- Department of Immunology, Institute for Biological Research “Siniša Stanković” - National Institute of Republic of Serbia, University of Belgrade, Belgrade, Serbia
Innate lymphoid cells type 3 (ILC3s) are the first line sentinels at the mucous tissues, where they contribute to the homeostatic immune response in a major way. Also, they have been increasingly appreciated as important modulators of chronic inflammatory and autoimmune responses, both locally and systemically. The proper identification of ILC3 is of utmost importance for meaningful studies on their role in immunity. Flow cytometry is the method of choice for the detection and characterization of ILC3. However, the analysis of ILC3-related papers shows inconsistency in ILC3 phenotypic definition, as different inclusion and exclusion markers are used for their identification. Here, we present these discrepancies in the phenotypic characterization of human and mouse ILC3s. We discuss the pros and cons of using various markers for ILC3 identification. Furthermore, we consider the possibilities for the efficient isolation and propagation of ILC3 from different organs and tissues for in-vitro and in-vivo studies. This paper calls upon uniformity in ILC3 definition, isolation, and propagation for the increased possibility of confluent interpretation of ILC3’s role in immunity.
1 Introduction
Group 3 innate lymphoid cells (ILC3s) have been identified as critical regulators of intestinal homeostasis and immune responses to both commensal and pathogenic bacteria in the gut. Subsequent studies showed that ILC3s are present in a variety of tissues and organs, including the lungs, skin, and lymphoid tissues. ILC3s are now recognized as important players in the immune response against different pathogens and are suggested to interfere with the development of inflammatory and autoimmune diseases (1, 2). The discovery of ILC3 has opened new avenues for the study of the role of the innate immune system in health and disease. Overall, the discovery of these cells represents a significant milestone in our understanding of the innate immune system.
ILCs were first identified in the early 2010s by researchers studying the immune system in the intestinal mucosa. Before being classified as a distinct cell population, they had been identified in mice and humans as cells that lack antigen-specific T-cell or B-cell receptors (TCR or BCR), expressing natural killer (NK) cell markers and producing various cytokines in response to stimulation. There are three populations of ILCs, namely, ILC1, ILC2, and ILC3, broadly corresponding to Th1, Th2, and Th17/Treg cells, respectively. Initially, ILC3s were described as RORγt-dependent NKp46+ NK cells that produce IL-22 and provide mucosal immune defense in the intestine of mice (3–5). At the same time, IL-2-producing NKp44+ NK cells were detected in human mucosa-associated lymphoid tissues, such as tonsils and Peyer’s patches (6). Due to the almost simultaneous discovery by several research groups, IL-22-producing natural cytotoxicity receptor-expressing (NCR+) cells have been referred to as NK22 (6, 7), NCR22 (8), NKR-Lti (9), or ILC22 (10). Given the similarities between IL-22-producing NCR+ ILC and NK cells, researchers sought to elucidate their developmental pathways. Although RORγt+ ILCs were originally thought to be a subpopulation of NK cells, cell fate-mapping studies showed that their developmental pathways were distinct. This conclusion is supported by the fact that NK cells do not express RORγt during development (8, 9). Further distinctions between ILC3 and NK cells are observed at the mature stage. Like T helper (Th) 17 cells, ILC3s express RORγt and aryl hydrocarbon receptor (AhR) and produce a high amount of IL-22 in response to IL-23. On the other hand, NK cells from peripheral or cord blood cultured under Th17 conditions with or without AhR ligands are unable to produce IL-22 (6). Additionally, under Th17-polarizing conditions, IL-22-producing NCR+ ILC cells do not produce IL-17 (6). In light of this, it has been suggested that ILC3s and NK cells undergo distinct differentiation pathways.
Since ILC3s express RORγt, another explanation was that they may develop from lymphoid tissue inducer (LTi) cells, previously known to also express RORγt (9). LTi cells are involved in the formation of lymphoid tissue during fetal development in mice and humans. While LTi cells express IL-17 and IL-22, NKp46+ ILCs express only IL-22 (11, 12). Moreover, LTi cells are distinguished from (NKp46+/−) ILCs by expressing markers such as CD4, CD25, and CCR6 on their surface (13, 14). In contrast to LTi, which are only found during fetal development, “adult LTi,” called LTi-like cells, can be found in the lamina propria of the intestine and in the tonsils, and they arise from bone marrow precursors (15). Although LTi-like cells and LTi are very similar in their expression profile, only the former express OX40L and CD30L (16). Evidence that NCR+ ILCs are a distinct lineage from LTi was provided by studies showing that RORγt+α4β7+ cells develop into CD4+ and CD4− LTi, whereas RORγt+α4β7− give rise to IL-22-producing NKp46+ cells in mice (12). Therefore, the term ILC3 was introduced by Spits and colleagues to describe the group 3 ILCs which are distinct from LTi (17). However, it is still not clear if NCR+ ILC3s have a different developmental pathway from LTi. It is generally presumed that LTi cells arise in the fetal liver and are primarily involved in developing lymphoid tissues, while NCR+ ILC3s develop from bone marrow lymphoid progenitors in the gut and lymphoid tissue. Accordingly, RORγt+CD34+α4β7+ hematopoietic progenitor cells have been defined in humans as IL-22+ ILC3-specific precursors (18). Still, there was a report that LTi cells gave rise to ILC3 in fetal lymph nodes (11), thus suggesting that the development of LTi and ILC3 is not strictly divergent. Furthermore, NCR+ ILC3 can be generated from RORγt+ precursors stemming from LTi-like cells. Indeed, in a study on mice, it was shown that NKp46+RORγt+ cells differentiate directly from LTi-like cells (NKp46−RORγt+) (9). Along the same line, it was demonstrated that human RORγt+NKp46+ cells can be generated in vitro from LTi-like cells obtained from adult tonsils (19).
As in mice, the development of ILC in humans begins with the common lymphoid progenitor (CLP), characterized as lineage(Lin)−CD34+CD45RA+CD10+KIT–, from which all lymphocyte subsets, including ILC, T, and B cells can develop. Intermediate stages of NK development are also thought to differentiate from this progenitor, at least in secondary lymphoid tissues, such as tonsils and lymph nodes (20). ILC differentiation further leads to the early tonsil progenitor cells (EToP), with two distinct phenotypes, EToP1 (Lin−CD34+CD10+KIT−) or EToP2 (Lin−CD34+CD10+KIT+) (21, 22). EToP2 can be divided into two groups based on ST2 (IL-33R, IL1r1) expression and on their ability to form ILC (21). Unlike ST2− EToP, which can give rise to T cells and dendritic cells (DCs) in vitro, ST2+ EToPs give rise exclusively to ILC, both in vitro and in vivo, when isolated from pediatric tonsils and injected into immunodeficient NGS mice (22), and cannot give rise to non-ILC cells. Thus, these cells are the earliest human common ILC progenitors identified to date (22). Two other restricted progenitors were identified, one of which is a Lin–CD34–CD7+IL-7R+KIT+ ILC progenitor (ILCP), found in cord blood, fetal liver, peripheral blood, and other human tissues (23). This progenitor is common for ILC3, LTi, and ILC2 cells. Another progenitor, identified downstream of the ILCP, is CD34–KIT+CD56+, termed restricted ILCP (rILCP) (24), and can give rise to NK cells, ILC1, and ILC3, but not to ILC2 cells (25). This may partially explain the ILC plasticity observed in different tissues and under different conditions. Single-cell RNA sequencing of human ILC3 and ILC1 from the tonsils and intestine and bioinformatics analysis (RNA velocity) recently enabled the identification of a cluster of cells that represent a spectrum of cell conversions between ILC3 and ILC1, with a pronounced bias toward ILC1. Cell transfers into humanized mice demonstrated in-vivo conversion of ILC3 to ILC1, which occurred preferentially in TGF-β-expressing tissues, such as the spleen (26). As observed in mice, the formation of mature human ILC3 subsets not only occurs during fetal development but can also occur after birth locally in tissues, such as adult tonsils and intestine (19). Different developmental pathways and marked plasticity even in adult ILC complicate their classification into subtypes.
2 Literature survey results
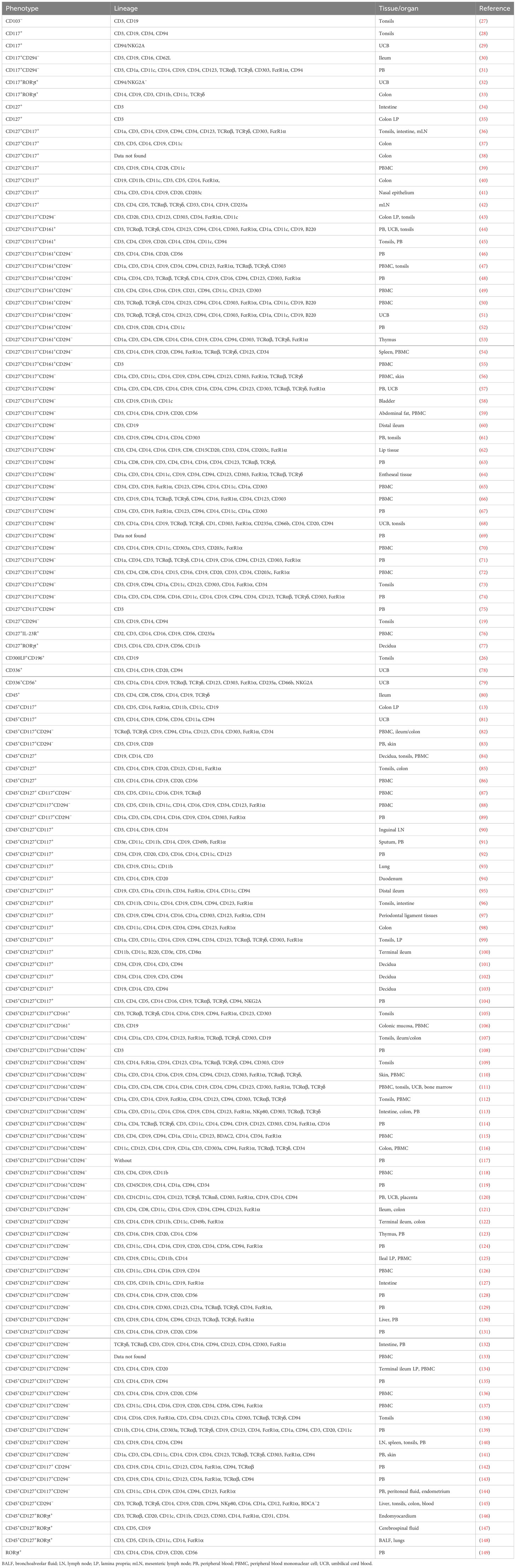
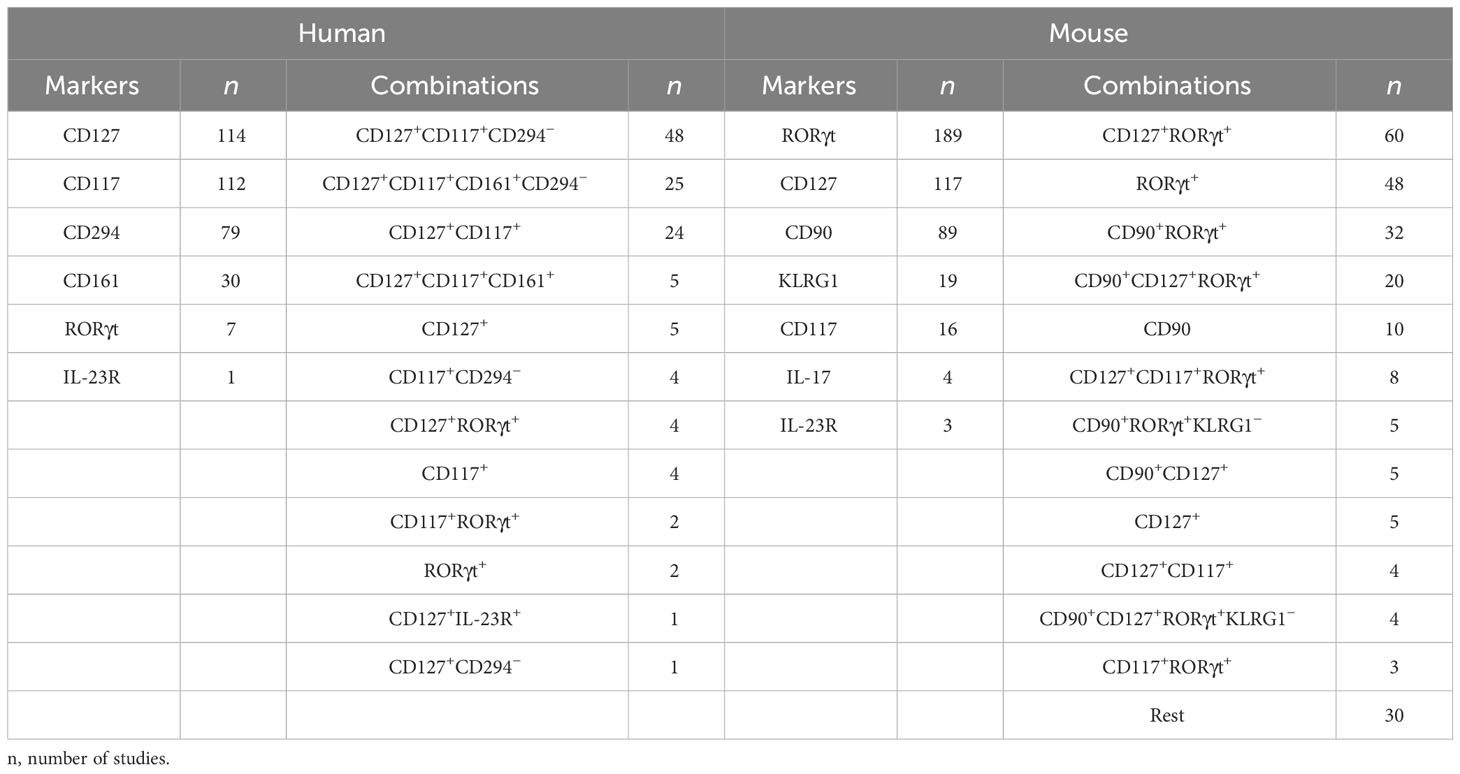
A detailed survey of papers on ILC3 was performed using the PubMed database (https://pubmed.ncbi.nlm.nih.gov/) until 15 June 2023. Searching for “ILC3 mouse” and “ILC3 human” retrieved more than 300 articles each. Papers containing data on ILC3 identification by flow cytometry (121 and 226 for human and mouse ILC3, respectively) were analyzed for the following information: phenotypic markers used for the definition of ILC3, lineage markers used for excluding immune cells other than ILC3, and organ/tissue of ILC3 origin. The obtained information was compiled in Tables 1, 2, dedicated to human and mouse ILC3, respectively. The overview of the tables shows inconsistency in the markers used for the identification of ILC3, as well as of those used in lineage cocktails for the exclusion of non-ILC3 cells. The most commonly used inclusion markers for human ILC3 are CD117, CD127, and CD161, while CD294 is the most frequently used exclusion marker (Figure 1A). The most frequently used marker combinations for the identification of human ILC3 are 1) CD127+CD117+CD294−, 2) CD127+CD117+CD161+CD294−, and 3) CD127+CD117+ (Figure 1B). The most commonly used markers for mouse ILC3 are RORγt, CD127, and CD90 (Figure 1C), while the prevalent combinations are CD127+RORγt+ and CD90+RORγt+ (Figure 1D). A detailed list of the markers and their combinations is given in Table 3. Interestingly, RORγt is frequently used as a single marker for ILC3 identification in mice and is by far the predominantly used marker for ILC3 in mouse, but not in human samples (vide infra). The adequacy of the phenotypic markers used for defining ILC3 in relation to the tissue/organ of their origin is discussed in the following chapters.
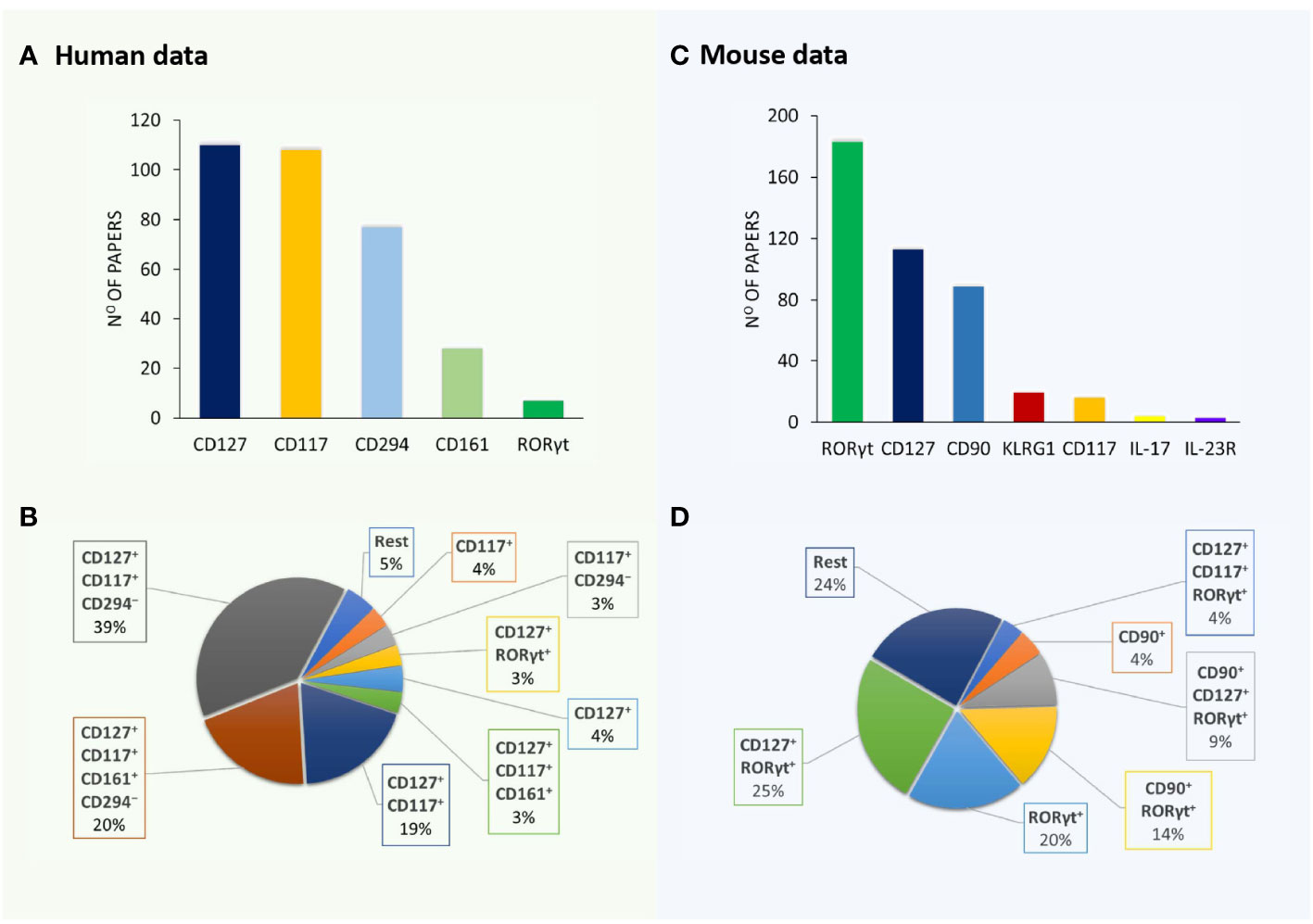
Figure 1 Markers and their combinations used for the identification of ILC3. Visual representation of data from Tables 1, 2. The number of papers in which specific markers were used for the identification of human (A) and mouse (B) ILC3s. The most frequently used marker combinations for the identification of human (C) and mouse (D) ILC3s (% of all papers examined).
3 Lineage cocktails
CD3 is a marker that is targeted by almost all lineage cocktails. Additionally, in some of the mouse studies, CD3 is the only negative marker used for ILC3 identification. The logic behind this strategy is clear when RORγt is used as the key positive ILC3 marker. As RORγt is expressed by T cells and ILC3 (346), the latter can be identified as CD3−RORγt+ cells. It is known that human bone marrow and blood neutrophils, as well as mouse lung neutrophils, can express RORγt (347, 348). However, neutrophils are easily distinguished from the lymphocytic populations by flow cytometry using forward and side scatter. Also, there is a report on a RORγt-expressing DC subset in the spleen (349), while the existence of a similar subset was not confirmed in the intestine (98). Thus, CD3 and RORγt can be used as the basic discriminators of ILC3 in the murine intestine, but not the spleen. As for human samples, it seems that RORγt can be expressed by all ILC subsets, as well as by NK cells (24), thus making it an unsuitable marker for ILC3 discrimination.
Other T-cell markers can be used instead of CD3, such as TCRαβ and TCRγδ. Lineage cocktails used in some studies contained both CD3- and TCR-specific antibodies, even though this combination seems redundant having in mind that CD3 already defines all TCR-positive cells. Additionally, CD4 and CD8a were used as lineage markers in some studies. While there are no data on the expression of CD8 in ILC3, there are studies showing that the LTi ILC3 subset can express CD4 (160, 350), thus making CD4 inadequate as a lineage marker. Also, there are data on the expression of RORγt by CD3+CD4−CD8− cells in patients with systemic lupus erythematosus and psoriasis (351, 352), which contribute to the view that CD3, and not CD4 and CD8, should be used in lineage cocktails. CD5-specific antibodies are also frequently used in mice, as this marker is specific for T cells and a subset of B cells.
The second most commonly used lineage markers are B-cell markers, including B220 (CD45RA) and CD19. CD45RA is an isoform of CD45, a molecule expressed on all types of hematopoietic cells except mature erythrocytes and platelets (353). Although it is expressed in mouse DC (354), naive T cells (355), and T cells undergoing apoptosis (356, 357), it is considered a pan-B-cell marker in mice, present on pre-B cells and at all stages of B-cell maturation (358), making it a suitable negative marker for phenotyping ILC. CD19 is a molecule belonging to the immunoglobulin superfamily, and its expression is specific to normal and neoplastic B cells, as well as follicular DC (359). Both B220 and CD19 molecules are suitable B-cell markers in mice, whereas in humans, B220 is detected on ILC3 in the blood and cord blood, with heterogeneous expression in tonsillar, intestinal, and lung ILC3 (360). Therefore, CD19, and not B220, should be used as a B-cell-related lineage component for human ILC detection.
The most challenging distinction is the one between NK and ILC3 cells. NK cells and ILC share the same progenitor, while the mature forms of ILC also share some markers with NK cells. For instance, ILC1, some ILC3, and NK cells express T-bet. Therefore, it is important to distinguish these cells when detecting ILC3. NK1.1-specific antibodies are frequently used in mice lineage cocktails, while CD94 (Klrd1) is preferentially used in human samples to exclude NK cells, although the use of NKG2A has also been reported in some studies. Additionally, CD56 is used to exclude NK cells and other immune cells able to express this molecule, such as T cells, DC, and monocytes (361). Some researchers also use markers specific for lymphocyte subpopulations, such as CD49b for some T cells (Tr1, NKT), Klrg1 for effector memory CD8+ T and NK cells, or CD27 for activated effector T cells, NK cells, and activated B cells. One important issue is that different mouse strains carry different NK-related markers. For example, the Balb/c strain lacks NK1.1 (362), so CD49b can be used for the determination of NK cells.
CD11b, CD11c, Gr1, Ly6G, Ly6C, and Ly6B are markers that are used in lineage cocktails to eliminate monocytes, DC, granulocytes, macrophages, NK cells, and subsets of T and B cells. Additionally, F4/80 is a relevant marker of murine macrophages. FcϵRIα is also used to detect epidermal Langerhans cells, eosinophils, mast cells, basophils, and various antigen-presenting cells. However, there are reports on ILC3 expressing CD11b and CD11c (27), thus compromising their use in lineage cocktails. Finally, Ter119 is routinely used to exclude erythrocytes from analyses.
Different lineage cocktails can be used while studying ILC3. It is on the researchers to make the best choice for a particular species and tissue in which ILC3s are being detected. It seems that the uncritical use of redundant markers in the lineage cocktails does not improve the efficacy of ILC3 detection, while the use of markers that are potentially expressed by ILC3 increases the risk of excluding some ILC3 subpopulations from the analysis. So, the advice on lineage cocktails might be the following: keep it simple and efficient.
4 Markers used for the discrimination of ILC3
Various markers are used, alone or in combination, to identify ILC3. CD45 is a marker of hematopoietic cells and is, therefore, expressed by ILC3. The use of anti-CD45 antibodies is essential when identifying ILC3 in the intestine, lungs, and other organs and tissues where there is plenty of non-hematopoietic cells. There, gating on CD45+ cells as one of the first steps in flow cytometry analysis is a prerequisite to prevent contamination with non-hematopoietic cells and to get precise data on ILC3. On the other hand, identification of ILC3 in the peripheral blood, spleen, tonsils, or lymph nodes does not require the use of anti-CD45 antibodies, as there are few non-hematopoietic cells, if any. Still, it is clear from Tables 1, 2 that some authors do not use CD45-specific antibodies where necessary, and the results of such studies should be carefully considered.
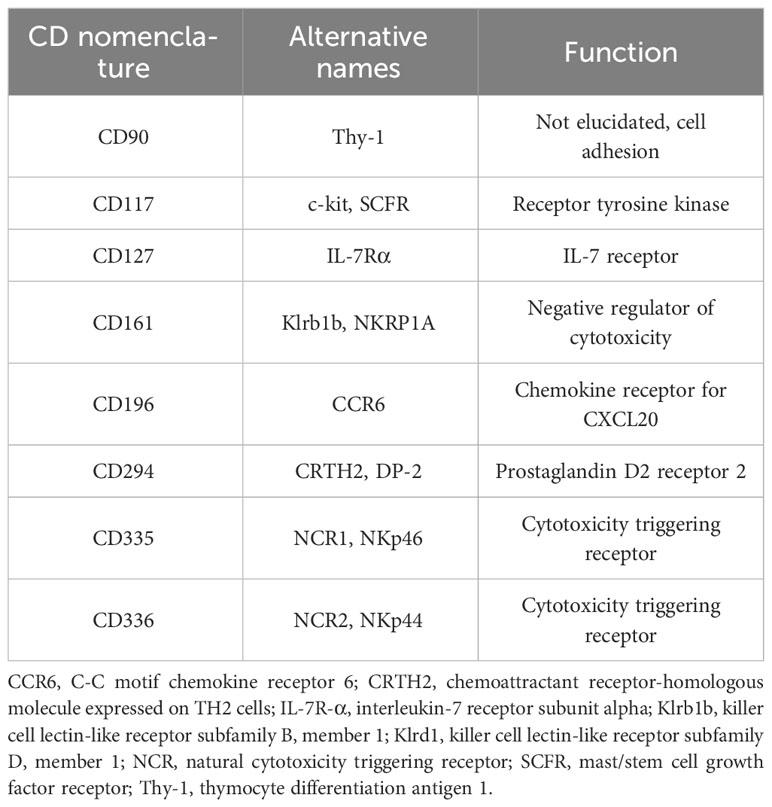
Table 4 presents the most common markers used in ILC3-related studies. Both CD nomenclature and alternative names are used in research papers, thus confusing readers who are not specialized in the ILC3 field. While alternative names have the advantage of functional comprehension and historical connotation, CD designation offers a more systematic approach. Thus, it seems productive to use CD nomenclature as much as possible, or at least to provide information on CD designation in the papers that use alternative names.
4.1 CD127
According to the literature (Figure 1; Table 3), CD127 is the most commonly used ILC3 marker in humans and the second most common in mice. This marker is an α subunit of the IL-7 receptor. IL-7 is important for the early development of T and B cells from bone marrow precursors and for the development of T cells in the thymus (363). As IL-7 is the major growth factor of ILC, but not NK cells, CD127 is expressed by the former (364), but not by the latter, and is thus a valuable marker for the distinction of these two cell types. In addition to ILC, CD127 is expressed in naive (365), memory (366), and regulatory T cells (367). Therefore, using T-cell-specific antibodies in the lineage cocktail is necessary to be able to use CD127 as a reliable marker of ILC. This marker is of particular interest for discriminating between NK and ILC1 cells, as both cell types are known to express T-bet. Still, keeping in mind that NK cells do not express RORγt, CD127 does not appear to be essential for the discrimination between ILC3 and NK cells. It seems that it is reasonable to use CD127 in ILC3 research when analyzing all ILC subsets, while it is not essential for ILC3 subset identification.
4.2 CD117
This marker is used in ILC3 studies in humans almost as frequently as CD127 is, yet its use in mouse studies is not so frequent (Figure 1; Table 3). CD117 is a tyrosine kinase, also known as c-Kit, and it is the receptor for the SCF. CD117 plays an important role in early hematopoiesis and its expression is lost during cellular differentiation (368). However, in addition to mature mast cells and melanocytes, CD117 is also expressed on recently activated human CD8+ T cells (369) and circulating B cells (370). Within the ILC, CD117 is expressed on ILC2 and ILC3 (17). Since CD117 is not exclusively expressed on ILC3, additional markers should be used to distinguish between ILC2 and ILC3, and its use may not be crucial for detecting ILC3.
4.3 CD294
This marker is frequently used in human but not in mouse studies. CD294, also known as CRTH2, is a prostaglandin D2 receptor expressed on Th2 cells and ILC2 in humans, but not in mice. It is also present in both human and mouse eosinophils and basophils (371), while it is not expressed on ILC3 (17). Thus, when phenotyping ILC3, it serves to exclude the ILC2 population. CD294 is most often used in combination with CD127 and CD117, as based on the expression of these three markers in lineage-negative cells, all three ILC subsets can be distinguished. However, when only detecting the ILC3 population, it does not seem necessary to use all of these markers.
4.4 CD90
This marker is very frequently used in mouse but not in human studies, although the mouse and human CD90 proteins are highly similar. Despite this, CD90 expression is negligible in human cells and, thus, cannot be used as a marker for ILC (372). CD90 is a glycophosphatidylinositol-anchored cell surface protein and a member of the immunoglobulin superfamily. While its function is still not fully elucidated, it has been shown to interact with integrins and is therefore presumably important for cell adhesion (274). Within immune cells, CD90 is expressed in T cells, NK cells, and ILCs. It is found on the surface of all subtypes of ILC (373). In Rag1−/− mice, the anti-CD90 antibody is commonly used to deplete ILC (374). Although CD90 is considered a pan-ILC marker, a recent study showed that CD90 is not constitutively expressed in intestinal ILC (375). Additionally, having in mind that CD90 is expressed in all ILC subsets, CD90 does not appear to be the best choice for the detection of ILC3. Furthermore, based on their results of different expressions of CD90 on ILC, Schroeder et al. (375) suggested that CD127 is a much more reliable ILC marker than CD90.
4.5 CD161
CD161 is used as an additional marker to determine ILC3 in humans, but not in mice. CD161 is a human homolog of NK1.1 and is expressed in NK cells, T cells, NKT cells, monocytes, and DCs (376). It is used in combination with CD127 and CD117, with or without CD294 (Figure 1; Table 3). Aside from ILC3, CD161 is also expressed on ILC2 (377). Therefore, this molecule cannot be used as a basic, but only as a supplementary marker for ILC3 phenotyping.
4.6 RORγt
RORγt is by far the most frequently used marker for the discrimination of ILC3 in mouse studies, but not in humans. This molecule is expressed by T cells and ILC3 (346). Human bone marrow and blood neutrophils, as well as mouse lung neutrophils and splenic DCs, can also express RORγt, as previously mentioned (347–349). However, in human secondary lymphoid tissues, including the tonsils, lymph nodes, and spleen, RORγt is expressed in all ILC subsets, as well as in NK cells. In these tissues, a novel Lin−CD34+CD45RA+CD117+IL-1R1+RORγt+ cell population was found, capable of differentiating into all major ILC populations (24). The expression of RORγt was also found in human fetal gut and adult peripheral blood ILC2 cells (377). Although there are no data showing the expression of RORγt in adult human intestinal ILC1 and ILC2, Cogswell et al. (378) have demonstrated the expression of RORγt in ILC1, ILC3, and NK cells in the colon of rhesus macaques, which additionally compromises the use of RORγt in the identification of ILC3 in humans. However, in all of the abovementioned studies, the expression of RORγt was much higher in ILC3 than in other ILC subsets. Thus, RORγt is the crucial marker for the identification of ILC3 in mice and a supplementary one in humans.
5 Molecules important for the identification of ILC3 subsets
5.1 CD196
CD196 or CCR6 is a G protein-coupled receptor expressed on DCs, CD4+ T cells, B cells, and ILCs. CCL20 is a high-affinity ligand for CCR6, and pro-inflammatory cytokines have been shown to induce its expression (379). CCL20 is highly present in inflamed tissues and drives the recruitment of CCR6-expressing immune cells (380). Within ILC3, CCR6 is expressed on LTi and LTi-like cells (381) and is an important marker for identifying these cells. While CCR6-expressing ILC3s are important for the formation of lymphoid organs during development, in adult mice, these cells are usually aggregated with DCs, B cells, and stromal cells in cryptopatches and isolated lymphoid follicles (382). Thus, CD196 is a valuable marker for the identification of ILC3 subpopulations.
5.2 CD335
CD335 or NKp46 is a member of the immunoglobulin superfamily and one of the NCRs of NK cells. While there are three NCRs in humans, namely, NKp30, NKp44, and NKp46, the only NCR described in mice is NKp46 (383). It is expressed on the surface of non-activated and activated NK cells (384), a small population of T lymphocytes, ILC1, and a subset of ILC3, called NCR+ ILC3 (385). Although the functional role of NKp46 on ILC3 is poorly understood, this molecule is essential for the identification of ILC3 subsets.
5.3 CD336
CD336 or NKp44 is another NCR and is expressed only in humans. Like NKp46, it belongs to the immunoglobulin superfamily, but there is no homology between these two molecules (386). While NKp30 and NKp46 are constitutively expressed in human NK cells, NKp44 is expressed only upon activation (384), and the use of anti-NKp44 antibodies has been shown to stimulate cytokine production by human ILC3 (387). In addition to NK cells, NKp44 is also expressed on ILC1 and ILC3 (388) and is thus helpful in defining ILC3 subsets.
6 ILC3 subsets in humans
Three different ILC3 subtypes can be distinguished in humans (Figure 2). LTi cells are the subtype that is most distinct by their developmental pathway and functional properties, and some even consider them to be a separate cell population (385). However, when it comes to their identification by flow cytometry, there is a clear lack of appropriate markers (385). Two other important subtypes are distinguished in humans based on the expression of CD336 (NKp44) and are referred to as NKp44− ILC3 and NKp44+ ILC3 (38, 48, 387, 389). Still, LTi cells are also defined as NKp44−. The distinction between NKp44− ILC3 and LTi in humans can only be performed through analysis of CD45 expression. While NKp44− ILC3s have high CD45 expression, LTi cells express intermediate levels of CD45 (385). Thus, we are in need of a novel marker that would make a clear distinction between NKp44− ILC3 and LTi. An additional subpopulation of NKp44+ ILC3 that loses the ability to express RORγt but keeps the expression of T-bet, and thus becomes closer to the ILC1 phenotype, can be identified as “ex-ILC3” cells (390).
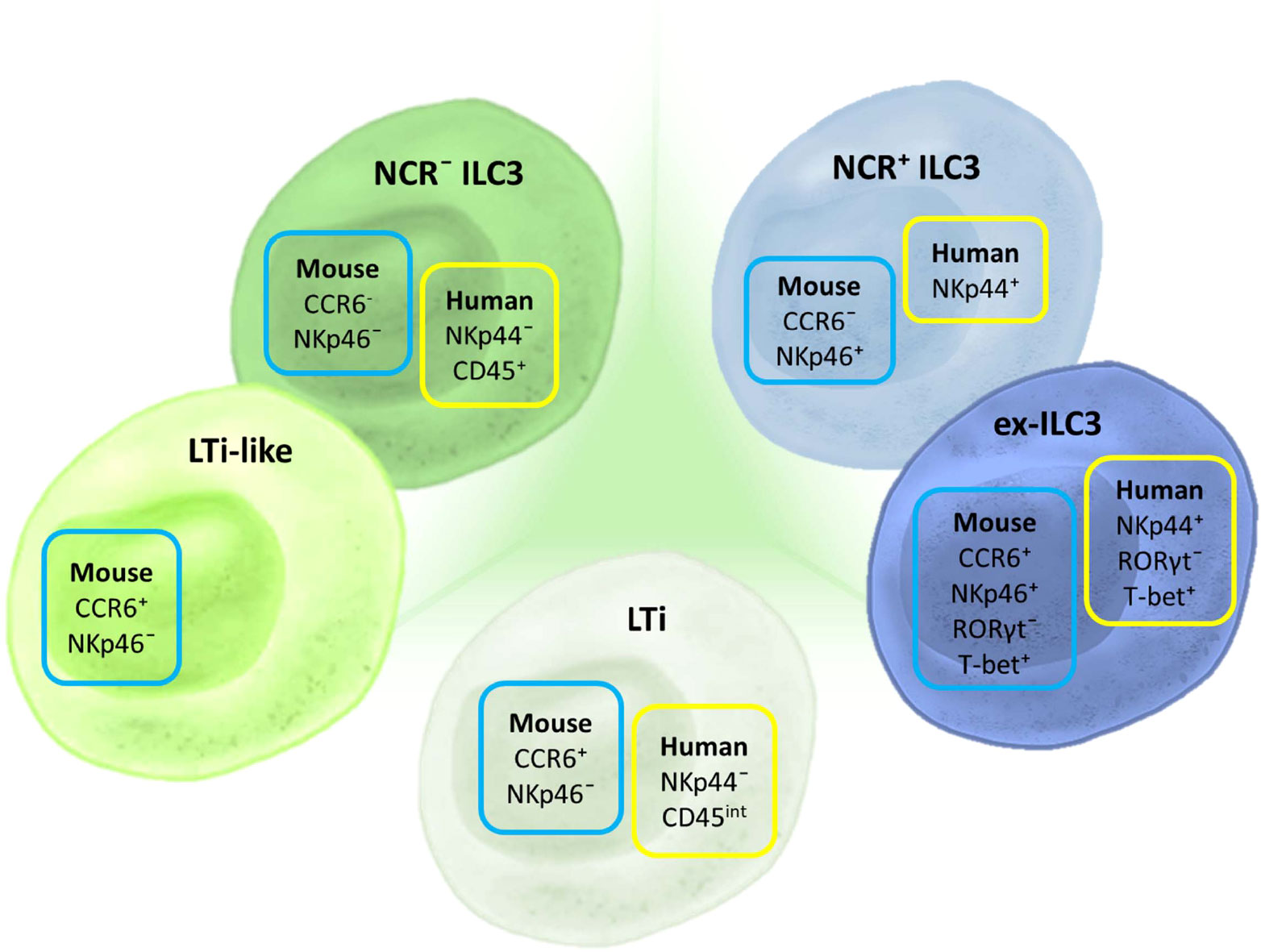
Figure 2 Markers for ILC3 subset discrimination in humans and mice. The basic classification of ILC3 includes three subtypes in humans and mice: LTi, NCR+, and NCR− ILC3. In mice, an additional subpopulation can be identified—LTi-like cells, although it is still not clear if this subpopulation is distinct from NCR− ILC3. Different NCRs are used for characterizing human and mouse ILC3s: NKp44 and NKp46, respectively. CCR6 is an additional marker used for ILC3 classification in mice. Aside from these basic subtypes, there is a defined group of ILCs that shares certain markers and properties of both ILC3 and ILC1. These cells are considered to have converted from ILC3 toward ILC1 and are thus termed “ex-ILC3.” They are distinguished from NCR+ ILC3 by the expression of RORγt and T-bet.
ILC3s in humans are most often found in mucosal tissue and are most abundant in the intestine, where they exhibit several functions (30). For instance, it was shown that after gastrointestinal transplantation, the recipients’ ILC rapidly infiltrated the graft (95). Increased frequencies of human NKp44+ ILC3 were observed in the graft, which has been associated with successful intestinal transplants as it reduces the risk of rejection (99). A similar observation was made in leukemic patients after hematopoietic stem cell transplantation (95). These observations have further strengthened the hypothesis that NKp44+ ILC3s are essential for maintaining homeostasis and protecting the gastrointestinal tract. However, there are conflicting data from some cancer studies that the role of IL-22-producing NKp44+ ILC3 may be detrimental to patients. For example, patients with hepatocarcinoma had more NKp44+ ILC3s than healthy controls (48). On the other hand, NKp44– ILC3s have been described to exacerbate inflammation, especially in inflammatory bowel disease, due to their secretion of IFN-γ and IL-17A (122, 390, 391). Additionally, it was shown that upon activation, human ILC3s were able to induce antigen-specific CD4+ memory T-cell responses (392). Both NKp44+ ILC3 and NKp44– cells can produce IL-17, but the ability to produce IL-22 is mainly restricted to NKp44+ ILC3 (387). By producing IL-22, NKp44+ ILC3s promote tissue integrity, maintain barrier functions, and promote homeostasis (238), particularly in the gastrointestinal tract. IL-22 secretion by NKp44+ ILC3 is activated after various stimuli such as food intake (241, 314), danger signals (111, 393), and changes in the cytokine milieu (6, 7). In addition, NKp44+ ILC3 can interact with other cells through the expression of neuroregulatory receptors which allows them to directly interact with glial cells (246) and goblet cells (394).
The attempt to classify ILC3 subgroups becomes more complex when different tissues are observed. For example, single-cell RNA sequencing analysis has shown that three populations can be identified in the tonsils based on the expression of NKp44, human leukocyte antigen D-related (HLA-DR), and CD62L, and each of these subpopulations has a different cytokine profile (395). HLA-DR+ ILC3s have also been observed in the human intestine (138, 396). Apart from this, NKp44+ ILC3s can be further divided into two populations based on the expression of neuropilin-1 (NRP-1). NRP-1+ ILC3s produce more cytokines than NRP-1− ILC3, and these cells have been detected only in lymphoid tissues and inflamed lung tissue (105). Overall, these data make it necessary to find a consensus in identifying and classifying ILC3s into specific subtypes.
It was generally presumed that all human ILC3s express a common transcription factor RORγt, which implied that RORγt is essential for their functions. However, in patients with RORγt deficiency, IL-22+ but not IL-17+ ILC3s were found. Similarly, in-vitro experiments showed that chemical inhibition of RORγt did not affect the ability of ILC3 to produce IL-22 (40). This suggests that other transcription factors such as AhR or GATA3 may regulate IL-22+ ILC3, as studies in mice have found that these transcription factors are implicated in the regulation of IL-22 production by ILC3 (161, 397).
7 ILC3 subsets in mice
Discrimination among ILC3 subgroups in mice is mainly based on the expression of two cell markers: CD196 (CCR6) and CD335 (NKp46) (Figure 2). There are three basic populations of murine ILC3: NCR+ ILC3 (CCR6−NKp46+), NCR− ILC3 (CCR6−NKp46+), and LTi cells (CCR6+NKp46−). LTi cells identified postnatally are referred to as LTi-like. Additionally, there is a subpopulation of NCR+ ILC3 that is called “ex-ILC3,” which is characterized by diminished RORγt and upregulated T-bet expression.
LTi cells are vital for the formation of secondary lymphoid organs throughout the prenatal period as they produce lymphotoxin-α1β2, which activates lymphoid tissue organizer cells in future lymph node and Peyer’s patch (PP) sites (15, 398). Post-birth, LTi-like cells also play a role in the development of cryptopatches, small lymphoid aggregates in the gut, which can grow into isolated lymphoid follicles (ILFs) in response to microbial signals (398). CCL20, the major CCR6 ligand, is constitutively expressed within the intestinal PP, ILF, and mesenteric lymph nodes (mLNs), confirming the importance of CCR6-mediated signals for secondary lymphoid organogenesis. These signals also help keep leukocytes at sites critical for immune surveillance (399). Accordingly, CCR6+ LTi-like ILC3s are the prevailing ILC3 subset in the PP, mLN, and colon lamina propria (243). In the intestine, LTi-like cells are also important for maintaining local gut homeostasis, through their interactions with both the innate and adaptive immune responses. For instance, LTi-like cells can produce IL-22 and IL-17, which are able to increase the expression of intestinal antimicrobial peptides (350, 400).
Additionally, LTi-like cells, unlike CCR6− ILC3, constitutively express MHC II in the gut while lacking the expression of co-stimulatory molecules CD40, CD80, and CD86, compared with DCs (369). This is one of the features that allows them to play a part in maintaining tolerance through the suppression of commensal antigen-specific T effector cells, enhanced generation of memory T cells, and promotion of mucosal antibody responses [reviewed by Zhong et al. (14)]. LTi-like cells can also be separated into CD4+ and CD4− subsets, even though gene expression analysis showed that there were minimal differences between the two groups—probably not enough to allow functional significance (288). Aside from confirming that CCR6 transcripts were expressed at higher levels in LTi-like cells compared with other ILC3s, the same was shown for an additional chemokine receptor, CXCR5 (288). Finally, fate-mapping studies have demonstrated that while the LTi lineages develop from ILC progenitors that have never expressed the transcription factor PLZF, all other ILC3 subtypes are generated from progenitors that do express it (401). GATA3 is another transcription factor that was shown to be necessary for the development of non-LTi ILCs, while it was not required for the generation of LTi-like cells (402). Additional precursor single-cell analyses (403) and the use of multitranscription factor reporter mice (404) confirmed the distinct origins of ILC and LTi lineages. Based on the differences in gene expression needed for their development, LTi-like cells could be considered a separate group of ILCs, despite sharing similarities in phenotype and function with NKp46+ ILC3.
NKp46+CCR6− ILC3, unlike CCR6+ LTi-like cells, are marked by T-bet expression. Their postnatal development from NKp46−CCR6−RORγt+ progenitors, often labeled as double negative (DN) ILC3, is in part guided by commensal microbiota signals and IL-23, while c-Maf-controlled graded T-bet expression ensures the expression of NKp46 (247, 405). To maintain their phenotype, NKp46+ ILC3s also require Notch signaling, which can be regulated by both AhR (406) and T-bet (407), while it was shown that TGF-β impairs their development (290).
In the gut, NKp46+ ILC3s are predominantly found in the small intestine lamina propria (243), and their localization within the lamina propria villi is influenced by CXCR6 (160). NKp46+ ILC3s also make up most of the ILC3s in the cecum and are necessary for its homeostasis (289). NKp46 has mostly been studied as one of the NCRs that are expressed by NK cells and whose activation by various pathogen-derived ligands can trigger NK cell-mediated cytotoxicity accomplished by the secretion of perforin or proinflammatory cytokines such as IFN-γ and TNF or through activation-induced cell death. Unlike NK cells, it seems that intestinal NKp46+ ILC3s are not able to perform these functions, while they do produce IL-22 to fight intestinal inflammation (3), similar to LTi-like cells. Interestingly, questions about the redundancy of the NKp46+ subgroup of ILC3s have been brought up in some experimental settings. For instance, NKp46+ ILC3s were shown to be inessential in the fight with a Citrobacter rodentium infection, when compared with LTi-like ILC3s (256). More specifically, IL-22 was necessary for the resistance to C. rodentium, but IL-22 production by NKp46+ ILC3 was not (289). However, NKp46+ ILC3s also have the potential to secrete GM-CSF and were crucial in promoting the accumulation and activation of inflammatory monocytes in an anti-CD40-induced colitis model (256).
A fate-mapping study by Viant et al. (290) demonstrated a notable proportion of NKp46− ILC3 used to express NKp46, and this phenomenon was predominantly observed in mucosal tissues but not in the bone marrow. Furthermore, an RORγt fate-mapping study revealed that, in some inflammatory conditions, NKp46+ ILC3 can significantly lower or lose RORγt expression, while T-bet expression increases. At the same time, these “ex-ILC3s” start producing IFN-γ and upregulate perforin and granzyme B expression, becoming more akin to ILC1 (9). Following Salmonella typhimurium infection, these cells were the main source of IFN-γ in the small intestine (405), which implies an adaptive role in microbial defense. On the other hand, NKp46+ ILC3 plasticity is regulated by c-Maf, which suppresses their conversion into the ILC1-like state by restraining T-bet expression and supporting RORγt activity (219, 247). Finally, the transcriptional profile of NKp46+ ILC3 was shown to have features in between those of NKp46− LTi-like ILC3 and NKp46+ ILC1 (while sharing more transcripts with NKp46+ ILC1). This might be the basis of their plasticity potential, which is probably shaped by their specific microenvironment (288).
8 ILC3 sorting and culture
Our literature survey shows that a consensus about the minimal or optimal panel of surface markers for ILC3 sorting is still missing. However, in most sorting protocols, the live/dead discrimination and doublet exclusion seem to be mandatory. Some authors suggest that before cell sorting, enrichment of CD90+ (233) or Lin− cells (408) should take place. The most efficient sorting of mouse ILC3 occurs in samples obtained from mice that carry a fluorescently labeled reporter RORγt. In this case, the sorting protocol might include only a CD3 staining, after which ILC3s are sorted as CD3−RORγt+ cells (the exclusion of Th17 cells is achieved). However, most of the protocols include additional surface markers—ILC3 can be sorted as CD45intCD90highRORγt+ (275) or as Lin−CD45+CD90.2+RORγt+ (270). In the studies not using RORγt reporter mice, investigators tend to use as many markers as they can for the determination of ILC3. For example, Zhou et al. (96) sorted an enriched population of ILC3 as Lin−CD45lowCD90.2highCD127+KLRG1−. However, other groups used fewer denominators for the enrichment of ILC3, for example, the CD3− and CD90highCD45low gating (254) or complete lineage-negative population coupled with CD90high and CD45low markers (238, 269). Some groups use KLRG1 as an additional discriminative factor for ILC3 enrichment in the following panel Lin−CD90.2+KLRG1− (330) or Lin−CD45+CD90.2+KLRG1− (128). As recent data indicate that surface CD90 can be downregulated when ILC3s are functionally active, the usage of CD90 for ILC3 sorting is questionable (375). CD127 and ST2 are also used to determine ILC3 for sorting in addition to Lin−CD45+CD4−. The same study uses the NKp46 marker to sort-purify NCR+ and NCR− ILC3s (122). In all articles that we have analyzed so far, the percentage of acquired RORγt+ cells after ILC3 sorting (without the use of knock-in fluorescent labeling of RORγt) was not determined. Therefore, cells sorted in such a way can be referred to as enriched ILC3s, or just ILCs (as they are composed out of all three ILC types). The recommendation for future studies where these cells are used in vitro would be to evaluate the ILC distribution in post-sorted samples before engaging in any treatment or stimulation.
Human ILC3 cell sorting tends to be more difficult, as their surface markers may be different in peripheral blood mononuclear cells (PBMCs) and within the tissues. Also, unlike in mice, human ILC2 and ILC3 cannot exclusively be defined by transcription factors GATA3 and RORγt (409); therefore, there is no need for confirmation in post-sort samples. In general, ILC3 sorting from the PBMC or colon was performed according to the surface markers Lin−CD45+CD127+CRTH2−CD117+ (410, 411). However, when ILC3s are sorted from the lungs, additional markers were included such as CD16−NKG2A− and CD161+ (411).
ILC3 sorting is usually done in order to perform single-cell RNA sequencing, in-vitro culture, or cell transfer. In-vitro culture of enriched human ILC3 is a prolonged process that requires several growth factors (SCF, IL-7, Flt-3L, IL-2, IL-15). The success of ILC3 propagation in vitro can depend upon a layer of cells to support ILC3 proliferation (such as mesenchymal stem cells) (79). Mouse-enriched ILC3s are usually cultured with SCF and IL-7 and on occasion with the addition of IL-1β, IL-12, or IL-23. In-vitro culture and stimulation of ILC3 with PMA/ionomycin/brefeldin is done in order to detect ILC3-derived cytokines. However, this can lead to the downregulation of ILC3-specific surface markers, thus interfering with flow cytometry analysis of post-treatment ILC3 (412). For performing a cell transfer, ILC3s are usually used freshly sorted from cell suspension derived from intestinal lamina propria or the lungs. Lamina propria is most often chosen as the source of ILC3 because of the abundance of ILC3s in the innate immune cell compartment. As the numbers of obtained ILC3 from tissues are very low, researchers use from 100,000 to 500,000 cells per transfer (233, 267). Having in mind the required precision in ILC3 phenotyping, sorting, and culture conditions and the paucity of tissue ILC3, as well as the limited availability of organs/tissues for their isolation, the use of ILC3 for human cell therapy seems to be complex and unrealistic. Still, alternative strategies, such as propagating tissue-like ILC3 from CD34+ hematopoietic progenitors (79), could overcome the above-stated obstacles on the road toward ILC3-based therapy.
9 Conclusions
The ILC3 research field is still relatively novel, and ILC3 identification by flow cytometry is not yet standardized. The expanding complexity of phenotypic determinants of ILC3 and the tissue specificities of ILC3 contribute to the discrepancies in protocols used by different research groups. Through analysis of the current state of phenotypic characterization of ILC3 by flow cytometry, we conclude that markers for the identification of ILC3 should be chosen in relation to the study species and design, as well as to the tissue of origin. The following are our suggestions for the identification of ILC3 by flow cytometry.
9.1 Mice: CD3−RORγt+
RORγt is the most reliable marker for ILC3 in mice. It is enough to use CD3 as the lineage marker in combination with RORγt in most of the studies. Additional lineage markers seem redundant as long as samples are not obtained from the lungs or spleen when neutrophil or myeloid/DC exclusion is needed. CD127 is required as an additional positive marker only if ILC1 and ILC2 are identified in parallel. CD90 should not be used as an exclusive ILC3 marker.
9.2 Humans: CD3−CD127+CD117+CD294−
CD127 in combination with T-cell-specific lineage markers is a proper way to distinguish human ILC from NK cells. CD294 is an ILC2-specific marker and is a good marker to discriminate between ILC2 and ILC3. As for the distinction of ILC1, CD117 is expressed on ILC2 and ILC3, but not on ILC1. The addition of CD161 is not necessary. It can be used instead of CD127, but in that case, additional lineage markers for myeloid cells are required. RORγt cannot be used for distinguishing ILC3 from other innate lymphocytes in humans.
9.3 General
There is no need to use CD45 when analyzing cells isolated from the blood, lymph nodes, spleen, or tonsils, but it is necessary to use it in quantitative analysis of tissue-residing ILC3.
In a number of studies analyzed in this review, we observe a superfluous use of lineage cocktail markers. Although unnecessary, such overapplication of markers does not jeopardize proper ILC3 identification. On the other hand, there are studies in which ILC3 is identified by an insufficient number of markers or inappropriate marker combinations. This raises the question of the accuracy of the results and the conclusions of such studies. We hope that this paper will help researchers interested in ILC3-related studies to tailor their research design to the best of their specific needs.
Author contributions
ĐMil: Conceptualization, Funding acquisition, Writing – original draft, Writing – review & editing. IK: Visualization, Writing – original draft, Writing – review & editing. SS: Visualization, Writing – original draft. BJ: Writing – original draft, Writing – review & editing. DMic: Writing – original draft, Writing – review & editing. IS: Conceptualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Ministry of Science, Technological Development, and Innovations, Republic of Serbia, contract No. 451-03-47/2023-01/200007, and Science Fund, Republic of Serbia, Ideas Program, project GUTtoAID, contract No. 7742898.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Miljković Đ, Jevtić B, Stojanović I, Dimitrijević M. ILC3, a central innate immune component of the gut-brain axis in multiple sclerosis. Front Immunol (2021) 12:657622. doi: 10.3389/fimmu.2021.657622
2. Stojanović I, Saksida T, Miljković Đ, Pejnović N. Modulation of intestinal ILC3 for the treatment of type 1 diabetes. Front Immunol (2021) 12:653560. doi: 10.3389/fimmu.2021.653560
3. Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity (2008) 29(6):958–70. doi: 10.1016/j.immuni.2008.11.001
4. Luci C, Reynders A, Ivanov II, Cognet C, Chiche L, Chasson L, et al. Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat Immunol (2009) 10(1):75–82. doi: 10.1038/ni.1681
5. Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, et al. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol (2009) 10(1):83–91. doi: 10.1038/ni.1684
6. Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JK, et al. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature (2009) 457(7230):722–5. doi: 10.1038/nature07537
7. Cella M, Otero K, Colonna M. Expansion of human NK-22 cells with IL-7, IL-2, and IL-1beta reveals intrinsic functional plasticity. Proc Natl Acad Sci U.S.A. (2010) 107(24):10961–6. doi: 10.1073/pnas.1005641107
8. Satoh-Takayama N, Lesjean-Pottier S, Vieira P, Sawa S, Eberl G, Vosshenrich CA, et al. IL-7 and IL-15 independently program the differentiation of intestinal CD3-NKp46+ cell subsets from Id2-dependent precursors. J Exp Med (2010) 207(2):273–80. doi: 10.1084/jem.20092029
9. Vonarbourg C, Mortha A, Bui VL, Hernandez PP, Kiss EA, Hoyler T, et al. Regulated expression of nuclear receptor RORγt confers distinct functional fates to NK cell receptor-expressing RORγt(+) innate lymphocytes. Immunity (2010) 33(5):736–51. doi: 10.1016/j.immuni.2010.10.017
10. Spits H, Di Santo JP. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat Immunol (2011) 12(1):21–7. doi: 10.1038/ni.1962
11. Cupedo T, Crellin NK, Papazian N, Rombouts EJ, Weijer K, Grogan JL, et al. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nat Immunol (2009) 10(1):66–74. doi: 10.1038/ni.1668
12. Sawa S, Lochner M, Satoh-Takayama N, Dulauroy S, Bérard M, Kleinschek M, et al. RORγt+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol (2011) 12(4):320–6. doi: 10.1038/ni.2002
13. Hepworth MR, Fung TC, Masur SH, Kelsen JR, McConnell FM, Dubrot J, et al. Immune tolerance. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria-specific CD4+ T cells. Science (2015) 348(6238):1031–5. doi: 10.1126/science.aaa4812
14. Zhong C, Zheng M, Zhu J. Lymphoid tissue inducer-A divergent member of the ILC family. Cytokine Growth Factor Rev (2018) 42:5–12. doi: 10.1016/j.cytogfr.2018.02.004
15. van de Pavert SA. Lymphoid Tissue inducer (LTi) cell ontogeny and functioning in embryo and adult. BioMed J (2021) 44(2):123–32. doi: 10.1016/j.bj.2020.12.003
16. Kim MY, Anderson G, White A, Jenkinson E, Arlt W, Martensson IL, et al. OX40 ligand and CD30 ligand are expressed on adult but not neonatal CD4+CD3- inducer cells: evidence that IL-7 signals regulate CD30 ligand but not OX40 ligand expression. J Immunol (2005) 174(11):6686–91. doi: 10.4049/jimmunol.174.11.6686
17. Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells–a proposal for uniform nomenclature. Nat Rev Immunol (2013) 13(2):145–9. doi: 10.1038/nri3365
18. Montaldo E, Teixeira-Alves LG, Glatzer T, Durek P, Stervbo U, Hamann W, et al. Human RORγt(+)CD34(+) cells are lineage-specified progenitors of group 3 RORγt(+) innate lymphoid cells. Immunity (2014) 41(6):988–1000. doi: 10.1016/j.immuni.2014.11.010
19. Crellin NK, Trifari S, Kaplan CD, Cupedo T, Spits H. Human NKp44+IL-22+ cells and LTi-like cells constitute a stable RORC+ lineage distinct from conventional natural killer cells. J Exp Med (2010) 207(2):281–90. doi: 10.1084/jem.20091509
20. Scoville SD, Freud AG, Caligiuri MA. Modeling human natural killer cell development in the era of innate lymphoid cells. Front Immunol (2017) 8:360. doi: 10.3389/fimmu.2017.00360
21. Freud AG, Yokohama A, Becknell B, Lee MT, Mao HC, Ferketich AK, et al. Evidence for discrete stages of human natural killer cell differentiation in vivo. J Exp Med (2006) 203(4):1033–43. doi: 10.1084/jem.20052507
22. Scoville SD, Freud AG, Caligiuri MA. Cellular pathways in the development of human and murine innate lymphoid cells. Curr Opin Immunol (2019) 56:100–6. doi: 10.1016/j.coi.2018.11.003
23. Lim AI, Li Y, Lopez-Lastra S, Stadhouders R, Paul F, Casrouge A, et al. Systemic human ILC precursors provide a substrate for tissue ILC differentiation. Cell (2017) 168(6):1086–1100.e10. doi: 10.1016/j.cell.2017.02.021
24. Scoville SD, Mundy-Bosse BL, Zhang MH, Chen L, Zhang X, Keller KA, et al. A progenitor cell expressing transcription factor RORγt generates all human innate lymphoid cell subsets. Immunity (2016) 44(5):1140–50. doi: 10.1016/j.immuni.2016.04.007
25. Chen L, Youssef Y, Robinson C, Ernst GF, Carson MY, Young KA, et al. CD56 expression marks human group 2 innate lymphoid cell divergence from a shared NK cell and group 3 innate lymphoid cell developmental pathway. Immunity (2018) 49(3):464–476.e4. doi: 10.1016/j.immuni.2018.08.010
26. Cella M, Gamini R, Sécca C, Collins PL, Zhao S, Peng V, et al. Subsets of ILC3-ILC1-like cells generate a diversity spectrum of innate lymphoid cells in human mucosal tissues. Nat Immunol (2019) 20(8):980–91. doi: 10.1038/s41590-019-0425-y
27. Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, Newberry RD, et al. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity (2013) 38(4):769–81. doi: 10.1016/j.immuni.2013.02.010
28. Victor AR, Nalin AP, Dong W, McClory S, Wei M, Mao C, et al. IL-18 drives ILC3 proliferation and promotes IL-22 production via NF-κB. J Immunol (2017) 199(7):2333–42. doi: 10.4049/jimmunol.1601554
29. Damele L, Amaro A, Serio A, Luchetti S, Pfeffer U, Mingari MC, et al. EZH1/2 inhibitors favor ILC3 development from human HSPC-CD34+ Cells. Cancers (Basel) (2021) 13(2):319. doi: 10.3390/cancers13020319
30. Hoorweg K, Peters CP, Cornelissen F, Aparicio-Domingo P, Papazian N, Kazemier G, et al. Functional differences between human NKp44(-) and NKp44(+) RORC(+) innate lymphoid cells. Front Immunol (2012) 3:72. doi: 10.3389/fimmu.2012.00072
31. Munneke JM, Björklund AT, Mjösberg JM, Garming-Legert K, Bernink JH, Blom B, et al. Activated innate lymphoid cells are associated with a reduced susceptibility to graft-versus-host disease. Blood (2014) 124(5):812–21. doi: 10.1182/blood-2013-11-536888
32. Damele L, Montaldo E, Moretta L, Vitale C, Mingari MC. Effect of tyrosin kinase inhibitors on NK cell and ILC3 development and function. Front Immunol (2018) 9:2433. doi: 10.3389/fimmu.2018.02433
33. Longman RS, Diehl GE, Victorio DA, Huh JR, Galan C, Miraldi ER, et al. CX3CR1+ mononuclear phagocytes support colitis-associated innate lymphoid cell production of IL-22. J Exp Med (2014) 211(8):1571–83. doi: 10.1084/jem.20140678
34. Talayero P, Mancebo E, Calvo-Pulido J, Rodríguez-Muñoz S, Bernardo I, Laguna-Goya R, et al. Innate lymphoid cells groups 1 and 3 in the epithelial compartment of functional human intestinal allografts. Am J Transplant (2016) 16(1):72–82. doi: 10.1111/ajt.13435
35. Powell N, Lo JW, Biancheri P, Vossenkämper A, Pantazi E, Walker AW, et al. Interleukin 6 increases production of cytokines by colonic innate lymphoid cells in mice and patients with chronic intestinal inflammation. Gastroenterology (2015) 149(2):456–67.e15. doi: 10.1053/j.gastro.2015.04.017
36. Bernink JH, Krabbendam L, Germar K, de Jong E, Gronke K, Kofoed-Nielsen M, et al. Interleukin-12 and -23 control plasticity of CD127(+) group 1 and group 3 innate lymphoid cells in the intestinal lamina propria. Immunity (2015) 43(1):146–60. doi: 10.1016/j.immuni.2015.06.019
37. Castellanos JG, Woo V, Viladomiu M, Putzel G, Lima S, Diehl GE, et al. Microbiota-induced TNF-like ligand 1A drives group 3 innate lymphoid cell-mediated barrier protection and intestinal T cell activation during colitis. Immunity (2018) 49(6):1077–1089.e5. doi: 10.1016/j.immuni.2018.10.014
38. Seidelin JB, Bahl MI, Licht TR, Mead BE, Karp JM, Johansen JV, et al. Acute experimental barrier injury triggers ulcerative colitis-specific innate hyperresponsiveness and ulcerative colitis-type microbiome changes in humans. Cell Mol Gastroenterol Hepatol (2021) 12(4):1281–96. doi: 10.1016/j.jcmgh.2021.06.002
39. Le Coz C, Nolan BE, Trofa M, Kamsheh AM, Khokha MK, Lakhani SA, et al. Cytotoxic T-lymphocyte-associated protein 4 haploinsufficiency-associated inflammation can occur independently of T-cell hyperproliferation. Front Immunol (2018) 9:1715. doi: 10.3389/fimmu.2018.01715
40. Withers DR, Hepworth MR, Wang X, Mackley EC, Halford EE, Dutton EE, et al. Transient inhibition of RORγt therapeutically limits intestinal inflammation by reducing TH17 cells and preserving group 3 innate lymphoid cells. Nat Med (2016) 22(3):319–23. doi: 10.1038/nm.4046
41. Latek M, Lacwik P, Molinska K, Blauz A, Lach J, Rychlik B, et al. Effect of an intranasal corticosteroid on quality of life and local microbiome in young children with chronic rhinosinusitis: A randomized clinical trial. JAMA Pediatr (2023) 177(4):345–52. doi: 10.1001/jamapediatrics.2022.6172
42. Rethacker L, Boy M, Bisio V, Roussin F, Denizeau J, Vincent-Salomon A, et al. Innate lymphoid cells: NK and cytotoxic ILC3 subsets infiltrate metastatic breast cancer lymph nodes. Oncoimmunology (2022) 11(1):2057396. doi: 10.1080/2162402X.2022.2057396
43. Castleman MJ, Dillon SM, Purba CM, Cogswell AC, Kibbie JJ, McCarter MD, et al. Commensal and Pathogenic Bacteria Indirectly Induce IL-22 but Not IFNγ Production From Human Colonic ILC3 via Multiple Mechanisms. Front Immunol (2019) 10:649. doi: 10.3389/fimmu.2019.00649
44. Eken A, Yetkin MF, Vural A, Okus FZ, Erdem S, Azizoglu ZB, et al. Fingolimod alters tissue distribution and cytokine production of human and murine innate lymphoid cells. Front Immunol (2019) 10:217. doi: 10.3389/fimmu.2019.00217
45. Komlósi ZI, Kovács N, van de Veen W, Kirsch AI, FAhRner HB, Wawrzyniak M, et al. Human CD40 ligand-expressing type 3 innate lymphoid cells induce IL-10-producing immature transitional regulatory B cells. J Allergy Clin Immunol (2018) 142(1):178–194.e11. doi: 10.1016/j.jaci.2017.07.046
46. Ssekamatte P, Nakibuule M, Nabatanzi R, Egesa M, Musubika C, Bbuye M, et al. Type 2 diabetes mellitus and latent tuberculosis infection moderately influence innate lymphoid cell immune responses in Uganda. Front Immunol (2021) 12:716819. doi: 10.3389/fimmu.2021.716819
47. Kindstedt E, Koskinen Holm C, Palmqvist P, Sjöström M, Lejon K, Lundberg P. Innate lymphoid cells are present in gingivitis and periodontitis. J Periodontol (2019) 90(2):200–7. doi: 10.1002/JPER.17-0750
48. Heinrich B, Ruf B, Subramanyam V, Myojin Y, Lai CW, Craig AJ, et al. Checkpoint inhibitors modulate plasticity of innate lymphoid cells in peripheral blood of patients with hepatocellular carcinoma. Front Immunol (2022) 13:849958. doi: 10.3389/fimmu.2022.849958
49. Darboe A, Nielsen CM, Wolf AS, Wildfire J, Danso E, Sonko B, et al. Age-related dynamics of circulating innate lymphoid cells in an african population. Front Immunol (2020) 11:594107. doi: 10.3389/fimmu.2020.594107
50. Eken A, Cansever M, Somekh I, Mizoguchi Y, Zietara N, Okus FZ, et al. Genetic deficiency and biochemical inhibition of ITK affect human th17, treg, and innate lymphoid cells. J Clin Immunol (2019) 39(4):391–400. doi: 10.1007/s10875-019-00632-5
51. Haliloglu Y, Ozcan A, Erdem S, Azizoglu ZB, Bicer A, Ozarslan OY, et al. Characterization of cord blood CD3+ TCRVα7.2+ CD161high T and innate lymphoid cells in the pregnancies with gestational diabetes, morbidly adherent placenta, and pregnancy hypertension diseases. Am J Reprod Immunol (2022) 88(1):e13555. doi: 10.1111/aji.13555
52. Geier CB, Kraupp S, Bra D, Eibl MM, Farmer JR, Csomos K, et al. Reduced numbers of circulating group 2 innate lymphoid cells in patients with common variable immunodeficiency. Eur J Immunol (2017) 47(11):1959–69. doi: 10.1002/eji.201746961
53. Nagasawa M, Germar K, Blom B, Spits H. Human CD5+ Innate lymphoid cells are functionally immature and their development from CD34+ Progenitor cells is regulated by id2. Front Immunol (2017) 8:1047. doi: 10.3389/fimmu.2017.01047
54. Audia S, Moulinet T, Ciudad-Bonté M, Samson M, Facy O, Ortega-Deballon P, et al. Altered distribution and function of splenic innate lymphoid cells in adult chronic immune thrombocytopenia. J Autoimmun (2018) 93:139–44. doi: 10.1016/j.jaut.2018.07.015
55. Eken A, Cansever M, Okus FZ, Erdem S, Nain E, Azizoglu ZB, et al. ILC3 deficiency and generalized ILC abnormalities in DOCK8-deficient patients. Allergy (2020) 75(4):921–32. doi: 10.1111/all.14081
56. Petrasca A, Hambly R, Molloy O, Kearns S, Moran B, Smith CM, et al. Innate lymphoid cell (ILC) subsets are enriched in the skin of patients with hidradenitis suppurativa. PloS One (2023) 18(2):e0281688. doi: 10.1371/journal.pone.0281688
57. Nagasawa M, Heesters BA, Kradolfer CMA, Krabbendam L, Martinez-Gonzalez I, de Bruijn MJW, et al. KLRG1 and NKp46 discriminate subpopulations of human CD117+CRTH2- ILC biased toward ILC2 or ILC3. J Exp Med (2019) 216(8):1762–76. doi: 10.1084/jem.20190490
58. Riding AM, Loudon KW, Guo A, Ferdinand JR, Lok LSC, Richoz N, et al. Group 3 innate lymphocytes make a distinct contribution to type 17 immunity in bladder defence. iScience (2022) 25(7):104660. doi: 10.1016/j.isci.2022.104660
59. Sattler FR, Mert M, Sankaranarayanan I, Mack WJ, Galle-Treger L, Gonzalez E, et al. Feasibility of quantifying change in immune white cells in abdominal adipose tissue in response to an immune modulator in clinical obesity. PloS One (2020) 15(9):e0237496. doi: 10.1371/journal.pone.0237496
60. Pucci Molineris M, González Polo V, Rumbo C, Fuxman C, Lowestein C, Nachman F, et al. Acute cellular rejection in small-bowel transplantation impairs NCR+ innate lymphoid cell subpopulation 3/interleukin 22 axis. Transpl Immunol (2020) 60:101288. doi: 10.1016/j.trim.2020.101288
61. Campana S, De Pasquale C, Barberi C, Oliveri D, Sidoti Migliore G, Galletti B, et al. Circulating ILC precursors expressing CD62L exhibit a type 2 signature distinctly decreased in psoriatic patients. Eur J Immunol (2021) 51(7):1792–8. doi: 10.1002/eji.202048893
62. Simmerman E, Qin X, Marshall B, Perry L, Cai L, Wang T, et al. Innate lymphoid cells: a paradigm for low SSI in cleft lip repair. J Surg Res (2016) 205(2):312–7. doi: 10.1016/j.jss.2016.06.081
63. Pearson C, Thornton EE, McKenzie B, Schaupp AL, Huskens N, Griseri T, et al. ILC3 GM-CSF production and mobilisation orchestrate acute intestinal inflammation. Elife (2016) 5:e10066. doi: 10.7554/eLife.10066
64. Cuthbert RJ, Fragkakis EM, Dunsmuir R, Li Z, Coles M, Marzo-Ortega H, et al. Brief report: group 3 innate lymphoid cells in human enthesis. Arthritis Rheumatol (2017) 69(9):1816–22. doi: 10.1002/art.40150
65. Wang S, Li J, Wu S, Cheng L, Shen Y, Ma W, et al. Type 3 innate lymphoid cell: a new player in liver fibrosis progression. Clin Sci (Lond) (2018) 132(24):2565–82. doi: 10.1042/CS20180482
66. Carvelli J, Piperoglou C, Bourenne J, Farnarier C, Banzet N, Demerlé C, et al. Imbalance of circulating innate lymphoid cell subpopulations in patients with septic shock. Front Immunol (2019) 10:2179. doi: 10.3389/fimmu.2019.02179
67. Wang SQ, Shen Y, Li J, Liu Y, Cheng LS, Wu SD, et al. Entecavir-induced interferon-λ1 suppresses type 2 innate lymphoid cells in patients with hepatitis B virus-related liver cirrhosis. J Viral Hepat (2021) 28(5):795–808. doi: 10.1111/jvh.13476
68. Bennstein SB, Scherenschlich N, Weinhold S, Manser AR, Noll A, Raba K, et al. Transcriptional and functional characterization of neonatal circulating Innate Lymphoid Cells. Stem Cells Transl Med (2021) 10(6):867–82. doi: 10.1002/sctm.20-0300
69. Braudeau C, Amouriaux K, Néel A, Herbreteau G, Salabert N, Rimbert M, et al. Persistent deficiency of circulating mucosal-associated invariant T (MAIT) cells in ANCA-associated vasculitis. J Autoimmun (2016) 70:73–9. doi: 10.1016/j.jaut.2016.03.015
70. Xuan X, Zhou J, Tian Z, Lin Y, Song J, Ruan Z, et al. ILC3 cells promote the proliferation and invasion of pancreatic cancer cells through IL-22/AKT signaling. Clin Transl Oncol (2020) 22(4):563–75. doi: 10.1007/s12094-019-02160-5
71. Heinrich B, Gertz EM, Schäffer AA, Craig A, Ruf B, Subramanyam V, et al. The tumour microenvironment shapes innate lymphoid cells in patients with hepatocellular carcinoma. Gut (2022) 71(6):1161–75. doi: 10.1136/gutjnl-2021-325288
72. Ercolano G, Wyss T, Salomé B, Romero P, Trabanelli S, Jandus C. Distinct and shared gene expression for human innate versus adaptive helper lymphoid cells. J Leukoc Biol (2020) 108(2):723–37. doi: 10.1002/JLB.5MA0120-209R
73. Ahn YO, Weeres MA, Neulen ML, Choi J, Kang SH, Heo DS, et al. Human group3 innate lymphoid cells express DR3 and respond to TL1A with enhanced IL-22 production and IL-2-dependent proliferation. Eur J Immunol (2015) 45(8):2335–42. doi: 10.1002/eji.201445213
74. Ruiter B, Patil SU, Shreffler WG. Vitamins A and D have antagonistic effects on expression of effector cytokines and gut-homing integrin in human innate lymphoid cells. Clin Exp Allergy (2015) 45(7):1214–25. doi: 10.1111/cea.12568
75. Min HK, Moon J, Lee SY, Lee AR, Lee CR, Lee J, et al. Expanded IL-22+ Group 3 innate lymphoid cells and role of oxidized LDL-C in the pathogenesis of axial spondyloarthritis with dyslipidaemia. Immune Netw (2021) 21(6):e43. doi: 10.4110/in.2021.21.e43
76. Wu Y, Yue J, Wu J, Zhou W, Li D, Ding K, et al. Obesity may provide pro-ILC3 development inflammatory environment in asthmatic children. J Immunol Res (2018) 2018:1628620. doi: 10.1155/2018/1628620
77. Xu Y, Romero R, Miller D, Silva P, Panaitescu B, Theis KR, et al. Innate lymphoid cells at the human maternal-fetal interface in spontaneous preterm labor. Am J Reprod Immunol (2018) 79(6):e12820. doi: 10.1111/aji.12820
78. Einenkel R, Ehrhardt J, Zygmunt M, Muzzio DO. Oxygen regulates ILC3 antigen presentation potential and pregnancy-related hormone actions. Reprod Biol Endocrinol (2022) 20(1):109. doi: 10.1186/s12958-022-00979-2
79. Bennstein SB, Weinhold S, Degistirici Ö, Oostendorp RAJ, Raba K, Kögler G, et al. Efficient in vitro generation of IL-22-secreting ILC3 from CD34+ Hematopoietic progenitors in a human mesenchymal stem cell niche. Front Immunol (2021) 12:797432. doi: 10.3389/fimmu.2021.797432
80. Ciccia F, Guggino G, Rizzo A, Saieva L, Peralta S, Giardina A, et al. Type 3 innate lymphoid cells producing IL-17 and IL-22 are expanded in the gut, in the peripheral blood, synovial fluid and bone marrow of patients with ankylosing spondylitis. Ann Rheum Dis (2015) 74(9):1739–47. doi: 10.1136/annrheumdis-2014-206323
81. Kiekens L, Wahlen S, Persyn E, De Vos Z, Taghon T, Vandekerckhove B, et al. T-BET drives the conversion of human type 3 innate lymphoid cells into functional NK cells. Front Immunol (2022) 13:975778. doi: 10.3389/fimmu.2022.975778
82. Forkel M, van Tol S, Höög C, Michaëlsson J, Almer S, Mjösberg J. Distinct alterations in the composition of mucosal innate lymphoid cells in newly diagnosed and established crohn's disease and ulcerative colitis. J Crohns Colitis (2019) 13(1):67–78. doi: 10.1093/ecco-jcc/jjy119
83. Villanova F, Flutter B, Tosi I, Grys K, Sreeneebus H, Perera GK, et al. Characterization of innate lymphoid cells in human skin and blood demonstrates increase of NKp44+ ILC3 in psoriasis. J Invest Dermatol (2014) 134(4):984–91. doi: 10.1038/jid.2013.477
84. Pelosi A, Alicata C, Tumino N, Ingegnere T, Loiacono F, Mingari MC, et al. An anti-inflammatory microRNA signature distinguishes group 3 innate lymphoid cells from natural killer cells in human decidua. Front Immunol (2020) 11:133. doi: 10.3389/fimmu.2020.00133
85. Pascual-Reguant A, Köhler R, Mothes R, Bauherr S, Hernández DC, Uecker R, et al. Multiplexed histology analyses for the phenotypic and spatial characterization of human innate lymphoid cells. Nat Commun (2021) 12(1):1737. doi: 10.1038/s41467-021-21994-8
86. Wang SC, Yang KD, Lin CY, Huang AY, Hsiao CC, Lin MT, et al. Intravenous immunoglobulin therapy enhances suppressive regulatory T cells and decreases innate lymphoid cells in children with immune thrombocytopenia. Pediatr Blood Cancer (2020) 67(2):e28075. doi: 10.1002/pbc.28075
87. Chen ZX, Liu HQ, Wu ZH, He JL, Zhong HJ. Type 3 innate lymphoid cells as an indicator of renal dysfunction and serum uric acid in hyperuricemia. Adv Clin Exp Med (2023) 32(3):307–13. doi: 10.17219/acem/154625
88. Huang JC, Schleisman M, Choi D, Mitchell C, Watson L, Asquith M, et al. Preliminary report on interleukin-22, GM-CSF, and IL-17F in the pathogenesis of acute anterior uveitis. Ocul Immunol Inflammation (2021) 29(3):558–65. doi: 10.1080/09273948.2019.1686156
89. Kawka L, Felten R, Schleiss C, Fauny JD, Le Van Quyen P, Dumortier H, et al. Alteration of innate lymphoid cell homeostasis mainly concerns salivary glands in primary Sjögren's syndrome. RMD Open (2023) 9(2):e003051. doi: 10.1136/rmdopen-2023-003051
90. Rodríguez-Carrio J, Hähnlein JS, Ramwadhdoebe TH, Semmelink JF, Choi IY, van Lienden KP, et al. Brief report: altered innate lymphoid cell subsets in human lymph node biopsy specimens obtained during the at-risk and earliest phases of rheumatoid arthritis. Arthritis Rheumatol (2017) 69(1):70–6. doi: 10.1002/art.39811
91. Ham J, Kim J, Sohn KH, Park IW, Choi BW, Chung DH, et al. Cigarette smoke aggravates asthma by inducing memory-like type 3 innate lymphoid cells. Nat Commun (2022) 13(1):3852. doi: 10.1038/s41467-022-31491-1
92. Zhang Z, Cheng L, Zhao J, Li G, Zhang L, Chen W, et al. Plasmacytoid dendritic cells promote HIV-1-induced group 3 innate lymphoid cell depletion. J Clin Invest (2015) 125(9):3692–703. doi: 10.1172/JCI82124
93. De Grove KC, Provoost S, Verhamme FM, Bracke KR, Joos GF, Maes T, et al. Characterization and quantification of innate lymphoid cell subsets in human lung. PloS One (2016) 11(1):e0145961. doi: 10.1371/journal.pone.0145961
94. Graves CL, Li J, LaPato M, Shapiro MR, Glover SC, Wallet MA, et al. Intestinal epithelial cell regulation of adaptive immune dysfunction in human type 1 diabetes. Front Immunol (2017) 7:679. doi: 10.3389/fimmu.2016.00679
95. Gómez-Massa E, Lasa-Lázaro M, Gil-Etayo FJ, Ulloa-Márquez E, Justo I, Loinaz C, et al. Donor helper innate lymphoid cells are replaced earlier than lineage positive cells and persist long-term in human intestinal grafts - a descriptive study. Transpl Int (2020) 33(9):1016–29. doi: 10.1111/tri.13609
96. Zhou L, Zhou W, Joseph AM, Chu C, Putzel GG, Fang B, et al. Group 3 innate lymphoid cells produce the growth factor HB-EGF to protect the intestine from TNF-mediated inflammation. Nat Immunol (2022) 23(2):251–61. doi: 10.1038/s41590-021-01110-0
97. Li C, Liu J, Pan J, Wang Y, Shen L, Xu Y. ILC1s and ILC3 exhibit inflammatory phenotype in periodontal ligament of periodontitis patients. Front Immunol (2021) 12:708678. doi: 10.3389/fimmu.2021.708678
98. Lyu M, Suzuki H, Kang L, Gaspal F, Zhou W, Goc J, et al. ILC3 select microbiota-specific regulatory T cells to establish tolerance in the gut. Nature (2022) 610(7933):744–51. doi: 10.1038/s41586-022-05141-x
99. Kang J, Loh K, Belyayev L, Cha P, Sadat M, Khan K, et al. Type 3 innate lymphoid cells are associated with a successful intestinal transplant. Am J Transplant (2021) 21(2):787–97. doi: 10.1111/ajt.16163
100. Zhou L, Chu C, Teng F, Bessman NJ, Goc J, Santosa EK, et al. Innate lymphoid cells support regulatory T cells in the intestine through interleukin-2. Nature (2019) 568(7752):405–9. doi: 10.1038/s41586-019-1082-x
101. Vacca P, Montaldo E, Croxatto D, Loiacono F, Canegallo F, Venturini PL, et al. Identification of diverse innate lymphoid cells in human decidua. Mucosal Immunol (2015) 8(2):254–64. doi: 10.1038/mi.2014.63
102. Croxatto D, Micheletti A, Montaldo E, Orecchia P, Loiacono F, Canegallo F, et al. Group 3 innate lymphoid cells regulate neutrophil migration and function in human decidua. Mucosal Immunol (2016) 9(6):1372–83. doi: 10.1038/mi.2016.10
103. Vacca P, Pesce S, Greppi M, Fulcheri E, Munari E, Olive D, et al. PD-1 is expressed by and regulates human group 3 innate lymphoid cells in human decidua. Mucosal Immunol (2019) 12(3):624–31. doi: 10.1038/s41385-019-0141-9
104. Croft CA, Thaller A, Marie S, Doisne JM, Surace L, Yang R, et al. Notch, RORC and IL-23 signals cooperate to promote multi-lineage human innate lymphoid cell differentiation. Nat Commun (2022) 13(1):4344. doi: 10.1038/s41467-022-32089-3
105. Shikhagaie MM, Björklund ÅK, Mjösberg J, Erjefält JS, Cornelissen AS, Ros XR, et al. Neuropilin-1 is expressed on lymphoid tissue residing LTi-like group 3 innate lymphoid cells and associated with ectopic lymphoid aggregates. Cell Rep (2017) 18(7):1761–73. doi: 10.1016/j.celrep.2017.01.063
106. Mitsialis V, Wall S, Liu P, Ordovas-Montanes J, Parmet T, Vukovic M, et al. Single-cell analyses of colon and blood reveal distinct immune cell signatures of ulcerative colitis and crohn's disease. Gastroenterology (2020) 159(2):591–608.e10. doi: 10.1053/j.gastro.2020.04.074
107. Konya V, Czarnewski P, Forkel M, Rao A, Kokkinou E, Villablanca EJ, et al. Vitamin D downregulates the IL-23 receptor pathway in human mucosal group 3 innate lymphoid cells. J Allergy Clin Immunol (2018) 141(1):279–92. doi: 10.1016/j.jaci.2017.01.045
108. Kroeze A, van Hoeven V, Verheij MW, Turksma AW, Weterings N, van Gassen S, et al. Presence of innate lymphoid cells in allogeneic hematopoietic grafts correlates with reduced graft-versus-host disease. Cytotherapy (2022) 24(3):302–10. doi: 10.1016/j.jcyt.2021.10.011
109. Coorens M, Rao A, Gräfe SK, Unelius D, Lindforss U, Agerberth B, et al. Innate lymphoid cell type 3-derived interleukin-22 boosts lipocalin-2 production in intestinal epithelial cells via synergy between STAT3 and NF-κB. J Biol Chem (2019) 294(15):6027–41. doi: 10.1074/jbc.RA118.007290
110. Bernink JH, Ohne Y, Teunissen MBM, Wang J, Wu J, Krabbendam L, et al. c-Kit-positive ILC2s exhibit an ILC3-like signature that may contribute to IL-17-mediated pathologies. Nat Immunol (2019) 20(8):992–1003. doi: 10.1038/s41590-019-0423-0
111. Hazenberg MD, Haverkate NJE, van Lier YF, Spits H, Krabbendam L, Bemelman WA, et al. Human ectoenzyme-expressing ILC3: immunosuppressive innate cells that are depleted in graft-versus-host disease. Blood Adv (2019) 3(22):3650–60. doi: 10.1182/bloodadvances.2019000176
112. Mazzurana L, Forkel M, Rao A, Van Acker A, Kokkinou E, Ichiya T, et al. Suppression of Aiolos and Ikaros expression by lenalidomide reduces human ILC3-ILC1/NK cell transdifferentiation. Eur J Immunol (2019) 49(9):1344–55. doi: 10.1002/eji.201848075
113. Krämer B, Goeser F, Lutz P, Glässner A, Boesecke C, Schwarze-Zander C, et al. Compartment-specific distribution of human intestinal innate lymphoid cells is altered in HIV patients under effective therapy. PloS Pathog (2017) 13(5):e1006373. doi: 10.1371/journal.ppat.1006373
114. Gelmez MY, Cinar S, Cetin EA, Ozcit-Gürel G, Babuna-Kobaner G, Erdugan M, et al. Inflammatory status might direct ILC and NK cells to IL-17 expressing ILC3 and NK subsets in Behcet's disease. Immunol Lett (2021) 235:1–8. doi: 10.1016/j.imlet.2021.04.008
115. Cole S, Murray J, Simpson C, Okoye R, Tyson K, Griffiths M, et al. Interleukin (IL)-12 and IL-18 synergize to promote MAIT cell IL-17A and IL-17F production independently of IL-23 signaling. Front Immunol (2020) 11:585134. doi: 10.3389/fimmu.2020.585134
116. Creyns B, Jacobs I, Verstockt B, Cremer J, Ballet V, Vandecasteele R, et al. Biological therapy in inflammatory bowel disease patients partly restores intestinal innate lymphoid cell subtype equilibrium. Front Immunol (2020) 11:1847. doi: 10.3389/fimmu.2020.01847
117. Yu H, Wei Y, Dong Y, Chen P. Regulation of notch signaling pathway to innate lymphoid cells in patients with acute myocardial infarction. Immunol Invest (2023) 52(2):241–55. doi: 10.1080/08820139.2022.2158856
118. Mohr A, Trésallet C, Monot N, Bauvois A, Abiven D, Atif M, et al. Tissue infiltrating LTi-like group 3 innate lymphoid cells and T follicular helper cells in graves' and hashimoto's thyroiditis. Front Immunol (2020) 11:601. doi: 10.3389/fimmu.2020.00601
119. Zhang L, Lin Q, Jiang L, Wu M, Huang L, Quan W, et al. Increased circulating innate lymphoid cell (ILC)1 and decreased circulating ILC3 are involved in the pathogenesis of Henoch-Schonlein purpura. BMC Pediatr (2022) 22(1):201. doi: 10.1186/s12887-022-03262-w
120. Mendes J, Rodrigues-Santos P, Areia AL, Almeida JS, Alves V, Santos-Rosa M, et al. Type 2 and type 3 innate lymphoid cells at the maternal-fetal interface: implications in preterm birth. BMC Immunol (2021) 22(1):28. doi: 10.1186/s12865-021-00423-x
121. Goc J, Lv M, Bessman NJ, Flamar AL, Sahota S, Suzuki H, et al. Dysregulation of ILC3 unleashes progression and immunotherapy resistance in colon cancer. Cell (2021) 184(19):5015–5030.e16. doi: 10.1016/j.cell.2021.07.029
122. Chang Y, Kim JW, Yang S, Chung DH, Ko JS, Moon JS, et al. Increased GM-CSF-producing NCR- ILC3s and neutrophils in the intestinal mucosa exacerbate inflammatory bowel disease. Clin Transl Immunol (2021) 10(7):e1311. doi: 10.1002/cti2.1311
123. de Sousa TR, Sgnotto FDR, Fagundes BO, Duarte AJDS, Victor JR. Non-atopic neonatal thymic innate lymphoid cell subsets (ILC1, ILC2, and ILC3) identification and the modulatory effect of igG from dermatophagoides pteronyssinus (Derp)-atopic individuals. Front Allergy (2021) 2:650235. doi: 10.3389/falgy.2021.650235
124. Schielke L, Zimmermann N, Hobelsberger S, Steininger J, Strunk A, Blau K, et al. Metabolic syndrome in psoriasis is associated with upregulation of CXCL16 on monocytes and a dysbalance in innate lymphoid cells. Front Immunol (2022) 13:916701. doi: 10.3389/fimmu.2022.916701
125. Chu C, Moriyama S, Li Z, Zhou L, Flamar AL, Klose CSN, et al. Anti-microbial functions of group 3 innate lymphoid cells in gut-associated lymphoid tissues are regulated by G-protein-coupled receptor 183. Cell Rep (2018) 23(13):3750–8. doi: 10.1016/j.celrep.2018.05.099
126. Cruz-Zárate D, Cabrera-Rivera GL, Ruiz-Sánchez BP, Serafín-López J, Chacón-Salinas R, López-Macías C, et al. Innate Lymphoid Cells Have Decreased HLA-DR Expression but Retain Their Responsiveness to TLR Ligands during Sepsis. J Immunol (2018) 201(11):3401–10. doi: 10.4049/jimmunol.1800735
127. Teng F, Goc J, Zhou L, Chu C, Shah MA, Eberl G, et al. A circadian clock is essential for homeostasis of group 3 innate lymphoid cells in the gut. Sci Immunol (2019) 4(40):eaax1215. doi: 10.1126/sciimmunol.aax1215
128. Wang X, Cai J, Lin B, Ma M, Tao Y, Zhou Y, et al. GPR34-mediated sensing of lysophosphatidylserine released by apoptotic neutrophils activates type 3 innate lymphoid cells to mediate tissue repair. Immunity (2021) 54(6):1123–1136.e8. doi: 10.1016/j.immuni.2021.05.007
129. Hu C, Xu B, Wang X, Wan WH, Lu J, Kong D, et al. Gut microbiota-derived short-chain fatty acids regulate group 3 innate lymphoid cells in HCC. Hepatology (2023) 77(1):48–64. doi: 10.1002/hep.32449
130. Siegler JJ, Correia MP, Hofman T, Prager I, Birgin E, Rahbari NN, et al. Human ILC3 exert TRAIL-mediated cytotoxicity towards cancer cells. Front Immunol (2022) 13:742571. doi: 10.3389/fimmu.2022.742571
131. Pan L, Chen X, Liu X, Qiu W, Liu Y, Jiang W, et al. Innate lymphoid cells exhibited IL-17-expressing phenotype in active tuberculosis disease. BMC Pulm Med (2021) 21(1):318. doi: 10.1186/s12890-021-01678-1
132. Qi J, Crinier A, Escalière B, Ye Y, Wang Z, Zhang T, et al. Single-cell transcriptomic landscape reveals tumor specific innate lymphoid cells associated with colorectal cancer progression. Cell Rep Med (2021) 2(8):100353. doi: 10.1016/j.xcrm.2021.100353
133. Hou M, Liu S. Innate lymphoid cells are increased in systemic lupus erythematosus. Clin Exp Rheumatol (2019) 37(4):676–9.
134. Tang Y, Tan SA, Iqbal A, Li J, Glover SC. STAT3 Genotypic Variant rs744166 and Increased Tyrosine Phosphorylation of STAT3 in IL-23 Responsive Innate Lymphoid Cells during Pathogenesis of Crohn's Disease. J Immunol Res (2019) 2019:9406146. doi: 10.1155/2019/9406146
135. Fu W, Wang W, Zhang J, Zhao Y, Chen K, Wang Y, et al. Dynamic change of circulating innate and adaptive lymphocytes subtypes during a cascade of gastric lesions. J Leukoc Biol (2022) 112(4):931–8. doi: 10.1002/JLB.5MA0422-505R
136. Guo C, Zhou M, Zhao S, Huang Y, Wang S, Fu R, et al. Innate lymphoid cell disturbance with increase in ILC1 in systemic lupus erythematosus. Clin Immunol (2019) 202:49–58. doi: 10.1016/j.clim.2019.03.008
137. Soare A, Weber S, Maul L, Rauber S, Gheorghiu AM, Luber M, et al. Cutting edge: homeostasis of innate lymphoid cells is imbalanced in psoriatic arthritis. J Immunol (2018) 200(4):1249–54. doi: 10.4049/jimmunol.1700596
138. Björklund ÅK, Forkel M, Picelli S, Konya V, Theorell J, Friberg D, et al. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nat Immunol (2016) 17(4):451–60. doi: 10.1038/ni.3368
139. Camelo A, Rosignoli G, Ohne Y, Stewart RA, Overed-Sayer C, Sleeman MA, et al. IL-33, IL-25, and TSLP induce a distinct phenotypic and activation profile in human type 2 innate lymphoid cells. Blood Adv (2017) 1(10):577–89. doi: 10.1182/bloodadvances.2016002352
140. Bar-Ephraim YE, Cornelissen F, Papazian N, Konijn T, Hoogenboezem RM, Sanders MA, et al. Cross-tissue transcriptomic analysis of human secondary lymphoid organ-residing ILC3 reveals a quiescent state in the absence of inflammation. Cell Rep (2017) 21(3):823–33. doi: 10.1016/j.celrep.2017.09.070
141. Teunissen MBM, Munneke JM, Bernink JH, Spuls PI, Res PCM, Te Velde A, et al. Composition of innate lymphoid cell subsets in the human skin: enrichment of NCR(+) ILC3 in lesional skin and blood of psoriasis patients. J Invest Dermatol (2014) 134(9):2351–60. doi: 10.1038/jid.2014.146
142. Aglas-Leitner F, Juillard P, Juillard A, Byrne SN, Hawke S, Grau GE, et al. Circulating CCR6+ILC proportions are lower in multiple sclerosis patients. Clin Transl Immunol (2022) 11(12):e1426. doi: 10.1002/cti2.1426
143. Aglas-Leitner FT, Juillard P, Juillard A, Byrne SN, Hawke S, Grau GE, et al. Mass cytometry reveals cladribine-induced resets among innate lymphoid cells in multiple sclerosis. Sci Rep (2022) 12(1):20411. doi: 10.1038/s41598-022-24617-4
144. Sugahara T, Tanaka Y, Hamaguchi M, Fujii M, Shimura K, Ogawa K, et al. Reduced innate lymphoid cells in the endometrium of women with endometriosis. Am J Reprod Immunol (2022) 87(1):e13502. doi: 10.1111/aji.13502
145. Raabe J, Kaiser KM, ToVinh M, Finnemann C, Lutz P, Hoffmeister C, et al. Identification and characterisation of a hepatic IL-13 producing ILC3-like population potentially involved in liver fibrosis. Hepatology (2023) 78(3):787–802. doi: 10.1097/HEP.0000000000000350
146. Bracamonte-Baran W, Chen G, Hou X, Talor MV, Choi HS, Davogustto G, et al. Non-cytotoxic cardiac innate lymphoid cells are a resident and quiescent type 2-commited population. Front Immunol (2019) 10:634. doi: 10.3389/fimmu.2019.00634
147. Baban B, Braun M, Khodadadi H, Ward A, Alverson K, Malik A, et al. AMPK induces regulatory innate lymphoid cells after traumatic brain injury. JCI Insight (2021) 6(1):e126766. doi: 10.1172/jci.insight.126766
148. Oherle K, Acker E, Bonfield M, Wang T, Gray J, Lang I, et al. Insulin-like growth factor 1 supports a pulmonary niche that promotes type 3 innate lymphoid cell development in newborn lungs. Immunity (2020) 52(2):275–294.e9. doi: 10.1016/j.immuni.2020.01.005
149. Rodríguez OL, Lugo DA, Cabrera M, Sánchez MA, Zerpa O, Tapia FJ. Innate lymphoid cells in peripheral blood of patients with American Cutaneous Leishmaniasis. Exp Dermatol (2021) 30(7):982–7. doi: 10.1111/exd.14351
150. Mxinwa V, Dludla PV, Nyambuya TM, Nkambule BB. Circulating innate lymphoid cell subtypes and altered cytokine profiles following an atherogenic high-fat diet. Innate Immun (2021) 27(7-8):525–32. doi: 10.1177/17534259211053634
151. Kawano Y, Edwards M, Huang Y, Bilate AM, Araujo LP, Tanoue T, et al. Microbiota imbalance induced by dietary sugar disrupts immune-mediated protection from metabolic syndrome. Cell (2022) 185(19):3501–3519.e20. doi: 10.1016/j.cell.2022.08.005
152. von Burg N, Chappaz S, Baerenwaldt A, Horvath E, Bose Dasgupta S, Ashok D, et al. Activated group 3 innate lymphoid cells promote T-cell-mediated immune responses. Proc Natl Acad Sci U.S.A. (2014) 111(35):12835–40. doi: 10.1073/pnas.1406908111
153. Wu C, He S, Liu J, Wang B, Lin J, Duan Y, et al. Type 1 innate lymphoid cell aggravation of atherosclerosis is mediated through TLR4. Scand J Immunol (2018) 87(5):e12661. doi: 10.1111/sji.12661
154. Hoorweg K, Narang P, Li Z, Thuery A, Papazian N, Withers DR, et al. A stromal cell niche for human and mouse type 3 innate lymphoid cells. J Immunol (2015) 195(9):4257–63. doi: 10.4049/jimmunol.1402584
155. Niu X, Daniel S, Kumar D, Ding EY, Savani RC, Koh AY, et al. Transient neonatal antibiotic exposure increases susceptibility to late-onset sepsis driven by microbiota-dependent suppression of type 3 innate lymphoid cells. Sci Rep (2020) 10(1):12974. doi: 10.1038/s41598-020-69797-z
156. Zhao D, Yang B, Ye C, Zhang S, Lv X, Chen Q. Enteral nutrition ameliorates the symptoms of Crohn's disease in mice via activating special pro-resolving mediators through innate lymphoid cells. Innate Immun (2021) 27(7-8):533–42. doi: 10.1177/17534259211057038
157. Yu Y, Tsang JC, Wang C, Clare S, Wang J, Chen X, et al. Single-cell RNA-seq identifies a PD-1hi ILC progenitor and defines its development pathway. Nature (2016) 539(7627):102–6. doi: 10.1038/nature20105
158. Shen W, Hixon JA, McLean MH, Li WQ, Durum SK. IL-22-expressing murine lymphocytes display plasticity and pathogenicity in reporter mice. Front Immunol (2016) 6:662. doi: 10.3389/fimmu.2015.00662
159. Warren KJ, Poole JA, Sweeter JM, DeVasure JM, Dickinson JD, Peebles RS Jr, et al. Neutralization of IL-33 modifies the type 2 and type 3 inflammatory signature of viral induced asthma exacerbation. Respir Res (2021) 22(1):206. doi: 10.1186/s12931-021-01799-5
160. Satoh-Takayama N, Serafini N, Verrier T, Rekiki A, Renauld JC, Frankel G, et al. The chemokine receptor CXCR6 controls the functional topography of interleukin-22 producing intestinal innate lymphoid cells. Immunity (2014) 41(5):776–88. doi: 10.1016/j.immuni.2014.10.007
161. Zhong C, Cui K, Wilhelm C, Hu G, Mao K, Belkaid Y, et al. Group 3 innate lymphoid cells continuously require the transcription factor GATA-3 after commitment. Nat Immunol (2016) 17(2):169–78. doi: 10.1038/ni.3318
162. Li Z, Hodgkinson T, Gothard EJ, Boroumand S, Lamb R, Cummins I, et al. Epidermal Notch1 recruits RORγ(+) group 3 innate lymphoid cells to orchestrate normal skin repair. Nat Commun (2016) 7:11394. doi: 10.1038/ncomms11394
163. Dutton EE, Camelo A, Sleeman M, Herbst R, Carlesso G, Belz GT, et al. Characterisation of innate lymphoid cell populations at different sites in mice with defective T cell immunity. Wellcome Open Res (2017) 2:117. doi: 10.12688/wellcomeopenres.13199.3
164. Miranda MCG, Oliveira RP, Torres L, Aguiar SLF, Pinheiro-Rosa N, Lemos L, et al. Frontline Science: Abnormalities in the gut mucosa of non-obese diabetic mice precede the onset of type 1 diabetes. J Leukoc Biol (2019) 106(3):513–29. doi: 10.1002/JLB.3HI0119-024RR
165. Godinho-Silva C, Domingues RG, Rendas M, Raposo B, Ribeiro H, da Silva JA, et al. Light-entrained and brain-tuned circadian circuits regulate ILC3 and gut homeostasis. Nature (2019) 574(7777):254–8. doi: 10.1038/s41586-019-1579-3
166. Gao X, Shen X, Liu K, Lu C, Fan Y, Xu Q, et al. The transcription factor thPOK regulates ILC3 lineage homeostasis and function during intestinal infection. Front Immunol (2022) 13:939033. doi: 10.3389/fimmu.2022.939033
167. Fuchs A, Ghosh S, Chang SW, Bochicchio GV, Turnbull IR. Pseudomonas aeruginosa pneumonia causes a loss of type-3 and an increase in type-1 innate lymphoid cells in the gut. J Surg Res (2021) 265:212–22. doi: 10.1016/j.jss.2021.03.043
168. Yan J, Yu J, Yuan S, Tang W, Ma W, Yang X, et al. Musculin is highly enriched in Th17 and IL-22-producing ILC3 and restrains pro-inflammatory cytokines in murine colitis. Eur J Immunol (2021) 51(4):995–8. doi: 10.1002/eji.202048573
169. Hu L, Hu J, Chen L, Zhang Y, Wang Q, Yang X. Interleukin-22 from type 3 innate lymphoid cells aggravates lupus nephritis by promoting macrophage infiltration in lupus-prone mice. Front Immunol (2021) 12:584414. doi: 10.3389/fimmu.2021.584414
170. Jones R, Cosway EJ, Willis C, White AJ, Jenkinson WE, Fehling HJ, et al. Dynamic changes in intrathymic ILC populations during murine neonatal development. Eur J Immunol (2018) 48(9):1481–91. doi: 10.1002/eji.201847511
171. Cai J, Lu H, Su Z, Mi L, Xu S, Xue Z. Dynamic changes of NCR- type 3 innate lymphoid cells and their role in mice with bronchopulmonary dysplasia. Inflammation (2022) 45(2):497–508. doi: 10.1007/s10753-021-01543-7
172. Fiancette R, Finlay CM, Willis C, Bevington SL, Soley J, Ng STH, et al. Reciprocal transcription factor networks govern tissue-resident ILC3 subset function and identity. Nat Immunol (2021) 22(10):1245–55. doi: 10.1038/s41590-021-01024-x
173. Cuff AO, Male V. Conventional NK cells and ILC1 are partially ablated in the livers of Ncr1 iCreTbx21 fl/fl mice. Wellcome Open Res (2017) 2:39. doi: 10.12688/wellcomeopenres.11741.2
174. Javadzadeh SM, Keykhosravi M, Tehrani M, Asgarian-Omran H, Rashidi M, Hossein-Nattaj H, et al. Evaluation of innate lymphoid cells (ILC) population in the mouse model of colorectal cancer. Iran J Immunol (2022) 19(4):339–48. doi: 10.22034/IJI.2022.92467.2152
175. Gray J, Oehrle K, Worthen G, Alenghat T, Whitsett J, Deshmukh H. Intestinal commensal bacteria mediate lung mucosal immunity and promote resistance of newborn mice to infection. Sci Transl Med (2017) 9(376):eaaf9412. doi: 10.1126/scitranslmed.aaf9412
176. Okubo Y, Tokumaru S, Yamamoto Y, Miyagawa SI, Sanjo H, Taki S. Generation of a common innate lymphoid cell progenitor requires interferon regulatory factor 2. Int Immunol (2019) 31(8):489–98. doi: 10.1093/intimm/dxz019
177. Yamamoto Y, Yoshizawa K, Takamoto M, Soejima Y, Sanjo H, Taki S. Circulating T cells and resident non-T cells restrict type 2 innate lymphoid cell expansion in the small intestine. Biochem Biophys Res Commun (2022) 618:93–9. doi: 10.1016/j.bbrc.2022.06.007
178. Leon-Coria A, Kumar M, Workentine M, Moreau F, Surette M, Chadee K. Muc2 mucin and nonmucin microbiota confer distinct innate host defense in disease susceptibility and colonic injury. Cell Mol Gastroenterol Hepatol (2021) 11(1):77–98. doi: 10.1016/j.jcmgh.2020.07.003
179. Walker JA, Clark PA, Crisp A, Barlow JL, Szeto A, Ferreira ACF, et al. Polychromic reporter mice reveal unappreciated innate lymphoid cell progenitor heterogeneity and elusive ILC3 progenitors in bone marrow. Immunity (2019) 51(1):104–118.e7. doi: 10.1016/j.immuni.2019.05.002
180. Aparicio-Domingo P, Romera-Hernandez M, Karrich JJ, Cornelissen F, Papazian N, Lindenbergh-Kortleve DJ, et al. Type 3 innate lymphoid cells maintain intestinal epithelial stem cells after tissue damage. J Exp Med (2015) 212(11):1783–91. doi: 10.1084/jem.20150318
181. Wanke F, Tang Y, Gronke K, Klebow S, Moos S, Hauptmann J, et al. Expression of IL-17F is associated with non-pathogenic Th17 cells. J Mol Med (Berl) (2018) 96(8):819–29. doi: 10.1007/s00109-018-1662-5
182. Castro-Dopico T, Fleming A, Dennison TW, Ferdinand JR, Harcourt K, Stewart BJ, et al. GM-CSF calibrates macrophage defense and wound healing programs during intestinal infection and inflammation. Cell Rep (2020) 32(1):107857. doi: 10.1016/j.celrep.2020.107857
183. Sim S, Lee DH, Kim KS, Park HJ, Kim YK, Choi Y, et al. Micrococcus luteus-derived extracellular vesicles attenuate neutrophilic asthma by regulating miRNAs in airway epithelial cells. Exp Mol Med (2023) 55(1):196–204. doi: 10.1038/s12276-022-00910-0
184. Deng F, Hu JJ, Lin ZB, Sun QS, Min Y, Zhao BC, et al. Gut microbe-derived milnacipran enhances tolerance to gut ischemia/reperfusion injury. Cell Rep Med (2023) 4(3):100979. doi: 10.1016/j.xcrm.2023.100979
185. Frascoli M, Jeanty C, Fleck S, Moradi PW, Keating S, Mattis AN, et al. Heightened immune activation in fetuses with gastroschisis may be blocked by targeting IL-5. J Immunol (2016) 196(12):4957–66. doi: 10.4049/jimmunol.1502587
186. Dalli J, Colas RA, Arnardottir H, Serhan CN. Vagal regulation of group 3 innate lymphoid cells and the immunoresolvent PCTR1 controls infection resolution. Immunity (2017) 46(1):92–105. doi: 10.1016/j.immuni.2016.12.009
187. Schaeuble K, Britschgi MR, Scarpellino L, Favre S, Xu Y, Koroleva E, et al. Perivascular fibroblasts of the developing spleen act as LTα1β2-dependent precursors of both T and B zone organizer cells. Cell Rep (2017) 21(9):2500–14. doi: 10.1016/j.celrep.2017.10.119
188. Tripathi D, Radhakrishnan RK, Sivangala Thandi R, Paidipally P, Devalraju KP, Neela VSK, et al. IL-22 produced by type 3 innate lymphoid cells (ILC3) reduces the mortality of type 2 diabetes mellitus (T2DM) mice infected with Mycobacterium tuberculosis. PloS Pathog (2019) 15(12):e1008140. doi: 10.1371/journal.ppat.1008140
189. de Araújo EF, Preite NW, Veldhoen M, Loures FV, Calich VLG. Pulmonary paracoccidioidomycosis in AhR deficient hosts is severe and associated with defective Treg and Th22 responses. Sci Rep (2020) 10(1):11312. doi: 10.1038/s41598-020-68322-6
190. Huang J, Fu L, Huang J, Zhao J, Zhang X, Wang W, et al. Group 3 innate lymphoid cells protect the host from the uropathogenic escherichia coli infection in the bladder. Adv Sci (Weinh) (2022) 9(6):e2103303. doi: 10.1002/advs.202103303
191. Liu N, He J, Fan D, Gu Y, Wang J, Li H, et al. Circular RNA circTmem241 drives group III /innate lymphoid cell differentiation via initiation of Elk3 transcription. Nat Commun (2022) 13(1):4711. doi: 10.1038/s41467-022-32322-z
192. Darby M, Roberts LB, Mackowiak C, Chetty A, Tinelli S, Schnoeller C, et al. ILC3-derived acetylcholine promotes protease-driven allergic lung pathology. J Allergy Clin Immunol (2021) 147(4):1513–1516.e4. doi: 10.1016/j.jaci.2020.10.038
193. Zhao D, Cai C, Zheng Q, Jin S, Song D, Shen J, et al. Vancomycin pre-treatment impairs tissue healing in experimental colitis: Importance of innate lymphoid cells. Biochem Biophys Res Commun (2017) 483(1):237–44. doi: 10.1016/j.bbrc.2016.12.160
194. Shi Z, Takeuchi T, Nakanishi Y, Kato T, Beck K, Nagata R, et al. A Japanese herbal formula, daikenchuto, alleviates experimental colitis by reshaping microbial profiles and enhancing group 3 innate lymphoid cells. Front Immunol (2022) 13:903459. doi: 10.3389/fimmu.2022.903459
195. Stehle C, Rückert T, Fiancette R, Gajdasik DW, Willis C, Ulbricht C, et al. T-bet and RORα control lymph node formation by regulating embryonic innate lymphoid cell differentiation. Nat Immunol (2021) 22(10):1231–44. doi: 10.1038/s41590-021-01029-6
196. Hamaguchi M, Okamura T, Fukuda T, Nishida K, Yoshimura Y, Hashimoto Y, et al. Group 3 innate lymphoid cells protect steatohepatitis from high-fat diet induced toxicity. Front Immunol (2021) 12:648754. doi: 10.3389/fimmu.2021.648754
197. Kawano R, Okamura T, Hashimoto Y, Majima S, Senmaru T, Ushigome E, et al. Erythritol ameliorates small intestinal inflammation induced by high-fat diets and improves glucose tolerance. Int J Mol Sci (2021) 22(11):5558. doi: 10.3390/ijms22115558
198. Canesso MCC, Lemos L, Neves TC, Marim FM, Castro TBR, Veloso ÉS, et al. The cytosolic sensor STING is required for intestinal homeostasis and control of inflammation. Mucosal Immunol (2018) 11(3):820–34. doi: 10.1038/mi.2017.88
199. Dong Y, Fan H, Zhang Z, Jiang F, Li M, Zhou H, et al. Berberine ameliorates DSS-induced intestinal mucosal barrier dysfunction through microbiota-dependence and Wnt/β-catenin pathway. Int J Biol Sci (2022) 18(4):1381–97. doi: 10.7150/ijbs.65476
200. Hatfield JK, Brown MA. Group 3 innate lymphoid cells accumulate and exhibit disease-induced activation in the meninges in EAE. Cell Immunol (2015) 297(2):69–79. doi: 10.1016/j.cellimm.2015.06.006
201. Ebihara T, Song C, Ryu SH, Plougastel-Douglas B, Yang L, Levanon D, et al. Runx3 specifies lineage commitment of innate lymphoid cells. Nat Immunol (2015) 16(11):1124–33. doi: 10.1038/ni.3272
202. Verrier T, Satoh-Takayama N, Serafini N, Marie S, Di Santo JP, Vosshenrich CA. Phenotypic and functional plasticity of murine intestinal NKp46+ Group 3 innate lymphoid cells. J Immunol (2016) 196(11):4731–8. doi: 10.4049/jimmunol.1502673
203. Saez de Guinoa J, Jimeno R, Farhadi N, Jervis PJ, Cox LR, Besra GS, et al. CD1d-mediated activation of group 3 innate lymphoid cells drives IL-22 production. EMBO Rep (2017) 18(1):39–47. doi: 10.15252/embr.201642412
204. Liu B, Ye B, Yang L, Zhu X, Huang G, Zhu P, et al. Long noncoding RNA lncKdm2b is required for ILC3 maintenance by initiation of Zfp292 expression. Nat Immunol (2017) 18(5):499–508. doi: 10.1038/ni.3712
205. Lu Y, Zhang X, Bouladoux N, Kaul SN, Jin K, Sant'Angelo D, et al. Zbtb1 controls NKp46+ ROR-gamma-T+ innate lymphoid cell (ILC3) development. Oncotarget (2017) 8(34):55877–88. doi: 10.18632/oncotarget.19645
206. Li Z, Buttó LF, Buela KA, Jia LG, Lam M, Ward JD, et al. Death receptor 3 signaling controls the balance between regulatory and effector lymphocytes in SAMP1/yitFc mice with crohn's disease-like ileitis. Front Immunol (2018) 9:362. doi: 10.3389/fimmu.2018.00362
207. Yin S, Yu J, Hu B, Lu C, Liu X, Gao X, et al. Runx3 mediates resistance to intracellular bacterial infection by promoting IL12 signaling in group 1 ILC and NCR+ILC3. Front Immunol (2018) 9:2101. doi: 10.3389/fimmu.2018.02101
208. Hou P, Zhou X, Yu L, Yao Y, Zhang Y, Huang Y, et al. Exhaustive exercise induces gastrointestinal syndrome through reduced ILC3 and IL-22 in mouse model. Med Sci Sports Exerc (2020) 52(8):1710–8. doi: 10.1249/MSS.0000000000002298
209. Mincham KT, Snelgrove RJ. OMIP-086: Full spectrum flow cytometry for high-dimensional immunophenotyping of mouse innate lymphoid cells. Cytometry A (2022) 103(2):110–6. doi: 10.1002/cyto.a.24702
210. Gogoleva VS, Kuprash DV, Grivennikov SI, Tumanov AV, Kruglov AA, Nedospasov SA. LTα, TNF, and ILC3 in peyer's patch organogenesis. Cells (2022) 11(12):1970. doi: 10.3390/cells11121970
211. Schroeder JH, Meissl K, Hromadová D, Lo JW, Neves JF, Howard JK, et al. T-bet controls cellularity of intestinal group 3 innate lymphoid cells. Front Immunol (2021) 11:623324. doi: 10.3389/fimmu.2020.623324
212. Schroeder JH, Roberts LB, Meissl K, Lo JW, Hromadová D, Hayes K, et al. Sustained post-developmental T-bet expression is critical for the maintenance of type one innate lymphoid cells in vivo. Front Immunol (2021) 12:760198. doi: 10.3389/fimmu.2021.760198
213. de Araújo EF, Loures FV, Preite NW, Feriotti C, Galdino NA, Costa TA, et al. AhR ligands modulate the differentiation of innate lymphoid cells and T helper cell subsets that control the severity of a pulmonary fungal infection. Front Immunol (2021) 12:630938. doi: 10.3389/fimmu.2021.630938
214. Liu B, Liu N, Zhu X, Yang L, Ye B, Li H, et al. Circular RNA circZbtb20 maintains ILC3 homeostasis and function via Alkbh5-dependent m6A demethylation of Nr4a1 mRNA. Cell Mol Immunol (2021) 18(6):1412–24. doi: 10.1038/s41423-021-00680-1
215. Liu Y, Song Y, Lin D, Lei L, Mei Y, Jin Z, et al. NCR- group 3 innate lymphoid cells orchestrate IL-23/IL-17 axis to promote hepatocellular carcinoma development. EBioMedicine (2019) 41:333–44. doi: 10.1016/j.ebiom.2019.02.050
216. Serafini N, Jarade A, Surace L, Goncalves P, Sismeiro O, Varet H, et al. Trained ILC3 responses promote intestinal defense. Science (2022) 375(6583):859–63. doi: 10.1126/science.aaz8777
217. Michaudel C, Bataille F, Maillet I, Fauconnier L, Colas C, Sokol H, et al. Ozone-induced aryl hydrocarbon receptor activation controls lung inflammation via interleukin-22 modulation. Front Immunol (2020) 11:144. doi: 10.3389/fimmu.2020.00144
218. Melo-Gonzalez F, Kammoun H, Evren E, Dutton EE, Papadopoulou M, Bradford BM, et al. Antigen-presenting ILC3 regulate T cell-dependent IgA responses to colonic mucosal bacteria. J Exp Med (2019) 216(4):728–42. doi: 10.1084/jem.20180871
219. Tizian C, Lahmann A, Hölsken O, Cosovanu C, Kofoed-Branzk M, Heinrich F, et al. c-Maf restrains T-bet-driven programming of CCR6-negative group 3 innate lymphoid cells. Elife (2020) 9:e52549. doi: 10.7554/eLife.52549
220. Xia P, Liu J, Wang S, Ye B, Du Y, Xiong Z, et al. WASH maintains NKp46+ ILC3 cells by promoting AHR expression. Nat Commun (2017) 8:15685. doi: 10.1038/ncomms15685
221. Bergmann H, Roth S, Pechloff K, Kiss EA, Kuhn S, Heikenwälder M, et al. Card9-dependent IL-1β regulates IL-22 production from group 3 innate lymphoid cells and promotes colitis-associated cancer. Eur J Immunol (2017) 47(8):1342–53. doi: 10.1002/eji.201646765
222. Saksida T, Paunović V, Koprivica I, Mićanović D, Jevtić B, Jonić N, et al. Development of type 1 diabetes in mice is associated with a decrease in IL-2-producing ILC3 and foxP3+ Treg in the small intestine. Molecules (2023) 28(8):3366. doi: 10.3390/molecules28083366
223. Xie J, Tian S, Liu J, Huang S, Yang M, Yang X, et al. Combination therapy with indigo and indirubin for ulcerative colitis via reinforcing intestinal barrier function. Oxid Med Cell Longev (2023) 2023:2894695. doi: 10.1155/2023/2894695
224. Chan L, Mehrani Y, Minott JA, Bridle BW, Karimi K. The potential of dendritic-cell-based vaccines to modulate type 3 innate lymphoid cell populations. Int J Mol Sci (2023) 24(3):2403. doi: 10.3390/ijms24032403
225. Lewis JM, Monico PF, Mirza FN, Xu S, Yumeen S, Turban JL, et al. Chronic UV radiation-induced RORγt+ IL-22-producing lymphoid cells are associated with mutant KC clonal expansion. Proc Natl Acad Sci U.S.A. (2021) 118(37):e2016963118. doi: 10.1073/pnas.2016963118
226. Fachi JL, Sécca C, Rodrigues PB, Mato FCP, Di Luccia B, Felipe JS, et al. Acetate coordinates neutrophil and ILC3 responses against C. difficile through FFAR2. J Exp Med (2020) 217(3):e20190489. doi: 10.1084/jem.20190489
227. Peng V, Cao S, Trsan T, Bando JK, Avila-Pacheco J, Cleveland JL, et al. Ornithine decarboxylase supports ILC3 responses in infectious and autoimmune colitis through positive regulation of IL-22 transcription. Proc Natl Acad Sci U.S.A. (2022) 119(45):e2214900119. doi: 10.1073/pnas.2214900119
228. Kim SH, Cho BH, Kiyono H, Jang YS. Microbiota-derived butyrate suppresses group 3 innate lymphoid cells in terminal ileal Peyer's patches. Sci Rep (2017) 7(1):3980. doi: 10.1038/s41598-017-02729-6
229. Li S, Bostick JW, Ye J, Qiu J, Zhang B, Urban JF Jr, et al. Aryl hydrocarbon receptor signaling cell intrinsically inhibits intestinal group 2 innate lymphoid cell function. Immunity (2018) 49(5):915–928.e5. doi: 10.1016/j.immuni.2018.09.015
230. Kang L, Zhang X, Ji L, Kou T, Smith SM, Zhao B, et al. The colonic macrophage transcription factor RBP-J orchestrates intestinal immunity against bacterial pathogens. J Exp Med (2020) 217(4):e20190762. doi: 10.1084/jem.20190762
231. Bando JK, Gilfillan S, Song C, McDonald KG, Huang SC, Newberry RD, et al. The tumor necrosis factor superfamily member RANKL suppresses effector cytokine production in group 3 innate lymphoid cells. Immunity (2018) 48(6):1208–1219.e4. doi: 10.1016/j.immuni.2018.04.012
232. Lo BC, Gold MJ, Hughes MR, Antignano F, Valdez Y, Zaph C, et al. The orphan nuclear receptor RORα and group 3 innate lymphoid cells drive fibrosis in a mouse model of Crohn's disease. Sci Immunol (2016) 1(3):eaaf8864. doi: 10.1126/sciimmunol.aaf8864
233. Tait Wojno ED, Beamer CA. Isolation and identification of innate lymphoid cells (ILCs) for immunotoxicity testing. Methods Mol Biol (2018) 1803:353–70. doi: 10.1007/978-1-4939-8549-4_21
234. Geiger TL, Abt MC, Gasteiger G, Firth MA, O'Connor MH, Geary CD, et al. Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. J Exp Med (2014) 211(9):1723–31. doi: 10.1084/jem.20140212
235. Xu W, Cherrier DE, Chea S, Vosshenrich C, Serafini N, Petit M, et al. An id2RFP-reporter mouse redefines innate lymphoid cell precursor potentials. Immunity (2019) 50(4):1054–1068.e3. doi: 10.1016/j.immuni.2019.02.022
236. Liu Q, Kim MH, Friesen L, Kim CH. BATF regulates innate lymphoid cell hematopoiesis and homeostasis. Sci Immunol (2020) 5(54):eaaz8154. doi: 10.1126/sciimmunol.aaz8154
237. Sepahi A, Liu Q, Friesen L, Kim CH. Dietary fiber metabolites regulate innate lymphoid cell responses. Mucosal Immunol (2021) 14(2):317–30. doi: 10.1038/s41385-020-0312-8
238. Yu HB, Yang H, Allaire JM, Ma C, Graef FA, Mortha A, et al. Vasoactive intestinal peptide promotes host defense against enteric pathogens by modulating the recruitment of group 3 innate lymphoid cells. Proc Natl Acad Sci U.S.A. (2021) 118(41):e2106634118. doi: 10.1073/pnas.2106634118
239. Jonckheere AC, Seys SF, Steelant B, Decaesteker T, Dekoster K, Cremer J, et al. Innate lymphoid cells are required to induce airway hyperreactivity in a murine neutrophilic asthma model. Front Immunol (2022) 13:849155. doi: 10.3389/fimmu.2022.849155
240. Zhou J, Hou P, Yao Y, Yue J, Zhang Q, Yi L, et al. Dihydromyricetin Improves High-Fat Diet-Induced Hyperglycemia through ILC3 Activation via a SIRT3-Dependent Mechanism. Mol Nutr Food Res (2022) 66(16):e2101093. doi: 10.1002/mnfr.202101093
241. Seillet C, Luong K, Tellier J, Jacquelot N, Shen RD, Hickey P, et al. The neuropeptide VIP confers anticipatory mucosal immunity by regulating ILC3 activity. Nat Immunol (2020) 21(2):168–77. doi: 10.1038/s41590-019-0567-y
242. Emgård J, Kammoun H, García-Cassani B, Chesné J, Parigi SM, Jacob JM, et al. Oxysterol sensing through the receptor GPR183 promotes the lymphoid-tissue-inducing function of innate lymphoid cells and colonic inflammation. Immunity (2018) 48(1):120–132.e8. doi: 10.1016/j.immuni.2017.11.020
243. Melo-Gonzalez F, Hepworth MR. Identification and functional characterization of murine group 3 innate lymphoid cell (ILC3) subsets in the intestinal tract and associated lymphoid tissues. Methods Mol Biol (2020) 2121:37–49. doi: 10.1007/978-1-0716-0338-3_4
244. Linley H, Ogden A, Jaigirdar S, Buckingham L, Cox J, Priestley M, et al. CD200R1 promotes IL-17 production by ILC3, by enhancing STAT3 activation. Mucosal Immunol (2023) 16(2):167–79. doi: 10.1016/j.mucimm.2023.01.001
245. Sudan R, Fernandes S, Srivastava N, Pedicone C, Meyer ST, Chisholm JD, et al. LRBA deficiency can lead to lethal colitis that is diminished by SHIP1 agonism. Front Immunol (2022) 13:830961. doi: 10.3389/fimmu.2022.830961
246. Ibiza S, García-Cassani B, Ribeiro H, Carvalho T, Almeida L, Marques R, et al. Glial-cell-derived neuroregulators control type 3 innate lymphoid cells and gut defence. Nature (2016) 535(7612):440–3. doi: 10.1038/nature18644
247. Parker ME, Barrera A, Wheaton JD, Zuberbuehler MK, Allan DSJ, Carlyle JR, et al. c-Maf regulates the plasticity of group 3 innate lymphoid cells by restraining the type 1 program. J Exp Med (2020) 217(1):e20191030. doi: 10.1084/jem.20191030
248. Nakagawa S, Matsumoto M, Katayama Y, Oguma R, Wakabayashi S, Nygaard T, et al. Staphylococcus aureus virulent PSMα Peptides induce keratinocyte alarmin release to orchestrate IL-17-dependent skin inflammation. Cell Host Microbe (2017) 22(5):667–677.e5. doi: 10.1016/j.chom.2017.10.008
249. Chun E, Lavoie S, Fonseca-Pereira D, Bae S, Michaud M, Hoveyda HR, et al. Metabolite-sensing receptor ffar2 regulates colonic group 3 innate lymphoid cells and gut immunity. Immunity (2019) 51(5):871–884.e6. doi: 10.1016/j.immuni.2019.09.014
250. Bielecki P, Riesenfeld SJ, Hütter JC, Torlai Triglia E, Kowalczyk MS, Ricardo-Gonzalez RR, et al. Skin-resident innate lymphoid cells converge on a pathogenic effector state. Nature (2021) 592(7852):128–32. doi: 10.1038/s41586-021-03188-w
251. Zhou J, Yue J, Yao Y, Hou P, Zhang T, Zhang Q, et al. Dihydromyricetin protects intestinal barrier integrity by promoting IL-22 expression in ILC3 through the AMPK/SIRT3/STAT3 signaling pathway. Nutrients (2023) 15(2):355. doi: 10.3390/nu15020355
252. Crittenden S, Cheyne A, Adams A, Forster T, Robb CT, Felton J, et al. Purine metabolism controls innate lymphoid cell function and protects against intestinal injury. Immunol Cell Biol (2018) 96(10):1049–59. doi: 10.1111/imcb.12167
253. Seo GY, Shui JW, Takahashi D, Song C, Wang Q, Kim K, et al. LIGHT-HVEM signaling in innate lymphoid cell subsets protects against enteric bacterial infection. Cell Host Microbe (2018) 24(2):249–260.e4. doi: 10.1016/j.chom.2018.07.008
254. Huang J, Lee HY, Zhao X, Han J, Su Y, Sun Q, et al. Interleukin-17D regulates group 3 innate lymphoid cell function through its receptor CD93. Immunity (2021) 54(4):673–686.e4. doi: 10.1016/j.immuni.2021.03.018
255. Vojkovics D, Kellermayer Z, Gábris F, Schippers A, Wagner N, Berta G, et al. Differential effects of the absence of nkx2-3 and MAdCAM-1 on the distribution of intestinal type 3 innate lymphoid cells and postnatal SILT formation in mice. Front Immunol (2019) 10:366. doi: 10.3389/fimmu.2019.00366
256. Song C, Lee JS, Gilfillan S, Robinette ML, Newberry RD, Stappenbeck TS, et al. Unique and redundant functions of NKp46+ ILC3 in models of intestinal inflammation. J Exp Med (2015) 212(11):1869–82. doi: 10.1084/jem.20151403
257. Goto Y, Obata T, Kunisawa J, Sato S, Ivanov II, Lamichhane A, et al. Innate lymphoid cells regulate intestinal epithelial cell glycosylation. Science (2014) 345(6202):1254009. doi: 10.1126/science.1254009
258. Fung TC, Bessman NJ, Hepworth MR, Kumar N, Shibata N, Kobuley D, et al. Lymphoid-tissue-resident commensal bacteria promote members of the IL-10 cytokine family to establish mutualism. Immunity (2016) 44(3):634–46. doi: 10.1016/j.immuni.2016.02.019
259. Silver JS, Kearley J, Copenhaver AM, Sanden C, Mori M, Yu L, et al. Inflammatory triggers associated with exacerbations of COPD orchestrate plasticity of group 2 innate lymphoid cells in the lungs. Nat Immunol (2016) 17(6):626–35. doi: 10.1038/ni.3443
260. Paustian AMS, Paez-Cortez J, Bryant S, Westmoreland S, Waegell W, Kingsbury G. Continuous IL-23 stimulation drives ILC3 depletion in the upper GI tract and, in combination with TNFα, induces robust activation and a phenotypic switch of ILC3. PloS One (2017) 12(8):e0182841. doi: 10.1371/journal.pone.0182841
261. Yang C, Kwon DI, Kim M, Im SH, Lee YJ. Commensal microbiome expands Tγδ17 cells in the lung and promotes particulate matter-induced acute neutrophilia. Front Immunol (2021) 12:645741. doi: 10.3389/fimmu.2021.645741
262. Bhatt B, Zeng P, Zhu H, Sivaprakasam S, Li S, Xiao H, et al. Gpr109a limits microbiota-induced IL-23 production to constrain ILC3-mediated colonic inflammation. J Immunol (2018) 200(8):2905–14. doi: 10.4049/jimmunol.1701625
263. Yang FC, Chiu PY, Chen Y, Mak TW, Chen NJ. TREM-1-dependent M1 macrophage polarization restores intestinal epithelium damaged by DSS-induced colitis by activating IL-22-producing innate lymphoid cells. J BioMed Sci (2019) 26(1):46. doi: 10.1186/s12929-019-0539-4
264. Poholek CH, Dulson SJ, Zajac AJ, Harrington LE. IL-21 controls ILC3 cytokine production and promotes a protective phenotype in a mouse model of colitis. Immunohorizons (2019) 3(6):194–202. doi: 10.4049/immunohorizons.1900005
265. Di Luccia B, Gilfillan S, Cella M, Colonna M, Huang SC. ILC3 integrate glycolysis and mitochondrial production of reactive oxygen species to fulfill activation demands. J Exp Med (2019) 216(10):2231–41. doi: 10.1084/jem.20180549
266. Bauché D, Joyce-Shaikh B, Jain R, Grein J, Ku KS, Blumenschein WM, et al. LAG3+ Regulatory T cells restrain interleukin-23-producing CX3CR1+ Gut-resident macrophages during group 3 innate lymphoid cell-driven colitis. Immunity (2018) 49(2):342–352.e5. doi: 10.1016/j.immuni.2018.07.007
267. Seo GY, Giles DA, Kronenberg M. Bacterial infection allows for functional examination of adoptively transferred mouse innate lymphoid cell subsets. Methods Mol Biol (2020) 2121:129–40. doi: 10.1007/978-1-0716-0338-3_12
268. He L, Zhou M, Li YC. Vitamin D/vitamin D receptor signaling is required for normal development and function of group 3 innate lymphoid cells in the gut. iScience (2019) 17:119–31. doi: 10.1016/j.isci.2019.06.026
269. Fachi JL, Pral LP, Dos Santos JAC, Codo AC, de Oliveira S, Felipe JS, et al. Hypoxia enhances ILC3 responses through HIF-1α-dependent mechanism. Mucosal Immunol (2021) 14(4):828–41. doi: 10.1038/s41385-020-00371-6
270. Li Y, Ge J, Zhao X, Xu M, Gou M, Xie B, et al. Cell autonomous expression of BCL6 is required to maintain lineage identity of mouse CCR6+ ILC3s. J Exp Med (2023) 220(4):e20220440. doi: 10.1084/jem.20220440
271. Xiong L, Wang S, Dean JW, Oliff KN, Jobin C, Curtiss R 3rd, et al. Group 3 innate lymphoid cell pyroptosis represents a host defence mechanism against Salmonella infection. Nat Microbiol (2022) 7(7):1087–99. doi: 10.1038/s41564-022-01142-8
272. Peng V, Xing X, Bando JK, Trsan T, Di Luccia B, Collins PL, et al. Whole-genome profiling of DNA methylation and hydroxymethylation identifies distinct regulatory programs among innate lymphocytes. Nat Immunol (2022) 23(4):619–31. doi: 10.1038/s41590-022-01164-8
273. Chang D, Zhang H, Ge J, Xing Q, Guo X, Wang X, et al. A cis-element at the Rorc locus regulates the development of type 3 innate lymphoid cells. Front Immunol (2023) 14:1105145. doi: 10.3389/fimmu.2023.1105145
274. Hu P, Leyton L, Hagood JS, Barker TH. Thy-1-integrin interactions in cis and trans mediate distinctive signaling. Front Cell Dev Biol (2022) 10:928510. doi: 10.3389/fcell.2022.928510
275. Krzywinska E, Sobecki M, Nagarajan S, Zacharjasz J, Tambuwala MM, Pelletier A, et al. The transcription factor HIF-1α mediates plasticity of NKp46+ innate lymphoid cells in the gut. J Exp Med (2022) 219(2):e20210909. doi: 10.1084/jem.20210909
276. Hou Q, Huang J, Xiong X, Guo Y, Zhang B. Role of nutrient-sensing receptor GPRC6A in regulating colonic group 3 innate lymphoid cells and inflamed mucosal healing. J Crohns Colitis (2022) 16(8):1293–305. doi: 10.1093/ecco-jcc/jjac020
277. Chen L, He Z, Slinger E, Bongers G, Lapenda TLS, Pacer ME, et al. IL-23 activates innate lymphoid cells to promote neonatal intestinal pathology. Mucosal Immunol (2015) 8(2):390–402. doi: 10.1038/mi.2014.77
278. Ma Z, Li C, Xue L, Zhang S, Yang Y, Zhang H, et al. Linggan Wuwei Jiangxin formula ameliorates airway hyperresponsiveness through suppression of IL-1β and IL-17A expression in allergic asthmatic mice especially with diet-induced obesity. Ann Transl Med (2021) 9(8):682. doi: 10.21037/atm-21-1189
279. Kim HY, Lee HJ, Chang YJ, Pichavant M, Shore SA, Fitzgerald KA, et al. Interleukin-17-producing innate lymphoid cells and the NLRP3 inflammasome facilitate obesity-associated airway hyperreactivity. Nat Med (2014) 20(1):54–61. doi: 10.1038/nm.3423
280. He J, Shen X, Fu D, Yang Y, Xiong K, Zhao L, et al. Human periodontitis-associated salivary microbiome affects the immune response of diabetic mice. J Oral Microbiol (2022) 14(1):2107814. doi: 10.1080/20002297.2022.2107814
281. Nettleford SK, Zhao L, Qian F, Herold M, Arner B, Desai D, et al. The essential role of selenoproteins in the resolution of citrobacter rodentium-induced intestinal inflammation. Front Nutr (2020) 7:96. doi: 10.3389/fnut.2020.00096
282. Di Censo C, Marotel M, Mattiola I, Müller L, Scarno G, Pietropaolo G, et al. Granzyme A and CD160 expression delineates ILC1 with graded functions in the mouse liver. Eur J Immunol (2021) 51(11):2568–75. doi: 10.1002/eji.202149209
283. Valle-Noguera A, Gómez-Sánchez MJ, Girard-Madoux MJH, Cruz-Adalia A. Optimized protocol for characterization of mouse gut innate lymphoid cells. Front Immunol (2020) 11:563414. doi: 10.3389/fimmu.2020.563414
284. Huang S, Wang X, Xie X, Su Y, Pan Z, Li Y, et al. Dahuang Mudan decoction repairs intestinal barrier in chronic colitic mice by regulating the function of ILC3. J Ethnopharmacol (2022) 299:115652. doi: 10.1016/j.jep.2022.115652
285. Chen MJ, Feng Y, Gao L, Lin MX, Wang SD, Tong ZQ. Composite sophora colon-soluble capsule ameliorates DSS-induced ulcerative colitis in mice via gut microbiota-derived butyric acid and NCR+ ILC3. Chin J Integr Med (2023) 29(5):424–33. doi: 10.1007/s11655-022-3317-1
286. Mielke LA, Groom JR, Rankin LC, Seillet C, Masson F, Putoczki T, et al. TCF-1 controls ILC2 and NKp46+RORγt+ innate lymphocyte differentiation and protection in intestinal inflammation. J Immunol (2013) 191(8):4383–91. doi: 10.4049/jimmunol.1301228
287. Xie X, Zhao M, Huang S, Li P, Chen P, Luo X, et al. Luteolin alleviates ulcerative colitis by restoring the balance of NCR-ILC3/NCR+ILC3 to repairing impaired intestinal barrier. Int Immunopharmacol (2022) 112:109251. doi: 10.1016/j.intimp.2022.109251
288. Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK, et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol (2015) 16(3):306–17. doi: 10.1038/ni.3094
289. Rankin LC, Girard-Madoux MJ, Seillet C, Mielke LA, Kerdiles Y, Fenis A, et al. Complementarity and redundancy of IL-22-producing innate lymphoid cells. Nat Immunol (2016) 17(2):179–86. doi: 10.1038/ni.3332
290. Viant C, Rankin LC, Girard-Madoux MJ, Seillet C, Shi W, Smyth MJ, et al. Transforming growth factor-β and Notch ligands act as opposing environmental cues in regulating the plasticity of type 3 innate lymphoid cells. Sci Signal (2016) 9(426):ra46. doi: 10.1126/scisignal.aaf2176
291. Goverse G, Labao-Almeida C, Ferreira M, Molenaar R, Wahlen S, Konijn T, et al. Vitamin A controls the presence of RORγ+ Innate lymphoid cells and lymphoid tissue in the small intestine. J Immunol (2016) 196(12):5148–55. doi: 10.4049/jimmunol.1501106
292. Robinette ML, Bando JK, Song W, Ulland TK, Gilfillan S, Colonna M. IL-15 sustains IL-7R-independent ILC2 and ILC3 development. Nat Commun (2017) 8:14601. doi: 10.1038/ncomms14601
293. Liu B, Ye B, Zhu X, Huang G, Yang L, Zhu P, et al. IL-7Rα glutamylation and activation of transcription factor Sall3 promote group 3 ILC development. Nat Commun (2017) 8(1):231. doi: 10.1038/s41467-017-00235-x
294. Robinette ML, Cella M, Telliez JB, Ulland TK, Barrow AD, Capuder K, et al. Jak3 deficiency blocks innate lymphoid cell development. Mucosal Immunol (2018) 11(1):50–60. doi: 10.1038/mi.2017.38
295. Abou-Samra E, Hickey Z, Aguilar OA, Scur M, Mahmoud AB, Pyatibrat S, et al. NKR-P1B expression in gut-associated innate lymphoid cells is required for the control of gastrointestinal tract infections. Cell Mol Immunol (2019) 16(11):868–77. doi: 10.1038/s41423-018-0169-x
296. Gronke K, Hernández PP, Zimmermann J, Klose CSN, Kofoed-Branzk M, Guendel F, et al. Interleukin-22 protects intestinal stem cells against genotoxic stress. Nature (2019) 566(7743):249–53. doi: 10.1038/s41586-019-0899-7
297. Li Y, Xie HQ, Zhang W, Wei Y, Sha R, Xu L, et al. Type 3 innate lymphoid cells are altered in colons of C57BL/6 mice with dioxin exposure. Sci Total Environ (2019) 662:639–45. doi: 10.1016/j.scitotenv.2019.01.139
298. Yamano T, Dobeš J, Vobořil M, Steinert M, Brabec T, Ziętara N, et al. Aire-expressing ILC3-like cells in the lymph node display potent APC features. J Exp Med (2019) 216(5):1027–37. doi: 10.1084/jem.20181430
299. Qi H, Li Y, Yun H, Zhang T, Huang Y, Zhou J, et al. Lactobacillus maintains healthy gut mucosa by producing L-Ornithine. Commun Biol (2019) 2:171. doi: 10.1038/s42003-019-0424-4
300. Ke K, Chen TH, Arra M, Mbalaviele G, Swarnkar G, Abu-Amer Y. Attenuation of NF-κB in intestinal epithelial cells is sufficient to mitigate the bone loss comorbidity of experimental mouse colitis. J Bone Miner Res (2019) 34(10):1880–93. doi: 10.1002/jbmr.3759
301. Gao Y, Bian Z, Xue W, Li Q, Zeng Y, Wang Y, et al. Human IL-23R cytokine-binding homology region-fc fusion protein ameliorates psoriasis via the decrease of systemic th17 and ILC3 cell responses. Int J Mol Sci (2019) 20(17):4170. doi: 10.3390/ijms20174170
302. Wang Q, Robinette ML, Billon C, Collins PL, Bando JK, Fachi JL, et al. Circadian rhythm-dependent and circadian rhythm-independent impacts of the molecular clock on type 3 innate lymphoid cells. Sci Immunol (2019) 4(40):eaay7501. doi: 10.1126/sciimmunol.aay7501
303. Kim HJ, Lee SH, Hong SJ. Antibiotics-induced dysbiosis of intestinal microbiota aggravates atopic dermatitis in mice by altered short-chain fatty acids. Allergy Asthma Immunol Res (2020) 12(1):137–48. doi: 10.4168/aair.2020.12.1.137
304. Bank U, Deiser K, Plaza-Sirvent C, Osbelt L, Witte A, Knop L, et al. c-FLIP is crucial for IL-7/IL-15-dependent NKp46+ ILC development and protection from intestinal inflammation in mice. Nat Commun (2020) 11(1):1056. doi: 10.1038/s41467-020-14782-3
305. Zhu Y, Shi T, Lu X, Xu Z, Qu J, Zhang Z, et al. Fungal-induced glycolysis in macrophages promotes colon cancer by enhancing innate lymphoid cell secretion of IL-22. EMBO J (2021) 40(11):e105320. doi: 10.15252/embj.2020105320
306. Zhang W, Zhou Q, Liu H, Xu J, Huang R, Shen B, et al. Bacteroides fragilis strain ZY-312 facilitates colonic mucosa regeneration in colitis via motivating STAT3 signaling pathway induced by IL-22 from ILC3 secretion. Front Immunol (2023) 14:1156762. doi: 10.3389/fimmu.2023.1156762
307. Chu S, Ma L, Yang X, Xiao B, Liang Y, Zheng S, et al. NCR negative group 3 innate lymphoid cell (NCR- ILC3) participates in abnormal pathology of lung in cigarette smoking-induced COPD mice. Immun Inflammation Dis (2023) 11(3):e816. doi: 10.1002/iid3.816
308. Ma B, Zhou Y, Hu Y, Duan H, Sun Z, Wang P, et al. Mapping resident immune cells in the murine ocular surface and lacrimal gland by flow cytometry. Ocul Immunol Inflammation (2023) 31(4):748–59. doi: 10.1080/09273948.2023.2182327
309. Zhao X, Liang W, Wang Y, Yi R, Luo L, Wang W, et al. Ontogeny of RORγt+ cells in the intestine of newborns and its role in the development of experimental necrotizing enterocolitis. Cell Biosci (2022) 12(1):3. doi: 10.1186/s13578-021-00739-6
310. Lehmann FM, von Burg N, Ivanek R, Teufel C, Horvath E, Peter A, et al. Microbiota-induced tissue signals regulate ILC3-mediated antigen presentation. Nat Commun (2020) 11(1):1794. doi: 10.1038/s41467-020-15612-2
311. Irshad S, Flores-Borja F, Lawler K, Monypenny J, Evans R, Male V, et al. RORγt+ Innate lymphoid cells promote lymph node metastasis of breast cancers. Cancer Res (2017) 77(5):1083–96. doi: 10.1158/0008-5472.CAN-16-0598
312. Seshadri S, Allan DSJ, Carlyle JR, Zenewicz LA. Bacillus anthracis lethal toxin negatively modulates ILC3 function through perturbation of IL-23-mediated MAPK signaling. PloS Pathog (2017) 13(10):e1006690. doi: 10.1371/journal.ppat.1006690
313. Giacomin PR, Moy RH, Noti M, Osborne LC, Siracusa MC, Alenghat T, et al. Epithelial-intrinsic IKKα expression regulates group 3 innate lymphoid cell responses and antibacterial immunity. J Exp Med (2015) 212(10):1513–28. doi: 10.1084/jem.20141831
314. Talbot J, Hahn P, Kroehling L, Nguyen H, Li D, Littman DR. Feeding-dependent VIP neuron-ILC3 circuit regulates the intestinal barrier. Nature (2020) 579(7800):575–80. doi: 10.1038/s41586-020-2039-9
315. Sasaki T, Moro K, Kubota T, Kubota N, Kato T, Ohno H, et al. Innate lymphoid cells in the induction of obesity. Cell Rep (2019) 28(1):202–217.e7. doi: 10.1016/j.celrep.2019.06.016
316. Muraoka WT, Korchagina AA, Xia Q, Shein SA, Jing X, Lai Z, et al. Campylobacter infection promotes IFNγ-dependent intestinal pathology via ILC3 to ILC1 conversion. Mucosal Immunol (2021) 14(3):703–16. doi: 10.1038/s41385-020-00353-8
317. Wang B, Lim JH, Kajikawa T, Li X, Vallance BA, Moutsopoulos NM, et al. Macrophage β2-integrins regulate IL-22 by ILC3 and protect from lethal citrobacter rodentium-induced colitis. Cell Rep (2019) 26(6):1614–1626.e5. doi: 10.1016/j.celrep.2019.01.054
318. Snyder LM, Doherty CM, Mercer HL, Denkers EY. Induction of IL-12p40 and type 1 immunity by Toxoplasma gondii in the absence of the TLR-MyD88 signaling cascade. PloS Pathog (2021) 17(10):e1009970. doi: 10.1371/journal.ppat.1009970
319. Van Maele L, Carnoy C, Cayet D, Ivanov S, Porte R, Deruy E, et al. Activation of Type 3 innate lymphoid cells and interleukin 22 secretion in the lungs during Streptococcus pneumoniae infection. J Infect Dis (2014) 210(3):493–503. doi: 10.1093/infdis/jiu106
320. Lo BC, Canals Hernaez D, Scott RW, Hughes MR, Shin SB, Underhill TM, et al. The transcription factor RORα Preserves ILC3 lineage identity and function during chronic intestinal infection. J Immunol (2019) 203(12):3209–15. doi: 10.4049/jimmunol.1900781
321. Stier MT, Goleniewska K, Cephus JY, Newcomb DC, Sherrill TP, Boyd KL, et al. STAT1 represses cytokine-producing group 2 and group 3 innate lymphoid cells during viral infection. J Immunol (2017) 199(2):510–9. doi: 10.4049/jimmunol.1601984
322. Yang J, Cornelissen F, Papazian N, Reijmers RM, Llorian M, Cupedo T, et al. IL-7-dependent maintenance of ILC3s is required for normal entry of lymphocytes into lymph nodes. J Exp Med (2018) 215(4):1069–77. doi: 10.1084/jem.20170518
323. Liébana-García R, Olivares M, Francés-Cuesta C, Rubio T, Rossini V, Quintas G, et al. Intestinal group 1 innate lymphoid cells drive macrophage-induced inflammation and endocrine defects in obesity and promote insulinemia. Gut Microbes (2023) 15(1):2181928. doi: 10.1080/19490976.2023.2181928
324. Chan IH, Jain R, Tessmer MS, Gorman D, Mangadu R, Sathe M, et al. Interleukin-23 is sufficient to induce rapid de novo gut tumorigenesis, independent of carcinogens, through activation of innate lymphoid cells. Mucosal Immunol (2014) 7(4):842–56. doi: 10.1038/mi.2013.101
325. Wu X, Khatun A, Kasmani MY, Chen Y, Zheng S, Atkinson S, et al. Group 3 innate lymphoid cells require BATF to regulate gut homeostasis in mice. J Exp Med (2022) 219(11):e20211861. doi: 10.1084/jem.20211861
326. Friedrich C, Mamareli P, Thiemann S, Kruse F, Wang Z, Holzmann B, et al. MyD88 signaling in dendritic cells and the intestinal epithelium controls immunity against intestinal infection with C. rodentium. PloS Pathog (2017) 13(5):e1006357. doi: 10.1371/journal.ppat.1006357
327. Kim M, Gu B, Madison MC, Song HW, Norwood K, Hill AA, et al. Cigarette smoke induces intestinal inflammation via a th17 cell-neutrophil axis. Front Immunol (2019) 10:75. doi: 10.3389/fimmu.2019.00075
328. Snyder LM, Belmares-Ortega J, Doherty CM, Denkers EY. Impact of MyD88, Microbiota, and Location on Type 1 and Type 3 Innate Lymphoid Cells during Toxoplasma gondii Infection. Immunohorizons (2022) 6(9):660–70. doi: 10.4049/immunohorizons.2200070
329. Lin YD, Arora J, Diehl K, Bora SA, Cantorna MT. Vitamin D is required for ILC3 derived IL-22 and protection from citrobacter rodentium infection. Front Immunol (2019) 10:1. doi: 10.3389/fimmu.2019.00001
330. Teufel C, Horvath E, Peter A, Ercan C, Piscuoglio S, Hall MN, et al. mTOR signaling mediates ILC3-driven immunopathology. Mucosal Immunol (2021) 14(6):1323–34. doi: 10.1038/s41385-021-00432-4
331. Chea S, Perchet T, Petit M, Verrier T, Guy-Grand D, Banchi EG, et al. Notch signaling in group 3 innate lymphoid cells modulates their plasticity. Sci Signal (2016) 9(426):ra45. doi: 10.1126/scisignal.aaf2223
332. Li S, Heller JJ, Bostick JW, Lee A, Schjerven H, Kastner P, et al. Ikaros inhibits group 3 innate lymphoid cell development and function by suppressing the aryl hydrocarbon receptor pathway. Immunity (2016) 45(1):185–97. doi: 10.1016/j.immuni.2016.06.027
333. Suo C, Fan Z, Zhou L, Qiu J. Perfluorooctane sulfonate affects intestinal immunity against bacterial infection. Sci Rep (2017) 7(1):5166. doi: 10.1038/s41598-017-04091-z
334. Aguiar SLF, Miranda MCG, Guimarães MAF, Santiago HC, Queiroz CP, Cunha PDS, et al. High-salt diet induces IL-17-dependent gut inflammation and exacerbates colitis in mice. Front Immunol (2018) 8:1969. doi: 10.3389/fimmu.2017.01969
335. Mikami Y, Scarno G, Zitti B, Shih HY, Kanno Y, Santoni A, et al. NCR+ ILC3 maintain larger STAT4 reservoir via T-BET to regulate type 1 features upon IL-23 stimulation in mice. Eur J Immunol (2018) 48(7):1174–80. doi: 10.1002/eji.201847480
336. Almeida FF, Tognarelli S, Marçais A, Kueh AJ, Friede ME, Liao Y, et al. A point mutation in the Ncr1 signal peptide impairs the development of innate lymphoid cell subsets. Oncoimmunology (2018) 7(10):e1475875. doi: 10.1080/2162402X.2018.1475875
337. Deng T, Suo C, Chang J, Yang R, Li J, Cai T, et al. ILC3-derived OX40L is essential for homeostasis of intestinal Tregs in immunodeficient mice. Cell Mol Immunol (2020) 17(2):163–77. doi: 10.1038/s41423-019-0200-x
338. Li J, Shi W, Sun H, Ji Y, Chen Y, Guo X, et al. Activation of DR3 signaling causes loss of ILC3 and exacerbates intestinal inflammation. Nat Commun (2019) 10(1):3371. doi: 10.1038/s41467-019-11304-8
339. Yordanova IA, Lamatsch M, Kühl AA, Hartmann S, Rausch S. Eosinophils are dispensable for the regulation of IgA and Th17 responses in Giardia muris infection. Parasite Immunol (2021) 43(3):e12791. doi: 10.1111/pim.12791
340. Cho SX, Rudloff I, Lao JC, Pang MA, Goldberg R, Bui CB, et al. Characterization of the pathoimmunology of necrotizing enterocolitis reveals novel therapeutic opportunities. Nat Commun (2020) 11(1):5794. doi: 10.1038/s41467-020-19400-w
341. Chang J, Ji X, Deng T, Qiu J, Ding Z, Li Z, et al. Setd2 determines distinct properties of intestinal ILC3 subsets to regulate intestinal immunity. Cell Rep (2022) 38(11):110530. doi: 10.1016/j.celrep.2022.110530
342. Sano T, Huang W, Hall JA, Yang Y, Chen A, Gavzy SJ, et al. An IL-23R/IL-22 circuit regulates epithelial serum amyloid A to promote local effector th17 responses. Cell (2015) 163(2):381–93. doi: 10.1016/j.cell.2015.08.061
343. Tan C, Hong G, Wang Z, Duan C, Hou L, Wu J, et al. Promoting effect of L-fucose on the regeneration of intestinal stem cells through AHR/IL-22 pathway of intestinal lamina propria monocytes. Nutrients (2022) 14(22):4789. doi: 10.3390/nu14224789
344. Riba A, Olier M, Lacroix-Lamandé S, Lencina C, Bacquié V, Harkat C, et al. Early life stress in mice is a suitable model for Irritable Bowel Syndrome but does not predispose to colitis nor increase susceptibility to enteric infections. Brain Behav Immun (2018) 73:403–15. doi: 10.1016/j.bbi.2018.05.024
345. Park HJ, Lee SW, Van Kaer L, Hong S. CD1d-dependent iNKT cells control DSS-induced colitis in a mouse model of IFNγ-mediated hyperinflammation by increasing IL22-secreting ILC3 cells. Int J Mol Sci (2021) 22(3):1250. doi: 10.3390/ijms22031250
346. Eberl G. RORγt, a multitask nuclear receptor at mucosal surfaces. Mucosal Immunol (2017) 10(1):27–34. doi: 10.1038/mi.2016.86
347. Taylor PR, Roy S, Leal SM Jr, Sun Y, Howell SJ, Cobb BA, et al. Activation of neutrophils by autocrine IL-17A-IL-17RC interactions during fungal infection is regulated by IL-6, IL-23, RORγt and dectin-2. Nat Immunol (2014) 15(2):143–51. doi: 10.1038/ni.2797
348. Savers A, Rasid O, Parlato M, Brock M, Jouvion G, Ryffel B, et al. Infection-Mediated Priming of Phagocytes Protects against Lethal Secondary Aspergillus fumigatus Challenge. PloS One (2016) 11(4):e0153829. doi: 10.1371/journal.pone.0153829
349. Brown CC, Gudjonson H, Pritykin Y, Deep D, Lavallée VP, Mendoza A, et al. Transcriptional basis of mouse and human dendritic cell heterogeneity. Cell (2019) 179(4):846–863.e24. doi: 10.1016/j.cell.2019.09.035
350. Sonnenberg GF, Monticelli LA, Elloso MM, Fouser LA, Artis D. CD4(+) lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity (2011) 34(1):122–34. doi: 10.1016/j.immuni.2010.12.009
351. Crispín JC, Oukka M, Bayliss G, Cohen RA, Van Beek CA, Stillman IE, et al. Expanded double negative T cells in patients with systemic lupus erythematosus produce IL-17 and infiltrate the kidneys. J Immunol (2008) 181(12):8761–6. doi: 10.4049/jimmunol.181.12.8761
352. Ueyama A, Imura C, Fusamae Y, Tsujii K, Furue Y, Aoki M, et al. Potential role of IL-17-producing CD4/CD8 double negative αβ T cells in psoriatic skin inflammation in a TPA-induced STAT3C transgenic mouse model. J Dermatol Sci (2017) 85(1):27–35. doi: 10.1016/j.jdermsci.2016.10.007
353. Thomas ML. The leukocyte common antigen family. Annu Rev Immunol (1989) 7:339–69. doi: 10.1146/annurev.iy.07.040189.002011
354. Nakano H, Yanagita M, Gunn MD. CD11c(+)B220(+)Gr-1(+) cells in mouse lymph nodes and spleen display characteristics of plasmacytoid dendritic cells. J Exp Med (2001) 194(8):1171–8. doi: 10.1084/jem.194.8.1171
355. Merkenschlager M, Fisher AG. CD45 isoform switching precedes the activation-driven death of human thymocytes by apoptosis. Int Immunol (1991) 3(1):1–7. doi: 10.1093/intimm/3.1.1
356. Renno T, Attinger A, Rimoldi D, Hahne M, Tschopp J, MacDonald HR. Expression of B220 on activated T cell blasts precedes apoptosis. Eur J Immunol (1998) 28(2):540–7. doi: 10.1002/(SICI)1521-4141(199802)28:02<540::AID-IMMU540>3.0.CO;2-Y
357. Oka S, Mori N, Matsuyama S, Takamori Y, Kubo K. Presence of B220 within thymocytes and its expression on the cell surface during apoptosis. Immunology (2000) 100(4):417–23. doi: 10.1046/j.1365-2567.2000.00063.x
358. Melchers F, ten Boekel E, Seidl T, Kong XC, Yamagami T, Onishi K, et al. Repertoire selection by pre-B-cell receptors and B-cell receptors, and genetic control of B-cell development from immature to mature B cells. Immunol Rev (2000) 175:33–46. doi: 10.1111/j.1600-065X.2000.imr017510.x
359. Wang K, Wei G, Liu D. CD19: a biomarker for B cell development, lymphoma diagnosis and therapy. Exp Hematol Oncol (2012) 1(1):36. doi: 10.1186/2162-3619-1-36
360. Meininger I, Carrasco A, Rao A, Soini T, Kokkinou E, Mjösberg J. Tissue-specific features of innate lymphoid cells. Trends Immunol (2020) 41(10):902–17. doi: 10.1016/j.it.2020.08.009
361. Van Acker HH, Capsomidis A, Smits EL, Van Tendeloo VF. CD56 in the immune system: more than a marker for cytotoxicity? Front Immunol (2017) 8:892. doi: 10.3389/fimmu.2017.00892
362. Carlyle JR, Mesci A, Ljutic B, Belanger S, Tai LH, Rousselle E, et al. Molecular and genetic basis for strain-dependent NK1.1 alloreactivity of mouse NK cells. J Immunol (2006) 176(12):7511–24. doi: 10.4049/jimmunol.176.12.7511
363. Sheikh A, Abraham N. Interleukin-7 receptor alpha in innate lymphoid cells: more than a marker. Front Immunol (2019) 10:2897. doi: 10.3389/fimmu.2019.02897
364. Artis D, Spits H. The biology of innate lymphoid cells. Nature (2015) 517(7534):293–301. doi: 10.1038/nature14189
365. Alves NL, van Leeuwen EM, Remmerswaal EB, Vrisekoop N, Tesselaar K, Roosnek E, et al. A new subset of human naive CD8+ T cells defined by low expression of IL-7R alpha. J Immunol (2007) 179(1):221–8. doi: 10.4049/jimmunol.179.1.221
366. Huster KM, Busch V, Schiemann M, Linkemann K, Kerksiek KM, Wagner H, et al. Selective expression of IL-7 receptor on memory T cells identifies early CD40L-dependent generation of distinct CD8+ memory T cell subsets. Proc Natl Acad Sci U.S.A. (2004) 101(15):5610–5. doi: 10.1073/pnas.0308054101
367. Liu W, Putnam AL, Xu-Yu Z, Szot GL, Lee MR, Zhu S, et al. CD127 expression inversely correlates with FoxP3 and suppressive function of human CD4+ T reg cells. J Exp Med (2006) 203(7):1701–11. doi: 10.1084/jem.20060772
368. Lennartsson J, Rönnstrand L. Stem cell factor receptor/c-Kit: from basic science to clinical implications. Physiol Rev (2012) 92(4):1619–49. doi: 10.1152/physrev.00046.2011
369. Frumento G, Zuo J, Verma K, Croft W, Ramagiri P, Chen FE, et al. CD117 (c-kit) is expressed during CD8+ T cell priming and stratifies sensitivity to apoptosis according to strength of TCR engagement. Front Immunol (2019) 10:468. doi: 10.3389/fimmu.2019.00468
370. Onkanga IO, Hamilton R, Mwinzi PNM, Schneider T, Ondigo BN, Sang H, et al. Expression of CD117 (c-kit) on circulating B cells in pediatric schistosomiasis. Infect Immun (2022) 90(8):e0016022. doi: 10.1128/iai.00160-22
371. Oliver ET, Chichester K, Devine K, Sterba PM, Wegner C, Vonakis BM, et al. Effects of an oral CRTh2 antagonist (AZD1981) on eosinophil activity and symptoms in chronic spontaneous urticaria. Int Arch Allergy Immunol (2019) 179(1):21–30. doi: 10.1159/000496162
372. Sauzay C, Voutetakis K, Chatziioannou A, Chevet E, Avril T. CD90/thy-1, a cancer-associated cell surface signaling molecule. Front Cell Dev Biol (2019) 7:66. doi: 10.3389/fcell.2019.00066
373. Corral D, Charton A, Krauss MZ, Blanquart E, Levillain F, Lefrançais E, et al. ILC precursors differentiate into metabolically distinct ILC1-like cells during Mycobacterium tuberculosis infection. Cell Rep (2022) 39(3):110715. doi: 10.1016/j.celrep.2022.110715
374. Sonnenberg GF, Monticelli LA, Alenghat T, Fung TC, Hutnick NA, Kunisawa J, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science (2012) 336(6086):1321–5. doi: 10.1126/science.1222551
375. Schroeder JH, Beattie G, Lo JW, Zabinski T, Powell N, Neves JF, et al. CD90 is not constitutively expressed in functional innate lymphoid cells. Front Immunol (2023) 14:1113735. doi: 10.3389/fimmu.2023.1113735
376. Poggi A, Rubartelli A, Moretta L, Zocchi MR. Expression and function of NKRP1A molecule on human monocytes and dendritic cells. Eur J Immunol (1997) 27(11):2965–70. doi: 10.1002/eji.1830271132
377. Mjösberg JM, Trifari S, Crellin NK, Peters CP, van Drunen CM, Piet B, et al. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat Immunol (2011) 12(11):1055–62. doi: 10.1038/ni.2104
378. Cogswell A, Ferguson N, Barker E. Presence of inflammatory group I and III innate lymphoid cells in the colon of simian immunodeficiency virus-infected rhesus macaques. J Virol (2020) 94(9):e01914–19. doi: 10.1128/JVI.01914-19
379. Starner TD, Barker CK, Jia HP, Kang Y, McCray PB Jr. CCL20 is an inducible product of human airway epithelia with innate immune properties. Am J Respir Cell Mol Biol (2003) 29(5):627–33. doi: 10.1165/rcmb.2002-0272OC
380. Kulkarni N, Pathak M, Lal G. Role of chemokine receptors and intestinal epithelial cells in the mucosal inflammation and tolerance. J Leukoc Biol (2017) 101(2):377–94. doi: 10.1189/jlb.1RU0716-327R
381. Ignacio A, Breda CNS, Camara NOS. Innate lymphoid cells in tissue homeostasis and diseases. World J Hepatol (2017) 9(23):979–89. doi: 10.4254/wjh.v9.i23.979
382. Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi Y, Littman DR. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat Immunol (2004) 5(1):64–73. doi: 10.1038/ni1022
383. Glasner A, Isaacson B, Viukov S, Neuman T, Friedman N, Mandelboim M, et al. Increased NK cell immunity in a transgenic mouse model of NKp46 overexpression. Sci Rep (2017) 7(1):13090. doi: 10.1038/s41598-017-12998-w
384. Sivori S, Vitale M, Morelli L, Sanseverino L, Augugliaro R, Bottino C, et al. p46, a novel natural killer cell-specific surface molecule that mediates cell activation. J Exp Med (1997) 186(7):1129–36. doi: 10.1084/jem.186.7.1129
385. Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate lymphoid cells: 10 years on. Cell (2018) 174(5):1054–66. doi: 10.1016/j.cell.2018.07.017
386. Bottino C, Biassoni R, Millo R, Moretta L, Moretta A. The human natural cytotoxicity receptors (NCR) that induce HLA class I-independent NK cell triggering. Hum Immunol (2000) 61(1):1–6. doi: 10.1016/s0198-8859(99)00162-7
387. Glatzer T, Killig M, Meisig J, Ommert I, Luetke-Eversloh M, Babic M, et al. RORγt+ innate lymphoid cells acquire a proinflammatory program upon engagement of the activating receptor NKp44. Immunity (2013) 38(6):1223–35. doi: 10.1016/j.immuni.2013.05.013
388. Cella M, Miller H, Song C. Beyond NK cells: the expanding universe of innate lymphoid cells. Front Immunol (2014) 5:282. doi: 10.3389/fimmu.2014.00282
389. Simoni Y, Fehlings M, Kløverpris HN, McGovern N, Koo SL, Loh CY, et al. Human innate lymphoid cell subsets possess tissue-type based heterogeneity in phenotype and frequency. Immunity (2017) 46(1):148–61. doi: 10.1016/j.immuni.2016.11.005
390. Bernink JH, Peters CP, Munneke M, te Velde AA, Meijer SL, Weijer K, et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol (2013) 14(3):221–9. doi: 10.1038/ni.2534
391. Geremia A, Arancibia-Cárcamo CV, Fleming MP, Rust N, Singh B, Mortensen NJ, et al. IL-23-responsive innate lymphoid cells are increased in inflammatory bowel disease. J Exp Med (2011) 208(6):1127–33. doi: 10.1084/jem.20101712
392. Rao A, Strauss O, Kokkinou E, Bruchard M, Tripathi KP, Schlums H, et al. Cytokines regulate the antigen-presenting characteristics of human circulating and tissue-resident intestinal ILCs. Nat Commun (2020) 11(1):2049. doi: 10.1038/s41467-020-15695-x
393. Jarade A, Di Santo JP, Serafini N. Group 3 innate lymphoid cells mediate host defense against attaching and effacing pathogens. Curr Opin Microbiol (2021) 63:83–91. doi: 10.1016/j.mib.2021.06.005
394. Pian Y, Chai Q, Ren B, Wang Y, Lv M, Qiu J, et al. Type 3 Innate Lymphoid Cells Direct Goblet Cell Differentiation via the LT-LTβR Pathway during Listeria Infection. J Immunol (2020) 205(3):853–63. doi: 10.4049/jimmunol.2000197
395. Simoni Y, Newell EW. Dissecting human ILC heterogeneity: more than just three subsets. Immunology (2018) 153(3):297–303. doi: 10.1111/imm.12862
396. Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature (2013) 498(7452):113–7. doi: 10.1038/nature12240
397. Kiss EA, Diefenbach A. Role of the aryl hydrocarbon receptor in controlling maintenance and functional programs of RORγt(+) innate lymphoid cells and intraepithelial lymphocytes. Front Immunol (2012) 3:124. doi: 10.3389/fimmu.2012.00124
398. Sécca C, Bando JK, Fachi JL, Gilfillan S, Peng V, Di Luccia B, et al. Spatial distribution of LTi-like cells in intestinal mucosa regulates type 3 innate immunity. Proc Natl Acad Sci U.S.A. (2021) 118(23):e2101668118. doi: 10.1073/pnas.2101668118
399. Ito T, Carson WF4, Cavassani KA, Connett JM, Kunkel SL. CCR6 as a mediator of immunity in the lung and gut. Exp Cell Res (2011) 317(5):613–9. doi: 10.1016/j.yexcr.2010.12.018
400. Takatori H, Kanno Y, Watford WT, Tato CM, Weiss G, Ivanov II, et al. Lymphoid tissue inducer-like cells are an innate source of IL-17 and IL-22. J Exp Med (2009) 206(1):35–41. doi: 10.1084/jem.20072713
401. Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature (2014) 508(7496):397–401. doi: 10.1038/nature13047
402. Zhong C, Zheng M, Cui K, Martins AJ, Hu G, Li D, et al. Differential expression of the transcription factor GATA3 specifies lineage and functions of innate lymphoid cells. Immunity (2020) 52(1):83–95.e4. doi: 10.1016/j.immuni.2019.12.001
403. Ishizuka IE, Chea S, Gudjonson H, Constantinides MG, Dinner AR, Bendelac A, et al. Single-cell analysis defines the divergence between the innate lymphoid cell lineage and lymphoid tissue-inducer cell lineage. Nat Immunol (2016) 17(3):269–76. doi: 10.1038/ni.3344
404. Kasal DN, Bendelac A. Multi-transcription factor reporter mice delineate early precursors to the ILC and LTi lineages. J Exp Med (2021) 218(2):e20200487. doi: 10.1084/jem.20200487
405. Klose CS, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, d'Hargues Y, et al. A T-bet gradient controls the fate and function of CCR6-RORγt+ innate lymphoid cells. Nature (2013) 494(7436):261–5. doi: 10.1038/nature11813
406. Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M, et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat Immunol (2011) 13(2):144–51. doi: 10.1038/ni.2187
407. Rankin LC, Groom JR, Chopin M, Herold MJ, Walker JA, Mielke LA, et al. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat Immunol (2013) 14(4):389–95. doi: 10.1038/ni.2545
408. Cossarizza A, Chang HD, Radbruch A, Abrignani S, Addo R, Akdis M, et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies (third edition). Eur J Immunol (2021) 51(12):2708–3145. doi: 10.1002/eji.202170126
409. Simoni Y, Fehlings M, Kløverpris HN, McGovern N, Koo SL, Loh CY, et al. Human innate lymphoid cell subsets possess tissue-type based heterogeneity in phenotype and frequency. Immunity (2018) 48(5):1060. doi: 10.1016/j.immuni.2018.04.028
410. Bonne-Année S, Bush MC, Nutman TB. Differential modulation of human innate lymphoid cell (ILC) subsets by IL-10 and TGF-β. Sci Rep (2019) 9(1):14305. doi: 10.1038/s41598-019-50308-8
411. Mazzurana L, Czarnewski P, Jonsson V, Wigge L, Ringnér M, Williams TC, et al. Tissue-specific transcriptional imprinting and heterogeneity in human innate lymphoid cells revealed by full-length single-cell RNA-sequencing. Cell Res (2021) 31(5):554–68. doi: 10.1038/s41422-020-00445-x
Keywords: innate lymphoid cells, phenotype, flow cytometry, humans, mice
Citation: Koprivica I, Stanisavljević S, Mićanović D, Jevtić B, Stojanović I and Miljković Đ (2023) ILC3: a case of conflicted identity. Front. Immunol. 14:1271699. doi: 10.3389/fimmu.2023.1271699
Received: 02 August 2023; Accepted: 02 October 2023;
Published: 17 October 2023.
Edited by:
Carolina Jancic, National Scientific and Technical Research Council (CONICET), ArgentinaReviewed by:
Mohamad El-Zaatari, University of Michigan, United StatesLuke Roberts, The University of Manchester, United Kingdom
Copyright © 2023 Koprivica, Stanisavljević, Mićanović, Jevtić, Stojanović and Miljković. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Đorđe Miljković, ZGpvcmRqZW1AaWJpc3MuYmcuYWMucnM=
†These authors have contributed equally to this work
 Ivan Koprivica
Ivan Koprivica Suzana Stanisavljević
Suzana Stanisavljević Dragica Mićanović
Dragica Mićanović Bojan Jevtić
Bojan Jevtić Ivana Stojanović
Ivana Stojanović Đorđe Miljković
Đorđe Miljković