
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 04 September 2023
Sec. Inflammation
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1197752
This article is part of the Research Topic Inflammation in Respiratory and Neurological Diseases and the immune-interaction of the lung-brain axis View all 17 articles
 Hui Zhao1,2†
Hui Zhao1,2† Lan Wang1†
Lan Wang1† Yi Yan1,3†
Yi Yan1,3† Qin-Hua Zhao1
Qin-Hua Zhao1 Jing He1
Jing He1 Rong Jiang1
Rong Jiang1 Ci-Jun Luo1
Ci-Jun Luo1 Hong-Ling Qiu1
Hong-Ling Qiu1 Yu-Qing Miao2
Yu-Qing Miao2 Su-Gang Gong1*
Su-Gang Gong1* Ping Yuan1*
Ping Yuan1* Wen-Hui Wu1*
Wen-Hui Wu1*Pulmonary fibrosis (PF) and pulmonary hypertension (PH) have common pathophysiological features, such as the significant remodeling of pulmonary parenchyma and vascular wall. There is no effective specific drug in clinical treatment for these two diseases, resulting in a worse prognosis and higher mortality. This study aimed to screen the common key genes and immune characteristics of PF and PH by means of bioinformatics to find new common therapeutic targets. Expression profiles are downloaded from the Gene Expression Database. Weighted gene co-expression network analysis is used to identify the co-expression modules related to PF and PH. We used the ClueGO software to enrich and analyze the common genes in PF and PH and obtained the protein–protein interaction (PPI) network. Then, the differential genes were screened out in another cohort of PF and PH, and the shared genes were crossed. Finally, RT-PCR verification and immune infiltration analysis were performed on the intersection genes. In the result, the positive correlation module with the highest correlation between PF and PH was determined, and it was found that lymphocyte activation is a common feature of the pathophysiology of PF and PH. Eight common characteristic genes (ACTR2, COL5A2, COL6A3, CYSLTR1, IGF1, RSPO3, SCARNA17 and SEL1L) were gained. Immune infiltration showed that compared with the control group, resting CD4 memory T cells were upregulated in PF and PH. Combining the results of crossing characteristic genes in ImmPort database and RT-PCR, the important gene IGF1 was obtained. Knocking down IGF1 could significantly reduce the proliferation and apoptosis resistance in pulmonary microvascular endothelial cells, pulmonary smooth muscle cells, and fibroblasts induced by hypoxia, platelet-derived growth factor-BB (PDGF-BB), and transforming growth factor-β1 (TGF-β1), respectively. Our work identified the common biomarkers of PF and PH and provided a new candidate gene for the potential therapeutic targets of PF and PH in the future.
Pulmonary hypertension (PH), defined as a mean pulmonary artery pressure (mPAP) >20 mmHg at rest (1), is a major global health issue, with an estimated prevalence of ~1% in the population worldwide and 10% in individuals aged >65 years (1). PH is often accompanied by pulmonary fibrosis (PF). In idiopathic pulmonary fibrosis (IPF), 8%–15% of patients have PH at diagnosis, and the prevalence is raised to 30%–50% in the advanced stage and >60% at the end stage of the disease. PH significantly complicates the course of PF and heralds an unfavorable disease prognosis (2, 3). As a well-known fatal lung disease, the average life expectancy of IPF is 3–5 years after diagnosis (4, 5), and the mortality climbs even higher when pulmonary vascular remodeling is involved (6–8). Currently, although several drugs targeting group 1 PH [pulmonary arterial hypertension (PAH)] have been studied in patients with PF associated with PH, the results are inconsistent, with some studies indicative of ineffective or even harmful impact (9–11), which unfortunately left lung transplantation the only durable treatment for PH secondary to PF (6). Therefore, finding a new common therapeutic target for PF-PH is particularly important.
The pathological feature of PF associated with PH involves an overexuberant fibroproliferative process gradually destructing the normal lung architecture and sustained remodeling in the pulmonary vasculature. Even though the underlying pathophysiology is complex and unique, accumulating evidence indicates that the development of fibrosis and vascular obliteration (both in PAH and secondary PH) share several similar mechanisms involving epigenetics, metabolism, inflammation, immunity, DNA damage, and oxidative stress (6, 12–14). We have recently reported that targeting checkpoint kinases 1/2 exhibited a promising antifibrotic and antiproliferative activity in PF and pulmonary vascular remodeling (13, 14). However, the pathological origins of PF and PH remain undetermined, and understanding at the systems level and developing novel therapeutic strategies remain highly recommended.
With rapid advances in sequencing technology (15), it is available to measure the expression of thousands of genes in various diseases, which helps people understand the pathogenesis of diseases in depth from the gene level (16). In this study, we identified co-expression modules and shared genes of PF and PH by using publicly available gene expression data from the NCBI Gene Expression Omnibus (GEO) database. The results revealed that PF and PH shared 313 genes in discovery cohorts, and they were mainly enriched in lymphocyte activation. We further validated that IGF1 was particularly noteworthy, which may serve as the key modulator and potential therapeutic target in PF and PH in the future.
The lung tissues from six PF-PH and five healthy subjects were obtained from patients undergoing an open lung biopsy or lung transplant in Shanghai Pulmonary Hospital in the period from 2020 to 2022. The diagnosis of PF-PH was established from the European Society of Cardiology and the European Respiratory Society guidelines (1). All of the experimental procedures using human tissues were approved and supervised by the Ethics Committee of Shanghai Pulmonary Hospital (numbers: K22-137Y). Written informed consent was obtained from all participants.
We used “pulmonary arterial hypertension” or “idiopathic pulmonary fibrosis,” the two typical diseases for pulmonary vascular remodeling and PF, as keywords to search in the GEO database (https://www.ncbi.nlm.nih.gov/geo/) and obtain the expression profiles. Database inclusion criteria were as follows. First, the sequencing methods of the selected database are expression profiling by array, and the dataset must contain both control and disease groups. Second, the tissue sources used for sequencing were all human lung tissue. Third, the number of samples should be sufficient to ensure the accuracy of each analysis, and the database must provide either processed or raw data that can be used for reanalysis. Finally, we selected GSE53845, GSE113439, GSE110147, and GSE15197 from the GEO database. Subsequently, we used R 4.2.0 software to convert probes in GSE53845 and GSE113439 into gene symbols. We provide all involved R programs in Supplementary Table 1.
PF- and PH-related modules were gained by weighted gene co-expression network analysis (WGCNA) (17), and WGCNA was carried out using SangerBox (sangerbox.com). The gene cluster map and the correlation heat map between modules and phenotypes were obtained.
We selected the modules highly correlated with PF and PH and screened the overlapping shared genes in the modules that are positively associated with PF and PH. ClueGO (18) explored the potential role of these shared genes in PF and PH and made a biological analysis of these shared genes. A p-value <0.05 was considered significant. Protein–protein interaction (PPI) network analysis used STRING (string-db.org), and the “MCODE” algorithm in Cytoscape software was used to realize visualization.
To further identify potential functional genes for PF and PH, differential expression analysis was performed using GEO2R between patients and healthy controls in GSE110147 or GSE15197. The screening threshold of differentially expressed genes (DEGs) was set at a log2 |fold change (FC)| >1 and a p-value <0.05. Overlapping DEGs in PF and PH databases were acquired by Venn. A set of immunity-related genes was achieved from the ImmPort database (19) and crossed with identified common genes by Venn mapping.
The normalized GSE53845 and GSE113439 expression data were analyzed by CIBERSORT (https://cibersort.stanford.edu/) to evaluate enrichment for immune invasions and to obtain an immunocyte infiltration matrix. Furthermore, we displayed the percentage of each immunocyte in the samples in a histogram, constructed a heatmap of 22 kinds of immunocytes, and compared the levels of 22 immunocytes between patients and controls using SangerBox. Spearman’s rank correlation analysis was used to analyze the relationship between immunocytes and feature genes. A p-value <0.05 was considered statistically significant.
Lung tissue samples were obtained from patients with PF-PH who underwent a lung transplant and healthy donors (controls) and stored at −80°C until use. Total RNA was extracted from the lung tissue using TRIzol reagent. RNA purity was determined using the NanoDrop 2000 spectrophotometer at 260/280 nm (ratio = 1.9–2.1). Then, reverse transcription using the PrimeScript™ RT reagent Kit (TaKaRa, RR0371, Japan) was performed according to the manufacturer’s instructions. RT-PCR was then performed with the SYBR Green-based RT-PCR (TOYOBO, QPK201, Japan). For RT-PCR analysis, the expression level of genes was represented as a fold change using the 2−△△Ct method, and a p-value <0.05 was considered statistically significant. Primer sequences are listed in Supplementary Table S2.
Human pulmonary microvascular endothelial cells (PMECs), human pulmonary artery smooth muscle cells (PASMCs), and human fibroblasts were purchased from ScienceCell (Shanghai, China) and cultured in endothelial cell medium, smooth muscle cell medium, and Dulbecco’s modified Eagle’s medium, respectively, and incubated with 5% CO2 at 37°C. Hypoxia-induced PMECs were acquired in anoxic incubators with 5% O2 at 37°C. PASMCs and fibroblasts were induced to proliferate with 10 ng/mL platelet-derived growth factor-BB (PDGF-BB) (PeproTech, USA) and 5 ng/mL transforming growth factor-β1 (TGF-β1) (PeproTech, USA), respectively.
Cells were transfected with 20 μM siRNA IGF1 (Genomeditch, Shanghai, China) using Lipo2000 Transfection Reagent (Invitrogen, USA). The control groups were treated with equal concentrations of negative control sequences to eliminate nonspecific effects.
Cell Counting Kit-8 (CCK-8, Dojindo, Japan) was used to measure cell proliferation. Cells were inoculated into a 96-well plate at a density of 3 × 103 cells per well. After 48 h of siRNA IGF1 treatment under the stimulation of proliferation induction, cells were treated with 10 μL CCK-8 solution at 37°C for 60 min and then tested. The absorbance at 450 nm was then detected, with a relative proliferation level obtained by normalization from five independent experiments.
In this study, 2 × 103 treated cells were planted in each well of a 96-well plate. After 48 h of treatment, cell proliferation was measured by EdU Kit (BeyoClick™, China). Hoechst (blue) labeled the nucleus and EdU (green) labeled the proliferating cells, and these were observed and counted under the fluorescence microscope.
The Annexin V apoptosis kit (Dojindo, Japan) was used to detect the apoptosis of cells. In this study, 2 × 105 cells were planted into each well of a 6-well plate with or without treatment. After 48 h, cells were digested with trypsin, and cells were washed with phosphate buffered saline (PBS) three times. Then, 500 μL of 1× Annexin V binding solution was added to the washed cells to prepare a cell suspension, and 3 μL of Annexin V-FITC and PI were added, respectively. The samples were mixed thoroughly and were left in the dark for 15 min. Finally, the apoptosis was analyzed by flow cytometry.
The cells were analyzed for fluorescence by Calcein-AM/PI staining (Calcein-AM/PI Double Stain Kit, YESEN, China). Cells were seeded in 24-well plates (2 × 105 cells) for 48 h with or without treatment. Dyeing solution was added, and the cells were incubated at 37°C for 30 min. Finally, the cells were analyzed using a fluorescence microscope.
In order to further verify the ability of protein IGF1 to target drugs, molecular docking was carried out. Firstly, a database was selected in The NCGC Pharmaceutical Collection, which contained 7,929 FDA-approved drugs. The crystal structure of 2OJ9 was downloaded from the PDB database as the receptor protein structure for molecular docking. Secondly, the Discovery Studio LibDock program was used for molecular docking screening, and the top 20 compounds of LibDock score were selected for further study. Subsequently, for the top 20 compounds, the ligands and protein needed for molecular docking were prepared by AutoDock Vina software (http://vina.scripps.edu/), and for the target protein, the crystal structure obtained from the PDB database (https://www.rcsb.org/) needed pretreatment. Finally, the target structure was molecularly docked with the active ingredient structure, and the affinity value of the target structure is the binding ability of the target structure and the active ingredient structure using vina in pyrx software (https://pyrx.sourceforge.io/).
SangerBox (sangerbox.com) was used for all statistical analyses. Differences between the two groups were compared using parametric tests for normally distributed variables. All of the statistical differences were p-value <0.05.
Based on the screening criteria mentioned in the method, we selected four GEO datasets numbered GSE53845, GSE110147, GSE113439, and GSE15197. Furthermore, GSE53845 (20) and GSE113439 (21) served as discovery queues for WGCNA. A concise description of these samples was summarized in Supplementary Table S3. Additionally, GSE110147 and GSE15197 served as validation queues for DEG analysis.
We identified eight relevant modules for GSE53845 through WGCNA, with each color representing a different module. A heat map of the module–trait relationship was then drawn based on the correlation coefficients to assess the association of each module with disease (Figures 1A, C). Among them, the “red” module (r = 0.81, p = 4.9e-12) had a high correlation with PF and was selected as the module with the highest positive correlation with PF, including 1,260 genes. Similarly, in GSE113439, module “pink” (r = 0.89, p = 1.8e-9) was the module with the highest positive correlation with PH, including 5,013 genes (Figures 1B, D).
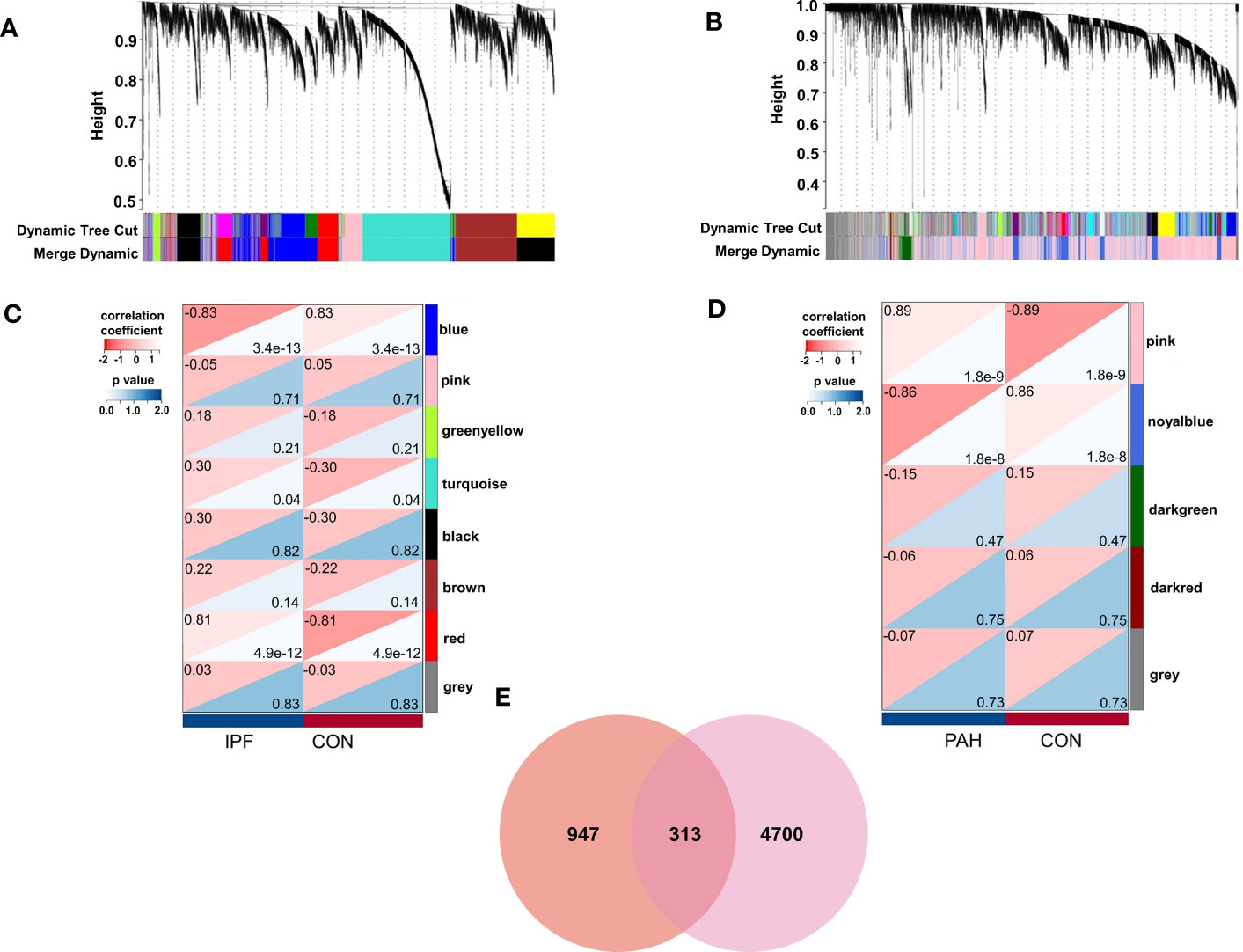
Figure 1 Weighted gene co-expression network analysis (WGCNA). (A) The cluster dendrogram of co-expression genes in PF. (B) The cluster dendrogram of co-expression genes in PH. (C) Module–trait relationships in PF. (D) Module–trait relationships in PH. (E) Shared genes overlap between the red module of PF and the pink module of PH. IPF, idiopathic pulmonary fibrosis; PAH, pulmonary arterial hypertension; CON, control.
Among PF and PH, 313 genes in the most relevant positive correlation module overlapped and were defined as gene set 1 (Figure 1E). To explore the potential function of gene set 1, Gene Ontology (GO) enrichment was analyzed using ClueGO. The first significantly enriched GO terms for Biological Process (BP) are “lymphocyte activation,” which accounted for 53.12% of the total GO terms (Figures 2A, B ), confirming that this pathway may be essential in both PF and PH. Then, we constructed a protein-level PPI network for dataset 1 (Figures 3A-D). Four clusters were extracted using MCODE analysis, and cluster 1 contains 60 nodes and 759 edges (score = 25.73). We performed functional enrichment analysis on the genes of the four clusters respectively and selected the first 10 immune-related pathways (Figure 3E). GO enrichment analysis showed that the genes in cluster 1 were mainly related to the lymphocyte activation pathway; this was consistent with the enrichment of dataset 1. Therefore, this gene cluster belongs to the gene part shared by PF in PH.
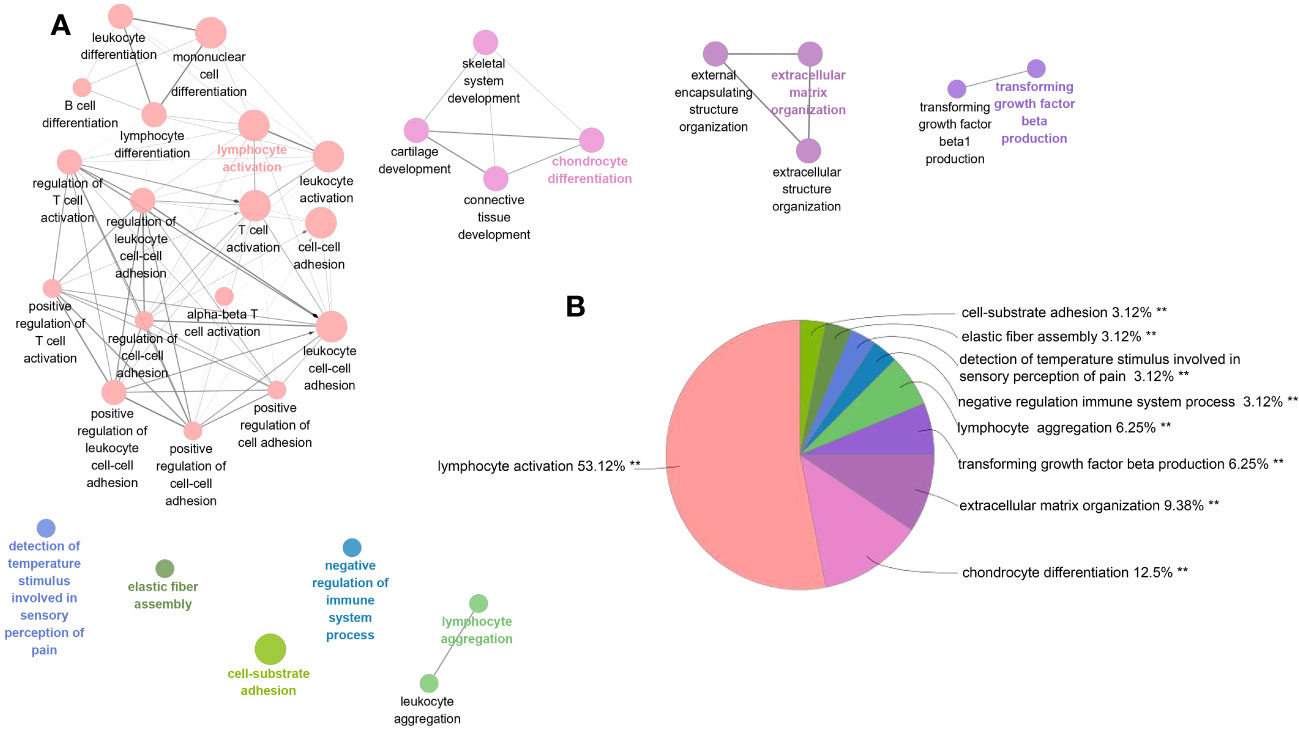
Figure 2 ClueGO enrichment analysis of dataset 1. (A) The interaction network of GO terms generated by the Cytoscape plug-in ClueGO. The significant term of each group is highlighted. (B) Proportion of each GO terms group in the total. GO, gene ontology. **p < 0.05.
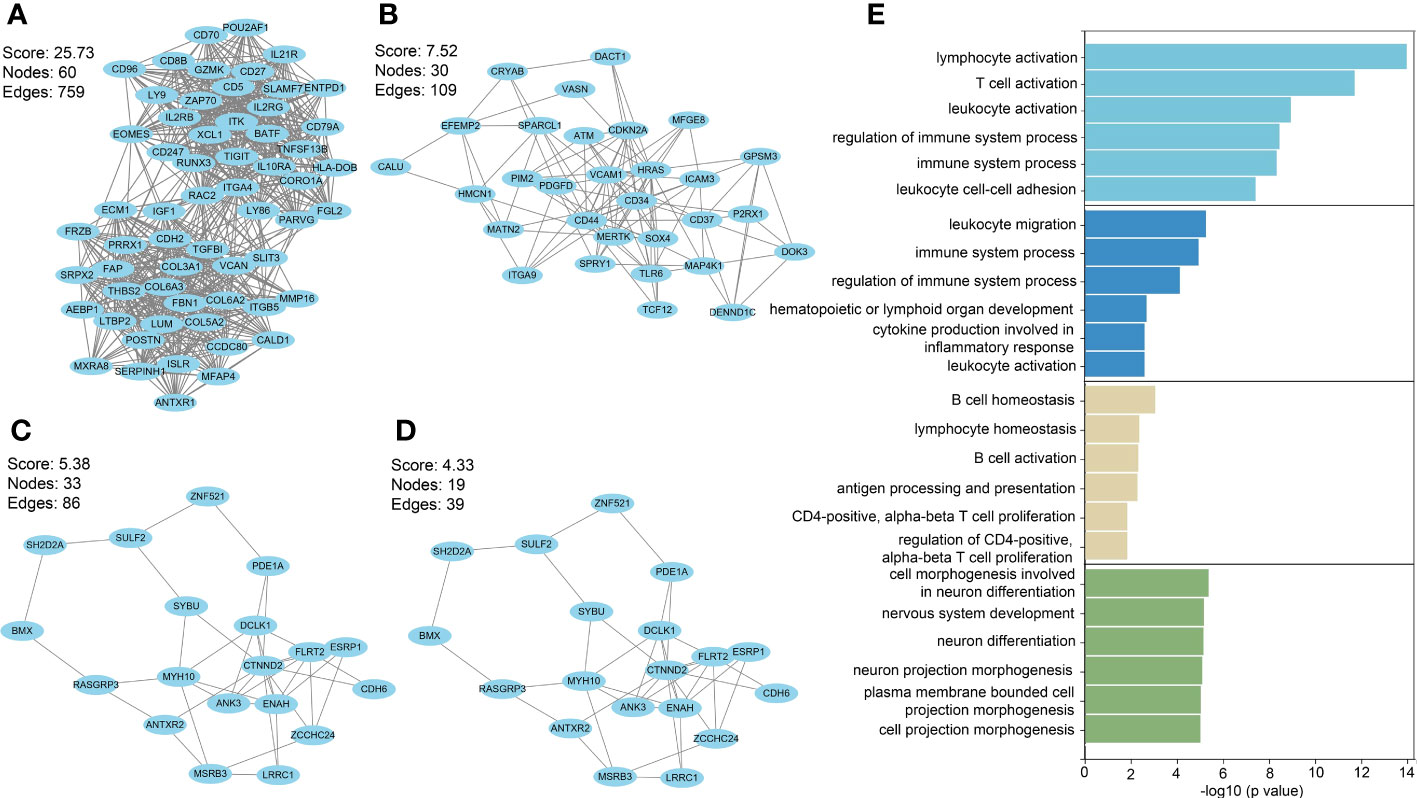
Figure 3 (A–D) PPI network analysis was performed on gene set 1, and four clusters were identified. (E) GO analysis was performed on each cluster.
We performed DEG analyses of GSE110147 and GSE15197 to validate our results. Among the 2,579 DEGs in GSE110147, 1,213 were significantly downregulated and 1,366 were significantly upregulated. In GSE15197, 3,497 DEGs were identified, including 1,328 downregulated and 2,169 upregulated genes. Further cross-analysis of GSE110147 and GSE15197 revealed that there were 32 co-downregulated genes and 165 co-upregulated genes in PF and PH, defined as gene set 2 and expressed by Venn diagram (Figures 4A, B). There were eight overlapped genes, ACTR2, COL5A2, COL6A3, CYSLTR1, IGF1, RSPO3, SCARNA17 and SEL1L, in GS1 and GS2 (Figure 4C). Notably, COL5A2, IGF1, and COL6A3 are contained in cluster 1, and RSPO3 is contained in cluster 3. At the same time, the expression level of the overlapped genes in the four datasets was visualized (Figures 4E-H).
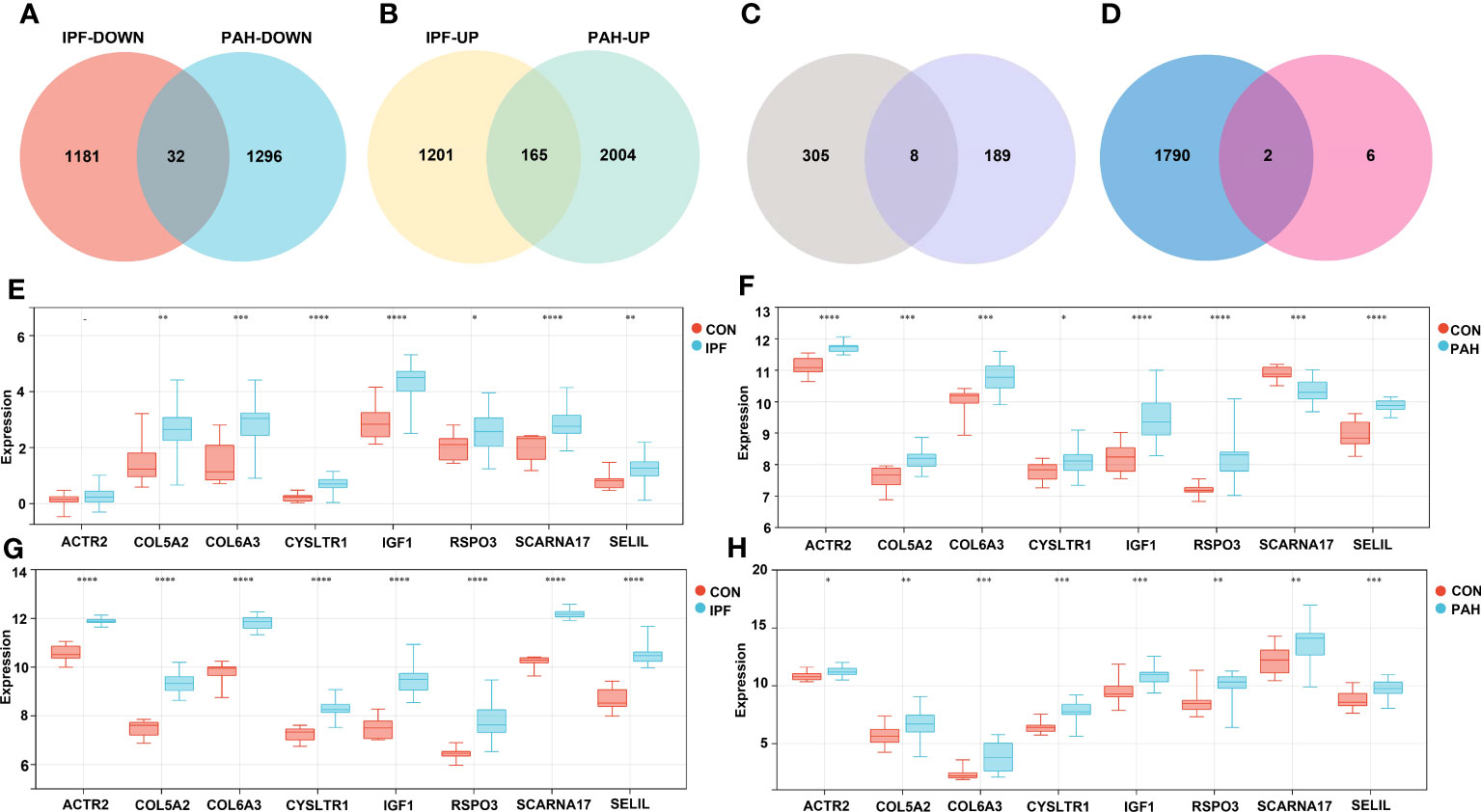
Figure 4 Validation Group: (A, B) Differential gene analysis of PF dataset GSE110147 and PH dataset GSE15197, respectively: downregulated intersection gene and upregulated intersection gene. (C) The intersection gene set 1 and gene set 2 were selected, and there were eight key genes. (D) The intersection of immune-related genes with the eight key genes was retrieved in the ImmPort database. Analysis of expression levels of eight key genes in the verification group of the discovery group: GSE53845 (E), GSE113439 (F), GSE110147 (G), and GSE15197 (H). IPF, idiopathic pulmonary fibrosis; PAH, pulmonary arterial hypertension; CON, control. **p < 0.01; ***p < 0.001; ****p < 0.0001.
In the enrichment assay, immune-related biological functions were significantly enriched in PF and PH. Therefore, we further performed CIBERSORT to predict immune cell infiltration between patients with the disease and control groups. The percentage of each of the 22 immune cells in each sample was shown in the bar and heat graphs of GSE53845 (Figures 5A, B) and GSE113439 (Figures 5D, E). The box diagram of the differences in immunocyte infiltration when samples were classified into disease and control groups showed significant differences of T cells CD4 memory resting, T cells CD4 memory activated, T cells regulatory (Tregs), T cells gamma delta, natural killer (NK) cells resting, Macrophages M1, Macrophages M2, and Neutrophils in PF patients compared with those in controls (Figure 5C). Compared with the control group, there were significant differences in T cells CD8, T cells CD4 memory activated, T cells follicular helper, NK cell activated, Mast cells resting, Eosinophils, and Neutrophils in patients with PH (Figure 5F). As can be seen from the results, T cells CD4 memory activated was a common significantly different immune feature to PF and PH. In addition, IGF1 and CYSLTR1 were the shared genes between 1,792 immune-related genes retrieved from the ImmPort database and eight common genes (Figure 4D). IGF1 and CYSLTR1 were also related to several immune cells (Figure 6).
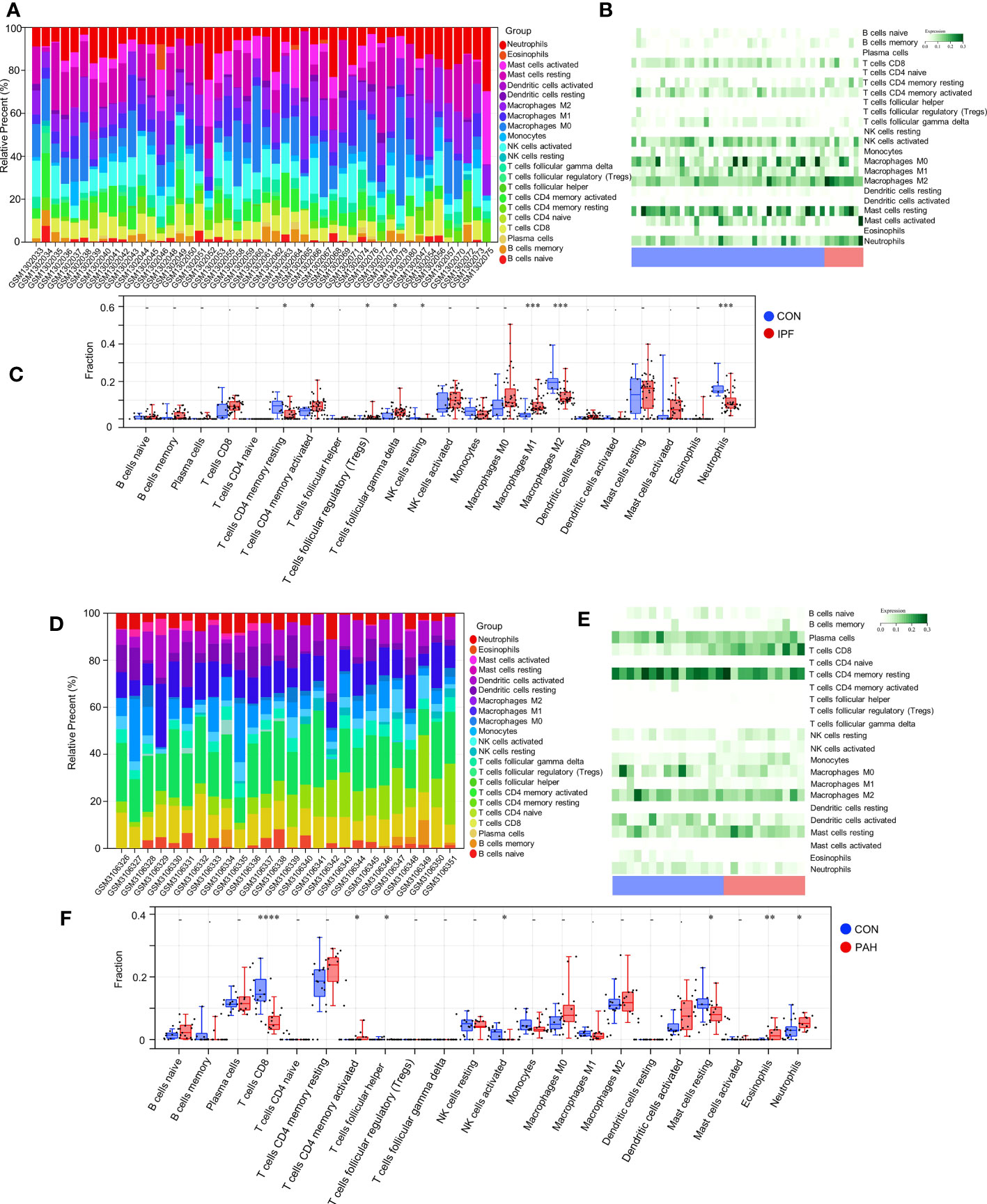
Figure 5 Immune infiltrates between PF and normal controls: (A) Relative percentage of 22 immune cells; (B) Heat map of 22 immune cells; (C) Difference in immune infiltrates between PH and normal controls. Immune infiltrates between PH and normal controls: (D) Relative percentages of 22 immune cells; (E) Heat maps of 22 immune cells; (F) Differences in immune infiltrates between PH and normal controls. IPF, idiopathic pulmonary fibrosis; PAH, pulmonary arterial hypertension; CON, control. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
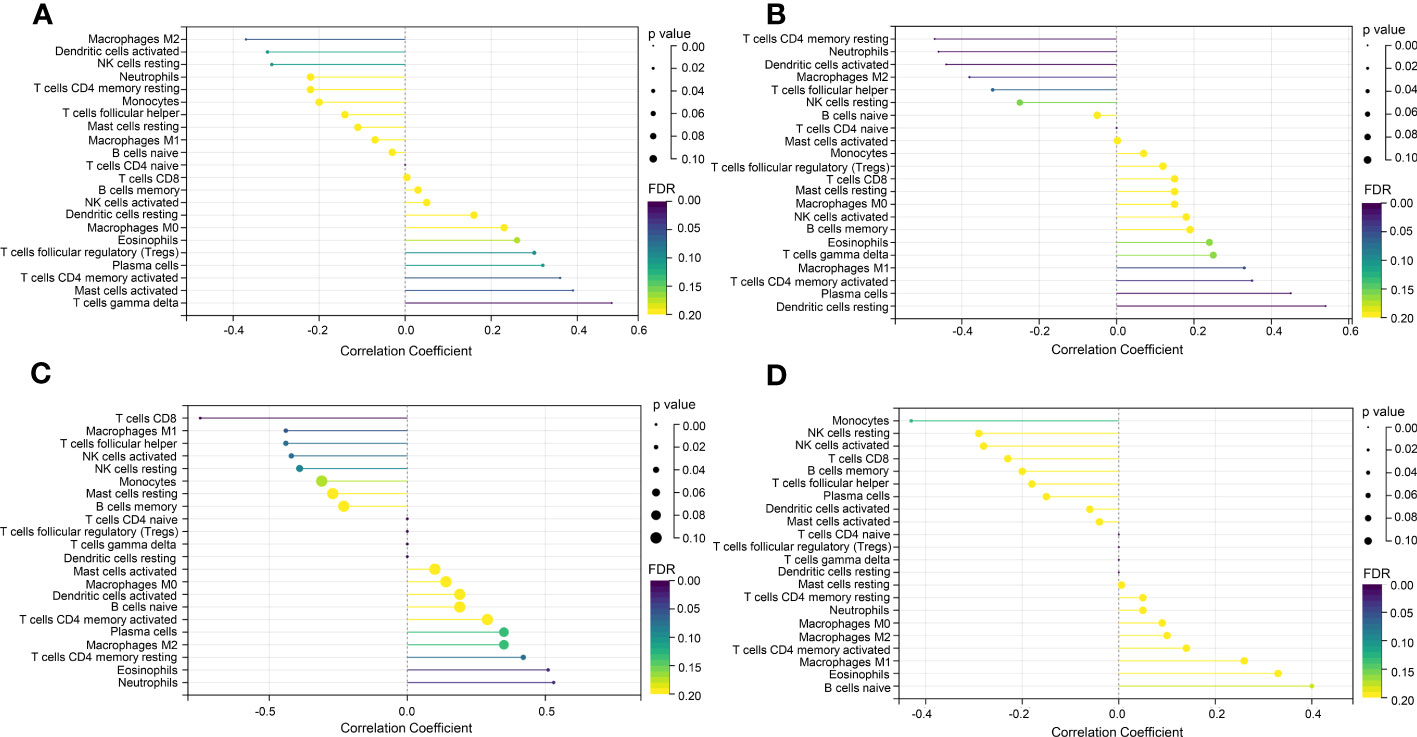
Figure 6 Correlation between IGF1 and CYSLTR1 and immune cells. (A) Correlation between IGF1 and immune cells (in GSE53845). (B) Correlation between CYSLTR1 and immune cells (in GSE53845). (C) Correlation between IGF1 and immune cells (in GSE113439). (D) Correlation between CYSLTR1 and immune cells (in GSE113439). The length of the line represents the correlation between genetic biomarkers and immune cells. The size and color of the points represent p-values and FDR, respectively, and the smaller the point, the greater the difference.
To further demonstrate the expression of overlapping genes in PF-PH patients, we performed RT-PCR in six lung tissue samples from PF-PH patients and five healthy donors. The results showed that IGF1 and COL5A2 expression was distinctly increased in PF-PH samples compared with healthy samples (Figure 7), which is consistent with previous findings in DEGs of PF and PH.
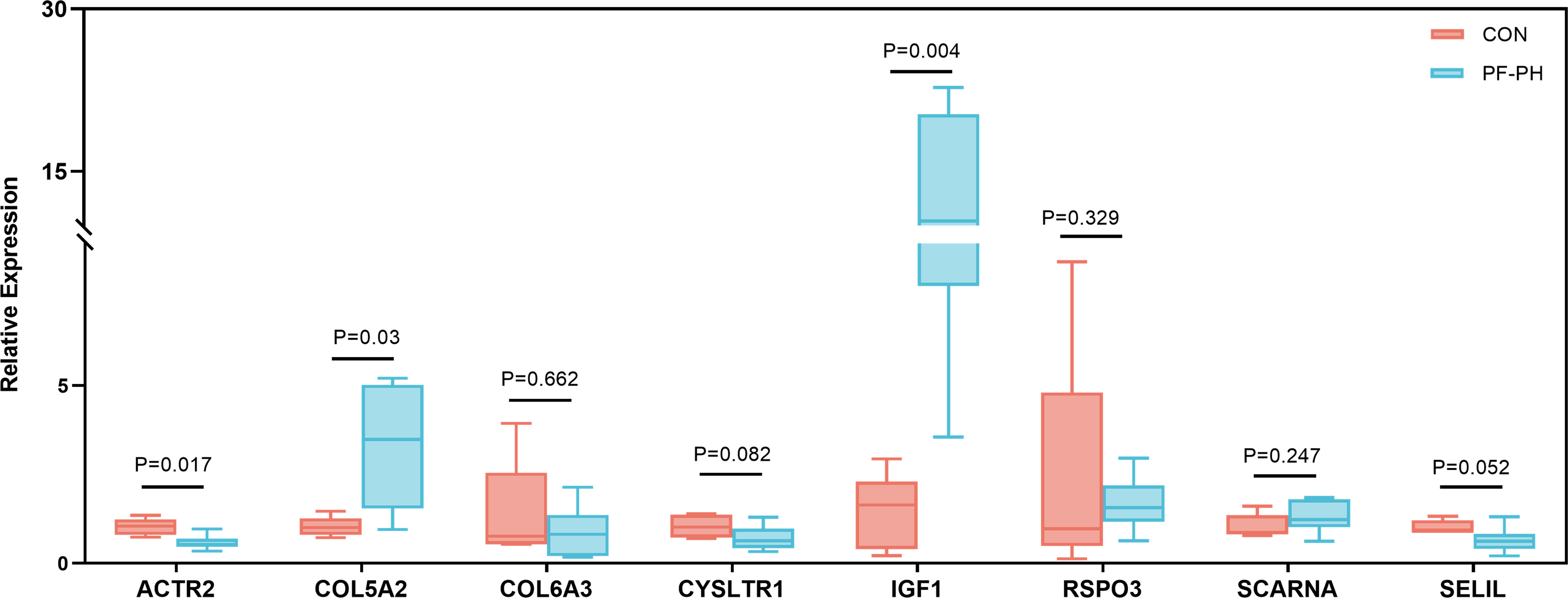
Figure 7 RT-PCR for the expression of common genes of PF and PH in PF-PH patients and healthy donors. PF, pulmonary fibrosis; PH, pulmonary hypertension; CON, control.
In view of the upregulation of IGF1 in lung tissue of patients with PF-PH, we used siRNA IGF1 to reduce the expression of IGF in PMECs, PASMCs, and fibroblasts to further study the effect of IGF1 on cell function. Hypoxia, PDGF-BB, and TGF-β1 were used to induce the proliferation of PMECs, PASMCs, and fibroblasts, respectively. As shown in Figures 8A–C, the cell proliferation was increased obviously after stimulation and significantly inhibited after adding siRNA IGF1. In addition, we used EdU Kit to verify the cell proliferation, and the results were consistent (Figures 8D–F). Then, using flow cytometry and AM/PI Kit, we identified that siRNA IGF1 could reduce the apoptosis rate and death rate promoted by hypoxia, PDGF-BB, and TGF-β1 in PMECs (Figures 9A, D), PASMCs (Figures 9B, E), and fibroblasts (Figures 9C, F), respectively.
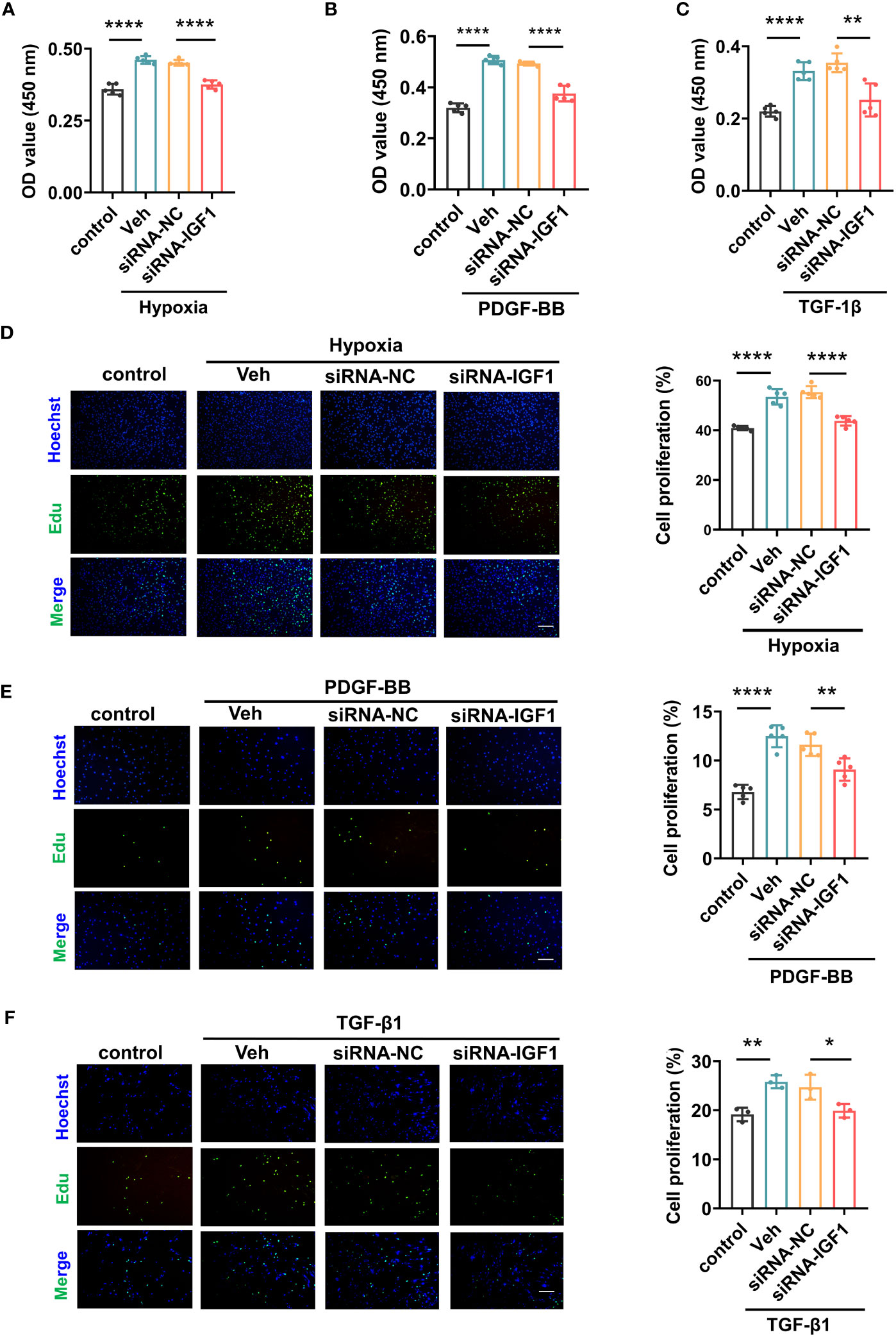
Figure 8 Effects of IGF1 on the proliferation of PMECs, PASMCs, and fibroblasts. The CCK-8 Kit test proliferation of PMECs (A), PASMCs (B), and fibroblasts (C) with siRNA IGF1. The EdU Kit test proliferation and analysis results of PMECs (D), PASMCs (E), and fibroblasts (F). All data are presented as the mean ± SEM. Scale bar 100 μm. *p < 0.05; **p < 0.01; ****p < 0.0001. Veh, vehicle; PDGF-BB, platelet-derived growth factor-BB; TGF-β1, transforming growth factor-β1.
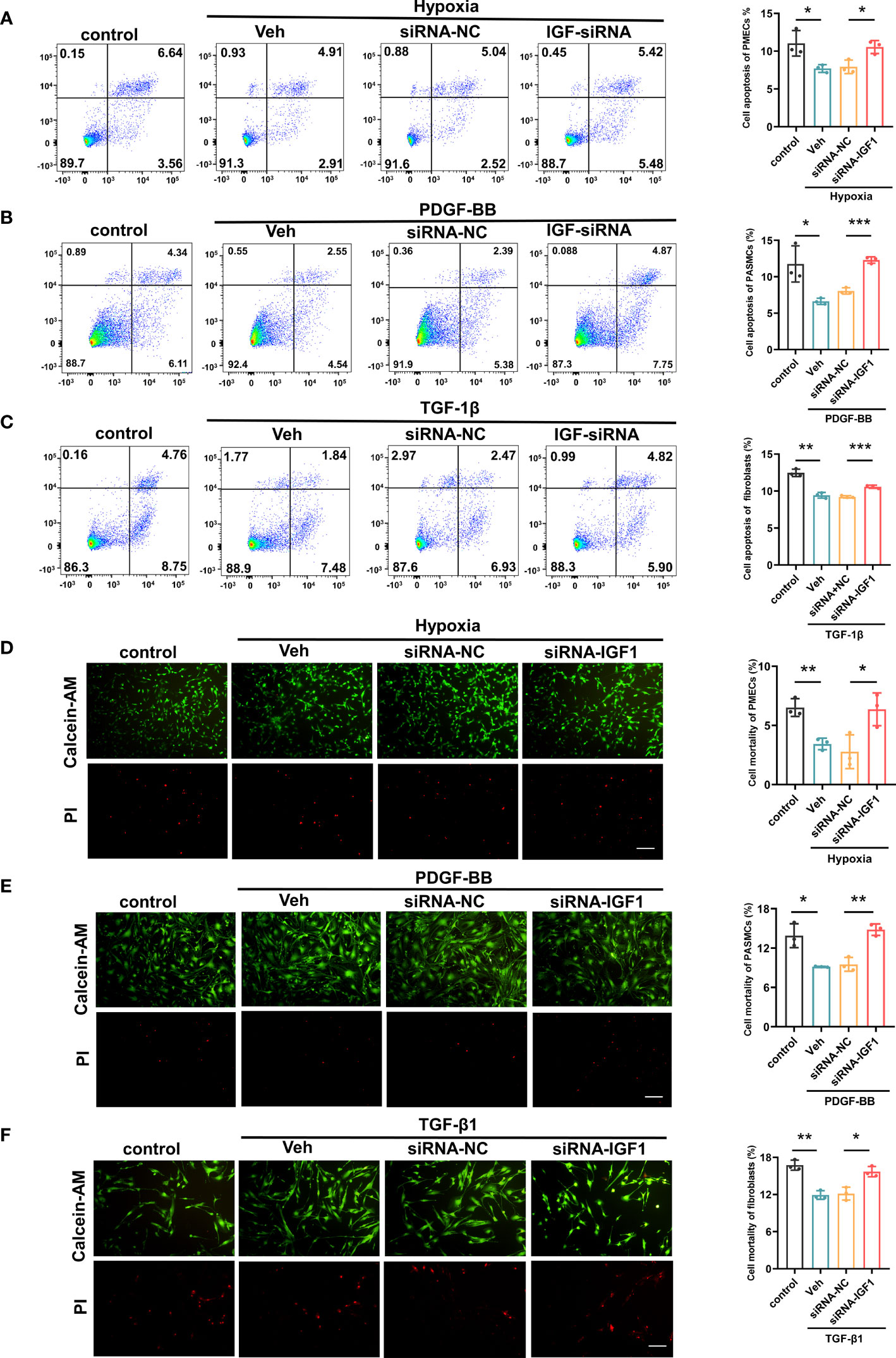
Figure 9 Effects of IGF1 on the apoptosis and mortality of PMECs, PASMCs, and fibroblasts. The flow cytometry test apoptosis of PMECs (A), PASMCs (B), and fibroblasts (C) with siRNA IGF1. The AM/PI Kit test mortality and analysis results of PMECs (D), PASMCs (E), and fibroblasts (F). All data are presented as the mean ± SEM. Scale bar 100 μm. *p < 0.05; **p < 0.01; ***p < 0.001. Veh, vehicle; PDGF-BB, platelet-derived growth factor-BB; TGF-β1, transforming growth factor-β1.
The LibDock docking results showed that 2,720 compounds in 7,929 were successfully docked to protein IGF1. The higher the LibDock score, the closer the interaction between molecules and proteins. We selected the top 20 compounds of LibDock score (Table 1) to enter the next research. Subsequently, the lower the binding ability, the more stable the binding between the ligand and the receptor (the lower the binding energy, the better the binding) in vina docking results. Therefore, among the first 20 compounds, we screened out the compounds with binding energy less than -7.0 kcal/mol by vina (22), namely, deslanoside (Figure 10A), flavitan (Figure 10C), ivermectin (Figure 10E), and posaconazole (Figure 10G), which have strong binding activity with IGF1. The results also show the interaction between these drugs and IGF1. Deslanoside forms hydrogen bonds (HBs) with ARG1062, GLU985, and ARG973 of IGF1 and forms alkyl bonds with ARG1062 and ARG1054 (Figure 10B). Six HBs were formed between flavitan and IGF1, namely, SER1059, SER1063, ARG1054, GLU1115, GLU985, and GLU974, and also formed alkyl with ILE1130 and pi-alkyl with ARG1062 (Figure 10D). Ivermectin forms HBs with GLU985 and ASP1056 of IGF1 and alkyl with ARG1054 (Figure 10F). Posaconazole forms HBs with ILE1136, ARG1062, and SER1059 of IGF1, alkyl with ARG1054, pi-pi stacked with TYR1131, and halogen with GLU974 and GLU985 (Figure 10H).
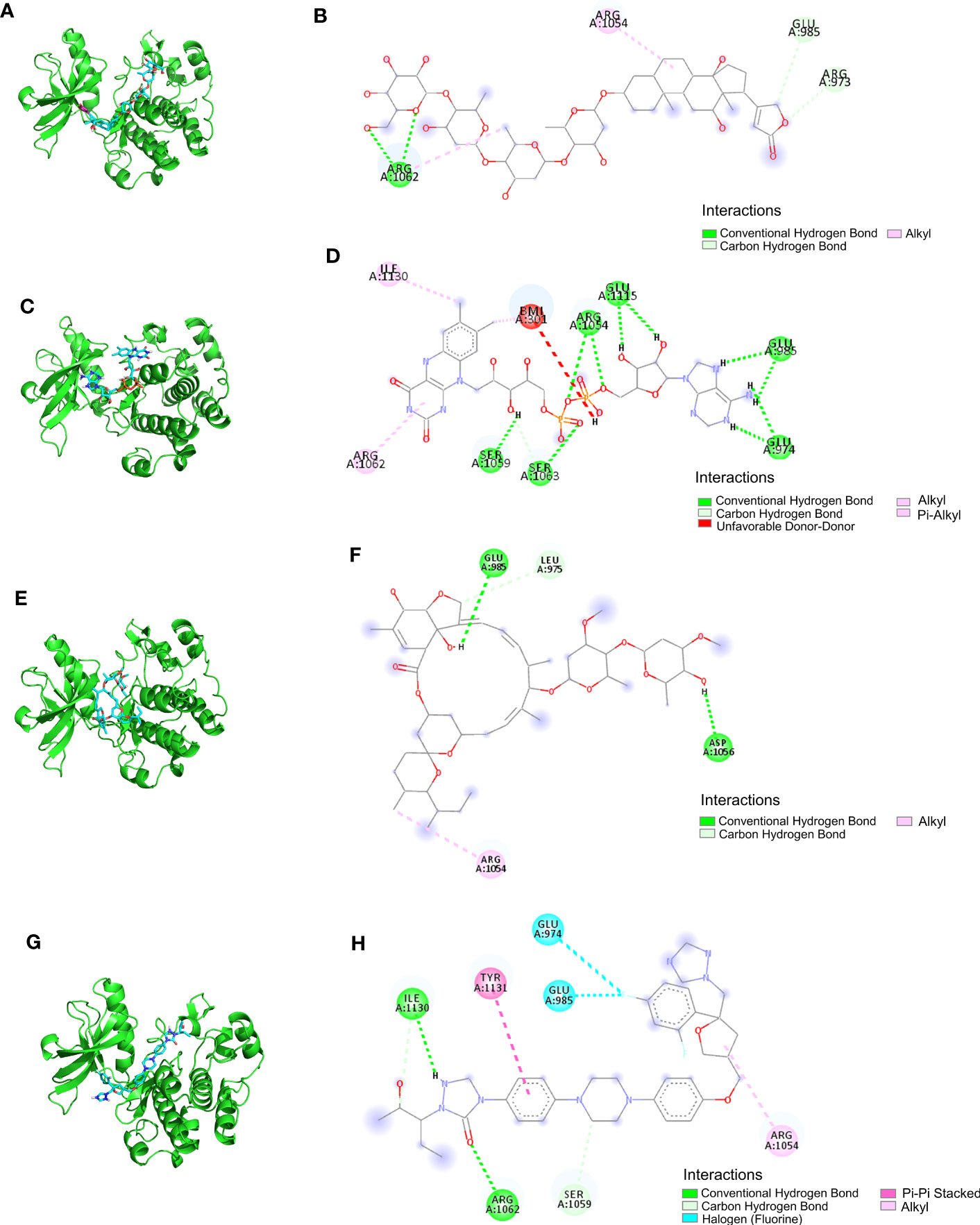
Figure 10 The ability of IGF1 to target deslanoside (A, B), flavitan (C, D), ivermectin (E, F), and posaconazole (G, H).
The recent approval of the first PAH therapy for treating PF-associated PH represents an encouraging advancement, which indicates that despite the heterogeneity in patients, shared molecular mechanisms contribute to the perpetuation of the fibrotic process and pulmonary vascular remodeling (23). In this study, we conducted WGCNA to find clusters (modules) of highly relevant genes, explored the relationship between gene networks and the two diseases (17), and then obtained the DEGs, which refer to the specific genes expressed between healthy subjects and patients with PF or pulmonary vascular remodeling. After integration, the final “important” intersecting genes were discovered, probably the potential vital genes and therapeutic targets of PF-associated PH.
Drawing support from WGCNA, we first described several intersection genes of PF and pulmonary vascular remodeling, most of which are enriched in lymphocyte activation. Then, we performed CIBERSORT analytical tool (24) to specifically analyze the essential fractions of 22 subpopulations of immune cells and observed that activated memory CD4+ T cells were upregulated in the lung of both PF and PH patients compared with control. Further evaluation and validation allowed us to identify IGF1 as a potential immune-related critical gene for pulmonary vascular remodeling and fibrosis.
Immunity is one of the critical factors involved in the pathogenesis of PH and PF besides genetic predisposition (25, 26), epigenetic regulation (27, 28), metabolic derangement (26), and environment insults (28, 29). Both innate immunity and adaptive immunity contribute to all forms of PF and pulmonary artery vascular remodeling. Pulmonary vasculopathy in patients with PH is characterized by varying degrees of perivascular inflammatory infiltration, including T lymphocytes, B lymphocytes, macrophages, dendritic cells, and mast cells (30). Meanwhile, those immune cells are also important players in fibroblast biology and fibrogenesis (31, 32). In our study, we first demonstrated that the shared genes between PF and PH are mainly functionally enriched in lymphocyte activation, which refers to a series of cellular biochemical changes and processes of lymphocytes under the stimulation of antigen or mitogen, including the transmission of the cell activation signal and the activation of a variety of related enzymes. According to their migration, surface molecules, and functions, they can be divided into T lymphocytes (also known as T cells), B lymphocytes (also known as B cells), and NK cells.
Using CIBERSORT, we predicted that CD4+ T cell activation was a common immune feature to PF and PH. CD4+ T cells can differentiate into several effector subsets, such as Th1, Th2, Th17, Tregs, and Tr1 cells, depending on factors in the local environment, such as cytokines and T cell receptor (TCR) signaling strength (33). Previous studies have already described a specific depletion of CD4+ T cells in peripheral venous blood of patients with PH (34) and PF (35); this significant downregulation is associated with poor outcomes in patients with PF (35) and PH (36). While in the perivascular (37) and fibrosis area (38), a significantly increased helper T cell (CD4) prevalence could be found, such as Th17, which produces several cytokines promoting pulmonary vascular remodeling (36) and fibrogenesis (38). Unlike Th17, Treg cells, especially their CD4+/CD25+/Foxp3+ phenotype, are essential for maintaining immune system homeostasis, especially in long-term inflammatory responses. They inhibit the activity of other T cells, especially CD8+ cytotoxic T cells, limit the immune response (39), reduce endothelial injury, and then impede PH-mediated vascular remodeling (40, 41) by upregulating COX-2, PTGIS, HO-1, and PD-L1 in the media layer of the pulmonary arteriole (40). A previous study identified Treg-associated genes in the progression of PH, and the gene signature based on these genes was revealed to be a novel indicator to distinguish PH from controls (42). The imbalanced Th1/Th2 immune response has been considered central to the pathogenesis of PF (43); Treg impairment is also dedicated to the development of PF (44). Global impairment of CD4+CD25+FOXP3+ Tregs has been identified in the peripheral blood and bronchoalveolar lavage fluid in IPF patients and is strongly correlated with disease severity (45). Tregs can attenuate fiber recruitment and PF by inhibiting the CXC chemokine ligand (CXCL)12 (46). Recently, Ichikawa et al. (47) found that depletion of CD69hiCD103hiFoxp3+ Tregs resulted in substantially higher levels of lung fibrosis in mice during chronic exposure to Aspergillus fumigatus because tissue-resident CD44hiCD69hi CD4+ T cells had increased expression of fibrosis-related genes. Nevertheless, a profibrosis effect of CD4+CD25hiFoxp3+ Tregs has been shown in bleomycin-induced PF (48) and other types of fibrosis animals (31), so the potential role of Tregs in PF remains uncertain and can be controversial.
To further identify the hub gene, we validated 313 shared genes of PF and PH in two GEO datasheets by differential gene expression analysis. Then, we confirmed their mRNA expression in the lung tissues of PF-PH patients. Ultimately, the IGF1 gene with high functional significance was selected as a central shared gene related to immunity in PF and PH. It is a single-chain polypeptide with a high sequence homology to pro-insulin and has been reported as a critical factor for developing T and B cells from pluripotent precursors (49). IGF1 interacts with the IGF1 receptor, whose signaling regulates T-cell proliferation, apoptosis, and Treg cell function by inducing phosphorylation of phosphoinositide 3-kinase/Akt pathway (50, 51). Meanwhile, IGF1 is also highly expressed in lung fibroblasts (52, 53), pulmonary artery endothelial cells, and PASMCs (54). In vivo, IGF1 can increase the levels of phosphorylated AKT at residue position S473 in PASMCs and elevate the expression of endothelin-1 (ET-1) in pulmonary artery endothelial cells (54). In neonatal mice, hypoxia exposure can modulate DNA methylation in the IGF1 promoter region, upregulating its levels in the lungs (54). Smooth muscle cell (SMC)-specific deletion of IGF1 reduced the proliferation of PASMCs and attenuated hypoxia-induced pulmonary vascular remodeling, right ventricular hypertrophy, and right ventricular systolic pressure (55). Consistent with our analysis, IGF1 has been shown to be overexpressed in fibrotic fibroblasts, especially class 1 and IGF-1Ea variants, and promoting fibroblast proliferation and extracellular matrix deposition (53).
Blocking the IGF1 pathway by a monoclonal antibody against the IGF1 receptor exhibited a protective effect on bleomycin-induced lung injury in animal models (56). All of these indicate that IGF1 was probably involved in the pathogenesis of fibrogenesis and PH. As IGF1 has long been implicated in various tumorigeneses, including pancreatic cancer, breast cancer, Ewing sarcoma, and melanoma, several small molecules and monoclonal antibodies targeting IGF1 signaling have been developed (57, 58). Further translational research is needed to provide direct evidence proving that inhibition of IGF1 could become a potential new therapeutic target for PF and PH.
Among the eight shared genes we discovered, COL5A2 is another gene that has been successfully validated. Studies have shown that COL5A2 is related to fibrosis. Foxf2 interacted with Smad6, downregulated COL5A2 transcription, and reduced fibrosis (59). Moreover, C57BL/6 mice presented with increased tissue elastance and a nonspecific interstitial pneumonia histologic pattern in the lung, combined with the thickening of the small and medium intrapulmonary arteries, increased fibrosis, and increased COL5A2 gene expression (60). In addition, COL5A2 expression linked to cell morphogenesis, angiogenesis, and blood vessel development (61). Gene co-expression network revealed that COL5A2 shows predictive potential in myocardial infarction, which may be a new candidate marker for identifying and treating ischemic cardiovascular diseases (62).
The results of molecular docking show that four important compounds (deslanoside, flavitan, ivermectin, and posaconazole) have strong binding activity with the target protein IGF1, and the main forms of interaction between components and targets are HBs, alkyl interaction, π-π stacking, and halogen bonding. They make that compound have a strong binding force with the target protein IGF1.
Deslanoside is widely used to treat a variety of heart diseases, such as arrhythmia and hypotension, mainly by inhibiting Na/K-ATPase. Recently, in vitro and in vivo studies have shown that deslanoside reduces the proliferation rate of tumor cells, causes G2/M cell cycle arrest, induces cell apoptosis, reduces colony formation, and inhibits migration and invasion, thus exerting anticancer activity (63). Flavin is an important component of flavin adenine dinucleotide (FAD) and riboflavin. FAD was effective in recycling glutathione disulfide in vivo and promoted alveologenesis but did not impact alveolar fluid clearance nor attenuate fibrosis following high fraction inspired oxygen exposure (64). However, ATP content was increased, and the content of free fatty acids and reactive oxygen species were decreased by FAD in vivo and in vitro (65). FAD can inhibit pathological cardiac hypertrophy and fibrosis through activating short-chain acyl-CoA dehydrogenase, thus preventing them from developing into heart failure (65). Riboflavin also works as an antioxidant by scavenging free radicals. Administration of lipopolysaccharide (LPS) resulted in marked cellular changes including interstitial edema, hemorrhage, infiltration of PMNs, which were reversed by riboflavin administration (66). Ivermectin is one of the most important drugs for the control of parasitic infection, which has antiviral activity against many DNA and RNA viruses, including coronavirus disease-2019 (COVID-19) (67). With regard to its anti-inflammatory properties, ivermectin has been proven to significantly reduce the recruitment of immune cells and the production of cytokines (68) and inhibit TNF-α and IL-6 induced by LPS (69). Posaconazole is being used for invasive pulmonary aspergillosis at present (70). In addition, posaconazole accumulates in lung tissue, especially in macrophages (71). This characteristic allows for the long duration of action commonly observed in epithelial cells (71).
We acknowledge that this study has some limitations. First, the datasets used were all derived from transcriptomics. The use, mutual verification, and supplement of other omics must be addressed. Further analysis and verification of other omics will be needed after this research. Second, the sample size in RT-PCR validation was small because obtaining lung samples from patients and controls is challenging. Third, heterogeneity of PF and PH has not been considered in this study. Fourth, further demonstration of machinery by future studies is necessary.
In conclusion, we proposed that lymphocyte activation might be a vital pathway affecting the pathogenesis of PF and PH and identified IGF1 as the critical immune-related common gene. These data findings provide a basis for future research and new potential therapeutic targets for the future treatment of PF and PH.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
The studies involving humans were approved by the Ethics Committee of Shanghai Pulmonary Hospital (numbers: K22-137Y). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
HZ, LW, and YY investigated the literature research, got the data, and analyzed the data. Q-HZ and JH wrote the article. RJ and C-JL modified the figures. H-LQ and Y-QM revised the article. W-HW, PY, and S-GG conceived the idea of the study, designed the steps of the study, and directed the data analysis. All authors contributed to the article and approved the submitted version.
This work was supported by the Program of National Natural Science Foundation of China (81900050, 82000059), the Program of Natural Science Foundation of Shanghai (22ZR1452400), the Pujiang Talent Program (22PJD064), the Special Youth Program of Clinical Research in Health Industry of Shanghai Health and Wellness Committee (20204Y0384) and the Program of Shanghai Pulmonary Hospital (fk18003, fkly20005).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1197752/full#supplementary-material
1. Humbert M, Kovacs G, Hoeper MM, Badagliacca R, Berger RMF, Brida M, et al. ESC/ERS Guidelines for the diagnosis and treatment of pulmonary hypertension. Eur Respir J (2022) 61:2200879. doi: 10.1183/13993003.00879-2022
2. Nathan SD, Barbera JA, Gaine SP, Harari S, Martinez FJ, Olschewski H, et al. Pulmonary hypertension in chronic lung disease and hypoxia. Eur Respir J (2019) 53:1801914. doi: 10.1183/13993003.01914-2018
3. Raghu G, Amatto VC, Behr J, Stowasser S. Comorbidities in idiopathic pulmonary fibrosis patients: a systematic literature review. Eur Respir J (2015) 46:1113–30. doi: 10.1183/13993003.02316-2014
4. Pahal P, Sharma S. Idiopathic Pulmonary Artery Hypertension. In: StatPearls. Treasure Island (FL: StatPearls Publishing Copyright © 2022, StatPearls Publishing LLC. (2022).
5. Martinez FJ, Collard HR, Pardo A, Raghu G, Richeldi L, Selman M, et al. Idiopathic pulmonary fibrosis. Nat Rev Dis Primers (2017) 3:17074. doi: 10.1038/nrdp.2017.74
6. Rajagopal K, Bryant AJ, Sahay S, Wareing N, Zhou Y, Pandit LM, et al. Idiopathic pulmonary fibrosis and pulmonary hypertension: Heracles meets the Hydra. Br J Pharmacol (2021) 178:172–86. doi: 10.1111/bph.15036
7. George PM, Patterson CM, Reed AK, Thillai M. Lung transplantation for idiopathic pulmonary fibrosis. Lancet Respir Med (2019) 7:271–82. doi: 10.1016/S2213-2600(18)30502-2
8. Lettieri CJ, Nathan SD, Barnett SD, Ahmad S, Shorr AF. Prevalence and outcomes of pulmonary arterial hypertension in advanced idiopathic pulmonary fibrosis. Chest (2006) 129:746–52. doi: 10.1378/chest.129.3.746
9. Collum SD, Amione-Guerra J, Cruz-Solbes AS, DiFrancesco A, Hernandez AM, Hanmandlu A, et al. Pulmonary hypertension associated with idiopathic pulmonary fibrosis: current and future perspectives. Can Respir J (2017) 2017:1430350. doi: 10.1155/2017/1430350
10. Milara J, Ballester B, Morell A, Ortiz JL, Escrivá J, Fernández E, et al. JAK2 mediates lung fibrosis, pulmonary vascular remodelling and hypertension in idiopathic pulmonary fibrosis: an experimental study. Thorax (2018) 73:519–29. doi: 10.1136/thoraxjnl-2017-210728
11. Chen NY, Collum SD, Luo F, Weng T, Le TT, Hernamdez A,M, et al. Macrophage bone morphogenic protein receptor 2 depletion in idiopathic pulmonary fibrosis and Group III pulmonary hypertension. Am J Physiol Lung Cell Mol Physiol (2016) 311:L238–54. doi: 10.1152/ajplung.00142.2016
12. Rajkumar R, Konishi K, Richards TJ, Ishizawar DC, Wiechert AC, Kaminski N, et al. Genomewide RNA expression profiling in lung identifies distinct signatures in idiopathic pulmonary arterial hypertension and secondary pulmonary hypertension. Am J Physiol Heart Circ Physiol (2010) 298:H1235–48. doi: 10.1152/ajpheart.00254.2009
13. Wu WH, Bonnet S, Shimauchi T, Toro V, Grobs Y, ROmanet C, et al. Potential for inhibition of checkpoint kinases 1/2 in pulmonary fibrosis and secondary pulmonary hypertension. Thorax (2022) 77:247–58. doi: 10.1136/thoraxjnl-2021-217377
14. Bourgeois A, Bonnet S, Breuils-Bonnet S, Habbout K, Paradis R, Tremblay E, et al. Inhibition of CHK 1 (Checkpoint kinase 1) elicits therapeutic effects in pulmonary arterial hypertension. Arterioscler Thromb Vasc Biol (2019) 39:1667–81. doi: 10.1161/ATVBAHA.119.312537
15. Govindarajan R, Duraiyan J, Kaliyappan K, Palanisamy M. Microarray and its applications. J Pharm Bioallied Sci (2012) 4:S310–2. doi: 10.4103/0975-7406.100283
16. Yao M, Zhang C, Gao C, Wang Q, Dai M, Yue R, et al. Exploration of the shared gene signatures and molecular mechanisms between systemic lupus erythematosus and pulmonary arterial hypertension: evidence from transcriptome data. Front Immunol (2021) 12:658341. doi: 10.3389/fimmu.2021.658341
17. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinf (2008) 9:559. doi: 10.1186/1471-2105-9-559
18. Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics (2009) 25:1091–3. doi: 10.1093/bioinformatics/btp101
19. Lu Y, Li K, Hu Y, Wang X. Expression of immune related genes and possible regulatory mechanisms in Alzheimer's disease. Front Immunol (2021) 12:768966. doi: 10.3389/fimmu.2021.768966
20. DePianto DJ, Chandriani S, Abbas AR, Jia G, N'Diaye EN, Caplazi P, et al. Heterogeneous gene expression signatures correspond to distinct lung pathologies and biomarkers of disease severity in idiopathic pulmonary fibrosis. Thorax (2015) 70:48–56. doi: 10.1136/thoraxjnl-2013-204596
21. Mura M, Cecchini MJ, Joseph M, Granton JT. Osteopontin lung gene expression is a marker of disease severity in pulmonary arterial hypertension. Respirology (2019) 24:1104–10. doi: 10.1111/resp.13557
22. Wang R, Lee YG, Dhandapani S, Baek NI, Kim KP, Cho YE, et al. 8-paradol from ginger exacerbates PINK1/Parkin mediated mitophagy to induce apoptosis in human gastric adenocarcinoma. Pharmacol Res (2023) 187:106610. doi: 10.1016/j.phrs.2022.106610
23. Nathan SD, Waxman A, Rajagopal S, Case A, Johri S, DuBrock H, et al. Inhaled treprostinil and forced vital capacity in patients with interstitial lung disease and associated pulmonary hypertension: a post-hoc analysis of the INCREASE study. Lancet Respir Med (2021) 9:1266–74. doi: 10.1016/S2213-2600(21)00165-X
24. Zeng H, Liu X, Zhang Y. Identification of potential biomarkers and immune infiltration characteristics in idiopathic pulmonary arterial hypertension using bioinformatics analysis. Front Cardiovasc Med (2021) 8:624714. doi: 10.3389/fcvm.2021.624714
25. Gu S, Kumar R, Lee MH, Mickael C, Graham BB. Common genetic variants in pulmonary arterial hypertension. Lancet Respir Med (2019) 7:190–1. doi: 10.1016/S2213-2600(18)30448-X
26. Xu W, Janocha AJ, Erzurum SC. Metabolism in pulmonary hypertension. Annu Rev Physiol (2021) 83:551–76. doi: 10.1146/annurev-physiol-031620-123956
27. Yan Y, He YY, Jiang X, Wang Y, Chen JW, Zhao JH, et al. DNA methyltransferase 3B deficiency unveils a new pathological mechanism of pulmonary hypertension. Sci Adv (2020) 6:eaba2470. doi: 10.1126/sciadv.aba2470
28. Vaz M, Hwang SY, Kagiampakis I, Phallen J, Patil A, O'Hagan HM, et al. Chronic cigarette smoke-induced epigenomic changes precede sensitization of bronchial epithelial cells to single-step transformation by KRAS mutations. Cancer Cell (2017) 32:360–376.e6. doi: 10.1016/j.ccell.2017.08.006
29. Zhu L, Liu F, Hao Q, Feng T, Chen Z, Luo S, et al. Dietary geranylgeranyl pyrophosphate counteracts the benefits of statin therapy in experimental pulmonary hypertension. Circulation (2021) 143:1775–92. doi: 10.1161/CIRCULATIONAHA.120.046542
30. Stacher E, Graham BB, Hunt JM, Gandjeva A, Groshong SD, McLaughlin VV, et al. Modern age pathology of pulmonary arterial hypertension. Am J Respir Crit Care Med (2012) 186:261–72. doi: 10.1164/rccm.201201-0164OC
31. Kolahian S, Fernandez IE, Eickelberg O, Hartl D. Immune mechanisms in pulmonary fibrosis. Am J Respir Cell Mol Biol (2016) 55:309–22. doi: 10.1165/rcmb.2016-0121TR
32. Sharma SK, MacLean JA, Pinto C, Kradin RL. The effect of an anti-CD3 monoclonal antibody on bleomycin-induced lymphokine production and lung injury. Am J Respir Crit Care Med (1996) 154:193–200. doi: 10.1164/ajrccm.154.1.8680680
33. Brummelman J, Pilipow K, Lugli E. The single-cell phenotypic identity of human CD8(+) and CD4(+) T cells. Int Rev Cell Mol Biol (2018) 341:63–124. doi: 10.1016/bs.ircmb.2018.05.007
34. Hautefort A, Girerd B, Montani D, Cohen-Kaminsky S, Price L, Lambrecht BN, et al. T-helper 17 cell polarization in pulmonary arterial hypertension. Chest (2015) 147:1610–20. doi: 10.1378/chest.14-1678
35. Gilani SR, Vuga LJ, Lindell KO, Gibson KF, Xue J, Kaminski N, et al. CD28 down-regulation on circulating CD4 T-cells is associated with poor prognoses of patients with idiopathic pulmonary fibrosis. PloS One (2010) 5:e8959. doi: 10.1371/journal.pone.0008959
36. Tian W, Jiang SY, Jiang X, Tamosiuniene R, Kim D, Guan T, et al. The role of regulatory T cells in pulmonary arterial hypertension. Front Immunol (2021) 12:684657. doi: 10.3389/fimmu.2021.684657
37. Savai R, Pullamsetti SS, Kolbe J, Bieniek E, Voswinckel R, Fink L, et al. Immune and inflammatory cell involvement in the pathology of idiopathic pulmonary arterial hypertension. Am J Respir Crit Care Med (2012) 186:897–908. doi: 10.1164/rccm.201202-0335OC
38. Celada LJ, Kropski JA, Herazo-Maya JD, Luo W, Creecy A, Abad AT, et al. PD-1 up-regulation on CD4(+) T cells promotes pulmonary fibrosis through STAT3-mediated IL-17A and TGF-β1 production. Sci Transl Med (2018) 10:eaar8356. doi: 10.1126/scitranslmed.aar8356
39. Tomaszewski M, Bębnowska D, Hrynkiewicz R, Dworzyński J, Niedźwiedzka-Rystwej P, Kopeć G, et al. Role of the immune system elements in pulmonary arterial hypertension. J Clin Med (2021) 10:3757. doi: 10.3390/jcm10163757
40. Tamosiuniene R, Manouvakhova O, Mesange P, Saito T, Qian J, Sanyal M, et al. Dominant role for regulatory T cells in protecting females against pulmonary hypertension. Circ Res (2018) 122:1689–702. doi: 10.1161/CIRCRESAHA.117.312058
41. Tamosiuniene R, Tian W, Dhillon G, Wang L, Sung YK, Gera L, et al. Regulatory T cells limit vascular endothelial injury and prevent pulmonary hypertension. Circ Res (2011) 109:867–79. doi: 10.1161/CIRCRESAHA.110.236927
42. Liu Y, Shi JZ, Jiang R, Liu SF, He YY, van der Vorst EPC, et al. Regulatory T cell-related gene indicators in pulmonary hypertension. Front Pharmacol (2022) 13:908783. doi: 10.3389/fphar.2022.908783
43. Heukels P, Moor CC, von der Thüsen JH, Wijsenbeek MS, Kool M. Inflammation and immunity in IPF pathogenesis and treatment. Respir Med (2019) 147:79–91. doi: 10.1016/j.rmed.2018.12.015
44. Vignali DA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol (2008) 8:523–32. doi: 10.1038/nri2343
45. Kotsianidis I, Nakou E, Bouchliou I, Tzouvelekis A, Spanoudakis E, Steiropoulos P, et al. Global impairment of CD4+CD25+FOXP3+ regulatory T cells in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med (2009) 179:1121–30. doi: 10.1164/rccm.200812-1936OC
46. Garibaldi BT, D'Alessio FR, Mock JR, Files DC, Chau E, Eto Y, et al. Regulatory T cells reduce acute lung injury fibroproliferation by decreasing fibrocyte recruitment. Am J Respir Cell Mol Biol (2013) 48:35–43. doi: 10.1165/rcmb.2012-0198OC
47. Ichikawa T, Hirahara K, Kokubo K, Kiuchi M, Aoki A, Morimoto Y, et al. CD103(hi) T(reg) cells constrain lung fibrosis induced by CD103(lo) tissue-resident pathogenic CD4 T cells. Nat Immunol (2019) 20:1469–80. doi: 10.1038/s41590-019-0494-y
48. Birjandi SZ, Palchevskiy V, Xue YY, Nunez S, Kern R, Weigt SS, et al. CD4(+)CD25(hi)Foxp3(+) cells exacerbate bleomycin-induced pulmonary fibrosis. Am J Pathol (2016) 186:2008–20. doi: 10.1016/j.ajpath.2016.03.020
49. Murphy WJ, Durum SK, Longo DL. Human growth hormone promotes engraftment of murine or human T cells in severe combined immunodeficient mice. Proc Natl Acad Sci U.S.A. (1992) 89:4481–5. doi: 10.1073/pnas.89.10.4481
50. Mirdamadi Y, Bommhardt U, Goihl A, Guttek K, Zouboulis CC, Quist S, et al. Insulin and Insulin-like growth factor-1 can activate the phosphoinositide-3-kinase /Akt/FoxO1 pathway in T cells in vitro. Dermatoendocrinol (2017) 9:e1356518. doi: 10.1080/19381980.2017.1356518
51. Bilbao D, Luciani L, Johannesson B, Piszczek A, Rosenthal N. Insulin-like growth factor-1 stimulates regulatory T cells and suppresses autoimmune disease. EMBO Mol Med (2014) 6:1423–35. doi: 10.15252/emmm.201303376
52. Oherle K, Acker E, Bonfield M, Wang T, Gray J, Lang I, et al. Insulin-like growth factor 1 supports a pulmonary niche that promotes type 3 innate lymphoid cell development in newborn lungs. Immunity (2020) 52:275–294.e9. doi: 10.1016/j.immuni.2020.01.005
53. Bloor CA, Knight RA, Kedia RK, Spiteri MA, Allen JT. Differential mRNA expression of insulin-like growth factor-1 splice variants in patients with idiopathic pulmonary fibrosis and pulmonary sarcoidosis. Am J Respir Crit Care Med (2001) 164:265–72. doi: 10.1164/ajrccm.164.2.2003114
54. Yang Q, Sun M, Ramchandran R, Raj JU. IGF-1 signaling in neonatal hypoxia-induced pulmonary hypertension: Role of epigenetic regulation. Vascul Pharmacol (2015) 73:20–31. doi: 10.1016/j.vph.2015.04.005
55. Sun M, Ramchandran R, Chen J, Yang Q, Raj JU. Smooth muscle insulin-like growth factor-1 mediates hypoxia-induced pulmonary hypertension in neonatal mice. Am J Respir Cell Mol Biol (2016) 55:779–91. doi: 10.1165/rcmb.2015-0388OC
56. Choi JE, Lee SS, Sunde DA, Huizar I, Haugk KL, Thannickal VJ, et al. Insulin-like growth factor-I receptor blockade improves outcome in mouse model of lung injury. Am J Respir Crit Care Med (2009) 179:212–9. doi: 10.1164/rccm.200802-228OC
57. Weigel B, Malempati S, Reid JM, Voss SD, Cho SY, Chen HX, et al. Phase 2 trial of cixutumumab in children, adolescents, and young adults with refractory solid tumors: a report from the Children's Oncology Group. Pediatr Blood Cancer (2014) 61:452–6. doi: 10.1002/pbc.24605
58. Kundranda M, Gracian AC, Zafar SF, Meiri E, Bendell J, Algül H, et al. Randomized, double-blind, placebo-controlled phase II study of istiratumab (MM-141) plus nab-paclitaxel and gemcitabine versus nab-paclitaxel and gemcitabine in front-line metastatic pancreatic cancer (CARRIE). Ann Oncol (2020) 31:79–87. doi: 10.1016/j.annonc.2019.09.004
59. Chen G, Liu L, Sun J, Zeng L, Cai H, He Y. Foxf2 and Smad6 co-regulation of collagen 5A2 transcription is involved in the pathogenesis of intrauterine adhesion. J Cell Mol Med (2020) 24:2802–18. doi: 10.1111/jcmm.14708
60. Teodoro WR, de Jesus Queiroz ZA, Dos Santos LA, Catanozi S, Dos Santos Filho A, Bueno C, et al. Proposition of a novel animal model of systemic sclerosis induced by type V collagen in C57BL/6 mice that reproduces fibrosis, vasculopathy and autoimmunity. Arthritis Res Ther (2019) 21:278. doi: 10.1186/s13075-019-2052-2
61. Meng XY, Shi MJ, Zeng ZH, Chen C, Liu TZ, Wu QJ, et al. The role of COL5A2 in patients with muscle-invasive bladder cancer: A bioinformatics analysis of public datasets involving 787 subjects and 29 cell lines. Front Oncol (2018) 8:659. doi: 10.3389/fonc.2018.00659
62. Azuaje F, Zhang L, Jeanty C, Puhl SL, Rodius S, Wagner DR. Analysis of a gene co-expression network establishes robust association between Col5a2 and ischemic heart disease. BMC Med Genomics (2013) 6:13. doi: 10.1186/1755-8794-6-13
63. Liu M, Huang Q, A J, Li L, Li X, Zhang Z, et al. The cardiac glycoside deslanoside exerts anticancer activity in prostate cancer cells by modulating multiple signaling pathways. Cancers (Basel) (2021) 13:5809. doi: 10.3390/cancers13225809
64. Grant GJ, Mimche PN, Paine R 3rd, Liou TG, Qian WJ, Helms MN. Enhanced epithelial sodium channel activity in neonatal Scnn1b mouse lung attenuates high oxygen-induced lung injury. Am J Physiol Lung Cell Mol Physiol (2021) 321:L29–l41. doi: 10.1152/ajplung.00538.2020
65. Ma Z, Qin X, Zhong X, Liao Y, Su Y, Liu X, et al. Flavine adenine dinucleotide inhibits pathological cardiac hypertrophy and fibrosis through activating short chain acyl-CoA dehydrogenase. Biochem Pharmacol (2020) 178:114100. doi: 10.1016/j.bcp.2020.114100
66. Al-Harbi NO, Imam F, Nadeem A, Al-Harbi MM, Korashy HM, Sayed-Ahmed MM, et al. Riboflavin attenuates lipopolysaccharide-induced lung injury in rats. Toxicol Mech Methods (2015) 25:417–23. doi: 10.3109/15376516.2015.1045662
67. Formiga FR, Leblanc R, de Souza Rebouças J, Farias LP, de Oliveira RN, Pena L. Ivermectin: an award-winning drug with expected antiviral activity against COVID-19. J Control Release (2021) 329:758–61. doi: 10.1016/j.jconrel.2020.10.009
68. Yan S, Ci X, Chen N, Chen C, Li X, Chu X, et al. Anti-inflammatory effects of ivermectin in mouse model of allergic asthma. Inflammation Res (2011) 60:589–96. doi: 10.1007/s00011-011-0307-8
69. Zhang X, Song Y, Ci X, An N, Ju Y, Li H, et al. Ivermectin inhibits LPS-induced production of inflammatory cytokines and improves LPS-induced survival in mice. Inflammation Res (2008) 57:524–9. doi: 10.1007/s00011-008-8007-8
70. Vanderbeke L, Janssen NAF, Bergmans D, Bourgeois M, Buil JB, Debaveye Y, et al. Posaconazole for prevention of invasive pulmonary aspergillosis in critically ill influenza patients (POSA-FLU): a randomised, open-label, proof-of-concept trial. Intensive Care Med (2021) 47:674–86. doi: 10.1007/s00134-021-06431-0
Keywords: pulmonary fibrosis, pulmonary hypertension, WGCNA, differential gene analysis, T cells CD4
Citation: Zhao H, Wang L, Yan Y, Zhao Q-H, He J, Jiang R, Luo C-J, Qiu H-L, Miao Y-Q, Gong S-G, Yuan P and Wu W-H (2023) Identification of the shared gene signatures between pulmonary fibrosis and pulmonary hypertension using bioinformatics analysis. Front. Immunol. 14:1197752. doi: 10.3389/fimmu.2023.1197752
Received: 31 March 2023; Accepted: 14 August 2023;
Published: 04 September 2023.
Edited by:
Maria Pini, Alira Health, United StatesReviewed by:
Rami M. Elshazli, Horus University, EgyptCopyright © 2023 Zhao, Wang, Yan, Zhao, He, Jiang, Luo, Qiu, Miao, Gong, Yuan and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Hui Wu, d2VuaHVpNTYyMTAwNkAxMjYuY29t; Ping Yuan, cGFuZHl5dWFuQHRvbmdqaS5lZHUuY24=; Su-Gang Gong, Z29uZ3N1Z2FuZ0B0b25namkuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.