- 1Department of Clinical Laboratory, The First People’s Hospital of Kunshan, Suzhou, China
- 2Department of Biochemistry and Molecular Biology, School of Life Sciences, Central South University, Changsha, China
Tuberculosis (TB) is a major infectious disease induced by Mycobacterium tuberculosis (M. tb) which causes the world’s dominant fatal bacterial contagious disease. Increasing studies have indicated that exosomes may be a novel option for the diagnosis and treatment of TB. Exosomes are nanovesicles (30-150 nm) containing lipids, proteins and non-coding RNAs (ncRNAs) released from various cells, and can transfer their cargos and communicate between cells. Furthermore, exosomal ncRNAs exhibit diagnosis potential in bacterial infections, including TB. Additionally, differential exosomal ncRNAs regulate the physiological and pathological functions of M. tb-infected cells and act as diagnostic markers for TB. This current review explored the potential biological roles and the diagnostic application prospects of exosomal ncRNAs, and included recent information on their pathogenic and therapeutic functions in TB.
1 Introduction
Tuberculosis (TB) is a severe infectious disease that is still the major cause of death from infection, regardless of worldwide progress in health and diseases (1, 2). Recent estimates from a new World Health Organization report show that ~1/4 of the international population are infected with Mycobacterium tuberculosis (M. tb), which leads to 1.4 million deaths per year (3, 4). Pathogenically, M. tb can survive for a long time in macrophages of tubercle granulomas in the human host (5). Macrophages are key components of the host innate immune responses to M. tb that could eliminate mycobacteria via different mechanisms, including apoptosis, immune-inflammatory responses and phagocytic activity (6).
The diagnosis and analysis of TB are mainly based on TB culture or PCR. Usually, sputum smear microscopy is the most extensively used; however, regardless of its high specificity, its low sensitivity restricts its diagnostic value (7). However, TB cultures take nearly 42 days to detect identifiable growth, thus the long culture period limits its clinical diagnosis (8). At present, the Xpert MTB/RIF and ultra-version tests detect genetic material from M. tb as sensitively as microbial cultures (9). These assays are rapid but expensive and widely unavailable (10, 11), and call for better diagnostic technologies for M. tb. Therefore, sensitive and specific diagnostic assays are important in controlling and preventing infections from spreading, and novel biomarkers are urgently required due to these problems of current TB diagnostics (12, 13).
In recent years, exosomes, small vesicles derived from various cells, have shown great potential as diagnostic markers and in treatment depending on the cargo inside. Furthermore, it has been indicated that exosomal non-coding RNAs (ncRNAs) are critical regulators involved in the immune defense of M. tb infection. Exosomal ncRNAs regulate the host to resist M. tb infection by dominating the relevant signaling pathways in the infection process (14). With the development of sequencing technology, a large number of exosomal ncRNAs have been identified in different biological processes of TB. More and more studies have demonstrated that exosomal ncRNAs regulate host gene expression at the level of transcription and post-transcription, which is closely related to the adaptation of TB to the host environment and the generation of pathogenicity (15). These exosomal ncRNAs interact with each other, as well as other components, including proteins and DNA, thus affecting the occurrence and development of TB.
The present study comprehensively reviews the function and molecular mechanisms of exosomal ncRNAs in the physiological and pathological process of M. tb infection.
2 The biological functions of exosomes
Exosomes are 30-150 nm microvesicles, have the same topology as their origin cells and exhibit enrichment of selected proteins, lipids and nucleic acids (16, 17). Exosomes can be distinguished from other extracellular structures (ectosomes and apoptotic blebs) due to their size and the protein factors intercalated in their membranes (18). Exosomes originate from almost all cell types, are separated from nearly all human biofluids and carry functional molecules, including nucleic acids (mRNA, ncRNAs and DNA), proteins, metabolites and lipid modulators (19–22). Exosomes are pivotal in cell-cell communication and transfer biological information by shuttling their cargo to either local or distant cells, and thereby, modulate the function of the recipient cells (Figure 1) (23–25). Exosomes are regarded as novel diagnostic biomarkers under various pathological environments, including cancers and infections (26–29). Exosomal contents are employed as signatures of various cancers and infectious diseases (30).
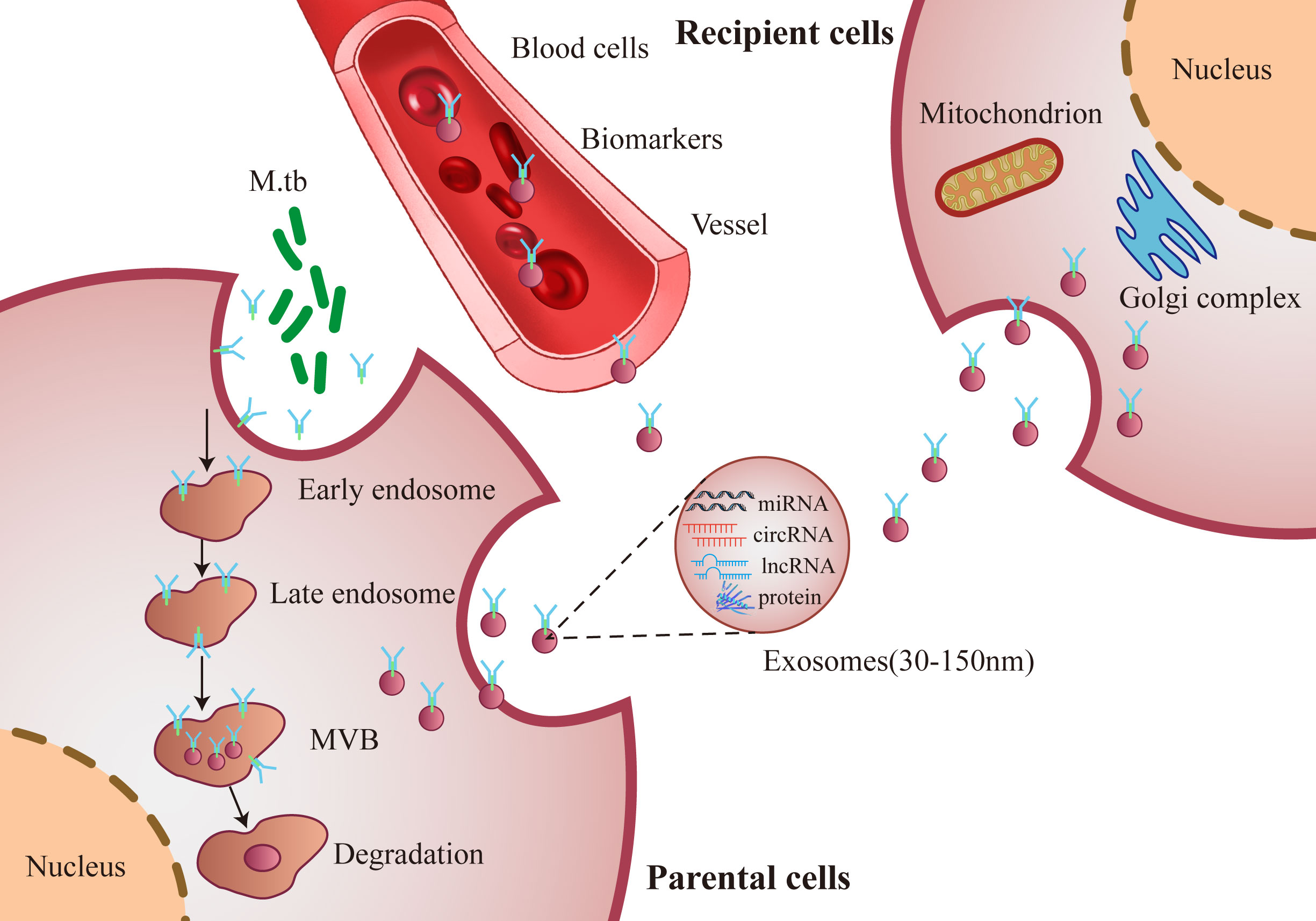
Figure 1 Schematic diagram of secretion synthesis and transmission of exosomes. M. tb infected host cells forms and secretes exosomes, which transmit biological information and regulate the functions between cells through exosomes components, such as miRNA, circRNA, lncRNA and proteins.
Although exosomes involved in cell-to-cell communication and immune regulation are a topic of intense research, most reports divert to finding aberrations in exosomal protein and RNA contents related to disease occurrence and advancement, which are initially associated with cancer or infection (31). In particular, microRNAs (miRNAs/miRs) are transferred by exosomes that participate in the initiation of various respiratory system diseases, including TB, in which they are critical in inflammation and pathogenesis (32, 33). The miRNA profiles expressed differently in different pathological environments suggest that microvesicles may be related to the occurrence of diseases (34). It is well-known that exosomes induce a series of immune responses in M. tb infection.
It has been reported that M. tb could induce macrophages to secrete exosomes with pathogen-associated molecular patterns, and these exosomes are in turn transferred into uninfected macrophages to be activated through the Toll-like receptor and myeloid differentiation factor 88-dependent pathway (35, 36). Notably, exosomes from T cells are transferred into dendritic cells (DCs) and induce more resistant DC antiviral responses via the cyclic GMP-AMP synthase/stimulator of interferon response cGAMP interactor-1 cytosolic DNA-sensing pathway and via the expression of interferon regulatory factor-3-dependent interferon-regulated genes (37). Notably, exosomes may be a candidate vector for vaccines or treatment. Exosomes carrying mycobacterial antigens can protect mice against M. tb infection, indicating the potential of exosomes in serving as a novel cell-free vaccine targeting M. tb infection (38). Exosomes from M. tb-infected bone marrow-derived macrophages could induce autophagy for in vitro M. tb killing and also decreased the mycobacterial burden in the lungs of mice with low tissue damage (39). These findings indicate that exosomes not only induce the immune response but can also be used as a vaccine. However, the detailed immunological function of exosomes during M. tb infection is still to be revealed.
The present review describes the discovered roles of exosomes in TB disease, the translation work in vaccine development and how to govern the circling of these dynamic vesicles for diagnostic aims. In addition, exosomal RNAs (especially miRNAs) may be a perfect platform to generate diagnostic biomarkers because the exosomes-regulated resistance against RNA degradation is durable (40).
3 The functions of exosomal ncRNAs
NcRNAs are non-protein coding molecules with <200 nucleotides, and comprise a myriad of RNA classes, including miRNAs, circular RNAs (circRNAs; circs) and long non-coding RNAs (lncRNAs), with diverse functions, including numerous small transcripts without related functions (41, 42). Exosomes derived from cells in different physiological states contain various ncRNAs, and the composition and species of ncRNAs differ markedly.
Expression assays demonstrate that ncRNAs are accurately modulated in space and time, and a disturbance in gene expression modes can lead to pathological phenotypes, including various cancers and infectious diseases (43). Among ncRNAs, miRNAs are the most-studied and participate in post-transcriptional modulation. MiRNAs are largely conserved ncRNAs that could modulate gene expression by base-pairing. Therefore, variations in transcriptional output from these molecules reflect anomalies in global transcription in cells. Additionally, miRNAs can be delivered into or out of cells by vesicles, which could be detected in body fluids (44). In addition, miRNA profiles can be related to disease development or division among diseases with identical symptoms (45, 46), demonstrating their clinical significance.
Accumulating studies have demonstrated that exosomal ncRNAs are associated with various diseases, including diabetes, coronary artery disease, cancers and infectious diseases (47–49). For instance, Lai et al. reported that exosomal lncRNA SOX2 overlapping transcript (SOX2-OT) promoted ovarian cancer progression, and SOX2-OT, miR-181b-5p and stearoyl-CoA desaturase 1 may serve as potential targets for the treatment of ovarian cancer (50). However, in breast cancer, Zhao et al. found that exosomal transfer of miR-181b-5p conferred senescence-mediated doxorubicin resistance by modulating BCL2 associated transcription factor 1 (51). These studies indicate that exosomal ncRNAs are altered in diseases, and the regulatory targets involved by exosomal ncRNAs are also different.
Exosomes transport ncRNAs to communicate phenotypic characteristics between cells (52). Some potential biomarkers (human miRNAs, mRNAs and mycobacterial RNAs in exosomes) can be used for the identification of TB (53). A new issue in molecular biology is that fluids (e.g., blood or urine) are used to substitute invasive biopsy in disease prognostics and diagnosis. These body fluids carry large protein-covering lipoprotein composites (e.g., exosomes), and DNA and RNA molecules, and are actively derived by living cells (54). The genetic substances covered in exosomes are stable and undegradable and can be used to noninvasively detect both chronic and infectious disorders (55, 56). However, there is no profound exosomal RNA-sequencing (RNA-seq) in human clinical samples of patients with active TB (ATB) or latent TB infection (LTBI).
4 The function of exosomal miRNAs in M. tb infection
Exosomes mediate the exchange of intricate intercellular messages such as miRNAs (57). miRNAs are small 18-22-nt RNAs that are pivotal in mediating gene expression and translation. miRNAs impact most biological functions, and their dysregulation is related to several pathologies (58). Functional miRNAs may be embedded in exosomes and delivered to target cells, regulating the roles of recipient cells by affecting their transcriptome and/or proteome (59). Additionally, exosomal miRNAs can exist stably in body fluids, and are associated with information of maternal tissues or cells depending on miRNA expression and composition (60, 61).
MiRNAs are involved in the mediation of inflammatory procedures amid M. tb infection (62, 63). M. tb infection triggers various physiological responses in infected cells, resulting in host immune abnormality and metabolic re-patterning (64). This regulation of host cell reactions allows bacteria to isolate vital host factors to meet their material and energy demand to facilitate intracellular survival (65). These steps may be managed by subversing host miRNA networks involved in the mediation of carbon, nitrogen and lipid metabolism in the infected cells (66, 67).
Exosomes are critical extracellular antigen sources and can promote T cell immunity after M. tb infection in mice (68). Generally, exosomes are promising for diagnosis, treatment or vaccine invention, and exosome-based treatment for patients with TB should be further studied (69). Recent research shows that exosomal miRNAs are potential diagnostic tools for TB. The present review summarizes and elaborates the information on the pathogenic and therapeutic functions and roles as diagnostic biomarkers of exosomal miRNAs in TB (Table 1).
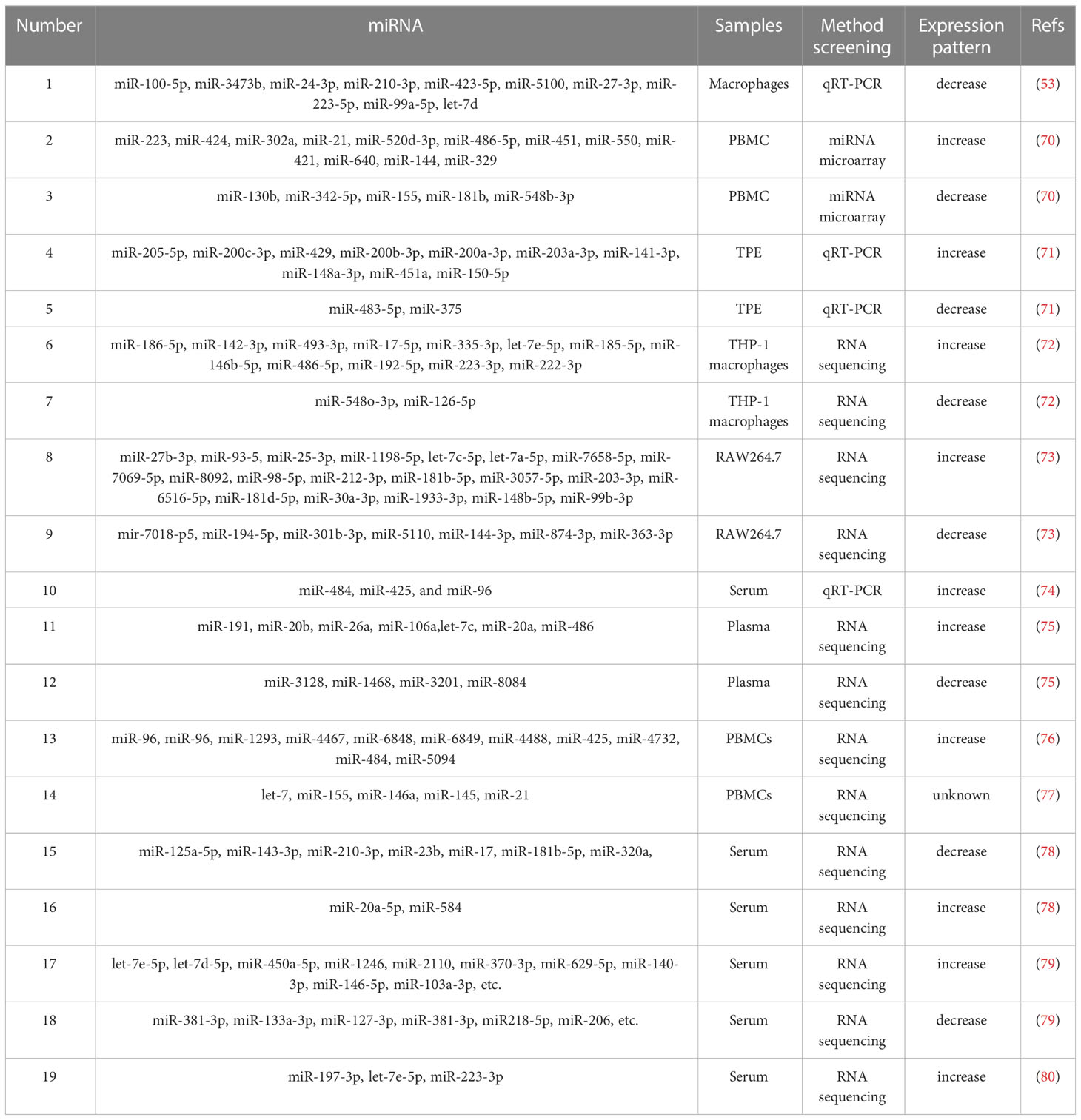
It has been reported that 57 exosomal miRNAs were found in M. tb-infected macrophages (e.g., Mmu-223 and 486-5p) and most of these miRNAs were decreased (53). The suppression of these exosomal miRNAs was evaluated by the quantified miRNAs for mRNA targets using miRDB and functional Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis, and the results indicated that potential gene targets for these miRNAs included those associated with immune surveillance and inflammation. These results imply the richness of cell miRNAs after M. tb infection and reveal immune mechanisms induced by the pathogen. Wang et al. detected 17 exosomal miRNAs differentially expressed (DE) in peripheral blood mononuclear cells (PBMCs) in patients with ATB, patients with LTBI and healthy controls, respectively, and these exosomal miRNAs were involved in functions in hematopoietic cell differentiation and the transition from LTB to ATB (70). Among these 17 differential miRNAs, six miRNAs (miR-21, miR-223, miR-302a, miR-424, miR-451 and miR-486-5p) were upregulated and miR-130b exhibited reduced expression in patients with ATB; and four miRNAs (miR-144, miR-365, miR-133a and miR-424) were upregulated and three miRNAs (miR-500, miR-661 and miR-892b) were downregulated in patients with ATB (70). However, miR-424 was upregulated in patients with ATB compared with patients with LTBI and healthy controls (70). Therefore, mycobacterial infection of macrophages led to general inhibition of miRNA incorporation into exosomes and exosomal miRNA released from M. tb-infected macrophages may have a potential function in diagnosis during mycobacterial infection.
Wang et al. reported that miR-148a-3p, -451a and -150-5p were all upregulated with different fold changes in pleural effusions (PEs) of tuberculosis and other benign lesions using RNA-seq and reverse transcription-quantitative PCR (RT-qPCR). These different miRNA profiles may support the use as biomarkers for differential diagnosis of PEs with more verification based on larger cohorts (71). As reported, 495 exosomal miRNAs related to TB infection were found using whole transcriptome high-throughput sequencing, and it was identified that miR-185-5p, miR-146b-5p and miR-17-5p were increased in exosomes (72). Interestingly, miR-185-5p was markedly increased in all the following samples, including bacille Calmette-Guerin (BCG)-infected monocytes from PBMCs and exosomes from patients with TB (72). These differential miRNAs are excellent biomarkers for the infection of TB. Zhan et al. found 20 upregulated and seven downregulated exosomal miRNAs in M. tb-infected macrophages, of which miR-27b-3p, miR-93-5p, miR-25-3p, miR-1198-5p, let-7c-5p and let-7a-5p were considerably upregulated based on high-throughput sequencing (73). A bioinformatics experiment implied that these DE exosomal miRNAs were engaged in several bioprocesses and pathways, and the target genes of the top six miRNAs in the upregulated group were positively related to apoptosis modulation (73). The miRNA expression profile in macrophage exosomes differed after BCG infection, and the DE miRNAs participated in multiple bioprocesses and pathways (73).
Alipoor et al. identified miR-484, miR-425 and miR-96 in serum-derived exosomes of patients with TB by RT-qPCR, and these three miRNAs were markedly increased in the serum, suggesting that these exosomal miRNAs have diagnostic potential in ATB, and their diagnostic value could be improved through combination with conventional diagnostic markers (74). Hu et al. identified six exosomal miRNAs (miR-20a, miR-20b, miR-26a, miR-106a, miR-191 and miR-486) that were increased, and four miRNAs (miR-3128, miR-1468, miR-3201 and miR-8084) that were decreased in patients with TB using RT-qPCR (75). These exosomal miRNAs combined with health records could facilitate clinical discovery of TB meningitis and pulmonary TB (PTB) (75). Alipoor et al. detected 11 exosomal miRNAs (miR-1224, miR-1293, miR-425, miR-4467, miR-4732, miR-484, miR-5094, miR-6848, miR-6849, miR-96 and miR-4488) that were upregulated in BCG-infected monocyte-derived macrophages (MDMs), and these miRNAs were engaged in some key pathways, such as central C metabolism, fatty acid and sugar metabolism, amino acid metabolism, bacterial invasion pathways, and cell pathways (76). These exosomal miRNAs reflect the host-pathogen interaction and subversion of host metabolic processes following infection (76). Mortaz et al. reported that the infection of MDMs with BCG led to the release of a number of exosomal miRNAs, including let-7 family members, and miR-155, miR-146a, miR-145 and miR-21, all of which could target critical immune genes and pathways in BCG-infected MDMs (77). However, these results need to be verified and the presence of these miRNAs in the blood should be tested further to estimate their specificity and selectivity as a diagnostic tool in patients with TB.
Guio et al. found three downregulated miRNAs (miR-143-3p, miR-210-3p and miR-20a-5p) and one upregulated miRNA (miR-20a-5p) for LTB, and three decreased miRNAs (miR-23b, miR-17 and miR-181b-5p) and one increased miRNA (miR-584) for ATB using small RNA-seq, and only two miRNAs were shared by the two types of TB (miR-125a-5p and miR-203a) (78). These exclusive miRNAs are promising regulators of common or exclusive KEGG pathways related to infectious disorders, cancers and immunology. Lyu et al. identified 250 exosomal miRNAs, including 85 specifically expressed miRNAs in serum exosomes of patients with LTBI or TB and healthy controls using small RNA-seq (79). Among the 250 DE miRNAs, 49 upregulated (e.g., miR-146-5p, miR-103a-3p and miR-103b) and 21 downregulated (e.g., miR-381-3p, miR-133a-3p and miR-127-3p) miRNAs were detected for LTBI, and 37 upregulated (e.g., miR-629-5p, miR-140-3p and miR-151a-3p) and 10 downregulated (e.g., miR-381-3p, miR-128-3p and miR-218-5p) miRNAs were identified for TB (79). In addition, they identified 18 and 67 specifically expressed miRNAs in the LTBI (e.g., let-7e-5p, let-7d-5p, miR-450A-5p) and TB (e.g., miR-1246, miR-2110 and miR-370-3p) groups. These findings provide a critical reference and better understanding about miRNAs and repetitive region-obtained small RNAs in exosomes amid M. tb infection, and promote the generation of potential molecular targets for the diagnosis of LTB or ATB (79). In addition, Lyu et al. used RNA-seq for a limited RNA library to test divergent exosomal miRNA modes in sera of healthy individuals, and patients with LTBI and TB, and revealed six exosomal miRNAs, and three continually increased miRNAs, including miR-3184-5p, miR-140-3p and miR-423-3p, were detected as potential regulators in TB advancement (79). Additionally, they both assessed the DE miRNA mode with DE mRNA modes. They reported relevant data on the potential roles of exosomes in the whole M. tb contagion and suggested that the identified exosomal miRNAs may be used as biomarkers for diagnosis. Interestingly, various literatures show that the types and abundance of exosomal ncRNAs in active and latent tuberculosis are very different, and there are some specific ncRNAs related to tuberculosis progress. Therefore, exosomal ncRNAs can be used to distinguish active and latent tuberculosis.
Carranza et al. found three upregulated miRNAs (miR-let-7e-5p, miR-197-3p and -223-3p) to be sensitive separators between controls and patients with TB for both drug-resistant-TB and multidrug-resistant (MDR)-TB groups using a multivariate analysis (80). MiR-let-7e-5p was upregulated in the MDR-TB group without type 2 diabetes mellitus (T2DM), indicating that miR-let7e-5p is a possible biomarker for the detection and treatment of MDR-TB without T2DM (64). MiRNAs in exosomes, which are exhaled, have been suggested as potential biomarkers for individuals with respiratory tract infections such as TB (81).
These exosomal miRNA sequencing profiles reveal the molecular mechanism of regulating target genes, and of regulation of host physiology and pathology in the process of TB infection by differential miRNAs. These findings provide a comprehensive understanding of miRNAs and repetitive region-derived small RNAs in exosomes during the M. tb infectious process and facilitate the development of potential molecular targets for the detection/diagnosis of TB.
5 The roles of other exosomal ncRNAs in M. tb infection
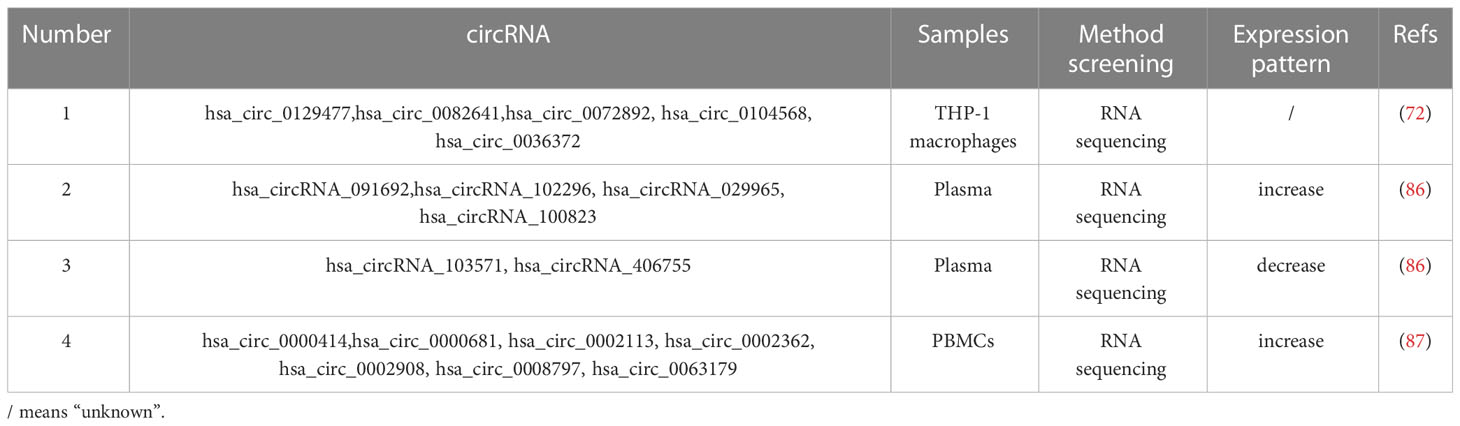
CircRNAs are new classes of internal ncRNAs with tissue- and cell-specific expression profiles, and are covalently shut down and extensively expressed in eukaryotes (82). CircRNAs act as miRNA or protein inhibitors (‘sponges’), regulate protein function or self-translate to serve critical biological roles (83, 84). Previous study has revealed that exosomes could carry and protect circRNAs in various body fluids. Exosomal circRNAs in cancers could function at target cells or organs by transporting exosomes, and then participate in the regulation of tumor growth and metastasis (85). Since exosomes are present in diverse body fluids and exosomal circRNAs are highly stable, exosomal circRNAs are potential biomarkers for the diagnosis and prognosis of early and minimally invasive cancer (Table 2) (88).
After the exosomal ncRNA profile was analyzed in M. tb H37Ra- and M. bovis BCG-infected macrophages, circ_0129477, circ_0082641, circ_0072892, circ_0104568 and circ_0036372 were detected and the possible downstream regulatory pathway of these differential circRNAs was revealed (72). The circRNA-miRNA network of interaction implies that a single miRNA can be a target of several circRNAs, while multiple miRNAs can also be targeted by a single circRNA. Yi et al. reported that hsa_circRNA_103571 was markedly decreased in patients with ATB and was involved in the ras pathway, mediation of actin cytoskeleton, and the T- and B-cell receptor pathway, and suggested that circRNA_103571 may be a potential biomarker for ATB identification (86). Qian et al. explored circRNA expression in the peripheral blood of patients with TB using RNA-seq and microarray analysis, and found seven increased circRNAs (circ_0000414, circ_0000681, circ_0002113, circ_0002362, circ_0002908, circ_0008797 and circ_0063179), suggesting that circRNAs may be used as marker molecules to diagnose ATB (87). These studies suggest that the exosomal circRNA signature in TB infection may offer possible targets for the clinical diagnosis of TB. Nevertheless, the roles of circRNAs as biomarkers should be verified in a large sample cluster.
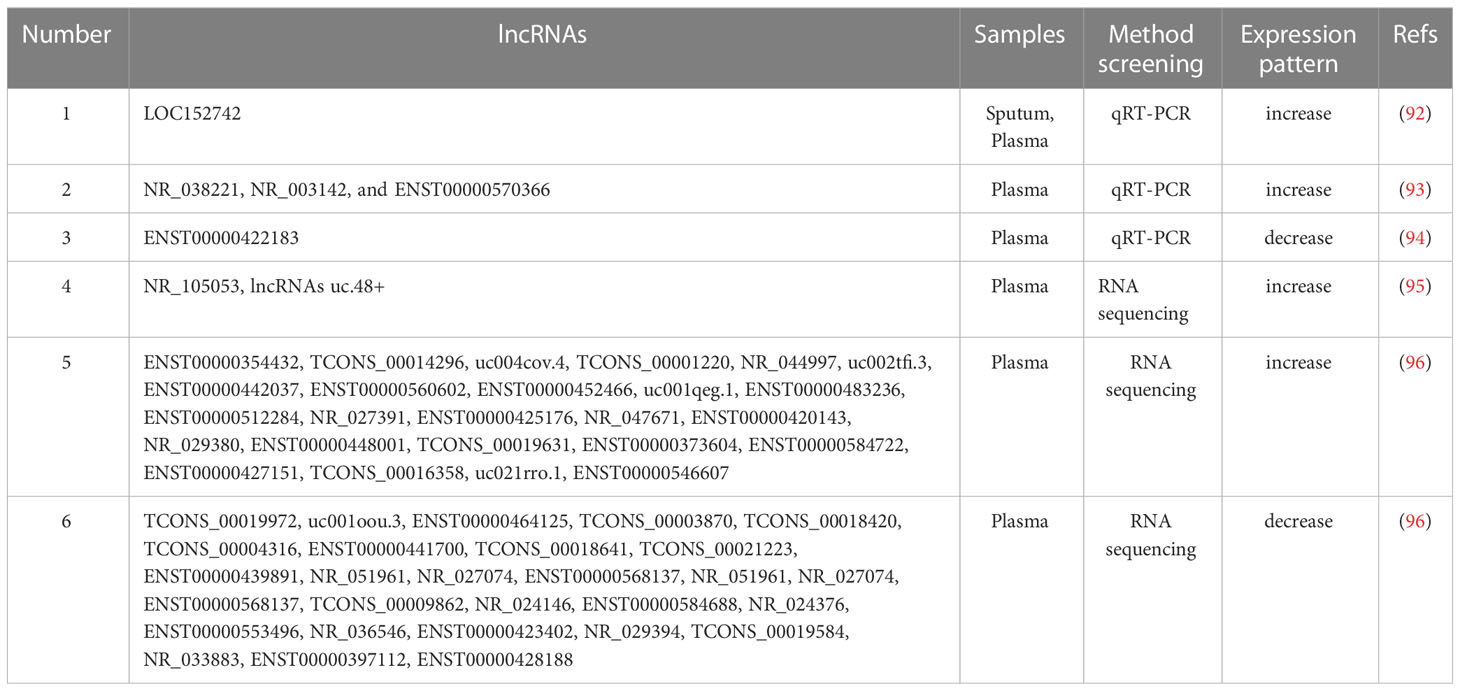
LncRNAs comprise transcripts longer than 200 nucleotides. LncRNAs function as competitive endogenous RNAs by competitively occupying the shared binding sequences of miRNAs, thus sequestering the miRNAs and changing the expression of their downstream target genes (89). Exosomal lncRNAs may also act as biomarkers in the diagnosis of cancers and infectious diseases (90, 91). The present review summarizes all lncRNAs related to TB in following studies (Table 3).
Evidences demonstrate that lncRNA expression levels are abnormal in PBMCs of patients with TB, implying that lncRNAs are associated with the pathology of TB (97–100). LOC152742 levels in plasma of patients with ATB are higher than those in patients with previous episodes of TB and BCG-vaccinated individuals, suggesting that LOC152742 may be a potential biomarker for ATB discovery and treatment (92). Some small lncRNA sets are highly sensitive and specific in diagnosis. Four DE lncRNAs (NR_038221, NR_003142, ENST-00000570366 and ENST-00000422183) can efficiently separate patients with PTB from controls, with an area under the curve of 0.845 (93). Two lncRNAs (ENST-00000354432 and ENST-00000427151) in the plasma are potential biomarkers for TB diagnosis (94). LncRNAs NR_105053 and uc.48+ were increased in the plasma and may be potential biomarkers to differentiate untreated and cured TB subjects (95). Li et al. found 351 upregulated lncRNAs and 841 downregulated lncRNAs in the serum exosomes of patients with ATB, and NONHSAT-101518.2, NONHSAT-067134.2, NONHSAT-148822.1 and NONHSAT-078957.2 were downregulated in the plasma, suggesting that they may be potential biomarkers for ATB diagnosis (96).
Lv et al. identified mycobacterial transcripts in the exosomes derived from infected macrophages and in serum exosomes of patients with TB. To the best of our knowledge, this was the first report to recognize bacterial RNA in exosomes (101). Gutkin et al. investigated M. tb genes in serum exosomes using gene sequencing, and found 2 M. tb genes (rrs and rrl) in LTBI and 3 RNA genes (rrs, rrl and Rv2917) in ATB samples (102).
Although exosomal ncRNAs are closely related to TB infection and regulate the corresponding miRNA or mRNA to ultimately mediate transcription or the proteome, their specific regulatory mechanisms and use as diagnostic markers of TB infection need to be further clarified and confirmed in large-scale clinical trials.
6 Clinical potential application of exosomal ncRNAs in M. tb infection
Exosomes released from M. tb infected macrophages contain pathogen-associated molecular patterns (PAMPs), mycobacteria components lipoarabinomannan and the 19-kDa lipoprotein (103). Moreover, exosomes isolated from M. bovis BCG- and M. tb-infected macrophages in mice, stimulate the production of TNF-alpha and IL-12 revealing that exosomes promote intercellular communication during an immune response to intracellular pathogens, and exosomes containing PAMPs is an important mechanism of immune surveillance (104). In addition, there was a similar TH1 immune response but a more limited TH2 response in exosome-vaccinated mice compared to BCG-vaccinated mice suggesting that exosomes might serve as a novel cell-free vaccine against an M. tb infection (30). These documents show that exosomes cloud be used as an immunotherapy or vaccine to prevent tuberculosis infection in clinic (Figure 2). This study is mainly aimed at exosomes as a biomarker for the diagnosis of tuberculosis infection, especially for ncRNAs in exosomes.
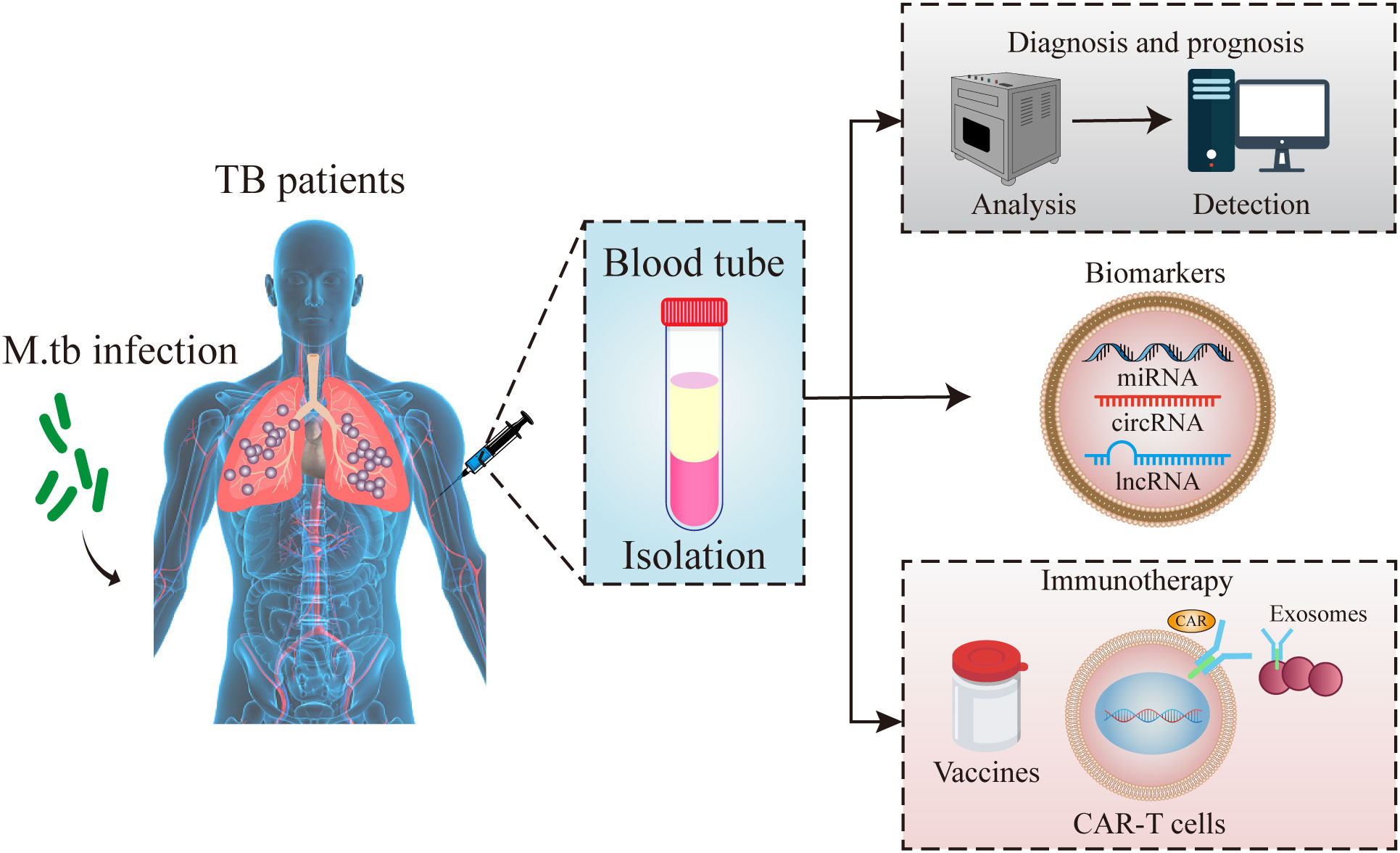
Figure 2 Schematic diagram of exosomes functions. Exosomes in peripheral blood of tuberculosis patients could be used for the diagnostic markers, vaccines and immunotherapy for tuberculosis infection.
Screening approaches based on biomarkers (specially quantified in blood specimens) help detect subjects with a higher risk of developing ATB, or at early stages of disease (105, 106). To satisfy these requirements, some clinical studies have recognized molecular signatures using PCR in blood samples, which were demonstrated to be effective in prospective cohorts. Such studies utilize gene or protein expression screening approaches (microarrays, RT-qPCR and RNA-seq) to find consistent changes related to specific clinical features, typically comparing patients with TB with patients with LTBI or healthy individuals, and verifying a short-list of chosen candidate markers in follow-up research of TB groups to forecast who will suffer pulmonary disease (107–109). Numerous signatures have been suggested that were more sensitive and specific than the existing screening assays (110–113). In a similar manner, signatures of genes or proteins that are expressed differently in response to TB therapy can assist in clinical follow-up assessment or support prognostic assays of new TB cases (114).
As for cancers and infectious diseases, miRNAs are considered as a probable source of such biomarkers, bringing about numerous discovery reports, which either screen subject-vs-control cohorts or aim to confirm exosomal ncRNAs found in experiments (115). As aforementioned, M. tb infection induces the abnormal expression of genes (including ncRNAs) involved in immune, inflammation, autophagy and apoptosis pathways that can be potential diagnostic or prognostic biomarkers of disease and therapy response. Pedersen et al. have attempted to find known ncRNA signatures that can be assayed in accessible specimens, focusing on circulating ncRNAs, which were measured in blood (PBMCs or serum/plasma) (116). The expected ncRNA signature should specifically find M. tb-infected subjects, and may separate an active infection from LTBI.
The amount of exosomal biomolecules in the blood is difficult to detect. Compared with proteins and lipids, exosomal ncRNAs can be amplified by RT-qPCR, and thus, is more frequently used in the clinic and assayed (117). Due to inclusion in exosomes, ncRNA degradation can be prevented and ncRNAs may be a source of stable RNAs that can be used as disease biomarkers (118). Their enrichment and high stability in exosomes may permit their noninvasive detection in body fluids (119, 120). Therefore, ncRNAs in exosomes may be used in diagnosis, as biomarkers and for therapy in TB infection (Figure 2).
7 Conclusions
In M. tb infection, exosomes from the infected immune cells exert inherent immune regulation effects on anti-TB immunity. Therefore, exploring potential specific components and biological functions of exosomes is helpful in developing novel diagnosis and therapy strategies. Different ncRNA molecules have been found in exosomes after M. tb infection, which shed new light on the potential role of exosomal ncRNAs as novel TB biomarkers for developing the next generation of TB diagnostic strategies. Exosomes have shown strong potential in delivering vaccine components (proteins, peptides and RNA) in different infectious diseases, showing the potential to provide a more effective vaccine strategy for TB.
Author contributions
YL, JWu, and NW conceived the work and wrote the manuscript. JWa and LT designed and revised the manuscript. XY and YJ discussed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 82002111), the Medical Scientific Research Project of Jiangsu Provincial Health Commission (No. Z2019001), the Science and Technology Project of Suzhou (No. KJXW2018068), the Suzhou Science and Technology Development Plan Project (SKY2022078).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Acharya B, Acharya A, Gautam S, Ghimire SP, Mishra G, Parajuli N, et al. Advances in diagnosis of tuberculosis: An update into molecular diagnosis of mycobacterium tuberculosis. Mol Biol Rep (2020) 47(5):4065–75. doi: 10.1007/s11033-020-05413-7
2. Rajaei E, Hadadi M, Madadi M, Aghajani J, Ahmad MM, Farnia P, et al. Outdoor air pollution affects tuberculosis development based on geographical information system modeling. BioMed Biotechnol Res J (2018) 2:39. doi: 10.4103/bbrj.bbrj_5_18
3. Sinigaglia A, Peta E, Riccetti S, Venkateswaran S, Manganelli R, Barzon L. Tuberculosis-associated MicroRNAs: From pathogenesis to disease biomarkers. Cells (2020) 9(10):2160. doi: 10.3390/cells9102160
4. Bouzeyen R, Javid B. Therapeutic vaccines for tuberculosis: An overview. Front Immunol (2022) 13:878471. doi: 10.3389/fimmu.2022.878471
5. Mi J, Liang Y, Liang J, Gong W, Wang S, Zhang J, et al. The research progress in immunotherapy of tuberculosis. Front Cell Infect Microbiol (2021) 11:763591. doi: 10.3389/fcimb.2021.763591
6. Zaheen A, Bloom BR. Tuberculosis in 2020 - new approaches to a continuing global health crisis. N Engl J Med (2020) 382(14):e26. doi: 10.1056/NEJMp2000325
7. Ortiz-Brizuela E, Menzies D, Behr MA. Testing and treating mycobacterium tuberculosis infection. Med Clin North Am (2022) 106(6):929–47. doi: 10.1016/j.mcna.2022.08.001
8. Flynn JL, Chan J. Immune cell interactions in tuberculosis. Cell (2022) 185(25):4682–702. doi: 10.1016/j.cell.2022.10.025
9. Mohammadzadeh R, Ghazvini K, Farsiani H, Soleimanpour S. Mycobacterium tuberculosis extracellular vesicles: Exploitation for vaccine technology and diagnostic methods. Crit Rev Microbiol (2021) 47(1):13–33. doi: 10.1080/1040841X.2020.1830749
10. Abdulgader SM, Okunola AO, Ndlangalavu G, Reeve BWP, Allwood BW, Koegelenberg CFN, et al. Diagnosing tuberculosis: What do new technologies allow us to (Not) do? Respiration (2022) 101(9):797–813. doi: 10.1159/000525142
11. Krug S, Parveen S, Bishai WR. Host-directed therapies: Modulating inflammation to treat tuberculosis. Front Immunol (2021) 12:660916. doi: 10.3389/fimmu.2021.660916
12. Kim JS, Kim YR, Yang CS. Host-directed therapy in tuberculosis: Targeting host metabolism. Front Immunol (2020) 11:1790. doi: 10.3389/fimmu.2020.01790
13. Frydrychowicz M, Kolecka-Bednarczyk A, Madejczyk M, Yasar S, Dworacki G. Exosomes-structure, biogenesis and biological role in non-small-cell lung cancer. Scand J Immunol (2015) 81:2–10. doi: 10.1111/sji.12247
14. Liang S, Ma J, Gong H, Shao J, Li J, Zhan Y, et al. Immune regulation and emerging roles of noncoding RNAs in mycobacterium tuberculosis infection. Front Immunol (2022) 13:987018. doi: 10.3389/fimmu.2022.987018
15. Tang XH, Guo T, Gao XY, Wu XL, Xing XF, Ji JF, et al. Exosome-derived noncoding RNAs in gastric cancer: Functions and clinical applications. Mol Cancer (2021) 20(1):99. doi: 10.1186/s12943-021-01396-6
16. Zhang L, Yu D. Exosomes in cancer development, metastasis, and immunity. Biochim Biophys Acta Rev Cancer (2019) 1871(2):455–68. doi: 10.1016/j.bbcan.2019.04.004
17. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science (2020) 367(6478):eaau6977. doi: 10.1126/science.aau6977
18. Zhu L, Sun HT, Wang S, Huang SL, Zheng Y, Wang CQ, et al. Isolation and characterization of exosomes for cancer research. J Hematol Oncol (2020) 13(1):152. doi: 10.1186/s13045-020-00987-y
19. Kok VC, Yu CC. Cancer-derived exosomes: Their role in cancer biology and biomarker development. Int J Nanomed (2020) 15:8019–36. doi: 10.2147/IJN.S272378
20. Meldolesi J. Exosomes and ectosomes in intercellular communication. Curr Biol (2018) 28(8):R435–44. doi: 10.1016/j.cub.2018.01.059
21. Zhang W, Jiang X, Bao J, Wang Y, Liu H, Tang L. Exosomes in pathogen infections: A bridge to deliver molecules and link functions. Front Immunol (2018) 9:90. doi: 10.3389/fimmu.2018.00090
22. Kourembanas S. Exosomes: Vehicles of intercellular signaling, biomarkers, and vectors of cell therapy. Annu Rev Physiol (2015) 77:13–27. doi: 10.1146/annurev-physiol-021014-071641
23. Dai J, Su Y, Zhong S, Cong L, Liu B, Yang J, et al. Exosomes: Key players in cancer and potential therapeutic strategy. Signal Transduct Target Ther (2020) 5(1):145. doi: 10.1038/s41392-020-00261-0
24. Schorey JS, Harding CV. Extracellular vesicles and infectious diseases: New complexity to an old story. J Clin Invest (2016) 126:1181–9. doi: 10.1172/JCI81132
25. Zhang Y, Bi J, Huang J, Tang Y, Du S, Li P. Exosome: A review of its classification, isolation techniques, storage, diagnostic and targeted therapy applications. Int J Nanomed (2020) 15:6917–34. doi: 10.2147/IJN.S264498
26. Zöller M. Exosomes in cancer disease. Methods Mol Biol (2016) 1381:111–49. doi: 10.1007/978-1-4939-3204-7_7
27. Novais AA, Chuffa LGA, Zuccari DAPC, Reiter RJ. Exosomes and melatonin: Where their destinies intersect. Front Immunol (2021) 12:692022. doi: 10.3389/fimmu.2021.692022
28. Jiang H, Zhao H, Zhang M, He Y, Li X, Xu Y, et al. Hypoxia induced changes of exosome cargo and subsequent biological effects. Front Immunol (2022) 13:824188. doi: 10.3389/fimmu.2022.824188
29. Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res (2012) 40(Database issue):D1241e4. doi: 10.1093/nar/gkr828
30. Yang TT, Liu CG, Gao SC, Zhang Y, Wang PC. The serum exosome derived microRNA-135a, -193b, and -384 were potential alzheimer’s disease biomarkers. BioMed Environ Sci (2018) 31:87–96. doi: 10.3967/bes2018.011
31. Wang J, Wang Y, Tang L, Garcia RC. Extracellular vesicles in mycobacterial infections: Their potential as molecule transfer vectors. Front Immunol (2019) 10:1929. doi: 10.3389/fimmu.2019.01929
32. Jin Y, Xing J, Xu K, Liu D, Zhuo Y. Exosomes in the tumor microenvironment: Promoting cancer progression. Front Immunol (2022) 13:1025218. doi: 10.3389/fimmu.2022.1025218
33. Booton R, Lindsay MA. Emerging role of microRNAs and long noncoding RNAs in respiratory disease. Chest (2014) 146:193–204. doi: 10.1378/chest.13-2736
34. Sessa R, Hata A. Role of microRNAs in lung development and pulmonary diseases. Pulm Circ (2013) 3:315–28. doi: 10.4103/2045-8932.114758
35. Noonin C, Thongboonkerd V. Exosome-inflammasome crosstalk and their roles in inflammatory responses. Theranostics (2021) 11(9):4436–51. doi: 10.7150/thno.54004
36. Liu M, Wang Z, Ren S, Zhao H. Exosomes derived from mycobacterium tuberculosis-infected MSCs induce a pro-inflammatory response of macrophages. Aging (Albany NY) (2021) 13:11595–609. doi: 10.18632/aging.202854
37. Hui WW, Hercik K, Belsare S, Alugubelly N, Clapp B, Rinaldi C, et al. Salmonella enterica serovar typhimurium alters the extracellular proteome of macrophages and leads to the production of proinflammatory exosomes. Infect Immun (2018) 86:e00386–17. doi: 10.1128/IAI.00386-17
38. Torralba D, Baixauli F, Villarroya-Beltri C, Fernández-Delgado I, Latorre-Pellicer A, Acín-Pérez R, et al. Priming of dendritic cells by DNA-containing extracellular vesicles from activated T cells through antigen-driven contacts. Nat Commun (2018) 9:2658. doi: 10.1038/s41467-018-05077-9
39. Cheng Y, Schorey JS. Exosomes carrying mycobacterial antigens can protect mice against mycobacterium tuberculosis infection. Eur J Immunol (2013) 43:3279–90. doi: 10.1002/eji.201343727
40. Cheng Y, Schorey JS. Extracellular vesicles deliver Mycobacterium RNA to promote host immunity and bacterial killing. EMBO Rep (2019) 20:e46613. doi: 10.15252/embr.201846613
41. Weil PP, Hensel KO, Weber D, Postberg J. An expanding universe of noncoding RNAs between the poles of basic science and clinical investigations. Epigenomics (2016) 8(3):317–9. doi: 10.2217/epi.15.65
42. Jiang Y, Zhao L, Wu Y, Deng S, Cao P, Lei X, et al. The role of NcRNAs to regulate immune checkpoints in cancer. Front Immunol (2022) 13:853480. doi: 10.3389/fimmu.2022.853480
43. Maracaja-Coutinho V, Paschoal AR, Caris-Maldonado JC, Borges PV, Ferreira AJ, Durham AM. Noncoding RNAs databases: Current status and trends. Methods Mol Biol (2019) 1912:251–85. doi: 10.1007/978-1-4939-8982-9_10
44. Sampath P, Periyasamy KM, Ranganathan UD, Bethunaickan R. Monocyte and macrophage miRNA: Potent biomarker and target for host-directed therapy for tuberculosis. Front Immunol (2021) 12:667206. doi: 10.3389/fimmu.2021.667206
45. Wang L, Xiong Y, Fu B, Guo D, Zaky MY, Lin X, et al. MicroRNAs as immune regulators and biomarkers in tuberculosis. Front Immunol (2022) 13:1027472. doi: 10.3389/fimmu.2022.1027472
46. Mori MA, Ludwig RG, Garcia-Martin R, Brandão BB, Kahn CR. Extracellular miRNAs: From biomarkers to mediators of physiology and disease. Cell Metab (2019) 30(4):656–73. doi: 10.1016/j.cmet.2019.07.011
47. Xu YX, Pu SD, Li X, Yu ZW, Zhang YT, Tong XW, et al. Exosomal ncRNAs: Novel therapeutic target and biomarker for diabetic complications. Pharmacol Res (2022) 178:106135. doi: 10.1016/j.phrs.2022.106135
48. Li J, Wu X, Ma H, Sun G, Ding P, Lu S, et al. New developments in non-exosomal and exosomal ncRNAs in coronary artery disease. Epigenomics (2022) 14(21):1355–72. doi: 10.2217/epi-2022-0201
49. Li L, Bi Y, Diao S, Li X, Yuan T, Xu T, et al. Exosomal LncRNAs and hepatocellular carcinoma: From basic research to clinical practice. Biochem Pharmacol (2022) 200:115032. doi: 10.1016/j.bcp.2022.115032
50. Lai Y, Dong L, Jin H, Li H, Sun M, Li J. Exosome long non-coding RNA SOX2-OT contributes to ovarian cancer malignant progression by miR-181b-5p/SCD1 signaling. Aging (Albany NY) (2021) 13(20):23726–38. doi: 10.18632/aging.203645
51. Zhao S, Pan T, Deng J, Cao L, Vicencio JM, Liu J, et al. Exosomal transfer of miR-181b-5p confers senescence-mediated doxorubicin resistance via modulating BCLAF1 in breast cancer. Br J Cancer (2022) 128(4):665–77. doi: 10.1038/s41416-022-02077-x
52. Li C, YQ NI, Xu H, QY X, Zhao Y, JK Z, et al. Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Signal Transduct Target Ther (2021) 6(1):383. doi: 10.1038/s41392-021-00779-x
53. Singh PP, Li L, Schorey JS. Exosomal RNA from Mycobacterium tuberculosis infected cells is functional in recipient macrophages. Traffic (2015) 16(6):555–71. doi: 10.1111/tra.12278
54. Kishikawa T, Otsuka M, Ohno M, Yoshikawa T, Takata A, Koike K. Circulating RNAs as new biomarkers for detecting pancreatic cancer. World J Gastroenterol (2015) 21(28):8527–40. doi: 10.3748/wjg.v21.i28.8527
55. Leung RK, Wu YK. Circulating microbial RNA and health. Sci Rep (2015) 5:16814. doi: 10.1038/srep16814
56. Matamala JM, Arias-Carrasco R, Sanchez C, Uhrig M, Bargsted L, Matus S, et al. Genome-wide circulating microRNA expression profiling reveals potential biomarkers for amyotrophic lateral sclerosis. Neurobiol Aging (2018) 64:123–38. doi: 10.1016/j.neurobiolaging.2017.12.020
57. Miller IV, Grunewald TG. Tumour-derived exosomes: Tiny envelopes for big stories. Biol Cell (2015) 107:287–305. doi: 10.1111/boc.201400095
58. Alipoor SD, Adcock IM, Garssen J, Mortaz E, Varahram M, Mirsaeidi M, et al. The roles of miRNAs as potential biomarkers in lung diseases. Eur J Pharmacol (2016) 791:395–404. doi: 10.1016/j.ejphar.2016.09.015
59. Alipoor SD, Mortaz E, Garssen J, Movassaghi M, Mirsaeidi M, Adcock IM. Exosomes and exosomal miRNA in respiratory diseases. Mediators Inflammation (2016) 2016:5628404. doi: 10.1155/2016/5628404
60. Gurung S, Perocheau D, Touramanidou L, Baruteau J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun Signal (2021) 19(1):47. doi: 10.1186/s12964-021-00730-1
61. Preethi KA, Selvakumar SC, Ross K, Jayaraman S, Tusubira D, Sekar D. Liquid biopsy: Exosomal microRNAs as novel diagnostic and prognostic biomarkers in cancer. Mol Cancer (2022) 21(1):54. doi: 10.1186/s12943-022-01525-9
62. Lin Y, Zhang Y, Yu H, Tian R, Wang G, Li F. Identification of unique key genes and miRNAs in latent tuberculosis infection by network analysis. Mol Immunol (2019) 112:103–14. doi: 10.1016/j.molimm.2019.04.032
63. Furci L, Schena E, Miotto P, Cirillo DM. Alteration of human macrophages microRNA expression profile upon infection with mycobacterium tuberculosis. Int J Mycobacteriol (2013) 2:128–34. doi: 10.1016/j.ijmyco.2013.04.006
64. Kundu M, Basu J. The role of microRNAs and long non-coding RNAs in the regulation of the immune response to Mycobacterium tuberculosis infection. Front Immunol (2021) 12:687962. doi: 10.3389/fimmu.2021.687962
65. Mehrotra P, Jamwal SV, Saquib N, Sinha N, Siddiqui Z, Manivel V, et al. Pathogenicity of Mycobacterium tuberculosis is expressed by regulating metabolic thresholds of the host macrophage. PloS Pathog (2014) 10:e1004265. doi: 10.1371/journal.ppat.1004265
66. Eisenreich W, Heesemann J, Rudel T, Goebel W. Metabolic host responses to infection by intracellular bacterial pathogens. Front Cell Infect Microbiol (2013) 3:24. doi: 10.3389/fcimb.2013.00024
67. Enriquez AB, Izzo A, Miller SM, Stewart EL, Mahon RN, Frank DJ, et al. Advancing adjuvants for Mycobacterium tuberculosis therapeutics. Front Immunol (2021) 12:740117. doi: 10.3389/fimmu.2021.740117
68. Smith VL, Cheng Y, Bryant BR, Schorey JS. Exosomes function in antigen presentation during an in vivo mycobacterium tuberculosis infection. Sci Rep (2017) 7:43578. doi: 10.1038/srep43578
69. Hadifar S, Fateh A, Yousefi MH, Siadat SD, Vaziri F. Exosomes in tuberculosis: Still terra incognita? J Cell Physiol (2019) 234(3):2104–11. doi: 10.1002/jcp.27555
70. Wang C, Yang S, Sun G, Tang X, Lu S, Neyrolles O, et al. Comparative miRNA expression profiles in individuals with latent and active tuberculosis. PloS One (2011) 6:e25832. doi: 10.1371/journal.pone.0025832
71. Wang Y, Xu YM, Zou YQ, Lin J, Huang B, Liu J, et al. Identification of differential expressed PE exosomal miRNA in lung adenocarcinoma, tuberculosis, and other benign lesions. Med (Baltimore) (2017) 96(44):e8361. doi: 10.1097/MD.0000000000008361
72. Kaushik AC, Wu Q, Lin L, Li H, Zhao L, Wen Z, et al. Exosomal ncRNAs profiling of mycobacterial infection identified miRNA-185-5p as a novel biomarker for tuberculosis. Brief Bioinform (2021) 22(6):bbab210. doi: 10.1093/bib/bbab210
73. Zhan X, Yuan W, Zhou Y, Ma R, Ge Z. Small RNA sequencing and bioinformatics analysis of RAW264.7-derived exosomes after Mycobacterium bovis bacillus calmette-guérin infection. BMC Genomics (2022) 23(1):355. doi: 10.1186/s12864-022-08590-w
74. Alipoor SD, Tabarsi P, Varahram M, Movassaghi M, Dizaji MK, Folkerts G, et al. Serum exosomal miRNAs are associated with active pulmonary tuberculosis. Dis Markers (2019) 2019:1907426. doi: 10.1155/2019/1907426
75. Hu X, Liao S, Bai H, Wu L, Wang M, Wu Q, et al. Integrating exosomal microRNAs and electronic health data improved tuberculosis diagnosis. EBioMedicine (2019) 40:564–73. doi: 10.1016/j.ebiom.2019.01.023
76. Alipoor SD, Mortaz E, Tabarsi P, Farnia P, Mirsaeidi M, Garssen J, et al. Bovis bacillus calmette-guerin (BCG) infection induces exosomal miRNA release by human macrophages. J Transl Med (2017) 15(1):105. doi: 10.1186/s12967-017-1205-9
77. Mortaz E, Alipoor SD, Tabarsi P, Adcock IM, Garssen J, Velayati AA. The analysis of exosomal micro-RNAs in peripheral blood mononuclear cell-derived macrophages after infection with bacillus calmette-guérin by RNA sequencing. Int J Mycobacteriol (2016) 5(Suppl 1):S184–5. doi: 10.1016/j.ijmyco.2016.09.045
78. Guio H, Aliaga-Tobar V, Galarza M, Pellon-Cardenas O, Capristano S, Gomez HL, et al. Comparative profiling of circulating exosomal small RNAs derived from Peruvian patients with tuberculosis and pulmonary adenocarcinoma. Front Cell Infect Microbiol (2022) 12:909837. doi: 10.3389/fcimb.2022.909837
79. Lyu L, Zhang X, Li C, Yang T, Wang J, Pan L, et al. Small RNA profiles of serum exosomes derived from individuals with latent and active tuberculosis. Front Microbiol (2019) 10:1174. doi: 10.3389/fmicb.2019.01174
80. Carranza C, Herrera MT, Guzmán-Beltrán S, Salgado-Cantú MG, Salido-Guadarrama I, Santiago E, et al. A dual marker for monitoring MDR-TB treatment: Host-derived miRNAs and m. tuberculosis derived RNA sequences in serum. Front Immunol (2021) 12:760468. doi: 10.3389/fimmu.2021.760468
81. Gallo A, Tandon M, Alevizos I, Illei GG. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PloS One (2012) 7:e30679. doi: 10.1371/journal.pone.0030679
82. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet (2019) 20:675–91. doi: 10.1038/s41576-019-0158-7
83. Chen X, Yang T, Wang W, Xi W, Zhang T, Li Q, et al. Circular RNAs in immune responses and immune diseases. Theranostics (2019) 9:588–607. doi: 10.7150/thno.29678
84. Wang S, Dong Y, Gong A, Kong H, Gao J, Hao X, et al. Exosomal circRNAs as novel cancer biomarkers: Challenges and opportunities. Int J Biol Sci (2021) 17(2):562–73. doi: 10.7150/ijbs.48782
85. Li R, Jiang J, Shi H, Qian H, Zhang X, Xu W. CircRNA: A rising star in gastric cancer. Cell Mol Life Sci (2020) 77(9):1661–80. doi: 10.1007/s00018-019-03345-5
86. Yi Z, Gao K, Li R, Fu Y. Dysregulated circRNAs in plasma from active tuberculosis patients. J Cell Mol Med (2018) 22(9):4076–4084.7. doi: 10.1111/jcmm.13684
87. Qian Z, Liu H, Li M, Shi J, Li N, Zhang Y, et al. Potential diagnostic power of blood circular RNA expression in active pulmonary tuberculosis. EBioMedicine (2018) 27:18–26. doi: 10.1016/j.ebiom.2017.12.007
88. Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer (2019) 18(1):116. doi: 10.1186/s12943-019-1041-z
89. Lakshmi S, Hughes TA, Priya S. Exosomes and exosomal RNAs in breast cancer: A status update. Eur J Cancer (2021) 144:252–68. doi: 10.1016/j.ejca.2020.11.033
90. Li Y, Yin Z, Fan J, Zhang S, Yang W. The roles of exosomal miRNAs and lncRNAs in lung diseases. Signal Transduct Target Ther (2019) 4:47. doi: 10.1038/s41392-019-0080-7
91. Vosough P, Khatami SH, Hashemloo A, Tajbakhsh A, Karimi-Fard F, Taghvimi S, et al. Exosomal lncRNAs in gastrointestinal cancer. Clin Chim Acta (2022) 540:117216. doi: 10.1016/j.cca.2022.117216
92. Wang L, Xie B, Zhang P, Ge Y, Wang Y, Zhang D. LOC152742 as a biomarker in the diagnosis of pulmonary tuberculosis infection. J Cell Biochem (2019) 120(6):8949–55. doi: 10.1002/jcb.27452
93. Chen ZL, Wei LL, Shi LY, Li M, Jiang TT, Chen J, et al. Screening and identification of lncRNAs as potential biomarkers for pulmonary tuberculosis. Sci Rep (2017) 7(1):16751. doi: 10.1038/s41598-017-17146-y
94. He J, Ou Q, Liu C, Shi L, Zhao C, Xu Y, et al. Differential expression of long non-coding RNAs in patients with tuberculosis infection. Tuberculosis (Edinb) (2017) 107:73–9. doi: 10.1016/j.tube.2017.08.007
95. Fang Y, Zhao J, Wang X, Wang X, Wang L, Liu L, et al. Identification of differentially expressed lncRNAs as potential plasma biomarkers for active tuberculosis. Tuberculosis (Edinb) (2021) 128:102065. doi: 10.1016/j.tube.2021.102065
96. Li ZB, Han YS, Wei LL, Shi LY, Yi WJ, Chen J, et al. Screening and identification of plasma lncRNAs uc.48+ and NR_105053 as potential novel biomarkers for cured pulmonary tuberculosis. Int J Infect Dis (2020) 92:141–50. doi: 10.1016/j.ijid.2020.01.005
97. Fu Y, Gao K, Tao E, Li R, Yi Z. Aberrantly expressed long non-coding RNAs in CD8(+) T cells response to active tuberculosis. J Cell Biochem (2017) 118(12):4275–84. doi: 10.1002/jcb.26078
98. Wang Y, Zhong H, Xie X, Chen CY, Huang D, Shen L, et al. Long noncoding RNA derived from CD244 signaling epigenetically controls CD8+ T-cell immune responses in tuberculosis infection. Proc Natl Acad Sci U.S.A. (2015) 112(29):E3883–92. doi: 10.1073/pnas.1501662112
99. Yi Z, Li J, Gao K, Fu Y. Identification of differentially expressed long non-coding RNAs in CD4+ T cells response to latent tuberculosis infection. J Infect (2014) 69(6):558–68. doi: 10.1016/j.jinf.2014.06.016
100. Deng G, Ji N, Shi X, Zhang W, Qin Y, Sha S, et al. Effects of Mycobacterium tuberculosis Rv1096 on mycobacterial cell division and modulation on macrophages. Microb Pathog (2020) 141:103991. doi: 10.1016/j.micpath.2020.103991
101. Lv L, Li C, Zhang X, Ding N, Cao T, Jia X, et al. RNA Profiling analysis of the serum exosomes derived from patients with active and latent Mycobacterium tuberculosis infection. Front Microbiol (2017) 8:1051. doi: 10.3389/fmicb.2017.01051
102. Gutkin A, Uziel O, Beery E, Nordenberg J, Pinchasi M, Goldvaser H, et al. Tumor cells derived exosomes contain hTERT mRNA and transform nonmalignant fibroblasts into telomerase positive cells. Oncotarget (2016) 7:59173–88. doi: 10.18632/oncotarget.10384
103. Bhatnagar S, Shinagawa K, Castellino FJ, Schorey JS. Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response. Vitro vivo. Blood (2007) 110:3234–44. doi: 10.1182/blood-2007-03-079152
104. Singh PP, LeMaire C, Tan JC, Zeng E, Schorey JS. Exosomes released from m. tuberculosis infected cells can suppress IFN-γ mediated activation of naïve macrophages. PloS One (2011) 6:e18564. doi: 10.1371/journal.pone.0018564
105. Fiore-Gartland A, Carpp LN, Naidoo K, Thompson E, Zak DE, Self S, et al. Considerations for biomarker-targeted intervention strategies for tuberculosis disease prevention. Tuberculosis (2018) 109:61–8. doi: 10.1016/j.tube.2017.11.009
106. Levin M, Kaforou M. Predicting active tuberculosis progression by RNA analysis. Lancet (2016) 387:2268–70. doi: 10.1016/S0140-6736(16)00165-3
107. Sigal GB, Segal MR, Mathew A, Jarlsberg L, Wang M, Barbero S, et al. Biomarkers of tuberculosis severity and treatment effect: A directed screen of 70 host markers in a randomized clinical trial. EBioMedicine (2017) 25:112–21. doi: 10.1016/j.ebiom.2017.10.018
108. Anderson ST, Kaforou M, Brent AJ, Wright VJ, Banwell CM, Chagaluka G, et al. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N Engl J Med (2014) 370:1712–23. doi: 10.1056/NEJMoa1303657
109. Leong S, Zhao Y, Joseph NM, Hochberg NS, Sarkar S, Pleskunas J, et al. Existing blood transcriptional classifiers accurately discriminate active tuberculosis from latent infection in individuals from south India. Tuberculosis (2018) 109:41–51. doi: 10.1016/j.tube.2018.01.002
110. Maertzdorf J, McEwen G, Weiner J, Tian S, Lader E, Schriek U, et al. Concise gene signature for point-of-care classification of tuberculosis. EMBO Mol Med (2016) 8:86–95. doi: 10.15252/emmm.201505790
111. Sambarey A, Devaprasad A, Mohan A, Ahmed A, Nayak S, Swaminathan S, et al. Unbiased identification of blood-based biomarkers for pulmonary tuberculosis by modeling and mining molecular interaction networks. EBioMedicine (2017) 15:112–26. doi: 10.1016/j.ebiom.2016.12.009
112. Suliman S, Thompson EG, Sutherland J, Weiner J 3rd, Ota MOC, Shankar S, et al. Four-gene pan-African blood signature predicts progression to tuberculosis. Am J Respir Crit Care Med (2018) 197(9):1198–208. doi: 10.1164/rccm.201711-2340OC
113. Sweeney TE, Braviak L, Tato CM, Khatri P. Genome-wide expression for diagnosis of pulmonary tuberculosis: A multicohort analysis. Lancet Respir Med (2016) 4:213–24. doi: 10.1016/S2213-2600(16)00048-5
114. Zak DE, Penn-Nicholson A, Scriba TJ, Thompson E, Suliman S, Amon LM, et al. A blood RNA signature for tuberculosis disease risk: A prospective cohort study. Lancet (2016) 387:2312–22. doi: 10.1016/S0140-6736(15)01316-1
115. Thompson EG, Du Y, Malherbe ST, Shankar S, Braun J, Valvo J, et al. Host blood RNA signatures predict the outcome of tuberculosis treatment. Tuberculosis (2017) 107:48–58. doi: 10.1016/j.tube.2017.08.004
116. Pedersen JL, Bokil NJ, Saunders BM. Developing new TB biomarkers, are miRNA the answer? Tuberculosis (2019) 118:101860. doi: 10.1016/j.tube.2019.101860
117. Cheng L, Sharples RA, Scicluna BJ, Hill AF. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J Extracell Vesicles (2014) 3:1–14. doi: 10.3402/jev.v3.23743
118. Ge Q, Zhou Y, Lu J, Bai Y, Xie X, Lu Z. miRNA in plasma exosome is stable under different storage conditions. Molecules (2014) 19:1568–75. doi: 10.3390/molecules19021568
119. Dejima H, Iinuma H, Kanaoka R, Matsutani N, Kawamura M. Exosomal microRNA in plasma as a non-invasive biomarker for the recurrence of non-small cell lung cancer. Oncol Lett (2017) 13:1256–63. doi: 10.3892/ol.2017.5569
Keywords: exosomes, non-coding RNAs, Mycobacterium tuberculosis, tuberculosis, biomarkers
Citation: Wang J, Li Y, Wang N, Wu J, Ye X, Jiang Y and Tang L (2023) Functions of exosomal non-coding RNAs to the infection with Mycobacterium tuberculosis. Front. Immunol. 14:1127214. doi: 10.3389/fimmu.2023.1127214
Received: 19 December 2022; Accepted: 13 March 2023;
Published: 22 March 2023.
Edited by:
Louisy Santos, Rio de Janeiro State University, BrazilReviewed by:
Erick Oliveira, Juiz de Fora Federal University, BrazilJoyoti Basu, Bose Institute, India
Esmaeil Mortaz, Shahid Beheshti University of Medical Sciences, Iran
Copyright © 2023 Wang, Li, Wang, Wu, Ye, Jiang and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lijun Tang, dGxqeGllQGNzdS5lZHUuY24=; Jianjun Wang, d2FuZ2ppYW5qdW4wNTIwQDE2My5jb20=
†These authors have contributed equally to this work
 Jianjun Wang
Jianjun Wang Yujie Li1†
Yujie Li1† Nan Wang
Nan Wang Xiaojian Ye
Xiaojian Ye Lijun Tang
Lijun Tang