
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 17 October 2023
Sec. Comparative Immunology
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1276357
This article is part of the Research TopicCommunity Series in Recent Advances in Drosophila Cellular and Humoral Innate Immunity, volume IIView all 10 articles
A correction has been applied to this article in:
Corrigendum: Interactions between innate immunity and insulin signaling affect resistance to infection in insects
An active immune response is energetically demanding and requires reallocation of nutrients to support resistance to and tolerance of infection. Insulin signaling is a critical global regulator of metabolism and whole-body homeostasis in response to nutrient availability and energetic needs, including those required for mobilization of energy in support of the immune system. In this review, we share findings that demonstrate interactions between innate immune activity and insulin signaling primarily in the insect model Drosophila melanogaster as well as other insects like Bombyx mori and Anopheles mosquitos. These studies indicate that insulin signaling and innate immune activation have reciprocal effects on each other, but that those effects vary depending on the type of pathogen, route of infection, and nutritional status of the host. Future research will be required to further understand the detailed mechanisms by which innate immunity and insulin signaling activity impact each other.
Innate immunity and insulin/insulin-like signaling (IIS) are both integral for homeostasis and anti-pathogen defense in insects. An immune response is critical for protecting an organism from invading pathogens that pirate resources and reproduce within the host (1, 2). The IIS pathway functions as a nutrient-sensing pathway that regulates cell and tissue growth, as well as whole-organism metabolism (3, 4). Innate immunity and IIS activity have been thought of as independent processes, but developments over the past decade have demonstrated them to be connected in mobilizing energy stores required for an effective immune response (5). In this review, we will discuss what we know of the physiological consequences and genetic mechanisms underlying interactions between insulin and immune pathways in insects, emphasizing primary examples from Drosophila melanogaster and integrating data from other insect models.
The innate immune system and IIS pathway are highly conserved across insect orders. Comparative genomics has revealed considerable conservation of orthologs in immune genes across almost all insects whose genomes have been sequenced (6). Similarly, insulin like-peptides (ILPs) that share structural and functional homology to mammalian insulin, have also been identified in major insect orders including Orthoptera, Diptera, Hymenoptera, Coleoptera, and Hemiptera (7–11). Concurrently, the development of genetic technologies from RNAi to CRISPR/Cas9 has enabled us to begin to understand how these pathways operate and how they impact each other across diverse insects (12–17), paving the way for future studies of how these systems interact to maintain organismal homeostasis in the presence and absence of infection.
Mounting an immune response is an energetically costly process, requiring systemic physiological shifts in metabolism and energetic stores (16, 18–21). A broad range of pathogenic infections, from bacterial, viral, parasitic, or fungal, can lead to decreases in energetic stores like glycogen and triglycerides (12, 22–26). In insects, the fat body is a multifunctional tissue that can secrete antimicrobial peptides in response to immune stimulus and is also a site for the storage and breakdown of carbohydrates and lipids (27). Energetic stores like glycogen can be critical for supplying energy to activate the immune response. Glycogen is the polymeric form of glucose, and can be broken down in tissues like the fat body to be released into the hemolymph as free glucose by the enzyme glycogen phosphorylase (glyp) (28). D. melanogaster exhibit decreased glycogen levels and increased circulating glucose levels after infection with Streptococcus pneumoniae, with increased gene expression of glyp stimulated by the extracellular release of adenosine (25, 29). Phagocytic immune cells uptake the circulating glucose during the acute phase of S. pneumoniae infection, enabling phagocytosis. D. melanogaster deficient for the adenosine receptor or glyp exhibit lower levels of glycogen and higher mortality after S. pneumoniae infection (25, 29).
A metabolomics study on larvae of the silkworm Bombyx mori infected with the entomophathogenic fungi Beauveria bassiana found major shifts in hemolymph levels of metabolites like lipids, carbohydrates, and amino acids after infection (30). Fungal infection also caused higher levels of glucose and an associated reduction in trehalose (30). Trehalose is a disaccharide comprised of two glucose molecules that can be readily liberated into free glucose by the enzyme trehalase. Trehalose is the primary sugar found in insect hemolymph and is used for energetic needs like flight and metabolic homeostasis (28, 31). Although Xu et al. (30) did not measure trehalase activity, Praveena et al. (32) found that trehalase activity progressively diminishes in the hemolymph of B. mori larvae over five days of B. beauveria infection while they remain consistent in uninfected controls. These authors hypothesize that trehalase activity reduces over the course of the infection because the early hydrolysis of trehalose to glucose consumes the substrate pool available for conversion. The data suggest that during a fungal infection, trehalose may be converted to glucose to allocate energy to fighting the infection.
The breakdown of glycogen and trehalose provides free glucose for immune cell function (29). However, the role of triglyceride breakdown during infection is less well established. It has been recently demonstrated that D. melanogaster larvae exhibit reduced triglyceride stores during an active immune response, and shift from lipid storage to synthesizing phosphatidylcholine (PC) and phosphatidylethanolamine (PE) (16), which are two of the major phospholipids that comprise secretory vesicles and cell and organelle membranes (33). Active humoral immune responses drive high levels of AMP gene expression (e.g., 34), which could put a heavy burden for the endoplasmic reticulum (ER) to synthesize and secrete the encoded effectors into circulation. This shift to PE and PC synthesis during chronic immune activation has been hypothesized to support the secretion of immune effectors during the immune response.
Cellular stresses, including those imposed by immune reactions, stimulate the unfolded protein response (UPR) to allow the endoplasmic reticulum (ER) to regulate proper folding and secretion of proteins (35, 36). One marker of UPR in the ER is the upregulation of the transcription factor X-box binding protein 1 (XBP1), which positively regulates phospholipid synthesis and enzymes that increase the size of the ER (37). Both infection (34, 38) and chronic immune activation in the fat body (16) have been demonstrated to increase gene expression of XBP1. This elevated expression of ER-stress response genes is also associated with increased expansion of the ER lumen in infected mated females (38) and in uninfected larvae with an overactive immune response (16). These results suggest that the innate immune response induces ER stress, which increases phospholipid synthesis to support secretion of immune effectors. The infection-induced reduction of triglyceride stores observed across several D. melanogaster studies (12, 19, 23, 39) could arise from a need to mobilize energetic stores to support ER homeostasis by way of upregulating phospholipid synthesis.
Sustained long-term infections can result in energetic wasting. In a healthy animal, insulin signaling regulates glucose homeostasis (40, 41). Phosphorylation of the IIS protein Akt leads to uptake of extracellular glucose (42) and promotes glycogen and triglyceride synthesis (43) by positively regulating the genes glycogen synthase and acetyl coenzyme-A (Figure 1; 44–46). Dionne et al. (12) demonstrated that Drosophila melanogaster infected with the bacterium Mycobacterium marinum exhibit impaired IIS activity over several days of infection. Reduction in phosphorylation of Akt leads to reduced expression of glycogen synthase and acetyl coenzyme-A. Consequently, flies infected with M. marinum become hyperglycemic and progressively lose glycogen and triglyceride stores until they ultimately die from the infection.
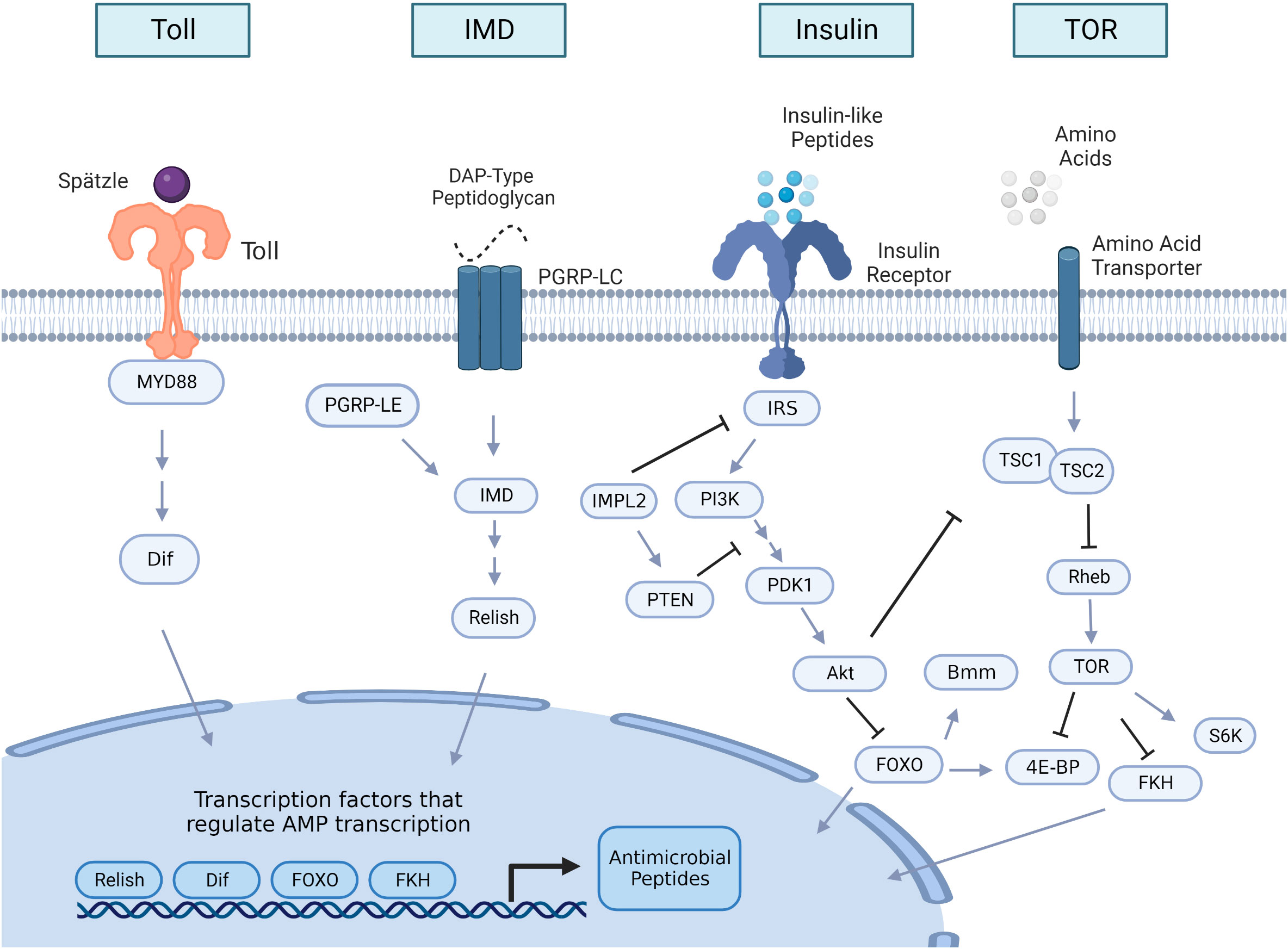
Figure 1 Insulin and innate immune pathway signaling overview in insects. The Toll and the immune deficiency (IMD) pathways are the two major innate immune signaling pathways that are induced upon bacterial or fungal infection, leading to the production of antimicrobial peptides (AMPs). The Toll receptor is activated by the cleaved form of the cytokine Spätzle, which is processed in a proteolytic cascade after the detection of Gram-positive bacteria or fungi. Once Toll is active, it initiates an intracellular cascade that leads to activation of the nuclear factor κB (NF-κB) transcription factor Dif. When activated, Dif translocate to the nucleus to initiate AMP transcription. IMD signaling is initiated by peptidoglycan recognition protein LC (PGRP-LC) or peptidoglycan recognition protein LE (PGRP-LE) binding DAP-type peptidoglycan shed by Gram-negative bacteria and some Gram-positive bacteria. This binding triggers a cascade that activates the NF-kB transcription factor Relish to translocate to the nucleus to transcribe AMPs. Insulin signaling activity begins with the production of insulin-like peptides (ILPs), which are predominantly secreted from insulin-producing cells and from other tissue sources like the fat body or muscle. An intracellular cascade occurs after ILPS bind to the insulin receptor, activating the protein Akt. Once Akt is active, it impacts multiple downstream processes within the cell, including inhibition of nuclear translocation of the forkhead family transcription factor, Forkhead box-O (FOXO), which can regulate transcription of AMPs, and the lipase brummer (bmm). Akt activation also stimulates the target of rapamycin (TOR) pathway by TSC1/TSC2 suppression. TOR is also activated by amino acid sensing. Downstream targets of TOR activation include repression of the forkhead box family member forkhead (FKH), which also regulates AMP transcription, activation of the protein kinase S6K, which is critical for growth processes, and the inhibition of translational regulator 4E-BP (eukaryotic initiation factor 4 binding protein). Arrows in the figure denote positive interactions and bars denote negative interactions. Double arrows indicate that additional steps in the pathway are not illustrated. IRS, insulin receptor substrate; IMPL2, ecdysone-inducible gene L2; PI3K; phosphatidylinositol 3-kinase; PTEN, phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase; PDK1 (phosphoinositide-dependent protein kinase); TSC, tuberous sclerosis tumor suppressor Rheb, Ras homolog enriched in brain; S6K, ribosomal S6 kinase.
In examples such as this (12, 23) it can be difficult to determine whether the metabolic change reflects a manipulation of the host by the parasite for the parasite’s own nutritional benefit, or whether the wasting reflects a collateral cost of sustained immune reactions. In the case of bacteria in the genus Mycobacterium, the bacteria primarily depend on lipids like cholesterol as their source of carbon (47, 48) and a large portion of the genome for this genus is devoted to lipid metabolism (49). It is possible that wasting phenotypes observed in this infection could be a result of pathogen nutrient acquisition. This can be distinguished with appropriately designed experiments. For example, one human study investigated metabolic shifts induced by infection with Mycobacterium tuberculosis (Mtb), the causative agent of tuberculosis (TB). These authors measured metabolic shifts in blood samples from patients that were healthy, patients infected with Mtb and given short-term antibiotic treatment, and Mtb-infected patients without treatment (50). They found that Mtb infection, regardless of treatment, significantly shifted metabolic profiles compared to healthy patients, and they observed no significant differences in metabolites between TB patients treated with antibiotics or not, despite reduced pathogen load in patients given antibiotics (50). These authors suggest that their observation that antibiotic treatment does not impact infection-induced metabolic shifts implies that the host consumes metabolites during an active immune response. This study does not rule out the possibility that virulence factors produced by the pathogen could continue to impact host metabolism even after pathogen loads are reduced by antibiotic treatment. However, it can serve as an example of how to begin to study whether metabolic consequences of infection are due to host energetic reallocation or pathogen nutrient acquisition.
Both the immune system and insulin signaling are heavily influenced by dietary nutrition (51–55), thus any variation in the composition of diet can influence an organism’s ability to resist infection and regulate global metabolism. When Drosophila melanogaster are reared on high glucose (0.57 M), they experience reduced resistance to a systemic infection by the bacterial pathogen Providencia rettgeri (56). Similarly, those reared on high sucrose (1 M) experience increased mortality following infection by the pathogen Pseudomonas aeruginosa (52). High sucrose diets also impair immunological melanization in larval hemolymph (52) and reduce phagocytosis of fungal spores by D. melanogaster larvae (53).
IIS activity might mediate the higher post-infection mortality observed in flies fed high-sugar diets. Musselman et al. (52) found through an RNA-seq analysis that uninfected larvae with knockdown of the insulin receptor (InR) in the fat body exhibit elevated expression of AMPs while larvae with constitutive activation of InR in the fat body exhibit reduced AMP expression. Knockdown of InR in the adult fat body increases survivorship of systemic P. aerugionsa infection in flies fed a high-sugar (0.7 M) diet (52). These data suggest that reduction in insulin signaling could increase infection survival on high-sugar diets, potentially due to elevated expression of immune effectors. In that study, however, they did not specifically measure AMP expression during an infection in flies with InR knockdown so it remains to be determined whether InR knockdown promotes survival of infection by elevating AMP expression.
These systemic infections performed by Unckless et al. (56) and Musselman et al. (52) were performed on Drosophila reared on high dietary sugar throughout larval development. High-sugar diets cause developmental delays, lipidemia, hyperglycemia, and reduced body size in both larvae and adults (51, 52). Thus, the consequences of a high-sugar diet on adult defense could be due to dietary effects on larval development that may affect immune capacity in the adult stage. For example, D. melanogaster larvae fed high yeast diets exhibit increased expression of antimicrobials as adults (57). Drosophila larvae fed low-protein diets have lower counts of phagocytic immune cells (58) and are more susceptible to systemic Pseudomonas entomophila infection in the adult stage (59). Mosquito larvae fed low protein diets were more susceptible to Plasmodium falciparum (60) and Sindbis virus infections (61). Infection in the D. melanogaster larval stage can also affect adult fitness, including reduced lifespan (62) and smaller body size (63). Future research should distinguish between the immediate metabolic effects and developmental effects of sugar overnutrition. One strategy for doing this is to rear juveniles on the same rearing medium then transfer adults to varied experimental diets after development is complete. This approach could be directly compared to cohorts of insects that are reared on varied diets high in dietary sugar to determine the relative contributions of developmental history and immediate metabolic dysregulation on immune function.
Some studies suggest a potentially protective effect of dietary sugar during enteric infection. For example, D. melanogaster fed high-glucose diets (0.55 M) exhibit increased survival after an enteric Vibrio cholerae infection (64). Feeding sugar to the mosquito Aedes aegypti also increases expression of antiviral genes in the intestinal tract and protects mosquitos from oral arbovirus infections (65). High-sugar diets have been demonstrated to elevate reactive oxygen species levels (66) and increase expression of antimicrobial peptides (67) in the midgut of D. melanogaster. These data suggest high-sugar diets may alter gut immunity, potentially as a consequence of dysregulation of the endogenous microbiota (68), which would not occur in the hemocoel during a systemic infection. These data draw attention to important differences in the consequences of dietary sugar that may depend on whether the infection is acquired systemically or orally.
The dependency of infection-related phenotypes on dietary nutrition raises an important concern for comparisons across studies performed by different research groups. There is very little standardization of diets used for rearing D. melanogaster in infection studies (69, 70). For example, the diet used for the enteric infection referenced in (64) was a holidic (chemically-defined) diet, which is known to delay development relative to sugar-yeast based diets that are standard in D. melanogaster studies (71, 72), and the definition of “high” sugar varied nearly 2-fold between Musselman et al. (52) and Unckless et al. (56). Potentially, differences in dietary composition between studies could affect metabolism and infection phenotypes even when diet is not itself an explicit focus of the experiment.
Transcriptomic studies of infected insects often show differential expression of genes in gene ontology (GO) categories related to carbohydrate and lipid metabolism (34, 73–75). The IIS pathway is a critical regulator of carbohydrate and lipid metabolism (4, 76, 77). Thus, infection-induced shifts in carbohydrate and lipid metabolism could be mediated by alterations in IIS activity driven by the immune system.
The Toll signaling pathway (Figure 1) is one of two major innate immune signaling pathways in insects (78). Toll signaling is homologous to Toll-like signaling in vertebrates (79) and leads to activation of the NF-κB transcription factor Dif. Gram-positive bacterial and fungal pathogens primarily activate Toll signaling (80, 81). Activation of Toll in the D. melanogaster fat body either through genetic manipulation or infection suppresses insulin signaling as measured by reduced phosphorylation of Akt (19, 39, 82). Since the insulin pathway is critical for regulating growth and lipid metabolism (76, 77), inhibition of IIS activity by Toll activation during development results in reduced body size and depletion of triglyceride stores (19, 82).
Suzawa et al. (82) showed that infection of D. melanogaster larvae with the Gram-positive bacterium Enterococcus faecalis reduces production of insulin-like peptide 6 (dILP6), which is an ILP secreted from the fat body that regulates larval growth (83, 84). Overexpression of Dif in the larval fat body also reduces dILP6 mRNA transcripts, and the infection-induced reduction in dILP6 and growth inhibition can be rescued by RNAi knockdown of Dif (82). Roth et al. (39) further found that activation of Toll in the larval fat body inhibits PDK1 from phosphorylating Akt at residue T342, resulting in growth inhibition and reduced triglyceride storage (39). Body size and triglyceride storage were rescued by forced phosphorylation of Akt site T342 in the fat body of larvae with constitutively activated Toll (39). These results clearly place alteration of dILP6-production and larval insulin signaling downstream of the Toll signaling pathway. However, further experiments are required to test whether Dif directly regulates dILP6 expression or whether insulin signaling is secondarily regulated by proteins whose expression is directly controlled by Dif.
The immune deficiency (IMD) pathway (Figure 1) is the second main arm of the insect innate immune system (73). IMD signaling is responsive to DAP-type peptidoglycan shed by Gram-negative bacteria as well as some Bacillus species (81) and activates the NF-κB transcription factor Relish to induce transcription of antimicrobial peptides and other immune response genes (Figure 1; 81, 85). There is mixed evidence of IMD activity impacting insulin signaling and mobilization of energetic stores in D. melanogaster. DiAngelo et al. (19) found that constitutive activation of Relish in the fat body did not significantly impact Akt phosphorylation or triglyceride content (19). Although infection with the Gram-negative bacterium Escherichia coli reduced Akt phosphorylation, the suppression was lost when Toll pathway mutants were infected with E. coli, indicating that the attenuation of IIS was due to cross activation of the Toll pathway and not due to IMD signaling (19). In contrast, Davoodi et al. (86) found that constitutive activation of the IMD protein in the fat body significantly reduces phosphorylation of Akt and S6K throughout the body. S6K is a serine kinase downstream of Akt (Figure 1) that is critical for regulating larval growth (87). IMD activation in the fat body also reduced total triglyceride and trehalose levels in 3rd-instar larvae (86). Those larvae weighed less, had delayed development, and had reduced adult eclosion rates. These phenotypes are similar to those of insulin-signaling mutants and flies that overexpress Toll in the fat body (19, 88). One major difference between these two studies is the fly genotypes used to constitutively activate the IMD signaling pathway. Davoodi et al. (86) overexpressed the IMD protein where DiAngelo et al. (19) forced constitutive expression of the transcription factor Relish. The IMD molecule is an adaptor protein that acts upstream of Relish (Figure 1; 81), and, when activated, it triggers an intracellular signaling cascade that results in Relish activation (89). Potentially, there may be branching of the IMD pathway such that targets downstream of the IMD protein that disrupts insulin signaling and alter metabolic stores may not be regulated by Relish.
Davoodi et al. (86), also performed a transcriptomic analysis on larval fat body with constitutive IMD protein activation and identified modified expression of genes that regulate metabolic processes. Genes involved in the negative regulation of IIS were upregulated, including impl2 and downstream targets of IIS like 4E-BP (86). Lipid synthesis and gluconeogenesis genes were conversely downregulated in larvae with constitutive expression of the IMD pathway (86). These transcriptional data suggest that the IMD pathway, like Toll signaling, can influence metabolism.
Toll and IMD signaling are both critical for systemic immunity, however, the IMD pathway is primarily responsible for regulating gut immunity, including in response to endogenous microbiota (90). Impaired IMD signaling in the gut has been demonstrated to affect insulin signaling activity and metabolic stores. Kamareddine et al. (91) found a significant depletion of lipids in the fat body and an accumulation of lipid droplets in the anterior region of the midguts of several IMD mutants of D. melanogaster (Dredd, key, RelE20, PGRP-LC and PGRP-LE) in the absence of a systemic infection. Despite having depleted lipids in the fat body, whole-body triglyceride content and glucose levels were higher in the IMD mutants. These mutants also exhibited lower levels of Ilp3 transcripts and reduced phosphorylated Akt in the intestine (91). Davoodi et al. (86) similarly observed that uninfected imd mutants have higher whole-body triglyceride levels and reduced expression of Ilp2, Ilp3, and Ilp5, although only in males. These results demonstrate that IMD signaling plays a critical role in regulating metabolic homeostasis even in the absence of systemic infection, potentially through actions in the gut or on the endogenous gut microbiota.
The insect digestive tract is a complex organ composed of multiple cell types that vary in function including immune defense, absorption of nutrients, and lipid metabolism (92). Enteroendocrine (EE) cells in the midgut epithelium produce hormones, regulate stem cell activity, and express insulin-like peptides (93). Kamareddine et al. (91) found that Relish knockdown in EE cells reduced Ilp3 transcripts in the gut, increased EE lipid content, and reduced phosphorylation of Akt, while Relish knockdown in the fat body yielded no effect on Akt phosphorylation, triglyceride levels, or glucose content (91). Previous studies have demonstrated a role for commensal D. melanogaster gut microbiota like Acetobactor pomorum in regulating host metabolism, particularly via the microbial metabolic byproduct acetate (94, 95). Provision of acetate to germ-free flies increases nuclear localization of Relish and restores Akt phosphorylation (91). Germ-free wildtype flies have reduced IMD activity and exhibit metabolic phenotypes similar to those of flies with IMD knockdown in EE cells (91). Reciprocally, IIS activity and metabolic homeostasis are restored by overexpression of Relish in the EE cells of germ-free flies (91).
These data corroborate findings from an earlier microarray study that demonstrate microbiota-induced expression of host gene expression is Relish-dependent (96). Particularly, that study found that conventionally-reared flies exhibit upregulation of 285 genes in the gut relative to flies reared germ-free, and that GO categories for metabolic genes were enriched. When those authors then compared the transcriptomes of germ-free and conventionally reared Relish mutants, they found that expression of 151 of those 285 genes upregulated in response to microbiota was altered, including the insulin signaling genes PI3K, InR, and thor (96). Several other studies similarly demonstrate microbiota-Relish-dependent regulation of host gene expression in the gut. For example, oral infection with Erwinia carotovora also causes shifts in midgut gene expression that are Relish-dependent, including both immune and metabolic gene regulators (90). Relish mutants do not exhibit expression of microbiota-regulated genes observed in wildtype flies, regardless of whether they are germ-free or associated with microbiota (97). These data suggest a model whereby the host IMD pathway regulates the gut microbiota to maintain metabolic homeostasis, including through altered production of IIS activity by microbiota-derived metabolites such as acetate. However, the mechanistic details of signal integration under this model are unclear, and future experiments should study how downstream effectors of the IMD pathway interact with components of IIS to shape global metabolic regulation.
Since the IIS pathway is a critical global regulator of metabolism (76), it is also vital for regulating communication among tissues, and there are several studies that demonstrate innate immune activation also stimulating inter-organ communication in response to infection in Drosophila (98–100; see 101 for a review on inter-organ communication during an immune response). For example, glutamate derived from the muscle stimulates reallocation of lipids from the fat body to improve survivorship during oral infection with Pseudomonas entomophila (100). While not often directly investigated, there is evidence to suggest tissue communication during an immune response occurs in part by altered IIS activity. One study found that both systemic infection and overexpression of Toll and IMD in the fat body increased triglyceride and lipid droplet size in the midgut (99). This increase in lipid accumulation in the midgut was also associated with increased midgut gene expression of negative IIS regulators like Impl2 and thor (99), which suggests that Toll and IMD activation in the fat body can suppress IIS activity in the midgut in response to innate immune activity in the fat body. In this study, lipid accumulation in the midgut was critical for surviving systemic Photorhabdus luminsecens infection (99). It is an exciting avenue of research to consider the extent to which innate immune activity and IIS activity may communicate across tissues to drive organism-level responses and increase survivorship during infection.
Several studies in D. melanogaster have characterized the effect that suppressing IIS activity has on infection outcome. The insulin receptor substrate chico is important for mediating the downstream cascade of IIS activity after insulin-like peptides bind to the insulin receptor (Figure 1, 76, 88). Chico mutants experienced increased survival of systemic infection by Pseudomonas aeruginosa and Enterococcus faecalis compared to wildtype flies (102). This higher rate of survivorship post-infection was not associated with increased levels of infection-induced gene expression of the AMP-encoding genes Diptericin, Attacin, and Drosomycin, although chico mutants do exhibit increased expression of thor in response to infection (102). Thor is an inhibitor of 5’-cap-dependent mRNA translation (103) and can prioritize translation of AMPs during an infection (104). Systemic infection with several different bacteria induces higher expression of thor (34, 104, 105) and thor mutants are highly susceptible to bacterial and fungal infection (104–106). Potentially, suppression of insulin signaling could be indirectly protective against infection due to downstream effects on cap-independent translation of proteins like AMPs.
A subsequent study conducted by McCormack et al. (107) found that chico mutants sustained lower pathogen loads after systemic infection with E. coli and Photorhabdus luminescens, although survivorship proportion was similar to wildtype flies. The effects on immune system activity were mixed, with reduced infection-induced transcription of the AMP genes Diptericin, Cecropin A1, and Drosomycin, and even reduced phagocytic capacity (107). However, the chico mutants exhibited increased phenoloxidase activity and melanization compared to wildtype. Melanization is important for insect wound healing and to clear pathogens at an injury site (108) and has been demonstrated to increase survivorship after bacterial and fungal infections (109). Potentially, increased phenoloxidase activity could also contribute to the increased survivorship of E. faecalis infection observed by Libert et al. (102) since melanization is also important for surviving E. faecalis infection (109). Similarly, higher phenoloxidase and melanization activity has been suggested to increase survivorship after P. luminescens infection (110, 111). However, melanization activity is not critical for surviving E. coli infection (112), thus improved phenoloxidase activity alone does not fully explain improved survivorship for chico mutants after E. coli infections observed by Libert et al. (102).
A more recent study assaying the effect of disrupted IIS activity on infection survival in D. melanogaster further demonstrated pathogen-specific infection outcomes. Davoodi et al. (86) delivered systemic and oral infections with five different bacterial pathogens (Vibrio cholerae, Enterococcus faecalis, Pseudomonas sneebia, Providencia rettgeri, and a virulent strain of Serratia marcescens) to triple mutants of insulin-like peptides 2, 3, and 5. P. sneebia systemic infections resulted in 100% mortality for both wildtype and ILP mutants, while systemic infection for all other bacteria tested resulted in higher proportion of death in ilp2,3,5 mutants. Pathogen loads were higher for ilp2,3,5 mutants only after systemic infection with P. rettgeri and E. faecalis. In contrast, ilp2,3,5 mutants showed higher survival of oral infection with P. sneebia, V. cholerae and S. marcescens, although only V. cholerae loads were reduced in ilp2,3,5 mutants (86). When IIS activity is increased due to mutation in the IIS pathway antagonist impl2, flies suffered higher mortality and increased pathogen load than wildtype after oral V. cholerae infection (86). Collectively, these data suggest that functional IIS activity increases susceptibility to oral infections. D. melanogaster with oral infection of V. cholerae exhibit reduced activation of the IIS pathway (95), which further suggest that reduction in IIS activity could be a protective mechanism against enteric V. cholerae infections.
The combination of results across all studies demonstrates that the effects of IIS signaling on immunity varies with the specific pathogen and route of infection in D. melanogaster. Future experiments using tissue-specific approaches to manipulate IIS could elucidate how specific organs contribute to the interaction between IIS and immunity. Additionally, conditional genetic manipulations could be deployed to differentiate immediate immunological effects of altered insulin signaling from pleiotropic consequences of developmental effects due to constitutively altered IIS.
There is also evidence that IIS affects immunity in mosquitoes. For example, several studies demonstrate that Plasmodium infection results in IIS activation and host immune suppression in Anopheles stephensi mosquitoes (e.g., 113, 114). P. falciparum infection induces the transcription of insulin-like peptide 1, 3, 4, and 5 in A. stephensi (10, 114). Artificial feeding of insulin-like peptide 3 (ILP3) and insulin-like peptide 4 (ILP4) to A. stephensi results in reduced expression of NF-kB-dependent genes in the midgut, including those encoding the antimicrobial peptides Gambicin, Cecropin, and Defensin (113, 115) and encapsulation-related proteins APL1, TEP1, LRIM1 (115). Similarly, feeding ILP4 or human insulin to A. stephensi at levels expected to be present in a bloodmeal (170 pM) induces IIS pathway activation and increases prevalence Plasmodium falciparum (113, 115, 116), where prevalence is defined as the proportion of mosquitos within the experimental population that become infected with Plasmodium. Reciprocally, inhibition of IIS activity with the PI3K inhibitor LY294002 or morpholinos that target ILP3 and ILP4 reduces the prevalence of P. falciparum infection and increases the expression level of immune effector genes in the mosquito midgut (113, 114). Together, these results demonstrate that insulin produced endogenously or obtained from a bloodmeal can suppress mosquito immune activity and could benefit infecting Plasmodium.
In a reciprocal experiment, Hauck et al. (117) demonstrated that overexpression of a negative regulator of Akt, phosphatase and tensin homolog (PTEN, Figure 1), in the A. stephensi midgut can reduce prevalence of Plasmodium infection. The proportion of PTEN-overexpressing mosquitos that became infected with P. falciparum was significantly lower than in wildtype controls and the transgenic mosquitoes developed fewer oocysts (117). Despite reduced prevalence of Plasmodium in the midgut, there was no effect on infection-induced expression of the AMP gene Defensin or in genes responsible for controlling P. falciparum, including NOS, TEP1, APL1, LRIM1 (117). PTEN overexpression enhanced expression of genes that promote autophagy, which is associated with midgut integrity and improves infection resistance, and midguts that overexpressed PTEN were less permeable than midguts that overexpress Akt (117). Thus, enhanced midgut integrity may provide another mechanism for reduced prevalence of Plasmodium in IIS-suppressed mosquitoes.
Nevertheless, an independent set of studies suggested an opposite phenomenon. Corby-Harris et al. (13) generated transgenic A. stephensi lines that drive constitutive activation of Akt under the control of the midgut carboxypeptidase promoter to assay the effects that midgut Akt activation has on Plasmodium development. In contrast to the previously cited studies, they found that activation of Akt significantly reduced prevalence of oocysts in the midgut and the proportion of mosquitos infected with P. falciparum (13), with Akt overexpression impacting mitochondrial function and the production of nitric oxide species (NOS) (13, 118). NOS has been associated with killing Plasmodium berghei and Plasmodium falciparum (119–121). Suppression of NOS in mosquitos that overexpress Akt increases their susceptibility to P. falciparum infection, demonstrating that the Akt-induced suppression of Plasmodium prevalence is mediated at least in part by elevated NOS levels (122). Overexpression of Akt in the midgut also results in abnormal mitochondria morphology and significantly reduced number of mitochondria in the midgut (122), with mitochondrial dysfunction evident in reduced activity of Mitochondrial Complexes I, II-II, and V. Additionally, Akt-overexpression in the fat body of mosquitos systemically infected with E. coli or Bacillus subtilis led to higher induction of antimicrobial peptides and higher survivorship of infection (118). These findings suggest that activation of IIS pathway in the fat body and midgut can induce an immune response to limit Plasmodium falciparum development and control bacterial infections.
These inconsistency between studies showing that insulin suppresses the immune response and increases P. falciparum prevalence (113, 115–117) versus those showing that overexpression of Akt increases immunity and suppresses Plasmodium establishment (13, 118) may result from differences in IIS activity or expression pattern. Surachetpong et al. (116) demonstrated that feeding human insulin to A. stephensi increased ROS production and NOS synthesis but that the elevated NOS synthesis was not associated with reduced P. falciparum prevalence (116). Potentially, transgenic mosquitos that overexpress Akt in the midgut or fat body may synthesize higher levels of NOS and ROS than is produced in wildtype mosquitos fed insulin. This could be tested in future studies. Additionally, insulin may have other immune-suppressive effects on mosquito physiology independent of Akt activation. More refined experimentation, including tissue-specific transcriptomic contrasts between mosquitoes fed on insulin compared to those overexpressing Akt could help elucidate the cause of the differences between the studies.
Interestingly, insulin treatment of vertebrate macrophages similarly suppresses inflammation and innate immunity regulated by NF-κB transcription factors (123–126). For example, mouse macrophages primed with insulin prior to immune challenge with lipopolysaccharide (LPS) exhibited attenuated activity of Toll-like Receptor 4, significantly reduced nuclear localization of NF-κB, and reduced gene expression of Tumor Necrosis Factor alpha (TNFα) (126). When insulin-primed cells were treated with a PI3K inhibitor, insulin-mediated suppression of immune activity was alleviated (126). These data further suggest that insulin-mediated suppression of NF-κB activity is evolutionary conserved between vertebrates and invertebrates.
Target of Rapamycin (TOR) is a nutrient sensing pathway that is jointly regulated by IIS activity and the abundance of free amino acids (Figure 1; 76, 127, 128). The Tsc1/Tsc2 complex negatively regulates TOR, and is inhibited by Akt (76). Thus, Akt phosphorylation activates the TOR complex to induce downstream cellular processes that promote growth (76). Inhibition of TOR can potentiate immunity in both mosquitos and Drosophila (129–131). For example, inhibition of A. stephensi TOR with rapamycin increased expression of genes like those encoding the NF-kB transcription factor Rel2, antimicrobial peptides Attacin and Cecropin, the complement-like protein thioester-containing protein 1 (TEP1), and the CLIP domain serine protease SPCLIP1 (130). TOR inhibition also reduced infection prevalence of Plasmodium berghei in A. stephensi (130). Similarly, suppressing TOR using rapamycin treatment in D. melanogaster increased expression of the antimicrobial peptides Diptericin and Metchnikowin (129), and also improved survival of systemic infection against the Gram-negative bacterium Pseudomonas aeruginosa (132).
Suppression of TOR through genetic manipulation has similar effects on immune function as rapamycin treatment (129, 132). For example, both uninfected TOR mutants and ubiquitous overexpression of the negative TOR regulator Tsc1/Tsc2 increased Diptericin and Metchnikowin expression in D. melanogaster under conditions of high protein availability (129). RagA is a GTPase that positively stimulates TOR activity (133), and flies that expressed a dominant negative form of RagA (which suppresses TOR activity) exhibited increased survivorship after P. aeruginosa infection (132). Rheb is a GTP-binding protein that positively regulates TOR activity (134, 135). Overexpression of Rheb reduced expression of the AMP genes Diptericin and Metchnikowin (129). Interestingly, genetic suppression of TOR by overexpression of Tsc1/Tsc2 increased susceptibility to systemic infection with the human pathogen Burkholderia cepacia, with flies exhibiting higher mortality and pathogen burden (136). TOR associates with two complexes, TOR complex 1 (TORC1) which is responsive to rapamycin and Tsc1/Tsc2 activity, and the less-studied TOR complex 2 (TORC2), which does not interact with Tsc1/Tsc2 or rapamycin treatment (137). When TORC2 is genetically suppressed via mutations lacking TORC2-specific components SIN1 and Rictor, flies exhibit increased survivorship and lower pathogen loads after B. cepacia (136). It is not clear to what extent TORC2 activity effects survivorship of other pathogens or its effects on immune capacity such as AMP expression.
TOR activity has also been recently demonstrated to play a role in D. melanogaster survival after oral infection. Deshpande et al. (131) discovered that active TOR signaling is necessary to survive an enteric infection with the entomopathogenic bacteria Pseudomonas entomophila. Oral infection with P. entomophila increases TOR activity and elevates expression of lipid synthesis genes like fatty acid synthase 1 and Lipin. Suppression of TOR via rapamycin treatment decreases infection survivorship without affecting bacterial load or AMP gene transcription (131). The higher mortality post-infection observed in flies with suppressed TOR activity was associated with significant depletion of lipid stores, specifically in the fat body and anterior region of the gut. This reduction in lipid after infection is associated with decrease in expression of lipid synthesis genes and an excessive loss in lipid stores (131), and suggests that TOR can regulate lipid homeostasis for tolerance of enteric infection.
Forkhead box proteins are a family of transcription factors whose activity is suppressed by IIS (Figure 1) (138, 139). The forkhead box-O (FOXO) protein in particular is involved in various physiological functions including apoptosis, stress responses and metabolic homeostasis (140–142), which are conserved from invertebrates to vertebrates (139, 143–146). Activated Akt phosphorylates FOXO, preventing FOXO from localizing to the nucleus to drive transcription of target genes including those that mobilize metabolic stores (76). FOXO is responsive to varied stress stimuli including starvation, diapause, hypoxia, and infection (12, 147–151) and regulates processes like lipolysis and autophagy in response to these stressors (149, 152).
Even in the absence of infection, starvation induces FOXO-dependent expression of antimicrobial peptides in the gut, trachea, fat body, and epidermis of D. melanogaster that is independent of Toll and IMD signaling (153). Similarly, the transcription factor Forkhead (FKH) drives AMP transcription in Drosophila in the absence of infection as a function of inhibited TOR signaling, again independent of Toll and IMD signaling (129). Starving larvae of the silkworm Bombyx mori larvae also exhibit decreased Akt phosphorylation and elevated expression of the AMP genes CecropinB6, Attacin1 and Defensin B (154). AMP gene promoters in varied insects frequently contain binding sites for forkhead-family transcription factors (153–156). These may allow infection-independent FKH/FOXO-regulated expression of these genes, and there may also be synergism between forkhead family and NF-kB family transcription factors. Future study should emphasize the extent to which forkhead-family transcription factors synergize with canonical immune signaling during an active infection.
Inhibition of FOXO results in increased nutrient storage and activation of FOXO results in nutrient mobilization and release. As could then be expected, D. melanogaster mutants for FOXO experience reduced loss of glycogen and a partial rescue of metabolic wasting compared to wildtype flies during a systemic Mycobacterium marinum infection (12). FOXO-deficient flies have increased survival of infection with M. marinum despite having pathogen load similar to that of wildtype flies, which suggests that the progressive loss of nutrient stores associated with activated FOXO increases mortality.
FOXO additionally has also been implicated in contributing to IMD-mediated effects on lipid metabolism in the absence of infection. Molaei et al. (149) found Relish mutants to have lower whole-body levels of triglyceride. Knockdown of Relish in the fat body had no effect on triglyceride content under normal rearing conditions, but when knockdown flies were fasted, they exhibited a significant reduction in triglyceride levels that was not seen in starved, wildtype flies (149). This drastic reduction in triglyceride levels in a Relish knockdown background was also associated with elevated expression of the lipase gene brummer (bmm). These authors determined that the depletion of lipids in starved Relish-deficient flies was due to FOXO-dependent regulation of bmm.
FOXO activates transcription of the lipase bmm (Figure 1) to positively regulate lipolysis (157, 158). Reducing FOXO activity in Relish mutants restores wildtype levels of fasting-dependent bmm gene expression and triglyceride storage in the fat body (149). In flies that are starved or fully fed, Relish can bind to the Bmm locus (149). Relish has been suggested to modify histone acetylation during starvation conditions demonstrated by enrichment of histone 3 lysine 9 acetylation (H3K9ac) in relish mutants at the site where Relish binds the Bmm locus (149) Together, these data suggest that Relish may antagonize FOXO-dependent regulation of lipolysis to prevent rapid loss of lipids during starvation. Future experiments could elucidate whether Relish-FOXO antagonism mediates the shifts in lipid metabolism observed during infection (12, 23, 131).
FOXO also plays tissue-specific roles during oral infections. During oral infection of D. melanogaster with Serratia marcescens, FOXO becomes activated and localized to the nucleus in gut epithelial cells, where it contributes to infection-induced expression of antimicrobial peptides (155). Even in the absence of infection, overexpression of FOXO in D. melanogaster intestines increases transcript levels of antimicrobial peptide genes Attacin, Drosomycin, Diptericin, and Defensin in gut cells (155). FOXO mutants experience reduced survivorship and higher bacteria load in response to oral infection with S. marcescens compared to wildtype, presumably as a consequence of the impaired AMP expression (155).
Recent evidence in B. mori demonstrates a role for FOXO in controlling infection by the orally-acquired baculovirus Bombyx mori nuclear polyhedrosis virus (BmNPV). B. mori cell lines infected with BmNPV exhibit increased phosphorylation of Akt (159, 160) and reduced FOXO gene expression (17). Phosphoenolpyruvate carboxykinase (PEPCK) is a downstream target of FOXO that is known for its role in regulating gluconeogenesis (161), and more recently for its antiviral role in B. mori (17, 162). B. mori cells overexpressing PEPCK exhibited a reduction in replication of BmNPV and elevated expression of the autophagy-related protein ATG8 (162). PGRP2-2 negatively regulates PTEN, which is itself a negative regulator of IIS (160). Thus PGRP2-2 is a positive regulator of IIS, and consequently a negative regulator of FOXO and PEPCK. PGRP2-2 knockdown and FOXO overexpression reduce replication of BmNPV in a B. mori cell line (160) and increase expression of PEPCK and autophagy-related genes like ATG6, ATG7, and ATG8 (17). Pharmaceutical inhibition of Akt phosphorylation in B. mori cells using a PI3K inhibitor also suppresses viral replication (160). These results suggest that FOXO plays a critical role in this antiviral immune response, and viruses like BmNPV can impact expression of its host IIS to evade the immune response.
Evidence from D. melanogaster also suggests FOXO mediates anti-viral immunity. Constitutive activation of FOXO can reduce the viral load of the Cricket Paralysis virus in D. melanogaster S2 cells (148). FOXO mutants are also more susceptible to RNA virus infections (148). FOXO regulates stress responses like autophagy and apoptosis that can be critical for combatting viral infection, providing a potential mechanism for FOXO-mediated resistance to viruses. The anti-viral effects of FOXO have thus far been predominantly evaluated in cell culture. Future experiments may determine whether FOXO is involved in tissue-specific antiviral responses and how those may impact host survivorship in vivo.
In this review, we have discussed examples of known interactions between innate immune and IIS pathways in the model insect D. melanogaster as well as in insects of economic and public health relevance like B. mori and A. stephensi. Figure 2 summarizes known interactions between Toll and IMD and IIS/TOR and questions to consider for further investigation. Supplementary Table 1 contains a list of published experiments in D. melanogaster, A. stephensi, and B. mori that have evaluated how different pathogens and routes of infection (i.e., oral vs. systemic) impact IIS/TOR activity and immunological phenotypes. Supplementary Table 2 summarizes several published studies in D. melanogaster that tested how manipulating the expression of innate immune genes affects IIS activity and IIS-dependent metabolism. The literature reviewed demonstrates a key role of insulin signaling in responding to oral, parasitic, viral, and systemic infections (12, 17, 19, 39, 115, 155). It also reveals critical differences between tissues, pathogens, life stages, and routes of infection in how insulin-immunity interactions play out. Different adaptive strategies of IIS regulation could be deployed by the host depending on the type of pathogenic infection. IIS and TOR upregulation could promote tolerance of pathogens that stimulate wasting of nutrient stores. For example, Akt and TOR are activated by oral infection with P. entomophila, which promotes lipid synthesis and alleviates infection-induced wasting to increase survivorship (131). But for infections that may not pose a threat of host wasting, suppression of IIS and promotion of FOXO-dependent signaling to regulate lipid metabolism and transcription of immune effectors may be more advantageous. Akt activation is inhibited after oral infection with S. marcescens, which activates FOXO to potentially help upregulate transcription of AMPs that may act as a defense at the gut epithelium (155). Potentially, FOXO-dependent regulation of AMPs in the gut epithelial barrier may enable a localized response that is more efficient than dumping antimicrobials into the gut lumen, where they would be at lower effective concentrations and could cause dysbiosis of the resident microbiota. Additionally, pathogens and parasites may manipulate their host metabolism to gain nutritional resources or may cause host metabolic changes as a consequence of nutrient consumption. Future experiments are necessary to explore to what extent infection-induced shifts to IIS activity serve as an adapted host response to infection versus as a mechanism for pathogens to exploit host physiology for their own gains.
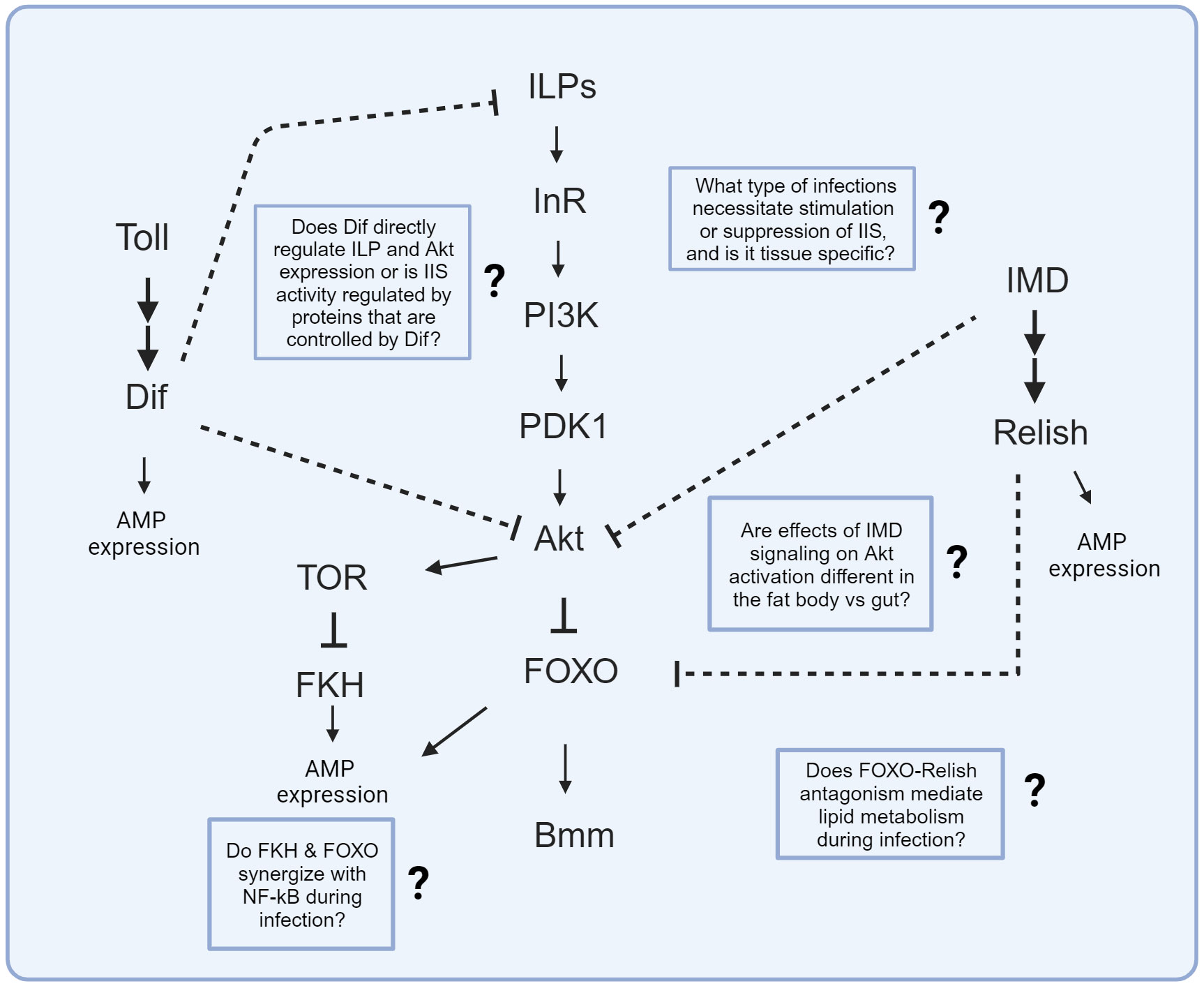
Figure 2 Known signaling interactions between innate immune and IIS/TOR pathways. Arrows depict known activation while bars represent known suppression in molecular interactions. Dashed bars represent known suppression between innate immune and IIS pathways, but the specific molecular interaction is unknown. Text boxes with question marks indicate outstanding questions regarding these signaling interactions that could be the study of future research.
Studies that make use of genetic manipulation have been critical in advancing our understanding of how IIS and innate immune activity interact. While infection studies in D. melanogaster IIS mutants have shown varying effects on infection outcome (86, 102, 107), overexpression experiments driving Toll and IMD signaling in the D. melanogaster fat body demonstrate an antagonistic response of innate immune signaling on IIS activation (16, 19, 39, 82, 86). Future experiments using tissue-specific or conditionally inducible knockdowns and overexpressions (e.g. 163) will further reveal how IIS affects immune function, including disentangling direct antagonisms from pleiotropic developmental effects and distinguishing tissue autonomy from interorgan communication. Additionally, experiments coupling pharmacological intervention and genetic manipulation of IIS could help elucidate the contributions of TOR- and IIS- mediated effects on immunity. IIS, metabolism, and innate immunity are interrelated in a feedback network, and carefully conceived experiments will be required to disentangle the mechanisms that connect them.
Genetic manipulation of insulin signaling in mosquitos has revealed that activation of IIS can increase production of immune effectors, leading to reduced prevalence of Plasmodium infection (117, 118, 122). Overexpression of negative regulators of IIS similarly contributes to reduced Plasmodium infectivity (117). It is thus possible that either upregulation or downregulation of IIS could create unfavorable environments for parasite development, increasing the potential for control of disease transmission via genetic manipulation of the mosquito vector. However, direct feeding of insulin to mosquitoes promotes Plasmodium falciparum infection success, and the parasite can induce expression of ILPs to suppress the mosquito immune response. The discrepancy between direct feeding of insulin versus genetic manipulation of IIS activity in mosquitos remains to be understood and brings an appropriate note of caution in interpreting individual experiments. Despite these nuances in experimental design, studies that demonstrate how feeding insulin or treating mosquitos with pharmaceutical inhibitors alter metabolic and immune function in mosquitos can inform novel strategies to target for vector pest management.
Common themes of interaction between IIS and immunity have emerged from the body of work described throughout this review. Infection may stimulate or suppress IIS activity to initiate nutrient mobilization in support of an immune response or to preserve metabolic stores. Potentially, we could expect an adapted metabolic response to support varying metabolic needs that could arise when a host is challenged with diverse types of pathogens. Understanding how IIS activity and the innate immune system interact within insects is an exciting avenue of research with compelling foundation but clearly much left to learn.
AMD: Conceptualization, Investigation, Project administration, Visualization, Writing – original draft, Writing – review & editing. BPL: Investigation, Resources, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by NIH grant AI141385. AMD is supported by fellowships from the Ford Foundation and the State University of New York (SUNY) system.
We are thankful to members of the Lazzaro lab for advice and comments on the manuscript, including Katie Gordon and Scott Keith. Additional thanks to Tory Hendry, Angela Poole, Sri Lakshmi Sravani Devarakonda, Taylar Mouton, and Dorian Jackson for additional comment and feedback on the manuscript. The main figures were created with BioRender.com.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1276357/full#supplementary-material
1. Danilova N. The evolution of immune mechanisms. J Exp Zool Part B: Mol Dev Evol (2006) 306B:496–520. doi: 10.1002/jez.b.21102
2. Wang X, Zhang Y, Zhang R, Zhang J. The diversity of pattern recognition receptors (PRRs) involved with insect defense against pathogens. Curr Opin Insect Sci (2019) 33:105–10. doi: 10.1016/j.cois.2019.05.004
3. Badisco L, Van Wielendaele P, Vanden Broeck J. Eat to reproduce: a key role for the insulin signaling pathway in adult insects. Front Physiol (2013) 4:202. doi: 10.3389/fphys.2013.00202
4. Nässel DR, Liu Y, Luo J. Insulin/IGF signaling and its regulation in Drosophila. Gen Comp Endocrinol (2015) 221:255–66. doi: 10.1016/j.ygcen.2014.11.021
5. Hotamisligil GS. Inflammation, metaflammation and immunometabolic disorders. Nature (2017) 542:177–85. doi: 10.1038/nature21363
6. Viljakainen L. Evolutionary genetics of insect innate immunity. Brief Funct Genomics (2015) 14:407–12. doi: 10.1093/bfgp/elv002
7. Lagueux M, Lwoff L, Meister M, Goltzene F, Hoffmann JA. cDNAs from neurosecretory cells of brains of Locusta migratoria (Insecta, Orthoptera) encoding a novel member of the superfamily of insulins. Eur J Biochem (1990) 187:249–54. doi: 10.1111/j.1432-1033.1990.tb15302.x
8. Ishizaki H. Molecular characterization of the brain secretory peptides, prothoracicotropic hormone (PTTH) and bombyxin, of the silkmoth Bombyx mori. Proc Jpn Acad Ser B Phys Biol Sci (2004) 80:195–203. doi: 10.2183/pjab.80.195
9. Wu Q, Brown MR. Signaling and function of insulin-like peptides in insects. Annu Rev Entomol (2006) 51:1–24. doi: 10.1146/annurev.ento.51.110104.151011
10. Marquez AG, Pietri JE, Smithers HM, Nuss A, Antonova Y, Drexler AL, et al. Insulin-like peptides in the mosquito Anopheles stephensi: Identification and expression in response to diet and infection with Plasmodium falciparum. Gen Comp Endocrinol (2011) 173:303–12. doi: 10.1016/j.ygcen.2011.06.005
11. Ihle KE, Baker NA, Amdam GV. Insulin-like peptide response to nutritional input in honey bee workers. J Insect Physiol (2014) 69:49–55. doi: 10.1016/j.jinsphys.2014.05.026
12. Dionne MS, Pham LN, Shirasu-Hiza M, Schneider DS. Akt and foxo dysregulation contribute to infection-induced wasting in. Drosophila Curr Biol (2006) 16:1977–85. doi: 10.1016/j.cub.2006.08.052
13. Corby-Harris V, Drexler A, Jong LW, Antonova Y, Pakpour N, Ziegler R, et al. Activation of Akt signaling reduces the prevalence and intensity of malaria parasite infection and lifespan in Anopheles stephensi mosquitoes. PloS Pathog (2010) 6:e1001003. doi: 10.1371/journal.ppat.1001003
14. Sun D, Guo Z, Liu Y, Zhang Y. Progress and prospects of CRISPR/Cas systems in insects and other arthropods. Front Physiol (2017) 8:608. doi: 10.3389/fphys.2017.00608
15. Christiaens O, Niu J, Nji Tizi Taning C. RNAi in insects: A revolution in fundamental research and pest control applications. Insects (2020) 11:415. doi: 10.3390/insects11070415
16. Martínez BA, Hoyle RG, Yeudall S, Granade ME, Harris TE, Castle JD, et al. Innate immune signaling in Drosophila shifts anabolic lipid metabolism from triglyceride storage to phospholipid synthesis to support immune function. PloS Genet (2020) 16:e1009192. doi: 10.1371/journal.pgen.1009192
17. Kang X, Wang Y, Liang W, Tang X, Zhang Y, Wang L, et al. Bombyx mori nucleopolyhedrovirus downregulates transcription factor BmFoxO to elevate virus infection. Dev Comp Immunol (2021) 116:103904. doi: 10.1016/j.dci.2020.103904
18. Sheldon BC, Verhulst S. Ecological immunology: costly parasite defences and trade-offs in evolutionary ecology. Trends Ecol Evol (1996) 11:317–21. doi: 10.1016/0169-5347(96)10039-2
19. DiAngelo JR, Bland ML, Bambina S, Cherry S, Birnbaum MJ. The immune response attenuates growth and nutrient storage in Drosophila by reducing insulin signaling. Proc Natl Acad Sci (2009) 106:20853–8. doi: 10.1073/pnas.0906749106
20. Adamo SA, Bartlett A, Le J, Spencer N, Sullivan K. Illness-induced anorexia may reduce trade-offs between digestion and immune function. Anim Behav (2010) 79:3–10. doi: 10.1016/j.anbehav.2009.10.012
21. Povey S, Cotter SC, Simpson SJ, Wilson K. Dynamics of macronutrient self-medication and illness-induced anorexia in virally infected insects. J Anim Ecol (2014) 83:245–55. doi: 10.1111/1365-2656.12127
22. Schilder RJ, Marden JH. Metabolic syndrome and obesity in an insect. Proc Natl Acad Sci (2006) 103:18805–9. doi: 10.1073/pnas.0603156103
23. Chambers MC, Song KH, Schneider DS. Listeria monocytogenes infection causes metabolic shifts in Drosophila melanogaster. PloS One (2012) 7:e50679. doi: 10.1371/journal.pone.0050679
24. Yang H, Hultmark D. Drosophila muscles regulate the immune response against wasp infection via carbohydrate metabolism. Sci Rep (2017) 7:15713. doi: 10.1038/s41598-017-15940-2
25. Bajgar A, Dolezal T. Extracellular adenosine modulates host-pathogen interactions through regulation of systemic metabolism during immune response in. Drosophila PloS Pathog (2018) 14:e1007022. doi: 10.1371/journal.ppat.1007022
26. Vincent CM, Simoes da Silva CJ, Wadhawan A, Dionne MS. Origins of metabolic pathology in Francisella-infected Drosophila. Front Immunol (2020) 11:1419. doi: 10.3389/fimmu.2020.01419
27. Li S, Yu X, Feng Q. Fat body biology in the last decade. Annu Rev Entomol (2019) 64:315–33. doi: 10.1146/annurev-ento-011118-112007
28. Arrese EL, Soulages JL. Insect fat body: Energy, metabolism, and regulation. Annu Rev Entomol (2010) 55:207–25. doi: 10.1146/annurev-ento-112408-085356
29. Krejčová G, Danielová A, Nedbalová P, Kazek M, Strych L, Chawla G, et al. Drosophila macrophages switch to aerobic glycolysis to mount effective antibacterial defense. eLife (2019) 14(4):e1007022. doi: 10.7554/eLife.50414
30. Xu Y-J, Luo F, Gao Q, Shang Y, Wang C. Metabolomics reveals insect metabolic responses associated with fungal infection. Anal Bioanal Chem (2015) 407:4815–21. doi: 10.1007/s00216-015-8648-8
31. Thompson SN. Trehalose – The insect ‘blood’ sugar. Adv Insect Physiol (2003) 31:205–85. doi: 10.1016/S0065-2806(03)31004-5
32. Praveena M, Savithri G. Analysis of digestive enzymes in different breeds of Bombyx mori in response to Beauveria bassiana infection. Int J Entomol Res (2021) 6:3:129–34.
33. Strahl T, Thorner J. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim Biophys Acta (BBA) Mol Cell Biol Lipids (2007) 1771:353–404. doi: 10.1016/j.bbalip.2007.01.015
34. Troha K, Im JH, Revah J, Lazzaro BP, Buchon N. Comparative transcriptomics reveals CrebA as a novel regulator of infection tolerance in D. melanogaster. PloS Pathog (2018) 14:e1006847. doi: 10.1371/journal.ppat.1006847
35. Rutkowski DT, Kaufman RJ. A trip to the ER: coping with stress. Trends Cell Biol (2004) 14:20–8. doi: 10.1016/j.tcb.2003.11.001
36. Sriburi R, Jackowski S, Mori K, Brewer JW. XBP1 : a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol (2004) 167:35–41. doi: 10.1083/jcb.200406136
37. Grootjans J, Kaser A, Kaufman RJ, Blumberg RS. The unfolded protein response in immunity and inflammation. Nat Rev Immunol (2016) 16:469–84. doi: 10.1038/nri.2016.62
38. Gupta V, Frank AM, Matolka N, Lazzaro BP. Inherent constraints on a polyfunctional tissue lead to a reproduction-immunity tradeoff. BMC Biol (2022) 20:127. doi: 10.1186/s12915-022-01328-w
39. Roth SW, Bitterman MD, Birnbaum MJ, Bland ML. Innate immune signaling in Drosophila blocks insulin signaling by uncoupling PI(3,4,5)P 3 production and Akt activation. Cell Rep (2018) 22:2550–6. doi: 10.1016/j.celrep.2018.02.033
40. Kwak S-J, Hong S-H, Bajracharya R, Yang S-Y, Lee K-S, Yu K. Drosophila adiponectin receptor in insulin producing cells regulates glucose and lipid metabolism by controlling insulin secretion. PloS One (2013) 8:e68641. doi: 10.1371/journal.pone.0068641
41. Ugrankar R, Theodoropoulos P, Akdemir F, Henne WM, Graff JM. Circulating glucose levels inversely correlate with Drosophila larval feeding through insulin signaling and SLC5A11. Commun Biol (2018) 1:1–15. doi: 10.1038/s42003-018-0109-4
42. Huang X, Liu G, Guo J, Su Z. The PI3K/AKT pathway in obesity and type 2 diabetes. Int J Biol Sci (2018) 14:1483–96. doi: 10.7150/ijbs.27173
43. DiAngelo JR, Birnbaum MJ. Regulation of fat cell mass by insulin in Drosophila melanogaster. Mol Cell Biol (2009) 29:6341–52. doi: 10.1128/MCB.00675-09
44. Cross DAE, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature (1995) 378:785–9. doi: 10.1038/378785a0
45. Xu C, Kim N-G, Gumbiner BM. Regulation of protein stability by GSK3 mediated phosphorylation. Cell Cycle (2009) 8:4032–9. doi: 10.4161/cc.8.24.10111
46. Armstrong AR, Drummond-Barbosa D. Insulin signaling acts in adult adipocytes via GSK-3β and independently of FOXO to control Drosophila female germline stem cell numbers. Dev Biol (2018) 440:31–9. doi: 10.1016/j.ydbio.2018.04.028
47. Wipperman MF, Sampson NS, Thomas ST. Pathogen roid rage: Cholesterol utilization by Mycobacterium tuberculosis. Crit Rev Biochem Mol Biol (2014) 49:269–93. doi: 10.3109/10409238.2014.895700
48. Ghith A, Bruning JB, Bell SG. The oxidation of cholesterol derivatives by the CYP124 and CYP142 enzymes from Mycobacterium marinum. J Steroid Biochem Mol Biol (2023) 231:106317. doi: 10.1016/j.jsbmb.2023.106317
49. Singh G, Singh G, Jadeja D, Kaur J. Lipid hydrolizing enzymes in virulence: Mycobacterium tuberculosis as a model system. Crit Rev Microbiol (2010) 36:259–69. doi: 10.3109/1040841X.2010.482923
50. Ding Y, Raterink R-J, Marín-Juez R, Veneman WJ, Egbers K, van den Eeden S, et al. Tuberculosis causes highly conserved metabolic changes in human patients, mycobacteria-infected mice and zebrafish larvae. Sci Rep (2020) 10:11635. doi: 10.1038/s41598-020-68443-y
51. Musselman LP, Fink JL, Narzinski K, Ramachandran PV, Hathiramani SS, Cagan RL, et al. A high-sugar diet produces obesity and insulin resistance in wild-type Drosophila. DMM Dis Models Mech (2011) 4(6):842–9. doi: 10.1242/dmm.007948
52. Musselman LP, Fink JL, Grant AR, Gatto JA, Tuthill BF, Baranski TJ. A complex relationship between immunity and metabolism in Drosophila diet-induced insulin resistance. Mol Cell Biol (2017) 38(2). doi: 10.1128/mcb.00259-17
53. Yu S, Zhang G, Jin LH. A high-sugar diet affects cellular and humoral immune responses in Drosophila. Exp Cell Res (2018) 368:215–24. doi: 10.1016/j.yexcr.2018.04.032
54. Ponton F, Morimoto J, Robinson K, Kumar SS, Cotter SC, Wilson K, et al. Macronutrients modulate survival to infection and immunity in Drosophila. J Anim Ecol (2020) 89:460–70. doi: 10.1111/1365-2656.13126
55. Regan JC, Froy H, Froy H, Walling CA, Moatt JP, Nussey DH. Dietary restriction and insulin-like signalling pathways as adaptive plasticity: A synthesis and re-evaluation. Funct Ecol (2020) 34:107–28. doi: 10.1111/1365-2435.13418
56. Unckless RL, Rottschaefer SM, Lazzaro BP. The complex contributions of genetics and nutrition to immunity in Drosophila melanogaster. PloS Genet (2015) 11:1005030. doi: 10.1371/journal.pgen.1005030
57. Fellous S, Lazzaro BP. Larval food quality affects adult (but not larval) immune gene expression independent of effects on general condition. Mol Ecol (2010) 19:1462–8. doi: 10.1111/j.1365-294X.2010.04567.x
58. Bakopoulos D, Beadle LF, Esposito KM, Mirth CK, Warr CG, Johnson TK. Insulin-like signalling influences the coordination of larval hemocyte number with body Size in Drosophila melanogaster. G3 Genes|Genomes|Genetics (2020) 10:2213–20. doi: 10.1534/g3.120.401313
59. Savola E, Montgomery C, Waldron FM, Monteith KM, Vale P, Walling C. Testing evolutionary explanations for the lifespan benefit of dietary restriction in fruit flies (Drosophila melanogaster). Evolution (2021) 75:450–63. doi: 10.1111/evo.14146
60. Linenberg I, Christophides GK, Gendrin M. Larval diet affects mosquito development and permissiveness to Plasmodium infection. Sci Rep (2016) 6:38230. doi: 10.1038/srep38230
61. Muturi EJ, Kim C-H, Alto BW, Berenbaum MR, Schuler MA. Larval environmental stress alters Aedes aEgypti competence for Sindbis virus. Trop Med Int Health (2011) 16:955–64. doi: 10.1111/j.1365-3156.2011.02796.x
62. Duneau DF, Lazzaro BP. Persistence of an extracellular systemic infection across metamorphosis in a holometabolous insect. Biol Lett (2018) 14:20170771. doi: 10.1098/rsbl.2017.0771
63. Futerman PH, Layen SJ, Kotzen ML, Franzen C, Kraaijeveld AR, Godfray HCJ. Fitness effects and transmission routes of a microsporidian parasite infecting Drosophila and its parasitoids. Parasitology (2006) 132:479–92. doi: 10.1017/S0031182005009339
64. Galenza A, Hutchinson J, Campbell SD, Hazes B, Foley E. Glucose modulates Drosophila longevity and immunity independent of the microbiota. Biol Open (2016) 5:165–73. doi: 10.1242/bio.015016
65. Almire F, Terhzaz S, Terry S, McFarlane M, Gestuveo RJ, Szemiel AM, et al. Sugar feeding protects against arboviral infection by enhancing gut immunity in the mosquito vector Aedes aEgypti. PloS Pathog (2021) 17:e1009870. doi: 10.1371/journal.ppat.1009870
66. Zhang X, Jin Q, Jin LH. High sugar diet disrupts gut homeostasis though JNK and STAT pathways in Drosophila. Biochem Biophys Res Commun (2017) 487:910–6. doi: 10.1016/j.bbrc.2017.04.156
67. Bonfini A, Dobson AJ, Duneau D, Revah J, Liu X, Houtz P, et al. Multiscale analysis reveals that diet-dependent midgut plasticity emerges from alterations in both stem cell niche coupling and enterocyte size. eLife (2021) 10:e64125. doi: 10.7554/eLife.64125
68. Vandehoef C, Molaei M, Karpac J. Dietary adaptation of microbiota in Drosophila requires NF-κB-dependent control of the translational regulator 4E-BP. Cell Rep (2020) 31:107736. doi: 10.1016/j.celrep.2020.107736
69. Lüersen K, Röder T, Rimbach G. Drosophila melanogaster in nutrition research - The importance of standardizing experimental diets. Genes Nutr (2019) 14(3). doi: 10.1186/s12263-019-0627-9
70. Lesperance DNA, Broderick NA. Meta-analysis of diets used in drosophila microbiome research and introduction of the drosophila dietary composition calculator (DDCC). G3 Genes|Genomes|Genetics (2020) 10:2207–11. doi: 10.1534/g3.120.401235
71. Piper MD, Blanc E, Leitão-Gonçalves R, Yang M, He X, Linford NJ, et al. A holidic medium for Drosophila melanogaster. Nat Methods (2014) 11:100–5. doi: 10.1038/nmeth.2731
72. Piper MD. Using artificial diets to understand the nutritional physiology of Drosophila melanogaster. Curr Opin Insect Sci (2017) 23:104–11. doi: 10.1016/j.cois.2017.07.014
73. De Gregorio E. The Toll and Imd pathways are the major regulators of the immune response in. Drosophila EMBO J (2002) 21:2568–79. doi: 10.1093/emboj/21.11.2568
74. Li G, Qian H, Luo X, Xu P, Yang J, Liu M, et al. Transcriptomic analysis of resistant and susceptible Bombyx mori strains following BmNPV infection provides insights into the antiviral mechanisms. Int J Genomics (2016) 2016:e2086346. doi: 10.1155/2016/2086346
75. Caragata EP, Pais FS, Baton LA, Silva JBL, Sorgine MHF, Moreira LA. The transcriptome of the mosquito Aedes fluviatilis (Diptera: Culicidae), and transcriptional changes associated with its native Wolbachia infection. BMC Genomics (2017) 18:6. doi: 10.1186/s12864-016-3441-4
76. Grewal SS. Insulin/TOR signaling in growth and homeostasis: A view from the fly world. Int J Biochem Cell Biol (2009) 41:1006–10. doi: 10.1016/j.biocel.2008.10.010
77. Chatterjee N, Perrimon N. What fuels the fly: Energy metabolism in Drosophila and its application to the study of obesity and diabetes. Sci Adv (2021) 7:eabg4336. doi: 10.1126/sciadv.abg4336
78. Imler J-L, Zheng L. Biology of Toll receptors: lessons from insects and mammals. J Leukocyte Biol (2004) 75:18–26. doi: 10.1189/jlb.0403160
79. Medzhitov R. Toll-like receptors and innate immunity | Nature Reviews Immunology. Nat Rev Immunol (2001) 1(2):135–45. doi: 10.1038/35100529
80. Khush RS, Leulier F, Lemaitre B. Drosophila immunity: two paths to NF-κB. Trends Immunol (2001) 22:260–4. doi: 10.1016/S1471-4906(01)01887-7
81. Buchon N, Silverman N, Cherry S. Immunity in Drosophila melanogaster — from microbial recognition to whole-organism physiology. Nat Rev Immunol (2014) 14:796–810. doi: 10.1038/nri3763
82. Suzawa M, Muhammad NM, Joseph BS, Bland ML. The Toll signaling pathway targets the insulin-like peptide Dilp6 to inhibit growth in Drosophila. Cell Rep (2019) 28:1439–1446.e5. doi: 10.1016/j.celrep.2019.07.015
83. Okamoto N, Yamanaka N, Yagi Y, Nishida Y, Kataoka H, O’Connor MB, et al. A fat body-derived IGF-like peptide regulates postfeeding growth in Drosophila. Dev Cell (2009) 17:885–91. doi: 10.1016/j.devcel.2009.10.008
84. Slaidina M, Delanoue R, Gronke S, Partridge L, Léopold P. A Drosophila insulin-like peptide promotes growth during nonfeeding states. Dev Cell (2009) 17:874–84. doi: 10.1016/j.devcel.2009.10.009
85. Keehnen NLP, Rolff J, Theopold U, Wheat CW. Chapter one - insect antimicrobial defences: A brief history, recent findings, biases, and a way forward in evolutionary studies. In: Ligoxygakis P, editor. Advances in Insect Physiology Insect Immunity. Cambridge, Massachusetts: Academic Press (2017). p. 1–33. doi: 10.1016/bs.aiip.2017.02.003
86. Davoodi S, Galenza A, Panteluk A, Deshpande R, Ferguson M, Grewal S, et al. The immune deficiency pathway regulates metabolic homeostasis in Drosophila. J Immunol (2019) 202:2747–59. doi: 10.4049/jimmunol.1801632
87. Miron M, Lasko P, Sonenberg N. Signaling from Akt to FRAP/TOR targets both 4E-BP and S6K in Drosophila melanogaster. Mol Cell Biol (2003) 23:9117–26. doi: 10.1128/MCB.23.24.9117-9126.2003
88. Böhni R, Riesgo-Escovar J, Oldham S, Brogiolo W, Stocker H, Andruss BF, et al. Autonomous control of cell and organ Size by CHICO, a Drosophila homolog of vertebrate IRS1–4. Cell (1999) 97:865–75. doi: 10.1016/S0092-8674(00)80799-0
89. Paquette N, Broemer M, Aggarwal K, Chen L, Husson M, Ertürk-Hasdemir D, et al. Caspase mediated cleavage, IAP binding and ubiquitination: Linking three mechanisms crucial for Drosophila NF-κB signaling. Mol Cell (2010) 37:172. doi: 10.1016/j.molcel.2009.12.036
90. Buchon N, Broderick NA, Poidevin M, Pradervand S, Lemaitre B. Drosophila intestinal response to bacterial infection: activation of host defense and stem cell proliferation. Cell Host Microbe (2009) 5:200–11. doi: 10.1016/j.chom.2009.01.003
91. Kamareddine L, Robins WP, Berkey CD, Mekalanos JJ, Watnick PI. The Drosophila immune deficiency pathway modulates enteroendocrine function and host metabolism. Cell Metab (2018) 28(3):449–62.e5. doi: 10.1016/j.cmet.2018.05.026
92. Miguel-Aliaga I, Jasper H, Lemaitre B. Anatomy and physiology of the digestive tract of drosophila melanogaster. Genetics (2018) 210:357–96. doi: 10.1534/genetics.118.300224
93. Amcheslavsky A, Song W, Li Q, Nie Y, Bragatto I, Ferrandon D, et al. Enteroendocrine cells support intestinal stem cell-mediated homeostasis in. Drosophila Cell Rep (2014) 9:32–9. doi: 10.1016/j.celrep.2014.08.052
94. Shin SC, Kim SH, You H, Kim B, Kim AC, Lee KA, et al. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science (2011) 334:670–4. doi: 10.1126/science.1212782
95. Hang S, Purdy AE, Robins WP, Wang Z, Mandal M, Chang S, et al. The acetate switch of an intestinal pathogen disrupts host insulin signaling and lipid metabolism. Cell Host Microbe (2014) 16:592–604. doi: 10.1016/j.chom.2014.10.006
96. Broderick NA, Buchon N, Lemaitre B. Microbiota-induced changes in drosophila melanogaster host gene expression and gut morphology. mBio (2014) 5(3). doi: 10.1128/mBio.01117-14
97. Combe BE, Defaye A, Bozonnet N, Puthier D, Royet J, Leulier F. Drosophila microbiota modulates host metabolic gene expression via IMD/NF-κB signaling. PloS One (2014) 9(7):e104120. doi: 10.1371/journal.pone.0094729
98. Thomson TC, Schneemann A, Johnson J. Oocyte destruction is activated during viral infection. genesis (2012) 50:453–65. doi: 10.1002/dvg.22004
99. Harsh S, Heryanto C, Eleftherianos I. Intestinal lipid droplets as novel mediators of host–pathogen interaction in Drosophila. Biol Open (2019) 8:bio039040. doi: 10.1242/bio.039040
100. Zhao X, Karpac J. Glutamate metabolism directs energetic trade-offs to shape host-pathogen susceptibility in Drosophila. Cell Metab (2021) 33:2428–2444.e8. doi: 10.1016/j.cmet.2021.10.003
101. Yu S, Luo F, Xu Y, Zhang Y, Jin LH. Drosophila innate immunity involves multiple signaling pathways and coordinated communication between different tissues. Front Immunol (2022) 13:2022.905370. doi: 10.3389/fimmu.2022.905370
102. Libert S, Chao Y, Zwiener J, Pletcher SD. Realized immune response is enhanced in long-lived puc and chico mutants but is unaffected by dietary restriction. Mol Immunol (2008) 5(3):810–7. doi: 10.1016/j.molimm.2007.06.353
103. Bai H, Post S, Kang P, Tatar M. Drosophila longevity assurance conferred by reduced insulin receptor substrate Chico partially requires d4eBP. PloS One (2015) 10:e0134415. doi: 10.1371/journal.pone.0134415
104. Vasudevan D, Clark NK, Sam J, Cotham VC, Ueberheide B, Marr MT, et al. The GCN2-ATF4 signaling pathway activates 4E-BP to bias mRNA translation and boost antimicrobial peptide synthesis in response to bacterial infection. Cell Rep (2017) 21:2039–47. doi: 10.1016/j.celrep.2017.10.096
105. Bernal A, Kimbrell DA. Drosophila Thor participates in host immune defense and connects a translational regulator with innate immunity. Proc Natl Acad Sci U.S.A. (2000) 97:6019–24. doi: 10.1073/pnas.100391597
106. Levitin A, Marcil A, Tettweiler G, Laforest MJ, Oberholzer U, Alarco AM, et al. Drosophila melanogaster thor and response to Candida albicans infection. Eukaryotic Cell (2007) 6:658–63. doi: 10.1128/EC.00346-06
107. McCormack S, Yadav S, Shokal U, Kenney E, Cooper D, Eleftherianos I. The insulin receptor substrate Chico regulates antibacterial immune function in Drosophila. Immun Ageing (2016) 13(15). doi: 10.1186/s12979-016-0072-1
108. Tang H. Regulation and function of the melanization reaction in Drosophila. Fly (2009) 3:105–11. doi: 10.4161/fly.3.1.7747
109. Binggeli O, Neyen C, Poidevin M, Lemaitre B. Prophenoloxidase activation is required for survival to microbial infections in. Drosophila PloS Pathog (2014) 10:e1004067. doi: 10.1371/journal.ppat.1004067
110. Shokal U, Eleftherianos I. Thioester-containing protein-4 regulates the Drosophila immune signaling and function against the pathogen Photorhabdus. J Innate Immun (2016) 9:83–93. doi: 10.1159/000450610
111. Shokal U, Kopydlowski H, Eleftherianos I. The distinct function of Tep2 and Tep6 in the immune defense of Drosophila melanogaster against the pathogen Photorhabdus. Virulence (2017) 8:1668–82. doi: 10.1080/21505594.2017.1330240
112. Ayres JS, Schneider DS. A signaling protease required for melanization in Drosophila affects resistance and tolerance of infections. PloS Biol (2008) 6:e305. doi: 10.1371/journal.pbio.0060305
113. Pakpour N, Corby-Harris V, Green GP, Smithers HM, Cheung KW, Riehle MA, et al. Ingested human insulin inhibits the mosquito NF-κB-dependent immune response to Plasmodium falciparum. Infect Immun (2012) 80:2141–9. doi: 10.1128/IAI.00024-12
114. Pietri JE, Pietri EJ, Potts R, Riehle MA, Luckhart S. Plasmodium falciparum suppresses the host immune response by inducing the synthesis of insulin-like peptides (ILPs) in the mosquito Anopheles stephensi. Dev Comp Immunol (2015) 53:134–44. doi: 10.1016/j.dci.2015.06.012
115. Pietri JE, Pakpour N, Napoli E, Song G, Pietri E, Potts R, et al. Two insulin-like peptides differentially regulate malaria parasite infection in the mosquito through effects on intermediary metabolism. Biochem J (2016) 473:3487–503. doi: 10.1042/BCJ20160271
116. Surachetpong W, Pakpour N, Cheung KW, Luckhart S. Reactive oxygen species-dependent cell signaling regulates the mosquito immune response to Plasmodium falciparum. Antioxid Redox Signaling (2011) 14:943–55. doi: 10.1089/ars.2010.3401
117. Hauck ES, Antonova-Koch Y, Drexler A, Pietri J, Pakpour N, Liu D, et al. Overexpression of phosphatase and tensin homolog improves fitness and decreases Plasmodium falciparum development in Anopheles stephensi. Microbes Infect (2013) 15:775–87. doi: 10.1016/j.micinf.2013.05.006
118. Hun LV, Cheung KW, Brooks E, Zudekoff R, Luckhart S, Riehle MA. Increased insulin signaling in the Anopheles stephensi fat body regulates metabolism and enhances the host response to both bacterial challenge and Plasmodium falciparum infection. Insect Biochem Mol Biol (2021) 139:103669. doi: 10.1016/j.ibmb.2021.103669
119. Kumar S, Christophides GK, Cantera R, Charles B, Han YS, Meister S, et al. The role of reactive oxygen species on Plasmodium melanotic encapsulation in Anopheles Gambiae. Proc Natl Acad Sci (2003) 100:14139–44. doi: 10.1073/pnas.2036262100
120. Peterson TML, Luckhart S. A mosquito 2-Cys peroxiredoxin protects against nitrosative and oxidative stresses associated with malaria parasite infection. Free Radic Biol Med (2006) 40:1067–82. doi: 10.1016/j.freeradbiomed.2005.10.059
121. Molina-Cruz A, DeJong RJ, Charles B, Gupta L, Kumar S, Jaramillo-Gutierrez G, et al. Reactive oxygen species modulate Anopheles Gambiae immunity against bacteria and. Plasmodium. J Biol Chem (2008) 283:3217–23. doi: 10.1074/jbc.M705873200
122. Luckhart S, Giulivi C, Drexler AL, Antonova-Koch Y, Sakaguchi D, Napoli E, et al. Sustained activation of Akt elicits mitochondrial dysfunction to block Plasmodium falciparum infection in the mosquito host. PloS Pathog (2013) 9:e1003180. doi: 10.1371/journal.ppat.1003180
123. Aljada A, Ghanim H, Saadeh R, Dandona P. Insulin inhibits NFκB and MCP-1 expression in human aortic endothelial cells. J Clin Endocrinol Metab (2001) 86:450–3. doi: 10.1210/jcem.86.1.7278
124. Dandona P, Aljada A, Mohanty P, Ghanim H, Hamouda W, Assian E, et al. Insulin inhibits intranuclear nuclear factor κB and stimulates IκB in mononuclear cells in obese subjects: evidence for an anti-inflammatory effect? J Clin Endocrinol Metab (2001) 86:3257–65. doi: 10.1210/jcem.86.7.7623
125. Ghanim H, Mohanty P, Deopurkar R, Sia CL, Korzeniewski K, Abuaysheh S, et al. Acute modulation of toll-like receptors by insulin. Diabetes Care (2008) 31:1827–31. doi: 10.2337/dc08-0561
126. Pal S, Nath P, Das D, Hajra S, Maitra S. Cross-talk between insulin signalling and LPS responses in mouse macrophages. Mol Cell Endocrinol (2018) 476:57–69. doi: 10.1016/j.mce.2018.04.009
127. Oldham S, Montagne J, Radimerski T, Thomas G, Hafen E. Genetic and biochemical characterization of dTOR, the Drosophila homolog of the target of rapamycin. Genes Dev (2000) 14:2689–94. doi: 10.1101/gad.845700
128. Parisi F, Riccardo S, Daniel M, Saqcena M, Kundu N, Pession A, et al. Drosophila insulin and target of rapamycin (TOR) pathways regulate GSK3 beta activity to control Myc stability and determine Myc expression in vivo. BMC Biol (2011) 9:65. doi: 10.1186/1741-7007-9-65
129. Varma D, Bülow MH, Pesch Y-Y, Loch G, Hoch M. Forkhead, a new cross regulator of metabolism and innate immunity downstream of TOR in Drosophila. J Insect Physiol (2014) 69:80–8. doi: 10.1016/j.jinsphys.2014.04.006
130. Feng Y, Chen L, Gao L, Dong L, Wen H, Song X, et al. Rapamycin inhibits pathogen transmission in mosquitoes by promoting immune activation. PloS Pathog (2021) 17:e1009353. doi: 10.1371/journal.ppat.1009353
131. Deshpande R, Lee B, Qiao Y, Grewal SS. TOR signalling is required for host lipid metabolic remodelling and survival following enteric infection in. Drosophila Dis Models Mech (2022) 15:dmm049551. doi: 10.1242/dmm.049551
132. Lee J-E, Rayyan M, Liao A, Edery I, Pletcher SD. Acute dietary restriction acts via TOR, PP2A, and Myc signaling to boost innate immunity in Drosophila. Cell Rep (2017) 20:479–90. doi: 10.1016/j.celrep.2017.06.052
133. Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan K-L. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol (2008) 10:935–45. doi: 10.1038/ncb1753
134. Yamagata K, Sanders LK, Kaufmann WE, Yee W, Barnes CA, Nathans D, et al. rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel Ras-related protein. J Biol Chem (1994) 269:16333–9. doi: 10.1016/S0021-9258(17)34012-7
135. Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol (2003) 5:566–71. doi: 10.1038/ncb996
136. Allen VW, O’Connor RM, Ulgherait M, Zhou CG, Stone EF, Hill VM, et al. period-regulated feeding behavior and TOR signaling modulate survival of infection. Curr Biol: CB (2016) 26:184–94. doi: 10.1016/j.cub.2015.11.051
137. Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell (2002) 10:457–68. doi: 10.1016/s1097-2765(02)00636-6
138. Cantley LC. The phosphoinositide 3-kinase pathway. Science (2002) 296:1655–7. doi: 10.1126/science.296.5573.1655
139. Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell (2004) 117:421–6. doi: 10.1016/S0092-8674(04)00452-0
140. Gross DN, van den Heuvel APJ, Birnbaum MJ. The role of FoxO in the regulation of metabolism. Oncogene (2008) 27:2320–36. doi: 10.1038/onc.2008.25
141. Teleman AA, Hietakangas V, Sayadian AC, Cohen SM. Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab (2008) 7:21–32. doi: 10.1016/j.cmet.2007.11.010
142. Eijkelenboom A, Burgering BMT. FOXOs: signalling integrators for homeostasis maintenance. Nat Rev Mol Cell Biol (2013) 14:83–97. doi: 10.1038/nrm3507
143. Wang B, Moya N, Niessen S, Hoover H, Mihaylova MM, Shaw RJ, et al. A hormone-dependent module regulating energy balance. Cell (2011) 145:596–606. doi: 10.1016/j.cell.2011.04.013
144. Webb AE, Brunet A. FOXO transcription factors: key regulators of cellular quality control. Trends Biochem Sci (2014) 39:159–69. doi: 10.1016/j.tibs.2014.02.003
145. Tsuchiya K, Ogawa Y. Forkhead box class O family member proteins: The biology and pathophysiological roles in diabetes. J Diabetes Investig (2017) 8:726–34. doi: 10.1111/jdi.12651
146. Gui T, Burgering BMT. FOXOs: masters of the equilibrium. FEBS J (2021) 289(24):7918 –39. doi: 10.1111/febs.16221
147. Kramer JM, Davidge JT, Lockyer JM, Staveley BE. Expressionof Drosophila FOXO regulates growth and can phenocopy starvation. BMC Dev Biol (2003) 3:5. doi: 10.1186/1471-213X-3-5
148. Spellberg MJ, Marr MT. FOXO regulates RNA interference in Drosophila and protects from RNA virus infection. Proc Natl Acad Sci (2015) 112:14587–92. doi: 10.1073/pnas.1517124112
149. Molaei M, Vandehoef C, Karpac J. NF-κB shapes metabolic adaptation by attenuating Foxo-mediated lipolysis in. Drosophila Dev Cell (2019) 49:802–810.e6. doi: 10.1016/j.devcel.2019.04.009
150. Barretto EC, Polan DM, Beevor-Potts AN, Lee B, Grewal SS. Tolerance to hypoxia is promoted by FOXO regulation of the innate immunity transcription factor NF-κB/Relish in Drosophila. Genetics (2020) 215:1013–25. doi: 10.1534/genetics.120.303219
151. Li Y-N, Ren X-B, Liu Z-C, Ye B, Zhao Z-J, Fan Q, et al. Insulin-like peptide and FoxO mediate the trehalose catabolism enhancement during the diapause termination period in the Chinese Oak Silkworm (Antheraea pernyi). Insects (2021) 12:784. doi: 10.3390/insects12090784
152. Demontis F, Perrimon N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell (2010) 143:813–25. doi: 10.1016/j.cell.2010.10.007
153. Becker T, Loch G, Beyer M, Zinke I, Aschenbrenner AC, Carrera P, et al. FOXO-dependent regulation of innate immune homeostasis. Nature (2010) 463:369–73. doi: 10.1038/nature08698
154. Zhang J, Yang W, Xu J, Yang W, Li Q, Zhong Y, et al. Regulation of antimicrobial peptide genes via insulin-like signaling pathway in the silkworm Bombyx mori. Insect Biochem Mol Biol (2018) 103:12–21. doi: 10.1016/j.ibmb.2018.10.002
155. Fink C, Hoffmann J, Knop M, Li Y, Isermann K, Roeder T. Intestinal FoxO signaling is required to survive oral infection in. Drosophila Mucosal Immunol (2016) 9:927–36. doi: 10.1038/mi.2015.112
156. Zhong X, Chowdhury M, Li C-F, Yu X-Q. Transcription factor forkhead regulates expression of antimicrobial peptides in the tobacco hornworm, Manduca sexta. Sci Rep (2017) 7(2688). doi: 10.1038/s41598-017-02830-w
157. Kang P, Chang K, Liu Y, Bouska M, Birnbaum A, Karashchuk G, et al. Drosophila Kruppel homolog 1 represses lipolysis through interaction with dFOXO. Sci Rep (2017) 7:16369. doi: 10.1038/s41598-017-16638-1
158. Yoshinari Y, Kosakamoto H, Kamiyama T, Hoshino R, Matsuoka R, Kondo S, et al. The sugar-responsive enteroendocrine neuropeptide F regulates lipid metabolism through glucagon-like and insulin-like hormones in Drosophila melanogaster. Nat Commun (2021) 12:4818. doi: 10.1038/s41467-021-25146-w
159. Jin S, Cheng T, Guo Y, Lin P, Zhao P, Liu C, et al. Bombyx mori epidermal growth factor receptor is required for nucleopolyhedrovirus replication. Insect Mol Biol (2018) 27:464–77. doi: 10.1111/imb.12386
160. Jiang L, Liu W, Guo H, Dang Y, Cheng T, Yang W, et al. Distinct functions of Bombyx mori peptidoglycan recognition protein 2 in immune responses to bacteria and viruses. Front Immunol (2019) 11:776. doi: 10.3389/fimmu.2019.00776
161. Méndez-Lucas A, Duarte JAG, Sunny NE, Satapati S, He T, Fu X, et al. PEPCK-M expression in mouse liver potentiates, not replaces, PEPCK-C mediated gluconeogenesis. J Hepatol (2013) 59:105–13. doi: 10.1016/j.jhep.2013.02.020
162. Guo H, Xu G, Wang B, Xia F, Sun Q, Wang Y, et al. Phosphoenolpyruvate carboxykinase is involved in antiviral immunity against Bombyx mori nucleopolyhedrovirus. Dev Comp Immunol (2019) 92:193–8. doi: 10.1016/j.dci.2018.11.015
Keywords: innate immunity, insulin signaling, metabolism, immunometabolism, humoral immunity, Drosophila, mosquito, Bombyx
Citation: Darby AM and Lazzaro BP (2023) Interactions between innate immunity and insulin signaling affect resistance to infection in insects. Front. Immunol. 14:1276357. doi: 10.3389/fimmu.2023.1276357
Received: 11 August 2023; Accepted: 03 October 2023;
Published: 17 October 2023.
Edited by:
Susanna Valanne, Tampere University, FinlandReviewed by:
Adam Bajgar, University of South Bohemia in České Budějovice, CzechiaCopyright © 2023 Darby and Lazzaro. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrea M. Darby, YW1kNDM5QGNvcm5lbGwuZWR1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.