- 1School of Marine Sciences, Ningbo University, Ningbo, Zhejiang, China
- 2Agriculture Ministry Key Laboratory of Healthy Freshwater Aquaculture, Zhejiang Institule of Freshwater Fisheries, Huzhou, Zhejiang, China
- 3Key Laboratory of Aquacultural Biotechnology, Ministry of Education, Ningbo University, Ningbo, China
- 4National Engineering Research Laboratory of Marine Biotechnology and Engineering, Ningbo University, Ningbo, Zhejiang, China
Background: Macrophage colony-stimulating factor 2 (MCSF-2) is an important cytokine that controls how cells of the monocyte/macrophage lineage proliferate, differentiate, and survive in vertebrates. Two isoforms of MCSF have been identified in fish, each exhibiting distinct gene organization and expression patterns. In this study, we investigated a goldfish MCSF-2 gene in terms of its immunomodulatory and functional properties.
Methods: In this study, goldfish were acclimated for 3 weeks and sedated with TMS prior to handling. Two groups of fish were used for infection experiments, and tissues from healthy goldfish were collected for RNA isolation. cDNA synthesis was performed, and primers were designed based on transcriptome database sequences. Analysis of gfMCSF-2 sequences, including nucleotide and amino acid analysis, molecular mass prediction, and signal peptide prediction, was conducted. Real-time quantitative PCR (qPCR) was used to analyze gene expression levels, while goldfish head kidney leukocytes (HKLs) were isolated using standard protocols. The expression of gfMCSF-2 in activated HKLs was investigated, and recombinant goldfish MCSF-2 was expressed and purified. Western blot analysis, cell proliferation assays, and flow cytometric analysis of HKLs were performed. Gene expression analysis of transcription factors and pro-inflammatory cytokines in goldfish head kidney leukocytes exposed to rgMCSF-2 was conducted. Statistical analysis using one-way ANOVA and Dunnett’s post hoc test was applied.
Results: We performed a comparative analysis of MCSF-1 and MCSF-2 at the protein and nucleotide levels using the Needleman-Wunsch algorithm. The results revealed significant differences between the two sequences, supporting the notion that they represent distinct genes rather than isoforms of the same gene. Sequence alignment demonstrated high sequence identity with MCSF-2 homologs from fish species, particularly C. carpio, which was supported by phylogenetic analysis. Expression analysis in various goldfish tissues demonstrated differential expression levels, with the spleen exhibiting the highest expression. In goldfish head kidney leukocytes, gfMCSF-2 expression was modulated by chemical stimuli and bacterial infection, with upregulation observed in response to lipopolysaccharide (LPS) and live Aeromonas hydrophila. Recombinant gfMCSF-2 (rgMCSF-2) was successfully expressed and purified, showing the ability to stimulate cell proliferation in HKLs. Flow cytometric analysis revealed that rgMCSF-2 induced differentiation of sorted leukocytes at a specific concentration. Moreover, rgMCSF-2 treatment upregulated TNFα and IL-1β mRNA levels and influenced the expression of transcription factors, such as MafB, GATA2, and cMyb, in a time-dependent manner.
Conclusion: Collectively, by elucidating the effects of rgMCSF-2 on cell proliferation, differentiation, and the modulation of pro-inflammatory cytokines and transcription factors, our findings provided a comprehensive understanding of the potential mechanisms underlying gfMCSF-2-mediated immune regulation. These results contribute to the fundamental knowledge of MCSF-2 in teleosts and establish a foundation for further investigations on the role of gfMCSF-2 in fish immune responses.
1 Introduction
MCSF, also named colony stimulating factor 1 (CSF-1), is a growth factor that has multiple effects on cells such as monocytes, macrophages, and bone marrow progenitors (1, 2). Indeed, MCSF plays an essential role in controlling the maturation, shape, survival, and function of tissue macrophages and osteoclasts (3). All of MCSF’s effects are triggered by one high-affinity transmembrane receptor (MCSF-R), which is generated by the c-fms protooncogene (4). Through binding to its receptor, MCSF upregulates the expression of additional markers of macrophage differentiation and promotes the expansion of macrophage progenitor populations (5). By aiding in its chemotactic, phagocytic, and destructive activities, MCSF supports monocyte and macrophage defense mechanisms (6). Therefore, it is clear that MCSF plays a crucial role in regulation of downstream immunological responses, early defense against invading pathogens, tissue homeostasis, and tissue repair based on its effects on this myeloid lineage (3).
In mammals, there are three different biologically active MCSF isoforms, a heavily glycosylated form that associates with the extracellular matrix, a membrane-bound form that can be released by the action of proteases, and a soluble form (7–9). Endothelial cells are responsible for producing the soluble form of MCSF that circulates in the blood, while other forms of MCSF are produced locally in the tissues (10, 11). Moreover, many cell lines produce MCSF in vitro, either naturally or after being induced. In vivo, MCSF has been hypothesized to enhance M2 polarization because of homeostatic expression and the overall M2-like phenotype of resident macrophage populations under normal conditions (12, 13). Several diseases, including inflammatory and autoimmune conditions, as well as viral and bacterial infections, have been linked to MCSF in mammals (14–16).
Macrophage-like cells may be found in almost all multicellular creatures and their roles have been substantially preserved throughout evolution. While MCSFs have been extensively studied and documented in mammals, this is not the case for lower vertebrates (17). Goldfish (Carassius auratus) was the first non-mammalian species to have their MCSF gene identified and characterized (18). Subsequent discoveries included grouper (Epinephelus coioides) (19, 20), zebrafish (Danio rerio), rainbow trout (Oncorhynchus mykiss) (21, 22), Japanese flounder (Paralichthys olivaceus) (17). Although numerous studies have been conducted on MCSFs (19, 21, 23–25), only a few have pointed to a role for teleost MCSFs in fish immune systems (20, 25). Furthermore, limited functional studies have been conducted, in particular the effects of MCSF on monocyte differentiation and proliferation in goldfish (26). In addition, recombinant trout MCSF may boost CXCR3 expression in head kidney macrophages and promote proliferation of kidney head leukocytes (21). Recent studies in Japanese flounder (Paralichthys olivaceus) have looked at the immunomodulatory and physiologic effects of MCSF (17). Despite the fact that Wang and co-authors found two MCSF genes in fish with distinct gene organization and expression patterns (21), it is surprising that previous studies have focused more on MCSF-1 compared to MCSF-2.
Currently, studies concerning the functional characterization of MCSF-2 in teleosts are limited. Therefore, the present study will, examine the immunomodulatory ability and functional properties of MCSF-2 in teleosts, and specifically in the goldfish.
2 Materials and methods
2.1 Fish
Goldfish (Carassius auratus L.) of the Crucian carp variety were obtained from a local aquatic market in Ningbo and housed in the aquatic facilities of the Laboratory of Aquatic Animal Diseases and Immunology, University of Ningbo. Fish were acclimated for a minimum of 3 weeks, fed daily with pellets, and maintained at room temperature under a simulated natural photoperiod using a circulating water system. Randomly selected male or female goldfish, approximately 2 years old, measuring between 10 and 15 cm in length, were used for the experiments. Before handling, the fish were sedated with a solution of TMS (tricaine methane sulphonate) at 40 to 50 mg/L in water. The experimental protocol, identified as Protocol # NBU20210046, was approved by the Ningbo University Council of Animal Care, and all studies were conducted in accordance with the established regulations.
2.2 Bacterial infection
To prepare the A. hydrophila strain for infection experiments, TSA plates were used to incubate during 24 h at 28°C, the bacterial culture. The culture was then transferred to TSB medium and shaken for 12 h at 28°C. The sediment was rinsed three times with phosphate-buffered saline (PBS) after centrifugation at 1,000 g for 5 minutes at 4°C. Two groups of fish were used for the infection assay: a control group and an experimental group. The control group received 100 µL of sterile PBS, while the experimental group received 100 µL of sterile PBS containing A. hydrophila at a concentration of 1 × 108 CFU/mL.
2.3 RNA isolation and cDNA synthesis
Tissues including spleen (Sp), kidney (Ki), brain (Br), heart (He), liver (Li), muscle (Mu), intestine (In) and gills (Gi) were taken from 6 healthy goldfish. Following homogenization, tissues were extracted using Trizol reagent (Omega Biotech, China) to isolate RNA. After being checked for purity and concentration, RNA samples were stored at -80°C and analyzed using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, USA). A Prime-Script RT Reagent Kit with gDNA Eraser (Takara, Japan) was used to reverse transcribe RNA and make first-strand cDNA. Until needed, cDNA samples were kept at -20°C.
2.4 Gene cloning, and sequence analysis
The primers were designed according to the sequences obtained from the previously generated transcriptome database (27). All primers employed in cloning and expression of gfMCSF-2 are provided in Supplementary Table 1 (Table S1). For cDNA template preparation, we extracted RNA from goldfish kidney, followed by utilizing the PerfectStart Green qPCR SuperMix (TransGen, China). To amplify the product, we followed the thermocycling parameters consisting of a first step of denaturation at 95°C for 2 min, followed by 30 rounds of amplification at 95°C for 30 s, 55 for 45 s, 72°C for 2 min for 30 s, a last step of extension at 72°C for 12 min. Sequence study of gfMCSF-2 nucleotides and predicted amino acids, we used the NCBI BLAST program and the ExPASy server Molecular Biology (http://us.expasy.org). The Compute pI/Mw tool at ExPASy (http://www.expasy.ch/) was used to predict the molecular mass and isoelectric point of the putative gfMCSF-2, and SignalP was utilized to predict the signal peptide. The alignment of several sequences was performed by CLUSTAL-W (http://www.genome.jp/tools-bin/clustalw), and images were generated using ESPript 3.0 (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi). The p-distance method of the MEGA 11 program, with values expressed as percentages, was employed to perform the gfMCSF-2 phylogenetic analysis, 10,000 bootstrap replicates.
2.5 Real-time quantitative PCR analysis
Goldfish specific MCSF-2 primers were created with Primer Express software (Applied Biosystems, USA). Tissues and cell groups from six different goldfish (n = 6) were analyzed using the ABI QuantStudio 5 instrument (Thermo Fisher Scientific, USA) for Q-PCR. Thermocycling settings were 95°C for 10 min, 40 cycles of 15 s at 95°C, and 60°C for 1 min. GraphPad Prism (GraphPad Software, USA) was utilized for statistical analysis and graphing purposes. Each gene’s transcription levels were quantified using a standard curve and standardized to EF-1α. Expression variations were computed by comparing the experimental group’s mean expression level to the control groups.
2.6 Isolation of goldfish head kidney leukocytes
HKLs were isolated from goldfish using standard protocols (28). Briefly, the kidneys were excised and kept in ice-cold NMGFL-15 medium. The tissues were then homogenized, and the cell solution was suspended on top of 51% Percoll using a 400 g centrifuge at 4°C for 25 min to remove debris. At the 51% Percoll/medium junction, the cells were collected and washed twice with incomplete medium before resuspending the HKLs in complete medium. Viable HKLs were counted using the trypan blue exclusion method before use in experiments.
2.7 Analysis of MCSF-2 expression in activated goldfish HKLs
To prepare heat-killed and alive bacteria, the methods previously described were used (29). Six goldfish were used for this study (n = 6), and leukocytes from their kidneys were utilized to investigate gfMCSF-2 expression. Leukocytes were cultured at a density of 2 x 106 cells/mL for a volume of 500 μL in each well on a 24 plate, then exposed to either 1 x 107 CFU/mL of live A. hydrophila, 1 x 107 CFU/mL of heat-killed A. hydrophila, or 10 µg/mL of LPS (Sigma L2630). A total of 1 x 106 cells were used in 500 µL of complete NMGFL-15 media for each treatment group. After the cells were challenged, they were kept alive at 26 °C for 6 and 12 h. The TranScript Uni All-in-One First-Strand cDNA synthesis Supermix (Transgen, China) was used to synthesize cDNA from various cell types’ RNA. The reference gene EF-1α was used to determine gfMCSF-2’s relative expression levels.
2.8 Prokaryotic expression and scale up production of recombinant goldfish recombinant MCSF-2
Goldfish MCSF-2 ORF excluding singnal peptide was amplified by PCR using gene-specific primers that included 5’-end BamH I and Hind III insertions (Table S1). Restriction enzymes BamH I and Hind III were used to thoroughly digest the PCR result. (Thermo Fisher Scientific, USA), then ligated to the pET32a (+) vector digested with BamH I/Hind III. This resulted in the creation of the recombinant plasmid pET32a-gfMCSF-2, which were subsequently converted into competent E. coli (BL21/DE3) cells (TransGen, China). To express the RgMCSF-2 proteins, 0.1 mM IPTG induction was carried out at 16°C overnight, and recombinants proteins expression was analyzed by SDS-PAGE. Purification of the recombinant protein was accomplished by using Ni-NTA Sefinose Resin (Sangon, China) as directed by the manufacturer. As a control protein in subsequent experiments, the pET32a (+) vector containing a thioredoxin tag without an insert was expressed and purified in the same manner. Subsequently, a ProteoSpin Endotoxin removal column (Norgen Biotek, USA) was used to purify the recombinant protein after it was dialyzed overnight at 4°C in 1 x PBS. Micro BCA Protein Assay Kit (Beyotime, China) was used to determine protein content.
2.9 Purification of recombinant proteins
The recombinant proteins of the genes studied were transformed into E. coli Transetta (DE3) (TransGen, China). After plates were kept at 37°C for one night, cells were grown in LB+AMP broth until the OD600 hit 0.6–0.8. A final concentration of 0.1 mM IPTG was added to the cell culture, which was then kept at 37°C for 3–5 hours, pelleted, and kept at -80°C overnight or until use. The pellet was dissolved with ultra-sonication for 10 minutes after being mixed with Solution I (150 mM NaCl, 1 mM EDTA, 2 M Urea, Triton (1X) 0.5%, pH=7.9). After centrifugation, Ultra-sonication was repeated for 10 minutes after the addition of solution II (150 mM NaCl, 20 mM Tris-HCL, 30 mM Imidazole, 8 M Urea, Triton (1X) 0.2%, pH= 7.9). To acquire the supernatants, we centrifuged the samples at 7,500 rpm for 20 minutes at 4°C after the prior procedure. For protein separation, a Nickel-nitrilotriacetic acid (Ni-NTA) column (Sangon Biotech, China) was used. Solution II was used to clean the Ni-NTA column, and the sample was left in the column overnight at 4°C while it was constantly shaken. The sample was passed through the Ni-NTA column and washed the next day with solution III (NaCl 150 mM, Tris-HCL 20 mM, Imidazole 20 mM, Urea 6 M, Triton (1X) 0,1%) to remove any residual material. Proteins were recovered from the column after being eluted with Elution solution (10 mM NaCl, 2 mM Tris-HCL, 100 mM Imidazole, 1.5 M Urea).
2.10 Renaturation of purified recombinant proteins
Subsequently, the denatured proteins were renatured overnight in 10 times the volume of renaturation buffer (5 mM EDTA, 2 mM oxidized glutathione, 4 mM reduced glutathione, 50 mM sodium borate, pH = 8.5). To eliminate the excess urea and imidazole molecules, the renatured protein was added to a 20 kDa dialysis bag and stored overnight at 4°C in 1 X PBS. The following day, PEG20000 [HO(CH2CH2O)nH] was used to remove excess buffer, and protein was collected in new tubes. Western blot analysis confirmed the presence of protein, and the concentration was determined using a BCA Protein Assay Kit (Beyotime, China) and stored at -80°C for future use.
2.11 Western blot analysis
The expressed and purified recombinant rgMCSF-2 and recombinant thioredoxin (rTrx) were analyzed by western blot. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was used to separate the samples, and then they were transferred to a nitrocellulose membrane at 200 mA for 2 h. Primary antibody [anti-His (1:3,000)] was incubated with the membrane at 4°C overnight after being blocked with 5% skim milk in TBST buffer (0.05% Tween-20 in TBS, pH 7.4) for 2 hours at room temperature. The membrane was rinsed and incubated for 1 hour the next day, with a secondary antibody [HRP-conjugated goat anti-mouse IgG (1:5,000)] for 1 h at room temperature. Detection of the recombinant proteins was conducted by Tanon 5200 Chemiluminescent Imaging System (Tanon, China).
2.12 Detection of cell proliferation of HKLs with MTT assay
Goldfish HKLs suspension (1 x 105 cells/mL) was plated in 96-well dishes and treated with rgMCSF-2 at various concentrations (0.1, 0.25, 0.5, 0.75 and 1 μg/mL) for 24 h and 48 h. As a control, rTrx was added at 5 μg/mL. After centrifugation at 230 g for 5 min, MTT was added to each well, and the plates were kept in the incubator for an extra 4 h. The formazon salts were dissolved by adding DMSO, and the optical density was measured at 540 nm using a microplate reader (Bio-rad, USA). Three independent experiments were conducted.
2.13 Flow cytometric analysis of HKLs
After being isolated, HKLs from goldfish were seeded at densities of 1×106 cells/well on a 6-well plate. Cells were exposed to concentrations of rgMCSF-2 ranging from 0.5 to 1 μg/mL. As a control, we employed rTrx at a final concentration of 5 μg/mL. The samples were then analyzed using a Fluorescence-activated cell sorter (FACS) Calibur flow cytometer (Becton Dickinson, USA) 24 h and 48 h after treatment, while the plates were incubated at 20°C. Cellular forward (size) and side (internal complexity) scatter light patterns were measured across all treatment groups (n = 3).
2.14 Transcription factor and pro-inflammatory cytokine gene expression in goldfish leukocytes triggered by rgMCSF-2
To evaluate the effect of rgMCSF-2 on goldfish leukocytes, six different fish were used to create primary cultures that were then exposed to either 5 μg/mL of rgMCSF-2 or 5 μg/mL of rTrx (as a control). TRIzol reagent was used to extract RNA from the cells after they had been incubated at 26°C for 6 h and 12 h. Quantitative PCR was used to analyzed the mRNA levels of a variety of transcription factor genes and pro-inflammatory cytokine relative to the housekeeping gene EF-1α. In a total volume of 500 μL of complete NMGFL-15 media, 1 × 106 cells were used for each treatment group.
2.15 Statistical analysis
All experimental data were analyzed using one-way ANOVA, and then compared to the control and treatment groups using Dunnett’s post hoc test P < 0.05 was used as the threshold for significance.
3 Results
3.1 Sequence analysis and characterization of goldfish MCSF-2
The goldfish MCSF-2 nucleotide sequence has been deposited to the Genbank under the Accession No. OQ459355. The goldfish MCSF-2 cDNA transcript exhibited a distinct ORF comprising 804 base pairs (bp) and encoding a protein of 267 aa. Notably, it possessed a signal peptide of aa (Figure S1). Comparative analysis of goldfish MCSF-1 and MCSF-2 coding region nucleotide sequences was performed using Clustal multiple sequence alignment (Figure S2). These findings highlight the differential characteristics of the gfMCSF isoforms, emphasizing the unique attributes of gfMCSF-2 in relation to its precursor gfMCSF-1. Additionally, sequence alignment of MCSF-1 and MCSF-2 protein sequences indicated obvious difference between the two proteins (Figure S3). These results strongly suggest that MCSF-1 and MCSF-2 represent distinct proteins with notable variations.
A conservative CSF-1 superfamily domain was predicted and observed between positions 38 and 155 aa. The theoretical molecular weight (Mw) of full gfMCSF-2 protein was 30.57 kDa, and its isoelectronic point (pI) was 6.14. Based on multiple sequence alignment, gfMCSF-2 has the highest sequence identity with MCSF-2 homologs from C. carpio (94%), S. rhinocerous (92%), C. idella (89%), and the lowest identity (18-20%) with those from mammals, and aves (Table S2). In addition, the secondary structure of gfMCSF-2 is similar to that of human MCSF-2 (Figure S4). In order to investigate the phylogenetic relationships of gfMCSF-2, we retrieved homologous amino acid sequences from various teleost fishes through the NCBI database. These sequences were utilized to construct a phylogenetic tree encompassing multiple vertebrate species. The MEGA 11 software and the p-distance method were employed to generate a neighboring joining tree. The resulting phylogenetic analysis, depicted in Figure 1, demonstrated the closest evolutionary relationship between gfMCSF-2 and the MCSF-2 of C. carpio. Additionally, when examining the phylogenetic analysis of goldfish MCSF-1 and MCSF-2 alongside other species, we observed a clear distinction between two distinct groups, as illustrated in Figure S5. This observation implies a genetic consistency between these species throughout their evolutionary history, as reflected in the phylogenetic tree.
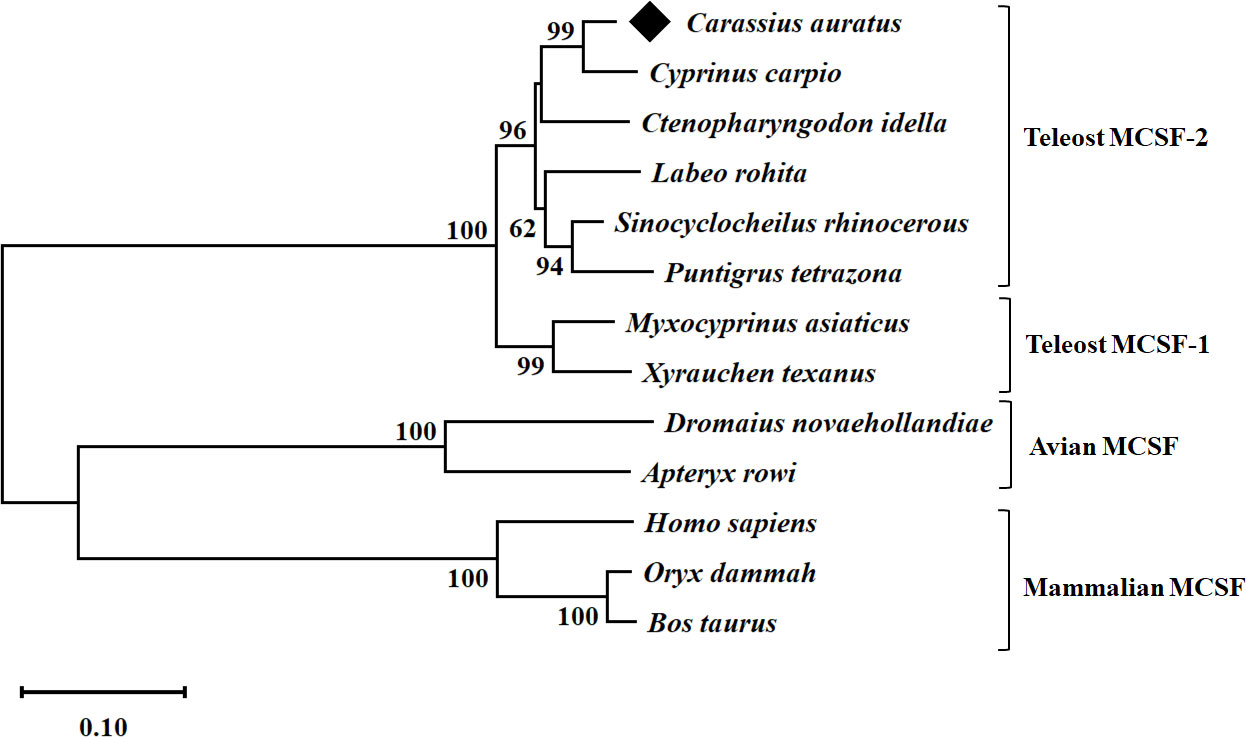
Figure 1 Phylogenetic analysis of goldfish MCSF-2 from different species. The evolutionary history was inferred using the p-distance method. The optimal tree is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (10,000 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method and are in the units of the number of base substitutions per site. All ambiguous positions were removed for each sequence pair (pairwise deletion option). Evolutionary analyses were conducted in MEGA 11.
3.2 MCSF-2 expression in healthy goldfish tissues
qRT-PCR was carried out in eight different tissues of healthy goldfish, with the muscle tissue serving as the reference tissue for the analysis. As shown in Figure 2, the mRNA expression of gfMCSF-2 was found in all tested organs. The spleen had the highest mRNA level of MCSF-2, according to the analysis of gfMCSF-2 expression, followed by the kidney and the gills (Figure 2). The mRNA levels in the muscle, intestine, and heart were found to be relatively lower than other tissues (Figure 2).
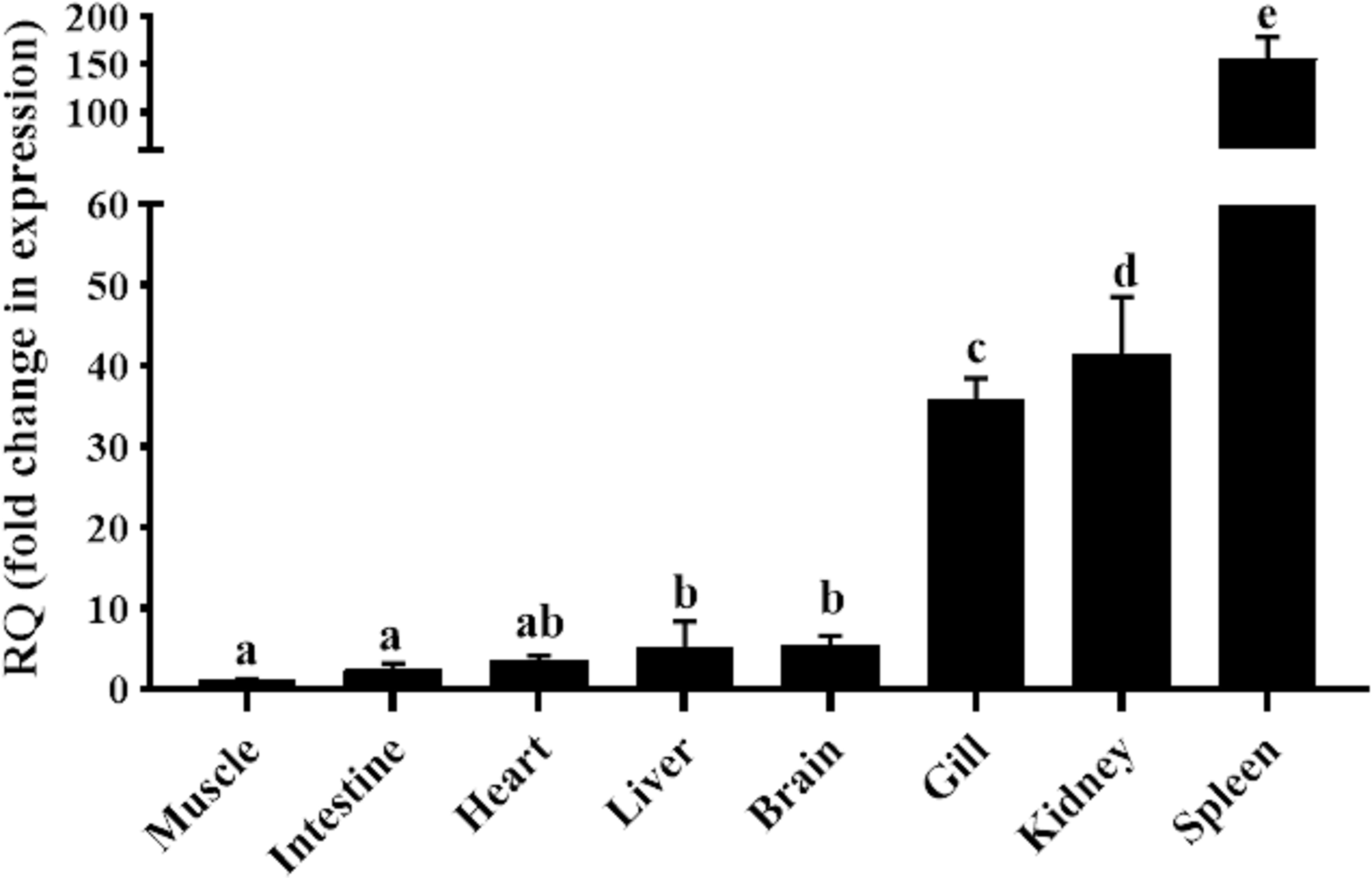
Figure 2 Expression analysis of MCSF-2 in tissues obtained from normal goldfish. Analysis of the relative tissue expression was done using tissues from six fish (n = 6), qPCR was done in triplicate for each tissue. The expression of MCSF-2 was relative to endogenous control gene, EF-1α. All results were normalized to lowest expression tissue (muscle). Statistical analysis was performed using one-way ANOVA followed by Dunnett’s post-hoc test. Different letters above each bar denote statistically different (P < 0.05), and the same letter indicates no statistical differences between groups.
3.3 Analysis of goldfish MCSF-2, TNFα and IL-1β expression in HKLs treated with chemical stimuli and bacteria
As for TNFα and IL-1β which were used in this experiment as a positive control, a significant increase of these cytokines’ mRNAs level was observed after treatment of goldfish HKLs with the various stimuli at 6 h and 12 h when compared with the control groups (Figure 3).
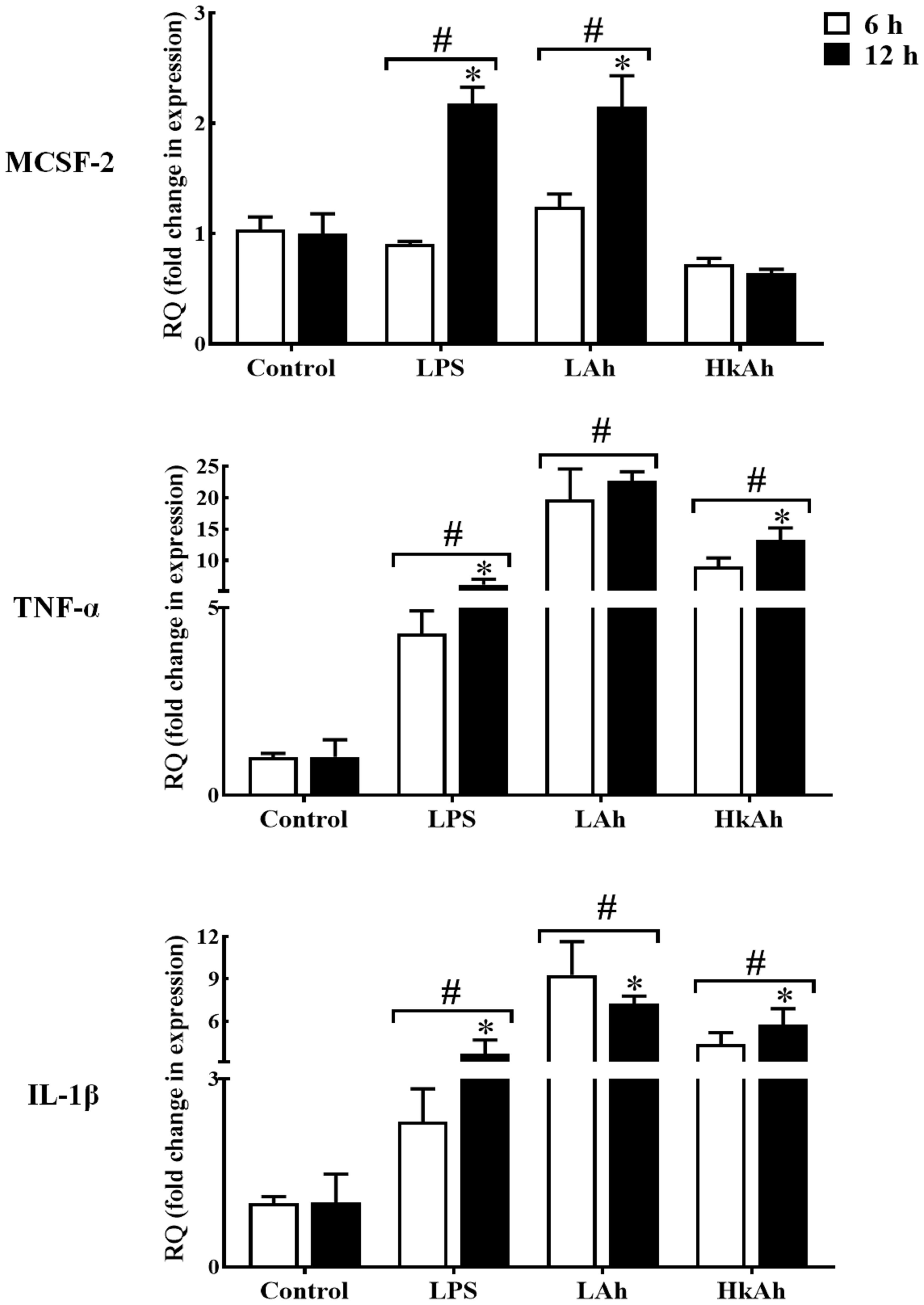
Figure 3 Quantitative expression of goldfish MCSF-2, TNF-α and IL-1β in leukocytes treated with different stimuli. The expression of MCSF-2, TNF-α and IL-1β in HKLs treated with either rTrx as a control, or lipopolysaccharide (LPS), heat-killed and live A. hydrophila. The expression of MCSF-2, TNF-α and IL-1β were examined relative to the endogenous control gene, elongation factor 1 alpha (EF-1α). The pound sign (#) indicates the significant difference at P < 0.05 compared to the control at 6 h or 12 h time point. The asterisk (*) denotes the significant difference at P < 0.05 between 6 h and 12 h treated groups.
3.4 Recombinant goldfish MCSF-2 expression and purification
We expressed rgMCSF-2 in E. coli and purified it using the Ni-NTA Sefinose Resin protein purification system to investigate the functional activities of gfMCSF-2. A single band of approximately 50.27 kDa was observed in SDS-PAGE analysis of Commassie blue stained rgMCSF-2, which included the N-terminal six-histidine tag and Trx fusion protein (Figure S6). Figure S6 shows that the recombinant protein was also verified by Western blotting using an anti-His antibody.
3.5 Effects of rgMCSF-2 treatment on cell proliferation of goldfish HKLs
Cultured HKLs were exposed to either rTrx as a control at a final concentration of 5 μg/mL or rgMCSF-2 at concentrations of 0.1, 0.25, 0.5, 0.75, and 1 μg/mL to determine whether rgMCSF-2 might stimulate the proliferation of immune cells. Treatment with rgMCSF-2 for 24 h resulted in mild cell proliferation at 0.75 and 1 μg/mL, while treatment for 48 h showed significant cell proliferation at all concentrations tested except 0.1 μg/mL (Figure 4).
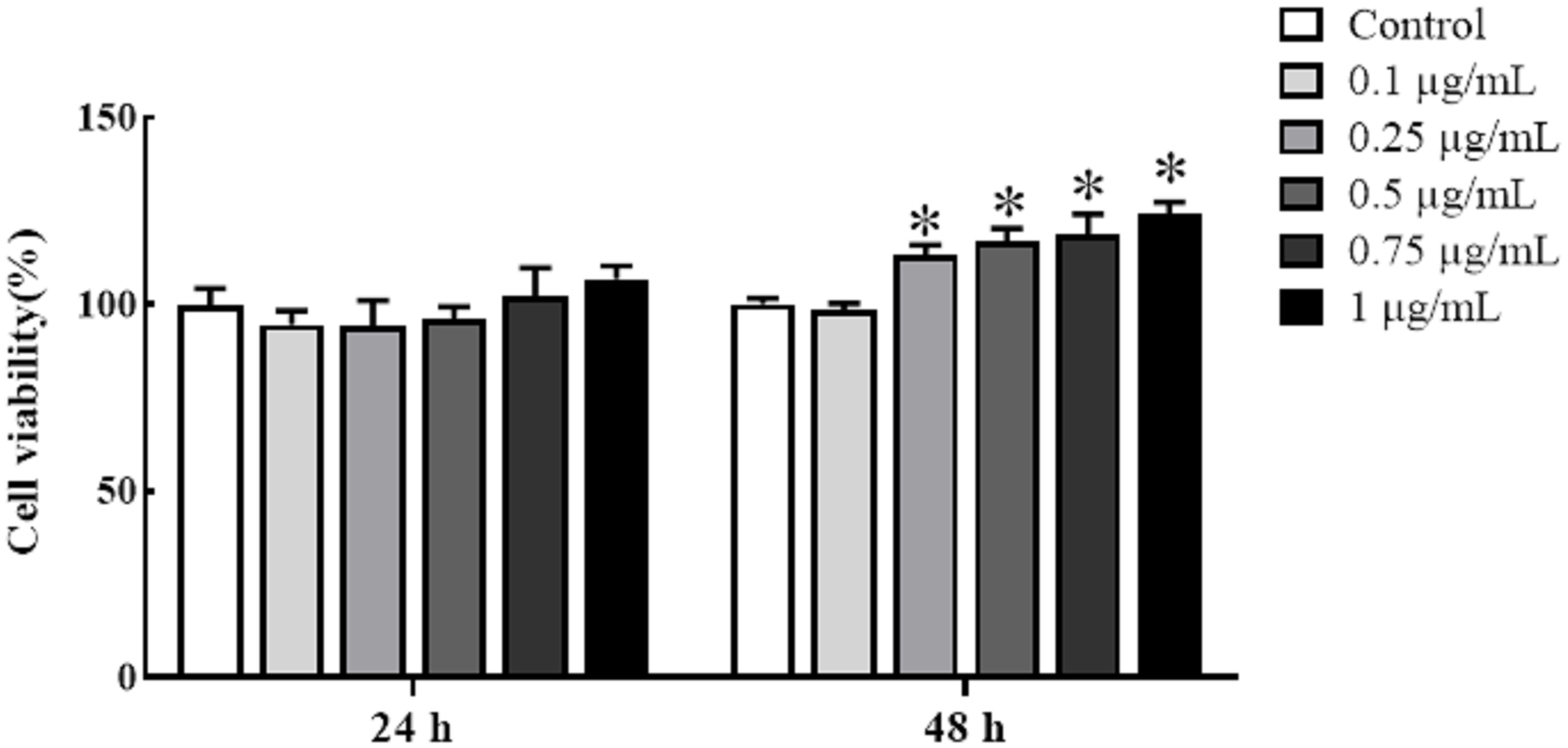
Figure 4 Effects of rgMCSF-2 on the proliferation of goldfish leucocytes using MTT assay. Goldfish head kidney leucocytes were treated with or without rgMCSF-2 at different concentrations for 24 h and 48 h. The proliferation of the cells was determined by MTT assay. Data are presented as means ± SEM (n = 3). Significance is denoted by (*) compared to the reference sample (P < 0.05).
3.6 Flow cytometric results of goldfish HKLs
Sorted goldfish leucocytes were treated with recombinant goldfish MCSF-2 at concentrations of 0.1, 0.25, 0.5, 0.75, and 1 μg/mL. Over the course of two days, the influence of rgMCSF-2 on cell differentiation in isolated subpopulations of sorted leukocytes was evaluated by monitoring these cells with flow cytometry every 24 h. The differentiation of sorted goldfish leukocytes was unaffected after treatment at the concentrations of 0.1, 0.25, and 0.5 μg/mL of rgMCSF-2. Sorted leukocytes, however, were induced to differentiate when treated with rgMCSF-2 at a concentration of 1 μg/mL (Figure 5).
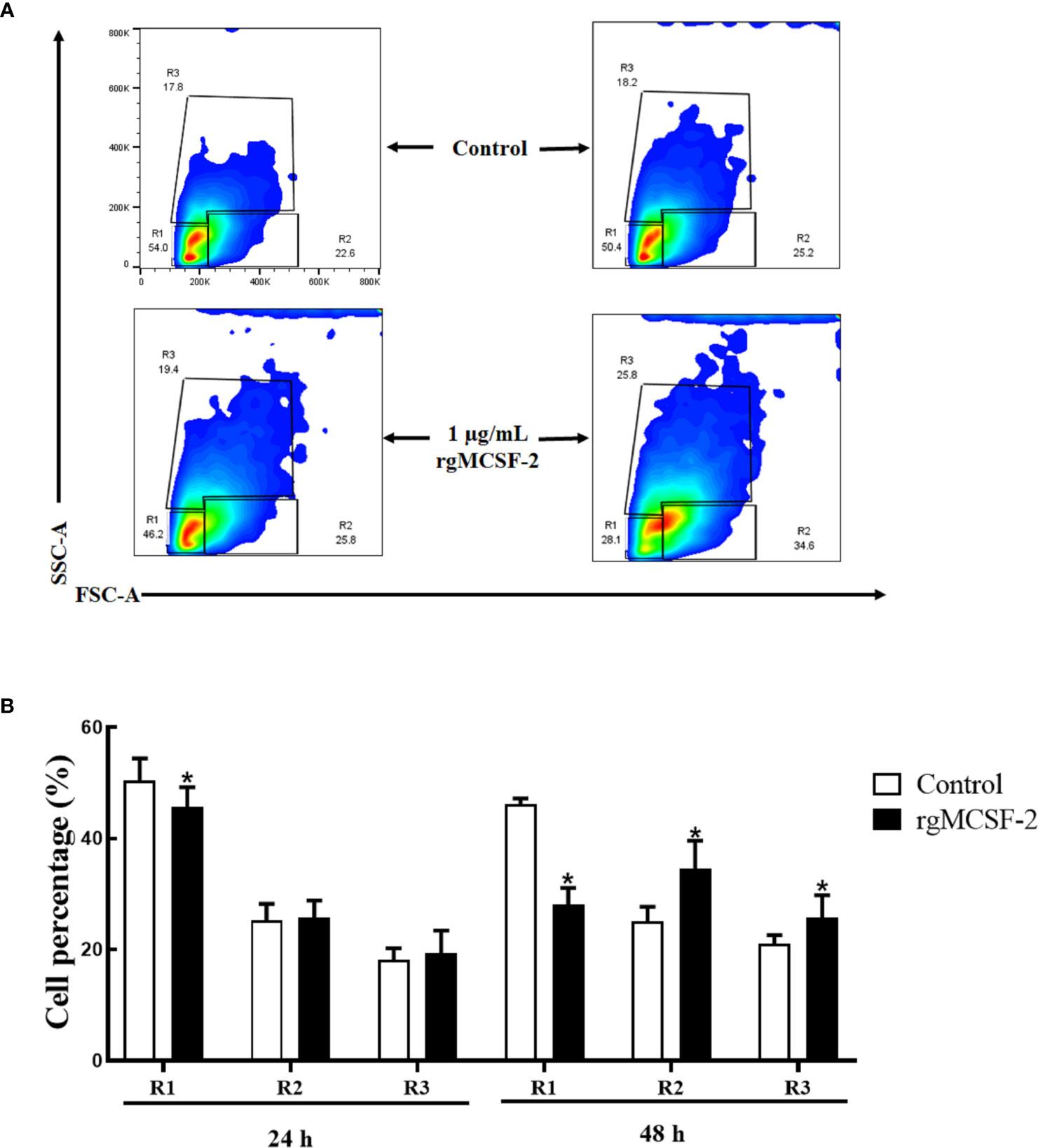
Figure 5 Flow cytometric analysis showing the differentiation of goldfish progenitors, after treatment with 1 µg/mL rgMCSF-2. (A) Analysis shows the results obtained at 24 h and 48 h taken from three individual fish. (B) Histogram shows the percentage of cells in R1, R2, and R3 after treatment with 1 µg/mL rTrx as a control and of rgMCSF-2 for 24 h and 48 h. Data are presented as means ± SEM (n = 3). Significance is denoted by (*) compared to the reference sample (P < 0.05).
3.7 Proinflammatory cytokine analysis in rgMCSF-2 treated goldfish HKLs
Goldfish leukocyte cultures were treated with rgMCSF-2, and we measured the induction of TNFα, IL-1β and IFNγ protein levels and gene expression. The pro-inflammatory cytokines TNFα (Figure 6A) and IL-1β (Figure 6B) had their mRNA levels significantly upregulated after 6 h and 12 h, respectively, of treatment with 5 μg/mL rgMCSF-2 in goldfish HKLs. IFNγ mRNA production, however, was not significantly affected by rgMCSF-2 (Figure 6C).
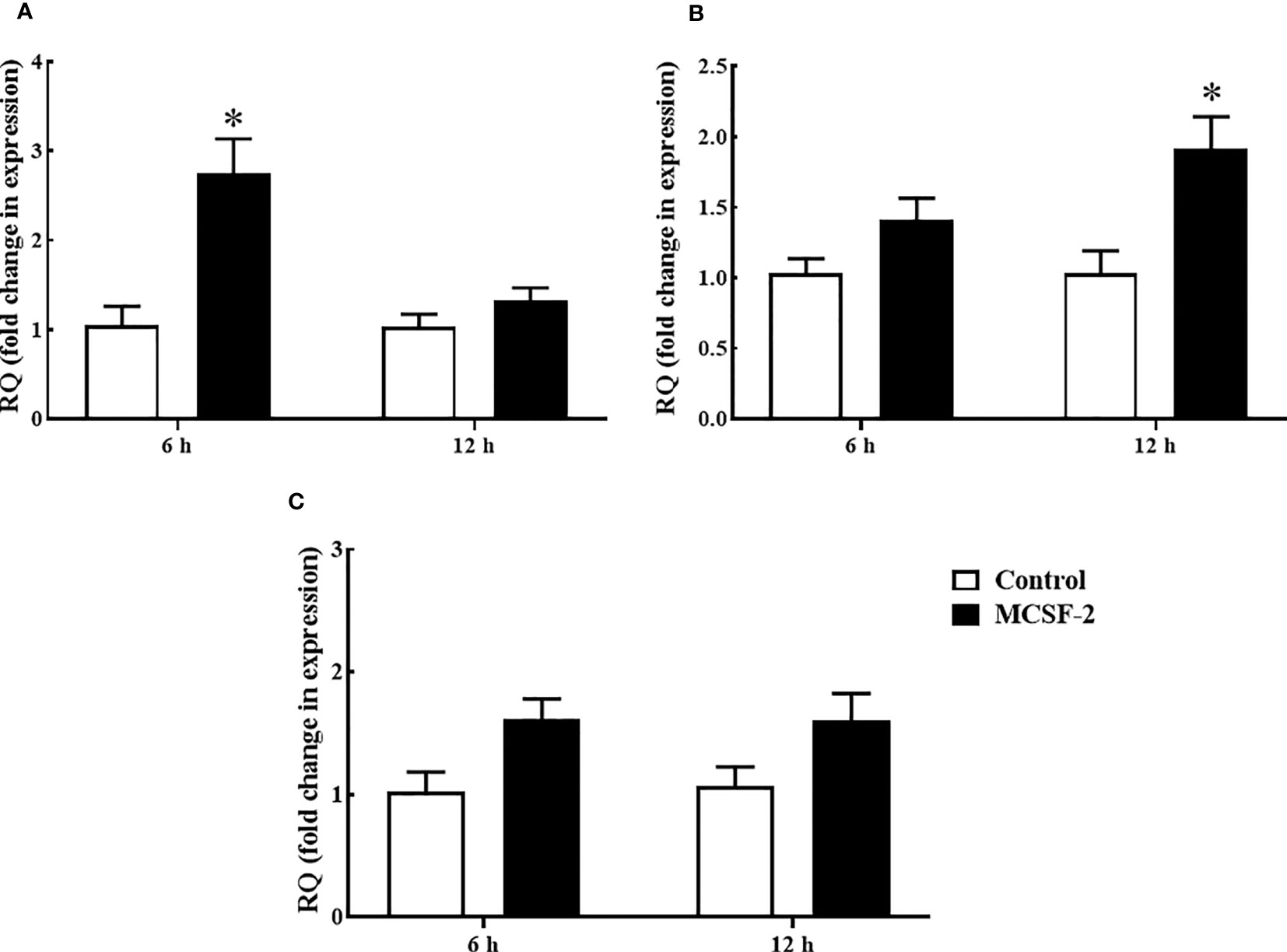
Figure 6 Quantitative expression analysis of goldfish proinflammatory cytokines TNF-α (A), IL-1β (B) and IFNγ (C) in HKLs treated with 5 µg/mL rgMCSF-2 for 6 h and 12 h. The expression of goldfish MCSF-2 was examined against the endogenous control gene, elongation factor 1 alpha (EF-1α). Expression values were normalized to those seen in rTrx -treated cells. Results are the mean ± S.E.M. of primary leukocyte cultures established from six individual fish (n = 6). The asterisk (*) denotes the significant difference at P < 0.05 between the control and MCSF2-treated groups.
3.8 Transcription factor analysis in rgMCSF-2 treated goldfish HKLs
Next, we looked at whether or not rgMCSF-2 had a role in controlling transcriptional factors. After 6 h of treatment with 5 μg/mL rgMCSF-2, HKLs showed elevated mRNA levels of transcriptional regulators MafB, GATA2, and cMyb (Figure 7A). After 12 h of treatment, mRNA levels of transcriptional regulators MafB and GATA2 were also dramatically increased in HKLs (Figure 7B). In contrast, there were no obvious changes in cJun, Egr1, PU.1, or Runx1 mRNA levels over time (Figure 7).
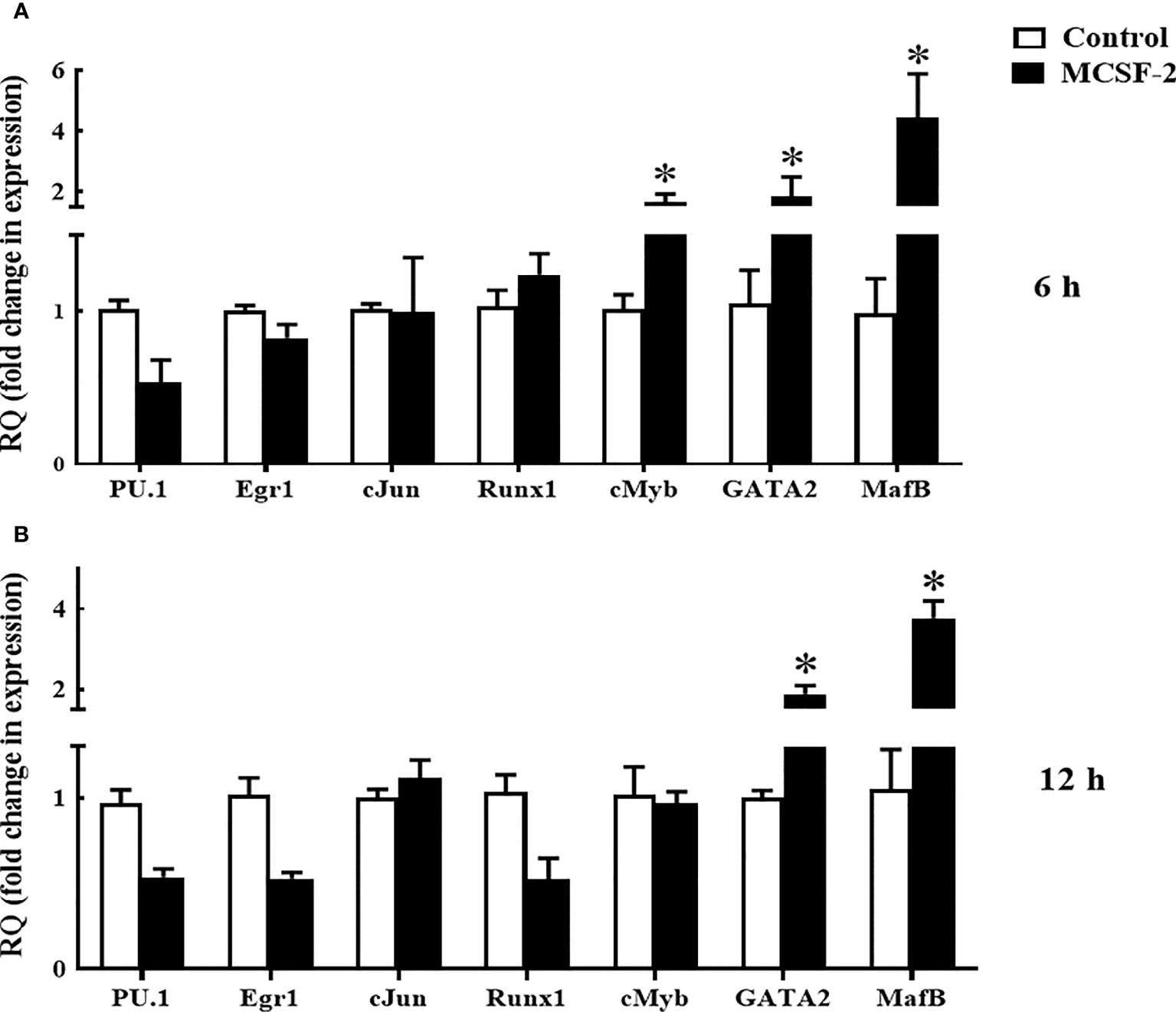
Figure 7 Quantitative expression analysis of goldfish transcriptions factors in HKLs treated with 5 µg/mL rgMCSF-2 for 6 h (A) and 12 h (B). The expression of goldfish MCSF-2 was examined against the endogenous control gene, elongation factor 1 alpha (EF-1α). Expression values were normalized to those seen in rTrx -treated cells. Results are the mean ± S.E.M. of primary leukocyte cultures established from six individual fish (n = 6). The asterisk (*) denotes the significant difference at P < 0.05 between the control and MCSF2-treated groups.
4 Discussion
MCSF is a multifunctional cytokine that promotes the survival, proliferation, and differentiation of macrophages and their precursors, also it plays a significant role in the antimicrobial immune response in a wide variety of species, including mammals, birds, and teleosts (4, 30). Wang and coauthors found two MCSF genes in fish, both of which are differentially expressed and have distinct gene structures (21). Although MCSF has also been identified in teleosts, the majority of the research to date has concentrated on MCSF-1. This research described the expression and functional properties of the goldfish MCSF-2 gene.
From our previously generated transcriptome database, the goldfish MCSF-2 gene was cloned and isolated. The gfMCSF-2 cDNA transcripts contained an ORF of 804 bp encoding 267 aa, which is nearly identical to the MCSF-2 genes of rainbow trout (276 aa) and zebrafish (284 aa) (21), indicating that the structural features of MCSF-2 have been conserved in vertebrates throughout evolution. The observed differences in both protein and nucleotide sequences provide valuable insights into the evolutionary divergence and genetic variation of MCSF-1 and MCSF-2. The substantial gaps in the alignment results suggest the presence of insertions or deletions, indicating structural differences between the two proteins. These variations may contribute to distinct functional properties or regulatory mechanisms associated with gfMCSF-2. The phylogenetic tree clearly demonstrated that gfMCSF-2 is a member of the teleost MCSF-2 family, showing a high level of cluster with the MCSF-2 proteins from other fishes. Following the same pattern as previous research (17, 21), gfMCSF-2 was clustered with fish from the class Actinopterygii, and more specifically with Cyprinus carpio, indicating that MCSF-2 in teleosts has been evolutionarily conserved.
Gills, kidneys, and spleen had relatively higher gfMCSF-2 expression, while the heart, intestine, and muscle had the lower mRNA level. These findings matched those of previous studies on MCSF-2 tissue distribution in O. mykiss and D. rerio, which found that MCSF-2 expression was typically highest in the kidney and spleen (21). Therefore, kidney is where most bony fishes’ hematopoiesis and monocyte/macrophage formation takes place, the spleen also being a particularly macrophage-rich organ (31).
To investigate MCSF-2 activators in fish, we first treated HKLs isolated from goldfish primary kidneys with LPS and fish pathogen A. hydrophila for 6 h and 12 h. Our results showed that the gfMCSF-2 mRNA level was increased in the LPS and live A. hydrophila groups compared to the control group after 12 h. Also, the results of the LPS and live A. hydrophila treatments between 6 and 12 h were significantly different. However, gfMCSF-2 mRNA expression was reduced after treatment with heat-killed A. hydrophila, although this change was not statistically significant at 6 h and 12 h. These findings are in line with previous studies in higher vertebrates that have demonstrated the stimulatory effect of LPS on MCSF production. For example, in human monocytes, LPS stimulation leads to increased production of MCSF. Similarly, in murine macrophages, LPS treatment induces the secretion of MCSF (5). This highlights the role of LPS as a potent inducer of MCSF production in higher vertebrates. In mammals, LPS has been shown to induce the expression of MCSF in various cell types (32). In an experimental rabbit model, Eichinger and colleagues found that MCSF was significantly up-regulated following valvular surgery and Staphylococcus aureus infection (33). Mice exposed to Leishmania donovani amastigote antigens have their serum MCSF production stimulated (34). Furthermore, the induction of MCSF by LPS is associated with the recruitment and activation of macrophages, which play a crucial role in the innate immune response. MCSF serves as a chemoattractant for monocytes and macrophages, promoting their differentiation and activation (11). This recruitment and activation of macrophages are essential for the clearance of pathogens and the initiation of an appropriate immune response. Unfortunately, there is no relative research reports on MCSF-2 regarding immune defense against pathogenic infection in teleost. Therefore, our results show that HKLs of goldfish treated with live A. hydrophila and LPS significantly increased the expression of gfMCSF-2, implying that MCSF-2 may be involved in fish resistance to bacterial and viral infections, as well as in mammalian defense.
Mammalian MCSF was discovered to be a hematopoietic growth factor that promotes the growth, survival and differentiation of monocytes, macrophages, and their progenitors in bone (35–37). In teleost, goldfish MCSF-1 elicited functional responses including increased macrophage differentiation, proliferation from hematopoietic phagocytosis, chemotaxis, precursors, and production of antimicrobial reactive oxygen and nitrogen intermediates (18, 23, 26). Administration of MCSF-1 to goldfish in vivo increased the quantity of bloodstream-circulating monocytes (26). However, the role of MCSF-2 in teleost immune-related cells has not yet been fully reported. Hence, our study investigated the potential effects of rgMCSF-2 on immune cell proliferation and differentiation in goldfish. We found that treatment with rgMCSF-2 resulted in both cell proliferation and differentiation. Our study investigated the potential effects of rgMCSF-2 on immune cell proliferation and differentiation in goldfish. We found that treatment with rgMCSF-2 resulted in both cell proliferation and differentiation. In terms of cell proliferation, our results demonstrated a time-dependent effect of rgMCSF-2 treatment. After 24 h of treatment, mild cell proliferation was observed at concentrations of 0.75 and 1 μg/mL, while significant cell proliferation was evident at all concentrations tested except 0.1 μg/mL after 48 h. These findings suggest that rgMCSF-2 has the capacity to stimulate immune cell proliferation in goldfish. Similar stimulatory effects of MCSF on cell proliferation have been reported in higher vertebrates. In mammals, MCSF has been shown to promote the proliferation of mononuclear phagocytes, including macrophages and osteoclasts (11). For instance, in murine macrophages, MCSF treatment induces their proliferation and enhances their survival (13, 37). These studies support the notion that the proliferative effects of MCSF-2 on immune cells are conserved across vertebrates. Regarding cell differentiation, our results indicated that treatment with rgMCSF-2 at concentrations of 0.1, 0.25, and 0.5 μg/mL did not significantly affect the differentiation of sorted goldfish leukocytes. However, at a concentration of 1 μg/mL, rgMCSF-2 induced the differentiation of sorted leukocytes. This observation suggests that rgMCSF-2 can drive the differentiation of specific immune cell subpopulations in goldfish. The capacity of MCSF to induce cell differentiation has been well-documented in higher vertebrates. In mammals, MCSF is a critical factor for the differentiation of monocytes into macrophages and their subsequent activation (11). Moreover, MCSF is involved in osteoclastogenesis, promoting the differentiation of precursor cells into functional osteoclasts (13, 37). These findings indicate the conserved role of MCSF-2 in regulating immune cell differentiation and function across different vertebrate species.
Furthermore, it has been discovered that MCSF-1 upregulates goldfish antimicrobial responses by inducing proinflammatory gene expression (23). However, there are no relevant research reports regarding the role of MCSF-2 in regulating pro-inflammatory responses in teleosts. To explore the function of rgMCSF-2 in the stimulation of pro-inflammatory cytokine production, rgMCSF-2 was given to goldfish HKLs, and mRNA and protein levels of IFNγ, IL-1β, and TNFα were measured. After treating goldfish HKLs with 5 μg/mL of rgMCSF-2, the pro-inflammatory cytokines TNFα and IL-1β mRNA levels increased significantly. These findings mirrored those from studies in mammals, which found that MCSF can trigger the production of various cytokines, including TNFα (38), and those from studies on teleost, which found that MCSF-1 can stimulate the expression of pro-inflammatory genes including TNFα and IL-1β in goldfish and Japanese flounder (17, 23). Consequently, these findings point to the possibility that MCSF-2 in fish is essential for pro-inflammatory responses by activating TNFα and IL-1β.
In order to gain a better understanding of hematopoietic cells in teleosts and goldfish in particular, we looked at the effect rgMCSF-2 has on the differentiation of progenitor cell populations by measuring the mRNA levels of relevant transcription factors. Thus, myeloid transcription factor MafB, GATA2, and cMyb mRNA levels were significantly upregulated in HKLs after treatment with rgMCSF-2, as is typical for cell lines derived from monocytes and macrophages (39). MafB has been reported to stimulate the differentiation of myeloblasts into monocytes and macrophages (40–43) and to be highly expressed in monocytes/macrophages (29). Despite the dearth of research on the function of MafB in teleost myelopoiesis, the goldfish model revealed the presence of a MafB transcript and demonstrated that MafB mRNA increased with macrophage development in the goldfish primary renal macrophage culture system (44). Activation of myeloid genes, as well as the maintenance and expansion of multipotent progenitor cells, are all processes in which GATA2 plays a role as a transcription factor (39). An evolutionarily conserved function for cMyb in hematopoiesis is shown by the fact that it is a marker of definitive HSCs in embryonic zebrafish (45) and is required for the formation of all blood cells (46). These findings further support our prior hypothesis that rgMCSF-2 plays an important role in myeloid lineage cell proliferation and differentiation.
In conclusion, we provided insights into the functionality of gfMCSF-2 by demonstrating that rgMCSF-2 not only promoted the proliferation and differentiation of primary head kidney cells but also increased the expression of proinflammatory cytokines such as TNFα and IL-1β in HKLs. Significant increases in the mRNA levels of the transcriptional regulators MafB, GATA2, and cMyb were also triggered by rgMCSF-2. Overall, our results indicated that gfMCSF-2 may have a significant function in goldfish progenitor cell growth, differentiation, and immunological response. Thus, our findings have laid the groundwork for the functional characterization of MCSF-2 in teleosts.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
All fish experiments were conducted in accordance with the guidelines set by Council of Animal Care of Ningbo University under the Protocol # NBU20210046.
Author contributions
MG, ZB designed the research, completed most experiemnts and wrote the manuscript. MG, XY analyzed the data and provided further input. JJ and JX generated the research idea and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Zhejiang Provincial Natural Science Foundation of China (No. LQ21C190002), the National Natural Science Foundation of China (No. 42106105), the Ningbo Public Welfare Science and Technology Project (No. 2021S061), the Ningbo 2025 Major Project of Science Technology and Innovation (No. 2021Z003), and the Ningbo Yongjiang Talent Introduction Programme (No. 2021B-029-C). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1235370/full#supplementary-material
References
1. Fontana MF, de Melo GL, Anidi C, Hamburger R, Kim CY, Lee SY, et al. Macrophage colony stimulating factor derived from CD4+ T cells contributes to control of a blood-borne infection. PloS Pathog (2016) 12(12):e1006046. doi: 10.1371/journal.ppat.1006046
2. Trus E, Basta S, Gee K. Who’s in charge here? macrophage colony stimulating factor and granulocyte macrophage colony stimulating factor: Competing factors in macrophage polarization. Cytokine (2020) 127:154939. doi: 10.1016/j.cyto.2019.154939
3. Rieger AM, Hanington PC, Belosevic M, Barreda DR. Control of CSF-1 induced inflammation in teleost fish by a soluble form of the CSF-1 receptor. Fish Shellfish Immunol (2014) 41(1):45–51. doi: 10.1016/j.fsi.2014.03.035
4. Chitu V, Stanley ER. Regulation of embryonic and postnatal development by the CSF-1 receptor. Curr Top Dev Biol (2017) 123:229–75. doi: 10.1016/bs.ctdb.2016.10.004
5. Sweet MJ, Hume DA. CSF-1 as a regulator of macrophage activation and immune responses. Arch Immunol Ther Exp (2003) 51(3):169–78.
6. Roca FJ, Sepulcre MP, López-Castejón G, Meseguer J, Mulero V. The colony-stimulating factor-1 receptor is a specific marker of macrophages from the bony fish gilthead seabream. Mol Immunol (2006) 43(9):1418–23. doi: 10.1016/j.molimm.2005.07.028
7. Gow DJ, Sester DP, Hume DA. CSF-1, IGF-1, and the control of postnatal growth and development. J Leukoc Biol (2010) 88(3):475–81. doi: 10.1189/jlb.0310158
8. Droin N, Solary E. CSF1R, CSF-1, and IL-34, a “ménage à trois” conserved across vertebrates. J Leukoc Biol (2010) 87(5):745–7. doi: 10.1189/jlb.1209780
9. Sinha SK, Miikeda A, Fouladian Z, Mehrabian M, Edillor C, Shih D, et al. Local M-CSF (macrophage colony-stimulating factor) expression regulates macrophage proliferation and apoptosis in atherosclerosis. Arterioscler Thromb Vasc Biol (2021) 41(1):220–33. doi: 10.1161/ATVBAHA.120.315255
10. Ryan GR, Dai XM, Dominguez MG, Tong W, Chuan F, Chisholm O, et al. Rescue of the colony-stimulating factor 1 (CSF-1)–nullizygous mouse (Csf1 op/Csf1 op) phenotype with a CSF-1 transgene and identification of sites of local CSF-1 synthesis. Blood Am Soc Hematol (2001) 98(1):74–84. doi: 10.1182/blood.V98.1.74
11. Pixley FJ, Stanley ER. CSF-1 regulation of the wandering macrophage: complexity in action. Trends Cell Biol (2004) 14(11):628–38. doi: 10.1016/j.tcb.2004.09.016
12. Davies LC, Jenkins SJ, Allen JE, Taylor PR. Tissue-resident macrophages. Nat Immunol (2013) 14(10):986–95. doi: 10.1038/ni.2705
13. Hamilton TA, Zhao C, Pavicic PG Jr., Datta S. Myeloid colony-stimulating factors as regulators of macrophage polarization. Front Immunol (2014) 5:554. doi: 10.3389/fimmu.2014.00554
14. Higgins DM, Sanchez-Campillo J, Rosas-Taraco AG, Higgins JR, Lee EJ, Orme IM, et al. Relative levels of M-CSF and GM-CSF influence the specific generation of macrophage populations during infection with Mycobacterium tuberculosis. J Immunol (2008) 180(7):4892–900. doi: 10.4049/jimmunol.180.7.4892
15. Garcia S, Hartkamp LM, Malvar-Fernandez B, van Es IE, Lin H, Wong J, et al. Colony-stimulating factor (CSF) 1 receptor blockade reduces inflammation in human and murine models of rheumatoid arthritis. Arthritis Res Ther (2016) 18(1):1–4. doi: 10.1186/s13075-016-0973-6
16. Collison J. CSF1 drives pain in arthritis. Nat Rev Rheumatol (2018) 14(10):562–2. doi: 10.1038/s41584-018-0087-4
17. Gu H, Wang B, He J, Hu Y. Macrophage colony stimulating factor (MCSF) of Japanese flounder (Paralichthys olivaceus): Immunoregulatory property, anti-infectious function, and interaction with MCSF receptor. Dev Comp Immunol (2021) 116:103920. doi: 10.1016/j.dci.2020.103920
18. Hanington PC, Wang T, Secombes CJ, Belosevic M. Growth factors of lower vertebrates: characterization of goldfish (Carassius auratus L.) macrophage colony-stimulating factor-1. J Biol Chem (2007) 282(44):31865–72. doi: 10.1074/jbc.M706278200
19. Dan XM, Zhong ZP, Li YW, Luo XC, Li AX. Cloning and expression analysis of grouper (Epinephelus coioides) M-CSFR gene post Cryptocaryon irritans infection and distribution of M-CSFR+ cells. Fish Shellfish Immunol (2013) 35(2):240–8. doi: 10.1016/j.fsi.2013.04.019
20. Mo ZQ, Li YW, Zhou L, Li AX, Luo XC, Dan XM. Grouper (Epinephelus coioides) IL-34/MCSF2 and MCSFR1/MCSFR2 were involved in mononuclear phagocytes activation against Cryptocaryon irritans infection. Fish Shellfish Immunol (2015) 43(1):142–9. doi: 10.1016/j.fsi.2014.12.023
21. Wang T, Hanington PC, Belosevic M, Secombes CJ. Two macrophage colony-stimulating factor genes exist in fish that differ in gene organization and are differentially expressed. J Immunol (2008) 181(5):3310–22. doi: 10.4049/jimmunol.181.5.3310
22. Wang T, Kono T, Monte MM, Kuse H, Costa MM, Korenaga H, et al. Identification of IL-34 in teleost fish: differential expression of rainbow trout IL-34, MCSF1 and MCSF2, ligands of the MCSF receptor. Mol Immunol (2013) 53(4):398–409. doi: 10.1016/j.molimm.2012.09.008
23. Grayfer L, Hanington PC, Belosevic M. Macrophage colony-stimulating factor (CSF-1) induces pro-inflammatory gene expression and enhances antimicrobial responses of goldfish (Carassius auratus L.) macrophages. Fish Shellfish Immunol (2009) 26(3):406–13. doi: 10.1016/j.fsi.2008.12.001
24. Honda T, Nishizawa T, Uenobe M, Kohchi C, Kuroda A, Ototake M, et al. Molecular cloning and expression analysis of a macrophage-colony stimulating factor receptor-like gene from rainbow trout, Oncorhynchus mykiss. Mol Immunol (2005) 42(1):1–8. doi: 10.1016/j.molimm.2004.07.002
25. Katzenback BA, Belosevic M. Colony-stimulating factor-1 receptor protein expression is a specific marker for goldfish (Carassius auratus L.) macrophage progenitors and their differentiated cell types. Fish Shellfish Immunol (2012) 32(3):434–45. doi: 10.1016/j.fsi.2011.12.003
26. Hanington PC, Hitchen SJ, Beamish LA, Belosevic M. Macrophage colony stimulating factor (CSF-1) is a central growth factor of goldfish macrophages. Fish Shellfish Immunol (2009) 26(1):1–9. doi: 10.1016/j.fsi.2008.09.020
27. Li C, Jiang J, Xie J, Yang W, Wang Y. Transcriptome profiling and differential expression analysis of the immune-related genes during the acute phase of infection with Mycobacterium marinum in the goldfish (Carassius auratus L.). Aquaculture (2021) 533:736198. doi: 10.1016/j.aquaculture.2020.736198
28. Yang X, Wei H, Qin L, Zhang S, Wang X, Zhang A, et al. Reciprocal interaction between fish TGF-β1 and IL-1β is responsible for restraining IL-1β signaling activity in grass carp head kidney leukocytes. Dev Comp Immunol (2014) 47(2):197–204. doi: 10.1016/j.dci.2014.07.023
29. Xie J, Hodgkinson JW, Li C, Kovacevic N, Belosevic M. Identification and functional characterization of the goldfish (Carassius auratus L.) high mobility group box 1 (HMGB1) chromatin-binding protein. Dev Comp Immunol (2014) 44(1):245–53. doi: 10.1016/j.dci.2013.12.015
30. Wu Z, Harne R, Chintoan-Uta C, Hu TJ, Wallace R, MacCallum A, et al. Regulation and function of macrophage colony-stimulating factor (CSF1) in the chicken immune system. Dev Comp Immunol (2020) 105:103586. doi: 10.1016/j.dci.2019.103586
31. Zapata A, Diez B, Cejalvo T, Gutierrez-de Frias C, Cortés A. Ontogeny of the immune system of fish. Fish Shellfish Immunol (2006) 20(2):126–36. doi: 10.1016/j.fsi.2004.09.005
32. Furman I, Baudet C, Brachet P. Differential expression of M-CSF, LIF, and TNF-α genes in normal and malignant rat glial cells: Regulation by lipopolysaccharide and vitamin D. J Neurosci Res (1996) 46(3):360–6. doi: 10.1002/(SICI)1097-4547(19961101)46:3<360::AID-JNR9>3.0.CO;2-I
33. Eichinger WB, Grammer JB, Zhao B, Brückner J, Mendler N, Lange R, et al. Transcriptional regulation of α5β1 integrin, fibronectin, VCAM-1, MCSF-1/c-fms, and MCP-1 in atrioventricular valves after valvular surgery and Staphylococcus aureus bacteremia. Cytokine (2005) 31(6):465–72. doi: 10.1016/j.cyto.2005.07.005
34. Singal P, Singh PP. Leishmania donovani amastigote components-induced colony-stimulating factors production. Parasitol Int (2005) 54(1):9–20. doi: 10.1016/j.parint.2004.08.001
35. Tushinski RJ, Oliver IT, Guilbert LJ, Tynan PW, Warner JR, Stanley ER. Survival of mononuclear phagocytes depends on a lineage-specific growth factor that the differentiated cells selectively destroy. Cell (1982) 28(1):71–81. doi: 10.1016/0092-8674(82)90376-2
36. Sherr CJ, Stanley ER. Colony-stimulating factor 1 (macrophage colony-stimulating-factor). Pept Growth factors their receptors I (1990) 95(1):667–98. doi: 10.1007/978-1-4612-3210-0_15
37. Fixe P, Praloran V. M-CSF: haematopoietic growth factor or inflammatory cytokine? Cytokine (1998) 10(1):32–7. doi: 10.1006/cyto.1997.0249
38. Evans R, Kamdar SJ, Fuller JA, Krupke DM. The potential role of the macrophage colony-stimulating factor, CSF-1, in inflammatory responses: characterization of macrophage cytokine gene expression. J Leukoc Biol (1995) 58(1):99–107. doi: 10.1002/jlb.58.1.99
39. Katzenback BA, Karpman M, Belosevic M. Distribution and expression analysis of transcription factors in tissues and progenitor cell populations of the goldfish (Carassius auratus L.) in response to growth factors and pathogens. Mol Immunol (2011) 48(9-10):1224–35. doi: 10.1016/j.molimm.2011.03.007
40. Sarrazin S, Mossadegh-Keller N, Fukao T, Aziz A, Mourcin F, Vanhille L, et al. MafB restricts M-CSF-dependent myeloid commitment divisions of hematopoietic stem cells. Cell (2009) 138(2):300–13. doi: 10.1016/j.cell.2009.04.057
41. Gemelli C, Montanari M, Tenedini E, Zanocco Marani T, Vignudelli T, Siena M, et al. Virally mediated MafB transduction induces the monocyte commitment of human CD34+ hematopoietic stem/progenitor cells. Cell Death Differ (2006) 13(10):1686–96. doi: 10.1038/sj.cdd.4401860
42. Kelly LM, Englmeier U, Lafon I, Sieweke MH, Graf T. MafB is an inducer of monocytic differentiation. EMBO J (2000) 19(9):1987–97. doi: 10.1093/emboj/19.9.1987
43. Aziz A, Soucie E, Sarrazin S, Sieweke MH. MafB/c-Maf deficiency enables self-renewal of differentiated functional macrophages. Science (2009) 326(5954):867–71. doi: 10.1126/science.117605
44. Barreda DR, Hanington PC, Walsh CK, Wong P, Belosevic M. Differentially expressed genes that encode potential markers of goldfish macrophage development in vitro. Dev Comp Immunol (2004) 28(7-8):727–46. doi: 10.1016/j.dci.2003.11.005
45. Bertrand JY, Kim AD, Teng S, Traver D. CD41+ cmyb+ precursors colonize the zebrafish pronephros by a novel migration route to initiate adult hematopoiesis. Development (2008) 135:1853–62. doi: 10.1242/dev.015297
Keywords: fish, macrophage colony-stimulating factor 2, progenitors, hematopoiesis, cytokine
Citation: Gouife M, Ban Z, Yue X, Jiang J and Xie J (2023) Molecular characterization, gene expression and functional analysis of goldfish (Carassius auratus L.) macrophage colony stimulating factor 2. Front. Immunol. 14:1235370. doi: 10.3389/fimmu.2023.1235370
Received: 06 June 2023; Accepted: 13 July 2023;
Published: 01 August 2023.
Edited by:
Xinjiang Lu, Zhejiang University, ChinaReviewed by:
Qingguo Meng, Nanjing Normal University, ChinaAvinash Premraj, Management of Scientific Centers and Presidential Camels, United Arab Emirates
Copyright © 2023 Gouife, Ban, Yue, Jiang and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiasong Xie, eGllamlhc29uZ0BuYnUuZWR1LmNu
 Moussa Gouife
Moussa Gouife Ziqi Ban1
Ziqi Ban1 Jiasong Xie
Jiasong Xie