
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Immunol., 15 March 2023
Sec. Viral Immunology
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1129190
This article is part of the Research TopicChanges in T cell populations and cytokine production in SARS-CoV-2 infected individuals; their role in prognosisView all 20 articles
 Gerasimos Eleftheriotis1
Gerasimos Eleftheriotis1 Efthymios P. Tsounis2
Efthymios P. Tsounis2 Ioanna Aggeletopoulou2,3
Ioanna Aggeletopoulou2,3 Periklis Dousdampanis4
Periklis Dousdampanis4 Christos Triantos2
Christos Triantos2 Athanasia Mouzaki3
Athanasia Mouzaki3 Markos Marangos1
Markos Marangos1 Stelios F. Assimakopoulos1*
Stelios F. Assimakopoulos1*Although coronavirus disease 2019 (COVID-19) is primarily associated with mild respiratory symptoms, a subset of patients may develop more complicated disease with systemic complications and multiple organ injury. The gastrointestinal tract may be directly infected by SARS-CoV-2 or secondarily affected by viremia and the release of inflammatory mediators that cause viral entry from the respiratory epithelium. Impaired intestinal barrier function in SARS-CoV-2 infection is a key factor leading to excessive microbial and endotoxin translocation, which triggers a strong systemic immune response and leads to the development of viral sepsis syndrome with severe sequelae. Multiple components of the gut immune system are affected, resulting in a diminished or dysfunctional gut immunological barrier. Antiviral peptides, inflammatory mediators, immune cell chemotaxis, and secretory immunoglobulins are important parameters that are negatively affected in SARS-CoV-2 infection. Mucosal CD4+ and CD8+ T cells, Th17 cells, neutrophils, dendritic cells, and macrophages are activated, and the number of regulatory T cells decreases, promoting an overactivated immune response with increased expression of type I and III interferons and other proinflammatory cytokines. The changes in the immunologic barrier could be promoted in part by a dysbiotic gut microbiota, through commensal-derived signals and metabolites. On the other hand, the proinflammatory intestinal environment could further compromise the integrity of the intestinal epithelium by promoting enterocyte apoptosis and disruption of tight junctions. This review summarizes the changes in the gut immunological barrier during SARS-CoV-2 infection and their prognostic potential.
Although the primary target of severe acute respiratory syndrome coronavirus-2 (SARS−CoV−2) is type II alveolar epithelial cells, the virus can also infect gastrointestinal mucosal cells by binding to angiotensin-converting enzyme 2 (ACE2) and its cofactor, transmembrane serine protease 2 (TMPRSS2), both of which are widely expressed on the surface of enterocytes. Notably, ACE2 expression is higher in intestinal cells of the ileum and colon than in lung cells (1). In experimental models, intranasal inoculation of SARS-CoV-2 resulted in disruption of gut barrier integrity as a consequence of systemic release of proinflammatory mediators (2). Therefore, the virus can cause either direct damage to ACE-2-expressing intestinal epithelial cells or indirect damage through a systemic hyperinflammatory response (2, 3). Recombination is a common mechanism in coronaviruses that allows them to resist selective pressure and adapt to new habitats. Recombination events with other gut-targeting coronaviruses could potentially enhance SARS-CoV-2 virulence and tropism for the gastrointestinal tract (4). Indeed, up to 24% of COVID-19 patients develop gastrointestinal symptoms, including diarrhea, abdominal discomfort, nausea, vomiting, and loss of appetite (3, 5). Notably, COVID-19 patients may even develop severe duodenitis and present with gastrointestinal bleeding requiring red blood cell transfusion (6). Immunohistochemical staining of these biopsies was positive for SARS-CoV-2 spike protein, suggesting that duodenitis developed as a result of direct enterocyte invasion by SARS-CoV-2; in situ hybridization also provided evidence of active viral replication (6). SARS-CoV-2 infection is associated with multifactorial impairment of the gut barrier, as it has deleterious effects on all of its critical aspects of defense, which consist of a balance between the gut microbiota (biological barrier), intestinal epithelial cells and their junctions (mechanical barrier), and gut-associated immune cells, immunoglobulins, and cytokine production (immune barrier). Previous studies have shown that the integrity of the intestinal barrier is significantly impaired in COVID-19 patients, as evidenced by various surrogate markers. Giron et al. (7) demonstrated that severe COVID-19 is associated with higher plasma levels of zonulin, indicating profound disruption of tight junction homeostasis, as well as increased levels of lipopolysaccharide (LPS)-binding protein (LBP) and β-glucan, which are reliable markers of bacterial and fungal translocation, respectively. Importantly, serum markers of tight junction permeability and microbial translocation were significantly associated with circulating proinflammatory mediators such as IL-6, suggesting that systemic inflammation is triggered to some extent by gut barrier disruption. In addition, intestinal fatty acid binding protein (I-FABP), a protein synthesized by mature enterocytes responsible for fatty acid turnover and used as a biomarker of intestinal injury, was measured in the urine of 283 patients hospitalized for COVID-19 (8, 9). Urinary I-FABP levels were significantly elevated compared with controls and remained high in patients’ samples two weeks after hospitalization; levels were even higher in patients with critical illness than in milder cases (8). Another study found that serum levels of zonula occludins-1 (ZO-1), a marker of structural and functional integrity of the paracellular barrier, were significantly elevated in patients with COVID-19 pneumonia but were not predictive of progression to severe respiratory failure (10). In addition, numerous studies have shown significant changes in the composition of the gut microbiota in patients with COVID-19. A recent meta-analysis detailed the changes in gut microbiota composition during SARS-CoV-2 infection (11). In several studies, changes in the gut microbiome were also closely associated with the clinical severity of COVID-19, suggesting a prognostic role in such patients (12–24). The present review focuses specifically on COVID-19 associated changes in the gut immunological barrier and their prognostic potential.
Fecal calprotectin has also been studied in detail in COVID-19 patients because it is produced predominantly by neutrophils that migrate to and are activated in the intestine, making it a reliable marker of bowel inflammation in other conditions such as inflammatory bowel disease (25). Ojetti et al. (26) showed that high fecal calprotectin levels in SARS-CoV-2 infected individuals are an independent risk factor for the development of COVID-19 pneumonia, while data from other studies indicate that fecal calprotectin levels are positively correlated with serum IL-6, degree of hypoxemia, and days of hospitalization (27–29). Although there is no correlation between gastrointestinal symptoms and fecal calprotectin, an increase in this parameter has a better predictive value for progression of severe disease compared with C-reactive protein (30). This finding possibly suggests that the increase in fecal calprotectin associated with SARS-CoV-2 infection is due in part to chemotaxis of immune cells into to the gastrointestinal tract and hypoxic intestinal damage rather than intestinal inflammation and destruction of enterocytes (29, 30). In addition, the role of serum calprotectin was also investigated and was found to be an effective marker for predicting the future status of SARS-CoV-2-infected individuals (31). The strong correlation of serum calprotectin with poor clinical outcomes highlights the potential value of this marker in identifying COVID-19 patients at high risk for disease progression (31).
Data are also available on changes in immune cells in the gastrointestinal mucosa of patients infected with SARS-CoV-2. Mass cytometric analysis of intestinal tissue from deceased individuals with COVID-19 revealed leukocytic infiltration consisting of monocytes, CD11b+ macrophages, CD11c+ dendritic cells (DCs), natural killer (NK) cells, and B cells (32). Similar results were obtained from lung tissue, suggesting that these organs are the epicenter of the immune response during SARS-CoV-2 infection (32). Moreover, analysis of duodenal biopsies after the onset of COVID-19 symptoms in patients with macroscopically normal mucosa who underwent endoscopy for other reasons (e.g., upper abdominal pain) revealed increased numbers of CD68+, CD14+ macrophages, CD11c+ DCs, mucosal CD4+ T cells, and intraepithelial CD8+ T cells (33). The aforementioned intraepithelial lymphocytes were antigen-experienced and exhibited a CD8+ effector cell phenotype (CD45RA+, CD27-) (33). An increase in intraepithelial lymphocytes in intestinal biopsies was still observed one month after SARS-CoV-2 infection (34). Exhaustion/depletion of CD4+ T cells, a hallmark of HIV infection, is also observed in SARS-CoV-2 infection. The resulting dysregulation of CD4+ T cells in the gut may contribute to intestinal epithelial barrier dysfunction and leaky gut, which promotes systemic inflammation (35). Accordingly, IL-17 producing Th17 cells are overactivated in SARS-CoV-2 infection (36).
Invasion of SARS-CoV-2 into intestinal cells leads to increased expression of type I and III IFN and other proinflammatory cytokines, such as IL-8 and IL-12 (37, 38). All types of IFN can activate the JAK/STAT pathway. Type I IFN can be secreted by many cell types, especially plasmacytoid DCs. SARS-CoV-2 has developed several strategies to evade immune surveillance by attenuating type I and III IFN responses (39). This effect is mediated by a SARS-CoV-2 membrane protein that inhibits the formation of a multiprotein complex responsible for the phosphorylation and subsequent activation of IFN regulatory factor (IRF) 3 (39). IRF3 activation is a prerequisite for IFN transcription and synthesis. The SARS-CoV-2 accessory protein ORF9b is another molecule that blocks the IRF3 activation pathway (40). ORF9b also inhibits IFN gene expression by interacting with the stimulator of IFN genes (STING); STING can recruit TANK binding kinase 1 (TBK1), one of the IRF3 phosphorylators (40). In addition, ORF9b targets the translocase of outer mitochondrial membrane 70 (TOMM70), which is located on the mitochondrial membrane and functions as a receptor for mitochondrial antiviral signaling protein (MAVS). MAVS is also involved in the IRF3 phosphorylation pathway. Therefore, ORF9b downregulates type I IFN production by interfering with the interaction between TOMM70 and MAVS (41, 42). However, overexpression of TOMM70 can overcome ORF9b-mediated inhibition and restore IFN-β expression (41).
After pretreatment of human intestinal cell lines with IFN-β and human colon organoids with IFN-β1 and type III IFN, respectively, a protective effect against SARS-CoV-2 infection was observed, resulting in a significantly milder infection (43, 44). However, the antiviral activity of type III IFN against SARS-CoV-2 in the gut is stronger and more durable (45). Of note, infection of colon organoids with SARS-CoV-2 resulted in upregulation of type III IFN but not type I IFN, despite the ability of these organoids to produce both types in response to viral infection (44, 46). Conversely, depletion of the type III IFN receptor resulted in increased SARS-CoV-2 infectivity, viral genome replication, and virion production (44, 45).
Although neutralizing autoantibodies to type I IFN have been previously demonstrated in humans and are a universal finding, particularly in autoimmune polyendocrine syndrome type 1 (APS-1), they have not been associated with increased prevalence or severity in viral infections (47–50). SARS-CoV-2 infection appears to be an exception; in an international cohort study, 19 of 22 APS-1 patients with COVID-19 were hospitalized, and 11 of 22 required mechanical ventilation (51). Pre-existing neutralizing IFN type I antibodies in the serum of previously healthy individuals represented a major risk factor for severe COVID-19 (51). Moreover, their prevalence increases with age; during SARS-CoV-2 infection, 10.2% of patients with life-threatening disease, almost exclusively men, had pre-existing neutralizing IFN-I autoantibodies and low or undetectable serum IFN-α levels, whereas no such antibodies were detected in patients with mild infection (52, 53). In addition, COVID-19 patients with type I IFN antibodies had significantly higher viral loads than patients without these antibodies (54). Similar results were obtained in other studies investigating the presence of neutralizing IFN-I antibodies in patients with COVID-19 in the intensive care unit (55, 56). Such autoantibodies were detected in 9.5% to 18% of these patients; 87% to 92.3% of them were men (55, 57). In contrast, non-neutralizing IFN-I antibodies are common in critically ill non-COVID-19 patients and do not affect clinical outcome (58). The aforementioned data provide an additional explanation for the increased likelihood of severe disease in elderly men; notably, this group of individuals accounts for approximately 20% of COVID-19 deaths (53).
Furthermore, impairment of IFN-I-dependent immunity caused by any mechanism can lead to severe COVID-19 symptoms. Loss-of-function variants of genes such as IRF3, IRF7, IFN-α receptor, and Toll-like receptor 3 (TLR3) were detected in 3.5% of individuals with life-threatening COVID-19 and no history of other severe infections, whereas no patient with mild or asymptomatic disease carried these variants; all of these variants resulted in disproportionately low IFN I production in response to SARS-CoV-2 (59).
Following intranasal infection with SARS-CoV-2, cytokines such as IL-4, IL-1β, TNF-α, IL-17A, and other inflammatory mediators are initially produced in gastrointestinal tissues (2). In parallel, upregulation of the anti-inflammatory IL-10 and inhibition of the pro-inflammatory IL-1β and IFN-γ can be induced by inoculation in the digestive tract (2). A gut-on-a-chip model of SARS-CoV-2 infection provided further evidence for the release of cytokines in the digestive tract (60). In particular, the IL-6 and TNF genes and C-X-C motif chemokine ligand 10 (CXCL10), a chemoattractant for NK cells and T cells and a monocyte inducer, were significantly upregulated (60). Gene set enrichment analysis in pluripotent stem cells derived from small intestinal epithelial cells or intestinal organoids from SARS-CoV-2 infection models also revealed increased expression of IL-1β, IL-6, CXCL10, C-C motif chemokine ligand 5 (CCL5, chemoattractant for monocytes), and significant upregulation of IL-6 and the nuclear factor-κB (NF-κB) pathway (61, 62). Quantification of cytokines in the stool of patients hospitalized for COVID-19 revealed higher IL-8, IL-18, and lower IL-10 levels (63, 64).
In contrast, treatment of human colon tissue samples with short-chain fatty acids (SCFAs), metabolites capable of reducing pro-inflammatory mediators such as IL-6, IL-12, and IFN-γ, showed no effect on cell permeability; however, treatment with SCFAs showed a modest, albeit significant, effect in reducing the expression of the type III IFN receptor, interferon lambda receptor 1 (IFNLR1), and the serine protease TMPRSS2 (56, 65). TMPRSS2 is a membrane-bound protein that has been shown to promote SARS-CoV-2 infection in enteroids by supporting virus-enterocyte fusion (66). Of note, depletion of the gut microbiota after antibiotic administration did not affect mortality in a mouse model of SARS-CoV-2 infection although colonic concentrations of IL-17 and CXCL2 were significantly increased (67). In contrast, administration of remdesivir to SARS-CoV-2-infected intestinal epithelial cells resulted in reduced induction of the IL-1β, IL-6, CXCL10, and CCL5 genes (62).
Another protein inversely correlated with serum IL-6 levels during SARS-CoV-2 infection is soluble mucosal addressin cell adhesion molecule (sMAdCAM), which is expressed by gut endothelial venules to induce migration of immune cells into the intestine (68, 69). At the same time, sMAdCAM levels were lower in COVID-19 patients compared to healthy controls or convalescent subjects, suggesting that normalization of sMAdCAM levels may signify the restoration of mucosal homeostasis and highlighting its role as an important systemic and gut homing parameter that needs to be monitored for better therapeutic guidance and prophylactic intervention in COVID-19 (68, 69).
Regarding the antiviral response of the gastrointestinal tract, Paneth cells and neutrophils are also capable of producing the immunomodulatory proteins, defensins. Defensins are important members of the antimicrobial peptide (AMP) family with diverse immunoregulatory functions and a broad spectrum of antimicrobial and antiviral effects. These proteins act as chemoattractants and activators for immature DCs, monocytes, and naive T cells (70, 71). During SARS-CoV-2 infection, α-defensin 5, a lectin-like protein that can recognize lipids and glycoproteins, shields the ACE2 receptor and prevents SARS-CoV-2 binding (72). Although SARS-CoV-2 has a higher affinity for the ACE2 receptor than α-defensin 5 (72), intestinal α-defensin 5 has a protective effect because it is highly abundant in the digestive tract. Consequently, α-defensin 5 levels are elevated before infection (72). Indeed, administration of α-defensin 5 to a cell line model after infection resulted in no antiviral response, whereas pretreatment with α-defensin 5 showed a beneficial effect (73). Of note, β-defensin 1 production is increased in later stages of SARS-CoV-2 infection due to intestinal hypoxia mediated by hypoxia-inducible factor 1α (74).
The immunologic barrier of the gut is also strengthened by secretory immunoglobulin A (sIgA) produced by mucosal lymphoid tissues, including gut-associated lymphoid tissue (GALT). Dimeric sIgAs are the predominant mucosal antibodies and form an essential component of the immunologic barrier (75). Commensal microorganisms play a central role in controlling IgA class switching and effective antibody production. Indeed, the number of functional IgA-secreting B cells is drastically reduced in germ-free animal models (76, 77). The predominance of sIgA in the intestine is likely another explanation for the attenuated gut inflammation compared with lung tissue; IgA dimers are able to inactivate toxins or pathogens without inducing inflammation because they cannot bind and activate complement (78). An in vitro study examining the neutralizing ability of IgG and IgA from B cells of COVID-19 convalescent subjects found that dimeric IgA was much more effective than IgA monomers or IgG in neutralizing SARS-CoV-2 (79).
The constant interaction of immune cells with the gut microbiome maintains the balance between tolerance to beneficial bacteria and eradication of pathogenic species (80). A complex, dynamic, and bilateral interaction between the gut microbiome and COVID-19 has been described (81). The gut microbiome of patients with SARS-CoV-2 infection exhibits significant alterations, possibly due to a severe systemic inflammatory response. The mechanisms underlying COVID-19-related dysbiosis are still unclear. However, interactions between the ACE2 receptor and SARS-CoV-2 have been associated with alterations in the composition of the gut microbiota by impairing the secretion AMPs. The function of the amino acid transporter B0AT1, which mediates intestinal uptake of tryptophan, is dependent on the ACE2 pathway (82). Tryptophan modulates the production of AMPs via the mammalian target of rapamycin (mTOR) pathway (83). Therefore, the deficiency of tryptophan caused by ACE-2 blockade may decrease the production of AMPs and disrupt the intraluminal microbial species. Commensal bacteria also play a critical role in mucosal homeostasis by modulating the expression of ACE2 in the gut (84). Secretion of proinflammatory cytokines, particularly TNF-α, during respiratory tract infections has a dynamic anorexigenic effect via hypothalamic activity. The decrease in fiber and caloric intake disrupts the composition of the gut microbiota and the synthesis of its metabolites, which in turn strongly influence the transcriptional “training” of innate immune cells (85).
A recent meta-analysis detailed the changes in the gut microbiota during SARS-CoV-2 infection (11). At the phylum level, dysbiosis is characterized by a reduction in the ratio of Firmicutes to Bacteroidetes. In particular, COVID-19 is associated with fewer butyrate-producing bacterial species, including Faecalibacterium and Roseburia (11, 15, 86). The genus Roseburia is closely associated with colon motility and mucosal tissue integrity and has a crucial anti-inflammatory effect by regulating IL-10 synthesis (87). Several other beneficial genera, including Eubacterium, Alistipes, and Bifidobacterium, are also reduced in COVID-19 patients (11). Bifidobacterium strains mediate robust antimicrobial and antiviral activity, which is balanced by promotion of Treg-mediated responses and induction of tolerogenic DC phenotypes (88).
Alterations in the gut microbiome have also been closely associated with clinical severity of COVID-19 in several studies, suggesting a prognostic role in such patients (12–24). Bacterial genera with significant prognostic value included Eubacterium, Ruminococcus, Faecalibacterium, Bacteroides, Lactobacillus, Clostridium, Roseburia, and Bifidobacterium (12–24). An increase in the dominant genus Enterococcus and a decrease in the families Ruminococcaceae and Lachnospiraceae have been reported in severe COVID-19 cases admitted to the intensive care unit (21).
The changes in the gut microbiota in patients with COVID-19 should be considered as a dynamic process (81). Emerging evidence suggests that the regulatory functions of the gut microbiota effectively support recovery from SARS-CoV-2 infection. The main features of intestinal immune barrier disruption during SARS-CoV-2 infection are shown in Figure 1.
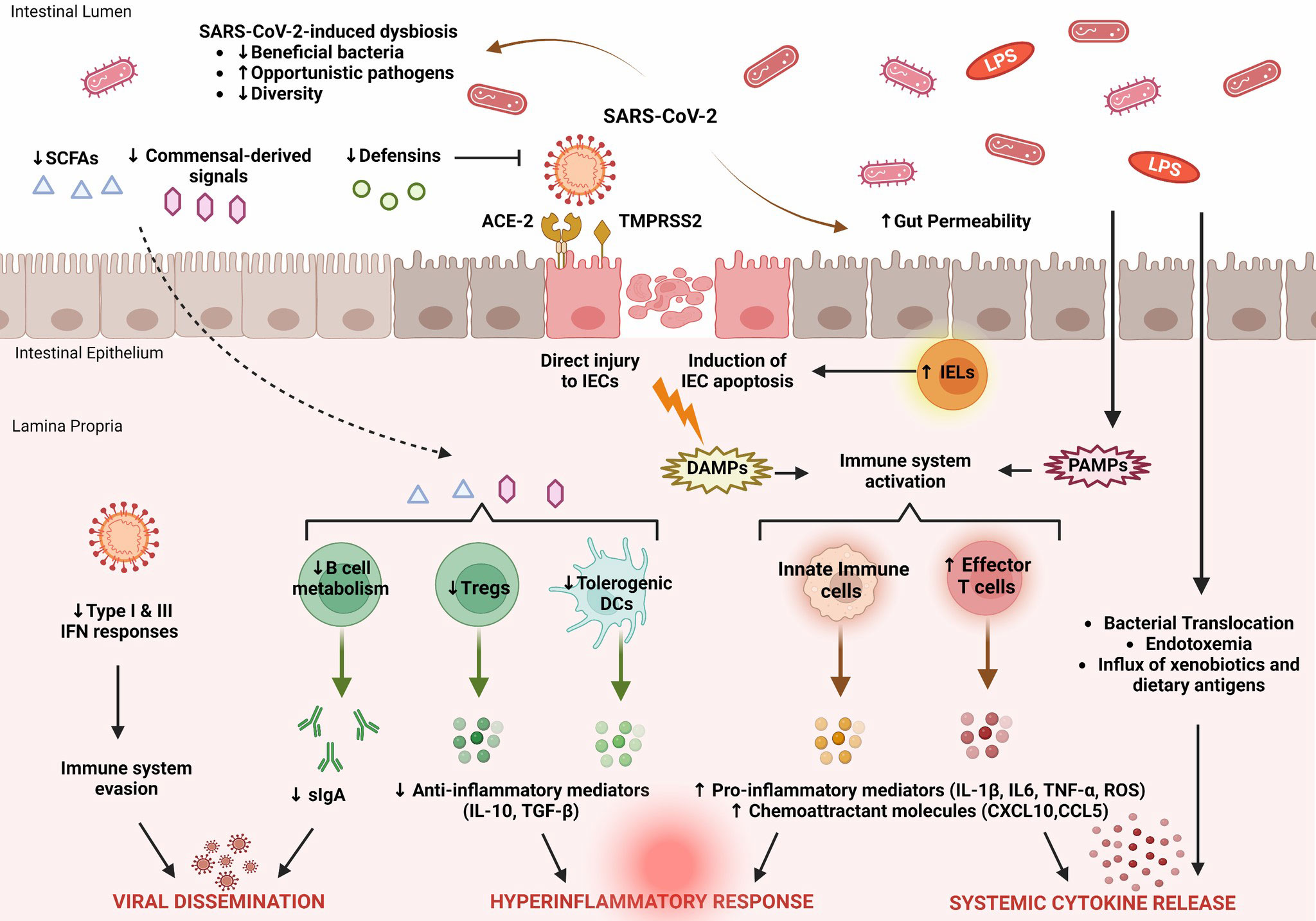
Figure 1 Key features of intestinal immune barrier disruption during SARS-CoV-2 infection. SARS-CoV-2 infection is associated with profound alterations in the intestinal microflora, manifested by decreased species diversity, depletion of symbiotic microorganisms, and prevalence of pathogenic species. Signals and metabolites derived from the intestinal flora, such as short-chain fatty acids (SCFAs), play an important role in controlling mucosal immunity by promoting T regulatory cell (Treg) responses and the activity of tolerogenic dendritic cells (DCs). This immunoregulatory environment, rich in anti-inflammatory mediators (IL-10, TGF-β), is significantly impaired by SARS-CoV-2. As a result, B cell metabolism and maturation are severely impaired, leading to exhaustion of effective plasma cells that produce secretory dimeric immunoglobulin A (sIgA), which is essential for viral containment. The proliferation of SARS-CoV-2 is also facilitated by its ability to evade recognition by the immune system by interfering with type I and type III IFN signaling. SARS-CoV-2 exerts either direct cytopathic effects on intestinal epithelial cells (IECs) expressing ACE2 and TMPRSS2 receptors or indirect immune-mediated injury. During COVID-19, the expression of several antimicrobial peptides, including defensins, is dysregulated, which increases the infectivity of SARS-CoV-2. In addition, recruitment of intraepithelial lymphocytes (IELs) accelerates IEC apoptosis. The release of damage-associated molecular patterns (DAMPs) due to cell injury and the influx of pathogen-associated molecular patterns (PAMPs) as a result of increased gut permeability lead to immune activation. Macrophages/monocytes, neutrophils, and other cells of the innate immune system secrete large amounts of proinflammatory mediators (IL-1β, IL-6, TNF-α, ROS) and chemokines (CCL5, CXCL10) that cause recruitment of additional immune cells and prime effector T cells. In parallel, disruption of the intestinal barrier facilitates bacterial translocation, endotoxemia, and dissemination of other gut-derived stimuli that contribute to systemic hyperinflammatory responses and cytokine release syndrome, leading to severe COVID-19.
SARS-CoV-2 affects multiple systems, including the gastrointestinal tract and gut barrier integrity. COVID-19 is associated with multifaceted disruption of the various components of the mucosal immune barrier, and the extent of these changes reflects the severity of the underlying disease. In particular, SARS-CoV-2 is able to evade the innate immune response by disrupting interferon signaling. The expression pattern of several cytokines in the mucosal compartment is severely affected, essentially leading to the recruitment and activation of additional immune cells that support this proinflammatory milieu. In addition, the production and release of antimicrobial peptides and secretory IgA, which are important regulators of intestinal immune integrity, are impaired. As a result, profound alterations of the gut microbiome and metabolome occur, characterized by depletion of symbiotic species and dominance of pathogenic microorganisms. The main features of SARS-CoV-2-induced dysregulation of the intestinal immune barrier are shown in Figure 1.
The prognostic value of various parameters related to the gut immunological barrier was evaluated. A strong association was described between certain prognostic factors and disease severity, poor prognosis, hospitalization, or mortality due to SARS-CoV-2. Table 1 summarizes important parameters related to the gut immunologic barrier that have been studied as prognostic markers for severity and progression of SARS-CoV-2 infection. It is unlikely that a single index of gut immunologic barrier function can independently predict progression of COVID-19. Alternatively, the development and validation of a prognostic scoring system that incorporates the most robust immunologic parameters and combines them with additional epidemiologic, clinical, and laboratory data may provide the best results. Further prospective studies with larger numbers of participants are warranted to identify markers of gut barrier dysfunction that could help identify high-risk COVID-19 patients who require early or enhanced support.
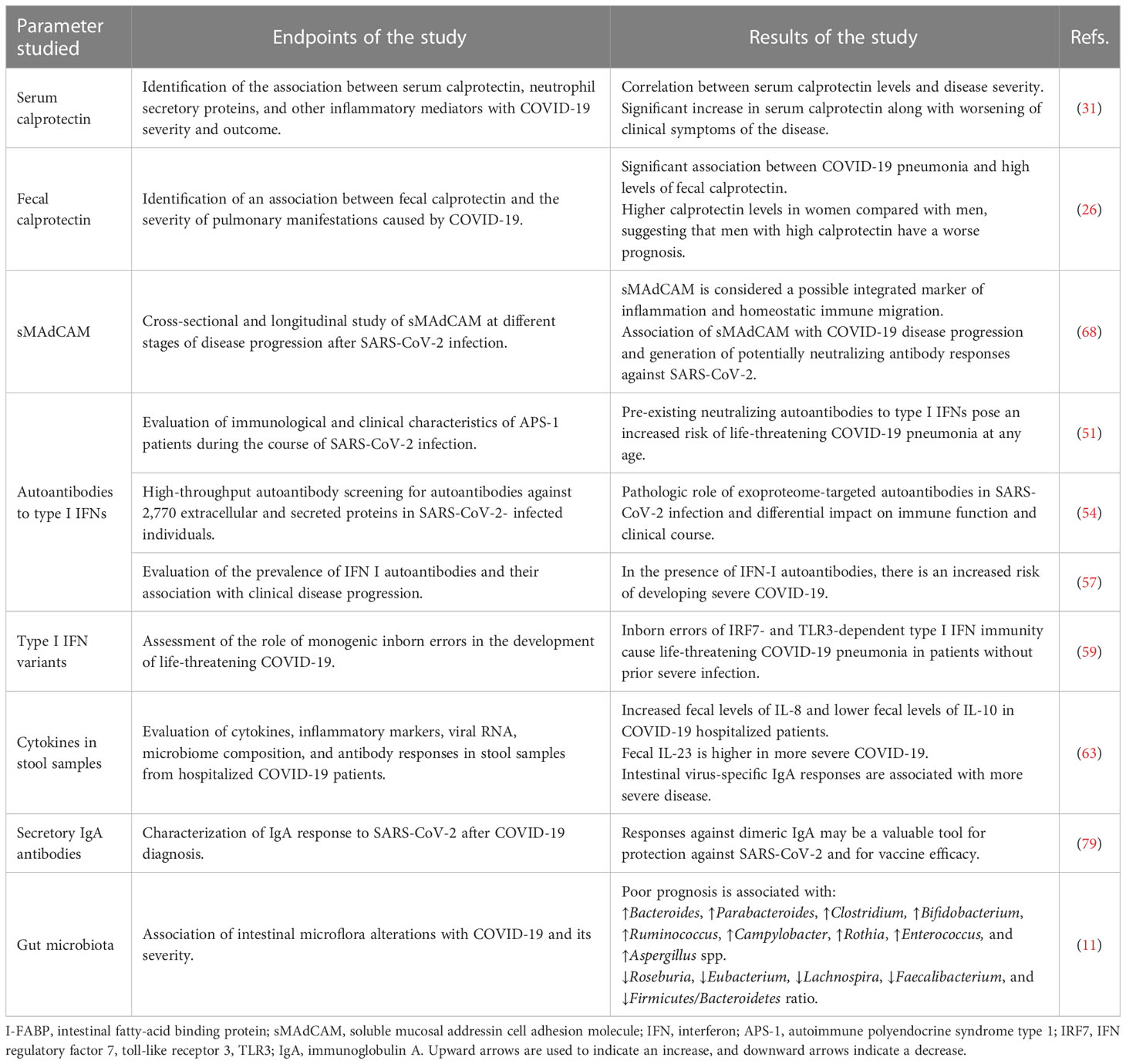
Table 1 Key gut immunologic barrier parameters that serve as prognostic markers for severity of SARS-CoV-2 infection and poor prognosis.
SARS-CoV-2 infection is associated with significant disruption of intestinal immunological homeostasis and impairs mucosal immune cell function and production of signaling molecules. SARS-CoV-2-induced gut dysbiosis could drive many of these immunological changes through commensal-derived signals and metabolites that maintain a continuous dialog between the mucosal immune system and the gut microflora. Conversely, dysregulation of intestinal immune cells and overproduction of proinflammatory cytokines could compromise the integrity of intestinal epithelial cells (apoptosis induction) and their connections (disruption of tight junctions), further promoting gut barrier dysfunction. These alterations contribute to the breakdown of intestinal barrier integrity, which may subsequently lead to translocation of microbes and endotoxins from the intestinal lumen into the systemic circulation, promoting a hyperinflammatory response associated with distant organ dysfunction and the development of a “viral sepsis syndrome” (89). The importance of gut immunologic barrier alterations in COVID-19 is underscored by several studies demonstrating their prognostic potential. Features of intestinal immune barrier failure occur early in the course of infection and correlate well with the severity of COVID-19, suggesting that immune barrier dysfunction is not only a bystander but an active participant in fueling exuberant immune responses and systemic inflammation. Further clinical studies are needed to explore the role of appropriate biomarker-based immunologic therapies in improving gut barrier function, which could lead to an expansion of therapeutic options against COVID-19.
SFA, conceptualization; GE, PD, and CT, review of bibliography; GE, ET, and IA, preparation of draft; ET and IA, preparation of figure; GE, PD, CT, MM, SFA, and AM, preparation of revised version of manuscript, review and editing of manuscript; all authors were responsible for revising the manuscript for important intellectual content. All authors contributed to the article and approved the submitted version.
The publication fees of this manuscript has been financed by the Research Council of the University of Patras.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Guimarães Sousa S, Kleiton de Sousa A, Maria Carvalho Pereira C, Sofia Miranda Loiola Araújo A, de Aguiar Magalhães D, Vieira de Brito T, et al. Sars-Cov-2 infection causes intestinal cell damage: Role of interferon's imbalance. Cytokine (2022) 152:155826. doi: 10.1016/j.cyto.2022.155826
2. Jiao L, Li H, Xu J, Yang M, Ma C, Li J, et al. The gastrointestinal tract is an alternative route for sars-Cov-2 infection in a nonhuman primate model. Gastroenterology (2021) 160(5):1647–61. doi: 10.1053/j.gastro.2020.12.001
3. Kariyawasam JC, Jayarajah U, Riza R, Abeysuriya V. Seneviratne SL gastrointestinal manifestations in covid-19. Trans R Soc Trop Med Hygiene (2021) 115(12):1362–88. doi: 10.1093/trstmh/trab042
4. Focosi D, Maggi FJV. Recombination in coronaviruses, with a focus on sars-Cov-2. Viruses (2022) 14(6):1239. doi: 10.3390/v14061239
5. Dorrell RD, Dougherty MK, Barash EL, Lichtig AE, Clayton SB, Jensen ETJJO. Gastrointestinal and hepatic manifestations of covid-19: A systematic review and meta-analysis. JGH Open (2021) 5(1):107–15. doi: 10.1002/jgh3.12456
6. Neuberger M, Jungbluth A, Irlbeck M, Streitparth F, Burian M, Kirchner T, et al. Duodenal tropism of sars-Cov-2 and clinical findings in critically ill covid-19 patients. Infection (2022) 50(5):1111–20. doi: 10.1007/s15010-022-01769-z
7. Giron LB, Dweep H, Yin X, Wang H, Damra M, Goldman AR, et al. Plasma markers of disrupted gut permeability in severe covid-19 patients. Front Immunol (2021) 12:686240. doi: 10.3389/fimmu.2021.686240
8. Saia RS, Giusti H, Luis-Silva F, Pedroso KJB, Auxiliadora-Martins M, Morejón KML, et al. Clinical investigation of intestinal fatty acid-binding protein (I-fabp) as a biomarker of sars-Cov-2 infection. Int J Infect Dis (2021) 113:82–6. doi: 10.1016/j.ijid.2021.09.051
9. Pelsers MM, Namiot Z, Kisielewski W, Namiot A, Januszkiewicz M, Hermens WT, et al. Intestinal-type and liver-type fatty acid-binding protein in the intestine. Tissue Distribution Clin Utility. Clin Biochem (2003) 36(7):529–35. doi: 10.1016/s0009-9120(03)00096-1
10. Assimakopoulos SF, Mastronikolis S, DE Lastic AL, Aretha D, Papageorgiou D, Chalkidi T, et al. Intestinal barrier biomarker Zo1 and endotoxin are increased in blood of patients with covid-19-Associated pneumonia. In Vivo (2021) 35(4):2483–8. doi: 10.21873/invivo.12528
11. Farsi Y, Tahvildari A, Arbabi M, Vazife F, Sechi LA, Shahidi Bonjar AH, et al. Diagnostic, prognostic, and therapeutic roles of gut microbiota in covid-19: A comprehensive systematic review. Front Cell Infect Microbiol (2022) 182. doi: 10.3389/fcimb.2022.804644
12. Tang L, Gu S, Gong Y, Li B, Lu H, Li Q, et al. Clinical significance of the correlation between changes in the major intestinal bacteria species and covid-19 severity. Eng (Beijing) (2020) 6(10):1178–84. doi: 10.1016/j.eng.2020.05.013
13. Zuo T, Zhang F, Lui GCY, Yeoh YK, Li AYL, Zhan H, et al. Alterations in gut microbiota of patients with covid-19 during time of hospitalization. Gastroenterology (2020) 159(3):944–55.e8. doi: 10.1053/j.gastro.2020.05.048
14. Zuo T, Liu Q, Zhang F, Lui GC, Tso EY, Yeoh YK. Depicting sars-Cov-2 faecal viral activity in association with gut microbiota composition in patients with covid-19. Gut (2021) 70(2):276–84. doi: 10.1136/gutjnl-2020-322294
15. Cao J, Wang C, Zhang Y, Lei G, Xu K, Zhao N, et al. Integrated gut virome and bacteriome dynamics in covid-19 patients. Gut Microbes (2021) 13(1):1–21. doi: 10.1080/19490976.2021.1887722
16. Lv L, Jiang H, Chen Y, Gu S, Xia J, Zhang H, et al. The faecal metabolome in covid-19 patients is altered and associated with clinical features and gut microbes. Anal Chim Acta (2021) 1152:338267. doi: 10.1016/j.aca.2021.338267
17. Yeoh YK, Zuo T. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with covid-19. Gut (2021) 70(4):698–706. doi: 10.1136/gutjnl-2020-323020
18. Prasad R, Patton MJ, Floyd JL, Fortmann S, DuPont M, Harbour A, et al. Plasma microbiome in covid-19 subjects: An indicator of gut barrier defects and dysbiosis. Int J Mol Sci (2022) 23(16):9141. doi: 10.3390/ijms23169141
19. Moreira-Rosário A, Marques C, Pinheiro H, Araújo JR, Ribeiro P, Rocha R, et al. Gut microbiota diversity and c-reactive protein are predictors of disease severity in covid-19 patients. Front Microbiol (2021) 12:705020. doi: 10.3389/fmicb.2021.705020
20. Wu Y, Cheng X. Altered oral and gut microbiota and its association with sars-Cov-2 viral load in covid-19 patients during hospitalization. NPJ Biofilms Microbiomes (2021) 7(1):61. doi: 10.1038/s41522-021-00232-5
21. Gaibani P, D'Amico F, Bartoletti M, Lombardo D, Rampelli S, Fornaro G, et al. The gut microbiota of critically ill patients with covid-19. Front Cell Infect Microbiol (2021) 11:670424. doi: 10.3389/fcimb.2021.670424
22. Zuo T, Liu Q, Zhang F, Yeoh YK, Wan Y, Zhan H, et al. Temporal landscape of human gut rna and DNA virome in sars-Cov-2 infection and severity. Microbiome (2021) 9(1):91. doi: 10.1186/s40168-021-01008-x
23. Khan M, Mathew BJ, Gupta P, Garg G, Khadanga S, Vyas AK, et al. Gut dysbiosis and il-21 response in patients with severe covid-19. Microorganisms (2021) 9(6):1292. doi: 10.3390/microorganisms9061292
24. Hegazy M, Ahmed Ashoush O, Tharwat Hegazy M, Wahba M, Lithy RM, Abdel-Hamid HM, et al. Beyond probiotic legend: Essap gut microbiota health score to delineate sars-Cov-2 infection severity. Br J Nutr (2022) 127(8):1180–9. doi: 10.1017/s0007114521001926
25. Ayling RM, Kok K. Fecal calprotectin. Adv Clin Chem (2018) 87:161–90. doi: 10.1016/bs.acc.2018.07.005
26. Ojetti V, Saviano A, Covino M, Acampora N, Troiani E, Franceschi F. Covid-19 and intestinal inflammation: Role of fecal calprotectin. Dig Liver Dis (2020) 52(11):1231–3. doi: 10.1016/j.dld.2020.09.015
27. O'Moráin N, Stack R, Doherty J, Tosetto M, Garcia Leon A, Mallon P, et al. Faecal calprotectin as a potential biomarker of disease severity in sars-Cov-2 infection. J Infect (2022) 85(4):436–80. doi: 10.1016/j.jinf.2022.06.016
28. Effenberger M, Grabherr F, Mayr L, Schwaerzler J, Nairz M, Seifert M, et al. Faecal calprotectin indicates intestinal inflammation in covid-19. Gut (2020) 69(8):1543–4. doi: 10.1136/gutjnl-2020-321388
29. Adriana DN, Sugihartono T, Nusi IA, Setiawan PB, Purbayu H, Maimunah U, et al. Role of fecal calprotectin as a hypoxic intestinal damage biomarker in covid-19 patients. Gut Pathog (2022) 14(1):34. doi: 10.1186/s13099-022-00507-y
30. Shokri-Afra H, Alikhani A, Moradipoodeh B, Noorbakhsh F, Fakheri H, Moradi-Sardareh H. Elevated fecal and serum calprotectin in covid-19 are not consistent with gastrointestinal symptoms. Sci Rep (2021) 11(1):22001. doi: 10.1038/s41598-021-01231-4
31. Shokri-Afra H, Moradi M, Musavi H, Moradi-Sardareh H, Moradi Poodeh B, Kazemi Veisari A, et al. Serum calprotectin can indicate current and future severity of covid-19. J Clin Lab Anal (2023) 37(1):e24809. doi: 10.1002/jcla.24809
32. Wang C, Xu J, Wang S, Pan S, Zhang J, Han Y, et al. Imaging mass cytometric analysis of postmortem tissues reveals dysregulated immune cell and cytokine responses in multiple organs of covid-19 patients. Front Microbiol (2020) 11:600989. doi: 10.3389/fmicb.2020.600989
33. Lehmann M, Allers K, Heldt C, Meinhardt J, Schmidt F, Rodriguez-Sillke Y, et al. Human small intestinal infection by sars-Cov-2 is characterized by a mucosal infiltration with activated Cd8(+) T cells. Mucosal Immunol (2021) 14(6):1381–92. doi: 10.1038/s41385-021-00437-z
34. Livanos AE, Jha D, Cossarini F, Gonzalez-Reiche AS, Tokuyama M, Aydillo T, et al. Intestinal host response to sars-Cov-2 infection and covid-19 outcomes in patients with gastrointestinal symptoms. Gastroenterology (2021) 160(7):2435–50.e34. doi: 10.1053/j.gastro.2021.02.056
35. Peng X, Ouyang J, Isnard S, Lin J, Fombuena B, Zhu B, et al. Sharing Cd4+ T cell loss: When covid-19 and hiv collide on immune system. Front Immunol (2020) 11:596631. doi: 10.3389/fimmu.2020.596631
36. Martonik D, Parfieniuk-Kowerda A, Rogalska M, Flisiak R. The role of Th17 response in covid-19. Cells (2021) 10(6):1550. doi: 10.3390/cells10061550
37. Chu H, Chan JF, Wang Y, Yuen TT, Chai Y, Shuai H, et al. Sars-Cov-2 induces a more robust innate immune response and replicates less efficiently than sars-cov in the human intestines: An ex vivo study with implications on pathogenesis of covid-19. Cell Mol Gastroenterol Hepatol (2021) 11(3):771–81. doi: 10.1016/j.jcmgh.2020.09.017
38. Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, et al. Sars-Cov-2 productively infects human gut enterocytes. Science (2020) 369(6499):50–4. doi: 10.1126/science.abc1669
39. Zheng Y, Zhuang MW, Han L, Zhang J, Nan ML, Zhan P, et al. Severe acute respiratory syndrome coronavirus 2 (Sars-Cov-2) membrane (M) protein inhibits type I and iii interferon production by targeting rig-I/Mda-5 signaling. Signal Transduct Target Ther (2020) 5(1):299. doi: 10.1038/s41392-020-00438-7
40. Han L, Zhuang MW, Deng J, Zheng Y, Zhang J, Nan ML, et al. Sars-Cov-2 Orf9b antagonizes type I and iii interferons by targeting multiple components of the rig-I/Mda-5-Mavs, Tlr3-trif, and cgas-sting signaling pathways. J Med Virol (2021) 93(9):5376–89. doi: 10.1002/jmv.27050
41. Jiang HW, Zhang HN, Meng QF, Xie J, Li Y, Chen H, et al. Sars-Cov-2 Orf9b suppresses type I interferon responses by targeting Tom70. Cell Mol Immunol (2020) 17(9):998–1000. doi: 10.1038/s41423-020-0514-8
42. Kreimendahl S, Rassow J. The mitochondrial outer membrane protein Tom70-mediator in protein traffic, membrane contact sites and innate immunity. Int J Mol Sci (2020) 21(19):7262. doi: 10.3390/ijms21197262
43. Shuai H, Chu H, Hou Y, Yang D, Wang Y, Hu B, et al. Differential immune activation profile of sars-Cov-2 and sars-cov infection in human lung and intestinal cells: Implications for treatment with ifn-B and ifn inducer. J Infect (2020) 81(4):e1–e10. doi: 10.1016/j.jinf.2020.07.016
44. Stanifer ML, Kee C, Cortese M, Zumaran CM, Triana S, Mukenhirn M, et al. Critical role of type iii interferon in controlling sars-Cov-2 infection in human intestinal epithelial cells. Cell Rep (2020) 32(1):107863. doi: 10.1016/j.celrep.2020.107863
45. Metz-Zumaran C, Kee C, Doldan P, Guo C, Stanifer ML, Boulant S. Increased sensitivity of sars-Cov-2 to type iii interferon in human intestinal epithelial cells. J Virol (2022) 96(7):e0170521. doi: 10.1128/jvi.01705-21
46. Pervolaraki K, Rastgou Talemi S, Albrecht D, Bormann F, Bamford C, Mendoza JL, et al. Differential induction of interferon stimulated genes between type I and type iii interferons is independent of interferon receptor abundance. PloS Pathog (2018) 14(11):e1007420. doi: 10.1371/journal.ppat.1007420
47. Meager A, Visvalingam K, Peterson P, Möll K, Murumägi A, Krohn K, et al. Anti-interferon autoantibodies in autoimmune polyendocrinopathy syndrome type 1. PloS Med (2006) 3(7):e289. doi: 10.1371/journal.pmed.0030289
48. Meyer S, Woodward M, Hertel C, Vlaicu P, Haque Y, Kärner J, et al. Aire-deficient patients harbor unique high-affinity disease-ameliorating autoantibodies. Cell (2016) 166(3):582–95. doi: 10.1016/j.cell.2016.06.024
49. Bruserud Ø, Oftedal BE, Landegren N, Erichsen MM, Bratland E, Lima K, et al. A longitudinal follow-up of autoimmune polyendocrine syndrome type 1. J Clin Endocrinol Metab (2016) 101(8):2975–83. doi: 10.1210/jc.2016-1821
50. Orlova EM, Sozaeva LS, Kareva MA, Oftedal BE, Wolff ASB, Breivik L, et al. Expanding the phenotypic and genotypic landscape of autoimmune polyendocrine syndrome type 1. J Clin Endocrinol Metab (2017) 102(9):3546–56. doi: 10.1210/jc.2017-00139
51. Bastard P, Orlova E, Sozaeva L, Lévy R, James A, Schmitt MM, et al. Preexisting autoantibodies to type I ifns underlie critical covid-19 pneumonia in patients with aps-1. J Exp Med (2021) 218(7):e20210554. doi: 10.1084/jem.20210554
52. Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann HH, Zhang Y, et al. Autoantibodies against type I ifns in patients with life-threatening covid-19. Science (2020) 370(6515):eabd4585. doi: 10.1126/science.abd4585
53. Bastard P, Gervais A, Le Voyer T, Rosain J, Philippot Q, Manry J, et al. Autoantibodies neutralizing type I ifns are present in ~4% of uninfected individuals over 70 years old and account for ~20% of covid-19 deaths. Sci Immunol (2021) 6(62):eabl4340. doi: 10.1126/sciimmunol.abl4340
54. Wang EY, Mao T, Klein J, Dai Y, Huck JD, Jaycox JR, et al. Diverse functional autoantibodies in patients with covid-19. Nature (2021) 595(7866):283–8. doi: 10.1038/s41586-021-03631-y
55. Solanich X, Rigo-Bonnin R, Gumucio VD, Bastard P, Rosain J, Philippot Q, et al. Pre-existing autoantibodies neutralizing high concentrations of type I interferons in almost 10% of covid-19 patients admitted to intensive care in Barcelona. J Clin Immunol (2021) 41(8):1733–44. doi: 10.1007/s10875-021-01136-x
56. Pascoal LB, Rodrigues PB, Genaro LM, Gomes A, Toledo-Teixeira DA, Parise PL, et al. Microbiota-derived short-chain fatty acids do not interfere with sars-Cov-2 infection of human colonic samples. Gut Microbes (2021) 13(1):1–9. doi: 10.1080/19490976.2021.1874740
57. Goncalves D, Mezidi M, Bastard P, Perret M, Saker K, Fabien N, et al. Antibodies against type I interferon: Detection and association with severe clinical outcome in covid-19 patients. Clin Transl Immunol (2021) 10(8):e1327. doi: 10.1002/cti2.1327
58. Koning R, Bastard P, Casanova JL, Brouwer MC, van de Beek D. Autoantibodies against type I interferons are associated with multi-organ failure in covid-19 patients. Intensive Care Med (2021) 47(6):704–6. doi: 10.1007/s00134-021-06392-4
59. Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, et al. Inborn errors of type I ifn immunity in patients with life-threatening covid-19. Science (2020) 370(6515):eabd4570. doi: 10.1126/science.abd4570
60. Guo Y, Luo R, Wang Y, Deng P, Song T, Zhang M, et al. Sars-Cov-2 induced intestinal responses with a biomimetic human gut-on-Chip. Sci Bull (Beijing) (2021) 66(8):783–93. doi: 10.1016/j.scib.2020.11.015
61. Mithal A, Hume AJ, Lindstrom-Vautrin J, Villacorta-Martin C, Olejnik J, Bullitt E, et al. Human pluripotent stem cell-derived intestinal organoids model sars-Cov-2 infection revealing a common epithelial inflammatory response. Stem Cell Rep (2021) 16(4):940–53. doi: 10.1016/j.stemcr.2021.02.019
62. Yamada S, Noda T, Okabe K, Yanagida S, Nishida M, Kanda Y. Sars-Cov-2 induces barrier damage and inflammatory responses in the human ipsc-derived intestinal epithelium. J Pharmacol Sci (2022) 149(3):139–46. doi: 10.1016/j.jphs.2022.04.010
63. Britton GJ, Chen-Liaw A, Cossarini F, Livanos AE, Spindler MP, Plitt T, et al. Limited intestinal inflammation despite diarrhea, fecal viral rna and sars-Cov-2-Specific iga in patients with acute covid-19. Sci Rep (2021) 11(1):13308. doi: 10.1038/s41598-021-92740-9
64. Tao W, Zhang G, Wang X, Guo M, Zeng W, Xu Z, et al. Analysis of the intestinal microbiota in covid-19 patients and its correlation with the inflammatory factor il-18. Med Microecol (2020) 5:100023. doi: 10.1016/j.medmic.2020.100023
65. Parada Venegas D, de la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, et al. Short chain fatty acids (Scfas)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol (2019) 10:277. doi: 10.3389/fimmu.2019.00277
66. Zang R, Gomez Castro MF, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, et al. Tmprss2 and Tmprss4 promote sars-Cov-2 infection of human small intestinal enterocytes. Sci Immunol (2020) 5(47):eabc3582. doi: 10.1126/sciimmunol.abc3582
67. Rodrigues PB, Gomes GF, Angelim M, Souza GF, Muraro SP, Toledo-Teixeira DA, et al. Impact of microbiota depletion by antibiotics on sars-Cov-2 infection of K18-Hace2 mice. Cells (2022) 11(16):2572. doi: 10.3390/cells11162572
68. Jagtap D, Bhor VM, Bhowmick S, Kasarpalkar N, Sagvekar P, Kulkarni B, et al. Smadcam: Il-6 ratio influences disease progression and anti-viral responses in sars-Cov-2 infection. Front Immunol (2021) 12:619906. doi: 10.3389/fimmu.2021.619906
69. Singh AK, Kasarpalkar N, Bhowmick S, Paradkar G, Talreja M, Shah K, et al. Opposing roles for smadcam and il-15 in covid-19 associated cellular immune pathology. J Leukoc Biol (2022) 111(6):1287–95. doi: 10.1002/jlb.3covbcr0621-300r
70. Territo MC, Ganz T, Selsted ME, Lehrer R. Monocyte-chemotactic activity of defensins from human neutrophils. J Clin Invest (1989) 84(6):2017–20. doi: 10.1172/jci114394
71. Yang D, Chen Q, Chertov O, Oppenheim JJ. Human neutrophil defensins selectively chemoattract naive T and immature dendritic cells. J Leukoc Biol (2000) 68(1):9–14. doi: 10.1189/jlb.68.1.9
72. Wang C, Wang S, Li D, Wei DQ, Zhao J, Wang J. Human intestinal defensin 5 inhibits sars-Cov-2 invasion by cloaking Ace2. Gastroenterology (2020) 159(3):1145–7.e4. doi: 10.1053/j.gastro.2020.05.015
73. Xu C, Wang A, Marin M, Honnen W, Ramasamy S, Porter E, et al. Human defensins inhibit sars-Cov-2 infection by blocking viral entry. Viruses (2021) 13(7):1246. doi: 10.3390/v13071246
74. Singhal R, Shah YM. Oxygen battle in the gut: Hypoxia and hypoxia-inducible factors in metabolic and inflammatory responses in the intestine. J Biol Chem (2020) 295(30):10493–505. doi: 10.1074/jbc.REV120.011188
75. Underdown BJ, Schiff JM. Immunoglobulin a: Strategic defense initiative at the mucosal surface. Annu Rev Immunol (1986) 4:389–417. doi: 10.1146/annurev.iy.04.040186.002133
76. Macpherson AJ, Martinic MM, Harris N. The functions of mucosal T cells in containing the indigenous commensal flora of the intestine. Cell Mol Life Sci (2002) 59(12):2088–96. doi: 10.1007/s000180200009
77. Brandtzaeg P, Farstad IN, Johansen FE, Morton HC, Norderhaug IN, Yamanaka T. The b-cell system of human mucosae and exocrine glands. Immunol Rev (1999) 171(1):45–87. doi: 10.1111/j.1600-065x.1999.tb01342.x
78. Cerutti A. The regulation of iga class switching. Nat Rev Immunol (2008) 8(6):421–34. doi: 10.1038/nri2322
79. Wang Z, Lorenzi JCC, Muecksch F, Finkin S, Viant C, Gaebler C, et al. Enhanced sars-Cov-2 neutralization by dimeric iga. Sci Transl Med (2021) 13(577):eabf1555. doi: 10.1126/scitranslmed.abf1555
80. Assimakopoulos SF, Triantos C, Maroulis I, Gogos C. The role of the gut barrier function in health and disease. Gastroenterol Res (2018) 11(4):261. doi: 10.14740/gr1053w
81. Farsi Y, Tahvildari A, Arbabi M, Vazife F, Sechi LA, Shahidi Bonjar AH, et al. Diagnostic, prognostic, and therapeutic roles of gut microbiota in covid-19: A comprehensive systematic review. Front Cell Infect Microbiol (2022) 12:804644. doi: 10.3389/fcimb.2022.804644
82. Hashimoto T, Perlot T, Rehman A, Trichereau J, Ishiguro H, Paolino M, et al. Ace2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature (2012) 487(7408):477–81. doi: 10.1038/nature11228
83. Zhao Y, Chen F, Wu W, Sun M, Bilotta AJ, Yao S, et al. Gpr43 mediates microbiota metabolite scfa regulation of antimicrobial peptide expression in intestinal epithelial cells Via activation of mtor and Stat3. Mucosal Immunol (2018) 11(3):752–62. doi: 10.1038/mi.2017.118
84. Edwinson A, Yang L, Chen J, Grover M. Colonic expression of Ace2, the sars-Cov-2 entry receptor, is suppressed by commensal human microbiota. Gut Microbes (2021) 13(1):1984105. doi: 10.1080/19490976.2021.1984105
85. Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E. Dysbiosis and the immune system. Nat Rev Immunol (2017) 17(4):219–32. doi: 10.1038/nri.2017.7
86. Gu S, Chen Y, Wu Z, Chen Y, Gao H, Lv L, et al. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1n1 influenza. Clin Infect Dis (2020) 71(10):2669–78. doi: 10.1093/cid/ciaa709
87. Tamanai-Shacoori Z, Smida I, Bousarghin L, Loreal O, Meuric V, Fong SB, et al. Roseburia spp.: A marker of health? Future Microbiol (2017) 12(2):157–70. doi: 10.2217/fmb-2016-0130
88. Konieczna P, Akdis CA, Quigley EM, Shanahan F, O'Mahony L. Portrait of an immunoregulatory bifidobacterium. Gut Microbes (2012) 3(3):261–6. doi: 10.4161/gmic.20358
Keywords: COVID-19, gut barrier, cytokines, interferon, defensins, microbiome, cytokine storm
Citation: Eleftheriotis G, Tsounis EP, Aggeletopoulou I, Dousdampanis P, Triantos C, Mouzaki A, Marangos M and Assimakopoulos SF (2023) Alterations in gut immunological barrier in SARS-CoV-2 infection and their prognostic potential. Front. Immunol. 14:1129190. doi: 10.3389/fimmu.2023.1129190
Received: 21 December 2022; Accepted: 06 March 2023;
Published: 15 March 2023.
Edited by:
Rupesh K. Srivastava, All India Institute of Medical Sciences, IndiaReviewed by:
Stéphane Isnard, McGill University Health Center, CanadaCopyright © 2023 Eleftheriotis, Tsounis, Aggeletopoulou, Dousdampanis, Triantos, Mouzaki, Marangos and Assimakopoulos. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Stelios F. Assimakopoulos, c2Fzc2ltQHVwYXRyYXMuZ3I=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.