- 1Department of Occupational Medicine and Environmental Toxicology, Nantong Key Laboratory of Environmental Toxicology, School of Public Health, Nantong University, Nantong, China
- 2College of International Studies, Southwest University, Chongqing, China
- 3Nantong Institute of Liver Diseases, Nantong Third People’s Hospital Affiliated Nantong Hospital of Nantong University, Nantong, China
m6A is the most prevalent internal modification of eukaryotic mRNA, and plays a crucial role in tumorigenesis and various other biological processes. Lung cancer is a common primary malignant tumor of the lungs, which involves multiple factors in its occurrence and progression. Currently, only the demethylases FTO and ALKBH5 have been identified as associated with m6A modification. These demethylases play a crucial role in regulating the growth and invasion of lung cancer cells by removing methyl groups, thereby influencing stability and translation efficiency of mRNA. Furthermore, they participate in essential biological signaling pathways, making them potential targets for intervention in lung cancer treatment. Here we provides an overview of the involvement of m6A demethylase in lung cancer, as well as their potential application in the diagnosis, prognosis and treatment of the disease.
1 Introduction
Lung cancer is a highly prevalent and devastating form of primary lung malignancy, contributing to a significant portion of cancer-related deaths worldwide. It accounts for approximately 18% of all cancer fatalities. Unfortunately, the survival rates for lung cancer remain alarmingly low, with five‐year relative survival rates of 23% (1). Understanding the diverse histopathological classifications is crucial for comprehending the complexity of this disease. In terms of histopathologically, lung cancer can be categorized into non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). NSCLC is the most common type, and it accounts for approximately 85% of all cases, while SCLC represents about 15%. Within the subtypes of NSCLC, lung adenocarcinoma is the most prevalent histological variant, making up around 40% of NSCLC cases. Lung squamous cell carcinoma closely follows, comprising approximately 35% of NSCLC cases (2).
Epigenetic modifications play a crucial role in the initiation and progression of various tumours. These modifications mainly involve regulation of gene function and expression through processes such as DNA methylation, regulation of non-coding RNA, histone modification, and chromatin remodelling. By modifying these epigenetic marks, tumor cells can manipulate gene activity and disrupt normal cellular processes, thereby contributing to the development and advancement of cancer (3). Among the various epigenetic modifications, one of the most extensively studied is N6-adenylate methylation (m6A), which is a common internal modification in eukaryotic mRNA molecules. It represents around 60% of all known RNA modifications identified in mammals (4, 5). m6A takes place when methylation occurs at the sixth position of adenylate (A) nucleotides within the RNA molecule (6). The discovery of m6A modification provides new insights into the intricate regulation of gene expression in cancer cells. This modification serves as a dynamic and reversible mark that can impact multiple aspects of mRNA metabolism, such as splicing, stability, localization, and translation efficiency. Through m6A modification, tumor cells possess the ability to precisely regulate the expression of crucial genes that are involved in pathways essential for cell survival, proliferation, invasion, and metastasis (7). In addition, there is evidence indicating that dysregulation of m6A modification machinery and aberrant m6A patterns are commonly observed in various cancer types, can affect the occurrence and development of breast cancer, ovarian cancer, gastric cancer, lung cancer, colon cancer and other cancers (8–12). These changes can affect the behaviour of tumour cells, leading to disease advancement and resistance to treatment. Consequently, some researchers have concentrated their efforts on uncovering the specific mechanisms involved in m6A modification and its functional implications in the development of tumors (13, 14).
The occurrence of m6A modification involves the participation of multiple enzymes, including Writers, Readers and Erasers. During the transcription of DNA into RNA, the sixth N of adenosine is methylated and modified by methyltransferases such as METTL3, METTL14 and WTAP (15–17). These enzyme are referred to as Writers, specifically methylases. RNA base sites often require specific enzymes to recognize them after methylation. These enzymes, known as Readers, primarily include YTH domain proteins, nuclear heterogeneous riboproteins (hnRNPs) and eukaryotic initiation factors (eIFs) (18–20). Readers can recognize the bases where m6A methylation occurs and have the functions that include participating in mRNA degradation, downstream translation, and accelerating the rate of mRNA nuclear export. m6A demethylases are called Erasers, mainly including Fat mass and obesity-associated protein (FTO) and human AlkB homolog 5 (ALKBH5). FTO, a member of the Alkb protein family, is the first m6A demethylase that acts on mRNA in an iron-dependent manner (21). It primarily localizes in the nucleus and exhibits a punctate pattern in the nucleoplasm, partially co-localizing with splicing or splicing-related spot factor SART1 (22). Under physiological conditions, FTO demonstrates the highest affinity for m6A as a substrate. Changes or dysfunction in FTO expression may contribute to the occurrence and development of various tumors, where it can act as either a tumour suppressor gene or oncogene. FTO has been found to play a critical role in tumor cell proliferation, metastasis and apoptosis. ALKBH5, another important m6A demethylase, is capable of demethylating mRNA in the nucleus. It possesses an alanine-rich region at the N-terminus and a unique coiled-coil structure (23). Current studies have revealed dysregulation of ALKBH5 expression in various cancers, including lung, breast, and gastric cancer. ALKBH5 can exert both carcinogenic and tumor-suppressive effects, depending on the specific cancer type. Its involvement in cancer is closely related to death, migration, invasion, and metastasis (12, 24, 25).
In general, m6A demethylases catalyze the removal of methylation at the N6 position of adenylate in mRNA, thereby modulating the epigenetic information of mRNA. Upregulation of their expression decreases m6A modification, impacting the stability and translation efficiency of mRNA. m6A modification has been implicated in the promotion of lung cancer growth and progression, affecting the proliferation, invasion, and metastasis of lung cancer cells. Aberrant m6A modification may result from abnormal expression of writers and erasers. When exposed to external factors such as environmental pollutants, m6A modification abnormalities may occur, influencing the onset and progression of lung cancer and other cancers. Additionally, other proteins and signaling pathways involved in m6A modification may also play a role in lung cancer. m6A modification can influence the characteristics and maintenance of lung cancer stem cells, thereby facilitating tumor initiation (26, 27). It can also impact the transcription and translation processes in lung cancer cells, thereby regulating key signaling pathways and promoting tumor progression and metastasis. Moreover, m6A modification can modulate the expression of genes associated with lung cancer by altering mRNA stability, thereby affecting crucial processes such as cell cycle control and apoptosis (10, 28). Notably, lung cancer is a multifaceted disease influenced by various contributors, including genetic mutations, epigenetic alterations, and environmental factors. Thus, further investigations are needed to comprehensively elucidate the precise mechanisms underlying m6A modification in lung cancer.
Understanding the significance of m6A modification in cancer is crucial for the advancement of targeted treatment strategies and the discovery of potential biomarkers. By unraveling the intricate interplay between m6A writers, erasers, and readers, researchers can gain insights into the molecular mechanisms driving tumor progression. Recent studies have highlighted the importance of FTO and ALKBH5, as erasers involved in m6A RNA demethylation, in the development of lung cancer. These demethylases have been found to be closely associated with occurrence and development of the cancer. Furthermore, we will discuss future directions of research in this field and explore the potential clinical applications of targeting of FTO and ALKBH5 in the treatment of lung cancer. The relationship between erasers and lung cancer is shown in Figures 1, 2.
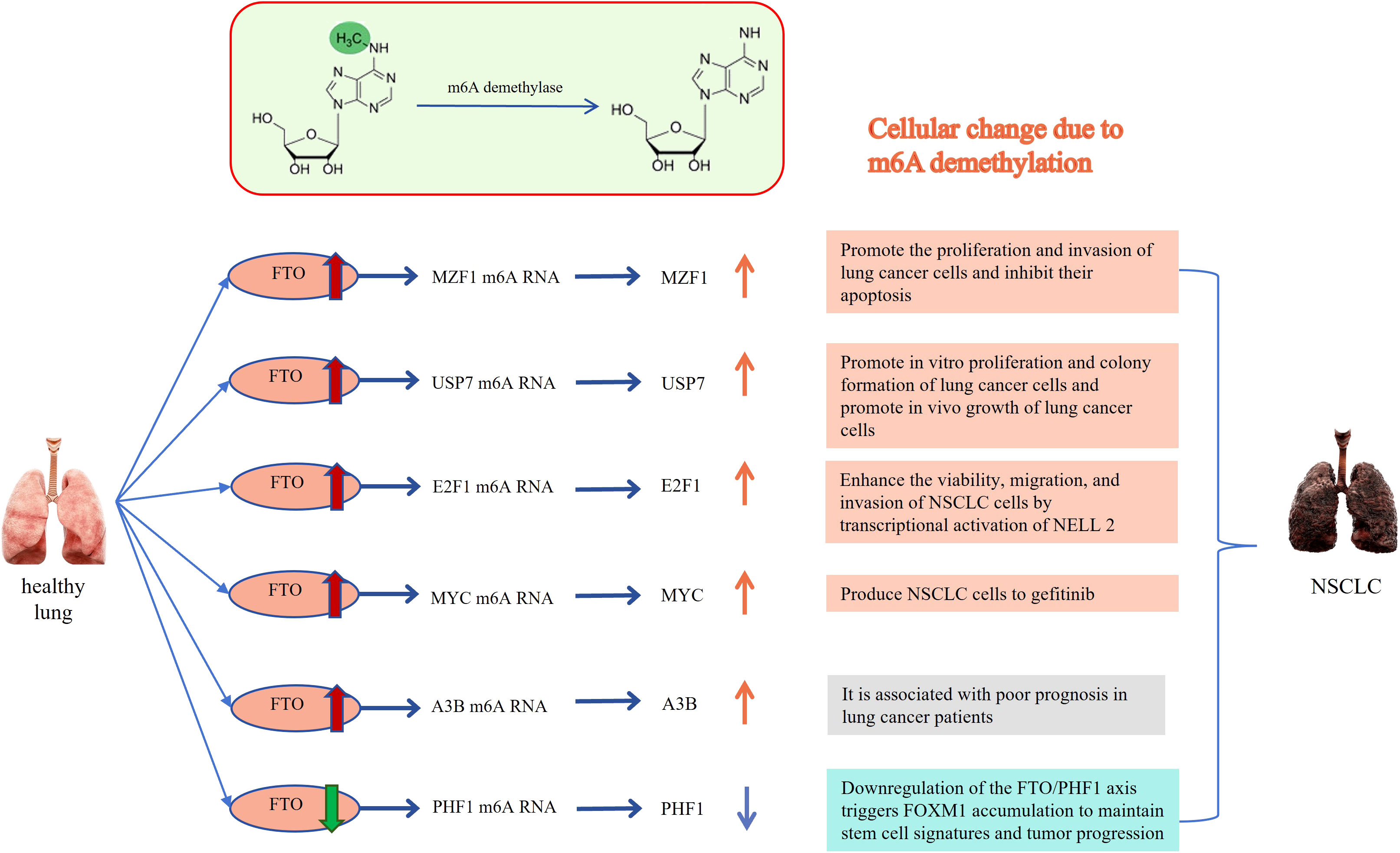
Figure 1 Relationship between FTO and lung cancer: The m6A methylated RNA can be demethylated by FTO, resulting in the occurrence of cytological behaviors that contribute to the development of NSCLC. MZF1, Myeloid Zinc Finger Protein 1; USP7, ubiquitin-specific protease-7; E2F1, E2F transcription factor-1; MYC, v-myc avian myelocytomatosis viral oncogene homolog; A3B, APOBEC3B; PHF1, Human Plant Homeodomain (PHD) finger protein 1. The Red and orange arrows represent increased m6A demethylase levels or m6A modification levels of RNA; The green and blue arrows represent decreased m6A demethylase levels or m6A modification levels of RNA.
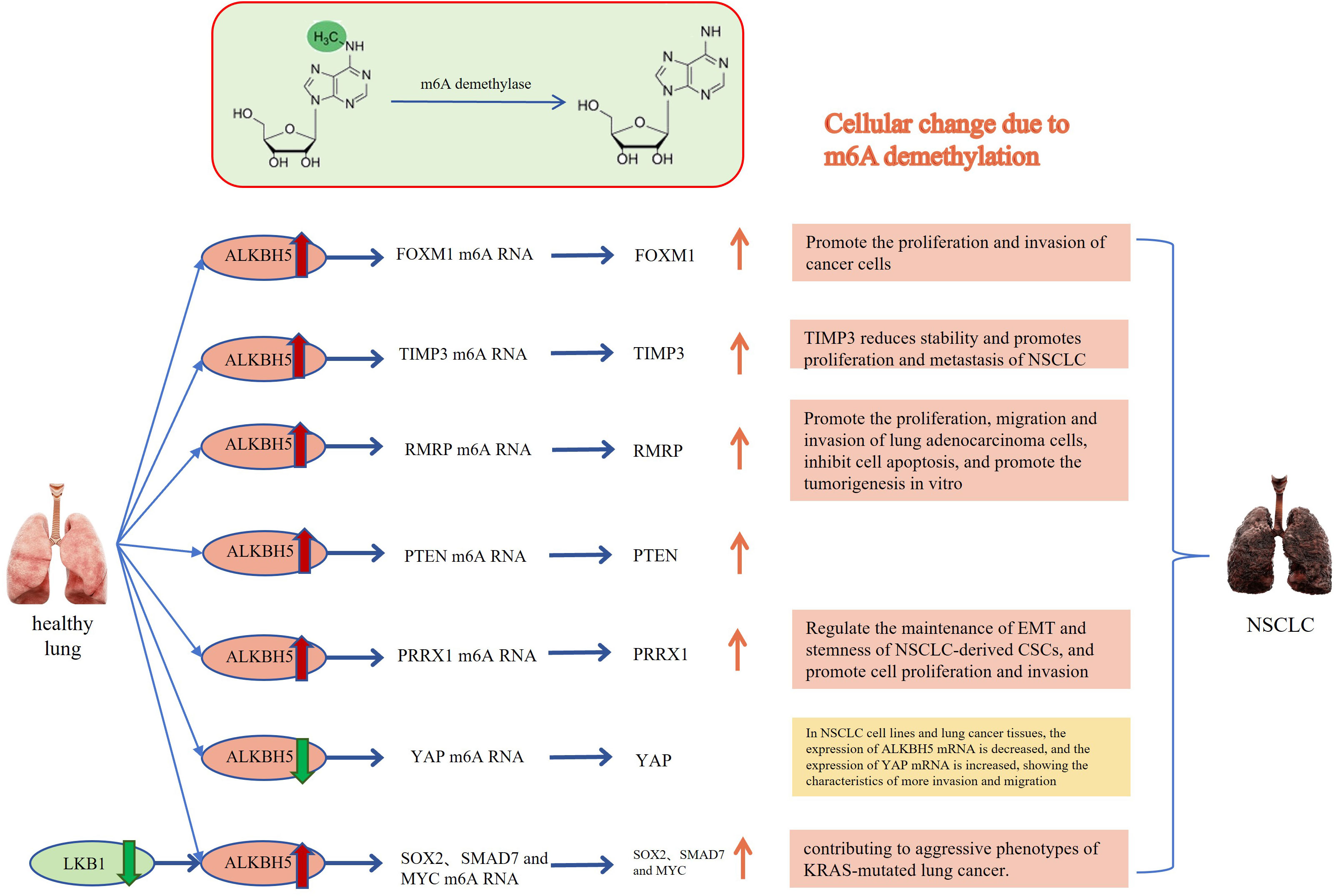
Figure 2 Relationship between ALKBH5 and lung cancer: The m6A methylated RNA undergoes demethylation through the action of ALKBH5, leading to the occurrence of cytological behaviors that promote the development of NSCLC. FOXM1, Forkhead box M1; TIMP3, TIMP metallopeptidase inhibitor 3; RMRP, RNA Component of Mitochondrial RNA Processing Endoribonuclease; PTEN, Phosphatase and tensin homolog deleted on chromosome 10; PRRX1, Paired related homeobox 1; YAP, Yes-associated protein; SOX2, SRY-box transcription factor 2; SMAD7, SMAD family member 7. The Red and orange arrows represent increased m6A demethylase levels or m6A modification levels of RNA; The green and blue arrows represent decreased m6A demethylase levels or m6A modification levels of RNA.
2 The impact of m6A demethylases on lung cancer progression
m6A, as the most prevalent mRNA modification, has methyltransferases like METTL3 being responsible for adding methyl groups to specific mRNA sites, while m6A demethylases FTO and ALKBH5 are responsible for removing methyl groups from m6A. The interaction between methyltransferases and demethylases helps maintain a balanced level of m6A. When m6A levels are stable, mRNA transcripts with appropriate m6A levels can undergo proper splicing, transport, translation, or degradation. Imbalanced m6A regulation can lead to RNA metabolism defects (29, 30). FTO and ALKBH5 are known to be upregulated in lung cancer tissues. They have been found to play a crucial in tumor cell proliferation, apoptosis, invasion, and migration, and their functions are dependent on the presence of m6A modifications (31, 32). These demethylases also contribute to the regulation of tumor progression in lung cancer (33). The possible mechanisms by which m6A demethylation affects the development and progression of lung cancer are listed in Table 1.
2.1 The role of demethylase overexpression in cell proliferation and apoptosis
The research on the relationship between demethylase and lung cancer, as well as its mechanism is still in the early stages. However, increasing evidence suggests that m6A demethylases FTO and ALKBH5 are frequently overexpressed in lung cancer and significantly associated with the tumor’s prognosis. As research progresses, the involvement of m6A modification in lung cancer is gradually being confirmed. Numerous studies have demonstrated that the demethylation process in lung cancer cells plays a crucial role in regulating the expression of key genes involved in essential cellular processes such as cell proliferation and apoptosis. Disturbances in DNA methylation patterns can disrupt the normal regulation of these genes, leading to increased proliferation of lung cancer cells and decreased apoptosis. These alterations contribute to progression and aggressiveness of tumors (46, 47). In lung cancer, abnormal DNA demethylation can occur at specific genomic regions, resulting in the activation of oncogenes or the silencing of tumor suppressor genes (48). Through the removal of the methyl groups from the DNA molecule, demethylation can lead to the overexpression of genes that promote cell proliferation, giving cancer cells a growth advantage (10). Moreover, the suppression of genes responsible for apoptosis can enable cancer cells to evade programmed cell death, aiding in their survival and expansion (28).
Li et al. conducted a study that revealed a significant association between high expression of m6A demethylase FTO and cancer development. The overexpression of FTO could promote cell proliferation and colony formation (34, 49, 50). Both mRNA and protein levels of FTO were observed to be overexpressed in human NSCLC tissues and cell lines. Conversely, the loss of FTO function resulted in a reduced proliferation rate of cancer cells. Mechanistically, FTO can decrease m6A levels through its demethylase activity, enhance the stability of ubiquitin-specific protease (USP7) mRNA, and promote the growth of lung cancer cells (49, 50). Another study demonstrated that FTO can promote the proliferation and invasion of LUSC by reducing the m6A level in myeloid zinc finger 1 (zinc finger 1, MZF1) mRNA transcription and maintaining the mRNA stability (38). Additionally, some studies have shown that circular RNAs can act as miRNA sponges, promoting cancer cell proliferation and participating in the process of NSCLC. For instance, Hsa_circ_0072309 can sponge miR-607, leading to the upregulation of its target gene FTO and promoting the occurrence of NSCLC (44). Another m6A demethylase also ALKBH5 also plays a key role in promoting or inhibiting cell proliferation and apoptosis. FOXM1 is involved in important cellular processes such as cell proliferation, cell differentiation, cell cycle regulation, angiogenesis and metastasis. It is often upregulated in human malignant tumors, indicating a poor prognosis (51, 52). Studies have shown that upregulation of ALKBH5 in cancer cells can enhance the translation efficiency of FOXM1 mRNA, leading to increased FOXM1 protein expression and promoting the proliferation and invasion of cancer cells (24). Similarly, Zhu et al. demonstrated in their studies that ALKBH 5 promotes the proliferation of NSCLC cells and reduces apoptosis in vitro. Knockdown of ALKBH5 in vivo inhibited tumor growth primarily by destabilizing TIMP 3 mRNA. ALKBH5 interacts with the methylation site of TIMP3 in the 3’UTR to facilitate protein production (10). Additionally, studies have shown that knockout ALKBH5 can induce G1 phase arrest, inhibit the cancer cell proliferation, and increase the number of apoptotic cells (43). ALKBH5 also can indirectly regulating autophagy. For instance, UBE2C, a ubiquitin-conjugating enzyme, selectively inhibits autophagy in NSCLC. Destroying UBE2C-mediated autophagy inhibition weakens the proliferation and clonogenicity of NSCLC. ALKBH5 stabilizes UBE2C transcription by reducing the m6A methylation level in its mRNA (28). Indeed, m6A demethylation plays a significant role in lung cancer cell proliferation, providing important insights into the understanding and treatment of lung cancer. The biological function and specific mechanism leading to cancer development and progression of m6A demethylation are not fully understood, and further research is required for validation.
2.2 m6A demethylases-mediated regulation of invasion and migration in lung cancer cells
Demethylation is closely related to the invasion and migration of lung cancer cells as well. Studies have shown that demethylation can affect the metastatic ability of lung cancer cells, thus facilitating the metastasis and invasion of these cells. Demethylation plays a role in gene expression regulation, particularly in relation to cell metastasis by transcription factors and signaling molecules. One example is the activation of cell migration by FTO through mRNA demethylation. This activation can contribute to the progression of lung cancer and affect the prognosis of patients (35). Liu et al.’s study demonstrated that the higher levels of FTO are associated with worse prognosis in LUSC patients. FTO acts as an m6A mRNA demethylase, promoting cell migration and invasion (38). Another study found that FTO can inhibit the M6A modification of E2F1 in vivo, leading to increased expression of E2F1. E2F1, in turn, enhances the survival, migration, and invasion of NSCLC cells by activating NELL2 transcriptionally. Furthermore, FTO can promote the formation and metastasis of NSCLC through the FTO/E2F1/NELL2 axis in vitro (36). As for ALKBH5, some studies have found that ALKBH5 can promote cell migration and invasion. In lung cancer patients, the levels of ALKBH5 protein expression have been observed to be positively associated with tumor size, TNM staging, and clinical staging. Research by Yu et al. indicated that patients with higher ALKBH5 gene expression had a poorer prognosis (53). ALKBH5 upregulates the expression of RMRP through demethylation, and the upregulation of RMRP promotes the proliferation and invasion of lung adenocarcinoma cells while inhibiting cell apoptosis. However, other researchers have discovered that overexpression of ALKBH5 effectively reverses the proliferation, colony formation and migration of kras mutant lung cancer cells that are regulated by LKB1 (47).
The impact of demethylation on the phenotypic transition of cells, such as the transition from epithelial cells to mesenchymal cells, and its effect on the invasion and migration of lung cancer cells have yielded different conclusions, including both promotion and inhibition of lung cancer cell migration by ALKBH. Experiments conducted by Guo et al. demonstrated that downregulation of ALKBH5 using miR-381 and siRNA specifically impedes the EMT of NSCLC cells, thereby inhibiting their migration and invasive growth (28). Liu et al. also found that ALKBH 5 is highly expressed in NSCLC-derived CSCs (cancer stem cells), and downregulation of ALKBH5 significantly reduces Sox2 expression, leading to increased levels of E-cadherin protein, inhibition of EMT, and suppression of tumor invasive development and metastasis (54). However, some researchers have come to the opposite conclusion. Studies have shown that TGF-β induces EMT in NSCLC and regulate cell migration and invasion. Interestingly, overexpression of ALKBH 5 can inhibit the metastasis of NSCLC cells stimulated by TGF-β in vivo (45). Jin et al. also believe that ALKBH5 acts as an inhibitor of NSCLC cell migration and invasion, ALKBH5 regulates the miR-107/LATS2 axis in a HURP-dependent manner, thereby reducing the expression and activity of YAP, thereby inhibiting tumor growth and metastasis (48). The possible explanation for this phenomenon is that when the gene interacts with a tumor suppressor gene it promotes cancer cell growth and metastasis while when it interacts with an oncogene it inhibits cancer cell growth. In addition, lung cancer is a highly heterogeneous disease, and biological diversity and subtype differences in lung cancer need to be considered. The specific reasons for the differences still need to be explored. Notably, patients with Trp53 gene mutations tend to have a poorer prognosis when FTO is expressed at high levels, while patients with the wild-type gene don’t exhibit the same trend (39). This suggests that the observed phenomenon may be associated with specific genotype mutations, highlighting the need for further research. Overall, demethylation is a critical factor in the invasion and migration of lung cancer cells, playing a role in their metastatic potential. By influencing gene expression, the phenotypic transition of cells and other processes, demethylation can promote the acquisition of invasive and migratory characteristics in lung cancer cells. Further research focusing on the identification of specific genes and molecular pathways affected by demethylation in the context of lung cancer metastasis could offer valuable insights for targeted therapies to address metastatic disease.
3 Potential applications of m6A demethylase in the diagnosis, prognosis assessment and treatment of lung cancer
3.1 Application of m6A demethylase in the lung cancer diagnosis
Recent studies have shown that significant application value of m6A demethylase in diagnosing lung cancer. Specifically, by comparing the expression levels of m6A demethylase in lung cancer tissue and normal lung tissue, researchers have observed significant alterations in its expression, either upregulation or downregulation, indicating its potential as a diagnostic marker for lung cancer. These findings have garnered considerable attention in the field of lung cancer diagnosis. Among the m6A demethylases, FTO and ALKBH5 may be associated with lung cancer, and their relationship has been gradually studied. These enzymes have demonstrated potential in developing risk assessment models that enhance the accuracy of diagnosing lung adenocarcinoma (LUAD), a prevalent histological subtype of lung cancer. By incorporating methylation-related enzymes, such as FTO and ALKBH5, into these risk assessment models, researchers have achieved improved prediction accuracy for LUAD diagnosis (55). Moreover, evaluating the expression levels of m6A demethylases in blood or tissue samples from lung cancer patients holds the potential for early diagnosis, pathological classification, and staging of lung cancer staging (47). The application of m6A demethylases in lung cancer diagnosis is an exciting area of research with significant clinical implications. By incorporating these enzymes into diagnostic algorithms and utilizing their association with tumorigenesis, healthcare professionals may enhance the accuracy and efficiency of lung cancer diagnosis (56). Additionally, the ability to detect m6A demethylase expression in various sample types, including blood and tissue, may offer a non-invasive and easily accessible approach for the early detection and monitoring of lung cancer (57, 58). Indeed, additional research is required to validate and enhance the diagnostic potential of m6A demethylases in lung cancer. Large-scale clinical studies involving diverse patient populations are necessary to establish robust diagnostic models and determine the specific thresholds for demethylase expression levels in different stages and subtypes of lung cancer (59, 60). These investigations will aid in developing standardized diagnostic protocols and facilitate the integration of m6A demethylases into routine clinical practice for accurate lung cancer diagnosis. In summary, recent studies have demonstrated the potential value of m6A demethylases in diagnosing lung cancer. The differential expression of these enzymes in lung cancer tissue and their potential integration into risk assessment models underscore their diagnostic potential (61). Moreover, assessing m6A demethylase expression levels in blood or tissue samples shows promise for early diagnosis, pathological typing, and lung cancer staging (62). Further research and validation are needed to fully realize the diagnostic capabilities of m6A demethylases, leading to improved lung cancer diagnosis and patient care.
3.2 Application of m6A demethylase in the lung cancer prognosis assessment
m6A demethylases have shown significant potential in the prognostic assessment of lung cancer. The expression levels of these enzymes in lung cancer tissues have been strongly correlated with patient survival rates and overall prognosis (63). Abnormally expression of demethylases, either too high or too low, may serve as indicators of poor clinical outcomes. Studies have indeed reported that high expression of ALKBH5 and FTO is associated with a favourable prognosis in lung cancer patients (64). Elevated levels of these demethylases may indicate better outcomes for patient (65). Furthermore, the expression levels of m6A demethylases can be used to develop prognostic models to assess the risk of poor prognosis in patients. Recent research has focused on the construction of such models using factors including FTO and other genes. For instance, Zhang et al. developed a model, and found that a two-gene model combining FTO and METTL3 was more effective in guiding prognostic assessment of lung cancer (66). Additionally, researchers have utilized methylation-related enzymes such as FTO and ALKBH5 to develop risk assessment models for LUAD patients. These models divide LUAD patients into high-risk and low-risk categories based on the constructed models, and correlations have been observed with various clinical factors including TNM staging, lymph node staging, gender, and tumour stage (55). This suggests that the risk assessment model based on m6A demethylases can provide valuable insights into the prognosis and clinical characteristics of LUAD patients. These findings highlight the potential of m6A demethylases as prognostic markers in lung cancer. Incorporating m6A demethylase expression levels into prognostic models or risk assessment models can improve the accuracy of prognostic evaluation and assist in clinical decision-making. By considering the expression levels of these demethylases along with other relevant clinical factors, healthcare professionals can better predict patient outcomes and tailor treatment strategies accordingly. It is important to note that further research and validation are necessary to establish standardized prognostic models incorporating m6A demethylases in lung cancer (67, 68). Large-scale studies involving diverse patient populations must be conducted to confirm the associations between demethylase expression, prognostic risk, and clinical characteristics (69). These efforts will contribute to developing reliable prognostic tools that can guide patient management and improve prognostic assessments in lung cancer. m6A demethylase, along with other m6A regulatory factors, collaborates to regulate methylation in the body. Relying solely on ALKBH5 and FTO is not reliable for diagnosing and predicting prognosis in lung cancer patients. Therefore, it is more meaningful to collectively detect various regulatory factors than focusing on individual ones, as it can better reflect the actual conditions of lung cancer patients (70, 71). In clinical practice, monitoring the expression level of m6A demethylase in lung cancer patients is expected to offer personalized treatment recommendations and an accurate assessment of efficacy and prognosis. Numerous studies have indicated that m6A demethylase has the potential as a diagnostic and prognostic marker for lung cancer.
3.3 Application of m6A demethylation drugs in lung cancer treatment
Lung cancer, particularly NSCLC, continues to be the primary cause of cancer-associated mortality globally, constituting 85% of newly diagnosed cases (72). With the in-depth study of m6A demethylase mechanism in lung cancer, the potential application of m6A demethylation drugs in treating lung cancer is becoming increasingly evident. Significant progress has been made in the laboratory and clinical trials small molecule inhibitors and activators targeting demethylases such as FTO and ALKBH5 have made certain research progress. At present, the most commonly used m6A demethylase drugs are mainly focused on m6A demethylase inhibitors, which can regulate the activity of m6A demethylase by binding to its active site. FTO inhibitors included FB23 and FB23-2, rhein, Meclofenamic acid, fluorescein, R-2-hydroxyglutarate (R-2HG), et al. (73–77). In recent years, 44/ZLD115, Xanthine derivatives and other FTO inhibitors have been developed. Among them, 44/ZLD115 shows good anti-leukemia activity in xenograft mice and is a very promising FTO inhibitor (78, 79). With the continuous progress of scientific research, the development of ALKBH inhibitors is also ongoing. For example,2- [(1-hydroxy-2-oxo-2-phenylethyl) sulfanyl] acetic acid (3) and 4-[(furan-2-yl) methyl] amino-1,2-diazinane-3,6-dione (6). These two novel ALKBH5 inhibitors could selectively inhibit the growth of leukemia cell lines (80). These drugs have shown promise in affecting tumor cell growth, invasion and metastasis by regulating the activity of m6A demethylase. However, the current development of m6A demethylase inhibitors is mainly focused on various types of leukemia, breast cancer, bowel cancer and other cancers, but they have shown promising characteristics, while m6A demethylase inhibitors for lung cancer still need to be developed by researchers.
A research team reported that the synthesis of an FTO inhibitor, which demonstrated its ability to inhibit cancer progression via the inhibition of cell invasion, migration, and EMT (epithelial-mesenchymal transition). Additionally, the inhibitor showed potential in inhibiting angiogenesis, a critical process for tumor growth and metastasis (27). Therapeutic resistance to multiple small molecules, including chemotherapeutics and targeted agents, is a significant factor contributing to poor prognosis in NSCLC (71, 81). Gefitinib, an essential drugs for treating lung cancer, has been found to be associated with demethylase-related resistance in NSCLC. Specifically, studies have reported that there is an association between the methylation level of WIF1 in cfDNA (Cell-free DNA) and the insensitivity of gefitinib in the treatment of lung cancer. In patients with more advanced disease, the DNA methylation levels of the WIF1 promoter are significantly elevated (41). Co-administration of GE (gefitinib) with MA (meclofenamic acid) has been demonstrated to enhance the sensitivity of drug-resistant NSCLC cells to treatment (42). This effect is attributed to the inhibition of BCRP and MRP7 expression levels through the FTO/m6A/MYC axis. These findings suggest that combining drugs can potentially overcome treatment resistance in NSCLC. Moreover, exosomes derived from gefitinib-resistant (GR) cells can play a role in intercellular transmission of gefitinib resistant through the FTO/YTHDF2/ABCC10 axis. These findings confirm the Feasibility of targeting FTO-m6A axis to prevent or delay the acquisition of gefitinib resistance in NSCLC (40).
In the future, m6A demethylation drugs have the potential to become a vital component of the comprehensive treatment strategy for lung cancer. By combining these drugs with traditional treatments such as surgery, radiotherapy, and chemotherapy, it is expected that the survival rate and quality of life for lung cancer patients can be improved. It is important to highlight that the research and development of drugs targeting m6A demethylation still encounter certain challenges. Firstly, there is a need to enhance the effectiveness of these drugs and minimize the occurrence of side effects by further refining their the selectivity and specificity (82). In addition, owing to the considerable heterogeneity of lung cancer, variations in the response of different patients to m6A demethylating drugs may exist (83). Hence, it is imperative to study biomarkers that can predict patient response to drugs and prognosis in order to achieve precision medicine (84). Overall, m6A demethylases hold significant potential for applications in lung cancer diagnosis, prognosis assessment, and therapy. Future research should focus on gaining a deeper understanding of the mechanism of action of m6A demethylase, expediting the development of drugs, and providing more effective and personalized treatment options for lung cancer patients (85). Additionally, interdisciplinary cooperation and clinical trials will play an essential role in advancing the application of M6A demethylase in lung cancer.
4 Environment specific factor for abnormal m6A modification in lung cancer
Research has indicated that prolonged exposure to environments with heavy metals increases the risk of lung cancer in humans. For instance, exposure to beryllium (Be) and arsenic (As) compounds, both in vivo and in vitro is strongly associated with the occurrence and development of lung cancer (86). m6A demethylase also plays a role in the interaction between heavy metals and lung cancer. For example, FTO protein is highly expressed in tumor samples of NSCLC patients and it can mediate the reduction of m6A modification induced by arsenic in A3B, resulting in increased expression of A3B (87). Similarly, ALKBH5 can regulate the m6A methylation level of PTEN mRNA and reduce the stability of PTEN mRNA and promoting the cadmium-induced malignant transformation of human bronchial epithelial cells, as well as enhancing proliferation, migration and invasion of cancer cells (88). Furthermore, ALKBH5 may be involved in silica-induced pulmonary fibrosis, which may be through the miR-320a-3p/FOXM1 axis or by directly targeting FOXM1 (89). However, there is limited knowledge regarding how m6A demethylases contribute to the pathogenesis of lung cancer and their relationship with heavy metal exposure. Studying and understanding these mechanisms is of great significance as it can provide new ideas and methods for the treatment and prevention lung cancer.
5 Discussion
In recent years, m6A demethylase, a novel RNA-modifying enzyme, has emerged as a prominent molecular mechanisms in lung cancer research. Studies have demonstrated that m6A demethylase affects the stability, transcription, and translation of target genes, subsequently impacting the proliferation, migration, invasion and other processes of lung cancer cells. It also interacts with other molecules such as MZF1, USP7, FOXM1, forming a complex molecular regulatory network. Additionally, m6A demethylase can regulate certain MicroRNAs, including miR-107, miR-607 (44, 48), thereby affecting various physiological functions in the body. Furthermore, researchers have discovered that m6A demethylase may also have an impact on cancer metastasis. For example, m6A modification can regulate the malignancy of breast cancer lung metastasis cells, and overexpression of FTO can significantly inhibit the lung colonization of BT-549LMF3 cells (26). All these findings highlight the close association between m6A demethylase and the malignant progression of lung cancer. Increasing evidence suggests that m6A demethylase holds significant potential in the diagnosis, prognosis evaluation, and treatment of lung cancer. However, the current research is unable to fully elucidate the complete functionality of m6A demethylase. m6A methylation can be likened to a double-edged sword since its overexpression may contribute to certain tumor types, while the absence of m6A modification may drive the progression of other tumors. The inconsistent results observed among researchers can be attributed to various factors. Therefore, conducting more multicenter, large-scale studies is imperative in order to delve deeper into this topic and establish a solid foundation for the effective treatment of human tumors. Advancing the field of m6A demethylases requires a thorough investigation into their underlying mechanisms. By unravelling the complex molecular pathways and regulatory networks involved in the malignant transformation of lung cancer cells, researchers can lay the foundation for development of more precise and targeted treatments (90). Gaining a comprehensive understanding of the specific role of m6A demethylases within these networks will provide valuable theoretical insights and offer practical support for the design and implementation of effective therapeutic strategies. Additionally, investigating molecular network regulation models involving m6A demethylases in lung cancer holds great potential. By unraveling the intricate interactions between m6A demethylases and other critical genes implicated in the progression of lung cancer, researchers can identify novel targets and pathways that can be targeted for therapeutic purposes (91, 92). This approach has the potential to revolutionize the treatment landscape by enabling the development of innovative combination therapies or the identification of specific molecular signatures that can guide personalized treatment approaches for individuals with lung cancer.
Although it will be some time before m6A demethylase can be applied to clinical practice, the current study has shown its translational value. This requires large-scale clinical samples and cell model studies to obtain sufficient data support. In addition, accurate and meticulous analysis of the large amount of RNA modification data is also a difficult point that needs to be broken through. The development of targeted drugs for different stages of lung cancer may be challenging and promising.
6 Conclusion
Overall, m6A modification is one of the most common RNA modifications. It not only plays an important role in various cell biological processes, but also participates in the occurrence and development of cancers, such as lung cancer, acute myeloid leukemia and breast cancer, etc. Therefore, m6A-modified related molecules are considered as potential tumor diagnostic markers and therapeutic targets. Conducting in-depth research on the mechanisms of m6A demethylase function and its intricate network interactions in lung cancer will greatly enhance our understanding of the disease and o provide opportunities for targeted treatments. Through rigorous scientific inquiry, researchers can establish the groundwork for the development of more effective therapies to fight against lung cancer and well-being of patients affected by this condition.
Author contributions
MY: Methodology, Conceptualization, Data curation, Formal Analysis, Investigation, Writing – original draft. WJ: Methodology, Supervision, Writing – review & editing. XY: Data curation, Formal Analysis, Writing – review & editing. KT: Writing – review & editing, Formal Analysis, Visualization. XM: Formal Analysis, Visualization, Writing – review & editing. SY: Writing – review & editing, Conceptualization, Supervision. LC: Conceptualization, Supervision, Writing – review & editing. XZ: Supervision, Writing – review & editing, Funding acquisition, Methodology, Project administration.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (82173554); the Natural Science Foundation of Jiangsu Province (BK20201444); Qing Lan Project for Excellent Young Key Teachers of Colleges and Universities of Jiangsu Province (2020).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer ZD declared a shared affiliation with the author WJ to the handling editor at the time of review.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin (2023) 73(1):17–48. doi: 10.3322/caac.21763
2. Leon G, MacDonagh L, Finn SP, Cuffe S, Barr MP. Cancer stem cells in drug resistant lung cancer: Targeting cell surface markers and signaling pathways. Pharmacol Ther (2016) 158:71–90. doi: 10.1016/j.pharmthera.2015.12.001
3. Sun L, Zhang H, Gao P. Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell (2022) 13(12):877–919. doi: 10.1007/s13238-021-00846-7
4. Xu J, Liu Y, Liu J, Xu T, Cheng G, Shou Y, et al. The identification of critical m(6)A RNA methylation regulators as Malignant prognosis factors in prostate adenocarcinoma. Front Genet (2020) 11:602485. doi: 10.3389/fgene.2020.602485
5. Chen Y, Zhou W, Gong Y, Ou X. N6-methyladenosine-related long noncoding RNA is a potential biomarker for predicting pancreatic cancer prognosis. Tumor Discovery (2022) 1(2):165. doi: 10.36922/td.v1i2.165
6. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell (2017) 169(7):1187–200. doi: 10.1016/j.cell.2017.05.045
7. Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. BioMed Pharmacother (2019) 112:108613. doi: 10.1016/j.biopha.2019.108613
8. Chang G, Shi L, Ye Y, Shi H, Zeng L, Tiwary S, et al. YTHDF3 induces the translation of m(6)A-enriched gene transcripts to promote breast cancer brain metastasis. Cancer Cell (2020) 38(6):857–71 e7. doi: 10.1016/j.ccell.2020.10.004
9. Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res (2020) 48(7):3816–31. doi: 10.1093/nar/gkaa048
10. Zhu Z, Qian Q, Zhao X, Ma L, Chen P. N(6)-methyladenosine ALKBH5 promotes non-small cell lung cancer progress by regulating TIMP3 stability. Gene (2020) 731:144348. doi: 10.1016/j.gene.2020.144348
11. Yang P, Wang Q, Liu A, Zhu J, Feng J. ALKBH5 holds prognostic values and inhibits the metastasis of colon cancer. Pathol Oncol Res (2020) 26(3):1615–23. doi: 10.1007/s12253-019-00737-7
12. Zhang J, Guo S, Piao HY, Wang Y, Wu Y, Meng XY, et al. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem (2019) 75(3):379–89. doi: 10.1007/s13105-019-00690-8
13. Muller S, Glass M, Singh AK, Haase J, Bley N, Fuchs T, et al. IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res (2019) 47(1):375–90. doi: 10.1093/nar/gky1012
14. Yang X, Zhang S, He C, Xue P, Zhang L, He Z, et al. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol Cancer (2020) 19(1):46. doi: 10.1186/s12943-020-1146-4
15. Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B, et al. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol Cancer (2020) 19(1):106. doi: 10.1186/s12943-020-01220-7
16. Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell (2016) 62(3):335–45. doi: 10.1016/j.molcel.2016.03.021
17. Wei W, Sun J, Zhang H, Xiao X, Huang C, Wang L, et al. Circ0008399 interaction with WTAP promotes assembly and activity of the m(6)A methyltransferase complex and promotes cisplatin resistance in bladder cancer. Cancer Res (2021) 81(24):6142–56. doi: 10.1158/0008-5472.CAN-21-1518
18. Cho S, Lee G, Pickering BF, Jang C, Park JH, He L, et al. mTORC1 promotes cell growth via m(6)A-dependent mRNA degradation. Mol Cell (2021) 81(10):2064–75. e8. doi: 10.1016/j.molcel.2021.03.010
19. Li S, Qi Y, Yu J, Hao Y, He B, Zhang M, et al. Nuclear Aurora kinase A switches m(6)A reader YTHDC1 to enhance an oncogenic RNA splicing of tumor suppressor RBM4. Signal Transduct Target Ther (2022) 7(1):97. doi: 10.1038/s41392-022-00905-3
20. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature (2014) 505(7481):117–20. doi: 10.1038/nature12730
21. Ning J, Wang F, Bu J, Zhu K, Liu W. Down-regulated m6A reader FTO destabilizes PHF1 that triggers enhanced stemness capacity and tumor progression in lung adenocarcinoma. Cell Death Discovery (2022) 8(1):354. doi: 10.1038/s41420-022-01125-y
22. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol (2011) 7(12):885–7. doi: 10.1038/nchembio.687
23. Purslow JA, Nguyen TT, Egner TK, Dotas RR, Khatiwada B, Venditti V. Active site breathing of human alkbh5 revealed by solution NMR and accelerated molecular dynamics. Biophys J (2018) 115(10):1895–905. doi: 10.1016/j.bpj.2018.10.004
24. Chao Y, Shang J, Ji W. ALKBH5-m(6)A-FOXM1 signaling axis promotes proliferation and invasion of lung adenocarcinoma cells under intermittent hypoxia. Biochem Biophys Res Commun (2020) 521(2):499–506. doi: 10.1016/j.bbrc.2019.10.145
25. Zhang C, Zhi WI, Lu H, Samanta D, Chen I, Gabrielson E, et al. Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget (2016) 7(40):64527–42. doi: 10.18632/oncotarget.11743
26. Chen F, Chen Z, Guan T, Zhou Y, Ge L, Zhang H, et al. N(6) -methyladenosine regulates mRNA stability and translation efficiency of KRT7 to promote breast cancer lung metastasis. Cancer Res (2021) 81(11):2847–60. doi: 10.1158/0008-5472.CAN-20-3779
27. Xie G, Wu XN, Ling Y, Rui Y, Wu D, Zhou J, et al. A novel inhibitor of N (6)-methyladenosine demethylase FTO induces mRNA methylation and shows anti-cancer activities. Acta Pharm Sin B (2022) 12(2):853–66. doi: 10.1016/j.apsb.2021.08.028
28. Guo J, Wu Y, Du J, Yang L, Chen W, Gong K, et al. Deregulation of UBE2C-mediated autophagy repression aggravates NSCLC progression. Oncogenesis (2018) 7(6):49. doi: 10.1038/s41389-018-0054-6
29. Zhang H, Shi X, Huang T, Zhao X, Chen W, Gu N, et al. Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res (2020) 48(11):6251–64. doi: 10.1093/nar/gkaa347
30. Wang J, Wang K, Liu W, Cai Y, Jin H. m6A mRNA methylation regulates the development of gestational diabetes mellitus in Han Chinese women. Genomics (2021) 113(3):1048–56. doi: 10.1016/j.ygeno.2021.02.016
31. Zhao T, Wang J, Wu Y, Han L, Chen J, Wei Y, et al. Increased m6A modification of RNA methylation related to the inhibition of demethylase FTO contributes to MEHP-induced Leydig cell injury(☆). Environ Pollut (2021) 268(Pt A):115627. doi: 10.1016/j.envpol.2020.115627
32. Hu Y, Gong C, Li Z, Liu J, Chen Y, Huang Y, et al. Demethylase ALKBH5 suppresses invasion of gastric cancer via PKMYT1 m6A modification. Mol Cancer (2022) 21(1):34. doi: 10.1186/s12943-022-01522-y
33. Feng M, Xie X, Han G, Zhang T, Li Y, Li Y, et al. YBX1 is required for maintaining myeloid leukemia cell survival by regulating BCL2 stability in an m6A-dependent manner. Blood (2021) 138(1):71–85. doi: 10.1182/blood.2020009676
34. Li J, Han Y, Zhang H, Qian Z, Jia W, Gao Y, et al. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem Biophys Res Commun (2019) 512(3):479–85. doi: 10.1016/j.bbrc.2019.03.093
35. Ding Y, Qi N, Wang K, Huang Y, Liao J, Wang H, et al. FTO facilitates lung adenocarcinoma cell progression by activating cell migration through mRNA demethylation. Onco Targets Ther (2020) 13:1461–70. doi: 10.2147/OTT.S231914
36. Wang Y, Li M, Zhang L, Chen Y, Zhang S. m6A demethylase FTO induces NELL2 expression by inhibiting E2F1 m6A modification leading to metastasis of non-small cell lung cancer. Mol Ther Oncolytics (2021) 21:367–76. doi: 10.1016/j.omto.2021.04.011
37. Shen W, Pu J, Zuo Z, Gu S, Sun J, Tan B, et al. The RNA demethylase ALKBH5 promotes the progression and angiogenesis of lung cancer by regulating the stability of the LncRNA PVT1. Cancer Cell Int (2022) 22(1):353. doi: 10.1186/s12935-022-02770-0
38. Liu J, Ren D, Du Z, Wang H, Zhang H, Jin Y. m(6)A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem Biophys Res Commun (2018) 502(4):456–64. doi: 10.1016/j.bbrc.2018.05.175
39. Zhao Z, Wan J, Guo M, Wang Y, Yang Z, Zhou F, et al. Expression and prognostic significance of m6A-related genes in TP53-mutant non-small-cell lung cancer. J Clin Lab Anal (2022) 36(1):e24118. doi: 10.1002/jcla.24118
40. Xiao P, Liu YK, Han W, Hu Y, Zhang BY, Liu WL. Exosomal Delivery of FTO Confers Gefitinib Resistance to Recipient Cells through ABCC10 Regulation in an m6A-dependent Manner. Mol Cancer Res (2021) 19(4):726–38. doi: 10.1158/1541-7786.MCR-20-0541
41. Shen Z, Chen C, Sun J, Huang J, Liu S. The status of WIF1 methylation in cell-free DNA is associated with the insusceptibility for gefitinib in the treatment of lung cancer. J Cancer Res Clin Oncol (2021) 147(8):2239–48. doi: 10.1007/s00432-021-03640-4
42. Chen H, Jia B, Zhang Q, Zhang Y. Meclofenamic acid restores gefinitib sensitivity by downregulating breast cancer resistance protein and multidrug resistance protein 7 via FTO/m6A-demethylation/c-myc in non-small cell lung cancer. Front Oncol (2022) 12:870636. doi: 10.3389/fonc.2022.870636
43. Tsuchiya K, Yoshimura K, Iwashita Y, Inoue Y, Ohta T, Watanabe H, et al. m(6)A demethylase ALKBH5 promotes tumor cell proliferation by destabilizing IGF2BPs target genes and worsens the prognosis of patients with non-small-cell lung cancer. Cancer Gene Ther (2022) 29(10):1355–72. doi: 10.1038/s41417-022-00451-8
44. Mo WL, Deng LJ, Cheng Y, Yu WJ, Yang YH, Gu WD. Circular RNA hsa_circ_0072309 promotes tumorigenesis and invasion by regulating the miR-607/FTO axis in non-small cell lung carcinoma. Aging (Albany NY) (2021) 13(8):11629–45. doi: 10.18632/aging.202856
45. Sun Z, Su Z, Zhou Z, Wang S, Wang Z, Tong X, et al. RNA demethylase ALKBH5 inhibits TGF-beta-induced EMT by regulating TGF-beta/SMAD signaling in non-small cell lung cancer. FASEB J (2022) 36(5):e22283. doi: 10.1096/fj.202200005RR
46. Li H, Zhang Y, Guo Y, Liu R, Yu Q, Gong L, et al. ALKBH1 promotes lung cancer by regulating m6A RNA demethylation. Biochem Pharmacol (2021) 189:114284. doi: 10.1016/j.bcp.2020.114284
47. Zhang D, Ning J, Okon I, Zheng X, Satyanarayana G, Song P, et al. Suppression of m6A mRNA modification by DNA hypermethylated ALKBH5 aggravates the oncological behavior of KRAS mutation/LKB1 loss lung cancer. Cell Death Dis (2021) 12(6):518. doi: 10.1038/s41420-021-00435-x
48. Jin D, Guo J, Wu Y, Yang L, Wang X, Du J, et al. m(6)A demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs-mediated YAP expression and inhibiting miR-107/LATS2-mediated YAP activity in NSCLC. Mol Cancer (2020) 19(1):40. doi: 10.1186/s12943-020-01161-1
49. Sun Y, Cao L, Sheng X, Chen J, Zhou Y, Yang C, et al. WDR79 promotes the proliferation of non-small cell lung cancer cells via USP7-mediated regulation of the Mdm2-p53 pathway. Cell Death Dis (2017) 8(4):e2743. doi: 10.1038/cddis.2017.162
50. Wang Y, Wen L, Zhao SH, Ai ZH, Guo JZ, Liu WC. FoxM1 expression is significantly associated with cisplatin-based chemotherapy resistance and poor prognosis in advanced non-small cell lung cancer patients. Lung Cancer (2013) 79(2):173–9. doi: 10.1016/j.lungcan.2012.10.019
51. Liao GB, Li XZ, Zeng S, Liu C, Yang SM, Yang L, et al. Regulation of the master regulator FOXM1 in cancer. Cell Commun Signal (2018) 16(1):57. doi: 10.1186/s12964-018-0266-6
52. Malapelle U, Morra F, Ilardi G, Visconti R, Merolla F, Cerrato A, et al. USP7 inhibitors, downregulating CCDC6, sensitize lung neuroendocrine cancer cells to PARP-inhibitor drugs. Lung Cancer (2017) 107:41–9. doi: 10.1016/j.lungcan.2016.06.015
53. Yu H, Zhang Z. ALKBH5-mediated m6A demethylation of lncRNA RMRP plays an oncogenic role in lung adenocarcinoma. Mamm Genome (2021) 32(3):195–203. doi: 10.1007/s00335-021-09872-6
54. Liu X, Wang Z, Yang Q, Hu X, Fu Q, Zhang X, et al. RNA Demethylase ALKBH5 Prevents Lung Cancer Progression by Regulating EMT and Stemness via Regulating p53. Front Oncol (2022) 12:858694. doi: 10.3389/fonc.2022.858694
55. Zhuang Z, Chen L, Mao Y, Zheng Q, Li H, Huang Y, et al. Diagnostic, progressive and prognostic performance of m(6)A methylation RNA regulators in lung adenocarcinoma. Int J Biol Sci (2020) 16(11):1785–97. doi: 10.7150/ijbs.39046
56. Li N, Zhan X. Identification of pathology-specific regulators of m(6)A RNA modification to optimize lung cancer management in the context of predictive, preventive, and personalized medicine. EPMA J (2020) 11(3):485–504. doi: 10.1007/s13167-020-00220-3
57. Liu WR, Zhang B, Chen C, Li Y, Ye X, Tang DJ, et al. Detection of circulating genetically abnormal cells in peripheral blood for early diagnosis of non-small cell lung cancer. Thorac Cancer (2020) 11(11):3234–42. doi: 10.1111/1759-7714.13654
58. Yang Q, Chen M, Gu J, Niu K, Zhao X, Zheng L, et al. Novel biomarkers of dynamic blood PD-L1 expression for immune checkpoint inhibitors in advanced non-small-cell lung cancer patients. Front Immunol (2021) 12:665133. doi: 10.3389/fimmu.2021.665133
59. Li TT, Gao X, Gao L, Gan BL, Xie ZC, Zeng JJ, et al. Role of upregulated miR-136-5p in lung adenocarcinoma: A study of 1242 samples utilizing bioinformatics analysis. Pathol Res Pract (2018) 214(5):750–66. doi: 10.1016/j.prp.2018.02.017
60. Ye Z, Zhang X, Luo Y, Li S, Huang L, Li Z, et al. Prognostic values of vimentin expression and its clinicopathological significance in non-small cell lung cancer: A meta-analysis of observational studies with 4118 cases. PloS One (2016) 11(9):e0163162. doi: 10.1371/journal.pone.0163162
61. Li Y, Qi D, Zhu B, Ye X. Analysis of m6A RNA methylation-related genes in liver hepatocellular carcinoma and their correlation with survival. Int J Mol Sci (2021) 22(3):1474. doi: 10.3390/ijms22031474
62. Zhang M, Yang C, Ruan X, Liu X, Wang D, Liu L, et al. CPEB2 m6A methylation regulates blood-tumor barrier permeability by regulating splicing factor SRSF5 stability. Commun Biol (2022) 5(1):908. doi: 10.1038/s42003-022-03878-9
63. Monzon AZ, Franco-Lopez Á, Culebras JM. Benefits and harms of screening: Overdiagnosis and anticipatory medicine – A secondary publication. Tumor Discovery (2022) 1(2):228. doi: 10.36922/td.v1i2.228
64. Zhu J, Wang M, Hu D. Deciphering N(6)-methyladenosine-related genes signature to predict survival in lung adenocarcinoma. BioMed Res Int (2020) 2020:2514230. doi: 10.1155/2020/2514230
65. Wang L, Feng X, Jiao Z, Gan J, Meng Q. Characterization of the prognostic and diagnostic values of ALKBH family members in non-small cell lung cancer. Pathol Res Pract (2022) 231:153809. doi: 10.1016/j.prp.2022.153809
66. Zhang K, Han Z, Zhao H, Liu S, Zeng F. An integrated model of FTO and METTL3 expression that predicts prognosis in lung squamous cell carcinoma patients. Ann Transl Med (2021) 9(20):1523. doi: 10.21037/atm-21-4470
67. Robles AI, Arai E, Mathe EA, Okayama H, Schetter AJ, Brown D, et al. An Integrated Prognostic Classifier for Stage I Lung Adenocarcinoma Based on mRNA, microRNA, and DNA Methylation Biomarkers. J Thorac Oncol (2015) 10(7):1037–48. doi: 10.1097/JTO.0000000000000560
68. Xie Y, Xiao G, Coombes KR, Behrens C, Solis LM, Raso G, et al. Robust gene expression signature from formalin-fixed paraffin-embedded samples predicts prognosis of non-small-cell lung cancer patients. Clin Cancer Res (2011) 17(17):5705–14. doi: 10.1158/1078-0432.CCR-11-0196
69. Liu J, Mei J, Li S, Wu Z, Zhang Y. Establishment of a novel cell cycle-related prognostic signature predicting prognosis in patients with endometrial cancer. Cancer Cell Int (2020) 20:329. doi: 10.1186/s12935-020-01428-z
70. Shao L, Hong W, Zheng L, He C, Zhang B, Xie F, et al. Joint serum tumor markers serve as survival predictive model of erlotinib in the treatment of recurrent non-small cell lung cancer. Zhongguo Fei Ai Za Zhi (2014) 17(5):391–400. doi: 10.3779/j.issn.1009-3419.2014.05.05
71. Zhou S, Huang R, Cao Y. Detection of epidermal growth factor receptor mutations in peripheral blood circulating tumor DNA in patients with advanced non-small cell lung cancer: A PRISMA-compliant meta-analysis and systematic review. Med (Baltimore) (2020) 99(40):e21965. doi: 10.1097/MD.0000000000021965
72. Vansteenkiste J, Wauters E, Reymen B, Ackermann CJ, Peters S, De Ruysscher D. Current status of immune checkpoint inhibition in early-stage NSCLC. Ann Oncol (2019) 30(8):1244–53. doi: 10.1093/annonc/mdz175
73. Huang Y, Su R, Sheng Y, Dong L, Dong Z, Xu H, et al. Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell (2019) 35(4):677–91 e10. doi: 10.1016/j.ccell.2019.03.006
74. Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m(6)A/MYC/CEBPA signaling. Cell (2018) 172(1-2):90–105 e23. doi: 10.1016/j.cell.2017.11.031
75. Wang T, Hong T, Huang Y, Su H, Wu F, Chen Y, et al. Fluorescein derivatives as bifunctional molecules for the simultaneous inhibiting and labeling of FTO protein. J Am Chem Soc (2015) 137(43):13736–9. doi: 10.1021/jacs.5b06690
76. Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res (2015) 43(1):373–84. doi: 10.1093/nar/gku1276
77. Chen B, Ye F, Yu L, Jia G, Huang X, Zhang X, et al. Development of cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J Am Chem Soc (2012) 134(43):17963–71. doi: 10.1021/ja3064149
78. Tanaka K, Suda A, Uesugi M, Futaki S, Imanishi M. Xanthine derivatives inhibit FTO in an L-ascorbic acid-dependent manner. Chem Commun (Camb) (2023) 59(72):10809–12. doi: 10.1039/D3CC02484A
79. Xiao P, Duan Z, Liu Z, Chen L, Zhang D, Liu L, et al. Rational design of RNA demethylase FTO inhibitors with enhanced antileukemia drug-like properties. J Med Chem (2023) 66(14):9731–52. doi: 10.1021/acs.jmedchem.3c00543
80. Selberg S, Seli N, Kankuri E, Karelson M. Rational design of novel anticancer small-molecule RNA m6A demethylase ALKBH5 inhibitors. ACS Omega (2021) 6(20):13310–20. doi: 10.1021/acsomega.1c01289
81. Lim ZF, Ma PC. Emerging insights of tumor heterogeneity and drug resistance mechanisms in lung cancer targeted therapy. J Hematol Oncol (2019) 12(1):134. doi: 10.1186/s13045-019-0818-2
82. Mao W, Cai Y, Chen D, Jiang G, Xu Y, Chen R, et al. Statin shapes inflamed tumor microenvironment and enhances immune checkpoint blockade in non-small cell lung cancer. JCI Insight (2022) 7(18):e161940. doi: 10.1172/jci.insight.161940
83. Peterson BS. Editorial: Biomarkers in precision medicine for mental illnesses. J Child Psychol Psychiatry (2020) 61(12):1279–81. doi: 10.1111/jcpp.13357
84. Milne S, Sin DD. Biomarkers in chronic obstructive pulmonary disease: the gateway to precision medicine. Clin Chest Med (2020) 41(3):383–94. doi: 10.1016/j.ccm.2020.06.001
85. Laranjeira ABA, Hollingshead MG, Nguyen D, Kinders RJ, Doroshow JH, Yang SX. DNA damage, demethylation and anticancer activity of DNA methyltransferase (DNMT) inhibitors. Sci Rep (2023) 13(1):5964. doi: 10.1038/s41598-023-32509-4
86. Boffetta P, Fryzek JP, Mandel JS. Occupational exposure to beryllium and cancer risk: a review of the epidemiologic evidence. Crit Rev Toxicol (2012) 42(2):107–18. doi: 10.3109/10408444.2011.631898
87. Gao M, Qi Z, Feng W, Huang H, Xu Z, Dong Z, et al. m6A demethylation of cytidine deaminase APOBEC3B mRNA orchestrates arsenic-induced mutagenesis. J Biol Chem (2022) 298(2):101563. doi: 10.1016/j.jbc.2022.101563
88. Li L, Zhou M, Chen B, Wang Q, Pan S, Hou Y, et al. ALKBH5 promotes cadmium-induced transformation of human bronchial epithelial cells by regulating PTEN expression in an m6A-dependent manner. Ecotoxicol Environ Saf (2021) 224:112686. doi: 10.1016/j.ecoenv.2021.112686
89. Sun W, Li Y, Ma D, Liu Y, Xu Q, Cheng D, et al. ALKBH5 promotes lung fibroblast activation and silica-induced pulmonary fibrosis through miR-320a-3p and FOXM1. Cell Mol Biol Lett (2022) 27(1):26. doi: 10.1186/s11658-022-00329-5
90. Shen S, Zhang R, Jiang Y, Li Y, Lin L, Liu Z, et al. Comprehensive analyses of m6A regulators and interactive coding and non-coding RNAs across 32 cancer types. Mol Cancer (2021) 20(1):67. doi: 10.1186/s12943-021-01362-2
91. Xie Q, Li Z, Luo X, Wang D, Zhou Y, Zhao J, et al. piRNA-14633 promotes cervical cancer cell Malignancy in a METTL14-dependent m6A RNA methylation manner. J Transl Med (2022) 20(1):51. doi: 10.1186/s12967-022-03257-2
Keywords: lung cancer, m6A demethylase, FTO, ALKBH5, immunotherapy
Citation: Yu M, Ji W, Yang X, Tian K, Ma X, Yu S, Chen L and Zhao X (2023) The role of m6A demethylases in lung cancer: diagnostic and therapeutic implications. Front. Immunol. 14:1279735. doi: 10.3389/fimmu.2023.1279735
Received: 18 August 2023; Accepted: 13 November 2023;
Published: 29 November 2023.
Edited by:
Zichao Luo, National University of Singapore, SingaporeReviewed by:
Zhen Dong, Southwest University, ChinaQi Wu, Shanghai Children’s Hospital, China
Matias I Hepp, Universidad Católica de la Santísima Concepción, Chile
Copyright © 2023 Yu, Ji, Yang, Tian, Ma, Yu, Chen and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shali Yu, NzY5MzUzOUBudHUuZWR1LmNu; Xinyuan Zhao, emhhb3hpbnl1YW5AbnR1LmVkdS5jbg==; Lin Chen, eGlhb2JlaTIyN0BzaW5hLmNvbQ==
†These authors have contributed equally to this work
 Mengjiao Yu1†
Mengjiao Yu1† Wenqian Ji
Wenqian Ji Shali Yu
Shali Yu Xinyuan Zhao
Xinyuan Zhao