- 1Research Center for Child Mental Development, Kanazawa University, Kanazawa, Japan
- 2Division of Socio-Cognitive-Neuroscience, United Graduate School of Child Development, Osaka University, Kanazawa University, Hamamatsu University School of Medicine, Chiba University and University of Fukui, Kanazawa, Japan
Bone marrow stromal cell antigen-1 (BST-1/CD157) is an immune/inflammatory regulator that functions as both nicotinamide adenine dinucleotide-metabolizing ectoenzyme and cell-surface signaling receptor. BST-1/CD157 is expressed not only in peripheral tissues, but in the central nervous system (CNS). Although its pathophysiological significance in the CNS is still unclear, clinical genetic studies over a decade have begun revealing relationships between BST-1/CD157 and neuropsychiatric diseases including Parkinson’s disease, autism spectrum disorders, sleep disorders, depressive disorders and restless leg syndrome. This review summarizes the accumulating evidence for the involvement of BST-1/CD157 in these disorders.
Introduction
Bone marrow stromal antigen-1 (BST-1/CD157) is a cell-surface membrane molecule that promotes pre-B lymphocyte growth (1, 2). BST-1/CD157, along with its paralogue CD38, constitutes a nicotinamide adenine dinucleotidase (NADase)/ADP-ribosyl cyclase family (2–10). These two enzymes catalyze the synthesis of cyclic ADP-ribose (cADPR) from NAD+ and thereby regulate the intracellular Ca2+ homeostasis (8–10). Also, BST-1/CD157 has a base-exchange activity for nicotinamide riboside and nicotinic acid riboside (11). In addition to these enzymatic activities, BST-1/CD157 as well as CD38 serves as a cell-membrane receptor that transmits signals for cell polarization, migration, and diapedesis (12).
BST-1/CD157 is expressed by myeloid lineage cells including neutrophils, eosinophils, basophils and macrophages in the peripheral blood, and by B-cell and myeloid precursors in the bone marrow (2, 10, 12–17). Its expression has also been reported in other tissues, such as peripheral mesothelium (18), vascular endothelium (19, 20) and Peyer’s patches (21). BST-1/CD157 thus plays diverse roles in humoral immune responses, leukocyte transmigration, and the maintenance of hematopoietic, intestinal and vascular endothelial stem cells (2, 12–21).
More importantly, BST-1/CD157 holds much pathogenetic and clinical significance in various diseases including autoimmune diseases, hematologic malignancies and solid tumors (10, 17). Nurse-like cells cloned from bone marrow and synovial tissues of patients with rheumatoid arthritis promoted survival of peripheral B cells, which was significantly blocked by anti-BST-1/CD157 antibody; and recombinant soluble BST-1/CD157 showed a similar survival effect (2, 22). It has been also demonstrated that BST-1/CD157 is involved in the progression and differentiation of leukemia (23–25), metastasis of ovarian carcinoma cells (26–28), malignant mesothelioma (29, 30) and glioma (31), and thus could be used as diagnostic or prognostic markers. Particularly, BST-1/CD157 has been regarded as a target for immunotherapy of acute myeloid leukemia (23–25). Despite the advances in the study of these diseases, it remains unclear whether BST-1/CD157 is involved in the pathogenesis of neuropsychiatric disorders in humans.
In this review, I survey the past studies on the BST-1/CD157 gene and discuss over its implications in neuropsychiatric disorders.
Structure of the human BST-1/CD157 gene and its expression in the nervous system
The human BST-1/CD157 gene maps to the short arm of chromosome 4 (4p15.32), where its paralogue CD38 gene is also located. The major transcript for BST-1/CD157 is encoded by nine exons that encompass over 35 kb in this chromosomal region (Figure 1).
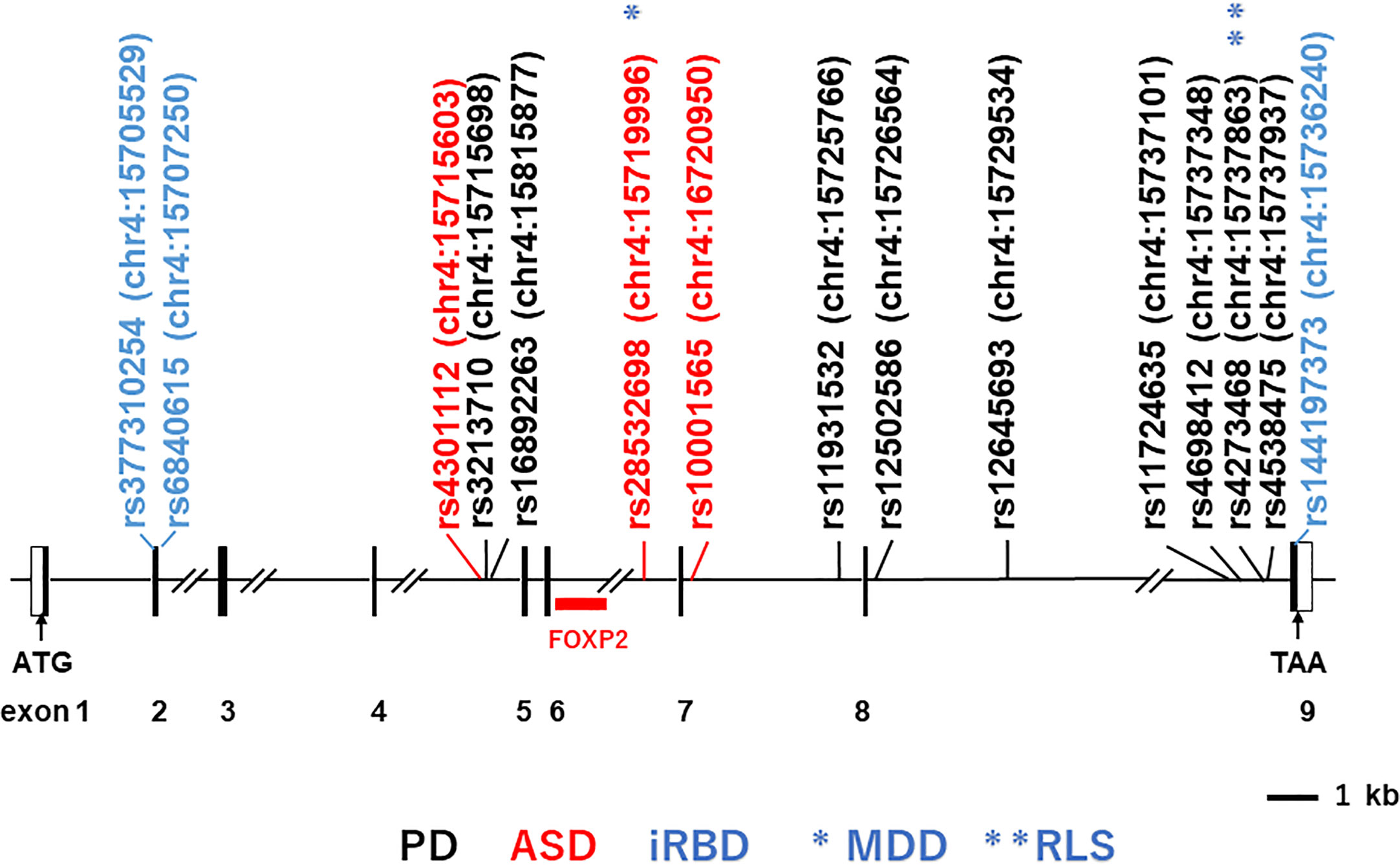
Figure 1 Structure of the human BST-1/CD157 gene and locations of main single-nucleotide-polymorphisms (SNPs). Depicted is the exon-intron organization based on GenBank accession numbers NM_004334 and NC_000004. Black and open boxes represent protein-coding regions and untranslated regions, respectively. The locations of the SNPs on human chromosome 4 (chr4) are indicated in parentheses; numbers after colons represent genomic positions based on the human genome assembly the UCSC GRCh38/hg38 genome browser (http://www.genome.ucsc.edu/cgi-bin/hgGateway?db=hg38). SNPs in black, red and blue stand for those reported to be associated with Parkinson’s disease (PD; representative ones), autism spectrum disorder (ASD) and isolated REM sleep behavior disorder (iRBD), respectively. Single asterisk and double asterisks (in blue) represent association with major depressive disorder (MDD) and restless leg syndrome (RLS), respectively.
Although BST-1/CD157 exists widely in both lymphoid and non-lymphoid tissues including blood, bone marrow, thymus, spleen, lymph nodes, lung, liver, gut, uterus, and vascular endothelial cells (8, 10, 17), little is known about its expression in the nervous system. RNA blot hybridization analysis in earlier studies did not detect BST-1/CD157 mRNA in human and mouse brains (1, 3). According to the Human Protein Atlas (32, 33), BST-1/CD157 mRNA is detectable in the normal human brain at low levels without regional specificity. Our immunohistochemical staining detected BST-1/CD157-immuoreactivity in the amygdala and somatosensory cortex of mice (34, 35). To date, changes in BST-1/CD157 expression in inflamed CNS have not fully been examined.
Parkinson’s disease
Parkinson’s disease (PD) is a common and complex neurological disorder that exhibits classical motor dysfunctions, including bradykinesia, resting tremor and gait disturbance, and non-motor features, such as psychiatric symptoms, sleep disorder and cognitive impairment (36). Epidemiological studies have revealed that both genetic and environmental factors are attributable to PD (36–41).
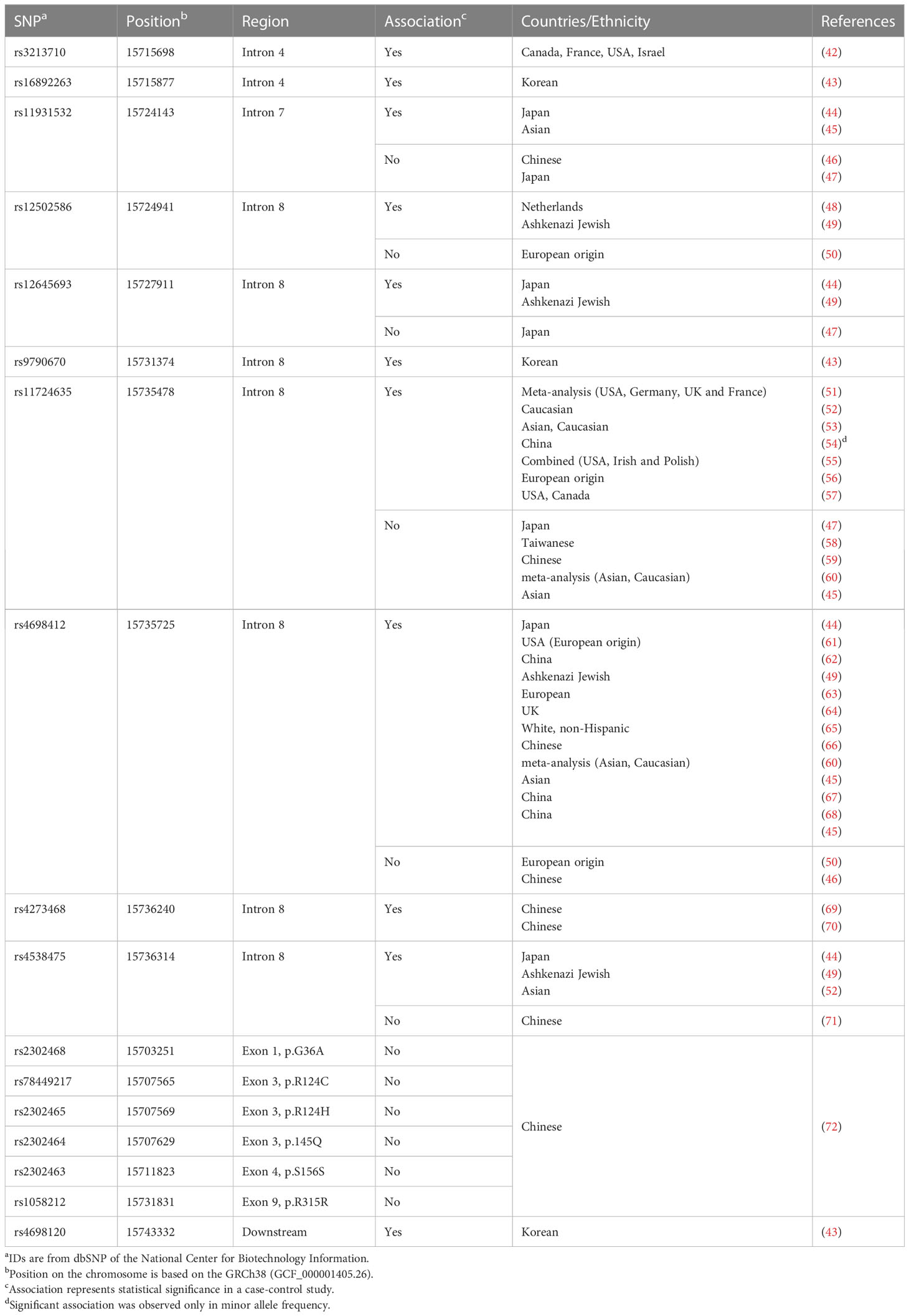
The initial genome-wide association study (GWAS) in a Japanese population reported rs11931532, rs12645693, rs4698412 and rs4538475 in the BST-1/CD157 gene as risk SNPs for sporadic late-onset PD (Figure 1; Table 1) (44). Afterwards, studies in various ethnicities have identified nearly ten PD-associated SNPs (Table 1). Among them, two SNPs, rs11724635 and rs4698412 (Figure 1; Table 1), have been examined most repeatedly. The statistically significant association of rs1573458 has been observed in six subsequent studies (Figure 1; Table 1) (51–57), but not in Asian and Caucasian cohorts (45, 47, 58–60).
The association of rs4698412 has been confirmed in eleven subsequent studies in populations with different ethnic backgrounds (45, 49, 60–68), but was not in European (50) and Chinese cohorts (46).
In search of PD-associated SNPs in exons, Wang et al. re-sequenced all the 9 exons of the BST-1/CD157 gene in a Chinese cohort. Of 524 PD cases and 527 controls, 6 non-synonymous SNPs were identified in exons 1, 3, 4, 7, and 9; but their association was insignificant (72). Thus, all PD-associated SNPs identified so far are located in introns, making it difficult to define a causal relationship between these SNPs and the pathogenesis of PD. In addition, all the SNPs in this review represent common variation in normal population, with their minor allele frequency being more than 10%. Hence any of them alone could not be an appropriate diagnostic or prognostic biomarker for PD. It is worth examining, however, whether these SNPs could be integrated effectively into polygenic risk score analysis (73) in combination with SNPs of IL-6, TNF-α and many other PD-related genes (41).
Autism spectrum disorder and other diseases
Autism spectrum disorder (ASD) is a neurodevelopmental disorder characterized by social communication deficits and restricted repetitive behaviors with a strong genetic inheritability as well as other environmental causes (74–76). An initial notable report was on a patient with both autistic symptoms and asthma (77). In this case, an 84-kb deletion between the BST-1/CD157 and CD38 genes resulted in an in-frame BST-1/CD157 and CD38 fusion transcript (77). One hypothetical explanation is that disruption of the CD38 gene in the vicinity reduced cyclic ADP-ribose formation, resulting in dysfunctional calcium (Ca2+)-induced Ca2+-release for the secretion of oxytocin, a neurohypophyseal hormone for social behavior and recognition (78–80); however, the functional consequence of this fusion transcript is unknown.
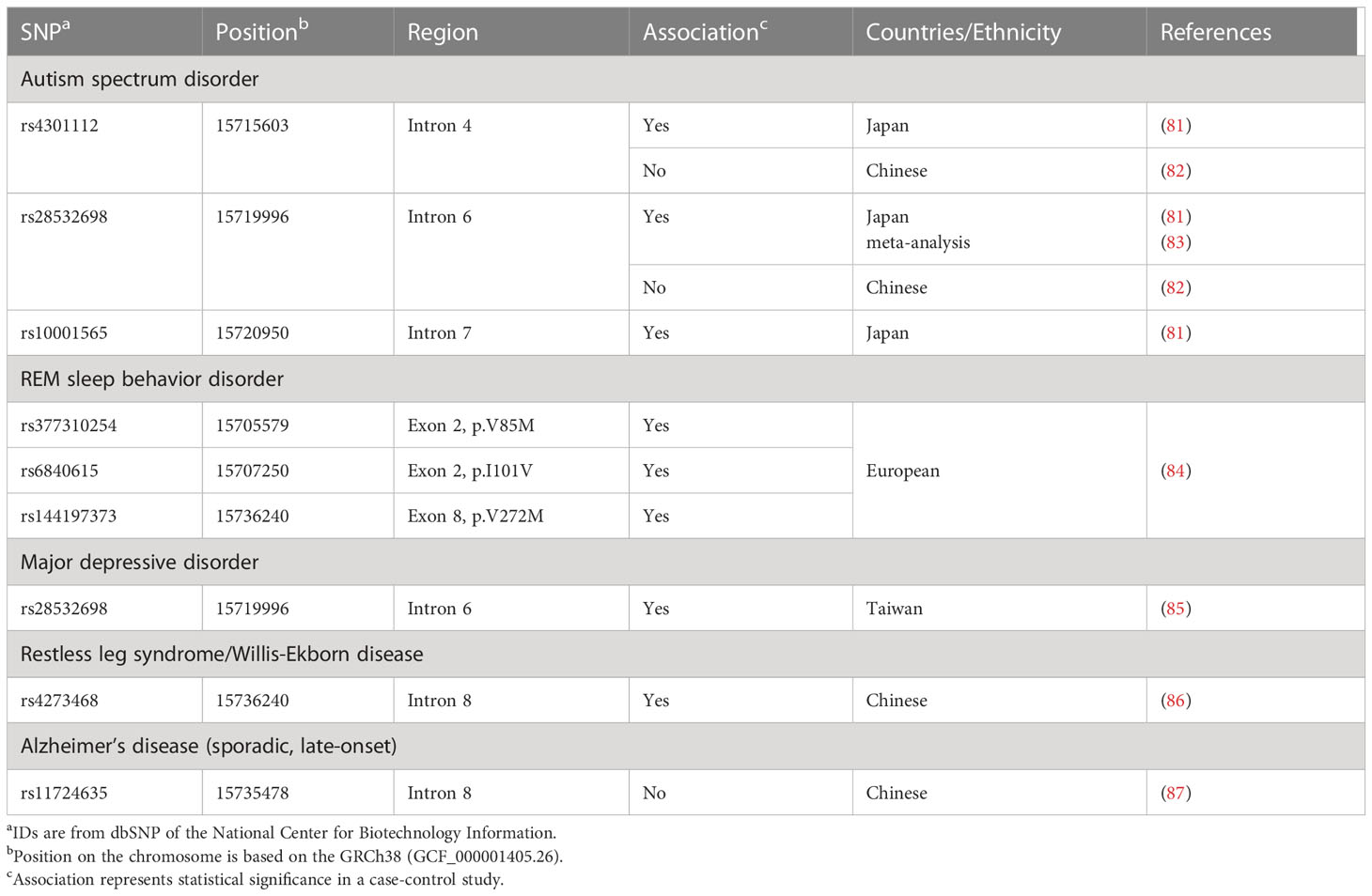
We subsequently reported association between 3 SNPs (rs4301112, rs28532698, and rs10001565) located in the BST-1/CD157 gene with ASD (Figure 1; Table 2) (81). This case-control study in a Japanese population tested genetic association between 93 SNPs in the BST-1/CD157 gene and ASD, and found out these possible risk SNPs. These SNPs are located separately from Parkinson’s disease-associated ones. As they are in high linkage disequilibrium (81), it is likely that the results represent single underlying pathogenetic process.
Bioinformatic analysis of the BST-1/CD157 gene using the HaploReg program (88, 89) predicts that genetic variations at these three SNPs may be associated with altered binding of neural development-related transcription factors: histone deacetylase C2 (HDAC2) (90), POU class 6 homeobox 1 (POU6F1) (91), and hes-related family bHLH transcription factor with YRPW motif 1 (HEY1s) (92), respectively. In addition, in the UCSC (GRCh37/hg19) track “Transcription Factor ChIP-seq (161 factors) from ENCODE (93) with Factorbook Motifs”, the region between rs4301112 and rs10001565 [chr4:15717226–15722573 (corresponding to chr4:15715603–15720950 in GRCh38/hg38)] includes potential binding sites for c-Jun, STAT3 (signal transducer and activator of transcription 3), FOXP2 (forkhead box protein P2) (Figure 1), PolR2a (Polκ RNA polymerase II polypeptide A), Elf-1 (E74-like factor 1), HNF4G (hepatocyte nuclear factor 4 gamma), HNF4A (hepatocyte nuclear factor 4 alpha), JunD, and C/EBPβ (CCAAT/enhancer binding protein beta). These potential regulatory sites are overlapped with a peak of H3K27Ac Mark track, where acetylation of lysine 27 of the H3 histone protein is assumed to regulate brain development at the level of transcription (94, 95). In particular, FOXP2 seems important because its genetic abnormalities have been implicated in speech and language disorders (96, 97). A chromosomal translocation disrupting the FOXP2 gene and an amino-acid substitution in its forkhead domain have been demonstrated in patients with severe developmental disorders of speech and language (96). FOXP2 mRNA is expressed in the developing human brain, in good concordance with anomalous sites identified by brain imaging in adult speech and language disorders (97). It is thus tempting to postulate that BST-1/CD157 expression is mediated by FOXP2 during the early brain development.
In other genes, these factors as well as FOXP2 are known to repress transcription through binding to cis-regulatory elements (90–92, 94, 95, 97). Currently, however, there is no data for their binding to cis-regulatory elements in the BST-1/CD157 gene. Also, it remains unknown whether genetic variation(s) in the BST-1/CD157 gene can change their repressive effect. I would hypothesize that nucleotide substitution(s) reduce binding affinities, weaker repressive effects on transcription and thereby dysregulate (possibly upregulate) expression of BST-1/CD157. As in the increase in CD38 and decrease in NAD+ (98–100), disruption of the NAD+ homeostasis would result in sustained immune/inflammatory reactions (Figure 2).
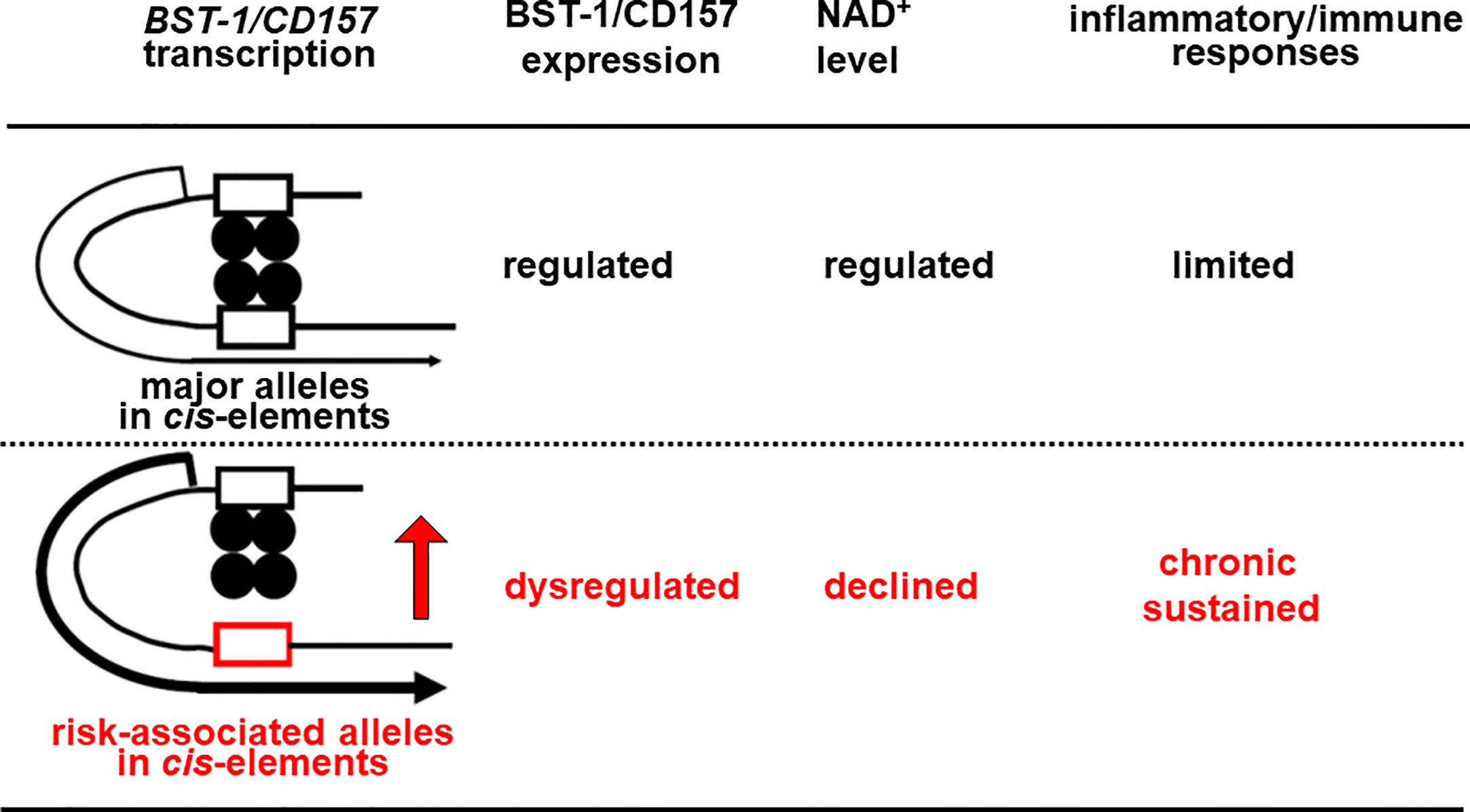
Figure 2 Hypothetical scheme for Bst-1/CD157-mediated inflammatory/immune regulation in the CNS. Nucleotide substitution(s) may lower binding affinities of transcription factors (closed circles) to in cis-regulatory regions (open boxes), decrease repressive effects on transcription and thereby upregulate the expression of the BST-1/CD157 gene, presumably in myeloid cells migrated from the periphery and/or microglia. This would disrupt the NAD+ homeostasis in the CNS, resulting in sustained immune/inflammatory reaction.
It is now well known that sustained immune/inflammatory activation is observed in the brain of the patients with developmental disorders and neurodegenerative diseases (101). Vargas et al. reported activation of microglia and astrocytes in autistic patients (102). Thus, it would be worth examining whether BST-1/CD157 is involved in such pathological state.
Interestingly, the ASD-associated SNP rs28532698 also showed association with major depressive disorder (MDD) in a Taiwan population (Figure 1; Table 2) (85). Huang et al. found that rs4273468 increased the risk of idiopathic restless leg syndrome (RLS)/Willis-Ekbom disease (WED) patients in a southeastern Chinese population (86). Although rs4273468 is also associated with PD (Figure 1; Table 2), relationship between this common sleep related movement disorder and PD remains unknown (103). Also, Mufti et al. reported that rare coding SNPs in the BST-1/CD157 gene, together with rare noncoding variants in the LAMP3 (lysosomal associated membrane protein 3) gene, was associated with isolated REM sleep behavior disorder (iRBD; Table 2) (84). All these non-synonymous variants (p.V85M, p.I101V, and p.V272M) seem to be loss-of-function variants with a potential effect on the protein structure and stability.
Shared genetic architecture and phenotypic traits
As above, SNPs in the BST-1/CD157 gene have been reported to be associated with at least five different neuropsychiatric diseases: Parkinson’s disease, ASD, iBRD, MDD and RLS. This multiple association could be regarded as genetic pleiotropy in which one genetic variant has influence on more than one phenotype (104, 105). Although both common and rare genetic variants are known to show genetic pleiotropy, this phenomenon is more frequently demonstrated in common variants than in rare variants (105). In consistent, with the exception of the exonic SNPs in iBRD, most risk alleles are common ones with frequencies > 1% in general human populations.
In the current conception, many common variants, each of which has a small effect size, in sum could be genetic risk of psychiatric neuropsychiatric disorders; in contrast, rare variants possess a large effect size, and one or small number of such variants are sufficient to cause disorders (104, 106). In most case-control studies of BST-1/CD157 SNPs, odds ratios have been estimated less than 2, suggesting that the BST-1/CD157 variations identified so far have a small effect size in the pathogenesis of common polygenic neuropsychiatry disorders.
The most common phenotypic trait among the five disorders is anxiety. In mice deficient in the BST-1/CD157 gene, Lopatina et al. reported anxiety-related and depression-like behaviors without apparent motor dysfunction, along with communication impairment (34, 35, 107, 108). These behaviors were alleviated by the treatment with anxiolytic agents, such as benzodiazepines (109), monoamine oxidase B inhibitors (109) and oxytocin (34, 107, 110). CD157 was weakly expressed in the amygdala and c-Fos-immunoreactivity, an indirect marker of neuronal excitability, which was less evident in BST-1/CD157-knockout (BST-1/CD157 -/-) mice than in wild-type mice (34). These observations in mice suggest that altered BST-1/CD157 expression in a certain brain region might affect mental state.
Conclusion and perspectives
In the past decade, an increasing number of genetic studies have suggested that the BST-1/CD157 gene could be a risk locus for several different neuropsychiatric disorders including PD and ASD. Future studies should define the nature of shared influences of BST-1/CD157 between psychiatric disorders and other diseases and phenotypic traits, especially immune/inflammatory dysfunction. The existing data, however, indicate nothing more than correlation between genetic variation and diagnoses. While the role of BST-1/CD157 variation in the genetic architecture of neuropsychiatric diseases has become clearer, the underlying molecular mechanisms remain elusive. At the same time, the physiological functions of BST-1/CD157 in the brain are still unclear. It is necessary to analyze BST-1/CD157 expression and their regulatory processes in the both developing and inflamed brain in detail.
Moreover, influences of BST-1/CD157 in the periphery on the CNS should be explored more extensively. A flurry of recent reports has documented microbiome-gut-brain axis (111, 112). Changes in gut microbiota has been shown to modulate anxiety (113, 114), depression (113, 114) and core symptoms of ASD (115, 116). Given its regulatory roles in the immune/inflammatory reactions (2, 12, 13, 117) and in the renewal of intestinal stem cells (21), it is conceivable that altered BST-1/CD157 activity may dysregulate conditions of the gut and enteric nervous system and thus result in mental disorders.
Author contributions
SY conceived, wrote and revised the manuscript. The author confirms being the sole contributor of this review article and has approved it for publication.
Funding
Our studies cited in this report was supported in part by the Collaborative Research Program of the Collaborative Research Network for Asian Children with Developmental Disorders: MEXT Policy Initiative FY2021, under joint research conducted through the initiative.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ASD, autism spectrum disorder; BST-1, bone marrow stromal cell antigen-1; CNS, central nervous system; GWAS, genome-wide association study; iRBD, isolated REM sleep behavior; MDD, major depressive disorder; NAD, nicotinamide adenine dinucleotide; PD, Parkinson’s disease; RLS; restless leg syndrome; SNP, single-nucleotide polymorphism.
References
1. Kaisho T, Ishikawa J, Oritani K, Inazawa J, Tomizawa H, Muraoka O, et al. BST-1, a surface molecule of bone marrow stromal cell lines that facilitates pre-B-cell growth. Proc Natl Acad Sci USA (1994) 91(12):5325–9. doi: 10.1073/pnas.91.12.5325
2. Ishihara K, Hirano T. BST-1/CD157 regulates the humoral immune responses in vivo. Chem Immunol (2000) 75:235–55. doi: 10.1159/000058772
3. Itoh M, Ishihara K, Tomizawa H, Tanaka H, Kobune Y, Ishikawa J, et al. Molecular cloning of murine BST-1 having homology with CD38 and aplysia ADP-ribosyl cyclase. Biochem Biophys Res Commun (1994) 203(2):1309–17. doi: 10.1006/bbrc.1994.2325
4. Kishimoto H, Hoshino S, Ohori M, Kontani K, Nishina H, Suzawa M, et al. Molecular mechanism of human CD38 gene expression by retinoic acid. identification of retinoic acid response element in the first intron. J Biol Chem (1998) 273(25):15429–34. doi: 10.1074/jbc.273.25.15429
5. Ferrero E, Saccucci F, Malavasi F. The human CD38 gene: polymorphism, CpG island, and linkage to the CD157 (BST-1) gene. Immunogenetics (1999) 49(7-8):597–604. doi: 10.1007/s002510050654
6. Lee HC. Cyclic ADP-ribose and NAADP: structures, metabolism and functions. New York, NY: Springer (2002).
7. Guse AH. Second messenger function and the structure-activity relationship of cyclic adenosine diphosphoribose (cADPR). FEBS J (2005) 272(18):4590–7. doi: 10.1111/j.1742-4658.2005.04863.x
8. Malavasi F, Deaglio S, Funaro A, Ferrero E, Horenstein AL, Ortolan E, et al. Evolution and function of the ADP ribosyl cyclase/CD38 gene family in physiology and pathology. Physiol Rev (2008) 88(3):841–86. doi: 10.1152/physrev.00035.2007
9. Lee HC. Cyclic ADP-ribose and nicotinic acid adenine dinucleotide phosphate (NAADP) as messengers for calcium mobilization. J Biol Chem (2012) 287(38):31633–40. doi: 10.1074/jbc.R112.349464
10. Quarona V, Zaccarello G, Chillemi A, Brunetti E, Singh VK, Ferrero E, et al. CD38 and CD157: a long journey from activation markers to multifunctional molecules. Cytomet B Clin Cytom (2013) 84(4):207–17. doi: 10.1002/cyto.b.21092
11. Yaku K, Palikhe S, Izumi H, Yoshida T, Hikosaka K, Hayat F, et al. BST1 regulates nicotinamide riboside metabolism via its glycohydrolase and base-exchange activities. Nat Commun (2021) 12(1):6767. doi: 10.1038/s41467-021-27080-3
12. Malavasi F, Deaglio S, Ferrero E, Funaro A, Sancho J, Ausiello CM, et al. CD38 and CD157 as receptors of the immune system: a bridge between innate and adaptive immunity. Mol Med (2006) 12(11-12):334–41. doi: 10.2119/2006-00094.Malavasi
13. Funaro A, Ortolan E, Ferranti B, Gargiulo L, Notaro R, Luzzatto L, et al. CD157 is an important mediator of neutrophil adhesion and migration. Blood (2004) 104(13):4269–78. doi: 10.1182/blood-2004-06-2129
14. Podesta M, Zocchi E, Pitto A, Usai C, Franco L, Bruzzone S, et al. Extracellular cyclic ADP-ribose increases intracellular free calcium concentration and stimulates proliferation of human hemopoietic progenitors. FASEB J (2000) 14(5):680–90. doi: 10.1096/fasebj.14.5.680
15. Cerletti M, Jang YC, Finley LW, Haigis MC, Wagers AJ. Short-term calorie restriction enhances skeletal muscle stem cell function. Cell Stem Cell (2012) 10(5):515–9. doi: 10.1016/j.stem.2012.04.002
16. Mouchiroud L, Houtkooper RH, Auwerx J. NAD+ metabolism: a therapeutic target for age-related metabolic disease. Crit Rev Biochem Mol Biol (2013) 48(4):397–408. doi: 10.3109/10409238.2013.789479
17. Ortolan E, Augeri S, Fissolo G, Musso I, Funaro A. CD157: from immunoregulatory protein to potential therapeutic target. Immunol Lett (2019) 205:59–64. doi: 10.1016/j.imlet.2018.06.007
18. Ross JA, Ansell I, Hjelle JT, Anderson JD, Miller-Hjelle MA, Dobbie JW. Phenotypic mapping of human mesothelial cells. Adv Perit Dial. (1998) 14:25–30.
19. Wakabayashi T, Naito H, Suehiro JI, Lin Y, Kawaji H, Iba T, et al. CD157 marks tissue-resident endothelial stem cells with homeostatic and regenerative properties. Cell Stem Cell (2018) 22(3):384–97.e6. doi: 10.1016/j.stem.2018.01.010
20. Wakabayashi T, Naito H. Cellular heterogeneity and stem cells of vascular endothelial cells in blood vessel formation and homeostasis: insights from single-cell RNA sequencing. Front Cell Dev Biol (2023) 11:1146399. doi: 10.3389/fcell.2023.1146399
21. Yilmaz OH, Katajisto P, Lamming DW, Gultekin Y, Bauer-Rowe KE, Sengupta S, et al. mTORC1 in the paneth cell niche couples intestinal stem-cell function to calorie intake. Nature (2012) 486(7404):490–5. doi: 10.1038/nature11163
22. Shimaoka Y, Attrep JF, Hirano T, Ishihara K, Suzuki R, Toyosaki T, et al. Nurse-like cells from bone marrow and synovium of patients with rheumatoid arthritis promote survival and enhance function of human b cells. J Clin Invest. (1998) 102(3):606–18. doi: 10.1172/JCI3162
23. Vaisitti T, Arruga F, Guerra G, Deaglio S. Ectonucleotidases in blood malignancies: a tale of surface markers and therapeutic targets. Front Immunol (2019) 10:2301. doi: 10.3389/fimmu.2019.02301
24. Yakymiv Y, Augeri S, Fissolo G, Peola S, Bracci C, Binaschi M, et al. CD157: from myeloid cell differentiation marker to therapeutic target in acute myeloid leukemia. Cells (2019) 8(12):1580. doi: 10.3390/cells8121580
25. Yakymiv Y, Augeri S, Bracci C, Marchisio S, Aydin S, D'Ardia S, et al. CD157 signaling promotes survival of acute myeloid leukemia cells and modulates sensitivity to cytarabine through regulation of anti-apoptotic Mcl-1. Sci Rep (2021) 11(1):21230. doi: 10.1038/s41598-021-00733-5
26. Morone S, Lo-Buono N, Parrotta R, Giacomino A, Nacci G, Brusco A, et al. Overexpression of CD157 contributes to epithelial ovarian cancer progression by promoting mesenchymal differentiation. PloS One (2012) 7(8):e43649. doi: 10.1371/journal.pone.0043649
27. Lo Buono N, Morone S, Giacomino A, Parrotta R, Ferrero E, Malavasi F, et al. CD157 at the intersection between leukocyte trafficking and epithelial ovarian cancer invasion. Front Biosci (Landmark Ed) (2014) 19(2):366–78. doi: 10.2741/4213
28. Ortolan E, Ferrero E, Funaro A. Re: CD157 in ovarian carcinoma: how does it help us? J Natl Cancer Inst (2010) 102(22):1741. doi: 10.1093/jnci/djq394
29. Ortolan E, Giacomino A, Martinetto F, Morone S, Lo Buono N, Ferrero E, et al. CD157 enhances malignant pleural mesothelioma aggressiveness and predicts poor clinical outcome. Oncotarget (2014) 5(15):6191–205. doi: 10.18632/oncotarget.2186
30. Augeri S, Capano S, Morone S, Fissolo G, Giacomino A, Peola S, et al. Soluble CD157 in pleural effusions: a complementary tool for the diagnosis of malignant mesothelioma. Oncotarget (2018) 9(32):22785–801. doi: 10.18632/oncotarget.25237
31. Chen X, Wu W, Wang Y, Zhang B, Zhou H, Xiang J, et al. Development of prognostic indicator based on NAD+ metabolism related genes in glioma. Front Surg (2023) 10:1071259. doi: 10.3389/fsurg.2023.1071259
32. Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. tissue-based map of the human proteome. Science (2015) 347(6220):1260419. doi: 10.1126/science.1260419
33. Sjöstedt E, Zhong W, Fagerberg L, Karlsson M, Mitsios N, Adori C, et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science (2020) 367(6482):1090. doi: 10.1126/science.aay5947
34. Lopatina O, Yoshihara T, Nishimura T, Zhong J, Akther S, Fakhrul AA, et al. Anxiety- and depression-like behavior in mice lacking the CD157/BST1 gene, a risk factor for Parkinson's disease. Front Behav Neurosci (2014) 8:133. doi: 10.3389/fnbeh.2014.00133
35. Higashida H, Liang M, Yoshihara T, Akther S, Fakhrul A, Stanislav C, et al. An immunohistochemical, enzymatic, and behavioral study of CD157/BST-1 as a neuroregulator. BMC Neurosci (2017) 18(1):35. doi: 10.1186/s12868-017-0350-7
36. Kalia LV, Lang AE. Parkinson's disease. Lancet (2015) 386(9996):896–912. doi: 10.1016/S0140-6736(14)61393-3
37. Zimprich A. Genetics of Parkinson's disease and essential tremor. Curr Opin Neurol (2011) 24(4):318–23. doi: 10.1097/WCO.0b013e3283484b87
38. Yokoyama SH H. CD157/BST-1 and neuropsychiatric disorders. MESSENGER (2014) 3(12):21–6. doi: 10.1166/msr.2014.1034
39. Dulski J, Uitti RJ, Ross OA, Wszolek ZK. Genetic architecture of Parkinson's disease subtypes - review of the literature. Front Aging Neurosci (2022) 14:1023574. doi: 10.3389/fnagi.2022.1023574
40. Mai AS, Yau CE, Tseng FS, Foo QXJ, Wang DQ, Tan EK. Linking autism spectrum disorders and parkinsonism: clinical and genetic association. Ann Clin Transl Neurol (2023) 10(4):484–96. doi: 10.1002/acn3.51736
41. Yi M, Li J, Jian S, Li B, Huang Z, Shu L, et al. Quantitative and causal analysis for inflammatory genes and the risk of Parkinson's disease. Front Immunol (2023) 14:1119315. doi: 10.3389/fimmu.2023.1119315
42. Rudakou U, Yu E, Krohn L, Ruskey JA, Asayesh F, Dauvilliers Y, et al. Targeted sequencing of Parkinson's disease loci genes highlights SYT11, FGF20 and other associations. Brain (2021) 144(2):462–72. doi: 10.1093/brain/awaa401
43. Chung SJ, Jung Y, Hong M, Kim MJ, You S, Kim YJ, et al. Alzheimer's disease and Parkinson's disease genome-wide association study top hits and risk of Parkinson's disease in Korean population. Neurobiol Aging (2013) 34(11):2695 e1–7. doi: 10.1016/j.neurobiolaging.2013.05.022
44. Satake W, Nakabayashi Y, Mizuta I, Hirota Y, Ito C, Kubo M, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson's disease. Nat Genet (2009) 41(12):1303–7. doi: 10.1038/ng.485
45. Li J, Luo J, Liu L, Fu H, Tang L. The association between CD157/BST1 polymorphisms and the susceptibility of Parkinson's disease: a meta-analysis. Neuropsychiatr Dis Treat (2019) 15:1089–102. doi: 10.2147/NDT.S190935
46. Tan EK, Kwok HH, Tan LC, Zhao WT, Prakash KM, Au WL, et al. Analysis of GWAS-linked loci in Parkinson's disease reaffirms PARK16 as a susceptibility locus. Neurology (2010) 75(6):508–12. doi: 10.1212/WNL.0b013e3181eccfcd
47. Miyake Y, Tanaka K, Fukushima W, Kiyohara C, Sasaki S, Tsuboi Y, et al. Lack of association between BST1 polymorphisms and sporadic Parkinson's disease in a Japanese population. J Neurol Sci (2012) 323(1-2):162–6. doi: 10.1016/j.jns.2012.09.008
48. Simon-Sanchez J, van Hilten JJ, van de Warrenburg B, Post B, Berendse HW, Arepalli S, et al. Genome-wide association study confirms extant PD risk loci among the Dutch. Eur J Hum Genet (2011) 19(6):655–61. doi: 10.1038/ejhg.2010.254
49. Liu X, Cheng R, Verbitsky M, Kisselev S, Browne A, Mejia-Sanatana H, et al. Genome-wide association study identifies candidate genes for Parkinson's disease in an ashkenazi Jewish population. BMC Med Genet (2011) 12:104. doi: 10.1186/1471-2350-12-104
50. Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, et al. Genome-wide association study reveals genetic risk underlying Parkinson's disease. Nat Genet (2009) 41(12):1308–12. doi: 10.1038/ng.487
51. International Parkinson Disease Genomics C, Nalls MA, Plagnol V, Hernandez DG, Sharma M, Sheerin UM, et al. Imputation of sequence variants for identification of genetic risks for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet (2011) 377(9766):641–9. doi: 10.1016/S0140-6736(10)62345-8
52. Lill CM, Roehr JT, McQueen MB, Kavvoura FK, Bagade S, Schjeide BM, et al. Comprehensive research synopsis and systematic meta-analyses in Parkinson's disease genetics: the PDGene database. PloS Genet (2012) 8(3):e1002548. doi: 10.1371/journal.pgen.1002548
53. Sharma M, Ioannidis JP, Aasly JO, Annesi G, Brice A, Van Broeckhoven C, et al. Large-Scale replication and heterogeneity in Parkinson disease genetic loci. Neurology (2012) 79(7):659–67. doi: 10.1212/WNL.0b013e318264e353
54. Liu J, Xiao Q, Wang Y, Xu ZM, Wang Y, Yang Q, et al. Analysis of genome-wide association study-linked loci in Parkinson's disease of mainland China. Mov Disord (2013) 28(13):1892–5. doi: 10.1002/mds.25599
55. Soto-Ortolaza AI, Heckman MG, Labbe C, Serie DJ, Puschmann A, Rayaprolu S, et al. GWAS risk factors in Parkinson's disease: LRRK2 coding variation and genetic interaction with PARK16. Am J Neurodegener Dis (2013) 2(4):287–99.
56. Nalls MA, Pankratz N, Lill CM, Do CB, Hernandez DG, Saad M, et al. Large-Scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat Genet (2014) 46(9):989–93. doi: 10.1038/ng.3043
57. Hayete B, Wuest D, Laramie J, McDonagh P, Church B, Eberly S, et al. A Bayesian mathematical model of motor and cognitive outcomes in Parkinson's disease. PloS One (2017) 12(6):e0178982. doi: 10.1371/journal.pone.0178982
58. Chen ML, Lin CH, Lee MJ, Wu RM. BST1 rs11724635 interacts with environmental factors to increase the risk of Parkinson's disease in a Taiwanese population. Parkinsonism Relat Disord (2014) 20(3):280–3. doi: 10.1016/j.parkreldis.2013.11.009
59. Chang KH, Wu YR, Chen YC, Fung HC, Lee-Chen GJ, Chen CM. STK39, but not BST1, HLA-DQB1, and SPPL2B polymorphism, is associated with Han-Chinese Parkinson's disease in Taiwan. Med (Baltimore). (2015) 94(41):e1690. doi: 10.1097/MD.0000000000001690
60. Wang S, Xu YF, Ding XY, Liu ZR, Ding Y, Jin B, et al. Association between bone marrow stromal cell antigen 1 gene polymorphisms and the susceptibility to Parkinson's disease: a meta-analysis. Neurosci Lett (2015) 599:120–4. doi: 10.1016/j.neulet.2015.05.026
61. Hamza TH, Zabetian CP, Tenesa A, Laederach A, Montimurro J, Yearout D, et al. Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson's disease. Nat Genet (2010) 42(9):781–5. doi: 10.1038/ng.642
62. Chang XL, Mao XY, Li HH, Zhang JH, Li NN, Burgunder JM, et al. Association of GWAS loci with PD in China. Am J Med Genet B Neuropsychiatr Genet (2011) 156B(3):334–9. doi: 10.1002/ajmg.b.31167
63. Saad M, Lesage S, Saint-Pierre A, Corvol JC, Zelenika D, Lambert JC, et al. Genome-wide association study confirms BST1 and suggests a locus on 12q24 as the risk loci for Parkinson's disease in the European population. Hum Mol Genet (2011) 20(3):615–27. doi: 10.1093/hmg/ddq497
64. Consortium UKPsD, Wellcome Trust Case Control C, Spencer CC, Plagnol V, Strange A, Gardner M, Paisan-Ruiz C, Band G, et al. Dissection of the genetics of Parkinson's disease identifies an additional association 5' of SNCA and multiple associated haplotypes at 17q21. Hum Mol Genet (2011) 20(2):345–53. doi: 10.1093/hmg/ddq469
65. Pankratz N, Beecham GW, DeStefano AL, Dawson TM, Doheny KF, Factor SA, et al. Meta-analysis of Parkinson's disease: identification of a novel locus, RIT2. Ann Neurol (2012) 71(3):370–84. doi: 10.1002/ana.22687
66. Guo JF, Li K, Yu RL, Sun QY, Wang L, Yao LY, et al. Polygenic determinants of Parkinson's disease in a Chinese population. Neurobiol Aging. (2015) 36(4):1765 e1– e6. doi: 10.1016/j.neurobiolaging.2014.12.030
67. Shen YT, Wang JW, Wang M, Zhi Y, Li JY, Yuan YS, et al. BST1 rs4698412 allelic variant increases the risk of gait or balance deficits in patients with Parkinson's disease. CNS Neurosci Ther (2019) 25(4):422–9. doi: 10.1111/cns.13099
68. Cheng WW, Zhu Q, Zhang HY. Identifying risk genes and interpreting pathogenesis for Parkinson's disease by a multiomics analysis. Genes (Basel). (2020) 11(9): 1100. doi: 10.3390/genes11091100
69. Wang J, Shete S. Testing departure from Hardy-Weinberg proportions. Methods Mol Biol (2012) 850:77–102. doi: 10.1007/978-1-61779-555-8_6
70. Shi C, Zheng Z, Wang Q, Wang C, Zhang D, Zhang M, et al. Exploring the effects of genetic variants on clinical profiles of Parkinson's disease assessed by the unified Parkinson's disease rating scale and the Hoehn-Yahr stage. PloS One (2016) 11(6):e0155758. doi: 10.1371/journal.pone.0155758
71. Zhu LH, Luo XG, Zhou YS, Li FR, Yang YC, Ren Y, et al. Lack of association between three single nucleotide polymorphisms in the Park9, Park16, and BST1 genes and Parkinson's disease in the northern Han Chinese population. Chin Med J (Engl). (2012) 125(4):588–92. doi: 10.3760/cma.j.issn.0366-6999.2012.04.006
72. Wang C, Feng X, Xie S, Gu Z, Chan P. Exonic sequencing revealed no causative mutation in the BST1 gene in patients with Parkinson's disease. Neurobiol Aging. (2013) 34(11):2695 e9– e10. doi: 10.1016/j.neurobiolaging.2013.05.024
73. Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet (2018) 19(9):581–90. doi: 10.1038/s41576-018-0018-x
74. American Psychiatric Association D-TF. Diagnostic and statistical manual of mental disorders: DSM-5™. 5th ed. Washington, DC: American Psychiatric Publishing, Inc (2013).
75. Lai MC, Lombardo MV, Baron-Cohen S. Autism. Lancet (2014) 383(9920):896–910. doi: 10.1016/S0140-6736(13)61539-1
76. Lord C, Elsabbagh M, Baird G, Veenstra-Vanderweele J. Autism spectrum disorder. Lancet (2018) 392(10146):508–20. doi: 10.1016/S0140-6736(18)31129-2
77. Ceroni F, Sagar A, Simpson NH, Gawthrope AJ, Newbury DF, Pinto D, et al. A deletion involving CD38 and BST1 results in a fusion transcript in a patient with autism and asthma. Autism Res (2014) 7(2):254–63. doi: 10.1002/aur.1365
78. Jin D, Liu HX, Hirai H, Torashima T, Nagai T, Lopatina O, et al. CD38 is critical for social behaviour by regulating oxytocin secretion. Nature (2007) 446(7131):41–5. doi: 10.1038/nature05526
79. Munesue T, Yokoyama S, Nakamura K, Anitha A, Yamada K, Hayashi K, et al. Two genetic variants of CD38 in subjects with autism spectrum disorder and controls. Neurosci Res (2010) 67(2):181–91. doi: 10.1016/j.neures.2010.03.004
80. Higashida H, Yokoyama S, Huang JJ, Liu L, Ma WJ, Akther S, et al. Social memory, amnesia, and autism: brain oxytocin secretion is regulated by NAD+ metabolites and single nucleotide polymorphisms of CD38. Neurochem Int (2012) 61(6):828–38. doi: 10.1016/j.neuint.2012.01.030
81. Yokoyama S, Al Mahmuda N, Munesue T, Hayashi K, Yagi K, Yamagishi M, et al. Association study between the CD157/BST1 gene and autism spectrum disorders in a Japanese population. Brain Sci (2015) 5(2):188–200. doi: 10.3390/brainsci5020188
82. Mo W, Liu J, Zhang Z, Yu H, Yang A, Qu F, et al. A study of single nucleotide polymorphisms in CD157, AIM2 and JARID2 genes in Han Chinese children with autism spectrum disorder. Nord J Psychiatry (2018) 72(3):179–83. doi: 10.1080/08039488.2017.1410570
83. Mpoulimari I, Zintzaras E. Synthesis of genetic association studies on autism spectrum disorders using a genetic model-free approach. Psychiatr Genet (2022) 32(3):91–104. doi: 10.1097/YPG.0000000000000316
84. Mufti K, Yu E, Rudakou U, Krohn L, Ruskey JA, Asayesh F, et al. Novel associations of BST1 and LAMP3 with REM sleep behavior disorder. Neurology (2021) 96(10):e1402–e12. doi: 10.1212/WNL.0000000000011464
85. Chen DT, Cheng SW, Chen T, Chang JP, Hwang BF, Chang HH, et al. Identification of genetic variations in the NAD-related pathways for patients with major depressive disorder: a case-control study in Taiwan. J Clin Med (2022) 11(13): 3622. doi: 10.3390/jcm11133622
86. Huang Y, Wang P, Luo Q, Ma J. Association of BST1 polymorphism with idiopathic restless legs syndrome in Chinese population. Sleep Breath. (2021) 25(4):1987–93. doi: 10.1007/s11325-021-02326-y
87. Zhu XC, Cao L, Tan MS, Jiang T, Wang HF, Lu H, et al. Association of Parkinson's disease GWAS-linked loci with Alzheimer's disease in Han Chinese. Mol Neurobiol (2017) 54(1):308–18. doi: 10.1007/s12035-015-9649-5
88. Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res (2012) 40(Database issue):D930–4. doi: 10.1093/nar/gkr917
89. Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res (2016) 44(D1):D877–81. doi: 10.1093/nar/gkv1340
90. Hagelkruys A, Lagger S, Krahmer J, Leopoldi A, Artaker M, Pusch O, et al. A single allele of Hdac2 but not Hdac1 is sufficient for normal mouse brain development in the absence of its paralog. Development (2014) 141(3):604–16. doi: 10.1242/dev.100487
91. Zhang L, Ju X, Cheng Y, Guo X, Wen T. Identifying Tmem59 related gene regulatory network of mouse neural stem cell from a compendium of expression profiles. BMC Syst Biol (2011) 5:152. doi: 10.1186/1752-0509-5-152
92. Ghahramani Seno MM, Hu P, Gwadry FG, Pinto D, Marshall CR, Casallo G, et al. Gene and miRNA expression profiles in autism spectrum disorders. Brain Res (2011) 1380:85–97. doi: 10.1016/j.brainres.2010.09.046
93. Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature (2012) 489(7414):57–74. doi: 10.1038/nature11247
94. Kim HJ, Rosenfeld MG. Epigenetic control of stem cell fate to neurons and glia. Arch Pharm Res (2010) 33(10):1467–73. doi: 10.1007/s12272-010-1001-z
95. Sheikh BN. Crafting the brain - role of histone acetyltransferases in neural development and disease. Cell Tissue Res (2014) 356(3):553–73. doi: 10.1007/s00441-014-1835-7
96. Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature (2001) 413(6855):519–23. doi: 10.1038/35097076
97. Lai CS, Gerrelli D, Monaco AP, Fisher SE, Copp AJ. FOXP2 expression during brain development coincides with adult sites of pathology in a severe speech and language disorder. Brain (2003) 126(Pt 11):2455–62. doi: 10.1093/brain/awg247
98. Hogan KA, Chini CCS, Chini EN. The multi-faceted ecto-enzyme CD38: roles in immunomodulation, cancer, aging, and metabolic diseases. Front Immunol (2019) 10:1187. doi: 10.3389/fimmu.2019.01187
99. Piedra-Quintero ZL, Wilson Z, Nava P, Guerau-de-Arellano M. CD38: an immunomodulatory molecule in inflammation and autoimmunity. Front Immunol (2020) 11:597959. doi: 10.3389/fimmu.2020.597959
100. Zeidler JD, Kashyap S, Hogan KA, Chini EN. Implications of the NADase CD38 in COVID pathophysiology. Physiol Rev (2022) 102(1):339–41. doi: 10.1152/physrev.00007.2021
101. Salter MW, Stevens B. Microglia emerge as central players in brain disease. Nat Med (2017) 23(9):1018–27. doi: 10.1038/nm.4397
102. Vargas DL, Nascimbene C, Krishnan C, Zimmerman AW, Pardo CA. Neuroglial activation and neuroinflammation in the brain of patients with autism. Ann Neurol (2005) 57(1):67–81. doi: 10.1002/ana.20315
103. Ferini-Strambi L, Carli G, Casoni F, Galbiati A. Restless legs syndrome and Parkinson disease: a causal relationship between the two disorders? Front Neurol (2018) 9:551. doi: 10.3389/fneur.2018.00551
104. Giangrande EJ, Weber RS, Turkheimer E. What do we know about the genetic architecture of psychopathology? Annu Rev Clin Psychol (2022) 18:19–42. doi: 10.1146/annurev-clinpsy-081219-091234
105. Andreassen OA, Hindley GFL, Frei O, Smeland OB. New insights from the last decade of research in psychiatric genetics: discoveries, challenges and clinical implications. World Psychiatry (2023) 22(1):4–24. doi: 10.1002/wps.21034
106. Gratten J, Wray NR, Keller MC, Visscher PM. Large-Scale genomics unveils the genetic architecture of psychiatric disorders. Nat Neurosci (2014) 17(6):782–90. doi: 10.1038/nn.3708
107. Lopatina OL, Furuhara K, Ishihara K, Salmina AB, Higashida H. Communication impairment in ultrasonic vocal repertoire during the suckling period of Cd157 knockout mice: transient improvement by oxytocin. Front Neurosci (2017) 11:266. doi: 10.3389/fnins.2017.00266
108. Higashida H, Hashii M, Tanaka Y, Matsukawa S, Higuchi Y, Gabata R, et al. CD38, CD157, and RAGE as molecular determinants for social behavior. Cells (2019) 9(1):62. doi: 10.3390/cells9010062
109. Kasai S, Yoshihara T, Lopatina O, Ishihara K, Higashida H. Selegiline ameliorates depression-like behavior in mice lacking the CD157/BST1 gene, a risk factor for Parkinson's disease. Front Behav Neurosci (2017) 11:75. doi: 10.3389/fnbeh.2017.00075
110. Mizuno A, Cherepanov SM, Kikuchi Y, Fakhrul AA, Akther S, Deguchi K, et al. Lipo-oxytocin-1, a novel oxytocin analog conjugated with two palmitoyl groups, has long-lasting effects on anxiety-related behavior and social avoidance in CD157 knockout mice. Brain Sci (2015) 5(1):3–13. doi: 10.3390/brainsci5010003
111. Benakis C, Martin-Gallausiaux C, Trezzi JP, Melton P, Liesz A, Wilmes P. The microbiome-gut-brain axis in acute and chronic brain diseases. Curr Opin Neurobiol (2020) 61:1–9. doi: 10.1016/j.conb.2019.11.009
112. Butler MI, Cryan JF, Dinan TG. Man and the microbiome: a new theory of everything? Annu Rev Clin Psychol (2019) 15:371–98. doi: 10.1146/annurev-clinpsy-050718-095432
113. Lach G, Schellekens H, Dinan TG, Cryan JF. Anxiety, depression, and the microbiome: a role for gut peptides. Neurotherapeutics (2018) 15(1):36–59. doi: 10.1007/s13311-017-0585-0
114. Peirce JM, Alvina K. The role of inflammation and the gut microbiome in depression and anxiety. J Neurosci Res (2019) 97(10):1223–41. doi: 10.1002/jnr.24476
115. Tan Q, Orsso CE, Deehan EC, Kung JY, Tun HM, Wine E, et al. Probiotics, prebiotics, synbiotics, and fecal microbiota transplantation in the treatment of behavioral symptoms of autism spectrum disorder: a systematic review. Autism Res (2021) 14(9):1820–36. doi: 10.1002/aur.2560
116. Lukens JR, Eyo UB. Microglia and neurodevelopmental disorders. Annu Rev Neurosci (2022) 45:425–45. doi: 10.1146/annurev-neuro-110920-023056
Keywords: anxiety, autism spectrum disorder, BST-1, CD157, neuroimmune dysfunction, Parkinson’s disease, single-nucleotide polymorphism
Citation: Yokoyama S (2023) Genetic polymorphisms of bone marrow stromal cell antigen-1 (BST-1/CD157): implications for immune/inflammatory dysfunction in neuropsychiatric disorders. Front. Immunol. 14:1197265. doi: 10.3389/fimmu.2023.1197265
Received: 30 March 2023; Accepted: 15 May 2023;
Published: 29 May 2023.
Edited by:
Takashi Nakagawa, University of Toyama, JapanReviewed by:
Fabio Malavasi, University of Turin, ItalyAdriana Sumoza-Toledo, Universidad Veracruzana, Mexico
Copyright © 2023 Yokoyama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shigeru Yokoyama, c2hpZ2VydXlAbWVkLmthbmF6YXdhLXUuYWMuanA=
 Shigeru Yokoyama
Shigeru Yokoyama