
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 03 January 2024
Sec. Autoimmune and Autoinflammatory Disorders: Autoinflammatory Disorders
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1286776
This article is part of the Research Topic Epithelial Immune Microenvironment and Inflammatory Skin Diseases View all 8 articles
 Lichen Zhang1,2†
Lichen Zhang1,2† Rongrong Chai1,2†
Rongrong Chai1,2† Zongguang Tai1,2
Zongguang Tai1,2 Fengze Miao1,2
Fengze Miao1,2 Xinwei Shi1,2
Xinwei Shi1,2 Zhongjian Chen1,2*
Zhongjian Chen1,2* Quangang Zhu1,2*
Quangang Zhu1,2*Inflammatory skin diseases are a group of diseases caused by the disruption of skin tissue due to immune system disorders. Histone modification plays a pivotal role in the pathogenesis and treatment of chronic inflammatory skin diseases, encompassing a wide range of conditions, including psoriasis, atopic dermatitis, lupus, systemic sclerosis, contact dermatitis, lichen planus, and alopecia areata. Analyzing histone modification as a significant epigenetic regulatory approach holds great promise for advancing our understanding and managing these complex disorders. Additionally, therapeutic interventions targeting histone modifications have emerged as promising strategies for effectively managing inflammatory skin disorders. This comprehensive review provides an overview of the diverse types of histone modification. We discuss the intricate association between histone modification and prevalent chronic inflammatory skin diseases. We also review current and potential therapeutic approaches that revolve around modulating histone modifications. Finally, we investigated the prospects of research on histone modifications in the context of chronic inflammatory skin diseases, paving the way for innovative therapeutic interventions and improved patient outcomes.
The epithelial tissues, comprising the outermost layer of an organism, serve as a protective barrier against environmental stressors, including physical, chemical, and microbial agents. Epithelial tissues are not only a physical barrier; they are also immunological organs that are activated in response to the attack of foreign agents and trigger different sets of transcriptional cascades that stimulate a specific type of immune response (1). The epithelial immune microenvironment is an important component in implementing this process; however, an aberration in this microenvironment may contribute to inflammatory skin diseases (2, 3).
Inflammatory skin diseases encompass a range of skin disorders characterized by the infiltration of inflammatory cells and a significant increase in inflammatory cytokines (4). The most prevalent inflammatory skin diseases include psoriasis, atopic dermatitis (AD), lupus, systemic sclerosis (SSc), contact dermatitis, lichen planus (LP), and alopecia areata (AA) The underlying causes of these diseases remain unclear. Genetic susceptibility has been proposed to partially explain the immune imbalances observed in patients with inflammatory skin conditions. Research indicates that defects in the filaggrin gene may increase the risk of allergic sensitization, atopic eczema, and allergic rhinitis (5). Another study discovered that the TYK2 gene’s loss-of-function missense variant rs34536443 is associated with decreased susceptibility to various autoimmune diseases in individuals with psoriasis (6). However, investigations involving monozygotic twins have shown that genetic factors do not comprehensively explain the underlying causes of inflammatory skin conditions (7, 8).
Epigenetic modifications, characterized by reversible alterations in gene expression, independent of DNA sequence changes, are manifested during various stages of development or in response to environmental stimuli. These modifications, commonly known as the “second code,” have gained increasing attention due to their significance in inflammatory skin diseases, specifically immune activation, T-cell polarization, and impairment of skin barrier function (9–13). In eukaryotic cells, the chromosomal DNA is organized into a compact structure known as chromatin (14). The chromatin comprises nucleosome subunits, consisting of a histone octamer made up of two copies each of the core histones H2A, H2B, H3, and H4, wrapped by 147 base pairs of DNA (15, 16). The nucleosomes are then condensed further into a higher-order chromatin structure when the linker histone H1 binds to the linker DNA. This condensation enables the storage of a significant amount of DNA, approximately six feet long when stretched from end to end, within a single cell (17–19). Dynamic chromatin remodeling significantly regulates various DNA-dependent biological processes, including RNA transcription, DNA replication, DNA repair, and chromosome condensation and segregation (20).
The chromatin structure can be classified into two categories according to its association with gene transcription. In a heterochromatic state, robust DNA-protein interactions induce highly condensed chromatin, characterized by limited interaction between transcription factors and the genome, which results in gene repression. Conversely, in a euchromatic state, DNA-protein interactions are diminished, resulting in loosely packed chromatin with an accessible structure that facilitates transcription factor binding to DNA, stimulating gene activation (21, 22). The core histones within the histone octamer exhibit a predominantly globular structure, except for their N-terminal “tails” that extend from the nucleosome. These N-terminal “tails” are rich in basic lysine and arginine residues and are subject to various modifications. Subsequently, these modifications result in conformational changes within the chromatin, enabling it to adopt relaxed or condensed states that either stimulate or inhibit transcription, respectively (23, 24). Histone modification has been shown to interact with the immune microenvironment and promote the occurrence and development of inflammatory skin diseases. A study reported that the α-KG–H3K9me3–BBOX1 axis is critical in the metabolic reprogramming of cluster of differentiation (CD)147, which is pivotal in glycolysis reprogramming during the pathogenesis of psoriasis (25). One of the histone deacetylases, SIRT1, potentially inhibits the apoptosis of keratinocytes induced by UVB in cutaneous lupus erythematosus (26). Another histone deacetylase, HDAC1, exhibits dysregulation in patients with AA and acne vulgaris (27). Some aberrant histone acetylation and methylation modifications were also found in PBMCs of patients with pemphigus vulgaris. These modifications may contribute to the pathological immune responses in the disease (28).
In this review, we described different types of histone modifications, analyzed their relationship with common chronic inflammatory skin diseases, and reviewed the therapeutic approaches currently employed for treating these conditions in association with histone modification. Additionally, we evaluated the prospects of utilizing histone modification in treating inflammatory skin diseases.
Histones typically contain amino acids with basic side chains that carry a positive charge, while genomic DNA carries a negative charge, creating an attractive force between the two (29). The process of histone acetylation primarily occurs at lysine residues, which neutralizes the positive charge of histones and reduces the interaction between nucleosomes and DNA. Consequently, histone acetylation results in a heterochromatic state, whereas histone deacetylation tends to result in a euchromatic state, significantly affecting the initiation and elongation of gene transcription (30, 31). The acetylation and deacetylation of histones are primarily regulated by histone acetyltransferases (HAT) and histone deacetylases (HDACs). HATs are protein complexes that facilitate the transfer of acetyl groups, including acetyl-CoA, onto the amino tails of histones (32). HDACs are categorized into four distinct classes: Class I (HDACs 1-3 and 8), Class II (HDACs 4-7, 9, and 10), and Class IV (HDAC11). HDACs require Zn2+ for their enzymatic activity, while Class III (Sirt1-7) HDACs are NAD+-dependent. These HDAC classes remove acetyl marks from histones and other protein substrates (17, 33). Table 1 shows the classification and family of the enzymes involved in histone acetylation. Histone acetylation plays a significant role in the pathogenesis and progression of inflammatory diseases. Among the HAT proteins, KAT2A has been identified as one of them. The expression of KAT2A has been positively correlated with the immunopathology of inflammatory joint diseases in both patients with rheumatoid arthritis and mice with experimental arthritis. This correlation is attributed to the promotion of Il1b and Nlrp3 transcription through histone H3K9ac modification and the inhibition of NRF2 transcription repressor activity by KAT2A. Additionally, another type of HAT, CBP/p300, enhances cytokine expression by increasing the levels of H3K27a (34). Another type of HAT, known as CBP/p300, facilitates the expression of cytokines by upregulating the abundance of H3K27ac (35). Given the role of histone acetylation in modulating inflammatory responses, it is plausible to consider manipulating histone acetylation levels as a potential therapeutic approach for managing inflammatory disorders. Figure 1 shows how histone acetylation influence inflammatory reaction.
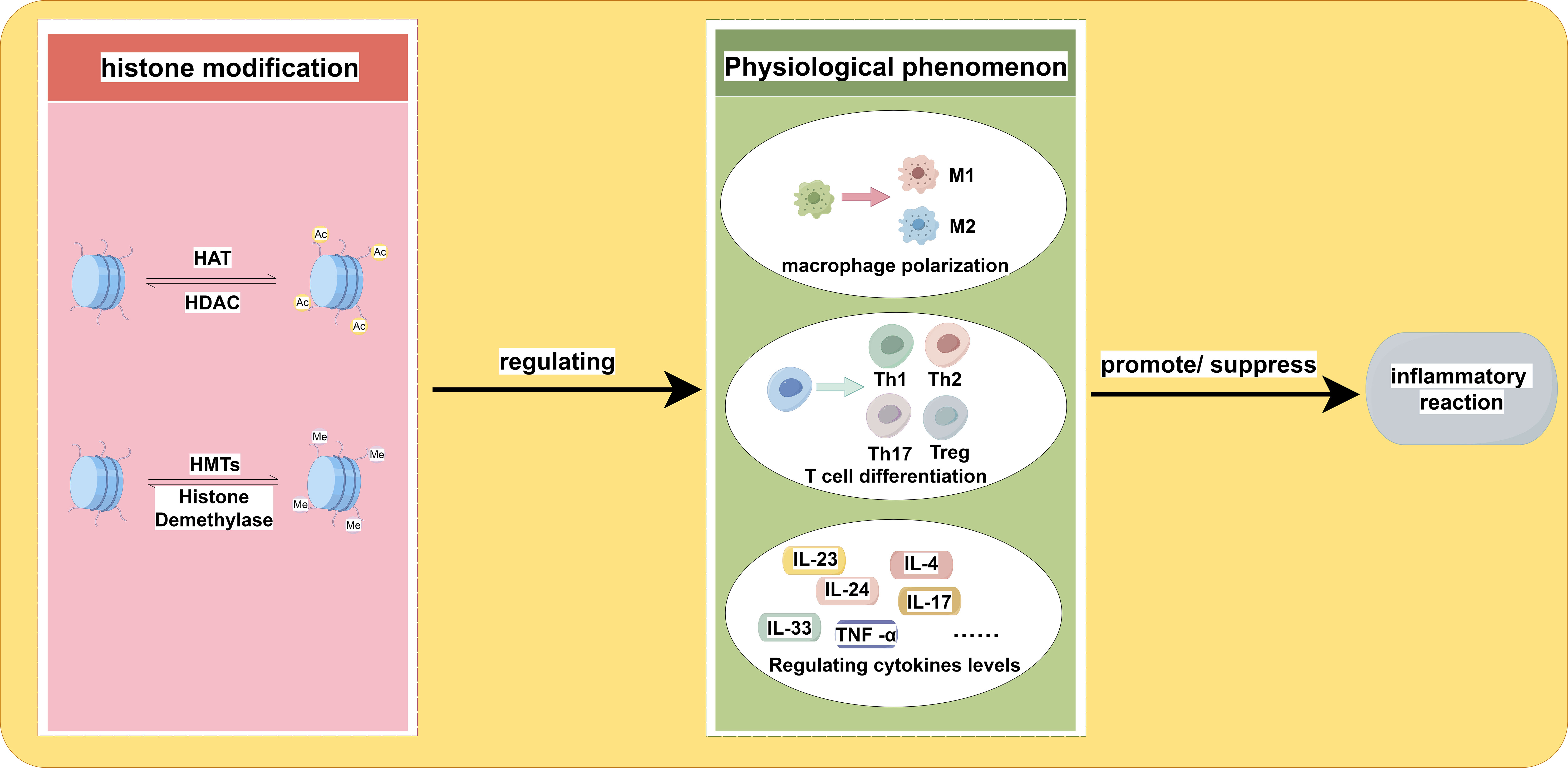
Figure 1 The impact of histone acetylation and histone methylation on the inflammatory response is examined. The up-regulation and down-regulation of these two types of histone modification play a role in macrophage polarization, T cell differentiation, and variation in cytokine levels. The specific locations and types of these modifications may lead to different directions of variation, ultimately influencing the regulation of the inflammatory response. The adjustment of histone modifications is mediated by relevant enzymes.
Histone methylation occurs at specific lysine or arginine residues located on histone’s N-terminal “tails” of histone (36). Compared to acetylation, methylation is a more intricate and nuanced process as it does not alter the charge in histone methylation, and both lysine and arginine residues can undergo multiple methylation states (37). Lysine residues, specifically H2BK5, H3K4, H3K9, H3K27, H3K36, H3K79, and H4K20, can undergo monomethylation, dimethylation, or trimethylation on the ϵ-amino groups of lysine residues. On the other hand, arginine residues, specifically H3R2, H3R8, H3R17, H3R26, and H4R3, can undergo monomethylation, symmetrical dimethylation, or asymmetrical dimethylation on their guanidinyl groups (23). Both the positioning of modified residues and the quantity of methyl groups are crucial for the functional consequences of histone methylation (38). The trimethylation of H3K4 (H3K4me3) at gene promoters and H3K36me3 across gene bodies results in open regions within chromatin and transcription activation. Conversely, H3K27me3 and H3K9me3 are commonly associated with gene silencing (17). In some conditions, monomethylation, including H3K9me1 and H3K27me1, is involved in the activation of transcription, while trimethylation at the same sites is associated with repression (17). Table 1 displays the classification and family of the enzymes involved in histone methylation. The polarization of macrophages can be influenced by various types and levels of histone methylation, which in turn can impact the occurrence and progression of inflammation. The function of M1 macrophages is to produce pro-inflammatory cytokines, including TNF-α, IL-1β, IL-6, IL-12, and IL-23, through the expression of transcription factors, mainly the NF-κB. Macrophages adopt the M1 phenotype in response to pro-inflammatory cytokines secreted in response to persistent and severe inflammation or infection. However, if the M1 phase continues, it causes chronic inflammation, tissue damage, and loss of organ functions. Two kinds of histone methylation, H3K9me3 and H3K36me2, can negatively regulate the M1 phenotype (39, 40). On the contrary, a decrease of H3K27me3 may promote the M1 macrophage phenotype (41). Histone methylation is also very important in T-cell responses. The differentiation from naive CD4+ T cells to Th17 cells causes upregulation of H3K4me3 in the RORC promoter while decreasing H3K27me3. In contrast, the differentiation of Th1 cells is anticipated to contribute to the upregulation of H3K27me3 at the regulatory regions of RORC (42). These findings provide evidence suggesting that histone methylation plays a pivotal role in the inflammatory response. Figure 1 shows how histone methylation influence inflammatory reaction.
The most studied histone modifications include acetylation and methylation, while other significant modifications include phosphorylation, ubiquitination, sumoylation, biotinylation, and ADP-ribosylation. Histone phosphorylation at serine and threonine residues contributes to chromatin relaxation (43) in various cellular processes, including transcription, mitosis, DNA repair, and apoptosis. Additionally, it enhances the effectiveness of histone acetylation (44, 45). Ubiquitin and small ubiquitin-like modifiers (SUMO) are conserved small proteins that undergo covalent attachment to an ϵ-amino group of a lysine residue on the histone (46, 47). Ubiquitination and sumoylation are associated with DNA double-strand break repair (48, 49). Histone biotinylation also occurs at lysine residues and may be involved in transcriptional repression (19, 50). ADP-ribosylation, catalyzed by ADP-ribose transferases using NAD+ as a cofactor, adds an ADP-ribose unit to an N-terminal chain. This process occurs in lysine, arginine, glutamic acid, and aspartic acid residues and is associated with DNA damage (51). Figure 2 indicates different kinds of histone modifications and their position in N-terminal “tails”. The enrichment of H3 serine 28 phosphorylation was observed at induced genes in mouse macrophages stimulated with bacterial lipopolysaccharide, serving as a mediator for MSKs in the regulation of inflammatory response (52). USP38, a histone deubiquitinase, exhibits specificity in removing monoubiquitin from H2B at lysine 120. This process plays a role in coordinating inflammatory responses with the histone H3K4 modifier KDM5B (53). The process of histone ADP-riboyslation, mediated by PARP1, enhances the expression of inflammatory cytokines in microglia by promoting the accessibility of promoter DNA (54). The investigation of the association between inflammation and other kinds of histone modifications remains infrequently explored in scholarly literature.
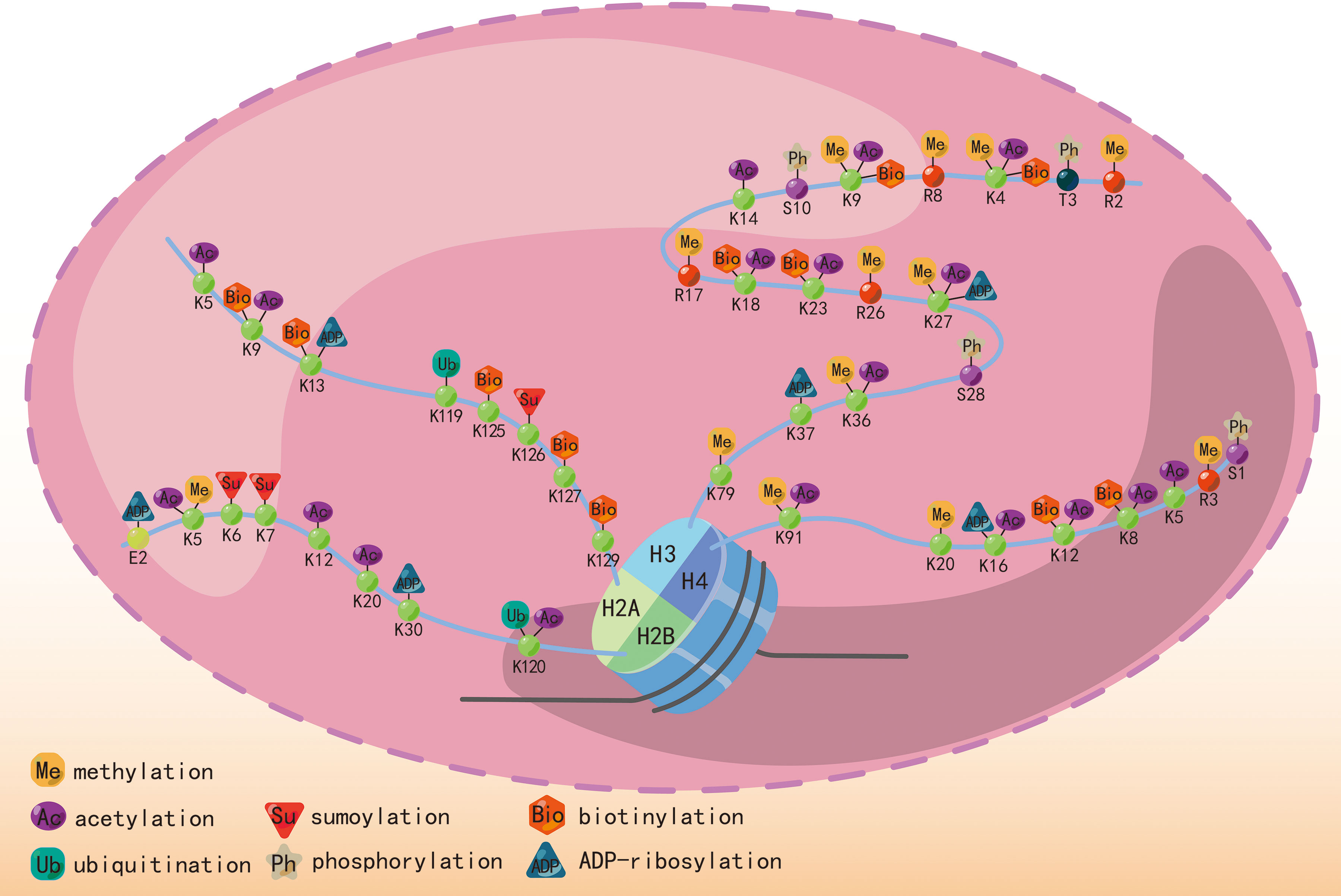
Figure 2 Different histone modifications, including methylation, acetylation, ubiquitination, phosphorylation, and sumoylation, and their position in the N-terminal “tails”.
Psoriasis, a prevalent chronic inflammatory skin disease, exhibits a significant genetic predisposition and autoimmune pathogenic characteristics (55, 56). The presentation of psoriasis in patients varies in terms of morphology, distribution, severity, and course, although scaling papules and plaques are frequently observed. This condition is categorized as a “chronic, non-communicable, painful, disfiguring, and disabling disease for which there is no cure,” affecting a substantial population of over 60 million adults and children (57, 58). Psoriasis is characterized as an inflammatory skin disease mediated by Th17 cells. In the study conducted by Ovejero-Benito et al. (59), individuals with psoriasis had elevated levels of H3K4 compared to healthy individuals. Additionally, significant alterations in H3K27 were observed between individuals who responded positively to biological drugs and those who did not, specifically at the 3-month follow-up. H3K27 has been identified as a regulator of Th17 cell differentiation, while H3K4 has been associated with Th17 plasticity (60, 61). Additionally, a decrease in H3K9me2 in keratinocytes may contribute to the upregulation of IL-23, thereby playing a role in developing chronic inflammatory diseases dependent on the IL-23/IL-17 axis (62). A study of Rasheed et al. showed that the expression of Class III HDACs, specifically SIRT1, was reduced while SIRT6 was increased in psoriatic skin (63). Another study also observed an increase in HDAC1 and a decrease in SIRT1 levels in skin tissues (64). The transcription cofactor p300 is a histone acetyltransferase with histone acetylase activity. Liao et al. (65) reported that the transcription factor Wilms Tumor 1 regulates the expression of IL-1β by facilitating the binding of P300 to the IL-1β promoter, thereby contributing to the development of psoriasis. These publications demonstrated that histone acetylation and methylation play an important role in the occurrence and development of psoriasis. Table 2 shows studies published on histone modification variation in psoriasis.
AD is characterized by the presence of recurring eczematous lesions and severe itchiness and discomfort, resulting in sleep deprivation, reduced self-esteem, and impaired academic and occupational performance. Globally, AD affects approximately 7%−10% of adults and up to 25% of children with a significant genetic influence (80, 81). This allergic disease is primarily mediated by Th2 cells. Kwon’s study suggested that the interaction between mast cells, keratinocytes, and dermal fibroblast cells can potentially contribute to AD development. The presence of CXCL13 in the exosomes of mast cells in a pathological model was observed to enhance the expression of HDAC6 in both skin mast cells and dermal fibroblast cells.
Additionally, HDAC6 was found to negatively regulate MiR-9, thereby influencing the expression of SIRT1. The downregulation or inhibition of HDAC6 and SIRT1 demonstrated a suppressive effect on AD, as discussed in this article (82). Mesenchymal stem cells can interact with both the innate and adaptive immune systems and inhibit the activation of immune cells. These cells have been observed to be protective against AD, and this function has been linked to an increase in the expression levels of HDAC3 (83). Furthermore, the expression of miR-335, a potent inducer of keratinocyte differentiation, is abnormally diminished in AD-lesioned skin. This reduction is epigenetically regulated by histone deacetylases (84). The influence of histone modification to AD is mainly concentrated on acetylation. Publications on the histone methylation to AD are seldom to see. Table 2 indicates studies published on histone modification variation in AD.
Scleroderma, or SSc, is an infrequent autoimmune disease affecting multiple systems, characterized by dysregulated innate and adaptive immunity resulting in extensive systemic fibrosis (85). Skin thickening has been extensively studied as a symptom of SSc because it facilitates diagnosis, and there has been a proven correlation between increased skin involvement and more severe internal organ damage, an unfavorable prognosis, and heightened disability (86). EZH2 is involved in T cell differentiation, EC-mediated angiogenesis, myofibroblast transformation, and tissue fibrosis in SSc (87–89). It has been reported that both EZH2 and H3K27me3 were elevated in SSc dermal fibroblasts and endothelial cells compared with healthy controls (90). JMJD3, a type of histone demethylase, is upregulated in SSc fibroblasts. This upregulation decreases H3K27me3 levels at the FRA2 promoter, which stimulates FRA2 expression and ultimately promotes SSc fibroblast activation (91). Ciechomska et al. reported that epigenetic alterations in monocytes can influence the pathogenesis of SSc by exerting either promotive or repressive effects on myofibrogenic differentiation (92). Histone acetylation also plays a significant role in SSc. In a study by Shin, ChIP-qPCR results showed an increase in H3K27 acetylation in SSc fibroblasts (93). The variation of histone modification is significant in fibroblasts and monocytes, which may be a valuable approach to understanding the development and changes of SSc. Table 2 displays the studies published on histone modification variation in SSc.
Systemic lupus erythematosus (SLE) is a chronic systemic autoimmune disease that may affect multiple tissues and organ systems, including cutaneous, renal, cardiopulmonary, musculoskeletal, neural, and hematologic systems (94). It is observed that nearly all patients with SLE experience cutaneous manifestations at some point during the progression of their disease (95). Currently, the treatment of SLE involves using glucocorticoids and immunosuppressive agents; however, the long-term prognosis for SLE remains unfavorable (96). Additionally, the regulation of SLE is significantly influenced by H3K27me3. Compared with healthy controls, lupus patients showed elevated levels of H3K27me3 in CD4+ T cells, a process facilitated by EZH2, a histone methyltransferase (97). Luo et al. observed a substantial increase in H3K27me3 at the HPK1 promoter in Tfh cells of SLE patients (98). In SLE mice, glomerular cells showed significantly upregulated HDAC6 and HDAC9 expression compared with healthy mice. Targeted inhibition of HDAC6 effectively reversed SLE-associated abnormalities by modulating the proportions of cells in the late pro- and early pre-B cell fractions and altering T cell differentiation, as indicated by increased Treg cells and decreased thymic DN T cells (99). Zhou et al. (100) showed a significant augmentation in H3 acetylation and H3K4me2 within the CD4+ T cells of individuals diagnosed with lupus. The anomalous alterations in histone modifications of CD4+ T cells potentially play a role in the pathogenesis of lupus by upregulating CD70 expression. More evidence suggested that histone modification take part in the abnormal differentiation of T cell in SLE individuals, which lead to the changes in pathological characteristics. The results of this study provided evidence that histone acetylation and methylation both play a role in regulating the immune microenvironment in SLE. Table 2 lists the studies published on histone modification variation in SLE.
Contact dermatitis, a prevalent inflammatory skin disease, is caused by direct exposure to environmental chemical substances that have the potential to irritate or cause allergenic reactions (101). These chemical substances facilitate the differentiation of T lymphocytes into Th2 cells, resulting in elevated levels of inflammatory cytokines and blood IgE (102). Additionally, the upregulation of IL-4 hinders the functionality of Th1 cells, resulting in reduced cell-mediated immunity and exacerbating the inflammatory response (103). Occupational contact dermatitis, a prominent subset of contact dermatitis, primarily affects individuals regularly exposed to water or irritating substances in their occupational settings (104). Hairdressers, construction workers, and healthcare professionals are particularly vulnerable to developing occupational contact dermatitis. Sonday et al. conducted a study involving 697 health workers in two tertiary hospitals in Southern Africa and reported that 12.3% of them exhibited a likelihood of contact dermatitis within the past 12 months (105). Additionally, contact with cosmetics and medical supplies was identified as a common source of contact dermatitis (106–108). A knockout experiment conducted in mice showed that HMGB1 exhibits anti-inflammatory properties in keratinocytes of contact dermatitis mice. The reduction in IL-24 expression is accomplished by inhibiting H3K4me3 binding to the promoter region of IL-24 (109). Additionally, another study indicated that the absence of Utx, a histone demethylase, could contribute to various T cell abnormalities and worsen the symptoms of contact dermatitis (110). Gaining insight into alterations in histone modification levels associated with contact dermatitis holds the potential to facilitate the development of novel preventive and therapeutic approaches aimed at enhancing the occupational and residential conditions of certain populations.
LP is a chronic mucocutaneous disease characterized by the presence of reticular, purple papules and plaques on the skin, as well as white papules and erosions on mucous membranes, and progresses through a chronic relapsing course (111–113). LP primarily affects the skin and oral mucosa, affecting approximately 1%-2% of the general population (111). LP is more prevalent in middle-aged or older adults and women (114). LP is characterized by dysfunctional T cells that initiate epithelial cell apoptosis. Immune infiltration, primarily consisting of T cells, is a common occurrence in LP. Most cytotoxic clones observed in lichen planus lesions are CD8+ T cells, while most non-cytotoxic clones are CD4+ (115). In addition to contributing to the development of autoimmunity, these cytotoxic CD8+ T cells are essential for promoting apoptosis of oral mucosal basal cells (116). The World Health Organization (WHO) recognizes oral LP as a potentially malignant disorder (115). Some studies have found that histone acetylation take part in the process of LP. A study conducted on 66 patients with oral LP and 23 patients with cutaneous LP showed that H3K9 histone acetylation is more prevalent in both lesions, with no significant distinction between them (117). Another study reported that H3K9 histone acetylation acts as an epigenetic marker for the recurrence of oral LP (118). Jun et al. observed a decrease in histone H3 acetylation and an increase in HDAC activity in CD4+ T cells of oral LP patients, which may impact the synthesis of inflammatory cytokines. The level of histone H3 acetylation showed a negative association with IL-4 and MCP-1 production, while the expression of HDAC6 mRNA exhibited a positive correlation with MCP-1 production (119). Nevertheless, the relationship between LP and other kinds of histone modifications are required to be researched.
Alopecia areata (AA) is a prevalent autoimmune disorder that manifests as temporary and non-scarring hair loss. The affected regions can vary from small areas of baldness to complete hair loss on the scalp and face and, in some cases, extend to body hair (120). The global prevalence of AA is estimated at approximately 2%, with 0.27% prevalence in China (121). AA commonly affects a patient’s physical appearance and significantly impacts their health-related quality of life (121–123). Research indicates that individuals with AA may have a more than twofold increased risk of experiencing depression or anxiety disorders compared to the general population (124). AA is a disease that primarily affects the hair follicles; however, systemic immune activation and dysregulation of serum cytokine levels may be associated with this condition (125). AA patients typically exhibit perifollicular and intrafollicular infiltration, with CD4+ Th1 cells predominantly located near the hair follicle and CD8+ Tc1 cells within the follicular epithelium (126). Zhao et al. compared the epigenetic profiles of peripheral blood mononuclear cell (PBMC) samples from 25 AA patients and 20 healthy controls. AA patients exhibit a significant elevation in histone H3 acetylation levels and a significant reduction in histone H3 lysine-4 methylation levels compared with healthy controls. Additionally, there is a positive correlation between AA disease severity and the acetylation levels of histone H3. This study also observed an upregulation in the expression of p300, HDAC1, MLL, SETD7, EHMT2, KDM4C, and KDM5A, as well as a downregulation in the expression of HDAC2, HDAC7, KDM1A, KDM4A, and KDM4B. These results showed that epigenetic modification of PBMC may be involved in the pathogenesis of alopecia areata (127). In a study comprising a cohort of 25 individuals with patchy AA, 26 patients with acne vulgaris, and 25 healthy controls, the enzyme-linked immunosorbent assay (ELISA) data showed a decreased level of histone deacetylase 1 (HDAC1) in both AA and acne vulgaris patients (128). Additionally, the inhibition of SIRT1 enhanced the synthesis of Th1 cytokines (IFN-γ and TNF-α) IFN-inducible chemokines (CXCL9 and CXCL10). It facilitated T cell migration in hair follicles’ outer root sheath cells. Conversely, the activation of SIRT1 suppressed the autoreactive inflammatory reactions. The antagonistic impact of SIRT1 on the immune response was mediated via the deacetylation of NF-κB and the phosphorylation of STAT3. Thus, SIRT1 downregulation may contribute to AA development (129). The variation of histone acetylation is observed in AA patients, which provides new ideas for the treatment of AA.
The occurrence and progression of inflammatory skin diseases are frequently accompanied by gut microbiota alterations, which serve multiple functions (130–133). The gut microbiota produces a wide range of short-chain fatty acids (SCFAs) through the fermentation of non-digestible carbohydrates, including dietary fiber (134). SCFAs are fatty acids with carbon backbones ranging from one to six, and the predominant SCFAs found in the intestines, with concentrations exceeding 100 mM, are propionate, acetate, and butyrate (134, 135). SCFAs are produced by two predominant bacterial groups, with acetate and propionate primarily generated by the Bacteroidetes phylum, while butyrate is mainly produced by the Firmicutes phylum (136). An animal model study showed that mice with intestinal microbiota exhibited higher histone acetylation levels than germ-free mice. Conversely, when SCFAs were administered through drinking water, histone acetylation in germ-free mice increased significantly to a higher level compared with the microbiota group. This study proves that gut microbiota regulates histone modification through SCFA metabolism (137). Another study showed that SCFAs produced by Propionibacterium acnes can inhibit the activity of HDAC, thereby contributing to the development of inflammatory responses (27). Short-chain fatty acids (SCFAs) are produced through the metabolic processes of gut bacteria and exert an impact on the development of inflammatory skin conditions by inhibiting histone deacetylase (HDAC). This observation underscores the intricate nature of the etiology of inflammatory skin diseases and presents a novel therapeutic avenue for their management. In contrast, imiquimod reduces the suppressive activity of Treg, resulting in the upregulation of IL-17 and IL-6 and the downregulation of IL-10 and FOXP3. Consequently, this effect helps develop a psoriasis-like skin inflammation model. One specific SCFA, butyrate, can reverse these processes by inhibiting HDACs (138). SCFAs represent a promising therapeutic target for inflammatory skin diseases due to their involvement in the regulation of histone modification. Krautkramer et al. (137)reported that administering SCFAs through water to germ-free mice can replicate gut colonization and its impact on histone modification. The combined treatment of sodium butyrate, one of the SCFAs, and the EGFR inhibitor PD153035 has enhanced keratinocyte differentiation (139). Another study showed that administering either injected or topically applied sodium butyrate onto the ears of mice sensitized to 2,4,6-trinitro-1-chlorobenzene significantly reduced the contact hypersensitivity reaction (140). As histone deacetylase inhibitors, butyrate and propionate upregulate miRNAs that suppress AID expression, thereby modulating autoantibody responses in lupus-prone MRL/Faslpr/lpr mice (141). Luo et al. reported that butyrate can suppress IL-33 expression in keratinocytes infected with S. aureus by inhibiting HDAC3 (142). These findings suggest that the modulation of SCFAs on histone modification can potentially treat inflammatory skin diseases; however, additional research is required on autoimmune disorder therapies.
HDAC inhibitors modulate gene expression by inhibiting the removal of acetyl groups from the N-terminal tails of histones through binding to HDAC (13, 143). Extensive research has shown the potential of HDAC inhibitors in antineoplastic therapy (144–146). Additionally, the antiproliferative and anti-inflammatory properties of HDAC inhibitors have prompted an active investigation into their efficacy for treating inflammatory skin diseases (13). Trichostatin A (TSA) is one of this context’s most extensively studied HDAC inhibitors. A study by Kim showed that TSA treatment resulted in a decreased IL-4 production and an increased T reg cells population, effectively suppressing AD-like skin lesions in NC/Nga mice treated with DNFB under SPF conditions (147). Additionally, TSA can reverse the aberrant expression of multiple genes associated with the immunopathogenesis of SLE patients (148). Mohammadi’s (149) study showed that sodium valproate inhibits HDAC in SLE patients, resulting in immunomodulatory effects on macrophages. Additionally, a study on 10 AD patients and 6 healthy individuals showed that the HDAC inhibitor belinostat restored epidermal miR-335 expression and rescued the impaired skin barrier in AD (84). Souliotis et al. reported that administering vorinostat resulted in hyperacetylation of histone H4, chromatin decondensation, improved DNA repair efficiency, and decreased apoptosis among patients with SLE. Consequently, genes involved in DNA damage repair and signaling pathways were significantly under-expressed, while genes associated with apoptosis were significantly over-expressed (150). Additionally, panobinostat treatment in SLE mice significantly reduced autoreactive plasma-cell numbers, autoantibodies, and nephritis, while other immune parameters remained largely unaffected. The findings indicate that panobinostat has the potential to be a therapeutic option for autoimmune conditions driven by B-cells without causing significant long-term negative effects on B-cell memory (151). Clinical applications for systemic lupus erythematosus were demonstrated by suberoylanilide hydroxamic acid in two distinct mouse model experiments (152, 153). Different types of HDAC inhibitors have been shown to have an impact on inflammatory skin diseases, thereby offering novel support for the management of such conditions.
Bromodomain proteins, a group of evolutionarily conserved motifs involved in protein-protein interactions, have been found to recognize acetylated lysine residues on histones and play a significant role in chromatin remodeling (154). Bromodomain proteins are characterized by the presence of four left-handed bundle α-helices, which are arranged in a manner that creates two distinct interhelical αZ-αA and αB-αC loop regions. These loop regions contribute to the formation of a hydrophobic pocket. The primary function of this hydrophobic pocket is to facilitate the recognition of acetylation modifications on lysine residues (155). The bromodomain and extra-terminal domain (BET) family of proteins is most widely studied among bromodomains in cellular biology. BET proteins consist of two tightly packed BRDs, BD1 and BD2, and an extra-terminal domain (ET) located at the carboxy terminus. The ET region is also highly conserved, consisting of 80 amino acids (156). This domain’s primary role is to interact with various cellular proteins, including histone-lysine N-methyltransferase and Jumonji domain-containing 6 (157, 158). As is shown in Figure 3, known as “chromatin readers,” BET proteins play a crucial role in numerous biological processes, including chromosomal architecture, DNA replication, DNA damage repair, and transcriptional regulation (156). JQ1, one of the BET inhibitors, has exhibited significant efficacy in suppressing multiple inflammatory and autoimmune diseases. JQ1 mitigates lupus in MRL-lpr mice by suppressing BAFF, pro-inflammatory cytokines, and autoimmunity. This finding suggests the therapeutic potential of JQ1 in treating lupus disease (159). Another study involving mice and cell culture showed that BRD4 inhibitors exhibited superior antifibrotic effects compared to other BET inhibitions (160). Additionally, it has been reported that OTX015 and ABBV075 can reduce the severity of imiquimod-induced psoriasis in mice. ABBV075 achieves the same pharmacodynamics at a dosage one-tenth of that required for OTX015 (161). Sato et al. (162) designed and synthesized various pyrido-benzodiazepinone derivatives, among which one exhibited the most effective therapeutic effect in treating imiquimod-induced psoriasis mice. Currently, the primary emphasis of clinical investigation concerning BET inhibitors lies in their application for anti-tumor therapy. Nevertheless, considering their involvement in transcription subsequent to histone modification, it is anticipated that these pharmaceutical agents will find utility in the management of inflammatory skin disorders.
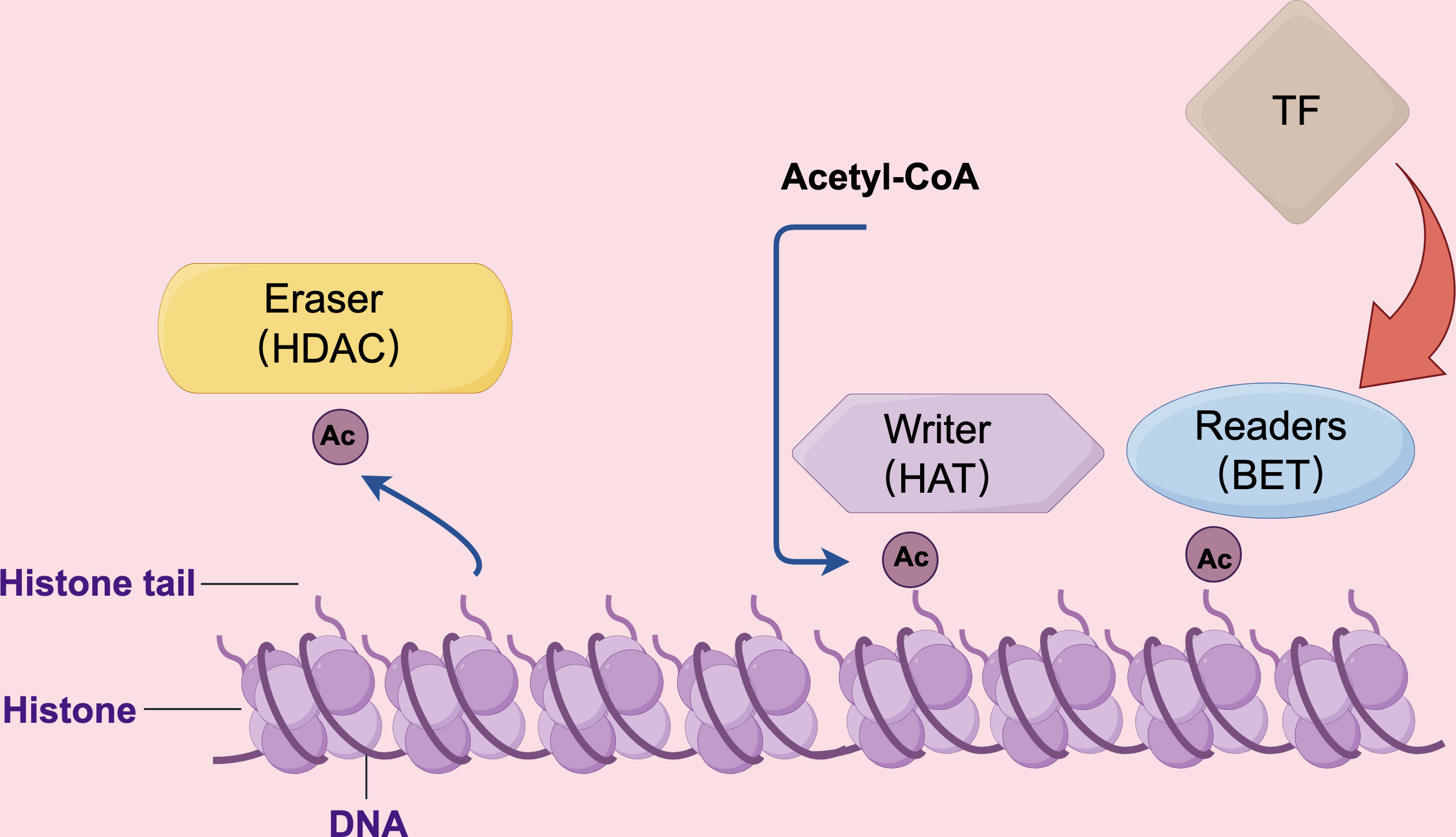
Figure 3 The HAT, HDAC, and BET proteins perform distinct roles in the regulation of gene transcription. HAT proteins function as “writers” by modifying lysine residues with acetyl groups using acetyl CoA. Conversely, BET proteins act as “readers” by binding to acetylated lysine residues and facilitating the recruitment of transcription factors. HDAC proteins, on the other hand, function as “erasers” by deacetylating lysine residues, thereby reducing the frequency of transcription events.
The epithelial immune microenvironment of the skin is a dynamic network composed of keratinocytes, immune cell subpopulations, cytokines, and metabolites. The disorder of the epithelial immune microenvironment may contribute to inflammation reactions. Inflammation has long been recognized as a defensive mechanism triggered by infections or injuries characterized by rubor, calor, swelling, and pain. It is an immune cell-mediated biological response that occurs when the body is stimulated by microorganisms, pathogens, damaged cells, and various internal and external environmental factors (163). Inflammation serves as a crucial defense mechanism in preserving the well-being of the host. It stimulates the activation of the immune system to eliminate the pathogen, initiates the healing process, reduces the severity of tissue damage and infection caused by the stimulus, and ultimately counteracts the threat while restoring homeostasis (164). Conversely, inflammation frequently coincides with heightened acidity in the affected area. Prolonged persistence of unresolved inflammation may result in the development of chronic inflammation and tissue deterioration, which can further increase the prevalence of inflammatory disorders and skin diseases.
Inflammation is an intricate and sequential phenomenon requiring synchronized involvement of various cellular and tissue components. The intercellular communication within inflammatory systems is primarily orchestrated by cytokines, encompassing protein, peptide, or glycoprotein messenger molecules (165). While inflammation shares similarities with the nervous system regarding sensory and effector pathways, inflammatory cells can migrate to any location within any tissue (166). Consequently, there may be inconsistencies in the etiology and distribution of inflammatory diseases, occasionally resulting in systemic abnormalities. The inflammatory cascade comprises cytokines, chemokines, growth factors, and other peptides synthesized by skin keratinocytes and immune cells. The expression of these components is controlled by epigenetic modifications (13). The influence of histone modification on inflammatory reaction often focuses on two sides: immunocyte and epithelial cells. Section two demonstrates that the modulation of histone modification can impact the polarization of macrophages. Additionally, histone modification exerts an influence on the differentiation and function of T cells. These mechanisms have the potential to either facilitate or impede inflammatory responses, thereby influencing the development of inflammatory skin conditions. It is worth noting that both histone methylation and histone acetylation play a role in this regard. For instance, the administration of VPA has been shown to inhibit the activity of HDAC, modulate macrophage polarization, increase the expression of anti-inflammatory cytokines, and enhance the immune response against inflammation in patients with SLE (149). In a separate study, the inhibition of SIRT1 was found to stimulate the secretion of Th1 cytokines, IFN-inducible chemokines, and T cell migration in ORS cells, thereby contributing to the development of AA (129). The significance of epithelial cells in the regulation of immunity has gained increasing recognition (167). Keratinocytes, being the principal epithelial cell in the skin, exert influence on immune responses through two mechanisms: direct expression of cytokines and antimicrobial peptides, as well as indirect functions as a barrier separating the environment from classical immunocytes (12). The reduction of H3K9me2 in keratinocytes within psoriatic lesions demonstrates a positive association with IL-23 expression, potentially contributing to the development of psoriasis (65). The inhibition of HDAC3 by butyrate leads to a decrease in IL-33 expression and subsequently alleviates skin inflammation in a mouse model resembling atopic dermatitis (142). The aforementioned findings suggest that the modulation of keratinocytes through epigenetic mechanisms has the potential to impact the development and progression of inflammatory skin diseases.
However, the complexity of this regulatory mechanism arises from the fact that similar epigenetic modifications can elicit diverse responses in various cell types (168, 169). Histone acetylation induces a state of open chromatin that facilitates the binding of transcription proteins, potentially leading to elevated expression of pro-inflammatory cytokines or suppression of inflammatory mediators in various cell types. Typically, HDAC inhibitors are considered crucial in anti-inflammatory approaches; however, Sanford et al. reported contradictory findings (170). SCFAs, acting as HDAC inhibitors, possess anti-inflammatory properties in myeloid-derived cells; they may disrupt the epidermis’ tolerance to inflammatory stimuli. Applying SCFAs on the skin surface and their subcutaneous administration may yield contrasting outcomes. Another study on cell cultures revealed that SCFAs can exhibit pro-inflammatory effects under specific circumstances (27). The elimination of Sirt2 was observed to exacerbate psoriasiform skin inflammation, whereas the reintroduction of Sirt2 through genetic means reduced disease severity (171).
The consideration of drug side effects, particularly those related to histone modification, is crucial in the treatment of inflammatory skin diseases. It has been reported that hematologic toxicity represents a significant concern in certain HDAC inhibitor drugs (172). Prolonged treatments with HDAC inhibitors have been associated with various adverse events, including leucopenia, neutropenia, thrombocytopenia, anemia, peripheral sensory neuropathy, fatigue, vomiting, and myalgia (173, 174). Therefore, a comprehensive evaluation of the side effects associated with HDAC inhibitor therapy is required (175–177). Developing new HDAC inhibitors aims to achieve good efficacy while minimizing side effects. HDAC inhibitors can be categorized into four groups: hydroxamates, benzamide derivatives, cyclic peptides, and aliphatic acids. Benzamide derivatives are believed to possess lower toxicity than hydroxamates due to their isotype-selective nature as opposed to being pan-HDAC inhibitors (178). Bollmann et al. have successfully developed a novel and highly selective HDAC inhibitor called YAK540. The combined administration of YAK540 and the proteasome inhibitor bortezomib shows potential as a promising strategy against leukemias, as it enhances the anticancer efficacy while minimizing the overall toxicity associated with HDAC inhibitors (179). A novel drug delivery system utilizing oxygen-containing nanosomes has been developed to efficiently transport HDAC inhibitors to dormant HIV-infected cells. Incorporating oxygen nanosomes in this study alleviates drug toxicity and regulates the rate of drug release (180). The administration of BET inhibitors also presents side effects. The most commonly observed treatment-related adverse events include thrombocytopenia, anemia, fatigue, and gastrointestinal complications (181). These side effects arise due to the ability of BET inhibitors to target any proteins containing bromodomains (182). Therefore, a BET inhibitor with enhanced selectivity and reduced side effects has been developed compared to pan BET inhibitors. GSK778 and dBET57 exhibit BD1-selectivity, while GSK046, ABBV-744, and SJ432 exhibit BD2-selectivity. Conversely, AZD5153 is a bivalent BET inhibitor specifically targeting both BD1 and BD2 of Brd4 (183–187). Although most of these studies do not pertain to inflammatory skin disease, it is anticipated that novel inflammatory skin disease epigenetic therapies with low toxicity and high efficacy will be developed soon.
Our assay aims at summarize the relationship between inflammatory skin disease and one type of epigenetics: histone modification. For it is a huge family of epigenetic modification and various function can be achieve by this kind of phenomena. Therapeutic method and potential therapeutic method related to histone modification was introduced by this assay. Many recently published literature has also been reviewed in this assay. In conclusion, histone modification is a crucial biological process in various inflammatory skin diseases. The studies on variations in histone modification during the pathogenesis of these diseases continue accumulating, and our understanding of their onset and progression is progressively expanding. Developing novel therapeutic approaches targeting histone modification, such as next-generation drugs and combination therapy methods, has become a pivotal area of research interest.
LZ: Conceptualization, Writing – original draft. RC: Visualization, Writing – original draft. ZT: Conceptualization, Writing – original draft. FM: Visualization, Writing – review & editing. XS: Writing – review & editing. QZ: Funding acquisition, Writing – review & editing. ZC: Funding acquisition, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work is supported by Science and Technology Commission of Shanghai Municipality (20DZ2255200, 21S21900900, and 22S21902700).
The Figures 1, 3 was drawn by Figdraw.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Dainichi T, Matsumoto R, Mostafa A, Kabashima K. Immune control by TRAF6-mediated pathways of epithelial cells in the EIME (Epithelial immune microenvironment). Front Immunol (2019) 10:1107. doi: 10.3389/fimmu.2019.01107
2. Dainichi T, Kitoh A, Otsuka A, Nakajima S, Nomura T, Kaplan DH, et al. The epithelial immune microenvironment (EIME) in atopic dermatitis and psoriasis. Nat Immunol (2018) 19(12):1286–98. doi: 10.1038/s41590-018-0256-2
3. Chen XY, Wang ZY, Zhou Y, Ye LR, Man XY. Keratinoctye-neuro-immune-units (KNICUs): collaborative impact on the initiation and maintenance of psoriasis. Front Med (Lausanne) (2023) 10:1191057. doi: 10.3389/fmed.2023.1191057
4. Wu H, Yin H, Chen H, Sun M, Liu X, Yu Y, et al. A deep learning, image based approach for automated diagnosis for inflammatory skin diseases. Ann Transl Med (2020) 8(9):581. doi: 10.21037/atm.2020.04.39
5. van den Oord RA, Sheikh A. Filaggrin gene defects and risk of developing allergic sensitisation and allergic disorders: systematic review and meta-analysis. BMJ (2009) 339:b2433. doi: 10.1136/bmj.b2433
6. Yuan S, Wang L, Zhang H, Xu F, Zhou X, Yu L, et al. Mendelian randomization and clinical trial evidence supports TYK2 inhibition as a therapeutic target for autoimmune diseases. EBioMedicine (2023) 89:104488. doi: 10.1016/j.ebiom.2023.104488
7. Hedrich CM, Mäbert K, Rauen T, Tsokos GC. DNA methylation in systemic lupus erythematosus. Epigenomics (2017) 9(4):505–25. doi: 10.2217/epi-2016-0096
8. Xing J, Wang Y, Zhao X, Li J, Hou R, Niu X, et al. Variants in PRKCE and KLC1, potential regulators of type I psoriasis. Clin Cosmet Investig Dermatol (2022) 15:1237–45. doi: 10.2147/ccid.S371719
9. Nedoszytko B, Reszka E, Gutowska-Owsiak D, Trzeciak M, Lange M, Jarczak J, et al. Genetic and epigenetic aspects of atopic dermatitis. Int J Mol Sci (2020) 21(18):1–18. doi: 10.3390/ijms21186484
10. Moltrasio C, Romagnuolo M, Marzano AV. Epigenetic mechanisms of epidermal differentiation. Int J Mol Sci (2022) 23(9):1–19. doi: 10.3390/ijms23094874
11. Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat Rev Genet (2018) 19(2):81–92. doi: 10.1038/nrg.2017.80
12. Sawada Y, Gallo RL. Role of epigenetics in the regulation of immune functions of the skin. J Invest Dermatol (2021) 141(5):1157–66. doi: 10.1016/j.jid.2020.10.012
13. Yi JZ, McGee JS. Epigenetic-modifying therapies: An emerging avenue for the treatment of inflammatory skin diseases. Exp Dermatol (2021) 30(8):1167–76. doi: 10.1111/exd.14334
14. Gardner KE, Allis CD, Strahl BD. Operating on chromatin, a colorful language where context matters. J Mol Biol (2011) 409(1):36–46. doi: 10.1016/j.jmb.2011.01.040
15. Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell (1999) 98(3):285–94. doi: 10.1016/s0092-8674(00)81958-3
16. Audia JE, Campbell RM. Histone modifications and cancer. Cold Spring Harb Perspect Biol (2016) 8(4):a019521. doi: 10.1101/cshperspect.a019521
17. Ghosh K, O'Neil K, Capell BC. Histone modifiers: Dynamic regulators of the cutaneous transcriptome. J Dermatol Sci (2018) 89(3):226–32. doi: 10.1016/j.jdermsci.2017.12.006
18. Ma F, Zhang CY. Histone modifying enzymes: novel disease biomarkers and assay development. Expert Rev Mol Diagn (2016) 16(3):297–306. doi: 10.1586/14737159.2016.1135057
19. Pestinger V, Wijeratne SS, Rodriguez-Melendez R, Zempleni J. Novel histone biotinylation marks are enriched in repeat regions and participate in repression of transcriptionally competent genes. J Nutr Biochem (2011) 22(4):328–33. doi: 10.1016/j.jnutbio.2010.02.011
20. Wolffe AP, Hayes JJ. Chromatin disruption and modification. Nucleic Acids Res (1999) 27(3):711–20. doi: 10.1093/nar/27.3.711
21. Mendenhall EM, Bernstein BE. Chromatin state maps: new technologies, new insights. Curr Opin Genet Dev (2008) 18(2):109–15. doi: 10.1016/j.gde.2008.01.010
22. Borrelli E, Nestler EJ, Allis CD, Sassone-Corsi P. Decoding the epigenetic language of neuronal plasticity. Neuron (2008) 60(6):961–74. doi: 10.1016/j.neuron.2008.10.012
23. Araki Y, Mimura T. The histone modification code in the pathogenesis of autoimmune diseases. Mediators Inflamm (2017) 2017:2608605. doi: 10.1155/2017/2608605
24. Bach SV, Hegde AN. The proteasome and epigenetics: zooming in on histone modifications. Biomol Concepts (2016) 7(4):215–27. doi: 10.1515/bmc-2016-0016
25. Engin B, Huseynova C, Ak T, Ayla AY, Can G, Uğurlu S. Screening of antigliadin and antitissue transglutaminase antibodies in patients with chronic plaque psoriasis: a case-control study. Turk J Med Sci (2023) 53(2):544–51. doi: 10.55730/1300-0144.5615
26. Zhou X, Qiu Y, Mu K, Li Y. Decreased SIRT1 protein may promote HMGB1 translocation in the keratinocytes of patients with cutaneous lupus erythematosus. Indian J Dermatol Venereol Leprol (2023) 1-8. doi: 10.25259/ijdvl_814_2022
27. Sanford JA, O'Neill AM, Zouboulis CC, Gallo RL. Short-chain fatty acids from cutibacterium acnes activate both a canonical and epigenetic inflammatory response in human sebocytes. J Immunol (2019) 202(6):1767–76. doi: 10.4049/jimmunol.1800893
28. Zhao M, Huang W, Zhang Q, Gao F, Wang L, Zhang G, et al. Aberrant epigenetic modifications in peripheral blood mononuclear cells from patients with pemphigus vulgaris. Br J Dermatol (2012) 167(3):523–31. doi: 10.1111/j.1365-2133.2012.11007.x
29. Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem (2007) 76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114
30. Yang XJ, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene (2007) 26(37):5310–8. doi: 10.1038/sj.onc.1210599
31. Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev (1998) 12(5):599–606. doi: 10.1101/gad.12.5.599
32. Kimura A, Matsubara K, Horikoshi M. A decade of histone acetylation: marking eukaryotic chromosomes with specific codes. J Biochem (2005) 138(6):647–62. doi: 10.1093/jb/mvi184
33. Duman M, Martinez-Moreno M, Jacob C, Tapinos N. Functions of histone modifications and histone modifiers in Schwann cells. Glia (2020) 68(8):1584–95. doi: 10.1002/glia.23795
34. Zhang Y, Gao Y, Ding Y, Jiang Y, Chen H, Zhan Z, et al. Targeting KAT2A inhibits inflammatory macrophage activation and rheumatoid arthritis through epigenetic and metabolic reprogramming. MedComm (2020) 2023) 4(3):e306. doi: 10.1002/mco2.306
35. Compton SE, Kitchen-Goosen SM, DeCamp LM, Lau KH, Mabvakure B, Vos M, et al. LKB1 controls inflammatory potential through CRTC2-dependent histone acetylation. Mol Cell (2023) 83(11):1872–86.e5. doi: 10.1016/j.molcel.2023.04.017
36. Zhang Y, Reinberg D. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev (2001) 15(18):2343–60. doi: 10.1101/gad.927301
37. Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res (2011) 21(3):381–95. doi: 10.1038/cr.2011.22
38. Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, et al. High-resolution profiling of histone methylations in the human genome. Cell (2007) 129(4):823–37. doi: 10.1016/j.cell.2007.05.009
39. Eames HL, Saliba DG, Krausgruber T, Lanfrancotti A, Ryzhakov G, Udalova IA. KAP1/TRIM28: an inhibitor of IRF5 function in inflammatory macrophages. Immunobiology (2012) 217(12):1315–24. doi: 10.1016/j.imbio.2012.07.026
40. Xu G, Liu G, Xiong S, Liu H, Chen X, Zheng B. The histone methyltransferase Smyd2 is a negative regulator of macrophage activation by suppressing interleukin 6 (IL-6) and tumor necrosis factor α (TNF-α) production. J Biol Chem (2015) 290(9):5414–23. doi: 10.1074/jbc.M114.610345
41. Kruidenier L, Chung CW, Cheng Z, Liddle J, Che K, Joberty G, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature (2012) 488(7411):404–8. doi: 10.1038/nature11262
42. Ghannam S, Pène J, Moquet-Torcy G, Jorgensen C, Yssel H. Mesenchymal stem cells inhibit human Th17 cell differentiation and function and induce a T regulatory cell phenotype. J Immunol (2010) 185(1):302–12. doi: 10.4049/jimmunol.0902007
43. Rossetto D, Avvakumov N, Côté J. Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics (2012) 7(10):1098–108. doi: 10.4161/epi.21975
44. Fan L, Xu L, Tian S, Zheng X. Identification of a novel histone phosphorylation prognostic signature in hepatocellular carcinoma based on bulk and single-cell RNA sequencing. Front Endocrinol (Lausanne) (2022) 13:965445. doi: 10.3389/fendo.2022.965445
45. Sawicka A, Seiser C. Sensing core histone phosphorylation - a matter of perfect timing. Biochim Biophys Acta (2014) 1839(8):711–8. doi: 10.1016/j.bbagrm.2014.04.013
46. Sekiguchi M, Matsushita N. DNA damage response regulation by histone ubiquitination. Int J Mol Sci (2022) 23(15):1–14. doi: 10.3390/ijms23158187
47. da Silva Z, Glanzner WG, Currin L, de Macedo MP, Gutierrez K, Guay V, et al. DNA damage induction alters the expression of ubiquitin and SUMO regulators in preimplantation stage pig embryos. Int J Mol Sci (2022) 23(17):1–16. doi: 10.3390/ijms23179610
48. Tang M, Li S, Chen J. Ubiquitylation in DNA double-strand break repair. DNA Repair (Amst) (2021) 103:103129. doi: 10.1016/j.dnarep.2021.103129
49. Ryu HY, Hochstrasser M. Histone sumoylation and chromatin dynamics. Nucleic Acids Res (2021) 49(11):6043–52. doi: 10.1093/nar/gkab280
50. Hasim S, Tati S, Madayiputhiya N, Nandakumar R, Nickerson KW. Histone biotinylation in Candida albicans. FEMS Yeast Res (2013) 13(6):529–39. doi: 10.1111/1567-1364.12056
51. Karch KR, Langelier MF, Pascal JM, Garcia BA. The nucleosomal surface is the main target of histone ADP-ribosylation in response to DNA damage. Mol Biosyst (2017) 13(12):2660–71. doi: 10.1039/c7mb00498b
52. Josefowicz SZ, Shimada M, Armache A, Li CH, Miller RM, Lin S, et al. Chromatin kinases act on transcription factors and histone tails in regulation of inducible transcription. Mol Cell (2016) 64(2):347–61. doi: 10.1016/j.molcel.2016.09.026
53. Zhao Z, Su Z, Liang P, Liu D, Yang S, Wu Y, et al. USP38 couples histone ubiquitination and methylation via KDM5B to resolve inflammation. Adv Sci (Weinh) (2020) 7(22):2002680. doi: 10.1002/advs.202002680
54. Martínez-Zamudio RI, Ha HC. PARP1 enhances inflammatory cytokine expression by alteration of promoter chromatin structure in microglia. Brain Behav (2014) 4(4):552–65. doi: 10.1002/brb3.239
55. Rendon A, Schäkel K. Psoriasis pathogenesis and treatment. Int J Mol Sci (2019) 20(6):1–28. doi: 10.3390/ijms20061475
56. Kamiya K, Kishimoto M, Sugai J, Komine M, Ohtsuki M. Risk factors for the development of psoriasis. Int J Mol Sci (2019) 20(18):1–14. doi: 10.3390/ijms20184347
57. Langley RG, Krueger GG, Griffiths CE. Psoriasis: epidemiology, clinical features, and quality of life. Ann Rheum Dis (2005) 64 Suppl 2(Suppl 2):ii18–23. doi: 10.1136/ard.2004.033217
58. Griffiths CEM, Armstrong AW, Gudjonsson JE, Barker J. Psoriasis. Lancet (2021) 397(10281):1301–15. doi: 10.1016/s0140-6736(20)32549-6
59. Ovejero-Benito MC, Reolid A, Sánchez-Jiménez P, Saiz-Rodríguez M, Muñoz-Aceituno E, Llamas-Velasco M, et al. Histone modifications associated with biological drug response in moderate-to-severe psoriasis. Exp Dermatol (2018) 27(12):1361–71. doi: 10.1111/exd.13790
60. Liu Z, Cao W, Xu L, Chen X, Zhan Y, Yang Q, et al. The histone H3 lysine-27 demethylase Jmjd3 plays a critical role in specific regulation of Th17 cell differentiation. J Mol Cell Biol (2015) 7(6):505–16. doi: 10.1093/jmcb/mjv022
61. Bending D, Newland S, Krejcí A, Phillips JM, Bray S, Cooke A. Epigenetic changes at Il12rb2 and Tbx21 in relation to plasticity behavior of Th17 cells. J Immunol (2011) 186(6):3373–82. doi: 10.4049/jimmunol.1003216
62. Li H, Yao Q, Mariscal AG, Wu X, Hülse J, Pedersen E, et al. Epigenetic control of IL-23 expression in keratinocytes is important for chronic skin inflammation. Nat Commun (2018) 9(1):1420. doi: 10.1038/s41467-018-03704-z
63. Rasheed H, El-Komy M, Hegazy RA, Gawdat HI, AlOrbani AM, Shaker OG. Expression of sirtuins 1, 6, tumor necrosis factor, and interferon-γ in psoriatic patients. Int J Immunopathol Pharmacol (2016) 29(4):764–8. doi: 10.1177/0394632016662475
64. Hwang YJ, Na JI, Byun SY, Kwon SH, Yang SH, Lee HS, et al. Histone deacetylase 1 and sirtuin 1 expression in psoriatic skin: A comparison between guttate and plaque psoriasis. Life (Basel) (2020) 10(9):1–8. doi: 10.3390/life10090157
65. Liao Y, Su Y, Wu R, Zhang P, Feng C. Overexpression of Wilms tumor 1 promotes IL-1β expression by upregulating histone acetylation in keratinocytes. Int Immunopharmacol (2021) 96:107793. doi: 10.1016/j.intimp.2021.107793
66. Chen C, Yi X, Liu P, Li J, Yan B, Zhang D, et al. CD147 facilitates the pathogenesis of psoriasis through glycolysis and H3K9me3 modification in keratinocytes. Res (Wash D C) (2023) 6:167. doi: 10.34133/research.0167
67. Huang S, Zhen Y, Yin X, Yang Z, Li X, Wang R, et al. KMT2C induced by FABP5P3 aggravates keratinocyte hyperproliferation and psoriasiform skin inflammation by upregulating the transcription of PIK3R3. J Invest Dermatol (2023) 143(1):37–47.e8. doi: 10.1016/j.jid.2022.06.025
68. Masalha M, Ben-Dov IZ, Ram O, Meningher T, Jacob-Hirsch J, Kassem R, et al. H3K27Ac modification and gene expression in psoriasis. J Dermatol Sci (2021) 103(2):93–100. doi: 10.1016/j.jdermsci.2021.07.003
69. Zhang T, Yang L, Ke Y, Lei J, Shen S, Shao S, et al. EZH2-dependent epigenetic modulation of histone H3 lysine-27 contributes to psoriasis by promoting keratinocyte proliferation. Cell Death Dis (2020) 11(10):826. doi: 10.1038/s41419-020-03028-1
70. Xia X, Cao G, Sun G, Zhu L, Tian Y, Song Y, et al. GLS1-mediated glutaminolysis unbridled by MALT1 protease promotes psoriasis pathogenesis. J Clin Invest (2020) 130(10):5180–96. doi: 10.1172/jci129269
71. Kim Y, Eom S, Kim K, Lee YS, Choe J, Hahn JH, et al. Transglutaminase II interacts with rac1, regulates production of reactive oxygen species, expression of snail, secretion of Th2 cytokines and mediates in vitro and in vivo allergic inflammation. Mol Immunol (2010) 47(5):1010–22. doi: 10.1016/j.molimm.2009.11.017
72. Poppenberg KE, Tutino VM, Tarbell E, Jarvis JN. Broadening our understanding of genetic risk for scleroderma/systemic sclerosis by querying the chromatin architecture surrounding the risk haplotypes. BMC Med Genomics (2021) 14(1):114. doi: 10.1186/s12920-021-00964-5
73. Wasson CW, Abignano G, Hermes H, Malaab M, Ross RL, Jimenez SA, et al. Long non-coding RNA HOTAIR drives EZH2-dependent myofibroblast activation in systemic sclerosis through miRNA 34a-dependent activation of NOTCH. Ann Rheum Dis (2020) 79(4):507–17. doi: 10.1136/annrheumdis-2019-216542
74. Tsou PS, Khanna D, Sawalha AH. Identification of cysteine-rich angiogenic inducer 61 as a potential antifibrotic and proangiogenic mediator in scleroderma. Arthritis Rheumatol (2019) 71(8):1350–9. doi: 10.1002/art.40890
75. van der Kroef M, Castellucci M, Mokry M, Cossu M, Garonzi M, Bossini-Castillo LM, et al. Histone modifications underlie monocyte dysregulation in patients with systemic sclerosis, underlining the treatment potential of epigenetic targeting. Ann Rheum Dis (2019) 78(4):529–38. doi: 10.1136/annrheumdis-2018-214295
76. Tian J, Huang T, Chen J, Wang J, Chang S, Xu H, et al. SIRT1 slows the progression of lupus nephritis by regulating the NLRP3 inflammasome through ROS/TRPM2/Ca(2+) channel. Clin Exp Med (2023) 23(7):3465–78. doi: 10.1007/s10238-023-01093-2
77. Mei Y, Xin Y, Li X, Yin H, Xiong F, Yang M, et al. Aberrant expression of JMJD3 in SLE promotes B-cell differentiation. Immunobiology (2023) 228(2):152347. doi: 10.1016/j.imbio.2023.152347
78. Zhao M, Feng D, Hu L, Liu L, Wu J, Hu Z, et al. 3D genome alterations in T cells associated with disease activity of systemic lupus erythematosus. Ann Rheum Dis (2023) 82(2):226–34. doi: 10.1136/ard-2022-222653
79. Zhou W, Wang J, Wang X, Wang B, Zhao Z, Fu J, et al. Degradation of HDAC10 by autophagy promotes IRF3-mediated antiviral innate immune responses. Sci Signal (2022) 15(765):eabo4356. doi: 10.1126/scisignal.abo4356
80. Weidinger S, Beck LA, Bieber T, Kabashima K, Irvine AD. Atopic dermatitis. Nat Rev Dis Primers (2018) 4(1):1. doi: 10.1038/s41572-018-0001-z
81. Løset M, Brown SJ, Saunes M, Hveem K. Genetics of atopic dermatitis: from DNA sequence to clinical relevance. Dermatology (2019) 235(5):355–64. doi: 10.1159/000500402
82. Kwon Y, Choi Y, Kim M, Jeong MS, Jung HS, Jeoung D. HDAC6 and CXCL13 mediate atopic dermatitis by regulating cellular interactions and expression levels of miR-9 and SIRT1. Front Pharmacol (2021) 12:691279. doi: 10.3389/fphar.2021.691279
83. Kim M, Lee SH, Kim Y, Kwon Y, Park Y, Lee HK, et al. Human adipose tissue-derived mesenchymal stem cells attenuate atopic dermatitis by regulating the expression of MIP-2, miR-122a-SOCS1 axis, and Th1/Th2 responses. Front Pharmacol (2018) 9:1175. doi: 10.3389/fphar.2018.01175
84. Liew WC, Sundaram GM, Quah S, Lum GG, Tan JSL, Ramalingam R, et al. Belinostat resolves skin barrier defects in atopic dermatitis by targeting the dysregulated miR-335:SOX6 axis. J Allergy Clin Immunol (2020) 146(3):606–20.e12. doi: 10.1016/j.jaci.2020.02.007
85. Allanore Y, Simms R, Distler O, Trojanowska M, Pope J, Denton CP, et al. Systemic sclerosis. Nat Rev Dis Primers (2015) 1:15002. doi: 10.1038/nrdp.2015.2
86. Tieu A, Chaigne B, Dunogué B, Dion J, Régent A, Casadevall M, et al. Autoantibodies versus skin fibrosis extent in systemic sclerosis: A case-control study of inverted phenotypes. Diagnostics (Basel) (2022) 12(5):1–13. doi: 10.3390/diagnostics12051067
87. Dreger H, Ludwig A, Weller A, Stangl V, Baumann G, Meiners S, et al. Epigenetic regulation of cell adhesion and communication by enhancer of zeste homolog 2 in human endothelial cells. Hypertension (2012) 60(5):1176–83. doi: 10.1161/hypertensionaha.112.191098
88. Xiao X, Senavirathna LK, Gou X, Huang C, Liang Y, Liu L. EZH2 enhances the differentiation of fibroblasts into myofibroblasts in idiopathic pulmonary fibrosis. Physiol Rep (2016) 4(17):1–15. doi: 10.14814/phy2.12915
89. Tumes DJ, Onodera A, Suzuki A, Shinoda K, Endo Y, Iwamura C, et al. The polycomb protein Ezh2 regulates differentiation and plasticity of CD4(+) T helper type 1 and type 2 cells. Immunity (2013) 39(5):819–32. doi: 10.1016/j.immuni.2013.09.012
90. Tsou PS, Campbell P, Amin MA, Coit P, Miller S, Fox DA, et al. Inhibition of EZH2 prevents fibrosis and restores normal angiogenesis in scleroderma. Proc Natl Acad Sci USA (2019) 116(9):3695–702. doi: 10.1073/pnas.1813006116
91. Bergmann C, Brandt A, Merlevede B, Hallenberger L, Dees C, Wohlfahrt T, et al. The histone demethylase Jumonji domain-containing protein 3 (JMJD3) regulates fibroblast activation in systemic sclerosis. Ann Rheum Dis (2018) 77(1):150–8. doi: 10.1136/annrheumdis-2017-211501
92. Ciechomska M, O'Reilly S, Przyborski S, Oakley F, Bogunia-Kubik K, van Laar JM. Histone demethylation and toll-like receptor 8-dependent cross-talk in monocytes promotes transdifferentiation of fibroblasts in systemic sclerosis via Fra-2. Arthritis Rheumatol (2016) 68(6):1493–504. doi: 10.1002/art.39602
93. Shin JY, Beckett JD, Bagirzadeh R, Creamer TJ, Shah AA, McMahan Z, et al. Epigenetic activation and memory at a TGFB2 enhancer in systemic sclerosis. Sci Transl Med (2019) 11(497):1–23. doi: 10.1126/scitranslmed.aaw0790
94. Nagy N, Papp G, Gáspár-Kiss E, Diószegi Á, Tarr T. Changes in clinical manifestations and course of systemic lupus erythematosus and secondary antiphospholipid syndrome over three decades. Biomedicines (2023) 11(4):1–10. doi: 10.3390/biomedicines11041218
95. Chan ES, Herlitz LC, Jabbari A. Ruxolitinib attenuates cutaneous lupus development in a mouse lupus model. J Invest Dermatol (2015) 135(7):1912–5. doi: 10.1038/jid.2015.107
96. Thomas G, Mancini J, Jourde-Chiche N, Sarlon G, Amoura Z, Harlé JR, et al. Mortality associated with systemic lupus erythematosus in France assessed by multiple-cause-of-death analysis. Arthritis Rheumatol (2014) 66(9):2503–11. doi: 10.1002/art.38731
97. Tsou PS, Coit P, Kilian NC, Sawalha AH. EZH2 modulates the DNA methylome and controls T cell adhesion through junctional adhesion molecule A in lupus patients. Arthritis Rheumatol (2018) 70(1):98–108. doi: 10.1002/art.40338
98. Luo S, Xie Y, Huang J, Liu J, Zhang Q. Decreased jumonji domain-containing 3 at the promoter downregulates hematopoietic progenitor kinase 1 expression and cytoactivity of T follicular helper cells from systemic lupus erythematosus patients. J Immunol Res (2022) 2022:3690892. doi: 10.1155/2022/3690892
99. Regna NL, Vieson MD, Luo XM, Chafin CB, Puthiyaveetil AG, Hammond SE, et al. Specific HDAC6 inhibition by ACY-738 reduces SLE pathogenesis in NZB/W mice. Clin Immunol (2016) 162:58–73. doi: 10.1016/j.clim.2015.11.007
100. Zhou Y, Qiu X, Luo Y, Yuan J, Li Y, Zhong Q, et al. Histone modifications and methyl-CpG-binding domain protein levels at the TNFSF7 (CD70) promoter in SLE CD4+ T cells. Lupus (2011) 20(13):1365–71. doi: 10.1177/0961203311413412
101. Chamani S, Mobasheri L, Rostami Z, Zare I, Naghizadeh A, Mostafavi E. Heavy metals in contact dermatitis: A review. J Trace Elem Med Biol (2023) 79:127240. doi: 10.1016/j.jtemb.2023.127240
102. Han SC, Kang JI, Choi YK, Boo HJ, Yoon WJ, Kang HK, et al. Intermittent fasting modulates immune response by generating tregs via TGF-β Dependent mechanisms in obese mice with allergic contact dermatitis. Biomol Ther (Seoul) (2023), 1–10. doi: 10.4062/biomolther.2023.053
103. Chai W, Zhang X, Lin M, Chen Z, Wang X, Wang C, et al. Allergic rhinitis, allergic contact dermatitis and disease comorbidity belong to separate entities with distinct composition of T-cell subsets, cytokines, immunoglobulins and autoantibodies. Allergy Asthma Clin Immunol (2022) 18(1):10. doi: 10.1186/s13223-022-00646-6
104. van den Oord LJ, Naber RJ, van Manen BCT, Rustemeyer T. The prevalence of contact dermatitis among surgical assistants, anaesthesia assistants and anaesthesiologists working at the Amsterdam University Medical Centre. Contact Dermatitis (2023) 89(3):171–77. doi: 10.1111/cod.14371
105. Sonday Z, Baatjies R, Mwanga HH, Jeebhay MF. Prevalence of work-related skin symptoms and associated factors among tertiary hospital workers exposed to cleaning agents in Southern Africa. Contact Dermatitis (2023) 89(3):178–89. doi: 10.1111/cod.14374
106. Barbaud A, Kurihara F, Raison-Peyron N, Milpied B, Valois A, Assier H, et al. Allergic contact dermatitis from essential oil in consumer products: Mode of uses and value of patch-tests with an essential oil series. Results of a French study of the DAG (dermato-allergy group of the French Society of Dermatology). Contact Dermatitis (2023) 89(3):190–97. doi: 10.1111/cod.14377
107. Coppola CM, Tobin JG, Lawrence JP. Allergic contact dermatitis to Dermabond Prineo after abdominal wound closure for anterior lumbar interbody fusion: case report. J Spine Surg (2023) 9(2):191–4. doi: 10.21037/jss-22-93
108. Ojo DE, Martinez VH, Sheean AJ, Hartzler RU. Documented contact allergy impacts risk for surgical adhesive-associated contact dermatitis after shoulder arthroplasty. Arthrosc Sports Med Rehabil (2023) 5(3):e839–e42. doi: 10.1016/j.asmr.2023.04.013
109. Senda N, Yanai H, Hibino S, Li L, Mizushima Y, Miyagaki T, et al. HMGB1-mediated chromatin remodeling attenuates Il24 gene expression for the protection from allergic contact dermatitis. Proc Natl Acad Sci USA (2021) 118(1):1–7. doi: 10.1073/pnas.2022343118
110. Inoue T, Omori-Miyake M, Maruyama S, Okabe M, Kuwahara M, Honda H, et al. The loss of H3K27 histone demethylase Utx in T cells aggravates allergic contact dermatitis. J Immunol (2021) 207(9):2223–34. doi: 10.4049/jimmunol.2001160
111. Schinner J, Cunha T, Mayer JU, Hörster S, Kind P, Didona D, et al. Skin-infiltrating T cells display distinct inflammatory signatures in lichen planus, bullous pemphigoid and pemphigus vulgaris. Front Immunol (2023) 14:1203776. doi: 10.3389/fimmu.2023.1203776
112. Le Cleach L, Chosidow O. Clinical practice. Lichen planus. N Engl J Med (2012) 366(8):723–32. doi: 10.1056/NEJMcp1103641
113. Sugerman PB, Savage NW, Walsh LJ, Zhao ZZ, Zhou XJ, Khan A, et al. The pathogenesis of oral lichen planus. Crit Rev Oral Biol Med (2002) 13(4):350–65. doi: 10.1177/154411130201300405
114. Rotaru DI, Sofineti D, Bolboacă SD, Bulboacă AE. Diagnostic criteria of oral lichen planus: A narrative review. Acta Clin Croat (2020) 59(3):513–22. doi: 10.20471/acc.2020.59.03.16
115. Sugerman PB, Savage NW. Oral lichen planus: causes, diagnosis and management. Aust Dent J (2002) 47(4):290–7. doi: 10.1111/j.1834-7819.2002.tb00540.x
116. Zhang J, Peng G, Chi H, Yang J, Xie X, Song G, et al. CD8 + T-cell marker genes reveal different immune subtypes of oral lichen planus by integrating single-cell RNA-seq and bulk RNA-sequencing. BMC Oral Health (2023) 23(1):464. doi: 10.1186/s12903-023-03138-0
117. Gonzaga AKG, Lopes M, Squarize CH, Castilho RM, de Medeiros AMC, Rocha KBF, et al. Expression profile of DNA repair proteins and histone H3 lys-9 acetylation in cutaneous and oral lichen planus. Arch Oral Biol (2020) 119:104880. doi: 10.1016/j.archoralbio.2020.104880
118. Dillenburg CS, Martins MAT, Almeida LO, Meurer L, Squarize CH, Martins MD, et al. Epigenetic modifications and accumulation of DNA double-strand breaks in oral lichen planus lesions presenting poor response to therapy. Med (Baltimore) (2015) 94(30):e997. doi: 10.1097/md.0000000000000997
119. Shen J, Yin C, Jiang X, Wang X, Yang S, Song G. Aberrant histone modification and inflammatory cytokine production of peripheral CD4+ T cells in patients with oral lichen planus. J Oral Pathol Med (2019) 48(2):136–42. doi: 10.1111/jop.12790
120. Islam N, Leung PS, Huntley AC, Gershwin ME. The autoimmune basis of alopecia areata: a comprehensive review. Autoimmun Rev (2015) 14(2):81–9. doi: 10.1016/j.autrev.2014.10.014
121. Muntyanu A, Gabrielli S, Donovan J, Gooderham M, Guenther L, Hanna S, et al. The burden of alopecia areata: A scoping review focusing on quality of life, mental health and work productivity. J Eur Acad Dermatol Venereol (2023), 1490–1520. doi: 10.1111/jdv.18926
122. Ito T, Kamei K, Yuasa A, Matsumoto F, Hoshi Y, Okada M, et al. Health-related quality of life in patients with alopecia areata: Results of a Japanese survey with norm-based comparisons. J Dermatol (2022) 49(6):584–93. doi: 10.1111/1346-8138.16364
123. Ohyama M, Kamei K, Yuasa A, Anderson P, Milligan G, Sakaki-Yumoto M. Economic burden of alopecia areata: A study of direct and indirect cost in Japan using real-world data. J Dermatol (2023) 50(10):1246–54. doi: 10.1111/1346-8138.16888
124. Singam V, Patel KR, Lee HH, Rastogi S, Silverberg JI. Association of alopecia areata with hospitalization for mental health disorders in US adults. J Am Acad Dermatol (2019) 80(3):792–4. doi: 10.1016/j.jaad.2018.07.044
125. Waśkiel-Burnat A, Osińska M, Salińska A, Blicharz L, Goldust M, Olszewska M, et al. The role of serum Th1, Th2, and Th17 cytokines in patients with alopecia areata: clinical implications. Cells (2021) 10(12):1–14. doi: 10.3390/cells10123397
126. Żeberkiewicz M, Rudnicka L, Malejczyk J. Immunology of alopecia areata. Cent Eur J Immunol (2020) 45(3):325–33. doi: 10.5114/ceji.2020.101264
127. Zhao M, Liang G, Wu X, Wang S, Zhang P, Su Y, et al. Abnormal epigenetic modifications in peripheral blood mononuclear cells from patients with alopecia areata. Br J Dermatol (2012) 166(2):226–73. doi: 10.1111/j.1365-2133.2011.10646.x
128. Abdelkader HA, Amin I, Rashed LA, Samir M, Ezzat M. Histone deacetylase 1 in patients with alopecia areata and acne vulgaris: An epigenetic alteration. Australas J Dermatol (2022) 63(2):e138–e41. doi: 10.1111/ajd.13784
129. Hao L, Nam KH, Lee GJ, Kim D, Shin JM, Lee Y, et al. SIRT1 downregulation provokes immune-inflammatory responses in hair follicle outer root sheath cells and may contribute to development of alopecia areata. J Dermatol Sci (2023) 111(1):2–9. doi: 10.1016/j.jdermsci.2023.05.005
130. Zekey E, Tunçez Akyürek F, Tunçez A, Akyürek F, Doğan ME. The relationship of serum trimethylamine N-oxide levels with carotid intima-media thickness and disease activity in psoriasis patients. Dermatol Pract Concept (2023) 13(2):1–10. doi: 10.5826/dpc.1302a116
131. Yang P, Xu R, Chen F, Chen S, Khan A, Li L, et al. Fungal gut microbiota dysbiosis in systemic lupus erythematosus. Front Microbiol (2023) 14:1149311. doi: 10.3389/fmicb.2023.1149311
132. Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol (2012) 129(2):434–40, 40.e1-2. doi: 10.1016/j.jaci.2011.10.025
133. Stec A, Maciejewska M, Zaremba M, Paralusz-Stec K, Michalska M, Rudnicka L, et al. The clinical significance of serum biomarkers of the intestinal barrier in systemic sclerosis: A cross-sectional study. J Pers Med (2023) 13(4):1–11. doi: 10.3390/jpm13040678
134. Ganapathy V, Thangaraju M, Prasad PD, Martin PM, Singh N. Transporters and receptors for short-chain fatty acids as the molecular link between colonic bacteria and the host. Curr Opin Pharmacol (2013) 13(6):869–74. doi: 10.1016/j.coph.2013.08.006
135. Postler TS, Ghosh S. Understanding the holobiont: how microbial metabolites affect human health and shape the immune system. Cell Metab (2017) 26(1):110–30. doi: 10.1016/j.cmet.2017.05.008
136. Levy M, Thaiss CA, Elinav E. Metabolites: messengers between the microbiota and the immune system. Genes Dev (2016) 30(14):1589–97. doi: 10.1101/gad.284091.116
137. Krautkramer KA, Kreznar JH, Romano KA, Vivas EI, Barrett-Wilt GA, Rabaglia ME, et al. Diet-microbiota interactions mediate global epigenetic programming in multiple host tissues. Mol Cell (2016) 64(5):982–92. doi: 10.1016/j.molcel.2016.10.025
138. Schwarz A, Philippsen R, Schwarz T. Induction of regulatory T cells and correction of cytokine disbalance by short-chain fatty acids: implications for psoriasis therapy. J Invest Dermatol (2021) 141(1):95–104.e2. doi: 10.1016/j.jid.2020.04.031
139. Leon Carrion S, Sutter CH, Sutter TR. Combined treatment with sodium butyrate and PD153035 enhances keratinocyte differentiation. Exp Dermatol (2014) 23(3):211–4. doi: 10.1111/exd.12333
140. Schwarz A, Bruhs A, Schwarz T. The short-chain fatty acid sodium butyrate functions as a regulator of the skin immune system. J Invest Dermatol (2017) 137(4):855–64. doi: 10.1016/j.jid.2016.11.014
141. Casali P, Shen T, Xu Y, Qiu Z, Chupp DP, Im J, et al. Estrogen reverses HDAC inhibitor-mediated repression of aicda and class-switching in antibody and autoantibody responses by downregulation of miR-26a. Front Immunol (2020) 11:491. doi: 10.3389/fimmu.2020.00491
142. Luo CH, Lai AC, Chang YJ. Butyrate inhibits Staphylococcus aureus-aggravated dermal IL-33 expression and skin inflammation through histone deacetylase inhibition. Front Immunol (2023) 14:1114699. doi: 10.3389/fimmu.2023.1114699
143. Shen C, Li M, Duan Y, Jiang X, Hou X, Xue F, et al. HDAC inhibitors enhance the anti-tumor effect of immunotherapies in hepatocellular carcinoma. Front Immunol (2023) 14:1170207. doi: 10.3389/fimmu.2023.1170207
144. Palamaris K, Moutafi M, Gakiopoulou H, Theocharis S. Histone deacetylase (HDAC) inhibitors: A promising weapon to tackle therapy resistance in melanoma. Int J Mol Sci (2022) 23(7):1–20. doi: 10.3390/ijms23073660
145. Jannoo R, Walker W, Kanamarlapudi V. Targeting and sensitization of breast cancer cells to killing with a novel interleukin-13 receptor α2-specific hybrid cytolytic peptide. Cancers (Basel) (2023) 15(10):1–19. doi: 10.3390/cancers15102772
146. Hong Y, Song M, Lan Y, Wang J, Lu S, Zhang Y, et al. Efficacy and safety of programmed cell death receptor 1 inhibition-based regimens in patients with pediatric Malignancies: the real-world study in China. Front Immunol (2023) 14:1182751. doi: 10.3389/fimmu.2023.1182751
147. Kim TH, Jung JA, Kim GD, Jang AH, Cho JJ, Park YS, et al. The histone deacetylase inhibitor, trichostatin A, inhibits the development of 2,4-dinitrofluorobenzene-induced dermatitis in NC/Nga mice. Int Immunopharmacol (2010) 10(10):1310–5. doi: 10.1016/j.intimp.2010.08.004
148. Mishra N, Brown DR, Olorenshaw IM, Kammer GM. Trichostatin A reverses skewed expression of CD154, interleukin-10, and interferon-gamma gene and protein expression in lupus T cells. Proc Natl Acad Sci USA (2001) 98(5):2628–33. doi: 10.1073/pnas.051507098
149. Mohammadi S, Saghaeian-Jazi M, Sedighi S, Memarian A. Sodium valproate modulates immune response by alternative activation of monocyte-derived macrophages in systemic lupus erythematosus. Clin Rheumatol (2018) 37(3):719–27. doi: 10.1007/s10067-017-3922-0
150. Souliotis VL, Vougas K, Gorgoulis VG, Sfikakis PP. Defective DNA repair and chromatin organization in patients with quiescent systemic lupus erythematosus. Arthritis Res Ther (2016) 18(1):182. doi: 10.1186/s13075-016-1081-3
151. Waibel M, Christiansen AJ, Hibbs ML, Shortt J, Jones SA, Simpson I, et al. Manipulation of B-cell responses with histone deacetylase inhibitors. Nat Commun (2015) 6:6838. doi: 10.1038/ncomms7838
152. Li N, Zhao D, Kirschbaum M, Zhang C, Lin CL, Todorov I, et al. HDAC inhibitor reduces cytokine storm and facilitates induction of chimerism that reverses lupus in anti-CD3 conditioning regimen. Proc Natl Acad Sci USA (2008) 105(12):4796–801. doi: 10.1073/pnas.0712051105
153. Reilly CM, Mishra N, Miller JM, Joshi D, Ruiz P, Richon VM, et al. Modulation of renal disease in MRL/lpr mice by suberoylanilide hydroxamic acid. J Immunol (2004) 173(6):4171–8. doi: 10.4049/jimmunol.173.6.4171
154. Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov (2014) 13(5):337–56. doi: 10.1038/nrd4286
155. Wen Q, Liu H, Lou K, Zhang X, Chao W, Xin J, et al. Essential role of bromodomain proteins in renal cell carcinoma (Review). Mol Med Rep (2023) 28(1):1–11. doi: 10.3892/mmr.2023.13026
156. Gargano D, Segatto M, Di Bartolomeo S. Regulation of cell plasticity by bromodomain and extraterminal domain (BET) proteins: A new perspective in glioblastoma therapy. Int J Mol Sci (2023) 24(6):1–26. doi: 10.3390/ijms24065665
157. Liu W, Ma Q, Wong K, Li W, Ohgi K, Zhang J, et al. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell (2013) 155(7):1581–95. doi: 10.1016/j.cell.2013.10.056
158. Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol (2011) 31(13):2641–52. doi: 10.1128/mcb.01341-10
159. Wei S, Sun Y, Sha H. Therapeutic targeting of BET protein BRD4 delays murine lupus. Int Immunopharmacol (2015) 29(2):314–9. doi: 10.1016/j.intimp.2015.10.036
160. Vichaikul S, Gurrea-Rubio M, Amin MA, Campbell PL, Wu Q, Mattichak MN, et al. Inhibition of bromodomain extraterminal histone readers alleviates skin fibrosis in experimental models of scleroderma. JCI Insight (2022) 7(9):1–15. doi: 10.1172/jci.insight.150871
161. Wu X, Shi Z, Hsu DK, Chong J, Huynh M, Mendoza L, et al. A monocyte-keratinocyte-derived co-culture assay accurately identifies efficacies of BET inhibitors as therapeutic candidates for psoriasiform dermatitis. J Dermatol Sci (2020) 100(1):31–8. doi: 10.1016/j.jdermsci.2020.08.005
162. Sato M, Kondo T, Kohno Y, Seto S. Discovery of benzo[f]pyrido[4,3-b][1,4]oxazepin-10-one derivatives as orally available bromodomain and extra-terminal domain (BET) inhibitors with efficacy in an in vivo psoriatic animal model. Bioorg Med Chem (2021) 34:116015. doi: 10.1016/j.bmc.2021.116015
163. Chae BJ, Lee KS, Hwang I, Yu JW. Extracellular acidification augments NLRP3-mediated inflammasome signaling in macrophages. Immune Netw (2023) 23(3):e23. doi: 10.4110/in.2023.23.e23
164. Zhao Z, Zhang L, Ocansey DKW, Wang B, Mao F. The role of mesenchymal stem cell-derived exosome in epigenetic modifications in inflammatory diseases. Front Immunol (2023) 14:1166536. doi: 10.3389/fimmu.2023.1166536
165. Terheyden H, Stadlinger B, Sanz M, Garbe AI, Meyle J. Inflammatory reaction - communication of cells. Clin Oral Implants Res (2014) 25(4):399–407. doi: 10.1111/clr.12176
166. Nathan C. Nonresolving inflammation redux. Immunity (2022) 55(4):592–605. doi: 10.1016/j.immuni.2022.03.016
167. Zhang LJ, Sen GL, Ward NL, Johnston A, Chun K, Chen Y, et al. Antimicrobial peptide LL37 and MAVS signaling drive interferon-β Production by epidermal keratinocytes during skin injury. Immunity (2016) 45(1):119–30. doi: 10.1016/j.immuni.2016.06.021
168. Korsten S, Vromans H, Garssen J, Willemsen LEM. Butyrate protects barrier integrity and suppresses immune activation in a caco-2/PBMC co-culture model while HDAC inhibition mimics butyrate in restoring cytokine-induced barrier disruption. Nutrients (2023) 15(12):1–27. doi: 10.3390/nu15122760
169. Turgeon N, Gagné JM, Blais M, Gendron FP, Boudreau F, Asselin C. The acetylome regulators Hdac1 and Hdac2 differently modulate intestinal epithelial cell dependent homeostatic responses in experimental colitis. Am J Physiol Gastrointest Liver Physiol (2014) 306(7):G594–605. doi: 10.1152/ajpgi.00393.2013
170. Sanford JA, Zhang LJ, Williams MR, Gangoiti JA, Huang CM, Gallo RL. Inhibition of HDAC8 and HDAC9 by microbial short-chain fatty acids breaks immune tolerance of the epidermis to TLR ligands. Sci Immunol (2016) 1(4):1–10. doi: 10.1126/sciimmunol.aah4609
171. Hao L, Park J, Jang HY, Bae EJ, Park BH. Inhibiting protein kinase activity of pyruvate kinase M2 by SIRT2 deacetylase attenuates psoriasis. J Invest Dermatol (2021) 141(2):355–63.e6. doi: 10.1016/j.jid.2020.06.024
172. Candelaria M, Gallardo-Rincón D, Arce C, Cetina L, Aguilar-Ponce JL, Arrieta O, et al. A phase II study of epigenetic therapy with hydralazine and magnesium valproate to overcome chemotherapy resistance in refractory solid tumors. Ann Oncol (2007) 18(9):1529–38. doi: 10.1093/annonc/mdm204
173. Lassen U, Molife LR, Sorensen M, Engelholm SA, Vidal L, Sinha R, et al. A phase I study of the safety and pharmacokinetics of the histone deacetylase inhibitor belinostat administered in combination with carboplatin and/or paclitaxel in patients with solid tumours. Br J Cancer (2010) 103(1):12–7. doi: 10.1038/sj.bjc.6605726
174. Dizon DS, Blessing JA, Penson RT, Drake RD, Walker JL, Johnston CM, et al. A phase II evaluation of belinostat and carboplatin in the treatment of recurrent or persistent platinum-resistant ovarian, fallopian tube, or primary peritoneal carcinoma: a Gynecologic Oncology Group study. Gynecol Oncol (2012) 125(2):367–71. doi: 10.1016/j.ygyno.2012.02.019
175. Roy R, Ria T, RoyMahaPatra D, Sk UH. Single inhibitors versus dual inhibitors: role of HDAC in cancer. ACS Omega (2023) 8(19):16532–44. doi: 10.1021/acsomega.3c00222
176. Jenner MW, Pawlyn C, Davies FE, Menzies T, Hockaday A, Olivier C, et al. The addition of vorinostat to lenalidomide maintenance for patients with newly diagnosed multiple myeloma of all ages: results from 'Myeloma XI', a multicentre, open-label, randomised, phase III trial. Br J Haematol (2023) 201(2):267–79. doi: 10.1111/bjh.18600
177. Tretyakov BA, Filatova NV, Mumyatova VA, Gadomsky SY, Terent'ev AA. Pyridine derivative of succinic acid hydroxylamide enhances the cytotoxic effect of cisplatin and actinomycin D. Bull Exp Biol Med (2023) 175(1):23–6. doi: 10.1007/s10517-023-05803-4
178. Pan D-S, Yang Q-J, Fu X, Shan S, Zhu J-Z, Zhang K, et al. Discovery of an orally active subtype-selective HDAC inhibitor, chidamide, as an epigenetic modulator for cancer treatment. MedChemComm (2014) 5(12):1789–96. doi: 10.1039/C4MD00350K
179. Bollmann LM, Skerhut AJ, Asfaha Y, Horstick N, Hanenberg H, Hamacher A, et al. The novel class IIa selective histone deacetylase inhibitor YAK540 is synergistic with bortezomib in leukemia cell lines. Int J Mol Sci (2022) 23(21):1–21. doi: 10.3390/ijms232113398
180. Hong J, Choi Y, Lee G, Kim J, Jang Y, Yoon CH, et al. Nanosome-mediated delivery of hdac inhibitors and oxygen molecules for the transcriptional reactivation of latent Hiv-infected Cd4(+) T cells. Small (2023) 19(37):e2301730. doi: 10.1002/smll.202301730
181. Piha-Paul SA, Hann CL, French CA, Cousin S, Braña I, Cassier PA, et al. Phase 1 study of molibresib (GSK525762), a bromodomain and extra-terminal domain protein inhibitor, in NUT carcinoma and other solid tumors. JNCI Cancer Spectr (2020) 4(2):pkz093. doi: 10.1093/jncics/pkz093
182. To KKW, Xing E, Larue RC, Li PK. BET bromodomain inhibitors: novel design strategies and therapeutic applications. Molecules (2023) 28(7):1–38. doi: 10.3390/molecules28073043
183. Faivre EJ, McDaniel KF, Albert DH, Mantena SR, Plotnik JP, Wilcox D, et al. Selective inhibition of the BD2 bromodomain of BET proteins in prostate cancer. Nature (2020) 578(7794):306–10. doi: 10.1038/s41586-020-1930-8
184. Gilan O, Rioja I, Knezevic K, Bell MJ, Yeung MM, Harker NR, et al. Selective targeting of BD1 and BD2 of the BET proteins in cancer and immunoinflammation. Science (2020) 368(6489):387–94. doi: 10.1126/science.aaz8455
185. Ali SR, Jordan M, Nagarajan P, Amit M. Nerve density and neuronal biomarkers in cancer. Cancers (Basel) (2022) 14(19):1–15. doi: 10.3390/cancers14194817
186. Nowak RP, DeAngelo SL, Buckley D, He Z, Donovan KA, An J, et al. Plasticity in binding confers selectivity in ligand-induced protein degradation. Nat Chem Biol (2018) 14(7):706–14. doi: 10.1038/s41589-018-0055-y
Keywords: histone modification, inflammatory skin disease, therapy, epigenetic, inflammatory reaction
Citation: Zhang L, Chai R, Tai Z, Miao F, Shi X, Chen Z and Zhu Q (2024) Noval advance of histone modification in inflammatory skin diseases and related treatment methods. Front. Immunol. 14:1286776. doi: 10.3389/fimmu.2023.1286776
Received: 01 September 2023; Accepted: 08 December 2023;
Published: 03 January 2024.
Edited by:
Dongqing Li, Chinese Academy of Medical Sciences & Peking Union Medical College, ChinaReviewed by:
Rong Zeng, Chinese Academy of Medical Sciences and Peking Union Medical College, ChinaCopyright © 2024 Zhang, Chai, Tai, Miao, Shi, Chen and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Quangang Zhu, cWd6aHVAMTI2LmNvbQ==; Zhongjian Chen, YWFqaWFuODE4QDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.