
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 17 October 2023
Sec. Cytokines and Soluble Mediators in Immunity
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1281845
This article is part of the Research Topic Harnessing the Potential of the Common Gamma-Chain Family of Cytokines in Immunotherapy and Vaccine Development View all 6 articles
Background: Observational studies have reported an association between circulating cytokines and sepsis. However, the precise causal relationship between these factors remains unclear. The objective of this study was to explore the causal link between circulating cytokines and sepsis using genetic data within the framework of Mendelian Randomization (MR).
Methods: We performed a two-sample MR analysis to investigate this causality relationship in individuals of European ancestry. The publicly available genome-wide association studies (GWAS) statistics were used. We selected eligible instrumental single nucleotide polymorphisms (SNPs) that were significantly related to the circulating cytokines. Multiple MR analysis approaches were carried out, which included inverse variance weighted (IVW), Weighted Median, MR-Egger, Weighted Mode, Simple Mode, and MR pleiotropy residual sum and outlier (MR-PRESSO) methods.
Results: We found evidence to support the causal role of genetically predicted circulating levels on decreased risk of sepsis, including RANTES (OR = 0.920, 95% CI: 0.849-0.997, P = 0.041) and basic fibroblast growth factor (basic-FGF) (OR = 0.869, 95% CI: 0.766-0.986, P = 0.029). Additionally, MR analysis positive causal association of between beta-nerve growth factor (β-NGF) and sepsis (OR = 1.120, 95% CI: 1.037-1.211, P = 0.004). The results of MR-Egger, Weighted Median, Weighted Mode, and Simple Mode methods were consistent with the IVW estimates. Sensitivity analysis showed no horizontal pleiotropy to bias the causal estimates.
Conclusion: This MR study provides first novel evidence that genetically predicted causal association of circulating levels of RANTES, basic-FGF, and β-NGF with altered sepsis risk. The findings shed light on the potential involvement of these cytokines in sepsis pathogenesis. Although requiring additional confirmation, the results contribute new insights into cytokine mediators in sepsis and suggest promising future research directions.
Sepsis, a systemic inflammatory response syndrome triggered by the body in response to pathogens, is a critical condition that demands prompt and effective treatment. Failure to intervene in a timely manner can result in the dysfunction of multiple organs and a mortality rate ranging from 30% to 50% (1). Epidemiological studies have shed light on the global prevalence of sepsis, with approximately 17 million cases reported in 2017, showing a notable decrease of 18.8% compared to the 60.2 million cases reported in 1990 (2). The alarming impact of sepsis is further highlighted by estimations of 11 million sepsis-related deaths worldwide in 2017, making up a staggering 19.7% of all deaths that year (2). These statistics underscore the urgent need for heightened awareness, improved prevention strategies, and enhanced treatment options to combat this life-threatening condition.
Inflammation lies at the heart of sepsis pathophysiology (3). While an inflammatory response is vital to combat infection initially, excessive inflammation can inflict damage on the host (4). This explains the early use of glucocorticoids in sepsis to temper harmful hyperinflammation (5, 6). However, as sepsis progresses, the inflammatory response wanes while immunosuppression comes to the fore (7). Prolonged glucocorticoid administration then risks precipitating immune paralysis (8). The inflammatory cascade in sepsis is driven by the release of potent cytokines including TNF-α, interleukin (IL)-1, and IL-6, as well as other mediators like chemokines, complement, and reactive oxygen species (4, 9–12). The ensuing dysregulated immune response and tissue injury arise from complex interplay between inflammatory pathways (13). Though research continues to unravel these nuanced relationships, the precise causal mechanisms remain enigmatic. Recent advances harnessing models and technology have offered fresh insights, while clinical studies illuminated potential anti-inflammatory therapeutic targets (14, 15). In summary, despite copious evidence linking inflammatory mediators to sepsis pathogenesis, the intricate inflammatory imbalance underpinning sepsis progression remains incompletely understood.
Mendelian randomization (MR) is an epidemiological technique that utilizes genetic variants as instrumental variables (IVs) to assess causal relationships between modifiable exposures and disease outcomes (16). The random assortment of alleles at conception results in a random distribution of genotypes in the population. Therefore, genetic variants can be leveraged as unconfounded proxies for exposures of interest. By examining the association between genetic variants linked to exposures and disease risk, MR can infer causality while minimizing biases from reverse causation and confounding that afflict conventional observational studies (17).
In this study, we aimed to employ MR to explore the causal nature of the relationship between circulating cytokines and sepsis. By using genetic instruments as proxies for circulating cytokines, we can assess whether inflammation has a causal role in sepsis development or merely represents an epiphenomenon. Elucidating these causal pathways will provide greater biological insight and may uncover novel therapeutic targets for this deadly syndrome.
In our two-sample MR study (Figure 1), single nucleotide polymorphisms (SNPs) were exploited as instrument variables (IVs). To ensure the data validity, we chose SNPs through three major assumptions: (1) IVs should be significantly associated with the exposure factors (‘‘Relevance assumption’’); (2) IVs affect the outcomes only via exposure factors rather than the other pathways, which implies no horizontal pleiotropy (‘‘Exclusivity assumption’’); (3) IVs were not relevant to any confounding factors (‘‘Independence assumption’’) (18).
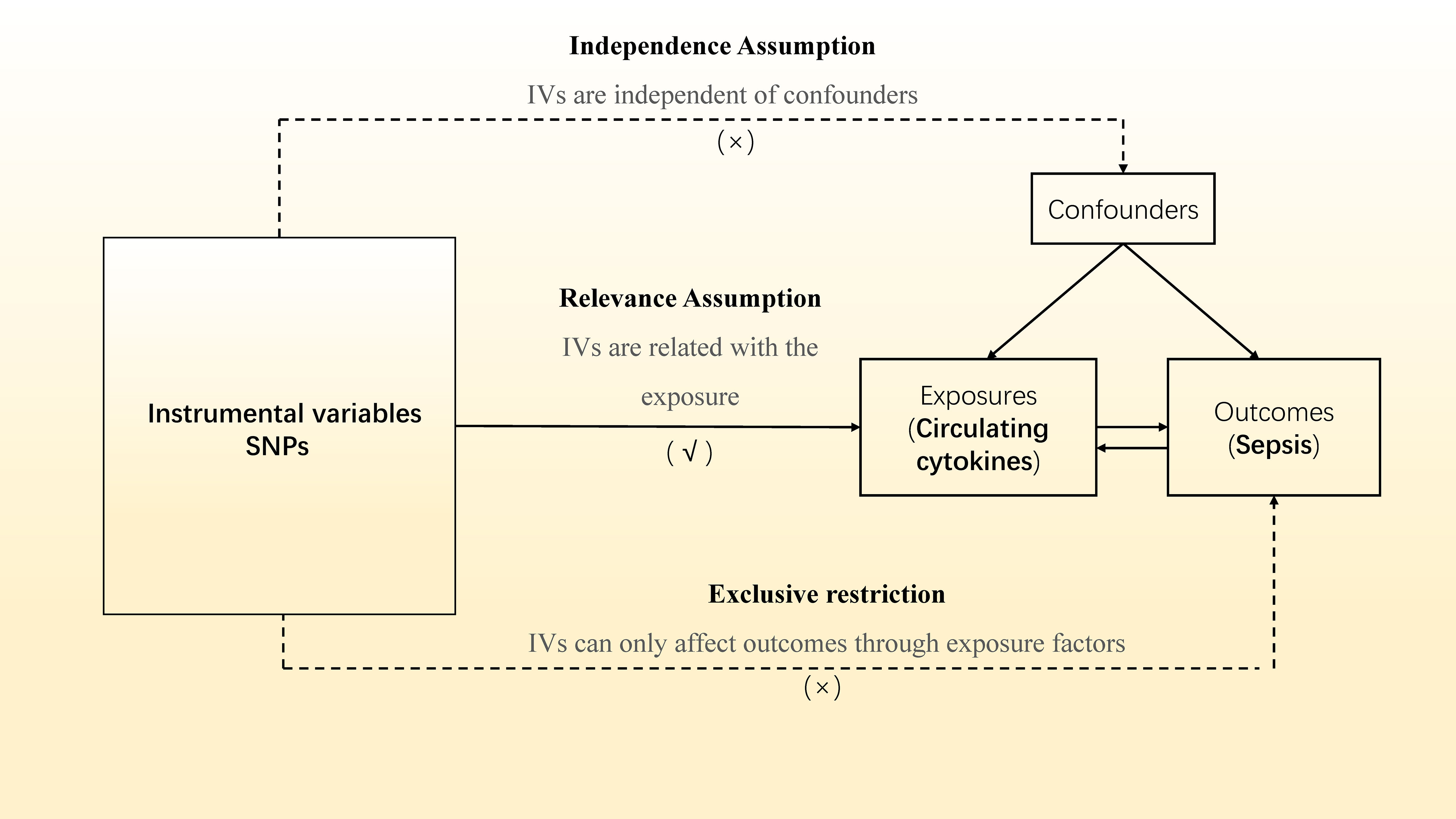
Figure 1 Overview of the current Mendelian randomization (MR) study. SNPs, single nucleotide polymorphisms; IVs, instrumental variables. A √ symbol indicates that the criteria was achieved; a × symbol indicates that the criteria was not achieved.
SNPs on circulating cytokines and sepsis was selected from the latest genome-wide association study (GWAS), as presented in Supplementary Table 1.
Figure 1 depicts the study’s overview in detail. Summary data from the most thorough and extensive cytokine GWAS were used for the genetic tool of cytokines. The cytokine GWAS meta-analysis included 8,293 Finns from three distinct population-based cohorts: the Young Finns Cardiovascular Risk Study, FINRISK1997, and FINRISK2002 studies (19). Participants were chosen at random from five distinct geographic regions and between the ages of 25 and 74 during the survey’s administration in Finland. The subjects’ EDTA plasma, heparin plasma, and blood were tested for cytokine levels. Only observations within each cytokine’s detectable range and cytokines with more than 90% of their values missing were removed from the study (7 out of 48). Written informed permission was given by each subject.
To explore the causal effect of exposure of various circulating cytokines on the risk of sepsis, we selected datasets for sepsis as the outcome from the summary statistics of the GWAS from UK Biobank (n = 11,643 for sepsis case, n = 474,841 for control). Notably, sepsis cases were identified by International Classification of Diseases (ICD), 10th edition, codes A02, A39, A40, and A41, aligning with definitions used in recent research (20–22). Cases were included when these codes appeared in either the primary or secondary diagnostic position within hospital episode statistics data, or similar datasets from devolved nations, as provided by UK Biobank. Then, using GWAS summary statistics, we used two-sample MR methods to deduce the causative link between inflammatory factors and sepsis. Since samples of inflammatory regulators and sepsis were acquired from various consortiums, there was no overlap.
We performed a set of methods to filter valid SNPs that suit the three core MR assumptions. Firstly, the independent SNPs strongly linked to different circulating cytokines were selected (23, 24). SNPs with P-value < 5×10-6 were considered to be significantly associated with circulating cytokines to obtain more SNPs as IVs. Secondly, we adopted the clumping process to evaluate the linkage disequilibrium (LD) among the SNPs (r2< 0.001 and clumping distance = 10,000 kb). The SNPs with LD were removed to avoid biased results. Thirdly, we searched all the screened SNPs on PhenoScanner V2 (http://www.phenoscanner.medschl.cam.ac.uk/) (25). PhenoScanner V2 provides the phenotypes information of SNPs, which can be used to determine whether the SNPs only affect the outcomes through exposure. The SNPs related to the confounding factors, such as smoking, diabetes and worries, were excluded to eliminate the bias. Finally, we harmonized the exposure and outcome datasets to remove the non-concordant SNPs. The remaining SNPs were used as the genetic IVs.
Moreover, the F statistics for the SNPs were calculated by the following equation: F = R2 × (N–2)/(1–R2). R2 was the proportion of variance. N represented the sample size. Weak instruments were identified by IVs with an F statistic less than 10 (F < 10) and excluded from the analysis (26).
After selecting the valid SNPs, we adopted inverse variance weighted (IVW) as the main way to estimate the MR analysis. IVW assesses the overall causal impact of exposure on the outcomes. It is the most accurate way to evaluate causality if all the selected SNPs are valid (27). We also applied complementary methods to analyze causal association, including Weighted Median, MR Egger, Weighted Mode, and Simple Mode methods. The Weighted Median method will generate a more potent effect when more than half of the SNPs are valid (28). MR Egger provides accurate effect estimates even if all the SNPs are invalid (29).
We further conducted the MR-Egger regression and the MR Pleiotropy Residual Sum and Outlier (MR-PRESSO) test to evaluate the possible horizontal pleiotropy (30, 31). In the MR-Egger regression, the intercept term indicates the average pleiotropic effect of IVs (31). We used Cochran’s Q statistic and MR-egger regression to test the heterogeneities. Additionally, the leave-one-out analysis was utilized to assess the robustness and consistency of the results.
All the analyses were performed with the packages “Two Sample MR” and “MRPRESSO” in R version 4.2.1. P < 0.05 is statistically significant.
After the series of filters mentioned in the method, 4-16 SNPs were left as IVs for circulating cytokines (Supplementary Table 1). All the selected SNPs were robust instruments, as confirmed by the F-statistic values being more than 10.
Next, we adopted these SNPs to analyze the causal link. The MR estimates between circulating cytokines and sepsis of different methods are presented in (Supplementary Table 2). Specifically, the preliminary results of IVW revealed negative causal effect of two cytokines on sepsis, including RANTES (regulated on activation, normal T-cell expressed and secreted (CCL5)) [OR = 0.920, 95% CI: 0.849-0.997, P = 0.041] and basic fibroblast growth factor (basic-FGF) [OR = 0.869, 95% CI: 0.766-0.986, P = 0.029], and positive causal effect of beta-nerve growth factor (β-NGF) and sepsis [OR = 1.120, 95% CI: 1.037-1.211, P = 0.004] (Table 1 and Figure 2). In addition, the MR-Egger, Weighted Median method showed consistent results. The scatter plots demonstrated the specific effects of each method per outcome database (Supplementary Figures 1, 3, 5).

Figure 2 Forest plot for the causal effect of circulating cytokines on the risk of sepsis derived from IVW. OR, odds ratio; CI, confdence interval; IVW, inverse variance weighting; CTACK, cutaneous T-cell attracting (CCL27); RANTES, regulated on activation, normal T-cell expressed and secreted (CCL5); TRAIL TNF-related apoptosis-inducing ligand. The * symbol represents a p-value of less than 0.05, which is statistically significant. Bold values represent p-values less than 0.05, which is statistically significant.
The MR-Egger regression and MR-PRESSO global test results revealed no horizontal pleiotropy (Table 2; Supplementary Table 3). More importantly, Cochran’s Q statistic and MR-egger regression showed no heterogeneity between the individual SNPs (P > 0.05). Additionally, the p-values of the MR PRESSO global test for circulating cytokines on sepsis were all greater than 0.05. The leave-one-out analysis further confirmed that the causal estimates of circulating cytokines were given (Supplementary Figures 2, 4, 6).
Apart from RANTES, basic-FGF and β-NGF, the other 38 cytokines (e.g., GRO-α, Trail, MIG, IL-17) did not show any association with the risk of sepsis in either IVW primary MR analysis or in other secondary analyses (Supplementary Table 2). In the heterogeneity assay, most of the cytokines were significantly non-heterogeneous, except for hepatocyte growth factor (P = 0.022). MR-egger regression did not show pleiotropy in p values for all cytokines (Table 2). An additional solidity test, the MR-PRESSO assay, did not show any abnormal values for the significant MR results (Supplementary Table 3).
To further explore the causal effect of sepsis on the circulating cytokines, we selected independent SNPs as IVs for sepsis. The F-statistic values were all higher than 10, which confirmed that all the selected SNPs were valid instruments. The results of the IVW analysis confirmed no causal effect of sepsis on all circulating cytokines (Table 3). Furthermore, most results revealed no heterogeneities based on the results of Cochran’s Q statistic, except for macrophage inflammatory protein 1b (P = 0.009). Horizontal pleiotropy was not detected in the results of several cytokines based on the results of MR-Egger intercept (Pintercept > 0.05) and MR-PRESSO global test (Pglobal test > 0.05). The detailed data are shown in Table 4; Supplementary Table 4.
In the present study, we utilized the two-sample MR method to investigate the potential causal associations between circulating levels of 41 cytokines and the risk of sepsis. We found suggestive evidence that the genetically predicted circulating levels of RANTES, basic-FGF and β-NGF were associated with sepsis. These findings provide new insights into the pathogenesis of sepsis.
RANTES/CCL5 is a chemokine that plays an important role in inflammation by recruiting leukocytes including monocytes, memory T cells, eosinophils and NK cells to sites of inflammation (32). Little is known about the role of RANTES in sepsis, and the studies that are currently available have inconsistent results regarding the role of RANTES. In basic research, Ness et al. found in a mouse sepsis model that the use of CCL5 increased sepsis-induced lethality in wild-type mice, while neutralization of CCL5 improved survival (33), in contrast to a recent study by Xie et al. who demonstrated that RANTES levels were significantly elevated in septic mice (34). In some small-sample clinical observational studies it was indicated that RANTES concentrations were significantly lower in the plasma of septic neonates and could be used as a biomarker for predicting neonatal infections (35–37). However, Cavaillon et al. found that RANTES levels were significantly higher in sepsis survivors than in non-survivors; moreover, RANTES levels did not correlate with any other cellular levels (including IL-6, IL-8, monocyte chemotactic protein-1 [MCP-1], etc.), and negatively correlated with acute physiologic assessment and chronic health evaluation II (APACHE II) scores, with low levels being significantly predictive of poor outcome (38). These findings do not entirely align with our own research. The specific role of RANTES in sepsis is currently unclear, as various studies have produced conflicting results and there are doubts about its clinical significance. These discrepancies may be due to variations in the animal models used, the timing and dosage of RANTES administration, and other factors that can affect its effects. Additionally, the limited sample size in current clinical studies may have contributed to individual differences. Nevertheless, our MR analysis suggests that circulating RANTES may have a potential protective effect against sepsis, although further studies are needed to validate these findings. Importantly, our inverse MR analysis did not identify a causal relationship between sepsis and basal levels of RANTES. Mechanistically, RANTES plays a crucial role in recruiting immune cells, such as monocytes, to sites of infection in order to eliminate pathogens and provide protection against infections (32). Additionally, it is capable of facilitating T cell proliferation and differentiation (39). Furthermore, elevated levels of RANTES may counteract immunosuppression in sepsis, particularly in advanced stages, by supporting the recovery of T lymphocytes (40). Given these functions, further research is necessary to fully understand the exact mechanisms through which RANTES contributes to the development of sepsis.
Basic-FGF, also known as FGF-2, is a member of the fibroblast growth factor family (41). It is a potent mitogen involved in wound healing, angiogenesis and embryonic development. Basic-FGF stimulates the proliferation, differentiation and migration of many cell types involved in inflammation and tissue repair, including endothelial cells, fibroblasts, smooth muscle cells and keratinocytes (42). In addition to its effects on cell growth and motility, basic-FGF also regulates the production of inflammatory mediators, extracellular matrix components and proteinases (43). Through its diverse biological activities, basic-FGF plays a key role in mediating inflammatory responses, angiogenesis and wound healing (44). Currently, there is only one clinical study that compares blood FGF-2 concentrations in 118 healthy control subjects with 18 sepsis patients. This study found that the median FGF-2 concentration in sepsis patients was significantly lower than that in healthy controls (25.7 vs. 37.7, P =0.0057) (45). Other studies have focused on basic research. For example, Pan et al. found that FGF-2 had a therapeutic effect on sepsis-associated acute lung injury by improving capillary leakage and reducing inflammatory response through cellular and animal experiments (46). Similarly, Sun et al. suggested that FGF-2 inhibited coagulant activity in septic mice, reduced lung and liver injuries, and improved survival, highlighting the role of FGF-2 in ameliorating sepsis-induced coagulation abnormalities (47). Moreover, in sepsis-induced cardiac injury, delivery of basic-FGF via nanoparticles as a carrier has been shown to treat sepsis-induced cardiac injury and protect cardiomyocytes from oxidative and inflammatory damage (48). Although an observational study found that basic-FGF levels were lower in sepsis patients compared to the normal population, analysis of prognosis was absent. Evidence from experimental studies supports our finding of a potential protective effect of basic-FGF in sepsis. These data suggest that basic-FGF may be a potential therapeutic target for sepsis, but further studies are warranted to confirm the underlying biological mechanisms. Specifically, large-scale clinical studies analyzing the prognostic value of basic-FGF levels in sepsis patients are needed. Additionally, more research is required to elucidate the complex immunomodulatory effects of basic-FGF in sepsis through in vitro and animal models. Gaining a better understanding of how basic-FGF regulates inflammatory pathways and mediates organ damage and recovery will be crucial in determining its viability as a sepsis treatment. Overall, basic-FGF shows promise as a therapeutic target in sepsis, but clinical and mechanistic validation is still lacking.
β-NGF, also known as nerve growth factor beta (NGFB), is a vital component of the nerve growth factor family. It is synthesized by multiple types of cells and plays a crucial role in the development, maintenance, and survival of neurons in the central and peripheral nervous systems (49). By binding to tropomyosin receptor kinase A (TrkA) and p75 neurotrophin receptor (p75NTR), β-NGF activates signaling cascades that regulate neuronal differentiation, neurite outgrowth, and nociception (50). In inflammation, β-NGF elicits hypersensitivity to pain and heat by increasing neurotransmitter release from damaged sensory neurons (51). It also stimulates endothelial cells to enhance adhesion molecule expression and facilitate leukocyte infiltration (52). Additionally, β-NGF triggers mast cell degranulation and histamine release, while activating production of inflammatory mediators like substance P and calcitonin gene-related peptide (CGRP) (53). Overall, β-NGF plays a crucial role as a mediator of neuroinflammation and development of inflammatory hyperalgesia, exerting pro-inflammatory effects on both nerve and immune cells. To date, there have been no direct studies examining the relationship between β-NGF and sepsis. However, Boucly et al. discovered that serum β-NGF levels can serve as a predictor of death or lung transplantation in patients with pulmonary arterial hypertension (54). Additionally, a study by Hepburn et al. revealed that β-NGF plays a key regulatory role in the immune response to S. aureus infection by enhancing phagocytosis and superoxide-dependent killing, stimulating proinflammatory cytokine production, and promoting calcium-dependent neutrophil recruitment (55). These findings provide indirect support for our result that genetically predicted higher β-NGF levels increase sepsis risk. This association may be due to the fact that β-NGF is connected to excessive inflammatory response and cellular damage in sepsis. Further research is still needed to directly investigate the mechanistic links between β-NGF and sepsis pathogenesis.
In summary, the role of RANTES in sepsis has produced inconsistent results in previous research. However, our MR analysis suggests that RANTES may have a protective effect, although further validation is necessary. Mechanistically, RANTES recruits immune cells to the infection sites, promotes T cell proliferation, and may counteract immunosuppression in late-stage sepsis. Future research should focus on determining its effects on inflammatory pathways in sepsis and confirming its prognostic value through extensive clinical studies. Moreover, basic-FGF shows promise as a therapeutic target, as experimental studies have demonstrated its protective effects against sepsis-associated organ damage and coagulation abnormalities. However, there is a lack of clinical research, and further work is needed to validate the prognostic value and mechanisms of basic-FGF in sepsis using human samples. Understanding its immunomodulatory functions may provide support for the potential use of basic-FGF as a treatment. Finally, it is suggested that β-NGF may contribute to excessive inflammation and cellular injury in sepsis, although there is currently no direct evidence linking it to sepsis pathogenesis. Moving forward, it is crucial for studies to investigate the direct involvement of β-NGF in sepsis development through clinical evaluations and mechanistic experiments.
This study has numerous strengths. Firstly, it employs MR to establish causal inferences while minimizing confounding bias, giving it a significant advantage over conventional observational studies. Secondly, it explores a wide range of 41 cytokines, offering a comprehensive overview of potential cytokine mediators in sepsis. Thirdly, it uncovers new connections between RANTES, basic-FGF, β-NGF, and sepsis risk, providing valuable insights into novel pathogenic mechanisms. Lastly, it proposes logical directions for future research to validate and build upon these findings.
However, it is important to acknowledge the limitations of this study. Firstly, our selection of IVs utilized a relaxed significance threshold of P < 5 × 10-6, which introduces the potential for false-positive variants and subsequent bias. However, it is worth noting that the IVs consistently demonstrated F-statistics greater than 10, indicating a weak instrumental bias is less likely. Secondly, the focus on a single ethnicity (European) limits the generalizability of our findings to other populations. Thirdly, while efforts were made to mitigate confounding, it is impossible to completely rule out the presence of pleiotropy. Lastly, we did not examine potential downstream mechanisms that could elucidate the connection between the identified cytokines and the pathogenesis of sepsis.
This MR study provides first novel evidence that genetically predicted causal association of circulating levels of RANTES, basic FGF, and β-NGF with altered sepsis risk. The findings shed light on the potential involvement of these cytokines in sepsis pathogenesis. Further experimental research is warranted to validate the observed associations and elucidate the underlying biological mechanisms linking RANTES, basic-FGF, β-NGF to sepsis development. Clinical studies with diverse patient populations are also needed to confirm the prognostic utility of these cytokines. Although requiring additional confirmation, the results contribute new insights into cytokine mediators in sepsis and suggest promising future research directions.
The original contributions presented in the study are included in the article/Supplementary Material. All data used in the current study are publicly available GWAS summary data.
SL: Conceptualization, Funding acquisition, Methodology, Writing – original draft, Writing – review & editing. XM: Data curation, Software, Visualization, Writing – original draft. WH: Software, Visualization, Writing – original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Key Project of the Affiliated Hospital of North Sichuan Medical College (2023ZD008).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1281845/full#supplementary-material
Supplementary Figure 1 | Scatter plot of β-NGF levels on sepsis.
Supplementary Figure 2 | Leave-one-out plot of β-NGF levels on sepsis.
Supplementary Figure 3 | Scatter plot of RANTES levels on sepsis.
Supplementary Figure 4 | Leave-one-out plot of RANTES levels on sepsis.
Supplementary Figure 5 | Scatter plot of basic-FGF levels on sepsis.
Supplementary Figure 6 | Leave-one-out plot of basic-FGF levels on sepsis.
GWAS, genome-wide association studies; SNPs, single nucleotide polymorphisms; IVs, instrumental variables; OR, odds ratio; CI, confdence interval; IVW, inverse variance weighting; MR-PRESSO, MR pleiotropy residual sum and outlier; CTACK, cutaneous T-cell attracting (CCL27); RANTES, regulated on activation, normal T-cell expressed and secreted (CCL5); TRAIL TNF-related apoptosis-inducing ligand; TNF-α, tumor necrosis factor-alpha; IL, interleukin; ICD, International Classification of Diseases; MCP-1, monocyte chemotactic protein-1; CGRP, calcitonin gene-related peptide; basic-FGF, basic fibroblast growth factor; β-NGF, beta-nerve growth factor.
1. Rhodes A, Evans LE, Alhazzani W, Levy MM, Antonelli M, Ferrer R, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock: 2016. Intensive Care Med (2017) 43(3):304–77. doi: 10.1007/s00134-017-4683-6
2. Rudd KE, Johnson SC, Agesa KM, Shackelford KA, Tsoi D, Kievlan DR, et al. Global, regional, and national sepsis incidence and mortality, 1990-2017: analysis for the Global Burden of Disease Study. Lancet (2020) 395(10219):200–11. doi: 10.1016/S0140-6736(19)32989-7
3. Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, et al. The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA (2016) 315(8):801–10. doi: 10.1001/jama.2016.0287
4. Oberholzer A, Oberholzer C, Moldawer LL. Sepsis syndromes: understanding the role of innate and acquired immunity. Shock (2001) 16(2):83–96. doi: 10.1097/00024382-200116020-00001
5. Moran JL, Graham PL, Rockliff S, Bersten AD. Updating the evidence for the role of corticosteroids in severe sepsis and septic shock: a Bayesian meta-analytic perspective. Crit Care (2010) 14(4):R134. doi: 10.1186/cc9182
6. Annane D, Bellissant E, Bollaert PE, Briegel J, Confalonieri M, De Gaudio R, et al. Corticosteroids in the treatment of severe sepsis and septic shock in adults: a systematic review. JAMA (2009) 301(22):2362–75. doi: 10.1001/jama.2009.815
7. Hotchkiss RS, Monneret G, Payen D. Immunosuppression in sepsis: a novel understanding of the disorder and a new therapeutic approach. Lancet Infect Dis (2013) 13(3):260–8. doi: 10.1016/S1473-3099(13)70001-X
8. Otto GP, Sossdorf M, Claus RA, Rödel J, Menge K, Reinhart K, et al. The late phase of sepsis is characterized by an increased microbiological burden and death rate. Crit Care (2011) 15(4):R183. doi: 10.1186/cc10332
9. Remick DG. Pathophysiology of sepsis. Am J Pathol (2007) 170(5):1435–44. doi: 10.2353/ajpath.2007.060872
10. Rittirsch D, Flierl MA, Ward PA. Harmful molecular mechanisms in sepsis. Nat Rev Immunol (2008) 8(10):776–87. doi: 10.1038/nri2402
11. Víctor VM, Espulgues JV, Hernández-Mijares A, Rocha M. Oxidative stress and mitochondrial dysfunction in sepsis: a potential therapy with mitochondria-targeted antioxidants. Infect Disord Drug Targets (2009) 9(4):376–89. doi: 10.2174/187152609788922519
12. Lee S, Nakahira K, Dalli J, Siempos II, Norris PC, Colas RA, et al. NLRP3 Inflammasome Deficiency Protects against Microbial Sepsis via Increased Lipoxin B4 Synthesis. Am J Respir Crit Care Med (2017) 196(6):713–26. doi: 10.1164/rccm.201604-0892OC
13. Bosmann M, Ward PA. The inflammatory response in sepsis. Trends Immunol (2013) 34(3):129–36. doi: 10.1016/j.it.2012.09.004
14. Buras JA, Holzmann B, Sitkovsky M. Animal models of sepsis: setting the stage. Nat Rev Drug Discovery (2005) 4(10):854–65. doi: 10.1038/nrd1854
15. Marshall JC. Why have clinical trials in sepsis failed? Trends Mol Med (2014) 20(4):195–203. doi: 10.1016/j.molmed.2014.01.007
16. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol (2013) 37(7):658–65. doi: 10.1002/gepi.21758
17. Emdin CA, Khera AV, Kathiresan S. Mendelian randomization. JAMA (2017) 318(19):1925–6. doi: 10.1001/jama.2017.17219
18. Davey Smith G, Holmes MV, Davies NM, Ebrahim S. Mendel's laws, Mendelian randomization and causal inference in observational data: substantive and nomenclatural issues. Eur J Epidemiol (2020) 35(2):99–111. doi: 10.1007/s10654-020-00622-7
19. Ahola-Olli AV, Würtz P, Havulinna AS, Aalto K, Pitkänen N, Lehtimäki T, et al. Genome-wide association study identifies 27 loci influencing concentrations of circulating cytokines and growth factors. Am J Hum Genet (2017) 100(1):40–50. doi: 10.1016/j.ajhg.2016.11.007
20. Zekavat SM, Lin SH, Bick AG, Liu A, Paruchuri K, Wang C, et al. Hematopoietic mosaic chromosomal alterations increase the risk for diverse types of infection. Nat Med (2021) 27(6):1012–24. doi: 10.1038/s41591-021-01371-0
21. You J, Bi X, Zhang K, Xie D, Chai Y, Wen S, et al. Causal associations between gut microbiota and sepsis: A two-sample Mendelian randomization study. Eur J Clin Invest (2023):e14064. doi: 10.1111/eci.14064
22. Hamilton FW, Thomas M, Arnold D, Palmer T, Moran E, Mentzer AJ, et al. Therapeutic potential of IL6R blockade for the treatment of sepsis and sepsis-related death: A Mendelian randomisation study. PloS Med (2023) 20(1):e1004174. doi: 10.1371/journal.pmed.1004174
23. Luo J, le Cessie S, Blauw GJ, Franceschi C, Noordam R, van Heemst D. Systemic inflammatory markers in relation to cognitive function and measures of brain atrophy: a Mendelian randomization study. Geroscience (2022) 44(4):2259–70. doi: 10.1007/s11357-022-00602-7
24. Chen H, Zhang Y, Li S, Tao Y, Gao R, Xu W, et al. The association between genetically predicted systemic inflammatory regulators and polycystic ovary syndrome: A mendelian randomization study. Front Endocrinol (Lausanne) (2021) 12:731569. doi: 10.3389/fendo.2021.731569
25. Kamat MA, Blackshaw JA, Young R, Surendran P, Burgess S, Danesh J, et al. PhenoScanner V2: an expanded tool for searching human genotype-phenotype associations. Bioinformatics (2019) 35(22):4851–3. doi: 10.1093/bioinformatics/btz469
26. Burgess S, Thompson SG, CRP CHD Genetics Collaboration. Avoiding bias from weak instruments in Mendelian randomization studies. Int J Epidemiol (2011) 40(3):755–64. doi: 10.1093/ije/dyr036
27. Bowden J, Del Greco MF, Minelli C, Zhao Q, Lawlor DA, Sheehan NA, et al. Improving the accuracy of two-sample summary-data Mendelian randomization: moving beyond the NOME assumption. Int J Epidemiol (2019) 48(3):728–42. doi: 10.1093/ije/dyy258
28. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol (2016) 40(4):304–14. doi: 10.1002/gepi.21965
29. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol (2015) 44(2):512–25. doi: 10.1093/ije/dyv080
30. Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet (2018) 50(5):693–8. doi: 10.1038/s41588-018-0099-7
31. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol (2017) 32(5):377–89. doi: 10.1007/s10654-017-0255-x
32. Appay V, Rowland-Jones SL. RANTES: a versatile and controversial chemokine. Trends Immunol (2001) 22(2):83–7. doi: 10.1016/S1471-4906(00)01812-3
33. Ness TL, Carpenter KJ, Ewing JL, Gerard CJ, Hogaboam CM, Kunkel SL. CCR1 and CC chemokine ligand 5 interactions exacerbate innate immune responses during sepsis. J Immunol (2004) 173(11):6938–48. doi: 10.4049/jimmunol.173.11.6938
34. Xie C, Wang P, Wu H, Hu X, Nie T, Li X, et al. Protective effect of the novel cyclic peptide ASK0912 on mice with sepsis induced by Acinetobacter baumannii. BioMed Pharmacother (2023) 164:114965. doi: 10.1016/j.biopha.2023.114965
35. Frimpong A, Owusu EDA, Amponsah JA, Obeng-Aboagye E, van der Puije W, Frempong AF, et al. Cytokines as potential biomarkers for differential diagnosis of sepsis and other non-septic disease conditions. Front Cell Infect Microbiol (2022) 12:901433. doi: 10.3389/fcimb.2022.901433
36. Ng PC, Li K, Leung TF, Wong RP, Li G, Chui KM, et al. Early prediction of sepsis-induced disseminated intravascular coagulation with interleukin-10, interleukin-6, and RANTES in preterm infants. Clin Chem (2006) 52(6):1181–9. doi: 10.1373/clinchem.2005.062075
37. Sugitharini V, Prema A, Berla Thangam E. Inflammatory mediators of systemic inflammation in neonatal sepsis. Inflammation Res (2013) 62(12):1025–34. doi: 10.1007/s00011-013-0661-9
38. Cavaillon JM, Adib-Conquy M, Fitting C, Adrie C, Payen D. Cytokine cascade in sepsis. Scand J Infect Dis (2003) 35(9):535–44. doi: 10.1080/00365540310015935
39. Appay V, Brown A, Cribbes S, Randle E, Czaplewski LG. Aggregation of RANTES is responsible for its inflammatory properties. Characterization of nonaggregating, noninflammatory RANTES mutants. J Biol Chem (1999) 274(39):27505–12. doi: 10.1074/jbc.274.39.27505
40. Bozza FA, Salluh JI, Japiassu AM, Soares M, Assis EF, Gomes RN, et al. Cytokine profiles as markers of disease severity in sepsis: a multiplex analysis. Crit Care (2007) 11(2):R49. doi: 10.1186/cc5783
41. Ornitz DM, Itoh N. The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip Rev Dev Biol (2015) 4(3):215–66. doi: 10.1002/wdev.176
42. Werner S, Grose R. Regulation of wound healing by growth factors and cytokines. Physiol Rev (2003) 83(3):835–70. doi: 10.1152/physrev.2003.83.3.835
43. Sörensen I, Adams RH, Gossler A. DLL1-mediated Notch activation regulates endothelial identity in mouse fetal arteries. Blood (2009) 113(22):5680–8. doi: 10.1182/blood-2008-08-174508
44. Broadley KN, Aquino AM, Woodward SC, Buckley-Sturrock A, Sato Y, Rifkin DB, et al. Monospecific antibodies implicate basic fibroblast growth factor in normal wound repair. Lab Invest (1989) 61(5):571–5.
45. Koga T, Sumiyoshi R, Furukawa K, Sato S, Migita K, Shimizu T, et al. Interleukin-18 and fibroblast growth factor 2 in combination is a useful diagnostic biomarker to distinguish adult-onset Still's disease from sepsis. Arthritis Res Ther (2020) 22(1):108. doi: 10.1186/s13075-020-02200-4
46. Pan X, Xu S, Zhou Z, Wang F, Mao L, Li H, et al. Fibroblast growth factor-2 alleviates the capillary leakage and inflammation in sepsis. Mol Med (2020) 26(1):108. doi: 10.1186/s10020-020-00221-y
47. Sun Y, Ye F, Li D, Yang H, Xu T, Zhong X, et al. Fibroblast growth factor 2 (FGF2) ameliorates the coagulation abnormalities in sepsis. Toxicol Appl Pharmacol (2023) 460:116364. doi: 10.1016/j.taap.2023.116364
48. Li X, Hong G, Zhao G, Pei H, Qu J, Chun C, et al. Red blood cell membrane-camouflaged PLGA nanoparticles loaded with basic fibroblast growth factor for attenuating sepsis-induced cardiac injury. Front Pharmacol (2022) 13:881320. doi: 10.3389/fphar.2022.881320
49. Huang EJ, Reichardt LF. Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci (2001) 24:677–736. doi: 10.1146/annurev.neuro.24.1.677
50. Otten U, März P, Heese K, Hock C, Kunz D, Rose-John S. Cytokines and neurotrophins interact in normal and diseased states. Ann N Y Acad Sci (2000) 917:322–30. doi: 10.1111/j.1749-6632.2000.tb05398.x
51. Woolf CJ, Safieh-Garabedian B, Ma QP, Crilly P, Winter J. Nerve growth factor contributes to the generation of inflammatory sensory hypersensitivity. Neuroscience (1994) 62(2):327–31. doi: 10.1016/0306-4522(94)90366-2
52. El Baassiri M G, Dosh L, Haidar H, Gerges A, Baassiri S, Leone A, et al. Nerve growth factor and burn wound healing: Update of molecular interactions with skin cells. Burns (2023) 49(5):989–1002. doi: 10.1016/j.burns.2022.11.001
53. Weihrauch T, Limberg MM, Gray N, Schmelz M, Raap U. Neurotrophins: neuroimmune interactions in human atopic diseases. Int J Mol Sci (2023) 24(7):6105. doi: 10.3390/ijms24076105
54. Boucly A, Tu L, Guignabert C, Rhodes C, De Groote P, Prévot G, et al. Cytokines as prognostic biomarkers in pulmonary arterial hypertension. Eur Respir J (2023) 61(3):2201232. doi: 10.1164/ajrccm-conference.2023.207.1_MeetingAbstracts.A2513
Keywords: Mendelian randomization, cytokines, sepsis, causality, genome-wide association study
Citation: Lin S, Mao X and He W (2023) Causal association of circulating cytokines with sepsis: a Mendelian randomization study. Front. Immunol. 14:1281845. doi: 10.3389/fimmu.2023.1281845
Received: 24 August 2023; Accepted: 05 October 2023;
Published: 17 October 2023.
Edited by:
Annalisa Marcuzzi, University of Ferrara, ItalyReviewed by:
Georgia Damoraki, National and Kapodistrian University of Athens, GreeceCopyright © 2023 Lin, Mao and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shan Lin, ZHIuc2hhbmxpbkBmb3htYWlsLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.