- 1Department of Epidemiology and Biostatistics, School of Public Health, Anhui Medical University, Hefei, Anhui, China
- 2Department of Infectious Diseases, The First Affiliated Hospital of Anhui Medical University, Hefei, Anhui, China
- 3Department of Public Health, Medical Department, Qinghai University, Xining, China
- 4Department of Rheumatology and Immunology, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui, China
Th17 cells play a key role in immunity against Mycobacterium tuberculosis (MTB), and this study aimed to explore the association of Th17 pathway gene polymorphisms with pulmonary tuberculosis (PTB) susceptibility in a Chinese population. A total of 10 single nucleotide polymorphisms in Th17 pathway genes (IL-17A gene rs2275913, rs3748067, rs8193036, rs3819024, IL-17F gene rs7741835, rs763780, IL-21 gene rs907715, rs2055979, IL-23R gene rs11805303, and rs7518660) were genotyped in 456 PTB patients and 466 controls using SNPscan technique. The IL-23R rs11805303 CC genotype, C allele frequencies were significantly lower in PTB patients than in controls, and the rs11805303 variant was significantly associated with the reduced risk of PTB in a recessive model. There were no significant associations between IL-17A, IL-17F, and IL-21 gene variations and PTB risk. In IL-17A gene, rs2275913, rs3748067, and rs3819024 variants were associated with drug resistance in PTB patients. In IL-17F gene, rs7741835 variant affected drug resistance, and rs763780 variant was associated with hypoproteinemia in PTB patients. In addition, the lower frequencies of the TT genotype, T allele of rs2055979 were found in PTB patients with drug-induced liver injury. Haplotype analysis showed that IL-23R CG haplotype frequency was significantly lower in PTB patients than in controls, while the TG haplotype frequency was higher. In conclusion, IL-23R rs11805303 polymorphism may contribute to the genetic underpinnings of PTB in the Chinese population, and the IL-17A, IL-17F, and IL-21 genetic variations are associated with several clinical manifestations of PTB patients.
Introduction
Tuberculosis (TB) is a common chronic infectious disease caused by Mycobacterium tuberculosis (MTB) and remains a major cause of morbidity and mortality in several developing countries. The global incidence of TB was estimated to be 9.9 million in 2021 (1). Approximately one-fourth of the general population was thought to be infected with MTB, while only 5–10% of these individuals eventually developed active TB during their lifetime (2, 3). This implied that the development of TB involved complex interactions, and bacteria, host genetic variation, and environmental factors played important roles (4). Previous studies have identified various host genes associated with pulmonary TB (PTB) susceptibility; unfortunately, the genetic factors influencing PTB development are not fully understood (5, 6). Therefore, identifying the role of host genetic variations in the development of PTB would facilitate further understanding of the pathogenesis of PTB and guide treatment strategies.
It has been suggested that a successful immune response against MTB infection depends on the activation of CD4+T lymphocytes (7). The Th17 cells, known as novel members of the CD4+T lymphocyte family, can induce interleukin (IL)-21 and IL-23R expression to promote cell proliferation and maturation and secrete several cytokines such as IL-17A, IL-17F, IL-22 (8, 9). Th17 cells play a key role in immunity against PTB, and previous studies have found that reduced Th17 response is related to severe outcomes of MTB infection (10). In addition, the secretion of Th17-related cytokines may contribute to the immune response of the human host against MTB (11). For example, the production of IL-17A can eliminate the primary infection and establish an effective memory response, and IL-17A can augment autophagy in MTB-infected monocytes from individuals with strong immunity against the bacterium (12, 13). A recent study found that the production of IL-17F using an in vitro model of human primary cell cultures was stimulated by MTB-Ag and demonstrated that individuals that mount an effective immune response against MTB secreted the highest concentrations of IL-17F (7). This suggested that IL-17F played a protective role in the immune response of the host against mycobacteria.
Several studies have shown that genetic variations in Th17 pathway genes (including IF-17A, IF-17F, IL-21, and IL-23R) are associated with infectious disease susceptibility and clinical manifestations (14, 15). However, the reports of studies on Th17 pathway gene variation and PTB susceptibility have been inconsistent (16). In addition, only a few studies have examined the association between these gene variants and the clinical manifestations of PTB. Therefore, it is important to further evaluate the roles of Th17 pathway gene variations in the pathogenesis of PTB through genetic susceptibility association studies. This study was conducted to investigate the possible roles of the IL-17A, IL-17F, IL-21, and IL-23R gene variation in PTB susceptibility and their clinical manifestations.
Materials and methods
Study participants
A total of 456 PTB patients were enrolled from Anhui Chest Hospital in this investigation. They included 194 females and 262 males with an average age of 43.37 ± 13.90 years. The diagnosis of PTB was based on the following: suspicious clinical symptoms, chest radiography, sputum and/or bronchoalveolar lavage fluid MTB culture, microscopy for acid-fast bacilli, and the effect of anti-TB treatment. Patients were excluded from this study if they had diseases such as cancer, acquired immunodeficiency syndrome, hepatitis, or immune deficiency. Meanwhile, 466 healthy individuals with the same ethnic background and no history of TB, cancer, and acquired immunodeficiency syndrome were enrolled from the same area as controls. The controls comprised 203 males and 263 females (average age: 45.61 ± 17.68 years), and were asymptomatic with negative sputum smear and culture and had normal chest radiographs.
This study was approved by the ethics committee of Anhui Medical University (20200250), and written informed consent was obtained from every participant before enrolment in the study. We collected peripheral blood samples and relevant data, including demographic characteristics and clinical manifestations (pulmonary infection, leukopenia, fever, drug resistance, drug-induced liver injury (DILI), and sputum smear status) from all the participants.
Single nucleotide polymorphism (SNP) selection and genotyping
Two approaches were used for SNP selection: literature retrieval and tag SNP selection. Previous studies on the association between the Th17 pathway gene (IL-17A, IL-17F, IL-21, and IL-23R) polymorphisms and human diseases were reviewed to identify SNPs related to human disease susceptibility. The genotype data of IL-17A, IL-17F, IL-21, and IL-23R in CHB were obtained from Ensembl Genome Browser 85 and CHBS_1000g, and we used the HaploView 4.0 software (Cambridge, MA, USA) to select the tag SNPs, which captured all the common SNPs located in the chromosome locus transcribed into these genes and their flanking 2 000 bp region. In addition, all SNPs included in this study had to meet the following two criteria: minor allele frequency of ≥ 0.05 in CHB and r2 threshold of > 0.8. We finally selected 10 SNPs (IL-17A gene rs2275913, rs3748067, rs8193036, rs3819024, IL-17F gene rs7741835, rs763780, IL-21 gene rs907715, rs2055979, IL-23R gene rs11805303, and rs7518660) for genotyping.
Peripheral blood was drawn from the medial cubital vein, collected in EDTA-containing tubes, and preserved at a temperature of -20°C. The Flexi Gene-DNA Kit (Qiagen, Valencia, CA) was used to extract genomic DNA from the peripheral blood. With the technical support of the Center for Genetic and Genomic Analysis, Genesky Biotechnologies (Inc., Shanghai), genetic polymorphism was detected using SNPscan technique.
Statistical analysis
All statistical analyses were conducted using SPSS 23.0 (Armonk, NY: IBM Corp, USA). The Chi-squared (χ2) test was used to confirm that the genotype distribution of all the SNPs in normal controls was consistent with the Hardy-Weinberg Equilibrium. The frequency distributions of genotypes and alleles in PTB patients and normal controls were compared using the chi-square test (χ2), and odds ratios with 95% confidence intervals were calculated using logistic regression analysis. The SHEsis software was used to perform Haplotype analysis (17), and the relationship between the Th17 pathway gene variations and susceptibility to PTB under two genetic models (dominant and recessive models) was also examined. A two-tailed P-value < 0.05 was considered as statistical significance.
Results
Association of Th17 pathway gene polymorphisms with susceptibility to PTB
The allele-genotype frequency distribution of the Th17 pathway gene polymorphisms (IL-17A gene rs2275913, rs3748067, rs8193036, rs3819024, IL-17F gene rs7741835, rs763780, IL-21 gene rs907715, rs2055979, IL-23R gene rs11805303, and rs7518660) are shown in Table 1. The genotype frequencies of these SNPs in normal controls were determined by the Hardy-Weinberg Equilibrium test, and we found that all SNPs reached genetic equilibrium. In IL-23R gene, the rs11805303 CC genotype frequency was significantly lower in the PTB patients than in the controls (CC vs. TT: P = 0.021), and the patients carrying the C allele had lower susceptibility to PTB than the T allele carriers (C vs. T: P = 0.024). In addition, the rs11805303 variant was associated with the reduced risk of PTB under the recessive model (CC vs. CT+TT: P = 0.037).
The genotype and allele frequencies of the IL-17A gene rs2275913, rs3748067, rs8193036, and rs3819024 variants did not affect susceptibility to PTB (all P > 0.05). Similarly, our results found no associations between the IL-17F gene rs7741835, rs763780, IL-21 gene rs907715, and rs2055979 variants and PTB susceptibility (all P > 0.05).
Influence of Th17 pathway gene polymorphisms on clinical manifestations of PTB
During PTB development, patients have various clinical manifestations, including fever, drug resistance, DILI, pulmonary infection, and hypoproteinemia, which influence treatment and affect prognosis. These clinical manifestations are generally affected by host genetic variation; hence, this study determined whether the polymorphisms of Th17 pathway genes influence the development of these clinical manifestations (Table 2). We found that the AA genotype and A allele carrier of IL-17A rs2275913 markedly increased the risk of drug resistance (P = 0.001, P = 0.003, respectively), and a higher frequency of the rs3748067 TT genotype was associated with drug resistance in PTB patients (P = 0.016). On the other hand, the IL-17A rs3819024 GG genotype and G allele frequencies were significantly increased in PTB patients with drug resistance relative to the patients without this clinical manifestation (P = 0.013, P = 0.017, respectively).
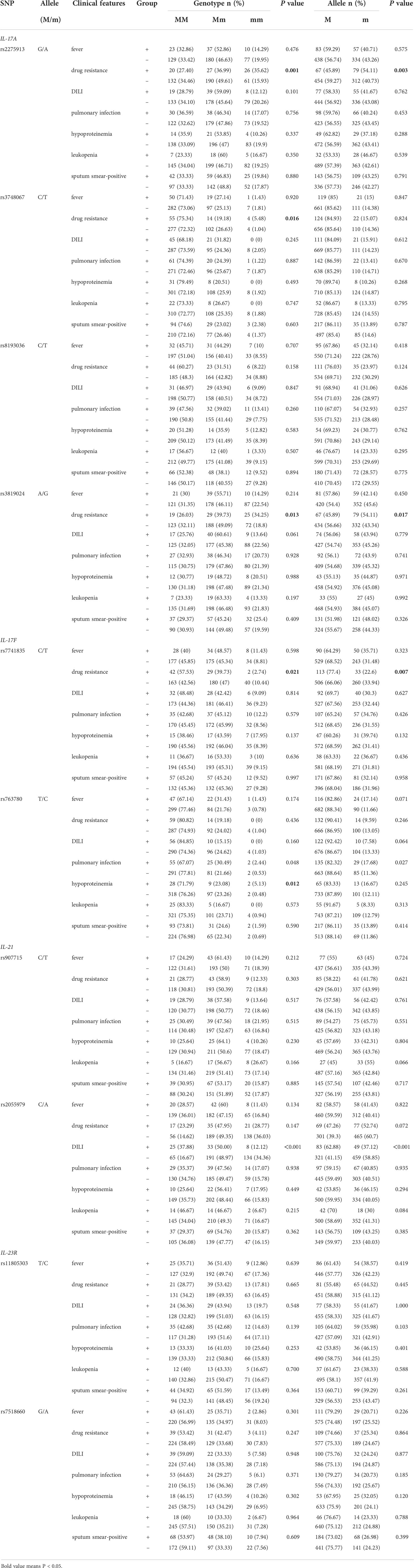
Table 2 Association between Th17 pathway genes polymorphisms and the clinical manifestations of PTB patients.
For the IL-17F gene, the rs7741835 TT genotype and T allele frequencies in PTB patients with drug resistance were significantly lower than those in the patients without drug resistance (P = 0.021, P = 0.007, respectively), and the rs763780 C allele frequency was increased in PTB patients with hypoproteinemia (P = 0.012). Regarding the IL-21 gene, PTB patients carrying the TT genotype and T allele of rs2055979 showed reduced susceptibility to DILI (P < 0.001, P < 0.001, respectively). However, the IL-23R rs11805303 and rs7518660 variants had no relationship with the clinical manifestations of PTB patients (all P > 0.05).
Haplotype analysis
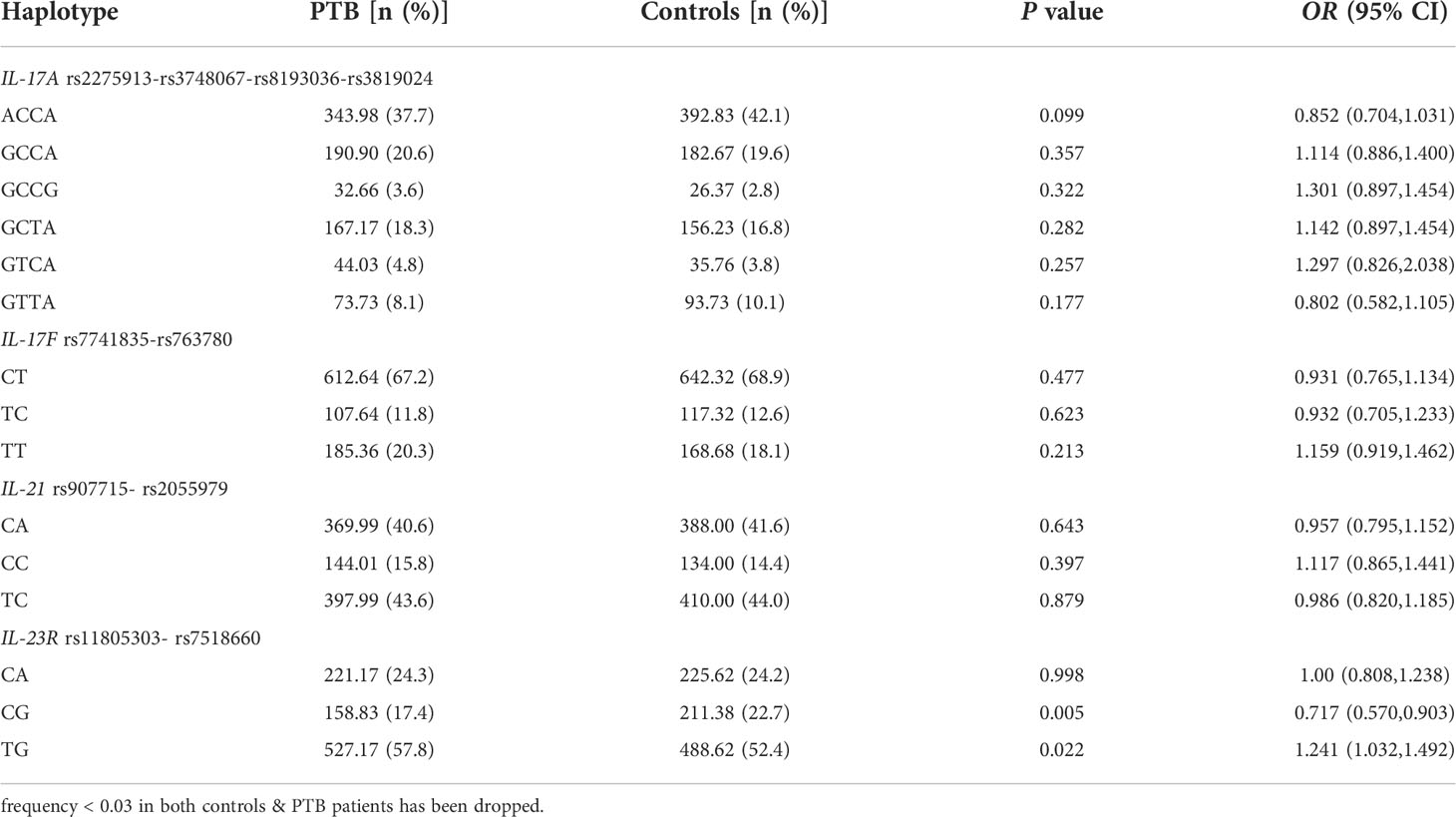
We detected the main haplotypes of the Th17 pathway genes (IL-17A, IL-17F, IL-21, IL-23R) determined by the SHEsis software. The main haplotypes we eventually detected were as follows: IL-17A ACCA, GCCA, GCCG, GCTA, GTCA, GTTA, IL-17F CT, TC, TT, IL-21 CA, CC, TC, IL-23R CA, CG, and TG.
The haplotype distributions in the PTB patients and controls are summarized in Table 3. The results suggested that the IL-23R CG haplotype frequency was significantly lower in PTB patients than in normal controls (P = 0.005), while the TG haplotype frequency was significantly higher (P = 0.022). However, there were no significant associations between the IL-17A, IL-17F, and IL-21 haplotype frequencies and PTB risk.
Discussion
PTB is a major public health problem but the factors underlying the human immune response to mycobacterium are still largely unknown. Identifying prospective genetic biomarkers of PTB is important because it may improve our understanding of its pathogenesis and facilitate early diagnosis and prompt clinical treatment. It is widely known that Th17 cells are involved in adaptive immunity against MTB, and Th17-related cytokines are essential regulators of anti-TB immune responses (18, 19). The contribution of Th17 pathway gene polymorphisms to PTB has been studied, with some studies reporting conflicting results. For example, several reports by studies on IL-17A rs2275913, rs3748067, rs3819024, and PTB susceptibility are inconsistent (20, 21). Hence, our study verified the associations between selected SNPs and PTB risk. In addition, the roles of IL-17F, IL-21, and IL-23R gene variations in the development of PTB among the Chinese Han population have been poorly studied, and we explored the genetic polymorphisms of IL-17F rs7741835, rs763780, IL-21 rs907715, rs2055979, IL-23R rs11805303, and rs7518660 and their associations with PTB.
Cytokine secretion is induced by the interaction between different immune cells and bacteria, and IL-17 acts as a pro-inflammatory cytokine by recruiting granulocytes to the sites of infection (22, 23). IL-17A and IL-17F, which belong to the IL-17 family, have similar biological functions and can induce target cells to produce multiple inflammatory cytokines, metalloproteinases, and chemokines, resulting in neutrophil recruitment, activation, and exudation to trigger inflammation (24). Previous studies have demonstrated that the IL-17A concentration increases in TB and is associated with its severity. The IL-17A concentration increased in mouse models and human PBMC cultures in vitro after stimulation with MTB in previous studies (24–26). In another study, the authors found that IL-17F secretion increased in the population with an effective immune response against MTB (7). A recent meta-analysis suggested that IL-17A rs2275913 polymorphisms may be associated with a reduced risk of PTB in Caucasians, and rs3748067 polymorphism was considered a risk factor for PTB in Asians (21). On the other hand, IL-17A rs3819024 and IL-17F rs763780 polymorphisms did not influence PTB susceptibility (21). As a functional SNP of IL-17A, Wang et al. found that rs8193036 could regulate gene expressions by influencing the binding activity of transcription factors, and rs8193036 T frequency was associated with active PTB (27). However, another study described no relationship between rs8193036 and PTB risk (28). In this study, we did not find any effects of IL-17A rs2275913, rs3748067, rs8193036, rs3819024, IL-17F rs7741835, and rs763780 polymorphisms on PTB susceptibility. This was consistent with the results of several previous studies (28, 29) and helped improve our understanding of the roles of IL-17A and IL-17F gene variations in PTB development. Our results were different from those of some studies due to the different sample sizes, ethnicities, genotyping methods, and study design, among others (27, 30). Hence, studies with larger samples and multiple ethnicities are needed.
IL-23 is a key proinflammatory cytokine in the innate and adaptive immune system that was required for long-term control of MTB, and the activity of this cytokine is mediated by its binding to the IL23R complex (31, 32). An important function of IL-23 mediated by the IL-23R complex is to promote the differentiation of T-cells to Th17, thereby increasing the release of other cytokines such as IL-17 and TNF, which are critical for the progression of PTB (12, 33). Several studies have confirmed that several SNPs of the IL-23R gene are associated with susceptibility to infectious disease, such as HBeAg-positive chronic hepatitis B (24). The study by Jiang et al. revealed that IL-23R rs7518660 was associated with PTB in Chinese Uygurs (34). In contrast, our study did not find a statistical association between rs7518660 and PTB susceptibility in the Chinese Han population. These inconsistent results were largely due to ethnic differences, and more studies were needed to confirm these results. In this study, we also found that the rs11805303 CC genotype and C allele of IL-23R were related to PTB. In addition, two haplotype frequencies were also abnormal in patients with PTB; this result supported the hypothesis that the IL-23R rs11805303 variant influenced susceptibility to PTB. Previous studies have shown that the IL-21 signaling pathway plays an important role in T-cell response to MTB infection by enhancing CD8+T cell activation and promoting T-cell accumulation in the lung and secretion of T-cell cytokines (35, 36). The level of expression of IL-21 in the peripheral blood of PTB patients also significantly decreases (35). Previous studies have analyzed the association between IL-21 gene variation and the risk of infectious diseases (37, 38), but its role in PTB has not been explored. This is the first study to analyze the associations between IL-21 gene rs907715 and rs2055979 polymorphisms and PTB; however, no significant association was found.
Studies have confirmed that host gene variations influence the clinical manifestations and prognosis of PTB patients. Wang et al. found that IL-17A rs3819024 and IL-17F rs763780 are weakly associated with the prognosis of PTB (27). Our previous study revealed that the CYP27A1 rs17470271 and rs933994 T alleles were significantly associated with leukopenia and drug resistance in PTB patients, respectively (39). In this study, we provided evidence of the associations between multiple SNPs (IL-17A rs2275913, rs3748067, rs3819024, and IL-17F rs7741835) and drug resistance in patients with PTB. The IL-17F rs763780 and IL-21 rs2055979 variants were associated with the development of hypoproteinemia and DILI in PTB patients, respectively. Based on these findings, it is reasonable to assume that these SNPs can be used to predict several clinical manifestations of PTB patients, which, in turn, will guide therapeutic schedules for patients.
In summary, this study suggests that IL-23R rs11805303 polymorphisms are associated with a decreased susceptibility to PTB, and IL-17A, IL-17F, and IL-21 gene polymorphisms do not involve the genetic background of PTB in Chinese. We found genetic evidence of significant relationships between IL-17A, IL-17F, and IL-21 genetic variations and several clinical manifestations, including drug resistance and hypoproteinemia in PTB patients. Some limitations of this study need to be considered. Firstly, the possible influence of some confounding factors, such as environmental factors and treatment regimen, in this study was not excluded. Secondly, the sample was relatively small, and larger samples should be used in future studies. Further studies with larger sample sizes that involve different ethnic groups should be conducted to further reveal the roles of Th17 pathway genes in PTB development.
Data availability statement
The original contributions presented in the study are included in the article. Further inquiries can be directed to the corresponding authors.
Ethics statement
This study was approved by the ethics committee of Anhui Medical University (20200250). The patients/participants provided their written informed consent to participate in this study.
Author contributions
T-PZ and H-FP designed the study. H-ML conducted the experiment. L-JW performed the statistical analyses. L-JW and QH participated in sample collection. H-ML drafted the manuscript. T-PZ and H-FP contributed to the manuscript revision. All authors approved the final submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (82003515).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. World Health Organization. Global tuberculosis report (2021). Available at: https://www.who.int/tb/publications/global_report/en/.
2. Houben RM, Dodd PJ. The global burden of latent tuberculosis infection: a re-estimation using mathematical modelling. PloS Med (2016) 13:e1002152. doi: 10.1371/journal.pmed.1002152
3. Lönnroth K, Raviglione M. Global epidemiology of tuberculosis: prospects for control. Semin Respir Crit Care Med (2008) 29:481–91. doi: 10.1055/s-0028-1085700
4. Liu Q, Chen X, Dai X. The association of cytokine gene polymorphisms with tuberculosis susceptibility in several regional populations. Cytokine. (2022) 156:155915. doi: 10.1016/j.cyto.2022.155915
5. Lee SW, Chuang TY, Huang HH, Liu CW, Kao YH, Wu LS. VDR and VDBP genes polymorphisms associated with susceptibility to tuberculosis in a han Taiwanese population. J Microbiol Immunol Infect (2016) 49(5):783–7. doi: 10.1016/j.jmii.2015.12.008
6. Shin JG, Park BL, Kim LH, Namgoong S, Kim JO, Chang HS, et al. Association study of polymorphisms in interferon-gamma receptor genes with the risk of pulmonary tuberculosis. Mol Med Rep (2015) 12(1):1568–78. doi: 10.3892/mmr.2015.3544
7. Khader SA, Bell GK, Pearl JE, Fountain JJ, Rangel-Moreno J, Cilley GE, et al. IL-23 and IL-17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during mycobacterium tuberculosis challenge. Nat Immunol (2007) 8(4):369–77. doi: 10.1038/ni1449
8. Allison SJ. Autoimmune disease: Egress of intestinal TH17 cells in autoimmune renal disease. Nat Rev Nephrol (2017) 13(2):61.
9. Feng H, Yin J, Han YP, Zhou XY, Chen S, Yang L, et al. Sustained changes of treg and Th17 cells during interferon-α therapy in patients with chronic hepatitis b. Viral Immunol (2015) 28(8):412–7. doi: 10.1089/vim.2015.0024
10. Chen X, Zhang M, Liao M, Graner MW, Wu C, Yang Q, et al. Reduced Th17 response in patients with tuberculosis correlates with IL-6R expression on CD4+ T cells. Am J Respir Crit Care Med (2010) 181(7):734–42. doi: 10.1164/rccm.200909-1463OC
11. Shen H, Chen ZW. The crucial roles of Th17-related cytokines/signal pathways in m. tuberculosis infection. Cell Mol Immunol (2018) 15:216–25. doi: 10.1038/cmi.2017.128
12. Khader SA, Cooper AM. IL-23 and IL-17 in tuberculosis. Cytokine. (2008) 41(2):79–83. doi: 10.1016/j.cyto.2007.11.022
13. Tateosian NL, Pellegrini JM, Amiano NO, Rolandelli A, Casco N, Palmero DJ, et al. IL17A augments autophagy in mycobacterium tuberculosisinfected monocytes from patients with active tuberculosis in association with the severity of the disease. Autophagy. (2017) 13:1191–204. doi: 10.1080/15548627.2017.1320636
14. Ren W, Wu Z, Ma R, Liu Z, Wang Y, Wu L, et al. Polymorphisms in the IL-17 gene (rs2275913 and rs763780) are associated with hepatitis b virus infection in the han Chinese population. Genet Test Mol Biomark (2017) 21(5):286–91. doi: 10.1089/gtmb.2016.0177
15. Tayefinasrabadi H, Mohebbi SR, Hosseini SM, Azimzadeh P, Pourhoseingholi MA, Ghaemi A, et al. Association of interleukin-17 gene polymorphisms with susceptibility to chronic hepatitis b virus infection and clearance in Iranian population. Microb Pathog (2020) 144:104195. doi: 10.1016/j.micpath.2020.104195
16. Wang M, Xu G, Lü L, Xu K, Chen Y, Pan H, et al. Genetic polymorphisms of IL-17A, IL-17F, TLR4 and miR-146a in association with the risk of pulmonary tuberculosis. Sci Rep (2016) 6:28586. doi: 10.1038/srep28586
17. Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res (2009) 19(4):519–23. doi: 10.1038/cr.2009.33
18. Lyadova IV, Panteleev AV. Th1 and Th17 cells in tuberculosis: Protection, pathology, and biomarkers. Mediators Inflamm (2015) 2015:854507. doi: 10.1155/2015/854507
19. Scriba TJ, Kalsdorf B, Abrahams DA, Isaacs F, Hofmeister J, Black G, et al. Distinct, specific IL-17- and IL-22-producing CD4+ T cell subsets contribute to the human anti-mycobacterial immune response. J Immunol (2008) 180(3):1962–70. doi: 10.4049/jimmunol.180.3.1962
20. Eskandari-Nasab E, Moghadampour M, Tahmasebi A, Asadi-Saghandi A. Interleukin-17 a and f gene polymorphisms affect the risk of tuberculosis: An updated meta-analysis. Indian J Tuberc (2018) 65(3):200–7. doi: 10.1016/j.ijtb.2017.08.027
21. Yu ZG, Wang BZ, Li J, Ding ZL, Wang K. Association between interleukin-17 genetic polymorphisms and tuberculosis susceptibility: An updated meta-analysis. Int J Tuberc Lung Dis (2017) 21(12):1307–13. doi: 10.5588/ijtld.17.0345
22. Etna MP, Giacomini E, Severa M, Coccia EM. Pro- and anti-inflammatory cytokines in tuberculosis: a two-edged sword in TB pathogenesis. Semin Immunol (2014) 26(6):543–51. doi: 10.1016/j.smim.2014.09.011
23. Bulat-Kardum LJ, Etokebe GE, Lederer P, Balen S, Dembic Z. Genetic polymorphisms in the toll-like receptor 10, interleukin (IL)17A and IL17F genes differently affect the risk for tuberculosis in Croatian population. Scand J Immunol (2015) 82(1):63–9. doi: 10.1111/sji.12300
24. Ju H, Liu H, Tian ZB, Jiang YP, Zhang CP, Liu XS. Association of polymorphisms in key Th-17 immune response genes with HBeAg-positive chronic hepatitis b susceptibility and response to PEG-IFNa-2α. Virology. (2017) 509:35–41. doi: 10.1016/j.virol.2017.05.011
25. Peng MY, Wang ZH, Yao CY, Jiang LN, Jin QL, Wang J, et al. Interleukin 17-producing gamma delta T cells increased in patients with active pulmonary tuberculosis. Cell Mol Immunol (2008) 5(3):203–8. doi: 10.1038/cmi.2008.25
26. Freches D, Korf H, Denis O, Havaux X, Huygen K, Romano M. Mice genetically inactivated in interleukin-17A receptor are defective in long-term control of mycobacterium tuberculosis infection. Immunology. (2013) 140(2):220–31. doi: 10.1111/imm.12130
27. Wang W, Deng G, Zhang G, Yu Z, Yang F, Chen J, et al. Genetic polymorphism rs8193036 of IL17A is associated with increased susceptibility to pulmonary tuberculosis in Chinese han population. Cytokine. (2020) 127:154956. doi: 10.1016/j.cyto.2019.154956
28. Yu ZQ, Wang WF, Dai YC, Chen XC, Chen JY. Interleukin-22 receptor 1 is expressed in multinucleated giant cells: A study on intestinal tuberculosis and crohn’s disease. World J Gastroenterol (2019) 25(20):2473–88. doi: 10.3748/wjg.v25.i20.2473
29. Du J, Han J, Li X, Zhang Y, Li H, Yang S. StIL-17 gene polymorphisms in the development of pulmonary tuberculosis. Int J Clin Exp Pathol (2015) 8:3225–9.
30. Peng R, Yue J, Han M, Zhao Y, Liu L, Liang L. The IL-17F sequence variant is associated with susceptibility to tuberculosis. Gene. (2013) 515(1):229–32. doi: 10.1016/j.gene.2012.11.017
31. Khader SA, Guglani L, Rangel-Moreno J, Gopal R, Junecko BA, Fountain JJ, et al. IL-23 is required for long-term control of mycobacterium tuberculosis and b cell follicle formation in the infected lung. J Immunol (2011) 187(10):5402–7. doi: 10.4049/jimmunol.1101377
32. Parham C, Chirica M, Timans J, Vaisberg E, Travis M, Cheung J, et al. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J Immunol (2002) 168(11):5699–708. doi: 10.4049/jimmunol.168.11.5699
33. Stein CM, Guwatudde D, Nakakeeto M, Peters P, Elston RC, Tiwari HK, et al. Heritability analysis of cytokines as intermediate phenotypes of tuberculosis. J Infect Dis (2003) 187(11):1679–85. doi: 10.1086/375249
34. Jiang D, Wubuli A, Hu X, Ikramullah S, Maimaiti A, Zhang W, et al. The variations of IL-23R are associated with susceptibility and severe clinical forms of pulmonary tuberculosis in Chinese uygurs. BMC Infect Dis (2015) 15:550. doi: 10.1186/s12879-015-1284-2
35. Kumar NP, Banurekha VV, Nair D, Babu S. Diminished plasma levels of common γ-chain cytokines in pulmonary tuberculosis and reversal following treatment. PloS One (2017) 12(4):e0176495. doi: 10.1371/journal.pone.0176495
36. Booty MG, Barreira-Silva P, Carpenter SM, Nunes-Alves C, Jacques MK, Stowell BL, et al. IL-21 signaling is essential for optimal host resistance against mycobacterium tuberculosis infection. Sci Rep (2016) 6:36720. doi: 10.1038/srep36720
37. Shoraka S, Mohebbi SR, Hosseini SM, Hosseini Razavi A, Hatami Y, Sharifian A, et al. Association between interleukin-21 and interleukin-21 receptor gene polymorphisms with susceptibility to chronic hepatitis b virus infection and HBV spontaneous clearance in Iranian population. Microb Pathog (2019) 128:263–7. doi: 10.1016/j.micpath.2019.01.008
38. Shoraka S, Mohebbi SR, Hosseini SM, Hosseini Razavi A, Ghaemi A, Kazemian S, et al. Interleukin-21 rs2055979 and interleukin-21 receptor rs3093390 genetic variants and hepatitis c virus chronic infection. Gastroenterol Hepatol Bed Bench (2017) 10(Suppl1):S154–60.
Keywords: pulmonary tuberculosis, Th17 pathway genes, IL-17A, IL-17F, IL-21, IL-23R, single nucleotide polymorphisms
Citation: Li H-M, Wang L-J, Huang Q, Pan H-F and Zhang T-P (2022) Exploring the association between Th17 pathway gene polymorphisms and pulmonary tuberculosis. Front. Immunol. 13:994247. doi: 10.3389/fimmu.2022.994247
Received: 14 July 2022; Accepted: 28 October 2022;
Published: 21 November 2022.
Edited by:
Nicola Ivan Lorè, San Raffaele Scientific Institute (IRCCS), ItalyReviewed by:
Muki Shey, University of Cape Town, South AfricaUtpal Sengupta, The Leprosy Mission Trust India, India
Copyright © 2022 Li, Wang, Huang, Pan and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tian-Ping Zhang, emhhbmd0aWFucGluZ0B1c3RjLmVkdS5jbg==; Hai-Feng Pan, cGFuaGFpZmVuZ0BhaG11LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
 Hong-Miao Li
Hong-Miao Li Li-Jun Wang2†
Li-Jun Wang2† Hai-Feng Pan
Hai-Feng Pan Tian-Ping Zhang
Tian-Ping Zhang