- 1Department of Rheumatology and Immunology, The First Affiliated Hospital of USTC, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China
- 2Department of Nosocomial Infection Management, The First Affiliated Hospital of Anhui Medical University, Hefei, China
- 3Department of Infectious Diseases, The First Affiliated Hospital of Anhui Medical University, Hefei, China
- 4Department of Public Health, Medical Department, Qinghai University, Xining, China
- 5Department of Epidemiology and Biostatistics, School of Public Health, Anhui Medical University, Hefei, China
Objective: The aim of the current study was to investigate the contributing role of gene variation and transcription levels among the m6A methyltransferases METTL3, METTL14, and WTAP in pulmonary tuberculosis (PTB).
Methods: A case-control study including 461 PTB patients and 467 normal controls was designed for genotyping. Three SNPs in METTL3 (rs1061027, rs1139130, rs1061026), three SNPs in METTL14 (rs62328061, rs4834698, rs1064034), and two SNPs in WTAP (rs1853259, rs11752345) were genotyped via the SNPscan™ technique. METTL3, METTL14, and WTAP transcription levels were determined in 78 PTB patients and 86 controls via quantitative real-time reverse-transcription PCR.
Results: Frequencies of the METTL14 rs62328061 GG genotype, WTAP rs11752345 CT genotype, and T allele were significantly increased in PTB patients compared to controls. An increased risk of rs62328061 was detected in a recessive model, and a decreased risk of rs11752345 was detected in a dominant model in the PTB group. METTL3 gene variation was not associated with PTB risk. The METTL3 rs1139130 GG genotype was significantly increased with drug resistance, and the G allele was significantly decreased with drug-induced liver injury in PTB patients. A reduced frequency of the METTL14 rs62328061 G allele was associated with leukopenia, a reduced frequency of the WTAP rs11752345 T allele was associated with sputum smear positivity, and a higher frequency of the METTL14 rs4834698 TC genotype was evident in PTB patients with hypoproteinemia. Compared to controls, METTL3, METTL14, and WTAP transcription levels in PTB patients were significantly decreased, and the level of WTAP was increased in PTB patients with drug resistance. METTL3 level was negatively associated with erythrocyte sedimentation rate and aspartate aminotransferase, and METTL14 level was negatively correlated with alanine aminotransferase and aspartate aminotransferase.
Conclusion: METTL14 rs62328061 and WTAP rs11752345 variants were associated with the genetic background of PTB, and METTL3, METTL14, and WTAP levels were abnormally decreased, suggesting that these m6A methyltransferases may play important roles in PTB.
Introduction
Tuberculosis (TB) is a common infectious disease caused by Mycobacterium tuberculosis (MTB), and it remains one of the most serious public health problems worldwide (1). A World Health Organization update reported that there was an estimated 9.9 million new incident TB patients globally in 2021 (2). It is well known that people infected with MTB can develop several possible outcomes, such as MTB clearance, primary TB, latent TB infection, and active TB. Studies indicate that approximately 10% of individuals infected with MTB will eventually progress to active TB (3, 4). The occurrence and development of TB is mainly influenced by complex interactions between the MTB strain, the external environment, and genetic factors (5, 6). Host genetics are the essential factor determining disease susceptibility and outcomes after MTB infection in many case-control studies, animal model studies, and twin and family studies (7, 8). Continuing studies on the roles of genetic variants in TB susceptibility would contribute to the development of advantageous approaches to TB prevention, diagnosis, and treatment, and many genetic variations have been shown to be associated with susceptibility to TB (9, 10).
N6-methyladenosine (m6A) is formed via methylation of the sixth N atom on the adenine base, which is reportedly the most abundant post-transcriptional modification of RNA in eukaryotes, particularly messenger RNA (mRNA) (11). m6A regulates post-transcriptional mRNA levels in a dynamic and reversible manner (12). m6A modification is regulated by several key regulators, including RNA methyltransferases (METTL3, METTL14, and WTAP), demethylases, and m6A-binding proteins (13). m6A modification is involved in many important biological processes, and it is closely related to the occurrence and development of many diseases.
Epigenetic modification, including DNA methylation and noncoding RNAs, reportedly contributes to pulmonary TB (PTB) progression (14, 15). In recent years m6A methylation was also found in the MTB genome (16). Therefore, m6A methylation may be involved in the pathogenesis of PTB. Strong evidence indicates that abnormal expression of m6A key regulators may result in abnormal RNA m6A modification, leading to a variety of diseases (17, 18). In addition, some functional single nucleotide polymorphisms (SNPs) in specific regions of m6A key regulator genes may influence m6A methylation, thereby affecting disease development. For example, several SNPs in METTL3 and METTL14 genes have been associated with neuroblastoma, Wilms’ tumor, acute lymphoblastic leukemia, and autoimmune thyroid disease (19–22). In a previous study, genetic variations in m6A demethylases FTO could affect susceptibility to PTB (23), but few studies have investigated associations between variation in key m6A modification regulatory genes and PTB risk. The current epidemiological study was conducted to identify novel m6A methyltransferases SNPs associated with susceptibility to PTB. The study assessed associations between three m6A methyltransferase genes (METTL3, METTL14, and WTAP) SNPs, as well as their transcription levels, and PTB risk.
Materials and methods
Study subjects
In this case-control study, 461 PTB patients and 467 normal controls were consecutively enrolled, and associations between m6A methyltransferase gene polymorphisms and PTB susceptibility were analyzed. Then, 78 PTB patients and 86 normal controls were enrolled to detect m6A methyltransferase transcription levels. All PTB patients were recruited from the Department of Tuberculosis at Anhui Chest Hospital, whereas normal controls were selected from a health center in the same area. PTB patients were diagnosed by specialists based on indicative clinical symptoms, chest radiography, sputum and/or bronchoalveolar lavage fluid MTB culture, microscopy for acid fast bacilli, and effects of anti-TB treatment. The exclusion criteria included HIV positivity, hepatitis, malignancy, and immunodeficiency. Individuals without a history of TB, malignant tumor, HIV or other infectious diseases were included in the study as normal controls. The control group was required to be asymptomatic with negative sputum smears and cultures, and normal chest radiographs. All subjects were of Chinese Han ethnicity.
The study was approved by the Medical Ethics Committee of Anhui Medical University (approval number 20200250). After obtaining informed consent, peripheral blood samples and relevant information were collected from each participant with the help of professional physicians. The data collected included basic demographic characteristics and some clinical data such as fever, drug resistance, drug-induced liver injury (DILI), pulmonary infection, leukopenia, sputum smears, total bilirubin, aspartate aminotransferase (AST), alanine aminotransferase (ALT), and erythrocyte sedimentation rate (ESR).
SNP selection
The three m6A methyltransferases METTL3, METTL14, and WTAP were included in the analyses, and the functional SNPs in their corresponding genes were chosen based on previous studies (24). Existing studies investigating associations between METTL3, METTL14, and WTAP gene polymorphisms and human diseases were systematically reviewed, and the SNPs related to human diseases were identified. Ensembl Genome Browser 85 and CHBS_1000g software were then used to obtain genotype data on these genes derived from Han Chinese people living in Beijing, and the tag SNPs of these genes were selected via the pairwise option of HaploView 4.0 software (Cambridge, MA, USA). The tag SNP selection criteria were: (1) a minor allele frequency ≥ 5% in CHB_1000g, and an r2 threshold > 0.8; (2) the common SNPs located within the chromosome locus transcribed into METTL3, METTL14, or WTAP and their flanking 2000 bp regions were captured; (3) potentially functional SNPs located within the 5’ untranslated region, 3’ untranslated region, and exon were preferentially selected. Lastly, three SNPs in the METTL3 gene (rs1061027, rs1139130, rs1061026), three SNPs in the METTL14 gene (rs62328061, rs4834698, rs1064034), and two SNPs in the WTAP gene (rs1853259, rs11752345) were selected.
DNA extraction and genotyping
Samples of approximately 5 mL of peripheral early morning fasting blood were collected from all subjects, and genomic DNA was extracted via a Flexi Gene-DNA Kit (Qiagen, Valencia, CA) for genotyping. Genotyping of all DNA samples for the selected SNPs was conducted via the Three SNPs in METTL3 (rs1061027, rs1139130, rs1061026), three SNPs in METTL14 (rs62328061, rs4834698, rs1064034), and two SNPs in WTAP (rs1853259, rs11752345) were genotyped via the SNPscan™ technique, with technical support from the Center for Genetic and Genomic Analysis, Genesky Biotechnologies Inc. (Shanghai). Only participants with 100% genotyping success for the selected SNPs were included in the final analysis.
Quantitative real-time reverse-transcription PCR
Peripheral blood mononuclear cells (PBMCs) were isolated from 3 mL anticoagulant peripheral blood, and total RNA was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA). A NanoDrop 2000 spectrophotometer (Thermo Scientific, USA) was used to determine total RNA concentrations. Total RNA was then reverse transcribed into cDNA by the PrimeScript™ RT Reagent Kit (Takara Bio Inc., Japan). In this experiment a 20-μL reverse transcription reaction system with a maximum of 1 μg total RNA was used, and the amount of total RNA needed in the reaction system was determined based on the RNA concentration.
METTL3, METTL14, and WTAP mRNA levels in PBMCs were detected via quantitative real-time reverse transcription (qRT) PCR with SYBR Green (SYBR Premix Ex Taq II, Takara Bio Inc., Japan). qRT-PCRs were conducted using a QuantStudio 12K Flex Real-Time PCR System (Applied Biosystems, Foster City, CA, USA), and the cycle conditions of reactions were 95°C for 1 min, followed by 42 cycles at 95°C for 10 sec, 60°C for 30 sec, and 72°C for 1 min. Relative transcription levels of METTL3, METTL14, and WTAP were calculated via comparisons with the housekeeping gene (internal control) β-actin in the same sample, and the 2-△△Ct method was used to express levels (25).
Statistical analysis
Whether the genotypes distribution of all SNPs in normal controls were in Hardy-Weinberg equilibrium was assessed via the Chi-square test. Associations between m6A methyltransferase gene polymorphisms and PTB susceptibility were estimated via odds ratios (OR) and 95% confidence intervals (CI) with logistic regression analyses. Two genetic models (dominant and recessive) were used to analyze associations between these SNPs and PTB risk, and haplotype analysis was conducted via SHEsis software (26). Transcription levels of METTL3, METTL14, and WTAP are expressed as median and quartile intervals, and the Mann-Whitney U test and the Kruskal-Wallis H test were respectively used to evaluate differences in these m6A methyltransferase levels between two groups and among three groups. Correlations between these m6A methyltransferase levels and experimental indexes in PTB patients were analyzed via the Spearman rank correlation coefficient test. All statistical analyses were conducted with SPSS 23.0 (SPSS Inc., IL, USA), and two-sided p values of < 0.05 were deemed to indicate statistical significance.
Results
Associations between m6A methyltransferase gene polymorphisms and PTB susceptibility
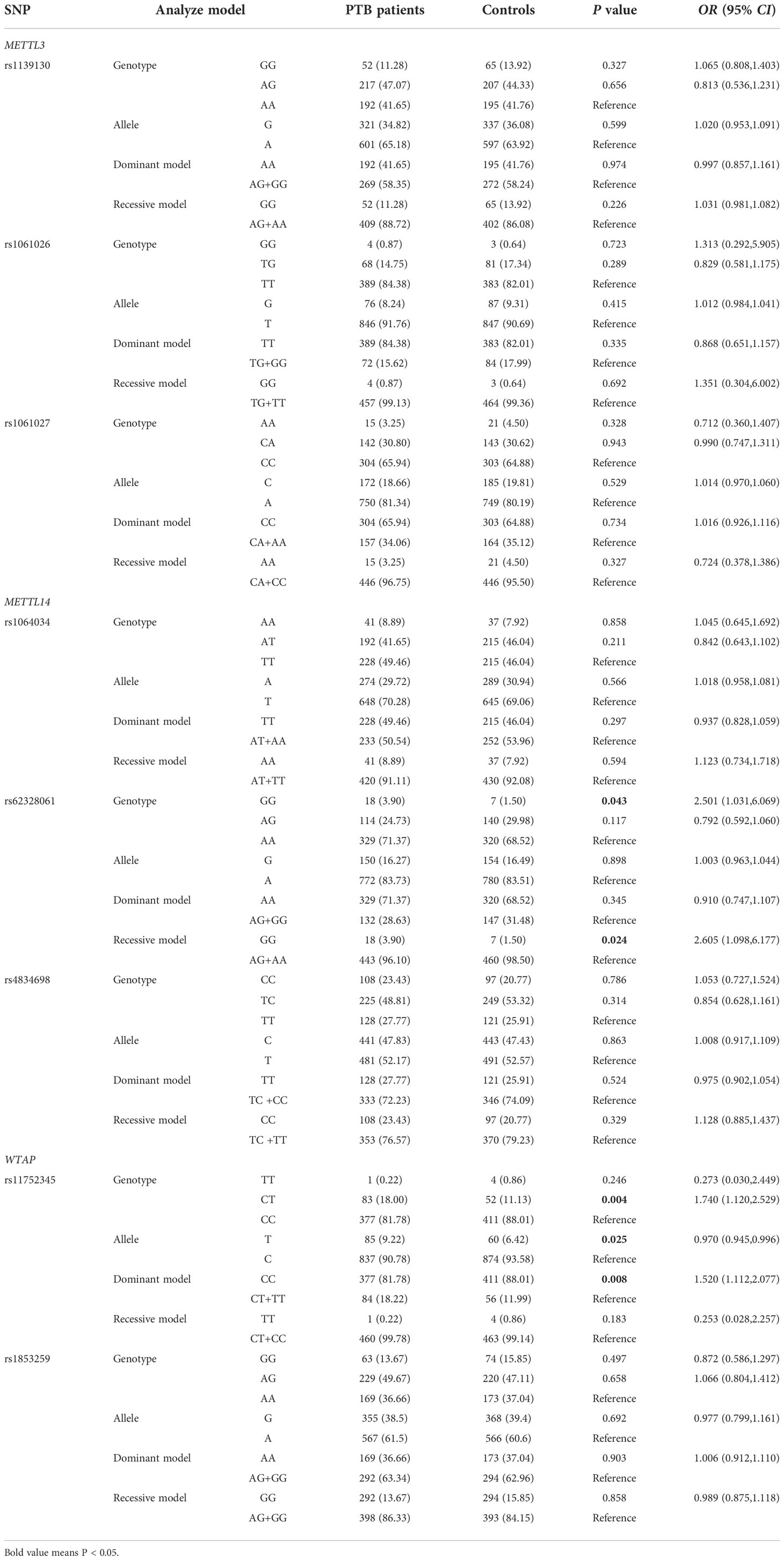
The PTB group genotyped included 194 females and 267 males, with a mean age of 45.56 ± 17.75 years. The normal control group genotyped included 265 females and 202 males, with a mean age of 43.38 ± 13.86 years. The genotype distributions of all SNPs in normal controls were in Hardy-Weinberg equilibrium, and the allele and genotype frequencies of these SNPs are shown in Table 1.
In the METTL14 gene, the frequency of the rs62328061 GG genotype was significantly higher in PTB patients than in normal controls, and the genotype was associated with PTB susceptibility in the recessive model (GG versus AA p = 0.043; GG versus AG+AA p = 0.024). There was no significant association between the rs1064034 variant and PTB risk. In comparisons of WTAP rs11752345 variant genotype and allele frequencies between the PTB group and control group, the CT genotype and T allele frequencies were significantly increased in PTB patients (CT versus CC p = 0.004; T versus C p = 0.025). There was a decreased risk of the rs11752345 variant in the dominant model (CC versus CT+TT p = 0.008). PTB risk was not significantly associated with the WTAP rs1853259 variant, or with the METTL3 rs1139130, rs1061026, or rs1061027 variants (all p > 0.05).
A case-only analysis was performed to analyze associations between METTL3, METTL14, and WTAP gene variants and common clinical features of PTB (Table 2). METTL3 gene rs1139130 GG genotype frequency was significantly positively associated with drug resistance (p = 0.017), whereas rs1139130 G allele frequency was significantly negatively associated with DILI (p = 0.041) in PTB patients. The METTL14 gene G allele frequency of the rs62328061 variant was significantly associated with a decreased risk of leukopenia (p = 0.016), and a higher frequency of the rs4834698 TC genotype was observed in PTB patients with hypoproteinemia (p = 0.042). A decreased frequency of the WTAP rs11752345 T allele was significantly associated with sputum smear positivity (p = 0.041). There were no significant associations between other SNPs and the clinical features of PTB.
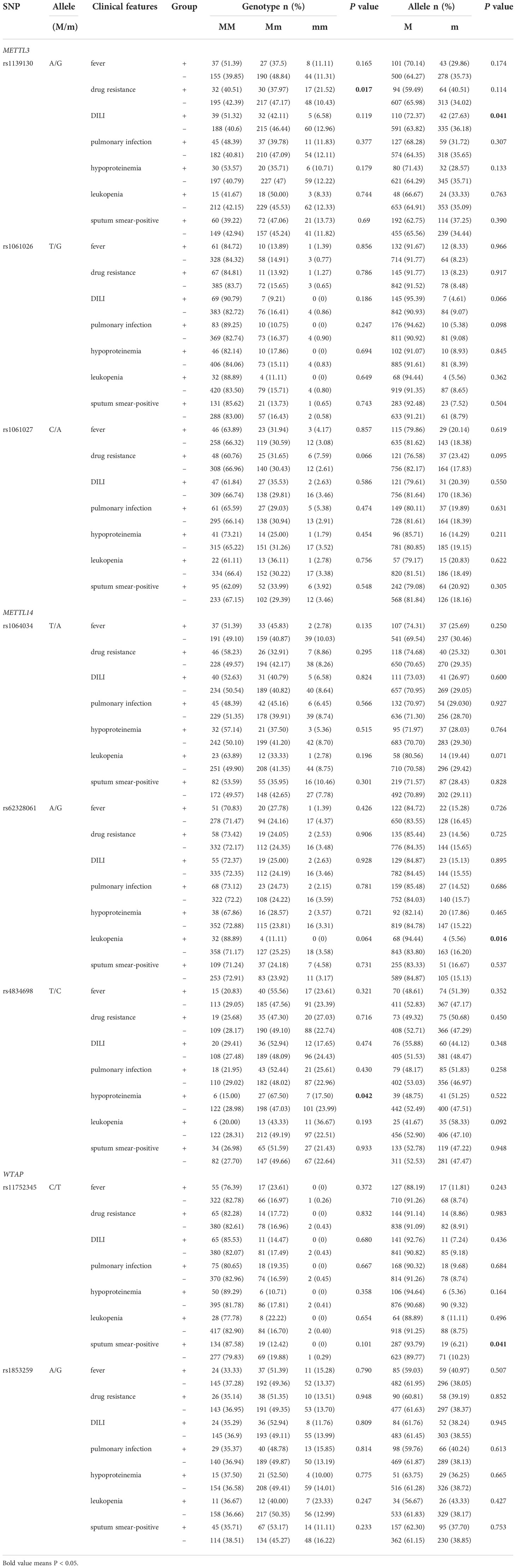
Table 2 Associations between METTL3, METTL14, and WTAP genes polymorphisms and clinical features of PTB patients.
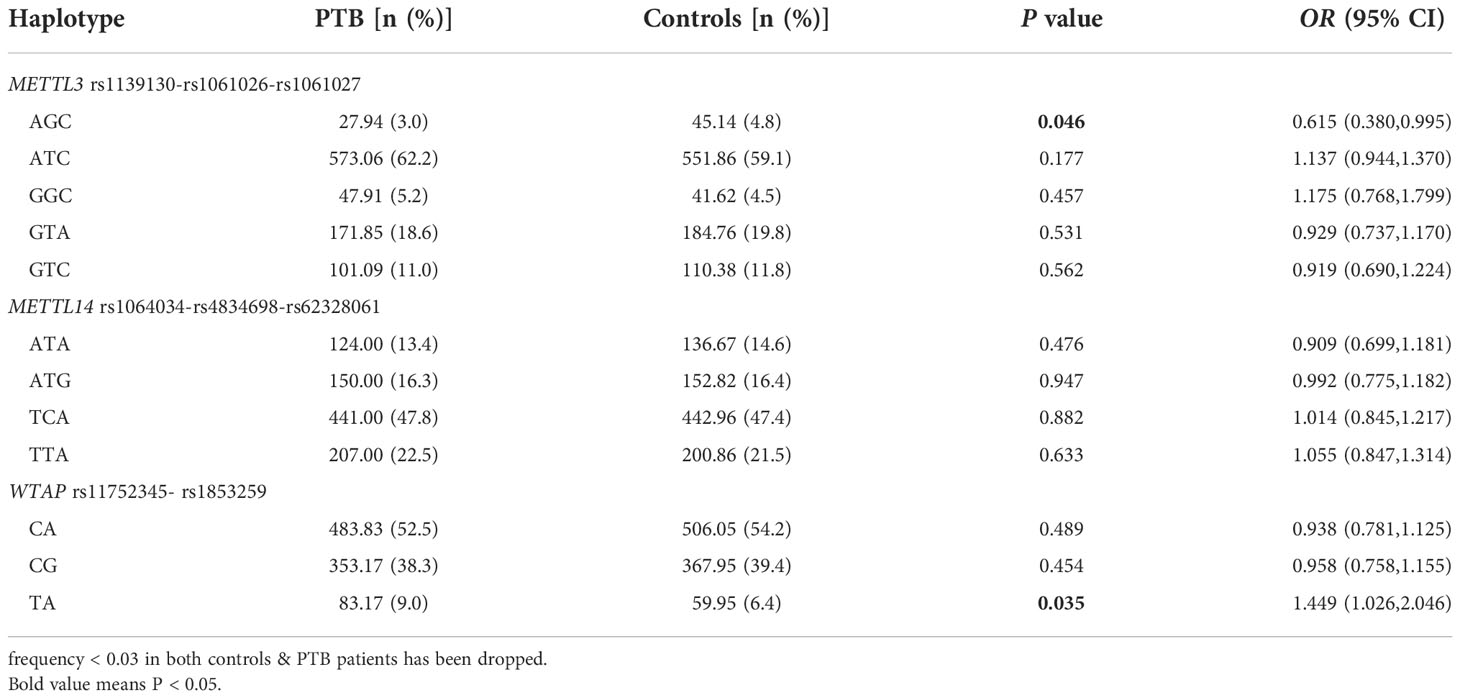
Haplotype analysis
The main haplotypes of METTL3, METTL14, and WTAP genes were detected via SHEsis software, and the frequency distributions of these haplotypes, including five for METTL3 (AGC, ATC, GGC, GTA, GTC), four for METTL14 (ATA, ATG, TCA, TTA), and three for WTAP (CA, CG, TA) are shown in Table 3. Compared with normal controls, the frequency of the METTL3 AGC haplotype was significantly lower in PTB patients (p = 0.046), and the frequency of the WTAP TA haplotype was significantly higher (p = 0.035).
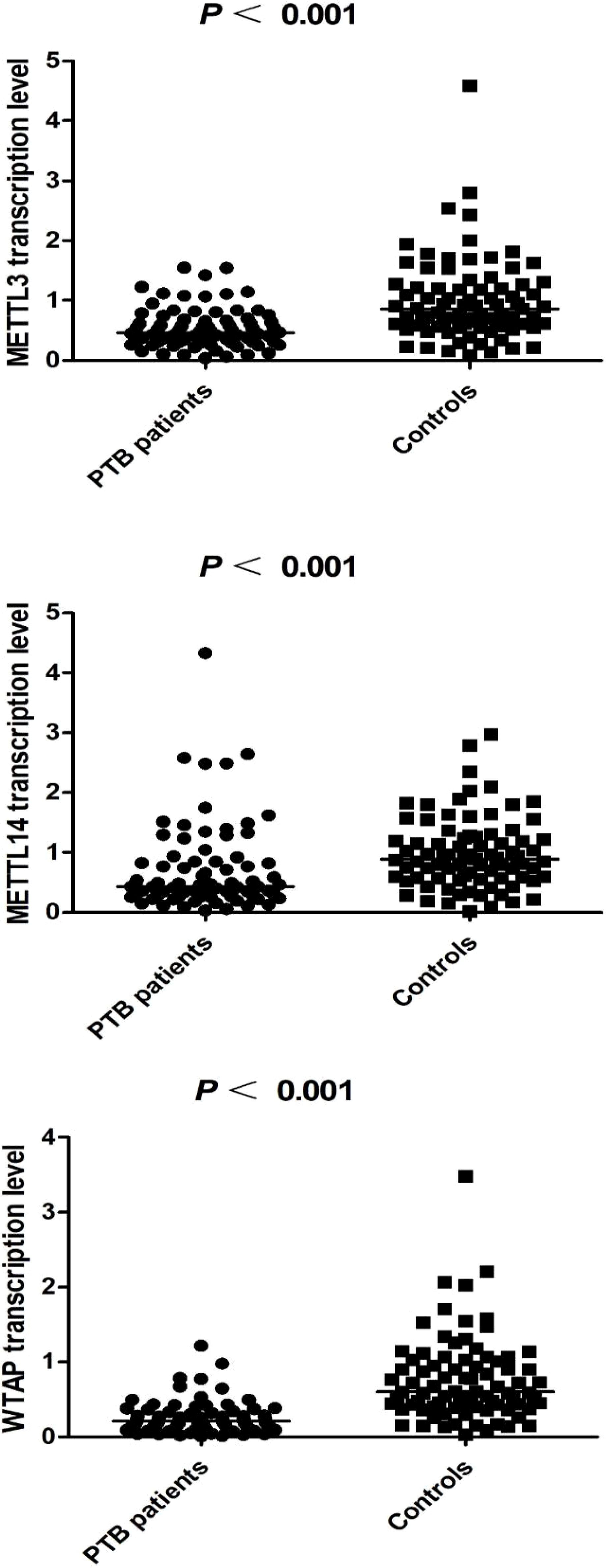
m6A methyltransferase transcription levels in the PTB group and the control group
The transcription levels of METTL3, METTL14, and WTAP in PBMCs were significantly lower in PTB patients than in normal controls (all p < 0.001) (Figure 1). Associations between METTL3, METTL14, and WTAP mRNA levels and multiple clinical features were investigated in PTB patients. WTAP transcription level was significantly greater in PTB patients with drug resistance than in PTB patients without drug resistance (p = 0.030) (Table 4). METTL3 transcription level was negatively associated with ESR (p = 0.035) and AST (p = 0.022) in PTB patients, and METTL14 transcription level was negatively associated with ALT (p = 0.049) and AST (p = 0.004) (Table 5).
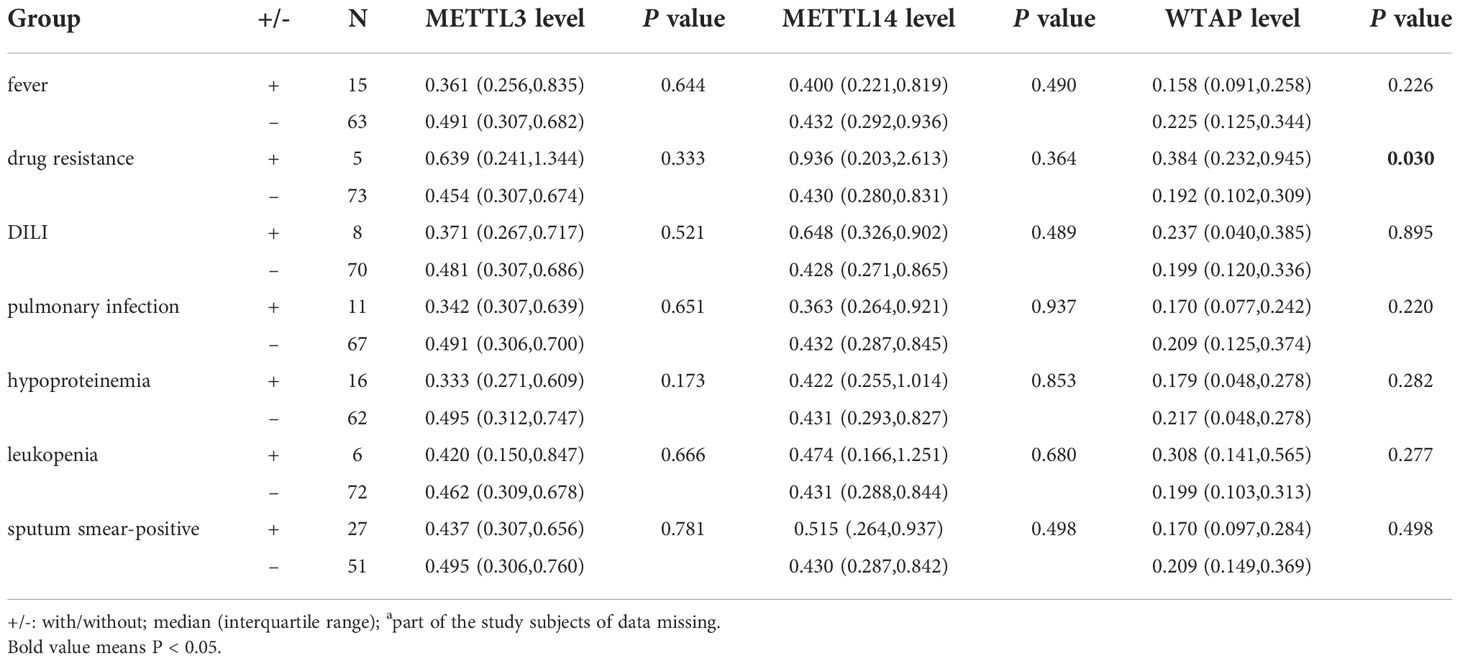
Table 4 Association between METTL3, METTL14, and WTAP transcription levels and several clinical features in PTB patients.
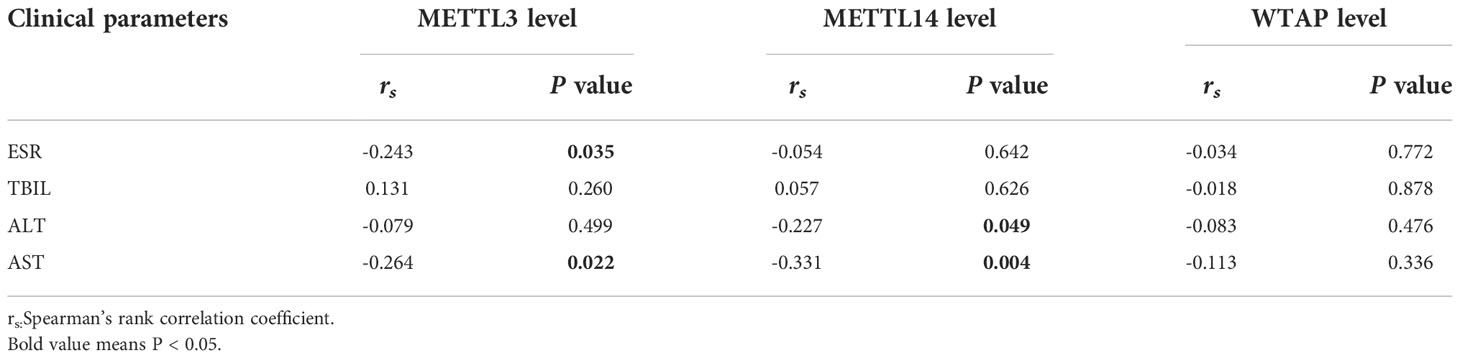
Table 5 The correlation between METTL3, METTL14, and WTAP transcription levels and ESR, TBIL, ALT, AST of PTB patients.
Associations between m6A methyltransferase gene polymorphisms and their transcription levels in PTB patients
The Associations between genes variation in m6A methyltransferase METTL3, METTL14, and WTAP and their transcription levels were assessed in 64 PTB patients, and no significant differences were detected (all p > 0.05) (Table 6).
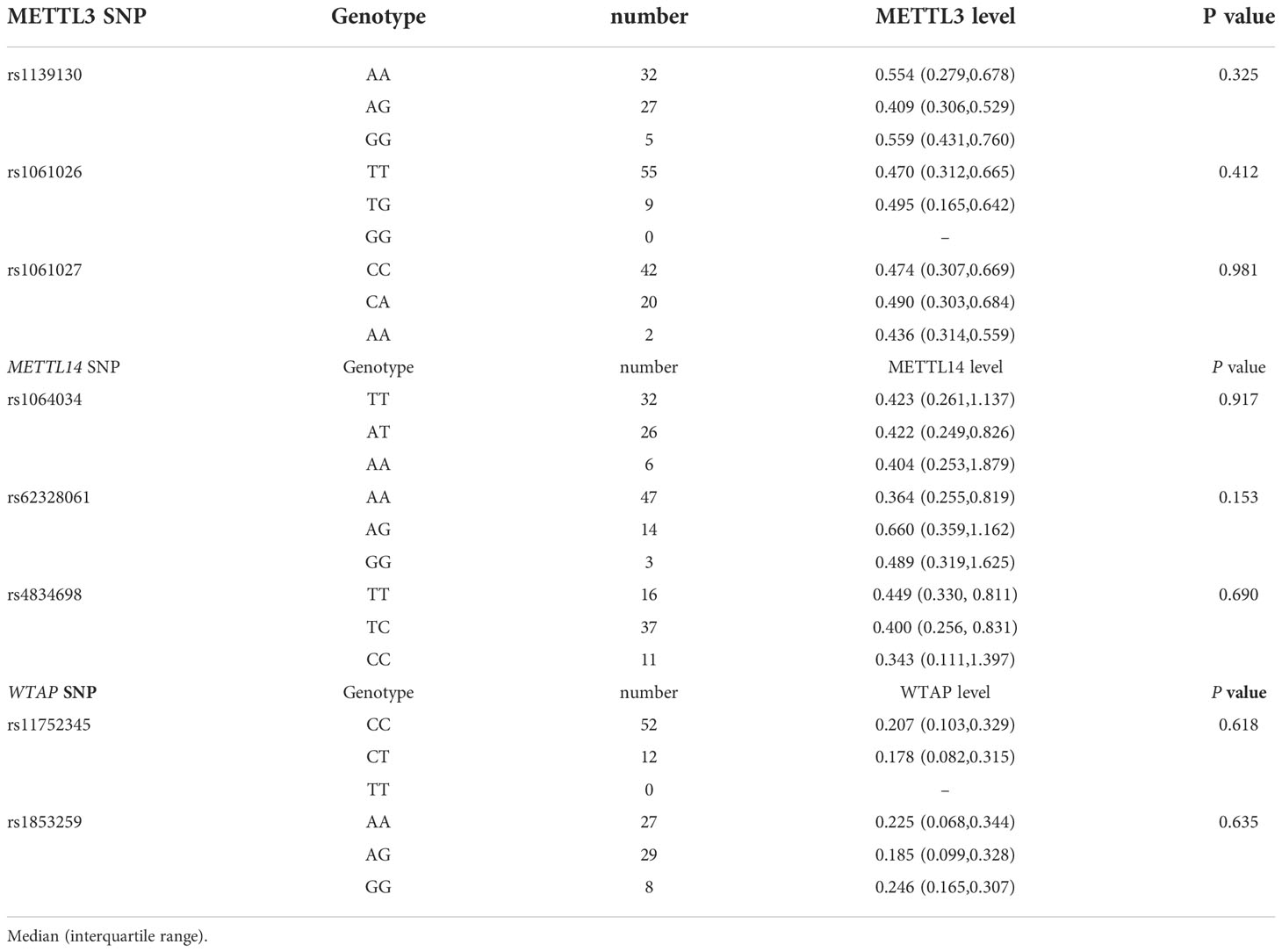
Table 6 Association between METTL3, METTL14, and WTAP genes polymorphisms with their transcription levels in PTB patients.
Discussion
Many novel genetic variants associated with susceptibility to PTB have been identified via genome-wide association studies and candidate gene studies, but unearthing the full range of PTB susceptibility variations remains a challenge. Given the important role of m6A modification in the development of multiple diseases, potential associations between genetic variation of m6A-modified genes and disease susceptibility have also attracted increased attention in recent years (27, 28). The classic m6A methyltransferase complex mainly consists of METTL3, METTL14, and WTAP, which mediate m6A methylation of mRNA. METTL3 is a vital methyltransferase as an S-adenosylmethionine-binding subunit, whereas METTL14 is an RNA-binding scaffold for substrate recognition. WTAP interacts with METTL3 and METTL14, then localizes them into nuclear speckles (29, 30). Notably the SNPs located in these m6A methyltransferases are reportedly associated with disease susceptibility via m6A methylation and related biological processes (19, 20, 31). To the best of our knowledge, the current study is the first to investigate associations between polymorphic variants of the m6A methyltransferase genes METTL3, METTL14, and WTAP and PTB susceptibility. The study provides strong evidence that the METTL14 gene variant rs62328061 and the WTAP gene variant rs11752345 are associated with the risk of PTB, and that METTL3, METTL14, and WTAP levels are decreased in PTB patients.
To date, studies on genetic variation of METTL3 and disease susceptibility have mainly focused on cancers. Lin et al. (32) suggested that the combination of rs1139130, rs1263801, rs1061026, and rs1061027 variants in the METTL3 gene could reduce the risk of Wilms’ tumor in children. Another study reported that these four SNPs were also related to higher susceptibility to neuroblastoma (19). In contrast with these findings, rs1061027, rs1139130, and rs1061026 variants were not significantly associated with PTB susceptibility in the present study. Compared with single SNPs, association studies based on haplotypes of multiple markers can significantly enhance the mapping and characterization of disease susceptibility genes (33). In this study, we assessed whether the main haplotypes consisting of the aforementioned three polymorphisms were associated with PTB, and the AGC haplotype was significantly associated with a lower risk of PTB. These results suggest that there may be weak associations between METTL3 gene variation of and PTB susceptibility, and these variants may interact with each other to modify the PTB risk. This should be confirmed in future studies with larger sample sizes. In addition, rs1139130 polymorphism was also significantly associated with drug resistance and DILI in PTB patients. This further proved that METTL3 gene variation was involved in the pathogenesis of PTB, and indicated that this SNP could be used to predict adverse reactions in patients, thus contributing to the formulation of more appropriate treatment measures.
In previous studies, METTL14 gene rs62328061 polymorphism was significantly associated with reduced susceptibility to neuroblastoma (34), and rs1064034, rs298982 polymorphisms were significantly associated with reduced susceptibility to Wilms’ tumor (21). In the current study, the rs62328061 GG genotype was associated with an increased risk of PTB, but there was no significant association between the rs1064034 variant and PTB susceptibility. These results provided key indications of an association between METTL14 gene variation and PTB susceptibility. In PTB patients, a decreased frequency of the rs62328061 G allele was associated with leukopenia, and an increased frequency of the rs4834698 TC genotype was associated with hypoproteinemia. This confirmed the important role of METTL14 gene variation in the development of PTB, and further functional studies are needed to investigate the molecular mechanisms involved in the effects of rs62328061 on PTB risk. The present study also investigated associations between WTAP gene polymorphisms and the risk of PTB, and the WTAP rs11752345 CT genotype and T allele frequencies were significantly increased in PTB patients. Moreover, the rs11752345 T allele was closely related to sputum smear in PTB patients. These results suggested that the CT genotype/T allele of rs11752345 may increase the risk of PTB, and the T allele may be useful for distinguishing different phenotypes in PTB patients.
Increasing studies indicate that transcription levels of m6A methyltransferases are closely related to the pathogenesis and progression of many diseases. Dysregulation of METTL3 is considered an important factor affecting the progression of various malignant tumors such as endometrial cancer (35) and bladder cancer (36). Another study detected lower METTL14 expression in colorectal cancer (37). Functional m6A methyltransferase SNPs may affect m6A methyltransferase expression and thus modify disease susceptibility. Hence, we designed a case-control study to determine METTL3, METTL14, and WTAP transcription levels in PTB patients to investigate the possible mechanism of SNP-mediated PTB susceptibility. Transcription levels of METTL3, METTL14, and WTAP were significantly decreased in PTB patients compared to controls. This suggested that METTL3, METTL14, and WTAP may be involved in PTB occurrence, and their levels may be used as auxiliary indicators for PTB diagnosis. This must be verified in the future by well-designed studies. WTAP level was also significantly associated with drug resistance, ALT, and AST, and METTL3 level was negatively associated with ESR and AST in PTB patients. These findings contribute to improving our understanding of the role of m6A methyltransferases in PTB development. Lastly, we analyzed potential associations between m6A methyltransferase gene variants and their transcription levels in PTB patients. No significant associations were detected, possibly due to the small sample size.
The major strengths of this study include its novelty and rational design, but the study also had several limitations. It was focused on the analysis of genetic factors in PTB, but it did not assess the effects of environmental factors on PTB, or interactions between environmental factors and genetic variation. Second, associations between gene variation and the prognosis of PTB were not analyzed. Lastly, the study lacked a functional investigation into the effects of m6A methyltransferase on PTB pathogenesis.
In summary, the current study indicates that polymorphisms of METTL14 rs62328061 and WTAP rs11752345 may contribute to increased PTB susceptibility, and several SNPs in METTL3, METTL14, and WTAP genes were associated with the clinical features including drug resistance, DILI, and leukopenia in PTB patients. The study demonstrated the important roles of alterations in METTL3, METTL14, and WTAP in PTB, and these m6A methyltransferases in the development of PTB. In order to further validate these findings and determine the underlying biological mechanisms involving these m6A methyltransferases in PTB, more studies incorporating larger sample sizes and different ethnic populations are warranted.
Data availability statement
The data presented in the study are deposited in the dbSNP (1063446) . Further inquiries can be directed to the corresponding authors.
Ethics statement
This study was approved by the Ethical Committee of Anhui Medical University. The patients/participants provided their written informed consent to participate in this study.
Author contributions
T-PZ and H-ML designed the study. H-ML conducted the experiment. RL performed the statistical analyses. L-JW and QH participated in the collection of samples. H-ML and T-PZ drafted the manuscript. All author contributed to manuscript revision. All the authors approved the final submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (82003515).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Kaufmann SHE. New vaccines against tuberculosis. Bundesgesundheitsbla (2020) 63:56–64. doi: 10.1007/s00103-019-03065-y
2. World Health Organization. Global tuberculosis report (2021). Available at: https://www.who.int/tb/publications/global_report/en/.
3. Ji G, Zhang M, Liu Q, Wu S, Wang Y, Chen G, et al. Functional polymorphism in the NFE2L2 gene associated with tuberculosis susceptibility. Front Immunol (2021) 12:660384. doi: 10.3389/fimmu.2021.660384
5. Caws M, Thwaites G, Dunstan S, Hawn TR, Lan NTN, Thuong NTT, et al. The influence of host and bacterial genotype on the development of disseminated disease with mycobacterium tuberculosis. PloS Pathog (2008) 4:e1000034. doi: 10.1371/journal.ppat.1000034
6. Gagneux S. Host-pathogen coevolution in human tuberculosis. Philos Trans R Soc Lond B Biol Sci (2012) 367:850–9. doi: 10.1098/rstb.2011.0316
7. Orlova M, Schurr E. Human genomics of mycobacterium tuberculosis infection and disease. Curr Genet Med Rep (2017) 5:125–31. doi: 10.1007/s40142-017-0124-7
8. Zhang TP, Chen SS, Zhang GY, Shi SJ, Wei L, Li HM. Association of vitamin d pathway genes polymorphisms with pulmonary tuberculosis susceptibility in a Chinese population. Gnes Nutr (2021) 16:6. doi: 10.1186/s12263-021-00687-3
9. Harishankar M, Selvaraj P, Bethunaickan R. Influence of genetic polymorphism towards pulmonary tuberculosis susceptibility. Front Med (Lausanne) (2018) 5:213. doi: 10.3389/fmed.2018.00213
10. Ortega E, Hernández-Bazán S, Sánchez-Hernández B, Licona-Limón I, Fuentes-Dominguez J. TLR4 single nucleotide polymorphisms in affect susceptibility to tuberculosis in Mexican population from the state of veracruz. J Immunol Res (2020) 2020:2965697. doi: 10.1155/2020/2965697
11. Cao G, Li HB, Yin Z, Flavell RA. Recent advances in dynamic m6A RNA modification. Open Biol (2016) 6:160003. doi: 10.1098/rsob.160003
12. Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N(6)-methyladenosine modification in cancers: Current status and perspectives. Cell Res (2018) 28:507–17. doi: 10.1038/s41422-018-0034-6
13. He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer (2019) 18:176. doi: 10.1186/s12943-019-1109-9
14. Wang M, Kong W, He B, Li Z, Song H, Shi P, et al. Vitamin d and the promoter methylation of its metabolic pathway genes in association with the risk and prognosis of tuberculosis. Clin Epigenet (2018) 10:118. doi: 10.1186/s13148-018-0552-6
15. Shell SS, Prestwich EG, Baek SH, Shah RR, Sassetti CM, Dedon PC, et al. DNA Methylation impacts gene expression and ensures hypoxic survival of mycobacterium tuberculosis. PloS Pathog (2013) 9:e1003419. doi: 10.1371/journal.ppat.1003419
16. Gong Z, Wang G, Zeng J, Stojkoska A, Huang H, Xie J. Differential DNA methylomes of clinical MDR, XDR and XXDR mycobacterium tuberculosis isolates revealed by using single-molecule real-time sequencing. J Drug Target (2021) 29:69–77. doi: 10.1080/1061186X.2020.1797049
17. Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN, et al. METTL3 facilitates tumor progression via an m6A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer (2019) 18:112. doi: 10.1186/s12943-019-1038-7
18. Orouji E, Peitsch WK, Orouji A, Houben R, Utikal J. Oncogenic role of an epigenetic reader of m6A RNA modification: YTHDF1 in merkel cell carcinoma. Cancers (Basel) (2020) 12:202. doi: 10.3390/cancers12010202
19. Bian J, Zhuo Z, Zhu J, Yang Z, Jiao Z, Li Y, et al. Association between METTL3 gene polymorphisms and neuroblastoma susceptibility: A nine-centre case-control study. J Cell Mol Med (2020) 24:9280–6. doi: 10.1111/jcmm.15576
20. Song RH, Liu XR, Gao CQ, Du P, Zhang JA. METTL3 gene polymorphisms contribute to susceptibility to autoimmune thyroid disease. Endocrine (2021) 72:495–504. doi: 10.1007/s12020-020-02503-1
21. Zhuo Z, Hua RX, Zhang H, Lin H, Fu W, Zhu J, et al. METTL14 gene polymorphisms decrease wilms tumor susceptibility in Chinese children. BMC Cancer (2021) 21(1):1294. doi: 10.1186/s12885-021-09019-5
22. Luo A, Yang L, Li M, Cai M, Huang A, Liu X, et al. Genetic variants in METTL14 are associated with the risk of acute lymphoblastic leukemia in southern Chinese children: A five-center case-control study. Cancer Manag Res (2021) 13:9189–200. doi: 10.2147/CMAR.S335925
23. Feng Y, Wang F, Pan H, Qiu S, Lü J, Wu L, et al. Obesity-associated gene FTO rs9939609 polymorphism in relation to the risk of tuberculosis. BMC Infect Dis (2014) 14:592. doi: 10.1186/s12879-014-0592-2
24. Li HM, Wang LJ, Tang F, Pan HF, Zhang TP. Association of leptin and leptin receptor genes variants and pulmonary tuberculosis susceptibility, clinical manifestations in a Chinese population. Microb Pathog (2022) 165:105499. doi: 10.1016/j.micpath.2022.105499
25. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc (2008) 3:1101–8.27. doi: 10.1038/nprot.2008.73
26. Li Z, Zhang Z, He Z, Tang W, Li T, Zeng Z, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis. Cell Res (2009) 19:519–23. doi: 10.1038/cr.2009.33
27. Ruan X, Tian M, Kang N, Ma W, Zeng Y, Zhuang G, et al. Genome-wide identification of m6A-associated functional SNPs as potential functional variants for thyroid cancer. Am J Cancer Res (2021) 11:5402–14.
28. Chen M, Lin W, Yi J, Zhao Z. Exploring the epigenetic regulatory role of m6A-associated SNPs in type 2 diabetes pathogenesis. Pharmgenom Pers Med (2021) 14:1369–78. doi: 10.2147/PGPM.S334346
29. Meyer KD, Jaffrey SR. Rethinking m6A readers, writers, and erasers. Annu Rev Cell Dev Biol (2017) 33:319–42. doi: 10.1146/annurev-cellbio-100616-060758
30. Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res (2014) 24:177–89. doi: 10.1038/cr.2014.3
31. Zhuo ZJ, Hua RX, Chen Z, Zhu J, Wang M, Yang Z, et al. WTAP gene variants confer hepatoblastoma susceptibility: A seven-center case-control study. Mol Ther Oncol (2020) 18:118–25. doi: 10.1016/j.omto.2020.06.007
32. Lin A, Zhou M, Hua RX, Zhang J, Zhou H, Li S, et al. METTL3 polymorphisms and wilms tumor susceptibility in Chinese children: A five-center case control study. J Gene Med (2020) 22:e3255. doi: 10.1002/jgm.3255
33. Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest (2008) 118:1590–605. doi: 10.1172/JCI34772
34. Zhuo Z, Lu H, Zhu J, Hua RX, Li Y, Yang Z, et al. METTL14 gene polymorphisms confer neuroblastoma susceptibility: An eight-center case-control study. Mol Ther Nucleic Acids (2020) 22:17–26. doi: 10.1016/j.omtn.2020.08.009
35. Liu J, Eckert MA, Harada BT, Liu SM, Lu Z, Yu K, et al. M(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat Cell Biol (2018) 20:1074–83. doi: 10.1038/s41556-018-0174-4
36. Jin H, Ying X, Que B, Wang X, Chao Y, Zhang H, et al. N6-methyladenosine modification of ITGA6 mRNA promotes the development and progression of bladder cancer. EBioMedicine (2019) 47:195–207. doi: 10.1016/j.ebiom.2019.07.068
Keywords: m6A methyltransferase, single nucleotide polymorphisms, pulmonary tuberculosis, METTL3, METTL14, WTAP
Citation: Zhang T-P, Li R, Wang L-J, Huang Q and Li H-M (2022) Roles of the m6A methyltransferases METTL3, METTL14, and WTAP in pulmonary tuberculosis. Front. Immunol. 13:992628. doi: 10.3389/fimmu.2022.992628
Received: 12 July 2022; Accepted: 15 August 2022;
Published: 07 December 2022.
Edited by:
Jianping Xie, Southwest University, ChinaCopyright © 2022 Zhang, Li, Wang, Huang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong-Miao Li, eXVxaWNhaTE5OTBAcXEuY29t
†These authors have contributed equally to this work and share first authorship
 Tian-Ping Zhang
Tian-Ping Zhang Rui Li2†
Rui Li2† Qian Huang
Qian Huang Hong-Miao Li
Hong-Miao Li