- 1Department of Cell Biology, College of Basic Medical Sciences, Army Medical University (Third Military Medical University), Chongqing, China
- 2Department of Wound Infection and Drug, State Key Laboratory of Trauma, Burn and Combined Injury, Daping Hospital, Army Medical University (Third Military Medical University), Chongqing, China
- 3Department of Oncology, The Second Affiliated Hospital of Zunyi Medical University, Zunyi, Guizhou, China
Interferon regulatory factor (IRF) 7 was originally identified as master transcriptional factor that produced IFN-I and regulated innate immune response, subsequent studies have revealed that IRF7 performs a multifaceted and versatile functions in multiple biological processes. In this review, we provide a comprehensive overview on the current knowledge of the role of IRF7 in immunity and autoimmunity. We focus on the latest regulatory mechanisms of IRF7 in IFN-I, including signaling pathways, transcription, translation, and post-translational levels, the dimerization and nuclear translocation, and the role of IRF7 in IFN-III and COVID-19. In addition to antiviral immunity, we also discuss the role and mechanism of IRF7 in autoimmunity, and the further research will expand our understanding of IRF7.
1 Introduction
Interferon regulatory factors (IRFs) are a family of master transcription factors, and although IRFs are recognized as transcriptional regulators of type I IFNs (IFN-I) and IFN-inducible genes, this family is now characterized as key factors in the regulation of many different processes, such as immunity, oncogenesis, metabolism, cell differentiation and apoptosis (1).
The IRF family consists of nine members, IRF1–9, previously reported in mammals. In addition, IRF10 has been identified in birds, dogs and teleost fish, and IRF11 is only found in teleost fish (2–4).
The IRF family has a conserved N-terminal region, and all members possess a helix-turn-helix DNA-binding domain (DBD), which contains five tryptophan repeats and recognizes the core DNA sequence of the 5’-GAAA-3’ tetranucleotide contained within the IFN-stimulated response elements (ISREs). The C-terminal regions of IRFs are diverse and related to distinct functions and contain two types of IRF-associated domains (IADs). The IAD mediates homo and heteromeric interactions with other IRF members, transcription factors, or cofactors to recognize DNA sequences and regulate gene transcription (5, 6).
IRF7 is a lymphoid-specific factor that is predominantly expressed in the cytoplasm of the spleen, thymus, and peripheral blood lymphocytes, such as B cells, plasmacytoid dendritic cells (pDCs) and monocytes. Although IRF7 was originally identified in Epstein-Barr virus (EBV) infection and characterized as a transcriptional regulator of IFN-I and IFN-stimulated gene (ISG), recent studies have revealed that IRF7 exerts a broad range of activities in different biological processes (7). In this review, as documented elsewhere, the structure of IRF7 is briefly summarized, however, we focus on the current knowledge about the regulation and function of IRF7.
2 The structure of IRF7
The IRF7 gene is located on chromosome 11p15.5 in humans and 7 F5 in mice, and four splicing variants of IRF7A, IRF7B, IRF7C, and IRF7H have been identified. IRF7A is the major splicing variant. Human IRF7A cDNA encodes a protein of 503 amino acid (aa) residues and mouse IRF7 consists of 457 aa residues. The nucleic acid homology of human and mouse IRF7 reaches 72.86% (8–10).
IRF7 (IRF7A) has a distinctive multiple domain structure in its C-terminal region, except for the DBD in the N-terminal region, present in all IRF members. With the deletion mutations of IRF7, a constitutive activation domain (CAD) is located between 151–246 aa adjacent to the conserved DBD (1-150 aa), which maintains the activity of IRF7, and a nuclear localization sequence may be present in the region between 1 and 246 aa. The region located between 278 and 305 aa contributed to the virus activated domain (VAD), which is indispensable for the activation of IRF7, and the sequence collaborates with the C-terminal signal response domain for maximal response to virus infection, and nuclear translocation may be controlled by the region. The inhibitory domain (ID) located between 341 and 467 aa interferes with the transactivation function of IRF7, but primarily within the region of 416–467 aa, and this sequence contains an efficient nuclear export signal (NES), deletion of the region of 417 and 440 aa, IRF7 failed to nuclear translocation. The second inhibitory domain is located between 341-415 aa, which may affect the DNA binding and/or transactivation activity of IRF7. The signal response domain (SRD) or accessory inducibility region located at the C-terminal end between 468 and 503 aa mediates IRF7 dimerization and contains a serine-rich domain, and the phosphorylation of Ser 477 and Ser 479 is vital for IRF7 because the substitution of S477 and S479 completely abrogated cytoplasmic to nuclear translocation. Nuclear export of IRF7 requires sequences within VAD, ID and phosphorylation of S477 and S479 (11, 12) (Figure 1). IRF7 is rich in the PEST (proline (P), glutamic acid (E), serine (S), and threonine (T)) sequence due to its very short half-life, and its stability is controlled by the ubiquitin–proteasome system (9, 13).
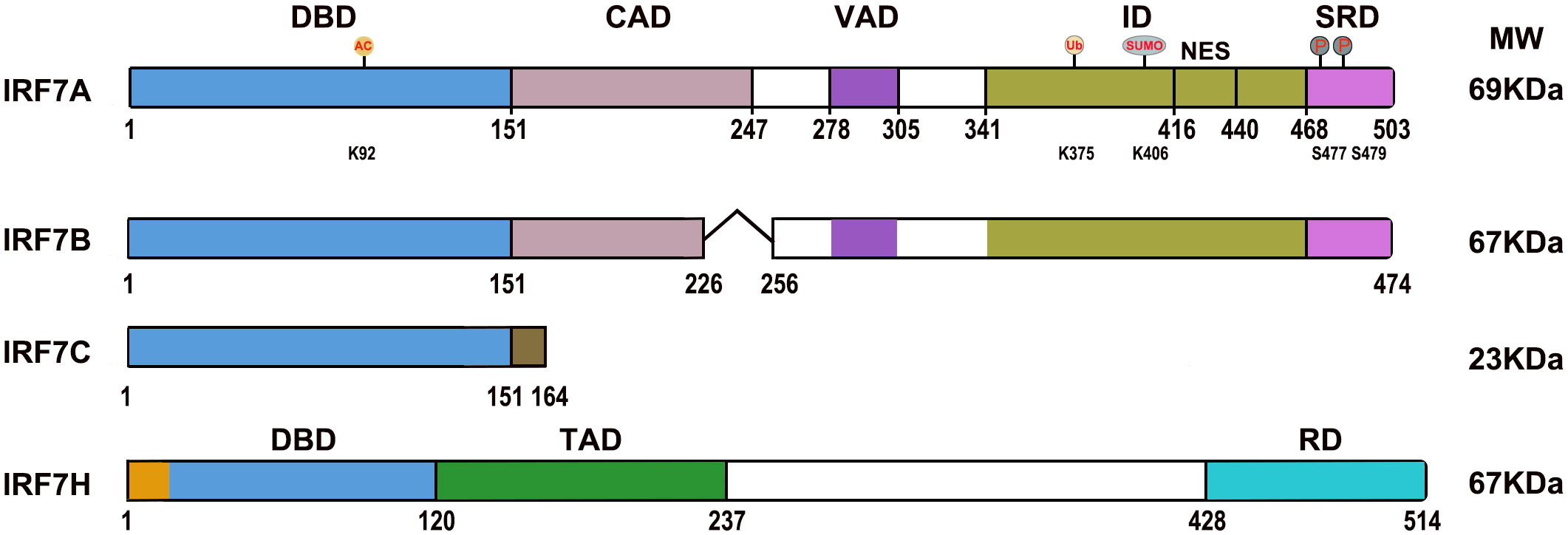
Figure 1 Diagrams of the IRF7 isoform domains. DBD, DNA binding domain; CAD, Constitutive activation domain; VAD, Virus activated domain; ID, Inhibitory domain; NES, Nuclear export signal; SRD, Signal response domain; TAD, Transactivation domain; RD, Regulatory domain. “˄” represents deleted regions. AC, Acetylation; Ub, Ubiquitination; SUMO, SUMOylation; P, Phosphorylation; K92, lysine 92; K375, lysine 375; K406, lysine 406; S477, Serine 477; S479, Serine 479.
IRF7B lacks 29 aa (from G227 to T255) in the middle of the CAD difference from IRF7A. IRF7C consists of 164 amino acid residues and unique 13 aa sequence at the C-terminus difference from IRF 7A; it is not only a dominant negative regulator that blocks the activation of IFN for IRF7A and IRF7B but is also associated with EBV transformation of human primary B cells as well as EBV type III latency and plays a role in the development of lymphoma (10, 12, 14). The spliced variant IRF7H encodes 514 aa and differs from IRF7A by 18 aa at the N-terminal region, displays functional similarity to IRF3, plays an important role in regulating the expression of the IFNA gene (8) (Figure 1).
3 The function of IRF7
IRF7 has a wide range of functions. Herein, we present a review regarding its function and regulation in immunity. We also discuss the latest relevant research in autoimmunity and oncogenesis toward understanding IRF7 beyond its role in immunity.
3.1 IRF7 in immunity
IRF7 is involved in the regulation of mouse and human IFN-I/III, especially IFN-I.
3.1.1 IRF7 and IFN-I
3.1.1.1 IRF7-mediated IFN-I signaling pathway
IFN-I, mainly IFN-α and IFN-β, is a critical pleiotropic cytokine for immunity against the viral response and has been characterized as triggering antiviral states in cells and potentiating adaptive immune responses. IRF7 is the master regulator of IFN-I immune responses, not only regulating further expression of IFN-β but also triggering IFN-α production (15). The specific knockout of IRF7 in mouse pDCs almost loses the ability to produce IFN-α (16, 17), and the deficiency of IRF7 in humans also significantly inhibits the production of IFN-α (18).
The production of IFN-I has been widely reported. Briefly, after viral infection and others, IRF7 in the cytosol is activated by distinct types of innate pattern recognition receptors (PRRs), and the PRRs associated with IRF7 can be classified as cytosolic and transmembrane signaling. The phosphorylation of IRF7 and IRF3 occurs mainly by innate immune cells contacting virus-specific antigenic substances (DNA, RNA) through PRRs and then by intracellular signaling molecules (MAVS, STING, TBK, IKKϵ, etc.). Dimerized IRF7 or IRF3/IRF7 can translocate into the nucleus to initiate expression of the IFN-I gene (Figure 2). Honda and Taniguchi reported that the homodimer of IRF7 or the heterodimer of IRF7/IRF3, rather than the homodimer of IRF3, is more important for the production of IFN-I under viral infection (19).
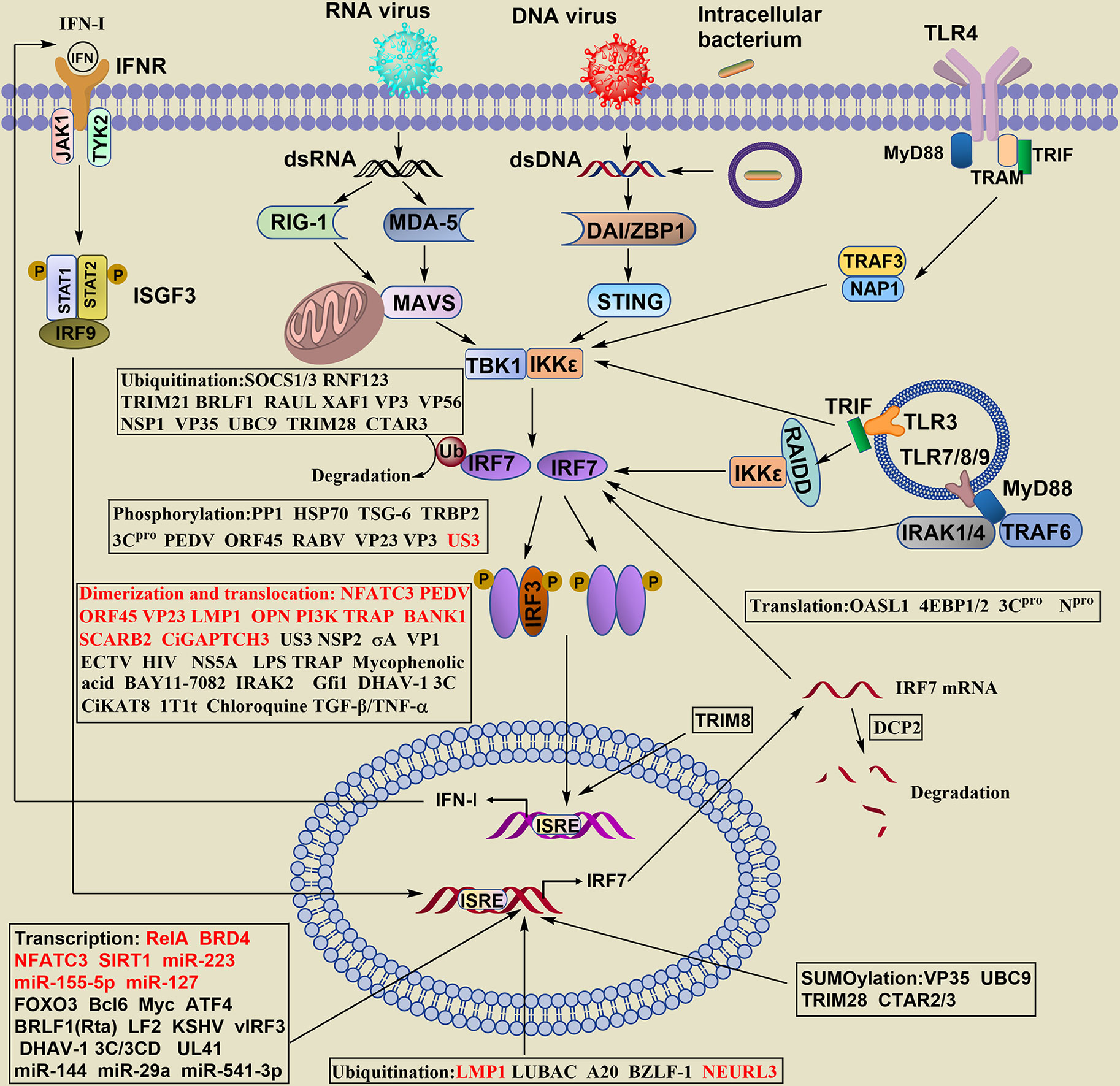
Figure 2 Regulation of IRF7 signaling pathway in the production of IFN- I. The stability of IRF7 can be regulated at the levels of transcription, translation, posttranslation, epigenetics, the dimerization and nuclear translocation. The regulatory levels are highlighted in boxes, words in red of the boxes indicate positive regulators, and words in black show negative regulators.
For cytoplasmic signals, RIG-I or MDA5 recognize viral RNA, DNA sensors also known as DAI (DNA-dependent activator of IRFs) or ZBP1 (Z-DNA-binding protein 1), including DDX (dead-box polypeptide) 41, MRE11 (meiotic recombinant 11 homolog A), IFI (IFN-γ induction) 16 and cGAS (loop CMP-AMP synthase), to detect cytosolic DNA. The induction of IFN-α in cytosolic DNA is required for both IRF3 and IRF7.
Transmembrane Toll-like receptor (TLR) signaling can be divided into MyD88-dependent and TRIF-dependent (MyD88-independent) pathways. TLR7/8/9 is expressed in the membrane of endosomes and phagosomes and employs a MyD88-dependent pathway to induce IFN-I. IRF7 is activated and translocated to the nucleus via the signaling cascade of MyD88-IRAK4-IRAK1-TRAF6, leading to the production of large amounts of IFN-I in pDCs.
TLR3 is mainly localized to endosomes and uses the adaptor protein TRIF to recruit TANK-binding kinase (TBK)1 to endosomes and phagosomes, activating IRF3 and IRF7 to induce the expression of IFN-I. Additionally, the adaptor molecule adaptor molecule RIP associated ICH-1/CED-3-homologous protein with a death domain (RAIDD) interacts with IKKϵ and IRF7 but not TBK1, mediates IRF7 phosphorylation and triggers IFN-I production (20).
TLR4 is located on the cell surface and is the only member in the TLR family that transduces signals by two pathways. The MyD88-dependent pathway requires IRF3 rather than IRF7. Endocytosed TLR4 activates the adaptor protein TRAM-TRIF, recruits TRAF3, NAP1 and TBK1/IKKϵ, and activates IRF7 to induce IFN-β and IFN-α4 but not other IFN-α genes.
IRF7 is required for IFN priming at an early stage, which restricts not only the acute infection of herpesvirus families but also chronic gamma herpesvirus infection. IRF7 suppresses the establishment of latency and viral reactivation in the peritoneal cavity and attenuates viral reactivation in the spleen, promoting the expression of select MHV68-restircting ISGs in peritoneal cells (21).
IRF7 is also required for IFN-I amplification at later stages, and the secondary, larger wave of IFN-I expression depends on the positive regulatory loop between IRF7 and IFN-I. This pathway regulates the transcription of IRF7 (Figure 2).
3.1.1.2 The regulation of IRF7 in IFN-I
For the key role of IRF7 in IFN-I production, the balance between its activation and repression needs to be delicately maintained. The regulation of IRF7 is complex and involves many positive and negative feedback mechanisms. The stability of IRF7 can be regulated by various mechanisms at the transcriptional, translational, posttranslational, epigenetic levels, such as phosphorylation, ubiquitination, SUMOylation, acetylation, et al., and the dimerization and nuclear translocation of IRF7 (Figure 2).
3.1.1.2.1 Transcriptional level
The transcription of IRF7 has two distinct pathways: IFN-triggered and IFN-independent signaling pathways. IFN signals through its receptor to induce the phosphorylation of STAT1 and STAT2, which results in the formation of ISGF3 and then promotes the transcription of IRF7 by binding directly to the IRF7 interferon (IFN)-stimulated response element (ISRE) and IRF-binding element (IRFE), which is IFN-triggered signaling (22, 23). Virus-induced formation of a virus-activated factor complex (formed by IRF7, IRF3 and p300/CREB-binding protein) directly binds to the IRF7 ISRE and IRFE and stimulates the intrinsic transcriptional activity of IRF7, and this induction is independent of the IFN-triggered pathway (24, 25). Some factors can regulate IRF7 transcription positively or negatively.
Phosphorylated RelA and bromodomain containing (BRD)4 induced by respiratory syncytial virus (RSV) can bind to the IRF7 promoter, triggering the IRF7-RIG-1 amplification loop for IFN-I/III expression (26, 27). Transcription factor nuclear factor of activated T cells (NFATC) 3 selectively binds to the autoinhibitory domain (373-443 aa) of IRF7, promotes transcription and nuclear translocation, and enhances CpG DNA–induced IFN-α production in pDCs (28).
FOXO3 is a negative regulator of IRF7 transcription, and the ternary complex consisting of FOXO3, nuclear corepressor 2 (NCOR2) and histone deacetylase 3 (HDAC3) on the Irf7 promoter enhances the closed chromatin structure and represses Irf7 expression and Ifnb1 production (29). B-cell lymphoma (Bcl)6, interacting with NCOR2 and HDAC3, binds to IRF7 loci and restrains its transcription (30). Myc forms a repressor complex together with NOCR2 and HDAC3 to reduce the expression of IRF7 through histone deacetylation (31). Activating transcription factor (ATF) 4 inhibits the transcription of IRF7 by regulating its promoter. However, IRF7 increases the expression and function of ATF4 directly, and cross-regulation between IFN and integrated stress responses is mediated by the reverse correlation between ATF4 and IRF7 (16).
Viral derived proteins also inhibit the transcription of IRF7, such as Epstein-Barr virus immediate early protein BRLF1 (Rta) and LF2, Kaposi´s sarcoma-associated herpesvirus (KSHV) viral homologs of IRF3 (vIRF3), duck hepatitis A virus type 1 (DHAV-1) 3C and 3CD protein, and duck enteritis virus tegument protein UL41 (17, 32–35).
Elevated expression of the decapping enzyme Dcp2 induced by viral infection and double-stranded RNA treatment inhibited IRF7 mRNA stability and protein accumulation (36).
MicroRNAs regulate IRF7 expression by modulating mRNA stability. miR-223 and miR-155-5p enhance IRF7 expression by targeting FOXO3 (37, 38), miR-127 promotes IRF7 expression by inhibiting the signal-dependent turnover of the Bcl6 coregulator (30), miR-144 negatively regulates IRF7 activity by suppressing the TRAF6-IRF7 signaling axis (39), and miR29a inhibits the expression of IRF7 and TRAF6 (40). miR-541-3p promotes the replication of porcine reproductive and respiratory syndrome virus (PRRSV) 2 by negatively regulating the transcription of IFN-I by targeting IRF7 (41).
3.1.1.2.2 Methylation
As a major epigenetic modification, CpG hypermethylation in the promoter region can lead to the inactivation of specific gene expression and function. Zhang Y et al. checked the 5-methylcytosine level of the CpG island infected by infectious bursal disease virus (IBDV) and found that CpG island methylation in the IRF7 promoter regions was substantially decreased (42). Rezaei R analyzed the methylation status of CpG sites of the IRF7 promoter in peripheral blood mononuclear cells (PBMCs) of systemic sclerosis (SSc) patients and found that hypomethylated CpG2 was associated with increased disease risk (43). The zinc finger CXXC family epigenetic regulator CXXC5 maintains the hypomethylation states of the CpG-containing island (CGI) within the Irf7 gene by recruiting the DNA demethylase Tet2 (44). Cigarette smoke condensate decreases the expression of IRF7 mRNA with increased CpG methylation of its promoter region in PBMCs (45).
3.1.1.2.3 Translational level
Although the regulation of transcription levels can control gene expression from the source, the slower nuclear pathways for mRNA synthesis and transport affect the effect. Translation control of gene expression enables cells to respond quickly to external and internal triggers or clues, and translational control of IRF7 is vital to the induction of IFN-I production.
By binding to the 5’ untranslated region (UTR) of IRF7, 2ʹ-5ʹ-oligoadenylate synthase-like protein 1 (OASL1) prevents the loaded 43S preinitiation complex from passing the UTR and inhibits the translation of Irf7 mRNA. The translational repressors 4E-BP1 and 4E-BP2 also suppress the translation of Irf7 mRNA by recognizing the 5’UTR, negatively regulating the production of IFN-I (46, 47).
The 3C protein of enterovirus 71 mediates the cleavage of IRF7 at the Q189-S190 site in the CAD domain and inhibits IRF7 function (48).
Npro of classical swine fever virus (CSFV) interacts with IRF7 through its Zn-binding domain and negatively regulates IRF7 protein expression but not mRNA expression in pDCs (49).
3.1.1.2.4 Posttranslational modification of IRF7
Posttranslational modifications (PTMs) are important epigenetic mechanisms regulating various biological processes, with the key advantages being that PTMs regulate faster and require less energy than protein regulation. These modifications include phosphorylation, acetylation, ubiquitylation, SUMOylation, and neddylation.
After pathogenic infection, IRF7 changes from an inactive to activated state by phosphorylation, and the activation of IRF7 requires two phosphorylation events. IRF7 needs at least one phosphorylation event at S477, S479, S483 and S487 in addition to phosphorylation of both S471 and S472 (50).
The phosphorylation of IRF7 is linked to PRR pathways. TLR7/8 antagonists reduce Helicobacter pylori infection-mediated IRF7 phosphorylation (51). Kinases such as IKKϵ, TBK1, IRAK1 and IKKα are responsible for the phosphorylation of IRF7 in a cell type-specific manner (52, 53).
In addition to the direct signaling kinases that regulate the phosphorylation of IRF7, other molecules may act by regulating these signaling molecules.
The phosphatase protein phosphatase (PPP)1 can target four key phosphorylation sites (Ser471, -472, -477 and -479) of IRF7, attenuate the phosphorylation level and DNA-binding activity stimulated by IKKϵ, and inhibit IRF7-mediated IFN-I immune responses (54). HSP70 interferes with IKKϵ-mediated IRF7 phosphorylation via simple competition for IRF7 binding (55), and the anti-inflammatory factor TNF-a–stimulated gene 6 (TSG-6) downregulates IRF7 activity by suppressing its phosphorylation mediated by CD44 (56). TARBP2 promotes the K48-linked polyubiquitination and proteasome-dependent degradation of TRAF6 and weakens the interaction between TRAF6 and IRF7, thus inhibiting the phosphorylation of IRF7 (57).
Viral proteins can interact with IRF7 and suppress its phosphorylation, dimerization and nuclear translocation. For example, Seneca Valley Virus (SVV) protein 3Cpro (58), the 1-55 region of porcine epidemic diarrhea virus (PEDV) membrane protein (59), Kaposi´s sarcoma-associated herpesvirus (KSHV) immediate-early proteins ORF45 (60), the negative single stranded RNA rabies virus (RABV) phosphoprotein P (61–63), and Marek´s disease virus VP23 protein (64), enterovirus D68 VP3 (65) et al. Unique-short kinase (Us)3 of Marek´s disease virus or duck enteritis virus can induce IRF7 phosphorylation by interacting with its activation domain but suppress dimerization and nuclear translocation (66, 67).
The acetylation of transcription factors can affect DNA binding affinity, stability, degradation and protein-protein interactions (68). IRF7 is subject to acetylation by the histone acetyltransferase (HAT) of the PCAF (p300/CBP Associated Factor)/GCN5 family. The unique lysine residue target for acetylation is located in the DNA binding domain at lysine 92 (K92), and acetylation inhibits IRF7 DNA binding and decreases transcriptional activity (69) (Figure 1).
The acetyltransferase KAT8 in grass carp (CiKAT8) directly interacts with IRF3/7 via the acetyltransferase domain (CiKAT8-∆264-487), blocks the interaction between IRF3/7 and ISRE, and inhibits IFN-1 expression (70).
Liquid–liquid phase separation (LLPS) is a key mechanism for transcriptional regulation. NAD-dependent protein deacetylase sirtuin-1 (SIRT1) directly deacetylates K39 and K77 on IRF3, and K45 and K92 on IRF7 are located in the DBD, promoting LLPS of IRF3/7 and enhancing IFN-1 transcription (71).
Ubiquitination can either activate or inactivate/degrade IRF7. Nondegradative ubiquitination is involved in the activation of IRF7 and is a prerequisite for its phosphorylation. Epstein-Barr virus latent membrane protein (LMP) 1 promotes the phosphorylation and nuclear translocation of IRF7 through ubiquitination (72, 73). IRF7 binds to the ISRE of the LMP1 promoter and forms a positive regulatory loop between IRF7 and LMP1 (74). The linear ubiquitin assembly complex (LUBAC) of the LMP1 downstream target, antiapoptotic factor deubiquitinase A20 induced by LMP1, and EB virus BZLF-1 negatively regulate the transcriptional activity of IRF7 via LMP1-stimulated IRF7 ubiquitination (75–77). The E3 ubiquitin ligase NEURL3 ubiquitinates the K63-linked polyubiquitin chain on IRF7 lysine 375 (K375) (Figure 1), impairs the association of IRF7 with histone deacetylase (HDAC)1, and enhances the transcription of IFN-I and ISGs (78).
In addition to activating IRF7, ubiquitin-mediated degradation of IRF7 is an effective mechanism to regulate its activity. Suppressor of cytokine signaling (SOCS) 1/3 mediates the degradation of IRF7 by lysine 48-linked polyubiquitination through the SH2 domain and suppresses IFN-I production (79). Ring finger protein (RNF)123 promotes TLR-3- and IRF7-mediated IFN-I expression by ubiquitination and proteasomal degradation of SOCS1 (80). TRIM21 (Ro52) cooperates with the Fas-associated death domain (FADD) to enhance TRIM21 ubiquitin ligase activity, promote the ubiquitination and degradation of IRF7, and repress its phosphorylation and transcriptional activities. FADD and TRIM21 constitute a negative feedback loop of the IFN-α pathway (81). RAUL (RTA-associated ubiquitin ligase) directly catalyzes lysine 48-linked polyubiquitination of IRF7, promotes ubiquitin-proteasome dependent proteolysis (82). and the XAF1-XIAP-KLHL22 axis inhibits IFN-I induction through CUL3-KLHL22, which directly ubiquitinates IRF7 (83).
In addition to inhibiting the phosphorylation and nuclear translocation of IRF7, enterovirus D68 VP3 disturbs the TRAF6-triggered K63-ubiquitination of IRF7 and blocks its activation (65). KSHV BRLF1 (Rta) promotes polyubiquitination and proteosome-mediated degradation of IRF7 (84), Rotavirus nonstructural protein NSP1, acting as a putative E3 ubiquitin ligase, induces proteasome-mediated degradation of IRF7 and suppresses the expression of IFN-β (85).Grass carp reovirus (GCRV) VP56 promotes the K48-linked ubiquitination of phosphorylated IRF7, inhibits IFN expression (86).
Small ubiquitin-like modifier (SUMO) is a ubiquitin-like small protein that can be conjugated onto target proteins to increase the functional repertoire of the eukaryotic proteome (87). TLR and RIG-I/MDA-5 signaling mediate the SUMOylation of IRF7 but not the IFN-activated JAK/STAT pathway in response to vesicular stomatitis virus (VSV) or encephalomyocarditis virus (EMCV) infection. SUMOylation is independent of phosphorylation, and lysine 406 (K406) of IRF7 is the SUMO conjugation site (88) (Figure 1). The encoded protein VP35 by filoviruses, such as Ebola virus and Marburg virus, not only interacts with IKK-ϵ and TBK-1 to block the activation of IRF7 but also promotes PIAS1-mediated SUMOylation of IRF7 to inhibit the transcription of the IFN gene (89, 90). The LMP1-mediated SUMO conjugating enzyme UBC9 not only promotes SUMOylation but also negatively regulates the chromatin binding of IRF7. TRIM28, the IRF7-specific SUMO E3 ligase, increased the SUMOylation of IRF7 during viral infections, inhibiting its transcriptional activity (91). Bentz G et al. showed that LMP1 C-terminal activating region (CTAR)3, in cooperation with CTAR2, induces the SUMOylation of IRF7 and decreases its turnover rate and transcriptional activity (88, 92).
Neddylation is a ubiquitin-like posttranslational protein modification that is indispensable for the production of RNA virus-induced IFN-α. IRF7 was identified as the neddylation substrate, and neddylation was first detected in zebrafish (93). Neddylation cannot promote RNA virus-induced IRF7 expression but enhances the transcriptional activity and nuclear translocation of IRF7 (94).
3.1.1.2.5 Dimerization and nuclear translocation of IRF7
After phosphorylation, dimerized IRF7 in the nucleus can bind to the promoter regions of target genes to activate transcription with the help of other coactivators, and the regulation of dimerization and translocation of IRF7 is a critical step in the production of IFN-I.
In addition to NFATC3, PEDV membrane protein, ORF45, VP23 and LMP1 promote the dimerization and nuclear translocation of IRF7, and other viral proteins, such as Us3, nonstructural protein(NSP) 2 of H1N1 influenza A virus, avian reovirus σA protein and the structural protein VP1 of chicken anemia virus, decrease the dimerization and nuclear translocation of IRF7 (95–97).
Viral infection affects the nuclear transport of IRF7. Infection with Ectromelia virus (ECTV) decreases the nuclear translocation of NF-κB, IRF3 and IRF7 in murine GM-CSF-derived bone marrow cells (98). HIV suppresses AKT phosphorylation to inhibit the translocation of IRF7 into the pDC nucleus (99). In HCV-positive hepatoma cells, stimulation with the TLR7 ligand increases IRF7 nuclear translocation (100). In contrast, in hepatocytes, HCV infection disturbs the phosphorylation of STAT1, blocks the nuclear translocation of IRF7 through the NS5A protein and inhibits the expression of IFN-α (101, 102). LPS, the major component of the outer membrane of gram-negative bacteria, suppresses virus-mediated phosphorylation and nuclear translocation of IRF3/IRF7 (103).
The components of the signal transduction pathway can regulate the nuclear translocation of IRF7. Intracellular osteopontin (OPN) and PI3K selectively promote IRF7 nuclear translocation and subsequent type I IFN production (104, 105). Tartrate-resistant acid phosphatase (TRAP) decreases the phosphorylation of OPN and then blocks the nuclear translocation of IRF7 and p65 (106). Mycophenolic acid, the metabolic product of mycophenolate mofetil, inhibits IRF7 nuclear translocation and IFN-α production by suppressing PI3K-AKT activity (107). An inhibitor of the IKK complex, BAY11, (E)-3-(4-methylphenylsulfonyl)-2-propenenitrile (BAY11-7082), has the ability to repress the nuclear translocation of IRF7 and IFN-α production (108). IL-1R-associated kinase (IRAK) 2 decreases the nuclear translocation of IRF7 in response to stimulation with TLR9 ligands (109). The transcriptional repressor growth factor independence 1 (Gfi1) prevents spontaneous SLE by negatively regulating TLR7 signaling and nuclear translocation of IRF7 in DCs (110). B-cell scaffold with ankyrin repeats (BANK)1 can increase the expression and nuclear translocation of IRF7 upon TLR7 stimulation in B cells and promote IgG production in autoimmune disease (111). Scavenger receptor class B member 2 (SCARB2) deficiency in pDCs impairs nuclear translocation of IRF7 and decreases endosomal translocation of TLR9 (112). Some small molecules affect the nuclear translocation of IRF7 in pDCs. 1T1t, the small molecule CXCR4 ligand, inhibits TLR-7 mediated IFN-α production through blocking the phosphorylation and nuclear translocation of IRF7 (113). Chloroquine, an endosomal inhibitor, blocks TLR signaling, decreases the expression and nuclear translocation of IRF7 and production of IFN-α (114, 115). TGF-β and TNF- α synergistically impaired IFN- α production of TLR-activated pDC through blocking the expression and nuclear translocation of IRF7 (116).
Additionally, cord blood pDCs do not function as their adult counterparts, especially in terms of the defect in IFN production, due to the deficiency in IRF7 nuclear translocation (117). Toxoplasma gondii inhibits virus-induced nuclear translocation of IRF7 via the tyrosine kinase ROP16 in pDCs (118). Ctenopharyngodon Idella (Ci) GAPTCH3 directly interacts with CiSTING and enhances the phosphorylation and nuclear translocation of CiIRF7 (119), and the DHAV-1 3C protein inhibits the nuclear translocation of IRF7 (35).
In the nucleus, TRIM8 stabilizes phosphorylated IRF7 and protects it from peptidyl-prolyl isomerase Pin1-induced degradation in primary pDCs (120).
3.1.2 IRF7 and IFN-IIIs
IFN-IIIs are also called lambda IFNs (IFNλs), produced by cells of hematopoietic origin or epithelia at barrier surfaces, and include four members in humans (IFNλ1 or IL-29, IFNλ2 or IL-28A, IFNλ3 or IL-28B, IFNλ4) and two in mice (IFNλ2 or IL-28A, IFNλ3 or IL-28B). IFN-III genes are located on chromosome 19 in humans and chromosome 7 in mice, though IFN-IIIs share homology, expression patterns, antiviral functions and signaling cascades with IFN-Is, some features distinguish the two IFNs: (i) the initiation time, IFN-IIIs are the earliest and predominant IFNs during virus infection, mediate front-line antiviral defense without activating inflammation, while IFN-Is come up later to enhance viral resistance and proinflammatory responses (121, 122). (ii) the distinct heterodimeric receptors, the IFNAR receptor complex (IFNAR1 and IFNAR2) is ubiquitously expressed, and is bound to IFN-Is, while the IFNLR receptor complex (IFNLR1 and IL10Rβ) is confined expressed on epithelial cells and a subset of myeloid lineage leukocytes, and is bound to IFN-IIIs (122). (iii) the signaling pathway, in addition to activating the same JAK1 as IFN-Is, IFN-IIIs also activate JAK2, and MAPKs are required for the antiviral activity of IFN-IIIs but not IFN-Is (123); contrary to mitochondrial MAVS for activate IFN-Is, peroxisome MAVS induces IFN-IIIs expression only.(iv) the signaling kinetics of ISG induction, IFN-Is mediate rapid and short-lived expression of ISGs with higher magnitude, while IFN-IIIs induce persistent expression of ISGs with lower magnitude (124).
In addition to IFN-I, IRF7 is also involved in IFN-III, IFNλ1 gene is regulated by virus-activated IRF3 and IRF7 like IFN-β gene is, while IFNλ2/3 gene is controlled by IRF7 like IFN-α gene is (125), and IRF7-mediated IFN-III plays a major role in antiviral protection of epithelial barriers due to restricted expression of the receptors (126, 127).
IRF7 mediates the induction of IFN-III caused by viral infection. Senecavirus A (SVA) is recognized by RIG-1/MDA-5 receptors in porcine kidney PK-15 cells and then activates downstream IRF7- but not IRF3-mediated signaling pathways, causing the upregulation of IFN-λ1, IFN-λ3 and related ISGs (126). 5-aza-2-deoxycytidine (5-AZA-CdR), a DNA-demethylating agent, induces IFN-III but not IFN-I production by the MDA5/MAVS/IRF7-dependent or JAK-dependent “viral mimicry” pathway (128). Rotavirus UK-like UKtc NSP1 reduces IRF7-dependent transcription at high IRF7 concentrations and inhibits IFN-III induction in intestinal epithelial cells (129).
With a synergistic interaction between IRF7 and p65 at the TA-repeat polymorphism (rs72258881) of the IFN-λ3 promoter, IRF7 is sensitive to changes in DNA phasing and mediates the transcription of IFN-λ3 (130).
Clinically, the upstream gene of IRF7 is reduced in patients with atopic dermatitis with a history of eczema herpeticum (ADEH+) compared with healthy subjects and patients with ADEH- after HSV-1 exposure, thus affecting the expression of IFN-IIIs (131). Med23, a subunit of the Mediator multiprotein complex, is specific to HSV-1 and regulates the induction of IFN-λ by interacting with and enhancing the activity of IRF7 (132).
3.1.3 IRF7 and COVID-19
The role of IRF7 in COVID-19 is conflicting. On the one hand, IRF7 is protective against viral infection. Campbell TM et al. reported that IRF7-deficient patients are prone to severe respiratory viral infections, with influenza and COVID-19, due to impaired type I and III IFN expression in both pDCs and respiratory epithelial cells (133–135). Patients with severe COVID-19 at early time points show decreased levels of type I and III IFN (136). TLR3- and TLR7-dependent production of IFN-I by pDCs and respiratory epithelial cells is essential for host defense against SARS-CoV-2 (137). Zhang et al. sequenced the genome or exome of 659 patients with COVID-19 to test 118 rare nonsynonymous variants of 13 human loci that underlie life-threatening influenza pneumonia. Autosomal-recessive and autosomal-dominant deficiencies of IRF7 are involved in the TLR3- and IRF7-dependent induction and amplification of IFN-I (138), and TLR7 together with IRF7 combat COVID-19 by the large amounts of IFN-I produced by pDCs (139). However, Povysil G et al. declared that they did not observe the enrichment of predicted loss-of-function (pLOF) variants in severe cases relative to population controls or mild COVID-19 cases postulated by Zhang et al. (140). The high expression of SCV2-miR-ORF1ab-2-5p, one of the four unique microRNA-like small RNAs encoded by SARS-CoV-2, inhibits the host IFN response by targeting IRF7 and IRF9 (141).
However, IRF7 may exacerbate the progression of COVID-19. IFN-I, IRF7 and ISGs are highly expressed in the oropharyngeal cells of SARS-CoV-2-positive patients (142), and strong IFN- I responses in patients with severe COVID-19 (143), for IFN- I may aggravate TNF- and IL-1-driven inflammation. With the analysis of the most frequent comorbidities in COVID-19, the authors found that the hub protein IRF7 is upregulated in COVID-19 patients, which is associated with the pathogenesis of diabetes mellitus and lung cancer (144). IRF7 is strongly hypomethylated in SARS-CoV-2 individuals, and IRF7 DNA methylation signatures may differentiate patients with SARS-CoV-2 infection from uninfected individuals and predict COVID-19 disease severity (145). The role of IFN-1 in COVID-19 may depend on mild versus severe and early versus late disease.
3.2 IRF7 and autoimmune diseases
Autoimmune diseases refer to diseases caused by the body’s immune response to its own antigen and damage to its particular tissue or system, which can be classified as systemic autoimmune disease (SAD) and organ-specific autoimmune disease. Upregulation of IFN-I is a hallmark of SAD, and the continuous increase in IFN-I/III may be accompanied by clinical manifestations and disease activity (146, 147). As the master regulator of IFN-I/III, IRF7 has a dual role as a protector and cause of autoimmune diseases. In published articles, decreased expression was observed in multiple sclerosis (MS)/experimental autoimmune encephalomyelitis (EAE) and rheumatoid arthritis (RA), while increased expression of IRF7 was observed in patients with systemic lupus erythematosus (SLE), systemic sclerosis (SSc), autoimmune pancreatitis (AIP), autoimmune thyroid diseases (AITD) and diabetes compared to healthy controls (Table 1).
3.2.1 IRF7 and MS/EAE
MS is a myelin-specific chronic inflammatory autoimmune disease. IRF7 is reduced in pDCs from patients with relapsing-remitting MS compared with healthy controls (148). In EAE, an animal model for MS, IRF7 deficiency resulted in more severe EAE, more infiltrating macrophages and T cells, and elevated levels of CCL2, CXCL10, IL-1β and IL17. The decreased expression of IRF7 represents a destructive function in MS/EAE (149).
3.2.2 IRF7 and RA
RA is characterized by persistent synovitis, systemic inflammation, and autoantibodies. In an arthritis model, IRF7 deficiency exacerbates the clinical severity, proinflammatory cytokines are increased, anti-inflammatory cytokine IFN-β is decreased, and IRF7 regulates the expression of IFN-β in murine macrophages but not in stromal fibroblast-like synoviocytes (150).
3.2.3 IRF7 and SLE
SLE is characterized by the production of a variety of autoantibodies, complement activation and immune complex deposition, resulting in tissue and organ damage. Accumulating evidence implies that IRF7 is a susceptibility locus for SLE. IRF7 mRNA expression is significantly increased in SLE patients, and genetic polymorphisms near/in IRF7 have been substantiated to be related to the onset of SLE. Single nucleotide polymorphisms (SNPs) (rs191491376, rs1131665, rs1061501, rs4963128, rs702966, rs2246614) were found to be associated with SLE susceptibility, although some conflicting results were reported for the genetic heterogeneity between these populations (151–158). SLE patients treated with autologous stem cell transplantation show that high expression of IRF7 is correlated with recurrent lupus disease activity (159). For epigenetic modification, significant hypomethylation is observed in the IRF7 methylated site (160, 161). As mentioned above, TRAP and Gfi1 prevent susceptibility to SLE by regulating the nuclear transport of IRF7 (106, 110).
3.2.4 IRF7 and SSc
SSc is a complex multisystem autoimmune disease that is characterized by widespread skin and internal organ fibrosis, immune system dysregulation, and vasculopathy. GWAS confirmed IRFs are genetic susceptibility loci in SSc (162). IRF5 SNP rs2004640 and rs2280714 are identified as a risk factor for SSc in whites and Asians., however rs4728142 is associated with lower IRF5 gene expression, longer survival and milder interstitial lung disease of SSc patients. The expression of IRF8 is decreased in SSc patients (163), IRF8 SNP rs11642873, rs2280381 and rs11117432 exhibit the strongest association with SSc risk (164–166). IRF4 SNP rs9328192 shows protective effect for SSc.
In addition to IRF5/IRF8/IRF4, IRF7 is regarded as a top predicted transcription factor of patients with SSc because increased expression is observed in peripheral blood cells (167, 168). The functional SNP rs1131665 leads to the activation of IRF7, and is associated with SSc risk for the presence of anticentromere autoantibodies (169). The methylation of IRF7 is associated with SSc, among the methylation status of 16 CpG sites at the promoter region of the IRF7 gene, CpG2 is significantly hypomethylated in SSc PBMCs and associated with increased disease risk, a significant difference in IRF7 mRNA expression between CpG8 methylated and unmethylated SSc patients, with four times higher in those who had a methylated CpG8 site than an unmethylated site (43). The mechanism may be that the overexpression of IRF7 in SSc fibrotic skin forms complexes with Smad3, the key component of TGF-β signaling for collagen production and fibrosis, and these complexes mediate fibrosis in dermal fibroblasts (170).
3.2.5 IRF7 and AIP
IRF7 mediates AIP and human type 1 AIP (171). The authors previously reported that IFN-α and IL-33 produced by pDCs drive experimental AIP and human type 1 AIP (172–174). Now they disclosed that IRF7 activation and nuclear translocation is detected in AIP and human type 1 AIP, blockade of IRF7 signaling pathways decreased chronic fibroinflammatory responses via the suppression of pDC-mediated IFN-I and IL-33, the IRF7-IFN-I-IL-33 axis mediates the development of AIP.
3.2.6 IRF7 and AITD
AITD is an organ-specific autoimmune disorder with immune attack on the thyroid and includes Graves’ disease (GD) and Hashimoto’s thyroiditis (HT), clinical hyperthyroidism and hypothyroidism (175, 176). IRF7 is a susceptibility gene for AITD, especially for GD and Graves’ ophthalmopathy. In a Chinese Han cohort, the IRF7 SNPs rs1131665 and rs1061502 were associated with increased susceptibility to GD, while rs1061501 was correlated with ophthalmopathy in GD patients (177). The function and mechanism of IRF7 in AITDs have not been clarified.
3.2.7 IRF7 and diabetes
Type 1 diabetes (T1D) is characterized by autoimmune destruction of pancreatic β-islet cells, and fulminant type 1 diabetes (FT1D) is a subtype of idiopathic diabetes characterized by the absence of both insulitis and diabetes-related antibodies due to the destruction of pancreatic beta cells (178). IRF7 activated by TLR9 can bind to the Foxp3 core promoter and promote its transcriptional activity. The hypermethylated Foxp3 promoter blocks IRF7 binding to Foxp3, impairing the development and function of Tregs in FT1D patients, and deficient Tregs are prone to the development of FT1D (179). Using an IRF7-/- bone marrow chimera model, Lang PA et al. found that the reduced expression of CD8+ T cells in pancreatic β-islet cells decreased the induction of T1D after LCMV infection (180).
In β cells, IFN-α promotes the nuclear translocation of STAT1 and IRF7, and the STAT1-IRF7-MHC I complex axis creates positive feedback through IRF7-STAT2 cascade amplifying signals and promotes the proliferation of CD8+ T cells, accelerating the process of T1D (181).
The IRF7-driven inflammatory network (IDIN) is enriched for viral response genes, and the human chromosome 13q32 locus controlling the IDIN was associated with the risk of T1D at the single nucleotide polymorphism rs9585056. Combined analyses of gene networks and DNA sequence variation implicated IRF7 network genes and their regulatory locus in the pathogenesis of T1D (182).
IRF7 deficiency prevents diet-induced obesity and insulin resistance (183). IRF7 transactivates MCP-1 by binding to its promoter in adipocytes, promoting the development of type 2 diabetes (184).
4 Conclusion and prospects
IFN signaling plays a causal role in host defense against infectious pathogenic organisms, the dysregulation is widely associated with autoimmune diseases, interferonopathy, infection, cancer and others, therefore, selective regulation of the IFN signaling may provide a therapeutic strategy. pDCs are the major producers of IFN- I, making them an appealing target for the treatment of autoimmune diseases. Litifilimab, a selected antibody against Blood dendritic cell antigen (BDCA) 2, a receptor is exclusively expressed on pDCs, shows efficacy in the treatment of SLE and cutaneous lupus erythematosus by decreasing the expression of IFN- I and ISGs (185, 186). IFN- I antagonists, such as anti-IFNα (sifalimumab, rontalizumab), anti- IFNαR(anifrolumab) appear effective in autoimmune disease, and can reduce ISG expression, the expression and nuclear translocation of IRF7 can be suppressed by the IFNα/β-Ab treatment (187–190). JAK inhibitors are approved for autoimmune, allergic diseases and most recently COVID-19 due to their potent efficacy in reducing IFN- I -driven inflammation. Tofacitinib, a pan-JAK inhibitor, used in the treatment of rheumatoid arthritis, ulcerative colitis et al, inhibits IFN- I production by suppressing transcription and nuclear translocation of IRF7, and affects DCs activities. Fedratinib, a JAK2 inhibitor, on one hand, can block new HIV-1 replication in acute HIV- I infection, on the other hand, can upregulate IRF7 transcription and phosphorylation, induce HIV-1 reactivation by an IFN-independent manner (191–193). As mentioned above, IRF7 is the main regulatory factor of IFN-I/III, small molecules can affect the transcription, translation, post translational regulation and nuclear transport of IRF7, thereby regulating the production of IFN. Therefore, IRF7 is expected to become an attractive therapeutic target for IFN-associated diseases.
IRF7 has multiple functions, as the vital step of the signaling pathway in IFN-I/III induction, the function and regulatory mechanism of IRF7 is important, which may help in understanding how to protect the host to reduce viral infection and maintain body balance. On the other hand, due to its tissue- and cell-specific and important role in autoimmune diseases, the relationship between IFN-I antagonists and IRF7, as well as the regulation of IRF7, is crucial for understanding the development of autoimmune diseases.
Although the role of IRF7 as a regulator of immune cell function has been extensively investigated, many questions remain unanswered and require explanation, such as the duality of IRF7’s role in conferring protection or exacerbation in different diseases, and the sophisticated signaling pathways require further elucidation. On the other hand, the role and mechanisms of IRF7 in IFN-III and autoimmunity remain to be explored. In addition, the expression, function and regulation of IRF7 in nonimmune cells remain unexplored, and more functions and mechanisms of IRF7 will be discovered with more in-depth studies in nonimmune cells. A deeper understanding of the precise functions and molecular mechanisms of IRF7 will be important for disease treatment.
Author contributions
The manuscript was conceptualized by QG and WM. WM and QG wrote the manuscript. The figures were designed by GH and drawn by GH and ZW. LW summarized the tables. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by National Natural Science Foundation of China (81772717), the Guizhou Provincial Basic Research Program (Natural Science) [ (2023) 525].
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Zhao G-N, Jiang D-S, Li H. Interferon regulatory factors: at the crossroads of immunity, metabolism, and disease. Biochim Biophys Acta (BBA) - Mol Basis Dis (2015) 1852(2):365–78. doi: 10.1016/j.bbadis.2014.04.030
2. Huang B, Qi ZT, Xu Z, Nie P. Global characterization of interferon regulatory factor (Irf) genes in vertebrates: glimpse of the diversification in evolution. BMC Immunol (2010) 11:22. doi: 10.1186/1471-2172-11-22
3. Suzuki Y, Yasuike M, Kondo H, Aoki T, Hirono I. Molecular cloning and expression analysis of interferon regulatory factor 10 (Irf10) in Japanese flounder, paralichthys olivaceus. Fish Shellfish Immunol (2011) 30(1):67–76. doi: 10.1016/j.fsi.2010.09.010
4. Tamura T, Yanai H, Savitsky D, Taniguchi T. The irf family transcription factors in immunity and oncogenesis. Annu Rev Immunol (2008) 26:535–84. doi: 10.1146/annurev.immunol.26.021607.090400
5. Savitsky D, Tamura T, Yanai H, Taniguchi T. Regulation of immunity and oncogenesis by the irf transcription factor family. Cancer immunology immunother: CII (2010) 59(4):489–510. doi: 10.1007/s00262-009-0804-6
6. Yanai H, Negishi H, Taniguchi T. The irf family of transcription factors: inception, impact and implications in oncogenesis. Oncoimmunology (2012) 1(8):1376–86. doi: 10.4161/onci.22475
7. Qing F, Liu Z. Interferon regulatory factor 7 in inflammation, cancer and infection. Front Immunol (2023) 14:1190841. doi: 10.3389/fimmu.2023.1190841
8. Au WC, Moore PA, LaFleur DW, Tombal B, Pitha PM. Characterization of the interferon regulatory factor-7 and its potential role in the transcription activation of interferon a genes. J Biol Chem (1998) 273(44):29210–7. doi: 10.1074/jbc.273.44.29210
9. Ning S, Pagano JS, Barber GN. Irf7: activation, regulation, modification and function. Genes Immun (2011) 12(6):399–414. doi: 10.1038/gene.2011.21
10. Zhang L, Pagano JS. Irf-7, a new interferon regulatory factor associated with epstein-barr virus latency. Mol Cell Biol (1997) 17(10):5748–57. doi: 10.1128/mcb.17.10.5748
11. Lin R, Mamane Y, Hiscott J. Multiple regulatory domains control irf-7 activity in response to virus infection. J Biol Chem (2000) 275(44):34320–7. doi: 10.1074/jbc.M002814200
12. Zhang L, Pagano JS. Structure and function of irf-7. J Interferon Cytokine research: Off J Int Soc Interferon Cytokine Res (2002) 22(1):95–101. doi: 10.1089/107999002753452700
13. Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, et al. Distinct and essential roles of transcription factors irf-3 and irf-7 in response to viruses for ifn-alpha/beta gene induction. Immunity (2000) 13(4):539–48. doi: 10.1016/s1074-7613(00)00053-4
14. Zhao Y, Xu D, Jiang Y, Zhang L. Dual functions of interferon regulatory factors 7c in epstein-barr virus-mediated transformation of human B lymphocytes. PloS One (2010) 5(3):e9459. doi: 10.1371/journal.pone.0009459
15. Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, et al. Irf-7 is the master regulator of type-I interferon-dependent immune responses. Nature (2005) 434(7034):772–7. doi: 10.1038/nature03464
16. Liang Q, Deng H, Sun CW, Townes TM, Zhu F. Negative regulation of irf7 activation by activating transcription factor 4 suggests a cross-regulation between the ifn responses and the cellular integrated stress responses. J Immunol (Baltimore Md: 1950) (2011) 186(2):1001–10. doi: 10.4049/jimmunol.1002240
17. Bentz GL, Liu R, Hahn AM, Shackelford J, Pagano JS. Epstein-barr virus brlf1 inhibits transcription of irf3 and irf7 and suppresses induction of interferon-beta. Virology (2010) 402(1):121–8. doi: 10.1016/j.virol.2010.03.014
18. Zhou B, Yang W, Li W, He L, Lu L, Zhang L, et al. Zdhhc2 is essential for plasmacytoid dendritic cells mediated inflammatory response in psoriasis. Front Immunol (2021) 11:607442. doi: 10.3389/fimmu.2020.607442
19. Honda K, Taniguchi T. Irfs: master regulators of signalling by toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol (2006) 6(9):644–58. doi: 10.1038/nri1900
20. Maney SK, Xu HC, Huang J, Pandyra AA, Ehlting C, Aguilar-Valenzuela R, et al. Raidd mediates tlr3 and irf7 driven type I interferon production. Cell Physiol Biochem (2016) 39(4):1271–80. doi: 10.1159/000447832
21. Johnson KE, Aurubin CA, Jondle CN, Lange PT, Tarakanova VL. Interferon regulatory factor 7 attenuates chronic gammaherpesvirus infection. J Virol (2020) 94(24):e01554-20. doi: 10.1128/jvi.01554-20
22. Marié I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J (1998) 17(22):6660–9. doi: 10.1093/emboj/17.22.6660
23. Sato M, Hata N, Asagiri M, Nakaya T, Taniguchi T, Tanaka N. Positive feedback regulation of type I ifn genes by the ifn-inducible transcription factor irf-7. FEBS Lett (1998) 441(1):106–10. doi: 10.1016/s0014-5793(98)01514-2
24. Ning S, Huye LE, Pagano JS. Regulation of the transcriptional activity of the irf7 promoter by a pathway independent of interferon signaling. J Biol Chem (2005) 280(13):12262–70. doi: 10.1074/jbc.M404260200
25. Yang H, Lin CH, Ma G, Baffi MO, Wathelet MG. Interferon regulatory factor-7 synergizes with other transcription factors through multiple interactions with P300/cbp coactivators. J Biol Chem (2003) 278(18):15495–504. doi: 10.1074/jbc.M212940200
26. Fang L, Choudhary S, Tian B, Boldogh I, Yang C, Ivanciuc T, et al. Ataxia telangiectasia mutated kinase mediates nf-kappab serine 276 phosphorylation and interferon expression via the irf7-rig-I amplification loop in paramyxovirus infection. J Virol (2015) 89(5):2628–42. doi: 10.1128/JVI.02458-14
27. Tian B, Yang J, Zhao Y, Ivanciuc T, Sun H, Garofalo RP, et al. Brd4 couples nf-kappab/rela with airway inflammation and the irf-rig-I amplification loop in respiratory syncytial virus infection. J Virol (2017) 91(6):e00007-17. doi: 10.1128/JVI.00007-17
28. Bao M, Wang Y, Liu Y, Shi P, Lu H, Sha W, et al. Nfatc3 promotes irf7 transcriptional activity in plasmacy–toid dendritic cells. J Exp Med (2016) 213(11):2383–98. doi: 10.1084/jem.20160438
29. Litvak V, Ratushny AV, Lampano AE, Schmitz F, Huang AC, Raman A, et al. A foxo3-irf7 gene regulatory circuit limits inflammatory sequelae of antiviral responses. Nature (2012) 490(7420):421–5. doi: 10.1038/nature11428
30. Xu F, Kang Y, Zhuang N, Lu Z, Zhang H, Xu D, et al. Bcl6 sets a threshold for antiviral signaling by restraining irf7 transcriptional program. Sci Rep (2016) 6:18778. doi: 10.1038/srep18778
31. Kim TW, Hong S, Lin Y, Murat E, Joo H, Kim T, et al. Transcriptional repression of ifn regulatory factor 7 by myc is critical for type I ifn production in human plasmacytoid dendritic cells. J Immunol (Baltimore Md: 1950) (2016) 197(8):3348–59. doi: 10.4049/jimmunol.1502385
32. Gao L, Liu R, Yang F, Li X, Liu C, Qi X, et al. Duck enteritis virus inhibits the cgas-sting DNA-sensing pathway to evade the innate immune response. J Virol (2022) 96(24):e0157822. doi: 10.1128/jvi.01578-22
33. Joo CH, Shin YC, Gack M, Wu L, Levy D, Jung JU. Inhibition of interferon regulatory factor 7 (Irf7)-mediated interferon signal transduction by the kaposi’s sarcoma-associated herpesvirus viral irf homolog virf3. J Virol (2007) 81(15):8282–92. doi: 10.1128/jvi.00235-07
34. Wu L, Fossum E, Joo CH, Inn KS, Shin YC, Johannsen E, et al. Epstein-barr virus lf2: an antagonist to type I interferon. J Virol (2009) 83(2):1140–6. doi: 10.1128/JVI.00602-08
35. Xia X, Cheng A, Wang M, Ou X, Sun D, Zhang S, et al. Dhav 3cd targets irf7 and rig-I proteins to block the type I interferon upstream signaling pathway. Vet Res (2023) 54(1):5. doi: 10.1186/s13567-023-01134-4
36. Li Y, Dai J, Song M, Fitzgerald-Bocarsly P, Kiledjian M. Dcp2 decapping protein modulates mrna stability of the critical interferon regulatory factor (Irf) irf-7. Mol Cell Biol (2012) 32(6):1164–72. doi: 10.1128/MCB.06328-11
37. Chen L, Song Y, He L, Wan X, Lai L, Dai F, et al. Microrna-223 promotes type I interferon production in antiviral innate immunity by targeting forkhead box protein O3 (Foxo3). J Biol Chem (2016) 291(28):14706–16. doi: 10.1074/jbc.M115.700252
38. Yang D, Wang X, Gao H, Chen B, Si C, Wang S. Downregulation of mir-155-5p facilitates enterovirus 71 replication through suppression of type I ifn response by targeting foxo3/irf7 pathway. Cell Cycle (Georgetown Tex) (2020) 19(2):179–92. doi: 10.1080/15384101.2019.1704512
39. Lopez CB, Rosenberger CM, Podyminogin RL, Diercks AH, Treuting PM, Peschon JJ, et al. Mir-144 attenuates the host response to influenza virus by targeting the traf6-irf7 signaling axis. PloS Pathog (2017) 13(4):e1006305. doi: 10.1371/journal.ppat.1006305
40. Shah AU, Cao Y, Siddique N, Lin J, Yang Q. Mir29a and mir378b influence cpg-stimulated dendritic cells and regulate cgas/sting pathway. Vaccines (2019) 7(4):197. doi: 10.3390/vaccines7040197
41. Shi X, Yang Y, Zhang X, Chang X, Chen J, Wang C, et al. Mir-541-3p promoted porcine reproductive and respiratory syndrome virus 2 (Prrsv-2) replication by targeting interferon regulatory factor 7. Viruses (2022) 14(1):126. doi: 10.3390/v14010126
42. Zhang Y, Yu Y, Ou C, Ma J, Wang Q, Du S, et al. Alleviation of infectious-bursal-disease-virus-induced bursal injury by betaine is associated with DNA methylation in il-6 and interferon regulatory factor 7 promoter. Poult Sci (2019) 98(10):4457–64. doi: 10.3382/ps/pez280
43. Rezaei R, Mahmoudi M, Gharibdoost F, Kavosi H, Dashti N, Imeni V, et al. Irf7 gene expression profile and methylation of its promoter region in patients with systemic sclerosis. Int J rheumatic Dis (2017) 20(10):1551–61. doi: 10.1111/1756-185x.13175
44. Ma S, Wan X, Deng Z, Shi L, Hao C, Zhou Z, et al. Epigenetic regulator cxxc5 recruits DNA demethylase tet2 to regulate tlr7/9-elicited ifn response in pdcs. J Exp Med (2017) 214(5):1471–91. doi: 10.1084/jem.20161149
45. Suzuki F, Maeyama JI, Kubota A, Nishimune A, Horiguchi S, Takii T, et al. Effect of cigarette smoke on mucosal vaccine response with activation of plasmacytoid dendritic cells: the outcomes of in vivo and in vitro experiments. Vaccine (2023) 41(8):1447–56. doi: 10.1016/j.vaccine.2023.01.019
46. Colina R, Costa-Mattioli M, Dowling RJ, Jaramillo M, Tai LH, Breitbach CJ, et al. Translational control of the innate immune response through irf-7. Nature (2008) 452(7185):323–8. doi: 10.1038/nature06730
47. Lee MS, Kim B, Oh GT, Kim YJ. Oasl1 inhibits translation of the type I interferon-regulating transcription factor irf7. Nat Immunol (2013) 14(4):346–55. doi: 10.1038/ni.2535
48. Lei X, Xiao X, Xue Q, Jin Q, He B, Wang J. Cleavage of interferon regulatory factor 7 by enterovirus 71 3c suppresses cellular responses. J Virol (2013) 87(3):1690–8. doi: 10.1128/JVI.01855-12
49. Fiebach AR, Guzylack-Piriou L, Python S, Summerfield A, Ruggli N. Classical swine fever virus N(Pro) limits type I interferon induction in plasmacytoid dendritic cells by interacting with interferon regulatory factor 7. J Virol (2011) 85(16):8002–11. doi: 10.1128/JVI.00330-11
50. Dalskov L, Narita R, Andersen LL, Jensen N, Assil S, Kristensen KH, et al. Characterization of distinct molecular interactions responsible for irf3 and irf7 phosphorylation and subsequent dimerization. Nucleic Acids Res (2020) 48(20):11421–33. doi: 10.1093/nar/gkaa873
51. Lee CYQ, Chan YT, Cheok YY, Tan GMY, Tang TF, Cheong HC, et al. Helicobacter pylori infection elicits type I interferon response in human monocytes via toll-like receptor 8 signaling. J Immunol Res (2022) 2022:3861518. doi: 10.1155/2022/3861518
52. Marsili G, Perrotti E, Remoli AL, Acchioni C, Sgarbanti M, Battistini A. Ifn regulatory factors and antiviral innate immunity: how viruses can get better. J Interferon Cytokine research: Off J Int Soc Interferon Cytokine Res (2016) 36(7):414–32. doi: 10.1089/jir.2016.0002
53. Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an ikk-related pathway. Sci (New York NY) (2003) 300(5622):1148–51. doi: 10.1126/science.1081315
54. Wang L, Zhao J, Ren J, Hall KH, Moorman JP, Yao ZQ, et al. Protein phosphatase 1 abrogates irf7-mediated type I ifn response in antiviral immunity. Eur J Immunol (2016) 46(10):2409–19. doi: 10.1002/eji.201646491
55. Lee KJ, Lee H, Joo CH. Negative regulation of ikkepsilon-mediated irf7 phosphorylation by hsp70. J Immunol (Baltimore Md: 1950) (2020) 204(9):2562–74. doi: 10.4049/jimmunol.1900297
56. Kui L, Chan GC, Lee PP. Tsg-6 downregulates ifn-alpha and tnf-alpha expression by suppressing irf7 phosphorylation in human plasmacytoid dendritic cells. Mediators Inflammation (2017) 2017:7462945. doi: 10.1155/2017/7462945
57. Ling T, Weng GX, Li J, Li C, Wang W, Cao L, et al. Tarbp2 inhibits irf7 activation by suppressing traf6-mediated K63-linked ubiquitination of irf7. Mol Immunol (2019) 109:116–25. doi: 10.1016/j.molimm.2019.02.019
58. Xue Q, Liu H, Zhu Z, Yang F, Ma L, Cai X, et al. Seneca valley virus 3c(Pro) abrogates the irf3- and irf7-mediated innate immune response by degrading irf3 and irf7. Virology (2018) 518:1–7. doi: 10.1016/j.virol.2018.01.028
59. Li S, Zhu Z, Yang F, Cao W, Yang J, Ma C, et al. Porcine epidemic diarrhea virus membrane protein interacted with irf7 to inhibit type I ifn production during viral infection. J Immunol (Baltimore Md: 1950) (2021) 206(12):2909–23. doi: 10.4049/jimmunol.2001186
60. Zhu FX, King SM, Smith EJ, Levy DE, Yuan Y. A kaposi’s sarcoma-associated herpesviral protein inhibits virus-mediated induction of type I interferon by blocking irf-7 phosphorylation and nuclear accumulation. Proc Natl Acad Sci United States America (2002) 99(8):5573–8. doi: 10.1073/pnas.082420599
61. Wang C, Lv L, Wu Q, Wang Z, Luo Z, Sui B, et al. The role of interferon regulatory factor 7 in the pathogenicity and immunogenicity of rabies virus in a mouse model. J Gen Virol (2021) 102(10):001665. doi: 10.1099/jgv.0.001665
62. Vidy A, El Bougrini J, Chelbi-Alix MK, Blondel D. The nucleocytoplasmic rabies virus P protein counteracts interferon signaling by inhibiting both nuclear accumulation and DNA binding of stat1. J Virol (2007) 81(8):4255–63. doi: 10.1128/jvi.01930-06
63. Brzozka K, Finke S, Conzelmann KK. Inhibition of interferon signaling by rabies virus phosphoprotein P: activation-dependent binding of stat1 and stat2. J Virol (2006) 80(6):2675–83. doi: 10.1128/JVI.80.6.2675-2683.2006
64. Gao L, Li K, Zhang Y, Liu Y, Liu C, Zhang Y, et al. Inhibition of DNA-sensing pathway by marek’s disease virus vp23 protein through suppression of interferon regulatory factor 7 activation. J Virol (2019) 93(4):e01934-18. doi: 10.1128/JVI.01934-18
65. Kang J, Huang M, Li J, Zhang K, Zhu C, Liu S, et al. Enterovirus D68 vp3 targets the interferon regulatory factor 7 to inhibit type I interferon response. Microbiol Spectr (2023) 11(3):e0413822. doi: 10.1128/spectrum.04138-22
66. Du X, Zhou D, Zhou J, Xue J, Wang G, Cheng Z. Marek’s disease virus serine/threonine kinase us3 facilitates viral replication by targeting irf7 to block ifn-beta production. Vet Microbiol (2022) 266:109364. doi: 10.1016/j.vetmic.2022.109364
67. Liu R, Gao L, Yang F, Li X, Liu C, Qi X, et al. Duck enteritis virus protein kinase us3 inhibits DNA sensing signaling by phosphorylating interferon regulatory factor 7. Microbiol Spectr (2022) 10(6):e0229922. doi: 10.1128/spectrum.02299-22
68. Park JM, Jo SH, Kim MY, Kim TH, Ahn YH. Role of transcription factor acetylation in the regulation of metabolic homeostasis. Protein Cell (2015) 6(11):804–13. doi: 10.1007/s13238-015-0204-y
69. Caillaud A, Prakash A, Smith E, Masumi A, Hovanessian AG, Levy DE, et al. Acetylation of interferon regulatory factor-7 by P300/creb-binding protein (Cbp)-associated factor (Pcaf) impairs its DNA binding. J Biol Chem (2002) 277(51):49417–21. doi: 10.1074/jbc.M207484200
70. Li M, Hu J, Mao H, Li D, Jiang Z, Sun Z, et al. Grass carp (Ctenopharyngodon idella) kat8 inhibits ifn 1 response through acetylating irf3/irf7. Front Immunol (2021) 12:808159. doi: 10.3389/fimmu.2021.808159
71. Qin Z, Fang X, Sun W, Ma Z, Dai T, Wang S, et al. Deactylation by sirt1 enables liquid-liquid phase separation of irf3/irf7 in innate antiviral immunity. Nat Immunol (2022) 23(8):1193–207. doi: 10.1038/s41590-022-01269-0
72. Huye LE, Ning S, Kelliher M, Pagano JS. Interferon regulatory factor 7 is activated by a viral oncoprotein through rip-dependent ubiquitination. Mol Cell Biol (2007) 27(8):2910–8. doi: 10.1128/MCB.02256-06
73. Zhang L, Pagano JS. Interferon regulatory factor 7 mediates activation of tap-2 by epstein-barr virus latent membrane protein 1. J Virol (2001) 75(1):341–50. doi: 10.1128/JVI.75.1.341-350.2001
74. Ning S, Hahn AM, Huye LE, Pagano JS. Interferon regulatory factor 7 regulates expression of epstein-barr virus latent membrane protein 1: A regulatory circuit. J Virol (2003) 77(17):9359–68. doi: 10.1128/jvi.77.17.9359-9368.2003
75. Hahn AM, Huye LE, Ning S, Webster-Cyriaque J, Pagano JS. Interferon regulatory factor 7 is negatively regulated by the epstein-barr virus immediate-early gene, bzlf-1. J Virol (2005) 79(15):10040–52. doi: 10.1128/JVI.79.15.10040-10052.2005
76. Ning S, Pagano JS. The A20 deubiquitinase activity negatively regulates lmp1 activation of irf7. J Virol (2010) 84(12):6130–8. doi: 10.1128/JVI.00364-10
77. Wang L, Wang Y, Zhao J, Ren J, Hall KH, Moorman JP, et al. The linear ubiquitin assembly complex modulates latent membrane protein 1 activation of nf-Kb and interferon regulatory factor 7. J Virol (2017) 91(4):e01138-16. doi: 10.1128/jvi.01138-16
78. Qi F, Zhang X, Wang L, Ren C, Zhao X, Luo J, et al. E3 ubiquitin ligase neurl3 promotes innate antiviral response through catalyzing K63-linked ubiquitination of irf7. FASEB journal: Off Publ Fed Am Societies Exp Biol (2022) 36(8):e22409. doi: 10.1096/fj.202200316R
79. Yu CF, Peng WM, Schlee M, Barchet W, Eis-Hubinger AM, Kolanus W, et al. Socs1 and socs3 target irf7 degradation to suppress tlr7-mediated type I ifn production of human plasmacytoid dendritic cells. J Immunol (Baltimore Md: 1950) (2018) 200(12):4024–35. doi: 10.4049/jimmunol.1700510
80. Huang S, Cui M, Huang J, Wu Z, Cheng A, Wang M, et al. Rnf123 mediates ubiquitination and degradation of socs1 to regulate type I interferon production during duck tembusu virus infection. J Virol (2023) 97(4):e0009523. doi: 10.1128/jvi.00095-23
81. Young JA, Sermwittayawong D, Kim HJ, Nandu S, An N, Erdjument-Bromage H, et al. Fas-associated death domain (Fadd) and the E3 ubiquitin-protein ligase trim21 interact to negatively regulate virus-induced interferon production. J Biol Chem (2011) 286(8):6521–31. doi: 10.1074/jbc.M110.172288
82. Yu Y, Hayward GS. The ubiquitin E3 ligase raul negatively regulates type I interferon through ubiquitination of the transcription factors irf7 and irf3. Immunity (2010) 33(6):863–77. doi: 10.1016/j.immuni.2010.11.027
83. Liu BQ, Liu RB, Li WP, Mao XT, Li YN, Huang T, et al. Xaf1 prevents hyperproduction of type I interferon upon viral infection by targeting irf7. EMBO Rep (2023) 24(1):e55387. doi: 10.15252/embr.202255387
84. Yu Y, Wang SE, Hayward GS. The kshv immediate-early transcription factor rta encodes ubiquitin E3 ligase activity that targets irf7 for proteosome-mediated degradation. Immunity (2005) 22(1):59–70. doi: 10.1016/j.immuni.2004.11.011
85. Barro M, Patton JT. Rotavirus nsp1 inhibits expression of type I interferon by antagonizing the function of interferon regulatory factors irf3, irf5, and irf7. J Virol (2007) 81(9):4473–81. doi: 10.1128/JVI.02498-06
86. Zhang C, Lu LF, Li ZC, Zhou XY, Zhou Y, Chen DD, et al. Grass carp reovirus vp56 represses interferon production by degrading phosphorylated irf7. Fish shellfish Immunol (2020) 99:99–106. doi: 10.1016/j.fsi.2020.02.004
87. Celen AB, Sahin U. Sumoylation on its 25th anniversary: mechanisms, pathology, and emerging concepts. FEBS J (2020) 287(15):3110–40. doi: 10.1111/febs.15319
88. Kubota T, Matsuoka M, Chang TH, Tailor P, Sasaki T, Tashiro M, et al. Virus infection triggers sumoylation of irf3 and irf7, leading to the negative regulation of type I interferon gene expression. J Biol Chem (2008) 283(37):25660–70. doi: 10.1074/jbc.M804479200
89. Basler CF, Amarasinghe GK. Evasion of interferon responses by ebola and marburg viruses. J Interferon Cytokine research: Off J Int Soc Interferon Cytokine Res (2009) 29(9):511–20. doi: 10.1089/jir.2009.0076
90. Chang TH, Kubota T, Matsuoka M, Jones S, Bradfute SB, Bray M, et al. Ebola zaire virus blocks type I interferon production by exploiting the host sumo modification machinery. PloS Pathog (2009) 5(6):e1000493. doi: 10.1371/journal.ppat.1000493
91. Liang Q, Deng H, Li X, Wu X, Tang Q, Chang TH, et al. Tripartite motif-containing protein 28 is a small ubiquitin-related modifier E3 ligase and negative regulator of ifn regulatory factor 7. J Immunol (Baltimore Md: 1950) (2011) 187(9):4754–63. doi: 10.4049/jimmunol.1101704
92. Bentz GL, Shackelford J, Pagano JS. Epstein-barr virus latent membrane protein 1 regulates the function of interferon regulatory factor 7 by inducing its sumoylation. J Virol (2012) 86(22):12251–61. doi: 10.1128/JVI.01407-12
93. Yu G, Liu X, Tang J, Xu C, Ouyang G, Xiao W. Neddylation facilitates the antiviral response in zebrafish. Front Immunol (2019) 10:1432. doi: 10.3389/fimmu.2019.01432
94. Zhao M, Zhang Y, Yang X, Jin J, Shen Z, Feng X, et al. Myeloid neddylation targets irf7 and promotes host innate immunity against rna viruses. PloS Pathog (2021) 17(9):e1009901. doi: 10.1371/journal.ppat.1009901
95. Gao L, Liu R, Luo D, Li K, Qi X, Liu C, et al. Avian reovirus Σa protein inhibits type I interferon production by abrogating interferon regulatory factor 7 activation. J Virol (2023) 97(1):e0178522. doi: 10.1128/jvi.01785-22
96. Liu X, Xi D, Xu A, Wang Y, Song T, Ma T, et al. Chicken anemia virus vp1 negatively regulates type I interferon via targeting interferon regulatory factor 7 of the DNA-sensing pathway. Poult Sci (2022) 102(1):102291. doi: 10.1016/j.psj.2022.102291
97. Zhang B, Liu M, Huang J, Zeng Q, Zhu Q, Xu S, et al. H1n1 influenza a virus protein ns2 inhibits innate immune response by targeting irf7. Viruses (2022) 14(11):2411. doi: 10.3390/v14112411
98. Szulc-Dabrowska L, Struzik J, Ostrowska A, Guzera M, Toka FN, Bossowska-Nowicka M, et al. Functional paralysis of gm-csf-derived bone marrow cells productively infected with ectromelia virus. PloS One (2017) 12(6):e0179166. doi: 10.1371/journal.pone.0179166
99. Dhamanage AS, Thakar MR, Paranjape RS. Hiv-1-mediated suppression of ifn-alpha production is associated with inhibition of irf-7 translocation and pi3k/akt pathway in plasmacytoid dendritic cells. AIDS Res Hum Retroviruses (2019) 35(1):40–8. doi: 10.1089/AID.2018.0136
100. Chang S, Kodys K, Szabo G. Impaired expression and function of toll-like receptor 7 in hepatitis C virus infection in human hepatoma cells. Hepatology (2010) 51(1):35–42. doi: 10.1002/hep.23256
101. Chowdhury JB, Kim H, Ray R, Ray RB. Hepatitis C virus ns5a protein modulates irf-7-mediated interferon-alpha signaling. J Interferon Cytokine research: Off J Int Soc Interferon Cytokine Res (2014) 34(1):16–21. doi: 10.1089/jir.2013.0038
102. Raychoudhuri A, Shrivastava S, Steele R, Dash S, Kanda T, Ray R, et al. Hepatitis C virus infection impairs irf-7 translocation and alpha interferon synthesis in immortalized human hepatocytes. J Virol (2010) 84(21):10991–8. doi: 10.1128/JVI.00900-10
103. Juang YT, Au WC, Lowther W, Hiscott J, Pitha PM. Lipopolysaccharide inhibits virus-mediated induction of interferon genes by disruption of nuclear transport of interferon regulatory factors 3 and 7. J Biol Chem (1999) 274(25):18060–6. doi: 10.1074/jbc.274.25.18060
104. Guiducci C, Ghirelli C, Marloie-Provost MA, Matray T, Coffman RL, Liu YJ, et al. Pi3k is critical for the nuclear translocation of irf-7 and type I ifn production by human plasmacytoid predendritic cells in response to tlr activation. J Exp Med (2008) 205(2):315–22. doi: 10.1084/jem.20070763
105. Shinohara ML, Lu L, Bu J, Werneck MB, Kobayashi KS, Glimcher LH, et al. Osteopontin expression is essential for interferon-alpha production by plasmacytoid dendritic cells. Nat Immunol (2006) 7(5):498–506. doi: 10.1038/ni1327
106. An J, Briggs TA, Dumax-Vorzet A, Alarcon-Riquelme ME, Belot A, Beresford M, et al. Tartrate-resistant acid phosphatase deficiency in the predisposition to systemic lupus erythematosus. Arthritis Rheumatol (Hoboken NJ) (2017) 69(1):131–42. doi: 10.1002/art.39810
107. Shigesaka M, Ito T, Inaba M, Imai K, Yamanaka H, Azuma Y, et al. Mycophenolic acid, the active form of mycophenolate mofetil, interferes with irf7 nuclear translocation and type I ifn production by plasmacytoid dendritic cells. Arthritis Res Ther (2020) 22(1):264. doi: 10.1186/s13075-020-02356-z
108. Miyamoto R, Ito T, Nomura S, Amakawa R, Amuro H, Katashiba Y, et al. Inhibitor of ikappab kinase activity, bay 11-7082, interferes with interferon regulatory factor 7 nuclear translocation and type I interferon production by plasmacytoid dendritic cells. Arthritis Res Ther (2010) 12(3):R87. doi: 10.1186/ar3014
109. Wan Y, Kim TW, Yu M, Zhou H, Yamashita M, Kang Z, et al. The dual functions of il-1 receptor-associated kinase 2 in tlr9-mediated ifn and proinflammatory cytokine production. J Immunol (Baltimore Md: 1950) (2011) 186(5):3006–14. doi: 10.4049/jimmunol.1003217
110. Desnues B, Macedo AB, Ordonez-Rueda D, Roussel-Queval A, Malissen B, Bruhns P, et al. The transcriptional repressor gfi1 prevents lupus autoimmunity by restraining tlr7 signaling. Eur J Immunol (2016) 46(12):2801–11. doi: 10.1002/eji.201646573
111. Wu YY, Kumar R, Iida R, Bagavant H, Alarcon-Riquelme ME. Bank1 regulates igg production in a lupus model by controlling tlr7-dependent stat1 activation. PloS One (2016) 11(5):e0156302. doi: 10.1371/journal.pone.0156302
112. Guo H, Zhang J, Zhang X, Wang Y, Yu H, Yin X, et al. Scarb2/limp-2 regulates ifn production of plasmacytoid dendritic cells by mediating endosomal translocation of tlr9 and nuclear translocation of irf7. J Immunol (Baltimore Md: 1950) (2015) 194(10):4737–49. doi: 10.4049/jimmunol.1402312
113. Smith N, Rodero MP, Bekaddour N, Bondet V, Ruiz-Blanco YB, Harms M, et al. Control of tlr7-mediated type I ifn signaling in pdcs through cxcr4 engagement-a new target for lupus treatment. Sci Adv (2019) 5(7):eaav9019. doi: 10.1126/sciadv.aav9019
114. Martinson JA, Montoya CJ, Usuga X, Ronquillo R, Landay AL, Desai SN. Chloroquine modulates hiv-1-induced plasmacytoid dendritic cell alpha interferon: implication for T-cell activation. Antimicrob Agents Chemother (2010) 54(2):871–81. doi: 10.1128/AAC.01246-09
115. Osawa Y, Iho S, Takauji R, Takatsuka H, Yamamoto S, Takahashi T, et al. Collaborative action of nf-kappab and P38 mapk is involved in cpg DNA-induced ifn-alpha and chemokine production in human plasmacytoid dendritic cells. J Immunol (Baltimore Md: 1950) (2006) 177(7):4841–52. doi: 10.4049/jimmunol.177.7.4841
116. Sisirak V, Vey N, Goutagny N, Renaudineau S, Malfroy M, Thys S, et al. Breast cancer-derived transforming growth factor-beta and tumor necrosis factor-alpha compromise interferon-alpha production by tumor-associated plasmacytoid dendritic cells. Int J Cancer J Int du Cancer (2013) 133(3):771–8. doi: 10.1002/ijc.28072
117. Danis B, George TC, Goriely S, Dutta B, Renneson J, Gatto L, et al. Interferon regulatory factor 7-mediated responses are defective in cord blood plasmacytoid dendritic cells. Eur J Immunol (2008) 38(2):507–17. doi: 10.1002/eji.200737760
118. Pierog PL, Zhao Y, Singh S, Dai J, Yap GS, Fitzgerald-Bocarsly P. Toxoplasma gondii inactivates human plasmacytoid dendritic cells by functional mimicry of il-10. J Immunol (Baltimore Md: 1950) (2018) 200(1):186–95. doi: 10.4049/jimmunol.1701045
119. Li M, Liu C, Xu X, Liu Y, Jiang Z, Li Y, et al. Grass carp (Ctenopharyngodon idella) gpatch3 initiates ifn 1 expression via the activation of sting-irf7 signal axis. Dev Comp Immunol (2020) 112:103781. doi: 10.1016/j.dci.2020.103781
120. Maarifi G, Smith N, Maillet S, Moncorgé O, Chamontin C, Edouard J, et al. Trim8 is required for virus-induced ifn response in human plasmacytoid dendritic cells. Sci Adv (2019) 5(11):eaax3511. doi: 10.1126/sciadv.aax3511
121. Galani IE, Triantafyllia V, Eleminiadou EE, Koltsida O, Stavropoulos A, Manioudaki M, et al. Interferon-Lambda Mediates Non-Redundant Front-Line Antiviral Protection against Influenza Virus Infection without Compromising Host Fitness. Immunity (2017) 46(5):875–90 e6. doi: 10.1016/j.immuni.2017.04.025
122. Lazear HM, Schoggins JW, Diamond MS. Shared and distinct functions of type I and type iii interferons. Immunity (2019) 50(4):907–23. doi: 10.1016/j.immuni.2019.03.025
123. Pervolaraki K, Stanifer ML, Munchau S, Renn LA, Albrecht D, Kurzhals S, et al. Type I and type iii interferons display different dependency on mitogen-activated protein kinases to mount an antiviral state in the human gut. Front Immunol (2017) 8:459. doi: 10.3389/fimmu.2017.00459
124. Ye L, Schnepf D, Staeheli P. Interferon-lambda orchestrates innate and adaptive mucosal immune responses. Nat Rev Immunol (2019) 19(10):614–25. doi: 10.1038/s41577-019-0182-z
125. Osterlund PI, Pietila TE, Veckman V, Kotenko SV, Julkunen I. Ifn regulatory factor family members differentially regulate the expression of type iii ifn (Ifn-lambda) genes. J Immunol (Baltimore Md: 1950) (2007) 179(6):3434–42. doi: 10.4049/jimmunol.179.6.3434
126. Peng K, Deng L, Wei J, Zhao J, Deng H, Tao Q, et al. Transcriptome analyses of senecavirus a-infected pk-15 cells: rig-I and irf7 are the important factors in inducing type iii interferons. Front Microbiol (2022) 13:846343. doi: 10.3389/fmicb.2022.846343
127. Wells AI, Coyne CB. Type iii interferons in antiviral defenses at barrier surfaces. Trends Immunol (2018) 39(10):848–58. doi: 10.1016/j.it.2018.08.008
128. Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen Shu Y, et al. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell (2015) 162(5):961–73. doi: 10.1016/j.cell.2015.07.056
129. Iaconis G, Jackson B, Childs K, Boyce M, Goodbourn S, Blake N, et al. Rotavirus nsp1 inhibits type I and type iii interferon induction. Viruses (2021) 13(4):589. doi: 10.3390/v13040589
130. Roy S, Guha Roy D, Bhushan A, Bharatiya S, Chinnaswamy S. Functional genetic variants of the ifn-lambda3 (Il28b) gene and transcription factor interactions on its promoter. Cytokine (2021) 142:155491. doi: 10.1016/j.cyto.2021.155491
131. Bin L, Edwards MG, Heiser R, Streib JE, Richers B, Hall CF, et al. Identification of novel gene signatures in patients with atopic dermatitis complicated by eczema herpeticum. J Allergy Clin Immunol (2014) 134(4):848–55. doi: 10.1016/j.jaci.2014.07.018
132. Griffiths SJ, Koegl M, Boutell C, Zenner HL, Crump CM, Pica F, et al. A systematic analysis of host factors reveals a med23-interferon-lambda regulatory axis against herpes simplex virus type 1 replication. PloS Pathog (2013) 9(8):e1003514. doi: 10.1371/journal.ppat.1003514
133. Campbell TM, Liu Z, Zhang Q, Moncada-Velez M, Covill LE, Zhang P, et al. Respiratory viral infections in otherwise healthy humans with inherited irf7 deficiency. J Exp Med (2022) 219(7):e20220202. doi: 10.1084/jem.20220202
134. Ciancanelli MJ, Abel L, Zhang SY, Casanova JL. Host genetics of severe influenza: from mouse mx1 to human irf7. Curr Opin Immunol (2016) 38:109–20. doi: 10.1016/j.coi.2015.12.002
135. Ciancanelli MJ, Huang SX, Luthra P, Garner H, Itan Y, Volpi S, et al. Infectious disease. Life-threatening influenza and impaired interferon amplification in human irf7 deficiency. Sci (New York NY) (2015) 348(6233):448–53. doi: 10.1126/science.aaa1578
136. Rajamanickam A, Kumar NP, Pandiaraj AN, Selvaraj N, Munisankar S, Renji RM, et al. Restoration of dendritic cell homeostasis and type I/type iii interferon levels in convalescent covid-19 individuals. BMC Immunol (2022) 23(1):51. doi: 10.1186/s12865-022-00526-z
137. Zhang Q, Bastard P, Effort CHG, Cobat A, Casanova JL. Human genetic and immunological determinants of critical covid-19 pneumonia. Nature (2022) 603(7902):587–98. doi: 10.1038/s41586-022-04447-0
138. Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, et al. Inborn errors of type I ifn immunity in patients with life-threatening covid-19. Sci (New York NY) (2020) 370(6515):eabd4570. doi: 10.1126/science.abd4570
139. Smith CIE, Zain R, Österborg A, Palma M, Buggert M, Bergman P, et al. Do reduced numbers of plasmacytoid dendritic cells contribute to the aggressive clinical course of covid-19 in chronic lymphocytic leukaemia? Scandinavian J Immunol (2022) 95(4):e13153. doi: 10.1111/sji.13153
140. Povysil G, Butler-Laporte G, Shang N, Wang C, Khan A, Alaamery M, et al. Rare loss-of-function variants in type I ifn immunity genes are not associated with severe covid-19. J Clin Invest (2021) 131(14):e147834. doi: 10.1172/jci147834
141. Zhu Y, Zhang Z, Song J, Qian W, Gu X, Yang C, et al. Sars-cov-2-encoded mirnas inhibit host type I interferon pathway and mediate allelic differential expression of susceptible gene. Front Immunol (2021) 12:767726. doi: 10.3389/fimmu.2021.767726
142. Scagnolari C, Pierangeli A, Frasca F, Bitossi C, Viscido A, Oliveto G, et al. Differential induction of type I and iii interferon genes in the upper respiratory tract of patients with coronavirus disease 2019 (Covid-19). Virus Res (2021) 295:198283. doi: 10.1016/j.virusres.2020.198283
143. Lee JS, Shin EC. The type I interferon response in covid-19: implications for treatment. Nat Rev Immunol (2020) 20(10):585–6. doi: 10.1038/s41577-020-00429-3
144. Nain Z, Barman SK, Sheam MM, Syed SB, Samad A, Quinn JMW, et al. Transcriptomic studies revealed pathophysiological impact of covid-19 to predominant health conditions. Brief Bioinform (2021) 22(6):bbab197. doi: 10.1093/bib/bbab197
145. Konigsberg IR, Barnes B, Campbell M, Davidson E, Zhen Y, Pallisard O, et al. Host methylation predicts sars-cov-2 infection and clinical outcome. Commun Med (Lond) (2021) 1(1):42. doi: 10.1038/s43856-021-00042-y
146. Chasset F, Dayer JM, Chizzolini C. Type I interferons in systemic autoimmune diseases: distinguishing between afferent and efferent functions for precision medicine and individualized treatment. Front Pharmacol (2021) 12:633821. doi: 10.3389/fphar.2021.633821
147. Hall JC, Rosen A. Type I interferons: crucial participants in disease amplification in autoimmunity. Nat Rev Rheumatol (2010) 6(1):40–9. doi: 10.1038/nrrheum.2009.237
148. Deckx N, Willekens B, Wens I, Eijnde BO, Goossens H, Van Damme P, et al. Altered molecular expression of tlr-signaling pathways affects the steady-state release of il-12p70 and ifn-alpha in patients with relapsing-remitting multiple sclerosis. Innate Immun (2016) 22(4):266–73. doi: 10.1177/1753425916642615
149. Salem M, Mony JT, Løbner M, Khorooshi R, Owens T. Interferon regulatory factor-7 modulates experimental autoimmune encephalomyelitis in mice. J Neuroinflamm (2011) 8:181. doi: 10.1186/1742-2094-8-181
150. Sweeney SE, Corr M, Kimbler TB. Role of interferon regulatory factor 7 in serum-transfer arthritis: regulation of interferon-beta production. Arthritis rheumatism (2012) 64(4):1046–56. doi: 10.1002/art.33454
151. Niewold TB, Kelly JA, Kariuki SN, Franek BS, Kumar AA, Kaufman KM, et al. Irf5 haplotypes demonstrate diverse serological associations which predict serum interferon alpha activity and explain the majority of the genetic association with systemic lupus erythematosus. Ann rheumatic Dis (2012) 71(3):463–8. doi: 10.1136/annrheumdis-2011-200463
152. Lin LH, Ling P, Liu MF. The potential role of interferon-regulatory factor 7 among Taiwanese patients with systemic lupus erythematosus. J Rheumatol (2011) 38(9):1914–9. doi: 10.3899/jrheum.101004
153. Li P, Cao C, Luan H, Li C, Hu C, Zhang S, et al. Association of genetic variations in the stat4 and irf7/kiaa1542 regions with systemic lupus erythematosus in a northern han chinese population. Hum Immunol (2011) 72(3):249–55. doi: 10.1016/j.humimm.2010.12.011
154. Salloum R, Franek BS, Kariuki SN, Rhee L, Mikolaitis RA, Jolly M, et al. Genetic variation at the irf7/phrf1 locus is associated with autoantibody profile and serum interferon-alpha activity in lupus patients. Arthritis rheumatism (2010) 62(2):553–61. doi: 10.1002/art.27182
155. Akahoshi M, Nakashima H, Sadanaga A, Miyake K, Obara K, Tamari M, et al. Promoter polymorphisms in the irf3 gene confer protection against systemic lupus erythematosus. Lupus (2008) 17(6):568–74. doi: 10.1177/0961203308089340
156. Harley JB, Alarcón-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in itgam, pxk, kiaa1542 and other loci. Nat Genet (2008) 40(2):204–10. doi: 10.1038/ng.81
157. Fu Q, Zhao J, Qian X, Wong JLH, Kaufman KM, Yu CY, et al. Association of a functional irf7 variant with systemic lupus erythematosus. Arthritis Rheumatism (2011) 63(3):749–54. doi: 10.1002/art.30193
158. Xu WD, Zhang YJ, Xu K, Zhai Y, Li BZ, Pan HF, et al. Irf7, a functional factor associates with systemic lupus erythematosus. Cytokine (2012) 58(3):317–20. doi: 10.1016/j.cyto.2012.03.003
159. Sweeney SE. Hematopoietic stem cell transplant for systemic lupus erythematosus: interferon regulatory factor 7 activation correlates with the ifn signature and recurrent disease. Lupus (2011) 20(9):975–80. doi: 10.1177/0961203310394897
160. Imgenberg-Kreuz J, Carlsson Almlöf J, Leonard D, Alexsson A, Nordmark G, Eloranta ML, et al. DNA methylation mapping identifies gene regulatory effects in patients with systemic lupus erythematosus. Ann rheumatic Dis (2018) 77(5):736–43. doi: 10.1136/annrheumdis-2017-212379
161. Joseph S, George NI, Green-Knox B, Treadwell EL, Word B, Yim S, et al. Epigenome-wide association study of peripheral blood mononuclear cells in systemic lupus erythematosus: identifying DNA methylation signatures associated with interferon-related genes based on ethnicity and sledai. J Autoimmun (2019) 96:147–57. doi: 10.1016/j.jaut.2018.09.007
162. Thoreau B, Chaigne B, Renaud A, Mouthon L. Pathophysiology of systemic sclerosis. Presse Med (2021) 50(1):104087. doi: 10.1016/j.lpm.2021.104087
163. Ototake Y, Yamaguchi Y, Asami M, Komitsu N, Akita A, Watanabe T, et al. Downregulated irf8 in monocytes and macrophages of patients with systemic sclerosis may aggravate the fibrotic phenotype. J Invest Dermatol (2021) 141(8):1954–63. doi: 10.1016/j.jid.2021.02.015
164. Gorlova O, Martin JE, Rueda B, Koeleman BP, Ying J, Teruel M, et al. Identification of novel genetic markers associated with clinical phenotypes of systemic sclerosis through a genome-wide association strategy. PloS Genet (2011) 7(7):e1002178. doi: 10.1371/journal.pgen.1002178
165. Terao C, Ohmura K, Kawaguchi Y, Nishimoto T, Kawasaki A, Takehara K, et al. Pld4 as a novel susceptibility gene for systemic sclerosis in a Japanese population. Arthritis rheumatism (2013) 65(2):472–80. doi: 10.1002/art.37777
166. Arismendi M, Giraud M, Ruzehaji N, Dieude P, Koumakis E, Ruiz B, et al. Identification of nf-kappab and plcl2 as new susceptibility genes and highlights on a potential role of irf8 through interferon signature modulation in systemic sclerosis. Arthritis Res Ther (2015) 17(1):71. doi: 10.1186/s13075-015-0572-y
167. Assassi S, Swindell WR, Wu M, Tan FD, Khanna D, Furst DE, et al. Dissecting the heterogeneity of skin gene expression patterns in systemic sclerosis. Arthritis Rheumatol (Hoboken NJ) (2015) 67(11):3016–26. doi: 10.1002/art.39289
168. Tan FK, Zhou X, Mayes MD, Gourh P, Guo X, Marcum C, et al. Signatures of differentially regulated interferon gene expression and vasculotrophism in the peripheral blood cells of systemic sclerosis patients. Rheumatol (Oxford England) (2006) 45(6):694–702. doi: 10.1093/rheumatology/kei244
169. Carmona FD, Gutala R, Simeón CP, Carreira P, Ortego-Centeno N, Vicente-Rabaneda E, et al. Novel identification of the irf7 region as an anticentromere autoantibody propensity locus in systemic sclerosis. Ann rheumatic Dis (2012) 71(1):114–9. doi: 10.1136/annrheumdis-2011-200275
170. Wu M, Skaug B, Bi X, Mills T, Salazar G, Zhou X, et al. Interferon regulatory factor 7 (Irf7) represents a link between inflammation and fibrosis in the pathogenesis of systemic sclerosis. Ann rheumatic Dis (2019) 78(11):1583–91. doi: 10.1136/annrheumdis-2019-215208
171. Minaga K, Watanabe T, Arai Y, Shiokawa M, Hara A, Yoshikawa T, et al. Activation of interferon regulatory factor 7 in plasmacytoid dendritic cells promotes experimental autoimmune pancreatitis. J Gastroenterol (2020) 55(5):565–76. doi: 10.1007/s00535-020-01662-2
172. Arai Y, Yamashita K, Kuriyama K, Shiokawa M, Kodama Y, Sakurai T, et al. Plasmacytoid dendritic cell activation and ifn-A Production are prominent features of murine autoimmune pancreatitis and human igg4-related autoimmune pancreatitis. J Immunol (Baltimore Md: 1950) (2015) 195(7):3033–44. doi: 10.4049/jimmunol.1500971
173. Watanabe T, Minaga K, Kamata K, Kudo M, Strober W. Mechanistic insights into autoimmune pancreatitis and igg4-related disease. Trends Immunol (2018) 39(11):874–89. doi: 10.1016/j.it.2018.09.005
174. Watanabe T, Yamashita K, Arai Y, Minaga K, Kamata K, Nagai T, et al. Chronic fibro-inflammatory responses in autoimmune pancreatitis depend on ifn-A and il-33 produced by plasmacytoid dendritic cells. J Immunol (Baltimore Md: 1950) (2017) 198(10):3886–96. doi: 10.4049/jimmunol.1700060
175. Antonelli A, Ferrari SM, Corrado A, Di Domenicantonio A, Fallahi P. Autoimmune thyroid disorders. Autoimmun Rev (2015) 14(2):174–80. doi: 10.1016/j.autrev.2014.10.016
176. Lee HJ, Li CW, Hammerstad SS, Stefan M, Tomer Y. Immunogenetics of autoimmune thyroid diseases: A comprehensive review. J Autoimmun (2015) 64:82–90. doi: 10.1016/j.jaut.2015.07.009
177. Yao Q, An X, Zhang J, Mu K, Li L, Song R, et al. Irf7 gene variations confer susceptibility to autoimmune thyroid diseases and graves’ Ophthalmopathy. Int J Endocrinol (2019) 2019:7429187. doi: 10.1155/2019/7429187
178. Imagawa A, Hanafusa T, Miyagawa J, Matsuzawa Y. A novel subtype of type 1 diabetes mellitus characterized by a rapid onset and an absence of diabetes-related antibodies. Osaka iddm study group. New Engl J Med (2000) 342(5):301–7. doi: 10.1056/nejm200002033420501
179. Wang Z, Zheng Y, Hou C, Yang L, Li X, Lin J, et al. DNA methylation impairs tlr9 induced foxp3 expression by attenuating irf-7 binding activity in fulminant type 1 diabetes. J Autoimmun (2013) 41:50–9. doi: 10.1016/j.jaut.2013.01.009
180. Lang PA, Cervantes-Barragan L, Verschoor A, Navarini AA, Recher M, Pellegrini M, et al. Hematopoietic cell-derived interferon controls viral replication and virus-induced disease. Blood (2009) 113(5):1045–52. doi: 10.1182/blood-2007-10-117861
181. Jiang H, Li Y, Shen M, Liang Y, Qian Y, Dai H, et al. Interferon-alpha promotes mhc I antigen presentation of islet beta cells through stat1-irf7 pathway in type 1 diabetes. Immunology (2022) 166(2):210–21. doi: 10.1111/imm.13468
182. Heinig M, Petretto E, Wallace C, Bottolo L, Rotival M, Lu H, et al. A trans-acting locus regulates an anti-viral expression network and type 1 diabetes risk. Nature (2010) 467(7314):460–4. doi: 10.1038/nature09386
183. Wang XA, Zhang R, Zhang S, Deng S, Jiang D, Zhong J, et al. Interferon regulatory factor 7 deficiency prevents diet-induced obesity and insulin resistance. Am J Physiol Endocrinol Metab (2013) 305(4):E485–95. doi: 10.1152/ajpendo.00505.2012
184. Kuroda M, Nishiguchi M, Ugawa N, Ishikawa E, Kawabata Y, Okamoto S, et al. Interferon regulatory factor 7 mediates obesity-associated mcp-1 transcription. PloS One (2020) 15(5):e0233390. doi: 10.1371/journal.pone.0233390
185. Furie R, Werth VP, Merola JF, Stevenson L, Reynolds TL, Naik H, et al. Monoclonal antibody targeting bdca2 ameliorates skin lesions in systemic lupus erythematosus. J Clin Invest (2019) 129(3):1359–71. doi: 10.1172/JCI124466
186. Werth VP, Furie RA, Romero-Diaz J, Navarra S, Kalunian K, van Vollenhoven RF, et al. Trial of anti-bdca2 antibody litifilimab for cutaneous lupus erythematosus. New Engl J Med (2022) 387(4):321–31. doi: 10.1056/NEJMoa2118024
187. Goulden B, Isenberg D. Anti-ifnαr mabs for the treatment of systemic lupus erythematosus. Expert Opin Biol Ther (2021) 21(4):519–28. doi: 10.1080/14712598.2021.1841164
188. Paredes JL, Niewold TB. Type I interferon antagonists in clinical development for lupus. Expert Opin Investig Drugs (2020) 29(9):1025–41. doi: 10.1080/13543784.2020.1797677
189. Psarras A, Emery P, Vital EM. Type I interferon–mediated autoimmune diseases: pathogenesis, diagnosis and targeted therapy. Rheumatology (2017) 56(10):1662–75. doi: 10.1093/rheumatology/kew431
190. Takauji R, Iho S, Takatsuka H, Yamamoto S, Takahashi T, Kitagawa H, et al. Cpg-DNA-induced ifn-alpha production involves P38 mapk-dependent stat1 phosphorylation in human plasmacytoid dendritic cell precursors. J leukocyte Biol (2002) 72(5):1011–9. doi: 10.1189/jlb.72.5.1011
191. Ezeonwumelu IJ, Garcia-Vidal E, Felip E, Puertas MC, Oriol-Tordera B, Gutierrez-Chamorro L, et al. Irf7 expression correlates with hiv latency reversal upon specific blockade of immune activation. Front Immunol (2022) 13:1001068. doi: 10.3389/fimmu.2022.1001068
192. Kubo S, Yamaoka K, Kondo M, Yamagata K, Zhao J, Iwata S, et al. The jak inhibitor, tofacitinib, reduces the T cell stimulatory capacity of human monocyte-derived dendritic cells. Ann rheumatic Dis (2014) 73(12):2192–8. doi: 10.1136/annrheumdis-2013-203756
Keywords: IRF7, IFN-I, IFN-III, COVID-19, immunity, autoimmunity
Citation: Ma W, Huang G, Wang Z, Wang L and Gao Q (2023) IRF7: role and regulation in immunity and autoimmunity. Front. Immunol. 14:1236923. doi: 10.3389/fimmu.2023.1236923
Received: 08 June 2023; Accepted: 25 July 2023;
Published: 10 August 2023.
Edited by:
Uday Kishore, United Arab Emirates University, United Arab EmiratesReviewed by:
Jean-philippe Herbeuval, UMR8601 Laboratoire de Chimie et Biochimie Pharmacologiques et Toxicologiques, FranceYunhao Tan, AbbVie, United States
Copyright © 2023 Ma, Huang, Wang, Wang and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiangguo Gao, cWlhbmdndW9nYW9AMTYzLmNvbQ==
 Wei Ma1,2
Wei Ma1,2 Qiangguo Gao
Qiangguo Gao