- 1CAS Center for Excellence in Biotic Interactions, College of Life Science, University of Chinese Academy of Sciences, Beijing, China
- 2College of Life Science, Agricultural University of Hebei, Baoding, China
Iron plays a crucial role in the biochemistry and development of nearly all living organisms. Iron starvation of pathogens during infection is a striking feature utilized by a host to quell infection. In mammals and some other animals, iron is essentially obtained from diet and recycled from erythrocytes. Free iron is cytotoxic and is readily available to invading pathogens. During infection, most pathogens utilize host iron for their survival. Therefore, to ensure limited free iron, the host’s natural system denies this metal in a process termed nutritional immunity. In this fierce battle for iron, hosts win over some pathogens, but others have evolved mechanisms to overdrive the host barriers. Production of siderophores, heme iron thievery, and direct binding of transferrin and lactoferrin to bacterial receptors are some of the pathogens’ successful strategies which are highlighted in this review. The intricate interplay between hosts and pathogens in iron alteration systems is crucial for understanding host defense mechanisms and pathogen virulence. This review aims to elucidate the current understanding of host and pathogen iron alteration systems and propose future research directions to enhance our knowledge in this field.
1 Introduction
Iron is vital for animals, plants, and microorganisms because of its redox potential. Iron is required as a cofactor for many important biological functions like mitochondrial respiration, oxygen transport, DNA repair and synthesis and the citric acid cycle. Similarly, iron is crucial for many enzymes like cytochromes, aconitases, polymerases, and oxidoreductases (1–3). Therefore, iron is maintained in a critical equilibrium by different protein regulators because both low and high levels of iron can compromise cellular activities. In normal conditions, iron is tightly regulated by absorption, recycling, and intracellular storage (4). Conversely, excessive or unused iron and other transition metals are highly toxic and damaging to cells and to vital organs as they can generate free radicals by the Fenton-Haber Weiss reaction (5, 6). Thereby, iron and other transition metals are very critically monitored to maintain healthy physiological conditions within the body.
High levels of iron have been associated with different pathophysiological conditions like neurodegenerative disorders, cancers, hormonal abnormalities, diabetes, liver and heart disease, and immune system dysfunctions (7, 8). In contrast low levels can lead to iron deficiency, anemia, compromised activation and proliferation of immune cells, and other pathological conditions, so an equilibrium state of iron is mandatory (9, 10). Around 3 to 5 gm of iron is present in normal human adults. In most organisms, the antimicrobial immune pathways and the proliferative capability of many pathogens are limited by the bioavailability of iron (11). When humans and other organisms face pathogens, their bodies’ iron is regulated very strictly by different mechanisms (12, 13) because iron availability is vital for pathogenesis. In most cases the iron level is downregulated by the host to deny available iron to the invading pathogens; in this way, pathogens are iron-starved to death (14). In some cases, the iron level is upregulated aimed to pose the toxic effects of iron to stop the pathogen’s growth (15).
Interestingly, iron also influences the host’s immune response to infection. Iron deficiency has been associated with impaired immune function, making individuals more susceptible to infections. Conversely, excessive iron levels can promote inflammation and contribute to the severity of certain infections, such as malaria (16). The initial importance of host iron withholding during infection comes to the observation in 1946 when the host iron level was significantly declined within 48 hours by Staphylococcus aureus inoculation (17). Similarly, iron sequestration and the elevation of different iron regulators have been found during the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection that causes COVID-19, a globally devastating disease (18).
As we know, iron is required for the vital activities of mammals and other organisms (1, 2, 19), therefore, in the majority of vertebrates, iron is stored intracellularly and extracellularly within storage proteins or complexed with cofactors of myoglobin or hemoglobin. In reaction to infectious agents, the host decreases the accessibility of iron in both the intracellular and extracellular spaces by utilizing iron-binding proteins and iron regulatory proteins such as transferrin (Tsf), ferritin, calprotectin, lipocalin 2 (LCN2), etc., to limit the availability of iron to invading pathogens as shown in Figure 1. In addition, to make it more difficult for invading pathogens to access host iron, some factors like neutral pH, production of iron-binding proteins and aerobic environment of serum provides additional barriers. Thereby, these numerous barriers make it very difficult for invading pathogens to survive and proliferate (20, 21), And hence, vertebrate hosts have evolved several strategies of iron limitation to prevent bacterial proliferation—a process called nutritional immunity. It is a bacteriostatic strategy of the host immune system that limits many essential metals from pathogens.
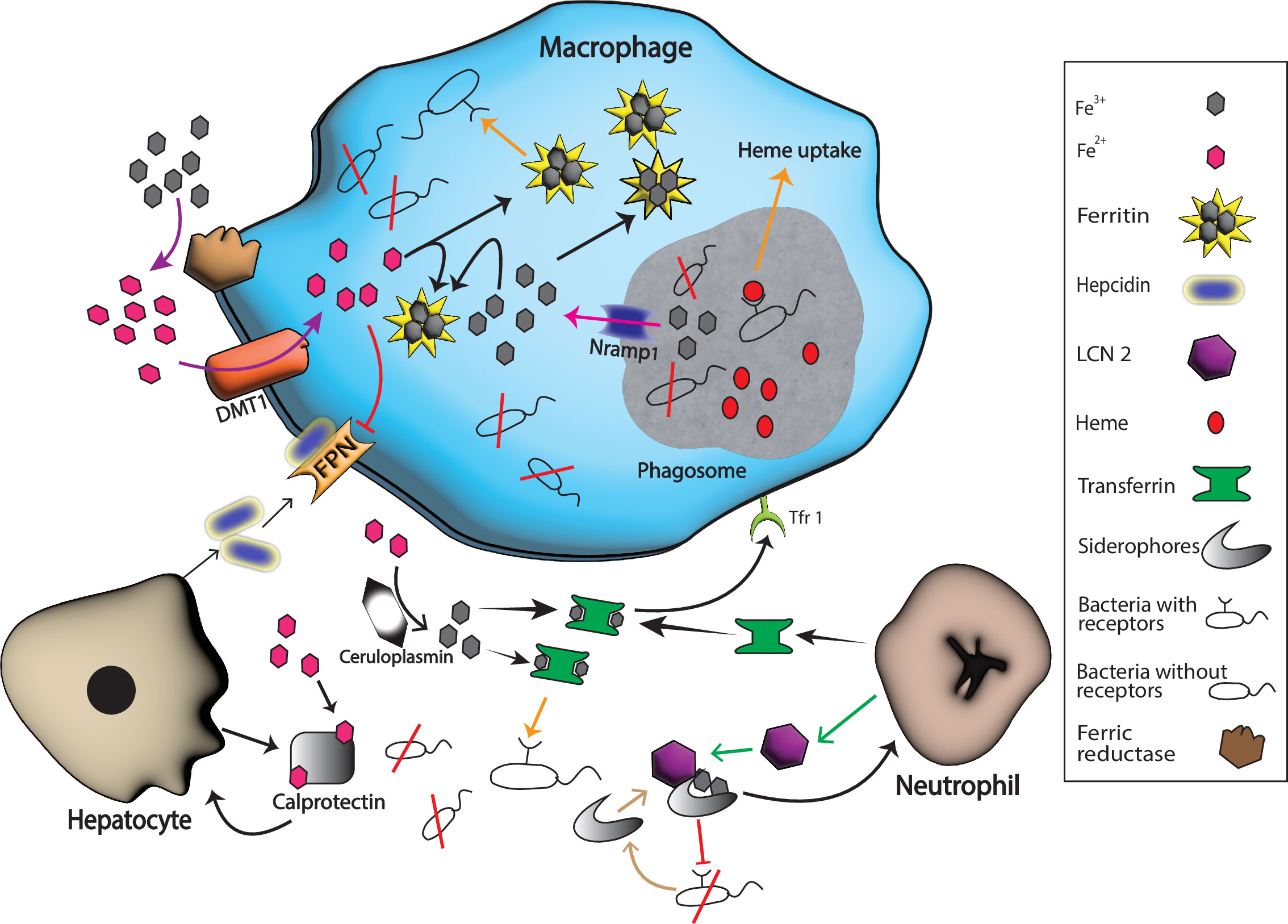
Figure 1 Host and pathogen mechanisms for iron uptake. Hosts produce various substances with a purpose to starve pathogens of iron in response to different inflammatory signals. For example, neutrophil produces transferrin and LCN 2 that binds extracellular Fe3+ and bacterial siderophores, respectively while hepatocytes boost the production of calprotectin and hepcidin. Calprotectin binds extracellular Fe2+ while hepcidin helps in the degradation of ferroportin 1 (FPN), the known iron exporter, and hence extracellular iron is limited by these actions. Nramp1 production on the surface of phagosomes helps in the export of iron form this compartment, and ferritin stores the intracellular Fe3+ that helps in the starvation of intracellular pathogens. Many bacteria escape these barriers because they have specific extracellular receptors for host iron-rich molecules that successfully bind these molecules and fulfill their need for iron for survival (represented by orange arrows). Black arrows represent host iron sequestration mechanisms, while orange arrows represent bacterial iron uptake.
However, pathogens have also evolved strategies to overcome a host’s iron restrictions. For instance, certain bacteria produce siderophores which are small molecules that scavenge iron from host proteins and transport it back to the pathogen. This allows the bacteria to acquire iron even in iron-limited environments (22, 23). Once pathogens enter the host, their survival and proliferation depend on the available host iron for completing different synthetic and metabolic reactions (24). Because of the importance of iron to both host and pathogens, there is competition for iron between host and pathogen. Meanwhile, pathogens employ various iron acquisition mechanisms, such as siderophores and specialized receptors, to obtain iron from the host, as shown in Figure 1 (11, 25, 26). In this review, some of the important impacts of iron regulators that hosts put into action during pathogen attack in an attempt to starve the pathogens to death by sequestration or transport of iron, an important trace element for the survival of pathogens, are highlighted; as are pathogens’ responsive strategies to escape this iron starvation. Understanding the interplay between these key players is crucial for developing strategies to combat iron-related diseases and infections.
2 Host iron regulation during pathogen attack
The regulation of iron plays a crucial role in the immune response during a pathogen attack. As we know, iron is an essential nutrient for both host and pathogens, so its availability can impact the outcome of an infection. Pathogens require iron for their growth and survival, while the host employs various strategies to limit its availability to pathogens, thereby restricting their ability to cause infection. During an infection, the host activates a series of defense mechanisms that aim to sequester and limit the access of iron to pathogens as follows;
2.1 Transferrin
Transferrins (Tsf) are glycoproteins found in the fluids of vertebrates and invertebrates, having an important role in iron transport and metabolism that bind two iron molecules with high affinity in the extracellular matrix and keep low concentration of iron in blood and other bodily fluids and transport it to various organs (27, 28). Tsf binds approximately all iron released to the plasma from the diet to prevent the formation of free radicals and ensure its transport to target cells (29). It is known that Tsf can inhibit the growth of certain microorganisms due to its strong affinity for iron and hence is involved in the innate immune system of different organisms by sequestering iron from them. There are three homologs of Tsf in insects, i.e. Tsf1, Tsf2, and Tsf3 (30, 31). Different insects like Drosophila melanogaster boost the production of Tsf1 when exposed to pathogenic bacteria. Tsf1 is similar to Tsf in mammals (14, 32, 33). However, there is no evidence of the defensive or immune functions of Tsf2 and Tsf3 (31, 34).
Furthermore, Tsf1 transports iron from the gut to the fat body via the hemolymph. The fat body is an important adipose-like tissue in insects that plays a crucial role in various physiological processes, including energy storage, metabolism, and immunity (35, 36). However, the mechanism of relocation of iron from the gut to the fat body needs to be understood. Recently, iron has been detected at high levels in the fat body while at a low level in other tissues during infection, and it is proposed that iron is relocated by Tsf via hemolymph from other tissues toward the fat body in a purpose of limiting the access of the invading pathogens to iron (37). However, the precise mechanism behind the Tsf1 increase cannot be determined, and additional attention is needed in exploring mechanisms contributing to its production and relocation during infection.
In mammals, there are four different types of Tsf, i.e. serum Tsf, lactoferrin (LF), melanotransferrin, and inhibitor of carbonic anhydrase (ICA) (38). Serum Tsf are present in all vertebrates while LF is present in the secretory fluids of mammals. LF can be distinguished from serum Tsf by its different sequence, its higher isoelectric point, and different functions and locations in the body (28). For instance, serum Tsf and LF are two secreted transferrins known to be involved in nutritional immunity by hiding iron to help protect the host from pathogens (39), while the immune functions of melanotransferrin and ICA are not reported. Iron is transported from the blood into the cells by serum Tsf via Tsf receptors. In addition, Tsf receptor expression is downregulated by cytokine signaling in case of intracellular infection in order to stop the import of iron-bound Tsf to cells (40, 41). There is a contrasting regulation of Tsf levels in humans and mice upon infection. Downregulation of Tsf at the site of infection in humans may be due to iron being relocated by serum Tsf to other tissues because many bacterial species can utilize human Tsf for iron acquisition (11, 42). In mice, the disparity in Tsf regulation could be attributed to the distinct pathogen-host interactions and also due to differences in murine protein (43).
Lactoferrin is the major iron-binding protein in human milk and is also found in some specific granules of neutrophils and exocrine gland secretions (44). LF plays an important role in antimicrobial activities as it lowers or stops the proliferative and adhesive capabilities of microbes (45). As LF is present in biological fluids, it lowers the iron concentration in these fluids upon infection and destabilizes the membrane of pathogens. Moreover, in our body, many proteins help regulate iron, but LF is the only regulator that acts as the first line of defense against pathogens entering the body through the mucosa (46, 47). In addition, the hepatic production of LF is increased by IFN-γ, TNF-α, and IL-6 in response to infection that results in the sequestration of extracellular iron, as shown in Figure 2 (11).
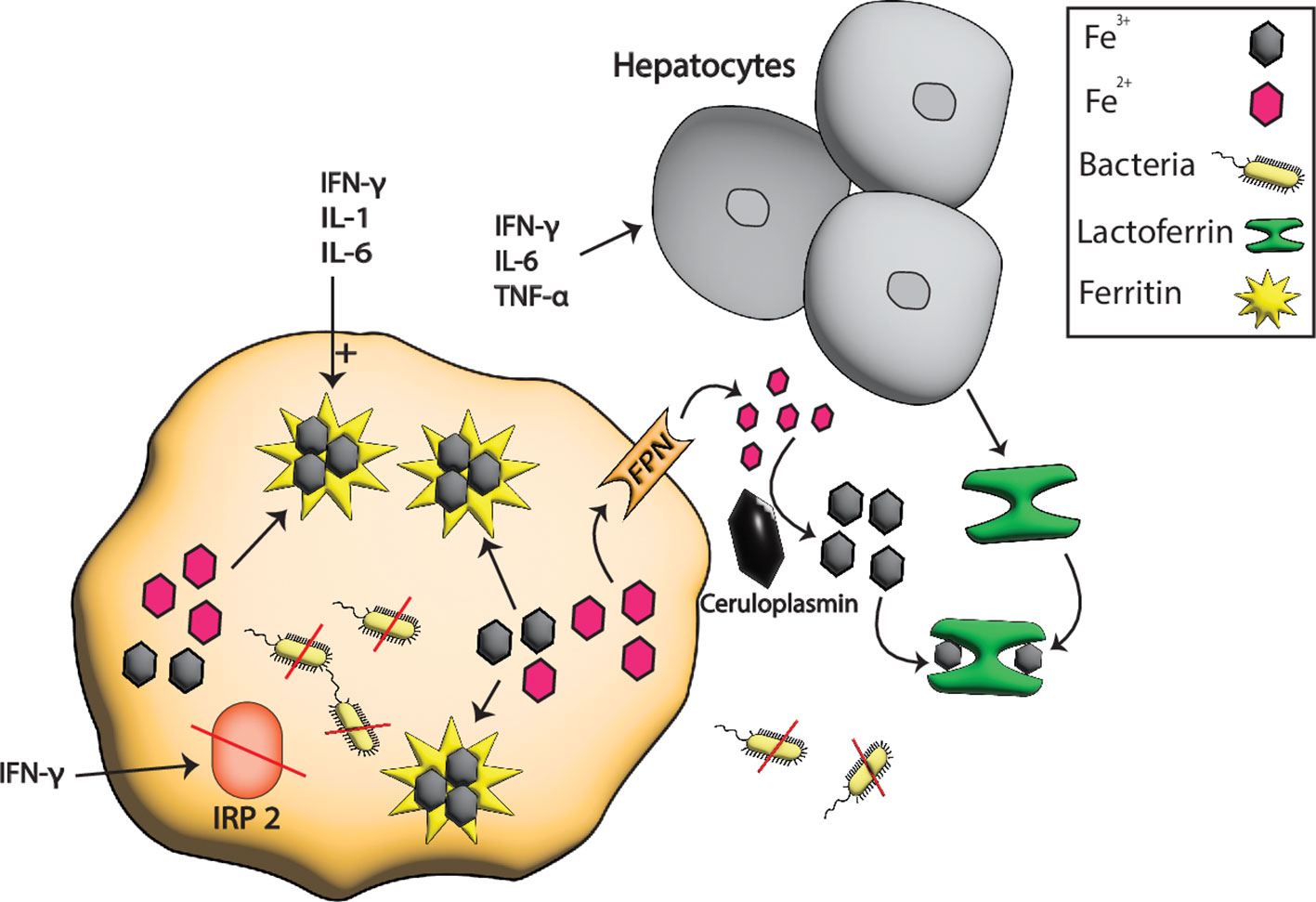
Figure 2 Iron sequestration by LF and ferritin during pathogenesis. At the host-pathogen interface, the hepatocytes boost the production of serum Tsf especially LF in response to different inflammatory signals such as IFN-γ, TNF-α, and IL-6 that sequester the extracellular iron from pathogens. In contrast, the intracellular iron is stored in ferritin whose production is boosted by TNF-α, IL-6, and IL1. IFN-γ also degrades the ferritin repressor IRP2 which further enhances the production of ferritins resulting in more iron storage and hence the starvation of intracellular pathogens due to low or no availability of iron.
Most of the iron in healthy individuals is intracellular and is bound to the cofactor of cytochromes, ferritin, FeS proteins, and heme erythrocytes (5, 48). After the destruction of erythrocytes, heme and the bounded iron are released into the circulation and are taken up by serum ferroxidase ceruloplasmin (21, 49). LF has the ceruloplasmin binding affinity and once bounded, iron transfer between them is possible. Hence, this transfer prevents the formation of free radicals and the iron acquisition by pathogenic bacteria (50). Moreover, LF can bind iron at acidic pH and can be an effective scavenger of iron in acidotic infection. This can happen in conditions such as diabetic ketoacidosis, sepsis, or certain types of tissue damage. Acidosis not only impairs the immune response but also provides a favorable environment for the growth of iron-dependent microorganisms. Therefore, LF’s ability to sequester iron under acidic conditions is especially important in combating infections in these contexts (51).
Besides the role of LF in host immunity by hiding iron, it also functions in cell differentiation and proliferation. However, the precise mechanisms through which LF influences cell proliferation and differentiation are still being investigated. It is hypothesized that LF may interact with specific cell surface receptors, triggering signaling pathways that regulate gene expression (52). In addition, it is commonly upregulated upon the infection of many microorganisms, so it can be used as a biomarker of infection (53). The iron scavenging activity of LF also has a significant role in different neurodegenerative diseases (5, 54, 55), inflammatory diseases (56), and allergic responses (57). Due to the emergence of multidrug-resistant (MDR) bacterial strains, much research has diverted to using innate molecules as alternatives. Interestingly, LF shows no resistance and has antimicrobial potential against staphylococcus infection and mucormycosis through iron chelation (53, 58, 59), so it can be exploited in combination with other antibiotics as an anti-infective agent.
2.2 Ferritin
Ferritins are complex three-dimensional hollow spherical structures that accommodate large quantities of iron in a nontoxic and bioavailable form in its hollow center. They have a mol. wt of about 450 kDa and can store around 4500 ferric iron (Fe3+) atoms in their cores. Ferritins are considered as major iron storage protein that is found in plants, animals, and prokaryotes (60, 61). Ferritins, a type of iron-binding protein complex, have been extensively studied for their crucial role in various physiological processes. Recent research has provided evidence to support the significant involvement of higher levels of ferritins in immunity, inflammation, signal transduction, and angiogenesis. By binding and sequestering excess iron, ferritin prevents pathogens from accessing this essential nutrient, limiting their ability to establish an infection. This mechanism is particularly important in the initial stages of infection when the immune system is still mobilizing its defenses (62, 63).
Ferritins are found in both insects and mammals, where they exhibit a common structure consisting of one heavy chain (FTH) and one light chain (FTL). The FTH and FTL serve distinct functions, contributing to the oxidation and nucleation of the iron core, respectively (60, 64). Whereas in plants and bacteria only FTH is present, which helps in the oxidation of only two iron atoms (65). FTH is responsible for detoxification by ferroxidase activity that converts toxic ferrous iron (Fe2+) to nontoxic Fe3+. Thereby, this activity of FTH helps ferritin sequester iron in its hollow core as hydrous ferric oxide (66). While FTL subunits function in iron nucleation, mineralization, and ferritin protein stability. Together, these chains ensure the proper regulation and storage of iron, safeguarding cellular function and preventing oxidative damage (67). Moreover, FTH expression abnormally increases in different tissues during malignancies and inflammation (68–70).
Ferritin plays an important role in host nutritional immunity, inflammation, and hypoxia (63). In humans and other mammals’ oral cavity, ferritin expression is very high because the oral cavity is exposed to different bacterial species that depend on iron for proliferation, and hence the increased expression of ferritin to sequester iron from bacteria (71, 72). Depending on the context, Ferritin can act as a pro-inflammatory and anti-inflammatory molecule. It can promote inflammation by inducing the production of inflammatory cytokines and chemokines, which attract immune cells to the site of infection. This helps to mount an effective immune response against invading pathogens (73). Conversely, ferritin can also exert anti-inflammatory effects by sequestering iron and preventing its participation in oxidative stress reactions. Iron-mediated oxidative stress can exacerbate inflammation and tissue damage. By binding excess iron, ferritin helps to reduce oxidative stress and limit inflammation, thereby promoting tissue repair and resolution of the immune response (74).
During inflammatory or pathological conditions, ferritin expression is stimulated by pro-inflammatory cytokines such as tumor necrosis factor alpha (TNF-α), interleukin 1 (IL-1), and interleukin 6 (IL-6) through nuclear factor (NF)-κB pathway. In addition, interferon-gamma (IFN-γ) and lipopolysaccharide degrade iron-responsive protein 2 (IRP2), a repressor of ferritin expression, via a nitric-oxide-dependent pathway that initiates the expression of ferritin in macrophages as shown in Figure 2 (70, 75, 76). Moreover, IL-6 is also an important player in pathological responses because it enhances the expression of FTH and FTL in the hepatocytes (77). All these signaling elevate the level of ferritins that bind and sequester intracellular iron, that lowers the iron level, which is then scarcely available for pathogens.
Ferritin heavy chain is expressed mostly against inflammatory responses while FTL is sensitive to elevated iron levels (78). Ferritins/FTH are also highly expressed in glial cells in mice and Drosophila to protect neurons from iron-mediated ferroptotic damage. Ferritin provides an antioxidant defense system to protect neurons from iron-mediated cytotoxicity (79). The main source of ferritin secretion in insects are intestinal cells and is abundant in the fat body, midgut, and hemolyph (37, 80). The synthesis and storage of ferritin differ between mammals and Drosophila. In mammals, ferritin primarily originates from hepatocytes or in case of infection from macrophages (60). Recent studies have provided evidence supporting the extraction of small quantities of iron from ferritin by Enterobacteriaceae under conditions of oxidative stress regulation by siderophore-independent mechanisms, enough to promote bacteria survival. However, the exact mechanisms behind this process are still being investigated (81). Therefore, understanding the complex mechanisms by which ferritin participates in these biological processes is crucial for developing targeted therapies and interventions. It was also hypothesized that ferritin can act as an iron exporter and cells can secrete ferritin. The expression of many receptors on the cell surface like Tsf receptor 1, SCARA5, and TIM2 further support this hypothesis. The expression of these receptors are also used as an effective drug delivery system (82, 83).
2.3 Hepcidin and ferroportin 1
Hepcidin, initially identified as a cysteine-rich antimicrobial peptide, has been found to play a crucial role in iron regulation during inflammation and infection. It is primarily synthesized in hepatocytes and released from the body through urine, where it was first discovered (84). Hepcidin is also synthesized in other parts, including heart, adipose tissue, kidney, liver, myeloid cells, monocytes, and splenic macrophages (85). This peptide exhibits dual functions, acting as both a potent antimicrobial agent and a key regulator of iron homeostasis. Its significance lies in its ability to modulate iron metabolism in response to various physiological and pathological conditions (86). By inhibiting iron transport across the intestinal epithelium and blocking iron release from macrophages and hepatocytes, hepcidin effectively limits the availability of iron during periods of inflammation and infection. This regulation helps prevent the growth and proliferation of pathogenic microorganisms that depend on iron for survival (87). Hepcidin also plays a crucial role in the pathogenesis of iron-related disorders such as anemia of inflammation and hereditary hemochromatosis (88, 89). Understanding the intricate mechanisms of hepcidin synthesis and its regulatory functions is of great importance in the field of iron metabolism and holds promise for the development of novel therapeutic strategies for various iron-related disorders.
Hepcidin is a very important iron regulator found only in vertebrates. No data related to hepcidin-iron regulation is available in other organisms including insects. However, in insects, divalent metal transporter-1 (DMT1) homolog Malvoli, multicopper oxidase-1, and ferritin have been found to play crucial roles in iron import, export, and storage (90–92). It is reported that Salmonella typhimurium infection induces hepcidin formation via the estrogen-related receptor γ (ERR), thereby counteracting nitric oxide (NO) and nuclear factor erythroid 2-related factor-2 (Nrf2) mediated iron export from macrophages exerting antimicrobial effect on S. typhimurium. Conversely, it also results in an increased supply of the metal for intracellular microbes (93, 94). Induction of hepcidin also results in the development of hypoferremia and eventually, anemia which are well-known mechanisms occurring upon inflammation and infection (89). Interestingly, a study conducted on fish also reported the induction of hepcidin in response to infection and inflammation. In this particular study, fish were infected with Streptococcus iniae, a bacterium known to cause infections in aquatic animals. The researchers found that the level of hepcidin mRNA increased significantly by a staggering 4500 times (95).
The primary mechanism against infection and inflammation involves the activation of macrophages. With the activation of macrophages, pro-inflammatory cytokines are released, especially IL-1 and IL-6 which induces the production of hepcidin. In addition, myeloid cells can also induce hepcidin production via the activation of TRL4 receptors present on macrophages and neutrophils during inflammation, leading to iron modulation and starvation of infectious agents (96). The increased hepcidin binds FPN, a well-known cellular iron exporter, and is then internalized and degraded by lysosomal machinery (86, 97). With the reduction in FPN due to hepcidin-aided degradation, iron export from cells to plasma reduces, limiting extracellular iron to pathogens, as shown in Figure 3. This limited iron export has important implications in the context of pathogen control, as it restricts the availability of extracellular iron to invading pathogens.
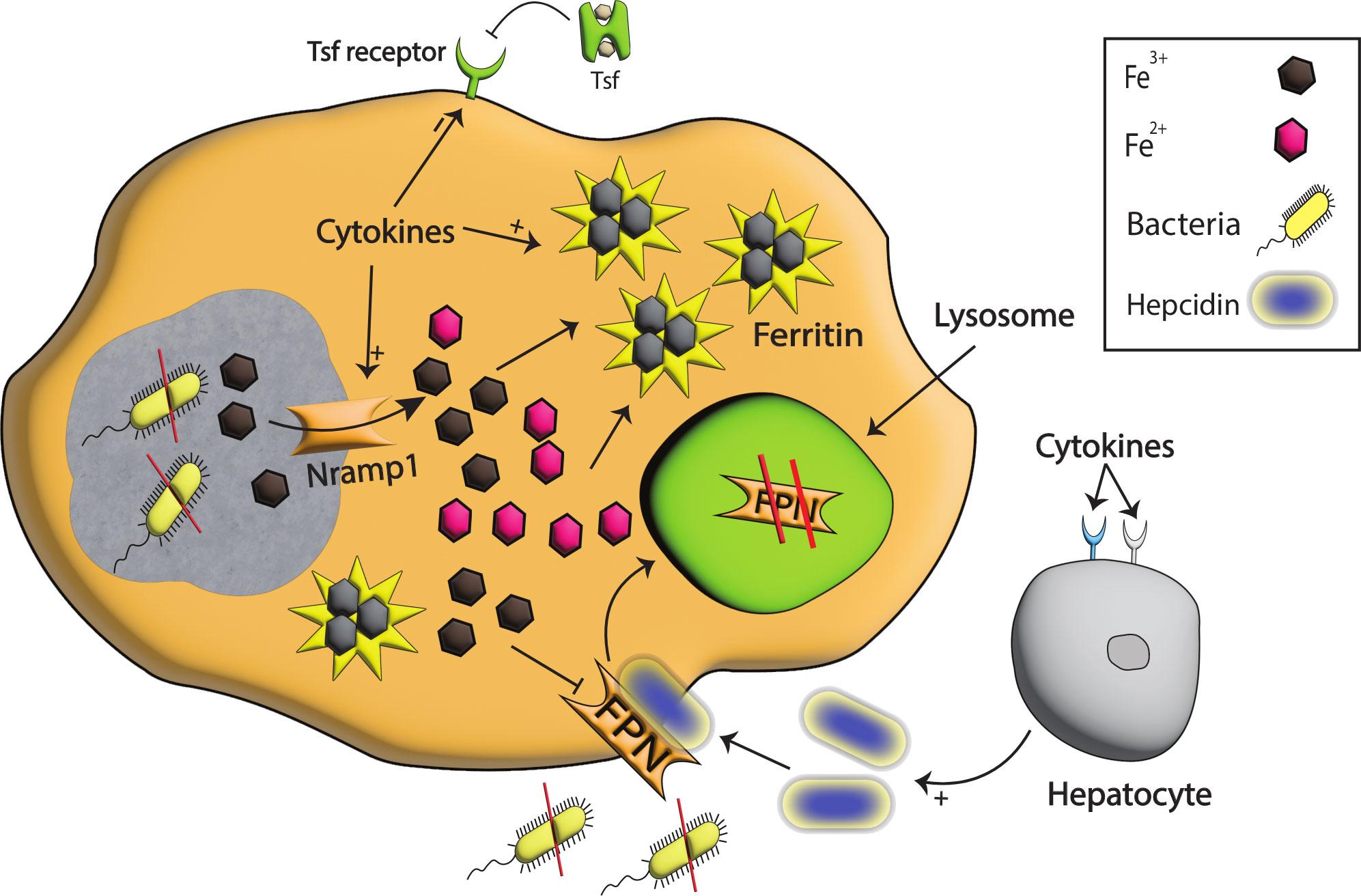
Figure 3 Iron modulation by Nramp1, Hepcidin, and FPN during infection. With the detection of bacterial species, pro-inflammatory cytokines are produced that signal hepcidin production. The binding of hepcidin to FPN results in the degradation of FPN in the lysosomes, and as a consequence iron egress from the cell is reduced, and extracellular pathogens are starved. Pro-inflammatory cytokines also enhance the production of Nramp1, a well-known exporter of iron on phagosomes that helps export iron from phagosomes, stored intracellularly in ferritin, and in this way, intracellular pathogens are starved to death. Moreover, the tsf receptor on the cell surface is reduced, restricting iron import to cells and further limiting intracellular iron availability.
2.4 Nramp1
The natural resistance-associated macrophage protein (Nramp) family contains evolutionarily conserved divalent metal ion transporters, which play important roles in regulating intracellular divalent ion transport and can be found in animals, plants, and bacteria. In animals, Nramp proteins are present in different tissues and organs. They are particularly abundant in cells of the immune system, where they are involved in the transport of divalent metal ions across cellular membranes. This is important for the proper functioning of immune cells, as divalent metal ions are essential for many immune processes (98–100). Nramp1, a member of the Nramp family, is crucial in protecting against certain intracellular pathogens. This protein is also being significant in regulating iron levels during host-pathogen interactions. Notably, Nramp1’s mRNA expression is significantly upregulated in polymorphonuclear leukocytes and macrophages in humans when exposed to inflammatory signals. This suggests that Nramp1 is actively involved in enhancing innate immunity responses. Its ability to respond to inflammatory signals and its high expression in key immune cells highlights the importance of Nramp1 in the host’s defense against pathogens (101, 102).
In addition, Nramp1 mutation is associated with a significant increase in susceptibility to a range of intracellular pathogens, including Salmonella, Leishmania spp., and Mycobacteria. This heightened susceptibility can be attributed to the loss of function of Nramp1, which leads to the increased availability of iron and other divalent metals to these pathogens (103–105). In human, Nramp1 is required for resistance against intracellular bacteria especially those residing inside phagosomes by restricting the availability of Fe2+ and Mn2+. For instance, pro-inflammatory cytokines and pattern recognition receptor (PRR) production decrease the production of transferrin receptors on phagocytes which in turn enhance the expression of Nramp1. As a result, iron is exported to the cytoplasm, leading to iron starvation of pathogens in this compartment as shown in Figure 3 (106). Moreover, macrophages are also responsible for the phagocytosis of red blood cells which contain a high quantity of iron, and this iron is transported from phagosomes by Nramp1 after phagocytosis (107). The expression of Nramp1 also induces lipocalin 2 and nitric oxide production that are linked with iron-mediated immunity (108, 109).
Nramp 2 known as DMT1 helps uptake iron across the brush border membrane of intestinal epithelial cells (110). DMT1 also helps combat pathogens as its expression elevates during viral infection (111). Recent studies have shown the role of DMT1 in the immune response of sea cucumbers. Upon bacterial invasion, sea cucumbers exhibit a significant increase in DMT1 expression. This suggests that DMT1 is crucial in modulating iron metabolism as part of the defense response (112). In addition, there is recent evidence on the role of DMT-1 in bacterial infection. In Salmoenlla infection DMT-1 expression controlled iron delivery to intracellular bacteria and induced the siderophore scavenger lipocalin-2 (113). However, the upregulation of DMT1 expression indicates its importance but further studies are required to investigate the precise mechanisms by which DMT1 modulates iron metabolism and its impact on bacterial iron starvation. Such knowledge will not only provide insights into the host defense strategies but may also have implications for the development of novel therapeutic approaches targeting bacterial infections.
In plants, Nramp family proteins are located on the plasma membrane, and they help in uptaking iron from the soil, especially during iron deficiency (99, 114). Plant Nramps exhibit sequence similarity with mouse Nramp1. It has been observed that some of the Nramp genes play a role in plant immunity. In the case of Arabidopsis thaliana, the AtNramp3 gene is upregulated in leaves following infection. However, it remains unclear whether AtNramp3 is involved in regulating iron homeostasis during infection. Further research is needed to determine the specific role of AtNramp3 in this context (115). Nramp1 homologs have been identified in many bacterial species like Escherichia coli and Mycobacterium tuberculosis and have been described as divalent metal iron transporters (116, 117). While malvolio is a Nramp1 homolog in insects and functions in the absorption of dietary iron (118, 119), no data is present about iron regulation by malvolio during infection.
2.5 Lipocalin-2
Lipocalin-2 (LCN2) plays a central role in protecting and regulating against intracellular infection by scavenging iron (120). LCN2, also known as neutrophil gelatinase-associated lipocalin (NGAL) or siderocalin, belongs to bacteriostatic factors, produced by neutrophil secondary granules and later also reported to be synthesized by macrophages and epithelial cells (121, 122). It has been found to possess bactericidal, anti-stress, and anti-inflammatory effects, making it a crucial component of the immune response. Its expression is induced by pro-inflammatory cytokine signals, i.e. TNF-α, IFN-γ, IL-1β, IL-17, NF-κB, and JAK-STAT signaling pathways (123, 124).
While LCN2 has various functions, it is best characterized by its ability to inhibit the proliferation of bacteria that rely on siderophores for host iron uptake. Siderophores are specialized proteins produced by bacterial species during infection. These proteins have a high affinity for ferric iron, allowing them to scavenge this essential nutrient from the host (125, 126). However, LCN2 acts as a potent defense mechanism against siderophore-dependent bacteria. It binds to the siderophores, preventing them from delivering iron to the bacterial cells. This effectively limits iron availability for bacterial growth, inhibiting their proliferation and survival within the host, as shown in Figure 4. Because of its significant contribution and production, LCN2 has been used as a biomarker in anti-bacterial and anti-inflammatory responses (127, 128).
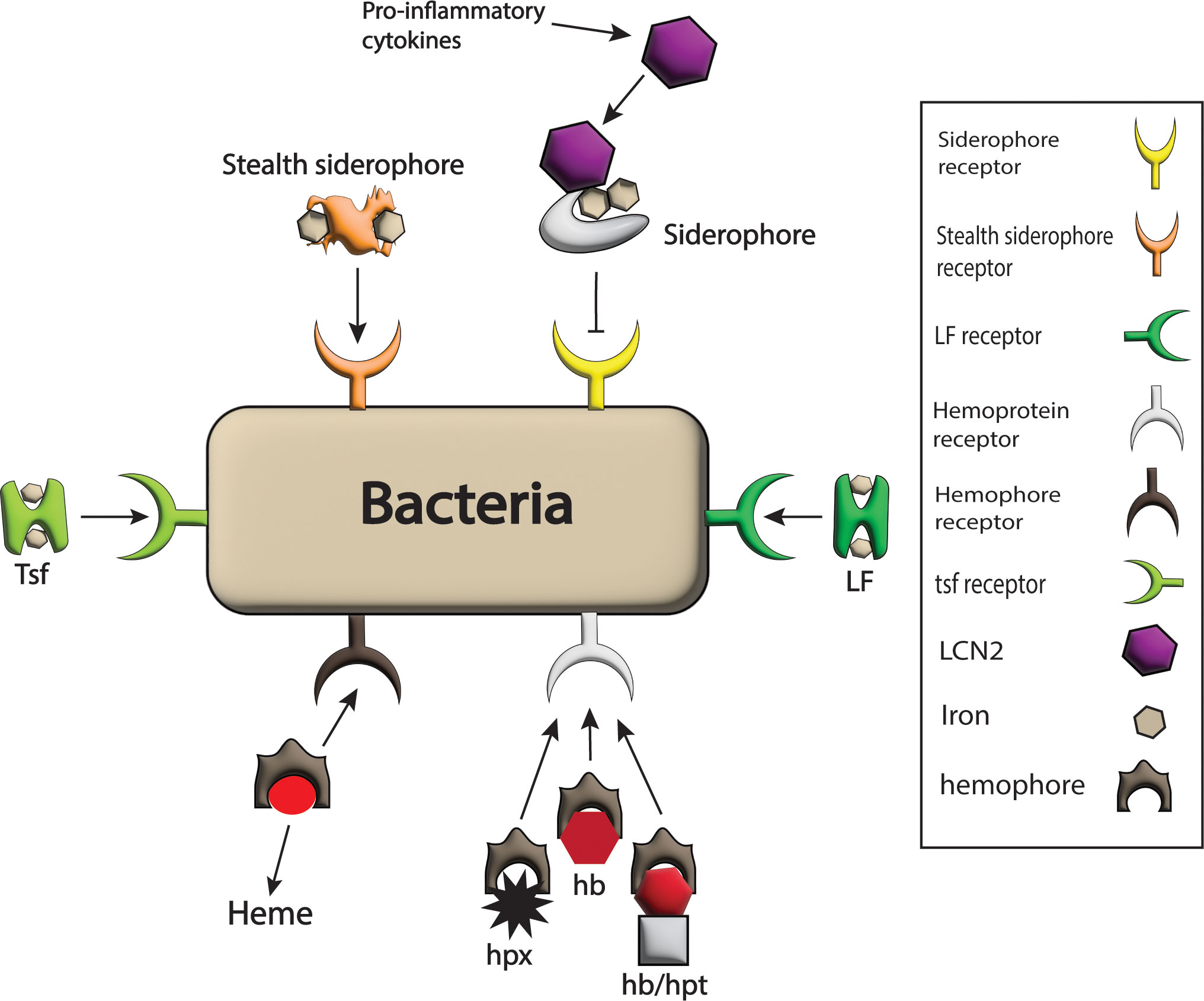
Figure 4 Bacterial strategies against host barriers for iron uptake. The bacterial system expresses different receptors on its surface such as LF receptor, hemophores receptor, Tsf receptor, stealth siderophores receptor, siderophores receptors, and hemoprotein receptor for the acquisition of iron-rich proteins like LF, heme, Tsf, stealth siderophores, siderophores and hpx, hb and hb/hpt complex respectively and hence iron requirements of pathogens are fulfilled. Here only siderophores are blocked from bacteria by the host by the production of LCN2 in response to pro-inflammatory cytokines that bind siderophores.
Accordingly, LCN2 knock-out mice are more susceptible to enterobactin producing bacterial infections (129, 130). Moreover, LCN2 can stop the proliferation of M. tuberculosis in vitro and cultured cell lines of macrophages (131, 132), suggesting its role in iron regulation in infection. In animals, one of the crucial molecules 2,5 dihydroxbenzoic acid (2,5-DHBA), which is homologous to siderophores in structure and is being scavenged by some bacterial species, plays an important role in iron homeostasis. During bacterial invasion, this molecule is also downregulated by TLR-mediated pro-inflammatory signaling (133, 134), but the role of 2,5-DHBA downregulation in the innate immune response is still unclear. However, due to the homology of 2,5-DHBA with siderophores, there is a possibility that during bacterial invasion, LCN2 binds 2,5-DHBA to prevent its acquisition by bacteria. Moreover, further research is needed to fully understand the implications of 2,5-DHBA downregulation and the role of LCN2 in the innate immune response. The exact mechanisms by which LCN2 interacts with 2,5-DHBA and its impact on bacterial invasion are still not precise.
2.6 Calprotectin
Calprotectin (CT) is a vital component of the innate immune response due to its ability to employ a strategy involving bivalent metal ions withholding. CT is a host-defense bactericidal protein that is primarily expressed by neutrophils and epithelial cells during bacterial invasion, making it an important player in the host defense system (135). CT, previously known for its role in scavenging zinc and manganese during microbial invasion, has recently been found to have a broader function in sequestering Fe2+ from extracellular pathogens (136, 137). The role of Fe2+ in the host innate immune response has been overlooked, but studies have shown its abundance at the infection site (138). Notably, CT at an infection site poses an innate immune response by taking Fe2+, which has recently attracted great attention (139).
Calprotectin is advantageous to the host in response to those pathogens that have the capability of up-taking Fe2+ for survival. CT is the only known Fe2+ scavenging protein released by neutrophils at the site of infection. CT is also a very critical host metal ions binding protein having two sites for the acquisition of divalent metal ions for antimicrobial effect (139, 140). Some studies have demonstrated that CT can also be used as a biomarker for the diagnosis of different inflammatory and bacterial infections (141–143). It’s proposed that CT and LF/Tsf are part of the first line of host defense against microbial infections and help to limit bacterial multiplication, which is completed at least partly by binding divalent metals like iron to deprive the pathogen (39, 139).
The role of CT in Fe2+ sequestration during infection has been questionable for a long time in nutritional immunity, because Fe2+ is very unstable under aerobic and oxidative conditions and converts to Fe3+ and CT has shown negligible affinity to Fe3+. The precise mechanism by which calprotectin withholds Fe2+ is still under investigation. However, it is believed that CT binds to free iron ions in the extracellular environment, forming stable complexes that are inaccessible to microorganisms (139, 144, 145). In addition, CT has the capability of maintaining Fe2+ oxidation state and can convert Fe3+ to Fe2+ in aerobic conditions (146). These uncertainties have been tackled in a study conducted on the opportunistic infectious bacteria Pseudomonas aeruginosa which has very high Fe2+ requirements. This study shows CT-mediated Fe2+ sequestration in both aerobic and anaerobic environments that hinder P. aeruginosa proliferation (147). Due to the vital role of CT in infection and inflammation by scavenging Fe2+, understanding the complex roles of CT and its potential therapeutic applications in the elimination of Fe2+-dependent pathogens is crucial in advancing our knowledge in the field of host-pathogen interactions.
2.7 Ceruloplasmin
Ceruloplasmin (CP) is a ferroxidase belonging to the multicopper oxidase (MCO) family that stores and carries copper in blood and also functions in iron metabolism, mainly synthesized in hepatocytes and to a lesser extent in lymphocytes and macrophages. CP acts as ferroxidase converting Fe2+ to Fe3+ which is essential for its incorporation into Tsf, a major iron regulator in nutritional immunity. Both lower and higher levels of CP are linked with different kinds of body disorders (148, 149), and some studies have found upregulation of CP during bacterial invasion (150), suggesting it plays a role not only in iron metabolism but also in iron-mediated nutritional immunity. However, understanding the precise molecular mechanisms by which CP modulates iron-mediated immunity is crucial. This could involve investigating the interaction between CP and immune cells, as well as identifying the signaling pathways involved in ceruloplasmin-mediated immune regulation. Moreover, in insects MCO1 is thought to be involved in gut immunity while it also has been reported to function in the oxidation of Fe2+ to Fe3+ (92, 151) which may have a close association with nutritional immunity by iron sequestration that needs further investigation.
2.8 Haptoglobin and hemopexin
Haptoglobin (hpt) and hemopexin (hpx) are produced in the liver that binds hemoglobin and heme, respectively. These iron-rich proteins are cleared by macrophages from the circulation by phagocytosis, or these complexes are transported to the liver, and the iron is recycled. Hemoglobin and heme in the free state are toxic to tissues;however, they are safe and protected from bacterial uptake when complexed with hpt and hpx (152). While hpt and hpx are primarily known for their involvement in hemoglobin and heme binding, their potential roles in iron transport and distribution within the body warrant investigation. Understanding how these proteins contribute to iron delivery to specific tissues and cells could have implications for the development of targeted therapies for iron-related disorders. Since host iron regulation is very strict and free iron is very roughly available for the invading pathogens, heme is being utilized by some bacterial pathogens to acquire iron (153, 154). Thereby, the expression of hpt and hpx is induced by interleukin-22 (IL-22) at the infectious site, and the induced hpx is recognized to limit heme iron availability to the infectious bacteria E. coli. Still, knowledge about hpt iron regulation in this regard is limited (155). However, hpt lowers iron and hemoglobin levels during S. aureus septic model induction and that has a significant effect on survival, though in vivo studies are limited that can unravel the underlying mechanisms of hemoglobin regulation during pathogen interface (156).
2.9 Iron regulatory proteins
Iron regulatory proteins (IRPs) have been explored in the setting of infection. Altered IRP1 and IRP2 expression have been first described upon scrapie infection in the brain of mice (157). Subsequently, Trichomonas vaginalis attracted interest because it can manipulate iron delivery by an iron regulatory binding protein homolog (158). Lastly, IRPs were shown to restrict bacterial iron access and promote iron scavenging via LCN2 in macrophages (159). In iron overload and limited conditions, IRPs detach or bind iron-responsive elements (IRE) on key genes involved in iron metabolism, and thus reduce iron export/import, utilization, and storage according to the underlying pathophysiological conditions or body needs (160, 161). Similarly, IRP1 has been reported to induce the production of Tsf receptors in host cells when challenged with Toxoplasma gondii. This indicates that IRP 1 plays a significant role in host innate immunity by modulating iron availability and potentially restricting the parasite’s growth. Further research in this area will help deepen our understanding of the host-parasite interaction and may lead to new interventions against T. gondii infection (162).
In short, all the mechanisms at cellular levels are post-transcriptionally under the control of IRP 1 and IRP 2 proteins. In inflammation, infection, iron-limited or excess conditions these proteins help regulate the key genes involved in iron homeostasis. Iron is an essential nutrient for host cells and invading pathogens (163). During infection, hosts put forward different mechanisms to deprive pathogens of iron. But pathogens also have evolved different mechanisms to circumvent the host barriers in order to snatch iron from the host.
3 Bacterial iron acquisition against host barriers
Iron is an essential nutrient for the growth and survival of bacteria, but it is often tightly regulated and sequestered by the host as a defense mechanism. In mammals, strict iron regulation leads to minimal levels of free iron in both intracellular and extracellular spaces. Notably, many pathogens have a high dependency on iron for survival and pathogenicity; therefore, they have developed different mechanisms to acquire iron from the host or steal it from host deposition sites (164). Many sites of iron in the host are potentially available for invading pathogens. For instance, pathogens have evolved mechanisms to acquire iron from heme, hpx, hemoglobin, LF, and ferritin, as shown in Figure 4.
Many pathogenic microorganisms have evolved the ability to produce siderophores to acquire iron from the host. Different bacteria produce different and very specific siderophores via varying iron binding capacities which are selectively ingested via specific receptors. Siderophores serve as high-affinity iron chelators that scavenge iron from the host’s iron-binding proteins or other sources (165, 166). Special transcription repressors control siderophores expression called ferric uptake regulator (FUR) that binds siderophores genes in iron excess conditions consequently repressing siderophores production. Conversely, under iron-deficient conditions, FUR is inhibited from binding to the DNA binding sites of siderophores genes which results in the expression of siderophores (167, 168). By utilizing siderophores, pathogens gain a competitive advantage in acquiring iron, which is often limited in the host’s environment. This iron piracy mechanism allows them to evade the host’s immune system and proliferate within the host. Siderophores are not only produced by bacteria, but fungi and plants can also produce them during low iron availability (169, 170).
During infection, thehost tightly regulates iron concentrations by producing different iron regulators to prevent its utilization by pathogens (171). Bacteria sense such low iron concentrations, and in response, siderophores are produced to acquire iron with a higher affinity than that of host-produced products. More than 500 siderophores different in structure have been identified. Once siderophores bind Fe3+, the complexes are then reclaimed by bacterial cells, where they are docked to the special surface receptors called Fep proteins and internalized for utilization by different mechanisms depending on the type of bacteria (172–174). The siderophores produced by one bacteria might not get back to the same bacteria and hence might not be advantageous to the producer. This may be advantageous to other bacteria of the same species or species with the same siderophores requirements (22). These mechanisms of bacterial siderophore production and acquisition to hijack host iron systems require clearer understanding.
Catecholates, hydroxamates, and carboxylates are the three main siderophore families classified based on structure (175). Conversely, to protect from iron thievery by siderophores, especially catecholates, the host produces LCN2 that can bind and acquire iron from these siderophores (Figure 4) (125). However, many bacteria have evolved mechanisms to evade LCN2 inhibition either by expressing stealth siderophores or competitive antagonists that augment the microbial pathogenicity by iron acquisition (166, 169). Additionally, siderophores also scavenge other metals like zinc and copper, protecting some bacteria against oxidative stress (176). Understanding the mechanisms of siderophore production and iron acquisition by pathogens is crucial for developing strategies to disrupt this process and combat infections. Researchers are exploring approaches such as developing siderophore analogs or inhibitors that can disrupt iron uptake and render pathogens more susceptible to host defenses or existing antibiotics. For instance, a novel antibiotic called cefiderocol is already in clinical use, that binds extracellular free iron and is taken up via the siderophore transporters thereby circumvention potential resistance pathways in gram negative bacteria (177). Similarly, by targeting siderophore-mediated iron acquisition pathways, it may be possible to develop novel therapeutic interventions against infectious diseases.
Another mechanism by which some bacteria uptake host iron is by the production of hemophores. Hemophores are specialized proteins that can bind free heme, hpx, hb, and hemoglobin/haptoglobin (hb/hpt) complex. The bacterial cell surface has specialized receptors that can recognize all these substrates, where the complexes are internalized and degraded for the liberation of iron. Heme, being cytotoxic, is accessed by many bacteria by producing exotoxins that can lyse erythrocytes, and the heamoglobin-bound heme is then released (24, 178). In some bacteria heme and heme-associated proteins are directly attached to the bacterial surface, where some specialized proteins internalize them and iron is extracted (164, 179). Though most intracellular and extracellular pathogens utilize the same mechanisms, some intracellular pathogens employ different mechanisms to hijack a host’s iron. For instance, S. enterica and M. tuberculosis reduce iron export by downregulating FPN expression which allows more intracellular iron for these bacteria (160, 180). In addition, Francisella tularensis expresses cell receptors called Tsf receptor 1, FupA, and FslE that successfully acquire iron from Tsf, direct binding of Fe2+,and siderophore-mediated Fe2+ uptake respectively (181). Another mechanism was observed in a recent study which shows that Leishmania donovani can cleave poly(rC)-binding proteins that load iron in ferritin, in macrophages which results in lower loading of iron in ferritin and subsequently its higher availability to the pathogen (182). Ehrlichia chaffeensis, the causative agent of human monocytic ehrlichiosis, induces ferritinophagy by producing a protein called Etf-3, which increases the cellular labile iron pool for its proliferation (183).
In addition, some infectious bacterial species have specific membrane receptors, capable of directly binding Tsf and LF which are the key players of nutritional immunity. After binding, iron ions are extracted from Tsf and LF by these special membrane proteins, shifted to periplasm, and then bounded by ferric binding proteins and transported to the cytoplasm where they are utilized by the bacterial cells (42, 184). By employing these strategies, pathogens successfully acquire iron from different sites within the host, ensuring their survival and enhancing their pathogenic potential. Understanding these mechanisms provides valuable insights into the host-pathogen interaction and can open avenues for developing targeted approaches to disrupt pathogen iron acquisition, potentially mitigating infections and related diseases.
4 Conclusion
In conclusion, iron plays a multifaceted role in infection. Both bacteria and host have mechanisms to acquire and withhold iron, respectively. In this iron battle, the host efficiently starves and eliminates a number of pathogens; however, some bacteria use more than one system for iron uptake, e.g. the production of different types of siderophores gives some bacteria better chances of survival than others (165). We need a better understanding of how immune cells interact with each other and with pathogens to learn how to modify the iron metabolism in a way that will help us to manipulate host responses to infection and pathogenicity. Moreover, by which pathways host restrict iron availability to extra- and intracellular pathogens are poorly understood. The alteration in either host or pathogen iron sequestration and acquisition pathways pharmacologically may hold key for discovering new approaches that will help prevent or treat infections.
In recent years, significant advancement has been made in developing therapeutic and preventive measures by studying the interconnections between host and pathogens. Taking control over iron is a central part of infection, thus controlling this element could influence the infection in favor of host. For this purpose, siderophores can be modified to be used as iron chelators, or the currently used iron chelators can be improved, aiding the immune system for pathogens’ starvation. Recently, siderophores have been actively exploited as antibiotic carriers to pathogenic bacterial cells, however further investigation is required to enhance the efficacy of this system. Furthermore, all types of siderophores need exploitation to be used as an effective antibiotic-siderophore conjugates delivery system to bacterial cells that will help eliminate a broad range of resistant pathogens. With the increasing knowledge of the metabolic requirements and mechanisms of up taking iron in many pathogens, drug development and modulation of immune pathways are becoming more active research areas that may result in the elimination of antibiotic drug resistance. Future research directions should investigate the role of iron regulatory proteins, host iron sequestration strategies, pathogen iron acquisition systems, and the intricate interplay between host and pathogen in iron alteration processes. By unraveling these mechanisms, we can gain valuable insights into host defense mechanisms and pathogen virulence, ultimately leading to the development of targeted therapeutic approaches.
Author contributions
ML: Conceptualization, Supervision, Writing – review & editing. IU: Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Class B Breeding Program of Special Projects for Leading Science and Technology of the Chinese Academy of Sciences (XDPB16), the Fundamental Research Funds for the Central Universities, Beijing Municipal Natural Science Foundation (7202129), the National Natural Science Foundation of China (31571042), the Key Basic Research Project of Applied Basic Research Program of Hebei Province (18966315D), and One Hundred Outstanding Creative Talents Support Program of Hebei (BR2-218).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Volani C, Doerrier C, Demetz E, Haschka D, Paglia G, Lavdas AA, et al. Dietary iron loading negatively affects liver mitochondrial function. Metallomics (2017) 9(11):1634–44. doi: 10.1039/C7MT00177K
2. Oexle H, Gnaiger E, Weiss G. Iron-dependent changes in cellular energy metabolism: influence on citric acid cycle and oxidative phosphorylation. Biochim Biophys Acta (BBA)-Bioenergetics (1999) 1413(3):99–107. doi: 10.1016/S0005-2728(99)00088-2
3. Ni S, Yuan Y, Kuang Y, Li X. Iron metabolism and immune regulation. Front Immunol (2022) 13. doi: 10.3389/fimmu.2022.816282
4. Duan G, Li J, Duan Y, Zheng C, Guo Q, Li F, et al. Mitochondrial iron metabolism: the crucial actors in diseases. Molecules (2023) 28(1):29. doi: 10.3390/molecules28010029
5. Kell DB. Iron behaving badly: inappropriate iron chelation as a major contributor to the aetiology of vascular and other progressive inflammatory and degenerative diseases. BMC Med Genomics (2009) 2(1):1–79. doi: 10.1186/1755-8794-2-2
6. Sposi NM. Oxidative stress and iron overload in B-thalassemia: an overview. In: Beta thalassemia. London, United Kingdom: IntechOpen (2019).
7. Fraga CG, Oteiza PI. Iron toxicity and antioxidant nutrients. Toxicology (2002) 180(1):23–32. doi: 10.1016/S0300-483X(02)00379-7
8. Peng Y, Chang X, Lang M. Iron homeostasis disorder and alzheimer’s disease. Int J Mol Sci (2021) 22(22):12442. doi: 10.3390/ijms222212442
9. Camaschella C. New insights into iron deficiency and iron deficiency anemia. Blood Rev (2017) 31(4):225–33. doi: 10.1016/j.blre.2017.02.004
10. Abuga KM, Nairz M, MacLennan CA, Atkinson SH. Severe anaemia, iron deficiency, and susceptibility to invasive bacterial infections. Wellcome Open Res (2023) 8:48. doi: 10.12688/wellcomeopenres.18829.1
11. Parrow NL, Fleming RE, Minnick MF. Sequestration and scavenging of iron in infection. Infect Immun (2013) 81(10):3503–14. doi: 10.1128/IAI.00602-13
12. Nairz M, Dichtl S, Schroll A, Haschka D, Tymoszuk P, Theurl I, et al. Iron and innate antimicrobial immunity—Depriving the pathogen, defending the host. J Trace Elements Med Biol (2018) 48:118–33. doi: 10.1016/j.jtemb.2018.03.007
13. Verbon EH, Trapet PL, Stringlis IA, Kruijs S, Bakker PA, Pieterse CM. Iron and immunity. Annu Rev Phytopathol (2017) 55:355–75. doi: 10.1146/annurev-phyto-080516-035537
14. Iatsenko I, Marra A, Boquete J-P, Peña J, Lemaitre B. Iron sequestration by transferrin 1 mediates nutritional immunity in drosophila melanogaster. Proc Natl Acad Sci (2020) 117(13):7317–25. doi: 10.1073/pnas.1914830117
15. Kao J-K, Wang S-C, Ho L-W, Huang S-W, Chang S-H, Yang R-C, et al. Chronic iron overload results in impaired bacterial killing of thp-1 derived macrophage through the inhibition of lysosomal acidification. PloS One (2016) 11(5):e0156713. doi: 10.1371/journal.pone.0156713
16. Mitterstiller A-M, von Raffay L, Nairz M. Iron Deficiency, Anemia, and the Immune System. In: Nutritional Anemia. Switzerland: Springer (2022). p. 235–48.
17. Cartwright G, Lauristen M, Humphreys S, Jones P, Merrill I, Wintrobe M. The anemia associated with chronic infection. Sci (Washington) (1946) 103:72–3. doi: 10.1126/science.103.2664.72
18. Girelli D, Marchi G, Busti F, Vianello A eds. Iron Metabolism in Infections: Focus on Covid-19. In: Seminars in Hematology. WB Saunders, United States: Elsevier.
19. Muckenthaler MU, Rivella S, Hentze MW, Galy B. A red carpet for iron metabolism. Cell (2017) 168(3):344–61. doi: 10.1016/j.cell.2016.12.034
20. Kehl-Fie TE, Skaar EP. Nutritional immunity beyond iron: A role for manganese and zinc. Curr Opin Chem Biol (2010) 14(2):218–24. doi: 10.1016/j.cbpa.2009.11.008
21. Skaar EP. The battle for iron between bacterial pathogens and their vertebrate hosts. PloS Pathog (2010) 6(8):e1000949. doi: 10.1371/journal.ppat.1000949
22. Kramer J, Özkaya Ö, Kümmerli R. Bacterial siderophores in community and host interactions. Nat Rev Microbiol (2020) 18(3):152–63. doi: 10.1038/s41579-019-0284-4
23. Page MG. The role of iron and siderophores in infection, and the development of siderophore antibiotics. Clin Infect Dis (2019) 69(Supplement_7):S529–S37. doi: 10.1093/cid/ciz825
25. Correnti C, Strong RK. Iron sequestration in immunity. Metals in Cells. United Kingdom: John Wiley (2016), 16–349.
26. Zughaier SM, Cornelis P. Role of iron in bacterial pathogenesis. Front Cell Infect Microbiol (2018) 8:344. doi: 10.3389/fcimb.2018.00344
27. Lambert LA. Molecular evolution of the transferrin family and associated receptors. Biochim Biophys Acta (BBA)-General Subj (2012) 1820(3):244–55. doi: 10.1016/j.bbagen.2011.06.002
28. Silva AM, Moniz T, de Castro B, Rangel M. Human transferrin: an inorganic biochemistry perspective. Coordination Chem Rev (2021) 449:214186. doi: 10.1016/j.ccr.2021.214186
29. Orkin SH, Nathan DG, Ginsburg D, Look AT, Fisher DE, Lux S. Nathan and oski’s hematology and oncology of infancy and childhood E-book. WB Saunders, United States: Elsevier Health Sciences (2014).
30. Yoshiga T, Georgieva T, Dunkov BC, Harizanova N, Ralchev K, Law JH. Drosophila melanogaster transferrin: cloning, deduced protein sequence, expression during the life cycle, gene localization and up-regulation on bacterial infection. Eur J Biochem (1999) 260(2):414–20. doi: 10.1046/j.1432-1327.1999.00173.x
31. Tiklová K, Senti K-A, Wang S, Gräslund A, Samakovlis C. Epithelial septate junction assembly relies on melanotransferrin iron binding and endocytosis in drosophila. Nat Cell Biol (2010) 12(11):1071–7. doi: 10.1038/ncb2111
32. Weber JJ, Park YJ, Gorman M. Secreted insect transferrin-1 with strong and reversible iron-binding has potentially tissue specific roles in immunity and iron transport. FASEB J (2020) 34(S1):1–. doi: 10.1096/fasebj.2020.34.s1.05330
33. Hrdina A, Iatsenko I. The roles of metals in insect–microbe interactions and immunity. Curr Opin Insect Sci (2022) 49:71–7. doi: 10.1016/j.cois.2021.12.004
34. Mandilaras K, Missirlis F. Genes for iron metabolism influence circadian rhythms in drosophila melanogaster. Metallomics (2012) 4(9):928–36. doi: 10.1039/c2mt20065a
35. Levy F, Bulet P, Ehret-Sabatier L. Proteomic analysis of the systemic immune response of drosophila. Mol Cell Proteomics (2004) 3(2):156–66. doi: 10.1074/mcp.M300114-MCP200
36. Arrese EL, Soulages JL. Insect fat body: energy, metabolism, and regulation. Annu Rev Entomol (2010) 55:207–25. doi: 10.1146/annurev-ento-112408-085356
37. Xiao G, Liu Z-H, Zhao M, Wang H-L, Zhou B. Transferrin 1 functions in iron trafficking and genetically interacts with ferritin in drosophila melanogaster. Cell Rep (2019) 26(3):748–58. e5. doi: 10.1016/j.celrep.2018.12.053
38. Lambert LA, Perri H, Halbrooks PJ, Mason AB. Evolution of the transferrin family: conservation of residues associated with iron and anion binding. Comp Biochem Physiol Part B: Biochem Mol Biol (2005) 142(2):129–41. doi: 10.1016/j.cbpb.2005.07.007
39. Ong ST, Ho JZS, Ho B, Ding JL. Iron-withholding strategy in innate immunity. Immunobiology (2006) 211(4):295–314. doi: 10.1016/j.imbio.2006.02.004
40. Rouault TA. How mammals acquire and distribute iron needed for oxygen-based metabolism. PloS Biol (2003) 1(3):e79. doi: 10.1371/journal.pbio.0000079
41. Jennifer B, Berg V, Modak M, Puck A, Seyerl-Jiresch M, Künig S, et al. Transferrin receptor 1 is a cellular receptor for human heme-albumin. Commun Biol (2020) 3(1):1–13. doi: 10.1038/s42003-020-01294-5
42. Chan C, Ng D, Fraser ME, Schryvers AB. Structural and functional insights into iron acquisition from lactoferrin and transferrin in gram-negative bacterial pathogens. BioMetals (2022) 36(3):683–702. doi: 10.1007/s10534-022-00466-6.
43. Ganz T. Iron in innate immunity: starve the invaders. Curr Opin Immunol (2009) 21(1):63–7. doi: 10.1016/j.coi.2009.01.011
44. Adlerova L, Bartoskova A, Faldyna M. Lactoferrin: A review. Veterinarni Medicina (2008) 53(9):457–68. doi: 10.17221/1978-VETMED
45. Sienkiewicz M, Jaśkiewicz A, Tarasiuk A, Fichna J. Lactoferrin: an overview of its main functions, immunomodulatory and antimicrobial role, and clinical significance. Crit Rev Food Sci Nutr (2022) 62(22):6016–33. doi: 10.1080/10408398.2021.1895063
46. Legrand D, Pierce A, Elass E, Carpentier M, Mariller C, Mazurier J. Lactoferrin structure and functions. Bioactive Components Milk (2008) 606:163–94. doi: 10.1007/978-0-387-74087-4_6
47. Abd El-Hack ME, Abdelnour SA, Kamal M, Khafaga AF, Shakoori AM, Bagadood RM, et al. Lactoferrin: antimicrobial impacts, genomic guardian, therapeutic uses and clinical significance for humans and animals. Biomed Pharmacother (2023) 164:114967. doi: 10.1016/j.biopha.2023.114967
48. Kell DB, Pretorius E. Serum ferritin is an important inflammatory disease marker, as it is mainly a leakage product from damaged cells. Metallomics (2014) 6(4):748–73. doi: 10.1039/C3MT00347G
49. White KN, Conesa C, Sánchez L, Amini M, Farnaud S, Lorvoralak C, et al. The transfer of iron between ceruloplasmin and transferrins. Biochim Biophys Acta (BBA)-General Subj (2012) 1820(3):411–6. doi: 10.1016/j.bbagen.2011.10.006
50. Kell DB, Pretorius E. No effects without causes: the iron dysregulation and dormant microbes hypothesis for chronic, inflammatory diseases. Biol Rev (2018) 93(3):1518–57. doi: 10.1111/brv.12407
51. Baker HM, Baker EN. A structural perspective on lactoferrin function. Biochem Cell Biol (2012) 90(3):320–8. doi: 10.1139/o11-071
52. Cao X, Ren Y, Lu Q, Wang K, Wu Y, Wang Y, et al. Lactoferrin: A glycoprotein that plays an active role in human health. Front Nutr (2023) 9:1018336. doi: 10.3389/fnut.2022.1018336
53. Kell DB, Heyden EL, Pretorius E. The biology of lactoferrin, an iron-binding protein that can help defend against viruses and bacteria. Front Immunol (2020) 11:1221. doi: 10.3389/fimmu.2020.01221
54. Siqueiros-Cendón T, Arévalo-Gallegos S, Iglesias-Figueroa BF, García-Montoya IA, Salazar-Martínez J, Rascón-Cruz Q. Immunomodulatory effects of lactoferrin. Acta Pharmacol Sin (2014) 35(5):557–66. doi: 10.1038/aps.2013.200
55. Schirmbeck GH, Sizonenko S, Sanches EF. Neuroprotective role of lactoferrin during early brain development and injury through lifespan. Nutrients (2022) 14(14):2923. doi: 10.3390/nu14142923
56. Chen C, Lu M, Zhang Z, Qin L. The role of lactoferrin in atherosclerosis. BioMetals (2023) 36(3):509–19. doi: 10.1007/s10534-022-00441-1
57. Van de Graaf E, Out T, Kobesen A, Jansen H. Lactoferrin and secretory iga in the bronchoalveolar lavage fluid from patients with a stable asthma. Lung (1991) 169(1):275–83. doi: 10.1007/BF02714163
58. Hussan JR, Irwin SG, Mathews B, Swift S, Williams DL, Cornish J. Optimal dose of lactoferrin reduces the resilience of in vitro staphylococcus aureus colonies. PloS One (2022) 17(8):e0273088. doi: 10.1371/journal.pone.0273088
59. Singh A, Ahmad N, Varadarajan A, Vikram N, Singh T, Sharma S, et al. Lactoferrin, a potential iron-chelator as an adjunct treatment for mucormycosis–a comprehensive review. Int J Biol Macromol (2021) 187:988–98. doi: 10.1016/j.ijbiomac.2021.07.156
60. Arosio P, Ingrassia R, Cavadini P. Ferritins: A family of molecules for iron storage, antioxidation and more. Biochim Biophys Acta (BBA)-General Subj (2009) 1790(7):589–99. doi: 10.1016/j.bbagen.2008.09.004
61. Piperno A, Pelucchi S, Mariani R. Hereditary hyperferritinemia. Int J Mol Sci (2023) 24(3):2560. doi: 10.3390/ijms24032560
62. Wang W, Knovich MA, Coffman LG, Torti FM, Torti SV. Serum ferritin: past, present and future. Biochim Biophys Acta (BBA)-General Subj (2010) 1800(8):760–9. doi: 10.1016/j.bbagen.2010.03.011
63. Mahroum N, Alghory A, Kiyak Z, Alwani A, Seida R, Alrais M, et al. Ferritin–from iron, through inflammation and autoimmunity, to covid-19. J Autoimmun (2022) 126:102778. doi: 10.1016/j.jaut.2021.102778
64. Moreira AC, Mesquita G, Gomes MS. Ferritin: an inflammatory player keeping iron at the core of pathogen-host interactions. Microorganisms (2020) 8(4):589. doi: 10.3390/microorganisms8040589
65. Harrison PM, Arosio P. The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta (BBA)-Bioenergetics (1996) 1275(3):161–203. doi: 10.1016/0005-2728(96)00022-9
66. Laghaei R, Evans DG, Coalson RD. Metal binding sites of human H-chain ferritin and iron transport mechanism to the ferroxidase sites: A molecular dynamics simulation study. Proteins: Structure Function Bioinf (2013) 81(6):1042–50. doi: 10.1002/prot.24251
67. Beard JL, Connor JR. Iron status and neural functioning. Annu Rev Nutr (2003) 23(1):41–58. doi: 10.1146/annurev.nutr.23.020102.075739
68. Chekhun S, Lukyanova NY, Shvets Y, Burlaka A, Buchynska L. Significance of ferritin expression in formation of Malignant phenotype of human breast cancer cells. Exp Oncol (2014) 36,№ 3):179–83.
69. Huang H, Qiu Y, Huang G, Zhou X, Zhou X, Luo W. Value of ferritin heavy chain (Fth1) expression in diagnosis and prognosis of renal cell carcinoma. Med Sci Monit: Int Med J Exp Clin Res (2019) 25:3700. doi: 10.12659/MSM.914162
70. Kernan KF, Carcillo JA. Hyperferritinemia and inflammation. Int Immunol (2017) 29(9):401–9. doi: 10.1093/intimm/dxx031
71. Hou J, Yamada S, Kajikawa T, Ozaki N, Awata T, Yamaba S, et al. Role of ferritin in the cytodifferentiation of periodontal ligament cells. Biochem Biophys Res Commun (2012) 426(4):643–8. doi: 10.1016/j.bbrc.2012.09.008
72. Huang W, Li W, Zhu W, Liu J, Hou J, Meng H. Ferritin expression in the periodontal tissues of primates. Eur J Histochem: EJH (2019) 63(3):136–43. doi: 10.4081/ejh.2019.3046
73. Gómez-Pastora J, Weigand M, Kim J, Wu X, Strayer J, Palmer AF, et al. Hyperferritinemia in critically ill covid-19 patients–is ferritin the product of inflammation or a pathogenic mediator? Clin Chim Acta; Int J Clin Chem (2020) 509:249. doi: 10.1016/j.cca.2020.06.033
74. Fan Y, Zhang J, Cai L, Wang S, Liu C, Zhang Y, et al. The effect of anti-inflammatory properties of ferritin light chain on lipopolysaccharide-induced inflammatory response in murine macrophages. Biochim Biophys Acta (BBA)-Mol Cell Res (2014) 1843(11):2775–83. doi: 10.1016/j.bbamcr.2014.06.015
75. Ruddell RG, Hoang-Le D, Barwood JM, Rutherford PS, Piva TJ, Watters DJ, et al. Ferritin functions as a proinflammatory cytokine via iron-independent protein kinase C zeta/nuclear factor kappab–regulated signaling in rat hepatic stellate cells. Hepatology (2009) 49(3):887–900. doi: 10.1002/hep.22716
76. Kim S, Ponka P. Effects of interferon-Γ and lipopolysaccharide on macrophage iron metabolism are mediated by nitric oxide-induced degradation of iron regulatory protein 2. J Biol Chem (2000) 275(9):6220–6. doi: 10.1074/jbc.275.9.6220
77. Miller LL, Miller SC, Torti SV, Tsuji Y, Torti FM. Iron-independent induction of ferritin H chain by tumor necrosis factor. Proc Natl Acad Sci (1991) 88(11):4946–50. doi.org/10.1073/pnas.88.11.4946
78. Torti FM, Torti SV. Regulation of ferritin genes and protein. Blood J Am Soc Hematol (2002) 99(10):3505–16. doi: 10.1182/blood.V99.10.3505
79. Mukherjee C, Kling T, Russo B, Miebach K, Kess E, Schifferer M, et al. Oligodendrocytes provide antioxidant defense function for neurons by secreting ferritin heavy chain. Cell Metab (2020) 32(2):259–72. e10. doi: 10.1016/j.cmet.2020.05.019
80. Lu ZJ, Xie YX, Yu HZ, Toufeeq S, Wang J, Huang YL, et al. Identification and functional analysis of an iron-binding protein, ferritin heavy chain subunit, from the swallowtail butterfly, papilio xuthus. Arch Insect Biochem Physiol (2019) 102(1):e21592. doi: 10.1002/arch.21592
81. Gehrer CM, Hoffmann A, Hilbe R, Grubwieser P, Mitterstiller A-M, Talasz H, et al. Availability of ferritin-bound iron to enterobacteriaceae. Int J Mol Sci (2022) 23(21):13087. doi: 10.3390/ijms232113087
82. Tu Z, Timashev P, Chen J, Liang XJ. Ferritin-based drug delivery system for tumor therapy. BMEMat (2023) 1(2):e12022. doi: 10.1002/bmm2.12022
83. Li L, Fang CJ, Ryan JC, Niemi EC, Lebrón JA, Björkman PJ, et al. Binding and uptake of H-ferritin are mediated by human transferrin receptor-1. Proc Natl Acad Sci (2010) 107(8):3505–10. doi: 10.1073/pnas.0913192107
84. Barton JC, Acton RT. Hepcidin, iron, and bacterial infection. Vitamins Hormones (2019) 110:223–42. doi: 10.1016/bs.vh.2019.01.011
85. Collins JF, Wessling-Resnick M, Knutson MD. Hepcidin regulation of iron transport. J Nutr (2008) 138(11):2284–8. doi: 10.3945/jn.108.096347
86. Michels K, Nemeth E, Ganz T, Mehrad B. Hepcidin and host defense against infectious diseases. PloS Pathog (2015) 11(8):e1004998. doi: 10.1371/journal.ppat.1004998
87. Nemeth E, Ganz T. Hepcidin and iron in health and disease. Annu Rev Med (2023) 74:261–77. doi: 10.1146/annurev-med-043021-032816
88. Brissot P, Cavey T, Ropert M, Guggenbuhl P, Loréal O. Genetic hemochromatosis: pathophysiology, diagnostic and therapeutic management. La Presse Médicale (2017) 46(12):e288–e95. doi: 10.1016/j.lpm.2017.05.037
89. Nemeth E, Ganz T. Hepcidin-ferroportin interaction controls systemic iron homeostasis. Int J Mol Sci (2021) 22(12):6493. doi: 10.3390/ijms22126493
90. Bettedi L, Aslam MF, Szular J, Mandilaras K, Missirlis F. Iron depletion in the intestines of malvolio mutant flies does not occur in the absence of a multicopper oxidase. J Exp Biol (2011) 214(6):971–8. doi: 10.1242/jeb.051664
91. Tang X, Zhou B. Ferritin is the key to dietary iron absorption and tissue iron detoxification in drosophila melanogaster. FASEB J (2013) 27(1):288–98. doi: 10.1096/fj.12-213595
92. Lang M, Braun CL, Kanost MR, Gorman MJ. Multicopper oxidase-1 is a ferroxidase essential for iron homeostasis in drosophila melanogaster. Proc Natl Acad Sci (2012) 109(33):13337–42. doi: 10.1073/pnas.1208703109
93. Kim D-K, Jeong J-H, Lee J-M, Kim KS, Park S-H, Kim YD, et al. Inverse agonist of estrogen-related receptor Γ Controls salmonella typhimurium infection by modulating host iron homeostasis. Nat Med (2014) 20(4):419–24. doi: 10.1038/nm.3483
94. Nairz M, Schleicher U, Schroll A, Sonnweber T, Theurl I, Ludwiczek S, et al. Nitric oxide–mediated regulation of ferroportin-1 controls macrophage iron homeostasis and immune function in salmonella infection. J Exp Med (2013) 210(5):855–73. doi: 10.1084/jem.20121946
95. Shike H, Lauth X, Westerman ME, Ostland VE, Carlberg JM, Van Olst JC, et al. Bass hepcidin is a novel antimicrobial peptide induced by bacterial challenge. Eur J Biochem (2002) 269(8):2232–7. doi: 10.1046/j.1432-1033.2002.02881.x
96. Peyssonnaux C, Zinkernagel AS, Datta V, Lauth X, Johnson RS, Nizet V. Tlr4-dependent hepcidin expression by myeloid cells in response to bacterial pathogens. Blood (2006) 107(9):3727–32. doi: 10.1182/blood-2005-06-2259
97. Wojciechowska M, Wisniewski O, Kolodziejski P, Krauss H. Role of hepcidin in physiology and pathophysiology. Emerging experimental and clinical evidence. J Physiol Pharmacol (2021) 72(1):10.26402. doi: 10.26402/jpp.2021.1.03
98. Blackwell JM, Searle S, Goswami T, Miller EN. Understanding the multiple functions of nramp1. Microbes Infect (2000) 2(3):317–21. doi: 10.1016/S1286-4579(00)00295-1
99. Chen H-M, Wang Y-M, Yang H-L, Zeng Q-Y, Liu Y-J. Nramp1 promotes iron uptake at the late stage of iron deficiency in poplars. Tree Physiol (2019) 39(7):1235–50. doi: 10.1093/treephys/tpz055
100. Cellier M, Prive G, Belouchi A, Kwan T, Rodrigues V, Chia W, et al. Nramp defines a family of membrane proteins. Proc Natl Acad Sci (1995) 92(22):10089–93. doi: 10.1073/pnas.92.22.10089
101. Govoni G, Gauthier S, Billia F, Iscove NN, Gros P. Cell-specific and inducible nramp1 gene expression in mouse macrophages in vitro and in vivo. J Leukocyte Biol (1997) 62(2):277–86. doi: 10.1002/jlb.62.2.277
102. Dwiyanti R, Muthaher AA, Achmad D, Arsyadi G, Nelwan BJ, Cangara MH, et al. Analysis of natural resistance-associated macrophage protein-1 (Nramp-1) level based on death, comorbidities and severity of covid-19 patients: A cross-sectional study. Ann Med Surg (2023) 85(6):2587. doi: 10.1097/MS9.0000000000000830
103. Ardiyana M, Gunawan A, Murtini S, Sartika T, Sumantri C. Polymorphisms and associations of the nramp-1 and inos genes on newcastle disease and salmonella enteritidis resistances in sensi-1 agrinak chickens. Trop Anim Sci J (2020) 43(2):95–102. doi: 10.5398/tasj.2020.43.2.95
104. Laranjeira-Silva MF, Hamza I, Pérez-Victoria JM. Iron and heme metabolism at the leishmania–host interface. Trends Parasitol (2020) 36(3):279–89. doi: 10.1016/j.pt.2019.12.010
105. Loomis WP, Johnson ML, Brasfield A, Blanc M-P, Yi J, Miller SI, et al. Temporal and anatomical host resistance to chronic salmonella infection is quantitatively dictated by nramp1 and influenced by host genetic background. PloS One (2014) 9(10):e111763. doi: 10.1371/journal.pone.0111763
106. Banerjee S, Datta R. Leishmania infection triggers hepcidin-mediated proteasomal degradation of nramp1 to increase phagolysosomal iron availability. Cell Microbiol (2020) 22(12):e13253. doi: 10.1111/cmi.13253
107. Sohn Y-S, Ghoti H, Breuer W, Rachmilewitz E, Attar S, Weiss G, et al. The role of endocytic pathways in cellular uptake of plasma non-transferrin iron. Haematologica (2012) 97(5):670. doi: 10.3324/haematol.2011.054858
108. Fritsche G, Nairz M, Libby SJ, Fang FC, Weiss G. Slc11a1 (Nramp1) impairs growth of salmonella enterica serovar typhimurium in macrophages via stimulation of lipocalin-2 expression. J Leukocyte Biol (2012) 92(2):353–9. doi: 10.1189/jlb.1111554
109. Fritsche G, Nairz M, Theurl I, Mair S, Bellmann-Weiler R, Barton HC, et al. Modulation of macrophage iron transport by nramp1 (Slc11a1). Immunobiology (2008) 212(9-10):751–7. doi: 10.1016/j.imbio.2007.09.014
110. Anderson GJ, Vulpe CD. Mammalian iron transport. Cell Mol Life Sci (2009) 66(20):3241–61. doi: 10.1007/s00018-009-0051-1
111. El Younis C, Zhang Y. Expression of divalent metal transporter 1 (Dmt1) and ferroportin 1 (Fp1) in the duodenum of hepatitis C virus (Hcv)-infected patients: 277. Off J Am Coll Gastroenterol | ACG (2010) 105:S103–S4. doi: 10.14309/00000434-201010001-00277
112. Huang B, Lv Z, Li Y, Li C. Identification and functional characterization of natural resistance-associated macrophage protein 2 from sea cucumber apostichopus japonicus. Dev Comp Immunol (2021) 114:103835. doi: 10.1016/j.dci.2020.103835
113. Grander M, Hoffmann A, Seifert M, Demetz E, Grubwieser P, Pfeifhofer-Obermair C, et al. Dmt1 protects macrophages from salmonella infection by controlling cellular iron turnover and lipocalin 2 expression. Int J Mol Sci (2022) 23(12):6789. doi: 10.3390/ijms23126789
114. Spielmann J, Vert G. The many facets of protein ubiquitination and degradation in plant root iron-deficiency responses. J Exp Bot (2021) 72(6):2071–82. doi: 10.1093/jxb/eraa441
115. Segond D, Dellagi A, Lanquar V, Rigault M, Patrit O, Thomine S, et al. Nramp genes function in arabidopsis thaliana resistance to erwinia chrysanthemi infection. Plant J (2009) 58(2):195–207. doi: 10.1111/j.1365-313X.2008.03775.x
116. Makui H, Roig E, Cole ST, Helmann JD, Gros P, Cellier MF. Identification of the escherichia coli K-12 nramp orthologue (Mnth) as a selective divalent metal ion transporter. Mol Microbiol (2000) 35(5):1065–78. doi: 10.1046/j.1365-2958.2000.01774.x
117. Bosma EF, Rau MH, van Gijtenbeek LA, Siedler S. Regulation and distinct physiological roles of manganese in bacteria. FEMS Microbiol Rev (2021) 45(6):fuab028. doi: 10.1093/femsre/fuab028
118. Folwell JL, Barton CH, Shepherd D, Immunolocalisation of the D. Melanogaster nramp homologue malvolio to gut and malpighian tubules provides evidence that malvolio and Nramp2 are orthologous. J Exp Biol (2006) 209(10):1988–95. doi: 10.1242/jeb.02193
119. Mehlferber EC, Benowitz KM, Roy-Zokan EM, McKinney EC, Cunningham CB, Moore AJ. Duplication and sub/neofunctionalization of malvolio, an insect homolog of nramp, in the subsocial beetle nicrophorus vespilloides. G3: Genes Genomes Genet (2017) 7(10):3393–403. doi: 10.1534/g3.117.300183
120. Sousa Geros A, Simmons A, Drakesmith H, Aulicino A, Frost JN. The battle for iron in enteric infections. Immunology (2020) 161(3):186–99. doi: 10.1111/imm.13236
121. Gombart AF, Kwok SH, Anderson KL, Yamaguchi Y, Torbett BE, Koeffler HP. Regulation of neutrophil and eosinophil secondary granule gene expression by transcription factors C/Ebpϵ and pu. 1. Blood J Am Soc Hematol (2003) 101(8):3265–73. doi: 10.1182/blood-2002-04-1039
122. Sheldon JR, Himmel LE, Kunkle DE, Monteith AJ, Maloney KN, Skaar EP. Lipocalin-2 is an essential component of the innate immune response to acinetobacter baumannii infection. PloS Pathog (2022) 18(9):e1010809. doi: 10.1371/journal.ppat.1010809
123. Al Jaberi S, Cohen A, D’Souza C, Abdulrazzaq YM, Ojha S, Bastaki S, et al. Lipocalin-2: structure, function, distribution and role in metabolic disorders. Biomed Pharmacother (2021) 142:112002. doi: 10.1016/j.biopha.2021.112002
124. Chan YR, Liu JS, Pociask DA, Zheng M, Mietzner TA, Berger T, et al. Lipocalin 2 is required for pulmonary host defense against klebsiella infection. J Immunol (2009) 182(8):4947–56. doi: 10.4049/jimmunol.0803282
125. Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK. The neutrophil lipocalin ngal is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell (2002) 10(5):1033–43. doi: 10.1016/s1097-2765(02)00708-6
126. Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature (2004) 432(7019):917–21. doi: 10.1038/nature03104
127. Thanh T, Casals-Pascual C, Ny NH, Ngoc N, Geskus R, Nhu LT, et al. Value of lipocalin 2 as a potential biomarker for bacterial meningitis. Clin Microbiol Infect (2021) 27(5):724–30. doi: 10.1016/j.cmi.2020.07.006
128. Abella V, Scotece M, Conde J, Gómez R, Lois A, Pino J, et al. The potential of lipocalin-2/Ngal as biomarker for inflammatory and metabolic diseases. Biomarkers (2015) 20(8):565–71. doi: 10.3109/1354750X.2015.1123354
129. Berger T, Togawa A, Duncan GS, Elia AJ, You-Ten A, Wakeham A, et al. Lipocalin 2-deficient mice exhibit increased sensitivity to escherichia coli infection but not to ischemia-reperfusion injury. Proc Natl Acad Sci (2006) 103(6):1834–9. doi: 10.1073/pnas.0510847103
130. Cramer EP, Dahl SL, Rozell B, Knudsen KJ, Thomsen K, Moser C, et al. Lipocalin-2 from both myeloid cells and the epithelium combats klebsiella pneumoniae lung infection in mice. Blood J Am Soc Hematol (2017) 129(20):2813–7. doi: 10.1182/blood-2016-11-753434
131. Saiga H, Nishimura J, Kuwata H, Okuyama M, Matsumoto S, Sato S, et al. Lipocalin 2-dependent inhibition of mycobacterial growth in alveolar epithelium. J Immunol (2008) 181(12):8521–7. doi: 10.4049/jimmunol.181.12.8521
132. Johnson EE, Srikanth CV, Sandgren A, Harrington L, Trebicka E, Wang L, et al. Siderocalin inhibits the intracellular replication of mycobacterium tuberculosis in macrophages. FEMS Immunol Med Microbiol (2010) 58(1):138–45. doi: 10.1111/j.1574-695X.2009.00622.x
133. Devireddy LR, Hart DO, Goetz DH, Green MR. A mammalian siderophore synthesized by an enzyme with a bacterial homolog involved in enterobactin production. Cell (2010) 141(6):1006–17. doi: 10.1016/j.cell.2010.04.040
134. Liu Z, Reba S, Chen W-D, Porwal SK, Boom WH, Petersen RB, et al. Regulation of mammalian siderophore 2, 5-dhba in the innate immune response to infection. J Exp Med (2014) 211(6):1197–213. doi: 10.1084/jem.20132629
135. Hadley RC, Nolan EM. Preparation and iron redox speciation study of the fe (Ii)-binding antimicrobial protein calprotectin. In: Calcium-Binding Proteins of the Ef-Hand Superfamily. Humana Press, New York, NY: Springer (2019). p. 397–415. doi: 10.1007/978-1-4939-9030-6_25
136. Baker TM, Nakashige TG, Nolan EM, Neidig ML. Magnetic circular dichroism studies of iron (Ii) binding to human calprotectin. Chem Sci (2017) 8(2):1369–77. doi: 10.1039/C6SC03487J
137. Zygiel EM, Obisesan AO, Nelson CE, Oglesby AG, Nolan EM. Heme protects pseudomonas aeruginosa and staphylococcus aureus from calprotectin-induced iron starvation. J Biol Chem (2021), 296. doi: 10.1074/jbc.RA120.015975
138. Hunter RC, Asfour F, Dingemans J, Osuna BL, Samad T, Malfroot A, et al. Ferrous iron is a significant component of bioavailable iron in cystic fibrosis airways. MBio (2013) 4(4):e00557–13. doi: 10.1128/mBio.00557-13
139. Nakashige TG, Zhang B, Krebs C, Nolan EM. Human calprotectin is an iron-sequestering host-defense protein. Nat Chem Biol (2015) 11(10):765–71. doi: 10.1038/nchembio.1891
140. Zygiel EM, Nolan EM. Exploring iron withholding by the innate immune protein human calprotectin. Accounts Chem Res (2019) 52(8):2301–8. doi: 10.1021/acs.accounts.9b00250
141. Kotsiou OS, Papagiannis D, Papadopoulou R, Gourgoulianis KI. Calprotectin in lung diseases. Int J Mol Sci (2021) 22(4):1706. doi: 10.3390/ijms22041706
142. Terrin G, Passariello A, Manguso F, Salvia G, Rapacciuolo L, Messina F, et al. Serum calprotectin: an antimicrobial peptide as a new marker for the diagnosis of sepsis in very low birth weight newborns. Clin Dev Immunol (2011), 2011. doi: 10.1155/2011/291085
143. Wei L, Liu M, Xiong H. Role of calprotectin as a biomarker in periodontal disease. Mediators Inflammation (2019), 2019. doi: 10.1155/2019/3515026
144. Radin JN, Kelliher JL, Párraga Solórzano PK, Kehl-Fie TE. The two-component system arlrs and alterations in metabolism enable staphylococcus aureus to resist calprotectin-induced manganese starvation. PloS Pathog (2016) 12(11):e1006040. doi: 10.1371/journal.ppat.1006040
145. Pantopoulos K, Porwal SK, Tartakoff A, Devireddy L. Mechanisms of mammalian iron homeostasis. Biochemistry (2012) 51(29):5705–24. doi: 10.1021/bi300752r
146. Nakashige TG, Nolan EM. Human calprotectin affects the redox speciation of iron. Metallomics (2017) 9(8):1086–95. doi: 10.1039/C7MT00044H
147. Zygiel EM, Nelson CE, Brewer LK, Oglesby-Sherrouse AG, Nolan EM. The human innate immune protein calprotectin induces iron starvation responses in pseudomonas aeruginosa. J Biol Chem (2019) 294(10):3549–62. doi: 10.1074/jbc.RA118.006819
148. Hellman NE, Gitlin JD. Ceruloplasmin metabolism and function. Annu Rev Nutr (2002) 22(1):439–58. doi: 10.1146/annurev.nutr.22.012502.114457
149. Gaware V, Kotade K, Dhamak K, Somawanshi S. Ceruloplasmin its role and significance: a review. International J Biomedical Research (2010) 1(4):153–62. doi: 10.7439/ijbr.v1i4.69
150. Liu H, Peatman E, Wang W, Abernathy J, Liu S, Kucuktas H, et al. Molecular responses of ceruloplasmin to edwardsiella ictaluri infection and iron overload in channel catfish (Ictalurus punctatus). Fish Shellfish Immunol (2011) 30(3):992–7. doi: 10.1016/j.fsi.2010.12.033
151. Gorman MJ, Dittmer NT, Marshall JL, Kanost MR. Characterization of the multicopper oxidase gene family in anopheles Gambiae. Insect Biochem Mol Biol (2008) 38(9):817–24. doi: 10.1016/j.ibmb.2008.07.001
152. Schaer DJ, Vinchi F, Ingoglia G, Tolosano E, Buehler PW. Haptoglobin, hemopexin, and related defense pathways—Basic science, clinical perspectives, and drug development. Front Physiol (2014) 5:415. doi: 10.3389/fphys.2014.00415
153. Hammer ND, Skaar EP. Molecular mechanisms of staphylococcus aureus iron acquisition. Annu Rev Microbiol (2011) 65:129–47. doi: 10.1146/annurev-micro-090110-102851
154. Richard KL, Kelley BR, Johnson JG. Heme uptake and utilization by gram-negative bacterial pathogens. Front Cell Infect Microbiol (2019) 9:81. doi: 10.3389/fcimb.2019.00081
155. Sakamoto K, Kim Y-G, Hara H, Kamada N, Caballero-Flores G, Tolosano E, et al. Il-22 controls iron-dependent nutritional immunity against systemic bacterial infections. Sci Immunol (2017) 2(8). doi: 10.1126/sciimmunol.aai8371
156. Remy KE, Cortés-Puch I, Sun J, Feng J, Lertora JJ, Risoleo T, et al. Haptoglobin therapy has differential effects depending on severity of canine septic shock and cell-free hemoglobin level. Transfusion (2019) 59(12):3628–38. doi: 10.1111/trf.15567
157. Kim B-H, Jun Y-C, Jin J-K, Kim J-I, Kim N-H, Leibold EA, et al. Alteration of iron regulatory proteins (Irp1 and Irp2) and ferritin in the brains of scrapie-infected mice. Neurosci Lett (2007) 422(3):158–63. doi: 10.1016/j.neulet.2007.05.061
158. Figueroa-Angulo EE, Calla-Choque JS, Mancilla-Olea MI, Arroyo R. Rna-binding proteins in trichomonas vaginalis: atypical multifunctional proteins involved in a posttranscriptional iron regulatory mechanism. Biomolecules (2015) 5(4):3354–95. doi: 10.3390/biom5043354
159. Nairz M, Ferring-Appel D, Casarrubea D, Sonnweber T, Viatte L, Schroll A, et al. Iron regulatory proteins mediate host resistance to salmonella infection. Cell Host Microbe (2015) 18(2):254–61. doi: 10.1016/j.chom.2015.06.017
160. Cronin SJ, Woolf CJ, Weiss G, Penninger JM. The role of iron regulation in immunometabolism and immune-related disease. Front Mol Biosci (2019) 6:116. doi: 10.3389/fmolb.2019.00116
161. Gao M, Zhao T, Zhang C, Li P, Wang J, Han J, et al. Ferritinophagy-mediated iron competition in rutis: tug-of-war between upec and host. Biomed Pharmacother (2023) 163:114859. doi: 10.1016/j.biopha.2023.114859
162. Gail M, Gross U, Bohne W. Transferrin receptor induction in toxoplasma gondii-infected hff is associated with increased iron-responsive protein 1 activity and is mediated by secreted factors. Parasitol Res (2004) 94(3):233–9. doi: 10.1007/s00436-004-1209-2
163. Maio N, Zhang D-L, Ghosh MC, Jain A, SantaMaria AM, Rouault TA eds. Mechanisms of Cellular Iron Sensing, Regulation of Erythropoiesis and Mitochondrial Iron Utilization. In: Seminars in Hematology. WB Saunders, United States: Elsevier. doi: 10.1053/j.seminhematol.2021.06.001
164. Murdoch CC, Skaar EP. Nutritional immunity: the battle for nutrient metals at the host–pathogen interface. Nat Rev Microbiol (2022) 20(11):657–70. doi: 10.1038/s41579-022-00745-6
165. Ratledge C, Dover LG. Iron metabolism in pathogenic bacteria. Annu Rev Microbiol (2000) 54(1):881–941. doi: 10.1146/annurev.micro.54.1.881
166. Golonka R, San Yeoh B, Vijay-Kumar M. The iron tug-of-war between bacterial siderophores and innate immunity. J Innate Immun (2019) 11(3):249–62. doi: 10.1159/000494627
167. Pasqua M, Visaggio D, Lo Sciuto A, Genah S, Banin E, Visca P, et al. Ferric uptake regulator fur is conditionally essential in pseudomonas aeruginosa. J Bacteriol (2017) 199(22):e00472–17. doi: 10.1128/jb.00472-17
168. Hassan HM, Troxell B. Transcriptional regulation by ferric uptake regulator (Fur) in pathogenic bacteria. Front Cell Infect Microbiol (2013) 3:59. doi: 10.3389/fcimb.2013.00059
169. Hider RC, Kong X. Chemistry and biology of siderophores. Natural Product Rep (2010) 27(5):637–57. doi: 10.1039/B906679A
170. Kotsiliti E. Siderophore cross-feeding between fungi and salmonella. Nat Rev Gastroenterol Hepatol (2023) 20(2):65–. doi: 10.1038/s41575-022-00731-6
171. Nairz M, Weiss G. Iron in infection and immunity. Mol Aspects Med (2020) 75:100864. doi: 10.1016/j.mam.2020.100864
172. Hannauer M, Barda Y, Mislin GL, Shanzer A, Schalk IJ. The ferrichrome uptake pathway in pseudomonas aeruginosa involves an iron release mechanism with acylation of the siderophore and recycling of the modified desferrichrome. J Bacteriol (2010) 192(5):1212–20. doi: 10.1128/JB.01539-09
173. Matzanke BF, Anemüller S, Schünemann V, Trautwein AX, Hantke K. Fhuf, part of a siderophore– reductase system. Biochemistry (2004) 43(5):1386–92. doi: 10.1021/bi0357661
174. Brickman TJ, McIntosh MA. Overexpression and purification of ferric enterobactin esterase from escherichia coli. Demonstration of enzymatic hydrolysis of enterobactin and its iron complex. J Biol Chem (1992) 267(17):12350–5. doi: 10.1016/S0021-9258(19)49846-3
175. Marchetti M, De Bei O, Bettati S, Campanini B, Kovachka S, Gianquinto E, et al. Iron metabolism at the interface between host and pathogen: from nutritional immunity to antibacterial development. Int J Mol Sci (2020) 21(6):2145. doi: 10.3390/ijms21062145
176. Behnsen J, Raffatellu M. Siderophores: more than stealing iron. MBio (2016) 7(6):e01906–16. doi: 10.1128/mBio.01906-16
177. Syed YY. Cefiderocol: A review in serious gram-negative bacterial infections. Drugs (2021) 81(13):1559–71. doi: 10.1007/s40265-021-01580-4
178. Debarbieux L, Wandersman C. Hemophore-dependent heme acquisition systems. Iron Transport Bacteria (2004) 15:38–47. doi: 10.1128/9781555816544.ch3
179. Skaar EP, Schneewind O. Iron-regulated surface determinants (Isd) of staphylococcus aureus: stealing iron from heme. Microbes Infect (2004) 6(4):390–7. doi: 10.1016/j.micinf.2003.12.008
180. Nairz M, Schroll A, Sonnweber T, Weiss G. The struggle for iron–a metal at the host–pathogen interface. Cell Microbiol (2010) 12(12):1691–702. doi: 10.1111/j.1462-5822.2010.01529.x
181. Pérez NM, Ramakrishnan G. The reduced genome of the francisella tularensis live vaccine strain (Lvs) encodes two iron acquisition systems essential for optimal growth and virulence. PloS One (2014) 9(4):e93558. doi: 10.1371/journal.pone.0093558
182. Sen S, Bal SK, Yadav S, Mishra P, Rastogi R, Mukhopadhyay CK. Intracellular pathogen leishmania intervenes in iron loading into ferritin by cleaving chaperones in host macrophages as an iron acquisition strategy. J Biol Chem (2022) 298(12). doi: 10.1016/j.jbc.2022.102646
183. Yan Q, Zhang W, Lin M, Teymournejad O, Budachetri K, Lakritz J, et al. Iron robbery by intracellular pathogen via bacterial effector–induced ferritinophagy. Proc Natl Acad Sci (2021) 118(23):e2026598118. doi: 10.1073/pnas.2026598118
Keywords: hepatocytes, nutritional immunity, pathogens, siderophores, transferrin
Citation: Ullah I and Lang M (2023) Key players in the regulation of iron homeostasis at the host-pathogen interface. Front. Immunol. 14:1279826. doi: 10.3389/fimmu.2023.1279826
Received: 18 August 2023; Accepted: 03 October 2023;
Published: 24 October 2023.
Edited by:
Hajo Haase, Technical University of Berlin, GermanyReviewed by:
Guenter Weiss, Innsbruck Medical University, AustriaElizabeth A. Novak, University of Pittsburgh, United States
Copyright © 2023 Ullah and Lang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Minglin Lang, langml@ucas.ac.cn
 Inam Ullah
Inam Ullah Minglin Lang
Minglin Lang