
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 21 November 2023
Sec. Cancer Immunity and Immunotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1292861
This article is part of the Research Topic Personalized Immunotherapy: Advancing processes to extend patient collectives View all 12 articles
Immunotherapy has revolutionized the conventional treatment approaches for colorectal cancer (CRC), offering new therapeutic prospects for patients. Liquid biopsy has shown significant potential in early screening, diagnosis, and postoperative monitoring by analyzing circulating tumor cells (CTC) and circulating tumor DNA (ctDNA). In the era of immunotherapy, liquid biopsy provides additional possibilities for guiding immune-based treatments. Emerging technologies such as mass spectrometry-based detection of neoantigens and flow cytometry-based T cell sorting offer new tools for liquid biopsy, aiming to optimize immune therapy strategies. The integration of liquid biopsy with immunotherapy holds promise for improving treatment outcomes in colorectal cancer patients, enabling breakthroughs in early diagnosis and treatment, and providing patients with more personalized, precise, and effective treatment strategies.
Colorectal cancer (CRC) ranks as the second leading cause of cancer-related mortality worldwide (1), with increasing incidence and mortality rates. Most patients with metastatic CRC receive systemic drug therapy, which can prolong survival and improve symptoms but generally falls short of achieving a cure, making long-term survival challenging (2). In recent years, immunotherapy, represented by immune checkpoint inhibitors (ICIs), has revolutionized the traditional treatment approaches for CRC (3–5).
Immunotherapy has emerged as a promising approach for treating various cancers, including CRC. However, a challenge in the field of immunotherapy is the accurate assessment of treatment response and monitoring the effectiveness of immune interventions. Some biomarkers have been identified as predictors of the anti-tumor efficacy of ICIs, but there remains a need for clinically useful biomarkers. Traditional response assessment criteria, such as tissue biopsies, fail to capture the complex dynamics of the immune system and tumor microenvironment(TME) (6, 7), highlighting the urgent need for novel detection methods to monitor the efficacy of immunotherapy in real time and enable timely treatment adjustments (8–10).
With the rapid advancements in cell isolation and genetic testing technologies, liquid biopsy, which involves minimally invasive acquisition of tumor material, has gained recognition for its importance in precision oncology (11–13). It allows real-time monitoring of tumor progression, recurrence, or treatment response at the molecular level (14, 15). Circulating tumor cells (CTCs) and circulating tumor DNA (ctDNA) have emerged as representative liquid biopsy biomarkers (16).
In this review, we will first discuss the current biomarkers used for immune monitoring in CRC. Secondly, we will analyze the recent research progress in liquid biopsy, specifically focusing on ctDNA and CTCs, as adjuncts for CRC treatment. Finally, we will discuss the potential of novel technologies to address the challenges of immune therapy monitoring by providing solutions for liquid biopsy in the context of adjuvant immunotherapy.
Currently, the treatment modalities for CRC include endoscopic and surgical resection, systemic adjuvant chemotherapy, radiotherapy, targeted therapy, and immunotherapy (1, 17). Over the past five years, the discovery of ICIs and the successful use of ICIs have revolutionized the treatment paradigm for CRC. ICIs have brought new opportunities for the treatment of CRC (18–21). In 2017, the U.S. Food and Drug Administration (FDA) approved the use of immune therapy drugs for the treatment of metastatic colorectal cancer (mCRC) (22–25). Pembrolizumab, an anti- programmed death receptor 1(PD-1) monoclonal antibody, has been established as the first-line treatment standard for microsatellite-high/deficient mismatch repair (MSI-H/dMMR) mCRC (5). Immunotherapy is gradually becoming an essential component of precision treatment for mCRC.
With the continuous development of medical science and technological advancements, biomarkers play an increasingly important role in clinical applications. These biomarkers provide crucial information to assist physicians in the diagnosis, treatment, and monitoring of diseases. Some biomarkers, such as programmed cell death ligand 1 (PD-L1), tumor mutational burden (TMB), and microsatellite stability, have been identified as predictors of the anti-tumor efficacy of ICIs. However, there remains a gap in the clinical demand for effective biomarkers (26).
Microsatellite stability is currently the most relevant biomarker for immunotherapy sensitivity in CRC and is typically evaluated through solid tissue specimens (27, 28). MSI is a condition of genetic instability caused by defects in DNA repair mechanisms and is commonly observed in a subset of CRC patients. However, despite the promising prospects of MSI-H/dMMR as a biomarker for immunotherapy in CRC, there is variability in the reported overall response rates (ORR) in MSI-H mCRC patients, ranging from 30% to 70% (29–32). This suggests that a significant number of MSI-H mCRC patients do not benefit from immunotherapy (33). Conversely, a small subset of microsatellite-stable (MSS) CRC patients exhibit a response to immunotherapy. One contributing factor to this phenomenon is diagnostic errors caused by the detection methods (34).
Currently available methods for detecting microsatellite instability include immunohistochemistry (IHC), polymerase chain reaction (PCR), and next-generation sequencing (NGS). Immunohistochemistry detects the expression of four mismatch repair genes (MLH1, MSH2, MSH6, and PMS2) in the nuclei of tumor cells, and the absence of one or more of these proteins is defined as dMMR, otherwise known as proficient mismatch repair (pMMR) (35). Detection of MSI status is accomplished through immunohistochemistry on tissue specimens, which has the limitations of subjectivity and a lack of uniform standards (36).
TMB is associated with the treatment response to immunotherapy, and elevated plasma TMB levels (≥28 Mut/Mb) have shown predictable responses to the combination therapy of PD-L1 inhibitor durvalumab and CTLA4 inhibitor tremelimumab in MSS CRC patients (37). TMB has been approved by the U.S. FDA as a diagnostic biomarker for the use of pembrolizumab or dostarlimab in cancer immunotherapy (38, 39). Furthermore, studies have shown that high TMB (TMB ≥8 Mut/Mb) in CRC patients is associated with longer overall survival (OS) and better prognosis compared to low TMB (34, 40). However, the use of TMB as a sole predictor of immunotherapy response in CRC remains controversial. Limitations of using TMB as a predictive biomarker for immunotherapy response in CRC were observed in the KEYNOTE 177 trial (41). TMB assessment requires tumor tissue specimens as the gold standard, and tumor heterogeneity poses limitations to its precise estimation (42). Additionally, similar to any other gene or genomic biomarker, TMB may undergo changes in CRC following standard cytotoxic drug treatments (43).
Moreover, PD-L1 expression levels serve as important indicators of the immune status in cancer patients, which reflects the tumor’s response to immunotherapy (44–46). In certain solid tumors such as non-small cell lung cancer, melanoma, and renal cell carcinoma, PD-L1 expression has been proposed as a predictive biomarker for immunotherapy response (47–49). High PD-L1 expression is associated with a better response to immunotherapy. Tumor cells induce immune evasion by upregulating the expression of PD-L1, which binds to PD-1 on the surface of T cells, leading to T cell inactivation (Figure 1). ICIs block the interaction between PD-1 and PD-L1, thereby reactivating the body’s anti-tumor immune response (47). CRC patients have been reported to exhibit positive PD-L1 expression (50, 51). Although high PD-L1 expression is associated with a favorable prognosis in CRC patients (52–54), current clinical data suggest that the use of PD-L1 expression alone cannot accurately predict the immunotherapy response in CRC (55).
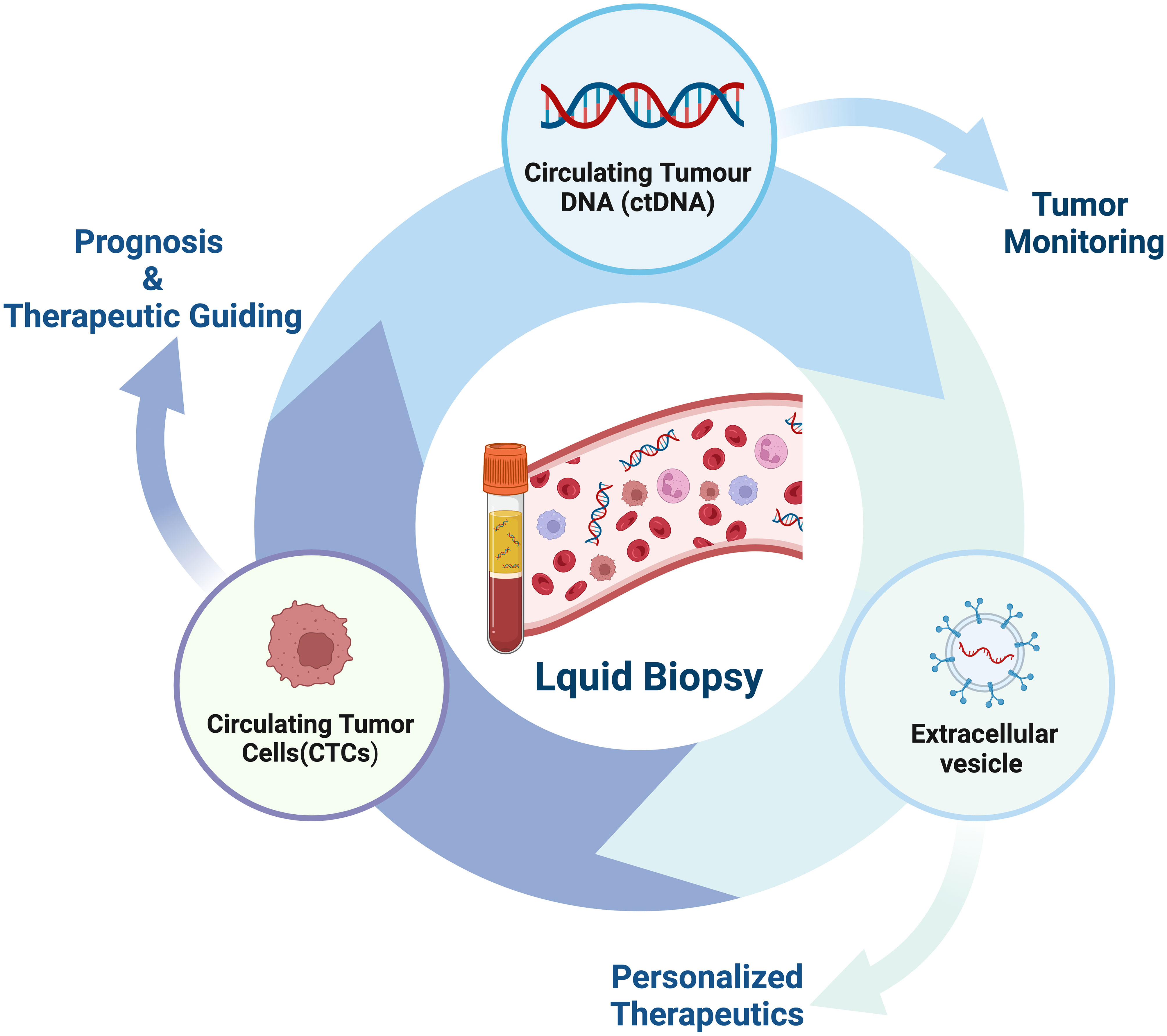
Figure 1 Programmed cell death protein 1 (PD-1) plays a crucial role in the initiation and effector phases of the anti-tumor immune response. T cell activation is a fundamental process in immune response, involving antigen presentation by dendritic cells and recognition by the T cell receptor (TCR). Once activated, T cells migrate to the tumor site to eliminate malignant cells. However, the tumor or bystander cells such as macrophages may upregulate PD-L1, which hinders T cell function by inducing inhibitory intracellular signaling.
However, despite the widespread application of certain biomarkers, we still face various challenges and limitations. To overcome these limitations, researchers are actively searching for more suitable detection methods and therapies to enhance the reliability and effectiveness of biomarkers in clinical practice. Through continuous exploration and innovation, we hope to open up new fields and approaches that will bring greater breakthroughs in disease prevention, diagnosis, and treatment. Therefore, the quest for more accurate and reliable biomarkers has become a hot topic in medical research, offering new opportunities and hopes for improving patient health outcomes.
Liquid biopsy has opened up a new avenue for cancer patients in terms of prognostic evaluation, detection of minimal residual disease (MRD), treatment selection, resistance mechanisms and monitoring, as well as early cancer diagnosis (56–61) (Figure 2). The fundamental principle of liquid biopsy is the non-invasive detection and assessment of tumors using circulating cell-free DNA (cfDNA), RNA, or tumor cells present in bodily fluids such as blood, urine, and cerebrospinal fluid (62–66). CTCs and ctDNA are important components and are generally considered the foundation of liquid biopsy. ctDNA is formed by apoptotic and necrotic tumor cells, which release fragmented DNA into the bloodstream and harbor genetic alterations of the original tumor cells (67, 68). CTCs are cancer cells that spontaneously detach from primary or metastatic tumors and circulate in the bloodstream (69). They serve as “seeds” of the tumor and can contribute to recurrence through liver metastasis, lymphatic dissemination, and angiogenesis (Figure 3).
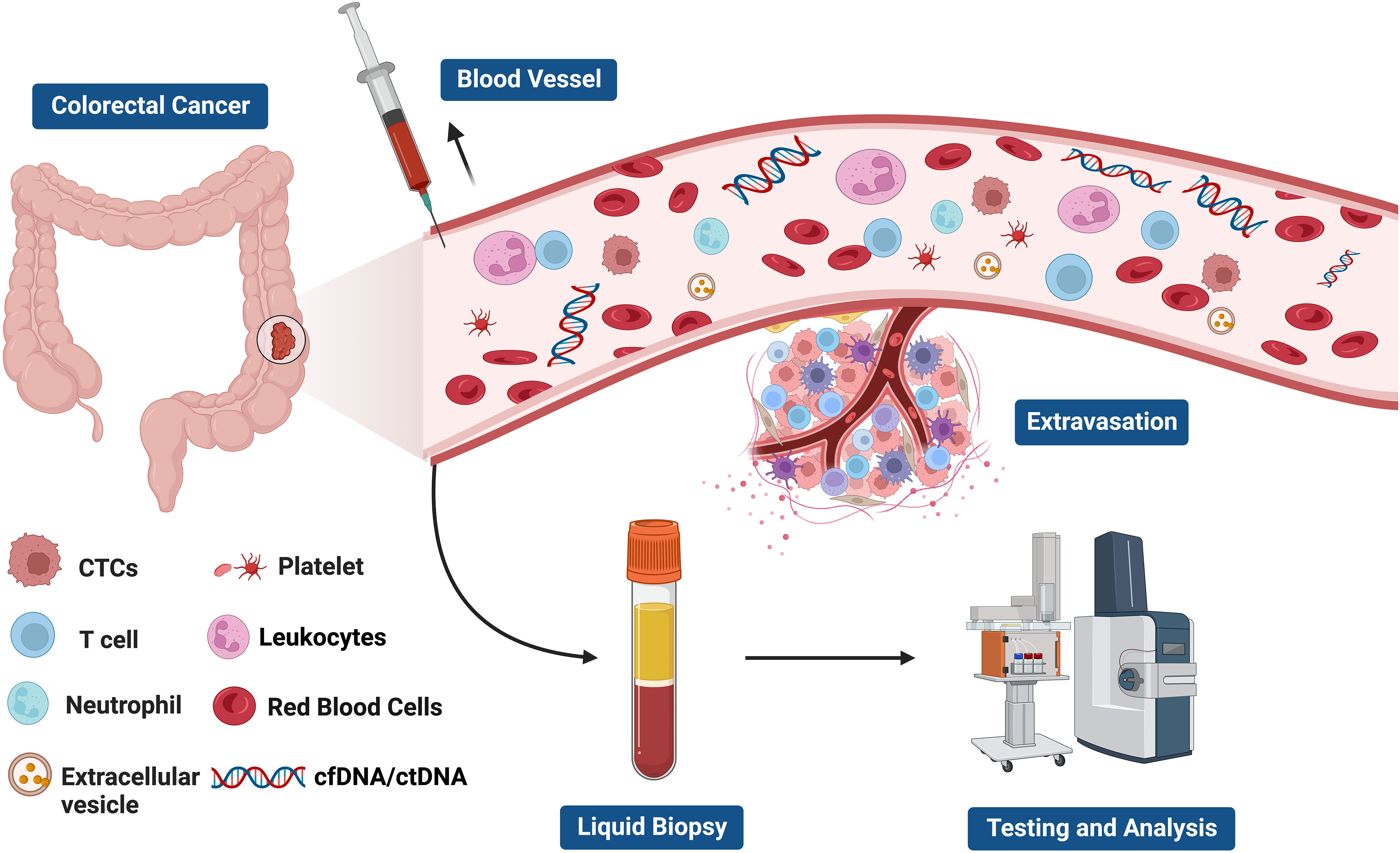
Figure 2 The current clinical applications of liquid biopsy. Liquid biopsy provides a non-invasive approach to assess the dynamic changes and treatment response of tumors by analyzing components such as circulating tumor DNA (ctDNA), circulating tumor cells (CTCs), and extracellular vesicles. These components can be obtained from blood samples and detected using highly sensitive analytical techniques. The information derived from liquid biopsy aids in guiding personalized treatment strategies, including the selection of appropriate drugs, monitoring treatment efficacy, and providing treatment guidance. Liquid biopsy holds promising applications in tumor management, offering patients more accurate and effective treatment choices.
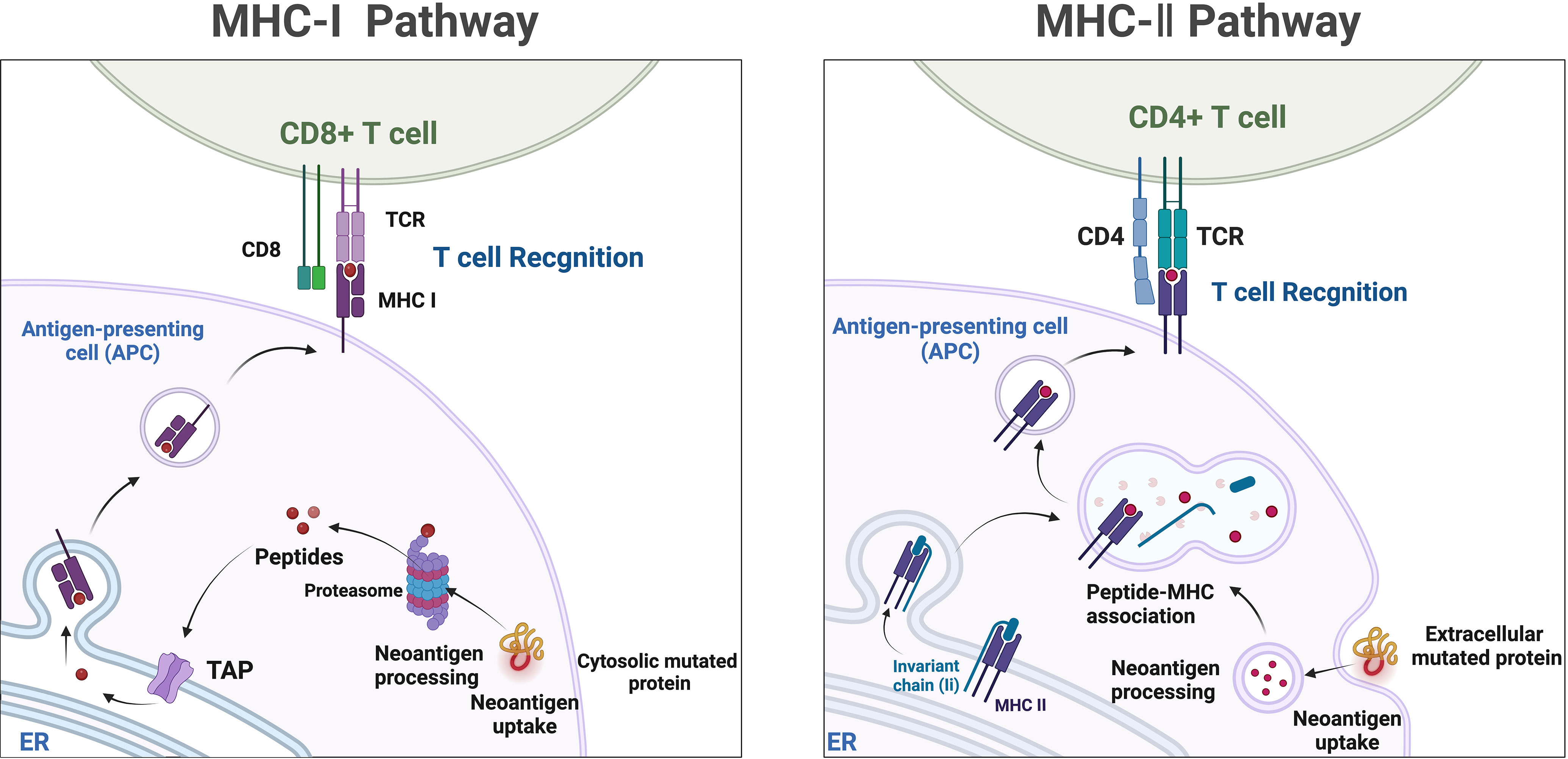
Figure 3 The components of liquid biopsy in colorectal cancer(CRC). circulating tumor cells (CTCs) and Circulating tumor DNA (ctDNA) are the main constituents of current liquid biopsy approaches. ctDNA, extracellular vesicle, and CTCs are shed directly from tumor masses or metastatic lesions into the bloodstream. After collection of blood samples, further analysis of these components provides a comprehensive tumor characterization.
Certain characteristics of CTCs, such as the expression of surface markers or genetic mutations, are associated with the prognosis of cancer patients. Changes in CTC counts are correlated with shortened disease-free survival (DFS), progression-free survival (PFS) and OS (70, 71). Increased levels of ctDNA may indicate disease progression (72, 73). By regularly monitoring changes in CTCs and ctDNA, the effectiveness of treatment and the dynamic changes of the tumor can be assessed (59).
MRD refers to the presence of extremely low levels of cancer cells or cancer-associated genetic material after completion of treatment (74). Early detection of MRD can be achieved through the detection of CTCs and ctDNA (75–77). According to the latest results from the GALAXY observational study presented at the 2023 ASCO conference, the detection of MRD through ctDNA testing at 4 weeks post-surgery is the strongest prognostic risk factor for DFS in stage II to IV CRC patients, regardless of BRAF V600E or MSI status (78, 79).
In early-stage CRC patients, the presence of ctDNA positivity after curative surgery is associated with a higher risk of disease recurrence (74, 80–85). A study demonstrated that ctDNA positivity after adjuvant chemotherapy is associated with poorer DFS, and ctDNA detection precedes radiological relapse by a median of 11.5 months (86). The DYNAMIC trial (87) investigated whether a ctDNA-guided approach could reduce the use of adjuvant therapy without compromising the risk of recurrence compared to the standard approach in stage II CRC. Among the 455 randomly assigned patients, 302 were assigned to the ctDNA-guided management group, and 153 were assigned to the standard management group. The median follow-up time was 37 months. The proportion of patients receiving adjuvant chemotherapy was lower in the ctDNA-guided group compared to the standard management group (15% vs. 28%). The ctDNA-guided management was non-inferior to standard management in terms of 2-year disease-free survival rates (93.5% vs. 92.4%). The 3-year disease-free survival rate was 86.4% in ctDNA-positive patients receiving adjuvant chemotherapy and 92.5% in ctDNA-negative patients not receiving adjuvant chemotherapy. The ctDNA-guided approach can reduce the use of adjuvant chemotherapy without compromising disease-free survival in the treatment of stage II CRC. In various cohorts of non-mCRC and resected colorectal liver metastasis patients, the proportion of disease recurrence has consistently exceeded 80% in patients with detectable ctDNA who did not receive adjuvant therapy (74, 88, 89). A study (90)demonstrated that preoperative ctDNA could be detected in 108 out of 122 (88.5%) patients with stage I to III CRC. Longitudinal ctDNA analysis identified 14 out of 16 (87.5%) recurrences after definitive treatment. Furthermore, at postoperative day 30, ctDNA-positive patients were more likely to experience recurrence compared to ctDNA-negative patients. Another study (91) evaluated the prognostic impact of postoperative ctDNA in stage I-III CRC patients and found that ctDNA status was the most significant and independent factor in predicting recurrence-free survival (RFS). Postoperative plasma samples from 108 patients underwent NGS quality control, with 17 (15.7%) classified as ctDNA-positive and 91 classified as ctDNA-negative. Among these 17 ctDNA-positive patients, 2 were stage II, and 15 were stage III. The recurrence rate for ctDNA-positive patients was 76.5% (13/17), significantly higher than the 16.5% (15/91) in ctDNA-negative patients. Kaplan-Meier survival curves showed significantly poorer recurrence-free survival (RFS) for ctDNA-positive patients compared to ctDNA-negative patients. The study results also demonstrated a sensitivity of 49.6% and specificity of 94.7% for ctDNA alone in predicting 2-year RFS. A predictive model combining ctDNA with clinical-pathological risk factors, referred to as CTCP prediction model, exhibited better RFS predictive value than ctDNA alone in stage I-III CRC patients and increased the sensitivity for 2-year RFS to 87.5%. The predictive value of this model was also externally validated. Additionally, ctDNA can be utilized for monitoring locally advanced rectal cancer (LARC) patients who achieve complete response after neoadjuvant therapy and adopt an “watch-and-wait” strategy (92, 93).
Precision therapy involves customizing drug treatments based on the individual characteristics of tumors (94). Liquid biopsy provides molecular profiling information of tumors, such as gene mutations and chromosomal rearrangements, to select appropriate targeted therapy drugs. In CRC, analysis of the molecular features of individual CTCs has revealed significant heterogeneity in the presence of EGFR mutations and other genetic mutations associated with EGFR inhibition (such as KRAS and PIK3CA mutations) among patients and between patients, which explains the different response rates to EGFR-targeted therapy (95). By analyzing mutations in ctDNA, patients who may benefit from targeted EGFR therapy or BRAF and MEK inhibitors can be identified (96–98). In ctDNA-positive CRC patients, plasma testing for RAS status demonstrated a sensitivity of 92.9% and specificity of 87.7% (99). The CHRONOS trial confirmed the importance of evaluating RAS status using ctDNA in metastatic CRC patients (100, 101). The study found that patients who were mutation-negative in ctDNA had good clinical responses to anti-EGFR retreatment. An ongoing randomized Phase III trial (102) is expected to reveal that liquid biopsy-based retreatment with anti-EGFR monoclonal antibodies achieves approximately one-third objective responses in mCRC patients, prospectively demonstrating the effective management of patients through genetic profiling using liquid biopsy.
Similar to predicting response to chemotherapy and/or targeted therapy, liquid biopsy based on ctDNA can guide immunotherapy. While immune therapy has prolonged PFS in patients with MSI-H CRC, it is interesting to note that in the KEYNOTE-177 study, approximately 30% of patients showed no response to pembrolizumab (32). Using ctDNA monitoring to identify non-responders at an early stage can provide an opportunity for physicians to switch to chemotherapy or consider the addition of anti-CTLA-4 agents (103). The mutational burden in ctDNA is associated with the efficacy of immunotherapy and serves as a direct reflection of tumor burden (104–110). Liquid biopsy utilizes ctDNA released into the bloodstream, providing a non-invasive alternative. However, similar to TMB, the MSI status is also influenced by spatial and temporal heterogeneity, making it challenging to monitor its therapeutic value through liquid biopsy (111). A study used liquid biopsy to detect the MSI status in ctDNA and found a high concordance with results from traditional tissue biopsy, effectively predicting immunotherapy sensitivity and clinical outcomes in patients (112). Another study demonstrated that liquid biopsy could monitor changes in MSI status at different time points, providing important information on treatment response and disease progression in patients (74). Furthermore, recent studies have proposed that the concentration of cfDNA can serve as a predictive biomarker for immune therapy response (113–116). cfDNA can be detected in MSI-H CRC patients who respond well to immunotherapy (117, 118). Moreover, dynamic changes in ctDNA have been shown to predict the efficacy of other immunotherapies, including chimeric antigen receptor T-cell (CAR-T) therapy (119, 120). Analysis of tumor-derived structural alterations through shallow whole-genome sequencing revealed a decrease in ctDNA levels in patients who responded well to CAR-T cell therapy, while an increase was observed in patients who did not achieve a treatment response. The abundance of CAR-T cell construct-derived DNA in peripheral blood may be correlated with the dynamic changes in ctDNA and can be used in combination (121).
Several clinical trials focusing on liquid biopsy in the context of immunotherapy are currently underway. The ongoing ARETHUSA trial (NCT03519412) is investigating the use of ctDNA-based TMB assessment as a predictive marker for immunotherapy response following pretreatment with temozolomide in MGMT-methylated mCRC (122). It is worth noting that there are ongoing efforts to identify the optimal approach for TMB analysis (123). The use of ctDNA for predicting response to immunotherapy has shown promise in the INSPIRE study, a prospective Phase II trial that conducted serial ctDNA assessments in 94 patients with advanced solid tumors receiving pembrolizumab (124). It was found that in 42% of patients, an increase in ctDNA and tumor volume was observed at 6 weeks, accurately predicting lack of response with 100% specificity. During immunotherapy, 16% of patients exhibited ctDNA clearance, with a median follow-up exceeding 25 months and an OS of 100%. At the start of the third treatment cycle, 98% of patients had an increase in ctDNA, indicating lack of objective response. This may enable the avoidance of ineffective treatment in a subset of patients. Zhang et al. characterized the prognostic and predictive impact of ctDNA in patients with 16 different solid tumor types enrolled in Phase I/II trials of single-agent durvalumab or combination therapy with tremelimumab (125). Higher pretreatment variant allele frequency (VAF) was associated with poorer survival but not with ORR. In contrast, reductions in VAF during treatment were associated with prolonged PFS, OS, and ORR, suggesting the predictive benefit of ctDNA during the treatment course. In ongoing clinical trials across various tumor types, including CRC, the dynamics of ctDNA, as measured by changes in VAF percentage and/or ctDNA clearance, have emerged as important biomarkers.
The application of liquid biopsy-guided adjuvant therapy for CRC is still in the research stage and requires further clinical validation and optimization. However, it has the advantages of non-invasiveness, repeatability, and real-time monitoring, and is expected to become one of the important auxiliary tools for personalized treatment of CRC.
Significant progress has been made in the study of CTCs and ctDNA using traditional liquid biopsy methods, which have played a powerful auxiliary role in tumor treatment (126–128). Immunotherapy has shown remarkable efficacy in various types of cancer, but it may impact the results of liquid biopsy. Therefore, it is necessary to reassess the traditional liquid biopsy criteria to accommodate the needs of immunotherapy (129, 130). Emerging detection technologies have provided support for liquid biopsy in optimizing treatment strategies, thus contributing to further advancements in the field of immunotherapy.
T-cell subset isolation is a method used to separate and purify T cells from a mixed population of cells. Flow cytometry can analyze various indicators such as T-cell subgroups, functional status, and expression of immune checkpoint molecules in blood samples. It can aid in evaluating a patient’s immune status and predicting the response to immune therapy (131–135).
The peripheral blood TCR repertoire serves as an important biomarker for the selection of ICIs therapy (136). TCR sequencing enables the study of the immune response mechanism of T cells. Longitudinal monitoring of the dynamic therapeutic evaluation of the TCR repertoire in ctDNA in peripheral blood provides insights into the co-evolution of tumors and immune components during ICIs treatment. TCR repertoire diversity, early conversion of peripheral T cells, and overall remodeling of the T-cell repertoire are associated with clonal regulation during ICIs treatment and are linked to anti-tumor immune responses. The presence of persistently exhausted TCR clones in peripheral blood is associated with adverse reactions to immune therapy (137–142). By combining flow cytometry and gene sequencing techniques, TCR sequences can be rapidly and accurately detected to understand T-cell clonal expansion and diversity.
Peripheral blood immune cell biomarkers, as one of the easily accessible biomarkers, can assess treatment response in the early stages and facilitate adjustments in early management (143–145). Studies have shown that the quantity and function of Tregs cells change in patients receiving immune therapy and are associated with poor prognosis (146–150). Studies using flow cytometry and RNA analysis have found that the percentage of circulating CD4+ and CD8+ T cells is associated with inflammatory tumors, indicating the significant role of these biomarkers in anti-tumor responses (151). Additionally, circulating T-cell lymphocyte subpopulations have been identified as biomarkers for mCRC (152). Decreased proportions of CD4+ cells and Tregs during treatment with folinic acid, 5-fluorouracil, and irinotecan (FOLFIRI) plus bevacizumab are associated with improved survival rates (153). Systemic immune inflammation index, ratios of different immune cells, and ratios of immune cells to platelets are also biomarkers for prognosis and prediction in CRC patients, including platelet-to-lymphocyte ratio (PLR) and neutrophil-to-lymphocyte ratio (NLR) (154–156). Recent studies have discovered novel circulating non-tumor cells and their biomarkers and extracellular matrix components, which have clinical application value in diagnosis, prognosis, and treatment response (157). Some studies suggest that circulating tumor endothelial cells (CTECs) from the tumor may play a prognostic role in CRC, with higher predictive value than CTCs (158–160). Similarly, in patients with mCRC receiving treatment with bevacizumab and chemotherapy, CECs and CD276-positive CTECs based on flow cytometry significantly increase (161). Studies have also shown that CXCR4-positive CECs are associated with longer PFS and OS, providing predictive value for mCRC patients receiving bevacizumab treatment (162–164).
Furthermore, single-cell sequencing technology (scRNA-seq) allows the study of gene expression and genetic variations at the individual cell level (165). The first immunotranscriptomic study based on scRNA-seq was conducted on CD4+ T cells infiltrating CRC. In this study, the impact of the tumor immune microenvironment (TIME) on specific gene expression (LAYN, MAGEH1, and CCR8) in tumor-infiltrating Tregs cells was confirmed, and these gene expressions were found to be correlated with immune therapy response, tumor-suppressive activity, and prognosis (166).
The gene characteristics of peripheral blood immune cells have received attention in the field of immune therapy and precision medicine. By combining T-cell subset isolation and liquid biopsy, comprehensive monitoring tools for immune therapy can be obtained, leading to a better understanding of tumor immune response and treatment outcomes, as well as optimization of treatment strategies.
Mass spectrometry-based liquid-phase detection is a novel technique that allows for the molecular-level monitoring of chemical components within cells and organisms, providing deeper insights into biological information. Neoantigens are novel antigenic epitopes generated by genetic mutations and serve as important targets in personalized immunotherapy (129). Neoantigens can be produced through proteasome-mediated endogenous protein degradation, and the resulting mutated peptides are subsequently transported to the endoplasmic reticulum (ER) via antigen processing-associated transporter (TAP), where they may be loaded onto MHC-I. MHC-II dimers assemble in the ER and associate with the invariant chain (Ii). The Ii-MHC-II complex can be transported directly from the cell surface or, at times, indirectly endocytosed into the MHC-II compartment (MIIC). Within the MIIC, a series of endolysosomal proteases degrade Ii, releasing it and enabling MHC-II to bind specific peptide segments derived from mutated proteins within the endocytic pathway. These peptide-MHC (pMHC) complexes are subsequently transported to the cell surface, where they are recognized by T cells (167) (Figure 4). Neoantigens possess potential high specificity and targeting, but they are predominantly patient-specific, making it challenging to categorize their utility, and they are often prominent in cancer patient populations. Currently, immune therapies, ICIs, tumor-specific vaccines, and neoantigen-based tumor-infiltrating lymphocytes (TILs) play increasingly important roles in cancer treatment (168). Studies have observed that certain CRC patients with MSI-H may benefit from ICIs treatment due to the presence of neoantigens (169). One of the main obstacles faced in personalized neoantigen immunotherapy is the accessibility of tumor biopsies. Thus far, the identification of neoantigens has typically involved genomic analysis of various tumor biopsies (170). Although this approach is time-consuming, invasive, and has a low positivity rate, it is more common in challenging cases requiring repeated sampling or when samples are limited, particularly in cases of frequent occurrence and metastatic cancers. Specifically, the presence of natural neoantigens at the top of immune checkpoints can enhance the effectiveness of significant inhibitors (171, 172). Given the current situation, liquid biopsies can serve as a viable alternative approach to identify potential neoantigens as immune therapeutic targets, applicable to numerous cancers. Although the limitations of detecting genomic mutations in plasma samples lie in detecting low allele frequencies, the reliability of genetic information obtained from liquid biopsies has been demonstrated (173). Therefore, based on current research on liquid biopsies, valuable insights can be provided for the use of neoantigens in treatment selection. Mass spectrometry-based liquid-phase detection allows for efficient identification and quantification of protein compositions within tumor cells, enabling the discovery of novel tumor-specific antigens by monitoring neoantigens in serum (174). Neoantigen-based immunotherapeutic approaches, such as ICIs, tumor-specific vaccines, and TILs, have become increasingly important in cancer treatment (168). Not all MSI-H CRC patients benefit from ICIs treatment; however, certain MSI-H colorectal cancer patients may benefit from ICIs treatment due to the presence of highly immunogenic neoantigens (169, 173).
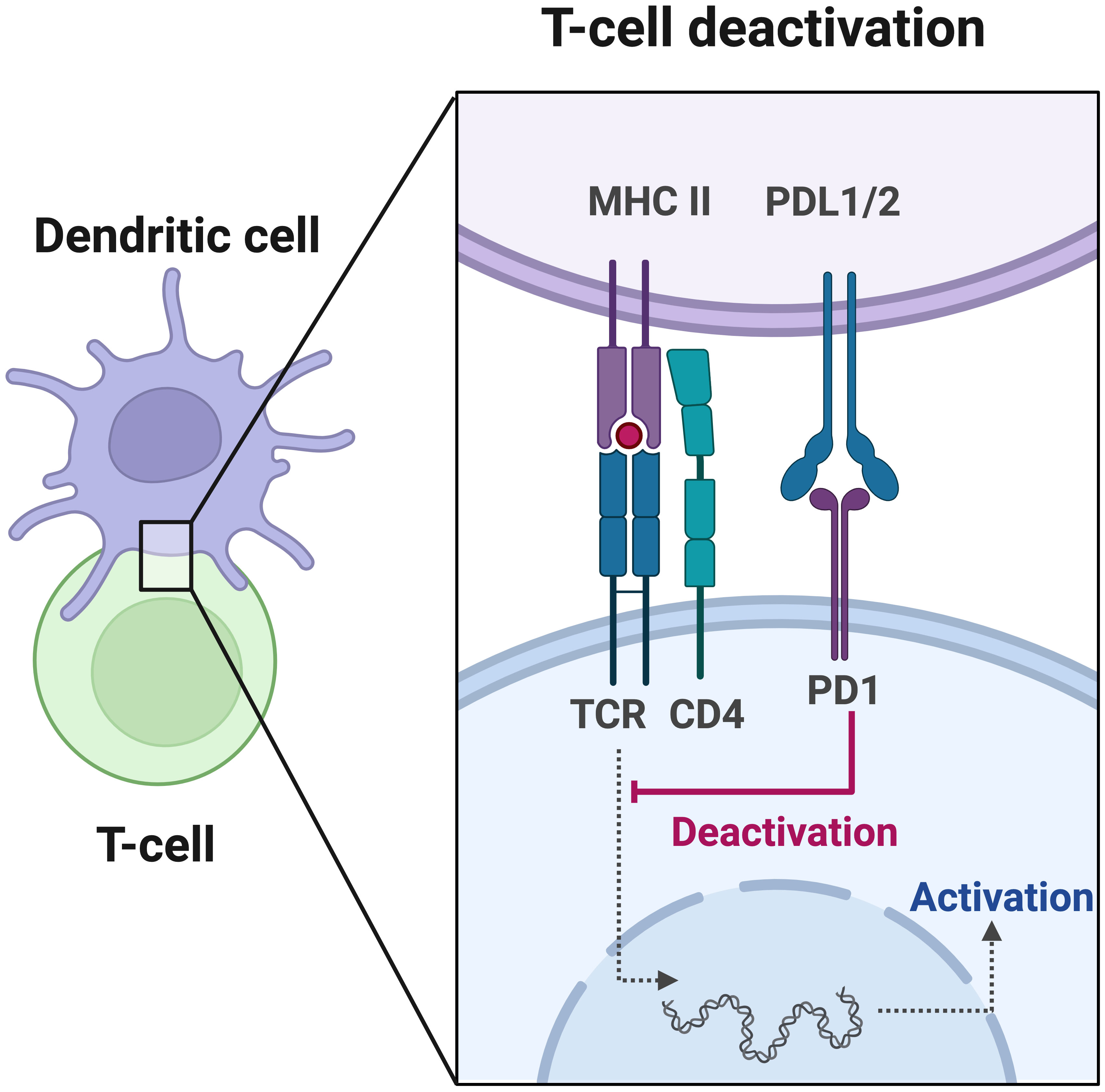
Figure 4 Neoantigens, which are mutated peptides generated through proteasome-mediated endogenous protein degradation, can subsequently be transported to the endoplasmic reticulum (ER) via antigen processing-associated transporter (TAP) and may be loaded onto MHC-I. In the ER, MHC-II dimers assemble and associate with the invariant chain (Ii). The Ii-MHC-II complex can be transported directly from the cell surface or, in some cases, indirectly endocytosed into the MHC-II compartment (MIIC). Within the MIIC, a series of endolysosomal proteases degrade Ii, releasing it and enabling MHC-II to bind specific peptide segments derived from mutated proteins within the endocytic pathway. These pMHC complexes are subsequently transported to the cell surface, where they are recognized by T cells.
Non-mutated neoantigens (NM-neoAgs) are immunogenic protein fragments generated through translational modifications or protein degradation of apoptotic tumor cells (175–181). These unique fragments do not exist in normal cells and are more easily processed and cross-presented by antigen-presenting cells (182). Studies utilizing mass spectrometry techniques and memory T cells as probes have identified NM-neoAgs in serum and found a strong correlation between high levels of NM-neoAgs and the efficacy of immunotherapy. Following induction chemotherapy, the response of NM-neoAgs-specific effector T cells (CD4+ and CD8+ T cells) increases and is further enhanced after immunotherapy, closely associated with patients’ survival rates and decreased expression levels of PD-1 (182). NM-neoAgs can target tumors with lower mutational burdens, contributing to the development of effective T cell-based immunotherapies for various cancer patients (182), and expanding the potential targets of liquid biopsy.
In summary, neoantigens and NM-neoAgs are tumor cell-specific antigens with tremendous potential in personalized immunotherapy. New detection methods such as flow cytometry and mass spectrometry techniques provide powerful tools for evaluating the efficacy of immunotherapy, thereby offering more effective treatment strategies for patients.
Liquid biopsy, as a non-invasive detection method, has emerged as a promising approach for early screening, diagnosis, postoperative monitoring, treatment response assessment, and evaluation of tumor resistance (183). With advancements in mass spectrometry-based detection of neoantigens and T cell sorting techniques such as flow cytometry, liquid biopsy has gained support as an adjunctive tool in the field of immunotherapy, providing opportunities for optimizing treatment strategies. However, despite significant progress, liquid biopsy remains in the exploratory and developmental stage, facing various challenges and complexities.
These include issues including the typically low concentrations of analytes collected from samples (184, 185), lack of standardization and uniformity for liquid biopsy biomarkers, and a dearth of widely accepted clinical practice guidelines (186, 187),related to false-positive results (188), variations in sensitivity among studies (82, 189), limitations in detection sensitivity and specificity (186, 190), and susceptibility to interference (184, 189, 191, 192). Overcoming these challenges and advancing liquid biopsy requires the development of highly sensitive and specific detection methods, standardization of experimental procedures and validation methods, and the application of artificial intelligence and machine learning algorithms for data analysis and interpretation. Additionally, the exploration of new biomarkers and the conduct of large-scale multicenter studies and clinical trials are essential to enhance the accuracy of early diagnosis and treatment prediction (193–196).
Despite the challenges that remain, the potential of liquid biopsy-assisted immunotherapy in transforming the field of immunotherapy is undeniable. Looking ahead, in the era of immunotherapy, liquid biopsy-assisted immunotherapy has the potential to fundamentally change the field and provide patients with more precise, effective, and personalized treatment strategies. Continued research, clinical trials, and technological advancements will play a crucial role in fully harnessing liquid biopsy as a valuable tool for guiding immunotherapy and improving future patient outcomes.
SY: Conceptualization, Data curation, Formal analysis, Writing – original draft. YH: Investigation, Methodology, Writing – review & editing. MY: Resources, Software, Writing – original draft. KJ: Conceptualization, Data curation, Writing – original draft, Writing – review & editing. HL: Investigation, Software, Validation, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Natural Science Foundation of China [grant no. 82104445 to HRL], Zhejiang Provincial Science and Technology Projects (grant no. LGF22H160046 to HRL), Jinhua Municipal Science and Technology Projects (grants no. 2021-3-040 to KTJ, and 2021-3-046 to HRL).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
2. Pitroda SP, Chmura SJ, Weichselbaum RR. Integration of radiotherapy and immunotherapy for treatment of oligometastases. Lancet Oncol (2019) 20:e434–42. doi: 10.1016/s1470-2045(19)30157-3
3. Schmidt C. The benefits of immunotherapy combinations. Nature (2017) 552:S67–s69. doi: 10.1038/d41586-017-08702-7
4. Yao S, Lan H, Han Y, Mao C, Yang M, Zhang X, et al. From organ preservation to selective surgery: How immunotherapy changes colorectal surgery? Surg Open Sci (2023) 15:44–53. doi: 10.1016/j.sopen.2023.07.024
5. André T, Shiu K-K, Kim TW, Jensen BV, Jensen LH, Punt C, et al. Pembrolizumab in microsatellite-instability–high advanced colorectal cancer. New Engl J Med (2020) 383:2207–18. doi: 10.1056/NEJMoa2017699
6. Raza A, Khan AQ, Inchakalody VP, Mestiri S, Yoosuf Z, Bedhiafi T, et al. Dynamic liquid biopsy components as predictive and prognostic biomarkers in colorectal cancer. J Exp Clin Cancer Res (2022) 41:99. doi: 10.1186/s13046-022-02318-0
7. Lokhandwala T, Bittoni MA, Dann RA, D'Souza AO, Johnson M, Nagy RJ, et al. Costs of diagnostic assessment for lung cancer: A medicare claims analysis. Clin Lung Cancer (2017) 18:e27–34. doi: 10.1016/j.cllc.2016.07.006
8. Marron TU, Ryan AE, Reddy SM, Kaczanowska S, Younis RH, Thakkar D, et al. Considerations for treatment duration in responders to immune checkpoint inhibitors. J Immunother Cancer (2021) 9. doi: 10.1136/jitc-2020-001901
9. Meric-Bernstam F, Larkin J, Tabernero J, Bonini C. Enhancing anti-tumour efficacy with immunotherapy combinations. Lancet (2021) 397:1010–22. doi: 10.1016/s0140-6736(20)32598-8
10. Ott PA, Hodi FS, Kaufman HL, Wigginton JM, Wolchok JD. Combination immunotherapy: a road map. J Immunother Cancer (2017) 5:16. doi: 10.1186/s40425-017-0218-5
11. Ye Q, Ling S, Zheng S, Xu X. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer (2019) 18:114. doi: 10.1186/s12943-019-1043-x
12. Diaz LA Jr., Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol (2014) 32:579–86. doi: 10.1200/jco.2012.45.2011
13. Alix-Panabières C, Pantel K. Clinical applications of circulating tumor cells and circulating tumor DNA as liquid biopsy. Cancer Discovery (2016) 6:479–91. doi: 10.1158/2159-8290.Cd-15-1483
14. Heidrich I, Ačkar L, Mossahebi Mohammadi P, Pantel K. Liquid biopsies: Potential and challenges. Int J Cancer (2021) 148:528–45. doi: 10.1002/ijc.33217
15. Saini A, Pershad Y, Albadawi H, Kuo M, Alzubaidi S, Naidu S, et al. Liquid biopsy in gastrointestinal cancers. Diagnostics (Basel) (2018) 8. doi: 10.3390/diagnostics8040075
16. Ivanova E, Ward A, Wiegmans AP, Richard DJ. Circulating tumor cells in metastatic breast cancer: from genome instability to metastasis. Front Mol Biosci (2020) 7:134. doi: 10.3389/fmolb.2020.00134
17. Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut (2017) 66:683–91. doi: 10.1136/gutjnl-2015-310912
18. Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer (2012) 12:252–64. doi: 10.1038/nrc3239
19. Hodi FS, Chiarion-Sileni V, Gonzalez R, Grob JJ, Rutkowski P, Cowey CL, et al. Nivolumab plus ipilimumab or nivolumab alone versus ipilimumab alone in advanced melanoma (CheckMate 067): 4-year outcomes of a multicentre, randomised, phase 3 trial. Lancet Oncol (2018) 19:1480–92. doi: 10.1016/s1470-2045(18)30700-9
20. Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, et al. Updated analysis of KEYNOTE-024: pembrolizumab versus platinum-based chemotherapy for advanced non-small-cell lung cancer with PD-L1 tumor proportion score of 50% or greater. J Clin Oncol (2019) 37:537–46. doi: 10.1200/jco.18.00149
21. Motzer RJ, Rini BI, McDermott DF, Arén Frontera O, Hammers HJ, Carducci MA, et al. Nivolumab plus ipilimumab versus sunitinib in first-line treatment for advanced renal cell carcinoma: extended follow-up of efficacy and safety results from a randomised, controlled, phase 3 trial. Lancet Oncol (2019) 20:1370–85. doi: 10.1016/s1470-2045(19)30413-9
22. Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med (2015) 372:2509–20. doi: 10.1056/NEJMoa1500596
23. Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol (2018) 36:773–9. doi: 10.1200/jco.2017.76.9901
24. Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz HJ, Morse MA, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol (2017) 18:1182–91. doi: 10.1016/s1470-2045(17)30422-9
25. Marcus L, Lemery SJ, Keegan P, Pazdur R. FDA approval summary: pembrolizumab for the treatment of microsatellite instability-high solid tumors. Clin Cancer Res (2019) 25:3753–8. doi: 10.1158/1078-0432.Ccr-18-4070
26. Xu Y, Fu Y, Zhu B, Wang J, Zhang B. Predictive biomarkers of immune checkpoint inhibitors-related toxicities. Front Immunol (2020) 11:2023. doi: 10.3389/fimmu.2020.02023
27. Yoshino T, Arnold D, Taniguchi H, Pentheroudakis G, Yamazaki K, Xu RH, et al. Pan-Asian adapted ESMO consensus guidelines for the management of patients with metastatic colorectal cancer: a JSMO-ESMO initiative endorsed by CSCO, KACO, MOS, SSO and TOS. Ann Oncol (2018) 29:44–70. doi: 10.1093/annonc/mdx738
28. Benson AB, Venook AP, Al-Hawary MM, Arain MA, Chen YJ, Ciombor KK, et al. Colon cancer, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw (2021) 19:329–59. doi: 10.6004/jnccn.2021.0012
29. Lenz HJ, Van Cutsem E, Luisa Limon M, Wong KYM, Hendlisz A, Aglietta M, et al. First-line nivolumab plus low-dose ipilimumab for microsatellite instability-high/mismatch repair-deficient metastatic colorectal cancer: the phase II checkMate 142 study. J Clin Oncol (2022) 40:161–70. doi: 10.1200/jco.21.01015
30. Cohen R, Bennouna J, Meurisse A, Tournigand C, de la Fouchardière C, Tougeron D, et al. RECIST and iRECIST criteria for the evaluation of nivolumab plus ipilimumab in patients with microsatellite instability-high/mismatch repair-deficient metastatic colorectal cancer: the GERCOR NIPICOL phase II study. J Immunother Cancer (2020) 8. doi: 10.1136/jitc-2020-001499
31. Asaoka Y, Ijichi H, Koike K. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med (2015) 373:1979. doi: 10.1056/NEJMc1510353
32. Le DT, Kim TW, Van Cutsem E, Geva R, Jäger D, Hara H, et al. Phase II open-label study of pembrolizumab in treatment-refractory, microsatellite instability-high/mismatch repair-deficient metastatic colorectal cancer: KEYNOTE-164. J Clin Oncol (2020) 38:11–9. doi: 10.1200/jco.19.02107
33. Huyghe N, Baldin P, Van den Eynde M. Immunotherapy with immune checkpoint inhibitors in colorectal cancer: what is the future beyond deficient mismatch-repair tumours? Gastroenterol Rep (Oxf) (2020) 8:11–24. doi: 10.1093/gastro/goz061
34. Cohen R, Hain E, Buhard O, Guilloux A, Bardier A, Kaci R, et al. Association of primary resistance to immune checkpoint inhibitors in metastatic colorectal cancer with misdiagnosis of microsatellite instability or mismatch repair deficiency status. JAMA Oncol (2019) 5:551–5. doi: 10.1001/jamaoncol.2018.4942
35. Ganesh K, Stadler ZK, Cercek A, Mendelsohn RB, Shia J, Segal NH, et al. Immunotherapy in colorectal cancer: rationale, challenges and potential. Nat Rev Gastroenterol Hepatol (2019) 16:361–75. doi: 10.1038/s41575-019-0126-x
36. Amato M, Franco R, Facchini G, Addeo R, Ciardiello F, Berretta M, et al. Microsatellite instability: from the implementation of the detection to a prognostic and predictive role in cancers. Int J Mol Sci (2022) 23:23. doi: 10.3390/ijms23158726
37. Chen EX, Jonker DJ, Loree JM, Kennecke HF, Berry SR, Couture F, et al. Effect of combined immune checkpoint inhibition vs best supportive care alone in patients with advanced colorectal cancer: the canadian cancer trials group CO.26 study. JAMA Oncol (2020) 6:831–8. doi: 10.1001/jamaoncol.2020.0910
38. Passaro A, Stenzinger A, Peters S. Tumor mutational burden as a pan-cancer biomarker for immunotherapy: the limits and potential for convergence. Cancer Cell (2020) 38:624–5. doi: 10.1016/j.ccell.2020.10.019
39. Sha D, Jin Z, Budczies J, Kluck K, Stenzinger A, Sinicrope FA. Tumor mutational burden as a predictive biomarker in solid tumors. Cancer Discovery (2020) 10:1808–25. doi: 10.1158/2159-8290.Cd-20-0522
40. Innocenti F, Ou FS, Qu X, Zemla TJ, Niedzwiecki D, Tam R, et al. Mutational analysis of patients with colorectal cancer in CALGB/SWOG 80405 identifies new roles of microsatellite instability and tumor mutational burden for patient outcome. J Clin Oncol (2019) 37:1217–27. doi: 10.1200/jco.18.01798
41. Bortolomeazzi M, Keddar MR, Montorsi L, Acha-Sagredo A, Benedetti L, Temelkovski D, et al. Immunogenomics of colorectal cancer response to checkpoint blockade: analysis of the KEYNOTE 177 trial and validation cohorts. Gastroenterology (2021) 161:1179–93. doi: 10.1053/j.gastro.2021.06.064
42. Gandara DR, Paul SM, Kowanetz M, Schleifman E, Zou W, Li Y, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med (2018) 24:1441–8. doi: 10.1038/s41591-018-0134-3
43. Pich O, Muiños F, Lolkema MP, Steeghs N, Gonzalez-Perez A, Lopez-Bigas N. The mutational footprints of cancer therapies. Nat Genet (2019) 51:1732–40. doi: 10.1038/s41588-019-0525-5
44. Peng QH, Wang CH, Chen HM, Zhang RX, Pan ZZ, Lu ZH, et al. CMTM6 and PD-L1 coexpression is associated with an active immune microenvironment and a favorable prognosis in colorectal cancer. J Immunother Cancer (2021) 9. doi: 10.1136/jitc-2020-001638
45. Omura Y, Toiyama Y, Okugawa Y, Yin C, Shigemori T, Kusunoki K, et al. Prognostic impacts of tumoral expression and serum levels of PD-L1 and CTLA-4 in colorectal cancer patients. Cancer Immunol Immunother (2020) 69:2533–46. doi: 10.1007/s00262-020-02645-1
46. Masugi Y, Nishihara R, Yang J, Mima K, da Silva A, Shi Y, et al. Tumour CD274 (PD-L1) expression and T cells in colorectal cancer. Gut (2017) 66:1463–73. doi: 10.1136/gutjnl-2016-311421
47. Hou W, Zhou X, Yi C, Zhu H. Immune check point inhibitors and immune-related adverse events in small cell lung cancer. Front Oncol (2021) 11:604227. doi: 10.3389/fonc.2021.604227
48. Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. (2018) 560:382–6. doi: 10.1038/s41586-018-0392-8
49. Wang X, Teng F, Kong L, Yu J. PD-L1 expression in human cancers and its association with clinical outcomes. Onco Targets Ther (2016) 9:5023–39. doi: 10.2147/ott.S105862
50. Patel SP, Kurzrock R. PD-L1 expression as a predictive biomarker in cancer immunotherapy. Mol Cancer Ther (2015) 14:847–56. doi: 10.1158/1535-7163.Mct-14-0983
51. Ahn AR, Kim KM, Jang KY, Moon WS, Ha GW, Lee MR, et al. Correlation of PIK3CA mutation with programmed death ligand-1 (PD-L1) expression and their clinicopathological significance in colorectal cancer. Ann Transl Med (2021) 9:1406. doi: 10.21037/atm-21-2315
52. Yang L, Xue R, Pan C. Prognostic and clinicopathological value of PD-L1 in colorectal cancer: a systematic review and meta-analysis. Onco Targets Ther (2019) 12:3671–82. doi: 10.2147/ott.S190168
53. Li Y, He M, Zhou Y, Yang C, Wei S, Bian X, et al. The prognostic and clinicopathological roles of PD-L1 expression in colorectal cancer: A systematic review and meta-analysis. Front Pharmacol (2019) 10:139. doi: 10.3389/fphar.2019.00139
54. Wang S, Yuan B, Wang Y, Li M, Liu X, Cao J, et al. Clinicopathological and prognostic significance of PD-L1 expression in colorectal cancer: a meta-analysis. Int J Colorectal Dis (2021) 36:117–30. doi: 10.1007/s00384-020-03734-4
55. Hou W, Yi C, Zhu H. Predictive biomarkers of colon cancer immunotherapy: Present and future. Front Immunol (2022) 13:1032314. doi: 10.3389/fimmu.2022.1032314
56. Pantel K, Alix-Panabières C. Circulating tumour cells in cancer patients: challenges and perspectives. Trends Mol Med (2010) 16:398–406. doi: 10.1016/j.molmed.2010.07.001
57. Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic - implementation issues and future challenges. Nat Rev Clin Oncol (2021) 18:297–312. doi: 10.1038/s41571-020-00457-x
58. Bidard FC, Peeters DJ, Fehm T, Nolé F, Gisbert-Criado R, Mavroudis D, et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: a pooled analysis of individual patient data. Lancet Oncol (2014) 15:406–14. doi: 10.1016/s1470-2045(14)70069-5
59. Alix-Panabières C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer (2014) 14:623–31. doi: 10.1038/nrc3820
60. Alix-Panabières C, Pantel K. Liquid biopsy: from discovery to clinical application. Cancer Discovery (2021) 11:858–73. doi: 10.1158/2159-8290.Cd-20-1311
61. Markou A, Tzanikou E, Lianidou E. The potential of liquid biopsy in the management of cancer patients. Semin Cancer Biol (2022) 84:69–79. doi: 10.1016/j.semcancer.2022.03.013
62. Diehl F, Schmidt K, Durkee KH, Moore KJ, Goodman SN, Shuber AP, et al. Analysis of mutations in DNA isolated from plasma and stool of colorectal cancer patients. Gastroenterology (2008) 135:489–98. doi: 10.1053/j.gastro.2008.05.039
63. De Mattos-Arruda L, Mayor R, Ng CKY, Weigelt B, Martínez-Ricarte F, Torrejon D, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun (2015) 6:8839. doi: 10.1038/ncomms9839
64. Kimura H, Fujiwara Y, Sone T, Kunitoh H, Tamura T, Kasahara K, et al. EGFR mutation status in tumour-derived DNA from pleural effusion fluid is a practical basis for predicting the response to gefitinib. Br J Cancer (2006) 95:1390–5. doi: 10.1038/sj.bjc.6603428
65. Wang Y, Springer S, Mulvey CL, Silliman N, Schaefer J, Sausen M, et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med (2015) 7:293ra104. doi: 10.1126/scitranslmed.aaa8507
66. Mouliere F, El Messaoudi S, Pang D, Dritschilo A, Thierry AR. Multi-marker analysis of circulating cell-free DNA toward personalized medicine for colorectal cancer. Mol Oncol (2014) 8:927–41. doi: 10.1016/j.molonc.2014.02.005
67. Muluhngwi P, Valdes R Jr., Fernandez-Botran R, Burton E, Williams B, Linder MW. Cell-free DNA diagnostics: current and emerging applications in oncology. Pharmacogenomics (2019) 20:357–80. doi: 10.2217/pgs-2018-0174
68. Spindler KG. Methodological, biological and clinical aspects of circulating free DNA in metastatic colorectal cancer. Acta Oncol (2017) 56:7–16. doi: 10.1080/0284186x.2016.1253861
69. Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem (2013) 59:110–8. doi: 10.1373/clinchem.2012.194258
70. Qi Y, Wang W. Clinical significance of circulating tumor cells in squamous cell lung cancer patients. Cancer biomark (2017) 18:161–7. doi: 10.3233/cbm-160090
71. Castello A, Carbone FG, Rossi S, Monterisi S, Federico D, Toschi L, et al. Circulating tumor cells and metabolic parameters in NSCLC patients treated with checkpoint inhibitors. Cancers (Basel) (2020) 12. doi: 10.3390/cancers12020487
72. Yu H, Han L, Yuan J, Sun Y. Circulating tumor cell free DNA from plasma and urine in the clinical management of colorectal cancer. Cancer biomark (2020) 27:29–37. doi: 10.3233/cbm-182344
73. Kobayashi S, Nakamura Y, Taniguchi H, Odegaard JI, Nomura S, Kojima M, et al. Impact of preoperative circulating tumor DNA status on survival outcomes after hepatectomy for resectable colorectal liver metastases. Ann Surg Oncol (2021) 28:4744–55. doi: 10.1245/s10434-020-09449-8
74. Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med (2016) 8:346ra92. doi: 10.1126/scitranslmed.aaf6219
75. Mauri G, Vitiello PP, Sogari A, Crisafulli G, Sartore-Bianchi A, Marsoni S, et al. Liquid biopsies to monitor and direct cancer treatment in colorectal cancer. Br J Cancer (2022) 127:394–407. doi: 10.1038/s41416-022-01769-8
76. Montagut C, Vidal J. Liquid biopsy for precision adjuvant chemotherapy in colon cancer. N Engl J Med (2022) 386:2330–1. doi: 10.1056/NEJMe2204625
77. McDonald BR, Contente-Cuomo T, Sammut SJ, Odenheimer-Bergman A, Ernst B, Perdigones N, et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci Transl Med (2019) 11. doi: 10.1126/scitranslmed.aax7392
78. Kotani D, Oki E, Nakamura Y, Yukami H, Mishima S, Bando H, et al. Molecular residual disease and efficacy of adjuvant chemotherapy in patients with colorectal cancer. Nat Med (2023) 29:127–34. doi: 10.1038/s41591-022-02115-4
79. Coombes RC, Page K, Salari R, Hastings RK, Armstrong A, Ahmed S, et al. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clin Cancer Res (2019) 25:4255–63. doi: 10.1158/1078-0432.Ccr-18-3663
80. Kasi PM, Fehringer G, Taniguchi H, Starling N, Nakamura Y, Kotani D, et al. Impact of circulating tumor DNA-based detection of molecular residual disease on the conduct and design of clinical trials for solid tumors. JCO Precis Oncol (2022) 6:e2100181. doi: 10.1200/po.21.00181
81. Tie J, Cohen JD, Wang Y, Christie M, Simons K, Lee M, et al. Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol (2019) 5:1710–7. doi: 10.1001/jamaoncol.2019.3616
82. Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med (2008) 14:985–90. doi: 10.1038/nm.1789
83. Faulkner LG, Howells LM, Pepper C, Shaw JA, Thomas AL. The utility of ctDNA in detecting minimal residual disease following curative surgery in colorectal cancer: a systematic review and meta-analysis. Br J Cancer (2023) 128:297–309. doi: 10.1038/s41416-022-02017-9
84. Reinert T, Henriksen TV, Christensen E, Sharma S, Salari R, Sethi H, et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol (2019) 5:1124–31. doi: 10.1001/jamaoncol.2019.0528
85. Tie J, Cohen JD, Lo SN, Wang Y, Li L, Christie M, et al. Prognostic significance of postsurgery circulating tumor DNA in nonmetastatic colorectal cancer: Individual patient pooled analysis of three cohort studies. Int J Cancer (2021) 148:1014–26. doi: 10.1002/ijc.33312
86. Tarazona N, Gimeno-Valiente F, Gambardella V, Zuñiga S, Rentero-Garrido P, Huerta M, et al. Targeted next-generation sequencing of circulating-tumor DNA for tracking minimal residual disease in localized colon cancer. Ann Oncol (2019) 30:1804–12. doi: 10.1093/annonc/mdz390
87. Tie J, Cohen JD, Lahouel K, Lo SN, Wang Y, Kosmider S, et al. Circulating tumor DNA analysis guiding adjuvant therapy in stage II colon cancer. N Engl J Med (2022) 386:2261–72. doi: 10.1056/NEJMoa2200075
88. Tie J, Cohen JD, Wang Y, Li L, Christie M, Simons K, et al. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: a prospective biomarker study. Gut (2019) 68:663–71. doi: 10.1136/gutjnl-2017-315852
89. Parikh AR, Van Seventer EE, Siravegna G, Hartwig AV, Jaimovich A, He Y, et al. Minimal residual disease detection using a plasma-only circulating tumor DNA assay in patients with colorectal cancer. Clin Cancer Res (2021) 27:5586–94. doi: 10.1158/1078-0432.Ccr-21-0410
90. Taniguchi H, Nakamura Y, Kotani D, Yukami H, Mishima S, Sawada K, et al. CIRCULATE-Japan: Circulating tumor DNA-guided adaptive platform trials to refine adjuvant therapy for colorectal cancer. Cancer Sci (2021) 112:2915–20. doi: 10.1111/cas.14926
91. Gao Z, Huang D, Chen H, Yang Y, An K, Ding C, et al. Development and validation of postoperative circulating tumor DNA combined with clinicopathological risk factors for recurrence prediction in patients with stage I-III colorectal cancer. J Transl Med (2023) 21:63. doi: 10.1186/s12967-023-03884-3
92. Habr-Gama A, Perez RO, Nadalin W, Sabbaga J, Ribeiro U Jr., Silva e Sousa AH Jr., et al. Operative versus nonoperative treatment for stage 0 distal rectal cancer following chemoradiation therapy: long-term results. Ann Surg (2004) 240:711–7; discussion 717-8. doi: 10.1097/01.sla.0000141194.27992.32
93. Habr-Gama A, Sabbaga J, Gama-Rodrigues J, São Julião GP, Proscurshim I, Bailão Aguilar P, et al. Watch and wait approach following extended neoadjuvant chemoradiation for distal rectal cancer: are we getting closer to anal cancer management? Dis Colon Rectum (2013) 56:1109–17. doi: 10.1097/DCR.0b013e3182a25c4e
94. Singh T, Fatehi Hassanabad M, Fatehi Hassanabad A. Non-small cell lung cancer: Emerging molecular targeted and immunotherapeutic agents. Biochim Biophys Acta Rev Cancer (2021) 1876:188636. doi: 10.1016/j.bbcan.2021.188636
95. Gasch C, Bauernhofer T, Pichler M, Langer-Freitag S, Reeh M, Seifert AM, et al. Heterogeneity of epidermal growth factor receptor status and mutations of KRAS/PIK3CA in circulating tumor cells of patients with colorectal cancer. Clin Chem (2013) 59:252–60. doi: 10.1373/clinchem.2012.188557
96. Li W, Liu JB, Hou LK, Yu F, Zhang J, Wu W, et al. Liquid biopsy in lung cancer: significance in diagnostics, prediction, and treatment monitoring. Mol Cancer (2022) 21:25. doi: 10.1186/s12943-022-01505-z
97. Cremolini C, Rossini D, Dell'Aquila E, Lonardi S, Conca E, Del Re M, et al. Rechallenge for patients with RAS and BRAF wild-type metastatic colorectal cancer with acquired resistance to first-line cetuximab and irinotecan: A phase 2 single-arm clinical trial. JAMA Oncol (2019) 5:343–50. doi: 10.1001/jamaoncol.2018.5080
98. Martinelli E, Martini G, Famiglietti V, Troiani T, Napolitano S, Pietrantonio F, et al. Cetuximab rechallenge plus avelumab in pretreated patients with RAS wild-type metastatic colorectal cancer: the phase 2 single-arm clinical CAVE trial. JAMA Oncol (2021) 7:1529–35. doi: 10.1001/jamaoncol.2021.2915
99. Bachet JB, Bouché O, Taieb J, Dubreuil O, Garcia ML, Meurisse A, et al. RAS mutation analysis in circulating tumor DNA from patients with metastatic colorectal cancer: the AGEO RASANC prospective multicenter study. Ann Oncol (2018) 29:1211–9. doi: 10.1093/annonc/mdy061
100. Sartore-Bianchi A, Pietrantonio F, Lonardi S, Mussolin B, Rua F, Fenocchio E, et al. Phase II study of anti-EGFR rechallenge therapy with panitumumab driven by circulating tumor DNA molecular selection in metastatic colorectal cancer: The CHRONOS trial. J Clin Oncol (2021) 39:3506–6. doi: 10.1200/JCO.2021.39.15_suppl.3506
101. Parseghian CM, Loree JM, Morris VK, Liu X, Clifton KK, Napolitano S, et al. Anti-EGFR-resistant clones decay exponentially after progression: implications for anti-EGFR re-challenge. Ann Oncol (2019) 30:243–9. doi: 10.1093/annonc/mdy509
102. Metastatic Colorectal Cancer (RAS-wildtype) After Response to First-line Treatment With FOLFIR Plus Cetuximab (AIO-KRK-0114). Available at: https://clinicaltrials.gov/study/NCT02934529.
103. Kasi PM, Budde G, Krainock M, Aushev VN, Koyen Malashevich A, Malhotra M, et al. Circulating tumor DNA (ctDNA) serial analysis during progression on PD-1 blockade and later CTLA-4 rescue in patients with mismatch repair deficient metastatic colorectal cancer. J Immunother Cancer (2022) 10. doi: 10.1136/jitc-2021-003312
104. Lee JH, Menzies AM, Carlino MS, McEvoy AC, Sandhu S, Weppler AM, et al. Longitudinal monitoring of ctDNA in patients with melanoma and brain metastases treated with immune checkpoint inhibitors. Clin Cancer Res (2020) 26:4064–71. doi: 10.1158/1078-0432.Ccr-19-3926
105. Lee JH, Long GV, Boyd S, Lo S, Menzies AM, Tembe V, et al. Circulating tumour DNA predicts response to anti-PD1 antibodies in metastatic melanoma. Ann Oncol (2017) 28:1130–6. doi: 10.1093/annonc/mdx026
106. Braune J, Keller L, Schiller F, Graf E, Rafei-Shamsabadi D, Wehrle J, et al. Circulating tumor DNA allows early treatment monitoring in BRAF- and NRAS-mutant Malignant melanoma. JCO Precis Oncol (2020) 4:20–31. doi: 10.1200/po.19.00174
107. Valpione S, Gremel G, Mundra P, Middlehurst P, Galvani E, Girotti MR, et al. Plasma total cell-free DNA (cfDNA) is a surrogate biomarker for tumour burden and a prognostic biomarker for survival in metastatic melanoma patients. Eur J Cancer (2018) 88:1–9. doi: 10.1016/j.ejca.2017.10.029
108. Wong SQ, Raleigh JM, Callahan J, Vergara IA, Ftouni S, Hatzimihalis A, et al. Circulating tumor DNA analysis and functional imaging provide complementary approaches for comprehensive disease monitoring in metastatic melanoma. JCO Precis Oncol (2017) 1:1–14. doi: 10.1200/po.16.00009
109. Chang GA, Tadepalli JS, Shao Y, Zhang Y, Weiss S, Robinson E, et al. Sensitivity of plasma BRAFmutant and NRASmutant cell-free DNA assays to detect metastatic melanoma in patients with low RECIST scores and non-RECIST disease progression. Mol Oncol (2016) 10:157–65. doi: 10.1016/j.molonc.2015.09.005
110. Ito K, Schöder H, Teng R, Humm JL, Ni A, Wolchok JD, et al. Prognostic value of baseline metabolic tumor volume measured on (18)F-fluorodeoxyglucose positron emission tomography/computed tomography in melanoma patients treated with ipilimumab therapy. Eur J Nucl Med Mol Imaging (2019) 46:930–9. doi: 10.1007/s00259-018-4211-0
111. He WZ, Hu WM, Wang F, Rong YM, Yang L, Xie QK, et al. Comparison of mismatch repair status between primary and matched metastatic sites in patients with colorectal cancer. J Natl Compr Canc Netw (2019) 17:1174–83. doi: 10.6004/jnccn.2019.7308
112. Diaz LA Jr., Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature (2012) 486:537–40. doi: 10.1038/nature11219
113. Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, et al. Development of tumor mutation burden as an immunotherapy biomarker: utility for the oncology clinic. Ann Oncol (2019) 30:44–56. doi: 10.1093/annonc/mdy495
114. Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther (2017) 16:2598–608. doi: 10.1158/1535-7163.Mct-17-0386
115. Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science (2015) 348:124–8. doi: 10.1126/science.aaa1348
116. Cabel L, Riva F, Servois V, Livartowski A, Daniel C, Rampanou A, et al. Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: a proof-of-concept study. Ann Oncol (2017) 28:1996–2001. doi: 10.1093/annonc/mdx212
117. Willis J, Lefterova MI, Artyomenko A, Kasi PM, Nakamura Y, Mody K, et al. Validation of microsatellite instability detection using a comprehensive plasma-based genotyping panel. Clin Cancer Res (2019) 25:7035–45. doi: 10.1158/1078-0432.Ccr-19-1324
118. Tieng FYF, Abu N, Lee LH, Ab Mutalib NS. Microsatellite instability in colorectal cancer liquid biopsy-current updates on its potential in non-invasive detection, prognosis and as a predictive marker. Diagnostics (Basel) (2021) 11. doi: 10.3390/diagnostics11030544
119. Goodman AM, Holden KA, Jeong AR, Kim L, Fitzgerald KD, Almasri E, et al. Assessing CAR T-cell therapy response using genome-wide sequencing of cell-free DNA in patients with B-cell lymphomas. Transplant Cell Ther (2022) 28:30.e1–7. doi: 10.1016/j.jtct.2021.10.007
120. Frank MJ, Hossain NM, Bukhari A, Dean E, Spiegel JY, Claire GK, et al. Monitoring of circulating tumor DNA improves early relapse detection after axicabtagene ciloleucel infusion in large B-cell lymphoma: results of a prospective multi-institutional trial. J Clin Oncol (2021) 39:3034–43. doi: 10.1200/jco.21.00377
121. Mika T, Thomson J, Nilius-Eliliwi V, Vangala D, Baraniskin A, Wulf G, et al. Quantification of cell-free DNAfor the analysis of CD19-CAR-T cells during lymphoma treatment. Mol Ther Methods Clin Dev (2021) 23:539–50. doi: 10.1016/j.omtm.2021.10.009
122. Crisafulli G, Sartore-Bianchi A, Lazzari L, Pietrantonio F, Amatu A, Macagno M, et al. Temozolomide treatment alters mismatch repair and boosts mutational burden in tumor and blood of colorectal cancer patients. Cancer Discovery (2022) 12:1656–75. doi: 10.1158/2159-8290.Cd-21-1434
123. Stenzinger A, Allen JD, Maas J, Stewart MD, Merino DM, Wempe MM, et al. Tumor mutational burden standardization initiatives: Recommendations for consistent tumor mutational burden assessment in clinical samples to guide immunotherapy treatment decisions. Genes Chromosomes Cancer (2019) 58:578–88. doi: 10.1002/gcc.22733
124. Bratman SV, Yang SYC, Iafolla MAJ, Liu Z, Hansen AR, Bedard PL, et al. Personalized circulating tumor DNA analysis as a predictive biomarker in solid tumor patients treated with pembrolizumab. Nat Cancer (2020) 1:873–81. doi: 10.1038/s43018-020-0096-5
125. Zhang Q, Luo J, Wu S, Si H, Gao C, Xu W, et al. Prognostic and predictive impact of circulating tumor DNA in patients with advanced cancers treated with immune checkpoint blockade. Cancer Discovery (2020) 10:1842–53. doi: 10.1158/2159-8290.Cd-20-0047
126. Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature (2017) 545:446–51. doi: 10.1038/nature22364
127. Malla M, Loree JM, Kasi PM, Parikh AR. Using circulating tumor DNA in colorectal cancer: current and evolving practices. J Clin Oncol (2022) 40:2846–57. doi: 10.1200/jco.21.02615
128. Stadler JC, Belloum Y, Deitert B, Sementsov M, Heidrich I, Gebhardt C, et al. Current and future clinical applications of ctDNA in immuno-oncology. Cancer Res (2022) 82:349–58. doi: 10.1158/0008-5472.Can-21-1718
129. Mirza S, Bhadresha K, Mughal MJ, McCabe M, Shahbazi R, Ruff P, et al. Liquid biopsy approaches and immunotherapy in colorectal cancer for precision medicine: Are we there yet? Front Oncol (2022) 12:1023565. doi: 10.3389/fonc.2022.1023565
130. Oshi M, Katsuta E, Yan L, Ebos JML, Rashid OM, Matsuyama R, et al. A novel 4-gene score to predict survival, distant metastasis and response to neoadjuvant therapy in breast cancer. Cancers (Basel) (2020) 12. doi: 10.3390/cancers12051148
131. Perfetto SP, Ambrozak D, Nguyen R, Chattopadhyay PK, Roederer M. Quality assurance for polychromatic flow cytometry using a suite of calibration beads. Nat Protoc (2012) 7:2067–79. doi: 10.1038/nprot.2012.126
132. Wei C, Wang M, Gao Q, Yuan S, Deng W, Bie L, et al. Dynamic peripheral blood immune cell markers for predicting the response of patients with metastatic cancer to immune checkpoint inhibitors. Cancer Immunol Immunother (2023) 72:23–37. doi: 10.1007/s00262-022-03221-5
133. Nixon AB, Schalper KA, Jacobs I, Potluri S, Wang IM, Fleener C. Peripheral immune-based biomarkers in cancer immunotherapy: can we realize their predictive potential? J Immunother Cancer (2019) 7:325. doi: 10.1186/s40425-019-0799-2
134. Maecker HT, McCoy JP, Nussenblatt R. Standardizing immunophenotyping for the human immunology project. Nat Rev Immunol (2012) 12:191–200. doi: 10.1038/nri3158
135. Wu Q, Liu Z, Gao Z, Luo Y, Li F, Yang C, et al. KLF5 inhibition potentiates anti-PD1 efficacy by enhancing CD8(+) T-cell-dependent antitumor immunity. Theranostics (2023) 13:1381–400. doi: 10.7150/thno.82182
136. Rossi G, Russo A, Tagliamento M, Tuzi A, Nigro O, Vallome G, et al. Precision medicine for NSCLC in the era of immunotherapy: new biomarkers to select the most suitable treatment or the most suitable patient. Cancers (Basel) (2020) 12. doi: 10.3390/cancers12051125
137. Hogan SA, Courtier A, Cheng PF, Jaberg-Bentele NF, Goldinger SM, Manuel M, et al. Peripheral blood TCR repertoire profiling may facilitate patient stratification for immunotherapy against melanoma. Cancer Immunol Res (2019) 7:77–85. doi: 10.1158/2326-6066.Cir-18-0136
138. Valpione S, Galvani E, Tweedy J, Mundra PA, Banyard A, Middlehurst P, et al. Immune-awakening revealed by peripheral T cell dynamics after one cycle of immunotherapy. Nat Cancer (2020) 1:210–21. doi: 10.1038/s43018-019-0022-x
139. Poran A, Scherer J, Bushway ME, Besada R, Balogh KN, Wanamaker A, et al. Combined TCR repertoire profiles and blood cell phenotypes predict melanoma patient response to personalized neoantigen therapy plus anti-PD-1. Cell Rep Med (2020) 1:100141. doi: 10.1016/j.xcrm.2020.100141
140. Wang X, Muzaffar J, Kirtane K, Song F, Johnson M, Schell MJ, et al. T cell repertoire in peripheral blood as a potential biomarker for predicting response to concurrent cetuximab and nivolumab in head and neck squamous cell carcinoma. J Immunother Cancer (2022) 10. doi: 10.1136/jitc-2022-004512
141. Huang AC, Postow MA, Orlowski RJ, Mick R, Bengsch B, Manne S, et al. T-cell invigoration to tumour burden ratio associated with anti-PD-1 response. Nature (2017) 545:60–5. doi: 10.1038/nature22079
142. Oliveira G, Stromhaug K, Klaeger S, Kula T, Frederick DT, Le PM, et al. Phenotype, specificity and avidity of antitumour CD8(+) T cells in melanoma. Nature (2021) 596:119–25. doi: 10.1038/s41586-021-03704-y
143. Relecom A, Merhi M, Inchakalody V, Uddin S, Rinchai D, Bedognetti D, et al. Emerging dynamics pathways of response and resistance to PD-1 and CTLA-4 blockade: tackling uncertainty by confronting complexity. J Exp Clin Cancer Res (2021) 40:74. doi: 10.1186/s13046-021-01872-3
144. Kim KH, Cho J, Ku BM, Koh J, Sun JM, Lee SH, et al. The first-week proliferative response of peripheral blood PD-1(+)CD8(+) T cells predicts the response to anti-PD-1 therapy in solid tumors. Clin Cancer Res (2019) 25:2144–54. doi: 10.1158/1078-0432.Ccr-18-1449
145. Li S, Zhang C, Pang G, Wang P. Emerging blood-based biomarkers for predicting response to checkpoint immunotherapy in non-small-cell lung cancer. Front Immunol (2020) 11:603157. doi: 10.3389/fimmu.2020.603157
146. Kavanagh B, O'Brien S, Lee D, Hou Y, Weinberg V, Rini B, et al. CTLA4 blockade expands FoxP3+ regulatory and activated effector CD4+ T cells in a dose-dependent fashion. Blood (2008) 112:1175–83. doi: 10.1182/blood-2007-11-125435
147. Long SA, Buckner JH. CD4+FOXP3+ T regulatory cells in human autoimmunity: more than a numbers game. J Immunol (2011) 187:2061–6. doi: 10.4049/jimmunol.1003224
148. Deng L, Zhang H, Luan Y, Zhang J, Xing Q, Dong S, et al. Accumulation of foxp3+ T regulatory cells in draining lymph nodes correlates with disease progression and immune suppression in colorectal cancer patients. Clin Cancer Res (2010) 16:4105–12. doi: 10.1158/1078-0432.Ccr-10-1073
149. Wolf D, Wolf AM, Rumpold H, Fiegl H, Zeimet AG, Muller-Holzner E, et al. The expression of the regulatory T cell-specific forkhead box transcription factor FoxP3 is associated with poor prognosis in ovarian cancer. Clin Cancer Res (2005) 11:8326–31. doi: 10.1158/1078-0432.Ccr-05-1244
150. Farinha P, Al-Tourah A, Gill K, Klasa R, Connors JM, Gascoyne RD. The architectural pattern of FOXP3-positive T cells in follicular lymphoma is an independent predictor of survival and histologic transformation. Blood (2010) 115:289–95. doi: 10.1182/blood-2009-07-235598
151. Manjarrez-Orduño N, Menard LC, Kansal S, Fischer P, Kakrecha B, Jiang C, et al. Circulating T cell subpopulations correlate with immune responses at the tumor site and clinical response to PD1 inhibition in non-small cell lung cancer. Front Immunol (2018) 9:1613. doi: 10.3389/fimmu.2018.01613
152. Roselli M, Formica V, Cereda V, Jochems C, Richards J, Grenga I, et al. The association of clinical outcome and peripheral T-cell subsets in metastatic colorectal cancer patients receiving first-line FOLFIRI plus bevacizumab therapy. Oncoimmunology (2016) 5:e1188243. doi: 10.1080/2162402x.2016.1188243
153. Bencsikova B, Budinska E, Selingerova I, Pilatova K, Fedorova L, Greplova K, et al. Circulating T cell subsets are associated with clinical outcome of anti-VEGF-based 1st-line treatment of metastatic colorectal cancer patients: a prospective study with focus on primary tumor sidedness. BMC Cancer (2019) 19:687. doi: 10.1186/s12885-019-5909-5
154. Passardi A, Scarpi E, Cavanna L, Dall'Agata M, Tassinari D, Leo S, et al. Inflammatory indexes as predictors of prognosis and bevacizumab efficacy in patients with metastatic colorectal cancer. Oncotarget. (2016) 7:33210–9. doi: 10.18632/oncotarget.8901
155. Chang J, Lin G, Ye M, Tong D, Zhao J, Zhu D, et al. Decreased mean platelet volume predicts poor prognosis in metastatic colorectal cancer patients treated with first-line chemotherapy: results from mCRC biomarker study. BMC Cancer (2019) 19:15. doi: 10.1186/s12885-018-5252-2
156. Dell'Aquila E, Cremolini C, Zeppola T, Lonardi S, Bergamo F, Masi G, et al. Prognostic and predictive role of neutrophil/lymphocytes ratio in metastatic colorectal cancer: a retrospective analysis of the TRIBE study by GONO. Ann Oncol (2018) 29:924–30. doi: 10.1093/annonc/mdy004
157. Galindo-Pumariño C, Collado M, Herrera M, Peña C. Tumor microenvironment in metastatic colorectal cancer: the arbitrator in patients' Outcome. Cancers (Basel) (2021) 13. doi: 10.3390/cancers13051130
158. Manzoni M, Comolli G, Torchio M, Mazzini G, Danova M. Circulating endothelial cells and their subpopulations: role as predictive biomarkers in antiangiogenic therapy for colorectal cancer. Clin Colorectal Cancer (2015) 14:11–7. doi: 10.1016/j.clcc.2014.12.002
159. Rahbari NN, Schölch S, Bork U, Kahlert C, Schneider M, Rahbari M, et al. Prognostic value of circulating endothelial cells in metastatic colorectal cancer. Oncotarget (2017) 8:37491–501. doi: 10.18632/oncotarget.16397
160. Cima I, Kong SL, Sengupta D, Tan IB, Phyo WM, Lee D, et al. Tumor-derived circulating endothelial cell clusters in colorectal cancer. Sci Transl Med (2016) 8:345ra89. doi: 10.1126/scitranslmed.aad7369
161. Gootjes EC, Kraan J, Buffart TE, Bakkerus L, Zonderhuis BM, Verhoef C, et al. CD276-positive circulating endothelial cells do not predict response to systemic therapy in advanced colorectal cancer. Cells (2020) 9. doi: 10.3390/cells9010124
162. Ronzoni M, Manzoni M, Mariucci S, Loupakis F, Brugnatelli S, Bencardino K, et al. Circulating endothelial cells and endothelial progenitors as predictive markers of clinical response to bevacizumab-based first-line treatment in advanced colorectal cancer patients. Ann Oncol (2010) 21:2382–9. doi: 10.1093/annonc/mdq261
163. Malka D, Boige V, Jacques N, Vimond N, Adenis A, Boucher E, et al. Clinical value of circulating endothelial cell levels in metastatic colorectal cancer patients treated with first-line chemotherapy and bevacizumab. Ann Oncol (2012) 23:919–27. doi: 10.1093/annonc/mdr365
164. Matsusaka S, Mishima Y, Suenaga M, Terui Y, Kuniyoshi R, Mizunuma N, et al. Circulating endothelial progenitors and CXCR4-positive circulating endothelial cells are predictive markers for bevacizumab. Cancer (2011) 117:4026–32. doi: 10.1002/cncr.25977
165. Tieng FYF, Lee LH, Ab Mutalib NS. Deciphering colorectal cancer immune microenvironment transcriptional landscape on single cell resolution - A role for immunotherapy. Front Immunol (2022) 13:959705. doi: 10.3389/fimmu.2022.959705
166. De Simone M, Arrigoni A, Rossetti G, Gruarin P, Ranzani V, Politano C, et al. Transcriptional landscape of human tissue lymphocytes unveils uniqueness of tumor-infiltrating T regulatory cells. Immunity (2016) 45:1135–47. doi: 10.1016/j.immuni.2016.10.021
167. Xie N, Shen G, Gao W, Huang Z, Huang C, Fu L. Neoantigens: promising targets for cancer therapy. Signal Transduct Target Ther (2023) 8:9. doi: 10.1038/s41392-022-01270-x
168. Klebanoff CA, Rosenberg SA, Restifo NP. Prospects for gene-engineered T cell immunotherapy for solid cancers. Nat Med (2016) 22:26–36. doi: 10.1038/nm.4015
169. Dudley JC, Lin MT, Le DT, Eshleman JR. Microsatellite instability as a biomarker for PD-1 blockade. Clin Cancer Res (2016) 22:813–20. doi: 10.1158/1078-0432.Ccr-15-1678
170. Yarchoan M, Johnson BA 3rd, Lutz ER, Laheru DA, Jaffee EM. Targeting neoantigens to augment antitumour immunity. Nat Rev Cancer (2017) 17:209–22. doi: 10.1038/nrc.2016.154
171. Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science (2017) 357:409–13. doi: 10.1126/science.aan6733
172. Kreiter S, Vormehr M, van de Roemer N, Diken M, Löwer M, Diekmann J, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature (2015) 520:692–6. doi: 10.1038/nature14426
173. Parkhurst MR, Robbins PF, Tran E, Prickett TD, Gartner JJ, Jia L, et al. Unique neoantigens arise from somatic mutations in patients with gastrointestinal cancers. Cancer Discovery (2019) 9:1022–35. doi: 10.1158/2159-8290.Cd-18-1494
174. Sellars MC, Wu CJ, Fritsch EF. Cancer vaccines: Building a bridge over troubled waters. Cell (2022) 185:2770–88. doi: 10.1016/j.cell.2022.06.035
175. Hanada K, Yewdell JW, Yang JC. Immune recognition of a human renal cancer antigen through post-translational protein splicing. Nature (2004) 427:252–6. doi: 10.1038/nature02240
176. Cobbold M, de la Peña H, Norris A, Polefrone JM, Qian J, English AM, et al. MHC class I-associated phosphopeptides are the targets of memory-like immunity in leukemia. Sci Transl Med (2013) 5:203ra125. doi: 10.1126/scitranslmed.3006061
177. Malaker SA, Penny SA, Steadman LG, Myers PT, Loke JC, Raghavan M, et al. Identification of glycopeptides as posttranslationally modified neoantigens in leukemia. Cancer Immunol Res (2017) 5:376–84. doi: 10.1158/2326-6066.Cir-16-0280
178. Oliveira-Ferrer L, Legler K, Milde-Langosch K. Role of protein glycosylation in cancer metastasis. Semin Cancer Biol (2017) 44:141–52. doi: 10.1016/j.semcancer.2017.03.002
179. Brentville VA, Metheringham RL, Gunn B, Symonds P, Daniels I, Gijon M, et al. Citrullinated vimentin presented on MHC-II in tumor cells is a target for CD4+ T-cell-mediated antitumor immunity. Cancer Res (2016) 76:548–60. doi: 10.1158/0008-5472.Can-15-1085
180. Marijt KA, Blijleven L, Verdegaal EME, Kester MG, Kowalewski DJ, Rammensee HG, et al. Identification of non-mutated neoantigens presented by TAP-deficient tumors. J Exp Med (2018) 215:2325–37. doi: 10.1084/jem.20180577
181. Yewdell JW, Nicchitta CV. The DRiP hypothesis decennial: support, controversy, refinement and extension. Trends Immunol (2006) 27:368–73. doi: 10.1016/j.it.2006.06.008
182. Grimaldi A, Cammarata I, Martire C, Focaccetti C, Piconese S, Buccilli M, et al. Combination of chemotherapy and PD-1 blockade induces T cell responses to tumor non-mutated neoantigens. Commun Biol (2020) 3:85. doi: 10.1038/s42003-020-0811-x
183. Mehnert JM, Monjazeb AM, Beerthuijzen JMT, Collyar D, Rubinstein L, Harris LN. The challenge for development of valuable immuno-oncology biomarkers. Clin Cancer Res (2017) 23:4970–9. doi: 10.1158/1078-0432.Ccr-16-3063
184. Kilgour E, Rothwell DG, Brady G, Dive C. Liquid biopsy-based biomarkers of treatment response and resistance. Cancer Cell (2020) 37:485–95. doi: 10.1016/j.ccell.2020.03.012
185. Biswas D, Ganeshalingam J, Wan JCM. The future of liquid biopsy. Lancet Oncol (2020) 21:e550. doi: 10.1016/s1470-2045(20)30687-2
186. Neumann MHD, Bender S, Krahn T, Schlange T. ctDNA and CTCs in liquid biopsy - current status and where we need to progress. Comput Struct Biotechnol J (2018) 16:190–5. doi: 10.1016/j.csbj.2018.05.002
187. Ding Y, Li W, Wang K, Xu C, Hao M, Ding L. Perspectives of the application of liquid biopsy in colorectal cancer. BioMed Res Int (2020) 2020:6843180. doi: 10.1155/2020/6843180
188. Morganti S, Tarantino P, Ferraro E, D'Amico P, Duso BA, Curigliano G. Next generation sequencing (NGS): A revolutionary technology in pharmacogenomics and personalized medicine in cancer. Adv Exp Med Biol (2019) 1168:9–30. doi: 10.1007/978-3-030-24100-1_2
189. Schøler LV, Reinert T, Ørntoft MW, Kassentoft CG, Árnadóttir SS, Vang S, et al. Clinical implications of monitoring circulating tumor DNA in patients with colorectal cancer. Clin Cancer Res (2017) 23:5437–45. doi: 10.1158/1078-0432.Ccr-17-0510
190. Cabel L, Proudhon C, Gortais H, Loirat D, Coussy F, Pierga JY, et al. Circulating tumor cells: clinical validity and utility. Int J Clin Oncol (2017) 22:421–30. doi: 10.1007/s10147-017-1105-2
191. Reinert T, Schøler LV, Thomsen R, Tobiasen H, Vang S, Nordentoft I, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut (2016) 65:625–34. doi: 10.1136/gutjnl-2014-308859
192. Zhou H, Zhu L, Song J, Wang G, Li P, Li W, et al. Liquid biopsy at the frontier of detection, prognosis and progression monitoring in colorectal cancer. Mol Cancer (2022) 21:86. doi: 10.1186/s12943-022-01556-2
193. Liu Y, Zugazagoitia J, Ahmed FS, Henick BS, Gettinger SN, Herbst RS, et al. Immune cell PD-L1 colocalizes with macrophages and is associated with outcome in PD-1 pathway blockade therapy. Clin Cancer Res (2020) 26:970–97. doi: 10.1158/1078-0432.Ccr-19-1040
194. Lin SY, Chang SC, Lam S, Irene Ramos R, Tran K, Ohe S, et al. Prospective molecular profiling of circulating tumor cells from patients with melanoma receiving combinatorial immunotherapy. Clin Chem (2020) 66:169–77. doi: 10.1373/clinchem.2019.307140
195. Dou X, Hua Y, Chen Z, Chao F, Li M. Extracellular vesicles containing PD-L1 contribute to CD8+ T-cell immune suppression and predict poor outcomes in small cell lung cancer. Clin Exp Immunol (2022) 207:307–17. doi: 10.1093/cei/uxac006
Keywords: liquid biopsy, immunotherapy, circulating tumor DNA (ctDNA), Circulating tumor cells (CTC), colorectal cancer, neoantigen, mass spectrometry, flow cytometry
Citation: Yao S, Han Y, Yang M, Jin K and Lan H (2023) Integration of liquid biopsy and immunotherapy: opening a new era in colorectal cancer treatment. Front. Immunol. 14:1292861. doi: 10.3389/fimmu.2023.1292861
Received: 12 September 2023; Accepted: 03 November 2023;
Published: 21 November 2023.
Edited by:
Patrick Schmidt, National Center for Tumor Diseases (NCT), GermanyReviewed by:
Veronika Vymetalkova, Institute of Experimental Medicine (ASCR), CzechiaCopyright © 2023 Yao, Han, Yang, Jin and Lan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ketao Jin, amlua2V0YW8yMDAxQHpqdS5lZHUuY24=; Huanrong Lan, bGFuaHIyMDE4QDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.