
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Immunol. , 22 November 2023
Sec. Vaccines and Molecular Therapeutics
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1289032
This article is part of the Research Topic Emerging Talents in Vaccines and Molecular Therapeutics: 2022 View all 9 articles
 Nadim Sharif1*
Nadim Sharif1* Nazmul Sharif2
Nazmul Sharif2 Afsana Khan3
Afsana Khan3 Irma Domínguez Azpíroz4,5,6
Irma Domínguez Azpíroz4,5,6 Raquel Martínez Diaz4,7,8
Raquel Martínez Diaz4,7,8 Isabel De la Torre Díez9
Isabel De la Torre Díez9 Anowar Khasru Parvez1
Anowar Khasru Parvez1 Shuvra Kanti Dey1*
Shuvra Kanti Dey1*Introduction: Rotavirus infection is a major cause of mortality among children under 5 years in Bangladesh. There is lack of integrated studies on rotavirus prevalence and genetic diversity during 1973 to 2023 in Bangladesh.
Methods: This meta-analysis was conducted to determine the prevalence, genotypic diversity and seasonal distribution of rotavirus during pre-vaccination period in Bangladesh. This study included published articles on rotavirus A, rotavirus B and rotavirus C. We used Medline, Scopus and Google Scholar for published articles. Selected literatures were published between 1973 to 2023.
Results: This study detected 12431 research articles published on rotavirus. Based on the inclusion criteria, 29 of 75 (30.2%) studies were selected. Molecular epidemiological data was taken from 29 articles, prevalence data from 29 articles, and clinical symptoms from 19 articles. The pooled prevalence of rotavirus was 30.1% (95% CI: 22%-45%, p = 0.005). Rotavirus G1 (27.1%, 2228 of 8219) was the most prevalent followed by G2 (21.09%, 1733 of 8219), G4 (11.58%, 952 of 8219), G9 (9.37%, 770 of 8219), G12 (8.48%, 697 of 8219), and G3 (2.79%, 229 of 8219), respectively. Genotype P[8] (40.6%, 2548 of 6274) was the most prevalent followed by P[4] (12.4%, 777 of 6274) and P[6] (6.4%, 400 of 6274), respectively. Rotavirus G1P[8] (19%) was the most frequent followed by G2P [4] (9.4%), G12P[8] (7.2%), and G9P[8], respectively. Rotavirus infection had higher odds of occurrence during December and February (aOR: 2.86, 95% CI: 2.43-3.6, p = 0.001).
Discussion: This is the first meta-analysis including all the studies on prevalence, molecular epidemiology, and genetic diversity of rotavirus from 1973 to 2023, pre-vaccination period in Bangladesh. This study will provide overall scenario of rotavirus genetic diversity and seasonality during pre-vaccination period and aids in policy making for rotavirus vaccination program in Bangladesh. This work will add valuable knowledge for vaccination against rotavirus and compare the data after starting vaccination in Bangladesh.
Group A rotavirus is the main (family Reoviridae) pathogen of gastroenteritis among children under five years in Bangladesh (1–3). About 300 000 deaths related to rotavirus infection are reported worldwide every year (1). Rotavirus associated death has been reduced in the developed countries due to the implementation of rotavirus vaccination program (4–6). However, the health burden of rotavirus associated morbidity and mortality are still high in developing countries like Bangladesh without vaccination against rotavirus (7–9). The estimated incidence of rotavirus is 10,000 cases per 100,000 children under five years in Bangladesh. According to WHO, severe health conditions are reported from 25-30% of rotavirus infected children (8, 9). Every year about 2500-3000 children under five die of rotavirus infection in Bangladesh. The TRIVAC model by Pan American Health Organization indicates that vaccination against rotavirus can prevent about 3 million cases and 4000 fatalities among children under five during next 10 years in Bangladesh (7–10).
Rotavirus is a nonenveloped, double stranded RNA (dsRNA, segmented) virus with a genome of about 18.55 kilo base pairs (11). Rotavirus encodes for six structural proteins namely, VP1 to VP6 and six nonstructural proteins namely, NSP1 to NSP6 (10, 11). Among ten species of rotavirus (referred to as A, B, C, D, E, F, G, H, I and J), rotavirus A is mainly involved with majority of human cases (90%). Two genotypes of rotavirus are named as G genotype from glycoprotein (VP7) and P genotype from protease (VP4) (11, 12). Nearly, 36-types and 51 P-types have been reported worldwide since 1973 (10). Until 2000s, the most commonly reported G-types were G1‐G4 (11–17). After 2000s, genotype G9 transmitted rapidly became one of important genotypes worldwide, whereas genotype G12 became prevalent in the Americas and developing countries in Asia. Among the P-type, P[4], P[6] and P[8] remain the most common worldwide (12–17). Globally rotavirus genotypes G1P[8], G2P[4], G3P[8], G4P[8] and G9P[8] have contributed to the majority of the reported cases.(12-17) The genotypic diversity of rotavirus has changed in Bangladesh over the years (16, 17).
Four rotavirus A vaccines are prequalified by WHO and available internationally namely, RotaTeq (bovine-human reassorted vaccine), Rotarix (human rotavirus), Rotavac (bovine-human reassorted vaccine of G9P11), and RotaSiil (bovine-human reassorted vaccine) Besides, several other vaccines are available and licensed for use in India (Rotasiil, pentavalent human bovine reassortant vaccine), Vietnam (Rotavin-M1, human genotyp G1P[8]) and China (Lanzhou lamb, genotype G10P[12]of lamb rotavirus) (10). Though rotavirus vaccination is not included in the national immunization program in Bangladesh, international rotavirus vaccines are available in Bangladesh (9, 10, 13). The efficacy of rotavirus vaccines namely, Rotarix and RotaTeq is high in developed countries (13). After the introduction of rotavirus vaccination, the cases and fatalities among children under five reduced significantly in the developed countries. However, in developing and under-developed countries the vaccines have lower efficacy. A recent Rotarix trial in Bangladesh has revealed that vaccine efficacy is decreasing and it is below 50%. This lower efficacy may be attributed to the circulation of heterotypic strains and high number of mutational and reassortment events of homotypic rotaviruses (9, 10). However, integrated studies on the prevalence and epidemiology of rotavirus are lacking in Bangladesh. Knowing the epidemiology and genetic diversity of rotavirus can contribute in evaluation of effects of rotavirus vaccines in Bangladesh. The main aim of this study is to determine the prevalence and molecular epidemiology of rotavirus in Bangladesh during pre-vaccination period, from 1973 to 2023.
Epidemiology of rotavirus is defined as the distribution and determinants of the outbreaks in different population and strategies taken to minimize the health effect among them. The clinical feature was defined including both the signs and symptoms during and after the rotavirus infection. Transmission of rotavirus is defined as the transfer of the virus from source to human and carrier to human. This study included epidemiological, clinical and virological research published in Bangladesh on rotavirus. The rotavirus positive case was determined by ELISA-based method before the invention of RT-PCR laboratory-confirmed RT-PCR test. The standards of the Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) Statement and Cochrane Collaboration were followed to conduct the study (18).
The study was performed by following different steps including, identification of precise objectives and search strategies, appropriate research articles, inclusion of manuscripts, collection of data, analysis, and summarization and inclusion of the findings, exclusion of irrelevant studies. We included previous original research articles focusing epidemiological studies, case studies, surveillance work, outbreak investigation, hospital-based surveillance and online databases. No previous study on the topic with strict parameters for the quality assessment is available. As a result, we relied on the quality reports of the selected articles by the authors.
We performed searches for collecting relevant articles in MEDLINE (through PubMed), Web of Science, EMBASE, Scopus, The New England Journal of Medicine (NEJM) and The Lancet with no restriction on language. The place of the search strategy was fixed for Bangladesh. All published manuscripts and scientific writings till December 31, 2022 were included in this study. The notable search term included Rotavirus, RVA, RVB, RVC, Epidemiology of rotavirus, Epidemiology of rotavirus in Bangladesh, Clinical features of rotavirus, Sign and Symptoms of rotavirus, Clinical characteristics of rotavirus, Cases of rotavirus, Transmission of rotavirus, Molecular epidemiology of rotavirus, Genetic diversity, Bangladesh, Children under five, and combination of these search terms. Distinguish searches were made for each term in every website and database.
Further, we performed search on the grey literature and Google Scholar database. These sources included data and articles from CDC (Centers for Disease Control and Prevention, USA), ECDC (European Centers for Disease Control and Prevention) WHO (the World Health Organization), Epicentre, ProMed, IEDCR of Bangladesh, ICDDR, B in Bangladesh. We searched for the yearly update of the data in these websites and databases. Further, articles and data were searched and included from different preprint databases including SSRN, bioRxiv, medRxiv, and AAS Open Research. Additionally, we manually analyzed the first twenty pages on the Google Scholar search system for each search term. We tried to determine the epidemiology of rotavirus by including prevalence over the years, genetic diversity, incidence, case reports, clinical history, case fatality rate, seasonality and vaccination.
Two authors, NS and NS conducted the evaluation of eligible scientific studies. After completion of searching of all the databases and websites, the potential articles were selected by removing the duplicates and screening out the eligible ones by NS and AKP. Articles with specific and relevant topics were selected for all cities and location in Bangladesh, all ethnicity, age group, sex and clinical features for full-text analyzing. We also included the articles focusing on the molecular epidemiology of rotavirus involving animals and human or only animal or only human published during 1973 and 2022. We excluded modelling and prediction studies, correlation of environmental factors studies, review articles and non-relevant studies to our objectives. Further, critical evaluation on the quality of the selected articles, screening out duplicated articles, and removing correspondence or comment of duplicated data were performed by NS, SKD and AK, separately. The seasonal exclusion criteria were implemented only on the studies on seasonality of rotavirus not on environmental impact.
The risk of bias was measured using The Systematic Review Centre for Laboratory Animal Experimentation (SYRCLE) assessment tool (19). The SYRCLE is consisted of 10 parameters to assess different biases in this study. The parameter included attrition bias, detection bias, reporting bias, performance bias, selection bias and other biases. The bias for each parameter was measured by using the possible outcomes as yes, no, and unclear, representing low, high, and unclear bias, respectively (19). Data on environmental factors were collected from different websites including, the official website of Bangladesh Meteorological Department (http://live4.bmd.gov.bd/satelite/v/sat_infrared/), meteoblue (www.meteoblue.com), AccuWeather (www.accuweather.com) and WeatherOnline (www.weatheronline.co.uk). Data on sociodemographic factors were included from the analyzed studies and database of national surveillance system on rotavirus in Bangladesh.
A confirmed case of rotavirus was defined by following the WHO case definition and final classification guideline, the presence of rotavirus is confirmed by an enzyme immunoassay (EIA) or reverse transcriptase polymerase chain reaction (PCR)-based methods. The type of the used PCR assay will be determined on the basis of local guidelines and availability of the test method.
Total number of cases, circulating genotypes and seasonality were determined for each decade by summation of the reported cases per genotypes. Pooled statistical analyses were conducted for determining the overall prevalence of rotavirus and genotypic distribution by using SAS version 9.4. Logistic regression analysis was conducted to determine the impact of sociodemographic factors and environmental factors on the odds of rotavirus infection.
This study detected 12431 research articles on the prevalence and molecular epidemiology of rotavirus in Bangladesh (Figure 1). Firstly, among 12431 articles, 231 were screened and initially considered for analyzing abstract. Secondly, from 231 articles we selected 96 articles for full text investigation and excluded 135 articles. Thirdly, after analyzing the full texts, we excluded 67 studies and included 29 (30.2%) studies for further analysis. Among the selected manuscripts, we extracted molecular epidemiological data from 29 articles, prevalence data from 29 articles, clinical symptoms from 19 articles (Figure 1). Finally, 23 studies were included in the meta-analysis.
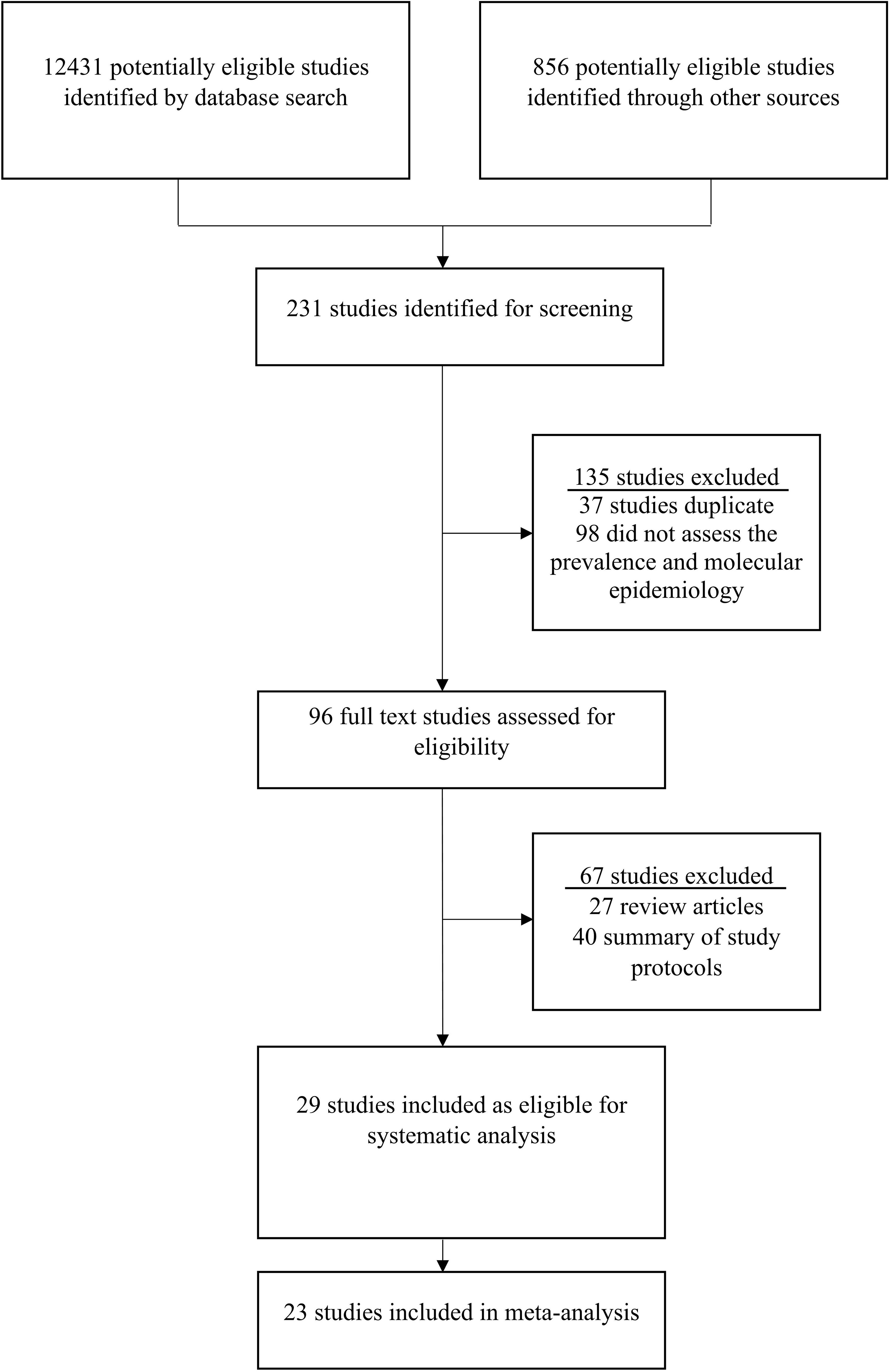
Figure 1 Procedure of study selection on rotavirus in Bangladesh. The standards of the Preferred Reporting Items for Systematic Review and Meta-Analysis (PRISMA) Statement was followed to select the studies.
Prevalence of rotavirus infection varied from the first report to the last report of rotavirus during the last 50 years in Bangladesh. The pooled prevalence of rotavirus infection was 30.1% (95% CI: 22%-45%, p = 0.005) in Bangladesh before the introduction of mass vaccination. Among these 29 studies during 1973-2023, rotavirus infection was reported from 25 (86.2%) studies (20–50). Rotavirus A (23 of 25, 92% studies) was the most predominant followed by rotavirus B (1 of 25, 4% study) and C (1 of 25, 4% study). Rotavirus A was reported from about 99.93% (45671 of 45679) of the cases (Table 1). Most of the cases (80%) were reported among children. Co-prevalence of pathogenic bacteria, virus and parasites were reported in 7 studies. Male was the most commonly affected group. In majority of the studies, ELISA and reverse-transcriptase polymerase chain reaction was used to detect the presence of rotavirus (Table 1). Before 2010, studies on rotavirus were conducted in only three cities including Dhaka, Mymensingh and Matlab.
Rotavirus infection was detected across the country. The first report was from Matlab, Chandpur during 1977-1979 with lower prevalence and bacterial co-infection (20). Only few studies were conducted in selected cities in Bangladesh during 1975-1999 (20–25). Larger prevalence (>20%) of rotavirus among children was detected in Matlab during 1978-1979 and 1987-1989 followed by Dhaka and Mymensingh during 1988 and 1989 (Figure 2). However, cases of rotavirus were not reported from any other part of the country during 1975-1999. The number of studies increased during 2000-2023 in Bangladesh. About 22 studies were conducted on the prevalence and epidemiology of rotavirus on human during 2000-2023 (Figure 3) (26–49). Majority of the studies were reported from the three cities including Dhaka, Mymensingh and Matlab until 2010 in Bangladesh. Two surveillance studies during 2012-2015 and 2012-2017 included rotavirus cases from 8 divisional cities in Bangladesh and reported higher prevalence of about 64% (Figure 3). Besides these divisional cities studies on rotavirus have been conducted only in limited areas including Sirajganj, Savar and Tangail. Presence of other enteric viruses and bacterial co-infection have been reported from Dhaka, Chittagong, Savar, Tangail, and Sirajganj during 2010-2019 (43, 44).
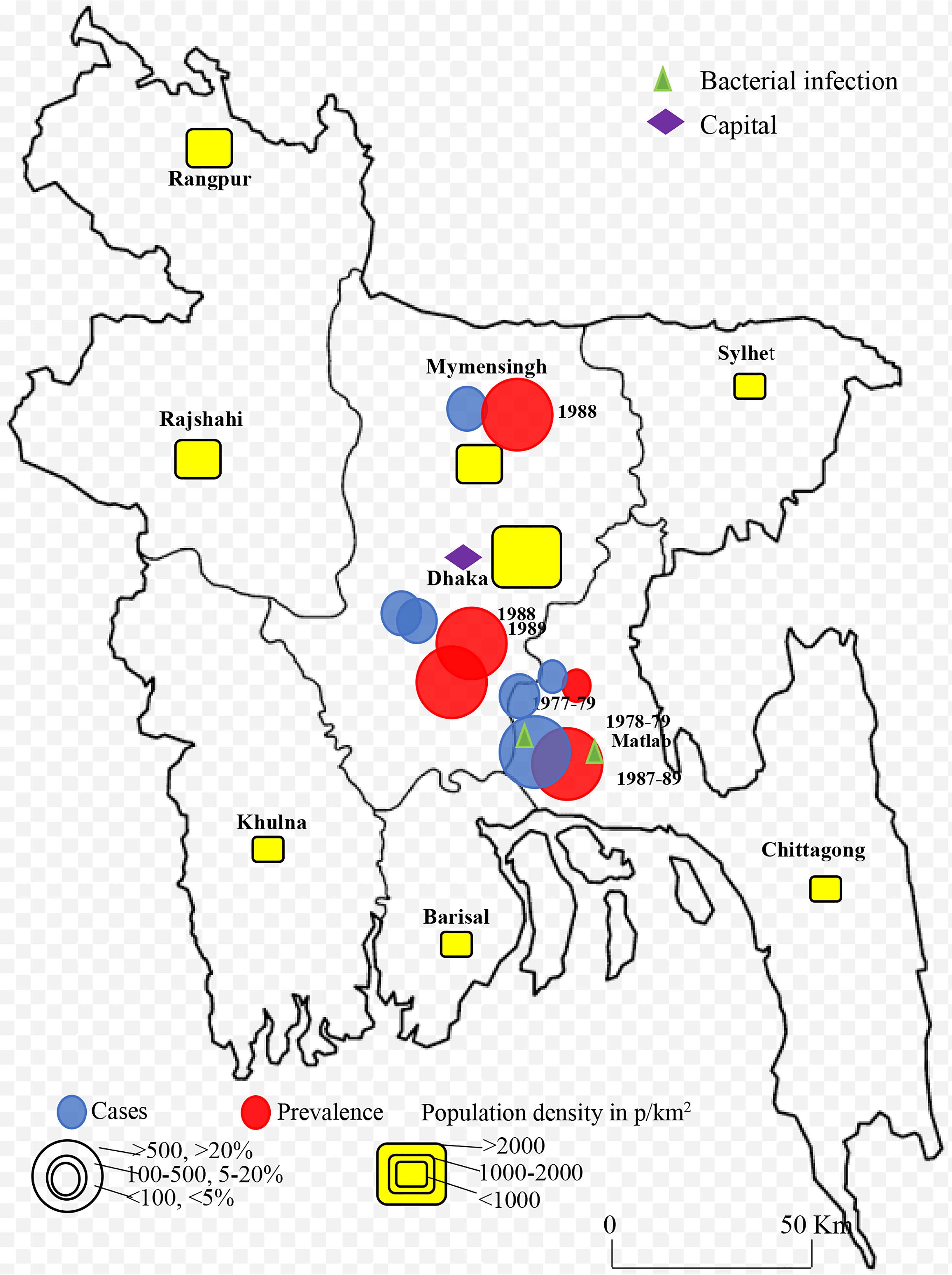
Figure 2 Spatial and temporal distribution of rotavirus cases and prevalence in Bangladesh during 1973-1999 and B. 2000-2023. Blue color circle indicated cases, red color circle indicated prevalence yellow color box indicated population density in p/km2.
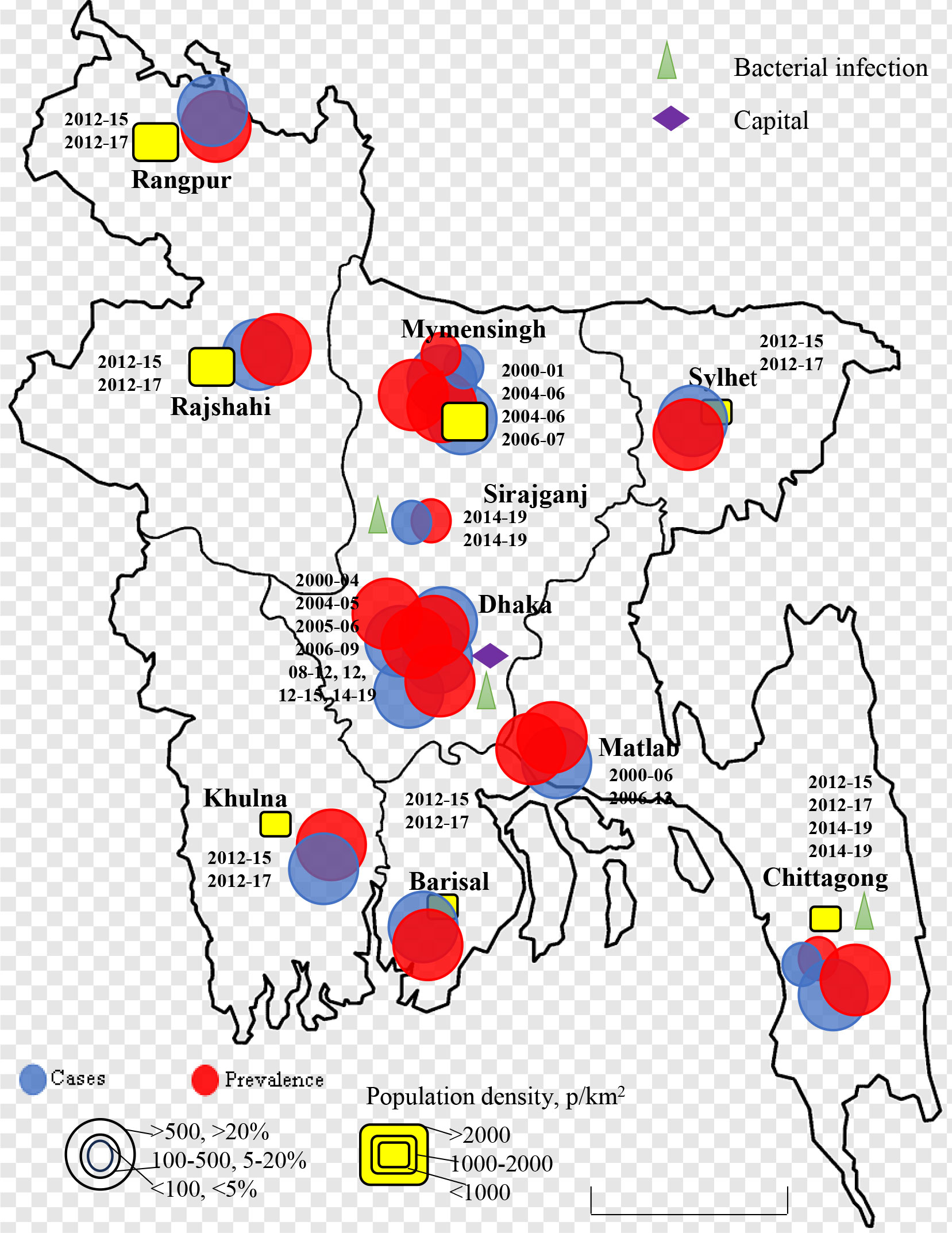
Figure 3 Spatial and temporal distribution of rotavirus cases and prevalence in Bangladesh during 2000-2023. Blue color circle indicated cases, red color circle indicated prevalence yellow color box indicated population density in p/km2.
Data on genetic diversity of rotavirus from all the reported works were included. Rotavirus G-typing was found in 19 articles, P-typing in 13 articles and G [P] typing in 13 articles. G-typing was conducted for 8219 samples. Rotavirus G1 (27.1%, 2228 of 8219) was the most prevalent followed by G2 (21.09%, 1733 of 8219), G4 (11.58%, 952 of 8219), G9 (9.37%, 770 of 8219), G12 (8.48%, 697 of 8219), and G3 (2.79%, 229 of 8219), respectively. About 2.5% (200 of 8219) of the isolates were mixed and 16.7% were non-typable G type (Table 2A). The number of studies on P typing were less than G typing. Among the reported P-type in Bangladesh, P[8] (40.6%, 2548 of 6274) was the most prevalent followed by P[4] (12.4%, 777 of 6274) and P[6] (6.4%, 400 of 6274), respectively. About 35.5% samples were non-typable and 2.3% were mixed P type in Bangladesh during 1973-2023 (Table 2B). Significantly diverse genotypes of rotavirus have been reported in Bangladesh. Among the regularly reported genotypes of rotavirus, G1P[8] (19.1%, 1199 of 6274) was the most frequent followed by G2P[4] (9.4%, 590 of 6274), G12P[8] (7.2%, 451 of 6274), G9P[8] (6.97%, 437 of 6274), G12P[6] (3.5%, 219 of 6274), G4P[8] (2.9%, 183 of 6274), G9P[4] (1.8%, 111 of 6274) and G9P[6] (1.5%, 94 of 6274), respectively (Table 2C). Other irregularly irregularly reported genotypes of rotavirus in Bangladesh included, G1P[4], G1P[6], G2P[6], G2P[8], G3P[3], G3P[15], G3P[8], G4P[4], G4P[6], G4P[49], G6P[11], G8P[1], G10P[1], G10P[11], G10P[15], G11P[25], and G12P[4] (Table 2C). Yearly distribution of genotypes of rotavirus showed significant changes during the study period. During 1985-1989, G2 (25%) was the most prevalent followed by G1 (22%) and G4 (19%) and during the next 10 years the prevalence of non-typable G type (45%) increased significantly. However, during 2000-04 and 2004-09, the prevalence of G1 (33%), G2 (32%) and G9 (16%) increased. In the next ten years (2010-2019), a sharp increase in the prevalence of G12 (18%) was observed and G1 (40%) remained the most prevalent genotype in Bangladesh. Prevalence of genotype G4 decreased from 20% to 2% from 1985 to 2019 (Figure 4A).
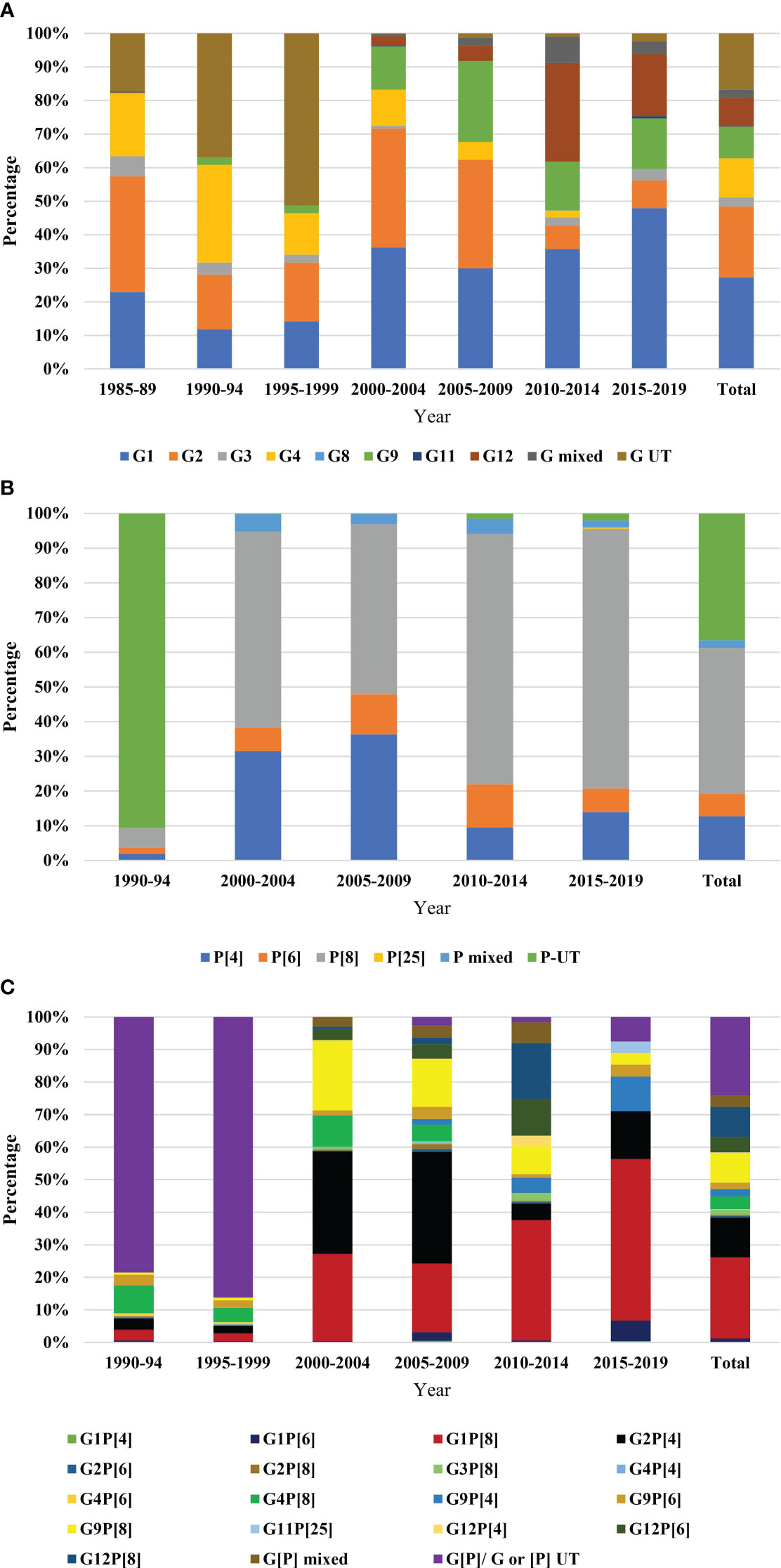
Figure 4 Year-wise genotypic distribution of rotavirus (A) G-type, data were represented from 1985 to 2019. Five years interval was used. The bar in the right side represented total diversity during 1985-2019, (B) P-type, data were represented from 1990 to 2019. Five years interval was used. The bar in the right side represented total diversity during 1990-2019, and (C) G[P]-types, data were represented from 1990 to 2019. Five years interval was used. The bar in the right side represented total diversity during 1990-2019.
Majority of the isolates were un-typable (90%) for P-type during 1990-94. The prevalence of P[8] rotavirus increased from 7% to 72% during 1990 to 2019 and remained the most reported P type in Bangladesh. A sharp decrease in the prevalence of P[4] from 31% to 14% was reported from 2000 to 2019 (Figure 4B).
Among the circulating strains, 80-90% were un-typable during 1990-1999. Genotype G4P[8] was prevalent (7%) during 1990-99, the prevalence increased in 2000-05 (10%) and decreased to 1% during 2005-2019. During 2000-04, genotype G2P [4] (32%) was the most prevalent followed by G1P[8] (28%), G9P[8] (23%) and G4P[8] (10%), respectively. During the next five years (2005-09), the prevalence of G2P[4] increased to 38%. After 2009, genotype G1P[8] (50%) became the most predominant and the diversity of genotypes increased by 80% during 2010-2019 (Figure 4C).
Regional distribution of rotavirus revealed that G1 was most prevalent in Dhaka (42.3%, 942 of 2228) followed by Matlab (38%, 848 of 2228) and Mymensingh (5.9%, 132 of 2228), respectively. Genotype G2 and G4 were also most prevalent in Dhaka (48.7% and 69%, respectively) and Matlab (39% and 28%, respectively). Diversity of G genotype was significantly higher in Dhaka than any other cities in Bangladesh. Among the sequenced P types, P[4], P[6] and P[8] were also prevalent in Dhaka (about 50%) followed by Matlab (about 30%) and Mymensingh (about 10%). About 45% (95% CI: 32-63%) of reported G1P[8], G2P[4], G12P[8], and G9P[8] were documented in Dhaka, followed by 31% (95% CI: 22-43%) in Matlab. Genotypes G1P[8], G2P[4], G2P[6], G9P[4] G12P[8], and G9P[8] were also reported in minor frequency from Mymensingh, Rajshahi, Sylhet, Chittagong, Rangpur, Khulna and Barisal (Table 2C).
About 19 of the studies included clinical symptoms of rotavirus infected people. Diarrhea was the most common symptoms followed by dehydration, vomiting, fever and abdominal pain, respectively. About 89.5% (17 of 19) of the studies reported diarrhea followed by dehydration (63.1, 12 of 19), vomiting (57.9%, 11 of 19), fever (57.9%, 11 of 19) and abdominal pain (36.8, 7 of 19), respectively (Table 3). Hospitalization was required for 83% (95% CI: 62-98%, p = 0.001) of the reported cases of rotavirus in Bangladesh. Diarrhea (97.3%, 95% CI: 87-99%, p = 0.04) with characteristics watery stools of rotavirus infection (70.5%, 95% CI: 63-89%, p=0.005) was the most prevalent symptoms followed by vomiting (56.5%, 95% CI: 45-89%, p = 0.006), dehydration (28.8%, 95% CI: 24-75%, p = 0.002), and fever (16.5%, 95% CI: 8-54%, p = 0.003), respectively (Table 3). Only three of the studies reported death of children infected with rotavirus with mortality rate varying from 0.1-1.6%. Rotavirus positive children had higher risk of developing severe dehydration (aOR: 2.81, 95% CI: 2.45-3.36, p = 0.005), diarrhea (aOR: 1.12, 95% CI: 0.74-1.54, p = 0.01), and watery stool (aOR: 1.49, 95% CI: 1.19-1.71, p = 0.003) (Figure 5). However, rotavirus negative children with diarrhea had higher risk of developing fever (aOR: 1.22, 95% CI: 0.94-1.43, p = 0.005) and abdominal pain (aOR: 1.03, 95% CI: 0.79-1.23, p = 0.01) (Figure 5).
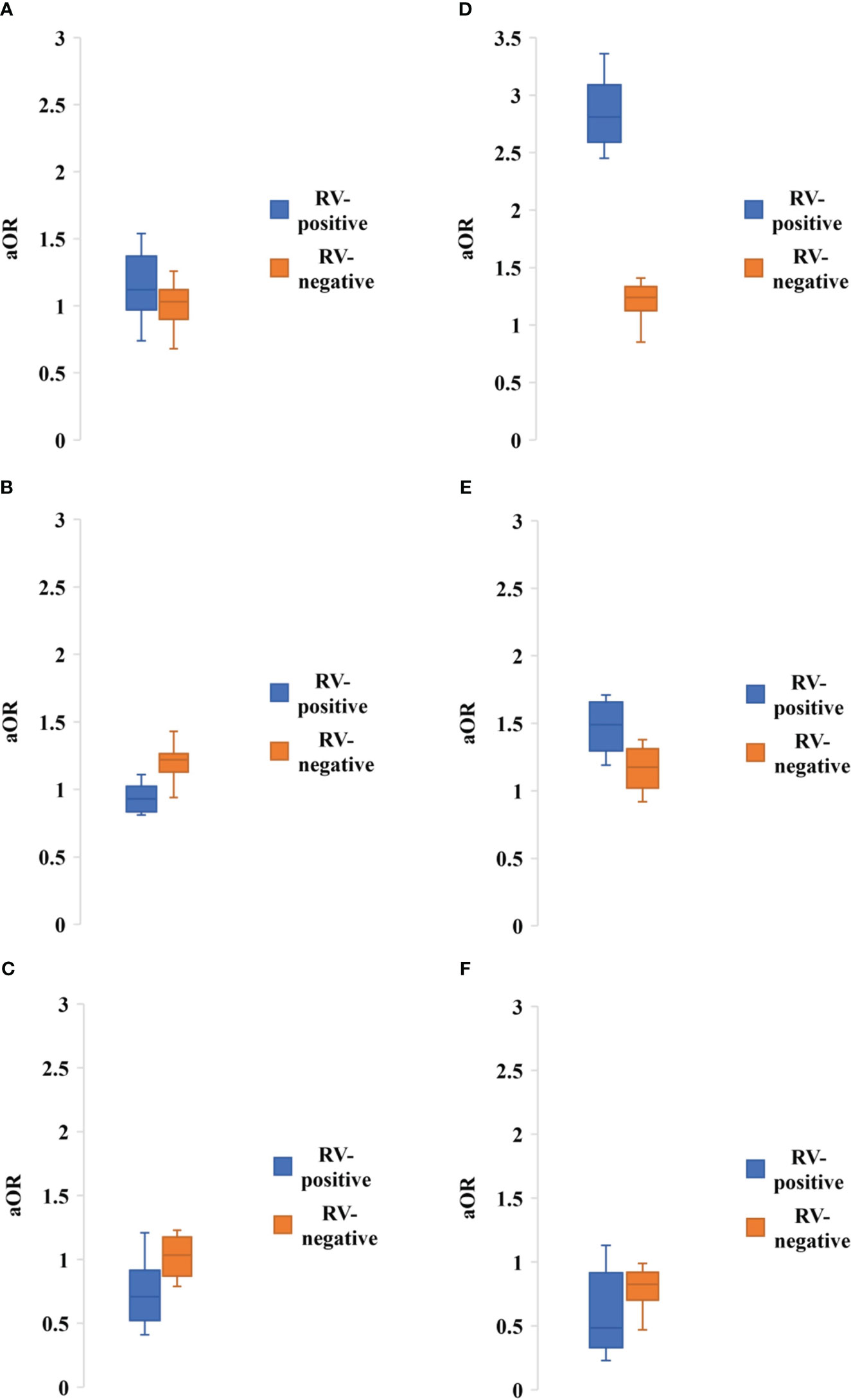
Figure 5 Adjusted odds ratio of having symptoms of (A) Diarrhea, (B) Severe dehydration, (C) Fever, (D) Watery stool, (E) Abdominal pain, (F) Hospitalization among people with rotavirus positive vs negative infection. Blue bar indicated rotavirus positive and brown bar indicated rotavirus negative cases.
Seasonality data of rotavirus infection was extracted from 13 studies during 1978-2023 in Bangladesh (Figure 6). Rotavirus infection in the study regions was detected all around the year. The highest peak of infection (43%, 95% CI: 39-58%, p = 0.001) was reported during December to February (winter season with average temperature 19-21°C) followed by March to May (21%, 95% CI: 17-31%, p = 0.002) (Figure 6). The pattern of the seasonal peak was consistent in all the included studies (Table 4). Impact of different environmental factors on the rotavirus incidence was determined. Incidence of rotavirus had higher odds of occurrence during December and February (aOR: 2.86, 95% CI: 2.43-3.6, p = 0.001) (Figure 7). We also found that higher odds of rotavirus infection at average temperature <25°C (aOR: 1.85, 95% CI: 1.61-1.94, p = 0.002), average UV index <7 (aOR: 2.11, 95% CI: 1.89-2.31, p = 0.004) and rainfall <40 mm (aOR: 1.86, 95% CI: 1.57-1.91, p = 0.005) (Figure 7).
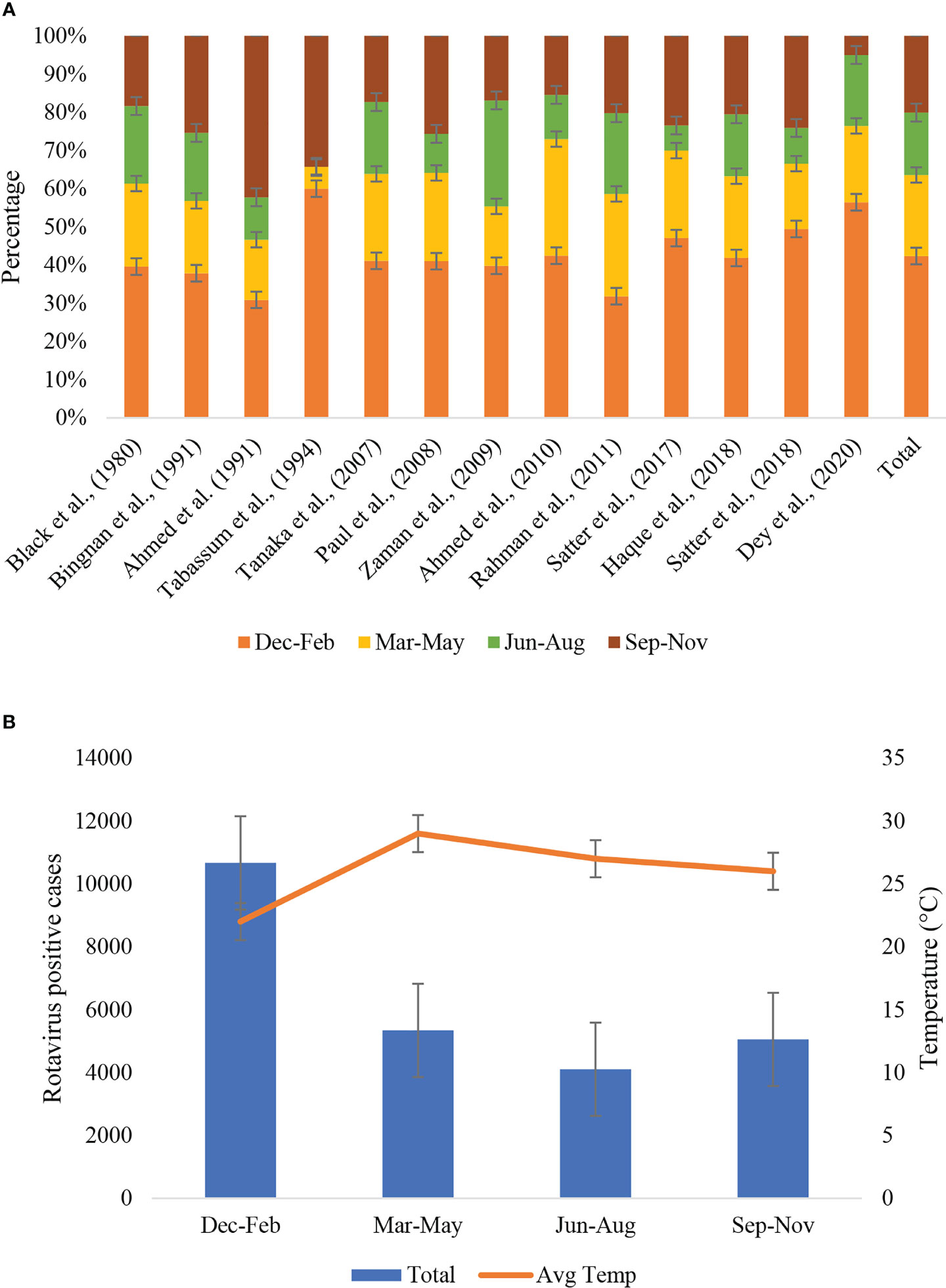
Figure 6 (A) Seasonal distribution of rotavirus infection described in different studies during 1973-2023 in Bangladesh. According to the variation of average temperature, four seasonal groups were included, December to February, March to May, June to August and September to November. The last bar in the right side represented cumulative seasonality of rotavirus infection. (B) Seasonal distribution of rotavirus positive cases and trends of average temperature in Bangladesh during 1973-2023.
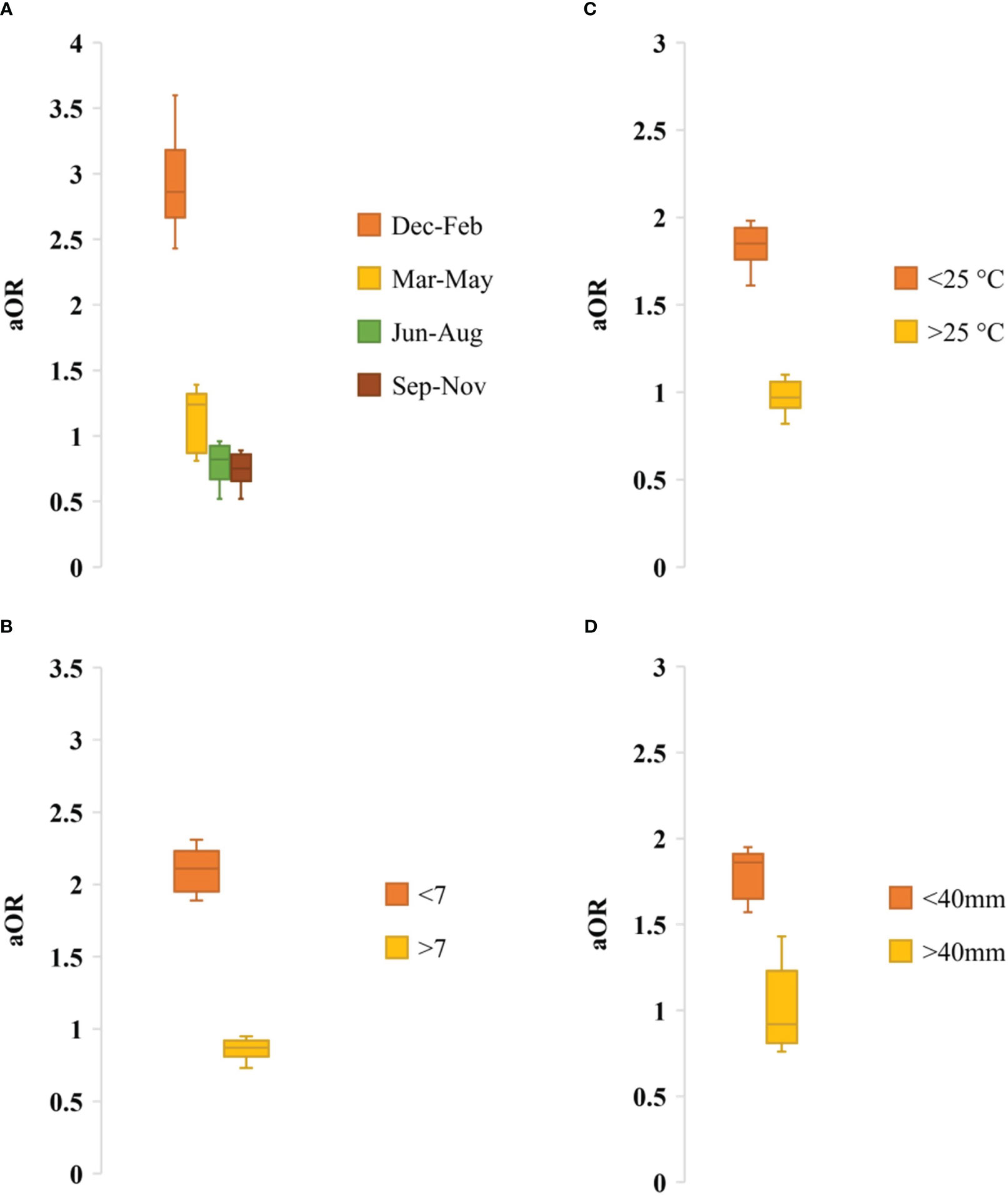
Figure 7 Adjusted odds ratio of rotavirus infection by (A) Seasonality, (B) UV index, (C) Average temperature and (D) Average rainfall.
Impact of socio-demographic factors associated with rotavirus infection was evaluated. Children aged <2 years had higher risk of rotavirus infection (aOR: 1.76, 95% CI: 1.23-2.32, p = 0.001), male children had higher risk of rotavirus infection (aOR: 1.48, 95% CI: 1.14-1.92, p = 0.003), children in rural areas had higher risk of infection (aOR: 1.4, 95% CI: 0.98-1.65, p = 0.01) than in urban areas in Bangladesh. In the seasonal analysis, we also found that rotavirus infection was prevalent during winter (aOR: 2.37, 95% CI: 1.91-3.21, p = 0.02) than other seasons (Figure 8).
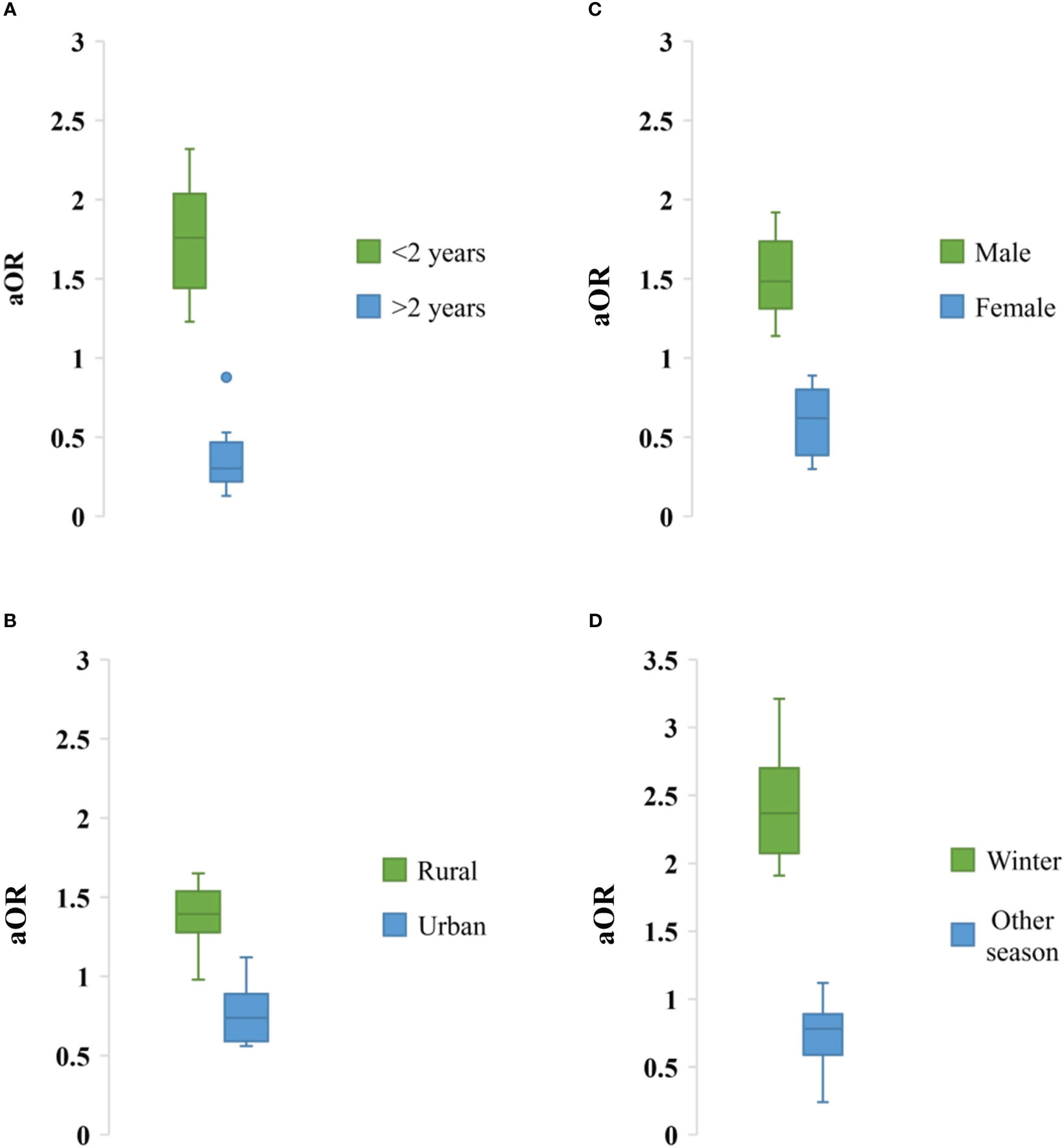
Figure 8 Association of different sociodemographic factors (A) Age, (B) Residence, (C) Sex and (D) Weather with odds of rotavirus infection in Bangladesh during 1973-2023.
Rotavirus has remained one of the major causes of children mortality and hospitalization in Bangladesh (43–45). However, rotavirus vaccination has not been included in the national immunization program in Bangladesh (9). As a result, the prevalence of rotavirus among children in Bangladesh is still high (40, 41, 43, 44). Before inclusion of rotavirus vaccine on national immunization program, integrated knowledge on prevalence, genotypic diversity, symptoms and seasonality of rotavirus can contribute significantly in the public health policy making. However, there is a lack of integrated study on rotavirus in Bangladesh. We conducted this study to provide an integrated insight of rotavirus infection in Bangladesh during pre-vaccination period. This is the first report of meta-analysis of rotavirus in Bangladesh including all data from 1973 to 2023. We found high prevalence of rotavirus (30.1%, 95% CI: 22%-45%, p = 0.005) in Bangladesh, that is higher than previous meta-analysis and surveillance study conducted in Turkey, China, worldwide data (50–65). However, a meta-study found higher prevalence of rotavirus in Iran. Prevalence of rotavirus ranged from 2%-64% in the included studies in Bangladesh. Further, we found higher prevalence of rotavirus among children aged <5 years (38.2%, 95% CI: 24%-51%, p = 0.001) than any other meta studies conducted previously (55, 56, 58–69). These data suggest that prevalence of rotavirus is relatively higher than most of the countries and a consistent health problem in Bangladesh (40–45, 65–72). This study also suggested a higher prevalence of rotavirus among people of all age groups. Other epidemiological findings including male to female ratio of (male was mostly infected) rotavirus infection was in good agreement with previous studies in Bangladesh, China, Turkey, India and Pakistan (50–56, 62–69). Most of the studies reported circulation of rotavirus A (99.9% cases) among Bangladeshi population, which is in good agreement with previous studies (60–69).
Recent studies after 2005 found higher prevalence (ranging from 23% to 64%) of rotavirus among children under 5 years in Bangladesh than previous studies. Usage of reverse transcriptase polymerase chain reaction and molecular sequencing were applied more commonly after 2000 in Bangladesh for the detection and molecular characterization of rotavirus. Only six of the studies reported co-circulation of bacterial pathogens among diarrheal children with rotavirus in Bangladesh. Among the reported coinfecting bacterial pathogens, V. cholerae, Shigella spp, and Salmonella spp were most commonly found in different studies with prevalence ranging from 1% to 32%. These findings are highly similar with previous studies in Bangladesh (43, 44, 70–72). We found high genotypic diversity of rotavirus A in Bangladesh. About 15 G-types and 8 P-types of rotaviruses have been documented. Genotype G1, G2 and G4 (60% combinedly) were the most prevalent rotaviruses in Bangladesh. Among the P-genotypes P[8] was the most frequent (40%) followed by P[4] (12%). These findings are in good agreement with pre-vaccination data in developing and developed countries (51, 55, 58–69). As mass vaccination is yet to be introduced, the prevalence and genotypic diversity remained unchanged over the years and places in Bangladesh. Further, genotypic diversity and heterogeneity due to mutation and recombination contributing to numerous un-typable G-types and P-types, which can contribute in lowering the vaccine efficacy in Bangladesh. Among the typable isolates, 28 genotypic combinations were documented, which is higher than the previous reports from a country (58–69). Rotavirus G1P[8] (19%) was the most prevalent and G2P[4], G9P[8] and G12P[8] were regularly reported among children with diarrhea in Bangladesh since 1978. This genotypic distribution is in good agreement with reported genotypic diversity data in Ethiopia, India, Iran and South Korea (51, 53–68). However, we detected relatively lower prevalence of G3 (2.8%) and G4 (11.6%) in Bangladesh than in other countries (60–68). This pattern of genotypic distribution reflects lack of vaccination contributed to the continuous prevalence of G1, G2, G9 and G12 in Bangladesh. These findings are highly similar with previous studies (51, 53–68).
We also found that only limited studies were conducted in Dhaka, Mymensingh and Matlab before 2000 in Bangladesh. The number of studies and localities increased during 2000-2010. Majority of the studies reported rotavirus from Dhaka, Matlab, Mymensingh (91%) and extended to Sylhet, Chittagong, Rangpur, Khulna and Barisal during 2010-2023. This study reports that published documents represent mostly hospitalized patients in selected areas (20–50). In future, more studies including samples from both home staying and hospitalized children with diarrhea from all over the country should be conducted. Hospital oriented surveillance during 2012-2017 included divisional cities and large number of samples. Molecular surveillance is lacking significantly to represent actual diversity of rotavirus in Bangladesh.
Clinical symptoms of rotavirus positive people were similar with previous reports (40–44). Diarrhea was the most prevalent symptoms followed by dehydration and vomiting. We found fever and abdominal pain inconsistently reported from the infected children in Bangladesh. Studies reporting co-infection of other bacterial pathogens have found higher frequency of fever, abdominal pain and vomiting than only rotavirus infection. Hospitalization was required for majority of the rotavirus infected children (80-100%) in Bangladesh. These findings are in good agreement with previous studies in Bangladesh (40–44).
We found significant impact of environmental factors and seasonality on rotavirus infection in Bangladesh. Rotavirus infection was detected all around the year with a sharp increase of cases during the winter season (December to February) with average air temperature 19-21°C. Further, we found that risk of rotavirus infection was high at lower temperature, lower UV index and lower amount of rainfall. These findings reflect seasonality and association of increased rotavirus infection with environmental parameters in Bangladesh (21, 23, 24, 29, 30, 32, 35, 36, 40–44). In some of the previous studies two peaks of rotavirus, one in winter and another in summer have been reported (40–44). However, in this integrated study we found a significant frequency of cases (about 48%) were documented during winter. Our findings are completely supported by previous studies in the developing countries. Previous studies have reported abundance of rotavirus infection among male children aged <5 years. However, lack of statistical analysis to evaluate the relationship between socio-demographic factors and rotavirus infection were found. In this study, we found that male (aOR: 1.48, 95% CI: 1.14-1.92, p = 0.003) children aged <2 years (aOR: 1.76, 95% CI: 1.23-2.32, p = 0.001) living in rural areas had higher risk of rotavirus infection in Bangladesh. These findings will add knowledge on the impact of sociodemographic factors on rotavirus infection. Our findings are supported strongly by previous findings (40–44, 51, 53–68).
The main limitation of this study was not including any data on rotavirus vaccination due to lack of studies. Further, we could not include mutational data of Bangladeshi rotavirus strains due to lack of studies. This study could be improved if data from all the regions were available and included for longer periods.
This is the first meta-analysis including all the studies on prevalence, molecular epidemiology, and genetic diversity of rotavirus from 1973 to 2023 in Bangladesh. This study created an integrated overview of rotavirus infection in Bangladesh during 1973 to 2023. Vaccine coverage of rotavirus is still low in Bangladesh as rotavirus vaccines are not included in the EPI program. As a result, high prevalence of rotavirus (30.1%, 95% CI: 22%-45%, p = 0.005) in Bangladesh contributing to majority of health problems and mortality among children aged <5 years till now. We also found great genetic diversity of rotaviruses in the study regions with high prevalence of G1P[8], G2P[4], G9P[8] and G12P[8]. Introduction of existing rotavirus vaccines and development of more effective vaccines to prevent the health burden associated with rotavirus infection are immediately needed. This study will provide overall scenario of rotavirus infection during pre-vaccination period and aid in policy making for rotavirus vaccination program in Bangladesh.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
NadS: Conceptualization, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. NazS: Formal Analysis, Investigation, Methodology, Software, Writing – review & editing. AK: Data curation, Formal Analysis, Investigation, Software, Writing – review & editing. IA: Data curation, Formal Analysis, Investigation, Software, Writing – review & editing. RM: Data curation, Formal Analysis, Investigation, Software, Writing – review & editing. ID: Data curation, Funding acquisition, Investigation, Resources, Software, Validation, Visualization, Writing – review & editing. AP: Data curation, Investigation, Methodology, Software, Writing – review & editing. SD: Conceptualization, Data curation, Investigation, Methodology, Project administration, Software, Supervision, Writing – review & editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Kotloff KL, Platts-Mills JA, Nasrin D, Roose A, Blackwelder WC, Levine MM. Global burden of diarrheal diseases among children in developing countries: Incidence, etiology, and insights from new molecular diagnostic techniques. Vaccine (2017) 35:6783–9. doi: 10.1016/j.vaccine.2017.07.036
2. Aliabadi N, Antoni S, Mwenda JM, Weldegebriel G, Biey JN, Cheikh D, et al. Global impact of rotavirus vaccine introduction on rotavirus hospitalisations among children under 5 years of age, 2008–16: findings from the Global Rotavirus Surveillance Network. Lancet Global Health (2019) 7:e893–903. doi: 10.1016/S2214-109X(19)30207-4
3. Coşkun USŞ, Kasap T. Frequency of rotavirus and adenovirus in pediatric patients with acute gastroenteritis. J Contemp Med (2019) 9:85–8. doi: 10.16899/gopctd.459823
4. Milivojevic V, Milosavljevic T. Burden of gastroduodenal diseases from the global perspective. Curr Treat Options Gastroenterol (2020) 18:148–57. doi: 10.1007/s11938-020-00277-z
5. Kotloff KL. The burden and etiology of diarrheal illness in developing countries. Pediatr Clinics (2017) 64:799–814. doi: 10.1016/j.pcl.2017.03.006
6. Hartman S, Brown E, Loomis E, Russell HA. Gastroenteritis in children. Am Family Physician (2019) 99:159–65.
7. Tilmanne A, Lepage P, Vandenberg O, Martiny D, Hallin M, Quach C. Rotavirus: the guard dies, but it does not surrender. Infect Dis (2019) 51:67–70. doi: 10.1080/23744235.2018.1508885
8. Dennehy PH. Rotavirus infection: a disease of the past? Infect Dis Clinics (2015) 29:617–35. doi: 10.1016/j.idc.2015.07.002
9. Pecenka C, Parashar U, Tate JE, Khan JA, Groman D, Chacko S, et al. Impact and cost-effectiveness of rotavirus vaccination in Bangladesh. Vaccine (2017) 35(32):3982–7. doi: 10.1016/j.vaccine.2017.05.087
10. Sadiq A, Bostan N, Yinda KC, Naseem S, Sattar S. Rotavirus: Genetics, pathogenesis and vaccine advances. Rev Med Virol (2018) 28(6):e2003. doi: 10.1002/rmv.2003
11. Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res (2018) 46:D708–17. doi: 10.1093/nar/gkx932
12. Patton JT. Rotavirus diversity and evolution in the post-vaccine world. Discov Med (2012) 13:85.
13. Troeger C, Khalil IA, Rao PC, Cao S, Blacker BF, Ahmed T, et al. Rotavirus vaccination and the global burden of rotavirus diarrhea among children younger than 5 years. JAMA Pediatrics (2018) 172(10):958–65. doi: 10.1001/jamapediatrics.2018.1960
14. Zeng Y, Li T, Zhao B, Lai F, Tang X, Qiao Y, et al. Molecular epidemiology of group A rotavirus in outpatient diarrhea infants and children in Chongqing, China, 2011-2015. J Med Virol (2019) 91:1788–96. doi: 10.1002/jmv.25530
15. Liu N, Xu Z, Li D, Zhang Q, Wang H, Duan ZJ. Update on the disease burden and circulating strains of rotavirus in China: a systematic review and meta-analysis. Vaccine (2014) 32:4369–75. doi: 10.1016/j.vaccine.2014.06.018
16. Kawai K, O’Brien MA, Goveia MG, Mast TC, El Khoury AC. Burden of rotavirus gastroenteritis and distribution of rotavirus strains in Asia: a systematic review. Vaccine (2012) 30:1244–54. doi: 10.1016/j.vaccine.2011.12.092
17. Miles MG, Lewis KD, Kang G, Parashar UD, Steele AD. A systematic review of rotavirus strain diversity in India, Bangladesh, and Pakistan. Vaccine (2012) 30:A131–9. doi: 10.1016/j.vaccine.2011.10.002
18. Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. Updating guidance for reporting systematic reviews: development of the PRISMA 2020 statement. J Clin Epidemiol (2021) 134:103–12. doi: 10.1016/j.jclinepi.2021.02.003
19. Zeng X, Zhang Y, Kwong JS, Zhang C, Li S, Sun F, et al. The methodological quality assessment tools for preclinical and clinical studies, systematic review and meta-analysis, and clinical practice guideline: a systematic review. J Evidence-Based Med (2015) 8(1):2–10. doi: 10.1111/jebm.12141
20. Black R, Huq I, Merson M, Alim AR, Yunus MD. Incidence and severity of rotavirus and Escherichia coli diarrhoea in rural Bangladesh: implications for vaccine development. Lancet (1981) 317(8212):141–3. doi: 10.1016/S0140-6736(81)90719-4
21. Black RE, Merson MH, Rahman AM, Yunus M, Alim AA, Huq I, et al. A two-year study of bacterial, viral, and parasitic agents associated with diarrhea in rural Bangladesh. J Infect Dis (1980) 142(5):660–4. doi: 10.1093/infdis/142.5.660
22. Ahmed MU, Taniguchi KO, Kobayashi NO, Urasawa TO, Wakasugi F, Islam MU, et al. Characterization by enzyme-linked immunosorbent assay using subgroup-and serotype-specific monoclonal antibodies of human rotavirus obtained from diarrheic patients in Bangladesh. J Clin Microbiol (1989) 27(7):1678–81. doi: 10.1128/jcm.27.7.1678-1681.1989
23. Tabassum S, Shears P, Hart CA. Genomic characterization of rotavirus strains obtained from hospitalized children with diarrhoea in Bangladesh. J Med Virol (1994) 43(1):50–6. doi: 10.1002/jmv.1890430110
24. Fun BN, Unicomb L, Rahim Z, Banu NN, Podder G, Clemens J, et al. Rotavirus-associated diarrhea in rural Bangladesh: two-year study of incidence and serotype distribution. J Clin Microbiol (1991) 29(7):1359–63. doi: 10.1128/jcm.29.7.1359-1363.1991
25. Bern C, Unicomb L, Gentsch JR, Banul N, Yunus M, Sack RB, et al. Rotavirus diarrhea in Bangladeshi children: correlation of disease severity with serotypes. J Clin Microbiol (1992) 30(12):3234–8. doi: 10.1128/jcm.30.12.3234-3238.1992
26. Sanekata T, Ahmed MU, Kader A, Taniguchi K, Kobayashi N. Human group B rotavirus infections cause severe diarrhea in children and adults in Bangladesh. J Clin Microbiol (2003) 41(5):2187–90. doi: 10.1128/JCM.41.5.2187-2190.2003
27. Rahman M, Banik S, Faruque AS, Taniguchi K, Sack DA, Van Ranst M, et al. Detection and characterization of human group C rotaviruses in Bangladesh. J Clin Microbiol (2005) 43(9):4460–5. doi: 10.1128/JCM.43.9.4460-4465.2005
28. Rahman M, Sultana R, Ahmed G, Nahar S, Hassan ZM, Saiada F, et al. Prevalence of G2P [4] and G12P [6] rotavirus, Bangladesh. Emerg Infect Dis (2007) 13(1):18. doi: 10.3201/eid1301.060910
29. Tanaka G, Faruque AS, Luby SP, Malek MA, Glass RI, Parashar UD. Deaths from rotavirus disease in Bangladeshi children: estimates from hospital-based surveillance. Emerg Infect Dis (2007) 26(11):1014–8. doi: 10.1097/INF.0b013e318125721c
30. Paul SK, Kobayashi N, Nagashima S, Ishino M, Watanabe S, Alam MM, et al. Phylogenetic analysis of rotaviruses with genotypes G1, G2, G9 and G12 in Bangladesh: evidence for a close relationship between rotaviruses from children and adults. Arch Virol (2008) 153:1999–2012. doi: 10.1007/s00705-008-0212-9
31. Paul SK, Ahmed MU, Hossain MA, Mahmud MC, Bhuiyan MR, Saha SK, et al. Molecular characterization of group A human rotavirus among hospitalized children and adults in Bangladesh: finding of emerging G12 strain. Mymensingh Med Journal: MMJ (2010) 19(1):16–26.
32. Zaman K, Yunus MD, Faruque AS, El Arifeen S, Hossain I, Azim T, et al. Surveillance of rotavirus in a rural diarrhoea treatment centre in Bangladesh, 2000–2006. Vaccine (2009) 27:F31–4. doi: 10.1016/j.vaccine.2009.08.063
33. Dey SK, Thongprachum A, Islam AR, Phan GT, Rahman M, Mizuguchi M, et al. Molecular analysis of G3 rotavirus among infants and children in Dhaka City, Bangladesh after 1993. Infect Genet Evol (2009) 9(5):983–6. doi: 10.1016/j.meegid.2009.06.016
34. Islam MS, Alam MM, Ahmed MU, Saifuzzaman AB, Kobayashi N, Kayesh ME, et al. Molecular epidemiologic study on rotavirus infection in human and birds in association with gastroenteritis. Bangladesh J Veterinary Med (2009) 7(1):233–7.
35. Ahmed K, Ahmed S, Mitui MT, Rahman A, Kabir L, Hannan A, et al. Molecular characterization of VP7 gene of human rotaviruses from Bangladesh. Virus Genes. (2010) 40:347–56. doi: 10.1007/s11262-010-0463-x
36. Rahman M, Alamgir AS, Saiada F, Hassan Z, Faruque AS, Cravioto A, et al. Co-circulation of G1, G2 and G9 rotaviruses in hospitalized patients in Bangladesh during 2006-2009. Hum Vaccines (2011) 7(9):929–33. doi: 10.4161/hv.7.9.15988
37. Afrad MH, Hassan Z, Farjana S, Moni S, Barua S, Das SK, et al. Changing profile of rotavirus genotypes in Bangladesh, 2006–2012. BMC Infect Dis (2013) 13(1):1–7. doi: 10.1186/1471-2334-13-320
38. Sarker MH, Das SK, Ahmed S, Ferdous F, Das J, Farzana FD, et al. Changing characteristics of rotavirus diarrhea in children younger than five years in urban Bangladesh. PloS One (2014) 9(8):e105978. doi: 10.1371/journal.pone.0105978
39. Tanmoy AM, Ahmed AN, Arumugam R, Hossain B, Marzan M, Saha S, et al. Rotavirus surveillance at a WHO-coordinated invasive bacterial disease surveillance site in Bangladesh: a feasibility study to integrate two surveillance systems. PloS One (2016) 11(4):e0153582. doi: 10.1371/journal.pone.0153582
40. Satter SM, Gastanaduy PA, Islam K, Rahman M, Rahman M, Luby SP, et al. Hospital-based surveillance for rotavirus gastroenteritis among young children in Bangladesh: defining the potential impact of a rotavirus vaccine program. Pediatr Infect Dis J (2017) 36(2):168. doi: 10.1097/INF.0000000000001381
41. Haque W, Haque J, Barai D, Rahman S, Moni S, Hossain ME, et al. Distribution of rotavirus genotypes in Dhaka, Bangladesh, 2012–2016: re-emergence of G3P [8] after over a decade of interval. Vaccine (2018) 36(43):6393–400. doi: 10.1016/j.vaccine.2018.08.081
42. Satter SM, Aliabadi N, Gastañaduy PA, Haque W, Mamun A, Flora MS, et al. An update from hospital-based surveillance for rotavirus gastroenteritis among young children in Bangladesh, July 2012 to June 2017. Vaccine (2018) 36(51):7811–5. doi: 10.1016/j.vaccine.2018.05.032
43. Dey SK, Sharif N, Sarkar OS, Sarkar MK, Talukder AA, Phan T, et al. Molecular epidemiology and surveillance of circulating rotavirus among children with gastroenteritis in Bangladesh during 2014–2019. PloS One (2020) 15(11):e0242813. doi: 10.1371/journal.pone.0242813
44. Sharif N, Nobel NU, Sakib N, Liza SM, Khan ST, Billah B, et al. Molecular and epidemiologic analysis of diarrheal pathogens in children with acute gastroenteritis in Bangladesh during 2014–2019. Pediatr Infect Dis J (2020) 39(7):580–5. doi: 10.1097/INF.0000000000002637
45. Sarkar S, Esona MD, Gautam R, Castro CJ, Ng TF, Haque W, et al. Outbreak of diarrhoea in piglets caused by novel rotavirus genotype G4P [49] in north-western district of Bangladesh, February 2014. Transbound Emerg Dis (2020) 67(1):442–9. doi: 10.1111/tbed.13343
46. Islam A, Hossain ME, Haider N, Rostal MK, Mukharjee SK, Ferdous J, et al. Molecular characterization of group A rotavirus from rhesus macaques (Macaca mulatta) at human–wildlife interfaces in Bangladesh. Transbound Emerg Dis (2020) 67(2):956–66. doi: 10.1111/tbed.13431
47. Islam A, Hossain ME, Rostal MK, Ferdous J, Islam A, Hasan R, et al. Epidemiology and molecular characterization of rotavirus A in fruit bats in Bangladesh. EcoHealth (2020) 17:398–405. doi: 10.1007/s10393-020-01488-7
48. Hossain MB, Rahman MS, Watson OJ, Islam A, Rahman S, Hasan R, et al. Epidemiology and genotypes of group A rotaviruses in cattle and goats of Bangladesh, 2009-2010. Infect Genet Evol (2020) 79:104170. doi: 10.1016/j.meegid.2020.104170
49. Unicomb LE, Podder G, Gentsch JR, Woods PA, Hasan KZ, Faruque AS, et al. Evidence of high-frequency genomic reassortment of group A rotavirus strains in Bangladesh: emergence of type G9 in 1995. J Clin Microbiol (1999) 37(6):1885–91. doi: 10.1128/JCM.37.6.1885-1891.1999
50. Selim A, Kabir AR, Aminur R, Maleeha H, Soofia K, Abdul H. Severity of rotavirus diarrhea in children: one year experience in a children hospital of Bangladesh. Iranian J Pediatrics (2009) 19(2):108–16.
51. Ahmed MU, Urasawa S, Taniguchi K, Urasawa T, Kobayashi N, Wakasugi F, et al. Analysis of human rotavirus strains prevailing in Bangladesh in relation to nationwide floods brought by the 1988 monsoon. J Clin Microbiol (1991) 29(10):2273–9. doi: 10.1128/jcm.29.10.2273-2279.1991
52. Ahmed MU, Kobayashi N, Wakuda M, Sanekata T, Taniguchi K, Kader A, et al. Genetic analysis of group B human rotaviruses detected in Bangladesh in 2000 and 2001. J Med Virol (2004) 72(1):149–55. doi: 10.1002/jmv.10546
53. Giri S, Hemavathy RP, Arumugam R, Sherchand JB, Thu HM, Galagoda G, et al. Molecular epidemiology of rotaviruses in the south-east Asian region from 2009 to 2015. Vaccine (2018) 36:7851–5. doi: 10.1016/j.vaccine.2018.02.092
54. Sadiq A, Bostan N, Bokhari H, Matthijnssens J, Yinda KC, Raza S, et al. Molecular characterization of human group A rotavirus genotypes circulating in Rawalpindi, Islamabad, Pakistan during 2015-2016. PloS One (2019) 14(7):e0220387. doi: 10.1371/journal.pone.0220387
55. Ouermi D, Soubeiga D, Nadembega WMC, Sawadogo PM, Zohoncon TM, Obiri-Yeboah D, et al. Molecular epidemiology of rotavirus in children under five in Africa (2006-2016): A systematic review. Pak J Biol Sci (2017) 20(2):59–69. doi: 10.3923/pjbs.2017.59.69
56. Tcheremenskaia O, Marucci G, De Petris S, Ruggeri FM, Dovecar D, Sternak SL, et al. Molecular epidemiology of rotavirus in Central and Southeastern Europe. J Clin Microbiol (2007) 45(7):2197–204. doi: 10.1128/JCM.00484-07
57. Grimprel E, Rodrigo C, Desselberger U. Rotavirus disease: impact of coinfections. Pediatr Infect Dis J (2008) 27:S3–S10. doi: 10.1097/INF.0b013e31815eedfa
58. Jagai JS, Sarkar R, Castronovo D, Kattula D, McEntee J, Ward H, et al. Seasonality of rotavirus in South Asia: a meta-analysis approach assessing associations with temperature, precipitation, and vegetation index. PloS One (2012) 7:e38168. doi: 10.1371/journal.pone.0038168
59. Ameta P, Nayak VH, Goyal SC. Prevalence and Seasonal distribution of Rotavirus Diarrhea in hospitalized children less than 5 years old in South Rajasthan. Int J Biomed Res (2015) 6:214–8. doi: 10.7439/ijbr.v6i3.1672
60. Güzel M, Akpınar O, Kılıç MB. Prevalence of rotavirus-associated acute gastroenteritis cases in early childhood in Turkey: meta-analysis. Children (2020) 7(10):159. doi: 10.3390/children7100159
61. Yu J, Lai S, Geng Q, Ye C, Zhang Z, Zheng Y, et al. Prevalence of rotavirus and rapid changes in circulating rotavirus strains among children with acute diarrhea in China, 2009–2015. J Infect (2019) 78(1):66–74. doi: 10.1016/j.jinf.2018.07.004
62. Arakaki L, Tollefson D, Kharono B, Drain PK. Prevalence of rotavirus among older children and adults with diarrhea: A systematic review and meta-analysis. Vaccine (2021) 39(33):4577–90. doi: 10.1016/j.vaccine.2021.06.073
63. Monavari SH, Hadifar S, Mostafaei S, Miri A, Keshavarz M, Babaei F, et al. Epidemiology of rotavirus in the Iranian children: A systematic review and meta-analysis. J Global Infect Dis (2017) 9(2):66. doi: 10.4103/0974-777X.205173
64. Gupta S, Chaudhary S, Bubber P, Ray P. Epidemiology and genetic diversity of group A rotavirus in acute diarrhea patients in pre-vaccination era in Himachal Pradesh, India. Vaccine (2019) 37(36):5350–6. doi: 10.1016/j.vaccine.2019.07.037
65. Leshem E, Lopman B, Glass R, Gentsch J, Bányai K, Parashar U, et al. Distribution of rotavirus strains and strain-specific effectiveness of the rotavirus vaccine after its introduction: a systematic review and meta-analysis. Lancet Infect Dis (2014) 14(9):847–56. doi: 10.1016/S1473-3099(14)70832-1
66. Damtie D, Melku M, Tessema B, Vlasova AN. Prevalence and genetic diversity of rotaviruses among under-five children in Ethiopia: A systematic review and meta-analysis. Viruses (2020) 12(1):62. doi: 10.3390/v12010062
67. Zhou N, Lv D, Wang S, Lin X, Bi Z, Wang H, et al. Continuous detection and genetic diversity of human rotavirus A in sewage in eastern China, 2013–2014. Virol J (2016) 13:1–8. doi: 10.1186/s12985-016-0609-0
68. Jeong S, Kim W. A systematic review of genetic diversity of human rotavirus circulating in South Korea. Infect Genet Evol (2014) 28:462–9. doi: 10.1016/j.meegid.2014.08.020
69. Sharif N, Parvez AK, Haque A, Talukder AA, Ushijima H, Dey SK. Molecular and epidemiological trends of human bocavirus and adenovirus in children with acute gastroenteritis in Bangladesh during 2015 to 2019. J Med Virol (2020) 92(12):3194–201. doi: 10.1002/jmv.25812
70. Dey SK, Sharif N, Billah B, Siddique TT, Islam T, Parvez AK, et al. Molecular epidemiology and genetic diversity of norovirus infection in children with acute gastroenteritis in Bangladesh, 2014–2019. J Med Virol (2021) 93(6):3564–71. doi: 10.1002/jmv.26772
71. Sharif N, Ahmed SN, Sharif N, Alzahrani KJ, Alsuwat MA, Alzahrani FM, et al. High prevalence of norovirus GII. 4 Sydney among children with acute gastroenteritis in Bangladesh, 2018–2021. J Infect Public Health (2023) 16(7):1015–22. doi: 10.1016/j.jiph.2023.05.002
Keywords: rotavirus, genotype, epidemiology, diversity, Bangladesh, pre-vaccination
Citation: Sharif N, Sharif N, Khan A, Azpíroz ID, Diaz RM, Díez IDlT, Parvez AK and Dey SK (2023) Prevalence and genetic diversity of rotavirus in Bangladesh during pre-vaccination period, 1973-2023: a meta-analysis. Front. Immunol. 14:1289032. doi: 10.3389/fimmu.2023.1289032
Received: 05 September 2023; Accepted: 06 November 2023;
Published: 22 November 2023.
Edited by:
Susu M. Zughaier, Qatar University, QatarReviewed by:
David A. Montero, University of Concepcion, ChileCopyright © 2023 Sharif, Sharif, Khan, Azpíroz, Diaz, Díez, Parvez and Dey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuvra Kanti Dey, c2h1dnJhZGV5QHlhaG9vLmNvbQ==; Nadim Sharif, bmFkaW1ibWJAbGl2ZS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.