
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 24 January 2023
Sec. Vaccines and Molecular Therapeutics
Volume 14 - 2023 | https://doi.org/10.3389/fimmu.2023.1058327
This article is part of the Research Topic Insights in Vaccines and Molecular Therapeutics: 2022 View all 13 articles
 Fengqing Wang1,2
Fengqing Wang1,2 Qing Zhang3*
Qing Zhang3* Fanqing Zhang1
Fanqing Zhang1 En Zhang1
En Zhang1 Mei Li1
Mei Li1 Shiwei Ma1
Shiwei Ma1 Jianming Guo1
Jianming Guo1 Zhibiao Yang1
Zhibiao Yang1 Jianguo Zhu1*
Jianguo Zhu1*Porcine epidemic diarrhea virus (PEDV) mainly infects the intestinal epithelial cells of pigs, causing porcine epidemic diarrhea (PED). In particular, the virus causes severe diarrhea, dehydration, and death in neonatal piglets. Maternal immunity effectively protects neonatal piglets from PEDV infection; however, maternal antibodies can only prevent PEDV attachment and entry into target cells, but have no effects on intracellular viruses. Intracellular antibodies targeting virus-encoded proteins are effective in preventing viral infection. We previously identified four single chain variable fragments (scFvs), ZW1-16, ZW3-21, ZW1-41, and ZW4-16, which specifically targeted the PEDV N protein and significantly inhibited PEDV replication and up-regulated interferon-λ1 (IFN-λ1) expression in host cells. In our current study, the four scFvs were subcloned into replication-defective adenovirus vectors to generate recombinant adenoviruses rAdV-ZW1-16, rAdV-ZW3-21, rAdV-ZW1-41, and rAdV-ZW4-16. ScFvs were successfully expressed in Human Embryonic Kidney 293 (HEK293) cells and intestinal porcine epithelial cell line J2 (IPEC-J2) and were biosafe for piglets as indicated by body temperature and weight, scFv excretion in feces, IFN-γ and interleukin-4 (IL-4) expression in jejunum, and pathological changes in porcine tissue after oral administration. Western blotting, immunofluorescence, and immunohistochemical analyses showed that scFvs were expressed in porcine jejunum. The prophylactic effects of rAdV-ZW, a cocktail of the four rAdV-scFvs, on piglet diarrhea caused by PEDV was investigated. Clinical symptoms in piglets orally challenged with PEDV, following a two-time treatment with rAdV-ZW, were significantly reduced when compared with PEDV-infected piglets treated with phosphate buffered saline (PBS) or rAdV-wild-type. Also, no death and jejunal lesions were observed. ScFv co-localization with the PEDV N protein in vivo was also observed. Next, the expression of pro-inflammatory serum cytokines such as tumor necrosis factor-α (TNF-α), IL-6, IL-8, IL-12, and IFN-λ was assessed by enzyme-linked immunosorbent assay (ELISA), which showed that scFvs significantly suppressed PEDV-induced pro-inflammatory cytokine expression and restored PEDV-inhibited IFN-λ expression. Therefore, our study supported a promising role for intracellular scFvs targeting the PEDV N protein to prevent and treat diarrhea in PEDV-infected piglets.
Porcine epidemic diarrhea (PED) is caused by porcine epidemic diarrhea virus (PEDV) and is one of the most lethal diseases in piglets; neonatal piglet mortality rates can reach 90%–100% and cause serious economic losses to the global pig breeding industry (1–4). Diseased piglets are clinically characterized by lethargy, loss of appetite, watery diarrhea, dehydration, and even death (1, 2). PED prevention measures have included enhanced biosecurity on pig farms and vaccinations (5, 6). However, more PEDV variant strains have been recently reported, and similarly, when PED outbreaks are caused by variant PEDV strains, traditional vaccines are largely ineffective in controlling infection and spread (7–9).
The neonatal piglet immune system is immature and takes 3 weeks to produce antibodies, therefore, maternal immunity is vital for protecting piglets against PEDV infection (10). Piglets receive maternal antibodies, mainly IgG and secretory IgA, from colostrum and milk of sows immunized with the PEDV vaccine, which provides piglets with passive immune protection against systemic infection and intestinal infection, respectively (10, 11). Although maternal antibodies prevent PEDV attaching to and entering target cells, they are ineffective against viruses inside host cells (12, 13). Therefore, new intracellular small molecules that specifically recognize and bind to PEDV must be generated to effectively prevent intracellular virus replication and control PED.
Single chain variable fragments (scFvs) are small molecule antibodies where the heavy chain variable region and the light chain variable region of the immunoglobulin molecule are linked by a short flexible peptide, commonly comprising glycine and serine residues (14, 15). When compared with monoclonal antibodies, scFvs not only retain intact monoclonal antibody specificity but also have low molecular weights, which provide stronger tumor penetration and higher molecular imaging sensitivity. Therefore, scFvs are ideal tools for tumor molecular imaging, disease therapy, and tracking targeted proteins in vivo (16–18).
PEDV is an enveloped, single-stranded positive-sense RNA virus, which belongs to the Alphacoronavirus genus in the Coronaviridae family (6). With an RNA genome of approximately 28 kb, the virus is composed of seven open reading frames (ORFs), a 5´ untranslated region (5′-UTR), and a 3´-UTR. The ORFs encode four structural proteins, including a spike (S) protein, an envelope (E) protein, a membrane (M) protein, and a nucleocapsid (N) protein, and three non-structural proteins, including replicase 1a, replicase 1b, and an accessory protein (19). Oral scFv administration to neonatal piglets targets the PEDV S protein and forms protection, which helps prevent and control PEDV infection (20). However, the S protein is frequently mutated, which challenges preventive and treatment strategies for PEDV (21, 22). As a structural protein, the N protein is the most abundant viral protein expressed in PEDV-infected cells and forms a ribonucleoprotein complex with the viral genomic RNA (23). During virion assembly, the N-RNA complex is incorporated into the virion by the interaction of N protein with M protein (24). It is highly conserved and plays important roles in PEDV replication (25–27). In addition, it involves regulate interferon (IFN) gene expression in host cells (28). Therefore, intracellular expressed scFvs targeting the PEDV N protein may exert effective inhibitory effects on viruses inside host cells.
Most scFvs bind only to extracellular pathogens, but not pathogens inside cells. Intracellular expressed scFvs that specifically bind to target proteins are important tools for studying protein molecular functions and preventing and treating disease. Adenovirus expression systems are widely used in gene therapy because they do not integrate foreign genes into host cell genomes, and they exhibit effective transduction abilities (29, 30). Adenovirus vectors are used in vaccine development to express antigens (31, 32) and in cancer therapy to express small molecules that target antigens such as scFvs and siRNAs (33–35). Recombinant adenoviruses can be administered intramuscularly, intravenously, intranasally or orally. Therefore, the adenovirus-mediated delivery of antigen-targeting scFvs into the body could provide protective effects and generate resistance to pathogen infection.
We previously reported that four scFvs, ZW1-16, ZW3-21, ZW1-41, and ZW4-16, specifically targeted the PEDV N protein, significantly suppressing PEDV replication, and upregulating IFN-λ1 expression in host cells (36). Herein, to investigate scFv effects on PEDV-induced diarrhea in piglets, four recombinant adenoviruses expressing scFvs against the PEDV N protein were generated. Recombinant adenovirus biosafety was assessed in vivo, and their distribution in piglet jejunums investigated. We also examined the protective effect of a combined scFv application in 3-day-old piglets with diarrhea caused by PEDV infection. Diarrhea severity in the oral recombinant adenovirus rAdV-scFv group was improved and sera IFN-λ expression upregulated when compared with control groups. Overall, adenovirus vector-mediated scFvs targeting of the PEDV N protein demonstrated effective preventive effects toward PEDV-induced diarrhea in piglets, therefore scFv-based therapeutic agents should be investigated to prevent and treat PED.
The HEK293 cell line kindly provided by Prof. Xuanming Yang (Shanghai Jiao Tong University, Shanghai, China) or African green monkey kidney cell line (Vero E6) was cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco, Carlsbad, CA, USA) and 10% fetal bovine serum (FBS; Gibco). The IPEC-J2 was a gift from Prof. Jianxiong Xu (Shanghai Jiao Tong University, Shanghai, China) and cultured in DMEM/F12 and 10% FBS. Cell cultures contained 100 units/mL penicillin and 100 μg/mL streptomycin (Gibco). The PEDV (SH2012-5, GenBank accession number: MG837011.1) was propagated in Vero E6 cells.
ScFvs targeting the PEDV N protein were previously generated and described (36). Briefly, a porcine recombinant scFv library was constructed and four scFvs targeting the PEDV N protein were generated. ScFvs were amplified using upstream primers containing the restriction enzyme SalI (Thermo Fisher Scientific, Waltham, MA, USA) and downstream primers with HindIII (Table 1). PCR thermal cycling conditions were: 95°C for 5 min; 35 cycles of 95°C for 30 s, 62°C for 30 s, and 72°C for 55 s; and 72°C for 5 min. PCR products were purified and cloned into the shuttle vector pAdTrack-CMV (Addgene, Watertown, MA, USA). The linearized recombinant vector pAdTrack-CMV-scFv, digested with the restriction enzyme PmeI, was transformed into BJ5183 competent cells which contained the adenoviral backbone vector pAdEasy-1. Recombinant adenovirus plasmids in BJ5183 cells were selected using streptomycin and kanamycin, and termed pAdEasy-scFv (Addgene). After this, pAdEasy-scFvs were linearized with PacI and transfected into HEK293 cells using TurboFect Transfection Reagent (Thermo Fisher Scientific). Until fluorescence and cytopathic effects appeared, cells were freeze-thawed three times to generate the recombinant adenovirus rAdV-scFv. A recombinant rAdV-wild-type adenovirus was used as a control.
HEK293 or IPEC-J2 cells were seeded in 6-well plates and cultured at 37°C in 5% CO2 until 80% confluence. Cells were then infected with the recombinant adenovirus rAdV-scFv or rAdV-wild-type at a multiplicity of infection of 1 and 100, respectively. At 36 h post-infection, cells were lysed in RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China) and scFv expression analyzed by western blotting. Total proteins were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes (Santa Cruz Biotechnology, Santa Cruz, CA, USA). After blocking in 5% skim milk, membranes were incubated with a primary mouse monoclonal anti-HA antibody (CMCTAG, Milwaukee, WI, USA) overnight at 4°C, and then incubated with a horseradish peroxidase (HRP)-conjugated Affinipure goat anti-mouse IgG (H + L) antibody (Jackson ImmunoResearch, West Grove, PA, USA) at 37°C for 1 h. Protein bands were revealed using Immobilon Western Chemiluminescent HRP Substrate (Merck-Novagen, Darmstadt, Germany).
Twenty-seven 3-day-old piglets without the PEDV vaccine were purchased from a commercial non-PEDV epidemic pig farm (Wujiang Tianyu Biological Technology Co., Ltd, Suzhou, China). Piglets were randomly divided into nine piglets/groups. Group 1 piglets were orally given 2 mL 1010TCID50 (median tissue culture infective dose) recombinant adenovirus rAdV-ZW (equal quantities of rAdV-ZW1-16, rAdV-ZW3-21, rAdV-ZW1-41, and rAdV-ZW4-16). Piglets in groups 2 and 3 were orally given 2 mL 1010TCID50 recombinant adenovirus rAdV-wild-type and 2 mL PBS, respectively. Three piglets from each group were humanely sacrificed on days 2, 14, and 18 after oral administration, respectively. A jejunum section was excised and lysed in RIPA lysis buffer to evaluate scFv expression by western blotting. Additionally, a jejunum section was fixed in paraformaldehyde and paraffin-embedded tissue sections were prepared to analyze scFv expression using indirect immunofluorescence and immunohistochemistry assays (see below).
Nine 3-day-old non-PEDV infected piglets were purchased, randomly divided into three groups, and treated as described. Before oral recombinant adenovirus administration, piglet weights were recorded. Rectum temperatures were also measured and recorded every day after oral administration. Feces were collected at 1 day intervals to measure scFv levels using indirect enzyme-linked immunosorbent assay (ELISA). The following method (37) was used: 1 mL PBS buffer containing 20 mM phenylmethylsulfonyl fluoride (PMSF, Beyotime Biotechnology) was added to 0.5 g feces. After intense vortexing, the mixture was shaken at 200 × g for 1 h at 4°C and centrifuged at 8,000 × g for 10 min to collect the supernatant which was filtered through a 0.45 μm filter and used to coat a 96-well plate. The plate was incubated overnight at 4°C and then blocked in 4% bovine serum albumin (BSA) at 37°C for 2 h. A mouse monoclonal anti-HA antibody was added to each well and incubated at 37°C for 2 h. A secondary HRP-conjugated Affinipure goat anti-mouse IgG (H + L) antibody was added to each well and incubated at 37°C for 1 h. After terminating the reaction, the optical density at 450 nm was measured using a Multiskan™ FC Microplate Photometer (Thermo Fisher Scientific). All piglets were weighed on day 10 after oral administration and percentage weight gains calculated.
Piglets were monitored for 10 days and then humanely sacrificed. Jejunums were collected and scFv expression analyzed by western blotting and indirect immunofluorescence and immunohistochemistry assays. Hearts, livers, spleens, lungs, kidneys, stomachs, small intestines, and large intestines were excised and fixed in paraformaldehyde. Pathological sections were generated and tissue morphology observed using hematoxylin-eosin (H&E) staining. IFN-γ and interleukin-4 (IL-4) quantification in jejunum samples was performed as described below.
Jejunums from piglets orally treated with recombinant adenovirus were collected and frozen in liquid nitrogen on day 10 and then ground in physiological saline solution. After centrifugation at 1,000 × g for 10 min, the supernatant was collected and IFN-γ and IL-4 levels measured using commercial ELISA kits according to manufacturer’s recommendations (Shanghai Enzyme-linked Biotechnology Co., Ltd., Shanghai, China).
Sera were collected on day 3 after PEDV challenge and IL-6, IL-8, IL-12, TNF-α, and IFN-λ levels quantified using commercial ELISA kits (Shanghai Enzyme-linked Biotechnology Co., Ltd.).
An indirect immunofluorescence assay was performed to evaluate scFv infection efficiency and distribution in jejunum samples. Jejunal sections were dewaxed, rehydrated, and subjected to antigen retrieval. After blocking in 3% BSA at room temperature for 30 min, sections were incubated with a primary mouse monoclonal anti-HA antibody overnight at 4°C, followed by incubation with a Cy3-conbjugated Affinipure goat anti-mouse IgG (H + L) secondary antibody at room temperature for 1 h. After cell nuclei were stained with 4´,6-diamidino-2-phenylindole (DAPI), sections were analyzed using a Nikon Eclipse microscope (Nikon, Tokyo, Japan). PEDV N protein expression was detected by staining sections with a rabbit polyclonal anti-PEDV N protein antibody and then a HRP-conjugated Affinipure goat anti-rabbit IgG (H + L) secondary antibody. Sections were observed under a Nikon Eclipse microscope (Nikon) following incubation with fluorescein isothiocyanate (FITC)-tyramide signal amplification.
After dewaxing, rehydrating, and antigen retrieval, endogenous peroxidases were blocked in 3% hydrogen peroxide for 25 min. Jejunal sections were blocked in 3% BSA and probed with a primary mouse monoclonal anti-HA antibody overnight at 4°C. After incubation with a Cy3-conjugated Affinipure goat anti-mouse IgG (H + L) secondary antibody, samples were assessed using a Dako REAL EnVision Detection System (Dako, Glostrup, Denmark) at room temperature for 5 min. Sections were washed in ddH2O to terminate reactions and stained in Harris hematoxylin for 3 min. Then, samples were dehydrated, cleared, mounted, and observed and imaged under a microscope (Nikon).
As shown in Table 2, 15 × 3-day-old piglets were randomly divided into five groups with three piglets/group. Group I and IV piglets were orally treated with 2 mL 1010TCID50 rAdV-ZW. Group II piglets orally received 2 mL 1010TCID50 rAdV-wild-type. Group III and V piglets were treated orally with 2 mL PBS (controls). All piglets were retreated at 2 day intervals. At 6 h after the second oral recombinant adenovirus administration, groups I–III piglets were orally challenged with 1 mL 5×105TCID50 PEDV, while groups IV and V piglets were orally challenged with 1 mL PBS. Piglets were monitored for 7 days, and clinical signs and symptoms, including body weight, body temperature, and diarrhea, were evaluated. Average weight gain and rectum temperatures were recorded every day during studies. Fecal rectal samples were used to assess diarrhea severity. Fecal consistency was evaluated using a scoring scale of 0–3: 0 = solid feces, normal; 1 = pasty feces, slight diarrhea; 2, semi-liquid feces, moderate diarrhea; and 3, liquid feces, severe diarrhea (38, 39).
Fecal samples were collected every day after the PEDV challenge. Viral RNA was extracted using the E.Z.N.A. Stool RNA Kit (Omega Bio-tek, Norcross, GA, USA) and served as a template for complementary DNA (cDNA) synthesis using the PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) (Takara Bio, Shiga, Japan). PEDV M gene expression was analyzed by reverse transcription (RT)-quantitative PCR (qPCR) to quantify virus production in feces (36).
Data were presented as the mean ± standard error of the mean (SEM). Statistical analyses were performed using Origin 8.0 software (OriginLab, Northampton, USA) and GraphPad Prism v8.0 software (GraphPad Software Inc., San Diego, CA, USA). P < 0.05 was considered statistically significant.
Four scFvs, ZW1-16, ZW3-21, ZW1-41, and ZW4-16, targeting the PEDV nucleocapsid (N) protein were generated from a porcine recombinant scFv library (36). Genes of the scFvs were amplified, cloned into a replication-defective Ad5 vector, and packaged into HEK293 cells (Supplementary Figure 1). A green fluorescent protein (GFP) gene incorporated into the adenoviral backbone allows direct observation of the efficiency of viral infection (40). Green fluorescence was observed in cells infected with rAdV-ZW1-16, rAdV-ZW3-21, rAdV-ZW1-41, rAdV-ZW4-16, and rAdV-wild-type (Figure 1A). Next, the expression of four scFvs was analyzed by western blotting. The prominent bands of ZW1-16, ZW3-21, and ZW4-16 were approximately 30 kDa, and that of ZW1-41 was approximately 35 kDa (Figure 1B). The sizes of scFvs were consistent with our previous data (36). To measure scFv expression in porcine cells, IPEC-J2 cells were infected with rAdV-scFvs; four major bands at approximately 30-35 kDa were detected (Figure 1C). Thus, four rAdV-scFvs encoding scFv genes were successfully constructed and expressed in IPEC-J2 cells.
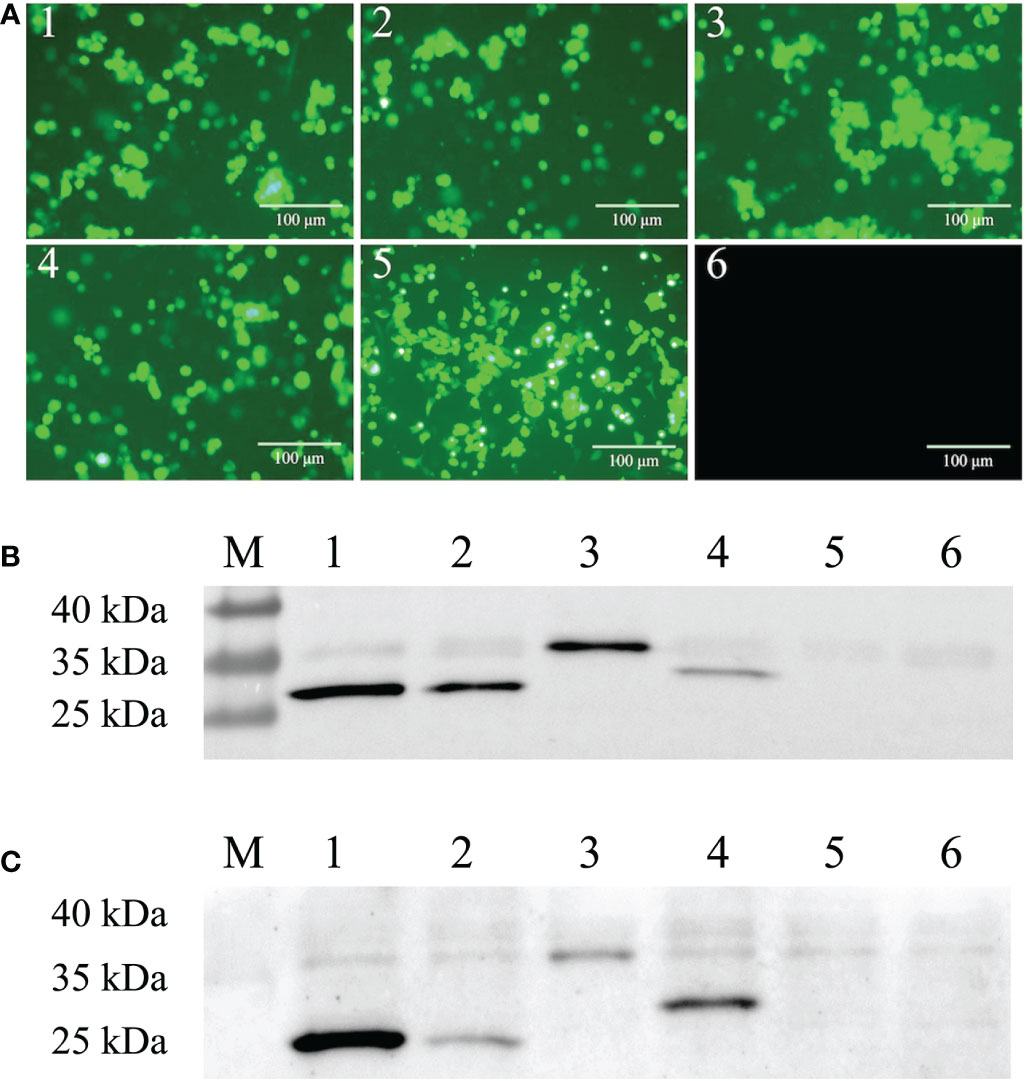
Figure 1 Construction of recombinant adenovirus rAdV-scFvs and expression in cells. (A) Fluorescent images of rAdV-scFv-infected HEK293 cells, rAdV-wild-type infected HEK293 cells, and uninfected HEK293 cells. HEK293 cells were infected with rAdV-ZW1-16, rAdV-ZW3-21, rAdV-ZW1-41, rAdV-ZW4-16, and rAdV-wild-type (1–5) and observed under fluorescence microscopy; uninfected HEK293 cells were used as MOCK controls (6). Scale bar = 100 μm. (B, C) Western blots showing scFv expression in HEK293 (B) and IPEC-J2 cells (C) infected with rAdV-scFvs and rAdV-wild type, respectively. Lane M, protein molecular weight marker; lanes 1–4, whole cell lysates from infected cells expressing rAdV-ZW1-16, rAdV-ZW3-21, rAdV-ZW1-41, and rAdV-ZW4-16, respectively; lane 5, whole cell lysate from infected cells plus rAdV-wild-type; lane 6, uninfected cell lysate.
To explore the biosafety of recombinant adenovirus mediated the scFvs in vivo, 3-day-old piglets were orally administered rAdV-ZW (equal quantities of rAdV-ZW1-16, rAdV-ZW3-21, rAdV-ZW1-41, and rAdV-ZW4-16), rAdV-wild-type, or PBS. During studies, no adverse clinical effects were observed. Rectal temperatures were measured every day and were always approximately 38.5°C (Figure 2A). No significant differences in body temperatures among groups were identified. Piglets were weighed and weight gain rates during studies were calculated. The average percentage weight gain was approximately 40%, with no statistically significant differences (Figure 2C). Fecal samples were collected every day to measure scFv levels using indirect ELISA. Fecal samples from all groups were negative for scFv levels, indicating that adenovirus-mediated scFvs were not environmentally released via defecation (Figure 2B). To assess if rAdV-ZW induced Th1 and Th2 immune responses, IFN-γ and IL-4 expression levels in jejunum samples were analyzed by ELISA. No significant differences in IFN-γ and IL-4 expression were identified among groups (Figures 2D, E). Additionally, histological morphology analyses of heart, liver, spleen, lung, kidney, stomach, small intestine, and large intestine revealed no abnormalities (Figure 3). Collectively, adenovirus-mediated anti-PEDV N scFv expression in the jejunum was biosafe for piglets.
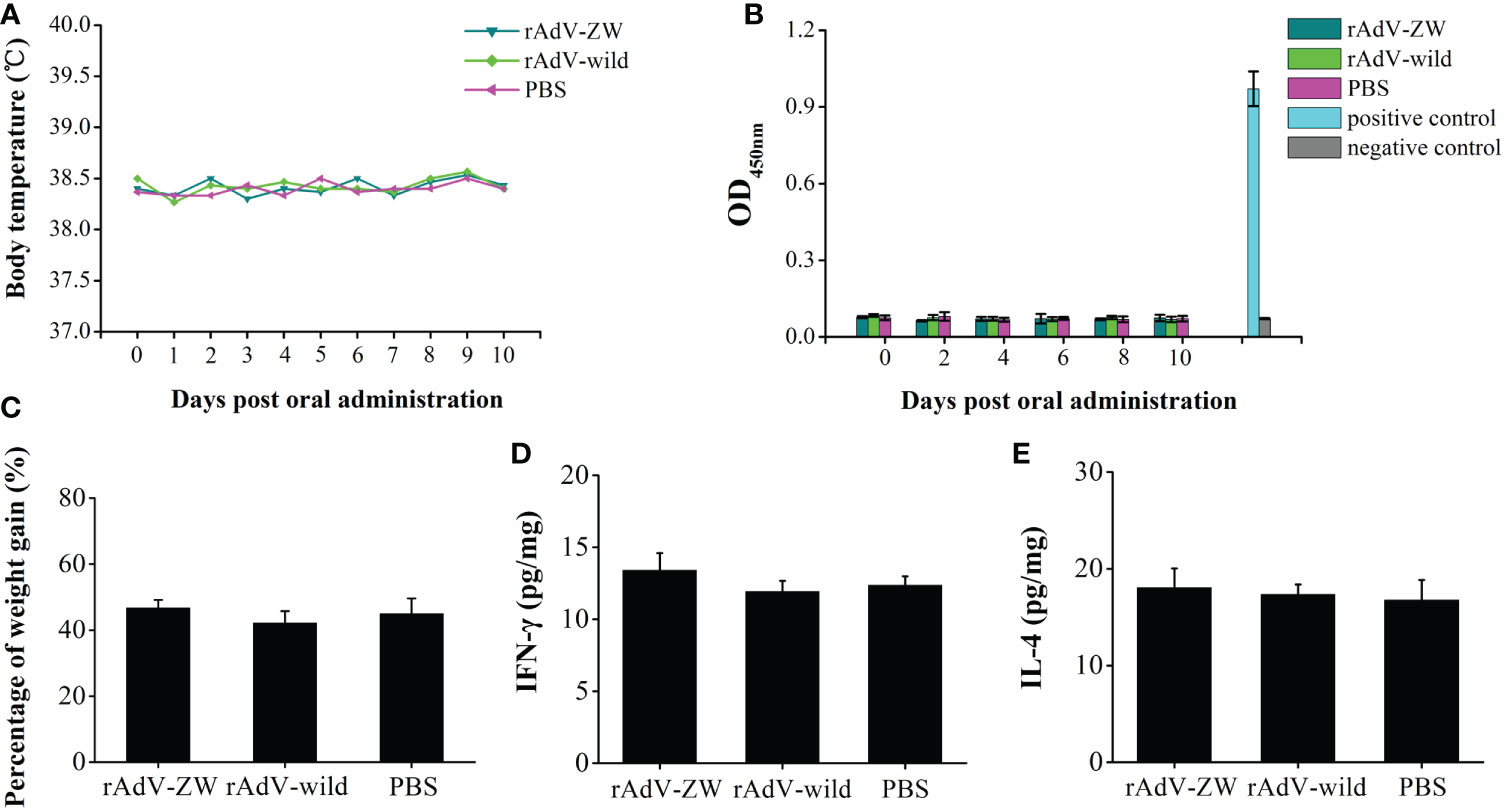
Figure 2 Biosafety rAdV-ZW testing. Three-day-old piglets were orally administrated 2 mL 1010TCID50 rAdV-ZW, 2 mL 1010TCID50 rAdV-wild-type, and 2 mL PBS. (A) The rAdV-ZW effects on rectal temperature. After administration, piglet temperatures were measured daily. (B) Viral shedding determined by ELISA. After administration, feces samples were collected at one day intervals. (C) Piglet percentage weight gain after rAdV-ZW administration. Jejunal (D) IFN-γ and (E) IL-4 expression by ELISA at day 10 after oral administration. Values are presented as the mean ± standard error of the mean (SEM).
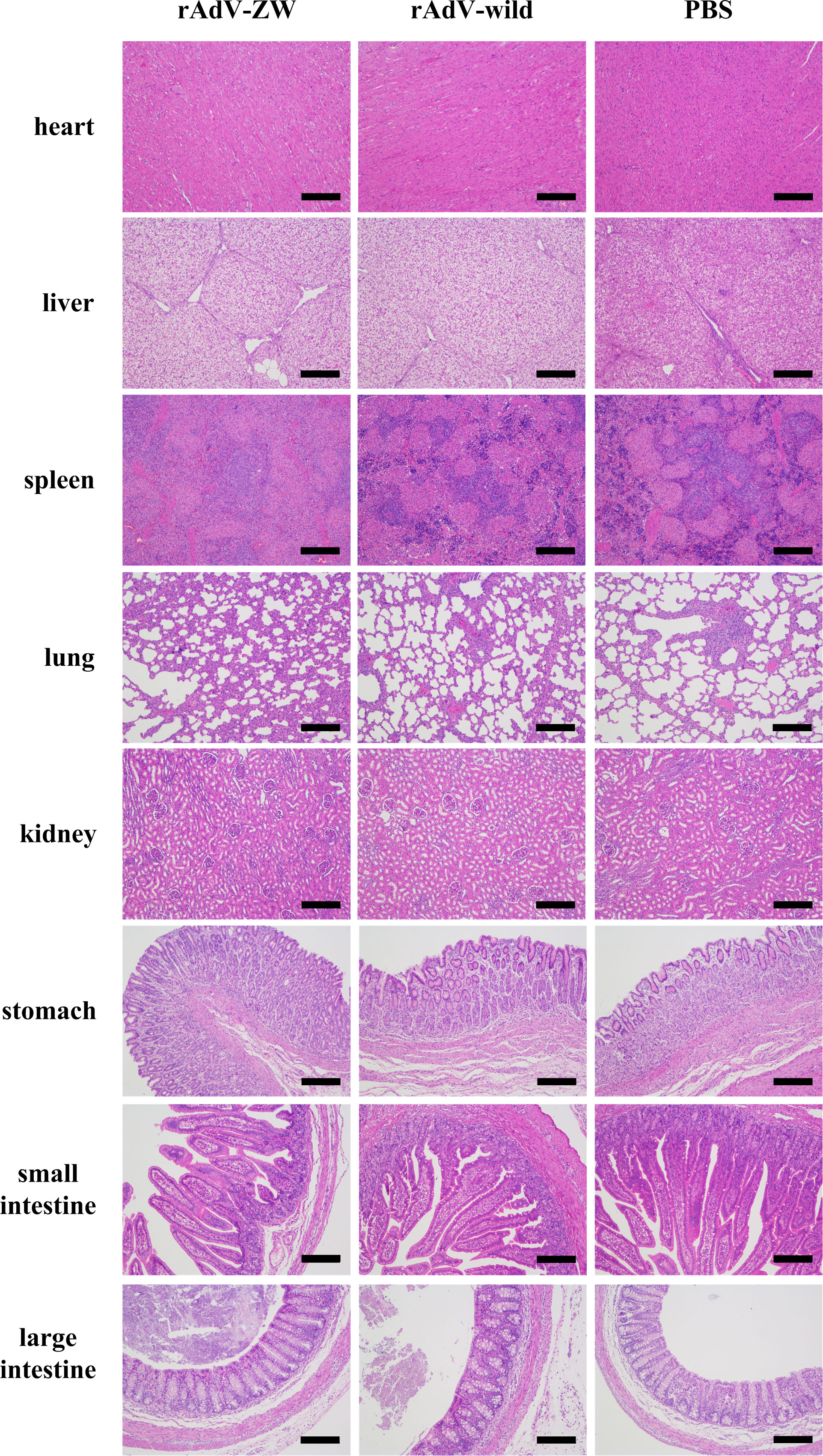
Figure 3 Histopathological analyses of different tissues after orally administrating rAdV-ZW, rAdV-wild-type, or PBS to piglets. Hematoxylin & eosin (H&E) staining of heart, liver, spleen, lung, kidney, stomach, small intestine, and large intestine. Scale bar = 200 μm.
Previous reports revealed that scFv cocktails exhibited better therapeutic effect than single scFvs (41, 42). In our study, a four-scFv cocktail of ZW1-16, ZW3-21, ZW1-41, and ZW4-16 was used. To analyze scFv expression in the jejunum, piglets were orally administered recombinant adenovirus rAdV-ZW or rAdV-wild-type. On days 2, 10, 14, and 18 after oral administration, jejunums were collected and analyzed by western blotting, immunofluorescence, and immunohistochemistry. As shown in Figure 4A, scFvs were successfully expressed in the jejunum of the rAdV-ZW group, with protein sizes consistent with in vitro data (Figures 1B, C). No specific protein bands were identified in orally administered rAdV-wild-type and PBS piglets. Immunofluorescence analysis showed that scFv fluorescence signals were distributed in epithelial cells mainly on jejunum villi surfaces at day 2 after rAdV-ZW oral administration (Figure 4Ba). Notably, scFvs were expressed in cells of the entire jejunum mucosa, including epithelium, underlying lamina propria, muscularis mucosa, and submucosa, at day 10 after oral administration (Figure 4Bb). Over time, the signal of scFv expression gradually weakened (Figures 4Bc, Bd). The expression of scFvs in jejunum could still be detected at day 18 after oral administration (Figure 4Bd). Additionally, immunohistochemical analyses was used to determine scFv expression and distribution in the jejunum of piglets mediate by recombinant adenovirus. ScFv expression on day 10 were higher than on days 2, 14, and 18 in the rAdV-ZW oral administration group (Figure 4C). These immunohistochemical data were consistent with immunofluorescence data. No fluorescence signals were observed in jejunum tissue from rAdV-wild-type and PBS groups. Collectively, western blotting, immunofluorescence, and immunohistochemistry data confirmed that scFvs were successfully expressed in piglet jejunums.
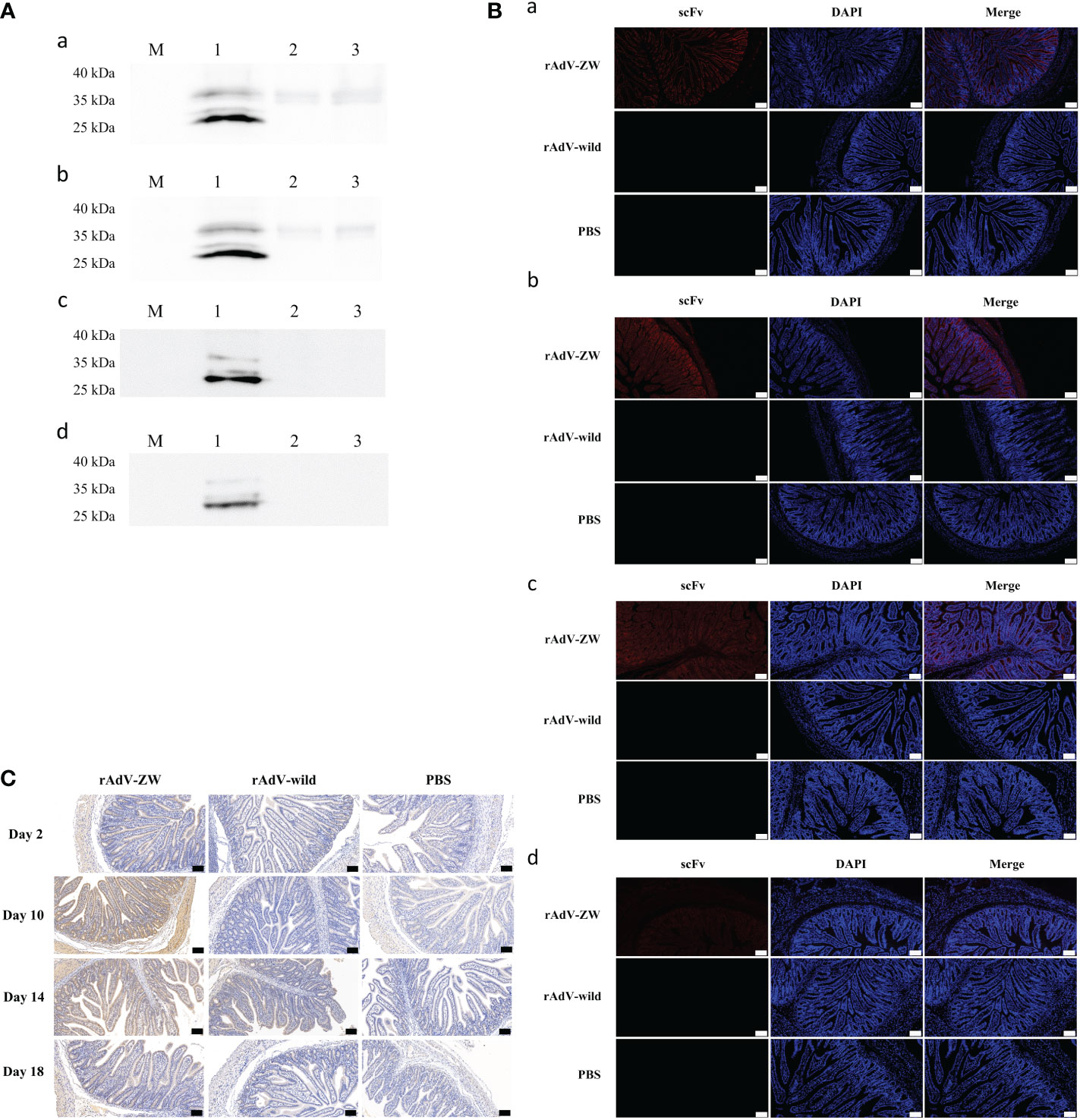
Figure 4 Jejunal expression of four scFvs. After rAdV-ZW, rAdV-wild-type or PBS oral administration, jejunums were collected on days 2 (Aa, Ba), 10 (Ab, Bb), 14 (Ac, Bc), and 18 (Ad, Bd). (A) Western blotting of scFv expression in jejunum tissue showing several bands at approximately 30-35 kDa. Lanes 1–3, jejunum tissue lysate from piglets administrated rAdV-ZW (lane 1), rAdV-wild-type (lane 2), and PBS (lane 3). (B) Immunofluorescence analysis showing scFv expression in jejunum tissue. Jejunum paraffin sections were incubated with a primary mouse monoclonal anti-HA antibody to detect HA-tagged scFvs, and then stained with DAPI to observe nuclei. Red fluorescence represents scFvs and blue represents nuclei. Scale bar = 100 μm. (C) Immunohistochemical jejunum tissue staining to analyze scFv expression. Brown staining indicates scFvs and blue represents hematoxylin-stained nuclei. Scale bar = 100 μm.
To explore scFv protective effects, piglets were orally administered rAdV-ZW and challenged with the prevalent PEDV strain following the indicated scheme (Table 2 and Figure 5A). Clinical symptoms were observed and recorded. Group II and III piglets had slight diarrhea at 1-day post-challenge (dpc). Over time, diarrhea symptoms became more significant. All group II and III piglets developed severe diarrhea at 2 or 3 dpc (Figure 5B). As shown in Figure 5B, group I piglets had slight diarrhea at 2 dpc and moderate diarrhea at 3 or 4 dpc, but no severe diarrhea occurred and piglets had no diarrhea at 7 dpc. Body temperatures in group II and III piglets increased gradually and almost reached 40°C on day 3 after challenge (Figure 5C). Body temperatures in group I piglets increased slightly after challenge, but then gradually returned to normal (Figure 5C). Thus, the degree of PEDV-induced diarrhea of piglets prevented with rAdV-ZW in group I was reduced.
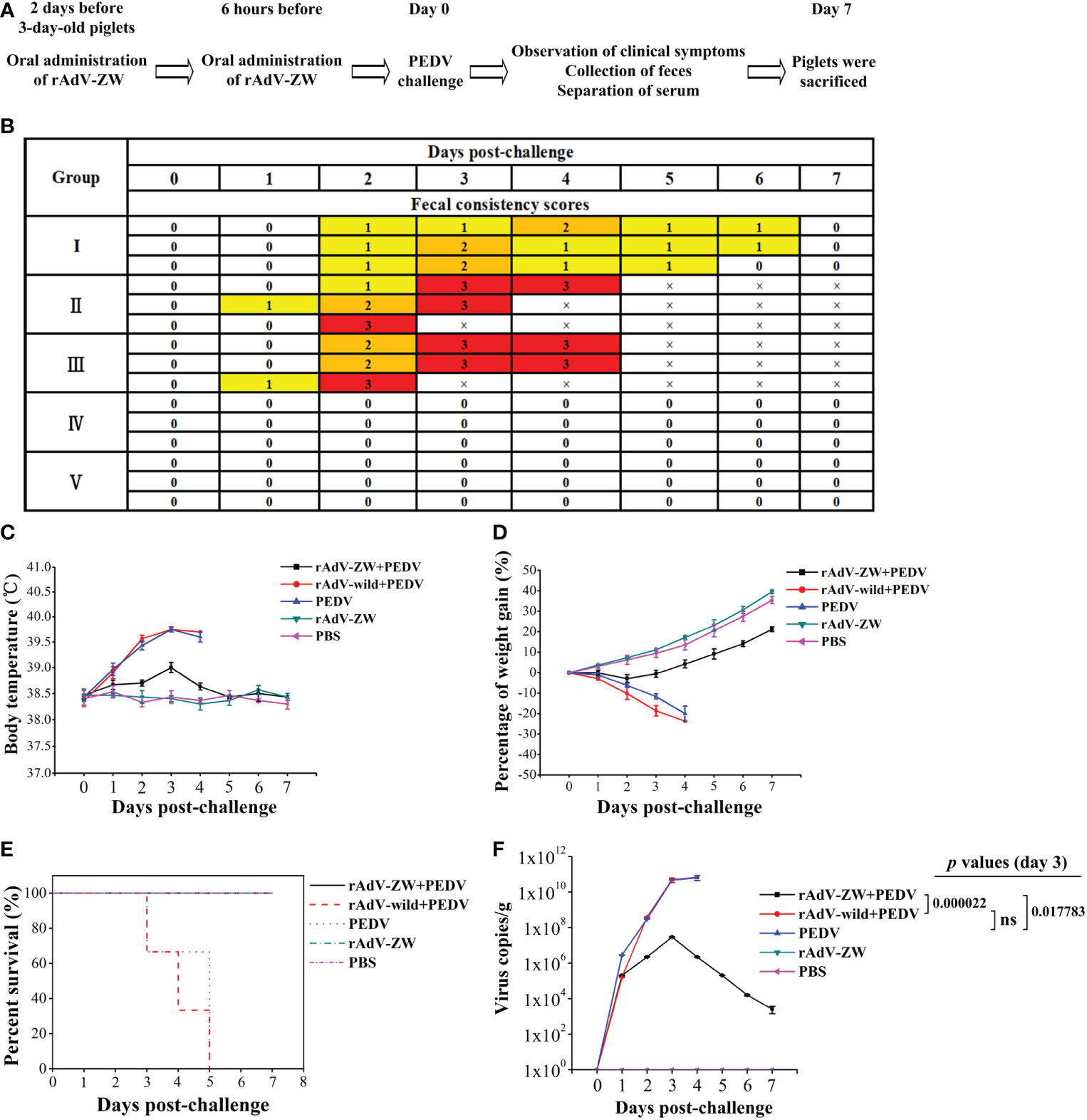
Figure 5 Evaluating the protective effects of scFvs on PEDV-induced diarrhea in piglets. (A) Experimental design showing the oral administration of rAdV-ZW, PEDV challenge, and how we evaluated protective effects in piglets. (B) After PEDV challenge, fecal consistency scores were measured to evaluate the effects of rAdV-ZW on diarrhea severity. Fecal consistency was evaluated using a 0-3 scoring scale: 0 = solid feces; 1 = pasty feces; 2 = semi-liquid feces; and 3 = liquid feces. (C) Effects of rAdV-ZW on rectal temperatures of piglets challenged with PEDV. Rectal temperatures, induced by PEDV in piglets via the oral administration of rAdV-wild-type or PBS, were significantly increased; the highest temperature was approximately 40°C, and rAdV-ZW administration delayed and attenuated increased rectal temperatures induced by PEDV in piglets. (D) Piglet weight gain percentages during studies. Piglets were weighed before PEDV challenge and at 7 dpc, with weight gain percentages shown. PEDV-infected piglets upon oral rAdV-wild-type or PBS administration lost approximately 20% of their body weight. (E) Piglets were observed for 7 days after challenge to assess the effects of rAdV-ZW on survival rates. After PEDV infection, piglets orally administered rAdV-wild-type or PBS died, while those orally administered rAdV-ZW survived. (F) PEDV feces quantification; fecal samples were collected daily and viral RNA extracted to analyze PEDV production by RT-qPCR using specific PEDV M gene primers. Values were presented as the mean ± standard error of the mean (SEM). Ns = not significant (P > 0.05).
Piglets in control groups IV and V had a normal mental state, normal appetites, and gained 35%–40% of their body weight (Figure 5D). After PEDV challenge, group II and III piglets were depressed and their appetite decreased. By the time of death, they had lost approximately 20% of their body weight. The mental state of group I piglets was hardly affected; body weights were almost unchanged in the first 3 days after challenge; they gradually increased from the fourth day and increased by approximately 20% at 7 dpc (Figure 5D). Statistical survival rate analyses showed that all piglets in groups II and III died at 3–5 dpc, whereas survival rate in the other three groups was 100% (Figure 5E). Therefore, scFvs targeting the PEDV N protein alleviated diarrhea in piglets, which was caused by PEDV infection.
Fecal samples were collected every day (Table 2 and Figure 5A) to determine viral shedding by RT-qPCR. As shown in Figure 5F, PEDV virions were detected in feces from groups I–III on the first day after challenge, while viral RNA levels in groups II and III increased sharply, reaching 1010 copies/g at 3 and 4 dpc. Viral RNA levels in group I increased gradually and peaked at 107 copies/g at 3 dpc, but these levels were significantly lower in groups II and III (P < 0.05). Over time, viral RNA levels in group I gradually decreased to 103 copies/g at 7 dpc. No PEDV virions were detected in feces from groups IV and V (Figure 5F). These results showed that scFvs targeting the PEDV N protein significantly inhibited virus replication in PEDV-infected piglets.
In our previous study, we reported that the four scFvs co-localized with the wild-type N protein in PEDV-infected cells (36). To determine scFv and wild-type N protein expression and distribution in vivo, jejunums from piglets orally administered rAdV-ZW and challenged with the prevalent PEDV strain (Table 2 and Figure 5A) were collected, fixed, and paraffinized. Both scFv and PEDV N protein expression were analyzed using indirect immunofluorescence assays (Figure 6); scFvs were highly expressed in the jejunum in groups I and IV, while no fluorescence (red) was observed in groups II, III, and V. Interestingly, PEDV was evenly distributed in the whole small intestine in group II and III piglets. However, when piglets were orally administrated rAdV-ZW in advance and then challenged with PEDV, PEDV was mainly distributed on jejunum villi surfaces, and scFvs were distinctly co-localized with PEDV N proteins in the jejunum. Together, these data suggested that PEDV infected epithelial cells on jejunum villi surfaces after viral challenge, and scFvs expressed in jejunum epithelial cells specifically bound PEDV N proteins, resulting in inhibited PEDV replication and decreased intact PEDV virions, with PEDV unable to infect more cells.
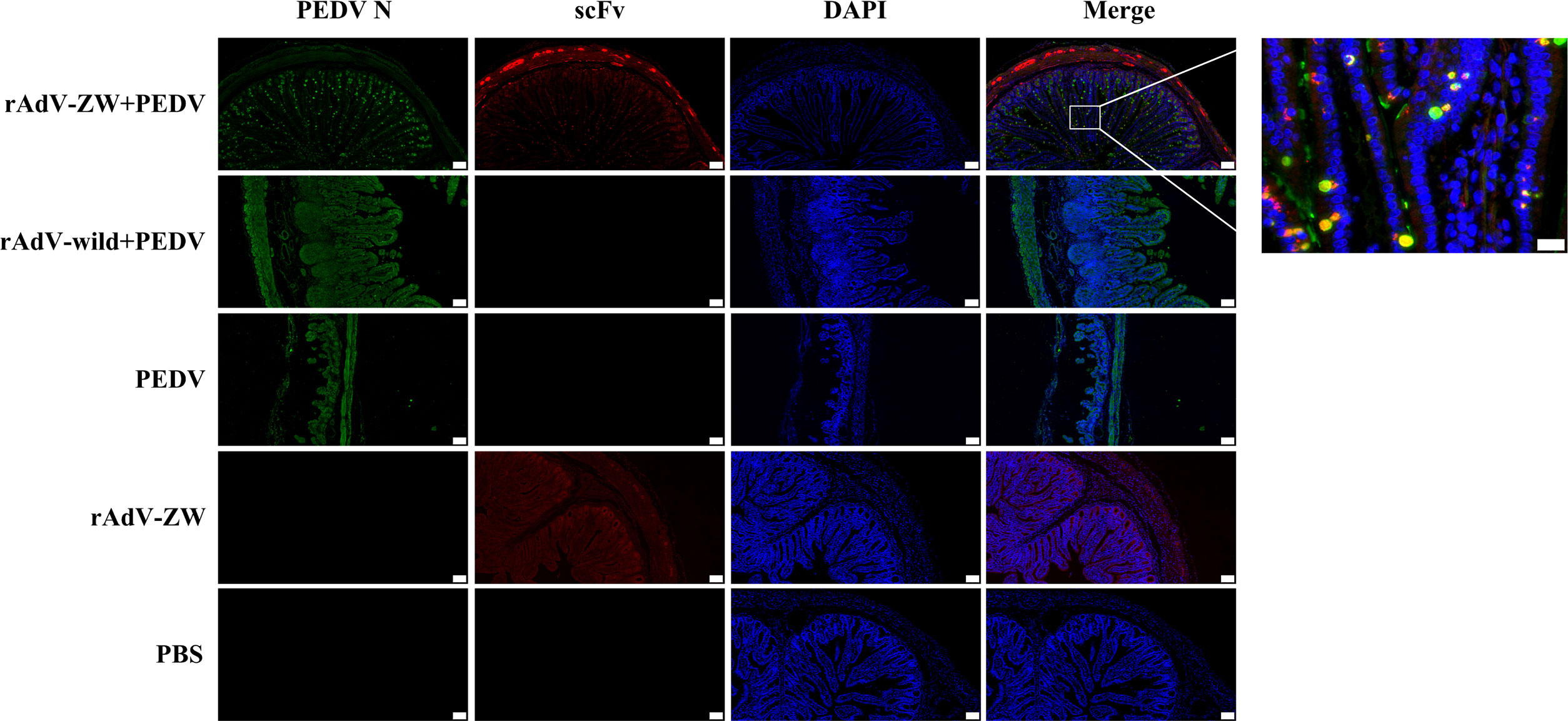
Figure 6 ScFv and PEDV N protein expression in jejunum by immunofluorescence. Jejunum samples were collected from piglets orally administrated rAdV-ZW after challenge with PEDV. Paraffin sections were stained with FITC, CY3, and DAPI and observed at 100× magnification (left). Scale bar = 100 μm. (Right) (630×) Enlarged framed area in (left). Scale bar = 20 μm. Green represents the PEDV N protein; red represents scFvs, and blue indicates DAPI-stained nuclei. In merged images, orange indicates ZW co-localization (including ZW1-16, ZW3-21, ZW1-41, and ZW4-16) with the PEDV N protein.
Histological analyses showed that jejunum villi were considerably atrophied, with thinning intestinal walls, mesenteric hyperemia, necrosis, and intestinal villus epithelial cell shedding, with intestinal mucosa structures severely damaged in group II and III piglets (Figure 7A). The jejunum villi were mildly atrophied, and the intestinal mucosa structure was slightly damaged of piglets prevented by the scFvs against the PEDV N protein before infection with PEDV in group I. Jejunum morphology in control groups IV and V was normal. These pathological observations showed that jejunum damage caused by PEDV infection was significantly alleviated when scFvs directed against the PEDV N protein were expressed in the jejunum.
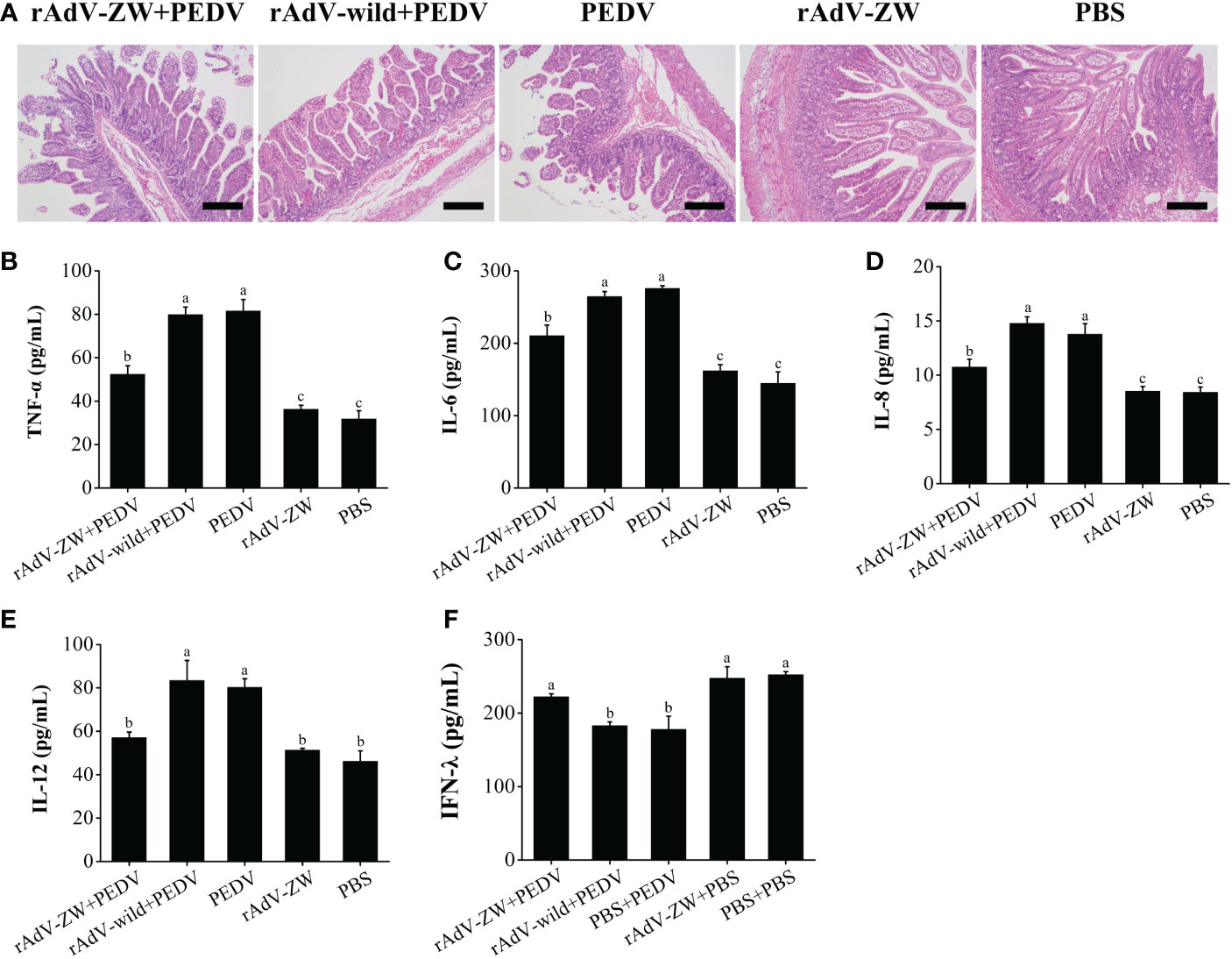
Figure 7 Histomorphological observations in jejunum and cytokine production levels in piglets. (A) Jejunum tissue underwent H&E staining to assess the effects of rAdV-ZW on jejunum pathological changes in piglets infected with PEDV. H&E staining showed that rAdV-ZW reduced jejunal damage severity. Scale bar = 200 μm. (B–E) Pro-inflammatory sera cytokine expression in piglets. At 3 dpc, TNF-α, IL-6, IL-8 and IL-12 serum levels were assayed using indirect ELISA. (B) TNF-α expression, (C) IL-6 expression, (D) IL-8 expression, (E) IL-12 expression, and (F) IFN-λ expression profiles in piglet sera at 3 dpc. Values were presented as the mean ± standard error of the mean (SEM). Values indicated by different letters are significantly different (P < 0.05). No significant differences between values are indicated by identical letters.
Serum pro-inflammatory cytokine levels, including TNF-α, IL-6, IL-8, and IL-12 were evaluated by ELISA (Figures 7B–E). Serum TNF-α, IL-6, IL-8, and IL-12 levels in group II and III piglets infected were significantly increased when compared with control groups IV and V (P < 0.05). TNF-α, IL-6, and IL-8 serum expression levels in PEDV-infected group I piglets treated with rAdV-ZW were significantly lower than in groups II and III (P < 0.05), although levels were significantly higher than in control groups IV and V (P < 0.05). Serum IL-12 levels in PEDV-infected group I piglets treated with rAdV-ZW were significantly decreased when compared with groups II and III (P < 0.05), with no significant differences between groups I, IV, and V (P > 0.05).
We previous showed that IFN-λ1 expression levels were significantly up-regulated after the PEDV N protein was blocked by the four scFvs in PEDV-infected IPEC-J2 cells (36). To examine if IFN-λ expression level in piglets infected with PEDV changed after scFvs were delivered to the jejunum, serum was collected and processed using indirect ELISA. As shown in Figure 7F, IFN-λ serum expression in PEDV-infected group II and III piglets was significantly down-regulated when compared with control groups IV and V (P < 0.05), whereas expression was up-regulated in PEDV-infected group I piglets treated with rAdV-ZW (P < 0.05). Thus, the four scFvs eliminated the antagonistic effects of PEDV infection toward IFN-λ expression by blocking PEDV N protein expression in vivo.
In recent years, vaccination has become a most effective approach controlling PEDV; traditional strategies have mainly included PEDV inactivated vaccines, live attenuated vaccines, and genetically engineered vaccines (43, 44). However, because inactivated vaccines cannot be replicated, pigs must be immunized multiple times with large doses. Also, attenuated live vaccines may revert to wild-type PEDV strains, and most genetically engineered vaccines currently developed use the PEDV S protein as antigen, however, the mutational probability of the S gene in variant strains is very high, resulting in the prevention and control effect of PED caused by PEDV variant strain is not ideal (8, 22, 45). Therefore, novel effective strategies are required against PEDV.
Some scFvs were previously evaluated for their efficacy in detecting, preventing, and treating disease (18, 46, 47). The scFvs targeting the PEDV S protein were effective in controlling piglet diarrhea (20). The PEDV N protein is a structural protein and highly conserved when compared with the S protein. It has key functions in the viral life cycle and is involved in interactions between the virus and host cells. Coronavirus N protein contributes to viral RNA synthesis by interacting with nonstructural protein 3 (nsp3), a component of the replication-transcription complex (48). It was shown that the PEDV N protein promoted viral replication (25) and different regulatory factors in host cells could suppress viral replication by degrading the N protein (49, 50). Because the protein has multiple functions, specific protein targeting by scFvs is an ideal way to inhibit viral infection. We constructed a recombinant scFv library from the spleens of pigs immunized with purified recombinant PEDV N protein, and identified four scFvs, ZW1-16, ZW3-21, ZW1-41, and ZW4-16, which specifically bound to the PEDV N protein (36). ScFvs co-localized with the wild-type PEDV N protein, significantly suppressed PEDV replication, and observably upregulated IFN-λ1 expression in PEDV-infected cells (36).
Adenovirus is a double-stranded DNA virus with an icosahedral capsid and no envelope. Adenovirus vectors are effective for in vivo gene delivery systems (51, 52); human adenovirus type 5 is the most commonly used vector for vaccine preparations and gene therapy (40, 53). In our study, the adenoviral plasmid pAdEasy-1 was replication defective and contained most of the human adenovirus type 5 genome, except for E1 and E3 genes, and was used as the backbone to generate the recombinant adenovirus rAdV-scFv. Four recombinant adenoviruses were successfully packaged into HEK293 cells and scFvs successfully expressed in rAdV-scFv-infected IPEC-J2 cells.
Previous studies have shown that a cocktail of several scFvs targeting different molecular epitopes could enhance sensitivity of scFv-based disease detection and improve disease prevention potency when compared with individual scFvs (54, 55). Therefore, we combined four scFvs against the PEDV N protein to explore their efficacy in preventing PEDV infection. We evaluated rAdV-scFvs biosafety by measuring piglet body temperatures, body weights, scFv levels in feces, IFN-γ and IL-4 expression in the jejunum, and pathological changes in eight tissues after oral administration. No significant differences were observed in body temperatures, weight gain rates, and IFN-γ and IL-4 expression levels in jejunum when compared with control piglets. No scFvs were detected in feces and no lesions were identified in tissue sections of rAdV-ZW-treated piglets.
Recombinant adenoviruses deliver genes into the body in several ways. Orally administered adenovirus vaccines may induce mucosal immune responses in animals (56, 57). Because PEDV mainly infects porcine small-intestinal epithelial cells, in our study, rAdV-scFvs directly delivered four scFvs to piglet intestinal tissues via oral administration. We investigated scFv distribution in the jejunum using western blotting, immunofluorescence, and immunohistochemical analysis at days 2 and 10 after oral administration - expression of the four scFvs was detected at both time points. Immunohistofluorescence and immunohistochemical analyses showed scFvs were mainly expressed in cells on jejunum villi surfaces on day 2 and distributed to the whole jejunum by day 10.
After confirming scFvs were expressed in porcine jejunum tissue and biosafe for piglets, we evaluated the prophylactic effects of rAdV-ZW on diarrhea caused by PEDV. Immunofluorescence analyses showed that scFvs were co-localized with the PEDV N protein in jejunum epithelial cells in piglets challenged with PEDV, consistent with vero E6 cells in a previous study (36). Surprisingly, after challenge, PEDV was almost uniformly distributed in all jejunum cells in piglets administered rAdV-wild or PBS, whereas, PEDV was mainly detected in jejunum villi cells surfaces in piglets administered rAdV-ZW. We speculated that scFvs expressed in epithelial cells bound to the N protein of intracellular PEDV, inhibiting PEDV replication, not forming complete virions in assembly processes, and not generating enough viruses to infect other cells. Consequently, epithelial cells infected by PEDV were mainly present on villi surfaces.
After PEDV challenge, piglets administrated rAdV-wild or PBS showed elevated body temperatures and severe diarrhea, and became thinner during studies, while piglets treated with rAdV-ZW showed reduced clinical symptoms with a slight increase in body temperature, moderate diarrhea, and a body weight gain of approximately 20%. Thus, scFvs already expressed in intestinal epithelial cells provided effective protection against viral infection. PEDV infection in piglets treated with rAdV-wild or PBS induced the severe destruction of jejunal villi and 100% mortality in both groups. In contrast, jejunal villi in piglets orally administrated rAdV-ZW were slightly damaged, and all piglets survived. Therefore, a four-scFv cocktail prevented PEDV infection in piglets.
The PEDV N protein promotes viral replication and accelerates higher titer viral growth (25, 26). Several studies reported that PEDV replication was suppressed when the N protein was degraded (49, 58, 59). Our scFvs targeting the PEDV N protein significantly suppressed PEDV replication in PEDV-infected cells, as described in our previous study (36). Previous studies indicated that PEDV infection in piglets caused fecal viral RNA shedding (39, 60). In our study, fecal samples were analyzed to assess PEDV RNA shedding titers; no PEDV was detected in feces from control animals administered rAdV-ZW or PBS and not exposed to PEDV. Viral RNA excretion occurred in the other groups after PEDV infection; the virus shedding titer in the rAdV-ZW group was significantly lower than in rAdV-wild-type and PBS groups at 3 dpc (P < 0.05). Therefore, scFvs inhibited PEDV proliferation in vivo by blocking the PEDV N protein.
Viral infection elicits host immune responses, leading to changes in cytokine expression levels. Expression of the pro-inflammatory cytokines TNF-α, IL-6, IL-8, and IL-12 was up-regulated in PEDV-infected IPEC-J2 cells (61, 62), while PEDV infection increased TNF-α, IL-6, and IL-12 serum expression levels in piglets (63). TNF-α, IL-6, IL-8, and IL-12 serum expression levels in piglets on day 3 after PEDV challenge were significantly increased when compared with control IV and V groups (P < 0.05). After challenge, serum cytokine expression in piglets treated with rAdV-ZW was distinctly decreased when compared with group III piglets (P < 0.05), while rAdV-wild-type had no obvious effects on expression. Therefore, the N protein had important roles during proinflammatory cytokine production induced by PEDV, while scFvs effectively antagonized cytokine production.
PEDV mainly infects porcine small intestinal epithelial cells and proliferates in cells leading to atrophy and small intestinal villi shedding. Type III IFN has important roles in mucosal immunity which exhibits potent antiviral activity (64). Porcine IFN-λ1 and IFN-λ3 inhibit PEDV infection in IPEC-J2 cells (65, 66). To generate a cellular environment conducive to viral growth, PEDV has evolved several mechanisms to counteract or evade antiviral effects. Among PEDV proteins, 11, including nsp1, nsp3, nsp5, nsp8, nsp14, nsp15, nsp16, ORF3, E, M, and N suppressed IFN-λ1 promoter activities (67). Recent studies reported that the PEDV N protein antagonized IFN-λ3 production by blocking nuclear translocation of nuclear factor-κB (68). In our previous study, IFN-λ1 expression was up-regulated in PEDV-infected IPEC-J2 cells expressing all four scFvs against the PEDV N protein (36). As shown in Supplementary Figure 2, IFN-λ1 promoter activity was increased by intracellular scFv expression in a concentration-dependent manner. We also analyzed type III IFN serum expression levels; IFN-λ expression was dramatically downregulated after PEDV infection (P < 0.05). When piglets were administered rAdV-ZW before PEDV infection, IFN-λ expression was significantly upregulated (P < 0.05). These results were consistent with our previous study which showed the effects of all four scFvs against the PEDV N protein on type III IFN production in IPEC-J2 cells. Therefore, we hypothesize that our four scFvs expressed in the jejunum antagonized the negative regulatory effects of the PEDV N protein toward IFN-λ production by blocking the N protein in vivo.
In summary, four scFvs specifically targeting the PEDV N protein were cloned into adenovirus vectors and successfully expressed in vitro and in vivo. Recombinant adenoviruses were biosafe for piglets, and adenovirus vector-mediated scFvs effectively expressed in jejunum tissue were co-localized with the PEDV N protein in jejunal epithelial cells in PEDV-infected piglets. Further animal studies showed that a rAdV-scFv cocktail generated effective protective effects against PEDV infection. After scFv administration, IFN-λ serum expression levels in infected piglets were up-regulated, consistent with our previous study and providing new mechanistic insights on how the PEDV N protein antagonizes type III IFN expression. Our results provide encouraging antiviral potential of anti-N scFvs and a basis for developing scFv-based drugs to control PEDV-induced diarrhea in piglets. Additionally, the optimal level of recombinant adenovirus and the optimal time of PEDV challenge after oral administration of recombinant adenovirus are necessary to investigate to achieve optimal protection for piglets and the investigation is under the way.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by Animal Ethics Committee of Shanghai Jiao Tong University, School of Agriculture and Biology.
FW, QZ, FZ, and JZ designed the research. JZ, QZ and ZY applied for funding. FW, EZ, ML, SM, and JG performed experiments. FW and QZ performed data analysis and manuscript writing. JZ supervised the project. All authors contributed to the article and approved the submitted version.
The authors acknowledge the Key Project of Science and Technology for Agriculture of Shanghai (Hu Nong Ke Gong Zi (2015) No. 1–8) (To Jianguo Zhu), Natural Science Foundation of Shanghai (21ZR1433900), Natural Science Foundation of China (31472211), and China Postdoctoral Science Foundation (.2020TQ0197) for supporting this work.
We thank the School of Agriculture and Biology, Shanghai Jiao Tong University for assistance with animal experiments.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2023.1058327/full#supplementary-material
1. Stevenson GW, Hoang H, Schwartz KJ, Burrough ER, Sun D, Madson D, et al. Emergence of porcine epidemic diarrhea virus in the united states: Clinical signs, lesions, and viral genomic sequences. J Vet Diagn Invest (2013) 25:649–54. doi: 10.1177/1040638713501675
2. Pensaert MB, de Bouck P. A new coronavirus-like particle associated with diarrhea in swine. Arch Virol (1978) 58:243–7. doi: 10.1007/BF01317606
3. Reveles-Felix S, Carreon-Napoles R, Mendoza-Elvira S, Quintero-Ramirez V, Garcia-Sanchez J, Martinez-Bautista R, et al. Emerging strains of porcine epidemic diarrhoea virus (PEDV) in Mexico. Transbound Emerg Dis (2020) 67:1035–41. doi: 10.1111/tbed.13426
4. He WT, Bollen N, Xu Y, Zhao J, Dellicour S, Yan Z, et al. Phylogeography reveals association between swine trade and the spread of porcine epidemic diarrhea virus in China and across the world. Mol Biol Evol (2022) 39:msab364. doi: 10.1093/molbev/msab364
5. Stewart SC, Dritz SS, Woodworth JC, Paulk C, Jones CK. A review of strategies to impact swine feed biosecurity. Anim Health Res Rev (2020) 21:61–8. doi: 10.1017/S146625231900015X
6. Jung K, Saif LJ, Wang Q. Porcine epidemic diarrhea virus (PEDV): An update on etiology, transmission, pathogenesis, and prevention and control. Virus Res (2020) 286:198045. doi: 10.1016/j.virusres.2020.198045
7. Gao Q, Zheng Z, Wang H, Yi S, Zhang G, Gong L. The new porcine epidemic diarrhea virus outbreak may mean that existing commercial vaccines are not enough to fully protect against the epidemic strains. Front Vet Sci (2021) 8:697839. doi: 10.3389/fvets.2021.697839
8. Li Z, Ma Z, Li Y, Gao S, Xiao S. Porcine epidemic diarrhea virus: Molecular mechanisms of attenuation and vaccines. Microb Pathog (2020) 149:104553. doi: 10.1016/j.micpath.2020.104553
9. Crawford K, Lager KM, Kulshreshtha V, Miller LC, Faaberg KS. Status of vaccines for porcine epidemic diarrhea virus in the united states and Canada. Virus Res (2016) 226:108–16. doi: 10.1016/j.virusres.2016.08.005
10. Langel SN, Paim FC, Lager KM, Vlasova AN, Saif LJ. Lactogenic immunity and vaccines for porcine epidemic diarrhea virus (PEDV): Historical and current concepts. Virus Res (2016) 226:93–107. doi: 10.1016/j.virusres.2016.05.016
11. Leidenberger S, Schroder C, Zani L, Auste A, Pinette M, Ambagala A, et al. Virulence of current German PEDV strains in suckling pigs and investigation of protective effects of maternally derived antibodies. Sci Rep (2017) 7:10825. doi: 10.1038/s41598-017-11160-w
12. Li C, Li W, Lucio de Esesarte E, Guo H, van den Elzen P, Aarts E, et al. Cell attachment domains of the porcine epidemic diarrhea virus spike protein are key targets of neutralizing antibodies. J Virol (2017) 91:e00273–17. doi: 10.1128/JVI.00273-17
13. Wen Z, Xu Z, Zhou Q, Li W, Wu Y, Du Y, et al. A heterologous 'Prime-boost' anti-PEDV immunization for pregnant sows protects neonatal piglets through lactogenic immunity against PEDV. Lett Appl Microbiol (2019) 69:258–63. doi: 10.1111/lam.13197
14. Hoedemaeker FJ, Signorelli T, Johns K, Kuntz DA, Rose DR. A single chain fv fragment of p-Glycoprotein-Specific monoclonal antibody C219. design, expression, and crystal structure at 2.4 a resolution. J Biol Chem (1997) 272:29784–9. doi: 10.1074/jbc.272.47.29784
15. Lilley GG, Dolezal O, Hillyard CJ, Bernard C, Hudson PJ. Recombinant single-chain antibody peptide conjugates expressed in escherichia coli for the rapid diagnosis of HIV. J Immunol Methods (1994) 171:211–26. doi: 10.1016/0022-1759(94)90041-8
16. Duranti C, Carraresi L, Sette A, Stefanini M, Lottini T, Crescioli S, et al. Generation and characterization of novel recombinant anti-hERG1 scFv antibodies for cancer molecular imaging. Oncotarget (2018) 9:34972–89. doi: 10.18632/oncotarget.26200
17. Islam A, Pishesha N, Harmand TJ, Heston H, Woodham AW, Cheloha RW, et al. Converting an anti-mouse CD4 monoclonal antibody into an scFv positron emission tomography imaging agent for longitudinal monitoring of CD4(+) T cells. J Immunol (2021) 207:1468–77. doi: 10.4049/jimmunol.2100274
18. Rafiq S, Yeku OO, Jackson HJ, Purdon TJ, van Leeuwen DG, Drakes DJ, et al. Targeted delivery of a PD-1-Blocking scFv by CAR-T cells enhances anti-tumor efficacy in vivo. Nat Biotechnol (2018) 36:847–56. doi: 10.1038/nbt.4195
19. Song D, Park B. Porcine epidemic diarrhoea virus: A comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes (2012) 44:167–75. doi: 10.1007/s11262-012-0713-1
20. Zhang F, Chen Y, Ke Y, Zhang L, Zhang B, Yang L, et al. Single chain fragment variable (scFv) antibodies targeting the spike protein of porcine epidemic diarrhea virus provide protection against viral infection in piglets. Viruses (2019) 11:58. doi: 10.3390/v11010058
21. Liu J, Cong G, Shi H, Wang Y, Gao R, Chen J, et al. Neutralizing monoclonal antibody fails to recognize porcine epidemic diarrhea virus with two regions deleted in spike protein. Vet Microbiol (2020) 251:108917. doi: 10.1016/j.vetmic.2020.108917
22. Diep NV, Norimine J, Sueyoshi M, Lan NT, Yamaguchi R. Novel porcine epidemic diarrhea virus (PEDV) variants with Large deletions in the spike (S) gene coexist with PEDV strains possessing an intact s gene in domestic pigs in Japan: A new disease situation. PloS One (2017) 12:e0170126. doi: 10.1371/journal.pone.0170126
23. de Haan CA, Rottier PJ. Molecular interactions in the assembly of coronaviruses. Adv Virus Res (2005) 64:165–230. doi: 10.1016/S0065-3527(05)64006-7
24. McBride R, van Zyl M, Fielding BC. The coronavirus nucleocapsid is a multifunctional protein. Viruses-Basel (2014) 6:2991–3018. doi: 10.3390/v6082991
25. Su M, Shi D, Xing X, Qi S, Yang D, Zhang J, et al. Coronavirus porcine epidemic diarrhea virus nucleocapsid protein interacts with p53 to induce cell cycle arrest in s-phase and promotes viral replication. J Virol (2021) 95:e0018721. doi: 10.1128/JVI.00187-21
26. Liwnaree B, Narkpuk J, Sungsuwan S, Jongkaewwattana A, Jaru-Ampornpan P. Growth enhancement of porcine epidemic diarrhea virus (PEDV) in vero E6 cells expressing PEDV nucleocapsid protein. PloS One (2019) 14:e0212632. doi: 10.1371/journal.pone.0212632
27. Xu J, Mao J, Han X, Shi F, Gao Q, Wang T, et al. Porcine epidemic diarrhea virus inhibits HDAC1 expression to facilitate its replication Via binding of its nucleocapsid protein to host transcription factor Sp1. J Virol (2021) 95:e0085321. doi: 10.1128/JVI.00853-21
28. Ding Z, Fang LR, Jing HY, Zeng SL, Wang D, Liu LZ, et al. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J Virol (2014) 88:8936–45. doi: 10.1128/Jvi.00700-14
30. Wold WS, Toth K. Adenovirus vectors for gene therapy, vaccination and cancer gene therapy. Curr Gene Ther (2013) 13:421–33. doi: 10.2174/1566523213666131125095046
31. Wu S, Huang J, Zhang Z, Wu J, Zhang J, Hu H, et al. Safety, tolerability, and immunogenicity of an aerosolised adenovirus type-5 vector-based COVID-19 vaccine (Ad5-nCoV) in adults: Preliminary report of an open-label and randomised phase 1 clinical trial. Lancet Infect Dis (2021) 21:1654–64. doi: 10.1016/S1473-3099(21)00396-0
32. Zhu FC, Guan XH, Li YH, Huang JY, Jiang T, Hou LH, et al. Immunogenicity and safety of a recombinant adenovirus type-5-Vectored COVID-19 vaccine in healthy adults aged 18 years or older: A randomised, double-blind, placebo-controlled, phase 2 trial. Lancet (2020) 396:479–88. doi: 10.1016/S0140-6736(20)31605-6
33. Qian J, Yang M, Feng Q, Pan XY, Yang LL, Yang JL. Inhibition of glioma by adenovirus KGHV500 encoding anti-p21Ras scFv and carried by cytokine-induced killer cells. Exp Biol Med (Maywood) (2021) 246:1228–38. doi: 10.1177/1535370220986769
34. Dai F, Zhang PB, Feng Q, Pan XY, Song SL, Cui J, et al. Cytokine-induced killer cells carrying recombinant oncolytic adenovirus expressing p21Ras scFv inhibited liver cancer. J Cancer (2021) 12:2768–76. doi: 10.7150/jca.51434
35. Guo J, Gao J, Li Z, Gong Y, Man X, Jin J, et al. Adenovirus vector-mediated Gli1 siRNA induces growth inhibition and apoptosis in human pancreatic cancer with smo-dependent or smo-independent hh pathway activation in vitro and in vivo. Cancer Lett (2013) 339:185–94. doi: 10.1016/j.canlet.2013.06.010
36. Wang F, Wang M, Zhang L, Cheng M, Li M, Zhu J. Generation and functional analysis of single chain variable fragments (scFvs) targeting the nucleocapsid protein of porcine epidemic diarrhea virus. Appl Microbiol Biotechnol (2022) 106:995–1009. doi: 10.1007/s00253-021-11722-z
37. Tabrizi NM, Amani J, Ebrahimzadeh M, Nazarian S, Kazemi R, Almasian P. Preparation and evaluation of chitosan nanoparticles containing CtxB antigen against vibrio cholera. Microb Pathog (2018) 124:170–7. doi: 10.1016/j.micpath.2018.08.037
38. Kelly D, O'Brien JJ, McCracken KJ. Effect of creep feeding on the incidence, duration and severity of post-weaning diarrhoea in pigs. Res Vet Sci (1990) 49:223–8. doi: 10.1016/S0034-5288(18)31082-8
39. Hou Y, Lin CM, Yokoyama M, Yount BL, Marthaler D, Douglas AL, et al. Deletion of a 197-Amino-Acid region in the n-terminal domain of spike protein attenuates porcine epidemic diarrhea virus in piglets. J Virol (2017) 91:e00227–17. doi: 10.1128/JVI.00227-17
40. He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U.S.A. (1998) 95:2509–14. doi: 10.1073/pnas.95.5.2509
41. Nejatollahi F, Jaberipour M, Asgharpour M. Triple blockade of HER2 by a cocktail of anti-HER2 scFv antibodies induces high antiproliferative effects in breast cancer cells. Tumour Biol (2014) 35:7887–95. doi: 10.1007/s13277-014-1854-8
42. Froude JW, Herbert AS, Pelat T, Miethe S, Zak SE, Brannan JM, et al. Post-exposure protection in mice against Sudan virus by a two antibody cocktail. Viruses (2018) 10:286. doi: 10.3390/v10060286
43. Hou X, Jiang X, Jiang Y, Tang L, Xu Y, Qiao X, et al. Oral immunization against PEDV with recombinant lactobacillus casei expressing dendritic cell-targeting peptide fusing COE protein of PEDV in piglets. Viruses (2018) 10:106. doi: 10.3390/v10030106
44. Gerdts V, Zakhartchouk A. Vaccines for porcine epidemic diarrhea virus and other swine coronaviruses. Vet Microbiol (2017) 206:45–51. doi: 10.1016/j.vetmic.2016.11.029
45. Antas M, Olech M, Szczotka-Bochniarz A. Molecular characterization of porcine epidemic diarrhoea virus (PEDV) in Poland reveals the presence of swine enteric coronavirus (SeCoV) sequence in s gene. PloS One (2021) 16:e0258318. doi: 10.1371/journal.pone.0258318
46. Roth KDR, Wenzel EV, Ruschig M, Steinke S, Langreder N, Heine PA, et al. Developing recombinant antibodies by phage display against infectious diseases and toxins for diagnostics and therapy. Front Cell Infect Microbiol (2021) 11:697876. doi: 10.3389/fcimb.2021.697876
47. Chen F, Ma K, Madajewski B, Zhuang L, Zhang L, Rickert K, et al. Ultrasmall targeted nanoparticles with engineered antibody fragments for imaging detection of HER2-overexpressing breast cancer. Nat Commun (2018) 9:4141. doi: 10.1038/s41467-018-06271-5
48. Cong Y, Ulasli M, Schepers H, Mauthe M, V'Kovski P, Kriegenburg F, et al. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J Virol (2020) 94:e01925–19. doi: 10.1128/JVI.01925-19
49. Kong N, Shan T, Wang H, Jiao Y, Zuo Y, Li L, et al. BST2 suppresses porcine epidemic diarrhea virus replication by targeting and degrading virus nucleocapsid protein with selective autophagy. Autophagy (2020) 16:1737–52. doi: 10.1080/15548627.2019.1707487
50. Wang H, Kong N, Jiao Y, Dong S, Sun D, Chen X, et al. EGR1 suppresses porcine epidemic diarrhea virus replication by regulating IRAV to degrade viral nucleocapsid protein. J Virol (2021) 95:e0064521. doi: 10.1128/JVI.00645-21
51. Crystal RG. Adenovirus: The first effective in vivo gene delivery vector. Hum Gene Ther (2014) 25:3–11. doi: 10.1089/hum.2013.2527
52. Greber UF, Gomez-Gonzalez A. Adenovirus - a blueprint for gene delivery. Curr Opin Virol (2021) 48:49–56. doi: 10.1016/j.coviro.2021.03.006
53. Dong J, Li W, Dong AN, Mao SY, Shen LJ, Li S, et al. Gene therapy for unresectable hepatocellular carcinoma using recombinant human adenovirus type 5. Med Oncol (2014) 31:95. doi: 10.1007/s12032-014-0095-4
54. van Dorsten RT, Wagh K, Moore PL, Morris L. Combinations of single chain variable fragments from HIV broadly neutralizing antibodies demonstrate high potency and breadth. Front Immunol (2021) 12:734110. doi: 10.3389/fimmu.2021.734110
55. Pacifico MD, Pearl RA, Kupsch JM. The use of a cocktail of single chain fv antibody fragments to improve the in vitro and in vivo targeting of melanoma. J Exp Clin Canc Res (2006) 25:45–53.
56. Do VT, Jang J, Park J, Dao HT, Kim K, Hahn TW. Recombinant adenovirus carrying a core neutralizing epitope of porcine epidemic diarrhea virus and heat-labile enterotoxin b of escherichia coli as a mucosal vaccine. Arch Virol (2020) 165:609–18. doi: 10.1007/s00705-019-04492-7
57. Xie Y, Gao P, Li Z. A recombinant adenovirus expressing P12A and 3C protein of the type O foot-and-Mouth disease virus stimulates systemic and mucosal immune responses in mice. BioMed Res Int (2016) 2016:7849203. doi: 10.1155/2016/7849203
58. Dong SJ, Kong N, Zhang Y, Li YW, Sun DG, Qin WZ, et al. TARDBP inhibits porcine epidemic diarrhea virus replication through degrading viral nucleocapsid protein and activating type I interferon signaling. J Virol (2022) 96:e0007022. doi: 10.1128/jvi.00070-22
59. Wang H, Chen X, Kong N, Jiao Y, Sun D, Dong S, et al. TRIM21 inhibits porcine epidemic diarrhea virus proliferation by proteasomal degradation of the nucleocapsid protein. Arch Virol (2021) 166:1903–11. doi: 10.1007/s00705-021-05080-4
60. Lin CM, Ghimire S, Hou Y, Boley P, Langel SN, Vlasova AN, et al. Pathogenicity and immunogenicity of attenuated porcine epidemic diarrhea virus PC22A strain in conventional weaned pigs. BMC Vet Res (2019) 15:26. doi: 10.1186/s12917-018-1756-x
61. Lin H, Li B, Chen L, Ma Z, He K, Fan H. Differential protein analysis of IPEC-J2 cells infected with porcine epidemic diarrhea virus pandemic and classical strains elucidates the pathogenesis of infection. J Proteome Res (2017) 16:2113–20. doi: 10.1021/acs.jproteome.6b00957
62. Wang S, Wu J, Wang F, Wang H, Wu Z, Wu S, et al. Expression pattern analysis of antiviral genes and inflammatory cytokines in PEDV-infected porcine intestinal epithelial cells. Front Vet Sci (2020) 7:75. doi: 10.3389/fvets.2020.00075
63. Jung K, Miyazaki A, Saif LJ. Immunohistochemical detection of the vomiting-inducing monoamine neurotransmitter serotonin and enterochromaffin cells in the intestines of conventional or gnotobiotic (Gn) pigs infected with porcine epidemic diarrhea virus (PEDV) and serum cytokine responses of gn pigs to acute PEDV infection. Res Vet Sci (2018) 119:99–108. doi: 10.1016/j.rvsc.2018.06.009
64. Stanifer ML, Guo CC, Doldan P, Boulant S. Importance of type I and III interferons at respiratory and intestinal barrier surfaces. Front Immunol (2020) 11:608645. doi: 10.3389/fimmu.2020.608645
65. Li L, Fu F, Xue M, Chen W, Liu J, Shi H, et al. IFN-lambda preferably inhibits PEDV infection of porcine intestinal epithelial cells compared with IFN-alpha. Antiviral Res (2017) 140:76–82. doi: 10.1016/j.antiviral.2017.01.012
66. Liu YS, Liu Q, Jiang YL, Yang WT, Huang HB, Shi CW, et al. Surface-displayed porcine IFN-Lambda3 in lactobacillus plantarum inhibits porcine enteric coronavirus infection of porcine intestinal epithelial cells. J Microbiol Biotechnol (2020) 30:515–25. doi: 10.4014/jmb.1909.09041
67. Zhang Q, Ke H, Blikslager A, Fujita T, Yoo D. Type III interferon restriction by porcine epidemic diarrhea virus and the role of viral protein Nsp1 in IRF1 signaling. J Virol (2018) 92:e01677–17. doi: 10.1128/JVI.01677-17
Keywords: porcine epidemic diarrhea virus, nucleocapsid protein, single chain variable fragment, recombinant adenovirus, protective effects
Citation: Wang F, Zhang Q, Zhang F, Zhang E, Li M, Ma S, Guo J, Yang Z and Zhu J (2023) Adenovirus vector-mediated single chain variable fragments target the nucleocapsid protein of porcine epidemic diarrhea virus and protect against viral infection in piglets. Front. Immunol. 14:1058327. doi: 10.3389/fimmu.2023.1058327
Received: 30 September 2022; Accepted: 04 January 2023;
Published: 24 January 2023.
Edited by:
Lee Mark Wetzler, Boston University, United StatesReviewed by:
Roopali Rajput, Jamia Hamdard University, IndiaCopyright © 2023 Wang, Zhang, Zhang, Zhang, Li, Ma, Guo, Yang and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing Zhang, cWluZ3poYW5nQHNqdHUuZWR1LmNu; Jianguo Zhu, emh1X2pnQHNqdHUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.