
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
CLINICAL TRIAL article
Front. Immunol. , 27 November 2023
Sec. Vaccines and Molecular Therapeutics
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1292568
 Margherita Rosati1
Margherita Rosati1 Evangelos Terpos2
Evangelos Terpos2 Philip Homan3
Philip Homan3 Cristina Bergamaschi1
Cristina Bergamaschi1 Sevasti Karaliota1,4
Sevasti Karaliota1,4 Ioannis Ntanasis-Stathopoulos2
Ioannis Ntanasis-Stathopoulos2 Santhi Devasundaram1
Santhi Devasundaram1 Jenifer Bear1
Jenifer Bear1 Robert Burns1
Robert Burns1 Tina Bagratuni2
Tina Bagratuni2 Ioannis P. Trougakos5
Ioannis P. Trougakos5 Meletios A. Dimopoulos2
Meletios A. Dimopoulos2 George N. Pavlakis6
George N. Pavlakis6 Barbara K. Felber1*
Barbara K. Felber1*Introduction: Cytokines and chemokines play an important role in shaping innate and adaptive immunity in response to infection and vaccination. Systems serology identified immunological parameters predictive of beneficial response to the BNT162b2 mRNA vaccine in COVID-19 infection-naïve volunteers, COVID-19 convalescent patients and transplant patients with hematological malignancies. Here, we examined the dynamics of the serum cytokine/chemokine responses after the 3rd BNT162b2 mRNA vaccination in a cohort of COVID-19 infection-naïve volunteers.
Methods: We measured serum cytokine and chemokine responses after the 3rd dose of the BNT162b2 mRNA (Pfizer/BioNtech) vaccine in COVID-19 infection-naïve individuals by a chemiluminescent assay and ELISA. Anti-Spike binding antibodies were measured by ELISA. Anti-Spike neutralizing antibodies were measured by a pseudotype assay.
Results: Comparison to responses found after the 1st and 2nd vaccinations showed persistence of the coordinated responses of several cytokine/chemokines including the previously identified rapid and transient IL-15, IFN-γ, CXCL10/IP-10, TNF-α, IL-6 signature. In contrast to the transient (24hrs) effect of the IL-15 signature, an inflammatory/anti-inflammatory cytokine signature (CCL2/MCP-1, CCL3/MIP-1α, CCL4/MIP-1β, CXCL8/IL-8, IL-1Ra) remained at higher levels up to one month after the 2nd and 3rd booster vaccinations, indicative of a state of longer-lasting innate immune change. We also identified a systemic transient increase of CXCL13 only after the 3rd vaccination, supporting stronger germinal center activity and the higher anti-Spike antibody responses. Changes of the IL-15 signature, and the inflammatory/anti-inflammatory cytokine profile correlated with neutralizing antibody levels also after the 3rd vaccination supporting their role as immune biomarkers for effective development of vaccine-induced humoral responses.
Conclusion: These data revealed that repeated SARS-Cov-2 BNT162b2 mRNA vaccination induces both rapid transient as well as longer-lasting systemic serum cytokine changes associated with innate and adaptive immune responses.
Clinical trial registration: Clinicaltrials.gov, identifier NCT04743388.
Cytokines and chemokines play an important role in shaping innate and adaptive immunity in response to infection and vaccination. Dissecting early innate vaccine signatures may predict immunogenicity to help optimize the efficacy of mRNA and other vaccine strategies. Several studies identified immune signatures early after Yellow Fever, HIV-Ade5, HIV ALVAC, Ebola rVSV-ZEBOV, and trivalent influenza (TIV) vaccination (1–7). Understanding the effect of COVID-19 BNT162b2 mRNA (Pfizer/BioNtech) (8) vaccine on innate and trained immunity responses [reviewed in (9)] is of great interest and contributes to the modulation of adaptive immunity. We and others reported on early effects after BNT162b2 mRNA vaccination in humans using systems serology (10, 11), multi-omic approaches and flow-based assays (12–14). Systems serology identified immunological parameters predictive of beneficial response of the BNT162b2 mRNA vaccine in COVID-19 infection-naïve volunteers (10), COVID-19 convalescent patients (10) and transplant patients with hematological malignancies (11). We identified a systemic signature, detectable at 24hrs after the 1st vaccination, comprising IL-15, IFN-γ, and CXCL10/IP-10, which was expanded to include tumor necrosis factor alpha (TNF-α) and IL-6 at 24hrs after the 2nd vaccination (10). We further reported a correlation of changes of IL-15 and IFN-γ levels with binding antibody titers in BNT162b2 mRNA vaccine recipients (10). In contrast to vaccination of a cohort of healthy participants, patients with hematological malignancies who had a lower anti-Spike response, also had a diminished systemic cytokine response (IFN-γ, IL-15 and CXCL10/IP-10), and this correlated with the lower anti-Spike antibody levels (11). On the other hand, in patients who failed to develop antibodies, the innate systemic response showed a lack of the IL-15/IFN-γ signature with responses dominated by CCL8/IL-8 and CCL3/MIP-1α (11). Thus, we concluded that successful development of an anti-Spike immune response was associated with a robust transient activation of the IL-15 signature (10, 11). Similar to our findings after BTN162b2 mRNA vaccination, Andersen-Nissen et al. (4) reported that a different vaccine using a non-replicating vaccinia vector expressing HIV antigens (ALVAC-HIV) induced a signature of serum cytokines featuring IFN-γ, IL-15, and CXCL10/IP-10 in humans. Multi-omic approaches further reported an involvement of myeloid cells in response to the BNT162b2 mRNA vaccination in humans and mice (12–15).
In this report, we examined the dynamics of the serum cytokine/chemokine responses after the 3rd BNT162b2 mRNA vaccination in a cohort of COVID-19 infection-naïve volunteers. In addition to the previously identified cytokine/chemokine responses (11), we detected transient induction of CXCL13, a biomarker for germinal center activation and a key regulator of B cells (16), found only after the 3rd vaccination. In addition a set of pro-inflammatory and anti-inflammatory cytokines was detectable for up to one month post the 2nd and the 3rd vaccination. Together, these data revealed that repeated BNT162b2 mRNA vaccination induced both rapid/transient as well as longer-lasting cytokine/chemokine changes. Changes of some biomarkers, including IL-15, also correlated with vaccine-induced neutralizing antibody responses supporting an association of innate and adaptive immune responses.
This is a prospective study (NCT04743388) that evaluates the kinetics of antibodies against SARS-CoV-2 as well as the kinetics of serum cytokines and chemokines associated with the immune response in volunteers receiving the BNT162b2 vaccine against SARS-Cov-2 (Comirnaty™), initiated on January 4, 2021, in Greece (10). Volunteer donors (Table 1) were tested in the period January to December 2021 in the vaccine center of Alexandra General Hospital in Athens, Greece. Major inclusion criteria for participation in this study included: (i) age above 18 years; (ii) ability to sign the informed consent form, and (iii) eligibility for vaccination, according to the national (Greek) program for COVID-19 vaccination (i.e., individuals who had no serious allergy problem and especially they have not been hospitalized due to a serious allergic reaction (anaphylaxis). Major exclusion criteria included the presence of: (i) autoimmune disorder under immunosuppressive therapy; (ii) active malignant disease and (iii) end-stage renal disease, as previously described (10). Individuals with documented (PCR, ELISA) prior COVID-19 infection were excluded from this report. The study recruited volunteers for vaccination 1 (n=63), vaccination 2 (n=73) and vaccination 3 (n=44). The vaccinations were administered at day 1, day 22, and month 9. The present study included frequent serum collections but did not include collection of PBMC. All study procedures were carried out in accordance with the declaration of Helsinki (18th World Medical Association Assembly), its subsequent amendments, the Greek regulations and guidelines, as well as the good clinical practice guidelines (GCP) as defined by the International Conference of Harmonization. The study was also approved by the local ethic committee of Alexandra General (no 15/23 December 2020).
Serum was collected within 4 hours from blood collection as detailed in Figure 1A and stored at -80°C until the day of measurement. Sera collected after 1st and 2nd vaccination were part of a previous report (10). An in-house ELISA assay measuring IgG (19, 20) was used to detect Ab endpoint titers against the complete trimeric ancestral WA1/2020 Spike protein (21). The Spike pseudotyped pHIV-1NLΔEnv-Nanoluc assay (22, 23) was described (17, 19) measuring eight 4-fold serial dilutions starting at 1:50. Neutralization assays were performed using 8 dilutions of heat-inactivated sera (1:40 to 1: 655,360) in triplicates in HEK293T/ACE2wt cells and luciferase levels in the cell lysates were measured. Neutralization was measured against the WA1 and Omicron BA.1 Spike as well as against the BA.5, BQ.1.1, XBB, and XBB.1.5 Spike proteins and these data were previously reported (17, 18). Ab titers and 50% Inhibitory Dose (ID50) of NAb were calculated using GraphPad Prism version 9.4.1 for MacOS X (GraphPad Software, Inc, La Jolla, CA).
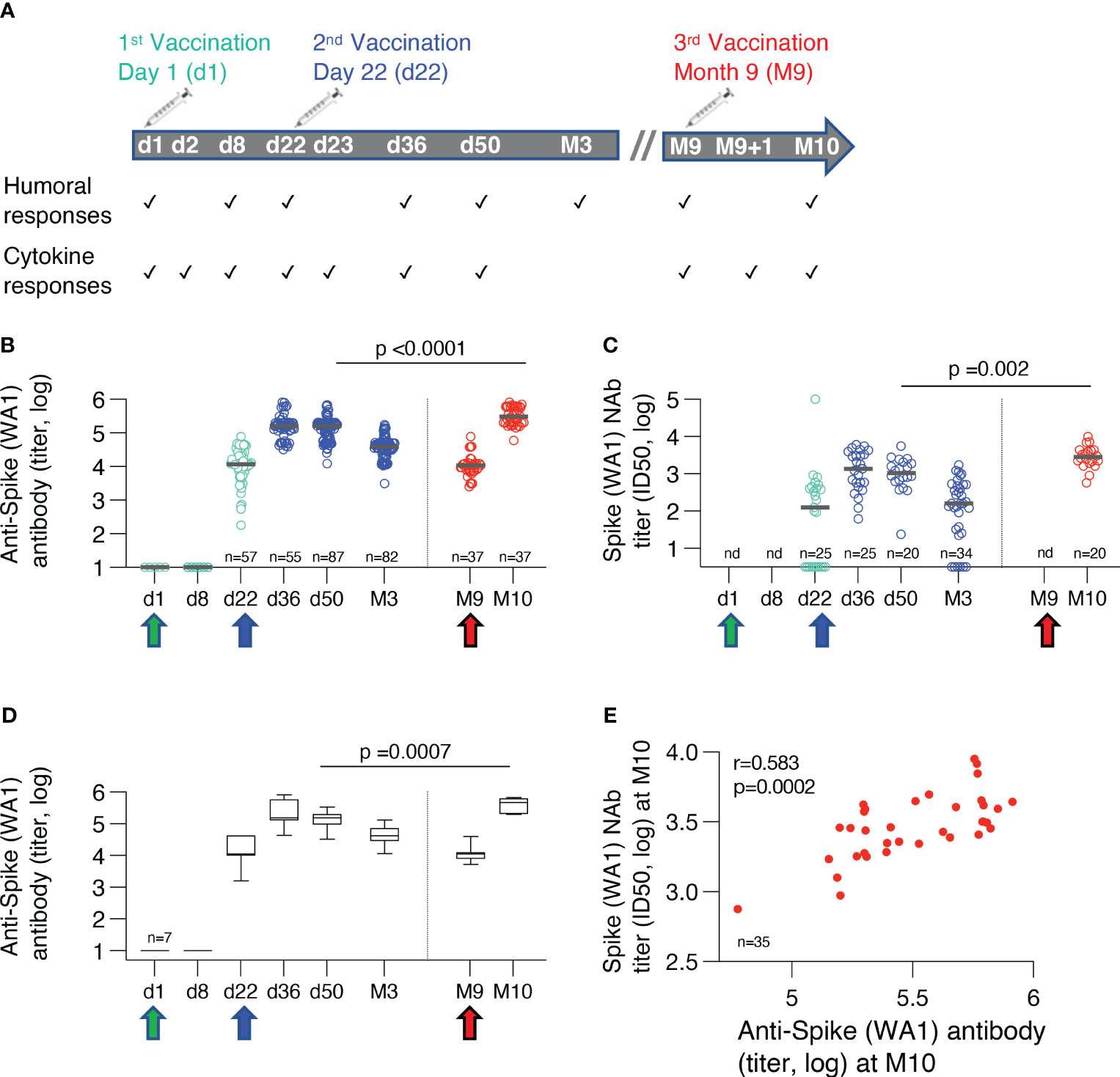
Figure 1 BNT162b2 mRNA cohorts and humoral immune responses. (A) Time points of measurements of adaptive and innate immune responses over the course of three BNT162b2 mRNA vaccinations. (B) Binding antibodies against ancestral trimeric Spike WA1 were measured by ELISA. Antibody titers from some individual time points have been reported previously (10, 17, 18). The Ab titers were compared at one month after the 2nd (n=87) and the 3rd vaccination (n=36). The p-value is from unpaired, non-parametric t test, Mann-Whitney. Median values are shown with black bars. (C) Neutralizing antibodies (NAb) were assessed using HIV-1NLΔEnv-NanoLuc derived pseudotype virus assay carrying Wuhan-Hu-1 Spike mutant D614G. The NAb titers were compared at one month after the 2nd (n=25) and the 3rd vaccination (n=20). The p-value is from unpaired, non-parametric t test, Mann-Whitney. nd, not done. Median values are shown with black bars. (D) Binding antibodies against ancestral trimeric Spike WA1 were measured by ELISA in a subset of vaccine recipients for whom sequential samples were available for all the time points. Median values are shown with black bars. (E) Correlation of neutralizing (WA1 D614G) and binding Ab titers to WA1 measured at one month after the booster vaccination (Spearman r=0.583; p=0.002).
Cytokine/chemokine levels (pg/ml) of 53 biomarkers were measured in sera using the chemiluminescent assay V-PLEX Human Biomarker Assay kit (Meso Scale Diagnostics LLC, Maryland, US), as listed in Table 2. Biomarkers which did not show changes at the time points analyzed or were below the threshold of the assay are listed. CXCL13 was measured by commercial ELISA (Invitrogen cat# EHCXCL13) using 1:2 diluted serum samples. Biomarker levels falling below the detection limit/standard range were removed if absent in more than 50% of the samples or adjusted to 50% of the lowest standard point or detection limits.
Biomarker analysis was performed with a workflow written in R and through a user interface developed on the Foundry Platform (Palantir Technologies). Heatmaps of 26 analytes represent the log2 fold change in concentration between time points. Analytes with a False Discovery Rate (FDR) <0.05 were considered significantly changed. Significant changes in analyte concentrations after Vaccine 2 (d22,d23,d36,d50) and Vaccine 3(M9, M9+d1, M10) a was determined using non-parametric Anova (Friedman test). The pairwise inter-relationship between analytes were identified by calculating the Spearman correlation between analytes using the log2 fold-changes in concentration after vaccine 2 (d23 vs d22) and vaccine 3 (M9+d1 vs M9) across each patient. Analyte pairs with a Spearman’s correlation coefficient < 0.005 were considered significant relationships. Univariate correlations for analytes were identified by calculating the spearman correlation between the analytes log2 fold-change in concentration after vaccine 2 (d23 vs d22) and vaccine 3 (M9+d1 vs M9) and the corresponding NAb levels for each patient at the vaccine time point. Analytes with a Spearman’s correlation coefficient < 0.05 were considered significant.
COVID-19 infection-naïve individuals were enrolled to receive three Pfizer/BioNtech BNT162b2 mRNA vaccinations between January and November 2021 (NCT04743388), administered at day 1 (n=63), day 22 (n=73) and at month 9 (M9, n=44). Cohorts receiving the two priming vaccinations and the 3rd booster vaccination had similar age and gender distribution (Table 1). Sequential serum samples were collected on the days of vaccination (d1, d22, M9, respectively) and after each vaccination which included 24hrs later (d2, d23, M9 + 1d, respectively), as well as on d36, d50, month 3 (M3) after the 2nd vaccination and one month (M10) after the 3rd vaccination (Figure 1A).
Humoral immune responses were measured against the ancestral trimeric WA1 Spike by ELISA. Neutralizing Ab was measured using the HIV-derived pseudotype virus assay against WA1 Spike. The cross-sectional humoral response analysis showed robust responses after the 2nd vaccination followed by the expected contraction of about 1 log in endpoint Ab titers over the 8 months of follow-up. Of note, this analysis showed that the 3rd vaccination at M9 resulted in significantly higher Spike-specific binding (Figure 1B) and neutralizing antibodies (NAb) (Figure 1C) compared to the 2nd vaccination. Similar to the cross-sectional evaluation, we made a sequential analysis of a subset of vaccinees (n=7) for whom samples were available for all the time points between the 2nd and 3rd vaccinations (Figure 1D). This analysis corroborated the cross-sectional analysis (Figure 1B) showing significantly higher humoral responses at M10 compared to d50, comparing the responses at one month after each vaccination. Together, the 3rd vaccination provided a significant enhancement of the humoral immunity. The previously identified correlation of binding and neutralizing Ab was also maintained after the 3rd vaccination (Figure 1E). Overall, the 3rd vaccination provided a beneficial increase in humoral immunity.
Next, we performed a sequential analysis of serum cytokine/chemokine levels after each BNT162b2 mRNA administration both at the day of vaccination and 24hrs later (see Figure 1A). Sera were analyzed using the Meso Scale Discovery platform (V-plex, 54-Plex kit) and an ELISA assay (Invitrogen) to measure CXCL13. We previously reported effects in 58 participants after the 1st and 2nd vaccination using a smaller panel of analytes (10). The current analysis included a larger dataset with 7 additional biomarkers, including CXCL13, IL-10 and IL-27 (Table 2) after the 2nd and 3rd vaccination, including 62 participants after the 1st vaccination, 72 participants after the 2nd vaccination (of which 62 were also analyzed after the 1st vaccination) and 44 participants after the 3rd vaccination (of which 10 were also analyzed after vaccinations 1 and 2) (Table 1).
The fold changes in cytokine/chemokine levels comparing the values at the day of vaccine administration and 24hrs later were compared after the 1st (d2 vs d1), 2nd (d23 vs d22) and 3rd (M9 + 1d vs M9) vaccinations (Figures 2A, B). Of the 54 analytes, 26 had significant changes over the 24-hr-follow-up, 11 analytes had no or marginal changes and 17 were below the threshold of detection (Table 2). Biomarkers were grouped by hierarchical clustering based on average fold change at each vaccination (Figure 2B). Biomarkers in cluster 1 showed no or low responses after the 2nd vaccination but strong responses after the 1st and 3rd vaccinations; cluster 2 showed strong responses both after the 2nd and 3rd vaccinations, whereas cluster 3 showed a stronger response after the 2nd vaccination.
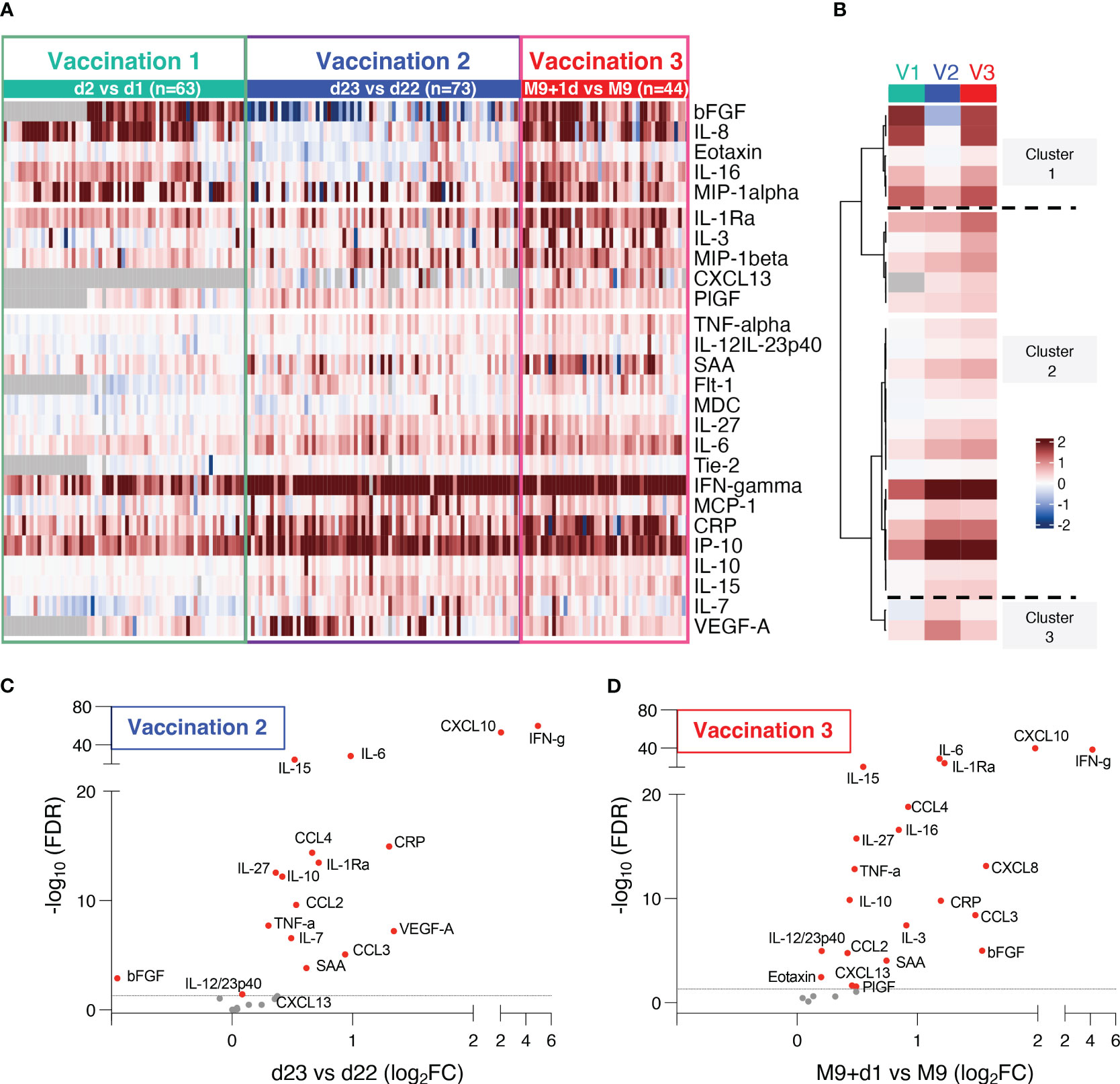
Figure 2 Serum cytokine and chemokine levels after BNT162b2 mRNA vaccination. (A, B) The heatmap (A) shows comparison of log2 fold changes of 26 cytokines and chemokines measured after the 1st and 2nd and 3rd vaccination. Changes compared samples collected 24hrs after vaccination to the respective day of vaccination (d2 vs d1, d23 vs d22, M9+d1 vs M9). Grey cells denote missing values for five analytes not measured for 22 participants at d1 and d2. The analysis after the 1st and 2nd vaccinations includes additional data from 7 analytes (CXCL13, bFGF, Flt-1, IL-10, IL-27, PIGF, Tie-2) which were not previously reported (10). (B) Mean log2 fold changes for each analyte and each group from data shown in (A, C, D) Volcano plots depict differentially expressed analytes after the 2nd vaccination (d23 vs d22) and (D) the 3rd vaccination (M9d+1 vs M9) using adjusted p values. Red dots indicate significant changes (FDR<0.05) capturing 17 analytes after the 2nd (C) and 21 analytes after the 3rd vaccination (D). Horizontal broken line represents FDR-adjusted p-values=0.05.
The differentially expressed biomarkers are shown in volcano plots after the 2nd vaccination (Figure 2C) and the 3rd vaccination (Figure 2D). Of the 26 analytes, significant changes were found in 17 analytes after the 2nd vaccination, and in 21 analytes after the 3rd vaccination. Absolute levels of individual biomarkers after the 2nd and 3rd vaccination are shown (Figure 3). Consistent with the rapid response after the 2nd vaccination, activation of several cytokines including IL-15, IFN-γ, CXCL10/IP-10, IL-6, TNF-α, CCL4/MIP-1β, IL-1Ra, IL-10, IL-27 were detected also after the 3rd vaccination.
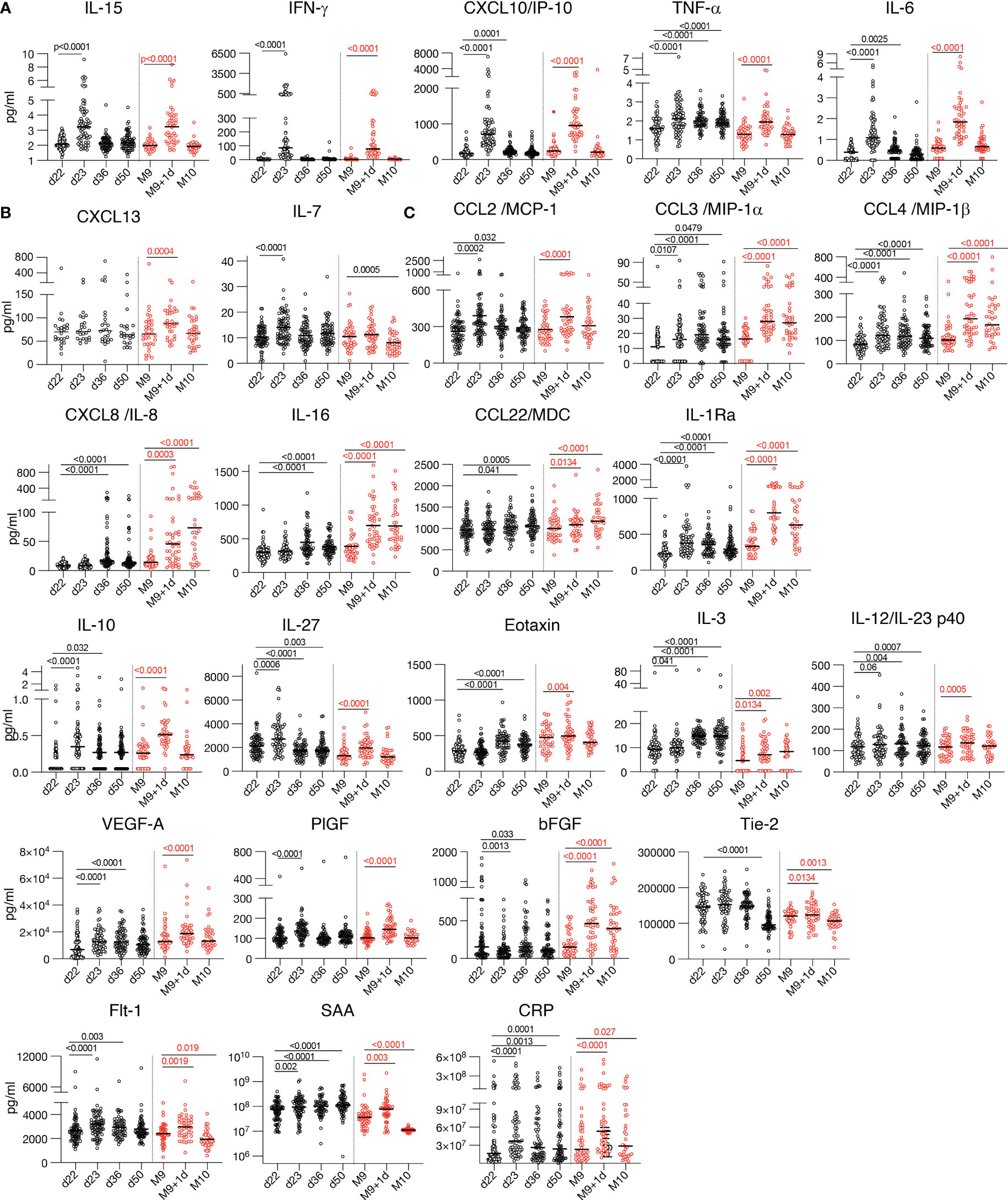
Figure 3 Cytokine and chemokine levels over time. Cytokine and chemokine measurements are shown over time using the Meso Scale Discovery assay and the ELISA for CXCL13 at the day of vaccination (d22, M9) and at one day after the 2nd vaccination (d23) and the 3rd vaccination (M9 + 1d). Additional measurements at 2 and 4 weeks (d36 and d50) after the 2nd vaccination and 4 weeks (M10) after the 3rd vaccination were plotted. Values (pg/ml) from individual vaccine recipients are shown after the 2nd vaccination (black symbols and after the 3rd vaccination (red symbols) and median values are shown (black lines). The p values (GraphPad Prism) are from Anova (Friedman test) for the 58 participants having 4 over time paired samples available after the 2nd vaccination and the 36 participants having 3 over time samples after the 3rd vaccination, respectively. The plots show the absolute values of all measurements: d22, d23 (n=72); d36 (n=62); d50 (n=64); M9 and M9 + 1d (n=44) and M10 (n=36). (A-C) Serum levels of 17 selected analytes among 26 analytes showing changes after the 2nd and 3rd vaccination are plotted over time: (A) cytokines belonging to the previously identified IL-15 signature; (B) Germinal center activation biomarkers CXCL13 and IL-7; (C) cytokine with pro-inflammatory, anti-inflammatory or dual function.
Additionally, we further found a significant transient increase of CXCL13 only after the 3rd vaccination (Figures 2, 3B). CXCL13 is a biomarker for germinal center activation and a key regulator of B cells (16) [reviewed in (24)]. This finding is in agreement with the significantly higher humoral immune response found after the 3rd vaccination (Figures 1B, C), suggesting further stimulation of B-cell development.
We also found significant increases of several other chemokines and cytokines produced by myeloid cells [reviewed in (25, 26)], including CCL2/MCP-1, CCL3/MIP-1α, CCL4/MIP-1β, CXCL8/IL-8, CCL22/MDC, and the anti-inflammatory cytokine IL-1Ra (Figures 2, 3). A strong transient activation of IL-1Ra was also found. The IL-1/IL-1Ra regulatory axis plays a role in the adaptive immune response development (27, 28) with the pro-inflammatory IL-1 acting as master cytokine in inflammation (29–31). Although no systemic levels of the IL-1 cytokines, IL-1α or IL-1β, were found one day later (Table 2), we cannot exclude faster and transient changes of these biomarkers.
IL-12/23p40, involved in the development and modulation of adaptive responses (32), was also significantly increased (Figures 2, 3). Changes in the levels of the IL-12/23p40 chain, which is part of heterodimeric IL-12p70 and IL-23 cytokines, could reflect their respective expression. Although ex vivo stimulation of PBMC from vaccine recipients with Spike peptide resulted in IL-12p70 expression (8), systemic serum levels of IL-12p70 as well as of IL-23 remained below the threshold of detection in mRNA/LNP vaccinated humans (Table 2).
The 3rd vaccination also induced a significant increase of biomarkers having a pro-inflammatory role including CXCL10/IP-10, IL-16; anti-inflammatory function (IL-1Ra, IL-10, IL-3, Eotaxin) or both (IL-27) (Figures 2, 3) involved in shaping the innate and adaptive immune response. Molecules released in response to inflammation [reviewed in (33)] included acute phase proteins serum amyloid A (SAA) and C-reactive protein CRP (Figure 3). Significant changes were found in the angiogenic cytokines VEGF-A, Flt-1, bFGF, as well as PIGF and Tie-2, two cytokines which were also reported to have macrophage activating function (34) (Figure 3).
Of note, a set of analytes including CXCL8/IL-8, IL-16, Eotaxin and bFGF which showed no changes after the 2nd vaccination, showed significant increases within 24hrs both after the 1st and the 3rd vaccinations (Figures 2A, B, 3). This discrepancy may be attributed, at least in part, to the short time between 1st and 2nd vaccination which could influence response kinetics of these biomarkers.
In addition to the cross-sectional analysis of the transient changes over 24hrs after each vaccination, a sequential analysis (Figures 4A, B) was performed. Biomarker changes were recorded from day 1 (day 1 of 1st vaccination) to 3 weeks later (d22, day of 2nd vaccination), to day 36 (2 weeks after the 2nd vaccination), to day 50 (4 weeks after the 2nd vaccination), to 8 months later at M9 (day of 3rd vaccination);, and to M10 (4 weeks after the 3rd vaccination). A subset of patients (n=10) had samples available for the complete time range starting at day 1 of the study, reported at M9 and M10.
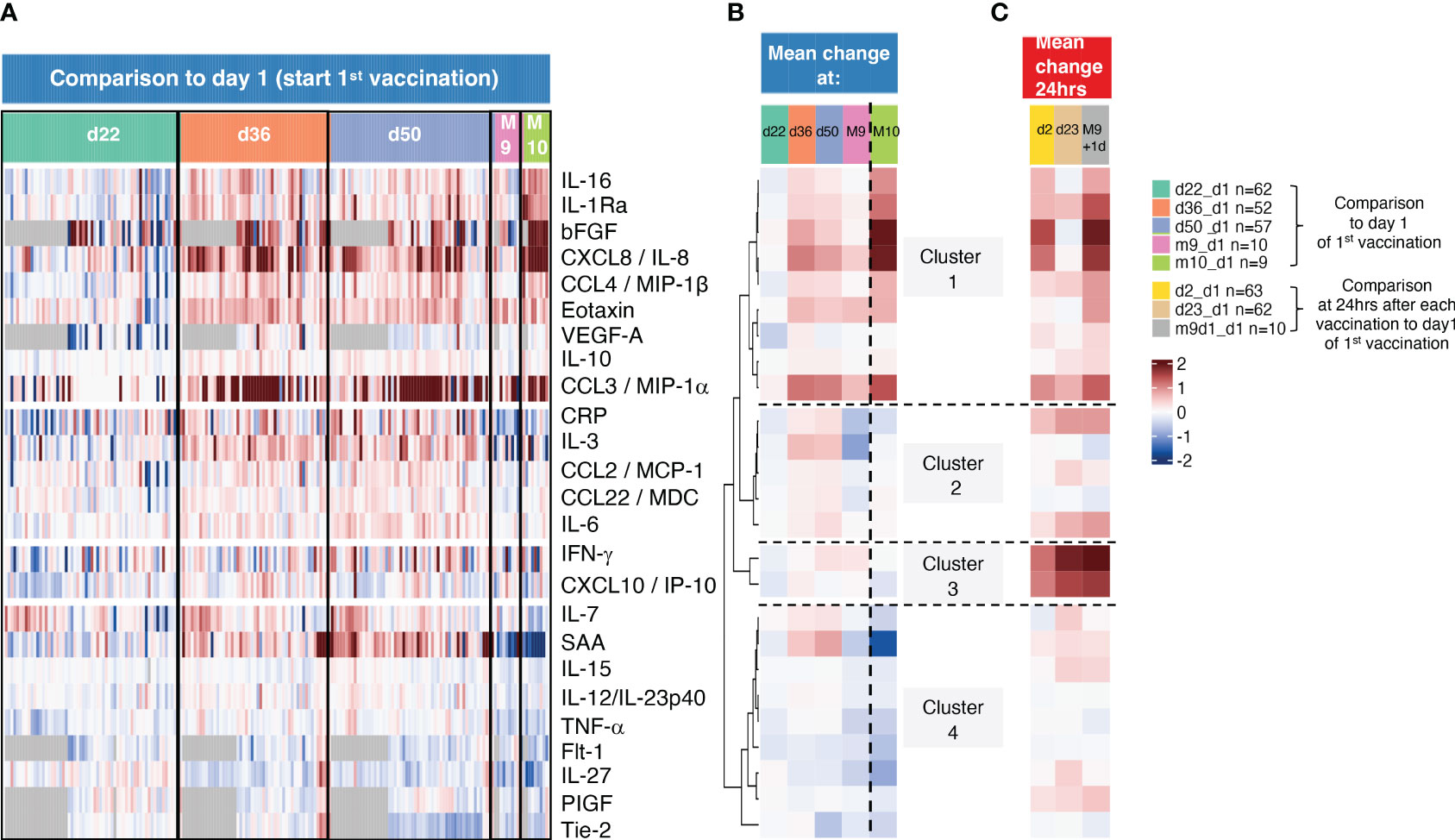
Figure 4 Sequential analysis of biomarkers with longer-lasting changes. (A) The heatmap shows log2 fold changes comparing all sequential measurements to the day 1 measurements before the 1st vaccination. d22 samples were collected before 2nd vaccination, d36, d50, and M9 before the 3rd vaccination and M10, one month after the 3rd vaccination. Grey cells denote missing values for five analytes not measured for 22 participants. (B, C) Mean log2 fold changes for each analyte and for (B) data obtained d22, d36, d50, M9 and M10 from (B, C) and data obtained 24hrs after the 1st, 2nd, and the 3rd vaccination (d2, d23, m9+d1).
Sequential changes are summarized (Figure 4B). By d22, all biomarkers reached baseline, as we previously observed (10). A set of biomarkers including primarily myeloid cell associated inflammatory and anti-inflammatory cytokines (CXCL8/IL-8, IL-16, CCL3/MIP-1α, CCL4/MIP-1β, IL-1Ra, Eotaxin, IL-10 and the angiogenic cytokines bFGF and VEGF-A) in clusters 1 and 2 continued to remain upregulated at d36 and d50. However, their levels were greatly reduced reaching baseline by M9 (8 months after the 2nd vaccination), except CXCL8/IL-8, CCL3/MIP-1α, Eotaxin which declined to a lesser extent. The levels of these biomarkers also remained upregulated by M10, 4 weeks after the 3rd vaccination (compare d50 and M10), reflecting their response kinetics after the 2nd vaccination. The responses of biomarkers in cluster 2 also continued to be higher (d36, d50) but to a lower extent. In contrast, biomarkers in clusters 3 and 4 were no longer detectable at these time points in agreement with their rapid transient responses (Figures 2, 3).
To compare longer-term changes to the transient changes, samples collected at 24hrs after each vaccination (d2:1st vaccination, d23:2nd vaccination, M9 + 1:3rd vaccination) were also included in the sequential analysis comparing changes to day 1 (day 1 of 1st vaccination) (Figure 4C). These data showed that the biomarkers in cluster 1 increased only after the 1st (d2) and the 3rd (M9 + 1d) vaccinations, whereas clusters 2-4 showed a stronger increase after the 2nd and the 3rd vaccinations with cluster 3 (IFN-γ, CXCL10/IP10) showing the highest responses, as expected (Figures 2, 3). Comparison of samples from 24hrs after each vaccination (Figure 4C) and over time responses (Figure 4B) illustrated the more durable presence of a subset of biomarkers in clusters 1 and 2.
Together, these data demonstrated both a transient increase for some cytokines, as well as a longer lasting and more pronounced presence of a subset of biomarkers, persisting up to one month after the 2nd and 3rd vaccinations, These biomarkers primarily affect the regulation of the immune chemotaxis and myeloid cell function.
To assess the inter-relationship of the vaccine-induced effects on different serum cytokines and chemokines, a pairwise correlation analyses was performed using the log2 fold changes obtained after the 3rd vaccination (M9 + 1d vs M9; Figure 5A). A correlation matrix of the measurements was calculated to identify significant relationships using a cut-off Spearman coefficient with a p value <0.005. The data were compared to a similar analysis performed after the 2nd vaccination (10) (d23 vs d22; Figure 5B). Several inter-relationships among the cytokines were identified. Importantly, the 3rd vaccination largely recapitulated our previous findings after the 2nd vaccination (10). Comparison between the 2nd and 3rd vaccination showed that cytokine inter-relations after the 3rd vaccination were slightly weaker, depicted with broader ellipses, and that some associations did not meet the significance requirements (Figure 5A vs 5B). The fact that several biomarkers did not exhibit significant associations may reflect the lack of these interactions in general or it may be due to the time point analyzed (24hrs after the vaccination).
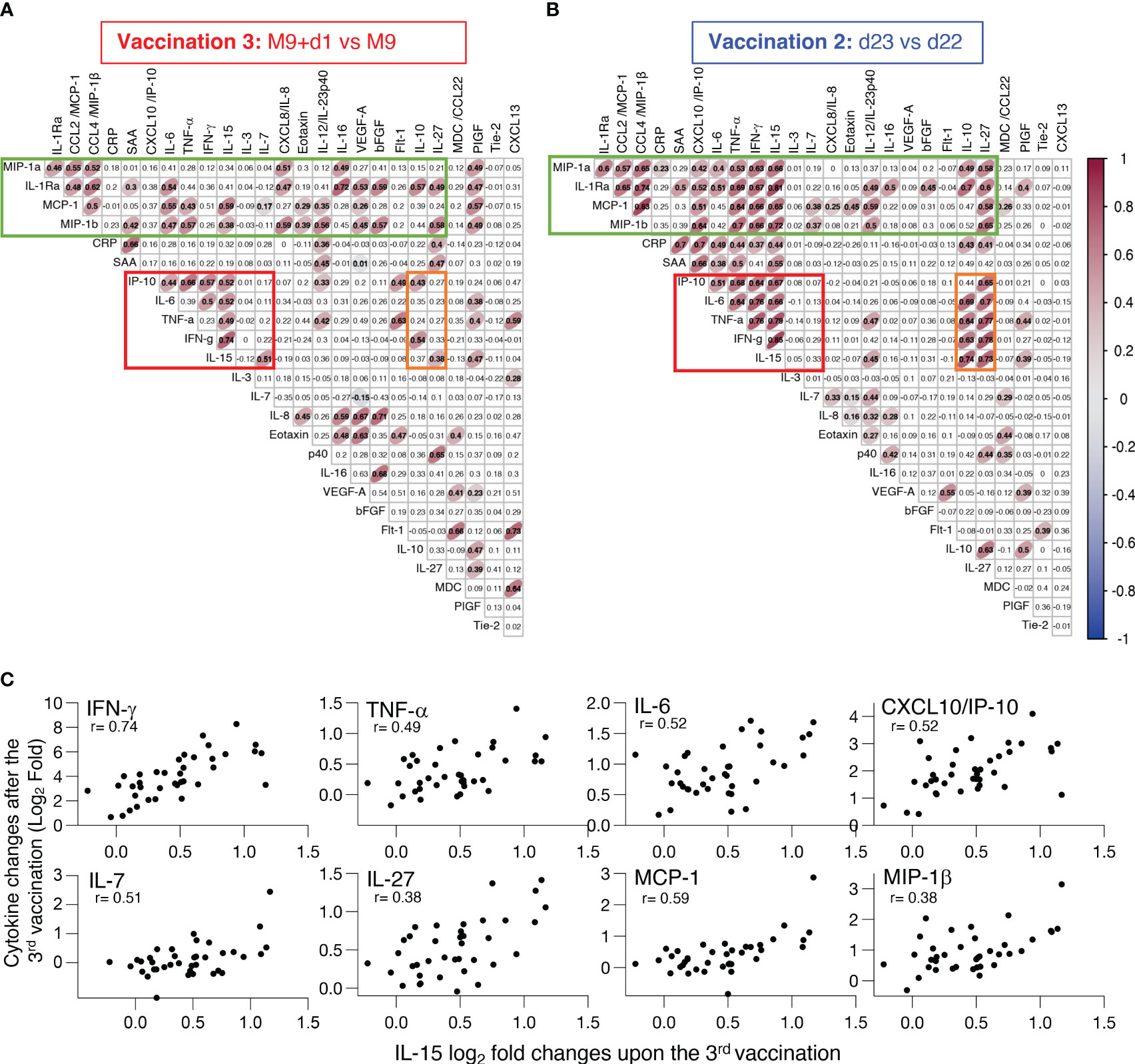
Figure 5 Inter-relationship of the vaccine-induced effects on different serum cytokines and chemokines. (A, B) Pairwise correlations were calculated between the log2 fold changes after (A) 3rd vaccination (M9 + 1d vs M9) and (B) 2nd vaccination (d23 vs d22) for the 26 biomarkers that were affected by the vaccinations using the Spearman correlation coefficient. Significant correlations (p-value < 0.005) are represented by ellipses whose color and shape correspond to the value of the Spearman correlation coefficient with red color (Figure 2) indicating a positive correlation. The red boxes identify the cluster of positive associations featuring IFN-γ, IL-15, TNF-α, IL-6 and IP-10/CXCL10. The green boxes denote the cluster of concerted interactions of inflammatory and anti-inflammatory cytokines. The orange boxes denote the cluster of IL-10 and IL-27 associated with the IL-15 cluster. (C) Scatter plots comparing the log2 fold change after the booster vaccination (M9+d1 vs M9) shown in panel A for analytes (IFN-γ, TNF-α, IL-6, CXCL10/IP-10, IL-7, IL-27, MCP-1, MIP-1β) significantly correlated to IL-15. Each dot represents the compared analytes log2 fold change for a single vaccine recipient. r and p values are shown in plots.
Despite this, a concerted and highly significant effect among IL-15, IFN-γ, TNF-α, IL-6 and IP-10/CXCL10 was observed at 24hrs after the 3rd vaccination (Figure 5A, red box) comparable to the reported interrelation after the 2nd vaccination (10) (Figure 5B, red box). These data confirmed that the concerted cytokine increase induced after the 1st vaccination and enhanced after the 2nd vaccination (10) was recapitulated after the 3rd vaccination (Figure 5). The correlating cytokine pairs included IL-15 and IFN-γ, IL-15 and TNF-α, IL-15 and IL-6, IL-15 and IP-10/CXCL10; individual correlation plots are shown in Figure 5C. IL-10 and IL-27 were reported to play a role with IL-15 in NK cell biology (35–37) and were associated with all the components of the IL-15 cluster after the 2nd vaccination (Figure 5B, orange box), these relations were weaker or lost after the 3rd vaccination (Figure 5A, orange box). In addition, the extended panel of cytokine/chemokines used in this work revealed additional concerted interactions of the IL-15 signature cytokines with IL-10 and IL-27 after the 2nd vaccination (Figure 5B) that were decreased after the 3rd vaccination (Figure 5A).
The concerted inter-relationship of other myeloid cell-produced cytokines comprising of CCL3/MIP-1α, CCL4/MIP-1β, CCL2/MCP-1, TNF-α also included IL-1Ra, IL-6, and CXCL8/IL-8, IL-16. This concerted release appeared more pronounced after the 3rd vaccination (Figures 5A, B, green boxes). This signature also associated with PIGF, a factor reported to activate monocytes, resulting in increased expression of TNF-α, IL-1Ra, CCL2/MCP-1, CXCL8/IL-8, and CCL4/MIP-1β (38). We also found that IL-15 associated with different components of this signature as well as with PIGF. This association is broader after the 3rd vaccination compared to the data obtained after the 2nd vaccination, although the significance was lower (depicted with wider ellipses). These data indicated a robust BNT162b2 mRNA vaccine-induced activation of myeloid cells. Interestingly, the analysis after the 3rd vaccination further showed an association of TNF-α and CXCL13 (Figure 5A), a relation attributed to B cell recruitment (39, 40).
We also noted a link between CRP and SAA changes both after the 2nd and 3rd vaccination, reflecting a response to inflammation [reviewed in (33)]. On the other hand, although a series of angiogenic cytokines (VEGF-A, bFGF, Tie-1, Flt-1), reflecting processes of endothelial injury and repair, greatly increased after vaccination, no concerted interactions were found under the condition of this analysis.
In summary, the 3rd vaccination resulted in a coordinated release of several biomarkers, including the IL-15 signature cytokines, which are likely responsible for the recruitment and mobilization of different immune cell subsets, supporting regulated priming and activation of immune responses.
To identify biomarkers of efficient humoral responses to vaccination, we examined the relationships between alterations in these cytokines (log2 fold changes after the 2nd vaccination d23 vs d22 and after the 3rd vaccination M9 vs M9+d1) and the levels of anti-Spike (WA1) NAb, measured after the respective vaccinations. Correlations found after the 2nd vaccination (Figure 6A) were compared to those after the 3rd vaccination (Figure 6B). We found significant univariate correlations of both IL-15 and IFN-γ changes after both vaccination and NAb levels. Together these results show that the IL-15/IFN-γ signature continues to serve as immune biomarker for effective development of vaccine-induced humoral responses.
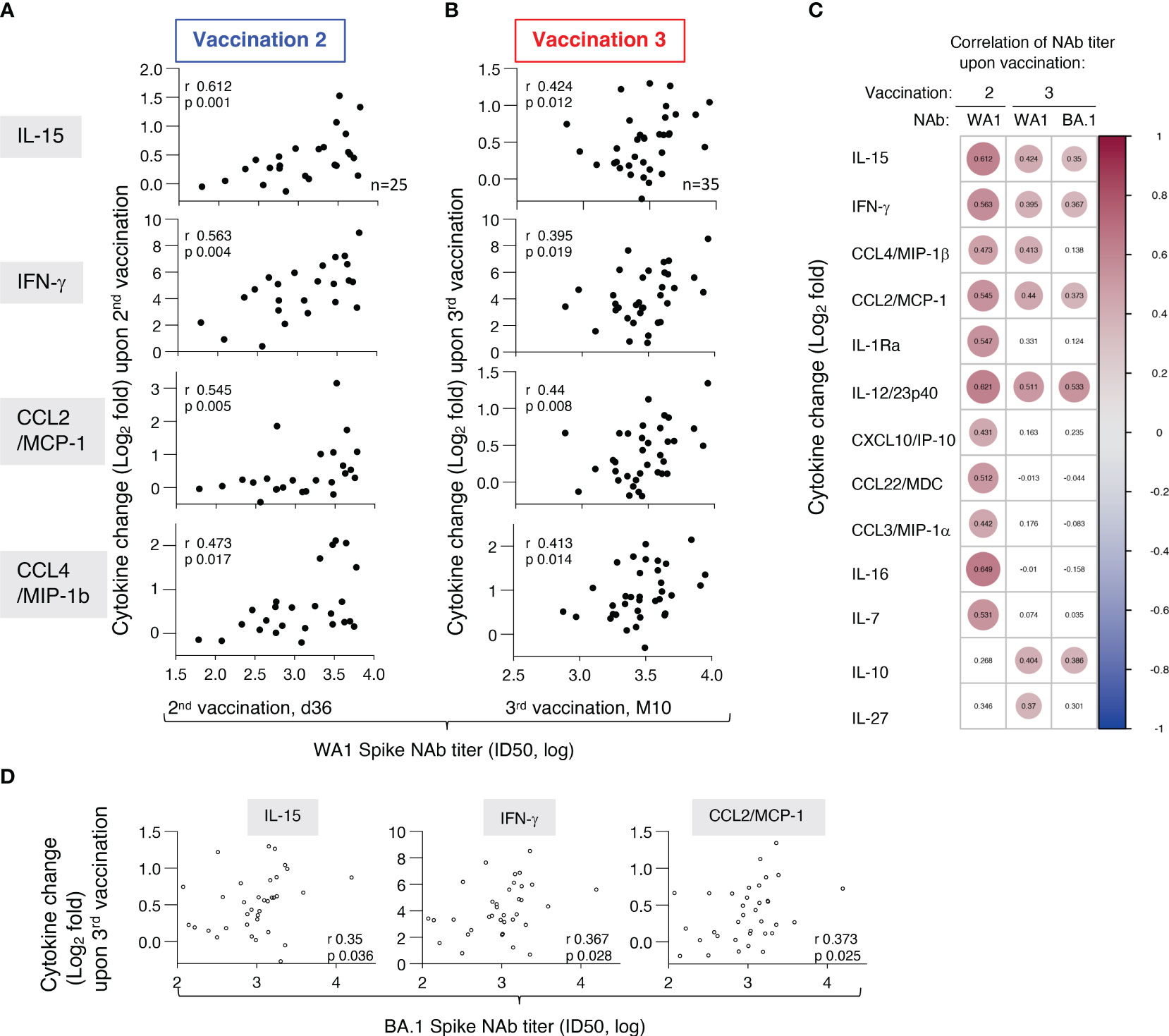
Figure 6 Association of biomarkers and vaccine-induced humoral immune responses. Univariate correlations of log2 fold changes of selected cytokines (IL-15, IFN-γ, CCL2/MCP-1, CCL4/MIP-1β, IL-1Ra) after the (A) 2nd vaccination (d23 vs d22) and (B) 3rd vaccination (M9 + 1d vs M9) and levels of anti-Spike (WA1) NAb, measured at d36 and M10, respectively. Spearman r and p values are given (calculated in R). Note the different scales of the X-axis after the 2nd and 3rd vaccination, with a tighter spread of NAb responses after the 3rd vaccination. (C) Pairwise Spearman correlations for analytes with positive correlations between cytokine fold changes and NAb titers against WA1 after the 2nd and 3rd vaccination and NAb titers against BA.1 after the 3rd vaccination. Significant correlations (p-value < 0.05) are represented by circles whose size and coloring correspond to the value of the Spearman correlation r values (calculated in R). (D) Univariate correlations of log2 fold changes of selected cytokines (IL-15, IFN-γ, CCL2/MCP-1) after the 3rd vaccination (M9 + 1d vs M9) and levels of anti-Spike BA.1 NAb, measured at M10. Spearman r and p values are given (calculated in R).
Additional significant correlations were identified between NAb, both after the 2nd and the 3rd vaccination, and with changes with CCL4/MIP-1β, CCL2/MCP-1 (Figures 6A, B) and IL-1Ra as well as with IL-12/23p40 (Figure 6C). We further found correlations of NAb levels and the pro-inflammatory IL-10, IL-7, IL-16, and IL-27 which play a role in adaptive immune response development. Some of these Spearman correlations were only found after the 3rd vaccination [IL-10 (r=0.404, p 0.016); IL-27 (r=0.37, p 0.029)], while others were only found after the 2nd vaccination [IL-16 (r=0.649, p 0.0004), IL-7 (r= 0.531, p 0.006)] (Figure 6C). These correlations support our previous observation of correlation of some of these cytokines (CCL3/MIP-1α, CCL4/MIP-1β, CCL22/MDC, IL-1Ra) and anti-Spike humoral responses (10). Interestingly, these positive correlations included several cytokines belonging to the co-expressed cluster as reported in Figure 5 and were re-established after the 3rd vaccination. The correlation with NAb after the 3rd vaccination, although weaker due to the significantly lower titers (18), further extended to some of the same cytokines and NAb titers against the more closely related Omicron BA.1 variant [IFN-γ (r=0.367, p 0.028); IL-15 (r=0.35, p 0.036), CCL2/MCP-1 (r=0.373, p 0.025), IL-10 (r= 0.386, p 0.02); IL-12/23p40 (r= 0.533, p 0.0008)] (Figures 6C, D). In contrast, the cytokine/chemokine responses did not correlate with NAb titers against the more divergent Spike variants BA.5, BQ.1.1, XBB, and XBB.1.5. Of note, this is due to the fact that the WA1 mRNA vaccine induced NAb showed significantly lower response rates and magnitudes against these Spike variants (17) than WA1 and BA.1. Thus, the cytokine/chemokine correlations observed were most pronounced towards the vaccine-matched WA1 NAb titers.
In conclusion, our analysis showed correlations between increases of the vaccine-induced cytokine levels with NAb levels after the 2nd and 3rd vaccinations. We did not find such associations using the respective baseline measurements (d1, d22 or M9) of the different biomarkers and these values did not serve as predictor of the vaccine outcome. Thus, the correlations reported here reflected the effects of vaccination on the dynamics of the cytokine/chemokine responses and their effect on the humoral response. These findings emphasize the significant contributions of innate responses to the BNT162b2 mRNA vaccination in shaping adaptive immunity and also revealed the persistence of a coordinated responses of several cytokine/chemokines after the 3rd vaccination.
In this report, we show the induction of higher binding and neutralizing Ab after the 3rd vaccination, indicating a stronger adaptive immune response compared to the 2nd vaccination. This observation could reflect a benefit of the longer rest period of eight months between the vaccinations. Applying systems serology, we studied the effects of the BNT162b2 mRNA COVID-19 vaccine on key plasma cytokines and chemokines measured one day after each vaccination and up to one month after the 2nd and 3rd vaccinations. The systemic effects of the 3rd vaccination were compared to those of the 1st and 2nd vaccinations, showing induction of a distinct cytokine response featuring the rapid (24hrs) transient induction of IL-15 signature, including IFN-γ, TNF-a, IL-6, CXCL10/IP-10 also after the 3rd vaccination. We further identified a longer lasting inflammatory/anti-inflammatory signature (>1 month). Importantly, we also found induction of biomarkers for germinal center activation including CXCL13, IL-6, IL-7, supporting the robust humoral immune response after the 3rd vaccination.
We measured the changes in biomarker levels and the inter-relationship of these changes identifying clusters of biomarkers with concerted changes. This analysis revealed two major clusters including a cluster of IL-15, IFN-γ, CXCL10/IP-10, IL-6, and TNF-α and a cluster of pro-and anti-inflammatory cytokines including CCL2/MCP-1, CCL3/MIP-1α, CCL4/MIP-1β, TNF-α, IL-1Ra, IL-6, CXCL8/IL-8, IL-16. While a rapid transient change within 24hrs after vaccination was found for the IL-15 cluster, several members of the inflammatory/anti-inflammatory cytokine cluster (CXCL8/IL-8, IL-16, CCL22/MDC, Eotaxin) showed more long-lasting elevated levels, detectable up to one month after the 2nd and 3rd vaccination.
Correlation of several immunological parameters, including IL-15 and IFN-γ with neutralizing antibody titers indicated their positive association with humoral immune response development. In addition, the discovery of systemic CXCL13 induction indicated a stronger effect of lymph node activation after the 3rd vaccination. CXCL13, also known as B-cell attracting chemokine-1 (BCA-1), plays a critical role in activating T follicular helper (Tfh) cells, which are essential contributors to B cell proliferation, differentiation, and high-affinity antibody synthesis and are required for germinal center (GC) formation and maintenance (41, 42). Other biomarkers supporting LN activity are IL-7, IL-21, IL-6, IL-4 and IL-23, cytokines involved in the development and modulation of adaptive responses and Tfh function [ (43); reviewed in (41, 42, 44)]. Systemic IL-2, IL-4, and IL-23 levels remained below the threshold of detection (Table 2), whereas the transient increase of IL-7 after the 3rd vaccination did not reach significance, which is in contrast to its significant increase after the 2nd vaccination. IL-6, readily induced after each mRNA vaccination, was reported to contribute to Tfh cell differentiation in mice (43). Together, the increases of CXCL13, IL-6 and IL-7 further support activation of LN events. In addition, others reported that additional vaccinations resulted in increase and expansion of the memory B-cells which could be in-line with increased GC activation (45).
We also observed that CXCL8/IL-8, IL-16 and bFGF which showed significant systemic increases after the 1st vaccination, failed to show any changes within 24hrs after the 2nd vaccination, but were readily increased within 24hrs after the 3rd vaccination. One possible explanation for this could be that the 3-week rest between the 1st and 2nd vaccination may have been too short. A more extended interval between the two vaccinations could potentially facilitate the development of a more robust adaptive immune responses. Indeed, the 3rd vaccination resulted in significantly higher humoral responses. The length of time between vaccinations has been intensely debated, supporting the notion that a prolonged rest period could allow for stronger adaptive immune response development (46–48). The short time interval between the first two BNT162b2 mRNA vaccinations of just three weeks was due to the urgency to combat the first period of COVID-19 epidemic. In future, a 0-2-6-months and a 0-6-12-months vaccine regimen should be compared to the present schedule, for SARS-CoV-2-naïve persons, so that an optimal vaccination regimen can be selected. Based on the available data it appears that repeated vaccinations within a short period, like within three weeks, may be sub-optimal.
Our analysis of early responses was performed with sera collected 24hrs post vaccination. We cannot exclude faster or delayed kinetics of cytokine responses, which could affect our interpretation of the dataset. This could apply for biomarkers that scored below the threshold of the assay as well as positive biomarkers, which could have peak responses either before or past 24hrs and thereby also affecting the correlations or lack thereof after a certain vaccination. These issues could only be addressed by more frequent sampling or by testing the effects of mRNA vaccine in an animal model, which also would allow sampling of tissues such as lymph nodes, in addition to blood.
We found that the IL-15 cluster showed a transient rapid response after the 3rd mRNA vaccination. The vaccine effect was comparable to the effect of the 2nd vaccination. Importantly, changes of several members of this cluster including IL-15, IFN-γ, CXCL10/IP-10 positively correlated with neutralizing antibody titers. These data showed that our observation after the 1st vaccination of the concerted change of IL-15, IFN-γ, CXCL10/IP-10, which was enriched by TNF-α and IL-6 after the 2nd vaccination (10) was recapitulated after the 3rd vaccination. Together with the data from the HIV-ALVAC platform by Andersen-Nissen et al. (4), these findings underscore an important early role of IL-15 as part of development of an effective vaccine. Of note, although individuals with COVID-19 infection prior to the 1st vaccination were excluded from this study, we had reported that such individuals showed increased transient IFN-γ, IP-10/CXCL10, TNF-α, and IL-6 responses compared to infection-naïve persons, but only after the 1st vaccination (10). Thus, these data further support our findings of similar mRNA vaccine induced transient changes of the IL-15 signature cytokines upon the 2nd and 3rd vaccination.
IL-15 and components of this cluster have been reported to play a role in the development of innate and adaptive immune responses. IL-15, a heterodimeric cytokine (49–51), plays a role in proliferation, survival and function of many lymphocytes [reviewed in (52)] and is produced by dendritic cells and monocytes/macrophages (53, 54) as well as from endothelial cells and stroma cells in some tissues. IL-15 contributes to the development of immune responses correlating with vaccine efficacy (55–61), affects durability (62–64) and cytotoxic activity (65–67) of the immune response. IL-15 activates IL-10 production by NK cells (35, 36) and directly stimulates lymphocytes to produce IFN-γ [reviewed in (68)]. Like the other components of the IL-15 cluster, we also found correlation of IL-10 changes with NAb levels. Macrophages release IL-15, IL-6, and TNF-α as part of the inflammatory response (69). Both IL-6 and TNF-α are biomarkers of trained immunity [reviewed in (70)] and may contribute to the stronger immune response after repeated mRNA vaccination. CXCL10/IP-10 is released in response to IFN-γ, promoting chemotaxis of CXCR3+ cells (71, 72). An interplay between macrophage-produced CXCL10/IP-10 and B-cell produced IL-6 driving B differentiation and association with antibody production was shown (73). Serum levels of CXCL10/IP-10 were identified as part of an innate signature associated with higher vaccine-induced antibody titers (6, 7). These data underscore our reported role of IL-15 in inducing both IFN-γ directly and DC produced CXCL10/IP-10 and CXCL9/MIG through the IFN type-2 pathway in the mouse model (67). The systemic increase of IL-15, IFN-γ, CXCL10/IL-10, TNF-α and IL-6 and associated biomarkers and their concerted increase after BNT162b2 mRNA vaccination are in good agreement with other reports (12–14). Others reported associations of myeloid cells, NK cells and monocytes as well as NAb levels with changes in levels of IFN-γ, IL-6 and CXCL10/IL-10 (14) and a correlation of IFN-γ with monocyte and DC (12) and systemic increase of CXCL10 and IL-10 (12, 13). In further support of IL-15 as key early mediator of activation of innate responses, treatment of humans with recombinant hetIL-15 was reported to induce IFN-γ, IL-6, IL-27, CXCL10/IP-10, CCL3/MIP-1α, CCL4/MIP-1β (74). Indeed, our work also showed a concerted increase of IL-15 and these biomarkers after the 2nd and/or 3rd vaccinations. In non-human primate studies, we further identified an increase of IL-15 and IL-7, which in turn acts in lymph nodes to promote CXCL13 production (75). While PBMC were not collected as part of this vaccine study, our systems serology study point to the importance of analyzing the events in lymph nodes. Such studies however need to be performed in an animal model such as non-human primates and this was not part of this project.
We also found a concerted increase of a cluster of biomarkers (CCL2/MCP-1, CCL3/MIP-1α, CCL4/MIP-1β, CCL22/MDC, CXCL8/IL-8, IL-16) associated with myeloid cells after the 3rd vaccination. We previously reported correlations of changes in CCL2/MCP-1, CCL3/MIP-1α, CCL4/MIP-1β, and CCL22/MDC after the 2nd vaccination and anti-Spike antibody levels. After the 3rd vaccination, several of these biomarker changes (CCL2/MCP-1, CCL4/MIP-1β) continued to correlate with NAb levels. Correlation of CCL2/MCP-1 with NK and monocytes as well as NAb levels after the 2nd vaccination was recently reported (14). Thus, our data showing repeated activation after mRNA vaccination of biomarkers associated with myeloid cells [(10, 11) and this report] and the detailed transcriptomics and flow work in humans reported by others (12–14) support activation of a network of chemokines and cytokines affecting innate and adaptive cellular and humoral immune responses.
These results highlight the important role of innate responses to the BNT162b2 mRNA vaccination in shaping adaptive immunity. The persistent coordinated response of several cytokine/chemokines also after the 3rd vaccination provides further insight into our understanding on their contribution to innate and adaptive immune response development. Whereas recent reports showed pre-vaccination transcriptional state of the immune system (76) or CXCL10/IP-10 levels (13) as predictive for antibody responses, our systemic cytokine analysis performed pre-vaccination (day 1) and at day 22 and month 9 was correlating with antibody response levels. In contrast, it was the dynamics of biomarker changes after each vaccination which was established as predictor of anti-Spike Ab responses. We found a correlation of a distinct set of cytokine changes (IL-15, IFN-γ, CXCL10/IP-10, CCL3/MIP-1α, CCL4/MIP-1β) after BNT162b2 mRNA vaccination of antigen-naïve individuals after 1st, 2nd, and the 3rd vaccination (this report). Patients with hematological malignancies showing diminished systemic cytokine response (IFN-γ, IL-15 and CXCL10/IP-10) also had correlating but lower anti-Spike antibody levels (11).
Monitoring biomarker changes up to one month after the 2nd and 3rd vaccinations, we were able to detect longer-lasting changes for some cytokines. In contrast to the IL-15 signature and several other biomarkers, significant higher levels of an inflammatory/anti-inflammatory cytokine cluster were still detected showing longer-lasting increases. Interestingly, we noted previously that some of these cytokines were also still detected on day 8 post the 1st vaccination but declined to baseline levels by day 22, the day of the 2nd vaccination. Thus, our data showed that after the 2nd vaccination and the 3rd vaccination, biomarkers associated with this inflammatory/anti-inflammatory signature were produced over longer time or have longer half-lives, which resulted in their prolonged presence. It is possible that this feature is associated with or favors the development of trained immunity [reviewed in (9)].
Together, our results show coordinated responses to the BNT162b2 mRNA vaccine and highlight the important role of a network of innate responses, centering on IL-15, in shaping adaptive immunity after vaccination. This study suggests that understanding the role of these biomarkers could also help the refinement of other regimens to increase vaccine efficacy.
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding author.
The studies involving humans were approved by Ethics Committee of Alexandra General Hospital (Ref No. 15/23 December 2020). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
MR: Writing – review & editing, Data curation, Formal Analysis, Investigation, Visualization. ET: Conceptualization, Supervision, Writing – review & editing, Funding acquisition, Investigation. PH: Formal Analysis, Writing – review & editing, Data curation, Visualization. CB: Writing – review & editing, Investigation. SK: Writing – review & editing, Investigation. IN-S: Writing – review & editing, Investigation. SD: Writing – review & editing, Investigation. JB: Writing – review & editing, Investigation. RB: Writing – review & editing, Investigation. TB: Writing – review & editing, Investigation. IT: Writing – review & editing, Investigation. MD: Supervision, Writing – review & editing, Conceptualization, Funding acquisition, Investigation. GP: Conceptualization, Formal Analysis, Writing – review & editing, Data curation, Funding acquisition, Supervision, Visualization, Writing – original draft. BF: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Supervision, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by funding from the Intramural Research Program, National Institutes of Health, National Cancer Institute, Center for Cancer Research to BF. The funders had no role in the experimental design, collection of data or writing the paper.
We thank Y. Wang and J. Inglefield, Clinical Services Program, Frederick National Laboratory for Cancer Research for MSD assay performance; D. Esposito (Protein Expression lab, Frederick National Laboratory for Cancer Research, NCI) for SARS-CoV-2 protein; M. Angel for participating in initial steps of the analysis; A. Kane for graphic work support; M. Moussa and D. Stellas for discussion; T. Jones for editorial assistance.
Author ET has received honoraria from Astra/Zeneca and Pfizer. Authors PH and SK were employed by the company Leidos Biomedical Research, Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Gaucher D, Therrien R, Kettaf N, Angermann BR, Boucher G, Filali-Mouhim A, et al. Yellow fever vaccine induces integrated multilineage and polyfunctional immune responses. J Exp Med (2008) 205(13):3119–31. doi: 10.1084/jem.20082292
2. Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol (2009) 10(1):116–25. doi: 10.1038/ni.1688
3. Zak DE, Andersen-Nissen E, Peterson ER, Sato A, Hamilton MK, Borgerding J, et al. Merck Ad5/HIV induces broad innate immune activation that predicts CD8(+) T-cell responses but is attenuated by preexisting Ad5 immunity. Proc Natl Acad Sci U.S.A. (2012) 109(50):E3503–12. doi: 10.1073/pnas.1208972109
4. Andersen-Nissen E, Fiore-Gartland A, Ballweber Fleming L, Carpp LN, Naidoo AF, Harper MS, et al. Innate immune signatures to a partially-efficacious HIV vaccine predict correlates of HIV-1 infection risk. PloS Pathog (2021) 17(3):e1009363. doi: 10.1371/journal.ppat.1009363
5. Wimmers F, Donato M, Kuo A, Ashuach T, Gupta S, Li C, et al. The single-cell epigenomic and transcriptional landscape of immunity to influenza vaccination. Cell (2021) 184(15):3915–35 e21. doi: 10.1016/j.cell.2021.05.039
6. Goncalves E, Bonduelle O, Soria A, Loulergue P, Rousseau A, Cachanado M, et al. Innate gene signature distinguishes humoral versus cytotoxic responses to influenza vaccination. J Clin Invest (2019) 129(5):1960–71. doi: 10.1172/JCI125372
7. Rechtien A, Richert L, Lorenzo H, Martrus G, Hejblum B, Dahlke C, et al. Systems vaccinology identifies an early innate immune signature as a correlate of antibody responses to the ebola vaccine rVSV-ZEBOV. Cell Rep (2017) 20(9):2251–61. doi: 10.1016/j.celrep.2017.08.023
8. Sahin U, Muik A, Derhovanessian E, Vogler I, Kranz LM, Vormehr M, et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature (2020) 586(7830):594–9. doi: 10.1038/s41586-020-2814-7
9. Verbeke R, Hogan MJ, Lore K, Pardi N. Innate immune mechanisms of mRNA vaccines. Immunity (2022) 55(11):1993–2005. doi: 10.1016/j.immuni.2022.10.014
10. Bergamaschi C, Terpos E, Rosati M, Angel M, Bear J, Stellas D, et al. Systemic IL-15, IFN-gamma, and IP-10/CXCL10 signature associated with effective immune response to SARS-CoV-2 in BNT162b2 mRNA vaccine recipients. Cell Rep (2021) 36:109504. doi: 10.1016/j.celrep.2021.109504
11. Bergamaschi C, Pagoni M, Rosati M, Angel M, Tzannou I, Vlachou M, et al. Reduced antibodies and innate cytokine changes in SARS-CoV-2 BNT162b2 mRNA vaccinated transplant patients with hematological malignancies. Front Immunol (2022) 13:899972. doi: 10.3389/fimmu.2022.899972
12. Arunachalam PS, Scott MKD, Hagan T, Li C, Feng Y, Wimmers F, et al. Systems vaccinology of the BNT162b2 mRNA vaccine in humans. Nature (2021) 596(7872):410–6. doi: 10.1038/s41586-021-03791-x
13. Ryan FJ, Norton TS, McCafferty C, Blake SJ, Stevens NE, James J, et al. A systems immunology study comparing innate and adaptive immune responses in adults to COVID-19 mRNA and adenovirus vectored vaccines. Cell Rep Med (2023) 4(3):100971. doi: 10.1016/j.xcrm.2023.100971
14. Takano T, Morikawa M, Adachi Y, Kabasawa K, Sax N, Moriyama S, et al. Distinct immune cell dynamics correlate with the immunogenicity and reactogenicity of SARS-CoV-2 mRNA vaccine. Cell Rep Med (2022) 3(5):100631. doi: 10.1016/j.xcrm.2022.100631
15. Li C, Lee A, Grigoryan L, Arunachalam PS, Scott MKD, Trisal M, et al. Mechanisms of innate and adaptive immunity to the Pfizer-BioNTech BNT162b2 vaccine. Nat Immunol (2022) 23(4):543–55. doi: 10.1038/s41590-022-01163-9
16. Havenar-Daughton C, Lindqvist M, Heit A, Wu JE, Reiss SM, Kendric K, et al. CXCL13 is a plasma biomarker of germinal center activity. Proc Natl Acad Sci U.S.A. (2016) 113(10):2702–7. doi: 10.1073/pnas.1520112113
17. Devasundaram S, Terpos E, Rosati M, Ntanasis-Stathopoulos I, Bear J, Burns R, et al. XBB.1.5 neutralizing antibodies upon bivalent COVID-19 vaccination are similar to XBB but lower than BQ.1.1. Am J Hematol (2023) 98(5):E123–E6. doi: 10.1002/ajh.26887
18. Rosati M, Terpos E, Bear J, Burns R, Devasundaram S, Ntanasis-Stathopoulos I, et al. Low spike antibody levels and impaired BA.4/5 neutralization in patients with multiple myeloma or waldenstrom's macroglobulinemia after BNT162b2 booster vaccination. Cancers (Basel) (2022) 14(23):5816. doi: 10.3390/cancers14235816
19. Rosati M, Agarwal M, Hu X, Devasundaram S, Stellas D, Chowdhury B, et al. Control of SARS-CoV-2 infection after Spike DNA or Spike DNA+Protein co-immunization in rhesus macaques. PloS Pathog (2021) 17(9):e1009701. doi: 10.1371/journal.ppat.1009701
20. Rosati M, Terpos E, Ntanasis-Stathopoulos I, Agarwal M, Bear J, Burns R, et al. Sequential Analysis of Binding and Neutralizing Antibody in COVID-19 Convalescent Patients at 14 months after SARS-CoV-2 infection. Front Immunol (2021) 12:793953. doi: 10.3389/fimmu.2021.793953
21. Esposito D, Mehalko J, Drew M, Snead K, Wall V, Taylor T, et al. Optimizing high-yield production of SARS-CoV-2 soluble spike trimers for serology assays. Protein Expr Purif (2020) 174:105686. doi: 10.1016/j.pep.2020.105686
22. Robbiani DF, Gaebler C, Muecksch F, Lorenzi JCC, Wang Z, Cho A, et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature (2020) 584(7821):437–42. doi: 10.1038/s41586-020-2456-9
23. Schmidt F, Weisblum Y, Muecksch F, Hoffmann HH, Michailidis E, Lorenzi JCC, et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. J Exp Med (2020) 217(11):e20201181. doi: 10.1084/jem.20201181
24. Bekele Feyissa Y, Chiodi F, Sui Y, Berzofsky JA. The role of CXCL13 in antibody responses to HIV-1 infection and vaccination. Front Immunol (2021) 12:638872. doi: 10.3389/fimmu.2021.638872
25. Mantovani A, Sica A, Allavena P, Garlanda C, Locati M. Tumor-associated macrophages and the related myeloid-derived suppressor cells as a paradigm of the diversity of macrophage activation. Hum Immunol (2009) 70(5):325–30. doi: 10.1016/j.humimm.2009.02.008
26. Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol (2004) 25(12):677–86. doi: 10.1016/j.it.2004.09.015
27. Ritvo PG, Klatzmann D. Interleukin-1 in the response of follicular helper and follicular regulatory T cells. Front Immunol (2019) 10:250. doi: 10.3389/fimmu.2019.00250
28. Tahtinen S, Tong AJ, Himmels P, Oh J, Paler-Martinez A, Kim L, et al. IL-1 and IL-1ra are key regulators of the inflammatory response to RNA vaccines. Nat Immunol (2022) 23(4):532–42. doi: 10.1038/s41590-022-01160-y
29. Arend WP. The balance between IL-1 and IL-1Ra in disease. Cytokine Growth Factor Rev (2002) 13(4-5):323–40. doi: 10.1016/S1359-6101(02)00020-5
30. Mantovani A, Dinarello CA, Molgora M, Garlanda C. Interleukin-1 and related cytokines in the regulation of inflammation and immunity. Immunity (2019) 50(4):778–95. doi: 10.1016/j.immuni.2019.03.012
31. Dinarello CA. Overview of the IL-1 family in innate inflammation and acquired immunity. Immunol Rev (2018) 281(1):8–27. doi: 10.1111/imr.12621
32. Trinchieri G, Pflanz S, Kastelein RA. The IL-12 family of heterodimeric cytokines: new players in the regulation of T cell responses. Immunity (2003) 19(5):641–4. doi: 10.1016/S1074-7613(03)00296-6
33. Mantovani A, Garlanda C. Humoral innate immunity and acute-phase proteins. N Engl J Med (2023) 388(5):439–52. doi: 10.1056/NEJMra2206346
34. Kong X, Bu J, Chen J, Ni B, Fu B, Zhou F, et al. PIGF and Flt-1 on the surface of macrophages induces the production of TGF-beta1 by polarized tumor-associated macrophages to promote lung cancer angiogenesis. Eur J Pharmacol (2021) 912:174550. doi: 10.1016/j.ejphar.2021.174550
35. Clark SE, Burrack KS, Jameson SC, Hamilton SE, Lenz LL. NK cell IL-10 production requires IL-15 and IL-10 driven STAT3 activation. Front Immunol (2019) 10:2087. doi: 10.3389/fimmu.2019.02087
36. Park JY, Lee SH, Yoon SR, Park YJ, Jung H, Kim TD, et al. IL-15-induced IL-10 increases the cytolytic activity of human natural killer cells. Mol Cells (2011) 32(3):265–72. doi: 10.1007/s10059-011-1057-8
37. Zwirner NW, Ziblat A. Regulation of NK cell activation and effector functions by the IL-12 family of cytokines: the case of IL-27. Front Immunol (2017) 8:25. doi: 10.3389/fimmu.2017.00025
38. Selvaraj SK, Giri RK, Perelman N, Johnson C, Malik P, Kalra VK. Mechanism of monocyte activation and expression of proinflammatory cytochemokines by placenta growth factor. Blood (2003) 102(4):1515–24. doi: 10.1182/blood-2002-11-3423
39. Armas-Gonzalez E, Dominguez-Luis MJ, Diaz-Martin A, Arce-Franco M, Castro-Hernandez J, Danelon G, et al. Role of CXCL13 and CCL20 in the recruitment of B cells to inflammatory foci in chronic arthritis. Arthritis Res Ther (2018) 20(1):114. doi: 10.1186/s13075-018-1611-2
40. Mandik-Nayak L, Huang G, Sheehan KC, Erikson J, Chaplin DD. Signaling through TNF receptor p55 in TNF-alpha-deficient mice alters the CXCL13/CCL19/CCL21 ratio in the spleen and induces maturation and migration of anergic B cells into the B cell follicle. J Immunol (2001) 167(4):1920–8. doi: 10.4049/jimmunol.167.4.1920
41. Winer H, Rodrigues GOL, Hixon JA, Aiello FB, Hsu TC, Wachter BT, et al. IL-7: comprehensive review. Cytokine (2022) 160:156049. doi: 10.1016/j.cyto.2022.156049
42. Spolski R, Leonard WJ. IL-21 and T follicular helper cells. Int Immunol (2010) 22(1):7–12. doi: 10.1093/intimm/dxp112
43. Choi YS, Eto D, Yang JA, Lao C, Crotty S. Cutting edge: STAT1 is required for IL-6-mediated Bcl6 induction for early follicular helper cell differentiation. J Immunol (2013) 190(7):3049–53. doi: 10.4049/jimmunol.1203032
44. Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity (2014) 41(4):529–42. doi: 10.1016/j.immuni.2014.10.004
45. Yang L, Van Beek M, Wang Z, Muecksch F, Canis M, Hatziioannou T, et al. Antigen presentation dynamics shape the antibody response to variants like SARS-CoV-2 Omicron after multiple vaccinations with the original strain. Cell Rep (2023) 42(4):112256. doi: 10.1016/j.celrep.2023.112256
46. Amirthalingam G, Bernal JL, Andrews NJ, Whitaker H, Gower C, Stowe J, et al. Serological responses and vaccine effectiveness for extended COVID-19 vaccine schedules in England. Nat Commun (2021) 12(1):7217. doi: 10.1038/s41467-021-27410-5
47. Grunau B, ASamoah-Boaheng M, Lavoie PM, Karim ME, Kirkham TL, Demers PA, et al. A higher antibody response is generated with a 6- to 7-week (vs standard) severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccine dosing interval. Clin Infect Dis (2022) 75(1):e888–e91. doi: 10.1093/cid/ciab938
48. Grunau B, Goldfarb DM, ASamoah-Boaheng M, Golding L, Kirkham TL, Demers PA, et al. Immunogenicity of extended mRNA SARS-CoV-2 vaccine dosing intervals. JAMA (2022) 327(3):279–81. doi: 10.1001/jama.2021.21921
49. Bergamaschi C, Bear J, Rosati M, Kelly Beach R, Alicea C, Sowder R, et al. Circulating interleukin-15 (IL-15) exists as heterodimeric complex with soluble IL-15 receptor alpha (IL-15Ralpha) in human serum. Blood (2012) 120:e1–8. doi: 10.1182/blood-2011-10-384362
50. Chertova E, Bergamaschi C, Chertov O, Sowder R, Bear J, Roser JD, et al. Characterization and favorable in vivo properties of heterodimeric soluble IL-15/IL-15Ra cytokine compared to IL-15 monomer. J Biol Chem (2013) 288:18093–103. doi: 10.1074/jbc.M113.461756
51. Bergamaschi C, Rosati M, Jalah R, Valentin A, Kulkarni V, Alicea C, et al. Intracellular interaction of interleukin-15 with its receptor alpha during production leads to mutual stabilization and increased bioactivity. J Biol Chem (2008) 283(7):4189–99. doi: 10.1074/jbc.M705725200
52. Bergamaschi C, Stravokefalou V, Stellas D, Karaliota S, Felber BK, Pavlakis GN. Heterodimeric IL-15 in cancer immunotherapy. Cancers (Basel) (2021) 13(4):837. doi: 10.3390/cancers13040837
53. Colpitts SL, Stonier SW, Stoklasek TA, Root SH, Aguila HL, Schluns KS, et al. Transcriptional regulation of IL-15 expression during hematopoiesis. J Immunol (2013) 191(6):3017–24. doi: 10.4049/jimmunol.1301389
54. Cui G, Hara T, Simmons S, Wagatsuma K, Abe A, Miyachi H, et al. Characterization of the IL-15 niche in primary and secondary lymphoid organs in vivo. Proc Natl Acad Sci U.S.A. (2014) 111(5):1915–20. doi: 10.1073/pnas.1318281111
55. Barrenas F, Hansen SG, Law L, Driscoll C, Green RR, Smith E, et al. Interleukin-15 response signature predicts RhCMV/SIV vaccine efficacy. PloS Pathog (2021) 17(7):e1009278. doi: 10.1371/journal.ppat.1009278
56. Huntington ND, Alves NL, Legrand N, Lim A, Strick-Marchand H, Mention JJ, et al. IL-15 transpresentation promotes both human T-cell reconstitution and T-cell-dependent antibody responses in vivo. Proc Natl Acad Sci U.S.A. (2011) 108(15):6217–22. doi: 10.1073/pnas.1019167108
57. Moore AC, Kong WP, Chakrabarti BK, Nabel GJ. Effects of antigen and genetic adjuvants on immune responses to human immunodeficiency virus DNA vaccines in mice. J Virol (2002) 76(1):243–50. doi: 10.1128/JVI.76.1.243-250.2002
58. Oh S, Berzofsky JA, Burke DS, Waldmann TA, Perera LP. Coadministration of HIV vaccine vectors with vaccinia viruses expressing IL-15 but not IL-2 induces long-lasting cellular immunity. Proc Natl Acad Sci U.S.A. (2003) 100(6):3392–7. doi: 10.1073/pnas.0630592100
59. Valentin A, von Gegerfelt A, Rosati M, Miteloudis G, Alicea C, Bergamaschi C, et al. Repeated DNA therapeutic vaccination of chronically SIV-infected macaques provides additional virological benefit. Vaccine (2010) 28(8):1962–74. doi: 10.1016/j.vaccine.2009.10.099
60. Ramanathan MP, Kutzler MA, Kuo YC, Yan J, Liu H, Shah V, et al. Coimmunization with an optimized IL15 plasmid adjuvant enhances humoral immunity via stimulating B cells induced by genetically engineered DNA vaccines expressing consensus JEV and WNV E DIII. Vaccine (2009) 27(32):4370–80. doi: 10.1016/j.vaccine.2009.01.137
61. Kutzler MA, Robinson TM, Chattergoon MA, Choo DK, Choo AY, Choe PY, et al. Coimmunization with an optimized IL-15 plasmid results in enhanced function and longevity of CD8 T cells that are partially independent of CD4 T cell help. J Immunol (2005) 175(1):112–23. doi: 10.4049/jimmunol.175.1.112
62. Rubinstein MP, Lind NA, Purton JF, Filippou P, Best JA, McGhee PA, et al. IL-7 and IL-15 differentially regulate CD8+ T-cell subsets during contraction of the immune response. Blood (2008) 112(9):3704–12. doi: 10.1182/blood-2008-06-160945
63. Schluns KS, Williams K, Ma A, Zheng XX, Lefrancois L. Cutting edge: requirement for IL-15 in the generation of primary and memory antigen-specific CD8 T cells. J Immunol (2002) 168(10):4827–31. doi: 10.4049/jimmunol.168.10.4827
64. Li J, Valentin A, Ng S, Beach RK, Alicea C, Bergamaschi C, et al. Differential effects of IL-15 on the generation, maintenance and cytotoxic potential of adaptive cellular responses induced by DNA vaccination. Vaccine (2015) 33(9):1188–96. doi: 10.1016/j.vaccine.2014.12.046
65. Watson DC, Moysi E, Valentin A, Bergamaschi C, Devasundaram S, Fortis SP, et al. Treatment with native heterodimeric IL-15 increases cytotoxic lymphocytes and reduces SHIV RNA in lymph nodes. PloS Pathog (2018) 14(2):e1006902. doi: 10.1371/journal.ppat.1006902
66. Ng SSM, Nagy BA, Jensen SM, Hu X, Alicea C, Fox BA, et al. Heterodimeric IL-15 treatment enhances tumor infiltration, persistence and effector functions of adoptively transferred tumor-specific T cells in the absence of lymphodepletion. Clin Cancer Res (2017) 23:2817–30. doi: 10.1158/1078-0432.CCR-16-1808
67. Bergamaschi C, Pandit H, Nagy BA, Stellas D, Jensen SM, Bear J, et al. Heterodimeric IL-15 delays tumor growth and promotes intratumoral CTL and dendritic cell accumulation by a cytokine network involving XCL1, IFN-gamma, CXCL9 and CXCL10. J Immunother Cancer (2020) 8(1):e000599. doi: 10.1136/jitc-2020-000599
68. Perera PY, Lichy JH, Waldmann TA, Perera LP. The role of interleukin-15 in inflammation and immune responses to infection: implications for its therapeutic use. Microbes Infect (2012) 14(3):247–61. doi: 10.1016/j.micinf.2011.10.006
69. McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med (2011) 365(23):2205–19. doi: 10.1056/NEJMra1004965
70. Al B, Suen TK, Placek K, Netea MG. Innate (learned) memory. J Allergy Clin Immunol (2023) 152(3):551–6. doi: 10.1016/j.jaci.2023.06.014
71. Liu M, Guo S, Stiles JK. The emerging role of CXCL10 in cancer (Review). Oncol Lett (2011) 2(4):583–9. doi: 10.3892/ol.2011.300
72. Groom JR, Luster AD. CXCR3 in T cell function. Exp Cell Res (2011) 317(5):620–31. doi: 10.1016/j.yexcr.2010.12.017
73. Xu W, Joo H, Clayton S, Dullaers M, Herve MC, Blankenship D, et al. Macrophages induce differentiation of plasma cells through CXCL10/IP-10. J Exp Med (2012) 209(10):1813–23, S1-2. doi: 10.1084/jem.20112142
74. Conlon K, Watson DC, Waldmann TA, Valentin A, Bergamaschi C, Felber BK, et al. Phase I study of single agent NIZ985, a recombinant heterodimeric IL-15 agonist, in adult patients with metastatic or unresectable solid tumors. J Immunother Cancer (2021) 9(11):e003388. doi: 10.1136/jitc-2021-003388
75. Pandit H, Valentin A, Angel M, Deleage C, Bergamaschi C, Bear J, et al. Step-dose IL-7 treatment promotes systemic expansion of T cells and alters immune cell landscape in blood and lymph nodes. iScience (2023) 26(2):105929. doi: 10.1016/j.isci.2023.105929
Keywords: Innate immunity, IL-15, IFN-g, CXCL10/IP10, TNF-a, CXCL13, myeloid cell biomarkers, anti-spike neutralizing antibody
Citation: Rosati M, Terpos E, Homan P, Bergamaschi C, Karaliota S, Ntanasis-Stathopoulos I, Devasundaram S, Bear J, Burns R, Bagratuni T, Trougakos IP, Dimopoulos MA, Pavlakis GN and Felber BK (2023) Rapid transient and longer-lasting innate cytokine changes associated with adaptive immunity after repeated SARS-CoV-2 BNT162b2 mRNA vaccinations. Front. Immunol. 14:1292568. doi: 10.3389/fimmu.2023.1292568
Received: 11 September 2023; Accepted: 14 November 2023;
Published: 27 November 2023.
Edited by:
Elke Bergmann-Leitner, Walter Reed Army Institute of Research, United StatesReviewed by:
Sivaram Gunisetty, Emory University, United StatesCopyright © 2023 Rosati, Terpos, Homan, Bergamaschi, Karaliota, Ntanasis-Stathopoulos, Devasundaram, Bear, Burns, Bagratuni, Trougakos, Dimopoulos, Pavlakis and Felber. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Barbara K. Felber, QmFyYmFyYS5mZWxiZXJAbmloLmdvdg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.