
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 20 September 2022
Sec. Parasite Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.998756
This article is part of the Research Topic Host Immune Response and Regulation to Parasitic Infections: Therapeutic Approaches and Defence Strategies View all 9 articles
 Luoluo Wang1†
Luoluo Wang1† Jian Wu2†
Jian Wu2† Runzhou Liu1†
Runzhou Liu1† Wenjun Chen3
Wenjun Chen3 Zhichang Pang1
Zhichang Pang1 Fan Zhou1
Fan Zhou1 Lu Xia2,4
Lu Xia2,4 Jia Huang1
Jia Huang1 Tao Pan5
Tao Pan5 Xin-zhuan Su2*
Xin-zhuan Su2* Xiaoyun Wang1*
Xiaoyun Wang1*N6-Methyladenosine (m6A), the most abundant mammalian mRNA modification, has been reported to modulate various viral infections. Although it has been confirmed that RNA modifications can also modulate the replication and development of different parasites, the role of the RNA epitranscriptome in the regulation of host response post parasite infection remains to be elucidated. Here we report host spleen m6A epitranscriptome landscapes induced by different strains of the malaria parasite Plasmodium yoelii. We found that malaria parasite infection dramatically changes host spleen m6A mRNA modification and gene expression. Additionally, malaria parasite infection reprograms host immune response pathways by regulating the m6A modification enzymes. Collectively, our study is the first characterization of host spleen m6A methylome triggered by malaria parasite infections, and our data identify m6A modifications as significant transcriptome-wide marks during host-parasite interactions. We demonstrate that host mRNA methylation machinery can sense and respond to malaria parasite infections, and provide new insights into epitranscriptomic mechanisms underlying parasite-induced pathogenesis.
N6-Methyladenosine (m6A), the most pervasive mRNA modification in mammals, can affect all aspects of mRNA biology, including RNA stability, splicing, decay, and translation (1, 2). The biological functions of m6A have been associated with stem cell differentiation, immune response, cancer, and others (3). As the most abundant mRNA modification in mammals, m6A epitranscriptomic mapping and biology have been extensively studied in the past few years (4). The epitranscriptome of m6A in host cells could be altered after pathogen infections, and there are growing pieces of evidence showing that m6A modifications play important roles in various pathogen infections including host responses to infections of SARS-CoV-2 (5), HIV (6), flavivirus (7), influenza A virus (8), hepatitis B virus (9), human metapneumovirus (10), adenovirus (11), enterovirus 71 (12). The expression of m6A-modified genes in host cells could be altered after virus infections. Thus, m6A is now considered one of the predominant epitranscriptome marks in host-pathogen interactions (13). With the advance in sequencing technologies, transcriptome-wide mappings of m6A and other RNA modifications after infections of pathogens have been reported, laying the foundation for a better understanding of m6A functions and corresponding mechanisms.
Malaria is a deadly mosquito-borne disease affecting hundreds of millions of people every year and remains a significant global health challenge (14, 15). The parasites mostly reside in red blood cells (RBCs), hiding from direct contact or recognition by host immune cells. In recent years, RNA epitranscriptome has been recognized as a new posttranscriptional modulator of gene expression during malaria parasite development, rendering RNA epigenetic marks as novel vaccine candidates (16–19). For example, m6A modification was associated with P. falciparum stage-specific fine-tuning of the transcriptional cascade and could shape the parasite transcriptome profile during blood-stage development (16). More importantly, a recent study showed that NOP2/Sun RNA Methyltransferase 2 (NSUN2)-mediated mRNA m5C modifications could regulate mRNA transcript stability and sexual differentiation in Plasmodium yoelii and Plasmodium falciparum (20). Additionally, DNMT2-mediated tRNA cytosine methylation in P. falciparum is a key regulator for the response to drug treatment and sexual commitment (21). In addition to Plasmodium, m6A enrichment was reported in 342 transcripts of Trypanosoma brucei including transcripts encoding variant surface glycoproteins that are essential for the survival of the parasites (22). In Toxoplasma gondii, m6A has been reported as a critical mRNA modification widespread across multiple stages of the parasite’s life cycle and is essential for parasite viability and development (23, 24). Emerging evidence has provided new insights into the roles of RNA epitranscriptome in parasite biology and the treatment of the associated diseases (25).
Although the role of m6A modification in the development of malaria and other parasites has been demonstrated, the host m6A epitranscriptomic modifications and the functional consequences post malaria parasite infection remain elusive to date. In this study, we investigate genome-wide host m6A RNA modifications after malaria parasite infections and show that host RNA m6A machinery can sense and respond to malaria parasite infections including regulation of immune response-associated pathways.
Freshly thawed parasites (P. y. nigeriensis N67, P. y. nigeriensis N67C, P. y. yoelii YM, and P. y. yoelii 17XNL) were injected into C57BL/6 mice (aged 6-8 weeks) to initiate infections. An inoculum containing 1×106 iRBCs suspended in 100 μL phosphate buffer saline from the donor mice was injected into experimental mice intraperitoneally. Four days later, the spleens and red blood cells (with malaria parasites inside) from infected or uninfected mice were rapidly separated and freshly frozen on dry ice and stored at –80°C. All animal procedures in this study were performed following the protocol approved (approval number LMVR11E) by the Institutional Animal Care and Use Committee at the National Institute of Allergy and Infectious Diseases following the guidelines of the Public Health Service Policy on Humane Care and Use of Laboratory Animals and AAALAC.
Total RNA was isolated from the mice spleens and red blood cells using TRIzol (Ambion). Total RNA concentration and purity were quantified using NanoDrop 2000 Spectrophotometer (Thermo Scientific). All RNA samples were kept at –80°C until used. To obtain mRNA from mouse spleens, total RNA was purified using PolyATtract® mRNA Isolation System IV (PRZ5310, Promega) followed by the RiboMinus transcriptome isolation kit for human/mouse (K155002, Invitrogen). To obtain mRNA from the malaria parasites, total RNA was purified using PolyATtract® mRNA Isolation System IV (PRZ5310, Promega) followed by the RiboMinus™ Eukaryote Kit (A1083708, Invitrogen).
The purified mRNAs (150 ng) were digested with 2 U nuclease P1 (N8630, Sigma) in 30 μL of buffer containing 25 mM NaCl and 2.5 mM ZnCl2 for 2 h at 37°C, followed by the addition of 2 μL FastAP Thermosensitive Alkaline Phosphatase (EF0651, Thermo Scientific) and incubation at 37°C for 4 h. The samples were then filtered through a 0.22 μm PVDF filter (Millipore) and transferred into mass spectrometry tubes. Ten μL of each sample was injected into a C18 reverse phase column coupled online to Agilent 6460 LC-MS/MS spectrometer in positive electrospray ionization mode. The nucleosides were quantified by using retention time and the nucleoside to base ion mass transitions (268-to-136 for A; 282-to-150 for m6A). Quantification was performed by comparing with the standard curve obtained from pure nucleoside standards running with the same batch of samples. The m6A/A ratio was calculated based on the calibrated concentrations.
MeRIP-seq experiments were carried out according to procedures in our previous report (26). Briefly, poly(A)-selected and ribodepletion-treated mRNA samples (1.5 μg) were used for RNA fragmentation followed by immunoprecipitation with EpiMark® N6-Methyladenosine Enrichment Kit (E1610S, New England Biolabs). RNAs were eluted from protein G magnetic beads in 100 μL of Buffer RLT (79216, Qiagen) followed by precipitation. The RNAs were dissolved in 12 μL RNase-free water. Input RNAs and immunoprecipitated RNAs were used for library preparation using the TruSeq Stranded mRNA kit (RS-122-2101, Illumina) according to the manufacturer’s instructions. The concentration and quality of libraries were measured using the Agilent 2100 bioanalyzer. RNA sequencing was carried out at the University of Chicago Genomics Facility on an Illumina HiSeq2500 platform that generates 100-bp paired-end reads.
After removing adapters and low-quality bases using Cutadapt (v1.15), the Fastq files were aligned to the reference genome (mm10 and VSV) using Hisat2 (v2.1.0) (27). Reads mapped to tRNA and rRNA were removed and each sample obtained ~30 million useful reads for the following analysis.
Stringtie (v1.3.3b) (28) was used to calculate the FPKM of each gene to represent their mRNA expression level. The differentially expressed genes were identified by a negative binomial model using the DEseq2 package (29), combining information from all replicates. Significantly differentially expressed genes must meet all the following criteria: q-value ≤ 0.01, log2 (fold-change) ≥ 1 or ≤ -1. KEGG enrichment analyses of differential genes were conducted by R package clusterProfiler (v3.8.1) (30).
The m6A peak calling method was modified from published report (31). In brief, the longest isoform of each gene was scanned using a 100-bp sliding window with 10-bp steps. We excluded windows with read counts less than 1/20 of the top window in both the input and m6A-IP sample to reduce bias from potentially inaccurate gene structure annotation and the arbitrary use of the longest isoform. The read counts in each window were normalized by the median count of all windows of that gene. The differential windows between the input and IP samples were identified with a negative binomial model by using the edgeR package (32), combing information from all replicates. A positive window was called if the false discovery rate (FDR) < 0.01 and log2 (enrichment score) ≥ 1, and overlapped positive windows were merged. The following four numbers were calculated to obtain the enrichment score of each peak (or window): reads count of the IP sample in the current peak/window (a); median reads count of the IP sample in all 100-bp windows on the current mRNA (b); reads count of the input sample in the current peak/window (c); and median reads count of the input sample in all 100-bp windows on the current mRNA (d). The enrichment score of each window was calculated as (a × d)/(b × c). Sequence motifs on m6A peaks were identified using HOMER (33).
The PrimeScript RT Reagent Kit (RR047A, Takara) was used to synthesize cDNA with 1 μg of total RNA from the mouse spleen. The cDNAs were 20-fold diluted for real-time RT-PCR reaction using Hieff qPCR SYBR Green Master Mix (11202ES08, YEASEN). The fluorescence intensity of the amplification process was monitored using CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad). Primers used for RT-qPCR are listed in Table 1. Actin was used as the internal control, and the relative expression levels of target mRNAs were calculated using the 2−ΔΔCt method.
Protein samples were extracted using RIPA buffer (Thermo Fisher Scientific) with freshly added 1 mM PMSF. The lysate was supplemented with NuPage Sample Buffer (Thermo Fisher no. NP0008) and denatured at 75°C for 10 min. The samples were subjected to polyacrylamide gel electrophoresis, transferred onto a polyvinylidene difluoride membrane, and then blocked for 1 h at room temperature in 1% milk with 0.1% Tween-20 in PBS. Primary antibodies included rabbit anti-METTL14 antibody (HPA038002, Sigma, 1: 3,000), rabbit anti-METTL3 antibody (ab195352, Abcam, 1: 1,000), rabbit anti-ALKBH5 antibody (HPA007196, Sigma, 1: 1,000), mouse anti-FTO antibody (ab92821, Abcam, 1: 1,000), mouse anti-beta actin antibody (ab6276, Abcam, 1: 5,000). The membranes were probed with primary antibodies for 2 h at RT, washed 3X with blocking buffer, and with secondary antibodies conjugated to HRP (1:10,000). The HRP signal was developed using Amersham™ ECL™ Western Blotting Detection Reagents (GE Healthcare). The intensities of the protein bands were quantified using the Image Lab Software.
Data are presented as mean ± standard deviation (SD). Asterisks represent significant differences between samples, as determined by the Mann-Whitney test or Student’s t test (P < 0.05).
To investigate host response to malaria parasite infection regulated by m6A modification, we infected mice with four P. yoelii strains, including N67, N67C, YM, and 17XNL in parallel with noninfected controls. These four strains of P. yoelii trigger dramatically different disease phenotypes in mice (14). Four days post-infection, the spleens and blood samples were collected from mice for total RNA extraction and mRNA purification. We measured total m6A/A ratios of mRNA using liquid chromatography-tandem mass spectrometry (LC-MS/MS), and investigated mRNA m6A epitranscriptome profiles using m6A-MeRIP sequencing and data analysis (Figure 1A). LC-MS/MS data showed that the average m6A levels in all P. yoelii-infected host spleen mRNAs (0.52~0.55%) were significantly higher than that in noninfected mRNA (0.23%, Figure 1B). In addition, m6A levels in four P. yoelii strains were diverse (Figure 1C), with the highest level of m6A modification in 17XNL (0.56%) and the lowest level in N67C (0.23%).
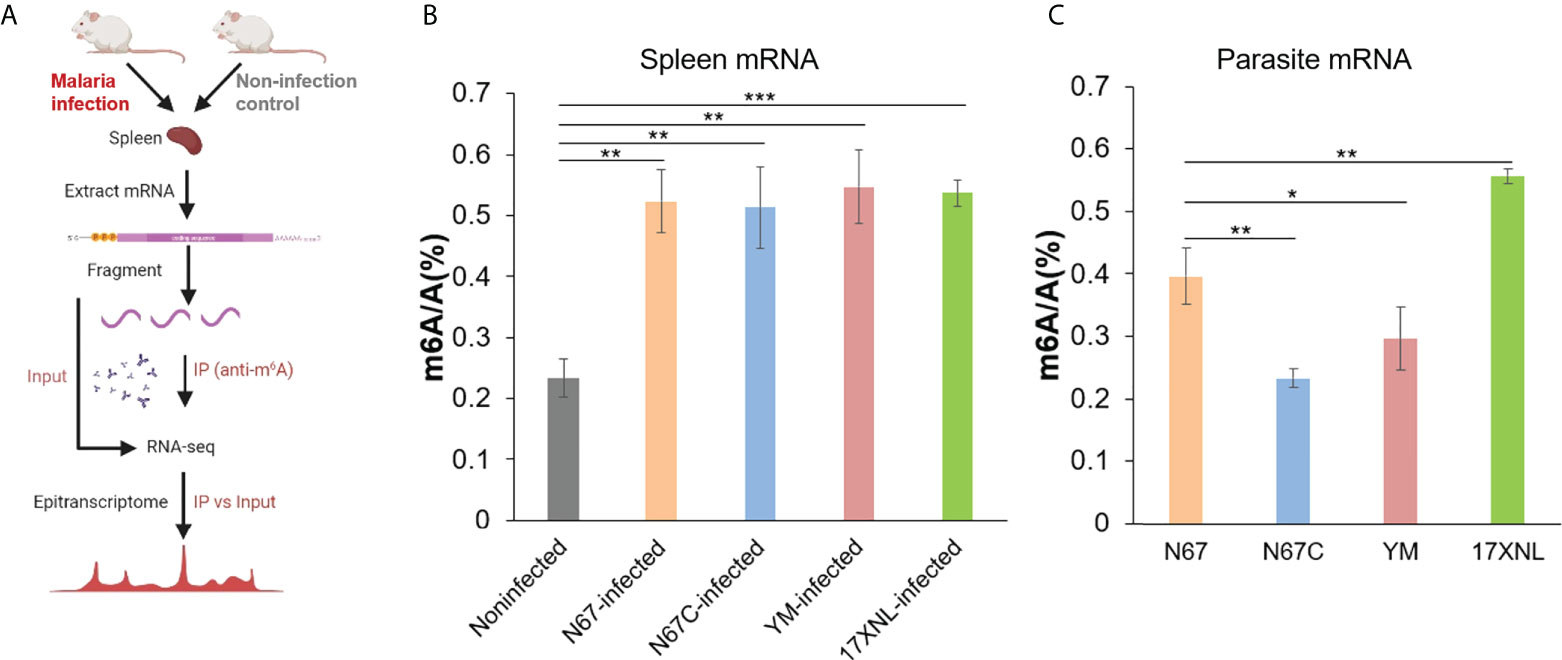
Figure 1 Malaria parasite infections in mice alter host spleen mRNA m6A methylation levels. (A) Schematic of the experimental design and MeRIP-seq protocol used in this study to identify differential m6A methylation profiles in mice spleens following infection with Plasmodium yoelii parasites. (B) LC-MS/MS quantification of m6A level in poly(A)-selected and RiboMinus-treated mRNAs isolated from the spleens of noninfected, P. y. nigeriensis N67-, P. y. nigeriensis N67C-, P. y. yoelii YM-, or P. y. yoelii 17XNL- infected mice. (C) LC-MS/MS quantification of m6A level in poly(A)-selected and RiboMinus-treated mRNAs isolated from N67, N67C, YM and 17XNL parasites. Values are the means ± SD, n = 3. *P < 0.05; **P < 0.01; ***P < 0.001, Student’s t test.
Although m6A modification has been extensively studied in mammals, the level and function of m6A modification in species such as parasites and other infectious pathogens are limited. Especially for the malaria parasites, the m6A RNA epitranscriptome was recognized as an important posttranscriptional regulator of gene expression for the parasite development very recently (16–19); however, the profiles of m6A modification levels in different malaria strains and spleens of infected-hosts remain unclear. Here, we showed that m6A modification levels in malaria parasite could be variable among strains and m6A modification levels in the host could be altered post malaria infection. Considering m6A epitranscriptome profiles induced by other pathogens in previous studies and the dramatic changes in mRNA m6A level post malaria parasite infection, we next performed m6A-MeRIP sequencing to obtain transcriptome-wide maps of m6A modifications. For the sequencing in this study, we focused on the spleen mRNA m6A profiles induced by N67 (N67Infected) and N67C (N67CInfected) in comparison with noninfected control due to their unique disease phenotypes and their highly similar genome sequences.
From the m6A MeRIP-seq data analysis, we identified 14319, 16835, and 6986 m6A peaks for N67Infected, N67CInfected, and noninfected spleens, respectively (Figure 2A). Approximately 80% of differential peaks were induced by malaria parasite infection, and only 20% of the m6A peaks in the infected samples (22.58% for N67Infected and 19.29% for N67CInfected) overlapped with noninfected samples (Figure 2B). Correspondingly, 4581 and 5284 m6A peak-containing genes were mapped to mouse spleens infected with N67- and N67C-, respectively (Figure 2C). Among them, 3848 m6A peak-containing genes were shared by N67- and N67C-infected samples, and 733 and 1436 m6A peak-containing genes were unique for N67- and N67C-infected samples, respectively (Figure 2D). We next performed epitranscriptome analysis to investigate the sequence features of m6A modification post malaria parasite infections. The known m6A motif RRACH (where R represents G or A; H represents A, C, or U), particularly GGACU, was enriched in the identified peaks in the three groups of samples (Figure 2E). The peak distribution analysis showed that N67-/N67C- infection lead to an obvious increase in m6A peak density in the regions of 5’UTR and CDS, but a decrease in the 3’UTR region (Figure 2F). A detailed presentation of m6A peak distribution confirmed that malaria parasite infection induced a dramatic increase of mRNA m6A modification around 5’UTR, start codon, and CDS (Figures 2G–I). These results demonstrate that malaria parasite infections in mice can dramatically alter host spleen mRNA m6A methylation levels and m6A methylome profiles.
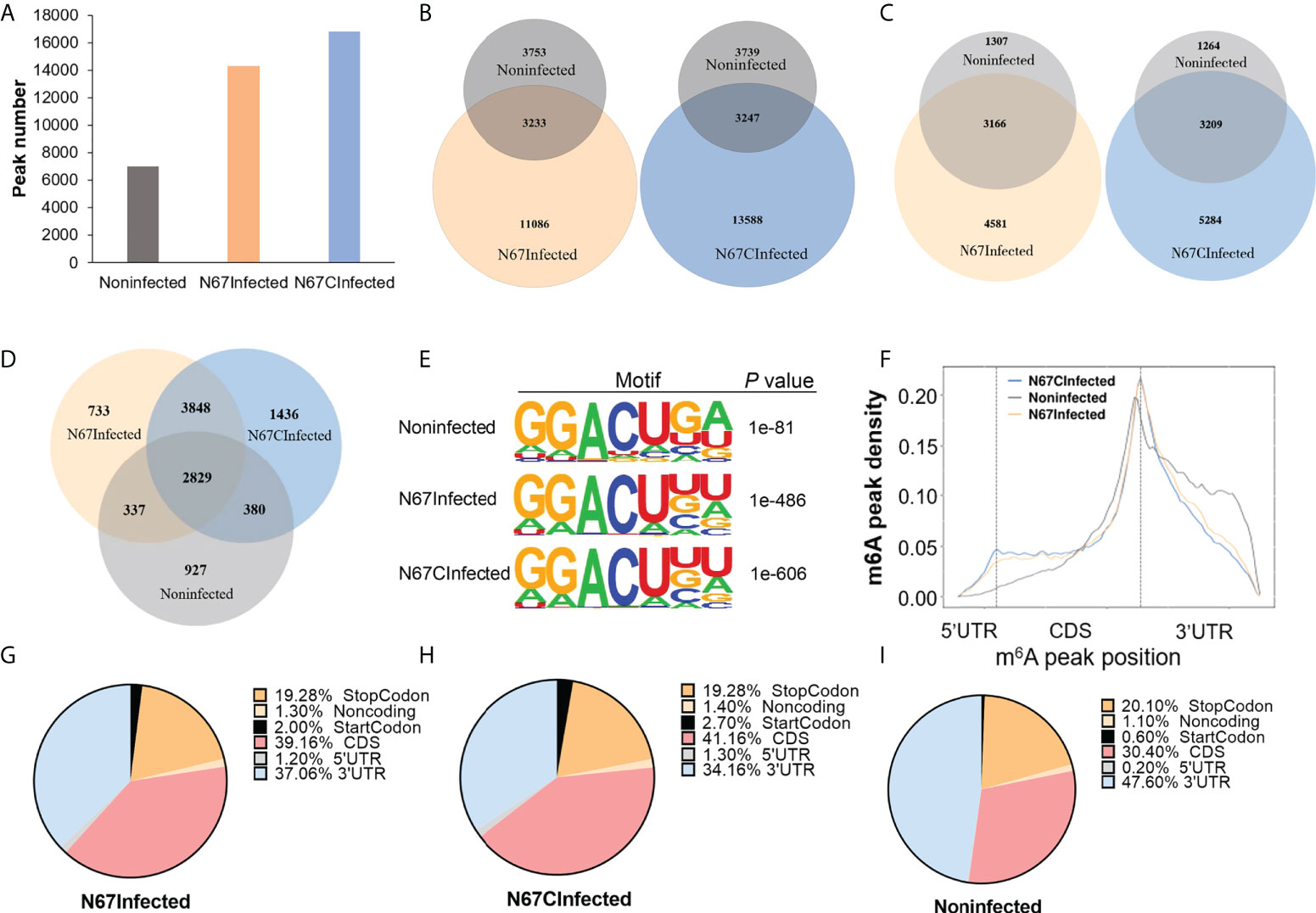
Figure 2 Malaria parasite infections in mice alter host spleen mRNA m6A methylome profiles. (A) m6A peak number identified from MeRIP in N67-, N67C-infected or noninfected spleens. (B) Venn diagrams showing the differences and overlaps of m6A peaks identified from MeRIP between N67- or N67C-infected and noninfected spleens, respectively. (C) Venn diagrams showing the differences and overlaps of m6A-containing genes between N67- or N67C-infected and noninfected spleens, respectively. (D) Venn diagram showing the differences and overlaps of m6A-containing genes among N67-, N67C-infected and noninfected spleens. (E) Consensus motifs and corresponding P values of m6A peaks identified by HOMER from m6A-seq analysis. (F) m6A peak distribution across the mRNA regions (5’UTR, CDS, 3’UTR). m6A peaks were mapped to the corresponding gene and plotted according to their position. (G) Pie chart showing the proportion of m6A peaks in different mRNA regions of N67-infected mice spleens. (H) Pie chart showing the proportion of m6A peaks in different mRNA regions of N67C-infected mice spleens. (I) Pie chart showing the proportion of m6A peaks in different mRNA regions of noninfected mice spleens.
Next, we analyzed overall gene expression patterns in mice spleens post malaria parasite infection using Input samples. Principal component analysis showed that both Input samples and IP samples for N67Infected (Figure S1A) and N67C-infected samples (Figure S1B) could be separated from noninfected samples. Volcano plots and heap maps showed that large members of genes were differentially expressed between N67Infected and noninfected samples (Figure S2A, Figure S3A) or between N67CInfected and noninfected samples (Figure S2B, Figure S3A). Although the gene expression patterns were similar between N67Infected and N67CInfected samples, there were still hundreds of differentially expressed genes (Figures S2C, S3B). Gene Ontology analysis using differentially expressed genes showed that a substantial group of pathways related to innate immune responses were enriched upon malaria infection both for N67- (Figure S4A) and N67C-infected (Figure S4B) samples, which was consistent with a previous report (14).
To decipher the role of m6A modifications in the host response to malaria parasite infection, we further performed an in-depth m6A epitranscriptome analysis using m6A-IP samples. First, the transcripts with various numbers of m6A peaks are different in three groups. For example, among 7747, 8493, and 4473 m6A-containing transcripts in three groups, 3972, 3994, and 2782 transcripts have a single m6A peak in N67Infected, N67CInfected and noninfected samples, respectively (Figure 3A). We observed that the ratio for both N67-/noninfected and N67C-/noninfected was about 1.4 for one peak-containing transcript, while there were higher ratios of transcripts that were multi-methylated (e.g., 2.4 and 3.0 for N67Infected and N67CInfected samples, respectively, Figure 3A). Moreover, both N67Infected and N67CInfected samples had a broader distribution of m6A peak/exon ratio compared to the noninfected samples (Figure 3B). We also analyzed the expression level of transcripts with m6A modifications, and the results showed that the abundance of m6A-containing transcripts was significantly lower in N67- and N67C- infected samples than in noninfected samples (Figure 3C). Overall, malaria infection-induced m6A modifications showed a negative effect on host gene expression, considering the higher level of m6A modification in N67- or N67C- infected samples compared to noninfected samples. We further analyzed the expression level of genes with malaria infection-induced m6A peaks enriched in different mRNA regions (Figures 3D, E). The results showed that m6A peaks enriched in CDS had a negative effect on gene expression, while m6A peaks enriched in 3’UTR had a positive effect on gene expression. Our results suggested that malaria-induced m6A modification in 3’UTR may increase mRNA stability leading to higher mRNA level.
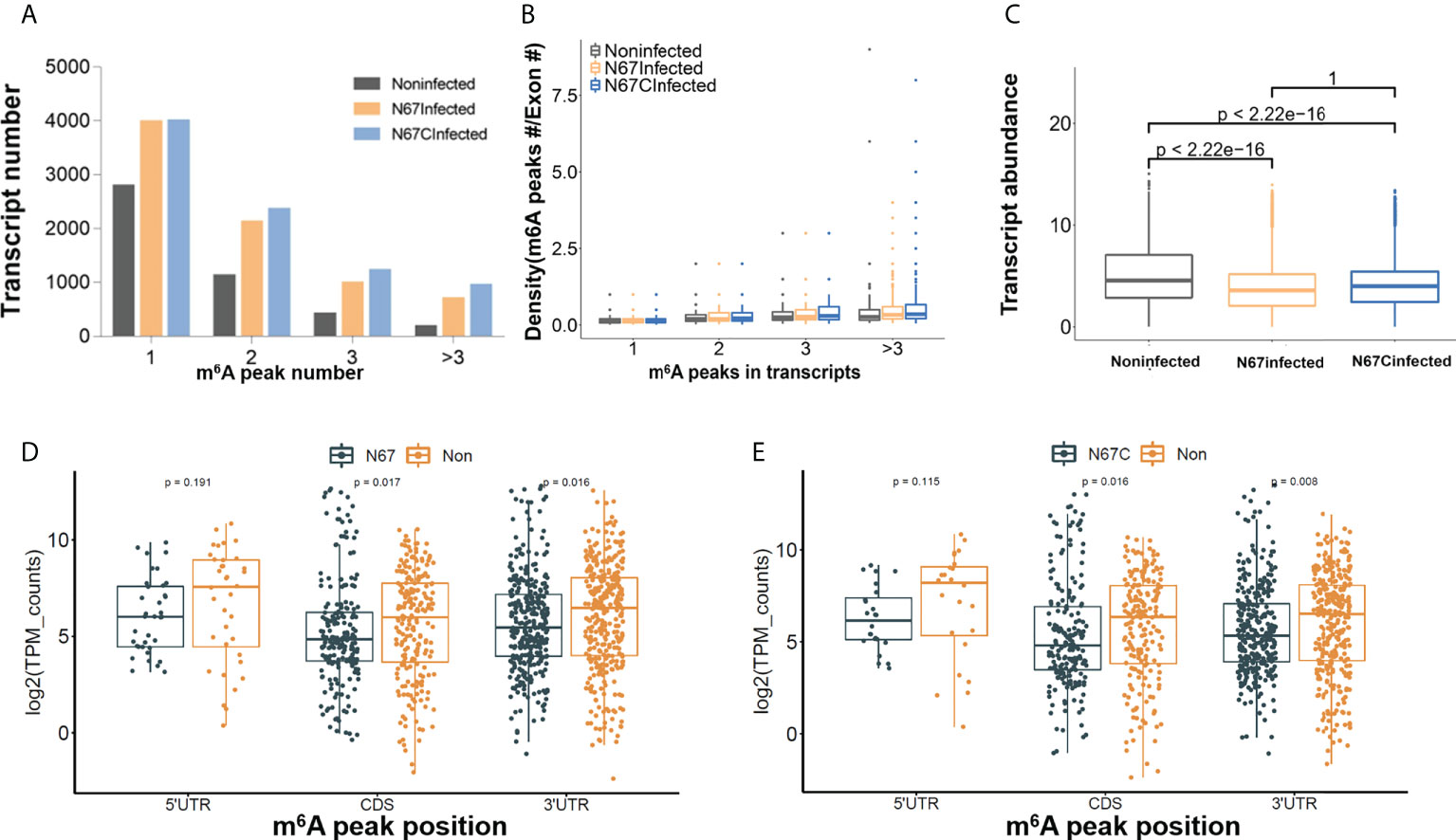
Figure 3 Malaria parasite infection-induced m6A modifications impact host gene expression patterns. (A) Total number of transcripts containing different m6A sites in N67- or N67C-infected or noninfected spleens, respectively. (B) Plots of m6A peaks within exons in N67- or N67C-infected or noninfected spleens, respectively. (C) Box plots showing the abundance of m6A-containing transcripts in N67-, N67C-infected or noninfected spleens, respectively. Transcript abundance was shown in log2 transform. (D) Boxplot showing expression level of genes with peaks enriched in 5’UTR, CDS and 3’UTR in N67Infected and noninfected samples. (E) Boxplots showing expression level of genes with peaks enriched in 5’UTR, CDS and 3’UTR in N67CInfected and noninfected samples. R package ggboxplot was used for plots and Kruskal-Wallis test was used to show the significance between groups.
The overall effect of m6A modifications showed a negative effect on host spleen gene expression (Figure 3C), some immune response-related genes were still shown to be positively regulated by m6A modification. We have performed RT-qPCR and validated our sequencing results with specific primers targeting interferon pathway-related genes (Irf7, Ifngr2, Ifnar2, Irf1, Ifit2) that were previously reported in anti-parasite defense (14, 15). Those immune response-related genes gained m6A peaks but the expression levels were still upregulated in N67- (Figure 4A) or N67CInfected (Figure 4B) samples compared to noninfected control. For example, Ifit2 is an interferon-stimulated gene with well-established antimicrobial activity through binding to and enhancing the translation efficiency of host mRNAs by suppressing ribosome pausing (34). Representative read coverage plots were shown for Ifit2 (Figure 4C), this gene gained an obvious m6A peak in 3’UTR in N67CInfected samples and the expression level was dramatically up-regulated by N67C infection (Figure 4B), consistent with the data that m6A enriched in 3’UTR had a positive effect on gene expression (Figure 3E). A recent study showed that the translation of interferon-induced genes could be enhanced after m6A modification as the antiviral mechanism (35). Thus, it is conceivable that m6A modification on these genes may have a similar role in the immune response to malaria infections.
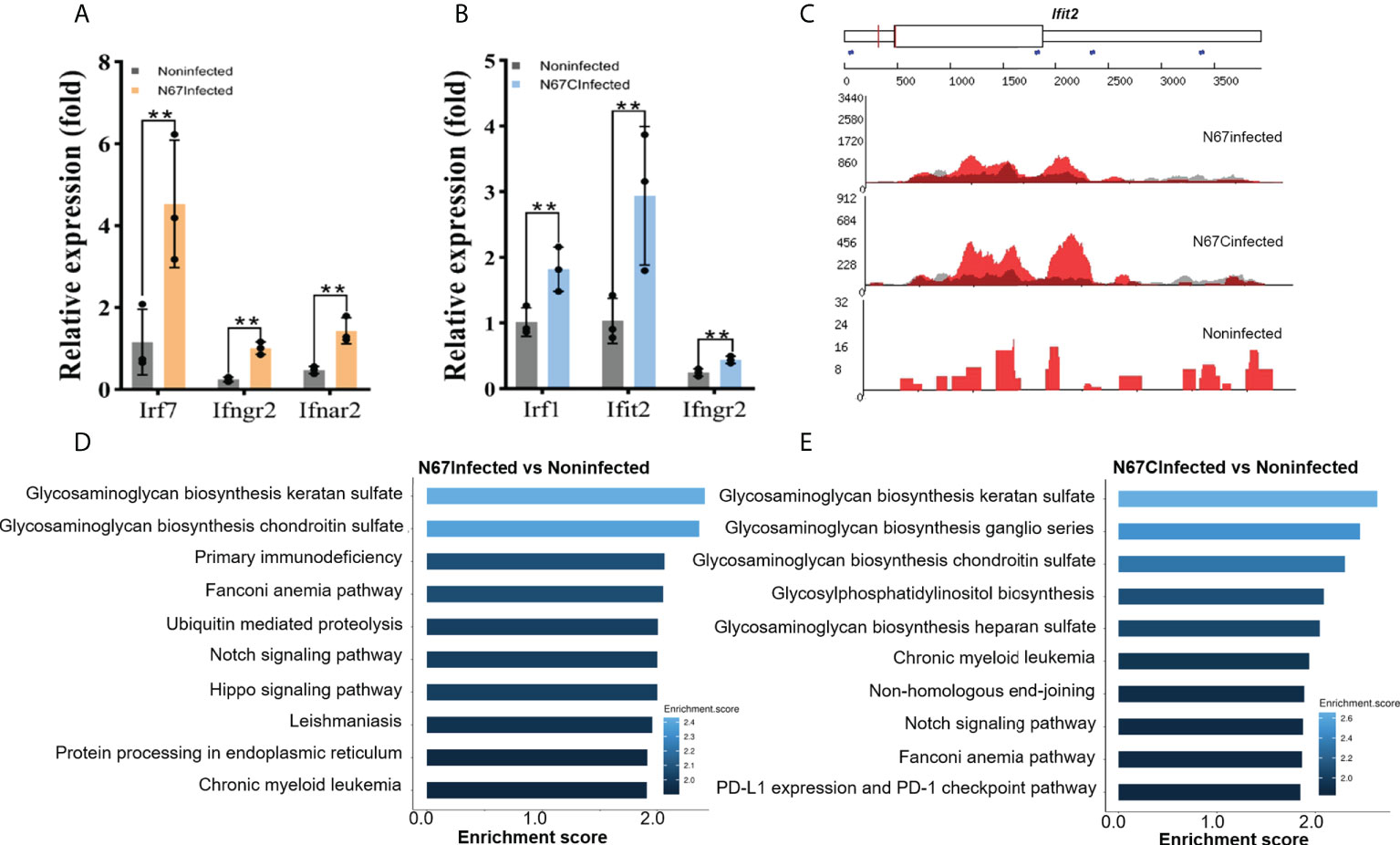
Figure 4 Malaria parasite infection-induced m6A modifications reprogram host immune response pathways. (A) Transcript expression of interferon stimulated genes from N67Infected and noninfected spleens, respectively. (B) Transcript expression of interferon stimulated genes from N67CInfected and noninfected spleens, respectively. Values are the means ± SD, n=3. **P < 0.01, Student’s t test. (C) Integrative genomics viewer (IGV) plots of m6A-IP (red) and Input (grey) showing m6A peaks of Ifit2 in N67-, N67C-infected or noninfected spleens. The Y axis represents the normalized m6A signal along the gene. (D) Biological processes of differentially expressed genes with m6A peaks in N67-infected mice spleens compared to noninfected mice spleens. (E) Biological processes of differentially expressed genes with m6A peaks in N67C-infected mice spleens compared to noninfected mice spleens.
We further performed Gene Ontology analysis using m6A-marked genes. Interestingly, we found that glycosaminoglycans (GAGs) biosynthesis pathways were significantly enriched for m6A-modified genes in both N67Infected (Figure 4D) and N67CInfected samples (Figure 4E). GAGs are complex carbohydrates ubiquitously present on the cell surface and in the extracellular matrix, and increasing evidence indicates a key role for GAGs in the invasion of various parasitic pathogens such as Toxoplasma, Plasmodium, and Trypanosoma parasites (36). Therefore, our results suggest that GAGs might be the prime targets of malaria parasites through the mechanism of m6A modifications, which requires further investigation.
Since the two parasite strains N67 and N67C can cause different disease phenotypes though have similar genome sequences (>99% identity) (37), we tried to explore the potential differences at m6A epitranscriptome level. We performed Gene Ontology analysis using m6A-modified genes which are unique for N67 (733 genes) or N67C (1436 genes). The pathways with the highest confidence were “establishment of protein localization to organelle” and “morphogenesis of branching epithelium” for the N67 strain and N67C strain, respectively (Figures 5A, B). Whether m6A modification plays strain-specific roles in regulating different phenotypes through those strain-unique m6A-modified genes and associated pathways requires further investigation. We also performed KEGG analysis using malaria infection-gained m6A-modified genes (Table 2) and found that most of the enriched pathways from KEGG analysis were related to pathogen infections (Figure 5C), leading to the support that malaria parasite infection-induced m6A modifications indeed influence host immune responses.
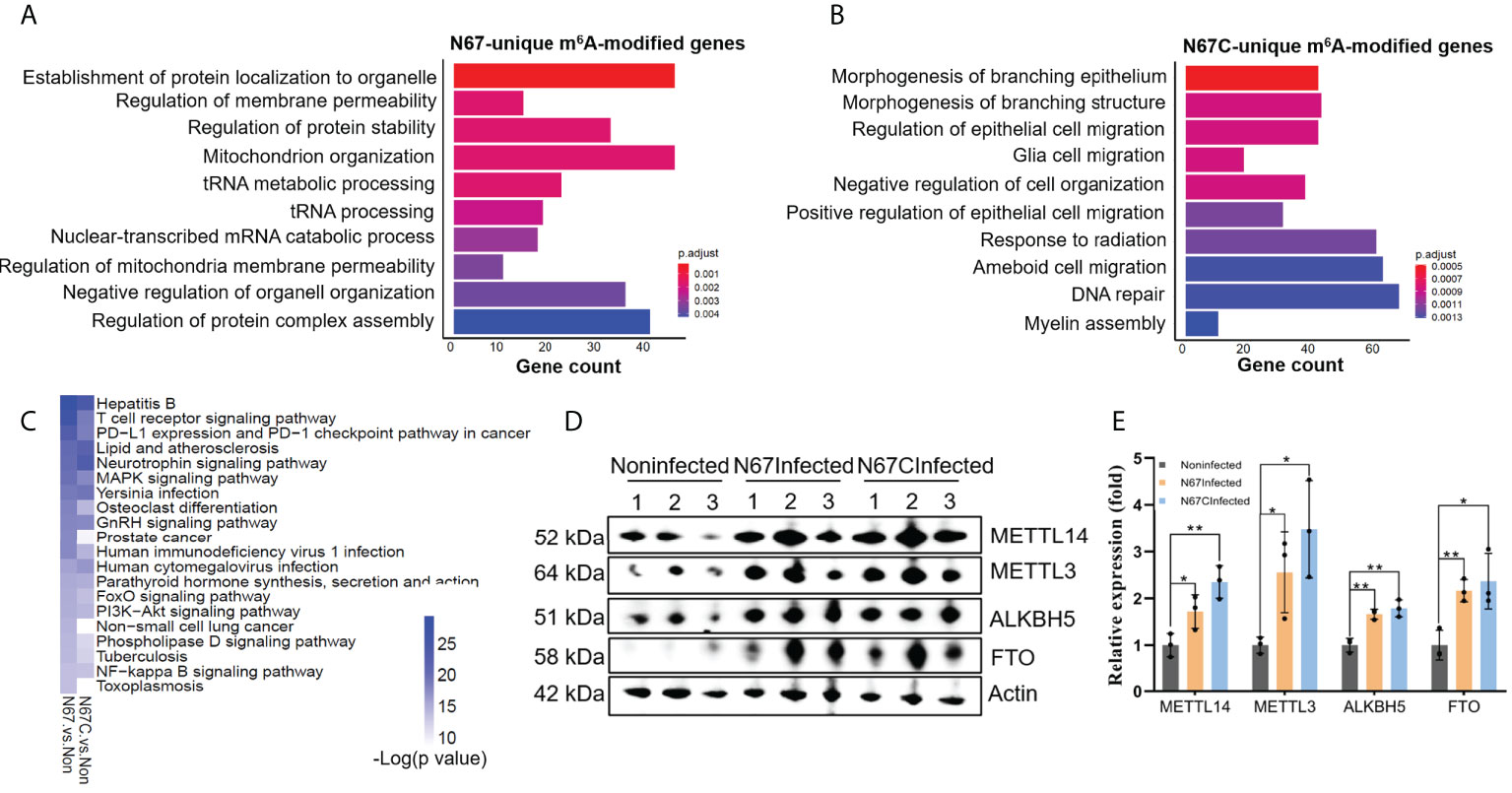
Figure 5 Strain-specific immune response pathways regulated by m6A modifications. (A) Biological processes of 733 N67-unique m6A-modified genes. Shown are the top 10 enriched pathways. (B) Biological processes of 1436 N67C-unique m6A-modified genes. Shown are the top 10 enriched pathways. (C) KEGG overlapping pathways of genes gained m6A peaks post malaria infections. Shown are the top twenty enriched pathways with low p-value for N67/noninfected and N67C/noninfected comparisons. (D) Western blots showing expression levels of m6A writer proteins METTL3, METTL14, and eraser proteins FTO, ALKBH5 in spleens of N67-, N67C-infected or noninfected mice. (E) Relative expression levels of m6A methylation machinery proteins. Signal intensity relative to β-actin was determined using Image (J). Values are the means ± SD, n=3. *P < 0.05, **P < 0.01, Student’s t test.
We tried to explore the potential mechanism of m6A modifications in regulating host immune response to malaria infection. In mammals, dynamic m6A modification is maintained by the enzyme complex containing the METTL3 and METTL14 proteins, and two eraser enzymes of FTO and ALKBH5 (26). To obtain a mechanistic understanding of m6A changes in malaria parasite-infected mice, we measured the protein levels of m6A writer proteins METTL3 and METTL14, and the m6A eraser proteins FTO and ALKBH5 using western blot (Figures 5D, E, Figure S5). We found that both m6A writer proteins and m6A eraser proteins were highly overexpressed in the N67-/N67C-infected samples compared to that in noninfected ones. In our study, the simultaneous overexpression of the m6A writer complex and the erasers in N67- and N67C-infected samples suggests that mRNA m6A modification plays a key role in the regulation of host immune function and response to parasite infection.
In summary, we have characterized the dynamic m6A mRNA methylation profiles in mice infected with different P. yoelii strains. Our results show that m6A is a crucial mechanism of post-transcriptional regulation during malaria parasite infections. Malaria parasite infection dramatically changes the host m6A mRNA modification profile and gene expression in the spleen by regulating the m6A modification enzymes. Future studies include delineating specific contributions and signaling mechanisms of key molecules that regulate host m6A methylome during malaria parasite infections, which may help develop vaccines or drugs to combat malaria. For instance, small molecule drugs targeting enzymes of the m6A machinery or genes modified by malaria-induced m6A-machinery may activate or enhance host immune responses to control malaria infections.
The RNA-seq and m6A-seq data generated by this study have been deposited in the NCBI GEO database under the accession number GSE150546.
All animal procedures in this study were performed following the protocol approved (approval number LMVR11E) by the Institutional Animal Care and Use Committee at the National Institute of Allergy and Infectious Diseases following the guidelines of the Public Health Service Policy on Humane Care and Use of Laboratory Animals and AAALAC.
X-zS and XW designed the research project. LW, JW, WC, ZP, FZ, LX, and XW performed the experiments. LW, RL, JH, and XW contributed to the data analysis. LW, JW, TP, X-zS, and XW wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China (32070615, 81902093), the National Institutes of Health of the USA (K01 DK111764), the fellowship of the China Postdoctoral Science Foundation (2020M672676), and the Guangdong Project of Basic and Applied Basic Research (20201910240000150), and partially by the Division of Intramural Research at National Institute of Allergy and Infectious Diseases (NIAID) in National Institutes of Health (NIH), USA. XW is a recipient of Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2019).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.998756/full#supplementary-material
1. Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell (2015) 161:1388–99. doi: 10.1016/j.cell.2015.05.014
2. Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet (2014) 15:293–306. doi: 10.1038/nrg3724
3. Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell (2017) 169:1187–200. doi: 10.1016/j.cell.2017.05.045
4. Meyer KD, Jaffrey SR. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol (2014) 15:313–26. doi: 10.1038/nrm3785
5. Liu J, Xu YP, Li K, Ye Q, Zhou HY, Sun H, et al. The m(6)A methylome of SARS-CoV-2 in host cells. Cell Res (2021) 31:404–14. doi: 10.1038/s41422-020-00465-7
6. Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, et al. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol (2016) 1:16011. doi: 10.1038/nmicrobiol.2016.11
7. Gokhale NS, McIntyre ABR, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, et al. N6-methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe (2016) 20:654–65. doi: 10.1016/j.chom.2016.09.015
8. Courtney DG, Kennedy EM, Dumm RE, Bogerd HP, Tsai K, Heaton NS, et al. Epitranscriptomic enhancement of influenza a virus gene expression and replication. Cell Host Microbe (2017) 22:377–386 e5. doi: 10.1016/j.chom.2017.08.004
9. Imam H, Khan M, Gokhale NS, McIntyre ABR, Kim GW, Jang JY, et al. N6-methyladenosine modification of hepatitis b virus RNA differentially regulates the viral life cycle. Proc Natl Acad Sci U.S.A. (2018) 115:8829–34.
10. Lu M, Zhang Z, Xue M, Zhao BS, Harder O, Li A, et al. N(6)-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat Microbiol (2020) 5:584–98. doi: 10.1038/s41564-019-0653-9
11. Price AM, Hayer KE, McIntyre ABR, Gokhale NS, Abebe JS, Della Fera AN, et al. Direct RNA sequencing reveals m(6)A modifications on adenovirus RNA are necessary for efficient splicing. Nat Commun (2020) 11:6016. doi: 10.1038/s41467-020-19787-6
12. Hao H, Hao S, Chen H, Chen Z, Zhang Y, Wang J, et al. N6-methyladenosine modification and METTL3 modulate enterovirus 71 replication. Nucleic Acids Res (2019) 47:362–74. doi: 10.1093/nar/gky1007
13. Winkler R, Gillis E, Lasman L, Safra M, Geula S, Soyris C, et al. m(6)A modification controls the innate immune response to infection by targeting type I interferons. Nat Immunol (2019) 20:173–82. doi: 10.1038/s41590-018-0275-z
14. Wu J, Tian L, Yu X, Pattaradilokrat S, Li J, Wang M, et al. Strain-specific innate immune signaling pathways determine malaria parasitemia dynamics and host mortality. Proc Natl Acad Sci U.S.A. (2014) 111:E511–20. doi: 10.1073/pnas.1316467111
15. Wu J, Xia L, Yao X, Yu X, Tumas KC, Sun W, et al. The E3 ubiquitin ligase MARCH1 regulates antimalaria immunity through interferon signaling and T cell activation. Proc Natl Acad Sci U.S.A. (2020) 117:16567–78. doi: 10.1073/pnas.2004332117
16. Baumgarten S, Bryant JM, Sinha A, Reyser T, Preiser PR, Dedon PC, et al. Transcriptome-wide dynamics of extensive m(6)A mRNA methylation during plasmodium falciparum blood-stage development. Nat Microbiol (2019) 4:2246–59. doi: 10.1038/s41564-019-0521-7
17. Sinha A, Baumgarten S, Distiller A, McHugh E, Chen P, Singh M, et al. Functional characterization of the m(6)A-dependent translational modulator PfYTH.2 in the human malaria parasite. mBio (2021) 12:e00661-21. doi: 10.1128/mBio.00661-21
18. Govindaraju G, Kadumuri RV, Sethumadhavan DV, Jabeena CA, Chavali S, Rajavelu A. N(6)-adenosine methylation on mRNA is recognized by YTH2 domain protein of human malaria parasite plasmodium falciparum. Epigenet Chromatin (2020) 13:33. doi: 10.1186/s13072-020-00355-7
19. Govindaraju G, Chavali S, Rajavelu A. Plasmodium falciparum YTH2 domain binds to m6A-containing mRNA and regulates translation. mBio (2021) 12:e0136721. doi: 10.1128/mBio.01367-21
20. Liu M, Guo G, Qian P, Mu J, Lu B, He X, et al. 5-methylcytosine modification by plasmodium NSUN2 stabilizes mRNA and mediates the development of gametocytes. Proc Natl Acad Sci U.S.A. (2022) 119:e2110713119. doi: 10.1073/pnas.2110713119
21. Hammam E, Sinha A, Baumgarten S, Nardella F, Liang J, Miled S, et al. Malaria parasite stress tolerance is regulated by DNMT2-mediated tRNA cytosine methylation. mBio (2021) 12:e0255821. doi: 10.1128/mBio.02558-21
22. Viegas IJ, de Macedo JP, Serra L, De Niz M, Temporao A, Silva Pereira S, et al. N(6)-methyladenosine in poly(A) tails stabilize VSG transcripts. Nature (2022) 604:362–70. doi: 10.1038/s41586-022-04544-0
23. Holmes MJ, Padgett LR, Bastos MS, Sullivan WJ Jr. m6A RNA methylation facilitates pre-mRNA 3'-end formation and is essential for viability of toxoplasma gondii. PloS Pathog (2021) 17:e1009335. doi: 10.1371/journal.ppat.1009335
24. Farhat DC, Bowler MW, Communie G, Pontier D, Belmudes L, Mas C, et al. A plant-like mechanism coupling m6A reading to polyadenylation safeguards transcriptome integrity and developmental gene partitioning in toxoplasma. Elife (2021) 10:e68312. doi: 10.7554/eLife.68312.sa2
25. Maran SR, de Lemos Padilha Pitta JL, Dos Santos Vasconcelos CR, McDermott SM, Rezende AM, Silvio Moretti N. Epitranscriptome machinery in trypanosomatids: New players on the table? Mol Microbiol (2021) 115:942–58. doi: 10.1111/mmi.14688
26. Wang X, Li Y, Chen W, Shi H, Eren AM, Morozov A, et al. Transcriptome-wide reprogramming of N(6)-methyladenosine modification by the mouse microbiome. Cell Res (2019) 29:167–70. doi: 10.1038/s41422-018-0127-2
27. Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods (2015) 12:357–60. doi: 10.1038/nmeth.3317
28. Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol (2015) 33:290–5. doi: 10.1038/nbt.3122
29. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol (2014) 15:550. doi: 10.1186/s13059-014-0550-8
30. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an r package for comparing biological themes among gene clusters. OMICS (2012) 16:284–7. doi: 10.1089/omi.2011.0118
31. Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc (2013) 8:176–89. doi: 10.1038/nprot.2012.148
32. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics (2010) 26:139–40. doi: 10.1093/bioinformatics/btp616
33. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and b cell identities. Mol Cell (2010) 38:576–89. doi: 10.1016/j.molcel.2010.05.004
34. Tran V, Ledwith MP, Thamamongood T, Higgins CA, Tripathi S, Chang MW, et al. Influenza virus repurposes the antiviral protein IFIT2 to promote translation of viral mRNAs. Nat Microbiol (2020) 5:1490–503. doi: 10.1038/s41564-020-0778-x
35. McFadden MJ, McIntyre ABR, Mourelatos H, Abell NS, Gokhale NS, Ipas H, et al. Post-transcriptional regulation of antiviral gene expression by N6-methyladenosine. Cell Rep (2021) 34:108798. doi: 10.1016/j.celrep.2021.108798
36. Kamhi E, Joo EJ, Dordick JS, Linhardt RJ. Glycosaminoglycans in infectious disease. Biol Rev Camb Philos Soc (2013) 88:928–43. doi: 10.1111/brv.12034
Keywords: RNA epitranscriptome, N6-methyladenosine, malaria infection, immune response, plasmodium yoelii
Citation: Wang L, Wu J, Liu R, Chen W, Pang Z, Zhou F, Xia L, Huang J, Pan T, Su X-z and Wang X (2022) Epitranscriptome profiling of spleen mRNA m6A methylation reveals pathways of host responses to malaria parasite infection. Front. Immunol. 13:998756. doi: 10.3389/fimmu.2022.998756
Received: 20 July 2022; Accepted: 05 September 2022;
Published: 20 September 2022.
Edited by:
Arif Jamal Siddiqui, University of Hail, Saudi ArabiaReviewed by:
Manish Goyal, Boston University, United StatesCopyright © 2022 Wang, Wu, Liu, Chen, Pang, Zhou, Xia, Huang, Pan, Su and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xin-zhuan Su, eHN1QG5pYWlkLm5paC5nb3Y=; Xiaoyun Wang, d2FuZ3h5QHNjbnUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.