- 1SC Ematologia, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milan, Italy
- 2Dipartimento di Oncologia e Oncoematologia, Università degli Studi di Milano, Milan, Italy
Autoimmune hemolytic anemia (AIHA) is due to autoantibodies with or without complement activation and involves cellular and cytokine dysregulation. Here, we investigated cytokine single-nucleotide polymorphisms (SNPs) of TNF-α, TGF-β1, IL-10, IL-6, and IFN-γ, along with their serum levels. The former were related to hematological parameters, therapy, and clinical outcome. The study included 123 consecutive patients with primary AIHA [77 warm AIHA and 46 cold agglutinin disease (CAD)], followed up for a median of 49 months. Results show that the allelic frequency of TNF-α -308 G/A polymorphisms was significantly lower in patients versus controls. Moreover, the genotypic frequency of TNF-α -308G/A and TGF-β gene codon 25 G/C genotypes was significantly lower in patients versus controls. Considering cytokine SNP genotypes associated with different gene expression levels, TNF-α high gene expression was significantly more frequent in patients, TGF-β and IL-10 high gene expression was higher in patients with more severe anemia, and TGF-β high gene expression was higher in patients with active disease. Considering treatment, TNF-α and TGF-β high gene expression was more frequent in multitreated patients and particularly in CAD. It may be speculated that this genetic predisposition to a stronger inflammatory response may result in a greater immune dysregulation and in a relapsed/refractory disease. Regarding cytokine serum levels, TNF-α and TGF-β were significantly lower, and IL-10 and IL-6 were significantly higher in patients versus controls, underlying the complex interplay between genetic background and disease features.
Introduction
Autoimmune hemolytic anemia (AIHA) is characterized by the increased destruction of red blood cells (RBCs) by anti-RBC autoantibodies, with or without complement activation. According to the autoantibody Ig class and thermal characteristics (thermal range of reaction with autologous erythrocytes), AIHAs can be classified as warm (wAIHA), cold [cold agglutinin disease (CAD)], mixed, and atypical forms (1–3).
The pathogenesis of AIHA is complex and involves autoantibodies, monocyte/macrophages, complement, and dysregulation of humoral immunity (B-lymphocytes and long-lived plasma cells) as well as T cells (2, 4–6). The latter include cytotoxic, helper, and regulatory subsets. In particular, T regulatory cells (Treg) are key players in immunologic tolerance, producing interleukin (IL)-10 and transforming growth factor β (TGF-β). Moreover, the IL-10/IL-12 disequilibrium as well as the altered T helper 17/Treg ratio may play a role in the onset and/or maintenance of autoimmunity (2). Many other abnormalities of immunoregulatory cytokines have been described in serum and culture experiments in AIHA (7–9). However, cytokine levels are affected by several incidental factors, including overt and subclinical infections, inflammatory triggers, comorbidities, and AIHA clinical phases and therapy. These confounders may alter the interpretation of cytokine landscape in the pathogenesis of AIHA. There is growing interest in the possible role of the genetic background in the regulation of immune effectors and cytokines in several autoimmune diseases, including outcome and response to biological drugs (10–14). Specifically, single-nucleotide polymorphisms (SNPs) in the regulatory sequences of cytokine genes may provide further insights into cytokine patterns beyond incidental confounders.
We investigated cytokine SNPs of several cytokines, including TNF-α promoter, TGF-β1 gene codon, IL-10 promoter, IL-6 promoter, and IFN-γ genes, along with their serum levels, in AIHA. The results were related to hematological parameters, therapy, and clinical outcome.
Methods
Study population
The study population included 123 consecutive patients with primary AIHA, enrolled from 2001 to 2021 diagnosed and followed at our center. The study protocol was approved by the Ethical Committee, patients gave informed consent, and the study was conducted according to the Declaration of Helsinki. Primary AIHA was defined by hemolytic anemia and positive DAT, in the absence of underlying lymphoproliferative, infectious, autoimmune, or neoplastic diseases (15). Complete clinical examination, blood counts, hemolytic markers (reticulocytes, total and unconjugated bilirubin, LDH, and haptoglobin), and DAT were recorded at enrollment. Seventy-seven patients were classified as wAIHA (DAT+ for IgG only or IgG plus C3d) and 46 were classified as CAD (DAT+ for C3d only, with cold agglutinins titer ≥64), according to DAT and to the thermal characteristics of the autoantibody. Infectious and thrombotic complications were graded according to the Common terminology criteria CTCAE version 5.
Cytokine genotyping
DNA was extracted from whole blood using the salting-out standard techniques. Cytokine genotyping by PCR-sequence specific primer (PCR-SSP) amplification was done for five cytokine genes SNPs: TNF-α, TGF-β1, IL-10, IL-6, and IFN-γ. The Cytokine Genotyping Tray (One Lambda, Canoga Park, CA) has been used to evaluate the TNF-α promoter -308G>A SNP (rs1800629); two TGF-β1 SNPs, codon 10 + 869C>T (rs1800470) and codon 25 + 915G>C (rs1800471); three IL-10 promoter SNPs, -1082A>G (rs1800896), -819 T>C (rs1800871), and -592A>C (rs1800872); the IL-6 promoter -174C>G (rs1800795); and IFN-γ +874T>A (rs2430561). These SNPs were selected on the basis of previous data in the literature on various autoimmune diseases, including AIHA (16–22). Allele and genotype frequency results were compared with those described in the Italian population by Poli et al. (2002) (23) and Uboldi de Capei et al. (2003) (24). The high-, intermediate-, and low-expression categories were defined according to available literature and the manufacturer instructions of the kit used.
Cytokine serum levels
Peripheral blood serum samples were collected and the following cytokines have been tested in 64 patients using high-sensitivity ELISA kits: IL-6, IL-10, IFN-γ, TNF-α (High Sensitivity Elisa kits, eBioscience, Inc., Vienna Austria), and TGF-β (Immunological Sciences, Rome, Italy). Patient cytokine serum levels have been compared with median values of 40 healthy controls.
Statistical analysis
Analysis of variance was performed by using mean, median, ranges, and standard errors. Student t-test has been used for continuous variables. For the comparison of categorical variables, Chi-squared or Fisher’s exact tests have been used appropriately.
Results
Clinical and laboratory characteristics of patients
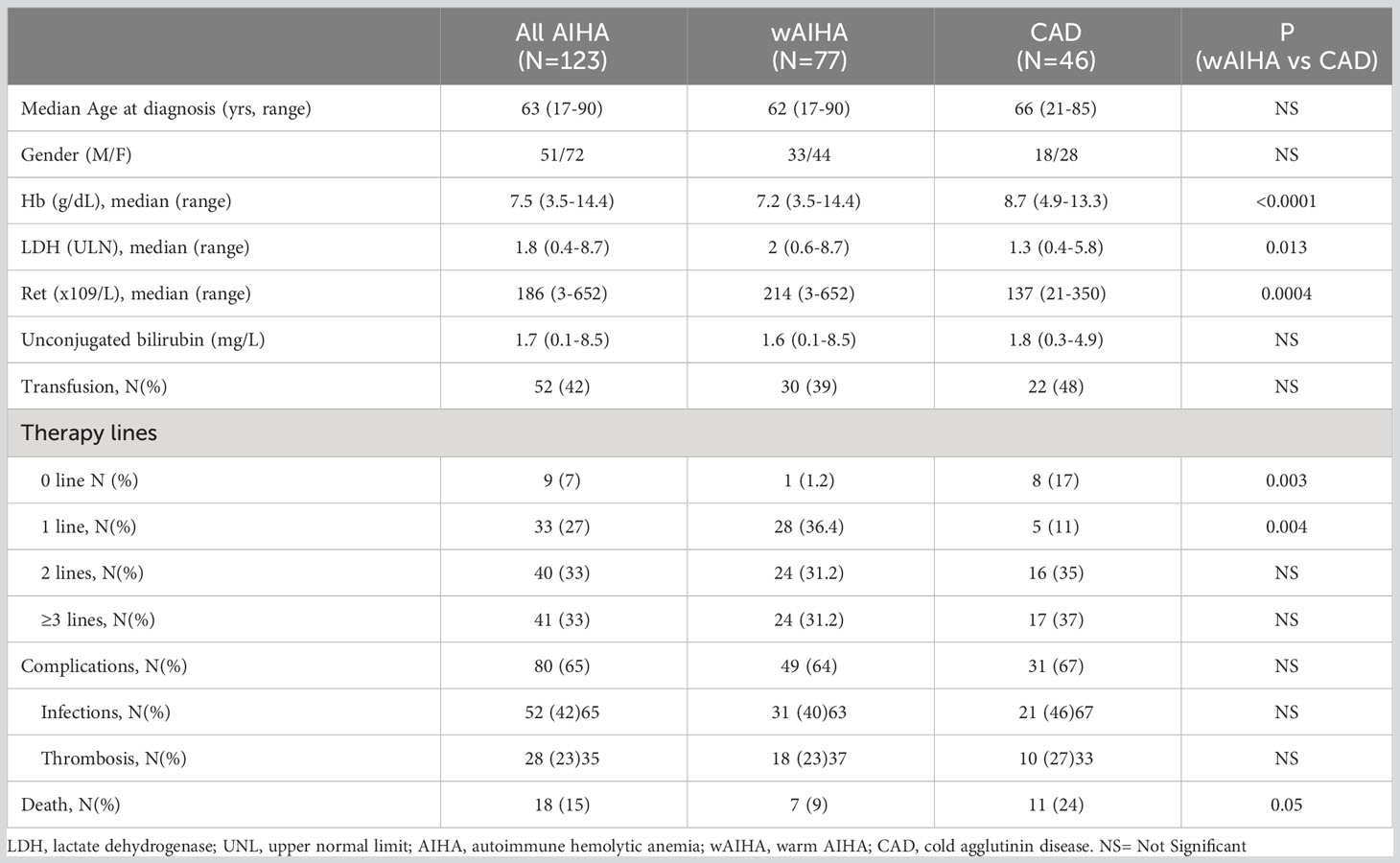
Table 1 shows the clinical and laboratory characteristics of the 123 patients studied. At diagnosis, wAIHA patients showed a more severe anemia compared to CAD ones (p < 0.0001) with higher LDH (p = 0.013) and absolute reticulocytes (p = 0.0004). During a median follow-up of 49 months (1–306), 52 patients required transfusions and 114 were treated (33 patients, 1 therapy line; 40 patients, 2 lines; and 41 patients, ≥3 lines). Nine patients did not require treatment (one wAIHA and eight CAD). The following complications were observed: 28 thrombotic events (18 in wAIHA and 10 in CAD), 52 infectious episodes ≥grade 3 (31 in wAIHA and 21 in CAD), and 18 patients (15%) died (7 wAIHA and 11 CAD), for causes not directly related to AIHA.
Allele frequencies
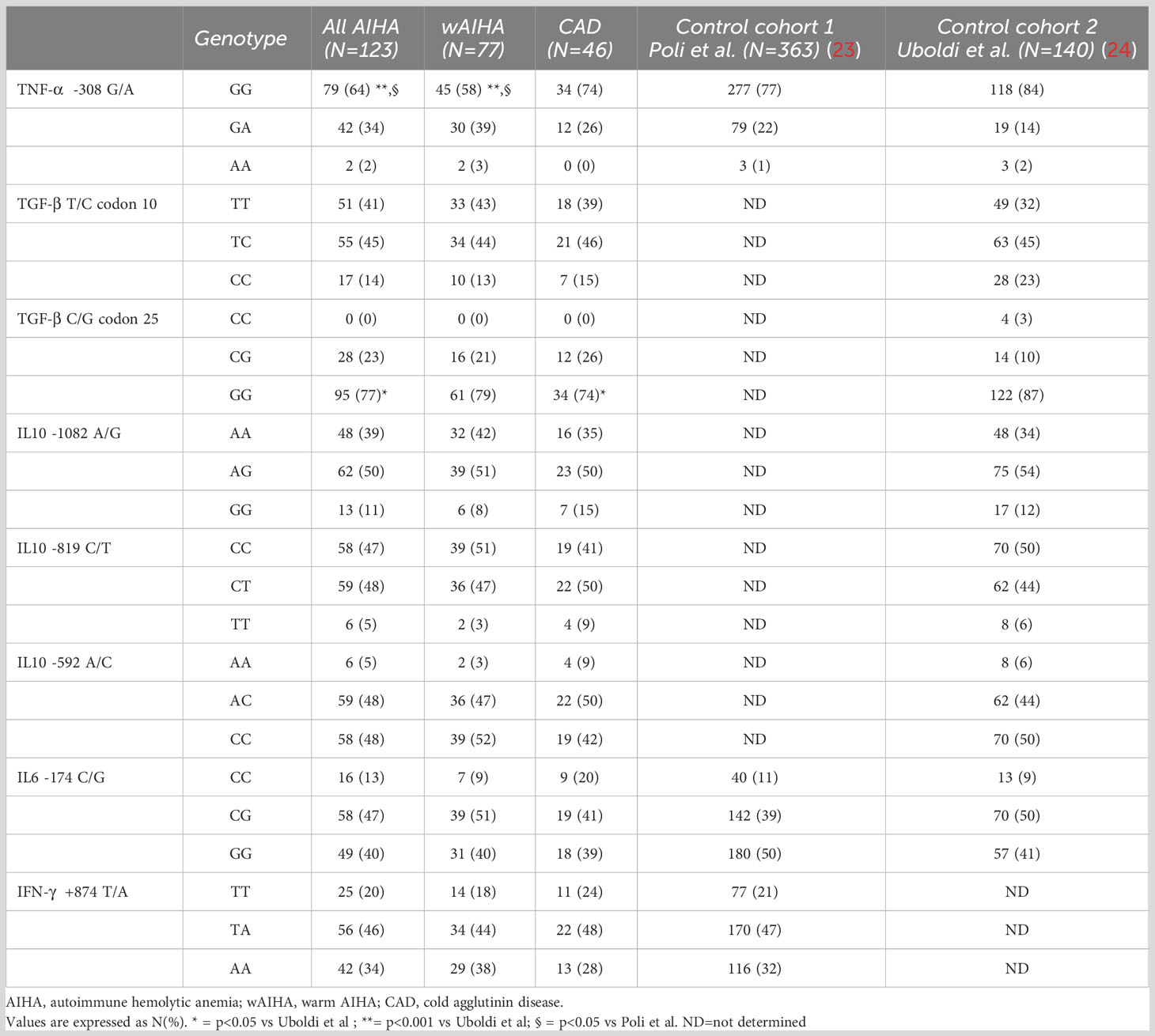
We compared our frequency results with two different control cohorts, namely, control cohort 1 and control cohort 2 (23, 24). The allelic frequency of the TNF-α -308 G/A polymorphism was significantly lower in patients compared with both controls (Supplementary Table 1). In detail, the frequency of TNF-α -308G allele was 81% in all AIHAs and 77% in wAIHA, compared with 88% (control cohort 1) and 91% (control cohort 2). Non-significant differences between patients and controls were observed considering the other SNPs. No differences in allele distribution of any other cytokine SNPs (TGF-β1, IL-10, IL-6, and IFN-γ) were observed between wAIHA and CAD.
Genotype frequency
Table 2 shows the genotypic frequency of the cytokine gene SNPs. The frequency of the TNF-α -308G/A and the TGF-β gene codon 25 G/C genotypes were significantly lower in patients compared with controls. Specifically, the frequency of the TNF-α -308 GG genotype was 64% in all AIHAs and 58% in wAIHA, compared with 77% (control cohort 1) and 84% (control cohort 2). The frequency of the TGF-β gene codon 25 GG genotype was 77% in all AIHAs, 79% in wAIHA, and 74% in CAD versus 87% in controls.
By investigating cytokine SNP genotypes associated with different gene expression levels (high versus intermediate versus low producers), the TNF-α high gene expression was significantly more frequent both in all AIHAs and in wAIHA patients compared with controls (36% and 41% versus 23% and 16%, respectively) (Supplementary Table 2). No other differences were observed considering other cytokine SNPs (TGF-β1, IL-10, IL-6, and IFN-γ).
Cytokine SNP analysis according to clinical and hematological features
The frequency of TGF-β (T/T-G/G and T/C-G/G, 80% vs. 59%, p = 0.03) and IL-10 (GCC-GCC, 16% vs. 5%, p = 0.02) high gene expression level genotypes was higher in patients with more severe anemia (Hb ≤7.5 g/dL, i.e., median of all AIHA) versus the remaining AIHA population. Likewise, the frequency of IFN-γ high plus intermediate gene expression level genotypes (T/T plus T/A) was higher within the group with Hb levels ≤7.5 g/dL at diagnosis (74% vs. 57%, p = ns) (Table 3). The frequency of TGF-β high gene expression level genotypes was higher in patients with LDH greater than 1.8 xULN (i.e., the median of all AIHA, 80% vs. 58%, p = 0.05). Finally, considering treatment, TNF-α -308 high gene expression level genotypes were more frequent among CAD patients requiring three or more therapy lines (33% versus 20%). Likewise, TGF-β high gene expression level genotypes were more frequent in all multitreated AIHA patients (75% versus 60%) and CAD subgroup (76% versus 54%) (Supplementary Table 3).
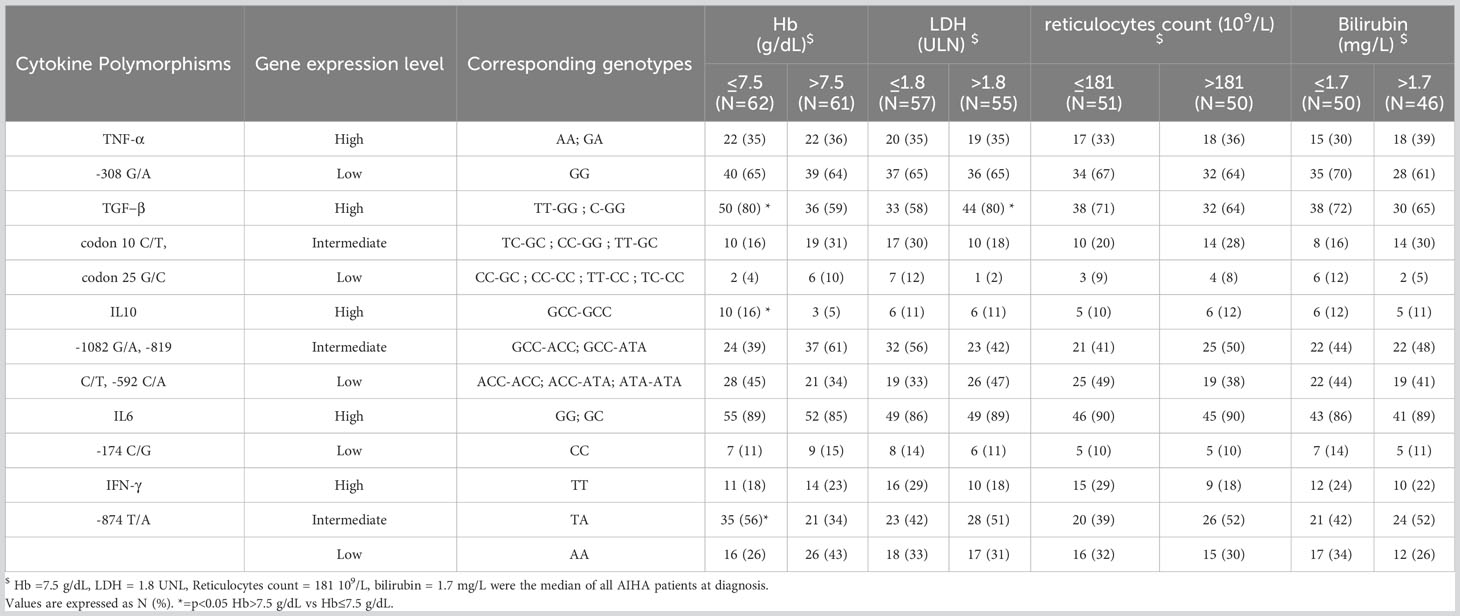
Table 3 Gene expression levels of cytokine single nucleotide polymorphism in AIHA patients according to anemia and markers of hemolysis.
Cytokine serum levels
As shown in Figure 1, TNF-α and TGF-β serum levels were significantly lower in patients compared with controls (median 0.23 pg/mL, range 0.01–2.65 vs. 1.5, 0.1–2.9, p < 0.001 for TNF-α; 2,284 pg/mL, 1,076–5,908 vs. 3,531 pg/mL, 924–6086, p = 0.006 for TGF-β). Moreover, IL-10 and IL-6 serum levels were significantly higher in AIHA patients versus controls (median 1.67 pg/mL, 0.05–33.74 vs. 0.4 pg/mL, 0.02–2.7, p < 0.001 for IL-10; 1.26 pg/mL, 0.11–12.9 vs. 0.7 pg/mL, 0.2–4.7, p = 0.001 for IL-6). Finally, IFN-γ was slightly reduced versus controls (median 0.4 pg/mL, 0.01–11.40 vs. 0.7 pg/mL, 0.02–3.2), although not significantly. A slight association was found between gene expression genotypes and cytokine serum levels, i.e., high gene expression genotypes displayed higher cytokine levels: IL-6 (median 1.29 versus 1.26 pg/mL, not significant) and IFN-γ (median 0.7 versus 0.32 pg/mL, not significant). At variance, an inverse relationship between high gene expression genotypes and cytokine serum levels was found for TNF-α (median 0.21 versus 0.24 pg/mL, not significant), TGF-β (median 2,192 versus 2,425 pg/mL, not significant), and IL-10 (median 0.9 versus 2.08 pg/mL, not significant).
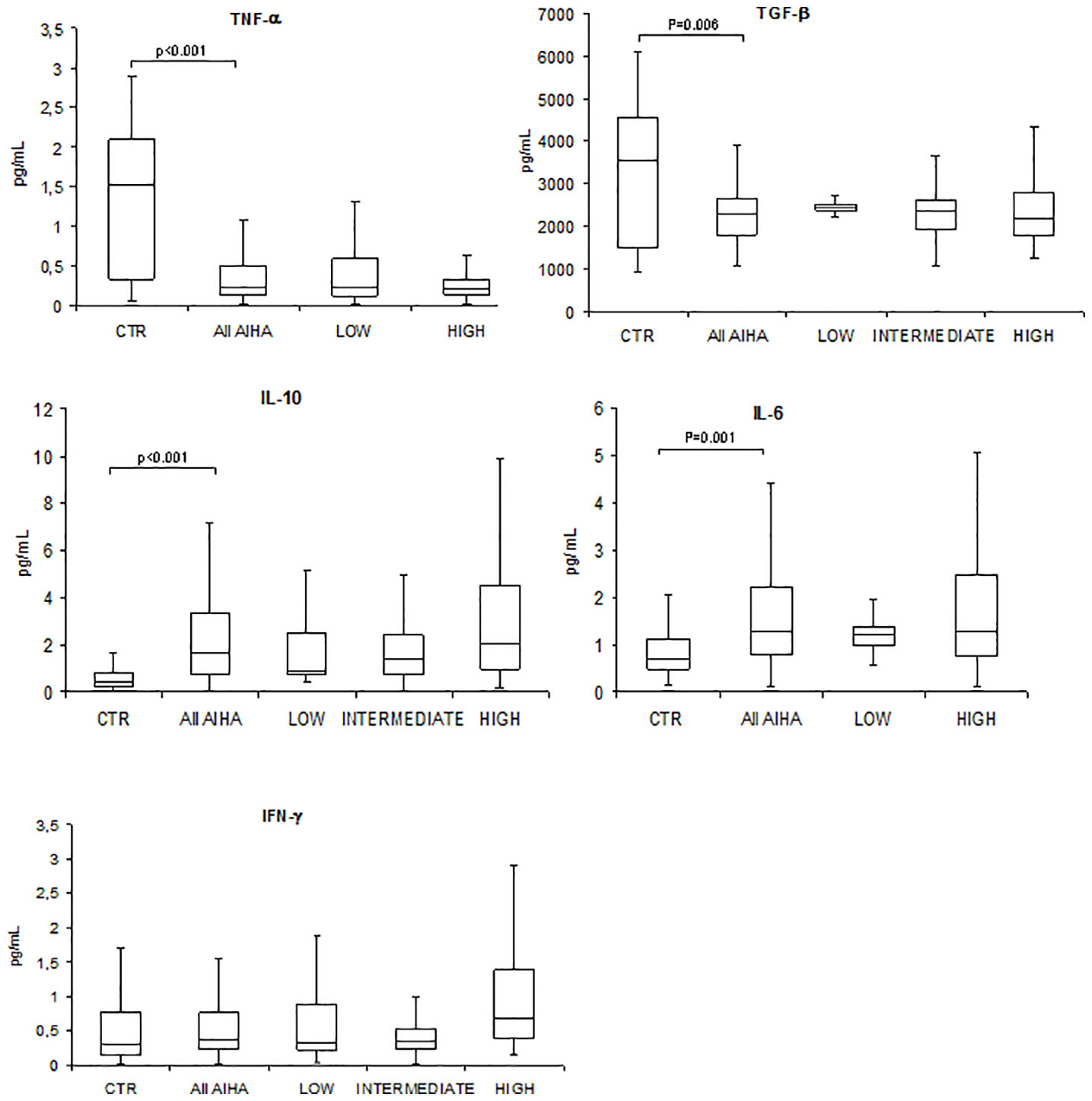
Figure 1 Cytokine serum levels in autoimmune hemolytic anemia patients (All AIHA) versus controls, and according to high/intermediate/low gene expression level. Tumor necrosis factor alpha (TNF-α), transforming growth factor beta (TGF-β), interleukin (IL)-10, IL-6, and interferon gamma (IFN-γ).
Discussion
Here, we investigated several cytokine polymorphisms in AIHA and evaluated their relationship with cytokine serum levels and with disease severity. Overall, the allele frequency of some SNPs of TNF-α and the TNF-α and TGF-β genotypes were less frequently found in patients compared with controls (23, 24). Regarding TNF-α, our results are in agreement with those of Pavkovic et al., who found reduced G- and consequently increased A- alleles of TNF-α-308G/A in AIHA versus controls (16). At variance, no difference in genotype distribution, as compared to healthy controls, was reported by D’Abronzo et al. (25) who however investigated a small number of patients. Furthermore, in our study, the TNF-α genotype associated with high gene expression was more frequent in patients compared with controls, as well as in patients with more severe disease and generally in multitreated patients. It may be speculated that this genetic predisposition to a stronger inflammatory response may result in a greater immune dysregulation and in a relapsed/refractory disease. Alternatively, the association between TNF-α genotype and AIHA may be driven by the rarity of AA genotype. Surprisingly, TNF-α serum levels were found to be lower in AIHA than in controls, in agreement with several previous reports from our group (26–28). This apparent discrepancy may reflect the several incidental factors, such as overt and subclinical infections, inflammatory triggers, and treatments that may influence cytokine serum levels notwithstanding genetic background. Moreover, it may be assumed that cytokine serum levels represent the net result between production and utilization by cellular effectors. Thus, low levels may indicate a greater uptake and consumption in the context of a massive inflammatory/autoimmune process as observed in more severe disease. This has already been hypothesized for in vitro cultures assessing cytokine production and the effect of exogenous cytokines on autoantibody production: in the active disease, production of TGF-β was increased, and addition of TGF-β increased antibody production and binding to RBC. This leads to the speculation that overproduction of TGF-β may downregulate cellular responses (7). TGF-β is a pleiotropic cytokine regulating a wide variety of cell functions, with primarily anti-inflammatory, antimitotic, and profibrotic properties (29). Moreover, it could impair the generation of Th1 cells through an indirect IFN-γ-mediated mechanism and is a crucial inducer of T-helper 17 and Tregs. The role of TGF-β in autoimmunity is complex, depending on the phase of the disease and the main effector mechanisms involved (cellular versus humoral), but generally favors autoimmune phenomena. Here, we found that TGF-β high producer genotypes were more frequent in more hemolytic and multitreated AIHA patients, along with lower serum levels. As observed for TNF-α, the discrepancy between cytokine serum levels and high producer genotypes may be the result of cytokine utilization by effector cells.
For other cytokines, the picture is less clear-cut. Concerning IL-10, which is both a Th2 and anti-inflammatory cytokine, its serum levels were increased in patients versus controls, along with a greater frequency of high gene expression genotype in more severe cases. The increased IL-10 production may be an attempt to counteract the over-inflammatory autoimmune response, resulting in an excess free/unutilized cytokine in serum. Finally, serum levels of the Th2 and inflammatory cytokine IL-6 were found increased in patients consistently with the main humoral pathogenic mechanism of AIHA, although without a clear relationship with SNPs. The slight association found for IL-6 and IFN-γ cytokine serum levels and high gene expression genotype may possibly indicate cell autonomous activity of the cytokine, which contributes to an autocrine signaling. At variance, the absence of this association for TNF-α, TGF-β, and IL-10 suggests more complex regulatory pathways between genotype and phenotype.
In conclusion, the analysis of cytokine gene SNPs highlighted the role of the TNF-α− and TGF-β-mediated inflammatory response as a genetic background that may influence the severity and refractoriness of AIHA. Cytokine serum levels are one side of the coin, reflect the balance between production and utilization, and may be more influenced by incidental environmental factors. Overall, the results pinpoint the complexity of the cytokine network, where redundancy and pleiotropism of the various cytokines may generate opposite effects depending on the existing milieu.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Ethic Committee Milano Area 2. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
BF and WB designed the study, followed patients, analyzed data, wrote the paper, and revised the paper for important intellectual content. AZ performed biological studies, collected and analyzed data, wrote the paper, and revised the paper for important intellectual content. LP followed patients and critically revised the manuscript. CV and AM collected samples and critically revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by Italian Ministry of Health - Current research, Fondazione IRCCS Ca’ Granda Policlinico Milano, project n. RC 175/01.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1221582/full#supplementary-material
References
1. Petz LD, Garratty G. Immune Hemolytic Anemias. 2nd ed. Churchill Livingstone: Philadelphia (2004).
2. Barcellini W, Zaninoni A, Giannotta JA, Fattizzo B. New insights in autoimmune hemolytic anemia: from pathogenesis to therapy stage 1. J Clin Med (2020) 9(12):3859. doi: 10.3390/jcm9123859
3. Berentsen S, Barcellini W. Autoimmune hemolytic anemias. N Engl J Med (2021) 385(15):1407–19. doi: 10.1056/NEJMra2033982
4. Fagiolo E. Immunological tolerance loss vs. erythrocyte self antigens and cytokine network disregulation in autoimmune hemolytic anemia. Autoimmun Rev (2004) 3(2):53–9. doi: 10.1016/S1568-9972(03)00085-5
5. Gilsanz F, de la Serna J, Moltó L, Alvarez-Mon M. Hemolytic anemia in chronic large granular lymphocytic leukemia of natural killer cells: cytotoxicity of natural killer cells against autologous red cells is associated with hemolysis. Transfusion (1996) 36:463–6. doi: 10.1046/j.1537-2995.1996.36596338025.x
6. Mahévas M, Michel M, Vingert B, Moroch J, Boutboul D, Audia S, et al. Emergence of long-lived autoreactive plasma cells in the spleen of primary warm auto-immune hemolytic anemia patients treated with rituximab. J Autoimmun (2015) 62:22–30. doi: 10.1016/j.jaut.2015.05.006
7. Barcellini W, Clerici G, Montesano R, Taioli E, Morelati F, Rebulla P, et al. In vitro quantification of anti-red blood cell antibody production in idiopathic autoimmune haemolytic anaemia: effect of mitogen and cytokine stimulation. Br J Haematol (2000) 111:452–60. doi: 10.1046/j.1365-2141.2000.02380.x
8. Xu L, Zhang T, Liu Z, Li Q, Xu Z, Ren T. Critical role of Th17 cells in development of autoimmune hemolytic anemia. Exp Hematol (2012) 40(12):994–1004.e4. doi: 10.1016/j.exphem.2012.08.008
9. Toriani-Terenzi C, Fagiolo E. IL-10 and the cytokine network in the pathogenesis of human autoimmune hemolytic anemia. Ann N Y Acad Sci (2005) 1051:29–44. doi: 10.1196/annals.1361.044
10. Wan YN, Wang YJ, Yan JW, Li XP, Tao JH, Wang BX, et al. The effect of TGF-β1 polymorphism on systemic sclerosis: a systematic review and pooled analysis of available literature. Rheumatol Int (2013) 33:2859–65. doi: 10.1007/s00296-013-2826-9
11. Linares-Pineda TM, Cañadas-Garre M, Sánchez-Pozo A, Calleja-Hernández MA. Gene polymorphisms as predictors of response to biological therapies in psoriasis patients. Pharmacol Res (2016) 113:71–80. doi: 10.1016/j.phrs.2016.07.020
12. Bakr NM, Hashim NA, El-Baz HAE, Khalaf EM, Elharoun AS. Polymorphisms in proinflammatory cytokines genes and susceptibility to Multiple Sclerosis. Mult Scler Relat Disord (2021) 47:102654. doi: 10.1016/j.msard.2020.102654
13. Wang R, Zhang X, Wang S. Differential genotypes of TNF-α and IL-10 for immunological diagnosis in discoid lupus erythematosus and oral lichen planus: A narrative review. Front Immunol (2022) 13:967281. doi: 10.3389/fimmu.2022.967281
14. Lee YH, Song GG. Association between the interferon-γ +874 T/A polymorphism and susceptibility to systemic lupus erythematosus and rheumatoid arthritis: A meta-analysis. Int J Immunogenet (2022) 49:365–71. doi: 10.1111/iji.12599
15. Jäger U, Barcellini W, Broome CM, Gertz MA, Hill A, Hill QA, et al. Diagnosis and treatment of autoimmune hemolytic anemia in adults: Recommendations from the First International Consensus Meeting. Blood Rev (2020) 41:100648. doi: 10.1016/j.blre.2019.100648
16. Pavkovic M, Petlichkovski A, Angelovic R, Genadieva-Stavric S, Cevreska L, Stojanovic A. Tumor necrosis factor gene polymorphisms in adult patients with autoimmune hemolytic anemia. Int J Lab Hematol (2017) 39(3):e74–6. doi: 10.1111/ijlh.12624
17. Wilson AG, Gordon C, di Giovine FS, de Vries N, van de Putte LB, Emery P, et al. A genetic association between systemic lupus erythematosus and tumor necrosis factor alpha. Eur J Immunol (1994) 24(1):191–5. doi: 10.1002/eji.1830240130
18. Stadtlober NP, Flauzino T, Santos LFDRF, Iriyoda TMV, Costa NT, Lozovoy MAB, et al. TGFB1 +869 T > C (rs1800470) variant is independently associated with susceptibility, laboratory activity, and TGF-β1 in patients with systemic lupus erythematosus. Autoimmunity (2021) 54(8):569–75. doi: 10.1080/08916934.2021.1975680
19. Iriyoda TMV, Flauzino T, Costa NT, Lozovoy MAB, Reiche EMV, Simão ANC. TGFB1 (rs1800470 and rs1800469) variants are independently associated with disease activity and autoantibodies in rheumatoid arthritis patients. Clin Exp Med (2022) 22(1):37–45. doi: 10.1007/s10238-021-00725-9
20. Mokhtar GM, El-Beblawy NM, Adly AA, Elbarbary NS, Kamal TM, Hasan EM. Cytokine gene polymorphism [tumor necrosis factor-alpha (-308), IL-10 (-1082), IL-6 (-174), IL-17F, 1RaVNTR] in pediatric patients with primary immune thrombocytopenia and response to different treatment modalities. Blood Coagul Fibrinolysis (2016) 27(3):313–23. doi: 10.1097/MBC.0000000000000431
21. Robledo G, Dávila-Fajardo CL, Márquez A, Ortego-Centeno N, Callejas Rubio JL, de Ramón Garrido E, et al. Association between -174 interleukin-6 gene polymorphism and biological response to rituximab in several systemic autoimmune diseases. DNA Cell Biol (2012) 31(9):1486–91. doi: 10.1089/dna.2012.1684
22. Lee YH, Bae SC. Association between interferon-γ +874 T/A polymorphism and susceptibility to autoimmune diseases: a meta-analysis. Lupus (2016) 25(7):710–8. doi: 10.1177/0961203315624557
23. Poli F, Piccolo G, Scalamogna M. Genetic polymorphisms influencing therapy and susceptibility to rejection in organ allograft recipients. BioDrugs (2002) 16(1):11–7. doi: 10.2165/00063030-200216010-00002
24. Uboldi de Capei M, Dametto E, Fasano ME, Rendine S, Curtoni ES. Genotyping for cytokine polymorphisms: allele frequencies in the Italian population. Eur J Immunogenet (2003) 30(1):5–10. doi: 10.1046/j.1365-2370.2003.00361.x
25. D’Abronzo LS, Barros MM, Bordin JO, Fig-ueiredo MS. Analysis of polymorphisms of TNF-α, LT-α, IL-10, IL-12 and CTLA-4 inpatients with warm autoimmune haemoly-tic anaemia. Int J Lab Hematol (2012) 34:356–61. doi: 10.1111/j.1751-553X.2012.01400.x
26. Barcellini W, Zaja F, Zaninoni A, Imperiali FG, Battista ML, Di Bona E, et al. Low-dose rituximab in adult patients with idiopathic autoimmune hemolytic anemia: clinical efficacy and biologic studies. Blood (2012) 119(16):3691–7. doi: 10.1182/blood-2011-06-363556
27. Fattizzo B, Zaninoni A, Giannotta JA, Binda F, Cortelezzi A, Barcellini W. Reduced 25-OH vitamin D in patients with autoimmune cytopenias, clinical correlations and literature review. Autoimmun Rev (2016) 15(7):770–5. doi: 10.1016/j.autrev.2016.03.015
28. Fattizzo B, Michel M, Zaninoni A, Giannotta J, Guillet S, Frederiksen H, et al. Efficacy of recombinant erythropoietin in autoimmune hemolytic anemia: a multicenter international study. Haematologica (2021) 106(2):622–5. doi: 10.3324/haematol.2020.250522
Keywords: warm autoimmune hemolytic anemia, cold agglutinin disease, cytokine polymorphism, interleukin (IL)-6, IL-10, Interferon (IFN)-γ, tumor necrosis factor (TNF)-α, transforming growth factor (TGF)-β
Citation: Zaninoni A, Fattizzo B, Pettine L, Vercellati C, Marcello AP and Barcellini W (2023) Cytokine polymorphisms in patients with autoimmune hemolytic anemia. Front. Immunol. 14:1221582. doi: 10.3389/fimmu.2023.1221582
Received: 12 May 2023; Accepted: 23 October 2023;
Published: 10 November 2023.
Edited by:
Arnaud Zaldumbide, Leiden University Medical Center (LUMC), NetherlandsReviewed by:
Jaal Jan Zwaginga, Leiden University Medical Center (LUMC), NetherlandsDominic John Ciavatta, University of North Carolina at Chapel Hill, United States
Copyright © 2023 Zaninoni, Fattizzo, Pettine, Vercellati, Marcello and Barcellini. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anna Zaninoni, YW5uYS56YW5pbm9uaUBwb2xpY2xpbmljby5taS5pdA==
 Anna Zaninoni
Anna Zaninoni Bruno Fattizzo
Bruno Fattizzo Loredana Pettine
Loredana Pettine Cristina Vercellati
Cristina Vercellati Anna P. Marcello1
Anna P. Marcello1 Wilma Barcellini
Wilma Barcellini