- 1Peter Medawar Building for Pathogen Research, Nuffield Department of Medicine, University of Oxford, Oxford, United Kingdom
- 2National Institute for Health and Care Research Health Protection Research Unit in Emerging and Zoonotic Infections, University of Liverpool, Liverpool, United Kingdom
- 3Peter Medawar Building for Pathogen Research, Department of Paediatrics, University of Oxford, Oxford, United Kingdom
The factors determining whether infection will occur following exposure to SARS-CoV-2 remain elusive. Certain SARS-CoV-2-exposed individuals mount a specific T-cell response but fail to seroconvert, representing a population that may provide further clarity on the nature of infection susceptibility and correlates of protection against SARS-CoV-2. Exposed seronegative individuals have been reported in patients exposed to the blood-borne pathogens Human Immunodeficiency virus and Hepatitis C virus and the sexually transmitted viruses Hepatitis B virus and Herpes Simplex virus. By comparing the quality of seronegative T-cell responses to SARS-CoV-2 with seronegative cellular immunity to these highly divergent viruses, common patterns emerge that offer insights on the role of cellular immunity against infection. For both SARS-CoV-2 and Hepatitis C, T-cell responses in exposed seronegatives are consistently higher than in unexposed individuals, but lower than in infected, seropositive patients. Durability of T-cell responses to Hepatitis C is dependent upon repeated exposure to antigen – single exposures do not generate long-lived memory T-cells. Finally, exposure to SARS-CoV-2 induces varying degrees of immune activation, suggesting that exposed seronegative individuals represent points on a spectrum rather than a discrete group. Together, these findings paint a complex landscape of the nature of infection but provide clues as to what may be protective early on in SARS-CoV-2 disease course. Further research on this phenomenon, particularly through cohort studies, is warranted.
Introduction
Exposure to viral pathogens does not guarantee infection. The clearest examples of this phenomenon are in the failure of test subjects in human challenge studies to consistently become infected (1–3). Variation in host susceptibility has been linked to host genetics, inoculum viral load, and prior exposure to related pathogens (2–6). Among those individuals who are exposed but fail to become infected, a small but well-documented population generate pathogen-specific T-cell responses in the absence of viraemia or antibodies (7–9). The earliest reports of this phenomenon occurred in the late 1980s concerning apparent Human Immunodeficiency virus (HIV) resistance in at-risk individuals (10–14). These patients, despite exposure to HIV and measurable cellular immunity, failed to develop an antibody response and were therefore classified as “exposed seronegative” (ESN). In the 30 years since initial reports in HIV, the phenomenon has been appreciated to occur following exposure to Hepatitis C virus (HCV), Hepatitis B virus (HBV), and Herpes Simplex virus 2 (HSV-2) (8, 15–18), and recently Severe Acute Respiratory Syndrome coronavirus 2 (SARS-CoV-2), a pathogen to which most of the global population has been exposed (7, 19, 20). SARS-CoV-2, a member of the Coronaviridae family, is the causative agent of coronavirus disease 2019 (COVID-19). Canonical immunity to SARS-CoV-2 is well characterised despite its recent emergence: a delay in innate immune activation resulting from viral evasion of interferon (IFN) responses enables infection to occur (21). Both humoral and cellular mechanisms are essential for viral control; weak or delayed adaptive responses can lead to severe or fatal COVID-19, with immunopathology and cytokine hyperactivation characteristic of end-stage disease (21).
The causes and consequences of the ESN phenomenon following exposure to SARS-CoV-2, as well as other viruses, are the focus of this review. We outline the circumstances in which seronegative cellular immunity occurs and examine the quality of the T-cell response. In addition, we address the role of viral exposure on response durability, and whether this offers protection against infection. Finally, potential mechanisms are discussed, as well as gaps in current knowledge that future research must fill.
Definitions and terminology
For this review, an ESN is defined as an individual who mounts a cellular immune response following viral exposure without generating detectable virus-specific antibody. Instances where infection has been prevented by the innate immune system alone, as reported elsewhere (22, 23), are out of scope for this review due to not inducing a T-cell response. In the literature, the terminology for ESNs is largely consistent within but not between viruses and include exposed seronegative(s), exposed uninfected, infected seronegative, immune seronegative, and highly exposed persistently seronegative. These terms will be collectively referred to as ESN.
At-risk demographics
The dynamics of exposure, such as inoculum viral load, exposure frequency, and exposure duration, may influence cellular responses in ESNs. To make robust comparisons between SARS-CoV-2 and other viruses, we examine cellular immunity in two demographics at high risk of exposure – close contacts of seropositive individuals, and healthcare workers (HCWs). This enables the analysis of common and differing patterns of immunity to unrelated viruses through similar modes of exposure.
Close and household contacts
Close contacts of SARS-CoV-2-infected individuals represent a population exposed to SARS-CoV-2 over short time periods. Wang et al. (2021) assessed cellular immunity in 90 seropositive individuals and 69 seronegative contacts who had been within 1.5m of a patient for over one hour or in the same household for over 24 hours (20). The authors observed higher CD8+ and CD4+ T-cell activation in both cohorts compared to unexposed controls, as measured by IFNγ production following stimulation with Spike (S), Membrane (M), Nucleocapsid (NP), and Envelope peptides. Gallais et al. (2021) also identified T-cell responses in close contacts of SARS-CoV-2-infected family members (24).
Family members of HCV-infected individuals face viral exposure through sexual and in-utero transmission. Kamal et al. (2004) identified 14 seronegative sexual partners of HCV-infected individuals who generated IFNγ responses to HCV, although at lower magnitudes than seropositive resolvers (25). HCV is a positive-sense RNA virus in the Flaviviridae family (26). Canonically, adaptive immune responses appear one to two months after infection (27), and both CD4+ and CD8+ T-cells are associated with control (26). In individuals exposed to HCV via a family member, Scognamiglio et al. (1999) identified CD8+ T-cells targeting both structural and non-structural proteins (NSPs) (16). There was no correlation between magnitude of response and mode of exposure, suggesting that similar cellular responses are generated following HCV exposure through different routes.
HCWs represent a population where SARS-CoV-2 exposure in clinical settings enables the study of ESNs. da Silva Antunes et al. (2021) recruited 26 PCR+ HCWs, 32 seronegative HCWs at high risk of exposure (treated as ESNs here), and 33 community controls (28). ESNs demonstrated higher T-cell responses than unexposed individuals. Notably, the T-cell activation markers HLA-DR and CD38 were upregulated in PCR+ HCWs, but not in ESNs, leading authors to conclude that cellular responses in ESNs were generated by exposure rather than infection and waning of antibody. Considerable overlap in levels of HLA-DR/CD38 expression was observed between seropositive and ESN HCWs. A study of SARS-CoV-2 ESN HCWs in Swadling et al. (2022) found a significant correlation between the magnitude of the T-cell response and transcript levels of IFI17, an IFN-inducible marker of early infection (7). Swadling et al. concluded that transient infection had occurred, but was aborted by early T-cell responses (7). However, ESNs with elevated IFI17 transcripts only represented 10% of ESNs, and mean IFI17 expression was lower in ESNs than seropositive HCWs (7). ESNs may therefore represent not one discrete group, but a spectrum of immune engagement, from subclinical exposure to transient infection.
Ogbe et al. (2020) identified SARS-CoV-2-specific T-cell responses in exposed HCWs: three of 10 generated IFNγ to S, M and NP, whilst all generated cellular responses to M and NP by proliferation assay, indicating the potentially higher sensitivity of this assay (19). T-cell responses in ESNs were of greater magnitude than unexposed controls for CD4+ but not CD8+ cells, although both CD4+ and CD8+ cells targeted a greater number of antigens in ESNs compared to controls. The presence of T-cells targeting multiple antigens in ESNs is supported by da Silva Antunes et al. (2021), where cellular immunity targeting S as well as the rest of the proteome was higher in ESNs compared to controls (28).
Kubitschke et al. (2007) identified T-cell immunity in seronegative HCWs exposed to HCV-contaminated needles (29). CD4+ responses occurred in four of 10 individuals within eight weeks of injury but were absent after 2.5 years. Heller et al. (2013) assayed cellular immunity in HCWs exposed to HCV via needlestick, cut, or mucosal exposure, and identified HCV-specific proliferation and IFNγ production in 48% (n=30) and 42% (n=26) of individuals, respectively (30). Three-quarters of responses were directed towards NSPs. Responses peaked four to six weeks after exposure, unlike canonical HCV infection where immunity generally peaks at seven to 14 weeks (31). The authors suggested this may reflect boosting of pre-existing cellular memory. However, responses were transient and returned to baseline within months, unlike canonical long-lived memory responses (32). The dynamics of cellular immunity in HCV ESNs appears distinct from seropositive infection in its more rapid induction and reduced longevity.
Finally, Clerici et al. (1994) studied eight ESN HCWs following HIV+ needlestick injury. Four to eight weeks after injury, production of interleukin (IL)-2 by T-cells specific for the env glycoprotein was observed in six of eight ESNs, compared to only one of nine unexposed controls (33). However, two ESNs seroconverted at six and 19 months after sampling, reflecting early-stage infection rather than exposure.
Duration and dose
Viruses that have been well-studied for decades provide valuable information on the roles of exposure frequency and response durability in ESNs. Thurairajah et al. (2011) studied seronegative injection drug users exposed to HCV with differing injection behaviours (non-injectors in rehabilitation, infrequent injectors, and continuing injectors) (34). Continuing injectors had stronger and more numerous T-cell responses to HCV compared to non-injectors and healthy controls. Furthermore, individuals who had last injected over 12 months ago had a lower proportion of positive responses than those who had injected in the last six months. These data indicate that ongoing exposure to virus is one factor in the maintenance of T-cell responses in HCV ESNs.
Animal studies also provide insight into the role of antigen exposure for HCV. Shata et al. (2003) demonstrated that two chimpanzees exposed at six month intervals to increasing doses of HCV generated transient T-cell responses (35). 12 months later, both chimpanzees were exposed to a tenfold greater dose of the virus and became infected. The chimpanzee with consistently stronger T-cell responses cleared infection whilst the other developed chronic disease. Interpretation of this result is limited by the small cohort size. Furthermore, macaques exposed to infectious doses of simian immunodeficiency virus seroconverted but generated weak cellular responses, whilst those exposed to sub-infectious doses generated cellular responses only (36). These findings suggest that dose may factor into which arm of adaptive immunity dominates upon viral exposure. Similar challenge studies in primates or humans exposed to differing doses of SARS-CoV-2 would be necessary to make conclusions about the role of dose in SARS-CoV-2 ESNs.
T-cell responses in seronegative household contacts exposed to SARS-CoV-2 suggest that prolonged exposure may not be essential for cellular immunity (7, 19, 28). The durability of these responses is unknown due to the short timescale since virus emergence as well as the confounding influence of vaccination against SARS-CoV-2. Future studies on SARS-CoV-2 ESNs would benefit from sampling high-risk seronegative individuals, but although NP-targeting immunoassays could be used, these studies will be hamstrung by vaccination.
Target antigens
Determining which antigens are targeted in SARS-CoV-2 ESNs provides insight into mechanisms of response. T-cells targeting the replication-transcription complex (RTC) of SARS-CoV-2 were described by Swadling et al. (2022) in ESNs (7). The RTC is comprised of the RNA polymerase NSP12, a co-factor NSP7, and the helicase NSP13 (37). Its expression early in the SARS-CoV-2 replication cycle makes the RTC a target for rapidly-induced T-cell responses (7). The authors identified fivefold-higher RTC-specific T-cell responses in ESNs compared to unexposed controls. Furthermore, cellular immunity in ESNs preferentially targeted the RTC over structural proteins compared to seropositive individuals. However, the authors did not assay cellular responses to other NSPs. In a study of six ESN sexual partners of HSV-2-infected individuals by Posavad et al. (2010), T cell responses in ESNs were skewed towards peptides expressed early in the virus replication cycle, whereas HSV-2 seropositive individuals more frequently generated responses to structural proteins present in virions (8). The authors speculated that this skew in ESNs reflected early T-cell engagement with infected cells before the production of infectious virions. Together, these data support a model whereby rapid T-cell responses targeting early translated NSPs may prevent infection from gaining a foothold.
Cytokine profile
In a cohort of 52 household contacts of SARS-CoV-2-infected individuals, Kundu et al. (2022) identified higher frequencies of IL-2-, but not IFNγ-, secreting T-cells in ESNs compared to individuals that later became infected (38). A similar study of household contacts from Brand et al. (2021) reported no T-cell recognition of SARS-CoV-2 epitopes in seronegative individuals – this was measured by a novel IFNγ assay, which may lack sensitivity for low magnitude responses in ESNs (39). Assays that measure IFNγ production alone may underestimate the prevalence of cellular immune responses in ESNs, highlighting the need for multiple sensitive immunophenotyping methods, such as flow cytometric or proliferation assays, to accurately quantify responses.
TH1-focussed cytokine production has been described in HBV ESNs (18). Sexual partners of infected individuals generated proliferative T-cell and IFNγ responses to HBV peptides. No ESNs generated TNFα or IL-10 responses, unlike seropositive individuals (40). Finally, IFNγ secretion was described in seronegative individuals exposed to Ebola virus (EBOV) (41). A study of EBOV close contacts (n=42) from Thom et al. (2021) identified two ESNs. However, responses in these ESNs were not present in further samples, potentially reflecting experimental artefacts. This further highlights the need for sensitive immunophenotyping assays to examine the true prevalence of ESNs.
Proposed mechanism
To prevent infection before seroconversion, a rapid cellular response appears critical. Chandran et al. (2021) assayed weekly nasopharyngeal swabs and blood samples from HCWs, and demonstrated that SARS-CoV-2 specific T-cell proliferation can occur before PCR positivity (42). These rapid responses may originate from pre-existing, cross-reactive T-cells specific for human coronaviruses (HCoVs). Cross-recognition of SARS-CoV-2 by HCoV-specific T-cells has been widely described (43–50), and T-cells from COVID-19 convalescents preferentially target conserved epitopes over SARS-CoV-2-specific epitopes (49). HCWs display higher levels of HCoV-specific T-cells than community controls (28), which may contribute to the abundance of ESNs amongst HCWs. The activation of cross-reactive T-cells by related viruses has been termed ‘heterologous immunity’ (51). This is distinct from autologous viral infection in that neutralising antibody responses to the heterologous virus may be suboptimal, allowing cellular memory to dominate.
The RTC is highly conserved between SARS-CoV-2 and HCoVs (7). Tetramer staining of T-cells with an HCoV-HKU1 homologue of the RTC component NSP7 showed strong responses in SARS-CoV-2 ESNs. Swadling et al. (2022) suggested that prior exposure to HCoV-HKU1 generates cross-reactive T-cells specific for NSP7, enabling rapid abortion of SARS-CoV-2 infection (7). A study of camel workers in Saudi Arabia identified both CD4+ and CD8+ responses to Middle-East Respiratory Syndrome coronavirus in four highly-exposed seronegative individuals, suggesting that the ESN phenomenon may be common to other human-infective coronaviruses (52).
It is unclear whether cross-reactive T-cells contribute to ESN immunity in HCV, HIV, HBV or HSV. Cross-reactivity between HCV and influenza A has been described, with HCV-seronegative individuals generating T-cell responses to a cross-reactive HCV epitope (53). However, human viruses with homology to HIV, HBV or HSV have not been described and are thus unlikely to be the driver of the ESN phenomenon for these viruses.
Correlates of protection from infection
Key to understanding correlates of protection against SARS-CoV-2 infection is deciphering the role of cellular versus humoral immunity. Seropositivity may not always be the most appropriate marker if cellular immunity is protective. This is particularly relevant for assessing vaccine-induced protection against disease where neutralizing antibody titres are a common endpoint, and particularly for SARS-CoV-2 where an arms race between booster vaccination and waning antibody titres has begun.
In a model whereby cellular immunity in ESNs is protective, one would reasonably expect that the magnitude of cellular response in ESNs would be greater than in seropositive individuals, to compensate for the lack of humoral immunity. Cellular immunity is able to clear SARS-CoV-2 infection in isolation; patients with X-linked agammaglobulinemia who cannot produce antibodies eventually clear SARS-CoV-2 infection, and mount higher magnitude CD8+ T-cell responses to SARS-CoV-2 compared to immunocompetent individuals (54). However, in Wang et al. (2021) the magnitude of the SARS-CoV-2-specific CD4+ T-cell response was twice as high in infected individuals compared to ESNs. This casts doubt on their role in protection against infection.
In influenza virus infection, cytotoxic T-cells target conserved non-structural proteins while antibodies target the divergent neuraminidase and hemagglutinin proteins and are thus strain-specific. In 1983, McMichael and colleagues demonstrated that individuals with cross-reactive T-cells targeting influenza A were able to clear infection in the absence of subtype-specific antibody (55). Later studies showed cross-reactive CD4+ and CD8+ T-cells are associated with milder disease in individuals lacking cross-reactive antibody (56, 57). Animal challenge models have shed light on whether cellular immunity following vaccination can confer protection against influenza. Vaccination of mice with a virus-like particle vaccine against influenza A virus promoted cross-reactive CD8+ T-cell-mediated protection against later challenge with a heterosubtypic strain, supporting the idea that cellular immunity in the absence of subtype-specific antibody can confer protection against infection (58). The applicability of cross-reactive T cell responses in influenza virus models to other virus families is unclear. However, the observations in these studies strongly support cellular immunity being considered in estimates of correlates of protection for viral infection.
Discussion
A model for the dynamics of adaptive immunity in infection versus exposure is shown in Figure 1. In canonical infection (Figure 1A), T-cells and antibodies reduce viral load and contribute to disease resolution. In ESNs (Figure 1B), T-cells proliferate alone and at lower levels than canonical infection. Viral load never reaches detectable levels and clinical disease does not occur. This may result from early proliferation of cross-reactive memory T-cells. Rather than being a discrete group, ESNs likely represent points along a spectrum of immune engagement, influenced by viral and host factors (Figure 1C). Discrepancies across studies likely reflect individuals at different points along this spectrum, dependent upon viral dose, existing cross-reactive immunity, and frequency of exposure.
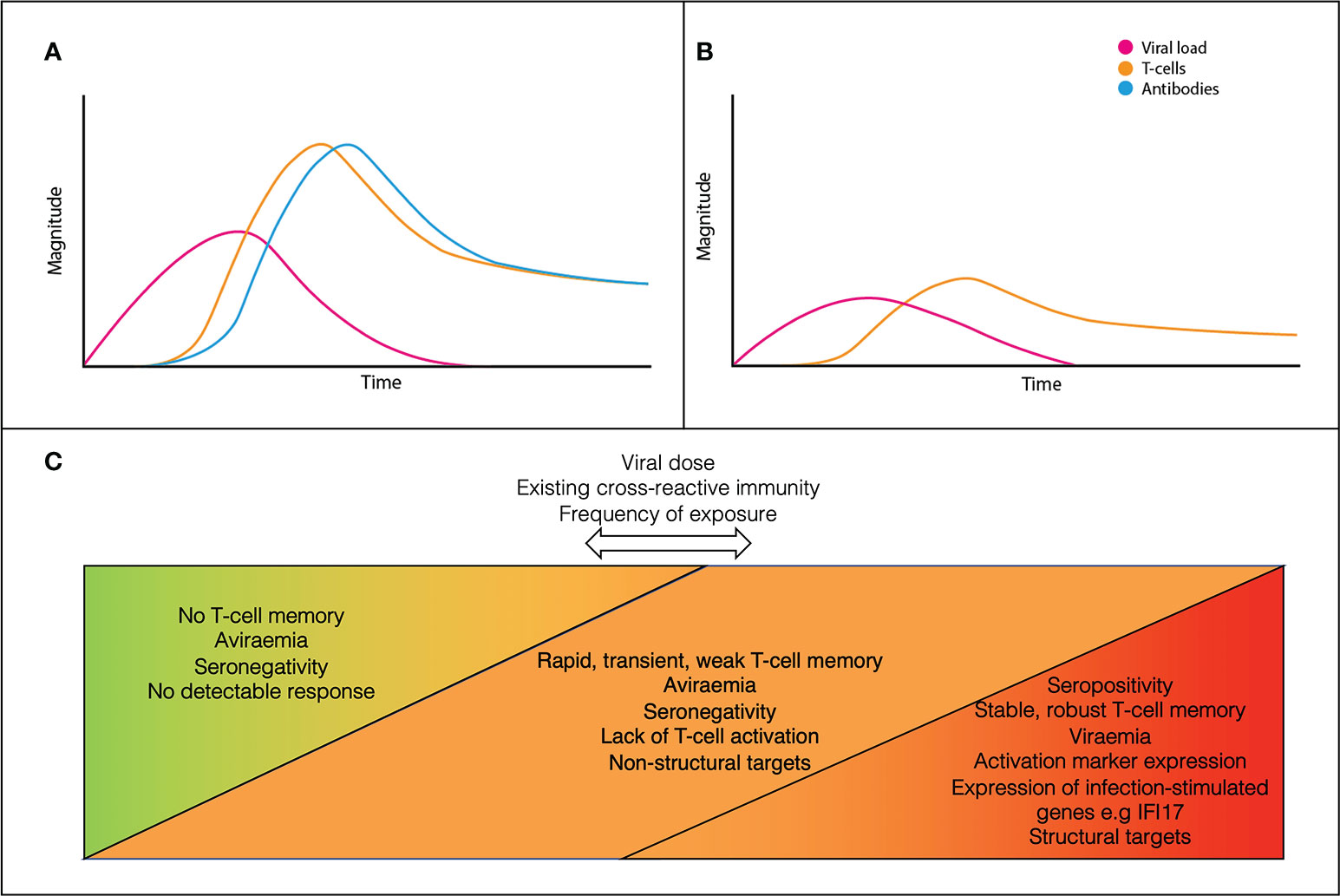
Figure 1 A conceptual model for the dynamics of canonical acute infection (A) vs ESN infection (B). ESNs (orange) represent points along a spectrum of immune engagement, influenced by viral dose, immune cross-reactivity, and frequency of exposure (C).
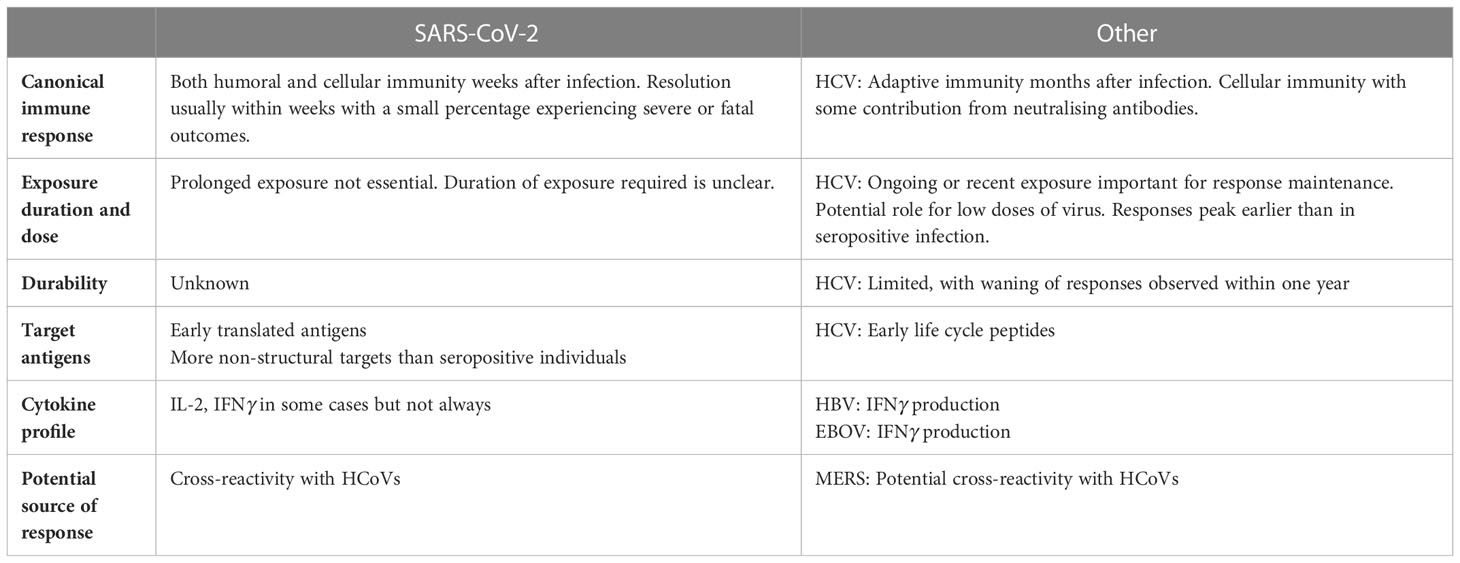
Table 1 displays a summary of findings for SARS-CoV-2 and other viruses. However, significant gaps in our understanding of this phenomenon remain. Specific areas that would provide further clarification include:
1. Repeated exposure and response durability. Frequent exposure appears critical in the durability of HCV-specific cellular responses. SARS-CoV-2 challenge studies in primates and humans would clarify the role of dose and exposure frequency in ESNs, durability of responses, and the extent to which cellular immune responses correlate with protection against infection.
2. Interaction with innate components. Although not covered here, cross-reactive T-cells likely act in coordination with innate immunity to prevent infection. Natural killer cells have been demonstrated to mediate resistance to HCV infection (59–61), and polymorphisms in immune mediator genes such as IL28B likely contribute to disease susceptibility (62). Future research into correlates of protection for SARS-CoV-2 should examine both innate and cellular components in seronegative infection, for example with the use of flow cytometric assays that enable precise dissection of immune components.
3. Other T-cell subsets. Many of the studies outlined in this review use whole blood samples representing circulating immunity. It is critical for future research to consider mucosal and tissue-resident cells to generate a complete picture wherever possible (63). This would require additional sampling such as nasopharyngeal swabs or respiratory samples.
Our understanding of the ESN phenomenon remains in its infancy yet offers opportunities for development. The remarkable heterogeneity in outcome following SARS-CoV-2 exposure makes understanding infection susceptibility crucial for prevention and treatment. Significant insight can be gained into correlates of protection against SARS-CoV-2 by further investigating this phenomenon and gaining a deeper understanding of the role of cellular immunity in protection against infection.
Author contributions
CJ conceived the idea and wrote the manuscript. JR assisted with finalising the manuscript. LT, PG and PK proofread the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Killingley B, Mann A, Kalinova M, Boyers A, Goonawardane N, Zhou J, et al. Safety, tolerability and viral kinetics during SARS-CoV-2 human challenge. Nature Medicine 28, 1031–41 (2022). doi: 10.21203/rs.3.rs-1121993/v1
2. Memoli MJ, Czajkowski L, Reed S, Athota R, Bristol T, Proudfoot K, et al. Validation of the wild-type influenza a human challenge model H1N1pdMIST: An A(H1N1)pdm09 dose-finding investigational new drug study. Clin Infect Dis (2015) 60:693–702. doi: 10.1093/cid/ciu924
3. Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature (1996) 382:722–5. doi: 10.1038/382722a0
4. Kelly JD, Frankfurter RG, Tavs JM, Barrie MB, McGinnis T, Kamara M, et al. Association of lower exposure risk with Paucisymptomatic/Asymptomatic infection, less severe disease, and unrecognized Ebola virus disease: A seroepidemiological study. Open Forum Infect Dis (2022) 9:ofac052. doi: 10.1093/ofid/ofac052
5. Asabe S, Wieland SF, Chattopadhyay PK, Roederer M, Engle RE, Purcell RH, et al. The size of the viral inoculum contributes to the outcome of hepatitis b virus infection. J Virol (2009) 83:9652–62. doi: 10.1128/JVI.00867-09
6. Cobey S, Hensley SE. Immune history and influenza virus susceptibility. Curr Opin Virol (2017) 22:105–11. doi: 10.1016/j.coviro.2016.12.004
7. Swadling L, Diniz MO, Schmidt NM, Amin OE, Chandran A, Shaw E, et al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature (2022) 601:110–7. doi: 10.1038/s41586-021-04186-8
8. Posavad CM, Remington M, Mueller DE, Zhao L, Magaret AS, Wald A, et al. Detailed characterization of T cell responses to herpes simplex virus-2 in immune seronegative persons. J Immunol (2010) 184:3250–9. doi: 10.4049/jimmunol.0900722
9. Miyazawa M, Lopalco L, Mazzotta F, Caputo SL, Veas F, Clerici M, et al. The ‘immunologic advantage’ of HIV-exposed seronegative individuals. AIDS (2009) 23:161–75. doi: 10.1097/QAD.0b013e3283196a80
10. Ranki A, Mattinen S, Yarchoan R, Broder S, Ghrayeb J, Lähdevirta J, et al. T-Cell response towards HIV in infected individuals with and without zidovudine therapy, and in HIV-exposed sexual partners. AIDS (1989) 3:63–70. doi: 10.1097/00002030-198902000-00002
11. Clerici M, Berzofsky JA, Shearer GM, Tacket CO. Exposure to human immunodeficiency virus (HIV) type I indicated by HIV-specific T helper cell responses before detection of infection by polymerase chain reaction and serum antibodies [corrected]. J Infect Dis (1991) 164:178–82. doi: 10.1093/infdis/164.1.178
12. Clerici M, Giorgi JV, Chou CC, Gudeman VK, Zack JA, Gupta P, et al. Cell-mediated immune response to human immunodeficiency virus (HIV) type 1 in seronegative homosexual men with recent sexual exposure to HIV-1. J Infect Dis (1992) 165:1012–9. doi: 10.1093/infdis/165.6.1012
13. Kelker HC, Seidlin M, Vogler M, Valentine FT. Lymphocytes from some long-term seronegative heterosexual partners of HIV-infected individuals proliferate in response to HIV antigens. AIDS Res Hum Retroviruses (1992) 8:1355–9. doi: 10.1089/aid.1992.8.1355
14. Rowland-Jones SL, Nixon DF, Aldhous MC, Gotch F, Ariyoshi K, Hallam N, et al. HIV-Specific cytotoxic T-cell activity in an HIV-exposed but uninfected infant. Lancet (1993) 341:860–1. doi: 10.1016/0140-6736(93)93063-7
15. Koziel MJ, Wong DKH, Dudley D, Houghton M, Walker BD. Hepatitis c virus-specific cytolytic T lymphocyte and T helper cell responses in seronegative persons. J Infect Dis (1997) 176:859–66. doi: 10.1086/516546
16. Scognamiglio P, Accapezzato D, Casciaro MA, Cacciani A, Artini M, Bruno G, et al. Presence of effector CD8+ T cells in hepatitis c virus-exposed healthy seronegative donors. J Immunol (1999) 162:6681–9. doi: 10.4049/jimmunol.162.11.6681
17. Psaros Einberg A, Brenndörfer ED, Frelin L, Hallberg L, Sällberg M, Fischler B. Neonatal exposure to hepatitis c virus antigens in uninfected children born to infected mothers. J Pediatr Gastroenterol Nutr (2018) 66:106–11. doi: 10.1097/MPG.0000000000001755
18. Wiegand J, Meya S, Schlaphoff V, Manns MP, Mössner J, Wedemeyer H, et al. HBV-specific T-cell responses in healthy seronegative sexual partners of patients with chronic HBV infection. J Viral Hepat (2010) 17:631–9. doi: 10.1111/j.1365-2893.2009.01220.x
19. Ogbe A, Kronsteiner B, Skelly DT, Pace M, Brown A, Adland E, et al. T Cell assays differentiate clinical and subclinical SARS-CoV-2 infections from cross-reactive antiviral responses. Nat Commun (2021) 12:2055. doi: 10.1038/s41467-021-21856-3
20. Wang Z, Yang X, Zhong J, Zhou Y, Tang Z, Zhou H, et al. Exposure to SARS-CoV-2 generates T-cell memory in the absence of a detectable viral infection. Nat Commun (2021) 12:1724. doi: 10.1038/s41467-021-22036-z
21. Sette A, Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell (2021) 184:861–80. doi: 10.1016/j.cell.2021.01.007
22. Bancroft GJ. The role of natural killer cells in innate resistance to infection. Curr Opin Immunol (1993) 5:503–10. doi: 10.1016/0952-7915(93)90030-V
23. Warshow UM, Riva A, Hegazy D, Thurairajah PH, Kaminski ER, Chokshi S, et al. Cytokine profiles in high risk injection drug users suggests innate as opposed to adaptive immunity in apparent resistance to hepatitis c virus infection. J Viral Hepat (2012) 19:501–8. doi: 10.1111/j.1365-2893.2011.01574.x
24. Gallais F, Velay A, Nazon C, Wendling M-J, Partisani M, Sibilia J, et al. Intrafamilial exposure to SARS-CoV-2 associated with cellular immune response without seroconversion, France. Emerg Infect Dis (2021) 27, 113–121. doi: 10.3201/eid2701.203611
25. Kamal SM, Amin A, Madwar M, Graham CS, He Q, Al Tawil A, et al. Cellular immune responses in seronegative sexual contacts of acute hepatitis c patients. J Virol (2004) 78:12252–8. doi: 10.1128/JVI.78.22.12252-12258.2004
26. Shoukry NH, Cawthon AG, Walker CM. Cell-mediated immunity and the outcome of hepatitis c virus infection. Annu Rev Microbiol (2004) 58:391–424. doi: 10.1146/annurev.micro.58.030603.123836
27. Cashman SB, Marsden BD, Dustin LB. The humoral immune response to HCV: Understanding is key to vaccine development. Front Immunol (2014) 5:550. doi: 10.3389/fimmu.2014.00550
28. da Silva Antunes R, Pallikkuth S, Williams E, Dawen Yu E, Mateus J, Quiambao L, et al. Differential T-cell reactivity to endemic coronaviruses and SARS-CoV-2 in community and health care workers. J Infect Dis (2021) 224:70–80. doi: 10.1093/infdis/jiab176
29. Kubitschke A, Bahr MJ, Aslan N, Bader C, Tillmann HL, Sarrazin C, et al. Induction of hepatitis c virus (HCV)-specific T cells by needle stick injury in the absence of HCV-viraemia. Eur J Clin Invest (2007) 37:54–64. doi: 10.1111/j.1365-2362.2007.01753.x
30. Heller T, Werner JM, Rahman F, Mizukoshi E, Sobao Y, Gordon AM, et al. Occupational exposure to hepatitis c virus: Early T-cell responses in the absence of seroconversion in a longitudinal cohort study. J Infect Dis (2013) 208:1020–5. doi: 10.1093/infdis/jit270
31. Thimme R, Oldach D, Chang K-M, Steiger C, Ray SC, Chisari FV. Determinants of viral clearance and persistence during acute hepatitis c virus infection. J Exp Med (2001) 194:1395–406. doi: 10.1084/jem.194.10.1395
32. Takaki A, Wiese M, Maertens G, Depla E, Seifert U, Liebetrau A, et al. Cellular immune responses persist and humoral responses decrease two decades after recovery from a single-source outbreak of hepatitis c. Nat Med (2000) 6:578–82. doi: 10.1038/75063
33. Clerici M, Levin JM, Kessler HA, Harris A, Berzofsky JA, Landay AL, et al. HIV-Specific T-helper activity in seronegative health care workers exposed to contaminated blood. JAMA (1994) 271:42–6. doi: 10.1001/jama.1994.03510250058035
34. Thurairajah PH, Hegazy D, Demaine A, Kaminski ER, Cramp ME. Loss of virus-specific T-cell responses in HCV exposed uninfected injection drug users with drug rehabilitation. J Infect Dis (2011) 203:847–53. doi: 10.1093/infdis/jiq121
35. Shata MT, Tricoche N, Perkus M, Tom D, Brotman B, McCormack P, et al. Exposure to low infective doses of HCV induces cellular immune responses without consistently detectable viremia or seroconversion in chimpanzees. Virology (2003) 314:601–16. doi: 10.1016/S0042-6822(03)00461-6
36. Clerici M, Clark EA, Polacino P, Axberg I, Kuller L, Casey NI, et al. T-Cell proliferation to subinfectious SIV correlates with lack of infection after challenge of macaques. AIDS (1994) 8:1391–5. doi: 10.1097/00002030-199410000-00004
37. Wilamowski M, Hammel M, Leite W, Zhang Q, Kim Y, Weiss KL, et al. Transient and stabilized complexes of Nsp7, Nsp8, and Nsp12 in SARS-CoV-2 replication. Biophys J (2021) 120:3152–65. doi: 10.1016/j.bpj.2021.06.006
38. Kundu R, Narean JS, Wang L, Fenn J, Pillay T, Fernandez ND, et al. Cross-reactive memory T cells associate with protection against SARS-CoV-2 infection in COVID-19 contacts. Nat Commun (2022) 13:80. doi: 10.1038/s41467-021-27674-x
39. Brand I, Gilberg L, Bruger J, Garí M, Wieser A, Eser TM, et al. Broad T cell targeting of structural proteins after SARS-CoV-2 infection: High throughput assessment of T cell reactivity using an automated interferon gamma release assay. Front Immunol (2021) 12:688436. doi: 10.3389/fimmu.2021.688436
40. Bozkaya H, Bozdayi M, Türkyilmaz R, Sarioglu M, Cetinkaya H, Cinar K, et al. Circulating IL-2, IL-10 and TNF-alpha in chronic hepatitis b: their relations to HBeAg status and the activity of liver disease. Hepatogastroenterology (2000) 47:1675–9.
41. Thom R, Tipton T, Strecker T, Hall Y, Akoi Bore J, Maes P, et al. Longitudinal antibody and T cell responses in Ebola virus disease survivors and contacts: an observational cohort study. Lancet Infect Dis (2021) 21:507–16. doi: 10.1016/S1473-3099(20)30736-2
42. Chandran A, Rosenheim J, Nageswaran G, Swaddling L, Pollara G, Gupta RK, et al. Rapid synchronous type 1 IFN and virus-specific T cell responses characterize first wave non-severe SARS-CoV-2 infections. Cell Rep Med. (2022) 3(3):100557. doi: 10.1016/j.xcrm.2022.100557
43. Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell (2020) 181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015
44. Mateus J, Grifoni A, Tarke A, Sidney J, Ramirez SI, Dan JM, et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science (2020) 370:89–94. doi: 10.1126/science.abd3871
45. Lee CH, Pinho MP, Buckley PR, Woodhouse IB, Ogg G, Simmons A, et al. Potential CD8+ T Cell Cross-Reactivity Against SARS-CoV-2 Conferred by Other Coronavirus Strains. Front Immunol. (2020) 11:579480. doi: 10.3389/fimmu.2020.579480
46. Lipsitch M, Grad YH, Sette A, Crotty S. Cross-reactive memory T cells and herd immunity to SARS-CoV-2. Nat Rev Immunol (2020) 709–13:1–5. doi: 10.1038/s41577-020-00460-4
47. Low JS, Vaqueirinho D, Mele F, Foglierini M, Jerak J, Perotti M, et al. Clonal analysis of immunodominance and cross-reactivity of the CD4 T cell response to SARS-CoV-2. Science (2021) 372, 1336–1341.
48. Loyal L, Braun J, Henze L, Kruse B, Dingeldey M, Reimer U, et al. Cross-reactive CD4+ T cells enhance SARS-CoV-2 immune responses upon infection and vaccination. Science (2021) 0:eabh1823. Preprint available at SSRN: https://ssrn.com/abstract=3792503 or http://dx.doi.org/10.2139/ssrn.3792503
49. Myklebust C, York J, Persgård Lund K, Grødeland G, Tveita A, Nissen-Meyer L, et al. Preexisting Cross-Reactive T Cells are Boosted and Comprise Significant Immunity in COVID-19 Recovered Patients. doi: 10.2139/ssrn.3792503
50. Tan AT, Linster M, Tan CW, Bert NL, Chia WN, Kunasegaran K, et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep (2021) 34. doi: 10.1016/j.celrep.2021.108728
51. Welsh RM, Selin LK. No one is naive: the significance of heterologous T-cell immunity. Nat Rev Immunol (2002) 2:417–26. doi: 10.1038/nri820
52. Alshukairi AN, Zheng J, Zhao J, Nehdi A, Baharoon SA, Layqah L, et al. High prevalence of MERS-CoV infection in camel workers in Saudi Arabia. mBio (2018) 9:e01985–18. doi: 10.1128/mBio.01985-18
53. Wedemeyer H, Mizukoshi E, Davis AR, Bennink JR, Rehermann B. Cross-reactivity between hepatitis c virus and influenza a virus determinant-specific cytotoxic T cells. J Virol (2001) 75:11392–400. doi: 10.1128/JVI.75.23.11392-11400.2001
54. Buckland MS, Galloway JB, Fhogartaigh CN, Meredith L, Provine NM, Bloor S, et al. Treatment of COVID-19 with remdesivir in the absence of humoral immunity: a case report. Nat Commun (2020) 11:6385. doi: 10.1038/s41467-020-19761-2
55. McMichael AJ, Gotch FM, Noble GR, Beare PAS. Cytotoxic T-cell immunity to influenza. N Engl J Med (1983) 309:13–7. doi: 10.1056/NEJM198307073090103
56. Sridhar S, Begom S, Bermingham A, Hoschler K, Adamson W, Carman W, et al. Cellular immune correlates of protection against symptomatic pandemic influenza. Nat Med (2013) 19:1305–12. doi: 10.1038/nm.3350
57. Wilkinson T, Li C, Chui C, Huang A, Perkins M, Liebner J, et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med (2012) 18:274–80. doi: 10.1038/nm.2612
58. Hemann EA, Kang S-M, Legge KL. Protective CD8 T cell–mediated immunity against influenza a virus infection following influenza virus–like particle vaccination. J Immunol (2013) 191:2486–94. doi: 10.4049/jimmunol.1300954
59. Warshow U, Sieberhagen C, Hegazy D, Kaminski E, Knapp S, Khakoo S, et al. P45 natural killer cell cytotoxicity is enhanced in injection drug users with apparent resistance to hepatitis c virus infection. Gut (2011) 60:A21–2. doi: 10.1136/gutjnl-2011-300857a.45
60. Sugden PB, Cameron B, Mina M, Lloyd AR, HITS investigators. Protection against hepatitis c infection via NK cells in highly-exposed uninfected injecting drug users. J Hepatol (2014) 61:738–45. doi: 10.1016/j.jhep.2014.05.013
61. Ow MM, Hegazy D, Warshow UM, Cramp ME. Enhanced natural killer cell activity is found in exposed uninfected recipients of hepatitis c-contaminated blood. J Viral Hepat (2018) 25:245–53. doi: 10.1111/jvh.12810
62. Knapp S, Warshow U, Ho KMA, Hegazy D, Little A-M, Fowell A, et al. A polymorphism in IL28B distinguishes exposed, uninfected individuals from spontaneous resolvers of HCV infection. Gastroenterology (2011) 141:320–325, 325.e1–2. doi: 10.1053/j.gastro.2011.04.005
Keywords: SARS-CoV-2, seronegative, T-cells, exposed, hepatitis C
Citation: Jay C, Ratcliff J, Turtle L, Goulder P and Klenerman P (2023) Exposed seronegative: Cellular immune responses to SARS-CoV-2 in the absence of seroconversion. Front. Immunol. 14:1092910. doi: 10.3389/fimmu.2023.1092910
Received: 08 November 2022; Accepted: 16 January 2023;
Published: 26 January 2023.
Edited by:
Javier Castillo-Olivares, University of Cambridge, United KigdomReviewed by:
Stephen Rawlings, Maine Health, United StatesCopyright © 2023 Jay, Ratcliff, Turtle, Goulder and Klenerman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cecilia Jay, cecilia.jay@linacre.ox.ac.uk
†Present address: Jeremy Ratcliff, Applied Biological Sciences, Johns Hopkins Applied Physics Laboratory, Laurel, MD
 Cecilia Jay
Cecilia Jay Jeremy Ratcliff1†
Jeremy Ratcliff1† Lance Turtle
Lance Turtle Philip Goulder
Philip Goulder Paul Klenerman
Paul Klenerman