- 1Translational Medicine Institute, Affiliated Tianjin Union Medical Center of Nankai University, Nankai University, Tianjin, China
- 2Department of Immunology, Nankai University School of Medicine, Nankai University, Tianjin, China
- 3State Key Laboratory of Medicinal Chemical Biology, Nankai University, Tianjin, China
Tumor-associated macrophages (TAMs) are present in almost all solid tumor tissues. 16They play critical roles in immune regulation, tumor angiogenesis, tumor stem cell activation, tumor invasion and metastasis, and resistance to therapy. However, it is unclear how TAMs perform these functions. With the application of single-cell RNA sequencing (scRNA-seq), it has become possible to identify TAM subpopulations associated with distinct functions. In this review, we discuss four novel TAM subpopulations in distinct solid tumors based on core gene signatures by scRNA-seq, including FCN1+, SPP1+, C1Q+ and CCL18+ TAMs. Functional enrichment and gene expression in scRNA-seq data from different solid tumor tissues found that FCN1+ TAMs may induce inflammation; SPP1+ TAMs are potentially involved in metastasis, angiogenesis, and cancer cell stem cell activation, whereas C1Q+ TAMs participate in immune regulation and suppression; And CCL18+ cells are terminal immunosuppressive macrophages that not only have a stronger immunosuppressive function but also enhance tumor metastasis. SPP1+ and C1Q+ TAM subpopulations can be further divided into distinct populations with different functions. Meanwhile, we will also present emerging evidence highlighting the separating macrophage subpopulations associated with distinct functions. However, there exist the potential disconnects between cell types and subpopulations identified by scRNA-seq and their actual function.
1 Introduction
Macrophages, which exist in almost all tissues and organs, not only contribute to immune regulation, tissue regeneration and remodeling (1) but also play critical roles in the occurrence and development of diseases (2–4). TAMs are important immune cells in the tumor microenvironment (TME). They determine tumor growth, metastasis and prognosis (5–7). They are closely related to poor prognosis and resistance to therapy (8, 9). These cells include TAMs from embryo-derived TRMs and inflammatory monocytes (10, 11). TAMs from monocytes can increase with tumor growth due to self-proliferation, recruitment, and differentiation from circulating inflammatory monocytes (7, 12), which are mediated by elevated secretion of cytokines by cancer and stromal cells in tumors and associated metastases (13–15).
Macrophages are divided into two different subpopulations, M1 and M2, based on in vitro culture (16–18). TAMs with M1- and M2-like phenotypes represent two extremes of TAM polarization. These TAMs display distinct functions in tumor tissues. M1-like TAMs, which express surface molecules such as CD68 and produce cytokines such as IL-1β, could act as the main forces in innate host defense; Whereas M2-like TAMs, which express immunosuppressive molecules such as CD163 and produce cytokines such as IL-10 and CCL18, are critical in promoting epithelial-mesenchymal transition (EMT), angiogenesis, and immunosuppression of tumors (7, 19, 20). However, according to present literatures, it is defective or even misleading for the M1/M2 dichotomy in cancer biology. Accumulating evidence has shown that some TAM subpopulations can express genes of both M1 and M2 macrophages (21). Studies have also found that TAMs have significant plasticity and heterogeneity, and are composed of multiple different subpopulations in TME (19). However, it is unclear how to distinguish these TAM subpopulations with different functions.
With the application of scRNA-seq, it has become possible to distinguish TAM subpopulations with distinct functions. scRNA-seq can not only discover relationships between the genes, and track the trajectories of different cell lineages, but also more importantly reveal different subpopulations, especially some rare cell populations. To distinguish TAM subpopulations with different functions, analyses can be conducted according to different compositions, functional enrichment, and differential gene expression.
One of the most frequently employed enrichment analysis tools for scRNA-seq data is DAVID website (http://david.niaid.nih.gov), which aims to provide a functional interpretation of large lists of genes derived from genomic studies (22). It includes the gene functional classification tool, functional annotation tool, gene ID conversion tool, gene name viewer, and NIAID pathogen genome browser (22). According to scRNA-seq data from different solid tumors in the current literatures, TAMs in solid tumor tissues can be mainly divided into four different kinds of TAMs, including FCN1+, SPP1+, C1Q+ and CCL18+ subpopulations. Here, we will review these macrophage subpopulations, which are related to the occurrence and development of tumors. The identified TAM subpopulations in tumor tissues can be potential prognostic biomarkers(s) and/or candidate therapeutic targets.
2 Origin of tumor-associated macrophages
With the application of scRNA-seq and modern lineage tracing techniques, a large body of evidence has shown that TAMs, which are derived from embryo-derived TRMs and inflammatory monocytes, can be found in tumors such as colorectal cancer, liver cancer, pancreatic cancer, lung cancer, and glioblastoma (23–25).
2.1 TAMs derived from embryo-derived TRMs
Recent data utilizing specific fate mapping technologies have provided evidence for the embryonic origin of tissue macrophages (26–28). These macrophages possess self-renewal and proliferation capacity. In most normal tissues, TRMs are mainly embryonic macrophages (29), which are necessary for the development of tissues and organs. Notably, solid tumors have requirements similar to those of developing organs and tissues in forming complex structures (30). Thus, macrophages in tumor tissues can also be derived from embryo-derived TRMs (31, 32). These TAMs from embryo-derived TRMs can contribute to the occurrence and development of cancers (33, 34).
2.2 TAMs derived from monocytes
In solid tumors, monocyte-derived macrophages (MDMs) are recruited by cytokines and chemokines and then polarized into TAMs, which are universally heterogeneous (13–15). MDMs can also self-renew and proliferate (35). For example, the proliferation of TAMs can be induced in the presence of granulocyte macrophage colony stimulating factor (GM-CSF) in liver cancer tissues (36). These monocyte-derived TAMs can switch from one phenotype to another. They display remarkable plasticity within the TME (37), which can result in distinct subpopulations. Tumor-associated factors, such as tumor hypoxia in the TME, contribute to the heterogeneity of monocyte-derived TAMs. Thus, monocyte-derived TAMs may consist of multiple subpopulations generated through distinct developmental pathways.
In addition, there is also a minor splenic contribution to monocyte-derived TAMs (38, 39). Although the bone marrow is the primary hematopoietic tissue and monocyte reservoir, the spleen is also an identified reservoir of monocytes, which can play a significant role in the inflammatory response (40). Thus, the spleen is also an important extra-medullary site that can continuously supply growing tumors with monocytes (39).
3 Novel TAM subpopulations in different solid tumors
There are many different gene signatures in TAM populations and their subpopulations. However, four classes of genes are generally used to recognize TAMs and their subpopulations, including macrophage-specific markers such as CD14 and MHCII, T cell immune checkpoint ligands on TAMs, such as PD-L1, PD-L2, CD80, and CD86 (41), surface immune suppressive molecules such as CD163, CD68 and MRC1 (CD206) (42), and specific core gene signatures such as FCN1, C1Q, SPP1 or CCL18 in different TAM subpopulations. TAMs in different solid tumors are mainly divided into four subpopulations based on core gene signatures by scRNA-seq, including two main subpopulations C1Q+ and SPP1+TAMs, and two minor subpopulations FCN1+ and CCL18+ TAMs (Figure 1). Notably, there are differences in the gene expression in each TAM subpopulation in different tumors, the same tumors in different patients, and even different stages of the same tumor although there exists a core gene signature. ScRNA-seq studies have also demonstrated that these TAMs have high phenotypic plasticity and heterogeneity in cancers (43–45).
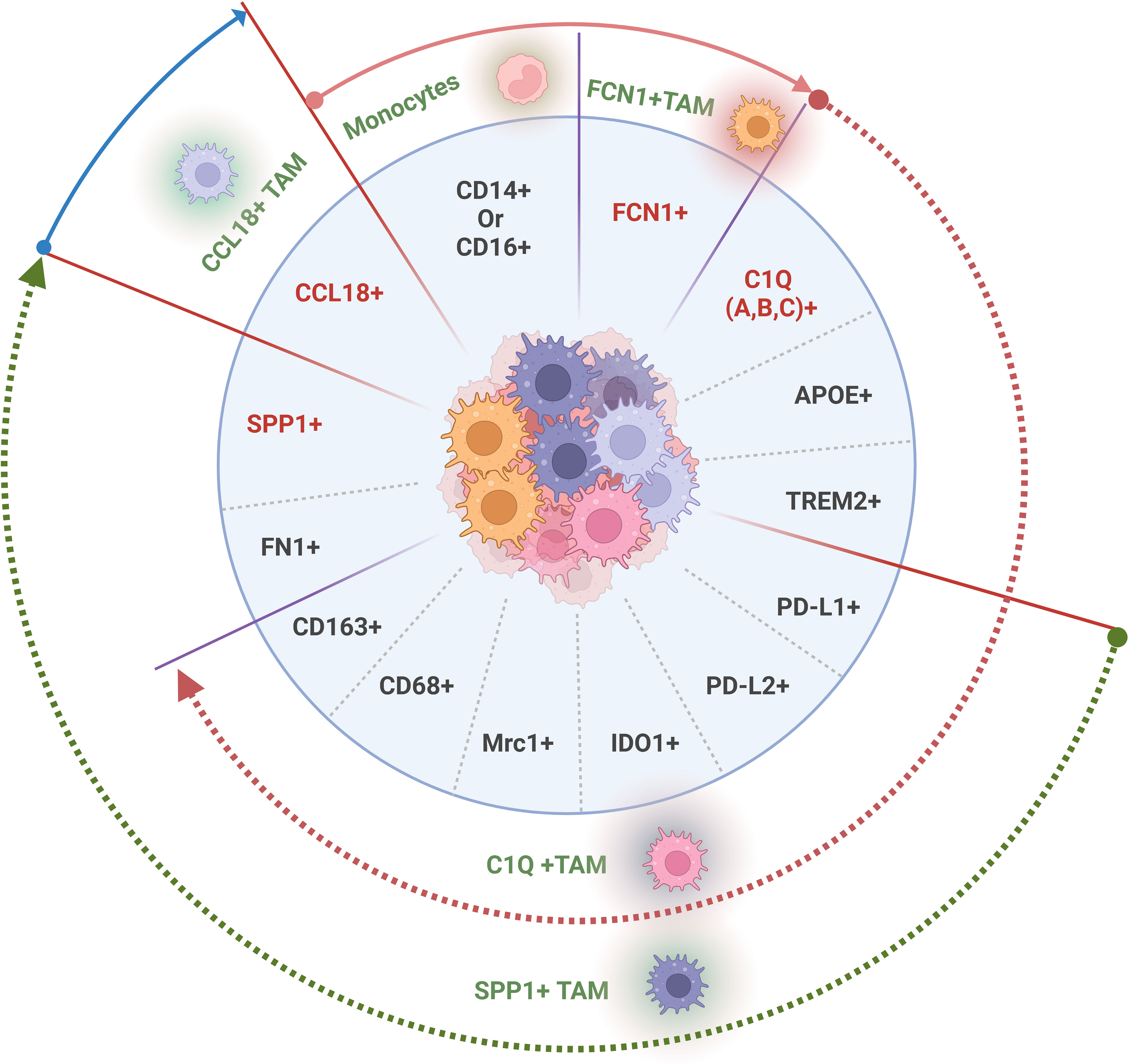
Figure 1 Tumor associated macrophage (TAM) subpopulations with distinct gene signatures in solid tumor tissues. The genes used to recognize TAM population and subpopulations, generally including T cell immune checkpoint ligands on the membrane surface such as PD-L1, PDL2, CD80, and CD86, surface immune suppressive molecules such as CD163, CD68 and MRC1 (CD206) and specific core gene signatures such as FCN1+, C1Q A/B/C+, SPP1+ or CCL18+ in different TAM subpopulations. Gene with red color, a core gene in each distinct TAM subpopulation.
3.1 SPP1+ TAM subpopulation
SPP1+TAM subpopulation is identified by specific expression of a core gene SPP1. SPP1+ TAM subpopulation also often expresses the following genes such as FN1, IL1RN, and other TAM genes IDO, Mrc1(CD206), PD-L1, PD-L2, CD68 and CD163. Notably, TAM associated CD68 (46) and/or CD206 (47) genes can be found only in the SPP1+ TAM subpopulation of some solid tumors but not in all solid tumors (48). In addition, SPP1+CD206+ TAM subpopulation also produces epithelial growth factor (EGF) (49).
SPP1+TAM subpopulation was initially found in colorectal cancers (CRC) (43), and later in lung and breast cancers (50, 51). In human CRCs, SPP1+ TAM subpopulations not only showed specific expression of SPP1 but also MARCO and VEGFA (43, 52, 53). SPP1 with glycolysis genes (GAPDH, ENO1, LDHA, ALDOA, and TPI1) was also expressed in TAMs in non-small cell lung cancer (NSCLC) (54). In lung squamous cell carcinoma, there also existed subpopulation of SPP1+TAMs. SPP1+ macrophages were significantly increased in the tumor microenvironment, which was related to the poor prognosis of patients with lung squamous cell carcinoma. SPP1+ macrophages (53) were also found in hepatocellular carcinoma. Macrophage subsets that express SPP1, TREM2 and FN1 anti-inflammatory TAMs were found in breast cancer (8, 55, 56). The patients with SPP1low TAMs had the best prognosis for cervical cancer, whereas the worst prognosis appeared in patients with SPP1high TAMs (57). Multi-omics analysis also revealed the distinct clinical significance of SPP1+ TAMs in cervical cancer (57). SPP1+, TAM subpopulations could also be detected in rental cancer. and in pancreatic cancer (58).
3.2 C1Q+ TAM subpopulation
The core gene signature of C1Q+TAM subpopulation is the expression of C1QA/B/C. C1Q+ TAMs are also characterized by the expression PD-L1, PD-L2, HAVCR2, LGALS9, and CEACAM1 (52). PD-L1 and PD-L2, which are checkpoint molecules, can prevent CD8+ and CD4+ Th1 immunity (59). C1Q+ TAMs can also highly express TREM2, MERTK, CD80 (43, 57, 60), SLC40A1, GPNMB (43, 52, 61) and other genes in TAMs, such as IDO, Mrc1 (CD206), CD68 and CD163. In addition, this population of macrophages can also produce pro-angiogenic cytokines, such as vascular endothelial growth factor (VEGF).
A subpopulation of TAMs expressing C1QA/B/C is present in multiple cancers, such as colorectal cancer (43), NSCLC (62), liver cancer (44), lung cancer (63), rental cancer (60, 64, 65), breast cancer (8, 50), pancreatic cancer (58) and cervical cancer (57). Single-cell and spatial analyses revealed immunosuppressive phenotypes of C1Q+APOE+ TAMs in CRCs (66). The patients with C1QChigh TAMs had the best prognosis for cervical cancer, whereas the worst prognosis could appear in patients with C1QClow TAMs (57).
3.3 FCN1+ TAM subpopulation
Another TAM subset, characterized by high expression of the core gene FCN1, is also identified in cancer tissues. This subpopulation of macrophages is a precursors of C1Q+TAM (43, 44). They are derived from inflammatory monocytes. Indeed, tumor-enriched FCN1+ monocyte-like cells showed a high similarity to blood CD14+ monocytes, representing a monocyte population migrating into tumors and harboring a tumor-specific transcriptional program (43, 45).
FCN1+ TAM subpopulation was also identified in human multiple solid cancers such as NSCLC (62), liver (44, 53), breast cancer (8), lung (45), CRCs (43) and pancreatic cancer (58). In human NSCLC, FCN1+ macrophages, together with typical FYN+ and STAT1+ macrophages, expressed genes related to increased inflammatory function (62).
3.4 CCL18+ TAM subpopulation
This population is characterized by the expression of a core gene CCL18. CCL18 is associated with the immunosuppressive nature of the tumor microenvironment. CCL18+ TAM subpopulation is also an important element in cancer immune evasion (67). Notably, both CCL18+ and SPP1+ macrophages highly express immunosuppressive M2-like genes (62), implying that they are the same type of TAMs. CCL18+ TAMs should be terminal SPP1+ macrophages.
CCL18+ TAM subpopulations can be found in many tumors such as NSCLC (62), breast cancer (68), CRCs (69, 70) hepatocellular carcinoma (53, 71), thyroid cancer (72) and intrahepatic cholangiocarcinoma (53).
Thus, four TAM subpopulations with different core genes such as C1Q+, SPP1+, FCN+ and CCL8+ TAM have been identified in distinct solid tumors by scRNA-seq data. However, there mainly are two TAM subpopulations in solid tumors, including C1Q+ and SPP1+ TAMs (43, 57, 73).
Notably, there also exist other classification methods based on single cell RNA sequences. For example, one study separated TAM into seven TAM subsets in tumor tissues, including inhibin beta A chain (INHBA)+ TAMs, complement C1q subcomponent subunit C (C1QC)+ TAMs, ubiquitin like protein ISG15 (ISG15)+ TAMs, NACHT, LRR and PYD domains containing protein 3 (NLRP3)+ TAMs, LYVE1+ TAMs, and sphingosine- 1-phosphate phosphatase 1 (SPP1)+ TAMs (74). TAMs were also separated into five TAM subsets in another study, including transcription factor HES-1 (HES1)+, complement component 1q (C1Q)hi, triggering receptor expressed on myeloid cells 2 (TREM2)+, IL4I1+ and proliferating TAMs (75). Other also showed seven TAM subsets in tumor tissues, including interferon-primed TAMs (IFN-TAMs), immune regulatory TAMs (reg-TAMs), inflammatory cytokine-enriched TAMs (inflam-TAMs), lipid-associated TAMs (LA-TAMs), pro-angiogenic TAMs (angio-TAMs), resident-tissue macrophage-like TAMs (RTM-TAMs) and proliferating TAMs (prolif-TAMs) (76).
4 Potential functions of novel TAM subpopulations
TAMs are generally associated with a poor prognosis and treatment resistance (8, 9, 77, 78). These TAMs play different roles in the occurrence and development of tumors, such as immunosuppression, angiogenesis, metastasis, tumor stem cell activation, inflammation, antigen-presenting and phagocytes (5–7). TAM subpopulations with distinct functions can be identified through functional enrichment and differential gene expression in scRNA-seq data (Figure 2).
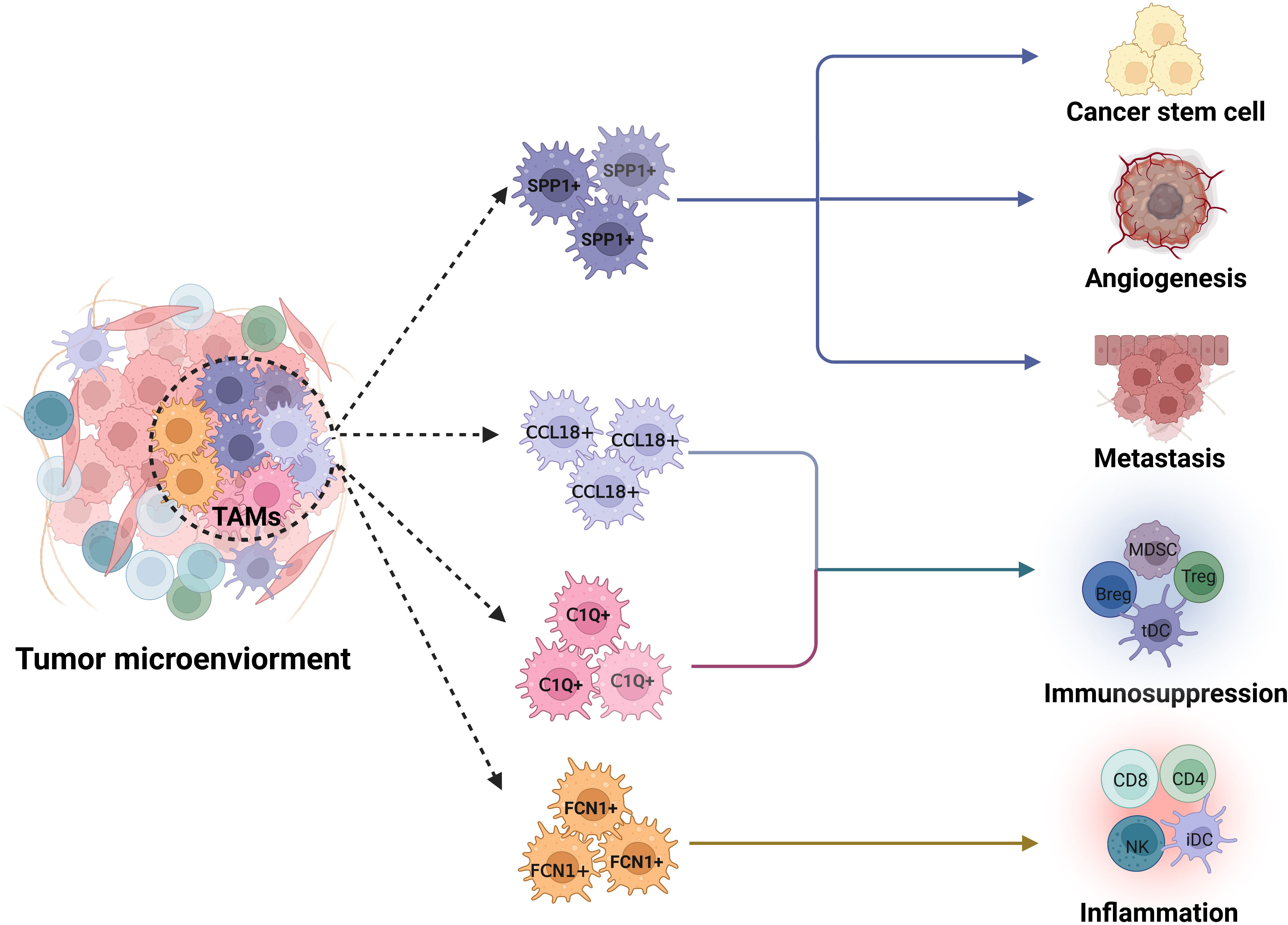
Figure 2 Potential functions of FCN1+, C1Q+, SPP1+ and CCL18+ TAM subpopulations in tumor microenvironments. SPP1+ tumor associated macrophage (TAM) subpopulation exerts multiple roles in tumor development such as tumor metastasis, angiogenesis and tumor stem cell activation; Whereas a main function of C1Q+TAM population is immunosuppression. CCL18 TAM subpopulation has stronger immunosuppressive function. FCN1+ TAM subpopulation is inflammatory macrophages, which can cause inflammation. Dotted line arrow, predicted TAM subpopulations based on core gene signatures.
4.1 SPP1+ TAM subpopulation
The number of SPP1+ macrophages significantly increases in the tumor microenvironment, which is correlated with poor prognosis (46, 79–85). SPP1+ TAM subpopulation has multiple roles in tumor development such as tumor metastasis (62, 86–91), angiogenesis (92–94), tumor stem cell activation and immunosuppression (95).
Tumor metastasis includes multiple steps, such as epithelial-to-mesenchymal transition, trans-endothelial migration, extracellular matrix remodeling, and formation of pre-metastatic niches. In the patients with tumors, the enrichment SPP1+ TAMs was associated with worse overall survival (86, 96). SPP1+ macrophages from tumor tissues showed high expression of MMP9, MMP12, MMP14, and MMP19, which could contribute to the degradation of the basement membrane for the invasion of tumor cells (82). There had also an interaction between SPP1 and CD44 in SPP1+ macrophages to promote metastasis. The interaction between SPP1+ macrophages and tumor epithelial cells activated downstream genes to mediate the activation of NF-κB, PI3K/Akt, VEGF, uPA, and MMPs, which could promote endothelial cell proliferation. The interaction of SPP1+ macrophages with FAP+ fibroblasts also appeared in colorectal cancer to promote metastasis (97). In SPP1+ macrophages, the main metabolic pathway was glycolysis, which could promote tumor metastasis via angiogenesis and matrix remodeling (62). SPP1+ macrophages were predominant in liver metastasis (86) and also potential biomarker for early lymph node metastasis (89). Colorectal cancer metastases in the liver could establish immunosuppressive spatial networking between tumor-associated SPP1+ macrophages and fibroblasts, which supported colorectal cancer growth in the immunosuppressed metastatic niche in the liver (98). Studies also found that SPP1+macrophages were metastasis accelerators of colorectal cancer (99). They were often found in mesenteric lymph node (MLN) with metastases. SPP1+ macrophages also promoted prostate tumor progression by increasing the incidence of prostate intraepithelial neoplasia (100).
For tumor angiogenesis, SPP1+ TAMs can be acted as “angiogenic switch.” They produce cytokines, such as VEGFA, platelet-derived growth factor (PDGF) and angiopoietin, to promote angiogenesis (93, 101). SPP1+ TAMs with high angiogenesis scores showed the expression of genes related to angiogenesis. There was also a strong enrichment of tumor angiogenesis and tumor vasculature pathways in SPP1+ TAMs (43). Under the influence of hypoxia, SPP1+TAMs could develop a pro-angiogenic phenotype by directly upregulating angiogenic molecules, such as VEGF-A (102) and angiogenic modulators, such as matrix metalloproteinase (MMP)7 (103). These cells could also interact with endothelial cells to promote angiogenic functions (94).
Cancer stem cells (CSCs) are subsets of tumor cells that play a key role in tumorigenesis. SPP1+TAMs could regulate CSC activation (104–106). For example, SPP1+TAMs promoted tumor growth and proliferation by secreting collagen (107) and insulin-like growth factor-1 (IGF-1) (108).
SPP1+ TAMs are also implicated in T cell suppression (97, 109), and play a role in immune evasion in the cancers such as colon cancer (52, 62). Upregulation of PD-L1 by SPP1 could mediate macrophage polarization and facilitate immune escape in lung adenocarcinoma (110). A dramatic increase in SPP1+ TAMs was positively correlated with FAP+ fibroblasts in CRC tissues, which could impair the immunotherapeutic effects (97). The patients with high SPP1 expression achieved less therapeutic benefit from an anti-PD-L1 therapy cohort (97). A tumor immune barrier (TIB) formed by the interaction of SPP1+ macrophages and cancer-associated fibroblasts (CAFs) was related to immunotherapy efficacy. Disruption of the TIB structure by blocking SPP1 should be considered a relevant therapeutic approach to enhance the therapeutic effect of immune checkpoint blockage (ICB) in HCC (111).
SPP1, an osteopontin, from SPP1+ macrophages is a multifunctional secreted phosphorylated glycoprotein (112). It is also present in other cells such as osteoblasts, fibroblasts, and in tumor cells (112). The correlations between levels of circulating SPP1 and/or increases in SPP1 expression on tumor cells had poor prognosis in many types of cancer (112). The interaction of SPP1 and FN1 in TAMs with certain integrins promoted tumorigenesis in CRCs (43). SPP1-CD44 signaling in the glioma perivascular niche also enhanced cancer stem cell phenotypes and promoted aggressive tumor growth via the γ-secretase-regulated intracellular domain of CD44 (113). SPP1 was a key gene in the lymph node metastasis and a potential predictor of poor prognosis in head and neck carcinoma (114). In addition, SPP1 is involved in resistance to chemoradiotherapy through the induction of EMT, autophagy, epigenetic alterations, aberrant glucose metabolism and reduction of drug uptake (79, 111, 115).
4.2 C1Q+ TAM subpopulation
The main functions of C1Q+ TAM subpopulation are immune regulation and immunosuppression (116–119). Indeed, TAMs with high C1Q A/B/C, HLA-DR, APOE, and TREM2 expression have a classically immunosuppressive phenotype (65, 116). These TAMs exert their immunosuppressive effects through surface molecules, cytokines, and metabolites. Surface molecules such as PD-L1, PDL2, CD80, and CD86, and cytokines such as IL-10 from TAMs could induce the differentiation of CD4+ T cells to Tregs (iTregs) (120, 121). A metabolite of tryptophan mediated by indoleamine 2, 3-dioxygenase (IDO) in C1Q+ TAMs also played an immunosuppressive function directly or via the aryl hydrocarbon receptor in Treg, NK, and DC cells (122). Arginine depletion in TAMs could cause “arginine starvation” in T cells to inhibit these cells (123). Increased levels of IDO also limited the proliferation of cytotoxic CD8 cells (124). C1Q+ macrophages could effectively suppress T cells (65, 125, 126). In RCC, the presence of C1Q+ macrophages correlated with exhausted T cells, forming a dysfunctional immune circuit (65, 127).
Notably, C1Q+ TAMs are also related to positive responses to ICB therapy in melanoma and lung-carcinoma patients (117–119). Indeed, C1Q+ cell density was correlated with inhibitory receptors PD-1 and LAG3 at the CD8+ T cell surface (128). These C1Q+ macrophages express additional immune checkpoint ligands, such as PD-L1, PDL-2 (127, 128) and others such as CD40L, CTLA4, LAG3, PD-1, and TIGIT (57). Thus, these C1Q+ TAMs may be a beneficial population for clinical applications (86). Notably, patients with C1Qhigh and SPP1low TAMs had the best prognosis, whereas the worst prognosis appeared in the patients with C1Qlow and SPP1high TAMs (57, 129). In lung cancer, increased CXCL-10 was described in C1Q+ TAM, which was related with an enrichment of the transcription factors IRF1, IRF7, and STAT1 (62). Interestingly, IRF1 was correlated with STAT1, HLA-DR, PD-1, and LAG-3 in metastases of colorectal cancer (130). In addition, C1Q+ TAMs were also involved in phagocytosis and antigen presentation (43).
C1Q+ macrophages to exert their functions also depend on C1Q, a recognition molecule of classical complement pathway, which can bind to immune complexes or other activators in the tumor microenvironment. Indeed, C1Q could regulate human macrophage polarization via interactions with LAIR1 (131), and modulate the cytokine expression while they digested lipid proteins, causing an M2-like polarization (132). C1Q also directly regulated T cell phenotype through internalization, binding to mitochondria, and regulation of mitochondrial metabolism (133, 134). However, it is incompletely clear how it controls immune activation, tolerance and exhaustion. In addition, C1Q could also interact directly with endothelial cells (EC) to promote neoangiogenesis, via still unknown cell surface receptors (128, 135).
4.3 FCN1+ TAM subpopulation
FCN1+ TAM subpopulation comprises inflammatory macrophages. This subpopulation of macrophages is associated with increased inflammatory function (62). Enriched FCN1+ TAMs were detected in tumor-adjacent tissues. These FCN1+ TAMs were considered an intermediate stage from monocytes to tumor macrophages (136). They were associated with angiogenesis. Subsequently, these cells can produce C1Q+ and SPP1+ TAM (43). In addition, FCN1 (ficolin-1) from FCN1+ TAMs can be as a novel macrophage infiltration-associated biomarker for the diagnosis of pediatric inflammatory bowel diseases (137).
4.4 CCL18+ TAM subpopulation
The effects of CCL18+ TAM subpopulation on tumor cells include tumor cell proliferation, migration induction, invasion, EMT, angiogenesis, and lymphangiogenesis (67). CCL18+ macrophages can be found in solid tumor tissues (53, 71) and in an immunosuppressive state in tumor tissues (70). They have a stronger tumor-promoting role than SPP1+ macrophages (53). Single-cell spatial transcriptomic analysis also identified highly metabolic CCL18+ TAMs in colorectal liver metastasis sites.
In the tumor, CCL18 chemokine produced by CCL18+ TAMs (67) was a marker of neoplastic diseases (67). Notably, elevated CCL18 levels in the serum and tumor are related to a worse prognosis in patients (53). Indeed, CCL18 is associated with immunosuppressive functions and cancer immune evasion. The CCL18 could recruit CD4+CD45RA+CD25- naïve T cells into the tumor niche, and then differentiated into Treg cells, as shown in gastric cancer (138) and breast cancer (139). Notably, the importance of CCL18 in neoplastic processes mainly includes a signal transduction from PITPNM3 (one of CCL18 receptors) in CCL18-dependent migration, invasion, and epithelial- EMT cancer cells (67). Studies with human umbilical vein endothelial cells (HUVECs) showed that CCL18 could cause the VEGF-independent migration and tube formation of these cells (140). CCL18 also had an influence on the proliferation of cancer cells, but this effect was dependent on the type of tumor (67). In breast phyllodes tumor, CCL18 also participated in myofibroblast differentiation (141). Thus, CCL18 should be a potential therapeutic target for anti-cancer therapy. However, there is very few studies on the effects on tumor development through blocking the activity of CCL18.
5 Differentiation of novel TAM subpopulations
TAMs have both embryonic and monocyte origins (142, 143). Genes related to immunosuppression and inflammation can be expressed by FCN1+ and C1Q+ TAMs (86, 87, 89, 97). Since monocyte-derived TAMs highly express genes related to inflammation and immunosuppression, FCN1+ and C1Q+ TAM subpopulations may be derived from monocytes. Whereas other two SPP1+ and CCL18+ TAM subpopulations express genes related to tissue remodeling, wound healing, metastasis and angiogenesis (67, 144). Notably, the genes related to tissue remodeling and wound healing also appear in embryo-derived TRMs (11, 24, 29, 142, 145). Thus, SPP1+ and CCL18+ TAMs might be derived from embryo-derived TRMs. Based on the scRNA-seq data, we suggest differentiation models of TAMs by tracking the development trajectories of distinct cell lineages (Figure 3). Monocyte-derived TAMs subsequently differentiate from peripheral blood CD14+ and/or CD16+-expressing monocytes to mature C1Q+ macrophages through FCN1+ inflammatory macrophages and pre-C1Q+macrophages; Whereas the differentiation of TAMs from embryo-derived RTMs to CCL18+ macrophages happens via pre-SPP1+ and mature SPP1+macrophages. Mature C1Q+ and SPP1+ macrophages can be further divided into the populations with distinct functions. However, others have also suggested that FCN1+ inflammatory macrophages can subsequently give rise to both C1Q+ and SPP1+ TAM populations (43).
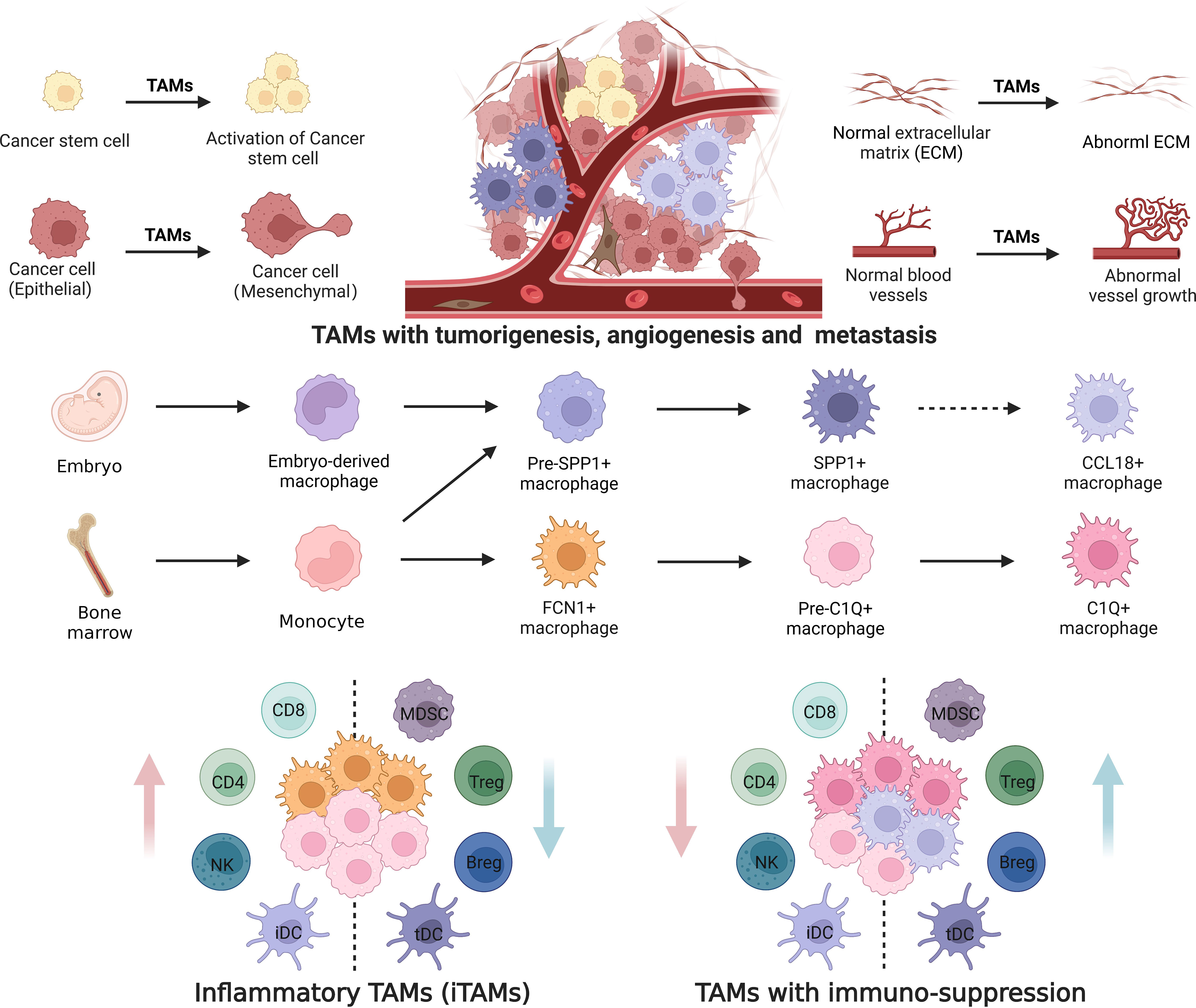
Figure 3 Differentiation of tumor associated macrophages with distinct functions. Tumor associated macrophages (TAMs) are both embryonic and monocyte origins. Monocytes derived TAMs highly express genes related to inflammatory and immunosuppression; While embryo-derived TAMs highly express genes related to tumorigenesis, metastasis and angiogenesis. FCN1+ and C1Q+ TAM subpopulations are derived from peripheral blood monocytes, whereas SPP1+ and CCL18+TAMs are derived from tissue-resident macrophages (pre-SPP1+ macrophages from embryo and monocyte derived macrophages). Differentiation of TAMs from peripheral blood monocytes to C1Q+ macrophages includes monocytes, FCN1+ macrophages, and pre-C1Q+ and mature C1Q+ macrophages. Differentiation of TAMs from tissue-resident macrophages into CCL18+ macrophages occurs via pre-SPP1+ and mature SPP1+. Furthermore, mature C1Q+ and SPP1+ macrophages can be divided into different populations with distinct functions. iDC, inflammatory dendritic cells; tDC, tolerant dendritic cells; MDSCs, myeloid-derived suppressor cells; NK, natural killer cells. Dotted line arrow: predicted pathway based on gene expression.
5.1 From FCN1+ to C1Q+TAMs
Monocyte subsets that highly expressed FCN1, S100A8,and S100A9 could subsequently produce C1Q+ TAM populations (43). C1Q+ TAMs include pre-mature and mature C1Q+ TAMs. Pre-C1Q+ macrophages possess pro-inflammatory and phagocytic phenotypes based on the TAM differentiation routine. C1Q+ and FCN1+ macrophages with a pro-inflammatory phenotype could be detected in NSCLC (62). C1Q+ TAMs with high phagocytic activity were also found in tumor tissues (125, 128). These TAMs highly also express genes involved in antigen presentation (43, 60). However, mature C1Q+ macrophages are the predominant contributors to immunosuppression. These C1Q+ macrophages express multiple immunosuppressive genes such as PD-L1, PD-L2, HAVCR2, LGALS9, and CEACAM1 (52). In addition, several different gene signature clusters in C1Q+ TAM subpopulations have been reported, such as C1Q+TAMs with genes C1QA/B/C, SLCO2B1, NRP1, SLAMF8, FCGR1A, MERTK, and SIGLEC1 (43); C1Q+TAMs with genes C1QA/B, APOE, TREM2, GPNMB and SLC40A1 (44), C1Q+TAMs with genes TREM2, CD81, MARCO, APOE, CALR, CD63 and SPP1 (63); and C1Q+TAMs with genes C1QB, APOE, FN1, CD276, TREM2, CHIT1, CCL18, MARCO, CD81 and NRP2 (50). Since C1Q+ TAMs mainly exert immunosuppressive functions, which are related to multiple immune cells, these TAMs with different gene signatures suggest the existence of multiple C1Q+ TAM populations with distinct immunosuppressive functions (Figure 3).
5.2 From SPP1+ to CCL18+ TAMs
SPP1+ macrophages not only promote tumorigenesis, but also metastasis and angiogenesis, which are possessed by embryo-derived TRMs (11, 24, 29, 142, 145), implying that this subpopulation is derived from embryo-derived TRMs. Several different gene signature clusters in SPP1+ TAM subpopulations have been reported, such as SPP1+TAMs with genes SPP1, BCL6, ADM, MARCO, FN1, AQP9, TIMP1, VEGFA, and IL1RN (57); SPP1+ TAMs with genes SEPP1,CD68, LYZ, MARCO, APOC1, SPP1 (82), SPP1+ TAMs with genes SPP1, MARCO, and VEGFA (43, 52); and SPP1+ TAMs with genes SPP1, AQP9, TNS3, FN1, C15ORF48, PHLDA1, and NDRG1 (43). These TAMs with different clusters suggest that SPP1+ TAM can be divided into different populations with distinct functions. In addition, SPP1+macrophages possessed anti-inflammatory phenotypes (62), which played a role in immune suppression and tumor evasion in colon cancer (97, 109).
CCL18+ macrophages belong to terminal TAMs. Increased immunosuppressive CCL18+ TAMs with a terminally differentiated state and metabolically energetic phenotype could be found in tumors. CCL18+ macrophages with high expression of CD163, MARCO, and CSF1R also exhibited stronger tumor-promoting effects than SPP1+ macrophages (53). These macrophages with an anti-inflammatory phenotype could also induce a worse prognosis (53, 62). The effects of CCL18 on tumor cells were similar to SPP1 TAM (67), implying that this CCL8+ macrophage subpopulation was originated from SPP1+ TAMs.
6 Relevance of TAM subpopulations by scRNA-seq with old M1 and M2 paradigm
Activated macrophages are usually divided into two categories, M1 and M2 (146). M1 is involved in pro-inflammatory responses, whereas M2 macrophages are mainly involved in anti-inflammatory responses. In tumor environments, M1 macrophages typically exert anti-tumor functions, including directly mediate cytotoxicity and antibody-dependent cell-mediated cytotoxicity to kill tumor cells; Whereas M2 macrophages can promote the occurrence and metastasis of tumor cells, inhibit T cell-mediated anti-tumor immune response, promote tumor angiogenesis, and lead to tumor progression (5). There are mainly two classes of TAM subpopulations identified by scRNA-seq in solid tumors, including C1Q+ and SPP1+ TAM subpopulations. C1Q+ subpopulation in solid tumors possesses the characteristics of both M1 and M2 functions. This subpopulation is potentially involved in immunosuppression but also exhibits pro-inflammatory phenotypes. C1Q+ TAM subpopulation (43, 44, 50, 63) also expresses common genes with both M1 and M2 macrophages (5, 20, 147). M2 TAMs are critical in promoting EMT, angiogenesis, and immunosuppression of tumors (7, 19, 20); Whereas SPP1+ subpopulation can also potentially perform immunosuppression and mediate metastasis and angiogenesis although SPP1+ TAM subpopulation also possesses other functions such as promotion of cell proliferation and tissue repair. This SPP1+ TAM subpopulation also shares some genes with M2 macrophages (43, 50, 51). Thus, although C1Q+ and SPP1+ TAM subpopulations are different from old M1 and M2 paradigm, there also exist some common characteristics such as genes and functions between C1Q+ and M1 TAM macrophages, and between SPP1+ and M2 TAM subpopulations.
7 Tumor-associated macrophages as a target against tumor
TAMs, the most abundant immune cells in the TME, not only influence cancer progression and metastasis but also tumor recurrence (148). Thus, it is a critical strategy to target macrophages against tumor, which not only ameliorate the tumor-associated immunosuppression but also elicit anti-tumor immune responses. Different targeting strategies for TAMs have been developed, such as small molecular inhibitors and immune checkpoint inhibitors and antibodies.
The differentiation and function of macrophages can be manipulated by targeting PI3Kγ, JAK-STAT, C/EBPα, PPARγ and JNK1/2 signaling pathway using small-molecule inhibitors (149–151), and targeting immune-metabolism pathways such as arginine, adenosine, glutamine, tryptophan, kynurenine and lactate which have all been implicated in TAM reprogramming (152–154). Notably, STING (stimulator of interferon genes) could regulate the polarization of tumor-associated macrophages to inhibit liver metastasis of colorectal cancer (155). Targeting tumor-associated macrophages with STING agonist improved the antitumor efficacy of osimertinib in a mouse model of EGFR-mutant lung cancer (156). STING agonist overcomed STAT3-mediated immunosuppression and adaptive resistance to PARP inhibition in ovarian cancer (157). The activation of STING signaling also enhanced anti-tumor immunity (158), and improved cancer immunotherapy against tumor (159). In addition, epigenetic regulators such as histone deacetylases could also modify macrophage phenotypes (160, 161). However, these agents are not specific to macrophages, which have been associated with adverse toxicity.
Immune checkpoint inhibitors and antibodies which target TAMs can be used in therapy against tumors. TAMs and/or their subpopulations can overexpress different immune-checkpoint molecules such as PD-L1, which contribute to T cell exhaustion (162). Li et al. developed a hydrogel loaded with Pexidartinib and anti-PD-1-conjugated platelets to prevent of tumor recurrence after surgery (163). CD47 could interact with the inhibitory receptor SIRPα on the macrophages to transmit the “don’t eat me” signal (164). Several antibodies or small molecules targeting CD47-SIRPα axis have entered clinical trials (165, 166). A bispecific single-domain antibody was also used in the treatment of malignant tumors, which could efficiently and specifically bind and neutralize CCL2 as well as CCL5. They significantly induced the polarization of TAMs, and reduced immunosuppression in the TME (167). CD206 blockade also enhanced antitumor immune response in syngeneic models and the mouse pancreatic tumor model (168). Blockade of complement receptor C5aR1 also reprogramed tumor-associated macrophages (169).
In addition, other strategies such as depletion of TAMs and genetic engineering of macrophages with chimeric antigen receptor (CAR), which allows them to recognize tumor antigens and perform tumor cell-specific phagocytosis have also been used in therapy against tumors (170). One attractive target for TAM depletion is the CSF1-CSF1R axis (170). More than 30 phase I/II trials of small-molecule inhibitors or blocking antibodies to CSF1R have been initiated (149).
Notably, with the finding of new macrophage subpopulations by single cell RNA sequencing, more precise strategies which can target different TAM subpopulations will be developed. However, to the success of TAM subpopulation targeted therapies, it will be important to determine the exact functions of TAM subpopulations. Future studies should offer sufficient functional and phenotypic markers in the different TAM subsets. This will pave the way for the future development of targeted drugs to macrophage subsets. Notably, it may be difficult to study how embryo-derived TRMs and monocyte-derived TRMs influence tumors owing to limited consensus markers for these TRMs (149).
8 Conclusion and perspectives
TAMs and their subpopulations are related to tumor growth and progression. These TAMs and their subpopulations can act as potential new targets for cancer immunotherapy. Owing to the plasticity and heterogeneity of TAMs, it is difficult to define TAM subpopulations and their functions. However, scRNA-seq has served as a powerful tool for identifying these different TAM subpopulations. Here, we review subpopulations of TAMs in solid tumors in the phenotypic and transcriptomic levels, as well as their differentiation at the single-cell level. This review not only offers important insights into redefining subpopulations with different functions but also offers the possibility of precisely targeting TAM subpopulations. The application of scRNA-seq will undoubtedly promote our understanding of the biological characteristics of tumors.
However, there exists the potential disconnect between cell types and subtypes identified by scRNA-seq and their actual function. Thus, although scRNA-seq data have suggested TAM subpopulations with distinct functions, further clarification of these subpopulations with different functions is necessary for precise therapy and understanding of TAM characteristics. Next step will be to establish the actual function per TAM subpopulation. To determine the exact function of these TAMs, TAM population and subpopulation can be deleted through specific cytotoxicity or conditioned knockout.
Author contributions
JW: Writing – original draft, Writing – review & editing. NZ: Writing – original draft, Writing – review & editing. XS: Supervision, Writing – review & editing. YG: Supervision, Writing – original draft. RY: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by NSFC grants (grant number 81901677, 91842302, 81970457 and 91629102); The Tianjin Science and Technology Commission (grant number, 18JCZDJC35300); A Ministry of Science and Technology (grant number, 2016YFC1303604); The State Key Laboratory of Medicinal Chemical Biology and the Fundamental Research Funds for the Central University, Nankai university (63191724).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Wynn TA, Vannella KM. Macrophages in tissue repair, regeneration, and fibrosis. Immunity (2016) 44:450–62. doi: 10.1016/j.immuni.2016.02.015
2. Wang LX, Zhang SX, Wu HJ, Rong XL, Guo J. M2b macrophage polarization and its roles in diseases. J Leukoc Biol (2019) 106:345–58. doi: 10.1002/JLB.3RU1018-378RR
3. Aegerter H, Lambrecht BN, Jakubzick CV. Biology of lung macrophages in health and disease. Immunity (2022) 55:1564–80. doi: 10.1016/j.immuni.2022.08.010
4. Guilliams M, Scott CL. Liver macrophages in health and disease. Immunity (2022) 55:1515–29. doi: 10.1016/j.immuni.2022.08.002
5. Pan Y, Yu Y, Wang X, Zhang T. Tumor-associated macrophages in tumor immunity. Front Immunol (2020) 11:583084. doi: 10.3389/fimmu.2020.583084
6. Cheng Y, Song S, Wu P, Lyu B, Qin M, Sun Y, et al. Tumor associated macrophages and TAMs-based anti-tumor nanomedicines. Adv Healthc Mater (2021) 10:e2100590. doi: 10.1002/adhm.202100590
7. Wu K, Lin K, Li X, Yuan X, Xu P, Ni P, et al. Redefining tumor-associated macrophage subpopulations and functions in the tumor microenvironment. Front Immunol (2020) 11:1731. doi: 10.3389/fimmu.2020.01731
8. Nalio Ramos R, Missolo-Koussou Y, Gerber-Ferder Y, Bromley CP, Bugatti M, Nunez NG, et al. Tissue-resident FOLR2(+) macrophages associate with CD8(+) T cell infiltration in human breast cancer. Cell (2022) 185:1189–207 e25. doi: 10.1016/j.cell.2022.02.021
9. Ngambenjawong C, Gustafson HH, Pun SH. Progress in tumor-associated macrophage (TAM)-targeted therapeutics. Adv Drug Delivery Rev (2017) 114:206–21. doi: 10.1016/j.addr.2017.04.010
10. Casanova-Acebes M, Dalla E, Leader AM, LeBerichel J, Nikolic J, Morales BM, et al. Tissue-resident macrophages provide a pro-tumorigenic niche to early NSCLC cells. Nature (2021) 595:578–84. doi: 10.1038/s41586-021-03651-8
11. Zhu Y, Herndon JM, Sojka DK, Kim KW, Knolhoff BL, Zuo C, et al. Tissue-resident macrophages in pancreatic ductal adenocarcinoma originate from embryonic hematopoiesis and promote tumor progression. Immunity (2017) 47:323–38 e6. doi: 10.1016/j.immuni.2017.07.014
12. Wang H, Tian T, Zhang J. Tumor-associated macrophages (TAMs) in colorectal cancer (CRC): from mechanism to therapy and prognosis. Int J Mol Sci (2021) 22. doi: 10.3390/ijms22168470
13. Sanford DE, Belt BA, Panni RZ, Mayer A, Deshpande AD, Carpenter D, et al. Inflammatory monocyte mobilization decreases patient survival in pancreatic cancer: a role for targeting the CCL2/CCR2 axis. Clin Cancer Res (2013) 19:3404–15. doi: 10.1158/1078-0432.CCR-13-0525
14. Ren G, Zhao X, Wang Y, Zhang X, Chen X, Xu C, et al. CCR2-dependent recruitment of macrophages by tumor-educated mesenchymal stromal cells promotes tumor development and is mimicked by TNFalpha. Cell Stem Cell (2012) 11:812–24. doi: 10.1016/j.stem.2012.08.013
15. Qian BZ, Li J, Zhang H, Kitamura T, Zhang J, Campion LR, et al. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature (2011) 475:222–5. doi: 10.1038/nature10138
16. Orecchioni M, Ghosheh Y, Pramod AB, Ley K. Macrophage Polarization: Different Gene Signatures in M1(LPS+) vs. Classically and M2(LPS-) vs. Alternatively Activated Macrophages. Front Immunol (2019) 10:1084. doi: 10.3389/fimmu.2019.01084
17. Cutolo M, Campitiello R, Gotelli E, Soldano S. The role of M1/M2 macrophage polarization in rheumatoid arthritis synovitis. Front Immunol (2022) 13:867260. doi: 10.3389/fimmu.2022.867260
18. Wang N, Liang H, Zen K. Molecular mechanisms that influence the macrophage m1-m2 polarization balance. Front Immunol (2014) 5:614. doi: 10.3389/fimmu.2014.00614
19. Locati M, Curtale G, Mantovani A. Diversity, mechanisms, and significance of macrophage plasticity. Annu Rev Pathol (2020) 15:123–47. doi: 10.1146/annurev-pathmechdis-012418-012718
20. Gao J, Liang Y, Wang L. Shaping polarization of tumor-associated macrophages in cancer immunotherapy. Front Immunol (2022) 13:888713. doi: 10.3389/fimmu.2022.888713
21. Gubin MM, Esaulova E, Ward JP, Malkova ON, Runci D, Wong P, et al. High-dimensional analysis delineates myeloid and lymphoid compartment remodeling during successful immune-checkpoint cancer therapy. Cell (2018) 175:1014–30 e19. doi: 10.1016/j.cell.2018.09.030
22. Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res (2007) 35:W169–75. doi: 10.1093/nar/gkm415
23. Klemm F, Maas RR, Bowman RL, Kornete M, Soukup K, Nassiri S, et al. Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell (2020) 181:1643–60 e17. doi: 10.1016/j.cell.2020.05.007
24. Bleriot C, Chakarov S, Ginhoux F. Determinants of resident tissue macrophage identity and function. Immunity (2020) 52:957–70. doi: 10.1016/j.immuni.2020.05.014
25. Bian Z, Gong Y, Huang T, Lee CZW, Bian L, Bai Z, et al. Deciphering human macrophage development at single-cell resolution. Nature (2020) 582:571–6. doi: 10.1038/s41586-020-2316-7
26. Chorro L, Sarde A, Li M, Woollard KJ, Chambon P, Malissen B, et al. Langerhans cell (LC) proliferation mediates neonatal development, homeostasis, and inflammation-associated expansion of the epidermal LC network. J Exp Med (2009) 206:3089–100. doi: 10.1084/jem.20091586
27. Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science (2010) 330:841–5. doi: 10.1126/science.1194637
28. Hoeffel G, Wang Y, Greter M, See P, Teo P, Malleret B, et al. Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J Exp Med (2012) 209:1167–81. doi: 10.1084/jem.20120340
29. Davies LC, Jenkins SJ, Allen JE, Taylor PR. Tissue-resident macrophages. Nat Immunol (2013) 14:986–95. doi: 10.1038/ni.2705
30. Egeblad M, Nakasone ES, Werb Z. Tumors as organs: complex tissues that interface with the entire organism. Dev Cell (2010) 18:884–901. doi: 10.1016/j.devcel.2010.05.012
31. Zhu Y, Herndon JM, Sojka DK, Kim KW, Knolhoff BL, Zuo C, et al. Tissue-resident macrophages in pancreatic ductal adenocarcinoma originate from embryonic hematopoiesis and promote tumor progression. Immunity (2017) 47:597. doi: 10.1016/j.immuni.2017.08.018
32. Loyher PL, Hamon P, Laviron M, Meghraoui-Kheddar A, Goncalves E, Deng Z, et al. Macrophages of distinct origins contribute to tumor development in the lung. J Exp Med (2018) 215:2536–53. doi: 10.1084/jem.20180534
33. Li W, Chang N, Li L. Heterogeneity and function of kupffer cells in liver injury. Front Immunol (2022) 13:940867. doi: 10.3389/fimmu.2022.940867
34. Wculek SK, Dunphy G, Heras-Murillo I, Mastrangelo A, Sancho D. Metabolism of tissue macrophages in homeostasis and pathology. Cell Mol Immunol (2022) 19:384–408. doi: 10.1038/s41423-021-00791-9
35. Pombo Antunes AR, Scheyltjens I, Lodi F, Messiaen J, Antoranz A, Duerinck J, et al. Single-cell profiling of myeloid cells in glioblastoma across species and disease stage reveals macrophage competition and specialization. Nat Neurosci (2021) 24:595–610. doi: 10.1038/s41593-020-00789-y
36. Wang J, Wang Y, Chu Y, Li Z, Yu X, Huang Z, et al. Tumor-derived adenosine promotes macrophage proliferation in human hepatocellular carcinoma. J Hepatol (2021) 74:627–37. doi: 10.1016/j.jhep.2020.10.021
37. Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest (2012) 122:787–95. doi: 10.1172/JCI59643
38. Shand FH, Ueha S, Otsuji M, Koid SS, Shichino S, Tsukui T, et al. Tracking of intertissue migration reveals the origins of tumor-infiltrating monocytes. Proc Natl Acad Sci USA (2014) 111:7771–6. doi: 10.1073/pnas.1402914111
39. Cortez-Retamozo V, Etzrodt M, Newton A, Rauch PJ, Chudnovskiy A, Berger C, et al. Origins of tumor-associated macrophages and neutrophils. Proc Natl Acad Sci USA (2012) 109:2491–6. doi: 10.1073/pnas.1113744109
40. Swirski FK, Nahrendorf M, Etzrodt M, Wildgruber M, Cortez-Retamozo V, Panizzi P, et al. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science (2009) 325:612–6. doi: 10.1126/science.1175202
41. Cassetta L, Pollard JW. Targeting macrophages: therapeutic approaches in cancer. Nat Rev Drug Discovery (2018) 17:887–904. doi: 10.1038/nrd.2018.169
42. Jayasingam SD, Citartan M, Thang TH, Mat Zin AA, Ang KC, Ch'ng ES. Evaluating the polarization of tumor-associated macrophages into M1 and M2 phenotypes in human cancer tissue: technicalities and challenges in routine clinical practice. Front Oncol (2019) 9:1512. doi: 10.3389/fonc.2019.01512
43. Zhang L, Li Z, Skrzypczynska KM, Fang Q, Zhang W, O'Brien SA, et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell (2020) 181:442–59 e29. doi: 10.1016/j.cell.2020.03.048
44. Zhang Q, He Y, Luo N, Patel SJ, Han Y, Gao R, et al. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell (2019) 179:829–45 e20. doi: 10.1016/j.cell.2019.10.003
45. Zilionis R, Engblom C, Pfirschke C, Savova V, Zemmour D, Saatcioglu HD, et al. Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity (2019) 50:1317–34 e10. doi: 10.1016/j.immuni.2019.03.009
46. Sun BY, Zhou C, Guan RY, Liu G, Yang ZF, Wang ZT, et al. Dissecting intra-tumoral changes following immune checkpoint blockades in intrahepatic cholangiocarcinoma via single-cell analysis. Front Immunol (2022) 13:871769. doi: 10.3389/fimmu.2022.871769
47. Filiberti A, Gmyrek GB, Berube AN, Carr DJJ. Osteopontin contributes to virus resistance associated with type I IFN expression, activation of downstream ifn-inducible effector genes, and CCR2(+)CD115(+)CD206(+) macrophage infiltration following ocular HSV-1 infection of mice. Front Immunol (2022) 13:1028341. doi: 10.3389/fimmu.2022.1028341
48. Melcher V, Graf M, Interlandi M, Moreno N, de Faria FW, Kim SN, et al. Macrophage-tumor cell interaction promotes ATRT progression and chemoresistance. Acta Neuropathol (2020) 139:913–36. doi: 10.1007/s00401-019-02116-7
49. Haque A, Moriyama M, Kubota K, Ishiguro N, Sakamoto M, Chinju A, et al. CD206(+) tumor-associated macrophages promote proliferation and invasion in oral squamous cell carcinoma via EGF production. Sci Rep (2019) 9:14611. doi: 10.1038/s41598-019-51149-1
50. Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell (2018) 174:1293–308 e36. doi: 10.1016/j.cell.2018.05.060
51. Kim N, Kim HK, Lee K, Hong Y, Cho JH, Choi JW, et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun (2020) 11:2285. doi: 10.1038/s41467-020-16164-1
52. Liu X, Xu X, Wu Z, Shan Q, Wang Z, Wu Z, et al. Integrated single-cell RNA-seq analysis identifies immune heterogeneity associated with KRAS/TP53 mutation status and tumor-sideness in colorectal cancers. Front Immunol (2022) 13:961350. doi: 10.3389/fimmu.2022.961350
53. Song G, Shi Y, Meng L, Ma J, Huang S, Zhang J, et al. Single-cell transcriptomic analysis suggests two molecularly subtypes of intrahepatic cholangiocarcinoma. Nat Commun (2022) 13:1642. doi: 10.1038/s41467-022-29164-0
54. Leader AM, Grout JA, Maier BB, Nabet BY, Park MD, Tabachnikova A, et al. Single-cell analysis of human non-small cell lung cancer lesions refines tumor classification and patient stratification. Cancer Cell (2021) 39:1594–609 e12. doi: 10.1016/j.ccell.2021.10.009
55. Zhang Y, Chen H, Mo H, Hu X, Gao R, Zhao Y, et al. Single-cell analyses reveal key immune cell subsets associated with response to PD-L1 blockade in triple-negative breast cancer. Cancer Cell (2021) 39:1578–93 e8. doi: 10.1016/j.ccell.2021.09.010
56. Pal B, Chen Y, Vaillant F, Capaldo BD, Joyce R, Song X, et al. A single-cell RNA expression atlas of normal, preneoplastic and tumorigenic states in the human breast. EMBO J (2021) 40:e107333. doi: 10.15252/embj.2020107333
57. Li X, Zhang Q, Chen G, Luo D. Multi-omics analysis showed the clinical value of gene signatures of C1QC(+) and SPP1(+) TAMs in cervical cancer. Front Immunol (2021) 12:694801. doi: 10.3389/fimmu.2021.694801
58. Raghavan S, Winter PS, Navia AW, Williams HL, DenAdel A, Lowder KE, et al. Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer. Cell (2021) 184:6119–37 e26. doi: 10.1016/j.cell.2021.11.017
59. Doroshow DB, Bhalla S, Beasley MB, Sholl LM, Kerr KM, Gnjatic S, et al. PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat Rev Clin Oncol (2021) 18:345–62. doi: 10.1038/s41571-021-00473-5
60. Long Z, Sun C, Tang M, Wang Y, Ma J, Yu J, et al. Single-cell multiomics analysis reveals regulatory programs in clear cell renal cell carcinoma. Cell Discovery (2022) 8:68. doi: 10.1038/s41421-022-00415-0
61. Li XY, Shen Y, Zhang L, Guo X, Wu J. Understanding initiation and progression of hepatocellular carcinoma through single cell sequencing. Biochim Biophys Acta Rev Cancer (2022) 1877:188720. doi: 10.1016/j.bbcan.2022.188720
62. Yang Q, Zhang H, Wei T, Lin A, Sun Y, Luo P, et al. Single-cell RNA sequencing reveals the heterogeneity of tumor-associated macrophage in non-small cell lung cancer and differences between sexes. Front Immunol (2021) 12:756722. doi: 10.3389/fimmu.2021.756722
63. Lavin Y, Kobayashi S, Leader A, Amir ED, Elefant N, Bigenwald C, et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell (2017) 169:750–65 e17. doi: 10.1016/j.cell.2017.04.014
64. Jaitin DA, Adlung L, Thaiss CA, Weiner A, Li B, Descamps H, et al. Lipid-associated macrophages control metabolic homeostasis in a trem2-dependent manner. Cell (2019) 178:686–98 e14. doi: 10.1016/j.cell.2019.05.054
65. Obradovic A, Chowdhury N, Haake SM, Ager C, Wang V, Vlahos L, et al. Single-cell protein activity analysis identifies recurrence-associated renal tumor macrophages. Cell (2021) 184:2988–3005 e16. doi: 10.1016/j.cell.2021.04.038
66. Wei J, Yu W, Chen J, Huang G, Zhang L, Chen Z, et al. Single-cell and spatial analyses reveal the association between gene expression of glutamine synthetase with the immunosuppressive phenotype of APOE+CTSZ+TAM in cancers. Mol Oncol (2023) 15:984–98. doi: 10.1002/1878-0261.13373
67. Korbecki J, Olbromski M, Dziegiel P. CCL18 in the progression of cancer. Int J Mol Sci (2020) 21. doi: 10.3390/ijms21217955
68. Huang X, Lai S, Qu F, Li Z, Fu X, Li Q, et al. CCL18 promotes breast cancer progression by exosomal miR-760 activation of ARF6/Src/PI3K/Akt pathway. Mol Ther Oncolytics (2022) 25:1–15. doi: 10.1016/j.omto.2022.03.004
69. Yue T, Liu X, Zuo S, Zhu J, Li J, Liu Y, et al. BCL2A1 and CCL18 are predictive biomarkers of cisplatin chemotherapy and immunotherapy in colon cancer patients. Front Cell Dev Biol (2021) 9:799278. doi: 10.3389/fcell.2021.799278
70. Wu Y, Yang S, Ma J, Chen Z, Song G, Rao D, et al. Spatiotemporal immune landscape of colorectal cancer liver metastasis at single-cell level. Cancer Discovery (2022) 12:134–53. doi: 10.1158/2159-8290.CD-21-0316
71. Song G, Shi Y, Zhang M, Goswami S, Afridi S, Meng L, et al. Global immune characterization of HBV/HCV-related hepatocellular carcinoma identifies macrophage and T-cell subsets associated with disease progression. Cell Discovery (2020) 6:90. doi: 10.1038/s41421-020-00214-5
72. Sun C, Wang P, Gao T, Chi J. CCL18 knockdown suppresses cell growth and migration in thyroid cancer. J Healthc Eng (2022) 2022:1548155. doi: 10.1155/2022/1548155
73. Ren X, Zhang L, Zhang Y, Li Z, Siemers N, Zhang Z. Insights gained from single-cell analysis of immune cells in the tumor microenvironment. Annu Rev Immunol (2021) 39:583–609. doi: 10.1146/annurev-immunol-110519-071134
74. Cheng S, Li Z, Gao R, Xing B, Gao Y, Yang Y, et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell (2021) 184:792–809 e23. doi: 10.1016/j.cell.2021.01.010
75. Mulder K, Patel AA, Kong WT, Piot C, Halitzki E, Dunsmore G, et al. Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease. Immunity (2021) 54:1883–900 e5. doi: 10.1016/j.immuni.2021.07.007
76. Ma RY, Black A, Qian BZ. Macrophage diversity in cancer revisited in the era of single-cell omics. Trends Immunol (2022) 43:546–63. doi: 10.1016/j.it.2022.04.008
77. Huang YH, Cai K, Xu PP, Wang L, Huang CX, Fang Y, et al. CREBBP/EP300 mutations promoted tumor progression in diffuse large B-cell lymphoma through altering tumor-associated macrophage polarization via FBXW7-NOTCH-CCL2/CSF1 axis. Signal Transduct Target Ther (2021) 6:10. doi: 10.1038/s41392-020-00437-8
78. Etzerodt A, Moulin M, Doktor TK, Delfini M, Mossadegh-Keller N, Bajenoff M, et al. Tissue-resident macrophages in omentum promote metastatic spread of ovarian cancer. J Exp Med (2020) 217:16–25. doi: 10.1084/jem.20191869
79. Matsubara E, Komohara Y, Esumi S, Shinchi Y, Ishizuka S, Mito R, et al. SPP1 derived from macrophages is associated with a worse clinical course and chemo-resistance in lung adenocarcinoma. Cancers (Basel) (2022) 14:3468–80. doi: 10.3390/cancers14184374
80. He C, Sheng L, Pan D, Jiang S, Ding L, Ma X, et al. Single-cell transcriptomic analysis revealed a critical role of SPP1/CD44-mediated crosstalk between macrophages and cancer cells in glioma. Front Cell Dev Biol (2021) 9:779319. doi: 10.3389/fcell.2021.779319
81. Gao W, Liu D, Sun H, Shao Z, Shi P, Li T, et al. SPP1 is a prognostic related biomarker and correlated with tumor-infiltrating immune cells in ovarian cancer. BMC Cancer (2022) 22:1367. doi: 10.1186/s12885-022-10485-8
82. Hao B, Zhang Z, Lu Z, Xiong J, Fan T, Song C, et al. Single-cell RNA sequencing analysis revealed cellular and molecular immune profiles in lung squamous cell carcinoma. Transl Oncol (2023) 27:101568. doi: 10.1016/j.tranon.2022.101568
83. Zhao K, Ma Z, Zhang W. Comprehensive analysis to identify SPP1 as a prognostic biomarker in cervical cancer. Front Genet (2021) 12:732822. doi: 10.3389/fgene.2021.732822
84. Liu L, Zhang R, Deng J, Dai X, Zhu X, Fu Q, et al. Construction of TME and Identification of crosstalk between Malignant cells and macrophages by SPP1 in hepatocellular carcinoma. Cancer Immunol Immunother (2022) 71:121–36. doi: 10.1007/s00262-021-02967-8
85. Cai X, Zhang H, Li T. The role of SPP1 as a prognostic biomarker and therapeutic target in head and neck squamous cell carcinoma. Int J Oral Maxillofac Surg (2022) 51:732–41. doi: 10.1016/j.ijom.2021.07.022
86. Liu Y, Zhang Q, Xing B, Luo N, Gao R, Yu K, et al. Immune phenotypic linkage between colorectal cancer and liver metastasis. Cancer Cell (2022) 40:424–37 e5. doi: 10.1016/j.ccell.2022.02.013
87. Wei J, Chen Z, Hu M, He Z, Jiang D, Long J, et al. Characterizing intercellular communication of pan-cancer reveals SPP1+ Tumor-associated macrophage expanded in hypoxia and promoting cancer Malignancy through single-cell RNA-seq data. Front Cell Dev Biol (2021) 9:749210. doi: 10.3389/fcell.2021.749210
88. Hoeft K, Schaefer GJL, Kim H, Schumacher D, Bleckwehl T, Long Q, et al. Platelet-instructed SPP1(+) macrophages drive myofibroblast activation in fibrosis in a CXCL4-dependent manner. Cell Rep (2023) 42:112131. doi: 10.1016/j.celrep.2023.112131
89. Dong B, Wu C, Huang L, Qi Y. Macrophage-related SPP1 as a potential biomarker for early lymph node metastasis in lung adenocarcinoma. Front Cell Dev Biol (2021) 9:739358. doi: 10.3389/fcell.2021.739358
90. Ozato Y, Kojima Y, Kobayashi Y, Hisamatsu Y, Toshima T, Yonemura Y, et al. Spatial and single-cell transcriptomics decipher the cellular environment containing HLA-G+ cancer cells and SPP1+ macrophages in colorectal cancer. Cell Rep (2023) 42:111929. doi: 10.1016/j.celrep.2022.111929
91. Kramerova I, Kumagai-Cresse C, Ermolova N, Mokhonova E, Marinov M, Capote J, et al. Spp1 (osteopontin) promotes TGFbeta processing in fibroblasts of dystrophin-deficient muscles through matrix metalloproteinases. Hum Mol Genet (2019) 28:3431–42. doi: 10.1093/hmg/ddz181
92. Chen P, Zhao D, Li J, Liang X, Li J, Chang A, et al. Symbiotic macrophage-glioma cell interactions reveal synthetic lethality in PTEN-null glioma. Cancer Cell (2019) 35:868–84 e6. doi: 10.1016/j.ccell.2019.05.003
93. Droho S, Rajesh A, Cuda CM, Perlman H, Lavine JA. CD11c+ macrophages are pro-angiogenic and necessary for experimental choroidal neovascularization. JCI Insight (2023) 34:108677. doi: 10.1172/jci.insight.168142
94. Jiang X, Zhang X, Jiang N, Sun Y, Li T, Zhang J, et al. The single-cell landscape of cystic echinococcosis in different stages provided insights into endothelial and immune cell heterogeneity. Front Immunol (2022) 13:1067338. doi: 10.3389/fimmu.2022.1067338
95. Tan Y, Zhao L, Yang YG, Liu W. The role of osteopontin in tumor progression through tumor-associated macrophages. Front Oncol (2022) 12:953283. doi: 10.3389/fonc.2022.953283
96. Taniguchi S, Elhance A, Van Duzer A, Kumar S, Leitenberger JJ, Oshimori N. Tumor-initiating cells establish an IL-33-TGF-beta niche signaling loop to promote cancer progression. Science (2020) 369:1124–31. doi: 10.1126/science.aay1813
97. Qi J, Sun H, Zhang Y, Wang Z, Xun Z, Li Z, et al. Single-cell and spatial analysis reveal interaction of FAP(+) fibroblasts and SPP1(+) macrophages in colorectal cancer. Nat Commun (2022) 13:1742. doi: 10.1038/s41467-022-29366-6
98. Sathe A, Mason K, Grimes SM, Zhou Z, Lau BT, Bai X, et al. Colorectal cancer metastases in the liver establish immunosuppressive spatial networking between tumor-associated SPP1+ Macrophages and fibroblasts. Clin Cancer Res (2023) 29:244–60. doi: 10.1158/1078-0432.CCR-22-2041
99. Liu X, Qin J, Nie J, Gao R, Hu S, Sun H, et al. ANGPTL2+cancer-associated fibroblasts and SPP1+macrophages are metastasis accelerators of colorectal cancer. Front Immunol (2023) 14:1185208. doi: 10.3389/fimmu.2023.1185208
100. Messex JK, Byrd CJ, Thomas MU, Liou GY. Macrophages cytokine spp1 increases growth of prostate intraepithelial neoplasia to promote prostate tumor progression. Int J Mol Sci (2022) 23:822–34. doi: 10.3390/ijms23084247
101. De Palma M, Biziato D, Petrova TV. Microenvironmental regulation of tumour angiogenesis. Nat Rev Cancer (2017) 17:457–74. doi: 10.1038/nrc.2017.51
102. Movahedi K, Laoui D, Gysemans C, Baeten M, Stange G, Van den Bossche J, et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res (2010) 70:5728–39. doi: 10.1158/0008-5472.CAN-09-4672
103. Henze AT, Mazzone M. The impact of hypoxia on tumor-associated macrophages. J Clin Invest (2016) 126:3672–9. doi: 10.1172/JCI84427
104. Zhang B, Ye H, Ren X, Zheng S, Zhou Q, Chen C, et al. Macrophage-expressed CD51 promotes cancer stem cell properties via the TGF-beta1/smad2/3 axis in pancreatic cancer. Cancer Lett (2019) 459:204–15. doi: 10.1016/j.canlet.2019.06.005
105. Chen P, Hsu WH, Han J, Xia Y, DePinho RA. Cancer stemness meets immunity: from mechanism to therapy. Cell Rep (2021) 34:108597. doi: 10.1016/j.celrep.2020.108597
106. Liguori M, Digifico E, Vacchini A, Avigni R, Colombo FS, Borroni EM, et al. The soluble glycoprotein NMB (GPNMB) produced by macrophages induces cancer stemness and metastasis via CD44 and IL-33. Cell Mol Immunol (2021) 18:711–22. doi: 10.1038/s41423-020-0501-0
107. Qiu S, Deng L, Liao X, Nie L, Qi F, Jin K, et al. Tumor-associated macrophages promote bladder tumor growth through PI3K/AKT signal induced by collagen. Cancer Sci (2019) 110:2110–8. doi: 10.1111/cas.14078
108. Quail DF, Bowman RL, Akkari L, Quick ML, Schuhmacher AJ, Huse JT, et al. The tumor microenvironment underlies acquired resistance to CSF-1R inhibition in gliomas. Science (2016) 352:aad3018. doi: 10.1126/science.aad3018
109. Chan JM, Quintanal-Villalonga A, Gao VR, Xie Y, Allaj V, Chaudhary O, et al. Signatures of plasticity, metastasis, and immunosuppression in an atlas of human small cell lung cancer. Cancer Cell (2021) 39:1479–96 e18. doi: 10.1016/j.ccell.2021.09.008
110. Zhang Y, Du W, Chen Z, Xiang C. Upregulation of PD-L1 by SPP1 mediates macrophage polarization and facilitates immune escape in lung adenocarcinoma. Exp Cell Res (2017) 359:449–57. doi: 10.1016/j.yexcr.2017.08.028
111. Liu Y, Xun Z, Ma K, Liang S, Li X, Zhou S, et al. Identification of a tumour immune barrier in the HCC microenvironment that determines the efficacy of immunotherapy. J Hepatol (2023) 78:770–82. doi: 10.1016/j.jhep.2023.01.011
112. Matsubara E, Yano H, Pan C, Komohara Y, Fujiwara Y, Zhao S, et al. The significance of SPP1 in lung cancers and its impact as a marker for protumor tumor-associated macrophages. Cancers (Basel). (2023) 15:108724. doi: 10.3390/cancers15082250
113. Pietras A, Katz AM, Ekstrom EJ, Wee B, Halliday JJ, Pitter KL, et al. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell (2014) 14:357–69. doi: 10.1016/j.stem.2014.01.005
114. Feng S, Yuan W, Sun Z, Guo X, Ling J, Chang A, et al. SPP1 as a key gene in the lymph node metastasis and a potential predictor of poor prognosis in head and neck carcinoma. J Oral Pathol Med (2022) 51:620–9. doi: 10.1111/jop.13333
115. Hao C, Lane J, Jiang WG. Osteopontin and cancer: insights into its role in drug resistance. Biomedicines (2023) 11:2461–70. doi: 10.3390/biomedicines11010197
116. Krishna C, DiNatale RG, Kuo F, Srivastava RM, Vuong L, Chowell D, et al. Single-cell sequencing links multiregional immune landscapes and tissue-resident T cells in ccRCC to tumor topology and therapy efficacy. Cancer Cell (2021) 39:662–77 e6. doi: 10.1016/j.ccell.2021.03.007
117. Dangaj D, Bruand M, Grimm AJ, Ronet C, Barras D, Duttagupta PA, et al. Cooperation between constitutive and inducible chemokines enables T cell engraftment and immune attack in solid tumors. Cancer Cell (2019) 35:885–900 e10. doi: 10.1016/j.ccell.2019.05.004
118. Qu Y, Wen J, Thomas G, Yang W, Prior W, He W, et al. Baseline frequency of inflammatory cxcl9-expressing tumor-associated macrophages predicts response to avelumab treatment. Cell Rep (2020) 32:108115. doi: 10.1016/j.celrep.2020.108115
119. House IG, Savas P, Lai J, Chen AXY, Oliver AJ, Teo ZL, et al. Macrophage-derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin Cancer Res (2020) 26:487–504. doi: 10.1158/1078-0432.CCR-19-1868
120. Zhou J, Li X, Wu X, Zhang T, Zhu Q, Wang X, et al. Exosomes released from tumor-associated macrophages transfer miRNAs that induce a Treg/Th17 cell imbalance in epithelial ovarian cancer. Cancer Immunol Res (2018) 6:1578–92. doi: 10.1158/2326-6066.CIR-17-0479
121. Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity (2014) 41:49–61. doi: 10.1016/j.immuni.2014.06.010
122. Labadie BW, Bao R, Luke JJ. Reimagining IDO pathway inhibition in cancer immunotherapy via downstream focus on the tryptophan-kynurenine-aryl hydrocarbon axis. Clin Cancer Res (2019) 25:1462–71. doi: 10.1158/1078-0432.CCR-18-2882
123. Geiger R, Rieckmann JC, Wolf T, Basso C, Feng Y, Fuhrer T, et al. L-arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell (2016) 167:829–42 e13. doi: 10.1016/j.cell.2016.09.031
124. Ye LY, Chen W, Bai XL, Xu XY, Zhang Q, Xia XF, et al. Hypoxia-induced epithelial-to-mesenchymal transition in hepatocellular carcinoma induces an immunosuppressive tumor microenvironment to promote metastasis. Cancer Res (2016) 76:818–30. doi: 10.1158/0008-5472.CAN-15-0977
125. Dong L, Chen C, Zhang Y, Guo P, Wang Z, Li J, et al. The loss of RNA N(6)-adenosine methyltransferase Mettl14 in tumor-associated macrophages promotes CD8(+) T cell dysfunction and tumor growth. Cancer Cell (2021) 39:945–57 e10. doi: 10.1016/j.ccell.2021.04.016
126. Chen LH, Liu JF, Lu Y, He XY, Zhang C, Zhou HH. Complement C1q (C1qA, C1qB, and C1qC) may be a potential prognostic factor and an index of tumor microenvironment remodeling in osteosarcoma. Front Oncol (2021) 11:642144. doi: 10.3389/fonc.2021.642144
127. Braun DA, Street K, Burke KP, Cookmeyer DL, Denize T, Pedersen CB, et al. Progressive immune dysfunction with advancing disease stage in renal cell carcinoma. Cancer Cell (2021) 39:632–48 e8. doi: 10.1016/j.ccell.2021.02.013
128. Roumenina LT, Daugan MV, Noe R, Petitprez F, Vano YA, Sanchez-Salas R, et al. Tumor cells hijack macrophage-produced complement C1q to promote tumor growth. Cancer Immunol Res (2019) 7:1091–105. doi: 10.1158/2326-6066.CIR-18-0891
129. Yao W, Liu H, Xu F, Cai Z, Hang L, Lu M, et al. C1QC is a prognostic biomarker with immune-related value in kidney renal clear cell carcinoma. Front Genet (2023) 14:1109991. doi: 10.3389/fgene.2023.1109991
130. Shao YJ, Ni JJ, Wei SY, Weng XP, Shen MD, Jia YX, et al. IRF1-mediated immune cell infiltration is associated with metastasis in colon adenocarcinoma. Med (Baltimore) (2020) 99:e22170. doi: 10.1097/MD.0000000000022170
131. Son M, Porat A, He M, Suurmond J, Santiago-Schwarz F, Andersson U, et al. C1q and HMGB1 reciprocally regulate human macrophage polarization. Blood (2016) 128:2218–28. doi: 10.1182/blood-2016-05-719757
132. Spivia W, Magno PS, Le P, Fraser DA. Complement protein C1q promotes macrophage anti-inflammatory M2-like polarization during the clearance of atherogenic lipoproteins. Inflammation Res (2014) 63:885–93. doi: 10.1007/s00011-014-0762-0
133. Ling GS, Crawford G, Buang N, Bartok I, Tian K, Thielens NM, et al. C1q restrains autoimmunity and viral infection by regulating CD8(+) T cell metabolism. Science (2018) 360:558–63. doi: 10.1126/science.aao4555
134. Revel M, Sautes-Fridman C, Fridman WH, Roumenina LT. C1q+ macrophages: passengers or drivers of cancer progression. Trends Cancer (2022) 8:517–26. doi: 10.1016/j.trecan.2022.02.006
135. Bulla R, Tripodo C, Rami D, Ling GS, Agostinis C, Guarnotta C, et al. C1q acts in the tumour microenvironment as a cancer-promoting factor independently of complement activation. Nat Commun (2016) 7:10346. doi: 10.1038/ncomms10346
136. Song H, Lou C, Ma J, Gong Q, Tian Z, You Y, et al. Single-cell transcriptome analysis reveals changes of tumor immune microenvironment in oral squamous cell carcinoma after chemotherapy. Front Cell Dev Biol (2022) 10:914120. doi: 10.3389/fcell.2022.914120
137. Chen X, Gao Y, Xie J, Hua H, Pan C, Huang J, et al. Identification of FCN1 as a novel macrophage infiltration-associated biomarker for diagnosis of pediatric inflammatory bowel diseases. J Transl Med (2023) 21:203. doi: 10.1186/s12967-023-04038-1
138. Hoves S, Krause SW, Schutz C, Halbritter D, Scholmerich J, Herfarth H, et al. Monocyte-derived human macrophages mediate anergy in allogeneic T cells and induce regulatory T cells. J Immunol (2006) 177:2691–8. doi: 10.4049/jimmunol.177.4.2691
139. Su S, Liao J, Liu J, Huang D, He C, Chen F, et al. Blocking the recruitment of naive CD4(+) T cells reverses immunosuppression in breast cancer. Cell Res (2017) 27:461–82. doi: 10.1038/cr.2017.34
140. Lin L, Chen YS, Yao YD, Chen JQ, Chen JN, Huang SY, et al. CCL18 from tumor-associated macrophages promotes angiogenesis in breast cancer. Oncotarget (2015) 6:34758–73. doi: 10.18632/oncotarget.5325
141. Nie Y, Huang H, Guo M, Chen J, Wu W, Li W, et al. Breast phyllodes tumors recruit and repolarize tumor-associated macrophages via secreting CCL5 to promote Malignant progression, which can be inhibited by CCR5 inhibition therapy. Clin Cancer Res (2019) 25:3873–86. doi: 10.1158/1078-0432.CCR-18-3421
142. Ginhoux F, Guilliams M. Tissue-resident macrophage ontogeny and homeostasis. Immunity (2016) 44:439–49. doi: 10.1016/j.immuni.2016.02.024
143. Hoeffel G, Ginhoux F. Ontogeny of tissue-resident macrophages. Front Immunol (2015) 6:486. doi: 10.3389/fimmu.2015.00486
144. Chen J, Yao Y, Gong C, Yu F, Su S, Chen J, et al. CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell (2011) 19:541–55. doi: 10.1016/j.ccr.2011.02.006
145. Yue Z, Nie L, Zhang P, Chen Q, Lv Q, Wang Q. Tissue-resident macrophage inflammaging aggravates homeostasis dysregulation in age-related diseases. Cell Immunol (2021) 361:104278. doi: 10.1016/j.cellimm.2020.104278
146. Yunna C, Mengru H, Lei W, Weidong C. Macrophage M1/M2 polarization. Eur J Pharmacol (2020) 877:173090. doi: 10.1016/j.ejphar.2020.173090
147. Boutilier AJ, Elsawa SF. Macrophage polarization states in the tumor microenvironment. Int J Mol Sci (2021) 22:1624. doi: 10.3390/ijms22136995
148. Song J, Xiao T, Li M, Jia Q. Tumor-associated macrophages: Potential therapeutic targets and diagnostic markers in cancer. Pathol Res Pract (2023) 249:154739. doi: 10.1016/j.prp.2023.154739
149. Barry ST, Gabrilovich DI, Sansom OJ, Campbell AD, Morton JP. Therapeutic targeting of tumour myeloid cells. Nat Rev Cancer (2023) 23:216–37. doi: 10.1038/s41568-022-00546-2
150. Liu X, Liu X, Mao W, Guo Y, Bai N, Jin L, et al. Tetrastigma polysaccharide reprogramming of tumor-associated macrophages via PPARgamma signaling pathway to play antitumor activity in breast cancer. J Ethnopharmacol (2023) 314:116645. doi: 10.1016/j.jep.2023.116645
151. Cheng Y, Zhong X, Nie X, Gu H, Wu X, Li R, et al. Glycyrrhetinic acid suppresses breast cancer metastasis by inhibiting M2-like macrophage polarization via activating JNK1/2 signaling. Phytomedicine (2023) 114:154757. doi: 10.1016/j.phymed.2023.154757
152. Mehla K, Singh PK. Metabolic regulation of macrophage polarization in cancer. Trends Cancer (2019) 5:822–34. doi: 10.1016/j.trecan.2019.10.007
153. Tao H, Zhong X, Zeng A, Song L. Unveiling the veil of lactate in tumor-associated macrophages: a successful strategy for immunometabolic therapy. Front Immunol (2023) 14:1208870. doi: 10.3389/fimmu.2023.1208870
154. Han S, Bao X, Zou Y, Wang L, Li Y, Yang L, et al. d-lactate modulates M2 tumor-associated macrophages and remodels immunosuppressive tumor microenvironment for hepatocellular carcinoma. Sci Adv (2023) 9:eadg2697. doi: 10.1126/sciadv.adg2697
155. Liu Y, Sun Q, Zhang C, Ding M, Wang C, Zheng Q, et al. STING-IRG1 inhibits liver metastasis of colorectal cancer by regulating the polarization of tumor-associated macrophages. iScience (2023) 26:107376. doi: 10.1016/j.isci.2023.107376
156. Lin Z, Wang Q, Jiang T, Wang W, Zhao JJ. Targeting tumor-associated macrophages with STING agonism improves the antitumor efficacy of osimertinib in a mouse model of EGFR-mutant lung cancer. Front Immunol (2023) 14:1077203. doi: 10.3389/fimmu.2023.1077203
157. Ding L, Wang Q, Martincuks A, Kearns MJ, Jiang T, Lin Z, et al. STING agonism overcomes STAT3-mediated immunosuppression and adaptive resistance to PARP inhibition in ovarian cancer. J Immunother Cancer (2023) 11:3896–907. doi: 10.1136/jitc-2022-005627
158. Liu X, Hogg GD, Zuo C, Borcherding NC, Baer JM, Lander VE, et al. Context-dependent activation of STING-interferon signaling by CD11b agonists enhances anti-tumor immunity. Cancer Cell (2023) 41:1073–90 e12. doi: 10.1016/j.ccell.2023.04.018
159. Wu YT, Fang Y, Wei Q, Shi H, Tan H, Deng Y, et al. Tumor-targeted delivery of a STING agonist improvescancer immunotherapy. Proc Natl Acad Sci USA (2022) 119:e2214278119. doi: 10.1073/pnas.2214278119
160. Guerriero JL, Sotayo A, Ponichtera HE, Castrillon JA, Pourzia AL, SChad S, et al. Class IIa HDAC inhibition reduces breast tumours and metastases through anti-tumour macrophages. Nature (2017) 543:428–32. doi: 10.1038/nature21409
161. Orillion A, Hashimoto A, Damayanti N, Shen L, Adelaiye-Ogala R, Arisa S, et al. Entinostat neutralizes myeloid-derived suppressor cells and enhances the antitumor effect of PD-1 inhibition in murine models of lung and renal cell carcinoma. Clin Cancer Res (2017) 23:5187–201. doi: 10.1158/1078-0432.CCR-17-0741
162. Horlad H, Ma C, Yano H, Pan C, Ohnishi K, Fujiwara Y, et al. An IL-27/Stat3 axis induces expression of programmed cell death 1 ligands (PD-L1/2) on infiltrating macrophages in lymphoma. Cancer Sci (2016) 107:1696–704. doi: 10.1111/cas.13065
163. Li Z, Ding Y, Liu J, Wang J, Mo F, Wang Y, et al. Depletion of tumor associated macrophages enhances local and systemic platelet-mediated anti-PD-1 delivery for post-surgery tumor recurrence treatment. Nat Commun (2022) 13:1845. doi: 10.1038/s41467-022-29388-0
164. Wang Y, Zhao C, Liu Y, Wang C, Jiang H, Hu Y, et al. Recent advances of tumor therapy based on the CD47-SIRPalpha axis. Mol Pharm (2022) 19:1273–93. doi: 10.1021/acs.molpharmaceut.2c00073
165. Zhang W, Huang Q, Xiao W, Zhao Y, Pi J, Xu H, et al. Advances in anti-tumor treatments targeting the CD47/SIRPalpha axis. Front Immunol (2020) 11:18. doi: 10.3389/fimmu.2020.00018
166. Bouwstra R, van Meerten T, Bremer E. CD47-SIRPalpha blocking-based immunotherapy: Current and prospective therapeutic strategies. Clin Transl Med (2022) 12:e943. doi: 10.1002/ctm2.943
167. Wang Y, Tiruthani K, Li S, Hu M, Zhong G, Tang Y, et al. mRNA delivery of a bispecific single-domain antibody to polarize tumor-associated macrophages and synergize immunotherapy against liver Malignancies. Adv Mater (2021) 33:e2007603. doi: 10.1002/adma.202007603
168. Jaynes JM, Sable R, Ronzetti M, Bautista W, Knotts Z, Abisoye-Ogunniyan A, et al. Mannose receptor (CD206) activation in tumor-associated macrophages enhances adaptive and innate antitumor immune responses. Sci Transl Med (2020) 12:663–71. doi: 10.1126/scitranslmed.aax6337
169. Zhang P, Gu Y, Wang J, Lv K, Lin C, Zhang H, et al. Complement receptor C5aR1 blockade reprograms tumor-associated macrophages and synergizes with anti-PD-1 therapy in gastric cancer. Int J Cancer (2023) 153:224–37. doi: 10.1002/ijc.34474
Keywords: tumor associated macrophages, cancer, single cell RNA sequencing, angiogenesis, metastasis
Citation: Wang J, Zhu N, Su X, Gao Y and Yang R (2024) Novel tumor-associated macrophage populations and subpopulations by single cell RNA sequencing. Front. Immunol. 14:1264774. doi: 10.3389/fimmu.2023.1264774
Received: 21 July 2023; Accepted: 30 November 2023;
Published: 29 January 2024.
Edited by:
Jun Zhang, Kumamoto University, JapanReviewed by:
Suchandan Sikder, James Cook University, AustraliaWang Huaitao, Kumamoto University, Japan
Copyright © 2024 Wang, Zhu, Su, Gao and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rongcun Yang, cnlhbmdAbmFua2FpLmVkdS5jbg==
†These authors have contributed equally to this work
 Juanjuan Wang1,2,3†
Juanjuan Wang1,2,3† Xiaomin Su
Xiaomin Su Rongcun Yang
Rongcun Yang