
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 04 September 2023
Sec. Autoimmune and Autoinflammatory Disorders : Autoimmune Disorders
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1247710
This article is part of the Research TopicAutoimmunity and Aging-related DisordersView all 6 articles
Objectives: Previous research has indicated a potential association between immune factors and osteoarthritis (OA), but the causal relationship between CD25 expression on immune cells and hip OA remains enigmatic. To shed light on this relationship, this study utilized the two-sample Mendelian Randomization (MR) method.
Methods: Leveraging genome-wide association studies (GWAS) data from the UK Biobank and arcOGEN, the investigation encompasses a substantial European cohort comprising 15,704 hip OA cases and 378,169 controls. Genetic insights into CD25 stem from a subgroup of 3,757 individuals with European ancestry, encompassing 77 CD25-related traits. Several MR methods were applied, and robustness was assessed through heterogeneity and sensitivity analysis.
Results: Among the 77 traits examined, 66 shared the same single nucleotide polymorphisms (SNPs) with hip OA. Of these, 7 CD25-related traits were found to be causally associated with hip OA (adjusted P><0.05), with F-statistics ranging from 33 to 122. These traits are specifically related to CD4+CD25+ T cells, exhibiting odds ratios (OR) and 95% confidence intervals (CI) less than 1. Notably, no causal link was discerned with the CD8+CD25+ T cell subset. Within absolute count (AC) and relative count (RC) trait types, a significant causal relationship was observed solely between CD4+CD25+ T cells and hip OA, without subtype localization. A more intricate examination of CD25 expression levels within the CD4+CD25+ T cell subset revealed a correlation with the CD39+ regulatory T (Treg) subset and hip OA, particularly within the CD39+ activated Treg subset. Furthermore, a notable causal relationship emerged between CD25 expression levels in the CD45RA- not Treg subset and hip OA. However, no significant causal link was established with any subsets of B cells.
Conclusion: The genetic prediction suggests that CD25, particularly within the realm of CD4+CD25+ T cells, may exert a protective influence against the development of hip OA. These findings provide a novel therapeutic approach for the prevention and treatment of hip OA.
Hip osteoarthritis (OA) is an intricate degenerative condition affecting the hip joint, primarily triggered by a combination of chronic inflammatory and metabolic factors. It is characterized by narrowing of the joint space, degeneration of articular cartilage, osteosclerosis of subchondral bone, osteophyte formation, and chronic inflammation (1, 2). The Global Burden of Disease Study 2019 underscores a gradual rise in the incidence of hip OA, attributed to the aging population, consequently heightening the risk of disability and inflicting substantial societal and patient-related burdens (3–5). Nevertheless, studies on the etiology and therapeutic avenues for hip OA have not kept pace with those for knee OA. This discrepancy may be attributed to the higher prevalence of knee OA, resulting in the direct extrapolation of etiological and clinical insights from knee OA studies (6). However, this may lead to the neglect of other causes that contribute independently to hip OA. While a comprehensive understanding of hip OA’s pathogenesis remains elusive, certain studies have validated the intricate connection between immune-related factors and the emergence of both knee and hip OA (7–10). In light of this context, we propose a hypothesis asserting the pivotal role of immune-related elements in the pathophysiology of hip OA, thereby offering a novel framework to explore its underlying etiological factors and potential avenues for therapeutic intervention.
CD25, the α chain of interleukin-2 receptor (IL-2R), is predominantly expressed on the surface of immune-related cells, including activated T cells, B cells, and NK cells (11) CD25 forms a high-affinity heterotrimeric receptor alongside β and γ chains, engaging in diverse immune responses upon interaction with the ligand IL-2 (12–14). Furthermore, CD25 on immune cells has been implicated in various immune and inflammatory diseases across multiple studies. Research has shown that low expression of CD4+CD25+ regulatory T cells (Tregs) in patients with rheumatoid arthritis suggests that immune suppression mediated by CD25+ Tregs is an important mechanism in the development of rheumatoid arthritis (15, 16). In a separate study exploring factors contributing to periodontitis, CD25 was found to promote the immune inhibitory effects of Tregs and B cells, thereby safeguarding periodontal tissues from inflammation-induced damage (11). The influence of CD25 within the tumor microenvironment has also been extensively investigated, emphasizing the potential of mitigating Treg-mediated immune suppression as a pivotal facet of tumor immunotherapy, positioning CD25 as a viable target for immunotherapeutic strategies (17–19). Moreover, CD25 plays an important role in mediating transplant tolerance and other autoimmune diseases such as atopic dermatitis, Hodgkin’s lymphoma, etc (14, 20). Although there is existing evidence suggesting the role of immune cells in the development of hip OA (21, 22), the precise impact of CD25 on immune cells in this context remains uncharted territory. The causal link between CD25 on immune cells and hip OA still lacks clarity, underscoring the need for comprehensive further research.
Within the realm of epidemiological research, conventional observational studies frequently introduce bias to research outcomes as a result of confounding variables and the potential for reverse causation, thereby diminishing their overall credibility. Conversely, the execution of randomized controlled trials (RCTs) encounters an array of practical limitations that render their implementation within clinical environments a formidable undertaking (23). Mendelian randomization (MR) emerges as an analytical methodology aimed at examining potential causal links between exposures and outcomes, utilizing data derived from genome-wide association studies (GWAS) (23–28). MR uses exposure-related genetic variations as instrumental variables (IVs) and follows the Mendelian gamete random allocation principle as well as the principle of free combination. This approach effectively mitigates the impact of confounding factors and reverse causation commonly encountered in conventional observational studies (29, 30). Thus, based on the available GWAS data on CD25 on immune cells and hip OA, the genetic variants of CD25 on immune cells were investigated as IVs in this study using a two-sample MR approach to investigate whether there is a causal association between CD25 on immune cells and hip OA.
In this research, a two-sample MR method was employed to conduct a causal association analysis. CD25 on immune cells served as the exposure variable, and single nucleotide polymorphisms (SNPs) that showed a significant association with CD25 on immune cells were employed as IVs. And the outcome of interest centered around hip OA. Heterogeneity Analysis and sensitivity analysis were conducted to ensure the robustness of the results.
A secondary analysis was conducted on existing data to identify dependable exposure and genetic variations available in publicly accessible databases. Consequently, no additional ethical approval was required. Considering that the frequency of fingerprint genetic variants varies among populations with different genetic backgrounds, leading to spurious associations between genetic variants and outcomes, we opted to focus our analytical framework on European populations within both samples.
To investigate the potential impact of CD25 on immune cells on hip OA, GWAS data for CD25 on immune cells and hip OA were obtained at the ieu open GWAS project website (https://gwas.mrcieu.ac.uk/datasets). The genetic variation for CD25 on immune cells was derived from a cohort of 3,757 Europeans (31). From this cohort, a total of 77 CD25-related traits were obtained, and all the data included in the analysis were obtained through Flow Cytometry. These 77 traits were categorized into three groups: Absolute count (AC), Relative count (RC), and Median Fluorescence Intensity (MFI). Among them, there were 14 traits in the AC group, representing the total number of cells in the analyzed subgroup. The RC group consisted of 28 traits, representing the proportion of cells in the analyzed subgroup compared to other cell groups. For example, CD25hi CD4+ T cell %CD4+ T cell is a relative value. The MFI group included 35 traits, which indirectly reflected the expression levels of CD25 protein in various subgroups of cells. The fluorescence intensity of CD25 expression can increase either by having more cells expressing CD25 or by having a higher expression level of CD25 in individual or specific subgroups of cells, even when the total cell count remains constant. Supplementary Table S1 presents the GWAS IDs of CD25-related traits, the name of the traits represented, the trait type, etc.
Moreover, genetic data on hip OA were obtained from the UK Biobank and Arthritis Research UK Osteoarthritis Genetics (arcOGEN) resources for a large European population, involving 15,704 cases and 378,169 controls, which included approximately 22 million SNPs, and the GWAS ID of hip OA is GCST007091 (32).
3 core assumptions must be met for SNPs to serve as IVs. First of all, correlation: the prerequisite of correlation necessitates that SNPs demonstrate a robust correlation with the exposure of interest. Notably, the strength of IVs, as measured by F values, should be set at a minimum threshold of ≥10. A value below 10 may potentially introduce significant bias into estimates of causal effects (33). Afterward, exclusivity: the principle of exclusivity mandates that IVs remain devoid of any direct association with the outcome. Their influence should be solely mediated through exposure, ensuring the absence of genetic pleiotropy. In the end, independence: the principle of independence dictates that IVs remain unlinked to confounding factors that exert an impact on both the exposure and the outcome (34). MR capitalizes on the stochastic segregation and recombination of genetic variants during gamete formation, thereby emulating a process of random assignment within a population. Importantly, this procedure typically remains uncorrelated with external environmental confounders, bolstering the integrity of MR analyses (35–37).
To ensure the authenticity and reliability regarding the causal relationship between CD25 on immune cells and hip OA, the following parameters were employed to screen the optimal IVs. Firstly, noteworthy SNPs were meticulously screened from the GWAS database of CD25-related traits. and the F statistic was set to F≥10, corresponding to the significance level of the “IV-exposure” association, commonly acknowledged as a robust screening threshold for IVs. In addition, a requirement for sufficient IV strength was upheld, with a screening threshold of P<5×10-8, effectively preempting potential biases originating from weak IVs. Secondly, the linkage disequilibrium (LD) coefficient was set as r2 of 0.001 and the maximum LD region span of 10,000kb was set. This meticulous parameterization ensured the independence of the selected SNPs and effectively mitigated any LD-related complications. Thirdly, leveraging information from the exposed and extracted SNPs, an explicit protocol was enacted. A minor allele frequency (MAF) threshold of 0.05 was established, and proxy SNPs were strategically employed for SNPs with missing data. SNPs devoid of alternative loci were systematically eliminated. Subsequently, the SNPs identified based on CD25-related traits were extracted from the GWAS database for hip OA. Finally, information from the above two datasets was pooled and excluded SNPs with palindromic structures and MAF > 0.42.
All data were statistically analyzed using R software version R-4.2.2. And all estimates were bilaterally ≤0.05 significant level. In this study, an array of methodologies was employed to infer the potential causal effects of CD25-related traits on hip OA, encompassing the Wald ratio, inverse-variance weighted (IVW), MR-Egger, simple mode, and weighted mode methods. If only one SNP was identified for a significant trait in both databases, only the Wald ratio approach was utilized for analysis. When there were two or more SNPs, the IVW approach was applied. For cases where there were three or more SNPs, the IVW and MR-Egger approaches were implemented. Assessment of SNP heterogeneity across the two samples was conducted via Cochran’s Q test, with significance established at P < 0.05 to indicate noteworthy heterogeneity. In cases where heterogeneity and pleiotropy were absent, precedence was given to the results derived from the IVW approach. In the presence of heterogeneity, the random effects model of IVW could be invoked. However, if pleiotropy was detected, preference was granted to the outcomes derived from MR-Egger calculations (38). To prevent type I errors that arise from multiple hypothesis testing, the Benjamini-Hochberg method of False Discovery Rate (FDR) was employed to adjust the P value and control the overall error rate. In the MR analysis, the beta value is commonly used to estimate the causal effect of genetic variation on the exposure and outcome. We convert the beta value to the odds ratio (OR) using the formula OR=exp(beta). The OR is a measure of association between the exposure and outcome, providing a more intuitive understanding of the actual implications of the results. If OR=1, it indicates that the exposure does not affect the odds of the outcome. If OR>1, it suggests that the exposure is associated with higher odds of the outcome. Conversely, if OR<1, it indicates that the exposure is associated with lower odds of the outcome.
The MR-Egger regression analysis was utilized to assess potential horizontal pleiotropy effects attributed to the encompassed SNPs. Should the intercept term of the MR-Egger method significantly deviate from 0, this would signal the presence of horizontal pleiotropy. Additionally, sensitivity analysis was performed using the Leave-one-out sensitivity test, whereby the outcomes were recalculated following the stepwise removal of individual SNPs. The objective was to ascertain whether the exclusion of any SNP yielded a substantial alteration in the results. The robustness of the MR findings was established if no SNP omission yielded a significant impact on the overall outcomes.
In addition, the chosen SNPs underwent a comprehensive cross-validation process using the website http://www.phenoscanner.medschl.cam, aimed at identifying any additional traits that could exert an influence on the study outcomes. SNPs found to exhibit associations with these additional traits were subsequently excluded, effectively minimizing the potential influence of confounding factors.
After a series of strict quality control, it was determined that among the 77 CD25-related traits, a total of 66 exhibited an intersection with SNPs sourced from the large-scale hip OA GWAS. However, only 7 of these CD25-related traits had a significantly adjusted P-value <0.05. Notably, the F-statistics for these 7 CD25-related traits ranged from 33 to 122, ensuring the strength of the IVs and avoiding bias due to weak instruments. The preliminary screened SNPs were obtained after removing LD effects, but one SNP (rs6584030) with palindromic structure and MAF > 0.42 was excluded. Comprehensive details about the SNPs, including SNP ID, beta value, standard error (se), P-value, effect allele (EA), and other allele, were systematically summarized for subsequent analysis, as shown in Supplementary Table S2.
As shown in Table 1, the transformation of beta values into OR via the formula OR=exp(beta) was executed, accompanied by the computation of 95% confidence interval (CI). A notable association emerges, indicating that 7 CD25-related traits are intricately linked with the susceptibility to hip OA, serving as protective elements against its development. All 7 CD25-related traits in the panel were Tregs, and they were classified into three different trait types: AC, RC, and MFI. AC and RC denote the quantity of a specific cellular subset within CD25+, whereas MFI represents the intensity of CD25 expression within a particular cellular subset of CD25+.
Among the above 7 traits, only CD25 on CD45RA- CD4 not Treg has two SNPs (rs181516190, rs61839660), with analysis conducted via the IVW method. In the heterogeneity evaluation, Cochran’s Q test showed a heterogeneity P-value of 0.877, indicating no heterogeneity. The remaining traits, characterized by just one SNP each, underwent analysis using the Wald ratio method, thereby obviating the need for a heterogeneity test. Furthermore, all 7 CD25-related trait SNPs were less than three, so no horizontal pleiotropy testing was needed. Upon comprehensive scrutiny within PhenoScanner, no direct links to the outcome were identified for all SNPs, and they exhibited no affiliation with potential confounding factors influencing the association between exposure and outcome. The subsets and molecular expression of CD25+ T cells and B cells in the results are illustrated in Figure 1.
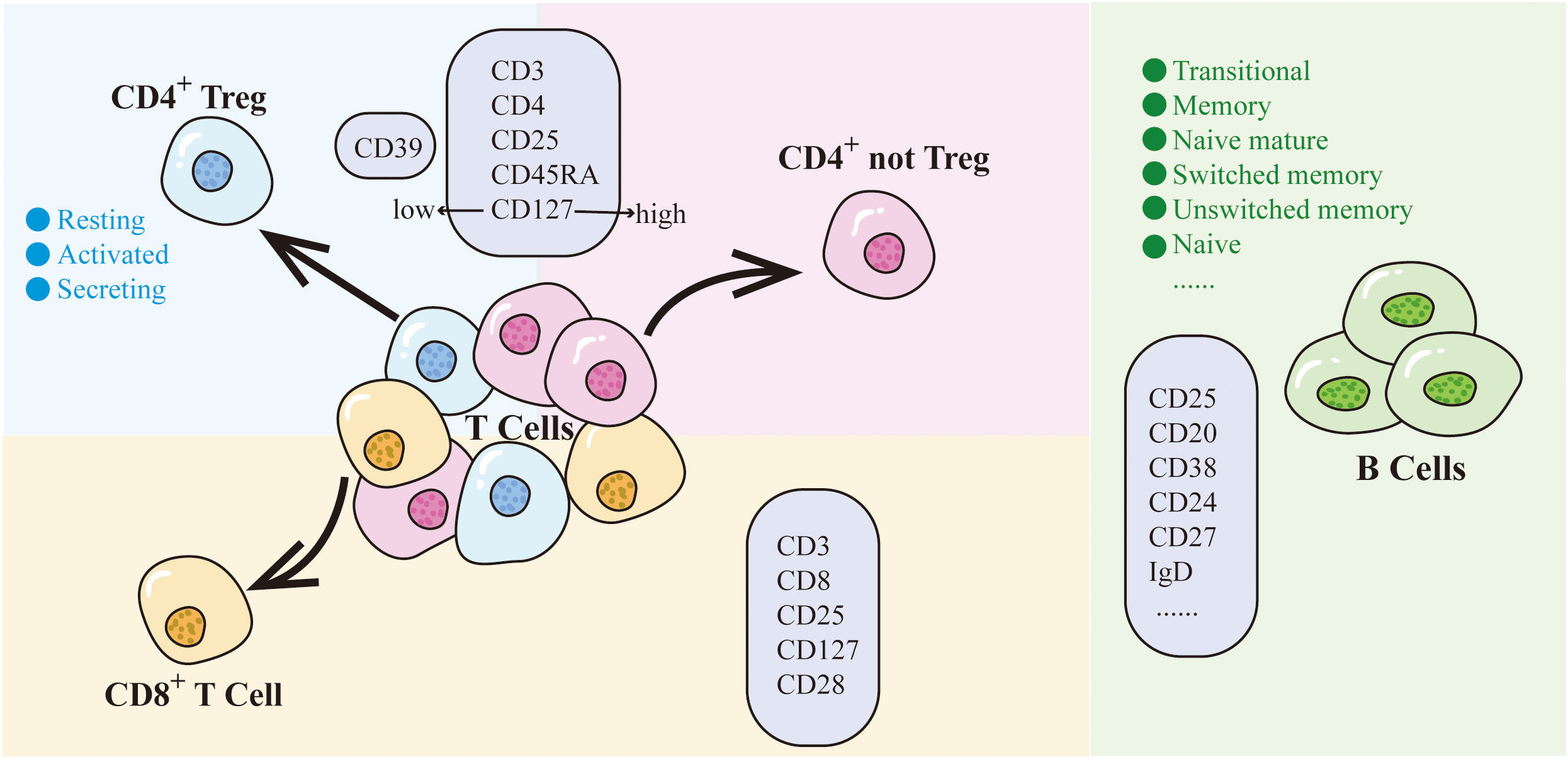
Figure 1 CD25 on T cells and B cells. Different colored backgrounds represent distinct cell subsets. T cells are divided into CD4+ subset and CD8+ subset. The CD4+ subset includes CD4+ Treg (light blue) and CD4+ not Treg (light pink). CD8+ T cells are depicted in light orange, while B cells are represented in green. The light purple box indicates the molecules expressed by cells within that particular subset.
Within the Treg panel, CD25+ cells can be classified into 2 main cell subpopulations based on the expression of CD4 and CD8 on the T cell surface: CD4+CD25+ and CD8+CD25+. The CD4+CD25+ traits within the Treg panel were further categorized into two distinct groups, Treg (CD25hiCD127low) and not Treg (CD25hiCD127hi), based on CD127 expression levels. Moreover, the CD4+CD25+ Treg subpopulation was systematically subdivided into 3 subtypes according to their cellular characteristics: activated (CD25+++CD45RA-), resting (CD25++CD45RA+), and secreting (CD25++CD45RA-). An additional layer of classification can be undertaken based on the expression of CD39. Similarly, the CD8+CD25+ traits can be divided into different subgroups depending on the expression of CD28 and CD127.
The findings demonstrated that CD4+CD25hi T cell AC (OR: 0.84, 95% CI: 0.76-0.93, P-adjusted=0.013) was significantly associated with hip OA. However, there was no significant causal association found between the AC traits of CD8+CD25+ cells and hip OA. Furthermore, the ensuing findings failed to indicate AC traits of CD4+CD25+ Treg (activated/resting/secreting) and CD4+CD25+ not Treg and hip OA were causally significantly associated with each other. Not only did AC show a significant association with hip OA within the Treg panel, but RC also exhibited a significant association with hip OA. Among the RC-related traits, CD4+CD25hi T cell %T cell (OR:0.88,95%CI:0.82-0.95, P-adjusted=0.013) and CD4+CD25hi T cell %CD4+ T cell (OR:0.90,95%CI:0.84-0.96, P-adjusted=0.013) indicated a significant causal relationship between the RC traits of CD25 and hip OA. However, no subpopulations within the RC traits of CD4+Treg (activated/resting/secreting) or CD4+ not Treg (CD45RA+/CD45RA-) were deemed significantly associated with hip OA. Additionally, no significant causal relationship was identified between any cell subpopulations within the RC traits of CD8+CD25+ cells and hip OA.
In addition to the significant causal relationships observed between the AC and RC traits of CD25 and hip OA, it is pertinent to highlight the link between the expression level of CD25 in immune cells and hip OA. We discovered traits within the CD4+CD25+ cell subpopulation that exhibit a causal association with hip OA. Specifically, the presence of CD25 on CD4+ T cell (OR: 0.84, 95% CI: 0.76-0.93, P-adjusted=0.013) was associated with a reduced risk of hip OA. In the realm of Tregs, CD25 on CD39+ CD4 Treg (OR: 0.80, 95% CI: 0.71-0.91, P-adjusted=0.024) emerged as a significant factor in hip OA. Further scrutiny revealed that within the CD39+ subgroup cells, CD25 on CD39+ activated CD4 Treg (OR: 0.79, 95% CI: 0.69-0.91, P-adjusted=0.024) displayed a close correlation with hip OA. Notably, among the not Treg subset, CD25 on CD45RA- CD4 not Treg (OR: 0.90, 95% CI: 0.85-0.96, P-adjusted=0.039) was associated with hip OA. However, no discernible causal relationship between any cell subpopulations within CD8+CD25+ and hip OA was discerned.
The presence of CD25 expression on B cells did not exhibit a causal link to hip OA. Within the B cell panel, a delineation into distinct subsets can be achieved based on CD24 versus CD38 expression, thereby encompassing transitional (CD24+CD38hi), memory (CD24+CD38−/dim), and naive mature (CD24−CD38−/dim) subsets. Further stratification based on CD27 versus IgD establishes additional subclasses, including switched memory (CD27+IgD−), unswitched memory (CD27+IgD+), naive (CD27−IgD+), and CD27−IgD− B cells. Additionally, classification using IgD versus CD38 lends itself to the identification of Bm1–Bm5 subgroups, and various other B cell subgroups can be discerned based on parameters like CD24 versus CD27, IgD versus CD24, and CD20 versus CD38. However, our findings demonstrated that CD25 expression on B cells, including all CD25+ B cell subgroups, was not causally related to hip OA.
Although OA has historically been considered a non-inflammatory degenerative disease, emerging evidence underscores the role of immune-related factors in its pathogenesis, with their active involvement in the inflammatory processes underlying OA. Therefore, the attenuation of anti-inflammatory effects and the enhancement of pro-inflammatory effects can both exacerbate the progression of inflammation, thereby driving the advancement of OA (7–10, 39, 40). CD25, a pivotal immune-related component, serves as the α chain of the IL-2R and is predominantly present on the surface of activated T cells, B cells, and NK cells. Functioning as a critical constituent of the IL-2R, CD25 competes for IL-2 binding, thereby reducing immune responses mediated by free IL-2. Up until now, no studies have used large-scale data to perform an in-depth analysis of the causal relationship between CD25 and hip OA. However, this study employs a two-sample MR analysis based on extensive GWAS data to establish a causal association between CD25 and hip OA, providing evidence for CD25 as a protective factor against hip OA. These findings are consistent with previous research on the role of CD25 in OA. For instance, an exploration into mesenchymal stem cell (MSC) therapy for OA revealed that treatment engendered the accumulation of CD4+CD25+ leukocyte subsets within the joints, effectively mitigating OA progression (41). Similarly, a distinct study demonstrated that OA patients exhibit lower proportions of CD4+CD25+ Tregs compared to healthy individuals, potentially contributing to increased pain and functional impairment (39). While previous studies have reported a significant increase in CD8+CD25+ T cells in patients with rheumatoid arthritis, which exacerbates the progression of inflammation and may be an important pathogenic mechanism (42). However, our findings do not provide evidence of a causal relationship between the CD8+CD25+ T cell subset and hip OA, suggesting the need for additional research to explore this association.
Based on the results of this MR study, it is evident that all 7 CD25-related traits exhibiting significant correlations with hip OA are specifically related to CD4+CD25+ T cells. Notably, among the AC and RC trait types, a significant causal relationship between CD4+CD25+ T cell subgroups and hip OA has been discerned, though the precise subgroup of cells responsible remains to be conclusively identified. Further analysis of CD25 expression levels revealed that both Treg and not Treg subgroups of CD4+CD25+ T cells play crucial roles in this association. The primary function of CD4+CD25+ Tregs is to maintain the body’s immune tolerance and suppress the immune response thus maintaining and regulating the body’s immune homeostasis, which is increasingly studied in inflammation, autoimmune diseases, and tumors (14, 16, 17). Consequently, an escalation in the count of CD25-expressing Tregs or elevated CD25 expression levels can enhance Tregs function, which is essential for suppressing the progression of immune inflammation. Remarkably, both the CD4+CD25+CD39+ Treg subset and its CD39+ activated Treg subgroup exhibit significant causal relationships with hip OA. CD39 is a cell membrane protein with enzymatic activities that hydrolyzes adenosine triphosphate (ATP) and adenosine diphosphate (ADP) into adenosine monophosphate (AMP), leading to energy depletion and the inhibition of immune reactions in various cells (43, 44). This enzymatic prowess of CD39 is likely instrumental in tempering the inflammatory cascade inherent in hip OA. The CD39+ activated Treg subgroup of CD39+ Tregs, not only exhibits high CD39 expression but also demonstrates the highest level of CD25 expression among all subgroups. This subgroup effectively suppresses immune reactions of hip OA, highlighting its immunosuppressive role in the disease. In contrast, within the not Treg subset, the presence of CD25 on CD45RA- CD4 T cell subset is significantly correlated with hip OA. Our conjecture posits that this subset, characterized by the absence of CD45RA expression, possibly encompasses non-naïve T cells that might be affiliated with effector T cells or memory T cells. However, due to the heightened CD25 expression, these cells engage in competitive binding with IL-2 and rapidly deploy immunosuppressive effects upon stimulation. Yet, the specific intricacies elucidating the immunosuppressive role of CD4+ not Tregs warrant comprehensive further exploration and analysis.
In short, all these traits are associated with the expression of CD25 on CD4+ T cells. An increased number of T cells expressing CD25 or an elevated expression level of CD25 both attenuate the inflammatory progression of hip OA. We hypothesize that this effect could be attributed to the heightened presence of CD25, which competitively intercepts IL-2, a pivotal factor essential for the survival of adjacent activated T cells. This interception, in turn, results in the suppression of their proliferation or the initiation of apoptosis. Additionally, it promotes the secretion of soluble negative immune molecules such as IL-10 and TGF-β by Tregs, enabling them to exert immunosuppressive effects. Consequently, these mechanisms collectively serve to dampen the inflammatory reaction and impede the progression of hip OA.
The 7 traits encompassing SNPs correspond to 2 genes, rs7078614, and rs61839660 are the IVs of IL2RA and rs181516190 is the IV of TBX20. IL2RA encodes the IL2R-α chain (CD25), which is crucial for the formation of the IL-2/IL-2R signaling pathway participation in several immune activities. It is an important functional gene related to immune-related factors (12, 45). Therefore, the polymorphisms in IL2RA are closely associated with hip OA.
TBX20, on the other hand, is also an encoding gene that produces a transcription factor belonging to the T-box family. The protein encoded by TBX20 is involved in regulating developmental processes, particularly in cardiac development, as a conserved master regulator of cardiac development and function (46, 47). While there are no specific reports on the involvement of TBX20 in hip OA, its polymorphisms suggest a potential link with the pathogenesis of hip OA and its potential as a therapeutic target. Yet, a comprehensive comprehension of the precise mechanisms necessitates further in-depth investigation.
This two-sample MR analysis aims to analyze the causal relationship between CD25 on immune cells and hip OA using available large-sample GWAS data, avoiding the effects of confounding factors and reverse causation and reducing the limitations of traditional observational studies. Furthermore, MR can also alleviate the representativeness and feasibility issues associated with RCTs. Nevertheless, there are some limitations in this study. Firstly, this study relies on publicly available GWAS data, which hinders the possibility of subgroup analysis considering factors like gender, age, and BMI to further explore the effects of other relevant factors on hip OA (26). Secondly, given that the dataset exclusively represents a European population, generalizing the findings to other ethnic groups should be done cautiously, necessitating further scrutiny (27). Thirdly, this study had a limited number of SNPs, and some of the CD25-related traits were associated with only one SNP. This could lead to an overestimation of the relationship between genetic variation and exposure, potentially influenced by the winner’s curse and leading to biased results (48). Finally, all CD25-related traits examined in this analysis were linked to the Treg panel, whereas prior research has indicated a significant role for CD25+ B cells in the inflammatory response as well (11, 49). Therefore, further research is necessary to investigate whether CD25+ B cells and CD25+ NK cells are related to hip OA. Fortunately, the F-statistic value of the IVs in this study was greater than 10, which prevents the potential bias resulting from weak IVs.
In conclusion, this two-sample MR study revealed a causal relationship between CD25 on immune cells and hip OA, particularly CD4+CD25+ T cells, providing genetic evidence for CD25 as a protective factor for hip OA. It provides a new therapeutic strategy for the treatment of hip OA, making CD25 a potential target for the prevention and treatment of hip OA.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethical approval was not required for the study involving humans in accordance with the local legislation and institutional requirements. Written informed consent to participate in this study was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and the institutional requirements.
SZ, HL, and YZ conceptualized and designed the study. SZ, HL, YZ, BG, ZR, ZL, and ZF performed material preparation, data collection, and analysis. SZ, HL, and YZ analyzed and interpreted the data, prepared the original draft, and reviewed and edited the final draft. All authors contributed to the article and approved the submitted version.
This work was supported by the following grants: the National Natural Science Foundation of China (No. 81902222,82172399), and the Central South University Graduate School-Enterprise Joint Innovation Project (NO. 2022XQLH186).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer BZ declared a shared parent affiliation with the authors to the handling editor at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1247710/full#supplementary-material
1. Aresti N, Kassam J, Nicholas N, Achan P. Hip osteoarthritis. BMJ (2016) 354:i3405. doi: 10.1136/bmj.i3405
2. Hoaglund FT, Steinbach LS. Primary osteoarthritis of the hip: etiology and epidemiology. J Am Acad Orthop Surg (2001) 9(5):320–7. doi: 10.5435/00124635-200109000-00005
3. Katz JN, Arant KR, Loeser RF. Diagnosis and treatment of hip and knee osteoarthritis: A review. JAMA (2021) 325(6):568–78. doi: 10.1001/jama.2020.22171
4. Diseases GBD, Injuries C. Global burden of 369 diseases and injuries in 204 countries and territories, 1990-2019: A systematic analysis for the global burden of disease study 2019. Lancet (2020) 396(10258):1204–22. doi: 10.1016/S0140-6736(20)30925-9
5. Glyn-Jones S, Palmer AJ, Agricola R, Price AJ, Vincent TL, Weinans H, et al. Osteoarthritis. Lancet (2015) 386(9991):376–87. doi: 10.1016/S0140-6736(14)60802-3
6. Murphy NJ, Eyles JP, Hunter DJ. Hip osteoarthritis: etiopathogenesis and implications for management. Adv Ther (2016) 33(11):1921–46. doi: 10.1007/s12325-016-0409-3
7. Assirelli E, Pulsatelli L, Dolzani P, Mariani E, Lisignoli G, Addimanda O, et al. Complement expression and activation in osteoarthritis joint compartments. Front Immunol (2020) 11:535010. doi: 10.3389/fimmu.2020.535010
8. Apinun J, Sengprasert P, Yuktanandana P, Ngarmukos S, Tanavalee A, Reantragoon R. Immune mediators in osteoarthritis: infrapatellar fat pad-infiltrating cd8+ T cells are increased in osteoarthritic patients with higher clinical radiographic grading. Int J Rheumatol (2016) 2016:9525724. doi: 10.1155/2016/9525724
9. de Lange-Brokaar BJ, Kloppenburg M, Andersen SN, Dorjee AL, Yusuf E, Herb-van Toorn L, et al. Characterization of synovial mast cells in knee osteoarthritis: association with clinical parameters. Osteoarthritis Cartilage (2016) 24(4):664–71. doi: 10.1016/j.joca.2015.11.011
10. Moradi B, Schnatzer P, Hagmann S, Rosshirt N, Gotterbarm T, Kretzer JP, et al. Cd4(+)Cd25(+)/highcd127low/(-) regulatory T cells are enriched in rheumatoid arthritis and osteoarthritis joints–analysis of frequency and phenotype in synovial membrane, synovial fluid and peripheral blood. Arthritis Res Ther (2014) 16(2):R97. doi: 10.1186/ar4545
11. Han Y, Yu C, Yu Y, Bi L. Cd25+ B cells produced il-35 and alleviated local inflammation during experimental periodontitis. Oral Dis (2022) 28(8):2248–57. doi: 10.1111/odi.13939
12. Li Y, Li X, Geng X, Zhao H. The il-2a receptor pathway and its role in lymphocyte differentiation and function. Cytokine Growth Factor Rev (2022) 67:66–79. doi: 10.1016/j.cytogfr.2022.06.004
13. Hernandez R, LaPorte KM, Hsiung S, Santos Savio A, Malek TR. High-dose il-2/cd25 fusion protein amplifies vaccine-induced Cd4(+) and Cd8(+) neoantigen-specific T cells to promote antitumor immunity. J Immunother Cancer (2021) 9(9):e002865. doi: 10.1136/jitc-2021-002865
14. Zammarchi F, Havenith K, Bertelli F, Vijayakrishnan B, Chivers S, van Berkel PH. Cd25-targeted antibody-drug conjugate depletes regulatory T cells and eliminates established syngeneic tumors via antitumor immunity. J Immunother Cancer (2020) 8(2):e000860. doi: 10.1136/jitc-2020-000860
15. Ju B, Zhu L, Wang J, Zheng J, Hao Z, Luo J, et al. The proportion and phenotypic changes of Cd4(+)Cd25(-)Foxp3(+) T cells in patients with untreated rheumatoid arthritis. BMC Immunol (2022) 23(1):41. doi: 10.1186/s12865-022-00517-0
16. Delavari S, Ghafourian M, Rajaei E, Mowla K, Ghadiri A. Evaluation of Cd4+/cd25+/high/cd127low/- regulatory T cells in rheumatoid arthritis patients. Iran J Immunol (2021) 18(3):179–87. doi: 10.22034/iji.2021.68100.1365
17. Peng Y, Tao Y, Zhang Y, Wang J, Yang J, Wang Y. Cd25: a potential tumor therapeutic target. Int J Cancer (2023) 152(7):1290–303. doi: 10.1002/ijc.34281
18. Solomon I, Amann M, Goubier A, Arce Vargas F, Zervas D, Qing C, et al. Cd25-T(Reg)-depleting antibodies preserving il-2 signaling on effector T cells enhance effector activation and antitumor immunity. Nat Cancer (2020) 1(12):1153–66. doi: 10.1038/s43018-020-00133-0
19. Villanueva MT. Anti-cd25 antibody tips the T cell balance. Nat Rev Drug Discovery (2021) 20(1):18. doi: 10.1038/d41573-020-00206-w
20. Ellery JM, Nicholls PJ. Alternate signalling pathways from the interleukin-2 receptor. Cytokine Growth Factor Rev (2002) 13(1):27–40. doi: 10.1016/s1359-6101(01)00023-5
21. Lopes EBP, Filiberti A, Husain SA, Humphrey MB. Immune contributions to osteoarthritis. Curr Osteoporos Rep (2017) 15(6):593–600. doi: 10.1007/s11914-017-0411-y
22. Scanzello CR, Goldring SR. The role of synovitis in osteoarthritis pathogenesis. Bone (2012) 51(2):249–57. doi: 10.1016/j.bone.2012.02.012
23. Li R, Guo Q, Zhao J, Kang W, Lu R, Long Z, et al. Assessing causal relationships between gut microbiota and asthma: evidence from two sample mendelian randomization analysis. Front Immunol (2023) 14:1148684. doi: 10.3389/fimmu.2023.1148684
24. Hu C, Li Y, Qian Y, Wu Z, Hu B, Peng Z. Kidney function and cardiovascular diseases: A large-scale observational and mendelian randomization study. Front Immunol (2023) 14:1190938. doi: 10.3389/fimmu.2023.1190938
25. Li W, Lu Q, Qian J, Feng Y, Luo J, Luo C, et al. Assessing the causal relationship between genetically determined inflammatory biomarkers and low back pain risk: A bidirectional two-sample mendelian randomization study. Front Immunol (2023) 14:1174656. doi: 10.3389/fimmu.2023.1174656
26. Tian D, Zhou Y, Chen Y, Wu Y, Wang H, Jie C, et al. Genetically predicted ankylosing spondylitis is causally associated with psoriasis. Front Immunol (2023) 14:1149206. doi: 10.3389/fimmu.2023.1149206
27. Zhao N, Guo P, Tang M, Yang F, Zhang T, Mao R. Evidence for a causal relationship between psoriasis and cutaneous melanoma: a bidirectional two-sample mendelian randomized study. Front Immunol (2023) 14:1201167. doi: 10.3389/fimmu.2023.1201167
28. Cho H, Okekunle A, Yie G, Youn J, Kang M, Jin T, et al. Association of coffee consumption with type 2 diabetes and glycemic traits: a mendelian randomization study. Nutr Res Pract (2023) 17(4):789–802. doi: 10.4162/nrp.2023.17.4.789
29. Ference BA, Holmes MV, Smith GD. Using mendelian randomization to improve the design of randomized trials. Cold Spring Harb Perspect Med (2021) 11(7):a040980. doi: 10.1101/cshperspect.a040980
30. Sekula P, Del Greco MF, Pattaro C, Kottgen A. Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol (2016) 27(11):3253–65. doi: 10.1681/ASN.2016010098
31. Orru V, Steri M, Sidore C, Marongiu M, Serra V, Olla S, et al. Complex genetic signatures in immune cells underlie autoimmunity and inform therapy. Nat Genet (2020) 52(10):1036–45. doi: 10.1038/s41588-020-0684-4
32. Tachmazidou I, Hatzikotoulas K, Southam L, Esparza-Gordillo J, Haberland V, Zheng J, et al. Identification of new therapeutic targets for osteoarthritis through genome-wide analyses of Uk biobank data. Nat Genet (2019) 51(2):230–6. doi: 10.1038/s41588-018-0327-1
33. Burgess S, Thompson SG, Collaboration CCG. Avoiding bias from weak instruments in mendelian randomization studies. Int J Epidemiol (2011) 40(3):755–64. doi: 10.1093/ije/dyr036
34. Hartwig FP, Borges MC, Horta BL, Bowden J, Davey Smith G. Inflammatory biomarkers and risk of schizophrenia: a 2-sample mendelian randomization study. JAMA Psychiatry (2017) 74(12):1226–33. doi: 10.1001/jamapsychiatry.2017.3191
35. Birney E. Mendelian randomization. Cold Spring Harb Perspect Med (2022) 12(4):a041302. doi: 10.1101/cshperspect.a041302
36. Bowden J, Holmes MV. Meta-analysis and mendelian randomization: a review. Res Synth Methods (2019) 10(4):486–96. doi: 10.1002/jrsm.1346
37. Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet (2014) 23(R1):R89–98. doi: 10.1093/hmg/ddu328
38. Sproviero W, Winchester L, Newby D, Fernandes M, Shi L, Goodday SM, et al. High blood pressure and risk of dementia: a two-sample mendelian randomization study in the uk biobank. Biol Psychiatry (2021) 89(8):817–24. doi: 10.1016/j.biopsych.2020.12.015
39. Nees TA, Zhang JA, Platzer H, Walker T, Reiner T, Tripel E, et al. Infiltration profile of regulatory T cells in osteoarthritis-related pain and disability. Biomedicines (2022) 10(9):2111. doi: 10.3390/biomedicines10092111
40. Li YS, Luo W, Zhu SA, Lei GH. T cells in osteoarthritis: alterations and beyond. Front Immunol (2017) 8:356. doi: 10.3389/fimmu.2017.00356
41. Lamers K, Baquero M, Karrow N, Hurtig M. Intra-articular xenogeneic mesenchymal stem cell-based therapy increases Cd4(+)Cd25(+) cells in synovial fluid. Vet Immunol Immunopathol (2020) 227:110085. doi: 10.1016/j.vetimm.2020.110085
42. Wang LQ, Wang Y, Wang R, Zhao RX, Meng Y, Ren LL, et al. [Changes and clinical significance of peripheral blood Cd8(+)Cd25(+)T cells in rheumatoid arthritis patients]. Zhonghua Yi Xue Za Zhi (2020) 100(20):1557–61. doi: 10.3760/cma.j.cn112137-20190829-01920
43. Timperi E, Barnaba V. Cd39 regulation and functions in T cells. Int J Mol Sci (2021) 22(15):8068. doi: 10.3390/ijms22158068
44. Moesta AK, Li XY, Smyth MJ. Targeting cd39 in cancer. Nat Rev Immunol (2020) 20(12):739–55. doi: 10.1038/s41577-020-0376-4
45. Qiao Y, Wang Y, Xu Y, Zhang J, Su Y, Cheng Y, et al. An association study of il2ra polymorphisms with cerebral palsy in a chinese population. BMC Med Genomics (2022) 15(1):208. doi: 10.1186/s12920-022-01350-5
46. Chen Y, Xiao D, Zhang L, Cai CL, Li BY, Liu Y. The role of tbx20 in cardiovascular development and function. Front Cell Dev Biol (2021) 9:638542. doi: 10.3389/fcell.2021.638542
47. Singh R, Kispert A. Tbx20, smads, and the atrioventricular canal. Trends Cardiovasc Med (2010) 20(4):109–14. doi: 10.1016/j.tcm.2010.09.004
48. Jiang T, Gill D, Butterworth AS, Burgess S. An empirical investigation into the impact of winner's curse on estimates from mendelian randomization. Int J Epidemiol (2022) 52(4):1209–19. doi: 10.1093/ije/dyac233
Keywords: hip OA, CD25, SNP, Mendelian randomization, causal relationship
Citation: Luo H, Zhu Y, Guo B, Ruan Z, Liu Z, Fan Z and Zhao S (2023) Causal relationships between CD25 on immune cells and hip osteoarthritis. Front. Immunol. 14:1247710. doi: 10.3389/fimmu.2023.1247710
Received: 26 June 2023; Accepted: 21 August 2023;
Published: 04 September 2023.
Edited by:
Qi Wang, Huazhong University of Science and Technology, ChinaReviewed by:
Bin Zhao, Second Xiangya Hospital, Central South University, ChinaCopyright © 2023 Luo, Zhu, Guo, Ruan, Liu, Fan and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shushan Zhao, emhhb3NodWlxdWFuQDEyNi5jb20=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.