
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol., 03 May 2023
Sec. Comparative Immunology
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1135588
This article is part of the Research TopicThe Epidemiologic Triads in Aquaculture: Host, Pathogen and EnvironmentView all 7 articles
A correction has been applied to this article in:
Corrigendum: Edwardsiella piscicida infection reshapes the intestinal microbiome and metabolome of big-belly seahorses: mechanistic insights of synergistic actions of virulence factors
 Lele Zhang1,2†
Lele Zhang1,2† Fang Wang3†
Fang Wang3† Longwu Jia1,2
Longwu Jia1,2 Hansheng Yan1,2
Hansheng Yan1,2 Longkun Gao1,2
Longkun Gao1,2 Yanan Tian1,2
Yanan Tian1,2 Xiaolei Su1,2
Xiaolei Su1,2 Xu Zhang1,2
Xu Zhang1,2 Chunhui Lv1,2
Chunhui Lv1,2 Zhenhao Ma1,2
Zhenhao Ma1,2 Yuanyuan Xue1,2
Yuanyuan Xue1,2 Qiang Lin4
Qiang Lin4 Kai Wang1,2*
Kai Wang1,2*Uncovering the mechanism underlying the pathogenesis of Edwardsiella piscicida-induced enteritis is essential for global aquaculture. In the present study, we identified E. piscicida as a lethal pathogen of the big-belly seahorse (Hippocampus abdominalis) and revealed its pathogenic pattern and characteristics by updating our established bacterial enteritis model and evaluation system. Conjoint analysis of metagenomic and metabolomic data showed that 15 core virulence factors could mutually coordinate the remodeling of intestinal microorganisms and host metabolism and induce enteritis in the big-belly seahorse. Specifically, the Flagella, Type IV pili, and Lap could significantly increase the activities of the representative functional pathways of both flagella assembly and bacterial chemotaxis in the intestinal microbiota (P < 0.01) to promote pathogen motility, adherence, and invasion. Legiobactin, IraAB, and Hpt could increase ABC transporter activity (P < 0.01) to compete for host nutrition and promote self-replication. Capsule1, HP-NAP, and FarAB could help the pathogen to avoid phagocytosis. Upon entering epithelial cells and phagocytes, Bsa T3SS and Dot/Icm could significantly increase bacterial secretion system activity (P < 0.01) to promote the intracellular survival and replication of the pathogen and the subsequent invasion of the neighboring tissues. Finally, LPS3 could significantly increase lipopolysaccharide biosynthesis (P < 0.01) to release toxins and kill the host. Throughout the pathogenic process, BopD, PhoP, and BfmRS significantly activated the two-component system (P < 0.01) to coordinate with other VFs to promote deep invasion. In addition, the levels of seven key metabolic biomarkers, Taurine, L-Proline, Uridine, L-Glutamate, Glutathione, Xanthosine, and L-Malic acid, significantly decreased (P < 0.01), and they can be used for characterizing E. piscicida infection. Overall, the present study systematically revealed how a combination of virulence factors mediate E. piscicida-induced enteritis in fish for the first time, providing a theoretical reference for preventing and controlling this disease in the aquaculture of seahorses and other fishes.
Edwardsiella spp. are the most common and serious gram-negative zoonotic pathogens affecting humans and animals worldwide (1, 2); in particular, they cause more than 90% of fish deaths in aquaculture (3). Edwardsiella piscicida, an emerging and important intestinal pathogen in fish (1), causes substantial biomass and economic losses in aquaculture globally (1), seriously threatening the healthy development of fisheries. As E. piscicida possesses virulence factors (VFs) similar to many intestinal pathogens, such as Flagella, adherence, invasion, type III and type VI secretion systems (T3SS and T6SS), quorum sensing (QS) regulators, iron uptake, two-component systems (TCS), toxins, and invasion proteins (1–3), it could be used as an attractive model organism to study the role of a single VF or a combination of VFs in pathogenic process (1–3). Unfortunately, current researches have mainly focused on revealing the functional mechanisms of single VFs in E. piscicida-induced diseases in mammals (1, 4, 5); the related research in fish is limited. Therefore, exploring the potential mechanisms underlying the mutual interaction of VFs to induce deep infection may not only enrich the knowledge of E. piscicida-induced enteritis in fish but also provide theoretical references for development of optimal strategies for the control and prevention of such diseases.
More than one billion microorganisms colonizing the intestinal tract constitute the first barrier against pathogen invasion (6, 7) and are the driving forces of host health (8). As the most dominant and important part of the intestinal microorganisms, bacterial microbiota can act mutually with the immune system and metabolic function of the host (9, 10), as well as the VFs and antibiotic resistance ontologies (AROs) of opportunistic pathogens (11, 12), to maintain intestinal homeostasis (13). However, pathogenic infections can disrupt such homeostasis and cause diseases (11, 14, 15). Multi-omics analyses have been used to reveal pathogenesis mechanisms from the perspective of pathogen-intestinal microbiota-host metabolism in humans and mice (11, 16, 17). Recently, more researches have focused on exploring the relationship among intestinal microbiota, metabolites, and fish diseases by a single-omics approach (14, 18, 19). However, research on multi-omics applications in fish diseases (20) are scant, limiting the characterization of the effects and understanding of mechanisms of intestinal pathogens from a broader molecular level perspective.
As flagship species of the marine ecological environment (21), a drastic decline of the seahorse (Hippocampus spp.) population has been observed in the past decade (22). Therefore, establishment of seahorse aquaculture is recommended to provide an alternative source of seahorses (23). Owing to the unique characteristics such as simple intestinal structure and lack of gut-associated lymphatic tissue (24), seahorses easily succumb to bacterial enteritis under crowded farming conditions, causing considerable economic losses (25, 26). To date, over 20 bacterial pathogens, mainly Vibrio spp., have been identified, the majority of which can cause enteritis in seahorses (25–28). However, the lack of research on the pathogenesis of bacterial enteritis in seahorses restricts the establishment of effective measures to control and prevent such disease in seahorse aquaculture. In our previous study, we found that E. tarda could induce lethal enteritis in farmed lined seahorses (H. erectus) and established an experimental model to evaluate the pattern and rate of disease progression for the first time (25). Whether such a model is suitable for other seahorses and pathogenic species requires further validation. Our recent research found that E. piscicida could also induce lethal enteritis in farmed big-belly seahorses (H. abdominalis), posing a major threat to the aquaculture of this species (25). Uncover the pathogenesis of E. piscicida induced enteritis in seahorses will be meaningful for the healthy development of the aquaculture industry.
In the present study, we aimed to determine the pathogenic characteristics of E. piscicida-induced enteritis in the big-belly seahorses. We enriched and reassessed the current research model and its evaluation system, identified key VFs and AROs, explored their effects on intestinal microbiota and functions through metagenomic analysis, determined the changes in host metabolism, identified key metabolic biomarkers (KMBs) through metabolomic analysis, and illustrated the potential mechanism underlying E. piscicida-induced enteritis via conjoint analysis. We believe that this study will help enrich our knowledge of Edwardsiella-induced enteritis and develop appropriate prevention and control strategies for fish pathogens in aquaculture.
Big-belly seahorses were maintained and treated in accordance with the guidelines of Animal Ethics Experimentation approved by the Animal Care and Use Committee of Ludong University (document number: LDU-RB20210308NXY-9). Healthy big-belly seahorses were collected from the Wendeng Seahorse Center of Ludong University, Yantai 264025, Shandong Province, China. Male and female seahorses were maintained in ponds connected to a central circulation system with mechanical and biological filtration, ultraviolet sterilization, and a protein skimmer that continuously aerates water (salinity: 31.5 ± 0.5‰, temperature: 19 ± 0.5°C, pH: 8.2 ± 0.1) at Ludong University for 2 weeks before the experiments. Plastic plants were used as holdfasts. The seahorses were fed three times per day (08:00, 12:00, and 16:00) with frozen Mysis, and residual feed and feces were siphoned out 2 h after each feeding session.
To construct the research model, seven groups were set up (20 male and 20 female seahorses per group) to determine the appropriate combination of seahorse size (wet weight, g) and challenge dose of E. piscicida (cfu/mL) via intraperitoneal (IP) injection (14). In detail, Con represents the combination of 2.5–3.0 g and physiological saline solution, and EPs represent the E. piscicida challenge groups with the combinations 3.5–4.0 g and 1 × 105 cfu/mL, 3.5–4.0 g and 1 × 107 cfu/mL, 3.5–4.0 g and 1 × 109 cfu/mL, 4.1–4.5 g and 1 × 105 cfu/mL, and 4.1–4.5 g and 1 × 107 cfu/mL. After the IP injection of E. piscicida, the male and female seahorses were separately maintained in tanks (50 × 40 × 40 cm3) under the same culture conditions mentioned above, except that the circulating water was turned off. After 24 h, the seahorses were fed normally, feces were siphoned out, and seawater was supplemented to maintain normal water levels. The number of deaths was recorded daily to calculate survival rate, and the seahorses were switched to new tanks every week for 21 days. The appropriate combination for research model construction was determined according to the survival rate and pathological characteristics of the seahorses.
Using the optimal combination of seahorse size and challenge dose, 200 big-belly seahorses (male: female = 1:1) were equally separated into two groups (Con and EP groups) with 50 males and 50 females per group, and a new research model was constructed. Both growth-related and pathological parameters of the big-belly seahorses listed in the established evaluation system were recorded on days 0, 1, 5, 9, 15, and 21 as previously reported (25). In addition, the respiratory rate (RR) of randomly chosen seahorses per group was recorded daily. After anesthetization with 0.035% MS-222 (Sigma-Aldrich, Saint Louis, Missouri, USA) for 2 min, intestinal samples were collected on days 0, 1, 5, 9, 15, and 21 for further histological and quantitative real-time polymerase chain reaction (q-PCR) analysis. After 21 d, the pathological characteristics of E. piscicida-induced enteritis in big-belly seahorses were examined.
The established evaluation system was updated by redefining the scoring range and supplementing the scoring system with new parameters according to our previously established principles (25). Disease activity index (DAI) was determined to reveal the pattern and key pathogenic time points of E. piscicida-induced enteritis in big-belly seahorses. To further reveal the pathogenesis of E. piscicida-induced enteritis in big-belly seahorses, 60 seahorses (male:female = 1:1) were divided into two groups (Con: 10 males and 10 females; EP: 20 males and 20 females). After repeating the model construction steps, intestinal samples were collected at key pathogenic time points for integrated metagenomics and metabolomics analyses.
The intestine samples randomly collected from the seahorses in the Con and EP groups (n = 3) were fixed in Bouin’s solution. Within 24 h, the tissues were dehydrated in an alcohol-xylene series and embedded in paraffin. The samples from different intestinal segments were cut into 8-μm-thick sections and stained with hematoxylin and eosin (Beyotime, Tianjin, Hebei, China) (25).The sections were then examined under a light microscope (BX53; Olympus, Tokyo, Japan) at 400× magnification to visualize the pathogenic changes in the intestine.
Intestine samples (n = 5) randomly collected from the Con and EP groups on days 0, 1, 5, 9, 15, and 21 were used to extract total RNA using Trizol Reagent (9109, Takara, USA). cDNA was synthesized using the PrimeScript RT Reagent cDNA Amplification Kit with a gDNA Eraser (RR047A; Takara, USA). q-PCR analysis was performed to evaluate gene expression using a SYBR® Premix Ex Taq™ (Takara, Dalian, China) on a Bio-Rad CFX96 Touch machine (Bio-Rad, USA) according to a previously reported method (29). The sequences of the primers used are listed in Table S1. The relevant levels of target genes were determined using the 2−ΔΔCt method (30).
Fifteen seahorses from each group were randomly selected, and the frequency of their gill cover movements per minute was recorded to calculate the RR (31).
To eliminate the potential sex-related differences and obtain better matching results of metagenomic and metabolomic sequencing analyses, eight intestinal samples per group, one male and one female seahorses, were randomly collected and thoroughly ground in liquid nitrogen. Half of each sample was used for metagenomic sequencing, and the other half was used for metabolomic sequencing.
To reveal the effects of E. piscicida infection on the composition, diversity, structure, function, VF, and ARO of intestinal microbiota, four samples from each group were randomly selected for metagenomic sequencing, and a paired-end sequencing approach was used to obtain raw data (32). Thereafter, data quality control, removal of host genome reads, metagenomic assembly, encoding gene prediction, and non-redundancy gene set construction were performed. Detailed information regarding these steps can be found in the Supplementary Materials and Methods. The raw sequencing data of metagenome in this study are available in the Sequence Read Archive database of National Center for Biotechnology Information (NCBI) under accession number PRJNA916642.
To determine the effects of E. piscicida infection on host metabolism, liquid chromatography-mass spectrometry (LC-MS) analysis of all eight samples from each group was used for untargeted metabolomic sequencing. Chromatographic separation was performed on a Waters UPLC Acquity I-Class PLUS (Waters Corp., Milford, Connecticut, USA) system with mass spectrometric detection using a Waters UPLC Xevo G2-XS QTOF attachment (Waters Corp.). The samples were analyzed in both positive and negative ion modes. All raw data were collected using MassLynx software (version 4.2, Milford, Connecticut, USA) and entered into Progenesis QI (version 2.4, Munich, Bavaria, Germany) software for further analysis (33).
For the bioinformatic analysis, α-diversity analyses were conducted to evaluate the diversity, evenness, and richness of the intestinal microbiota, especially for the bacteria and their function, VFs and AROs, and metabolites at different levels by using GraphPad Prism (version 8.0, San Diego, California, USA), and β-diversity analyses were conducted to evaluate the structure of the intestinal microbiota, especially for the bacteria and their function, VFs and AROs, and metabolites at different levels by BMK Cloud platform (http://www.biocloud.net/). Linear discriminant analysis effect size (LEfSe) was used to screen biomarkers of intestinal microbiota using the criteria linear discriminant analysis (LDA) > 3 and P < 0.05 (34).
Network analysis was employed to visualize the potential connections between bacterial genera and VFs (35). After annotation using the Nr, KEGG, GO, eggNOG, VFDB, and CARD databases (36, 37), correlations between bacterial genera and KEGG functional pathways of intestinal microbiota, as well as VFs and KEGG functional pathways of intestinal microbiota, were assessed using Pearson correlation analysis (https://cloud.majorbio.com/page/tools/). The affiliations of VFs and AROs with bacterial species were determined using data from the non-redundant gene set construction. Potential metabolic biomarkers (PMBs) and KMBs were identified according to previously reported criteria (variable importance in projection [VIP] > 1.0, P < 0.05, and VIP > 1.5, P < 0.05) (33). The fold change (FC) of KMBs was calculated using the formula E9D/Con.
Experimental data are presented as the mean ± standard deviation and analyzed using an independent samples t-test in SPSS software (version 23.0, Chicago, Illinois, USA). Structural differences in the intestinal microbiota, function, VF levels, and AROs were determined using the PERMANOVA algorithm (http://www.biocloud.net/). Results with P < 0.05 were considered significant and those with P < 0.01 were considered highly significant.
The survival rate of big-belly seahorses showed a weight- and dose-dependent relationship (Figure 1A). The survival rate of the seahorses (4.1–4.5 g) injected with 1 × 105 cfu/mL of E. piscicida was the same as that of the Con group (100%), which is suitable for model construction. Whereas, in the other EP groups, the time to death and survival rate appeared to positively and negatively correlate with body weight and challenge concentration, respectively. After the reconstruction of the research model, typical pathological changes associated with bacterial enteritis, such as intestinal epithelial dissolution, focal bleeding, villus atrophy, separation between the lamina propria and submucosa, thickened lamina propria and muscularis mucosae, vascular distorted congestion, and a large amount of inflammatory necrosis in the muscle layer and serosal surface, were observed (Figure S1). Compared with those of the Con group, the body weight and length of seahorses in the EP group (4.1–4.5 g and 1 × 105cfu/mL) were reduced, especially on day 9 (P < 0.05) (Figures 1B, C). The gene expression of intestinal proinflammatory cytokines (IL-1;interferon1, INF1; tumor necrosis factor-α, TNF-α, interleukin-1β, IL-1β; IL-1β receptors), anti-inflammatory factors (IL-10 and IL-2), antimicrobial peptides (hepcidin; liver-expressed antimicrobial peptide, LEAP; piscidin; lysozyme), and Toll-like receptor 5 (TLR5) significantly increased on day 9 and decreased to different extents thereafter (Figures S2A, B). At the same time, the gene expression patterns of ZO-1 were similar to those of the immune genes (Figure S2C), whereas the other two tight junction genes claudin 5 and occludin showed the opposite expression patterns (Figures 1D, E). The RR of the EP group declined from days 2 to 9, especially from days 3 to 9 (P < 0.05), recovered thereafter, and remained at the level of the Con group (Figure 1F). As the RR and gene expression of both claudin 5 and occludin varied consistently along with the pathogenic process and other parameters, we updated our previously published evaluation system by adding them to the DAI scoring system (Table S2). We found that the DAI of the EP group increased from day 1, became significantly higher than that of the Con group from day 5 (P < 0.05), peaked on day 9, and decreased there after until day 21 (Figure 1G).
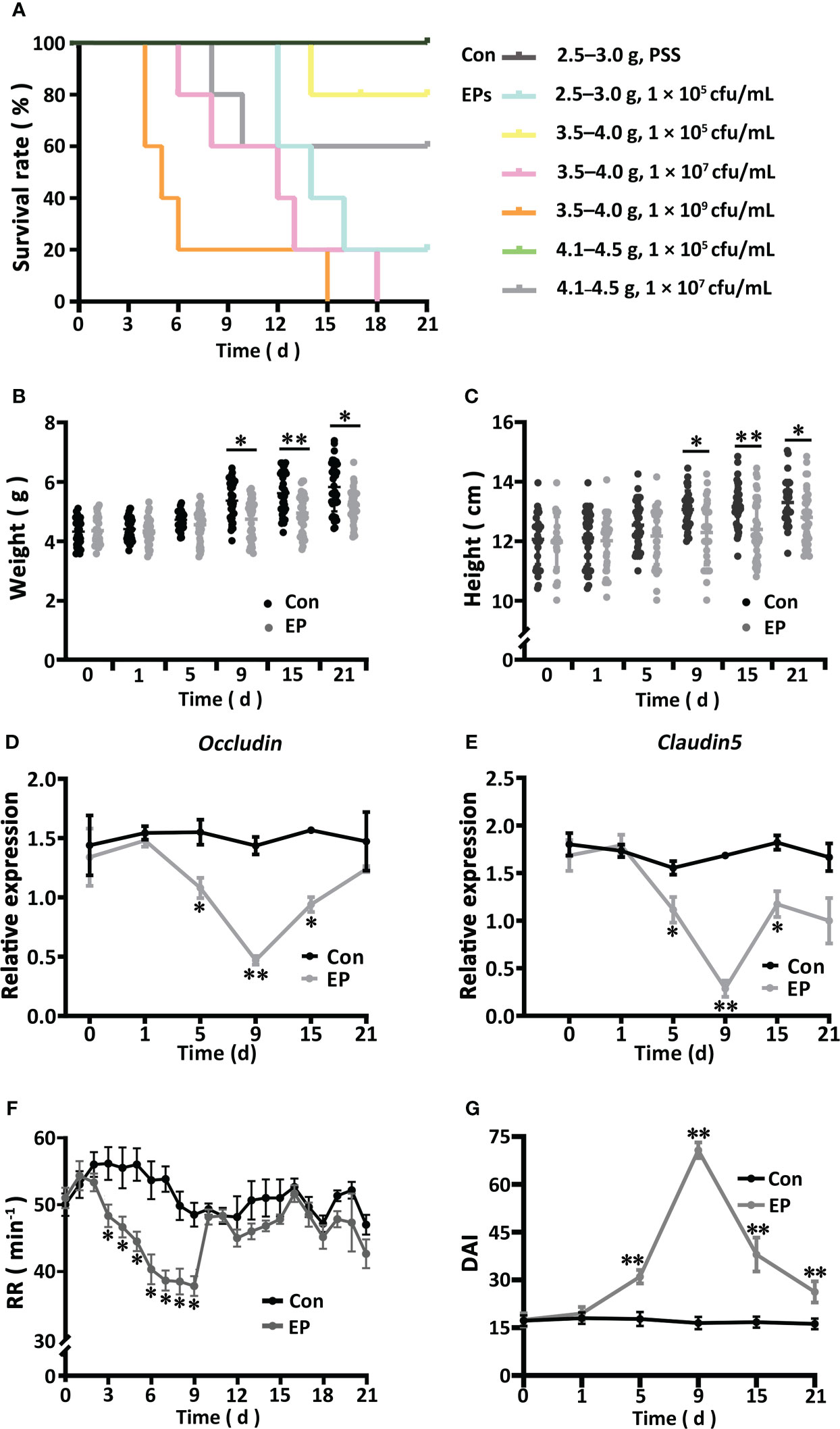
Figure 1 Effects of Edwardsiella piscicida infection on the survival (A), body weight (B), body height (C), gene expressions of Occludin (D) and Claudin 5 (E), RR (F), and DAI (G) of the big-belly seahorse (H. abdominalis). In (A), Con represents healthy controls (2.5–3.0 g, PSS), EPs represent E. piscicida-challenged groups (2.5–3.0 g, 1 × 105 cfu/mL; 3.5–4.0 g, 1 × 105 cfu/mL; 3.5–4.0 g, 1 × 107 cfu/mL; 3.5–4.0 g, 1 × 109 cfu/mL; 4.1–4.5 g, 1 × 105 cfu/mL; and 4.1–4.5 g, 1 × 107 cfu/mL). In (B–G), Con represents healthy controls (4.1–4.5 g, PSS), and EP represents E. piscicida- challenged group (4.1–4.5 g, 1 × 105 cfu/mL). PSS, physiological saline solution; RR, respiratory rate; DAI, disease activity index. *P < 0.05; **P < 0.01.
A total of 256,229 genes were identified via sequencing, belonging to 6 kingdoms, 95 phyla, 119 classes, 250 orders, 552 families, 1329 genera, and 3171 species (Figure 2A; Table S4). The number of sequences could reflect the structure and diversity of the microbial community because the corresponding rarefaction curves had already reached the saturation plateau (Figure S3A). The α-diversity and β-diversity of the intestinal microorganisms of the E9D group were significantly different from those of the Con and E21D groups at the species level (P < 0.01), whereas the difference between the Con and E21D groups was not significant (P > 0.05) (Figures S3B, C).
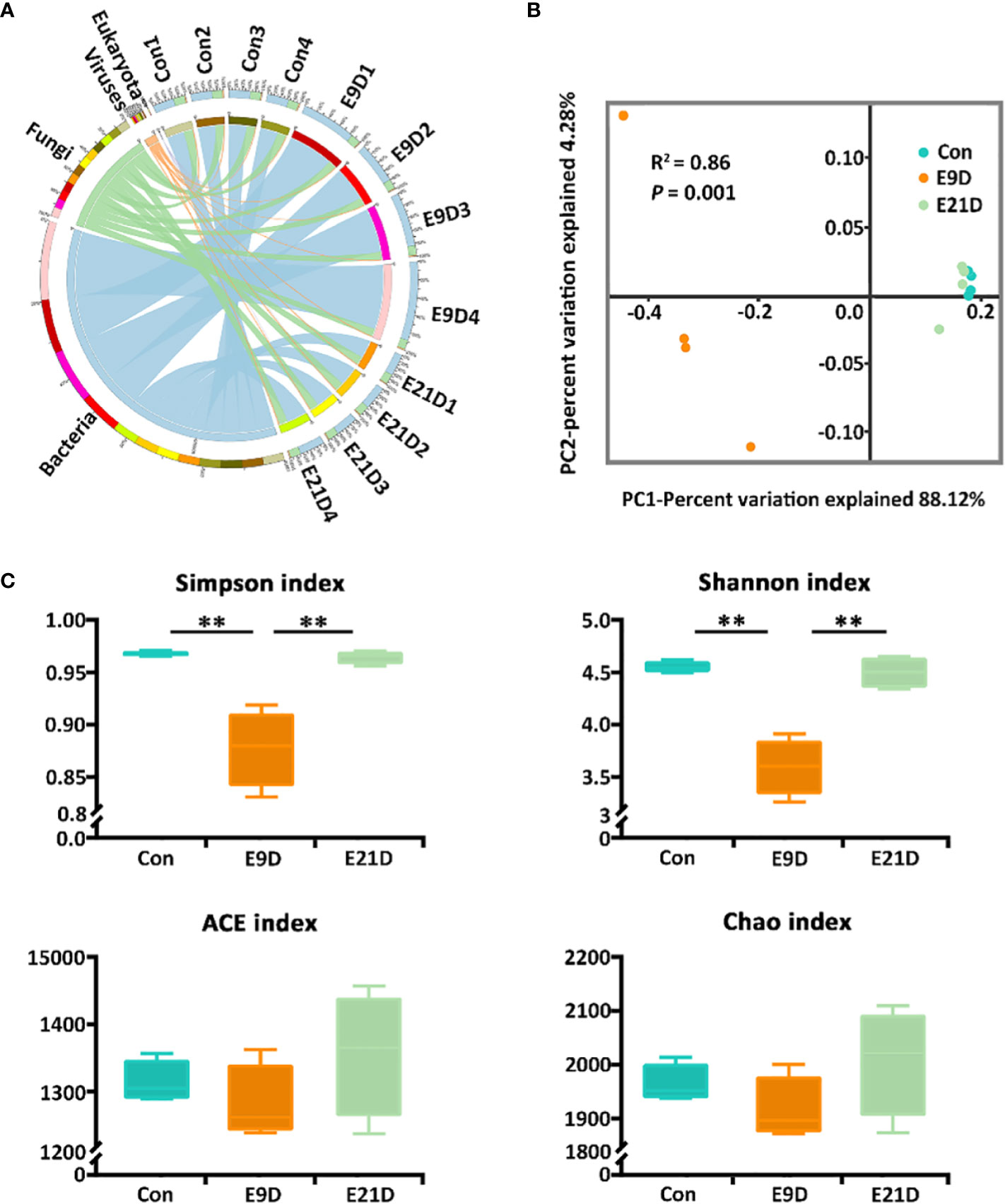
Figure 2 Effects of Edwardsiella piscicida infection on kingdom-level composition (A), structure (B), and diversity (C) of intestinal microbiota in big-belly seahorses. (B, C) represent bacterial intestinal microbiota at the species level. Con represents healthy controls (4.1–4.5 g, PSS); E9D and E21D represent the samples collected on days 9 and 21 of E. piscicida- challenged group (4.1–4.5 g, 1 × 105 cfu/mL), respectively (similarly hereinafter). **P < 0.01.
The intestinal microbiota composition at the kingdom level was numerically dominated by bacteria in each group (Figure 2A). As the strains used for stress were bacterial pathogens, all subsequent analyses were based on bacterial levels. There were 2066 shared bacterial species in the Con, E9D, and E21D groups, with 231, 227, and 380 endemic species, respectively (Figure S3D). As shown in Figure 2B, the β-diversity of the intestinal microbiota in the E9D group was significantly different from that of the Con and E21D groups at the species level (P < 0.01) but not between the Con and E21D groups (P > 0.05). The Shannon and Simpson indices of α-diversity in the E9D group were significantly lower than those of the Con and E21D groups (P < 0.01), but the ACE and Chao richness indices were not significantly different among the three groups (P > 0.05) (Figure 2C). This result indicates that pathogen infection altered the structure, and significantly reduced the evenness and diversity of intestinal microbiota in seahorses.
The composition of the intestinal microbiota at different levels changed after E. piscicida infection (Figure S4). The LEfSe analysis revealed significant differences in the phylogenetic distribution and biomarkers of the microbiota among the groups (P < 0.05) (Figure 3A). Notably, Edwardsiella and Edwardsiella piscicida accounted for 70.10% and 32.74% of the relative abundance in the identified bacteria, respectively (Table S5). Edwardsiella abundance was significantly positively correlated (P < 0.05, |r| > 0.7) with Chlamydia, Enterobacter, Yersinia, Pantoea, Salmonella, Xenorhabdus, Klebsiella, and Arthrobacter but negatively correlated (P < 0.05, |r| > 0.7) with Lactobacillus, Microbacterium, Enterococcus, Acinetobacter, Mycobacteroides, Flavobacterium, and Thalassococcus (Figure 3B).
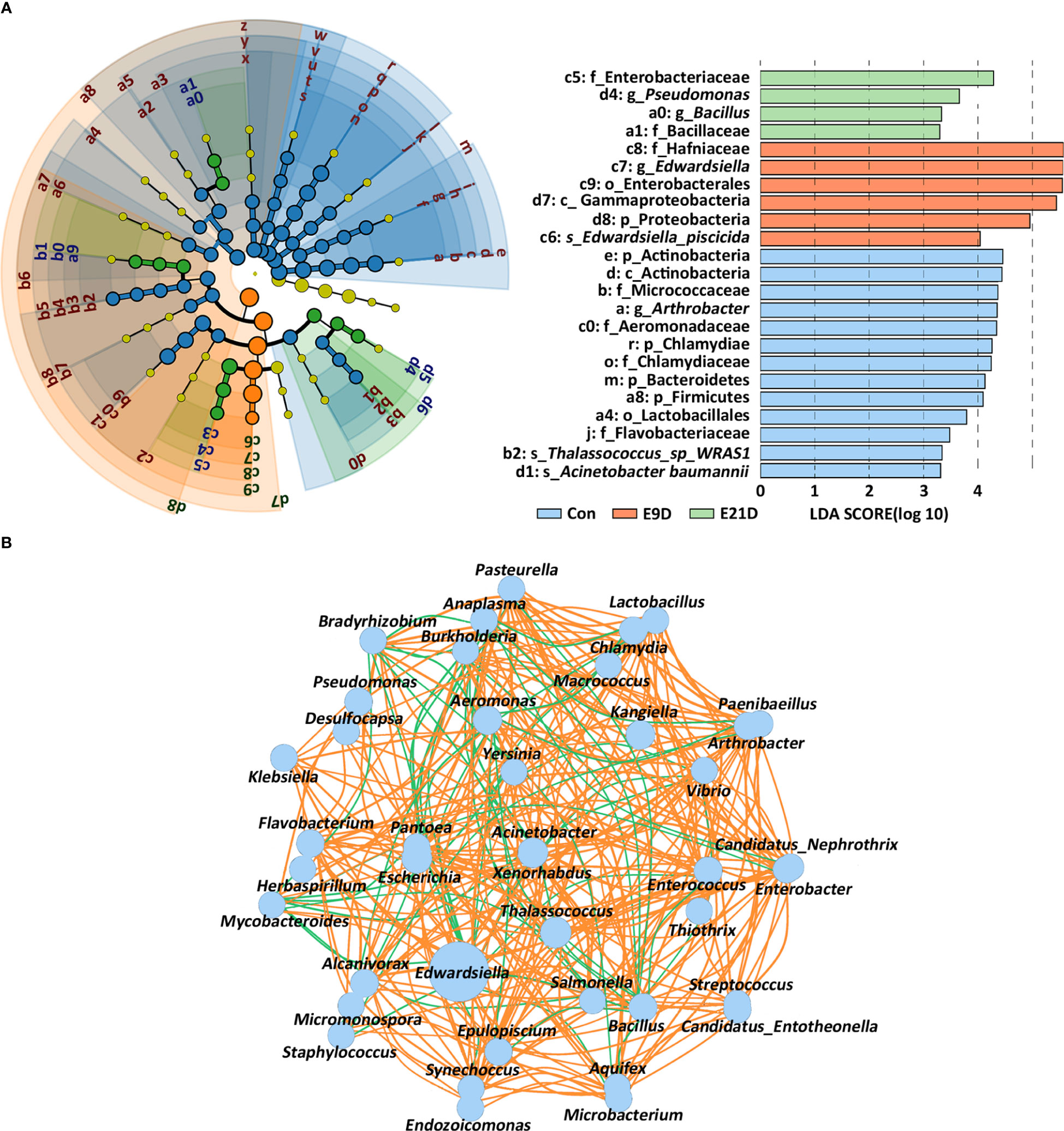
Figure 3 Effects of Edwardsiella piscicida infection on the expression of biomarkers (A) and interaction structure (B) of the intestinal microbiota in seahorses. The node size displays the abundance of the species. Intestinal microbiota biomarker screening criteria: P < 0.05, LDA > 3. The color of the line reflects the direction in the B-plot (P < 0.05, |r| > 0.7): orange, positive; green, negative.
The structure and activity of the functional pathways of the intestinal microbiota in the E9D group were significantly different from those of the Con and E21D groups at all three annotated KEGG, GO, and eggNOG levels (P < 0.01) (Figure 4A; Figure S5). As shown in Figure 4, compared with that of the Con group, the relative abundance of Edwardsiella, Chlamydia, Enterobacter, and Arthrobacter (P < 0.05) and the activities of 28 positively correlated functional pathways (P < 0.01, |r| > 0.7) were significantly increased (P < 0.01), whereas the relative abundance of Lactobacillus, Enterococcus, Microbacterium, Acinetobacter, Mycobacteroides, Aeromonas, and Burkholderia and the activities of 11 positively correlated functional pathways (P < 0.05, |r| > 0.7) were significantly decreased (P < 0.05) in the E9D group. Notably, the activities of eight functional pathways, including bacterial chemotaxis, flagellar assembly, lipopolysaccharide biosynthesis, phosphotransferase system, bacterial secretion system, QS, ABC transporters, and TCS, increased significantly (P < 0.01) in the E9D group (Figure 4), indicating the crucial role of these pathways during E. piscicida infection.
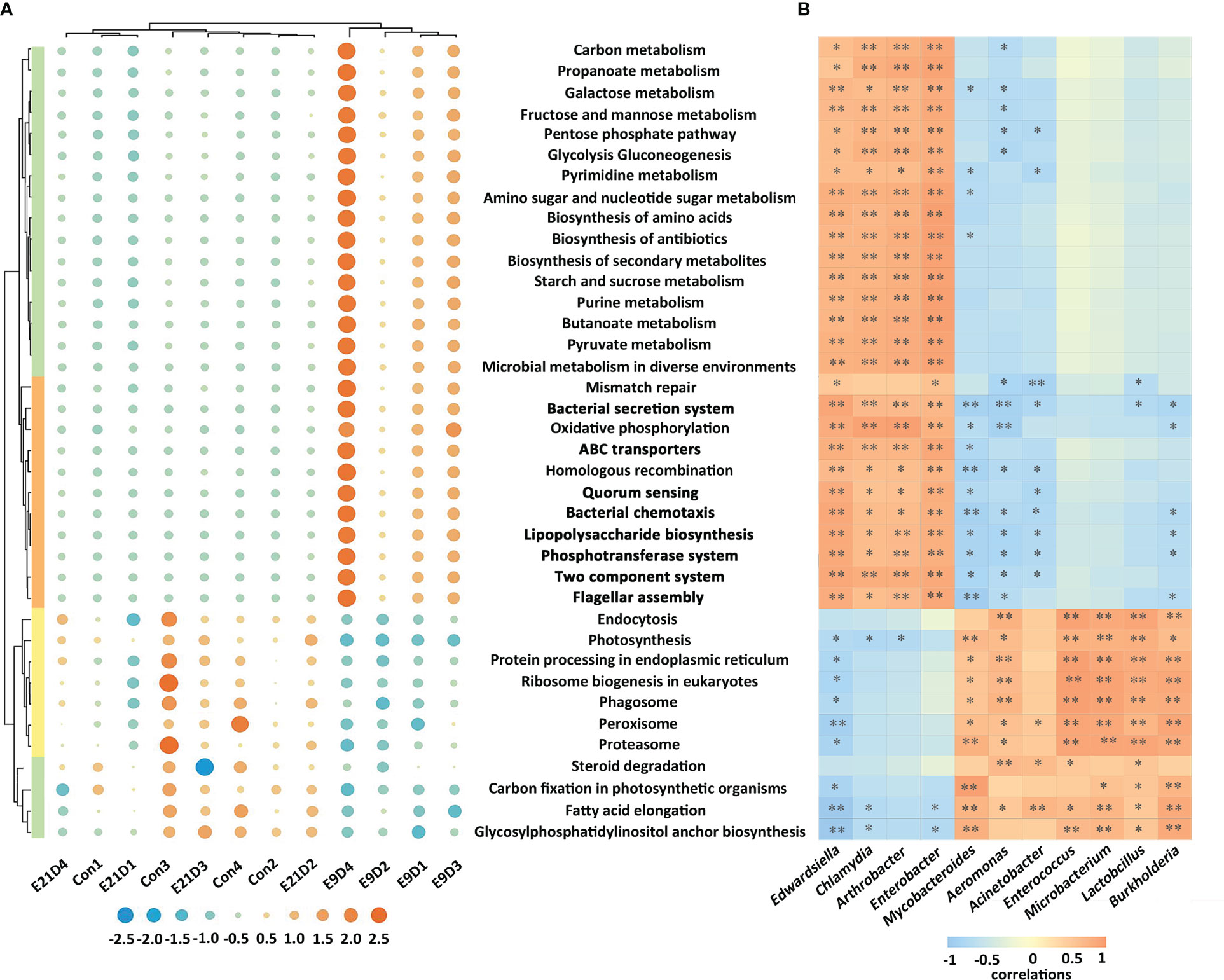
Figure 4 Effects of Edwardsiella piscicida infection on the functions (A) and its correlation with biomarkers (B) of intestinal microbiota in seahorses. In (A) the significance level of the KEGG functional activity difference of intestinal microbiota not in bold typeface was P < 0.05, whereas that in bold typeface was P < 0.01. The color column represents the KEGG level 1 functional classification (green: metabolism; orange: cellular process and environmental information processing; yellow: genetic information processing). The blue circles represent a decrease in relative abundance, whereas the orange circles represent an increase in abundance. In B, values > 0 represent a positive correlation, whereas values < 0 represent a negative correlation. Darker colors also indicate a stronger correlation. *P < 0.05; **P < 0.01.
Metagenomic analysis identified 10 categories, 181 VFs, and 478 virulence genes (Table S6; Figure S6A). The structure of VFs in the E9D group was significantly different from that of the Con and E21D groups (P < 0.01) (Figure S6B), which was similar to the results of the intestinal microbiota. Significant differences in α-diversity were found between the Con and E9D groups (P < 0.01) (Figure S6C; Table S6), consistent with the results of the intestinal microbiota and their functions.
Compared with that of the Con group, the relative abundance of 128 VFs in the E9D group significantly increased (P < 0.05), including 126 VFs that were extremely significantly increased (P < 0.01) (Figure 5A; Table S6). Edwardsiella could express 123 VFs covering all 10 categories (Table S6) and all six pathogenic processes. Significant positive correlations were observed among 50 VFs (P < 0.05, |r| > 0.7) (Figure 5B). The relative abundance of 15 of them, Flagella, Type IV pili, Lap, Bsa T3SS, Dot/Icm, FarAB, Capsule1, Hp-NAP, Legiobactin, IraAB, Hpt, LPS3, PhoP, BfmR, and BopD, was in the top 50 and complex nodes (P < 0.05) (Figure 5B; Table S6), indicating their pivotal role in regulating other VFs. As shown in Figure 5C, the top 50 most abundant VFs were significantly positively (P < 0.01, |r| > 0.7) and negatively (P < 0.05, |r| > 0.7) correlated with 28 significantly increased and 11 significantly decreased (P < 0.05) intestinal microbiota functions (Figure 4), respectively, indicating the importance of these VFs in regulating intestinal microbiota function. In addition, 48 AROs were annotated (Figure S7A; Table S7). The structure of AROs in the E9D group was significantly different from that in the Con and E21D groups (P < 0.05) (Figure S7B), which is similar to the results of the intestinal microbiota and VFs. The Chao and Shannon indices of α-diversity in the E9D group were significantly higher than those in the control group (P < 0.01) (Figure S7C). Six AROs with significantly increased abundance (P < 0.05) in the E9D group were identified (Figure 5D), five of which were expressed by Edwardsiella, adeF, msbA, Ecol_EFTu_PLV, Ecol_GlpT_FOF, and Hinf_PBP3_BCA (Table S7).

Figure 5 Effects of Edwardsiella piscicida infection on the composition and abundance of virulence factors (VFs) (A), the correlation between VFs (B), the correlation between VFs of relative abundance (TOP 50) and microbiota function (C), and abundance of antibiotic resistance ontologies (P < 0.05) (D) of the intestinal microbiota. The color in (A) represents the relative VF content (blue, decrease; orange, increase). The 15 VFs in bold typeface in (B) represent their core role in regulating other VFs and the functions of intestinal microbiota. The orange lines among VFs indicate significant positive correlations (P < 0.05, |r| > 0.7). Colored columns on the right of (C) represent the classification of VFs (blue, motility, adherence, and invasion; purple, effector delivery system; orange, iron uptake system; red, immune modulation; green, toxin; yellow, regulation). Values > 0 represent a positive correlation, whereas values < 0 represent a negative correlation. Darker colors also indicate a stronger correlation. *P < 0.05; **P < 0.01.
Metabolomic data revealed that the structure of metabolites of the E9D groups was different from that of the Con group (Figures S8A–C). A total of 17 categories, 4789 metabolites, and 1768 PMBs (VIP > 1.0 and P < 0.05; 623 upregulated and 1145 downregulated) were identified (Figures 6A, B, S8D; Table S8). At the same time, 491 KMBs (VIP > 1.5 and P < 0.05; 189 upregulated and 302 downregulated) were identified (Figure S8E), indicating that E. piscicida infection significantly affected metabolic processes in big-belly seahorses.
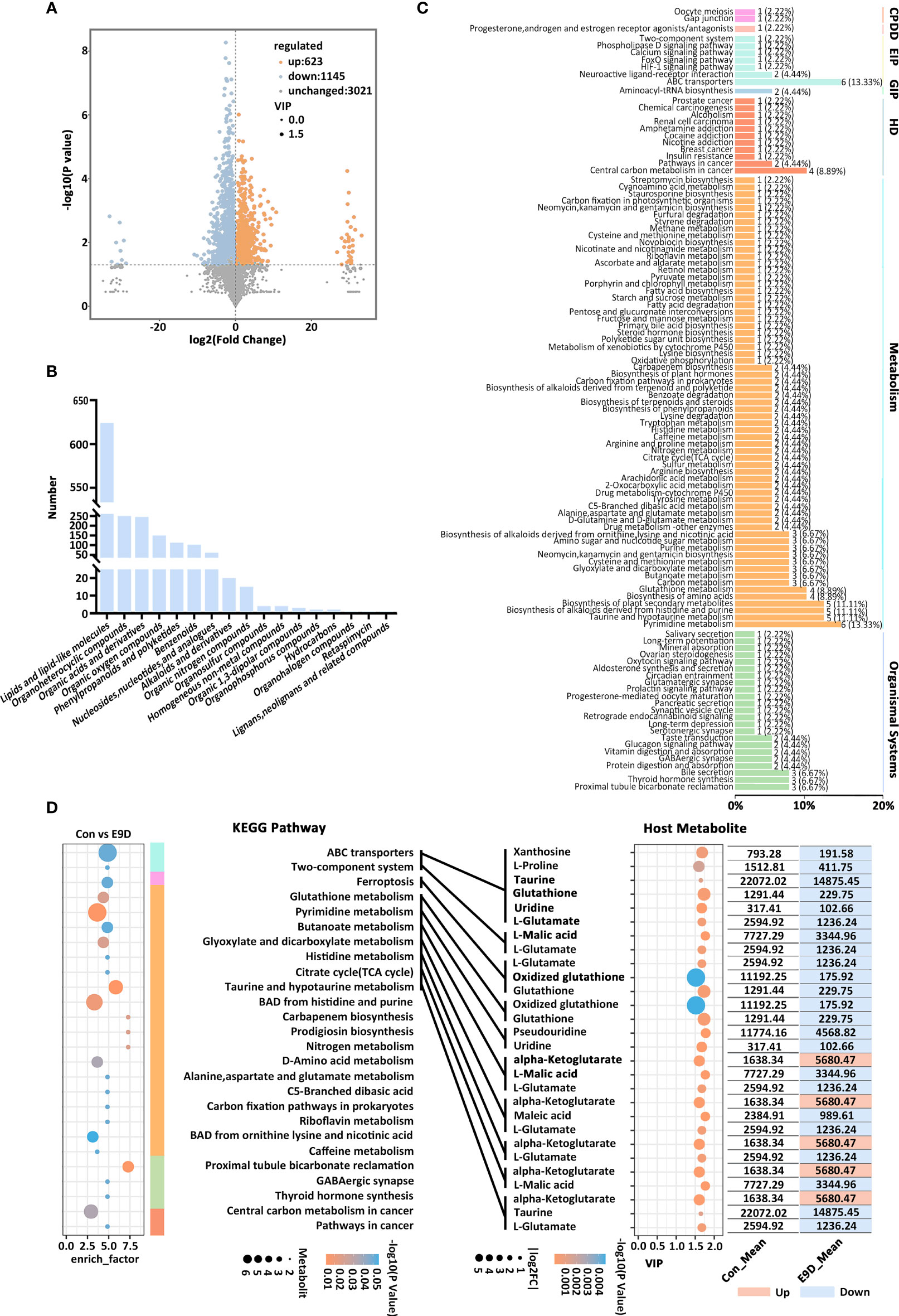
Figure 6 Effects of Edwardsiella piscicida infection on the composition (A), classification (B), KEGG functional classification of seahorse intestinal potential metabolite biomarkers (PMBs) (C), and the content of key metabolite biomarkers (KMBs) and their KEGG functions (D). CP, cellular processes; EIP, environmental information processing; GIP, genetic information processing; VIP, variable importance in projection. BAD, biosynthesis of alkaloids derived. The color column represents the KEGG level 1 functional classification in (D) (green, environmental information processing; blue, cellular processes; light blue, metabolism; orange, organismal systems; yellow, human diseases). The size the circle on the left side represents the levels of metabolites, whereas the size of the circle on the right side represents the |log2FC|.
Forty-eight out of the 491 KMBs (Table S9) were enriched in six major functional categories and 110 functional pathways (Figure 6C). The activities of 27 functional pathways from 5 major functional categories were also significantly different (P < 0.05) (Figure 6D left panel). Eleven KMBs (abundance TOP 15 and |log2FC | > 0.55) were identified and enriched in 10 significantly different functional pathways (P < 0.05) (Figure 6D). Compared with those of the Con group, KMBs associated with histidine metabolism, citrate cycle (TCA cycle), and taurine and hypotaurine metabolism pathways were mostly upregulated. In contrast, KMBs associated with ABC transporters, TCS, ferroptosis, glutathione metabolism, and pyrimidine metabolism pathways were all downregulated, whereas KMBs related to glyoxylate and dicarboxylate metabolism and butanoate metabolism pathway were mostly downregulated in the E9D group (Figure 6D). Notably, eight KMBs, alpha-Ketoglutarate, Taurine, Glutathione, Uridine, L-Glutamate, L-Malic acid, Glutathione, and Oxidized glutathione, were involved in regulating multiple functional pathways (Figure 6D right panel), suggesting essential roles during E. piscicida infection.
Centered on the eight closely related key functions of the intestinal microbiota, we found 34 KVFs significantly positively correlated with six of them (P < 0.01, |r| > 0.8), including15 were core VFs (CVFs) and the other 19 of the top 50 VFs in abundance. As shown in Figure 7A, 34 KVFs among the top 50 VFs with relatively complex nodes correlated with six key intestinal microbiota functions and seven KMBs after E. piscicida infection. For instance, in the E9D group, the relative abundance of VFs associated with motility, adhesion, and invasion of pathogens significantly and simultaneously increased with the functional activity of flagella assembly and bacterial chemotaxis (P < 0.01). The relative abundance of the effector delivery system-related VFs significantly and simultaneously increased with the functional activity of the bacterial secretion system (P < 0.01). The relative abundance of the iron uptake system-related VFs increased with the functional activity of ABC transporters of the intestinal microbiota (P < 0.01), whereas the levels of L-Proline, Taurine, Glutathione, Uridine, Xanthosine, and L-Glutamate significantly decreased (P < 0.05). Similarly, the relative abundance of toxin-related VFs significantly and simultaneously increased with the functional activity of LPS biosynthesis. The relative abundance of the regulation-related VFs significantly and simultaneously increased with the functional activity of TCS of the intestinal microbiota (P < 0.01), whereas the levels of KMBs L-Malic acid and L-Glutamate significantly decreased (P < 0.05) (Figures 4D left panel; 5C; 7A). In addition, seven of the eight KMBs with central roles were enriched and significantly negatively correlated with two key intestinal microbiota functions, TCS and ABC transporters (P < 0.05). The content of L-Malic acid and L-Glutamate related to TCS significantly decreased after E. piscicida infection (P < 0.05), as did the levels of L-Proline, Taurine, Glutathione, Uridine, Xanthosine, and L-Glutamate in relation to ABC transporters (P < 0.05) (Figure 7A).
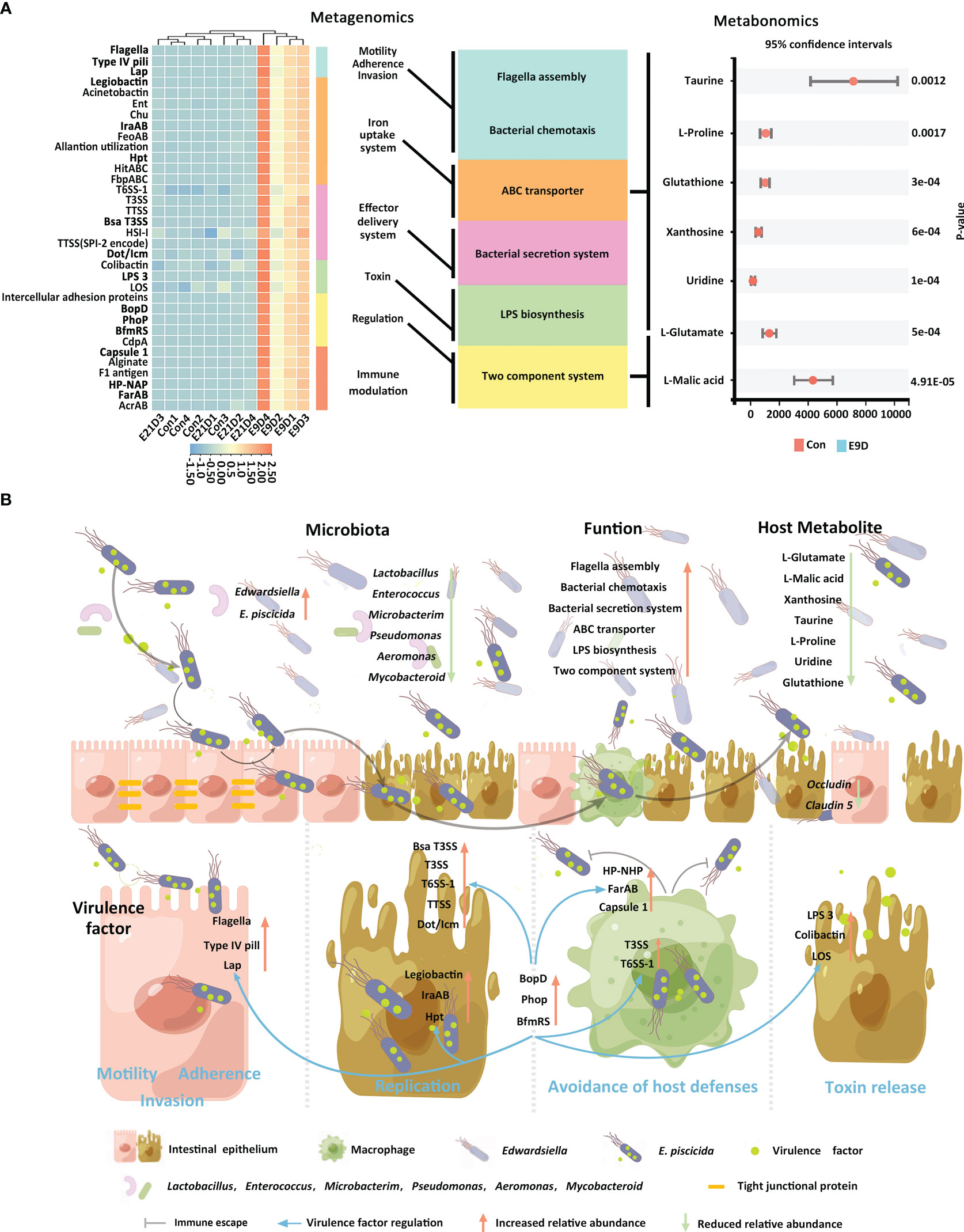
Figure 7 Correlation among core functions of the intestinal microbiota, key virulence factors, and key metabolites of the host (A). Significant positive correlations P < 0.01, |r| > 0.8. Schematic diagram of pathogenic mechanisms (B) of Edwardsiella piscicida infection. Edwardsiella piscicida infection could be divided into six stages: motility, adherence, invasion, replication, avoidance of host defenses, and toxin release. The KVFs Flagella, Type IV pili, and Lap allow pathogen approach, attachment, and binding to host epithelial cells, and invasion and internalization into the villus epithelial cells. After entering epithelial cells, legiobactin, IraAB, and Hpt help compete for more nutrients to improve pathogen survival and replication in a nutrient-deficient environment. Meanwhile, T3SS, T6SS-1, TTSS, HSI-I, VirB/VirD, and Dot/Icm promote pathogen replication and invasion to the adjacent epithelial cells and other tissues. Upon invasion into deeper tissues, pathogens could encounter phagocytes. HP-NAP, Capsule1, and FarAB could help pathogens avoid phagocytosis, whereas T3SS and T6SS-1 could help them survive and replicate within phagocytes. After release, they could destroy the intestinal barrier (characterized by the downregulations of Occludin and Claudin 5). After successful colonization, LPS3, Colibactin, and LOS could help release toxins, induce severe inflammatory responses, and finally lead to host death. Core VFs BopD, Phop, and BfmRS could coordinate with other VFs in responding to environmental changes to accelerate deep tissue infection. When a large number of E. piscicida is released into the intestinal lumen, it can induce enteritis by increasing the relative abundance of related pathogens and virulence factors and changing the composition structure, function, and host metabolism of the intestinal microbiota.
In this study, we found that E. piscicida infection inhibited growth and induced typical pathological features of enteritis in big-belly seahorses, similar to our previous findings on E. tarda-induced enteritis in lined seahorses (25). The RR of the big-belly seahorses was significantly reduced in the deep infection stage, indicating inhibition of basal metabolic activity after E. piscicida infection (31). Significant downregulation of occludin and claudin 5 expression on day 9 of E. piscicida infection might have led to alterations in intestinal permeability and increased bacterial paracellular flux (38). As a component of tight junctions, the expression of ZO-1 significantly decreases after pathogen infection in fish (39). Interestingly, ZO-1 expression in the present study was significantly upregulated in the deep infection stage, consistent with previous reports in mice (40), suggesting that ZO-1 is dispensable for barrier function and can act as a regulator of tight junctions to promote the repair of damaged mucosa (41). In addition, the DAI results suggest that the updated evaluation system can effectively reflect the pattern and intensity of disease progression. Different from E. tarda-induced enteritis in lined seahorses (25), more severe pathological features and sooner peaked DAI were found in E. piscicida-induced enteritis in the big-belly seahorses under similar challenge conditions. This finding may be attributed to the higher toxicity of E. piscicida or a relatively smaller body proportion of the trunk area of big-belly seahorses with pathogen infection than lined seahorses.
Systematic studies exploring relevant omics datasets will enable scientist to describe the complexity and characteristics of interactions in the host-pathogen network, and identify new targets or biomarkers for pathogenic infections (20, 42). In the present study, E. piscicida infection could alter the composition, structure, and abundance and significantly reduce the diversity of the intestinal microbiota of big-belly seahorses. These results are similar to previously reported pathogenic infections in lined seahorses, yellow seahorses (H. kuda), largemouth bronze gudgeon (Coreius guichenoti), and pearl gentian grouper (Epinephelus lanceolatus ♂ × E. fuscoguttatus ♀) (14, 16, 26, 43). Lipopolysaccharide (LPS) is a key VF and an important component of the outer membrane of gram-negative pathogens (44). Bacterial chemotaxis plays a crucial role at all stages of infection (45). Flagellar assembly is essential for bacterial pathogens to reach the optimal infection sites, promote biofilm formation, and adhere to host surfaces or cells (46). The bacterial secretion system can interfere with host immunity and disrupt or mimic host cellular processes, and help Edwardsiella avoid phagocytosis and replicate within phagocytes and epithelial cells of the intestine (2, 47), inducing severe systemic infections and killing the host (48). The phosphotransferase system is involved in stimulating biofilm formation, aggregating motility, and inducing pathogen colonization (49). ABC transporters, as nutritional VFs, help bacterial pathogens absorb nutrients, such as iron, vitamins, and metabolites, and grow and survive in nutrient-limited or harsh environments (50, 51). TCS is the primary control switch in signal transduction, physiology, cell-cell communication, adaptation to changing environments, and pathogenesis of bacterial pathogens (52). QS is a major regulator of natural competence, motility, and virulence, and controls the switch between the replicative and transmissive/virulent phases of Legionella pneumophila (53). In the present study, E. piscicida infection significantly increased the relative abundance of the opportunistic pathogens (Edwardsiella, Chlamydia, Enterobacter, and Arthrobacter) and the activities of their positively correlated 28 functional pathways, but decreased the abundance of the probiotic microbiota (Enterococcus, Microbacterium, Lactobacillus, and Burkholderia) and opportunistic pathogens (Aeromonas, Acinetobacter, and Mycobacteroides). This suggests that E. piscicida infection may enhance the competition for nutrients against autochonous microbiota and certain pathogenic bacteria by increasing the activities of functional pathways such as chemotaxis and QS, induce dysbiosis of the intestinal microbiota, and finally cause enteritis.
VFs are key features for the selective advantage of potentially pathogenic bacteria over common members of healthy gut microbiota (54). In the present study, we identified 123 VFs that were significantly increased in abundance after E. piscicida infection for the first time, of which 15 CVFs may play a key role in pathogenesis. Referring on previously reported VFs of other bacterial pathogens, E. piscicida may rely on Flagella, Type IV pili, and Lap for adhesion and infection (55, 56); Bsa T3SS and Dot/Icm for invasion, intracellular replication, and immune escape (57, 58); Legiobactin, IraAB, and Hpt for survival and growth (46, 59, 60); HP-NAP, Capsule1, and FarAB for evasion of host immune defenses and killing (61–63); and LPS3 for poisoning and killing the host (63). In addition, PhoP, BfmRs, and BopD may help E. piscicida sense environmental changes and coordinate the expression of other VFs (64–67). Finally, E. piscicida infection may reshape the intestinal microbiota and its functions and induce enteritis through the synergistic regulation and crosstalk between the 15 key VFs and other VFs. In addition, the five AROs of Edwardsiella may resist or evade the bactericidal effects of tetracycline, fluoroquinolone, elfamycin, fosfomycin, nitroimidazole, and beta-lactam antibiotics through mechanisms such as active efflux of antibiotics, transferring to other resistant bacteria, and biofilm formation (68, 69). More attention should be paid to these characteristics when designing prevention and control strategies against Edwardsiella infections.
The metabolite composition of organisms can provide substantial evidence for identifying biomarkers in pathological processes and analyzing pathogenic mechanisms (70). In this study, E. piscicida infection resulted in dysfunctions in metabolism, environmental information processing, and cellular processes, consistent with the results of E. tarda infection in tilapia (Oreochromis mossambicus) (71). The ABC transporters are associated with the absorption of nutrients, vitamins, and metabolites (51). TCS regulates magnesium ion levels, pH, and antimicrobial peptide-associated signaling (72), converting external signals into gene expression in a dose-dependent manner (73). Pyrimidine metabolism is essential for DNA and RNA synthesis (74). In this study, the functional activities of ABC transporters, TCS, and pyrimidine metabolism were significantly downregulated, suggesting the inhibition of host nutrient transport systems, environmental changes, immune-related signal transduction systems, and translation mechanisms during E. piscicida infection. Ferroptosis and glutathione metabolism are essential for pathogen defense in the host, and the downregulation of their activities could disrupt intestinal homeostasis (75, 76) and promote the invasion of E. piscicida. In addition, the TCA cycle, histidine metabolism, and taurine and hypotaurine metabolism were altered after E. piscicida infection, indicating disturbance in host energy metabolism (77) and immune defense mechanisms (78, 79). Collectively, these results suggest that E. piscicida infection affects energy metabolism and disrupts the defense mechanisms of big-belly seahorses by inhibiting nutrient transport, signal transduction, and translation to promote virulence regulation, replication, and transmission.
Pathogen infection can generally be divided into six stages: motility, adherence, invasion, replication, avoidance of host defenses, and toxin release (2), during which significant associations among VFs, intestinal microbiota, and metabolism are found (54, 80, 81). To the best of our knowledge, this is the first study to provide evidence that 34 KVFs are closely related to six key intestinal microbiota functions and seven host KMBs during E. piscicida infection in big-belly seahorses. Significant upregulation of KVFs associated with motility, adherence, and invasion may allow E. piscicida to adhere, bind to host intestinal epithelial cells, and invade or be internalized into host tissues and cells by increasing the activities of bacterial chemotaxis and flagellar assembly of the intestinal microbiota (45, 55). With the aggravation of local infection, the upregulation of KVFs associated with the effector delivery system and iron uptake system could help pathogens acquire nutrients for growth, survival, and proliferation under nutrient-limited and harsh host environments by enhancing the functional activity of pathogenic ABC transporters (51, 60). Meanwhile, local infection increases the activity of the bacterial secretion system, allowing the pathogen to survive and replicate in epithelial cells and spread to the adjacent epithelial cells and other tissues (1, 3). During deep invasion, E. piscicida may avoid phagocytosis by upregulating KVFs associated with immune regulation (82) or survive and replicate within phagocytes with the help of KVFs of the effector delivery system (1, 83). Through these two mechanisms, E. piscicida may successfully avoid attack by the host immune system and disrupt the host mucosal and microbial barriers. Moreover, significant increases of both the abundance of toxin KVFs and LPS biosynthesis results in the production of large amounts of LPS, leading to host tissue necrosis (2, 63). During the invasion process, the upregulation of KVFs may activate the TCS pathway, and thereby help signal transmission and activate appropriate VFs according to environmental conditions by regulating their transcriptional activity (1, 47, 52). The synergy of these processes increases the abundance and accelerates the deep infection of E. piscicida.
Taurine, L-Proline, and Uridine have antibacterial activity and can improve host resistance against pathogen invasion (6, 84, 85); therefore, they can be used as KMBs to characterize host resistance to infection by Edwardsiella spp. and other pathogens. L-Glutamate is essential for the host to maintain intestinal health and improve growth performance and survival (86). Glutathione has an antioxidant function and can be used to characterize the energy production of host mitochondria (87). Xanthosine and L-Malic acid can provide carbon and energy sources for the growth of pathogens (88, 89). In the present study, all seven KMBs were involved in multiple metabolic pathways and were significantly downregulated after infection, suggesting that E. piscicida can use metabolites from big-belly seahorses to promote its replication and infection while inhibiting the energy production and disease resistance of the host. These KMBs may act as typical metabolic features to characterize E. piscicida infection. In addition, KMBs related to ABC transporters and TCS pathway-related genes were significantly altered during deep infection, suggesting a key role for these two functions in that specific infection stage. Applying the analysis pattern employed in this study to investigate other infection stages may help identify more KMBs and better understand the underlying mechanisms of the pathogenic process. In summary, we drew a schematic figure to help interpret the mechanisms underlying E. piscicida infection-induced enteritis in big-belly seahorses (Figure 7B).
In this study, we updated the current research model and determined the pathogenic patterns and pathological characteristics of bacterial enteritis induced by E. piscicida infection in big-belly seahorses; elucidated the role of KVFs in regulating the diversity, structure, and function of intestinal microbiota; and determined the changes and KMBs of host intestinal metabolites for characterizing E. piscicida infection. Our results shed light on the pathogenetic mechanisms underlying E. piscicida-induced bacterial enteritis in big-belly seahorses. This may enrich the knowledge and provide theoretical references for preventing and controlling related diseases.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ebi.ac.uk/ena/browser/view/PRJNA916642.
The animal study was reviewed and approved by the Animal Care and Use Committee of Ludong University (document number: LDU-RB20210308NXY-9).
LZ and FW, conceptualization, methodology, investigation, formal analysis, data curation, visualization, resources, writing - original draft, writing- review and editing. LJ, investigation, formal analysis, data curation, visualization, and resources. HY and LG, visualization, data curation, and investigation. YT and XS, investigation and formal analysis. XZ, investigation. CL, sample collection and investigation. ZM, visualization. YX, resources. QL, writing-review and editing and conceptualization. KW, supervision, validation, funding acquisition, writing-review & editing, and conceptualization. All authors contributed to the preparation of article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (No. 42276118, 42230409), the Marine Economic Development Project (No. GDNRC [2022] 36), the Shandong Province Science and Technology Support Program for Outstanding Youth of Colleges and Universities (No. 2020KJF007), the Yantai Foundation for Development of Science and Technology (No. 2020LJRC120), the Strategic Priority Research Program of CAS (No. XDB42030204), the SCSIO Open Funding Project (No. LMB20200103) and the Shandong Province science and technology SMSE innovation capacity improvement Project (No. 2022TSGC1218).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1135588/full#supplementary-material
LPS, lipopolysaccharide; VF, virulence factor; CVF, core virulence factor; KVF, key virulence factor; KMB, key metabolic biomarker; PMB, potential metabolic biomarker; QS, quorum sensing; ARO, antibiotic resistance ontology; CFU, colony-forming unit; q-PCR, quantitative reverse transcription-polymerase chain reaction; LC-MS, liquid chromatography-mass spectrometry; DAI, disease activity index; TCS, two-component systems; T3SS/T6SS, type III/VI secretion systems; LEfSe, linear discriminant analysis effect size; IP, intraperitoneal; LDA, linear discriminant analysis; RR, respiratory rate; PSS, physiological saline solution; VIP, variable importance in projection; FC, fold change; CP, cellular processes; EIP, environmental information processing; GIP, genetic information processing; BAD, biosynthesis of alkaloids derived.
1. Leung KY, Wang Q, Yang Z, Siame BA. Edwardsiella piscicida: a versatile emerging pathogen of fish. Virulence (2019) 10(1):555–67. doi: 10.1080/21505594.2019.1621648
2. Leung KY, Siame BA, Tenkink BJ, Noort RJ, Mok YK. Edwardsiella tarda – virulence mechanisms of an emerging gastroenteritis pathogen. Microbes Infect (2012) 14(1):26–34. doi: 10.1016/j.micinf.2011.08.005
3. Machimbirike VI, Crumlish M, Dong HT, Santander J, Khunrae P, Rattanarojpong T. Edwardsiella ictaluri: a systemic review and future perspectives on disease management. Rev Aquacult (2022) 14(3):1–24. doi: 10.1111/raq.12665
4. Jiang JT, Wang WW, Sun F, Zhang YX, Liu Q, Yang DH. Bacterial infection reinforces host metabolic flux from arginine to spermine for NLRP3 inflammasome evasion. Cell Rep (2021) 34(10):108832. doi: 10.1016/j.celrep.2021.108832
5. Zeng ZX, Liu LY, Xiao SB, Lu JF, Liu YL, Li J, et al. Secreted in a type III secretion system-dependent manner, EsaH and EscE are the cochaperones of the T3SS needle protein EsaG of. Edwardsiella piscicida. mBio (2022) 13(4):e0125022. doi: 10.1128/mbio.01250-22
6. Stacy A, Andrade-Oliveira V, McCulloch JA, Hild B, Oh JH, Perez-Chaparro PJ, et al. Infection trains the host for microbiota-enhanced resistance to pathogens. Cell (2021) 184(3):615–27.e17. doi: 10.1016/j.cell.2020.12.011
7. Wang CY, Li QR, Ren JN. Microbiota-immune interaction in the pathogenesis of gut-derived infection. Front Immunol (2019) 10:1873. doi: 10.3389/fimmu.2019.01873
8. Raaijmakers JM, Kiers ET. Rewilding plant microbiomes. Science (2022) 378(6620):599–600. doi: 10.1126/science.abn6350
9. Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell (2006) 124(4):837–48. doi: 10.1016/j.cell.2006.02.017
10. Fan QX, Shi KP, Zhan M, Xu Q, Liu XB, Li ZJ, et al. Acute damage from the degradation of Ulva prolifera on the environmental microbiota, intestinal microbiota and transcriptome of Japanese flounder Paralichthys olivaceus. Environ Pollut (2022) 302:119022. doi: 10.1016/j.envpol.2022.119022
11. Kamada N, Kim YG, Sham HP, Vallance BA, Puente JL, Martens EC, et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science (2012) 336(6086):1325–9. doi: 10.1126/science.1222195
12. Ma X, Yang ZR, Xu TT, Qian MQ, Jiang XM, Zhan X, et al. Chlortetracycline alters microbiota of gut or faeces in pigs and leads to accumulation and migration of antibiotic resistance genes. Sci Total Environ (2021) 796:148976. doi: 10.1016/j.scitotenv.2021.148976
13. Qiao RX, Sheng C, Lu YF, Zhang Y, Ren HQ, Lemos B. Microplastics induce intestinal inflammation, oxidative stress, and disorders of metabolome and microbiome in zebrafish. Sci Total Environ (2019) 662:246–53. doi: 10.1016/j.scitotenv.2019.01.245
14. Zhang LL, Zou Q, Tian YN, Lv CH, Zheng SY, Jiang GJ, et al. Dysbiosis of both structure and function of intestinal microbiota in lined seahorses (Hippocampus erectus) as response to Edwardsiella tarda infection. J Trop Oceanogr (2022) 41(02):177–88. doi: 10.11978/2021074
15. Zhou CQ, Yang SW, Ka W, Gao P, Li Y, Long RJ, et al. Association of gut microbiota with metabolism in rainbow trout under acute heat stress. Front Microbiol (2022) 13:846336. doi: 10.3389/fmicb.2022.846336
16. Deng YQ, Zhang YQ, Chen HX, Xu LW, Wang Q, Feng J. Gut-liver immune response and gut microbiota profiling reveal the pathogenic mechanisms of Vibrio harveyi in pearl gentian grouper (Epinephelus lanceolatus ♂ × e. fuscoguttatus ♀). Front Immunol (2020) 11:607754. doi: 10.3389/fimmu.2020.607754
17. Franzosa EA, Sirota-Madi A, Avila-Pacheco J, Fornelos N, Haiser HJ, Reinker S, et al. Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol (2019) 4(2):293–305. doi: 10.1038/s41564-018-0306-4
18. Khurana H, Singh DN, Singh A, Singh Y, Lal R, Negi RK. Gut microbiome of endangered Tor putitora (Ham.) as a reservoir of antibiotic resistance genes and pathogens associated with fish health. BMC Microbiol (2020) 20(1):249. doi: 10.1186/s12866-020-01911-7
19. Jiang M, Chen ZG, Zheng J, Peng B. Metabolites-enabled survival of crucian carps infected by Edwardsiella tarda in high water temperature. Front Immunol (2019) 10:1991. doi: 10.3389/fimmu.2019.01991
20. Natnan M, Low CF, Chong CM, Bunawan H, Baharum S. Integration of omics tools for understanding the fish immune response due to microbial challenge. Front Mar Sci (2021) 8:668771. doi: 10.3389/fmars.2021.668771
21. Cohen FPA, Valenti WC, Planas M, Calado R. Seahorse aquaculture, biology and conservation: knowledge gaps and research opportunities. Rev Fish Sci Aquac (2017) 25:100–11. doi: 10.1080/23308249.2016.1237469
22. Er WCV, Christianus A, Harah M, Chong CM. Significance of water flow rate and period of nursing on the growth of juvenile seahorse, Hippocampus barbouri (Jordan and Richardson, 1908). J Surv Fish Sci (2017) 4:1–7. doi: 10.18331/SFS2017.4.1.1
23. Len YW, Christianus A, Worachananant S, Harah M, Chong CM. Estimation of live food consumption for Hippocampus barbouri and Hippocampus kuda. J Surv Fish Sci (2020) 7:43–54. doi: 10.18331/SFS2020.7.1.4
24. Tang L, Zhang YH, Wang X, Zhang CC, Qin G, Lin Q. Effects of chronic exposure to environmental levels of tributyltin on the lined seahorse (Hippocampus erectus) liver: analysis of bioaccumulation, antioxidant defense, and immune gene expression. Sci Total Environ (2021) 801:149646. doi: 10.1016/j.scitotenv.2021.149646
25. Wang XM, Wang F, Chen GZ, Yang B, Chen J, Fang Y, et al. Edwardsiella tarda induces enteritis in farmed seahorses (Hippocampus erectus): an experimental model and its evaluation. Fish Shellfish Immunol (2020) 98:391–400. doi: 10.1016/j.fsi.2020.01.049
26. Wang RP, Pan X, Xu YJ. Altered intestinal microbiota composition associated with enteritis in yellow seahorses Hippocampus kuda (Bleeker, 1852). Curr Microbiol (2020) 77(5):730–7. doi: 10.1007/s00284-019-01859-6
27. Evans D, Millar Z, Harding D, Pham PH, LePage V, Lumsden JS. Lipoid liver disease in Hippocampus erectus perry with Vibrio fortis-induced dermatitis and enteritis. J Fish Dis (2022) 45(8):1225–9. doi: 10.1111/jfd.13618
28. Jiang FY, Huang H, Yang N, Feng HM, Li Y, Han BB. Isolation, identification, and biological control in vitro of tail rot pathogen strain from Hippocampus kuda. PloS One (2020) 15(4):e0232162. doi: 10.1371/journal.pone.0232162
29. Zhang HX, Qin G, Zhang YH, Li SS, Lin Q. The leptin system and its expression at different nutritional and pregnant stages in lined seahorse (Hippocampus erectus). Biol Open (2016) 5(10):1508–15. doi: 10.1242/bio.020750
30. Zhang W, Tan BP, Deng JM, Yang QH, Chi SY, Pang AB, et al. PRR-mediated immune response and intestinal flora profile in soybean meal-induced enteritis of pearl gentian groupers, Epinephelus fuscoguttatus♀ × Epinephelus lanceolatus♂. Front Immunol (2022) 13:814479. doi: 10.3389/fimmu.2022.814479
31. Qin G, Zhang YH, Huang LM, Lin Q. Effects of water current on swimming performance, ventilation frequency, and feeding behavior of young seahorses (Hippocampus erectus). J Exp Mar Biol Ecol (2014) 461:337–43. doi: 10.1016/j.jembe.2014.09.001
32. Berlinberg AJ, Regner EH, Stahly A, Brar A, Reisz JA, Gerich ME, et al. Multi ‘omics analysis of intestinal tissue in ankylosing spondylitis identifies alterations in the tryptophan metabolism pathway. Front Immunol (2021) 12:587119. doi: 10.3389/fimmu.2021.587119
33. Wang JL, Zhang T, Shen XT, Liu J, Zhao DL, Sun YW, et al. Serum metabolomics for early diagnosis of esophageal squamous cell carcinoma by UHPLC-QTOF/MS. Metabolomics (2016) 12(7):116. doi: 10.1007/s11306-016-1050-5
34. Liu XM, Liu JN, Xiong K, Zhang CQ, Fang JK, Song J, et al. Effects of ocean acidification on molting, oxidative stress, and gut microbiota in juvenile horseshoe crab Tachypleus tridentatus. Front Physiol (2021) 12:813582. doi: 10.3389/fphys.2021.813582
35. Peng WJ, Huang JH, Yang JJ, Zhang ZY, Yu R, Fayyaz S, et al. Integrated 16S rRNA sequencing, metagenomics, and metabolomics to characterize gut microbial composition, function, and fecal metabolic phenotype in non-obese type 2 diabetic goto-kakizaki rats. Front Microbiol (2019) 10:3141. doi: 10.3389/fmicb.2019.03141
36. He ZH, Zhong YQ, Hou DQ, Hu XY, Fu ZB, Liu LY, et al. Integrated analysis of mRNA-seq and MiRNA-seq reveals the molecular mechanism of the intestinal immune response in Marsupenaeus japonicus under decapod iridescent virus 1 infection. Front Immunol (2021) 12:807093. doi: 10.3389/fimmu.2021.807093
37. Kim H, Kim M, Kim S, Lee YM, Shin SC. Characterization of antimicrobial resistance genes and virulence factor genes in an Arctic permafrost region revealed by metagenomics. Environ Pollut (2022) 294:118634. doi: 10.1016/j.envpol.2021.118634
38. Jang SE, Lim SM, Jeong JJ, Jang HM, Lee HJ, Han MJ, et al. Gastrointestinal inflammation by gut microbiota disturbance induces memory impairment in mice. Mucosal Immunol (2018) 11(2):369–79. doi: 10.1038/mi.2017.49
39. Ding WK, Shangguan YY, Zhu YQ, Sultan Y, Feng YY, Zhang B, et al. Negative impacts of microcystin-LR and glyphosate on zebrafish intestine: linked with gut microbiota and microRNAs? Environ Pollut (2021) 286:117685. doi: 10.1016/j.envpol.2021.117685
40. Ren SN, Wang CJ, Chen A, Lv WT, Gao RJ. The probiotic Lactobacillus paracasei ameliorates diarrhea cause by Escherichia coli O8via gut microbiota modulation1. Front Nutr (2022) 9:878808. doi: 10.3389/fnut.2022.878808
41. Kuo WT, Zuo L, Odenwald MA, Madha S, Singh G, Gurniak CB, et al. The tight junction protein ZO-1 is dispensable for barrier function but critical for effective mucosal repair. Gastroenterology (2021) 161(6):1924–39. doi: 10.1053/j.gastro.2021.08.047
42. Yuan B, Zhao LM, Zhuang ZX, Wang XR, Fu Q, Huang HB, et al. Transcriptomic and metabolomic insights into the role of the flgK gene in the pathogenicity of Pseudomonas plecoglossicida to orange-spotted grouper (Epinephelus coioides). Zool Res (2022) 43(6):952–65. doi: 10.24272/j.issn.2095-8137.2022.216
43. Li TT, Long M, Cheng J, Shen ZX, Gatesoupe FJ, Zhang XJ, et al. Alterations of the gut microbiome of largemouth bronze gudgeon (Coreius guichenoti) suffering from furunculosis. Sci Rep (2016) 6:30606. doi: 10.1038/srep30606
44. Gągała-Borowska I, Karwaciak I, Jaros D, Ratajewski M, Kokociński M, Jurczak T, et al. Cyanobacterial cell-wall components as emerging environmental toxicants-detection and holistic monitoring by cellular signaling biosensors. Sci Total Environ (2022) 807(Pt 2):150645. doi: 10.1016/j.scitotenv.2021.150645
45. Matilla MA, Krell T. The effect of bacterial chemotaxis on host infection and pathogenicity. FEMS Microbiol Rev (2018) 42(1):40–67. doi: 10.1093/femsre/fux052
46. Viswanathan VK, Edelstein PH, Pope CD, Cianciotto NP. The Legionella pneumophila iraAB locus is required for iron assimilation, intracellular infection, and virulence. Infect Immun (2000) 68(3):1069–79. doi: 10.1128/iai.68.3.1069-1079.2000
47. Caballero-Flores G, Pickard JM, Núñez G. Regulation of Citrobacter rodentium colonization: virulence, immune response and microbiota interactions. Curr Opin Microbiol (2021) 63:142–9. doi: 10.1016/j.mib.2021.07.003
48. Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science (2006) 312(5779):1526–30. doi: 10.1126/science.1128393
49. Gao TT, Ding MZ, Yang CH, Fan HY, Chai YR, Li Y. The phosphotransferase system gene ptsH plays an important role in MnSOD production, biofilm formation, swarming motility, and root colonization in Bacillus cereus 905. Res Microbiol (2019) 170(2):86–96. doi: 10.1016/j.resmic.2018.10.002
50. Murphy TF, Brauer AL, Johnson A, Kirkham C. ATP-binding cassette (ABC) transporters of the human respiratory tract pathogen, Moraxella catarrhalis: role in virulence. PloS One (2016) 11(7):e0158689. doi: 10.1371/journal.pone.0158689
51. Fu SL, Zhuang F, Guo L, Qiu YS, Xiong JL, Ye C, et al. Effect of baicalin-aluminum complexes on fecal microbiome in piglets. Int J Mol Sci (2019) 20(10):2390. doi: 10.3390/ijms20102390
52. Umasuthan N, Valderrama K, Vasquez I, Segovia C, Hossain A, Cao T, et al. A novel marine pathogen isolated from wild cunners (Tautogolabrus adspersus): comparative genomics and transcriptome profiling of Pseudomonas sp. strain J380. Microorganisms (2021) 9(4):812. doi: 10.3390/microorganisms9040812
53. Personnic N, Striednig B, Hilbi H. Legionella quorum sensing and its role in pathogen-host interactions. Curr Opin Microbiol (2018) 41:29–35. doi: 10.1016/j.mib.2017.11.010
54. Vich Vila A, Imhann F, Collij V, Jankipersadsing SA, Gurry T, Mujagic Z, et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci Transl Med (2018) 10(472):eaap8914. doi: 10.1126/scitranslmed.aap8914
55. Sevrin G, Massier S, Chassaing B, Agus A, Delmas J, Denizot J, et al. Adaptation of adherent-invasive E. coli to gut environment: impact on flagellum expression and bacterial colonization ability. Gut Microbes (2020) 11(3):364–80. doi: 10.1080/19490976.2017.1421886
56. Jagadeesan B, Koo OK, Kim KP, Burkholder KM, Mishra KK, Aroonnual A, et al. LAP, an alcohol acetaldehyde dehydrogenase enzyme in Listeria, promotes bacterial adhesion to enterocyte-like caco-2 cells only in pathogenic species. Microbiol (Reading) (2010) 156(Pt 9):2782–95. doi: 10.1099/mic.0.036509-0
57. Vander Broek CW, Chalmers KJ, Stevens MP, Stevens JM. Quantitative proteomic analysis of Burkholderia pseudomallei Bsa type III secretion system effectors using hypersecreting mutants. Mol Cell Proteomics (2015) 14(4):905–16. doi: 10.1074/mcp.M114.044875
58. Qiu JZ, Luo ZQ. Legionella and Coxiella effectors: strength in diversity and activity. Nat Rev Microbiol (2017) 15(10):591–605. doi: 10.1038/nrmicro.2017.67
59. Chico-Calero I, Suárez M, González-Zorn B, Scortti M, Slaghuis J, Goebel W, et al. Hpt, a bacterial homolog of the microsomal glucose-6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc Natl Acad Sci U.S.A. (2002) 99(1):431–6. doi: 10.1073/pnas.012363899
60. Cianciotto NP. An update on iron acquisition by Legionella pneumophila: new pathways for siderophore uptake and ferric iron reduction. Future Microbiol (2015) 10(5):841–51. doi: 10.2217/fmb.15.21
61. Su HC, Hu XJ, Xu WJ, Xu Y, Wen GL, Cao YC. Diversity, abundances and distribution of antibiotic resistance genes and virulence factors in the south China Sea revealed by metagenomic sequencing. Sci Total Environ (2022) 814:152803. doi: 10.1016/j.scitotenv.2021.152803
62. Bernard Md, D’Elios MM. The immune modulating activity of the Helicobacter pylori HP-NAP: friend or foe? Toxicon (2010) 56(7):1186–92. doi: 10.1016/j.toxicon.2009.09.020
63. Lee CR, Lee JH, Park M, Park KS, Bae IK, Kim YB, et al. Biology of Acinetobacter baumannii: pathogenesis, antibiotic resistance mechanisms, and prospective treatment options. Front Cell Infect Microbiol (2017) 7:55. doi: 10.3389/fcimb.2017.00055
64. Chakraborty S, Li M, Chatterjee C, Sivaraman J, Leung KY, Mok YK. Temperature and Mg2+ sensing by a novel PhoP-PhoQ two-component system for regulation of virulence in Edwardsiella tarda. J Biol Chem (2010) 285(50):38876–88. doi: 10.1074/jbc.M110.179150
65. Liou ML, Soo PC, Ling SR, Kuo HY, Tang CY, Chang KC. The sensor kinase BfmS mediates virulence in Acinetobacter baumannii. J Microbiol Immunol Infect (2014) 47(4):275–81. doi: 10.1016/j.jmii.2012.12.004
66. Guo HR, Zhao T, Huang C, Chen JY. The role of the two-component system PhoP/PhoQ in intrinsic resistance of Yersinia enterocolitica to polymyxin. Front Microbiol (2022) 13:758571. doi: 10.3389/fmicb.2022.758571
67. Creti R, Koch S, Fabretti F, Baldassarri L, Huebner J. Enterococcal colonization of the gastro-intestinal tract: role of biofilm and environmental oligosaccharides. BMC Microbiol (2006) 6:60. doi: 10.1186/1471-2180-6-60
68. Liu Y, Gao YN, Liu XH, Liu Q, Zhang YX, Wang QY, et al. Transposon insertion sequencing reveals T4SS as the major genetic trait for conjugation transfer of multi-drug resistance pEIB202 from Edwardsiella. BMC Microbiol (2017) 17(1):112. doi: 10.1186/s12866-017-1013-7
69. Santos L, Ramos F. Antimicrobial resistance in aquaculture: current knowledge and alternatives to tackle the problem. Int J Antimicrob Agents (2018) 52(2):135–43. doi: 10.1016/j.ijantimicag.2018.03.010
70. Rinschen MM, Ivanisevic J, Giera M, Siuzdak G. Identification of bioactive metabolites using activity metabolomics. Nat Rev Mol Cell Biol (2019) 20(6):353–67. doi: 10.1038/s41580-019-0108-4
71. Peng B, Ma YM, Zhang JY, Li H. Metabolome strategy against Edwardsiella tarda infection through glucose-enhanced metabolic modulation in tilapias. Fish Shellfish Immunol (2015) 45(2):869–76. doi: 10.1016/j.fsi.2015.06.004
72. Yuan J, Jin F, Glatter T, Sourjik V. Osmosensing by the bacterial PhoQ/PhoP two-component system. Proc Natl Acad Sci U.S.A. (2017) 114(50):E10792–E8. doi: 10.1073/pnas.1717272114
73. Mazé A, Benenson Y. Artificial signaling in mammalian cells enabled by prokaryotic two-component system. Nat Chem Bio (2020) 16(2):179–87. doi: 10.1038/s41589-019-0429-9
74. Chitrakar I, Kim-Holzapfel DM, Zhou WJ, French JB. Higher order structures in purine and pyrimidine metabolism. J Struct Biol (2017) 197(3):354–64. doi: 10.1016/j.jsb.2017.01.003
75. Wu GY, Fang YZ, Yang S, Lupton JR, Turner ND. Glutathione metabolism and its implications for health. J Nutr (2004) 134(3):489–92. doi: 10.1093/jn/134.3.489
76. Balihodzic A, Prinz F, Dengler MA, Calin GA, Jost PJ, Pichler M. Non-coding RNAs and ferroptosis: potential implications for cancer therapy. Cell Death Differ (2022) 29(6):1094–106. doi: 10.1038/s41418-022-00998-x
77. Wang C, Cui RN, Niu CY, Zhong XL, Zhu QC, Ji D, et al. Low-dose PCB126 exposure disrupts cardiac metabolism and causes hypertrophy and fibrosis in mice. Environ Pollut (2021) 290:118079. doi: 10.1016/j.envpol.2021.118079
78. Zhou J, Yao N, Wang SX, An DC, Cao KN, Wei JL, et al. Fructus Gardeniae-induced Gastrointestinal injury was associated with the inflammatory response mediated by the disturbance of vitamin B6, phenylalanine, arachidonic acid, taurine and hypotaurine metabolism. J Ethnopharmacol (2019) 235:47–55. doi: 10.1016/j.jep.2019.01.041
79. Vera-Aviles M, Vantana E, Kardinasari E, Koh NL, Latunde-Dada GO. Protective role of histidine supplementation against oxidative stress damage in the management of anemia of chronic kidney disease. Pharm (Basel) (2018) 11(4):111. doi: 10.3390/ph11040111
80. Canfora EE, Meex RCR, Venema K, Blaak EE. Gut microbial metabolites in obesity, NAFLD and T2DM. Nat Rev Endocrinol (2019) 15(5):261–73. doi: 10.1038/s41574-019-0156-z
81. Islam J, Tanimizu M, Shimizu Y, Goto Y, Ohtani N, Sugiyama K, et al. Development of a rational framework for the therapeutic efficacy of fecal microbiota transplantation for calf diarrhea treatment. Microbiome (2022) 10(1):31. doi: 10.1186/s40168-021-01217-4
82. Jang J, Cho MH, Chun JH, Cho MH, Park JC, Oh HB, et al. The poly-γ-D-glutamic acid capsule of Bacillus anthracis enhances lethal toxin activity. Infect Immun (2011) 79(9):3846–54. doi: 10.1128/iai.01145-10
83. Tan YP, Zheng J, Tung SL, Rosenshine I, Leung KY. Role of type III secretion in Edwardsiella tarda virulence. Microbiol (Reading) (2005) 151(Pt 7):2301–13. doi: 10.1099/mic.0.28005-0
84. Christgen SL, Becker DF. Role of proline in pathogen and host interactions. Antioxid Redox Signal (2019) 30(4):683–709. doi: 10.1089/ars.2017.7335
85. Munia NS, Hosen MA, Azzam KMA, Al-Ghorbani M, Baashen M, Hossain MK, et al. Synthesis, antimicrobial, SAR, PASS, molecular docking, molecular dynamics and pharmacokinetics studies of 5’-O-uridine derivatives bearing acyl moieties: POM study and identification of the pharmacophore sites. Nucleosides, Nucleotides Nucleic Acids (2022) 41(10):1036–83. doi: 10.1080/15257770.2022.2096898
86. Hou YQ, Wu GY. L-Glutamate nutrition and metabolism in swine. Amino Acids (2018) 50(11):1497–510. doi: 10.1007/s00726-018-2634-3
87. Rodrigues RR, Gurung M, Li ZP, García-Jaramillo M, Greer R, Gaulke C, et al. Transkingdom interactions between Lactobacilli and hepatic mitochondria attenuate western diet-induced diabetes. Nat Commun (2021) 12(1):101. doi: 10.1038/s41467-020-20313-x
88. Hansen MR, Dandanell G. Purification and characterization of RihC, a xanthosine-inosine-uridine-adenosine-preferring hydrolase from Salmonella enterica serovar typhimurium. Biochim Biophys Acta (2005) 1723(1-3):55–62. doi: 10.1016/j.bbagen.2005.01.012
Keywords: Edwardsiella piscicida, metagenome, metabolome, virulence factor, big-belly seahorse, pathogenesis
Citation: Zhang L, Wang F, Jia L, Yan H, Gao L, Tian Y, Su X, Zhang X, Lv C, Ma Z, Xue Y, Lin Q and Wang K (2023) Edwardsiella piscicida infection reshapes the intestinal microbiome and metabolome of big-belly seahorses: mechanistic insights of synergistic actions of virulence factors. Front. Immunol. 14:1135588. doi: 10.3389/fimmu.2023.1135588
Received: 01 January 2023; Accepted: 14 April 2023;
Published: 03 May 2023.
Edited by:
Chou Min Chong, Putra Malaysia University, MalaysiaCopyright © 2023 Zhang, Wang, Jia, Yan, Gao, Tian, Su, Zhang, Lv, Ma, Xue, Lin and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kai Wang, d2FuZ2t5dEAxNjMuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.