
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 14 March 2023
Sec. Cancer Immunity and Immunotherapy
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1162607
This article is part of the Research Topic Community Series in Post-Translational Modifications of Proteins in Cancer Immunity and Immunotherapy, volume II View all 12 articles
As the most abundant and conserved internal modification in eukaryote RNAs, N6-methyladenosine (m6A) is involved in a wide range of physiological and pathological processes. The YT521-B homology (YTH) domain-containing family proteins (YTHDFs), including YTHDF1, YTHDF2, and YTHDF3, are a class of cytoplasmic m6A-binding proteins defined by the vertebrate YTH domain, and exert extensive functions in regulating RNA destiny. Distinct expression patterns of the YTHDF family in specific cell types or developmental stages result in prominent differences in multiple biological processes, such as embryonic development, stem cell fate, fat metabolism, neuromodulation, cardiovascular effect, infection, immunity, and tumorigenesis. The YTHDF family mediates tumor proliferation, metastasis, metabolism, drug resistance, and immunity, and possesses the potential of predictive and therapeutic biomarkers. Here, we mainly summary the structures, roles, and mechanisms of the YTHDF family in physiological and pathological processes, especially in multiple cancers, as well as their current limitations and future considerations. This will provide novel angles for deciphering m6A regulation in a biological system.
In recent years, more than 170 different chemical RNA modifications have been identified, drawing more attention to the epitranscriptome (1). Among them, N6-methyladenosine (m6A), which adds a methyl group to the sixth nitrogen atom of adenine, is the most abundant internal transcriptome modification in eukaryotes (2, 3). By identifying the consensus motif “RRACH” (R = A/G; H = A/C/U), m6A usually occurs in the 3’ untranslated region (3’UTR) and coding sequence (CDS), especially in the vicinity of stop codons (4, 5). Accordingly, m6A modification regulates the metabolism of multiple types of RNAs and are ultimately participating in various pathophysiological processes.
The m6A methylation is dynamic and reversible, regulated by a series of m6A-modifying enzymes which can be classified into “writers”, methyltransferases that install m6A modifications, and “erasers”, demethylases that remove m6A from mRNA, as well as “readers” that recognize and bind to m6A-modified mRNA to mediate their ultimate fate. Methyltransferase complex (MTC) is the main “writers”, including methyltransferase like 3/14 (METTL3/14), Wilms’ tumor 1-associating protein (WTAP) (6, 7). They catalyze the formation of m6A methylation synergistically. Conversely, the fat mass and obesity-associated protein (FTO) and AlkB homolog 3/5 (ALKBH3/5) that belong to the “erasers” act as key proteins in m6A demethylation (8, 9). Moreover, “readers” are important m6A binding proteins such as YTHDFs, YTH domain-containing 1/2 (YTHDC1/2), heterogeneous nuclear ribonucleoproteins (HNRNP) family, insulin-like growth factor 2 mRNA-binding proteins (IGF2BP1/2/3), and eukaryotic initiation factor 3 (eIF3) (5, 10–16). They influence RNA splicing, export, translation, and decay, and then regulate diverse downstream signaling pathways.
The YTHDF family is the most studied “readers” of m6A, which includes YTHDF1, YTHDF2, and YTHDF3. They regulate the translation and stability of target mRNAs to alter the expression of downstream molecules, thus affecting diverse biological processes (10, 17). In this review, we summarize the structures and functions of the YTHDF family, especially the m6A-binding specificity. Moreover, we focus on its underline mechanisms in multiple physiological and pathological processes, especially in tumors, hoping to provide possible application value.
In “writers”, MTC is the main component that catalyzes the formation of m6A. Among them, METTL3 installs methyl groups in S-adenosylmethionine to RNA target sites, while METTL14 selects RNA adenine bases and stabilizes the catalytic process (6, 18, 19). WTAP, RBM15/15B, VIRMA, and ZC3H13 are also components of the MTC, directing complexes to nuclear speckles as well as RNA sites (7, 20–22). In addition to MTC, METTL16, ZCCHC4, and METTL5 also can catalyze m6A modification of specific RNAs (23–25). In contrast, FTO and ALKBH3/5 act as key “erasers” proteins in m6A demethylation (8, 9, 26). FTO and ALKBH5 target mRNA and are associated with obesity and spermatogenesis, respectively (9, 27). Whereas ALKBH3 removes m6A on tRNA (26).
Moreover, “readers” are required in m6A-regulated diverse downstream signaling pathways. For example, YTHDC1 promotes mRNA splicing in the nucleus as well as nuclear export (11, 12). Furthermore, YTHDC1 accelerates the function of XIST to silence the transcription of genes on the X chromosome (20). Interestingly, YTHDC2 promotes mRNA translation with a concomitant decrease in mRNA abundance and has ATPase and 3’ to 5’ RNA helicase activities (13, 28). In addition, the HNRNP family regulates the alternative splicing of mRNA through an “m6A-switch” mechanism (29–33). IGF2BPs stabilize target mRNAs in different ways under normal and stress conditions (15). And eIF3 binds m6A on the 5’UTR of mRNA and promotes mRNA translation in a cap-independent manner (16).
The YTHDF family was identified by selecting proteins containing the YTH domain and subsequently obtained in pull-down experiments using methylated RNA bait (5, 34, 35). Now, the features of the YTHDF family have been gradually unraveled. The YTH domains of YTHDFs have a hydrophobic pocket, which is critical to the recognition of m6A in the cytoplasm (36). But the role of each protein is different, for example, YTHDF1 promotes RNA translation, YTHDF2 facilitates RNA decay, and YTHDF3 exhibits a dual function depending on its binding partner (37). Thus, the YTHDF family is closely associated with many cancers and other biological processes (Figure 1).
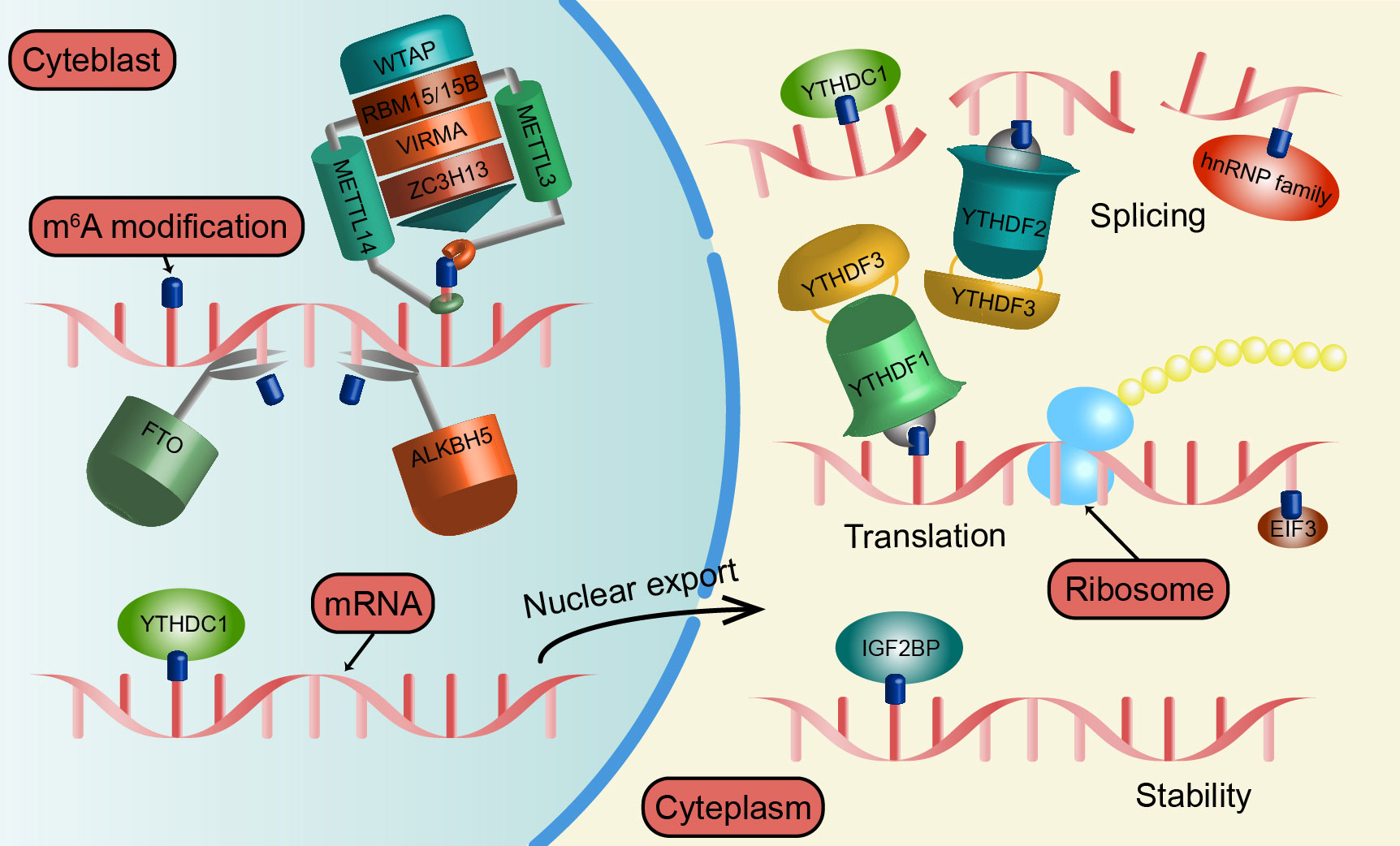
Figure 1 The regulation mechanism of m6A modification. METTL3, METTL14, WTAP, RBM15, VIRMA, and ZC3H13 all belong to the “writers” and catalyze the formation of m6A modification by constituting MTC. The “erasers” includes FTO and ALKBH5, which act as key proteins in m6A demethylation. YTHDF1/2/3, YTHDC1, IGF2BP, hnRNP family, and EIF3 as “readers” that bind to m6A and affect RNA splicing, output, translation, and decay.
The YTHDF family is composed of a C-terminal YTH domain and an N-terminal domain rich in P/Q/N (Pro/Gln/Asn). The YTH domain is the basis of recognizing m6A RNA specifically and its targeted position and consensus sequence are similar to the distribution pattern of m6A sites on mRNA (20, 38). YTH domain can also directly bind to N1-methyladenosine (m1A), but with a lower affinity than m6A (39). The prion-like low-complexity sequence regions (LCRs) of the N-terminal domain are associated with the liquid-liquid phase separation (LLPS) (40). The mRNA-YTHDF complexes are located in different membrane-less compartments in the cytoplasm, such as processing bodies (P-bodies), stress granules (SGs), or neuronal granules, which are the result of LLPS and can be enhanced by multivalent m6A modifications (41). Proteomic studies revealed that YTHDFs can be phosphorylated and myristoylated to regulate their expression and clustering (42). Additionally, the EGFR/SRC/ERK pathway stabilizes YTHDF2 protein by phosphorylating YTHDF2 at serine39 and threonine381 in glioblastoma cells (43). YTHDF2 can also be SUMOylated at site K571, thereby enhancing its binding affinity with m6A-modified mRNAs and accelerating cancer advancement (44). Therefore, targeting post-translational modifications represent a novel opportunity for YTHDFs to regulate their functions.
The crystal structures of the three YTH domains and their complexes with an m6A mononucleotide (or m6A oligoribonucleotides) have been revealed (45, 46). The YTH domains share a mixed α-helix-β-sheet fold, where the α-helices surround a barrel-shaped center arranged by the β-sheets. The surface of the YTH domain has a positively-charged groove in which m6A is tightly locked. Specifically, m6A is located in a hydrophobic pocket formed by three highly conserved aromatic residues called an aromatic cage. In the YTHDF-m6A complex, the m6A adenine moiety is sandwiched between the rings of two aromatic residues, paralleling them (Trp411 and Trp470 in YTHDF1, Trp432, and Trp491 in YTHDF2, Trp438, and Trp497 in YTHDF3). And the methyl group of m6A points to the ring of one aromatic residue (Trp465 in YTHDF1, Trp486 in YTHDF2, Trp492 in YTHDF3) (36, 47, 48). As well as aromatic residues, some amino acids (aa) of the YTH domain also interact with m6A. For example, the backbone NH of Tyr397 in YTHDF1 and Tyr418 in YTHDF2 form hydrogen bonds with the N3 of m6A. The carbonyl oxygen of Cys412 in YTHDF1, Cys433 in YTHDF2, and Cys439 in YTHDF3 bind to the N6 of m6A by hydrogen bonding. To sum up, the Π-Π interactions between the m6A adenine moiety and the aromatic cage, the cation-Π interactions between the methyl group and the aromatic cage, and a series of hydrogen bonds lay a foundation for m6A recognition (36) (Figure 2).
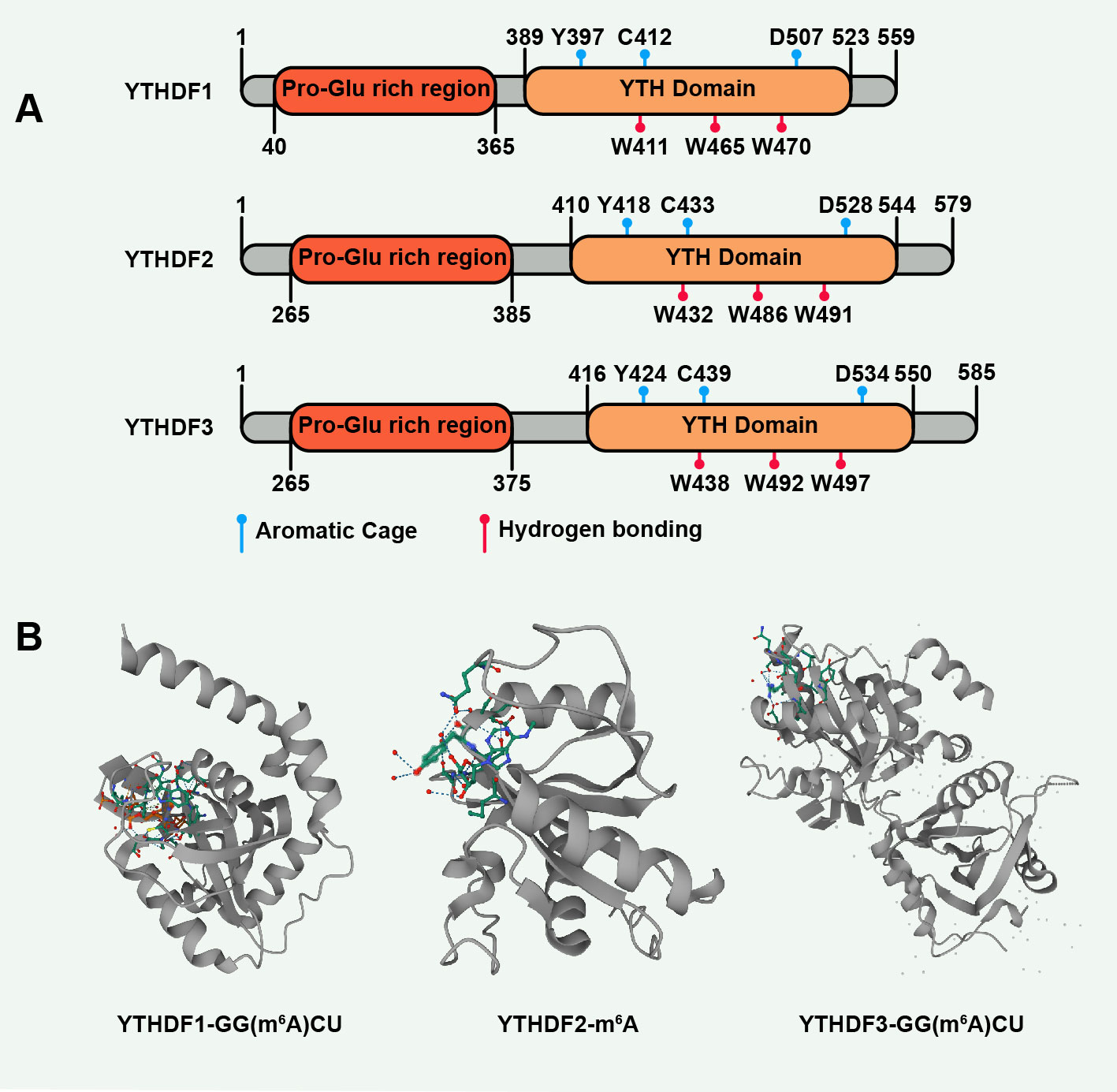
Figure 2 The structures of the YTHDF family, especially the YTH domain. (A) The YTH domain of YTHDFs: YTHDF1 (UniProt ID: Q9BYJ9), YTHDF2 (UniProt ID: Q9Y5A9), YTHDF3 (UniProt ID: Q7Z739). (B) Structures of YTHDFs in complex with m6A. YTHDF1 (PDB ID:4RCJ), YTHDF2 (PDB ID:4RDN), YTHDF3 (PDB ID:6ZOT). The secondary structures of proteins are shown in gray, and RNA molecules are shown in color.
Evidence confirms that the YTHDF family plays an integral role in the translation and degradation of m6A-modified mRNAs. YTHDF2 is the most explored YTHDFs and is generally expressed at much higher levels than YTHDF1 and YTHDF3 in most cells (42). YTHDF2 binds to m6A-modified mRNAs and recruits the CCR4-NOT deadenylase complex through its N-terminal 101-200 aa to initiate deadenylation, which is a prior condition of P-body localization and decay of targeted mRNAs (10, 49, 50). Additionally, m6A-modified mRNAs can also bind to YTHDF2 in an HRSP12-dependent manner, and subsequently cleaved by RNase P/MRP (endoribonucleases) (51, 52). In particular, HRSP12 bridges the N-terminal 100 aa of YTHDF2 and RNase P/MRP, contributing to the rapid degradation of mRNAs. And m6A-containing circular RNAs (circRNAs) are also degraded by this pathway. Interestingly, under heat shock stress, nuclear-translocated YTHDF2 protects m6A motifs in the 5’ untranslated region (5’UTR) of stress-induced transcripts and activates cap-independent translation initiation (53). The N-terminal of YTHDF1 (100-200 aa) is in charge of the translation of mRNAs with m6A modifications (54). YTHDF1 not only transports more mRNAs to translation machinery and promotes ribosome occupancy, but also enhances the translation-initiation rate by correlating eIF4G-mediated loop structure through interaction with eIF3 in a cap-dependent manner (17). YTHDF1 can also trigger translational elongation through interaction with elongation factors in some cancer cells (55–57). Apart from the above results, Li et al. found that YTHDF1 interacts with Argonaute 2 (AGO2) to stimulate the production of P-bodies for mRNA degradation (58). In addition, YTHDF3 augments m6A-mRNA translation by cooperating with YTHDF1 and interacting with the 40s/60s ribosome subunits (59). Besides that, YTHDF3 recruits eIF4G2 to m6A sites, driving translation initiation of circRNAs (60). YTHDF3 also promotes m6A-modified mRNA decay by working together with YTHDF2 (37). A recent study found that the effect of YTHDF3 in regulating targeted mRNA deadenylation during somatic cell reprogramming relies on the recruitment of the PAN2-PAN3 deadenylase complex (61).
Interestingly, the YTHDF family forms a classic functional model: upon entry into the cytoplasm, m6A-modified mRNAs are first bound by the YTHDF3 or YTHDF3-YTHDF1 complex and then recognized by YTHDF2, thereby regulating the different fates of the targeted mRNA (62). Nevertheless, it has recently been discovered that YTHDFs have redundant functions to a large extent (63). Those three YTHDFs share highly homologous structures (about 85% of aa sequence similarity) (64), similar RNA-binding properties (20), and a similar set of binding proteins, jointly regulating mRNA destiny in an m6A-dependent manner (65). Indeed, the distinct functions of YTHDFs depend on their expression levels, spatial locations, and post-translational modifications. Also, YTHDFs are affected by additional RNA-binding proteins that interact with YTHDFs, such as fragile X mental retardation protein (FMRP) (66, 67), and Proline-rich coiled-coil 2 A (Prrc2a) (68). Collectively, the role of YTHDFs in regulating gene expression is complex and requires further investigation.
Among the three YTHDFs, YTHDF2 is expressed and plays a pivotal role throughout mammalian gametogenesis. YTHDF2-knockout female mice are infertile while male mice are hypo fertile (65, 69). Specifically, YTHDF2 is intrinsically required for oocyte competence to support early zygotic development rather than MII oocytes formation and fertilization process (69). YTHDF2 regulates appropriate maternal transcript dosage during oocyte maturation by selectively mediating transcript destabilization. Additionally, YTHDF2 clears m6A-dependent matrix metallopeptidase transcripts to promote the adhesion and proliferation of spermatogonia during spermatogenesis (70). Knockout of YTHDF2 results in morphologically deformed and functionally impaired sperm, even severe loss (65, 71).
Intriguingly, unlike the previous view that maternal mRNAs clearance and maternal-to-zygotic transition (MZT) are dependent on YTHDF2, Kontur et al. found that individual YTHDFs deletion does not prevent embryonic development, whereas double mutations of YTHDF2/YTHDF3 disrupts oogenesis and triple YTHDF depletion causes lethality in zebrafish (72, 73). Despite evidence for the redundant functions of YTHDFs in early mouse embryonic development, depletion of YTHDF2 causes lethality at late embryonic development stages with embryos exhibiting severe neurological deficits (65, 74). Zheng et al. found that YTHDF3 reduction is an adaptive mechanism under a hypoxic environment in early embryonic development (75). Specifically, YTHDF3 binds to the m1A site of insulin-like growth factor 1 receptor (IGF1R) mRNA and degrades IGF1R mRNA, hindering migration and invasion of trophoblast.
Somatic cells are reprogrammed into induced pluripotent stem cells (iPSCs), which have unlimited proliferation and pluripotent differentiation potential similar to human embryonic stem cells (ESCs) (76). YTHDF2 and YTHDF3 play an essential role in this reprogramming process by clearing somatic mRNAs, especially Tead2, through distinct m6A-dependent deadenylation mechanisms (61). While YTHDF1 is capable of increasing the expression of the transcription factor Btg2 and promoting the reprogramming of induced neuronal cells (77). In terms of iPSCs functions, the YTHDF1/YTHDF2 orchestration is involved in METTL3-m6A-mediated maintenance of pluripotent state in porcine iPSCs by elevating JAK2 level, reducing SOSC3 expression, and provoking STAT3/KLF4/SOX2 signal axis (78). YTHDF1 upregulation depends on MATR3 and maintains a MATR3-mediated pluripotent state in human iPSCs by maintaining the expression of OCT4 and LIN28A transcripts (79). Importantly, YTHDF2 is overexpressed and disrupts the expression of a group of m6A-modified mRNAs associated with neurodevelopment, thereby blocking neural differentiation and promoting pluripotency in human iPSCs (80). Similarly, YTHDF3 reduces gene expression associated with the formation of three germ layers, and the absence of YTHDF3 impairs pluripotency in ESCs (81).
Several studies have revealed that the specification and characteristics of hematopoietic stem cells (HSCs) are significantly regulated by YTHDF2. The m6A-YTHDF2-mediated decay of Notch1 mRNA is critical for the generation of the earliest hematopoietic stem/progenitor cells (HSPCs) during the endothelial-to-hematopoietic transition (EHT) in both zebrafish and mice embryos (82, 83). Li et al. first reported that YTHDF2 specifically mediates the ex vivo expansion of human HSCs due to the regulation of the stability of multiple mRNAs essential for HSC self-renewal (84). Therefore, inhibition of YTHDF2 makes it possible to obtain a sufficient number of HSCs from human umbilical cord blood (hUCB), which facilitates the application of hUCB HSCs transplantation. Furthermore, YTHDF2 deletion also promotes the expansion and regeneration of HSCs by eliminating the decay of both WNT-targeted and survival-related genes under stress conditions (85). Interestingly, although YTHDF2 is dispensable for steady-state multilineage hematopoiesis, long-term deficiency of YTHDF2 dramatically impairs HSCs activity and blocks reconstitution of multilineage hematopoiesis (86). Given that hematopoietic-specific YTHDF2 deficiency-induced long-term HSCs impairment is consistent with the adverse consequences of inflammation in HSCs, the inflammation-induced increase in YTHDF2 may be a protective mechanism for the long-term integrity of HSCs. YTHDF3 is also involved in the regulation of HSCs. YTHDF3 binds m6A on the 5’UTR of CCND1 mRNA and cooperates with PABPC1 and EIF4G2 to promote the expression of CCND1, a positive regulator of HSCs reconstitution capacity (87). While YTHDF3 facilitates the translation of FOXM1 and ASXL1 transcripts and is critical for maintaining HSC properties under stress conditions (88).
YTHDF1 is indispensable for maintaining intestinal stem cells (ISCs) during regeneration after intestinal damage by driving a positive feedback loop of the YTHDF1/TCF4/WNT signaling axis (89). Similarly, YTHDF1 sustains the stemness of ISCs through a targeted translation of transcriptional-enhanced associate domain 1 (TEAD1) (90). In addition, YTHDF1 is also involved in the m6A-mediated self-renewal of mouse female germline stem cells (mFGSCs) (91).
YTHDFs play key roles in adipogenesis, particularly YTHDF2. YTHDF2 binds and degrades JAK1 mRNA to block the JAK1/STAT5/C/EBPβ pathway, thereby inhibiting the adipogenic differentiation of bone marrow stem cells (92). Similarly, YTHDF2-mediated silencing of the JAK2/STAT3/C/EBPβ pathway impedes adipogenesis (93). Indeed, YTHDF2 also impairs adipogenesis by degrading multiple target transcripts through methylation-dependent modifications. Cell cycle factors, including CCNA2, CDK2, and CCND1 promote cell cycle progression and mitotic clonal expression in adipocytes (94, 95). Epigallocatechin gallate (EGCG) and metformin reduce CCNA2 and CDK2 levels by increasing m6A modification in an FTO-YTHDF2-dependent manner (96, 97). Conversely, Zinc finger protein (Zfp217) binds and sequesters YTHDF2 to reduce m6A levels, thus reversing CCND1 mRNA degradation (98). YTHDF2 also reduces the content of FAM134B, fatty acids synthesis-related proteins such as FASN, and autophagy-related proteins, including ATG5 and ATG7, which inhibit adipogenesis (99–101). Furthermore, the liver Bmal1 regulates the circadian clock of lipid metabolism by controlling the abundance of m6A modifications on transcripts (102). Mechanistically, Bmal1 knockdown inhibits PPARα expression in an m6A-YTHDF2-dependent manner, which increases lipid accumulation. Moreover, AMPK upregulates CD36 levels through YTHDF2-dependent Parkin reduction, which enhances intestinal long-chain fatty acid uptake and induces obesity in high-fat diet mice (103).
Intriguingly, YTHDF1 inhibits ovine adipogenesis and promotes porcine adipogenesis by promoting the expression of PNPLA2 and MTCH2, respectively (62, 104). Chen et al. found that YTHDF1 restrains PPARγ expression in mice by promoting the translation of m6A-modified TRAF4 transcripts, while curcumin exerts an anti-obesity role by reducing the effect of ALKBH5 demethylation on TRAF4 m6A modification (105). In addition, YTHDF1 together with METTL3 amplifies the function of Rubicon that inhibits autophagy by stabilizing Rubicon mRNA, and further blocks the clearance of lipid droplets (LDs) in mouse nonalcoholic fatty liver disease (NAFLD) (106).
YTHDF1 mainly regulates axonal function as well as learning and memory, and YTHDF2 is mainly involved in neural development and differentiation. Functional axon regeneration under peripheral nervous system injury is supported by m6A-YTHDF1-derived increases in global protein translation (107). And YTHDF1 is a key player in enhancing Robo3.1 mRNA translation and guidance of pre-crossing commissural axons in the spinal cord, whereas YTHDF1 is inhibited by floor plate-induced signals in post-crossing axons guidance (108). Furthermore, dual depletion of YTHDF1/YTHDF3 affects spine morphology and excitatory synaptic transmission in hippocampal neurons (109). Further study revealed that YTHDF1 accelerates basal transmission and long-term potentiation of synapses by advancing neuronal stimulation-induced protein translation, thereby promoting learning and memory, especially long-term memory (110). In a Drosophila short-term memory experiment, memory-storing neurons require YTHDF to maintain normal memory function during aging (111). Furthermore, YTHDF1-mediated Dvl1 mRNA translation has a synergistic effect with YTHDF2-mediated Wnt5a mRNA degradation in inhibiting axon growth of cerebellar neurons (112).
During neural development, YTHDF2 is overexpressed and positively regulates early brain development by promoting the proliferation and differentiation of neural stem/progenitor cells (NSPCs) (74). Knockout of YTHDF2 significantly reduces cerebral cortical thickness and induces differentiated neurons to produce abnormal stress-sensitive neurites. Interestingly, YTHDF2-silenced NSPCs cannot differentiate into glial cells. Wu et al. showed that YTHDF2 competes with Prrc2a for binding to Olig2 mRNA, resulting in impaired oligodendrocyte specification and myelination (68). Moreover, YTHDF2 is detrimental to the extension and maintenance of retinal ganglion cell (RGC) dendritic arborization (113).
YTHDFs are also involved in a variety of brain disorders. For example, downregulated miR-421-3p in microglia after cerebral artery occlusion/reperfusion (MCAO/R) relieves the repression of YTHDF1, thereby promoting p65 mRNA translation, leading to aggravated inflammation and brain injury (114). Impairments of fine motor and cognitive function in young mice exposed to multiple sevoflurane are attributable to a specific decrease in YTHDF1 expression (115). Overexpression of YTHDF1 ameliorates diabetes-induced cognitive impairment (116). Additionally, elevated YTHDF2 under persistent light impedes cognitive behavior in mice by perturbing the stability of TrkappaB mRNA (117). And a recent case report found that most individuals with YTHDF3 haploinsufficiency show intellectual disability and/or developmental delay of variable degrees (118).
YTHDF1 promotes cardiomyocyte (CM) differentiation, whereas YTHDF3 does the opposite (81). YTHDF1, which is positively regulated by ALKBH5, also promotes CM proliferation in injury-induced cardiac regeneration by enhancing YAP mRNA translation (119). Xu et al. indicated that YTHDF2 degrades Myh7 mRNA to mitigate cardiac hypertrophy during heart failure development (120). Conversely, lncRNA MIAT-induced YTHDF2 high expression stimulates cardiac hypertrophy by downregulating CPT-1a levels in the PPARα pathway (121). Moreover, YTHDF1 and YTHDF2 promote ocular pathological angiogenesis via the METTL3-m6A-LRP6 axis and the FTO-m6A-FAK axis, respectively (122, 123). YTHDF1/YTHDF2 cooperation stimulates the atherogenic inflammatory cascade in the vascular endothelium by upregulating NLRP1 and downregulating KLF4 (124). Furthermore, loss of either YTHDF1 or YTHDF2 alleviates the proliferation of pulmonary arterial smooth muscle cells and pulmonary hypertension under hypoxia. Mechanistically, YTHDF1 promotes the translation of MAGED1 mRNA while YTHDF2 activates the PI3K/AKT signaling pathway by degrading PTEN mRNA (125, 126). And YTHDF3 knockout protects lung epithelial cells from inflammatory injury by inhibiting inflammatory cytokine secretion after hypoxia/reoxygenation (127).
YTHDFs play anti-viral roles in the life cycle of Epstein-Barr virus (EBV), Hepatitis B virus (HBV), Hepatitis C virus (HCV), Zika virus (ZIKV), and enterovirus 71 (EV71) (128–133). For example, the knockdown of each DF in EBV-infected cells promotes EBV lytic replication and reactivation. Mechanistically, YTHDF1 attracts ZAP, DDX17, and DCP2 forming RNA degradation complexes to accelerate the decapping of m6A-modified RNAs and degrade EBV cleavage gene transcripts (128). Furthermore, activation of caspases cleaves D166 and D367 sites on YTHDF2 upon EBV reactivation reduces YTHDF2 expression, thereby increasing caspase-8 protein levels and enhancing EBV replication (129). Alternatively, YTHDFs inhibit HCV infection by reducing viral particle production rather than blocking viral RNA replication (131). During the chronic HCV infection state, YTHDFs relocate to lipid droplets, bind to the m6A site in the HCV E1 region, and antagonize viral packaging caused by the binding of the viral core protein to the non-m6A site in the E1 region. In contrast, YTHDF2 promotes simian virus 40 (SV40) and influenza A virus (IAV) replication (134, 135). Moreover, YTHDF1 and YTHDF3 induce severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, YTHDF1 inhibits chikungunya virus (CHIKV) infection, and YTHDF2 functions opposite to that of YTHDF1 in both SARS-CoV-2 and CHIKV (136–138).
Notably, the regulation of YTHDFs in the transcription and replication of human immunodeficiency virus type 1 (HIV-1) and Kaposi’s sarcoma-associated herpesvirus (KSHV) remains controversial. Evidence suggests that YTHDFs hinder HIV-1 replication in target cells contradicting previous views that YTHDFs increase viral transcript and protein levels (139–141). Specifically, after HIV-1 infection into cells, YTHDFs impede HIV-1 reverse transcriptase by degrading incoming HIV-1 genomic RNA (gRNA) in an m6A-dependent manner, thereby limiting viral replication (139). Nevertheless, YTHDFs facilitate HIV-1 structural protein Gag synthesis and virus release, while forming a complex with HIV-1 Gag protein and viral and cellular RNAs in virus-producing cells (140). To ensure optimal HIV-1 infectivity, HIV-1 protease cleaves YTHDF3, which enters the virion in a nucleocapsid-dependent fashion (142). Additionally, Hesser et al. showed that YTHDF2 exerts pro- and anti-KSHV effects in iSLK and B cell lines, respectively (143). Instead, Tan et al. observed that YTHDF2 inhibits KSHV gene expression and virion production in iSLK cells (144). Together, the paradoxical phenomenon of YTHDFs in viral regulation may be explained by differences in cell types, viral life cycle stages, and experimental approaches.
The type I interferon (IFN) signaling pathway relies on the expression of IFN-stimulated genes (ISGs) to mediate a powerful innate antiviral immune response. YTHDF1-mediated upregulation of IFITM1, a subset of ISGs, initiates antiviral responses (145). Another study showed that YTHDF1 prevents viral double-stranded RNA (dsRNA)-driven IFN responses (146). YTHDF1 induces the IFN-mediated expression of ADAR1, which disrupts the secondary structure of dsRNA in an adenosine-to-inosine (A-to-I) RNA editing manner. Furthermore, YTHDF2 deletion enables increased levels of IFN-βand inflammatory factors, including interleukin-6 (IL-6) by stabilizing host antiviral transcripts (147, 148). YTHDF2 also binds and sequesters m6A-modified viral RNA, which protects viral RNA from RIG-I recognition, thereby inhibiting RIG-I activation and the downstream IFN signaling pathway (149, 150). In contrast, YTHDF2 is an essential cofactor for the IFN-α-induced degradation of m6A-methylated HBV RNA by ISG20 (151). Additionally, enterovirus 2A proteases cleave YTHDFs and limit antiviral responses during early viral infection (152). Among them, the cleavage of YTHDF3 dampens the IFN-I-stimulated JAK/STAT signaling pathway. Interestingly, only YTHDF3 attenuated ISGs expression in the absence of viral infection (153). Mechanistically, YTHDF3 rapidly translates forkhead box protein O3 (FOXO3) mRNA through cooperation with PABP1 and eIF4G2 in an m6A-independent way, thereby suppressing ISGs expression.
Inflammatory responses are also an important part of immunity. YTHDF1 counteracts the excessive and persistent development of inflammation in the septic response by promoting the expression of SOCS1, a negative regulator of macrophage-mediated inflammation (154). However, YTHDF1 knockout suppressed inflammatory lung or intestinal damage (155, 156). Macrophage-specific YTHDF1 knockdown may be a protective therapy against brain injury in severe sepsis rats with ECMO by enhancing adaptive immune function and alleviating inflammatory damage (157). YTHDF2 also negatively regulates inflammation. YTHDF2 inhibits the MAPK and NF-κB signaling pathways by downregulating the expression of MAP2K4, MAP4K4, STAT1, and PPAR-γ, and subsequently prevents macrophage polarization and proinflammatory cytokine secretion (158–160). And YTHDF2-dependent decay of KDM6B mRNA restricts H3K27me3 demethylation, which impedes transcription of proinflammatory cytokine genes (161).
Strikingly, the expression of YTHDFs has a strong relationship with the immune regulation of various tumors. The expression of YTHDF1 is not only the highest in normal immune cells but also dramatically correlated with tumor immune-infiltrated cells in cancer, especially CD8+ T cells, macrophages, and dendritic cells (DCs) (162). Han et al. revealed that YTHDF1 is an important target for anti-tumor immunotherapy (163). YTHDF1 depletion accelerates tumor antigen presentation and cross-priming of CD8+ T cells by retarding lysosomal cathepsin translation in DCs in an m6A-dependent manner. And the loss of YTHDF1 recruits DCs and activates IFN-γ receptor 1 and JAK/STAT1 signaling pathways, thereby promoting antitumor immunity in GC (164). Li et al. demonstrated that YTHDF1 hinders CD8+ T cell infiltration and increases immune checkpoint expression, such as PD-L1 and V-domain Ig suppressor of T cell activation (VISTA), in CRC (165). To this end, YTHDF1 consumption can be synergistic with anti-PD-1/PD-L1 immunotherapy for effective anti-tumor therapy. Similarly, YTHDF2-deficient tumors increased the sensitivity to anti-PD-1/PD-L1 immunotherapy by stabilizing PD-L1 mRNA in ICC (166). However, YTHDF2 participates in anti-tumor and anti-viral infection by regulating the maturation, proliferation, and effector functions of NK cells (167) (Figure 3).
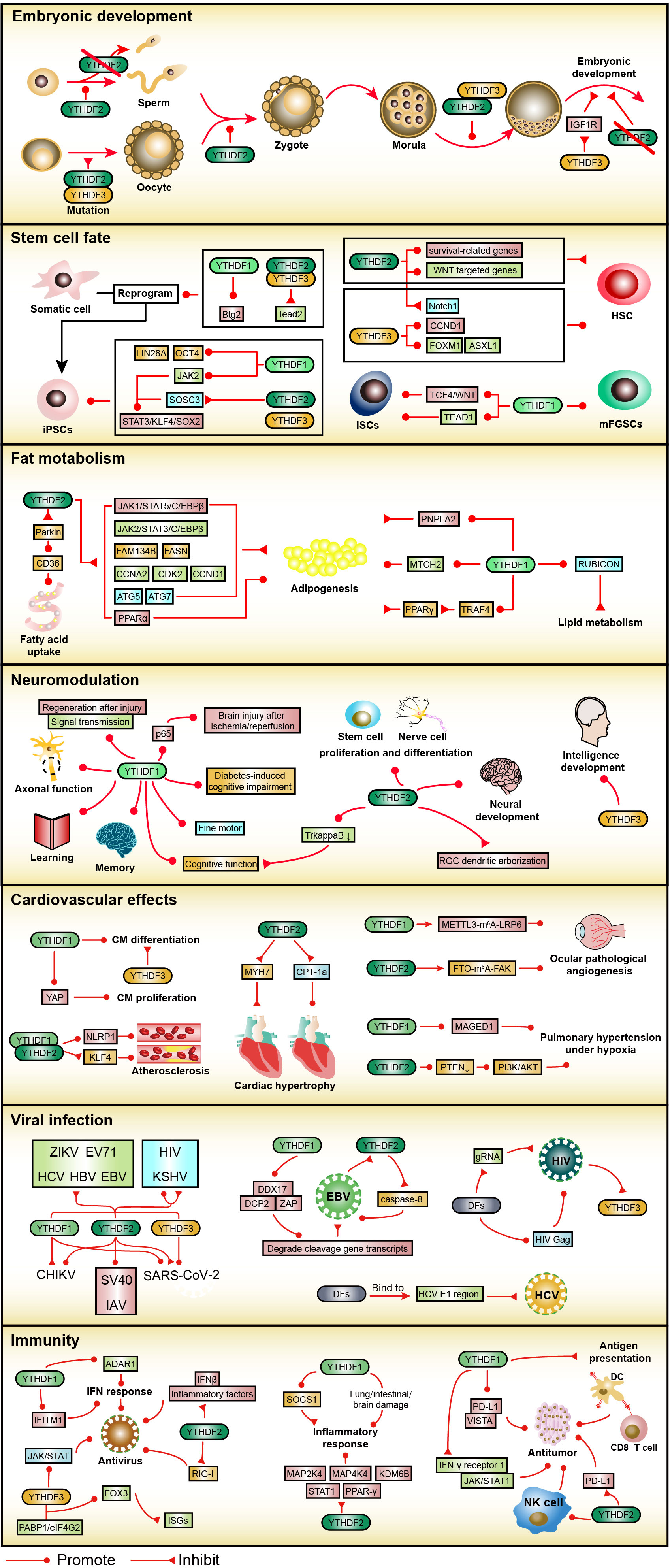
Figure 3 The roles of the YTHDF family in embryonic development, stem cell fate, fat metabolism, neuromodulation, cardiovascular effect, viral infection, and immunity. In embryonic development, YTHDF2 is essential for sperm, oocyte, zygote, and embryo formation. In stem cell fate, the YTHDF family promotes somatic cell reprogramming and the properties of iPSCs. In addition, YTHDF2 and YTHDF3 participate in the fate of HSC, and YTHDF1 in the fate of ISCs as well as mFGSCs. In fat metabolism, YTHDF1 and YTHDF2 regulate adipogenesis and fatty acid metabolism. In neuromodulation, YTHDF1 affects axonal function as well as learning and memory, YTHDF2 regulates neural development and differentiation, and YTHDF3 participates in intellectual development. In cardiovascular effect, YTHDF1 and YTHDF2 are closely related to the fate of CM, vascular endothelial cells, and pulmonary artery smooth muscle cells. In a viral infection, the YTHDF family is involved in the life cycle of several viruses, especially EBV, HCV, and HIV. In immunity, the YTHDF family plays an important role in antiviral immunity, inflammatory immunity, and anti-tumor immunity.
Studies have reported that YTHDF1 is an oncogene that is highly expressed and positively correlates with the pathology stage in hepatocellular carcinoma (HCC) (168, 169). YTHDF1 is also an independent factor for an unfavorable HCC prognosis. Lin et al. suggested that Snail induces epithelial-mesenchymal transition (EMT) to enhance the metastasis of HCC cells. Mechanistically, m6A-modified CDS facilitates translational elongation of the Snail mRNA in a YTHDF1/eEF2-dependent manner (55). In addition, the YTHDF1-mediated aggressive phenotypes are also associated with the activation of the AKT/GSK-3β/β-catenin pathway (170). Chi et al. perceived that the effect of YTHDF1 in enhancing HCC proliferation can be antagonized by hsa-miR-139-5p (171). YTHDF1 also promotes HCC cell growth by upregulating the PI3K/AKT/mTOR signaling pathway (172). Hu et al. showed that METTL3-m6A-YTHDF1-mediated RBM14 overexpression promotes Kupffer cell polarization and HCC progression (173). Furthermore, YTHDF1 is involved in the regulation of HCC under hypoxic stress. For example, hypoxia-inducible factor-1α (HIF-1α)-mediated upregulation of YTHDF1 promotes autophagy-associated genes ATG2A and ATG14 translation, thus aggravating HCC malignancy behavior (174). FOXO3 is a negative regulator of hypoxia-induced autophagy and mediates the sorafenib sensitivity in HCC (175). Importantly, YTHDF1 binds to METTL3-methylated m6A modification in the FOXO3 mRNA 3’UTR and increases its mRNA stability rather than translation. Moreover, under the sublethal heat stress from insufficient radiofrequency ablation (IRFA), YTHDF1 binds to the m6A site on the 5’UTR of EGFR mRNA and triggers EGFR translation, eventually resulting in HCC recurrence after IRFA (176).
Notably, YTHDF3 is also reported as a potential oncogene in HCC. YTHDF3 enhances HCC metastasis by maintaining ZEB1 mRNA stability in an m6A-dependent mechanism (177). YTHDF3/integrin subunit alpha 6 (ITGA6) is positively regulated by the lysine-specific demethylase 5B (KDM5B)/microRNA-448 axis and thereby enhances the self-renewal of HCC cells (178).
Intriguingly, YTHDF2 has a paradoxical effect on HCC in different studies. Zhong et al. professed that hypoxia-induced YTHDF2 downregulation reverses the repression of YTHDF2 on the ERK/MAPK signaling pathway, subsequently removing the inhibitory effect of YTHDF2 on the proliferation and growth of HCC cells (179). Mechanistically, YTHDF2 suppresses the activation of the ERK/MAPK signaling pathway by selectively recognizing the m6A site at the 3’UTR and triggering EGFR mRNA degradation. Hou et al. confirmed that YTHDF2 is significantly downregulated in HCC cells and YTHDF2 deficiency elicits inflammation, vascular abnormalization, and metastatic progression (180). Specifically, YTHDF2 destabilizes the mRNA of m6A-modified interleukin 11 (IL11) and serpin family E member 2 (SERPINE2) to exert an inhibitory effect. Conversely, YTHDF2 is also considered a tumor-promoting factor in HCC (181, 182). Yang et al. discovered that microRNA-145 targets the 3’UTR of YTHDF2 mRNA to attenuate its expression and thereby inhibits the proliferation of HCC cells (183). And YTHDF2 participates in METTL3-m6A-mediated HCC malignancy by shortening the half-life of the suppressor of cytokine signaling 2 (SOCS2) mRNA (184). Additionally, YTHDF2 increases the m6A levels in the 5’UTR of OCT4 mRNA in tandem with promoting OCT4 expression, eventually accelerating the HCC cancer stem cell (CSC) phenotype and metastasis (185). And PA2G4 depends on YTHDF2 to stabilize FYN mRNA and promote EMT-induced HCC metastasis (186). The discrepancy in the effect of YTHDF2 on HCC may be due to different cellular microenvironments or tumor heterogeneity (187).
In addition, YTHDF1 and YTHDF2 facilitate the advancement of intrahepatic cholangiocarcinoma (ICC) through increasing EGFR mRNA translation and IFIT2 mRNA decay, respectively (188, 189). Meanwhile, YTHDF2 silencing restrains ICC resistance to the exposure of cisplatin by reversing the degradation of cyclin-dependent kinase inhibitor 1B (CDKN1B) mRNA (190).
YTHDF1 mutations occur in approximately 7% of gastric cancer (GC) patients, and high expression of YTHDF1 is correlated with high-risk progression and poor prognosis in patients (191–193). YTHDF1 deficiency is capable to attenuate GC progression, including proliferation and metastasis in vitro and in vivo. Mechanistically, YTHDF1 relies on m6A modification to promote the translation of frizzled7 (FZD7) and USP14, which transmit WNT/β-catenin signaling and AKT/ERK signaling, respectively (192, 193). In addition, METTL3 promotes the malignancy behavior of GC through YTHDF1/eIF3a-dependent post-transcriptional translation of SPHK2 (194).
Zhang et al. showed that the knockdown of YTHDF2 inhibits GC cell proliferation and accelerates apoptosis in vitro (195). And lncRNA LINC00470 relies on YTHDF2 to degrade m6A-containing PTEN mRNA and thus promote GC advancement (196). Additionally, the HIF-1α-induced increase of lncRNA-CBSLR suppresses ferroptosis and chem-sensitive under hypoxic stress through the YTHDF2-CBS-ACSL4 axis (197). Specifically, CBSLR contributes to CBS mRNA destabilization by binding to the m6A site on the CDS of CBS mRNA by recruiting YTHDF2. However, Shen et al. found that YTHDF2 plays a suppressive role in GC by destabilizing FOXC2 mRNA (198).
Among the YTHDF family, YTHDF2 is the most studied protein in pancreatic cancer. YTHDF2 is elevated in pancreatic cancer and orchestrates the migration/proliferation dichotomy (199). Specifically, YTHDF2 prevents EMT, migration, and invasion by downregulating YAP signaling and enhances proliferation by activating AKT/GSK3B/CCND1 pathway. However, YTHDF2 downregulates the levels of PERP and PER1 mRNA to promote cell proliferation and migration in an m6A-dependent manner (200, 201). METTL3-m6A-YTHDF2-mediated decay of nucleobindin 1 (NUCB1) mRNA counteracts the effects of NUCB1 in halting pancreatic cancer growth and augmenting the antitumor with gemcitabine (GEM) (202). Conversely, another study showed that the rs142933486 G>T polymorphism in PIK3CB improves PIK3CB mRNA and protein levels by derailing m6A-YTHDF2-dependent degradation mechanisms, which is significantly associated with the poor prognosis of PTEN-deficient pancreatic cancer patients (203). And compared with PIK3CB[T], YTHDF2 mainly binds to PIK3CB[G]. Similarly, FTO reverses YTHDF2-regulated degradation of platelet-derived growth factor C (PDGFC) mRNA and promotes cell proliferation by reactivating the AKT signaling pathway (204). Notably, YTHDF1 is associated with the immune microenvironment and prognosis of pancreatic cancer (205–207). A recent study found that a novel antineoplastic drug, Olean-28,13β-lactam (B28), inhibits glutamine metabolism by reducing the expression of YTHDF1, which induces pancreatic cancer cell death (208). In addition, YTHDF3-mediated downregulation of lncRNA DICER1-AS1 reverses the repression of glycolysis by miR-5586-5p in pancreatic cancer (209).
In colorectal cancer (CRC), YTHDF1 may be a molecular target for diagnosis and treatment (210). Mechanistically, elevated YTHDF1 in CRC is mainly attributed to an increase in DNA copy number (211). The oncogene c-MYC, WNT signaling, and APC mutation can also upregulate YTHDF1 expression at the translational level (89, 212). Further studies found that YTHDF1 promotes tumorigenicity and CSC-like activity by amplifying the WNT/β-catenin pathway with little effect on normal intestinal development (211). And deletion of YTHDF1 in ISCs shrinks tumor size and prolongs the lifespan of CRC-formed mice substantially. YTHDF1 can promote CRC progression and metastasis by translating m6A-modified Rho/Rac guanine nucleotide exchange factor 2 (ARHGEF2) mRNA and activating RhoA signaling (213). Furthermore, circular RNA protein tyrosine kinase 2 (circPTK2) restores the miR-136-5p-mediated repression of YTHDF1 by competitively binding to miR-136-5p, resulting in the CRC advancement and chemoresistance (214). Chen et al. suggested that YTHDF1-mediated glutamine metabolism reduces the sensitivity of CRC cells to cisplatin (215). Specifically, YTHDF1 targets the m6A of glutaminase 1 (GLS1) mRNA 3’UTR to promote its translation. And METTL3 deletion inhibits LDHA mRNA translation by reducing the binding of YTHDF1 to LDHA mRNA CDS, thereby hindering glycolysis and promoting 5-fluorouracil sensitivity in CRC cells (216). Interestingly, the rs8100241 G>A mutation in ANKLE1 increases ANKLE1 levels in an m6A-YTHDF1-dependent fashion, thereby inhibiting proliferation and maintaining the genomic stability of CRC (217).
In addition, YTHDF2 often collaborates with “writers” and participates in CRC progression. For example, METTL3 downregulates YPEL5 in an m6A-YTHDF2-dependent manner and boosts CRC progression (218). METTL14 exerts an inhibitory effect in CRC by promoting the degradation of SYR-related high-mobility-group box 4 (SOX4) mRNA and long noncoding RNA XIST, which is dependent on YTHDF2 (219, 220). Han et al. deciphered that glutaminolysis inhibition increases ATF4 expression through FTO-mediated demethylation and YTHDF2-regulated decay, which further inactivates mTOR and promotes pro-survival autophagy of CRC cells (221). Moreover, in CRC, silencing of microRNA-6125 destabilizes GSK3β mRNA by upregulating the expression of YTHDF2, ultimately increasing WNT/β-catenin/Cyclin D1 pathway-related proteins and promoting CRC growth (222). Intriguingly, Zhou et al. found that HIF-1α-induced upregulation of lncRNA STEAP3-AS1 activates the WNT/β-catenin signaling pathway through overexpression of STEAP3, leading to CRC progression in a hypoxic environment (223). Specifically, after combining YTHDF2, STEAP3-AS1 prohibits STEAP3 mRNA from binding with YTHDF2, thus antagonizing STEAP3 mRNA decay.
Moreover, Ni et al. revealed that the long noncoding RNA GAS5-YAP-YTHDF3 axis forms a feedback loop in CRC (224). In detail, the downregulation of GAS5 enhances CRC proliferation and invasion by inhibiting phosphorylation and ubiquitin-mediated decay of YAP, which positively regulates YTHDF3. And YTHDF3 promotes the degradation of GAS5 mRNA by recognizing the m6A in GAS5 mRNA. Furthermore, YTHDF3 recruits eIF2AK2 and eIF3A on the 5’UTR of target mRNAs and promotes translation in oxaliplatin-resistant CRC (225).
The expression of YTHDF1 and YTHDF2 is markedly upregulated in tumor tissues of lung cancer series and possesses tumor-promoting activities (226). Shi et al. demonstrated that YTHDF1 is amplified and increases the translation of key regulators of the G0/G1 cell cycle transition, including CDK2, CDK4, and cyclin D1 mRNAs, intensifying non-small cell lung cancer (NSCLC) progression under normoxia conditions (227). In addition, microRNA-376c, delivered by endothelial cells through extracellular vesicles, inhibits the YTHDF1 and WNT/β-catenin pathway in NSCLC cells, resulting in the malignant progression of NSCLC cells (228). Nevertheless, under cisplatin-induced oxidative stress, YTHDF1 deficiency activates the antioxidant Nrf2-AKR1C1 axis by inhibiting the Keap1 mRNA transition, which resulted in cisplatin resistance and poor prognosis. Furthermore, the YTHDF1-m6A-enolase1 (ENO1) translation axis is a crucial pathway for stimulating glycolysis and tumorigenesis (229). In KRAS and TP53 co-mutated lung adenocarcinomas, YTHDF1 recognizes m6A modification and contributes to tumor proliferation and poor prognosis through the upregulation of cyclin B1 (230).
In addition, YTHDF2 promotes translation but not clearance of 6-phosphogluconate dehydrogenase (6PGD) mRNA in an m6A-dependent manner by interacting with eIF3a/b, which enhances the pentose phosphate pathway (PPP) flux for tumor growth (231). The transcriptional repressor ZBTB4 and the tumor suppressor DAPK2 are negatively regulated by YTHDF2 and significantly associates with smoking-induced lung cancer (232, 233). However, ALKBH5 attenuates YTHDF2-mediated downregulation of oncogenic drivers such as SOX2, SMAD7, and MYC, contributing to the progression of aggressive lung cancer with KRAS mutation/LKB1 loss (234). Furthermore, YTHDF2 produces a positive effect on lung adenocarcinoma progression through the mRNA decay of AXIN1, a negative regulator of the WNT/β-catenin pathway (235). YTHDF2 produces the same effect in a VIRMA-m6A-dependent fashion in lung adenocarcinoma and NSCLC by reducing BTG2 mRNA and DAPK3 mRNA stability, respectively (236, 237). Nevertheless, YTHDF2 induces sensitivity of lung adenocarcinoma to gefitinib via cleavage of circASK1 (238). Interestingly, YTHDF2 promotes proliferation and downregulates the FAM83D-TGFβ1-SMAD2/3 pathway to inhibit migration and invasion in lung adenocarcinoma cells (239). In lung squamous cell carcinoma, up-regulation of YTHDF2 under hypoxic conditions activates the mTOR/AKT signaling pathway and induces EMT to play a tumor-promoting role (240).
Interestingly, YTHDF1 and YTHDF2 regulate YAP expression by competitively binding to YTHDF3-m6A-YAP mRNA, thereby aggravating and attenuating the malignancy behavior of NSCLC, respectively (241). YTHDF1/3 recruits eIF3a/b to promote YAP mRNA translation, while YTHDF2/3 recruits AGO2 to promote YAP mRNA decay. And YTHDF3 indirectly increased YAP levels to empower NSCLC progression and drug resistance by enhancing MALAT1 mRNA stability (242).
YTHDF family plays a tumor-promoting role in bladder cancer. Specifically, METTL3 and YTHDF1 are closely related to malignant transformation and tumorigenesis in the presence of chemical carcinogens, with the m6A-methylated 3’UTR promoting oncogene CDCP1 translation (243). Moreover, YTHDF1/3 promotes aggressive phenotypes by translating ITGA6 mRNA, while YTHDF2 facilitates migration by degrading the mRNAs of the tumor suppressors SETD7 and KLF4 (244, 245).
YTHDF2 acts as a facilitator and is negatively regulated by miR-493-3p in prostate cancer (PCa) (246). Du et al. considered that KDM5A abrogates the inhibition of miR-495 on YTHDF2, and then upregulated YTHDF2 intensifies PCa progression by inducing m6A-MOB3B mRNA decay (247). In addition, YTHDF2 clears METTL3-mediated m6A-dependent mRNA of LHPP, NKX3-1, and USP4 (248, 249). The decrease of LHPP and NKX3-1 causes PCa proliferation and migration by inducing AKT phosphorylation. And downregulated USP4 promotes ARHGDIA expression by reducing ELAVL1 protein, thus accelerating invasion and metastasis of PCa. METTL14-mediated m6A modification of Thrombospondin 1 (THBS1) mRNA promotes PCa proliferation in a YTHDF2-dependent manner of transcriptome degradation (250).
In breast cancer, high expression of YTHDF1 and YTHDF3 is associated with gene copy number amplification and induces a poor prognosis (251, 252). YTHDF1 targets FOXM1 mRNA and positively regulates breast cancer progression (253). Additionally, hypoxia-mediated downregulation of miR-16-5p restored YTHDF1 expression, thereby promoting tumor glycolysis by enhancing PKM2 mRNA translation (254). Sun et al. demonstrated that YTHDF1 stabilizes E2F8 mRNA, which accelerates DNA damage repair and chemoresistance to adriamycin, cisplatin, and the PARP inhibitor olaparib in breast cancer cells (255). YTHDF1/eEF1-mediated translational elongation of KRT7 mRNA and YTHDF3-induced mRNAs translation of ST6GALNAC5, GJA1, and EGFR is involved in breast cancer lung and brain metastasis, respectively (57, 256). And YTHDF3 can be antagonized by miR-106b-5p (257). Moreover, YTHDF3 stabilizes ZEB1 mRNA to promote the invasion and migration of triple-negative breast cancer (TNBC) cells (258). Furthermore, YTHDF2 is upregulated in TNBC cells and prevents cell apoptosis (259, 260). YTHDF2 also targets the m6A site 5’UTR region of ATF3 mRNA to mitigate the resistance of breast cancer cells to tamoxifen (261).
YTHDF1 and YTHDF2 are considered oncogenes in ovarian cancer. YTHDF1 is recruited to the m6A site of EIF3C mRNA and stimulates EIF3C as well as overall protein translation (262). YTHDF1 also confers cisplatin-resistant ovarian cancer cells with CSC-like traits by promoting m6A-TRIM29 mRNA translation (263). Furthermore, FBW7 abrogates the mRNA degradation of YTHDF2 on pro-apoptotic gene BMF by inducing YTHDF2 decay, disrupting ovarian cancer progression (264). Moreover, YTHDF2 can be directly targeted and inhibited by miR-145 in ovarian cancer cells (265).
In cervical cancer (CC) cells, YTHDF1 accelerates m6A-augmented glycolysis and cancer progression by promoting translational elongation of pyruvate dehydrogenase kinase 4 (PDK4) mRNA and stabilization of hexokinase 2 (HK2) mRNA (56, 266). Specifically, the YTHDF1/eEF-2 complex binds the m6A site of PDK4 mRNA at the 5’UTR and YTHDF1 recognizes the m6A-modified 3’UTR of HK2 mRNA. Furthermore, YTHDF1 plays a tumor-promoting role by facilitating mitosis-associated RANBP2 mRNA translation in an m6A-mediated approach, while YTHDF2 exerts the same role by degrading the tumor suppressor GAS5 mRNA (267, 268). YTHDF2 deficiency suppresses the proliferation of CC cells, promotes apoptosis, and arrests the cells at the S phase (269). YTHDF2 can also facilitate EMT and cisplatin resistance in CC cells by stabilizing AXIN1 mRNA (270).
YTHDF1 and YTHDF2 modulate the negative regulator PHLPP2 and positive regulator mTORC2 of AKT respectively, which is unfavorable to the tumorigenicity of the AKT pathway in endometrial cancer (EC) (271). In addition, YTHDF2-mediated transcript degradation of IRS1 is accompanied by inhibition of the AKT/MMP9 signaling pathway, thereby impairing the activity of endometrial cells (272). And YTHDF2 deficiency activates the WNT signaling pathway by reducing the decay of HOXB13 mRNA, and thus promotes EC invasion and metastasis (273). Conversely, YTHDF2 degrades lncRNA FENDRR to enhance the expression of SOX4, which ultimately promotes EC cell proliferation and hinders apoptosis (274).
YTHDF1 and YTHDF2 were found to be highly overexpressed in glioblastoma (GBM) tissues compared to normal tissues (275). YTHDF1 is required for maintaining GBM CSC properties and promoting proliferation, migration, and chemoresistance (276). And Musashi-1(MSI1) is a GBM hyper-oncogenic regulator and positively regulates YTHDF1 expression. YTHDF1 also assists METTL3 in increasing levels of ADAR1 and thereby stimulates GBM cell growth (277). In addition, YTHDF2 is positively regulated by the EGFR/SRC/ERK pathway and facilitates the malignancy progression of GBM by degrading downstream transcripts, including LXRα, HIVEP2, UBXN1, and ASS1 mRNAs in an m6A-dependent manner (43, 278, 279). Among them, LXRα and ASS1 are related to cholesterol homeostasis and arginine metabolism, respectively. Strikingly, YTHDF2 recognizes m6A methylation to maintain MYC mRNA stability, thereby promoting the expression of the downstream effector IGFBP3, leading to GBM CSC growth (280). And this process occurs specifically in GBM CSCs but not in normal neural stem cells (NSCs). Chen et al. verified that YTHDF2 promotes temozolomide desensitization in GBM cells (281). Mechanistically, YTHDF2 activates PI3K/AKT and NF-κB signaling pathways by targeting the 3’UTR and downregulating the mRNAs stability of EPHB3 and TNFAIP3.
YTHDF1 is amplified in melanoma, and the combination of YTHDF1 and HNRNPA2B1 significantly increases the diagnostic validity (282). However, YTHDF1 inhibits ocular melanoma progression by facilitating HINT2 mRNA translation (283). YTHDF2 knockdown promotes tumor growth and reduces the sensitivity of anti-PD-1 therapy by enhancing the mRNAs stability of the intrinsic genes PD-1 (PDCD1), CXCR4, and SOX10 in an m6A-dependent fashion (284). Yu et al. discovered that histone lactylation promotes YTHDF2 expression in ocular melanoma, and YTHDF2 stimulates tumorigenesis by degrading m6A-modified PER1 and TP53 mRNAs (285). Similarly, YTHDF3 also promotes ocular melanoma progression by promoting CTNNB1 mRNA translation in an m6A-dependent manner (286).
The occurrence of Merkel cell carcinoma (MCC) is mostly attributed to the attack of the small T antigen of Merkel cell polyomavirus (MCPyV) (287). Meanwhile, overexpression of YTHDF1 improves the proliferative and clonogenic capacity of MCC cells by recruiting eIF3a/b to promote the translation initiation of small T antigen mRNA. Mechanistically, overexpression of YTHDF1 is caused by increased gene copy number.
Nguyen et al. first reported that YTHDF2 is identified as a novel acute myeloid leukemia1 (AML1) T translocation partner gene (288). Notably, YTHDF2 is highly expressed in different AML subtypes (289). And inhibition of YTHDF2 specifically impairs AML initiation and progression while expanding hematopoietic stem cells (HSCs) and maintaining normal hematopoietic function. In detail, YTHDF2 promotes the development and propagation of AML CSCs by degrading multiple m6A-modified mRNAs such as TNF receptor superfamily member 1b (TNFRSF1b) that are associated with the functional integrity of AML CSCs. Moreover, the AML1/ETO-HIF1α loop transactivates the YTHDF2 promoter to promote t (8, 21) AML cell proliferation (290). However, YTHDF2 may interfere with the glycolytic process of AML cells by destabilizing transcripts of phosphofructokinase platelet (PFKP) and lactate dehydrogenase B (LDHB) (291). Interestingly, the three YTHDFs can jointly degrade the associated transcripts and inhibit the differentiation of AML cells (63) (Figures 4–6) (Tables 1–3).
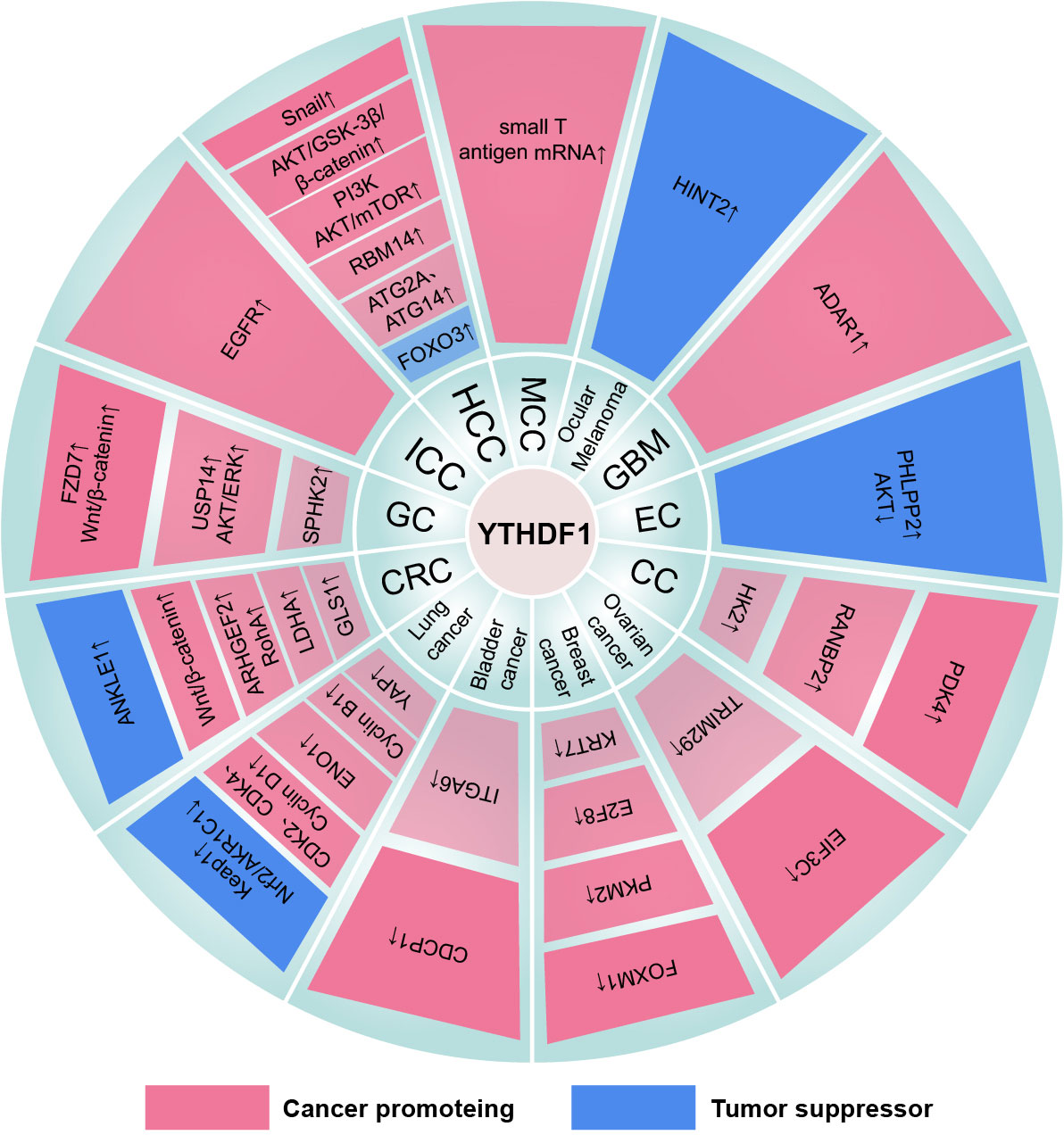
Figure 4 The mechanism of the YTHDF1 family in cancers. “↓” is the decrease of target mRNAs. “↑” is the increase of target mRNAs.
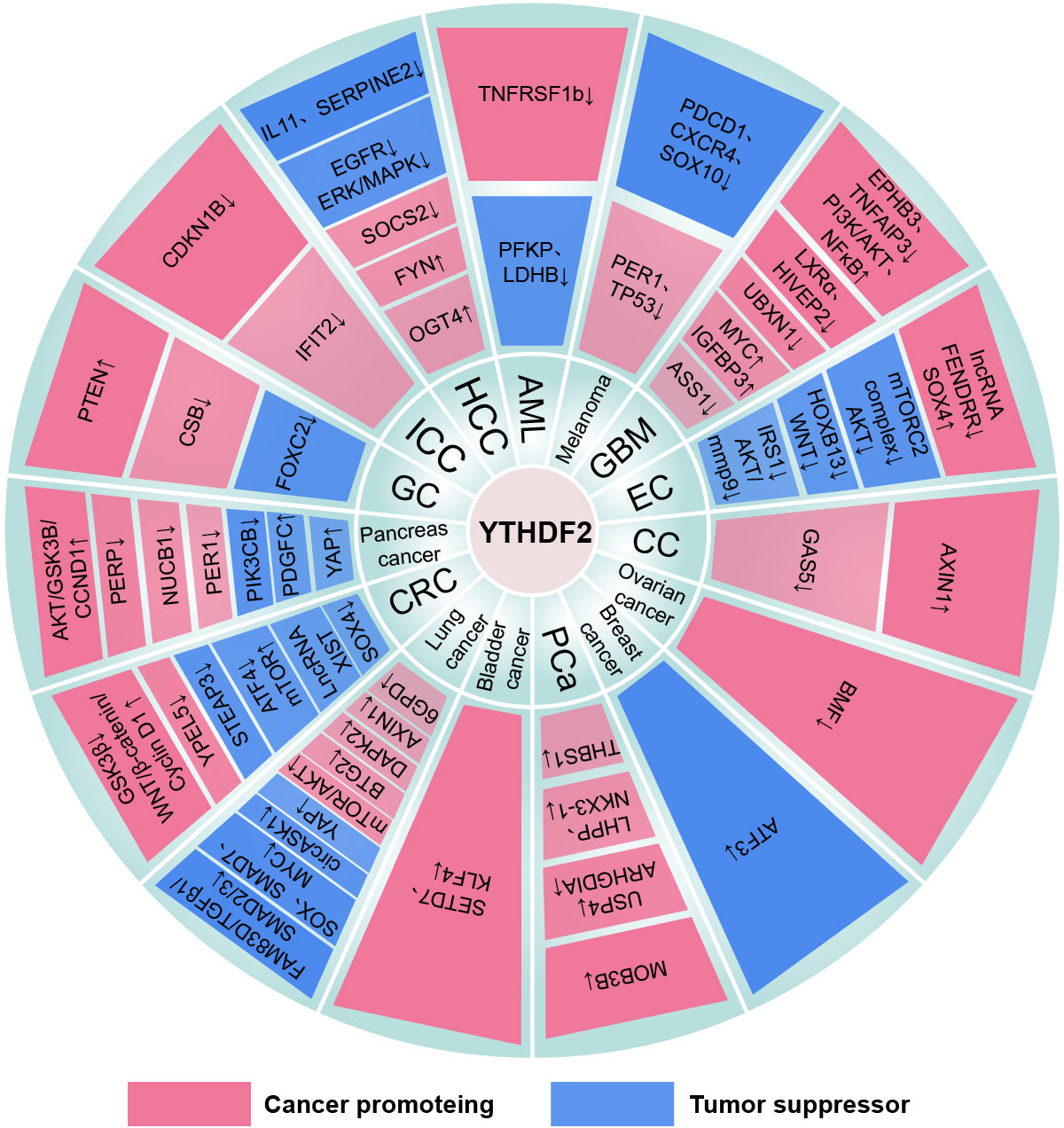
Figure 5 The mechanism of the YTHDF2 family in cancers. “↓” is the decrease of target mRNAs. “↑” is the increase of target mRNAs.
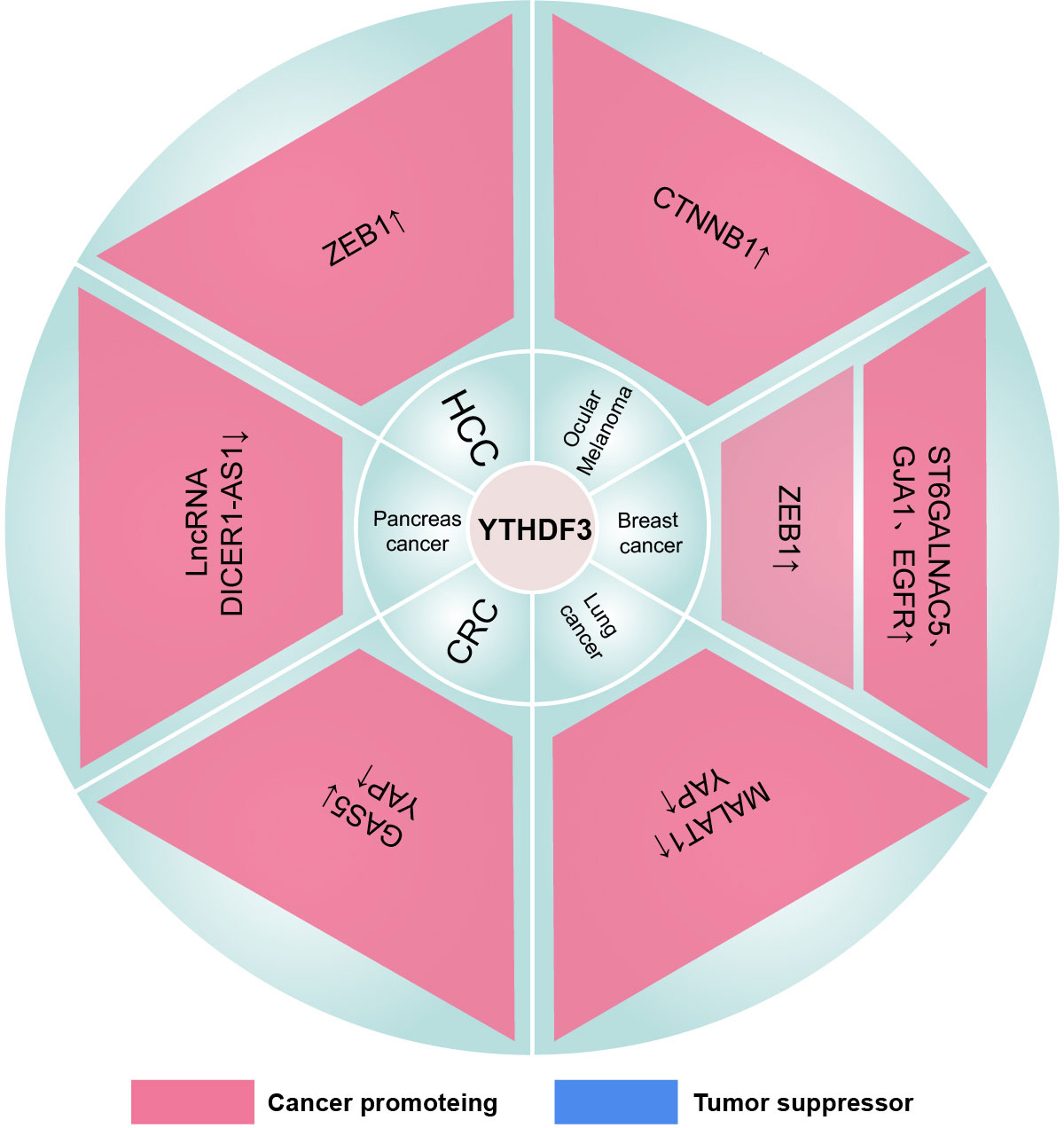
Figure 6 The mechanism of the YTHDF3 family in cancers. “↓” is the decrease of target mRNAs. “↑” is the increase of target mRNAs.
Although it has been revealed that the YTHDF family is involved in a variety of biological processes as the “readers” of m6A modification, there are still many mysteries about the YTHDF family that need to be discovered and solved in terms of structure, function, and treatment.
The discussion of the structure and function of YTHDFs is partially doubtful due to the limitations of technology and conditions. The reason why YTHDFs select the same or different target mRNAs and m6A sites on mRNAs, and why YTHDFs pair with different cooperating m6A regulators, has not been reached. In addition, YTHDFs can be localized in different cellular compartments and may re-enter the nucleus or transport out of the cell membrane, thus expanding the regulation of YTHDFs. The post-transcriptional modifications of YTHDFs and interactions of YTHDFs with other proteins also add to the structure and function complexity of YTHDFs. Therefore, the development of emerging technologies, the control of various conditions, and the change of different stimulus states are necessary to further investigations in the YTHDF family.
At present, many experiments have successfully constructed the YTHDF1/2/3 genetic KO mouse model using different techniques. First, the whole-body YTHDF1/2/3 KO mice are generated directly based on CRISPR/Cas9 by deleting a certain exon or inducing the premature appearance of a stop codon (87, 88, 110, 213). Second, the Cre/LoxP technique is used to generate cell-specific conditional YTHDF1/2/3 KO mice (74, 84, 108, 165, 167, 174, 227, 289). This represents an improvement in experimental research moving from in vitro to in vivo. However, the specific mutation of functional RNA binding sites of YTHDFs in mice needs to be further realized. In addition, one of the important purposes of experimental research is clinical transformation, so it is of great need to explore the application value of targeting YTHDFs in the clinic, especially in tumors. Many clinical-related studies have analyzed the expression profile of the m6A regulator in tumors and its association with the immune microenvironment, grading, staging, therapeutic effect, and prognosis. For example, the analysis of 162 HCC samples from the Zhou et al. and 177 HCC samples from the Nakagawa et al. showed that YTHDF1 was related to poor prognosis of HCC and YTHDF2 was related to HCC recurrence, respectively (169, 182). YTHDF1 was associated with a poor prognosis of GC in a study of 379 patients with GC (164). Interestingly, high expression of YTHDF1 and YTHDF2 was associated with a better prognosis in 603 cases of resected NSCLC, which might be due to increased tumor-infiltrating lymphocytes (TILs) and decreased co-inhibitor molecule PD-L1 (226). In addition, an assessment of single nucleotide polymorphisms (SNPs) in the YTHDF1 gene in 313 cases of hepatoblastoma showed that rs6090311 A>G was correlated with a reduced risk of hepatoblastoma (292). A similar SNPs assessment found that the YTHDF2 rs3738067 variant significantly increased glioma risk in 171 pediatric patients (293). Moreover, increasing evidence confirms the efficacy of bioinformatics analysis based on TCGA and other databases for the YTHDFs-associated model. To sum up, the expression of YTHDFs is significantly correlated with the grades and stages of various tumors and may be used as indicators to judge the occurrence and development of tumors. YTHDFs may act as independent prognostic factors for many tumors and affect survival-related indicators such as overall survival (OS), disease-free survival (DFS), and progression-free survival (PFS). At the therapeutic level, targeting YTHDFs can not only directly modulate the malignancy behavior of tumors, but also affect the sensitivity of chemotherapy and immunotherapy. Besides, YTHDFs also have the possibility of effective clinical application in non-cancer, including hematopoietic, anti-obesity, anti-viral, and anti-inflammatory.
However, studies of YTHDFs are still in the preclinical stage and many issues need attention. First, the clinical application of YTHDFs in different diseases, alone or in combination with other targets, requires further investigation. Second, the effectiveness of YTHDFs in diagnosing and predicting prognosis may vary across disease types, grades, and stages. Most importantly, the specific molecules targeting YTHDFs have not yet been developed. So how can YTHDFs be used in clinical treatment? The expression of YTHDFs can be regulated by other strategies. Targeting upstream or metabolic mechanisms of YTHDFs is an alternative approach to indirectly regulate the levels of YTHDFs (Figure 7). YTHDF2 has the capability of inhibiting the progression of HCC, and this effect can be antagonized by HIF-2α (180). Therefore, the HIF-2α antagonist (PT2385) can indirectly restore the effect of YTHDF2. And CDK1 inhibitors promote YTHDF2 proteolysis in AML (294). Furthermore, the delivery of target genes using viral vectors is also a feasible approach to target YTHDFs. YTHDF1 overexpression therapy can be achieved by injecting adeno-associated virus (AAV)-YTHDF1 into the hippocampus of diabetic cognitively impaired mice (116). In conclusion, clarifying the limitations of YTHDFs is conducive to better clinical transformation.
With multi-omics advancement, the roles of m6A modification have been gradually and seriously excavated. By binding to m6A, the YTHDF family plays an important role in the regulation of various physiological and pathological processes, including embryonic development, stem cell fate, fat metabolism, neuromodulation, cardiovascular effect, viral infection, immunity, and especially in tumors. In particular, YTHDFs regulate multiple tumor phenotypes such as proliferation, metastasis, metabolism, drug resistance, and immunity. Additionally, YTHDFs can be used as biomarkers for the diagnosis, treatment, and predictors of prognosis evaluation. On-going explorations of YTHDFs in modeling disease progression are still warranted for a better and deeper understanding of epigenetic modifications.
LC collected the related papers and drafted the manuscript. YG made the figures and revised the manuscript. SX edited and revised the manuscript. JG designed the framework and revised the manuscript. JY, MW, and TL revised the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by grants from the National Natural Science Foundation of China (81900518 to JG; 81702792 to SX; 82000088 to TL); the HUBEI Natural Science Foundation (2017CFB467 to MW); Tongji Hospital Clinical Research Flagship Program (grant no. 2019CR203), National Key Research and Development Program of China (grant no. 2022YFA110530 and 2019TFC1315905).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Boccaletto P, Stefaniak F, Ray A, Cappannini A, Mukherjee S, Purta E, et al. Modomics: A database of rna modification pathways. 2021 update. Nucleic Acids Res (2021) 50(D1):D231–D235. doi: 10.1093/nar/gkab1083
2. Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger rna from novikoff hepatoma cells. Proc Natl Acad Sci U S A (1974) 71(10):3971–5. doi: 10.1073/pnas.71.10.3971
3. Adams JM, Cory S. Modified nucleosides and bizarre 5'-termini in mouse myeloma mrna. Nature (1975) 255(5503):28–33. doi: 10.1038/255028a0
4. Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mrna methylation reveals enrichment in 3' utrs and near stop codons. Cell (2012) 149(7):1635–46. doi: 10.1016/j.cell.2012.05.003
5. Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, et al. Topology of the human and mouse M6a rna methylomes revealed by M6a-seq. Nature (2012) 485(7397):201–6. doi: 10.1038/nature11112
6. Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. A Mettl3-Mettl14 complex mediates mammalian nuclear rna N6-adenosine methylation. Nat Chem Biol (2014) 10(2):93–5. doi: 10.1038/nchembio.1432
7. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian wtap is a regulatory subunit of the rna N6-methyladenosine methyltransferase. Cell Res (2014) 24(2):177–89. doi: 10.1038/cr.2014.3
8. Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, et al. N6-methyladenosine in nuclear rna is a major substrate of the obesity-associated fto. Nat Chem Biol (2011) 7(12):885–7. doi: 10.1038/nchembio.687
9. Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, et al. Alkbh5 is a mammalian rna demethylase that impacts rna metabolism and mouse fertility. Mol Cell (2013) 49(1):18–29. doi: 10.1016/j.molcel.2012.10.015
10. Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N6-Methyladenosine-Dependent regulation of messenger rna stability. Nature (2014) 505(7481):117–20. doi: 10.1038/nature12730
11. Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, et al. Nuclear M(6)a reader Ythdc1 regulates mrna splicing. Mol Cell (2016) 61(4):507–19. doi: 10.1016/j.molcel.2016.01.012
12. Roundtree IA, Luo GZ, Zhang Z, Wang X, Zhou T, Cui Y, et al. Ythdc1 mediates nuclear export of N(6)-methyladenosine methylated mrnas. Elife (2017) 6:e31311. doi: 10.7554/eLife.31311
13. Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, et al. Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res (2017) 27(9):1115–27. doi: 10.1038/cr.2017.99
14. Geuens T, Bouhy D, Timmerman V. The hnrnp family: Insights into their role in health and disease. Hum Genet (2016) 135(8):851–67. doi: 10.1007/s00439-016-1683-5
15. Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, et al. Recognition of rna N(6)-methyladenosine by Igf2bp proteins enhances mrna stability and translation. Nat Cell Biol (2018) 20(3):285–95. doi: 10.1038/s41556-018-0045-z
16. Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, et al. 5' utr M(6)a promotes cap-independent translation. Cell (2015) 163(4):999–1010. doi: 10.1016/j.cell.2015.10.012
17. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine modulates messenger rna translation efficiency. Cell (2015) 161(6):1388–99. doi: 10.1016/j.cell.2015.05.014
18. Bhattarai PY, Kim G, Poudel M, Lim SC, Choi HS. Mettl3 induces Plx4032 resistance in melanoma by promoting M(6)a-dependent egfr translation. Cancer Lett (2021) 522:44–56. doi: 10.1016/j.canlet.2021.09.015
19. Chang YZ, Chai RC, Pang B, Chang X, An SY, Zhang KN, et al. Mettl3 enhances the stability of Malat1 with the assistance of hur Via M6a modification and activates nf-kappab to promote the malignant progression of idh-wildtype glioma. Cancer Lett (2021) 511:36–46. doi: 10.1016/j.canlet.2021.04.020
20. Patil DP, Chen CK, Pickering BF, Chow A, Jackson C, Guttman M, et al. M(6)a rna methylation promotes xist-mediated transcriptional repression. Nature (2016) 537(7620):369–73. doi: 10.1038/nature19342
21. Yue Y, Liu J, Cui X, Cao J, Luo G, Zhang Z, et al. Virma mediates preferential M(6)a mrna methylation in 3'utr and near stop codon and associates with alternative polyadenylation. Cell Discovery (2018) 4:10. doi: 10.1038/s41421-018-0019-0
22. Wen J, Lv R, Ma H, Shen H, He C, Wang J, et al. Zc3h13 regulates nuclear rna M(6)a methylation and mouse embryonic stem cell self-renewal. Mol Cell (2018) 69(6):1028–38.e6. doi: 10.1016/j.molcel.2018.02.015
23. Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, et al. The U6 snrna M(6)a methyltransferase Mettl16 regulates Sam synthetase intron retention. Cell (2017) 169(5):824–35.e14. doi: 10.1016/j.cell.2017.05.003
24. Ma H, Wang X, Cai J, Dai Q, Natchiar SK, Lv R, et al. N(6-)Methyladenosine methyltransferase Zcchc4 mediates ribosomal rna methylation. Nat Chem Biol (2019) 15(1):88–94. doi: 10.1038/s41589-018-0184-3
25. van Tran N, Ernst FGM, Hawley BR, Zorbas C, Ulryck N, Hackert P, et al. The human 18s rrna M6a methyltransferase Mettl5 is stabilized by Trmt112. Nucleic Acids Res (2019) 47(15):7719–33. doi: 10.1093/nar/gkz619
26. Ueda Y, Ooshio I, Fusamae Y, Kitae K, Kawaguchi M, Jingushi K, et al. Alkb homolog 3-mediated trna demethylation promotes protein synthesis in cancer cells. Sci Rep (2017) 7:42271. doi: 10.1038/srep42271
27. Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the fto gene is associated with body mass index and predisposes to childhood and adult obesity. Science (2007) 316(5826):889–94. doi: 10.1126/science.1141634
28. Wojtas MN, Pandey RR, Mendel M, Homolka D, Sachidanandam R, Pillai RS. Regulation of M(6)a transcripts by the 3'–>5' rna helicase Ythdc2 is essential for a successful meiotic program in the mammalian germline. Mol Cell (2017) 68(2):374–87.e12. doi: 10.1016/j.molcel.2017.09.021
29. Alarcon CR, Goodarzi H, Lee H, Liu X, Tavazoie S, Tavazoie SF. Hnrnpa2b1 is a mediator of M(6)a-dependent nuclear rna processing events. Cell (2015) 162(6):1299–308. doi: 10.1016/j.cell.2015.08.011
30. Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-Methyladenosine-Dependent rna structural switches regulate rna-protein interactions. Nature (2015) 518(7540):560–4. doi: 10.1038/nature14234
31. Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters rna structure to regulate binding of a low-complexity protein. Nucleic Acids Res (2017) 45(10):6051–63. doi: 10.1093/nar/gkx141
32. Wu B, Su S, Patil DP, Liu H, Gan J, Jaffrey SR, et al. Molecular basis for the specific and multivariant recognitions of rna substrates by human hnrnp A2/B1. Nat Commun (2018) 9(1):420. doi: 10.1038/s41467-017-02770-z
33. Huang XT, Li JH, Zhu XX, Huang CS, Gao ZX, Xu QC, et al. Hnrnpc impedes M(6)a-dependent anti-metastatic alternative splicing events in pancreatic ductal adenocarcinoma. Cancer Lett (2021) 518:196–206. doi: 10.1016/j.canlet.2021.07.016
34. Stoilov P, Rafalska I, Stamm S. Yth: A new domain in nuclear proteins. Trends Biochem Sci (2002) 27(10):495–7. doi: 10.1016/s0968-0004(02)02189-8
35. Zhang Z, Theler D, Kaminska KH, Hiller M, de la Grange P, Pudimat R, et al. The yth domain is a novel rna binding domain. J Biol Chem (2010) 285(19):14701–10. doi: 10.1074/jbc.M110.104711
36. Li F, Zhao D, Wu J, Shi Y. Structure of the yth domain of human Ythdf2 in complex with an M(6)a mononucleotide reveals an aromatic cage for M(6)a recognition. Cell Res (2014) 24(12):1490–2. doi: 10.1038/cr.2014.153
37. Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. Ythdf3 facilitates translation and decay of N(6)-Methyladenosine-Modified rna. Cell Res (2017) 27(3):315–28. doi: 10.1038/cr.2017.15
38. Luo S, Tong L. Molecular basis for the recognition of methylated adenines in rna by the eukaryotic yth domain. Proc Natl Acad Sci U S A (2014) 111(38):13834–9. doi: 10.1073/pnas.1412742111
39. Dai X, Wang T, Gonzalez G, Wang Y. Identification of yth domain-containing proteins as the readers for N1-methyladenosine in rna. Anal Chem (2018) 90(11):6380–4. doi: 10.1021/acs.analchem.8b01703
40. Gao Y, Pei G, Li D, Li R, Shao Y, Zhang QC, et al. Multivalent M(6)a motifs promote phase separation of ythdf proteins. Cell Res (2019) 29(9):767–9. doi: 10.1038/s41422-019-0210-3
41. Ries RJ, Zaccara S, Klein P, Olarerin-George A, Namkoong S, Pickering BF, et al. M(6)a enhances the phase separation potential of mrna. Nature (2019) 571(7765):424–8. doi: 10.1038/s41586-019-1374-1
42. Patil DP, Pickering BF, Jaffrey SR. Reading M(6)a in the transcriptome: M(6)a-binding proteins. Trends Cell Biol (2018) 28(2):113–27. doi: 10.1016/j.tcb.2017.10.001
43. Fang R, Chen X, Zhang S, Shi H, Ye Y, Shi H, et al. Egfr/Src/Erk-stabilized Ythdf2 promotes cholesterol dysregulation and invasive growth of glioblastoma. Nat Commun (2021) 12(1):177. doi: 10.1038/s41467-020-20379-7
44. Hou G, Zhao X, Li L, Yang Q, Liu X, Huang C, et al. Sumoylation of Ythdf2 promotes mrna degradation and cancer progression by increasing its binding affinity with M6a-modified mrnas. Nucleic Acids Res (2021) 49(5):2859–77. doi: 10.1093/nar/gkab065
45. Theler D, Dominguez C, Blatter M, Boudet J, Allain FH. Solution structure of the yth domain in complex with N6-methyladenosine rna: A reader of methylated rna. Nucleic Acids Res (2014) 42(22):13911–9. doi: 10.1093/nar/gku1116
46. Liao S, Sun H, Xu C. Yth domain: A family of N(6)-methyladenosine (M(6)a) readers. Genomics Proteomics Bioinf (2018) 16(2):99–107. doi: 10.1016/j.gpb.2018.04.002
47. Xu C, Liu K, Ahmed H, Loppnau P, Schapira M, Min J. Structural basis for the discriminative recognition of N6-methyladenosine rna by the human Yt521-b homology domain family of proteins. J Biol Chem (2015) 290(41):24902–13. doi: 10.1074/jbc.M115.680389
48. Li Y, Bedi RK, Moroz-Omori EV, Caflisch A. Structural and dynamic insights into redundant function of ythdf proteins. J Chem Inf Model (2020) 60(12):5932–5. doi: 10.1021/acs.jcim.0c01029
49. Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, et al. Ythdf2 destabilizes M(6)a-containing rna through direct recruitment of the Ccr4-not deadenylase complex. Nat Commun (2016) 7:12626. doi: 10.1038/ncomms12626
50. Zheng D, Ezzeddine N, Chen CY, Zhu W, He X, Shyu AB. Deadenylation is prerequisite for p-body formation and mrna decay in mammalian cells. J Cell Biol (2008) 182(1):89–101. doi: 10.1083/jcb.200801196
51. Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, et al. Endoribonucleolytic cleavage of M(6)a-containing rnas by rnase P/Mrp complex. Mol Cell (2019) 74(3):494–507 e8. doi: 10.1016/j.molcel.2019.02.034
52. Lee Y, Choe J, Park OH, Kim YK. Molecular mechanisms driving mrna degradation by M(6)a modification. Trends Genet (2020) 36(3):177–88. doi: 10.1016/j.tig.2019.12.007
53. Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic M(6)a mrna methylation directs translational control of heat shock response. Nature (2015) 526(7574):591–4. doi: 10.1038/nature15377
54. Rauch S, He C, Dickinson BC. Targeted M(6)a reader proteins to study epitranscriptomic regulation of single rnas. J Am Chem Soc (2018) 140(38):11974–81. doi: 10.1021/jacs.8b05012
55. Lin X, Chai G, Wu Y, Li J, Chen F, Liu J, et al. Rna M(6)a methylation regulates the epithelial mesenchymal transition of cancer cells and translation of snail. Nat Commun (2019) 10(1):2065. doi: 10.1038/s41467-019-09865-9
56. Li Z, Peng Y, Li J, Chen Z, Chen F, Tu J, et al. N(6)-methyladenosine regulates glycolysis of cancer cells through Pdk4. Nat Commun (2020) 11(1):2578. doi: 10.1038/s41467-020-16306-5
57. Chen F, Chen Z, Guan T, Zhou Y, Ge L, Zhang H, et al. N(6) -methyladenosine regulates mrna stability and translation efficiency of Krt7 to promote breast cancer lung metastasis. Cancer Res (2021) 81(11):2847–60. doi: 10.1158/0008-5472.CAN-20-3779
58. Li J, Chen K, Dong X, Xu Y, Sun Q, Wang H, et al. Ythdf1 promotes mrna degradation Via Ythdf1-Ago2 interaction and phase separation. Cell Prolif (2022) 55(1):e13157. doi: 10.1111/cpr.13157
59. Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, et al. Cytoplasmic M(6)a reader Ythdf3 promotes mrna translation. Cell Res (2017) 27(3):444–7. doi: 10.1038/cr.2017.10
60. Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular rnas driven by N(6)-methyladenosine. Cell Res (2017) 27(5):626–41. doi: 10.1038/cr.2017.31
61. Liu J, Gao M, Xu S, Chen Y, Wu K, Liu H, et al. Ythdf2/3 are required for somatic reprogramming through different rna deadenylation pathways. Cell Rep (2020) 32(10):108120. doi: 10.1016/j.celrep.2020.108120
62. Zhao YL, Liu YH, Wu RF, Bi Z, Yao YX, Liu Q, et al. Understanding M(6)a function through uncovering the diversity roles of yth domain-containing proteins. Mol Biotechnol (2019) 61(5):355–64. doi: 10.1007/s12033-018-00149-z
63. Zaccara S, Jaffrey SR. A unified model for the function of ythdf proteins in regulating M(6)a-modified mrna. Cell (2020) 181(7):1582–95 e18. doi: 10.1016/j.cell.2020.05.012
64. Hazra D, Chapat C, Graille M. M(6)a mrna destiny: Chained to the rhythm by the yth-containing proteins. Genes (Basel) (2019) 10(1):49. doi: 10.3390/genes10010049
65. Lasman L, Krupalnik V, Viukov S, Mor N, Aguilera-Castrejon A, Schneir D, et al. Context-dependent functional compensation between ythdf M(6)a reader proteins. Genes Dev (2020) 34(19-20):1373–91. doi: 10.1101/gad.340695.120
66. Edupuganti RR, Geiger S, Lindeboom RGH, Shi H, Hsu PJ, Lu Z, et al. N(6)-methyladenosine (M(6)a) recruits and repels proteins to regulate mrna homeostasis. Nat Struct Mol Biol (2017) 24(10):870–8. doi: 10.1038/nsmb.3462
67. Zhang F, Kang Y, Wang M, Li Y, Xu T, Yang W, et al. Fragile X mental retardation protein modulates the stability of its M6a-marked messenger rna targets. Hum Mol Genet (2018) 27(22):3936–50. doi: 10.1093/hmg/ddy292
68. Wu R, Li A, Sun B, Sun JG, Zhang J, Zhang T, et al. A novel M(6)a reader Prrc2a controls oligodendroglial specification and myelination. Cell Res (2019) 29(1):23–41. doi: 10.1038/s41422-018-0113-8
69. Ivanova I, Much C, Di Giacomo M, Azzi C, Morgan M, Moreira PN, et al. The rna M(6)a reader Ythdf2 is essential for the post-transcriptional regulation of the maternal transcriptome and oocyte competence. Mol Cell (2017) 67(6):1059–67.e4. doi: 10.1016/j.molcel.2017.08.003
70. Huang T, Liu Z, Zheng Y, Feng T, Gao Q, Zeng W. Ythdf2 promotes spermagonial adhesion through modulating mmps decay Via M(6)a/Mrna pathway. Cell Death Dis (2020) 11(1):37. doi: 10.1038/s41419-020-2235-4
71. Qi M, Sun H, Guo Y, Zhou Y, Gu X, Jin J, et al. M(6) a reader protein Ythdf2 regulates spermatogenesis by timely clearance of phase-specific transcripts. Cell Prolif (2022) 55(1):e13164. doi: 10.1111/cpr.13164
72. Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, Kuuspalu A, et al. M(6)a-dependent maternal mrna clearance facilitates zebrafish maternal-to-Zygotic transition. Nature (2017) 542(7642):475–8. doi: 10.1038/nature21355
73. Kontur C, Jeong M, Cifuentes D, Giraldez AJ. Ythdf M(6)a readers function redundantly during zebrafish development. Cell Rep (2020) 33(13):108598. doi: 10.1016/j.celrep.2020.108598
74. Li M, Zhao X, Wang W, Shi H, Pan Q, Lu Z, et al. Ythdf2-mediated M(6)a mrna clearance modulates neural development in mice. Genome Biol (2018) 19(1):69. doi: 10.1186/s13059-018-1436-y
75. Zheng Q, Gan H, Yang F, Yao Y, Hao F, Hong L, et al. Cytoplasmic M(1)a reader Ythdf3 inhibits trophoblast invasion by downregulation of M(1)a-methylated Igf1r. Cell Discovery (2020) 6:12. doi: 10.1038/s41421-020-0144-4
76. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell (2007) 131(5):861–72. doi: 10.1016/j.cell.2007.11.019
77. Choi H, Baek S, Cho B, Kim S, Kim J, Chang Y, et al. Epitranscriptomic N(6)-methyladenosine modification is required for direct lineage reprogramming into neurons. ACS Chem Biol (2020) 15(8):2087–97. doi: 10.1021/acschembio.0c00265
78. Wu R, Liu Y, Zhao Y, Bi Z, Yao Y, Liu Q, et al. M(6)a methylation controls pluripotency of porcine induced pluripotent stem cells by targeting Socs3/Jak2/Stat3 pathway in a Ythdf1/Ythdf2-orchestrated manner. Cell Death Dis (2019) 10(3):171. doi: 10.1038/s41419-019-1417-4
79. Pollini D, Loffredo R, Maniscalco F, Cardano M, Micaelli M, Bonomo I, et al. Multilayer and Matr3-dependent regulation of mrnas maintains pluripotency in human induced pluripotent stem cells. iScience (2021) 24(3):102197. doi: 10.1016/j.isci.2021.102197
80. Heck AM, Russo J, Wilusz J, Nishimura EO, Wilusz CJ. Ythdf2 destabilizes M(6)a-modified neural-specific rnas to restrain differentiation in induced pluripotent stem cells. RNA (2020) 26(6):739–55. doi: 10.1261/rna.073502.119
81. Wang S, Zhang J, Wu X, Lin X, Liu XM, Zhou J. Differential roles of Ythdf1 and Ythdf3 in embryonic stem cell-derived cardiomyocyte differentiation. RNA Biol (2020) 18(9):1354–63. doi: 10.1080/15476286.2020.1850628
82. Zhang C, Chen Y, Sun B, Wang L, Yang Y, Ma D, et al. M(6)a modulates haematopoietic stem and progenitor cell specification. Nature (2017) 549(7671):273–6. doi: 10.1038/nature23883
83. Lv J, Zhang Y, Gao S, Zhang C, Chen Y, Li W, et al. Endothelial-specific M(6)a modulates mouse hematopoietic stem and progenitor cell development Via notch signaling. Cell Res (2018) 28(2):249–52. doi: 10.1038/cr.2017.143
84. Li Z, Qian P, Shao W, Shi H, He XC, Gogol M, et al. Suppression of M(6)a reader Ythdf2 promotes hematopoietic stem cell expansion. Cell Res (2018) 28(9):904–17. doi: 10.1038/s41422-018-0072-0
85. Wang H, Zuo H, Liu J, Wen F, Gao Y, Zhu X, et al. Loss of Ythdf2-mediated M(6)a-dependent mrna clearance facilitates hematopoietic stem cell regeneration. Cell Res (2018) 28(10):1035–8. doi: 10.1038/s41422-018-0082-y
86. Mapperley C, van de Lagemaat LN, Lawson H, Tavosanis A, Paris J, Campos J, et al. The mrna M6a reader Ythdf2 suppresses proinflammatory pathways and sustains hematopoietic stem cell function. J Exp Med (2021) 218(3):e20200829. doi: 10.1084/jem.20200829
87. Zhang X, Cong T, Wei L, Zhong B, Wang X, Sun J, et al. Ythdf3 modulates hematopoietic stem cells by recognizing rna M6a modification on Ccnd1. Haematologica (2022) 107(10):2381–94. doi: 10.3324/haematol.2021.279739
88. Dang Q, Wu Q, Yu F, Sheng Y, Yu C, Song G, et al. M(6)a reader Ythdf3 protects hematopoietic stem cell integrity under stress by promoting the translation of Foxm1 and Asxl1 transcripts. Haematologica (2021) 107(8):1922–7. doi: 10.3324/haematol.2021.279300
89. Han B, Yan S, Wei S, Xiang J, Liu K, Chen Z, et al. Ythdf1-mediated translation amplifies wnt-driven intestinal stemness. EMBO Rep (2020) 21(4):e49229. doi: 10.15252/embr.201949229
90. Jiang D, Hou J, Qian Y, Gao Y, Gao X, Wei S. Ythdf1-regulated expression of Tead1 contributes to the maintenance of intestinal stem cells. Biochem Biophys Res Commun (2021) 557:85–9. doi: 10.1016/j.bbrc.2021.03.175
91. Zhao X, Tian GG, Fang Q, Pei X, Wang Z, Wu J. Comparison of rna M(6)a and DNA methylation profiles between mouse female germline stem cells and sto cells. Mol Ther Nucleic Acids (2021) 23:431–9. doi: 10.1016/j.omtn.2020.11.020
92. Yao Y, Bi Z, Wu R, Zhao Y, Liu Y, Liu Q, et al. Mettl3 inhibits bmsc adipogenic differentiation by targeting the Jak1/Stat5/C/Ebpbeta pathway Via an M(6)a-Ythdf2-Dependent manner. FASEB J (2019) 33(6):7529–44. doi: 10.1096/fj.201802644R
93. Wu R, Guo G, Bi Z, Liu Y, Zhao Y, Chen N, et al. M(6)a methylation modulates adipogenesis through Jak2-Stat3-C/Ebpbeta signaling. Biochim Biophys Acta Gene Regul Mech (2019) 1862(8):796–806. doi: 10.1016/j.bbagrm.2019.06.008
94. Wu R, Liu Y, Yao Y, Zhao Y, Bi Z, Jiang Q, et al. Fto regulates adipogenesis by controlling cell cycle progression Via M(6)a-Ythdf2 dependent mechanism. Biochim Biophys Acta Mol Cell Biol Lipids (2018) 1863(10):1323–30. doi: 10.1016/j.bbalip.2018.08.008
95. Liu Q, Zhao Y, Wu R, Jiang Q, Cai M, Bi Z, et al. Zfp217 regulates adipogenesis by controlling mitotic clonal expansion in a Mettl3-M(6)a dependent manner. RNA Biol (2019) 16(12):1785–93. doi: 10.1080/15476286.2019.1658508
96. Wu R, Yao Y, Jiang Q, Cai M, Liu Q, Wang Y, et al. Epigallocatechin gallate targets fto and inhibits adipogenesis in an mrna M(6)a-Ythdf2-Dependent manner. Int J Obes (Lond) (2018) 42(7):1378–88. doi: 10.1038/s41366-018-0082-5
97. Liao X, Liu J, Chen Y, Liu Y, Chen W, Zeng B, et al. Metformin combats obesity by targeting fto in an M(6)a-Ythdf2-Dependent manner. J Drug Target (2022) 30(9):983–91. doi: 10.1080/1061186X.2022.2071906
98. Song T, Yang Y, Wei H, Xie X, Lu J, Zeng Q, et al. Zfp217 mediates M6a mrna methylation to orchestrate transcriptional and post-transcriptional regulation to promote adipogenic differentiation. Nucleic Acids Res (2019) 47(12):6130–44. doi: 10.1093/nar/gkz312
99. Cai M, Liu Q, Jiang Q, Wu R, Wang X, Wang Y. Loss of M(6) a on Fam134b promotes adipogenesis in porcine adipocytes through M(6) a-Ythdf2-Dependent way. IUBMB Life (2019) 71(5):580–6. doi: 10.1002/iub.1974
100. Sun D, Zhao T, Zhang Q, Wu M, Zhang Z. Fat mass and obesity-associated protein regulates lipogenesis Via M(6) a modification in fatty acid synthase mrna. Cell Biol Int (2021) 45(2):334–44. doi: 10.1002/cbin.11490
101. Wang X, Wu R, Liu Y, Zhao Y, Bi Z, Yao Y, et al. M(6)a mrna methylation controls autophagy and adipogenesis by targeting Atg5 and Atg7. Autophagy (2020) 16(7):1221–35. doi: 10.1080/15548627.2019.1659617
102. Zhong X, Yu J, Frazier K, Weng X, Li Y, Cham CM, et al. Circadian clock regulation of hepatic lipid metabolism by modulation of M(6)a mrna methylation. Cell Rep (2018) 25(7):1816–28.e4. doi: 10.1016/j.celrep.2018.10.068
103. Wu W, Wang S, Liu Q, Shan T, Wang X, Feng J, et al. Ampk facilitates intestinal long-chain fatty acid uptake by manipulating Cd36 expression and translocation. FASEB J (2020) 34(4):4852–69. doi: 10.1096/fj.201901994R
104. Jiang Q, Sun B, Liu Q, Cai M, Wu R, Wang F, et al. Mtch2 promotes adipogenesis in intramuscular preadipocytes Via an M(6)a-Ythdf1-Dependent mechanism. FASEB J (2019) 33(2):2971–81. doi: 10.1096/fj.201801393RRR
105. Chen Y, Wu R, Chen W, Liu Y, Liao X, Zeng B, et al. Curcumin prevents obesity by targeting Traf4-induced ubiquitylation in M(6) a-dependent manner. EMBO Rep (2021) 22(5):e52146. doi: 10.15252/embr.202052146
106. Peng Z, Gong Y, Wang X, He W, Wu L, Zhang L, et al. Mettl3-M(6)a-Rubicon axis inhibits autophagy in nonalcoholic fatty liver disease. Mol Ther (2021) 30(2):932–46. doi: 10.1016/j.ymthe.2021.09.016
107. Weng YL, Wang X, An R, Cassin J, Vissers C, Liu Y, et al. Epitranscriptomic M(6)a regulation of axon regeneration in the adult mammalian nervous system. Neuron (2018) 97(2):313–25.e6. doi: 10.1016/j.neuron.2017.12.036
108. Zhuang M, Li X, Zhu J, Zhang J, Niu F, Liang F, et al. The M6a reader Ythdf1 regulates axon guidance through translational control of Robo3.1 expression. Nucleic Acids Res (2019) 47(9):4765–77. doi: 10.1093/nar/gkz157
109. Merkurjev D, Hong WT, Iida K, Oomoto I, Goldie BJ, Yamaguti H, et al. Synaptic N(6)-methyladenosine (M(6)a) epitranscriptome reveals functional partitioning of localized transcripts. Nat Neurosci (2018) 21(7):1004–14. doi: 10.1038/s41593-018-0173-6
110. Shi H, Zhang X, Weng YL, Lu Z, Liu Y, Lu Z, et al. M(6)a facilitates hippocampus-dependent learning and memory through Ythdf1. Nature (2018) 563(7730):249–53. doi: 10.1038/s41586-018-0666-1
111. Kan L, Ott S, Joseph B, Park ES, Dai W, Kleiner RE, et al. A neural M(6)a/Ythdf pathway is required for learning and memory in drosophila. Nat Commun (2021) 12(1):1458. doi: 10.1038/s41467-021-21537-1
112. Yu J, She Y, Yang L, Zhuang M, Han P, Liu J, et al. The M(6) a readers Ythdf1 and Ythdf2 synergistically control cerebellar parallel fiber growth by regulating local translation of the key Wnt5a signaling components in axons. Adv Sci (Weinh) (2021) 8(22):e2101329. doi: 10.1002/advs.202101329
113. Niu F, Han P, Zhang J, She Y, Yang L, Yu J, et al. The M(6)a reader Ythdf2 is a negative regulator for dendrite development and maintenance of retinal ganglion cells. Elife (2022) 11:e75827. doi: 10.7554/eLife.75827
114. Zheng L, Tang X, Lu M, Sun S, Xie S, Cai J, et al. Microrna-421-3p prevents inflammatory response in cerebral Ischemia/Reperfusion injury through targeting M6a reader Ythdf1 to inhibit P65 mrna translation. Int Immunopharmacol (2020) 88:106937. doi: 10.1016/j.intimp.2020.106937
115. Zhang L, Cheng Y, Xue Z, Li J, Wu N, Yan J, et al. Sevoflurane impairs M6a-mediated mrna translation and leads to fine motor and cognitive deficits. Cell Biol Toxicol (2021) 38(2):347–69. doi: 10.1007/s10565-021-09601-4
116. Li M, Zhong X, Zhao Z, Zeng Z, Yuan Q, Xiao X, et al. The expression of M6a enzymes in the hippocampus of diabetic cognitive impairment mice and the possible improvement of Ythdf1. Brain Res (2022) 1777:147766. doi: 10.1016/j.brainres.2021.147766
117. Yang Y, Feng Y, Hu Y, Liu J, Shi H, Zhao R. Exposure to constant light impairs cognition with fto inhibition and M(6)a-dependent trkappab repression in mouse hippocampus. Environ pollut (2021) 283:117037. doi: 10.1016/j.envpol.2021.117037
118. Terkelsen T, Brasch-Andersen C, Illum N, Busa T, Missirian C, Chandler K, et al. Mono-allelic loss of Ythdf3 and neurodevelopmental disorder: Clinical features of four individuals with 8q12.3 deletions. Clin Genet (2022) 101(2):208–13. doi: 10.1111/cge.14083
119. Han Z, Wang X, Xu Z, Cao Y, Gong R, Yu Y, et al. Alkbh5 regulates cardiomyocyte proliferation and heart regeneration by demethylating the mrna of Ythdf1. Theranostics (2021) 11(6):3000–16. doi: 10.7150/thno.47354
120. Xu H, Wang Z, Chen M, Zhao W, Tao T, Ma L, et al. Ythdf2 alleviates cardiac hypertrophy Via regulating Myh7 mrna decoy. Cell Biosci (2021) 11(1):132. doi: 10.1186/s13578-021-00649-7
121. Yang Y, Mbikyo MB, Zhang J, Zhang Y, Zhang N, Li Z. The lncrna miat regulates cpt-1a mediated cardiac hypertrophy through M(6)a rna methylation reading protein Ythdf2. Cell Death Discovery (2022) 8(1):167. doi: 10.1038/s41420-022-00977-8
122. Yao MD, Jiang Q, Ma Y, Liu C, Zhu CY, Sun YN, et al. Role of Mettl3-dependent N(6)-methyladenosine mrna modification in the promotion of angiogenesis. Mol Ther (2020) 28(10):2191–202. doi: 10.1016/j.ymthe.2020.07.022
123. Shan K, Zhou RM, Xiang J, Sun YN, Liu C, Lv MW, et al. Fto regulates ocular angiogenesis Via M(6)a-Ythdf2-Dependent mechanism. Exp Eye Res (2020) 197:108107. doi: 10.1016/j.exer.2020.108107
124. Chien CS, Li JY, Chien Y, Wang ML, Yarmishyn AA, Tsai PH, et al. Mettl3-dependent N(6)-methyladenosine rna modification mediates the atherogenic inflammatory cascades in vascular endothelium. Proc Natl Acad Sci U.S.A. (2021) 118(7):e2025070118. doi: 10.1073/pnas.2025070118
125. Hu L, Wang J, Huang H, Yu Y, Ding J, Yu Y, et al. Ythdf1 regulates pulmonary hypertension through translational control of Maged1. Am J Respir Crit Care Med (2021) 203(9):1158–72. doi: 10.1164/rccm.202009-3419OC
126. Qin Y, Qiao Y, Li L, Luo E, Wang D, Yao Y, et al. The M(6)a methyltransferase Mettl3 promotes hypoxic pulmonary arterial hypertension. Life Sci (2021) 274:119366. doi: 10.1016/j.lfs.2021.119366
127. Xiao K, Liu P, Yan P, Liu Y, Song L, Liu Y, et al. N6-methyladenosine reader yth N6-methyladenosine rna binding protein 3 or insulin like growth factor 2 mrna binding protein 2 knockdown protects human bronchial epithelial cells from Hypoxia/Reoxygenation injury by inactivating P38 mapk, akt, Erk1/2, and nf-kappab pathways. Bioengineered (2021) 13(5):11973–11986. doi: 10.1080/21655979.2021.1999550
128. Xia TL, Li X, Wang X, Zhu YJ, Zhang H, Cheng W, et al. N(6)-Methyladenosine-Binding protein Ythdf1 suppresses ebv replication and promotes ebv rna decay. EMBO Rep (2021) 22(4):e50128. doi: 10.15252/embr.202050128
129. Zhang K, Zhang Y, Maharjan Y, Sugiokto FG, Wan J, Li R. Caspases switch off the M(6)a rna modification pathway to foster the replication of a ubiquitous human tumor virus. mBio (2021) 12(4):e0170621. doi: 10.1128/mBio.01706-21
130. Imam H, Khan M, Gokhale NS, McIntyre ABR, Kim GW, Jang JY, et al. N6-methyladenosine modification of hepatitis b virus rna differentially regulates the viral life cycle. Proc Natl Acad Sci U S A (2018) 115(35):8829–34. doi: 10.1073/pnas.1808319115
131. Gokhale NS, McIntyre ABR, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, et al. N6-methyladenosine in flaviviridae viral rna genomes regulates infection. Cell Host Microbe (2016) 20(5):654–65. doi: 10.1016/j.chom.2016.09.015
132. Lichinchi G, Zhao BS, Wu Y, Lu Z, Qin Y, He C, et al. Dynamics of human and viral rna methylation during zika virus infection. Cell Host Microbe (2016) 20(5):666–73. doi: 10.1016/j.chom.2016.10.002
133. Hao H, Hao S, Chen H, Chen Z, Zhang Y, Wang J, et al. N6-methyladenosine modification and Mettl3 modulate enterovirus 71 replication. Nucleic Acids Res (2019) 47(1):362–74. doi: 10.1093/nar/gky1007
134. Tsai K, Courtney DG, Cullen BR. Addition of M6a to Sv40 late mrnas enhances viral structural gene expression and replication. PLoS Pathog (2018) 14(2):e1006919. doi: 10.1371/journal.ppat.1006919
135. Courtney DG, Kennedy EM, Dumm RE, Bogerd HP, Tsai K, Heaton NS, et al. Epitranscriptomic enhancement of influenza a virus gene expression and replication. Cell Host Microbe (2017) 22(3):377–86.e5. doi: 10.1016/j.chom.2017.08.004
136. Burgess HM, Depledge DP, Thompson L, Srinivas KP, Grande RC, Vink EI, et al. Targeting the M(6)a rna modification pathway blocks sars-Cov-2 and hcov-Oc43 replication. Genes Dev (2021) 35(13-14):1005–19. doi: 10.1101/gad.348320.121
137. Kim B, Arcos S, Rothamel K, Jian J, Rose KL, McDonald WH, et al. Discovery of widespread host protein interactions with the pre-replicated genome of chikv using vir-clasp. Mol Cell (2020) 78(4):624–40.e7. doi: 10.1016/j.molcel.2020.04.013
138. Liu J, Xu YP, Li K, Ye Q, Zhou HY, Sun H, et al. The M(6)a methylome of sars-Cov-2 in host cells. Cell Res (2021) 31(4):404–14. doi: 10.1038/s41422-020-00465-7
139. Tirumuru N, Zhao BS, Lu W, Lu Z, He C, Wu L. N(6)-methyladenosine of hiv-1 rna regulates viral infection and hiv-1 gag protein expression. Elife (2016) 5:e15528. doi: 10.7554/eLife.15528
140. Lu W, Tirumuru N, St Gelais C, Koneru PC, Liu C, Kvaratskhelia M, et al. N(6)-Methyladenosine-Binding proteins suppress hiv-1 infectivity and viral production. J Biol Chem (2018) 293(34):12992–3005. doi: 10.1074/jbc.RA118.004215
141. Kennedy EM, Bogerd HP, Kornepati AV, Kang D, Ghoshal D, Marshall JB, et al. Posttranscriptional M(6)a editing of hiv-1 mrnas enhances viral gene expression. Cell Host Microbe (2016) 19(5):675–85. doi: 10.1016/j.chom.2016.04.002
142. Jurczyszak D, Zhang W, Terry SN, Kehrer T, Bermudez Gonzalez MC, McGregor E, et al. Hiv protease cleaves the antiviral M6a reader protein Ythdf3 in the viral particle. PloS Pathog (2020) 16(2):e1008305. doi: 10.1371/journal.ppat.1008305
143. Hesser CR, Karijolich J, Dominissini D, He C, Glaunsinger BA. N6-methyladenosine modification and the Ythdf2 reader protein play cell type specific roles in lytic viral gene expression during kaposi's sarcoma-associated herpesvirus infection. PloS Pathog (2018) 14(4):e1006995. doi: 10.1371/journal.ppat.1006995
144. Tan B, Gao SJ. The rna epitranscriptome of DNA viruses. J Virol (2018) 92(22):e00696–18. doi: 10.1128/JVI.00696-18
145. McFadden MJ, McIntyre ABR, Mourelatos H, Abell NS, Gokhale NS, Ipas H, et al. Post-transcriptional regulation of antiviral gene expression by N6-methyladenosine. Cell Rep (2021) 34(9):108798. doi: 10.1016/j.celrep.2021.108798
146. Terajima H, Lu M, Zhang L, Cui Q, Shi Y, Li J, et al. N6-methyladenosine promotes induction of Adar1-mediated a-to-I rna editing to suppress aberrant antiviral innate immune responses. PLoS Biol (2021) 19(7):e3001292. doi: 10.1371/journal.pbio.3001292
147. Winkler R, Gillis E, Lasman L, Safra M, Geula S, Soyris C, et al. M(6)a modification controls the innate immune response to infection by targeting type I interferons. Nat Immunol (2019) 20(2):173–82. doi: 10.1038/s41590-018-0275-z
148. Xu J, Cai Y, Ma Z, Jiang B, Liu W, Cheng J, et al. The rna helicase Ddx5 promotes viral infection Via regulating N6-methyladenosine levels on the Dhx58 and nfkappab transcripts to dampen antiviral innate immunity. PloS Pathog (2021) 17(4):e1009530. doi: 10.1371/journal.ppat.1009530
149. Lu M, Xue M, Wang HT, Kairis EL, Ahmad S, Wei J, et al. Nonsegmented negative-sense rna viruses utilize n (6)-methyladenosine (M(6)a) as a common strategy to evade host innate immunity. J Virol (2021) 95(9):e01939–20. doi: 10.1128/JVI.01939-20
150. Kim GW, Imam H, Khan M, Siddiqui AN. (6)-methyladenosine modification of hepatitis b and c viral rnas attenuates host innate immunity Via rig-I signaling. J Biol Chem (2020) 295(37):13123–33. doi: 10.1074/jbc.RA120.014260
151. Imam H, Kim GW, Mir SA, Khan M, Siddiqui A. Interferon-stimulated gene 20 (Isg20) selectively degrades N6-methyladenosine modified hepatitis b virus transcripts. PLoS Pathog (2020) 16(2):e1008338. doi: 10.1371/journal.ppat.1008338
152. Kastan JP, Tremblay MW, Brown MC, Trimarco JD, Dobrikova EY, Dobrikov MI, et al. Enterovirus 2a(Pro) cleavage of the ythdf M(6)a readers implicates Ythdf3 as a mediator of type I interferon-driven Jak/Stat signaling. mBio (2021) 12(2):e00116–21. doi: 10.1128/mBio.00116-21
153. Zhang Y, Wang X, Zhang X, Wang J, Ma Y, Zhang L, et al. Rna-binding protein Ythdf3 suppresses interferon-dependent antiviral responses by promoting Foxo3 translation. Proc Natl Acad Sci U S A (2019) 116(3):976–81. doi: 10.1073/pnas.1812536116
154. Du J, Liao W, Liu W, Deb DK, He L, Hsu PJ, et al. N(6)-adenosine methylation of Socs1 mrna is required to sustain the negative feedback control of macrophage activation. Dev Cell (2020) 55(6):737–53.e7. doi: 10.1016/j.devcel.2020.10.023
155. Li SX, Yan W, Liu JP, Zhao YJ, Chen L. Long noncoding rna Snhg4 remits lipopolysaccharide-engendered inflammatory lung damage by inhibiting Mettl3 - mediated M(6)a level of Stat2 mrna. Mol Immunol (2021) 139:10–22. doi: 10.1016/j.molimm.2021.08.008
156. Zong X, Xiao X, Jie F, Cheng Y, Jin M, Yin Y, et al. Ythdf1 promotes Nlrp3 translation to induce intestinal epithelial cell inflammatory injury during endotoxic shock. Sci China Life Sci (2021) 64(11):1988–91. doi: 10.1007/s11427-020-1909-6
157. Xing Y, Cheng D, Shi C, Shen Z. The protective role of Ythdf1-knock down macrophages on the immune paralysis of severe sepsis rats with ecmo. Microvasc Res (2021) 137:104178. doi: 10.1016/j.mvr.2021.104178
158. Yu R, Li Q, Feng Z, Cai L, Xu Q. M6a reader Ythdf2 regulates lps-induced inflammatory response. Int J Mol Sci (2019) 20(6):1323. doi: 10.3390/ijms20061323
159. Huangfu N, Zheng W, Xu Z, Wang S, Wang Y, Cheng J, et al. Rbm4 regulates M1 macrophages polarization through targeting Stat1-mediated glycolysis. Int Immunopharmacol (2020) 83:106432. doi: 10.1016/j.intimp.2020.106432
160. Gu X, Zhang Y, Li D, Cai H, Cai L, Xu Q. N6-methyladenosine demethylase fto promotes M1 and M2 macrophage activation. Cell Signal (2020) 69:109553. doi: 10.1016/j.cellsig.2020.109553
161. Wu C, Chen W, He J, Jin S, Liu Y, Yi Y, et al. Interplay of M(6)a and H3k27 trimethylation restrains inflammation during bacterial infection. Sci Adv (2020) 6(34):eaba0647. doi: 10.1126/sciadv.aba0647
162. Hu J, Qiu D, Yu A, Hu J, Deng H, Li H, et al. Ythdf1 is a potential pan-cancer biomarker for prognosis and immunotherapy. Front Oncol (2021) 11:607224. doi: 10.3389/fonc.2021.607224
163. Han D, Liu J, Chen C, Dong L, Liu Y, Chang R, et al. Anti-tumour immunity controlled through mrna M(6)a methylation and Ythdf1 in dendritic cells. Nature (2019) 566(7743):270–4. doi: 10.1038/s41586-019-0916-x
164. Bai X, Wong CC, Pan Y, Chen H, Liu W, Zhai J, et al. Loss of Ythdf1 in gastric tumors restores sensitivity to antitumor immunity by recruiting mature dendritic cells. J Immunother Cancer (2022) 10(2):e003663. doi: 10.1136/jitc-2021-003663
165. Li T, Tan YT, Chen YX, Zheng XJ, Wang W, Liao K, et al. Methionine deficiency facilitates antitumour immunity by altering M(6)a methylation of immune checkpoint transcripts. Gut (2022) 72(3):501–11. doi: 10.1136/gutjnl-2022-326928
166. Qiu X, Yang S, Wang S, Wu J, Zheng B, Wang K, et al. M6a demethylase Alkbh5 regulates pd-L1 expression and tumor immunoenvironment in intrahepatic cholangiocarcinoma. Cancer Res (2021) 81(18):4778–93. doi: 10.1158/0008-5472.CAN-21-0468
167. Ma S, Yan J, Barr T, Zhang J, Chen Z, Wang LS, et al. The rna M6a reader Ythdf2 controls nk cell antitumor and antiviral immunity. J Exp Med (2021) 218(8):e20210279. doi: 10.1084/jem.20210279
168. Zhao X, Chen Y, Mao Q, Jiang X, Jiang W, Chen J, et al. Overexpression of Ythdf1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer biomark (2018) 21(4):859–68. doi: 10.3233/CBM-170791
169. Zhou Y, Yin Z, Hou B, Yu M, Chen R, Jin H, et al. Expression profiles and prognostic significance of rna N6-Methyladenosine-Related genes in patients with hepatocellular carcinoma: Evidence from independent datasets. Cancer Manag Res (2019) 11:3921–31. doi: 10.2147/CMAR.S191565
170. Bian S, Ni W, Zhu M, Song Q, Zhang J, Ni R, et al. Identification and validation of the N6-methyladenosine rna methylation regulator Ythdf1 as a novel prognostic marker and potential target for hepatocellular carcinoma. Front Mol Biosci (2020) 7:604766. doi: 10.3389/fmolb.2020.604766
171. Chi F, Cao Y, Chen Y. Analysis and validation of circrna-mirna network in regulating M(6)a rna methylation modulators reveals Circmap2k4/Mir-139-5p/Ythdf1 axis involving the proliferation of hepatocellular carcinoma. Front Oncol (2021) 11:560506. doi: 10.3389/fonc.2021.560506
172. Luo X, Cao M, Gao F, He X. Ythdf1 promotes hepatocellular carcinoma progression Via activating Pi3k/Akt/Mtor signaling pathway and inducing epithelial-mesenchymal transition. Exp Hematol Oncol (2021) 10(1):35. doi: 10.1186/s40164-021-00227-0
173. Hu J, Yang L, Peng X, Mao M, Liu X, Song J, et al. Mettl3 promotes M6a hypermethylation of Rbm14 Via Ythdf1 leading to the progression of hepatocellular carcinoma. Hum Cell (2022) 35(6):1838–55. doi: 10.1007/s13577-022-00769-3
174. Li Q, Ni Y, Zhang L, Jiang R, Xu J, Yang H, et al. Hif-1alpha-Induced expression of M6a reader Ythdf1 drives hypoxia-induced autophagy and malignancy of hepatocellular carcinoma by promoting Atg2a and Atg14 translation. Signal Transduct Target Ther (2021) 6(1):76. doi: 10.1038/s41392-020-00453-8
175. Lin Z, Niu Y, Wan A, Chen D, Liang H, Chen X, et al. Rna M(6) a methylation regulates sorafenib resistance in liver cancer through Foxo3-mediated autophagy. EMBO J (2020) 39(12):e103181. doi: 10.15252/embj.2019103181
176. Su T, Huang M, Liao J, Lin S, Yu P, Yang J, et al. Insufficient radiofrequency ablation promotes hepatocellular carcinoma metastasis through M(6) a mrna methylation dependent mechanism. Hepatology (2021) 74(3):1339–56. doi: 10.1002/hep.31766
177. Wang M, Yang Y, Yang J, Yang J, Han S. Circ_Kiaa1429 accelerates hepatocellular carcinoma advancement through the mechanism of M(6)a-Ythdf3-Zeb1. Life Sci (2020) 257:118082. doi: 10.1016/j.lfs.2020.118082
178. Guo JC, Liu Z, Yang YJ, Guo M, Zhang JQ, Zheng JF. Kdm5b promotes self-renewal of hepatocellular carcinoma cells through the microrna-448-Mediated Ythdf3/Itga6 axis. J Cell Mol Med (2021) 25(13):5949–62. doi: 10.1111/jcmm.16342
179. Zhong L, Liao D, Zhang M, Zeng C, Li X, Zhang R, et al. Ythdf2 suppresses cell proliferation and growth Via destabilizing the egfr mrna in hepatocellular carcinoma. Cancer Lett (2019) 442:252–61. doi: 10.1016/j.canlet.2018.11.006
180. Hou J, Zhang H, Liu J, Zhao Z, Wang J, Lu Z, et al. Ythdf2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol Cancer (2019) 18(1):163. doi: 10.1186/s12943-019-1082-3
181. Shao XY, Dong J, Zhang H, Wu YS, Zheng L. Systematic analyses of the role of the reader protein of n (6)-methyladenosine rna methylation, yth domain family 2, in liver hepatocellular carcinoma. Front Mol Biosci (2020) 7:577460. doi: 10.3389/fmolb.2020.577460
182. Nakagawa N, Sonohara F, Tanaka K, Sunagawa Y, Inokawa Y, Takami H, et al. Novel prognostic implications of yth domain family 2 in resected hepatocellular carcinoma. Oncol Lett (2021) 22(1):538. doi: 10.3892/ol.2021.12799
183. Yang Z, Li J, Feng G, Gao S, Wang Y, Zhang S, et al. Microrna-145 modulates N(6)-methyladenosine levels by targeting the 3'-untranslated mrna region of the N(6)-methyladenosine binding yth domain family 2 protein. J Biol Chem (2017) 292(9):3614–23. doi: 10.1074/jbc.M116.749689
184. Chen M, Wei L, Law CT, Tsang FH, Shen J, Cheng CL, et al. Rna N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through Ythdf2-dependent posttranscriptional silencing of Socs2. Hepatology (2018) 67(6):2254–70. doi: 10.1002/hep.29683
185. Zhang C, Huang S, Zhuang H, Ruan S, Zhou Z, Huang K, et al. Ythdf2 promotes the liver cancer stem cell phenotype and cancer metastasis by regulating Oct4 expression Via M6a rna methylation. Oncogene (2020) 39(23):4507–18. doi: 10.1038/s41388-020-1303-7
186. Sun S, Liu Y, Zhou M, Wen J, Xue L, Han S, et al. Pa2g4 promotes the metastasis of hepatocellular carcinoma by stabilizing fyn mrna in a Ythdf2-dependent manner. Cell Biosci (2022) 12(1):55. doi: 10.1186/s13578-022-00788-5
187. Liu GM, Zeng HD, Zhang CY, Xu JW. Identification of Mettl3 as an adverse prognostic biomarker in hepatocellular carcinoma. Dig Dis Sci (2021) 66(4):1110–26. doi: 10.1007/s10620-020-06260-z
188. Huang X, Zhu L, Wang L, Huang W, Tan L, Liu H, et al. Ythdf1 promotes intrahepatic cholangiocarcinoma progression Via regulating egfr mrna translation. J Gastroenterol Hepatol (2022) 37(6):1156–68. doi: 10.1111/jgh.15816
189. Xu QC, Tien YC, Shi YH, Chen S, Zhu YQ, Huang XT, et al. Mettl3 promotes intrahepatic cholangiocarcinoma progression by regulating Ifit2 expression in an M(6)a-Ythdf2-Dependent manner. Oncogene (2022) 41(11):1622–33. doi: 10.1038/s41388-022-02185-1
190. Huang CS, Zhu YQ, Xu QC, Chen S, Huang Y, Zhao G, et al. Ythdf2 promotes intrahepatic cholangiocarcinoma progression and desensitises cisplatin treatment by increasing Cdkn1b mrna degradation. Clin Transl Med (2022) 12(6):e848. doi: 10.1002/ctm2.848
191. Liu T, Yang S, Cheng YP, Kong XL, Du DD, Wang X, et al. The N6-methyladenosine (M6a) methylation gene Ythdf1 reveals a potential diagnostic role for gastric cancer. Cancer Manag Res (2020) 12:11953–64. doi: 10.2147/CMAR.S279370
192. Pi J, Wang W, Ji M, Wang X, Wei X, Jin J, et al. Ythdf1 promotes gastric carcinogenesis by controlling translation of Fzd7. Cancer Res (2021) 81(10):2651–65. doi: 10.1158/0008-5472.CAN-20-0066
193. Chen XY, Liang R, Yi YC, Fan HN, Chen M, Zhang J, et al. The M(6)a reader Ythdf1 facilitates the tumorigenesis and metastasis of gastric cancer Via Usp14 translation in an M(6)a-dependent manner. Front Cell Dev Biol (2021) 9:647702. doi: 10.3389/fcell.2021.647702
194. Huo FC, Zhu ZM, Zhu WT, Du QY, Liang J, Mou J. Mettl3-mediated M(6)a methylation of Sphk2 promotes gastric cancer progression by targeting Klf2. Oncogene (2021) 40(16):2968–81. doi: 10.1038/s41388-021-01753-1
195. Zhang J, Pi J, Liu Y, Yu J, Feng T. [Knockdown of yth N(6)-methyladenosine rna binding protein 2 (Ythdf2) inhibits proliferation and promotes apoptosis in mgc-803 gastric cancer cells]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi (2017) 33(12):1628–34.
196. Yan J, Huang X, Zhang X, Chen Z, Ye C, Xiang W, et al. Lncrna Linc00470 promotes the degradation of pten mrna to facilitate malignant behavior in gastric cancer cells. Biochem Biophys Res Commun (2020) 521(4):887–93. doi: 10.1016/j.bbrc.2019.11.016
197. Yang H, Hu Y, Weng M, Liu X, Wan P, Hu Y, et al. Hypoxia inducible lncrna-cbslr modulates ferroptosis through M6a-Ythdf2-Dependent modulation of cbs in gastric cancer. J Adv Res (2022) 37:91–106. doi: 10.1016/j.jare.2021.10.001
198. Shen X, Zhao K, Xu L, Cheng G, Zhu J, Gan L, et al. Ythdf2 inhibits gastric cancer cell growth by regulating Foxc2 signaling pathway. Front Genet (2020) 11:592042. doi: 10.3389/fgene.2020.592042
199. Chen J, Sun Y, Xu X, Wang D, He J, Zhou H, et al. Yth domain family 2 orchestrates epithelial-mesenchymal Transition/Proliferation dichotomy in pancreatic cancer cells. Cell Cycle (2017) 16(23):2259–71. doi: 10.1080/15384101.2017.1380125
200. Wang M, Liu J, Zhao Y, He R, Xu X, Guo X, et al. Upregulation of Mettl14 mediates the elevation of perp mrna N(6) adenosine methylation promoting the growth and metastasis of pancreatic cancer. Mol Cancer (2020) 19(1):130. doi: 10.1186/s12943-020-01249-8
201. Guo X, Li K, Jiang W, Hu Y, Xiao W, Huang Y, et al. Rna demethylase Alkbh5 prevents pancreatic cancer progression by posttranscriptional activation of Per1 in an M6a-Ythdf2-Dependent manner. Mol Cancer (2020) 19(1):91. doi: 10.1186/s12943-020-01158-w
202. Hua YQ, Zhang K, Sheng J, Ning ZY, Li Y, Shi WD, et al. Nucb1 suppresses growth and shows additive effects with gemcitabine in pancreatic ductal adenocarcinoma Via the unfolded protein response. Front Cell Dev Biol (2021) 9:641836. doi: 10.3389/fcell.2021.641836
203. Tian J, Zhu Y, Rao M, Cai Y, Lu Z, Zou D, et al. N(6)-methyladenosine mrna methylation of Pik3cb regulates akt signalling to promote pten-deficient pancreatic cancer progression. Gut (2020) 69(12):2180–92. doi: 10.1136/gutjnl-2019-320179
204. Tan Z, Shi S, Xu J, Liu X, Lei Y, Zhang B, et al. Rna N6-methyladenosine demethylase fto promotes pancreatic cancer progression by inducing the autocrine activity of pdgfc in an M(6)a-Ythdf2-Dependent manner. Oncogene (2022) 41(20):2860–72. doi: 10.1038/s41388-022-02306-w
205. Tang R, Zhang Y, Liang C, Xu J, Meng Q, Hua J, et al. The role of M6a-related genes in the prognosis and immune microenvironment of pancreatic adenocarcinoma. PeerJ (2020) 8:e9602. doi: 10.7717/peerj.9602
206. Xu F, Zhang Z, Yuan M, Zhao Y, Zhou Y, Pei H, et al. M6a regulatory genes play an important role in the prognosis, progression and immune microenvironment of pancreatic adenocarcinoma. Cancer Invest (2021) 39(1):39–54. doi: 10.1080/07357907.2020.1834576
207. Zhou Z, Zhang J, Xu C, Yang J, Zhang Y, Liu M, et al. An integrated model of N6-methyladenosine regulators to predict tumor aggressiveness and immune evasion in pancreatic cancer. EBioMedicine (2021) 65:103271. doi: 10.1016/j.ebiom.2021.103271
208. Wu S, Ai Y, Huang H, Wu G, Zhou S, Hong W, et al. A synthesized olean-28,13beta-Lactam targets Ythdf1-Gls1 axis to induce ros-dependent metabolic crisis and cell death in pancreatic adenocarcinoma. Cancer Cell Int (2022) 22(1):143. doi: 10.1186/s12935-022-02562-6
209. Hu Y, Tang J, Xu F, Chen J, Zeng Z, Han S, et al. A reciprocal feedback between N6-methyladenosine reader Ythdf3 and lncrna Dicer1-As1 promotes glycolysis of pancreatic cancer through inhibiting maturation of mir-5586-5p. J Exp Clin Cancer Res (2022) 41(1):69. doi: 10.1186/s13046-022-02285-6
210. Liu T, Li C, Jin L, Li C, Wang L. The prognostic value of M6a rna methylation regulators in colon adenocarcinoma. Med Sci Monit (2019) 25:9435–45. doi: 10.12659/MSM.920381
211. Bai Y, Yang C, Wu R, Huang L, Song S, Li W, et al. Ythdf1 regulates tumorigenicity and cancer stem cell-like activity in human colorectal carcinoma. Front Oncol (2019) 9:332. doi: 10.3389/fonc.2019.00332
212. Nishizawa Y, Konno M, Asai A, Koseki J, Kawamoto K, Miyoshi N, et al. Oncogene c-myc promotes epitranscriptome M(6)a reader Ythdf1 expression in colorectal cancer. Oncotarget (2018) 9(7):7476–86. doi: 10.18632/oncotarget.23554
213. Wang S, Gao S, Zeng Y, Zhu L, Mo Y, Wong CC, et al. N6-methyladenosine reader Ythdf1 promotes Arhgef2 translation and rhoa signaling in colorectal cancer. Gastroenterology (2022) 162(4):1183–96. doi: 10.1053/j.gastro.2021.12.269
214. Jiang Z, Hou Z, Liu W, Yu Z, Liang Z, Chen S. Circular rna protein tyrosine kinase 2 (Circptk2) promotes colorectal cancer proliferation, migration, invasion and chemoresistance. Bioengineered (2022) 13(1):810–23. doi: 10.1080/21655979.2021.2012952
215. Chen P, Liu XQ, Lin X, Gao LY, Zhang S, Huang X. Targeting Ythdf1 effectively re-sensitizes cisplatin-resistant colon cancer cells by modulating gls-mediated glutamine metabolism. Mol Ther Oncolytics (2021) 20:228–39. doi: 10.1016/j.omto.2021.01.001
216. Zhang K, Zhang T, Yang Y, Tu W, Huang H, Wang Y, et al. N(6)-Methyladenosine-Mediated ldha induction potentiates chemoresistance of colorectal cancer cells through metabolic reprogramming. Theranostics (2022) 12(10):4802–17. doi: 10.7150/thno.73746
217. Tian J, Ying P, Ke J, Zhu Y, Yang Y, Gong Y, et al. Ankle1 N(6) -Methyladenosine-Related variant is associated with colorectal cancer risk by maintaining the genomic stability. Int J Cancer (2020) 146(12):3281–93. doi: 10.1002/ijc.32677
218. Zhou D, Tang W, Xu Y, Xu Y, Xu B, Fu S, et al. Mettl3/Ythdf2 M6a axis accelerates colorectal carcinogenesis through epigenetically suppressing Ypel5. Mol Oncol (2021) 15(8):2172–84. doi: 10.1002/1878-0261.12898
219. Chen X, Xu M, Xu X, Zeng K, Liu X, Pan B, et al. Mettl14-mediated N6-methyladenosine modification of Sox4 mrna inhibits tumor metastasis in colorectal cancer. Mol Cancer (2020) 19(1):106. doi: 10.1186/s12943-020-01220-7
220. Yang X, Zhang S, He C, Xue P, Zhang L, He Z, et al. Mettl14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding rna xist. Mol Cancer (2020) 19(1):46. doi: 10.1186/s12943-020-1146-4
221. Han S, Zhu L, Zhu Y, Meng Y, Li J, Song P, et al. Targeting Atf4-dependent pro-survival autophagy to synergize glutaminolysis inhibition. Theranostics (2021) 11(17):8464–79. doi: 10.7150/thno.60028
222. Li H, Zhang N, Jiao X, Wang C, Sun W, He Y, et al. Downregulation of microrna-6125 promotes colorectal cancer growth through Ythdf2-dependent recognition of N6-Methyladenosine-Modified Gsk3beta. Clin Transl Med (2021) 11(10):e602. doi: 10.1002/ctm2.602
223. Zhou L, Jiang J, Huang Z, Jin P, Peng L, Luo M, et al. Hypoxia-induced lncrna Steap3-As1 activates Wnt/Beta-catenin signaling to promote colorectal cancer progression by preventing M(6)a-mediated degradation of Steap3 mrna. Mol Cancer (2022) 21(1):168. doi: 10.1186/s12943-022-01638-1
224. Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, et al. Long noncoding rna Gas5 inhibits progression of colorectal cancer by interacting with and triggering yap phosphorylation and degradation and is negatively regulated by the M(6)a reader Ythdf3. Mol Cancer (2019) 18(1):143. doi: 10.1186/s12943-019-1079-y
225. Zhao Y, Zhao H, Zhang D, Quan Q, Ge Y, Li L, et al. Ythdf3 facilitates Eif2ak2 and Eif3a recruitment on mrnas to regulate translational processes in oxaliplatin-resistant colorectal cancer. ACS Chem Biol (2022) 17(7):1778–88. doi: 10.1021/acschembio.2c00131
226. Tsuchiya K, Yoshimura K, Inoue Y, Iwashita Y, Yamada H, Kawase A, et al. Ythdf1 and Ythdf2 are associated with better patient survival and an inflamed tumor-immune microenvironment in non-Small-Cell lung cancer. Oncoimmunology (2021) 10(1):1962656. doi: 10.1080/2162402X.2021.1962656
227. Shi Y, Fan S, Wu M, Zuo Z, Li X, Jiang L, et al. Ythdf1 links hypoxia adaptation and non-small cell lung cancer progression. Nat Commun (2019) 10(1):4892. doi: 10.1038/s41467-019-12801-6
228. Zhou J, Xiao D, Qiu T, Li J, Liu Z. Loading microrna-376c in extracellular vesicles inhibits properties of non-small cell lung cancer cells by targeting Ythdf1. Technol Cancer Res Treat (2020) 19:1533033820977525. doi: 10.1177/1533033820977525. 1533033820977525.
229. Ma L, Xue X, Zhang X, Yu K, Xu X, Tian X, et al. The essential roles of M(6)a rna modification to stimulate Eno1-dependent glycolysis and tumorigenesis in lung adenocarcinoma. J Exp Clin Cancer Res (2022) 41(1):36. doi: 10.1186/s13046-021-02200-5
230. Lou X, Ning J, Liu W, Li K, Qian B, Xu D, et al. Ythdf1 promotes cyclin B1 translation through M(6)a modulation and contributes to the poor prognosis of lung adenocarcinoma with Kras/Tp53 Co-mutation. Cells (2021) 10(7):1669. doi: 10.3390/cells10071669
231. Sheng H, Li Z, Su S, Sun W, Zhang X, Li L, et al. Yth domain family 2 promotes lung cancer cell growth by facilitating 6-phosphogluconate dehydrogenase mrna translation. Carcinogenesis (2020) 41(5):541–50. doi: 10.1093/carcin/bgz152
232. Cheng C, Wu Y, Xiao T, Xue J, Sun J, Xia H, et al. Mettl3-mediated M(6)a modification of Zbtb4 mrna is involved in the smoking-induced emt in cancer of the lung. Mol Ther Nucleic Acids (2021) 23:487–500. doi: 10.1016/j.omtn.2020.12.001
233. Jin M, Li G, Liu W, Wu X, Zhu J, Zhao D, et al. Cigarette smoking induces aberrant N(6)-methyladenosine of Dapk2 to promote non-small cell lung cancer progression by activating nf-kappab pathway. Cancer Lett (2021) 518:214–29. doi: 10.1016/j.canlet.2021.07.022
234. Zhang D, Ning J, Okon I, Zheng X, Satyanarayana G, Song P, et al. Suppression of M6a mrna modification by DNA hypermethylated Alkbh5 aggravates the oncological behavior of kras Mutation/Lkb1 loss lung cancer. Cell Death Dis (2021) 12(6):518. doi: 10.1038/s41419-021-03793-7
235. Li Y, Sheng H, Ma F, Wu Q, Huang J, Chen Q, et al. Rna M(6)a reader Ythdf2 facilitates lung adenocarcinoma cell proliferation and metastasis by targeting the Axin1/Wnt/Beta-catenin signaling. Cell Death Dis (2021) 12(5):479. doi: 10.1038/s41419-021-03763-z
236. Zhang C, Sun Q, Zhang X, Qin N, Pu Z, Gu Y, et al. Gene amplification-driven rna methyltransferase Kiaa1429 promotes tumorigenesis by regulating Btg2 Via M6a-Ythdf2-Dependent in lung adenocarcinoma. Cancer Commun (Lond) (2022) 42(7):609–26. doi: 10.1002/cac2.12325
237. Xu Y, Chen Y, Yao Y, Xie H, Lu G, Du C, et al. Virma contributes to non-small cell lung cancer progression Via N(6)-Methyladenosine-Dependent Dapk3 post-transcriptional modification. Cancer Lett (2021) 522:142–54. doi: 10.1016/j.canlet.2021.08.027
238. Wang T, Liu Z, She Y, Deng J, Zhong Y, Zhao M, et al. A novel protein encoded by Circask1 ameliorates gefitinib resistance in lung adenocarcinoma by competitively activating Ask1-dependent apoptosis. Cancer Lett (2021) 520:321–31. doi: 10.1016/j.canlet.2021.08.007
239. Zhao T, Wang M, Zhao X, Weng S, Qian K, Shi K, et al. Ythdf2 inhibits the migration and invasion of lung adenocarcinoma by negatively regulating the Fam83d-Tgfbeta1-Smad2/3 pathway. Front Oncol (2022) 12:763341. doi: 10.3389/fonc.2022.763341
240. Xu P, Hu K, Zhang P, Sun ZG, Zhang N. Hypoxia-mediated Ythdf2 overexpression promotes lung squamous cell carcinoma progression by activation of the Mtor/Akt axis. Cancer Cell Int (2022) 22(1):13. doi: 10.1186/s12935-021-02368-y
241. Jin D, Guo J, Wu Y, Yang L, Wang X, Du J, et al. M(6)a demethylase Alkbh5 inhibits tumor growth and metastasis by reducing ythdfs-mediated yap expression and inhibiting mir-107/Lats2-Mediated yap activity in nsclc. Mol Cancer (2020) 19(1):40. doi: 10.1186/s12943-020-01161-1
242. Jin D, Guo J, Wu Y, Du J, Yang L, Wang X, et al. M(6)a mrna methylation initiated by Mettl3 directly promotes yap translation and increases yap activity by regulating the Malat1-Mir-1914-3p-Yap axis to induce nsclc drug resistance and metastasis. J Hematol Oncol (2019) 12(1):135. doi: 10.1186/s13045-019-0830-6
243. Yang F, Jin H, Que B, Chao Y, Zhang H, Ying X, et al. Dynamic M(6)a mrna methylation reveals the role of Mettl3-M(6)a-Cdcp1 signaling axis in chemical carcinogenesis. Oncogene (2019) 38(24):4755–72. doi: 10.1038/s41388-019-0755-0
244. Jin H, Ying X, Que B, Wang X, Chao Y, Zhang H, et al. N(6)-methyladenosine modification of Itga6 mrna promotes the development and progression of bladder cancer. EBioMedicine (2019) 47:195–207. doi: 10.1016/j.ebiom.2019.07.068
245. Xie H, Li J, Ying Y, Yan H, Jin K, Ma X, et al. Mettl3/Ythdf2 M(6) a axis promotes tumorigenesis by degrading Setd7 and Klf4 mrnas in bladder cancer. J Cell Mol Med (2020) 24(7):4092–104. doi: 10.1111/jcmm.15063
246. Li J, Meng S, Xu M, Wang S, He L, Xu X, et al. Downregulation of N(6)-methyladenosine binding Ythdf2 protein mediated by mir-493-3p suppresses prostate cancer by elevating N(6)-methyladenosine levels. Oncotarget (2018) 9(3):3752–64. doi: 10.18632/oncotarget.23365
247. Du C, Lv C, Feng Y, Yu S. Activation of the Kdm5a/Mirna-495/Ythdf2/M6a-Mob3b axis facilitates prostate cancer progression. J Exp Clin Cancer Res (2020) 39(1):223. doi: 10.1186/s13046-020-01735-3
248. Li J, Xie H, Ying Y, Chen H, Yan H, He L, et al. Ythdf2 mediates the mrna degradation of the tumor suppressors to induce akt phosphorylation in N6-Methyladenosine-Dependent way in prostate cancer. Mol Cancer (2020) 19(1):152. doi: 10.1186/s12943-020-01267-6
249. Chen Y, Pan C, Wang X, Xu D, Ma Y, Hu J, et al. Silencing of Mettl3 effectively hinders invasion and metastasis of prostate cancer cells. Theranostics (2021) 11(16):7640–57. doi: 10.7150/thno.61178
250. Wang Y, Chen J, Gao WQ, Yang R. Mettl14 promotes prostate tumorigenesis by inhibiting Thbs1 Via an M6a-Ythdf2-Dependent mechanism. Cell Death Discovery (2022) 8(1):143. doi: 10.1038/s41420-022-00939-0
251. Anita R, Paramasivam A, Priyadharsini JV, Chitra S. The M6a readers Ythdf1 and Ythdf3 aberrations associated with metastasis and predict poor prognosis in breast cancer patients. Am J Cancer Res (2020) 10(8):2546–54.
252. Li C, Zhang C, Zhang G, Chen B, Li X, Li K, et al. Ythdf1 amplification is correlated with worse outcome and lower immune cell infiltrations in breast cancer. Cancer biomark (2022) 35(2):127–42. doi: 10.3233/CBM-203103
253. Chen H, Yu Y, Yang M, Huang H, Ma S, Hu J, et al. Ythdf1 promotes breast cancer progression by facilitating Foxm1 translation in an M6a-dependent manner. Cell Biosci (2022) 12(1):19. doi: 10.1186/s13578-022-00759-w
254. Yao X, Li W, Li L, Li M, Zhao Y, Fang D, et al. Ythdf1 upregulation mediates hypoxia-dependent breast cancer growth and metastasis through regulating Pkm2 to affect glycolysis. Cell Death Dis (2022) 13(3):258. doi: 10.1038/s41419-022-04711-1
255. Sun Y, Dong D, Xia Y, Hao L, Wang W, Zhao C. Ythdf1 promotes breast cancer cell growth, DNA damage repair and chemoresistance. Cell Death Dis (2022) 13(3):230. doi: 10.1038/s41419-022-04672-5
256. Chang G, Shi L, Ye Y, Shi H, Zeng L, Tiwary S, et al. Ythdf3 induces the translation of M(6)a-enriched gene transcripts to promote breast cancer brain metastasis. Cancer Cell (2020) 38(6):857–71.e7. doi: 10.1016/j.ccell.2020.10.004
257. Liu M, Zhou S, Wang J, Zhang Q, Yang S, Feng J, et al. Identification of genes associated with survival of breast cancer patients. Breast Cancer (2019) 26(3):317–25. doi: 10.1007/s12282-018-0926-9
258. Lin Y, Jin X, Nie Q, Chen M, Guo W, Chen L, et al. Ythdf3 facilitates triple-negative breast cancer progression and metastasis by stabilizing Zeb1 mrna in an M(6)a-dependent manner. Ann Transl Med (2022) 10(2):83. doi: 10.21037/atm-21-6857
259. Wang S, Zou X, Chen Y, Cho WC, Zhou X. Effect of N6-methyladenosine regulators on progression and prognosis of triple-negative breast cancer. Front Genet (2020) 11:580036. doi: 10.3389/fgene.2020.580036
260. Einstein JM, Perelis M, Chaim IA, Meena JK, Nussbacher JK, Tankka AT, et al. Inhibition of Ythdf2 triggers proteotoxic cell death in myc-driven breast cancer. Mol Cell (2021) 81(15):3048–64.e9. doi: 10.1016/j.molcel.2021.06.014
261. Liu X, Yuan J, Zhang X, Li L, Dai X, Chen Q, et al. Atf3 modulates the resistance of breast cancer cells to tamoxifen through an N(6)-Methyladenosine-Based epitranscriptomic mechanism. Chem Res Toxicol (2021) 34(7):1814–21. doi: 10.1021/acs.chemrestox.1c00206
262. Liu T, Wei Q, Jin J, Luo Q, Liu Y, Yang Y, et al. The M6a reader Ythdf1 promotes ovarian cancer progression Via augmenting Eif3c translation. Nucleic Acids Res (2020) 48(7):3816–31. doi: 10.1093/nar/gkaa048
263. Hao L, Wang JM, Liu BQ, Yan J, Li C, Jiang JY, et al. M6a-Ythdf1-Mediated Trim29 upregulation facilitates the stem cell-like phenotype of cisplatin-resistant ovarian cancer cells. Biochim Biophys Acta Mol Cell Res (2021) 1868(1):118878. doi: 10.1016/j.bbamcr.2020.118878
264. Xu F, Li J, Ni M, Cheng J, Zhao H, Wang S, et al. Fbw7 suppresses ovarian cancer development by targeting the N(6)-methyladenosine binding protein Ythdf2. Mol Cancer (2021) 20(1):45. doi: 10.1186/s12943-021-01340-8
265. Li J, Wu L, Pei M, Zhang Y. Ythdf2, a protein repressed by mir-145, regulates proliferation, apoptosis, and migration in ovarian cancer cells. J Ovarian Res (2020) 13(1):111. doi: 10.1186/s13048-020-00717-5
266. Wang Q, Guo X, Li L, Gao Z, Su X, Ji M, et al. N(6)-methyladenosine Mettl3 promotes cervical cancer tumorigenesis and warburg effect through Ythdf1/Hk2 modification. Cell Death Dis (2020) 11(10):911. doi: 10.1038/s41419-020-03071-y
267. Wang H, Luo Q, Kang J, Wei Q, Yang Y, Yang D, et al. Ythdf1 aggravates the progression of cervical cancer through M(6)a-mediated up-regulation of Ranbp2. Front Oncol (2021) 11:650383. doi: 10.3389/fonc.2021.650383
268. Wang X, Zhang J, Wang Y. Long noncoding rna Gas5-As1 suppresses growth and metastasis of cervical cancer by increasing Gas5 stability. Am J Transl Res (2019) 11(8):4909–21.
269. Li Z, Luo Q, Wang H, Liu Y, Feng X, Li Z, et al. [Knockdown of yth N(6)-methyladenosine rna binding protein 2 (Ythdf2) inhibits cell proliferation and promotes apoptosis in cervical cancer cells]. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi (2020) 36(3):255–63.
270. Wu M, Chen G, Liao X, Xiao L, Zheng J. Ythdf2 interference suppresses the emt of cervical cancer cells and enhances cisplatin chemosensitivity by regulating Axin1. Drug Dev Res (2022) 83(5):1190–200. doi: 10.1002/ddr.21942
271. Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, et al. M(6)a mrna methylation regulates akt activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol (2018) 20(9):1074–83. doi: 10.1038/s41556-018-0174-4
272. Hong L, Pu X, Gan H, Weng L, Zheng Q. Ythdf2 inhibit the tumorigenicity of endometrial cancer Via downregulating the expression of Irs1 methylated with M(6)A. J Cancer (2021) 12(13):3809–18. doi: 10.7150/jca.54527
273. Zhang L, Wan Y, Zhang Z, Jiang Y, Lang J, Cheng W, et al. Fto demethylates M6a modifications in Hoxb13 mrna and promotes endometrial cancer metastasis by activating the wnt signalling pathway. RNA Biol (2021) 18(9):1265–78. doi: 10.1080/15476286.2020.1841458
274. Shen J, Feng XP, Hu RB, Wang H, Wang YL, Qian JH, et al. N-methyladenosine reader Ythdf2-mediated long noncoding rna fendrr degradation promotes cell proliferation in endometrioid endometrial carcinoma. Lab Invest (2021) 101(6):775–84. doi: 10.1038/s41374-021-00543-3
275. Du J, Hou K, Mi S, Ji H, Ma S, Ba Y, et al. Malignant evaluation and clinical prognostic values of M6a rna methylation regulators in glioblastoma. Front Oncol (2020) 10:208. doi: 10.3389/fonc.2020.00208
276. Yarmishyn AA, Yang YP, Lu KH, Chen YC, Chien Y, Chou SJ, et al. Musashi-1 promotes cancer stem cell properties of glioblastoma cells Via upregulation of Ythdf1. Cancer Cell Int (2020) 20(1):597. doi: 10.1186/s12935-020-01696-9
277. Tassinari V, Cesarini V, Tomaselli S, Ianniello Z, Silvestris DA, Ginistrelli LC, et al. Adar1 is a new target of Mettl3 and plays a pro-oncogenic role in glioblastoma by an editing-independent mechanism. Genome Biol (2021) 22(1):51. doi: 10.1186/s13059-021-02271-9
278. Chai RC, Chang YZ, Chang X, Pang B, An SY, Zhang KN, et al. Ythdf2 facilitates Ubxn1 mrna decay by recognizing Mettl3-mediated M(6)a modification to activate nf-kappab and promote the malignant progression of glioma. J Hematol Oncol (2021) 14(1):109. doi: 10.1186/s13045-021-01124-z
279. Miao YQ, Chen W, Zhou J, Shen Q, Sun Y, Li T, et al. N(6)-Adenosine-Methyltransferase-14 promotes glioma tumorigenesis by repressing argininosuccinate synthase 1 expression in an M6a-dependent manner. Bioengineered (2022) 13(1):1858–71. doi: 10.1080/21655979.2021.2018386
280. Dixit D, Prager BC, Gimple RC, Poh HX, Wang Y, Wu Q, et al. The rna M6a reader Ythdf2 maintains oncogene expression and is a targetable dependency in glioblastoma stem cells. Cancer Discovery (2021) 11(2):480–99. doi: 10.1158/2159-8290.CD-20-0331
281. Chen Y, Wang YL, Qiu K, Cao YQ, Zhang FJ, Zhao HB, et al. Ythdf2 promotes temozolomide resistance in glioblastoma by activation of the akt and nf-kappab signalling pathways Via inhibiting Ephb3 and Tnfaip3. Clin Transl Immunol (2022) 11(5):e1393. doi: 10.1002/cti2.1393
282. Li T, Gu M, Deng A, Qian C. Increased expression of Ythdf1 and Hnrnpa2b1 as potent biomarkers for melanoma: A systematic analysis. Cancer Cell Int (2020) 20:239. doi: 10.1186/s12935-020-01309-5
283. Jia R, Chai P, Wang S, Sun B, Xu Y, Yang Y, et al. M(6)a modification suppresses ocular melanoma through modulating Hint2 mrna translation. Mol Cancer (2019) 18(1):161. doi: 10.1186/s12943-019-1088-x
284. Yang S, Wei J, Cui YH, Park G, Shah P, Deng Y, et al. M(6)a mrna demethylase fto regulates melanoma tumorigenicity and response to anti-Pd-1 blockade. Nat Commun (2019) 10(1):2782. doi: 10.1038/s41467-019-10669-0
285. Yu J, Chai P, Xie M, Ge S, Ruan J, Fan X, et al. Histone lactylation drives oncogenesis by facilitating M(6)a reader protein Ythdf2 expression in ocular melanoma. Genome Biol (2021) 22(1):85. doi: 10.1186/s13059-021-02308-z
286. Xu Y, He X, Wang S, Sun B, Jia R, Chai P, et al. The M(6)a reading protein Ythdf3 potentiates tumorigenicity of cancer stem-like cells in ocular melanoma through facilitating Ctnnb1 translation. Oncogene (2022) 41(9):1281–97. doi: 10.1038/s41388-021-02146-0
287. Orouji E, Peitsch WK, Orouji A, Houben R, Utikal J. Oncogenic role of an epigenetic reader of M(6)a rna modification: Ythdf1 in merkel cell carcinoma. Cancers (Basel) (2020) 12(1):202. doi: 10.3390/cancers12010202
288. Nguyen TT, Ma LN, Slovak ML, Bangs CD, Cherry AM, Arber DA. Identification of novel Runx1 (Aml1) translocation partner genes Sh3d19, Ythdf2, and Znf687 in acute myeloid leukemia. Genes Chromosomes Cancer (2006) 45(10):918–32. doi: 10.1002/gcc.20355
289. Paris J, Morgan M, Campos J, Spencer GJ, Shmakova A, Ivanova I, et al. Targeting the rna M(6)a reader Ythdf2 selectively compromises cancer stem cells in acute myeloid leukemia. Cell Stem Cell (2019) 25(1):137–48.e6. doi: 10.1016/j.stem.2019.03.021
290. Chen Z, Shao YL, Wang LL, Lin J, Zhang JB, Ding Y, et al. Ythdf2 is a potential target of Aml1/Eto-Hif1alpha loop-mediated cell proliferation in T (Aml. Oncogene (2021) 40(22):3786–98. doi: 10.1038/s41388-021-01818-1
291. Qing Y, Dong L, Gao L, Li C, Li Y, Han L, et al. R-2-Hydroxyglutarate attenuates aerobic glycolysis in leukemia by targeting the Fto/M(6)a/Pfkp/Ldhb axis. Mol Cell (2021) 81(5):922–39.e9. doi: 10.1016/j.molcel.2020.12.026
292. Luo Z, Li G, Wang M, Zhu J, Yang Z, Li Y, et al. Ythdf1 Rs6090311 a>G polymorphism reduces hepatoblastoma risk: Evidence from a seven-center case-control study. J Cancer (2020) 11(17):5129–34. doi: 10.7150/jca.46120
293. He J, Yuan L, Lin H, Lin A, Chen H, Luo A, et al. Genetic variants in M(6)a modification core genes are associated with glioma risk in Chinese children. Mol Ther Oncolytics (2021) 20:199–208. doi: 10.1016/j.omto.2020.12.013
Keywords: M6A, YTHDF, biological process, cancer, clinical applications
Citation: Chen L, Gao Y, Xu S, Yuan J, Wang M, Li T and Gong J (2023) N6-methyladenosine reader YTHDF family in biological processes: Structures, roles, and mechanisms. Front. Immunol. 14:1162607. doi: 10.3389/fimmu.2023.1162607
Received: 09 February 2023; Accepted: 28 February 2023;
Published: 14 March 2023.
Edited by:
Xiangpeng Dai, Jilin University, ChinaReviewed by:
Zhijun Zhou, University of Oklahoma Health Sciences Center, United StatesCopyright © 2023 Chen, Gao, Xu, Yuan, Wang, Li and Gong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jun Gong, Z2p1bkBodXN0LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.