- 1College of Fisheries, Guangdong Ocean University, Zhanjiang, China
- 2Aquatic Animals Precision Nutrition and High Efficiency Feed Engineering Research Center of Guangdong Province, Zhanjiang, China
- 3Guangdong Provincial Laboratory of Southern Marine Science and Engineering, Zhanjiang, China
- 4Guangdong Provincial Key Laboratory of Pathogenic Biology and Epidemiology for Aquatic Economic Animals, Zhanjiang, China
In recent years, with global warming and increasing marine pollution, some novel marine viruses have become widespread in the aquaculture industry, causing huge losses to the aquaculture industry. Decapod iridescent virus 1 (DIV1) is one of the newly discovered marine viruses that has been reported to be detected in a variety of farmed crustacean and wild populations. Several previous studies have found that DIV1 can induce Warburg effect-related gene expression. In this study, the effects of DIV1 infection on intestinal health of shrimp were further explored from the aspects of histological, enzymatic activities, microorganisms and metabolites using Marsupenaeus japonicus as the object of study. The results showed that obvious injury in the intestinal mucosa was observed after DIV1 infection, the oxidative and antioxidant capacity of the shrimp intestine was unbalanced, the activity of lysozyme was decreased, and the activities of digestive enzymes were disordered, and secondary bacterial infection was caused. Furthermore, the increased abundance of harmful bacteria, such as Photobacterium and Vibrio, may synergized with DIV1 to promote the Warburg effect and induce metabolic reprogramming, thereby providing material and energy for DIV1 replication. This study is the first to report the changes of intestinal microbiota and metabolites of M. japonicus under DIV1 infection, demonstrating that DIV1 can induce secondary bacterial infection and metabolic reprogramming. Several bacteria and metabolites highly associated with DIV1 infection were screened, which may be leveraged for diagnosis of pathogenic infections or incorporated as exogenous metabolites to enhance immune response.
Highlights
● DIV1 infection disrupts the shrimp intestinal mechanical barrier.
● DIV1 infection resulted in imbalance of oxidative and antioxidant capacity, decreased immune enzyme activity, and disordered digestive enzyme activity in shrimp.
● DIV1 infection caused secondary bacterial infections, including Photobacterium and Vibrio.
● DIV1 can cooperate with harmful bacteria to induce metabolic reprogramming.
1 Introduction
Viruses are an important part of marine ecosystems. With the development of genetic technology, more and more new marine RNA and DNA viruses have been discovered (1, 2). Despite their tiny size, viruses play a large role in marine ecosystems and food webs. On the one hand, marine viruses can infect a variety of oceans organisms, lysing their cells and releasing carbon and other nutrients that impact the food web (3). On the other hand, marine viruses often contain host-derived metabolic genes (i.e., auxiliary metabolic genes; AMGs), which are hypothesized to increase viral replication and alter ecosystem-level productivity through reprogramming host metabolism (4). To date, several crustacean viruses have been found to induce metabolic reprogramming to promote survival and replication, including white spot syndrome virus (WSSV) (5–8), Taura syndrome virus (TSV) (9) and IHHNV (10). In recent years, with global warming and increasing marine pollution, some novel marine viruses have become widespread in the aquaculture industry, causing huge losses to the aquaculture industry (11). Decapod iridescent virus 1 (DIV1) is one of the newly discovered marine viruses. Since China initiated the DIV1 surveillance in 2017, DIV1 has been detected in shrimp culture ponds in several provinces (12). In addition to farmed shrimp, in 2020, Srisala et al. also detected DIV1 in wild populations of Penaeus monodon in the Indian Ocean (13). Up to now, DIV1 has been known as a highly lethal virus with global risk of transmission, capable of infecting freshwater and marine crustaceans, including Marsupenaeus japonicus (14).
Kuruma shrimp M. japonicus is widely distributed in the Indo-Western Pacific region and the East and South China seas (15). Due to its high economic value, strong environmental adaptability and suitability for long-distance transportation, it has now become one of the most farmed prawns in China (16, 17). Our previous study demonstrated that M. japonicus showed obvious clinical symptoms after DIV1 challenge, including empty stomach and intestine, atrophy of the hepatopancreas with yellowing, red body and soft shell. The shrimp mortality rate increased as the virus dose increased, and reached 100% mortality after at 76 hpi injected with 3.95 × 109 copies/μg DNA DIV1 inoculum (14). Through mRNA-seq and miRNA-seq analyses and their association analysis, we preliminarily revealed the hemocyte and intestinal immune response of M. japonicus to DIV1 infection (11, 14), and proposed that DIV1 can promote the Warburg effect by regulating host miRNA and mRNA expression. The Warburg effect was also observed in DIV1-infected Litopenaeus vannamei and Penaeus monodon. However, only the miRNA and mRNA expression profiles under DIV1 infection were revealed, the deep mechanism of the immune response needs further analysis, especially the intestinal immunity.
As an important immune and digestive organ of shrimp, the intestine and symbiotic microorganisms together constitute a complex ecosystem, which plays an important role in maintaining the function of the shrimp immune system. The intestinal immune system of shrimp is mainly composed of three parts, including the mechanical barrier formed by the tight junction and adhesion junction of intestinal mucosal cells, the innate immune barrier formed by intestinal hemocytes and their secreted immune factors, and the biological barrier formed by the intestinal microbiota and their secretions (18). The potential cooperation of bacteria and viruses in promoting disease development has received extensive attention from researchers in recent years. Numerous studies have shown that viral infection can alter the composition and function of the shrimp intestinal microbiota, induce secondary bacterial infection, and impair host immunity (19–21). However, similar to other invertebrates, shrimp lack specific immunity and cannot acquire antibodies through vaccination. Therefore, regulating host metabolic reprogramming by exogenous metabolites to enhance host immunity and ability to withstand environmental has become one of the best strategies for shrimp to resist viral infection (22–24). It will be interesting to explore the effect of metabolic reprogramming on host intestinal metabolome and microbiota and the consequence on immune response. However, there are no reports on the effects of DIV1 infection on the intestinal metabolome and microbiome of shrimp.
To gain a more comprehensive understanding of the interaction between DIV1 and shrimp, this study is the first to investigate the changes of intestinal microbiota and metabolites of M. japonicus under DIV1 infection through the integrated analysis of intestinal microbiome and metabolomics, demonstrating that DIV1 can induce secondary bacterial infection and metabolic reprogramming, and several highly related bacteria and metabolites highly associated with DIV1 infection were screened. The results are beneficial to provide a theoretical basis for virus control technology.
2 Materials and methods
2.1 Shrimp and rearing conditions
Healthy M. japonicus with an average body weight of 10.5 ± 1.6 g were randomly obtained from a local farming pond in East Island Marine Biological Research Base, Guangdong Ocean University (Zhanjiang, China). The shrimp were randomly sampled and tested by PCR to ensure that they were free from WSSV, IHHNV, and DIV1. The detection method was consistent with the previous study (25). Every 30 M. japonicus were acclimatized in 0.3-m3 tanks with aerated and filtered seawater (salinity 30‰, pH 7.5, temperature 28°C) for 7 days before the DIV1 challenge experiment. Commercial feed was provided to the shrimp three times per day at a feeding rate of 5% of their body weight, and nearly 90% of the seawater was exchanged per day.
2.2 DIV1 challenge and sample collection
After 7 days of acclimation, the healthy shrimp were divided into two groups: the PBS-injected group and the DIV1-infected group. Each group included three replicate tanks, and each tank containing 30 individuals. Base on LC50 test results from previous studies (14), we set the DIV1 injection concentration of this study as 3.95 × 109 copies/μg DNA. Each M. japonicus from the DIV1-infected group was intramuscularly injected with 50 μL of DIV1 inoculum, while each M. japonicus from the PBS-injected group was intramuscularly injected with 50 μL of phosphate-buffered saline (PBS; pH 7.4). The methods of viral inoculum preparation and quantification can be found in previous studies (26).
Based on previous research (14), the M. japonicus at twenty-four hour post-injection (hpi) were collected as samples under an aseptic condition. The intestines and their contents from 3 random individuals in the same group were combined as one sample and immediately frozen in liquid nitrogen before storing at -80°C until experimental analysis. In detail, each group had 6 samples were used for enzyme activity analysis, 6 samples were used for intestine microbiome analysis and 3 samples were used for intestinal metabolomics analysis. For histological analysis, the intestines of 6 shrimp per group were used to assess intestine tissue damage. Intestines of these shrimp were fixed in 10% formalin.
2.3 Histological analysis
After fixed in 10% formalin at 4°C for 24 h, the fixed intestine were dehydrated in gradient ethanol, hyalinized in xylene, and embedded in paraffin wax. Next, the paraffin blocks were sectioned at 5-μm thickness. The sections were collected on glass slides and stained with hematoxylin and eosin (H&E). The intestinal sections were examined by a microscope (Olympus, Nikon, Tokyo, Japan). The electronic images were further analyzed using ImageJ software to assess the dimensions of intestinal villus height and muscle thickness.
2.4 Enzyme activity analysis
Enzymatic biomarkers of functional responses in the intestine were measured using commercial detection kits (Jianglai Bioengineering Institute, Shanghai, China) according to protocols of the manufacturer. Prior to analysis, 6 samples in each group were homogenized in pre-chilled PBS (1:9 dilution) and then centrifuged for 10 min (4°C and 5,000 × g) to obtain the supernatant for further use. Superoxide dismutase (SOD), catalase (CAT) and lysozyme (LYZ) activities were used to reflect the nonspecific immunity in the shrimp intestine. α-Amylase (α-AMS), lipase (LPS) and trypsin (TPS) activities were used to reflect the digestive function of shrimp.
2.5 Intestinal microbiome analysis
Total bacterial DNA was extracted by the EasyPure® Marine Animal Genomic DNA Kit (TransGen Biotech, China) following the manufacturer’s directions. The concentration and purity of total DNA were determined by SimpliNano (GE Healthcare, United States) and 1% agarose gels. The primers pair 515F (5′-GTGCCAGCMGCCGCGG-3′) and 806R (5-′GGACTACNNGGGTATCTAAT-3′) were used to amplify the V4 hypervariable region of 16S rRNA gene, which was modified with a barcode tag with a random 6-base oligos. The PCR amplification reaction was performed with a 25 μL reaction mixture containing 12.5 μL rTaq mix (Takara Biotechnology (Dalian) Co., Ltd.), 1 μL of each primer, 1μL template and 9.5 μL ultrapure water. The reaction cycle parameters were as follows: an initial denaturation step of 95°C for 3 min; 30 serial cycles of a denaturation step of 95°C for 45 s, annealing at 55°C for 45 s, and extension at 72°C for 45 s; and a final extension step of 72°C for 10 min.
PCR amplification reaction system and parameters can refer to previous studies (27). After the PCR products were purified and mixed in equidensity ratios, sequencing libraries were constructed, and then sequenced by BGI (Shenzhen, China) with the Illumina Genome Analyzer technology. All raw sequencing data of intestinal microbiota was submitted to the Sequence Read Archive (SRA) (accession: PRJNA720257).
Sequences from raw data were analyzed and filtered by QIIME (v1.8.0). Sequence analysis was performed with UPARSE software (v7.0.1090), and the operational taxonomic units (OTUs) were defined with ≥ 97% similarity. Chimeric sequences were identified with UCHIME (v4.2.40). Alpha diversity was calculated using mothur software with five metrics, including the observed OTUs, Chao1, ACE, Shannon and Simpson indexes. Rarefaction curves were generated based on these metrics. The shared and unique OTUs between two groups were figured out by a Venn diagram. Beta diversity index based on the phylogenetic relationship between OTUs was used to calculate the Unifrac distance (weighted and unweighted Unifrac) and the results of Beta diversity were showed through PCoA and UPGMA Phylogenetic Dendrogram. A bar plot of the microbial community was constructed at the phylum, family and genus level respectively. To study the functional characteristics of bacterial communities, Kyoto Encyclopedia of Genes and Genomes (KEGG) functions were predicted using the PICRUSt software.
2.6 Intestinal metabolomics analysis
Six intestinal samples replicates of shrimp from each group were used for metabolomic analysis. All the samples were taken from the refrigerator at -80°C and thawed in the refrigerator at 4°C, and metabolite extraction was performed using methanol and 2-chlorobenzalanine. Twenty microlitres of each sample was taken for quality control (QC), and the rest was used for LC-MS detection. Liquid chromatography was accomplished in a Thermo Ultimate 3000 system equipped with an ACQUITY UPLC® BEH C18 (1.7 μm 2.1 × 100 mm, Waters, USA) column. Mass spectrometry was executed on a Q Exactive HF mass spectrometer (Thermo Fisher Scientific, USA). Data-dependent acquisition (DDA) MS/MS experiments were performed with HCD scans. Dynamic exclusion was implemented to remove some unnecessary information in the MS/MS spectra.
Peak-identification, peak-alignment and compound identification was conducted using Compound Discoverer software (v3.1). All the data were determined using quality control (QC) and quality assurance (QA). Partial least squares-discriminant analysis (PLS-DA) of the metabolomics data was performed using the R language ropls package. All the metabolites were classified according to Kyoto Encyclopedia of Genes and Genomes (KEGG) and Human Metabolome Database (HMDB). The PLS-DA model was used to determine the differential metabolites (DMs) between the PBS-injected group and the DIV1-infected group with the first principal component of variable importance in projection (VIP) values (VIP ≥ 1) combined with a q-value ≤ 0.05. The DMs were annotated with KEGG pathway analysis to further identified the change characteristics of the functional metabolites related to the immunity of the shrimp.
2.7 Correlation analysis of intestinal bacteria and DMs
Canonical correlation analysis and spearman correlation analysis were employed to reveal the correlation between significantly altered intestinal bacteria and intestinal DMs, and the results were shown with scatterplots and heat maps. The thresholds for correlation coefficients and p-values were not set. p < 0.05 was regarded as statistically significant, p < 0.01 was regarded as very significant, and p < 0.001 was regarded as extremely significant.
3 Result
3.1 Intestinal histological changes
Histological analysis of the intestinal sections from the PBS-injected group and the DIV1-infected group are shown in Figure 1. The H&E staining showed that no histological changes were observed in the PBS-injected group (Figures 1A). Contrastively, the microstructure of the intestine in the DIV1-infected group had lesions, some intestinal epithelial cells were detached from the basement membrane and destroyed (Figures 1D). Intestinal villus height and muscle thickness were measured to quantify the degree of intestinal damage. The results showed that the intestinal villus height in the PBS-injected group (65.02 ± 20.57 μm) was significantly higher than the DIV1-infected group (31.90 ± 4.48 μm) (p < 0.001), but the intestinal muscle thickness did not show significantly difference between the PBS-injected group (53.87 ± 15.50 μm) and the DIV1-infected group (51.79 ± 10.35 μm) (p > 0.05).
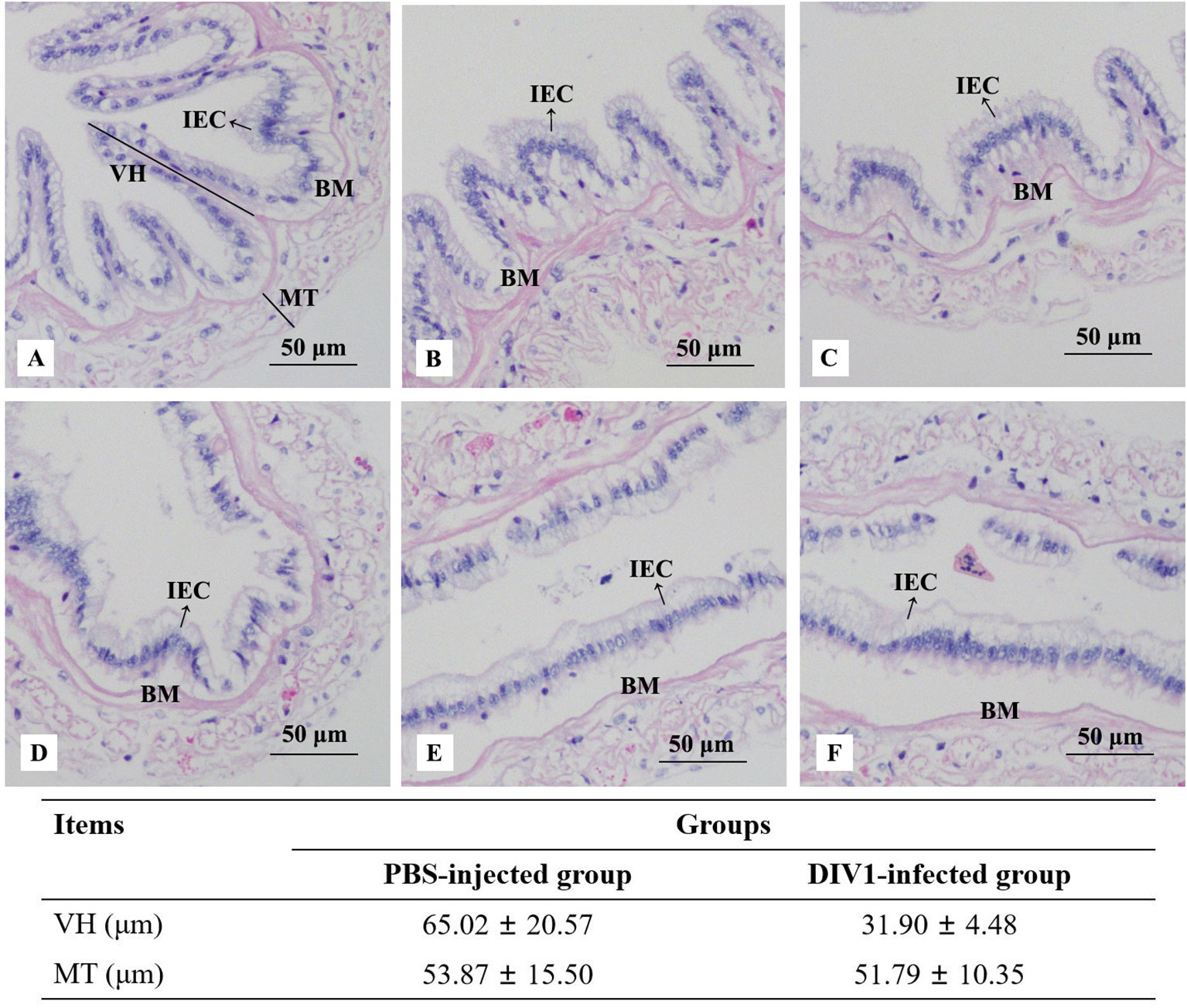
Figure 1 Photomicrographs of intestinal sections in the PBS-injected group (A-C) and the DIV1-infected group (D-F). IEC, intestinal epithelial cells; BM, basement membrane; VH, villus height; MT, muscle thickness.
3.2 Immune and digestive enzymes activity abnormalities
At 24 hpi, the activities of SOD, CAT and LYZ in the intestine were detected to evaluate the effect of DIV1 infection on the nonspecific immunity of M. japonicus (Figures 2A–C), and the activities of α-AMS, LPS and TPS in the intestine were detected to evaluate the digestive function (Figures 2D–F). Compared with the PBS-injected group, the activity of SOD, α-AMS and LPS were significantly increased (p < 0.001), while the activities of CAT, LYZ and TPS were significantly decreased (p < 0.001). It was worth noting that, after infection with DIV1, both the immune and digestive enzymes in the intestine were extremely significantly altered (p < 0.001).
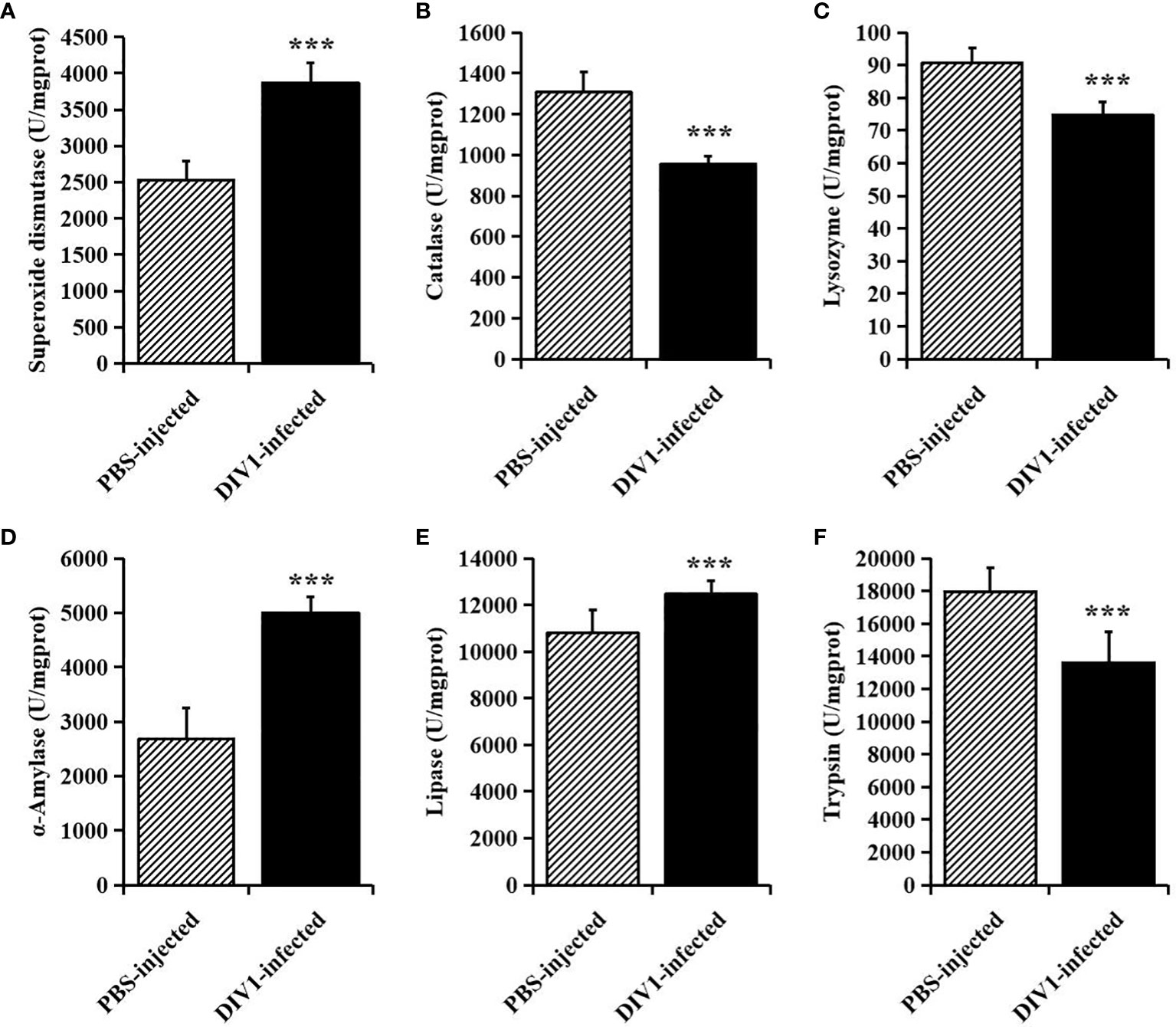
Figure 2 The activities of immune and digestive enzymes in the intestine of M. japonicus (mean ± SD). (A) Superoxide dismutase (SOD). (B) catalase (CAT). (C) lysozyme (LYZ). (D) α-Amylase. (E) lipase (LPS) and (F) trypsin (TPS). The statistically significant differences between the two groups were calculated by Student’s t-test (***p < 0.001).
3.3 Intestinal microbiota changes
3.3.1 Richness and diversity
A total of 711,264 high-quality sequences were generated from 12 intestinal samples. The clean reads ranged from 69,929 to 63,969, with an average of 59,272 clean reads per sample. After the alignment, the sequences of the PBS-injected group and the DIV1-infected group were clustered into 1,053 and 1,057 OTUs respectively with a 97% sequence similarity. Among them, there were 421 unique OTUs in the PBS-injected group, 425 unique OTUs in the DIV1-infected group, and 632 OTUs were the same in both the PBS-injected group and the DIV1-infected group (Figure 3A). A rarefaction curve analysis of the observed species per sample was sufficient (Figure 3B). To investigate the differences of species diversity and richness between two groups, the alpha diversity indexes were calculated, including observed OTUs, Chao1, ACE, Shannon and Simpson indexes, ranging from 270 to 715, 400.25 to 753.01, 424.50 to 765.22, 0.41 to 3.19 and 0.10 to 0.89, respectively (Table 1). Community richness indexes (observed OTUs, Chao1, ACE) were not significantly changed, while community diversity indexes (Shannon and Simpson) were significantly changed (p < 0.05). Beta diversity analysis was performed to comparative analyze the similarity and difference of intestinal microbial community in different groups. The PCoA with weighted and unweighted unifrac distance were further performed to confirm that intestinal bacteria in the PBS-injected group and the DIV1-infected group were clearly separated and samples from the same group were clustered closer (Figures 3C, D). UPGMA Phylogenetic Dendrogram with weighted and unweighted unifrac distance showed that all detected samples were divided into two main clades, and the intestinal microbial in the same group had a high degree of similarity (Figures 3E, F).
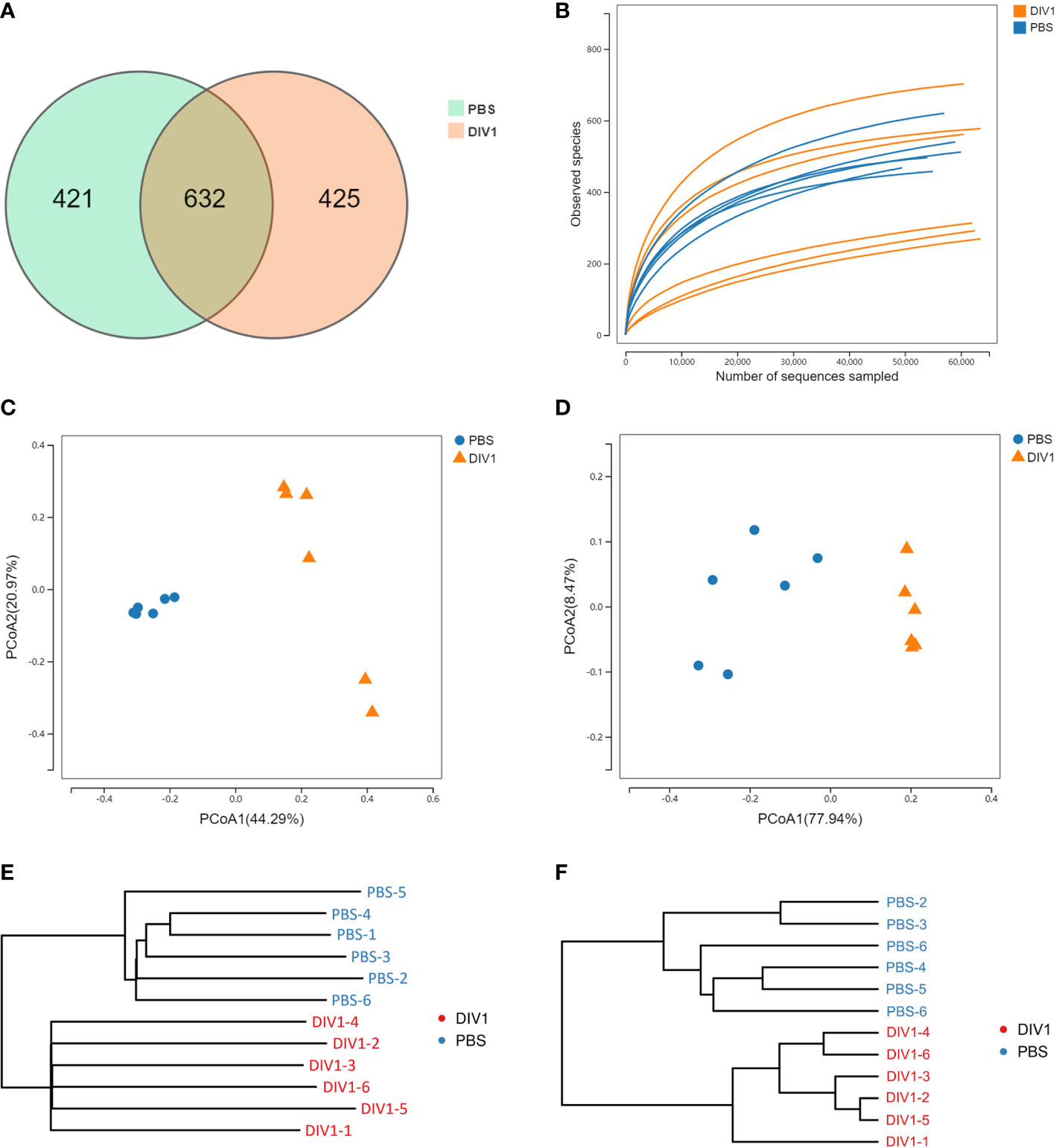
Figure 3 Intestinal microbial richness and diversity of M. japonicus after DIV1 infection. (A) Veen diagram showing the unique and shared OTUs of intestinal microbiota in the PBS-injected group and the DIV1-infected group. (B) Rarefaction curves of OTUs clustered at 97% sequence identity different samples. (C) PCoA plot with unweighted unifrac distance. (D) PCoA plot with weighted unifrac distance. (E) UPGMA Phylogenetic Dendrogram with unweighted unifrac distance. (F) UPGMA Phylogenetic Dendrogram with weighted unifrac distance.
3.3.2 Intestinal microbial composition
The taxa of dominant bacteria among the two groups were similar, while their abundance was altered significantly. At the phylum level, compared with the PBS-injected group, the relative abundance of Proteobacteria was significantly increased in the DIV1-infected group (p < 0.01), while the relative abundance of Actinobacteria and Cyanobacteria were significantly decreased (p < 0.01) (Figure 4A). At the family level, the relative abundance of Vibrionaceae and Sapropiraceae were significantly increased in the DIV1-infected group (p < 0.05), while the relative abundance of Corynebacteriaceae, Rhodobacteraceae and Enterobacteriaceae were significantly decreased (p < 0.05) (Figure 4B). At the genus level, differences were also observed. The relative abundance of Photobacterium was significantly decreased (p < 0.01), while the relative abundance of Corynebacterium, Sagittula and Morganella were significantly decreased (p < 0.05).
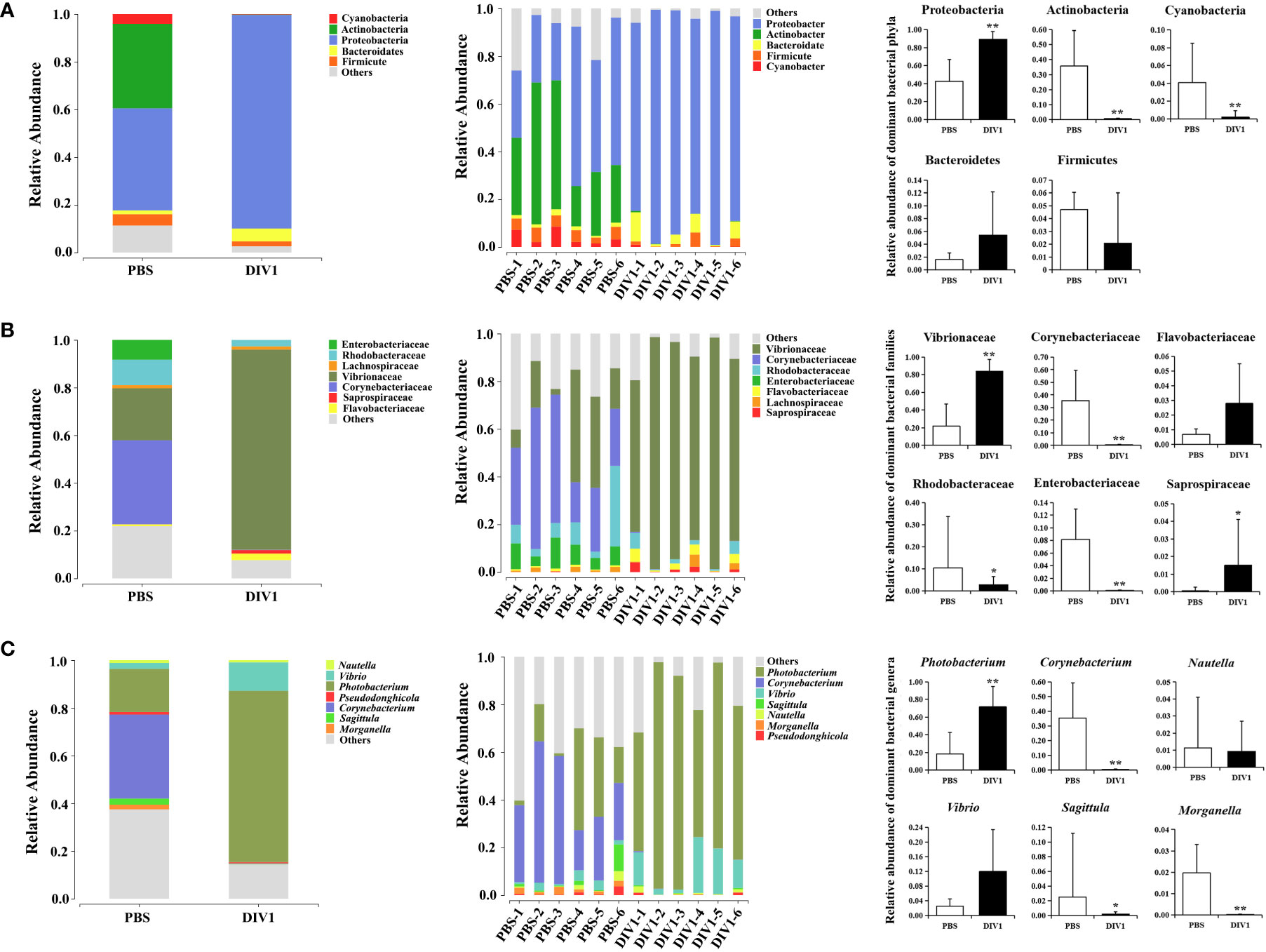
Figure 4 Structure and composition of the intestinal bacterial communities under DIV1 infection (mean ± SD). (A) Relative abundances of dominant bacterial phyla. (B) Relative abundances of dominant bacterial families. (C) Relative abundances of dominant bacterial genera.* Indicates a significant difference (*p < 0.05; **p < 0.01).
3.3.3 Changes in the intestinal bacterial phylotypes
LEfSe was employed to analyse the differential abundances of bacterial taxa in the two groups. In the cladogram, the families Vibrionaceae and Saprospiraceae were enriched in the DIV1-infected group, and Corynebacteriaceae, Rhodobacteraceae, Enterobacteriaceae and Lactobacillaceae were enriched in the PBS-injected group (Figure 5A). With an LDA score greater than 4.0, Photobacterium and Vibrio dominated in the DIV1-infected group, and Corynebacterium and Ruegeria dominated in the PBS-injected group (Figure 5B). The prediction function of the intestinal microbiota was analyzed using PICRUSt. Result showed that, in the KEGG level 2, the relative abundance of “carbohydrate metabolism”, “metabolism of cofactors and vitamins” and “amino acid metabolism” were the top 3 in the two groups (Figure 5C). It was worth noting that the relative abundance of “infectious diseases: bacterial” was significantly increased under DIV1 infection (p < 0.01). The distinct changes on a deeper resolution level within “Carbohydrate metabolism”, “Metabolism of cofactors and vitamins” and “Amino acid metabolism” were showed by the heatmap (Figure 5D). The results showed that the abundance value of KEGG level 3 term “alanine, aspartate and glutamate metabolism” and “valine, leucine and isoleucine biosynthesis” in the DIV1-infected group were significantly higher than the PBS-injected group (p < 0.01). The abundance value of some Warburg effect marker pathways and vitamin metabolism-related pathways were also significantly increased after DIV1 infection (p < 0.01), including “glycolysis/guconeogenesis”, “pyruvate metabolism”, “nicotinate and nicotinamide metabolism”, “vitamin B6 metabolism”, “riboflavin metabolism” and “pantothenate and CoA biosynthesis”.
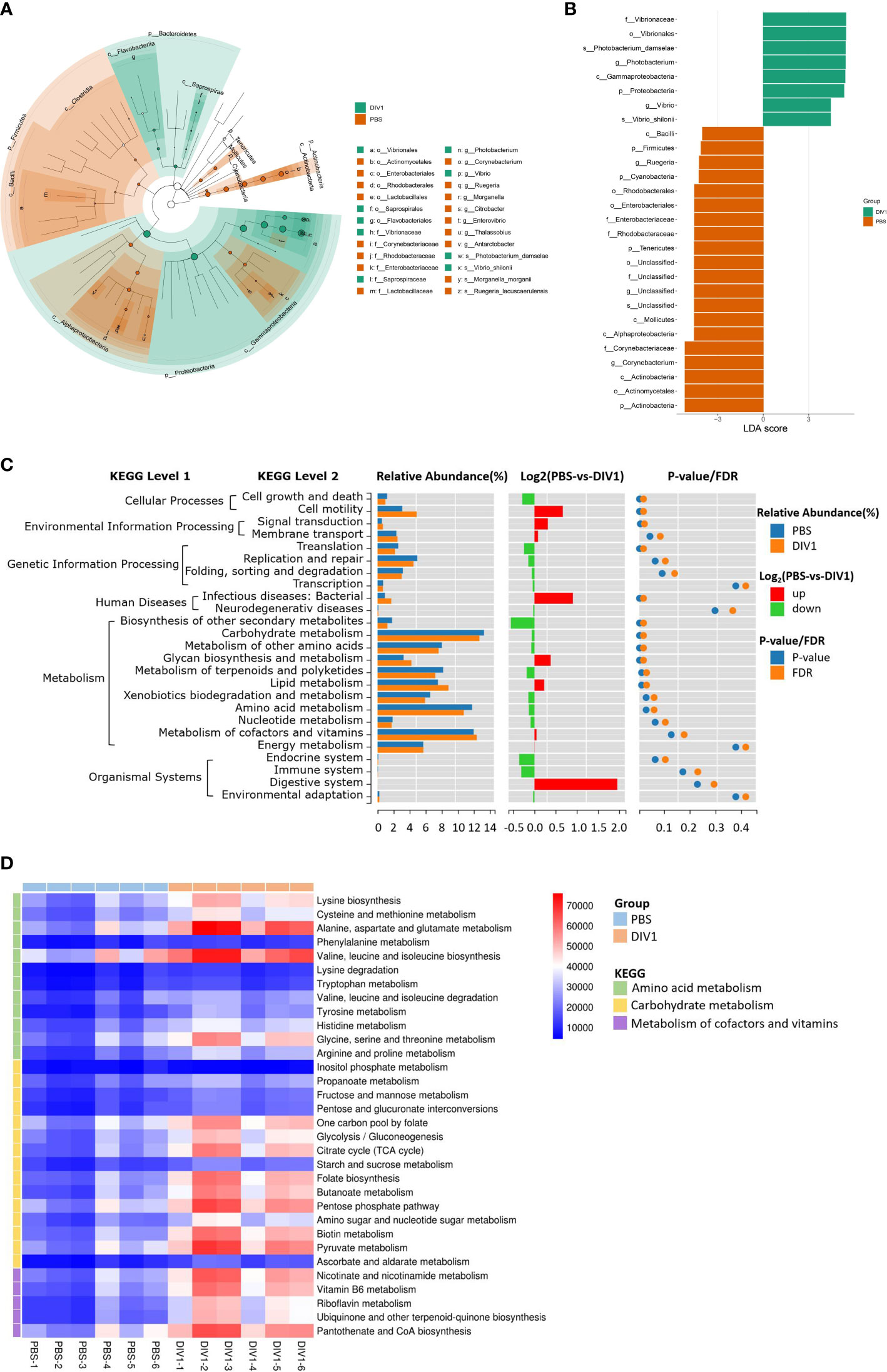
Figure 5 Intergroup variation, functional analysis of intestinal microbiota of M. japonicus after DIV1 infection. (A) LEfSe cladogram. (B) LDA score of LEfSe-PICRUSt. (C) Microbial metabolism prediction based on KEGG pathway analysis (KEGG level 2). (D) Abundance clustering heat map of predicted functions of intestinal microbiota in each sample (KEGG level 3).
3.4 Intestinal metabolic pattern alterations
3.4.1 Multivariate analysis of the metabolite profiles
Metabolomic analysis was conducted to explore the alterations in intestinal metabolic profiles after DIV1 infection. The PLS-DA score plot and permutation test showed a significant different between the two groups in both positive and negative ion mode (Figure 6), suggesting that DIV1 infection caused metabolic phenotype alterations in shrimp intestine. A total of 3,009 metabolites were identified in the shrimp intestine (including 2,096 metabolites were identified in the positive ion mode and 913 metabolites were identified in the negative ion mode). The classification results of the identified metabolites were shown in Figure 7. The largest category in the positive ion mode was “amino acids, peptides, and analogues” (253 metabolites), followed by “fatty acyls” (147 metabolites) and “benzene and derivatives” (139 metabolites) (Figure 7A), and the largest category in the negative ion mode was “amino acids, peptides, and analogues” (132 metabolites), followed by “fatty acyls” (58 metabolites) and “organic acids” (29 metabolites) (Figure 7B).
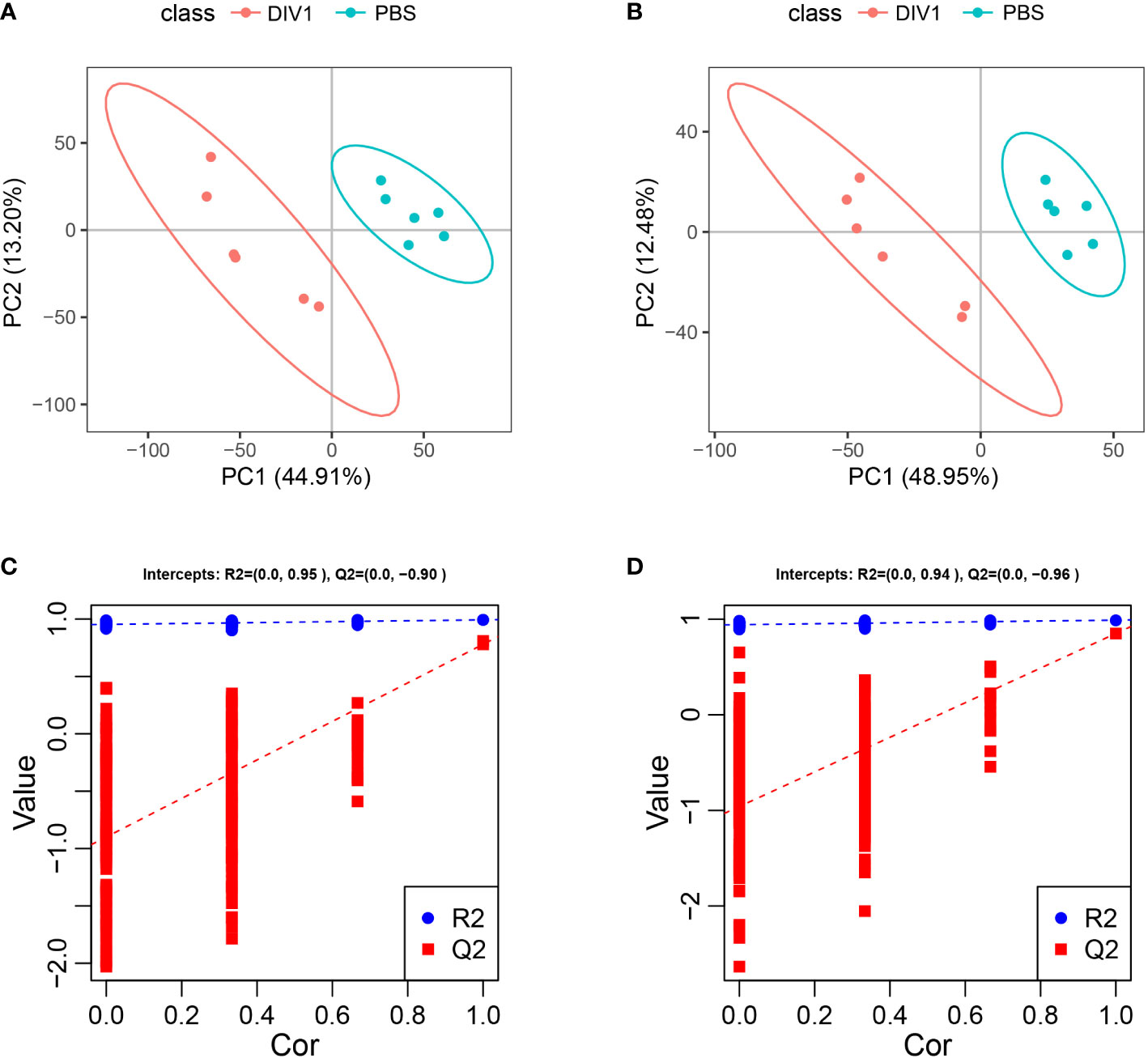
Figure 6 Derived PLS-DA score plots and corresponding permutation testing of PLS-DA from the LC-MS metabolite profiles in the intestine of M. japonicus after DIV1 infection. (A) PLS-DA score plot of positive ion mode. (B) PLS-DA score plot of negative ion mode. (C) Permutation testing of positive ion mode. (D) Permutation testing of negative ion mode.
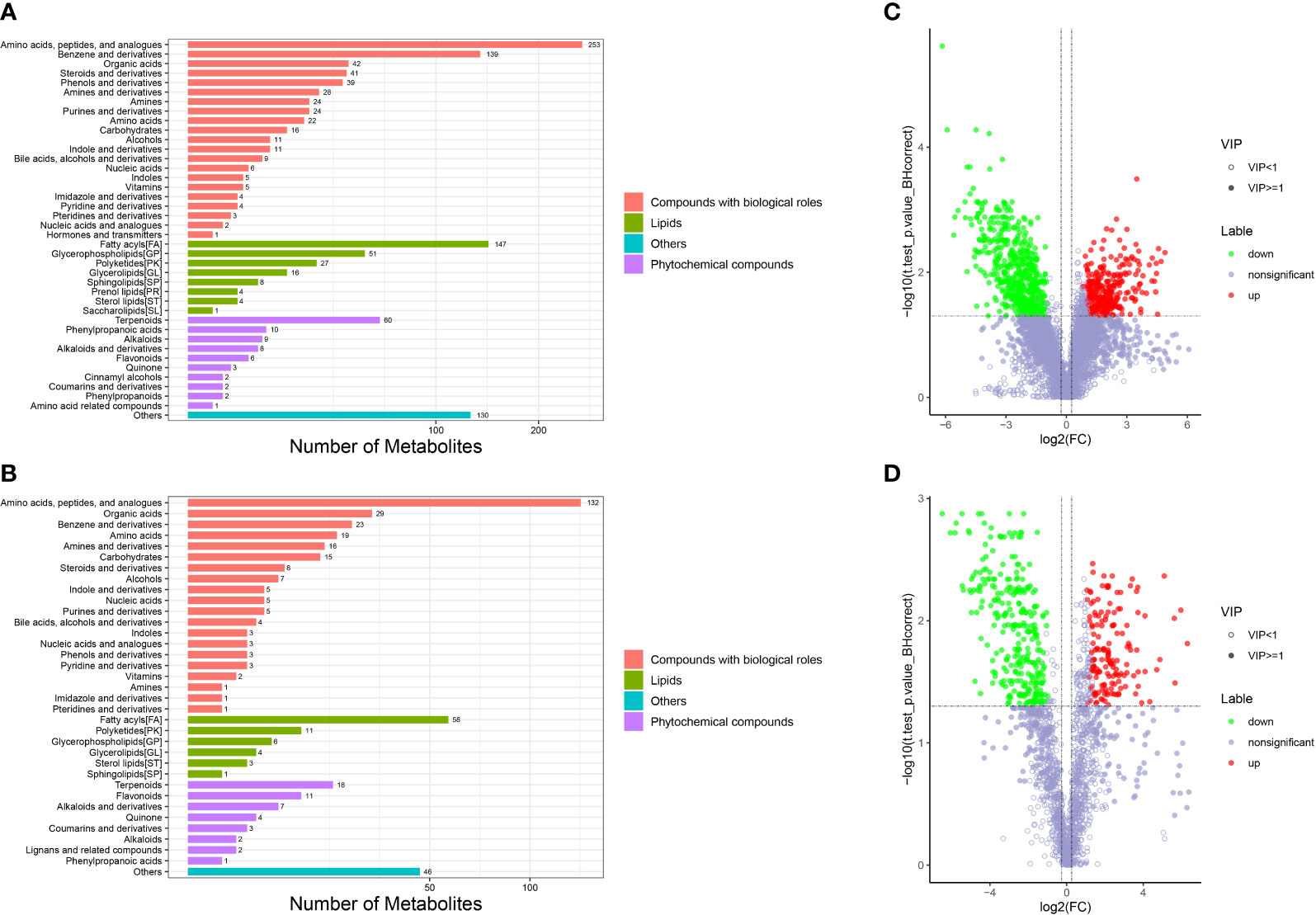
Figure 7 Taxonomic annotation of identified metabolites and volcano plots of DMs from the intestine of M. japonicus. (A) Taxonomic annotation of positive ion mode. (B) Taxonomic annotation of negative ion mode. Others means that the classification information is other categories, and the identified metabolites without classification information do not participate in the statistics. (C) Volcano plot of positive ion mode. (D) Volcano plot of negative ion mode.
3.4.2 Identification and functional annotation of the DMs
The DMs between the PBS-injected group and the DIV1-infected group were identified by the PLS-DA model with a cut-off VIP ≥ 1 and q-value ≤ 0.05. In the positive ion mode, a total of 868 DMs were obtained, including 312 up-regulated DMs and 556 down-regulated DMs, of which 419 DMs were identified in the database (Figure 7C). In the negative ion mode, a total of 454 DMs were obtained, including 158 up-regulated DMs and 296 down-regulated DMs, of which 209 DMs were identified in the database (Figure 7D).
For the KEGG pathway enrichment analysis, the top 20 KEGG pathway enrichments influenced by DIV1 infection in the positive ion mode and negative ion mode were shown in Figures 8A, B respectively. Among of them, four KEGG pathways related to vitamin metabolism were significantly enriched, including “vitamin digestion and absorption”, “retinol metabolism”, “vitamin B6 metabolism” and “pantothenate and CoA biosynthesis”. Three KEGG pathways related to amino acid metabolism were significantly enriched, including “tyrosine metabolism”, “phenylalanine metabolism” and “alanine, aspartate and glutamate metabolism”. Two Warburg effect marker pathways were significantly enriched, including “pyruvate metabolism” and “glycolysis/gluconeogenesis”. Both in positive ion mode and in negative ion mode, “primary bile acid biosynthesis”, “bile secretion”, “metabolic pathways”, “linoleic acid metabolism” and “arachidonic acid metabolism” were significantly enriched in KEGG pathway enrichment analysis.
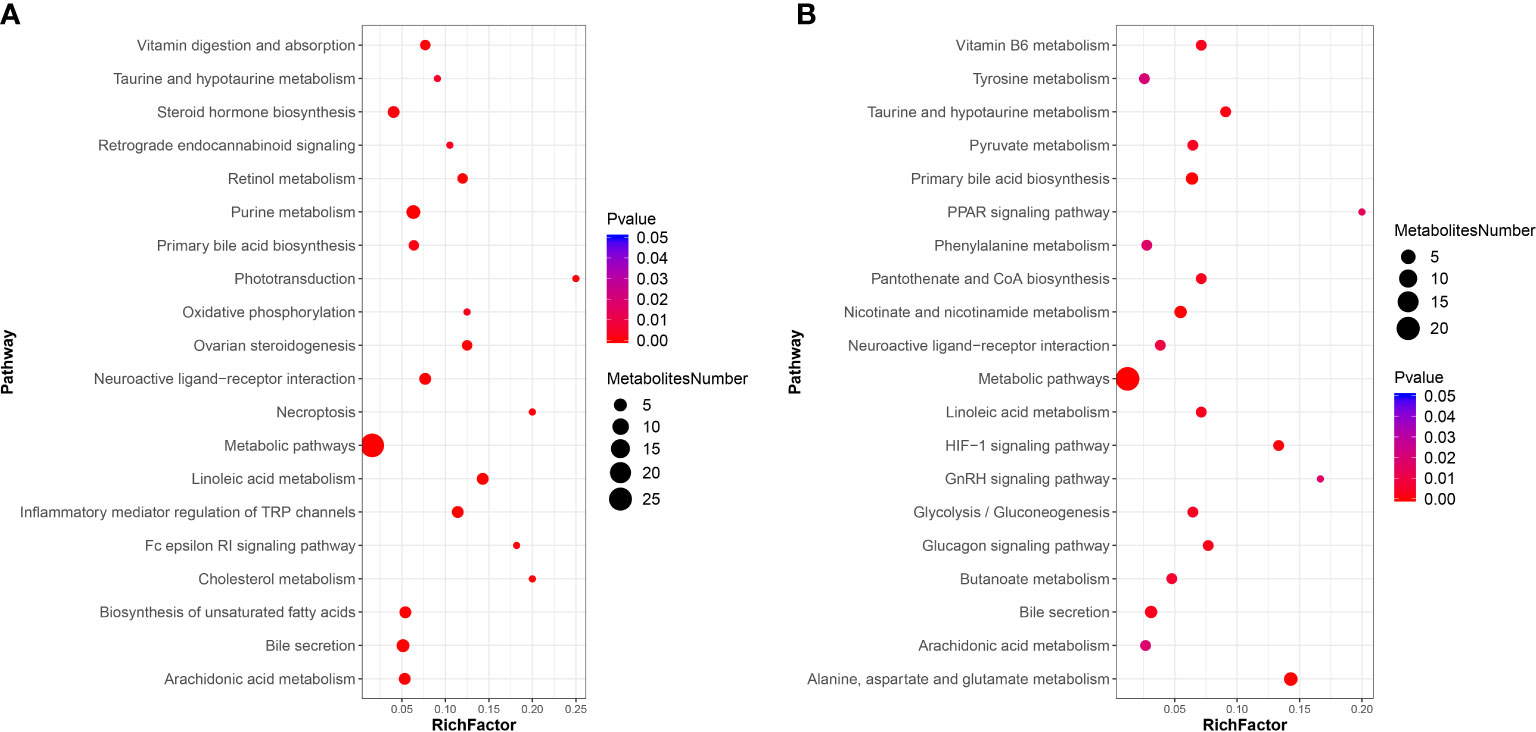
Figure 8 KEGG pathway enrichment analysis of DMs between the PBS-injected group and the DIV1-infected group (A) positive ion mode. (B) negative ion mode.
Further, in the positive ion mode, total of 41 known DMs were significantly enriched, including 25 up-regulated DMs and 16 down-regulated DMs (Table 2); in the negative ion mode, a total 24 DMs were significantly enriched, including 17 up-regulated DMs and 7 down-regulated DMs (Table 3). It was worth noting that seven types of lipid fatty acids were significantly changed after DIV1 infection, including arachidonic acid, eicosapentaenoic acid, docosapentaenoic acid, 9-oxo-10(e),12(e)-octadecadienoic acid, gamma-linolenic acid, 8(s)-hydroxy-(5z,9e,11z,14z) eicosatetraenoic acid and 16-hydroxyhexadecanoic acid. In addition, two Warburg effect marker metabolites were significantly up-regulated, including L-(+)-lactic acid and Pyruvic acid.
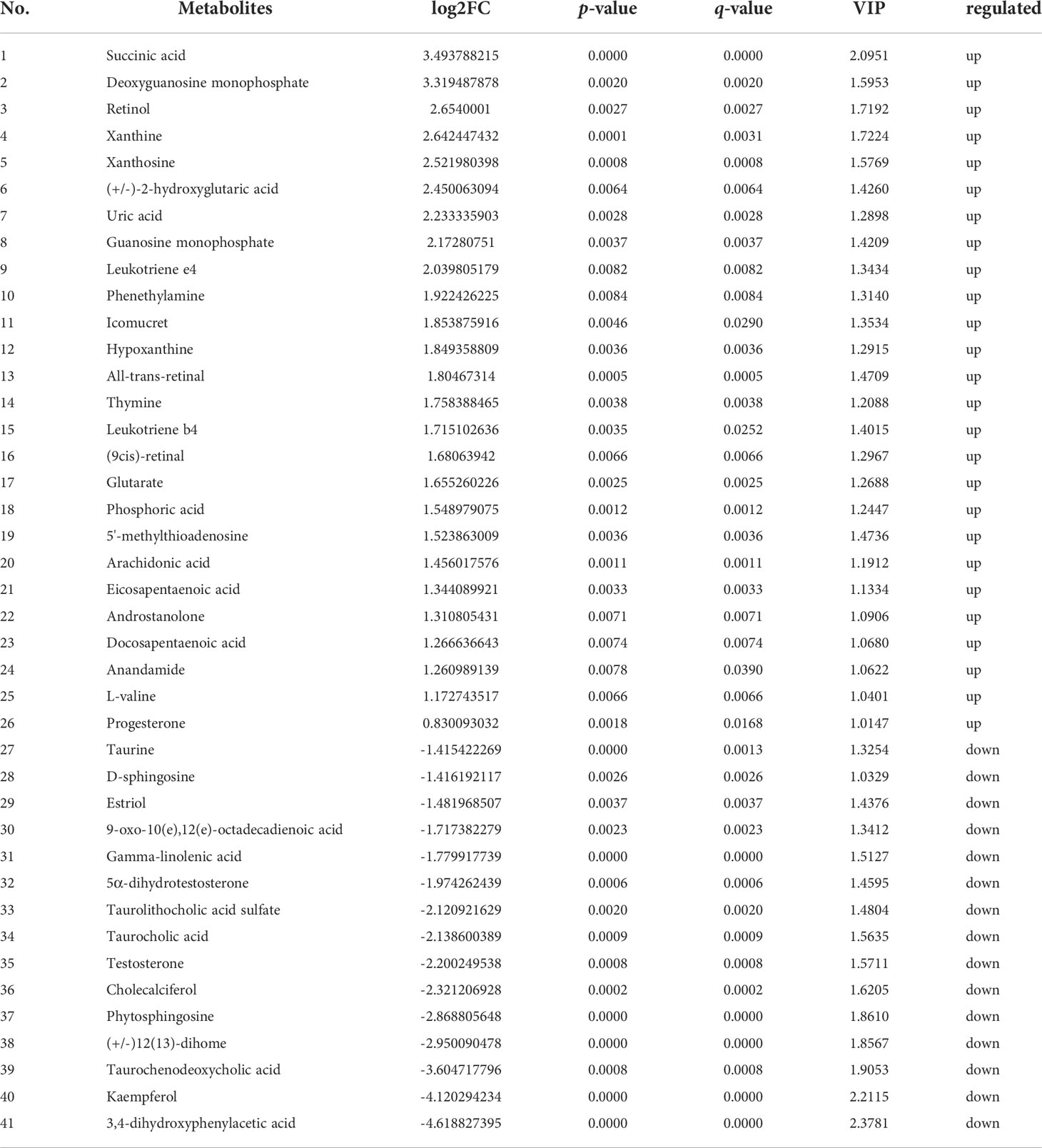
Table 2 41 known differential metabolites were significantly enriched in KEGG pathway enrichment analysis in positive ion mode.
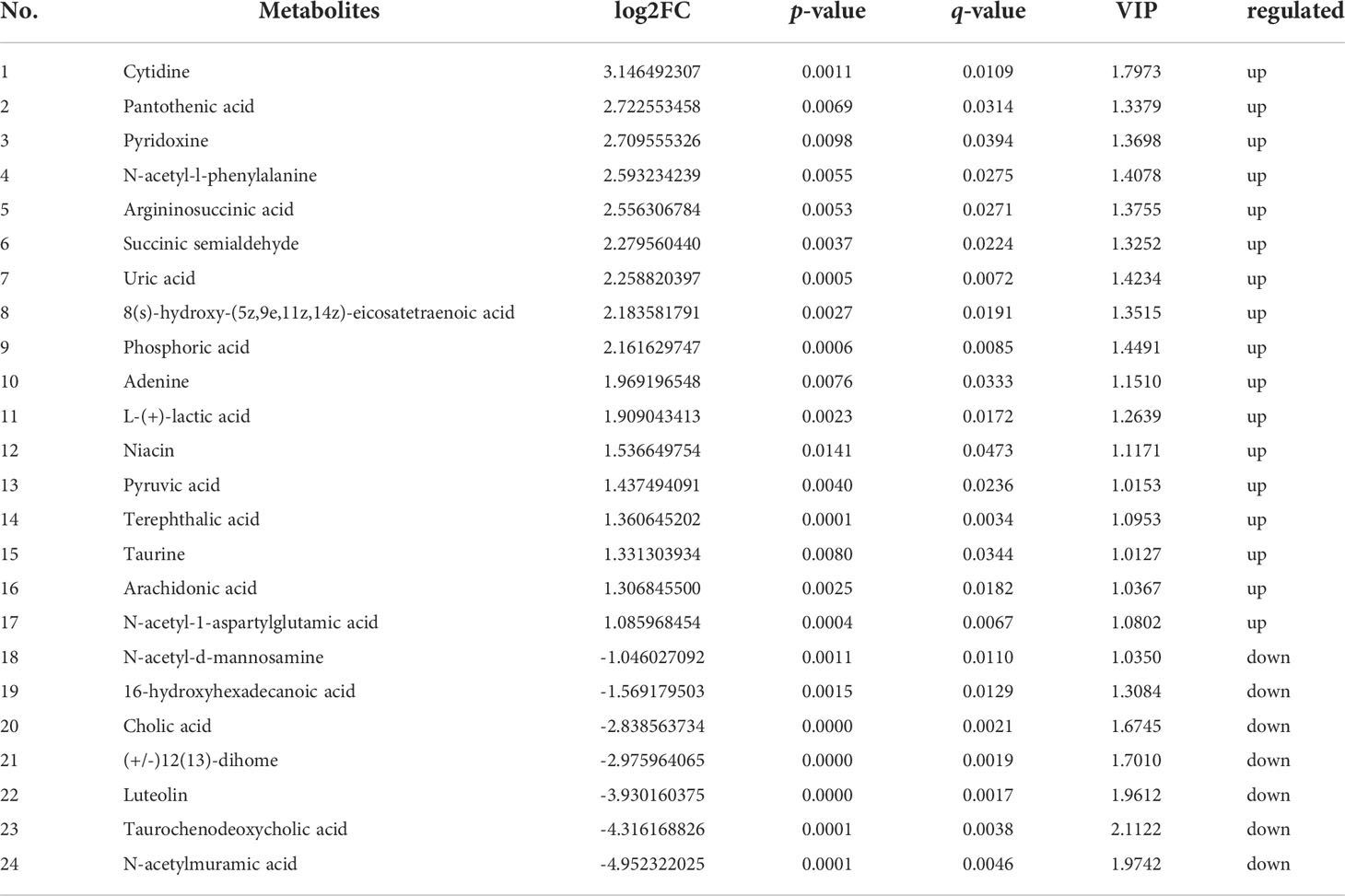
Table 3 24 known differential metabolites were significantly enriched in KEGG pathway enrichment analysis in negative ion mode.
3.5 Association between the altered metabolites and microbial
To reveal the relationships between intestinal microbial and DMs, scatterplots at the phylum, family, and genus levels were generated by canonical correlation analysis (Figure 9). The results showed that there was a strong correlation (R value > 0.75) between the host intestinal microbial and DMs at all taxonomic levels, and the samples from different groups had a large degree of dispersion. To further analyze the relationship between the nine marker DMs (including seven types of fatty acids and two Warburg effect marker metabolites) and host intestinal microbial, heat maps at the phylum, family, and genus levels were generated by spearman correlation analysis (Figure 10). The results showed that, at the phylum classification level, Proteobacteria was positively associated with arachidonic acid, 8(s)-hydroxy-(5z,9e,11z,14z) eicosatetraenoic acid, docosapentaenoic acid, eicosapentaenoic acid, pyruvic acid and L-(+)-lactic acid, and negatively associated with gamma-linolenic acid, 16-hydroxyhexadecanoic acid and 9-oxo-10(e),12(e)-octadecadienoic acid. The association of Cyanobacteria and Actinobacteria with marker metabolites was completely opposite to that of Proteobacteria, while neither Bacteriodetes nor Firmicutes were significantly associated with the marker metabolites (p > 0.05). It was worth noting that, Photobacterium and Vibrio in the Vibrionaceae that dominated the DIV1-infected group, were positively associated with Warburg effect marker metabolites L-(+)-lactic acid and pyruvic acid. In contrast, Corynebacterium in the Corynebacteriaceae that dominated the PBS-injected group, was negatively correlated with L-(+)-lactic acid and pyruvic acid. This phenomenon also appeared in the correlation with fatty acid metabolites.
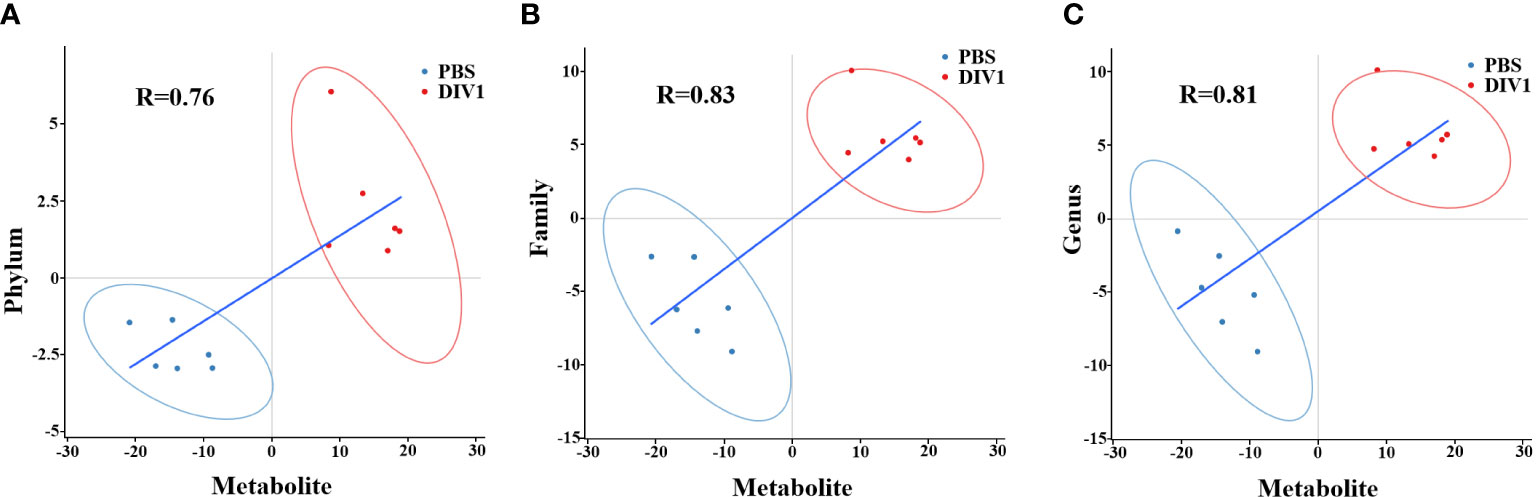
Figure 9 Scatter plots of correlation between DMs and hosts intestinal microbial. (A) Phylum classification level. (B) Family classification level. (C) Genus classification level.
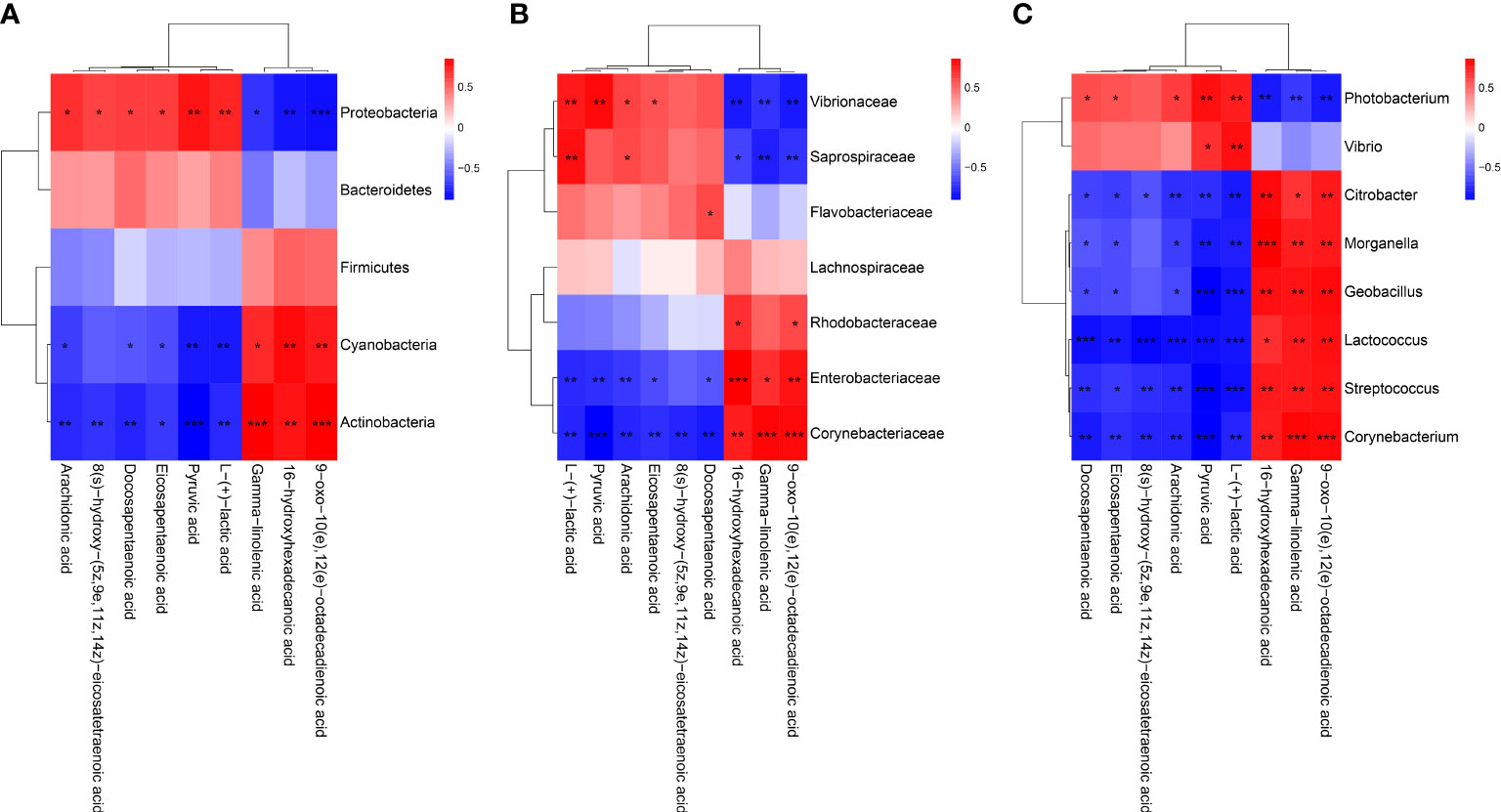
Figure 10 Heap maps of correlation between the nine marker DMs and the hosts intestinal microbial. (A) Phylum classification level. (B) Family classification level. (C) Genus classification level. Red indicateds a positive coreelation, and blue represents a negative correlation. * Indicates a significant difference (*p < 0.05; **p < 0.01; ***p < 0.001).
4 Discussion
Growing evidences suggested that viral infection or environmental stress can lead to changes in the structure and function of the host’s intestinal microbiota, and further affect the normal metabolism of the host and caused various adverse reactions (28–31). To date, there are no reports on the effects of DIV1 infection on the intestinal microbiota and metabolites of M. japonicus. The present study provides insights into the interaction of M. japonicus and DIV1 through the histological analysis, enzyme activity analysis and the integrated analysis of intestinal microbiome and metabolomics.
The results of histological analysis showed that severe intestinal mucosal damage was observed in the intestine of DIV1-infected shrimp, some intestinal epithelial cells were detached from the basement membrane and the intestinal villus height was significantly reduced. Similarly, Xue et al. found that WSSV-infected crayfish Procambarus clarkii exhibited worse intestinal histomorphology, with thinner intestinal walls and shorter intestinal villi compared to healthy crayfish (32). Liang et al. found that the intestinal epithelial cells of L. vannamei infected with Vibrio parahaemolyticus were completely detached from the basement membrane (33). Except for the intestinal mucosal damage, the disturbance of intestinal immunity and digestive function was also observed in the DIV1-infected shrimp. Enzyme activity analysis showed that all digestive enzymes and immune enzymes were significantly altered (p < 0.001), indicating that DIV1 infection severely affected the normal immune and digestive functions of the M. japonicus intestine. Studies have found that shrimp hemocytes can produce a large amount of free radicals and reactive oxygen species (ROS) during phagocytosis, thereby effectively killing the invading pathogens (34, 35). However, the mass accumulation of ROS in animals will cause serious cell damage, resulting in various diseases (36). SOD and CAT are two kinds of antioxidant enzymes, which can effectively remove free radicals and ROS, thereby protecting cells from oxidative stress (37–39). Under the action of SOD, ROS react with hydrogen ions to generate less toxic hydrogen peroxide, and then react with hydrogen ions under the action of CAT to finally produce harmless water (40). Therefore, only when the activities of SOD and CAT are coordinated, the free radicals and ROS in the organism can be maintained at a low level. However, the trends in the activity of SOD and CAT were not always consistent. In L. vannamei, intramuscular injection of 0.1 μg (g shrimp)−1 fluorescent red polyethylene microspheres (41) and experimental infection with Enterocytozoon hepatopenaei (42) both resulted in a significantly increase in SOD activity and a significant decrease in CAT activity. In Strongylocentrotus intermedius, the activity of SOD increased significantly and the activity of CAT decreased significantly with increasing temperature (43). In our study, the activities of SOD was significantly increased in the intestine of the M. japonicus under DIV1 infection, while the activities of CAT was significantly decreased. These results are supported by the previous findings, and suggested that DIV1 infection may cause a large amount of intestinal reactive oxygen species to be produced and inhibit the antioxidant capacity of the intestine to some extent. The specific mechanism needs to be further studied. LYZ has been shown to play an important role in shrimp resistance to viral and bacterial infections (44–46). The activity of LYZ can directly reflect the strength of shrimp immune function. After infection with DIV1, the activity of LYZ in the intestine of M. japonicus was significantly decreased, which indicated that the intestinal immune function was suppressed to some extent. α-AMS, LPS and TPS are three important digestive enzymes that also serve as markers to help diagnose diseases. Several studies had reported that the significantly increased α-AMS and LPS activities can be used as the basis for the diagnosis of mammalian enteritis and pancreatitis (47–51), while trypsin deficiency is closely related to enteritis (52–54). In this study, the intestinal activities of α-AMS and LPS were significantly increased after infection with DIV1, while the activities of TPS was significantly decreased. This suggested that DIV1 infection may cause damage to the shrimp intestine, causing enteritis and digestive dysfunction. In addition, this may also be one of the reasons for the decreased feeding rate of DIV1-infected shrimp and caused the symptoms of empty stomach and intestine (14, 25, 26, 55, 56).
In intestinal microbiome analysis, after DIV1 infection at 24 h, the intestine microbiome was significantly changed in the composition and diversity at the phylum, family and genus levels. In this study, the intestinal microbiota in M. japonicus were dominated by five phyla of Proteobacteria, Actinobacteria, Bacteroidetes, Firmicutes and Cyanobacteria in both the PBS-injected group and DIV1-infected group, which is consistent with the previous microbiome studies in L. vannamei. At the family level, the relative abundance of Vibrionaceae significantly increased in the intestine of DIV1-infected M. japonicus compared with healthy M. japonicus, which mainly reflected in the significant increase in the relative abundance of Photobacterium and Vibrio at the genus level. In addition, both the LEfSe cladogram and LDA score of LEfSe-PICRUSt showed that Photobacterium and Vibrio dominated in the DIV1-infected group. Several previous studies had shown that WSSV infection can cause an increase in the relative abundance of opportunistic pathogens in the intestine of shrimp, such as Aeromonas, Arcobacter, Vibrio and Trichococcus (19, 21, 32). Photobacterium had been shown to cause various diseases in fish and shrimp (57–61) causing huge losses in aquaculture. Vibrio, which is generally considered to be an opportunistic pathogen of shrimp and widely exists in the intestine of shrimp, can cause shrimp disease when the environment changes drastically (62–65). These results suggested that DIV1 infection can cause an increased abundance of opportunistic pathogens in the intestine, which may probably due to DIV1 infection disrupting the mechanical barrier of the intestine and impairing the ability of intestine to select microorganisms. The results of PICRUSt functional prediction showed that, in the KEGG level 2, the relative abundance of “carbohydrate metabolism”, “metabolism of cofactors and vitamins” and “amino acid metabolism” were the top 3 in the two groups. It meant that intestinal microbiota played an important role in regulating host vitamin metabolism, carbohydrate metabolism and amino acid metabolism. Notably, the relative abundance of “infectious diseases: bacterial” was significantly increased under DIV1 infection, which further suggested that DIV1 infection could cause secondary bacterial infection. In-depth analysis of the top 3 KEGG level 2 terms showed that the abundance value of amino acid metabolism, vitamin metabolism, glycolysis/gluconeogenesis and pyruvate metabolism were significantly increased after DIV1 infection. It meant that DIV1 can promote the Warburg effect by affecting the function of the host intestinal microbiota.
A recent study found that the “linoleic acid metabolism” and “arachidonic acid metabolism” were also the most disturbed pathways by HCoV-299E infection (66). Interestingly, exogenous supplement of linoleic acid or arachidonic acid in HCoV-229E-infected cells significantly suppressed HCoV-229E virus replication (66). Another study showed that linoleic acid could directly bound to WSSV to inhibited the viral replication and indirectly participate in the immune response against WSSV by activating the ERK-NF-κB signaling pathway to promote the expression of antimicrobial peptides and IFN-like gene Vago5 (67). Consistently, the intestinal metabolomics analysis in this study showed that lipid metabolism in the intestine of M. japonicus was remodeled after DIV1 infection, and a total of seven types of lipid fatty acids were significantly changed, including arachidonic acid, eicosapentaenoic acid, docosapentaenoic acid, 9-oxo-10(e), 12(e) -octadecadienoic acid, gamma-linolenic acid, 8(s)-hydroxy-(5z,9e,11z,14z) eicosatetraenoic acid and 16-hydroxyhexadecanoic acid. In addition, two Warburg effect marker metabolites were significantly up-regulated, including L-(+)-lactic acid and pyruvic acid. KEGG enrichment analysis indicated that the “Linoleic acid metabolism” and “arachidonic acid metabolism” pathways were the most affected by DIV1 infection. These results suggested that the lipid metabolic reprogramming of M. japonicus was significantly associated with DIV1 infection and replication. After infected with DIV1, two marker metabolites of the Warburg effect were significantly up-regulated in the shrimp intestine, including L-(+)-lactic acid and pyruvic acid, and significantly enriched in the “pyruvate metabolism” and “glycolysis/gluconeogenesis” pathways. In addition, some amino acid metabolism-related pathways were also significantly enriched under DIV1 infection, including “tyrosine metabolism”, “phenylalanine metabolism” and “alanine, aspartate and glutamate metabolism”. These phenomena also appeared in previous studies (11, 25, 26), which may be attributed to the material and energy requirements of DIV1 replication and suggested that DIV1 may contain AMGs for regulating host amino acid metabolism, carbohydrate metabolism and energy metabolism. It was worth noting that some pathways related to vitamin metabolism were significantly enriched, such as “retinol metabolism”, “vitamin B6 metabolism” and “pantothenate and CoA biosynthesis”. Retinol, also known as vitamin A (VA), plays an important and positive role in intestinal mucosal immunity and repair (68). The B vitamins can regulate the energy metabolism, nutrient accumulation and immune defense function of aquatic animals (69). Our previous study showed that DIV1 infection can regulate the vitamin metabolism of shrimp by affecting the expression of intestinal miRNA and mRNA (11, 26). In this study, from the metabolome level, it was further demonstrated that DIV1 infection can regulate vitamin metabolism in the intestine, thereby affecting the normal physiological function of M. japonicus.
Intestinal microbiota variation caused by viral infection or environmental stress can affect the metabolism and immunity of their host, and increase disease susceptibility (70, 71). To further explore the relationship between intestine microbiota variation and metabolites, the correlation analysis of dominant intestinal microbial and marker DMs was carried out. In this study, the increased levels of Photobacterium and Vibrio were positively correlated with changes in Warburg effect marker metabolites L-(+)-lactic acid and pyruvic acid. This indicated that Photobacterium and Vibrio may synergize with DIV1 to jointly promoted the Warburg effect, providing material and energy for the successful replication of DIV1. In addition, the abundances of Photobacterium and Vibrio were positively associated with arachidonic acid, eicosapentaenoic acid, docosapentaenoic acid and 8(s)-hydroxy-(5z,9e,11z,14z) eicosatetraenoic acid and negatively associated with 9-oxo-10(e),12(e)-octadecadienoic acid, gamma-linolenic acid and 16-hydroxyhexadecanoic acid. However, Corynebacterium, which dominated the PBS-injected group, was just the opposite of the above. It suggested that Photobacterium and Vibrio may be involved in the metabolic reprogramming of the shrimp intestine after DIV1 infection.
5 Conclusions
In conclusion, through the histological analysis, enzyme activity analysis and the integrated analysis of intestinal microbiome and metabolomics, the present study revealed that DIV1 infection can lead to the damage of the intestinal mechanical barrier of shrimp, the imbalance of oxidative and antioxidant capacity, the decrease of immune enzyme activities, and the disturbance of digestive enzyme activities. Futhermore, harmful bacteria may cooperate with DIV1 to promote the Warburg effect and induce metabolic reprogramming, thereby creating favorable conditions for the replication of DIV1. The speculation needs to be further verified. This study is the first to report the changes of intestinal microbiota and metabolites of M. japonicus under DIV1 infection, demonstrating that DIV1 can induce secondary bacterial infection and metabolic reprogramming (Figure 11).
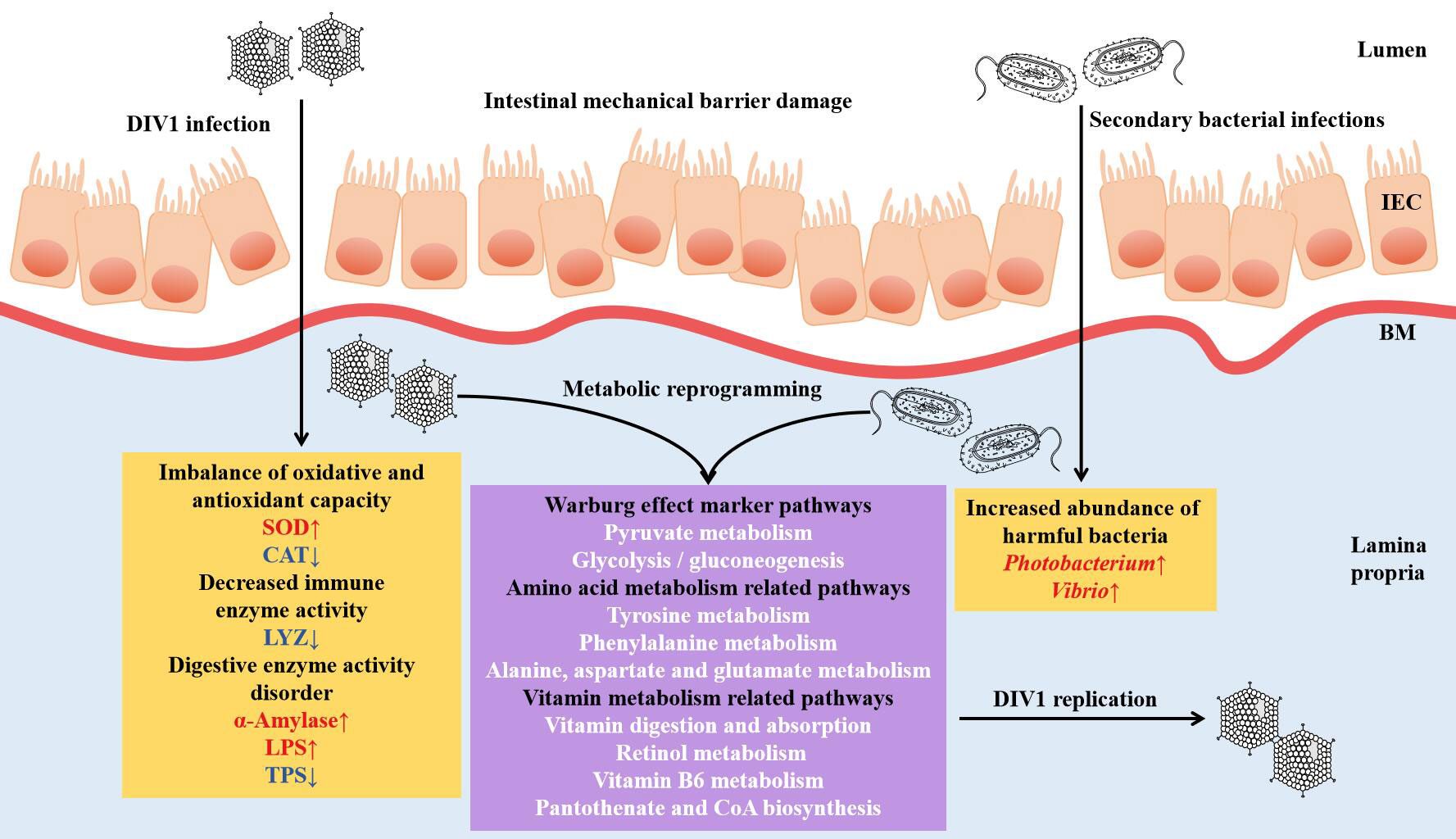
Figure 11 Proposed schematic diagram of secondary bacterial infection and metabolic reprogramming of M. japonicus induced by DIV1 infection. IEC, intestinal epithelial cells; BM, basement membrane; SOD, superoxide dismutase; CAT, catalase; LYZ, lysozyme; LPS, lipase; TPS, trypsin.
Data availability statement
The data presented in the study are deposited in the Sequence Read Archive repository, accession number PRJNA720257.
Author contributions
ZH, SZ, and CS contributed to conception and design of the study. ZH, YZ, ML, LD, and YW collected the samples and performed the experiments. ZH analyzed the sequencing data, organized all the results, wrote the original draft, and revised the manuscript. YZ helped to perform statistical analysis, conceptual analysis and manuscript revision. SZ performed the writing review and editing. CS contributed to project administration and funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the key research and development projects in Guangdong Province (Grant No. 2020B0202010009), the Science and Technology Program of Guangdong Province (Grant No. 2021B0202020003), and the project of the innovation team for the innovation and utilization of Economic Animal Germplasm in the South China Sea (Grant No. 2021KCXTD026).
Acknowledgments
Our acknowledgments go to all funders of this work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Gregory AC, Zayed AA, Conceição-Neto N, Temperton B, Bolduc B, Alberti A, et al. Marine DNA viral macro- and microdiversity from pole to pole. Cell (2019) 177(5):1109–1123.e14. doi: 10.1016/j.cell.2019.03.040
2. Zayed AA, Wainaina JM, Dominguez-Huerta G, Pelletier E, Guo J, Mohssen M, et al. Cryptic and abundant marine viruses at the evolutionary origins of earth’s RNA virome. Sci (New York NY) (2022) 376(6589):156–62. doi: 10.1126/science.abm5847
3. Suttle CA. Marine viruses–major players in the global ecosystem. Nat Rev Microbiol (2007) 5(10):801–12. doi: 10.1038/nrmicro1750
4. Hurwitz BL, U’Ren JM. Viral metabolic reprogramming in marine ecosystems. Curr Opin Microbiol (2016) 31:161–8. doi: 10.1016/j.mib.2016.04.002
5. Chen IT, Aoki T, Huang YT, Hirono I, Wang HC. White spot syndrome virus induces metabolic changes resembling the warburg effect in shrimp hemocytes in the early stage of infection. J Virol (2011) 85(24):12919–28. doi: 10.1128/JVI.05385-11
6. Hsieh YC, Chen YM, Li CY, Chang YH, Liang SY, Lin SY, et al. To complete its replication cycle, a shrimp virus changes the population of long chain fatty acids during infection via the pi3k-akt-mtor-hif1α pathway. Dev Comp Immunol (2015) 53(1):85–95. doi: 10.1016/j.dci.2015.06.001
7. Apún-Molina JP, Robles-Romo A, Alvarez-Ruiz P, Santamaria-Miranda A, Arjona O, Racotta IS. Influence of stocking density and exposure to white spot syndrome virus in biological performance, metabolic, immune, and bioenergetics response of whiteleg shrimp Litopenaeus vannamei. Aquaculture (2017) 479:528–37. doi: 10.1016/j.aquaculture.2017.06.027
8. Godoy-Lugo JA, Miranda-Cruz MM, Rosas-Rodríguez. JA, Adan-Bante NP, Soanez-Organis JG. Hypoxia inducible factor 1 regulates WSSV-induced glycolytic genes in the white shrimp Litopenaeus vannamei. Fish Shellfish Immunol (2019) 92:165–71. doi: 10.1016/j.fsi.2019.05.040
9. Zeng D, Chen X, Xie D, Zhao. Y, Yang C, Li Y, et al. Transcriptome analysis of pacific white shrimp (Litopenaeus vannamei) hepatopancreas in response to taura syndrome virus (TSV) experimental infection. Fish Sci Technol Guangxi (2014) 1:4–10. doi: 10.1371/journal.pone.0057515
10. Galvan-Alvarez D, Mendoza-Cano F, Hernandez-Lopez J, Sanchez-Paz A. Experimental evidence of metabolic disturbance in the white shrimp Penaeus vannamei induced by the infectious hypodermal and hematopoietic necrosis virus (IHHNV). J Invertebrate Pathol (2012) 111(1):60–7. doi: 10.1016/j.jip.2012.06.005
11. He Z, Zhong Y, Hou D, Hu X, Fu Z, Liu L, et al. Integrated analysis of mRNA-seq and miRNA-seq reveals the molecular mechanism of the intestinal immune response in Marsupenaeus japonicus under decapod iridescent virus 1 infection. Front Immunol (2022) 12:807093. doi: 10.3389/fimmu.2021.807093
12. Liao X, He J, Li C. Decapod iridescent virus 1: An emerging viral pathogen in aquaculture. Rev Aquacult (2022). doi: 10.1111/raq.12672
13. Srisala J, Sanguanrut P, Thaiue D, Laiphrom S, Siriwattano J, Khudet J, et al. Infectious myonecrosis virus (IMNV) and decapod iridescent virus 1 (DIV1) detected in captured, wild Penaeus monodon. Aquaculture (2021) 545:737262. doi: 10.1016/j.aquaculture.2021.737262
14. He Z, Zhao J, Chen X, Liao M, Xue Y, Zhou J, et al. The molecular mechanism of hemocyte immune response in Marsupenaeus japonicus infected with decapod iridescent virus 1. Front Microbiol (2021) 12:710845. doi: 10.3389/fmicb.2021.710845
15. Dall W, Hill BJ, Rothlisberg PC, Sharples DJ. The biology of the penaeidae. J Crustacean Biol (1990) 27:489. doi: 10.2307/1548534
16. He Z, Zhao J, Liao X, Chen X, Fu Z, Sun C, et al. The secondary bacterial infection caused by WSSV outbreaks impacts shrimp Marsupenaeus japonicus’ growth as well as its intestinal microbiota’s composition and function. Israeli J Aquacult Bamidgeh (2020) 72:1–17. 1227989. doi: 10.46989/001C.21687
17. Yuan R, Zheng J, Liu J, Jiang X. Reviews in genetics and breeding of Marsupenaeus japonicus. J Guangdong Ocean Univ (2016) 36(01):98–102.
18. Zheng X, Duan Y, Dong H, Zhang J. Progress on gut mucosal immunization of crustacean. Trans Oceanol Limnol (2016) 3:83–90. doi: 10.13984/j.cnki.cn37-1141.2016.03.012
19. Wang J, Huang Y, Xu K, Zhang X, Yan M. White spot syndrome virus (WSSV) infection impacts intestinal microbiota composition and function in Litopenaeus vannamei. Fish Shellfish Immunol (2018) 84:130–7. doi: 10.1016/j.fsi.2018.09.076
20. Chen H, Wang Y, Zhang J, Bao J. Intestinal microbiota in white spot syndrome virus infected red swamp crayfish (Procambarus clarkii) at different health statuses. Aquaculture (2021) 542:736826–6. doi: 10.1016/j.aquaculture.2021.736826
21. Niu G, Yan M, Li G, Lu P, Yu Z, Wang J. Infection with white spot syndrome virus affects the microbiota in the stomachs and intestines of kuruma shrimp. Sci Total Environ (2022) 839:156233. doi: 10.1016/j.scitotenv.2022.156233
22. Wang A, Yang Q, Tan B, Xiao W, Jia J, Dong X, et al. Effects of enzymolytic soybean meal on growth performance, serum biochemical indices, non-specific immunity and disease resistance of juvenile Litopenaeus vannamei. J Guangdong Ocean Univ (2018) 38(01):14–21.
23. Wu Y, Li R, Shen G, Huang F, Yang Q, Tan B, et al. Effects of dietary small peptides on growth, antioxidant capacity, nonspecific immunity and ingut microflora structure of Liopenaeus vannamei. J Guangdong Ocean Univ (2021) 41(05):1–9.
24. Wang Z, Zhang Y, Yao D, Zhao Y, Tran NT, Li S, et al. Metabolic reprogramming in crustaceans: A vital immune and environmental response strategy. Rev Aquacult (2021). doi: 10.1111/raq.12640
25. He Z, Chen X, Zhao J, Hou D, Fu Z, Zhong Y, et al. Establishment of infection mode and Penaeus monodon hemocytes transcriptomics analyses under decapod iridescent virus 1 (DIV1) challenge. Aquaculture (2021) 542:736816. doi: 10.1016/j.aquaculture.2021.736816
26. Liao X, Wang C, Wang B, Qin H, Hu S, Wang P, et al. Comparative transcriptome analysis of Litopenaeus vannamei reveals that triosephosphate isomerase-like genes play an important role during decapod iridescent virus 1 infection. Front Immunol (2020) 11:1904. doi: 10.3389/fimmu.2020.01904
27. Xiong J, Liu Y, Lin X, Zhang H, Zeng J, Hou J, et al. Geographic distance and pH drive bacterial distribution in alkaline lake sediments across Tibetan plateau. Environ Microbiol (2012) 14(9):2457–66. doi: 10.1111/j.1462-2920.2012.02799.x
28. Hsu BB, Gibson TE, Yeliseyev V, Liu Q, Lyon L, Bry L, et al. Dynamic modulation of the gut microbiota and metabolome by bacteriophages in a mouse model. Cell Host Microbe (2019) 25(6):803–814.e5. doi: 10.1016/j.chom.2019.05.001
29. Qiao R, Sheng C, Lu Y, Zhang Y, Ren H, Lemos B. Microplastics induce intestinal inflammation, oxidative stress, and disorders of metabolome and microbiome in zebrafish. Sci Total Environ (2019) 662:246–53. doi: 10.1016/j.scitotenv.2019.01.245
30. Duan Y, Xiong D, Wang Y, Li H, Dong H, Zhang J. Toxic effects of ammonia and thermal stress on the intestinal microbiota and transcriptomic and metabolomic responses of Litopenaeus vannamei. Sci Total Environ (2021) 754:141867. doi: 10.1016/j.scitotenv.2020.141867
31. Duan Y, Lu Z, Zeng S, Dan X, Mo Z, Zhang J, et al. Integration of intestinal microbiota and transcriptomic and metabolomic responses reveals the toxic responses of Litopenaeus vannamei to microcystin-LR. Ecotoxicol Environ Saf (2021) 228:113030. doi: 10.1016/j.ecoenv.2021.113030
32. Xue M, Jiang N, Fan Y, Yang T, Li M, Liu W, et al. White spot syndrome virus (WSSV) infection alters gut histopathology and microbiota composition in crayfish (Procambarus clarkii). Aquacult Rep (2022) 22:101006. doi: 10.1016/j.aqrep.2022.101006
33. Liang F, Li C, Hou T, Wen C, Kong S, Ma D, et al. Effects of chitosan-gentamicin conjugate supplement on non-specific immunity, aquaculture water, intestinal histology and microbiota of pacific white shrimp (Litopenaeus vannamei). Mar Drugs (2020) 18(8):419. doi: 10.3390/md18080419
34. Bogdan C, Röllinghoff M, Diefenbach A. Reactive oxygen and reactive nitrogen intermediates in innate and specific immunity. Curr Opin Immunol (2000) 12(1):64–76. doi: 10.1016/S0952-7915(99)00052-7
35. Duan Y, Zhang J, Dong H, Wang Y, Liu Q, Li H. Oxidative stress response of the black tiger shrimp Penaeus monodon to Vibrio parahaemolyticus challenge. Fish Shellfish Immunol (2015) 46(2):354–65. doi: 10.1016/j.fsi.2015.06.032
36. Yu BP. Cellular defenses against damage from reactive oxygen species. Physiol Rev (1994) 74(1):139–62. doi: 10.1152/physrev.1994.74.1.139
37. Zhu C, Li Y, Chen L, Li D, Li G, Deng S. Effects of nonylphenol (NP) on the activities of immunologic enzyme in blood serum of Macrobrachium rosenbergii. J Guangdong Ocean Univ (2012) 32(06):17–20.
38. Wu W, Chen L, Li Y, Zhu C. Effects of tributyltin on the activities of immunologic enzyme in blood serum of the Macrobrachium rosenbergill. J Guangdong Ocean Univ (2014) 34(03):17–21.
39. Chen Y, Li C, Huang X. Effects of microcystin on activities of immune enzymes in the white shrimp Litopenaeus vannamei. J Guangdong Ocean Univ (2015) 35(06):35–9.
40. Fridovich I. Oxygen toxicity: A radical explanation. J Exp Biol (1998) 201:1203–9. doi: 10.1242/jeb.201.8.1203
41. Hsieh SL, Wu YC, Xu RQ, Chen YT, Dong CD. Effect of polyethylene microplastics on oxidative stress and histopathology damages in Litopenaeus vannamei. Environ pollut (2021) 288:117800. doi: 10.1016/j.envpol.2021.117800
42. Subash P, Uma A, Ahilan B. Early responses in penaeus vannamei during experimental infection with Enterocytozoon hepatopenaei (EHP) spores by injection and oral routes. J Invertebrate Pathol (2022) 190:107740. doi: 10.1016/j.jip.2022.107740
43. Wang WP, Hang LS, Zhang XL, Liu P, Yang XY, Wang L, et al. Effects of thermal stress on the activities of antioxidant enzymes and mitochondrial structure and function in Strongylocentrotus intermedius. J Guangdong Ocean Univ (2022) 42(04):42–8.
44. Kaizu A, Fagutao FF, Kondo H, Aoki T, Hirono I. Functional analysis of c-type lysozyme in penaeid shrimp. J Biol Chem (2011) 286(52):44344–9. doi: 10.1074/jbc.M111.292672
45. Karthik V, Kamalakannan V, Thomas A, Sudheer NS, Singh IS, Narayanan RB. Functional characterization of a c-type lysozyme from Indian shrimp Fenneropenaeus indicus. Probiotics Antimicrobial Proteins (2014) 6(2):114–21. doi: 10.1007/s12602-013-9146-y
46. Liu HT, Wang J, Mao Y, Liu M, Niu SF, Qiao Y, et al. Identification and expression analysis of a new invertebrate lysozyme in kuruma shrimp (Marsupenaeus japonicus). Fish Shellfish Immunol (2016) 49:336–43. doi: 10.1016/j.fsi.2015.12.034
47. Tenner S, Baillie J, Dewitt J, Vege SS. American College of gastroenterology guideline: Management of acute pancreatitis. Am J Gastroenterol (2014) 109:1400–15. doi: 10.1038/ajg.2013.458
48. Muniraj T, Dang S, Pitchumoni CS. Pancreatitis or not? – elevated lipase and amylase in ICU patients. J Crit Care (2015) 30:1370–5. doi: 10.1016/j.jcrc.2015.08.020
49. Ali F, Hussain MR, Deep A, Abu-Sbeih H, Alrifai T, Myint PT, et al. Gastrointestinal, pancreatic, and hepatic toxicity profile of CTLA-4 immune checkpoint inhibitors alone and in combination with PD-1/PD-L1 inhibitors: A meta-analysis of clinical trials. J Clin Oncol (2019) 37:e14117–7. doi: 10.1200/JCO.2019.37.15_suppl.e14117
50. Al Kaabi S, Al Kaabi A, Al Nuaimi H. What is beyond salmonella gastroenteritis? A case of acute pancreatitis complicating salmonella infection in a child: A case report and literature review. BMC Pediatr (2021) 21(1):353. doi: 10.1186/s12887-021-02814-w
51. Choi S, Lee HJ, Seo AN, Bae HI, Kwon HJ, Cho CM, et al. Case report: Development of type 1 autoimmune pancreatitis in an adolescent with ulcerative colitis mimicking pancreatic cancer. Front Pediatr (2021) 9:791840. doi: 10.3389/fped.2021.791840
52. Delaney JP, Bonsack M. Acute radiation enteritis in rats: bile salts and trypsin. Surgery (1992) 112:587–92.
53. Krogdahl Å., Bakke-McKellep A, Baeverfjord G. Effects of graded levels of standard soybean meal on intestinal structure, mucosal enzyme activities, and pancreatic response in Atlantic salmon (Salmo salar l.). Aquacult Nutr (2003) 9:361–71. doi: 10.1046/j.1365-2095.2003.00264.x
54. Palliyeguru MCD, Rose S, Mackenzie A. Effect of trypsin inhibitor activity in soya bean on growth performance, protein digestibility and incidence of sub-clinical necrotic enteritis in broiler chicken flocks. Br Poultry Sci (2011) 52:359–67. doi: 10.1080/00071668.2011.577054
55. Chen X, Qiu L, Wang H, Zou P, Dong X, Li F, et al. Susceptibility of Exopalaemon carinicauda to the infection with shrimp hemocyte iridescent virus (SHIV 20141215), a strain of decapod iridescent virus 1 (DIV1). Viruses (2019) 11(4):387. doi: 10.3390/v11040387
56. Qiu L, Chen X, Zhao RH, Li C, Gao W, Zhang QL, et al. Description of a natural infection with decapod iridescent virus 1 in farmed giant freshwater prawn, Macrobrachium rosenbergii. Viruses (2019) 11(4):354. doi: 10.3390/v11040354
57. Vaseeharan B, Sundararaj S, Murugan T, Chen JC. Photobacterium damselae ssp. damselae associated with diseased black tiger shrimp Penaeus monodon fabricius in India. Lett Appl Microbiol (2007) 45(1):82–6. doi: 10.1111/j.1472-765X.2007.02139.x
58. Kanchanopas-Barnette P, Labella A, Alonso CM, Manchado M, Castro D, Borrego JJ. The first isolation of Photobacterium damselae subsp. damselae from Asian seabass lates calcarifer. Fish Pathol (2009) 44(1):47–50. doi: 10.3147/jsfp.44.47
59. Zhang X, Qin G, Bing X, Yan B, Bi K. Phenotypic and molecular characterization of Photobacterium damselae, a pathogen of the cultured tongue sole Cynoglossus semilaevis in China. New Z J Mar Freshw Res (2011) 45(1):1–13 doi: 10.1080/00288330.2010.531745
60. Singaravel V, Gopalakrishnan A, Dewangan NK, Kannan D, Shettu N, Martin GG. Photobacterium damselae subsp. damselae associated with bacterial myonecrosis and hepatopancreatic necrosis in broodstock pacific white leg shrimp, Litopenaeus vannamei (Boone 1931). Aquacult Int (2020) 28:1593–608. doi: 10.1007/s10499-020-00545-w
61. Wang Z, Shi C, Wang H, Wan X, Zhang Q, Song X, et al. A novel research on isolation and characterization of Photobacterium damselae subsp. damselae from pacific white shrimp, Penaeus vannamei, displaying black gill disease cultured in China. J Fish Dis (2020) 43(5):551–9 doi: 10.1111/jfd.13153
62. Saulnier D, Haffner P, Goarant C, Levy P, Ansquer D. Experimental infection models for shrimp vibriosis studies: a review. Aquaculture (2000) 191(1-3):133–44. doi: 10.1016/S0044-8486(00)00423-3
63. Zhou J, Fang W, Yang X, Zhou S, Hu L, Li X, et al. A nonluminescent and highly virulent Vibrio harveyi strain is associated with “bacterial white tail disease”. Litopenaeus Vannamei Shrimp PloS One (2012) 7(2):e29961. doi: 10.1371/journal.pone.0029961
64. Santhyia AV, Mulloorpeedikayil RG, Kollanoor RJ, Jeyaseelan PM. Molecular variations in Vibrio alginolyticus and v. harveyi in shrimp-farming systems upon stress. Braz J Microbiol: [Publ Braz Soc Microbiol] (2015) 46(4):1001–8. doi: 10.1590/S1517-838246420140410
65. Nguyen TV, Alfaro A, Arroyo BB, Leon JAR, Sonnenholzner S. Metabolic responses of penaeid shrimp to acute hepatopancreatic necrosis disease caused by Vibrio parahaemolyticus. Aquaculture (2021) 533:736174. doi: 10.1016/j.aquaculture.2020.736174
66. Yan B, Chu H, Yang D, Sze KH, Lai PM, Yuan S, et al. Characterization of the lipidomic profile of human coronavirus-infected cells: implications for lipid metabolism remodeling upon coronavirus replication. Viruses (2019) 11(1):73. doi: 10.3390/v11010073
67. Li C, Yang MC, Hong PP, Zhao XF, Wang JX. Metabolomic profiles in the intestine of shrimp infected by white spot syndrome virus and antiviral function of the metabolite linoleic acid in shrimp. J Immunol (Baltimore Md: 1950) (2021) 206(9):2075–87. doi: 10.4049/jimmunol.2001318
68. Sirisinha S. The pleiotropic role of vitamin a in regulating mucosal immunity. Asian Pacific J Allergy Immunol (2015) 33(2):71–89.
69. Mai K. Abances in methods of vitamin bs nutrition studies on aquatic animals. J Fish China (2000) 4:373–8.
70. Levy M, Blacher E, Elinav E. Microbiome, metabolites and host immunity. Curr Opin Microbiol (2017) 35:8–15. doi: 10.1016/j.mib.2016.10.003
Keywords: decapod iridescent virus 1, Marsupenaeus japonicus, intestinal microbiota, metabolomic, secondary bacterial infection, metabolic reprogramming
Citation: He Z, Zhong Y, Liao M, Dai L, Wang Y, Zhang S and Sun C (2022) Integrated analysis of intestinal microbiota and metabolomic reveals that decapod iridescent virus 1 (DIV1) infection induces secondary bacterial infection and metabolic reprogramming in Marsupenaeus japonicus. Front. Immunol. 13:982717. doi: 10.3389/fimmu.2022.982717
Received: 06 July 2022; Accepted: 31 August 2022;
Published: 16 September 2022.
Edited by:
Carlos Angulo, Centro de Investigación Biológica del Noroeste (CIBNOR), MexicoReviewed by:
Yo Okamura, University of Washington, United States;Li Li, Huazhong Agricultural University, ChinaCopyright © 2022 He, Zhong, Liao, Dai, Wang, Zhang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chengbo Sun, c3VuY2JAZ2RvdS5lZHUuY24=; Shuang Zhang, enNodWFuZ0BnZG91LmVkdS5jbg==
†These authors have contributed equally to this work
 Zihao He
Zihao He Yunqi Zhong
Yunqi Zhong Minze Liao
Minze Liao Linxin Dai1
Linxin Dai1 Yue Wang
Yue Wang Shuang Zhang
Shuang Zhang Chengbo Sun
Chengbo Sun