- 1Translational Healthcare Technologies Group, Centre for Inflammation Research, Queen’s Medical Research Institute, The University of Edinburgh, Edinburgh, United Kingdom
- 2Research & Development, Sanofi, Marcy L’Etoile, France
- 3Centre for Photonic and Physics, Bath University, Bath, United Kingdom
Introduction: Pulmonary-resident memory T cells (TRM) and B cells (BRM) orchestrate protective immunity to reinfection with respiratory pathogens. Developing methods for the in situ detection of these populations would benefit both research and clinical settings.
Methods: To address this need, we developed a novel in situ immunolabelling approach combined with clinic-ready fibre-based optical endomicroscopy (OEM) to detect canonical markers of lymphocyte tissue residency in situ in human lungs undergoing ex vivo lung ventilation (EVLV).
Results: Initially, cells from human lung digests (confirmed to contain TRM/BRM populations using flow cytometry) were stained with CD69 and CD103/CD20 fluorescent antibodies and imaged in vitro using KronoScan, demonstrating it’s ability to detect antibody labelled cells. We next instilled these pre-labelled cells into human lungs undergoing EVLV and confirmed they could still be visualised using both fluorescence intensity and lifetime imaging against background lung architecture. Finally, we instilled fluorescent CD69 and CD103/CD20 antibodies directly into the lung and were able to detect TRM/BRM following in situ labelling within seconds of direct intra-alveolar delivery of microdoses of fluorescently labelled antibodies.
Discussion: In situ, no wash, immunolabelling with intra-alveolar OEM imaging is a novel methodology with the potential to expand the experimental utility of EVLV and pre-clinical models.
Introduction
Human and non-human primate (NHP) studies have highlighted the protective role of lung-resident memory T cells (TRM) in controlling respiratory pathogens (1–3). Constituting the majority of T cells within the lung (4), TRM have a unique phenotype that differentiates them from other memory T cell subsets and their sequestration within the lung offers an ideal location to respond to respiratory infections (5). Requiring only cognate antigen for their activation (6), TRM are highly proliferative, producing polyfunctional progeny with superior effector function (7, 8). Activated CD4+ and CD8+ TRM in the lungs of patients with Mycobacterium tuberculosis (MTB) limit intracellular MTB replication in macrophages (9), whilst increased numbers of CD8+ TRM in BAL is associated with reduced symptoms and viral load following human respiratory syncytial virus (RSV) infection (1). In human influenza infection, CD4+ and CD8+ TRM confer heterosubtypic protection (10), with CD8+ TRM recognising universally conserved peptides expressed by influenza A, B and C viruses (11). Virus-specific pulmonary CD8+ TRM also have shown “innate like properties” that amplify inflammation and enhance neutrophil recruitment following noncognate bacterial infection to aid clearance, as demonstrated in murine models (12). While most actions of TRM benefit the host, following chronic exposure to allergens, TRM can also be detrimental and contribute significantly to pathology in experimental allergic asthma (13, 14), indicating monitoring the frequency and phenotype of TRM has relevance beyond respiratory infections.
Antigen-experienced lungs are also enriched with B cells expressing a resident memory phenotype (15). Murine parabiosis studies have demonstrated that pulmonary-resident memory B cells (BRM) require local antigen encounter and early CD40/CD40 ligand interactions with T cells for their formation and are phenotypically and functionally distinct from their systemic counterparts (5, 16). Murine BRM play a protective role in response to both viral influenza (16–18) and bacterial pneumococcal pneumonia (15) infections, providing rapid antibody secreting cells (ASC) capable of producing a range of class switched, cross-reactive antibodies to confer heterosubtypic protection (15–17).
Vaccination strategies designed to induce pulmonary resident memory lymphocyte populations have been successfully demonstrated in murine models (19). In NHP, pulmonary mucosal delivery of MTB vaccine is associated with enhanced protection over standard intradermal immunization via the induction of lung TRM (20). As TRM and BRM formation is dependent on pulmonary cognate antigen encounter (16, 21), it’s anticipated that pulmonary mucosal vaccination will also generate BRM. This can induce higher levels of local immunity than that generated by natural MTB infection (20). Strategies that establish antigen-specific TRM and BRM populations or enhance pre-existing populations have the potential to provide local long lasting heterosubtypic protection where it is most needed. Current methods for evaluating respiratory vaccine efficacy, however, rely mostly on peripheral blood sampling to reveal the level of humoral protection and circulating central memory T cell (TCM) and effector memory T cell (TEM) populations. However, as TRM and BRM do not circulate and limited blood biomarker surrogates exist, current methods for assessing local immunity are limited.
These studies highlight the pivotal role of TRM and BRM in orchestrating and directing protective responses to respiratory infections. The spatial and temporal characterisation of such pulmonary resident immune cells is thus of key importance, but most available methods require biopsy, tissue digestion and post-mortem studies (16, 22). In this study, we aimed to develop an in situ method for the direct detection of these cells in the human lung by exploiting recent advances in in situ-optical endomicroscopy, within the alveoli (23), which may provide a valuable tool for quantifying pulmonary resident memory populations following infection and vaccination.
The objective was to develop a technical platform for direct alveolar imaging of pulmonary TRM and BRM. We aimed to develop an in situ immunolabelling approach with a clinic-ready fibre-based optical endomicroscopy (OEM) system that combined both intensity imaging and fluorescence lifetime imaging (FLIM) to ensure contrast and specificity was achieved over endogenous elastin fluorescence of the lung. With NHP providing an ideal model for translational studies, this project also aimed to establish a cross-reactive antibody panel capable of detecting TRM and BRM in both humans and NHP species.
FLIM is a fluorescence imaging technique where the contrast is based on the lifetime of individual fluorophores rather than their emission spectra. The fluorescence lifetime (FLT) is defined as the average time that a molecule remains in an excited state prior to returning to the ground state by emitting a photon. The FLT of the fluorophore, rather than its intensity, is used to create the image in FLIM. FLT depends on the local micro-environment of the fluorophore, thus precluding any erroneous measurements in fluorescence intensity due to change in brightness of the light source, background light intensity or limited photo-bleaching (24).
A clinic-ready fibre-based FLIM system (KronoScan), incorporating a time-resolved spectrometer and an achromatic advanced confocal laser scanning microscope system was used coupled to a fully biocompatible multifunctional endoscopic fibre (Panoptes) with an outer diameter of 1.8 mm, which comprises an imaging fibre bundle packaged alongside two capillary channels for delivery and extraction of fluids
T cells recovered from bronchoalveolar lavage and lung tissue digests from ex vivo whole human lungs showed characteristic features of TRM with CD69+CD103+ CD8+ TRM being the most abundant population in the BAL and higher than in the lung tissue. A cross reactive panel of antibodies detected T cells bearing TRM markers in NHP lung, although at much lower levels than in human lung. Cells isolated from human lung digests co-expressing CD69 and CD103 could be visualised using fibre-based imaging via both fluorescence intensity and lifetime imaging when labelled with anti-CD69 and anti-CD103 antibodies, both in vitro and after instillation into human lungs. Using KronoScan with Panoptes (25), dual antibody labelled cells (expressing markers of tissue retention) were detected in situ within human lungs undergoing EVLV within seconds of direct in situ delivery.
Our study extends the methods available for live imaging of leukocytes at the host-environment interface of the lung and demonstrates the potential of specific immunolabelling for distal lung interrogation. The ex vivo human lung model, closely resembles the in vivo situation, enabling validation of imaging modalities and facilitates transfer to both research and clinical settings.
Methods
Tissue source
Human lungs from deceased subjects deemed non-suitable for transplantation were received from National Health Service Blood and Transplant (NHSBT), United Kingdom. Following explantation and perfusion, lungs were placed on ice and transported to the University of Edinburgh within 48 hours prior to ventilation/lavage. Whole NHP (cynomolgus macaque from Mauritius) lungs were obtained from Charles River Laboratories (Edinburgh, UK) and transported in Roswell Park Memorial Institute (RPMI) 1640 medium (11875-093, Gibco) prior to tissue digest. NHP lungs were non-perfused and obtained from healthy control animals not subjected to experimental manipulation.
Ex vivo lung ventilation
Human lungs undergoing EVLV were placed on a Dräger Savina 300 ventilator following placement of an endotracheal tube and ventilated with a tidal volume of 7 mL/kg (patient body mass) at a respiratory rate of 12 breaths per minute (Supplementary Figure 1A). Ventilation was volume controlled, air driven, and positive end expiratory pressure (PEEP) was set at 5 cm H2O. The airway pressure limit setting on the ventilator was increased to the maximum setting to prevent under ventilation.
Bronchoalveolar lavage
Bronchoalveolar lavage (BAL) was obtained from human lungs undergoing EVLV by instilling 100 mL phosphate-buffered saline (14190-094, PBS, Gibco) into the right middle lobe or selected lobes via a bronchoscope. Aspirated cells were passed through a 40 µm cell strainer (431750, Corning, USA) and counted using a NucleoCounter cell counter (Chemometec, Denmark). Cells were processed fresh or cryopreserved in liquid nitrogen using Recovery™ Cell Culture Freezing Medium (12648010, Thermo Fisher).
Lung tissue digest
Human (EVLV-derived samples from the distal lung) and NHP lung tissues were dissected with a scalpel and finely cut with scissors prior to incubation in 37°C DMEM (21969-035, Gibco) containing 1 mg/mL Collagenase IV (C5138, Sigma-Aldrich) and 0.1 mg/mL DNASE I (DN25 – Sigma Aldrich) for 1 hour with constant agitation. The sample was strained through a 70 µm cell strainer (22-363-549, Thermo Fisher) and spun at 350 x g for 5 minutes at room temperature. Red blood cell (RBC) lysis was performed using RBC lysis buffer (320301, Biolegend) for 10 minutes at room temperature prior to addition of DMEM and spun for another 5 minutes at 350 x g. Cells were counted using a NucleoCounter and processed for direct flow cytometry analysis or cryopreserved.
Peripheral blood mononuclear cells
Human peripheral blood mononuclear cells (PBMCs) were isolated from whole blood using Percoll gradients (26). PBMCs were then cryopreserved. Cryopreserved cynomolgus PBMCs were obtained from BioIVT (BioIVT, United Kingdom).
Flow cytometric analysis of lung tissue and BAL
Fresh lung digest and BAL cells were placed in 5 mL flow cytometry tubes. For cryopreserved cells, cells were gently warmed in 37°C water, placed in DMEM containing 10% foetal calf serum (FCS) and washed at 350 x g for 10 minutes prior to transfer to flow tubes. Cells were stained with live/dead marker Zombie UV (423107, Biolegend) in PBS for 30 minutes in the dark at room temperature. Cells were washed with FACS buffer (PBS + 2% FCS) and incubated with Fc block (564220, BD Biosciences) for 10 minutes prior to cell surface antibody staining containing BD Brilliant Horizon™ Staining Buffer (563794, BD Biosciences) (for details of flow cytometry antibodies used, see Supplementary Table 1). Following a 25-minute incubation in the dark at 4°C, cells were washed at 350 x g for 5 minutes and resuspended in Fixation Buffer (420801, Biolegend) for 20 minutes at room temperature before spinning at 350 x g for 5 minutes and resuspending in FACS buffer. For intracellular Ki-67 staining, cells were washed following cell surface antibody staining and processed using the Foxp3/Transcription Factor Staining Buffer Set (00-5523-00, Thermo Fisher Scientific) according to the manufacturer’s instructions. Ki-67 antibodies were used at dilutions recommended by the manufacturer (Supplementary Table 1). Samples were assessed using a 5L LSR Fortessa flow cytometer (BD Biosciences). Compensation was calculated using UltraComp eBeads™ (01-222-42, Thermo Fisher Scientific). Data analysis was performed using FlowJo software, version 10.7.2 (BD).
TRM/BRM immunofluorescence staining
Human and NHP lung tissues were placed in optimal cutting temperature compound (OCT, KMA-0100-00A, CellPath, UK) and snap frozen in liquid nitrogen. 10 µm sections were cut on to Super Frost microscope slides (12312148, Thermo Fisher) using a cryostat. Sections were air dried overnight at room temperature and then fixed in -20°C acetone for 10 minutes. Sections were air dried for <20 minutes prior to x2 washes with PBS. Multiplex immunofluorescence was performed on lung sections using a fully automated Bond-Rx Multiplex IHC Stainer (Leica Biosystems) and OPAL Multiplex IHC Detection Kit and counterstained with DAPI (Akoya Biosciences) according to a previously published protocol (27). TRM were stained with purified, cross-reactive CD4 (OKT4, Biolegend), CD8 (SK1, Biolegend), CD69 (FN50, Biolegend) and CD103 (2G5.1, Bio-Rad, UK) antibodies. BRM were stained with purified, cross-reactive CD20 (2H7, Biolegend), CD27 (M-T271, Biolegend) and CD69 (FN50, Biolegend) antibodies. Human lung (n=5) and NHP lung (n=3) sections were stained simultaneously using the same protocol and antibody concentrations. For full details, see online supplement.
Fluorescence intensity and lifetime imaging system (KronoScan) and fibre (Panoptes)
The clinic-ready fibre-based FLIM system (Supplementary Figures 1B–D) (KronoScan), incorporates a time-resolved spectrometer (with 32 lifetime channels) and achromatic advanced confocal laser scanning microscope system, using a supercontinuum laser (SuperK EVO, NKT Photonics, Denmark) filtered to excitation band 490 nm and 590 nm. 50 frame fluorescence intensity and lifetime video sequences were simultaneously recorded using 128 x 128-pixel images (350 x 350 µm) matching fibre bundle characteristics. Spectral emission ranges of 498-565 nm and 620-764 nm were selected for analysis. Lifetime decays were fitted using rapid lifetime determination method (28), which assumes a single exponential decay, enabling high-processing speeds and real-time imaging at 3 frames per second (20 µs exposure time).
Panoptes (25) is a fully biocompatible multi-functional endoscopic device, with an outer diameter of 1.6 mm. It comprises an imaging fibre bundle packaged alongside two capillary channels for delivery and extraction of fluids. The low-index contrast imaging fibre (0.3 numerical aperture (NA)) is derived from multi-mode telecommunications preforms (OM1 PCVD rods). It is fabricated by multi-stacking arrays of different sized cores to optimise pixel density and suppress core-to-core crosstalk (29). This high-resolution OEM & FLIM imaging fibre, which comprises 8100 cores, with 450 µm diagonal field of view, is deliverable via existing bronchoscopic and transthoracic platforms.
Fluorescence intensity and lifetime TRM/BRM fibre imaging in vitro
5x106 human lung digest cells or PBMCs were stained with 15 µL FITC-conjugated CD69 (FN50, Biolegend) and PE-Dazzle CD103 (BerACT8, Biolegend) to identify TRM, or 15 µL FITC-conjugated CD69 and PE-Dazzle-labelled CD20 (2H7, Biolegend) to identify BRM, for 20 minutes at 4°C in FACS buffer. Cells were spun at 350 x g for 5 minutes, resuspended in 100 µL PBS and transferred to a black 96-well plate. Fluorescence intensity and lifetime imaging was performed using OEM KronoScan Imaging system and Panoptes Multifunctional fibre. Videos were saved retrospectively when events were detected and analysed using the KronoScan Player (Firefinch Software Development, UK).
In situ detection of TRM/BRM
Using a bronchoscope, the Panoptes multifunctional fibre was navigated to the distal airways via autofluorescence intensity imaging to visualise airways and alveoli (Figure 1E). 5x106 pre-labelled human TRM or BRM were prepared as outlined in the previous section. For in situ TRM/BRM detection, 20 µL (2 µg) CD69 + 20 µL (0.5 µg) CD103 or 20 µL (0.8 µg) CD20 + CD69 was diluted in 500 µL PBS. Antibodies or pre-labelled cells were instilled into different alveolar segments (to prevent cross contamination) of human lungs undergoing EVLV via the delivery channel of Panoptes Multifunctional Fibre. Fluorescence intensity and lifetime imaging was performed after 5 minutes using KronoScan OEM.
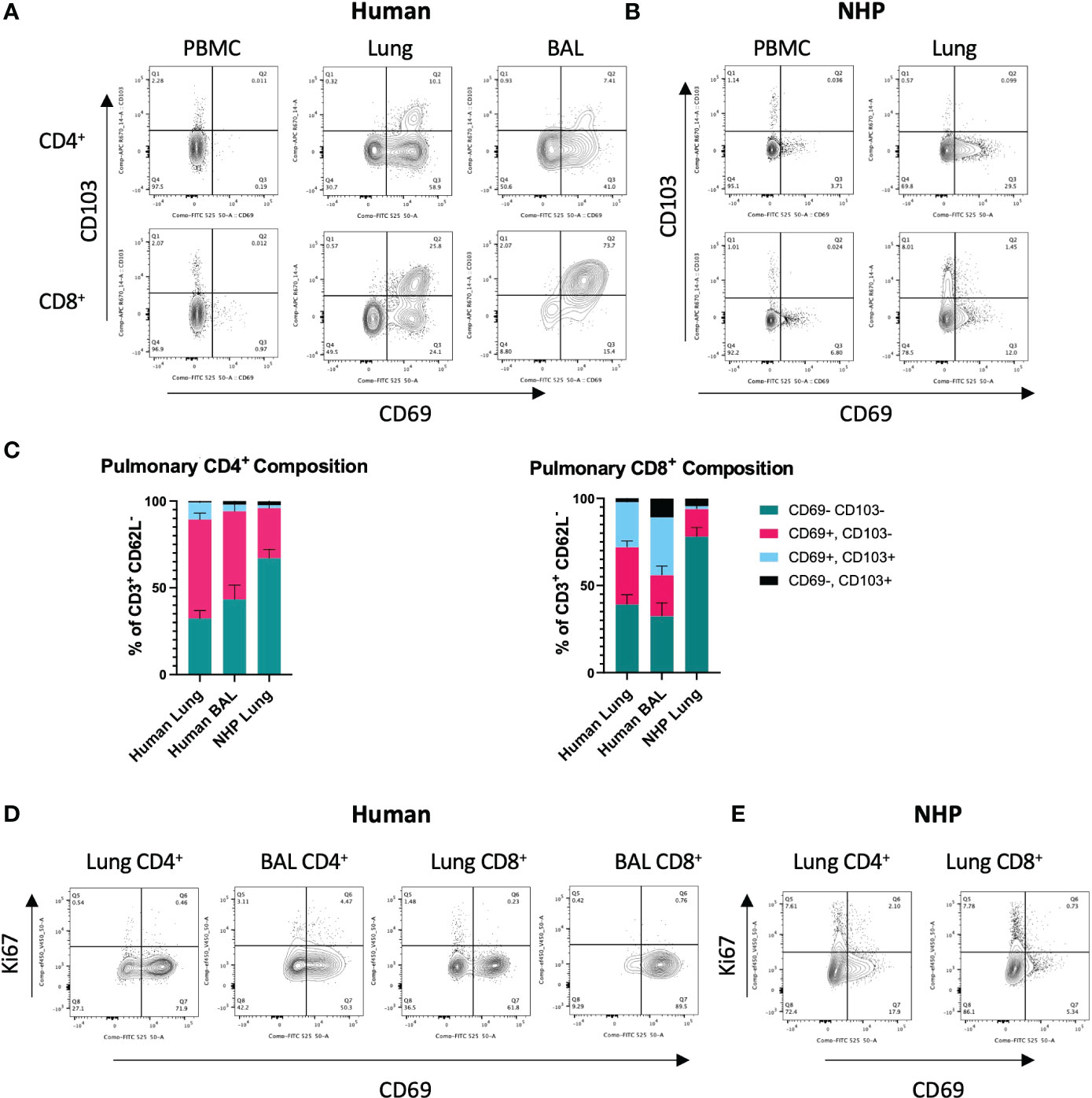
Figure 1 Characterisation of Human and NHP Pulmonary TRM.TRM from BAL and/or tissue digests were compared with peripheral T cells. (A) Identification of human pulmonary CD4+ and CD8+ TRM. TRM (live, CD3+, CD4+/CD8+, CD62-L-) were identified based on their expression of CD69 and CD103. (B) Identification of NHP CD4+ and CD8+ TRM. (C) Proportion of CD69+ and CD69+CD103+ TRM in human and NHP lung. (D) Ki-67 staining in human CD4+ and CD8+ TRM. (E) Ki-67 staining in NHP CD4+ and CD8+ TRM. Human and NHP samples were concatenated for analysis. Data represented as representative flow cytometry plots or compiled frequencies with mean ± SEM (C) from human lungs (n=15), BAL (n=9) and NHP lung (n=5).
Statistics
All data in text are represented as mean ± standard deviation (SD). All data in figures are represented as mean ± standard error of the mean (SEM). All graphs were generated using GraphPad Prism 9 (GraphPad Software, California, USA).
Results
Characterisation of human and NHP pulmonary TRM
To identify and characterise human and NHP pulmonary TRM, cells isolated from lung tissue were stained with cross-reactive antibodies for human and cynomolgus macaque and analysed by flow cytometry. Based on the existing knowledge (5), TRM were phenotyped as CD3+, CD4+/CD8+, CD62-L-, CD69+, ± CD103 (for TRM gating strategy, see Supplementary Figure 2). Exclusion of CD62-L+ (L-selectin) cells helped eliminate any contaminating circulating TCM present within the vasculature of the lung tissue.
CD4+ and CD8+ TRM were detected in human lung and BAL but were expectedly absent in peripheral blood (Figure 1A). The highest levels of CD103 expressing cells were seen in CD8+ TRM in the BAL where CD69+CD103+ TRM were the most abundant population (33.3% ± 22.9, Figures 1A, C). CD69+ CD4+ and CD8+ TRM were also detected in NHP lung, although at much lower levels than that found in human lung (Figures 1B, C). 57% ± 14.3 of CD4+ T cells in human lung were positive for CD69 compared to 28% ± 11 in NHP lung. 32.8% ± 14 of CD8+ T cells in human were positive for CD69 compared to 15.8% ± 11.7 in NHP lung. There were some similarities in the expression of CD69 and CD103 in human and NHP samples: CD69+ T cells were present at higher frequencies in lung versus peripheral blood (Figures 1A, B) and lung CD8+ T cells showed higher levels of CD103 expression than their CD4+ counterparts in both species (Figure 1C). However, the overall frequency of CD69+ TRM (± CD103) was lower in NHP than in human samples and co-expression of CD69 and CD103 was rarely observed in NHP (Figures 1A–C). Human lung contained 66.9% ± 17.6 and 58.7% ± 21.8 CD4+ and CD8+ TRM, respectively, compared to 30.6% ± 11.9 and 17.4% ± 12.3 in NHP lung. In NHP lung, only 1.74% ± 1.9 of CD4+ and 1.52% ± 0.8 of CD8+ cells co-expressed CD69 and CD103, compared to 9.9% ± 10.7 CD4+ and 25.9% ± 17.1 CD8+ cells within human lung. CD62L-CD69-CD103- TEM were the largest population observed in NHP lung, with 67% ± 17.5 of CD4+ T cells and 78% ± 21.6 of CD8+ T cells failing to express tissue-residency markers.
To assess rates of proliferation, intracellular levels of Ki-67 were measured in pulmonary TRM. Low levels of Ki-67 were observed in human CD4+ and CD8+ T cells isolated from lung and BAL (Figure 1D). Ki-67 levels were slightly higher in NHP lung, particularly in CD69- T cells, in line with their effector memory profile (Figure 1E).
Despite differences in expression of CD69 and CD103 between species, expression patterns of other markers associated with TRM phenotype, CD49a and programmed cell death protein 1 (PD-1), were similar on human and NHP TRM. CD49a was absent on peripheral blood T cells but expressed by human and NHP CD69+ TRM, particularly those co-expressing CD103 (Supplementary Figure 3). Increased levels of PD-1 were also observed on human and NHP TRM, with expression highest on CD103+ TRM and TRM isolated from BAL (Supplementary Figure 3).
Characterisation of human and NHP pulmonary BRM
Human and NHP lung cells were stained for pulmonary BRM markers. BRM were phenotyped as CD62-L-, B220+, CD20+, CD27+, CD69+ (5, 15) (for BRM gating strategy, see Supplementary Figure 4). Both human and NHP lungs contained CD27+CD69- memory B cells (BMEM) but a distinct population of CD27+CD69+ BRM was mostly observed in human BAL (Figures 2A, B). CD27+ BMEM were also detected in human and NHP peripheral blood, however these cells did not express CD69. Human lung and BAL contained similar frequencies of BMEM (41.1% and 40.6%, respectively) (Figure 2C). NHP lung contained significantly fewer BMEM, with only 9.5% present. Human BAL contained the highest proportion of BRM, with 25.6% of memory B cells expressing CD69 compared to 8.4% in human lung and only 1.4% in NHP lung (Figure 2C). Although BAL contained the highest proportion of BRM, numbers were far fewer than those found in the lung interstitium.
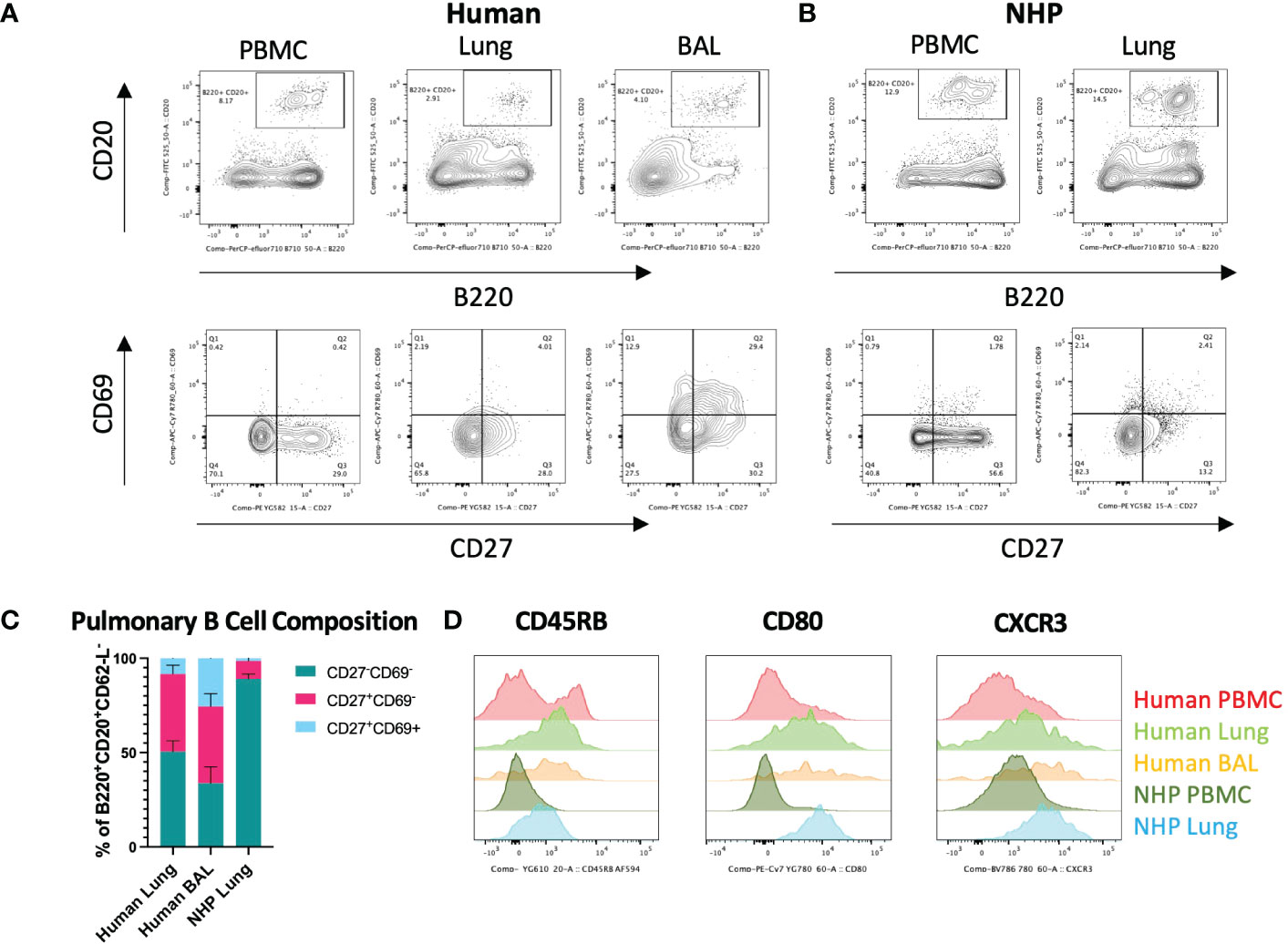
Figure 2 Characterisation of Human and NHP Pulmonary BRM. BRM were isolated from BAL and/or tissue digests and compared with peripheral B cells. (A) Identification of human pulmonary BRM. BRM (live, CD62L-, B220+, CD20+ (lower panels)) were identified based on their expression of CD27 and CD69. (B) Identification of NHP pulmonary BRM. (C) Proportion of BRM in human and NHP lung. (D) Human and NHP BRM surface marker expression. Human and NHP samples were concatenated for analysis. Surface marker expression was compared to B cells isolated from peripheral blood (PBMC). Data represented as representative flow cytometry plots, compiled frequencies with mean ± SEM or histograms from human lungs (n=9), BAL (n=7) and NHP lung (n=5).
Despite being fewer in frequency, NHP B cells from lung digests shared a similar surface marker phenotype to their human counterparts, expressing increased CD45RB, CD80 and CXCR3 levels compared to circulating B cells (Figure 2D).
TRM/BRM tissue immunofluorescence
Having identified the most robust markers and cross-reactive antibodies for detecting human and NHP TRM and BRM, frozen lung sections were stained for immunofluorescence imaging. Acetone-fixed frozen sections were used due to the sensitivities of CD69 and CD103 epitopes to formalin fixation. Tyramide signal amplification was also used as all primary antibodies were raised in mouse. Therefore, we investigated the localization of CD103+ and CD69+ CD4+ and CD8+ TRM and CD20+CD27+CD69+ BRM in human and NHP lungs.
CD69+ and CD69+CD103+ CD4+ and CD8+ TRM were identified within both human and NHP distal airway (Figure 3A – for individual fluorescent channels, see Supplementary Figures 5, 6). CD69-CD103- CD4+ and CD8+ T cells were also evident. CD20+CD27+CD69+ BRM were located within human and NHP distal lung, with less observed in NHP lung, mirroring flow cytometric results (Figure 3B). High frequencies of CD20+CD27-CD69- B cells were also observed in NHP lung, again mirroring flow cytometric results. No evidence of repair-associated memory depots (RAMD) or inducible bronchus-associated lymphoid tissue (iBALT) was apparent as shown by a lack of clusters of CD4+/CD8+ TRM or CD20+ B cells observed by immunofluorescence. DAPI staining following both staining protocols appeared diffuse, suggesting deterioration of the acetone-fixed, frozen tissue from the numerous wash and heat steps.
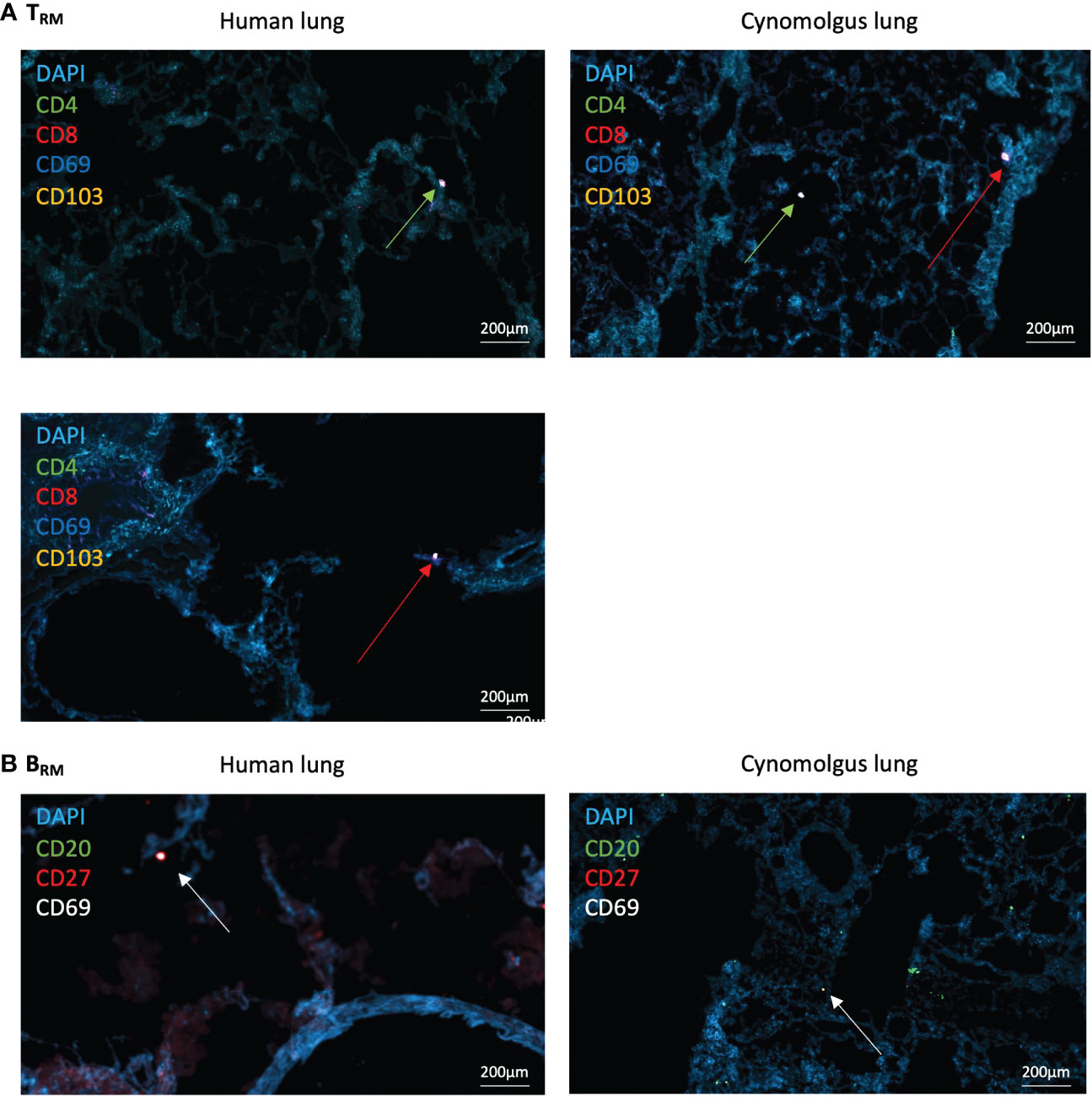
Figure 3 Human and NHP Immunofluorescence TRM/BRM Detection. Frozen lung sections or BAL cells were stained using tyramide signal amplification. (A) Human and NHP TRM detection in lung. CD4+ and CD8+ TRM were detected based on their expression of CD69 and CD103. (B) Human and NHP BRM detection in lung. BRM were detected based on their expression of CD20, CD27 and CD69. Images taken at x400 magnification. Green arrows represent CD4+ TRM and red arrows represent CD8+ TRM (A), and white arrows represent BRM (B).
Fluorescence intensity and lifetime imaging of human pulmonary TRM/BRM
As the current KronoScan system is optimised for two excitation channels, we were unable to employ the lineage-based gating strategies commonly used in flow cytometry and could not definitively label TRM in initial experiments. However, we assessed the potential of KronoScan to detect expression of CD69 and CD103, or CD69 and CD20, as the phenotypically distinguishing surface markers of TRM and BRM, respectively. Utilising the excitation wavelengths (490 nm and 590 nm) and two spectral windows (498-565 nm, “green”, and 620-764 nm, “red”) of KronoScan, FITC-conjugated CD69 and PE-Dazzle conjugated CD103 or CD20 were selected for fluorescence intensity and lifetime imaging. As a proof-of-concept experiment, human lung digest cells were stained with fluorescent antibodies for CD69 and CD103 or CD20, and subsequently imaged in vitro with KronoScan (Figure 4A). Human pulmonary CD69+ and CD69+CD103+ cells could be visualised via both fluorescence intensity and lifetime imaging (Figure 4B + Supplementary Video 1) as well as CD69+CD20+ cells (Figure 4C + Supplementary Video 2). No fluorescence intensity or lifetime signal was observed from unstained lung cells in either the green or red channels (Supplementary Figure 7A). As expected, no CD69 or CD103 signal was detected from PBMCs (as negative controls) (Supplementary Figure 7B + Supplementary Video 3), however CD20+ B cells were observed (Supplementary Figure 7C + Supplementary Video 4).
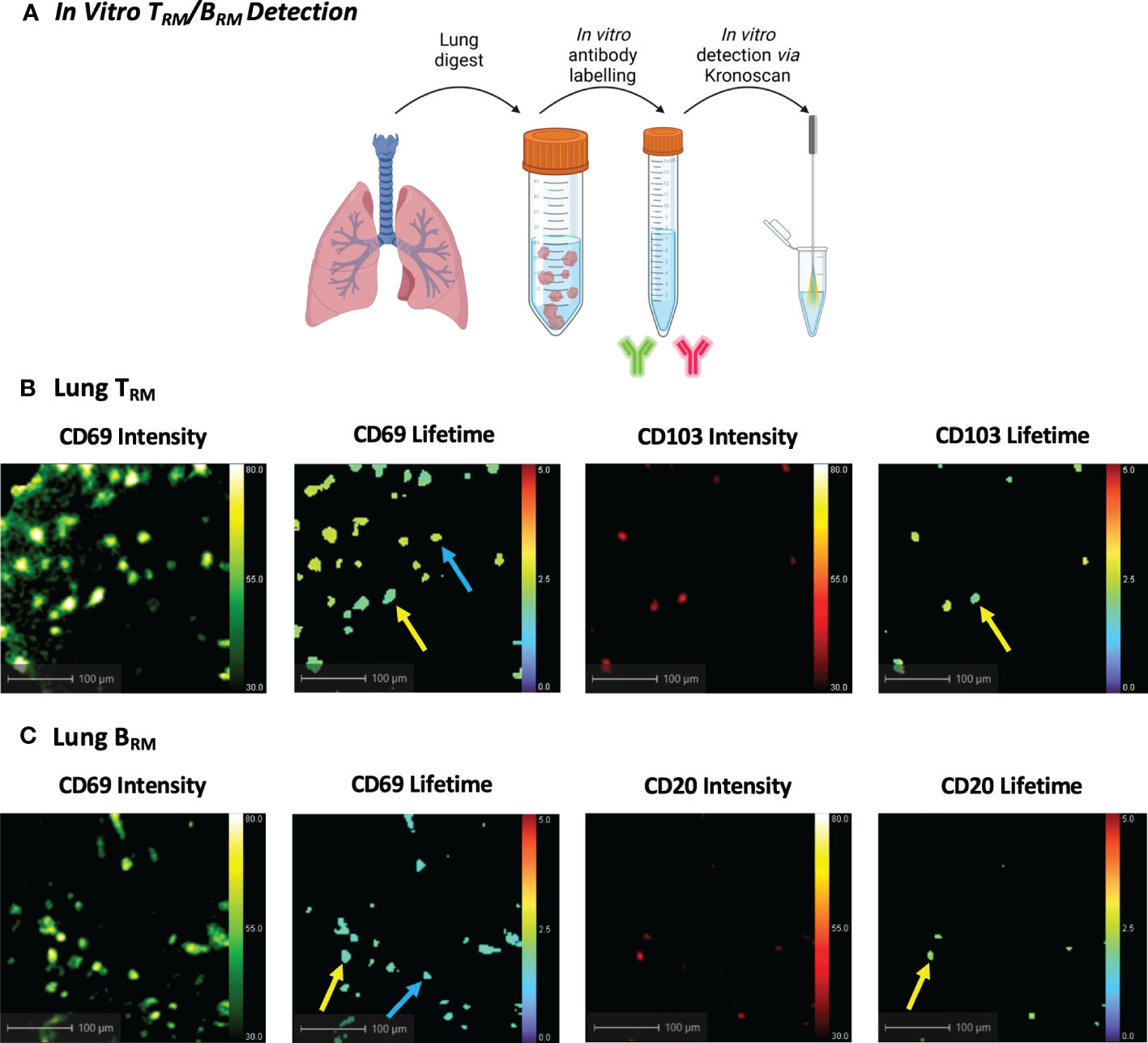
Figure 4 In Vitro Human TRM and BRM Detection using KronoScan and Panoptes. (A) Cells from human lung tissue digests were stained with TRM or BRM antibodies and imaged in vitro using KronoScan imaging system and Panoptes multifunctional fibre. (B) Human TRM detection. TRM were detected via expression of CD69 and CD103. Blue arrow indicates CD69+ cells and yellow arrow points CD69+CD103+ TRM. (C) Human BRM detection. Blue arrow indicates CD69+ cells and yellow arrow points CD69+ CD20+ BRM. Representative images from n=3 experiments.
In situ detection of human pulmonary TRM/BRM
To observe whether KronoScan and Panoptes could detect pulmonary CD69+/CD103+ and CD69+/CD20+ cells in situ, single cell suspensions from human lung digest were pre-stained for CD69 and CD103 or CD69 and CD20 and instilled into the alveolar space of lungs undergoing EVLV (Figure 5A). Fluorescence intensity imaging with 490 nm excitation was used to confirm that the Panoptes multifunctional fibre was within the alveolar space prior to instillation (Figure 5B + Supplementary Video 5). Alveolar lung architecture could be visualised via elastin autofluorescence of alveolar septae. No fluorescence intensity or lifetime signal was detected from lung tissue in the red channel. Upon instillation of labelled cells, CD69+ (Figure 5C + Supplementary Video 6), CD69+CD103+ cells (Figure 5D + Supplementary Video 7) and CD69+CD20+ cells (Figure 5E + Supplementary Video 8) could be visualised via an increase in fluorescence intensity counts as well as a lower lifetime signature of labelled cells that differentiated them from alveolar tissue and intrinsic unlabelled tissue cells.
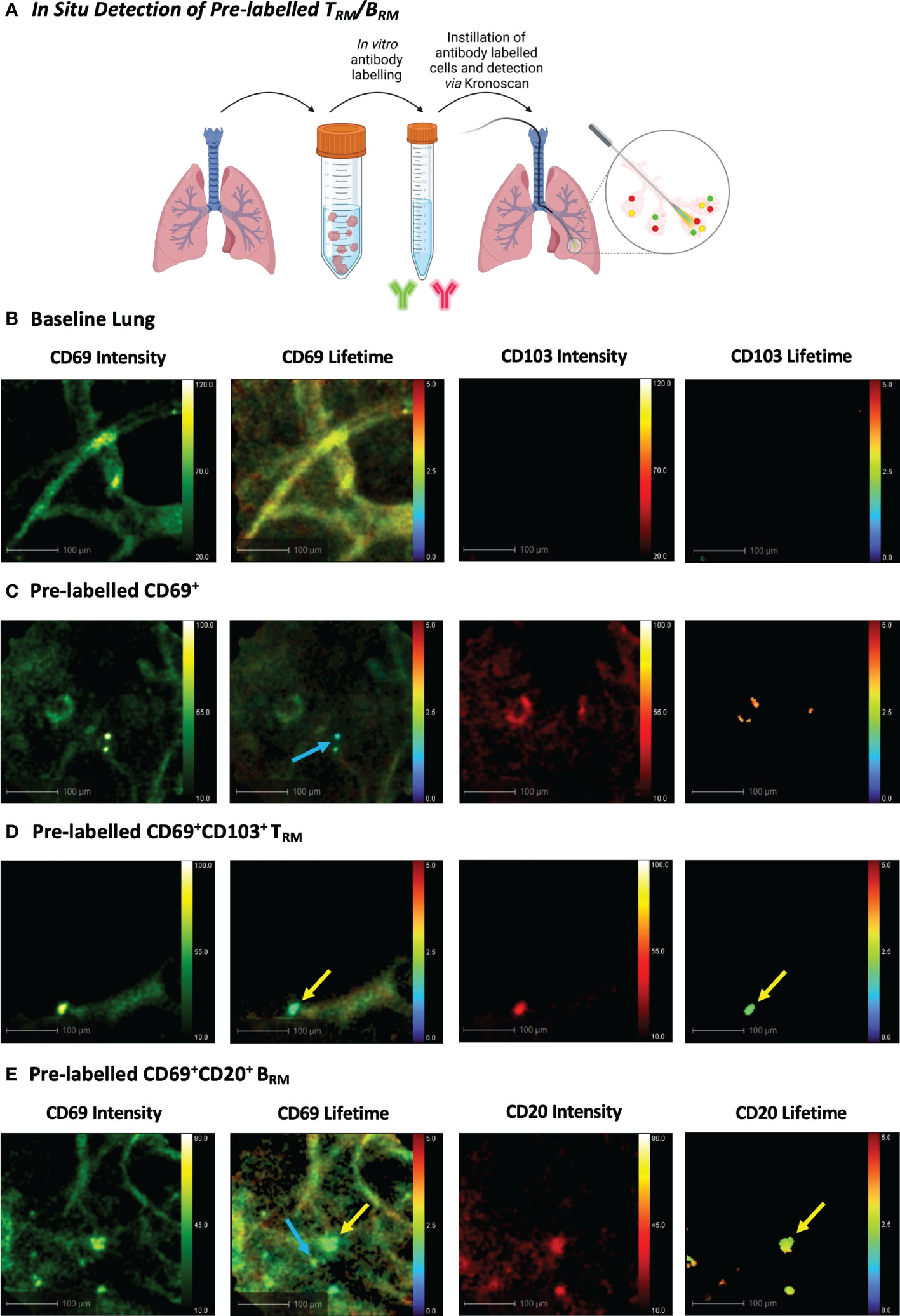
Figure 5 In Situ Detection of Pre-labelled TRM/BRM in Human Lung. (A) Pre-labelled human pulmonary TRM and BRM in suspension were instilled into the alveolar space and detected using KronoScan and Panoptes during human EVLV. (B) Baseline lung imaging prior to pre-labelled cell instillation. (C) In situ imaging of pre-labelled CD69+ cells. Lung tissue digest cells were stained with fluorescent CD69 and CD103 antibodies prior to instillation into the alveolar space using Panoptes. Cells were imaged with KronoScan. Blue arrow indicates CD69+ cells. (D) In situ imaging of pre-labelled CD69+CD103+ TRM. Yellow arrow indicates CD69+CD103+ TRM. (E) In situ imaging of pre-labelled BRM. Lung tissue digest cells were stained with fluorescent CD20 and CD69 antibodies prior to instillation to the alveolar space. Blue arrow indicates CD69+ cells and yellow arrow points CD69+CD20+ BRM. Representative images from n=3 experiments.
We next progressed to in situ labelling and detection of CD69+CD103+ and CD69+CD20+ cells following intra-alveolar delivery of microdoses (0.5–2 µg) of fluorescently labelled antibodies (Figure 6A). Baseline lung imaging confirmed that Panoptes was successfully navigated to the distal lung (Figure 6B). An instantaneous and rapid rise in fluorescence intensity counts confirmed successful delivery of the fluorescent antibody solution within the alveolar space (Supplementary Video 9). Within 120 seconds of CD69 and CD103 antibody delivery, CD69+ cells and CD69+CD103+ cells were visible via increased fluorescence intensity counts and a lifetime signature lower than that of lung tissue (Figures 6C, D + Supplementary Videos 10, 11). Pulmonary BRM were also visualised within the airways and alveolar space following instillation of CD69 and CD20 antibodies (Figure 6E + Supplementary Video 12).
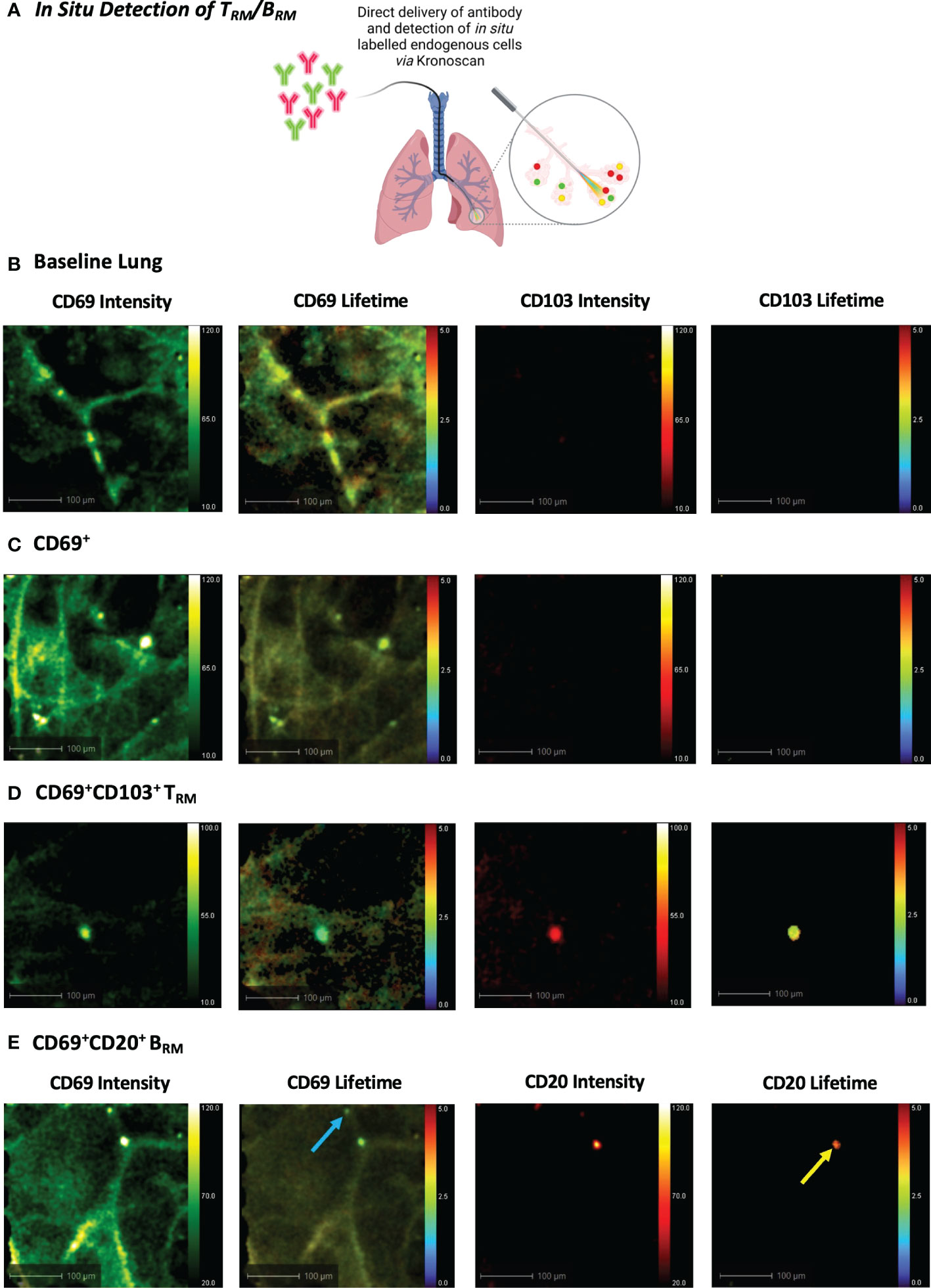
Figure 6 In Situ Detection of TRM/BRM in Human Lung following Antibody Delivery. (A) TRM or BRM antibodies were instilled into the alveolar space via Panoptes fibre and imaged with KronoScan. (B) Baseline lung imaging prior to antibody delivery. (C) In situ detection of CD69+ cells. CD69 and CD103 antibodies were instilled into the alveolar space to detect TRM. (D) In situ detection of CD69+CD103+ TRM. (E) In situ detection of BRM. CD20 and CD69 antibodies were instilled into the alveolar space to detect BRM. Blue arrow indicates CD69+ cells and yellow arrow points CD69+CD20+ BRM. Representative images from n=3 experiments.
Discussion
Using cross-reactive antibodies for CD69 and CD103, we identified pulmonary TRM in the lungs of humans and cynomolgus macaques. Both cell surface markers display key properties in the lung. CD69, although described as a marker of recent T cell activation induced through antigen stimulation and inflammatory cytokine exposure, supports effector T cell retention via the downregulation of sphingosine-1-phophsate receptor (S1PR1) and is considered as the main residency marker of pulmonary TRM (30, 31). CD103, preferentially expressed on pulmonary CD8+ TRM, promotes adherence to E-cadherin and contributes towards initial recruitment and persistence to aid surveillance (32). Moreover, human and NHP CD4+ and CD8+ CD69+ TRM expressed CD49a, particularly those co-expressing CD103. This integrin, specific for collagen IV, may contribute to the functionality of TRM in the lung by facilitating locomotion for surveillance and through its critical role in cell survival by limiting apoptosis (32). Increased levels of PD-1 were also detected on human and NHP TRM, particularly on CD8+ TRM and those isolated from human BAL. This marker may serve as a protective mechanism to prevent aberrant activation and excessive inflammation (33).
Although CD69+ T cells were identified in NHP lung with a higher frequency than in PBMC, there were notably fewer TRM than were found in human lung or BAL. High levels of pulmonary CD69+ TRM have been observed by others in Rhesus macaque, however CD69+CD103+ TRM frequencies were low, similar to our study, even post vaccination (34). We obtained lungs from healthy control animals bred in clean laboratory conditions, which differ greatly to human antigen experienced lungs. Human TRM frequencies are consistently higher than in mice maintained in clean conditions (35). In fact, CD69 expression in “dirty” pet store mice is more like that of humans (36). Limited pathogen exposure may therefore account for the reduced frequencies seen in this study. Moreover, TRM have been described to undergo “retrograde migration” to the mediastinal lymph node where they provide longer-lived regional memory (37).This, combined with limited pathogen exposure, may contribute to a reduction in number of pulmonary TRM in NHP macaques.
CD69 was also used to identify BRM in the lungs of humans and NHP, which has been shown to be a marker of residency rather than recent activation (15, 38). However, expression levels were very low and CD69+ BRM were only readily detectable in human BAL. Human and NHP lung B cells shared a similar phenotype to those reported in mice, with expression of CD80 and the chemokine receptor CXCR3 (17), suggesting their ability to rapidly differentiate into antibody secreting plasma cells (39). CD45RB, expressed by human BRM in the gut and tonsil, was also detected in our study on pulmonary B cells (40).
Human lung TRM weakly expressed Ki-67, indicating low level proliferation. Expression was similar to that found in blood, mirroring previous results (41). Murine studies suggest interstitial CD8+ TRM sustain airway TRM through a process of homeostatic proliferation (42), however we observed increased proliferation in human BAL. Proliferation was higher in NHP lung TRM. Of note, one human sample obtained from an individual who had suffered cardiac arrest out of hospital and aspirated expressed high levels of Ki-67 in lung and BAL TRM which may reflect significant lung injury and infection.
Pulmonary CD69+ CD103+ and CD69+ CD20+ human cells were successfully visualised in situ via fluorescence intensity and lifetime imaging using KronoScan and Panoptes OEM. The lung is highly autofluorescent in the green channel due to the presence of collagen and elastin (43) which can interfere with fluorescent signatures and mask positive signals when using fluorescence intensity imaging alone. In contrast to fluorescence intensity imaging, FLIM is insensitive to the concentration of probe bound to target, meaning only microdoses of antibody are required to visualise a target making FLIM particularly advantageous for lung endomicroscopy. Using both in combination allows for the imaging fibre to be successfully navigated to the alveolar space using fluorescence intensity imaging and FLIM used for the detection of labelled cells. This also allows for the use of commonly employed green channel fluorophores which may not be easily detected against background lung autofluorescence using fluorescence intensity imaging alone.
The direct instillation of fluorescently-tagged monoclonal antibodies into the lungs of living humans may raise ethical concerns. It will therefore be necessary to consider the functional properties of the antibodies and whether target engagement would directly affect ongoing local or systemic immune responses. Labelled humanised antibodies would be advantageous, whilst fluorescently labelled Fab or Fv antibody fragments lacking the Fc portion would prevent issues associated with antibody-dependent cellular cytotoxicity. In addition, as FLIM is largely independent of concentration, only very small amounts of antibody would be required. In this work the amount of antibody used (0.5 – 2 µg per experiment) is likely to fall within the microdose range and lack systemic effects when compared to the repeated mg/kg doses often used in immuno-oncology and is therefore likely to be well tolerated.
The lifetime signature of the fluorophores used in this study were lower than that of the lung tissue. Fluorescence lifetime is environmentally sensitive (43), meaning the temperature, oxygen saturation/hypoxia and pH within the lung will affect lifetime values and thus absolute lifetime values may differ from tissue to tissue. Thus, FLIM was used as a contrast tool for visualising fluorophores and labelled cells against the background lung rather than as an absolute measure of specific FLT values. The combination of high fluorescence intensity with a FLT value lower than that of background lung distinctly shows the presence of labelled TRM/BRM. The lifetime values of antibody-labelled cells are consistently lower than that of background lung tissue and are consistent between experiments (i.e., prelabelled or in situ labelling of both TRM and BRM).
FITC conjugated antibodies have the most spectral similarity to endogenous elastin and tissue autofluorescence, yet the FITC-CD69 was readily distinguishable with high specificity with KronoScan. The high concentration of local cell-specific labelling has been shown to diminish FLT as a function of antibody labelling concentration on the cell surface and is consistent with self-quenching effects expected at high densities of FITC molecules (44).
Due to limited fluorophore combinations on CD103 (clone 2G5) (and the spectral detection capabilities of KronoScan), we used the Ber-ACT8 CD103 clone. Although the Ber-ACT8 CD103 clone did not work in our cynomolgus model, it has been successfully used in Rhesus macaque models (34) and so has good translational potential. In the future, altering the band pass filters on KronoScan will enable the use of other fluorophore/cross reactive antibody combinations. Increasing the number of filter sets will also increase the number of fluorophore combinations that can be used simultaneously, allowing for more information to be collected (e.g., identifying and differentiating CD4+ or CD8+ TRM, or differentiating TRM and BRM subpopulations) and unmixed using both intensity and lifetime measurements.
In the wake of the COVID-19 pandemic, the realization that disease-relevant local immune responses in the lung are not reflected in the blood prompted a rallying call to immunologists to recognize that tissues, rather than blood, are where immune cells function (45). Understanding and investigating immune processes in situ is therefore essential. Successful in situ labelling of immune cells after direct delivery of small amounts (microdoses) of antibody by fluorescence intensity/lifetime imaging extends the methods available for molecular imaging and visualising cells in the distal lung following vaccination and infection. The large number of directly conjugated antibodies available for flow cytometry should make this technique readily adaptable to study the major immune cell types present in alveolar tissues. Direct antibody labelling could also be coupled with the use of activation based smart-probes or in situ imaging of pathogens in experimental settings to visualise infection-driven inflammatory interactions. A recent call for the development of intranasal vaccines capable of eliciting greater mucosal immunity may help to overcome the shortcomings of current COVID vaccines and prevent viral transmission (46). This highlights the potential benefits of increasing tissue-based immunity. Assessing local immunity with our in situ detection method could enhance the evaluation of such vaccines.
Fluorescent intensity-based OEM has shown proof of concept for real-time alveolar imaging, when combined with imaging agents for neutrophil activation (23), gram-negative (25) and gram-positive (47) bacteria. This is however the first report of alveolar fibre based FLIM imaging in the human lung, demonstrating that combining intensity and FLIM delivers contrast enhancement and multiplexing capability above and beyond existing OEM platforms. The multifunctional Panoptes fibre platform, which is designed as a single use disposable device, enables in situ delivery of microdoses of imaging agents and therapies.
We established a cross reactive antibody staining panel for assessing pulmonary TRM and BRM in human and cynomolgus macaque. Many of the antibodies used are cross reactive to other NHP species, including rhesus macaque, and so offer great translational research potential. This work reveals similarities and differences between human and NHP pulmonary resident memory lymphocyte subpopulations and demonstrates the potential of in situ pulmonary optical imaging for rapidly detecting immunologically diverse cells.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The studies involving human participants were reviewed and approved by London - Central Research Ethics Committee 16-LO-1883. Written informed consent was not provided because this study involves the use of human lungs that are unfit for transplant. Specialist Nurses for Organ Donation (SNODs) approach relatives of potential donors and obtain authorisation for the use of the donor’s organs and tissues for transplantation using the standard NHS Authorisation form. Using the same authorisation form, the SNODs also obtain authorisation for the use of the donor’s organs and tissues removed but subsequently found to be unsuitable for transplantation for other purposes (i.e. research studies, education, training). The use of lungs in this study has been approved by an independent ethics committee and NHSBT, the body responsible for organ donation and transplantation across the UK.
Author contributions
RO’C, KF, MC-R, FB, KD, and VP conceptualised the project. DH, RO’C, and KD provided methods. DH, RO’C, HS, TQ, EG, and BM performed the experiments. GW and JS provided materials and technical expertise. DH and HS performed the data analysis. DH wrote the manuscript. DH, RO’C, MC-R, FB, KD, and VP edited the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
We thank the Queen’s Medical Research Institute (QMRI) Flow Cytometry and Histology Facilities at the University of Edinburgh for assistance. We thank Charles River (Edinburgh, UK) for gifting lung tissue from cynomolgus macaque. All figures created with BioRender.com. This work was financially supported by Sanofi, EPSRC Healthcare Impact Partnership award (EP/S025987/1), CARB-X.
Conflict of interest
Authors DH, MC-R, FB and VP were employed by company Sanofi.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors declare that this study received funding from Sanofi. The funder had the following involvement in the study: study design, data analysis, decision to publish, and preparation of the manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1100161/full#supplementary-material
Supplementary Video 1 | In Vitro Fluorescence Imaging of Pulmonary TRM. Cells from human lung tissue digests were stained with TRM antibodies and imaged using KronoScan imaging system and Panoptes multifunctional fibre.
Supplementary Video 2 | In Vitro Fluorescence Imaging of Pulmonary BRM. Cells from human lung tissue digests were stained with BRM antibodies and imaged using KronoScan imaging system and Panoptes multifunctional fibre.
Supplementary Video 3 | Human PBMC CD69 CD103 In Vitro Fluorescence Imaging. Human PBMCs were stained with TRM antibodies and imaged using KronoScan imaging system and Panoptes multifunctional fibre.
Supplementary Video 4 | Human PBMC CD69 CD20 In Vitro Fluorescence Imaging. Human PBMCs were stained with BRM antibodies and imaged using KronoScan imaging system and Panoptes multifunctional fibre.
Supplementary Video 5 | Baseline Lung Fluorescence Imaging Prior to Pre-labelled Cell Instillation. KronoScan and Panoptes imaging fibre was navigated to the alveolar space prior to pre-labelled cell instillation.
Supplementary Video 6 | In Situ Fluorescence Imaging of Pre-labelled Human Pulmonary CD69+ cells. Lung tissue digest cells were stained with fluorescent CD69 and CD103 antibodies prior to instillation into the alveolar space using Panoptes. Cells were imaged with KronoScan.
Supplementary Video 7 | In Situ Fluorescence Imaging of Pre-labelled Human Pulmonary CD69+CD103+ TRM. Lung tissue digest cells were stained with fluorescent CD69 and CD103 antibodies prior to instillation into the alveolar space using Panoptes. Cells were imaged with KronoScan.
Supplementary Video 8 | In Situ Fluorescence Imaging of Pre-labelled Human Pulmonary CD69+CD20+ BRM. Lung tissue digest cells were stained with fluorescent CD69 and CD20 antibodies prior to instillation into the alveolar space using Panoptes. Cells were imaged with KronoScan.
Supplementary Video 9 | Baseline Lung Fluorescence Imaging Prior to Antibody Delivery. KronoScan and Panoptes imaging fibre was navigated to the alveolar space prior to fluorescent antibody delivery.
Supplementary Video 10 | In Situ Imaging of Human CD69+ Cells Following Antibody elivery. CD69 and CD103 antibodies were delivered to the alveolar space and subsequently imaged.
Supplementary Video 11 | In Situ Imaging of Human CD69+CD103+ TRM Following Antibody Delivery. CD69 and CD103 antibodies were delivered to the alveolar space and subsequently imaged.
Supplementary Video 12 | In Situ Fluorescence Imaging of Human Pulmonary BRM Following Antibody Delivery. CD69 and CD20 antibodies were delivered to the alveolar space and subsequently imaged.
References
1. Jozwik A, Habibi MS, Paras A, Zhu J, Guvenel A, Dhariwal J, et al. RSV-Specific airway resident memory CD8+ T cells and differential disease severity after experimental human infection. Nat Commun (2015) 6:1–15. doi: 10.1038/ncomms10224
2. Darrah PA, Zeppa JJ, Maiello P, Hackney JA, Wadsworth MH, Hughes TK, et al. Prevention of tuberculosis in macaques after intravenous BCG immunization. Nature (2020) 577:95–102. doi: 10.1038/s41586-019-1817-8
3. Ogongo P, Tezera LB, Ardain A, Nhamoyebonde S, Ramsuran D, Singh A, et al. Tissue-resident-like CD4+ T cells secreting IL-17 control mycobacterium tuberculosis in the human lung. J Clin Invest (2021) 131:e142014. doi: 10.1172/JCI142014
4. Snyder ME, Farber DL. Human lung tissue resident memory T cells in health and disease. Curr Opin Immunol (2019) 59:101–8. doi: 10.1016/j.coi.2019.05.011
5. Humphries DC, O’Connor RA, Larocque D, Chabaud-Riou M, Dhaliwal K, Pavot V. Pulmonary-resident memory lymphocytes: Pivotal orchestrators of local immunity against respiratory infections. Front Immunol (2021) 12:738955. doi: 10.3389/fimmu.2021.738955
6. de Bree GJ, van Leeuwen EMM, Out TA, Jansen HM, Jonkers RE, van Lier RAW. Selective accumulation of differentiated CD8+ T cells specific for respiratory viruses in the human lung. J Exp Med (2005) 202:1433–42. doi: 10.1084/jem.20051365
7. Pizzolla A, Nguyen THO, Sant S, Jaffar J, Loudovaris T, Mannering SI, et al. Influenza-specific lung-resident memory T cells are proliferative and polyfunctional and maintain diverse TCR profiles. J Clin Invest (2018) 128:721–33. doi: 10.1172/JCI96957
8. Pichyangkul S, Yongvanitchit K, Limsalakpetch A, Kum-Arb U, Im-Erbsin R, Boonnak K, et al. Tissue distribution of memory T and b cells in rhesus monkeys following influenza a infection. J Immunol (2015) 195:4378–86. doi: 10.4049/jimmunol.1501702
9. Yang Q, Zhang M, Chen Q, Chen W, Wei C, Qiao K, et al. Cutting edge: Characterization of human tissue-resident memory T cells at different infection sites in patients with tuberculosis. J Immunol (2020) 204:2331–6. doi: 10.4049/jimmunol.1901326
10. Pizzolla A, Wakim LM. Memory T cell dynamics in the lung during influenza virus infection. J Immunol (2019) 202:374–81. doi: 10.4049/jimmunol.1800979
11. Koutsakos M, Illing PT, Nguyen THO, Mifsud NA, Crawford JC, Rizzetto S, et al. Human CD8 + T cell cross-reactivity across influenza a, b and c viruses. Nat Immunol (2019) 20:613–25. doi: 10.1038/s41590-019-0320-6
12. Ge C, Monk IR, Pizzolla A, Wang N, Bedford JG, Stinear TP, et al. Bystander activation of pulmonary trm cells attenuates the severity of bacterial pneumonia by enhancing neutrophil recruitment. Cell Rep (2019) 29:4236–44. doi: 10.1016/j.celrep.2019.11.103
13. Turner DL, Goldklang M, Cvetkovski F, Paik D, Trischler J, Barahona J, et al. Biased generation and In situ activation of lung tissue-resident memory CD4 T cells in the pathogenesis of allergic asthma. J Immunol (2018) 200:1561–9. doi: 10.4049/jimmunol.1700257
14. Bošnjak B, Kazemi S, Altenburger LM, Mokrović G, Epstein MM. Th2-TRMs maintain life-long allergic memory in experimental asthma in mice. Front Immunol (2019) 10:840. doi: 10.3389/fimmu.2019.00840
15. Barker KA, Etesami NS, Shenoy AT, Arafa EI, Lyon de Ana C, Smith NM, et al. Lung-resident memory b cells protect against bacterial pneumonia. J Clin Invest (2021) 131:e141810. doi: 10.1172/JCI141810
16. Allie SR, Bradley JE, Mudunuru U, Schultz MD, Graf BA, Lund FE, et al. The establishment of resident memory b cells in the lung requires local antigen encounter. Nat Immunol (2019) 20:97–108. doi: 10.1038/s41590-018-0260-6
17. Onodera T, Takahashi Y, Yokoi Y, Ato M, Kodama Y, Hachimura S, et al. Memory b cells in the lung participate in protective humoral immune responses to pulmonary influenza virus reinfection. Proc Natl Acad Sci U.S.A. (2012) 109:2485–90. doi: 10.1073/pnas.1115369109
18. Rangel-Moreno J, Carragher DM, Misra RS, Kusser K, Hartson L, Moquin A, et al. B cells promote resistance to heterosubtypic strains of influenza via multiple mechanisms. J Immunol (2008) 180:454–63. doi: 10.4049/jimmunol.180.1.454
19. Knight FC, Wilson JT. Engineering vaccines for tissue-resident memory T cells. Adv Ther (Weinh) (2021) 4. doi: 10.1002/adtp.202000230
20. Dijkman K, Aguilo N, Boot C, Hofman SO, Sombroek CC, Vervenne RAW, et al. Pulmonary MTBVAC vaccination induces immune signatures previously correlated with prevention of tuberculosis infection. Cell Rep Med (2021) 2. doi: 10.1016/j.xcrm.2020.100187
21. McMaster SR, Wein AN, Dunbar PR, Hayward SL, Cartwright EK, Denning TL, et al. Pulmonary antigen encounter regulates the establishment of tissue-resident CD8 memory T cells in the lung airways and parenchyma. Mucosal Immunol (2018) 11:1071–8. doi: 10.1038/s41385-018-0003-x
22. Snyder ME, Finlayson MO, Connors TJ, Dogra P, Senda T, Bush E, et al. Generation and persistence of human tissue-resident memory T cells in lung transplantation. Sci Immunol (2019) 4:eaav5581. doi: 10.1126/sciimmunol.aav5581
23. Craven TH, Walton T, Akram AR, Scholefield E, McDonald N, Marshall ADL, et al. Activated neutrophil fluorescent imaging technique for human lungs. Sci Rep (2021) 11:976. doi: 10.1038/s41598-020-80083-w
24. Suhling K, Hirvonen LM, Levitt JA, Chung P-H, Tregidgo C, le Marois A, et al. Fluorescence lifetime imaging (FLIM): Basic concepts and some recent developments. Med Photonics (2015) 27:3–40. doi: 10.1016/j.medpho.2014.12.001
25. Parker HE, Stone JM, Marshall ADL, Choudhary TR, Thomson RR, Dhaliwal K, et al. Fibre-based spectral ratio endomicroscopy for contrast enhancement of bacterial imaging and pulmonary autofluorescence. BioMed Opt Express (2019) 10:1856–69. doi: 10.1364/BOE.10.001856
26. Dorward DA, Lucas CD, Doherty MK, Chapman GB, Scholefield EJ, Conway Morris A, et al. Novel role for endogenous mitochondrial formylated peptide-driven formyl peptide receptor 1 signalling in acute respiratory distress syndrome. Thorax (2017) 72:928–36. doi: 10.1136/thoraxjnl-2017-210030
27. Millar M. Mixed multiplex staining: Automated RNAscopeTM and OPALTM for multiple targets. In: Nielsen BS, Jones J, editors. Situ hybridization protocols. New York, NY: Springer US (2020). p. P. 277–298. doi: 10.1007/978-1-0716-0623-0
28. Li D, Arlt J, Tyndall D, Walker R, Richardson J, Henderson R, et al. Video-rate fluorescence lifetime imaging camera with CMOS single-photon avalanche diode arrays and high-speed imaging algorithm. J BioMed Opt (2011) 16:096012. doi: 10.1117/1.3625288
29. Stone JM, Wood HAC, Harrington K, Birks TA. Low index contrast imaging fibers. Opt Lett (2017) 42:1484–7. doi: 10.1364/OL.42.001484
30. Takamura S. Persistence in temporary lung niches: A survival strategy of lung-resident memory CD8+ T cells. Viral Immunol (2017) 30:438–50. doi: 10.1089/vim.2017.0016
31. Mackay LK, Braun A, Macleod BL, Collins N, Tebartz C, Bedoui S, et al. Cutting edge: CD69 interference with sphingosine-1-Phosphate receptor function regulates peripheral T cell retention. J Immunol (2015) 194:2059–63. doi: 10.4049/jimmunol.1402256
32. Reilly EC, Emo KL, Buckley PM, Reilly NS, Smith I, Chaves FA, et al. TRM integrins CD103 and CD49a differentially support adherence and motility after resolution of influenza virus infection. Proc Natl Acad Sci U.S.A. (2020) 117:12306–14. doi: 10.1073/pnas.1915681117
33. Kumar BV, Ma W, Miron M, Granot T, Guyer RS, Carpenter DJ, et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep (2017) 20:2921–34. doi: 10.1016/j.celrep.2017.08.078
34. Layton ED, Barman S, Wilburn DB, Yu KKQ, Smith MT, Altman JD, et al. T Cells specific for a mycobacterial glycolipid expand after intravenous bacillus calmette-guérin vaccination. J Immunol (2021) 206:1240–50. doi: 10.4049/jimmunol.2001065
35. Miron M, v. KB, Meng W, Granot T, Carpenter DJ, Senda T, et al. Human lymph nodes maintain TCF-1 hi memory T cells with high functional potential and clonal diversity throughout life. J Immunol (2018), 01(7):2132–40. doi: 10.4049/jimmunol.1800716
36. Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA, et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature (2016) 532. doi: 10.1038/nature17655
37. Stolley JM, Johnston TS, Soerens AG, Beura LK, Rosato PC, Joag V, et al. Retrograde migration supplies resident memory t cells to lung-draining LN after influenza infection. J Exp Med (2020) 217:e20192197. doi: 10.1084/jem.20192197
38. Tan H-X, Juno JA, Esterbauer R, Kelly HG, Wragg KM, Konstandopoulos P, et al. Lung-resident memory b cells established after pulmonary influenza infection display distinct transcriptional and phenotypic profiles. Sci Immunol (2022) 7:eabf5314. doi: 10.1126/sciimmunol.abf5314
39. Zuccarino-Catania GV, Sadanand S, Weisel FJ, Tomayko MM, Meng H, Kleinstein SH, et al. CD80 and PD-L2 define functionally distinct memory b cell subsets that are independent of antibody isotype. Nat Immunol (2014) 15:631–7. doi: 10.1038/ni.2914
40. Weisel NM, Weisel FJ, Farber DL, Borghesi LA, Shen Y, Ma W, et al. Comprehensive analyses of b-cell compartments across the human body reveal novel subsets and a gut-resident memory phenotype. Blood (2020), 136(24):2774–85. doi: 10.1182/blood.2019002782
41. Purwar R, Campbell J, Murphy G, Richards WG, Clark RA, Kupper TS. Resident memory T cells (T(RM)) are abundant in human lung: Diversity, function, and antigen specificity. PloS One (2011) 6:e16245–5. doi: 10.1371/journal.pone.0016245
42. Takamura S, Kato S, Motozono C, Shimaoka T, Ueha S, Matsuo K, et al. Interstitial-resident memory CD8+ T cells sustain frontline epithelial memory in the lung. J Exp Med (2019) 216:2736–47. doi: 10.1084/jem.20190557
43. Berezin MY, Achilefu S. Fluorescence lifetime measurements and biological imaging. Chem Rev (2010) 110:2641–84. doi: 10.1021/cr900343z
44. Deka C, Lehnert BE, Lehnert NM, Jones GM, Sklar LA, Steinkamp JA. Analysis of fluorescence lifetime and quenching of FITC-conjugated antibodies on cells by phase-sensitive flow cytometry. Cytometry (1996) 25:271–9. doi: 10.1002/(SICI)1097-0320(19961101)25:3<271::AID-CYTO8>3.0.CO;2-I
45. Farber DL. Tissues, not blood, are where immune cells function. Nature (2021) 593:506–9. doi: 10.1038/d41586-021-01396-y
46. Topol EJ, Iwasaki A. Operation nasal vaccine-lightning speed to counter COVID-19. Sci Immunol (2022) 7:eadd9947. doi: 10.1126/sciimmunol.add9947
Keywords: resident memory T cells, resident memory B cells, lung, optical endomicroscopy, fluorescence lifetime imaging
Citation: Humphries DC, O’Connor RA, Stewart HL, Quinn TM, Gaughan EE, Mills B, Williams GOS, Stone JM, Finlayson K, Chabaud-Riou M, Boudet F, Dhaliwal K and Pavot V (2023) Specific in situ immuno-imaging of pulmonary-resident memory lymphocytes in human lungs. Front. Immunol. 14:1100161. doi: 10.3389/fimmu.2023.1100161
Received: 16 November 2022; Accepted: 30 January 2023;
Published: 10 February 2023.
Edited by:
Nargis Khan, University of Calgary, CanadaReviewed by:
Pierre Guermonprez, Centre National de la Recherche Scientifique (CNRS), FranceBas G.J. Surewaard, University of Calgary, Canada
Ajitha Thanabalasuriar, McGill University, Canada
Copyright © 2023 Humphries, O’Connor, Stewart, Quinn, Gaughan, Mills, Williams, Stone, Finlayson, Chabaud-Riou, Boudet, Dhaliwal and Pavot. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kevin Dhaliwal, S2V2LkRoYWxpd2FsQGVkLmFjLnVr; Vincent Pavot, VmluY2VudC5QYXZvdEBzYW5vZmkuY29t
 Duncan C. Humphries
Duncan C. Humphries Richard A. O’Connor
Richard A. O’Connor Hazel L. Stewart
Hazel L. Stewart Tom M. Quinn1
Tom M. Quinn1 Gareth O.S. Williams
Gareth O.S. Williams Martine Chabaud-Riou
Martine Chabaud-Riou Florence Boudet
Florence Boudet Kevin Dhaliwal
Kevin Dhaliwal Vincent Pavot
Vincent Pavot