
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 17 October 2023
Sec. Molecular Innate Immunity
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1286820
This article is part of the Research Topic RNA Epigenetics in the Normal and Malignant Development of Immune Cells View all 4 articles
 Shenghai Shen1†
Shenghai Shen1† Li-Sheng Zhang1,2*
Li-Sheng Zhang1,2*The post-transcriptional RNA modifications impact the dynamic regulation of gene expression in diverse biological and physiological processes. Host RNA modifications play an indispensable role in regulating innate immune responses against virus infection in mammals. Meanwhile, the viral RNAs can be deposited with RNA modifications to interfere with the host immune responses. The N6-methyladenosine (m6A) has boosted the recent emergence of RNA epigenetics, due to its high abundance and a transcriptome-wide widespread distribution in mammalian cells, proven to impact antiviral innate immunity. However, the other types of RNA modifications are also involved in regulating antiviral responses, and the functional roles of these non-m6A RNA modifications have not been comprehensively summarized. In this Review, we conclude the regulatory roles of 2’-O-methylation (Nm), 5-methylcytidine (m5C), adenosine-inosine editing (A-to-I editing), pseudouridine (Ψ), N1-methyladenosine (m1A), N7-methylguanosine (m7G), N6,2’-O-dimethyladenosine (m6Am), and N4-acetylcytidine (ac4C) in antiviral innate immunity. We provide a systematic introduction to the biogenesis and functions of these non-m6A RNA modifications in viral RNA, host RNA, and during virus-host interactions, emphasizing the biological functions of RNA modification regulators in antiviral responses. Furthermore, we discussed the recent research progress in the development of antiviral drugs through non-m6A RNA modifications. Collectively, this Review conveys knowledge and inspiration to researchers in multiple disciplines, highlighting the challenges and future directions in RNA epitranscriptome, immunology, and virology.
Numerous emerging research areas, including cancer immunotherapy, gene therapy, regenerative medicine, and the pandemic, underscore the significance of investigating the innate immune system for advancing human healthcare and pharmaceutical innovation (1–4). As the fields of biomedical and biotechnology forge ahead at an unprecedented pace, our comprehension of this intricate regulatory network continually evolves, with established paradigms being supplemented from different perspectives and interdisciplinary insights emerging.
As the first line of defense against invaders, the innate immune system protects the body from the harmful effects of viral infections. A typical antiviral response can be succinctly outlined as follows: first and foremost, the innate immune system recognizes virus pathogen-associated molecular patterns (PAMPs) and triggers downstream signaling pathways. PAMPs typically include viral single-stranded (ss) or double-stranded (ds) RNAs, dsDNAs, and viral proteins (5, 6). The molecules in the immune system that detect PAMPs and transmit signals are pattern recognition receptors (PRRs). Common types of RNA-sensing PRRs include Toll-like receptors (TLRs), Retinoic Acid Inducible Gene-I (RIG-I)-like receptors (RLRs), NOD-like receptors (NLRs), C-type lectin receptors (CLRs), Protein Kinase R (PKR), and 2’-5’-Oligoadenylate Synthetases (OAS) (5, 7). These PRRs are one of the main targets for RNA viruses to evade immune surveillance (8–11). The characteristics and functions of the major PRRs are described in Table 1. Upon activation of signaling pathways by PRR, a large number of cytokines and chemokines (Activator protein-1 (AP-1), interferon regulatory factors (IRFs), etc.) are induced through pathways like NF-κB and MAPK, initiating the expression of downstream immunomodulatory and antiviral genes, with notable examples such as interferon (IFN) family and interleukin (IL) family (15). Concurrently, macrophages, natural killer (NK) cells, granulocytes, dendritic cells (DCs), and other immune cells are activated, recruited, and dynamically engaged in the antiviral immune response (5).
Due to the dynamic and rapid-response nature of the innate immune antiviral process, there has been considerable interest in exploring the realm of post-transcriptional regulation, a directly functional control layer of this process (16, 17). While extensive research has shed light on the roles of RNA splicing and non-coding (nc) RNA modulation in innate immunity regulation, there has been a growing focus on investigating the regulatory effects of RNA modifications (18–20). These chemical modifications have been demonstrated to be linked to the RNA sensing and activation of immune cells as early as 2005 (21). In recent years, many studies have reported diverse RNA modifications on viral and host RNAs and a complex network of interactions between RNA modifications and immune cells (22–25). Among these, N6-methyladenosine (m6A) stands out as the most abundant RNA modification in mammalian mRNA. The advancement of the base-resolution mapping tools targeting m6A, which are continually being refined, has facilitated the ongoing identification of its functional and mechanistic roles in diverse regulatory networks (26–32). Milestone studies continue to emerge, and the role of m6A in antiviral innate immunity regulation has been delving deeply, such as leading to immune evasion, influencing IFN production, and facilitating macrophage activation (33–35), with several related reviews published recently (34, 36).
Notably, in addition to m6A, many other RNA modifications have been demonstrated to possess a wide range of regulatory functions. Common types include 2’-O-Methylation (Nm), 5-methylcytidine (m5C), adenosine-inosine editing (A-to-I editing), pseudouridine (Ψ), N1-methyladenosine (m1A), N7-methylguanosine (m7G), N6,2’-O-dimethyladenosine (m6Am), and N4-acetylcytidine (ac4C). These non-m6A RNA modifications manifest diverse and intricate regulatory roles in controlling gene expression, modulating metabolic networks, and impacting the development of diseases (37), concomitant with the progress made in RNA epitranscriptome (38, 39). Intriguingly, these non-m6A RNA modifications are intimately linked with antiviral innate immunity, exerting biological and physicochemical regulatory functions upon both viral and host RNAs. Especially in recent years, the regulatory role of non-m6A RNA modifications in innate immunity has been heavily investigated, attracting considerable attention within translational medicine research (7, 20, 40).
Hence, in light of the present state of the cutting-edge investigation in non-m6A epitranscriptome and the gap of a summary related to innate immunity, here we comprehensively reviewed the regulatory role of the non-m6A RNA modifications in the antiviral innate immunity.
The role of diverse non-m6A RNA modifications in antiviral innate immune responses can be divided into the regulation of host RNA modification and viral RNA modification, impacting immune cell development and cytokine production, disguising endogenous RNA, and affecting viral development (36), with their chemical structures illustrated in Figure 1. From a dynamic perspective, the regulatory role of RNA modifications encompasses facilitating viral evasion from RNA sensing, infection-induced repercussions on cytokine production and signal transduction, and altering the epitranscriptome of immune cells to affect their function (37).
2’-O-methylation (Nm) modification is a highly conserved modification in which the 2’-hydroxyl group of nucleotides is methylated by 2’-O-methyltransferase (2’-O-MTase) co-transcriptionally or post-transcriptionally. Discovered initially to exist in ribosomal RNA (rRNA) and transfer RNA (tRNA), Nm has since been found to exhibit varying levels of abundance in diverse RNA species (41). Base-resolution sequencing techniques have been developed to detect Nm modifications, bringing breakthroughs in understanding the stoichiometric characteristics of this modification (42, 43). In human mRNAs, Nm modification near the cap structure is generally added by Cap methyltransferase 1 (CMTR1) or CMTR2, while Nm within internal positions could be installed by Fibrillarin (FBL) and FTSJ3 regulators (25, 44, 45). The involvement of a ribonucleoprotein (snoRNP) complex containing a small nucleolar RNA of the C/D box family (snoRNA) is essential in FBL-regulated Nm modifications (46, 47). Besides maintaining stability and promoting translation, the cap Nm of human mRNA commonly serves as a molecular signature of endogenous host RNA, and the lack of this modification may lead to autoimmune diseases (48). Internal mRNA Nm can hinder translation elongation by disrupting the interaction between translation components and tRNA decoding efficiency (49, 50); meanwhile, it is reported to stabilize mRNA and enhance its expression capacity (47). Furthermore, Nm plays essential roles in other types of RNA, such as rRNA Nm affecting ribosome heterogeneity and stability, and tRNA Nm maintaining translation accuracy and tRNA stability (48, 51). Pathologically, Nm exerts a regulatory influence on the occurrence of diseases such as cancer, autoimmune diseases, and epilepsy (52–55).
Apart from maintaining the stability of viral RNA by preventing the activity of host decapping and exoribonuclease protein, a paramount role of Cap Nm in antiviral innate immunity lies in facilitating viral RNA evasion from host RNA-sensing, thereby enabling immune evasion (Figure 2) (56). Since the Nm modification is installed at the first base (Cap 1 Nm) in eukaryotic host mRNA, numerous viruses employ this strategy to conceal themselves from detection by PRRs (57). By exhibiting Cap 1 Nm on their viral RNA, the viruses engage in a “subterfuge” that effectively evades recognition by RLRs, particularly RIG-I and MDA5 (58, 59), which disrupts signaling cascades, significantly suppresses the production of IFN, and impacting macrophage activation (60). In the case of RIG-I, this evasion effect can be attributed to steric hindrance resulting from the spatial obstruction caused by Nm when RIG-I binds to RNA, primarily facilitated by residue H830 (59, 61). On the other hand, although structural biology studies have inspired, the interaction mechanism between MDA5 and Nm still remains unclear. Recent studies suggest that Nm may reduce the catalytic activity switch of MDA5 (62). Moreover, viral RNA Cap 1 Nm also affects RNA sensing through TLR7 pathways, consequently impacting inflammatory responses (56). Although the inhibitory effect of Nm in bacterial tRNA on TLR activation has been studied to some extent, the relevant mechanisms and degree of inhibition in viruses are yet to be determined (63, 64).
Another escape strategy mediated by Cap Nm is to prevent the binding of viral RNA to IFN-induced protein with tetratricopeptide repeats 1 (IFIT1), an antiviral protein (65). IFIT1 is upregulated in response to viral infection, guided by interferon signals, and can recognize viral RNA lacking Cap Nm. It forms a complex with IFIT2 and IFIT3 and interacts with the eukaryotic initiation factor eIF3, inhibiting the transcription and translation of viral RNA, which is essential for IFN-induced antiviral response (60, 66). The internal RNA binding tunnel of IFIT1 exhibits a preference for RNA without Cap Nm, thus displaying a limited affinity for viral RNA with this modification (67). IFIT1B of the same family retains the ability to bind Cap1 Nm RNA, albeit with reduced affinity (68). Therefore, Cap Nm helps various viruses, such as West Nile virus, Zika virus, and human immunodeficiency virus (HIV), to escape the antiviral effect of IFN-induced antiviral factors (45, 69, 70). Coronaviruses lacking 2’-O-methyltransferase (2’-O MTase) exhibit high sensitivity to IFIT1 (71). Note that Cap Nm is only one of several strategies used by viruses to evade IFIT monitoring, while viral RNAs with Cap 1 Nm are still affected by IFIT1 at high concentrations; however, the viruses with only Cap 2 Nm are able to avoid IFIT1 effects completely, but this is rare among viruses (60, 67, 72, 73). Interestingly, a recent study reported that Cap 2 Nm in mammalian mRNAs shifts from Cap 1 Nm at a very slow methylation rate and mainly accumulates on host mRNAs, ensuring a low level of Cap 2 Nm in nascent viral RNAs and a certain level of immunostimulation (74).
Despite the incomplete comprehension of internal Nm modifications in viral RNA, recent studies have partially illuminated their regulatory role in innate immunity. Like internal Nm modifications in host mRNA, internal Nm modifications in viral RNA of various viruses can impair replication by affecting the elongation of the RNA polymerase (75). Interestingly, HIV is an exception to this. A recent report proved that internal Nm modifications induced by host 2’-O MTase promote HIV viral RNA replication (45). Furthermore, these modifications are implicated in immune evasion strategies employed by HIV, manifesting through their impact on MDA5 sensing and the antiviral activity of ISG20 (45, 76). While some investigations have shown that integrating Nm-modified adenosine into short RNAs can inhibit TLR7 activation, further exploration is warranted to determine if analogous mechanisms exist for other viral types (60, 77). Internal Nm modifications have been discovered in viral RNAs of many viruses, such as SARS-CoV-2, Ebola virus, and Dengue virus, but their functions remain unclear (75, 78, 79), awaiting more in-depth functional investigation.
In addition to the essential role in the development of immune cells such as macrophages and the expression of immune-related genes, host RNA Nm modification can also dynamically regulate the immune response through changes in Nm stoichiometry (80). The human Cap 1 2’-O MTase, CMTR1, exhibits an upregulated expression in response to interferon, thereby modifying the Nm status of specific antiviral ISG genes to enhance their expression and facilitate IFN-mediated antiviral response (25, 81). Some studies indicated the protective role of Nm modification on host RNA, such as increased vulnerability to viral infection in hosts lacking Nm-modified tRNA and resistance of Nm-modified host siRNA to targeting by poxviruses (82, 83). A recent study has also found that the introduction of Nm modifications on the RNA template significantly inhibits the synthesis of viral RNA (84). Moreover, viruses can manipulate the landscape of host RNA Nm modification. A typical example is that viruses can exploit FBL to modify host pre-rRNA’s Nm status, thereby attenuating protein synthesis. For instance, HIV infection disrupts FBL’s binding to nascent pre-rRNA, impairing ribosome biogenesis and function (85). Similarly, the Hendra virus orchestrates FBL methylation to influence proviral host genes and viral protein synthesis (44). Additionally, the disturbance of FBL has the potential to influence Nm modification level, consequently impeding the Type I IFN response and thereby facilitating viral infiltration into macrophages (86). Intriguingly, recent investigations have unveiled a decline in Nm sites within host mRNA upon SARS-CoV-2 infection, albeit the implications of this phenomenon on the host necessitate further study (78).
Comprehending how viruses synthesize Nm to circumvent immune responses is paramount in designing specialized antiviral medications. Three conventional ways for viral RNA to acquire Cap Nm have been identified, including utilizing host ‘writer’ enzymes (e.g., HIV), “cap snatching” from host mRNA (e.g., influenza virus), and encoding specialized enzymes and active sites for capping (e.g., SARS-COV-2) (25, 45, 87). The elucidation of these mechanisms propels advancements in translational medical research. Taking SARS-COV-2 as an example, several non-structural proteins (Nsps) are involved in viral RNA Nm modification, with pivotal participants including Nsp13, Nsp10, Nsp16, etc (88). Structural biology and biochemical studies of these 2’-O-MTases inspire drug development (89). Numerous drugs targeting these Nsp proteins have been conceptualized and developed (90–92). Similarly, drugs targeting the Nm synthesis mechanisms of viruses such as dengue, influenza, and Japanese encephalitis have also been reported (93–95). In addition, since viruses lacking the Cap Nm-deficient phenotype significantly attenuated virulence, these defective viruses demonstrate feasibility as a vaccine approach (96).
RNA m5C is a modification that occurs at the 5th position of cytosine residues and is widely distributed in eukaryotes RNA, with high enrichment in tRNA and other non-coding RNAs (37). With the advancement of related high-throughput sequencing technologies, the regulatory mechanisms of m5C and the corresponding writer, eraser, and reader regulatory proteins have been continually explored (97–99). Although much remains unknown, m5C writers have been relatively more well-studied. Two classes of writers have been identified: the DNA methyltransferase 2 (DNMT2) and the NOL1/NOP2/SUN domain (NSUN) family (100). Notably, these eukaryotic m5C writers deposit this modification selectively based on RNA types. For instance, NSUN2 and NSUN6 are responsible for m5C deposition on mRNA, NSUN1 and NSUN5 modify cytoplasmic ribosomal RNA, and mitochondrial RNA m5C is installed by NSUN3 and NSUN4 (100). This complexity directly leads to the diverse regulatory functions of m5C in innate immunity. In host RNA, the typical role of m5C is to maintain RNA stability. Furthermore, m5C impacts mRNA export through the ‘reader’ protein ALYREF, safeguards tRNA against stress-induced damage, influences protein synthesis rates, and actively partakes in ribosome biogenesis (20, 37, 101). Besides its involvement in embryonic development processes, m5C is intimately linked to various diseases, such as cancer and neurological disorders (20, 101).
Currently, there are no reports of viruses encoding their own m5C writer. However, they can utilize host writers to aid in their invasion (Figure 3). The most common case is the methylation of CpG islands on viral RNA mediated by DNMT2 and NSUN5 (102). Although the specific function is unclear, this may mediate viral heterogeneity (102). HIV has demonstrated its active recruitment of NSUN2 and DNMT for installing m5C on its viral RNA, thereby enhancing genome stability and facilitating replication, translation, and virus assembly efficiency (103, 104). Similar mechanisms have been observed in Mouse Leukemia Virus (MLV) and Alphaviruses, where a diminished NSUN2 level can reduce viral infection through downregulating m5C modifications (105, 106). Interestingly, the downregulation of NSUN2 in the Epstein-Barr (EB) virus increases viral RNA in vivo, as EB viral RNA degradation is m5C-dependent by RNase Angiogenin (107). In addition, viruses can employ m5C modifications on viral RNA to influence nuclear export and infectivity via interaction with host m5C readers like ALYREF (108, 109). Some studies have also reported the enrichment of m5C in the viral genome following infection; however, the specific function awaits further exploration (110).
In host cells, m5C is essential for maintaining immune homeostasis and immune cells like CD4 T Cells development (111, 112). In the innate antiviral response, m5C and its regulator have a sophisticated role. NSUN5 can bind to viral RNA and enhance the RNA-sensing function of RIG-I, functioning as a cardinal receptor (113). NSUN6 is involved in plasma cell formation (114). Meanwhile, DNMT2 responds to infection and relies on the dynamic installation of m5C to regulate the expression of antiviral genes, facilitating an efficient response (115, 116). NSUN1 inhibits HIV-1 replication and prolongs its latency by installing m5C on its transactivation response element RNA (117). It is worth mentioning that NSUN2, the most reported m5C writer associated with antiviral activity, exhibits functionally diverse regulatory roles. In response to viral infection, NSUN2 downregulates the levels of specific ncRNAs and alters their m5C levels, which directly and indirectly modulates the type I interferon response mediated by the RIG-I signaling pathway and enhances the antiviral response (118). In addition, NSUN2 can convert vault RNA (vtRNA) into smaller fragments through m5C installation (119, 120). Some viruses can induce high vtRNA expression, inhibiting the activation of PKR and subsequent IFN response, silencing the host antiviral immune response (121). Similarly, tRNA-derived non-coding fragments (tRFs) generated with the involvement of NSUN2 can be utilized by viruses for their immune escape with viral RNA replication (122–124). In addition, NSUN2 may interact with uridylated deaminase, which inhibits viral activity (102).
Although there are currently rare antiviral drugs targeting RNA m5C, due to substantial overlap with regulators of DNA 5-methylcytosine, a large number of antiviral drugs (e.g., azacytidine and decitabine) that target DNA methylation also exhibit effects on RNA m5C (125). Nonetheless, given that the dearth of regulatory mechanisms has only been filled in the last few years, the controversial distribution on the viral genome, and the bidirectional regulatory properties of the regulator in innate immunity, present a formidable challenge to the advancement of drugs targeting m5C (126).
A-to-I editing is an irreversible modification that widely exists in pre-mRNA, mRNA, and ncRNA, converting the amino group at position C6 of adenosine to a carbonyl group, resulting in inosine (37). In mammals, this modification is mediated by adenosine deaminases acting on RNA (ADARs) and adenosine deaminase acting on tRNA (ADAT), and these proteins have multiple dsRNA binding domains and a catalytic center (127). There are three types of ADARs in humans: ADAR1, ADAR2, and ADAR3 (which lacks catalytic activity in vitro and is considered a negative regulator of editing) (128). Among them, ADAR1 is responsible for the vast majority of modifications. Most ADAR’s actions occur within double-stranded RNAs formed by inverted Alu repeat elements scattered throughout the genome (128). This modification can disrupt RNA secondary structure and destabilize dsRNA. In the bioprocess, ADARs are involved in pre-mRNA processing (129). In host mRNA, inosine in non-coding regions can regulate specific gene expressions by adding or subtracting splicing donor or acceptor sites thus affecting alternative splicing, creating or destroying miRNA binding sites, and affecting mRNA stability (129–131). Within coding regions, A-to-I editing can regulate translation efficiency and generate new protein products (132). In ncRNA, ADARs play a role in the biogenesis of miRNA and circular RNA, and they can also adjust the miRNA targets (133). In addition, ADAR can change the secondary structure of lncRNA, thereby affecting its interaction with miRNA (20). Pathologically, A-to-I editing has been found to be associated with cancer, neurodegenerative diseases, and autoimmune diseases (127, 134, 135).
In host cells, ADAR1, especially its IFN-responsive isoform ADAR1 p150, is a critical IFN inhibitory factor in antiviral innate immunity, and its negative regulation is essential for suppressing abnormal antiviral responses and maintaining immune homeostasis (Figure 4) (136). Endogenous dsRNA may induce innate immune activation and cause autoimmune inflammatory diseases. ADAR1 possesses the ability to modify these immunogenic molecules through A-to-I editing, thereby influencing their interaction with dsRNA PRRs and effectively thwarting downstream sensor (e.g., MAVS) activation as well as ISG-IFN and pathways and inflammation (128). IFN signaling requires down-regulation of ADAR1-p110 during viral infection to execute effective antiviral activity (137). These mechanisms are also essential to prevent excessive immune responses to the viral RNA (138). Mutations in ADAR can be observed in many autoimmune diseases, such as Aicardi-Goutières syndrome (AGS) (139). In the RLR pathway, ADAR1 probably influences MDA5’s binding affinity to dsRNA by inducing structural alterations through installed inosine residues and imposing limitations on RIG-I RNA-sensing capacity via protein-protein interaction since this inhibition is not dependent on catalytic activity but rather RNA binding activity (140, 141). In the PKR pathway, ADAR1 inhibits the autophosphorylation activation of this dsRNA PRR, thereby preventing subsequent eIF2α-induced translation arrest (142). Some studies have also shown that ADAR affects the activation of the OAS-RNase L pathway and its induced RNA degradation, autophagy, and cell apoptosis (143). Moreover, ADAR1 has been found to interact with Z-DNA binding protein 1, limiting self-Z-RNA sensing and avoiding the type I IFN pathway (144, 145). Other studies demonstrated that ADAR1 can modulate immunity by directly editing IFN pathway components (128, 146, 147). Note that the details of how these editings affect PRRs and sensors need to be further investigated. In addition, host A-to-I editing regulates the development and activation of immune cells. A-to-I editing is essential for maintaining intracellular homeostasis in DCs and macrophages, and the loss of ADAR1 leads to cellular metabolic disorders (148). Furthermore, ADAR can affect macrophage polarization by inhibiting the biogenesis of miR-21 (147).
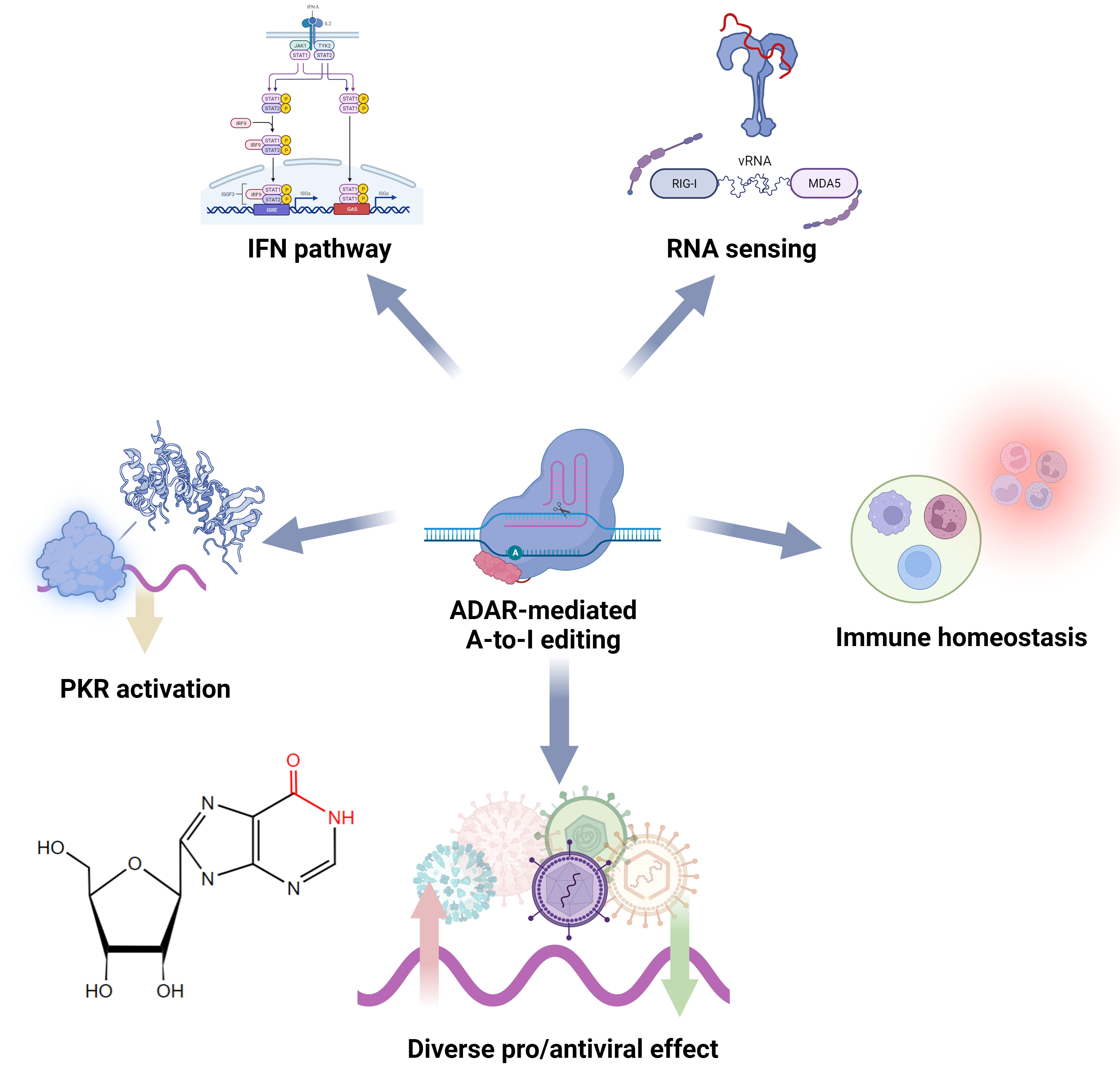
Figure 4 Functional roles of ADAR and A-to-I editing in the regulation of antiviral innate immunity.
Through editing the viral RNA, transcripts, template strands, and immune-responsive RNA, ADAR’s impact on viruses can be either proviral, antiviral, or even have no effect. This complexity primarily depends on the combination of the virus and host and the specificity of the editing sites. Like Nm, viruses can utilize/recruit host ADAR to edit viral RNA, effectively disguising it as “self” to evade detection by RNA-sensing (149). Notable examples include HIV, hepatitis C virus (HCV), Ebola virus, and measles virus (MeV), which utilize ADAR1 to edit their genomes/transcriptomes, thereby evading detection by RLR and other PRRs, and inhibiting IFN induction by suppressing IRF3 activation, thus blocking the immune response (150–154). Furthermore, some viruses have shown the ability to induce high expression of ADAR1 to raise the threshold for immune response (155). In addition, Dengue viruses were shown to modulate ADAR abundance by regulating the expression of microRNAs targeting ADAR1 (156). Different subtypes of ADAR1 (p150 and p110) may possess distinct modification sites and functions within the same virus. For instance, during influenza A virus infection, p150 hinders RLR signaling and assumes a pro-viral role; conversely, p110 appears to exert an antiviral effect by editing viral RNA to impact replication efficiency (157–160). On the other hand, ADAR2 has also been shown to promote the immune escape of the Borna disease virus by editing its genome (161). Interestingly, the linear correlation between PKR and IFN-β protein levels, as well as the antagonistic effect of PKR on ADAR’s immune inhibitory function in various viruses, suggests a balanced interaction between PKR and ADAR1 in antiviral immunity (154, 162). Escape mechanisms utilizing non-structural proteins and the transactivation response element were identified in viral infections by regulating PKR, interacting with the PKR activating compound of translation, and manipulating ADAR1 to inactivate PKR, which inhibited the suppression of viral replication and the subsequent formation of antiviral stress granules (163–167). Intriguingly, this “ loophole “ in ADAR that viruses can exploit appears to be an evolutionary invention for preventing aberrant antiviral responses (160, 168, 169). This is also evidenced by its complex regulatory network and low concentration level in the cytoplasm (170).
ADAR can achieve its proviral effects through other mechanisms as well. Viruses can exploit ADAR and influence viral RNA’s replication, transcription, and protein production efficiency through editing (171). A typical example is HIV, which hijacks ADAR1 and ADAR2 to achieve A-to-I editing, resulting in faster translation efficiency and significantly enhanced release of progeny viruses (152, 172). A study has demonstrated that ADAR1 is essential for efficiently replicating HIV-1 in T cells (173). It is worth noting that the stimulatory effect of ADAR1 on HIV-1 is achieved through both editing-dependent and editing-independent mechanisms (153). Moreover, ADAR1 can modify viral protein products, thereby impacting viral proliferation and infectivity. For instance, by modifying the RNA template of the hepatitis delta virus, it can induce the production of proteins that foster viral particle packaging (174, 175). ADAR can also provide a replication environment for viruses by disrupting endogenous RNA interference (176). In addition, ADAR1 has been shown to interact with the DICER protein and inhibit its cleavage of edited dsRNA (177). Viruses may recruit ADAR to suppress DICER, thereby escaping viral gene silencing (128). Recent studies have also found that ADAR editing accelerates the evolution of SARS-CoV-2 in humans and may be related to the infectivity of its spike protein (175, 178).
As an immune regulator, ADAR has been shown to have antiviral activity. Cells lacking ADAR exhibit increased susceptibility to various viruses, while overexpression of ADAR1 can inhibit viral replication (179). During different stages of viral infection, ADAR exhibits different proviral and antiviral effects that depend on the inflammatory response (180). Similar to its proviral regulatory, ADAR can also exert antiviral effects through site-specific editing. For instance, by editing HCV’s replicon, ADAR significantly curtails its replication; by editing MeV’s non-encapsidated defective interfering RNA, it diminishes infectivity; by editing long terminal repeat retrotransposons, it restricts their activity (181–183). ADAR can also reduce the expression of the encephalomyocarditis virus-encoded circRNA for antagonizing PKR activation by editing it to promote immune activation (184, 185). Additionally, ADAR1 can upregulate antiviral microRNA expression in response to infection (141). An interesting case is that since PKR activity favors HCV replication, the inhibitory effect of ADAR on PKR indirectly suppresses HCV (186). Furthermore, many studies have shown that, in the long run, the evolutionary effects of ADAR-induced editing pressure on viral genomes are likely detrimental (168, 187, 188). ADAR editing of the SARS-CoV-2 genome may reduce transmissibility (189). Thus, ADAR may be an “evolutionary weapon”. ADAR has also demonstrated an association with antiviral immune responses in many other cases, but whether ADAR directly involves these modulations remains unclear (186).
ADAR has been widely used in RNA editing therapy for diseases like cancer and metabolic disorders, as it exerts regulatory influence devoid of genome disruption (190). However, within the domain of antiviral drug development, despite the potential utility of investigating ADAR’s editing of viral genomes to inspire the development of potent vaccines, studies of related drugs are still sparse (191, 192). Numerous enigmas remain shrouded in ambiguity. Firstly, the mechanisms and biological effects of ADAR’s editing of viral genomes in the immune response are still inconclusive, and these effects may vary among different viruses. Viruses can also induce editing of host RNA during infection, such as the reduction of A-to-I editing of endogenous Alu RNA caused by SARS-CoV-2 (193). Further research is needed on the role of ADAR in virus-host interactions. Secondly, understanding how ADAR rapidly responds to viral infections, how host cells dynamically regulate the concentration of ADAR, and how this “double-edged sword” affects the diverse impact on dsRNAs are crucial for drug design. Research from a molecular evolution perspective may inspire. Molecular evolution studies may furnish valuable insights in this regard. Finally, ADAR has been shown to interact with other RNA modification regulators such as m6A readers and affect non-viral responsive RNAs like long interspersed element 1 (194, 195). Thus, more research perspectives with ADAR at the center of the immune regulatory network are essential for comprehending its antiviral prowess.
In addition to the earlier mentioned A-to-I editing, some other RNA editing modifications also contribute to the innate immune response. Ψ is the most abundant and widely distributed cellular RNA modification known as the “fifth nucleotide” (196). Ψ is the C5-glycoside isomer of uridine, and this conserved modification appears irreversible. The most notable function of Ψ is maintaining various RNAs’ structure and stability (197). This gives an understanding of the responsiveness of Ψ to stress (198). tRNA exhibits the most abundant Ψ sites, and these modifications are crucial for translation (198). In mRNA, Ψ affects pre-mRNA splicing and impact mRNA stability. One representative function is that when Ψ modification occurs at the stop codon, it inhibits translation termination of the mRNA (37). In addition, Ψ plays a role in small nuclear RNAs (snRNAs), in maintaining their structures and regulating RNA-protein interactions (199, 200). Research on the regulators of Ψ modification is still ongoing, and various writers (members in pseudouridine synthase) have been identified, but there have been no reports of erasers and readers. Recently, the quantitative sequencing technologies have been developed for mapping Ψ transcriptome-wide, which have advanced the study of this abundant modification’s role in biological processes and diseases context (201, 202).
Ψ has been detected in the genomes of many viruses, such as SARS-CoV-2 and influenza viruses (203, 204). Dated back to 2010, it was reported that Ψ on RNA can inhibit the activation of PKR and avoid degradation (Figure 5) (205). In viruses, Ψ modifications can help viral RNA escape detection by host PRRs. Studies have revealed that although RIG-I can discern RNA with Ψ, it cannot induce conformational changes that initiate downstream signaling (206). Moreover, Ψ has been demonstrated to obstruct the activation of TLRs, particularly TLR7 and TLR8 (21). Additionally, Ψ has also been reported to diminish the activity of OAS, thereby enabling modified RNA to be translated for an extended duration (207). Although many cases suggest the regulatory role of Ψ in antiviral RNA sensing, the specific mechanisms of immune evasion are still unclear. Considering the enhancing effect of Ψ on mRNA translation efficiency, Ψ on viral RNA also seems to promote viral gene expression; however, validation in vivo is lacking. Besides maintaining immune homeostasis, host RNA Ψ modification also plays a responsive role in antiviral immunity. A typical example is HIV-1 infection. The initiation of HIV reverse transcription is highly dependent on Ψ modifications on host tRNA, which stabilize the complex formed between tRNA and viral RNA (208). Furthermore, Ψ on 7SK snRNA regulates its stability and structure, influencing the formation of super elongation complexes during infection and indirectly inhibiting HIV-1 transcription. The loss of this modification promotes HIV-1 escape from latency and facilitates reverse transcription (209). In addition to affecting the development of immune cells, host Ψ has been shown to influence the activation of DCs and CD8+ T cells (21). Interestingly, many studies have observed an elevation in the abundance of host Ψ following viral infection, suggesting a combination of effects of host antiviral response and viral actions (204, 210). This may regulate antiviral gene expression by regulating RNA splicing and mRNA stability (204). A recent study also found that this stoichiometric regulation of Ψ was induced by the IFN pathway (211). Regardless, Ψ plays a role in host-virus interactions in the antiviral response, and the mechanisms need further investigation. In antiviral therapy, the most representative application of Ψ is to reduce immune response and improve translation efficiency in mRNA vaccine (7). This technology has been applied to commercialized vaccines (e.g. SARS-COV-2 vaccine) (212, 213).
Other forms of non-m6A RNA methylation also exert influence on the innate immune response. m7G, prevalent N7-methylated guanosine found in eukaryotic RNA, holds significant importance. Analogous to Nm, m7G serves as an integral component of the cap structure within eukaryotic RNA polymerase II (pol II) transcripts and is distributed internally across mRNA, tRNA, and rRNA (214). Cap m7C is a classic eukaryotic RNA structure that stabilizes transcripts and prevents degradation. Additionally, this structure regulates bioprocesses such as mRNA splicing, nuclear export, transcription elongation, and translation (215–218). Despite its late recognition as a crucial modification, numerous studies have demonstrated its regulatory role in mRNA translation efficiency, and related regulators have been continuously discovered, with quantitative sequencing techniques being developed (219–221). Similar to cap Nm, viruses can acquire cap m7G through various mechanisms. Common mechanisms include utilizing host Pol II to synthesize RNA with cap m7G, stealing cap from short host mRNA, and encoding their own capping methyltransferase (87). Cap m7G plays a crucial role in maintaining the stability of viral RNA, and notably, it helps viral RNA escape the action of 5’exonucleases and avoid degradation (215). In addition, cap m7G also promotes the translation and expression of viral RNA (222). These regulatory mechanisms for viral RNA cap m7G are highly analogous to those in the host mRNAs. Interestingly, the previously mentioned cap-snatching mechanism not only prevents the expression of host RNA but can also lead to gene fusion with the host, resulting in the production of immunomodulatory chimeric proteins after viral infection (223). Furthermore, a study reported hypermethylation of HIV-1’s Cap m7G directly affects infectivity (224). Compared to Cap Nm, the virus’s Cap m7G does not seem to promote its escape from RNA sensing recognition by RLR and other PRRs (61). As for internal m7G, it has not been reported to be found in viral RNA, and it is still unclear whether the landscape of the host internal m7G is responsive to viral infection. Antiviral drugs that interfere with viral RNA capping have been extensively developed based on the capping mechanisms of different viruses (225).
m1A, methylation of the N1 position of adenosine, is an abundant and conserved modification in eukaryotic non-coding RNAs. In recent years, we have witnessed the development of multiple single-base resolution sequencing technologies, which have sought to address the challenge of mapping m1A modifications transcriptome-wide (226–228). The m1A modification introduces steric hindrance, which affects base pairing and the spatial conformation of RNA. It also influences the interactions between RNA and proteins, other RNAs, or small molecules (37). These diverse functions are closely related to the location of m1A. In addition to being highly abundant in tRNA (affecting stability, translation efficiency, and accuracy) and rRNA (maintaining ribosome function), m1A has also been found to be distributed in mRNA (229). The functions of m1A on host mRNA are still poorly understood, although its site-specific involvement in translation regulation has been identified (228, 230). Interestingly, m1A is closely related to m6A. In addition to their convertibility to m6A, they share a variety of regulators like fat mass and obesity-associated (FTO) protein as an eraser (231, 232). In viral RNA, m1A has been identified and enriched in specific regions (203). However, its involvement in innate immunity is not yet clear. Viral infection has been shown to impact the expression levels of m1A writers and erasers on host mRNA, and some of them may have proviral effects (203). Intriguingly, upregulated m1A on host RNA inhibits the activity of the replication complex of SARS-COV-2 and thus achieves an antiviral effect, an effect that does not appear to be related to the steric hindrance caused by m1A (233). As for the impact on immune cells, research on m1A has mainly focused on cancer, such as its influence on immune cell infiltration (111). Nevertheless, this modification demonstrates research value in antiviral innate immunity. Further studies need to be conducted, especially on the mechanisms of regulators, the responsiveness of these modifications to viral infection, and the interactions with m6A during the antiviral response.
m6Am is another well-known mRNA modification, typically occurring at the first nucleotide after the cap structure in the 5’ UTR of eukaryotic mRNA, known as the Cap Nm position. When this nucleotide is adenosine, the phosphorylated CTD-interacting factor 1 (PCIF1) converts Am to m6Am (37). Similar to m6A and m1A, FTO mediates the demethylation of m6Am (234). The detailed role of m6Am in regulating mRNA metabolism is still under investigation. Existing studies have demonstrated its impact on mRNA stability and translation, but the conclusions from these studies are contradictory (235–237). m6Am plays a regulatory role in antiviral immune processes. Like Nm, m6Am installed by host PCIF1 on viral RNA can mediate immune evasion. The presence of m6Am on viral RNA prevents its detection by ISGs and weakens the antiviral effects of the IFN-β pathway (238). Unlike RIG-I or IFIT1 sensing, this effect does not depend on RIG-I or IFIT1 sensing. Additionally, viral m6Am can prevent viral RNA degradation by nucleases (234). Host m6Am also undergoes dynamic changes in antiviral responses, exhibiting both antiviral and proviral effects. HIV infection induces ubiquitination and degradation of host PCIF1, leading to a decrease in m6Am on cellular mRNA and regulating the host transcription factor ETS proto-oncogene 1 (ETS1) to promote viral replication (239). In this process, PCIF1 exerts its antiviral function by affecting ETS1 stability by installing m6Am. On the other hand, in SARS-CoV-2 infection, PCIF1 maintains the stability of angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2) mRNA by installing m6Am, promoting their expression and facilitating viral infection (240). In summary, m6Am, as a typical cap modification, has gained attention since the development of single-base resolution sequencing techniques (241). However, further research is needed to explore its functional connections and differences with Nm, as well as the role of PCIF1 in viral infection.
In addition to the regulatory modifications described above, some less-studied RNA modifications also function in antivirals. Although there is limited research on ac4C, this newly discovered, unique acetylation modification in eukaryotes has a vital role in bioprocesses and immune response. Initially discovered and demonstrated to regulate ribosome maturation and protein translation ability in tRNA and rRNA, ac4C was later found to be distributed in mRNA and enriched in the coding sequence (CDS) region using antibody-based sequencing methods (242). On mRNA, ac4C in the CDS region can maintain stability and promote translation, while the 5’ UTR has been found to potentially inhibit translation initiation by affecting the interaction between mRNA and tRNA/ribosomes (243, 244). ac4C has been found to be associated with diseases such as cancer, neurodegenerative disorders, and inflammation (242). In viral infections, the presence of ac4C on various viral RNAs plays a regulatory role. HIV-1 can utilize the host ac4C writer N-acetyltransferase 10 (NAT10) to add ac4C to viral RNA, increasing its stability and promoting replication (245). Inhibiting NAT10 can suppress the spread of HIV-1 without affecting cell viability. Interestingly, another investigation reported that NAT10 actually fosters HIV-1 latency while impeding Tat-mediated transcriptional processes (246). Further research is needed. NAT10 has also been found to recruit and add ac4C on viral RNA in Enterovirus, specifically recruiting host proteins to enhance viral RNA stability and translation (247). Additionally, changes in the abundance of ac4C in the 5’ UTR region of specific genes in the host have been observed after infection with the influenza A virus, which may be related to the virus-induced expression of NAT10 and its pro-viral effects (203, 248). Nevertheless, the specific functions of this modification distributed in the genomes of diverse viruses and the role of NAT10 in antiviral innate immunity await further investigation.
There are also several other low-abundance modifications detected on viral RNA, such as 5,2’-O-dimethylcytosine (m5Cm) (60). However, the functions of these modifications are still unknown. In addition, a number of other types of RNA editing also play a role in the regulation of antiviral innate immunity, although it is inconclusive whether these edits are categorized as RNA modifications. An example is the addition of uridine to the 3’ RNA terminus catalyzed by terminal uridyltransferases (TUTases), also known as uridylation and poly(U) tails. This editing occurs on almost all classes of RNAs and regulates processes such as mRNA decay, histone expression, and miRNA metabolism and targeting (249). Uridylation has been shown to have regulatory functions in innate immunity. TUTases can directly edit the genomes of various viruses, forming a “poly(U) tag” effect to promote immune response by mediating RNA exosome degradation, facilitating viral RNA decay, and targeting viral proteins for antiviral purposes (250, 251). Loss of TUTases leads to an increase in viral mRNA and protein levels. Furthermore, TUTases can be activated by TLR and regulate mRNA stability to promote the production of various cytokines, participating in immune responses (252). Another type of RNA editing involved in antiviral innate immunity is C-to-U deamidation editing mediated by the apolipoprotein B mRNA editing catalytic polypeptide-like (APOBEC) protein family. APOBEC proteins can significantly inhibit the replication of various retroviruses, endogenous viruses, and DNA viruses, including HIV-1, human T-cell leukemia virus type 1, hepatitis B virus, etc. (253). This viral restriction effect is mediated by multiple mechanisms, including inhibition of viral infection factor expression, disruption of viral particle assembly, excision of viral RNA bases, and direct editing of the genome of DNA viruses (253, 254). Moreover, APOBEC-mediated editing is essential for immune cell differentiation, development, and function (255–257). Many reviews have summarized the regulatory effects of APOBEC-mediated editing, so we will not review them in this work.
In this Review, we summarized the regulatory roles of eight typical non-m6A RNA modifications in antiviral innate immunity within the current scope of knowledge, including 2’-O-methylation (Nm), 5-methylcytidine (m5C), adenosine-inosine editing (A-to-I editing), pseudouridine (Ψ), N1-methyladenosine (m1A), N7-methylguanosine (m7G), N6,2’-O-dimethyladenosine (m6Am), and N4-acetylcytidine (ac4C). It is evident that compared to the extensively studied m6A modification, there is still a significant gap regarding the immune regulatory functions of non-m6A RNA modifications. The lack of clarity regarding the regulators of non-m6A RNA modifications has caused gaps in understanding the mechanisms underlying innate immune responses, particularly the utilization and induction patterns of viruses. In addition, low abundance, complexity of distribution, and diversity of responsive effects are all challenges for the study of non-m6A RNA modifications in antiviral immunity.
Considering the recently emerging base-resolution sequencing methods, the quantitative analysis of non-m6A RNA modifications, such as Ψ, Nm, m7G, m5C, m1A, and ac4C, has been enabled to resolve the challenges in studying the location and stoichiometry of these modifications on viral and host RNA, as well as the regulatory modes of RNA modification landscapes during host-virus interactions. These recent advances in base-resolution sequencing technology offer the super-resolution profiles of non-m6A RNA modifications transcriptome-wide; meanwhile, the breakthroughs in quantitative mapping tools could aid the comprehensive investigation of non-m6A RNA modifications in antiviral innate immunity through monitoring the dynamics of these RNA modifications.
Besides, some non-m6A RNA modifications have been demonstrated to facilitate the immune escape and replication of certain viruses; however, these non-m6A RNA modifications are also essential for the host to enhance the antiviral immune response. Thus, the manipulation of RNA modifications at specific sites within host RNA or viral RNA may have an impact on the development of antiviral drugs with therapeutic potential. Overall, the functional and mechanistic study of RNA modifications in antiviral innate immunity has led to entirely new perspectives, yielding in-depth insights into translational medicine and potentially benefiting related research in biological sciences, biomedical engineering, clinics, and the pharmaceutical industry.
SS: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review & editing. LZ: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This project was supported by Research Grant Council (RGC) grant ECS 26103623 (L-SZ).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Shanley LC, Mahon OR, Kelly DJ, Dunne A. Harnessing the innate and adaptive immune system for tissue repair and regeneration: Considering more than macrophages. Acta Biomater (2021) 133:208–21. doi: 10.1016/j.actbio.2021.02.023
2. Yamada T, Takaoka A. Innate immune recognition against SARS-CoV-2. Inflamm Regener (2023) 43:7. doi: 10.1186/s41232-023-00259-5
3. Gerada C, Ryan KM. Autophagy, the innate immune response and cancer. Mol Oncol (2020) 14:1913–29. doi: 10.1002/1878-0261.12774
4. Small S, Numan Y, Platanias LC. Innate immune mechanisms and immunotherapy of myeloid Malignancies. Biomedicines (2021) 9:1631. doi: 10.3390/biomedicines9111631
5. Xu Q, Tang Y, Huang G. Innate immune responses in RNA viral infection. Front Med (2021) 15:333–46. doi: 10.1007/s11684-020-0776-7
6. Iwama RE, Moran Y. Origins and diversification of animal innate immune responses against viral infections. Nat Ecol Evol (2023) 7:182–93. doi: 10.1038/s41559-022-01951-4
7. Liu A, Wang X. The pivotal role of chemical modifications in mRNA therapeutics. Front Cell Dev Biol (2022) 10:901510. doi: 10.3389/fcell.2022.901510
8. Zhang Q, Cao X. Epigenetic regulation of the innate immune response to infection. Nat Rev Immunol (2019) 19:417–32. doi: 10.1038/s41577-019-0151-6
9. Zhao H-J, Hu Y-F, Han Q-J, Zhang J. Innate and adaptive immune escape mechanisms of hepatitis B virus. World J Gastroenterol (2022) 28:881–96. doi: 10.3748/wjg.v28.i9.881
10. Yang B, Zhang X, Zhang D, Hou J, Xu G, Sheng C, et al. Molecular mechanisms of immune escape for foot-and-mouth disease virus. Pathogens (2020) 9:729. doi: 10.3390/pathogens9090729
11. Malik G, Zhou Y. Innate immune sensing of influenza A virus. Viruses (2020) 12:755. doi: 10.3390/v12070755
12. Zhang E, Lu M. Toll-like receptor (TLR)-mediated innate immune responses in the control of hepatitis B virus (HBV) infection. Med Microbiol Immunol (2015) 204:11–20. doi: 10.1007/s00430-014-0370-1
13. Rehwinkel J, Gack MU. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol (2020) 20:537–51. doi: 10.1038/s41577-020-0288-3
14. Munir M, Berg M. The multiple faces of proteinkinase R in antiviral defense. Virulence (2013) 4:85–9. doi: 10.4161/viru.23134
15. Suzzi S, Tsitsou-Kampeli A, Schwartz M. The type I interferon antiviral response in the choroid plexus and the cognitive risk in COVID-19. Nat Immunol (2023) 24:220–4. doi: 10.1038/s41590-022-01410-z
16. Guillemin A, Kumar A, Wencker M, Ricci EP. Shaping the innate immune response through post-transcriptional regulation of gene expression mediated by RNA-binding proteins. Front Immunol (2022) 12:796012. doi: 10.3389/fimmu.2021.796012
17. Burgess HM, Vink EI, Mohr I. Minding the message: tactics controlling RNA decay, modification, and translation in virus-infected cells. Genes Dev (2022) 36:108–32. doi: 10.1101/gad.349276.121
18. Gokhale NS, Smith JR, Van Gelder RD, Savan R. RNA regulatory mechanisms that control antiviral innate immunity. Immunol Rev (2021) 304:77–96. doi: 10.1111/imr.13019
19. Tagawa T, Serquin A, Kook I, Ziegelbauer J. Viral non-coding RNAs: Stealth strategies in the tug-of-war between humans and herpesviruses. Semin Cell Dev Biol (2021) 111:135–47. doi: 10.1016/j.semcdb.2020.06.015
20. Cui L, Ma R, Cai J, Guo C, Chen Z, Yao L, et al. RNA modifications: importance in immune cell biology and related diseases. Sig Transduct Target Ther (2022) 7:1–26. doi: 10.1038/s41392-022-01175-9
21. Karikó K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity (2005) 23:165–75. doi: 10.1016/j.immuni.2005.06.008
22. Yu P-L, Cao S-J, Wu R, Zhao Q, Yan Q-G. Regulatory effect of m(6)A modification on different viruses. J Med Virol (2021) 93:6100–15. doi: 10.1002/jmv.27246
23. Rajendren S, Karijolich J. The impact of RNA modifications on the biology of DNA virus infection. Eur J Cell Biol (2022) 101:151239. doi: 10.1016/j.ejcb.2022.151239
24. Li N, Rana TM. Regulation of antiviral innate immunity by chemical modification of viral RNA. Wiley Interdiscip Rev RNA (2022) 13:e1720. doi: 10.1002/wrna.1720
25. Sacco MT, Horner SM. Flipping the script: viral capitalization of RNA modifications. Brief Funct Genomics (2021) 20:86–93. doi: 10.1093/bfgp/elaa025
26. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang C-M, Li CJ, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell (2013) 49:18–29. doi: 10.1016/j.molcel.2012.10.015
27. Hu L, Liu S, Peng Y, Ge R, Su R, Senevirathne C, et al. m6A RNA modifications are measured at single-base resolution across the mammalian transcriptome. Nat Biotechnol (2022) 40:1210–9. doi: 10.1038/s41587-022-01243-z
28. Liu C, Sun H, Yi Y, Shen W, Li K, Xiao Y, et al. Absolute quantification of single-base m6A methylation in the mammalian transcriptome using GLORI. Nat Biotechnol (2023) 41:355–66. doi: 10.1038/s41587-022-01487-9
29. Xiao Y-L, Liu S, Ge R, Wu Y, He C, Chen M, et al. Transcriptome-wide profiling and quantification of N6-methyladenosine by enzyme-assisted adenosine deamination. Nat Biotechnol (2023) 41:993–1003. doi: 10.1038/s41587-022-01587-6
30. Wei J, Yu X, Yang L, Liu X, Gao B, Huang B, et al. FTO mediates LINE1 m6A demethylation and chromatin regulation in mESCs and mouse development. Science (2022) 376:968–73. doi: 10.1126/science.abe9582
31. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol (2011) 7:885–7. doi: 10.1038/nchembio.687
32. He PC, Wei J, Dou X, Harada BT, Zhang Z, Ge R, et al. Exon architecture controls mRNA m6A suppression and gene expression. Science (2023) 379(6633):677–82. doi: 10.1126/science.abj9090
33. Tong J, Wang X, Liu Y, Ren X, Wang A, Chen Z, et al. Pooled CRISPR screening identifies m6A as a positive regulator of macrophage activation. Sci Adv (2021) 7:eabd4742. doi: 10.1126/sciadv.abd4742
34. Lou X, Wang J-J, Wei Y-Q, Sun J-J. Emerging role of RNA modification N6-methyladenosine in immune evasion. Cell Death Dis (2021) 12:1–10. doi: 10.1038/s41419-021-03585-z
35. Winkler R, Gillis E, Lasman L, Safra M, Geula S, Soyris C, et al. m6A modification controls the innate immune response to infection by targeting type I interferons. Nat Immunol (2019) 20:173–82. doi: 10.1038/s41590-018-0275-z
36. McFadden MJ, Horner SM. N6-methyladenosine regulates host responses to viral infection. Trends Biochem Sci (2021) 46:366–77. doi: 10.1016/j.tibs.2020.11.008
37. Sun H, Li K, Liu C, Yi C. Regulation and functions of non-m6A mRNA modifications. Nat Rev Mol Cell Biol (2023) 24:714–31. doi: 10.1038/s41580-023-00622-x
38. Zhang Y, Lu L, Li X. Detection technologies for RNA modifications. Exp Mol Med (2022) 54:1601–16. doi: 10.1038/s12276-022-00821-0
39. Xu L, Seki M. Recent advances in the detection of base modifications using the Nanopore sequencer. J Hum Genet (2020) 65:25–33. doi: 10.1038/s10038-019-0679-0
40. Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer (2020) 20:303–22. doi: 10.1038/s41568-020-0253-2
41. Ayadi L, Galvanin A, Pichot F, Marchand V, Motorin Y. RNA ribose methylation (2′-O-methylation): Occurrence, biosynthesis and biological functions. Biochim Biophys Acta (BBA) - Gene Regul Mech (2019) 1862:253–69. doi: 10.1016/j.bbagrm.2018.11.009
42. Chen L, Zhang L-S, Ye C, Zhou H, Liu B, Gao B, et al. Nm-Mut-seq: a base-resolution quantitative method for mapping transcriptome-wide 2′-O-methylation. Cell Res (2023) 33:727–30. doi: 10.1038/s41422-023-00836-w
43. Dai Q, Moshitch-Moshkovitz S, Han D, Kol N, Amariglio N, Rechavi G, et al. Nm-seq maps 2′-O-methylation sites in human mRNA with base precision. Nat Methods (2017) 14:695–8. doi: 10.1038/nmeth.4294
44. Decle-Carrasco S, Rodriguez-Pina A, Rodriguez-Zapata L, Castano E. Current research on viral proteins that interact with fibrillarin. Mol Biol Rep (2023) 50:4631–43. doi: 10.1007/s11033-023-08343-2
45. Ringeard M, Marchand V, Decroly E, Motorin Y, Bennasser Y. FTSJ3 is an RNA 2 ’-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature (2019) 565:500–+. doi: 10.1038/s41586-018-0841-4
46. Monaco PL, Marcel V, Diaz J-J, Catez F. 2′-O-methylation of ribosomal RNA: towards an epitranscriptomic control of translation? Biomolecules (2018) 8:106. doi: 10.3390/biom8040106
47. Elliott BA, Ho H-T, Ranganathan SV, Vangaveti S, Ilkayeva O, Abou Assi H, et al. Modification of messenger RNA by 2′-O-methylation regulates gene expression in vivo. Nat Commun (2019) 10:3401. doi: 10.1038/s41467-019-11375-7
48. Dimitrova DG, Teysset L, Carré C. RNA 2′-O-methylation (Nm) modification in human diseases. Genes (2019) 10:117. doi: 10.3390/genes10020117
49. Hoernes TP, Clementi N, Faserl K, Glasner H, Breuker K, Lindner H, et al. Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res (2016) 44:852–62. doi: 10.1093/nar/gkv1182
50. Choi J, Indrisiunaite G, DeMirci H, Ieong K-W, Wang J, Petrov A, et al. 2′-O-methylation in mRNA disrupts tRNA decoding during translation elongation. Nat Struct Mol Biol (2018) 25:208–16. doi: 10.1038/s41594-018-0030-z
51. Vitali P, Kiss T. Cooperative 2 ’-O-methylation of the wobble cytidine of human elongator tRNA(Met)(CAT) by a nucleolar and a Cajal body-specific box C/D RNP. Genes Dev (2019) 33:741–6. doi: 10.1101/gad.326363.119
52. Zhou F, Aroua N, Liu Y, Rohde C, Cheng J, Wirth A-K, et al. A dynamic rRNA ribomethylome drives stemness in acute myeloid leukemia. Cancer Discov (2023) 13:332–47. doi: 10.1158/2159-8290.CD-22-0210
53. Zheng J, Wang C, Chang MR, Devarkar SC, Schweibenz B, Crynen GC, et al. HDX-MS reveals dysregulated checkpoints that compromise discrimination against self RNA during RIG-I mediated autoimmunity. Nat Commun (2018) 9:5366. doi: 10.1038/s41467-018-07780-z
54. Leschziner GD, Coffey AJ, Andrew T, Gregorio SP, Dias-Neto E, Calafato M, et al. Q8IYL2 is a candidate gene for the familial epilepsy syndrome of Partial Epilepsy with Pericentral Spikes (PEPS). Epilepsy Res (2011) 96:109–15. doi: 10.1016/j.eplepsyres.2011.05.010
55. Bao H, Chen X, Liu X, Wu W, Li Q, Xian J, et al. Box C/D snoRNA SNORD89 influences the occurrence and development of endometrial cancer through 2’-O-methylation modification of Bim. Cell Death Discov (2022) 8:1–10. doi: 10.1038/s41420-022-01102-5
56. Decombe A, El Kazzi P, Decroly E. Interplay of RNA 2′-O-methylations with viral replication. Curr Opin Virol (2023) 59:101302. doi: 10.1016/j.coviro.2023.101302
57. Hyde JL, Diamond MS. Innate immune restriction and antagonism of viral RNA lacking 2?-O methylation. Virology (2015) 479–480:66–74. doi: 10.1016/j.virol.2015.01.019
58. Zuest R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, et al. Ribose 2 ’-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol (2011) 12:137–U46. doi: 10.1038/ni.1979
59. Schuberth-Wagner C, Ludwig J, Bruder AK, Herzner A-M, Zillinger T, Goldeck M, et al. A conserved histidine in the RNA sensor RIG-I controls immune tolerance to N-1-2 ’ O-methylated self RNA. Immunity (2015) 43:41–51. doi: 10.1016/j.immuni.2015.06.015
60. Netzband R, Pager C. Epitranscriptomic marks: Emerging modulators of RNA virus gene expression. WILEY Interdiscip Rev RNA (2020) 11(3):e1576. doi: 10.1002/wrna.1576
61. Devarkar SC, Wang C, Miller MT, Ramanathan A, Jiang F, Khan AG, et al. Structural basis for m7G recognition and 2 ’-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc Natl Acad Sci USA (2016) 113:596–601. doi: 10.1073/pnas.1515152113
62. Andreou A, Papakyriakou A, Zervou M, Goulielmos G, Eliopoulos E. Is the association of the rare rs35667974 IFIH1 gene polymorphism with autoimmune diseases a case of RNA epigenetics? J Mol Evol (2023) 91:204–13. doi: 10.1007/s00239-022-10090-0
63. Rimbach K, Kaiser S, Helm M, Dalpke A, Eigenbrod T. 2 ’-O-methylation within bacterial RNA acts as suppressor of TLR7/TLR8 activation in human innate immune cells. J INNATE Immun (2015) 7:482–93. doi: 10.1159/000375460
64. Jockel S, Nees G, Sommer R, Zhao Y, Cherkasov D, Hori H, et al. The 2 ’-O-methylation status of a single guanosine controls transfer RNA-mediated Toll-like receptor 7 activation or inhibition. J Exp Med (2012) 209:235–41. doi: 10.1084/jem.20111075
65. Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, et al. 2 ’-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature (2010) 468:452–6. doi: 10.1038/nature09489
66. Diamond M. IFIT1: A dual sensor and effector molecule that detects non-2 ’-O methylated viral RNA and inhibits its translation. Cytokine Growth FACTOR Rev (2014) 25:543–50. doi: 10.1016/j.cytogfr.2014.05.002
67. Abbas YM, Laudenbach BT, Martínez-Montero S, Cencic R, Habjan M, Pichlmair A, et al. Structure of human IFIT1 with capped RNA reveals adaptable mRNA binding and mechanisms for sensing N1 and N2 ribose 2′-O methylations. Proc Natl Acad Sci (2017) 114:E2106–15. doi: 10.1073/pnas.1612444114
68. Kumar P, Sweeney T, Skabkin M, Skabkina O, Hellen C, Pestova T. Inhibition of translation by IFIT family members is determined by their ability to interact selectively with the 5 ’-terminal regions of cap0-, cap1-and 5 ’ ppp- mRNAs. Nucleic Acids Res (2014) 42:3228–45. doi: 10.1093/nar/gkt1321
69. Dong H, Zhang B, Shi P-Y. Flavivirus methyltransferase: A novel antiviral target. Antiviral Res (2008) 80:1–10. doi: 10.1016/j.antiviral.2008.05.003
70. Coutard B, Barral K, Lichiere J, Selisko B, Martin B, Aouadi W, et al. Zika virus methyltransferase: structure and functions for drug design perspectives. J Virol (2017) 91:e02202–16. doi: 10.1128/JVI.02202-16
71. Habjan M, Hubel P, Lacerda L, Benda C, Holze C, Eberl CH, et al. Sequestration by IFIT1 impairs translation of 2′O-unmethylated capped RNA. PLoS Pathog (2013) 9:e1003663. doi: 10.1371/journal.ppat.1003663
72. Young D, Andrejeva J, Li X, Inesta-Vaquera F, Dong C, Cowling V, et al. Human IFIT1 Inhibits mRNA Translation of Rubulaviruses but Not Other Members of the Paramyxoviridae Family. J Virol (2016) 90:9446–56. doi: 10.1128/JVI.01056-16
73. Hyde J, Gardner C, Kimura T, White J, Liu G, Trobaugh D, et al. A viral RNA structural element alters host recognition of nonself RNA. Science (2014) 343:783–7. doi: 10.1126/science.1248465
74. Despic V, Jaffrey S. mRNA ageing shapes the Cap2 methylome in mammalian mRNA. NATURE (2023) 614:358–+. doi: 10.1038/s41586-022-05668-z
75. Dong H, Chang D, Hua M, Lim S, Chionh Y, Hia F, et al. 2 ’-O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathog (2012) 8(4):e1002642. doi: 10.1371/journal.ppat.1002642
76. El Kazzi P, Rabah N, Chamontin C, Poulain L, Ferron F, Debart F, et al. Internal RNA 2 ’ O-methylation in the HIV-1 genome counteracts ISG20 nuclease-mediated antiviral effect. Nucleic Acids Res (2023) 51:2501–15. doi: 10.1093/nar/gkac996
77. Robbins M, Judge A, Liang L, McClintock K, Yaworski E, MacLachlan I. 2’-O-methyl-modified RNAs act as TLR7 antagonists. Mol Ther (2007) 15:1663–9. doi: 10.1038/sj.mt.6300240
78. Yang SL, DeFalco L, Anderson DE, Zhang Y, Aw JGA, Lim SY, et al. Comprehensive mapping of SARS-CoV-2 interactions in vivo reveals functional virus-host interactions. Nat Commun (2021) 12:5113. doi: 10.1038/s41467-021-25357-1
79. Martin B, Coutard B, Guez T, Paesen G, Canard B, Debart F, et al. The methyltransferase domain of the Sudan ebolavirus L protein specifically targets internal adenosines of RNA substrates, in addition to the cap structure. Nucleic Acids Res (2018) 46:7902–12. doi: 10.1093/nar/gky637
80. Nachmani D, Mendez L, Pandolfi P. Aberrant rRNA 2’-O-methylation causes bone marrow failure and defective immune function. Blood (2020) 136:11–2. doi: 10.1182/blood-2020-141809
81. Williams G, Gokhale N, Snider D, Horner S. The mRNA cap 2 ’-O-methyltransferase CMTR1 regulates the expression of certain interferon-stimulated genes. mSphere (2020) 5(3):e00202–20. doi: 10.1128/mSphere.00202-20
82. Angelova M, Dimitrova D, Da Silva B, Marchand V, Jacquier C, Achour C, et al. tRNA 2 ’-O-methylation by a duo of TRM7/FTSJ1 proteins modulates small RNA silencing in Drosophila. Nucleic Acids Res (2020) 48:2050–72. doi: 10.1093/nar/gkaa002
83. Backes S, Shapiro J, Sabin L, Pham A, Reyes I, Moss B, et al. Degradation of host microRNAs by poxvirus poly(A) polymerase reveals terminal RNA methylation as a protective antiviral mechanism. Cell Host Microbe (2012) 12:200–10. doi: 10.1016/j.chom.2012.05.019
84. Petushkov I, Esyunina D, Kulbachinskiy A. Effects of natural RNA modifications on the activity of SARS-CoV-2 RNA-dependent RNA polymerase. FEBS J (2023) 290:80–92. doi: 10.1111/febs.16587
85. Musinova YR, Sheval EV, Dib C, Germini D, Vassetzky YS. Functional roles of HIV-1 Tat protein in the nucleus. Cell Mol Life Sci (2016) 73:589–601. doi: 10.1007/s00018-015-2077-x
86. Li P, Liu Y, Song R, Zhao L, Yang J, Lu F, et al. RNA 2’-O-methyltransferase fibrillarin facilitates virus entry into macrophages through inhibiting type I interferon response. Front Immunol (2022) 13:793582. doi: 10.3389/fimmu.2022.793582
87. Decroly E, Ferron F, Lescar J, Canard B. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol (2012) 10:51–65. doi: 10.1038/nrmicro2675
88. Izadpanah A, Rappaport J, Datta PK. Epitranscriptomics of SARS-coV-2 infection. Front Cell Dev Biol (2022) 10:849298. doi: 10.3389/fcell.2022.849298
89. Viswanathan T, Arya S, Chan S, Qi S, Dai N, Misra A, et al. Structural basis of RNA cap modification by SARS-CoV-2. Nat Commun (2020) 11(1):1–7. doi: 10.1038/s41467-020-17496-8
90. Nguyen H, Thai N, Li M. Identifying inhibitors of NSP16-NSP10 of SARS-CoV-2 from large databases. J Biomol Struct Dyn (2022) 41(15):7045–54. doi: 10.1080/07391102.2022.2114941
91. Mohammad A, Alshawaf E, Marafie S, Abu-Farha M, Al-Mulla F, Abubaker J. Molecular simulation-based investigation of highly potent natural products to abrogate formation of the nsp10-nsp16 complex of SARS-coV-2. Biomolecules (2021) 11(4):573. doi: 10.3390/biom11040573
92. Perez-Lemus GR, Menéndez CA, Alvarado W, Byléhn F, de Pablo JJ. Toward wide-spectrum antivirals against coronaviruses: Molecular characterization of SARS-CoV-2 NSP13 helicase inhibitors. Sci Adv (2022) 8:eabj4526. doi: 10.1126/sciadv.abj4526
93. Tsukamoto Y, Hiono T, Yamada S, Matsuno K, Faist A, Claff T, et al. Inhibition of cellular RNA methyltransferase abrogates influenza virus capping and replication. Science (2023) 379:586–91. doi: 10.1126/science.add0875
94. Han S, Lee S. Inhibition of Japanese encephalitis virus (JEV) replication by specific RNA aptamer against JEV methyltransferase. Biochem AND Biophys Res Commun (2017) 483:687–93. doi: 10.1016/j.bbrc.2016.12.081
95. Jung J, Han S, Lee S. Development of RNA aptamer that inhibits methyltransferase activity of dengue virus. Biotechnol Lett (2018) 40:315–24. doi: 10.1007/s10529-017-2462-7
96. Li S, Dong H, Li X, Xie X, Zhao H, Deng Y, et al. Rational design of a flavivirus vaccine by abolishing viral RNA 2 ’-O methylation. J Virol (2013) 87:5812–9. doi: 10.1128/JVI.02806-12
97. Huang T, Chen W, Liu J, Gu N, Zhang R. Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat Struct Mol Biol (2019) 26:380–8. doi: 10.1038/s41594-019-0218-x
98. Yang X, Yang Y, Sun B-F, Chen Y-S, Xu J-W, Lai W-Y, et al. 5-methylcytosine promotes mRNA export — NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res (2017) 27:606–25. doi: 10.1038/cr.2017.55
99. Liu J, Huang T, Chen W, Ding C, Zhao T, Zhao X, et al. Developmental mRNA m5C landscape and regulatory innovations of massive m5C modification of maternal mRNAs in animals. Nat Commun (2022) 13:2484. doi: 10.1038/s41467-022-30210-0
100. Bohnsack KE, Höbartner C, Bohnsack MT. Eukaryotic 5-methylcytosine (m5C) RNA methyltransferases: mechanisms, cellular functions, and links to disease. Genes (2019) 10:102. doi: 10.3390/genes10020102
101. Gao Y, Fang J. RNA 5-methylcytosine modification and its emerging role as an epitranscriptomic mark. RNA Biol (2021) 18:117–27. doi: 10.1080/15476286.2021.1950993
102. Wnuk M, Slipek P, Dziedzic M, Lewinska A. The roles of host 5-methylcytosine RNA methyltransferases during viral infections. Int J Mol Sci (2020) 21:8176. doi: 10.3390/ijms21218176
103. Courtney DG, Tsai K, Bogerd HP, Kennedy EM, Law BA, Emery A, et al. Epitranscriptomic addition of m5C to HIV-1 transcripts regulates viral gene expression. Cell Host Microbe (2019) 26:217–27.e6. doi: 10.1016/j.chom.2019.07.005
104. Dev R, Ganji R, Singh S, Mahalingam S, Banerjee S, Khosla S. Cytosine methylation by DNMT2 facilitates stability and survival of HIV-1 RNA in the host cell during infection. Biochem J (2017) 474:2009–26. doi: 10.1042/BCJ20170258
105. Courtney D, Chalem A, Bogerd H, Law B, Kennedy E, Holley C, et al. Extensive epitranscriptomic methylation of A and C residues on murine leukemia virus transcripts enhances viral gene expression. mBio (2019) 10(3):e01209–19. doi: 10.1128/mBio.01209-19
106. Bhattacharya T, Yan L, Crawford J, Zaher H, Newton I, Hardy R. Differential viral RNA methylation contributes to pathogen blocking in Wolbachia-colonized arthropods. PLoS Pathog (2022) 18(3):e1010393. doi: 10.1371/journal.ppat.1010393
107. Henry B, Kanarek J, Kotter A, Helm M, Lee N. 5-methylcytosine modification of an Epstein-Barr virus noncoding RNA decreases its stability. RNA (2020) 26:1038–48. doi: 10.1261/rna.075275.120
108. Boyne JR, Colgan KJ, Whitehouse A. Recruitment of the Complete hTREX Complex Is Required for Kaposi’s Sarcoma–Associated Herpesvirus Intronless mRNA Nuclear Export and Virus Replication. PLoS Pathog (2008) 4:e1000194. doi: 10.1371/journal.ppat.1000194
109. Eckwahl M, Xu R, Michalkiewicz J, Zhang W, Patel P, Cai Z, et al. 5-methylcytosine RNA modifications promote retrovirus replication in an ALYREF reader protein-dependent manner. J Virol (2020) 94(13):e00544–20. doi: 10.1128/JVI.00544-20
110. Lu F, Zankharia U, Vladimirova O, Yi Y, Collman R, Lieberman P. Epigenetic landscape of HIV-1 infection in primary human macrophage. J Virol (2022) 96(7):e0016222. doi: 10.1128/jvi.00162-22
111. Zha L-F, Wang J-L, Cheng X. The effects of RNA methylation on immune cells development and function. FASEB J (2022) 36:e22552. doi: 10.1096/fj.202200716R
112. Guo G, Wang H, Shi X, Ye L, Yan K, Chen Z, et al. Disease Activity-Associated Alteration of mRNA m(5)C Methylation in CD4(+)T Cells of Systemic Lupus Erythematosus. Front Cell Dev Biol (2020) 8:430. doi: 10.3389/fcell.2020.00430
113. Sun B, Zeng H, Liang J, Zhang L, Hu H, Wang Q, et al. NSUN5 facilitates viral RNA recognition by RIG-I receptor. J Immunol (2020) 205:3408–18. doi: 10.4049/jimmunol.1901455
114. Wang W, Huang H, Jiang H, Tian C, Tang Y, Gan D, et al. A cross-tissue investigation of molecular targets and physiological functions of nsun6 using knockout mice. Int J Mol Sci (2022) 23(12):6584. doi: 10.3390/ijms23126584
115. Durdevic Z, Hanna K, Gold B, Pollex T, Cherry S, Lyko F, et al. Efficient RNA virus control in Drosophila requires the RNA methyltransferase Dnmt2. EMBO Rep (2013) 14:269–75. doi: 10.1038/embor.2013.3
116. Thiagarajan D, Dev R, Khosla S. The DNA methyltranferase Dnmt2 participates in RNA processing during cellular stress. Epigenetics (2011) 6(1):103–13. doi: 10.4161/epi.6.1.13418
117. Kong W, Biswas A, Zhou D, Fiches G, Fujinaga K, Santoso N, et al. Nucleolar protein NOP2/NSUN1 suppresses HIV-1 transcription and promotes viral latency by competing with Tat for TAR binding and methylation. PLoS Pathog (2020) 16(3):e1008430. doi: 10.1371/journal.ppat.1008430
118. Zhang Y, Zhang L, Dai Q, Chen P, Lu M, Kairis E, et al. 5-methylcytosine (m(5)C) RNA modification controls the innate immune response to virus infection by regulating type I interferons. Proc Natl Acad Sci USA (2022) 119(42):e2123338119. doi: 10.1073/pnas.2123338119
119. Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep (2013) 4:255–61. doi: 10.1016/j.celrep.2013.06.029
120. Sajini AA, Choudhury NR, Wagner RE, Bornelöv S, Selmi T, Spanos C, et al. Loss of 5-methylcytosine alters the biogenesis of vault-derived small RNAs to coordinate epidermal differentiation. Nat Commun (2019) 10:2550. doi: 10.1038/s41467-019-10020-7
121. Li F, Chen Y, Zhang Z, Ouyang J, Wang Y, Yan R, et al. Robust expression of vault RNAs induced by influenza A virus plays a critical role in suppression of PKR-mediated innate immunity. Nucleic Acids Res (2015) 43:10321–37. doi: 10.1093/nar/gkv1078
122. Deng J, Ptashkin RN, Chen Y, Cheng Z, Liu G, Phan T, et al. Respiratory syncytial virus utilizes a tRNA fragment to suppress antiviral responses through a novel targeting mechanism. Mol Ther (2015) 23:1622–9. doi: 10.1038/mt.2015.124
123. Ying S, Li P, Wang J, Chen K, Zou Y, Dai M, et al. tRF-Gln-CTG-026 ameliorates liver injury by alleviating global protein synthesis. Signal Transduct Target Ther (2023) 8:144. doi: 10.1038/s41392-023-01351-5
124. Yu X, Xie Y, Zhang S, Song X, Xiao B, Yan Z. tRNA-derived fragments: Mechanisms underlying their regulation of gene expression and potential applications as therapeutic targets in cancers and virus infections. Theranostics (2021) 11:461–9. doi: 10.7150/thno.51963
125. Schaefer M, Hagemann S, Hanna K, Lyko F. Azacytidine inhibits RNA methylation at DNMT2 target sites in human cancer cell lines. Cancer Res (2009) 69:8127–32. doi: 10.1158/0008-5472.CAN-09-0458
126. Huang A, Rieper L, Rieder D, Kimpel J, Lusser A. No evidence for epitranscriptomic m5C modification of SARS-CoV-2, HIV and MLV viral RNA. RNA (2023) 29:756–63. doi: 10.1261/rna.079549.122
127. Yuan J, Xu L, Bao H-J, Wang J, Zhao Y, Chen S. Biological roles of A-to-I editing: implications in innate immunity, cell death, and cancer immunotherapy. J Exp Clin Cancer Res (2023) 42:149. doi: 10.1186/s13046-023-02727-9
128. Quin J, Sedmík J, Vukić D, Khan A, Keegan LP, O’Connell MA. ADAR RNA modifications, the epitranscriptome and innate immunity. Trends Biochem Sci (2021) 46:758–71. doi: 10.1016/j.tibs.2021.02.002
129. Schoft VK, Schopoff S, Jantsch MF. Regulation of glutamate receptor B pre-mRNA splicing by RNA editing. Nucleic Acids Res (2007) 35:3723–32. doi: 10.1093/nar/gkm314
130. Nakahama T, Kawahara Y. Adenosine-to-inosine RNA editing in the immune system: friend or foe? Cell Mol Life Sci (2020) 77:2931–48. doi: 10.1007/s00018-020-03466-2
131. Stellos K, Gatsiou A, Stamatelopoulos K, Perisic Matic L, John D, Lunella FF, et al. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat Med (2016) 22:1140–50. doi: 10.1038/nm.4172
132. Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat Rev Mol Cell Biol (2016) 17:83–96. doi: 10.1038/nrm.2015.4
133. Eisenberg E, Levanon EY. A-to-I RNA editing — immune protector and transcriptome diversifier. Nat Rev Genet (2018) 19:473–90. doi: 10.1038/s41576-018-0006-1
134. Huang W, Sun Y-M, Pan Q, Fang K, Chen X-T, Zeng Z-C, et al. The snoRNA-like lncRNA LNC-SNO49AB drives leukemia by activating the RNA-editing enzyme ADAR1. Cell Discov (2022) 8:117. doi: 10.1038/s41421-022-00460-9
135. Tassinari V, La Rosa P, Guida E, Colopi A, Caratelli S, De Paolis F, et al. Contribution of A-to-I RNA editing, M6A RNA Methylation, and Alternative Splicing to physiological brain aging and neurodegenerative diseases. Mech Ageing Dev (2023) 212:111807. doi: 10.1016/j.mad.2023.111807
136. Hartner JC, Walkley CR, Lu J, Orkin SH. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol (2009) 10:109–15. doi: 10.1038/ni.1680
137. Li L, Qian G, Zuo Y, Yuan Y, Cheng Q, Guo T, et al. Ubiquitin-dependent turnover of adenosine deaminase acting on RNA 1 (ADAR1) is required for efficient antiviral activity of type I interferon. J Biol Chem (2016) 291:24974–85. doi: 10.1074/jbc.M116.737098
138. Wang H, Wang G, Zhang L, Zhang J, Zhang J, Wang Q, et al. ADAR1 suppresses the activation of cytosolic RNA-sensing signaling pathways to protect the liver from ischemia/reperfusion injury. Sci Rep (2016) 6:20248. doi: 10.1038/srep20248
139. R Gi, K Pr, F Gm, M Nm, G Sm, S M, et al. Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat Genet (2012) 44(11):1243–8. doi: 10.1038/ng.2414
140. Yang S, Deng P, Zhu Z, Zhu J, Wang G, Zhang L, et al. Adenosine deaminase acting on RNA 1 limits RIG-I RNA detection and suppresses IFN production responding to viral and endogenous RNAs. J Immunol (2014) 193:3436–45. doi: 10.4049/jimmunol.1401136
141. Liu G, Ma X, Wang Z, Wakae K, Yuan Y, He Z, et al. Adenosine deaminase acting on RNA-1 (ADAR1) inhibits hepatitis B virus (HBV) replication by enhancing microRNA-122 processing. J Biol Chem (2019) 294:14043–54. doi: 10.1074/jbc.RA119.007970
142. Lamers MM, van den Hoogen BG, Haagmans BL. ADAR1: “Editor-in-chief” of cytoplasmic innate immunity. Front Immunol (2019) 10:1763. doi: 10.3389/fimmu.2019.01763
143. Li Y, Banerjee S, Goldstein S, Dong B, Gaughan C, Rath S, et al. Ribonuclease L mediates the cell-lethal phenotype of double-stranded RNA editing enzyme ADAR1 deficiency in a human cell line. Elife (2017) 6:e25687. doi: 10.7554/eLife.25687
144. de Reuver R, Dierick E, Wiernicki B, Staes K, Seys L, De Meester E, et al. ADAR1 interaction with Z-RNA promotes editing of endogenous double-stranded RNA and prevents MDA5-dependent immune activation. Cell Rep (2021) 36:109500. doi: 10.1016/j.celrep.2021.109500
145. Jiao H, Wachsmuth L, Wolf S, Lohmann J, Nagata M, Kaya GG, et al. ADAR1 averts fatal type I interferon induction by ZBP1. Nature (2022) 607:776–83. doi: 10.1038/s41586-022-04878-9
146. Solomon O, Di Segni A, Cesarkas K, Porath HT, Marcu-Malina V, Mizrahi O, et al. RNA editing by ADAR1 leads to context-dependent transcriptome-wide changes in RNA secondary structure. Nat Commun (2017) 8:1440. doi: 10.1038/s41467-017-01458-8
147. Li J, Xie J, Liu S, Li X, Zhang D, Wang X, et al. ADAR1 attenuates allogeneic graft rejection by suppressing miR-21 biogenesis in macrophages and promoting M2 polarization. FASEB J (2018) 32:5162–73. doi: 10.1096/fj.201701449R
148. Baal N, Cunningham S, Obermann H-L, Thomas J, Lippitsch A, Dietert K, et al. ADAR1 is required for dendritic cell subset homeostasis and alveolar macrophage function. J Immunol (2019) 202:1099–111. doi: 10.4049/jimmunol.1800269
149. Domingo E. Paradoxical interplay of viral and cellular functions. VIRUSES-BASEL (2011) 3:272–7. doi: 10.3390/v3030272
150. Pujantell M, Franco S, Galvan-Femenia I, Badia R, Castellvi M, Garcia-Vidal E, et al. ADAR1 affects HCV infection by modulating innate immune response. Antiviral Res (2018) 156:116–27. doi: 10.1016/j.antiviral.2018.05.012
151. Khadka S, Williams C, Sweeney-Gibbons J, Basler C. Marburg and ebola virus mRNA 3 ’ Untranslated regions contain negative regulators of translation that are modulated by ADAR1 editing. J Virol (2021) 95(19):e0065221. doi: 10.1128/JVI.00652-21
152. Phuphuakrat A, Kraiwong R, Boonarkart C, Lauhakirti D, Lee T-H, Auewarakul P. Double-stranded RNA adenosine deaminases enhance expression of human immunodeficiency virus type 1 proteins. J Virol (2008) 82(21):10864–72. doi: 10.1128/jvi.00238-08
153. Doria M, Neri F, Gallo A, Farace MG, Michienzi A. Editing of HIV-1 RNA by the double-stranded RNA deaminase ADAR1 stimulates viral infection. Nucleic Acids Res (2009) 37:5848–58. doi: 10.1093/nar/gkp604
154. Li Z, Okonski K, Samuel C. Adenosine deaminase acting on RNA 1 (ADAR1) suppresses the induction of interferon by measles virus. J Virol (2012) 86:3787–94. doi: 10.1128/JVI.06307-11
155. Wang L, Sun Y, Song X, Wang Z, Zhang Y, Zhao Y, et al. Hepatitis B virus evades immune recognition via RNA adenosine deaminase ADAR1-mediated viral RNA editing in hepatocytes. Cell Mol Immunol (2021) 18:1871–82. doi: 10.1038/s41423-021-00729-1
156. Diosa-Toro M, Echavarria-Consuegra L, Flipse J, Fernandez G, Kluiver J, van den Berg A, et al. MicroRNA profiling of human primary macrophages exposed to dengue virus identifies miRNA-3614-5p as antiviral and regulator of ADAR1 expression. PLoS Negl Trop Dis (2017) 11(10):e0005981. doi: 10.1371/journal.pntd.0005981
157. Sarvestani ST, Tate MD, Moffat JM, Jacobi AM, Behlke MA, Miller AR, et al. Inosine-mediated modulation of RNA sensing by Toll-like receptor 7 (TLR7) and TLR8. J Virol (2014) 88:799–810. doi: 10.1128/JVI.01571-13
158. Cao Y, Cao R, Huang Y, Zhou H, Liu Y, Li X, et al. A comprehensive study on cellular RNA editing activity in response to infections with different subtypes of influenza a viruses. BMC Genomics (2018) 19(Suppl 1):925. doi: 10.1186/s12864-017-4330-1
159. Vogel O, Han J, Liang C, Manicassamy S, Perez J, Manicassamy B. The p150 Isoform of ADAR1 Blocks Sustained RLR signaling and Apoptosis during Influenza Virus Infection. PLoS Pathog (2020) 16(9):e1008842. doi: 10.1371/journal.ppat.1008842
160. Pfaller CK, Donohue RC, Nersisyan S, Brodsky L, Cattaneo R. Extensive editing of cellular and viral double-stranded RNA structures accounts for innate immunity suppression and the proviral activity of ADAR1p150. PLoS Biol (2018) 16:e2006577. doi: 10.1371/journal.pbio.2006577
161. Yanai M, Kojima S, Sakai M, Komorizono R, Tomonaga K, Makino A. ADAR2 is involved in self and nonself recognition of borna disease virus genomic RNA in the nucleus. J Virol (2020) 94(6):e01513–19. doi: 10.1128/JVI.01513-19
162. John L, Samuel C. Induction of stress granules by interferon and down-regulation by the cellular RNA adenosine deaminase ADAR1. Virology (2014) 454:299–310. doi: 10.1016/j.virol.2014.02.025
163. Okonski K, Samuel C. Stress granule formation induced by measles virus is protein kinase PKR dependent and impaired by RNA adenosine deaminase ADAR1. J Virol (2013) 87:756–66. doi: 10.1128/JVI.02270-12
164. Paget M, Cadena C, Ahmad S, Wang H, Jordan T, Kim E, et al. Stress granules are shock absorbers that prevent excessive innate immune responses to dsRNA. Mol Cell (2023) 83:1180–+. doi: 10.1016/j.molcel.2023.03.010
165. Cachat A, Alais S, Chevalier S, Journo C, Fusil F, Dutartre H, et al. ADAR1 enhances HTLV-1 and HTLV-2 replication through inhibition of PKR activity. Retrovirology (2014) 11:93. doi: 10.1186/s12977-014-0093-9
166. Chukwurah E, Handy I, Patel R. ADAR1 and PACT contribute to efficient translation of transcripts containing HIV-1 trans-activating response (TAR) element. Biochem J (2017) 474:1241–57. doi: 10.1042/BCJ20160964
167. Corbet G, Burke J, Parker R. ADAR1 limits stress granule formation through both translation-dependent and translation-independent mechanisms. J Cell Sci (2021) 134(17):jcs258783. doi: 10.1242/jcs.258783
168. Bar Yaacov D. Functional analysis of ADARs in planarians supports a bilaterian ancestral role in suppressing double-stranded RNA-response. PLoS Pathog (2022) 18(1):e1010250. doi: 10.1371/journal.ppat.1010250
169. Li T, Yang X, Li W, Song J, Li Z, Zhu X, et al. ADAR1 stimulation by IFN-alpha downregulates the expression of MAVS via RNA editing to regulate the anti-HBV response. Mol Ther (2021) 29:1335–48. doi: 10.1016/j.ymthe.2020.11.031
170. Ahmad S, Mu X, Yang F, Greenwald E, Park JW, Jacob E, et al. Breaching self-tolerance to alu duplex RNA underlies MDA5-mediated inflammation. Cell (2018) 172:797–810.e13. doi: 10.1016/j.cell.2017.12.016
171. Yuan L, Jia Q, Yang S, Idris N, Li Y, Wang Y, et al. ADAR1 promotes HBV replication through its deaminase domain. Front Biosci-Landmark (2020) 25:710–21. doi: 10.2741/4830
172. Doria M, Tomaselli S, Neri F, Ciafrè SA, Farace MG, Michienzi A, et al. ADAR2 editing enzyme is a novel human immunodeficiency virus-1 proviral factor. J Gen Virol (2011) 92:1228–32. doi: 10.1099/vir.0.028043-0
173. Cuadrado E, Booiman T, van Hamme J, Jansen M, van Dort K, Vanderver A, et al. ADAR1 facilitates HIV-1 replication in primary CD4(+) T cells. PLoS One (2015) 10(12):e0143613. doi: 10.1371/journal.pone.0143613
174. Poison AG, Bass BL, Casey JL. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature (1996) 380:454–6. doi: 10.1038/380454a0
175. Vlachogiannis NI, Verrou K-M, Stellos K, Sfikakis PP, Paraskevis D. The role of A-to-I RNA editing in infections by RNA viruses: Possible implications for SARS-CoV-2 infection. Clin Immunol (2021) 226:108699. doi: 10.1016/j.clim.2021.108699
176. Uhl S, Jang C, Frere J, Jordan T, Simon A, TenOever B. ADAR1 biology can hinder effective antiviral RNA interference. J Virol (2023) 97(4):e0024523. doi: 10.1128/jvi.00245-23
177. Montavon T, Baldaccini M, Lefevre M, Girardi E, Chane-Woon-Ming B, Messmer M, et al. Human DICER helicase domain recruits PKR and modulates its antiviral activity. PLoS Pathog (2021) 17(5):e1009549. doi: 10.1371/journal.ppat.1009549
178. Song Y, He X, Yang W, Wu Y, Cui J, Tang T, et al. Virus-specific editing identification approach reveals the landscape of A-to-I editing and its impacts on SARS-CoV-2 characteristics and evolution. Nucleic Acids Res (2022) 50:2509–21. doi: 10.1093/nar/gkac120
179. Samuel CE. ADARs, viruses and innate immunity. Curr Top Microbiol Immunol (2012) 353:10. doi: 10.1007/82_2011_148
180. Dong N, Dong C, Xiong S. Janus effects of ADAR1 on CVB3-induced viral myocarditis at different infection stages. Int J Cardiol (2016) 223:898–905. doi: 10.1016/j.ijcard.2016.08.315
181. Pfaller CK, Mastorakos GM, Matchett WE, Ma X, Samuel CE, Cattaneo R. Measles virus defective interfering RNAs are generated frequently and early in the absence of C protein and can be destabilized by adenosine deaminase acting on RNA-1-like hypermutations. J Virol (2015) 89:7735–47. doi: 10.1128/JVI.01017-15
182. Fischer S, Ruvkun G. Caenorhabditis elegans ADAR editing and the ERI-6/7/MOV10 RNAi pathway silence endogenous viral elements and LTR retrotransposons. Proc Natl Acad Sci USA (2020) 117:5987–96. doi: 10.1073/pnas.1919028117
183. Taylor DR, Puig M, Darnell MER, Mihalik K, Feinstone SM. New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through ADAR1. J Virol (2005) 79:6291–8. doi: 10.1128/JVI.79.10.6291-6298.2005
184. Liu C-X, Li X, Nan F, Jiang S, Gao X, Guo S-K, et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell (2019) 177:865–80.e21. doi: 10.1016/j.cell.2019.03.046
185. Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep (2015) 10:170–7. doi: 10.1016/j.celrep.2014.12.019
186. Pfaller C, George C, Samuel C. Adenosine deaminases acting on RNA (ADARs) and viral infections. Annu Rev Virol (2021) 8:239–64. doi: 10.1146/annurev-virology-091919-065320
187. Piontkivska H, Matos L, Paul S, Scharfenberg B, Farmerie W, Miyamoto M, et al. Role of host-driven mutagenesis in determining genome evolution of sigma virus (DMelSV; rhabdoviridae) in drosophila melanogaster. Genome Biol Evol (2016) 8:2952–63. doi: 10.1093/gbe/evw212
188. Rosani U, Bai C, Maso L, Shapiro M, Abbadi M, Domeneghetti S, et al. A-to-I editing of Malacoherpesviridae RNAs supports the antiviral role of ADAR1 in mollusks. BMC Evol Biol (2019) 19(1):149. doi: 10.1186/s12862-019-1472-6
189. Ringlander J, Fingal J, Kann H, Prakash K, Rydell G, Andersson M, et al. Impact of ADAR-induced editing of minor viral RNA populations on replication and transmission of SARS-CoV-2. Proc Natl Acad Sci USA (2022) 119(6):e2112663119. doi: 10.1073/pnas.2112663119
190. Booth BJ, Nourreddine S, Katrekar D, Savva Y, Bose D, Long TJ, et al. RNA editing: Expanding the potential of RNA therapeutics. Mol Ther (2023) 31:1533–49. doi: 10.1016/j.ymthe.2023.01.005
191. Khrustalev V, Khrustaleva T, Sharma N, Giri R. Mutational pressure in zika virus: local ADAR-editing areas associated with pauses in translation and replication. Front Cell Infect Microbiol (2017) 7:44. doi: 10.3389/fcimb.2017.00044
192. Peng X, Luo Y, Li H, Guo X, Chen H, Ji X, et al. RNA editing increases the nucleotide diversity of SARS-CoV-2 in human host cells. PLoS Genet (2022) 18(3):e1010130. doi: 10.1371/journal.pgen.1010130
193. Crooke PS, Tossberg JT, Porter KP, Aune TM. Reduced A-to-I editing of endogenous Alu RNAs in lung after SARS-CoV-2 infection1. Curr Res Immunol (2021) 2:52–9. doi: 10.1016/j.crimmu.2021.04.001
194. Terajima H, Lu M, Zhang L, Cui Q, Shi Y, Li J, et al. N-6-methyladenosine promotes induction of ADAR1-mediated A-to-I RNA editing to suppress aberrant antiviral innate immune responses. PLoS Biol (2021) 19(7):e3001292. doi: 10.1371/journal.pbio.3001292
195. Orecchini E, Doria M, Antonioni A, Galardi S, Ciafre S, Frassinelli L, et al. ADAR1 restricts LINE-1 retrotransposition. Nucleic Acids Res (2017) 45:155–68. doi: 10.1093/nar/gkw834
196. Li X, Ma S, Yi C. Pseudouridine: the fifth RNA nucleotide with renewed interests. Curr Opin Chem Biol (2016) 33:108–16. doi: 10.1016/j.cbpa.2016.06.014
197. Boo SH, Kim YK. The emerging role of RNA modifications in the regulation of mRNA stability. Exp Mol Med (2020) 52:400–8. doi: 10.1038/s12276-020-0407-z
198. Borchardt EK, Martinez NM, Gilbert WV. Regulation and function of RNA pseudouridylation in human cells. Annu Rev OF Genet (2020) 54:309–36. doi: 10.1146/annurev-genet-112618-043830
199. Roca X, Akerman M, Gaus H, Berdeja A, Bennett CF, Krainer AR. Widespread recognition of 5′ splice sites by noncanonical base-pairing to U1 snRNA involving bulged nucleotides. Genes Dev (2012) 26:1098–109. doi: 10.1101/gad.190173.112
200. Nguyen THD, Galej WP, Bai X-C, Oubridge C, Newman AJ, Scheres SHW, et al. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Å resolution. Nature (2016) 530:298–302. doi: 10.1038/nature16940
201. Dai Q, Zhang L-S, Sun H-L, Pajdzik K, Yang L, Ye C, et al. Quantitative sequencing using BID-seq uncovers abundant pseudouridines in mammalian mRNA at base resolution. Nat Biotechnol (2023) 41:344–54. doi: 10.1038/s41587-022-01505-w
202. Zhang M, Jiang Z, Ma Y, Liu W, Zhuang Y, Lu B, et al. Quantitative profiling of pseudouridylation landscape in the human transcriptome. Nat Chem Biol (2023) 19:1185–95. doi: 10.1038/s41589-023-01304-7
203. Furuse Y. RNA modifications in genomic RNA of influenza A virus and the relationship between RNA modifications and viral infection. Int J Mol Sci (2021) 22(17):9127. doi: 10.3390/ijms22179127
204. Karlebach G, Aronow B, Baylin SB, Butler D, Foox J, Levy S, et al. Betacoronavirus-specific alternate splicing. Genomics (2022) 114(2):110270. doi: 10.1016/j.ygeno.2022.110270
205. Anderson BR, Muramatsu H, Nallagatla SR, Bevilacqua PC, Sansing LH, Weissman D, et al. Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res (2010) 38:5884–92. doi: 10.1093/nar/gkq347
206. Durbin AF, Wang C, Marcotrigiano J, Gehrke L. RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. mBio (2016) 7(5):e00833–16. doi: 10.1128/mBio.00833-16
207. Anderson BR, Muramatsu H, Jha BK, Silverman RH, Weissman D, Kariko K. Nucleoside modifications in RNA limit activation of 2’-5’-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res (2011) 39:9329–38. doi: 10.1093/nar/gkr586
208. Galindo-Murillo R, Davis DR, Cheatham TE III. Probing the influence of hypermodified residues within the tRNA(3)(Lys) anticodon stem loop interacting with the A-loop primer sequence from HIV-1. Biochim ET Biophys ACTA-GENERAL Subj (2016) 1860:607–17. doi: 10.1016/j.bbagen.2015.11.009
209. Zhao Y, Karijolich J, Glaunsinger B, Zhou Q. Pseudouridylation of 7SK snRNA promotes 7SK snRNP formation to suppress HIV-1 transcription and escape from latency. EMBO Rep (2016) 17:1441–51. doi: 10.15252/embr.201642682
210. Martinez Campos C, Tsai K, Courtney DG, Bogerd HP, Holley CL, Cullen BR. Mapping of pseudouridine residues on cellular and viral transcripts using a novel antibody-based technique. RNA (2021) 27:1400–11. doi: 10.1261/rna.078940.121
211. Huang S, Zhang W, Katanski CD, Dersh D, Dai Q, Lolans K, et al. Interferon inducible pseudouridine modification in human mRNA by quantitative nanopore profiling. Genome Biol (2021) 22:330. doi: 10.1186/s13059-021-02557-y
212. Nance KD, Meier JL. Modifications in an emergency: the role of N1-methylpseudouridine in COVID-19 vaccines. ACS Cent Sci (2021) 7:748–56. doi: 10.1021/acscentsci.1c00197
213. Morais P, Adachi H, Yu Y-T. The critical contribution of pseudouridine to mRNA COVID-19 vaccines. Front Cell Dev Biol (2021) 9:789427. doi: 10.3389/fcell.2021.789427
214. Xia X, Wang Y, Zheng JC. Internal m7G methylation: A novel epitranscriptomic contributor in brain development and diseases. Mol Ther - Nucleic Acids (2023) 31:295–308. doi: 10.1016/j.omtn.2023.01.003
215. Galloway A, Cowling VH. mRNA cap regulation in mammalian cell function and fate. Biochim Biophys Acta (BBA) - Gene Regul Mech (2019) 1862:270–9. doi: 10.1016/j.bbagrm.2018.09.011
216. Pei Y, Shuman S. Interactions between fission yeast mRNA capping enzymes and elongation factor spt5 *. J Biol Chem (2002) 277:19639–48. doi: 10.1074/jbc.M200015200
217. Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, Emigh CA, et al. Dual roles for spt5 in pre-mRNA processing and transcription elongation revealed by identification of spt5-associated proteins. Mol Cell Biol (2003) 23:1368–78. doi: 10.1128/MCB.23.4.1368-1378.2003
218. Ramanathan A, Robb G, Chan S. mRNA capping: biological functions and applications. Nucleic Acids Res (2016) 44:7511–26. doi: 10.1093/nar/gkw551
219. Zhang L-S, Ju C-W, Liu C, Wei J, Dai Q, Chen L, et al. m7G-quant-seq: quantitative detection of RNA internal N7-methylguanosine. ACS Chem Biol (2022) 17:3306–12. doi: 10.1021/acschembio.2c00792
220. Zhao Z, Qing Y, Dong L, Han L, Wu D, Li Y, et al. QKI shuttles internal m7G-modified transcripts into stress granules and modulates mRNA metabolism. Cell (2023) 186:3208–26.e27. doi: 10.1016/j.cell.2023.05.047
221. Zhang L-S, Liu C, Ma H, Dai Q, Sun H-L, Luo G, et al. Transcriptome-wide mapping of internal N-7-methylguanosine methylome in mammalian mRNA. Mol Cell (2019) 74:1304–+. doi: 10.1016/j.molcel.2019.03.036
222. Yedavalli VSRK, Jeang K-T. Trimethylguanosine capping selectively promotes expression of Rev-dependent HIV-1 RNAs. Proc Natl Acad Sci USA (2010) 107:14787–92. doi: 10.1073/pnas.1009490107
223. Ho JSY, Angel M, Ma Y, Sloan E, Wang G, Martinez-Romero C, et al. Hybrid gene origination creates human-virus chimeric proteins during infection. CELL (2020) 181:1502–+. doi: 10.1016/j.cell.2020.05.035
224. Singh G, Seufzer B, Song Z, Zucko D, Heng X, Boris-Lawrie K. HIV-1 hypermethylated guanosine cap licenses specialized translation unaffected by mTOR. Proc Natl Acad Sci USA (2022) 119(1):e2105153118. doi: 10.1073/pnas.2105153118
225. Decroly E, Canard B. Biochemical principles and inhibitors to interfere with viral capping pathways. Curr Opin Virol (2017) 24:87–96. doi: 10.1016/j.coviro.2017.04.003
226. Shi J, Zhang Y, Tan D, Zhang X, Yan M, Zhang Y, et al. PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nat Cell Biol (2021) 23:424–36. doi: 10.1038/s41556-021-00652-7
227. Zhang L-S, Xiong Q-P, Peña Perez S, Liu C, Wei J, Le C, et al. ALKBH7-mediated demethylation regulates mitochondrial polycistronic RNA processing. Nat Cell Biol (2021) 23:684–91. doi: 10.1038/s41556-021-00709-7
228. Safra M, Sas-Chen A, Nir R, Winkler R, Nachshon A, Bar-Yaacov D, et al. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature (2017) 551:251–5. doi: 10.1038/nature24456
229. Xiong W, Zhao Y, Wei Z, Li C, Zhao R, Ge J, et al. N1-methyladenosine formation, gene regulation, biological functions, and clinical relevance. Mol Ther (2023) 31:308–30. doi: 10.1016/j.ymthe.2022.10.015
230. Li X, Xiong X, Zhang M, Wang K, Chen Y, Zhou J, et al. Base-resolution mapping reveals distinct m1A methylome in nuclear- and mitochondrial-encoded transcripts. Mol Cell (2017) 68:993–1005.e9. doi: 10.1016/j.molcel.2017.10.019
231. Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, et al. Differential m6A, m6Am, and m1A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol Cell (2018) 71:973–85.e5. doi: 10.1016/j.molcel.2018.08.011
232. Liu H, Zeng T, He C, Rawal VH, Zhou H, Dickinson BC. Development of mild chemical catalysis conditions for m(1)A-to-m(6) A rearrangement on RNA. ACS Chem Biol (2022) 17:1334–42. doi: 10.1021/acschembio.2c00178
233. Apostle A, Yin Y, Chillar K, Eriyagama AMDN, Arneson R, Burke E, et al. Effects of epitranscriptomic RNA modifications on the catalytic activity of the SARS-coV-2 replication complex. Chembiochem (2023) 24(8):e202300095. doi: 10.1002/cbic.202300095
234. Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature (2017) 541:371–5. doi: 10.1038/nature21022
235. Sun H, Zhang M, Li K, Bai D, Yi C. Cap-specific, terminal N6-methylation by a mammalian m6Am methyltransferase. Cell Res (2019) 29:80–2. doi: 10.1038/s41422-018-0117-4
236. Boulias K, Toczydłowska-Socha D, Hawley BR, Liberman N, Takashima K, Zaccara S, et al. Identification of the m6Am Methyltransferase PCIF1 Reveals the Location and Functions of m6Am in the Transcriptome. Mol Cell (2019) 75:631–43.e8. doi: 10.1016/j.molcel.2019.06.006
237. Sendinc E, Valle-Garcia D, Dhall A, Chen H, Henriques T, Navarrete-Perea J, et al. PCIF1 Catalyzes m6Am mRNA Methylation to Regulate Gene Expression. Mol Cell (2019) 75:620–30.e9. doi: 10.1016/j.molcel.2019.05.030
238. Tartell M, Boulias K, Hoffmann G, Bloyet L, Greer E, Whelan S. Methylation of viral mRNA cap structures by PCIF1 attenuates the antiviral activity of interferon-beta. Proc Natl Acad Sci USA (2021) 118(29):e2025769118. doi: 10.1073/pnas.2025769118
239. Zhang Q, Kang Y, Wang S, Gonzalez GM, Li W, Hui H, et al. HIV reprograms host m(6)Am RNA methylome by viral Vpr protein-mediated degradation of PCIF1. Nat Commun (2021) 12(1):5543. doi: 10.1038/s41467-021-25683-4
240. Wang L, Wang S, Wu L, Li W, Bray W, Clark AE, et al. PCIF1-mediated deposition of 5′-cap N6,2′-O-dimethyladenosine in ACE2 and TMPRSS2 mRNA regulates susceptibility to SARS-CoV-2 infection. Proc Natl Acad Sci (2023) 120:e2210361120. doi: 10.1073/pnas.2210361120
241. Sun H, Li K, Zhang X, Liu J, Zhang M, Meng H, et al. m6Am-seq reveals the dynamic m6Am methylation in the human transcriptome. Nat Commun (2021) 12:4778. doi: 10.1038/s41467-021-25105-5
242. Luo J, Cao J, Chen C, Xie H. Emerging role of RNA acetylation modification ac4C in diseases: Current advances and future challenges. Biochem Pharmacol (2023) 213:115628. doi: 10.1016/j.bcp.2023.115628
243. Arango D, Sturgill D, Alhusaini N, Dillman AA, Sweet TJ, Hanson G, et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell (2018) 175:1872–86.e24. doi: 10.1016/j.cell.2018.10.030
244. Arango D, Sturgill D, Yang R, Kanai T, Bauer P, Roy J, et al. Direct epitranscriptomic regulation of mammalian translation initiation through N4-acetylcytidine. Mol Cell (2022) 82:2912. doi: 10.1016/j.molcel.2022.06.022
245. Tsai K, Vasudevan A, Campos C, Emery A, Swanstrom R, Cullen B. Acetylation of cytidine residues boosts HIV-1 gene expression by increasing viral RNA stability. Cell Host Microbe (2020) 28:306–+. doi: 10.1016/j.chom.2020.05.011
246. Jean MJ, Power D, Kong W, Huang H, Santoso N, Zhu J. Identification of HIV-1 tat-associated proteins contributing to HIV-1 transcription and latency. Viruses (2017) 9:67. doi: 10.3390/v9040067
247. Hao H, Liu W, Miao Y, Ma L, Yu B, Liu L, et al. N4-acetylcytidine regulates the replication and pathogenicity of enterovirus 71. Nucleic Acids Res (2022) 50:9339–54. doi: 10.1093/nar/gkac675
248. Watanabe T, Kawakami E, Shoemaker JE, Lopes TJS, Matsuoka Y, Tomita Y, et al. Influenza virus-host interactome screen as a platform for antiviral drug development. Cell Host Microbe (2014) 16:795–805. doi: 10.1016/j.chom.2014.11.002
249. Zigáčková D, Vaňáčová Š. The role of 3′ end uridylation in RNA metabolism and cellular physiology. Philos Trans R Soc B: Biol Sci (2018) 373:20180171. doi: 10.1098/rstb.2018.0171
250. Gupta A, Li Y, Chen S, Papas B, Martin N, Morgan M. TUT4/7-mediated uridylation of a coronavirus subgenomic RNAs delays viral replication. Commun Biol (2023) 6(1):438. doi: 10.1038/s42003-023-04814-1
251. Le Pen J, Jiang H, Di Domenico T, Kneuss E, Kosalka J, Leung C, et al. Terminal uridylyltransferases target RNA viruses as part of the innate immune system. Nat Struct Mol Biol (2018) 25:778–+. doi: 10.1038/s41594-018-0106-9
252. Lin C, Shen Y, Chang C, Guo X, Young Y, Lai T, et al. Terminal uridyltransferase 7 regulates TLR4-triggered inflammation by controlling Regnase-1 mRNA uridylation and degradation. Nat Commun (2021) 12(1):3878. doi: 10.1038/s41467-021-24177-7
253. Harris RS, Dudley JP. APOBECs and virus restriction. Virology (2015) 479–480:131–45. doi: 10.1016/j.virol.2015.03.012
254. Xu WK, Byun H, Dudley JP. The role of APOBECs in viral replication. Microorganisms (2020) 8:1899. doi: 10.3390/microorganisms8121899
255. Pecori R, Di Giorgio S, Paulo Lorenzo J, Nina Papavasiliou F. Functions and consequences of AID/APOBEC-mediated DNA and RNA deamination. Nat Rev Genet (2022) 23:505–18. doi: 10.1038/s41576-022-00459-8
256. Squires KD, Monajemi M, Woodworth CF, Grant MD, Larijani M. Impact of APOBEC mutations on CD8(+) T cell recognition of HIV epitopes varies depending on the restricting HLA. JAIDS (2015) 70:172–8. doi: 10.1097/QAI.0000000000000689
Keywords: RNA modification, innate immunity, virus infection, 2’-O-methyltransferase, 5-methylcytidine, pseudouridine, RNA editing
Citation: Shen S and Zhang L-S (2023) The regulation of antiviral innate immunity through non-m6A RNA modifications. Front. Immunol. 14:1286820. doi: 10.3389/fimmu.2023.1286820
Received: 31 August 2023; Accepted: 04 October 2023;
Published: 17 October 2023.
Edited by:
Zhenhua Chen, Beckman Research Institute, United StatesReviewed by:
Lianjun Zhang, City of Hope National Medical Center, United StatesCopyright © 2023 Shen and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-Sheng Zhang, emhhbmdsc0B1c3QuaGs=
†ORCID: Shenghai Shen, orcid.org/0000-0001-8422-5423
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.