
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 12 October 2023
Sec. Nutritional Immunology
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1274401
This article is part of the Research TopicEffects of Medicinal Homologous Foods on Immunity through Intestinal FloraView all 8 articles
 Yanqun Cao1†
Yanqun Cao1† Khalid S. Ibrahim2,3†
Khalid S. Ibrahim2,3† Xing Li1†
Xing Li1† Aileen Wong2
Aileen Wong2 Yi Wu4
Yi Wu4 Xu-Dong Yu1
Xu-Dong Yu1 Xinzhi Zhou2
Xinzhi Zhou2 Zhoujin Tan4
Zhoujin Tan4 Zhiming He1
Zhiming He1 John A. Craft2*
John A. Craft2* Xinhua Shu1,2,5*
Xinhua Shu1,2,5*Background: Traditional Chinese Medicines have been used for thousands of years but without any sound empirical basis. One such preparation is the Qijudihuang pill (QP), a mixture of eight herbs, that has been used in China for the treatment of various conditions including age-related macular degeneration (AMD), the most common cause of blindness in the aged population. In order to explain the mechanism behind the effect of QP, we used an AMD model of high-fat diet (HFD) fed mice to investigate cholesterol homeostasis, oxidative stress, inflammation and gut microbiota.
Methods: Mice were randomly divided into three groups, one group was fed with control diet (CD), the other two groups were fed with high-fat-diet (HFD). One HFD group was treated with QP, both CD and the other HFD groups were treated with vehicles. Tissue samples were collected after the treatment. Cholesterol levels in retina, retinal pigment epithelium (RPE), liver and serum were determined using a commercial kit. The expression of enzymes involved in cholesterol metabolism, inflammation and oxidative stress was measured with qRT-PCR. Gut microbiota was analyzed using 16S rRNA sequencing.
Results: In the majority of the lipid determinations, analytes were elevated by HFD but this was reversed by QP. Cholesterol metabolism including the enzymes of bile acid (BA) formation was suppressed by HFD but again this was reversed by QP. BAs play a major role in signaling between host and microbiome and this is disrupted by HFD resulting in major changes in the composition of colonic bacterial communities. Associated with these changes are predictions of the metabolic pathway complexity and abundance of individual pathways. These concerned substrate breakdowns, energy production and the biosynthesis of pro-inflammatory factors but were changed back to control characteristics by QP.
Conclusion: We propose that the ability of QP to reverse these HFD-induced effects is related to mechanisms acting to lower cholesterol level, oxidative stress and inflammation, and to modulate gut microbiota.
Age-related macular degeneration (AMD) is an incurable visual disorder, which is the third highest cause of blindness in developed countries, with only cataracts and glaucoma as more prevalent eye diseases. Approximately 200 million individuals aged over 50 years are estimated to be affected (1). AMD is chronic in nature and worsens as it progresses from the early and intermediate stages to the late stage. Late AMD is subdivided into dry and wet forms. The dry form is most common and is characterized by the presence of abnormal deposits (drusen) underneath the retinal pigment epithelial (RPE) layer and RPE atrophy. Wet AMD is usually the cause of severe symptoms, such as vision loss, and can cause symptoms to develop in a much shorter timeframe, normally weeks to months. Wet AMD is defined by chronic neovascularization, when new blood vessels, induced by vascular endothelial growth factor (VEGF), grow under the RPE layer and break into the Bruch’s membrane, resulting in bleeding and sudden loss of vision (2). There is no effective treatment for dry AMD, though antioxidants have shown subtle benefits (3).In contrast, anti-VEGF therapy is effective for wet AMD; however, many patients experience incomplete responses, including persistent exudation, hemorrhage, and ongoing lesion fibrosis (4). AMD is a complex disease, associated with environmental and genetic risk factors, and multiple signaling pathways are involved (5).
Cholesterol plays a critical role in the maintenance of cellular structure and function and is associated with various disorders, including AMD (6). Cholesterol has been reported to enrich drusen and form sub-RPE crystals in patients with wet AMD, implicating dysregulation of cholesterol trafficking and metabolism (7, 8). Many studies have demonstrated that cholesterol metabolism and transport genes, apolipoprotein E (APOE), hepatic lipase (LIPC), CETP (cholesteryl ester transfer protein), and ATP Binding Cassette Subfamily A Member 1 (ABCA1) are associated with AMD pathogenesis and progression (9, 10). Knockout of cholesterol efflux-related genes (e.g., Apoe and Abca1) and cholesterol metabolism genes such as Cyp27a1 and Cyp46a1 cause retinal pathology in mice (11–14). We also found that the translocator protein, TSPO, mediated RPE cholesterol efflux, and loss of TSPO caused intracellular accumulation of cholesterol in human and mouse RPE cells (15, 16). High intake of dietary cholesterol has been reported to be associated with an increase in the risk of developing AMD (17–19). A high cholesterol-containing diet causes AMD pathological features in rabbits (20). All these findings suggest cholesterol is involved in AMD pathogenesis and that lowering cholesterol is a therapeutic strategy for patients with AMD.
Traditional Chinese Medicine has been used to treat both dry and wet AMD for thousands of years in China (2). Over 196 prescriptions have been described for treating AMD, among them, Qijudihuang pill (QP), which is regularly prescribed to patients with AMD. QP also shows therapeutic effects in other ocular disorders, such as diabetic retinopathy, retinitis pigmentosa, glaucoma, and dry eye disease (21, 22). QP is made of eight medicinal herbs, namely, Fructus lycii, Moutan Cortex, Dioscoreae Rhizoma, Corni Fructus, Poria, Alismatis Rhizoma, Flos Chrysanthemi Indici, and Rehmanniae Radix Praeparata. Using a network pharmacology approach, over 134 active compounds have been identified in QP and 72 candidate targets predicted, some of which were reported to have a role in retinal disorders (21). However, the underlying protective mechanisms of QP against ocular diseases, particularly AMD, are not explored.
In the present study, we treated high-fat diet-fed mice with QP and investigated changes in cholesterol metabolism, expression of antioxidant and inflammation genes, and gut microbiome.
For the preparation of QP, 12g of Fructus lycii, 10g of Moutan Cortex, 15g of Dioscoreae Rhizoma, 15g of Corni Fructus, 10g of Poria, 10g of Alismatis Rhizoma, 12g of Flos Chrysanthemi Indici, and 30g of Rehmanniae Radix Praeparata were pulverized together, put in 75ml distilled water, and boiled for 30 minutes with low heat. The liquid decoction was collected; the remaining material was again put in 75ml water and boiled for 30 minutes with low heat, and the decoction was collected. Both decoctions were mixed and concentrated to reach a final concentration of 1.10g/ml, and this was kept at 4°C.
All animal work was approved by the local animal welfare committee and followed the UK Home Office Animal Research guidance (Project License PP235815). Four-week-old male C57BL/6J mice were randomly allocated into three groups (eight animals/group). One group was fed with a control diet (CD) and the other two groups were fed for 13 weeks with a high-fat diet (HFD, 78.75% control diet to which 10% lard, 10% corn oil, 1% cholesterol, and 0.25% sodium cholate were added). The body weights of the mice were monitored weekly. After 13 weeks, the CD group and one HFD group were intra-gastrically treated daily with either physiological saline (0.4ml/animal) (CD group) or with the QP decoction (HFD+QP) at a dose of 14.69g/kg, which was calculated based on the clinical application (HFD group). After 30 days of treatment, animals were sacrificed and samples were collected.
Cholesterol was extracted from untreated, HFD, and HFD QP-treated serum, liver, retina, RPE/choroid with hexane: isopropanol (3:2, v/v). The extract was centrifuged, and the organic fraction was transferred into a new tube and dried under nitrogen gas. The dry lipid was dissolved in a cholesterol-working buffer and the total cholesterol was measured using an Amplex Red Cholesterol Assay kit (Thermo Fisher Scientific, UK) guided by the manufacturer’s protocol.
Total RNA was extracted from untreated, HFD, and HFD QP-treated mouse liver, retina, and RPE/choroid using TRIzol Reagent (Thermo Fisher Scientific, UK) based on the manufacturer’s protocol. CDNA was synthesized and targeted mRNA was detected using kits from Thermo Fisher Scientific, UK following the manufacturer’s instructions. Primers used for qRT-PCR are listed in Table S1.
Mouse cecum samples were collected and bacterial DNA was extracted using the QIAamp DNA Stool Mini Kit (QIAGEN, UK) based on the manufacturer’s instructions. Purified DNA from individual animals was used for PCR amplification and sequencing of 16S rRNA genes on an Illumina Nova-Seq with 2 x 300 base paired-end reads. Universal primers of the 16S rRNA genes were used to amplify the hypervariable regions, V3-V4 (V3F (338F) and V4R (806R)).
Sequences were processed with QIIME2 as described previously (23), except for using 230 as the truncation length in DADA2 (24). Reads and metadata have been submitted to the Sequence Read Archive (SRA) with Accession Number PRJNA1005835. The potential, functional metagenome was analyzed with the stand-alone version of PICRUSt2 (25). Raw abundance data was normalized for each pathway to generate Relative Abundance prior to analysis with ANOVA and Tukey’s post hoc correction to indicate significance between the three groups. This was conducted in R (v4.2.1) using the rstatix package, and results were plotted with ggplot2. The hierarchical clustering and heatmaps of the predicted proportion MetaCyc pathways and the Kyoto Encyclopedia of Genes and Genomes (KEGG) orthologues (KO) metagenomes were visualized by the SRplot tool (https://www.bioinformatics.com.cn/srplot).
Data on body weight, cholesterol level, and gene expression was analyzed using PRISM software (version 9) with one-way or two-way ANOVA followed by an appropriate post hoc test. Data was displayed as mean ± SD. 16S rRNA amplicon data was analyzed using Statistical Analysis of Metagenomic Profiles (STAMP) (26) by heatmap to explore the relationship between groups through ANOVA statistical test with Tukey-Kramer post hoc test after multiple test corrections. Statistical analysis of the abundance of taxa was conducted with Linear Discriminant Analysis (LDA) Effect Size (LEfSe) (27) using a Galaxy computational tool with settings of LDA at 3. LEfSe determines the features (genes, organisms, clades, and operational taxonomic units) of biologically relevant groupings. Wilcox test of two groups’ comparison (non-parametric) of (Bacteroidetes/Firmicutes ratio) with differences between at least two groups used SRplot online. A p-value of < 0.05 was considered significant.
During the 13-week initial feeding period, the body weight of the three groups continuously increased. Compared to the control group fed with a normal diet, animals fed with a high-fat diet had a significant increase in body weight from as early as 1 week of feeding that diet (Figure S1A). After the subsequent 30-day treatment with QP, the treated animals had similar body weight to the untreated HFD group, suggesting QP did not affect body weight (Figure S1B).
High-fat diet causes cholesterol accumulation in tissues and organs of animals and humans (23, 28). In the present study, we further confirmed animals fed with a high-fat diet had a significantly higher level of cholesterol in serum, liver, retina, and RPE/choroid. QP administration reversed the high-fat diet–induced effect on cholesterol levels in serum, liver, retina, and RPE/choroid (Figure 1). We also examined the expression of cholesterol homeostasis genes in the liver, RPE/choroid, and retina and found that cholesterol trafficking genes (Abca1 and Abcg1), cholesterol metabolism gene (Cyp27a1), and cholesterol transporting/metabolism-regulating gene (Nr1h3, encoding LXRα) had a significant decrease in expression in liver, RPE/choroid, and retina, compared to that of the control animals. QP treatment significantly increased the expression of these genes, compared to that of animals fed with a high-fat diet only. Expression of Cyp46a1 was markedly downregulated in liver and RPE/choroid of high-fat diet-fed animals, compared to that of control animals, and QP treatment increased its expression, compared to that of animals fed with high-fat diet alone. However, expression of Cyp46a1 was upregulated in the retina of high-fat diet-fed animals, but not changed in QP-treated animals. The cholesterol synthesis regulator, SCREBP2, had a significant increase in expression in the three tissues of animals fed with a high-fat diet, compared to control animals, whereas QP treatment significantly downregulated the expression of SCREBP2 in the three tissues compared to animals fed with high-fat diets (Figures 2, S2).
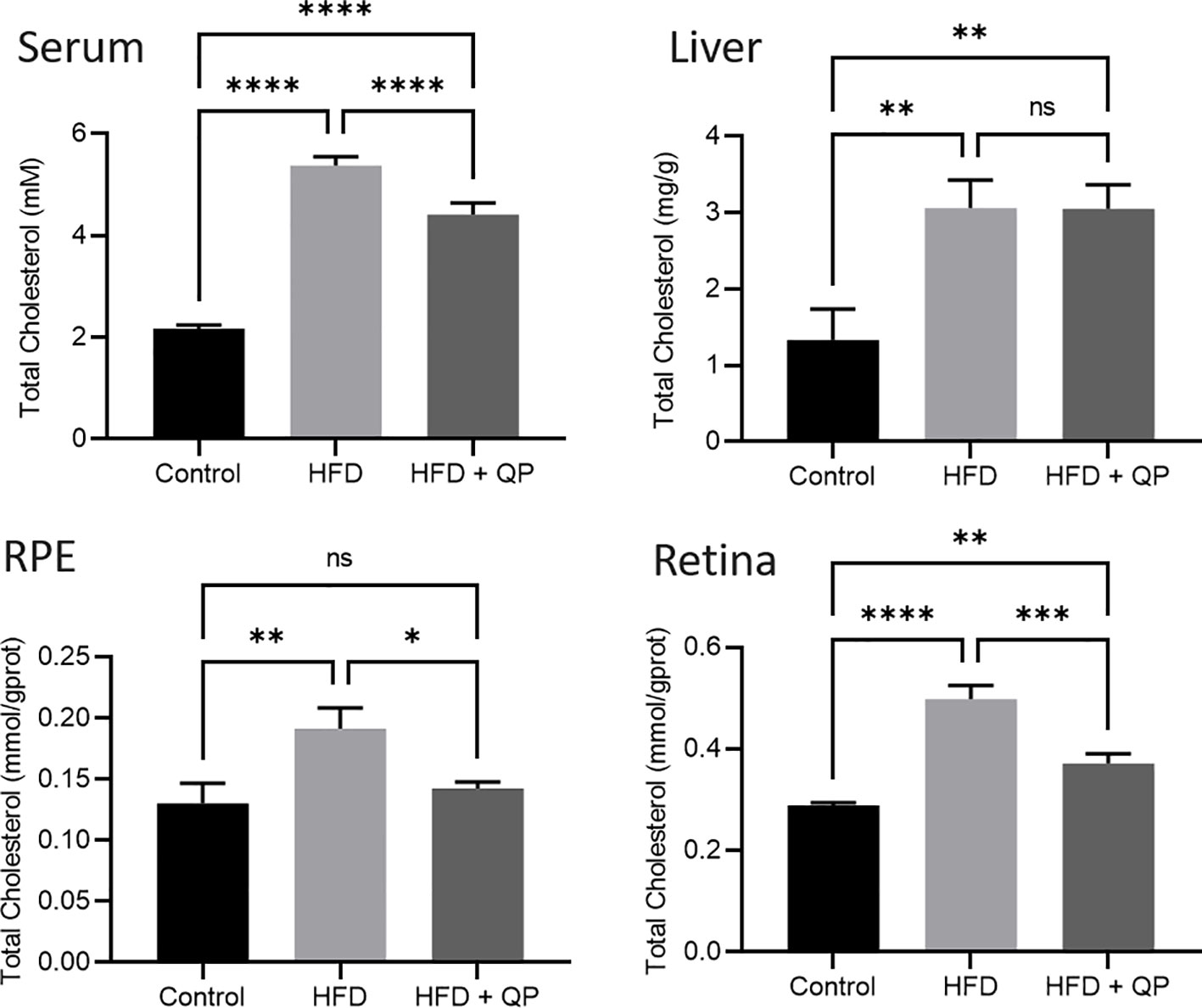
Figure 1 Effect of QP treatment on cholesterol level in serum, liver, RPE, and retina. Data was analyzed using one-way ANOVA followed by Tukey’s multiple comparisons test and presented as mean ± SD. HFD, high-fat diet-fed group; QP, Qijudihuang pill; RPE, retinal pigment epithelial cells. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001; ns, no significance. Eight samples/group.
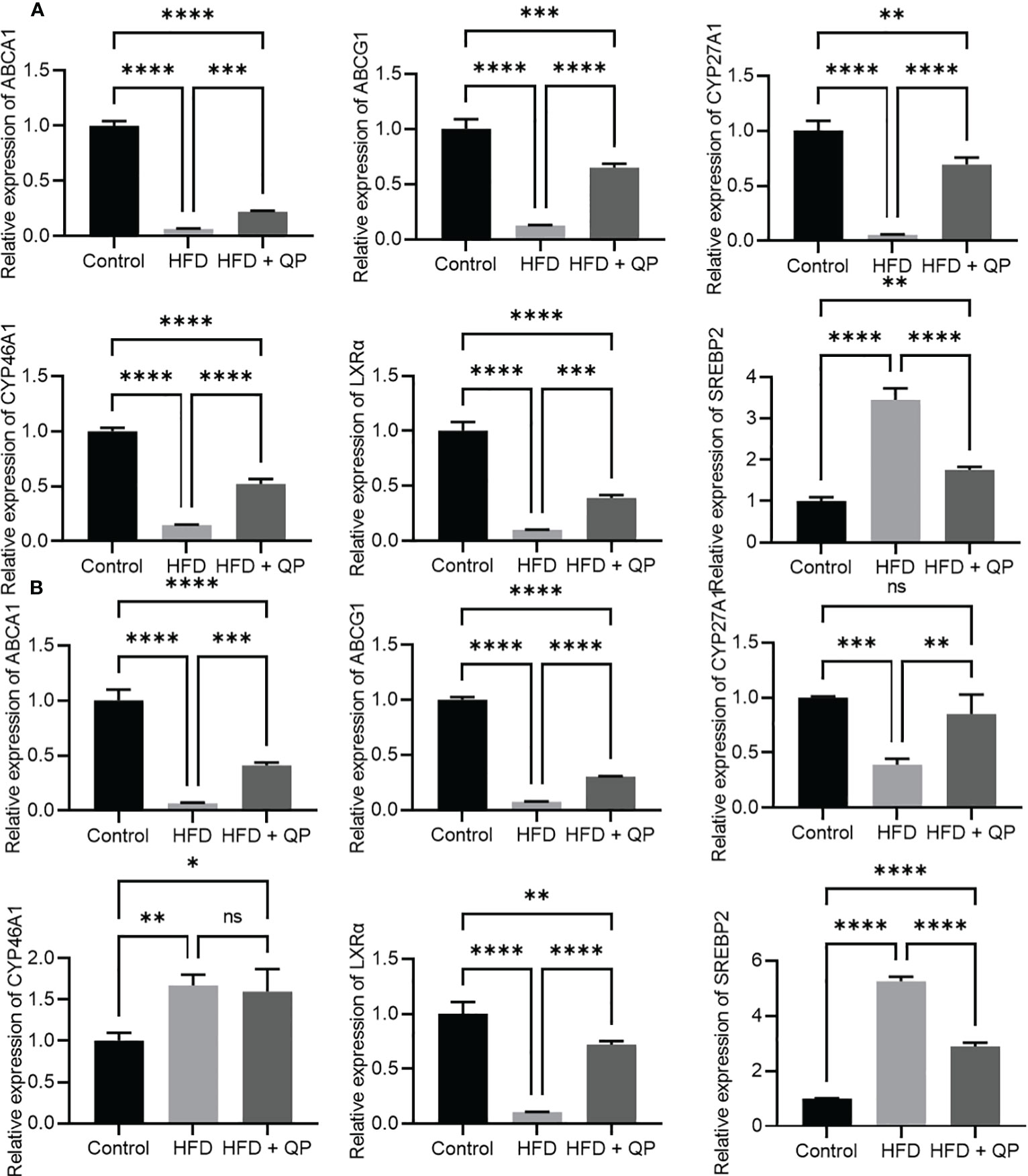
Figure 2 Effect of QP on expression of cholesterol homeostasis genes in RPE/choroid (A) and in the retina (B). Data was analyzed using one-way ANOVA followed by Tukey’s multiple comparisons test and presented as mean ± SD. HFD, high-fat diet-fed group; QP, Qijudihuang pill; RPE, retinal pigment epithelial cells. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001; ns, no significance. Eight samples/group.
It is well-documented that a high-fat diet induces oxidative stress and inflammation (29). In the present study, we found that the expression of antioxidant genes (Catalase, Gpx1, and Sod1) was markedly downregulated in RPE/choroid and retina of HFD animals compared to that of CD animals but QP treatment reversed this HFD-induced effect (Figure 3). Conversely, expression of pro-inflammatory cytokines (Il-1β and Tnfα) in RPE/choroid, retina, and liver of HFD mice was markedly upregulated compared to that of CD mice, and QP treatment significantly alleviated expression of the two cytokines in RPE/choroid, retina, and liver in the HFD+QP group (Figure 4).
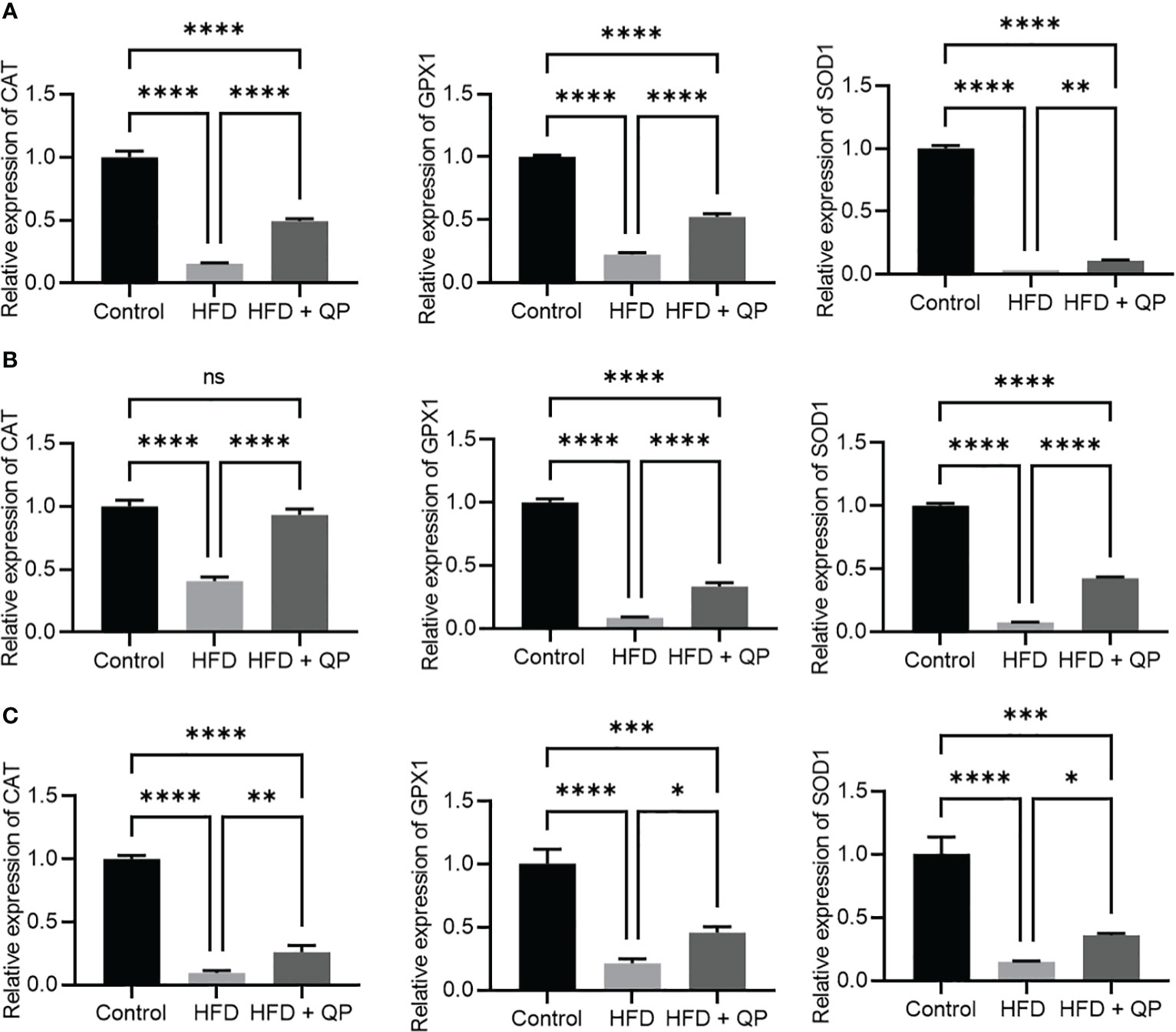
Figure 3 Effect of QP treatment on expression of antioxidant genes in RPE (A), retina (B), and liver (C). Data was analyzed using one-way ANOVA followed by Tukey’s multiple comparisons test and presented as mean ± SD. HFD, high-fat diet-fed group; QP, Qijudihuang pill; RPE, retinal pigment epithelial cells. * p<0.05; ** p<0.01; *** p<0.001; **** p<0.0001; ns, no significance; Eight samples/group, three repeats.
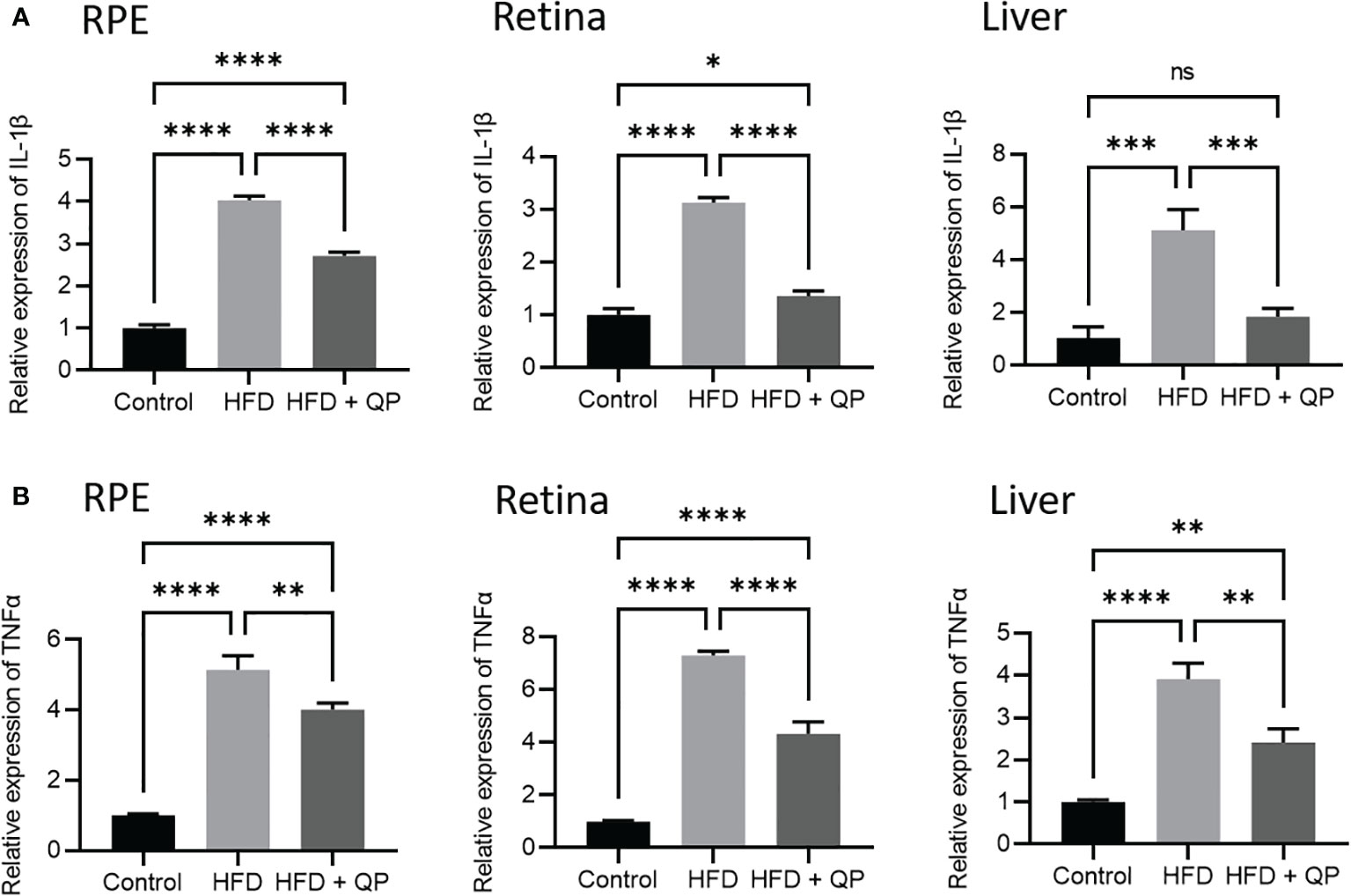
Figure 4 Effect of QP treatment on expression of IL-1β (A) and TNFα (B) in RPE, retina, and liver. Data was analyzed using one-way ANOVA followed by Tukey’s multiple comparisons test and presented as mean ± SD. HFD, high-fat diet-fed group; QP, Qijudihuang pill; RPE, retinal pigment epithelial cells. * p<0.05; ** p<0.01; *** p<0.001; **** p<0.0001; ns, no significance. Eight samples/group, three repeats.
Sequence read numbers and the numbers of non-chimeric, joined reads are shown in Table S2. Raw read numbers varied between 79700-114311, and after joining and elimination of chimeras, a range of 64.6-91.4% of input survived. Taxonomic profiles of the organisms present in each sample were obtained via QIIME2 packages with reference to the Green Genes database at 97% identity (Figure S3, Table S3). A total of 238 taxa from 8 phyla (Actinobacteria, Bacteroides, Firmicutes, Proteobacteria, Cyanobacteria, Fusobacteria, TM7, and Verrucomicrobia) from 120 genera with 97 identified species and 56 unclassified species are detailed in Table S3. Clear differences in bacterial communities between the groups were apparent by diversity analysis (Figure 5). Beta diversity indicated a clear separation of microbial communities between CD and HFD as well as between CD and HFD+QP but with some indication of the movement of HFD+QP towards CD. This is further shown in Emperor plots with alternative metrics of determination (Figure S3). Additionally, heat maps and hierarchical clustering show a separation between the CD and both high-fat groups (Figure S4). The ratio of Bacteroidetes compared to Firmicutes was markedly increased by the HFD compared to control but this ratio was much reduced in the HFD+QP group (Figure S4C).
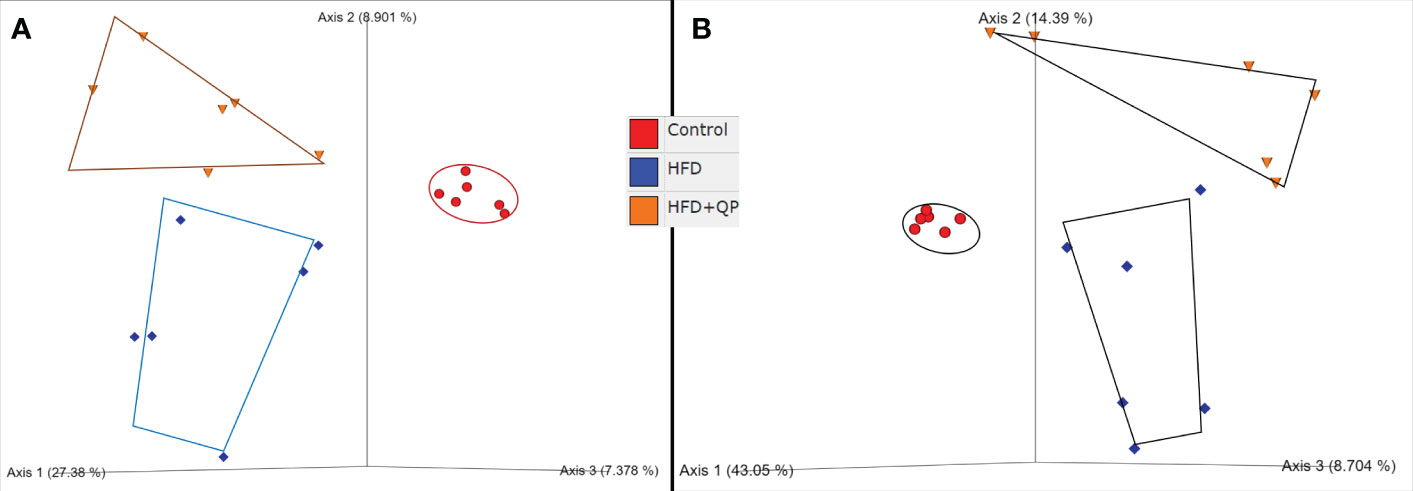
Figure 5 Beta diversity of bacterial communities in the three groups of mice by (A) Jaccard and (B) Bray-Curtis metrics as shown by Emperor plots. Six samples/group.
The differential abundances of taxa comparing the three experimental groups were determined using the Linear Discriminant Analysis (LDA) Effect Size (LEfSe) and Plot Cladogram (Figure 6). Mice fed with HFD showed profound changes in the abundance of taxa compared to taxa in mice given CD. Some taxa were present at significantly higher levels while others were present at much lower levels than found in CD. This is apparent in Figure 6 where 13 taxa are differentially increased in HFD mice while 19 are decreased. The pattern of changes and identity of taxa are shown in Figure S5 with the accompanying table. In contrast, HFD+QP had only two taxa with increased abundance (f_Aerococacceae and g_Clostridium) and these were also found when comparing HFD to CD. There was also a smaller list of taxa reduced in HFD+QP (13) of which 8 were common to the other CD comparison while 5 were unique. The complexities of the community changes became more apparent when comparing differential abundance in HFD to HFD+QP. In this case, the number of taxa with increased abundance in HFD was three (each of which was seen in the comparison to CD) and only one with reduced abundance relative to HFD+QP, again a taxa seen in CD (s_indistinctus).
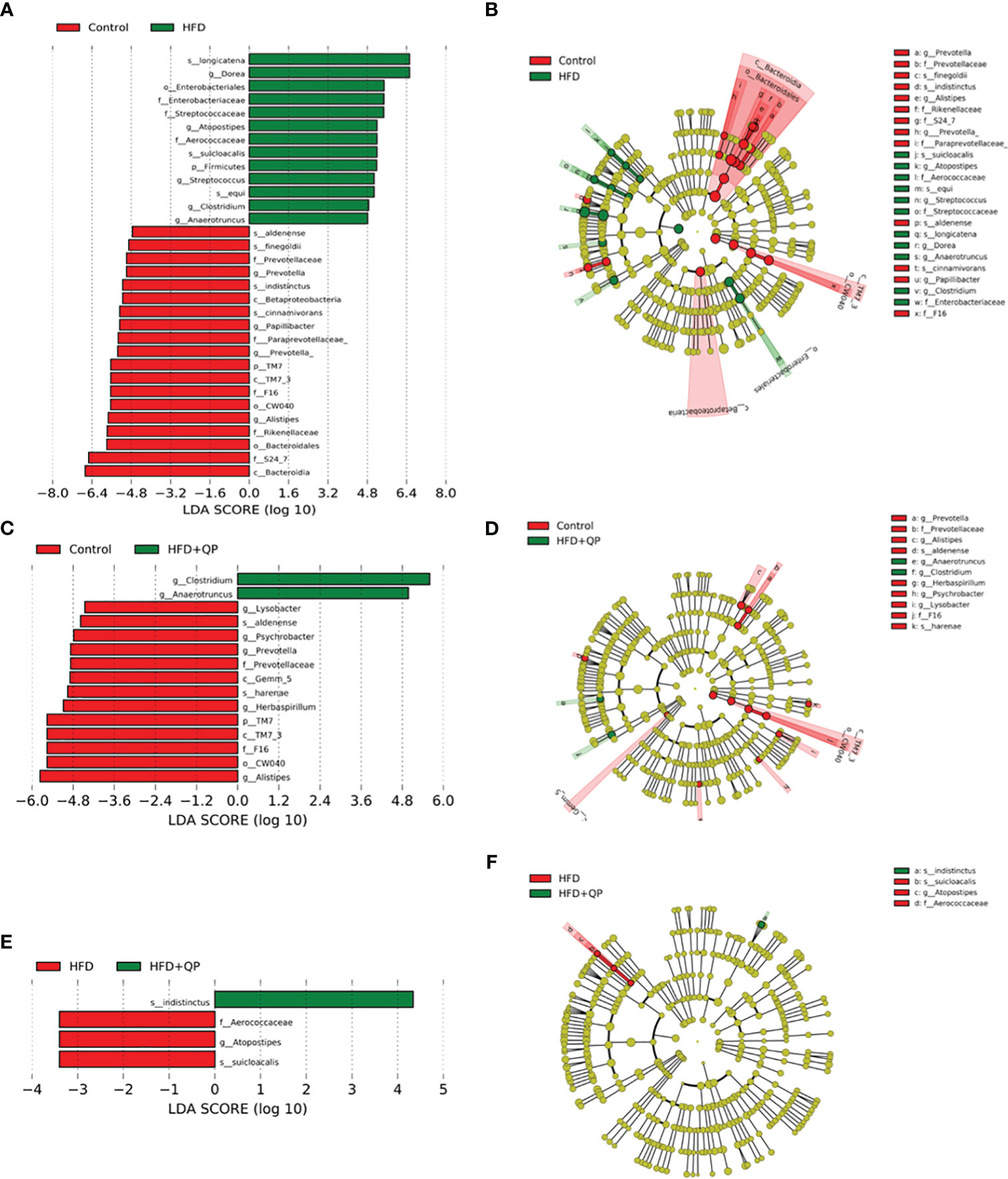
Figure 6 Differential abundance of bacteria in gut microbiome showing pairwise comparison of abundance using the LEfSe method. The figure shows features that are differentially abundant scaled with their effect size (A, C, E), and those differences are mapped to associated taxonomic trees (B, D, F). Effect size is indicated by the LDA score with the green colors showing taxa more abundant in HFD compared to the control group (A) and more abundant in HFD+QP compared to control (C) and HFD+QP relative to HFD (F). Negative LDA scores are shown in red indicating higher abundance in the comparison partner (B Control, D Control, and F HFD). The parallel, taxonomic representations utilize the same colors: red most abundant, green least abundant, and yellow indicates organisms without significant difference.
We used PICRUSt2 to provide insights into the potential metabolic pathways of the bacterial communities within each of the experimental groups. Analysis with PCoA revealed separation between each of the groups but with a clear movement of the HFD+QP towards CD (Figure 7A). Of the 415 MetaCyc pathways, 127 were significantly different between CD and HFD. In 55 of these pathways, there was no significant difference between CD and HFD+QP (Table S4). In some pathways (30), HFD increased pathway abundance from CD (Figure 7B) and this was reversed back to CD by the QP treatment, and in other cases (9) HFD suppressed abundance and this was increased back to CD by QP (Figure 7C).
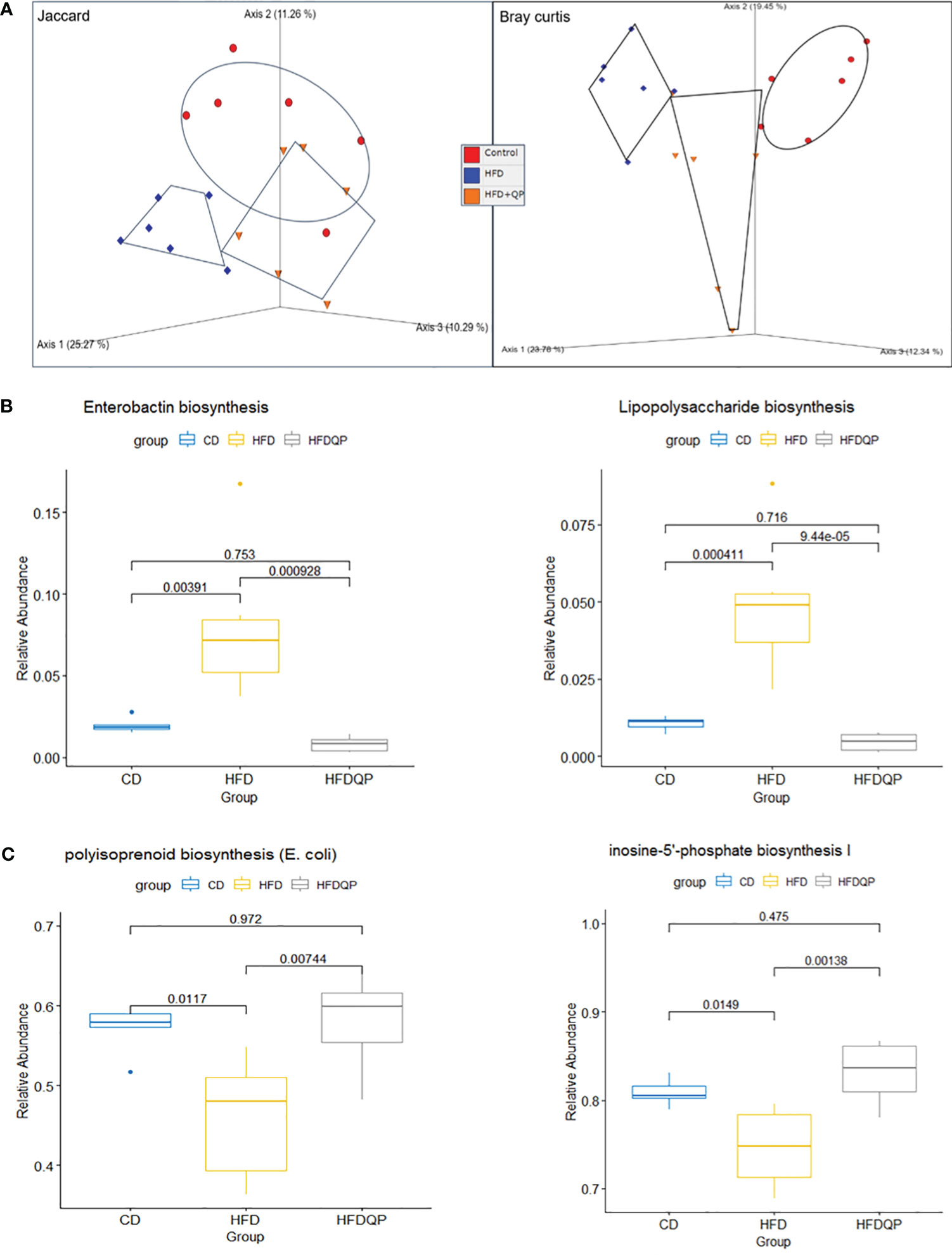
Figure 7 Functional analysis of the metagenome using PICRUSt2 with MetaCyc pathways. Beta diversity of pathways (A) by Jaccard and Bray Curtis metrics shown by PCoA. Relative Abundance of selected pathways that promote inflammation (B) or pathways of intermediary metabolism (C) affected by HFD and reversed by QP. CD, control diet; HFD, high-fat diet; HFDQP, high-fat diet with QP treatment.
The pathway changes were further studied by consideration of the Superclass ontologies specified by each of the MetaCyc classifications (Table S5). Of the 46 pathways elevated by HFD and reversed by QP, 19 were described by Superclass classification as “Biosynthesis”, 23 as “Degradation/Utilization/Assimilation”, and 4 as “Generation of Precursor Metabolites and Energy”. Of the “Biosynthesis” pathways, nine were involved with “Cofactor, Carrier, and Vitamin Biosynthesis”, mainly with compounds used in Electron Transport Chains, specifically menaquinones. Synthesis of lipopolysaccharides (LPS) and the phenolic siderophore, enterobactin were also notable. In the class “Degradation/Utilization/Assimilation”, “Aromatic Compound Degradation” (five) as well as “C1 Compound Utilization and Assimilation” (three), and “Carboxylic Acid Degradation” (three) along with degradation of amino acids and carbohydrates were represented. Of the other Superclass classifications, all representatives were described by the Superclass term “Generation of Precursor Metabolites and Energy”. Of the nine pathways decreased in HFD but reversed by QP all but one were described by “Biosynthesis”, the other being “Degradation/Utilization/Assimilation”. The representatives in “Biosynthesis” were involved in “Nucleoside and Nucleotide Biosynthesis” (three) or “Sugar Biosynthesis” (two). The example in Degradation was also involved in Purine metabolism.
Although QP has been used to treat AMD in China for a long period, the underlying functional mechanism has not been investigated. In the current study, we treated a high-fat diet-fed mouse model of AMD and found QP decreased HFD-induced elevated levels of cholesterol in the RPE/choroid, retina, liver, and serum, upregulated expression of antioxidant genes, and suppressed expression of pro-inflammatory genes in the RPE/choroid and retinas of high-fat diet-fed animals, as well as modulated gut microbial composition.
Cholesterol accumulates in the drusen of patients with AMD and can be enzymatically or non-enzymatically oxidized into oxysterols (6). Loss of oxysterol-producing enzymes or cholesterol transporters causes retinal degeneration in rodents. HFD exacerbates AMD pathological features in Apoe (involved in cholesterol efflux) knockout mice (31). HFD has also been shown to induce oxidative stress, inflammation, and dysbiosis, resulting in accelerated photoreceptor degeneration in rd10 mice (a retinitis pigmentosa mouse model) (32). In the present study, we also demonstrated that feeding with HFD caused increased cholesterol levels, decreased expression of antioxidant genes, and upregulated expression of inflammatory mediators in examined tissues and that these effects were reversed by QP treatment. Network pharmacology predicted that QP contains multiple active compounds and potential protein targets, of which the top ones are involved in inflammation (22). Therefore, it is supposed that there are multiple signaling pathways associated with QP’s protective effect in HFD-fed mice, which requires further investigation.
Excess cholesterol in the retina and RPE is reversibly transported back to the liver and converted to bile acids, principally by mitochondrial Cyp27A1 (33) and Cyp7A1 (34) located in the endoplasmic reticulum. The products are conjugated by taurine or glycine prior to transport into the gall bladder and their release into the duodenum is stimulated by feeding. The majority of the secreted BA is reabsorbed following deconjugation in the ileum to complete the BA enterohepatic circulation, while a small proportion arrives in the colon (35). Gut bacteria metabolize bile acids by a wide variety of reactions including deconjugation, re-conjugation, dihydroxylation, and oxidation (36, 37). The diversity of gut bacteria and their metabolic capacity to act on BAs determines effects on host systems via a multitude of BA-activated signal receptors that regulate hepatic cholesterol metabolism by affecting the expression of enzymes causing their formation, including the two Cyps (37).
When mice are provided with a high-fat diet there are multiple effects on cholesterol metabolism, including conversion to bile acids. These alter the spectrum of bacteria in the gut, and thus the signaling through bile acids, to affect the host including on the gut-brain axis (38). Indeed, HFD causes widespread changes in microbiota communities, leading to dysbiosis associated with obesity, type 2 diabetes, and AMD as well as a number of other health-compromising conditions (39–41). Those effects were observed in this study at the phylum level with the frequently observed increase of the ratio of Bacteroidetes compared to Firmicutes. The changes can also be seen in terms of beta diversity with the PCoA revealing a clear separation between the CD and HFD groups. Furthermore, analysis with the LEfSe tool revealed an HFD-associated increase in abundance of a variety of taxa previously commented on by ourselves and others (23, 42–44). At the same time, other taxa were decreased by this diet. For instance, Anaerotruncus was increased and has been associated with HCC and non-alcoholic steatohepatitis (45), whereas beneficial f-S27-4 was decreased (23, 43).
One objective of this project was to ascertain whether QP has an effect on gut microbiome and thus whether the observed changes play a mechanistic contribution to potential therapeutic effects. Even while the HFD was continued, QP, at least in part, reversed the effects observed with HFD alone. Thus, the spectrum of phyla reverted more towards that seen in CD as illustrated by a decrease of Bacteroidetes/Firmicutes, a movement of the HFD+QP group towards CD in PCoA plots, and a more restricted change in abundances of taxa in LEfSe, both for those that increased and those that decreased in abundance. The number and identity of taxa that were either increased or decreased in HFD+QP vs. CD were significantly diminished compared to when HFD alone was compared to CD.
Changes in the composition of microbial communities will have effects on global metabolic capacity and thus on interaction with the host. We examined the extent and nature of such potential changes with PICRUSt2. On a global scale, using diversity measures, clear changes induced by HFD and partially reversed by QP were apparent in PCoA plots, changes that closely parallel those seen when considering taxa alone. In terms of individual pathways increased by HFD, those generating pro-inflammatory effectors were notable. These included the synthesis of enterobactin, polymixin B, and lipopolysaccharide, each of which was increased by HFD but reversed when QP was included in the diet. We propose that the increase of IL-1B and TNF-a in the liver, RPE/choroid, and retina are also associated with these changes occurring in the gut microbiota due to HFDs. Likewise, we propose that the decrease of anti-oxidative enzymes catalase, GPX, and SOD1 is a response to changes in the abundance of bacterial taxa, their associated metabolic capacity, and nature. For instance, Enterobactin is a siderophore and derivative of 2,3 dihydroxy N-benzoylserine lactone that chelates primarily ferric iron and is produced by Gram-negative bacteria (46). Since iron is a requirement of host and microbes, an imbalance of iron uptake will impair host systems, and enterobactin has been shown to promote bacterial colonization (30) and interfere with host immune responses (47). Polymixins are microbially-produced antibiotics that illustrate two effects of the HFD. The production of polymixin antibiotics in the gut provides one mechanism leading to the remodeling of bacterial communities and indeed the original interest in these compounds arose as they provided a prospect for the treatment of multidrug-resistant organisms (48). Aligning with this mechanism of remodeling are bacteria, such as Pseudomonas, that have resistance to polymixin (49). However, the drugs also produced marked nephrotoxicity, illustrating the second point that the change of bacterial communities results in the formation of toxic compounds, causing harm to the host. The mode of action of polymixins is thought to be by interaction with phospholipids, causing membrane disruption including macrophages, and stimulating the production of IL-1β (50). An alternative theory suggests that polymixins induce oxidative stress caused by the formation of reactive oxygen species (51), which resonates with altered oxidative resistance in RPE. Resistant bacteria occur in Escherichia, Klebsiella, Salmonella, Shigella, Enterobacter, and Citrobacter (48), and have altered LPS structures that block the interaction with antibiotics (52). Other possible contributors to intestinal inflammation are changes to purinogenic pathways as found in inflammatory bowel disease (53). These pathways were found to be lowered by the HFD but raised to control levels when QP was administered.
While the complex effects of HFD on bacteria and their interaction with the host are starting to be understood, QP has not been previously explored at the levels described here. We have started to characterize the component chemicals present in the QP plant extract (in preparation) and established the principle components. How these might interact with the host and bacteria demands future investigation but one possibility is that they act with the promiscuous nuclear hormone receptor pregnane-X receptor (PXR) located in the liver and gut enterocytes. PXR regulates the expression of many Phase 1 and Phase 2 enzymes and thus affects the metabolism of cholesterol to BAs, for instance, the induction of SULT2A, which conjugates BAs prior to export, and of Mrp2 and Oatp2, the export components (54).
TCM preparations have widely shown lipid-lowering function in dyslipidemia (55, 56). For targeting cholesterol, TCM can inhibit intestinal absorption of cholesterol, suppress endogenous cholesterol synthesis, promote cholesterol reverse transport and excretion, and regulate the expression of cholesterol homeostasis-associated transcription factors (55, 56). Berberine, a major functional compound of many medicinal herbs, has been shown to lower blood cholesterol in atherogenic-diet-fed rats at least partly via the inhibition of intestinal cholesterol absorption (57). TCM Jiang-Zhi-Ning, containing four Chinese herbs, can significantly lower cholesterol, triglyceride, and low-density lipoprotein-cholesterol in hyperlipidemic rats, partly by inhibiting the expression of 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), the rate-limiting enzyme for cholesterol synthesis (58). TCM FufangZhenzhuTiaoZhi has been shown to decrease plasma cholesterol in hyperlipidemic rats via downregulating HMGCR expression and increasing expression and activity of 7-α hydroxylase (CYP7A1, the rate-limiting enzyme for bile acid synthesis) (59). In the present study, our data suggested that the effect of QP in lowering cholesterol is possibly through the promotion of cholesterol metabolism, transport, and excretion. It will be worth investigating whether QP inhibits cholesterol intestinal absorption and endogenous synthesis. Additionally, there are reports about the hepatotoxicity of TCM and natural products in animal models and human patients (60, 61), so it may be necessary to examine whether there is any potential toxicity of QP in animal models and patients with AMD.
This study aimed to investigate the relationship between HFD and AMD and start to unravel the mechanisms whereby QP may provide relief from the condition. HFD was shown to exacerbate lipid accumulation in the RPE and retina and associated inflammation and oxidative stress. Changes in the gut microbiota, their metabolic capacity, and signaling between the host and microbes will have contributed to these pathological conditions. We propose that the ability of QP to reverse these effects is related to mechanisms acting to lower cholesterol levels, oxidative stress, and inflammation, and to modulate gut microbiota; and these will be investigated in future studies.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was approved by The Animal Welfare Committee, Glasgow Caledonian University. The study was conducted in accordance with the local legislation and institutional requirements.
XS: Conceptualization, Funding acquisition, Writing – original draft, Writing – review & editing. YC: Investigation, Writing – original draft. KSI: Formal Analysis, Writing – original draft. XL: Investigation, Writing – original draft. AW: Investigation, Writing – original draft. YW: Investigation, Writing – original draft. XDY: Investigation, Writing – original draft. XZ: Formal Analysis, Writing – original draft. ZT: Project administration, Writing – original draft. ZH: Project administration, Writing – original draft. JAC: Formal Analysis, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was partially supported by the Locus Scholarship Programme of Hunan Province, China (2019-23 to XS), the TENOVUS Scotland (S20-02 to XS), the Chief Scientist Office/the RS Macdonald Charitable Trust (SNRF2021 to XS), and the Hunan Provincial Natural Science Foundation of China (2022JJ50167 to YC).
The author(s) declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1274401/full#supplementary-material
1. Wong WL, Su X, Li X, Cheung CMG, Klein R, Cheng CY, et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Global Health (2014) 2(2):e106–16. doi: 10.1016/S2214-109X(13)70145-1
2. Li Y, Li X, Li X, Zeng Z, Strang N, Shu X, et al. Non-neglectable therapeutic options for age-related macular degeneration: A promising perspective from traditional Chinese medicine. J Ethnopharmacol (2022) 282:114531. doi: 10.1016/j.jep.2021.114531
3. Cao Y, Li Y, Gkerdi A, Reilly J, Tan Z, Shu X. Association of nutrients, specific dietary patterns, and probiotics with age-related macular degeneration. Curr Medicinal Chem (2022) 29(40):pp.6141–6158. doi: 10.2174/0929867329666220511142817
4. Mettu PS, Allingham MJ, Cousins SW. Incomplete response to Anti-VEGF therapy in neovascular AMD: Exploring disease mechanisms and therapeutic opportunities. Prog retinal eye Res (2021) 82:100906. doi: 10.1016/j.preteyeres.2020.100906
5. Handa JT, Bowes Rickman C, Dick AD, Gorin MB, Miller JW, Toth CA, et al. A systems biology approach towards understanding and treating non-neovascular age-related macular degeneration. Nat Commun (2019) 10(1):3347. doi: 10.1038/s41467-019-11262-1
6. Zhang X, Alhasani RH, Zhou X, Reilly J, Zeng Z, Strang N, et al. Oxysterols and retinal degeneration. Br J Pharmacol (2021) 178(16):3205–19. doi: 10.1111/bph.15391
7. Curcio CA, Presley JB, Malek G, Medeiros NE, Avery DV, Kruth HS. Esterified and unesterified cholesterol in drusen and basal deposits of eyes with age-related maculopathy. Exp eye Res (2005) 81(6):731–41. doi: 10.1016/j.exer.2005.04.012
8. Pang CE, Messinger JD, Zanzottera EC, Freund KB, Curcio CA. The onion sign in neovascular age-related macular degeneration represents cholesterol crystals. Ophthalmology (2015) 122(11):2316–26. doi: 10.1016/j.ophtha.2015.07.008
9. DeAngelis MM, Owen LA, Morrison MA, Morgan DJ, Li M, Shakoor A, et al. Genetics of age-related macular degeneration (AMD). Hum Mol Genet (2017) 26(R1):R45–50. doi: 10.1093/hmg/ddx228
10. Fritsche LG, Igl W, Bailey JNC, Grassmann F, Sengupta S, Bragg-Gresham JL, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet (2016) 48(2):134–43. doi: 10.1038/ng.3448
11. Malek G, Johnson LV, Mace BE, Saloupis P, Schmechel DE, Rickman DW, et al. Apolipoprotein E allele-dependent pathogenesis: a model for age-related retinal degeneration. Proc Natl Acad Sci (2005) 102(33):11900–5. doi: 10.1073/pnas.0503015102
12. Omarova S, Charvet CD, Reem RE, Mast N, Zheng W, Huang S, et al. Abnormal vascularization in mouse retina with dysregulated retinal cholesterol homeostasis. J Clin Invest (2012) 122(8):3012–23. doi: 10.1172/JCI63816
13. Saadane A, Mast N, Trichonas G, Chakraborty D, Hammer S, Busik JV, et al. Retinal vascular abnormalities and microglia activation in mice with deficiency in cytochrome P450 46A1–mediated cholesterol removal. Am J Pathol (2019) 189(2):405–425. doi: 10.1016/j.ajpath.2018.10.013
14. Storti F, Klee K, Todorova V, Steiner R, Othman A, van der Velde-Visser S, et al. Impaired ABCA1/ABCG1-mediated lipid efflux in the mouse retinal pigment epithelium (RPE) leads to retinal degeneration. Elife (2019) 8:e45100. doi: 10.7554/eLife.45100
15. Biswas L, Zhou X, Dhillon B, Graham A, Shu X. Retinal pigment epithelium cholesterol efflux mediated by the 18 kDa translocator protein, TSPO, a potential target for treating age-related macular degeneration. Hum Mol Genet (2017) 26(22):4327–39. doi: 10.1093/hmg/ddx319
16. Farhan F, Almarhoun M, Wong A, Findlay AS, Bartholomew C, Williams MT, et al. Deletion of TSPO causes dysregulation of cholesterol metabolism in mouse retina. Cells (2021) 10(11):3066. doi: 10.3390/cells10113066
17. Mares-Perlman JA, Brady WE, Klein R, VandenLangenberg GM, Klein BE, Palta M. Dietary fat and age-related maculopathy. Arch Ophthalmol (1995) 113(6):743–8. doi: 10.1001/archopht.1995.01100060069034
18. SanGiovanni JP, Chew EY, Clemons TE, Davis MD, Ferris 3FL, Gensler GR, et al. The relationship of dietary lipid intake and age-related macular degeneration in a case-control study: AREDS Report No. 20. Arch Ophthalmol (Chicago Ill: 1960) (2007) 125(5):671–9. doi: 10.1001/archopht.125.5.671
19. Smith W, Mitchell P, Leeder SR. Dietary fat and fish intake and age-related maculopathy. Arch Ophthalmol (2000) 118(3):401–4. doi: 10.1001/archopht.118.3.401
20. Dasari B, Prasanthi JR, Marwarha G, Singh BB, Ghribi O. Cholesterol-enriched diet causes age-related macular degeneration-like pathology in rabbit retina. BMC Ophthalmol (2011) 11:1–11. doi: 10.1186/1471-2415-11-22
21. Liu F, Tian M. Study on the mechanism of Qiju Dihuang pill in the treatment of ophthalmic diseases based on systems pharmacology. Medicine (2022) 101(31):e30033. doi: 10.1097/MD.0000000000030033
22. Zhao X, Yu Z, Li D, Duan J. Using network pharmacology and molecular docking to explore the mechanism of qiju dihuang pill against dry eye disease. Comput Math Methods Med (2022) 2022:7316794. doi: 10.1155/2022/7316794
23. Ibrahim KS, Craft JA, Biswas L, Spencer J, Shu X. Etifoxine reverses weight gain and alters the colonic bacterial community in a mouse model of obesity. Biochem Pharmacol (2020) 180:114151. doi: 10.1016/j.bcp.2020.114151
24. Callahan B, McMurdie P, Rosen M, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods (2016) 13:581–3. doi: 10.1038/nmeth.3869
25. Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, et al. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol (2020) 38(6):685–8. doi: 10.1038/s41587-020-0548-6
26. Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics (2014) 30(21):3123–4. doi: 10.1093/bioinformatics/btu494
27. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol (2011) 12:1–18. doi: 10.1186/gb-2011-12-6-r60
28. Montani JP, Carroll JF, Dwyer TM, Antic V, Yang Z, Dulloo AG. Ectopic fat storage in heart, blood vessels and kidneys in the pathogenesis of cardiovascular diseases. Int J Obes (2004) 28(4):S58–65. doi: 10.1038/sj.ijo.0802858
29. Tan BL, Norhaizan ME. Effect of high-fat diets on oxidative stress, cellular inflammatory response and cognitive function. Nutrients (2019) 11(11):2579. doi: 10.3390/nu11112579
30. Pi H, Jones SA, Mercer LE, Meador JP, Caughron JE, Jordan L, et al. Role of catecholate siderophores in gram-negative bacterial colonization of the mouse gut. PloS One (2012) 7(11):e50020. doi: 10.1371/journal.pone.0050020
31. Skondra D, She H, Zambarakji HJ, Connolly E, Michaud N, Chan P, et al. Effects of ApoE deficiency, aging and high fat diet on laser-induced choroidal neovascularization and Bruch's membrane-RPE interface morphology. Invest Ophthalmol Visual Sci (2007) 48(13):1768–8.
32. Kutsyr O, Noailles A, Martínez-Gil N, Maestre-Carballa L, Martinez-Garcia M, Maneu V, et al. Short-term high-fat feeding exacerbates degeneration in retinitis pigmentosa by promoting retinal oxidative stress and inflammation. Proc Natl Acad Sci (2021) 118(43):e2100566118. doi: 10.1073/pnas.2100566118
33. Bjorkhem I. Mechanism of degradation of the steroid side chain in the formation of bile acids. J Lipid Res (1992) 33(4):455–71. doi: 10.1016/S0022-2275(20)41612-8
34. Myant NB, Mitropoulos KA. Cholesterol 7α-hydroxylase. J Lipid Res (1977) 18(2):135–53. doi: 10.1016/S0022-2275(20)41693-1
35. Martinez-Guryn K, Hubert N, Frazier K, Urlass S, Musch MW, Ojeda P, et al. Small intestine microbiota regulate host digestive and absorptive adaptive responses to dietary lipids. Cell Host Microbe (2018) 23(4):458–69.e5. doi: 10.1016/j.chom.2018.03.011
36. Lucas LN, Barrett K, Kerby RL, Zhang Q, Cattaneo LE, Stevenson D, et al. Dominant bacterial phyla from the human gut show widespread ability to transform and conjugate bile acids. MSystems (2021):e0080521. doi: 10.1128/mSystems.00805-21
37. Cai J, Rimal B, Jiang C, Chiang JY, Patterson AD. Bile acid metabolism and signaling, the microbiota, and metabolic disease. Pharmacol Ther (2022) 237:108238. doi: 10.1016/j.pharmthera.2022.108238
38. O’Mahony SM, Clarke G, Borre YE, Dinan TG, Cryan JF. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav Brain Res (2015) 277:32–48. doi: 10.1016/j.bbr.2014.07.027
39. Clemons TE, Milton RC, Klein R, Seddon JM, Ferris FL 3rd, Age-Related Eye Disease Study Research Group. Risk factors for the incidence of Advanced Age-Related Macular Degeneration in the Age-Related Eye Disease Study (AREDS) AREDS report no. 19. Ophthalmology (2005) 112(4):533–9. doi: 10.1016/j.ophtha.2004.10.047
40. Yang RL, Li W, Shi YH, Le GW. Lipoic acid prevents high-fat diet-induced dyslipidemia and oxidative stress: a microarray analysis. Nutrition (2008) 24(6):582–8. doi: 10.1016/j.nut.2008.02.002
41. Zisimopoulos A, Klavdianou O, Theodossiadis P, Chatziralli I. The role of the microbiome in age-related macular degeneration: A review of the literature. Ophthalmologica (2021) 244(3):173–8. doi: 10.1159/000515026
42. de La Serre CB, Ellis CL, Lee J, Hartman AL, Rutledge JC, Raybould HE. Propensity to high-fat diet-induced obesity in rats is associated with changes in the gut microbiota and gut inflammation. Am J Physiol Gastrointest Liver Physiol (2010) 299(2):G440–8. doi: 10.1152/ajpgi.00098.2010
43. Lagkouvardos I, Lesker TR, Hitch TCA, Gálvez EJC, Smit N, Neuhaus K, et al. Sequence and cultivation study of Muribaculaceae reveals novel species, host preference, and functional potential of this yet undescribed family. Microbiome (2019) 7(1):28. doi: 10.1186/s40168-019-0637-2
44. Stenkamp-Strahm CM, Nyavor YE, Kappmeyer AJ, Horton S, Gericke M, Balemba OB. Prolonged high fat diet ingestion, obesity, and type 2 diabetes symptoms correlate with phenotypic plasticity in myenteric neurons and nerve damage in the mouse duodenum. Cell Tissue Res (2015) 361(2):411–26. doi: 10.1007/s00441-015-2132-9
45. Ye JZ, Li YT, Wu WR, Shi D, Fang DQ, Yang LY, et al. Dynamic alterations in the gut microbiota and metabolome during the development of methionine-choline-deficient diet-induced nonalcoholic steatohepatitis. World J Gastroenterol (2018) 24(23):2468–81. doi: 10.3748/wjg.v24.i23.2468
46. Fischbach MA, Lin H, Liu DR, Walsh CT. How pathogenic bacteria evade mammalian sabotage in the battle for iron. Nat Chem Biol (2006) 2(3):132–8. doi: 10.1038/nchembio771
47. Saha P, Yeoh BS, Xiao X, Golonka RM, Abokor AA, Wenceslau CF, et al. Enterobactin induces the chemokine, interleukin-8, from intestinal epithelia by chelating intracellular iron. Gut Microbes (2020) 12(1):1–18. doi: 10.1080/19490976.2020.1841548
48. Ayoub Moubareck C. Polymyxins and bacterial membranes: a review of antibacterial activity and mechanisms of resistance. Membranes (2020) 10(8):p.181. doi: 10.3390/membranes10080181
49. Falagas ME, Kasiakou SK. Colistin: The revival of polymyxins for the management of multidrug-resistantgram-negative bacterial infections. Clin Infect Dis Off Publ Infect Dis Soc Am (2005) 40:1333–41. doi: 10.1086/429323
50. Kagi T, Naganuma R, Inoue A, Noguchi T, Hamano S, Sekiguchi Y, et al. The polypeptide antibiotic polymyxin B acts as a pro-inflammatory irritant by preferentially targeting macrophages. J Antibiot (Tokyo) (2022) 75(1):29–39. doi: 10.1038/s41429-021-00490-7
51. Trimble MJ, Mlynárčik P, Kolář M, Hancock RE. Polymyxin: alternative mechanisms of action and resistance. Cold Spring Harb Perspect Med (2016) 6(10):a025288. doi: 10.1101/cshperspect.a025288
52. Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol (2014) 5:643. doi: 10.3389/fmicb.2014.00643
53. Longhi MS, Moss A, Jiang ZG, Robson SC. Purinergic signaling during intestinal inflammation. J Mol Med (Berl) (2017) 95(9):915–25. doi: 10.1007/s00109-017-1545-1
54. Lv Y, Luo YY, Ren HW, Li CJ, Xiang ZX, Luan ZL. The role of pregnane X receptor (PXR) in substance metabolism. Front Endocrinol (Lausanne) (2022) 13:959902. doi: 10.3389/fendo.2022.959902
55. Bei WJ, Guo J, Wu HY, Cao Y. Lipid-regulating effect of traditional Chinese medicine: mechanisms of actions. Evidence-Based Complementary Altern Med (2012) 2012:970635. doi: 10.1155/2012/970635
56. Li Y, Wang X, Shen Z. Traditional Chinese medicine for lipid metabolism disorders. Am J Trans Res (2017) 9(5):2038.
57. Wang Y, Yi X, Ghanam K, Zhang S, Zhao T, Zhu X. Berberine decreases cholesterol levels in rats through multiple mechanisms, including inhibition of cholesterol absorption. Metabolism (2014) 63(9):1167–77. doi: 10.1016/j.metabol.2014.05.013
58. Chen J, Zhao H, Yang Y, Liu B, Ni J, Wang W. Lipid-lowering and antioxidant activities of Jiang-Zhi-Ning in traditional Chinese medicine. J Ethnopharmacol (2011) 134(3):919–30. doi: 10.1016/j.jep.2011.01.048
59. Guo J, Bei W, Hu Y, Tang C, He W, Liu X, et al. A new TCM formula FTZ lowers serum cholesterol by regulating HMG-CoA reductase and CYP7A1 in hyperlipidemic rats. J ethnopharmacol (2011) 135(2):299–307. doi: 10.1016/j.jep.2011.03.012
60. Duan J, Dong W, Xie L, Fan S, Xu Y, Li Y. Integrative proteomics-metabolomics strategy reveals the mechanism of hepatotoxicity induced by Fructus Psoraleae. J Proteomics (2020) 221:103767. doi: 10.1016/j.jprot.2020.103767
Keywords: traditional Chinese medicine, Qijudihuang pill, age-related macular degeneration, cholesterol, oxidative stress, inflammation, gut microbiota
Citation: Cao Y, Ibrahim KS, Li X, Wong A, Wu Y, Yu X-D, Zhou X, Tan Z, He Z, Craft JA and Shu X (2023) Chinese medicine, Qijudihuang pill, mediates cholesterol metabolism and regulates gut microbiota in high-fat diet-fed mice, implications for age-related macular degeneration. Front. Immunol. 14:1274401. doi: 10.3389/fimmu.2023.1274401
Received: 08 August 2023; Accepted: 21 September 2023;
Published: 12 October 2023.
Edited by:
Nemat Ali, King Saud University, Saudi ArabiaReviewed by:
Likui Feng, The Rockefeller University, United StatesCopyright © 2023 Cao, Ibrahim, Li, Wong, Wu, Yu, Zhou, Tan, He, Craft and Shu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John A. Craft, Si5BLkNyYWZ0QGdjdS5hYy51aw==; Xinhua Shu, WGluaHVhLlNodUBnY3UuYWMudWs=
†These authors share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.