- 1School of Life Science, Tianjin University, Tianjin, China
- 2China Animal Health and Epidemiology Center, Qingdao, Shandong, China
Introduction: The H5N8 influenza virus is a highly pathogenic pathogen for poultry and human. Vaccination is the most effective method to control the spread of the virus right now. The traditional inactivated vaccine, though well developed and used widely, is laborious during application and more interests are stimulated in developing alternative approaches.
Methods: In this study, we developed three hemagglutinin (HA) gene-based yeast vaccine. In order to explore the protective efficacy of the vaccines, the gene expression level in the bursa of Fabricius and the structure of intestinal microflora in immunized animals were analyzed by RNA seq and 16SrRNA sequencing, and the regulatory mechanism of yeast vaccine was evaluated.
Results: All of these vaccines elicited the humoral immunity, inhibited viral load in the chicken tissues, and provided partial protective efficacy due to the high dose of the H5N8 virus. Molecular mechanism studies suggested that, compared to the traditional inactivated vaccine, our engineered yeast vaccine reshaped the immune cell microenvironment in bursa of Fabricius to promote the defense and immune responses. Analysis of gut microbiota further suggested that oral administration of engineered ST1814G/H5HA yeast vaccine increased the diversity of gut microbiota and the increasement of Reuteri and Muciniphila might benefit the recovery from influenza virus infection. These results provide strong evidence for further clinical use of these engineered yeast vaccine in poultry.
Introduction
The H5N8 avian influenza viruses are highly pathogenic threats to poultry and public health (1, 2). The H5N8 subtype AIV has spread globally along with the migration of wild birds and human infection cases have been reported continually during the last decades (3–5). Vaccination is one of the most effective methods to prevent and control the pandemic influenza right now (6, 7). The traditional inactivated vaccine and protein subunit vaccine are widely used vaccines in China (8, 9). The inactivated vaccine for H5N8, although used for decades, has its critical limitations, including a lengthy manufacturing process, the need for lots of fertilized chicken eggs, and a laborious vaccination procedure (10, 11). Moreover, antigen drift is common phenomena in avian influenza H5N8 virus infection (12). The antigenicity changes will reduce the neutralization effect of the inactivated vaccine against the popular virus and the resistance of the vaccine to different subtypes of avian influenza, such as H5N6, has great differences. A safe, convenience and effective vehicle for rapidly developing vaccine against popular subtypes of avian influenza is urgently needed. Viral proteins such as HA have been demonstrated to be administrated as protein subunit vaccine to provide protective efficacy against influenza, with virus vector or yeasts (13). Adenovirus, Newcastle disease virus and fowlpox virus have been reported to be used as vehicles of the influenza protein subunit vaccine for their immunogenicity as virus (14). In this case, reverse genetic manipulation of recombinant virus with proper antigen is a complicated and critical step for successful vaccines (15, 16). Yeast surface display approach has been explored for vaccines against different viruses (17–20). There are several advantages of using yeast as vaccine vehicles (21). First, yeast can be easily modified to express new antigen protein on the surface of yeast or secreted to the culture medium. Second, the oral administration of yeast could stimulate the mucosal-associated immunity in the context of vaccination, as the properties of yeast wall cell composition could promote immune stimulation and the whole cell of yeast could be engulfed by professional antigen-presenting cells due to its diameter. Third, the antigen target protein expressed by yeast could be further processed by glycosylation modification, which may facilitate their recognition by host immune system and enhance the related immune responses. Besides, there are a few reports about yeast vaccine for the prophylactic and therapeutic cases recently. However, the molecular mechanism of protective efficacy provided by the yeast vaccine was not fully understood.
The concept of nucleic acid vaccine is to deliver target antigen DNA or RNA to cells of the immune system (22, 23). Use of mRNA COVID-19 vaccine has been permitted and the immunization programs have been carried out in several countries around the world since 2020 (24). In contrast to the inactivated vaccine and the protein subunit vaccine, nucleic acid vaccine is thought to be safer, cheaper and easily manufactured with stronger specific immune response. Consistent with the idea of nucleic acid vaccine, yeast is widely used in the delivery of functional DNA and mRNA molecules to monocytes and dendritic cells, which indicated yeast as a promising delivery vehicle of DNA and RNA vaccine. But there is no report about its feasibility as nucleic acid vaccine against the H5N8 avian influenza virus until now.
The intestinal microbial community structure of chicken is diverse and the community is composed of different bacteria, methanogenic archaea, fungi and viruses, which play an important role in inducing and regulating host responses to a variety of pathogens, such as AIV (25, 26). It is reported that the use of probiotics to alter the composition of intestinal microflora has a beneficial effect on immunity to influenza virus infection, mainly due to the activation of natural killer (NK) cells in the lung and spleen and the increased expression of various cytokines in the lung (27). Therefore, understanding the changes of chicken intestinal microbiota after immunization is very important to develop effective yeast vaccines to enhance immunity to the virus.
Herein, to further explore the molecular mechanism of yeast vaccine and evaluate the effect of H5HA-based specific yeast vaccine on chicken upon the H5N8 virus challenge, we employed Saccharomyces cerevisiae ST1814G strain to develop three types of yeast vaccine, namely, yeast surface display-based H5HA protein subunit vaccine, H5HA gene-based DNA and RNA combined nucleic acid vaccine, and a protein-nucleic acid combined H5HA vaccine. Our experimental results indicated that our engineered yeast vaccine elicited the humoral immunity and provided partial protective efficacy upon high dose of the H5N8 virus infection. Our mechanism studies suggested that engineered yeast vaccine could reshape the immune cell microenvironment and enhance the defense and immune response of bursa of Fabricus. On the other hand, the diversity of gut microbiota was increased after oral administration of our engineered ST1814G/H5HA yeast vaccine, which might further reduce the intestinal tract inflammation, protect intestinal barrier and regulate the metabolic processes to benefit for the recovery of chicken from influenza virus infection. Taken together, our experimental results indicate that the yeast vaccine developed in this study have great potentialities in the use of poultry in future.
Results
The design of recombinant protein vaccine, nucleic acid vaccine and protein-nucleic acid combined vaccine
We firstly constructed a recombinant S. cerevisiae ST1814G (MATa aga1 his3Δ200 leu2Δ0 lys2Δ0 trp1Δ63 ura3Δ0 met15Δ0) strain expressing 17-484 amino acids of H5HA protein excluding the signaling peptides, which was integrated into the S. cerevisiae chromosome IV according to the methods as our previous studies (28), and the HA protein was expressed correctly as expected as shown in Figure 1A. This genetically engineered yeast strain was used as recombinant protein vaccine (ST1814G/H5HA) and it also serves as yeast chassis for our next protein-nucleic acid combined vaccine.
Figure 1 Schematic diagram of vaccines preparation, chicken vaccination and virus challenge experiments. (A) Preparation of protein subunit vaccine. The plasmids were constructed as described in Materials and Methods and then transformed into Saccharomyces cerevisiae ST1814G strain to prepare ST1814G/H5HA vaccine. Western blot analysis showed that HA protein could be expressed normally in ST1814G/H5HA vaccine. (M: protein marker; CK: ST1814G yeast lysates, lane 1-2: ST1814G/H5HA yeast lysates. (B) Preparation of DNA-RNA nucleic acid vaccine. The plasmids were constructed as described in Materials and Methods and then transformed into Saccharomyces cerevisiae ST1814G strain or ST1814G/H5HA strain to prepare ST1814G/DNA-RNA vaccine or ST1814G/H5HA/DNA-RNA vaccine. (C) Both the eGFP protein and the mCherry protein could be efficiently expressed in stomach and intestine tissues of chickens. (D) Seven groups of two-week-old chickens were immunized as the schedule indicated and they were challenged intramuscularly with the H5N8 virus on the 6th day after the third immunization.
To develop an efficient nucleic acid vaccine system, we constructed a plasmid chassis of DNA and RNA combined nucleic acid vaccine, including a CMV promoter and IRES sequence-driven DNA nucleic acid element and a GAP promoter and IRES sequence-driven mRNA nucleic acid element (Figure 1B). The IRES element is supposed to increase levels of translation of recombinant antigen in mammalian cells and prevent translation in yeasts. The CMV promoter and GAP promoter were supposed to promote the transcription of target genes in mammalian cells and yeast, respectively. Based on ST1814G and ST1814G/H5HA yeast strain, we constructed an eGFP-mCherry vaccine (ST1814G/eGFP-mCherry), which was used for the efficacy test of the plasmid chassis, an H5HA-DNA-mRNA vaccine (ST1814G/DNA-mRNA), which was used as nucleic acid vaccines, and an H5HA protein-nucleic acid combined vaccine (ST1814G/H5HA/DNA-mRNA). As shown in Figure 1C, both the eGFP and the mCherry proteins could be efficiently expressed in stomach and intestine tissues of chickens. Therefore, the designed antigen, H5HA protein, should be efficiently expressed theoretically both in the ST1814G/DNA-mRNA nucleic acid vaccine and in the ST1814G/H5HA/DNA-mRNA protein-nucleic acid combined vaccine.
Stronger humoral immunity and efficient protective effect were elicited after vaccination with our engineered yeast vaccine
To better understand the efficacy of our engineered yeast vaccine, we next carried out the vaccination procedure and virus challenge experiments as indicated in Figure 1D and subsequently, the humoral immune responses were evaluated by ELISA assay as planned. As shown in Figures 2A–D, the antibody level of both the secreted IgA in anal swabs and the IgG in serum of the immunized groups, whatever the commercial inactivated vaccine or each of our designed engineered yeast vaccine, were higher than those in PBS treated group or ST1814G yeast treated group before the H5N8 virus challenge. Interestingly, compared to the yeast vaccine alone, our protein-nucleic acid vaccine (ST1814G/H5HA/DNA-mRNA) and recombinant protein vaccine combined with the commercial vaccine (ST1814G/H5HA/Inactivated) exhibited excellent abilities to elicit high levels of IgG and IgA, even after the H5N8 virus challenge. These results indicated our engineered yeast vaccine stimulate the humoral immune responses efficiently against the H5N8 virus infection. Consistent with the humoral immune responses, although it is lethal to the chickens of all the groups with our experimental dose, our yeast-based vaccine could prolong the survival time of infected chickens compared with the PBS treated or ST1814G yeast treated group, which began to die on the second day and over 50 percent of them was dead on the third day (Figure 2E).
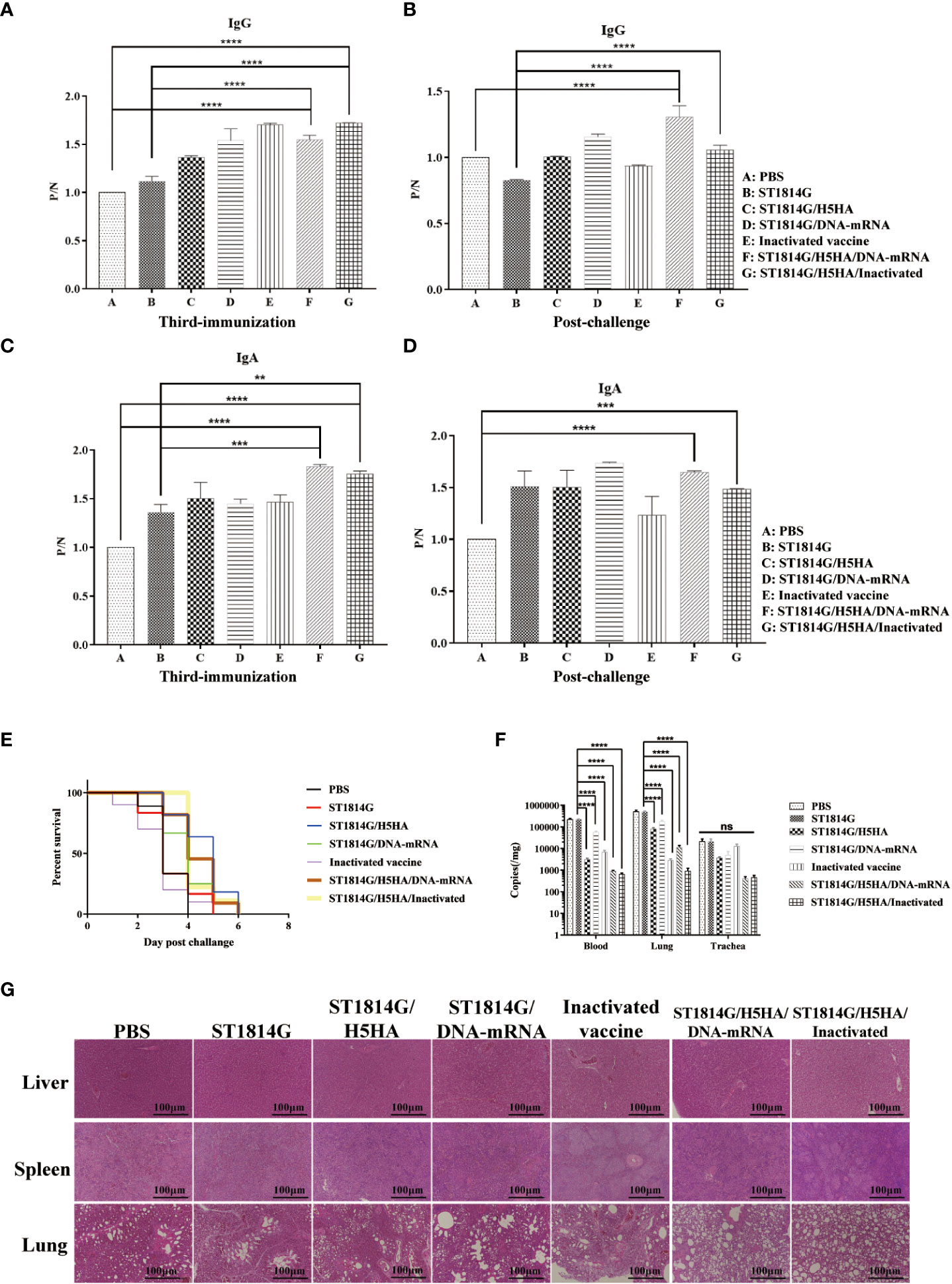
Figure 2 Protective effects of engineered yeast vaccine. (A–D) The level of both the secreted IgA in anal swabs and the IgG in serum of each group. The significance of differences was determined by two-way analysis of variance. (A, C), samples after the third immunizations; (B, D), samples post challenge), (**p < 0.01; ***p < 0.001; ****p < 0.001). (E) Survival rate of each group at the indicated time post infection. (F) Virus titer determination of each group in the blood and lung. Statistical analysis was analyzed by Student’s t-test. (****p < 0.001 n=3 chickens for each group). (G) H&E-stained section of the liver and spleen from control group showing spot of bleeding, the lung was showing a pattern of necrotizing alveolitis, the alveolar walls are necrotic and alveolar air spaces contained inflammatory cells in the control groups.
To further confirm the protective effect of our engineered yeast vaccine, we measured the virus titer in the blood, lung (Figure 2F) and trachea (Supplementary Figure 1A) of each group. As shown in Figure 2F, although combined use could not further decrease the viral load compared with the inactivated vaccine, yeast-based vaccines, both ST1814G/H5HA and ST1814G/DNA-mRNA, were significantly decreased the viral load in blood and lung tissue, which suggested our yeast vaccine could also provide comparable protective efficacy compared with the inactivated vaccine.
The histopathological examination showed that the H5N8 virus infection induced spot of bleeding in liver and spleen, the central vein of liver was congested with lymphocyte infiltration and the boundary between red pulp and white pulp of spleen was not clear, the lung showed a pattern of necrotizing alveolitis, the alveolar walls were necrotic and alveolar air spaces contained inflammatory cells in the control groups (Figure 2G). Meanwhile, the immunized groups presented only slight histopathological changes compared with the control groups, indicating that our engineered yeast vaccine could effectively alleviate the histopathological damage caused by the H5N8 infection in chickens.
Collectively, these results demonstrate that our engineered H5HA-based yeast vaccine could elicit the humoral immunity and provide protective efficacy for chickens after the H5N8 virus infection and we found that simultaneous administration of different types vaccines may be a better choice for the vaccination procedure.
The immune cell microenvironment of bursa of Fabriciu was reshaped to enhance the defense and immune response
Yeasts have been considered as a promising delivery platform for recombinant protein and nucleic acid vaccines for decades. To better understand the molecular mechanisms of the protective efficacy after immunized with yeast vehicles and the traditional inactivated vaccines, we next performed RNA sequencing using the bursa of Fabricius(BF) of chickens and the differentially-expressed genes (DEGs) were proceeded to enrichment analysis based on the Gene Ontology databases. With regards to our engineered yeast vaccine, lots of defense-related genes were significantly upregulated in chickens immunized with ST1814G/H5HA vaccine compared with those in the ST1814G yeast placebo (Figures 3A). The related biological processes included defense response, immune response and response to biotic stimulus, which might be responsible for the protective efficacy of our engineered yeast vaccine in chickens upon the H5N8 virus infection. For the traditional inactivated vaccine, enrichment analysis of upregulated genes also indicated that defense-related biological processes were activated in these chickens (Figure 3B). To further verify the influence of engineered yeast vaccine on the immune system of chicken, we next checked and verified the gene expression levels of immune system process (Supplementary Figures 1B, D). We found that, compared with the ST1814G group and the inactivated vaccine group, there were about 73 genes upregulated significantly, which play roles on positive regulation of the immune system process, and about 14 genes downregulated significantly, which play roles on negative regulation of the immune system process in the ST1814G/H5HA groups (Figure 3C).
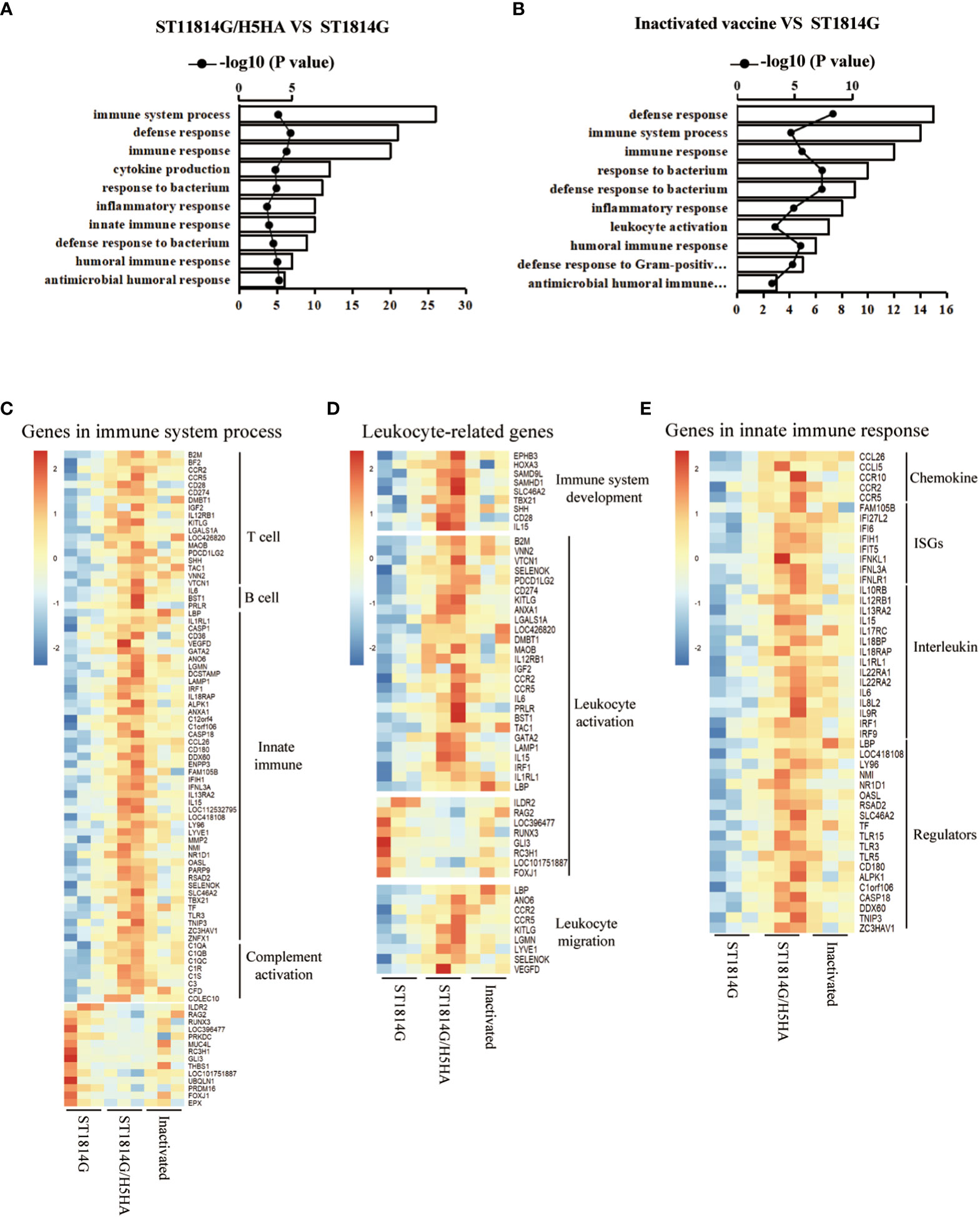
Figure 3 Effect of Vaccines on immune related genes of bursa of Fabricius in chickens. (A, B) Biological process analysis of the differentially-expressed genes (DEGs) based on the Gene Ontology databases. Abscissa: biological process; Ordinate: number of genes and –log10 (P value); (A) ST1814G/H5HA group vs ST1814G group; (B) Inactivated vaccine group vs ST1814G group. (C–E) Heatmaps of gene expression levels in BF for indicated gene lists. (C) Genes related to immune system process; (D) Genes related to leukocyte microenvironment of bursa of Fabricius; (E) Genes related to innate immune response.
The bursa of Fabricius is an important organ of chicken for the immune cell differentiation and proliferation and the microenvironments are crucial factors for the immune cell determination. As the immune system processes were significantly different in our engineered yeast vaccine group, we next checked the expression levels of leukocyte type-related genes to generally evaluate the change of the microenvironment of bursa of Fabricius. As shown in Figure 3D, compared with the ST1814G group and the inactivated vaccine group, the expression levels of genes regulating the immune system development and leukocyte migration were upregulated significantly in ST1814G/H5HA group. At the same time, the expression levels of genes, which were reported to positively regulate of lymphocyte, such as T cell and B cell, were also increased in ST1814G/H5HA group compared with the ST1814G group or the inactivated vaccine group. Additionally, the expression levels of genes in innate immune response, such as the interleukin receptors (ILRs), interferon ligands (IFNLs) and C-C Motif Chemokines receptors (CCLRs), which are important for the lymphocyte function, were also increased in the ST1814G/H5HA group (Figure 3E). These results suggested that the immune cell types produced in the bursa of Fabricius of ST1814G/H5HA group might be different compared with the ST1814G group and the inactivated vaccine group. Collectively, our above results indicated that the stimulation with our yeast-based influenza vaccine might reshape the immune cell microenvironment and it would elicit much more immune and defense responses compared with the traditional inactivated vaccine in chickens.
Yeast vaccine elevates the diversity of gut microbiota that might benefit the recovery from influenza virus infection
Microorganisms play important roles in the regulation of gut micro-ecology balance, especially during influenza virus infection. To better understand the molecular mechanism of the protective effect on the gut microbiota, we next performed 16S rRNA sequencing and investigated the diversity of gut microbiota in different groups after the H5N8 virus challenge. As shown in Figure 4A, the results indicated that there were much more species special sequences in the ST1814G/H5HA/DNA-RNA group and the ST1814G/H5HA/Inactivated group than the PBS group or the ST1814G group while the inactivated vaccine group did not show this advantage.
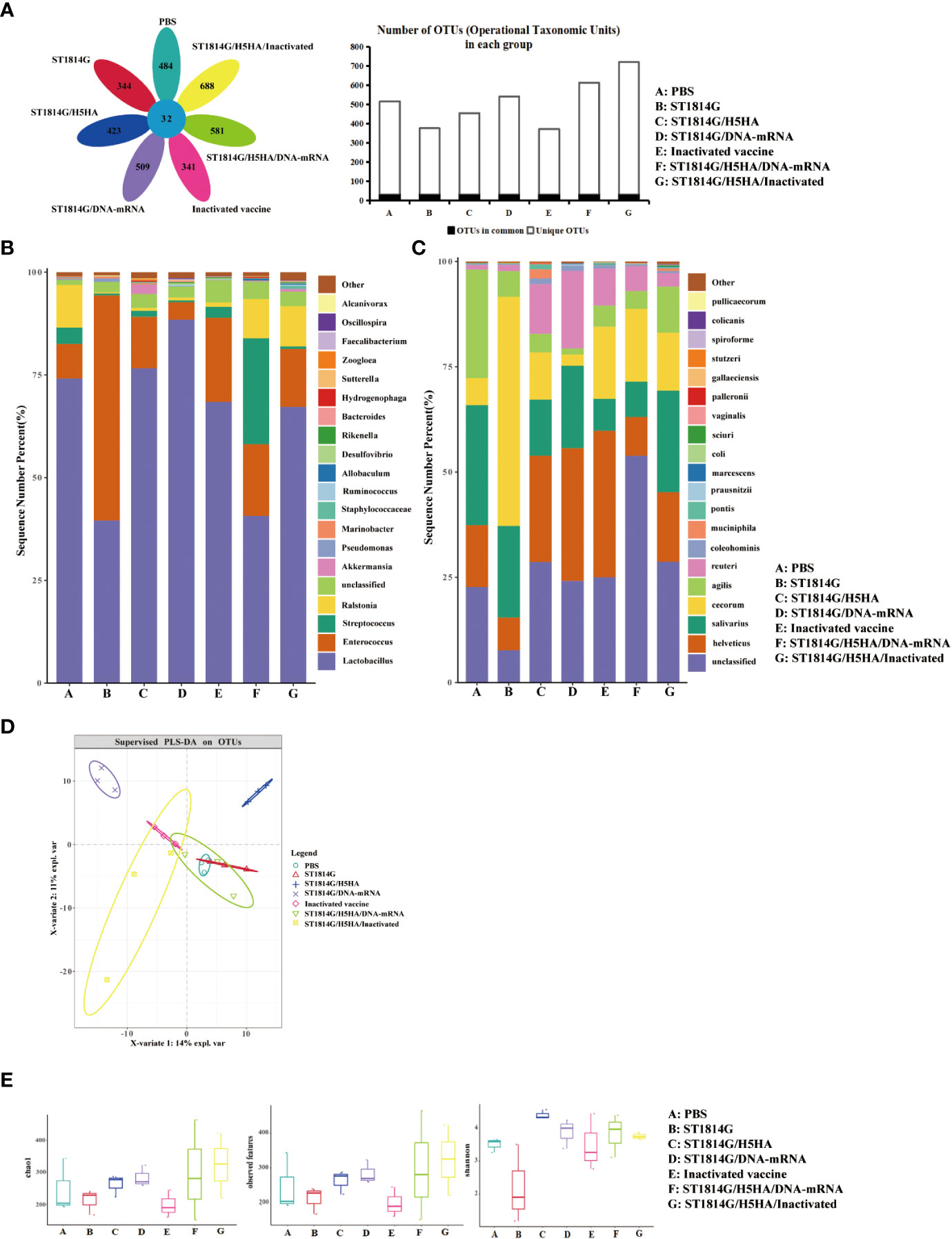
Figure 4 Effect of vaccines on intestinal microflora diversity post immunization. (A) Venn diagrams (left) and bar charts (right) of common or endemic species. (B) Relative content of bacterial genus in each group. (C) Relative content of bacterial species in each group. (D) Partial least-squares discrimination analysis (PLS-DA) revealed the diversity of chicken gut microbiota among the seven groups. Dots represent individual samples. (E) The analysis of Alpha diversity index, Chao1, Shannon and observed features, showed the diversity of chicken gut microbiota within each group post immunization. Dots represent individual samples.
Taxonomic analysis of the gut microbiota was performed to further explore the composition differences of species and genera in each group. As shown in Figures 4B, C, at genus level, the relative content of Lactobacillus was significantly higher in the ST1814G/DNA-RNA groups compared to the ST1814G group. Lactobacillus are supposed as probiotic bacteria (29), which plays roles on regulating gut microbiota, enhancing mucosal immune, and protecting gastric mucosa. At the same time, the relative content of Akkermansia increased significantly in ST1814G/H5HA group and ST1814G/H5HA/Inactivated group. Usually, Akkermansia is a marker of better growth status and diversity of gut microbiota. The difference with them is that the proportion of streptococcus in group ST1814G/H5HA/DNA-RNA was significantly increased. Streptococcus had critical contributions to the maintenance of intestinal homeostasis (30). This suggested that different combinations of vaccines affect the distribution of bacterial flora in the gut. At species level, the two species with large changes were Reuteri and Muciniphila (31–34). The relative content of Reuteri was significantly higher in the immunized groups, especially in the ST1814G/DNA-RNA group. Reuteri was reported to reshape the gut microbiota to reduce inflammation (35–37). The relative content of Muciniphila was higher in ST1814G/H5HA group and ST1814G/H5HA/Inactivated group, the combination of ST1814G/H5HA and inactivated vaccine led to a decrease in the proportion of Muciniphila but increased the number of microbiomes in the gut. Muciniphila is belived to play a protective role in the intestinal tract, as the decrease of Muciniphila could cause intestinal barrier damage, plasma endotoxin increase, low-grade inflammation and metabolic disorders (38). These results suggested that our yeast vaccines could improve the probiotic bacteria content of vaccinated chickens, such as Muciniphila, which might improve the microbiome in the gut together.
Partial least-squares discrimination analysis (PLS-DA) also showed that the distribution of bacterial communities in the ST1814G/H5HA group was different from the PBS group and the ST1814G group (Figure 4D), which were in accord with the increased relative abundance of Muciniphila as shown above. The alpha diversity analysis of each group showed that Shannon index was higher in the immunized groups, especially in the ST1814G/H5HA group, compared to the PBS group or the ST1814G group (Figure 4E), indicating that our engineered yeast vaccine could increase the gut microbiota diversity during influenza virus infection, which might be benefit for the recovery of the infected chickens.
Discussion
Vaccinations are the most effective manners to prevent the outbreak of avian influenza virus, such as the H5N8 virus. Although the traditional inactivated influenza vaccines are the most popular candidates in China right now, there are several reports about exploring yeast surface-displayed influenza vaccines during the past decades (39), which might be promising vaccine that greatly facilitate the vaccination procedure in poultry. However, the molecular mechanism of protective efficacy provided by the yeast vaccine was not fully understood.
In this study, we developed three yeast-based vaccines: yeast surface-displayed recombinant protein vaccine (ST1814G/H5HA), yeast-based DNA-RNA vaccine (ST1814G/DNA-RNA) and yeast-based protein-nucleic acid combined vaccine (ST1814G/H5HA/DNA-RNA). The vaccination program was designed to test the protective efficacy with the above vaccine alone or with the traditional inactivated vaccine together, which was aimed to explore better vaccine candidate. The results suggested that all of our engineered yeast vaccine could promote the humoral immune response and decrease the viral load in different tissues after high dose of the H5N8 virus challenge. Considering the special antibody response and the viral load experiments, simultaneous administration of ST1814G/H5HA/DNA-RNA or ST1814G/H5HA/Inactivated vaccines may be better candidates for the vaccination procedure in poultry. Unfortunately, the chicken survival experiments did not get a fully protective effect in the simultaneous administration groups or any other group, and it was highly possible that the H5N8 influenza virus was extremely pathogenic for the experiment chickens. Even so, the immunized groups survived longer than the PBS group or the ST1814G group, suggesting that our engineered yeast vaccine offered partial protective effects.
To better understand the molecular mechanism of yeast vaccine, we analyzed the gene expression patterns using the bursa of Fabricius, which is an important immune organ of chickens. The results suggested that our ST1814G/H5HA vaccine group greatly enhanced the defense and immune responses and further analysis suggested that the immune cell microenvironment of bursa of Fabricius was reshaped after our engineered yeast vaccination in chicken. The genes for immune system development, lymphocytes activation and leukocyte migration were partially upregulated in the ST1814G/H5HA group compared to the PBS group or the inactivated vaccine group. Meanwhile, the genes for innate immune response were also partially upregulated in the ST1814G/H5HA group. These results together suggested that the yeast vaccine not only stimulate the adaptive immune response, but also stimulate the innate immune response in bursa of Fabricius. The cross talks between innate immunity and adaptive immunity are extremely important for efficient protection upon pathogen challenge. Taken together, the complexity and crosstalk between the innate and adaptive immune system, stimulated by our engineered yeast vaccine but not the traditional inactivated vaccine, might explain how the yeast vaccine provided protective efficacy in vivo.
Gut microbiota have been supposed to provide the host with several functions that promote immune homeostasis, immune responses, and protection against pathogen colonization (40–42). To investigate the influence of our engineered yeast vaccine on the diversity of gut microbiota, we performed the taxonomic and alpha diversity analysis and the results suggested that yeast vaccine elevated the diversity of gut microbiota. The increased content of symbiotic bacterium, such as Reuteri which was in enhancing antiviral activity of chicken macrophages against AIV in chicken Macrophages and Muciniphila, previous research has shown that oral administration of A.muciniphila could decrease the levels of IL-6 and IL-1β in the lungs and the levels of IL-6 and TNF-α in the blood during the course of H7N9 infection, which suggested that inhibition of these cytokines was one of the mechanisms by A.muciniphila protects the host from influenza virus infection (43). Therefore, oral administration of our engineered yeast vaccine might use similar mechanisms to protect chickens from AIV.
Conclusion
We developed three types of the H5N8 HA-based yeast-produced avian influenza vaccines. Our experimental results demonstrated that all of the engineered yeast vaccine elicited humoral immunity in chicken, decreased the virus load in different tissues, and provided partial protection of chickens from high dose of lethal the H5N8 virus infection. It’s just that the combination of vaccines (ST1814G/H5HA/DNA-mRNA and ST1814G/H5HA/Inactivated) compared with yeast vaccine along could enhance more humoral immunity and obviously reduce viral titers in the blood and lungs. Mechanically, yeast vaccine promoted the defense and immune responses, reshaped the immune cell microenvironment in bursa of Fabricius. Although the proportion of probiotics in intestinal flora is different between yeast vaccine in separate and combined use, but all yeast vaccine promoted the immune homeostasis upon influenza virus infection. These yeast vaccines can be further used as influenza vaccine for preventing influenza outbreaks in poultry.
Materials and methods
Ethics statement
All animal studies were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of the People’s Republic of China. The animal study was reviewed and approved by the Institutional Animal Ethical Committee of Tianjin Institute of Pharmaceutical Research [Certificate Number: SYXK (Jin) 2016-0009, Tianjin, China].
Preparation of recombinant HA protein
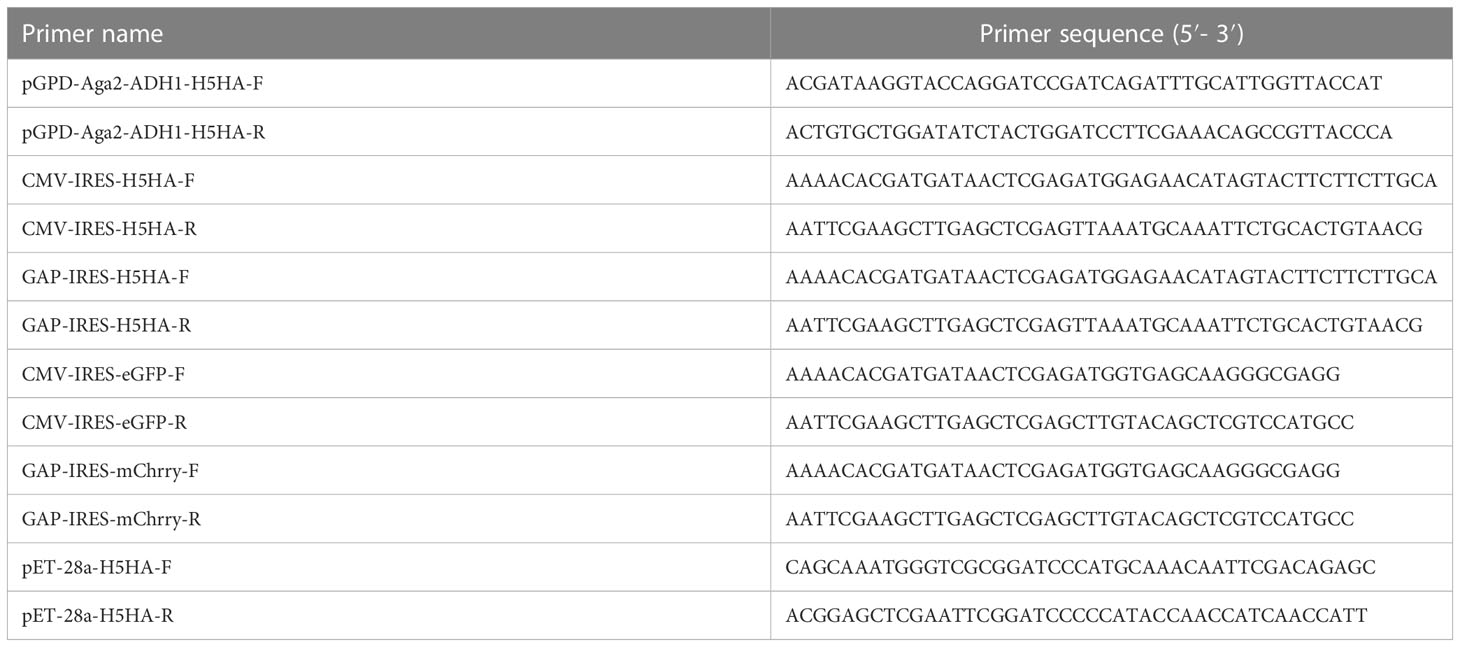
The 97-1131 nucleotides of the H5N8 HA gene were inserted into the restriction site B amHI of the pET-28a plasmid vector. The primers used in the study was showed in Table 1. The plasmid was transfected into Rossatta E.coli cell to induce the expression of HA protein by adding isopropyl-β-D-thiogalactoside (IPTG). The recombinant HA protein was further purified with Ni-NTA agarose beads (Thermo Scientific, 78605).
Vaccine preparation
Preparation of protein subunit vaccine
The 97-1131 nucleotides of the the H5N8 HA gene were inserted into the pGPD-AGA2-ADH1-POT plasmid vector. The plasmids, with terminal homologous arms as indicated, PMV-URR1, PMV-URR2, PMV-Leu and pGPD-AGA2-H5HA-ADH1-POT were digested with restriction enzymes BsmbI or BsaI, and then the fragments were ligated by T4 DNA ligase. The ligated products were transformed into Saccharomyces cerevisiae ST1814G strain (MATa aga1 his3Δ200 leu2Δ0 lys2Δ0 trp1Δ63 ura3Δ0 met15Δ0) and the positive clones were screened on SD/-Leu plate as previous described (44) (Figure 1A).
Preparation of nucleic acid vaccine
The 1-1704 nucleotides of the H5N8 HA gene were inserted into the pGAP-IRES plasmid vector and pCMV-IRES plasmid vector, respectively. The GAP promoter promotes the transcription of target genes in yeast and the IRES element prevents the translation in yeast, which function together as an RNA production combination. The CMV promoter promotes the transcription of target genes in mammalian cells and the IRES element increases the translation in mammalian cells, which function together as an DNA production combination. The plasmids pGAP-IRES-H5HA and pCMV-IRES-H5HA were digested with restriction enzyme BsaI and then the fragments were ligated by T4 DNA ligase. The ligated products were transformed into Saccharomyces cerevisiae ST1814G strain (MATa aga1 his3Δ200 leu2Δ0 lys2Δ0 trp1Δ63 ura3Δ0 met15Δ0) as previous described and the positive clones were screened on SD/-Trp plate (Figure 1B). For the eGFP-mCherry nucleic acid vaccine, the H5HA gene was replaced by eGFP or mCherry, respectively.
Inactivated vaccine
The commercial inactivated vaccine used in this experiment was Reassortant Avian Influenza Virus (H5+H7)Trivalent Vaccine (DaHuaNong, 19002346).
Chicken vaccination and virus challenge experiments
91 of 2-week-old chickens were randomly divided into 7 groups for the indicated vaccination program as Figure 1D. Serum samples, anal swab and intestinal contents were collected at the indicated time as the schedule indicated. Chickens were challenged with 106.8 TCID50/0.1 mL the H5N8 influenza virus at the 6th day after the third vaccination and the survival rate was recorded daily. 6 days after the challenge, blood, lung and trachea were collected to measure the viral RNA load. At the same time, the tissues of liver, spleen, lung and BF were prepared for tissue sections.
Enzyme-linked immunosorbent assay
Purified the H5N8 HA proteins were coated onto 96-well Maxisorp clear plates at 1 mg/mL in 50 mM Na2CO3, pH 9.6, overnight at 4°C. 500μL serum or PBS buffer-diluted anal swabs were incubated at 37°C for 40 minutes. Plates were washed 3 times with PBST (0.5% Tween-20), and then incubated with HRP-goat anti-chicken IgG (Solarbio, Beijing, China) or goat anti-chicken IgA (Abcam, USA). After 40 minutes incubation at 37°C, plates were washed 3 times with PBST (0.5% Tween-20) and 1-Step Ultra TMB-ELISA (Solarbio, Beijing, China) was added. Following 12 minutes incubation, reactions were stopped with 2 M sulfuric acid. The absorbance of each well at 405 nm was determined using ELISA Reader (Biored, USA).
Quantitative real-time PCR
Total RNA was extracted with TRIzol reagent (Yeasen Biotech, Shanghai, China). First-strand cDNA was synthesized from 1 μg total RNA using the Hifair®III 1st Strand cDNA Synthesis SuperMix (Yeasen Biotech, Shanghai, China), and qPCR was performed using the Hieff® qPCR SYBR Green Master Mix (Yeasen Biotech, Shanghai, China). The primers used in the study was showed in Table 2. The total volume of qPCR was 20μL, and the amplification step was 94 °C for 5 minutes, followed by 40 cycles of denaturation at 94°C for 10 seconds and extension at 60°C for 30 seconds, followed by dissociation curve analysis. ABI 7500 real-time PCR was used for numerical determination, and each sample was analyzed in triplicate. Data were calculated based on the 2−ΔΔCT method. Relative mRNA expression was normalized to that of β-actin.
RNA-seq and 16S rRNA seq
At the 6th day after the third vaccination, three immunized chickens were randomly selected in each group. The total RNA of bursa of Fabricius and the segments about 10centimeters from the cecum were sent to RNA-seq and 16S rRNA-seq, respectively.
In RNA-seq analysis, the raw fastq files were analyzed using FastQC (v0.11.9). Sequencing adaptors and low quality (base quality score<20) reads were trimmed by trim galore (v0.6.6). Reads shorter than 20nt were discarded. The trimmed reads were aligned to the Gallus (chicken) (GCF_016699485.2) by Hisat2 (v2.2.1) and the aligned data was further proceeded by Samtools (v1.3.1). The transcript was quantified by String Tie (v2.1.7). The gene counts were next analyzed by DESeq2 and the significant different genes were defined as absolute(log2FoldChange) >1.5 and pvalue < 0.05.
In the 16S rRNA analysis, the original sequencing data was filtered firstly, and the processed data was filtered to get effective data. Operational Taxonomic Units (OTU) clustering/denoising and species classification analysis were carried out based on the effective data to form the species abundance spectrum of OTU and other species classification levels. Based on the species abundance spectrum of OTU after homogenization of the data, the abundance and diversity index of OTU were analyzed, and the community structure of species annotation was statistically analyzed at each classification level.
Data availability statement
The data presented in the study are deposited in the GEO database, accession number GSE220714.
Ethics statement
The animal study was reviewed and approved by Institutional Animal Ethical Committee of Tianjin Institute of Pharmaceutical Research [Certificate Number: SYXK (Jin) 2016-0009, Tianjin, China].
Author contributions
Conceived and designed the experiments: JH, HL, RX, HuZ. Performed the experiments: RX, HuZ, HaZ, CL, DC, SL, ZL, HL. Analyzed the data: RX, HaZ, ZL, JH. Wrote the paper: JH, ZL, RX. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Tianjin Synthetic biotechnology innovation capability improvement project in China (TSBICIP-KJGG-014) and the key underprop project of Tianjin Municipal Science and Technology Bureau (20YFZCSN00340).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1125190/full#supplementary-material
Supplementary Figure 1 | Virus titer determination of each group in the trachea and Verification experiment of gene expression level in the Bursae of Fabricius post immunization. (A) Virus titer determination of each group in the trachea. Statistical analysis was analyzed by Student’s t-test. (ns, no significance. n=3 chickens for each group). The transcriptional levels of interleukin receptors (ILRs) (B), C-C motif chemokine receptors (CCLRs) (C), and interferon ligands (IFNLs) genes(D) in the Bursae of Fabricius of the immunized chickens were determined by RT-qPCR. Data were calculated based on the 2−ΔΔCT method, and relative mRNA expression was normalized to that of β-actin. The data in figures were obtained from three independent experiments and represent the averages ± SD. The significance of differences was determined by two-way analysis of variance (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.001 or ns, no significance).
References
1. Lewis NS, Banyard AC, Whittard E, Karibayev T, Al Kafagi T, Chvala I, et al. Emergence and spread of novel H5N8, H5N5 and H5N1 clade 2.3.4.4 highly pathogenic avian influenza in 2020. Emerg Microbes Infect (2021) 10(1):148–51. doi: 10.1080/22221751.2021.1872355
2. Lee DH, Torchetti MK, Winker K, Ip HS, Song CS, Swayne DE. Intercontinental spread of Asian-origin H5N8 to north America through beringia by migratory birds. J Virol (2015) 89(12):6521–4. doi: 10.1128/JVI.00728-15
3. Lee DH, Bertran K, Kwon JH, Swayne DE. Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3.4.4. J Vet Sci (2017) 18(S1):269–80. doi: 10.4142/jvs.2017.18.S1.269
4. Baek YG, Lee YN, Lee DH, Shin JI, Lee JH, Chung DH, et al. Multiple reassortants of H5N8 clade 2.3.4.4b highly pathogenic avian influenza viruses detected in south Korea during the winter of 2020-2021. Viruses (2021) 13(3). doi: 10.3390/v13030490
5. Pyankova OG, Susloparov IM, Moiseeva AA, Kolosova NP, Onkhonova GS, Danilenko AV, et al. Isolation of clade 2.3.4.4b A(H5N8), a highly pathogenic avian influenza virus, from a worker during an outbreak on a poultry farm, Russia, December 2020. Euro Surveill (2021) 26(24). doi: 10.2807/1560-7917.ES.2021.26.24.2100439
6. Pietrzak M, Maciola A, Zdanowski K, Protas-Klukowska AM, Olszewska M, Smietanka K, et al. An avian influenza H5N1 virus vaccine candidate based on the extracellular domain produced in yeast system as subviral particles protects chickens from lethal challenge. Antiviral Res (2016) 133:242–9. doi: 10.1016/j.antiviral.2016.08.001
7. Lee J, Kim DH, Noh J, Youk S, Jeong JH, Lee JB, et al. Live recombinant NDV-vectored H5 vaccine protects chickens and domestic ducks from lethal infection of the highly pathogenic H5N6 avian influenza virus. Front Vet Sci (2021) 8:773715. doi: 10.3389/fvets.2021.773715
8. Criado MF, Sa ESM, Lee DH, Salge CAL, Spackman E, Donis R, et al. Cross-protection by inactivated H5 prepandemic vaccine seed strains against diverse Goose/Guangdong lineage H5N1 highly pathogenic avian influenza viruses. J Virol (2020) 94(24). doi: 10.1128/JVI.00720-20
9. Belshe RB, Frey SE, Graham I, Mulligan MJ, Edupuganti S, Jackson LA, et al. Safety and immunogenicity of influenza a H5 subunit vaccines: effect of vaccine schedule and antigenic variant. J Infect Dis (2011) 203(5):666–73. doi: 10.1093/infdis/jiq093
10. Gao F, Liu X, Dang Y, Duan P, Xu W, Zhang X, et al. AddaVax-adjuvanted H5N8 inactivated vaccine induces robust humoral immune response against different clades of H5 viruses. Vaccines (Basel) (2022) 10(10). doi: 10.3390/vaccines10101683
11. Hatta M, Zhong G, Chiba S, Lopes TJS, Neumann G, Kawaoka Y. Effectiveness of whole, inactivated, low pathogenicity influenza A(H7N9) vaccine against antigenically distinct, highly pathogenic H7N9 virus. Emerg Infect Dis (2018) 24(10):1910–3. doi: 10.3201/eid2410.180403
12. Kosik I, Yewdell JW. Influenza hemagglutinin and neuraminidase: Yin(-)Yang proteins coevolving to thwart immunity. Viruses (2019) 11(4). doi: 10.3390/v11040346
13. Lei H, Jin S, Karlsson E, Schultz-Cherry S, Ye K. Yeast surface-displayed H5N1 avian influenza vaccines. J Immunol Res (2016) 2016:4131324. doi: 10.1155/2016/4131324
14. Niqueux E, Guionie O, Amelot M, Jestin V. Prime-boost vaccination with recombinant H5-fowlpox and Newcastle disease virus vectors affords lasting protection in SPF Muscovy ducks against highly pathogenic H5N1 influenza virus. Vaccine (2013) 31(38):4121–8. doi: 10.1016/j.vaccine.2013.06.074
15. Medina J, Boukhebza H, De Saint Jean A, Sodoyer R, Legastelois I, Moste C. Optimization of influenza a vaccine virus by reverse genetic using chimeric HA and NA genes with an extended PR8 backbone. Vaccine (2015) 33(35):4221–7. doi: 10.1016/j.vaccine.2015.06.112
16. Wen F, Ma JH, Yu H, Yang FR, Huang M, Zhou YJ, et al. Protective efficacy of a high-growth reassortant swine H3N2 inactivated vaccine constructed by reverse genetic manipulation. J Vet Sci (2014) 15(3):381–8. doi: 10.4142/jvs.2014.15.3.381
17. Kumar R, Kumar P. Yeast-based vaccines: New perspective in vaccine development and application. FEMS Yeast Res (2019) 19(2). doi: 10.1093/femsyr/foz007
18. Heinzelman P, Greenhalgh JC, Romero PA. Yeast surface display-based identification of ACE2 mutations that modulate SARS-CoV-2 spike binding across multiple mammalian species. Protein Eng Des Sel (2022) 35. doi: 10.1093/protein/gzab035
19. Trentelman JJA, Tomas-Cortazar J, Knorr S, Barriales D, Hajdusek O, Sima R, et al. Probing an ixodes ricinus salivary gland yeast surface display with tick-exposed human sera to identify novel candidates for an anti-tick vaccine. Sci Rep (2021) 11(1):15745. doi: 10.1038/s41598-021-92538-9
20. Zhang L, Yao L, Guo Y, Li X, Ma L, Sun R, et al. Oral SARS-CoV-2 spike protein recombinant yeast candidate prompts specific antibody and gut microbiota reconstruction in mice. Front Microbiol (2022) 13:792532. doi: 10.3389/fmicb.2022.792532
21. Han T, Sui J, Bennett AS, Liddington RC, Donis RO, Zhu Q, et al. Fine epitope mapping of monoclonal antibodies against hemagglutinin of a highly pathogenic H5N1 influenza virus using yeast surface display. Biochem Biophys Res Commun (2011) 409(2):253–9. doi: 10.1016/j.bbrc.2011.04.139
22. Shehata AA, Basiouni S, Ali A, Fawzy M, Hafez HM, Ulbert S, et al. Immunization of turkeys with a DNA vaccine expressing the haemagglutinin gene of low pathogenic avian influenza virus subtype H9N2. J Virol Methods (2020) 284:113938. doi: 10.1016/j.jviromet.2020.113938
23. Bahl K, Senn JJ, Yuzhakov O, Bulychev A, Brito LA, Hassett KJ, et al. Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol Ther (2022) 30(8):2874. doi: 10.1016/j.ymthe.2022.07.013
24. Kadali RAK, Janagama R, Peruru S, Malayala SV. Side effects of BNT162b2 mRNA COVID-19 vaccine: A randomized, cross-sectional study with detailed self-reported symptoms from healthcare workers. Int J Infect Dis (2021) 106:376–81. doi: 10.1016/j.ijid.2021.04.047
25. Yitbarek A, Taha-Abdelaziz K, Hodgins DC, Read L, Nagy E, Weese JS, et al. Gut microbiota-mediated protection against influenza virus subtype H9N2 in chickens is associated with modulation of the innate responses. Sci Rep (2018) 8(1):13189. doi: 10.1038/s41598-018-31613-0
26. Stefan KL, Kim MV, Iwasaki A, Kasper DL. Commensal microbiota modulation of natural resistance to virus infection. Cell (2020) 183(5):1312–24 e10. doi: 10.1016/j.cell.2020.10.047
27. Kawahara T, Takahashi T, Oishi K, Tanaka H, Masuda M, Takahashi S, et al. Consecutive oral administration of bifidobacterium longum MM-2 improves the defense system against influenza virus infection by enhancing natural killer cell activity in a murine model. Microbiol Immunol (2015) 59(1):1–12. doi: 10.1111/1348-0421.12210
28. Cao H, Hua D, Zhang H, Zhang H, Liu N, Feng Z, et al. Oral immunization of recombinant saccharomyces cerevisiae expressing fiber-2 of fowl adenovirus serotype 4 induces protective immunity against homologous infection. Vet Microbiol (2022) 271:109490. doi: 10.1016/j.vetmic.2022.109490
29. Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert consensus document. the international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol (2014) 11(8):506–14. doi: 10.1038/nrgastro.2014.66
30. Pagnini C, Saeed R, Bamias G, Arseneau KO, Pizarro TT, Cominelli F. Probiotics promote gut health through stimulation of epithelial innate immunity. Proc Natl Acad Sci U.S.A. (2010) 107(1):454–9. doi: 10.1073/pnas.0910307107
31. Zhang L, Liu J, Jin T, Qin N, Ren X, Xia X. Live and pasteurized akkermansia muciniphila attenuate hyperuricemia in mice through modulating uric acid metabolism, inflammation, and gut microbiota. Food Funct (2022) 13(23):12412–25. doi: 10.1039/d2fo02702j
32. Luo Y, Lan C, Li H, Ouyang Q, Kong F, Wu A, et al. Rational consideration of akkermansia muciniphila targeting intestinal health: advantages and challenges. NPJ Biofilms Microbiomes (2022) 8(1):81. doi: 10.1038/s41522-022-00338-4
33. Pei T, Hu R, Wang F, Yang S, Feng H, Li Q, et al. Akkermansia muciniphila ameliorates chronic kidney disease interstitial fibrosis via the gut-renal axis. Microb Pathog (2022) 174:105891. doi: 10.1016/j.micpath.2022.105891
34. Ross SM. Akkermansia muciniphila: A next-generation beneficial gut microbe and its role in obesity and diabetes. Holist Nurs Pract (2022) 36(6):372–4. doi: 10.1097/HNP.0000000000000556
35. Sinkiewicz G, Cronholm S, Ljunggren L, Dahlen G, Bratthall G. Influence of dietary supplementation with lactobacillus reuteri on the oral flora of healthy subjects. Swed Dent J (2010) 34(4):197–206.
36. Qi C, Tu H, Zhao Y, Zhou J, Chen J, Hu H, et al. Breast milk-derived limosilactobacillus reuteri prevents atopic dermatitis in mice via activating retinol absorption and metabolism in peyer's patches. Mol Nutr Food Res (2022):e2200444. doi: 10.1002/mnfr.202200444
37. Hu MX, He F, Guo YX, Mo LZ, Zhu X. Lactobacillus reuteri biofilms inhibit pathogens and regulate microbiota in In vitro fecal fermentation. J Agric Food Chem (2022) 70(38):11935–43. doi: 10.1021/acs.jafc.2c02372
38. Belzer C, de Vos WM. Microbes inside–from diversity to function: the case of akkermansia. ISME J (2012) 6(8):1449–58. doi: 10.1038/ismej.2012.6
39. Wasilenko JL, Sarmento L, Spatz S, Pantin-Jackwood M. Cell surface display of highly pathogenic avian influenza virus hemagglutinin on the surface of pichia pastoris cells using alpha-agglutinin for production of oral vaccines. Biotechnol Prog (2010) 26(2):542–7. doi: 10.1002/btpr.343
40. Mu Q, Edwards MR, Swartwout BK, Cabana Puig X, Mao J, Zhu J, et al. Gut microbiota and bacterial DNA suppress autoimmunity by stimulating regulatory b cells in a murine model of lupus. Front Immunol (2020) 11:593353. doi: 10.3389/fimmu.2020.593353
41. Schuijt TJ, van der Poll T, de Vos WM, Wiersinga WJ. The intestinal microbiota and host immune interactions in the critically ill. Trends Microbiol (2013) 21(5):221–9. doi: 10.1016/j.tim.2013.02.001
42. Czerucka D, Rampal P. Diversity of saccharomyces boulardii CNCM I-745 mechanisms of action against intestinal infections. World J Gastroenterol (2019) 25(18):2188–203. doi: 10.3748/wjg.v25.i18.2188
43. Hu X, Zhao Y, Yang Y, Gong W, Sun X, Yang L, et al. Akkermansia muciniphila improves host defense against influenza virus infection. Front Microbiol (2020) 11:586476. doi: 10.3389/fmicb.2020.586476
Keywords: influenza A virus, H5N8, Saccharomyces cerevisiae, vaccine, hemagglutinin (HA)
Citation: Xie R, Zhang H, Zhang H, Li C, Cui D, Li S, Li Z, Liu H and Huang J (2023) Hemagglutinin expressed by yeast reshapes immune microenvironment and gut microbiota to trigger diverse anti-infection response in infected birds. Front. Immunol. 14:1125190. doi: 10.3389/fimmu.2023.1125190
Received: 16 December 2022; Accepted: 22 March 2023;
Published: 18 April 2023.
Edited by:
Min Li, University of South China, ChinaReviewed by:
Wenbao Qi, South China Agricultural University, ChinaXingang Xu, Northwest A&F University, China
Copyright © 2023 Xie, Zhang, Zhang, Li, Cui, Li, Li, Liu and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hualei Liu, bGl1aHVhbGVpQGNhaGVjLmNu; Jinhai Huang, amluaGFpaEB0anUuZWR1LmNu
†These authors have contributed equally to this work
 Ruyu Xie1†
Ruyu Xie1† Zexing Li
Zexing Li Jinhai Huang
Jinhai Huang