
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 15 November 2023
Sec. Autoimmune and Autoinflammatory Disorders : Autoimmune Disorders
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1279139
This article is part of the Research Topic The Role of Metabolism in Immunological Skin Diseases View all 5 articles
Background: Chronic spontaneous urticaria (CSU) is defined by the spontaneous occurrence of wheals and/or angioedema for >6 weeks. The pathogenesis involves skin mast cells, but the complex causes of their activation remain to be characterized in detail.
Objectives: To explore disease-driving genes and biological pathways in CSU.
Methods: Two microarray data sets, e.g., GSE57178 and GSE72540, with mRNA information of skin from CSU patients, were downloaded from the Gene Expression Omnibus (GEO) database. An integrated bioinformatics pipeline including identification of differentially expressed genes (DEGs), functional enrichment analysis, protein-protein interaction (PPI) network analysis, co-expression and drug prediction analysis, and immune and stromal cells deconvolution analyses were applied to identify hub genes and key drivers of CSU pathogenesis.
Results: In total, we identified 92 up-regulated and 7 down-regulated genes in CSU lesions. These were significantly enriched in CSU-related pathways such as TNF, NF-κB, and JAK-STAT signaling. Based on PPI network modeling, four genes, i.e., IL-6, TLR-4, ICAM-1, and PTGS-2, were computationally identified as key pathogenic players in CSU. Immune infiltration analyses indicated that dendritic cells, Th2 cells, mast cells, megakaryocyte-erythroid progenitor, preadipocytes, and M1 macrophages were increased in lesional CSU skin.
Conclusion: Our results offer new insights on the pathogenesis of CSU and suggest that TNF, NF-κB, JAK-STAT, IL-6, TLR-4, ICAM-1, and PTGS-2 may be candidate targets for novel CSU treatments.
CSU is usually characterized by typically short-lived, itchy, and fleeting wheals, angioedema, or both, occurring spontaneously for longer than 6 weeks (1). CSU affects about 1-4% of the world population, across all ages, mostly young and middle-aged women (2). CSU carries a significant emotional and economic burden for the patient (3),often lasting a long time and affecting many areas of daily life. To date, only antihistamines and omalizumab are licensed for the treatment of CSU, but there are still many patients who do not use these therapies and need more effective treatment.
The development of novel and better treatments for CSU is very much needed. Possible pathophysiological mechanisms now known for CSU include autoantibodies and other mast cell activation signals that lead to skin MC degranulation, release of pro-inflammatory mediators, and recruitment of inflammatory cells to the skin. Beyond this, the pathogenesis of urticaria is poorly understood (4–6). Insights into the complexities and drivers of the pathogenesis of CSU can point to promising targets of newer and more efficacious targeted biological treatments for severe, refractory CSU.
Microarray analysis can detect thousands of gene expression changes in a short time (7). Bioinformatic analyses of microarray gene expression datasets can identify DEGs and relevant functional pathways (8–10). As of now, two CSU studies have assessed skin gene expression (11–13), but joint analyses of these microarray datasets, GSE57178 and GSE72540 are limited. One recent study of these datasets recently reported that the IL-6/miR-149-5p/ZBTB20-AS1 axis may be involved in mast cell activation in CSU (14). The lesional immune cell infiltrate and core genes, however have not been investigated by bioinformatics analyses.
In this study, we performed deep bioinformatic analyses of the merged CSU microarray datasets GSE57178 and GSE72540 available from the GEO, which contain cutaneous gene expression data from 16 CSU patients and 13 healthy individuals. The aim of this study was to identify key genes and immune cells related to the pathological mechanism of CSU.
Two microarray datasets (GSE57178 (11) and GSE72540 (12)) were downloaded from GEO (http://www.ncbi.nlm.nih.gov/geo). The details are shown in Table 1. GSE57178 contains the mRNA information of lesional skin (N=6) and non-lesional skin (N=7) of CSU patients, and of skin from healthy individuals (N=5). GSE72540 also contains information on lesional CSU skin (N=10), non-lesional CSU skin (N=13), and the skin of healthy control subjects (N=8). All included patients showed active CSU, i.e. urticaria activity score 7 (UAS7) ≥ 11 for at least 3 months, and were refractory to antihistamine treatment.
Background correction and data normalization were completed through the “Affy” package and the RMA algorithm.The limma package was applied to identify the DEGs between CSU skin and the skin of healthy controls. |LogFC| > 1 and P-value < 0.05 were defined as the cut-off.
GSEA (Gene Set Enrichment Analysis) is a calculation method to determine whether a set of genes defined in advance are differentially expressed in different samples (15). In order to explore whether most of the genes in the predefined gene set are highly expressed or poorly expressed in the sample, the H. All. V6.2. Symbols.gmt [Hallmarks] gene set database in GESA software is used for analysis. Significantly enriched biological process (BP) items of the DEGs were obtained by DAVID (version 6.8) and Metascape. Biological pathways analysis was completed by the functional enrichment analysis tool (Funrich) (16). The pathway enrichment analyses of DEGs were evaluated by two databases, including “KEGG pathway” and “Reactome”.
The PPI network of DEGs was predicted by STRING (version 11.0; http://string-db.org) and visualized by Cytoscape (version 3.7.2). Key modules were identified by using The Molecular Complex Detection (MCODE; version 1.5.1) of Cytoscape (MCODE scores > 5, degree cut‐off = 2, node score cut‐off = 0.2, max depth = 100 and k‐score = 2).
The cytoHubba plug-in in the Cytoscape software was used to analyze the topological attributes of nodes in the network, and the parameters are set to no weight. By sorting the score of each node, we identified the important nodes involved in protein interaction in the network. We selected six commonly used parameters to calculate the top ten genes and their respective rankings and obtained the common hub genes through the Venn diagram. GeneMANIA (http://www.genemania.org/) was used to explore the co-expression genes and functions of hub genes. The Drug-Gene Interaction database (DGIdb 3.0; http://www.dgidb.org/) was used to discover the existing targeted drugs for hub genes. All the network map was formed by Cytoscape.
xCell is an evaluation tool of immune cell infiltration based on gene expression data, which can identify potential immune and stromal cell subsets and calculate their relative abundance in tissues (17). We evaluated the immune cell infiltration of two microarray data by xCell. Finally, their common differential immune and stromal cells were obtained by Venn diagrams.
Spearman correlation analysis was used to explore the relationship between hub genes and infiltrating immune cells. The results were visualized by using the “ggplot2” software package.
Comparing lesional CSU skin and healthy control samples, 292 and 1221 DEGs were identified in GSE57178 and GSE72540, respectively, of which 99 were in both (Figure 1), 92 upregulated and 7 downregulated DEGs (Figure 1). The heat map shows that these DEGs can basically distinguish CSU lesion samples from healthy control samples (Figure 2).
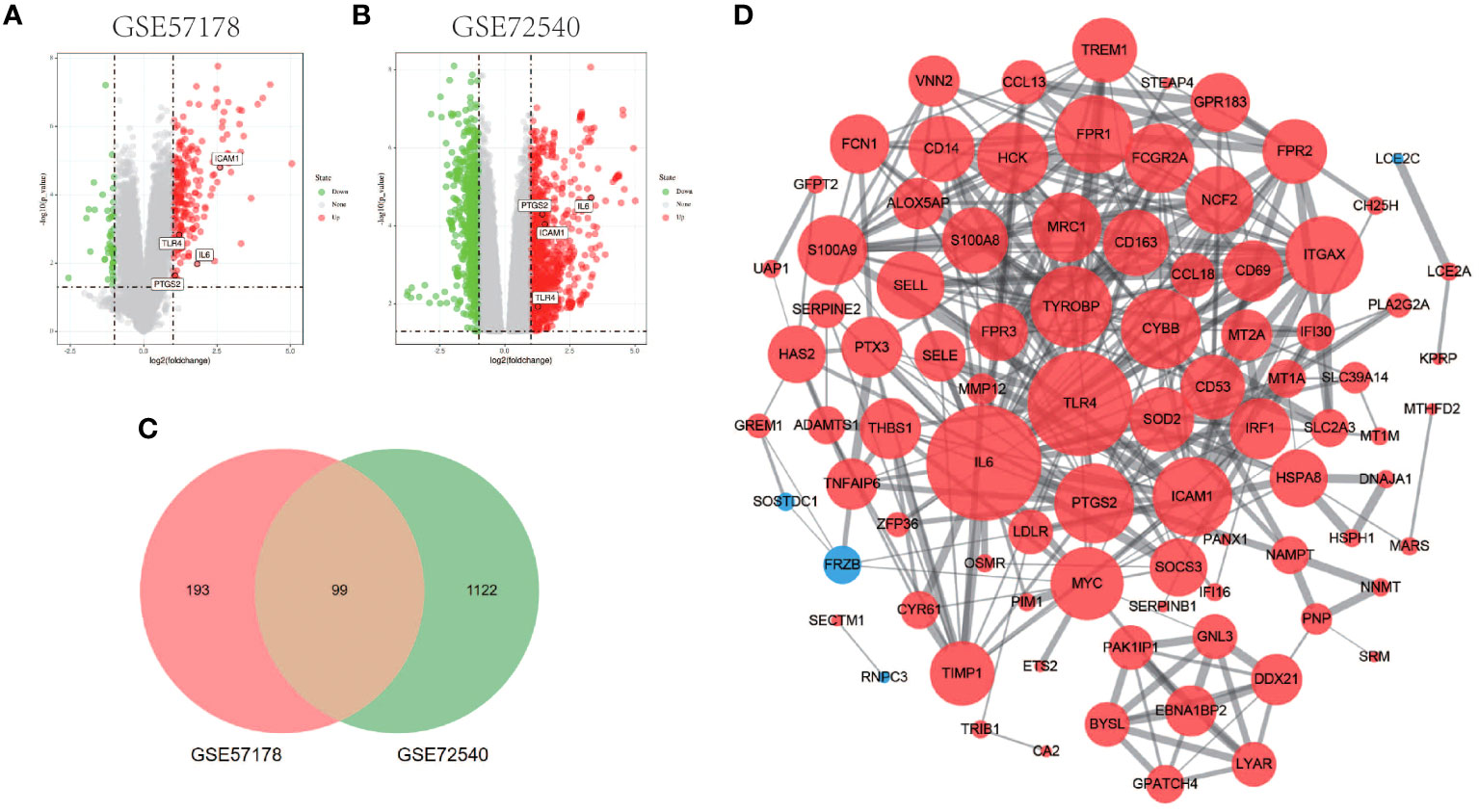
Figure 1 Volcano figure, Venn diagram, PPI network of common DEGs. (A, B) Volcano plot of DEGs. Among them, red indicates up-regulated genes and green indicates down-regulated genes. Gray dots represent genes with no significant difference. (C) GSE57178 and GSE72540 have 99 DEGs in common. (D) PPI network is constructed according to 99 common DEGs, in which up-regulated genes are represented by red circles and down-regulated genes by blue circles. The size of the gray node represents the size of the topological properties of the corresponding protein in the network, and the thickness of the line represents the strength of evidence of interaction between the two proteins.
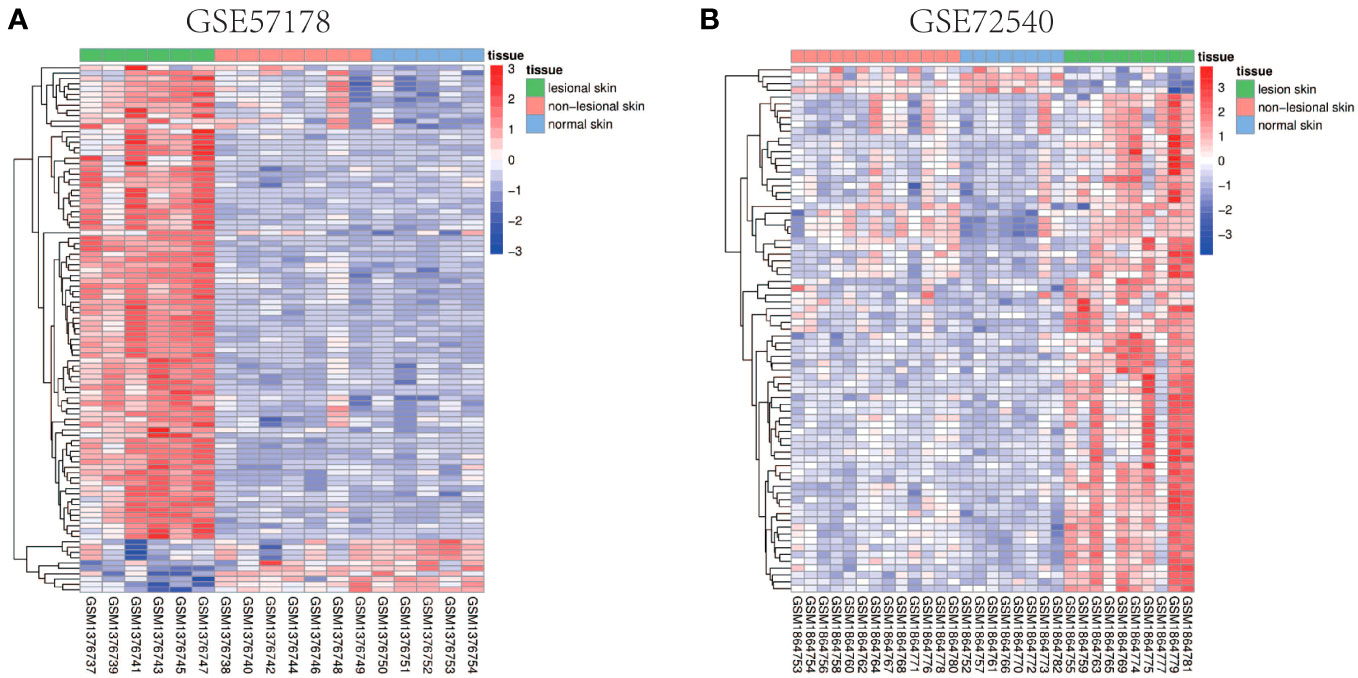
Figure 2 Heat map of common DEGs. (A, B) Heat maps of common DEGs between GSE57178 and GSE72540. Up-regulated genes are shown in red, down-regulated genes are shown in blue, and genes with little change are shown in white.
Gene set enrichment analyses showed that four common gene sets were significantly enriched in CSU and healthy controls samples (Figure 3), including TNF-mediated NF-κB pathway, IL-6/JAK/STAT3 signaling pathway, and responses to interferon-gamma. Biological processes enrichment analysis indicated that these were mainly linked to inflammation and leukocyte migration (Figure 4 and Table S1). Enrichment analyses of biological pathways demonstrated that DEGs in lesional vs non lesional skin were mainly enriched in IL-6-mediated signaling events and the biological process by which formyl peptide receptors bind to formyl peptides and many other ligands (Figure 5). Moreover, our KEGG analysis showed that 4 pathways were significantly different in lesional vs healthy skin of the control group, including Phagosome, Malaria, TNF signaling pathway, and Staphylococcus aureus infection. Reactome enrichment analyses revealed that the most significantly enriched pathways in CSU lesions vs non lesional skin were Immune System, Innate Immune System, Neutrophil degranulation, and Interleukin-4 and Interleukin-13 signaling (Figures 6A, B, Tables S2 and S3).

Figure 3 GSEA enrichment analysis. (A, B) The common gene set pathway obtained by GESA analysis of GSE57178 and GSE72540 respectively.
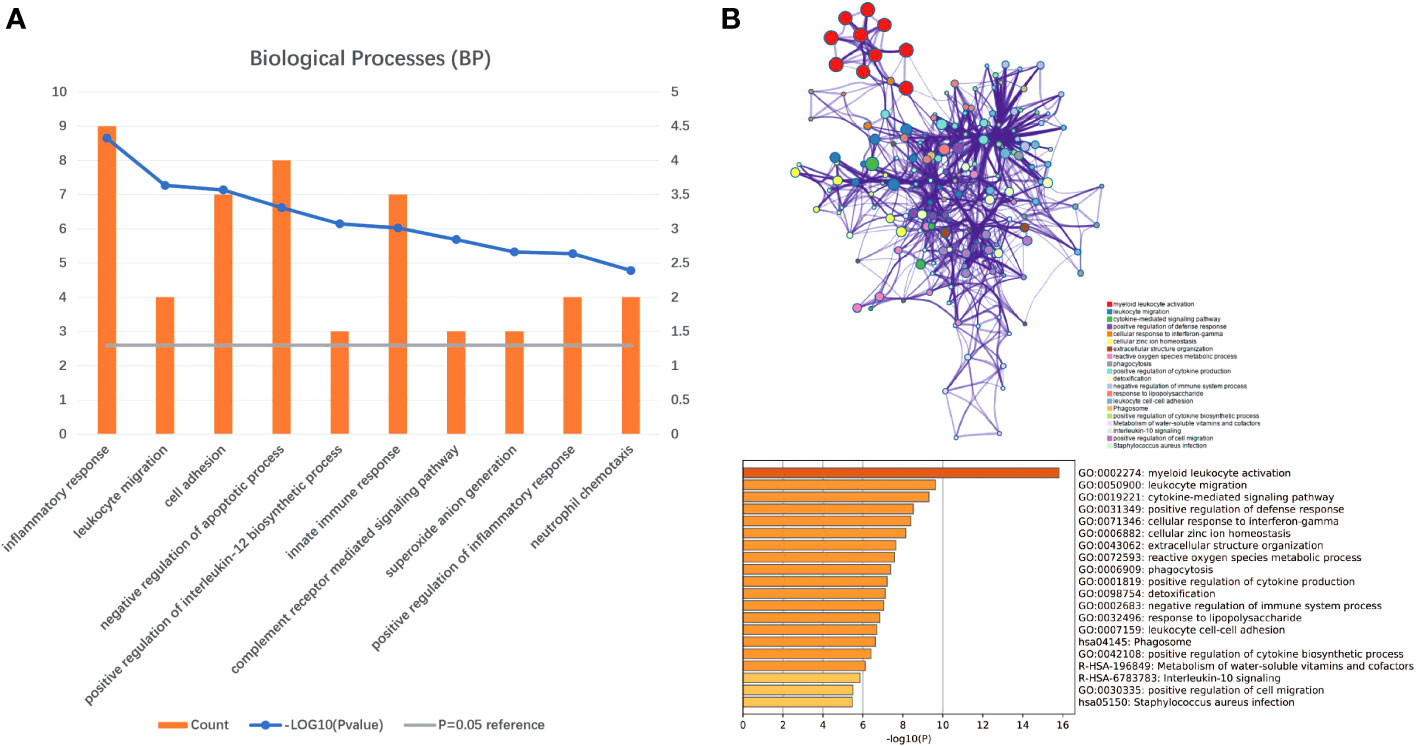
Figure 4 Significant biological processes were obtained through DAVID and Metascape. (A) The top 10 biological processes form DAVID. (B) The top 20 biological processes enriched by Metascape.
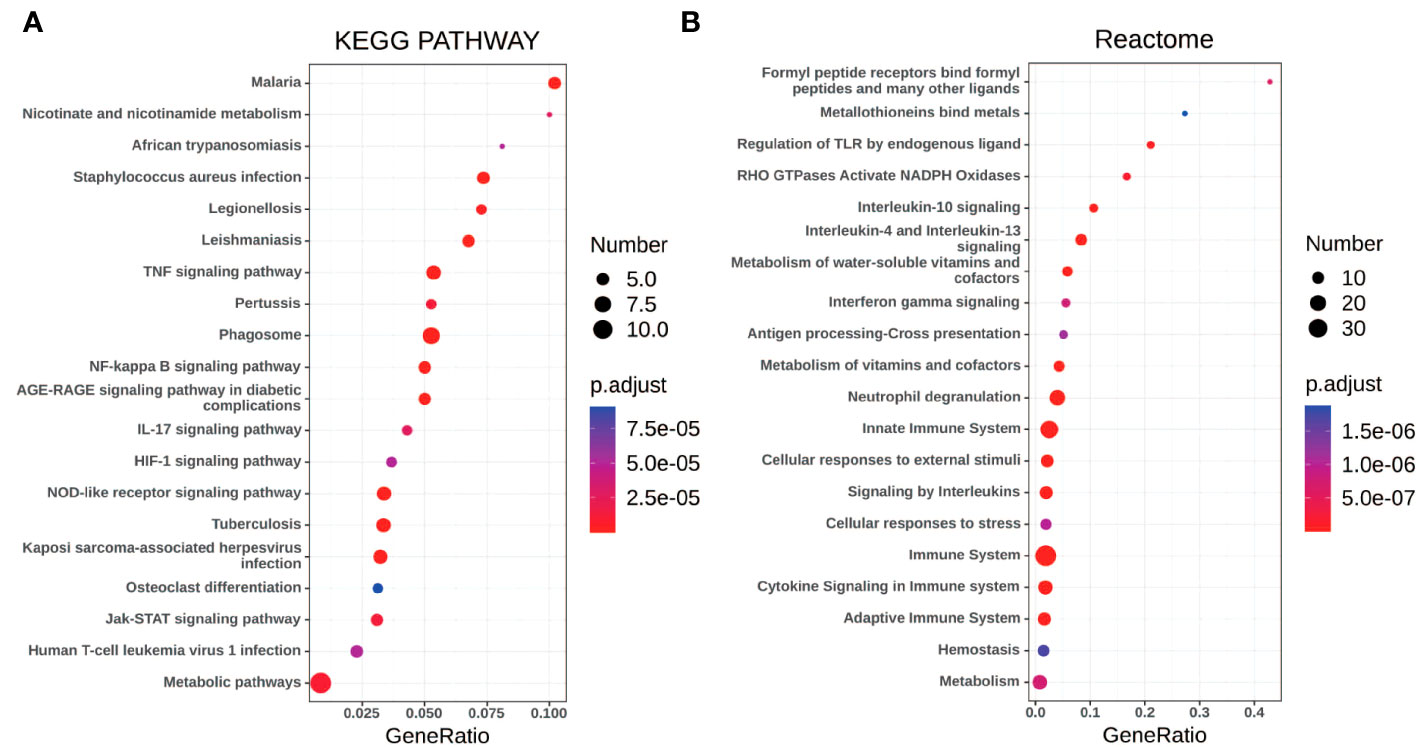
Figure 6 The pathway analysis of DEGs by KOBAS 3.0. (A, B) The pathway analysis of all the DEGs via KEGG and Reactome database. The size of the dot represents the number of enriched genes, the color represents the size of the P-value, the abscissa represents the proportion of the enriched genes to the total number of gene sets, and the ordinate represents the name of the pathway.
The PPI network of DEGs generated by Cytoscape consists of 87 nodes and 347 interaction pairs (Figure 1D). Two important gene modules were generated by MCODE plug-ins (Figures 7A, C). KEGG pathway enrichment analysis showed that three pathways are closely related to CSU, including TNF signaling pathway, NF-κB signaling pathway and JAK-STAT signaling pathway (Figure 7B, D).

Figure 7 KEGG pathway analysis of modular genes. (A, B) Module 1 and the KEGG pathway enrichment analysis. (C, D) Module 2 and the KEGG pathway enrichment analysis.
Using the 6 algorithms (MNC, Radiality, Stress, Degree, Closeness, and EPC) in the cytoHubba plugin, we obtained 4 lesional skin-expressed hub genes: IL-6, TLR-4, ICAM-1, and PTGS-2 (Figure 8A and Table 2). Then, a network of these 4 common hub genes and their co-expression genes were analyzed by GeneMANIA online platform. The analysis of co-expression network showed that hub genes and co-expression genes were mainly involved in the process of leukocyte migration and inflammatory response (Figure 8B). Furthermore, we obtained 12 drugs (FDA‐listed + Immunotherapies) that paired with these hub genes from the DGIdb (Figure 8C).
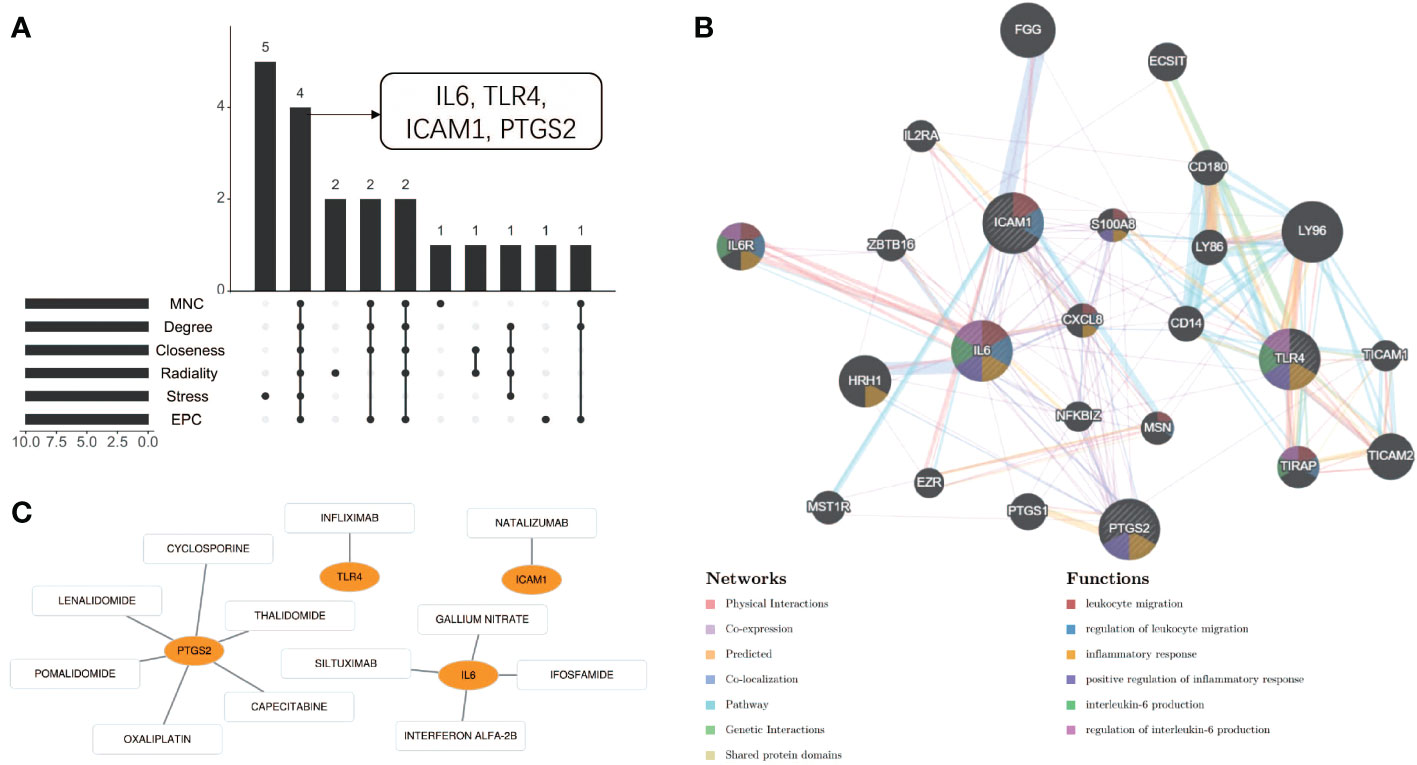
Figure 8 Co-expression analysis and drug-gene interaction analysis of hub genes. (A) Four common hub genes obtained by six algorithms. (B) The network diagram of Hub genes and their co-expressed genes. (C) Drug-gene interaction diagram.
To investigate the immune cell types that may be infiltrated in CSU lesions, cell enrichment analyses done with xCell showed marked differences between CSU and healthy controls samples in both datasets, GSE57178 and GSE72540. Eight cell types showed much higher expression in lesional CSU skin: immune cells, sensory nerves, dendritic cells (DC), preadipocytes, megakaryocyte-erythroid progenitor (MEP), mast cells, M1 macrophages, microvascular (mv) endothelial cells, and Th2 cells (Figure 9A). Specifically, both in GSE57178 and GSE72540, compared with normal healthy skin, CSU lesions showed higher infiltrations of DC, preadipocytes, Th2 cells, MEP, mast cells, and macrophages M1(Figure 9B, C), respectively.
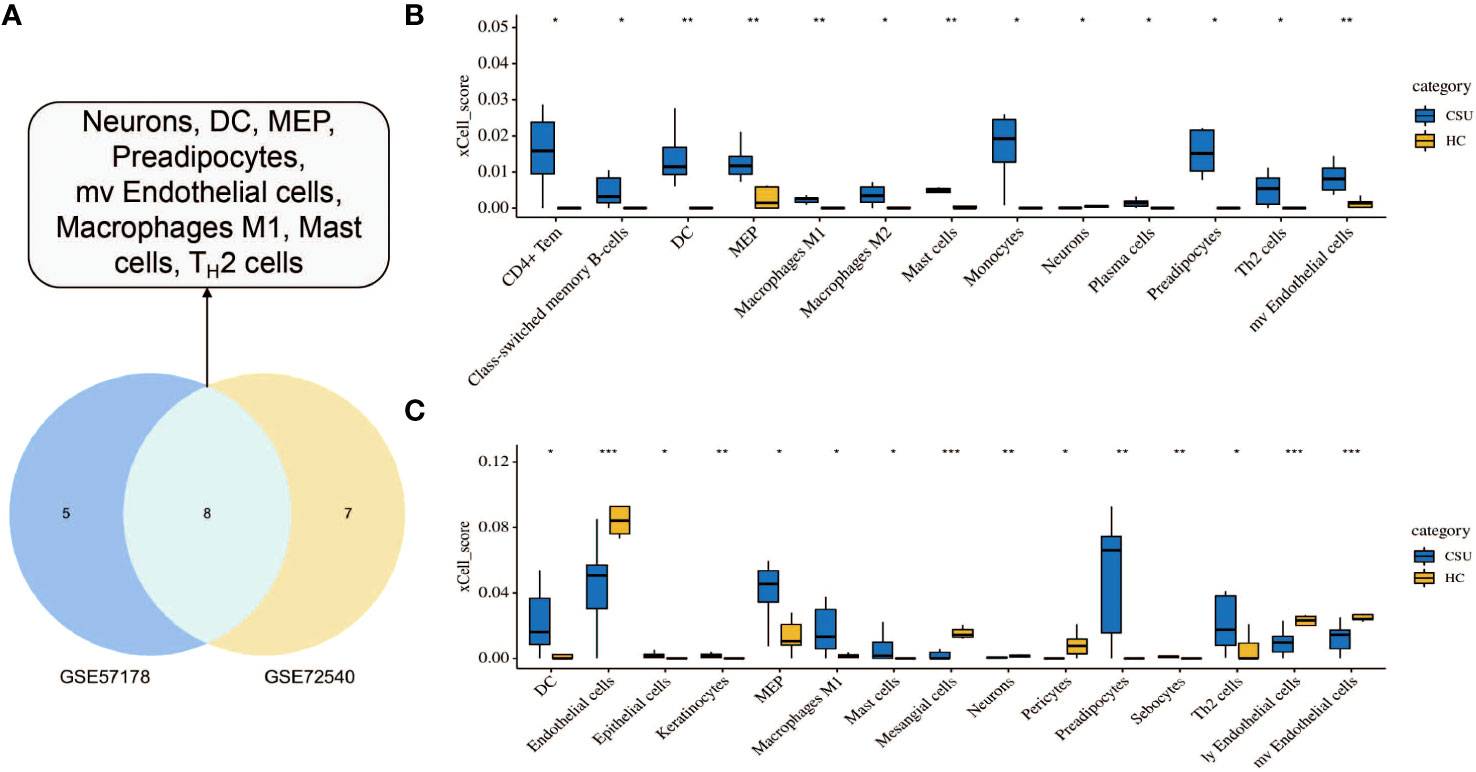
Figure 9 Immunological and stromal cell infiltration analysis. (A) Common differential cell types in GSE57178 and GSE72540. XCell scores of 64 cell types in (B) GSE57178 and (C) GSE72540.
Correlation analysis between hub genes and infiltrating immune cells in CSU lesions showed that 1) IL-6 (Figure 10A) was positively correlated with activated Th2 cells, preadipocytes, and mv Endothelial cells; 2) PTGS-2 (Figure 10B) was positively correlated with Th2 cells, neurons, and macrophages M1; 3) ICAM-1 (Figure 10C) was positively correlated with Th2 cells, mast cells, macrophages M1, mv Endothelial cells, MEP, and preadipocytes; 4) TLR-4 (Figure 10D) was positively correlated with mv Endothelial cells, MEP, macrophages M1, and preadipocytes.
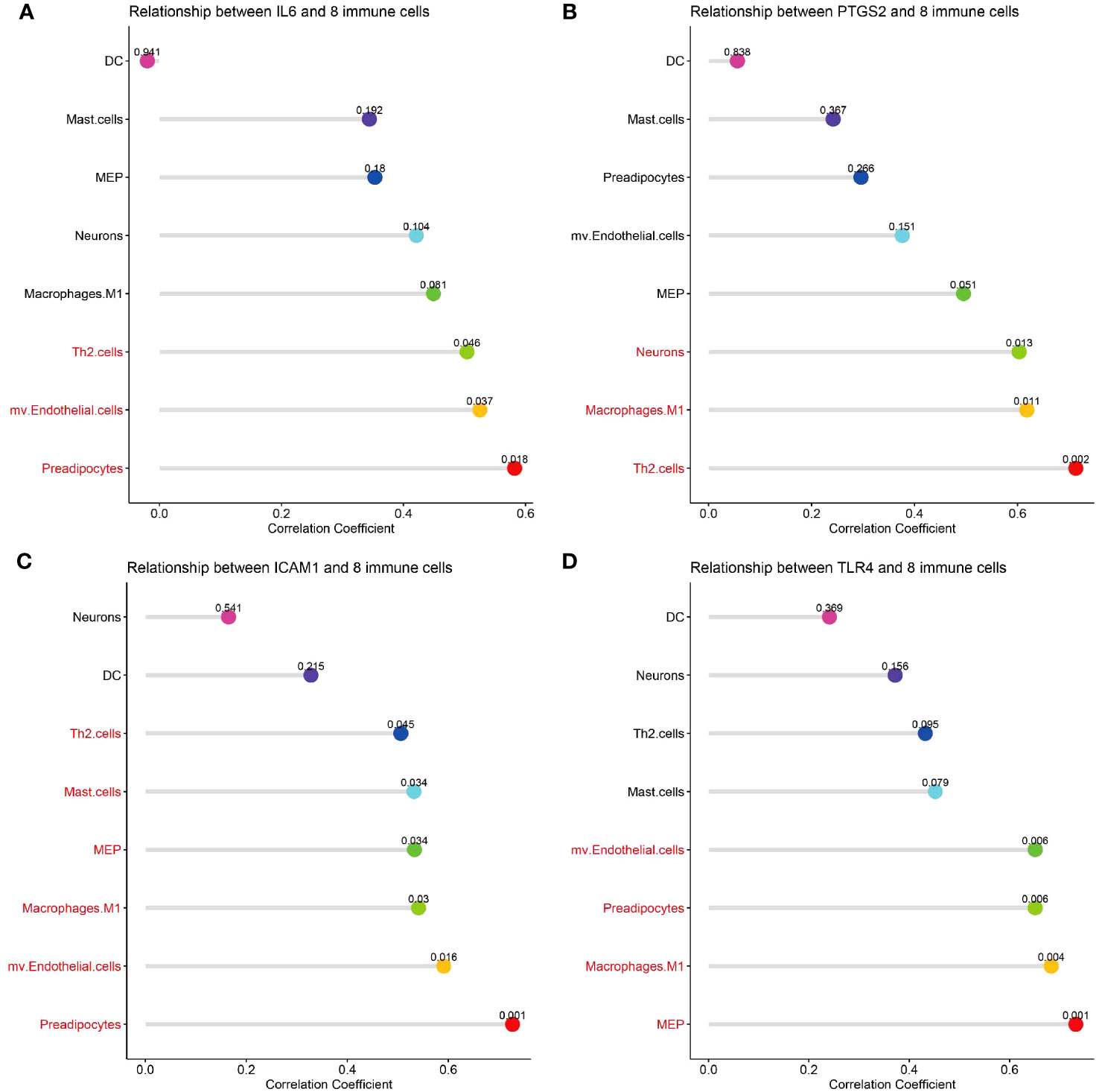
Figure 10 Correlation analysis of hub genes with immune and stromal cells. (A-D) The correlation between common differential cell types and IL-6, PTGS-2, ICAM-1, TLR-4. The terms marked with red indicate the cells that are related to the hub gene.
This study investigated potential disease driving gene signatures and functional networks in patients with CSU through microarray analysis. In total, 99 DEGs including 92 up-regulated genes and 7 down-regulated genes between CSU and healthy controls were identified. By enrichment analysis we identified core modules and three signaling pathways i.e. TNF, NF-κB, and JAK-STAT, that are closely related to the development of CSUs. In addition, we identified four hub genes, including IL-6, TLR-4, ICAM-1, and PTGS2.In addition, xCell is often used to assess local immune cell infiltration in skin biopsies of CSU patients. CSU lesions contained higher proportions of DCs, Th2 cells, MCs, megakaryocyte-erythroid progenitors, preadipocytes, and M1 macrophages compared to healthy skin. Finally, the above four hub genes IL-6, TLR-4, ICAM-1, and PTGS2 were found to be correlate with immune and stromal cell infiltration in CSU lesions.
CSU is not only associated with autoimmune processes but also systemic inflammatory responses (18). The release of various inflammatory mediators (histamine, protease, leukotriene and TNF, etc.) is closely related to the wheals of CSU. TNF, which also can be secreted by T cells, natural killer (NK) cells, monocytes, macrophages, basophils, eosinophils, keratinocytes, and fibroblasts, et al. (19). Dysregulation of the TNF pathway is a key feature and causative factor in many autoimmune and inflammatory diseases (20, 21). The formation of wheals in CSU is closely related to the activation of TNF signaling pathway, marked by 1) increased circulating concentrations of TNF-R, sTNF-R1, and sTNF-R2, 2) serum TNF level was positively correlated with disease activity in the samples of patients with CSU, 3) TNF is upregulated in the lesional and non-lesional skin of patients with CSU (22, 23). In addition, the use of TNF inhibitors (Etanercept, Adalimumab, Infliximab) for the treatment of CSU have also been validated in clinical care (24). Consistent with these findings, in the present study, we also observed that TNF is a hub gene of CSU. In sum, all these findings provide a theoretical basis for the potential of TNF targets in the treatment of CSU. Building on these foundations, larger and more rigorous trials are needed to make a stronger case that TNF for CSU is safe and effective.
CSU is also associated with chronic inflammation. And most chronic diseases, caused by lifestyle factors, appear to be linked to inflammation (25). As we all know, NF-κB is a pivotal pathway that mediates the pathogenesis of inflammatory skin diseases (26). When cells are subjected to various intracellular and extracellular stimuli, NF-κB dimers are released, which are further activated and translocated to the nucleus, leading to the production of inflammatory cytokines such as IL-6 and TNF (27, 28). In turn, these inflammatory cytokines promote the activation of NF-κB signalling, leading to a complex inflammatory cascade in vivo (27). In the present study, we have demonstrated that both IL-6 and TNF are hub genes of CSU. Besides, our enrichment analysis results showed that the NF-κB signaling pathway was significantly related to the occurrence and development of CSU. Based on these findings, we speculate that IL-6/TNF-mediated over-activation of the NF-κB signaling pathway is a new therapeutic target for CSU.
It has been shown in recent years that the JAK/STAT3 signalling pathway is associated with the pathogenesis of CSU. In the lesions of CSU patients, the high expression of OSMR gene can promote the expression of JAK/STAT3 signaling pathway related genes (29). They further observed that OSMR gene silencing can significantly inhibit the activity of JAK/STAT3 signal pathway, promote cell proliferation and migration, and further inhibit epithelial cell apoptosis in a chronic autoimmune urticaria mouse model. Other studies have also found that IL-9 and IL-10 can promote CSU through activation of the JAK/STAT3 signalling pathway (30). Our GSEA enrichment analysis based on both datasets showed that the IL-6/JAK/STAT3 pathway was also highly expressed in the CSU patient group. Additionally,the IL-6/JAK/STAT3 signalling pathway has been shown to be associated with mast cell degranulation and allergic reactions (31). All aforementioned findings indicate that blocking the IL-6/JAK/STAT3 signalling pathway may be a novel approach for the treatment of CSU.
IL-6 is an important regulator of inflammation and immune responses and is associated with a variety of autoimmune and chronic inflammatory diseases (32–34). Although the imbalance of inflammatory cells was detected in skin biopsies of CSU patients, the source of IL-6 in CSU has not been identified (6, 35). Previously, several studies have reported higher IL-6 levels in patients with CSU compared to those observed in the healthy group (36–39). IL-6 levels could be linked to CSU disease activity (40). In addition, IL-6 levels in CSU patients were reduced by omalizumab therapy.
Besides, IL-6/soluble IL-6 receptor (sIL-6R) complex can stimulate the synthesis of acute phase proteins (such as CRP) and promote the production of autoantibodies. On the other hand, it promotes the differentiation of Th2 and Th17 cells while inhibiting the differentiation of Th1 and Treg cells, ultimately leading to an imbalance of Th1/Th2 cells (32, 41). These findings suggest that IL-6 might contribute to CSU pathology, even though the utility of IL-6 as biomarker in CSU is still questionable (42).
Toll-like receptors (TLRs) are a group of pattern recognition receptors that recognise microbe-associated PAMPs and damage or tissue repair hazard-associated sub-DAMPs (43).. TLR-4 was one of the most important TLRs which play a key role in pathogen recognition and activation of various inflammatory signaling pathways (44). TLR-4 signaling also has been suggested to participate in allergic diseases as well as autoimmune responses (45, 46). TLR-4 expression was significantly upregulated in allergic rhinitis patients (47). Lately, although contradictory observations have also been published, TLR-4 polymorphisms have been reported to associate with atopy and asthma (48, 49). Interestingly, a decreased TLR-4 expression on CD14+ cells from chronic idiopathic urticaria patients was observed in ex vivo conditions (50). In the present study, for the first time, we identified TLR-4 as a hub gene in CSU. Taken together, these findings indicate that TLR-4 may be involved in the development of CSU. As for the specific mechanisms, clinical sample verification and laboratory data are still needed.
In CSU, although MCs initiate and remain in early-phase wealing response, cellular adhesion molecules (CAMs) are thought to sustain the late phase (51). CAMs are well documented in the selective recruitment and activation of inflammatory cells, which ultimately lead to tissue damage (52). Among the CAMs, ICAM-1 plays an important role in accelerating tissue oedema formation (53): 1) Overexpression of ICAM-1 in endothelial cells alters cell morphology and increases endothelial cell permeability or vascular leakage (54); 2) Moreover, antibody cross-linking specific for ICAM-1 also increased albumin passage from intact cremaster micro vessels (55). It has been demonstrated that ICAM-1 is the hallmark of allergic inflammation (56). Although it has been reported that ICAM-1 was not always significantly elevated in CU sera. ICAM-1, as a biomarker of endothelial dysfunction, is increased in sera and detected in spontaneous wheals of patients with CIU (35). Moreover, one study, which included a large number of CSU patients, showed a significant increase in soluble ICAM-1 level in sera (57). A recent study also demonstrated that specific knockout of ICAM-1 can inhibits the expression of IgE, ECP, CD62L, and CD11a, thereby reducing the adhesion between vascular endothelial cells in a mouse model of immune contact urticaria (58). In this study, we observed that ICAM-1 was a hub gene in CSU for the first time. Thus, the above previous findings combined with our data, all indicate that ICAM-1 plays an important role in tissue edema formation of CSU, where relevant clinical and laboratory evidence remains to be further explored.
In clinic, the effect of using H1 antihistamine alone in the treatment of CSU is not satisfactory (59). A reasonable explanation is that some inflammatory mediators can show similar functions to histamine, such as prostaglandins and coagulans, has been overlooked (60). Arachidonic acid forms prostaglandins and thromboxanes through the actions of the enzymes cyclooxygenases (COXs). Inducible COX-2 is associated with pseudoallergy and other inflammatory reactions (61). Besides, COX-2 is predominantly induced in activated leukocytes and inflammatory sites (62). Meanwhile, Nonsteroidal anti-inflammatory drug (NSAID)-sensitivity is very common in patients with chronic urticaria (63). The physiological and pathological processes of respiratory and skin reactions may involve the increase of cysteinyl leukotrienes (64). COX-2 selective inhibitor is a major alternative treatment to control pain and inflammation in these patients (65). In the present study, we further confirmed PTGS-2 as a hub gene in CSU through PPI network characterization. Thus COX-2 may be an important target for the treatment of CSUs.
The study found that CSU lesions contained a higher proportion of DCs, Th2 cells, mast cells, MEP, preadipocytes, and M1 macrophages compared with healthy skin. It was also discovered that increased expression of Th-2 cytokines (IL-4/IL-5) in CSU than in normal healthy controls (66). Mast cells are key effector cells in the pathogenesis of CSU, and when activated, they release inflammatory mediators such as histamine and PGD2, which in turn lead to an increase in vascular permeability, causing flare-ups, itching, angioedema (66). In CSU, IgE-mediated type I hypersensitivity is closely related to mast cell activation (67). A predominance of activated M2 macrophages was also observed in CSU patients with refractory for antihistamines (68). Thus, our experimental data are consistent with previous findings on the overview of immune cell infiltration in the CSU, which in turn demonstrates the accuracy of our analysis.
It’s worth noting that, so far there have been no reports on MEP or preadipocytes in CSU, and these potential connections were observed for the first time in the present study. Thus, the specific mechanism of MEP or preadipocyte involvement in the development of CSU still needs further verification and exploration. Moreover, in this study, we found that hub genes IL-6, TLR-4, ICAM-1, and PTGS-2 were positively correlated with immune and stromal cell infiltration in CSU lesions. These findings indicate that these hub genes also contribute to the infiltration of these immune cells to the lesions of patients with CSU. However, the specific mechanism of these DEGs on the immune invasion of CSU lesions remains to be further studied.
However, this study still has certain limitations. First, the sample size we analyzed is relatively small. Cause the amount of samples uploaded to public databases is currently valid, and we have analyzed the only two databases available until now. Second, all our data comes from publicly available databases. Third, the potential pathway involved in the CSU was not examined and verified. Future studies should be performed to identify the detailed pathway that participated in the progression of CSU. Nevertheless, our results provide a new insight into the pathological mechanism of CSU. These findings do require further substantiation such as by in vivo or in vitro experiments.
In summary, in the present study, a total of 4 hub genes (IL-6, TLR-4, ICAM-1, and PTGS-2) have been identified, which has the potential to be used as markers or as therapeutic targets for CSU. TNF signaling pathway, NF-κB signaling pathway, and JAK-STAT signaling pathway are thought to be closely related to the pathological mechanism of CSU. We observed higher proportions of DCs, Th2 cells, mast cells, MEP, preadipocytes, and M1 macrophages in CSU lesions. Finally, the positive correlation of hub genes IL-6, TLR-4, ICAM-1, and PTGS-2 and these immune cells infiltration in CSU lesions were found. Our findings will help to encourage further studies on the biological functions of these hub genes, immune infiltration profiles, as well as the relationship between them in CSU.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
WS: Software, Visualization, Writing – original draft, Writing – review & editing. YT: Conceptualization, Data curation, Formal Analysis, Writing – review & editing. YW: Data curation, Writing – review & editing. FH: Project administration, Supervision, Writing – review & editing. JJ: Data curation, Project administration, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported in part by the National Natural Science Foundation of China (82073434), Natural Science Foundation of Chongqing (cstc2021ycjh-bgzxm0291), The seventh batch of Suzhou health talents young top-notch talents (2020-088), Suzhou Minsheng Technology-Medical and Health Application Foundation (SYS2020135), and 2020 Medical Research Project of Jiangsu Provincial Health Commission (Z2020017).
A preprint of the study has been published (https://www.researchsquare.com/article/rs-137346/v1) (69).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1279139/full#supplementary-material
BP, Biological processes; COX-2, Cyclooxygenase 2; CRP, C-reactive protein; CSU, Chronic spontaneous urticarial; DCs, Dendritic cells; DEGs, Differentially expressed genes; DGIdb, Drug-Gene Interaction database; FC, Fold-changes; GEO, Gene Expression Omnibus; GSEA, Gene set enrichment analysis; ICAM-1, Intercellular adhesion molecule 1; IL-6, Interleukin 6; LS, Lesional skin; MEP, Megakaryocyte-erythroid progenitor; NL, Non-lesional skin; NS, Normal skin; PGD2, Prostaglandin D2; PPI, Protein-protein interaction; PTGS-2, Prostaglandin-endoperoxide synthase 2; sIL-6R, Soluble IL-6 receptor; TF, Tissue factor; TLR-4, Toll-like receptor 4; TLRs, Toll-like receptors; TNF, Tumor necrosis factor; TNFR1, TNF receptor 1; UAS7, urticaria activity score 7.
1. Zuberbier T, Abdul Latiff AH, Abuzakouk M, Aquilina S, Asero R, Baker D, et al. The international EAACI/GA(2)LEN/EuroGuiDerm/APAAACI guideline for the definition, classification, diagnosis, and management of urticaria. Allergy (2022) 77(3):734–66. doi: 10.1111/all.15090
2. Kolkhir P, Gimenez-Arnau AM, Kulthanan K, Peter J, Metz M, Maurer M. Urticaria. Nat Rev Dis Primers (2022) 8(1):61. doi: 10.1038/s41572-022-00389-z
3. Goncalo M, Gimenez-Arnau A, Al-Ahmad M, Ben-Shoshan M, Bernstein JA, Ensina LF, et al. The global burden of chronic urticaria for the patient and society. Br J Dermatol (2021) 184(2):226–36. doi: 10.1111/bjd.19561
4. Kolkhir P, Munoz M, Asero R, Ferrer M, Kocatürk E, Metz M, et al. Autoimmune chronic spontaneous urticaria. J Allergy Clin Immunol (2022) 149(6):1819–31. doi: 10.1016/j.jaci.2022.04.010
5. Maurer M, Zuberbier T, Metz M. The classification, pathogenesis, diagnostic workup, and management of urticaria: an update. Handb Exp Pharmacol (2022) 268:117–33. doi: 10.1007/164_2021_506
6. Zhou B, Li J, Liu R, Zhu L, Peng C. The role of crosstalk of immune cells in pathogenesis of chronic spontaneous urticaria. Front Immunol (2022) 13:879754. doi: 10.3389/fimmu.2022.879754
7. Lamot L, Vidovic M, Perica M, Bukovac LT, Harjacek M. Microarray and gene expression analysis. Lijec Vjesn (2015) 137(5-6):188–95.
8. Desany B, Zhang Z. Bioinformatics and cancer target discovery. Drug Discovery Today (2004) 9(18):795–802. doi: 10.1016/S1359-6446(04)03224-6
9. Yan Q. Bioinformatics databases and tools in virology research: an overview. In Silico Biol (2008) 8(2):71–85.
10. Oertelt S, Selmi C, Invernizzi P, Podda M, Gershwin ME. Genes and goals: an approach to microarray analysis in autoimmunity. Autoimmun Rev (2005) 4(7):414–22. doi: 10.1016/j.autrev.2005.05.004
11. Patel OP, Giorno RC, Dibbern DA, Andrews KY, Durairaj S, Dreskin SC. Gene expression profiles in chronic idiopathic (spontaneous) urticaria. Allergy Rhinol (Providence) (2015) 6(2):101–10. doi: 10.2500/ar.2015.6.0124
12. Gimenez-Arnau A, Curto-Barredo L, Nonell L, Puigdecanet E, Yelamos J, Gimeno R, et al. Transcriptome analysis of severely active chronic spontaneous urticaria shows an overall immunological skin involvement. Allergy (2017) 72(11):1778–90. doi: 10.1111/all.13183
13. Metz M, Torene R, Kaiser S, Beste MT, Staubach P, Bauer A, et al. Omalizumab normalizes the gene expression signature of lesional skin in patients with chronic spontaneous urticaria: A randomized, double-blind, placebo-controlled study. Allergy (2019) 74(1):141–51. doi: 10.1111/all.13547
14. Liu R, Lv D, Cao L, Liu Y, Wen X, Jiang Y, et al. The hub genes and their potential regulatory mechanisms in chronic spontaneous urticaria revealed by integrated transcriptional expression analysis. Exp Dermatol (2023) 32(6):840–51. doi: 10.1111/exd.14785
15. Subramanian A, Tamayo P, Mootha VK, Mukherjee K, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A (2005) 102(43):15545–50. doi: 10.1073/pnas.0506580102
16. Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CY, Williamson NA, et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics (2015) 15(15):2597–601. doi: 10.1002/pmic.201400515
17. Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol (2017) 18(1):220. doi: 10.1186/s13059-017-1349-1
18. Ferrer M. Immunological events in chronic spontaneous urticaria. Clin Transl Allergy (2015) 5:30. doi: 10.1186/s13601-015-0074-7
19. Church MK, Kolkhir P, Metz M, Maurer M. The role and relevance of mast cells in urticaria. Immunol Rev (2018) 282(1):232–47. doi: 10.1111/imr.12632
20. Jang DI, Lee AH, Shin HY, Song HR, Park JH, Kang TB, et al. The role of tumor necrosis factor alpha (TNF-alpha) in autoimmune disease and current TNF-alpha inhibitors in therapeutics. Int J Mol Sci (2021) 22(5):2719. doi: 10.3390/ijms22052719
21. Doss GP, Agoramoorthy G, Chakraborty C. TNF/TNFR: drug target for autoimmune diseases and immune-mediated inflammatory diseases. Front Biosci (Landmark Ed) (2014) 19(7):1028–40. doi: 10.2741/4265
22. Grzanka R, Damasiewicz-Bodzek A, Kasperska-Zajac A. Tumor necrosis factor-alpha and Fas/Fas ligand signaling pathways in chronic spontaneous urticaria. Allergy Asthma Clin Immunol (2019) 15:15. doi: 10.1186/s13223-019-0332-7
23. Hermes B, Prochazka AK, Haas N, Jurgovsky K, Sticherling M, Henz BM. Upregulation of TNF-alpha and IL-3 expression in lesional and uninvolved skin in different types of urticaria. J Allergy Clin Immunol (1999) 103(2 Pt 1):307–14. doi: 10.1016/S0091-6749(99)70506-3
24. Sand FL, Thomsen SF. TNF-alpha inhibitors for chronic urticaria: experience in 20 patients. J Allergy (Cairo) (2013) 2013:130905. doi: 10.1155/2013/130905
25. Bansal CJ, Bansal AS. Stress, pseudoallergens, autoimmunity, infection and inflammation in chronic spontaneous urticaria. Allergy Asthma Clin Immunol (2019) 15:56. doi: 10.1186/s13223-019-0372-z
26. Sur I, Ulvmar M, Toftgard R. The two-faced NF-kappaB in the skin. Int Rev Immunol (2008) 27(4):205–23. doi: 10.1080/08830180802130319
27. Jimi E, Fei H, Nakatomi C. NF-kappaB signaling regulates physiological and pathological chondrogenesis. Int J Mol Sci (2019) 20(24):6275. doi: 10.3390/ijms20246275
28. Liu T, Zhang L, Joo D, Sun SC. NF-kappaB signaling in inflammation. Signal Transduct Target Ther (2017) 2:17023–. doi: 10.1038/sigtrans.2017.23
29. Luo XY, Liu Q, Yang H, Tan Q, Gan LQ, Ren FL, et al. OSMR gene effect on the pathogenesis of chronic autoimmune Urticaria via the JAK/STAT3 pathway. Mol Med (2018) 24(1):28. doi: 10.1186/s10020-018-0025-6
30. Feng H, Feng J, Zhang Z, Xu Q, Hu M, Wu Y, et al. Role of IL-9 and IL-10 in the pathogenesis of chronic spontaneous urticaria through the JAK/STAT signalling pathway. Cell Biochem Funct (2020) 38(4):480–9. doi: 10.1002/cbf.3481
31. Hox V, O’Connell MP, Lyons JJ, Sackstein P, Dimaggio T, Jones N, et al. Diminution of signal transducer and activator of transcription 3 signaling inhibits vascular permeability and anaphylaxis. J Allergy Clin Immunol (2016) 138(1):187–99. doi: 10.1016/j.jaci.2015.11.024
32. Neurath MF, Finotto S. IL-6 signaling in autoimmunity, chronic inflammation and inflammation-associated cancer. Cytokine Growth Factor Rev (2011) 22(2):83–9. doi: 10.1016/j.cytogfr.2011.02.003
33. Tackey E, Lipsky PE, Illei GG. Rationale for interleukin-6 blockade in systemic lupus erythematosus. Lupus (2004) 13(5):339–43. doi: 10.1191/0961203304lu1023oa
34. Tanaka T, Narazaki M, Kishimoto T. Therapeutic targeting of the interleukin-6 receptor. Annu Rev Pharmacol Toxicol (2012) 52:199–219. doi: 10.1146/annurev-pharmtox-010611-134715
35. Caproni M, Volpi W, Macchia D, Giomi B, Manfred M, Campi P, et al. Infiltrating cells and related cytokines in lesional skin of patients with chronic idiopathic urticaria and positive autologous serum skin test. Exp Dermatol (2003) 12(5):621–8. doi: 10.1034/j.1600-0625.2003.00010.x
36. Metz M, Krull C, Maurer M. Histamine, TNF, C5a, IL-6, -9, -18, -31, -33, TSLP, neopterin, and VEGF are not elevated in chronic spontaneous urticaria. J Dermatol Sci (2013) 70(3):222–5. doi: 10.1016/j.jdermsci.2013.03.003
37. Gora A, Przybyl M, Swietochowska E, Machura E. Assessment of selected interleukins (IL-6, IL-17A, IL-18, IL-23) and chemokines (RANTES, IP-10) in children with acute and chronic urticaria. Ital J Pediatr (2022) 48(1):201. doi: 10.1186/s13052-022-01395-3
38. Grieco T, Porzia A, Paolino G, Chello C, Sernicola A, Faina V, et al. IFN-gamma/IL-6 and related cytokines in chronic spontaneous urticaria: evaluation of their pathogenetic role and changes during omalizumab therapy. Int J Dermatol (2020) 59(5):590–4. doi: 10.1111/ijd.14812
39. de Montjoye L, Choteau M, Herman A, Hendrickx E, Chéou P, Baeck M, et al. IL-6 and IL-1beta expression is increased in autologous serum skin test of patients with chronic spontaneous urticaria. Allergy (2019) 74(12):2522–4. doi: 10.1111/all.13928
40. Kolkhir P, Andre F, Church MK, Maurer M, Metz M. Potential blood biomarkers in chronic spontaneous urticaria. Clin Exp Allergy (2017) 47(1):19–36. doi: 10.1111/cea.12870
41. Ataie-Kachoie P, Pourgholami MH, Morris DL. Inhibition of the IL-6 signaling pathway: a strategy to combat chronic inflammatory diseases and cancer. Cytokine Growth Factor Rev (2013) 24(2):163–73. doi: 10.1016/j.cytogfr.2012.09.001
42. Asero R, Cugno M. Biomarkers of chronic spontaneous urticaria and their clinical implications. Expert Rev Clin Immunol (2021) 17(3):247–54. doi: 10.1080/1744666X.2021.1882304
43. Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity (2011) 34(5):637–50. doi: 10.1016/j.immuni.2011.05.006
44. Lim KH, Staudt LM. Toll-like receptor signaling. Cold Spring Harb Perspect Biol (2013) 5(1):a011247. doi: 10.1101/cshperspect.a011247
45. Zhang Y, Liu J, Wang C, Liu J, Lu W. Toll-like receptors gene polymorphisms in autoimmune disease. Front Immunol (2021) 12:672346. doi: 10.3389/fimmu.2021.672346
46. Kirtland ME, Tsitoura DC, Durham SR, Shamji MH. Toll-like receptor agonists as adjuvants for allergen immunotherapy. Front Immunol (2020) 11:599083. doi: 10.3389/fimmu.2020.599083
47. Radman M, Golshiri A, Shamsizadeh A, Zainodini N, Bagheri V, Arababadi MK, et al. Toll-like receptor 4 plays significant roles during allergic rhinitis. Allergol Immunopathol (Madr) (2015) 43(4):416–20. doi: 10.1016/j.aller.2014.04.006
48. Shukur W, Alyaqubi K, Dosh R, Al-Ameri A, Al-Aubaidy H, Al-Maliki R, et al. Association of Toll-like receptors 4 (TLR-4) gene expression and polymorphisms in patients with severe asthma. J Med Life (2021) 14(4):544–8. doi: 10.25122/jml-2021-0173
49. Rylander R, Michel O. Organic dust induced inflammation–role of atopy and TLR-4 and CD14 gene polymorphisms. Am J Ind Med (2005) 48(4):302–7. doi: 10.1002/ajim.20205
50. Azor MH, dos Santos JC, Futata EA, de Brito CA, Maruta CW, Rivitti EA, et al. Statin effects on regulatory and proinflammatory factors in chronic idiopathic urticaria. Clin Exp Immunol (2011) 166(2):291–8. doi: 10.1111/j.1365-2249.2011.04473.x
51. Caproni M, Giomi B, Volpi W, Melani L, Schincaglia E, Macchia D, et al. Chronic idiopathic urticaria: infiltrating cells and related cytokines in autologous serum-induced wheals. Clin Immunol (2005) 114(3):284–92. doi: 10.1016/j.clim.2004.10.007
52. Quarmby S, Kumar P, Kumar S. Radiation-induced normal tissue injury: role of adhesion molecules in leukocyte-endothelial cell interactions. Int J Cancer (1999) 82(3):385–95. doi: 10.1002/(SICI)1097-0215(19990730)82:3<385::AID-IJC12>3.0.CO;2-5
53. Yanaba K, Kaburagi Y, Takehara K, Steeber DA, Tedder TF, Sato S. Relative contributions of selectins and intercellular adhesion molecule-1 to tissue injury induced by immune complex deposition. Am J Pathol (2003) 162(5):1463–73. doi: 10.1016/S0002-9440(10)64279-4
54. Lawson C, Wolf S. ICAM-1 signaling in endothelial cells. Pharmacol Rep (2009) 61(1):22–32. doi: 10.1016/S1734-1140(09)70004-0
55. Sumagin R, Kuebel JM, Sarelius IH. Leukocyte rolling and adhesion both contribute to regulation of microvascular permeability to albumin via ligation of ICAM-1. Am J Physiol Cell Physiol (2011) 301(4):C804–813. doi: 10.1152/ajpcell.00135.2011
56. Canonica GW, Ciprandi G, Pesce GP, Buscaglia S, Paolieri F, Bagnasco M. ICAM-1 on epithelial cells in allergic subjects: a hallmark of allergic inflammation. Int Arch Allergy Immunol (1995) 107(1-3):99–102. doi: 10.1159/000236943
57. Puxeddu I, Panza F, Pratesi F, Bartaloni D, Casigliani Rabl S, Rocchi V, et al. CCL5/RANTES, sVCAM-1, and sICAM-1 in chronic spontaneous urticaria. Int Arch Allergy Immunol (2013) 162(4):330–4. doi: 10.1159/000354922
58. Lv G, Fan J. Silencing ICAM-1 reduces the adhesion of vascular endothelial cells in mice with immunologic contact urticaria. Gene (2020) 760:144965. doi: 10.1016/j.gene.2020.144965
59. Zuberbier T, Aberer W, Asero R, Bindslev-Jensen C, Brzoza Z, Canonica GW, et al. The EAACI/GA(2) LEN/EDF/WAO Guideline for the definition, classification, diagnosis, and management of urticaria: the 2013 revision and update. Allergy (2014) 69(7):868–87. doi: 10.1111/all.12313
60. Bristow MR, Ginsburg R, Kantrowitz NE, Baim DS, Rosenbaum JT. Coronary spasm associated with urticaria: report of a case mimicking anaphylaxis. Clin Cardiol (1982) 5(3):238–40. doi: 10.1002/clc.4960050307
61. Jawabrah Al-Hourani B, Sharma SK, Suresh M, Wuest F. Cyclooxygenase-2 inhibitors: a literature and patent review (2009 - 2010). Expert Opin Ther Pat (2011) 21(9):1339–432. doi: 10.1517/13543776.2011.593510
62. Kolaczkowska E, Goldys A, Kozakiewicz E, Lelito M, Plytycz B, van Rooijen N, et al. Resident peritoneal macrophages and mast cells are important cellular sites of COX-1 and COX-2 activity during acute peritoneal inflammation. Arch Immunol Ther Exp (Warsz) (2009) 57(6):459–66. doi: 10.1007/s00005-009-0053-6
63. Stevenson DD. Aspirin and NSAID sensitivity. Immunol Allergy Clin North Am (2004) 24(3):491–505. doi: 10.1016/j.iac.2004.03.001
64. Picado P. COX-2 specific inhibitors in NSAID-intolerant patients. Int J Immunopathol Pharmacol (2003) 16(2 Suppl):11–6.
65. Vane JR, Botting RM. Anti-inflammatory drugs and their mechanism of action. Inflammation Res (1998) 47 Suppl 2:S78–87. doi: 10.1007/s000110050284
66. Caproni M, Giomi B, Melani L, Volpi W, Antiga E, Torchia D, et al. Cellular infiltrate and related cytokines, chemokines, chemokine receptors and adhesion molecules in chronic autoimmune urticaria: comparison between spontaneous and autologous serum skin test induced wheal. Int J Immunopathol Pharmacol (2006) 19(3):507–15. doi: 10.1177/039463200601900306
67. Bracken SJ, Abraham S, MacLeod AS. Autoimmune theories of chronic spontaneous urticaria. Front Immunol (2019) 10:627. doi: 10.3389/fimmu.2019.00627
68. Criado RFJ, Criado PR, Pagliari C, Sotto MN, MaChado Filho CD, Bianco B. M2 macrophage polarization in chronic spontaneous urticaria refractory to antihistamine treatment. Allergol Int (2021) 70(4):504–6. doi: 10.1016/j.alit.2021.03.005
69. Su W, Huang B, Zhao Y, Zhang X, Chen L, Ji J, et al. (2020). Integrated bioinformatics approach to understand immune-related key genes and pathways in chronic spontaneous urticaria. PREPRINT (Version 1). Available at Research Square: https://doi.org/10.21203/rs.3.rs-137346/v1.
Keywords: chronic spontaneous urticaria, bioinformatics, differentially expressed genes, hub genes, pathway, immune infiltration, TNF, NF-κB
Citation: Su W, Tian Y, Wei Y, Hao F and Ji J (2023) Key genes and immune infiltration in chronic spontaneous urticaria: a study of bioinformatics and systems biology. Front. Immunol. 14:1279139. doi: 10.3389/fimmu.2023.1279139
Received: 17 August 2023; Accepted: 02 November 2023;
Published: 15 November 2023.
Edited by:
Cong Peng, Central South University, ChinaReviewed by:
Angelica Petraroli, Federico II University Hospital, ItalyCopyright © 2023 Su, Tian, Wei, Hao and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiang Ji, amlqaWFuZzIyMjJAMTI2LmNvbQ==; Fei Hao, aGFvZmVpNjJAMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.