
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 12 September 2023
Sec. Inflammation
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1250504
This article is part of the Research TopicSingle-cell analysis on the pathophysiology of autoimmune diseasesView all 16 articles
 Jaehwan Kim1,2,3*
Jaehwan Kim1,2,3* Jongmi Lee2
Jongmi Lee2 Xuan Li1
Xuan Li1 Norma Kunjravia1
Norma Kunjravia1 Darshna Rambhia1
Darshna Rambhia1 Inna Cueto1
Inna Cueto1 Katherine Kim2,3
Katherine Kim2,3 Vasuma Chaparala2
Vasuma Chaparala2 Younhee Ko3,4
Younhee Ko3,4 Sandra Garcet1,5
Sandra Garcet1,5 Wei Zhou6
Wei Zhou6 Junyue Cao6
Junyue Cao6 James G. Krueger1*
James G. Krueger1*Durable psoriasis improvement has been reported in a subset of psoriasis patients after treatment withdrawal of biologics blocking IL-23/Type 17 T-cell (T17) autoimmune axis. However, it is not well understood if systemic blockade of the IL-23/T17 axis promotes immune tolerance in psoriasis skin. The purpose of the study was to find translational evidence that systemic IL-17A blockade promotes regulatory transcriptome modification in human psoriasis skin immune cell subsets. We analyzed human psoriasis lesional skin 6 mm punch biopsy tissues before and after systemic IL-17A blockade using the muti-genomics approach integrating immune cell-enriched scRNA-seq (n = 18), microarray (n = 61), and immunohistochemistry (n = 61) with repository normal control skin immune cell-enriched scRNA-seq (n = 10) and microarray (n = 8) data. For the T17 axis transcriptome, systemic IL-17A blockade depleted 100% of IL17A+ T-cells and 95% of IL17F+ T-cells in psoriasis skin. The expression of IL23A in DC subsets was also downregulated by IL-17A blockade. The expression of IL-17-driven inflammatory mediators (IL36G, S100A8, DEFB4A, and DEFB4B) in suprabasal keratinocytes was correlated with psoriasis severity and was downregulated by IL-17A blockade. For the regulatory DC transcriptome, the proportion of regulatory semimature DCs expressing regulatory DC markers of BDCA-3 (THBD) and DCIR (CLEC4A) was increased in posttreatment psoriasis lesional skin compared to pretreatment psoriasis lesional skin. In addition, IL-17A blockade induced higher expression of CD1C and CD14, which are markers of CD1c+ CD14+ dendritic cell (DC) subset that suppresses antigen-specific T-cell responses, in posttreatment regulatory semimature DCs compared to pretreatment regulatory semimature DCs. In conclusion, systemic IL-17A inhibition not only blocks the entire IL-23/T17 cell axis but also promotes regulatory gene expression in regulatory DCs in human psoriasis skin.
Psoriasis is one of the most common organ-specific autoimmune diseases in the human population affecting 3.2% of the adult population (1–3). Psoriasis originates from the skin, but it progressively induces systemic inflammation that may accompany psoriatic arthritis, metabolic syndrome, diabetes, overt vascular inflammation, and cardiovascular disease (4–7). Increasing evidence indicates that the systemic impact of psoriasis shortens the lifespan of affected individuals by at least 3–5 years (8).
Psoriasis persists as a lifelong disease that rarely improves without treatment, but systemic administration of recent monoclonal antibodies targeting the IL-23/Type 17 T-cell (T17) autoimmune axis is highly effective for psoriasis treatment (3). In the current model of psoriasis immunopathogenesis, IL-23 from dendritic cells (DCs) triggers T-cells (T17 cells) to produce IL-17, and IL-17 induces inflammatory mediators in keratinocytes (KCs) such as IL-36γ. The inflammatory mediators from KCs amplify DC and T-cell recruitment and activation completing the feed-forward inflammatory loop.
Genomic medicine mapping of psoriasis-associated immune pathways in the human skin enabled the therapeutic success of IL-23/IL-17 antagonists. The high efficacy of most IL-23/IL-17 antagonists was identified through early phase IIb studies that incorporated total skin gene expression profiles on relatively small numbers of patients, often with <10 in a treatment cohort (9–11). Large phase II and III studies were needed mainly to develop safety profiles on a sufficient number of patients for new drug registration.
However, total skin gene expression profiling is confronting limitations to dissect the IL-23/T17 autoimmune axis for explaining recent clinical trial findings (1): systemic administration of an anti-IL-17A monoclonal antibody increased the expression of genes involved in skin homeostasis promotion and KC stem cell activation in psoriasis skin (10). (2) systemic administration of an anti-IL-17A or anti-IL-23p19 monoclonal antibody produced durable psoriasis improvement maintenance even after treatment withdrawal in a subset of psoriasis patients (9, 12–15). To understand immune tolerance promotion induced by IL-23/T17 autoimmune axis blockade, we need to segregate and compare the transcriptome of regulatory immune cell subsets in human psoriasis skin before and after blocking the IL-23/T17 autoimmune axis.
We recently used RT-PCR analyses of total skin from a randomized placebo-controlled clinical trial (ClinicalTrial.gov identifier: NCT03131570) to study psoriasis skin transcriptome modifications induced by systemic IL-17A blockade (10). Here, we used archived samples from this trial for further analysis, applying a multi-omics approach. We performed single cell RNA-sequencing (scRNA-seq) analyses of T-cell, DC, and KC subsets isolated from psoriasis skin. We also used whole psoriasis skin for microarray, RT-PCR, and IHC analyses (Figure 1). Our multi-omics approach provides novel translational evidence in human psoriasis skin that systemic anti-IL-17A monoclonal antibody administration not only blocks the entire feed-forward inflammatory amplification loop between T-cells, DCs, and KCs, but also promotes regulatory transcriptome modification in regulatory DCs.
23 adult psoriasis patients received anti-IL-17A monoclonal antibody (secukinumab) injections for more than 12 weeks in the phase II clinical trial (ClinicalTrial.gov identifier: NCT03131570). Patients received the anti-IL-17A monoclonal antibody at a dose of 300 mg with injections administered once weekly at baseline and at weeks 1, 2, 3, and 4 and then every 4 weeks. The clinical responses of patients and RT-PCR analyses of psoriasis skin were previously published (10).
For this study, psoriasis skin biopsy samples obtained from the clinical trial (10) were further analyzed by immune cell-enriched scRNA-seq (n = 18; GSE183047), microarray (n = 61; GSE226244), and immunohistochemistry (n = 61) together with our repository normal control skin immune cell-enriched scRNA-seq data (n = 10) (16) and microarray data (n = 8) (Supplementary Table 1). To minimize the batch effects, all the molecular experiments from the initial sample processing to cDNA library construction were performed by the same investigators under the same protocol.
Lesional biopsy specimens were harvested with a 6 mm punch biopsy from areas with representative psoriasis lesions. The individual skin sample information, including age, gender, psoriasis versus (vs.) control, and timepoint of biopsy is summarized in Supplementary Table 1. Psoriasis lesional skin biopsy samples before anti-IL-17A monoclonal antibody injections (pretreatment) and psoriasis lesional skin biopsy samples after 12 weeks of anti-IL-17 monoclonal antibody injections (posttreatment) were compared for this study.
Harvested skin samples were bisected immediately after the skin biopsy. Half of the two bisected skin tissue was incubated in a culture medium for single-cell RNA sequencing (scRNA-seq) experiments. Another half of the bisected skin tissue was snap-frozen and embedded in the Optimal Cutting Temperature (OCT) compound (Ted Pella, Redding, CA) for microarray and immunohistochemistry experiments.
Skin biopsy samples were incubated in culture medium for harvesting emigrating cells (16). To split the epidermis and dermis where most of the inflammatory cells were located, harvested skin tissue was immediately placed in 0.2% Dispase II (Sigma-Aldrich, St. Louis, MO) and incubated in a humidified incubator at 37°C and 5% CO2 for 3 hours. Then, the epidermis and dermis were separated with forceps and sliced into small pieces with #10 blade scalpels. The epidermis and dermis were separately incubated in RPMI-1640 medium with L-glutamine (Cytiva, Marlborough, MA) supplemented with 10% human albumin serum (Sigma-Aldrich, St. Louis, MO) in a humidified incubator at 37°C and 5% CO2. Nonplastic adherent cells that had emigrated out of the epidermis and dermis were harvested after 48 hours. The harvested cells from the epidermis and dermis were filtered through a 40-µm cell strainer (Corning, Glendale, AZ) and stored on ice. The cell numbers and viability were determined using a Countess automated cell counter (Invitrogen, Carlsbad, CA) and trypan blue staining (BioRad, Hercules, CA).
The 10x Genomics Chromium Single Cell 3′ Reagents Kit user guide (https://support.10xgenomics.com) was used to prepare the single-cell suspension. The appropriate volume of each sample was diluted to recover 10,000 cells. Subsequently, the single-cell suspension, gel beads, and oils were added to the 10X Genomics single-cell chip. After droplet generation, samples were transferred into PCR tubes and we performed reverse transcription using a ProFlex PCR Thermocycler System (Applied Biosystems, Foster City, CA). After reverse transcription, cDNA was recovered using a recovery agent, provided by 10X Genomics, followed by silane DynaBead clean-up as outlined in the user guide.
The raw sequencing data for individual samples were processed using the 10X Genomics standard sequencing protocol without modifications. The FASTQ files were aligned to the human genome reference sequence GRCh38. Cell Ranger was applied to FASTQ files to generate files containing a barcode table, a gene table, and a gene expression matrix. The number of reads, mean reads per cell, valid barcodes, sequencing saturation, and other scRNA-seq parameters are detailed in Supplementary Table 1. The scRNA-seq data have been deposited in NCBI’s Gene Expression Omnibus and are publicly accessible through GEO Series accession number GSE183047. There is no restriction on the use of the data.
We used the Seurat R package (version 4.0) installed in R (version 4.0.2) for the downstream single-cell analyses (17–19). Before data integration, single-cell data quality control was performed separately for each sample as previously described for human skin single-cell RNA sequencing analyses (16, 20–22). Genes expressed in <3 cells, and cells with <100 or >5,000 genes and a mitochondrial gene percentage of >25% were filtered out to eliminate partial cells and doublets. Ubiquitously expressed ribosomal protein-coding (RPS and RPL) and MALAT noncoding RNA, miRNA, and snoRNA genes were excluded from the analysis as we described in our prior single-cell analysis publication with the publicly available R scripts (16). Seurat objects were created followed by normalizing data, scaling data, and finding variable 2,000 genes.
We first merged single-cell data of samples with the identical reagent kit version and the identical sequencer. To harmonize merged groups into a single dataset reducing batch effects, correspondences between cells in merged datasets were identified by the FindIntegrationAnchors function, and the correspondences were used for data integration with the IntegratedData function as detailed by Stuart et al. (18).
Principal component analysis and graph-based clustering analysis were performed, and sixty principal components (PCs) were selected for Uniform Manifold Approximation and Projection for Dimension Reduction. With a resolution of 0.8, cells were clustered by the FindClusters function. The average gene expression of psoriasis vs. control cells within a cluster was calculated by the AverageExpression function.
When 10,000 cells are submitted to a microfluidic platform for droplet-based single-cell library construction, 2.3% to 4.6% of single-cell data could be technical artifacts caused by cell doublets formed during cell capture (Chromium Next GEM Single Cell 3′ Reagent Kits User Guide, 10X Genomics, USA). To remove clusters with possible doublets for downstream single-cell analysis, we excluded clusters expressing gene signatures of more than two different immune cell types (Supplementary Figure 1). We also excluded clusters of a minimal number of cells that were spatially separated from the analogous type of cells. To compare psoriasis and control cells within clusters representing each type of skin immune cells, we merged adjacent clusters of common immune cell subsets.
Skin biopsy samples were frozen and then mechanically disaggregated before RNA extraction. RNA was extracted with RNA isolation kits (Qiagen) according to the manufacturer’s protocol. A total of 100 ng of biotinylated cDNA was hybridized to the GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix). The expression values were obtained using the GCRMA algorithm (23), while normalization across samples was carried out using quantile normalization. The raw microarray data have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through accession number GSE226244.
Total RNA was extracted from frozen skin biopsies by the Qiagen miNeasy Mini Kit (Valencia, CA, USA). DNA was removed with on-column DNase digestion by the Qiagen RNase-free DNase Set (Valencia, CA, USA). The quality of extracted RNA was examined using Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA). Real-time PCR was performed on QuantStudio 7 Flex Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA) with TaqMan Array Cards (384-well plates preloaded with TaqMan assays, Thermo Fisher Scientific, Waltham, MA). All primers are listed in Supplementary Table 2.
For immunostaining, vertical sections were cut to a thickness of 6 μm (+/- 1 μm) from the frozen skin biopsy tissues embedded in the OCT compound. Frozen sections were dried at room temperature and then fixed for 2 minutes in acetone. Next, the samples were blocked with 10% normal serum of the species in which the secondary antibody was made, and then the samples were incubated overnight at 4°C with the appropriate primary antibody. Primary antibodies used in this study were all mouse antibodies: KRT16 (LSBio, Seattle, WA; clone 7A4, dilution 1:500), CD3 (BD Biosciences, San Jose, CA; clone SK7, dilution 1:100), and CD11c (BD Biosciences, San Jose, CA; clone Bly6, dilution 1:100). Biotin-labeled horse anti-mouse antibodies (Vector Laboratories, Burlingame, CA) were used to detect the primary antibodies. The staining signal was amplified with avidin-biotin complex (Vector Laboratories, Burlingame, CA) and developed using chromogen 3-amino-9-ethyl carbazole (Sigma-Aldrich, St. Louis, MO). Section images were acquired at ×10 magnification. The number of immunostaining-positive cell counts per section was manually counted using computer-assisted image analysis software (ImageJ V1.48, National Institute of Health, Bethesda, MD). 61 sections were quantified for CD3 immunostaining (n = 40 for pretreatment sections and n = 21 for posttreatment sections) and 60 sections were quantified for CD11c immunostaining (n = 40 for pretreatment sections and n = 20 for posttreatment sections).
Statistical analyses of scRNA-seq data at the level of total T-cells, DCs, or KCs were performed under the general framework of the Seurat R package (17–19). We used a Wilcoxon rank sum test to define differentially expressed genes. A value of p < 0.05 was considered statistically significant.
Statistical analyses of scRNA-seq data at the level of T-cell and KC subsets were performed under the trajectory inference framework of Monocle3 R package (24, 25). We used a spatial autocorrelation analysis called Moran’s I (25) with the graph-test function, which Cao et al. (24) showed to be effective in finding genes that are differentially expressed in single-cell RNA sequencing data. A p-value that has been adjusted for the False Discovery rate < 0.05 was considered statistically significant (Supplementary Tables 3, 4). Spearman’s rank correlation coefficient was calculated to test inter-correlations between the single-cell data.
For statistical analyses of microarray data, log2-transformed expression values were modeled using linear mixed-effects models. P-values from moderated t-tests were adjusted for multiple hypotheses across genes using the Benjamini-Hochberg procedure. A value of p < 0.05 and a fold change ratio > 2 was considered statistically significant.
For statistical analyses of RT-PCR data, RT-PCR results were modeled with the delta Ct (ΔΔCt) method for relative quantification to a housekeeping gene RPLP0 (Ribosomal Protein Lateral Stalk Subunit P0) and calibration with a reference sample Control01 under the general framework of pcr_analyze R package (26) as we described in our prior publication (10). Two-sided Wilcoxon rank-sum test was used to compare RT-PCR experiment results between group 1 and group 2. A value of p < 0.05 was considered statistically significant.
For statistical analyses of immunohistochemistry data, we used a Wilcoxon rank sum test to compare measures between groups. A value of p < 0.05 was considered statistically significant.
The study protocol and informed consent were approved by the Institutional Review Board of the Rockefeller University, New York, NY, USA. The study was conducted in accordance with Good Clinical Practice and the Declaration of Helsinki, and all subjects provided written informed consent before entering the study.
We first studied how the immune cell numbers are changed in psoriasis skin after systemic IL-17A blockade, and how the immune cell number changes are reflected in total skin transcriptome. KC hyperproliferation [KRT16 staining (27)], T-cell numbers (CD3 staining), and DC numbers (ITGAX (CD11c) staining) were decreased in psoriasis lesional skin after 12 weeks of anti-IL-17A monoclonal antibody injections (posttreatment) compared to pretreatment psoriasis lesional skin of the same patients (Figure 2A, p < 0.05).
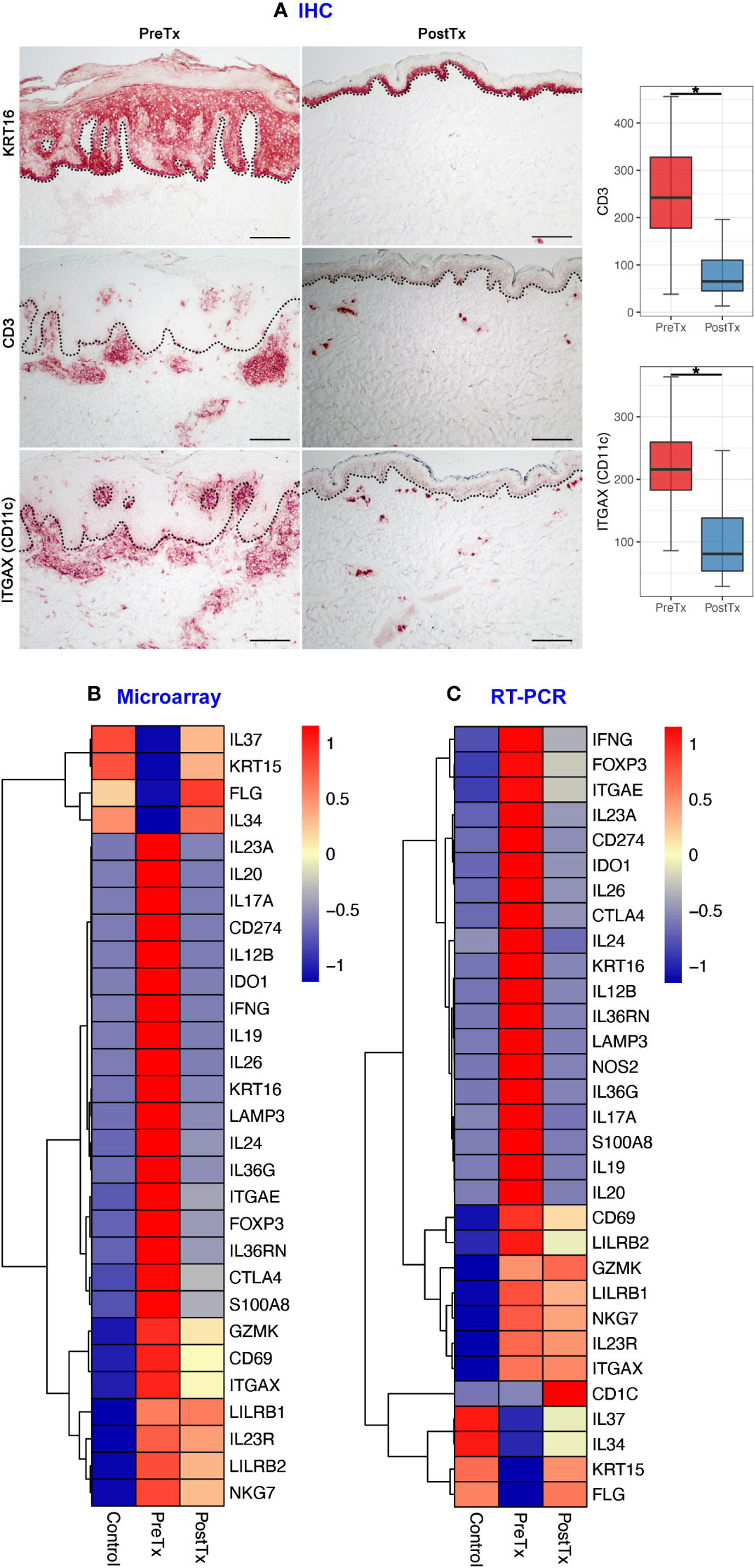
Figure 2 IL-17A blockade reduces T-cell signatures, dendritic cell signatures, and keratinocyte hyperproliferation signatures and increases regulatory molecular expression in psoriasis lesional skin at total skin transcriptome levels. (A) Immunohistochemistry of keratinocyte hyperproliferation (KRT16), CD3+ T-cells, and ITGAX (CD11c)+ dendritic cells (DCs) before (PreTx) and after (PostTx) 12 weeks of systemic anti-IL-17A monoclonal antibody (secukinumab) administration. Bar graphs display CD3+ T-cell (n = 40 for PreTx and n = 21 for PostTx) and ITGAX (CD11c)+ DC (n = 40 for PreTx and n = 20 for PostTx) number changes induced by systemic anti-IL-17A blockade. *p < 0.05. Black dotted lines delineate the junction between the epidermis and dermis (Scale bar = 200 μm). (B, C), Differentially expressed genes (DEGs) between PreTx and PostTx at total skin transcriptome levels. DEGs that are relevant to psoriasis pathogenesis and statistically significant in both microarray (B, fold change ratio > 2 and p < 0.05) and RT-PCR (C, p < 0.05) experiments are displayed.
Reflecting the decreased T-cell numbers by systemic IL-17A blockade, total skin transcriptome of T17 responses (IL17A, IL26, IL12B, and IL20) (3, 28), T-cell regulation (IL24 and CTLA4) (29, 30), resident memory T-cells (Trms) [ITGAE (CD103)] (31) and regulatory T-cells (Tregs) (FOXP3) (32) was decreased in posttreatment psoriasis lesional skin compared to pretreatment psoriasis lesional skin in both microarray and RT-PCR experiments (Figures 2B, C, p < 0.05).
Reflecting the decreased DC numbers by systemic IL-17A blockade, total skin transcriptome of T17 cell-stimulating DCs (IL23A) (3) and mature DCs (LAMP3 (DC-LAMP), CD274 (PD-L1) and IDO1) (16, 33, 34) were decreased in posttreatment psoriasis lesional skin compared to pretreatment psoriasis lesional skin in both microarray and RT-PCR experiments (Figures 2B, C, p < 0.05).
Reflecting the decreased KC hyperproliferation by systemic IL-17A blockade, total skin transcriptome of KC cytokines that stimulate DCs and T-cells in feed-forward mechanisms (S100A8 and IL36G) (3) was decreased in posttreatment psoriasis lesional skin compared to pretreatment psoriasis lesional skin in both microarray and RT-PCR experiments (Figures 2B, C, p < 0.05).
In contrast, total skin transcriptome of Treg-specific cytokine that mediates immune tolerance [IL34 (35–37)], an anti-inflammatory cytokine that inhibits innate immune signaling [IL37 (38, 39)], and KC stem cell marker of quiescence [KRT15 (40)] were increased in posttreatment psoriasis lesional skin compared to pretreatment psoriasis lesional skin in both microarray and RT-PCR experiments, albeit the decreased T-cell, DC, and KC numbers (Figures 2B, C, p < 0.05).
We next studied how systemic IL-17A blockade modified transcriptome of T17 cells and transcriptome of Tregs differently with single-cell analyses. Clustering analysis of 40,026 single cells from 28 samples of 20 subjects identified clusters of NK cells, CD161+ T-cells, CD8+ T-cells, CD4+ T-cells, Tregs, mature DCs, semimature DCs, melanocytes, KCs in different layers of Stratum (S.) corneum, S. granulosum, S. spinosum, S. basale, endothelial cells, and fibroblasts without subclustering (Figure 3A and Supplementary Figure 2). The identity of each cluster was determined by the expression of established cell type markers (Figure 3B). To characterize T17 cell transcriptome in psoriasis skin, we segregated T-cell (CD161+ T-cell, CD8+ T-cell, CD4+ T-cell, and Treg) clusters in the scRNA-seq data and tested if psoriasis T-cells express more T17 cell genes than control T-cells. The expression of T17 cell genes [KLRB1 (CD161) (41), IL17A, IL17F, and IL26 (42, 43)], a cytokine that represents a particularly T17-specific abnormality and positively associates with psoriasis severity (CXCL13) (44–47), and Treg genes (FOXP3, IL2RA (CD25), and TIGIT) was increased in pretreatment psoriasis lesional skin T-cells compared to control skin T-cells (Figure 4A, p < 0.05).
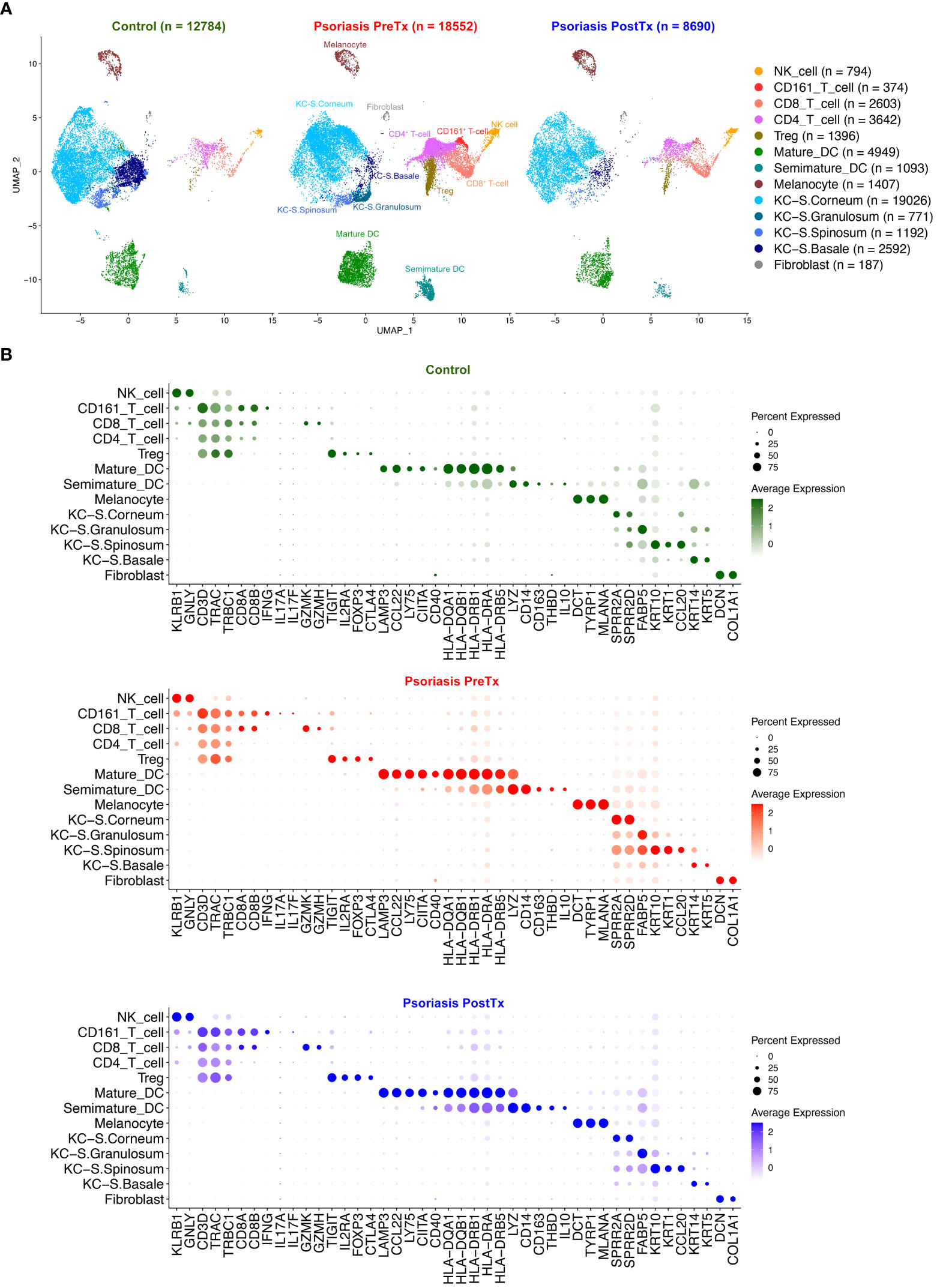
Figure 3 Human skin immune cell subset single-cell transcriptome comparison between psoriasis pretreatment (PreTx), psoriasis posttreatment (PostTx), and control (healthy volunteers). Psoriasis posttreatment is after anti-IL-17A monoclonal antibody (secukinumab) subcutaneous injections at a dose of 300 mg at baseline and weeks 1, 2, 3, 4, 8, and 12 (12 weeks of injections). (A) The Uniform Manifold Approximation and Projection plot (UMAP). (B) Dot plots displaying expression levels of cluster-defining genes.
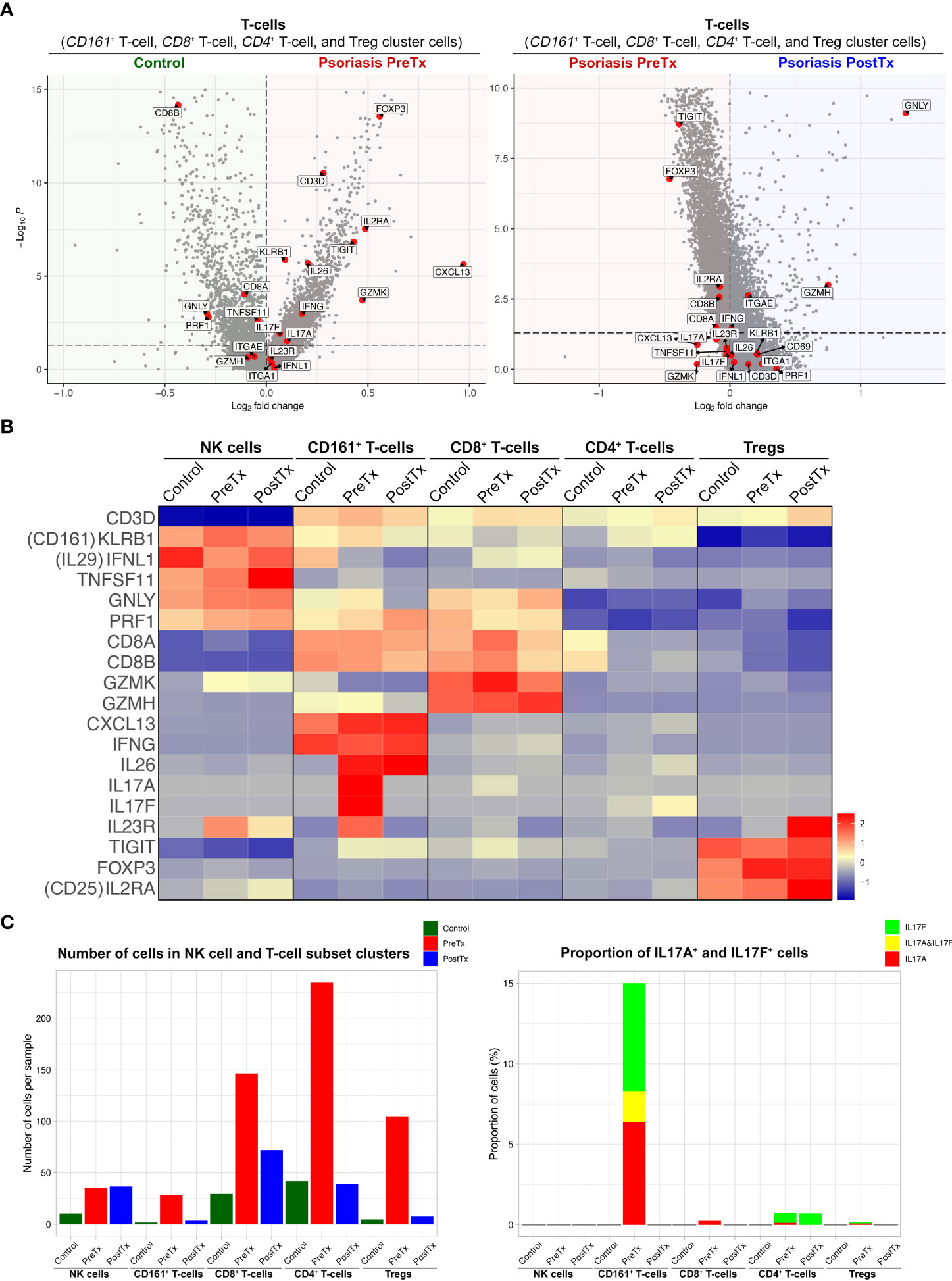
Figure 4 IL-17A blockade reduces Type 17 T-cell signatures and modifies regulatory T-cell signatures in psoriasis lesional skin at single-cell cluster levels. (A) Volcano plots displaying differentially expressed genes between control skin T-cells, psoriasis lesional skin T-cells before treatment (PreTx), and psoriasis lesional skin T-cells after 12 weeks of systemic anti-IL-17A monoclonal antibody (secukinumab) administration (PostTx). (B) Heatmap illustrating the average gene expression in clusters of NK cell and T-cell subsets, split by psoriasis PreTx, psoriasis PostTx, and control skin. (C) (Left) Number of cells per sample and (Right) proportion of IL17A+ and IL17F+ cells in NK cell and T-cell subset clusters, split by control, psoriasis PreTx, and psoriasis PostTx.
When we defined T17 cells as T-cell cluster cells expressing IL17A and/or IL17F, the majority of T17 cells were within the CD161+ T-cell cluster. 5.5% of total T-cells were CD161+ T-cell cluster cells and 15.0% of CD161+ T-cell cluster cells were T17 cells (Figure 4C). 76.5% of IL17A+ T-cells, 61.4% of IL17F+ T-cells, and 100% of IL17A+ IL17F+ T-cells were within the CD161+ T-cell cluster in pretreatment psoriasis lesional skin. In addition, 84.7% of CD161+ T-cell cluster cells expressed CXCL13, while only 1.6% of CD8+ T-cell cluster cells, 2.5% of CD4+ T-cell cluster cells, and 0.6% of Treg cluster cells expressed CXCL13 in pretreatment psoriasis lesional skin (Figure 4B).
After systemic IL-17A blockade (12 weeks of anti-IL-17A monoclonal antibody injections), the expression of IL17A in the CD161+ T-cell cluster was decreased to zero in psoriasis lesional skin (Figures 4B, C, and Supplementary Table 3; p < 0.05). Although anti-IL-17A monoclonal antibody does not directly target IL-17F, the expression of IL17F was also decreased in the CD161+ T-cell cluster in psoriasis lesional skin after IL-17A blockade (Figure 4B and Supplementary Table 3, p < 0.05). When all T-cell cluster cells were considered, IL-17A blockade decreased 95% of IL17F+ T-cells (Figure 4C).
We next studied how systemic IL-17A blockade modified transcriptome of different DC subsets that we previously described in our prior single-cell studies of human psoriasis skin (16). When we segregated DC subset (mature DC and semimature DC) clusters in the scRNA-seq data and compared their transcriptome, mature DCs in psoriasis skin were characterized by relatively high expression of MHC class II molecules (HLA-DRA and HLA-DPB1) and upregulated maturation markers [CD86 (48) and DC-LAMP (LAMP3) (49)] (16) (Figure 5A). In contrast, semimature DCs in psoriasis skin were characterized by relatively low expression of MHC class II molecules (HLA-DRA and HLA-DPB1) and upregulated inflammatory monocyte receptor markers [CD14 and CD64 (FCGR1A) (50)] (Figure 5B, p < 0.05). Semimature DCs were characterized as regulatory DCs expressing high levels of IL10, THBD (BDCA-3), LILRB2, and CLEC4A (DCIR) (51, 52) (Figure 5B, p < 0.05). We previously reported that both mature and semimature DCs in psoriasis lesional skin expressed high levels of IL23A compared to control skin (16) (Figure 5A). Our current study showed that the expression of IL23A in both mature and semimature DCs was decreased after systemic IL-17A blockade in posttreatment psoriasis lesional skin compared to pretreatment psoriasis lesional skin (Figure 5A, p < 0.05).
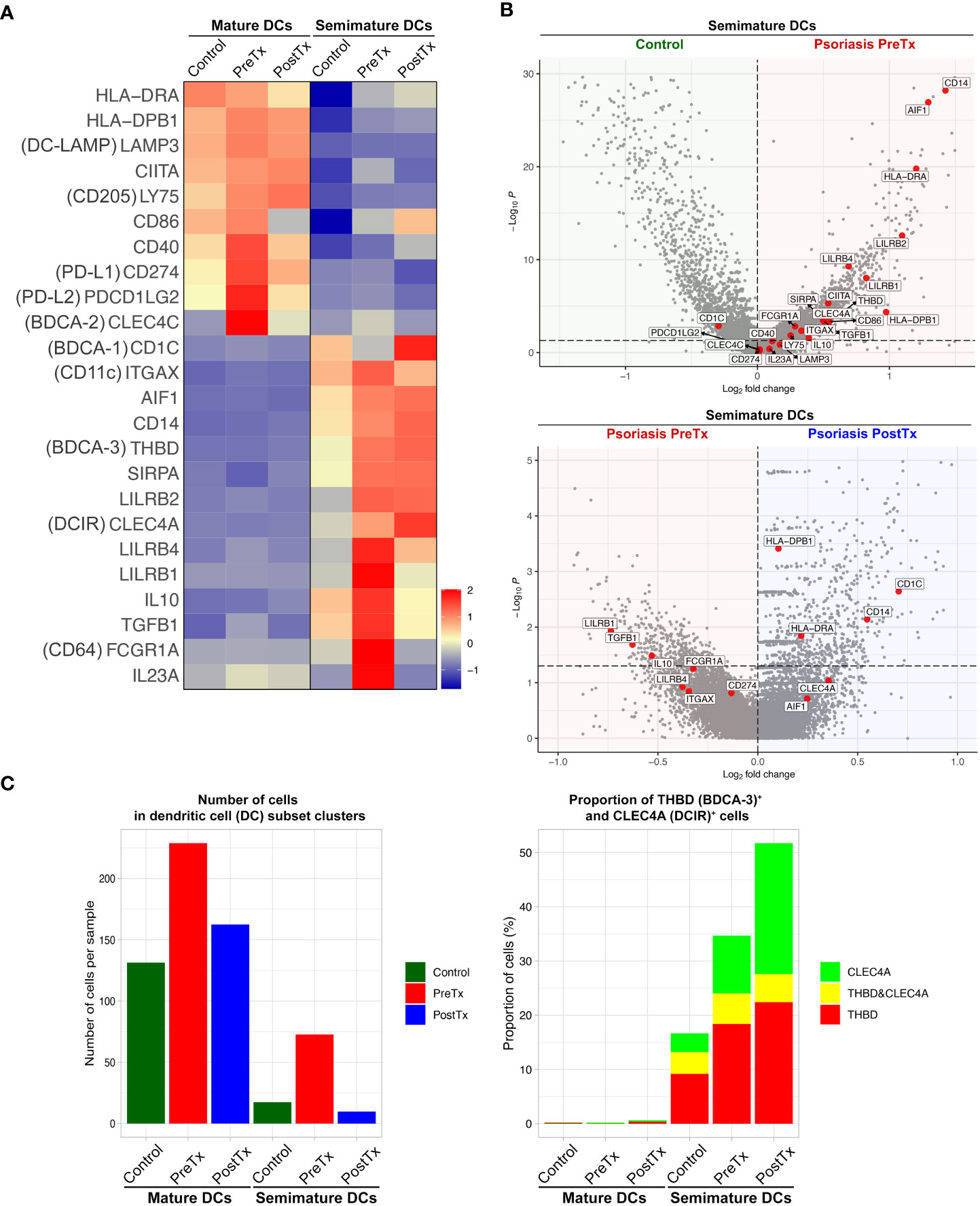
Figure 5 IL-17A blockade increases regulatory dendritic cell signatures in psoriasis lesional skin at single-cell cluster levels. (A) Heatmap illustrating the average gene expression in clusters of mature DCs and semimature DCs, split by control skin DCs, psoriasis lesional skin DCs before treatment (PreTx), and psoriasis lesional skin DCs after 12 weeks of systemic anti-IL-17A monoclonal antibody (secukinumab) administration (PostTx). (B) Volcano plots displaying differentially expressed genes between control skin DCs, psoriasis PreTx DCs, and psoriasis PostTx DCs. (C) (Left) Number of cells per sample and (Right) proportion of THBD (BDCA-3)+ and CLEC4A (DCIR)+ cells in DC subset clusters, split by control, psoriasis PreTx, and psoriasis PostTx.
Reflecting the decreased DC numbers after systemic IL-17A blockade in the immunohistochemistry experiment (Figure 2A), both mature and semimature DC numbers were decreased in posttreatment psoriasis lesional skin compared to pretreatment psoriasis lesional skin in scRNA-seq data (Figure 5C). Although semimature DC cell numbers were decreased after systemic IL-17A blockade, regulatory transcriptome expression of the semimature DCs was increased after systemic IL-17A blockade – 1) The proportion of semimature DCs expressing regulatory DC markers of BDCA-3 (THBD) (51) and DCIR (CLEC4A) (52) was increased in posttreatment psoriasis lesional skin compared to pretreatment psoriasis lesional skin (Figure 5C). 2) The expression of CD1C and CD14, which are genes of a CD1c+ CD14+ DC subset that suppresses antigen-specific T-cell responses (53, 54), was increased in posttreatment psoriasis lesional skin semimature DCs compared to pretreatment psoriasis lesional skin semimature DCs (Figure 5B, p < 0.05).
We next studied how systemic IL-17A blockade modified transcriptome of KCs in different layers of epidermis. We segregated KC clusters representing different layers of the epidermis (S. corneum, S. granulosum, S. spinosum, and S. basale) and tested if psoriasis KCs express more IL-17-driven inflammatory mediators than control KCs. The expression of inflammatory mediators induced by IL-17 in keratinocytes (IL36G, S100A8, DEFB4A, and DEFB4B (3)) was increased in pretreatment psoriasis lesional skin KCs compared to control skin KCs (Figure 6A, p < 0.05). After systemic IL-17A blockade, the expression of those inflammatory mediators (IL36G, S100A8, DEFB4A, and DEFB4B) was decreased in posttreatment psoriasis lesional skin KCs compared to pretreatment psoriasis lesional skin KCs (Figure 6A, p < 0.05).
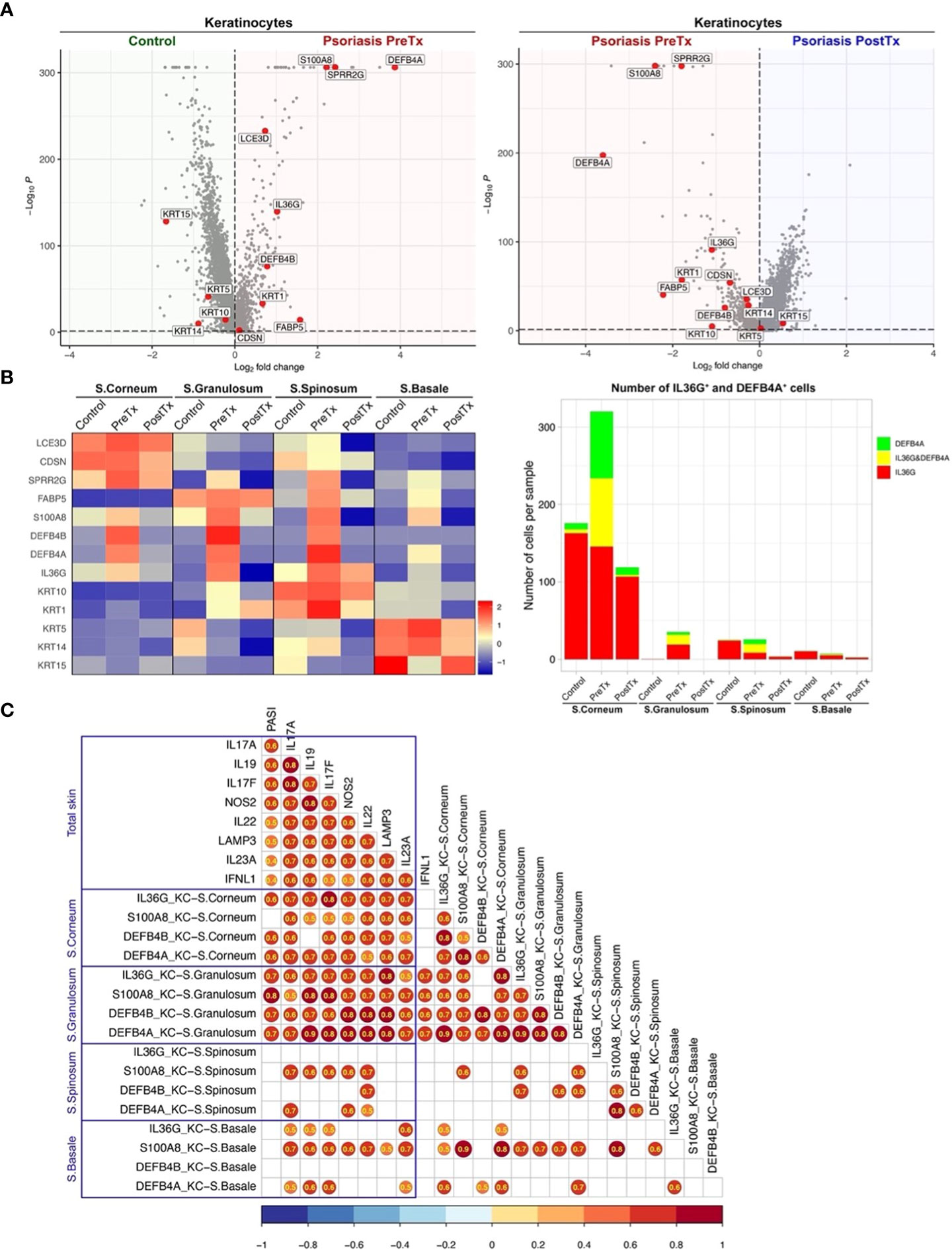
Figure 6 IL-17A blockade reduces IL-17-driven inflammatory mediator expression in suprabasal keratinocytes and increases keratinocyte stem cell marker expression in basal keratinocytes in psoriasis lesional skin at single-cell cluster levels. (A) Volcano plots displaying differentially expressed genes between control skin KCs, psoriasis lesional skin KCs before treatment (PreTx), and psoriasis lesional skin KCs after 12 weeks of systemic anti-IL-17A monoclonal antibody (secukinumab) administration (PostTx). (B) Heatmap illustrating the average gene expression in KC clusters representing different layers of epidermis (Stratum (S.) corneum, S. granulosum, S. spinosum, and S. basale), split by psoriasis PreTx, psoriasis PostTx, and control skin. Bar graphs displaying IL36G+ and DEFB4A+ cell numbers in KC clusters, split by control, psoriasis PreTx, and psoriasis PostTx. (C) Correlation plots displaying inter-correlations between Psoriasis Area-and-Severity Index (PASI), IL-23/T17 axis gene expression in total skin, and IL-17-driven KC inflammatory mediator expression in different epidermal layers. Circled numbers = correlation coefficients with p < 0.05.
When we compared KC transcriptome in different epidermal layers, the expression of IL-17-driven inflammatory mediators (IL36G, S100A8, DEFB4A, and DEFB4B) was localized to suprabasal layers (S. corneum, S. granulosum, and S. spinosum) in pretreatment psoriasis lesional skin (Figure 6B). A high proportion of IL36G expressing KCs co-expressed DEFB4A in suprabasal layers (37.7% in S. corneum, 39.9% in S. granulosum and 56.7% in S. spinosum). Reflecting that inflammation in the upper layers of the epidermis determines psoriasis disease severity (erythema, thickness, and scaling components of Psoriasis Area-and-Severity Index (PASI)), KC expression of IL36G, DEFB4A, and DEFB4B in S. corneum and S. granulosum was correlated with PASI (Figure 6C, p < 0.05). After IL-17A blockade, KC expression of IL36G, S100A8, DEFB4A, and DEFB4B in S. corneum and S. granulosum decreased in posttreatment psoriasis lesional skin compared to pretreatment psoriasis lesional skin (Figure 6B and Supplementary Table 4, p < 0.05). The number of IL36G and/or DEFB4A expressing KCs in S. corneum and S. granulosum was decreased by 3 times from pretreatment psoriasis lesional skin to posttreatment psoriasis lesional skin (Figure 6B).
In contrast to the inflammatory mediators induced by IL-17, the expression of the KC stem cell marker (KRT15) was decreased in pretreatment psoriasis lesional skin KCs compared to control skin KCs (Figure 6A, p < 0.05). After systemic IL-17A blockade, the expression of KRT15 was increased in posttreatment psoriasis lesional skin KCs compared to pretreatment psoriasis lesional skin KCs (Figure 6A, p < 0.05). The expression of KRT15 in pretreatment psoriasis lesional skin was localized to the basal layer in pretreatment psoriasis lesional skin (Figure 6B). After IL-17A blockade, KC expression of KRT15 in S. basale increased in posttreatment psoriasis lesional skin compared to pretreatment psoriasis lesional skin (Figure 6B and Supplementary Table 4, p < 0.05).
In this study, we applied multi-omics to human psoriasis skin providing a unique opportunity to compare the transcriptome of total skin (microarray and RT-PCR) and the transcriptome of skin immune cell subsets (scRNA-seq) generated from the same tissues with different techniques. When skin transcriptome changes by systemic IL-17A blockade were compared between total skin and skin immune cell subsets, IL-23/T17 axis cytokine expression was downregulated at both total skin levels and skin immune cell subset levels. The quantity of overall immune cells, including both pathogenic and regulatory subsets, was decreased in posttreatment psoriasis lesional skin compared to pretreatment psoriasis lesional skin (Figure 2A). Since the IL-23/T17 axis cytokine sources (T-cells, DCs, and KCs) were decreased in the predetermined volume of skin biopsy tissue (6 mm punch), total skin transcriptome of IL-23/T17 axis cytokines (IL17A and IL26 from T-cells, IL23A, IL12B, and IL20 from DCs, and S100A8 and IL36 from KCs) was decreased in posttreatment psoriasis lesional skin compared to pretreatment psoriasis lesional skin (Figure 2B).
Paired skin immune cell subset transcriptome analyses with scRNA-seq further elucidated which immune cell subsets were expressing the IL-23/T17 axis cytokines and how their transcriptome was affected by systemic IL-17A blockade. For T-cell subsets, our study showed that 1) CD161+ T-cell cluster was the major T17 cell cluster, but only 15.0% of CD161+ T-cell cluster cells were T17 cells. Single-cell clustering of immune cell subsets presented in this study was consistent with our previous single-cell paper (16), but T-cell clustering evolved as we increased the total number of single-cells for dimension reduction analyses from 23,220 to 40,026 (Figure 3). Our new dimension reduction and clustering analyses revealed that the majority of T17 cells in psoriasis skin were contained in the CD161+ T-cell cluster consistent with previous CD161+ T-cell studies (55) (Figure 4C). CD161 is a marker of all human T-cell subsets with the ability to produce IL-17, and it has been reported that IL-17-producing cells exclusively originate from naïve CD161+ T-cell precursors (56). In psoriasis, it has been reported that more CD161+ T-cells are present in psoriasis lesional skin compared to psoriasis nonlesional skin or normal skin (57, 58), and the greater frequency of CD161+ T-cells are present in prepsoriatic skin compared to normal skin, suggesting the role of CD161+ T-cells in the initial development of psoriatic lesions (57, 59). 2) CD161+ T-cell cluster was the major T-cell subset cluster expressing CXCL13, which is a cytokine that represents a particularly T17-specific abnormality and positively associates with psoriasis severity (44–47) (Figure 4B). In our single-cell dataset, 70.2% of CXCL13-expressing cells was T-cell cluster cells, and 73.2% of CXCL13-expressing T-cell cluster cells were CD161+ T-cell cluster cells.
For single-cell transcriptome modification of the IL-23/T17 cell axis induced by systemic IL-17A blockade, our study showed that 1) The expression of IL23A in dendritic cell subsets in psoriasis skin lesions was downregulated by systemic IL-17A blockade, potentially by blocking the entire feed-forward inflammatory amplification loop between T-cells, DCs, and KCs (Figure 5A), 2) Systemic IL-17A blockade removed 100% of IL17A-expressing T-cells. Although anti-IL-17A monoclonal antibody does not directly target IL-17F, systemic IL-17A blockade also removed 95% of IL17F-expressing T-cells in psoriasis skin, potentially mediated by IL-23 reduction in dendritic cells (Figure 4C). 2) The expression of IL-17-driven inflammatory mediators (IL36G, S100A8, DEFB4A, and DEFB4B) in KCs was localized to suprabasal KCs (Figure 6B). The expression of those IL-17-driven inflammatory mediators in suprabasal KCs was correlated with psoriasis severity (Figure 6C) and was downregulated by systemic IL-17A blockade (Figure 6B).
Mapping regulatory DC transcriptome is challenging because a master regulator of regulatory DCs is not identified. All regulatory DC markers of BDCA-3 (THBD) (51), DCIR (CLEC4A) (52), LILRB1 (60), LILRB2 (ILT4) (61), LILRB4 (ILT3) (61) and combination of CD1C and CD14 (53, 54) are surface markers that are not specific for DCs. Since the markers are expressed in other skin cells, total skin transcriptome changes of those markers did not represent transcriptome changes of regulatory DCs (Figure 2B). Instead, skin immune cell subset transcriptome analyses of the semimature DC cluster, which we previously reported as regulatory DCs in psoriasis skin (16), showed regulatory transcriptome promotion by systemic IL-17A blockade in regulatory DCs – 1) The proportion of semimature DCs expressing regulatory DC markers of BDCA-3 (THBD) (51) and DCIR (CLEC4A) (52) was increased in posttreatment psoriasis lesional skin compared to pretreatment psoriasis lesional skin (Figure 5C). 2) The expression of CD1C and CD14 [markers of CD1c+ CD14+ DC subset that suppresses antigen-specific T-cell responses (53, 54)] was increased in posttreatment psoriasis lesional skin semimature DCs compared to pretreatment psoriasis lesional skin semimature DCs (Figure 5B, p < 0.05).
Clinically, when moderate-to-severe psoriasis patients were treated with systemic anti-IL-17A monoclonal antibody (secukinumab) administration for 52 weeks and then stopped the treatment, 16.0% of them maintained response after treatment withdrawal over the next 52 weeks without need for retreatment (12, 13). Our study may explain how those patients restored immune tolerance to psoriasis autoantigens that may have prevented recurrence of psoriasis off the treatment. In addition to the co-modulation of IL-17A and IL-17F by direct and indirect effects on T-cells including IL-23 reduction in dendritic cells, systemic IL-17A blockade may have effectively modified regulatory transcriptome of regulatory DCs (Figures 5B, 6C) in those patients.
Our study has limitations: 1) Our single-cell analysis approach relied on immune cell emigration from the skin biopsies in culture. Whilst this is a recognized approach to isolate immune cells from the skin (16), some skin resident immune cells may not be migratory and may have been excluded in the immune cell subset analyses. 2) The unique challenges of scRNA-seq data analysis, including the sparsity of the scRNA-seq dataset and the dropout of lowly expressed genes, are widely acknowledged (62–67). In particular, differentially expressed gene analyses of scRNA-seq data are biased towards highly expressed genes (68), while the target genes investigated in the study (T17 axis cytokines) are lowly expressed genes in specific T-cell or DC subsets (16). To maximize the recovery of differentially expressed genes, we used a Wilcoxon rank sum test at total immune cell (T-cell, DC, or KC) levels and a spatial autocorrelation analysis (24, 25) at immune cell subset levels. However, we observed the discrepancies of IL23R status between the average expression visualized in the heatmap (Figure 4B) and the spatial autocorrelation test results (Supplementary Table 3) in Treg cluster analyses. The average expression of IL23R was higher in posttreatment Tregs compared to pretreatment Tregs (Figure 4B) but the difference was not statistically significant in the spatial autocorrelation test. 3) The transcriptome analyses were not validated by functional studies at protein levels. 4) Different body sites can harbor different microbes and may have different immune cell compositions, but the location of skin biopsies was not matched between comparison groups (Skin biopsy location of each sample is listed in Supplementary Table 1).
Our study provides an early demonstration that systemic anti-IL-17A monoclonal antibody administration blocks the entire IL-23/T17 cell axis depleting or modulating 100% of IL17A+ T17 cells and 95% of IL17F+ T17 cells at single-cell levels (Supplementary Figure 3). In addition, systemic anti-IL-17A monoclonal antibody administration promotes regulatory DCs, while strongly downregulating the expression of IL23A in dendritic cells in psoriasis skin lesions. Thus, we hypothesize that a monoclonal blockade of pathogenic T-cells, such as an IL-17A blockade or an IL-23p19 blockade, may induce expansion of regulatory immune cells subsets or expression of cytokines involved in skin homeostasis (9, 10, 69). To further test the hypothesis, we are currently conducting a clinical trial of short-term anti-IL-23p19 monoclonal antibody administration to induce long-term disease remission that incorporates scRNA-seq analyses of human skin before/after the monoclonal antibody administration (NCT04630652). We are hoping to better understand the regulatory immune cell promotion by blocking the IL-23/T17 cell axis and develop personalized medicine approaches to cure psoriasis without recurrence.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: GSE183047 and GSE226244 (GEO).
The study protocol and informed consent were approved by the Institutional Review Board of the Rockefeller University, New York, NY, USA. The study was conducted in accordance with Good Clinical Practice and the Declaration of Helsinki, and all subjects provided written informed consent before entering the study. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
All the authors have made substantial contributions to the acquisition, analysis, or interpretation of the data. In addition, all the authors have contributed to the drafting or editing of the manuscript and gave their approval for the version submitted. JK and JGK designed the clinical trial, experiments, and laboratory protocols. JK, NK, IC, KK, and VC contributed to patient consent and care. JL and XL conducted single-cell and microarray studies. DR conducted immunohistochemistry studies. JK, YK, SG, WZ, JC, and JGK interpreted the data. JK and JGK verified all the underlying data. All authors contributed to the article and approved the submitted version.
This work was supported, in part, by grant no. UL1TR001866 from the National Center for Advancing Translational Sciences (NCATS), National Institutes of Health (NIH) Clinical and Translational Science Award (CTSA) program. JK was supported by NIAMS K23 Career Development Award (K23AR080043). JK was also supported by the Group for Research and Assessment of Psoriasis and Psoriatic Arthritis (GRAPPA) pilot grant award.
JK has received research funds from AbbVie and Novartis. JGK. has received research support from Pfizer, Amgen, Janssen, Lilly, Merck, Novartis, Kadmon, Dermira, Boehringer, Innovaderm, Kyowa, BMS, Serono, BiogenIdec, Delenex, AbbVie, Sanofi, Baxter, Paraxel, Xenoport, and Kineta.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1250504/full#supplementary-material
Supplementary Figure 1 | The Uniform Manifold Approximation and Projection plot (UMAP) of the initial non-linear dimension reduction and clustering analysis (A) and the final clusters for the downstream differential expression testing (B).
Supplementary Figure 2 | Alluvial diagram visualizing how individual immune cells in single-cell RNA sequencing data are allocated across categorical dimensions. Left column: Patients -10 psoriasis patients and 10 control healthy volunteers. Middle column: Treatment - psoriasis pretreatment (Psoriasis_preTx), psoriasis posttreatment (Psoriasis_postTx) after weeks 12, 24, and 48 of systemic anti-IL-17A administration, and control, Right column: immune cell clusters – NK cell, CD161 T-cell, CD8 T-cell, CD4 T-cell, regulatory T-cell (Treg), mature dendritic cell (DC), semimature DC, melanocyte, Keratinocyte in S (Stratum) corneum, S. granulosum, S. spinosum, S. basale, and fibroblast clusters.
Supplementary Figure 3 | A summary of different transcriptomic impacts of anti-IL-17A blockade on human psoriasis skin at the levels of total skin and clusters of T-cell, dendritic cell, and keratinocyte subsets. DC = dendritic cell, KC = keratinocyte.
1. Rachakonda TD, Schupp CW, Armstrong AW. Psoriasis prevalence among adults in the United States. J Am Acad Dermatol (2014) 70:512–6. doi: 10.1016/j.jaad.2013.11.013
2. Armstrong AW, Mehta MD, Schupp CW, Gondo GC, Bell SJ, Griffiths CEM. Psoriasis prevalence in adults in the United States. JAMA Dermatol (2021) 157:940–6. doi: 10.1001/jamadermatol.2021.2007
3. Kim J, Krueger JG. Highly effective new treatments for psoriasis target the IL-23/type 17 T cell autoimmune axis. Annu Rev Med (2017) 68:255–69. doi: 10.1146/annurev-med-042915-103905
4. Sterry W, Strober BE, Menter A, International Psoriasis Council. Obesity in psoriasis: the metabolic, clinical and therapeutic implications. Report of an interdisciplinary conference and review. Br J Dermatol (2007) 157:649–55. doi: 10.1111/j.1365-2133.2007.08068.x
5. Gisondi P, Tessari G, Conti A, Piaserico S, Schianchi S, Peserico A, et al. Prevalence of metabolic syndrome in patients with psoriasis: a hospital-based case-control study. Br J Dermatol (2007) 157:68–73. doi: 10.1111/j.1365-2133.2007.07986.x
6. Cohen AD, Dreiher J, Shapiro Y, Vidavsky L, Vardy DA, Davidovici B, et al. Psoriasis and diabetes: a population-based cross-sectional study. J Eur Acad Dermatol Venereol (2008) 22:585–9. doi: 10.1111/j.1468-3083.2008.02636.x
7. Armstrong EJ, Harskamp CT, Armstrong AW. Psoriasis and major adverse cardiovascular events: a systematic review and meta-analysis of observational studies. J Am Heart Assoc (2013) 2:e000062. doi: 10.1161/JAHA.113.000062
8. Gelfand JM, Troxel AB, Lewis JD, Kurd SK, Shin DB, Wang X, et al. The risk of mortality in patients with psoriasis: results from a population-based study. Arch Dermatol (2007) 143:1493–9. doi: 10.1001/archderm.143.12.1493
9. Krueger JG, Ferris LK, Menter A, Wagner F, White A, Visvanathan S, et al. Anti-IL-23A mAb BI 655066 for treatment of moderate-to-severe psoriasis: Safety, efficacy, pharmacokinetics, and biomarker results of a single-rising-dose, randomized, double-blind, placebo-controlled trial. J Allergy Clin Immunol (2015) 136:116–124.e7. doi: 10.1016/j.jaci.2015.01.018
10. Kim J, Lee J, Hawkes JE, Li X, Kunjravia N, Rambhia D, et al. Secukinumab improves mild-to-moderate psoriasis: A randomized, placebo-controlled exploratory clinical trial. J Am Acad Dermatol (2023) 88:428–30. doi: 10.1016/j.jaad.2022.04.060
11. Harden JL, Johnson-Huang LM, Chamian MF, Lee E, Pearce T, Leonardi CL, et al. Humanized anti-IFN-γ (HuZAF) in the treatment of psoriasis. J Allergy Clin Immunol (2015) 135:553–6. doi: 10.1016/j.jaci.2014.05.046
12. Langley RG, Sofen H, Dei-Cas I, Reich K, Sigurgeirsson B, Warren RB, et al. Secukinumab long-term efficacy and safety in psoriasis through to year 5 of treatment: results of a randomized extension of the phase III ERASURE and FIXTURE trials. Br J Dermatol (2023) 188:198–207. doi: 10.1093/bjd/ljac040
13. Blauvelt A, Reich K, Warren RB, Szepietowski JC, Sigurgeirsson B, Tyring SK, et al. Secukinumab re-initiation achieves regain of high response levels in patients who interrupt treatment for moderate to severe plaque psoriasis. Br J Dermatol (2017) 177:879–81. doi: 10.1111/bjd.15656
14. Papp KA, Blauvelt A, Bukhalo M, Gooderham M, Krueger JG, Lacour J-P, et al. Risankizumab versus ustekinumab for moderate-to-severe plaque psoriasis. N Engl J Med (2017) 376:1551–60. doi: 10.1056/NEJMoa1607017
15. Gordon KB, Armstrong AW, Foley P, Song M, Shen Y-K, Li S, et al. Guselkumab efficacy after withdrawal is associated with suppression of serum IL-23-regulated IL-17 and IL-22 in psoriasis: VOYAGE 2 study. J Invest Dermatol (2019) 139:2437–2446.e1. doi: 10.1016/j.jid.2019.05.016
16. Kim J, Lee J, Kim HJ, Kameyama N, Nazarian R, Der E, et al. Single-cell transcriptomics applied to emigrating cells from psoriasis elucidate pathogenic versus regulatory immune cell subsets. J Allergy Clin Immunol (2021) 148:1281–92. doi: 10.1016/j.jaci.2021.04.021
17. Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell (2021) 184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048
18. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive integration of single-cell data. Cell (2019) 177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031
19. Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol (2015) 33:495–502. doi: 10.1038/nbt.3192
20. Der E, Ranabothu S, Suryawanshi H, Akat KM, Clancy R, Morozov P, et al. Single cell RNA sequencing to dissect the molecular heterogeneity in lupus nephritis. JCI Insight (2017) 2:e93009. doi: 10.1172/jci.insight.93009
21. Der E, Suryawanshi H, Morozov P, Kustagi M, Goilav B, Ranabothu S, et al. Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat Immunol (2019) 20:915–27. doi: 10.1038/s41590-019-0386-1
22. He H, Suryawanshi H, Morozov P, Gay-Mimbrera J, Del Duca E, Kim HJ, et al. Single-cell transcriptome analysis of human skin identifies novel fibroblast subpopulation and enrichment of immune subsets in atopic dermatitis. J Allergy Clin Immunol (2020) 145:1615–28. doi: 10.1016/j.jaci.2020.01.042
23. Wu Z, Irizarry RA. Preprocessing of oligonucleotide array data. Nat Biotechnol (2004) 22:656–8. doi: 10.1038/nbt0604-656b
24. Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, et al. The single-cell transcriptional landscape of mamMalian organogenesis. Nature (2019) 566:496–502. doi: 10.1038/s41586-019-0969-x
25. Moran PAP. Notes on continuous stochastic phenomena. Biometrika (1950) 37:17–23. doi: 10.1093/biomet/37.1-2.17
26. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (2001) 25:402–8. doi: 10.1006/meth.2001.1262
27. Leigh IM, Navsaria H, Purkis PE, McKay IA, Bowden PE, Riddle PN. Keratins (K16 and K17) as markers of keratinocyte hyperproliferation in psoriasis in vivo and in vitro. Br J Dermatol (1995) 133:501–11. doi: 10.1111/j.1365-2133.1995.tb02696.x
28. Wolk K, Witte K, Witte E, Raftery M, Kokolakis G, Philipp S, et al. IL-29 is produced by TH17 cells and mediates the cutaneous antiviral competence in psoriasis. Sci Transl Med (2013) 5:204ra129–204ra129. doi: 10.1126/scitranslmed.3006245
29. Chikuma S. CTLA-4, an essential immune-checkpoint for T-cell activation. Curr Top Microbiol Immunol (2017) 410:99–126. doi: 10.1007/82_2017_61
30. Zhang Y, Liu Y, Xu Y. Interleukin-24 regulates T cell activity in patients with colorectal adenocarcinoma. Front Oncol (2019) 9:1401. doi: 10.3389/fonc.2019.01401
31. Watanabe R, Gehad A, Yang C, Scott LL, Teague JE, Schlapbach C, et al. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Sci Transl Med (2015) 7:279ra39s. doi: 10.1126/scitranslmed.3010302
32. Bovenschen HJ, van de Kerkhof PC, van Erp PE, Woestenenk R, Joosten I, Koenen HJPM. Foxp3+ regulatory T cells of psoriasis patients easily differentiate into IL-17A-producing cells and are found in lesional skin. J Invest Dermatol (2011) 131:1853–60. doi: 10.1038/jid.2011.139
33. Peng Q, Qiu X, Zhang Z, Zhang S, Zhang Y, Liang Y, et al. PD-L1 on dendritic cells attenuates T cell activation and regulates response to immune checkpoint blockade. Nat Commun (2020) 11:4835. doi: 10.1038/s41467-020-18570-x
34. Mellor AL, Munn DH. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat Rev Immunol (2004) 4:762–74. doi: 10.1038/nri1457
35. Bézie S, Picarda E, Ossart J, Tesson L, Usal C, Renaudin K, et al. IL-34 is a Treg-specific cytokine and mediates transplant tolerance. J Clin Invest (2015) 125:3952–64. doi: 10.1172/JCI81227
36. Lelios I, Cansever D, Utz SG, Mildenberger W, Stifter SA, Greter M. Emerging roles of IL-34 in health and disease. J Exp Med (2020) 217:e20190290. doi: 10.1084/jem.20190290
37. Farrag DA, Asaad MK, Ghobrial CK. Evaluation of IL-34 in psoriasis and psoriatic arthritis patients: correlation with disease activity and severity. Egypt Rheumatol (2017) 39:25–31. doi: 10.1016/j.ejr.2016.05.008
38. Allaire JM, Poon A, Crowley SM, Han X, Sharafian Z, Moore N, et al. Interleukin-37 regulates innate immune signaling in human and mouse colonic organoids. Sci Rep (2021) 11:8206. doi: 10.1038/s41598-021-87592-2
39. Rønholt K, Nielsen AL-L, Johansen C, Vestergaard C, Fauerbye A, López-Vales R, et al. IL-37 expression is downregulated in lesional psoriasis skin. Immunohorizons (2020) 4:754–61. doi: 10.4049/immunohorizons.2000083
40. Garza LA, Yang C-C, Zhao T, Blatt HB, Lee M, He H, et al. Bald scalp in men with androgenetic alopecia retains hair follicle stem cells but lacks CD200-rich and CD34-positive hair follicle progenitor cells. J Clin Invest (2011) 121:613–22. doi: 10.1172/JCI44478
41. Cosmi L, De Palma R, Santarlasci V, Maggi L, Capone M, Frosali F, et al. Human interleukin 17-producing cells originate from a CD161+CD4+ T cell precursor. J Exp Med (2008) 205:1903–16. doi: 10.1084/jem.20080397
42. Dambacher J, Beigel F, Zitzmann K, De Toni EN, Göke B, Diepolder HM, et al. The role of the novel Th17 cytokine IL-26 in intestinal inflammation. Gut (2009) 58:1207–17. doi: 10.1136/gut.2007.130112
43. Donnelly RP, Sheikh F, Dickensheets H, Savan R, Young HA, Walter MR. Interleukin-26: an IL-10-related cytokine produced by Th17 cells. Cytokine Growth Factor Rev (2010) 21:393–401. doi: 10.1016/j.cytogfr.2010.09.001
44. Liu J, Chang H-W, Huang Z-M, Nakamura M, Sekhon S, Ahn R, et al. Single-cell RNA sequencing of psoriatic skin identifies pathogenic Tc17 cell subsets and reveals distinctions between CD8+ T cells in autoimmunity and cancer. J Allergy Clin Immunol (2021) 147:2370–80. doi: 10.1016/j.jaci.2020.11.028
45. Liu Y, Wang H, Cook C, Taylor MA, North JP, Hailer A, et al. Defining patient-level molecular heterogeneity in psoriasis vulgaris based on single-cell transcriptomics. Front Immunol (2022) 13:842651. doi: 10.3389/fimmu.2022.842651
46. Liu Y, Wang H, Taylor M, Cook C, Martínez-Berdeja A, North JP, et al. Classification of human chronic inflammatory skin disease based on single-cell immune profiling. Sci Immunol (2022) 7:eabl9165s. doi: 10.1126/sciimmunol.abl9165
47. Liu W, Zhou X, Wang A, Ma J, Bai Y. Increased peripheral helper T cells type 17 subset correlates with the severity of psoriasis vulgaris. Immunol Lett (2021) 229:48–54. doi: 10.1016/j.imlet.2020.11.005
48. Al-Ashmawy GMZ. “Dendritic cell subsets, maturation and function. ,” InTech (2018). doi: 10.5772/intechopen.79926
49. Kim T-G, Kim DS, Kim H-P, Lee M-G. The pathophysiological role of dendritic cell subsets in psoriasis. BMB Rep (2014) 47:60. doi: 10.5483/BMBRep.2014.47.2.014
50. Mehta H, Mashiko S, Angsana J, Rubio M, Hsieh Y-CM, Maari C, et al. Differential Changes in Inflammatory Mononuclear Phagocyte and T-Cell Profiles within Psoriatic Skin during Treatment with Guselkumab vs. Secukinumab. J Invest Dermatol (2021) 141:1707–1718.e9. doi: 10.1016/j.jid.2021.01.005
51. Chu C-C, Ali N, Karagiannis P, Di Meglio P, Skowera A, Napolitano L, et al. Resident CD141 (BDCA3)+ dendritic cells in human skin produce IL-10 and induce regulatory T cells that suppress skin inflammation. J Exp Med (2012) 209:935–45. doi: 10.1084/jem.20112583
52. Uto T, Fukaya T, Takagi H, Arimura K, Nakamura T, Kojima N, et al. Clec4A4 is a regulatory receptor for dendritic cells that impairs inflammation and T-cell immunity. Nat Commun (2016) 7:1–15. doi: 10.1038/ncomms11273
53. Bakdash G, Buschow SI, Gorris MAJ, Halilovic A, Hato SV, Sköld AE, et al. Expansion of a BDCA1+CD14+ Myeloid cell population in melanoma patients may attenuate the efficacy of dendritic cell vaccines. Cancer Res (2016) 76:4332–46. doi: 10.1158/0008-5472.CAN-15-1695
54. Heger L, Hofer TP, Bigley V, de Vries IJM, Dalod M, Dudziak D, et al. Subsets of CD1c+ DCs: dendritic cell versus monocyte lineage. Front Immunol (2020) 11:559166. doi: 10.3389/fimmu.2020.559166
55. Kim J, Moreno A, Krueger JG. The imbalance between Type 17 T-cells and regulatory immune cell subsets in psoriasis vulgaris. Front Immunol (2022) 13:1005115. doi: 10.3389/fimmu.2022.1005115
56. Maggi L, Santarlasci V, Capone M, Peired A, Frosali F, Crome SQ, et al. CD161 is a marker of all human IL-17-producing T-cell subsets and is induced by RORC. Eur J Immunol (2010) 40:2174–81. doi: 10.1002/eji.200940257
57. Peternel S, Kastelan M. Immunopathogenesis of psoriasis: focus on natural killer T cells. J Eur Acad Dermatol Venereol (2009) 23:1123–7. doi: 10.1111/j.1468-3083.2009.03292.x
58. Cameron AL, Kirby B, Fei W, Griffiths CE. Natural killer and natural killer-T cells in psoriasis. Arch Dermatol Res (2002) 294:363–9. doi: 10.1007/s00403-002-0349-4
59. Curry JL, Qin J-Z, Robinson J, Nickoloff BJ. Reactivity of resident immunocytes in normal and prepsoriatic skin using an ex vivo skin-explant model system. Arch Pathol Lab Med (2003) 127:289–96. doi: 10.5858/2003-127-0289-RORIIN
60. Young NT, Waller ECP, Patel R, Roghanian A, Austyn JM, Trowsdale J. The inhibitory receptor LILRB1 modulates the differentiation and regulatory potential of human dendritic cells. Blood (2008) 111:3090–6. doi: 10.1182/blood-2007-05-089771
61. Guttman-Yassky E, Zhou L, Krueger JG. The skin as an immune organ: Tolerance versus effector responses and applications to food allergy and hypersensitivity reactions. J Allergy Clin Immunol (2019) 144:362–74. doi: 10.1016/j.jaci.2019.03.021
62. Zimmerman KD, Espeland MA, Langefeld CD. A practical solution to pseudoreplication bias in single-cell studies. Nat Commun (2021) 12:738. doi: 10.1038/s41467-021-21038-1
63. Murphy AE, Skene NG. A balanced measure shows superior performance of pseudobulk methods in single-cell RNA-sequencing analysis. Nat Commun (2022) 13:7851. doi: 10.1038/s41467-022-35519-4
64. Gagnon J, Pi L, Ryals M, Wan Q, Hu W, Ouyang Z, et al. Recommendations of scRNA-seq differential gene expression analysis based on comprehensive benchmarking. Life (2022) 12:850. doi: 10.3390/life12060850
65. Hou W, Ji Z, Ji H, Hicks SC. A systematic evaluation of single-cell RNA-sequencing imputation methods. Genome Biol (2020) 21:218. doi: 10.1186/s13059-020-02132-x
66. Vallejos CA, Richardson S, Marioni JC. Beyond comparisons of means: understanding changes in gene expression at the single-cell level. Genome Biol (2016) 17:70. doi: 10.1186/s13059-016-0930-3
67. Mou T, Deng W, Gu F, Pawitan Y, Vu TN. Reproducibility of methods to detect differentially expressed genes from single-cell RNA sequencing. Front Genet (2019) 10:1331. doi: 10.3389/fgene.2019.01331
68. Squair JW, Gautier M, Kathe C, Anderson MA, James ND, Hutson TH, et al. Confronting false discoveries in single-cell differential expression. Nat Commun (2021) 12:5692. doi: 10.1038/s41467-021-25960-2
Keywords: psoriasis, multi-omics, single-cell RNA sequencing, IL-17, biologics impacts of IL-17A blockade on skin immune cell subsets
Citation: Kim J, Lee J, Li X, Kunjravia N, Rambhia D, Cueto I, Kim K, Chaparala V, Ko Y, Garcet S, Zhou W, Cao J and Krueger JG (2023) Multi-omics segregate different transcriptomic impacts of anti-IL-17A blockade on type 17 T-cells and regulatory immune cells in psoriasis skin. Front. Immunol. 14:1250504. doi: 10.3389/fimmu.2023.1250504
Received: 30 June 2023; Accepted: 28 August 2023;
Published: 12 September 2023.
Edited by:
Shiang-Jong Tzeng, National Taiwan University, TaiwanReviewed by:
Stephanie Schell, The Pennsylvania State University, United StatesCopyright © 2023 Kim, Lee, Li, Kunjravia, Rambhia, Cueto, Kim, Chaparala, Ko, Garcet, Zhou, Cao and Krueger. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jaehwan Kim, ZGVya2ltQHVjZGF2aXMuZWR1; James G. Krueger, amdrQHJvY2tlZmVsbGVyLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.