- 1ShanXi Key Laboratory of Stem Cells for Immunological Dermatosis, Institute of Dermatology, Taiyuan City Center Hospital, Taiyuan, China
- 2State Key Breeding Laboratory of Stem Cells for Immunological Dermatosis, Institute of Dermatology, Taiyuan City Center Hospital, Taiyuan, China
Psoriasis is a chronic inflammatory skin disease. The histopathological features of psoriasis include excessive proliferation of keratinocytes and infiltration of immune cells. The S100 proteins are a group of EF-hand Ca2+-binding proteins, including S100A2, -A7, -A8/A9, -A12, -A15, which expression levels are markedly upregulated in psoriatic skin. These proteins exert numerous functions such as serving as intracellular Ca2+ sensors, transduction of Ca2+ signaling, response to extracellular stimuli, energy metabolism, and regulating cell proliferation and apoptosis. Evidence shows a crucial role of S100 proteins in the development and progress of inflammatory diseases, including psoriasis. S100 proteins can possibly be used as potential therapeutic target and diagnostic biomarkers. This review focuses on the pathogenic role of S100 proteins in psoriasis.
Introduction
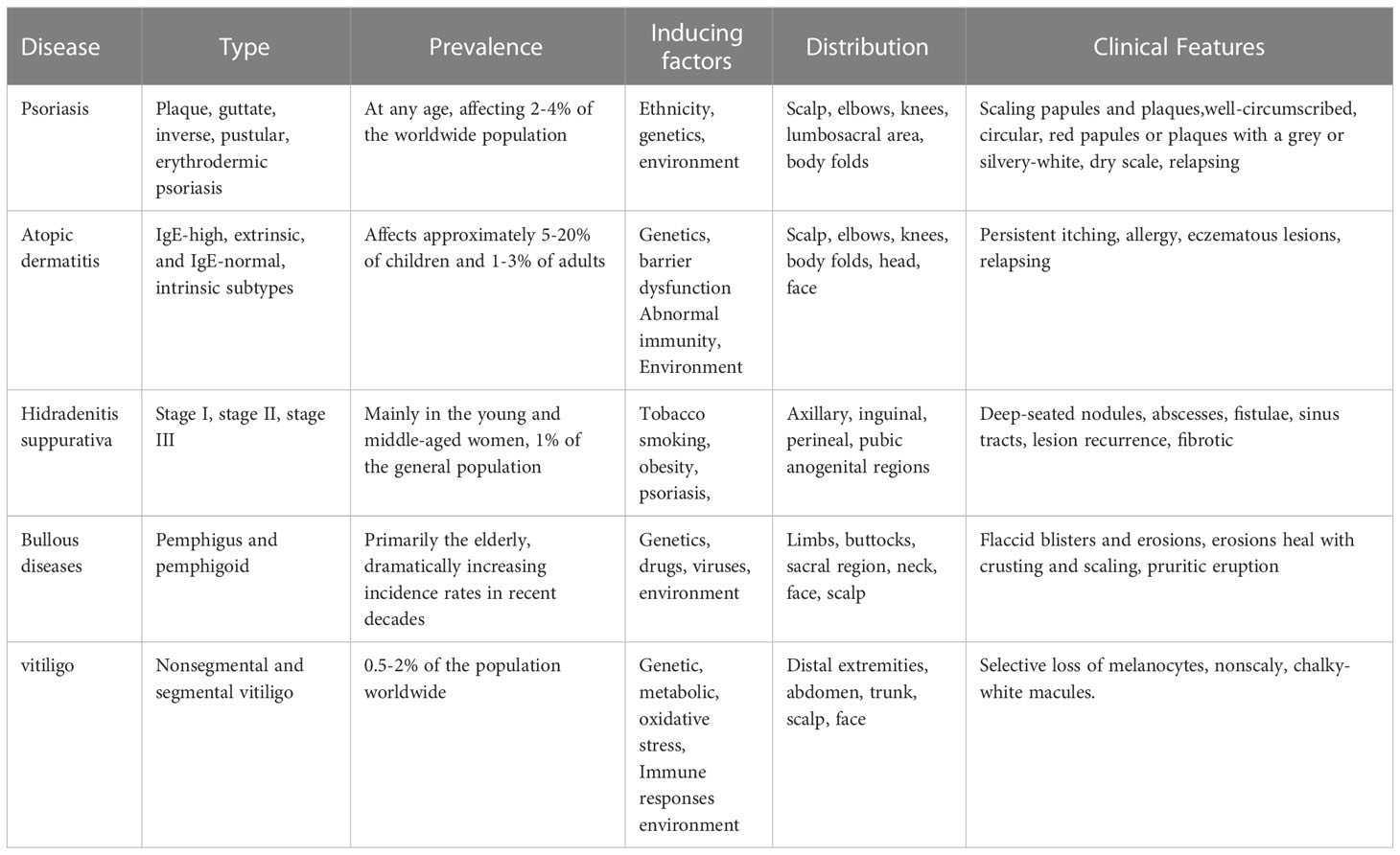
Psoriasis is a chronic, adaptive immune-mediated inflammatory skin disease with a strong genetic predisposition and autoimmune pathogenic features (1–4). Immune-mediated inflammatory dermatosis is characterized by complex multifactorial etiologies and is clinically very different from each other despite sharing a chronic inflammatory background (Table 1). Psoriasis presents a notable burden, with about 0.2 billion people worldwide suffering from psoriasis. The prevalence of psoriasis varies from country to country (5, 6). The pathogenesis of psoriasis is attributable to many factors, including ethnicity, genetics, gene variants, and environment (7, 8).
Psoriasis primarily comprises plaque psoriasis (the common form, approximately 90% of psoriatic cases are chronic plaque-type psoriasis), guttate psoriasis (after the streptococcal infection), inverse psoriasis (flexural psoriasis), pustular psoriasis (the rare and unstable), and erythrodermic psoriasis (systemic inflammation). Histopathologically, psoriasis is characterized by infiltration of immune cells, epidermal hyperproliferation, and abnormal keratinocyte (KC) differentiation (9). The recruitment of circulating leukocytes to the epidermis and the production of pro-inflammatory factors, such as TNF-α, IFN-γ, IL-6, IL-8, IL-23, IL-1β, and IL-17A play a crucial role in the development of psoriasis (10). In addition, psoriasis can be accompanied by comorbidities such as cardiovascular diseases, hyperlipidemia, hypertension, coronary artery disease, and diabetes (11–16).
Originally, psoriasis was thought to be due to dysregulation of keratinocyte proliferation (17), increasing evidence indicates a crucial role of the immune system in the pathogenesis of psoriasis (18). Psoriasis is caused by chronic interaction between KCs and activated immune cells (19). One of the most important features of psoriatic lesions is upregulation of expression levels of KC-originated antimicrobial peptides and proteins such as S100 protein subfamily, an important multifunctional player in inflammatory dermatoses (20, 21). The expression and secretion of S100 protein in keratinocytes, leads to the production of an array of pro-inflammatory cytokines, promoting dendritic cell maturation and CD4+ T cell proliferation (22), contributing to autoimmune system activation and psoriasis pathogenesis.
S100 protein family
The S100 protein family is a type of Ca2+-binding protein composed of a multigene family of low molecular mass proteins. The S100 proteins are expressed in a cell- and tissue-specific manner and engaged in multiple functions in various cell types and tissues (23, 24). The expression levels of S100 proteins are altered in various diseases such as cardiomyopathies, neurodegenerative, inflammatory disorders, and cancers (25–29). S100 proteins act as calcium sensors that regulate the function and subcellular distribution of specific target proteins participating in a variety of signaling pathways and playing a key role in diverse cellular processes such as cell proliferation, migration, differentiation, energy metabolism, apoptosis, etc. (30). In addition, S100 proteins have an extracellular activity in response to inflammatory stimuli (30–32).
The S100 protein is the largest subgroup within the superfamily of calcium-binding EF-hand motif. Genes of the S100 protein family are encoded in epidermal differentiation complex (EDC) and located within the cluster on human chromosome 1q21 (33). The members of the S100 protein family have a similar low molecular weight of 10-12 kDa and share 10% - 98% similarity in the amino acid sequences. S100 proteins are typically symmetric dimers, and each subunit contains two helix-loop-helix EF-hand motifs that are separated by a flexible hinge region and flanked by conserved hydrophobic residues at the amino- and carboxy-terminal ends (34) (Figure 1). The calcium-binding affinity of S100 proteins is low in the absence of target proteins, but the affinity is significantly increased by several orders of magnitude in the presence of specific target proteins (35, 36). The canonical C-terminal EF-hand motif binds to calcium with 100-fold higher than the N-terminal non-canonical EF-hand (37).
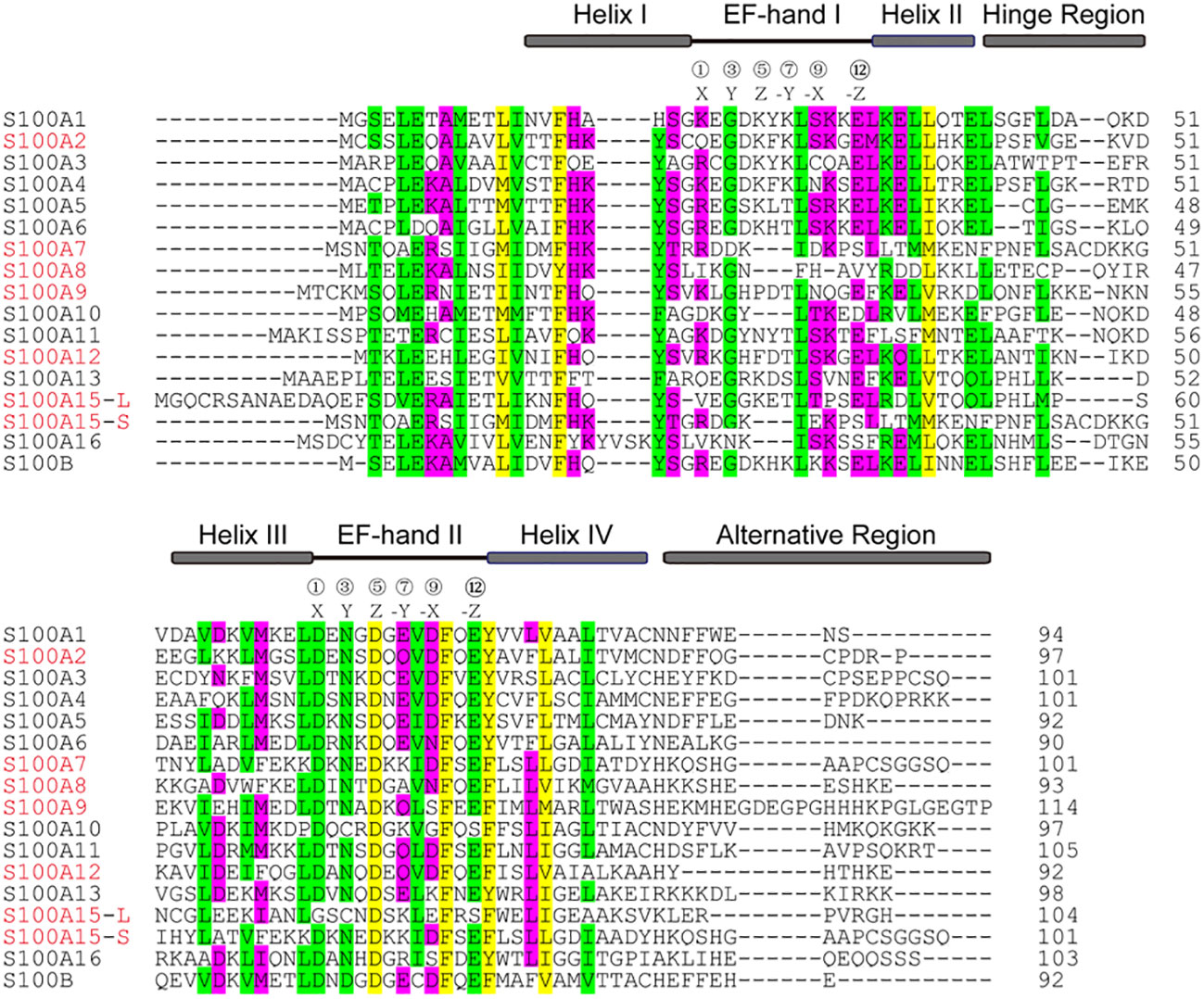
Figure 1 Alignment of the amino acid sequences of S100 from homo sapiens. The position of two EF-hands is indicated by black lines and the residues at positions 1, 3, 5, 7, 9, and 12 for Ca2+-coordination are labeled X, Y, Z, -Y, -X, -Z, respectively. Four α-helical segments, a hinge region, and an alternative region are indicated by grey boxes. The sequences highlighted in red are discussed in the text.
The 3D structures of S100 protein family have been resolved in three different states: Ca2+-free apo state, bound to Ca2+ and bound to its target protein (38). Upon calcium binding, the S100 protein undergoes a large conformational change that creates a special target protein recognition site and allows interaction with different target proteins, including cell surface receptors and other S100 proteins (39). Moreover, S100 proteins present as a non-covalent anti-parallel homo- or heterodimers form, and the target protein can bind to opposite ends of S100 protein dimer. Thus, S100 protein dimer is a cross-bridge between two target proteins.
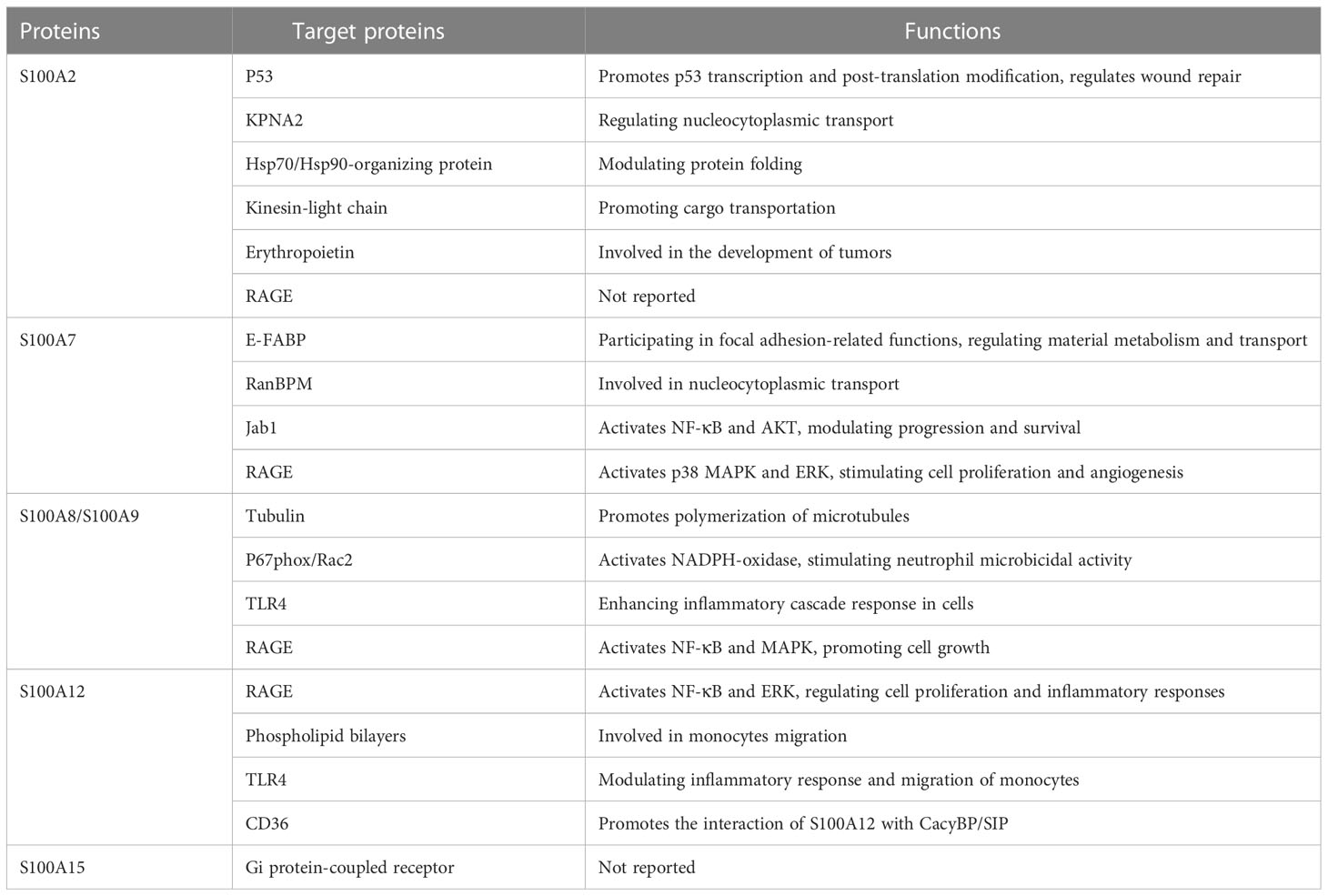
Because of multiple functions of the S100 proteins, many diseases, including psoriasis, are associated with altered expression levels of S100 proteins. Recent studies suggest the potential roles of S100 in keratinocyte proliferation, differentiation, and stress response. This review will focus on the expression and function of S100 protein-target protein interaction, and the signaling pathways of each S100 protein in psoriasis.
S100A2
S100A2 (S100 calcium-binding protein A2) is encoded by a gene located in human chromosome 1q21 (31), which was first identified as a tumor-suppressor gene by subtractive hybridization in human mammary epithelial cells (40). The three-dimensional crystal structure of a cysteine-deficient S100A2 in the calcium-free form is similar to other S100 proteins. S100A2 contains an N-terminal specific EF-hand loop by helix I and helix II, and a C-terminal classical EF-hand motif between helix III and helix IV, which are linked by a hinge region (41) (Figures 1, 2A, B). In addition to Ca2+, S100A2 also binds to Zn2+ with higher affinity, and the binding affinity of S100A2 to calcium is significantly reduced upon binding to zinc (42). Ca2+- and Zn2+-ions have opposite effects on the stability of S100A2, with Ca2+ acting as a protein stabilizer and Zn2+ as a protein destabilizer (43).
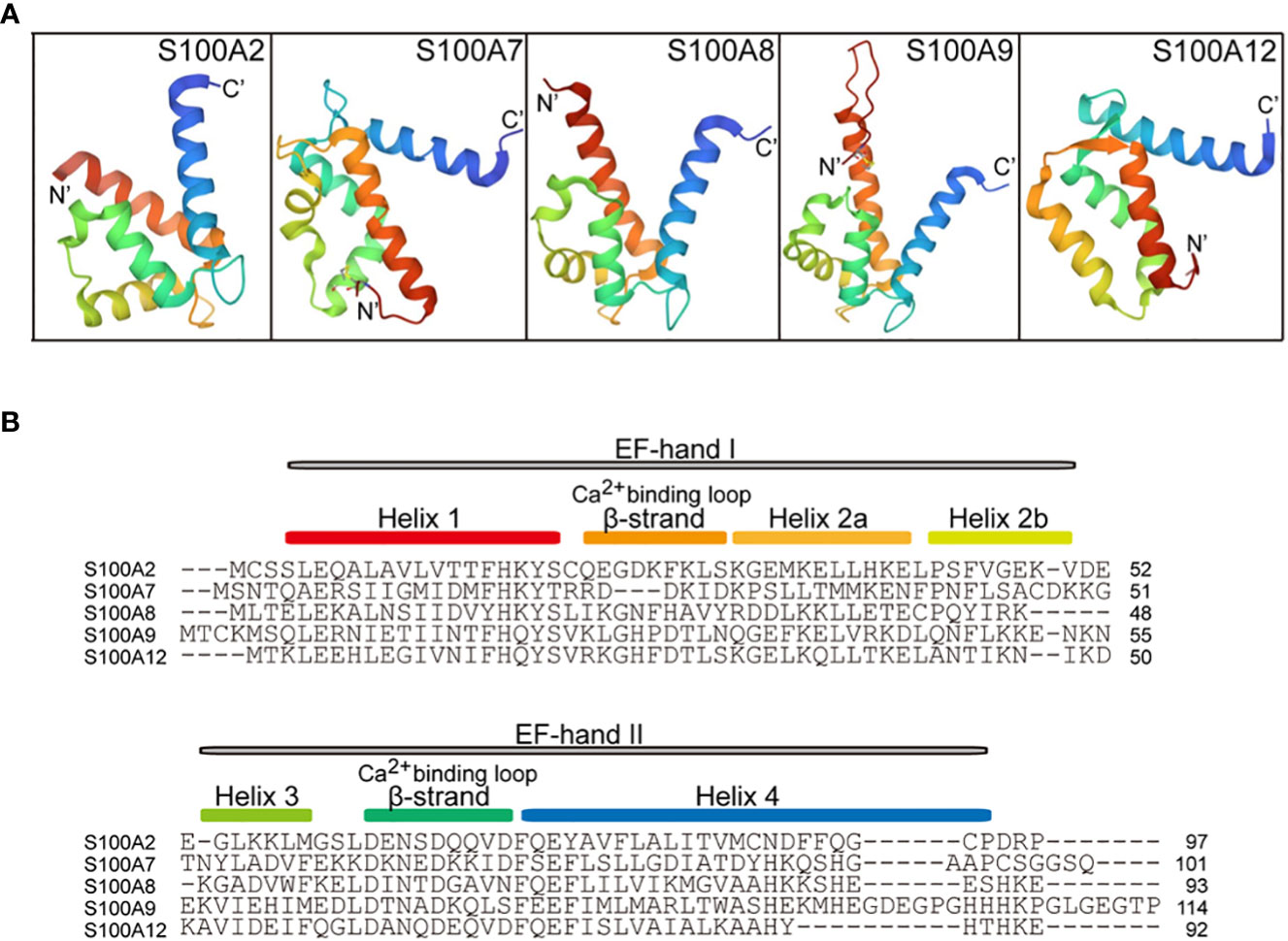
Figure 2 Cartoon representation of the tertiary structure in S100A proteins (PDB ID: 4DUQ, 2WOS, 1XK4, 6ZDY and 2WCF). (A), comparison of the crystal structure between S100A2, S100A7, S100A8, S100A9, and S100A12. The N-terminal and the C-terminal of S100 proteins indicate N’ and C’. (B), the primary secondary structure of S100A proteins. The red box indicates the helix 1 at the N-terminal, the Ca2+-binding loop 1, Helix 2a, Helix 2b, Helix 3, Ca2+-binding loop 2, and Helix 4 are indicated by the yellow box, orange box, yellow-orange box, green box, cyan box, and blue box, respectively. EF-hand regions are colored in grey boxes.
S100A2 is a member of the S100 EF-hand calcium-binding family found in mammary epithelial cells and other organs or tissues, including the lungs, kidneys, liver, and prostate gland (44). S100A2 has distinct functional roles in epithelial tissue of different origins (45). S100A2 is primarily expressed in the nucleus and moderately expressed in the cytoplasm of normal human keratinocytes (46). In the skin, S100A2 is co-localized with cytokeratin K14, an intermediate filament protein expressed primarily in basal proliferative KCs. Expression levels of S100A2 are correlated positively with the levels of cytokeratin K14. Expression levels of S100A2 gene is markedly upregulated in the involved skin of inflammatory dermatoses such as psoriasis and atopic dermatitis, and correlate positively with the severity of inflammatory skin disorders (47).
Many factors can regulate the distribution and expression levels of S100A2. For example, either oxidative stress or changes in intracellular Ca2+ levels enhance the translocation of S100A2 from the nucleus to the cytoplasm. Expression levels of the S100A2 gene can also be upregulated by multiple factors such as epidermal growth factor (EGF) (48), transforming growth factor β (TGF-β) (49), and interferon α (50). In keratinocyte cultures, EGF treatment significantly upregulated the expression levels of S100A2, while S100A2, but not EGF, is an effector of the regenerative hyperplasia pathway of epidermal differentiation (51). The binding of p53 to the binding site at the promoter of S100A2 can upregulate S100A2 expression (52–54). S100A2 protein is a direct transcriptional target of p73β and ΔNp63α, which both are required for the developmental and differentiation processes of keratinocytes (55). Moreover, ΔNp63α can interact with BRCA1, the breast/ovarian cancer susceptibility gene, upregulating S100A2 expression and enhancing tumor growth (56). Thus, multiple factors can regulate S100A2-mediated physiological and pathological reactions (47).
The S100A2 is an important regulator of keratinocyte differentiation, proliferation and wound healing. Epithelial-specific S100A2 transgenic mice exhibit increased proliferation and delayed skin wound repair (57). S100A2 and tumor suppressor factor p53, an important regulator in the wound healing process, can form a positive feedback loop to regulate the wound repair process. Moreover, S100A2 interacts with p53 to increase its transcriptional activity, and posttranslational modification of p53 increases its interaction with S100A2 (58). Electrophoresis and mass spectrometry assays showed a cross-link formation of S100A2 and S100A4 via copper-mediated oxidation of cysteine residues, resulting in increases in activation of NF-κB and secretion of TNF-α (59).
S100A2 interacts with KPNA2, a nucleocytoplasmic transport protein karyopherin α, to form a cotransport complex, transporting the tumor-associated transcription factor and regulating nucleocytoplasmic transport (60). Pulldown and coimmunoprecipitation assays revealed that the interaction between S100A2 and Hsp70/Hsp90-organizing protein or kinesin-light chain through tetratricopeptide domain modulates protein complex folding and KLC-cargo interaction (61). S100A2 can be recognized by and interact with erythropoietin, being involved in the development of tumors and other diseases (62).
RAGE (receptor for advanced glycation end products) is a member of an immunoglobulin protein family, including the extracellular part (composed of one variable like V-domain and two constants like C-type domains), transmembrane spanning helix, and a cytosolic domain (63). RAGE interacts with structurally diverse ligands, and plays an important role in human diseases. Although earlier studies did not show the interaction between S100A2 and RAGE, recent studies showed a micromolar affinity and strict calcium-dependent interaction between S100A2 and RAGE via the V-domain of RAGE (64) (Figure 3). However, further studies are needed to determine the functional significance of the interaction between S100A2 and RAGE.
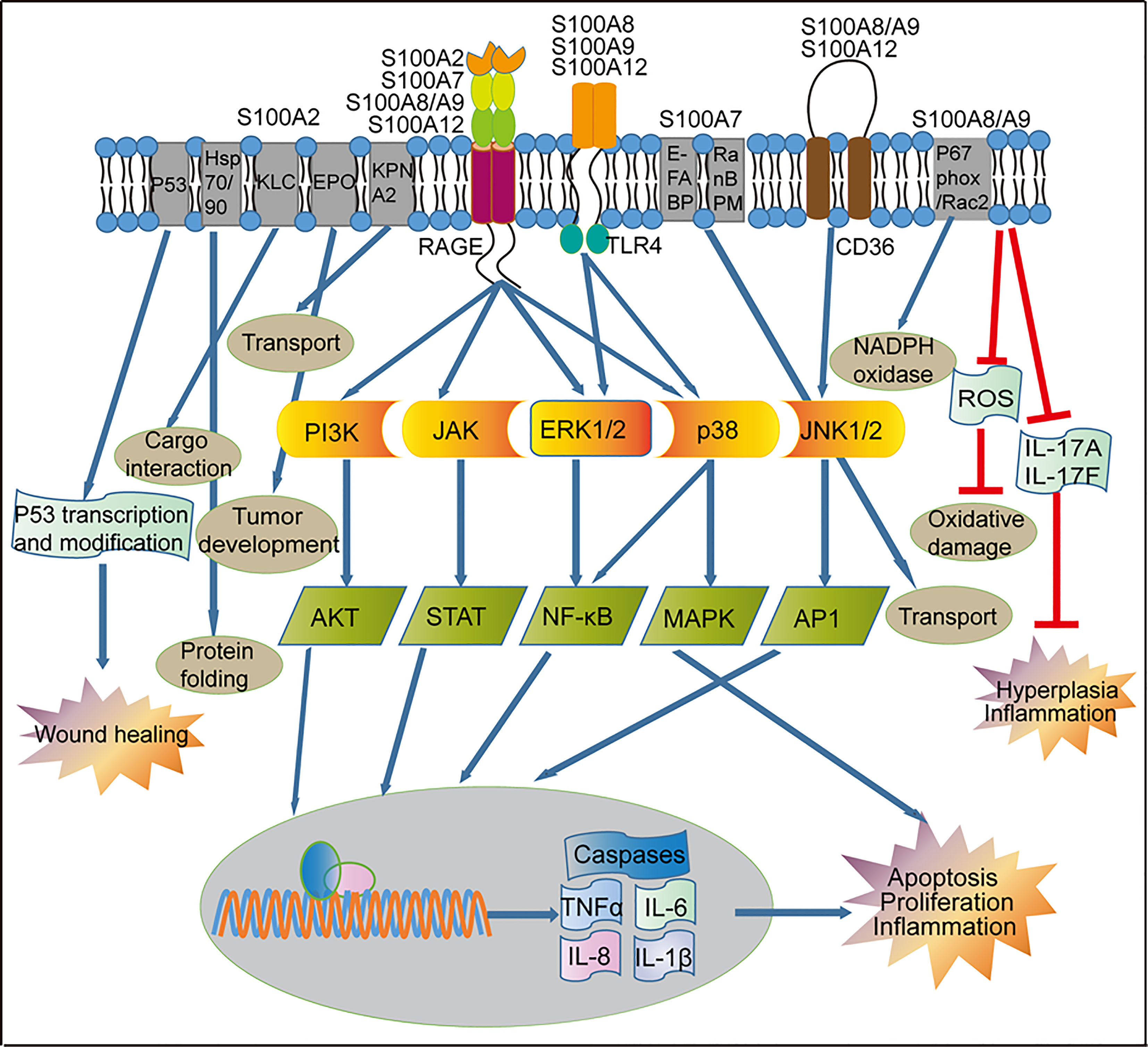
Figure 3 Regulation of signaling pathways by S100 proteins in psoriasis. Activation of RAGE by S100 proteins released from inflammatory cells can lead to the activation of the PI3K/AKT, JAK/STAT, and NF-κB pathways, which ultimately activate transcription of pro-inflammatory factors and other pathways that leads to proliferation and apoptosis. RAGE-mediated signaling can also activate the p38/MAPK signaling pathway in the immune cells, resulting in the up-regulated of genes involved in cell proliferation and inflammation. S100A8, S100A9 and S100A12 through the TLR4 receptor to activate NF-κB and MAPK. S100A8/A9 and S100A12 through the CD36 receptor activate the JNK1/2/AP1 signaling pathway and up-regulation of genes involved in cell inflammatory reactions. S100A2 and p53 can form a positive feedback loop to regulate the wound repair process in keratinocytes. S100A2 interacts with KPNA2 to form a cotransport complex, regulating nucleocytoplasmic transport in cancer cells, S100A2 interacts with Hsp70/Hsp90-organizing protein or kinesin-light chain, modulates protein complex folding and KLC-cargo interaction in fibroblasts. In keratinocytes, S100A7 promotes transport via interaction with E-FABP or RanBPM. In addition, S100A8/A9 reduces skin hyperplasia by inhibiting the production of IL-17A and IL-17F in the mouse keratinocytes. KLC, Kinesin-light chain; EPO, Erythropoietin; KPNA2, nucleocytoplasmic transport protein karyopherin α; E-FABP, epidermal fatty acid binding protein; RanBPM, Ran-binding protein M; RAGE, receptor for advanced glycation end products; TLR4, toll-like receptor 4; CD36, cluster of differentiation 36; PI3K, phosphatidylinositol 3-kinases; JAK, Janus kinase; ERK1/2, extracellular signaling-related kinase 1/2; JNK1/2, c-Jun N-terminal kinase; AKT, AGC kinase family; STAT, signal transducer and activator of transcription; NF-κB, nuclear factor kappa B, MAPK, mitogen-activated protein kinase; AP1, activator protein 1; TNF-α, tumor necrosis factor alpha; IL, interleukin.
S100A7
S100A7, also called psoriasin, was first found as a protein overexpressed in psoriasis-involved skin and later identified as a biomarker of psoriasis-involved skin (65–67). S100A7 is an EF-hand type calcium-binding protein with a molecular weight of 11.4 kDa localized in the cytoplasm of keratinocytes and distributed at the cell periphery in terminally differentiated keratinocytes (68). The three-dimensional crystal structure of S100A7 is typically a symmetric dimer containing four α helices (69). S100A7 contains two Ca2+-binding domains: an N-terminal non-canonical EF-hand domain and a C-terminal canonical EF-hand domain (Figures 1, 2). The amino acid and carboxyl ends are connected by a hinge region, which consists of 10-12 amino acid residues, and is crucial for target interactions. S100A7 lacks the ability of Ca2+-binding at the amino acid EF-hand motif, due to the absence of three amino acid residues and non-conserved substitution at position 12 of the EF-hand loop region (69). But S100A7 has a zinc-binding site composed of three histidines and an aspartate residue. The affinity of S100A7 for calcium is higher than that for zinc (70). Moreover, absence of zinc induces reorganization of the adjacent empty and distorted EF-hand loop in S100A7 structure, similar to a Ca2+-binding EF-hand (71). Contrary to most S100 proteins, S100A7 binding to calcium does not cause significant conformational changes.
Although S100A7 is constitutively expressed at relatively low levels in normal keratinocytes, its expression levels are dramatically increased in psoriatic lesions (72), suggesting its key role in response to inflammatory stimuli (73, 74) and the pathogenesis of psoriasis (68, 75). The altered expression of S100A7 is associated with keratinocyte differentiation and poor prognosis of psoriasis. Moreover, psoriasis-related cytokines and chemokines can upregulate S100A7 expression in normal and pathological conditions (76–81). S100A7 can also be produced by circulating cells, possibly contributing to systemic inflammation and psoriasis-associated comorbidities (10).
S100A7 exerts many functions, depending on its interaction with specific target proteins. Early studies suggested that epidermal fatty acid binding protein (E-FABP) is a candidate interaction target protein of S100A7 (82, 83). S100A7 and E-FABP form a complex participating in focal adhesion-related functions. Moreover, the S100A7/E-FABP complex binds to oleic acid to regulate oleic acid metabolism and transport. The S100A7/E-FABP complex is also involved in lipid transport and metabolism during epidermal barrier formation, and modulation of cell differentiation and migration in some dermatoses such as psoriasis (31, 68, 84).
Ran-binding protein M (RanBPM), a nucleoporin component of the nuclear pore complex, is another potential binding partner for S100A7. Both yeast two-hybrid and co-immunoprecipitation assays show an interaction of S100A7 with RanBPM, suggesting the involvement of S100A7/RanBPM complex in nucleocytoplasmic transport (85). Because expression levels of both nucleoporins Ran-binding protein 2 (RanBP2) and Ran-GTPase-activating protein 1 (RanGAP1) on the nuclear envelope are upregulated in the epidermis of psoriatic lesion, adequate expression of these nuclear envelope proteins is likely a prerequisite for nucleocytoplasmic transport in keratinocytes of the psoriatic epidermis (86).
Interaction of S100A7 with c-Jun activation domain-binding protein 1 (Jab1), a multifunctional signaling molecule, increases the activity of nuclear factor-κB (NF-κB) and phosphor-Akt (87, 88). The effects of S100A7 on NF-κB and phosphor-Akt pathway are dependent on the Jab1-binding domain and the interaction with Jab1 because mutation at the Jab1 binding domain of S100A7 does not stimulate phosphor-Akt (88). Treatment of keratinocytes with S100A7 also increases expression levels of both mRNA and protein of transglutaminase I and III via activation of MAPK signaling pathway. Moreover, psoriasis-involved skin displays higher expression levels of transglutaminase and alteration in the interaction between S100A7 and transglutaminase (89).
S100A7 can activate a variety of intracellular signaling pathways, and many S100A7 functions depend on the receptor for advanced glycation end products (RAGE) (90). Interaction of S100A7 with RAGE activates p38 MAPK (mitogen-activated protein kinase) and ERK (extracellular signal-regulated kinase) signaling pathways, leading to production of multiple inflammatory mediators involved in psoriasis, including IL-1α, IL-1β, IL-6, IL-8, TNF-α (91, 92) (Figure 3). S100A7 promotes cell proliferation and suppresses cell differentiation by inhibition of GATA3/Caspase14 (signal transducer and activator of transcription 3) signaling pathway (93, 94). Moreover, S100A7 can stimulate cell proliferation and angiogenesis through a RAGE-dependent up-regulation of endothelial growth factor (95). Recent study showed that lysine crotonylation at position 49 of S100A7 was suppressed in psoriatic lesions. Lysine crotonylation affects gene expression and epigenome, thereby affecting cell function and regulating immune responses (96, 97). Because of the important role of S100A7 in amplifying inflammatory process in psoriatic lesions, S100A7 becomes a potential diagnostic and therapeutic target for psoriasis.
S100A8/S100A9
S100A8 (MRP8) and S100A9 (MRP14) are Ca2+- and Zn2+-binding proteins, with damage-related molecular patterns (98). Human S100A8 and S100A9 are composed of 93 and 114 amino acid residues (Figures 1, 2B), respectively, and S100A9 has an isoform of 110 amino acid residues. The three-dimensional structure shows that both S100A8 and S100A9 have two helix-loop-helix motifs with charged amino acid residues (Figures 2A, B). The affinity of Ca2+-binding in the C-terminal EF-hand loop is higher than that in the N-terminal. S100A8 lacks the ability of Ca2+-binding in the N-terminal EF-hand motif, due to the absence of Ca2+-coordination necessary amino acid residues. S100A9 is different from other S100 proteins due to its long, very flexible C-terminal α-helix extension region (71).
S100A8 and S100A9 are predominantly expressed in myeloid cells (99, 100), epithelial cells (101), keratinocytes (102), and endothelial cells (103). S100A8 and S100A9 can form a stable homodimer or heterodimer by non-covalent bonding in the absence of calcium. S100A8 and S100A9 tend to form heterotetramers in the presence of calcium (104). The tetramer form is exposed to two high-affinity zinc ion binding sites (the crucial sites for functions) at the interface of the S100A8/S100A9 (calprotectin) complex (105, 106). S100A8 and S100A9 are the most abundant damage-associated molecular patterns in many inflammatory diseases. Expression levels of S100A8 and S100A9 are significantly elevated in psoriatic keratinocytes and leukocytes, and their expression levels correlate positively with the severity of psoriasis (107). The S100A8/S100A9 is a reliable biomarker for monitoring inflammatory disease activity (108–110).
S100A8/S100A9 complex plays a prominent role in regulation of inflammatory processes and immune response, including modulation of cell signaling transduction, differentiation, migration, apoptosis, regulation cytoskeleton, and so on (108, 111). S100A8/S100A9 complex promotes the polymerization of microtubules through direct interaction with tubulin, and modulates the cytoskeleton during the process of transendothelial migration (112). S100A8/S100A9 also stimulates neutrophil microbicidal activity by induction of reactive oxygen accumulation, resulting in activation of NADPH-oxidase (113). S100A8/S100A9-induced increase in NADPH-oxidase activity is via S100A9 transport of the cofactor arachidonic acid to the NADPH oxidase complex, and interaction of S100A8 with cytosolic phox proteins facilitating the enzyme assembly (114). Overexpression of S100A8/S100A9 induces cellular apoptosis (115), increases the production of pro-inflammatory cytokines and adhesion molecules, and enhances leukocyte adhesion and migration (116, 117). In addition, S100A8/S100A9 complex exerts bactericidal effects (118).
The role of S100A8 and S100A9 in the development of inflammation has been well demonstrated in murine models. For example, S100A8 deficiency induces a severe phenotype of psoriasis-like cutaneous inflammation, and rapid synchronous early resorption of the mouse embryo (119). Thus, S100A8 is an indispensable gene for mouse survival. Surprisingly, S100A9-deficient mice show slight decreases in the response of neutrophils to stimulation with chemoattractant (120) and slight aggravation of psoriasis symptoms compared with control mice (121), indicating functional differences between S100A8 and S100A9. In general, knock-out of the S100A8 or S100A9 leads to skin hyperplasia and aggravation of psoriatic symptoms in mouse models (121–123). S100A8 and S100A9 negatively regulate psoriatic skin inflammatory responses in mouse models. Combined with previous studies, it is suggested that S100A8 and S100A9 have both pro- and anti-inflammatory dualistic biological functions.
Several targets for S100A8/S100A9 have been identified, including Toll-like receptor 4 (TLR4) and myeloid differentiation factor 2 (MD 2). S100A8 interacts with TLR4-MD2 complex, enhancing inflammatory response in target cells (122) (Figure 3). Activation of TLR4 signaling pathway via S100A8/S100A9 appears to be crucial for inflammatory cascade response and systemic autoimmunity (123). In addition, S100A8/S100A9 complex interacts with p67phox and Rac2, increasing NADPH oxidase activity (114). S100A8/S100A9 complex is also involved in transcellular eicosanoid metabolism via interaction with scavenger receptor CD36 (124). Moreover, S100A8/S100A9 complex binds to endothelial cells via the S100A9 subunit interacting chiefly with heparin and heparan sulfate proteoglycans (125), contributing to the immobilization of the myeloid cell-derived S100A8/S100A9 complex on endothelium in human inflammatory diseases.
A line of evidence suggests the involvement of NF-κB signaling pathway in the action of S100A8 and S100A9. First, S100A8/S100A9 promotes tumor cell growth through the activation of the NF-κB signaling pathway in a RAGE-dependent manner (126), while S100A8/S100A9 can interact with RAGE (127). Second, amplification of pro-inflammatory cytokine response in macrophages by S100A8/S100A9 complex is via activation of MAPK and NF-κB signaling (128). Third, oxidative and carbonyl stresses can induce production of CML-motified (Nϵ-carboxymethyllysine) S100A8/S100A9. The latter binds to RAGE, leading to an increase in NF-κB-dependent pro-inflammatory gene expression, suggesting that CML-S100A8/S100A9 generated in inflammatory lesions can elicit a RAGE-dependent inflammatory response (129). Additionally, carboxylated glycans promotes the binding of S100A8/S100A9 to RAGE, resulting in activation of NF-κB signaling and cell proliferation (130) (Figure 3).
S100A12
S100A12, also named calgranulin C or EN-RAGE, is a pro-inflammatory protein, mainly expressed in keratinocytes of various inflammatory dermatoses (131, 132). In the psoriatic epidermis, S100A12 is expressed in the suprabasal epidermal layers (133), and its expression levels correlate positively with psoriasis severity (134). S100A12 has also been identified in neutrophils, macrophages, and lymphocytes, and functions as an innate immune defense against microorganisms. S100A12 can translocate from the cytosol to the membrane upon calcium activation.
S100A12 belongs to the S100 proteins family of EF-hand Ca2+-binding proteins, and its nucleotide sequence is localized within the epidermal differentiation complex on human chromosome 1q21. The predicted size of S100A12 is 92 amino acid residues with a molecular weight of 10.6 kDa (135) (Figures 1, 2B). The crystal structure of S100A12 reveals that monomeric subunits have four α-helices and two EF-hand motifs linked by a hinge domain (136, 137) (Figures 2A, B). The N-terminal domain of the target-binding site has two residues, Glu 5 and Glu 9, that are the most highly conserved in S100 proteins (136).
The crystal structure of Zn2+ or Ca2+/Cu2+-bound S100A12 shows that Zn2+ and Cu2+ share the same binding site. The type of ion binding to S100A12 determines its biological function (138). Binding of S100A12 to Cu2+ probably is essential in early immune reactions. Intracellular S100A12 is an anti-parallel homodimer form in the presence of zinc- and calcium- ions (138–142). Extracellular S100A12 is either a homodimer or hexamer form, with cytokine-like features.
Several mechanisms are involved in the action of S100A12. Interaction of S100A2 with target proteins regulates a variety of cellular processes and is linked to certain autoimmune responses (143). S100A12 recruits both mast cells and monocytes to inflammatory sites by its flexible hinge region during early inflammatory stages (144–146). Interaction of calcium-activated S100A12 with RAGE promotes cell proliferation and pro-inflammatory responses (147–149). The binding of S100A12 to the RAGE V domain increases cellular inflammatory response to oxidative stress (150) and participates in the pathogenesis of psoriasis (Figure 3). Moreover, S100A12 interacts with phospholipid bilayers by specific lipid and divalent ions in physiological responses (149), suggesting that it is involved in signaling events, such as migration of monocytes, located in the cell membrane (151). S100A12 binds more closely to negatively charged lipids in the presence of Ca2+ or Zn2+, both of which can change the conformation and enhance the thermal stability of S100A12 protein, further enhancing protein-membrane interaction (152). Furthermore, interaction of S100A12 with TLR4 activates TLR4 signaling pathway (Figure 3), resulting in enhanced inflammatory response and migration of monocytes (153). Additionally, interaction of S100A12 with CacyBP/SIP (Calcyclin binding protein/Siah-1 interacting protein) is via the CD36 (a class B scavenger) receptor (154, 155). Notably, RAGE upregulates while TLR4 downregulates the expression levels of CD36 (156, 157).
S100A12 exhibits pro-inflammatory cytokine-like activities and antimicrobial activity (139, 158, 159). The antimicrobial activity of S100A12 is mainly attributed to its ability to chelate metal ions during the nutritional immunity process (160–163). The binding of S100A12 to TLR4 activates TLR4 signaling pathway, and increases the expression levels of pro-inflammatory cytokines (164, 165). Carboxylated glycans on the V-domain of RAGE enhances its binding to S100A12, subsequently activating NF-κB/ERK signaling pathway (164, 166) and upregulating pro-inflammatory molecules (165). Thus, interaction of S100A12 with its specific target proteins increases pro-inflammatory immune response and induces inflammation (143).
S100A15
S100A15, also called koebnerisin or S100A7A, is recently identified as a member of the S100 protein family, and its amino acid sequences are 93% identical to S100A7 (167), suggesting the similarity of functions between S100A15 and S100A7. S100A15 protein is predictively composed of 101/104 amino acid residues, in which 13-48 residues contain the N-terminal non-canonical EF-hand domain, and 50-85 residues contain the canonical C-terminal EF-hand motif (168) (Figure 1). Interestingly, S100A15 is different from other S100 proteins because of its lack of acidic amino acids and the increased basic amino acids in the C-terminal (167).
The S100A15 gene has an alternative splicing site and two mRNA isoforms, i.e., the long isoform S100A15-L (104 amino acids) and the short isoform S100A15-S (101 amino acids) with exon 1 being spliced out (10, 90, 169). S100A15-L and S100A15-S variants are differentially expressed in normal, non-lesional and lesional skin of psoriasis. More S100A15-L than S100A15-S isoform is expressed in the skin. In comparison to normal individuals, expression levels of S100A15 are upregulated in both the uninvolved and involved skin of psoriatic subjects, with a more prominent upregulation in the involved skin.
S100A15 is an antimicrobial peptide and can increase the production and secretion of immunotropic cytokines such as TNF-α, IL-6, and IL-8 in keratinocytes, leading to the development of cutaneous inflammation (21, 170). S100A15 directly acts as a chemoattractant, promoting the infiltration of inflammatory cells and amplifying the pro-inflammatory cascade in the skin. Moreover, S100A15 act as a danger-associated molecular pattern or an alarmin factor by priming immune cells to produce pro-inflammatory mediators (168).
In contrast to S100A7, S100A15 is not able to either bind to Jab1 or interact with RAGE to activate respective signaling pathways. But it can interact with an as-yet-unknown Gi protein-coupled receptor. So far, little is known about the roles of S100A15 in multiple pathways involved in cell proliferation, migration and inflammation.
Conclusions
The S100 protein family consists of a series of homologous proteins that are involved in a wide range of intracellular processes, including inflammatory responses. The expression levels of several S100 proteins, such as S100A2, S100A7, S100A8/S100A9, S100A12, and S100A15, are significantly up-regulated in psoriasis-involved skin. The S100 protein family interacts with specific target proteins to regulate cell proliferation, differentiation, migration, signal transduction, apoptosis, energy metastasis, and more (Table 2). In addition, S100 protein family plays an important pathogenic role in inflammatory diseases such as psoriasis, and can be used as a potential biomarker to estimate the prognosis and severity of diseases. However, whether the S100 protein family can be used as drug target in the management of psoriasis and possibly other inflammatory dermatoses remains to be explored.
Author contributions
HL: data curation, visualization, writing-original draft preparation and funding acquisition; JL: reviewing and editing, funding acquisition; KZ: supervision, funding acquisition, reviewing and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81773336); the Shanxi Provincial Natural Science Foundation of China (No. 202203021211008) and the Shanxi Provincial Natural Science Foundation of China (No. 202103021224002).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Liang Y, Sarkar MK, Tsoi LC, Gudjonsson JE. Psoriasis: a mixed autoimmune and autoinflammatory disease. Curr Opin Immunol (2017) 49:1–8. doi: 10.1016/j.coi.2017.07.007
2. Rendon A, Schakel K. Psoriasis pathogenesis and treatment. Int J Mol Sci (2019) 20:1475. doi: 10.3390/ijms20061475
4. Nestle FO, Kaplan DH, Barker J. Psoriasis. N Engl J Med (2009) 361:496–509. doi: 10.1056/NEJMra0804595
5. Parisi R, Symmons DP, Griffiths CE, Ashcroft DM, Identification, Management of P, et al. Global epidemiology of psoriasis: a systematic review of incidence and prevalence. J Invest Dermatol (2013) 133:377–85. doi: 10.1038/jid.2012.339
6. Michalek IM, Loring B, John SM. A systematic review of worldwide epidemiology of psoriasis. J Eur Acad Dermatol Venereol (2017) 31:205–12. doi: 10.1111/jdv.13854
7. Kamiya K, Kishimoto M, Sugai J, Komine M, Ohtsuki M. Risk factors for the development of psoriasis. Int J Mol Sci (2019) 20:4347. doi: 10.3390/ijms20184347
8. Kuiper JJ, Prinz JC, Stratikos E, Kusnierczyk P, Arakawa A, Springer S, et al. EULAR study group on 'MHC-i-opathy': identifying disease-overarching mechanisms across disciplines and borders. Ann Rheum Dis (2023) 1–10. doi: 10.1136/ard-2022-222852
9. Lowes MA, Suarez-Farinas M, Krueger JG. Immunology of psoriasis. Annu Rev Immunol (2014) 32:227–55. doi: 10.1146/annurev-immunol-032713-120225
10. Batycka-Baran A, Hattinger E, Zwicker S, Summer B, Zack Howard OM, Thomas P, et al. Leukocyte-derived koebnerisin (S100A15) and psoriasin (S100A7) are systemic mediators of inflammation in psoriasis. J Dermatol Sci (2015) 79:214–21. doi: 10.1016/j.jdermsci.2015.05.007
11. Gerdes S, Mrowietz U, Boehncke WH. [Comorbidity in psoriasis]. Hautarzt (2016) 67:438–44. doi: 10.1007/s00105-016-3805-3
12. Prodanovich S, Kirsner RS, Kravetz JD, Ma F, Martinez L, Federman DG. Association of psoriasis with coronary artery, cerebrovascular, and peripheral vascular diseases and mortality. Arch Dermatol (2009) 145:700–3. doi: 10.1001/archdermatol.2009.94
13. Ahlehoff O, Gislason GH, Charlot M, Jorgensen CH, Lindhardsen J, Olesen JB, et al. Psoriasis is associated with clinically significant cardiovascular risk: a Danish nationwide cohort study. J Intern Med (2011) 270:147–57. doi: 10.1111/j.1365-2796.2010.02310.x
14. Armstrong EJ, Harskamp CT, Armstrong AW. Psoriasis and major adverse cardiovascular events: a systematic review and meta-analysis of observational studies. J Am Heart Assoc (2013) 2:e000062. doi: 10.1161/JAHA.113.000062
15. Samarasekera EJ, Neilson JM, Warren RB, Parnham J, Smith CH. Incidence of cardiovascular disease in individuals with psoriasis: a systematic review and meta-analysis. J Invest Dermatol (2013) 133:2340–6. doi: 10.1038/jid.2013.149
16. Xu T, Zhang YH. Association of psoriasis with stroke and myocardial infarction: meta-analysis of cohort studies. Br J Dermatol (2012) 167:1345–50. doi: 10.1111/bjd.12002
17. Di Cesare A, Di Meglio P, Nestle FO. The IL-23/Th17 axis in the immunopathogenesis of psoriasis. J Invest Dermatol (2009) 129:1339–50. doi: 10.1038/jid.2009.59
18. Benhadou F, Mintoff D, Del Marmol V. Psoriasis: keratinocytes or immune cells - which is the trigger? Dermatology (2019) 235:91–100. doi: 10.1159/000495291
19. Harden JL, Krueger JG, Bowcock AM. The immunogenetics of psoriasis: a comprehensive review. J Autoimmun (2015) 64:66–73. doi: 10.1016/j.jaut.2015.07.008
20. Morizane S, Gallo RL. Antimicrobial peptides in the pathogenesis of psoriasis. J Dermatol (2012) 39:225–30. doi: 10.1111/j.1346-8138.2011.01483.x
21. Batycka-Baran A, Maj J, Wolf R, Szepietowski JC. The new insight into the role of antimicrobial proteins-alarmins in the immunopathogenesis of psoriasis. J Immunol Res (2014) 2014:628289. doi: 10.1155/2014/628289
22. Takahashi T, Yamasaki K. Psoriasis and antimicrobial peptides. Int J Mol Sci (2020) 21:6791. doi: 10.3390/ijms21186791
23. Heizmann CW, Fritz G, Schafer BW. S100 proteins: structure, functions and pathology. Front Biosci (2002) 7:d1356–68. doi: 10.2741/A846
24. Donato R. Intracellular and extracellular roles of S100 proteins. Microsc Res Tech (2003) 60:540–51. doi: 10.1002/jemt.10296
25. Chan JK, Roth J, Oppenheim JJ, Tracey KJ, Vogl T, Feldmann M, et al. Alarmins: awaiting a clinical response. J Clin Invest (2012) 122:2711–9. doi: 10.1172/JCI62423
26. Kraus C, Rohde D, Weidenhammer C, Qiu G, Pleger ST, Voelkers M, et al. S100A1 in cardiovascular health and disease: closing the gap between basic science and clinical therapy. J Mol Cell Cardiol (2009) 47:445–55. doi: 10.1016/j.yjmcc.2009.06.003
27. Bresnick AR, Weber DJ, Zimmer DB. S100 proteins in cancer. Nat Rev Cancer (2015) 15:96–109. doi: 10.1038/nrc3893
28. Cristovao JS, Gomes CM. S100 proteins in alzheimer's disease. Front Neurosci (2019) 13:463. doi: 10.3389/fnins.2019.00463
29. Austermann J, Spiekermann C, Roth J. S100 proteins in rheumatic diseases. Nat Rev Rheumatol (2018) 14:528–41. doi: 10.1038/s41584-018-0058-9
30. Donato R, Cannon BR, Sorci G, Riuzzi F, Hsu K, Weber DJ, et al. Functions of S100 proteins. Curr Mol Med (2013) 13:24–57. doi: 10.2174/156652413804486214
31. Eckert RL, Broome AM, Ruse M, Robinson N, Ryan D, Lee K. S100 proteins in the epidermis. J Invest Dermatol (2004) 123:23–33. doi: 10.1111/j.0022-202X.2004.22719.x
32. Lai Y, Gallo RL. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol (2009) 30:131–41. doi: 10.1016/j.it.2008.12.003
33. Marenholz I, Heizmann CW, Fritz G. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature). Biochem Biophys Res Commun (2004) 322:1111–22. doi: 10.1016/j.bbrc.2004.07.096
34. Schafer BW, Heizmann CW. The S100 family of EF-hand calcium-binding proteins: functions and pathology. Trends Biochem Sci (1996) 21:134–40. doi: 10.1016/s0968-0004(96)80167-8
35. Wright NT, Cannon BR, Wilder PT, Morgan MT, Varney KM, Zimmer DB, et al. Solution structure of S100A1 bound to the CapZ peptide (TRTK12). J Mol Biol (2009) 386:1265–77. doi: 10.1016/j.jmb.2009.01.022
36. Markowitz J, Rustandi RR, Varney KM, Wilder PT, Udan R, Wu SL, et al. Calcium-binding properties of wild-type and EF-hand mutants of S100B in the presence and absence of a peptide derived from the c-terminal negative regulatory domain of p53. Biochemistry (2005) 44:7305–14. doi: 10.1021/bi050321t
37. Zimmer DB, Wright Sadosky P, Weber DJ. Molecular mechanisms of S100-target protein interactions. Microsc Res Tech (2003) 60:552–9. doi: 10.1002/jemt.10297
38. Otterbein LR, Kordowska J, Witte-Hoffmann C, Wang CL, Dominguez R. Crystal structures of S100A6 in the Ca(2+)-free and Ca(2+)-bound states: the calcium sensor mechanism of S100 proteins revealed at atomic resolution. Structure (2002) 10:557–67. doi: 10.1016/s0969-2126(02)00740-2
39. McClintock KA, Shaw GS. A novel S100 target conformation is revealed by the solution structure of the Ca2+-S100B-TRTK-12 complex. J Biol Chem (2003) 278:6251–7. doi: 10.1074/jbc.M210622200
40. Lee SW, Tomasetto C, Sager R. Positive selection of candidate tumor-suppressor genes by subtractive hybridization. Proc Natl Acad Sci U.S.A. (1991) 88:2825–9. doi: 10.1073/pnas.88.7.2825
41. Koch M, Diez J, Fritz G. Crystal structure of Ca2+ -free S100A2 at 1.6-a resolution. J Mol Biol (2008) 378:933–42. doi: 10.1016/j.jmb.2008.03.019
42. Koch M, Bhattacharya S, Kehl T, Gimona M, Vasak M, Chazin W, et al. Implications on zinc binding to S100A2. Biochim Biophys Acta (2007) 1773:457–70. doi: 10.1016/j.bbamcr.2006.12.006
43. Botelho HM, Koch M, Fritz G, Gomes CM. Metal ions modulate the folding and stability of the tumor suppressor protein S100A2. FEBS J (2009) 276:1776–86. doi: 10.1111/j.1742-4658.2009.06912.x
44. Wolf S, Haase-Kohn C, Pietzsch J. S100A2 in cancerogenesis: a friend or a foe? Amino Acids (2011) 41:849–61. doi: 10.1007/s00726-010-0623-2
45. Nagy N, Hoyaux D, Gielen I, Schafer BW, Pochet R, Heizmann CW, et al. The Ca2+-binding S100A2 protein is differentially expressed in epithelial tissue of glandular or squamous origin. Histol Histopathol (2002) 17:123–30. doi: 10.14670/HH-17.123
46. Zhang T, Woods TL, Elder JT. Differential responses of S100A2 to oxidative stress and increased intracellular calcium in normal, immortalized, and malignant human keratinocytes. J Invest Dermatol (2002) 119:1196–201. doi: 10.1046/j.1523-1747.2002.19520.x
47. Yoshioka M, Sawada Y, Saito-Sasaki N, Yoshioka H, Hama K, Omoto D, et al. High S100A2 expression in keratinocytes in patients with drug eruption. Sci Rep (2021) 11:5493. doi: 10.1038/s41598-021-85009-8
48. Lee SW, Tomasetto C, Swisshelm K, Keyomarsi K, Sager R. Down-regulation of a member of the S100 gene family in mammary carcinoma cells and reexpression by azadeoxycytidine treatment. Proc Natl Acad Sci U.S.A. (1992) 89:2504–8. doi: 10.1073/pnas.89.6.2504
49. Naz S, Ranganathan P, Bodapati P, Shastry AH, Mishra LN, Kondaiah P. Regulation of S100A2 expression by TGF-beta-induced MEK/ERK signalling and its role in cell migration/invasion. Biochem J (2012) 447:81–91. doi: 10.1042/BJ20120014
50. Foser S, Redwanz I, Ebeling M, Heizmann CW, Certa U. Interferon-alpha and transforming growth factor-beta co-induce growth inhibition of human tumor cells. Cell Mol Life Sci (2006) 63:2387–96. doi: 10.1007/s00018-006-6256-7
51. Stoll SW, Zhao X, Elder JT. EGF stimulates transcription of CaN19 (S100A2) in HaCaT keratinocytes. J Invest Dermatol (1998) 111:1092–7. doi: 10.1046/j.1523-1747.1998.00402.x
52. Kirschner RD, Sanger K, Muller GA, Engeland K. Transcriptional activation of the tumor suppressor and differentiation gene S100A2 by a novel p63-binding site. Nucleic Acids Res (2008) 36:2969–80. doi: 10.1093/nar/gkn132
53. van Dieck J, Fernandez-Fernandez MR, Veprintsev DB, Fersht AR. Modulation of the oligomerization state of p53 by differential binding of proteins of the S100 family to p53 monomers and tetramers. J Biol Chem (2009) 284:13804–11. doi: 10.1074/jbc.M901351200
54. Koch M, Diez J, Wagner A, Fritz G. Crystallization and calcium/sulfur SAD phasing of the human EF-hand protein S100A2. Acta Crystallogr Sect F Struct Biol Cryst Commun (2010) 66:1032–6. doi: 10.1107/S1744309110030691
55. Lapi E, Iovino A, Fontemaggi G, Soliera AR, Iacovelli S, Sacchi A, et al. S100A2 gene is a direct transcriptional target of p53 homologues during keratinocyte differentiation. Oncogene (2006) 25:3628–37. doi: 10.1038/sj.onc.1209401
56. Buckley NE, D'Costa Z, Kaminska M, Mullan PB. S100A2 is a BRCA1/p63 coregulated tumour suppressor gene with roles in the regulation of mutant p53 stability. Cell Death Dis (2014) 5:e1070. doi: 10.1038/cddis.2014.31
57. Pan SC, Li CY, Kuo CY, Kuo YZ, Fang WY, Huang YH, et al. The p53-S100A2 positive feedback loop negatively regulates epithelialization in cutaneous wound healing. Sci Rep (2018) 8:5458. doi: 10.1038/s41598-018-23697-5
58. van Dieck J, Teufel DP, Jaulent AM, Fernandez-Fernandez MR, Rutherford TJ, Wyslouch-Cieszynska A, et al. Posttranslational modifications affect the interaction of S100 proteins with tumor suppressor p53. J Mol Biol (2009) 394:922–30. doi: 10.1016/j.jmb.2009.10.002
59. Haase-Kohn C, Wolf S, Lenk J, Pietzsch J. Copper-mediated cross-linking of S100A4, but not of S100A2, results in proinflammatory effects in melanoma cells. Biochem Biophys Res Commun (2011) 413:494–8. doi: 10.1016/j.bbrc.2011.08.132
60. Han F, Zhang L, Liao S, Zhang Y, Qian L, Hou F, et al. The interaction between S100A2 and KPNA2 mediates NFYA nuclear import and is a novel therapeutic target for colorectal cancer metastasis. Oncogene (2022) 41:657–70. doi: 10.1038/s41388-021-02116-6
61. Shimamoto S, Takata M, Tokuda M, Oohira F, Tokumitsu H, Kobayashi R. Interactions of S100A2 and S100A6 with the tetratricopeptide repeat proteins, Hsp90/Hsp70-organizing protein and kinesin light chain. J Biol Chem (2008) 283:28246–58. doi: 10.1074/jbc.M801473200
62. Kazakov AS, Deryusheva EI, Sokolov AS, Permyakova ME, Litus EA, Rastrygina VA, et al. Erythropoietin interacts with specific S100 proteins. Biomolecules (2022) 12:120. doi: 10.3390/biom12010120
63. Schmidt AM, Vianna M, Gerlach M, Brett J, Ryan J, Kao J, et al. Isolation and characterization of two binding proteins for advanced glycosylation end products from bovine lung which are present on the endothelial cell surface. J Biol Chem (1992) 267:14987–97. doi: 10.1016/S0021-9258(18)42137-0
64. Leclerc E, Fritz G, Vetter SW, Heizmann CW. Binding of S100 proteins to RAGE: an update. Biochim Biophys Acta (2009) 1793:993–1007. doi: 10.1016/j.bbamcr.2008.11.016
65. Madsen P, Rasmussen HH, Leffers H, Honore B, Dejgaard K, Olsen E, et al. Molecular cloning, occurrence, and expression of a novel partially secreted protein "psoriasin" that is highly up-regulated in psoriatic skin. J Invest Dermatol (1991) 97:701–12. doi: 10.1111/1523-1747.ep12484041
66. Kulski JK, Lim CP, Dunn DS, Bellgard M. Genomic and phylogenetic analysis of the S100A7 (Psoriasin) gene duplications within the region of the S100 gene cluster on human chromosome 1q21. J Mol Evol (2003) 56:397–406. doi: 10.1007/s00239-002-2410-5
67. Wolf R, Voscopoulos CJ, FitzGerald PC, Goldsmith P, Cataisson C, Gunsior M, et al. The mouse S100A15 ortholog parallels genomic organization, structure, gene expression, and protein-processing pattern of the human S100A7/A15 subfamily during epidermal maturation. J Invest Dermatol (2006) 126:1600–8. doi: 10.1038/sj.jid.5700210
68. Broome AM, Ryan D, Eckert RL. S100 protein subcellular localization during epidermal differentiation and psoriasis. J Histochem Cytochem (2003) 51:675–85. doi: 10.1177/002215540305100513
69. Brodersen DE, Nyborg J, Kjeldgaard M. Zinc-binding site of an S100 protein revealed. two crystal structures of Ca2+-bound human psoriasin (S100A7) in the Zn2+-loaded and Zn2+-free states. Biochemistry (1999) 38:1695–704. doi: 10.1021/bi982483d
70. Vorum H, Madsen P, Rasmussen HH, Etzerodt M, Svendsen I, Celis JE, et al. Expression and divalent cation binding properties of the novel chemotactic inflammatory protein psoriasin. Electrophoresis (1996) 17:1787–96. doi: 10.1002/elps.1150171118
71. Heizmann CW, Ackermann GE, Galichet A. Pathologies involving the S100 proteins and RAGE. Subcell Biochem (2007) 45:93–138. doi: 10.1007/978-1-4020-6191-2_5
72. Alowami S, Qing G, Emberley E, Snell L, Watson PH. Psoriasin (S100A7) expression is altered during skin tumorigenesis. BMC Dermatol (2003) 3:1. doi: 10.1186/1471-5945-3-1
73. Algermissen B, Sitzmann J, LeMotte P, Czarnetzki B. Differential expression of CRABP II, psoriasin and cytokeratin 1 mRNA in human skin diseases. Arch Dermatol Res (1996) 288:426–30. doi: 10.1007/BF02505229
74. Jinquan T, Vorum H, Larsen CG, Madsen P, Rasmussen HH, Gesser B, et al. Psoriasin: a novel chemotactic protein. J Invest Dermatol (1996) 107:5–10. doi: 10.1111/1523-1747.ep12294284
75. D'Amico F, Trovato C, Skarmoutsou E, Rossi GA, Granata M, Longo V, et al. Effects of adalimumab, etanercept and ustekinumab on the expression of psoriasin (S100A7) in psoriatic skin. J Dermatol Sci (2015) 80:38–44. doi: 10.1016/j.jdermsci.2015.07.009
76. Johnston A, Gudjonsson JE, Aphale A, Guzman AM, Stoll SW, Elder JT. EGFR and IL-1 signaling synergistically promote keratinocyte antimicrobial defenses in a differentiation-dependent manner. J Invest Dermatol (2011) 131:329–37. doi: 10.1038/jid.2010.313
77. Rabeony H, Petit-Paris I, Garnier J, Barrault C, Pedretti N, Guilloteau K, et al. Inhibition of keratinocyte differentiation by the synergistic effect of IL-17A, IL-22, IL-1alpha, TNFalpha and oncostatin m. PloS One (2014) 9:e101937. doi: 10.1371/journal.pone.0101937
78. Yu N, Liu S, Yi X, Zhang S, Ding Y. Serum amyloid a induces interleukin-1beta secretion from keratinocytes via the NACHT, LRR and PYD domains-containing protein 3 inflammasome. Clin Exp Immunol (2015) 179:344–53. doi: 10.1111/cei.12458
79. Keermann M, Koks S, Reimann E, Abram K, Erm T, Silm H, et al. Expression of IL-36 family cytokines and IL-37 but not IL-38 is altered in psoriatic skin. J Dermatol Sci (2015) 80:150–2. doi: 10.1016/j.jdermsci.2015.08.002
80. Guttman-Yassky E, Lowes MA, Fuentes-Duculan J, Zaba LC, Cardinale I, Nograles KE, et al. Low expression of the IL-23/Th17 pathway in atopic dermatitis compared to psoriasis. J Immunol (2008) 181:7420–7. doi: 10.4049/jimmunol.181.10.7420
81. Liang SC, Tan XY, Luxenberg DP, Karim R, Dunussi-Joannopoulos K, Collins M, et al. Interleukin (IL)-22 and IL-17 are coexpressed by Th17 cells and cooperatively enhance expression of antimicrobial peptides. J Exp Med (2006) 203:2271–9. doi: 10.1084/jem.20061308
82. Hagens G, Roulin K, Hotz R, Saurat JH, Hellman U, Siegenthaler G. Probable interaction between S100A7 and e-FABP in the cytosol of human keratinocytes from psoriatic scales. Mol Cell Biochem (1999) 192:123–8. doi: 10.1023/A:1006894909694
83. Ruse M, Broome AM, Eckert RL. S100A7 (psoriasin) interacts with epidermal fatty acid binding protein and localizes in focal adhesion-like structures in cultured keratinocytes. J Invest Dermatol (2003) 121:132–41. doi: 10.1046/j.1523-1747.2003.12309.x
84. Dallaglio K, Marconi A, Truzzi F, Lotti R, Palazzo E, Petrachi T, et al. E-FABP induces differentiation in normal human keratinocytes and modulates the differentiation process in psoriatic keratinocytes. vitro Exp Dermatol (2013) 22:255–61. doi: 10.1111/exd.12111
85. Emberley ED, Gietz RD, Campbell JD, HayGlass KT, Murphy LC, Watson PH. RanBPM interacts with psoriasin in vitro and their expression correlates with specific clinical features in vivo in breast cancer. BMC Cancer (2002) 2:28. doi: 10.1186/1471-2407-2-28
86. Yasuda K, Sugiura K, Takeichi T, Ogawa Y, Muro Y, Akiyama M. Nuclear envelope localization of ran-binding protein 2 and ran-GTPase-activating protein 1 in psoriatic epidermal keratinocytes. Exp Dermatol (2014) 23:119–24. doi: 10.1111/exd.12324
87. Emberley ED, Niu Y, Leygue E, Tomes L, Gietz RD, Murphy LC, et al. Psoriasin interacts with Jab1 and influences breast cancer progression. Cancer Res (2003) 63:1954–61.
88. Emberley ED, Niu Y, Curtis L, Troup S, Mandal SK, Myers JN, et al. The S100A7-c-Jun activation domain binding protein 1 pathway enhances prosurvival pathways in breast cancer. Cancer Res (2005) 65:5696–702. doi: 10.1158/0008-5472.CAN-04-3927
89. Ling S, Xu B, Luo Y, Fang X, Liu X, Wang A, et al. Transglutaminase 3 attenuates skin inflammation in psoriasis by inhibiting NF-kappaB activation through phosphorylated STAT3-TET3 signaling. J Invest Dermatol (2022) 142:2968–77 e10. doi: 10.1016/j.jid.2022.03.035
90. Wolf R, Howard OM, Dong HF, Voscopoulos C, Boeshans K, Winston J, et al. Chemotactic activity of S100A7 (Psoriasin) is mediated by the receptor for advanced glycation end products and potentiates inflammation with highly homologous but functionally distinct S100A15. J Immunol (2008) 181:1499–506. doi: 10.4049/jimmunol.181.2.1499
91. Lei H, Li X, Jing B, Xu H, Wu Y. Human S100A7 induces mature Interleukin1alpha expression by RAGE-p38 MAPK-Calpain1 pathway in psoriasis. PloS One (2017) 12:e0169788. doi: 10.1371/journal.pone.0169788
92. Zheng Y, Niyonsaba F, Ushio H, Ikeda S, Nagaoka I, Okumura K, et al. Microbicidal protein psoriasin is a multifunctional modulator of neutrophil activation. Immunology (2008) 124:357–67. doi: 10.1111/j.1365-2567.2007.02782.x
93. Walsh DS, Borke JL, Singh BB, Do NN, Hsu SD, Balagon MV, et al. Psoriasis is characterized by altered epidermal expression of caspase 14, a novel regulator of keratinocyte terminal differentiation and barrier formation. J Dermatol Sci (2005) 37:61–3. doi: 10.1016/j.jdermsci.2004.10.003
94. Li T, Qi Z, Kong F, Li Y, Wang R, Zhang W, et al. S100A7 acts as a dual regulator in promoting proliferation and suppressing squamous differentiation through GATA-3/caspase-14 pathway in A431 cells. Exp Dermatol (2015) 24:342–8. doi: 10.1111/exd.12645
95. Shubbar E, Vegfors J, Carlstrom M, Petersson S, Enerback C. Psoriasin (S100A7) increases the expression of ROS and VEGF and acts through RAGE to promote endothelial cell proliferation. Breast Cancer Res Treat (2012) 134:71–80. doi: 10.1007/s10549-011-1920-5
96. Fellows R, Denizot J, Stellato C, Cuomo A, Jain P, Stoyanova E, et al. Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases. Nat Commun (2018) 9:105. doi: 10.1038/s41467-017-02651-5
97. Yang J, He Z, Chen C, Li S, Qian J, Zhao J, et al. Toxoplasma gondii infection inhibits histone crotonylation to regulate immune response of porcine alveolar macrophages. Front Immunol (2021) 12:696061. doi: 10.3389/fimmu.2021.696061
98. Edgeworth J, Gorman M, Bennett R, Freemont P, Hogg N. Identification of p8,14 as a highly abundant heterodimeric calcium binding protein complex of myeloid cells. J Biol Chem (1991) 266:7706–13. doi: 10.1016/S0021-9258(20)89506-4
99. Qin H, Lerman B, Sakamaki I, Wei G, Cha SC, Rao SS, et al. Generation of a new therapeutic peptide that depletes myeloid-derived suppressor cells in tumor-bearing mice. Nat Med (2014) 20:676–81. doi: 10.1038/nm.3560
100. Averill MM, Barnhart S, Becker L, Li X, Heinecke JW, Leboeuf RC, et al. S100A9 differentially modifies phenotypic states of neutrophils, macrophages, and dendritic cells: implications for atherosclerosis and adipose tissue inflammation. Circulation (2011) 123:1216–26. doi: 10.1161/CIRCULATIONAHA.110.985523
101. Henke MO, Renner A, Rubin BK, Gyves JI, Lorenz E, Koo JS. Up-regulation of S100A8 and S100A9 protein in bronchial epithelial cells by lipopolysaccharide. Exp Lung Res (2006) 32:331–47. doi: 10.1080/01902140600959580
102. Grimbaldeston MA, Geczy CL, Tedla N, Finlay-Jones JJ, Hart PH. S100A8 induction in keratinocytes by ultraviolet a irradiation is dependent on reactive oxygen intermediates. J Invest Dermatol (2003) 121:1168–74. doi: 10.1046/j.1523-1747.2003.12561.x
103. Perera C, McNeil HP, Geczy CL. S100 calgranulins in inflammatory arthritis. Immunol Cell Biol (2010) 88:41–9. doi: 10.1038/icb.2009.88
104. Vogl T, Leukert N, Barczyk K, Strupat K, Roth J. Biophysical characterization of S100A8 and S100A9 in the absence and presence of bivalent cations. Biochim Biophys Acta (2006) 1763:1298–306. doi: 10.1016/j.bbamcr.2006.08.028
105. Korndorfer IP, Brueckner F, Skerra A. The crystal structure of the human (S100A8/S100A9)2 heterotetramer, calprotectin, illustrates how conformational changes of interacting alpha-helices can determine specific association of two EF-hand proteins. J Mol Biol (2007) 370:887–98. doi: 10.1016/j.jmb.2007.04.065
106. Kehl-Fie TE, Chitayat S, Hood MI, Damo S, Restrepo N, Garcia C, et al. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of staphylococcus aureus. Cell Host Microbe (2011) 10:158–64. doi: 10.1016/j.chom.2011.07.004
107. Grantham HJ, Hussain AB, Reynolds NJ. Serum S100A8/A9 may act as biomarker of atherosclerosis severity in psoriasis. J Invest Dermatol (2022) 142:2848–50. doi: 10.1016/j.jid.2022.06.018
108. Austermann J, Zenker S, Roth J. S100-alarmins: potential therapeutic targets for arthritis. Expert Opin Ther Targets (2017) 21:739–51. doi: 10.1080/14728222.2017.1330411
109. Wang S, Song R, Wang Z, Jing Z, Wang S, Ma J. S100A8/A9 in inflammation. Front Immunol (2018) 9:1298. doi: 10.3389/fimmu.2018.01298
110. Vogl T, Eisenblatter M, Voller T, Zenker S, Hermann S, van Lent P, et al. Alarmin S100A8/S100A9 as a biomarker for molecular imaging of local inflammatory activity. Nat Commun (2014) 5:4593. doi: 10.1038/ncomms5593
111. Pruenster M, Vogl T, Roth J, Sperandio M. S100A8/A9: from basic science to clinical application. Pharmacol Ther (2016) 167:120–31. doi: 10.1016/j.pharmthera.2016.07.015
112. Vogl T, Ludwig S, Goebeler M, Strey A, Thorey IS, Reichelt R, et al. MRP8 and MRP14 control microtubule reorganization during transendothelial migration of phagocytes. Blood (2004) 104:4260–8. doi: 10.1182/blood-2004-02-0446
113. Simard JC, Simon MM, Tessier PA, Girard D. Damage-associated molecular pattern S100A9 increases bactericidal activity of human neutrophils by enhancing phagocytosis. J Immunol (2011) 186:3622–31. doi: 10.4049/jimmunol.1002956
114. Kerkhoff C, Nacken W, Benedyk M, Dagher MC, Sopalla C, Doussiere J. The arachidonic acid-binding protein S100A8/A9 promotes NADPH oxidase activation by interaction with p67phox and rac-2. FASEB J (2005) 19:467–9. doi: 10.1096/fj.04-2377fje
115. Li C, Chen H, Ding F, Zhang Y, Luo A, Wang M, et al. A novel p53 target gene, S100A9, induces p53-dependent cellular apoptosis and mediates the p53 apoptosis pathway. Biochem J (2009) 422:363–72. doi: 10.1042/BJ20090465
116. Viemann D, Strey A, Janning A, Jurk K, Klimmek K, Vogl T, et al. Myeloid-related proteins 8 and 14 induce a specific inflammatory response in human microvascular endothelial cells. Blood (2005) 105:2955–62. doi: 10.1182/blood-2004-07-2520
117. Koike A, Arai S, Yamada S, Nagae A, Saita N, Itoh H, et al. Dynamic mobility of immunological cells expressing S100A8 and S100A9 in vivo: a variety of functional roles of the two proteins as regulators in acute inflammatory reaction. Inflammation (2012) 35:409–19. doi: 10.1007/s10753-011-9330-8
118. Miyasaki KT, Bodeau AL, Murthy AR, Lehrer RI. In vitro antimicrobial activity of the human neutrophil cytosolic s-100 protein complex, calprotectin, against capnocytophaga sputigena. J Dent Res (1993) 72:517–23. doi: 10.1177/00220345930720020801
119. Passey RJ, Williams E, Lichanska AM, Wells C, Hu S, Geczy CL, et al. A null mutation in the inflammation-associated S100 protein S100A8 causes early resorption of the mouse embryo. J Immunol (1999) 163:2209–16. doi: 10.4049/jimmunol.163.4.2209
120. Hobbs JA, May R, Tanousis K, McNeill E, Mathies M, Gebhardt C, et al. Myeloid cell function in MRP-14 (S100A9) null mice. Mol Cell Biol (2003) 23:2564–76. doi: 10.1128/MCB.23.7.2564-2576.2003
121. Defrene J, Berrazouane S, Esparza N, Page N, Cote MF, Gobeil S, et al. Deletion of S100a8 and S100a9 enhances skin hyperplasia and promotes the Th17 response in imiquimod-induced psoriasis. J Immunol (2021) 206:505–14. doi: 10.4049/jimmunol.2000087
122. Vogl T, Tenbrock K, Ludwig S, Leukert N, Ehrhardt C, van Zoelen MA, et al. Mrp8 and Mrp14 are endogenous activators of toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat Med (2007) 13:1042–9. doi: 10.1038/nm1638
123. Loser K, Vogl T, Voskort M, Lueken A, Kupas V, Nacken W, et al. The toll-like receptor 4 ligands Mrp8 and Mrp14 are crucial in the development of autoreactive CD8+ T cells. Nat Med (2010) 16:713–7. doi: 10.1038/nm.2150
124. Kerkhoff C, Sorg C, Tandon NN, Nacken W. Interaction of S100A8/S100A9-arachidonic acid complexes with the scavenger receptor CD36 may facilitate fatty acid uptake by endothelial cells. Biochemistry (2001) 40:241–8. doi: 10.1021/bi001791k
125. Robinson MJ, Tessier P, Poulsom R, Hogg N. The S100 family heterodimer, MRP-8/14, binds with high affinity to heparin and heparan sulfate glycosaminoglycans on endothelial cells. J Biol Chem (2002) 277:3658–65. doi: 10.1074/jbc.M102950200
126. Ghavami S, Rashedi I, Dattilo BM, Eshraghi M, Chazin WJ, Hashemi M, et al. S100A8/A9 at low concentration promotes tumor cell growth via RAGE ligation and MAP kinase-dependent pathway. J Leukoc Biol (2008) 83:1484–92. doi: 10.1189/jlb.0607397
127. Boyd JH, Kan B, Roberts H, Wang Y, Walley KR. S100A8 and S100A9 mediate endotoxin-induced cardiomyocyte dysfunction via the receptor for advanced glycation end products. Circ Res (2008) 102:1239–46. doi: 10.1161/CIRCRESAHA.107.167544
128. Sunahori K, Yamamura M, Yamana J, Takasugi K, Kawashima M, Yamamoto H, et al. The S100A8/A9 heterodimer amplifies proinflammatory cytokine production by macrophages via activation of nuclear factor kappa b and p38 mitogen-activated protein kinase in rheumatoid arthritis. Arthritis Res Ther (2006) 8:R69. doi: 10.1186/ar1939
129. Andrassy M, Igwe J, Autschbach F, Volz C, Remppis A, Neurath MF, et al. Posttranslationally modified proteins as mediators of sustained intestinal inflammation. Am J Pathol (2006) 169:1223–37. doi: 10.2353/ajpath.2006.050713
130. Turovskaya O, Foell D, Sinha P, Vogl T, Newlin R, Nayak J, et al. RAGE, carboxylated glycans and S100A8/A9 play essential roles in colitis-associated carcinogenesis. Carcinogenesis (2008) 29:2035–43. doi: 10.1093/carcin/bgn188
131. Moroz OV, Antson AA, Grist SJ, Maitland NJ, Dodson GG, Wilson KS, et al. Structure of the human S100A12-copper complex: implications for host-parasite defence. Acta Crystallogr D Biol Crystallogr (2003) 59:859–67. doi: 10.1107/s0907444903004700
132. Roth J, Vogl T, Sorg C, Sunderkotter C. Phagocyte-specific S100 proteins: a novel group of proinflammatory molecules. Trends Immunol (2003) 24:155–8. doi: 10.1016/s1471-4906(03)00062-0
133. Mirmohammadsadegh A, Tschakarjan E, Ljoljic A, Bohner K, Michel G, Ruzicka T, et al. Calgranulin c is overexpressed in lesional psoriasis. J Invest Dermatol (2000) 114:1207–8. doi: 10.1046/j.1523-1747.2000.00005-2.x
134. Wilsmann-Theis D, Wagenpfeil J, Holzinger D, Roth J, Koch S, Schnautz S, et al. Among the S100 proteins, S100A12 is the most significant marker for psoriasis disease activity. J Eur Acad Dermatol Venereol (2016) 30:1165–70. doi: 10.1111/jdv.13269
135. Ilg EC, Troxler H, Burgisser DM, Kuster T, Markert M, Guignard F, et al. Amino acid sequence determination of human S100A12 (P6, calgranulin c, CGRP, CAAF1) by tandem mass spectrometry. Biochem Biophys Res Commun (1996) 225:146–50. doi: 10.1006/bbrc.1996.1144
136. Moroz OV, Antson AA, Murshudov GN, Maitland NJ, Dodson GG, Wilson KS, et al. The three-dimensional structure of human S100A12. Acta Crystallogr D Biol Crystallogr (2001) 57:20–9. doi: 10.1107/s090744490001458x
137. Moroz OV, Dodson GG, Wilson KS, Lukanidin E, Bronstein IB. Multiple structural states of S100A12: a key to its functional diversity. Microsc Res Tech (2003) 60:581–92. doi: 10.1002/jemt.10300
138. Moroz OV, Blagova EV, Wilkinson AJ, Wilson KS, Bronstein IB. The crystal structures of human S100A12 in apo form and in complex with zinc: new insights into S100A12 oligomerisation. J Mol Biol (2009) 391:536–51. doi: 10.1016/j.jmb.2009.06.004
139. Garcia AF, Garcia W, Nonato MC, Araujo AP. Structural stability and reversible unfolding of recombinant porcine S100A12. Biophys Chem (2008) 134:246–53. doi: 10.1016/j.bpc.2008.02.013
140. Wang Q, Aleshintsev A, Bolton D, Zhuang J, Brenowitz M, Gupta R. Ca(II) and Zn(II) cooperate to modulate the structure and self-assembly of S100A12. Biochemistry (2019) 58:2269–81. doi: 10.1021/acs.biochem.9b00123
141. Reis RA, Bortot LO, Caliri A. In silico assessment of S100A12 monomer and dimer structural dynamics: implications for the understanding of its metal-induced conformational changes. J Biol Inorg Chem (2014) 19:1113–20. doi: 10.1007/s00775-014-1149-y
142. Streicher WW, Lopez MM, Makhatadze GI. Modulation of quaternary structure of S100 proteins by calcium ions. Biophys Chem (2010) 151:181–6. doi: 10.1016/j.bpc.2010.06.003
143. Pietzsch J, Hoppmann S. Human S100A12: a novel key player in inflammation? Amino Acids (2009) 36:381–9. doi: 10.1007/s00726-008-0097-7
144. Yan WX, Armishaw C, Goyette J, Yang Z, Cai H, Alewood P, et al. Mast cell and monocyte recruitment by S100A12 and its hinge domain. J Biol Chem (2008) 283:13035–43. doi: 10.1074/jbc.M710388200
145. Miranda LP, Tao T, Jones A, Chernushevich I, Standing KG, Geczy CL, et al. Total chemical synthesis and chemotactic activity of human S100A12 (EN-RAGE). FEBS Lett (2001) 488:85–90. doi: 10.1016/s0014-5793(00)02392-9
146. Hofmann Bowman MA, Heydemann A, Gawdzik J, Shilling RA, Camoretti-Mercado B. Transgenic expression of human S100A12 induces structural airway abnormalities and limited lung inflammation in a mouse model of allergic inflammation. Clin Exp Allergy (2011) 41:878–89. doi: 10.1111/j.1365-2222.2011.03714.x
147. Chiou JW, Fu B, Chou RH, Yu C. Blocking the interactions between calcium-bound S100A12 protein and the V domain of RAGE using tranilast. PloS One (2016) 11:e0162000. doi: 10.1371/journal.pone.0162000
148. Xie J, Burz DS, He W, Bronstein IB, Lednev I, Shekhtman A. Hexameric calgranulin c (S100A12) binds to the receptor for advanced glycated end products (RAGE) using symmetric hydrophobic target-binding patches. J Biol Chem (2007) 282:4218–31. doi: 10.1074/jbc.M608888200
149. Nacken W, Sorg C, Kerkhoff C. The myeloid expressed EF-hand proteins display a diverse pattern of lipid raft association. FEBS Lett (2004) 572:289–93. doi: 10.1016/j.febslet.2004.07.024
150. Yao D, Brownlee M. Hyperglycemia-induced reactive oxygen species increase expression of the receptor for advanced glycation end products (RAGE) and RAGE ligands. Diabetes (2010) 59:249–55. doi: 10.2337/db09-0801
151. Yang Z, Tao T, Raftery MJ, Youssef P, Di Girolamo N, Geczy CL. Proinflammatory properties of the human S100 protein S100A12. J Leukoc Biol (2001) 69:986–94. doi: 10.1189/jlb.69.6.986
152. Garcia AF, Lopes JL, Costa-Filho AJ, Wallace BA, Araujo AP. Membrane interactions of S100A12 (Calgranulin c). PloS One (2013) 8:e82555. doi: 10.1371/journal.pone.0082555
153. Foell D, Wittkowski H, Kessel C, Luken A, Weinhage T, Varga G, et al. Proinflammatory S100A12 can activate human monocytes via toll-like receptor 4. Am J Respir Crit Care Med (2013) 187:1324–34. doi: 10.1164/rccm.201209-1602OC
154. Filipek A, Jastrzebska B, Nowotny M, Kuznicki J. CacyBP/SIP, a calcyclin and siah-1-interacting protein, binds EF-hand proteins of the S100 family. J Biol Chem (2002) 277:28848–52. doi: 10.1074/jbc.M203602200
155. Farokhzadian J, Mangolian Shahrbabaki P, Bagheri V. S100A12-CD36 axis: a novel player in the pathogenesis of atherosclerosis? Cytokine (2019) 122:154104. doi: 10.1016/j.cyto.2017.07.010
156. Tondera C, Laube M, Pietzsch J. Insights into binding of S100 proteins to scavenger receptors: class b scavenger receptor CD36 binds S100A12 with high affinity. Amino Acids (2017) 49:183–91. doi: 10.1007/s00726-016-2349-2
157. Zamora C, Canto E, Nieto JC, Angels Ortiz M, Juarez C, Vidal S. Functional consequences of CD36 downregulation by TLR signals. Cytokine (2012) 60:257–65. doi: 10.1016/j.cyto.2012.06.020
158. Moroz OV, Antson AA, Dodson EJ, Burrell HJ, Grist SJ, Lloyd RM, et al. The structure of S100A12 in a hexameric form and its proposed role in receptor signalling. Acta Crystallogr D Biol Crystallogr (2002) 58:407–13. doi: 10.1107/s0907444901021278
159. Rouleau P, Vandal K, Ryckman C, Poubelle PE, Boivin A, Talbot M, et al. The calcium-binding protein S100A12 induces neutrophil adhesion, migration, and release from bone marrow in mouse at concentrations similar to those found in human inflammatory arthritis. Clin Immunol (2003) 107:46–54. doi: 10.1016/s1521-6616(02)00043-8
160. Goyette J, Geczy CL. Inflammation-associated S100 proteins: new mechanisms that regulate function. Amino Acids (2011) 41:821–42. doi: 10.1007/s00726-010-0528-0
161. Haley KP, Delgado AG, Piazuelo MB, Mortensen BL, Correa P, Damo SM, et al. The human antimicrobial protein calgranulin c participates in control of helicobacter pylori growth and regulation of virulence. Infect Immun (2015) 83:2944–56. doi: 10.1128/IAI.00544-15
162. Jackson E, Little S, Franklin DS, Gaddy JA, Damo SM. Expression, purification, and antimicrobial activity of S100A12. J Vis Exp (2017) 123:55557. doi: 10.3791/55557
163. Shank JM, Kelley BR, Jackson JW, Tweedie JL, Franklin D, Damo SM, et al. The host antimicrobial protein calgranulin c participates in the control of campylobacter jejuni growth via zinc sequestration. Infect Immun (2018) 86:e00234–18. doi: 10.1128/IAI.00234-18
164. Kang JH, Hwang SM, Chung IY. S100A8, S100A9 and S100A12 activate airway epithelial cells to produce MUC5AC via extracellular signal-regulated kinase and nuclear factor-kappaB pathways. Immunology (2015) 144:79–90. doi: 10.1111/imm.12352
165. McEntee CP, Finlay CM, Lavelle EC. Divergent roles for the IL-1 family in gastrointestinal homeostasis and inflammation. Front Immunol (2019) 10:1266. doi: 10.3389/fimmu.2019.01266
166. Srikrishna G, Nayak J, Weigle B, Temme A, Foell D, Hazelwood L, et al. Carboxylated n-glycans on RAGE promote S100A12 binding and signaling. J Cell Biochem (2010) 110:645–59. doi: 10.1002/jcb.22575
167. Wolf R, Mirmohammadsadegh A, Walz M, Lysa B, Tartler U, Remus R, et al. Molecular cloning and characterization of alternatively spliced mRNA isoforms from psoriatic skin encoding a novel member of the S100 family. FASEB J (2003) 17:1969–71. doi: 10.1096/fj.03-0148fje
168. Kurpet K, Chwatko G. S100 proteins as novel therapeutic targets in psoriasis and other autoimmune diseases. Molecules (2022) 27:6640. doi: 10.3390/molecules27196640
169. Wolf R, Mascia F, Dharamsi A, Howard OM, Cataisson C, Bliskovski V, et al. Gene from a psoriasis susceptibility locus primes the skin for inflammation. Sci Transl Med (2010) 2:61ra90. doi: 10.1126/scitranslmed.3001108
Keywords: psoriasis, S100 proteins, interaction, signaling pathways, inflammation
Citation: Liang H, Li J and Zhang K (2023) Pathogenic role of S100 proteins in psoriasis. Front. Immunol. 14:1191645. doi: 10.3389/fimmu.2023.1191645
Received: 22 March 2023; Accepted: 22 May 2023;
Published: 06 June 2023.
Edited by:
Jeong Eun Kim, Hanyang University, Republic of KoreaReviewed by:
Dillon Mintoff, University of Malta, MaltaEvelyn Tsantikos, Monash University, Australia
Copyright © 2023 Liang, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kaiming Zhang, emhhbmdrYWltaW5nQHNpbmEuY29t
 Huifang Liang1,2
Huifang Liang1,2 Kaiming Zhang
Kaiming Zhang