- 1Department of Microbial Pathogens and Immunity, Rush University Medical Center, Chicago, IL, United States
- 2Department of Medical Microbiology and Immunology, Creighton University School of Medicine, Omaha, NE, United States
- 3Department of Biochemistry and Molecular Biology, School of Basic Medicine, Hubei University of Science and Technology, Xianning, Hubei, China
Cryptosporidium is a zoonotic apicomplexan parasite that infects the gastrointestinal epithelium and other mucosal surfaces in humans. It is an important opportunistic pathogen in AIDS patients and a leading cause of infectious diarrhea and diarrheal-related death in children worldwide. The intestinal epithelial cells provide the first line of defense against Cryptosporidium infection and play a central role in activating and regulating the host’s antiparasitic response. Increasing evidence suggests that long noncoding RNAs (lncRNAs) participate in host-pathogen interactions and play a regulatory role in the pathogenesis of diseases but the underlying molecular mechanisms are not fully understood. We previously identified a panel of host lncRNAs that are upregulated in murine intestinal epithelial cells following Cryptosporidium infection, including U90926. We demonstrate here that U90926 is acting in a pro-parasitic manner in regulating intestinal epithelial cell-autonomous antiparasitic defense. Inhibition of U90926 resulted in a decreased infection burden of the parasite while overexpression of U90926 showed an increase in infection burden in cultured murine intestinal epithelial cells. Induction of U90926 suppressed transcription of epithelial defense genes involved in controlling Cryptosporidium infection through epigenetic mechanisms. Specifically, transcription of Aebp1, which encodes the Aebp1 protein, a potent modulator of inflammation and NF-κB signaling, was suppressed by U90926. Gain- or loss-of-function of Aebp1 in the host’s epithelial cells caused reciprocal alterations in the infection burden of the parasite. Interestingly, Cryptosporidium carries the Cryptosporidium virus 1 (CSpV1), a double-stranded (ds) RNA virus coding two dsRNA fragments, CSpV1-dsRdRp and CSpV1-dsCA. Both CSpV1-dsRdRp and CSpV1-dsCA can be delivered into infected cells as previously reported. We found that cells transfected with in vitro transcribed CSpV1-dsCA or CSpV1-dsRdRp displayed an increased level of U90926, suggesting that CSpV1 is involved in the upregulation of U90926 during Cryptosporidium infection. Our study highlights a new strategy by Cryptosporidium to hijack a host lncRNA to suppress epithelial cell-autonomous antiparasitic defense and allow for a robust infection.
Introduction
Cryptosporidium is a protozoan parasite that infects the gastrointestinal epithelium and other mucosal surfaces in humans and animals (1). It is an important opportunistic pathogen in AIDS patients and is the second most common pathogen responsible for moderate to severe diarrhea in children under 2 years of age in developing countries (2). Infection with Cryptosporidium shows a significant association with mortality in children and appears to predispose them to lasting defects in body growth and cognitive ability (2–5). The majority of human Cryptosporidium infections are caused by two species: C. parvum and C. hominis (6, 7). Waterborne transmission grants the parasite the potential to cause large-scale outbreaks and epidemics in both developing and developed nations (8–10). After ingestion of Cryptosporidium oocysts from contaminated water or foods, the oocysts are excysted in the gastrointestinal tract to release infective sporozoites. The sporozoites then attach to the apical membrane of the intestinal epithelial cells and form an intracellular but extracytoplasmic vacuole where the organism remains and further develops (6, 11). Therefore, epithelial cells play a central role in activating and orchestrating both innate and adaptive anti-Cryptosporidium responses (12, 13).
Epithelial cells along the mucosal surface provide the first line of defense against luminal pathogen infections such as Cryptosporidium. These epithelial cells are an important component of digestive mucosal immunity and represent an integral component of a highly regulated communication network involving essential signals to and from the underlying cells of the gastrointestinal mucosa (14). Upon microbial challenge, gastrointestinal epithelial cells initiate a series of immune reactions including the release of inflammatory cytokines/chemokines and the production of antimicrobial molecules. These molecules then activate immune effector cells and mobilize them to the site of infection (15). Among these signaling pathways activated by C. parvum infection is the TLR4/nuclear factor kappa B (NF-κB) signaling pathway which results in the production and secretion of those cytokines/chemokines which may kill or inhibit parasite growth (16, 17). Nevertheless, the success of the parasite is the result of its ability to evade these host immune responses while still utilizing the host’s resources. The host and parasite partake in a co-evolutionary “arms race” in which the parasite evolves novel strategies to escape the immune response and the immune system simultaneously evolves strategies to kill the parasite. This dynamic competition results in the parasite employing innovative ways of utilizing host resources and machinery (18).
Increasing evidence suggests that a certain portion of the eukaryotic genome is transcribed as non-protein coding RNAs (ncRNAs) including long ncRNAs (lncRNAs) which are noncoding transcripts longer than 200 nt. Thousands of lncRNAs have been identified and the majority of them may be functional (19, 20). LncRNAs function to regulate gene transcription through specific interactions with cellular factors, including proteins, DNA, and other RNA molecules via nucleotide base pairing or the formation of structural domains generated by RNA folding (21–23). Studies have found many of these lncRNAs to be targets of inflammatory pathways and thus their expression is altered during inflammation or microbial infection (24, 25). Several lncRNAs have been shown to be induced in innate immune cells and may play a role in regulating the immune response (26–29). Furthermore, recent evidence suggests that microorganisms can utilize lncRNAs to regulate or evade the host’s immune response. During chlamydia trachomatis infection, the host lncRNA MIAT is upregulated and involved in not only resisting apoptosis but also the growth and development of chlamydia trachomatis (30). Toxoplasma gondii infection results in altered host lncRNA expression to target UNC93B1 expression, impairing the secretion of cytokines IL-12, TNF-α, IL-1β, and IFN-γ, ultimately inhibiting the host immune response (31).
Using an in vitro infection model of intestinal epithelial cells by C. parvum, a panel of host cell lncRNAs that are upregulated following C. parvum infection has been recently identified (25). Among the lncRNAs upregulated was U90926, previously also known as NR_033483, with its gene located on chromosome 5 near XR_880430.2 (32). This lncRNA can be induced in activated macrophages, is protective in endotoxic shock, and may encode a secreted peptide in macrophages (32). In this study, we demonstrate that U90926 displays an inhibitory effect on the innate immune response against C. parvum infection by transcriptionally repressing defense gene expression. Interestingly, the upregulation of U90926 itself seemed to be induced by a small double-stranded RNA virus found in Cryptosporidium called Cryptosporidium parvum virus (CSpV1) (33). Therefore, our data suggest that upon C. parvum infection, CSpV1 induces the upregulation of host lncRNA, U90926, which targets transcription of host defense genes and inhibits epithelial antiparasitic response. This study highlights a novel evasion strategy in which C. parvum hijacks a host’s lncRNA to suppress epithelial cell-autonomous antiparasitic defense to allow for a robust infection.
Materials and methods
C. parvum and cell lines
C. parvum oocysts of the Iowa strain were purchased from a commercial source (Bunch Grass Farm, Deary, ID). The mouse intestinal epithelial cell line (IEC4.1) was received as a kind gift from Dr. Pingchang Yang (McMaster University, Hamilton, Canada). Culture media were supplied with 10% FBS (Ambion) and antibiotics. Stable IEC4.1 cells with deficient in U90926 were generated through transfection of cells with the CRISPR/Cas9 KO(h) (U90926-CRISPR/Cas9 KO) and the HDR plasmid (U90926-HDR) as previously described (34).
Infection models and infection assays
Models of intestinal cryptosporidiosis using intestinal epithelial cell lines and 2D monolayers derived from 3D enteroids were employed as previously described (25, 35). Infection was done in culture medium (DMEM-F-12 with 100U/ml penicillin and 100µg/ml streptomycin) containing viable C. parvum oocysts after treatment with 1% sodium hypochlorite. Cells were then cultured for 4 hr at 37°C for attachment and invasion by the parasites. After extensive washing with DMEM-F-12 medium three times to remove free parasites. Cells were cultured for additional time periods. Intestinal epithelium and 3D enteroids were employed as previously described (35). Briefly, the small intestine was opened longitudinally and washed with ice-cold Ca2+ and Mg2+ free PBS before being cut into 1-2 mm fragments and washed with ice-cold Ca2+ and Mg2+ free PBS 3 times. The cut fragments were incubated in ice-cold 2 mM PBS/EDTA at 4° C for 30 min with gentle rotation followed by vigorous shake until the PBS solution was mostly opaque with dislodged crypts and villus particles. Large tissue fragments were removed through a 100-µm cell strainer (Becton-Dickinson Bioscience, Franklin Lakes, NJ). The pass-through was centrifuged 150 g for 5 min at 4°C and the pellet was collected as the intestinal epithelium. 2D monolayers derived from 3D enteroids were cultured as previously described (35–37) and exposed to C. parvum infection for 24-48 hr.
For in vivo experiments, a well-developed infection model of cryptosporidiosis in neonatal mice was used (38, 39). Briefly, 5 days after birth, mice received C. parvum oocysts by oral gavage (106 oocysts per mouse) to develop intestinal cryptosporidiosis. Mice that received PBS by oral gavage were used as controls. At 48- and 72 hr after C. parvum or PBS administration, animals were sacrificed, and ileum epithelium tissues were obtained for biochemical analysis and infection assessment as previously reported (25, 35, 40, 41). All research studies involving the use of animals were reviewed and approved by the Institutional Animal Care and Use Committees of the Rush University Medical Center and were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals.
Quantitative real-time PCR
For quantitative analysis of RNA and C. parvum RNA expression, qRT-PCR was performed as previously reported (35, 40–43), using the SYBR green PCR master mix (Applied Biosystems, Carlsbad, CA, USA). Briefly, total RNA was isolated and possible remaining DNA was removed using TRI reagent and treated with a DNase-free kit (Ambion, MA, USA). qRT-PCR was then performed using 25 ng of template cDNA from reverse transcription for each RNA gene of interest. The expression level of each RNA was calculated using the threshold cycles (ΔΔCT) method and normalized to glyceraldehyde-3-phosphate dehydrogenase (Gapdh). All sequences of PCR primers are listed in Table S1.
Transfections with siRNAs
Custom-designed RNA oligonucleotides against U90926 and a scrambled RNA (used as the control) were synthesized by IDT (Integrated DNA Technologies, Coralville, IA) and transfected into cells with the Lipofectamine RNAiMAX according to the manufacturer’s protocol (Invitrogen). Sequences of siRNAs were GAGGTTCTGTGAATTCTTTAA for U90926 and UUCUCCGAACGUGUCACGUUU for the control.
Chromatin immunoprecipitation and chromatin isolation by RNA purification
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (25, 28, 29) using the ChIP Assay Kit (Millipore Sigma) in accordance with the manufacturer’s protocol. An anti-H3K4m1 (Abcam) was used and the specific primers are listed in Table S1. Chromatin isolation by RNA purification (ChIRP) analysis was performed as previously reported (44). Briefly, glutaraldehyde cross-linked for chromatin isolation and a pool of tiling oligonucleotide probes with an affinity specific to the U90926 sequence were used. The sequences for each probe are listed in Table S1. The DNA sequences of the chromatin immunoprecipitates were confirmed by qRT-PCR using the same primer sets covering the gene promoter regions of interest as for ChIP analysis. A pool of scrambled oligonucleotide probes for LacZ (Table S1) was used as a control.
Luciferase reporter constructs and luciferase assay
The 266-1000 region of the U90926 gene, which contains five putative NF-kB binding sites, was cloned into the multiple cloning sites of the PNL1.1 NLUC vector (Fisher Scientific). The primers to amply the sequence are listed in Table S1. An empty vector was used as a control. Cultured IEC4.1 cells were transfected with the reporter construct overnight with or without C. parvum infection followed by an assessment of luciferase activity. The luciferase activity was normalized to the control β-galactosidase level and compared to the empty vector.
Statistical analysis
All values are given as mean ± SEM. The means of the groups were from at least three independent experiments and compared with Student’s t-test (unpaired) or the ANOVA test when appropriate. P values <0.05 were considered significant.
Results
Upregulation of U90926 expression in intestinal epithelium following C. parvum infection
The lab previously performed a genome-wide transcriptome analysis of C. parvum infected IEC4.1 cells, transformed by nontumorigenic intestinal epithelial cell line from neonatal mice, and identified a panel of host lncRNA genes whose expression levels are altered during infection (25). Among the lncRNAs that are upregulated is U90926, which is transcribed from the gene locus U90926 (25). To verify the upregulation of U90926 during C. parvum infection, qRT-PCR was performed in IEC4.1 cells following C. parvum infection at the 8, 24, and 48 hr time points. There was a significant increase in U90926 expression at 24 hrs post-infection (Figure 1A). Using a documented model of intestinal cryptosporidiosis in neonatal mice via oral administration of the parasite (35, 45), we detected infection via immunofluorescent staining, in the villus regions of the ileum from infected animals (Figure 1B). U90926 was also upregulated in isolated intestinal epithelium from neonatal mice infected with C. parvum at 48 and 72 hrs post-infection (Figure 1C). Using an ex vivo infection model employing 2D enteroid monolayers from neonatal mouse ileum (46), we detected a dose-dependent increase of U90926 expression in the infected monolayers (Figure 1D).
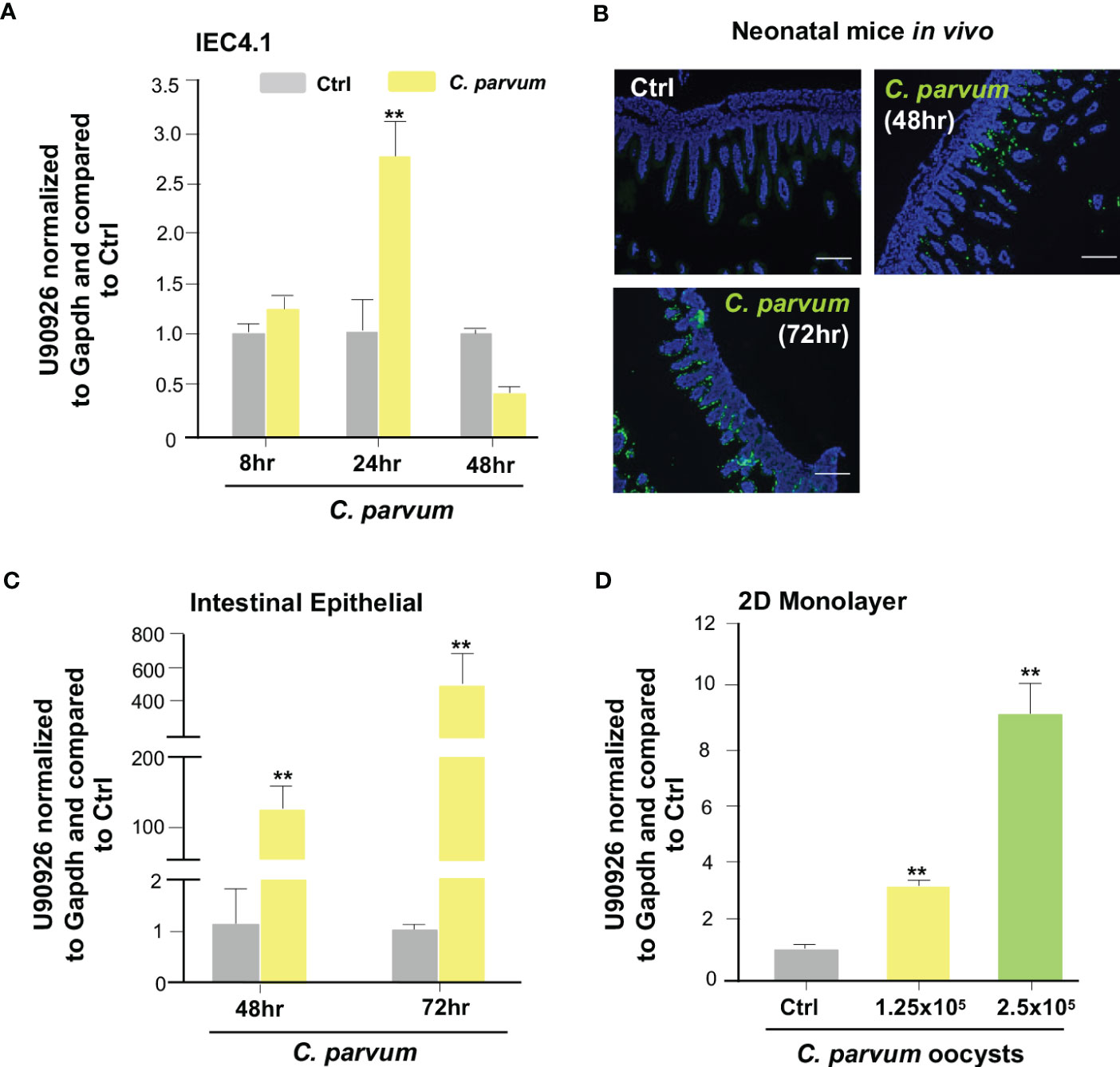
Figure 1 Upregulation of U90926 expression in intestinal epithelium following Cryptosporidium parvum infection. (A) Upregulation of U90926 in cultured intestinal epithelial cells (IEC4.1) following C. parvum infection. IEC4.1 cells were exposed to C. parvum infection for 8-48 hr and expression of U90926 was measured via qRT-PCR. (B) C. parvum infection of intestinal epithelium in neonatal mice. Indirect immunofluorescent staining of the ileum confirms C. parvum infection. (C) Upregulation of U90926 in murine intestinal epithelium following C. parvum infection in vivo. Neonates of mice (5 days old) were infected via oral administration for 48-72 hr. qRT-PCR was used to measure expression levels of U90926. (D) Upregulation of U90926 in 2D intestinal epithelial monolayers following C. parvum infection. 2D monolayers were exposed to C. parvum infection (at two different parasite oocyst doses: 1.25 X 105 and 2.5 X 105 oocysts/well) for 24 hrs and the expression of U90926 was measured via qRT-PCR. **p<0.001 vs cells of non-infected control (Ctrl).
Upregulation of U90926 may not be induced through the activation of immune signaling pathways in infected intestinal epithelial cells
We then investigated which host signaling pathways may be responsible for U90926 induction in intestinal epithelial cells following C. parvum infection. Previous studies have demonstrated multiple intracellular signals are activated during C. parvum infection, such as the NF-κB, JAK/STAT, JNK/MAPK, and type 1 IFN signal pathways (34, 40, 41, 47). IEC4.1 cells were stimulated with IFN-γ (10 ng/mL) and samples were collected at time points ranging from 1h to 8 hr post-treatment. Expression of U90926 was then measured via qRT-PCR. There was no change in U90926 when stimulated with IFN-γ (Figure 2A). IEC4.1 cells were also treated with IFN-α (50 u/mL and 25 u/mL) and there was a slight increase in U90926 expression at the 6 hr time point when cells were treated with IFN-α (50 u/mL), however, this modest change was not detected at any other time point or when cells were treated with a lower dose of IFN-α (25 u/mL) (Figures 2B, C). In addition, there was no change in U90926 expression in IEC4.1 cells stimulated with LPS or a JNK/MAPK activator anisomycin (48) (Figures 2D, E). Treatment with various Toll-like receptor (TLR) agonists, including TLR2, 4, 5, 7, and 9 all of which have been shown to be expressed on IEC4.1 cells (44), did not alter U90926 expression levels (Figure 2F). Of note, IEC4.1 cells displayed cellular response to these above stimuli when treated with the indicated doses and time course as reflected by the upregulation of selected genes following each stimulus [e.g., interferon-gamma induced GTPase (Igtp) for IFN-γ stimulation] (data not shown). The above data suggest that U90926 expression is increased during C. parvum infection but is not controlled by the tested host immune signaling pathways that are activated in infected IEC4.1 cells.
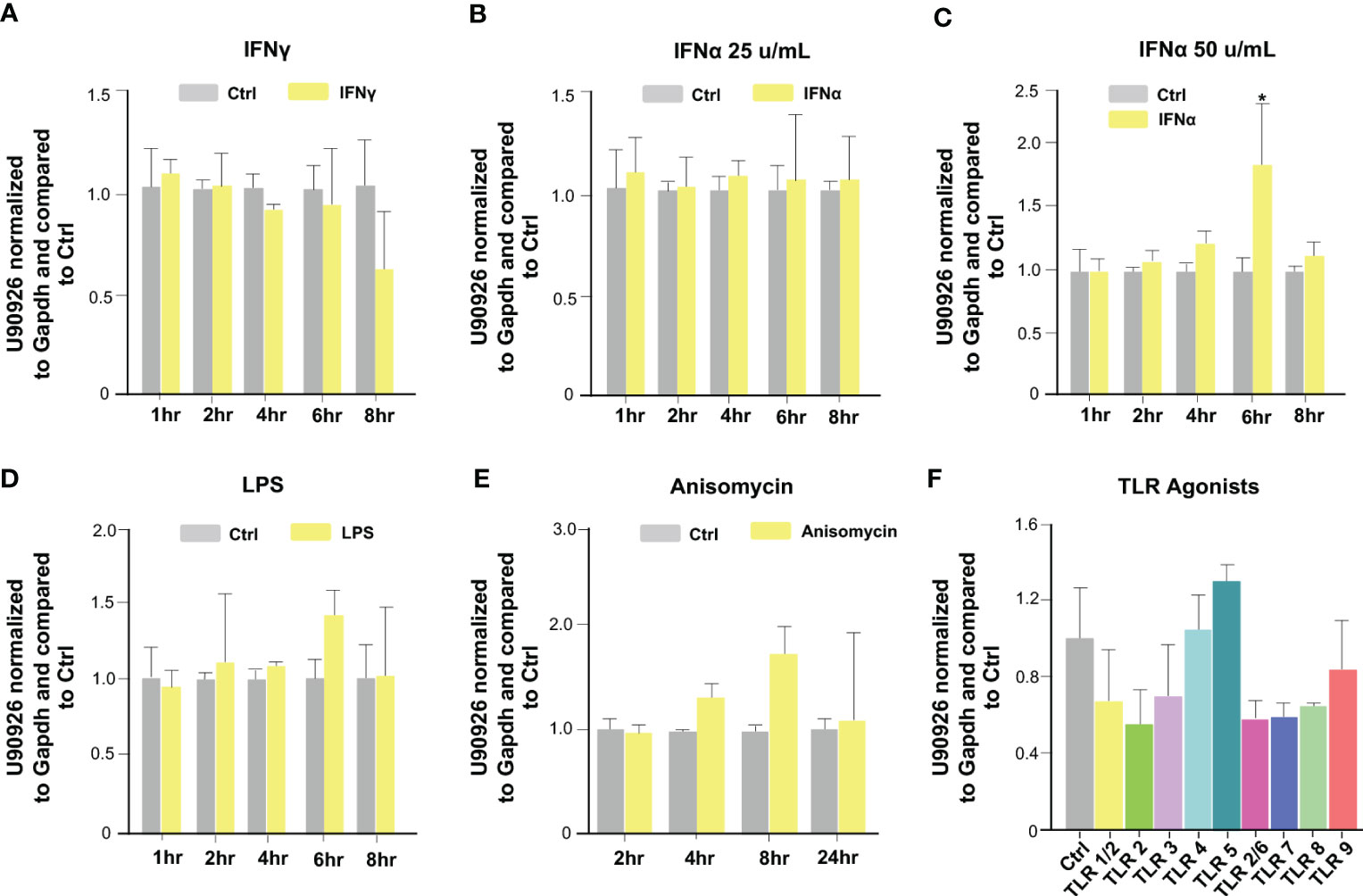
Figure 2 Upregulation of U90926 may not be induced through the activation of immune signaling pathways in infected intestinal epithelial cells. (A) Upregulation of U90926 was measured in IEC4.1 cells treated with IFN-γ (10 ng/mL), via qRT-PCR, at time points ranging from 1hr to 8hrs post-treatment. (B) IEC4.1 cells were treated with IFN-α (25 u/mL), and samples were collected at time points ranging from 1 to 8 hrs post-treatment. U90926 expression was measured via qRT-PCR. (C) IEC4.1 cells were treated with IFN-α (50 u/mL) and collected at time points ranging from 1hr to 8 hrs post-treatment and U90926 was measured via qRT-PCR. (D) Expression levels of U90926 were also measured in cells treated with LPS (12.5 ug/mL) and collected 1hr to 8hrs post-treatment. (E) IEC4.1 cells were treated with anisomycin (1 ug/mL) and U90926 expression was measured via qRT-PCR. (F) IEC4.1 cells were treated with TLR agonists for 16 hr and U90926 expression was measured via qPCR. Data represent means ± SEM from three independent experiments. *p<0.05 vs cells of non-infected control (Ctrl).
Impact of U90926 induction on parasite burden in intestinal epithelial cells
We sought to further explore the relationship between U90926 expression and C. parvum infection. We used RNA interference to knock down U90926 in host cells and measure its impact on C. parvum infection burden. C. parvum 18s (cp18s), Cryptosporidium parvum virus (CPV), and Hsp70 (cpHsp70) levels, reflecting parasite burden, were measured at 24 hrs after exposure to C. parvum (a time point reflecting activation of the cellular defenses protecting against infection), as well as at 2 hrs after exposure to the parasite (a time point reflecting the initial attachment and invasion of the parasite into host cells) (49). The designed siRNA to U90926 significantly decreased U90926 expression in both non-infected and C. parvum-infected IEC4.1 cells (Figure 3A). Interestingly, knock down of U90926 resulted in a decrease of C. parvum infection burden compared to cells treated with the nonspecific control siRNA after 24 hrs (Figure 3B) but no significant change in infection burden after 2h (Figure 3C). Additionally, U90926 CRISPR/Cas9 knockout cells (U90936-KO) were infected with C. parvum for 24 hrs and the infection burden was measured via qRT-PCR. U90926-KO cells displayed lower levels of C. parvum infection burden compared to IEC4.1 cells (Figure 3D). Complementarily, cells transfected with a plasmid expressing U90926 (U90926-OE) showed increased levels of U90926 in both C. parvum infected and non-infected cells (Figure 3E) and a slight increase in infection burden of the parasite after 24 hr (Figure 3F). These data indicate that U90926 may be playing a pro-parasitic role during C. parvum infection in IEC4.1 cells.
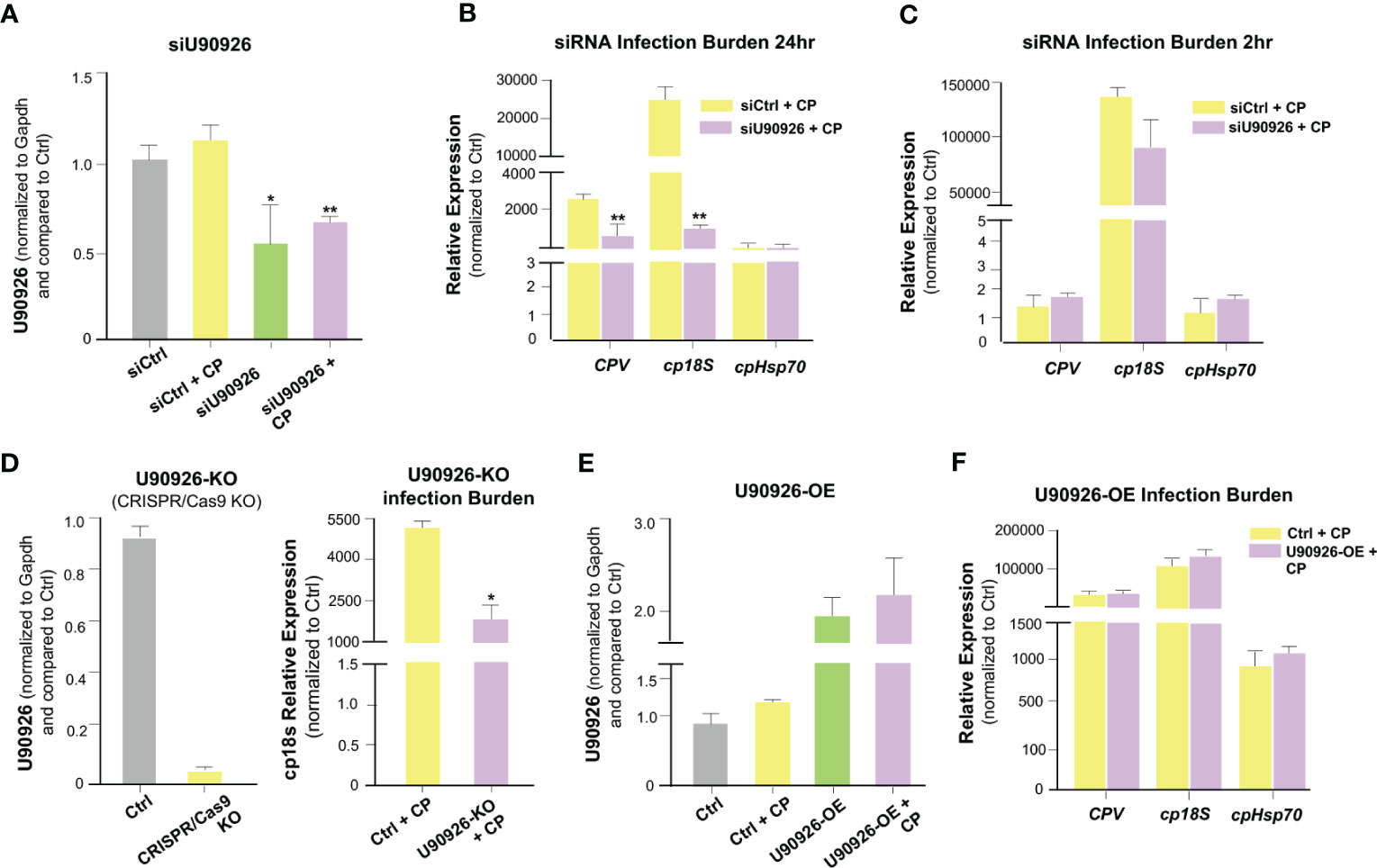
Figure 3 Impact of U90926 induction on parasite burden in intestinal epithelial cells. (A) Knockdown of U90926 with a designed siRNA in IEC4.1 cells. Cells were treated with siRNA to U90926 followed by infection with C. parvum (CP). U90926 expression was then measured via qRT-PCR and a nonspecific siRNA sequence was used as a control. (B, C) Impact of knockdown of U90926 on C. parvum infection burden. Cells were treated with knockdown U90926 siRNA and exposed to C. parvum infection for 24 hr (B) or 2 hr (C) followed by qRT-PCR to measure parasite infection burden. (D) U90926 CRISPR/Cas9 knockout cells (U90926-KO) were infected with C. parvum for 24 hrs and infection burden was measured via qRT-PCR. (E) IEC4.1 cells were transfected with a plasmid expressing U90926 RNA and infected with C. parvum. U90926 was measured via qRT-PCR. (F) Cells were transfected with a plasmid expressing U90926 RNA (U9926-OE) followed by infection with C. parvum. qRT-PCR was then used to measure infection burden. Data represent means ± SEM from three independent experiments. * p<0.05, **p<0.001 vs cells of non-infected control (Ctrl).
U90926 regulates the host’s gene expression profile in intestinal epithelial cells following C. parvum infection
Given its pro-parasitic role in intestinal epithelial cells, we sought to measure the effects of U90926 on the host’s gene expression profile in infected IEC4.1 cells. We used the siRNA interface to knock down U90926 during C. parvum infection and performed RNA-Seq analysis to compare the gene expression profiles in samples treated with an siRNA targeting U90926 (siU90926) and those treated with an siRNA control (siCtrl). A total of 1,338 genes were downregulated in the siU90926 samples following infection with C. parvum comparable to the 963 genes downregulated in the siCtrl-treated cells following infection (p<0.05). A total of 1,288 genes were significantly upregulated in the siU90926 treated samples following infection and 941 genes were significantly upregulated in the siCtrl samples infected (p<0.05). Among the 1,288 genes significantly upregulated, 576 were common in both the siU90926 samples and siCtrl samples, however, a subset of 712 genes were only upregulated in the siNR_033843 samples (Figure 4A). A full list of the upregulated and downregulated genes is provided in Table S2. Gene ontology analysis of these genes revealed a broad range of gene categories, distinct to each sample type. The most enriched molecular function pathways in the siU90926 treated samples infected with C. parvum compared to the infected siCtrl include DNA-binding transcription factor binding, transcription coregulator activity, and GTP binding (Figure 4B). Whereas in the siCtrl-treated samples infected with C. parvum, compared to siCtrl samples without infection, the most enriched pathways include ligase activity, double-stranded RNA binding, and ubiquitin-like protein ligase binding (Figure 4C). When we compared the expression levels of genes in samples treated with siCtrl, with or without C. parvum infection, we found that 260 genes were significantly altered, either downregulated (120) or upregulated (140) (adjusted p<0.05 with a fold change >1) (Figure 4D). Whereas when we compared infected samples treated with siU90926 and siCtrl samples with infection, there was a lower number of gene alterations, either downregulated (76) or upregulated (119) (adjusted p<0.05 with a fold change >1) (Figure 4E). The top 20 upregulated and downregulated genes are listed in Figure 4F. These top upregulated genes include Aebp1, Lox, and Slc27a and the top downregulated genes include Ccl5, Cyb5b, Kpna2, Mvd, and Phagalh (Figure 4F). Notably, the expression levels of a total of 207 genes were either upregulated (87) or downregulated (120) (adjusted p<0.05 with a fold change >1) between uninfected siU90926 treated cells and uninfected siCtrl treated cells (Figure S1), suggesting the impact of U90926 knockdown on the basal expression levels of those genes. All sequence data were described in accordance with MIAME guidelines and deposited in the ArrayExpress database (with the accession numbers: E-MTAB-12972). The expression levels of Aebp1, Lox, and Slc27a were all upregulated in IEC4.1 cells treated with U90926 siRNA followed by C. parvum infection for 24hrs compared to IEC4.1 cells treated with a control siRNA as further validated by qRT-PCR (Figure 4G). Additionally, a number of anti-defense genes were significantly downregulated in cells treated with siU90926 followed by C. parvum infection validated by qRT-PCR (Figure 4H).
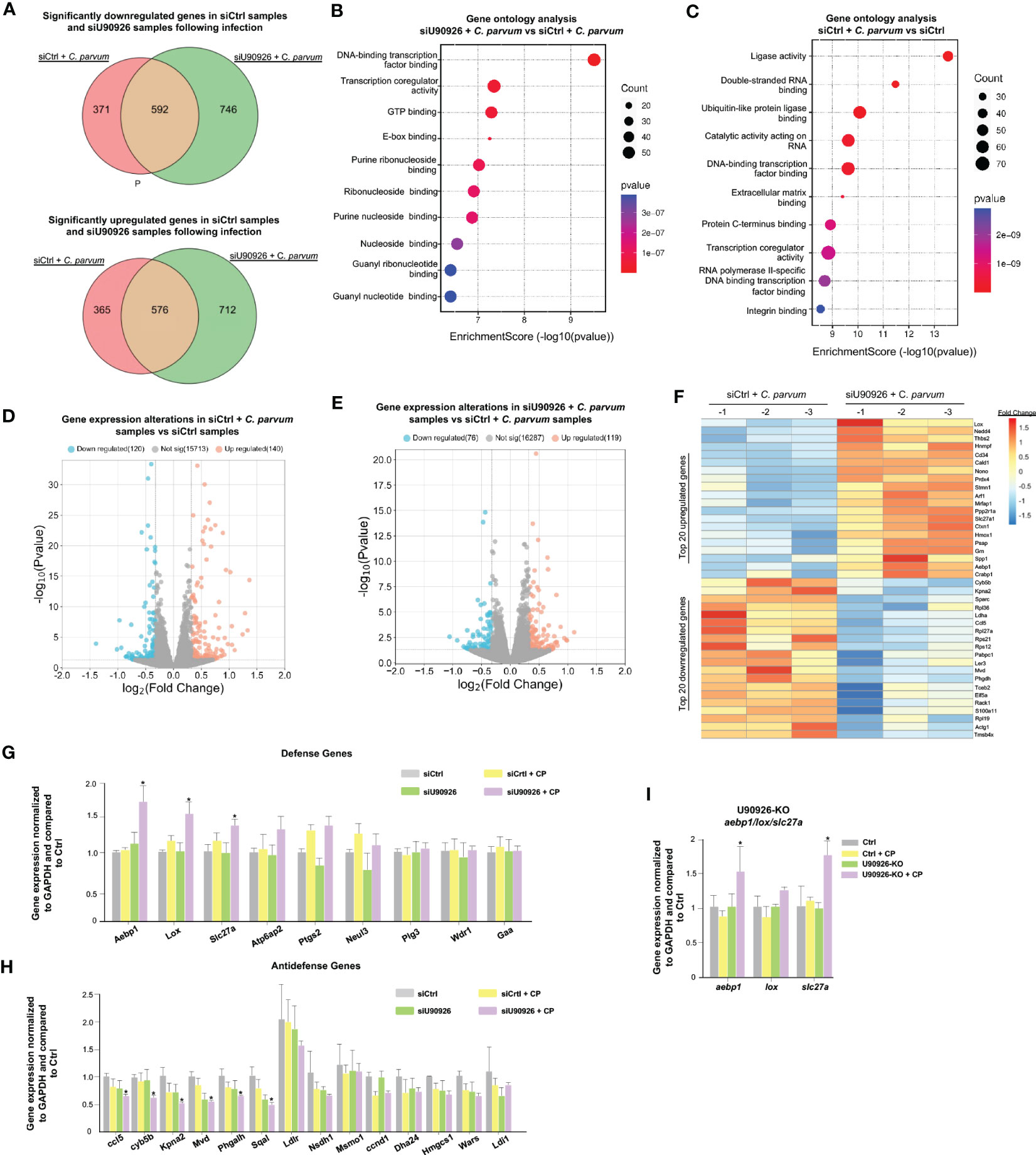
Figure 4 U90926 targets host defense gene expression during C. parvum infection. (A) Numbers of significantly downregulated and upregulated genes in siU90926 treated samples following C. parvum infection compared to siCtrl-treated samples following infection. (B) Gene ontology analysis of top 10 molecular functions in siU90926 treated samples following C. parvum infection compared to siCtrl-treated samples following infection. (C) Gene ontology analysis of top 10 molecular functions in siCtrl treated samples following C. parvum infection compared to siCtrl-treated samples. (D) Volcano plot of significantly altered genes in siCtrl-treated samples with infection compared to siCtrl-treated samples without infection (adjusted p<0.05 with a fold change >1). (E) Volcano plot of significantly altered genes in siU90926 treated samples with infection compared to siCtrl treated samples with infection (adjusted p<0.05 with a fold change >1). (F) Heatmap representing top 20 upregulated and top 20 downregulated genes in IEC cells treated with siRNA targeting U90926 followed by C. parvum infection. siRNA targeting U90926 was transfected into IEC cells and infected with C. parvum followed by genome-wide array analysis. The top 20 upregulated and top 20 downregulated genes are shown, presented as fold changes to the mean value of the log2 ratios in the noninfected control (n = 3). (G) Upregulation of host defense genes Aebp1, Lox, and Slc27a in IEC cells treated with siRNA targeting U90926 followed by C. parvum infection. siRNA targeting U90926 was transfected into IEC cells followed by C. parvum infection and expression levels of select defense genes were measured via qPCR and compared to a non-infected scrambled siRNA control. (H) Downregulation of host anti-defense genes in IEC cells treated with siRNA targeting U90926. A panel of anti-defense gene expression was measured via qRT-PCR and compared to non-infected samples transfected with a scrambled siRNA. (I) Upregulation of host defense genes Aebp1, Lox, and Slc27a in CRISPR/Cas9 U90926 knockout cells during C. parvum infection. Data represent means ± SEM from three independent experiments. *p<0.05 vs cells of scrambled siRNA control (Ctrl).
To further explore the relationship between U90926 and the host defense genes Aebp1, Slc27a, and Lox, U90926 CRISPR/Cas9 knockout cells were infected with C. parvum, and the expression levels of Aebp1, Slc27a1, and Lox were measured via qRT-PCR. The U90926-KO cells had an increase in Aebp1 and Slc27a expression, but no significant difference was observed in Lox expression (Figure 4I). Taken together, this finding supports that U90926 may play a pro-parasitic role during infection with C. parvum through the suppression of the host’s defense gene expression.
U90926 is recruited to the promoter regions of several host genes to regulate their transcription
We then sought to examine the impact NR_003483 might have on the transcriptional repression of Aebp1, Slc27a1, and Lox genes during C. parvum infection. More specifically, whether U90926 expression impacts the transcriptional active histone modifications associated with Aebp1, Slc27a, and Lox gene loci. By utilizing a ChIP assay, the histone modifications, such as H3K4m1 methylations, that are usually associated with gene transactivation can be examined (50). PCR primers were designed to cover the various regions of regulatory promoters of all three genes (Figure 5A). IEC4.1 cells infected with C. parvum had increased levels of H3K4m1 in the Aebp1 gene locus, however, this effect was greatly increased in U90926-KO IEC4.1 cells infected with C. parvum (Figure 5B). Similarly, U90926-KO cells had increased levels of H3K4m1 in the gene loci of Slc27a1 (Figure 5C) and Lox (Figure 5D). No change in H3K4m1 levels was observed in IEC4.1 cells at the Slc27a gene locus following C. parvum infection (Figure 5C) and there was a slight decrease in H3K4m1 levels at the Lox locus in IEC4.1 cells following C. parvum infection (Figure 5D).
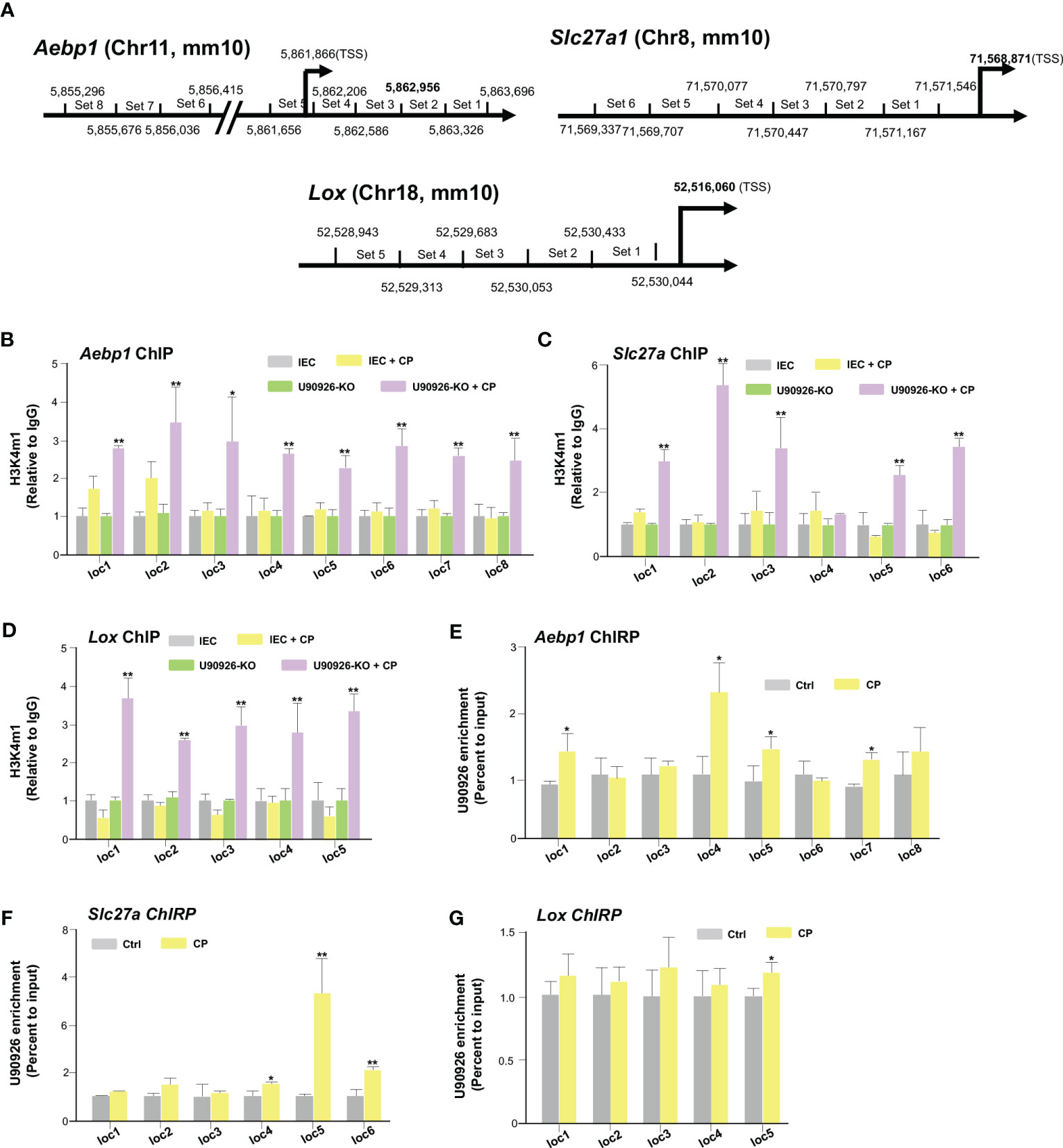
Figure 5 U90926 binds to the promoter regions of several host genes to regulate their transcription. (A) Diagram of primer sets of Aebp1, Slc27a, and Lox promoter regions. (B) Impact of U90926 on the enrichment of the activation marker H3K4m1 associated with Aebp1, (C) Slc27a, and (D) Lox gene loci in CRISPR/Cas9 U90926 knockout cells followed by Cryptosporidium infection for 24hrs. ChIP analysis was performed using the primer sets as designed. (E-G) U90926 recruitment to the Aebp1 (E), Slc27a (F), and (G) Lox gene loci. IEC4.1 cells were infected with Cryptosporidium for 24 hrs followed by ChIRP analysis using a pool of probes specific to U90926 and the PCR primer sets as designed. Data represent means ± SEM from three independent experiments. * p<0.05, **p<0.001 vs cells of non-infected control (Ctrl).
Furthermore, we examined how the transcription of Aebp1, Slc27a1, and Lox genes is selectively regulated by U90926 and whether this lncRNA is recruited directly to the gene loci. To analyze the occupancy of NR_003483 to the gene loci, ChIRP analysis was performed on IEC4.1 cells following C. parvum infection. A pool of biotinylated tiling oligonucleotide probes with an affinity for the U90926 sequence was used to precipitate the chromatin fragments through glutaraldehyde crossing-linking and chromatin isolation. The same PCR primers specific to various sequence regions of the Aebp1, Slc27a, and Lox gene loci for the above ChIP assay were used to identify the DNA sequence of the precipitated chromatin fragments (51). We detected an increased presence of U90926 in the gene loci of Aebp1 and Slc27a in IEC4.1 cells infected with C. parvum for 24 hrs (Figures 5E, F). There was also an increase in the occupancy of U90926 in the gene locus of Lox (Figure 5G). Taken together our data indicate that U90926 is recruited to the gene loci of Aebp1, Slc27a, and Lox and may play a role in the inhibition of their transcription.
Aebp1 is required for epithelial cell-autonomous anti-Cryptosporidium defense
Aebp1 is a ubiquitously expressed transcriptional repressor found in many different tissues and cells and is involved in several biological processes including adipogenesis, macrophage cholesterol homeostasis, and inflammation (52–54). Aebp1 has been shown to induce macrophage inflammatory responsiveness leading to enhanced expression of pro-inflammatory mediators (IL-6, TNF-α, MCP-1, and iNOS) via Aebp1’s potential role to promote NF-κB activity (54). To further explore the relationship between Aebp1 and C. parvum infection, we generated a CRISPR/Cas9 Aebp1 KO stable IEC4.1 cell line. The cells were verified as knockout cells via qRT-PCR with primers specific for Aebp1 (Figure S2A). These cells were infected with C. parvum for 2 or 24 hrs and the infection burden was quantified by measuring expression levels of C. parvum genes via qRT-PCR. Aebp1 KO cells (Aebp1KO 1-3 strains) showed no significant change in C. parvum infection burden after 2 hr of infection (a timepoint for C. parvum sporozoite attachment to and entry of host cells) however, after 24 hr infection, Aebp1 KO cells displayed significantly higher levels of C. parvum infection burden when compared to IEC4.1 cells infected with C. parvum (Figures 6A, B). Complementary, we generated CRISPR/Cas9 Aebp1 overexpression stable IEC4.1 cell line as were verified as overexpression cells via qRT-PCR with primers specific to Aebp1 (Figure S2B). These overexpression cells were infected with C. parvum for 2 or 24 hrs and infection burden was measured via qRT-PCR. In line with the knockout cell data, Aebp1 overexpressing cells (Aebp1OE 1 and 2 strains) showed a significant decrease in C. parvum infection burden after 24 hrs compared to the wild-type control. Interestingly, after a 2 hr infection, there was also a significant decrease in infection burden in Aebp1 overexpressing cells compared to the wild-type control cells (Figures 6C, D). These data highlight the protective role of Aebp1 during C. parvum infection.
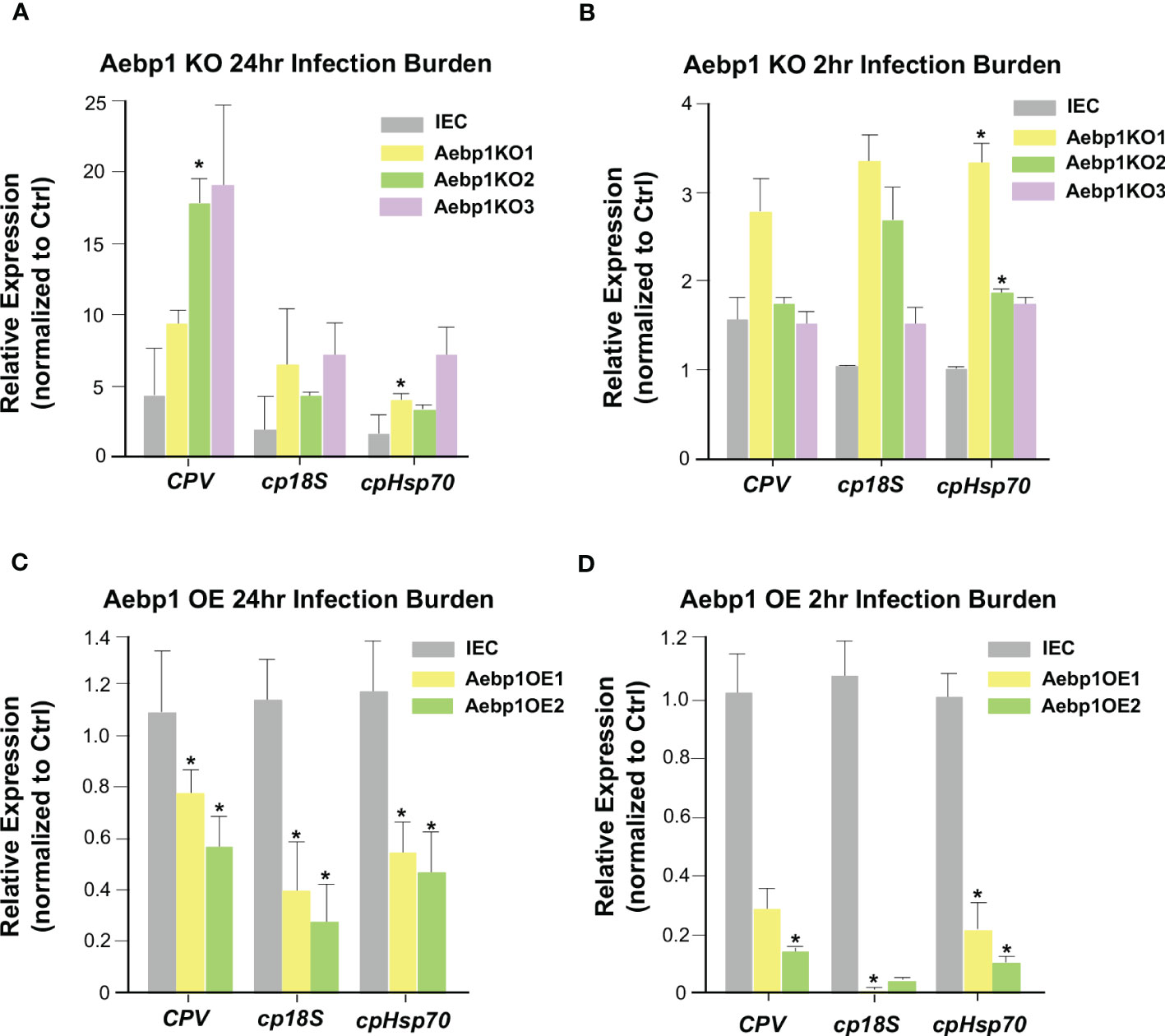
Figure 6 Aebp1 is required for epithelial cell-autonomous anti-Cryptosporidium defense. (A) Inhibition of Aebp1 results in increased Cryptosporidium infection. CRISPR/Cas9 Aebp1 knockout cells were generated (Aebp1KO1-3) and infected with C. parvum for 24 hrs and C. parvum infection burden was measured via qRT-PCR and compared to the IEC4.1 cell control. (B) CRISPR/Cas9 Aebp1 knockout cells showed no significant difference in C. parvum attachment/invasion compared to the IEC4.1 control. CRISPR/Cas9 Aebp1 knockout cells were infected with C. parvum for 2 hrs and C. parvum infection burden was measured via qRT-PCR and compared to the IEC4.1 control. (C) Aebp1 overexpression results in a decrease in C. parvum 24 hrs infection burden. CRISPR/Cas9 Aebp1 overexpression cells (Aebp1OE1-2) were infected with C. parvum for 24 hrs and C. parvum infection burden was measured via qPCR and compared to the IEC4.1 control. (D) Aebp1 overexpression results in a decrease in C. parvum 2 hrs infection burden. CRISPR/Cas9 Aebp1 overexpression cells were infected with C. parvum for 2 hrs and C. parvum infection burden was measured via qPCR and compared to the IEC4.1 control. Data represent means ± SEM from three independent experiments. * p<0.05 vs IEC control cells (IEC).
The CSpV1 of C. parvum induces the upregulation of U90926 during C. parvum infection
Proteins and RNAs of parasite origin can be selectively delivered into host cells following C. parvum infection (55, 56). In our previous studies, we identified a panel of RNA of parasite origin, as well as the dsRNAs of CSpv1, which are delivered into infected host cells (56, 57). CSpV1 is a dsRNA virus identified in Cryptosporidium oocysts from human stool (58, 59). CSpV1 contains two dsRNA segments (CSpV1-dsRaRp and CSpV1-dsCA) that are encapsulated separately and encode for two proteins: the RNA-dependent RNA polymerase (RdRp) and the capsid protein (CA), respectively. We reasoned that the delivery of RNAs of parasite origin is involved in C. parvum-induced U90926 expression. We measured U90926 expression levels in IEC4.1 cells transfected with plasmids expressing parasite RNAs that we previously demonstrated to be delivered into infected host cells (56). No obvious induction of U90926 expression was detected in cells transfected with the parasite RNAs tested (Figure S3). We then transfected IEC4.1 cells with in vitro transcribed CSpV1-dsCA or CSpV1-dsRdRp and examined U90926 expression levels via qRT-PCR. The expression levels of CSpV1-dsCA or CSpV1-dsRdRp were confirmed via qRT-PCR (Figure S4). Samples were compared to a scrambled siRNA control. The cells transfected with CSpV1-dsRdRp or CSpv1-dsCA had significantly higher expression levels of NR_003483 than control samples (Figure 7A). A pool of siRNAs targeting CSpV1-dsRdRp and CSpV1-dsCA was designed and transfected into IEC4.1 cells followed by infection with Cryptosporidium. There was no increase in U90926 in the IEC4.1 cells transfected with siRNA pool targeting CSpV1-dsRdRp and CSpV1-dsCA regardless of Cryptosporidium infection (Figure 7B), further supporting that CSpV1 is involved in the upregulation of U90926 expression during Cryptosporidium infection.
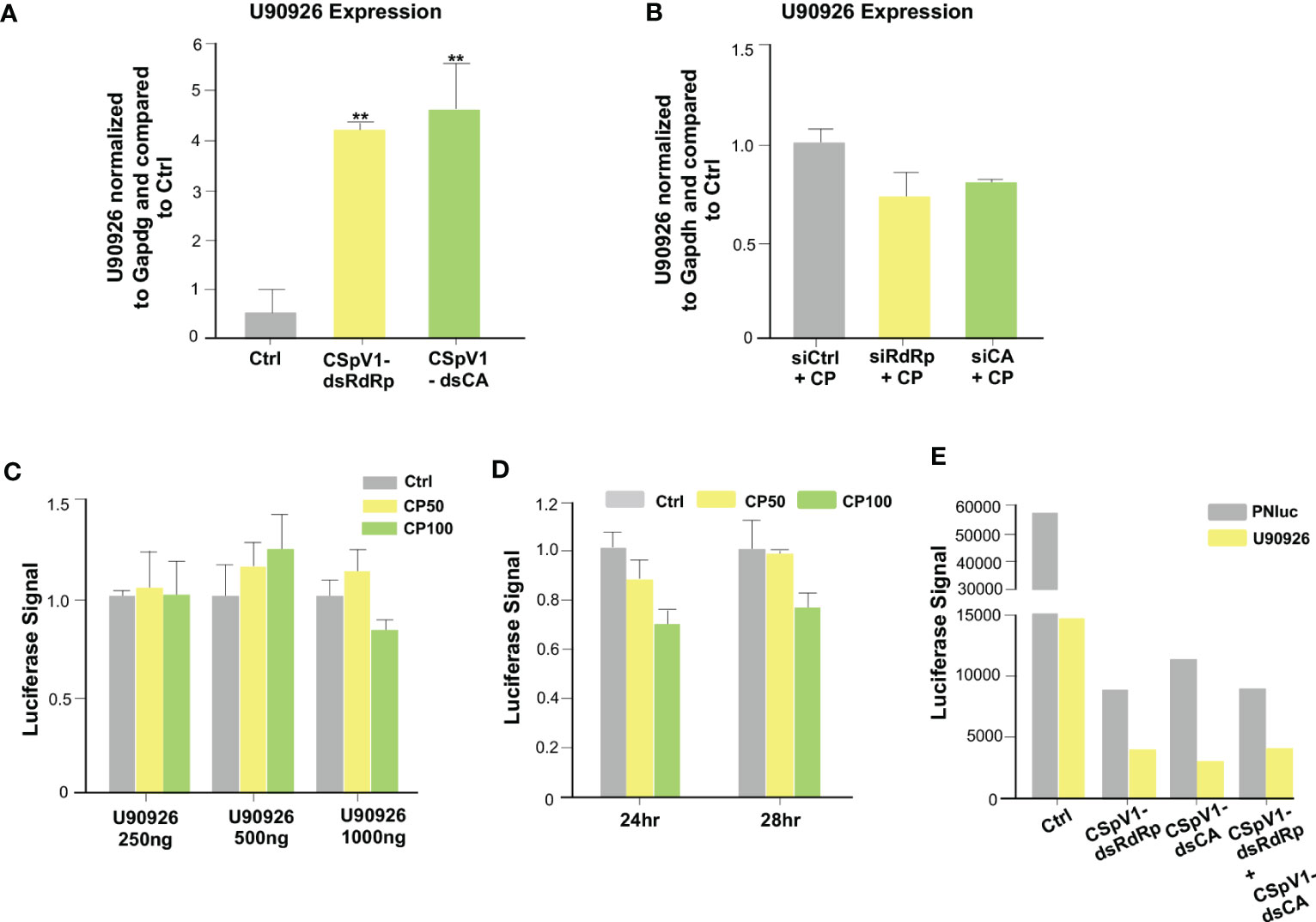
Figure 7 The CSpV1 of C. parvum induces the upregulation of U90926 during C. parvum infection. (A) Transfection of CSpV1-dsRdRp or CSpV1-dsCA increases U90926 expression. CSpV1-RdRp or CSpV1-CP dsRNA was transfected into IEC4.1 cells and U90926 expression was measured via qRT-PCR and compared to a scrambled dsRNA control. (B) Transfection of siRNAs to CSpV1-dsRdRp and CSpV1-dsCA suppresses U90926 upregulation during C. parvum infection. IEC4.1 cells were transfected with a pool of siRNAs targeting either CSpV1-RdRp or CSpV1-CA followed by infection with C. parvum and U90926 expression was measured via qRT-PCR and compared to a scrambled siRNA control. (C) Infection with C. parvum does not result in an increase in the U90926 luciferase construct signal. A luciferase construct carrying the promoter region of U90926 was transfected into IEC4.1 cells (250 ng, 500 ng, or 1000 ng) followed by C. parvum infection. The luciferase signal was measured and compared to a noninfected control. (D) No significant change in luciferase signal at 24 hrs or 28 hrs post-infection. The U90926 luciferase construct was transfected into IEC4.1 cells followed by C. parvum infection for 24 hrs or 28 hrs. The luciferase signal was measured and compared to a noninfected control. (E) Transfection of CSpV1-dsRdRp and/or CSpV1-dsCA does not increase the U90926 luciferase signal. The U90926 luciferase construct, CSpV1-dsRdRp, and/or CSpV1-dsCA were transfected into IEC cells and the luciferase signal was measured and compared to a scrambled siRNA control. Data represent means ± SEM from three independent experiments. **p<0.01 vs control cells (Ctrl).
To uncover the mechanism of CSpV1-induced upregulation of host lncRNA U90926, we generated a luciferase construct carrying the putative promoter region (1 kb upstream of start) of U90926. IEC cells were transfected with the U90926 luciferase construct at different concentrations and infected with Cryptosporidium (at two separate doses: 50 μl/well or 100 μl/well) and the luciferase signal was measured. Interestingly, there was no significant change in the luciferase signal regardless of the concentration of U90926 luciferase construct transfected into cells or Cryptosporidium concentration (Figure 7C). Additionally, there was no change in luciferase signal at 24 hr or 28 hr transfection infected with Cryptosporidium (Figure 7D). We also measured the luciferase signal when IEC4.1 cells were transfected with the U90926 luciferase construct and plasmids expressing CSpV1-dsRdRp or CSpV1-dsCA. We found no increase in the luciferase signal in the samples transfected with U90926 construct and plasmids expressing CSpV1-dsRdRp or CSpV1-dsCA when compared to control cells transfected with an empty vector (Figure 7E). Taken together, the data suggest that the upregulation of U90926 during Cryptosporidium infection seems to involve CSpV1, however, this may not be due to a transcriptional change in the U90926 promoter.
Discussion
Cell-autonomous immunity, defined as the ability of a host cell to eliminate an invasive/intracellular infectious agent at the cellular level, is a first line of defense against intracellular microbial pathogens (60). It relies on antimicrobial proteins, specialized degradative compartments, and programmed host cell death (60–63) and is mediated by tiered innate immune signaling networks that sense microbial pathogens and stimulate downstream pathogen elimination programming (61). Upon infection, Cryptosporidium sporozoites attach to the apical membrane surface of epithelial cells and form intracellular but extracytoplasmic vacuoles in which the organism remains and develops (6, 11). Intestinal epithelial cell-autonomous immunity is the host’s first defense against C. parvum infection.
In a previous study, the lab identified a panel of host cell lncRNAs that are upregulated in intestinal epithelial cells following infection with C. parvum (25). Among these host lncRNAs, U90926 was upregulated during infection in IEC4.1 cells, neonatal mice, and 2D intestinal epithelial monolayers. Interestingly, unlike other host lncRNAs identified in the panel whose expression is controlled by host immune signaling, such as the TLR/NF-κB signaling pathway (25, 64, 65), U90926 does not appear to be upregulated in IEC4.1 cells in response to treatment with IFN-γ, IFN-α, LPS, or anisomycin. Furthermore, the inhibition of U90926 via RNA interference in host intestinal epithelial cells resulted in a decreased infection burden of C. parvum and the overexpression of U90926 led to an increase in infection burden. This indicates that U90926 upregulation may be beneficial for the parasite during infection of intestinal epithelial cells through suppression of the host’s epithelial cell-autonomous defense mechanisms.
Our data show that U90926 targets several host genes during C. parvum infection, including Aebp1, Slc27a, and Lox. Inhibition of U90926 expression during C. parvum infection resulted in increased levels of all three genes, suggesting that U90926 is downregulating their expression during infection. lncRNAs function to regulate gene transcription through specific interactions with cellular factors including proteins, DNA, and other RNA molecules (21, 22). In the case of U90926, we found this lncRNA is recruited to the gene loci in the promoter regions of these genes and alters transcriptional active histone modifications.
The Aebp1 gene encodes for adipocyte enhancer-binding protein-1, a potent modulator of NF-κB signaling and inflammation (66, 67) whereas Lox is a cell surface endocytosis receptor for ox-LDL and recognizes pathogenic microorganisms (68). It is a proinflammatory scavenger protein and is structurally similar to a c-type lectin-like receptor (69, 70). Scl27a is an integral protein involved in the uptake of long-fatty acids into the cell. It has been reported to be important in intracellular signaling and the metabolism of innate immune cells (71–74). Among the three genes, Aebp1 showed the most drastic difference in expression in the U90926 knockout experiments. It has been shown to induce macrophage inflammatory responsiveness and play a key role in enhancing the expression of pro-inflammatory mediators by promoting NF-κB activity (52–54). In this study, we demonstrated that during C. parvum infection Aebp1 knockout cells showed a significantly higher C. parvum infection burden compared to control cells. This was complemented by Aebp1 overexpression cells exhibiting decreased C. parvum infection burden. Together, our data suggest that U90926 is induced during C. parvum infection and suppresses transcription of the Aebp1 gene. This, in turn, inhibits Aebp1’s cell-autonomous defense role in the intestinal epithelial cells and allows for a robust parasitic infection.
It is interesting that the small dsRNA virus, CSpV1, may be responsible for its upregulation. CSpV1 is a bisegmented dsRNA virus that is found in many Cryptosporidium spp. The two genome segments are packaged in separate virions and encode for an RNA-dependent RNA polymerase and a capsid protein (36, 58). CSpV1 seems to be transmitted vertically during the cell division of C. parvum cells as well as during gamete fusion of the C. parvum replication cycle. No horizontal extracellular transmission has been reported (37). This virus is thought to be associated with avirulent infections of its host and some authors have described C. parvum as having a “viral symbiont” (38). There have been few studies looking at CSpV1 and virulence with one study showing a correlation between levels of CSpV1 dsRNA and parasite fecundity (59). However, the cell biology of CSpV1 infection and the host factors that are involved in modulation, inhibition, or are affected by the virus are unknown. We previously demonstrated that several parasite RNAs and CSpV1-dsRNAs are delivered into infected host cells following Cryptosporidium infection (56, 57). Our current data indicate that the transfection of CSpV1-dsRdRp and CSpV1-dsCA into IEC4.1 cells leads to an increase in U90926 expression. This effect was inhibited when cells were treated with an siRNA pool targeting CSpV1-dsRdRp and CSpV1-dsCA. We speculate that CSpV1-dsRNAs are upregulating U90926 to target the host genes, including Aebp1, and inhibit the immune response to C. parvum infection.
In this study, we uncovered a novel strategy by C. parvum to evade the host’s immune response by hijacking host lncRNAs to suppress epithelial cell-autonomous antiparasitic defense. Our data also suggest a new role for Aebp1 in intestinal epithelial cell-autonomous defense. Further investigations are required to fully understand how CSpV1-dsRNAs induce U90926 expression and how Aebp1 may carry out its antiparasitic function. It will also be interesting to investigate whether U90926 plays a role during intestinal infection with other microbes.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below:E-MTAB-12972 (ArrayExpress; https://www.ebi.ac.uk/biostudies/arrayexpress).
Ethics statement
The animal study was reviewed and approved by The Institutional Animal Care and Use Committees (IACUC) of the Rush University Medical Center.
Author contributions
MG, ML, and X-MC designed experiments and wrote the manuscript. MG, ML, SD, KJ, and A-YG performed experiments. MG, ML, SW, and X-MC performed data analysis. A-YG and X-MC directed and supervised the study. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by funding from the National Institutes of Health (AI116323, AI136877, AI141325, and AI156370 to X-MC).
Acknowledgments
We thank Dr. Pingchang Yang (McMaster University, Hamilton, Canada) for sharing IEC4.1 cells and Dr. Lena Al-Harthi, Dr. Edward Barker, Dr. David L Williams, Dr. Jeffery Schneider (Department of Microbial Pathogens and Immunity, Rush University Medical Center) for their helpful suggestions and comments on the study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Author disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1205468/full#supplementary-material
References
1. O’Hara SP, Huang BQ, Chen X-M, Nelson J, LaRusso NF. Distribution of Cryptosporidium parvum sporozoite apical organelles during attachment to and internalization by cultured biliary epithelial cells. J Parasitol (2005) 91(5):995–9. doi: 10.1645/GE-495R.1
2. Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the global enteric multicenter study, GEMS): a prospective, case-control study. Lancet (2013) 382(9888):209–22. doi: 10.1016/S0140-6736(13)60844-2
3. Khalil IA, Troeger C, Rao PC, Blacker BF, Brown A, Brewer TG, et al. Morbidity, mortality, and long-term consequences associated with diarrhoea from Cryptosporidium infection in children younger than 5 years: a meta-analyses study. Lancet Global Health (2018) 6(7):e758–68. doi: 10.1016/S2214-109X(18)30283-3
4. Putignani L, Menichella D. Global distribution, public health and clinical impact of the protozoan pathogen Cryptosporidium. Interdiscip Perspect Infect Dis (2010) 2010:753512. doi: 10.1155/2010/753512
5. Pierce K, Kirkpatrick B. Update on human infections caused by intestinal protozoa. Curr Opin Gastroenterol (2009) 25(1):12–7. doi: 10.1097/mog.0b013e32831da7dd
6. Striepen B. Parasitic infections: time to tackle cryptosporidiosis. Nature (2013) 503:189–91. doi: 10.1038/503189a
7. Checkley W, White AC, Jaganath D, Arrowood MJ, Chalmers RM, Chen X-M, et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect Dis (2015) 15(1):85–94. doi: 10.1016/S1473-3099(14)70772-8
8. Widmer G, Carmena D, Kváč M, Chalmers RM, Kissinger JC, Xiao L, et al. Update on Cryptosporidium spp.: highlights from the seventh international Giardia and Cryptosporidium conference. Parasite (2020) 27:14. doi: 10.1051/parasite/2020011
9. O'Leary JK, Sleator RD, Lucey B. Cryptosporidium spp. diagnosis and research in the 21st century. Food Waterborne Parasitol (2021) 24:e00131. doi: 10.1016/j.fawpar.2021.e00131
10. Efstratiou A, Ongerth JE, Karanis P. Waterborne transmission of protozoan parasites: review of worldwide outbreaks - an update 2011-2016. Water Res (2017) 114:14–22. doi: 10.1016/j.watres.2017.01.036
11. Chen X-M, Keithly JS, Paya CV, LaRusso NF. Cryptosporidoisis. N Engl J Med (2002) 346(22):1723–31. doi: 10.1056/NEJMra013170
12. McDonald V, Korbel DS, Barakat FM, Choudhry N, Petry F. Innate immune responses against Cryptosporidium parvum infection. Parasite Immunol (2013) 35(2):55–64. doi: 10.1111/pim.12020
13. Ming Z, Zhou R, Chen X-M. Regulation of host epithelial responses to Cryptosporidium infection by microRNAs. Parasite Immunol (2017) 39(2):e12408. doi: 10.1111/pim.12408
14. Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol (2014) 14(3):141–53. doi: 10.1038/nri3608
15. Heidemann J, Domschke W, Kucharzik T, Maaser C. Intestinal microvascular endothelium and innate immunity in inflammatory bowel disease: a second line of defense? Infect Immun (2006) 74(10):5425–32. doi: 10.1128/IAI.00248-06
16. Laurent F, Kagnoff MF, Savidge TC, Naciri M, Eckmann L. Human intestinal epithelial cells respond to Cryptosporidium parvum infection with increased prostaglandin h synthase 2 expression and prostaglandin E2 and F2α production. Infect Immun (1998) 66(4):1787–90. doi: 10.1128/IAI.66.4.1787-1790.1998
17. Zhou R, Gong A-Y, Eischeid AN, Chen X-M. miR-27b targets KSRP to coordinate TLR4-mediated epithelial defense against Cryptosporidium parvum infection. PloS Pathog (2012) 8(5):e1002702. doi: 10.1371/journal.ppat.1002702
18. Walker DM, Oghumu S, Gupta G, McGwire BS, Drew ME, Satoskar AR. Mechanisms of cellular invasion by intracellular parasites. Cell Mol Life Sci (2014) 71(7):1245–63. doi: 10.1007/s00018-013-1491-1
19. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet (2009) 10(3):155–9. doi: 10.1038/nrg2521
20. Dinger ME, Amaral PP, Mercer TR, Mattick JS. Pervasive transcription of the eukaryotic genome: functional indices and conceptual implications. Brief Funct Genomics Proteomics (2009) 8(6):407–23. doi: 10.1093/bfgp/elp038
21. Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature (2009) 458(7235):223–7. doi: 10.1038/nature07672
22. Mathy NW, Chen X-M. Long non-coding RNAs (lncRNAs) and their transcriptional control of inflammatory responses. J Biol Chem (2017) 292(30):12375–82. doi: 10.1074/jbc.R116.760884
23. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem (2012) 81:145–66. doi: 10.1146/annurev-biochem-051410-092902
24. Peng X, Gralinski L, Armour CD, Ferris MT, Thomas MJ, Proll S, et al. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. mBio (2010) 1(5):e00206–10. doi: 10.1128/mBio.00206-10
25. Li M, Gong A-Y, Zhang X-T, Wang Y, Mathy NW, Martins GA, et al. Induction of a long noncoding RNA transcript, NR_045064, promotes defense gene transcription and facilitates intestinal epithelial cell responses against Cryptosporidium infection. J Immunol (2018) 201(12):3630–40. doi: 10.4049/jimmunol.1800566
26. Elling R, Chan J, Fitzgerald KA. Emerging role of long noncoding RNAs as regulators of innate immune cell development and inflammatory gene expression. Eur J Immunol (2016) 46(3):504–12. doi: 10.1002/eji.201444558
27. Atianand MK, Fitzgerald KA. Long non-coding RNAs and control of gene expression in the immune system. Trends Mol Med (2014) 20(11):623–31. doi: 10.1016/j.molmed.2014.09.002
28. Tong Q, Gong A-Y, Zhang X-T, Lin C, Ma S, Chen J, et al. LincRNA-Cox2 modulates TNF-α-induced transcription of Il12b gene in intestinal epithelial cells through regulation of mi-2/NuRD-mediated epigenetic histone modifications. FASEB J (2016) 30(3):1187–97. doi: 10.1096/fj.15-279166
29. Hu G, Gong A-Y, Wang Y, Ma S, Chen X, Chen J, et al. LincRNA-Cox2 promotes late inflammatory gene transcription in macrophages through modulating SWI/SNF-mediated chromatin remodeling. J Immunol (2016) 196(6):2799–808. doi: 10.4049/jimmunol.1502146
30. Luo F, Wen Y, Zhao L, Su S, Zhao Y, Lei W, et al. Chlamydia trachomatis induces lncRNA MIAT upregulation to regulate mitochondria-mediated host cell apoptosis and chlamydial development. J Cell Mol Med (2022) 26(1):163–77. doi: 10.1111/jcmm.17069
31. Liu W, Huang L, Wei Q, Zhang Y, Zhang S, Zhang W, et al. Microarray analysis of long non-coding RNA expression profiles uncovers a Toxoplasma-induced negative regulation of host immune signaling. Parasit Vectors (2018) 11(1):174. doi: 10.1186/s13071-018-2697-8
32. Sabikunnahar B, Caldwell S, Varnum S, Hogan T, Cooper A, Lahue KG, et al. Long noncoding RNA U90926 is induced in activated macrophages, is protective in endotoxic shock, and encodes a novel secreted protein. J Immunol (2023) 210(6):807–19. doi: 10.4049/jimmunol.2200215
33. Khramtsov NV, Woods KM, Nesterenko MV, Dykstra CC, Upton SJ. Virus-like, double-stranded RNAs in the parasitic protozoan Cryptosporidium parvum. Mol Microbiol (1997) 26(2):289–300. doi: 10.1046/j.1365-2958.1997.5721933.x
34. Ming Z, Gong A-Y, Wang Y, Zhang X-T, Li M, Mathy NW, et al. Involvement of Cryptosporidium parvum Cdg7_FLc_1000 RNA in the attenuation of intestinal epithelial cell migration via trans-suppression of host cell SMPD3. J Infect Dis (2017) 217(1):122–33. doi: 10.1093/infdis/jix392
35. Lacroix S, Mancassola R, Naciri M, Laurent F. Cryptosporidium parvum-specific mucosal immune response in C57BL/6 neonatal and gamma interferon-deficient mice: role of tumor necrosis factor alpha in protection. Infect Immun (2001) 69(2):1635–42. doi: 10.1128/IAI.69.3.1635-1642.2001
36. Nibert ML, Woods KM, Upton SJ, Ghabrial SA. Cryspovirus: a new genus of protozoan viruses in the family partitiviridae. Arch Virol (2009) 154(12):1959–65. doi: 10.1007/s00705-009-0513-7
37. Khramtsov NV, Upton SJ. Association of RNA polymerase complexes of the parasitic protozoan Cryptosporidium parvum with virus-like particles: heterogeneous system. J Virol (2000) 74(13):5788–95. doi: 10.1128/jvi.74.13.5788-5795.2000
38. Kniel KE, Higgins JA, Trout JM, Fayer R, Jenkins MC. Characterization and potential use of a Cryptosporidium parvum virus (CPV) antigen for detecting c. parvum oocysts. J Microbiol Methods (2004) 58:189–95. doi: 10.1016/j.mimet.2004.03.007
39. Wilke G, Funkhouser-Jones LJ, Wang Y, Ravindran S, Wang Q, Beatty WL, et al. A stem-cell-derived platform enables complete Cryptosporidium development in vitro and genetic tractability. Cell Host Microbe (2019) 26(1):123–134.e8. doi: 10.1016/j.chom.2019.05.007
40. Pantenburg B, Dann SM, Wang HC, Robinson P, Castellanos-Gonzalez A, Lewis DE, et al. Intestinal immune response to human Cryptosporidium sp. infection. Infect Immun (2008) 76(1):23–9. doi: 10.1128/IAI.00960-07
41. Deng M, Lancto CA, Abrahamsen MS. Cryptosporidium parvum regulation of human epithelial cell gene expression. Int J Parasitol (2004) 34(1):73–82. doi: 10.1016/j.ijpara.2003.10.001
42. Foulke-Abel J, In J, Kovbasnjuk O, Zachos NC, Ettayebi K, Blutt SE, et al. Human enteroids as an ex-vivo model of host–pathogen interactions in the gastrointestinal tract. Exp Biol Med (Maywood) (2014) 239(9):1124–34. doi: 10.1177/1535370214529398
43. Lantier L, Lacroix-Lamandé S, Potiron L, Metton C, Drouet F, Guesdon W, et al. Intestinal CD103+ dendritic cells are key players in the innate immune control of Cryptosporidium parvum infection in neonatal mice. PloS Pathog (2013) 9(12):e1003801. doi: 10.1371/journal.ppat.1003801
44. Price AE, Shamardani K, Lugo KA, Deguine J, Roberts AW, Lee BL, et al. A map of toll-like receptor expression in the intestinal epithelium reveals distinct spatial, cell type-specific, and temporal patterns. Immunity (2018) 49(3):560–575.e6. doi: 10.1016/j.immuni.2018.07.016
45. Kapel N, Benhamou Y, Buraud M, Magne D, Opolon P, Gobert J-G, et al. Kinetics of mucosal ileal gamma-interferon response during cryptosporidiosis in immunocompetent neonatal mice. Parasitol Res (1996) 82(8):664–7. doi: 10.1007/s004360050182
46. Zhang X-T, Gong A-Y, Wang Y, Chen X, Lim S-YS, Dolata CE, et al. Cryptosporidium parvum infection attenuates the ex vivo propagation of murine intestinal enteroids. Physiol Rep (2016) 4(24):e13060. doi: 10.14814/phy2.13060
47. Zhao G-H, Gong A-Y, Wang Y, Zhang X-T, Li M, Mathy NW, et al. Nuclear delivery of parasite Cdg2_FLc_0220 RNA transcript to epithelial cells during Cryptosporidium parvum infection modulates host gene transcription. Vet Parasitol (2018) 251:27–33. doi: 10.1016/j.vetpar.2017.12.015
48. Iordanov MS, Pribnow D, Magun JL, Dinh TH, Pearson JA, Chen SL, et al. Ribotoxic stress response: activation of the stress-activated protein kinase JNK1 by inhibitors of the peptidyl transferase reaction and by sequence-specific RNA damage to the alpha-sarcin/ricin loop in the 28S rRNA. Mol Cell Biol (1997) 17(6):3373–81. doi: 10.1128/MCB.17.6.3373
49. Chen XM, LaRusso NF. Mechanisms of attachment and internalization of Cryptosporidium parvum to biliary and intestinal epithelial cells. Gastroenterology (2000) 118(2):368–79. doi: 10.1016/s0016-5085(00)70219-8
50. Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J (2008) 27(2):406–20. doi: 10.1038/sj.emboj.7601967
51. Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell (2011) 44:667–78. doi: 10.1016/j.molcel.2011.08.027
52. Park JG, Muise A, He GP, Kim SW, Ro HS. Transcriptional regulation by the gamma5 subunit of a heterotrimeric G protein during adipogenesis. EMBO J (1999) 18(14):4004–12. doi: 10.1093/emboj/18.14.4004
53. Zhang L, Reidy SP, Nicholson TE, Lee H-J, Majdalawieh A, Webber C, et al. The role of AEBP1 in sex-specific diet-induced obesity. Mol Med (2005) 11(1-12):39–47. doi: 10.2119/2005-00021.Ro
54. Majdalawieh A, Zhang L, Fuki IV, Rader DJ, Ro H-S. Adipocyte enhancer-binding protein 1 is a potential novel atherogenic factor involved in macrophage cholesterol homeostasis and inflammation. Proc Natl Acad Sci USA (2006) 103(7):2346–51. doi: 10.1073/pnas.0508139103
55. Liao Q, Shen J, Liu J, Sun X, Zhao G, Chang Y, et al. Genome-wide identification and functional annotation of Plasmodium falciparum long noncoding RNAs from RNA-seq data. Parasitol Res (2014) 113(4):1269–81. doi: 10.1007/s00436-014-3765-4
56. Wang Y, Gong A-Y, Ma S, Chen X, Li Y, Su C-J, et al. Delivery of parasite RNA transcripts into infected epithelial cells during Cryptosporidium infection and its potential impact on host gene transcription. J Infect Dis (2017) 215(4):636–43. doi: 10.1093/infdis/jiw607
57. Deng S, He W, Gong AY, Li M, Wang Y, Xia Z, et al. Cryptosporidium uses CSpV1 to activate host type I interferon and attenuate antiparasitic defenses. Nat Commun (2023) 14(1):1456. doi: 10.1038/s41467-023-37129-0
58. Gallimore CI, Green J, Casemore DP, Brown DWG. Detection of a picobirnavirus associated with Cryptosporidium positive stools from humans. Arch Virol (1995) 140(7):1275–8. doi: 10.1007/BF01322752
59. Jenkins MC, Higgins J, Abrahante JE, Kniel KE, O’Brien C, Trout J, et al. Fecundity of cryptosporidium parvum is correlated with intracellular levels of the viral symbiont CPV. Int J Parasitol (2008) 38(8-9):1051–5. doi: 10.1016/j.ijpara.2007.11.005
60. Randow F, MacMicking JD, James LC. Cellular self-defense: how cell-autonomous immunity protects against pathogens. Science (2013) 340(6133):701–6. doi: 10.1126/science.1233028
61. Mostowy S, Shenoy AR. The cytoskeleton in cell-autonomous immunity: structural determinants of host defence. Nat Rev Immunol (2015) 15(9):559–73. doi: 10.1038/nri3877
62. Sridharan H, Upton JW. Programmed necrosis in microbial pathogenesis. Trends Microbiol (2014) 22(4):199–207. doi: 10.1016/j.tim.2014.01.005
63. Pasparakis M, Vandenabeele P. Necroptosis and its role in inflammation. Nature (2015) 517(7534):311–20. doi: 10.1038/nature14191
64. Deng S, Gong A-Y, Wang Y, Zhang X-T, Li M, Li J, et al. Induction of a long non-coding RNA, lncRNA-Chr1:1226, by Cryptosporidium infection primes intestinal epithelial cells for IFN-γ-mediated host antimicrobial gene transcription. J Immunol (2019) 202:190.12–2. doi: 10.4049/jimmunol.202.Supp.190.12
65. Lu Y, Liu X, Xie M, Liu M, Ye M, Li M, et al. The NF-κB–responsive long noncoding RNA FIRRE regulates posttranscriptional regulation of inflammatory gene expression through interacting with hnRNPU. J Immunol (2017) 199(10):3571–82. doi: 10.4049/jimmunol.1700091
66. Majdalawieh A, Zhang L, Ro H-S. Adipocyte enhancer-binding protein-1 promotes macrophage inflammatory responsiveness by up-regulating NF-κB via IκBα negative regulation. Mol Biol Cell (2007) 18(3):930–42. doi: 10.1091/mbc.e06-03-0217
67. Holloway RW, Bogachev O, Bharadwaj AG, McCluskey GD, Majdalawieh AF, Zhang L, et al. Stromal adipocyte enhancer-binding protein (AEBP1) promotes mammary epithelial cell hyperplasia via proinflammatory and hedgehog signaling. J Biol Chem (2012) 287(46):39171–81. doi: 10.1074/jbc.M112.404293
68. Li DY, Chen HJ, Staples ED, Ozaki K, Annex B, Singh BK, et al. Oxidized low-density lipoprotein receptor LOX-1 and apoptosis in human atherosclerotic lesions. J Cardiovasc Pharmacol Ther (2002) 7(3):147–53. doi: 10.1177/107424840200700304
69. Chen X-P, Zhang T-T, Du G-H. Lectin-like oxidized low-density lipoprotein receptor-1, a new promising target for the therapy of atherosclerosis? Cardiovasc Drug Rev (2007) 25(2):146–61. doi: 10.1111/j.1527-3466.2007.00009.x
70. Nakagawa T, Akagi M, Hoshikawa H, Chen M, Yasuda T, Mukai S, et al. Lectin-like oxidized low-density lipoprotein receptor 1 mediates leukocyte infiltration and articular cartilage destruction in rat zymosan-induced arthritis. Arthritis Rheum (2002) 46(9):2486–94. doi: 10.1002/art.10504
71. Ordovás L, Roy R, Zaragoza P, Rodellar C. Structural and functional characterization of the bovine solute carrier family 27 member 1 (SLC27A1) gene. Cytogenet Genome Res (2006) 115(2):115–22. doi: 10.1159/000095230
72. Barakat FM, McDonald V, Di Santo JP, Korbel DS. Roles for NK cells and an NK cell-independent source of intestinal gamma interferon for innate immunity to Cryptosporidium parvum infection. Infect Immun (2009) 77(11):5044–9. doi: 10.1128/IAI.00377-09
73. Zhou R, Hu G, Liu J, Gong A-Y, Drescher KM, Chen X-M. NF-kappaB p65-dependent transactivation of miRNA genes following Cryptosporidium parvum infection stimulates epithelial cell immune responses. PloS Pathog (2009) 5(12):e1000681. doi: 10.1371/journal.ppat.1000681
Keywords: lncRNA, Cryptosporidium, intestinal epithelium, defense, Aebp1, CSpV1, U90926
Citation: Graham ML, Li M, Gong A-Y, Deng S, Jin K, Wang S and Chen X-M (2023) Cryptosporidium parvum hijacks a host’s long noncoding RNA U90926 to evade intestinal epithelial cell-autonomous antiparasitic defense. Front. Immunol. 14:1205468. doi: 10.3389/fimmu.2023.1205468
Received: 13 April 2023; Accepted: 16 May 2023;
Published: 05 June 2023.
Edited by:
Silvia Amaral Gonçalves Da-Silva, Rio de Janeiro State University, BrazilReviewed by:
Alexandre Bello, Rio de Janeiro State University, BrazilPatrícia Santos, Federal University of Pernambuco, Brazil
Copyright © 2023 Graham, Li, Gong, Deng, Jin, Wang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xian-Ming Chen, eGlhbl9tX2NoZW5AcnVzaC5lZHU=
 Marion L. Graham1
Marion L. Graham1 Xian-Ming Chen
Xian-Ming Chen