- 1State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing, China
- 2Department of Respiratory Medicine, Center for Pathogen Biology and Infectious Diseases, Key Laboratory of Organ Regeneration and Transplantation of the Ministry of Education, The First Hospital of Jilin University, Changchun, China
Chlamydia trachomatis, one species of Chlamydia spp., has the greatest impact on human health and is the main cause of bacterial sexually transmitted diseases and preventable blindness among all Chamydia spp. species. The obligate intracellular parasitism and unique biphasic developmental cycle of C. trachomatis are the main barriers for the development of tools of genetic manipulation. The past decade has witnessed significant gains in genetic manipulation of C. trachomatis, including chemical mutagenesis, group II intron-based targeted gene knockout, fluorescence-reported allelic exchange mutagenesis (FRAEM), CRISPR interference (CRISPRi) and the recently developed transposon mutagenesis. In this review, we discuss the current status of genetic manipulations of C. trachomatis and highlights new challenges in the nascent field of Chlamydia genetics.
1 Introduction
Chlamydia are a group of gram-negative, obligate intracellular pathogens, and their broad host ranges from single-celled eukaryotes to cattle, sheep and humans (1). The main species capable of commonly infecting humans include Chlamydia trachomatis, Chlamydia pneumoniae, and Chlamydia psittaci. Among them, C. trachomatis has the greatest impact on human health. The eye infection of C. trachomatis can cause trachoma, which is the main cause of preventable blindness in developing countries of the world (2). According to the data of the World Health Organization (WHO) in June 2022, at least 125 million people are living in trachoma-endemic areas face the risk of trachoma blindness (2). Besides, genitourinary infection caused by C. trachomatis may lead to venereal lymphogranuloma or infertility (3).
C. trachomatis has a unique biphasic developmental cycle that alternates between two morphologically and functionally distinct developmental stages: the small, structurally stable, infectious elementary body (EB) and the large, metabolically vigorous, replicative reticulate body (RB) (1, 4). Infection begins with attachment and internalization of EBs to host cells. RBs replicate within a membrane bound compartment - the inclusion, early genes are transcribed and EBs differentiate into RBs (~6–8 hours post-infection) (1, 5, 6). Next, effectors that mediate nutrient acquisition and maintain the viability of the host cell are expressed. The bacteria divide by binary fission and the inclusion substantially expands (~8–16 hours post-infection) (6, 7). Last, RBs gradually re-differentiate back into EBs. By host cell lysis or by extrusion of intact inclusions, EBs are released to infect neighboring host cells at the end of the developmental cycle (~24–72 hours post-infection) (1, 8, 9).
Although the sequence of the first C. trachomatis genome was published in 1998 (10), the functional analysis of proteins of C. trachomatis has been limited by the lack of tools of genetic manipulation for a long time. A major barrier to the development of genetic manipulation is its dependence on a host replication and the unique biphasic developmental cycle. RB is only present in the host cell, and it is difficult for exogenous DNA to reach the bacterial cytoplasm through the four-layers of biofilm (11). Even EB can exist in the environment, it has a hard cell wall and low metabolic activity, so it is unlikely to reabsorb and integrate foreign DNA (11).
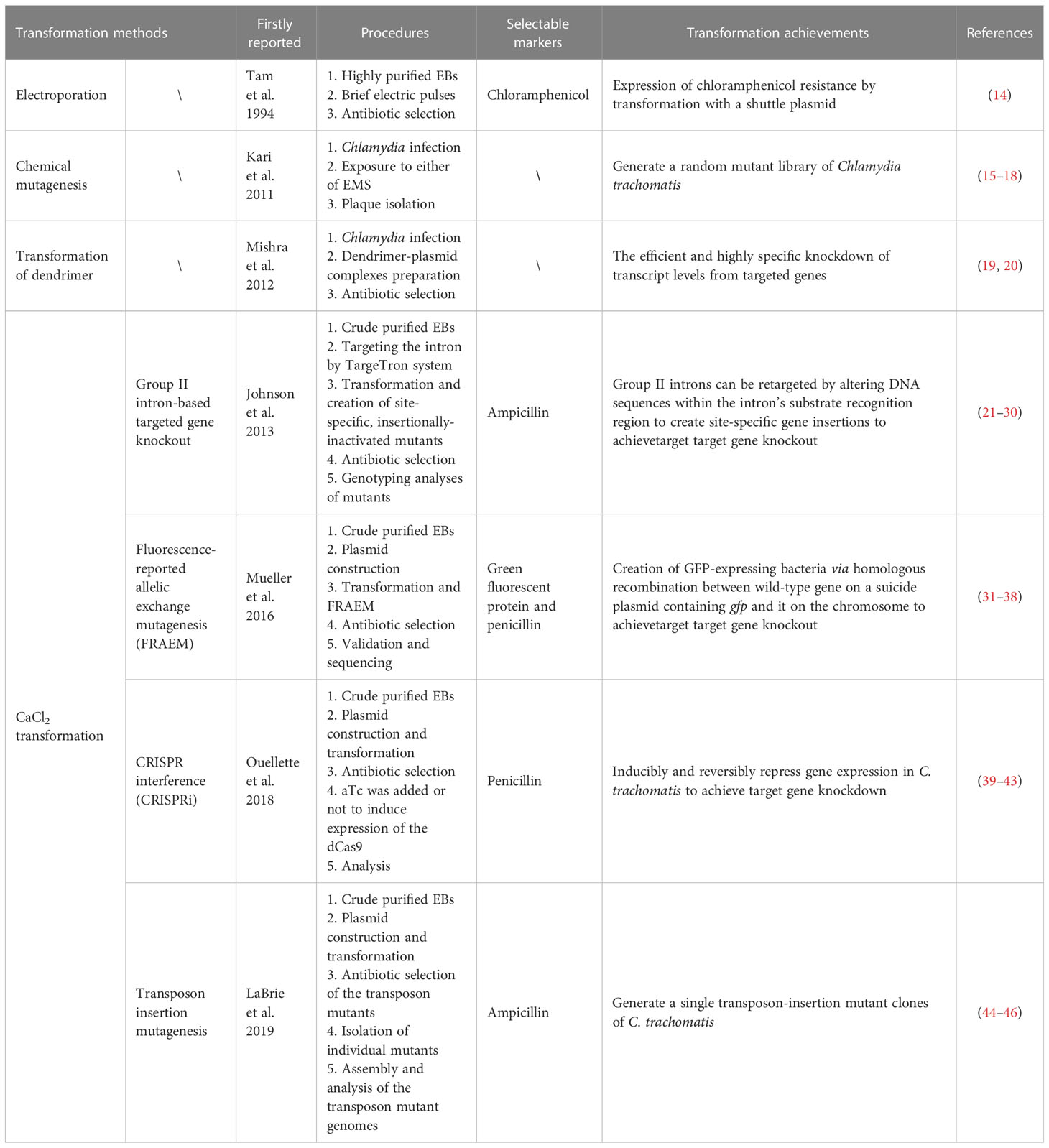
Fortunately, in the past decade, the development and application of several tools of genetic manipulation of C. trachomatis has made some progress, greatly expanding the current research on the biological characteristics of C. trachomatis and the function analysis of its virulence factors. The first step in almost all methods of genetic manipulation of Chlamydia is transformation. Four transformation methods have been reported: electroporation, chemically induced mutagenesis, polyamidoamine dendrimers (PAMAM dendrimers), and CaCl2 transformation (4, 12, 13). In this review, we categorize and summarize the tools of genetic manipulation that have been developed for C. trachomatis according to the methods of plasmid transformation (Figure 1; Table 1).
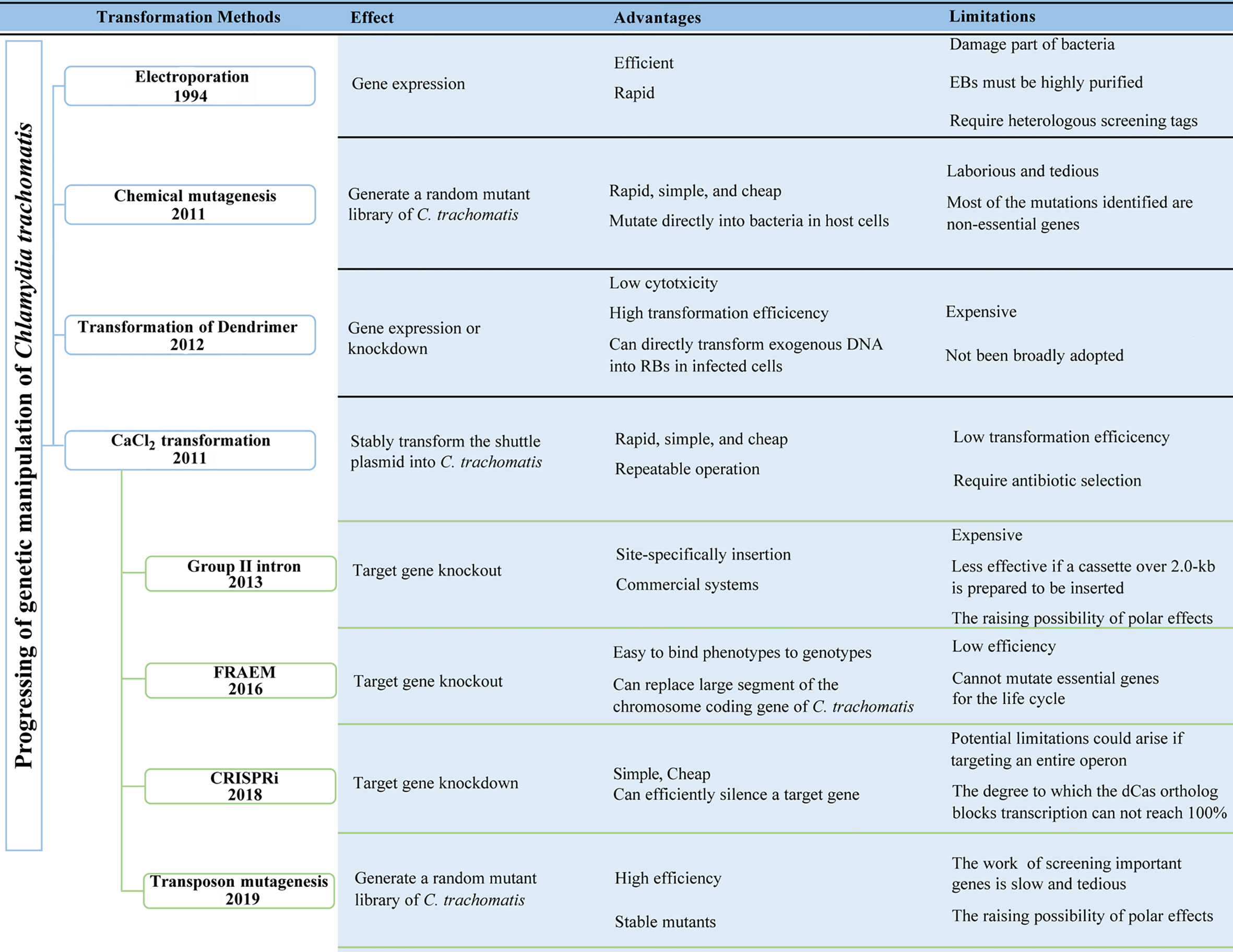
Figure 1 Progressing of the genetic manipulation of Chlamydia trachomatis. FRAEM, fluorescence-reported allelic exchange mutagenesis. CRISPRi, CRISPR interference.
2 Electroporation
Electroporation is universally effective in introducing heterologous DNA into obligate intracellular bacteria via brief electric pulses, which induce transient and reversible cell membrane permeabilization (47), and time constant and exponential decay pulse types, episomal DNA maintenance are the key features. Tam et al. successfully introduced the shuttle plasmin pPBW100 into C. trachomatis EBs by electroporation for the first time in 1994, and used it to infect McCoy cells and detected Chlamydia containing the chloramphenicol resistance gene in cell cultures (14). A similar electroporation method was used to mobilize an engineered vector into C. psittaci 6BC EBs and a efficiency was obtained with 10 µg of circular vector (1.9 ± 1.1×106, n=7) (48). For the successfully transformation, EBs must be highly purified and obtained by centrifugation through Renografin density gradients, and this complex procedure maybe limit the wide adoption of electroporation by other labs. Although the optimal transformation conditions for C. trachomatis electroporation have not been fully grasped, the study of Tam shows that exogenous DNA can be introduced into EBs by electroporation and the expression of heterologous screening tags can be achieve, which laid the foundation for the development of C. trachomatis electroporation technology in the future (14).
3 Chemical mutagenesis
Chemical mutagenesis is a technology that uses chemical mutagens such as base analogues, deamination agents, and alkylating agents to mutate DNA (49, 50).
C. trachomatis was subjected to low-level ethyl methanesulfonate (EMS) mutagenesis to generate chlamydiae that contained less than one mutation per genome in 2011 and a tryptophan synthase gene (trpB) null mutant incapable of avoiding the anti-microbial effect of IFN-γ–induced tryptophan starvation was isolated (15). Then mutagenesis in Chlamydia was performed by exposure of infected cells to either of the DNA alkylating compounds EMS or N-ethyl-N-nitrosourea (ENU), followed by plaque isolation of clonal strains in 2015 (4). In the study, Kokes et al. used ethyl methyl sulfonate (EMS) and N-ethyl-N-nitrosourea (ENU) to perform chemical mutagenesis on C. trachomatis to generate a mutant library, screening of mutants impaired in F-actin assembly and identifying InaC as an inclusion body membrane protein that binds host ARF and 14-3-3 proteins and regulates F-actin recombination and Golgi reorganization around vesicles (16).
Although studies have shown that it is possible to achieve either one mutation (15) or multiple mutations per genome of C. trachomatis (16–18) by adjusting different concentrations of mutagen, identifying and linking genotype and phenotype without a molecular signature is laborious and tedious, and most of the mutations identified are non-essential genes (51). Therefore, with the continuous development of genetic manipulation, the application of chemical mutagenesis will be gradually phased out.
4 Transformation of dendrimer
Polyamidoamine (PAMAM) dendrimers are hyperbranched polymers with low cytotoxicity. It can not only deliver small molecules to specific sites, but also effectively transfuse biological macromolecules such as oligonucleotides and plasmid DNA into cells (52). In addition, these dendrimers can be localized in Chlamydia inclusion bodies in Chlamydia-infected cells (53), indicating that PAMAM can directly transform exogenous DNA into RBs in infected monolayers (19, 52).
In 2012, Mishra et al. used PAMAM to transfer an antisense oligonucleotide into C. trachomatis and efficiently and specifically knockdown the transcription level of the target gene (19). Then in 2013, Kannan successfully used PAMAM to transfer a plasmid (pMW82: pL2-01-pL2-01P-GFP) into C. trachomatis and successfully detected green fluorescence in the initial transformed culture (20, 48). Despite plasmid replication and GFP expression being detected within the first infection cycle which indicating a high transformation efficiency, the dendrimer-based transformation method of Chlamydia has not been broadly adopted.
5 CaCl2 transformation
Since the first utilization of CaCl2 transformation method to stably transform the shuttle plasmid pBR325::L2 into C. trachomatis EBs by Wang et al. in 2011 (54). In the protocol, EBs were firstly incubated with plasmid DNA in CaCl2 buffer for 30 min at room temperature and then host cells resuspended in CaCl2 buffer were added, followed by an additional incubation for 20 min at room temperature. Due to its advantages of simple, rapid, cheap and repeatable operation, the CaCl2 transformation method is widely used as a general transformation method for Chlamydia. For the CaCl2 transformation, crude preparations of Chlamydia from host cell lysates exhibit more efficient than gradient purified EB preparations (54–56). Based on this method, group II intron-based targeted gene knockout (21–30), fluorescence-reported allelic exchange mutagenesis (FRAEM) (31–38), CRISPR interference (CRISPRi) (39–43) and transposon insertion mutagenesis (44–46) have been realized in C. trachomatis.
5.1 Group II intron-based targeted gene knockout
Group II introns are a class of self-splicing ribozymes capable of high-frequency movement between genes through a retrohoming (TargeTron system) with the help of intron-encoded protein (IEP, with RAN maturase, endonuclease, and reverse transcriptase activities) (21, 57). Based on this principle, the first targeted disruption of a gene on Chlamydia chromosome was performed by Johnson and Fisher in 2013 (21). In the study, a plasmid containing the coding sequence of β-lactamase was transformed successfully and site-specifically, insertionally inactivated incA of C. trachomatis L2 strain, confirming the requirement of this protein for homotypic fusion of Chlamydial inclusion (21, 25, 58).
Right now, group II intron integration technology has been used in commercial systems such as Sigma’s TargeTron gene knockout system, and it has been successfully used for gene knockout of other intracellular parasitic bacteria including Ehrlichia and Rickettsia (21, 59). However, this method requires bioinformatics analysis to determine the intron insertion site, so it is necessary to design several insertion sites at the same time to ensure the probability of gene knockout (21, 57, 60, 61), and it is also less effective if a cassette over 2.0-kb is prepared to be inserted. Another major limitation of this system is that intron insertions may have polar effects on the expression of neighboring genes if the knocked-out chlamydial genes exist within polycistronic operons.
5.2 FRAEM
In 2013, Wickstrum et al. developed an inducible gene expression system (shuttle plasmid pASK-GFP-L2) for Chlamydia, in which gene expression was controlled by Tet, developing a strategy for gene expression and/or complementation (62). Song et al. reported that pgp6 on the native pL2 plasmid of C. trachomatis is necessary for this plasmid maintenance (63). Then in 2016, Mueller et al. constructed a suicide plasmid pUS6 based on the inducible expression of pgp6 and permit rapid reverse genetics by FRAEM (31). This system can replace the chromosome coding gene of C. trachomatis with a 2.2-kb cassette encoding both GFP and β-lactamase, thus realizing the targeted knockout of C. trachomatis gene and permitting the monitoring of mutagenesis by fluorescence microscopy. They successfully constructed the trpA-deficient strain of C. trachomatis and found that the deficient strain was unable to grow in indole-containing medium (31). Later, they adapted FRAEM technology by leveraging a step-wise Cre-lox approach to excise selection marker genes from a deleted gene locus to eliminate the possibility of polar effects mediated by the inserted cassette (33).
Recently, Kenneth et al. present functional evidence that the region between C. trachomatis pgp6 and pgp7, containing four 22-bp tandem repeats in the endogenous plasmids, represents the chlamydial native plasmid origin of replication (32, 64). They constructed plasmid pKW-L2ori by mobilization of the entire region between these two genes from chlamydial native plasmid pL2 into a pUC19-based plasmid and proved that it could be maintained by C. trachomatis serovar D which contains a native chlamydial plasmid. Subsequently they proved that pKW can be utilized as a conditionally replicating plasmid sufficient for the generation of deletion mutants via allelic exchange (32).
Although FRAEM can specifically knockout gene of C. trachomatis, this strategy requires a low-frequency double-crossover event. Further optimized methods including using some heterologous site-specific recombinases [which have been reconstructed and applied to Coxiella burnetii (65)] could be applied to assist gene recombination in C. trachomatis.
5.3 CRISPRi
Since its release in 2012, the CRISPR/Cas9 system has been widely used due to its simple operation, low cost, and high efficiency (66). To repurpose the CRISPR system for transcription regulation, Matthew et al. have described an RNA-based method, CRISPR interference (CRISPRi), and they have shown that CRISPRi can efficiently silence a target gene with up to 99.9% in Escherichia coli (67). Until now, CRISPRi has been used for targeted silencing of transcription in intracellular bacteria including Mycobacterium tuberculosis (68), C. burnetii (69, 70).
In 2018, Ouellette successfully knocked down incA gene of C. trachomatis by using CRISPRi based on the catalytically inactive Cas9 variant (dCas9) of Staphylococcus aureus, proving that the system can be used to reversibly inhibit incA expression, in addition that they found the plasmid encoding the dCas9 from Staphylococcus pyogenes was not possible to successfully transform C. trachomatis with it (42). And in 2021, Ouellette et al. optimized and improved the missing expression of anhydrotetracycline (aTc) - inducible dCas9 orthologous genes and plasmid instability in the original system, and developed a second CRISPRi system based on the dCas12 system to expand the number of potential chromosomal targets (41). These two CRISPRi systems will allow for broad targeting of the C. trachomatis genome and for analysis of essential gene functions in C. trachomatis in a straightforward manner.
However, Wurihan et al. successfully transformed two plasmid encoding staphylococcal (S. aureus and S. pyogenes) dCas9 to C. trachomatis and found that conditional expression of the staphylococcal dCas9 strongly inhibits chlamydial growth in the absence of any specific guide RNA (gRNA) (40), suggesting that the staphylococcal dCas9 proteins in their current forms have limited utility for chlamydial research and strategies to overcome this problem should be developed.
5.4 Transposon mutagenesis
Transposon mutagenesis is an effective method for discovering specific genetic components associated with a given phenotype. The basic principle is that when a transposase drives an exogenous transposon integrating into the promoter region or coding region of an unknown gene randomly, the gene will be inactivated and a new mutant phenotype will be produced. Transposon mutagenesis has been applied to C. burnetii (71, 72), Rickettsia felis (73), Rickettsia prowazekii (74) and Ehrlichia chaffeensis (75) for the identification of virulence proteins.
In 2019, LaBrie et al. constructed a non-replicating plasmid termed pCMA to encode the widely utilized C9 Himar1 transposase (44). The pCMA plasmid was used in a C. trachomatis transformation procedure with β-lactams for selection and then a pool of 105 transposon mutant clones from 23 transformations was generated. Further experiments proved that a FAD-dependent monooxygenase (ct148) and a deubiquitinase (ct868) were important for infection, and identified CT339 as a ComEC (the DNA-uptake protein) homolog important for DNA uptake and lateral gene transfer (44). O’Neill et al. then describe the first application of a Transposon Directed Insertion Site sequencing (TraDIS) - based approach to C. trachomatis, offering a novel approach for saturation mutagenesis and thus identifying gene essentiality/functionality (45, 46). Later, transposon mutagenesis of Chlamydia muridarum was also development and 33 transposon mutants were generated from a total of 10 independent transformation experiments (76).
The development of transposon mutagenesis in C. trachomatis provides additional avenues for discovering the molecular mechanism underlying the pathogenesis of C. trachomatis and for a more thorough understanding of this important pathogen (44). A limitation of transposon mutagenesis is the raising possibility of polar effects mediated by the inserted transposon due to polycistronic operons existing within the chlamydial genes.
6 Summary and prospectives
The genetic intractability of C. trachomatis has severely limited molecular dissection of virulence factors associated with intracellular parasitism and pathogenic mechanisms that promote trachoma, venereal lymphogranuloma or infertility, because there was no methods for C. trachomatis virulence determinants inactivation and/or complementation. Great progress has been made in the development of genetic manipulation of Chlamydia in the past decade, and the application of the tools of genetic manipulation has significantly impeded progress in understanding the genetic basis of the pathogen’s unique lifestyle and virulence. Moreover, the increasing genetic tractability of C. trachomatis will enable the development of new pathogen countermeasures, such as rationally designed attenuated or subunit vaccines. But some problems still remain:
(1) Low transformation efficiencies remain an obstacle to further development of genetic tools. Reasons including suboptimal electroporation conditions/buffers, purity of host cell-derived organisms could be account for the poor efficiency.
(2) At present, the developed tools of genetic manipulation are mostly suitable for C. trachomatis and incapable for the commonly infecting humans pathogens include C. pneumoniae and C. psittaci.
Additional advances in genetic manipulation will be necessary to render Chlamydia significantly more genetically tractable. Ideally, a cell-free medium like C. burnetii (77, 78) for Chlamydia cultivation would address some issues. Omsland et al. developed a stage-specific metabolic and transcriptional activity of C. trachomatis in an axenic medium in 2012 (79), and host-free cultivation of Chlamydia may be achievable in the future. Improved electroporation conditions may be another avenue if the decreased chlamydial viability could be addressed. Overall, more rapid and definitive progress can be expected for this important and interesting intracellular parasite.
Author contributions
The manuscript was drafted by WW and DannL, and edited by JJ and DanL. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China [32000139] and the State Key Laboratory of Pathogen and Biosecurity (Academy of Military Medical Science) [SKLPBS2217].
Acknowledgments
We apologize to researchers whose work was not cited here due to space limitations.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Elwell C, Mirrashidi K, Engel J. Chlamydia cell biology and pathogenesis. Nat Rev Microbiol (2016) 14:385–400. doi: 10.1038/nrmicro.2016.30
2. Solomon AW, Burton MJ, Gower EW, Harding-Esch EM, Oldenburg CE, Taylor HR, et al. Trachoma. Nat Rev Dis Primers (2022) 8:32. doi: 10.1038/s41572-022-00359-5
3. Lujan AL, Croci DO, Gambarte Tudela JA, Losinno AD, Cagnoni AJ, Marino KV, et al. Glycosylation-dependent galectin-receptor interactions promote Chlamydia trachomatis infection. Proc Natl Acad Sci USA (2018) 115:E6000–9. doi: 10.1073/pnas.1802188115
4. Sixt BS, Valdivia RH. Molecular genetic analysis of Chlamydia species. Annu Rev Microbiol (2016) 70:179–98. doi: 10.1146/annurev-micro-102215-095539
5. Moore ER, Ouellette SP. Reconceptualizing the chlamydial inclusion as a pathogen-specified parasitic organelle: an expanded role for inc proteins. Front Cell Infect Microbiol (2014) 4:157. doi: 10.3389/fcimb.2014.00157
6. Gitsels A, Sanders N, Vanrompay D. Chlamydial infection from outside to inside. Front Microbiol (2019) 10:2329. doi: 10.3389/fmicb.2019.02329
7. Bayramova F, Jacquier N, Greub G. Insight in the biology of Chlamydia-related bacteria. Microbes Infect (2018) 20:432–40. doi: 10.1016/j.micinf.2017.11.008
8. Lee JK, Enciso GA, Boassa D, Chander CN, Lou TH, Pairawan SS, et al. Replication-dependent size reduction precedes differentiation in chlamydia trachomatis. Nat Commun (2018) 9:45. doi: 10.1038/s41467-017-02432-0
9. Yang C, Starr T, Song L, Carlson JH, Sturdevant GL, Beare PA, et al. Chlamydial lytic exit from host cells is plasmid regulated. mBio (2015) 6:e01648–15. doi: 10.1128/mBio.01648-15
10. Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, et al. Genome sequence of an obligate intracellular pathogen of humans: chlamydia trachomatis. Science (1998) 282:754–9. doi: 10.1126/science.282.5389.754
11. Omsland A, Sixt BS, Horn M, Hackstadt T. Chlamydial metabolism revisited: interspecies metabolic variability and developmental stage-specific physiologic activities. FEMS Microbiol Rev (2014) 38:779–801. doi: 10.1111/1574-6976.12059
12. McClure EE, Chavez ASO, Shaw DK, Carlyon JA, Ganta RR, Noh SM, et al. Engineering of obligate intracellular bacteria: progress, challenges and paradigms. Nat Rev Microbiol (2017) 15:544–58. doi: 10.1038/nrmicro.2017.59
13. Beare PA, Sandoz KM, Omsland A, Rockey DD, Heinzen RA. Advances in genetic manipulation of obligate intracellular bacterial pathogens. Front Microbiol (2011) 2:97. doi: 10.3389/fmicb.2011.00097
14. Tam JE, Davis CH, Wyrick PB. Expression of recombinant DNA introduced into Chlamydia trachomatis by electroporation. Can J Microbiol (1994) 40:583–91. doi: 10.1139/m94-093
15. Kari L, Goheen MM, Randall LB, Taylor LD, Carlson JH, Whitmire WM, et al. Generation of targeted Chlamydia trachomatis null mutants. Proc Natl Acad Sci USA (2011) 108:7189–93. doi: 10.1073/pnas.1102229108
16. Kokes M, Dunn JD, Granek JA, Nguyen BD, Barker JR, Valdivia RH, et al. Integrating chemical mutagenesis and whole-genome sequencing as a platform for forward and reverse genetic analysis of chlamydia. Cell Host Microbe (2015) 17:716–25. doi: 10.1016/j.chom.2015.03.014
17. Nguyen BD, Valdivia RH. Virulence determinants in the obligate intracellular pathogen Chlamydia trachomatis revealed by forward genetic approaches. Proc Natl Acad Sci USA (2012) 109:1263–8. doi: 10.1073/pnas.1117884109
18. Nguyen B, Valdivia R. A chemical mutagenesis approach to identify virulence determinants in the obligate intracellular pathogen chlamydia trachomatis. Methods Mol Biol (2014) 1197:347–58. doi: 10.1007/978-1-4939-1261-2_20
19. Mishra MK, Gerard HC, Whittum-Hudson JA, Hudson AP, Kannan RM. Dendrimer-enabled modulation of gene expression in chlamydia trachomatis. Mol Pharm (2012) 9:413–21. doi: 10.1021/mp200512f
20. Kannan RM, Gerard HC, Mishra MK, Mao G, Wang S, Hali M, et al. Dendrimer-enabled transformation of chlamydia trachomatis. Microb Pathog (2013) 65:29–35. doi: 10.1016/j.micpath.2013.08.003
21. Johnson CM, Fisher DJ. Site-specific, insertional inactivation of incA in Chlamydia trachomatis using a group II intron. PloS One (2013) 8:e83989. doi: 10.1371/journal.pone.0083989
22. Bauler LD, Hackstadt T. Expression and targeting of secreted proteins from chlamydia trachomatis. J Bacteriol (2014) 196:1325–34. doi: 10.1128/JB.01290-13
23. Bishop RC, Derre I. The Chlamydia trachomatis inclusion membrane protein CTL0390 mediates host cell exit via lysis through STING activation. Infect Immun (2022) 90:e0019022. doi: 10.1128/iai.00190-22
24. Faris R, McCullough A, Andersen SE, Moninger TO, Weber MM. The Chlamydia trachomatis secreted effector TmeA hijacks the n-WASP-ARP2/3 actin remodeling axis to facilitate cellular invasion. PloS Pathog (2020) 16:e1008878. doi: 10.1371/journal.ppat.1008878
25. Weber MM, Lam JL, Dooley CA, Noriea NF, Hansen BT, Hoyt FH, et al. Absence of specific Chlamydia trachomatis inclusion membrane proteins triggers premature inclusion membrane lysis and host cell death. Cell Rep (2017) 19:1406–17. doi: 10.1016/j.celrep.2017.04.058
26. Cortina ME, Bishop RC, DeVasure BA, Coppens I, Derre I. The inclusion membrane protein IncS is critical for initiation of the Chlamydia intracellular developmental cycle. PloS Pathog (2022) 18:e1010818. doi: 10.1371/journal.ppat.1010818
27. Cosse MM, Barta ML, Fisher DJ, Oesterlin LK, Niragire B, Perrinet S, et al. The loss of expression of a single type 3 effector (CT622) strongly reduces Chlamydia trachomatis infectivity and growth. Front Cell Infect Microbiol (2018) 8:145. doi: 10.3389/fcimb.2018.00145
28. Sixt BS, Bastidas RJ, Finethy R, Baxter RM, Carpenter VK, Kroemer G, et al. The Chlamydia trachomatis inclusion membrane protein CpoS counteracts STING-mediated cellular surveillance and suicide programs. Cell Host Microbe (2017) 21:113–21. doi: 10.1016/j.chom.2016.12.002
29. Carpenter V, Chen YS, Dolat L, Valdivia RH. The effector TepP mediates recruitment and activation of phosphoinositide 3-kinase on early Chlamydia trachomatis vacuoles. mSphere (2017) 2:e00207–17. doi: 10.1128/mSphere.00207-17
30. Pereira IS, Pais SV, Borges V, Borrego MJ, Gomes JP, Mota LJ. The type III secretion effector CteG mediates host cell lytic exit of chlamydia trachomatis. Front Cell Infect Microbiol (2022) 12:902210. doi: 10.3389/fcimb.2022.902210
31. Mueller KE, Wolf K, Fields KA. Gene deletion by fluorescence-reported allelic exchange mutagenesis in chlamydia trachomatis. mBio (2016) 7:e01817–15. doi: 10.1128/mBio.01817-15
32. Fields KA, Bodero MD, Scanlon KR, Jewett TJ, Wolf K. A minimal replicon enables efficacious, species-specific gene deletion in Chlamydia and extension of gene knockout studies to the animal model of infection using chlamydia muridarum. Infect Immun (2022) 90:e0045322. doi: 10.1128/iai.00453-22
33. Keb G, Hayman R, Fields KA. Floxed-cassette allelic exchange mutagenesis enables markerless gene deletion in Chlamydia trachomatis and can reverse cassette-induced polar effects. J Bacteriol (2018) 200:e00479-18. doi: 10.1128/JB.00479-18
34. Ghosh S, Ruelke EA, Ferrell JC, Bodero MD, Fields KA, Jewett TJ. Fluorescence-reported allelic exchange mutagenesis-mediated gene deletion indicates a requirement for Chlamydia trachomatis tarp during In vivo infectivity and reveals a specific role for the c terminus during cellular invasion. Infect Immun (2020) 88:e00841-19. doi: 10.1128/IAI.00841-19
35. Keb G, Ferrell J, Scanlon KR, Jewett TJ, Fields KA. Chlamydia trachomatis TmeA directly activates n-WASP to promote actin polymerization and functions synergistically with TarP during invasion. mBio (2021) 12:e02861-20. doi: 10.1128/mBio.02861-20
36. Bui DC, Jorgenson LM, Ouellette SP, Rucks EA. Eukaryotic SNARE VAMP3 dynamically interacts with multiple chlamydial inclusion membrane proteins. Infect Immun (2021) 89:e00409-20. doi: 10.1128/IAI.00409-20
37. Keb G, Fields KA. Markerless gene deletion by floxed cassette allelic exchange mutagenesis in chlamydia trachomatis. J Vis Exp (2020) 155. doi: 10.3791/60848-v
38. Wolf K, Rahnama M, Fields KA. Genetic manipulation of Chlamydia trachomatis: chromosomal deletions. Methods Mol Biol (2019) 2042:151–64. doi: 10.1007/978-1-4939-9694-0_11
39. Park JS, Helble JD, Lazarus JE, Yang G, Blondel CJ, Doench JG, et al. A FACS-based genome-wide CRISPR screen reveals a requirement for COPI in Chlamydia trachomatis invasion. iScience (2019) 11:71–84. doi: 10.1016/j.isci.2018.12.011
40. Wurihan W, Huang Y, Weber AM, Wu X, Fan H. Nonspecific toxicities of Streptococcus pyogenes and Staphylococcus aureus dCas9 in chlamydia trachomatis. Pathog Dis (2019) 77:ftaa005. doi: 10.1093/femspd/ftaa005
41. Ouellette SP, Blay EA, Hatch ND, Fisher-Marvin LA. CRISPR interference to inducibly repress gene expression in chlamydia trachomatis. Infect Immun (2021) 89:e0010821. doi: 10.1128/IAI.00108-21
42. Ouellette SP. Feasibility of a conditional knockout system for Chlamydia based on CRISPR interference. Front Cell Infect Microbiol (2018) 8:59. doi: 10.3389/fcimb.2018.00059
43. Wood NA, Blocker AM, Seleem MA, Conda-Sheridan M, Fisher DJ, Ouellette SP. The ClpX and ClpP2 orthologs of Chlamydia trachomatis perform discrete and essential functions in organism growth and development. mBio (2020) 11:e02016-20. doi: 10.1128/mBio.02016-20
44. LaBrie SD, Dimond ZE, Harrison KS, Baid S, Wickstrum J, Suchland RJ, et al. Transposon mutagenesis in Chlamydia trachomatis identifies CT339 as a ComEC homolog important for DNA uptake and lateral gene transfer. mBio (2019) 10:e01343-19. doi: 10.1128/mBio.01343-19
45. O’Neill CE, Skilton RJ, Forster J, Cleary DW, Pearson SA, Lampe DJ, et al. An inducible transposon mutagenesis approach for the intracellular human pathogen chlamydia trachomatis. Wellcome Open Res (2021) 6:312. doi: 10.12688/wellcomeopenres.16068.1
46. Skilton RJ, O’Neill C, Thomson NR, Lampe DJ, Clarke IN. Progress towards an inducible, replication-proficient transposon delivery vector for chlamydia trachomatis. Wellcome Open Res (2021) 6:82. doi: 10.12688/wellcomeopenres.16665.1
47. Wang P. Genetic transformation in Cryptococcus species. J Fungi (Basel) (2021) 7:56. doi: 10.3390/jof7010056
48. Binet R, Maurelli AT. Transformation and isolation of allelic exchange mutants of Chlamydia psittaci using recombinant DNA introduced by electroporation. Proc Natl Acad Sci U.S.A. (2009) 106:292–7. doi: 10.1073/pnas.0806768106
49. Bose JL. Chemical and UV mutagenesis. Methods Mol Biol (2016) 1373:111–5. doi: 10.1007/7651_2014_190
50. Chatterjee N, Walker GC. Mechanisms of DNA damage, repair, and mutagenesis. Environ Mol Mutagen (2017) 58:235–63. doi: 10.1002/em.22087
51. Bastidas RJ, Valdivia RH. Emancipating Chlamydia: advances in the genetic manipulation of a recalcitrant intracellular pathogen. Microbiol Mol Biol Rev (2016) 80:411–27. doi: 10.1128/MMBR.00071-15
52. Fox LJ, Richardson RM, Briscoe WH. PAMAM dendrimer - cell membrane interactions. Adv Colloid Interface Sci (2018) 257:1–18. doi: 10.1016/j.cis.2018.06.005
53. Mishra MK, Kotta K, Hali M, Wykes S, Gerard HC, Hudson AP, et al. PAMAM dendrimer-azithromycin conjugate nanodevices for the treatment of Chlamydia trachomatis infections. Nanomedicine (2011) 7:935–44. doi: 10.1016/j.nano.2011.04.008
54. Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PloS Pathog (2011) 7:e1002258. doi: 10.1371/journal.ppat.1002258
55. Kedzior M, Bastidas RJ. Forward and reverse genetic analysis of chlamydia. Methods Mol Biol (2019) 2042:185–204. doi: 10.1007/978-1-4939-9694-0_13
56. Mueller KE, Wolf K, Fields KA. Chlamydia trachomatis transformation and allelic exchange mutagenesis. Curr Protoc Microbiol (2017) 45:11A 3 1–11A 3 15. doi: 10.1002/cpmc.31
57. Lambowitz AM, Zimmerly S. Mobile group II introns. Annu Rev Genet (2004) 38:1–35. doi: 10.1146/annurev.genet.38.072902.091600
58. Weber MM, Noriea NF, Bauler LD, Lam JL, Sager J, Wesolowski J, et al. A functional core of IncA is required for Chlamydia trachomatis inclusion fusion. J Bacteriol (2016) 198:1347–55. doi: 10.1128/JB.00933-15
59. Noriea NF, Clark TR, Hackstadt T. Targeted knockout of the Rickettsia rickettsii OmpA surface antigen does not diminish virulence in a mammalian model system. mBio (2015) 6:e00323-15. doi: 10.1128/mBio.00323-15
60. Jeffrey BM, Suchland RJ, Eriksen SG, Sandoz KM, Rockey DD. Genomic and phenotypic characterization of in vitro-generated Chlamydia trachomatis recombinants. BMC Microbiol (2013) 13:142. doi: 10.1186/1471-2180-13-142
61. Perutka J, Wang W, Goerlitz D, Lambowitz AM. Use of computer-designed group II introns to disrupt Escherichia coli DExH/D-box protein and DNA helicase genes. J Mol Biol (2004) 336:421–39. doi: 10.1016/j.jmb.2003.12.009
62. Wickstrum J, Sammons LR, Restivo KN, Hefty PS. Conditional gene expression in Chlamydia trachomatis using the tet system. PloS One (2013) 8:e76743. doi: 10.1371/journal.pone.0076743
63. Song L, Carlson JH, Whitmire WM, Kari L, Virtaneva K, Sturdevant DE, et al. Chlamydia trachomatis plasmid-encoded Pgp4 is a transcriptional regulator of virulence-associated genes. Infect Immun (2013) 81:636–44. doi: 10.1128/IAI.01305-12
64. Thomas NS, Lusher M, Storey CC, Clarke IN. Plasmid diversity in chlamydia. Microbiol (Reading) (1997) 143(Pt 6):1847–54. doi: 10.1099/00221287-143-6-1847
65. Beare PA, Larson CL, Gilk SD, Heinzen RA. Two systems for targeted gene deletion in coxiella burnetii. Appl Environ Microbiol (2012) 78:4580–9. doi: 10.1128/AEM.00881-12
66. Doudna JA, Charpentier E. Genome editing. the new frontier of genome engineering with CRISPR-Cas9. Science (2014) 346:1258096. doi: 10.1126/science.1258096
67. Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell (2013) 152:1173–83. doi: 10.1016/j.cell.2013.02.022
68. Rock JM, Hopkins FF, Chavez A, Diallo M, Chase MR, Gerrick ER, et al. Programmable transcriptional repression in mycobacteria using an orthogonal CRISPR interference platform. Nat Microbiol (2017) 2:16274. doi: 10.1038/nmicrobiol.2016.274
69. Wachter S, Cockrell DC, Miller HE, Virtaneva K, Kanakabandi K, Darwitz B, et al. The endogenous Coxiella burnetii plasmid encodes a functional toxin-antitoxin system. Mol Microbiol (2022) 118:744–64. doi: 10.1111/mmi.15001
70. Fu M, Liu Y, Wang G, Wang P, Zhang J, Chen C, et al. A protein-protein interaction map reveals that the Coxiella burnetii effector CirB inhibits host proteasome activity. PloS Pathog (2022) 18:e1010660. doi: 10.1371/journal.ppat.1010660
71. Martinez E, Cantet F, Bonazzi M. Generation and multi-phenotypic high-content screening of Coxiella burnetii transposon mutants. J Vis Exp (2015) 99:e52851. doi: 10.3791/52851
72. Beare PA, Howe D, Cockrell DC, Omsland A, Hansen B, Heinzen RA. Characterization of a Coxiella burnetii ftsZ mutant generated by Himar1 transposon mutagenesis. J Bacteriol (2009) 191:1369–81. doi: 10.1128/JB.01580-08
73. Laukaitis HJ, Cooper TT, Suwanbongkot C, Verhoeve VI, Kurtti TJ, Munderloh UG, et al. Transposon mutagenesis of Rickettsia felis sca1 confers a distinct phenotype during flea infection. PloS Pathog (2022) 18:e1011045. doi: 10.1371/journal.ppat.1011045
74. Qin A, Tucker AM, Hines A, Wood DO. Transposon mutagenesis of the obligate intracellular pathogen rickettsia prowazekii. Appl Environ Microbiol (2004) 70:2816–22. doi: 10.1128/AEM.70.5.2816-2822.2004
75. Jaworski DC, Cheng C, Nair ADS, Ganta RR. Amblyomma americanum ticks infected with in vitro cultured wild-type and mutants of Ehrlichia chaffeensis are competent to produce infection in naive deer and dogs. Ticks Tick Borne Dis (2017) 8:60–4. doi: 10.1016/j.ttbdis.2016.09.017
76. Wang Y, LaBrie SD, Carrell SJ, Suchland RJ, Dimond ZE, Kwong F, et al. Development of transposon mutagenesis for chlamydia muridarum. J Bacteriol (2019) 201:e00366-19. doi: 10.1128/JB.00366-19
77. Omsland A, Beare PA, Hill J, Cockrell DC, Howe D, Hansen B, et al. Isolation from animal tissue and genetic transformation of Coxiella burnetii are facilitated by an improved axenic growth medium. Appl Environ Microbiol (2011) 77:3720–5. doi: 10.1128/AEM.02826-10
78. Omsland A, Cockrell DC, Howe D, Fischer ER, Virtaneva K, Sturdevant DE, et al. Host cell-free growth of the q fever bacterium coxiella burnetii. Proc Natl Acad Sci U.S.A. (2009) 106:4430–4. doi: 10.1073/pnas.0812074106
Keywords: intracellular bacterium, Chlamydia trachomatis, genetic manipulation, transformation, challenges
Citation: Wan W, Li D, Li D and Jiao J (2023) Advances in genetic manipulation of Chlamydia trachomatis. Front. Immunol. 14:1209879. doi: 10.3389/fimmu.2023.1209879
Received: 21 April 2023; Accepted: 14 June 2023;
Published: 28 June 2023.
Edited by:
Yanan Ma, Memorial Sloan Kettering Cancer Center, United StatesReviewed by:
Kunkun Wang, Fairbanks Memorial Hospital, United StatesJingyi Xie, University of Washington, United States
Weize Yuan, Massachusetts Institute of Technology, United States
Fang Li, University of Miami, United States
Dongdong Yang, University of Colorado Boulder, United States
Copyright © 2023 Wan, Li, Li and Jiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Jiao, jiaojun51920@sina.com
†These authors have contributed equally to this work
 Weiqiang Wan
Weiqiang Wan Danni Li
Danni Li Dan Li
Dan Li Jun Jiao
Jun Jiao