- 1Department of Immunopathology, Post Graduate Institute of Medical Education and Research, Chandigarh, India
- 2Department of Internal Medicine, Postgraduate Institute of Medical Education and Research, Chandigarh, India
- 3Allergy Immunology Unit, Department of Pediatrics, Advanced Pediatrics Centre, Post Graduate Institute of Medical Education and Research, Chandigarh, India
Introduction: Systemic sclerosis (SSc) is a chronic multisystem autoimmune rheumatic disease of unknown etiology. Several studies have established that SSc is triggered by a dynamic interplay between genetic factors and environmental stimuli. In the present study, we aimed to study the association of human leukocyte antigen (HLA) with familial and non-familial SSc patients [limited cutaneous SSc (lcSSc) and diffuse cutaneous SSc (dcSSc)] from North India.
Methods: The HLA-A, B, DRB1, and DQB1 genotyping of 150 (70 lcSSc and 80 dcSSc) adult-onset SSc patients and 150 age-gender-matched healthy controls were performed with sequence-specific oligonucleotide (SSO) typing kits using the luminex platform. HLA typing for HLA class I (A, B, and C) and II (DRB1, DQB1, and DPB1) in five North Indian families consisting of parent–child/sibling pairs affected with SSc or overlap syndrome was performed by Next Generation Sequencing (NGS) with Illumina MiniSeq.
Rseults: Among the non-familial SSc patients, HLA- DRB1*11 (P = 0.001, OR: 2.38, Pc = 0.01) was identified as a risk allele, and DRB1*12 (P = .0001, OR: 0.00, Pc = 0.001) as a protective allele. There was no statistical association found with HLA-DQB1*. Also, no significant association was observed between HLA antigens and different clinical subsets (lcSSc and dcSSc) of SSc. Two cases of familial SSc patients had the DRB1*11 allele. The DRB1*12 allele was absent in all the familial SSc patients.
Discussion: HLA DRB1*11 (risk allele) and DRB1*12 (protective allele) were found to be strongly associated with non-familial SSc patients and partially explain the disease’s familial clustering, supporting the susceptible genetic background theory for SSc development. The study also indicates the HLA allele as a common genetic risk factor in distinct autoimmune diseases contributing to overlap syndrome or polyautoimmunity.
1 Introduction
Systemic sclerosis (SSc) is a rare autoimmune rheumatic disease of unknown etiology characterized by excessive extracellular matrix deposition leading to fibrosis of the skin and visceral organs (1). Patients with SSc are classified into two subsets, limited cutaneous systemic sclerosis (lcSSc) and diffuse cutaneous systemic sclerosis (dcSSc), based on the extent of their skin involvement. SSc is heterogeneous in its clinical presentation that likely reflects different genetic or triggering factors (i.e., infectious agents such as cytomegalovirus and Epstein-Barr virus or environmental toxins such as organic solvents, toluene, xylene, trichloroethylene, and polyvinyl chloride) (2), which results in a chronic and self-amplifying process marked by vascular changes, inflammation, autoimmunity, and fibrosis. Endothelial cells, platelets, structural cells, immune cells, and other cell types are all involved in the disease process. These cells are activated by mediators such as transforming growth factor-β, interleukins, endothelin 1, serum autoantibodies, and reactive oxygen species (1, 2). The pathogenesis of SSc is still poorly understood. As per the literature, SSc develops in an individual with a “permissive” genetic background (2). Although SSc is not inherited in a Mendelian fashion, the relative risk of developing SSc is much higher (1.6%) in families with a history of SSc versus the general population (0.026%) (3). The familial history of SSc increases the risk of developing SSc in siblings (13- to 15-fold) and first-degree relatives (15- to 19-fold) compared with the general population (3). Hudson M et al. reported that 38% of SSc patients had at least one or other overlapping autoimmune disease (AID), suggesting AIDs share genetic risk factors and the role of genetic influences in SSc (4). The major histocompatibility complex (MHC) or human leukocyte antigen (HLA), located on chromosome 6p21.3, represents the densest and polymorphic region of the human genome and is observed to have the strongest genetic association with SSc (2). The MHC classes I and II have been identified as susceptible factors in SSc pathogenesis, and an evident ethnic variation is seen in these associations (2). Studies have reported different or the same HLA alleles as a risk factor or a protective factor in different ethnic populations (2). HLA polymorphism is also linked to susceptibility to a wide range of AIDs such as systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), autoimmune hepatitis (AIH), and so forth (4). Studies have reported HLA alleles as common risk factors in AIDs (4). From India to date, there is no report on the genetics of SSc. HLA associations can be unique for specific populations or common among different ethnic populations. Hence, in the present study, we wish to determine the HLA association with SSc patients having familial and non-familial inheritance and the genetic difference between lcSSc and dcSSc. This will be the first genetic report from India to decipher the role of HLA in SSc pathogenesis.
2 Materials and methods
2.1 Study population
This is a cross‐sectional study carried out in the Department of Immunopathology, and the SSc patients were recruited from the Department of Internal Medicine, Postgraduate Institute of Medical Research and Education, Chandigarh. One hundred fifty (female 138 and male 12) adult-onset (≥ 18 years) SSc patients, having a mean ± SD age of 40.3 ± 10.9 years, presenting with clinical signs and symptoms without familial history and fulfilling the ACR/EULAR criteria guidelines were enrolled from North India. They were categorized into two clinical subgroups according to the criteria of Le Roy et al. (5) as having lcSSc (n = 70) or dcSSc (n = 80). The controls consisted of 150 healthy unrelated individuals (female 138 and male 12), having a mean ± SD age of 41.5 ± 9.6 years with North Indian origin similar to the patients. Five North Indian families consisting of parent–child/Sibling pairs affected with SSc or overlap syndrome were also enrolled. This study was approved by the Institutional Ethics Committee (NK/3163/Ph.D./362), and written informed consent was obtained from each study subject.
2.2 ANA and autoantibodies tests
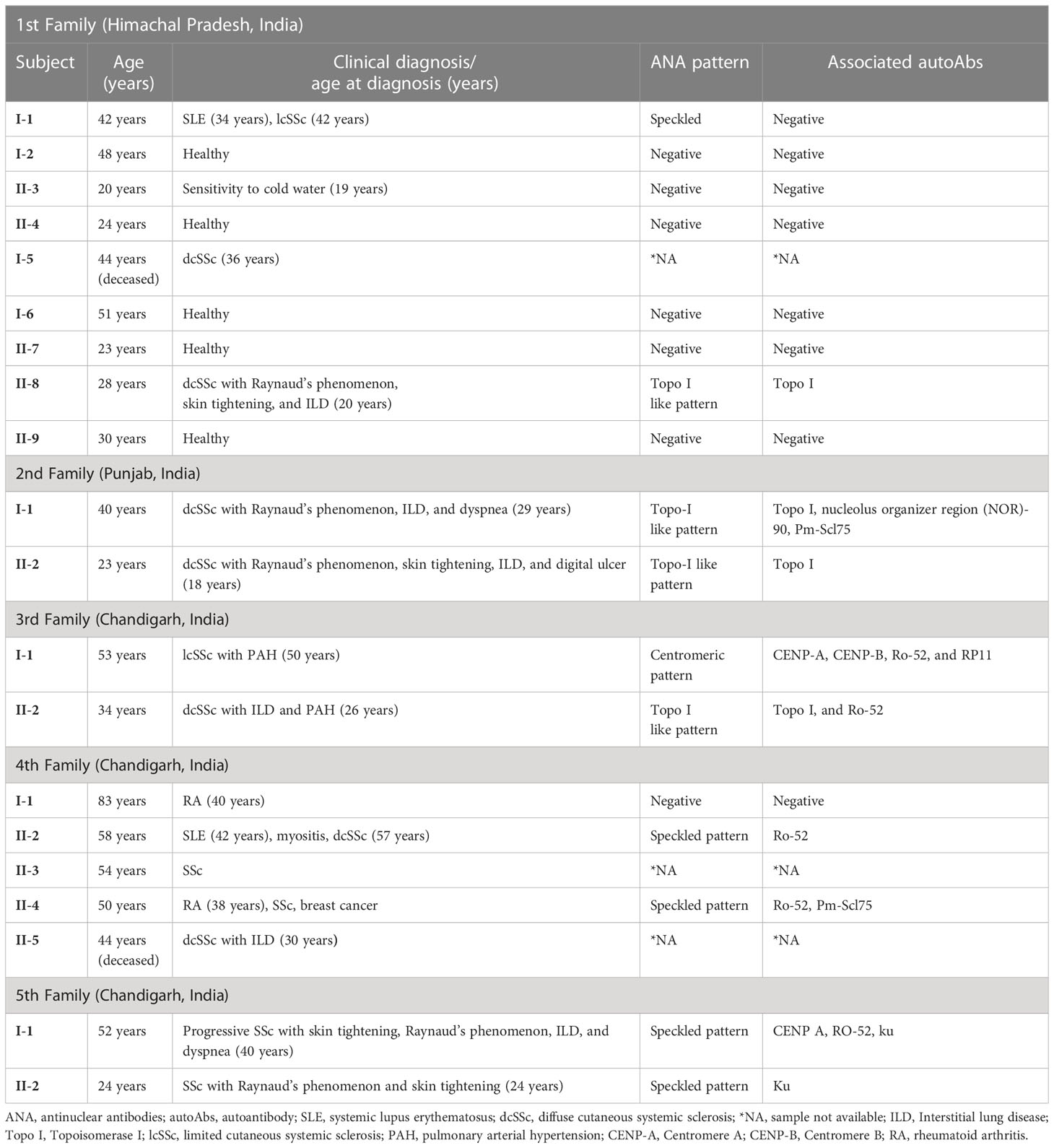
The sera from familial SSc patients were tested for antinuclear antibodies (ANA) by indirect immunofluorescence followed by line immunoblot assay for examining the specific autoantibodies, using the same reagents and protocol in our previously published article (6), which describes the ANA pattern and autoantibody profiling of North Indian SSc patients.
2.3 Genomic DNA extraction
Genomic DNA was extracted from whole blood using a commercially available kit (QIAamp DNA, Qiagen, Hilden, Germany). A260/A280 measurements assessed the concentration and purity of DNA on a spectrophotometer (Genova Nano, Jenway, UK).
2.4 HLA classes I (A and B) and II (DRB1 and DQB1) genotyping in non-familial SSc
The HLA-A, B, DRB1, DQB1 genotyping of 150 patients and 150 controls were performed with LIFE CODES SSO typing kits (Immucor, Norcross, GA, USA) using the Luminex platform (LX200, Austin, TX, USA). The HLA MatchIT DNA software was used in sequencing analysis and assigning HLA alleles.
2.5 HLA class I (A, B, and C) and II (DRB1, DQB1, and DPB1) genotyping in familial SSc
HLA typing for HLA classes I (A, B, and C) and II (DRB1, DQB1, and DPB1) for the familial study of SSc was performed by Next Generation Sequencing (NGS) with Illumina NovaSeq6000. HLA genotype was determined based on IMGT/HLA database release 3.42.0.
2.6 Statistical analysis
Phenotype frequencies were compared between patients and controls using the chi-square test or Fisher exact test. Bonferroni correction was applied to the p-values. Two-sided P-values less than 0.05 were considered significant. Analyses were performed using SPSS software (version 20.0).
3 Results
HLA types I and II association in patients with familial and non-familial SSc (lcSSc and dcSSc) from North India
3.1 HLA-A, HLA-B, HLA-DRB1, and HLA-DQB1 genotyping in non-familial SSc patients
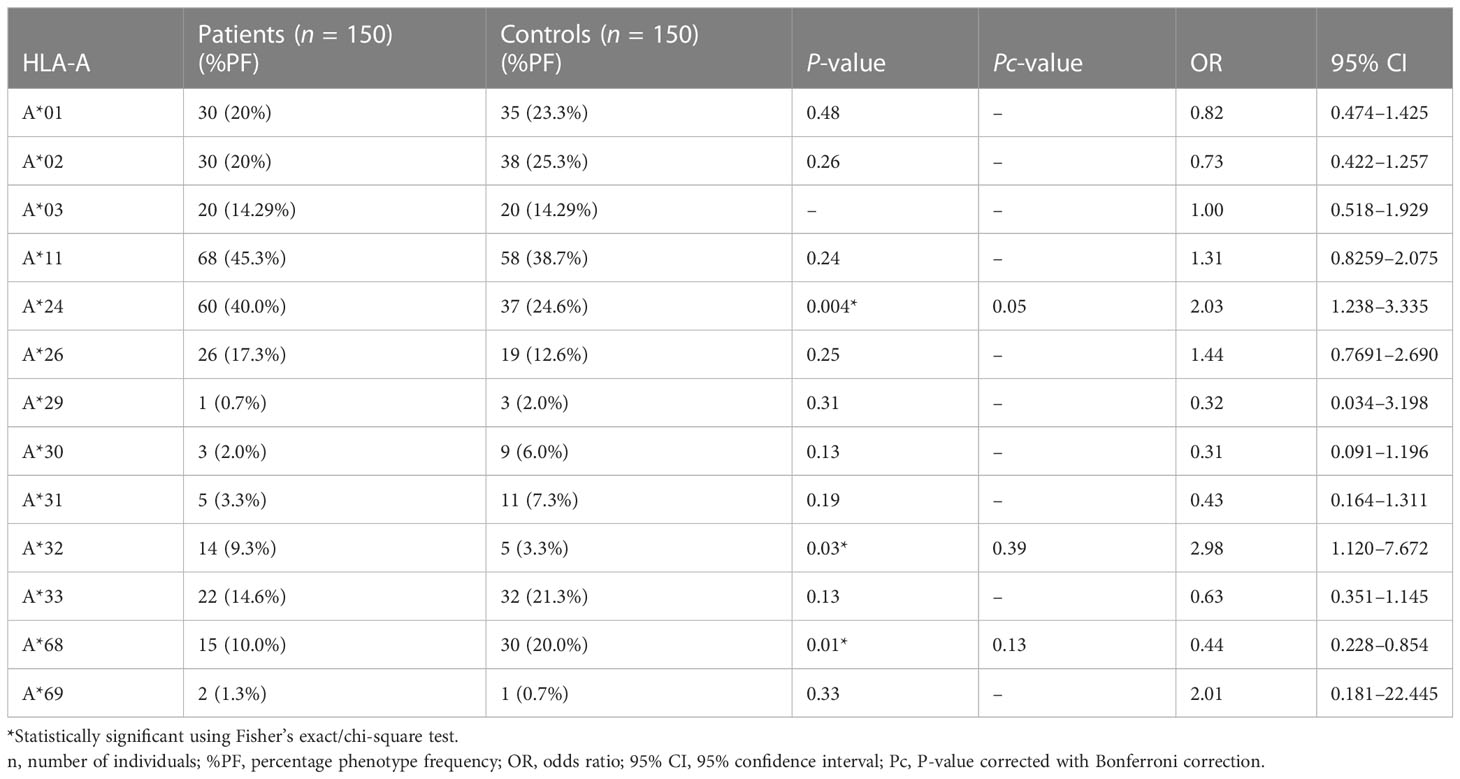
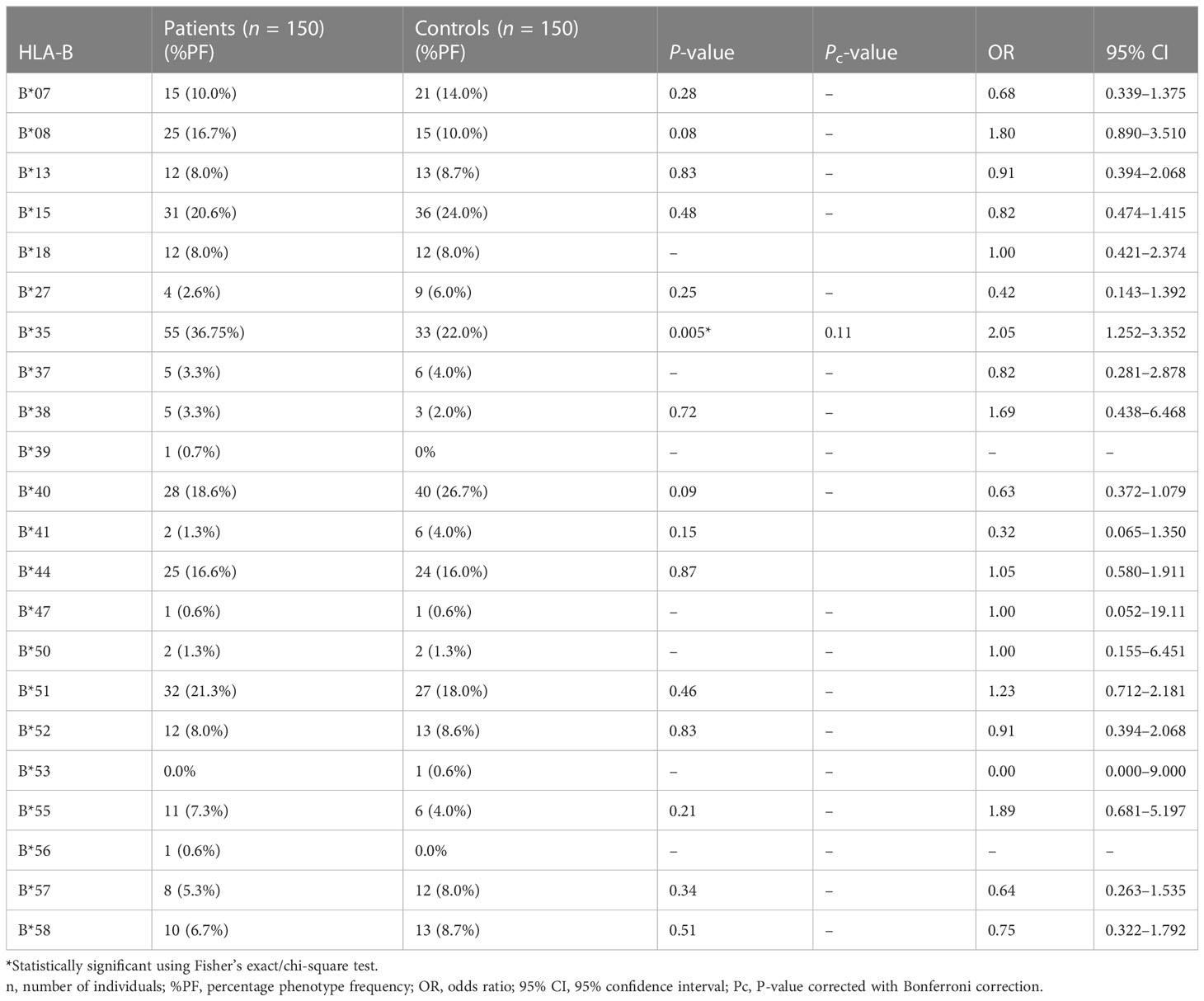
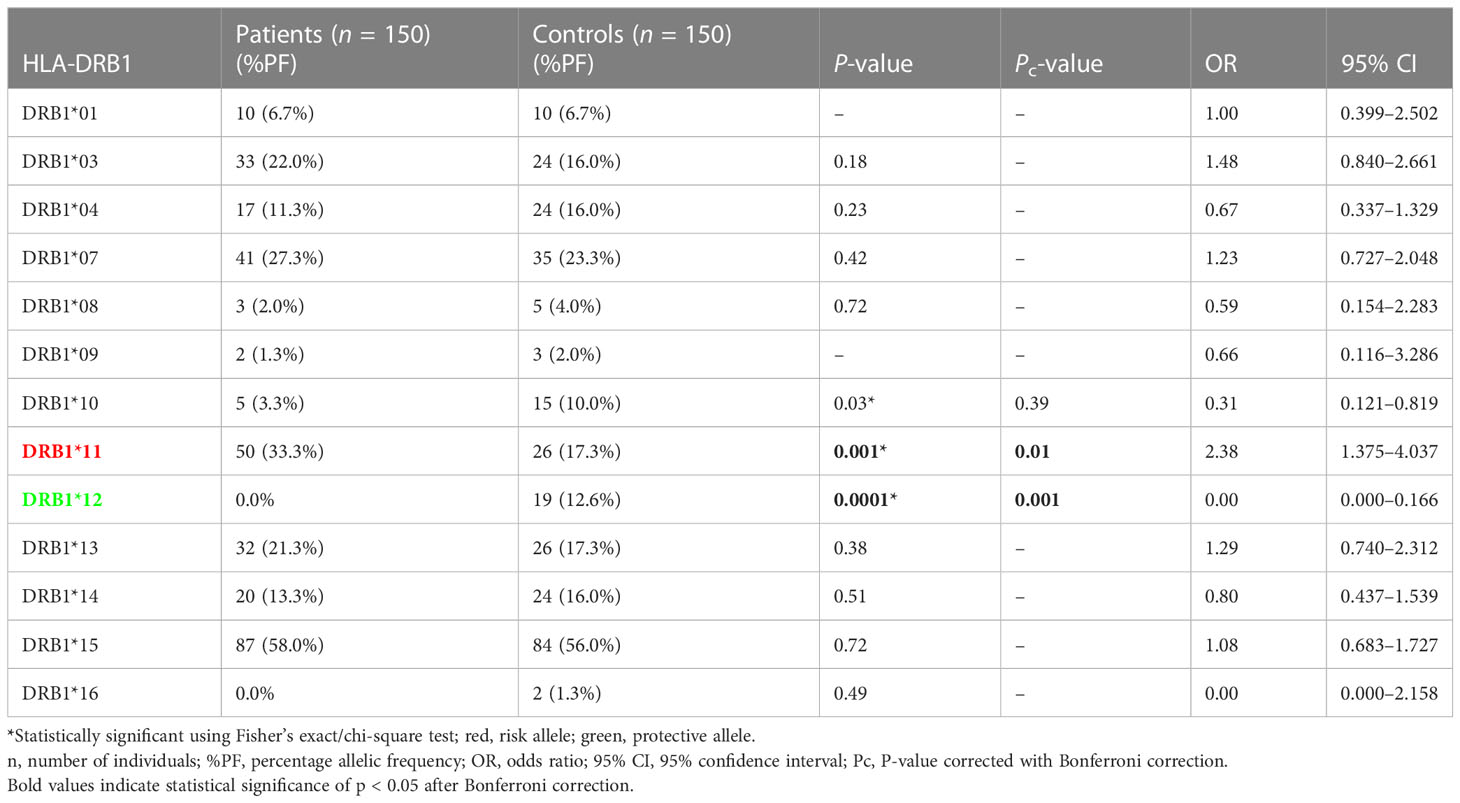
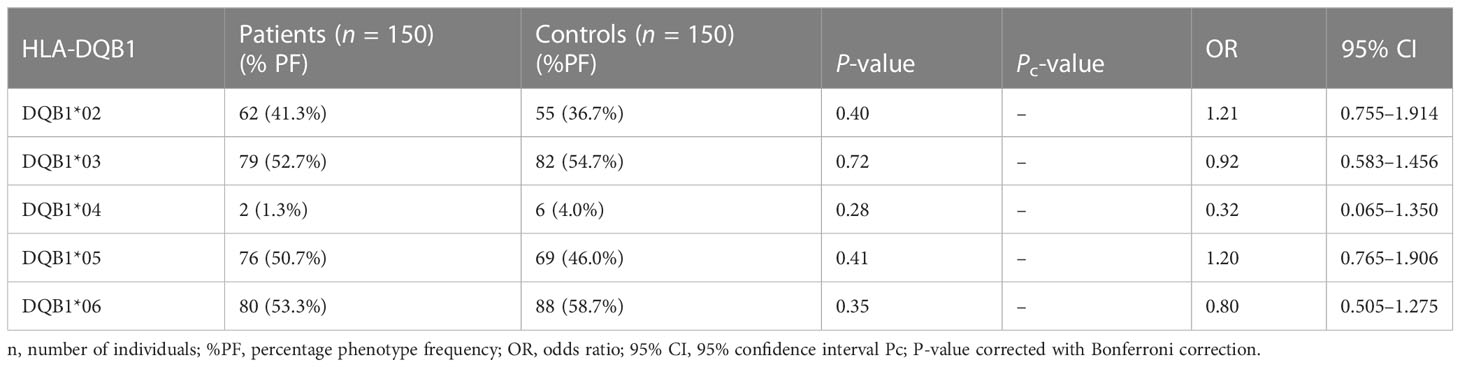
The results of genotyping for HLA-A, HLA-B, HLA-DRB1, and HLA-DQB1 in SSc patients (n = 150; 70 lcSSc and 80 dcSSc) and controls (n = 150) are summarized in Tables 1–8.
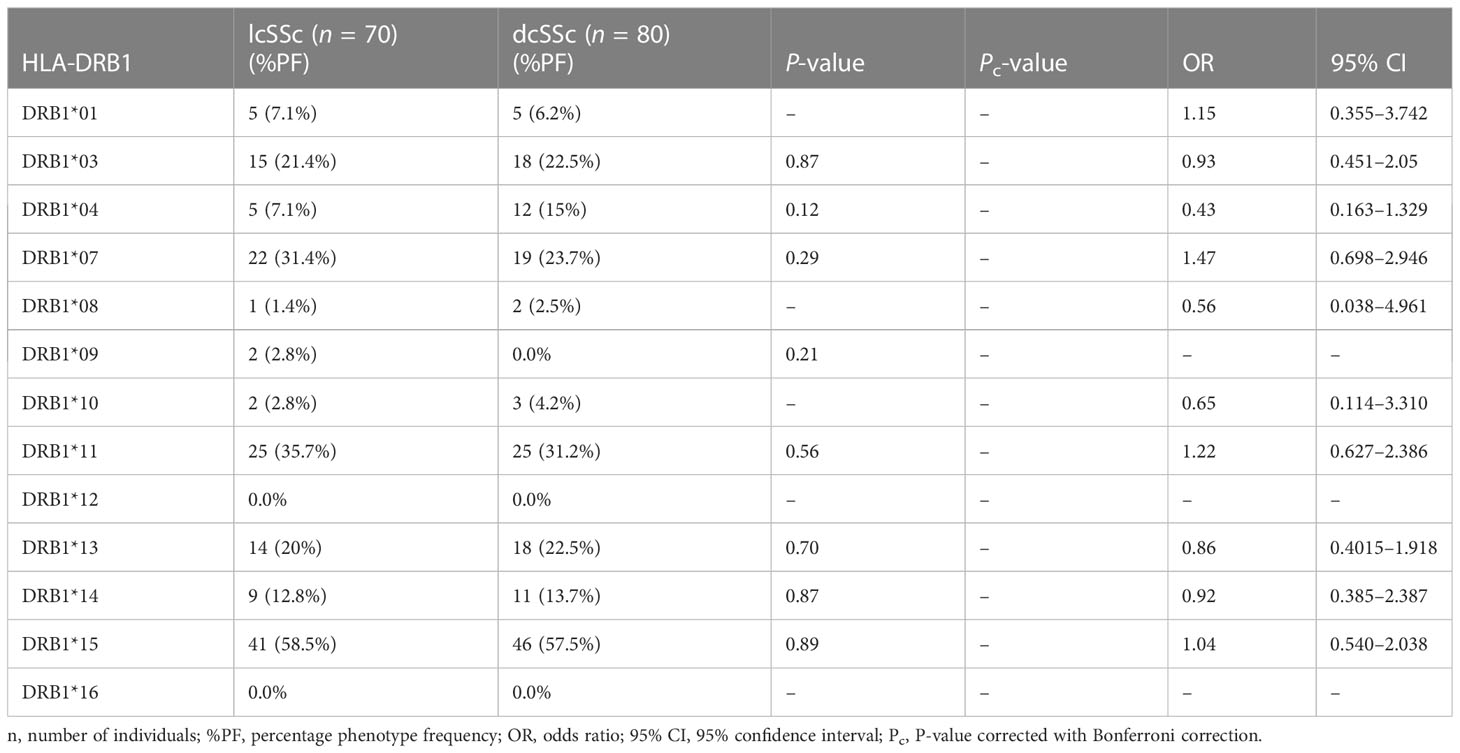
Phenotype (P = 0.001, OR: 2.38, CI: 1.375–4.037, Pc = 0.01) frequency of HLA-DRB1*11 was significantly high in SSc patients compared with controls suggesting their role in disease susceptibility.
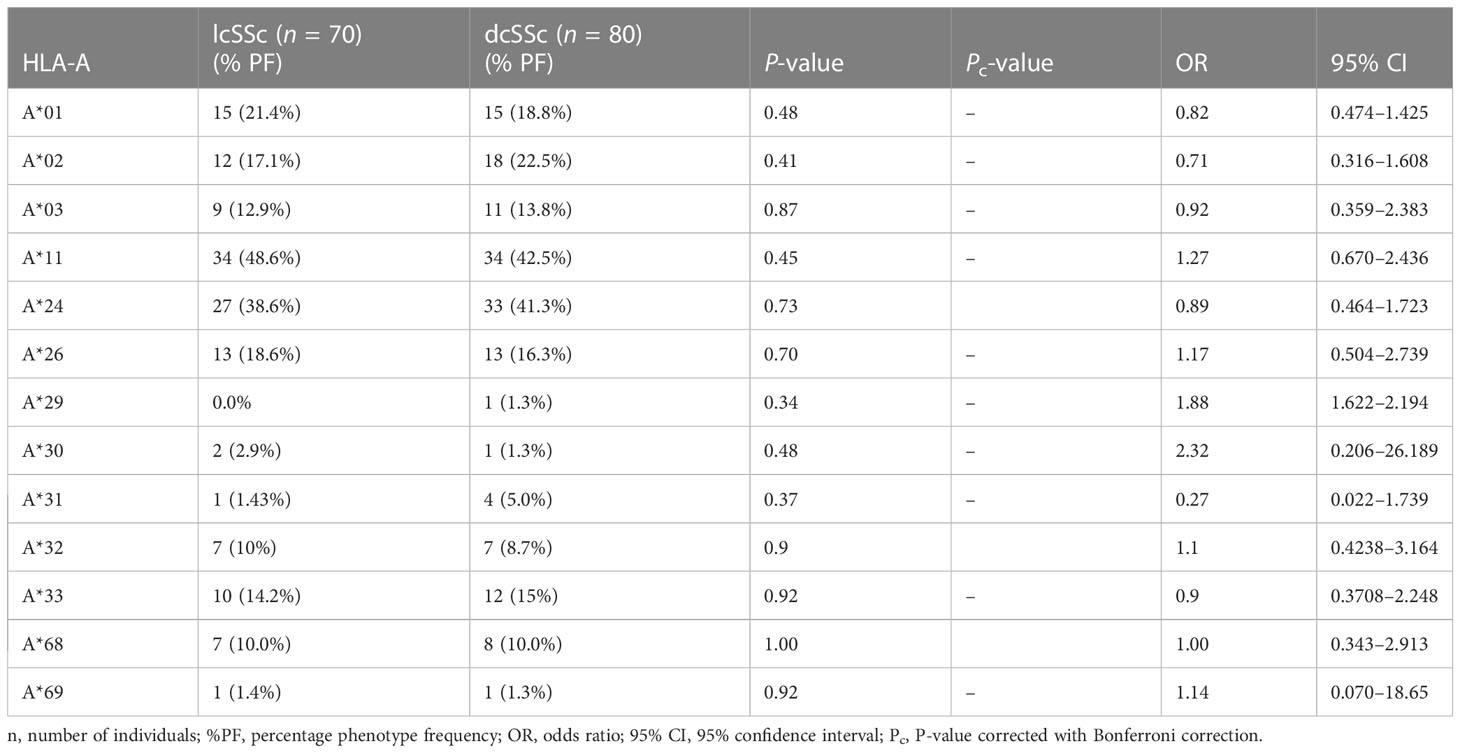
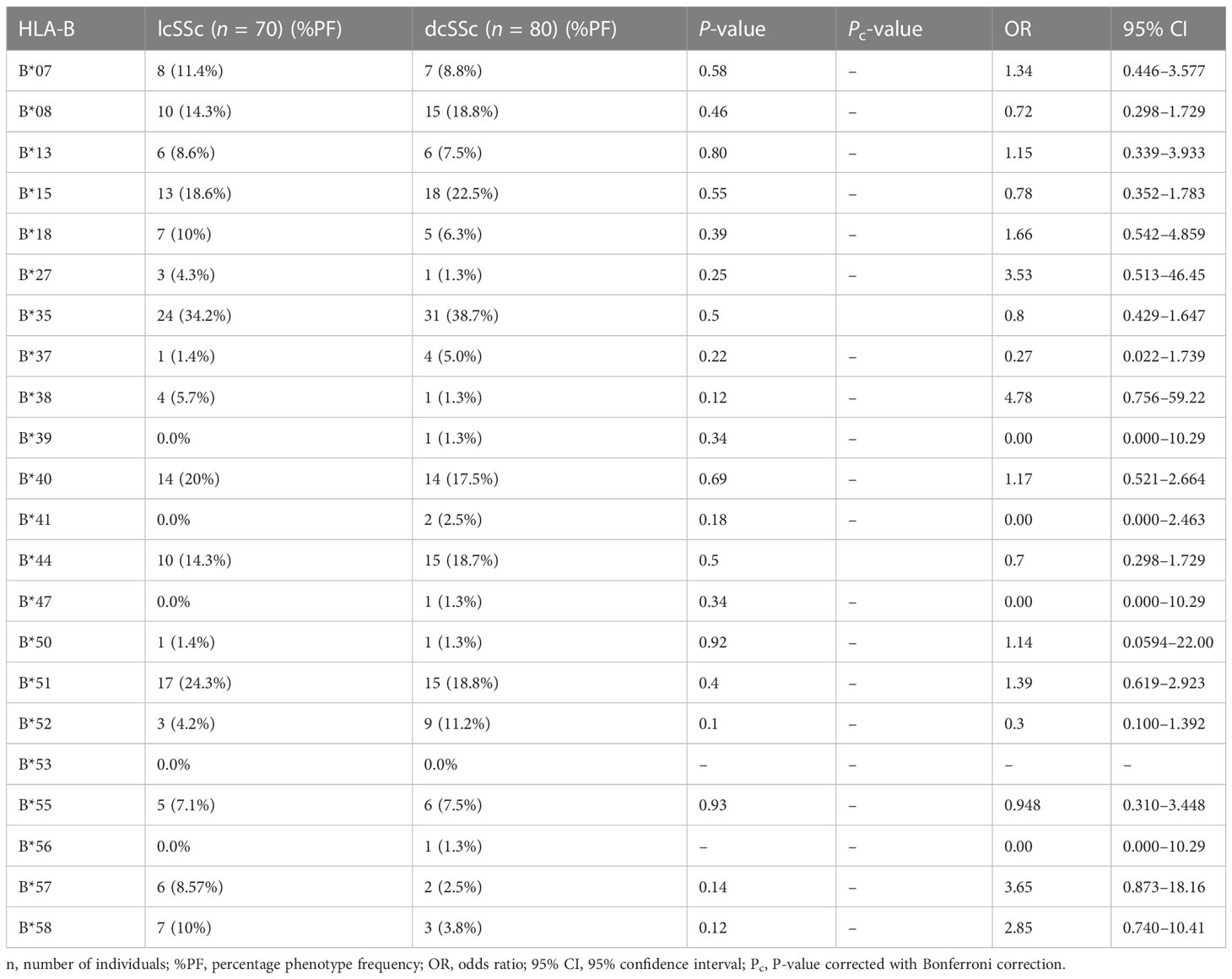
The phenotype frequencies of HLA-A*24 (P = 0.004, OR: 2.03, CI: 1.238–3.335, Pc = 0.05), HLA-A*32 (P = 0.03, OR: 2.98, CI: 1.120–7.672, Pc = 0.39), and HLA-B*35 (P = 0.005, OR: 2.05, CI: 1.252–3.352, Pc = 0.11), were observed to be high in SSc patients. However, after applying the Bonferroni correction, the P-value lost its significance.
Phenotype (P = .0001, OR: 0.00, CI: 0.000–0.166, Pc = 0.001) frequency of HLA-DRB1*12 was significantly less prevalent in SSc patients compared with controls suggesting a protective association.
The phenotype frequencies of HLA-A*68 (P = 0.01, OR: 0.44, CI: 0.228–0.854, Pc = 0.13) and HLA- DRB1*10 (P = 0.03, OR: 0.31, CI: 0.121–0.819, Pc = 0.39), were found to be less prevalent in SSc patients than controls; however, the association did not attain statistical significance after the Bonferroni correction.
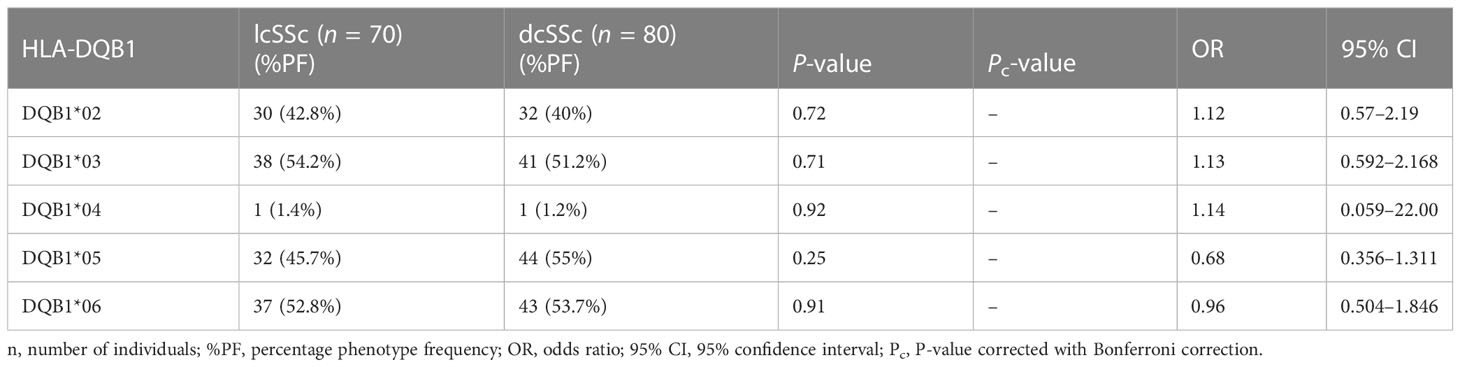
None of the HLA-DQB1 alleles were found to be associated with SSc patients compared with controls.
3.2 HLA-A, HLA-B, HLA-DRB1, and HLA-DQB1 phenotype frequency in lcSSc and dcSSc patients
Stratification of genotyping results into the limited and diffused form of SSc revealed no significant difference in the allele frequencies among the two groups (Tables 5–8).
3.3 HLA classes I (A, B, and C) and II (DRB1, DQB1, and DPB1) genotyping in familial SSc
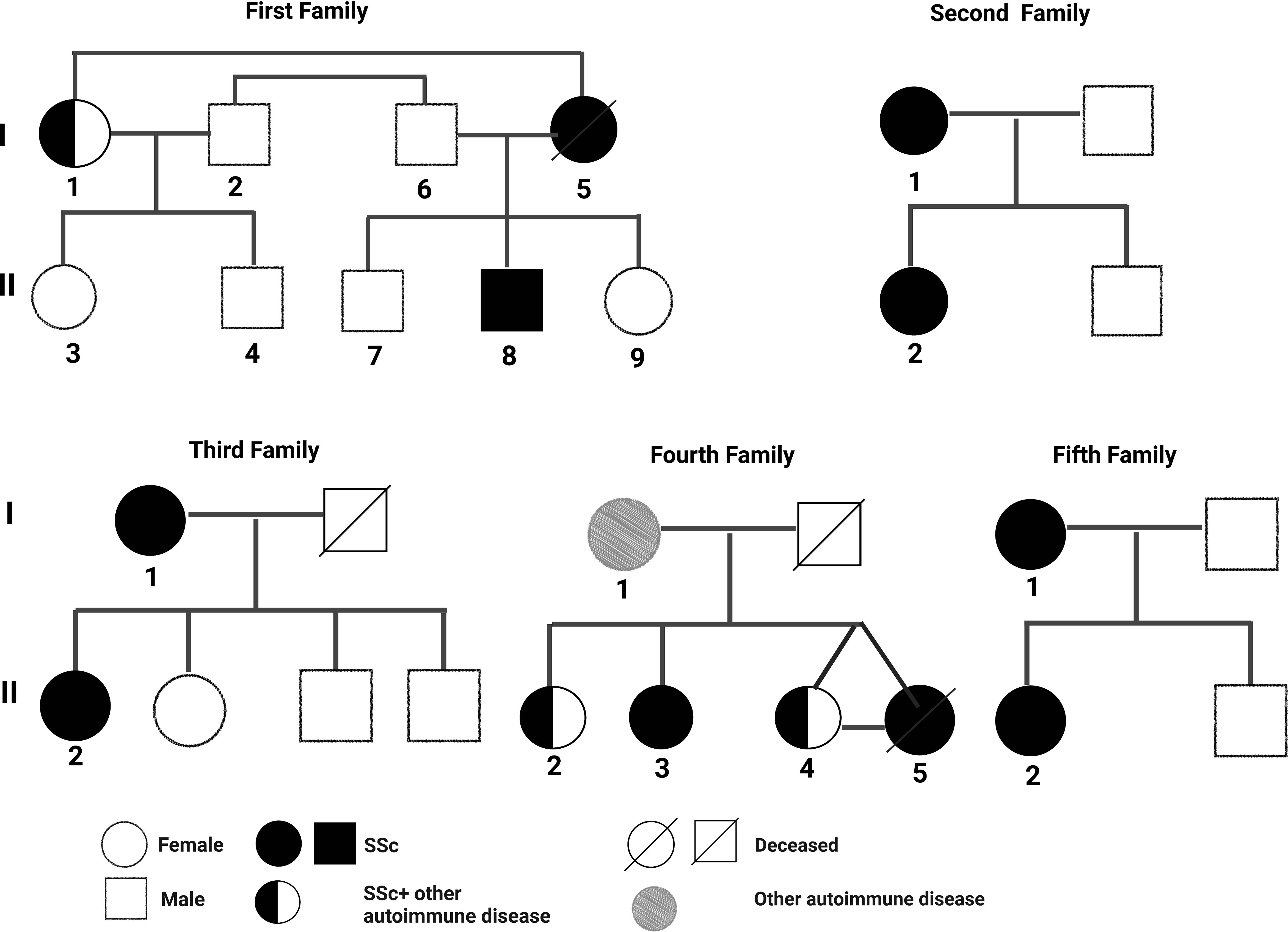
The pedigree, demographic-clinical details and HLA haplotype of five families are shown in Figure 1 and Tables 9, 10.
The risk allele HLA-DRB1*11 associated with the non-familial SSc patients was present in the SSc-affected family members of the second family. The protective allele HLA-DRB1*12 associated with the non-familial SSc patients was absent in all the SSc-affected family members.
4 Discussion
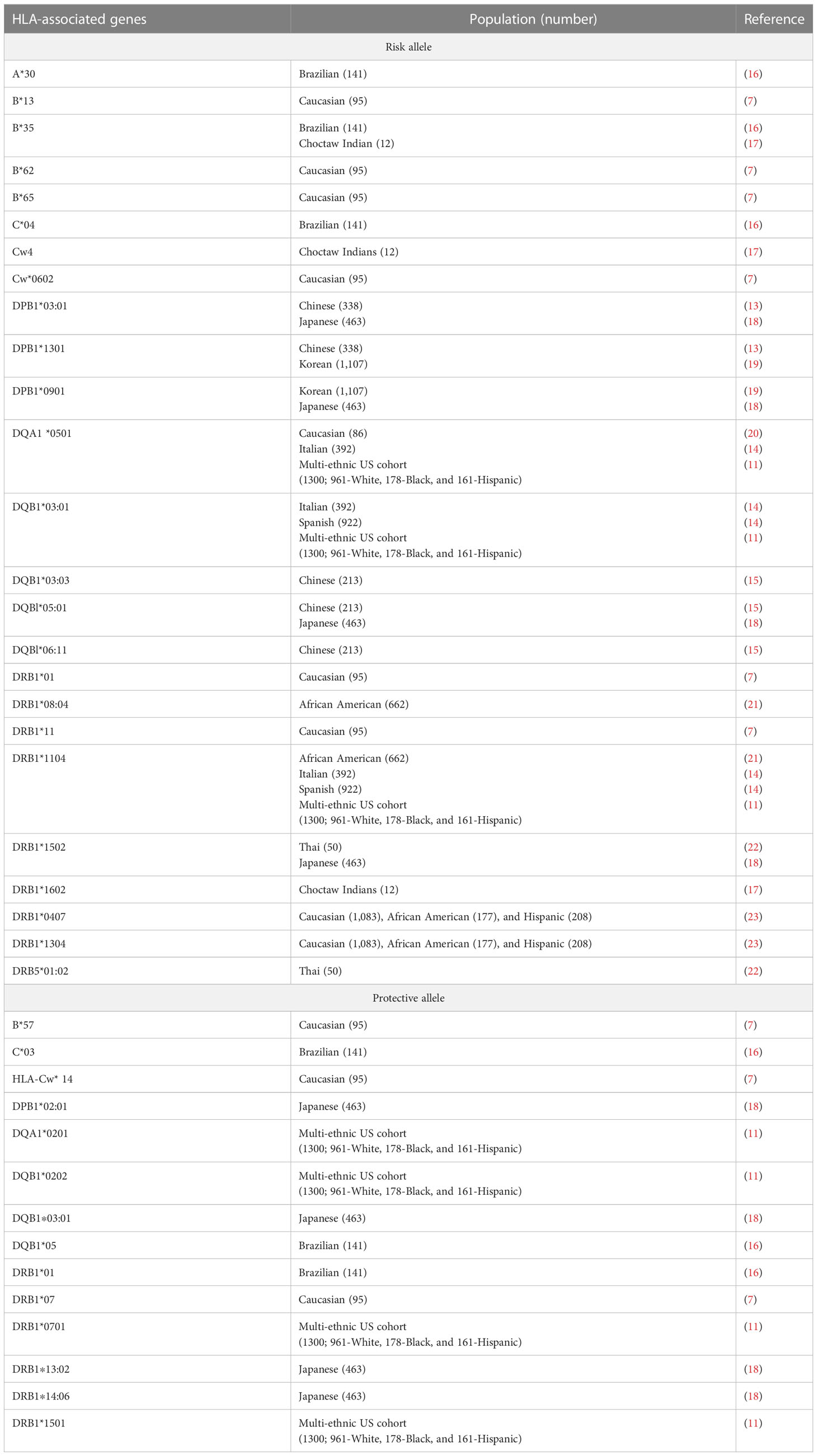
This is the first report on the HLA association with SSc from India. We investigated HLA types I and II association in patients with familial and non-familial SSc from North India. We also examined the HLA association within the subgroup of SSc (lcSSc vs. dcSSc). In this study, the HLA-DRB1*11 allele was significantly predisposing in SSc patients compared with the controls, while the HLA-DRB1*12 allele emerged as a protective allele. SSc’s most consistent genetic risk factor reported is the HLA-DRB1*11 alleles found in different ethnic populations. HLA-DRB1*11 has a strong genetic association among Caucasians, Spanish, and Greece populations (7–10). Also, in a large multicentric study involving three ethnic groups (Caucasian, Black, and Hispanic) in the United States, which covered 1,300 patients, the strongest association in the Caucasian and Hispanic subjects was with HLA-DRB1*11:04 haplotype; they also found DRB1*11:04 allele association with anti–topo-I positivity (11). In the present study, HLA-DRB1*11 showed the strongest association with SSc, further confirming HLA-DRB1*11 as a susceptibility allele for SSc. There are no previous reports on the association of the protective allele HLA-DRB1*12 with SSc in other ethnic populations. However, HLA-DRB1*12 is reported to be a protective allele against type 1 Diabetes (T1D) and Graves’ disease in the Taiwan population (12). Possibly genetic heterogeneity among ethnicities might be significantly impacting the complex trait of SSc. Previous studies of different populations already implicated ethnic differences in genetic association with SSc (2, 13), while some share the same genetic determinant as in the case of HLA-DRB*11 (7–10). Therefore, HLA-DRB1*12 is unique to our North Indian population of SSc. None of the HLA-DQB1 alleles were associated with North Indian SSc patients, unlike other ethnic populations (11, 14, 15). We also did not find any significant difference in the phenotype frequency among the two subsets of SSc (lcSSc vs. dcSSc), again, which could be due to ethnic variations. Table 11 shows the HLA association with SSc in the different ethnic populations.
The risk allele HLA-DRB1*11, found to be associated with the non-familial SSc cohort, was present only in the SSc-affected family members of the second family. The mother–daughter not only shared a risky HLA haplotype (HLA-DRB1*01) for SSc but also manifested a similar disease subset (dcSSc), and both were positive for SSc-specific topo-1 antibody. This is in line with prior studies indicating that the type of HLA can define the disease subset and autoantibodies (21, 24). The protective allele HLA-DRB1*12 found in non-familial SSc patients was absent in all the familial SSc patients investigated.
Interestingly, in the 3rd family, the daughter (subject II-1) had clinical signs and symptoms of dcSSc earlier at 26 years, and the mother (subject I-1) was diagnosed with lcSSc later at the age of 50 years. Also, both had a different set of multiple autoAbs. The possible reason could be the presence of the HLA-DPB1*13 allele in the daughter, making her more susceptible towards the SSc; the rest of the HLA haplotypes were the same in both mother and daughter. HLA-DPB1*13 allele is a known risk factor for SSc (13, 19) and is also associated with the topo-I autoantibody subset of SSc in the European American ancestral cohort (21). In our North Indian cohort of SSc, we found topo-I autoAb to be associated with the dcSSc and clinical complications such as Raynaud’s, ILD, and PAH (6). The daughter had a serological presence of topo-I autoAb. Altogether, the HLA-DPB1*13 allele in the daughter could be the possible reason for the existence of topo-I autoAb, causing early disease onset and development of a more severe subset of SSc compared with the mother.
Our study shows three cases in familial SSc patients where SSc was co-existing with other AIDs. Subject I-1 (1st family) had both SLE and lcSSc, subject II-2 (4th family) had SLE, myositis, APLA, and dcSSc, a case of polyautoimmunity, and subject II-4 (4th family) had RA and SSc. A meta-analysis study by Cruz-Tapias et al. identified HLA-DRB1*03 and HLA-DRB1*04 as common alleles conferring susceptibility to more than one AIDs: RA, SLE, AIH, MS (multiple sclerosis), and T1D (25). The presence of DRB1*04 in subject I-1 (1st family) and DRB1*03 and DRB1*04 in subject II-2,4 (4th family) might be the reason for the occurrence of distinct AIDs within an individual (polyautoimmunity) and within members of a nuclear family (familial autoimmunity), supporting the concept of a shared autoimmune genetic background among AIDs.
The findings partially explain the clustering of the disease within families, suggesting the role of non-HLA genes, environmental factors, and epigenetic influences in disease development.
Due to the resource-limited setting, we could not perform HLA-C and HLA-DPB1 genotyping and used a low-resolution detection kit for HLA genotyping in non-familial SSc patients, the major limitation of our study.
5 Conclusion
The HLA-DRB1*11 allele was found to be strongly associated with SSc risk in the non-familial cohort, whereas HLA-DRB1*12 was found to be protective; however, it partially explains the disease clustering in familial SSc patients, suggesting the role of many other factors such as non-HLA genes, epigenetics, and environmental factors, which needs to be further investigated. The present study also suggests HLA sharing in distinct AIDs resulting in polyautoimmunity and familial autoimmunity. Being the initial genetic report from India, this research provides a basis for future studies aiming to understand the complex etiopathogenesis of SSc and explore ethnic variations.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Figshare, https://figshare.com/articles/dataset/HLA_class_I_A_B_C_and_class_II_DRB1_DQB1_DPB1_genotyping_data_in_familial_systemic_sclerosis_by_Next-Generation_Sequencing/24005574.
Ethics statement
The studies involving human participants were reviewed and approved by Postgraduate Institute of Medical Research and Education, Institutional Ethics Committee (NK/3163/Ph.D./362). The patients/participants provided their written informed consent to participate in this study.
Author contributions
SM: executed the study, was involved in sample collection, processing and experiments, analyzed the data and wrote the manuscript. SKS: provided the clinical samples and assisted in the clinical classification of patients, and reviewed the manuscript. YK: analyzed the data and reviewed the manuscript. SS: provided the clinical samples and relevant clinical information and reviewed the manuscript. RA: analyzed the data and reviewed the manuscript. SA: contributed to the execution of ANA screening and immunoblotting of familial SSc patients. MK: contributed to the execution of HLA typing. HS: contributed to the execution of HLA typing. RM: responsible for study conception, design, drafting of the manuscript, and data analysis. All authors contributed to the article and approved the submitted version.
Funding
Indian Council of Medical Research, New Delhi (3/2/2/22/2018/Online Onco Fship/NCDIII).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ACR/EULAR, American College of Rheumatology/European League Against Rheumatism; AID, Autoimmune disease; AIH, Autoimmune hepatitis; ANA, Antinuclear antibodies; APLA, Antiphospholipid syndrome; AutoAb, Autoantibody; CENP A, Centromere A; CENP B, Centromere B; dcSSc, Diffuse cutaneous systemic sclerosis; ET-1, Endothelin-1; HLA, Human Leukocyte Antigen; ILD, Interstitial lung disease; lcSSc, Limited cutaneous systemic sclerosis; MHC, Major histocompatibility complex; MS, Multiple sclerosis; NGS, Next Generation Sequencing; PAH, Pulmonary arterial hypertension; RA, Rheumatoid arthritis; RP11, RNA polymerase III-11; SLE, Systemic lupus erythematosus; SSc, Systemic sclerosis; T1D, Type 1 Diabetes; Topo I, Topoisomerase I.
References
1. Denton CP, Khanna D. Systemic sclerosis. Lancet (2017) 390(10103):1685–99. doi: 10.1016/S0140-6736(17)30933-9
2. Pattanaik D, Brown M, Postlethwaite BC, Postlethwaite AE. Pathogenesis of systemic sclerosis. Front Immunol (2015) 6:272. doi: 10.3389/fimmu.2015.00272
3. Gorlova O, Martin JE, Rueda B, Koeleman BP, Ying J, Teruel M, et al. Identification of novel genetic markers associated with clinical phenotypes of systemic sclerosis through a genome-wide association strategy. PloS Genet (2011) 7(7):e1002178. doi: 10.1371/journal.pgen.1002178
4. Hudson M, Rojas-Villarraga A, Coral-Alvarado P, López-Guzmán S, Mantilla RD, Chalem P, et al. Polyautoimmunity and familial autoimmunity in systemic sclerosis. J Autoimmun (2008) 31(2):156–9. doi: 10.1016/j.jaut.2008.05.002
5. LeRoy EC, Black C, Fleischmajer R, Jablonska S, Krieg T, Medsger TA, et al. Scleroderma (systemic sclerosis): classification, subsets and pathogenesis. J Rheumatol (1988) 15(2):202–5.
6. Machhua S, Sharma SK, Kumar Y, Anand S, Handa S, Minz RW. Antinuclear antibody pattern and autoantibody profiling of systemic sclerosis patients in a tertiary referral center in north India. Pathol Int (2022) :1–10. doi: 10.1111/pin.13221
7. Gladman DD, Kung TN, Siannis F, Pellett F, Farewell VT, Lee P. HLA markers for susceptibility and expression in scleroderma. J Rheumatol (2005) 32(8):1481–7.
8. Loubière LS, Lambert NC, Madeleine MM, Porter AJ, Mullarkey ME, Pang JM, et al. HLA allelic variants encoding DR11 in diffuse and limited systemic sclerosis in Caucasian women. Rheumatology (2005) 44(3):318–22. doi: 10.1093/rheumatology/keh489
9. Simeón CP, Fonollosa V, Tolosa C, Palou E, Selva A, Solans R, et al. Association of HLA class II genes with systemic sclerosis in Spanish patients. J Rheumatol (2009) 36(12):2733–6. doi: 10.3899/jrheum.090377
10. Vlachoyiannopoulos PG, Dafni UG, Pakas I, Spyropoulou-Vlachou M, Stavropoulos-Giokas C, Moutsopoulos HM. Systemic scleroderma in Greece: low mortality and strong linkage with HLA-DRB1*1104 allele. Ann Rheum Dis (2000) 59(5):359–67. doi: 10.1136/ard.59.5.359
11. Arnett FC, Gourh P, Shete S, Ahn CW, Honey RE, Agarwal SK, et al. Major histocompatibility complex (MHC) class II alleles, haplotypes and epitopes which confer susceptibility or protection in systemic sclerosis: analyses in 1300 Caucasian, African-American and Hispanic cases and 1000 controls. Ann Rheum Dis (2010) 69(5):822–7. doi: 10.1136/ard.2009.111906
12. Tung YC, Fann CS, Chang CC, Chu CC, Yang WS, Hwu WL, et al. Comprehensive human leukocyte antigen genotyping of patients with type 1 diabetes mellitus in Taiwan. Pediatr Diabetes (2018) 19(4):699–706. doi: 10.1111/pedi.12645
13. Wang J, Guo X, Yi L, Guo G, Tu W, Wu W, et al. Association of HLA-DPB1 with scleroderma and its clinical features in Chinese population. PloS One (2014) 9(1):8–11. doi: 10.1371/journal.pone.0087363
14. Beretta L, Rueda B, Marchini M, Santaniello A, Simeón CP, Fonollosa V, et al. Analysis of class II human leucocyte antigens in Italian and Spanish systemic sclerosis. Rheumatology (2012) 51(1):52–9. doi: 10.1093/rheumatology/ker335
15. Zhou X, Yi L, Guo XJ, Chen E, Zou HJ, Jin L, et al. Association of HLA-DQB1*0501 with scleroderma and its clinical features in chinese population. Int J Immunopathol Pharmacol (2013) 26(3):747–51. doi: 10.1177/039463201302600318
16. Del Rio AP, Sachetto Z, Sampaio-Barros PD, Marques-Neto JF, Londe AC, Bertolo MB. HLA markers for poor prognosis in systemic sclerosis Brazilian patients. Dis Markers (2013) 35(2):73–8. doi: 10.1155/2013/301415
17. Arnett FC, Howard RF, Tan F, Moulds JM, Bias WB, Durban E, et al. Increased prevalence of systemic sclerosis in a native American tribe in Oklahoma: association with an Amerindian HLA haplotype. Arthritis Rheumatol (1996) 39(8):1362–70. doi: 10.1002/art.1780390814
18. Furukawa H, Oka S, Kawasaki A, Shimada K, Sugii S, Matsushita T, et al. Human leukocyte antigen and systemic sclerosis in japanese: the sign of the four independent protective alleles, DRB1 13:02, DRB1 14:06, DQB1 03:01, and DPB1 02:01. PloS One (2016) 11(4):1–13. doi: 10.1371/journal.pone.0154255
19. Zhou X, Lee JE, Arnett FC, Xiong M, Park MY, Yoo YK, et al. HLA-DPB1 and DPB2 are genetic loci for systemic sclerosis: a genome-wide association study in koreans with replication in north americans. Arthritis Rheumatol (2009) 60(12):3807–14. doi: 10.1002/art.24982
20. Lambert NC, Distler O, Müller-Ladner U, Tylee TS, Furst DE, Nelson JL. HLA-DQA1*0501 is associated with diffuse systemic sclerosis in Caucasian men. Arthritis Rheumatol (2000) 43(9):2005–10. doi: 10.1002/1529-0131(200009)43:9<2005::AID-ANR11>3.0.CO;2-#
21. Gourh P, Safran SA, Alexander T, Boyden SE, Morgan ND, Shah AA, et al. HLA and autoantibodies define scleroderma subtypes and risk in African and European americans and suggest a role for molecular mimicry. Proc Natl Acad Sci USA (2020) 117(1):552–62. doi: 10.1073/pnas.1906593116
22. Louthrenoo W, Kasitanon N, Wichainun R, Wangkaew S, Sukitawut W, Ohnogi Y, et al. Association of HLA-DRB1*15:02 and DRB5*01:02 allele with the susceptibility to systemic sclerosis in Thai patients. Rheumatol Int (2013) 33(8):2069–77. doi: 10.1007/s00296-013-2686-3
23. Nguyen B, Mayes MD, Arnett FC, del Junco D, Reveille JD, Gonzalez EB, et al. HLA-DRB1*0407 and *1304 are risk factors for scleroderma renal crisis. Arthritis Rheumatol (2011) 63(2):530–4. doi: 10.1002/art.30111
24. Kurteva EK, Boyadzhieva VV, Stoilov NR. Systemic sclerosis in mother and daughter with susceptible HLA haplotype and anti-topoisomerase I autoantibodies. Rheumatol Int (2020) 40(6):1001–9. doi: 10.1007/s00296-020-04516-5
Keywords: SSc, HLA, lcSSc, dcSSc, phenotype
Citation: Machhua S, Sharma SK, Kumar Y, Singh S, Aggarwal R, Anand S, Kumar M, Singh H and Minz RW (2023) Human leukocyte antigen association in systemic sclerosis patients: our experience at a tertiary care center in North India. Front. Immunol. 14:1179514. doi: 10.3389/fimmu.2023.1179514
Received: 04 March 2023; Accepted: 26 June 2023;
Published: 13 September 2023.
Edited by:
Veena Taneja, Mayo Clinic, United StatesReviewed by:
María Francisca González-Escribano, Virgen del Rocío University Hospital, SpainEdoardo Rosato, Sapienza University of Rome, Italy
Copyright © 2023 Machhua, Sharma, Kumar, Singh, Aggarwal, Anand, Kumar, Singh and Minz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ranjana Walker Minz, cndtaW56Lm1pbno4OEBnbWFpbC5jb20=
 Sanghamitra Machhua
Sanghamitra Machhua Shefali Khanna Sharma2
Shefali Khanna Sharma2 Surjit Singh
Surjit Singh Ranjana Walker Minz
Ranjana Walker Minz