
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 27 April 2023
Sec. Comparative Immunology
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1190590
The CpG oligodeoxynucleotides (CpG ODNs) reportedly possess the capacity to strengthen immunity in mammals. This experiment was conducted to evaluate the impact of dietary supplementation with 17 types of CpG ODNs on intestinal microbiota diversity, antioxidant capacity, and immune-related gene expression profiles of the shrimp Litopenaeus vannamei. Diets including 50 mg kg-1 CpG ODNs wrapped in egg whites were prepared and divided into 17 different groups, with 2 control groups (normal feed and feed with egg whites). These CpG ODNs supplemented diets and the control diets were fed to L. vannamei (5.15 ± 0.54 g) three times daily at 5%-8% shrimp body weight for three weeks. The results of consecutive detection of intestinal microbiota by 16S rDNA sequencing indicated that 11 of the 17 types of CpG ODNs significantly enhanced intestinal microbiota diversity, increased the populations of several probiotic bacteria, and activated possible mechanisms relevant to diseases. The immune-related genes expression and antioxidant capacity in hepatopancreas further demonstrated that the 11 types of CpG ODNs effectively improved the innate immunity of shrimp. Additionally, histology results showed that the CpG ODNs in the experiment did not damage the tissue structure of hepatopancreas. The results suggest that CpG ODNs could be used as a trace supplement to improve the intestinal health and immunity of shrimp.
Due to the rapid growth rate and the ability to tolerate wide range of temperatures and salinities, the Pacific white shrimp, Litopenaeus vannamei, has become one of the most profitable varieties in crustacean aquaculture (1, 2). However, the extended shrimp farming industry has sustained declining disease resistance, metabolic disorder, and frequent outbreaks of serious diseases, including viral an bacterial diseases, resulting in huge financial losses. Among all the shrimp bacterial diseases, vibriosis, which causes significant production losses, is of particular concern (3). Vaccines, such as subunit and recombinant treatments, are too costly for practical production on account of the individual size and farming scale of shrimp (4), while drugs such antibiotics carry the risk of excessive abuse, which can lead to a variety of drug-resistant bacteria, and disordered immune function, intestinal microbial flora imbalance, and even drug residues of aquatic animal (5). Therefore, it is necessary to develop dietary additives that can be used as immunoenhancement to enhance the innate immune capacity of shrimp (6).
CpG Oligonucleotides (CpG-ODNs) are synthetic short DNA with a CpG motif, which mimic bacterial genomic DNA and are significant potent activators of adaptive and innate immunity (7). CpG-ODNs are generally divided into four classes (A, B, C, and P) according to their structural differences, and each class of CpG-ODNs exhibits different immunostimulatory and immunomodulation effects (7). The A-class CpG-ODNs participate in the humoral immune response (8), the B-class CpG-ODNs relate to the cellular immune response (9), the C-class CpG-ODNs participate in both humoral and cellular immune responses (10), while the P-class CpG-ODNs affect hematology and plasma cytokines (11). Meanwhile, N-class CpG ODNs is a type of CpG-ODNs that cannot induce any immune stimulation on account of the backbone and the number, nature and spacing of the CpG motifs. For example, CpG 1720, CpG 2137 and CpG 2243 did not enhance the immunity of olive flounder (Paralichthys olivaceus) (12) and rainbow trout (Oncorhynchus mykiss) (13), respectively. CpG ODNs with similarly bacterial DNA motifs can typically initiate some innate immune responses by the recognition of molecules bearing conserved motifs, termed pathogen-associated (PAMPs), via pattern recognition receptors (PRRs) (14). It has been reported that CpG-ODNs are an immune response modifier in mammals (15). There are also slight differences in various animal models in terms of the immune stimulation effects caused by CpG ODNs. In murine and primate models, CpG-ODNs target Toll-like receptor (TLR) to trigger an immunostimulatory and immunomodulation cascade or stimulate an innate immune response, thereby contributing to the clearance of bacteria, parasites, and viruses and reducing the severity and duration of infection (16). In aquatic organisms, CpG ODNs can induce high resistance against viral hemorrhagic septicemia virus infection and intensive expression levels of Mx and ISG15 genes in Paralichthys olivaceus (12), produce remarkably high levels of O2− and H2O2 on macrophages and high anti-bactericidal activities in grass carp (17), and induce proliferation of spleen and head kidney cells, and peripheral blood leucocytes, from rainbow trout (18). The previous study verified that oral administration of CpG ODNs reduced the death rate of shrimps exposed to WSSV (19). However, it is unclear how CpG ODNs affect the intestines of shrimp. Therefore, it is necessary to investigate the mechanism of CpG-ODNs via intestinal absorption and recognition when stimulating innate immune response in L. vannamei.
The intestine is the main organ of shrimp digestion, absorption, and nutrient metabolism, as well as a significant component of its defense against pathogen invasion, which is mainly realized through the intestinal mucosal barrier. Due to the absence of an adaptive immune system, the intestinal mucosal barrier of L. vannamei plays a significant role in its innate immune system, which mainly consists of a mechanical, a chemical, and a microbial barrier (5, 20). The microbial barrier refers to the combination and adhesion of intestinal flora with intestinal mucosa, forming a microecosystem composed of membrane flora with certain regularity, and interdependent and interacting with the microspatial structure of the host. Many studies have shown that intestinal microbial community structure is closely related to the immunity and disease resistance of shrimp (21, 22). Therefore, the intestinal microbial community is a potential target for immune regulation of L. vannamei, and it is expected to achieve effective prevention and control of related diseases through immune regulation of intestinal microbiota structure.
In this study, 17 types of CpG ODNs (classified into A, B, C, P, N) were synthesized and added to shrimp feed. The variation of intestinal flora in shrimp was testified by 16S rDNA sequencing after continuous feeding to investigate the effects of each CpG ODN on the intestinal microbiota structure. The experiment of immune-related genes expression and antioxidant capacity in hepatopancreas was conducted to study the effects of CpG ODNs on nonspecific immune parameters. Additionally, hepatopancreas histomorphology verified whether CpG ODNs exert a negative effect on the vital organs of shrimp. These results provide novel insights into the immunoenhancement of CpG ODNs to improve shrimp immunity.
Seventeen types of CpG ODNs (Table S1), which were efficiently used in mammalian and aquatic animals, were synthesized, and inserted into plasmid pUC57 with 10 copies in series to form a series of CpG-rich DNA fragments with tandem CpG ODNs. Plasmids pUC57-CpG were massively extracted from Escherichia coli using a modified alkaline lysis method (23), then stored at -80 °C until further use.
The dietary composition of shrimps is shown in Table 1. A basal diet was purchased from Guangdong Zhengge Aquatic Science and Technology Co., Ltd., pulverized with a grinder, and sieved using a 300-mesh size nylon sieve. Different kinds of CpG ODNs dissolved in small amount of egg white were added to basal diet pulverized powder and stirred equably, then 40% water was added until stiff doughs containing 50 mg kg-1 CpG ODNs were obtained. The dough was pelletized by double knife noodle pressing machine (Weidi, China) with diameters of 1.0 mm. Basal diet and basal diet with small amount of egg white (about 5 g kg-1), made according to the same method, were set as control groups. The details of groups and samples are shown in Table S2. All diets were placed in a fume hood to air dry to about 10% moisture and stored at -20 °C in a freezer for further use.
The feeding experiment was conducted in Renhai Aquatic Science Technology Co., Ltd., Hainan Province, China. Healthy shrimps with an average initial body weight of 5.15 ± 0.54 g were acclimatized for 7 days prior to the experiment, in filtered aerated seawater with 24 ± 1 °C in temperature and 10 ± 1 ppt in salinity. A total of 1140 L. vannamei shrimps were randomly separated into nineteen 1200 L water tanks containing 800 L of sea water (randomly assigned 60 tails per tank). The shrimps were fed at 08:00, 14:00, and 20:00 each day. The initial ratio was 5-8% of their body weight, and the feeding experiment lasted for 14 days.
The whole hepatopancreas samples from three shrimp per tank at 0-, 7- and 14-days after feeding were collected, and immediately placed into liquid nitrogen to await further analysis of enzyme activity and gene expression profiles. The hepatopancreas tissue samples from three shrimp per tank at 14 days post feeding were pooled and placed in 4% paraformaldehyde solution for further histological experiments. For each tank, completed intestines from three shrimps at 0-, 7- and 14-days post feeding were dissected and immediately placed in liquid nitrogen until further analysis of intestinal microbiota.
Total RNA from pulverized L. vannamei hepatopancreas tissues was extracted using TRIzol Reagent (ThermoFisher, Carlsbad, CA, USA) and completely digested with DNase I (TaKaRa, Shiga, Japan) to remove potential genomic DNA contamination. The concentration and purity of obtained RNA were determined using NanoDrop One (ThermoFisher, Waltham, MA, USA), and its integrity was further verified by agarose gel electrophoresis analysis. The first-strand cDNA was synthesized using Reverse Transcriptase M-MLV (RNase H-) Kit (Takara, Shiga, Japan) with random primers using 1 μg total purified RNA.
Six pairs of primers (shown in Table 2), including interleukins (IL-1-β, IL-8, IL-10), inflammatory cytokines (TNF-α), anti-lipopolysaccharide factor (ALF) and target of rapamycin (TOR), were selected to analyze the immune-related gene expression during a 14-day feeding period. The assays were performed on a Light Cycler 480 (Roche, Mannheim, Germany) with 10 μL of 2×SYBR qPCR Master Mix (Everbright, Suzhou, China), 0.4 μL of forward primer (10 μM), 0.4 μL of reverse primer (10 μM), 1 μL diluted cDNA template (20 ng/μL) and 8.2 μL ddH2O using the following experiment set: 95 °C for 5 min, and 95 °C for 15 s and 35 cycles, 60 °C for 45 s. The β-actin gene was employed as an internal control (Table 2). All the qPCR reactions were repeated with two independent samples, and performed for three biological replicates. Relative mRNA expression was generated and calculated using 2-ΔΔCt method (24).
Hepatopancreas samples from the shrimps at 0-, 7- and 14- days after feeding were used to detect the activities of antioxidant enzymes, including superoxide dismutase (SOD), catalase (CAT), glutathione peroxidase (GPX), and glutathione S-transferase (GST). Superoxide Dismutase (SOD) assay kit, Catalase (CAT) assay kit, Glutathione Peroxidase (GSH-PX) assay kit and Glutathione S-transferase (GSH-ST) assay kit (Jiancheng, Nanjing, China) were used for these assays according to the manufacturer’s protocols.
The hepatopancreas tissue of L. vannamei at 14 days after feeding was immobilized in 4% paraformaldehyde fixative solution (Solarbio, Beijing, China) for 24 h and then dehydrated in various concentrations of alcohol (from 50% to 95%). The dehydrated tissues were then embedded in paraffin, and cut into thick slices with 4 μm in thick. The obtained tissues sections were stained with hematoxylin and eosin (H&E), then observed with a BX53M light microscope (Olympus, Tokyo, Japan).
DNA from the intestines of L. vannamei shrimps after 0-, 7- and 14- days of feeding, respectively, was extracted using a commercial DNA extraction kit (Omega, Guangzhou, China). The V3-V4 region of microbial 16S rDNA was amplified obtain an about 420 bp amplified fragment. The Illumina NovaSeq 6000 platform were used to obtain 2×250 bp paired-end sequence data. Paired-end reads were assigned to samples, truncated, and then merged using FLASH 1.2.8 for 16S or PEAR 0.9.6 for ITS2 (25). Raw reads were processed under specific filtering conditions to obtain the high-quality clean reads using fqtrim 0.94. Chimeric sequences were detected and filtered using V search software 2.3.4 (26). After dereplication using Divisive Amplicon Denoising Algorithm (DADA2), feature table and feature sequence of amplicon sequence variants (ASVs) were obtained. Alpha diversity and beta diversity were calculated according to the QIIME2 process, and visualized by R script. The sequence alignment of species annotation was performed by Blast against SILVA and NT-16S database (27). Based on the obtained species abundance statistics, the differences between the experimental and control groups were obtained and analyzed. A Mann-Whitney U test was used for comparison of the differences between the two groups of samples with biological replicates, while Kruskal–Wallis’s test was applied to compare multiple groups with biologically replicated samples (p<0.05). PICRUSt2 software was employed to predicts functional abundance based on marker gene sequences (28). The function of intestinal flora with relatively high abundance was annotated by PICRUSt2 (https://github.com/picrust/picrust2) based on COG (Clusters of Orthologous Groups) database. The differences between experimental and control groups were emphasized. BugBase (29) was employed to predict the phenotype of an intestinal microbial sample from each group.
Significant differences among different groups were tested by one-way analysis of variance (ANOVA) using SPSS 26.0 (IBM, Armonk, NY, USA). All data are presented in the form of means ± standard error (n = 3). Duncan’s multiple-range test was applied for multiple comparisons of group means. The histograms were implemented using OriginPro 2022b (Northampton, Massachusetts, USA). The difference was considered significant at a p value < 0.05.
As shown in Figure 1, after 14 days’ feeding, the expression of six immune-related genes exhibited amplitude and trends of different degrees of upregulation in most experimental groups, while some groups (CpG 1720, CpG 2137, CpG 21424, CpG 2243, CpG 21797, CpG 21425) consistent with the negative control groups did not reveal a stimulation effect for these genes. In the first seven days, at least 7 of 19 groups and at most 13 of 19 groups exhibited upregulation of all six detected genes. From seven to fourteen days, several groups, such as CpG 2395, CpG 1585, CpG 1681, CpG 8954, CpG 2143 and CpG M362, showed intense upregulation, while other groups exhibiting a tiny amount of upregulation (p<0.05).
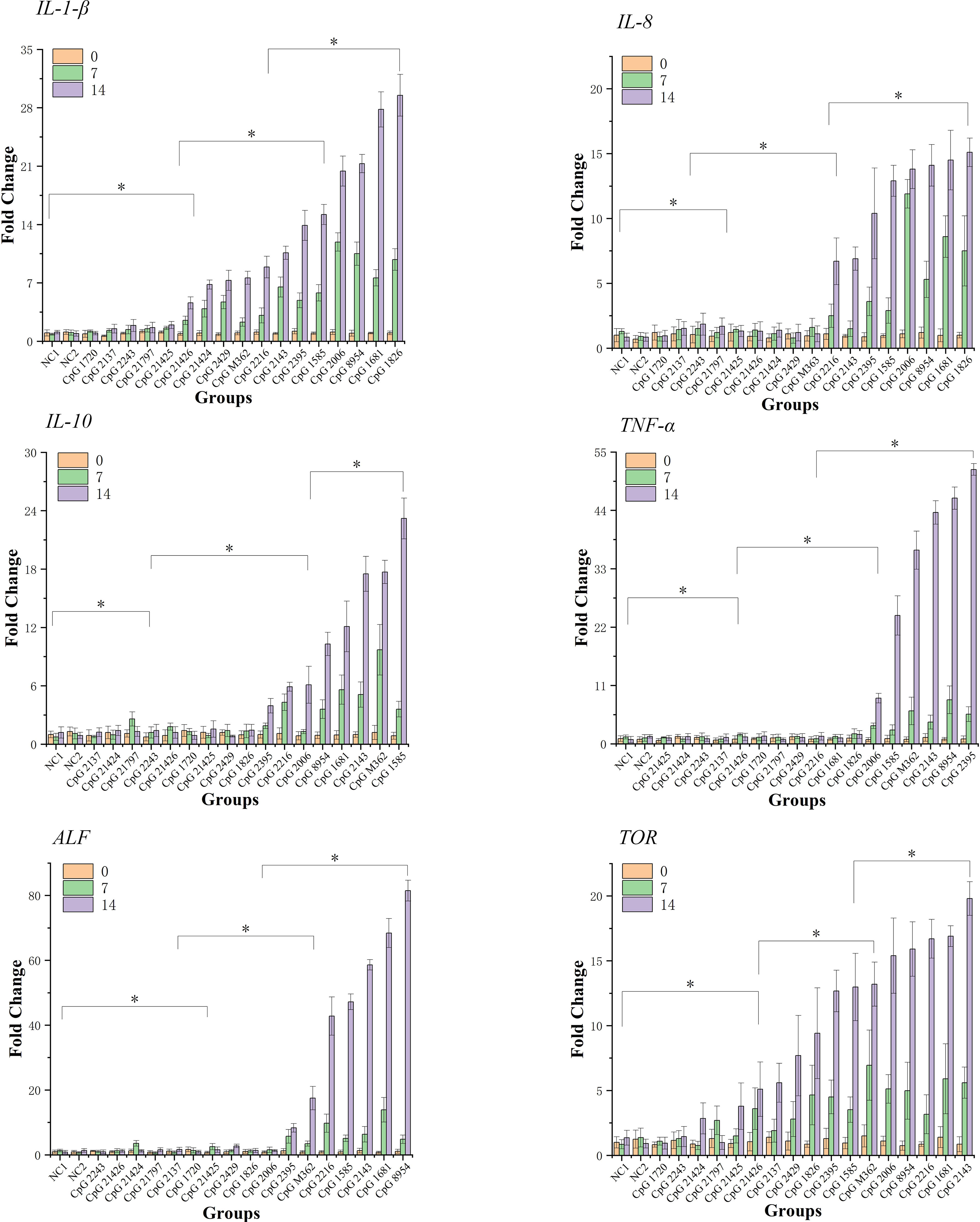
Figure 1 qPCR analysis of the expression of six immune-related genes detected in hepatopancreas of shrimps at 0-, 7- and 14-days. Data are means for three assays and presented as the means ± SD (p < 0.05). The symbol * indicates significant differences (p < 0.05).
Figure 2 illustrates the results of the four antioxidant enzymes activity (GST, CAT, GPX and SOD) analysis in hepatopancreas after 14 days’ feeding. Compared with the two negative control groups, of which the four antioxidant enzymes activities did not show any significant changes over 14 days, the C1, C2, C7, and C11 experimental groups exhibited significantly higher GST activity at 14 days than at 0 days (p<0.05); the C1, C2, C3, C4, and C7 experimental groups indicated significantly higher CAT activity at 14 days than at 0 days (p<0.05); the C1, C2, and C7 experimental groups showed significantly higher GPX activity at 14 days than at 0 days (p<0.05); and the C1, C2, C3, C4, C8, C9, and C15 experimental groups exhibited significantly higher SOD activity at 14 days than at 0 days (p<0.05). Additionally, no significant changes in antioxidant enzymes activity of the residual experimental groups were observed.
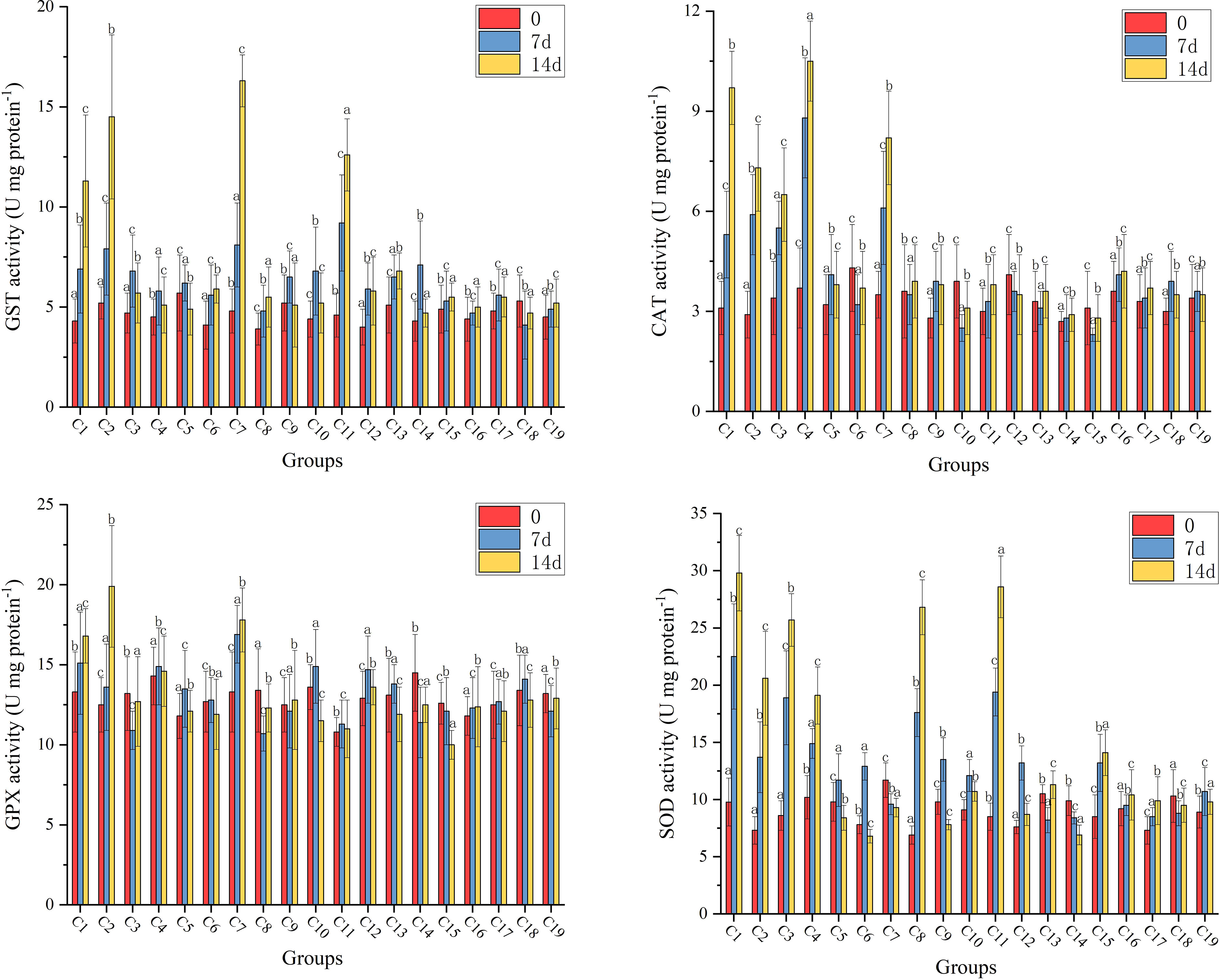
Figure 2 The results of the four antioxidant enzymes activity (GST, CAT, GPX and SOD) analysis in hepatopancreas of shrimps at 0-, 7- and 14-days. Data are means for three assays and presented as the means ± SD, different letters indicate significant differences (p < 0.05).
Hepatopancreas of Pacific white shrimp stained by H&E from each group were presented in Figure 3. The hepatopancreas tissue morphology of L. vannamei of all groups was characterized as normal. There were no hepatopancreas morphological alterations observed among all the experimental treatments. These results revealed that trace CpG ODNs in the diet had no obvious effect on the health status of the hepatopancreas tissue.
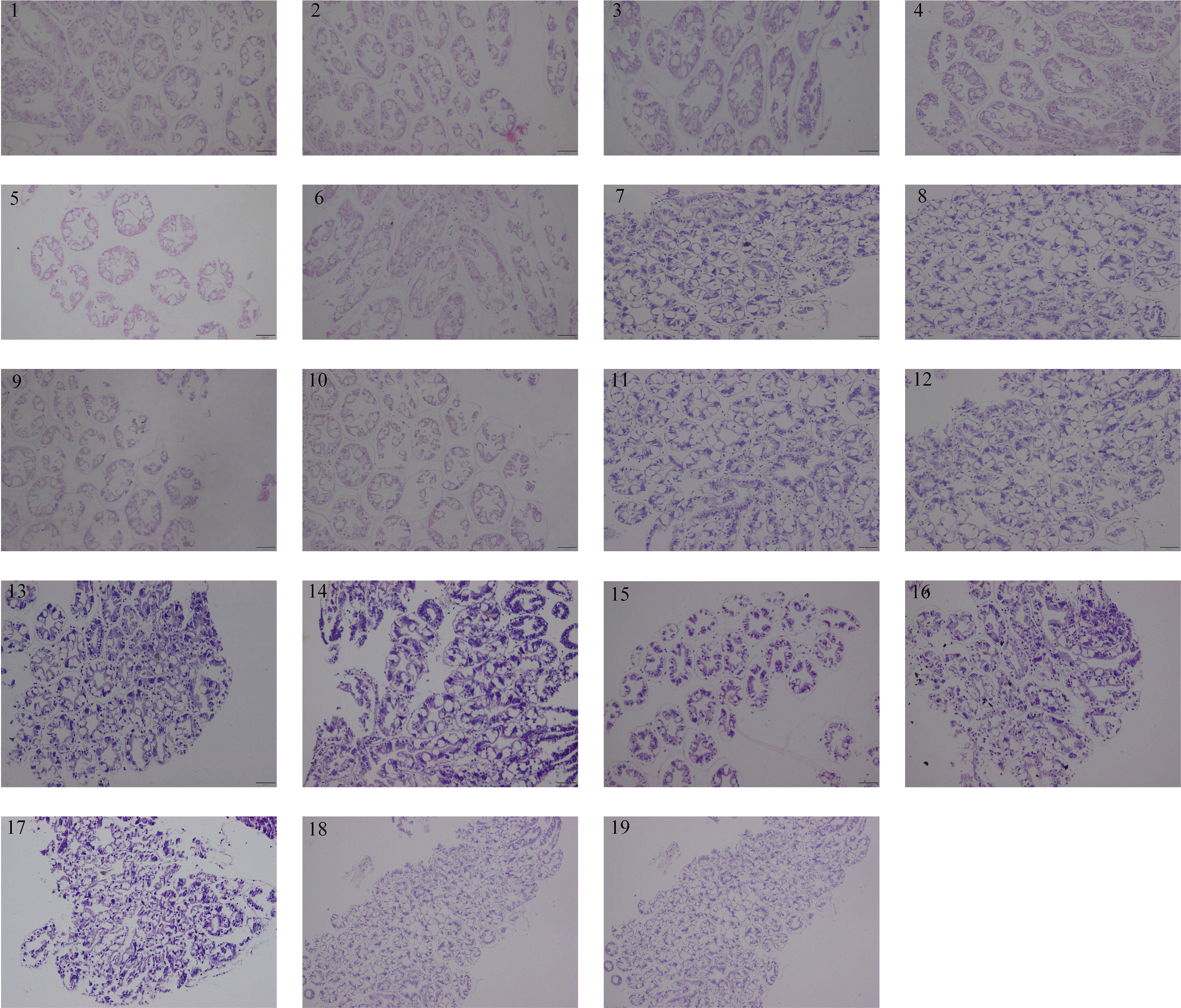
Figure 3 Hepatopancreas histomorphology of shrimps from each group at 14 days. All sections were stained with H&E. Scale bar 100 µm. 1. CpG ODN 1826, 2. CpG ODN 1681, 3. CpG ODN 2006, 4. CpG ODN M362, 5. CpG ODN 2137, 6. CpG ODN 8954, 7. CpG ODN 1585, 8. CpG ODN 2143, 9. CpG ODN 2395, 10. CpG ODN 2216, 11. CpG ODN 23617, 12. CpG ODN 2429, 13. CpG ODN 21424, 14. CpG ODN 21425, 15. CpG ODN 21426, 16. CpG ODN 2243, 17. CpG ODN 1720, 18. Negative control 1, 19. Negative control 2.
A total of 118,414,123 high-quality sequences were obtained from 171 experimental samples of the 19 groups. All the raw sequencing data were submitted to NCBI (PRJNA877472). ASV feature sequences were denoised by QIIME2, and randomly selected to calculate the values of alpha diversity (Supplementary: Figure S1). The community coverage of each sample was greater than 99% (Supplementary: Table S3), indicating that the identified sequence in the present research can represent most of the microorganism in each sample. Both Shannon and Simpson indices reflect the high community diversity, the Chao indices present high community richness, and the pielou-e indices exhibit Shannon’s evenness (Supplementary: Table S3). The results indicated that after two weeks of feeding, the alpha diversity of intestinal microbiota in the groups C1, C2, C3, C4, C6, C7, C8, C9, C10, C11, C12, C15 and C16 constantly increased, and was significantly higher than those in the two control groups, suggesting that the intestinal microbial abundances of shrimps in those groups were strengthened.
Together, beta and alpha diversity constitute the overall diversity or biological heterogeneity of certain microbiological communities. Principal component analysis (PCA), principal coordinates analysis (PCoA), an unweighted pair group method with arithmetic mean (UPGMA), and nonmetric multidimensional scaling (NMDS) were used to observe the differences between samples (Figure 4). The results indicated that the species diversity of intestinal microbial communities varied significantly in three samples (0-, 7- and 14-days) in the groups of C1, C2, C3, C4, C6, C7, C8, C9, C10, C11 and C15. This was due to the function of different CpG ODNs rather than other environmental factors.

Figure 4 The results of beta diversity from each group at 0, 7 and 14 days. (A) Principal component analysis (PCA), (B) Principal coordinates analysis (PCoA), (C) Nonmetric multidimensional scaling (NMDS), (D) Unweighted pair group method with arithmetic mean (UPGMA).
At the phylum level, the average intestinal microbiota community was dominated by Proteobacteria, Firmicutes, Planctomycetes, Actinobacteria, Bacteroidetes, Camplylobacterota and Verrucomicrobia (Figure 5A). Compared with the control groups, the relative abundances of Proteobacteria, Bacteroidetes, Planctomycetes and Firmicutes were significantly changed. Furthermore, there were more different phyla in the experimental groups (C1, C2, C4, C6, C7, C8, C9 and C15) than in the control groups, such as Nitrospirae, Gemmatimonadetes and Bedllovibrionota. At the genus level, the top 10 dominate species of the experimental group were Enterorhabdu, Candidatus Bacilloplasma, Vibrio, Caldilinea, Donghicola, flavobacterium, Ruegeria, Spongiimonas, Pir4_lineage and Formosa (Figure 5B). The relative abundance of Vibrio significantly decreased for C1, C2, C3, C4, C6, C7, C8, C9, C11, C13 and C15, while the relative abundances of Bacteroidota and Donghicola significantly increased (p<0.05) (Figure 5E). The distribution of the top five abundant intestinal flora of each sample at the phylum and the genus level, reflecting the composition proportion of dominant microbiological species in each group, and the distribution proportion of dominant microbiological species among different groups, were exhibited in Figures 5C, D, respectively. Figure 6A illustrates that the relative abundance of Candidatus Bacilloplasma and ZOR0006 under Firmicutes, and Vibrio under Proteobacteria of C1, C2, C3, C4, C6, C7, C8, C9, C10, C11, C15 decreased, while the relative abundance of Ruegeria and Alphaproteobacteria under Proteobacteria increased. The phylogenetic tree at the genus level analysis indicated that bacteria belonging to Proteobacteria were most, followed by Planctomycetota, Firmicutes, Bacteroidota, and Verrucomicrobiota, etc. (Figure 6B).
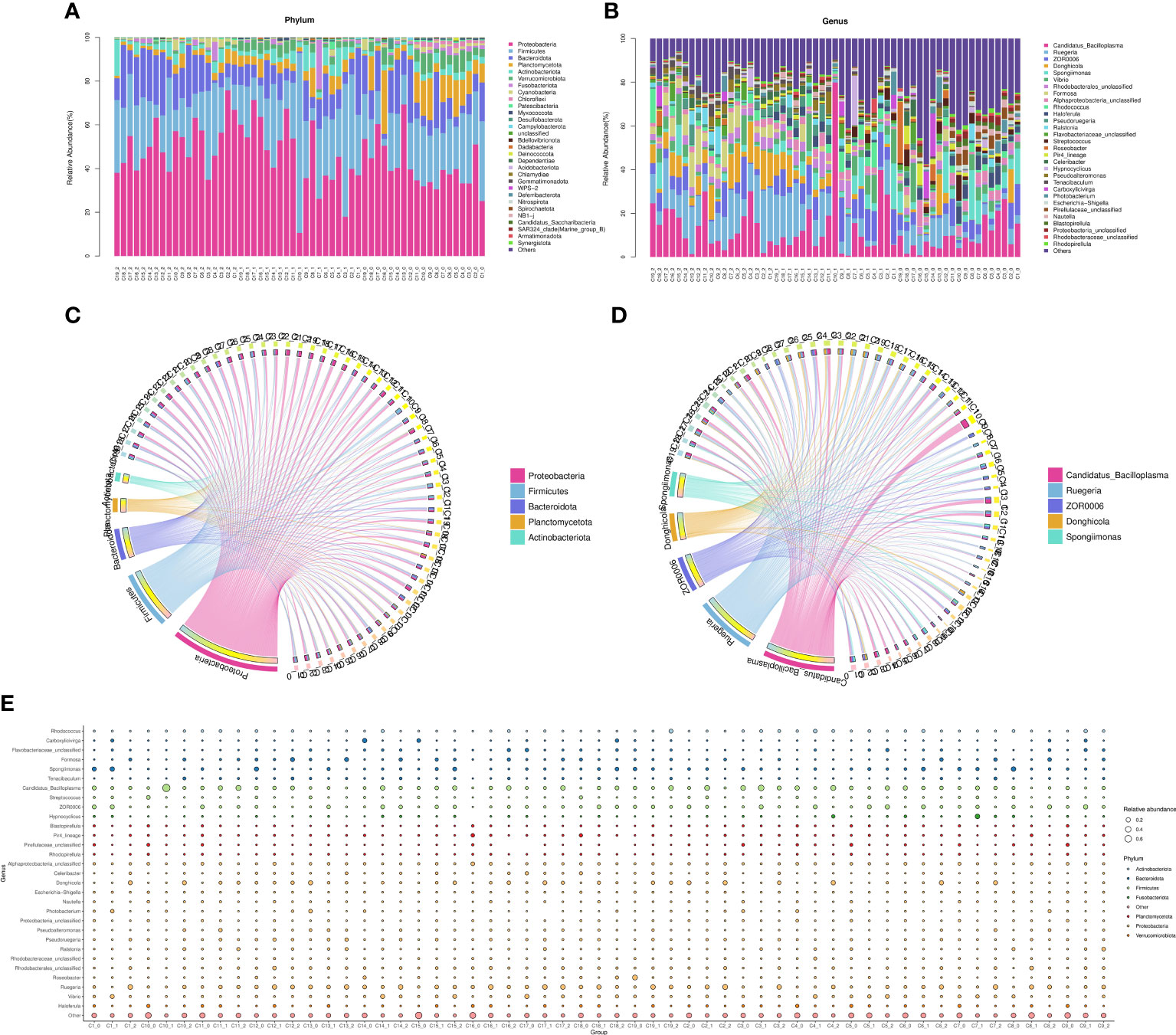
Figure 5 The microbial community composition in the intestine. (A) The distribution of the top 30 relative abundant species in each group at phylum level, (B) The distribution of the top 30 relative abundant species in each group at genus level, (C) The circus of top 5 relative abundant species in each group at phylum level, (D) The circus of top 5 relative abundant species in each group at genus level, (E) Species annotation information and relative abundance (circle size) at genus level in different sample groups.
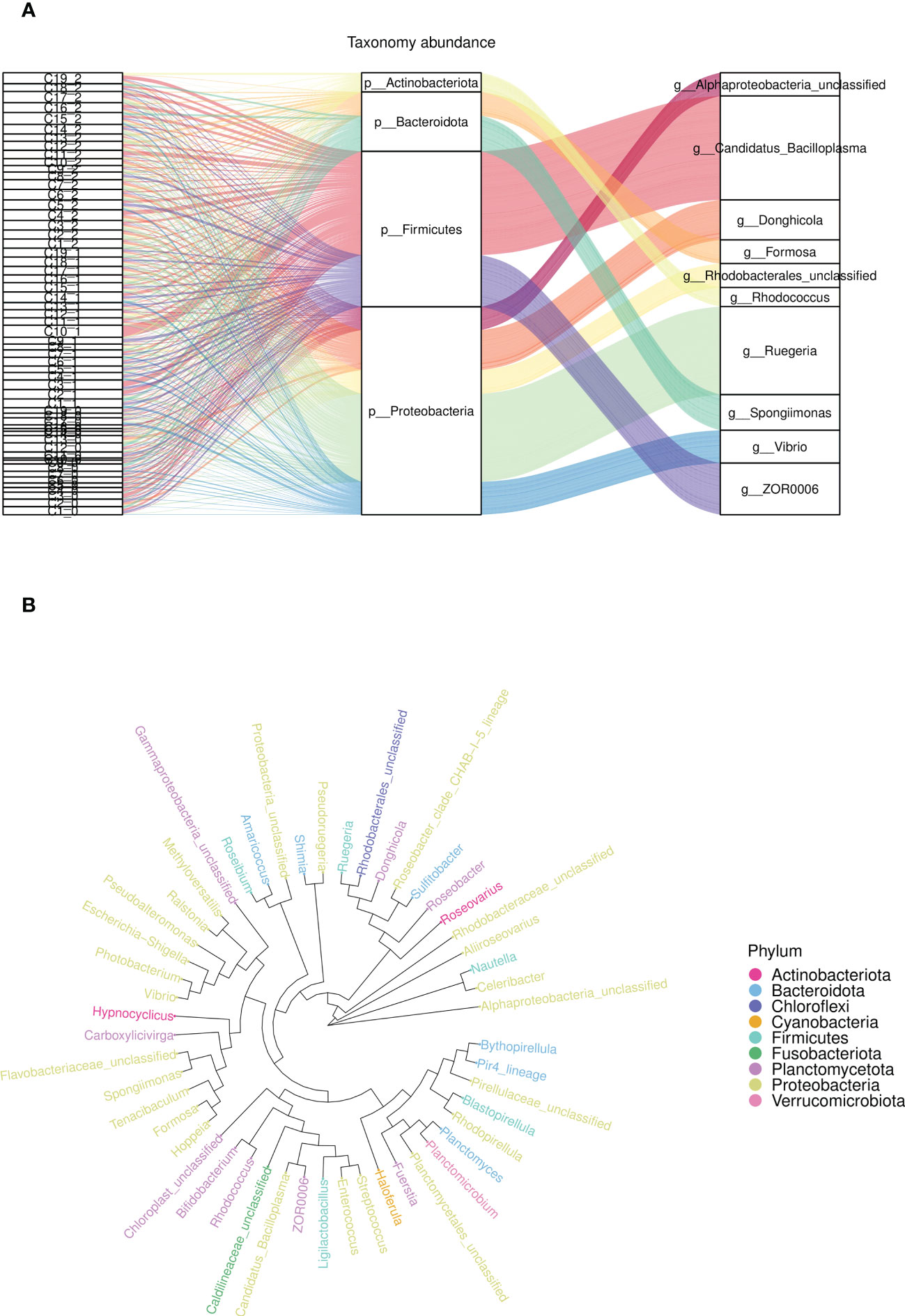
Figure 6 The relative abundance an phylogenetic tree of microbial community composition in the intestine. (A) The relative abundance of flora at phylum level (middle) and genus level (right) for different samples (left), (B) The phylogenetic tree of top 30 relative abundance species in each group at phylum level.
Bugbase was applied to analyze the variety in the presumptive phenotypes of the intestinal microbiota of shrimps in each group after feeding. The main predicted phenotypes were aerobic, anaerobic, contains mobile elements, facultatively anaerobic, forms biofilms, Gram-negative, Gram-positive, potentially pathogenic, and stress tolerant. The relative abundances of species in phylum level with different phenotypes in different groups were shown in Figure 7. The results indicated that the phenotypes of aerobic, contains mobile elements, forms biofilms, Gram-negative, potentially pathogenic, and stress tolerant were closely relevant with Proteobacteria and Tenericutes in each group. PICRUSt2 was used to analyze the variety in the presumptive functions of the intestinal microbiota of shrimps in each group after feeding. The results of pairwise comparison between all experimental groups and control groups were shown in Supplementary Figure S2, indicating that the pathways of Staphylococcus aureus infection, degradation of ethylbenzene, amoebiasis infection, taurine and hypo taurine metabolism, antigen processing and presentation, and pentose and glucuronic acid conversion were significantly strengthened. Additionally, the pathways of cysteine and methionine metabolism, lysine biosynthesis, and ansamycin biosynthesis were significantly attenuated.
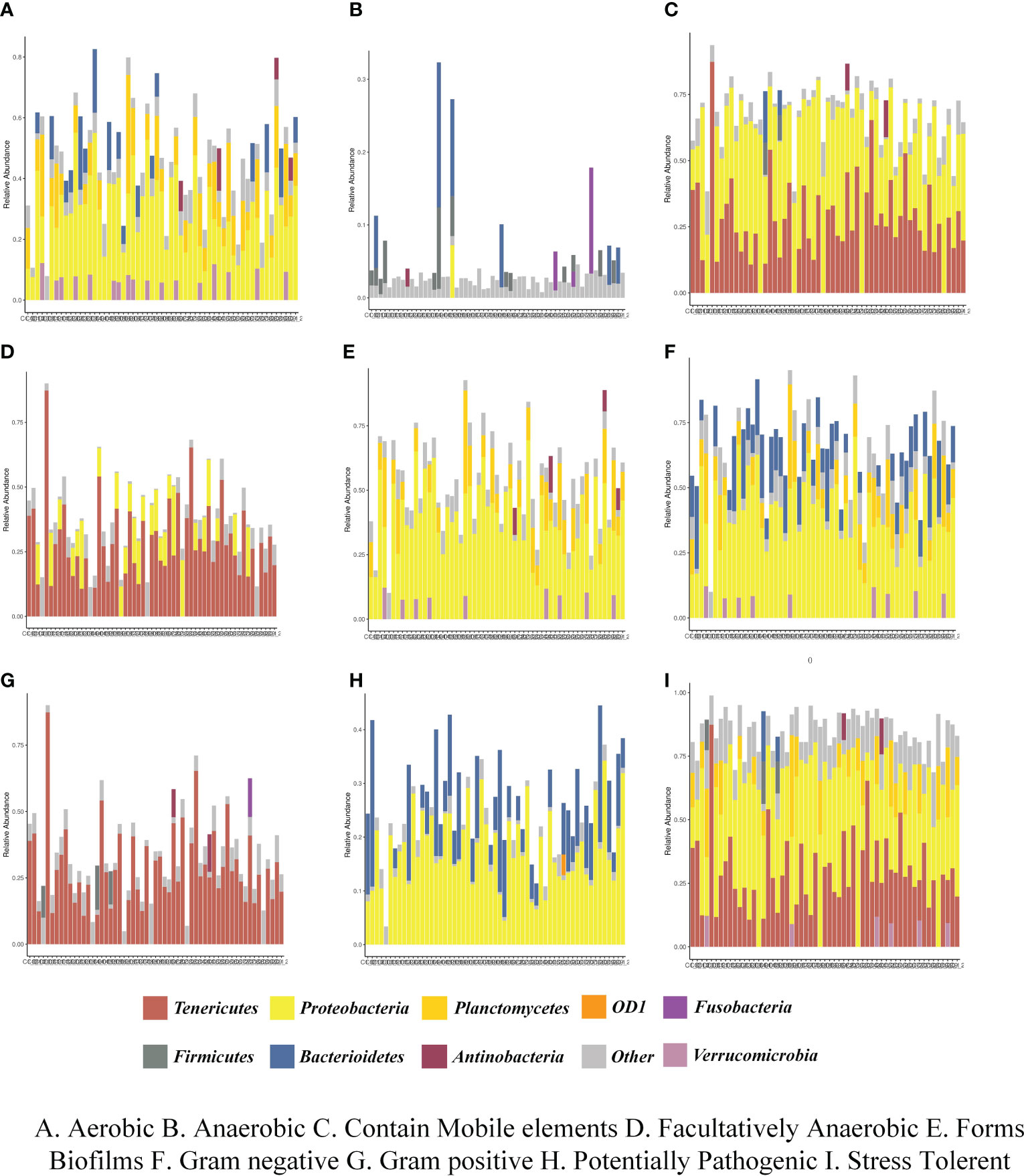
Figure 7 The relative abundances of species in phylum level with different phenotypes in different groups. (A) Aerobic, (B) Anaerobic, (C) Contain Mobile elements, (D) Facultatively Anaerobic, (E) Forms Biofilms, (F) Gram negative, (G) Gram positive, (H) Potentially Pathogenic, (I) Stress Tolerance.
The immunoprotected effects of CpG-ODNs have been reported in mice and other model organism. CpG-ODNs, as immunostimulatory agents, have been proven to stimulate immune cells and trigger immune responses to infections in many vertebral species, including humans (30), cattle and sheep (31), mice (32), poultry (33), and teleost fish (34, 35). There have also been studies on CpG-ODNs enhancing innate immunity in invertebrates, inducing autophagy through reactive oxygen species (ROS) in Eriocheir sinensis (36), enhancing hemolytic immune responses of Macrobrachium rosenbergii (37), and triggering hemocytes immune responses in Litopenaeus vannamei (38). In this study, different kinds of CpG ODNs produce discrepant immune-stimulated effects on account of their sequence divergence. Specifically, the expression of innate immune-related genes increased, the antioxidant capacity in hepatopancreas enhanced and the structure of intestinal flora improved.
The expression profiles of immune-related genes, including the IL-1β, IL-8, IL-10, TNF-α, ALF and TOR of L. vannamei fed with CpG 1826, CpG 1681, CpG 2006, CpG 8954, CpG M362, CpG 1585, and CpG 2143 exhibited significant upregulation. An interleukin (IL) system is essential for the proper regulation of T cell proliferation accompanied with an antigen encounter, as well as the modulation of apoptosis (39). Additionally, the IL family was well documented in invertebrates for its crucial roles in regulating innate immune system, such as in Chinese mitten crab (40) and in L. vannamei (41). The high expression of IL-1β, IL-8 and IL-10 stimulated by CpG ODNs indicated that the innate immunity of shrimps was effectively improved. TOR is highly conserved in eukaryotic organisms, and can integrate various biological processes such as translation, metabolism and autophagy of nutrient and growth factor regulatory proteins, and play a core regulatory role in cell proliferation, growth and metabolism (42). Autophagy has been demonstrated to play a pivotal function in shrimp innate immunity against invading pathogens (43). Similarly, there were significantly inductive effects for immune-related factors from CpG ODNs, including inflammatory cytokines, and anti-lipopolysaccharide factor, such as CpG 1681 and CpG 2006 in the spleen of grass carp Ctenopharyngodon idella (44), CpG M362 in murine plasmacytoid dendritic cells (45), CpG 1585 in serum of olive flounder (Paralichthys olivaceus) (46), CpG 8954 in pigs (47) and CpG 2143 in silver catfish (Rhamdia quelen) (48). The high expression of TNF-α and ALF in this study were consistent with the previously mentioned studies, which suggested that the immunity of shrimps was significantly enhanced as a result of orally administered CpG ODNs.
The increase in the antioxidant capacity in the hepatopancreas of L. vannamei illustrated that partial CpG ODNs could not only work at the genic level, but could also affect downstream protein expression. The antioxidant enzymes activity, including GST, CAT, GPX and SOD, plays a significant role in the innate immune system due to the absence of acquired immunity in L. vannamei. GST and GPX are important detoxification enzymes involved in apoptosis, cell proliferation, drug resistance and stress response (49). CAT and SOD are considered vital to survival because they eliminate ROS after pathogen invasion in the intestine (50). Early research only investigated the roles of CpG ODNs in hemocytes of shrimps, without a comprehensive or detailed evaluation (38). However, the positive effects on the antioxidant enzymes, together with immune-related genes in this study, proved that CpG ODNs could significantly enhance the innate immunity of L. vannamei administered orally.
The shrimp hepatopancreas comprises many tubules, mainly consists of basement membrane, cell layer, and hepatic canaliculus, and form the basic structural and functional units for absorption and digestion (51). The effective CpG-ODNs group in the study showed tight arrangement of tubules, complete basement membrane structure, and regular lumen shape, which improved the antioxidant properties of hepatopancreas, led to the repair of hepatopancreas damage, and maintained normal tissue and function. However, the increase in some inflammatory factors may also lead to corresponding histomorphological changes. The serious collapse and vacuolization of the hepatopancreas might be due to the dramatic increase in TNF-α after continuous CpG ODNs feeding, indicating that the immune response was strengthened.
The intestinal microbiota could participate in the regulation of multiple stress reactions, including high temperature, high pressure and other adverse conditions, by interacting with the host immune and digestive system (52). The roles of intestinal flora in the immunological stress of aquatic organisms have been widely studied, such as in Trachinotus ovatus and Scophthalmus maximus (53). CpG ODNs, which are in possession of similar bacterial DNA motifs, can be recognized by immune-related cells, such as phagocytes. However, there are fewer studies exploring the effects of CpG ODNs on the intestinal microbiota of aquatic organisms such as L. vannamei. In this study, there was strong evidence to suggest that different types of oral CpG ODNs administration modulate the structure and diversity of intestinal microbiota in L. vannamei, enhancing its innate immunity. The results indicated that the quantity and diversity of intestinal microorganisms significantly increased in partial CpG ODNs groups compared to the negative control groups, which was consistent with the regulation functions of the intestinal microbiota flora in terms of the nutritional composition changes or feed additives (54). In addition, the differentiate feature in the changes of the intestinal microbiota, both at the phylum and genus levels, of the effective dietary CpG ODNs groups was that pathogenic microorganisms decreased significantly, while beneficial microorganisms increased significantly. The dominant bacteria phylum were Proteobacteria, Firmicutes, Planctomycetes, Actinobacteria, and Bacteroidetes, which is consistent with previous results in fish and shrimp (54, 55). Furthermore, the relative abundances of Nitrospirae, Gemmatimonadetes and Bedllovibrionota were increased by the addition of partial CpG ODNs, supporting the point that the intestinal flora of shrimps became multitudinous. The variation tendency of microbial community at the genus level was consistent with those at the phylum level. Vibrio has been characterized as the dominant pathogenic bacterium in aquatic animals, and it can cause lethal diseases in shrimp (20, 22, 56). The relative abundance of the dominant genus Vibrio was significantly decreased in partial CpG ODNs groups in this study.
The results of functional annotation of key intestinal flora indicated that several pathways involved in immunity were strengthened, such as Staphylococcus aureus infection, amoebiasis infection and antigen processing and presentation. These innate immune responses were typically initiated by the recognition of PAMPs via PRRs due to the peculiar motifs of CpG ODNs. This indicated that CpG ONDs initiated the immunity of shrimps by activating related pathways and then improving other indicators. In addition, no studies have shown adverse effects from low concentrations of CpG ODN, which was also proved by the results of the hepatopancreas histomorphology of the treated shrimps in this study.
The present study showed that several dietary CpG ODNs can improve the intestinal health and immunity of shrimp without adverse effects, including CpG 2395, CpG 1585, CpG 1681, CpG 8954, CpG 2143, CpG 21424, CpG 21425, CpG 21426, CpG 2429, CpG 2216, and CpG M362. The enhancement covered immune-related genes, antioxidant enzymes and intestinal microbial diversity. These results indicated that the potential application of CpG ODNs improved the immunity of shrimp and laid a foundation for further investigation of its molecular series.
The data presented in the study are deposited in the NCBI repository, accession number PRJNA877472.
FH: Writing – original draft, conducted the experiments and wrote the manuscript. YW: conducted the experiments, data curation, analyzed the data. JH: conceived the study and designed the experiments, checked, and modified the manuscript. ZB: conceived the study and designed the experiments, checked, and modified the manuscript. All authors read and approved the final manuscript. MW: conceived the study and designed the experiments, checked, and modified the manuscript. All authors contributed to the article and approved the submitted version.
The work was supported by the Key Research and Development Program of Shandong Province (2021ZLGX03), the National Key Research and Development Program of China (2021YFD1200805), the Project of Sanya Yazhou Bay Science and Technology City Management Foundation (SKJC-2020-02-009), the Hainan Provincial Joint Project of Sanya Yazhou Bay Science and Technology City (320LH062), and the Startup Fund of Young Talents Project of Ocean University of China/Fundamental Research Funds for the Central Universities (202112019).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1190590/full#supplementary-material
1. Saoud IP, Davis DA, Rouse DB. Suitability studies of inland well waters for Litopenaeus vannamei culture. Aquaculture (2003) 217:373–83. doi: 10.1016/S0044-8486(02)00418-0
2. FAO. The state of agricultural commodity markets 2022. the geography of food and agricultural trade: policy approaches for sustainable development. (2022). Rome, FAO. doi: 10.4060/cc0471em
3. Munkongwongsiri N, Prachumwat A, Eamsaard W, Lertsiri K, Flegel TW, Stentiford GD, et al. Propionigenium And Vibrio species identified as possible component causes of shrimp white feces syndrome (WFS) associated with the microsporidian Enterocytozoon hepatopenaei. J Invertebr Pathol (2022) 192:107784. doi: 10.1016/j.jip.2022.107784
4. Lanh PT, Nguyen HM, Duong BTT, Hoa NT, Thom LT, Tam LT, et al. Generation of microalga Chlamydomonas reinhardtii expressing VP28 protein as oral vaccine candidate for shrimps against white spot syndrome virus (WSSV) infection. Aquaculture (2021) 540:736737. doi: 10.1016/j.aquaculture.2021.736737
5. Fu Z, Han F, Huang K, Zhang J, Qin JG, Chen L, et al. Combined toxic effects of thiamethoxam on intestinal flora, transcriptome and physiology of pacific white shrimp Litopenaeus vannamei. Sci Total Environ (2022) 830:154799. doi: 10.1016/j.scitotenv.2022.154799
6. Wu J, Tian S, Luo K, Zhang Y, Pan H, Zhang W, et al. Dietary recombinant human lysozyme improves the growth, intestinal health, immunity and disease resistance of pacific white shrimp Litopenaeus vannamei. Fish Shellfish Immunol (2022) 121:39–52. doi: 10.1016/j.fsi.2021.12.052
7. Dalpke AH, Heeg K. CpG-DNA as immune response modifier. Int J Med Microbiol (2004) 294:345–54. doi: 10.1016/j.ijmm.2004.07.005
8. Honda K, Ohba Y, Yanai H, Negishi H, Mizutani T, Takaoka A, et al. Spatiotemporal regulation of MyD88–IRF-7 signalling for robust type-I interferon induction. Nature (2005) 434:1035–40. doi: 10.1038/nature03547
9. Lai CY, Yu GY, Luo Y, Xiang R, Chuang T.H. Immunostimulatory activities of CpG-oligodeoxynucleotides in teleosts: toll-like receptors 9 and 21. Front Immunol (2019) 10:179. doi: 10.3389/fimmu.2019.00179
10. Li Y, Cao H, Wang N, Xiang Y, Lu Y, Zhao K, et al. A novel antagonist of TLR9 blocking all classes of immunostimulatory CpG-ODNs. Vaccine (2011) 29:2193–8. doi: 10.1016/j.vaccine.2010.10.042
11. Tosi I, Bureau F, Farnir F, Denoix JM, Lekeux P, Art T. Effects of a p-class CpG-ODN administered by intramuscular injection on plasma cytokines and on white blood cells of healthy horses. Vet Immunol Immunopathol (2018) 201:57–61. doi: 10.1016/j.vetimm.2018.05.004
12. Kang YJ, Kim KH. Effect of CpG-ODNs belonging to different classes on resistance of olive flounder (Paralichthys olivaceus) against viral hemorrhagic septicemia virus (VHSV) and miamiensis avidus (Ciliata; Scuticociliatia) infections. Aquaculture (2012) 324–325:39–43. doi: 10.1016/j.aquaculture.2011.11.008
13. Jørgensen JB, Zou J, Johansen A, Secombes CJ. Immunostimulatory CpG oligodeoxynucleotides stimulate expression of IL-1 β and interferon-like cytokines in rainbow trout macrophages via a chloroquine-sensitive mechanism. Fish Shellfish Immunol (2001) 11:673–82. doi: 10.1006/fsim.2001.0344
14. Heddes M, Altaha B, Niu Y, Reitmeier S, Kleigrewe K, Haller D, et al. The intestinal clock drives the microbiome to maintain gastrointestinal homeostasis. Nat Commun (2022) 13:6068. doi: 10.1038/s41467-022-33609-x
15. Moran MC, Bence AR, Vallecillo MFS, Lützelschwab CM, Rodriguez MG, Pardo R, et al. Polymeric antigen BLSOmp31 formulated with class b CpG-ODN in a nanostructure (BLSOmp31/CpG-ODN/Coa-ASC16) administered by parenteral or mucosal routes confers protection against Brucella ovis in balb/c mice. Res Vet Sci (2021) 135:217–27. doi: 10.1016/j.rvsc.2021.02.011
16. Manangeeswaran M, Lewkowicz AP, Israely T, Ireland DDC, Verthelyi D. CpG oligonucleotides protect mice from Alphavirus Encephalitis: role of NK cells, interferons, and TNF. Front Immunol (2020) 11:237. doi: 10.3389/fimmu.2020.00237
17. Meng Z, Shao J, Xiang L. CpG oligodeoxynucleotides activate grass carp (Ctenopharyngodon idellus) macrophages. Dev Comp Immunol (2003) 27:313–21. doi: 10.1016/S0145-305X(02)00104-0
18. Carrington AC, Collet B, Holland JW, Secombes CJ. CpG oligodeoxynucleotides stimulate immune cell proliferation but not specific antibody production in rainbow trout (Oncorhynchus mykiss). Vet Immunol Immunopathol (2004) 101:211–22. doi: 10.1016/j.vetimm.2004.04.022
19. Yi Q, Liu R, Sun R, Wang L, Zhou Z, Wang M, et al. The protection of CpG ODNs and yarrowia lipolytica harboring VP28 for shrimp Litopenaeus vannamei against white spot syndrome virus infection. Isj (2014) 11:119–31.
20. Jiao L, Dai T, Lu J, Tao X, Jin M, Sun P, et al. Excess iron supplementation induced hepatopancreas lipolysis, destroyed intestinal function in pacific white shrimp Litopenaeus vannamei. Mar pollut Bull (2022) 176:113421. doi: 10.1016/j.marpolbul.2022.113421
21. Holt CC, Bass D, Stentiford GD, van der Giezen M. Understanding the role of the shrimp gut microbiome in health and disease. J Invertebr Pathol (2020) 186:107387. doi: 10.1016/j.jip.2020.107387
22. Liang Q, Li Z, Ou M, Wu X, Qiao X, Wei W, et al. Hypoimmunity and intestinal bacterial imbalance are closely associated with blue body syndrome in cultured Penaeus vannamei. Aquaculture (2020) 522:735118. doi: 10.1016/j.aquaculture.2020.735118
23. Feliciello I, Chinali G. A modified alkaline lysis method for the preparation of highly purified plasmid DNA from Escherichia Coli. Anal Biochem (1993) 212:394–401. doi: 10.1006/abio.1993.1346
24. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods (2001) 25:402–8. doi: 10.1006/meth.2001.1262
25. Logue JB, Stedmon CA, Kellerman AM, Nielsen NJ, Andersson AF, Laudon H, et al. Experimental insights into the importance of aquatic bacterial community composition to the degradation of dissolved organic matter. ISME J (2016) 10:533–45. doi: 10.1038/ismej.2015.131
26. Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, et al. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems (2016) 1:e00009–15. doi: 10.1128/mSystems.00009-15
27. Takai K, Horikoshi K. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl Environ Microbiol (2000) 66:5066–72. doi: 10.1128/AEM.66.11.5066-5072.2000
28. Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol (2013) 31:814–21. doi: 10.1038/nbt.2676
29. Ward T, Larson J, Meulemans J, Hillmann B, Lynch J, Sidiropoulos D, et al. BugBase predicts organism-level microbiome phenotypes. bioRxiv (2017). doi: 10.1101/133462
30. Sabatel C, Radermecker C, Fievez L, Paulissen G, Chakarov S, Fernandes C, et al. Exposure to bacterial CpG DNA protects from Airway Allergic inflammation by expanding regulatory lung interstitial macrophages. Immunity (2017) 46:457–73. doi: 10.1016/j.immuni.2017.02.016
31. Nichani AK, Mena A, Popowych Y, Dent D, Townsend HGG, Mutwiri GK, et al. In vivo Immunostimulatory effects of CpG oligodeoxynucleotide in cattle and sheep. Vet Immunol Immunopathol (2004) 98:17–29. doi: 10.1016/j.vetimm.2003.10.001
32. Yeh D-W, Lai C-Y, Liu Y-L, Lu C-H, Tseng P-H, Yuh C-H, et al. CpG-oligodeoxynucleotides developed for grouper toll-like receptor (TLR) 21s effectively activate mouse and human TLR9s mediated immune responses. Sci Rep (2017) 7:17297. doi: 10.1038/s41598-017-17609-2
33. Gunawardana T, Ahmed KA, Goonewardene K, Popowich S, Kurukulasuriya S, Karunarathna R, et al. Synthetic CpG-ODN rapidly enriches immune compartments in neonatal chicks to induce protective immunity against bacterial infections. Sci Rep (2019) 9:341. doi: 10.1038/s41598-018-36588-6
34. Kang YJ, Kim KH. Therapeutic potential of CpG-ODN 1668 against scuticociliatosis in olive flounder (Paralichthys olivaceus). Aquaculture (2014) 430:17–20. doi: 10.1016/j.aquaculture.2014.03.035
35. Nguyen HT, Nguyen TTT, Wang Y-T, Wang P-C, Chen S-C. Effectiveness of formalin-killed vaccines containing CpG oligodeoxynucleotide 1668 adjuvants against Vibrio harveyi in orange-spotted grouper. Fish Shellfish Immunol (2017) 68:124–31. doi: 10.1016/j.fsi.2017.07.018
36. Sun M, Wang L, Jiang S, Liu R, Zhao D, Chen H, et al. CpG ODNs induced autophagy via reactive oxygen species (ROS) in Chinese mitten crab, Eriocheir sinensis. Dev Comp Immunol (2015) 52:1–9. doi: 10.1016/j.dci.2015.04.008
37. Sung H-H, Yang C-W, Lin Y-H, Chang P-T. The effect of two CpG oligodeoxynucleotides with different sequences on haemocytic immune responses of giant freshwater prawn, Macrobrachium rosenbergii. Fish Shellfish Immunol (2009) 26:256–63. doi: 10.1016/j.fsi.2008.11.017
38. Sun R, Qiu L, Yue F, Wang L, Liu R, Zhou Z, et al. Hemocytic immune responses triggered by CpG ODNs in shrimp Litopenaeus vannamei. Fish Shellfish Immunol (2013) 34:38–45. doi: 10.1016/j.fsi.2012.09.016
39. Cui X, Qian P, Rao T, Wei Y, Zhao F, Zhang H, et al. Cellular interleukin enhancer-binding factor 2, ILF2, inhibits Japanese encephalitis virus replication In Vitro. Viruses (2019) 11:559. doi: 10.3390/v11060559
40. Yang J, Wang L, Huang M, Wang L, Gai Y, Qiu L, et al. An interleukin-2 enhancer binding factor 2 homolog involved in immune response from Chinese mitten crab Eriocheir sinensis. Fish Shellfish Immunol (2011) 30:1303–9. doi: 10.1016/j.fsi.2011.03.014
41. Jiang X, Zhang X, Ren C, Ruan Y, Lu Y, Yuan L, et al. Interleukin-2 enhancer binding factor 2 (ILF2) in pacific white shrimp (Litopenaeus vannamei): alternatively spliced isoforms with different responses in the immune defenses against vibrio infection. Dev Comp Immunol (2021) 118:103975. doi: 10.1016/j.dci.2020.103975
42. Wang Y, Zhang H, Pang T, Zuo Z, Ren K. Rapamycin improves renal injury induced by iodixanol in diabetic rats by deactivating the mTOR/p70S6K signaling pathway. Life Sci (2020) 259:118284. doi: 10.1016/j.lfs.2020.118284
43. Wu W, Lin X, Wang C, Ke J, Wang L, Liu H. Transcriptome of white shrimp Litopenaeus vannamei induced with rapamycin reveals the role of autophagy in shrimp immunity. Fish Shellfish Immunol (2019) 86:1009–18. doi: 10.1016/j.fsi.2018.12.039
44. Su H, Yuan G, Su J. A specific CpG oligodeoxynucleotide induces protective antiviral responses against grass carp reovirus in grass carp Ctenopharyngodon idella. Dev Comp Immunol (2016) 60:218–27. doi: 10.1016/j.dci.2016.03.007
45. Lippitsch A, Chukovetskyi Y, Baal N, Bein G, Hackstein H. Unique high and homogenous surface expression of the transferrin receptor CD71 on murine plasmacytoid dendritic cells in different tissues. Cell Immunol (2017) 316:41–52. doi: 10.1016/j.cellimm.2017.03.005
46. Cha YJ, Lee CR, Kwon JY, Kang YJ. Protective effects of CpG-ODN 2007 administration against Edwardsiella tarda infection in olive flounder (Paralichthys olivaceus). Fish Shellfish Immunol (2017) 68:327–31. doi: 10.1016/j.fsi.2017.07.037
47. Dar A, Nichani A, Lai K, Potter A, Gerdts V, Babiuk LA, et al. All three classes of CpG ODNs up-regulate IP-10 gene in pigs. Res Vet Sci (2010) 88:242–50. doi: 10.1016/j.rvsc.2009.10.003
48. Canova R, Kirsten KS, de Figueiredo Soveral L, Frandoloso R, Kreutz LC. Oligodeoxynucleotides CpGs increase silver catfish (Rhamdia quelen) resistance to Aeromonas hydrophila challenge. Aquaculture (2017) 473:278–82. doi: 10.1016/j.aquaculture.2017.02.025
49. Butcherine P, Kelaher BP, Benkendorff K. Assessment of acetylcholinesterase, catalase, and glutathione s-transferase as biomarkers for imidacloprid exposure in penaeid shrimp. Aquat Toxicol (2022) 242:106050. doi: 10.1016/j.aquatox.2021.106050
50. Magara G, Sangsawang A, Pastorino P, Bellezza Oddon S, Caldaroni B, Menconi V, et al. First insights into oxidative stress and theoretical environmental risk of bronopol and detarox® AP, two biocides claimed to be ecofriendly for a sustainable aquaculture. Sci Total Environ (2021) 778:146375. doi: 10.1016/j.scitotenv.2021.146375
51. Zhou N, Wang Z, Yang L, Zhou W, Qin Z, Zhang H. Size-dependent toxicological effects of polystyrene microplastics in the shrimp Litopenaeus vannamei using a histomorphology, microbiome, and metabolic approach. Environ pollut (2023) 316:120635. doi: 10.1016/j.envpol.2022.120635
52. Lu Q, Stappenbeck TS. Local barriers configure systemic communications between the host and microbiota. Sci (80-.) (2022) 376:950–5. doi: 10.1126/science.abo2366
53. Liu M-J, Guo H-Y, Gao J, Zhu K-C, Guo L, Liu B-S, et al. Characteristics of microplastic pollution in golden pompano (Trachinotus ovatus) aquaculture areas and the relationship between colonized-microbiota on microplastics and intestinal microflora. Sci Total Environ (2023) 856:159180. doi: 10.1016/j.scitotenv.2022.159180
54. Zheng Y, Hou C, Yan Z, Chen J, Wang H, Tan B, et al. Effects of dietary zymosan-a on the growth performance and intestinal morphology, digestive capacity, and microbial community in Litopenaeus vannamei. Front Mar Sci (2022) 9:877865. doi: 10.3389/fmars.2022.877865
55. Dehler CE, Secombes CJ, Martin SAM. Environmental and physiological factors shape the gut microbiota of Atlantic salmon parr (Salmo salar L.). Aquaculture (2017) 467:149–57. doi: 10.1016/j.aquaculture.2016.07.017
Keywords: Litopenaeus vannamei, CpG oligodeoxynucleotides, intestinal microbiota, innate immunity, immune modulation
Citation: Hu F, Wang Y, Hu J, Bao Z and Wang M (2023) Comparative study of the impact of dietary supplementation with different types of CpG oligodeoxynucleotides (CpG ODNs) on enhancing intestinal microbiota diversity, antioxidant capacity, and immune-related gene expression profiles in Pacific white shrimp (Litopenaeus vannamei). Front. Immunol. 14:1190590. doi: 10.3389/fimmu.2023.1190590
Received: 21 March 2023; Accepted: 13 April 2023;
Published: 27 April 2023.
Edited by:
Jia Cai, Guangdong Ocean University, ChinaReviewed by:
Shengtao Guo, University of Chinese Academy of Sciences, ChinaCopyright © 2023 Hu, Wang, Hu, Bao and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mengqiang Wang, d2FuZ21lbmdxaWFuZ0BvdWMuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.