- 1Pathogen Molecular Genetics Section, Laboratory of Bacteriology, National Institute of Allergy and Infectious Diseases (NIAID), United States (US) National Institutes of Health (NIH), Bethesda, MD, United States
- 2Comparative Medicine Branch (CMB), NIAID, NIH, Bethesda, MD, United States
In contrast to the virulent human skin commensal Staphylococcus aureus, which secretes a plethora of toxins, other staphylococci have much reduced virulence. In these species, commonly the only toxins are those of the phenol-soluble modulin (PSM) family. PSMs are species-specific and have only been characterized in a limited number of species. S. xylosus is a usually innocuous commensal on the skin of mice and other mammals. Prompted by reports on the involvement of PSMs in atopic dermatitis (AD) and the isolation of S. xylosus from mice with AD-like symptoms, we here identified and characterized PSMs of S. xylosus with a focus on a potential involvement in AD phenotypes. We found that most clinical S. xylosus strains produce two PSMs, one of the shorter α- and one of the longer β-type, which were responsible for almost the entire lytic and pro-inflammatory capacities of S. xylosus. Importantly, PSMα of S. xylosus caused lysis and degranulation of mast cells at degrees higher than that of S. aureus δ-toxin, the main PSM previously associated with AD. However, S. xylosus did not produce significant AD symptoms in wild-type mice as opposed to S. aureus, indicating that promotion of AD by S. xylosus likely requires a predisposed host. Our study indicates that non-specific cytolytic potency rather than specific interaction underlies PSM-mediated mast cell degranulation and suggest that the previously reported exceptional potency of δ-toxin of S. aureus is due to its high-level production. Furthermore, they suggest that species that produce cytolytic PSMs, such as S. xylosus, all have the capacity to promote AD, but a high combined level of PSM cytolytic potency is required to cause AD in a non-predisposed host.
Introduction
Atopic dermatitis (AD) is an inflammatory skin disease that predominantly affects children and generally becomes less severe upon reaching adulthood (1). The pathophysiology of AD is a complex process, driven by environmental and genetic factors, barrier dysfunction, alterations in cell-mediated immune responses (e.g. pronounced Th2 immune responses in acute AD), and IgE-mediated hypersensitivity (2). Clinically, patients are presented with intense pruritus, chronic eczematous skin lesions, and epidermal thickening and hypertrophy (3). The inflammatory reactions in the epidermis are initiated by specialized antigen-presenting cells, such as Langerhans cells (4), which activate keratinocytes to release potent inflammatory mediators (4–7). These in turn (i) stimulate the expression of adhesion molecules in cells of the endothelium, thus attracting migration of macrophages and eosinophils to the site of inflammation (8), (ii) help induce a Th2-inflammatory phenotype (3, 9), and (iii) promote class-switching of B cells to produce Immunoglobulin E (IgE) (10, 11). Antigen-specific IgE molecules are recognized by the high-affinity receptor, FcϵRI, found on eosinophils, basophils and mast cells (12, 13) and upon their engagement of surface-bound IgE to multivalent antigens, many pro-inflammatory cellular processes are activated, including the rapid degranulation and release of preformed inflammatory mediators stored in intracellular granules (14), as well as the de novo synthesis and release of more pro-inflammatory mediators (15, 16). Mast cells are generally regarded as the main effector cell involved in that response (17).
While much is known about the mechanisms leading to skin inflammation in AD, we still do not have a complete understanding of the pathogenesis of AD, especially in the context of microbial infiltration in the skin. Staphylococcus aureus is a human pathogen associated with AD and AD severity (18, 19). More recently, a defined microbial factor produced by S. aureus could be linked to AD pathogenesis (20). This factor, δ-toxin, is a 26 amino-acid peptide toxin belonging to the family of phenol-soluble modulin (PSM) peptides (21, 22). PSMs are peptides of ~ 2 kDa (α-type) or 4 – 5 kDa (β-type) with cytotoxic and pro-inflammatory activities. They all contain amphiphilic α-helices, which give them a surfactant-like character that likely is responsible for their non-specific membrane-disturbing impact on a variety of cell types, leading to their lysis. Most staphylococci produce a species-specific repertoire of PSMs, frequently with only moderate or even lacking amino acid sequence similarity to those of other species. In many staphylococcal species other than S. aureus, PSMs are virtually the only secreted toxins. While in the initial study on the role of δ-toxin in AD (20), promotion of AD was driven exclusively by δ-toxin, a further study more generally implicated PSMs in mast cell degranulation as a hallmark of AD development (23). The reason for a potentially specific involvement of δ-toxin as opposed to other PSMs, which all share similar biological activities, has remained unknown.
Staphylococcus xylosus is a commensal of mice and other mammals, but rarely humans, where - like most other staphylococcal species that colonize mammals - it usually does not cause any harm (24). However, there are reports on skin infections in mice that are caused by S. xylosus (25, 26), in particular skin lesions with AD-like symptoms (27–31). We hypothesized that S. xylosus produces PSMs with pathogenic potential that may also trigger AD pathogenesis. Therefore, we here identified and characterized PSMs of S. xylosus, produced isogenic deletion mutants in the main detected psm genes, and used pure PSMs as well as supernatants of cultures of S. xylosus and its isogenic psm mutants to investigate the pathogenic potential of S. xylosus PSMs with a focus on AD pathogenesis. Our results not only give insight into what are most likely the main virulence determinants of S. xylosus, but give important general insight into the role of δ-toxin and PSMs in AD. Notably, they help explain why δ-toxin has an exceptional role in promoting AD.
Materials and methods
Ethics statement
Animal experiments and protocols were performed according to the regulations of the Division of Intramural Research Animal Care and Use Committee (DIR ACUC) of the National Institute of Allergy and Infectious Diseases (NIAID), animal study proposal LB1E. Animal work was conducted adhering to the institution’s guidelines for animal use and followed the guidelines and basic principles as outlined in the United States Public Health Service Policy on Humane Care and Use of Laboratory Animals, and the Guide for the Care and Use of Laboratory Animals by certified staff in an Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) international accredited facility. All animals were euthanized with CO2 at the end of the studies. Human neutrophils were isolated from venous blood of healthy volunteers in accordance with a protocol (No. 99-CC-0168) approved by the Institutional Review Board for Human Subjects, NIAID. Informed written consent was obtained from all volunteers.
Animals
Female 10- to 12-week-old SKH-1E mice (Charles River Laboratories) were used for mouse colonization/infection studies and C57BL/NCrl mice (Charles River Laboratories) were used for Evans blue sensitization experiments and for the collection of bone marrow for neutrophil and mast cell differentiation. All mice were bred and maintained under pathogen-free conditions in an AAALAC-accredited animal facility in the NIAID. Age-, sex- and littermate-matched mice were randomly assigned into treatment groups in each experiment.
Bacterial strains and constructs and growth conditions
The S. xylosus strains used in this study comprised of reference laboratory strains and a series of strains from a collection of skin isolates obtained from C57BL/6 mice [wild-type (WT) or associated genetically altered strains] with spontaneous ulcerative dermatitis at the NIH (Supplementary Table S1). For the identification of S. xylosus PSMs from culture filtrates, all isolates and reference strains from glycerol stocks were first streaked onto tryptic soy agar plates and incubated overnight at 37 °C. The following day, a single clone was inoculated into tryptic soy broth (TSB) and grown overnight with shaking at 37 °C and 180 rpm. Overnight cultures were then inoculated at 1:100 dilution into TSB and grown for 8 hours. At these time points, culture supernatants were collected and stored at -20 °C until needed. For in vitro experiments with culture filtrates, supernatant from 16-hour cultures were passed through 0.22-μm polyethersulphone (PES) membrane filters (Sigma-Aldrich).
Construction of isogenic gene deletion mutants in S. xylosus
To create markerless isogenic psm mutants in S. xylosus AG14, an allelic replacement procedure was employed (32). PCR primer pairs (Supplementary Table S2) were designed to amplify ~ 1-kb regions upstream and downstream of the psmα and psmβ1 genes in S. xylosus AG14 genomic DNA (Supplementary Table S2). Overlap PCR was performed with the upstream and downstream ~ 1-kb PCR fragments with primers Xylpsmα1 and Xylpsmα2 (Reverse) and Xylpsmβ1 and Xylpsmβ2 (Reverse) to create ~ 2 kb fragments, which lack the coding sequences for psmα and psmβ1, respectively, and then cloned into the SmaI restriction site of plasmid pIMAY (32) resulting in pIMAY-Δpsmα and pIMAY-Δpsmβ1. Plasmids were first transformed into S. carnosus TM300 and then into S. aureus PS187ΔΔ. Finally, the plasmids were transduced into S. xylosus AG14 with phage φ187 as described previously (33) and the allelic replacement procedure was performed essentially as described (32, 33) to create the single psmα and psmβ1 mutants. To create the double psmα and psmβ1 mutant, pIMAY-Δpsmβ1 was phage-transduced into the Δpsmα mutant, followed by allelic replacement. The fidelity of all constructs was confirmed by DNA sequencing and reversed-phase high pressure liquid chromatography/electrospray ionization mass spectrometry (RP-HPLC/ESI-MS) of culture filtrates.
N-terminal sequencing
PSMα of S. xylosus was subjected to N-terminal Edman sequencing performed by the Research Technology Branch of NIAID after boiling in 25% trifluoroacetic acid (TFA) for 2 h at 55°C for removal of potentially existing N-terminal N-formyl modification (34).
PSM detection and quantification
PSM production by S. xylosus strains was analyzed using a SOURCE 15RPC ST 4.6/100 column (Cytiva) using a linear gradient from 0.1% TFA in water to 0.1% TFA in acetonitrile over ~ 20 column volumes at a flow rate of 0.5 ml/min on an Agilent 1100 HPLC system connected to an Agilent Quadrupole LC/MS instrument with an ESI source. For purification of PSMα, we used a Zorbax SB-C18 5 μm 9.4 × 250 mm column (Agilent) and a GE AKTA Purifier system using the same buffers and a linear gradient from 0 to 100% buffer B in 20 column volumes. Further analysis of PSM production in S. xylosus strains was performed using our routine RP-HPLC/ESI-MS analysis method as for S. aureus or S. epidermidis by adapting the m/z values (two per PSM) for S. xylosus PSMs (35). This method was also used for the absolute quantification of PSMs in culture filtrates with standard curves obtained using known molar concentrations of synthetic PSMs.
Synthetic peptides
All PSM peptides were synthesized with an N-terminal N-formyl methionine modification at > 95% purity by Peptide 2.0: S. aureus PSMα3 (MEFVAKLFKFFKDLLGKFLGNN); S. aureus δ-toxin (MAQDIISTIGDLVKWIIDTVNKFTKK); S. xylosus PSMα (MSFIIDIIKKIVGLFKGE), S. xylosus PSMβ1 (MAEIVEAIGKAVSAGLSHDWATMGVSIAEVLGKGVDFVLGFFK). Peptides were dissolved in DMSO and stored at -20 °C.
Isolation of neutrophils from mouse bone marrow
The isolation of neutrophils from mouse bone marrow was performed as described previously (36). Briefly, 8- to 10-week-old C57Bl/6NCrl female mice were sacrificed followed by the collection of the tibiae and femora. The bones were flushed with phosphate-buffered saline (PBS) with 1% (v/v) bovine serum albumin (BSA) and the bone marrow was sedimented after brief centrifugation. The erythrocytes in the cell pellet were lysed after brief exposure to ACK lysis buffer (Lonza), and magnetic negative selection was used to purify neutrophils, which were finally resuspended in RPMI-1640 media without phenol red (Gibco) and 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (Invitrogen) [RPMI-H], to the desired concentration.
Isolation of mouse bone marrow-derived mast cells
The isolation of mouse bone marrow mast cells (mBMMCs) was performed as described previously (37). Briefly, after sedimentation of the erythrocyte-depleted bone marrow with ACK lysis buffer, the cells were resuspended in 10 ml of RPMI-1640 media (Gibco), supplemented with 1% (v/v) Penicillin-streptomycin-glutamine (Invitrogen), 2.5% (v/v) HEPES (Invitrogen), 1% (v/v) Non-essential amino acids (Invitrogen), 100 mM sodium pyruvate (Invitrogen), 50 mM β-mercaptoethanol (Sigma-Aldrich) and 10% (v/v) fetal bovine serum (Gibco) and supplemented with a final concentration of 0.4 μg/ml each of murine recombinant IL-3 (m-IL-3) and Stem Cell Factor (SCF) (PeproTech) at a final concentration of ~ 5 × 105 cells/ml. Approximately every 1-2 days for the first week, non-adherent cells were transferred to a new culturing vessel and diluted two-fold if a concentration of 1 × 106 cells/ml was reached. Thereafter, cells were diluted once every week with fresh media supplemented with m-IL-3 and SCF. After 3-4 weeks of propagation, the presence of mast cells was confirmed by flow cytometric analyses for the presence of both CD117 (c-Kit) and FcϵR surface markers.
Isolation of human neutrophils
Human neutrophils were isolated from heparinized blood from healthy donors as described previously (38). Briefly, erythrocytes and peripheral blood mononuclear cells were removed with Dextran (Sigma-Aldrich) and Ficoll-Paque PLUS (GE Healthcare), respectively. A final lysis step with sterile water removed contaminating erythrocytes and the resulting cell pellet of >90% neutrophils was resuspended in RPMI-H and enumerated by microscopy.
Cell lines
The human mast cell line LAD2 was derived from CD34+ cells following marrow aspiration of a patient with aggressive mastocytosis (39). These cells were cultured in StemPro-34 (Life technologies) supplemented with 2% (v/v) StemPro nutrient supplement (Life technologies), Penicillin-streptomycin-L-glutamine (Life technologies), and 100 ng/ml recombinant human stem cell factor (rhSCF) (Peprotech). Cells were diluted two-fold after a concentration of ~ 5 × 105 cells/ml was reached. Mast cell purity was confirmed by routine flow cytometric analyses for the presence of both CD117 (c-Kit) and FcϵR surface markers.
Measurements of erythrocyte lysis (hemolysis)
Whole blood from anesthetized mice was collected by a cardiac puncture procedure. After collection, bleeds were immediately transferred into heparinized tubes (Sarstedt), stored at 4°C and used within two days (as with freshly collected heparinized human blood). Hemolysis was measured by incubating 100 μl of synthetic PSMs or culture supernatants with 100 μl of 4% (v/v) washed erythrocytes for one hour at 37°C in 96 well tissue culture plates, as previously described (40). The levels of released hemoglobin were determined by scanning at OD540nm using a TECAN® Spark microplate reader. Next, the percentage of lysis was calculated by first subtracting readings from media-alone blanks from all test readings from and then by dividing the absorbance values of the test wells by those obtained with a Triton X-100 (Sigma-Aldrich) positive control indicating 100% lysis.
Real-time calcium imaging
Calcium flux in murine or human neutrophils was measured as previously described (36). Briefly, neutrophils were suspended at a concentration of 5 × 106 cells/ml in RPMI/H and incubated with ~ 2 mM Fluo3-AM (Invitrogen, F1242) for 20 minutes at 37°C. Cells were then washed three times with HEPES buffer, resuspended to a concentration of 5 × 105 cells/ml and dispensed into wells of a black 96-well plate (Costar) in 200-ml volumes. The plate was centrifuged at 240 × g for 5 minutes and calcium fluxes were monitored in a Spark® Multimode Microplate Reader (Tecan) by measuring fluorescence using excitation and emission wavelengths of 485 nm and 535 nm, respectively. Fluorescence signals from the non-stimulated controls were subtracted from stimulated samples to account for background.
Measurement of β-hexosaminidase release by mast cells
The detection of β-hexosaminidase from mBMMCs and human LAD2 cells was performed using p-nitrophenyl N-acetyl-β-D-glucosamide (PNAG) as previously described (41). Briefly, cells were grown in their corresponding media for six hours without IL-3 or SCF. mBMMCs and LAD2 cells were then sensitized overnight with mouse monoclonal anti-dinitrophenyl IgE (Sigma-Aldrich) and biotinylated anti human IgE (SeraCare), respectively. The following day, the cells were washed three times with sterile HEPES buffer (10 mM HEPES, Invitrogen), 137 mM sodium chloride (Sigma-Aldrich), 2.7 mM potassium chloride (Sigma-Aldrich), 0.4 mM sodium phosphate dibasic (Fisher), 5.6 mM glucose (Macron), 1.8 mM calcium chloride (Sigma-Aldrich), 1.3 mM magnesium sulfate (Sigma-Aldrich), 0.04% (w/v) bovine serum albumin, pH 7.4) and resuspended to concentrations at 1 × 106 cells/ml (LAD2 cells) or 5 × 106 cells/ml (mBMMCs). To wells of a 96 well plate, 90 μl of cells were added and first allowed to equilibrate at 37°C for 30 minutes prior to the addition of 10-μl samples. After co-incubation at 37°C for 20 minutes, plates were centrifuged and samples from each well were collected for LDH release (see below) and β-hexosaminidase release. Briefly, for the latter, plates were first centrifuged at 450 × g, at 4°C for 5 min and 50 μl of supernatant from each well was transferred to a new flat-bottomed 96 well plate. To the remaining cells, 50 μl of 1% (v/v) Triton X-100 was added to each well to yield 100% lysis and then 50 μl of each lysate was transferred to a new plate. Then, to all wells, 100 μl of PNAG (3.5 μg/ml) in citrate buffer (40 mM citric acid (Sigma-Aldrich), 20 mM sodium phosphate dibasic (Fisher, pH 4.5) was added and plates were incubated for 90 min at 37°C. The reactions were stopped with 50 μl of 400 mM glycine (pH 10.7, Sigma-Aldrich). Absorbances at 405nm and 620nm (background) were measured using a TECAN® Spark multiplate reader. The percentage of β-hexosaminidase released was determined by subtracting the 620nm values from the 405nm values and then dividing the supernatant values by 100% lysis values from the same well.
Lysis measurements of neutrophils and mast cells
For both human and mouse neutrophils, 100 μl of cells at a concentration of 2.5 × 105 cells per ml were incubated with 100 μl of sample for one hour at 37°C (20 min at 37°C for mast cells – see above) in 96 well tissue culture plates. Cell lysis was determined using a Cytotoxicity Detection Kit (Roche Applied Sciences) by release of lactate dehydrogenase (LDH), as described previously (38), and absorbance readings recorded in a TECAN® Spark microplate reader. After accounting for background by subtracting values of blanks, the percentage of lysis of each cell type was determined by dividing the absorbance values of the test wells by those obtained with a 1% (v/v) Triton X-100 (Sigma-Aldrich) positive control representing 100% lysis.
Mouse AD model
For animal infections, overnight cultures of bacterial strains were inoculated into TSB at 1:100 and allowed to grow for ~ 3 h at 37°C with shaking at 180 rpm. Bacteria were washed with sterile PBS and then adjusted to the desired CFU concentration. The murine AD model was performed essentially as previously described (20), with modifications. Briefly, 10- to 12-week-old SKH-1E female mice (Charles River Laboratories) were anesthetized and the back skin sterilized with an alcohol swab. A 1 cm × 1 cm piece of sterile gauze was placed on the dorsal area followed by the application of a 100-μl bacterial suspension containing 1 × 108 CFUs. Mice dorsa were bandaged with transparent occlusive dressing (Tegaderm™, 3M) to sustain bacterial contact with the skin. After seven days, mice were euthanized by CO2 and the occlusive dressing and gauze were removed. A 1 cm × 1 cm piece of the dorsal skin was excised in the area of gauze-skin contact and then subjected to histopathological analysis with hematoxylin and eosin (H and E) staining. The slides were examined by a veterinary pathologist and a score of inflammation ranging from 0 – 4 was used, with 4 indicating the greatest degree of inflammation.
Protein-fragment complementation assay
To assess mast cell degranulation in vivo, the PCA assay was performed as described previously (42) with the following modifications. Briefly, the ears of wild-type C57Bl/6NCrl mice were sensitized by intradermal injections of 20 ng of α-DNP-IgE (Sigma-Aldrich) resuspended in saline. After 15 hours of incubation, the mice were intradermally challenged with 20 μl containing 2% culture supernatants. Following inoculation, 100 μl of 5 mg/ml Evans blue dye (Sigma-Aldrich), diluted in saline, was intravenously injected into each mouse. After 30 minutes, the mice were sacrificed, and a biopsy was collected using a 5-mm diameter punch (Integra) from each ear. The punches were then placed in 200 μl formamide (Sigma-Aldrich) solution and incubated at 63°C overnight with shaking. Following this incubation, 150-μl formamide extracts were dispensed into wells of a clear flat-bottomed polypropylene 96 well plate and the absorbance values were measured at 600nm.
Statistical analysis
Graph Pad Prism (version 9.1.1) was used for statistical analyses. Unpaired, two-tailed Student’s t-tests were used when comparing two groups, and one-way or two-way ANOVA with Tukey’s post-tests when comparing more than two groups. ANOVAs were only used when data passed normality tests (Anderson–Darling, D’Agostino–Pearson, Shapiro–Wilk, Kolmogorov–Smirnov). Otherwise, non-parametric tests were used. All error bars depict the standard deviation.
Results
Identification of S. xylosus PSMs
To identify PSMs of S. xylosus, we used S. xylosus isolates obtained from spontaneous ulcerative dermatitis in mice and RP-HPLC/ESI-MS. In the characteristic elution range of PSMs, which elute exceptionally late during RP-HPLC due to their amphiphilic nature (21), we found three predominant peaks with deconvoluted masses of 2079.3 Da (peak 1), 2051.0 Da (peak 2), and 4479.8 Da (peak 3) in this order of elution (Figure 1A). Four out of six clinical isolates showed a similar pattern of production of peaks 1 and 3, one isolate only produced peak 2, and one isolate did not reveal PSM production. We also measured production of the peptides associated with these peaks in two S. xylosus standard strains, one of which showed a pattern similar to the clinical isolates with production of only peak 2 (strain C2a), and the other did not reveal production of PSM peptides (strain ATCC 24966) (Figure 1B).
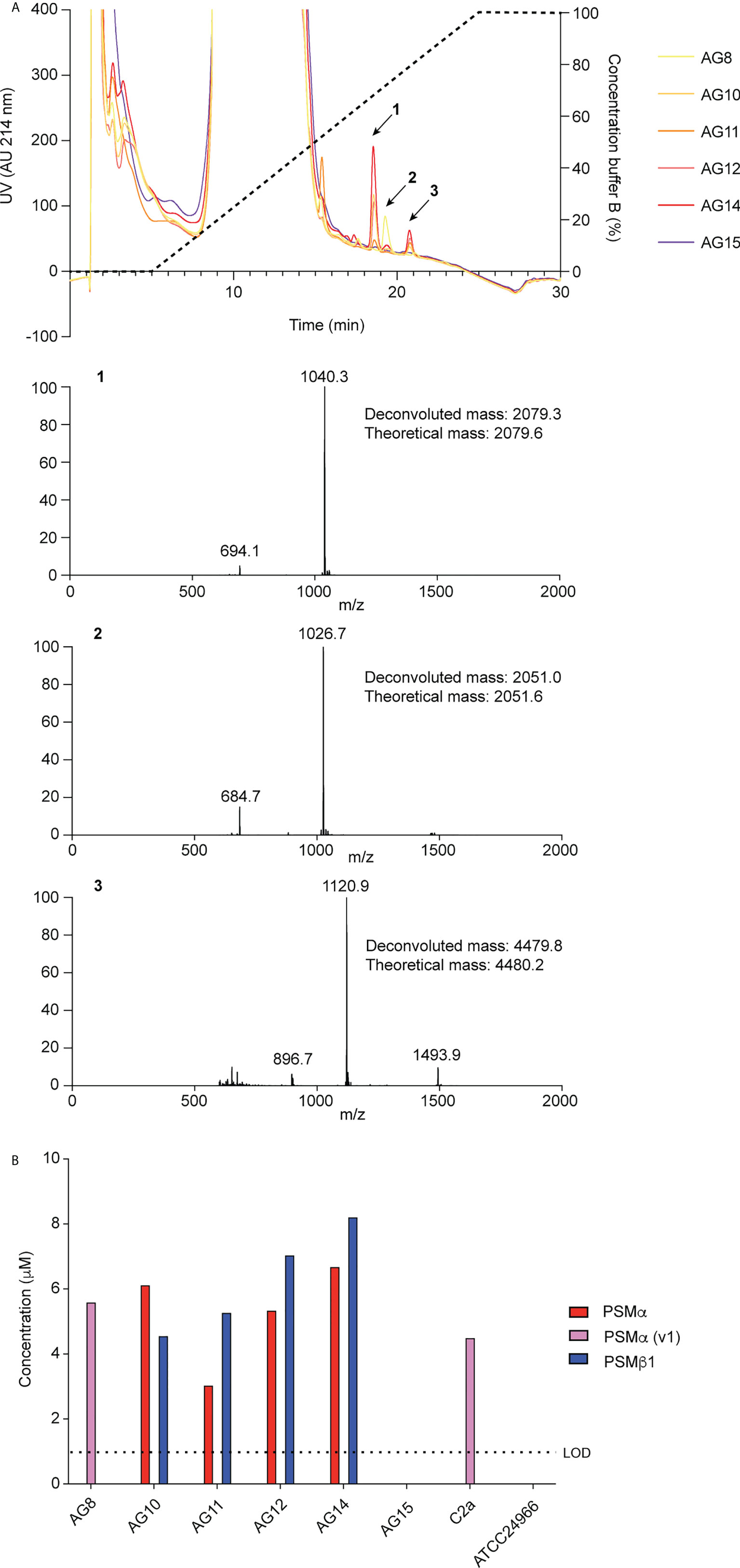
Figure 1 Detection of S. xylosus PSMs. (A) RP-HPLC/ESI-MS of culture filtrates of S. xylosus clinical isolates. Extracted mass spectra of peaks 1 to 3 are shown below the chromatogram. Results from deconvolution and theoretical average isotopic masses are noted. (B) Analysis of PSM production pattern by RP-HPLC/ESI-MS of culture filtrates collected from clinical S. xylosus isolates and reference strains. LOD, limit of detection.
PSMs can be classified in α-type PSMs, which have molecular weights (MWs) of ~ 2 kDa, and β-type PSMs, which have MWs of 4 to 5 kDa (22). Due to their higher MWs and similarity to known PSMs, β-type PSMs are usually annotated as such, or as hemolytic peptides, in staphylococcal genomes. In contrast, the identity of α -type PSMs cannot be deduced from genome sequences, as their length is below common annotation thresholds for open reading frames. Thus, the production of α-type PSMs in species where PSMs have not yet been characterized requires purification and N-terminal sequence conformation by Edman sequencing, in addition to the MW analysis provided by RP-HPLC/ESI-MS.
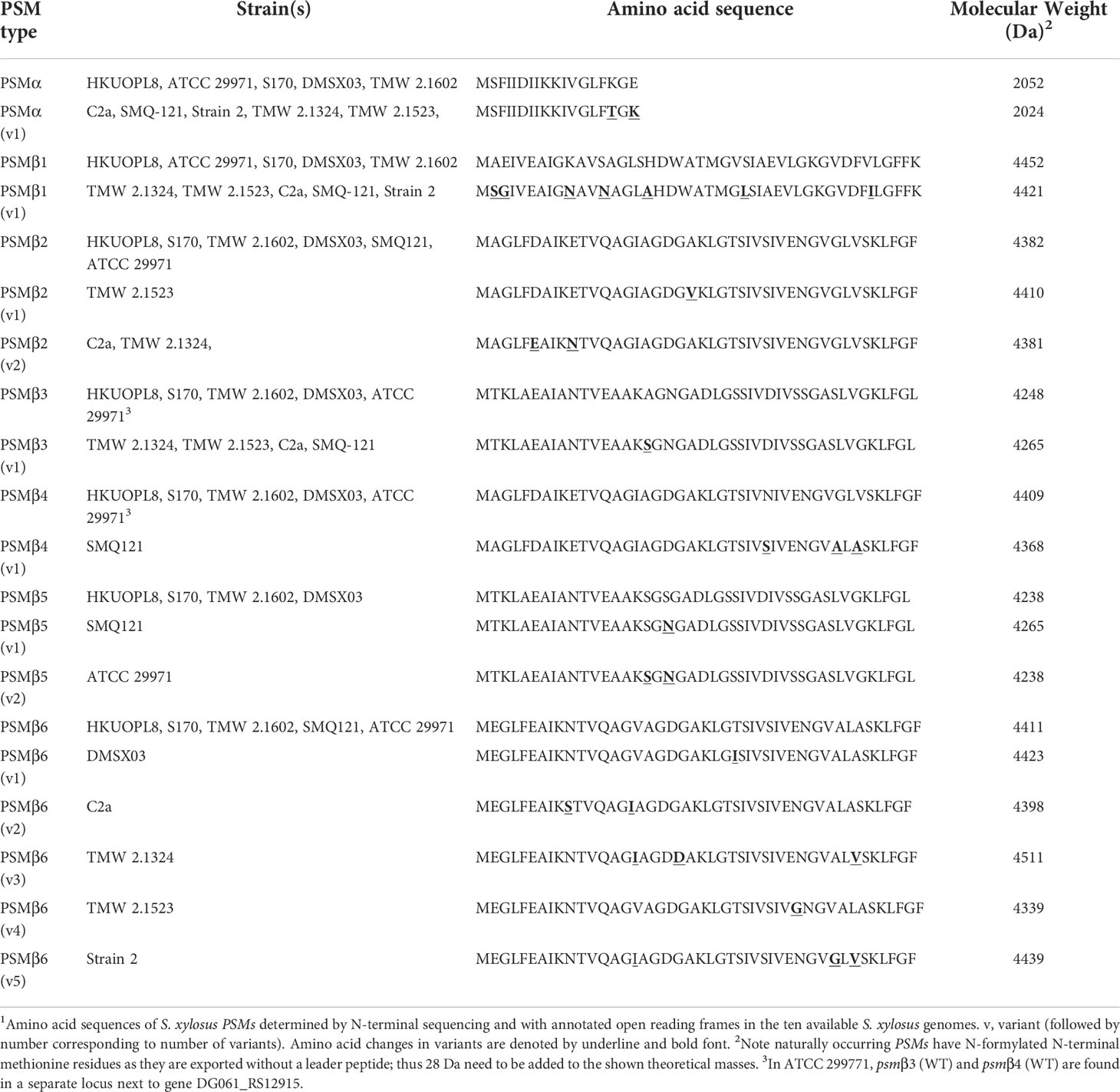
We therefore purified the main α-type PSM (peak 1) by RP-HPLC on a semi-preparative column. We assumed that the smaller α-type PSM peak 2 corresponded to an N-formylated version of peak 1, owing to their mass difference of 28 Da. PSMs are secreted as the primary translation product by a dedicated transporter (43), thus carrying an N-terminal N-formyl methionine like all primary bacterial translation products. However, some peptide molecules can undergo N-terminal deformylation by peptide deformylase (44), leading to a mass difference of 28 Da. Pairs of the same PSM peptides with a mass difference of 28 Da are therefore commonly found in PSM analyses and we thus assumed first that peaks 1 and 2 may represent N-formylated and N-deformylated versions of the same PSM peptide. N-terminal sequencing of the purified peptide corresponding to peak 1 yielded a sequence of MSFIIDIIKKIVGLFKGE, which represents the entire peptide and is in perfect agreement with the detected MW. The MW as determined by HPLC/MS and N-terminal sequence information, together with analysis of available S. xylosus genome sequences identified peak 1 as the peptide product of a gene situated between those encoding a GNAT family acetyltransferase and a dihydroxy-acid dehydratase in several S. xylosus genomes (Figure 2A). Following the nomenclature we established previously for staphylococcal PSMs (21), we called this peptide PSMα of S. xylosus.
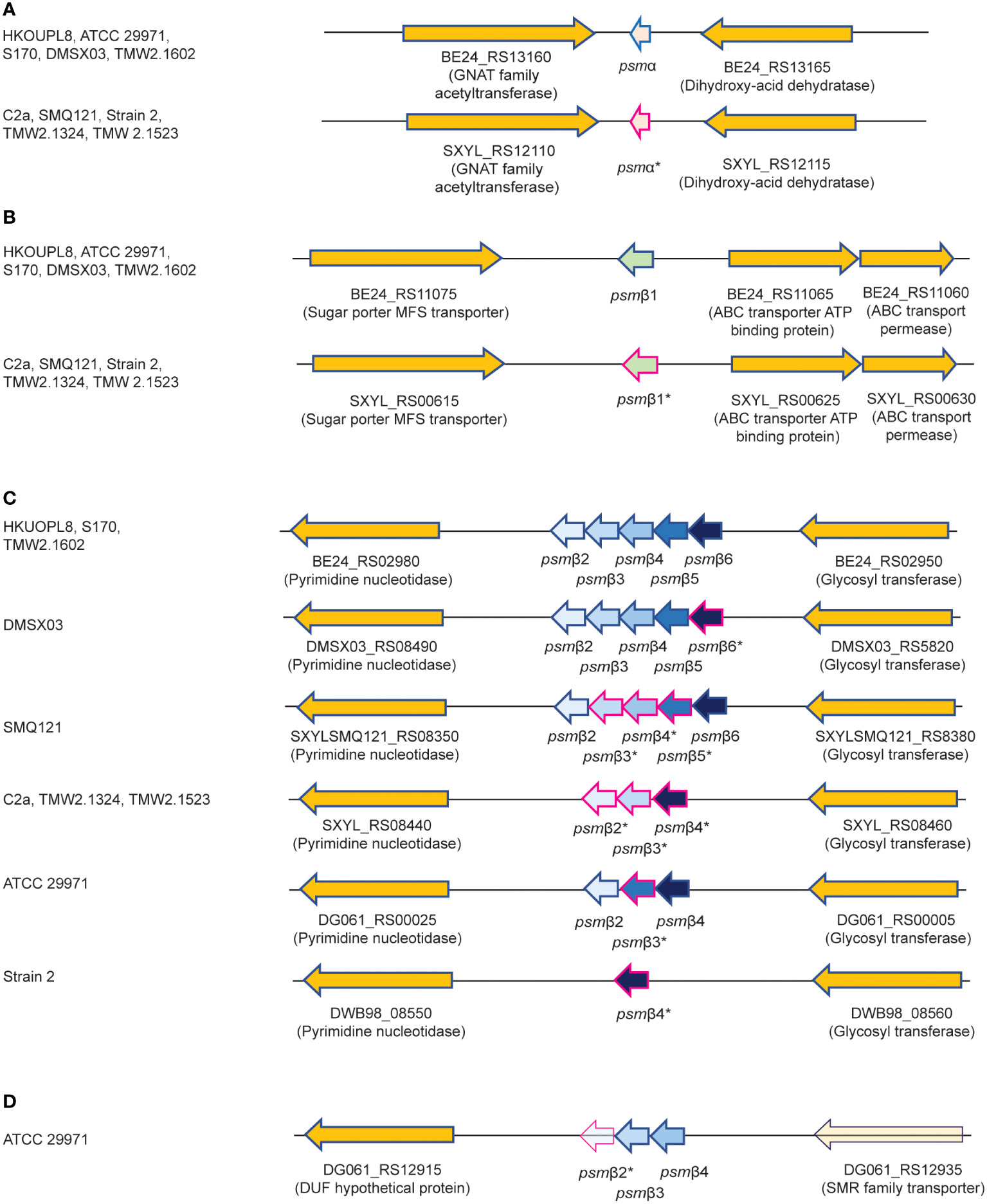
Figure 2 Amino acid sequences of S. xylosus PSMs and encoding genetic loci. (A) Genetic location of the psmα gene. (B) Genetic location of the psmβ1 gene. (C) Genetic location of the psmβ2-6 operon. Strain HKUOPL8 is used for reference (underlined) for the location of psmα, psmβ1 and psmβ2-6. (D) Genetic location of a second psmβ operon in ATCC 29971. *Genes encoding PSM variants are denoted by a fuchsia border around the arrow depicting the gene. For some PSMβ-type peptides, multiple variants exist but are not distinguished here. A complete list of predicted amino acid sequences of S. xylosus psm genes and variants can be found in Table 1.
We then analyzed all available S. xylosus genome sequences for sequences with similarity to that α-type peptide and for genes annotated as encoding PSMβ-like peptides, as well as sequences with similarity to PSMβ peptides found by this procedure. Results are shown in Figure 2 and deduced amino acid sequences in Table 1. Notably, we found that in several S. xylosus genomes a variant of the psmα gene is present, which encodes a peptide with a mass difference of 28 Da as compared to PSMα. As this peptide variant is found in the genome of strain C2a, where we also found peak 2 (rather than peak 1) using RP-HPLC/ESI-MS, together with the facts that (i) only PSMα (peak 1) or its variant (peak 2) but not both appear to be produced by different isolates, and (ii) N-formylated versions of the same PSM commonly elute later, not earlier, during RP-HPLC than N-deformylated forms, we concluded that peaks 1 and 2 stem from two different peptides, PSMα and PSMα (v1), whose mass difference only coincidentally is the same as that due to N-terminal formylation.
As for β-type peptides, we found a gene encoding a peptide whose calculated mass exactly corresponded to that of peak 3. We named this peptide PSMβ1 of S. xylosus (Figure 2B). According to genome analysis, this peptide also can occur as a peptide variant. Furthermore, we found a locus with considerable strain-to-strain variation and apparent gene duplication, encoding maximally five copies of a PSMβ peptide with moderate amino acid sequence variations (Figure 2C). Only in one strain, a second locus carrying additional psmβ genes was found at a different genomic location (Figure 2D). We named the corresponding encoded peptides PSMβ2 to PSMβ6 of S. xylosus. Re-examining our RP-HPLC/ESI-MS data, we found traces of signals corresponding to the expected masses of some of those peptides in the PSM elution range.
Altogether, our analysis revealed that S. xylosus produces a variety of PSM peptides with particularly pronounced variability as indicated by genome analysis. However, many of these peptides only appear to be produced at very low levels, and PSMα or its variant PSMα (v1) and PSMβ1 are the main PSMs produced by S. xylosus isolates. There appear to be two main clades of strains, one of which – to which most of our clinical isolates belong – produce PSMα and PSMβ1, while another clade predominantly produces a variant of PSMα, PSMα (v1), and barely PSMβ peptides. Complete absence of PSMs, as observed in one of our clinical isolates and one standard strain, is often found also in S. aureus where it is linked to mutation in the Agr quorum-sensing system, which strictly controls PSM production (45, 46). It is likely that this is also the case in the S. xylosus strains that do not produce PSMs. Lastly, one defining feature of S. xylosus is that it does not produce δ-toxin, a member of the PSM class produced in large amounts in other staphylococci, including S. aureus and S. epidermidis, for example (21, 47). This absence of a δ-toxin-like peptide in S. xylosus is in accordance with no obvious open reading frame being found within the apparent RNAIII gene of S. xylosus, where δ-toxin is usually encoded.
Phenotypic characterization of the two most abundant PSM peptides produced by clinical S. xylosus isolates
We focused on the two most abundant PSMs that are produced by the clinical isolates, PSMα and PSMβ1, and characterized their cytotoxic capacities toward human and mouse erythrocytes and neutrophils, as well as their pro-inflammatory capacities by determining calcium flux in human and mouse neutrophils. Calcium flux is the most commonly used readout for the pro-inflammatory capacity of PSMs (40, 48, 49).
PSMα of S. xylosus had strong cytolytic activity toward erythrocytes and neutrophils from humans and mice, at a level comparable to that of S. aureus PSMα3, a PSM known to have pronounced cytolytic activity (40) (Figures 3A, B). PSMβ1 had low cytolytic activity, as described generally for β-type PSMs of other staphylococci (40, 47, 50). The cytolytic potency of S. aureus δ-toxin, an α-type PSM, was higher than that of PSMβ1, but it did not reach the level of activity exerted by the strongly cytolytic S. xylosus PSMα or S. aureus PSMα3. Notably, there was no apparent species specificity of these effects.
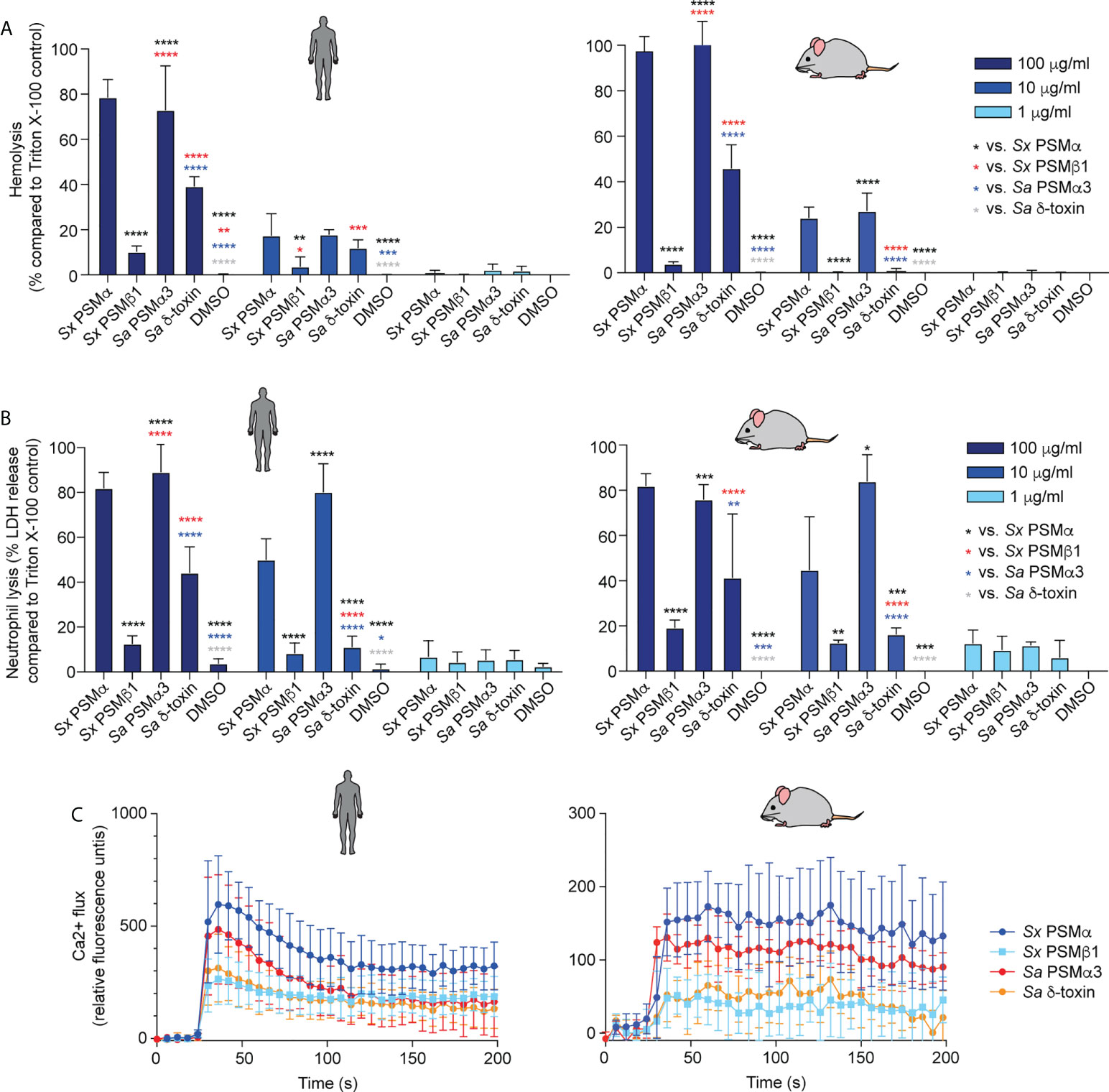
Figure 3 Cytolytic and pro-inflammatory capacities of main S. xylosus PSMs. Capacity of synthetic PSM peptides representative of predominant S. xylosus PSM species to lyse (A) human and mouse erythrocytes, (B) Human and mouse neutrophils and (C) induce calcium flux in human and mouse neutrophils. Synthetic S. aureus PSMα3 and δ-toxin were measured for comparison. For hemolysis and lactate dehydrogenase (LDH) release assays, triplicate measurements were performed with whole heparinized blood and purified neutrophils obtained from three donors, respectively. For calcium flux assays, duplicate measurements were performed from two donors and PSMs were added at a final concentration of 100 nM. Statistical analysis is by 2-way ANOVA with Tukey’s post-tests. *, p<0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; Error bars show the mean ± SD. Mouse and human schemes are from openclipart.org.
In contrast to the cytolytic activity of PSMs, their pro-inflammatory activity is receptor-dependent (48); thus, these two activities of PSMs are known not to be correlated. PSMα of S. xylosus showed the most pronounced pro-inflammatory activity as measured by calcium flux in neutrophils, followed by PSMα3 of S. aureus, and PSMβ1 of S. xylosus and S. aureus δ-toxin with similarly low activity, again with no apparent species specificity (Figure 3C).
These data showed that S. xylosus produces PSMs with pronounced cytolytic and pro-inflammatory activity, most notably PSMα, but there is no apparent species specificity of their effects.
Generation and analysis of isogenic psm mutants in S. xylosus
Focusing again on PSMα and PSMβ1 as the PSMs with the strongest expression in most of the analyzed S. xylosus clinical isolates, we produced single and combined isogenic deletion mutants in the psmα and psmβ1 loci of isolate AG14 as an isolate with high expression of these PSMs. PSM production analysis of the constructed Δpsmα, Δpsmβ1, and Δpsmαpsmβ1 mutants showed the expected expression pattern (Figure 4). Having established the absence of species specificity of S. xylosus PSM activities, we used human erythrocytes and neutrophils for these experiments.
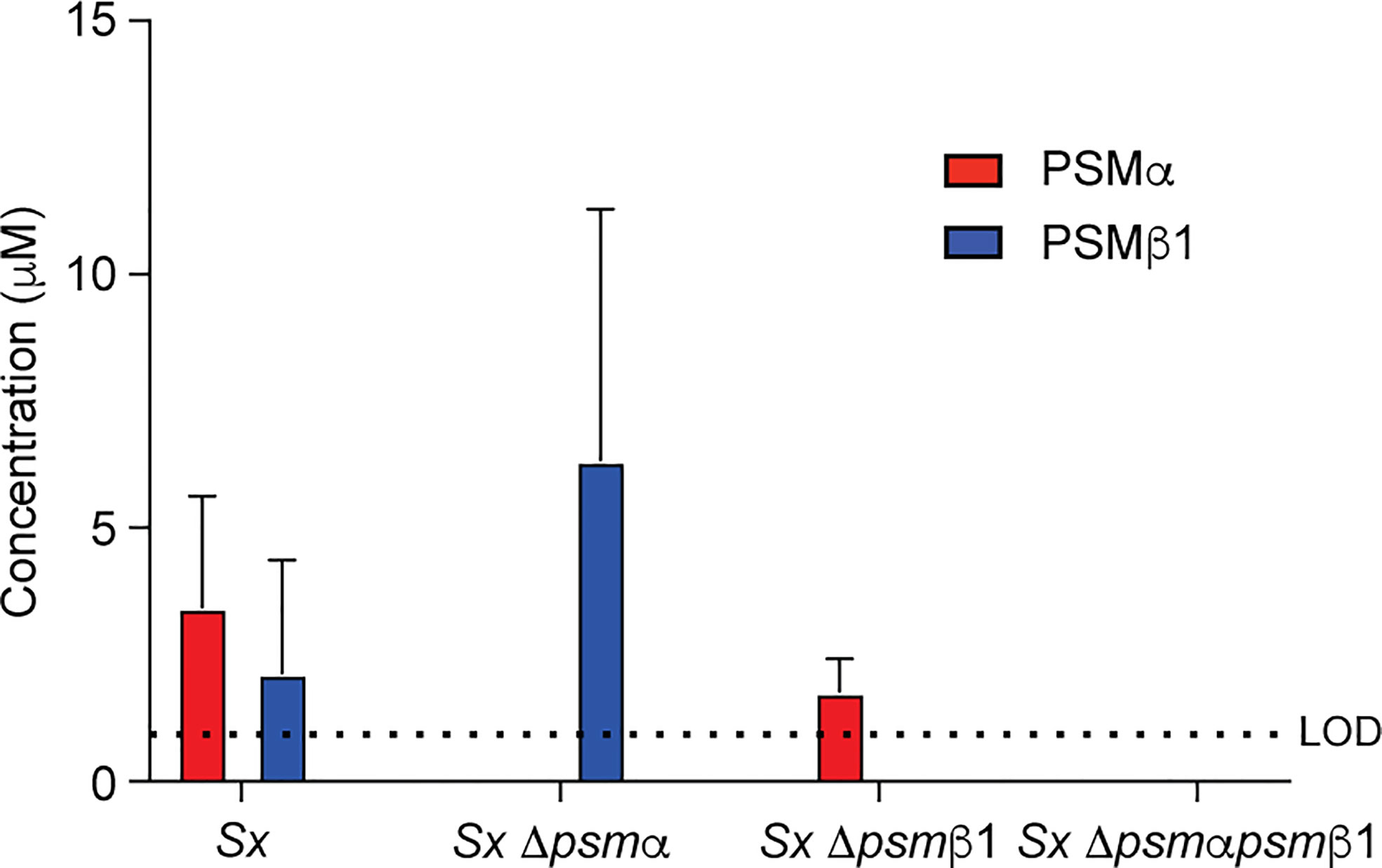
Figure 4 Construction of isogenic S. xylosus psm deletion and complementation strains. Analysis of PSM production pattern by RP-HPLC/ESI-MS of culture filtrates collected from constructed isogenic psm mutants of strain S. xylosus AG14.
PSMα and PSMβ1 both contributed considerably to cytolysis of erythrocytes and neutrophils in cell-free supernatants of harvested S. xylosus cultures (Figures 5A, B). Importantly, we could not detect any cytolytic activity when the genes for both these PSMs were deleted, emphasizing that they account for virtually the entire cytolytic activity of S. xylosus AG14, and by extrapolation based on PSM production patterns (see Figure 1B), of clinical S. xylosus strains in general. S. aureus δ-toxin was only responsible for part of the cytolytic activity of S. aureus, which is in accordance with previous findings (40) and to be expected by the fact that several additional cytolytic PSMs, as well as other cytolysins, are produced by S. aureus (51). Of note, as a likely result of the plethora of cytolysins produced by S. aureus, lytic activity toward both erythrocytes and neutrophils was considerably more pronounced in S. aureus than S. xylosus. Contrastingly, the capacity to induce calcium flux of these two species was about the same, with neither the δ-toxin nor PSMα of S. xylosus having a significant impact, while PSMβ1 appeared to be responsible for most of the pro-inflammatory effect of S. xylosus (Figures 5C).
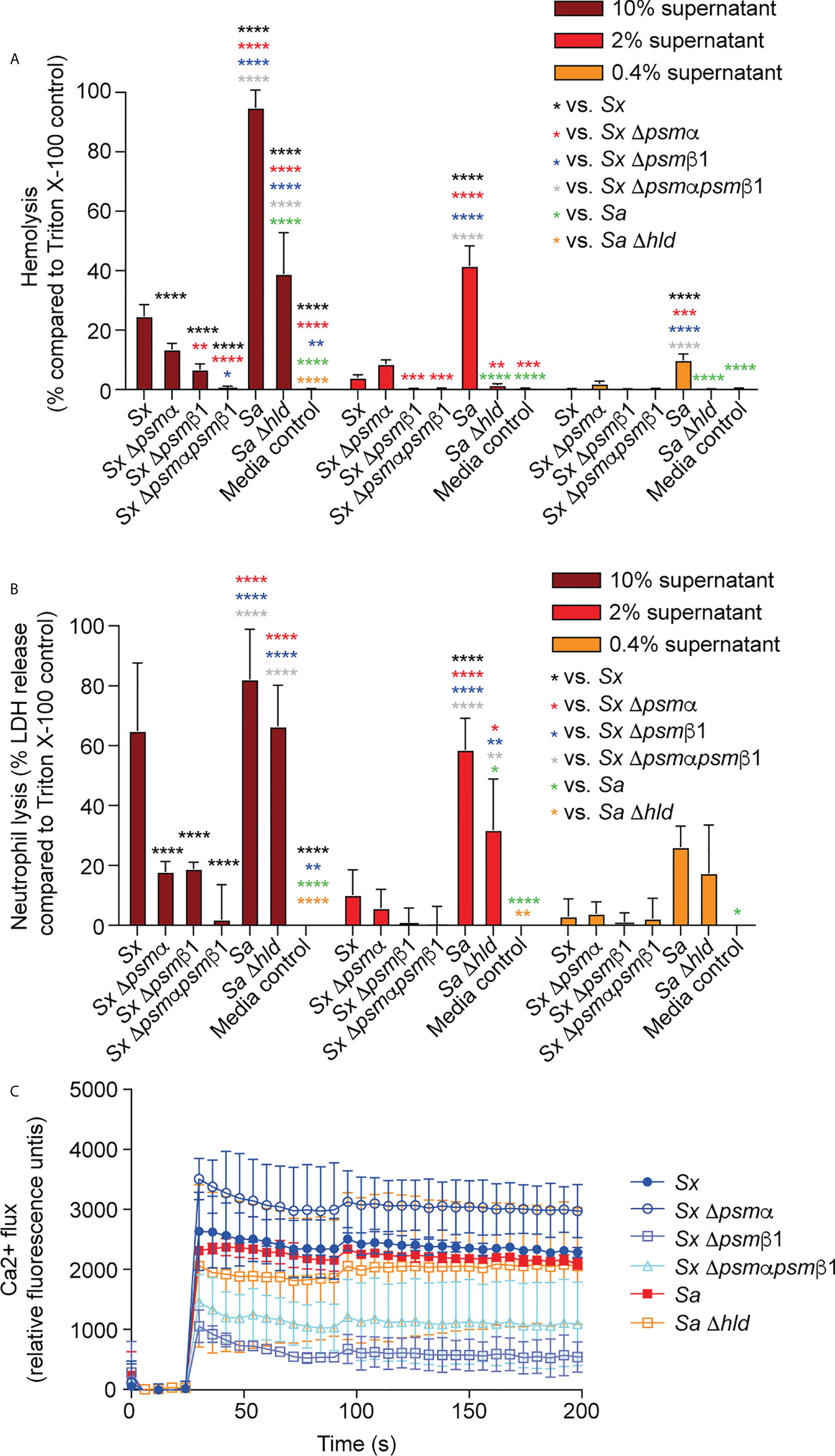
Figure 5 Cytolytic and pro-inflammatory capacities of S. xylosus psm deletion strains. Capacity of stationary-phase (16-h) culture filtrates from S. xylosus AG14 and isogenic psm mutants to induce lysis of (A) human erythrocytes, (B) human neutrophils and (C) induce calcium flux in human neutrophils. Culture filtrates from S. aureus LAC and its isogenic δ-toxin mutant were used as comparison. For hemolysis and lactate dehydrogenase (LDH) release assays, triplicate measurements were performed using whole heparinized blood and purified neutrophils obtained from three donors, respectively. For calcium flux assays, single measurements were performed from two donors. Statistical analysis is by 2-way ANOVA with Tukey’s post-tests. *, p<0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; Error bars show the mean ± SD.
Mast cell degranulation
While PSMs have been reported to represent important virulence determinants in a series of different S. aureus infection types, including skin (40, 52), blood (40), and bone (53) infections, of particular interest for the present investigation is their impact on atopic dermatitis. This is because the isolates we examined are from mice affected with dermatitis-like symptoms. Furthermore, there is discussion in the literature about whether mast cell degranulation as a main mechanism underlying AD pathogenesis is solely impacted by δ-toxin (20) or whether other PSMs also affect AD (23). We therefore examined the impact of S. xylosus PSMs on mast cell degranulation and also measured LDH release to determine whether degranulation is linked to cell lysis.
S. xylosus PSMα and PSMα3 of S. aureus caused strong, δ-toxin moderate, and S. xylosus PSMβ1 only minor mast cell lysis and degranulation (Figures 6A, B). Notably, lysis and degranulation by different PSMs were almost identical in degree and pattern. We also again included mouse mast cells in this experiment to verify the absence of species specificity on mast cells as a key cell type promoting AD symptoms. There were virtually no differences in the results obtained with human versus mouse mast cells.
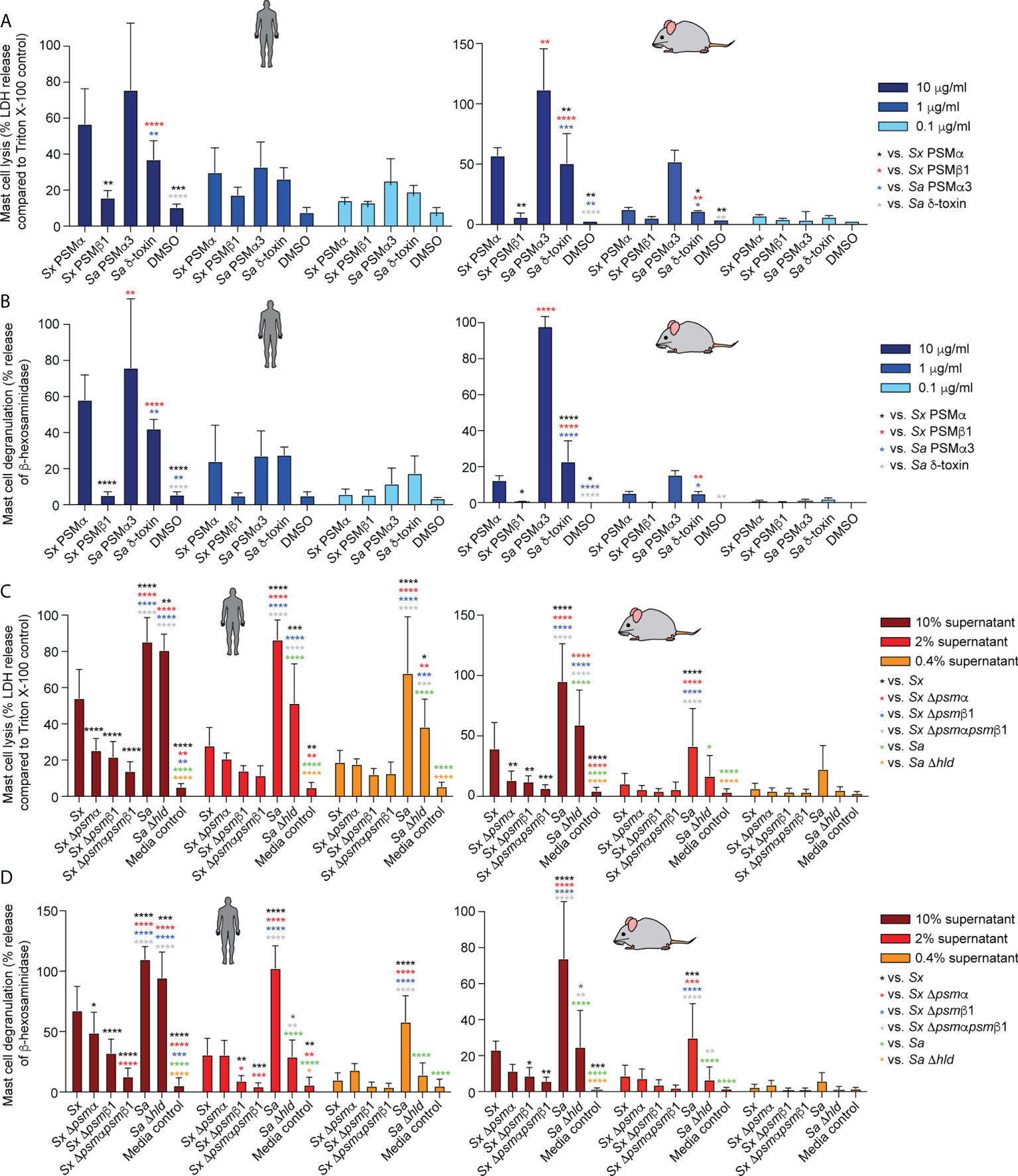
Figure 6 Mast cell degranulation by S. xylosus PSMs. Human and mouse mast cell degranulation and lysis by pure PSM peptides (A, B) and culture filtrates (C, D) of S. xylosus AG14 WT and isogenic psm mutants is shown. Statistical analysis is by 1-way ANOVA with Tukey’s post-tests. *, p<0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; Error bars show the mean ± SD. Mouse and human schemes are from openclipart.org.
As for the situation in the natural strain background, S. aureus supernatant caused considerably stronger lysis and degranulation than S. xylosus supernatant (Figures 6C, D). The δ-toxin had a strong contribution to mast cell degranulation and lysis, although there was still considerable potency of S. aureus supernatant to lyse mast cells and cause degranulation by S. aureus supernatant without δ-toxin. Most of S. xylosus’ capacity to lyse mast cells and cause degranulation was due to production of PSMα and PSMβ1, with production of both PSMs contributing to those effects. Again, experiments with mouse and human mast cells yielded similar results. As in our experiments with pure PSM peptides, the degree of lysis and degranulation generally was strongly correlated.
AD model and protein-fragment complementation assay
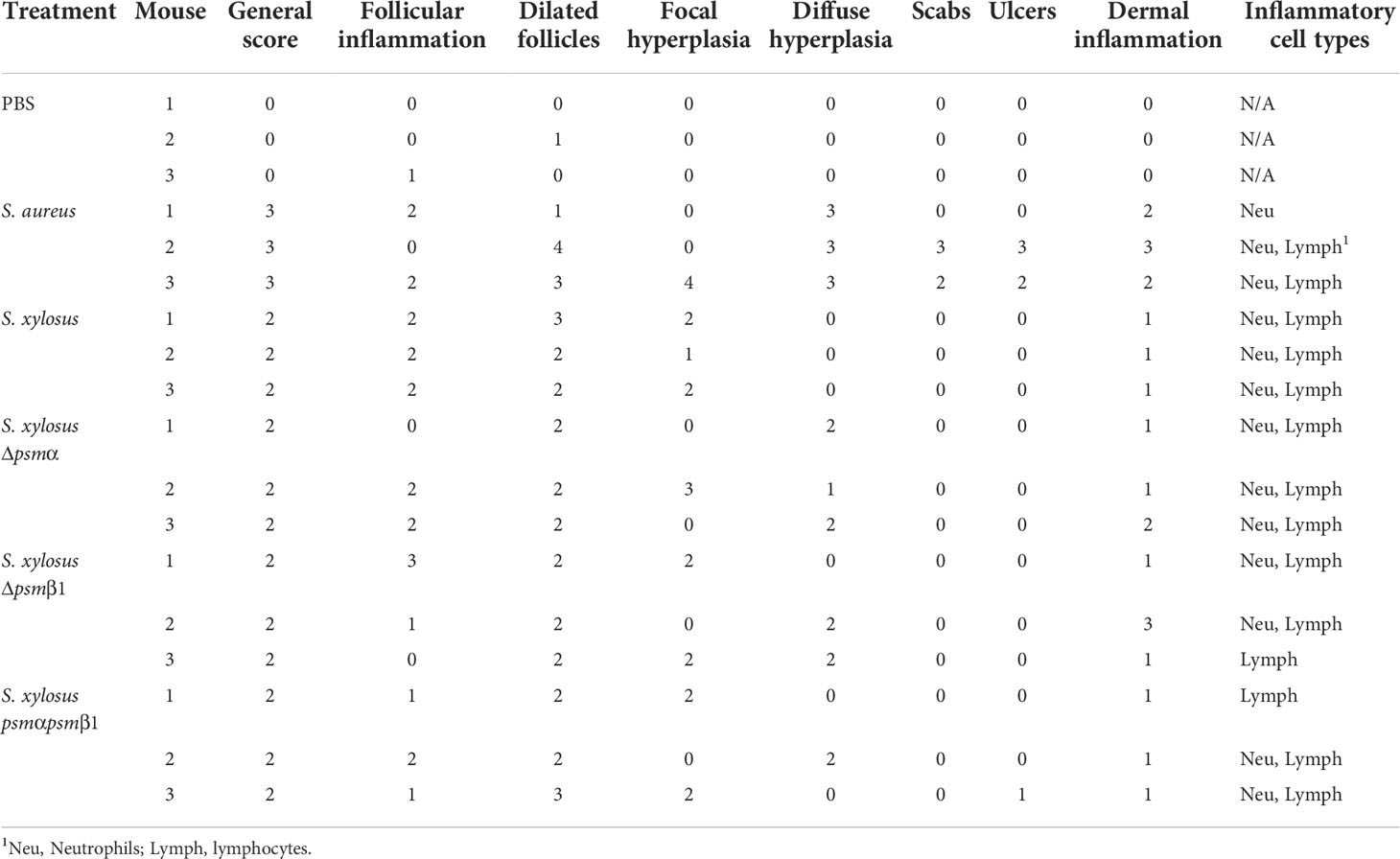
We employed an established model of AD in wild-type mice (20, 54, 55) to determine whether the results obtained in vitro with mast cells translate to in vivo pathogenicity. In accordance with the stronger degree of mast cell degranulation we had observed to be exerted by S. aureus, S. aureus caused visible scabbing on mouse skin, while S. xylosus did not (Figure 7A). Similarly, upon histological examination of infected skin tissue, S. aureus was found to cause, in all or some mice, follicular inflammation, dilated follicles, focal hyperplasia, diffuse hyperplasia, scabs, ulcers, dermal inflammation, and influx of neutrophils and lymphocytes (Figure 7B, Table 2). S. xylosus only caused these phenotypes to a very minor degree, and there was no difference to the degree of pathogenicity seen with S. xylosus psm mutants (Figure 7B, Table 2). Further confirming these results, in a protein-fragment complementation assay, which we performed to estimate the degree of in vivo mast cell degranulation, we observed significantly increased readings in mice injected with culture filtrates from S. aureus, but not with S. xylosus or its isogenic psm deletion mutants (Figure 7C).
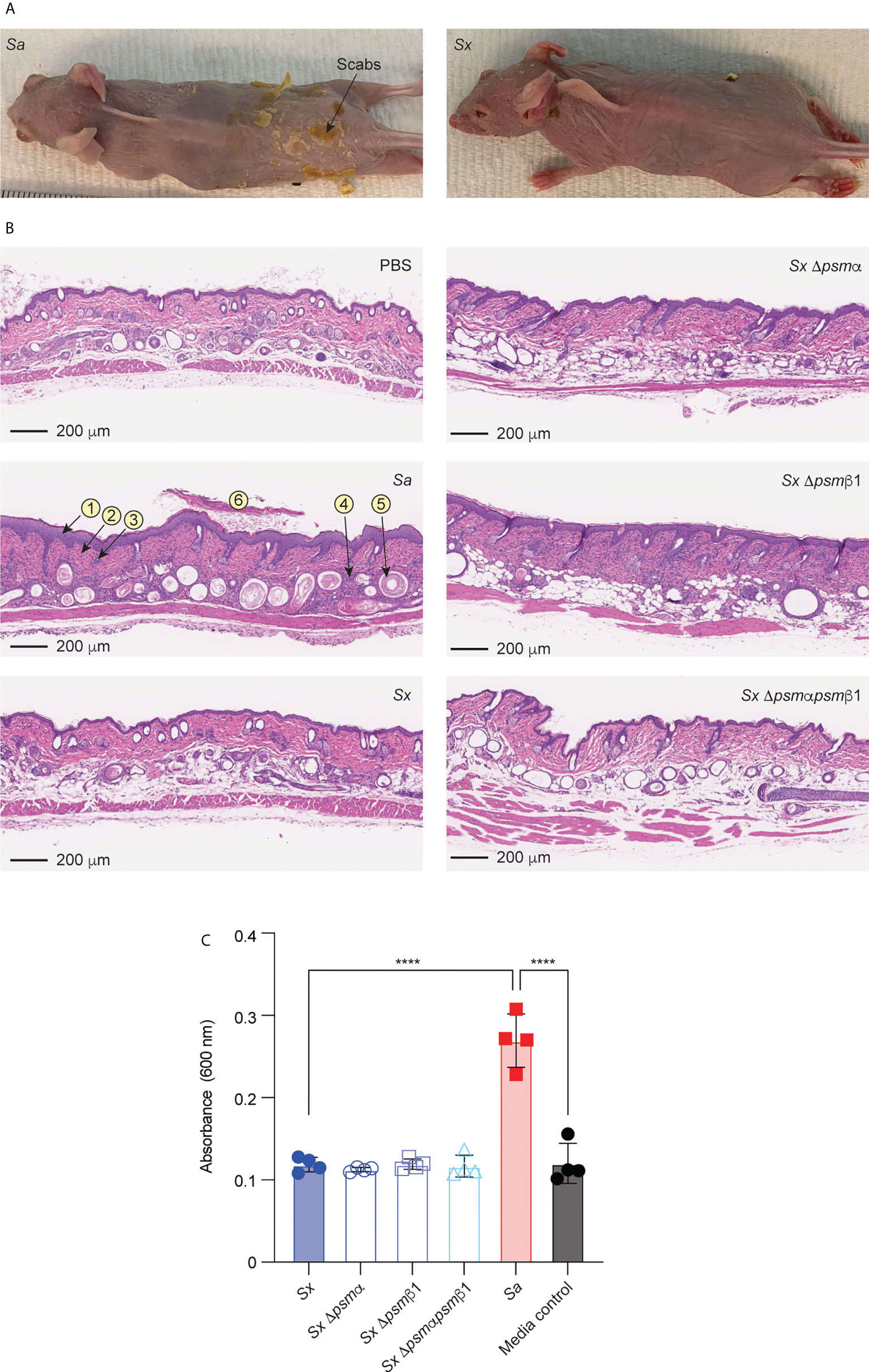
Figure 7 Mouse model of AD and in-vivo mast cell degranulation. (A) Representative pictures of mice infected on the skin with S. aureus LAC (Sa) and S. xylosus AG14 (Sx). (B) Histological evaluation of mice infected with S. aureus LAC, S. xylosus AG14 or isogenic S. xylosus psm mutants. See Table 2 for scores. Signs of inflammation and infection are marked in the S. aureus picture: 1, diffuse hyperplasia; 2, dermal inflammation; 3, focal hyperplasia; 4, follicular inflammation; 5, follicular dilation; 6, hyperkeratosis. These are absent or much less pronounced in all other pictures. (C) Mast cell degranulation in vivo as determined by a Protein-fragment complementation assay (PCA). Measurements were performed from both ears from two mice after injection with culture filtrates. Statistical analysis is by 1-way ANOVA with Tukey’s post-tests. ****, p < 0.0001; Error bars show the mean ± SD.
Discussion
In the present study, we identified and characterized PSM peptides of S. xylosus, a member of the genus Staphylococcus that has been reported to cause AD-like symptoms in mice (27–31). Our study was prompted by previous reports identifying members of the PSM family as virulence factors of the human pathogen S. aureus where they promote many infection types including AD (14, 20, 22, 40, 53). Most notably, the δ-toxin of S. aureus has been shown to cause AD-associated virulence phenotypes, such as mast cell degranulation, and AD symptoms in mouse models of AD (20). However, other studies have also implied other PSMs of S. aureus in mast cell degranulation (23). We thus performed this study not only to characterize what we assumed to be likely major virulence determinants of S. xylosus, because non-S. aureus staphylococci rarely produce toxins other than PSMs (56, 57), but also to gain insight into the role of δ-toxin versus other PSMs in causing AD.
We found that S. xylosus produces three main PSMs, with some strains producing one α- and one β-type PSM, and others mainly one α-type PSM, which is a variant of that found in the other strains. Importantly, no S. xylosus strain produced δ-toxin or a δ-toxin homologue. Genome analysis revealed the presence of further loci encoding β-type PSMs, but these genes are apparently only expressed in minor amounts. S. xylosus as a species overall showed pronounced heterogeneity regarding PSM production and amino sequence variations, while in other species that we have previously investigated, variants are rarer (21).
In accordance with what we know generally about PSMs (21, 22), the main S. xylosus PSMs promoted cytolysis of erythrocytes and neutrophils and calcium flux in neutrophils. The cytolytic activities of the two main S. xylosus PSMs were consistent in degree toward the different cell types, while the relative degree of pro-inflammatory activities differed from the cytolytic activity pattern, as expected from the fact that the cytolytic activities are not receptor-dependent while the pro-inflammatory activities are (48). Absence of production of both main S. xylosus PSMs in a constructed double mutant led to virtually complete reduction of those capacities. As for the cytolytic capacities this is expected from the general absence of cytolysins other than PSMs in these species. Like in S. aureus, where PSMs are by far the most pro-inflammatory secreted components (36), pro-inflammatory capacity of S. xylosus was also mainly due to PSMs.
Mast cell degranulation is a hallmark and key disease phenotype of AD (17). We therefore measured mast cell degranulation to estimate the potency of S. xylosus PSMs to contribute to AD development. Intriguingly, the potency of S. xylosus PSMα, as well as of S. aureus PSMα3, exceeded that of S. aureus δ-toxin, the main PSM previously associated with AD. Furthermore, the lytic activities toward mast cells correlated well with those toward erythrocytes and neutrophils as well as the abilities to promote mast cell degranulation. Nevertheless, cytolytic and mast cell degranulation potency of S. aureus supernatant was much higher than that of S. xylosus and to a large part due to δ-toxin.
The mechanism by which δ-toxin contributes to AD development has remained unknown, Nakamura et al. reported that interaction with the PSM receptor FPR2, which underlies the pro-inflammatory capacities of PSMs (48), is not involved (20). These authors also reported that sublytic concentrations of δ-toxin can cause mast cell degranulation, but that this is enhanced in the presence of anti-DNP IgE (20), suggesting that δ-toxin can trigger mast cell degranulation by an FcϵRI-dependent mechanism by engaging with the IgE-FcϵRI complex. On the other hand, in this and another study a correlation was found between mast cell degranulation and cytolytic potency of highly cytolytic α-type PSMs (20, 23), which is in accordance with our findings that show strong correlation of mast cell degranulation with general cytolytic capacity. This may indicate that lytic capacity determines δ-toxin’s degranulating activity on mast cells. While our findings imply a potential contribution of a mechanism that is based only on cytolysis, we planned our degranulation experiments always in the presence of IgE. Therefore, we are unable to comment further as to which mechanism has more significance. Notably, our findings indicate that the higher abundance of δ-toxin in S. aureus supernatants as compared to other PSMs (40, 47), rather than an exceptional potency to trigger mast cell degranulation, explain the increased corresponding potency of S. aureus as compared to S. xylosus.
While our results on mast cell degranulation indicate potency of S. xylosus to promote AD, albeit at a lower extent than S. aureus, we were not able to detect significant AD-like symptoms in an AD model using wild-type mice in which, contrastingly, S. aureus showed a clear impact. That we also could not detect an impact of psm genes of S. xylosus is likely due to already only very minor pathogenicity readouts we detected for S. xylosus wild-type. Given that AD-like symptoms in mice were reported exclusively in mice that were genetically or otherwise predisposed (27–31), similar to the mice from which the S. xylosus strains were isolated for this study, we believe that – in contrast to S. aureus – S. xylosus only has the potency to promote AD in the presence of other predisposing factors.
In conclusion, the findings of our study, which identified and characterized PSMs of S. xylosus, further substantiate the notion of widespread production in the genus Staphylococcus of cytolytic and pro-inflammatory peptides of the PSM family and underlines the previously emphasized species specificity of PSM production (21, 58). Furthermore, our findings have important implications for our understanding of PSM involvement in AD, since they suggest that the previously noted exceptional corresponding capacity of δ-toxin (20) is not due to specific characteristics of that peptide as compared to other PSMs, but its production levels. Because of that situation, a species like S. xylosus with absence of δ-toxin or other highly produced PSMs likely requires predisposing host factors for a PSM-mediated impact on AD development. Further exploration will be necessary to more completely understand the role of δ-toxin and other PSMs in AD development, such as to address questions that arise from the fact that in S. epidermidis for example, a δ-toxin homologue that is very similar to that of S. aureus is produced in high amounts (47); yet – despite some reports on the potential of S. epidermidis factors to trigger AD phenotypes (59) and S. epidermidis association with AD flares (18) - this species has not been associated with AD as clearly as S. aureus. Of potential importance in that regard, S. epidermidis does not produce high amounts of strongly lytic PSMs (47, 57). Thus, we speculate that the combined cytolytic potency of secreted PSMs determines the degree to which a given species or strain triggers mast cell degranulation and thereby, AD, a notion that will need to be substantiated by investigation with large numbers of different species and strains.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by Division of Intramural Research Animal Care and Use Committee (DIR ACUC) of the National Institute of Allergy and Infectious Diseases (NIAID).
Author contributions
AG and WE collected and provided S. xylosus skin isolates from mice. HJ, YZ, and MO performed RP-HPLC/ESI-MS analyses for identification and detection of PSMs. RH, GC and MO performed bioinformatical analyses. AV and JB created S. xylosus isogenic psm deletion and complementation strains. KR, RH, RL, TN, GC, and JB performed cellular experiments. KR, RH, RL, GC, and JB performed animal experiments. KR, GC and MO analyzed data. GC and MO designed the study. GC and MO supervised experiments. GC and MO wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases (NIAID), Laboratory of Bacteriology (project number ZIA AI000904 to MO) and the Comparative Medicine Branch, U.S. National Institutes of Health (NIH).
Acknowledgments
The authors thank Dr. Dean Metcalfe (Laboratory of Allergic Diseases, NIAID, NIH) for the kind gift of LAD2 cells and Dr. Jerrold Ward for thehistopathological evaluation of skin samples.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.999201/full#supplementary-material
References
1. Williams H, Flohr C. How epidemiology has challenged 3 prevailing concepts about atopic dermatitis. J Allergy Clin Immun (2006) 118(1):209–13. doi: 10.1016/j.jaci.2006.04.043
2. Boothe WD, Tarbox JA, Tarbox MB. Atopic dermatitis: Pathophysiology. Adv Exp Med Biol (2017) 1027:21–37. doi: 10.1007/978-3-319-64804-0_3
3. Weidinger S, Novak N. Atopic dermatitis. Lancet (2016) 387(10023):1109–22. doi: 10.1016/S0140-6736(15)00149-X
4. Nakajima S, Igyarto BZ, Honda T, Egawa G, Otsuka A, Hara-Chikuma M, et al. Langerhans cells are critical in epicutaneous sensitization with protein antigen via thymic stromal lymphopoietin receptor signaling. J Allergy Clin Immunol (2012) 129(4):1048–55 e6. doi: 10.1016/j.jaci.2012.01.063
5. Krishna S, Miller LS. Innate and adaptive immune responses against Staphylococcus aureus skin infections. Semin Immunopathol (2012) 34(2):261–80. doi: 10.1007/s00281-011-0292-6
6. Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol (2013) 13(3):159–75. doi: 10.1038/nri3399
7. Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol (2011) 30(1):16–34. doi: 10.3109/08830185.2010.529976
8. Liu FT, Goodarzi H, Chen HY. IgE, mast cells, and eosinophils in atopic dermatitis. Clin Rev Allergy Immunol (2011) 41(3):298–310. doi: 10.1007/s12016-011-8252-4
9. Dainichi T, Kitoh A, Otsuka A, Nakajima S, Nomura T, Kaplan DH, et al. The epithelial immune microenvironment (EIME) in atopic dermatitis and psoriasis. Nat Immunol (2018) 19(12):1286–98. doi: 10.1038/s41590-018-0256-2
10. Nakae S, Suto H, Iikura M, Kakurai M, Sedgwick JD, Tsai M, et al. Mast cells enhance T cell activation: importance of mast cell costimulatory molecules and secreted TNF. J Immunol (2006) 176(4):2238–48. doi: 10.4049/jimmunol.176.4.2238
11. Pawankar R, Okuda M, Yssel H, Okumura K, Ra C. Nasal mast cells in perennial allergic rhinitics exhibit increased expression of the fc epsilonRI, CD40L, IL-4, and IL-13, and can induce IgE synthesis in b cells. J Clin Invest (1997) 99(7):1492–9. doi: 10.1172/JCI119311
12. Kinet JP. The high-affinity IgE receptor (Fc epsilon RI): from physiology to pathology. Annu Rev Immunol (1999) 17:931–72. doi: 10.1146/annurev.immunol.17.1.931
13. Kraft S, Kinet JP. New developments in FcepsilonRI regulation, function and inhibition. Nat Rev Immunol (2007) 7(5):365–78. doi: 10.1038/nri2072
14. Johansson SGO. The history of IgE: From discovery to 2010. Curr Allergy Asthm R (2011) 11(2):173–7. doi: 10.1007/s11882-010-0174-3
15. Turner H, Kinet JP. Signalling through the high-affinity IgE receptor fc epsilon RI. Nature (1999) 402(6760):B24–30. doi: 10.1038/35037021
16. Rivera J, Fierro NA, Olivera A, Suzuki R. New insights on mast cell activation via the high affinity receptor for IgE. Adv Immunol (2008) 98:85–120. doi: 10.1016/S0065-2776(08)00403-3
17. Kawakami T, Ando T, Kimura M, Wilson BS, Kawakami Y. Mast cells in atopic dermatitis. Curr Opin Immunol (2009) 21(6):666–78. doi: 10.1016/j.coi.2009.09.006
18. Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res (2012) 22(5):850–9. doi: 10.1101/gr.131029.111
19. Leyden JJ, Marples RR, Kligman AM. Staphylococcus aureus in the lesions of atopic dermatitis. Br J Dermatol (1974) 90(5):525–30. doi: 10.1111/j.1365-2133.1974.tb06447.x
20. Nakamura Y, Oscherwitz J, Cease KB, Chan SM, Munoz-Planillo R, Hasegawa M, et al. Staphylococcus delta-toxin induces allergic skin disease by activating mast cells. Nature (2013) 503(7476):397–401. doi: 10.1038/nature12655
21. Cheung GY, Joo HS, Chatterjee SS, Otto M. Phenol-soluble modulins–critical determinants of staphylococcal virulence. FEMS Microbiol Rev (2014) 38(4):698–719. doi: 10.1111/1574-6976.12057
22. Peschel A, Otto M. Phenol-soluble modulins and staphylococcal infection. Nat Rev Microbiol (2013) 11(10):667–73. doi: 10.1038/nrmicro3110
23. Hodille E, Cuerq C, Badiou C, Bienvenu F, Steghens JP, Cartier R, et al. Delta hemolysin and phenol-soluble modulins, but not alpha hemolysin or panton-valentine leukocidin, induce mast cell activation. Front Cell Infect Microbiol (2016) 6:180. doi: 10.3389/fcimb.2016.00180
24. Nagase N, Sasaki A, Yamashita K, Shimizu A, Wakita Y, Kitai S, et al. Isolation and species distribution of staphylococci from animal and human skin. J Vet Med Sci (2002) 64(3):245–50. doi: 10.1292/jvms.64.245
25. Supre K, Haesebrouck F, Zadoks RN, Vaneechoutte M, Piepers S, De Vliegher S. Some coagulase-negative Staphylococcus species affect udder health more than others. J Dairy Sci (2011) 94(5):2329–40. doi: 10.3168/jds.2010-3741
26. Gozalo AS, Hoffmann VJ, Brinster LR, Elkins WR, Ding L, Holland SM. Spontaneous Staphylococcus xylosus infection in mice deficient in NADPH oxidase and comparison with other laboratory mouse strains. J Am Assoc Lab Anim Sci (2010) 49(4):480–6.
27. Won YS, Kwon HJ, Oh GT, Kim BH, Lee CH, Park YH, et al. Identification of Staphylococcus xylosus isolated from C57BL/6J-Nos2(tm1Lau) mice with dermatitis. Microbiol Immunol (2002) 46(9):629–32. doi: 10.1111/j.1348-0421.2002.tb02744.x
28. Bradfield JF, Wagner JE, Boivin GP, Steffen EK, Russell RJ. Epizootic fatal dermatitis in athymic nude mice due to Staphylococcus xylosus. Lab Anim Sci (1993) 43(1):111–3.
29. Kim Y, Lee YS, Yang JY, Lee SH, Park YY, Kweon MN. The resident pathobiont Staphylococcus xylosus in nfkbiz-deficient skin accelerates spontaneous skin inflammation. Sci Rep (2017) 7(1):6348. doi: 10.1038/s41598-017-05740-z
30. Russo M, Invernizzi A, Gobbi A, Radaelli E. Diffuse scaling dermatitis in an athymic nude mouse. Vet Pathol (2013) 50(4):722–6. doi: 10.1177/0300985812463408
31. Acuff NV, LaGatta M, Nagy T, Watford WT. Severe dermatitis associated with spontaneous Staphylococcus xylosus infection in rag(-/-)Tpl2(-/-) mice. Comp Med (2017) 67(4):344–9.
32. Monk IR, Shah IM, Xu M, Tan MW, Foster TJ. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio (2012) 3(2). doi: 10.1128/mBio.00277-11
33. Winstel V, Kuhner P, Rohde H, Peschel A. Genetic engineering of untransformable coagulase-negative staphylococcal pathogens. Nat Protoc (2016) 11(5):949–59. doi: 10.1038/nprot.2016.058
34. Shively JE, Hawke D, Jones BN. Microsequence analysis of peptides and proteins. III Artifacts Eff Impurities Anal Anal Biochem (1982) 120(2):312–2. doi: 10.1016/0003-2697(82)90352-9
35. Joo HS, Otto M. The isolation and analysis of phenol-soluble modulins of Staphylococcus epidermidis. Methods Mol Biol (2014) 1106:93–100. doi: 10.1007/978-1-62703-736-5_7
36. Nguyen TH, Cheung GYC, Rigby KM, Kamenyeva O, Kabat J, Sturdevant DE, et al. Rapid pathogen-specific recruitment of immune effector cells in the skin by secreted toxins. Nat Microbiol (2022) 7(1):62–72. doi: 10.1038/s41564-021-01012-9
37. Jensen BM, Swindle EJ, Iwaki S, Gilfillan AM. Generation, isolation, and maintenance of rodent mast cells and mast cell lines. Curr Protoc Immunol (2006) 3:23. doi: 10.1002/0471142735.im0323s74
38. Voyich JM, Braughton KR, Sturdevant DE, Whitney AR, Said-Salim B, Porcella SF, et al. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J Immunol (2005) 175(6):3907–19. doi: 10.4049/jimmunol.175.6.3907
39. Kirshenbaum AS, Akin C, Wu Y, Rottem M, Goff JP, Beaven MA, et al. Characterization of novel stem cell factor responsive human mast cell lines LAD 1 and 2 established from a patient with mast cell sarcoma/leukemia; activation following aggregation of FcepsilonRI or FcgammaRI. Leuk Res (2003) 27(8):677–82. doi: 10.1016/S0145-2126(02)00343-0
40. Wang R, Braughton KR, Kretschmer D, Bach TH, Queck SY, Li M, et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med (2007) 13(12):1510–4. doi: 10.1038/nm1656
41. Kuehn HS, Radinger M, Gilfillan AM. Measuring mast cell mediator release. Curr Protoc Immunol (2010) 7:38. doi: 10.1002/0471142735.im0738s91
42. Wershil BK, Wang ZS, Gordon JR, Galli SJ. Recruitment of neutrophils during IgE-dependent cutaneous late phase reactions in the mouse is mast cell-dependent. partial inhibition of the reaction with antiserum against tumor necrosis factor-alpha. J Clin Invest (1991) 87(2):446–53. doi: 10.1172/JCI115016
43. Chatterjee SS, Joo HS, Duong AC, Dieringer TD, Tan VY, Song Y, et al. Essential Staphylococcus aureus toxin export system. Nat Med (2013) 19(3):364–7. doi: 10.1038/nm.3047
44. Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev (1983) 47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983
45. Traber KE, Lee E, Benson S, Corrigan R, Cantera M, Shopsin B, et al. Agr function in clinical Staphylococcus aureus isolates. Microbiol (Reading) (2008) 154(Pt 8):2265–74. doi: 10.1099/mic.0.2007/011874-0
46. Queck SY, Jameson-Lee M, Villaruz AE, Bach TH, Khan BA, Sturdevant DE, et al. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol Cell (2008) 32(1):150–8. doi: 10.1016/j.molcel.2008.08.005
47. Cheung GY, Rigby K, Wang R, Queck SY, Braughton KR, Whitney AR, et al. Staphylococcus epidermidis strategies to avoid killing by human neutrophils. PloS Pathog (2010) 6(10):e1001133. doi: 10.1371/journal.ppat.1001133
48. Kretschmer D, Gleske AK, Rautenberg M, Wang R, Koberle M, Bohn E, et al. Human formyl peptide receptor 2 senses highly pathogenic Staphylococcus aureus. Cell Host Microbe (2010) 7(6):463–73. doi: 10.1016/j.chom.2010.05.012
49. Kretschmer D, Nikola N, Durr M, Otto M, Peschel A. The virulence regulator agr controls the staphylococcal capacity to activate human neutrophils via the formyl peptide receptor 2. J Innate Immun (2012) 4(2):201–12. doi: 10.1159/000332142
50. Da F, Joo HS, Cheung GYC, Villaruz AE, Rohde H, Luo X, et al. Phenol-soluble modulin toxins of Staphylococcus haemolyticus. Front Cell Infect Microbiol (2017) 7:206. doi: 10.3389/fcimb.2017.00206
51. Otto M. Staphylococcus aureus toxins. Curr Opin Microbiol (2014) 17:32–7. doi: 10.1016/j.mib.2013.11.004
52. Qi R, Joo HS, Sharma-Kuinkel B, Berlon NR, Park L, Fu CL, et al. Increased in vitro phenol-soluble modulin production is associated with soft tissue infection source in clinical isolates of methicillin-susceptible Staphylococcus aureus. J Infect (2016) 72(3):302–8. doi: 10.1016/j.jinf.2015.11.002
53. Cassat JE, Hammer ND, Campbell JP, Benson MA, Perrien DS, Mrak LN, et al. A secreted bacterial protease tailors the Staphylococcus aureus virulence repertoire to modulate bone remodeling during osteomyelitis. Cell Host Microbe (2013) 13(6):759–72. doi: 10.1016/j.chom.2013.05.003
54. Nakagawa S, Matsumoto M, Katayama Y, Oguma R, Wakabayashi S, Nygaard T, et al. Staphylococcus aureus virulent PSMalpha peptides induce keratinocyte alarmin release to orchestrate IL-17-Dependent skin inflammation. Cell Host Microbe (2017) 22(5):667–77.e5. doi: 10.1016/j.chom.2017.10.008
55. Nakamura Y, Takahashi H, Takaya A, Inoue Y, Katayama Y, Kusuya Y, et al. Staphylococcus agr virulence is critical for epidermal colonization and associates with atopic dermatitis development. Sci Transl Med (2020) 12(551). doi: 10.1126/scitranslmed.aay4068
56. Otto M. Virulence factors of the coagulase-negative staphylococci. Front Biosci (2004) 9:841–63. doi: 10.2741/1295
57. Otto M. Staphylococcus epidermidis–the ’accidental’ pathogen. Nat Rev Microbiol (2009) 7(8):555–67. doi: 10.1038/nrmicro2182
58. Otto M. Phenol-soluble modulins. Int J Med Microbiol (2014) 304(2):164–9. doi: 10.1016/j.ijmm.2013.11.019
Keywords: Staphylococcus xylosus, Staphylococcus aureus, phenol-soluble modulin, delta-toxin, virulence, cytolysin, atopic dermatitis
Citation: Reshamwala K, Cheung GYC, Hsieh RC, Liu R, Joo H-S, Zheng Y, Bae JS, Nguyen TH, Villaruz AE, Gozalo AS, Elkins WR and Otto M (2022) Identification and characterization of the pathogenic potential of phenol-soluble modulin toxins in the mouse commensal Staphylococcus xylosus. Front. Immunol. 13:999201. doi: 10.3389/fimmu.2022.999201
Received: 20 July 2022; Accepted: 25 August 2022;
Published: 15 September 2022.
Edited by:
René Köffel, University of Bern, SwitzerlandReviewed by:
Teruaki Nakatsuji, University of California, San Diego, United StatesBas G. J. Surewaard, University of Calgary, Canada
Copyright © 2022 Reshamwala, Cheung, Hsieh, Liu, Joo, Zheng, Bae, Nguyen, Villaruz, Gozalo, Elkins and Otto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Otto, bW90dG9AbmlhaWQubmloLmdvdg==; Gordon Y. C. Cheung, Y2hldW5nZ29AbmlhaWQubmloLmdvdg==
†These authors have contributed equally to this work
‡Present address: Hwang-Soo Joo, Department of Biotechnology, College of Engineering, Duksung Women’s University, Seoul, South Korea;Yue Zheng, Innovent Biologics Inc, Rockville, MD, United States;Justin S. Bae, Harvard University, Cambridge, MA, United States;Thuan H. Nguyen, University of Maryland, MD, United States
 Kunal Reshamwala
Kunal Reshamwala Gordon Y. C. Cheung
Gordon Y. C. Cheung Roger C. Hsieh
Roger C. Hsieh Ryan Liu
Ryan Liu Hwang-Soo Joo
Hwang-Soo Joo Yue Zheng1‡
Yue Zheng1‡ Justin S. Bae
Justin S. Bae Michael Otto
Michael Otto