- 1State Key Laboratory for Animal Disease Control and Prevention, Key Laboratory of Veterinary Parasitology of Gansu Province, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, Gansu, China
- 2Key Laboratory for Zoonosis Research of the Ministry of Education, Institute of Zoonosis, and College of Veterinary Medicine, Jilin University, Changchun, China
- 3Jiangsu Co-Innovation Center for the Prevention and Control of Important Animal Infectious Disease and Zoonoses, Yangzhou University, Yangzhou, China
Cysticercosis pisiformis, a highly prevalent parasitic disease worldwide, causes significant economic losses in the rabbit breeding industry. Previous investigations have identified a novel microRNA, designated as novel-miR1, within the serum of rabbit infected with Cysticercus pisiformis. In the present study, we found that C. pisiformis-derived novel-miR1 was released into the rabbit serum via exosomes. Through computational analysis using TargetScan, miRanda, and PITA, a total of 634 target genes of novel-miR1 were predicted. To elucidate the functional role of novel-miR1, a dual-luciferase reporter assay was utilized and demonstrated that novel-miR1 targets rabbit Toll-like receptor 2 (TLR2). Rabbit peripheral blood lymphocytes (PBLCs) were transfected with novel-miR1 mimic and mimic NC, and the in vitro experiments confirmed that novel-miR1 suppressed the expression of pro-inflammatory cytokines such as TNF-α, IL-1β, and IL-6 through the nuclear factor kappa B (NF-κB) pathway. In vivo experiments demonstrated that novel-miR1 was significantly upregulated during the 1–3 months following infection with C. pisiformis in rabbits. Notably, this upregulation coincided with a downregulation of TLR2, P65, pP65, TNF-α, IL-1β, and IL-6 in PBLCs. Collectively, these results indicate that the novel-miR1 derived from C. pisiformis inhibited the rabbits’ immune response by suppressing the NF-κB-mediated immune response. This immune modulation facilitates parasite invasion, survival, and establishment of a persistent infection.
1 Introduction
Cysticercus pisiformis is a widely prevalent larval form of tapeworm that primarily parasitizes the liver, omentum, and mesenteries of lagomorphs and rodents, leading to the development of cysticercosis pisiformis (1, 2). Among these hosts, rabbits are the most frequently affected and suffer significant consequences from cysticercosis pisiformis. This parasitic infection adversely affects rabbit reproductive rates and poses a serious threat to endangered rabbit species such as the teporingo (Romerolagus diazi) (3, 4). Moreover, it leads to decreased body weight and cholesterol levels, even leading to death (5). Importantly, C. pisiformis infection in rabbits impairs their immune responses, rendering them susceptible to secondary infections by other pathogens (6), which has substantial economic impacts in the rabbit breeding industry and impacts animal welfare.
Exosomes, extracellular vesicles ranging in size from 40 to 160 nm, are released by various cells, including both prokaryotes and eukaryotes (7). Numerous studies have reported that exosomes are mediators of vesicle transport, playing a pivotal role in parasite–host interactions (8–11). Trichinella spiralis excretory/secretory products have been shown to modulate programmed death 1-mediated M2 macrophage polarization (12). Exosomes derived from T. spiralis impact the nuclear factor kappa B (NF-κB) and mitogen-activated protein kinase (MAPK) signaling pathways, thereby inhibiting M1 macrophage polarization (13). Similarly, Taenia asiatica exosomes inhibit LoVo cell proliferation and autophagy via the Adenosine 5'-monophosphate (AMP)-activated protein kinase (AMPK) pathway (14). Notably, miRNAs are essential cargo within exosomes and are the key functional units (15, 16). Echinococcus multilocularis exosomes containing emu-miR-4989 induces UBE2N suppression (17). Fasciola hepatica releases fhe-miR-125b through exosomes to target the mammalian Argonaut protein (Ago-2) within macrophages during infection to downregulate the production of inflammatory cytokines (18). miR-1-3p and let-7-5p expressed by T. spiralis larvae-derived extracellular vesicles promote the polarization of bone marrow macrophages toward the M2b type while inhibiting fibroblast activation (19).
MicroRNAs (miRNAs) are small noncoding RNAs that play important roles in posttranscriptional gene silencing (20). They are capable of binding to the 3′-untranslated region (UTR), coding region, or 5′-UTR of the target mRNA to inhibit translation or facilitate mRNA degradation in a wide variety of physiological and pathological processes, including growth, proliferation, differentiation, development, metabolism, infection, immunity, and cell death (21, 22). Data from numerous studies have shown that miRNA expression profiles are altered during parasite infection and suggest that differentially expressed miRNAs play important roles in host immune responses (18, 23–25).
Toll-like receptors (TLRs) are pattern recognition receptors (PRRs) that play a crucial role in recognizing pathogen-associated molecular patterns (PAMPs) and activate intracellular signaling pathways, leading to the induction of inflammatory cytokine genes such as TNF-α, IL-6, IL-1β, and IL-12 (26, 27). Among the TLRs, TLR2 is involved in the recognition of various PAMPs derived from bacteria, fungi, parasites, and viruses (28). Upon recognition of PAMPs, including proteins, lipoproteins, lipids, nucleic acids, and endogenous ligands, TLR2 induces interferon (TIRF), leading to the activation of MAPKs through the MyD88 pathway or the Toll/IL-1R-related adaptor protein, resulting in the translocation of NF-κB and subsequent transcription and synthesis of pro-inflammatory cytokines (29).
Helminths, a class of pathogens, also elicit immune responses through TLR2. For instance, Clonorchis sinensis heat shock cognate B (CsHscB) activates TLR2 to induce the expression of IL-10, exerting immune regulatory activities (30). During T. spiralis infection, TLR2 is upregulated in the mouse small intestine, suggesting its crucial role in host defense mechanisms against this parasite (31). TLR2 is also important for the development of a Th1-dominant immune response in the mouse model of Taenia crassiceps cysticercosis (32). Cystic echinococcosis-derived antigens, including hydatid cyst fluid (HCF), germinal layer antigens (GL), somatic and excretory/secretory (ES) products of protoscoleces (PSC), have been shown to downregulate the expression of TLR2 in ovine peripheral blood leukocytes, potentially contributing to the establishment of chronic infection by suppressing host immunity (33).
Therefore, TLR2 plays important roles in the induction of innate immune responses. Despite the knowledge on the important roles played by parasite-derived miRNAs during infection, little is known about the role of TLR2 as a pattern recognition receptor in the context of C. pisiformis infection in rabbits. In our experiments, we observed a significant upregulation of novel-miR1, derived from C. pisiformis, in the serum of infected rabbits. Furthermore, TLR2 was predicted and verified as a target of novel-miR1. This study aims to investigate the mechanism underlying the interaction between novel-miR1 and TLR2 in rabbits infected with C. pisiformis. The findings will contribute valuable insights into the mechanism of C. pisiformis infection, thereby benefiting the diagnosis and treatment of cysticercosis pisiformis and providing strategies for the prevention and control of helminth infections.
2 Methods
2.1 Rabbit and C. pisiformis collection
Fifty-day-old New Zealand White rabbits weighing 1.5~2 kg were obtained from the Laboratory Animal Center of Lanzhou Veterinary Research (Lanzhou, China). The rabbits were allowed to adaptively acclimate for 1 week prior to the experiment. A total of six rabbits were orally infected with 1 ml of infective Taenia pisiformis eggs (1,000 eggs/ml), establishing the C. pisiformis infection group (CPi). As a negative control group (NC), three rabbits were administered 1 ml of phosphate-buffered saline (PBS) via oral gavage. All rabbits were reared with the same forage and clean water. Following the collection of required blood samples, necropsy was performed on the six rabbits to confirm C. pisiformis infection. The examination results are shown in Supplementary Table S1.
All rabbit experiments conducted in this study were approved by the Animal Ethics Committee of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (Permit No. LVRIAEC-2016-006), and the protocols used were strictly in accordance with the guidelines of animal welfare. Every effort was made to minimize any potential harm to the experimental rabbits in accordance with the study protocols.
2.2 Cells and media
Rabbit peripheral blood lymphocytes (PBLCs) were isolated from 5-ml fresh rabbit blood samples collected in tubes containing EDTA and separated using rabbit peripheral blood lymphocytes isolation kit based on Ficoll density gradient centrifugation (TBD Science, China, LTS10965). Cell viability was tested by trypan blue staining (TBD Science, China, TBD20180079) following the manufacturer’s instructions. The isolated PBLCs were cultured in RPMI 1640 medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS) (Gibco), penicillin (100 unit/ml) (Gibco), and streptomycin (100 µg/ml) (Gibco).
HEK293T cells were cultured in DMEM (Gibco) containing 10% FBS, penicillin (100 unit/ml), and streptomycin (100 µg/ml). All cells were cultured at 37°C in a 5% CO2 cell culture incubator.
2.3 Exosome isolation and identification
T. pisiformis larvae were harvested from the peritoneal cavities of CPi rabbits and cultured in 10 ml RPMI 1640 medium (Gibco, Grand Island, NY, USA) supplemented with 10% exosome-depleted FBS and antibiotics (100 U/ml penicillin, 100 μg/ml streptomycin) at 37°C in 5% CO2 cell culture incubator. After 12 h, the culture media were collected and replaced with fresh medium. The collected media were centrifuged at 2,000×g at 4°C for 10 min to remove cell debris, and the exosomes were isolated using the Minute™ Hi-Efficiency Exosome Isolation Reagent (Cat. No. EI-027; Invent Biotechnologies, Inc., Plymouth, MN, USA) following the manufacturer’s protocol. The exosome precipitate was resuspended in 500 μl phosphate-buffered saline (PBS) filtered through a 0.2-μM pore filter. The concentration of exosomes was determined using a Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). The particle size of the exosomes was analyzed using a NanoSight LM10 instrument (Nanosight, Wiltshire, UK). The exosome size and morphology were observed through transmission electron microscopy (TEM, Hitachi Ltd., Tokyo, Japan). Briefly, exosomes were adsorbed onto a 200-mesh formvar-coated copper grid (Agar Scientific Ltd., Stansted, UK) and incubated for 10 min at room temperature. After staining with 2% tungstophosphoric acid solution for 1 min, the exosomes were examined under an electron microscope (34).
2.4 Exosome labeling and confocal laser scanning microscope analysis
To visualize exosomal uptake by rabbit PBLCs, we labeled exosomes with DiD dye (ab275319, Abcam, Cambridge, MA, USA). A total of 6 µg of exosomes were mixed with 2 μl DiD dye (0.2%) and diluted to 1 ml PBS (35). As a negative control, only DiD dye mixed with PBS was used. The mixture was then incubated at 37°C for 30 min to allow for labeling of the exosomes. The labeled exosomes were subsequently washed with PBS and centrifuged at 120,000×g and 4°C for 1 h. The resulting precipitate was resuspended in 180 μl of PBS.
Subsequently, the labeled exosomes were added to three wells of a 12-well plate containing rabbit PBLCs and incubated for 12 h in a 5% CO2 incubator at 37°C. Following incubation, the cells were stained with 10 µg/ml of 4,6 diamidino-2-phenylindole (DAPI) (Beyotime, Shanghai, China) for 5 min. The stained PBLCs were added to a glass slide and covered with a coverslip for imaging under a confocal laser scanning microscope (Nikon, Tokyo, Japan) (36).
2.5 Novel-miR1/siRNA transfection and exosome treatment
PBLCs were isolated from fresh peripheral blood from healthy rabbits. The isolated cells were diluted to 2~10 × 106 cells/ml in RPMI 1640 medium supplemented with 10% FBS, penicillin (100 unit/ml), and streptomycin (100 µg/ml). Subsequently, 1 ml of the PBLC suspension was added to each well of a 12-well plate and cultured in a 37°C incubator with 5% CO2. For transfection experiments, 10 pmol/L of novel-miR1 mimic, mimic NC, inhibitor, inhibitor NC, siTLR2, and siRNA NC (RiboBio, Guangzhou, China) were transfected into the PBLCs using 2 μl of Lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific, USA) for a duration of 24 h. In exosome treatment experiments, 20 µg of total exosome protein or an equivalent volume of PBS was added per well and coincubated for 24 h.
2.6 Extraction of miRNAs and RNA
The miRNAs were separated from fresh blood samples or C. pisiformis-derived exosomes using an EasyPure miRNA Kit (TransGen, China) according to the manufacturer’s instructions. Total RNA was isolated from the PBLCs and C. pisiformis using TRIzol reagent (Thermo Fisher Scientific) following the manufacturer’s protocol.
2.7 Real-time qPCR analysis
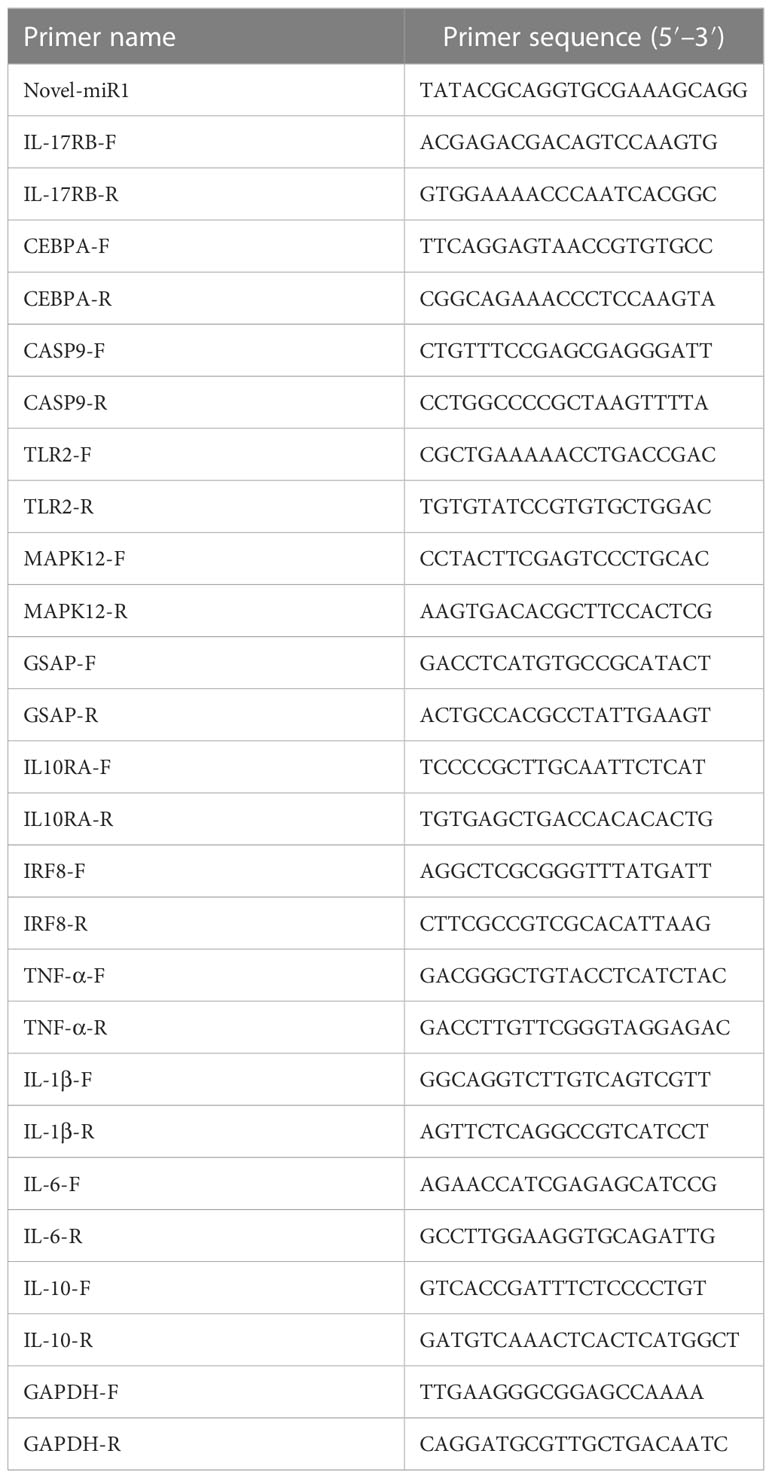
The expression of novel-miR1 and TLR2 in serum miRNAs or PBLC RNA in CPi and NC groups was analyzed using real-time quantitative PCR (qPCR). The miRNAs were reverse-transcribed into cDNA using the Mir-XTM miRNA First-Strand Synthesis Kit (Clontech, Mountain View, CA, USA). The cDNA first-strand synthesis of PBLC total RNA was performed using the HiScript® III 1st Strand cDNA Synthesis Kit (Vazyme, China). All cDNAs were used for real-time fluorescence qPCR detection using an ABI 7500 quantitative PCR instrument (Thermo Fisher Scientific). qPCR was performed in a 20-μl reaction volume, including 10 μl SYBR Advantage Premix (2×) (Takara, Japan), 2 μl cDNA, 0.4 μl ROX Dye (50×), 0.8 μl specific forward primer, 0.8 μl specific reversed primer, and 6 μl RNase-free water. qPCR was performed with the following: 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 34 s. The relative expressions were normalized to U6 small nuclear RNA (snRNA) or GAPDH. All primers were designed and produced by Sangon Biotech (Sangon Biotech, China) (Table 1). The relative expression of novel-miR1 was calculated using the 2−ΔΔCt method (37).
To demonstrate the source of novel-miR1 in rabbit serum, an absolute quantification qPCR was performed. Total RNA (1 μg) from all samples, including rabbit serum (CPi and NC), C. pisiformis, and C. pisiformis exosomes, was reverse-transcribed into cDNA using the Mir-XTM miRNA First-Strand Synthesis Kit. qPCR was conducted on an ABI 7500 using a TransStart Tip Green qPCR SuperMix Kit (TransGen Co., Beijing, China) with the following protocol: initial denaturation at 95°C for 30 s, followed by 40 cycles of 95°C for 15 s and 60°C for 34 s. The copy number of novel-miR1 in the samples was determined using a standard curve established with 10-fold serial dilutions (1 × 107 to 1 × 102 copies/μl) of the standard novel-miR1.
2.8 Function prediction of novel-miR1
To investigate the potential functional roles of novel-miR1, we employed bioinformatics tools to predict its target genes. The software TargetScan (http://www.targetscan.org/), miRanda (http://www.microrna.org/), and PITA (http://www.pita.org.fj/) were utilized, and the analysis was conducted on the Oryctolagus cuniculus genome (https://www.ncbi.nlm.nih.gov/genome/316?genome_assembly_id=1549366). The predicted miRNA target genes were determined by selecting the intersection set of results obtained from the three software programs. The functions of the identified target genes were analyzed by Gene Ontology (GO) functional annotation clustering analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. These analyses were performed using the online DAVID bioinformatics database functional annotation tool (https://david.ncifcrf.gov/).
2.9 Luciferase reporter assay
HEK293T cell lines were employed for the co-transfection experiments. The novel-miR1 mimic or mimic NC was cotransfected with dual-luciferase reporter plasmids (pmir-GLO-TLR2, pmir-GLO-TLR2-WT, or pmir-GLO-TLR2-Mut). The luciferase reporter assay was performed using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). For accurate comparisons, firefly luciferase activity was normalized to the Renilla luciferase activity. The effect of a novel-miR1 on the luciferase reporter constructs containing the TLR2 3′-UTR or the corresponding mutant was determined by comparing the reporter activity with the control. Each luciferase reporter assay was conducted in triplicate.
PBLCs proteins were extracted using RIPA lysis buffer supplemented with 1% protease inhibitor. Here, 30 µg total proteins were incubated at 100°C for 10 min. The protein samples were separated on a 10% SDS-PAGE gel alongside prestained protein markers. The separated proteins were transferred to a PVDF membrane and blocked with 5% skimmed milk at room temperature for 3 h. Rabbit anti-TLR2 monoclonal antibody (1:1,000; Abcam; cat. no. Ab209217), phospho-NF-κB p65 (Ser536) rabbit monoclonal antibody (1:1,000; Abcam; cat. no. ab239882), NF-κB p65 rabbit polyclonal antibody (1:1,000; Abcam; cat. no. Ab16502), rabbit anti-IL-6 polyclonal antibody (1:1,000; Bioss, Woburn, MA, USA; cat. no. bs-0782R), rabbit anti-TNF-α polyclonal antibody (1:1,000; Bioss; cat. no. bs-2081R), mouse anti-IL-1β monoclonal antibody (1:500; Santa Cruz Biotechnology, Dallas, TX, USA; cat. no. sc52012), and mouse anti-β-actin monoclonal antibody (1:2,000 dilution, Beyotime Biotechnology, China) were used as the primary antibodies in this study. The secondary antibodies used were goat anti-mouse IgG/HRP antibody (1:4,000 dilution, Beyotime Biotechnology) or goat anti-rabbit IgG/HRP antibody (1:4,000 dilution, Beyotime Biotechnology), as per the manufacturer’s instructions. The membrane was reacted with ultrasensitive ECL luminescence reagent (Beyotime Biotechnology) for color detection.
2.10 Cytokine analysis
Enzyme-linked immunosorbent assay (ELISA) was performed to quantify the levels of IL-6 (ml027844), TNF-α (ml028087), and IL-1β (ml027165). ELISA kits were obtained from Shanghai Enzyme-linked Biotechnology Co. Ltd. (Shanghai, China). PBLCs were isolated from the blood of CPi and NC rabbits and the cell culture supernatants were collected to measure the cytokine levels following the manufacturers’ instructions.
2.11 Statistical analysis
The Student’s t-test method was used to analyze the significance of the miRNA expression, ImageJ was used for Western blot grayscale analysis, and GraphPad 9 was used to create the relative expression diagram. p < 0.05 was considered statistically significant.
3 Results
3.1 Novel-miR1 derived from C. pisiformis exosomes was identified in infected rabbit serum
Exosomes derived from C. pisiformis were successfully isolated, and their measured size was about 100 nm (Figures 1A, B). To better understand the origin of the novel-miR1 in rabbit serum, absolute qPCR was used to detect the levels of miRNA in samples obtained from rabbit blood (CPi, NC), C. pisiformis, and C. pisiformis exosomes. The results revealed a significant enrichment of novel-miR1 within the C. pisiformis exosomes (Figure 1C). These findings indicate that novel-miR1 in infected rabbit serum is released into the bloodstream through C. pisiformis exosomes.
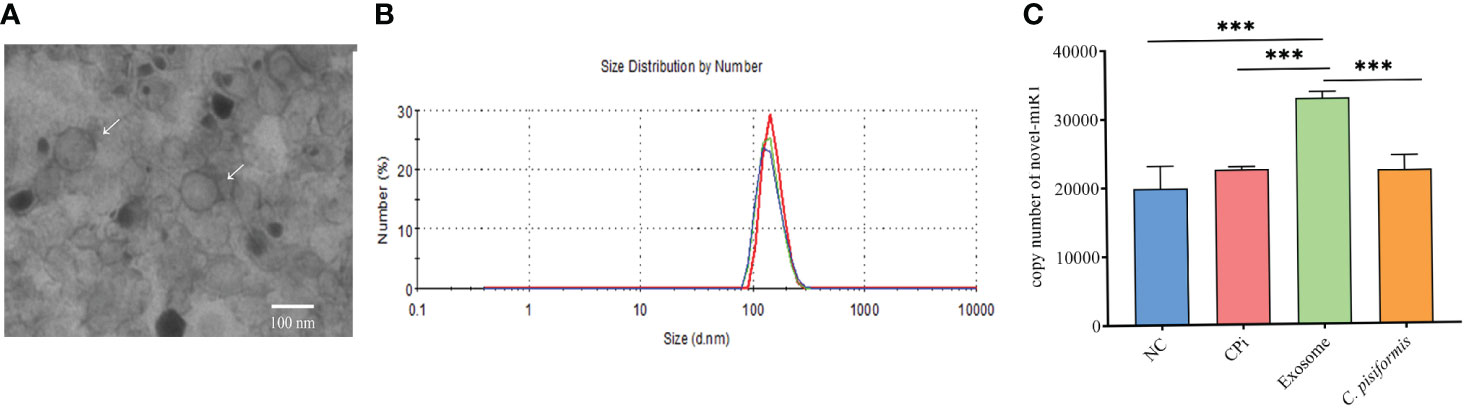
Figure 1 Identification and analysis of novel-miR1 source and target genes. (A) Morphological characterization of C. pisiformis exosomes through transmission electron microscope (TEM). (B) Size distribution of exosome of C. pisiformis by nanoparticle tracking analysis (NTA). (C) Quantification of novel-miR1 copy numbers in different samples. NC represents the negative control rabbit serum miRNA, CPi represents the C. pisiformis-infected rabbit serum total miRNA, exosomes are the C. pisiformis exosome total miRNA, and C. pisiformis represents the C. pisiformis total miRNA. Novel-miR1 copy numbers were determined by absolute quantitative PCR using 20 ng total miRNAs. *** indicates a significance level of p ≤ 0.001.
3.2 TLR2 is targeted by novel-miR1
A total of 634 target genes were predicted for novel-miR1 using TargetScan, miRNADA, and PITA (Figure 2A, Supplementary Table S2). GO functional annotation clustering analysis (Supplementary Table S3) revealed enrichment of novel-miR1 target genes in various biological processes (BPs), such as positive regulation of GTPase activity, intracellular signal transduction, protein autophosphorylation, protein stabilization, and ion transport. Molecular function (MF) analysis highlighted ATP binding, protein homodimerization activity, guanyl-nucleotide exchange factor activity, and transcription factor binding as enriched functions. Cellular component (CC) analysis showed localization to cytoplasm, nucleoplasm, integral component of plasma membrane, and lysosomal membrane (Supplementary Table S3). The top 10 terms for BP, MF, and CC are presented in Figure 2B. KEGG pathway analysis revealed the involvement of novel-miR1 target genes in 60 signaling pathways. Notably, these pathways included crucial signaling cascades such as Ras, MAPK, mTOR, Wnt, and ErbB. Moreover, the target genes were found to cluster in pathways related to pathogenic infection and immune responses, such as pathways in cancer, human immunodeficiency virus 1 infection, tuberculosis, Salmonella infection, human cytomegalovirus infection, hepatitis B, Kaposi sarcoma-associated herpesvirus infection, Influenza A, and T-cell receptor (Supplementary Table S4). Figure 2C illustrates the top 30 signaling pathways represented by the novel-miR1 target genes, highlighting the significant role of novel-miR1 in pathogenic infection.
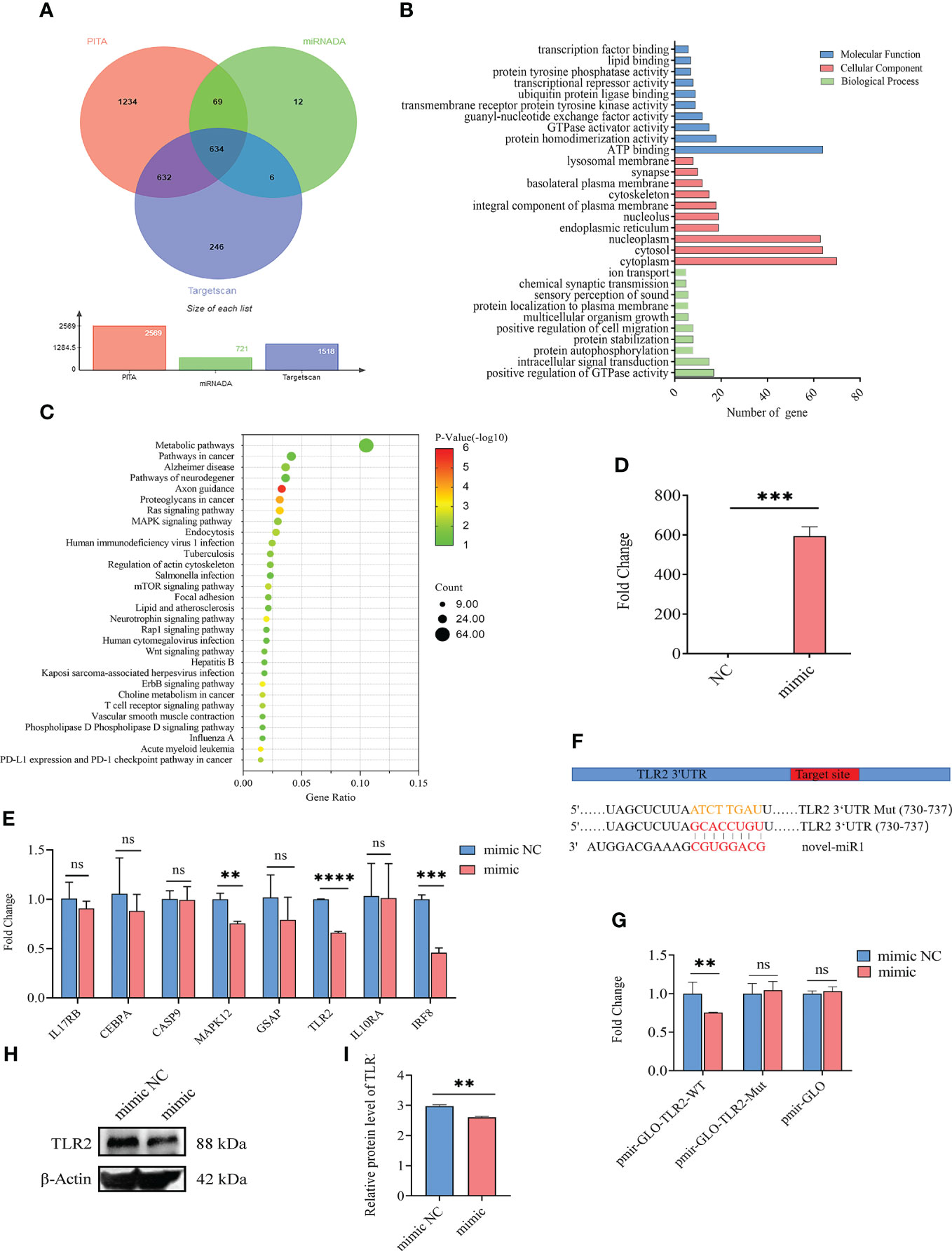
Figure 2 Targeting of TLR2 3′-UTR by novel-miR1. (A) Venn diagram showing the intersection of predicted target genes from different software programs, namely, PITA, miRanda, and TargetScan. (B) Gene Ontology (GO) molecular function annotations of the target genes of novel-miR1. The top 10 GO terms for biological processes, cellular component, and molecular function are shown based on their respective p-values. (C) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of the predicted target genes of novel-miR1. The size of dots indicates the number of genes associated with each pathway. (D) Overexpression of novel-miR1 in rabbit PBLCs. (E) Relative expression of novel-miR1 target genes in rabbit PBLCs after novel-miR1 treatment. (F) novel-miR1 binding TLR2 3′-UTR site and the sequences of luciferase reporter plasmids (pmir-GLO-TLR2-WT and pmir-GLO-TLR2-Mut). (G) Luciferase reporter assay results indicating the downregulation of TLR2 through binding to its target 3′-UTR. (H) Western blot assay results showing the downregulation of TLR2 protein in PBLCs by novel-miR1. (I) Western blot assay results demonstrating the downregulation of TLR2 protein in PBLCs by novel-miR1 (fold change was calculated by the gray values of the image). ns, p ≥ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001.
To further investigate the immunoregulatory function of novel-miR1 in rabbit serum, rabbit PBLCs were transfected with novel-miR1 mimic to achieve overexpression of novel-miR1 (850-fold change, Figure 2D). Eight immune-related target genes (IL17RB, CEBPA, CASP9, MAPK12, GSAP, TLR2, IL10RA, and IRF8) were selected for validation by qPCR analysis. Compared to the control group, novel-miR1 overexpression significantly inhibited the expression of MAPK12, TLR2, and IRF8 (p < 0.05), while IL17RB, CEBPA, and GSAP exhibited a tendency toward downregulation (Figure 2E).
Next, we verified the relationship between novel-miR1 and TLR2 and found that the seed sequence of novel-miR1 can bind to the 3′UTR of TLR2 (Gene ID: 100009578) using TargetScan software (Figure 2F). To verify the targeting relationship between novel-miR1 and TLR2, we conducted a dual luciferase reporter experiment by cotransfecting the vectors pmirGLO-TLR2-WT, pmirGLO-TLR2-Mut, and pmirGLO with novel-miR1 mimic/mimic NC into HEK293T cells. The results showed a decrease in firefly luciferase activity when pmirGLO-TLR2-WT was cotransfected with novel-miR1 mimics. However, differences were not found in the cells cotransfected with novel-miR1 mimics and pmirGLO-TLR2-Mut or novel-miR1 mimics and pmirGLO (Figure 2G). Additionally, Western blot analysis indicated that novel-miR1 inhibited the protein expression of TLR2 in PBLCs (Figures 2H, I).
3.3 Novel-miR1 inhibits the TLR2 signaling pathway and NF-κB activation
TLR2 is a pattern recognition receptor that plays a critical role in initiating innate inflammatory responses and promoting adaptive immune responses (38, 39). To investigate the effect of novel-miR1 targeting TLR2 in rabbit PBLCs, novel-miR1 was overexpressed in rabbit PBLCs via transfection with novel-miR1 mimic. Subsequently, the expression of cytokines related to the TLR2 signaling pathway and NF-κB protein was assessed. The experimental outline is shown in Figure 3.
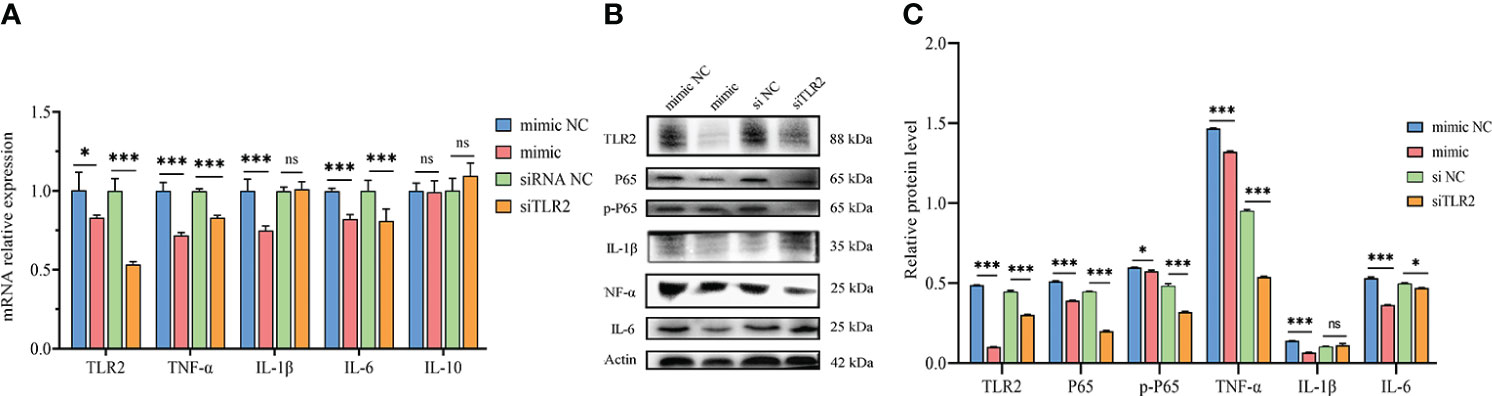
Figure 3 Inhibition of TLR2 signaling pathway and NF-κB activation by novel-miR1. (A) qPCR results demonstrate the reduction of TLR2, TNF-α, IL-1β, and IL-6 mRNA levels by novel-miR1. Each test was performed in triplicate. Fold change was calculated based on qPCR results. (B) Western blot assay results indicate the decrease in TLR2, P65, pP65, TNF-α, IL-1β, and IL-6 protein levels by novel-miR1. (C) Western blot assay results showing the decrease in protein levels of TLR2, P65, pP65, TNF-α, IL-1β, IL-6 (fold change was calculated by the gray values of the image). ns, p ≥ 0.05; *, p ≤ 0.05; ***, p ≤ 0.001.
The qPCR results indicated significant downregulation of TLR2, IL-6, TNF-α, and IL-1β mRNA expression in rabbit PBLCs. As a control, TLR2 siRNA was transfected into rabbit PBLCs, which confirmed that the mRNA expression of TLR2 was downregulated. Interestingly, the expression of IL-6 and TNF-α was significantly downregulated (Figure 3A). Consistent with the qPCR results, Westen blot results showed that the protein expression of TLR2 IL-6, TNF-α, and IL-1β was also downregulated. Additionally, we observed downregulation of P65 and phosphorylated-P65 (pP65) protein levels (Figures 3B, C), indicating inhibition of the NF-κB pathway activation due to reduced TLR2 expression. These results collectively suggest that novel-miR1 inhibits the NF-κB pathway signaling by targeting TLR2.
Next, C. pisiformis exosomes were marked with DiD dye and cocultured with rabbit PBLCs, which revealed that C. pisiformis exosomes can enter rabbit PBLCs (Figure 4A). Furthermore, qPCR results suggested that the expression of novel-miR1 and TLR2 was inhibited by C. pisiformis exosomes (Figure 4B). Similarly, Western blot results showed that C. pisiformis exosomes downregulated the expression of TLR2, P65, and pP65 proteins (Figures 4C, D).
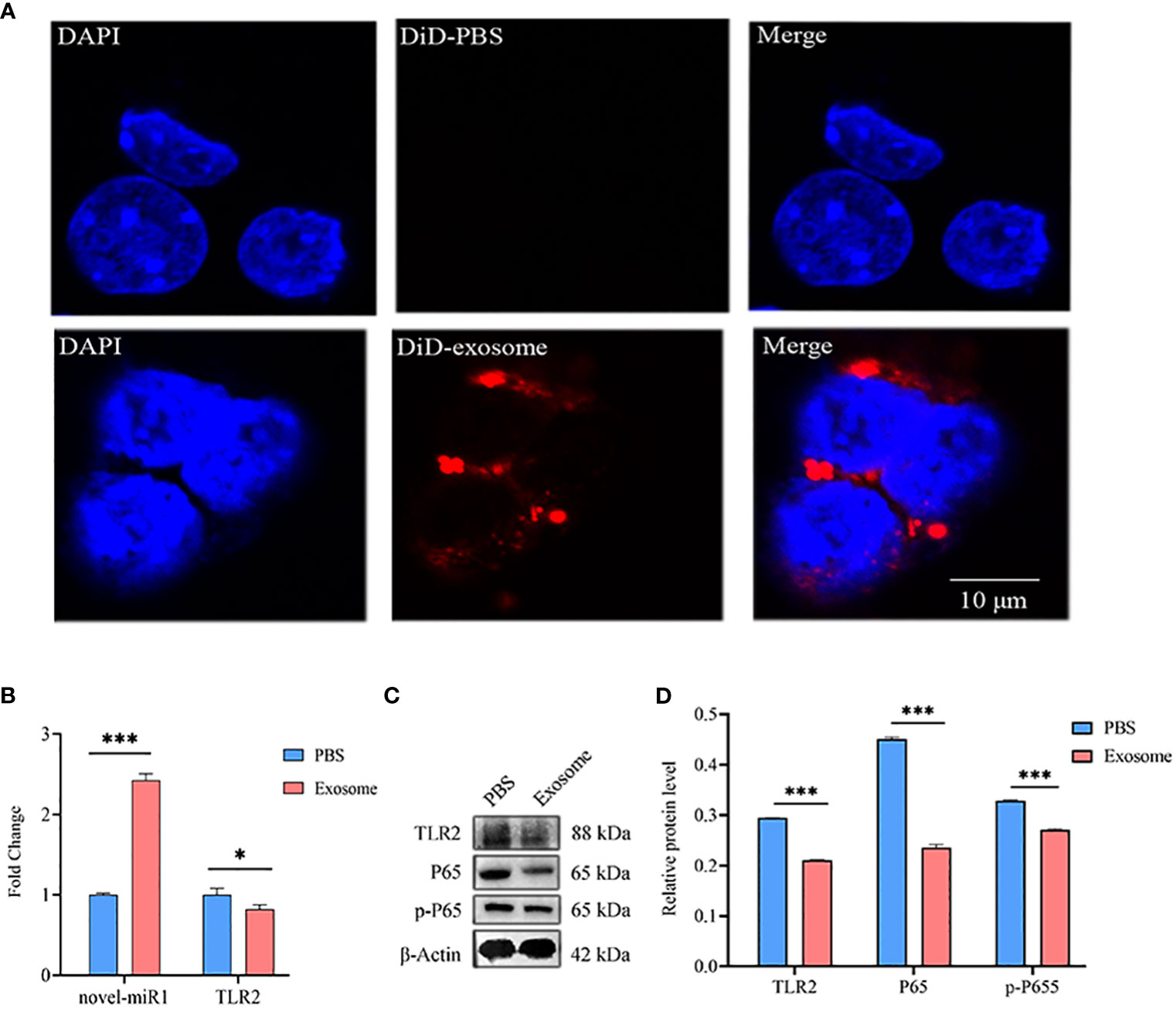
Figure 4 C. pisiformis exosomes inhibit the TLR2 signaling pathway and NF-κB activation. (A) C. pisiformis exosomes can enter rabbit PBLCs. (B) qPCR results demonstrate the increased expression of novel-miR1 and decreased TLR2 mRNA level in rabbit PBLCs after treatment with C. pisiformis exosomes. Each test was performed in triplicate; fold change was calculated based on qPCR results. (C) Western blot assay results suggest that treatment with C. pisiformis exosomes decreased TLR2, P65, and pP65 protein expression levels. (D) Western blot assay results indicating the reduction in protein expression levels of TLR2, P65, and pP65 by C. pisiformis exosomes (fold change was calculated by the gray values of the image). *, p ≤ 0.05; ***, p ≤ 0.001.
Lastly, rabbit PBLCs were isolated from the CPi and NC groups for detection and analysis. The results revealed consistent upregulation of novel-miR1 expression and downregulation of TLR2 mRNA during the initial 3 months (Figures 5A, B). Similarly, protein levels of TLR2, P65, pP65, IL-6, TNF-α, and IL-1β were downregulated (Figures 5C–E). In summary, these findings strongly support the release of novel-miR1 into the bloodstream through C. pisiformis exosomes in infected rabbits and demonstrate that novel-miR1 targets TLR2 to inhibit NF-κB pathway signaling.
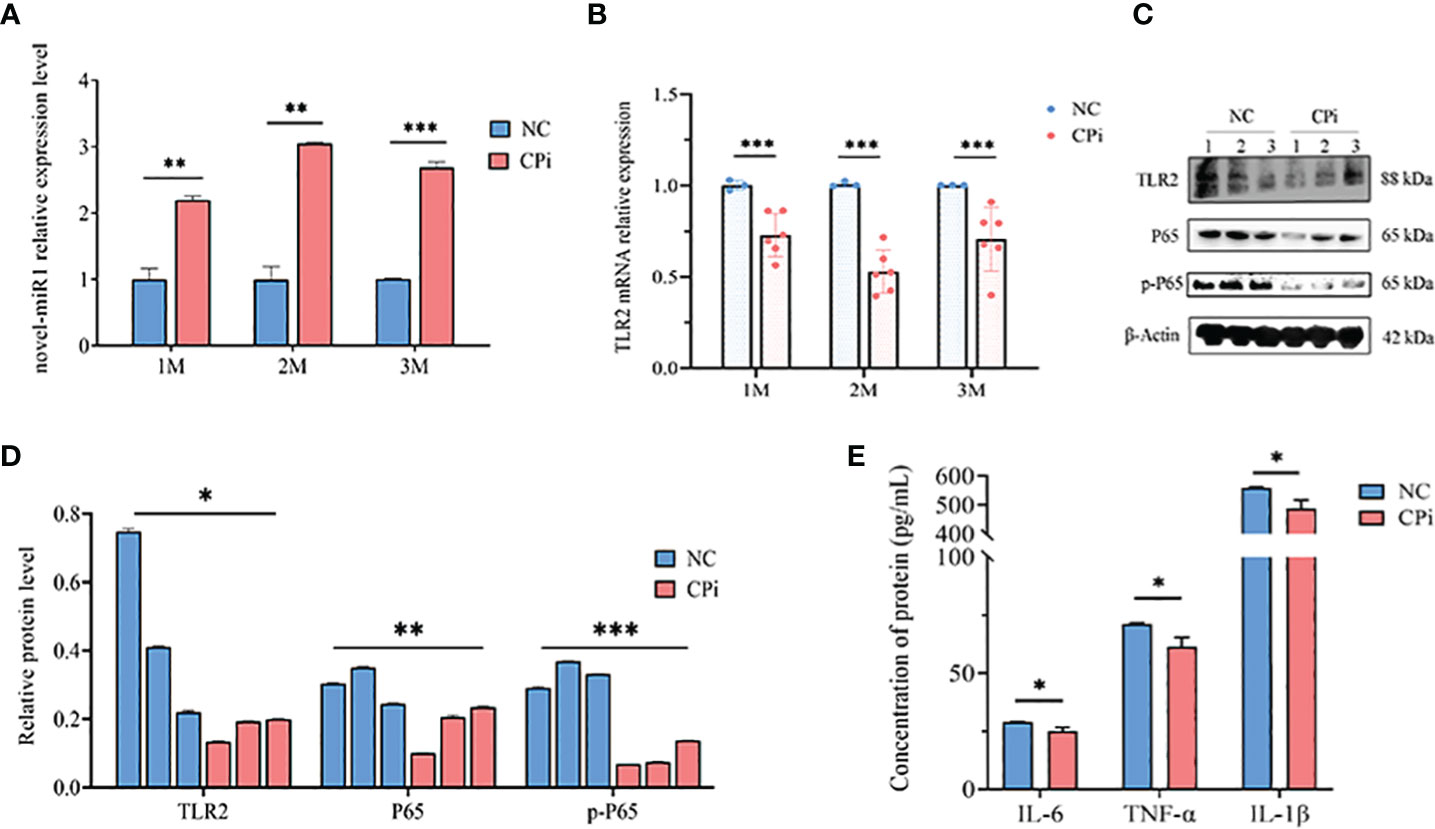
Figure 5 C. pisiformis inhibits the TLR2 signaling pathway and NF-κB activation. (A) Expression profile of novel-miR1 in rabbit PBLCs over a 3-month period. (B) Relative expression TLR2 mRNA in rabbit PBLCs during the initial 3-month period. (C) Western blot assay results indicate inhibition of TLR2, P65, and pP65 protein levels in C. pisiformis-infected rabbit PBLCs. (D) Western blot assay results demonstrating the inhibition of TLR2, P65, and pP65 protein levels in C. pisiformis-infected rabbit PBLCs (the fold change was calculated by the gray values of the image). (E) ELISA assay results indicate inhibition of IL-6, TNF-α, and IL-1β protein expression levels in C. pisiformis-infected rabbit PBLCs. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001.
4 Discussion
In our previous study, we observed a significant upregulation of novel-miR1 in rabbit serum through high-throughput sequencing of small RNA, which was further validated by qPCR. However, the sources of novel-miR1 within rabbit serum have not been well elucidated. In the present study, we conducted absolute quantification qPCR to assess total miRNA levels in samples from rabbit serum (CPi and NC), C. pisiformis, and C. pisiformis exosomes. The results indicated a significant enrichment of novel-miR1 in C. pisiformis exosomes. However, it is important to note that higher copy numbers observed in the NC group samples may be attributed to nonspecific sequence amplification.
To further elucidate the functions of novel-miR1, we employed transfection of novel-miR1 mimics and inhibitors into rabbit PBLCs. Interestingly, we were unable to detect any differences in novel-miR1 or novel-miR1 target gene expression in the groups transfected with novel-miR1 inhibitors (Supplementary Figures S1, S2). This finding suggests that novel-miR1 may not naturally exist within rabbit PBLCs and provides supporting evidence that rabbit serum novel-miR1 is released by C. pisiformis.
Our data revealed that the expression level of novel-miR1 increased up to 2 months and then decreased by the third month, while TLR2 mRNA expression followed an opposite trend, decreasing until the second month and subsequently increasing. The upregulation of novel-miR1 during the initial stages of infection and the subsequent decrease may be indicative of an ongoing enhancement of immune interactions between C. pisiformis and rabbits. After 2 months of infection, the immune response gradually weakens, facilitating continued parasitism by C. pisiformis. Additionally, the opposite expression trends observed between novel-miR1 and TLR2 provide evidence that TLR2 is a target gene of novel-miR1.
The findings in the present study highlight the significant immunoregulatory role of novel-miR1 by targeting rabbit PBLCs TLR2 during C. pisiformis infection. It has been reported that the miRNAs, as regulators of innate immunity, can regulate components of the TLR signaling pathway, including the TLRs themselves, associated signal proteins, regulatory molecules, transcription factors, and functional cytokines (40). For instance, mouse-derived mmu-miR-92a-2-5 can target TLR2 to inhibit Schistosoma japonicum-induced liver fibrosis and downregulate the expression of TLR2-related cytokines such as IL-4, IFN-γ, and TNF-α during S. japonicum infection in mice (41).
Further investigations have revealed that novel-miR1 can inhibit the expression of TNF-α-, IL-1β, and IL-6-related cytokines in rabbit PBLCs. Notably, novel-miR1 showed significant upregulation in rabbit PBLCs from 1 to 3 months during C. pisiformis infection in rabbits, while the corresponding expression of TLR2 was downregulated. TNF-α, IL-1β, and IL-6 play critical roles in regulating worm immune evasion. Cysteine protease inhibitors (cystatins) present in the ESPs of Fasciola hepatica were found to reduce the production of IL-6 and TNF-α in mouse macrophages (42). Similar findings have been reported in S. japonicum infections (43). Additionally, it has been reported that mice infected with Echinococcus multilocularis and treated with albendazole exhibited a reversal of elevated levels of IL-1β, IL-6, TNF-α, and IFN-γ in the liver (44). Inhibition of these inflammatory cytokines tends to regulate the host immune response toward Th2, and numerous studies have demonstrated the beneficial effects of helminths in inhibiting inflammatory cytokines for the management of auto-inflammatory/immune diseases (45–48).
Based on our study, we have identified that novel-miR1 has significant therapeutic potential for the treatment of allergic, inflammatory, and autoimmune disorders in rabbit. Furthermore, further investigation into the regulatory mechanisms governed by novel-miR1 holds potential for the prevention and treatment of human auto-inflammatory/immune diseases.
5 Conclusion
The results from the present study strongly indicate that C. pisiformis-derived exosomes serve as a means for the release of novel-miR1 into rabbit serum during parasite infection. Moreover, our results demonstrate that novel-miR1 exerts its regulatory effects by inhibiting the expression of TLR2, thereby modulating the production of pro-inflammatory factors via the NF-κB pathway. These observations support the notion that novel-miR1 plays a crucial role in facilitating parasite survival and establishment within the host (Figure 6).
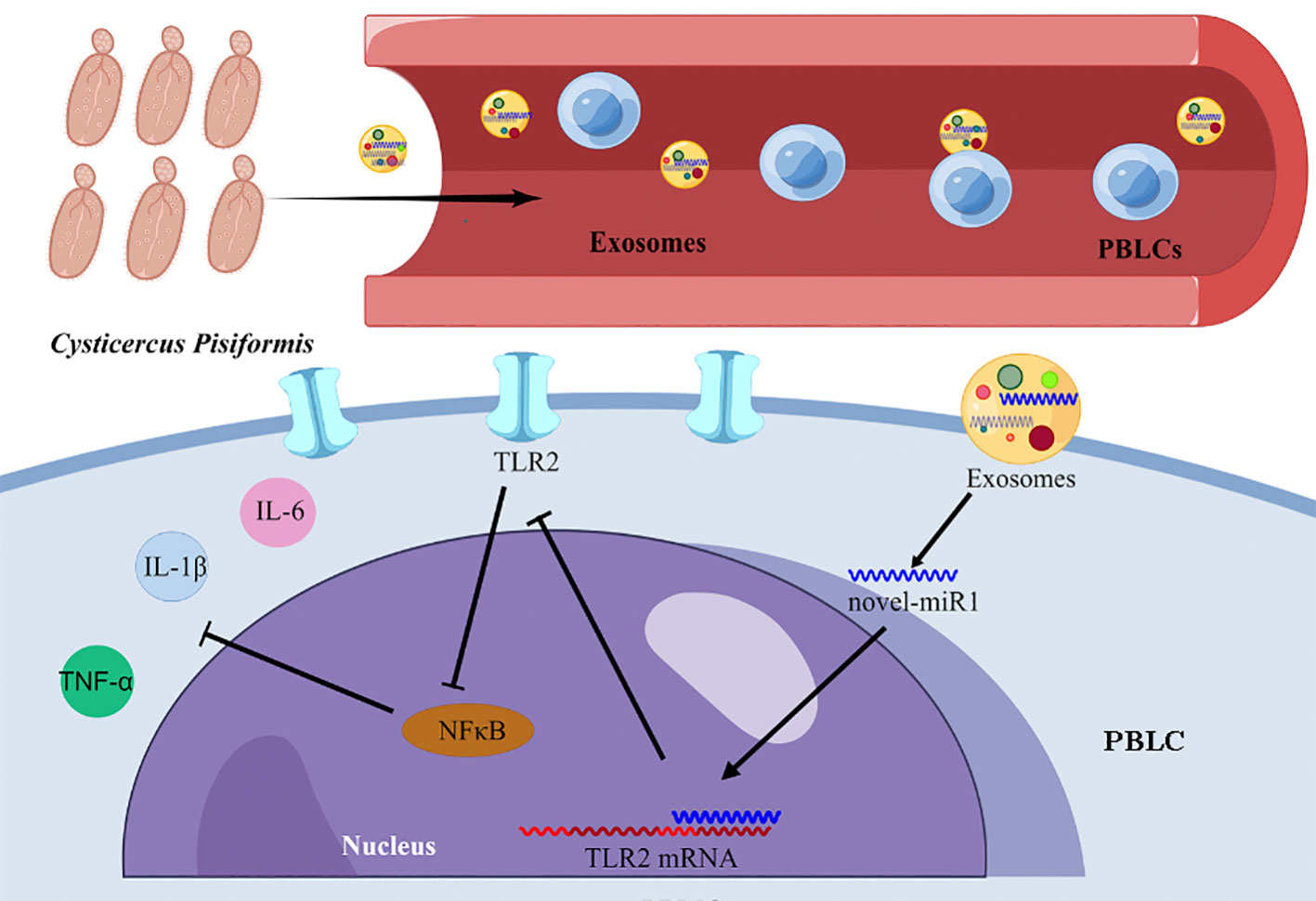
Figure 6 Graphical representation illustrating the mechanism of novel-miR1 targeting TLR2 in PBLCs to regulate the immune response in rabbits. The figure was created using Figdraw.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by Animal Ethics Committee of Lanzhou Veterinary Research Institute (SYXK 2020–0010).
Author contributions
GC and XNL conceived and designed the study, analyzed the data, drafted and critically revised the manuscript. GC, GP, LW performed the experiments. YL, LW, TL, HL, GP, SZ, XW, and XLL helped in the implementation of the study and performed and interpreted the computational analysis. All authors have read and approved the final manuscript.
Funding
This research was supported by grants from the National Natural Science Foundation of China (No. 32072889) and the National Key Research and Development Program of China (No.2017YFD0501303). The funders had no role in the design of the study, in the collection, analysis, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Acknowledgments
The authors wish to thank the Lanzhou Veterinary Research Institute for facilitating the implementation of this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1201455/full#supplementary-material
Supplementary Figure 1 | Relative expression of novel-miR1 in rabbit PBLCs treated with novel-miR1 inhibitors.
Supplementary Figure 2 | qPCR results showing the relative expression of TLR2, TNF-α, IL-1β, IL-6, and IL-10 mRNA in rabbit PBLCs treated with novel-miR1 inhibitors.
Supplementary Table 1 | Quantification of C. pisiformis in rabbits from CPi and NC groups.
Supplementary Table 2 | Prediction of target genes for novel-miR1 using Targetscan, miRNADA and PITA.
Supplementary Table 3 | Functional annotation clustering analysis of novel-miR1 target genes using GO.
Supplementary Table 4 | KEGG pathway analysis of novel-miR1 target genes.
References
1. Samorek-Pieróg M, Karamon J, Brzana A, Bilska-Zając E, Zdybel J, Cencek T. Molecular confirmation of massive Taenia pisiformis cysticercosis in one rabbit in Poland. Pathogens (2021) 10(8):1029. doi: 10.3390/pathogens10081029
2. Chen GL, Wang LQ, Liu TL, Li YP, Zhang SH, Li H, et al. Identification and expression profiling of circulating MicroRNAs in serum of Cysticercus pisiformis-infected rabbits. Genes (Basel) (2021) 12(10):1591. doi: 10.3390/genes12101591
3. Hallal-Calleros C, Morales-Montor J, Orihuela-Trujillo A, Togno-Peirce C, Murcia-Mejía C, Bielli A, et al. Taenia pisiformis cysticercosis induces decreased prolificacy and increased progesterone levels in rabbits. Vet Parasitol (2016) 229:50–3. doi: 10.1016/j.vetpar.2016.09.015
4. Stancampiano L, Ravagnan S, Capelli G, Militerno G. Cysticercosis by Taenia pisiformis in brown hare (Lepus europaeus) in northern Italy: epidemiologic and pathologic features. Int J Parasitol Parasites Wildl (2019) 9:139–43. doi: 10.1016/j.ijppaw.2019.04.004
5. Arias-Hernández D, García-Jiménez S, Domínguez-Roldan R, Murcia-Mejía C, Báez-Saldaña A, Hallal-Calleros C, et al. Effects of Taenia pisiformis infection and obesity on clinical parameters, organometry and fat distribution in Male rabbits. Pathogens (2020) 9(11):861. doi: 10.3390/pathogens9110861
6. Wang LQ, Liu TL, Liang PH, Zhang SH, Li TS, Li YP, et al. Characterization of exosome-like vesicles derived from Taenia pisiformis cysticercus and their immunoregulatory role on macrophages. Parasit Vectors (2020) 13(1):318. doi: 10.1186/s13071-020-04186-z
7. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science (2020) 367(6478):eaau6977. doi: 10.1126/science.aau6977
8. Liang PH, Mao L, Zhang SH, Guo XL, Liu GX, Wang LQ, et al. Identification and molecular characterization of exosome-like vesicles derived from the Taenia asiatica adult worm. Acta Trop (2019) 198:105036. doi: 10.1016/j.actatropica.2019.05.027
9. Zhang X, Gong W, Cao S, Yin J, Zhang J, Cao J, et al. Comprehensive analysis of non-coding RNA profiles of exosome-like vesicles from the protoscoleces and hydatid cyst fluid of Echinococcus granulosus. Front Cell Infect Microbiol (2020) 10:316. doi: 10.3389/fcimb.2020.00316
10. Wu Z, Wang L, Li J, Wang L, Wu Z, Sun X. Extracellular vesicle-mediated communication within host-parasite interactions. Front Immunol (2019) 9:3066. doi: 10.3389/fimmu.2018.03066
11. Twu O, Johnson PJ. Parasite extracellular vesicles: mediators of intercellular communication. PloS Pathog (2014) 10(8):e1004289. doi: 10.1371/journal.ppat.1004289
12. Wang Z, Hao C, Zhuang Q, Zhan B, Sun X, Huang J, et al. Excretory/Secretory products from Trichinella spiralis adult worms attenuated DSS-induced colitis in mice by driving PD-1-Mediated M2 macrophage polarization. Front Immunol (2020) 11:563784. doi: 10.3389/fimmu.2020.563784
13. Gao X, Yang Y, Liu X, Wang Y, Yang Y, Boireau P, et al. Extracellular vesicles derived from Trichinella spiralis prevent colitis by inhibiting M1 macrophage polarization. Acta Trop (2021) 213:105761. doi: 10.1016/j.actatropica.2020.105761
14. Liang PH, Li YP, Mao L, Liu TL, Zhang SH, Ehsan M, et al. Transcriptome analysis and autophagy investigation of LoVo cells stimulated with exosomes derived from t. asiatica adult worms. Microorganisms (2021) 9(5):994. doi: 10.3390/microorganisms9050994
15. Abou-El-Naga IF. Emerging roles for extracellular vesicles in schistosoma infection. Acta Trop (2022) 232:106467. doi: 10.1016/j.actatropica.2022.106467
16. Buck AH, Coakley G, Simbari F, McSorley HJ, Quintana JF, Le Bihan T, et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat Commun (2014) 5:5488. doi: 10.1038/ncomms6488
17. Cai MT, Ding JT, Li YT, He GT, Yang J, Liu TL, et al. Echinococcus multilocularis infection induces UBE2N suppression via exosomal emu-miR-4989. Acta Trop (2021) 223:106087. doi: 10.1016/j.actatropica.2021.106087
18. Tran N, Ricafrente A, To J, Lund M, Marques TM, Gama-Carvalho M, et al. Fasciola hepatica hijacks host macrophage miRNA machinery to modulate early innate immune responses. Sci Rep (2021) 11(1):6712. doi: 10.1038/s41598-021-86125-1
19. Wu J, Liao Y, Li DH, Zhu ZF, Zhang LC, Wu ZD, et al. Extracellular vesicles derived from Trichinella spiralis larvae promote the polarization of macrophages to M2b type and inhibit the activation of fibroblasts. Front Immunol (2022) 13:974332. doi: 10.3389/fimmu.2022.974332
20. Liu B, Li J, Cairns MJ. Identifying miRNAs, targets and functions. Brief Bioinform (2014) 15(1):1–19. doi: 10.1093/bib/bbs075
21. Li C, Wang Z, Chen S, Zhang J, Qu K, Liu C. MicroRNA-552 promotes hepatocellular carcinoma progression by downregulating WIF1. Int J Mol Med (2018) 42(6):3309–17. doi: 10.3892/ijmm.2018.3882
22. Kabekkodu SP, Shukla V, Varghese VK, D' Souza J, Chakrabarty S, Satyamoorthy K. Clustered miRNAs and their role in biological functions and diseases. Biol Rev Camb Philos Soc (2018) 93(4):1955–86. doi: 10.1111/brv.12428
23. He X, Pan W. Host-parasite interactions mediated by cross-species microRNAs. Trends Parasitol (2022) 38(6):478–488. doi: 10.1016/j.pt.2022.02.011
24. Zhou R, Gong AY, Eischeid AN, Chen XM. miR-27b targets KSRP to coordinate TLR4-mediated epithelial defense against Cryptosporidium parvum infection. PloS Pathog (2012) 8(5):e1002702. doi: 10.1371/journal.ppat.1002702
25. Jin X, Li Y, Yang X, Zheng Y. Modulatory effects of Echinococcus multilocularis emu-let-7-5p on the immunological functions of RAW264.7 macrophages. Front Vet Sci (2021) 8:663497. doi: 10.3389/fvets.2021.663497
26. Kawai T, Akira S. TLR signaling. Semin Immunol (2007) 19(1):24–32. doi: 10.1016/j.smim.2006.12.004
27. Zang R, Lian H, Zhong X, Yang Q, Shu HB. ZCCHC3 modulates TLR3-mediated signaling by promoting recruitment of TRIF to TLR3. J Mol Cell Biol (2020) 12(4):251–62. doi: 10.1093/jmcb/mjaa004
28. Hoppstädter J, Dembek A, Linnenberger R, Dahlem C, Barghash A, Fecher-Trost C, et al. Toll-like receptor 2 release by macrophages: an anti-inflammatory program induced by glucocorticoids and lipopolysaccharide. Front Immunol (2019) 10:1634. doi: 10.3389/fimmu.2019.01634
29. Rajasekaran S, Anuradha R, Bethunaickan R. TLR specific immune responses against helminth infections. J Parasitol Res (2017) 2017:6865789. doi: 10.1155/2017/6865789
30. Yan C, Fang F, Zhang YZ, Dong X, Wu J, Liu HL, et al. Recombinant CsHscB of carcinogenic liver fluke clonorchis sinensis induces IL-10 production by binding with TLR2. PloS Negl Trop Dis (2020) 14(10):e0008643. doi: 10.1371/journal.pntd.0008643
31. Wojtkowiak-Giera A, Derda M, Wandurska-Nowak E, Jagodziński PP, Kolasa-Wołosiuk A, Kosik-Bogacka D, et al. Changes in the expression of TLR2 during the intestinal phase of trichinellosis. J Vet Res (2020) 64(2):269–74. doi: 10.2478/jvetres-2020-0027
32. Reyes JL, González MI, Ledesma-Soto Y, Satoskar AR, Terrazas LI. TLR2 mediates immunity to experimental cysticercosis. Int J Biol Sci (2011) 7(9):1323–33. doi: 10.7150/ijbs.7.1323
33. Soleymani N, Taran F, Nazemshirazi M, Naghibi A, Torabi M, Borji H, et al. Dysregulation of ovine toll-like receptors 2 and 4 expression by hydatid cyst-derived antigens. Iran J Parasitol (2021) 16(2):219–28. doi: 10.18502/ijpa.v16i2.6271
34. Wang LQ, Liu TL, Chen GL, Li YP, Zhang SH, Mao L, et al. Exosomal microRNA let-7-5p from Taenia pisiformis cysticercus prompted macrophage to M2 polarization through inhibiting the expression of C/EBP δ. Microorganisms (2021) 9(7):1403. doi: 10.3390/microorganisms9071403
35. Deng Y, Ediriwickrema A, Yang F, Lewis J, Girardi M, Saltzman WM. A sunblock based on bioadhesive nanoparticles. Nat Mater (2015) 14(12):1278–85. doi: 10.1038/nmat4422
36. Li S, Yuan L, Su L, Lian Z, Liu C, Zhang F, et al. Decreased miR-92a-3p expression potentially mediates the pro-angiogenic effects of oxidative stress-activated endothelial cell-derived exosomes by targeting tissue factor. Int J Mol Med (2020) 46(5):1886–98. doi: 10.3892/ijmm.2020.4713
37. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods (2001) 25(4):402–8. doi: 10.1006/meth.2001.1262
38. Wang X, Ha T, Liu L, Hu Y, Kao R, Kalbfleisch J, et al. TLR3 mediates repair and regeneration of damaged neonatal heart through glycolysis dependent YAP1 regulated miR-152 expression. Cell Death Differ (2018) 25(5):966–82. doi: 10.1038/s41418-017-0036-9
39. Hua Z, Hou B. TLR signaling in b-cell development and activation. Cell Mol Immunol (2013) 10(2):103–6. doi: 10.1038/cmi.2012.61
40. He X, Jing Z, Cheng G. MicroRNAs: new regulators of toll-like receptor signalling pathways. BioMed Res Int (2014) 2014:945169. doi: 10.1155/2014/945169
41. Zhao Y, Dang Z, Chong S. Mmu-miR-92a-2-5p targets TLR2 to relieve Schistosoma japonicum-induced liver fibrosis. Int Immunopharmacol (2019) 69:126–35. doi: 10.1016/j.intimp.2019.01.007
42. Zhang K, Liu Y, Zhang G, Wang X, Li Z, Shang Y, et al. Molecular characteristics and potent immunomodulatory activity of Fasciola hepatica cystatin. Korean J Parasitol (2022) 60(2):117–26. doi: 10.3347/kjp.2022.60.2.117
43. Yang X, Liu J, Yue Y, Chen W, Song M, Zhan X, et al. Cloning, expression and characterisation of a type II cystatin from Schistosoma japonicum, which could regulate macrophage activation. Parasitol Res (2014) 113(11):3985–92. doi: 10.1007/s00436-014-4064-9
44. Weingartner M, Stücheli S, Jebbawi F, Gottstein B, Beldi G, Lundström-Stadelmann B, et al. Albendazole reduces hepatic inflammation and endoplasmic reticulum-stress in a mouse model of chronic Echinococcus multilocularis infection. PloS Negl Trop Dis (2022) 16(1):e0009192. doi: 10.1371/journal.pntd.0009192
45. Christen U, Hintermann E. Pathogens and autoimmune hepatitis. Clin Exp Immunol (2019) 195(1):35–51. doi: 10.1111/cei.13203
46. Lerner A, Arleevskaya M, Schmiedl A, Matthias T. Microbes and viruses are bugging the gut in celiac disease. are they friends or foes? Front Microbiol (2017) 8:1392. doi: 10.3389/fmicb.2017.01392
47. Gazzinelli-Guimaraes PH, Nutman TB. Helminth parasites and immune regulation. F1000Res (2018) 7:F1000 Faculty Rev–1685. doi: 10.12688/f1000research.15596.1
Keywords: rabbit, Cysticercosis pisiformis, novel-miR1, TLR2, NFκB
Citation: Chen G, Pu G, Wang L, Li Y, Liu T, Li H, Zhang S, Wang X, Liu X and Luo X (2023) Cysticercus pisiformis-derived novel-miR1 targets TLR2 to inhibit the immune response in rabbits. Front. Immunol. 14:1201455. doi: 10.3389/fimmu.2023.1201455
Received: 06 April 2023; Accepted: 26 June 2023;
Published: 25 July 2023.
Edited by:
Xiang Wu, Central South University, ChinaReviewed by:
Maria Elizbeth Alvarez Sanchez, Universidad Autónoma de la Ciudad de México, MexicoShuaiqin Huang, Central South University, China
Copyright © 2023 Chen, Pu, Wang, Li, Liu, Li, Zhang, Wang, Liu and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuenong Luo, bHVveHVlbm9uZ0BjYWFzLmNu
†These authors have contributed equally to this work
 Guoliang Chen
Guoliang Chen Guiting Pu
Guiting Pu Liqun Wang
Liqun Wang Yanping Li
Yanping Li Tingli Liu
Tingli Liu Hong Li
Hong Li Shaohua Zhang1
Shaohua Zhang1 Xuelin Wang
Xuelin Wang Xuenong Luo
Xuenong Luo