
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 04 October 2022
Sec. Inflammation
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.988668
This article is part of the Research Topic Acute-on-chronic Liver Failure: Systemic Inflammation and Immunosuppression View all 12 articles
 Ya-Chao Tao1,2
Ya-Chao Tao1,2 Yong-Hong Wang1,2
Yong-Hong Wang1,2 Meng-Lan Wang1,2
Meng-Lan Wang1,2 Wei Jiang1,2
Wei Jiang1,2 Dong-Bo Wu1,2
Dong-Bo Wu1,2 En-Qiang Chen1,2*
En-Qiang Chen1,2* Hong Tang1,2*
Hong Tang1,2*Background: Acute liver failure (ALF) and acute-on-chronic liver failure (ACLF) are the two most common subtypes of liver failure. They are both life-threatening clinical problems with high short-term mortality. Although liver transplantation is an effective therapeutic, its application is limited due to the shortage of donor organs. Given that both ACLF and ALF are driven by excessive inflammation in the initial stage, molecules targeting inflammation may benefit the two conditions. MicroRNAs (miRNAs) are a group of small endogenous noncoding interfering RNA molecules. Regulation of miRNAs related to inflammation may serve as promising interventions for the treatment of liver failure.
Aims: To explore the role and mechanism of miR-125b-5p in the development of liver failure.
Methods: Six human liver tissues were categorized into HBV-non-ACLF and HBV-ACLF groups. Differentially expressed miRNAs (DE-miRNAs) were screened and identified through high-throughput sequencing analysis. Among these DE-miRNAs, miR-125b-5p was selected for further study of its role and mechanism in lipopolysaccharide (LPS)/D-galactosamine (D-GalN) -challenged Huh7 cells and mice in vitro and in vivo.
Results: A total of 75 DE-miRNAs were obtained. Of these DE-miRNAs, miR-125b-5p was the focus of further investigation based on our previous findings and preliminary results. We preliminarily observed that the levels of miR-125b-5p were lower in the HBV-ACLF group than in the HBV-non-ACLF group. Meanwhile, LPS/D-GalN-challenged mice and Huh7 cells both showed decreased miR-125b-5p levels when compared to their untreated control group, suggesting that miR-125b-5p may have a protective role against liver injury, regardless of ACLF or ALF. Subsequent results revealed that miR-125b-5p not only inhibited Huh7 cell apoptosis in vitro but also relieved mouse ALF in vivo with evidence of improved liver histology, decreased alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels, and reduced tumor necrosis factor-α (TNF-α) and IL-1β levels. Based on the results of a biological prediction website, microRNA.org, Kelch-like ECH-associated protein 1 (Keap1) was predicted to be one of the target genes of miR-125b-5p, which was verified by a dual-luciferase reporter gene assay. Western blot results in vitro and in vivo showed that miR-125b-5p could decrease the expression of Keap1 and cleaved caspase-3 while upregulating the expression of nuclear factor (erythroid-derived 2)-like 2 (Nrf2) and heme oxygenase-1(HO-1).
Conclusion: Upregulation of miR-125b-5p can alleviate acute liver failure by regulating the Keap1/Nrf2/HO-1 pathway, and regulation of miR-125b-5p may serve as an alternative intervention for liver failure.
Liver failure is a life-threatening clinical problem with high short-term mortality. It can present as acute liver failure (ALF, without pre-existing chronic liver disease), acute-on-chronic liver failure (ACLF, an acute deterioration of underlying chronic liver disease) or an acute decompensation of an end-stage liver disease (1). Liver transplantation is an effective therapeutic option, irrespective of the etiology of liver failure. However, the application of liver transplantation is limited due to the shortage of donor organs (2). Thus, liver failure is still a severe clinical challenge, and other interventions assisting alleviating liver injury are being explored.
ALF and ACLF are the most widely discussed owing to their high incidence worldwide. Better knowledge of the pathophysiology of these diseases can provide insights into novel therapies. It has been reported that the development of both ALF and ACLF is driven by immune dysfunction and inflammatory imbalance, although the conditions are distinct clinical entities (3–5). The immune and inflammatory status of the diseases is dynamic, progressing from intensive inflammation to the development of immunoparalysis (3, 4, 6). In the initial phase of ALF, immune cells participating in the innate response are activated to produce proinflammatory mediators, which can stimulate a systemic inflammatory response. Patients with ACLF display an excessive innate immune response, which is characterized by leukocytosis, neutrophilia and lymphopenia, together with high levels of inflammatory mediators (7, 8). Initial systemic inflammatory response syndrome (SIRS) due to acute insult and/or subsequent secondary infection due to immunoparalysis can lead to extrahepatic organ failure (4, 9, 10). Thus, strategies modulating immune and proinflammatory mediators can be potential targets to alleviate liver failure.
MicroRNAs (miRNAs) are a class of small endogenous noncoding interfering RNA molecules and can induce gene silencing and translational repression by binding specific sequences in target mRNAs, thereby playing key roles in biological processes and in the development of various diseases (11). Accumulating studies have demonstrated that miRNAs are involved in the modulation of immunity and inflammation, and regulation of these miRNAs may be potential therapeutics for clinical problems (12–14). For instance, inhibition of miR-34b-5p could attenuate inflammation and apoptosis in acute lung injury, and thus miR‐34b‐5p and its target progranulin might be a potential intervention pathway for the treatment of acute lung injury (15). Thus, the regulation of miRNAs and their target genes may improve the outcome of various diseases by regulating immunity and inflammation.
In the present study, we identified miRNAs that were differentially expressed in human HBV-ACLF tissues compared to HBV-non-ACLF tissues. Then, we further explored the role of a certain miRNA in the development of liver failure, aiming to determine whether the miRNA may improve liver failure by regulating intensive inflammation, hoping to pave the way for miRNA-targeting therapies for liver failure.
Six patients with chronic HBV infection were enrolled. They all received antiviral therapy with nucleos(t)ide analogs. They were divided into HBV-non-ACLF (n=3) and HBV-ACLF (n=3) groups based on their liver histopathology and liver function. The histological assessments were performed using the METAVIR scoring system. Briefly, the degree of inflammation in the liver biopsies was assessed with the standard METAVIR histology activity index scoring system, defined as A0, no inflammation; A1, mild inflammation; A2, moderate inflammation, and A3, severe inflammation. The histological appearance of fibrosis was classified as F0 to F4, ranging from no fibrosis to cirrhosis (16).
HBV-non-ACLF referred to patients with chronic hepatitis B (CHB) who had abnormal liver function and mild-to-moderate inflammatory activity (≤A2) in the liver tissue due to HBV infection, but did not meet the diagnostic criteria of ACLF.
ACLF is diagnosed according to the recommendations of the Asian Pacific Association for the Study of the Liver (APASL) (1). Herein, patients who were previously diagnosed with CHB or cirrhosis belonged to the HBV-ACLF group if they manifested with jaundice (bilirubin>5 mg/dL), coagulopathy [prolonged international normalized ratio (INR)>1.5], encephalopathy, and ascites within 4 weeks. The histopathological findings of the HBV-ACLF group showed evident infiltration of inflammatory cells (A3), accompanied by the formation of pseudolobuli (F4). All HBV-ACLF patients later received liver transplantation.
Patients were excluded if they had any of the following conditions: (1) infections with other hepatitis viruses (including A, C, D, and E) or human immunodeficiency virus (HIV); (2) evidence of drug-induced liver injury, alcoholic liver disease, autoimmune liver diseases, severe systemic illnesses; (3) malignancies, such as hepatocellular carcinoma.
This study was carried out in accordance with the Declaration of Helsinki. All patients provided verbal informed consent. Detailed information about the study cohort is described in Table 1.
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The quantity and purity of total RNA were analyzed with an Agilent 2100 Bioanalyzer (Agilent, USA) with RIN> 6.5. Small RNAs of different length were separated using denaturing polyacrylamide gel electrophoresis (PAGE). Fragments between 18 and 30 nt in length were gel-purified and ligated to adaptors at both the 3′- and 5′-ends. The ligation products were subsequently reverse- transcribed into cDNA, and PCR amplification was performed using an Illumina sequencing kit (Illumina, USA) to generate a cDNA library according to previous studies (17, 18).
High-throughput sequencing was performed by Chengdu Life Baseline Technology Co., Ltd. As shown in Supplement 1, the raw read sequences were filtered to remove low-quality reads, 5′ adaptor contaminant reads, reads without 3’ adaptor sequences, reads containing poly (A) and adapter sequences, sequences <18 nt and sequences >32 nt. The clean reads were obtained and their length distributions were calculated using Fastx-Toolkit (19). Then, the clean reads were mapped and aligned to the human reference genome group and other small RNA databases using Bowtie2 software (20). The known miRNAs and novel miRNAs were identified and predicted using miRbase and miRDeep2, respectively (21, 22).
The miRNA expression levels between the two groups were compared to identify differentially expressed (DE)-miRNAs. The expression of miRNA was normalized using transcripts per million (TPM) as the following formula: TPM = (mapped read count/total clean read count) ×106 (23). DE-miRNAs were defined as |log2 (fold change)| >1 between two groups with a false discovery rate (FDR) of <0.05.
To understand the functions of these DE-miRNAs, their potential target genes were predicted by RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/) and miRanda (http://www.microrna.org/microrna/home.do) (24, 25). Only the target genes predicted by both methods were considered reliable targets for further analysis. Gene ontology (GO) functional enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed on these target genes (26, 27). GO analysis was conducted to provide functional annotation for predicted target genes of miRNAs by analyzing the classifications of Biological Process, Cellular Component and Molecular Function. Pathway analysis was based on KEGG, which is a database resource for understanding the high-level functions and utilities of biological systems. p<0.05 was regarded as the cutoff to select significantly enriched terms.
The human HCC cell line Huh7 was preserved in our laboratory. Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, D6429, Sigma, USA) containing 10% fetal bovine serum (12103C, Sigma, USA) and 1% penicillin/streptomycin under standard culture conditions (a humidified 5% carbon dioxide incubator at 37°C).
The miR-125b-5p overexpression vector and negative control (NC) vector were designed and provided by Heyuan Biotechnology (OBIO, Shanghai, China). After growth to 60%-70% confluence in six-well plates, Huh7 cells were transfected with miR-125b-5p vector or NC vector using Lipofectamine™ 2000 (11668019, Invitrogen, USA). On the following day, the cells were incubated with normal medium.
Apoptotic cells were assessed using an Annexin V- Fluorescein isothiocyanate (FITC) apoptosis detection kit (APOAF, Sigma, USA) according to the manufacturer’s protocol. Huh7 cells were collected and washed twice with PBS. Then, the cells were resuspended in 500 μL of 1× binding buffer and stained with 5 μL of Annexin V-FITC conjugate and 10 μL of PI solution. After incubation for 15 min in the dark at room temperature, stained cells were analyzed by flow cytometry (BD Accuri™ C6 flow cytometer).
Total RNA was extracted from Huh7 cells and liver tissues using TRIzol Reagent (15596026, Invitrogen, USA) according to the manufacturer’s instructions. Reverse transcription was performed using a First Strand cDNA Synthesis Kit (B300537, Sangon Biotech, China). The expression of miR-125b-5p was determined by quantitative real-time PCR using a miRNA qPCR detection kit (B532461, Sangon Biotech, China), as previously reported (28). The forward primer sequence for miR-125b-5p was CGTCCCTGAGA- CCCTAACTTGTGA. The reverse primers for miR-125b-5p were universal adaptor primers designed and provided by Sangon Biotech Company (Shanghai, China). The level of miR-125b-5p was calculated using the relative quantification 2-ΔΔCT method and normalized to the U6 transcript (Bio-Rad CFX Manager software), as previously described (29, 30)
Male C57BL/6J mice (6-8 weeks old, weighing 20-25 g) were purchased from Huaxi Laboratory Animal Center of Sichuan University (Chengdu, China). All mice were maintained under controlled conditions (24°C, 55% humidity and 12-h day/night rhythm) and given free access to water and food. The mice received humane care under guidance from the Institutional Review Board in accordance with the Animal Protection Art of Sichuan University. After 1 week of acclimation, the mice were prepared for further study.
The mouse model of ALF was established using lipopolysaccharide (LPS) and D-galactosamine (D-GalN) as previously described (31). In brief, C57BL/6J mice were given 700 mg/kg D-GalN (G0500, Sigma, USA) and 10 μg/kg LPS (Escherichia coli, 0111:B4, L2630, Sigma, USA) by intraperitoneal injection. Mice in the present study were randomly divided into four groups (n = 8/group): a normal control group, an LPS/D-GalN group, an LPS/D-GalN+ negative control (NC) group and an LPS/D-GalN+ miR-125b-5p group.
The miR-125b-5p overexpression vector or negative control vector was administered to mice via tail vein prior to the establishment of ALF as previously reported (32, 33). Seven hours after ALF model establishment, all mice were sacrificed, and serum and liver samples were harvested and stored for further analysis.
Liver tissue was obtained and fixed with 4% paraformaldehyde at 4°C for 48 h and then embedded in paraffin. After immobilization, samples were cut into sections and then stained with hematoxylin-eosin (HE) using a standard protocol. The sections were visualized under a light microscope, and representative images are presented.
Serum samples were collected from mice to detect alanine aminotransferase (ALT) and aspartate aminotransferase (AST) by an automatic biochemical analyzer.
The levels of serum tumor necrosis factor -α (TNF-α, EMC102a, NeoBioscience, China) and IL-1β (EMC001b, NeoBioscience, China) were detected using ELISA kits according to the manufacturer’s instructions.
Protein was extracted from Huh7 cells and liver tissues. The isolated proteins were separated by polyacrylamide gel electrophoresis and then transferred onto polyvinylidene fluoride (PVDF) membranes. Subsequently, the blots were exposed to primary antibodies against Kelch-like ECH-associated protein 1 (Keap1, #8047, CST, USA), nuclear factor (erythroid-derived 2)-like 2 (Nrf2, sc-365949, Santa Cruz, USA), heme oxygenase-1 (HO-1, #43966, CST, USA), and cleaved caspase-3 (#9661, CST, USA). Anti-GAPDH (TA-08), anti-β-tubulin (TA-10) and anti-β-actin (TA-09) were used as internal references, and were purchased from Beijing Zhong Shan-Golden Bridge Biological Technology Co., Ltd. The images of the gels were captured in a Bio-Rad Image Lab (ChemiDoc™ MP Imaging System, Bio-Rad, California, USA).
A biological prediction website, microRNA.org, was used to analyze the target genes of miR-125b-5p, and a dual-luciferase reporter gene assay was performed to detect whether miR-125b-5p extensively targeted Keap1. The full length of the 3′-untranslated region (3′UTR) (forward primer: GAGGAGTTGTGTTTGTGGAC; reverse primer: TGTAAAACGACGGCCAGT) of the Keap1 gene was amplified clonally. The wild-type (WT) vector of Keap1 (Keap1-WT) and a mutant type vector (Keap1-MUT) were constructed. Bioinformatics software was used to predict the binding sites between miR-125b-5p and Keap1. The vectors were cotransfected with miR-125b-5p mimic and mimic negative control into 293T cells according to the following groups: Keap1-WT+ miR-125b-5p mimic negative control (miR-NC), Keap1-WT+ miR-125b-5p mimic, Keap1-MUT+ miR-NC, and Keap1-MUT+ miR-125b-5p mimic. Firefly and Renilla luciferase activities were measured using a dual luciferase reporter gene detection kit. The experiment was repeated three times in each group.
Statistical analysis was performed using SPSS 20.0 software. Data are presented as the mean± deviation (SD) determined from a minimum of three independent experiments each performed with triplicate cultures. One-way ANOVA was performed to evaluate the level of significance, and the results were considered statistically significant if p<0.05.
Six liver tissues were obtained and categorized into HBV-non-ACLF and HBV-ACLF groups based on the severity of liver injury (Table 1, Figure 1A). The sequences of small RNAs ranged from 18-30 nt in length, of which the majority were 21-23 nt long, and the 22-nt small RNAs were the most abundant (Figure 1B). After removal of the adaptor, insert, poly (A) tail and short RNAs of <18 nt, a total of 59,160,359 and 49,723,211 clean reads were obtained for the two groups, respectively (Supplement 2). Small RNA sequences were searched against the GenBank, Rfam, and Repbase databases by Bowtie2 software, rRNA, snRNA, snoRNA, tRNA, sRNA, etc., were annotated and removed, and the unannotated RNAs were subjected to further analyses for miRNA identification (Figure 1C). We used the software miRbase and miRDeep2 to map the retained sequence reads to identify candidate miRNAs.
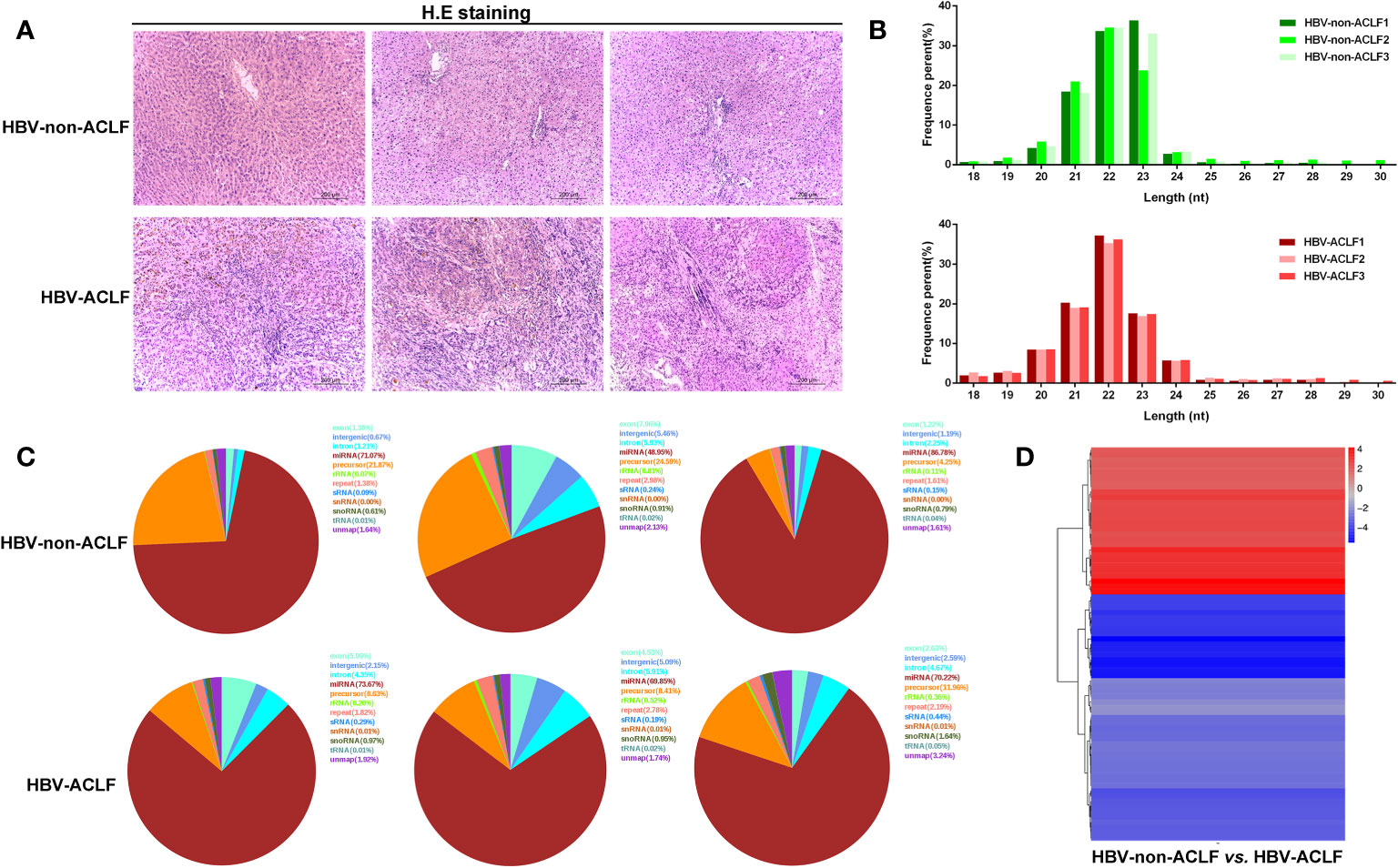
Figure 1 Results of high-throughput sequencing and identification of DE-miRNAs. (A) H&E staining of liver tissues from HBV-non-ACLF and HBV-ACLF groups; (B) Length distribution of the small RNA library; (C) Distribution of small RNAs among different categories; (D) Heatmap of DE-miRNAs constructed by comparing the levels of miRNAs between the two groups.
DE-miRNAs were obtained when they met the criteria of |log2 (fold change)|>1 between the two groups and FDR<0.05, and these DE-miRNAs are presented in a heatmap (Figure 1D). Compared with the HBV-non-ACLF group, the levels of 28 miRNAs were increased, while the levels of 47 miRNAs were decreased in the HBV-ACLF group. Some of the upregulated and downregulated miRNAs are shown in Tables 2 and 3, respectively.
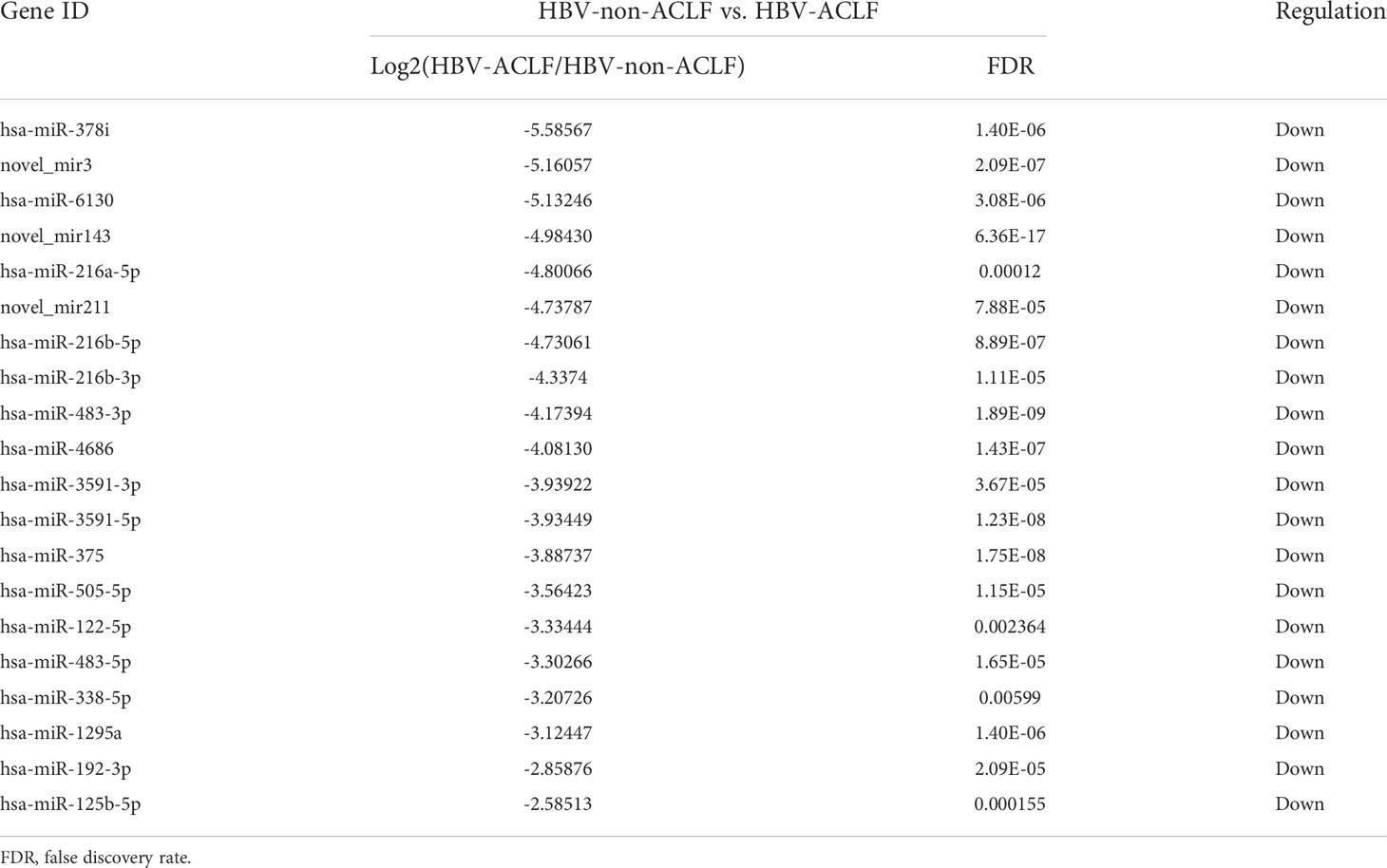
Table 3 Twenty of the downregulated DE-miRNAs in the HBV-ACLF group compared to the HBV-non-ACLF group.
To further understand the roles of miRNAs in the progression of liver injury, GO and KEGG analyses of putative target genes of these DE-miRNAs were performed. GO describes genes from three aspects, namely molecular function (MF), cellular component (CC) and biological process (BP).
Among the DE-miRNA target genes, “binding” was the most represented in the MF category, followed by “catalytic activity”, “nucleic acid binding transcription factor activity”, “antioxidant activity” and “transporter activity”. For the CC category, target genes mostly participated in the “cell”, “cell part”, “organelle”, “organelle part” and “membrane”. Genes involved in the “cellular process”, “single-organism process”, “biological process”, “metabolic process” and “regulation of biological process” were notably represented in the BP category (Figure 2A). In the KEGG analysis, target genes were primarily enriched in the “pathways in cancer”, “toxoplasmosis”, “pertussis”, “leishmaniasis” and “microRNAs in cancer” (Figure 2B).
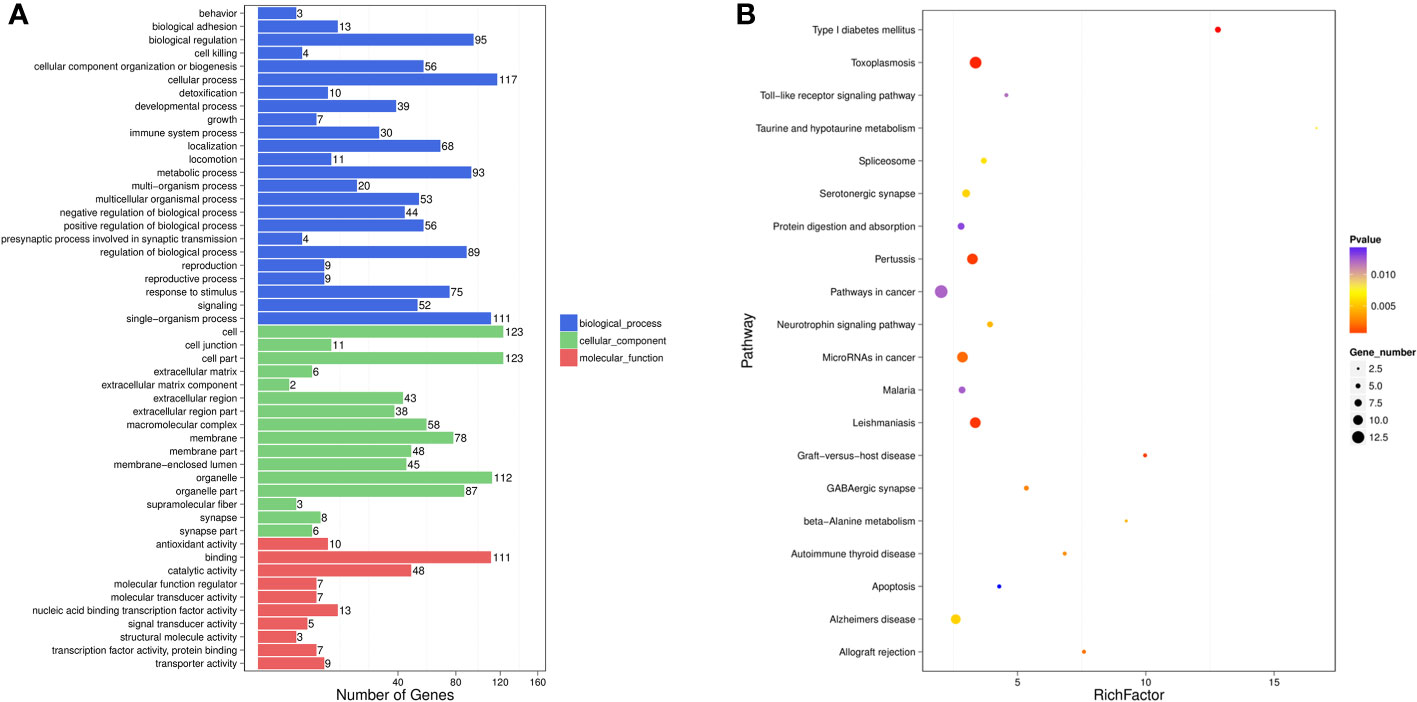
Figure 2 GO functional enrichment analysis (A) and KEGG pathway analysis (B) for DE-miRNAs obtained from the comparsion between HBV-non-ACLF and HBV-ACLF groups. Rich actor is the ratio of the DE gene number to the total gene number in a certain pathway. The size and color of the dots represent the gene number and range of the p value, respectively.
We previously proved that serum miR-125b-5p was associated with the severity of HBV-related liver damage, and high serum miR-125b-5p might serve as a predictor for poor outcomes in HBV-ACLF cases (28). In the present study, we observed that the levels of hepatic miR-125b-5p were decreased with the aggravation of HBV-induced liver injury based on miRNA array data. Meanwhile, we verified the expression of miR-125b-5p using 20 pairs of liver tissues from patients with HBV-non-ACLF and HBV-ACLF and confirmed that the levels of hepatic miR-125b-5p were lower in HBV-ACLF patients than in HBV-non-ACLF patients (p<0.001, Supplement 3A). In addition, the levels of miR-125b-5p were decreased in LPS/D-GalN-challenged Huh7 cells (p<0.01, Supplement 3B) and mice (p<0.001, Supplement 3C) compared to those in untreated control groups. Thus, it was well-founded to speculate that miR-125b-5p might play an important role in the development of liver failure, regardless of ACLF or ALF, and we focused on this miRNA for further study.
To determine the role of miR-125b-5p, Huh7 cells were transfected with miR-125b-5p overexpression vector or negative control vector. The expression of miR-125b-5p was significantly increased in the cells of the experimental group compared with the control group (p<0.001, Supplement 3D). Twenty-four hours after transfection with the vector, Huh7 cells were treated with LPS (20 ng/mL)/D-GalN (3 mol/mL) for 36 hours. Cell apoptosis was analyzed using annexin-V flow cytometry. As shown in Figures 3A, B, the apoptosis rates were significantly lower in cells transfected with miR-125b-5p vector before LPS/D-GalN addition than in cells only treated with LPS/D-GalN (29.41% ± 2.61% vs. 44.63% ± 5.45%, p<0.05) and in cells transfected with negative control vector (29.41% ± 2.61% vs. 50.39% ± 6.71%, p<0.01), demonstrating that miR-125b-5p may help to alleviate acute liver injury in vitro by inhibiting cell apoptosis.
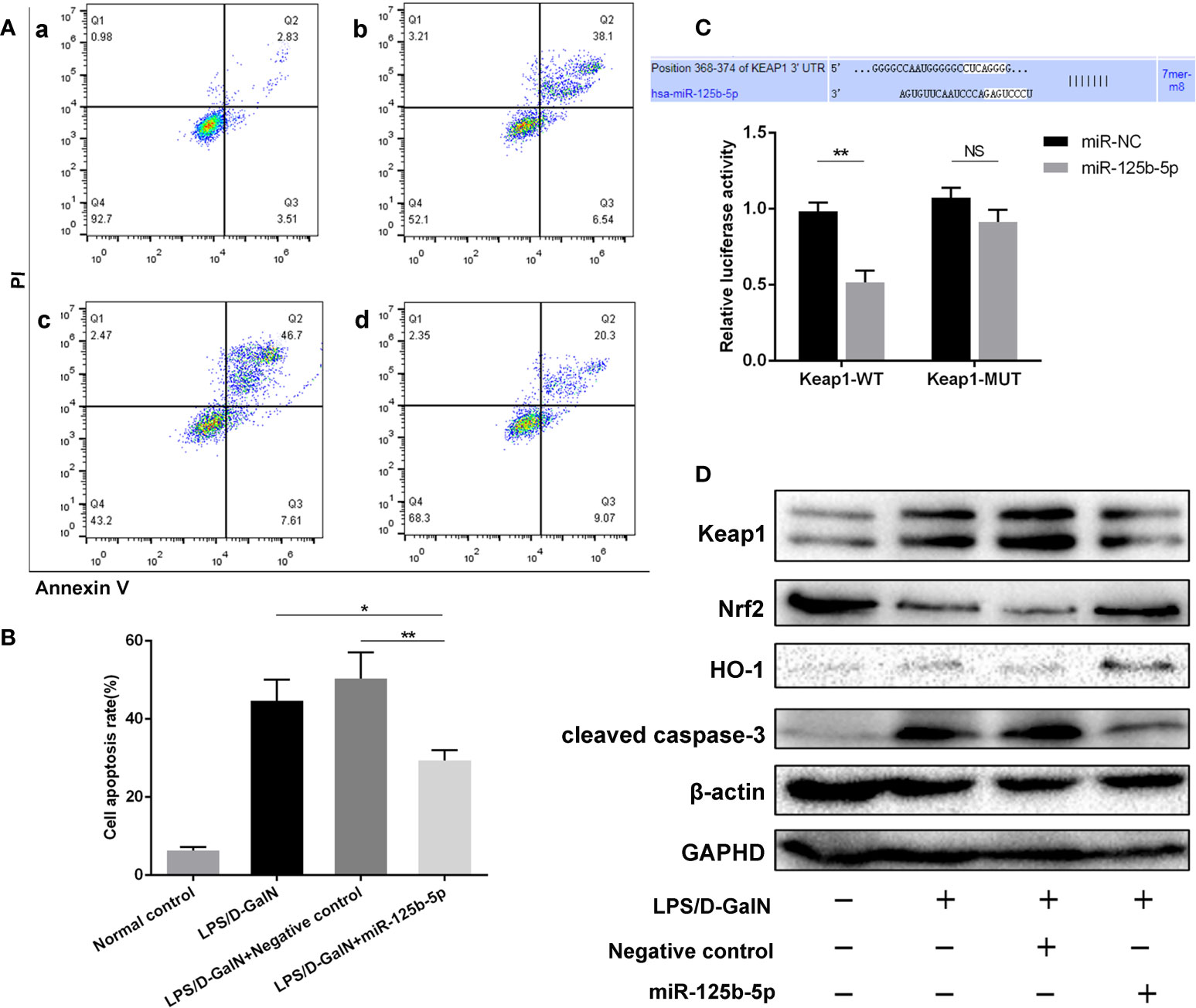
Figure 3 Role and mechanism of miR-125b-5p in injured Huh7 cells in vitro. (A) The cell apoptosis rate was calculated by flow cytometry. a: Normal control; b: LPS/D-GalN; c: LPS/D-GalN+ Negative control vector; d: LPS/D-GalN+ miR-125b-5p. (B) The cell apoptosis rate was calculated and compared. (C) A luciferase reporter assay was performed to identify the binding sites between human Keap1 and hsa-miR-125b-5p. (D) The expression levels of Keap1, Nrf2, HO-1 and cleaved caspase-3 were evaluated by western blotting. NS, no significance. *p< 0.05, **p < 0.01.
To further identify the protective role of miR-125b-5p in ALF in vivo, we established mouse ALF through intraperitoneal injection of LPS/D-GalN and assessed the improvements in liver histopathology and serum biochemical indicators after administration of the miR-125b-5p overexpression vector. The liver histopathological characteristics of the negative control groups showed no significant differences from those of the LPS/D-GalN-induced ALF groups, whereas administration of the miR-125b-5p vector improved the liver histopathology based on the retained liver structure, decreased inflammatory cell infiltration and reduced hemorrhage (Figure 4A). In addition, miR-125b-5p also reduced ALT and AST levels in mice treated with LPS/D-GalN (p<0.001, Figure 4B). Serum TNF-α and IL-1β levels were enhanced in the LPS/D-GalN-induced ALF group. In contrast, significant decreases in the two proinflammatory cytokines were observed in the miR-125b-5p treated group (p<0.001, Figure 4C).
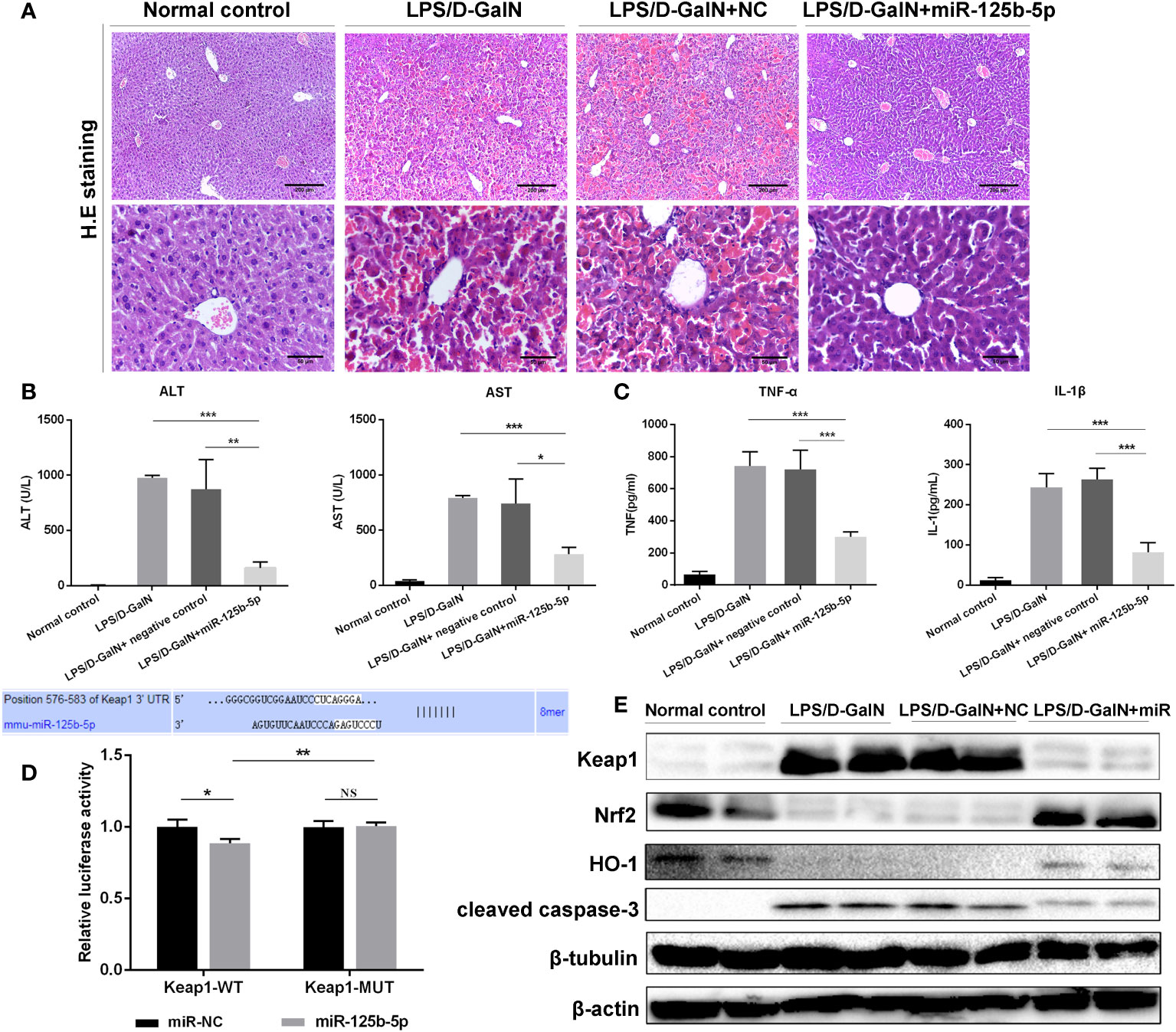
Figure 4 Role and mechanism of miR-125b-5p in LPS/D-GalN- induced ALF in vivo. (A) H&E staining of liver tissue. (B) Detection of serum ALF and AST. (C) Detection of two serum proinflammtory mediators, TNF-α and IL-1 β, using ELISA. (D) A luciferase reporter assay was performed to identify the binding sites between mouse Keap1 and mmu-miR-125b-5p. (E) The expression levels of Keap1, Nrf2, HO-1 and cleaved caspase-3 were evaluated by western blotting. NC, negative control. NS, no significance. *p<0.05, **p <0.01, ***p <0.001.
A dual-luciferase reporter gene assay was performed to detect whether miR-125b-5p targeted Keap1 through the potential binding site predicted by TargetScan. The luciferase assay activity levels were reduced in the miRNA-125b-5p group compared with the miR-NC group, demonstrating that the 3’−UTR of human Keap1 mRNA was the binding site of miRNA-125b-5p, and unchanged levels of luciferase activity in the presence of the mutated 3’−UTR of Keap1 further confirmed that Keap1 is the target of miR-125b-5p (Figure 3C). We then investigated whether the Keap1 protein and its involved signaling pathway were regulated by miR-125b-5p in vitro. Overexpression of miRNA-125b-5p significantly suppressed the protein expression of Keap1. The Keap1/Nrf2 pathway plays a key role in mitigating acute liver injury (34). Our study found that the expression levels of Nrf2 and one of its downstream proteins, HO-1, were upregulated, whereas cleaved caspase-3 was decreased in miRNA-125b-5p- overexpressing cells compared to the control groups (Figure 3D).
We further explored whether miR-125b-5p could improve ALF in vivo by targeting Keap1 and its downstream proteins. The dual-luciferase reporter gene assay confirmed the direct binding of miR-125b-5p with the 3’-UTR of mouse Keap1 but not with the mutated 3’-UTR (Figure 4D). Subsequently, we found elevated levels of Nrf2 and HO-1 in miR-125b-5p-treated mice (Figure 4E). The expression of cleaved caspase-3 was significantly decreased in mice administered the miR-125b-5p overexpression vector. Thus, miR-125b-5p can regulate Keap1, which leads to the activation of Nrf2 signaling and hence alleviates LPS/D-GalN-induced ALF.
Liver failure is a challenging condition with high mortality; thus, the development of novel therapeutic agents for liver failure is crucial. Both ACLF and ALF are driven by dysfunctional immunity and excessive inflammation. Hence, strategies targeting immunity and inflammation may provide new insight into the treatment of liver failure.
In the present study, we screened DE-miRNAs in human liver tissues from patients with HBV-non-ACLF and HBV-ACLF using high-throughput sequencing. To the best of our knowledge, this is the first study that systematically quantifies the expression features of miRNAs by high-throughput sequencing analysis in human HBV-related liver tissue.
A total of 75 DE-miRNAs were ultimately obtained, and subsequent GO analysis revealed that most target genes of these DE-miRNAs were involved in “binding”, “cell part”, “cellular process” and related categories. KEGG analysis showed that target genes predominantly participated in the “pathways in cancer”, “toxoplasmosis”, and “microRNAs in cancer”. Although the functions of these DE-miRNAs in the development of liver failure remain unclear, several lines of evidence from previous findings suggest that our results are biologically reasonable. For instance, in the present study, the level of hepatic miR-122-5p was downregulated with the aggravation of liver injury. Previous study proved that circulating miR-122-5p might function as a new biomarker for CHB patients with normal or nearly normal ALT levels (35), and miR-122-5p knockdown protected against acetaminophen-induced liver injury (36). Likewise, miR-192-5p was downregulated when liver injury worsened in the present study, while a previous study proved that mR-192-5p was upregulated in serum during liver injury, and downregulation of miR-192-5p might protect against liver cell death (37). Overall, it seems that DE-miRNAs may participate in the development of liver injury, and some of them may be potential targets for the treatment of liver failure.
Of these DE-miRNAs, miR-125b-5p was the focus of further investigation based on our previous findings and preliminary results. We previously documented that circulating miR-125b-5p might be a novel biomarker for liver injury, and high serum miR-125b-5p levels might predict a poor outcome in HBV-ACLF patients (28). In the present study, we preliminarily observed that the hepatic miR-125b-5p levels were lower in the HBV-ACLF group than in the HBV-non-ACLF group. Meanwhile, LPS/D-GalN-challenged mice and Huh7 cells both showed decreased miR-125b-5p levels compared to their untreated control groups. Therefore, it is reasonable to speculate that miR-125b-5p may have a protective role against liver injury, regardless of the presence of ACLF or ALF.
Subsequent experiments were performed to determine the role of miR-125b-5p in acute liver injury both in vitro and in vivo. We found a reduced apoptosis rate of injured Huh7 cells after transfection with the miR-125b-5p vector, demonstrating that miR-125b-5p was able to ameliorate acute liver injury. Later, using a mouse ALF model induced by LPS/D-GalN, we observed severely damaged liver histology and elevated ALT/AST levels in ALF mice, whereas upregulation of miR-125b-5p reversed the impacts of liver failure, in line with a previous study in which miR-125b-5p was proven to protect against paracetamol- and FAS-induced toxicity in hepatocytes (38).
We next explored the underlying mechanism of miR-125b-5p in relieving ALF. Patients with liver failure, either ALF or ACLF, display evidence of a proinflammatory state in the initial stage. Targeting factors that are involved in the inflammatory response may be an alternative therapeutic for liver failure. Immune dysfunction in ALF and ACLF shares many features with sepsis (3, 4, 39). During sepsis, miRNAs are crucial regulators of immune cell function and participate in the inflammatory response by regulating the production of inflammatory factors, the vascular barrier and endothelial function (40–42). Thus, dysregulated miRNAs may also be involved in the inflammatory response during liver failure. It has been reported that miR-125b-5p may directly or indirectly participate in inflammation (43–45). MiRNA-125b-5p elevation can restrain the inflammatory response and protect against sepsis-induced acute lung injury (43). Correspondingly, dramatic decreases in the levels of serum TNF-α and IL-1β in mice treated with miR-125b-5p were observed in our study; thus, we speculated that miR-125b-5p may alleviate ALF by suppressing inflammation.
Keap1, as a putative target of miR-125b-5p, was verified using a luciferase activity assay in the present study. There was a negative correlation between miR-125b-5p and Keap1 expression in Huh7 cells and liver tissues; thus, miR-125b-5p targeted and negatively regulated Keap1. The Keap1/Nrf2 signaling pathway plays a pivotal role in the maintenance of intracellular redox homeostasis and the regulation of inflammation. Under homeostatic conditions, Nrf2 is mainly retained by Keap1 in the cytoplasm. Upon oxidative stress, Keap1 is inactivated, and Nrf2 dissociates from Keap1. Nrf2 then translocates to the nucleus and binds to antioxidant response elements (AREs), leading to the expression of diverse antioxidant and metabolic genes, such as glutathione S-transferase (GST) and heme oxygenase 1 (HO-1) (46). HO-1 is a critical protective enzyme that inhibits the release of proinflammatory cytokines and activates the anti-inflammatory cytokines (47). The Nrf2/HO-1 axis has been found to reduce the levels of TNF-α, IL-6 and IL-1-β and play a major role in anti-inflammatory function (48).
In the present study, we found that LPS/D-GalN decreased hepatic miR-125b-5p levels to target Keap1 and inhibit the Nrf2 pathway, triggering liver inflammation and cell apoptosis. In contrast, upregulation of miR-125b-5p decreased the level of Keap1 and promoted the expression of Nrf2 and its target HO-1. Hence, miR-125b-5p protects against LPS/D-GalN- induced ALF by targeting Keap1 and disrupting the interaction between Nrf2 and Keap1, followed by the activation of the Nrf2/HO-1 axis, thereby decreasing the levels of TNF-α and IL-1-β and inhibiting cell apoptosis, which was in accordance with previous evidence that regulation of the Keap1/Nrf2/HO-1 signaling pathway could alleviate acute liver injury (34, 49, 50).
However, only miR-125b-5p was investigated herein, and further studies are needed to disclose the molecular mechanisms by which other miRNAs, individually or cooperatively, contribute to the development of liver injury. Moreover, it may be better to establish a miRNA-network diagram targeting inflammation in the development of liver failure, which may provide new insight into the pathophysiology of liver failure and lay a basis for the study of new therapeutics.
In conclusion, our study provides evidence that miR-125b-5p can alleviate LPS/D-GalN-induced ALF by regulating the Keap1/Nrf2/HO-1 signaling pathway, and therefore, regulation of miR-125b-5p and its downstream target Keap1 may serve as an alternative intervention for liver failure.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA885574.
The animal study was reviewed and approved by animal ethics committee of Sichuan University.
E-QC and HT proposed the study. Y-CT and Y-HW performed the research. Y-CT, M-LW, WJ, and D-BW contributed to the acquisition of data and drafting of the manuscript. All the authors contributed to the analysis and interpretation of data and approved the final version of the manuscript.
This work was supported by the 1.3.5 project for disciplines of excellence, West China Hospital, Sichuan University (NO.ZYGD20009) and the Science and Technological Supports Project of Sichuan Province, China (NO. 2019YFS0028).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.988668/full#supplementary-material
1. Sarin SK, Choudhury A, Sharma MK, Maiwall R, Al Mahtab M, Rahman S, et al. Acute-on-chronic liver failure: consensus recommendations of the Asian pacific association for the study of the liver (APASL): An update. Hepatol Int (2019) 13(4):353–90. doi: 10.1007/s12072-019-09946-3
2. Dogan S, Gurakar A. Liver transplantation update: 2014. Euroasian J Hepatogastroenterol (2015) 5(2):98–106. doi: 10.5005/jp-journals-10018-1144
3. Hensley MK, Deng JC. Acute on chronic liver failure and immune dysfunction: A mimic of sepsis. Semin Respir Crit Care Med (2018) 39(5):588–97. doi: 10.1055/s-0038-1672201
4. Dong V, Nanchal R, Karvellas CJ. Pathophysiology of acute liver failure. Nutr Clin Pract (2020) 35(1):24–9. doi: 10.1002/ncp.10459
5. Li J, Hu CH, Chen Y, Zhou MM, Gao ZJ, Fu MJ, et al. Characteristics of peripheral lymphocyte subsets in patients with acute-On-Chronic liver failure associated with hepatitis b. Front Med (Lausanne) (2021) 8:689865. doi: 10.3389/fmed.2021.689865
6. Laleman W, Claria J, Van der Merwe S, Moreau R, Trebicka J. Systemic inflammation and acute-on-Chronic liver failure: Too much, not enough. Can J Gastroenterol Hepatol (2018) p:1027152. doi: 10.1155/2018/1027152
7. Wu W, Yan H, Zhao H, Sun W, Yang Q, Sheng J, et al. Characteristics of systemic inflammation in hepatitis b-precipitated ACLF: Differentiate it from no-ACLF. Liver Int (2018) 38(2):248–57. doi: 10.1111/liv.13504
8. Weiss E, de la Grange P, Defaye M, Lozano JJ, Aguilar F, Hegde P, et al. Characterization of blood immune cells in patients with decompensated cirrhosis including ACLF. Front Immunol (2020) 11:619039. doi: 10.3389/fimmu.2020.619039
9. Khanam A, Kottilil S. Abnormal innate immunity in acute-on-Chronic liver failure: Immunotargets for therapeutics. Front Immunol (2020) 11:2013. doi: 10.3389/fimmu.2020.02013
10. Antoniades CG, Berry PA, Wendon JA, Vergani D. The importance of immune dysfunction in determining outcome in acute liver failure. J Hepatol (2008) 49(5):845–61. doi: 10.1016/j.jhep.2008.08.009
11. Vishnoi A, Rani S. MiRNA biogenesis and regulation of diseases: An overview. Methods Mol Biol (2017) 1509:1–10. doi: 10.1007/978-1-4939-6524-3_1
12. Nejad C, Stunden HJ, Gantier MP. A guide to miRNAs in inflammation and innate immune responses. FEBS J (2018) 285(20):3695–716. doi: 10.1111/febs.14482
13. Mikami Y, Philips RL, Sciume G, Petermann F, Meylan F, Nagashima H, et al. MicroRNA-221 and -222 modulate intestinal inflammatory Th17 cell response as negative feedback regulators downstream of interleukin-23. Immunity (2021) 54(3):514–25.e6. doi: 10.1016/j.immuni.2021.02.015
14. Fu B, Lin X, Tan S, Zhang R, Xue W, Zhang H, et al. MiR-342 controls mycobacterium tuberculosis susceptibility by modulating inflammation and cell death. EMBO Rep (2021) 22(9):e52252. doi: 10.15252/embr.202052252
15. Xie W, Lu Q, Wang K, Lu J, Gu X, Zhu D, et al. miR-34b-5p inhibition attenuates lung inflammation and apoptosis in an LPS-induced acute lung injury mouse model by targeting progranulin. J Cell Physiol (2018) 233(9):6615–31. doi: 10.1002/jcp.26274
16. Hu Y, Wu X, Ye Y, Ye L, Han S, Wang X, et al. Liver histology of treatment-naive children with chronic hepatitis b virus infection in shanghai China. Int J Infect Dis (2022) 123:112–8. doi: 10.1016/j.ijid.2022.08.017
17. Zhang Y, Xiang D, Hu X, Ruan Q, Wang L, Bao Z. Identification and study of differentially expressed miRNAs in aged NAFLD rats based on high-throughput sequencing. Ann Hepatol (2020) 19(3):302–12. doi: 10.1016/j.aohep.2019.12.003
18. Ouyang H, He X, Li G, Xu H, Jia X, Nie Q, et al. Deep sequencing analysis of miRNA expression in breast muscle of fast-growing and slow-growing broilers. Int J Mol Sci (2015) 16(7):16242–62. doi: 10.3390/ijms160716242
19. Zhao X, Chen Z, Zhou Z, Li Y, Wang Y, Zhou Z, et al. High-throughput sequencing of small RNAs and analysis of differentially expressed microRNAs associated with high-fat diet-induced hepatic insulin resistance in mice. Genes Nutr (2019) 14:6. doi: 10.1186/s12263-019-0630-1
20. Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods (2012) 9(4):357–9. doi: 10.1038/nmeth.1923
21. Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res (2014) 42(Database issue):D68–73. doi: 10.1093/nar/gkt1181
22. Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res (2012) 40(1):37–52. doi: 10.1093/nar/gkr688
23. Huang J, Li Y, Ma F, Kang Y, Liu Z, Wang J. Identification and characterization of microRNAs in the liver of rainbow trout in response to heat stress by high-throughput sequencing. Gene (2018) 679:274–81. doi: 10.1016/j.gene.2018.09.012
24. Wang J, Shao J, Li Y, Elzo MA, Jia X, Lai S. Genome-wide identification and characterization of perirenal adipose tissue microRNAs in rabbits fed a high-fat diet. Biosci Rep (2021) 41(4):BSR20204297. doi: 10.1042/BSR20204297
25. Ou N, Song Y, Xu Y, Yang Y, Liu X. Identification and verification of hub microRNAs in varicocele rats through high-throughput sequencing and bioinformatics analysis. Reprod Toxicol (2020) 98:189–99. doi: 10.1016/j.reprotox.2020.09.012
26. Gene Ontology C. The gene ontology: enhancements for 2011. Nucleic Acids Res (2012) 40(Database issue):D559–64. doi: 10.1093/nar/gkr1028
27. Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res (2017) 45(D1):D353–61. doi: 10.1093/nar/gkw1092
28. Tao YC, Wang ML, Wang M, Ma YJ, Bai L, Feng P. Quantification of circulating miR-125b-5p predicts survival in chronic hepatitis b patients with acute-on-chronic liver failure. Dig Liver Dis (2019) 51(3):412–8. doi: 10.1016/j.dld.2018.08.030
29. Ganjali S, Aghaee-Bakhtiari SH, Reiner Z, Sahebkar A. Differential expression of miRNA-223 in coronary in-stent restenosis. J Clin Med (2022) 11(3):849. doi: 10.3390/jcm11030849
30. Afonso MB, Rodrigues PM, Simao AL, Gaspar MM, Carvalho T, Borralho P, et al. miRNA-21 ablation protects against liver injury and necroptosis in cholestasis. Cell Death Differ (2018) 25(5):857–72. doi: 10.1038/s41418-017-0019-x
31. Wang H, Chen L, Zhang X, Xu L, Xie B, Shi H, et al. Kaempferol protects mice from d-GalN/LPS-induced acute liver failure by regulating the ER stress-Grp78-CHOP signaling pathway. BioMed Pharmacother (2019) 111:468–75. doi: 10.1016/j.biopha.2018.12.105
32. Tao YC, Wang ML, Wu DB, Luo C, Tang H, Chen EQ. Apolipoprotein A5 alleviates LPS/D-GalN-induced fulminant liver failure in mice by inhibiting TLR4-mediated NF-kappaB pathway. J Transl Med (2019) 17(1):151. doi: 10.1186/s12967-019-1900-9
33. Liu L, Wang P, Wang YS, Zhang YN, Li C, Yang ZY, et al. MiR-130a-3p alleviates liver fibrosis by suppressing HSCs activation and skewing macrophage to Ly6C(lo) phenotype. Front Immunol (2021) 12:696069. doi: 10.3389/fimmu.2021.696069
34. Wu CT, Deng JS, Huang WC, Shieh PC, Chung MI, Huang GJ. Salvianolic acid c against acetaminophen-induced acute liver injury by attenuating inflammation, oxidative stress, and apoptosis through inhibition of the Keap1/Nrf2/HO-1 signaling. Oxid Med Cell Longev 2019 (2019) p:9056845. doi: 10.1155/2019/9056845
35. Cheng JL, Zhao H, Yang SG, Chen EM, Chen WQ, Li LJ Plasma miRNA-122-5p and miRNA-151a-3p identified as potential biomarkers for liver injury among CHB patients with PNALT. Hepatol Int (2018) 12(3):277–87. doi: 10.1007/s12072-018-9871-0
36. Yang Z, Wu W, Ou P, Wu M, Zeng F, Zhou B, et al. MiR-122-5p knockdown protects against APAP-mediated liver injury through up-regulating NDRG3. Mol Cell Biochem (2021) 476(2):1257–67. doi: 10.1007/s11010-020-03988-0
37. Roy S, Benz F, Alder J, Bantel H, Janssen J, Vucur M, et al. Down-regulation of miR-192-5p protects from oxidative stress-induced acute liver injury. Clin Sci (Lond) (2016) 130(14):1197–207. doi: 10.1042/CS20160216
38. Yang D, Yuan Q, Balakrishnan A, Bantel H, Klusmann JH, Manns MP, et al. MicroRNA-125b-5p mimic inhibits acute liver failure. Nat Commun (2016) 7:11916. doi: 10.1038/ncomms11916
39. Claria J, Arroyo V, Moreau R. The acute-on-Chronic liver failure syndrome, or when the innate immune system goes astray. J Immunol (2016) 197(10):3755–61. doi: 10.4049/jimmunol.1600818
40. Kingsley SMK, Bhat BV. Role of microRNAs in sepsis. Inflammation Res (2017) 66(7):553–69. doi: 10.1007/s00011-017-1031-9
41. Zhong L, Simard MJ, Huot J. Endothelial microRNAs regulating the NF-kappaB pathway and cell adhesion molecules during inflammation. FASEB J (2018) 32(8):4070–84. doi: 10.1096/fj.201701536R
42. Lee LK, Medzikovic L, Eghbali M, Eltzschig HK, Yuan X. The role of MicroRNAs in acute respiratory distress syndrome and sepsis, from targets to therapies: A narrative review. Anesth Analg (2020) 131(5):1471–84. doi: 10.1213/ANE.0000000000005146
43. Jiang L, Ni J, Shen G, Xia Z, Zhang L, Xia S, et al. Upregulation of endothelial cell-derived exosomal microRNA-125b-5p protects from sepsis-induced acute lung injury by inhibiting topoisomerase II alpha. Inflammation Res (2021) 70(2):205–16. doi: 10.1007/s00011-020-01415-0
44. Luo X, Hu R, Zheng Y, Liu S, Zhou Z. Metformin shows anti-inflammatory effects in murine macrophages through dicer/microribonucleic acid-34a-5p and microribonucleic acid-125b-5p. J Diabetes Investig (2020) 11(1):101–9. doi: 10.1111/jdi.13074
45. Diao W, Lu L, Li S, Chen J, Zen K, Li L, et al. MicroRNA-125b-5p modulates the inflammatory state of macrophages via targeting B7-H4. Biochem Biophys Res Commun (2017) 491(4):912–8. doi: 10.1016/j.bbrc.2017.07.135
46. Lu MC, Ji JA, Jiang ZY, You QD. The Keap1-Nrf2-ARE pathway as a potential preventive and therapeutic target: An update. Med Res Rev (2016) 36(5):924–63. doi: 10.1002/med.21396
47. Ahmed SM, Luo L, Namani A, Wang XJ, Tang X. Nrf2 signaling pathway: Pivotal roles in inflammation. Biochim Biophys Acta Mol Basis Dis (2017) 1863(2):585–97. doi: 10.1016/j.bbadis.2016.11.005
48. Saha S, Buttari B, Panieri E, Profumo E, Saso L. An overview of Nrf2 signaling pathway and its role in inflammation. Molecules (2020) 25(22):5474. doi: 10.3390/molecules25225474
49. Lv H, Xiao Q, Zhou J, Feng H, Liu G, Ci X. Licochalcone a upregulates Nrf2 antioxidant pathway and thereby alleviates acetaminophen-induced hepatotoxicity. Front Pharmacol (2018) 9:147. doi: 10.3389/fphar.2018.00147
Keywords: acute liver failure, acute-on-chronic liver failure, microRNA-125b-5p, Kelch-like ECH-associated protein 1, high-throughput sequencing, inflammation
Citation: Tao Y-C, Wang Y-H, Wang M-L, Jiang W, Wu D-B, Chen E-Q and Tang H (2022) Upregulation of microRNA-125b-5p alleviates acute liver failure by regulating the Keap1/Nrf2/HO-1 pathway. Front. Immunol. 13:988668. doi: 10.3389/fimmu.2022.988668
Received: 07 July 2022; Accepted: 15 September 2022;
Published: 04 October 2022.
Edited by:
Xiaogang Xiang, Shanghai Jiao Tong University, ChinaReviewed by:
Wei Hou, Beijing Youan Hospital, Capital Medical University, ChinaCopyright © 2022 Tao, Wang, Wang, Jiang, Wu, Chen and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Tang, aHRhbmc2MTk4QGhvdG1haWwuY29t; En-Qiang Chen, Y2hlbmVucWlhbmcxOTgzQGhvdG1haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.