
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Immunol. , 26 May 2023
Sec. Molecular Innate Immunity
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1174695
This article is part of the Research Topic Metal Ions in Innate Immune Antimicrobial Responses: How the Bugs are Fighting Back View all 5 articles
 Brian R. Varghese1‡
Brian R. Varghese1‡ Kelvin G. K. Goh1‡
Kelvin G. K. Goh1‡ Devika Desai1
Devika Desai1 Dhruba Acharya1
Dhruba Acharya1 Collin Chee1
Collin Chee1 Matthew J. Sullivan1*†
Matthew J. Sullivan1*† Glen C. Ulett1,2*
Glen C. Ulett1,2*Streptococcus agalactiae, also known as group B Streptococcus, is an important human and animal pathogen. Zinc (Zn) is an essential trace element for normal bacterial physiology but intoxicates bacteria at high concentrations. Molecular systems for Zn detoxification exist in S. agalactiae, however the degree to which Zn detoxification may vary among different S. agalactiae isolates is not clear. We measured resistance to Zn intoxication in a diverse collection of clinical isolates of S. agalactiae by comparing the growth of the bacteria in defined conditions of Zn stress. We found significant differences in the ability of different S. agalactiae isolates to resist Zn intoxication; some strains such as S. agalactiae 18RS21 were able to survive and grow at 3.8-fold higher levels of Zn stress compared to other reference strains such as BM110 (6.4mM vs 1.68mM Zn as inhibitory, respectively). We performed in silico analysis of the available genomes of the S. agalactiae isolates used in this study to examine the sequence of czcD, which encodes an efflux protein for Zn that supports resistance in S. agalactiae. Interestingly, this revealed the presence of a mobile insertion sequence (IS) element, termed IS1381, in the 5′ region of czcD in S. agalactiae strain 834, which was hyper-resistant to Zn intoxication. Interrogating a wider collection of S. agalactiae genomes revealed identical placement of IS1381 in czcD in other isolates from the clonal-complex-19 (CC19) 19 lineage. Collectively, these results show a resistance spectrum among S. agalactiae isolates enables survival in varying degrees of Zn stress, and this phenotypic variability has implications for understanding bacterial survival in metal stress.
Streptococcus agalactiae colonizes the urogenital tract in a quarter to a third of all healthy women of child-bearing age (1). Of the approximately 30% of healthy women who are asymptomatic carriers of the bacteria, half of newborns of these mothers will be exposed to S. agalactiae (1, 2). Transmission of S. agalactiae from the mother to the neonate takes places vertically, during pregnancy or during labor (3, 4). S. agalactiae is a major cause of early- and late-onset disease in neonates (5), which can lead to meningitis (6, 7). S. agalactiae also causes various infections in adults, including skin and soft tissue infections, urinary tract infections (UTIs), pneumonia and bacteremia. The overall mortality rate of S. agalactiae infection in adults is estimated at 15% or more in the United States (3, 8). Thus, S. agalactiae is a usual asymptomatic colonizer of the female urogenital tract and causes a spectrum of disease in both newborns and adults.
There is currently no licensed vaccine for prevention of S. agalactiae infection, despite extensive research and development efforts over the past few decades (9, 10). Increased antibiotic usage during the past decade for infection-prevention strategies aimed at S. agalactiae (11) is a clinical practice associated with increasing antibiotic resistance as well as hyper-virulent strains of S. agalactiae, which is recognized as an emerging global threat (12–14). Thus, defining the mechanisms by which this pathogen causes disease in humans, including most notably, how S. agalactiae avoids the natural antimicrobial activities of the body’s defenses is of great importance. Such knowledge will ultimately deliver new opportunities to treat and prevent disease caused by this pathogen, such as by targeting newly defined virulence mechanisms or amplifying specific host defense pathways that are essential in the control of infection.
In bacteria, zinc (Zn) is an essential cofactor for metalloenzymes (15, 16) but is toxic at high concentrations, as can be encountered inside phagocytes (17, 18). In the host, bacterial pathogens internalize essential Zn (19, 20), but the host can restrict Zn availability as an antimicrobial strategy (21) and phagocytes can mobilise cellular Zn to expose internalized bacteria to higher metal concentrations that are antimicrobial (22–24). The innate immune response to S. agalactiae, for example, comprises a robust Zn mobilization response in macrophages (25). Host-driven Zn intoxication of bacteria may involve ablation of uptake of essential Mn (26), a compromised bacterial response to oxidative stress (27), or disrupted central carbon metabolism (28). Evidence of a role for metal intoxication of bacteria based on Zn as an antimicrobial of innate immunity has emerged only relatively recently, as reviewed elsewhere (17, 24).
To counteract actions of the innate immune response some bacteria can evade metal intoxication using mechanisms of detoxification that involve metal efflux (29). In S. agalactiae, a genetic system that manages Zn homeostasis by regulating metal import and export via a Zn-sensing transcriptional response regulator, encoded by sczA, and a Zn efflux transporter, encoded by czcD, has recently been described (25). The czcD-sczA Zn-management axis supports virulence of S. agalactiae by facilitating bacterial survival in the host during systemic infection (25). Zinc transport machinery in S. agalactiae appears to be expendable when the Zn-limiting pressure of calprotectin, a metal chelator abundant in neutrophils, is absent (30). In addition, regulatory cross-talk in S. agalactiae at the level of the copper-sensing transcriptional regulator CopY can support resistance to Zn intoxication (31). Knowledge of molecular management of Zn homeostasis in S. agalactiae is largely based on work with only a few reference strains, namely 874391 (25, 31), A909 and CJB111 (30). The degree to which different S. agalactiae clinical isolates may differ in sensitivity to Zn intoxication is unclear.
In this study, we assessed sixteen strains of S. agalactiae from diverse capsular serotypes, sequence types and isolation sources to compare relative resistance to Zn intoxication. S. agalactiae strains exhibited significant differences in Zn-resistance phenotypes and the implications for host-pathogen interactions are discussed.
The bacterial strains used in this study are outlined in Table S1. S. agalactiae were routinely grown at 37°C, with agitation at 200rpm in Todd-Hewitt Broth (THB) or on TH agar (1.5% w/v). To enumerate bacteria grown in liquid cultures we performed routine retrospective colony counts by plating dilutions of bacteria on tryptone soya agar containing 5% defibrinated horse blood (Thermo Fisher Scientific). Media were supplemented with antibiotics (spectinomycin (Sp) 100μg/mL; chloramphenicol (Cm) 10 μg/mL), as indicated.
To compare the growth of different bacterial strains we performed growth assays with 200μL culture volumes in 96-well plates (Greiner) that were sealed using Breathe-Easy membranes (Sigma-Aldrich); growth was measured as attenuance (D, at 600nm) using a ClarioSTAR multimode plate reader (BMG Labtech) in Well Scan mode using a 3mm 5x5 scan matrix with 5 flashes per scan point and path length correction of 5.88mm, with agitation at 300rpm and recordings taken every 30min. Media for growth assays was THB supplemented with Zn (supplied as ZnSO4) as indicated. For attenuance baseline correction, D600nm values were corrected against a cell-free blank of THB or THB with equivalent supplemental zinc. We also measured CFU/mL at 18h by performing retrospective colony counts to estimate the number of viable cells in each condition at the end of the assay period.
Plasmid DNA was isolated using miniprep kits (QIAGEN), with modifications for S. agalactiae as described elsewhere (32). Plasmids and primers are listed in Tables S2, S3, respectively. An isogenic czcD-deficient mutant of wild-type (WT) S. agalactiae 834, termed 834ΔczcD, harboring a deletion in the gene encoding the Zn efflux transporter CzcD, was constructed by allelic exchange using pHY304aad9 to replace czcD with a chloramphenicol (Cm) resistance marker as described previously (33, 34), using plasmid pGU2641 (25, 33, 34). The mutant was validated by PCR using primers external to the mutation site and DNA sequencing. Complementation with full-length czcD in trans was achieved using the E. coli-streptococcal shuttle vector pDL278-derivative, pGU2699 (25, 35, 36). qRT-PCR was used to analyse the capacity of Zn stress (1mM) to induce expression of czcD using mid-log-phase S. agalactiae cultures grown for 2.5 h prior to quantification of czcD, exactly as previously described (25).
The czcD gene from strain 874391 was used in a BLAST search to identify czcD homologues in the genomes of reference S. agalactiae strains (National Center for Biotechnology Information [NCBI] database) or clinical S. agalactiae strains (in-house database). Nucleotide sequences and protein sequences were then aligned using ClustalW within the MEGA X tool (37) and percentage identity matrices were generated. The czcD locus from 834 was used to probe a database of 143 S. agalactiae strains available on the NCBI database to identify other strains that possess the same IS1381 insertion within czcD. Molecular serotyping and sequence typing were performed using the PubMLST database (38). Sequence comparisons were generated using the Artemis Comparison Tool (39).
Statistical analyses were performed using GraphPad Prism 9 (GraphPad Prism Software Inc., La Jolla, California). Data shown represent at least three, independent biological repeats. Statistical significance was accepted at p values of ≤ 0.05.
We analysed the growth of 16 S. agalactiae strains using attenuance D600nm readings to estimate bacterial population densities over time in cultures grown for 18h in media conditions of defined Zn stress. Representative strains that were i) susceptible or ii) more resistant to Zn intoxication, namely A909 and 18RS21, respectively, are shown in Figure 1 and the remaining 14 strains are shown in Figure S1. The relative Zn resistance of different strains based on significant inhibitory concentration of Zn for each strain, according to relative recovery of viable cells (CFU/mL) at the end of the assay period are listed in Supplemental Table S1. These data show a marked variation in relative resistance to Zn intoxication among the S. agalactiae strains analyzed here. For example, S. agalactiae A909 was among the most Zn-susceptible strains; its growth was completely inhibited at ≥3.28mM Zn versus the control condition of THB without supplemental Zn [basal THB media contains 10.9 ± 0.07μM Zn (25)], according to D600nm readings (Figure 1A) and CFU/mL estimates that showed significantly less recovery of viable A909 at ≥1.68mM Zn after 18 h of culture versus control cultures without supplemental Zn (~5x108 vs ~5x107, p<0.01; Figure 1B). Increasing the concentration of Zn >3.28mM significantly enhanced the bactericidal effect. Other strains identified to exhibit susceptibility/low relative resistance to Zn intoxication included UPSA 714, UPSA 058, ABSA 729 and BM110. The growth of these strains was significantly inhibited at between 1.68-3.28mM Zn (p ≤ 0.05, one-way ANOVA; Supplementary Figure S1), noting variability between biological quadruplicates, with significantly fewer CFU/mL recovered after 18h of culture in 1.68-3.28mM Zn (p ≤ 0.01, one-way ANOVA).
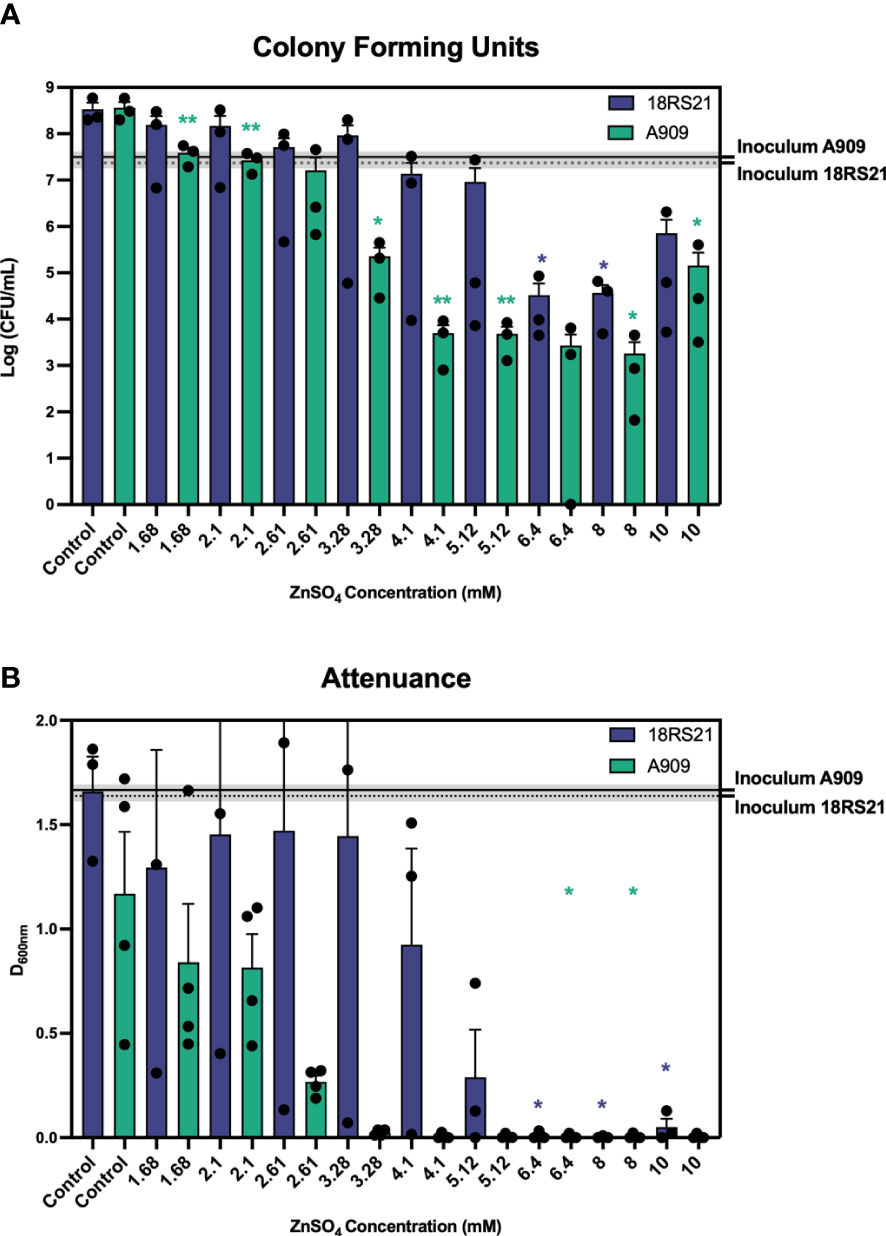
Figure 1 Survival of S. agalactiae in conditions of Zn stress showing differential resistance to Zn intoxication. S. agalactiae A909 and 18RS21 were grown for 18 hours prior to measuring CFU per mL (A) and attenuance (D600nm; B). Assays were performed in microtitre plates with starting inoculum (mean CFU/mL) represented as the black line with dashed lines and grey shading representative of the S.E.M of at least three independent experiments. Comparisons of means against the Control were performed using one-way repeated measures ANOVA; *p ≤ 0.05, **p ≤ 0.01.
A high degree of relative resistance to Zn intoxication was observed in multiple strains including 18RS21 (Figure 1), 807, 515 and NCTC 8181 (Supplementary Table S1). S. agalactiae 18RS21 exhibited the highest level of resistance to Zn with at least 6.4 mM required to significantly inhibit growth and reduce recovery of viable cells after 18h of culture compared to control cultures without supplemental Zn (Figure 1; p ≤ 0.05, one-way ANOVA; Supplementary Table S1). S. agalactiae CJB111 was significantly inhibited for growth in 4.1mM Zn and showed a 4-log10 reduction in CFU/mL (p ≤ 0.05, one-way ANOVA) (Supplementary Figure S1); similarly, S. agalactiae 834 showed a 99.99% reduction in CFU/mL at 5.12mM Zn (p ≤ 0.05, one-way ANOVA). Taken together, these results show a clear separation in levels of relative resistance to Zn intoxication among different S. agalactiae strains; where recovery of viable cells was significantly inhibited by ≤3.28mM Zn we classified a strain as exhibiting low relative Zn resistance, similarly for >3.28mM and ≤5.12mM a strain was classified as medium resistance, and ≥6.4mM as high relative resistance; (e.g., low relative resistance for A909, UPSA 714 compared to relatively resistant strains 18RS21, UPSA 807).
To further characterize relative resistance to Zn stress among the S. agalactiae strains, we generated growth curves for a sub-selection of five strains with Zn resistance phenotypes of high and low relative resistance classified using the cut-offs described above. Comparing these strains at a midpoint concentration of Zn, 4.1mM, further highlights the distinct phenotypes of Zn resistance between these strains; strain 834 exhibits significantly more growth in this level of Zn stress compared to other strains such as 2603V/R, 874391 and NCTC8181, for example (Figure 2). More detailed analysis of the growth curves of individual strains across a titration of Zn concentrations demonstrated other phenoptyic distinctions; for example, analysing strain A909 showed growth without supplemental Zn with a lag-phase of 1h, followed by log-phase from 1.5h to 4h prior to slower growth up to 12h (Supplementary Figure S2). In contrast, cultures of A909 that contained any supplemental Zn, even the lowest concentration tested (1.68mM) were significantly inhibited for growth. The lowest amount of supplemental Zn caused a delay of the bacteria entering log-phase until 3h, that lasted until 6h prior to stationary phase until 12h. Higher amounts of Zn completely ablated the growth of A909 (e.g., ≥4.1mM; Supplementary Figure S2). Analyzing S. agalactiae 834 demonstrated a log-phase from 3h to 5h followed by a decline phase up to 12h. Supplemental Zn caused incremental inhibition of growth of 834; even at the highest Zn concentration (6.4mM) growth was detectable even with an extended lag-phase of up to 6h (Supplementary Figure S2).
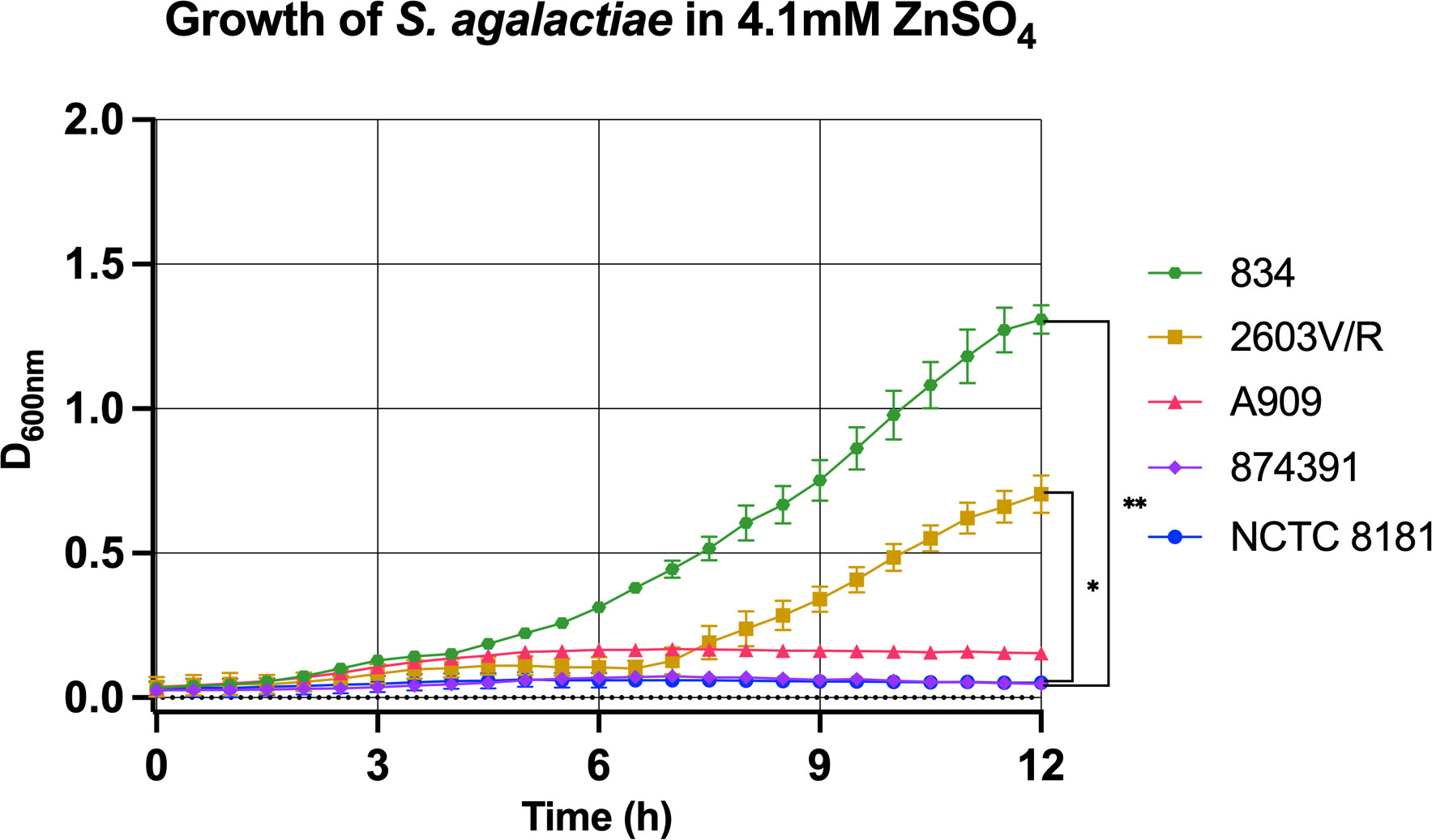
Figure 2 Distinct growth phenotypes of S. agalactiae in conditions of Zn stress. S. agalactiae strains, representative of distinct growth phenotypes, were grown for 12 hours in the presence of 4.1mM ZnSO4 with attenuance (D600nm) measured every 30 min and used to derive growth curves. Each data point represents an independent experiment and is shown with S.E.M. of at least n=3 experiments for each strain. Comparisons were performed using one-way repeated measures ANOVA to determine statistical significance; *p ≤ 0.05, **p ≤ 0.01.
Our results show that the Zn resistance phenotype varies significantly between different S. agalactiae strains. To investigate this further, we performed an in silico analysis of the czcD gene of the strains in this study, which have complete genomes available on the NCBI database. We found an overall high degree of sequence similarity to the genome-reference 874391 strain (Supplemental Table S2). Excepting single nucleotide polymorphisms (SNPs) detected in A909, 729 (position 93 A>T), 2603V/R and 1014 (position 228 T>C) and equating to 99.88% sequence identity (Supplementary Table S2), most other strains exhibited 100% sequence conservation in the czcD region. Conservation of amino acid sequence for these strains was 100% comparing to 874391 indicating redundancy in the SNPs among the strains. Strain 834 was an exception to the high degree of overall sequence similarity – this strain possessed only 49.20% identity to 874391 over the full length of the czcD open reading frame (Figure 3 and Supplementary Figure S3). This major difference could be accounted for by the presence of an 861 bp insertion in the 5′ region of czcD in the S. agalactiae 834 genome, 34bp after the ATG start site. The 861 bp insert was identical to a known mobile genetic element termed IS1381, first identified in other Streptococcus spp. (40). The makeup of IS1381 in this region comprised two open reading frames (ORFs) termed orfA and orfB (Figure 3), each encoding putative transposase genes. Further in silico analysis of a collection of one-hundred and forty-three complete S. agalactiae genomes available on NCBI database revealed six other strains that also contained an identical IS1381 insertion in the 5′ region of czcD. Interestingly, these six strains are all serotype III and of the clonal-complex-19 (CC-19) lineage (Supplementary Table S1). Despite the large insertion of IS1381 in the czcD gene, S. agalactiae 834 exhibited a higher degree of resistance to Zn stress compared to multiple other strains, including A909, 874391, UPSA 714, UPSA 058, ABSA 729 and BM110 (Figure 2; Supplementary Figure S1, Table S1). Interestingly, an analysis of the draft genome of 18RS21 revealed that it also possessed the same IS1381 insertion within czcD as 834 (Supplementary Figure S3). 18RS21 exhibited the highest degree of resistance to Zn stress among all the strains tested in this study. Notably, however, the genomes of other strains with high Zn resistance phenotypes, including 515, UPSA 807 and NCTC 8181 were examined and were shown to not carry IS1381 in czcD.
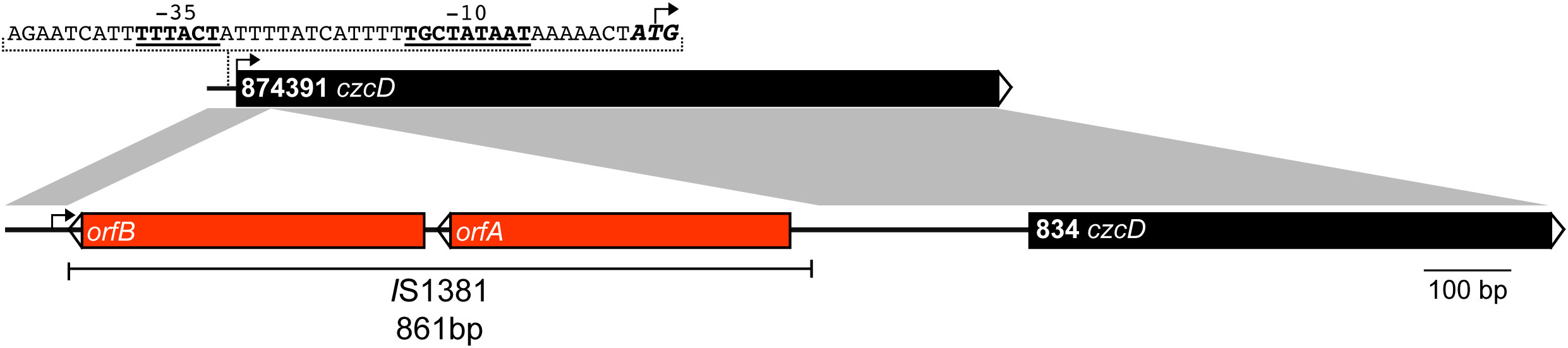
Figure 3 Physical map and linear comparison of czcD from S. agalactiae 874391 and 834. The black bars represent czcD, and the two red bars represent the orfA and orfB transposase genes within the IS1381 insertional element. The putative -35 and -10 transcription start sites (underlined and bold) are indicated, alongside the ATG start site of the gene (bold and italicised). The black arrow on each bar represents the ATG start site of czcD. The grey shaded regions represent a nucleotide identity of >99%.
To assess whether the IS1381 insertion element located in czcD of 834 might alter gene function in support of Zn resistance, we generated a czcD null mutant (i.e., lacking both IS1381 and czcD) and compared the growth of this mutant to WT 834. To do this, we used our previously designed pGU2461 plasmid that was used to delete full length czcD in S. agalactiae 874391. We also generated a complemented derivative of the null mutant, 834ΔczcD::pCzcD into which we cloned full length czcD from WT 874391 in trans. Interestingly, analysis of the growth curves demonstrated that WT 834, 834ΔczcD and 834ΔczcD::pCzcD were each able to grow well across the range of Zn concentrations tested, including 6.4mM Zn, without significant differences in the growth of the strains in any level of Zn tested (0mM-6.4mM), notably regardless of the presence of czcD. Each strain grew to high culture densities even in 6.4mM Zn conditions (Figure 4). In comparison, WT 874391 was able to grow to high culture densities only in lower concentrations of Zn (1.68mM-2.6mM), as consistent with prior assays (Supplementary Figure S2). There were also no significant differences in the growth curves of the strains in control conditions of no Zn. qRT-PCR analysis of WT 834, 834ΔczcD::pCzcD, 874391 and 807 to compare czcD expression in response to Zn stress showed significant differences in the response of these strains. The lowest expression ratio was detected in strain 834 and the highest response (five times higher than that of 834) was observed in 874391 (Supplementary Figure S4).
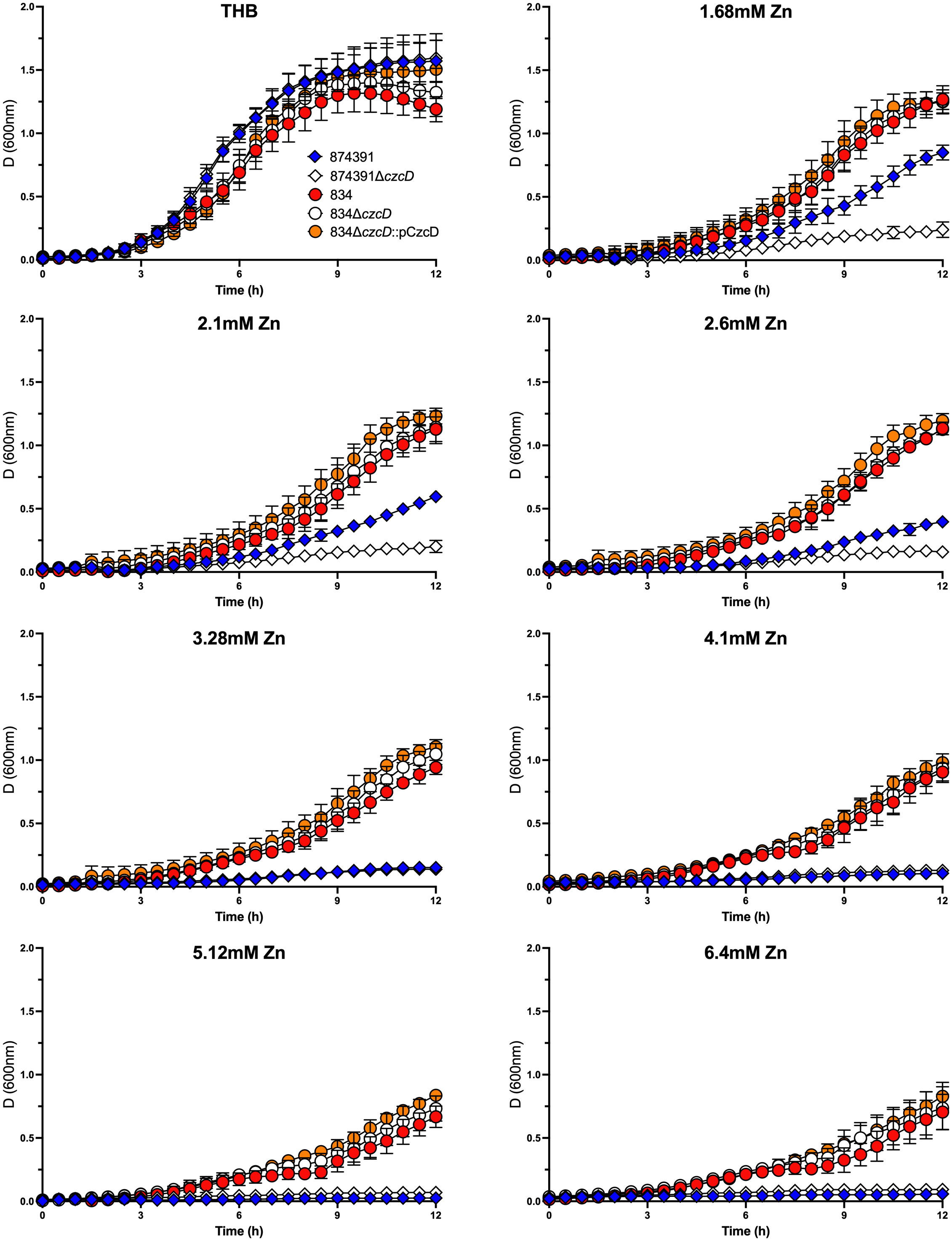
Figure 4 Growth kinetics of S. agalactiae 874391, 874391ΔczcD, 834, 834ΔczcD, and 834ΔczcD::PCzcD in the presence of increasing concentrations of Zn. The measurements of growth were derived from D600nm readings over 12 hours, shown as mean ± SD. Data were obtained from three independent experiments.
Two recent studies established that Zn exhibits antimicrobial activity against S. agalactiae (25, 41). In a genomic context, the S. agalactiae pan-genome, that is, the core genome that is shared by all isolates, constitutes approximately 80% of one genome, and the remainder comprises strain-specific or partially shared genes (42). Therefore, we tested a diversity of S. agalactiae strains, across different capsular serotypes, STs and clinical backgrounds to gain insight into the potential variability of phenotypes of susceptibility to Zn intoxication among a variety of S. agalactiae isolates. The central finding of this study is the sixteen S. agalactiae strains tested, that include several well-characterised, genome-sequenced reference strains exhibit a range of distinct phenotypes of Zn resistance with strains being classified as high or low-level Zn resistant. Such differential resistance to Zn intoxication among different S. agalactiae strains is phenotypically apparent in growth assays where strains can grow to varying degrees, including to high culture densities in defined levels of Zn stress. Additionally, these phenotypic distinctions are evident in time-kill assays in which some strains are killed by Zn stress conditions in which other more Zn resistant strains can survive and grow. This study also identifies a novel IS1381 insertion element within the Zn efflux transporter gene czcD that in S. agalactiae strain 834, along with czcD itself, appears to be redundant for the relative high resistance to Zn intoxication in this strain. Our finding of no significant difference in the growth curve of the 834ΔczcD null mutant compared to either the WT 834 or the 834ΔczcD::pCzcD complemented strain in Zn stress is surprising, given the contribution of CzcD in support of resistance against Zn stress in GBS strain 874391 (25). These data suggest that S. agalactiae 834 utilizes yet-to-be-characterized mechanisms, unrelated to czcD, to support resistance against Zn stress.
Our analysis of czcD transcriptional responses to 1 mM Zn amongst different strains was striking and negatively correlated with the relative Zn resistance phenotypes observed in comparing growth of strains 874391 and 834. For example, in 834, which has a high level of relative Zn resistance, czcD trancription was low following exposure to 1 mM Zn. Conversely, in 874391 that was determined to have low relative Zn resistance, czcD transcription was high, in identical growth conditions. This may be explained by differences in intracellular Zn content in these strains that were exposed to identical extracellular Zn levels. In future work it would be of interest to directly quantify intracellular Zn across a panel of S. agalactiae isolates with different relative Zn phenotypes. These data also support the notion that there are mechanisms separate to those driven by czcD that support resistance to Zn stress. A recent report that covR promotes resistance to Zn intoxication, and stp1/stk1 are essential for Zn resistance in GBS 874391 (31) highlights several genes beyond czcD that effect Zn detoxification. Additionally, cadD that mediates cadmium resistance in S. agalactiae also supports Zn resistance and promotes intracellular bacterial survival in macrophages and ascending infection during pregnancy (43). While we analyzed variation in czcD sequence for strains showing distinct phenotypes of Zn resistance in the current study, sequence analysis of other genes such as covR/S, stp1/stk1 and cadD would be interesting to explore potential correlations with variable Zn resistance. Taken together, this suggests that variable resistance to Zn intoxication among different S. agalactiae strains can involve mechanisms mediated by elements separate to CzcD to support resistance to Zn stress.
In the innate immune response to infection phagocytes can mobilise cellular Zn to expose bacteria that are located inside the phagocyte to metal concentrations that are antimicrobial (22–24). Recently, we showed that Zn mobilization occurs in the macrophage response to S. agalactiae (25). Some bacteria evade metal intoxication by mechanisms that involve metal efflux (29); and for S. agalactiae, Zn efflux pairs the SczA Zn-sensing transcriptional response regulator with the Zn efflux transporter CzcD (25). Zn stress in S. agalactiae also dysregulates transcription of numerous core metabolic pathways, including several for synthesis of purines, pyrimidines, riboflavin, and deoxynucleoside triphosphates. In this context, mediators additional to the SczA-CzcD Zn management axis regulate cellular management of intracellular Zn levels in S. agalactiae, including arcA, for example, which supports resistance to Zn intoxication (25). Other changes in S. agalactiae that occur because of Zn stress include broad dysregulation of cellular metal management aside from Zn itself; encompassing transcriptional shifts in the activities of putative manganese (Mn), iron (Fe), and nickel (Ni) transport loci, and major changes in the cellular pools of Mn and Fe (25). Thus, the roles of these metals in S. agalactiae strains that exhibit distinct Zn resistance phenotypes would be of interest. In this work, we focused on degrees of variation in resistance to Zn intoxication among different S. agalactiae strains and, subsequently, the presence of a novel IS1381 insertion element within czcD and how it might alter function. It will be of interest in the future to investigate the role of genes that influence Zn resistance other than czcD, including arcA, and genes for Mn, Fe, Ni (25) and Cd (43) transport in S. agalactiae strains such as 834 (Zn resistant) and A909 (Zn sensitive) under conditions of Zn stress.
A 2022 study by Francis et al. reported antimicrobial properties of Zn against a range of S. agalactiae strains, and suggested that isolation source, capsular serotype, and ST might contribute to susceptibility or resistance to Zn stress (41). Comparing the findings of that study with the current work shows consistency in relation to phenotypes of strains such as S. agalactiae A909 and COH1, for example, that are Zn susceptible and relatively resistant, respectively. Most of the strains tested however are unique to either of the two studies, precluding direct comparisons. Interestingly, the strains more susceptible to Zn intoxication in the current study, including BM110 and 874391 were of capsular type III; Francis et al. found that capsular type III strains of ST-19 exhibited the highest resistance to Zn intoxication (41). In our study, we cannot ascribe any clear role for capsular type in the phenotype of strains for Zn resistance, noting the number of isolates analysed in the current study was half the number of the study by Francis et al., which is a limitation of the present work. Analysis of more strains will be useful to further our understanding of the range of Zn resistance phenotypes in S. agalactiae.
A novel finding of this work is the identification of IS1381 in the 5′ region of czcD in S. agalactiae strain 834 that was relatively resistant to Zn intoxication. Insertion sequences can modify phenotypes by upregulating or inactivating proximal genes (44, 45). In S. agalactiae, some nonhemolytic strains were found to have an IS1381-variant integrated into an essential component of the toxin (46). Hence, we examined how the IS1381 element might affect the high Zn resistance observed in 834; to investigate the hypothesis that IS1381 was acting to increase the resistance of S. agalactiae 834 to high levels of Zn, we constructed a null mutant in 834ΔczcD and complement strain using full length czcD from WT 874391. Comparing the growth of these strains under conditions of Zn stress showed intriguingly that deletion of czcD and overexpression of prototypical czcD in trans did not significantly affect the growth of 834 in different concentrations of Zn. This result is surprising because the czcD gene encodes for a Zn efflux protein and is required by S. agalactiae for resistance against Zn stress (25); therefore, the null mutant was expected to be rendered more susceptible to Zn stress. However, a recent study used a transposon-insertion screen that identified a suite of genes that S. agalactiae uses to overcome Zn intoxication, that are separate to sczA-czcD Zn management (31). Therefore, it would be interesting to examine the function of these genes in 834 during growth in Zn stress to examine other factors which may contribute to this strain’s relative resistance to Zn. It is also noteworthy that IS1381 integrates at multiple sites across a bacterial genome, at seven other loci in the instance of S. agalactiae strain 834 including downstream of guaA involved in purine biosynthesis (47). It would be of interest to examine these additional sites for their contribution to surviving Zn intoxication.
We approached the measurement of resistance to Zn stress in S. agalactiae by using two measurements of attenuance and recovery of viable cells in cultures grown in THB, a widely utilized medium for β-haemolytic streptococci but one that does not closely represent the natural niches that S. agalactiae thrives in within humans and animals. Use of THB in this study provided new insight into the diversity of Zn resistance phenotypes among a defined collection of strains, and having established variations in Zn resistance among the strains in this study it would be of interest to extend the findings to chemically defined minimal medium, such as CDM, which more closely reflects a nutrient-limited host environment. It is of note that in growth assays of S. agalactiae in CDM, Zn exhibited elevated toxicity, since sub-millimolar levels (0.25mM) were sufficient to significantly inhibit wild-type S. agalactiae 874391 (25). It is also noteworthy, then, that the assays of Francis et al. used Brain Heart Infusion broth to compare phenotypes amongst S. agalactiae strains. Precisely how this medium differs from THB is yet to be determined, but collectively these observations imply that the specific growth conditions under which Zn intoxication assays are performed have a marked impact on the experimental findings.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Conceptualization: MS, GU and KG. Data curation: MS, KG, GU and BV. Formal analysis: MS, KG, GU and BV. Funding acquisition: MS, KG and GU. Investigation: MS, KG, GU, BV, DD, DA and CC. Methodology: MS, KG, GU, BV and DD. Project administration: GU and MS. Resources: GU. Supervision: GU, MS and KG. Validation: MS and KG. Writing – original draft: GU and BV. Writing – review & editing: MS, KG and GU. All authors contributed to the article and approved the submitted version.
This study was supported with an Ideas Grant from the National Health and Medical Research Council (2021475) to MS, KG and GU.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1174695/full#supplementary-material
1. Cho CY, Tang YH, Chen YH, Wang SY, Yang YH, Wang TH, et al. Group b streptococcal infection in neonates and colonization in pregnant women: an epidemiological retrospective analysis. J Microbiology Immunol Infection (2019) 52:265–72. doi: 10.1016/j.jmii.2017.08.004
2. Money D, Allen VM. The prevention of early-onset neonatal group b streptococcal disease. J Obstet Gynaecol Can (2016) 38:S326–35. doi: 10.1016/j.jogc.2016.09.042
3. Edwards MS, Baker CJ. 203 - streptococcus agalactiae (Group b streptococcus). In: Bennett JE, Dolin R, Blaser MJ, editors. Mandell, Douglas, and bennett's principles and practice of infectious diseases, Eighth Edition. Philadelphia: W.B. Saunders (2015). p. 2340–2348.e2.
4. Joubrel C, Tazi A, Six A, Dmytruk N, Touak G, Bidet P, et al. Group b streptococcus neonatal invasive infections, France 2007-2012. Clin Microbiol Infect (2015) 21:910–6. doi: 10.1016/j.cmi.2015.05.039
5. Rajagopal L. Understanding the regulation of group b streptococcal virulence factors. Future Microbiol (2009) 4:201–21. doi: 10.2217/17460913.4.2.201
6. Mullaney DM. Group b streptococcal infections in newborns. J Obstet Gynecol Neonatal Nurs (2001) 30:649–58. doi: 10.1111/j.1552-6909.2001.tb00012.x
7. Lin FY, Weisman LE, Troendle J, Adams K. Prematurity is the major risk factor for late-onset group b streptococcus disease. J Infect Dis (2003) 188:267–71. doi: 10.1086/376457
8. Farley MM. Group b streptococcal disease in nonpregnant adults. Clin Infect Dis (2001) 33:556–61. doi: 10.1086/322696
9. Johri AK, Paoletti LC, Glaser P, Dua M, Sharma PK, Grandi G, et al. Group b streptococcus: global incidence and vaccine development. Nat Rev Microbiol (2006) 4:932–42. doi: 10.1038/nrmicro1552
10. Nuccitelli A, Rinaudo CD, Maione D. Group b streptococcus vaccine: state of the art. Ther Adv Vaccines Immunotherapy (2015) 3:76–90. doi: 10.1177/2051013615579869
11. Centers for Disease Control. Revision of guidelines for the prevention of perinatal group b streptococcal disease. J Am Med Assoc (2002) 287:1106–7. doi: 10.1001/jama.287.9.1106-JWR0306-2-1
12. Da Cunha V, Davies MR, Douarre PE, Rosinski-Chupin I, Margarit I, Spinali S, et al. Streptococcus agalactiae clones infecting humans were selected and fixed through the extensive use of tetracycline. Nat Commun (2014) 5:4544. doi: 10.1038/ncomms5544
13. Teatero S, Ramoutar E, McGeer A, Li A, Melano RG, Wasserscheid J, et al. Clonal complex 17 group b streptococcus strains causing invasive disease in neonates and adults originate from the same genetic pool. Sci Rep (2016) 6:20047. doi: 10.1038/srep20047
14. Bisharat N, Crook DW, Leigh J, Harding RM, Ward PN, Coffey TJ, et al. Hyperinvasive neonatal group b streptococcus has arisen from a bovine ancestor. J Clin Microbiol (2004) 42:2161–7. doi: 10.1128/JCM.42.5.2161-2167.2004
15. Watly J, Potocki S, Rowinska-Zyrek M. Zinc homeostasis at the Bacteria/Host interface-from coordination chemistry to nutritional immunity. Chemistry (2016) 22:15992–6010. doi: 10.1002/chem.201602376
16. Osman D, Cavet JS. Copper homeostasis in bacteria. Adv Appl Microbiol (2008) 65:217–47. doi: 10.1016/S0065-2164(08)00608-4
17. Chandrangsu P, Rensing C, Helmann JD. Metal homeostasis and resistance in bacteria. Nat Rev Microbiol (2017) 15:338–50. doi: 10.1038/nrmicro.2017.15
18. German N, Doyscher D, Rensing C. Bacterial killing in macrophages and amoeba: do they all use a brass dagger? Future Microbiol (2013) 8:1257–64. doi: 10.2217/fmb.13.100
19. Bayle L, Chimalapati S, Schoehn G, Brown J, Vernet T, Durmort C. Zinc uptake by Streptococcus pneumoniae depends on both AdcA and AdcAII and is essential for normal bacterial morphology and virulence. Mol Microbiol (2011) 82:904–16. doi: 10.1111/j.1365-2958.2011.07862.x
20. Moulin P, Patron K, Cano C, Zorgani MA, Camiade E, Borezee-Durant E, et al. The Adc/Lmb system mediates zinc acquisition in Streptococcus agalactiae and contributes to bacterial growth and survival. J Bacteriol (2016) 198:3265–77. doi: 10.1128/JB.00614-16
21. Zygiel EM, Nolan EM. Transition metal sequestration by the host-defense protein calprotectin. Annu Rev Biochem (2018) 87:621–43. doi: 10.1146/annurev-biochem-062917-012312
22. Kapetanovic R, Bokil NJ, Achard ME, Ong CL, Peters KM, Stocks CJ, et al. Salmonella employs multiple mechanisms to subvert the TLR-inducible zinc-mediated antimicrobial response of human macrophages. FASEB J (2016) 30:1901–12. doi: 10.1096/fj.201500061
23. Achard MES, Stafford SL, Bokil NJ, Chartres J, Bernhardt PV, Schembri MA, et al. Copper redistribution in murine macrophages in response to Salmonella infection. Biochem J (2012) 444:51–7. doi: 10.1042/BJ20112180
24. Djoko KY, Ong CL, Walker MJ, McEwan AG. The role of copper and zinc toxicity in innate immune defense against bacterial pathogens. J Biol Chem (2015) 290:18954–61. doi: 10.1074/jbc.R115.647099
25. Sullivan MJ, Goh KGK, Ulett GC. Cellular management of zinc in group b streptococcus supports bacterial resistance against metal intoxication and promotes disseminated infection. mSphere (2021) 6(3):e00105–21. doi: 10.1128/mSphere.00105-21
26. McDevitt CA, Ogunniyi AD, Valkov E, Lawrence MC, Kobe B, McEwan AG, et al. A molecular mechanism for bacterial susceptibility to zinc. PloS Pathog (2011) 7:e1002357. doi: 10.1371/journal.ppat.1002357
27. Eijkelkamp BA, Morey JR, Ween MP, Ong CL, McEwan AG, Paton JC, et al. Extracellular zinc competitively inhibits manganese uptake and compromises oxidative stress management in streptococcus pneumoniae. PloS One (2014) 9:e89427. doi: 10.1371/journal.pone.0089427
28. Ong CL, Walker MJ, McEwan AG. Zinc disrupts central carbon metabolism and capsule biosynthesis in streptococcus pyogenes. Sci Rep (2015) 5:10799. doi: 10.1038/srep10799
29. Hassan KA, Pederick VG, Elbourne LD, Paulsen IT, Paton JC, McDevitt CA, et al. Zinc stress induces copper depletion in acinetobacter baumannii. BMC Microbiol (2017) 17:59. doi: 10.1186/s12866-017-0965-y
30. Burcham LR, Le Breton Y, Radin JN, Spencer BL, Deng L, Hiron A, et al. Identification of zinc-dependent mechanisms used by group b Streptococcus to overcome calprotectin-mediated stress. mBio (2020) 11(6):e02302–20. doi: 10.1128/mBio.02302-20
31. Sullivan MJ, Goh KGK, Ulett GC. Regulatory cross-talk supports resistance to zn intoxication in streptococcus. PloS Pathog (2022) 18:e1010607. doi: 10.1371/journal.ppat.1010607
32. Sullivan MJ, Ulett GC. Stable expression of modified green fluorescent protein in group b streptococci to enable visualization in experimental systems. Appl Environ Microbiol (2018) 84(18):e01262–18. doi: 10.1128/AEM.01262-18
33. Ipe DS, Ben Zakour NL, Sullivan MJ, Beatson SA, Ulett KB, Benjamin WHJ, et al. Discovery and characterization of human-urine utilization by asymptomatic-Bacteriuria-Causing streptococcus agalactiae. Infect Immun (2015) 84:307–19. doi: 10.1128/IAI.00938-15
34. Sullivan MJ, Leclercq SY, Ipe DS, Carey AJ, Smith JP, Voller N, et al. Effect of the Streptococcus agalactiae virulence regulator CovR on the pathogenesis of urinary tract infection. J Infect Dis (2017) 215:475–83. doi: 10.1093/infdis/jiw589
35. Dunny GM, Lee LN, LeBlanc DJ. Improved electroporation and cloning vector system for gram-positive bacteria. Appl Environ Microbiol (1991) 57:1194–201. doi: 10.1128/aem.57.4.1194-1201.1991
36. LeBlanc DJ, Lee LN, Abu-Al-Jaibat A. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid (1992) 28:130–45. doi: 10.1016/0147-619X(92)90044-B
37. Kumar S, Stecher G, Li M, Knyaz C, Tamura K, MEGA X. Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol (2018) 35:1547–9. doi: 10.1093/molbev/msy096
38. Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res (2018) 3:124. doi: 10.12688/wellcomeopenres.14826.1
39. Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. ACT: the Artemis comparison tool. Bioinformatics (2005) 21:3422–3. doi: 10.1093/bioinformatics/bti553
40. Sanchez-Beato AR, Garcia E, Lopez R, Garcia JL. Identification and characterization of IS1381, a new insertion sequence in streptococcus pneumoniae. J Bacteriol (1997) 179:2459–63. doi: 10.1128/jb.179.7.2459-2463.1997
41. Francis JD, Guevara MA, Lu J, Madhi SA, Kwatra G, Aronoff DM, et al. The antimicrobial activity of zinc against group b streptococcus is strain-dependent across diverse sequence types, capsular serotypes, and invasive versus colonizing isolates. BMC Microbiol (2022) 22:23. doi: 10.1186/s12866-021-02428-3
42. Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial "pan-genome". Proc Natl Acad Sci U. S. A (2005) 102:13950–5. doi: 10.1073/pnas.0506758102
43. Korir ML, Doster RS, Lu J, Guevara MA, Spicer SK, Moore RE, et al. Streptococcus agalactiae cadD alleviates metal stress and promotes intracellular survival in macrophages and ascending infection during pregnancy. Nat Commun (2022) 13:5392. doi: 10.1038/s41467-022-32916-7
44. Siguier P, Gourbeyre E, Chandler M. Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol Rev (2014) 38:865–91. doi: 10.1111/1574-6976.12067
45. Vandecraen J, Chandler M, Aertsen A, Van Houdt R. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit Rev Microbiol (2017) 43:709–30. doi: 10.1080/1040841X.2017.1303661
46. Spellerberg B, Pohl B, Haase G, Martin S, Weber-Heynemann J, Lutticken R. Identification of genetic determinants for the hemolytic activity of Streptococcus agalactiae by ISS1 transposition. J Bacteriol (1999) 181:3212–9. doi: 10.1128/JB.181.10.3212-3219.1999
Keywords: Streptococcus agalactiae, metal ions, metallobiology, zinc efflux, czcD
Citation: Varghese BR, Goh KGK, Desai D, Acharya D, Chee C, Sullivan MJ and Ulett GC (2023) Variable resistance to zinc intoxication among Streptococcus agalactiae reveals a novel IS1381 insertion element within the zinc efflux transporter gene czcD. Front. Immunol. 14:1174695. doi: 10.3389/fimmu.2023.1174695
Received: 27 February 2023; Accepted: 11 April 2023;
Published: 26 May 2023.
Edited by:
Jennifer Angeline Gaddy, Vanderbilt University Medical Center, United StatesReviewed by:
Mark Joseph Walker, The University of Queensland, AustraliaCopyright © 2023 Varghese, Goh, Desai, Acharya, Chee, Sullivan and Ulett. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Glen C. Ulett, Zy51bGV0dEBncmlmZml0aC5lZHUuYXU=; Matthew J. Sullivan, bWF0dGhldy5zdWxsaXZhbkB1ZWEuYWMudWs=
†Present address: Matthew J. Sullivan, School of Biological Sciences, University of East Anglia, Norwich, United Kingdom
‡These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.