
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 15 May 2023
Sec. Inflammation
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1168023
This article is part of the Research TopicInflammation in Respiratory and Neurological Diseases and the immune-interaction of the lung-brain axisView all 17 articles
Protein glycosylation is a widespread posttranslational modification that can impact the function of proteins. Dysregulated protein glycosylation has been linked to several diseases, including chronic respiratory diseases (CRDs). CRDs pose a significant public health threat globally, affecting the airways and other lung structures. Emerging researches suggest that glycosylation plays a significant role in regulating inflammation associated with CRDs. This review offers an overview of the abnormal glycoenzyme activity and corresponding glycosylation changes involved in various CRDs, including chronic obstructive pulmonary disease, asthma, cystic fibrosis, idiopathic pulmonary fibrosis, pulmonary arterial hypertension, non-cystic fibrosis bronchiectasis, and lung cancer. Additionally, this review summarizes recent advances in glycomics and glycoproteomics-based protein glycosylation analysis of CRDs. The potential of glycoenzymes and glycoproteins for clinical use in the diagnosis and treatment of CRDs is also discussed.
Protein glycosylation is a crucial posttranslational modification that affects more than 50% of known proteins. Glycosyltransferases (GTs) and glycoside hydrolases (GHs) are the two main types of glycoenzymes. The Carbohydrate-Active enZYmes Database (CAZy) (http://www.cazy.org) records 116 families of GTs (GT1-GT116) and 173 families of GHs (GH1-GH173), including 235 putative human GTs and 87 putative human GHs. GTs transfer sugar molecules from nucleotide sugar or lipid-linked sugar donors to hydroxyl groups of acceptors to form specific glycan structures and glycosidic linkages (1–3). For instance, sialyltransferases (STs), fucosyltransferases (FUTs), galactosyltransferases (GalTs) and MGAT3 (4, 5) are responsible for the formation of glycans that involve sialic acid (sialylation), fucose (fucosylation), galactose (galactosylation), and bisecting GlcNAc (6, 7), respectively. GHs constitute a superfamily of enzymes that hydrolyze glycosidic linkages during oligosaccharide maturation (3, 8), exhibiting broad and stringent substrate specificities (9). Common GHs include β-glucosidase, glucosidase II, N-acetyl-β-d-glucosaminidase, α-glucosidase, β-galactosidase, β-glucuronidase, α-mannosidase, β-mannosidase, α-fucosidase, and sialidase. O-linked β-N-acetylglucosamine glycosylation (O-GlcNAcylation), a distinctive form of protein glycosylation, involves the dynamic addition of N-acetylglucosamine from UDP-GlcNAc onto specific serine or threonine residues. O-GlcNAcylation cycling is mediated by O-GlcNAc transferase (OGT) and O-GlcNAcase (OGA) (10, 11).
Chronic respiratory diseases (CRDs), affecting the airways and other structures of the lungs, are a major threat to global public health. Common CRDs include chronic obstructive pulmonary disease (COPD), asthma, cystic fibrosis (CF), idiopathic pulmonary fibrosis (IPF), pulmonary arterial hypertension (PAH), non-CF bronchiectasis and lung cancer (12, 13). COPD obstructs the airways, making breathing difficult, while asthma is a chronic inflammatory condition caused by environmental and genetic factors, leading to airways narrowing and excess mucus production during attacks (14). CF is a multisystemic autosomal recessive disease caused by a mutation in the CF transmembrane conductance regulator (CFTR) gene, severely damaging the respiratory and digestive systems (15). Interstitial lung diseases (ILDs), such as IPF (16), are a heterogeneous family of lung disorders characterized by alveolar injury, inflammation, and fibrosis, while PAH is a rapidly progressive disease characterized by elevated pulmonary artery pressure. Non-CF bronchiectasis is a progressive lung disease resulting in permanently dilated airways. Lung cancers can develop from chronic irritation and inflammation (17). Persistent inflammation is a common feature of most CRDs, involving epithelial and immune cells and cytokines in the respiratory tract (18). Proinflammatory cytokines can regulate the glycosylation of cell surface-associated glycoproteins present in immune and epithelial cells (4). Therefore, inflammation-induced glycosylation changes may play a crucial role in the development of CRDs.
Glycoproteins play essential roles in a variety of physiological functions, such as protein folding, cell-cell interaction, cell adhesion, and ligand binding (19). Abnormal protein glycosylation has been observed in the pathological process of CRDs. In this review, we will focus on the role of glycosylation in CRDs by examining the involvement of glycoenzymes and aberrant glycosylation in the pathogenesis of different CRDs from three perspectives: (1) the altered expression of GTs and GHs, along with associated aberrant fucosylation, sialylation, and O-GlcNAcylation in CRDs; (2) advancements in glycomics and glycoproteomics that facilitate the exploration of CRDs; and (3) the potential clinical applications of glycoproteins and glycoenzymes as biomarkers and targets in CRDs.
Several experiments have shown that abnormal protein glycosylation is present in almost all CRDs, including COPD (20), asthma (21, 22), CF (23, 24), PAH (25), IPF (26), and lung cancer (27–29). For example, in COPD, an N-glycomics study found an abnormal N-glycosylation pattern in plasma proteins, with a decrease in low branching forms and increase in more complex forms (20). Aberrant fucosylation and sialylation of mucins were observed in asthmatic patients (30, 31). CF airway epithelial cells were characterized by increased fucosylation and decreased sialylation (23). In PAH, augmented expression and activity of OGT, the enzyme for O-GlcNAcylation, were observed, suggesting the role of abnormal O-GlcNAcylation in PAH development (32). Abnormal protein glycosylation is also observed in almost all cancers (33, 34), including lung cancer (27, 28). In IPF, the core fucosylation of TGF-β1, mediated by α-1,6-fucosyltransferase (FUT8), plays a crucial role in the transformation of pericytes into myofibroblasts (35). Changes in the protein glycosylation profile in body fluids, especially serum and sputum, may serve as biomarkers for early diagnosis of CRDs. For example, increased fucosylation levels of serum surfactant protein D (SP-D) were identified as a potential biomarker of COPD (36). Multiple integrated N-glycoproteomics analyses have been performed to screen new effective biomarkers for the early diagnosis of lung cancer and for monitoring lung cancer progression (37–40). Fucosylated alpha-1-acid glycoprotein (AGP), ceruloplasmin (CP), and paraoxonase 1 (PON1) in the serum may be potential biomarkers for lung cancer (41).
Protein glycosylation changes are prevalent in CRDs, making them a promising candidate for early diagnosis and therapeutic targets. Changes in protein glycosylation are often caused by aberrant expression and mutation of glycoenzymes, which affect glycosylation activity. Therefore, the analysis of glycoproteins and their related glycoenzymes has become a widely used approach for screening potential biomarkers and disease-associated regulators in various diseases, including CRDs.
CRDs are associated with persistent inflammation in the respiratory tract, where glycoproteins are believed to play a critical role in regulating inflammation-related functions such as cell adhesion, immunogenicity, and cell-to-cell and cell-to-substrate interactions. Proinflammatory cytokines, such as interleukin-1 (IL-1), IL-6, and tumor necrosis factor alpha (TNF-α), are important regulators of inflammation and can influence the expression of GTs, particularly STs and FUTs (42–45). This, in turn, affects downstream protein glycosylation changes, which contribute to the development of CRDs and other chronic inflammation diseases (46).
Various GTs are involved in the biosynthesis of different glycosylation types, and changes in GT expression can alter protein glycosylation patterns. Growing evidence suggests that alteration of GT expression is present in many pathophysiological conditions (19, 47). Dysregulation of GT expression has been observed in multiple cancers (19, 48), such as pancreatic cancer (49), colorectal cancer (50), and breast cancer (51), and it is associated with tumorigenesis, metastasis, and chemoresistance (50). Moreover, a higher frequency of GT variants has been detected in cancers with higher global mutation burdens, as revealed by an integrative pan-cancer analysis (52). Additionally, several GHs are overexpressed in various types of cancer (53), and they play a crucial role in prodrug therapy by activating prodrugs that target cancer and diabetes (53–55). Therefore, GTs and GHs are important factors in physiological and pathological conditions, and their dysregulated expression can lead to various diseases. Abnormal GTs and GHs could serve as potential biomarkers and therapeutic targets.
In CRDs, abnormal fucosylation, sialylation, and O-GlcNAcylation can be attributed to changes in the expression of glycoenzymes, which are responsible for the enzymatic reactions involved in protein glycosylation. The dysregulation of these glycoenzymes is a major factor for underling the aberrant glycosylation patterns observed in these diseases. Table 1 and Figure 1 provide a summary of the glycoenzyme-mediated glycosylation changes and associated dysfunction in CRDs. Specifically, we will review the impact of the variations in the expression of FUTs, STs, OGTs and GHs on protein glycosylation in these diseases.

Figure 1 Overview of the glycoenzyme-related protein glycosylation and the functional changes in CRDs. COPD, chronic obstructive pulmonary disease; CF, cystic fibrosis; IPF, idiopathic pulmonary fibrosis; PAH, pulmonary arterial hypertension; EMT, epithelial–mesenchymal transition; ST, sialyltransferase; FUT, fucosyltransferase; O-GlcNAcylation, O-linked β-N-acetylglucosamine glycosylation; OGT, O-GlcNAc transferase; ST6GAL, beta-galactoside alpha-2,6-sialyltransferase; ST3GAL, beta-galactoside alpha-2,3-sialyltransferase; NEU, neuraminidase; TGF, transforming growth factor; EGFR, epidermal growth factor receptor; JNK, c-Jun N-terminal protein kinase; IL, interleukin; STST3, signal transducer and activator of transcription 3; SAM68, SRC-associated in mitosis, 68 kDa; HCF-1, host cell factor-1; NO, nitric oxide; SP1, specificity protein 1; DC, dendritic cells; HA, hyaluronic acid; SAP, serum amyloid P; IGF1/PI3K/AKT, insulin-like growth factor 1/phosphotylinosital 3 kinase/protein kinase B; AEC, alveolar epithelial cell; SPARC, secreted protein acidic and rich in cysteine; EGF, epidermal growth factor.
Fucosylation is a process mediated by 11 N-linked and 2 O-linked fucosylation enzymes, collectively known as FUTs, which include FUT1-11 and protein O-fucosyltransferase 1 and 2 (POFUT1 and POFUT2). This process can be categorized as core fucosylation (α-1,6-fucosylation) or terminal fucosylation (α-1,2-fucosylation and α-1,3/4-fucosylation), depending on the location of fucose in N-glycan. FUT1 and FUT2 are responsible for α-1,2-linkage, while FUT3-7 and FUT9-11 catalyze α-1,3- and α-1,4-fucosylation, and FUT8 participates in core fucosylation. POFUT1 and POFUT2 transfer fucose from GDP-β-L-fucose to serine or threonine residues (41). It is increasingly evident that abnormal fucosylation occurs in cancer and inflammation (91, 92). Aberrant fucosylation of proteins can contribute to tumor proliferation, invasion, metastasis, and immune evasion (93, 94).
COPD, which encompasses emphysema and chronic bronchitis, is a progressive disease often linked to smoking and a heightened risk for lung cancer, particularly squamous cell carcinoma (95). Studies on COPD animal models and patients have shown that reduced levels of FUT8 and the corresponding reduction in core fucosylation may contribute to the pathogenesis of the disease. Fut8 knockout (Fut8-/-) mice exhibited emphysema-like changes in the lung, and Fut8 knockdown (Fut8+/-) mice exposed to cigarette smoke were found to be more susceptible to developing emphysema than wild-type mice (96). Knockout of FUT8 also resulted in alveolar destruction and loss of the core-fucosylated secreted protein acidic and rich in cysteine (SPARC), which impairs collagen binding (56). Decreased FUT8 activity has been correlated with poor lung function and exacerbation of COPD in patients (36, 96). FUT8 Thr267Lys mutation is also a risk factor for emphysema (97).
Studies have shown that treating airway smooth muscle cells in a lipopolysaccharide-induced COPD rat model with extracellular matrix components upregulates expression of cytokine factors like TGF-β1 and IL-6, but downregulates matrix metalloproteinase 9 (MMP-9) (98). In contrast, Fut8-/- mice overexpressed MMPs like MMP-12 and MMP-13 and showed a dysfunctional TGF-β1 due to lack of core fucosylation, leading to downregulation of the extracellular matrix (99). As TGF-β1 plays a significant role in lung remodeling (100), the abnormal expression of FUT8 and decline in core fucosylation may affect the cell-to-matrix communication via SPARC in COPD (56). Thus, FUT8 is a potential target for developing COPD therapies as it plays an important role in the COPD development and progression through its effects on the core fucosylation of various proteins, including TGF-β1 and SPARC.
Asthma is characterized by the hypersecretion of mucus that is primarily composed of glycoproteins. Studies in mice have shown that knockout of the FUT2 gene can reduce eosinophilic inflammation and airway hyperresponsiveness caused by house dust mites, a common trigger for asthma (22). Dysregulation of FUT2 and epithelial fucosylation have been associated with various chronic inflammatory diseases (92). The main component of mucus, MUC5AC, is typically fucosylated, and FUT2 exacerbates asthma through α-1,2-fucosylation of MUC5AC (59). FUT2 genotypes are also linked to asthma risk (101). FUT2 has three secretor genotypes: nonsecretors (homozygous for the loss-of-function of FUT2), heterozygous secretors, and homozygous secretors. Homozygous secretors of FUT2 may have more severe asthma exacerbations and poor lung function (72). Thus, FUT2 and α-1,2-fucosylation may play a crucial role in asthma. Glycosylated immunoglobulin (Ig) affects various allergic diseases, highlighting the importance of Ig glycosylation patterns in mediating allergies, including asthma. Moreover, the glycosylation of IgE, rather than IgG and IgA, has a dominant role in allergy (102).
CF is caused by defective CFTR, affecting chloride channels in mucus and sweat-producing cells. Phenotype analysis of the N-glycosylation of sputum proteins revealed that α-2,6-sialylation and α-1,6-core fucosylation are common structural features present in patients with CF (63). Abnormal glycosylation of mucins, especially MUC5B and MUC5AC, is reported in CF and other pulmonary conditions (61). However, aberrant O-glycosylation and N-glycosylation of mucins may result from bacterial infection and inflammation rather than CF pathogenesis, as both N-sialylation and N-fucosylation of mucins increase (63). In the small intestine of CF mouse models, increased levels of insoluble and soluble fucosylated mucins are observed (103), which may be due to upregulated FUT2 expression. The glycosylation patterns of CF airway epithelial cells alter with increases in α-1,3/4-fucosylation and decreases in α-1,2-fucosylation and sialylation (62). In CF, a different fucosylation phenotype was reported in mucus and airway epithelial cells, with increased α-1,2-fucosylation in mucus (103) and increased α-1,3/4-fucosylation in airway epithelial cells (23), indicating that different FUTs participate in the fucosylation of mucus and airway epithelial cells in asthma.
The exact etiology of IPF is still unknown, and the five-year survival rate is approximately 45% (104). Progress in improving overall survival in IPF has been limited since 2010 (104). Integrin α-3 (ITA3) mutation A349S has been discovered in ILD, leading to a gain-of-glycosylation (105). This mutation causes lung disorders by disturbing the biosynthesis of ITA3, a highly expressed integrin in lung epithelium that plays a key role in IPF and the epithelial-mesenchymal transition (EMT). The glycosylation of vacuolar H+-adenosine triphosphatase (V-ATPase) may promote collagen degradation and contribute to the progression of IPF (26). FUT8 upregulation is observed in a bleomycin-induced pulmonary fibrosis rat model, while glycyrrhizic acid can alleviate IPF by inhibiting FUT8-mediated core fucosylation of TGF-βR and WNT (67). Patients with IPF show upregulated expression of FUT8 and core fucosylation, which regulate the insulin-like growth factor 1(IGF1) signaling pathway in IPF (66). The upregulated expression of IGF1 is associated with the pathogenesis of IPF (106), and inhibition of core fucosylation alleviates IGF1-induced IPF (106). Therefore, FUT8 and core fucosylation may play a crucial role in the development and deterioration of IPF by regulating multiple signaling pathways, including those for IGF1, TGF-βR, and WNT. Interestingly, lower IPF expression is associated with increased levels of circulating FUT3 and FUT5 (65). Further research is needed to investigate the role of other FUTs in IPF besides FUT8.
Lung cancer is the leading cause of cancer-related mortality worldwide (107). It can be categorized into non-small cell lung cancer (NSCLC) or small cell lung cancer (SCLC), with NSCLC accounting for approximately 85% of cases and SCLC accounting for 15% (108). Lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) are the most common subtypes of NSCLC. A variety of changes in glycosylation have been observed in lung cancer, such as aberrant glycosylation of mucins and increased sialylation of proteins (29), which are potential biomarkers for tumor development and progression (28).
Abnormal protein fucosylation has also been found in lung cancer (41, 109), with FUT8 and POFUT1 proteins being upregulated in blood and tumor tissue of patients with lung cancer (110). These proteins have shown potential as biomarkers for early detection of lung cancer. In addition, the activity of α-1,3-FUTs increases in NSCLC tumor tissues, with upregulated mRNA expressions of FUT3, FUT6, and FUT7 and downregulated mRNA expression of FUT4 (111). Gene expression analysis has revealed that the mRNA expression of FUT2, FUT3, FUT6, and FUT8 are increased in NSCLC, whereas that of FUT1 is decreased (112). FUT1 expression has been correlated with treatment outcomes in LUAD patients receiving epidermal growth factor receptor tyrosine kinase inhibitor (EGFR-TKI) (73). FUT2 is overexpressed in LUAD and may facilitate autophagy and suppress apoptosis via the p53 and JNK pathways (74), as well as induce EMT through TGF-β/Smad signaling in LUAD (75). FUT4 expression shows a negative correlation with the overall survival in operable LUAD (113, 114), and may induce lung colonization and distant metastases of lung cancer cells (114). Autophagic tumor-associated macrophages have been found to promote TGF-β1 secretion and EMT in LUAD through the FUT4/p-ezrin pathway (76). Downregulated expression of FUT4 inhibits EGFR activation and MAPK and NF-κB signal pathways, resulting in reduced migration, invasion and EMT (77). FUT4 may also be related to multidrug resistance in lung cancer (115) and may participate in chemoresistance to cisplatin by suppressing FOXO1-induced apoptosis in lung cancer (116). Additionally, FUT4 expression is positively correlated with PD-1 expression, and may be involved in PD-1-mediated-immunosuppression (113). FUT4 and FUT7 may also participate in lung-to-brain metastasis of NSCLC cells (117). Reduced levels of FUT7 CpG DNA methylation have been found in lung cancer, especially in LUSC (118), and FUT7 promotes lung cancer proliferation by activation of the EGFR/AKT/mTOR signal pathway (119). FUT8, whose expression is upregulated in most cancers, regulates the core fucosylation of PD-1 (120, 121), PD-L2 (122), TGF-β (123), TNFR (124), EGFR (78, 125, 126), B7H3 (127), α3β1 integrin (128), and E-cadherin (129) as well as that of mucins (130). Upregulated expression of FUT8 in cancer-associated fibroblasts promotes the construction of an invasive tumor microenvironment in NSCLC through the core fucosylation of EGFR (78). FUT8-mediated core fucosylation of E-cadherin may also be involved in the EMT in lung cancer cells (131). Abnormal FUT8-mediated core fucosylation plays an important role in tumor proliferation, invasion and metastasis, and FUT8 may be a potential biomarker and therapeutic target in lung cancer and many other cancers.
Studies have reported an increase in glycosylation, particularly in O-GlcNAcylation, in the pulmonary vasculature of idiopathic PAH patients (46, 132). Additionally, there is evidence of a correlation between decreased O-GlcNAc levels and impaired angiogenesis and vascularization in idiopathic PAH (33). The glycosylation of IgG is also elevated in PAH patients (33), suggesting a potential role for glycosylation, especially O-GlcNAcylation, in the pathobiology of PAH.
Non-CF bronchiectasis, a progressive lung disease characterized by permanently dilated airways, may be driven by a complex cycle of infection and bronchial inflammation (133–135). Therefore, breaking this cycle is a promising therapeutic strategy (136). Research has shown that FUT2 polymorphism is closely associated with the prognosis of non-CF bronchiectasis (72). Patients with non-CF bronchiectasis with homozygous secretors of FUT2 had a poorer prognosis compared with those of nonsecretors and heterozygous secretors and exhibited lower lung function, more exacerbation, and a higher frequency of Pseudomonas aeruginosa-dominated infection (72). Therefore, FUT2 and α-1,2-fucosylation may serve as potential therapeutic targets for non-CF bronchiectasis.
Cell surface glycoproteins and glycolipids are commonly capped with sialic acids, which are covalently attached through sialylation by four known families of human STs: ST6GAL (ST6GAL1 and 2), ST6GALNAc (ST6GALNAc1-6), ST3GAL (ST3GAL1-6), and ST8SIA (ST8SIA1-6) (109, 132, 137, 138). ST6GAL1 and ST6GAL2 transfer sialic acid to a β-D-galactopyranosyl (Gal) residue (α-2,6-linked sialylation), while ST6GALNAc1-6 transfer sialic acid to a β-D-N-acetylgalactosaminyl (GalNAc) residue (α-2,6-linked sialylation). ST8SIA1-6, one the other hand, creates a linear α-2,8-polysialic acid on various glycoproteins. Different STs show tissue specificity, suggesting distinct sialylation traits in various tissues. Aberrant sialylation, commonly observed in cancer and nervous system diseases (139–141), is also found in the body fluids and tissue of patients with CRDs, especially with lung cancer (109, 142). To gain insight into the role of sialylation in CRDs, we reviewed the abnormal expression of STs and sialylation in CRDs below.
Research has shown that the sialylation of IgG and other glycoproteins increases in the blood of patients with COPD (143). The glycosylation changes in IgG could potentially serve as a promising biomarker to distinguish between COPD and lung cancer. Furthermore, the level of circulatory ST6GAL1 negatively correlates with the severity of acute airway inflammation, and the administration of recombinant ST6GAL1 to a murine model mimicking acute exacerbations of COPD can alleviate the inflammation symptoms (58). In patients with COPD, low levels of ST6GAL1 and α-2-6 sialylation are associated with poor prognosis, and this may relate to the regulatory effect of IL-6 expression/secretion by ST6GAL1 (57). Therefore, as a potential therapeutic approach, the administration of recombinant ST6GAL1could prevent the exacerbation of COPD.
Asthma is categorized as either type 2 (T2) or non-T2 based on the presence of T helper type 2 cells (Th2) and type 2 cytokines (144). The levels of ST6GAL1 and sialylated MUC4β have been shown to be increased in airway specimens from patients with T2 asthma (30), which may cause epithelial dysfunction. In contrast, the level of ST3GAL3-catalyzed sialylation of MUC5B is downregulated in most patients with asthma, and a reduced MUC5B level may be related to the severity of the disease (59). The sialylation pattern of mucins in asthma appears to play an important role in asthma pathogenesis. The sialylation of memory T helper and regulatory T cells alters in asthmatic children and is associated with asthma progression (145). Mice with a hepatocyte-specific knockout of ST6GAL1 (H-cKO) exhibit significantly increased morbidity due to T cell-dependent HDM-induced asthma. ST3GAL3 knockout (St3gal3−/−) and knockdown (St3gal3+/−) mice suffer more severe allergic eosinophilic airway inflammation. Therefore, sialylation could serve as a biomarker for the diagnosis and prognosis of asthma. The galactosylation and sialylation at Asn 297 of IgG decreased in the serum of adult allergic offspring from allergic mothers (21), and this reduction of IgG sialylation is associated with proinflammation (146). In summary, protein sialylation is altered in the airways, mucus, T cells, and sera of patients with asthma, and the related STs and sialylated proteins could potentially serve as biomarkers or therapeutic targets.
CF is a hereditary disorder cause by mutations of CFTR, mainly prevalent in white population from Europe, North America, and Australia (147). Approximately 90% of patients with CF have a phenylalanine deletion at codon 508 (ΔF508), which was associated with decreased α-2,3 sialylation on cell membranes (64). Knockout of CFTR (CFTR−/−) in piglets result in an increase in sialylated mucins in the airways (148). The aberrant sialylation of proteins, especially mucins, is associated with CF. Reduced sulfation and fucosylation and increased sialylation of MUC5B and MUC5AC are also reported in the sputum of patients with CF (61, 63). Bacterial infection can affect mucin O-glycosylation, and protein N-glycosylation in sputum, and the N-sialylation and N-fucosylation of sputum proteins were increased in these patients (63). The proinflammatory cytokine TNF induces the expression of ST3GAL4 in lung epithelial cells (149, 150), and the inflammatory cytokines IL-6 and IL-8 upregulate the expressions of ST6GAL2 and ST3GAL6 in CF epithelial cells (151), indicating that STs may play a crucial role in inflammation conditions, including CF.
Aberrant sialylation is a common feature in various cancers, including colorectal cancer (152), ovarian cancer (153), prostate cancer (153), and lung cancer (142). Sialylation is known to be involved in the regulation of tumor metastasis, cell survival, immune evasion, and multidrug resistance (140). A study on lung tumor N-glycoproteomics showed that 108 out of 303 quantified sialylated N-glycopeptides were differentially expressed, and differential Ig α-2,6-sialylation was also observed in lung tumor tissues (142). Moreover, hypersialylation and hyperfucosylation of saliva glycoproteins may be a hallmark of lung cancer in patients (109). The expressions of ST3GAL1, ST6GALNAc3, and ST8SIA6 are significantly reduced in lung cancer tissues and cells, whereas that of ST6GAL1 is significantly upregulated (80). ST6GAL1 may be involved in cell invasion and tumorigenesis of NSCLC via Notch1/Hes1/MMP signaling (80). ST3GAL4 may regulate the proliferation, invasion, and migration of NSCLC cells through α-2,3 sialylation of HSP60 (82). ST3GAL6 expression is downregulated in LUAD, which results in poor prognosis (79). Trp53R172H mutation increases the expression of ST6GALNAc1, which may promote tumor metastasis through sialylation of MUC5AC in LUAD (81). Thus, various aberrant ST expression profiles contribute to the abnormal sialylation patterns in lung cancer, which may partially contribute to its development and metastasis.
According to the Human Protein Atlas database (https://www.proteinatlas.org/), the expression of OGT is most prominent in the respiratory system compared to other organs, while OGA expression is average. The OGT/OGA pairs may actively work in the regulation of the respiratory system. O-GlcNAcylation participates in various fundamental cellular processes, including transcription, epigenetics, cell signaling dynamics, protein translation, stability, and turnover (10, 154). Dysfunction in O-GlcNAcylation is involved in several diseases, including neurodegenerative diseases (155, 156), diabetes (157, 158), and cancers (159, 160). Therefore, targeting O-GlcNAcylation could be critical in developing new treatments for these diseases (161), especially cancers (162).
Glycosylation, especially O-GlcNAcylation, increases in the pulmonary vasculature of patients with idiopathic PAH (IPAH) (32, 163), while decreased O-GlcNAc levels correlate with impaired angiogenesis and vascularization in IPAH (25), suggesting that O-GlcNAcylation may contribute to the pathobiology of PAH. OGT is overexpressed in the pulmonary vasculature of patients with IPAH, and its levels are negatively associated with the severity and prognosis of IPAH (32). In IPAH, the OGT/OGA pairs regulates cell proliferation by mediating the activation of host cell factor-1 (HCF-1) in IPAH (32) and can regulate the expression of vascular endothelial growth factor (VEGF) by O-GlcNAcylation of specificity protein 1 (SP1) (164). Nitric oxide (NO) deficiency is also implicated in the development PAH (165), as endothelial NO (eNOS) activity, which produces NO, is reduced by O-GlcNAcylation at residue 615 in PAH (71). Thus, targeting OGT may be a promising for the diagnosis and treatment of PAH.
In lung cancer, O-GlcNAcylation and expression of OGT are increased, potentially promoting tumorigenesis and cancer progression (166). O-GlcNAcylation can accelerate the KrasG12D-induced lung tumorigenesis (167). The nuclear RNA-binding protein, SAM68, has 11 identified O-GlcNAcylation sites (87). High levels of OGT and SAM68 result in poor prognosis of LUAD, and O-GlcNAcylated SAM68 may participate in modulating lung cancer aggressiveness (87). Overexpression of IL-8 can enhance protein O-GlcNAcylation, which may play an important role in the generation and maintenance of cancer stem cells in lung cancer (168). Elevated O-GlcNAcylation of p53/c-Myc induces cisplatin resistance, which could be an important mechanism of drug resistance (88). Upregulation of OGT is observed in SCLC and is associated with clinical outcomes of SCLC (89). OGT and O-GlcNAcylation may play a key role in the IL-6/STAT3 signaling-induced migration and invasion of lung cancer (169). altogether, OGT is typically overexpressed in lung cancer, resulting in hyper-O-GlcNAcylation of tumorigenesis and metastatic-related proteins. OGT has potential as a biomarker and drug target for lung cancer.
In addition to GTs, dysregulated expression of GHs can also contribute to the aberrant glycosylation in various diseases, especially in cancer (37, 38). GHs have been used in prodrug activation for cancer and diabetes treatment (38, 39). Interestingly, a higher incidence of congenital disorders of GHs has been observed due to the prevalence mutations in several GH genes in comparison to that in GT genes (40). GHs not only serve as critical enzymes in glycosylation but also play an essential role in prodrug design. The human sialidase family, also referred to as neuraminidases (NEUs), includes NEU1, NEU2, NEU3, and NEU4 (170). Among them, NEU1 is the predominant sialidase expressed in human airway epithelia and lung microvascular endothelia, which can inhibit endothelial cell migration (171, 172). Additionally, NEU1 has been implicated in various airway epithelia- and microvascular endothelia-related inflammatory reactions and diseases. NEU3 is also expressed in human airway epithelia and lung microvascular endothelia (171, 173).
NEU1 and NEU3 are the predominant sialidases in the lung microvascular endothelia (171). Increased expression of NEU1 is observed in the lungs of patients with IPF, and NEU1 may participate in the IPF pathogenesis by provoking lymphocytic infiltration and promoting accumulation of glycoprotein TGF-β1, type I and III collagen (68, 174). In mice, administration of NEU3 has been shown to induce lung fibrosis, with a gender-specific effect (175). NEU3 can stimulate extracellular accumulation of profibrotic cytokines IL-6 and IL-1β, and conversely, IL-6 can induce the expression of NEU3 in human peripheral blood mononuclear cells (70). Sialylation of serum amyloid P (SAP) is critical for its biological activity. It can inhibit the differentiation of monocytes into fibrocytes and promote high extracellular levels of IL-10. However, in patients with IPF, the sialidase NEU3 is highly expressed, leading to SAP desialylation in the sera (69). This desialylation may contribute to the dysregulated immune response and fibrotic remodeling observed in IPF. Maintaining SAP sialylation status could be a potential therapeutic strategy for modulating the pathogenesis of IPF. Therefore, inhibiting NEU1 and NEU3 could be a new therapeutic strategy for IPF.
The abnormal expression of certain GHs has been observed in lung cancer. For instance, β-glucuronidase is preferentially concentrated within areas of necrosis in lung tumor tissues (176). Glucosidase IIβ subunits are overexpressed in lung tumor tissues and promote cell growth and migration through receptor tyrosine kinase (RTK) signaling and the p53 pathway (177, 178). Inhibition of these subunits can induce autophagy and apoptosis in lung cancer cells (90). The expression of NEU1 is increased in NSCLC tumors with p53R273H mutation and is associated with poor prognosis (84). NEU1 is also correlated with the severity of drug resistance in DLKP, a lung cancer cell line (83). NEU3 may regulate the ERK pathway via EGFR and serve as a prognosis biomarker for EGFR-targeted therapies in NSCLC (85). Furthermore, FUCA2, a fucosidase, is upregulated in most tumors and predicts poor overall survival in pan-cancer, including LUAD (86). These observations suggest that several GHs may serve as potential biomarkers or drug targets for lung cancer.
There is a known correlation between diabetes mellitus (DM) and COPD, and studies have shown that patients with DM taking α-glucosidase inhibitor drugs have a higher morbidity of COPD (179, 180). In pediatric allergic asthma, the downregulation of NEU1 has been observed in the airway epithelial cells (181). In asthmatic mouse models, the interaction between hyaluronan and CD44 is crucial for the accumulation of antigen-specific Th2 cells (60, 182). NEU1 may also contribute to Th2 cell-mediated airway inflammation by influencing the glycosylation of CD44 (60, 182). Additional, abnormal activity levels of alpha-fucosidase, alpha-galactosidase, beta-galactosidase, alpha-glucosidase, beta-glucosidase, beta-glucuronidase, beta-hexosaminidase, and alpha-mannosidase have been were detected in the sera of patients with CF (183).
Bacterial infection and cigarette smoking are two major risk factors for CRDs. Smoke disrupts the heavily O-glycosylated MUC1 barrier that protects the airway, and N-acetyl-galactosaminyl transferase-6 (GALNT6), an enzyme that mediates the initial step of O-glycosylation, may be involved in the smoking-induced aberrant MUC1 glycosylation (184). Frequent respiratory infection is a symptom of many CRDs, such as COPD, CF, and non-CF bronchiectasis. Infection may change the glycosylation of lung epithelium cells, such as upregulation sialylation during Mycobacterium tuberculosis infection (185). Abnormal MUC1 glycosylation may be the major cause of persistent infection. Moreover, gender biases characterize most chronic inflammation diseases of airway (186, 187), including COPD (188, 189), asthma (190), CF (191), non-CF bronchiectasis (192),and lung cancer (188). Women usually suffer more severe symptoms and have a poorer prognosis for CRDs than men (187). One reason for this may be estrogen regulation of protein glycosylation by affecting the expressions of glycoenzymes. Estradiol can increase FUT4, FUT5, and FUT6 expressions as well as the total amount of fucosylation (193). Estrogen can affect the expressions of ST3GAL1 (194), ST6GAL1 (195), and ST6GAL3 (196), indicating a regulation of sialylation by estrogen. Therefore, estrogen may be a mediator in the progression of CRD.
In conclusion, alterations in protein glycosylation mediated by GTs play a crucial role in the pathogenesis of different CRDs. Various cytokines, such as TNF-α and IL-6, may participate in the regulation of GT expression leading to proinflammatory responses. Additionally, downstream regulatory molecules like TGF-β1 and mucins, which are glycoproteins, contribute significantly to the development of CRDs. The expression profile of GTs in CRDs is also affected by factors such as infection, smoking, and gender. Our review of glycoenzyme-mediated glycosylation changes in CRDs highlights the potential for identifying biomarkers and drug targets of various CRDs.
Traditional methods in molecular biology for screening target markers (28, 143, 145) and investigating physiological and pathological phenomena (36, 96, 97) often rely on studying the function and mechanism of one or a few proteins in tightly controlled experimental systems. However, such methods have limitations when it comes to analyzing large-scale samples and comprehensively capturing the complexity of biological systems. By contrast, omics strategies can provide a more inclusive molecular perspective of biological systems by analyzing diverse biological and clinical samples, which can lead to the identification of more effective biomarkers and a more comprehensive understanding of the underlying physiological and pathological mechanisms.
In recent years, glycomics and glycoproteomics strategies have been increasingly employed to study the protein glycosylation under various physiological and pathological conditions (197–199). Glycoproteomic analysis characterizes glycopeptides and provides information on glycoforms and their occupation sites (200), while glycomics profiles glycans, enabling the characterization of glycan structures and isomers (200, 201). These complementary strategies offer a comprehensive understanding of total glycosylation patterns. Lectin array and high-performance liquid chromatography (HPLC) with optical detection, commonly used for glycosylation analysis in respiratory-related diseases, have limited capabilities for glycan structure characterization (200–202). Mass spectrometry (MS)-based glycomics and glycoproteomics strategies have proven to be powerful for global glycosylation analysis in complex and large-scale biological samples (203–205) (206, 207). This section reviews recent developments in glycomics and glycoproteomics for the analysis of protein glycosylation in CRDs.
In recent years, there has been a growing interest in protein glycosylation and the use of MS technology for glycomics analysis in CRDs. A typical MS-based glycome experimental procedure involves sample pretreatment, glycan release, glycan purification and separation, glycan derivatization, MS analysis, and data interpretation. Efficient release of glycans from glycoproteins is a critical initial step in glycome analysis, which can be achieved by enzymes (e.g., peptide N-glycosidase F, peptide N-glycosidase A, endo-β-acetylglucosaminidase F, and O-glycosidases) or chemical methods (e.g., hydrazinolysis, β-elimination, and oxidation strategies) (201). MS-based characterization of released glycans can be performed in their native state or after chemical derivatization. Glycan derivatization can be achieved through derivation of the reducing end, hydroxyl group, and carboxyl group (201). For example, phenylhydrazine labeling (a reducing end derivation method) has been used for structural studies of fucosylated N-glycans by MALDI-MS in positive ion mode (208), and for efficient detection and discrimination of SA linkages when following alkyl esterification (209). MS-based systems commonly used for glycomic characterization include MALDI, electrospray ionization (ESI), capillary electrophoresis (CE), gas chromatography (GC) and liquid chromatography (LC) coupled to MS (143).
In the field of respiratory-related diseases, such as lung cancer, COPD, CF, and asthma, glycomics has gained increasing attention in recent years. Researchers have identified potential biomarkers for the diagnosis and differentiation of these diseases by studying fucosylated, sialylated, and galactosylated glycans with different structures. Borges’ research group (206, 207) found that α-2-6 sialylation, β-1-4 branching, β-1-6 branching, terminal, core and outer arm fucosylation markers were most effective in discriminating between lung cancer cases and controls, with the diagnostic performance being dependent on the cancer stage. Ma et al. (210) reported an increase in the abundance of fucosylated N-glycans from 40.9% to 48.3% in LUSC. Lattová et al. (211) reported a decrease in the amount of double-terminal core fucosylated glycans of the sialylation complex in LUAD. McQuiston et al. (212) found a significantly increased IgG1 N-glycan profile in lung transplant recipients with COPD and primary graft dysfunction. Glycomic analysis of mucins in CF revealed a significantly high abundance of sialylated glycans, and the total abundance of nonsulfated O-glycans correlated with the relative abundance of pathogens (213), whereas another study reported that submucosal gland mucins contained shorter and less branched glycans (214). The glycomics strategies utilized in the CRDs cases are concisely outline in Figure 2 (blue part).
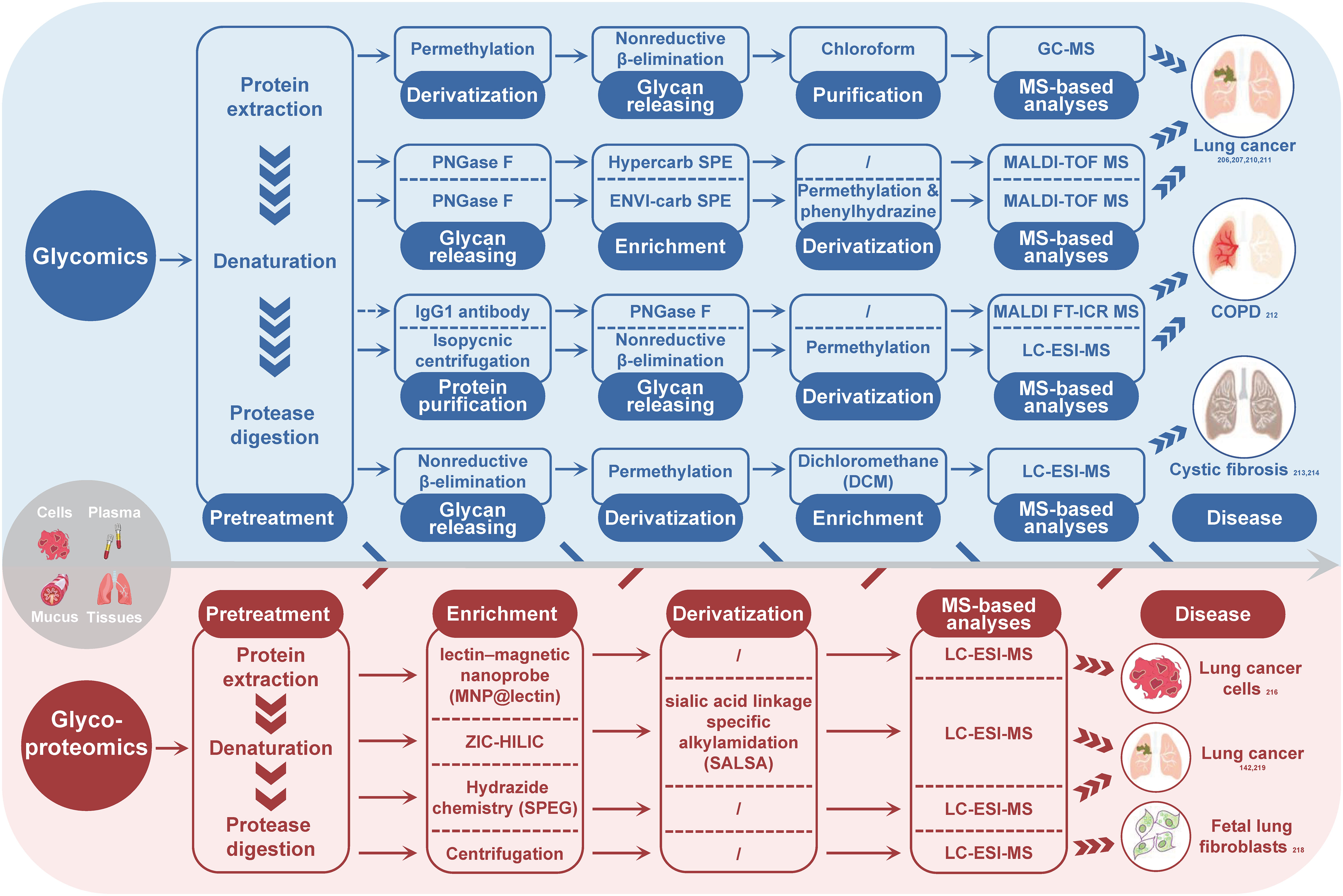
Figure 2 Summary of MS-based glycomics and glycoproteomics strategies used for glycosylation analysis in CRDs. The subscript number indicates the corresponding reference. PNGase F, peptide-N-glycosidase F; SPE, solid phase extraction; MS, mass spectrometry; GC-MS, gas chromatography-mass spectrometry; MALDI-TOF-MS, matrix-assisted laser desorption/ionization-time of flight mass spectrometry; MALDI FT-ICR MS, matrix-assisted laser desorption/ionization fourier transform-ion cyclotron resonance mass spectrometry; LC-ESI-MS, liquid chromatography-electronic spray ionization-mass spectrometry.
Glycomics enables the analysis of glycan phenotypes and provides information on the levels of different glycan subtype and associated glycoenzymens. In particular, glycan subtyping analysis can reveal the activity and expression of glycoenzymes, making it a valuable strategy for studying glycoenzyme-mediated functions. Studies on the human lung N-glycome have shown that biantennary complex-type N-glycans dominate (215), but dysregulation of the glycosylation-regulating mechanisms in CRDs can lead to changes in glycan structures, such as increased fucosylation (including terminal, core, and outer arm fucosylation), sialylation, and branching. The core fucosylation depends on FUT8 activity, while outer arm fucosylation relies on FUT3, FUT5, FUT6, and FUT11. ST6GalI glycosyltransferase is responsible for α-2-6 sialylation, and β-1-4 and β-1-6 branching of N-glycans are regulated by GnT-IVa and GnT-V enzymes, respectively (206, 207). However, because glycans can vary greatly in their length, branching, and connectivity, software for interpreting MS data from polysaccharides often requires manual intervention. The lack of generic annotation software has hindered the extensive investigation of the glycome in large scale sample sets from diverse disease, including the field of CRDs.
Glycoproteomics is a valuable strategy for identifying potential biomarkers and understanding physiological and pathological changes. Unlike glycomics, a typical experimental procedure of glycoproteomics involves protein digestion, glycopeptide enrichment, MS analysis and MS data interpretation by software tools (142, 216, 217). Glycopeptide enrichment is critical for efficient glycopeptide identification, and different enrichment methods have been developed, such as lectin enrichment, hydrophilic interaction chromatography, boronic chemistry, hydrazide chemistry, reductive amination chemistry, oxime click chemistry, and ultracentrifugation (218). Although MS-based site-specific glycosylation analysis methods have developed rapidly in recent years, only a few glycoproteomics studies on the intact glycopeptide level have been conducted in the context of CRDs, mainly on lung cancer and pulmonary fibroblasts. For instance, Waniwan et al. (216) developed a lectin-magnetic nanoprobe for glycopeptide enrichment and site-specific glycosylation analysis and identified over 2,000 glycopeptides in NSCLC cell lines. Yang et al. (142) conducted an N-glycoproteomic study using selective alkylamidation and multiple tandem mass tag (TMT)-tagged sialic acid linkages to specifically quantify glycoproteins in lung cancer tissues. Yang et al. (219) found that the expressions of 11 glycoproteins were upregulated in both LUAD and LUSC, while two glycoproteins (ELANE and IGFBP3) and six glycoproteins (ACAN, LAMC2, THBS1, LTBP1, PSAP, and COL1A2) were increased in either LUSC or LUAD. The most comprehensive study on lung fibroblasts was published by Takakura et al. in 2015 (218). They used glycoproteomics method to analyze the membrane fraction of human fetal lung fibroblasts and identified more than 272 glycoforms on 63 sites of 44 glycoproteins. Figure 2 (red part) summarizes the glycoproteoimcs strategies applied in the CRDs cases.
Although glycoproteomics is a valuable strategy for studying the physiology and pathology of diseases and discovering biomarkers, it has been rarely used in the field of CRDs, and there has been no large-scale clinical sample data output yet. Actually, in the field of glycoproteomics, several relatively mature software portfolios have been established for identification and quantification, such as pGlyco3 (220, 221) and pGlycoQuant (220, 221). These tools are state-of-the-art for site-specific glycome analysis, providing fast and precise identification and quantification of intact glycopeptides. Moreover, high-throughput glycopeptide enrichment technologies for large-scale samples are also available, such as the one developed by Jiang et al. (222), which allows for high-specificity and high-throughput glycopeptide parallel enrichment in a 96-well plate. With the emergence of more effective methods and accurate identification and quantitative tools in the field of glycoproteomics, it is expected that research on glycosylation in CRDs will reach new levels.
Protein glycosylation remodeling is a common feature of several pathological conditions, resulting from the dysregulated expression of glycoenzymes. Evaluating the expressions of glycoenzymes and protein glycosylation patterns could be a promising approach to creating diagnostic and prognostic biomarkers. Furthermore, targeting the dysregulated glycoenzymes and their associated changes could serve as a potential therapeutic strategy.
Protein glycosylation patterns vary across different CRDs due to the diverse profile of glycoenzymes (223, 224). Saliva and serum proteins with abnormal glycosylation have been identified as potential biomarkers for the diagnosis and prognosis of various CRDs (109, 225). Sputum proteins in progressive CRDs, such as COPD and CF, have also been observed to have aberrant glycosylation (63). Table 2 provides an overview of potential glycoprotein and glycoenzyme biomarkers.
For COPD, serum fucosylation levels of SP-D may serve as a diagnostic biomarker (36), while serum levels of ST6GAL1 may predict acute exacerbation of the disease (57). Glycosylated BPIFB1 in sputum may also act as a prognostic biomarker for COPD in smokers (226). In maternal asthma during pregnancy, the IgG glycosylation patterns may predict offspring asthma susceptibility (21). In IPF, circulating FUT3 levels are negatively associated with the risk of the disease and may serve as a biomarker. NEU3 may be involved in the IPF pathogenesis and a drug target, and serum NEU3 and sialylated SAP may act as biomarkers for IPF diagnosis and prognosis (69). FUT2 genotype in patients with non-CF bronchiectasis may also predict the disease outcomes (72). In lung cancer, various FUTs and STs expressed in tumor tissues have been identified as potential diagnostic and prognostic biomarkers (Tables 1, 2) (111, 112). Additionally, NEU3 is a promising biomarker for evaluating EGFR-targeted therapies in patients with NSCLC (85), and the expression level of GTs in lung cancer tumors may act as a biomarker for the diagnosis, prognosis, and treatment assessment (Table 2).
Glycoproteomics and glycomics are highly valuable strategies for the comprehensive analysis of glycoproteins and glycans in body fluids. By comparing the glycosylation profile of healthy and diseased individuals, researchers can identify specific changes in glycosylation patterns that are associated with particular diseases. This can be used to develop biomarkers for disease diagnosis, drug selection, and prognosis prediction, and to subtype patients and evaluate disease severity based on glycosylation patterns as illustrated in Figure 3. These techniques are also useful for monitoring the effects of therapy.
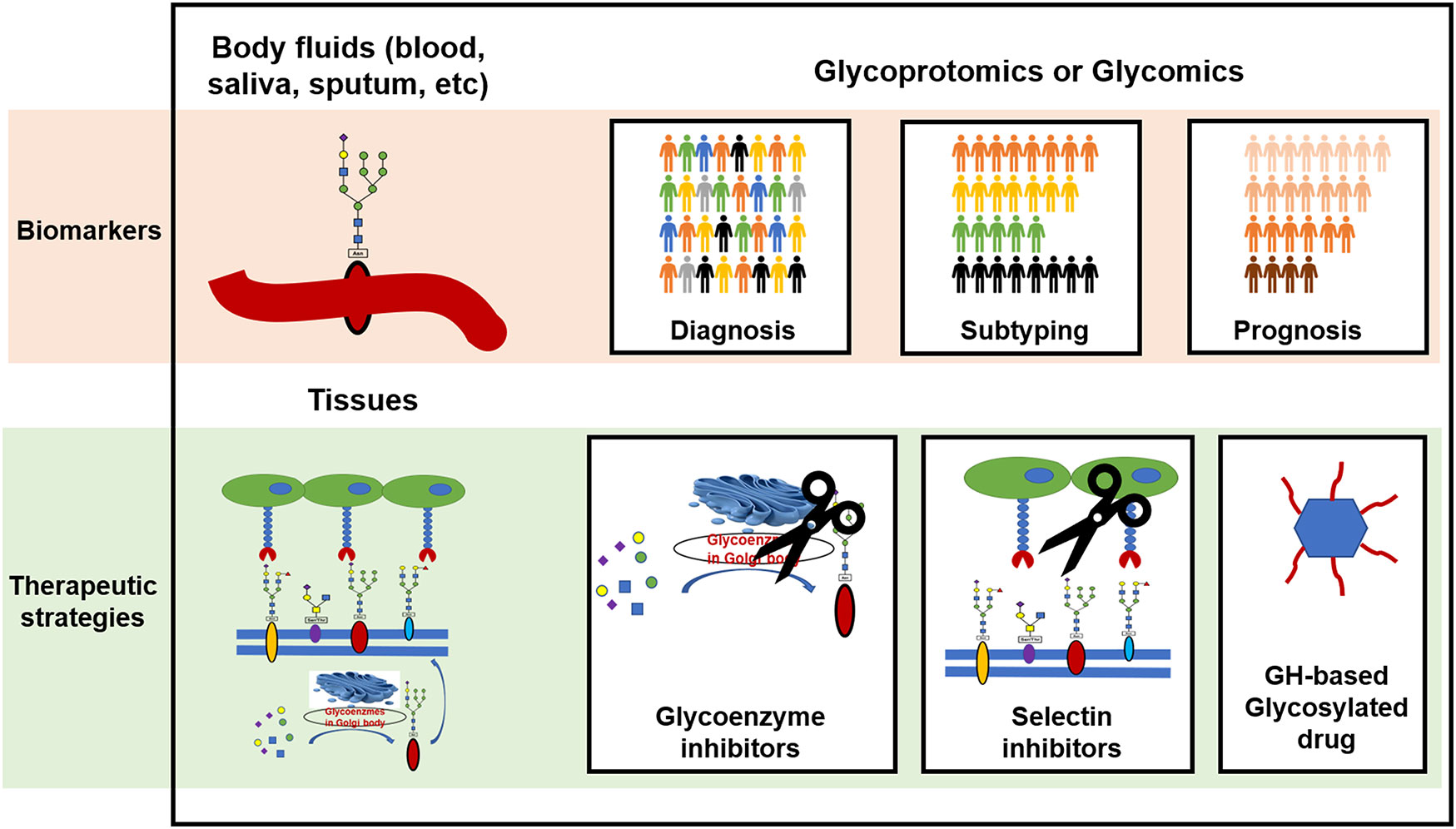
Figure 3 The clinical potential of protein glycosylation for the development of biomarkers and therapeutic strategies in CRDs. GH, glycoside hydrolase.
Aberrant protein glycosylation may serve as both a cause and a consequence of CRDs. These changes in glycosylation can influence the functions of the glycoproteins. Potential therapeutic strategies for CRDs include targeting glycoenzymes to correct the protein glycosylation status, blocking abnormal selectin-mediated cell-cell interactions, and clearing dysfunctional glycans as depicted in Figure 3.
One promising approach for treat diseases involves using specific glycoenzyme inhibitors. Aberrant expression of GTs and the related changes in glycosylation are reviewed in the second section of this review. Increased expression of FUTs and STs is characteristic of most patients with NSCLC, and patient-specific overexpressed FUTs and STs are promising targets for developing new therapies. Munkley J. reviewed the inhibitors of STs, including ST3GAL1, ST3GAL3, ST6GAL1, ST6GalNAc2, and ST8SIA3 (140). Increased expressions of ST3GAL4 (82), ST6GAL1 (80), and ST6GalNAc1 (81) were identified in most patients with NSCLC, and inhibiting these enzymes may suppress NSCLC cell metastasis. Further research is required to map the expression of all STs in various lung cancers, and the use of different ST inhibitors in combination therapy may be a new promising cancer therapy.
Overexpression of most FUTs, including FUT2-8, has been observed in lung cancers, especially NSCLC (78, 112, 114, 117, 228). In vitro studies have demonstrated that inhibiting or genetically depleting FUT2 (228) and FUT4 (114) can be effective therapies for these cancers. FUT8, a key regulator of the p53 signaling cascade, is a promising therapeutic target for cancer and inflammatory diseases, including Alzheimer’s disease (AD) (229). FUT8 and core fucosylation inhibition are prospective therapeutic strategies for cancer and inflammation. In addition to inhibitors, glycomimetics may also offer alternative therapeutic strategies. Keratan sulfate disaccharide L4 and derivatives show promise as potential drugs for treating emphysema and COPD (230).
Selectins, which are transmembrane glycoproteins found on endothelial cells (E-selectin), leukocytes (L-selectin) and platelets (P-selectin), play a critical role in mediating leucocyte-endothelial adhesion during inflammatory and immune reactions associated with tumorigenesis and metastasis (231). Blocking selectin-ligand interaction interactions is being investigated as an anti-metastasis therapy. P-selectin (232) and E-selectin (111) may be involved in NSCLC cell metastasis, and disrupting the selectin-ligand interactions could potentially serve as a complementary therapy to traditional anticancer therapy (140). Furthermore, selectin antagonists are explored as potential drug candidates for other respiratory inflammatory conditions, such as CF, asthma and COPD (23, 233, 234). Targeting selectins has the potential to be a promising immunomodulation intervention and combination therapy.
GHs can selectively hydrolyze glycosidic bonds and eliminate dysfunctional glycans. Glycosylated prodrugs, which have been widely used to reduce the side-effects of anticancer drugs, can be activated through targeted deglycosylation mediated by certain GHs. One such enzyme, β-glucuronidase, has been utilized to cleave the glycans of prodrugs to activate them (53, 235). Various GHs, including glucosidase II (90, 236), FUCA2 (86), NEU1 (83), and NEU3 (85), are overexpressed in lung cancer and have the potential to be utilized in the development of lung cancer-specific prodrugs.
CRDs encompass a range of inflammatory conditions affecting the respiratory tract. During inflammation reactions, various molecules involved in inflammation, such as p53 and selectins, undergo dysglycosylation. Targeting these glycoproteins represents a promising approach to anti-inflammatory and immunomodulation therapies. Moreover, the regulation of glycoenzymes and protein glycosylation by proinflammatory cytokines suggests a complex signaling pathway underlying the development and progression of CRDs.
Aberrant protein glycosylation plays a significant role in the pathogenesis of CRDs. Changes in the expressions of glycoenzymes in airway epithelial cells and mucus are responsible for variations in glycosylation patterns. Therefore, proteins with altered glycosylation patterns and various glycoenzymes present in the epithelial cells are potential targets for new monotherapies and combination therapies for CRDs. Abnormal protein glycosylation and glycoenzymes in body fluids, especially in sputum and serum, may serve as potential biomarkers for the diagnosis, prognosis, and treatment assessment of CRDs.
Glycoproteomics and glycomics are essential strategies for unveiling the protein glyco-codes, including glycosites, glycan structures, and glycosylation levels. This review focuses the glycoenzyme-protein glycosylation-CRD axis, highlighting two main aberrant terminal glycosylation modifications, fucosylation and sialylation, and their respective enzymes, FUTs and STs, in CRDs. The clinical potential of these glycoenzymes and glycosylated proteins has already been demonstrated in the diagnosis and treatment of CRDs.
The involvement of glycoenzyme-mediated glycosylation changes in CRD development and exacerbation suggests that targeting glycoenzymes and glycoproteins using chemical and biomacromolecular drugs may be a promising approach for CRD therapy. It is common to observe glycosylation changes such as increased fucosylation and sialylation. One potential avenue for treating different CRDs is inhibiting specific FUTs and STs. Furthermore, the study of GHs under different conditions can provide new insights into designing prodrugs that can minimize their side-effects. Despite some efforts to investigate glycoproteomic and glycomic changes in CRD research, there is still a considerable amount of work that needs to be done in the field of CRD development and diagnosis. A comprehensive understanding of the pathophysiological protein glycosylation in humans is urgently required. Therefore, a systematic mapping of such glycosylation patterns is essential to further advance our knowledge in this area.
WC conceived and designed the review; XX and SK contributed to the literature collection and the art work. XX, SK, and WC wrote and revised the manuscript. All authors contributed to the article and approved the submitted version.
This work is supported by grants from the National Natural Science Foundation of China Project (32271490 to WC).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem (2008) 77:521–55. doi: 10.1146/annurev.biochem.76.061005.092322
2. Na L, Li R, Chen X. Recent progress in synthesis of carbohydrates with sugar nucleotide-dependent glycosyltransferases. Curr Opin Chem Biol (2021) 61:81–95. doi: 10.1016/j.cbpa.2020.10.007
3. Jaroentomeechai T, Kwon YH, Liu Y, Young O, Bhawal R, Wilson JD, et al. A universal glycoenzyme biosynthesis pipeline that enables efficient cell-free remodeling of glycans. Nat Commun (2022) 13:6325. doi: 10.1038/s41467-022-34029-7
4. Reily C, Stewart TJ, Renfrow MB, Novak J. Glycosylation in health and disease. Nat Rev Nephrol (2019) 15:346–66. doi: 10.1038/s41581-019-0129-4
5. Nguyen NTB, Lin J, Tay SJ, Mariati, Yeo J, Nguyen-Khuong T, et al. Multiplexed engineering glycosyltransferase genes in cho cells Via targeted integration for producing antibodies with diverse complex-type n-glycans. Sci Rep (2021) 11:12969. doi: 10.1038/s41598-021-92320-x
6. Adjobimey T, Hoerauf A. Distinct n-linked immunoglobulin G glycosylation patterns are associated with chronic pathology and asymptomatic infections in human lymphatic filariasis. Front Immunol (2022) 13:790895. doi: 10.3389/fimmu.2022.790895
7. Nishima W, Miyashita N, Yamaguchi Y, Sugita Y, Re S. Effect of bisecting glcnac and core fucosylation on conformational properties of biantennary complex-type n-glycans in solution. J Phys Chem B (2012) 116:8504–12. doi: 10.1021/jp212550z
8. Davies G, Henrissat B. Structures and mechanisms of glycosyl hydrolases. Structure (1995) 3:853–9. doi: 10.1016/S0969-2126(01)00220-9
9. Stutz AE, Wrodnigg TM. Imino sugars and glycosyl hydrolases: historical context, current aspects, emerging trends. Adv Carbohydr Chem Biochem (2011) 66:187–298. doi: 10.1016/B978-0-12-385518-3.00004-3
10. Varki RD, Cummings JD, Esko HH, Freeze P, Stanley CR, Bertozzi GW, et al. (2009). Essentials of glycobiology. (NY: Cold Spring Harbor).
11. Stephen HM, Adams TM, Wells L. Regulating the regulators: mechanisms of substrate selection of the O-glcnac cycling enzymes ogt and oga. Glycobiology (2021) 31:724–33. doi: 10.1093/glycob/cwab005
12. Gaudet M, Plesa M, Mogas A, Jalaleddine N, Hamid Q, Al Heialy S. Recent advances in vitamin d implications in chronic respiratory diseases. Respir Res (2022) 23:252. doi: 10.1186/s12931-022-02147-x
13. Purghe B, Manfredi M, Ragnoli B, Baldanzi G, Malerba M. Exosomes in chronic respiratory diseases. BioMed Pharmacother (2021) 144:112270. doi: 10.1016/j.biopha.2021.112270
14. Kowalewicz-Kulbat M, Locht C. Bcg for the prevention and treatment of allergic asthma. Vaccine (2021) 39:7341–52. doi: 10.1016/j.vaccine.2021.07.092
15. Rafeeq MM, Murad HAS. Cystic fibrosis: current therapeutic targets and future approaches. J Transl Med (2017) 15:84. doi: 10.1186/s12967-017-1193-9
16. Danoff SK, Terry PB, Horton MR. A clinician’s guide to the diagnosis and treatment of interstitial lung diseases. South Med J (2007) 100:579–87. doi: 10.1097/SMJ.0b013e3180485c62
18. Aghasafari P, George U, Pidaparti R. A review of inflammatory mechanism in airway diseases. Inflammation Res (2019) 68:59–74. doi: 10.1007/s00011-018-1191-2
19. Schjoldager KT, Narimatsu Y, Joshi HJ, Clausen H. Global view of human protein glycosylation pathways and functions. Nat Rev Mol Cell Biol (2020) 21:729–49. doi: 10.1038/s41580-020-00294-x
20. Pavic T, Dilber D, Kifer D, Selak N, Keser T, Ljubicic D, et al. N-glycosylation patterns of plasma proteins and immunoglobulin G in chronic obstructive pulmonary disease. J Transl Med (2018) 16:323. doi: 10.1186/s12967-018-1695-0
21. Sodemann EB, Dahling S, Klopfleisch R, Boiarina E, Cataldo D, Alhasan MM, et al. Maternal asthma is associated with persistent changes in allergic offspring antibody glycosylation. Clin Exp Allergy (2020) 50:520–31. doi: 10.1111/cea.13559
22. Saku A, Hirose K, Ito T, Iwata A, Sato T, Kaji H, et al. Fucosyltransferase 2 induces lung epithelial fucosylation and exacerbates house dust mite-induced airway inflammation. J Allergy Clin Immunol (2019) 144:698–709 e9. doi: 10.1016/j.jaci.2019.05.010
23. Rhim AD, Stoykova L, Glick MC, Scanlin TF. Terminal glycosylation in cystic fibrosis (Cf): a review emphasizing the airway epithelial cell. Glycoconj J (2001) 18:649–59. doi: 10.1023/a:1020815205022
24. Scanlin TF, Glick MC. Terminal glycosylation and disease: influence on cancer and cystic fibrosis. Glycoconj J (2000) 17:617–26. doi: 10.1023/a:1011034912226
25. Vang S, Cochran P, Sebastian Domingo J, Krick S, Barnes JW. The glycobiology of pulmonary arterial hypertension. Metabolites (2022) 12:316. doi: 10.3390/metabo12040316
26. Lee MR, Lee GH, Lee HY, Kim DS, Chung MJ, Lee YC, et al. Bax inhibitor-1-Associated V-atpase glycosylation enhances collagen degradation in pulmonary fibrosis. Cell Death Dis (2014) 5:e1113. doi: 10.1038/cddis.2014.86
27. Kondo K, Harada Y, Nakano M, Suzuki T, Fukushige T, Hanzawa K, et al. Identification of distinct n-glycosylation patterns on extracellular vesicles from small-cell and non-Small-Cell lung cancer cells. J Biol Chem (2022) 298:101950. doi: 10.1016/j.jbc.2022.101950
28. Yang S, Xia J, Yang Z, Xu M, Li S. Lung cancer molecular mutations and abnormal glycosylation as biomarkers for early diagnosis. Cancer Treat Res Commun (2021) 27:100311. doi: 10.1016/j.ctarc.2021.100311
29. Lemjabbar-Alaoui H, McKinney A, Yang YW, Tran VM, Phillips JJ. Glycosylation alterations in lung and brain cancer. Adv Cancer Res (2015) 126:305–44. doi: 10.1016/bs.acr.2014.11.007
30. Zhou X, Kinlough CL, Hughey RP, Jin M, Inoue H, Etling E, et al. Sialylation of Muc4beta n-glycans by St6gal1 orchestrates human airway epithelial cell differentiation associated with type-2 inflammation. JCI Insight (2019) 4:e122475. doi: 10.1172/jci.insight.122475
31. KleinJan A. Stuck in the muc: infinity loop of fucosylation in allergic airway inflammation. J Allergy Clin Immunol (2019) 144:665–7. doi: 10.1016/j.jaci.2019.07.012
32. Barnes JW, Tian L, Heresi GA, Farver CF, Asosingh K, Comhair SA, et al. O-Linked beta-N-Acetylglucosamine transferase directs cell proliferation in idiopathic pulmonary arterial hypertension. Circulation (2015) 131:1260–8. doi: 10.1161/CIRCULATIONAHA.114.013878
33. Pinho SS, Reis CA. Glycosylation in cancer: mechanisms and clinical implications. Nat Rev Cancer (2015) 15:540–55. doi: 10.1038/nrc3982
34. Oliveira-Ferrer L, Legler K, Milde-Langosch K. Role of protein glycosylation in cancer metastasis. Semin Cancer Biol (2017) 44:141–52. doi: 10.1016/j.semcancer.2017.03.002
35. Sun W, Tang H, Gao L, Sun X, Liu J, Wang W, et al. Mechanisms of pulmonary fibrosis induced by core fucosylation in pericytes. Int J Biochem Cell Biol (2017) 88:44–54. doi: 10.1016/j.biocel.2017.05.010
36. Ito E, Oka R, Ishii T, Korekane H, Kurimoto A, Kizuka Y, et al. Fucosylated surfactant protein-d is a biomarker candidate for the development of chronic obstructive pulmonary disease. J Proteomics (2015) 127:386–94. doi: 10.1016/j.jprot.2015.07.011
37. Liu S, Wang H, Jiang X, Ji Y, Wang Z, Zhang Y, et al. Integrated n-glycoproteomics analysis of human saliva for lung cancer. J Proteome Res (2022) 21:1589–602. doi: 10.1021/acs.jproteome.1c00701
38. Tsai HY, Boonyapranai K, Sriyam S, Yu CJ, Wu SW, Khoo KH, et al. Glycoproteomics analysis to identify a glycoform on haptoglobin associated with lung cancer. Proteomics (2011) 11:2162–70. doi: 10.1002/pmic.201000319
39. Ueda K, Fukase Y, Katagiri T, Ishikawa N, Irie S, Sato TA, et al. Targeted serum glycoproteomics for the discovery of lung cancer-associated glycosylation disorders using lectin-coupled proteinchip arrays. Proteomics (2009) 9:2182–92. doi: 10.1002/pmic.200800374
40. Ahn JM, Sung HJ, Yoon YH, Kim BG, Yang WS, Lee C, et al. Integrated glycoproteomics demonstrates fucosylated serum paraoxonase 1 alterations in small cell lung cancer. Mol Cell Proteomics (2014) 13:30–48. doi: 10.1074/mcp.M113.028621
41. Jia L, Zhang J, Ma T, Guo Y, Yu Y, Cui J. The function of fucosylation in progression of lung cancer. Front Oncol (2018) 8:565. doi: 10.3389/fonc.2018.00565
42. Pabst M, Wu SQ, Grass J, Kolb A, Chiari C, Viernstein H, et al. Il-1beta and tnf-alpha alter the glycophenotype of primary human chondrocytes in vitro. Carbohydr Res (2010) 345:1389–93. doi: 10.1016/j.carres.2010.02.017
43. Higai K, Miyazaki N, Azuma Y, Matsumoto K. Interleukin-1beta induces sialyl Lewis X on hepatocellular carcinoma huh-7 cells Via enhanced expression of St3gal iv and fut vi gene. FEBS Lett (2006) 580:6069–75. doi: 10.1016/j.febslet.2006.09.073
44. Padro M, Mejias-Luque R, Cobler L, Garrido M, Perez-Garay M, Puig S, et al. Regulation of glycosyltransferases and Lewis antigens expression by il-1beta and il-6 in human gastric cancer cells. Glycoconj J (2011) 28:99–110. doi: 10.1007/s10719-011-9327-4
45. Jasper MJ, Care AS, Sullivan B, Ingman WV, Aplin JD, Robertson SA. Macrophage-derived lif and Il1b regulate Alpha(1,2)Fucosyltransferase 2 (Fut2) expression in mouse uterine epithelial cells during early pregnancy. Biol Reprod (2011) 84:179–88. doi: 10.1095/biolreprod.110.085399
46. Radovani B, Gudelj I. N-glycosylation and inflammation; the not-So-Sweet relation. Front Immunol (2022) 13:893365. doi: 10.3389/fimmu.2022.893365
47. Silsirivanit A. Glycosylation markers in cancer. Adv Clin Chem (2019) 89:189–213. doi: 10.1016/bs.acc.2018.12.005
48. Mereiter S, Balmana M, Campos D, Gomes J, Reis CA. Glycosylation in the era of cancer-targeted therapy: where are we heading? Cancer Cell (2019) 36:6–16. doi: 10.1016/j.ccell.2019.06.006
49. Gupta R, Leon F, Thompson CM, Nimmakayala R, Karmakar S, Nallasamy P, et al. Global analysis of human glycosyltransferases reveals novel targets for pancreatic cancer pathogenesis. Br J Cancer (2020) 122:1661–72. doi: 10.1038/s41416-020-0772-3
50. Fernandez-Ponce C, Geribaldi-Doldan N, Sanchez-Gomar I, Quiroz RN, Ibarra LA, Escorcia LG, et al. The role of glycosyltransferases in colorectal cancer. Int J Mol Sci (2021) 22:5822. doi: 10.3390/ijms22115822
51. Burchell JM, Beatson R, Graham R, Taylor-Papadimitriou J, Tajadura-Ortega V. O-Linked mucin-type glycosylation in breast cancer. Biochem Soc Trans (2018) 46:779–88. doi: 10.1042/BST20170483
52. Li Y, Lin Y, Aye L, Dong L, Zhang C, Chen F, et al. An integrative pan-cancer analysis of the molecular and biological features of glycosyltransferases. Clin Transl Med (2022) 12:e872. doi: 10.1002/ctm2.872
53. Martin H, Lazaro LR, Gunnlaugsson T, Scanlan EM. Glycosidase activated prodrugs for targeted cancer therapy. Chem Soc Rev (2022) 51:9694–716. doi: 10.1039/d2cs00379a
54. Zhang X, Han Y, Huang W, Jin M, Gao Z. The influence of the gut microbiota on the bioavailability of oral drugs. Acta Pharm Sin B (2021) 11:1789–812. doi: 10.1016/j.apsb.2020.09.013
55. Souza C, Pellosi DS, Tedesco AC. Prodrugs for targeted cancer therapy. Expert Rev Anticancer Ther (2019) 19:483–502. doi: 10.1080/14737140.2019.1615890
56. Wu TJ, Wang SH, Chen ES, Tsai HH, Chang YC, Tseng YH, et al. Loss of core-fucosylation of sparc impairs collagen binding and contributes to copd. Cell Mol Life Sci (2022) 79:348. doi: 10.1007/s00018-022-04381-4
57. Krick S, Helton ES, Easter M, Bollenbecker S, Denson R, Zaharias R, et al. St6gal1 and Alpha2-6 sialylation regulates il-6 expression and secretion in chronic obstructive pulmonary disease. Front Immunol (2021) 12:693149. doi: 10.3389/fimmu.2021.693149
58. Nasirikenari M, Lugade AA, Neelamegham S, Gao Z, Moremen KW, Bogner PN, et al. Recombinant sialyltransferase infusion mitigates infection-driven acute lung inflammation. Front Immunol (2019) 10:48. doi: 10.3389/fimmu.2019.00048
59. Raclawska DS, Ttofali F, Fletcher AA, Harper DN, Bochner BS, Janssen WJ, et al. Mucins and their sugars. critical mediators of hyperreactivity and inflammation. Ann Am Thorac Soc (2016) 13 Suppl 1:S98–9. doi: 10.1513/AnnalsATS.201511-743MG
60. Katoh S. Critical involvement of Cd44 in T helper type 2 cell-mediated eosinophilic airway inflammation in a mouse model of acute asthma. Front Immunol (2021) 12:811600. doi: 10.3389/fimmu.2021.811600
61. Schulz BL, Sloane AJ, Robinson LJ, Prasad SS, Lindner RA, Robinson M, et al. Glycosylation of sputum mucins is altered in cystic fibrosis patients. Glycobiology (2007) 17:698–712. doi: 10.1093/glycob/cwm036
62. Stoykova LI, Liu A, Scanlin TF, Glick MC. Alpha1,3fucosyltransferases in cystic fibrosis airway epithelial cells. Biochimie (2003) 85:363–7. doi: 10.1016/s0300-9084(03)00061-0
63. Venkatakrishnan V, Thaysen-Andersen M, Chen SC, Nevalainen H, Packer NH. Cystic fibrosis and bacterial colonization define the sputum n-glycosylation phenotype. Glycobiology (2015) 25:88–100. doi: 10.1093/glycob/cwu092
64. Dosanjh A, Muchmore EA. Expression of Deltaf508 cystic fibrosis transmembrane regulator (Cftr) decreases membrane sialylation. Open Respir Med J (2009) 3:79–84. doi: 10.2174/1874306400903010079
65. Nakanishi T, Cerani A, Forgetta V, Zhou S, Allen RJ, Leavy OC, et al. Genetically increased circulating Fut3 level leads to reduced risk of idiopathic pulmonary fibrosis: a mendelian randomisation study. Eur Respir J (2022) 59:2003979. doi: 10.1183/13993003.03979-2020
66. Sun W, Jing X, Yang X, Huang H, Luo Q, Xia S, et al. Regulation of the Igf1 signaling pathway is involved in idiopathic pulmonary fibrosis induced by alveolar epithelial cell senescence and core fucosylation. Aging (2021) 13:18852–69. doi: 10.18632/aging.203335
67. Gao L, Wang N, Jiang Y, Hu J, Ma B, Wu T. Glycyrrhizic acid inhibits core fucosylation modification modulated emt and attenuates bleomycin-induced pulmonary fibrosis. Evid Based Complement Alternat Med (2022) 2022:5943322. doi: 10.1155/2022/5943322
68. Luzina IG, Lockatell V, Hyun SW, Kopach P, Kang PH, Noor Z, et al. Elevated expression of Neu1 sialidase in idiopathic pulmonary fibrosis provokes pulmonary collagen deposition, lymphocytosis, and fibrosis. Am J Physiol Lung Cell Mol Physiol (2016) 310:L940–54. doi: 10.1152/ajplung.00346.2015
69. Chen W, Karhadkar TR, Ryu C, Herzog EL, Gomer RH. Reduced sialylation and bioactivity of the antifibrotic protein serum amyloid p in the sera of patients with idiopathic pulmonary fibrosis. Immunohorizons (2020) 4:352–62. doi: 10.4049/immunohorizons.2000043
70. Karhadkar TR, Chen W, Gomer RH. Attenuated pulmonary fibrosis in sialidase-3 knockout (Neu3(-/-)) mice. Am J Physiol Lung Cell Mol Physiol (2020) 318:L165–L79. doi: 10.1152/ajplung.00275.2019
71. Aulak KS, Barnes JW, Tian L, Mellor NE, Haque MM, Willard B, et al. Specific O-glcnac modification at ser-615 modulates enos function. Redox Biol (2020) 36:101625. doi: 10.1016/j.redox.2020.101625
72. Taylor SL, Woodman RJ, Chen AC, Burr LD, Gordon DL, McGuckin MA, et al. Fut2 genotype influences lung function, exacerbation frequency and airway microbiota in non-cf bronchiectasis. Thorax (2017) 72:304–10. doi: 10.1136/thoraxjnl-2016-208775
73. Zhuge J, Wang X, Li J, Wang T, Wang H, Yang M, et al. Construction of the model for predicting prognosis by key genes regulating egfr-tki resistance. Front Genet (2022) 13:968376. doi: 10.3389/fgene.2022.968376
74. Zhang Y, Yao E, Liu Y, Zhang Y, Ding M, Liu J, et al. Fut2 facilitates autophagy and suppresses apoptosis Via P53 and jnk signaling in lung adenocarcinoma cells. Cells (2022) 11:4031. doi: 10.3390/cells11244031
75. Deng G, Chen L, Zhang Y, Fan S, Li W, Lu J, et al. Fucosyltransferase 2 induced epithelial-mesenchymal transition Via tgf-Beta/Smad signaling pathway in lung adenocarcinaoma. Exp Cell Res (2018) 370:613–22. doi: 10.1016/j.yexcr.2018.07.026
76. Wang K, Chen X. Autophagic tumor-associated macrophages promote the endothelial mesenchymal transition in lung adenocarcinomas through the Fut4/P-ezrin pathway. J Thorac Dis (2021) 13:5973–85. doi: 10.21037/jtd-21-1519
77. Tian L, Shen D, Li X, Shan X, Wang X, Yan Q, et al. Ginsenoside Rg3 inhibits epithelial-mesenchymal transition (Emt) and invasion of lung cancer by down-regulating Fut4. Oncotarget (2016) 7:1619–32. doi: 10.18632/oncotarget.6451
78. Li F, Zhao S, Cui Y, Guo T, Qiang J, Xie Q, et al. Alpha1,6-fucosyltransferase (Fut8) regulates the cancer-promoting capacity of cancer-associated fibroblasts (Cafs) by modifying egfr core fucosylation (Cf) in non-small cell lung cancer (Nsclc). Am J Cancer Res (2020) 10:816–37.
79. Li J, Long Y, Sun J, Wu J, He X, Wang S, et al. Comprehensive landscape of the St3gal family reveals the significance of St3gal6-As1/St3gal6 axis on egfr signaling in lung adenocarcinoma cell invasion. Front Cell Dev Biol (2022) 10:931132. doi: 10.3389/fcell.2022.931132
80. Yuan Q, Chen X, Han Y, Lei T, Wu Q, Yu X, et al. Modification of Alpha2,6-sialylation mediates the invasiveness and tumorigenicity of non-small cell lung cancer cells in vitro and in vivo Via Notch1/Hes1/Mmps pathway. Int J Cancer (2018) 143:2319–30. doi: 10.1002/ijc.31737
81. Lakshmanan I, Chaudhary S, Vengoji R, Seshacharyulu P, Rachagani S, Carmicheal J, et al. St6galnac-I promotes lung cancer metastasis by altering Muc5ac sialylation. Mol Oncol (2021) 15:1866–81. doi: 10.1002/1878-0261.12956
82. Singh P, Joon A, Kumari M, Singh T, Bal A, Maan P, et al. Role of a disease-associated St3gal-4 in non-small cell lung cancer. Cell Biochem Biophys (2022) 80:781–93. doi: 10.1007/s12013-022-01091-3
83. Efeoglu E, Henry M, Clynes M, Meleady P. Label-free quantitative proteomics analysis of adriamycin selected multidrug resistant human lung cancer cells. Biomolecules (2022) 12:1401. doi: 10.3390/biom12101401
84. Lv T, Lv H, Fei J, Xie Y, Lian D, Hu J, et al. P53-R273h promotes cancer cell migration Via upregulation of neuraminidase-1. J Cancer (2020) 11:6874–82. doi: 10.7150/jca.44718
85. Forcella M, Oldani M, Epistolio S, Freguia S, Monti E, Fusi P, et al. Non-small cell lung cancer (Nsclc), egfr downstream pathway activation and tki targeted therapies sensitivity: effect of the plasma membrane-associated Neu3. PloS One (2017) 12:e0187289. doi: 10.1371/journal.pone.0187289
86. Zhong A, Chen T, Xing Y, Pan X, Shi M. Fuca2 is a prognostic biomarker and correlated with an immunosuppressive microenvironment in pan-cancer. Front Immunol (2021) 12:758648. doi: 10.3389/fimmu.2021.758648
87. Lin CH, Liao CC, Wang SY, Peng CY, Yeh YC, Chen MY, et al. Comparative O-glcnac proteomic analysis reveals a role of O-glcnacylated Sam68 in lung cancer aggressiveness. Cancers (Basel) (2022) 14:243. doi: 10.3390/cancers14010243
88. Luanpitpong S, Angsutararux P, Samart P, Chanthra N, Chanvorachote P, Issaragrisil S. Hyper-O-Glcnacylation induces cisplatin resistance Via regulation of P53 and c-myc in human lung carcinoma. Sci Rep (2017) 7:10607. doi: 10.1038/s41598-017-10886-x
89. Tang D, Li GS, Xu RX, Zhu SY, Luo J, Zheng JH, et al. Ogt demonstrated conspicuous clinical significance in cancers, from pan-cancer to small-cell lung cancer. J Oncol (2022) 2022:2010341. doi: 10.1155/2022/2010341
90. Khaodee W, Inboot N, Udomsom S, Kumsaiyai W, Cressey R. Glucosidase ii beta subunit (Gluiibeta) plays a role in autophagy and apoptosis regulation in lung carcinoma cells in a P53-dependent manner. Cell Oncol (Dordr) (2017) 40:579–91. doi: 10.1007/s13402-017-0349-1
91. Guo H, Abbott KL. Functional impact of tumor-specific n-linked glycan changes in breast and ovarian cancers. Adv Cancer Res (2015) 126:281–303. doi: 10.1016/bs.acr.2014.11.006
92. Goto Y, Uematsu S, Kiyono H. Epithelial glycosylation in gut homeostasis and inflammation. Nat Immunol (2016) 17:1244–51. doi: 10.1038/ni.3587
93. Shan M, Yang D, Dou H, Zhang L. Fucosylation in cancer biology and its clinical applications. Prog Mol Biol Transl Sci (2019) 162:93–119. doi: 10.1016/bs.pmbts.2019.01.002
94. Miyoshi E, Shinzaki S, Moriwaki K, Matsumoto H. Identification of fucosylated haptoglobin as a novel tumor marker for pancreatic cancer and its possible application for a clinical diagnostic test. Methods Enzymol (2010) 478:153–64. doi: 10.1016/S0076-6879(10)78006-X
95. Durham AL, Adcock IM. The relationship between copd and lung cancer. Lung Cancer (2015) 90:121–7. doi: 10.1016/j.lungcan.2015.08.017
96. Taniguchi N, Ohkawa Y, Maeda K, Kanto N, Johnson EL, Harada Y. N-glycan branching enzymes involved in cancer, alzheimer’s disease and copd and future perspectives. Biochem Biophys Res Commun (2022) 633:68–71. doi: 10.1016/j.bbrc.2022.09.027
97. Yamada M, Ishii T, Ikeda S, Naka-Mieno M, Tanaka N, Arai T, et al. Association of fucosyltransferase 8 (Fut8) polymorphism Thr267lys with pulmonary emphysema. J Hum Genet (2011) 56:857–60. doi: 10.1038/jhg.2011.118
98. Wang Z, Li R, Zhong R. Extracellular matrix promotes proliferation, migration and adhesion of airway smooth muscle cells in a rat model of chronic obstructive pulmonary disease Via upregulation of the Pi3k/Akt signaling pathway. Mol Med Rep (2018) 18:3143–52. doi: 10.3892/mmr.2018.9320
99. Wang X, Inoue S, Gu J, Miyoshi E, Noda K, Li W, et al. Dysregulation of tgf-Beta1 receptor activation leads to abnormal lung development and emphysema-like phenotype in core fucose-deficient mice. Proc Natl Acad Sci U.S.A. (2005) 102:15791–6. doi: 10.1073/pnas.0507375102
100. Halwani R, Al-Muhsen S, Al-Jahdali H, Hamid Q. Role of transforming growth factor-beta in airway remodeling in asthma. Am J Respir Cell Mol Biol (2011) 44:127–33. doi: 10.1165/rcmb.2010-0027TR
101. Azad MB, Wade KH, Timpson NJ. Fut2 secretor genotype and susceptibility to infections and chronic conditions in the alspac cohort. Wellcome Open Res (2018) 3:65. doi: 10.12688/wellcomeopenres.14636.2
102. Epp A, Sullivan KC, Herr AB, Strait RT. Immunoglobulin glycosylation effects in allergy and immunity. Curr Allergy Asthma Rep (2016) 16:79. doi: 10.1007/s11882-016-0658-x
103. Thomsson KA, Hinojosa-Kurtzberg M, Axelsson KA, Domino SE, Lowe JB, Gendler SJ, et al. Intestinal mucins from cystic fibrosis mice show increased fucosylation due to an induced Fucalpha1-2 glycosyltransferase. Biochem J (2002) 367:609–16. doi: 10.1042/BJ20020371
104. Zheng Q, Cox IA, Campbell JA, Xia Q, Otahal P, de Graaff B, et al. Mortality and survival in idiopathic pulmonary fibrosis: a systematic review and meta-analysis. ERJ Open Res (2022) 8:00591–2021. doi: 10.1183/23120541.00591-2021
105. Nicolaou N, Margadant C, Kevelam SH, Lilien MR, Oosterveld MJ, Kreft M, et al. Gain of glycosylation in integrin Alpha3 causes lung disease and nephrotic syndrome. J Clin Invest (2012) 122:4375–87. doi: 10.1172/JCI64100
106. Hernandez DM, Kang JH, Choudhury M, Andrianifahanana M, Yin X, Limper AH, et al. Ipf pathogenesis is dependent upon tgfbeta induction of igf-1. FASEB J (2020) 34:5363–88. doi: 10.1096/fj.201901719RR
107. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2021) 71:209–49. doi: 10.3322/caac.21660
108. Blandin Knight S, Crosbie PA, Balata H, Chudziak J, Hussell T, Dive C. Progress and prospects of early detection in lung cancer. Open Biol (2017) 7:170070. doi: 10.1098/rsob.170070
109. Gao Z, Xu M, Yue S, Shan H, Xia J, Jiang J, et al. Abnormal sialylation and fucosylation of saliva glycoproteins: characteristics of lung cancer-specific biomarkers. Curr Res Pharmacol Drug Discovery (2022) 3:100079. doi: 10.1016/j.crphar.2021.100079
110. Leng Q, Tsou JH, Zhan M, Jiang F. Fucosylation genes as circulating biomarkers for lung cancer. J Cancer Res Clin Oncol (2018) 144:2109–15. doi: 10.1007/s00432-018-2735-0
111. Ferreira IG, Carrascal M, Mineiro AG, Bugalho A, Borralho P, Silva Z, et al. Carcinoembryonic antigen is a sialyl Lewis x/a carrier and an E−Selectin ligand in Non−Small cell lung cancer. Int J Oncol (2019) 55:1033–48. doi: 10.3892/ijo.2019.4886
112. Park S, Lim JM, Chun JN, Lee S, Kim TM, Kim DW, et al. Altered expression of fucosylation pathway genes is associated with poor prognosis and tumor metastasis in Non−Small cell lung cancer. Int J Oncol (2020) 56:559–67. doi: 10.3892/ijo.2019.4953
113. Liu C, Li Z, Wang S, Fan Y, Zhang S, Yang X, et al. Fut4 is involved in pd-1-Related immunosuppression and leads to worse survival in patients with operable lung adenocarcinoma. J Cancer Res Clin Oncol (2019) 145:65–76. doi: 10.1007/s00432-018-2761-y
114. Lu HH, Lin SY, Weng RR, Juan YH, Chen YW, Hou HH, et al. Fucosyltransferase 4 shapes oncogenic glycoproteome to drive metastasis of lung adenocarcinoma. EBioMedicine (2020) 57:102846. doi: 10.1016/j.ebiom.2020.102846
115. Ding Y, Zhen Z, Nisar MA, Ali F, Din RU, Khan M, et al. Sesquiterpene lactones attenuate paclitaxel resistance Via inhibiting Malat1/Stat3/Fut4 axis and p-glycoprotein transporters in lung cancer cells. Front Pharmacol (2022) 13:795613. doi: 10.3389/fphar.2022.795613
116. Gao W, Liang J, Ye Y, Lu J, Lin T, Wang N, et al. Fut4sirna augments the chemosensitivity of non-small cell lung cancer to cisplatin through activation of Foxo1-induced apoptosis. BMC Cancer (2020) 20:895. doi: 10.1186/s12885-020-07324-z
117. Jassam SA, Maherally Z, Ashkan K, Pilkington GJ, Fillmore HL. Fucosyltransferase 4 and 7 mediates adhesion of non-small cell lung cancer cells to brain-derived endothelial cells and results in modification of the blood-Brain-Barrier: In vitro investigation of Cd15 and Cd15s in lung-to-Brain metastasis. J Neurooncol (2019) 143:405–15. doi: 10.1007/s11060-019-03188-x
118. Fang Y, Qu Y, Ji L, Sun H, Li J, Zhao Y, et al. Novel blood-based Fut7 DNA methylation is associated with lung cancer: especially for lung squamous cell carcinoma. Clin Epigenet (2022) 14:167. doi: 10.1186/s13148-022-01389-2
119. Liang JX, Gao W, Cai L. Fucosyltransferase vii promotes proliferation Via the Egfr/Akt/Mtor pathway in A549 cells. Onco Targets Ther (2017) 10:3971–8. doi: 10.2147/OTT.S140940
120. Okada M, Chikuma S, Kondo T, Hibino S, Machiyama H, Yokosuka T, et al. Blockage of core fucosylation reduces cell-surface expression of pd-1 and promotes anti-tumor immune responses of T cells. Cell Rep (2017) 20:1017–28. doi: 10.1016/j.celrep.2017.07.027
121. Zhang N, Li M, Xu X, Zhang Y, Liu Y, Zhao M, et al. Loss of core fucosylation enhances the anticancer activity of cytotoxic T lymphocytes by increasing pd-1 degradation. Eur J Immunol (2020) 50:1820–33. doi: 10.1002/eji.202048543
122. Xu Y, Gao Z, Hu R, Wang Y, Wang Y, Su Z, et al. Pd-L2 glycosylation promotes immune evasion and predicts anti-egfr efficacy. J Immunother Cancer (2021) 9:e002699. doi: 10.1136/jitc-2021-002699
123. Kuang M, Wu H, Hu L, Guo X, He D, Liu B, et al. Up-regulation of Fut8 inhibits tgf-Beta1-Induced activation of hepatic stellate cells during liver fibrogenesis. Glycoconj J (2021) 38:77–87. doi: 10.1007/s10719-021-09975-x
124. Lin S, Zhou L, Dong Y, Yang Q, Yang Q, Jin H, et al. Alpha-(1,6)-Fucosyltransferase (Fut8) affects the survival strategy of osteosarcoma by remodeling Tnf/Nf-Kappab2 signaling. Cell Death Dis (2021) 12:1124. doi: 10.1038/s41419-021-04416-x
125. Hoti N, Lih TS, Pan J, Zhou Y, Yang G, Deng A, et al. A comprehensive analysis of Fut8 overexpressing prostate cancer cells reveals the role of egfr in castration resistance. Cancers (Basel) (2020) 12:468. doi: 10.3390/cancers12020468
126. Kelel M, Yang RB, Tsai TF, Liang PH, Wu FY, Huang YT, et al. Fut8 remodeling of egfr regulates epidermal keratinocyte proliferation during psoriasis development. J Invest Dermatol (2021) 141:512–22. doi: 10.1016/j.jid.2020.07.030
127. Huang Y, Zhang HL, Li ZL, Du T, Chen YH, Wang Y, et al. Fut8-mediated aberrant n-glycosylation of B7h3 suppresses the immune response in triple-negative breast cancer. Nat Commun (2021) 12:2672. doi: 10.1038/s41467-021-22618-x
128. Zhao Y, Itoh S, Wang X, Isaji T, Miyoshi E, Kariya Y, et al. Deletion of core fucosylation on Alpha3beta1 integrin down-regulates its functions. J Biol Chem (2006) 281:38343–50. doi: 10.1074/jbc.M608764200
129. Liu D, Gao Z, Yue L. Fucosyltransferase 8 deficiency suppresses breast cancer cell migration by interference of the Fak/Integrin pathway. Cancer biomark (2019) 25:303–11. doi: 10.3233/CBM-190209
130. Cantero-Recasens G, Burballa C, Ohkawa Y, Fukuda T, Harada Y, Consortium IBDC, et al. The ulcerative colitis-associated gene Fut8 regulates the quantity and quality of secreted mucins. Proc Natl Acad Sci U.S.A. (2022) 119:e2205277119. doi: 10.1073/pnas.2205277119
131. Shao K, Chen ZY, Gautam S, Deng NH, Zhou Y, Wu XZ. Posttranslational modification of e-cadherin by core fucosylation regulates src activation and induces epithelial-mesenchymal transition-like process in lung cancer cells. Glycobiology (2016) 26:142–54. doi: 10.1093/glycob/cwv089
132. Montgomery AP, Xiao K, Wang X, Skropeta D, Yu H. Computational glycobiology: mechanistic studies of carbohydrate-active enzymes and implication for inhibitor design. Adv Protein Chem Struct Biol (2017) 109:25–76. doi: 10.1016/bs.apcsb.2017.04.003
133. Thornton CS, Somayaji R, Lim RK. Assessment of factors and interventions towards therapeutic adherence among persons with non-cystic fibrosis bronchiectasis. ERJ Open Res (2022) 8:00340–2022. doi: 10.1183/23120541.00340-2022
134. Goeminne P, Dupont L. Non-cystic fibrosis bronchiectasis: diagnosis and management in 21st century. Postgrad Med J (2010) 86:493–501. doi: 10.1136/pgmj.2009.091041
135. Schafer J, Griese M, Chandrasekaran R, Chotirmall SH, Hartl D. Pathogenesis, imaging and clinical characteristics of cf and non-cf bronchiectasis. BMC Pulm Med (2018) 18:79. doi: 10.1186/s12890-018-0630-8
136. Mandal P, Hill AT. Bronchiectasis: breaking the cycle of inflammation and infection. Lancet Respir Med (2013) 1:e5–6. doi: 10.1016/S2213-2600(13)70005-5
137. Harduin-Lepers A, Vallejo-Ruiz V, Krzewinski-Recchi MA, Samyn-Petit B, Julien S, Delannoy P. The human sialyltransferase family. Biochimie (2001) 83:727–37. doi: 10.1016/s0300-9084(01)01301-3
138. Huang RB, Cheng D, Liao SM, Lu B, Wang QY, Xie NZ, et al. The intrinsic relationship between structure and function of the sialyltransferase St8sia family members. Curr Top Med Chem (2017) 17:2359–69. doi: 10.2174/1568026617666170414150730
139. Lunemann JD, von Gunten S, Neumann H. Targeting sialylation to treat central nervous system diseases. Trends Pharmacol Sci (2021) 42:998–1008. doi: 10.1016/j.tips.2021.09.002
140. Munkley J. Aberrant sialylation in cancer: therapeutic opportunities. Cancers (Basel) (2022) 14:4248. doi: 10.3390/cancers14174248
141. Li F, Ding J. Sialylation is involved in cell fate decision during development, reprogramming and cancer progression. Protein Cell (2019) 10:550–65. doi: 10.1007/s13238-018-0597-5
142. Yang H, Tian Z. Sialic acid linkage-specific quantitative n-glycoproteomics using selective alkylamidation and multiplex tmt-labeling. Anal Chim Acta (2022) 1230:340391. doi: 10.1016/j.aca.2022.340391
143. Meszaros B, Jarvas G, Farkas A, Szigeti M, Kovacs Z, Kun R, et al. Comparative analysis of the human serum n-glycome in lung cancer, copd and their comorbidity using capillary electrophoresis. J Chromatogr B Analyt Technol BioMed Life Sci (2020) 1137:121913. doi: 10.1016/j.jchromb.2019.121913
144. Wangberg H, Woessner K. Choice of biologics in asthma endotypes. Curr Opin Allergy Clin Immunol (2021) 21:79–85. doi: 10.1097/ACI.0000000000000708
145. Yeh YL, Wu WC, Kannagi R, Chiang BL, Liu FT, Lee YL. Sialyl glycan expression on T cell subsets in asthma: a correlation with disease severity and blood parameters. Sci Rep (2019) 9:8947. doi: 10.1038/s41598-019-45040-2
146. Anthony RM, Nimmerjahn F. The role of differential igg glycosylation in the interaction of antibodies with fcgammars in vivo. Curr Opin Organ Transplant (2011) 16:7–14. doi: 10.1097/MOT.0b013e328342538f
148. Caballero I, Ringot-Destrez B, Si-Tahar M, Barbry P, Guillon A, Lantier I, et al. Evidence of early increased sialylation of airway mucins and defective mucociliary clearance in cftr-deficient piglets. J Cyst Fibros (2021) 20:173–82. doi: 10.1016/j.jcf.2020.09.009
149. Colomb F, Krzewinski-Recchi MA, Steenackers A, Vincent A, Harduin-Lepers A, Delannoy P, et al. Tnf up-regulates St3gal4 and sialyl-lewisx expression in lung epithelial cells through an intronic Atf2-responsive element. Biochem J (2017) 474:65–78. doi: 10.1042/BCJ20160602
150. Delmotte P, Degroote S, Lafitte JJ, Lamblin G, Perini JM, Roussel P. Tumor necrosis factor alpha increases the expression of glycosyltransferases and sulfotransferases responsible for the biosynthesis of sialylated and/or sulfated Lewis X epitopes in the human bronchial mucosa. J Biol Chem (2002) 277:424–31. doi: 10.1074/jbc.M109958200
151. Groux-Degroote S, Krzewinski-Recchi MA, Cazet A, Vincent A, Lehoux S, Lafitte JJ, et al. Il-6 and il-8 increase the expression of glycosyltransferases and sulfotransferases involved in the biosynthesis of sialylated and/or sulfated lewisx epitopes in the human bronchial mucosa. Biochem J (2008) 410:213–23. doi: 10.1042/BJ20070958
152. Holst S, Wuhrer M, Rombouts Y. Glycosylation characteristics of colorectal cancer. Adv Cancer Res (2015) 126:203–56. doi: 10.1016/bs.acr.2014.11.004
153. Lee WL, Wang PH. Aberrant sialylation in ovarian cancers. J Chin Med Assoc (2020) 83:337–44. doi: 10.1097/JCMA.0000000000000252
154. Yang X, Qian K. Protein O-glcnacylation: emerging mechanisms and functions. Nat Rev Mol Cell Biol (2017) 18:452–65. doi: 10.1038/nrm.2017.22
155. Yuzwa SA, Vocadlo DJ. O-Glcnac and neurodegeneration: biochemical mechanisms and potential roles in alzheimer’s disease and beyond. Chem Soc Rev (2014) 43:6839–58. doi: 10.1039/c4cs00038b
156. Zhu Y, Shan X, Yuzwa SA, Vocadlo DJ. The emerging link between O-glcnac and Alzheimer disease. J Biol Chem (2014) 289:34472–81. doi: 10.1074/jbc.R114.601351
157. Liu C, Dong W, Li J, Kong Y, Ren X. O-Glcnac modification and its role in diabetic retinopathy. Metabolites (2022) 12:725. doi: 10.3390/metabo12080725
158. Dias WB, Hart GW. O-Glcnac modification in diabetes and alzheimer’s disease. Mol Biosyst (2007) 3:766–72. doi: 10.1039/b704905f
159. Parker MP, Peterson KR, Slawson C. O-Glcnacylation and O-glcnac cycling regulate gene transcription: emerging roles in cancer. Cancers (Basel) (2021) 13:1666. doi: 10.3390/cancers13071666
160. Hu CW, Xie J, Jiang J. The emerging roles of protein interactions with O-glcnac cycling enzymes in cancer. Cancers (Basel) (2022) 14:5135. doi: 10.3390/cancers14205135
161. Zhu Y, Hart GW. Targeting O-glcnacylation to develop novel therapeutics. Mol Aspects Med (2021) 79:100885. doi: 10.1016/j.mam.2020.100885
162. Lu Q, Zhang X, Liang T, Bai X. O-Glcnacylation: an important post-translational modification and a potential therapeutic target for cancer therapy. Mol Med (2022) 28:115. doi: 10.1186/s10020-022-00544-y
163. Liu B, Zhu L, Yuan P, Marsboom G, Hong Z, Liu J, et al. Comprehensive identification of signaling pathways for idiopathic pulmonary arterial hypertension. Am J Physiol Cell Physiol (2020) 318:C913–C30. doi: 10.1152/ajpcell.00382.2019
164. Barnes JW, Tian L, Krick S, Helton ES, Denson RS, Comhair SAA, et al. O-Glcnac transferase regulates angiogenesis in idiopathic pulmonary arterial hypertension. Int J Mol Sci (2019) 20:6299. doi: 10.3390/ijms20246299
165. Lazar Z, Meszaros M, Bikov A. The nitric oxide pathway in pulmonary arterial hypertension: pathomechanism, biomarkers and drug targets. Curr Med Chem (2020) 27:7168–88. doi: 10.2174/0929867327666200522215047
166. Mi W, Gu Y, Han C, Liu H, Fan Q, Zhang X, et al. O-Glcnacylation is a novel regulator of lung and colon cancer malignancy. Biochim Biophys Acta (2011) 1812:514–9. doi: 10.1016/j.bbadis.2011.01.009
167. Taparra K, Wang H, Malek R, Lafargue A, Barbhuiya MA, Wang X, et al. O-Glcnacylation is required for mutant kras-induced lung tumorigenesis. J Clin Invest (2018) 128:4924–37. doi: 10.1172/JCI94844
168. Shimizu M, Tanaka N. Il-8-Induced O-glcnac modification Via Glut3 and gfat regulates cancer stem cell-like properties in colon and lung cancer cells. Oncogene (2019) 38:1520–33. doi: 10.1038/s41388-018-0533-4
169. Ge X, Peng X, Li M, Ji F, Chen J, Zhang D. Ogt regulated O-glcnacylation promotes migration and invasion by activating il-6/Stat3 signaling in nsclc cells. Pathol Res Pract (2021) 225:153580. doi: 10.1016/j.prp.2021.153580
170. Glanz VY, Myasoedova VA, Grechko AV, Orekhov AN. Sialidase activity in human pathologies. Eur J Pharmacol (2019) 842:345–50. doi: 10.1016/j.ejphar.2018.11.014
171. Cross AS, Hyun SW, Miranda-Ribera A, Feng C, Liu A, Nguyen C, et al. Neu1 and Neu3 sialidase activity expressed in human lung microvascular endothelia: Neu1 restrains endothelial cell migration, whereas Neu3 does not. J Biol Chem (2012) 287:15966–80. doi: 10.1074/jbc.M112.346817
172. Hyun SW, Liu A, Liu Z, Cross AS, Verceles AC, Magesh S, et al. The Neu1-selective sialidase inhibitor, C9-Butyl-Amide-Dana, blocks sialidase activity and Neu1-mediated bioactivities in human lung in vitro and murine lung in vivo. Glycobiology (2016) 26:834–49. doi: 10.1093/glycob/cww060
173. Lillehoj EP, Hyun SW, Feng C, Zhang L, Liu A, Guang W, et al. Human airway epithelia express catalytically active Neu3 sialidase. Am J Physiol Lung Cell Mol Physiol (2014) 306:L876–86. doi: 10.1152/ajplung.00322.2013
174. Luzina IG, Lillehoj EP, Lockatell V, Hyun SW, Lugkey KN, Imamura A, et al. Therapeutic effect of neuraminidase-1-Selective inhibition in mouse models of bleomycin-induced pulmonary inflammation and fibrosis. J Pharmacol Exp Ther (2021) 376:136–46. doi: 10.1124/jpet.120.000223
175. Pilling D, Sahlberg K, Karhadkar TR, Chen W, Gomer RH. The sialidase Neu3 promotes pulmonary fibrosis in mice. Respir Res (2022) 23:215. doi: 10.1186/s12931-022-02146-y
176. Bosslet K, Straub R, Blumrich M, Czech J, Gerken M, Sperker B, et al. Elucidation of the mechanism enabling tumor selective prodrug monotherapy. Cancer Res (1998) 58:1195–201.
177. Khaodee W, Udomsom S, Kunnaja P, Cressey R. Knockout of glucosidase ii beta subunit inhibits growth and metastatic potential of lung cancer cells by inhibiting receptor tyrosine kinase activities. Sci Rep (2019) 9:10394. doi: 10.1038/s41598-019-46701-y
178. Lei R, Zhou M, Zhang S, Luo J, Qu C, Wang Y, et al. Potential role of prkcsh in lung cancer: bioinformatics analysis and a case study of nano zno. Nanoscale (2022) 14:4495–510. doi: 10.1039/d1nr08133k
179. Wu SW, Ho YC, Luo CW, Chen HY, Su CH, Kuan YH. Oral treatment for diabetes using alpha-glucosidase inhibitors was a risk factor for chronic obstructive pulmonary disease: a cohort study. Int J Med Sci (2021) 18:778–84. doi: 10.7150/ijms.55361
180. Chen KY, Wu SM, Tseng CH, Lee KY, Lin YH, Liu HY, et al. Combination therapies with thiazolidinediones are associated with a lower risk of acute exacerbations in new-onset copd patients with advanced diabetic mellitus: a cohort-based case-control study. BMC Pulm Med (2021) 21:141. doi: 10.1186/s12890-021-01505-7
181. Lu LQ, Liao W. Screening and functional pathway analysis of genes associated with pediatric allergic asthma using a DNA microarray. Mol Med Rep (2015) 11:4197–203. doi: 10.3892/mmr.2015.3277
182. Katoh S, Maeda S, Fukuoka H, Wada T, Moriya S, Mori A, et al. A crucial role of sialidase Neu1 in hyaluronan receptor function of Cd44 in T helper type 2-mediated airway inflammation of murine acute asthmatic model. Clin Exp Immunol (2010) 161:233–41. doi: 10.1111/j.1365-2249.2010.04165.x
183. Hultberg B, Ceder O, Kollberg H. Acid hydrolases in sera and plasma from patients with cystic fibrosis. Clin Chim Acta (1981) 112:167–75. doi: 10.1016/0009-8981(81)90375-2
184. Zhang L, Gallup M, Zlock L, Chen YT, Finkbeiner WE, McNamara NA. Pivotal role of Muc1 glycosylation by cigarette smoke in modulating disruption of airway adherens junctions in vitro. J Pathol (2014) 234:60–73. doi: 10.1002/path.4375
185. Matos R, Fonseca KL, Mereiter S, Maceiras AR, Gomes J, Vilaplana C, et al. Mycobacterium tuberculosis infection up-regulates sialyl Lewis X expression in the lung epithelium. Microorganisms (2021) 9:99. doi: 10.3390/microorganisms9010099
186. Silveyra P, Fuentes N, Rodriguez Bauza DE. Sex and gender differences in lung disease. Adv Exp Med Biol (2021) 1304:227–58. doi: 10.1007/978-3-030-68748-9_14
187. Camp PG, Dimich-Ward H, Kennedy SM. Women and occupational lung disease: sex differences and gender influences on research and disease outcomes. Clin Chest Med (2004) 25:269–79. doi: 10.1016/j.ccm.2004.01.004
188. Siegfried JM. Sex and gender differences in lung cancer and chronic obstructive lung disease. Endocrinology (2022) 163:bqab254. doi: 10.1210/endocr/bqab254
189. Ntritsos G, Franek J, Belbasis L, Christou MA, Markozannes G, Altman P, et al. Gender-specific estimates of copd prevalence: a systematic review and meta-analysis. Int J Chron Obstruct Pulmon Dis (2018) 13:1507–14. doi: 10.2147/COPD.S146390
190. Postma DS. Gender differences in asthma development and progression. Gend Med (2007) 4 Suppl B:S133–46. doi: 10.1016/s1550-8579(07)80054-4
191. Harness-Brumley CL, Elliott AC, Rosenbluth DB, Raghavan D, Jain R. Gender differences in outcomes of patients with cystic fibrosis. J Womens Health (Larchmt) (2014) 23:1012–20. doi: 10.1089/jwh.2014.4985
192. Brooke-Hollidge A, Conway J, Lewis A. Gender differences in non-cystic fibrosis bronchiectasis severity and bacterial load: the potential role of hormones. Ther Adv Respir Dis (2021) 15:17534666211035311. doi: 10.1177/17534666211035311
193. Ercan A, Kohrt WM, Cui J, Deane KD, Pezer M, Yu EW, et al. Estrogens regulate glycosylation of igg in women and men. JCI Insight (2017) 2:e89703. doi: 10.1172/jci.insight.89703
194. Fan TC, Yeo HL, Hsu HM, Yu JC, Ho MY, Lin WD, et al. Reciprocal feedback regulation of St3gal1 and Gfra1 signaling in breast cancer cells. Cancer Lett (2018) 434:184–95. doi: 10.1016/j.canlet.2018.07.026
195. Engdahl C, Bondt A, Harre U, Raufer J, Pfeifle R, Camponeschi A, et al. Estrogen induces St6gal1 expression and increases igg sialylation in mice and patients with rheumatoid arthritis: a potential explanation for the increased risk of rheumatoid arthritis in postmenopausal women. Arthritis Res Ther (2018) 20:84. doi: 10.1186/s13075-018-1586-z
196. Peyrat JP, Recchi MA, Hebbar M, Pawlowski V, Hornez L, Dong-Lebouhris X, et al. Regulation of sialyltransferase expression by estradiol and 4-Oh-Tamoxifen in the human breast cancer cell mcf-7. Mol Cell Biol Res Commun (2000) 3:48–52. doi: 10.1006/mcbr.2000.0185
197. Kobeissy F, Kobaisi A, Peng W, Barsa C, Goli M, Sibahi A, et al. Glycomic and glycoproteomic techniques in neurodegenerative disorders and neurotrauma: towards personalized markers. Cells (2022) 11:581. doi: 10.3390/cells11030581
198. Ren W, Bian Q, Cai Y. Mass spectrometry-based n-glycosylation analysis in kidney disease. Front Mol Biosci (2022) 9:976298. doi: 10.3389/fmolb.2022.976298
199. Kailemia MJ, Xu G, Wong M, Li Q, Goonatilleke E, Leon F, et al. Recent advances in the mass spectrometry methods for glycomics and cancer. Anal Chem (2018) 90:208–24. doi: 10.1021/acs.analchem.7b04202
200. Lu H, Zhang Y, Yang P. Advancements in mass spectrometry-based glycoproteomics and glycomics. Nat Sci Rev (2016) 3:345–64. doi: 10.1093/nsr/nww019
201. Cao WQ, Liu MQ, Kong SY, Wu MX, Huang ZZ, Yang PY. Novel methods in glycomics: a 2019 update. Expert Rev Proteomics (2020) 17:11–25. doi: 10.1080/14789450.2020.1708199
202. Wu Y, Sha Q, Du J, Wang C, Zhang L, Liu BF, et al. Determination of n-glycans by high performance liquid chromatography using 6-Aminoquinolyl-N-Hydroxysuccinimidyl carbamate as the glycosylamine labeling reagent. J Chromatogr A (2018) 1535:114–22. doi: 10.1016/j.chroma.2018.01.016
203. Shajahan A, Heiss C, Ishihara M, Azadi P. Glycomic and glycoproteomic analysis of glycoproteins-a tutorial. Anal Bioanal Chem (2017) 409:4483–505. doi: 10.1007/s00216-017-0406-7
204. Gong Y, Qin S, Dai L, Tian Z. The glycosylation in sars-Cov-2 and its receptor Ace2. Signal Transduct Target Ther (2021) 6:396. doi: 10.1038/s41392-021-00809-8
205. Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M. Site-specific glycan analysis of the sars-Cov-2 spike. Science (2020) 369:330–3. doi: 10.1126/science.abb9983
206. Hu Y, Ferdosi S, Kapuruge EP, Diaz de Leon JA, Stücker I, Radoï L, et al. Diagnostic and prognostic performance of blood plasma glycan features in the women epidemiology lung cancer (Welca) study. J Proteome Res (2019) 18:3985–98. doi: 10.1021/acs.jproteome.9b00457
207. Ferdosi S, Rehder DS, Maranian P, Castle EP, Ho TH, Pass HI, et al. Stage dependence, cell-origin independence, and prognostic capacity of serum glycan fucosylation, B1-4 branching, B1-6 branching, and A2-6 sialylation in cancer. J Proteome Res (2018) 17:543–58. doi: 10.1021/acs.jproteome.7b00672
208. Lattová E, Skřičková J, Zdráhal Z. Applicability of phenylhydrazine labeling for structural studies of fucosylated n-glycans. Anal Chem (2019) 91:7985–90. doi: 10.1021/acs.analchem.9b01321
209. Jezková P, Skřičková J, Wimmer G Jr., Zelinková J, Zdráhal Z, Lattová E. Differentiation of sialyl linkages using a combination of alkyl esterification and phenylhydrazine derivatization: application for n-glycan profiling in the sera of patients with lung cancer. Anal Chem (2022) 94:6736–44. doi: 10.1021/acs.analchem.2c00105
210. Ma T, Wang Y, Jia L, Shu J, Yu H, Du H, et al. Increased expression of core-fucosylated glycans in human lung squamous cell carcinoma. RSC Adv (2019) 9:22064–73. doi: 10.1039/c9ra04341a
211. Lattová E, Skřičková J, Hausnerová J, Frola L, Křen L, Ihnatová I, et al. N-glycan profiling of lung adenocarcinoma in patients at different stages of disease. Mod Pathol (2020) 33:1146–56. doi: 10.1038/s41379-019-0441-3
212. McQuiston A, Scott D, Nord D, Langerude L, Pelaez A, Machuca T, et al. Pro-inflammatory Igg1 n-glycan signature correlates with primary graft dysfunction onset in copd patients. Transpl Immunol (2022) 71:101491. doi: 10.1016/j.trim.2021.101491
213. Batson BD, Zorn BT, Radicioni G, Livengood SS, Kumagai T, Dang H, et al. Cystic fibrosis airway mucus hyperconcentration produces a vicious cycle of mucin, pathogen, and inflammatory interactions that promotes disease persistence. Am J Respir Cell Mol Biol (2022) 67:253–65. doi: 10.1165/rcmb.2021-0359OC
214. Kato T, Radicioni G, Papanikolas MJ, Stoychev GV, Markovetz MR, Aoki K, et al. Mucus concentration-dependent biophysical abnormalities unify submucosal gland and superficial airway dysfunction in cystic fibrosis. Sci Adv (2022) 8:eabm9718. doi: 10.1126/sciadv.abm9718
215. Jia N, Byrd-Leotis L, Matsumoto Y, Gao C, Wein AN, Lobby JL, et al. The human lung glycome reveals novel glycan ligands for influenza a virus. Sci Rep (2020) 10:5320. doi: 10.1038/s41598-020-62074-z
216. Waniwan JT, Chen Y-J, Capangpangan R, Weng S-H, Chen Y-J. Glycoproteomic alterations in drug-resistant nonsmall cell lung cancer cells revealed by lectin magnetic nanoprobe-based mass spectrometry. J Proteome Res (2018) 17:3761–73. doi: 10.1021/acs.jproteome.8b00433
217. Alvarez MRS, Zhou Q, Grijaldo SJB, Lebrilla CB, Nacario RC, Heralde FM, et al. An integrated mass spectrometry-based glycomics-driven glycoproteomics analytical platform to functionally characterize glycosylation inhibitors. Molecules (2022) 27:3834. doi: 10.3390/molecules27123834
218. Takakura D, Tada M, Kawasaki N. Membrane glycoproteomics of fetal lung fibroblasts using Lc/Ms. Proteomics (2016) 16:47–59. doi: 10.1002/pmic.201500003
219. Yang S, Chen L, Chan DW, Li QK, Zhang H. Protein signatures of molecular pathways in non-small cell lung carcinoma (Nsclc): comparison of glycoproteomics and global proteomics. Clin Proteomics (2017) 14:31. doi: 10.1186/s12014-017-9166-9
220. Zeng WF, Cao WQ, Liu MQ, He SM, Yang PY. Precise, fast and comprehensive analysis of intact glycopeptides and modified glycans with Pglyco3. Nat Methods (2021) 18:1515–23. doi: 10.1038/s41592-021-01306-0
221. Kong S, Gong P, Zeng WF, Jiang B, Hou X, Zhang Y, et al. Pglycoquant with a deep residual network for quantitative glycoproteomics at intact glycopeptide level. Nat Commun (2022) 13:7539. doi: 10.1038/s41467-022-35172-x
222. Jiang B, Huang J, Yu Z, Wu M, Liu M, Yao J, et al. A multi-parallel n-glycopeptide enrichment strategy for high-throughput and in-depth mapping of the n-glycoproteome in metastatic human hepatocellular carcinoma cell lines. Talanta (2019) 199:254–61. doi: 10.1016/j.talanta.2019.02.010
223. Rudd PM, Dwek RA. Glycosylation: heterogeneity and the 3d structure of proteins. Crit Rev Biochem Mol Biol (1997) 32:1–100. doi: 10.3109/10409239709085144
224. Ashkani J, Naidoo KJ. Glycosyltransferase gene expression profiles classify cancer types and propose prognostic subtypes. Sci Rep (2016) 6:26451. doi: 10.1038/srep26451
225. Vasseur JA, Goetz JA, Alley WR Jr., Novotny MV. Smoking and lung cancer-induced changes in n-glycosylation of blood serum proteins. Glycobiology (2012) 22:1684–708. doi: 10.1093/glycob/cws108
226. Gao J, Ohlmeier S, Nieminen P, Toljamo T, Tiitinen S, Kanerva T, et al. Elevated sputum Bpifb1 levels in smokers with chronic obstructive pulmonary disease: a longitudinal study. Am J Physiol Lung Cell Mol Physiol (2015) 309:L17–26. doi: 10.1152/ajplung.00082.2015
227. Chen Y, Shen L, Chen B, Han X, Yu Y, Yuan X, et al. The predictive prognostic values of Cbfa2t3, Stx3, denr, Egln1, Fut4, and Pcdh7 in lung cancer. Ann Transl Med (2021) 9:843. doi: 10.21037/atm-21-1392
228. Zhou W, Ma H, Deng G, Tang L, Lu J, Chen X. Clinical significance and biological function of fucosyltransferase 2 in lung adenocarcinoma. Oncotarget (2017) 8:97246–59. doi: 10.18632/oncotarget.21896
229. Jin LW, di Lucente J, Ruiz Mendiola U, Tang X, Zivkovic AM, Lebrilla CB, et al. The role of Fut8-catalyzed core fucosylation in alzheimer’s amyloid-B oligomer-induced activation of human microglia. Glia (2023) 71:1346–59. doi: 10.1002/glia.24345
230. Ohkawa Y, Harada Y, Taniguchi N. Keratan sulfate-based glycomimetics using langerin as a target for copd: lessons from studies on Fut8 and core fucose. Biochem Soc Trans (2021) 49:441–53. doi: 10.1042/bst20200780
231. Borsig L, Lubli H. Cell adhesion during tumorigenesis and metastasis. In: Boffetta P., Hainaut P., (eds) Reference module in biomedical sciences (2017).
232. Gong L, Cai Y, Zhou X, Yang H. Activated platelets interact with lung cancer cells through p-selectin glycoprotein ligand-1. Pathol Oncol Res (2012) 18:989–96. doi: 10.1007/s12253-012-9531-y
233. Watz H, Bock D, Meyer M, Schierhorn K, Vollhardt K, Woischwill C, et al. Inhaled pan-selectin antagonist bimosiamose attenuates airway inflammation in copd. Pulm Pharmacol Ther (2013) 26:265–70. doi: 10.1016/j.pupt.2012.12.003
234. Romano SJ. Selectin antagonists: therapeutic potential in asthma and copd. Treat Respir Med (2005) 4:85–94. doi: 10.2165/00151829-200504020-00002
235. Compain G, Oumata N, Clarhaut J, Peraudeau E, Renoux B, Galons H, et al. A beta-Glucuronidase-Responsive albumin-binding prodrug for potential selective kinase inhibitor-based cancer chemotherapy. Eur J Med Chem (2018) 158:1–6. doi: 10.1016/j.ejmech.2018.08.100
Keywords: protein glycosylation, chronic respiratory diseases, abnormal glycoenzyme activity, glycoproteomics, glycobiomarkers
Citation: Xie X, Kong S and Cao W (2023) Targeting protein glycosylation to regulate inflammation in the respiratory tract: novel diagnostic and therapeutic candidates for chronic respiratory diseases. Front. Immunol. 14:1168023. doi: 10.3389/fimmu.2023.1168023
Received: 17 February 2023; Accepted: 02 May 2023;
Published: 15 May 2023.
Edited by:
Wen-Hui Wu, Tongji University, ChinaReviewed by:
Quanqing Zhang, University of California, Riverside, United StatesCopyright © 2023 Xie, Kong and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiqian Cao, d3FjYW9AZnVkYW4uZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.