
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 10 May 2023
Sec. Molecular Innate Immunity
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1162211
This article is part of the Research Topic Regulation and therapeutic development of type I interferon signaling View all 4 articles
 Shuai Yang1,2†
Shuai Yang1,2† Weishan Shen3†
Weishan Shen3† Jiajia Hu4†
Jiajia Hu4† Sihui Cai2
Sihui Cai2 Chenqiu Zhang2
Chenqiu Zhang2 Shouheng Jin2
Shouheng Jin2 Xiangdong Guan3*
Xiangdong Guan3* Jianfeng Wu3*
Jianfeng Wu3* Yaoxing Wu1,3*
Yaoxing Wu1,3* Jun Cui1,2*
Jun Cui1,2*Spatiotemporal separation of cellular components is vital to ensure biochemical processes. Membrane-bound organelles such as mitochondria and nuclei play a major role in isolating intracellular components, while membraneless organelles (MLOs) are accumulatively uncovered via liquid-liquid phase separation (LLPS) to mediate cellular spatiotemporal organization. MLOs orchestrate various key cellular processes, including protein localization, supramolecular assembly, gene expression, and signal transduction. During viral infection, LLPS not only participates in viral replication but also contributes to host antiviral immune responses. Therefore, a more comprehensive understanding of the roles of LLPS in virus infection may open up new avenues for treating viral infectious diseases. In this review, we focus on the antiviral defense mechanisms of LLPS in innate immunity and discuss the involvement of LLPS during viral replication and immune evasion escape, as well as the strategy of targeting LLPS to treat viral infectious diseases.
Spatiotemporal separation of cellular processes is necessary to ensure subcellular compartmentation and proper biological functions. Membrane-bound subcellular organelles are responsible for sequestering and compartmentalizing intracellular components in most cases (1); however, higher-order molecular condensates, also known as membraneless organelles (MLOs), have been recently revealed to participate in subcellular compartmentation through liquid-liquid phase separation (LLPS) (2–5). In contrast to classic organelles with lipid membranes, MLOs achieve specialized subregions through LLPS of biological polymers, such as proteins and nucleic acids, which allows subcellular enrichment of particular biomolecules to ensure their biological processes and biochemical reactions (3, 6–8). The pervasive roles of LLPS and MLOs during cellular processes have been greatly expanded in the past decade, including Cajal and promyelocytic leukemia (PML) bodies in the nucleus, as well as P-bodies (PBs) and stress granules (SGs) in the cytoplasm (9). Recent studies have focused on the particular involvement of biomolecular condensates in prompting interferon (IFN) relative antiviral biological processes (10, 11). Viruses employ strategies to form condensates for viral assembly and production, such as inclusion bodies (IBs) (12, 13). Therefore, unraveling the participation of LLPS during viral infection could open up new avenues for investigating the pathology and treatment of viral infectious diseases.
In this review, we focus on the involvement of LLPS as an antiviral defense mechanism during innate immunity and discuss the strategies involving LLPS used by viruses for replication and immune evasion. Additionally, we discuss the potential tactics of targeting the formation of LLPS to develop host-directed therapies as treatment options against viral infectious diseases.
The cellular membrane is responsible for dividing spaces for membranous organelles to perform specific biological functions (14–18). These membrane-bound organelles are convenient for constructing specific reaction systems and reaction environments, and reducing the influence of membrane proteins or reaction substances on the external environment. It has recently been revealed that biomolecules, including nucleic acids and proteins, can form membraneless compartments, also referred to as MLOs or LLPS (3, 19, 20). LLPS is a reversible physicochemical response in which large molecular components aggregate into a dense phase co-existing with a dilute phase. In 2009, Hyman’s team observed the formation of phase separation through the properties of P granules (21). In 2012, Li P. and colleagues found that multivalent proteins could undergo a rapid transition from small complexes to large polymeric assemblies, which increased the protein concentration (22). Moreover, Steven McKnight and colleagues found that RNA granules underwent LLPS condensate in a cell-free system (23). Since then, LLPS has become a new focus for targeting cellular processes.
As the global or regional concentration of macromolecules in solution increases, phase separation occurs from a low concentration dispersed state into a high concentration gel-like “liquid drop” state under appropriate conditions and these two states dynamically exchange. With the continued increase in the molecule concentration, the drop-like LLPS continues to transform into a colloidal form, which is termed solid-liquid phase separation. In a normal cell, the concentration of most proteins cannot reach the threshold of phase separation; however, under certain cellular processes, such as posttranslational modification, oligomerization, nucleic acid binding, or conformation change, several proteins can undergo phase separation at a low threshold concentration. In this context, cellular components can assemble more flexibly, which activates certain biochemical reactions and leads to the adoption of a continuum of material properties.
The LLPS system consists of two components: solutions and biomacromolecules. The multivalent forces between biomacromolecules include electrostatic interactions between charged residues, hydrogen bonds, hydrophobic interactions between weakly polar residues, superposition between aromatic residues, and cation superposition residues between positive charges and aromatic groups (3, 14, 22, 24, 25). LLPS is mainly induced by the following conditions: 1. Multivalent weak interactions between intrinsic disorder regions (IDR) or low-complexity regions of proteins (26); 2. scaffold proteins forming a phase separation through multivalent specific interaction networks, which allows enzymes or enzyme complexes to enter as “passengers”. Scaffold proteins drive LLPS and they are sufficient for spontaneous droplet and passenger molecules partition into condensates and influence the LLPS system (27); and 3. RNA-containing repetitive sequences, which are widely involved in the formation of membrane-free organelles rich in RNA/protein (28, 29). Additionally, LLPS processes are influenced by environmental parameters such as concentrations of components, the temperature of the system, salt, and pH (Figure 1) (30, 31).
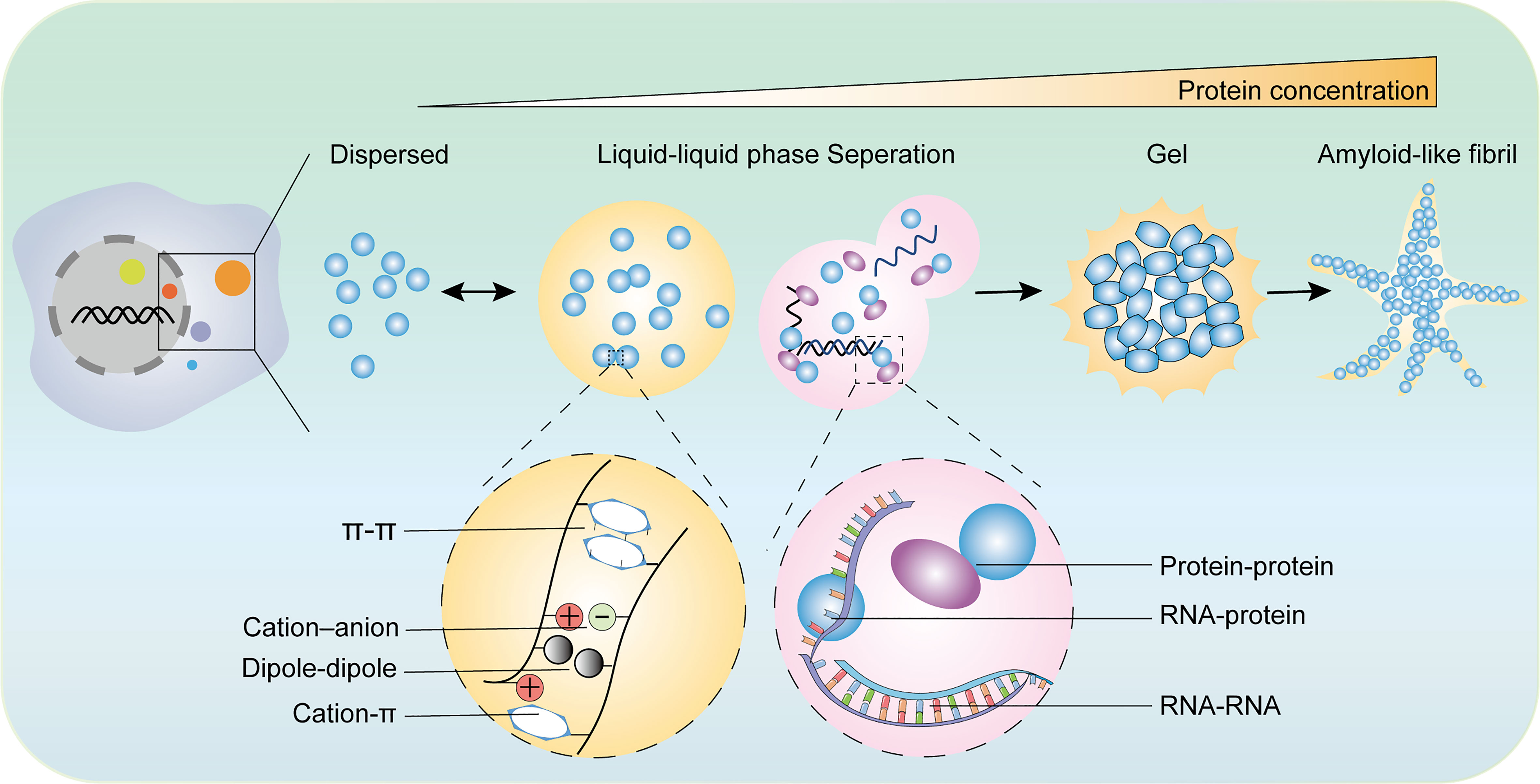
Figure 1 The diverse states of phase separated condensates and the driving forces of phase separation. The biomolecular condensates formed through LLPS are highly dynamic and exchange the components with surroundings. With the increase of protein concentration, the condensates of liquid-like condensates turn irreversibly to gels or amyloid fibrils. There are various types of multivalent interactions trigger phase transition including π–π interactions, cation–anion interactions, dipole−dipole interactions, cation–π interactions, and conventional multivalent interactions between protein and protein, protein and RNA, or RNA and RNA.
Biomolecular condensates specially recruit certain molecules while excluding others, which enables condensates to function as selective compartments. LLPS offers a space for participants to exchange the components with surroundings. The shape of LLPS is defined by the surface tension, and the surface tension tends to reduce the area of the interface until it reaches a minimum as a spherical shape. Therefore, LLPS tends to be spherical. The spherical droplets provide the equal chemical potentials of the proteins on either side of the boundary, as the mixing tendency is offset by interaction energy (32, 33). As a result, molecules in the condensates exhibit dynamic balance (34).
Biomacromolecules in solution tend to have less free energy, meaning that they diffuse evenly in the solution. The unit concentration can be increased as an effective strategy to ensure the timely participation of macromolecules in biological functions. In solutions with higher concentrations, the additional energy output can compensate for the entropy loss caused by the aggregation of biological macromolecules (20). Once the molecule concentration increases over the critical concentration, the solution undergoes phase separation to produce a dilute solution phase or even a colloidal phase (3, 6).
LLPS can be induced by multivalent interactions of biological macromolecules, such as intracellular protein-protein, protein–RNA, and RNA–RNA interactions (3, 35). Many studies have shown that proteins containing IDRs can interact with surrounding molecules and undergo LLPS, and IDR-containing proteins are more likely to undergo LLPS (4). Typically, the IDRs are highly enriched in specific proteins containing aromatic residues, charged residues, or hydrophilic residues. The amino acids within IDRs cannot fold tightly as a stable tertiary structure, which leads to a more flexible conformation and dynamic properties. Thus, they are closer to the surroundings and have more possibilities to interact with other biological molecules (36–38). The flexible conformation in IDRs perfectly meets the requirements of multivalent weak interactions in LLPS (30, 39). In addition to LLPS mediated by IDRs, LLPS can be triggered by molecular interactions, which are theoretically stronger and more specific for the complexity of biological processes. Researchers have also reported that RNA containing a large number of repetitive sequences can trigger intranuclear phase transitions in repeat expansion disorders, which can lead to neuromuscular diseases such as Huntington’s disease, muscular dystrophy, and amyotrophic lateral sclerosis (28, 40, 41).
LLPS participates in cellular processes, including genome remapping, gene expression, and the formation of nucleosome arrays, DNA damage foci, X-chromosome inactivation (XCI) centers, paraspeckles, stress granules, proteasomes, and autophagosomes (42–45). In the nucleus, the chromatin compartments can form MLOs, in which several structural chromosomal components, such as adenosine deaminase complexing protein 1 (ADCP1), heterochromatin protein 1 α HP1a, and Chromobox 2 (CBX2), are capable of undergoing LLPS. These chromatin compartmentations have been also shown to mediate the binding of transcription factors and their DNA promoters, resulting in changes in gene transcription. Indeed, the transcription co-activators bromodomain-containing protein 4 (BRD4) and mediator complex subunit 1 (MED1) can form droplets at the super enhancer region and gather to achieve the compartmentalization of the transcription process (27). The transcriptional activation domain of the transcription factors POU class 5 homeobox 1 (OCT4) and general control transcription factor 4 (GCN4) activates gene expression through LLPS with the transcriptional mediator complex mediator (28). The transcription factor YAP can mediate gene transcription through LLPS with a PDZ-binding motif (29). HP1α has been shown to occur in LLPS in cells and in vitro, which could promote chromatin conformation rearrangement and increase chromatin disorders (46). In the cytoplasm, LLPS participates in a wide range of biological processes, including maintaining cellular homeostasis, controlling immune responses, and promoting inflammasome protein degradation. The key autophagy-related proteins Atg13 and Atg17 can undergo LLPS to regulate the formation of autophagy (47). Speckle-type POZ protein (SPOP), the adapter protein of the proteasome pathway, was found to target DAXX (death-domain-associated protein) through LLPS, thus mediating the proteasome pathway degradation of the target protein (48). LLPS is also believed to regulate metabolic flow (49).
Abnormal LLPS is also related to the occurrence of diseases. Indeed, LLPS of Tau has been observed in neurons of patients with amyotrophic lateral sclerosis (17), Alzheimer’s disease (AD), Parkinson’s disease (PD), and frontotemporal dementia (FTD) (40, 50–52). Meanwhile, there is emerging evidence that LLPS is involved in cancer, viral diseases, and the anti-viral infection immune response (53–56). These studies indicate that LLPS plays a vital role in human health and diseases.
As obligatory intracellular parasites that co-evolve with their hosts over a long period, viruses have developed various strategies to drive and use LLPS in different steps of their lifecycles. Several virus-encoded proteins are characterized by a high degree of structural disorder and multivalence, as well as nucleic acid-binding capacity, which fulfills the classic prerequisites for LLPS. Viral protein-driven LLPS results in compartmentalization either in the cytoplasm or in the nucleus, and the formation of MLOs characterized by liquid-like features. These structures provide a specialized and isolated environment for the concentration of viral and cellular components to ensure the spatial organization and regulation of viral replication processes. Moreover, viral phase-separated compartments can prevent the activation of cell-intrinsic antiviral defenses by spatially excluding or sequestering innate immune components. Here, we have enumerated several examples of viral components with the capacity for LLPS and discussed their underlying mechanisms.
The biophysical mechanisms of LLPS by viral proteins concerning membraneless replication compartments have been well characterized among non-segmented negative-sense RNA viruses (nsNSV); these include several pathogens with high relevance to human diseases such as rabies virus (RABV), measles virus (MeV), respiratory syncytial virus (RSV), and Ebola virus (EBOV) (57). A defining feature of these viruses is the formation of cytoplasmic inclusions, referred to as viral inclusion bodies (IBs) or Negri bodies (NBs). FRAP experiments have revealed that viral IBs possess liquid-like properties shared by cellular MLOs, enabling them to fuse and fission, exchange materials with their surroundings, and respond to stimulation (58–60). Indeed, in vitro recombinant expression of viral nucleoprotein (N) and phosphoprotein (P) can reconstruct cellular minimal systems that recapitulate IB-like features (58–61). Both N and P proteins have the potential for LLPS, which is endowed with high levels of intrinsic disorder and multivalence. The RNA-binding capacity, P protein interaction, and oligomerization of N protein are essential for the IB formation of different viruses. Although the necessary structural elements of N and P vary according to virus species (as detailed in Table 1), the conserved IDRs in viral proteins mediate multiple protein-protein and protein-RNA interactions, which contribute to the multivalent interactions underlying LLPS (58, 60, 61, 65, 91, 92).
The IBs formed via N-P phase separation have often been described as specialized sites for viral transcription and replication, colocalizing with the viral RNA replication machinery including N, P, and the RNA-dependent RNA polymerase (RdRp) (62, 68). Phase separation of IBs increases the local concentration of viral components to support efficient replication. In particular, the functional sub-compartments within IBs, termed IB-associated granules (IBAGs), can recruit nascent viral mRNA and viral transcription anti-terminator M2-1, with the other IB components excluded (71). IBAGs have been proposed to function as viral mRNA sorting stations, in which the nascent viral mRNAs transiently concentrate after viral transcription and replication in other areas of IBs, followed by cytosolic export with M2-1 for translation (71). The viral IBs also selectively recruit several cellular factors with identified proviral effects to support viral replication and transcription, such as FAK and hsp70 in RABV (as detailed in Table 1) (63, 64). Viral IBs have also been found to participate in viral RNA encapsulation and RNP formation. RNA molecules preferentially localize to N-P protein droplets and trigger the production of nucleocapsid-like structures. Accordingly, in such cases, nucleocapsid assembly is significantly improved compared to non-phase-separated conditions (65). It has also been observed that RNPs are ejected from RABV NBs in a cytoskeleton-dependent manner before being further transported to the cytoplasm (58). Moreover, LLPS constitutes an indispensable part of virus antagonism against the host’s innate defense. It has also been shown that several viral proteins recruited into IBs display an inhibitory effect on the host IFN response (66, 93). Additionally, the formation of viral inclusions can spatially exclude cellular viral sensors and sequester key factors of the downstream pathways to prevent the initiation of antiviral signaling (58, 94–96). Collectively, the formation of viral IBs by LLPS participates in different stages of the viral lifecycle.
In contrast to the members of nsNSV, influenza A virus (IAV) contains a segmented, eight-partite RNA genome that replicates in the nucleus and is encapsulated into different types of vRNP complexes. Accumulating evidence indicates that LLPS may play a vital role in the spatio and temporal control of the IAV genome assembly. Following genome replication in the nucleus, the vRNPs are exported to the cytosol and present scattered distribution in the cytoplasm, colocalizing with the cellular GTPase Rab11, which is required for the biogenesis of vRNP hotspots (97). The vRNP/Rab11 condensates constitute viral inclusions, which display characteristics of liquid organelles. The formation and maintenance of phase separated viral inclusions depended on the interactions between viral and cellular proteins, whereas the RNA–RNA intersegment interactions among different types of vRNPs appear to be independent (97). Additionally, it has been demonstrated that the formation of IAV IBs is strictly spatially regulated and highly associated with membrane-bound organelles, developing in the vicinity of the endoplasmic reticulum exits sites (ERES) and depending on ER-Golgi vesicular cycling (97). It has been proposed that IAV IBs concentrate vRNPs that are transported to the cytosol at specific sites to allow the nucleation of vRNP–vRNP interactions for viral genome assembly and posterior delivery to the plasma membrane.
Viral protein-driven LLPS also contributes to the multiplication of positive-sense RNA viruses. However, RNA viruses triggered LLPS does not involve N-P-mediated interactions. In the case of SARS-CoV-2, as a representative, the nucleoprotein alone can undergo LLPS under specific conditions, independently of other viral proteins (54, 82, 83). Resembling the phase-separating proteins of nsNSV, the SARS-CoV-2 N protein exhibits a modular architecture, comprising an ordered N-terminal domain (NTD) and a C-terminal dimerization domain (CTD) flanked by IDRs, including an N-terminal IDR, a C-terminal IDR, and a central IDR (98). The essential roles of the central IDR, which has a serine/arginine (SR)-rich region and an adjacent leucine/glutamine (L/Q)-rich region, have been widely acknowledged in N phase separation (54, 82). More importantly, it has been shown that the introduction of RNA can dramatically enhance N protein phase separation (54, 82, 83), which is independent of RNA sequence specificity (82). Further research revealed that phosphorylation in the SR-rich region is involved in regulating the RNA-induced LLPS behavior of N, including its tendency for phase separation and the viscosity of the condensates (54). Regulation by phosphorylation modification in SR region confers the N protein with dual functions during viral replication, as hyperphosphorylated N protein promotes viral transcription, and hypophosphorylated N protein mediates RNA packaging (54, 83).
N phase separation plays a general role in SARS-CoV-2 genome packaging and virion assembly. Particles with shell-like architectures have been observed in soluble N-RNA complexes via negative-stain electron microscopy (99). Moreover, the soluble CTD of M proteins has been shown to interact with N and induce the formation of N condensates independently of RNA (54). A mixture of three components, including N, M, and RNA, spontaneously forms mutually exclusive condensates, with a central core of N-RNA condensation surrounded by a shell of N-M, which is easily reminiscent of a virion structure (54). In addition to its role in RNA packaging, recent advances have revealed that phosphorylated N proteins facilitate the synthesis of viral RNAs through the recruitment of the cellular RNA helicase DDX1 (84). Furthermore, N-induced condensates can act to sequester the host stress granule core protein G3BP1, indicating the role of N in suppressing innate immune responses (85).
Overall, LLPS is wildly exploited by RNA viruses during infection. Viral phase-separated compartments serve as the hub for various viral processes, including genome transcription, replication, and virion assembly. Additionally, the formation of viral inclusions could also prevent the activation of cell-intrinsic defenses by spatially excluding cellular sensors and sequestering key antiviral factors. These functions suggest that targeting LLPS may be a novel effective antiviral strategy to inhibit virus replication.
Compared to the wide involvement of LLPS during RNA virus replication, much less is known about the roles of LLPS in the lifecycle of double-stranded DNA (dsDNA) viruses. Recent advances in the research on herpesviruses have provided new insight into the functional importance of LLPS for dsDNA viruses. Generally, Herpesviridae is a large family of DNA viruses that is divided into three subfamilies, including alpha-Herpesviridae (e.g., HSV-1), beta-Herpesviridae (e.g., HCMV), and gamma-Herpesviridae (e.g., KSHV and MHV-68) (100). In contrast to RNA viruses, herpesviruses have evolved more complex strategies in their replication cycle and progeny virion production. Following viral invasion via membrane fusion at the cell surface, the capsid is uncoated and transported to a nuclear pore, thus releasing viral genomes into the nucleoplasm. The invading viral genomes can either initiate transcription and replication within the nucleus during lytic infection or stay static during latent infection (100). Recent studies have revealed that LLPS plays an essential role in different stages of the herpesvirus lifecycle, including latency maintenance, genome replication, and virion assembly.
The latency-associated nuclear antigen (LANA), encoded by Kaposi’s sarcoma-associated herpesvirus (KSHV), serves as a key regulator of viral latency. During latent infection, the KSHV genome changes its chromosome conformation and forms episomes, referred to as LANA-associated nuclear bodies (LANA-NBs), in a process that is strictly dependent on LANA LLPS. LANA binds to the terminal repeats (TRs) within viral template DNA through a structured C-terminal DNA binding domain, which leads to its oligomerization and reaching a concentration threshold for the induction of phase separation. In addition to DBD, LANA contains low-complexity (LC) domains that are responsible for various interactions involved in transcription regulation (87). A combination of LANA oligomerization, driven by the KSHV DNA template, and the multivalency of N-terminal LC domains eventually contributes to the formation of LANA-NB (86). Additionally, LANA oligomerization is necessary for the recruitment of the origin recognition complex protein (ORC2), as well as other nuclear factors, including the histone H3.3 chaperone DAXX, the polycomb-associated histone H3K27me3 methylase EZH2, and the histone H3K27me3, which is thought to maintain the chromatin organization of viral episomes (87). Notably, it has been observed that LANA-NBs undergo morphological changes in association with lytic reactivation, while LLPS disruption alters the KSHV genome conformation without inducing lytic reactivation, indicating a potential association between LLPS and lytic reactivation (86). However, the involvement of LANA-mediated LLPS in the transition from the latent to the lytic stage in the viral life cycle remains to be further explained.
The lytic infection cycle is initiated by viral genome replication with the simultaneous formation of viral replication compartments (VRCs). VRCs have been described as membraneless nuclear sub-compartments that provide a pro-replicative environment for viral replication. LLPS condensations induced by viral proteins are essential for VRC formation. The human cytomegalovirus (HCMV) UL112-113 protein, comprising a conserved N-terminal oligomerization domain and C-terminal IDRs, can independently induce biomolecular condensates with liquid-like properties (88) (101). The capacity of UL112-113 to drive LLPS relies on both self-oligomerization and multivalent interactions formed by IDRs (88). Additionally, UL112-113 LLPS is crucial for the recruitment of the viral DNA polymerase accessory factor UL44 at viral genomes to facilitate their replication (88). Similarly, the ICP4 protein of herpes simplex virus type-1 (HSV-1), an essential viral transcription factor, has also been identified as an IDP with LLPS properties that drives VRC formation (89). The ICP4 C-terminal activation domain (CTA) is an indispensable structural element for LLPS, while the N-terminal activation domain (NTA) and DNA binding domain (DBD) are likely to control the protein phase behavior (89). However, the biophysical properties of HSV-1 VRCs are not fully consistent with liquid-like nature of cellular condensates, and the cellular RNA polymerase II (RNA-PolII) within HSV-1 VRCs do not follow liquid-like diffusion kinetics when crossing the condensate interface, indicating additional mechanisms involved in the compartmentalization and molecule enrichment (102). Notably, the biophysical properties of VRCs change with the progression of infection. Mature VRCs exhibit irregular shape, higher viscosity and LLPS inhibitor resistance (88, 103). This transition can be blocked by inhibiting viral DNA replication, which implicates the involvement of viral genome replication in the maturation of VRCs as a condensate (103).
Following genome replication and nucleocapsid assembly within the nucleus, virion package occurs in the cytoplasm through a complicated multistage process including tegumentation and secondary envelopment (104). The formation of cytoplasmic virion assembly compartments (VACs) has been suggested to be essential for efficient viral replication at the stage of virion assembly via recruiting viral tegument proteins and host vesicles containing viral glycoproteins. However, the detailed mechanisms for VAC formation have not been completely elucidated. It has been shown that the formation and maintenance of VAC of HSV-1 and HCMV depend on the remodeling of the endomembrane system (105, 106). Recent research on MHV-68 (a γ-herpesvirus) also indicated the roles of LLPS in VAC formation. MHV-68 VACs was shown to exhibit liquid-like properties (107). And ORF52, a viral tegument protein with identified LLPS properties, has been found to underlie VAC formation. Moreover, nucleic acids participate in regulation of ORF52 phase separation. During the late stage of viral replication, cytoplasmic nascent RNA was co-aggregated with ORF52, promoting LLPS of ORF52 (107).
To summarize, akin to RNA viruses, LLPS is crucially at various stages of the herpesvirus lifecycle. Specifically, the initiation of LLPS by viral proteins induces the generation of viral condensates, such as VRCs, VACs, and latency-associated nuclear bodies, thus ensuring the spatial organization and regulation of various viral processes. The fundamental mechanism appears to be conserved among herpesviruses, offering novel perspectives for antiviral therapies.
The mammalian innate immune system serves as the first line of host defense. Pathogen-associated molecular pattern molecules (PAMPs) are recognized by pattern recognition receptors (PRRs) (108, 109). Upon viral infection, PRRs such as Toll-like receptors (TLRs) (110), retinoic acid-induced gene-I (RIG-I) like receptors (RLRs) (111), nucleotide-binding domain and leucine-rich repeat-containing receptors (NLRs) (112), cyclic GMP-AMP (cGAMP) synthase, and stimulator of interferon genes (STING) (113, 114), are employed to detect viral nucleic acids, hence leading to transcriptional expression of type I interferon (IFN-I) and hundreds of IFN-stimulated genes (ISGs), which amplify innate immune responses to effectively restrict viral replication (111). LLPS has also been shown to play a role in the regulation of innate immune signaling pathways (Figure 2). In the following section, we discuss the participation of LLPS during the innate immune response.
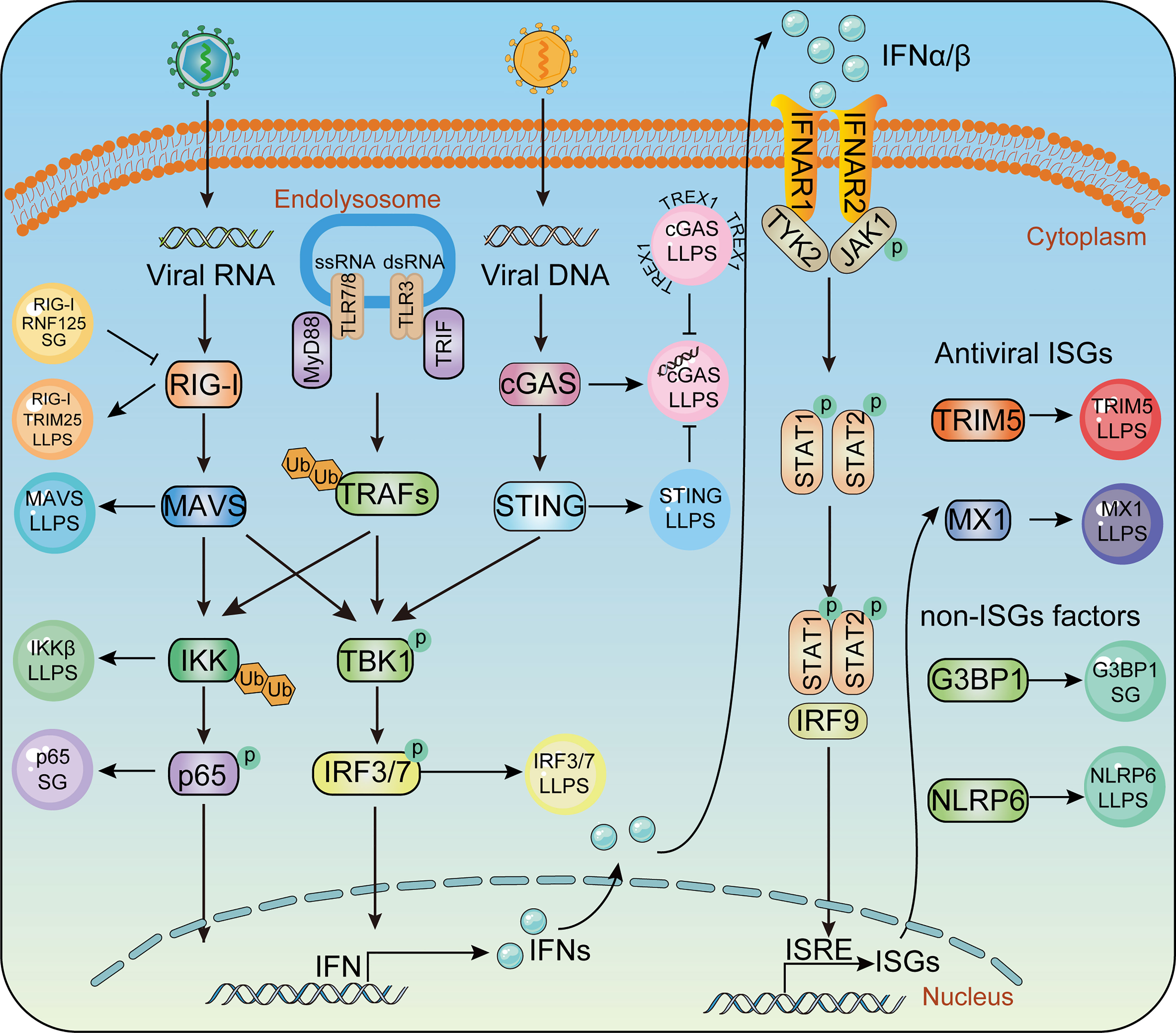
Figure 2 Phase separation in the innate immune pathway. Key molecules in innate immunity pathway undergoes LLPS to activate IFN signaling during virus infection, including TRIM25, MAVS, IRF3 in RLR signaling, cGAS, STING in cGAS-STING signaling and IKKβ, P65 in TLR pro-inflammatory signaling. Antiviral factors such as TRIM5, MX1, G3BP1, NLRP6 form liquid- like condensates and mediate antiviral immune responses.
RIG-I is the most famous RLR which specifically recognizes viral RNA (115, 116). Upon recognition of viral RNA, RIG-I undergoes K63-linked ubiquitination, conformational change, and tetramerization, thus allowing its interaction with the adaptor protein mitochondrial antiviral-signaling protein (MAVS). MAVS then anchors to mitochondria and undergoes prion-like aggregation, which in turn recruits and promotes the phosphorylation of TANK-binding kinase 1 (TBK1) and interferon regulatory factor 3 (IRF3). Phosphorylated IRF3 undergoes dimerization and nuclear translocation, which induces the expression of type I IFN. Recently, emerging evidence has revealed that LLPS participates in several stages of the RLR signaling pathway. TRIM25, a key E3 ligase promoting K63 ubiquitination of RIG-I and RIG-I activation, can form an LLPS state with RNA through its PRY/SPRY domains (117). MAVS, a key adaptor providing a scaffold to recruit downstream TBK1/IRF3 activation in the RLR pathway, has been proven to undergo LLPS after its K63-linked ubiquitination and oligomerization. The SARS-CoV-2 N protein has been found to inhibit the K63-linked polyubiquitination of MAVS and interfere with the LLPS of MAVS, thus inhibiting the activity of MAVS to facilitate immune evasion (108). IRF3, the pivotal transcriptional factor of IFN, has also been shown to undergo LLPS with interferon (IFN)-stimulated response element DNA and compartmentalized IRF7 in the nucleus, thus inducing the transcription of IFN. Deacetylation of IRF3 mediated by deacetylase SIRT1 is considered a prerequisite for IRF3/IRF7 LLPS, as hyper-acetylated IRF3 leads to a failure of IRF3 LLPS formation, with impairment of IFN induction and increased viral load and mortality in SIRT1 knockout mice (118). Therefore, we reason that LLPS contributes to the activation of the RLR-mediated IFN pathway.
The combination of DNA with cyclic GMP-AMP synthase (cGAS) leads to the generation of a secondary messenger loop GMP-AMP (cGAMP), which activates the innate immune response. The generation of the secondary messenger cGAMP binds to STING, which leads to STING transposition to the Golgi apparatus and the induction of IFN production through TBK1 and IRF3 (40, 114, 119–121).
At the initiation of cGAS-STING signaling, the combination of DNA with cGAS strongly induces the formation of cGAS LLPS droplets. The formation of cGAS-DNA LLPS condensates is affected by the disordered and positively charged region of cGAS, as well as the DNA length (40, 122, 123). Instead of directly controlling the activation of cGAS enzymes, the cGAS-DNA phase transition is thought to enhance its capability of DNA binding by inhibiting TREX1-mediated DNA degradation (124). TREx1 mutation specifically impairs the degradation of phase-separated DNA, which is associated with the serious autoimmune disease Aicardi-Goutières syndrome (124, 125). In addition to DNA, cGAS can undergo LLPS with RNA and spermine. Although neither RNA nor spermine can induce cGAS to generate cGAMP, RNA- or spermine-enhanced formation of cGAS can enhance cGAS activity, thus leading to downstream signaling and antiviral capability (121, 126).
Moderation of cGAMP in cells can induce STING transport to the Golgi apparatus and activate the antiviral natural immune pathway (121). As cGAMP is an effective activator of STING, it needs to be degraded to ensure a controlled signal transmission theoretically. Excessive accumulation of cGAMP in cells can also induce the phase separation of STING. The STING 309-342 fragment was identified as the IDR region mediating phase separation, and two conserved amino acid mutants, E336G/E337G, in this region impair the phase separation of STING. However, the cubic membrane structure of the endoplasmic reticulum generated by STING phase separation negatively regulates STING activation by spatially isolating STING-TBK1 from the transcription factor IRF3, thus preventing innate immune over-activation. Eventually, these cubic membrane structures could be decomposed by lysosomes or autolysosomes (127). Consistent with the above results, autoimmune disease-related mutants in STING exhibit significantly reduced phase separation capacity, suggesting that the downregulation of STING phase separators plays an abnormal role under pathological conditions (127, 128).
TLRs are the key PRRs in recognizing extracellular viral components and inducing IFN-I and pro-inflammatory cytokines. Currently, 11 TLR members have been identified in mammals, among which TLR3, TLR7, and TLR8 specifically recognize viral RNA, whereas TLR9 is a DNA recognition receptor (129). Upon activation, all TLRs, except for TLR3, recruit adaptor molecule myeloid differentiation factor 88 (MyD88) and activate transcription factors, including IRF3/7, nuclear factor-kappa B (NF-κB), and the activator protein 1 (AP-1) (130, 131). Ultimately, TLRs recognize molecular patterns leading to activation of the NF-κB.
The activation of NF-κB is a hallmark of most viral infections (132). Signaling cascades converging on the IKK complex can be activated during viral infection by viral proteins, virus-induced reactive oxygen species, and the release of viral nucleic acids. The IKK complex can induce NF-κB nuclear translocation and transcription of proinflammatory cytokines by promoting the phosphorylation and degradation of IκBα (133–136). Although the effects of NF-κB on viral replication depend on the viral species and the infected cell types, there is no dispute about the participation of NF-κB during viral infection. The IKK complex contains IKKα, IKKβ, and a regulatory subunit NEMO, which regulates the canonical NF-κB pathway. NEMO used to be considered to undergo head-to-head dimerization after binding to K63-linked or linear ubiquitination (137, 138). However, this model has been updated as NEMO has been recently shown to robustly undergo LLPS after binding to K63-linked or linear ubiquitination chains. The ubiquitin-binding (NUB) domain and the zinc-finger (ZF) domain of NEMO, which contribute to its Ub binding, are required for NEMO LLPS. Disease-associated mutations of NEMO, which impair its poly-ubiquitin binding and LLPS performance, lead to defects in NF-κB activation, indicating the importance of LLPS in NF-κB signaling (139).
ISGs contain a large proportion of antiviral factors that play direct roles in resistance against viral infection and replication. Accumulating evidence has shown that LLPS is also involved in the direct restriction or elimination of viruses by antiviral factors. Indeed, myxovirus resistance protein 1 (MX1), one of the most well-known antiviral ISGs, can inhibit multiple viruses by blocking the early steps of the viral replication cycle (140). Mx1 has been shown to form membraneless metastable (shape-changing) condensates in the cytoplasm. The human GFP Mx1 structure in the cytoplasm is considered a phase-separated membrane-free organelle that can include the VSV nucleocapsid (N) protein (10). TRIM5α, a well-known host factor that defends against invading retroviruses such as HIV-1, can also undergo LLPS, but the association between LLPS and the antiviral capability of TRIM5α remains to be elucidated (141, 142). LLPS also participates in the antiviral processes of non-ISGs to enhance the antiviral capability of ISGs. Cellular nucleic acid-binding protein (CNBP), which directly binds to the promoter of IFNβ in response to RNA virus infection, can promote IRF3 and IRF7 binding to IFN promoters for the maximal induction of IFN production (143). CNBP binds to SARS-CoV-2 viral RNA directly and competes with N protein, which restricts the formation of N protein-RNA LLPS of SARS-CoV-2 (144). G3BP1, a member of the heterogeneous nuclear RNA-binding protein, significantly enhances the recognition of cGAS to DNA and participates in virus clearance. Further research has shown that G3BP1 pre-assembles cGAS through LLPS to promote the activity of the cGAS enzyme. Additionally, G3BP1 interacts with RIG-I to enhance its binding to dsRNA and downstream signaling pathways (145, 146). NLRP6 is central to host defense by inducing the activation of inflammatory bodies and the production of interferon (147). Several studies have shown that NLRP6 undergoes LLPS when interacting with dsRNA in vitro and in cell model. The intrinsic disorder poly-lysine sequence of NLRP6 is important for multivalent interaction, phase separation, and activation of inflammatory bodies. In mice, NLRP6 deficiency or mutations in the LLPS region can lead to reduced activation of inflammasomes during infection with mouse hepatitis virus or rotavirus, indicating the anti-microbial immune function of NLRP6 LLPS (148).
Notably, m6A modification is the most common type of mRNA modification, which regulates gene expression. Three kinds of enzymes are involved in the m6A methylation of RNA: writers, erasers, and readers. Writers such as METTL3/14 and WTAP catalyze the m6A methylation of RNA, which is then recognized by readers such as YTHDF family proteins and participate in downstream translation and mRNA degradation, while erasers mediate the m6a demethylation modification. Targeting m6A not only mediates the control of the innate immune response but also blocks the replication of several viruses, including SARS-CoV-2 and influenza A virus (149, 150). The cytosolic m6A-binding proteins YTHDF1, YTHDF2, and YTHDF3 undergo phase separation (151–155); however, the association between the LLPS and the function of YTHDF proteins remains to be confirmed.
In conclusion, LLPS not only participates in the whole process of virus replication but also the antiviral process of the host cells, highlighting the importance of LLPS for viruses.
The preceding discussion has established the crucial role of LLPS in various aspects of the host antiviral response. To evade immune surveillance, viral proteins themselves also engage in LLPS as a means of circumventing host antiviral restrictions (Figure 3). This section presents a summary of several strategies used by viruses to evade host immunity via LLPS.
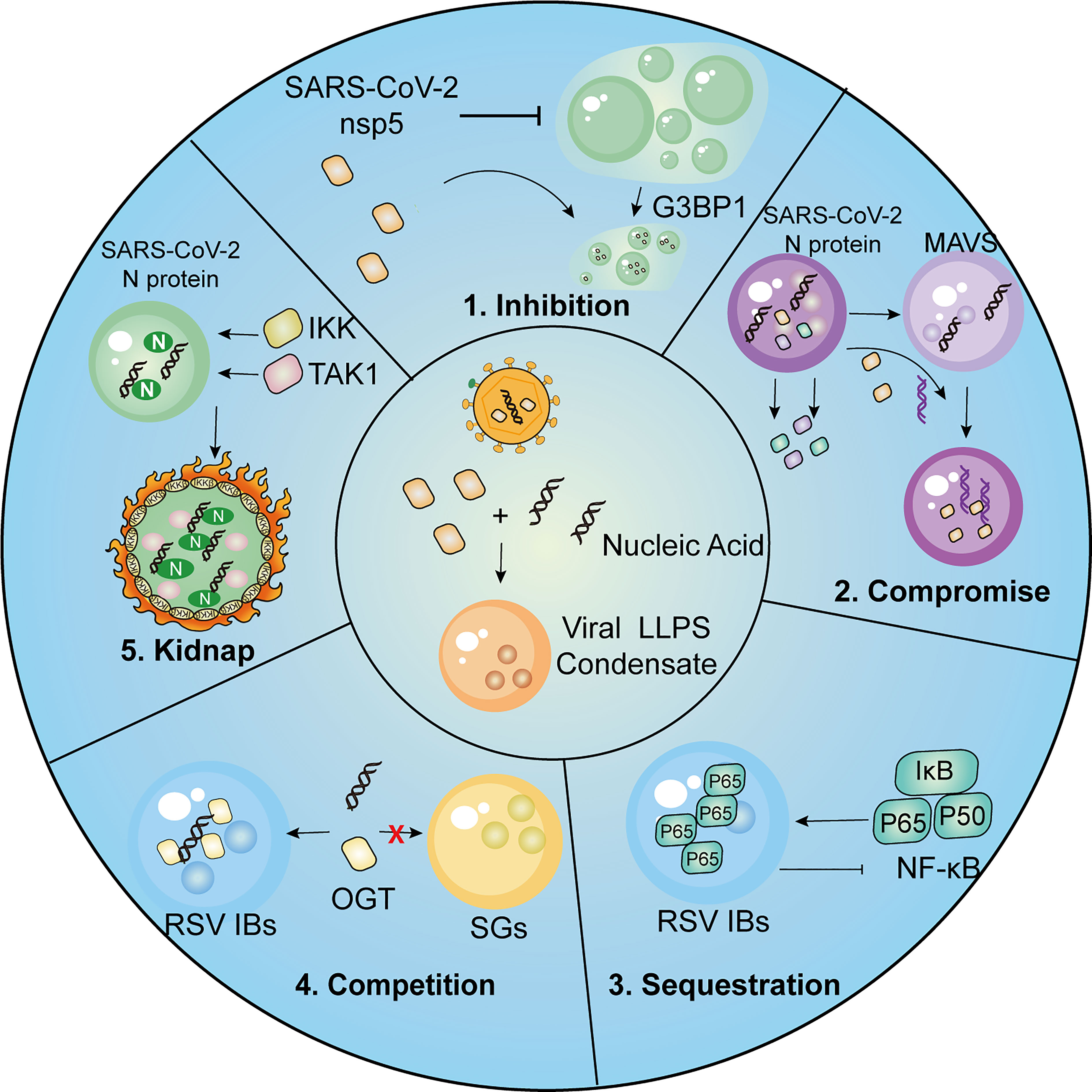
Figure 3 Summary of strategies employed by viruses for immune evasion.1, SARS-CoV-2 nsp5 inhibits SGs formation and promotes cleavage of G3BP1, resulting in inhibitory effect on IFN induction. 2, SARS-CoV-2 N protein undergoes LLPS thus compromising LLPS of MAVS and MAVS-dependent IFN induction. 3.The p65 subunit of NF-κB is trapped in the IBs of RSV, which leads to the restriction of its nuclear translocation and subsequent inhibitory effects on NF-κB signaling. 4. RSV-induced IBs compete OGT against SGs to suppress SG assembly. 5. SARS-CoV-2 N protein LLPS condensations recruit kinases IKKβ and TAK1 for NF-κB hyper-activation.
Viral proteins directly interact with the host factors and interfere with their roles in driving LLPS, which is regarded as a fundamental mechanism underlying the activation of host antiviral immunity. In particular, SGs, serving as cytoplasmic MLOs with highly sensitive to viral infections, are frequently antagonized by viruses. The initiation of the cellular stress response requires a core protein-RNA interaction network that drives LLPS and subsequent SG assembly. To counteract this vital host immune response, viruses employ a strategy to disrupt SG formation through competitive interaction with SG core proteins and interference with their mutual aggregation.
The Ebola virus (EBOV) multifunctional protein, VP35, has been shown to disrupt the aggregation of SG proteins, which is dose-dependently induced by exogenous stress. In cases that the level of VP35 is sufficient, the oligomerization of SG components is prevented during the late stages of EBOV infection (162). Notably, VP35-mediated SG deficiency is linked to its physical interaction with SG proteins, possibly through competitive binding, rather than its ability to contend for dsRNA (162). Similarly, nsp5, the SARS-CoV-2 main protease, can interact with G3BP1, inhibiting LLPS of G3BP1 and the SG formation. Instead of the direct cleavage of G3BP1 via nsp5 protease activity, SARS-CoV-2 nsp5 tends to competitively combine with SG components and interrupt G3BP1-triggered LLPS (85). As SG formation serves as the signaling hub to recruit antiviral factors such as RIG-I and MAVS, restriction of G3BP1 LLPS and SG formation would downregulate the IFN induction, resulting in the inhibition of cellular antiviral responses (145, 146).
In addition to direct competition with SG proteins, viral proteins can also indirectly inhibit host LLPS processes by limiting the interactions between cellular sensors and exogenous nucleic acids, which in turn allows viruses to evade and subvert the host immune surveillance and better establish infection within host cells.
Viral genomic DNA in the cytoplasm triggers the phase separation of the dsDNA sensor cGAS, which allows recognition of incoming pathogens and activation of antiviral signaling transduction. However, ORF52 and VP22, derived from KSHV and VZV, respectively, can interfere with cGAS-DNA phase separation (163). In vitro and in cell experiments revealed that the augment of ORF52 and VP22 displaced cGAS-DNA droplets, by simultaneously forming own liquid condensates with DNA (163). Additionally, ORF52-mediated restriction has been shown to impede the accumulation of cGAS substrates (ATP and GTP), thereby disrupting cGAS-induced signaling transduction (163). The capability of viral proteins to form multivalent interactions with DNA and undergo DNA-induced condensation plays an essential role in ORF52-mediated or VP22-mediated restriction of cGAS-DNA phase separation, which is determined by their IDRs (163). Another viral tegument protein, ORF9, from VZV, disrupts cGAS–DNA oligomers by undergoing DNA-dependent LLPS without associating with cGAS, which is similar to ORF52 and VP22 (163, 164).
Interestingly, RNA viruses such as SARS-CoV-2 could also impair MAVS function by restricting the formation of MAVS LLPS (159). In vitro, with the accumulation of N protein, the MAVS droplets are gradually displaced by N protein droplets, which relies on the dimerization domain of SARS-CoV-2 N protein (159). In addition to disrupting MAVS LLPS, SARS-CoV-2 N protein has been shown to directly interact with MAVS and inhibit Lys63-linked poly-ubiquitination and aggregation of MAVS, indicating multiple mechanisms by which SARS-CoV-2 can inhibit the MAVS-mediated interferon response via N protein (159).
Another evasion strategy that is widely employed by viruses is to sequester and shield key antiviral signaling molecules of the innate immune system within viral inclusions to ensure effective evasion and suppression of host innate immunity.
Respiratory syncytial virus (RSV) N protein within viral inclusion bodies (IBs) has been shown to sequester critical antiviral signaling molecules such as MAVS and MDA5, thus leading to the significant inhibition of RSV infection-induced IFN responses (94). The sequestration of MAVS and MDA5 by phase-separated N protein is further evidenced through their re-localization within IB-like structures by introducing RSV N and P proteins (94). Similarly, recruitment of the NF-κB subunit p65 within RSV IBs hinders its activation and subsequent nuclear translocation, thereby inhibiting downstream NF-κB signaling (165). RSV infection could also result in the sequestration of MAPK p38 within IBs and the subsequent interference with signal transduction through MAPK/MK2, which could be beneficial for virus replication (96). RABV multifunctional P protein targets cellular STAT proteins and sequesters them in the cytoplasm, which prevents the IFN-induced nuclear translocation of STATs and altogether inhibits JAK-STAT signaling in RABV-infected cells (166, 167). Similarly, infection with SFTSV leads to re-localization of the key signaling molecules to viral IBs, such as TBK1/IKKϵ and IRF3/IRF7 (168, 169).
The formation of viral IBs mediated by viral protein-driven LLPS results in spatial isolation of critical antiviral molecules from the cytoplasmic environment, leading to the blockage of host innate immunity.
Activation of antiviral responses requires the mobilization of various cellular resources. Therefore, viruses have also developed a novel mechanism for evading the immune system by competing with host cells for the cellular components of the antiviral immune system, which relies on viral proteins that promote phase separation.
As a concrete example, a KSHV inhibitor of cGAS (KicGAS) encoded by ORF52, forms condensates upon interacting with cytoplasmic dsDNA, thereby competitively inhibiting DNA-induced phase separation and subsequent activation of the dsDNA sensor cGAS (170). The N-terminal ordered domain has been shown to mediate KicGAS self-oligomerization, while the C-terminal IDR presumably mediates collective multivalent interactions with DNA. Both the N- and C-terminal domains of KicGAS are essential for KicGAS phase separation with DNA and the inhibition of cGAS (170).
Additionally, viral inclusions spatially separate cellular components from the cytoplasmic environment and preclude their roles in the cellular stress response, which presents a novel mechanism by which viruses contend for and occupy cellular resources to counteract antiviral immunity. For instance, the O-linked N-acetylglucosamine (OGN) transferase participates in regulating the stress response by catalyzing the post-translation modification of ribosomal proteins and supporting SG assembly (171, 172). It has been found that RSV infection results in the accumulation of OGT within viral inclusion bodies, leading to the inhibition of SG formation (96). Similarly, numerous necessary components for SG formation, including eIF4G, eIF3, PABP, and G3BP1, are also isolated within EBOV inclusions, which impedes the virus-induced SG assembly in EBOV-infected cells (162). Interestingly, the sequestration of these cellular proteins as well as the blockage of SG formation could be partly reversed upon stimulation of oxidative stress, with clear shrinkage and fragmentation of the viral IBs (96, 162), suggesting that sequestered proteins may be released into the cytoplasm due to the interruption of IB formation in response to exogenous stress. However, how viruses achieve the plunder of host factors within viral inclusions remains to be clarified as the formation of viral inclusion-like structures of nucleocapsid proteins was found to be insufficient to induce the relocation of SG proteins (162). In consideration of the RNA-binding capacity of SG proteins, the viral RNA colocalized with inclusion-bound granules may play a role in their competition (162).
Viruses hijack functional host factors and redirect them to virus-induced inclusions to implement their roles in viral life cycle replication, which is usually dependent on the interaction between host factors and phase-separating viral proteins.
It has been shown that hsp70, the major cellular heat shock protein, is frequently recruited by viruses and plays an indispensable role in various steps of viral replication. Indeed, RABV infection has been shown to induce enhanced expression of hsp70 and its recruitment to NBs, where viral transcription and replication occur (63). In contrast, specific inhibition of hsp70 synthesis significantly impaired viral transcription, viral protein accumulation, and virion production, indicative of a proviral effect of Hsp70 during RABV infection (63). Likewise, EBOV NP is known to recruit the cellular factor CAD into IBs, providing pyrimidines for EBOV RNA synthesis (78). The N protein of SARS-CoV-2 can also undergo liquid-liquid phase separation and recruit TAK1 and IKK complex, the key kinases of NF-κB signaling, which leads to the hyperactivation of NF-κB pro-inflammatory responses (173).
In addition to hijacking host factors to assist in viral replication, phase-separated viral proteins also recruit host-negative regulators to counteract host immune responses. For example, MeV-induced cytoplasmic IBs recruit WD repeat-containing protein 5 (WDR5) and promote viral replication (67). Depletion of WDR5 has been shown to enhance the induction of IFN-β during MeV infection (67). Additionally, the Tax1-binding protein 1 (TAX1BP1), which is directly involved in the negative regulation of NF-κB and IRF3 signaling pathways, has been identified as a cellular partner of RSV N protein (174). The depletion of TAX1BP1 resulted in enhanced antiviral and inflammatory responses and restricted RSV replication. These results suggest that RSV inhibits the host’s innate immune response through TAX1BP1 recruited by the RSV N protein (174).
LLPS constitutes an indispensable part of virus antagonism against the host’s innate defense. Viral proteins interfere with host cell functions by disrupting cellular LLPS processes to minimize host antiviral responses. Additionally, the phase separation of viral components and formation of viral MLOs lead to spatial and compartmental re-localization of host cellular factors, thus precluding the normal antiviral responses of host factors and facilitating viral replication.
As LLPS participates in multiple processes during virus infection, targeting the phase separation process may represent a new tactic for designing novel antiviral drugs. However, as both the host and virus share the LLPS system, which leads to double-edged sword effects, it remains to be seen whether LLPS needs to be upregulated or restricted when designing a treatment strategy. Thus, therapy development against infectious diseases necessitates targeting autophagy in a more selective way with specific molecular targets.
Targeting protein-protein interaction has long been considered a challenging task, but the number of successful cases has increased rapidly in the past decade. Recent studies have discovered successful chemical probes that dissolve viral condensates through interaction with viral proteins engaging in phase separation. A phase modulator is an optical modulator that can be used to control the optical phase of a laser beam and may target RNA or protein IDRs directly. LLPS of RNA and protein is widely involved in the assembly of organelles, and viral assembly of SARS-CoV-2 is also known to depend on LLPS (175). Treatment with 1,6- hexanediol (an LLPS inhibitor) can inhibit the formation of N protein phase separation, which not only weakens TAK1 and IKKβ in the process of virus infection but can also inhibit the inflammatory response and inflammatory factor release caused by COVID-19 infection, thus representing a potential anti-inflammatory strategy of targeting phase separation for treating COVID-19 (173). However, 1,6-hexanediol is highly cytotoxic and arrests phosphatase and kinase activities. A nontoxic alternative, propylene glycol, could provide an alternative to separate rotavirus replication factors (176). Additionally, many viruses achieve immune escape by hijacking host antiviral factors, among which cyclophilin A (CypA) is of concern because it can be hijacked by HIV, hepatitis C virus, and SARS, among others. Through an in-depth study of its mechanism, CypA has been found to bind to protein kinase R (PKR), which affects its ability to detect viruses (177). Therefore, in PKR knockout cells by CRISPR/Cas9, cyclophilin inhibitors have been shown to have a weak ability to prevent virus replication. Antiviral drugs targeting CypA can be used to treat many incurable viruses (177, 178). Targeting SARS-CoV-2-N protein LLPS is considered to be a promising treatment strategy, which does not restrict viral assembly but heightens MAVS-dependent IFN induction (159). Wang and colleagues designed and synthesized interference peptides NIP I-V with a DRI conformation, which were found to inhibit the formation of the interaction region of SARS-CoV-2 N protein dimer by blocking different DDs. In vitro and in vivo analyses demonstrated that NIP-V could disrupt SARS-CoV-2-N protein LLPS to alleviate the N protein-mediated suppression of the innate antiviral responses. Targeting N-RNA condensation with gallic catechin gallate (GCG) could be a potential treatment for COVID-19. The GCG in green tea polyphenols can interact with the virus N protein, thus inhibiting LLPS, achieving the effect of inhibiting SARS-CoV-2 replication and reducing virus transmission (179). Nanoantibodies are also believed to neutralize SARS-CoV-2 by blocking receptor interactions and LLPS (180, 181).
Post-translational modification (PTM) of LLPS phase-separated molecules is a critical factor affecting LLPS performance as PTMs can change the structure, charge, hydrophobicity, and other properties of phase-separated proteins. Several common PTMs (including phosphorylation, arginine methylation, arginine citrullination, acetylation, ubiquitination, and poly (ADP-ribosylation), could also regulate protein LLPS properties during viral infection, thus targeting PTMs of phase-separated proteins could contribute to anti-viral drug design (6, 20).
Small-molecule modulators of host kinases or phosphatases may regulate LLPS and serve as antiviral agents. Indeed, activators of the SR protein kinase 1 (SRPK1) may represent potential antivirals because the phosphorylation of the N protein SR region of SARS-CoV-2 could attenuate RNA-induced LLPS and viral RNA transcription (182). The RNA-binding protein TDP-43 is a key component of stress granules, and hyper-phosphorylated and ubiquitinated TDP-43 deposits act as endosomes in the brain and spinal cord of patients with motor neuron diseases (182). TDP-43 releases mtDNA into the cytoplasm through mPTP to activate signal transduction of the cGAS-STING pathway (183, 184). It has been demonstrated that cGAS and STING inhibitors can prevent inflammation induced by TDP-43. Acetylated TDP-43 colocalizes with SARS-CoV-2 N protein and simultaneously impairs the ability of TDP-43 to bind RNA (175). Ubiquitination promotes the aggregation of MAVS, and the SARS-CoV-2-N protein undergoes LLPS with RNA, which inhibits Lys63-linked poly-ubiquitination and suppresses the innate antiviral immune response (159). SIRT1-mediated DNA-binding domain (DBD) deacetylation of IRF3/IRF7 has been shown to inhibit LLPS and innate immunity, resulting in increased viral load and mortality in mice (118). Small-molecule inhibitors of protein acetyltransferase, such as C646, could be used to probe LLPS in viral infections (as detailed in Table 2) (185).
In addition to directly targeting the formation of LLPS, it is also possible to alter LLPS environmental factors to regulate the state of the target protein LLPS. The RNA binding domains (RBDs) of SARS-CoV-2 N-protein, RBD1 and RBD2, interact with different dsRNA. The addition of dsRNA reduces the condensation temperature, dependent on the RBD2 interaction, and regulates the inhibition of translation (186). It has been reported that in HPV-infected cells, the early protein 7 (E7) mRNA of the virus is modified by m6A and stabilized by the cell m6A reader IGF2BP1. Heat treatment promotes the aggregation of IGF2BP1 in the presence of m6A-modified E7 mRNA to form different heat-induced m6A E7 mRNA-IGF2BP1 particles, which are decomposed by the ubiquitin-proteasome system, and reverse HPV-related carcinogenesis in vitro and in vivo (187). During continued heat stress, the LLPS characteristics of heat-induced IGF2BP1 condensate suggested a liquid-to-solid phase transition of IGF2BP1 puncta. Several studies have suggested that high temperature and high humidity environments are conducive to reducing the transmission rate of COVID-19 (187). SGs are very sensitive to oxidative stress, osmotic stress, heat shock stress, and other cell stress conditions. FUS protein, a component of stress granules in cells, can form highly reversible amyloid fibrin through LLPS. FUS undergoes LLPS condensation with the SARS-CoV-2 N protein and may be involved in the host-mRNA processing of N; therefore, temperature may be related to some translation mechanisms around SG during stimulation (188). These studies show that controlling environmental factors, such as temperature, is an effective strategy to inhibit viral LLPS and virus replication, thus exploiting LLPS through adjusting the environmental factors might present tremendous potential as an adjuvant systematic therapy.
The application of LLPS during viral replication and host antiviral immunity has been greatly extended in the last several years. Several previous reports have observed viral structures such as IBs, Negri bodies from negative strand RNA viruses, and viral replication compartments from DNA viruses, which were recently regarded to arise from LLPS processes. These membraneless cellular viral factories formed by virus components contribute to viral replication, capsid assembly, virion generation, and immune restriction. In contrast, the host also uses the LLPS system to ensure the effective elimination of invading microorganisms. However, numerous questions remain unanswered regarding viral and host components of LLPS condensates. Further studies are required to investigate the detailed mechanisms and driving forces of the formation of host and viral LLPS condensation, the correlation between host and viral LLPS, the overall properties of host and viral molecular condensates, and the complete composition in viral LLPS MLOs. Considering the lack of a comprehensive understanding of host and viral LLPS condensates during infection, antiviral interventions targeting host and viral LLPS condensates are still at a very early stage. How to effectively restrict viral LLPS to limit viral replication and modulate LLPS of host antiviral components to ensure appropriate immune responses, even the precise control of specific molecules, remains a long-standing challenge in the implementation of phase separation-based therapies.
YW and JC conceived and designed the manuscript. SY, WS and YW drafted the manuscript. JH drew the plots. The manuscript was written by SY, WS and YW. All authors contributed to the article and approved the submitted version.
This research was supported by Guangdong Provincial Key R&D Program for Covid 19 (232020012620600001), National Natural Science Foundation of China (82272186), Natural Science Foundation of Guangdong Province, China (2021A1515012179), and Guangdong Clinical Research Center for Critical Care Medicine (2020B1111170005). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank all the members of Dr. Cui’s lab for the helpful discussion about our study. We thank the contributions and suggestions from Yishan Liu and Jiahui Yang during the proofreading and revision process.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol (2008) 9:112–24. doi: 10.1038/nrm2330
2. Alberti S, Hyman AA. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat Rev Mol Cell Biol (2021) 22:196–213. doi: 10.1038/s41580-020-00326-6
3. Banani SF, Lee HO, Hyman AA, Rosen MK. Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol (2017) 18:285–98. doi: 10.1038/nrm.2017.7
4. Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, et al. Protein phase separation: a new phase in cell biology. Trends Cell Biol (2018) 28:420–35. doi: 10.1016/j.tcb.2018.02.004
5. Lafontaine DLJ, Riback JA, Bascetin R, Brangwynne CP. The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol (2021) 22:165–82. doi: 10.1038/s41580-020-0272-6
6. Shin Y, Brangwynne CP. Liquid phase condensation in cell physiology and disease. Sci (2017) 357(6357):eaaf4382. doi: 10.1126/science.aaf4382
7. Gomes E, Shorter J. The molecular language of membraneless organelles. J Biol Chem (2019) 294:7115–27. doi: 10.1074/jbc.TM118.001192
8. Hyman AA, Weber CA, Julicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol (2014) 30:39–58. doi: 10.1146/annurev-cellbio-100913-013325
9. Sabari BR, Dall'Agnese A, Young RA. Biomolecular condensates in the nucleus. Trends Biochem Sci (2020) 45:961–77. doi: 10.1016/j.tibs.2020.06.007
10. Davis D, Yuan H, Liang FX, Yang YM, Westley J, Petzold C, et al. Human antiviral protein MxA forms novel metastable membraneless cytoplasmic condensates exhibiting rapid reversible tonicity-driven phase transitions. J Virol (2019) 93(22):e01014-19. doi: 10.1128/JVI.01014-19
11. Wang W, Chen J, Yu X, Lan HY. Signaling mechanisms of SARS-CoV-2 nucleocapsid protein in viral infection, cell death and inflammation. Int J Biol Sci (2022) 18:4704–13. doi: 10.7150/ijbs.72663
12. Etibor TA, Yamauchi Y, Amorim MJ. Liquid biomolecular condensates and viral lifecycles: review and perspectives. Viruses (2021) 13(3):366. doi: 10.3390/v13030366
13. Li H, Ernst C, Kolonko-Adamska M, Greb-Markiewicz B, Man J, Parissi V, et al. Phase separation in viral infections. Trends Microbiol (2022) 30:1217–31. doi: 10.1016/j.tim.2022.06.005
14. Nademi S, Dickhout JG. Protein misfolding in endoplasmic reticulum stress with applications to renal diseases. Adv Protein Chem Struct Biol (2019) 118:217–47. doi: 10.1016/bs.apcsb.2019.08.001
15. Banjade S, Rosen MK. Phase transitions of multivalent proteins can promote clustering of membrane receptors. Elife (2014) 3:e04123. doi: 10.7554/eLife.04123
16. Case LB, Ditlev JA, Rosen MK. Regulation of transmembrane signaling by phase separation. Annu Rev Biophys (2019) 48:465–94. doi: 10.1146/annurev-biophys-052118-115534
17. Liao YC, Fernandopulle MS, Wang G, Choi H, Hao L, Drerup CM, et al. RNA Granules hitchhike on lysosomes for long-distance transport, using annexin A11 as a molecular tether. Cell (2019) 179(1):147–164 e20. doi: 10.1016/j.cell.2019.08.050
18. Yamasaki A, Alam JM, Noshiro D, Hirata E, Fujioka Y, Suzuki K, et al. Liquidity is a critical determinant for selective autophagy of protein condensates. Mol Cell (2020) 77:1163–1175 e9. doi: 10.1016/j.molcel.2019.12.026
19. Cougot N, Cavalier A, Thomas D, Gillet R. The dual organization of p-bodies revealed by immunoelectron microscopy and electron tomography. J Mol Biol (2012) 420:17–28. doi: 10.1016/j.jmb.2012.03.027
20. Zhang H, Ji X, Li P, Liu C, Lou J, Wang Z, et al. Liquid-liquid phase separation in biology: mechanisms, physiological functions and human diseases. Sci China Life Sci (2020) 63:953–85. doi: 10.1007/s11427-020-1702-x
21. Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, et al. Germline p granules are liquid droplets that localize by controlled dissolution/condensation. Science (2009) 324:1729–32. doi: 10.1126/science.1172046
22. Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, et al. Phase transitions in the assembly of multivalent signalling proteins. Nature (2012) 483:336–40. doi: 10.1038/nature10879
23. Kato M, McKnight SL. The low-complexity domain of the FUS RNA binding protein self-assembles via the mutually exclusive use of two distinct cross-beta cores. Proc Natl Acad Sci U.S.A. (2021) 118(42):e2114412118. doi: 10.1073/pnas.2114412118
24. Lin YH, Forman-Kay JD, Chan HS. Theories for sequence-dependent phase behaviors of biomolecular condensates. Biochemistry (2018) 57:2499–508. doi: 10.1021/acs.biochem.8b00058
25. Murthy AC, Dignon GL, Kan Y, Zerze GH, Parekh SH, Mittal J, et al. Molecular interactions underlying liquid-liquid phase separation of the FUS low-complexity domain. Nat Struct Mol Biol (2019) 26:637–48. doi: 10.1038/s41594-019-0250-x
26. Wang Z, Zhang H. Phase separation, transition, and autophagic degradation of proteins in development and pathogenesis. Trends Cell Biol (2019) 29:417–27. doi: 10.1016/j.tcb.2019.01.008
27. Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, et al. Compositional control of phase-separated cellular bodies. Cell (2016) 166:651–63. doi: 10.1016/j.cell.2016.06.010
28. Zhou H, Song Z, Zhong S, Zuo L, Qi Z, Qu LJ, et al. Mechanism of DNA-induced phase separation for transcriptional repressor VRN1. Angew Chem Int Ed Engl (2019) 58:4858–62. doi: 10.1002/anie.201810373
29. Choi HJ, Lee JY, Kim K. Glutathionylation on RNA-binding proteins: a regulator of liquid−liquid phase separation in the pathogenesis of amyotrophic lateral sclerosis. Exp Mol Med (2023). doi: 10.1038/s12276-023-00978-2
30. Posey AE, Holehouse AS, Pappu RV. Phase separation of intrinsically disordered proteins. Methods Enzymol (2018) 611:1–30. doi: 10.1016/bs.mie.2018.09.035
31. Ruff KM, Roberts S, Chilkoti A, Pappu RV. Advances in understanding stimulus-responsive phase behavior of intrinsically disordered protein polymers. J Mol Biol (2018) 430:4619–35. doi: 10.1016/j.jmb.2018.06.031
32. Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell (2015) 57:936–47. doi: 10.1016/j.molcel.2015.01.013
33. Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, et al. A liquid-to-Solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell (2015) 162:1066–77. doi: 10.1016/j.cell.2015.07.047
34. Tong X, Tang R, Xu J, Wang W, Zhao Y, Yu X, et al. Liquid-liquid phase separation in tumor biology. Signal Transduct Target Ther (2022) 7:221. doi: 10.1038/s41392-022-01076-x
35. Elbaum-Garfinkle S, Brangwynne CP. Liquids, fibers, and gels: the many phases of neurodegeneration. Dev Cell (2015) 35:531–2. doi: 10.1016/j.devcel.2015.11.014
36. Feng Z, Jia B, Zhang M. Liquid-liquid phase separation in biology: specific stoichiometric molecular interactions vs promiscuous interactions mediated by disordered sequences. Biochemistry (2021) 60:2397–406. doi: 10.1021/acs.biochem.1c00376
37. Li J. Liquid-liquid phase separation in hair cell stereocilia development and maintenance. Comput Struct Biotechnol J (2023) 21:1738–45. doi: 10.1016/j.csbj.2023.02.040
38. Zhang X, Zheng R, Li Z, Ma J. Liquid-liquid phase separation in viral function. J Mol Biol (2023) 13:167955. doi: 10.1016/j.jmb.2023.167955
39. Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, Jahnel M, et al. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell (2018) 174:688–699 e16. doi: 10.1016/j.cell.2018.06.006
40. Du M, Chen ZJ. DNA-Induced liquid phase condensation of cGAS activates innate immune signaling. Science (2018) 361:704–9. doi: 10.1126/science.aat1022
41. Drino A, Schaefer MR. RNAs, phase separation, and membrane-less organelles: are post-transcriptional modifications modulating organelle dynamics? Bioessays (2018) 40:e1800085. doi: 10.1002/bies.201800085
42. Goode BL, Eskin JA, Wendland B. Actin and endocytosis in budding yeast. Genetics (2015) 199:315–58. doi: 10.1534/genetics.112.145540
43. Shan Z, Tu Y, Yang Y, Liu Z, Zeng M, Xu H, et al. Basal condensation of numb and pon complex via phase transition during drosophila neuroblast asymmetric division. Nat Commun (2018) 9:737. doi: 10.1038/s41467-018-03077-3
44. Yang P, Mathieu C, Kolaitis RM, Zhang P, Messing J, Yurtsever U, et al. G3BP1 is a tunable switch that triggers phase separation to assemble stress granules. Cell (2020) 181:325–345 e28. doi: 10.1016/j.cell.2020.03.046
45. Handwerger KE, Cordero JA, Gall JG. Cajal bodies, nucleoli, and speckles in the xenopus oocyte nucleus have a low-density, sponge-like structure. Mol Biol Cell (2005) 16:202–11. doi: 10.1091/mbc.e04-08-0742
46. Zhang L, Geng X, Wang F, Tang J, Ichida Y, Sharma A, et al. 53BP1 regulates heterochromatin through liquid phase separation. Nat Commun (2022) 13:360. doi: 10.1038/s41467-022-28019-y
47. Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell (2005) 16:2544–53. doi: 10.1091/mbc.e04-08-0669
48. Li XM, Wu HL, Xia QD, Zhou P, Wang SG, Yu X, et al. Novel insights into the SPOP E3 ubiquitin ligase: from the regulation of molecular mechanisms to tumorigenesis. BioMed Pharmacother (2022) 149:112882. doi: 10.1016/j.biopha.2022.112882
49. Knudsen C, Gallage NJ, Hansen CC, Moller BL, Laursen T. Dynamic metabolic solutions to the sessile life style of plants. Nat Prod Rep (2018) 35:1140–55. doi: 10.1039/C8NP00037A
50. Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J, et al. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science (2016) 352:595–9. doi: 10.1126/science.aad9964
51. Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell (2015) 163:123–33. doi: 10.1016/j.cell.2015.09.015
52. Sheu-Gruttadauria J, MacRae IJ. Phase transitions in the assembly and function of human miRISC. Cell (2018) 173:946–957 e16. doi: 10.1016/j.cell.2018.02.051
53. Zhu G, Xie J, Kong W, Xie J, Li Y, Du L, et al. Phase separation of disease-associated SHP2 mutants underlies MAPK hyperactivation. Cell (2020) 183:490–502 e18. doi: 10.1016/j.cell.2020.09.002
54. Lu S, Ye Q, Singh D, Cao Y, Diedrich JK, Yates JR, et al. The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated m protein. Nat Commun (2021) 12:502. doi: 10.1038/s41467-020-20768-y
55. Iserman C, Roden CA, Boerneke MA, Sealfon RSG, McLaughlin GA, Jungreis I, et al. Genomic RNA elements drive phase separation of the SARS-CoV-2 nucleocapsid. Mol Cell (2020) 80:1078–1091 e6. doi: 10.1016/j.molcel.2020.11.041
56. Perdikari TM, Murthy AC, Ryan VH, Watters S, Naik MT, Fawzi NL. SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. EMBO J. (2020) 39(24):e106478. doi: 10.15252/embj.2020106478
57. Amarasinghe GK, Ayllón MA, Bào Y, Basler CF, Bavari S, Blasdell KR, et al. Taxonomy of the order mononegavirales: update 2019. Arch Virol (2019) 164:1967–80. doi: 10.1007/s00705-019-04247-4
58. Nikolic J, Le Bars R, Lama Z, Scrima N, Lagaudriere-Gesbert C, Gaudin Y, et al. Negri bodies are viral factories with properties of liquid organelles. Nat Commun (2017) 8:58. doi: 10.1038/s41467-017-00102-9
59. Zhou Y, Su JM, Samuel CE, Ma D. Measles virus forms inclusion bodies with properties of liquid organelles. J Virol (2019) 93(21):e00948-19. doi: 10.1128/JVI.00948-19
60. Galloux M, Risso-Ballester J, Richard CA, Fix J, Rameix-Welti MA, Eleouet JF. Minimal elements required for the formation of respiratory syncytial virus cytoplasmic inclusion bodies In vivo and in vitro. mBio (2020) 11(5):e01202-20. doi: 10.1128/mBio.01202-20
61. Nevers Q, Scrima N, Glon D, Bars R, Decombe A, Garnier N, et al. Properties of rabies virus phosphoprotein and nucleoprotein biocondensates formed in vitro and in cellulo. PloS Pathog (2022) 18:e1011022. doi: 10.1371/journal.ppat.1011022
62. Lahaye X, Vidy A, Pomier C, Obiang L, Harper F, Gaudin Y, et al. Functional characterization of negri bodies (NBs) in rabies virus-infected cells: evidence that NBs are sites of viral transcription and replication. J Virol (2009) 83:7948–58. doi: 10.1128/JVI.00554-09
63. Lahaye X, Vidy A, Fouquet B, Blondel D. Hsp70 protein positively regulates rabies virus infection. J Virol (2012) 86:4743–51. doi: 10.1128/JVI.06501-11
64. Fouquet B, Nikolic J, Larrous F, Bourhy H, Wirblich C, Lagaudrière-Gesbert C, et al. Focal adhesion kinase is involved in rabies virus infection through its interaction with viral phosphoprotein p. J Virol (2015) 89:1640–51. doi: 10.1128/JVI.02602-14
65. Guseva S, Milles S, Jensen MR, Salvi N, Kleman JP, Maurin D, et al. Measles virus nucleo- and phosphoproteins form liquid-like phase-separated compartments that promote nucleocapsid assembly. Sci Adv (2020) 6(14):eaaz7095. doi: 10.1126/sciadv.aaz7095
66. Nakatsu Y, Takeda M, Ohno S, Shirogane Y, Iwasaki M, Yanagi Y. Measles virus circumvents the host interferon response by different actions of the c and V proteins. J Virol (2008) 82:8296–306. doi: 10.1128/JVI.00108-08
67. Ma D, George CX, Nomburg JL, Pfaller CK, Cattaneo R, Samuel CE. Upon infection, cellular WD repeat-containing protein 5 (WDR5) localizes to cytoplasmic inclusion bodies and enhances measles virus replication. J Virol (2018) 92(5):e01726–17. doi: 10.1128/JVI.01726-17
68. Carromeu C, Simabuco FM, Tamura RE, Farinha Arcieri LE, Ventura AM. Intracellular localization of human respiratory syncytial virus l protein. Arch Virol (2007) 152:2259–63. doi: 10.1007/s00705-007-1048-4
69. Weber E, Humbert B, Streckert HJ, Werchau H. Nonstructural protein 2 (NS2) of respiratory syncytial virus (RSV) detected by an antipeptide serum. Respiration (1995) 62:27–33. doi: 10.1159/000196385
70. Ghildyal R, Mills J, Murray M, Vardaxis N, Meanger J. Respiratory syncytial virus matrix protein associates with nucleocapsids in infected cells. J Gen Virol (2002) 83:753–7. doi: 10.1099/0022-1317-83-4-753
71. Rincheval V, Lelek M, Gault E, Bouillier C, Sitterlin D, Blouquit-Laye S, et al. Functional organization of cytoplasmic inclusion bodies in cells infected by respiratory syncytial virus. Nat Commun (2017) 8:563. doi: 10.1038/s41467-017-00655-9
72. Richard CA, Rincheval V, Lassoued S, Fix J, Cardone C, Esneau C, et al. RSV Hijacks cellular protein phosphatase 1 to regulate M2-1 phosphorylation and viral transcription. PloS Pathog (2018) 14:e1006920. doi: 10.1371/journal.ppat.1006920
73. Munday DC, Wu W, Smith N, Fix J, Noton SL, Galloux M, et al. Interactome analysis of the human respiratory syncytial virus RNA polymerase complex identifies protein chaperones as important cofactors that promote l-protein stability and RNA synthesis. J Virol (2015) 89:917–30. doi: 10.1128/JVI.01783-14
74. Radhakrishnan A, Yeo D, Brown G, Myaing MZ, Iyer LR, Fleck R, et al. Protein analysis of purified respiratory syncytial virus particles reveals an important role for heat shock protein 90 in virus particle assembly. Mol Cell Proteomics (2010) 9:1829–48. doi: 10.1074/mcp.M110.001651
75. Kolesnikova L, Mühlberger E, Ryabchikova E, Becker S. Ultrastructural organization of recombinant marburg virus nucleoprotein: comparison with marburg virus inclusions. J Virol (2000) 74:3899–904. doi: 10.1128/JVI.74.8.3899-3904.2000
76. Miyake T, Farley CM, Neubauer BE, Beddow TP, Hoenen T, Engel DA. Ebola Virus inclusion body formation and RNA synthesis are controlled by a novel domain of nucleoprotein interacting with VP35. J Virol (2020) 94(16):e02100-19. doi: 10.1128/JVI.02100-19
77. Biedenkopf N, Lier C, Becker S. Dynamic phosphorylation of VP30 is essential for Ebola virus life cycle. J Virol (2016) 90:4914–25. doi: 10.1128/JVI.03257-15
78. Brandt J, Wendt L, Bodmer BS, Mettenleiter TC, Hoenen T. The cellular protein CAD is recruited into Ebola virus inclusion bodies by the nucleoprotein NP to facilitate genome replication and transcription. Cells (2020) 9(5):1126. doi: 10.3390/cells9051126
79. Wendt L, Brandt J, Bodmer BS, Reiche S, Schmidt ML, Traeger S, et al. The Ebola virus nucleoprotein recruits the nuclear RNA export factor NXF1 into inclusion bodies to facilitate viral protein expression. Cells (2020) 9(1):187. doi: 10.3390/cells9010187
80. Fang J, Pietzsch C, Ramanathan P, Santos RI, Ilinykh PA, Garcia-Blanco MA, et al. Staufen1 interacts with multiple components of the Ebola virus ribonucleoprotein and enhances viral RNA synthesis. mBio (2018) 9(5):e01771–18. doi: 10.1128/mBio.01771-18
81. Chen J, He Z, Yuan Y, Huang F, Luo B, Zhang J, et al. Host factor SMYD3 is recruited by Ebola virus nucleoprotein to facilitate viral mRNA transcription. Emerg Microbes Infect (2019) 8:1347–60. doi: 10.1080/22221751.2019.1662736
82. Luo L, Li Z, Zhao T, Ju X, Ma P, Jin B, et al. SARS-CoV-2 nucleocapsid protein phase separates with G3BPs to disassemble stress granules and facilitate viral production. Sci Bull (Beijing) (2021) 66:1194–204. doi: 10.1016/j.scib.2021.01.013
83. Savastano A, Ibanez de Opakua A, Rankovic M, Zweckstetter M. Nucleocapsid protein of SARS-CoV-2 phase separates into RNA-rich polymerase-containing condensates. Nat Commun (2020) 11:6041. doi: 10.1038/s41467-020-19843-1
84. Wu CH, Chen PJ, Yeh SH. Nucleocapsid phosphorylation and RNA helicase DDX1 recruitment enables coronavirus transition from discontinuous to continuous transcription. Cell Host Microbe (2014) 16:462–72. doi: 10.1016/j.chom.2014.09.009
85. Zheng Y, Deng J, Han L, Zhuang MW, Xu Y, Zhang J, et al. SARS-CoV-2 NSP5 and n protein counteract the RIG-I signaling pathway by suppressing the formation of stress granules. Signal Transduct Target Ther (2022) 7:22. doi: 10.1038/s41392-022-00878-3
86. Vladimirova O, De Leo A, Deng Z, Wiedmer A, Hayden J, Lieberman PM. Phase separation and DAXX redistribution contribute to LANA nuclear body and KSHV genome dynamics during latency and reactivation. PloS Pathog (2021) 17:e1009231. doi: 10.1371/journal.ppat.1009231
87. De Leo A, Deng Z, Vladimirova O, Chen HS, Dheekollu J, Calderon A, et al. LANA oligomeric architecture is essential for KSHV nuclear body formation and viral genome maintenance during latency. PloS Pathog (2019) 15(1):e1007489. doi: 10.1371/journal.ppat.1007489
88. Caragliano E, Bonazza S, Frascaroli G, Tang J, Soh TK, Grunewald K, et al. Human cytomegalovirus forms phase-separated compartments at viral genomes to facilitate viral replication. Cell Rep (2022) 38:110469. doi: 10.1016/j.celrep.2022.110469
89. Seyffert M, Georgi F, Tobler K, Bourqui L, Anfossi M, Michaelsen K, et al. The HSV-1 transcription factor ICP4 confers liquid-like properties to viral replication compartments. Int J Mol Sci (2021) 22(9):4447. doi: 10.3390/ijms22094447
90. Zhou S, Fu Z, Zhang Z, Jia X, Xu G, Sun L, et al. Liquid-liquid phase separation mediates the formation of herpesvirus assembly compartments. J Cell Biol (2023) 222(1):e202201088. doi: 10.1083/jcb.202201088
91. Milles S, Jensen MR, Lazert C, Guseva S, Ivashchenko S, Communie G, et al. An ultraweak interaction in the intrinsically disordered replication machinery is essential for measles virus function. Sci Adv (2018) 4:eaat7778. doi: 10.1126/sciadv.aat7778
92. Bloyet LM, Brunel J, Dosnon M, Hamon V, Erales J, Gruet A, et al. Modulation of re-initiation of measles virus transcription at intergenic regions by PXD to NTAIL binding strength. PloS Pathog (2016) 12:e1006058. doi: 10.1371/journal.ppat.1006058
93. Zhang AP, Bornholdt ZA, Liu T, Abelson DM, Lee DE, Li S, et al. The ebola virus interferon antagonist VP24 directly binds STAT1 and has a novel, pyramidal fold. PloS Pathog (2012) 8:e1002550. doi: 10.1371/journal.ppat.1002550
94. Lifland AW, Jung J, Alonas E, Zurla C, Crowe JE Jr., Santangelo PJ. Human respiratory syncytial virus nucleoprotein and inclusion bodies antagonize the innate immune response mediated by MDA5 and MAVS. J Virol (2012) 86:8245–58. doi: 10.1128/JVI.00215-12
95. Jobe F, Simpson J, Hawes P, Guzman E, Bailey D. Respiratory syncytial virus sequesters NF-kappaB subunit p65 to cytoplasmic inclusion bodies to inhibit innate immune signaling. J Virol (2020) 94(22):e01380-20. doi: 10.1128/JVI.01380-20
96. Fricke J, Koo LY, Brown CR, Collins PL. p38 and OGT sequestration into viral inclusion bodies in cells infected with human respiratory syncytial virus suppresses MK2 activities and stress granule assembly. J Virol (2013) 87:1333–47. doi: 10.1128/JVI.02263-12
97. Alenquer M, Vale-Costa S, Etibor TA, Ferreira F, Sousa AL, Amorim MJ. Influenza a virus ribonucleoproteins form liquid organelles at endoplasmic reticulum exit sites. Nat Commun (2019) 10:1629. doi: 10.1038/s41467-019-09549-4
98. Huang Q, Yu L, Petros AM, Gunasekera A, Liu Z, Xu N, et al. Structure of the n-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry (2004) 43:6059–63. doi: 10.1021/bi036155b
99. Klein S, Cortese M, Winter SL, Wachsmuth-Melm M, Neufeldt CJ, Cerikan B, et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat Commun (2020) 11:5885. doi: 10.1038/s41467-020-19619-7
100. Gatherer D, Depledge DP, Hartley CA, Szpara ML, Vaz PK, Benko M, et al. ICTV virus taxonomy profile: herpesviridae 2021. J Gen Virol (2021) 102(10):001673. doi: 10.1099/jgv.0.001673
101. Schommartz T, Tang J, Brost R, Brune W. Differential requirement of human cytomegalovirus UL112-113 protein isoforms for viral replication. J Virol (2017) 91(17):e00254-17. doi: 10.1128/JVI.00254-17
102. McSwiggen DT, Hansen AS, Teves SS, Marie-Nelly H, Hao Y, Heckert AB, et al. Evidence for DNA-mediated nuclear compartmentalization distinct from phase separation. Elife (2019) 8:e47098. doi: 10.7554/eLife.47098
103. Caragliano E, Brune W, Bosse JB. Herpesvirus replication compartments: dynamic biomolecular condensates? Viruses (2022) 14(5):960. doi: 10.3390/v14050960
104. Mettenleiter TC, Klupp BG, Granzow H. Herpesvirus assembly: a tale of two membranes. Curr Opin Microbiol (2006) 9:423–9. doi: 10.1016/j.mib.2006.06.013
105. White S, Kawano H, Harata NC, Roller RJ. Herpes simplex virus organizes cytoplasmic membranes to form a viral assembly center in neuronal cells. J Virol (2020) 94(19):e00900-20. doi: 10.1128/JVI.00900-20
106. Sanchez V, Greis KD, Sztul E, Britt WJ. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J Virol (2000) 74:975–86. doi: 10.1128/JVI.74.2.975-986.2000
107. Zhou S, Fu Z, Zhang Z, Jia X, Xu G, Sun L, et al. Liquid–liquid phase separation mediates the formation of herpesvirus assembly compartments. J Cell Biol (2023) 222(1):e202201088. doi: 10.1083/jcb.202201088
108. Patel JR, Garcia-Sastre A. Activation and regulation of pathogen sensor RIG-I. Cytokine Growth Factor Rev (2014) 25:513–23. doi: 10.1016/j.cytogfr.2014.08.005
109. Sparrer KM, Gack MU. Intracellular detection of viral nucleic acids. Curr Opin Microbiol (2015) 26:1–9. doi: 10.1016/j.mib.2015.03.001
110. Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity (2010) 32:305–15. doi: 10.1016/j.immuni.2010.03.012
111. Taguchi T, Mukai K. Innate immunity signalling and membrane trafficking. Curr Opin Cell Biol (2019) 59:1–7. doi: 10.1016/j.ceb.2019.02.002
112. Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol (2004) 5:730–7. doi: 10.1038/ni1087
113. Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature (2008) 455:674–8. doi: 10.1038/nature07317
114. Motwani M, Pesiridis S, Fitzgerald KA. DNA Sensing by the cGAS-STING pathway in health and disease. Nat Rev Genet (2019) 20:657–74. doi: 10.1038/s41576-019-0151-1
115. Thoresen DT, Galls D, Gotte B, Wang W, Pyle AM. A rapid RIG-I signaling relay mediates efficient antiviral response. Mol Cell (2023) 83:90–104 e4. doi: 10.1016/j.molcel.2022.11.018
116. Thoresen D, Wang W, Galls D, Guo R, Xu L, Pyle AM. The molecular mechanism of RIG-I activation and signaling. Immunol Rev (2021) 304:154–68. doi: 10.1111/imr.13022
117. Haubrich K. RNA Binding regulates TRIM25-mediated RIG-I ubiquitylation. Preprint at bioRxiv (2020).
118. Qin Z, Fang X, Sun W, Ma Z, Dai T, Wang S, et al. Deactylation by SIRT1 enables liquid-liquid phase separation of IRF3/IRF7 in innate antiviral immunity. Nat Immunol (2022) 23:1193–207. doi: 10.1038/s41590-022-01269-0
119. Gao L, Liu R, Yang F, Li X, Liu C, Qi X, et al. Duck enteritis virus inhibits the cGAS-STING DNA-sensing pathway to evade the innate immune response. J Virol (2022) 96:e0157822. doi: 10.1128/jvi.01578-22
120. Xiao TS, Fitzgerald KA. The cGAS-STING pathway for DNA sensing. Mol Cell (2013) 51:135–9. doi: 10.1016/j.molcel.2013.07.004
121. Chen Q, Sun L, Chen ZJ. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat Immunol (2016) 17:1142–9. doi: 10.1038/ni.3558
122. Mehta S, Zhang J. Liquid-liquid phase separation drives cellular function and dysfunction in cancer. Nat Rev Cancer (2022) 22:239–52. doi: 10.1038/s41568-022-00444-7
123. Zhao M, Xia T, Xing JQ, Yin LH, Li XW, Pan J, et al. The stress granule protein G3BP1 promotes pre-condensation of cGAS to allow rapid responses to DNA. EMBO Rep (2022) 23(1):e53166. doi: 10.15252/embr.202153166
124. Zhou W, Mohr L, Maciejowski J, Kranzusch PJ. cGAS phase separation inhibits TREX1-mediated DNA degradation and enhances cytosolic DNA sensing. Mol Cell (2021) 81:739–755 e7. doi: 10.1016/j.molcel.2021.01.024
125. Ablasser A, Hemmerling I, Schmid-Burgk JL, Behrendt R, Roers A, Hornung V. TREX1 deficiency triggers cell-autonomous immunity in a cGAS-dependent manner. J Immunol (2014) 192:5993–7. doi: 10.4049/jimmunol.1400737
126. Wang L, Li S, Wang K, Wang N, Liu Q, Sun Z, et al. Spermine enhances antiviral and anticancer responses by stabilizing DNA binding with the DNA sensor cGAS. Immunity (2023) 56(2):272–288.e7. doi: 10.1016/j.immuni.2023.01.001
127. Yu X, Zhang L, Shen J, Zhai Y, Jiang Q, Yi M, et al. The STING phase-separator suppresses innate immune signalling. Nat Cell Biol (2021) 23:330–40. doi: 10.1038/s41556-021-00659-0
128. Meng F, Yu Z, Zhang D, Chen S, Guan H, Zhou R, et al. Induced phase separation of mutant NF2 imprisons the cGAS-STING machinery to abrogate antitumor immunity. Mol Cell (2021) 81:4147–4164 e7. doi: 10.1016/j.molcel.2021.07.040
129. Zheng W, Xu Q, Zhang Y, Gao XE,W, Zhang M, Zhai W, et al. Toll-like receptor-mediated innate immunity against herpesviridae infection: a current perspective on viral infection signaling pathways. Virol J (2020) 17:192. doi: 10.1186/s12985-020-01463-2
130. Kishida S, Sanjo H, Akira S, Matsumoto K, Ninomiya-Tsuji J. TAK1-binding protein 2 facilitates ubiquitination of TRAF6 and assembly of TRAF6 with IKK in the IL-1 signaling pathway. Genes Cells (2005) 10:447–54. doi: 10.1111/j.1365-2443.2005.00852.x
131. Lester SN, Li K. Toll-like receptors in antiviral innate immunity. J Mol Biol (2014) 426:1246–64. doi: 10.1016/j.jmb.2013.11.024
132. Santoro MG, Rossi A, Amici C. NF-kappaB and virus infection: who controls whom. EMBO J (2003) 22:2552–60. doi: 10.1093/emboj/cdg267
133. Wu P, Liu Y, Li X, Gu Y, Liu Y, Hu Y, et al. The regulation of the disease resistance, mTOR and NF-kB signaling pathway of aristichthys nobilis using rhodopseudomonas wastewater treatment. Dev Comp Immunol (2020) 104:103517. doi: 10.1016/j.dci.2019.103517
134. Almowallad S, Alqahtani LS, Mobashir M. NF-kB in signaling patterns and its temporal dynamics Encode/Decode human diseases. Life (Basel) (2022) 12(12):2012. doi: 10.3390/life12122012
135. Schlein LJ, Thamm DH. Review: NF-kB activation in canine cancer. Vet Pathol (2022) 59:724–32. doi: 10.1177/03009858221092017
136. Gilmore TD, NF-kappa B. KBF1, dorsal, and related matters. Cell (1990) 62:841–3. doi: 10.1016/0092-8674(90)90257-F
137. Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell (2006) 22:245–57. doi: 10.1016/j.molcel.2006.03.026
138. Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected]. Nat Cell Biol (2006) 8:398–406. doi: 10.1038/ncb1384
139. Du M, Ea CK, Fang Y, Chen ZJ. Liquid phase separation of NEMO induced by polyubiquitin chains activates NF-kappaB. Mol Cell (2022) 82:2415–2426 e5. doi: 10.1016/j.molcel.2022.03.037
140. Haller O, Staeheli P, Schwemmle M, Kochs G. Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends Microbiol (2015) 23:154–63. doi: 10.1016/j.tim.2014.12.003
141. Ganser-Pornillos BK, Pornillos O. Restriction of HIV-1 and other retroviruses by TRIM5. Nat Rev Microbiol (2019) 17:546–56. doi: 10.1038/s41579-019-0225-2
142. Tozawa T, Matsunaga K, Izumi T, Shigehisa N, Uekita T, Taoka M, et al. Ubiquitination-coupled liquid phase separation regulates the accumulation of the TRIM family of ubiquitin ligases into cytoplasmic bodies. PloS One (2022) 17:e0272700. doi: 10.1371/journal.pone.0272700
143. Chen Y, Lei X, Jiang Z, Fitzgerald KA. Cellular nucleic acid-binding protein is essential for type I interferon-mediated immunity to RNA virus infection. Proc Natl Acad Sci U.S.A. (2021) 118(26):e2100383118. doi: 10.1073/pnas.2100383118
144. Fitzgerald K, Chen Y, Lei X, Jiang Z, Humphries F, Mustone N, et al. CNBP restricts SARS-CoV2 by regulating IFN and disrupting RNA-protein condensates. Res Sq (2022) rs.3.rs-1576788. doi: 10.21203/rs.3.rs-1576788/v1
145. Yang W, Ru Y, Ren J, Bai J, Wei J, Fu S, et al. G3BP1 inhibits RNA virus replication by positively regulating RIG-i-mediated cellular antiviral response. Cell Death Dis (2019) 10:946. doi: 10.1038/s41419-019-2178-9
146. Kim SS, Sze L, Liu C, Lam KP. The stress granule protein G3BP1 binds viral dsRNA and RIG-I to enhance interferon-beta response. J Biol Chem (2019) 294:6430–8. doi: 10.1074/jbc.RA118.005868
147. Anand PK, Malireddi RK, Lukens JR, Vogel P, Bertin J, Lamkanfi M, et al. NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature (2012) 488:389–93. doi: 10.1038/nature11250
148. Shen C, Li R, Negro R, Cheng J, Vora SM, Fu TM, et al. Phase separation drives RNA virus-induced activation of the NLRP6 inflammasome. Cell (2021) 184:5759–5774 e20. doi: 10.1016/j.cell.2021.09.032
149. Wang Y, Wang Y, Gu J, Su T, Gu X, Feng Y. The role of RNA m6A methylation in lipid metabolism. Front Endocrinol (Lausanne) (2022) 13:866116. doi: 10.3389/fendo.2022.866116
150. Kisan A, Chhabra R. Modulation of gene expression by YTH domain family (YTHDF) proteins in human physiology and pathology. J Cell Physiol (2023) 238(1):5–31. doi: 10.1002/jcp.30907
151. Winkler R, Gillis E, Lasman L, Safra M, Geula S, Soyris C, et al. m(6)A modification controls the innate immune response to infection by targeting type I interferons. Nat Immunol (2019) 20(2):173–82. doi: 10.1038/s41590-018-0275-z
152. Burgess HM, Depledge DP, Thompson L, Srinivas KP, Grande RC, Vink EI, et al. Targeting the m(6)A RNA modification pathway blocks SARS-CoV-2 and HCoV-OC43 replication. Genes Dev (2021) 35(13-14):1005–19. doi: 10.1101/gad.348320.121
153. Zaccara S, Jaffrey SR. A unified model for the function of YTHDF proteins in regulating m(6)A-modified mRNA. Cell (2020) 181(7):1582–1595 e18. doi: 10.1016/j.cell.2020.05.012
154. Balacco DL, Soller M. The m(6)A writer: rise of a machine for growing tasks. Biochemistry (2019) 58(5):363–78. doi: 10.1021/acs.biochem.8b01166
155. Zhao L, Zhao Y, Liu Q, Huang J, Lu Y, Ping J. DDX5/METTL3-METTL14/YTHDF2 axis regulates replication of influenza a virus. Microbiol Spectr (2022) 10(3):e0109822. doi: 10.1128/spectrum.01098-22
156. Zhang Y, Ma Z, Wang Y, Boyer J, Ni G, Cheng L, et al. Streptavidin promotes DNA binding and activation of cGAS to enhance innate immunity. iScience (2020) 23:101463. doi: 10.1016/j.isci.2020.101463
157. Li S, Luo M, Wang Z, Feng Q, Wilhelm J, Wang X, et al. Prolonged activation of innate immune pathways by a polyvalent STING agonist. Nat BioMed Eng (2021) 5:455–66. doi: 10.1038/s41551-020-00675-9
158. Sato Y, Terawaki S, Oikawa D, Shimizu K, Okina Y, Ito H, et al. Involvement of heterologous ubiquitination including linear ubiquitination in alzheimer's disease and amyotrophic lateral sclerosis. Front Mol Biosci (2023) 10:1089213. doi: 10.3389/fmolb.2023.1089213
159. Wang S, Dai T, Qin Z, Pan T, Chu F, Lou L, et al. Targeting liquid-liquid phase separation of SARS-CoV-2 nucleocapsid protein promotes innate antiviral immunity by elevating MAVS activity. Nat Cell Biol (2021) 23:718–32. doi: 10.1038/s41556-021-00710-0
160. Jang M, Cai L, Udeani GO, Slowing KV, Thomas CF, Beecher CW, et al. Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science (1997) 275:218–20. doi: 10.1126/science.275.5297.218
161. Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature (2007) 450:712–6. doi: 10.1038/nature06261
162. Nelson EV, Schmidt KM, Deflubé LR, Doğanay S, Banadyga L, Olejnik J, et al. Ebola Virus does not induce stress granule formation during infection and sequesters stress granule proteins within viral inclusions. J Virol (2016) 90:7268–84. doi: 10.1128/JVI.00459-16
163. Xu G, Liu C, Zhou S, Li Q, Feng Y, Sun P, et al. Viral tegument proteins restrict cGAS-DNA phase separation to mediate immune evasion. Mol Cell (2021) 81:2823–2837.e9. doi: 10.1016/j.molcel.2021.05.002
164. Hertzog J, Zhou W, Fowler G, Rigby RE, Bridgeman A, Blest HT, et al. Varicella-zoster virus ORF9 is an antagonist of the DNA sensor cGAS. EMBO J (2022) 41:e109217. doi: 10.15252/embj.2021109217
165. Jobe F, Simpson J, Hawes P, Guzman E, Bailey D. Respiratory syncytial virus sequesters NF-κB subunit p65 to cytoplasmic inclusion bodies to inhibit innate immune signaling. J Virol (2020) 94(22):e01380–20. doi: 10.1128/JVI.01380-20
166. Vidy A, Chelbi-Alix M, Blondel D. Rabies virus p protein interacts with STAT1 and inhibits interferon signal transduction pathways. J Virol (2005) 79:14411–20. doi: 10.1128/JVI.79.22.14411-14420.2005
167. Brzózka K, Finke S, Conzelmann KK. Inhibition of interferon signaling by rabies virus phosphoprotein p: activation-dependent binding of STAT1 and STAT2. J Virol (2006) 80:2675–83. doi: 10.1128/JVI.80.6.2675-2683.2006
168. Hong Y, Bai M, Qi X, Li C, Liang M, Li D, et al. Suppression of the IFN-α and -β induction through sequestering IRF7 into viral inclusion bodies by nonstructural protein NSs in severe fever with thrombocytopenia syndrome bunyavirus infection. J Immunol (2019) 202:841–56. doi: 10.4049/jimmunol.1800576
169. Ning YJ, Wang M, Deng M, Shen S, Liu W, Cao WC, et al. Viral suppression of innate immunity via spatial isolation of TBK1/IKKϵ from mitochondrial antiviral platform. J Mol Cell Biol (2014) 6:324–37. doi: 10.1093/jmcb/mju015
170. Bhowmik D, Du M, Tian Y, Ma S, Wu J, Chen Z, et al. Cooperative DNA binding mediated by KicGAS/ORF52 oligomerization allows inhibition of DNA-induced phase separation and activation of cGAS. Nucleic Acids Res (2021) 49:9389–403. doi: 10.1093/nar/gkab689
171. Ohn T, Anderson P. The role of posttranslational modifications in the assembly of stress granules. Wiley Interdiscip Rev RNA (2010) 1:486–93. doi: 10.1002/wrna.23
172. Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol (2008) 10:1224–31. doi: 10.1038/ncb1783
173. Wu Y, Ma L, Cai S, Zhuang Z, Zhao Z, Jin S, et al. RNA-Induced liquid phase separation of SARS-CoV-2 nucleocapsid protein facilitates NF-kappaB hyper-activation and inflammation. Signal Transduct Target Ther (2021) 6:167. doi: 10.1038/s41392-021-00575-7
174. Descamps D, Peres de Oliveira A, Gonnin L, Madrieres S, Fix J, Drajac C, et al. Depletion of TAX1BP1 amplifies innate immune responses during respiratory syncytial virus infection. J Virol (2021) 95:e0091221. doi: 10.1128/JVI.00912-21
175. Perdikari TM, Murthy AC, Ryan VH, Watters S, Naik MT, Fawzi NL. SARS-CoV-2 nucleocapsid protein phase-separates with RNA and with human hnRNPs. EMBO J (2020) 39:e106478. doi: 10.15252/embj.2020106478
176. Wang Q, Sundar IK, Li D, Lucas JH, Muthumalage T, McDonough SR, et al. E-cigarette-induced pulmonary inflammation and dysregulated repair are mediated by nAChR alpha7 receptor: role of nAChR alpha7 in SARS-CoV-2 covid-19 ACE2 receptor regulation. Respir Res (2020) 21:154. doi: 10.1186/s12931-020-01396-y
177. Abdullah AA, Abdullah R, Nazariah ZA, Balakrishnan KN, Abdullah FFJ, Bala JA, et al. Cyclophilin a as a target in the treatment of cytomegalovirus infections. Antivir Chem Chemother (2018) 26:2040206618811413. doi: 10.1177/2040206618811413
178. Liao Y, Luo D, Peng K, Zeng Y. Cyclophilin a: a key player for etiological agent infection. Appl Microbiol Biotechnol (2021) 105:1365–77. doi: 10.1007/s00253-021-11115-2
179. Zhao M, Yu Y, Sun LM, Xing JQ, Li T, Zhu Y, et al. GCG inhibits SARS-CoV-2 replication by disrupting the liquid phase condensation of its nucleocapsid protein. Nat Commun (2021) 12:2114. doi: 10.1038/s41467-021-22297-8
180. Pymm P, Adair A, Chan LJ, Cooney JP, Mordant FL, Allison CC, et al. Nanobody cocktails potently neutralize SARS-CoV-2 D614G N501Y variant and protect mice. Proc Natl Acad Sci U.S.A. (2021) 118(19):e2101918118. doi: 10.1073/pnas.2101918118
181. El-Sherif DM, Abouzid M, Gaballah MS, Ahmed AA, Adeel M, Sheta SM. New approach in SARS-CoV-2 surveillance using biosensor technology: a review. Environ Sci pollut Res Int (2022) 29:1677–95. doi: 10.1007/s11356-021-17096-z
182. Yaron TM, Heaton BE, Levy TM, Johnson JL, Jordan TX, Cohen BM, et al. Host protein kinases required for SARS-CoV-2 nucleocapsid phosphorylation and viral replication. Sci Signal (2022) 15:eabm0808. doi: 10.1126/scisignal.abm0808
183. Yu CH, Davidson S, Harapas CR, Hilton JB, Mlodzianoski MJ, Laohamonthonkul P, et al. TDP-43 triggers mitochondrial DNA release via mPTP to activate cGAS/STING in ALS. Cell (2020) 183:636–649 e18. doi: 10.1016/j.cell.2020.09.020
184. Lee JD, Woodruff TM. TDP-43 puts the STING in ALS. Trends Neurosci (2021) 44:81–2. doi: 10.1016/j.tins.2020.12.001
185. Peng J, Ramatchandirin B, Wang Y, Pearah A, Namachivayam K, Wolf RM, et al. The P300 acetyltransferase inhibitor C646 promotes membrane translocation of insulin receptor protein substrate and interaction with the insulin receptor. J Biol Chem (2022) 298:101621. doi: 10.1016/j.jbc.2022.101621
186. Lupala CS, Ye Y, Chen H, Su XD, Liu H. Mutations on RBD of SARS-CoV-2 omicron variant result in stronger binding to human ACE2 receptor. Biochem Biophys Res Commun (2022) 590:34–41. doi: 10.1016/j.bbrc.2021.12.079
187. Wang L, Zhan G, Maimaitiyiming Y, Su Y, Lin S, Liu J, et al. m(6)A modification confers thermal vulnerability to HPV E7 oncotranscripts via reverse regulation of its reader protein IGF2BP1 upon heat stress. Cell Rep (2022) 41:111546. doi: 10.1016/j.celrep.2022.111546
Keywords: LLPS, virus replicaiton, innate immunity, immune evasion, Viral infection disease
Citation: Yang S, Shen W, Hu J, Cai S, Zhang C, Jin S, Guan X, Wu J, Wu Y and Cui J (2023) Molecular mechanisms and cellular functions of liquid-liquid phase separation during antiviral immune responses. Front. Immunol. 14:1162211. doi: 10.3389/fimmu.2023.1162211
Received: 09 February 2023; Accepted: 25 April 2023;
Published: 10 May 2023.
Edited by:
Cao-Qi Lei, Wuhan University, ChinaReviewed by:
Yong Du, Houston Methodist Research Institute, United StatesCopyright © 2023 Yang, Shen, Hu, Cai, Zhang, Jin, Guan, Wu, Wu and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yaoxing Wu, d3V5YW94NUBtYWlsLnN5c3UuZWR1LmNu; Xiangdong Guan, Z3VhbnhkQG1haWwuc3lzdS5lZHUuY24=; Jianfeng Wu, d3VqaWFuZkBtYWlsLnN5c3UuZWR1LmNu; Jun Cui, Y3VpajVAbWFpbC5zeXN1LmVkdS5jbg==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.