- Tianjin Key Laboratory of Digestive Diseases, Tianjin Institute of Digestive Diseases, Department of Gastroenterology and Hepatology, Tianjin Medical University General Hospital, Tianjin, China
Inflammatory bowel disease (IBD) is characterized by chronic and relapsing inflammation of gastrointestinal tract, with steadily increased incidence and prevalence worldwide. Although the precise pathogenesis remains unclear, gut microbiota, bile acids (BAs), and aberrant immune response play essential roles in the development of IBD. Lately, gut dysbiosis including certain decreased beneficial bacteria and increased pathogens and aberrant BAs metabolism have been reported in IBD. The bacteria inhabited in human gut have critical functions in BA biotransformation. Patients with active IBD have elevated primary and conjugated BAs and decreased secondary BAs, accompanied by the impaired transformation activities (mainly deconjugation and 7α-dehydroxylation) of gut microbiota. Probiotics have exhibited certain positive effects by different mechanisms in the therapy of IBD. This review discussed the effectiveness of probiotics in certain clinical and animal model studies that might involve in gut microbiota-BAs axis. More importantly, the possible mechanisms of probiotics on regulating gut microbiota-BAs axis in IBD were elucidated, which we focused on the elevated gut bacteria containing bile salt hydrolase or BA-inducible enzymes at genus/species level that might participate in the BA biotransformation. Furthermore, beneficial effects exerted by activation of BA-activated receptors on intestinal immunity were also summarized, which might partially explain the protect effects and mechanisms of probiotics on IBD. Therefore, this review will provide new insights into a better understanding of probiotics in the therapy targeting gut microbiota-BAs axis of IBD.
1 Introduction
Inflammatory bowel disease (IBD), which encompasses mainly two clinical phenotypes, was first described in 1895. The two phenotypes are ulcerative colitis (UC) and Crohn’s disease (CD) (1). The epidemiological stages of IBD’s evolution are Emergence, Acceleration in Incidence, Compounding Prevalence, and Prevalence Equilibrium. Western countries are in the third stage, newly industrialized regions are in the second stage, and developing countries are in the first stage as of the end of 2020. In addition, the rising global burden of IBD may be improved by comprehending the changing epidemiological patterns (2). Furthermore, it is of great importance to know the etiology and pathogenesis of IBD. The genetic agents, gut dysbiosis, environmental factors, immunological status, and permeability of gut barrier are closely related to the initiation and development of IBD, even though the precise causes and mechanisms still remain unclear (3). Among them, gut microbiota plays an irreplaceable role as its regulatory and metabolic functions. For instance, they participate in the bile acids (BAs) metabolism to produce unconjugated and secondary BAs for their possession of bile salt hydrolase (BSH) and bile acid–inducible enzymes (BAI) (4). The gut dysbiosis and bile acid dysmetabolism that existed in patients with IBD have been discovered by many studies, the levels of secondary BAs are lower, primary BAs are elevated because of the impairment of microbiota deconjugation and transformation activities, and the microbial BSH activity is decreased (5–7). Therefore, gut microbiota and BAs metabolism play important roles, which have been studied in IBD. In addition, it is widely believed that aberrant immune response against the gut microorganisms in genetically susceptible individuals is the cause of IBD. Immune cell trafficking and some cytokines, such as tumor necrosis factor (TNF), interleukin-10 (IL-10), IL-22, IL-6, IL-17, IL-12, and IL-23 are involved in the immunological pathogenesis of IBD (8). Probiotics, one of the adjuvant therapeutic methods of IBD, have been revealed to be effective in some clinical and animal studies. It can rebalance the aberrant gut microbiota by increasing beneficial microbes that may regulate BAs metabolism and simultaneously inhibiting pathogens (9). Chronic inflammatory, a gut condition in long-term IBD, has an increased risk of developing into intestinal cancer, which is perceived as colitis-associated cancer (CAC) (10). The abnormal BAs metabolism and gut dysbiosis also exist in CAC. The CAC mice model showed gut dysbiosis, decreased fecal BAs, and lessened transformation of primary to secondary BAs (11). Probiotic intervention exhibited inhibition of tumor formation and anti-inflammation effects in CAC mice to some extent (12, 13). Therefore, probiotics may adjust gut bacteria involved in BAs biotransformation in IBD and CAC.
For better understanding the probiotics, in the present review, we focused on the certain effects and probable functional roles of probiotics on regulating gut microbiota-BAs axis in IBD and CAC.
2 Microbial influence on BAs in IBD
2.1 BAs synthesis and circulation
In gastrointestinal tract, the biotransformation of intestinal BAs is central to the metabolic homeostasis (14). Disorders in BAs metabolism can result in dyslipidemia, cholestatic liver diseases, cardiovascular diseases, fatty liver diseases, diabetes, and so forth (15). Abnormal BAs metabolism detected in IBD may facilitate pro-inflammatory intestinal responses via its effects on immune cells and epithelial intestinal cells. The processes of BAs synthesis and circulation have been extensively reviewed elsewhere. BAs synthesize in hepatocytes through the classical and alternative pathways. Primary BAs include cholic acid (CA) and chenodeoxycholic acid (CDCA). Subsequently, CA and CDCA conjugate to either glycine (predominantly in humans) or taurine (mainly in mice) to form conjugated BAs. Then, these BAs are released into intestine after a meal, and approximately 95% of them recirculate through enterohepatic circulation in the distal ileum. The remaining about 5% BAs are transported into colon and further metabolized by the colonic microbiota. The bacteria with BSH activity can deconjugate BAs into their unconjugated forms, including CA and CDCA. Afterward, they are converted to secondary BAs, namely, ursodeoxycholic acid (UDCA), lithocholic acid (LCA), and deoxycholic acid (DCA) through the microbiota possessing enzyme action of 7α-dehydroxylation. In addition, iso-BAs are produced by oxidation and epimerization at C3, C7, and C12 positions of hydroxyl by bacteria with hydroxysteroid dehydrogenases (HSDH) (14, 16).
Crosstalk between BAs and gut microbiota affect metabolic phenotypes, immunological functions, and risk factors for many diseases including diabetes, obesity, non-alcoholic fatty liver disease, IBD, and various cancers (7). In addition to participating in lipid absorption and cholesterol homeostasis, regulating their own biosynthesis, and maintaining a healthy gut microbiota, BAs can also balance carbohydrate metabolism, insulin sensitivity, and innate immunity. The imbalance of BAs production, function, or reabsorption is related to distinct gastrointestinal diseases, such as intestinal inflammation and carcinogenesis, and gastrointestinal motility (5, 14). In IBD patients, although many studies have demonstrated the existence of BAs malabsorption or reduced BAs recycling, both are commonly neglected. Due to decreased microbial abundance in the distal ileum and colon, patients with active IBD had elevated conjugated BAs accumulation and decreased secondary BAs. In addition, the transformation activities that included deconjugation, 7α-dehydroxylation, and desulphation of gut microbiota were impaired (5, 17, 18). Analogously, the same results were observed in many colitis rodent models. In the trinitrobenzene sulfonic acid (TNBS)–induced colitis model, on account of decreased expression of BA transporters, BAs amassing in feces were increased, which resulted in the suppression of BAs recycling (19). Rats with dextran sodium sulfate (DSS)–induced colitis also exhibited accumulation of CA in feces (20). Of note, the microbiome of pediatric patients with IBD was overtly depleted in their BAs production potential (18). Furthermore, in colectomy-treated patients with UC, their pouches had reduced levels of DCA and LCA and lessened genes required to convert primary to secondary BAs (21). With respect to CAC, a study discovered that fecal BAs were lessened, accompanied by decreased transformation of primary to secondary BAs. Moreover, downregulated gut-liver farnesoid X receptor–fibroblast growth factor 15 (FXR-FGF15) axis was also revealed in the CAC mice model (11).
2.2 BAs metabolism: Regulatory role of gut microbiota
The synthesis and metabolism of BAs are prominently modified by gut bacteria with diverse enzymes (22). BSH and BAI expressed by certain gut bacteria are the two main enzymes that involve in the deconjugation of primary BAs and subsequent transformation into secondary BAs (4). The major BAs biotransformation in the human gut comprises deconjugation, 7α-dehydroxylation, oxidation and epimerization, desulfatation, and esterification (23). BAs biotransformation is a collaborative effort through the gut microbiota and host. This biotransformation process may play a role in bile detoxification, thus reducing BAs toxicity, and may have beneficial effects on bacteria that carry functional genes. The gut microbiota modulates BAs generation and signalling by the biotransformation of gut BAs to unconjugated and secondary forms that have influence on host health and diseases (24). The major bacterial genera of gut microbiota participated in gut microbiota-BAs axis include Bifidobacterium, Lactobacillus, Bacteroides, Clostridium, and Listeria in BAs deconjugation; Clostridium and Eubacterium in 7α-dehydroxylation; Clostridium, Bacteroides, Escherichia, Eubacterium, Egghertella, Ruminococcu, and Peptostreptococcus in oxidation and epimerization; Fusobacterium, Clostridium, Peptococcus, and Pseudomonas in desulfatation; Lactobacillus, Bacteroides, and Eubacterium in esterification (25). BAs and gut microbiota can influence each other, gut microbiota modulates the size and constitute of the BAs pool, which in turn regulate microbiota composition. Hydrophobic BAs at high concentrations can generate direct antimicrobial activities mainly via membrane damage. BAs could also indirectly shape the composition of gut microbiota through BAs receptors. Gram-positive microorganisms are often more sensitive to BAs than Gram-negative microorganisms (7). Nevertheless, BAs could induce the proliferation of some gut microbes. In the context of high fat diet, elevated taurine-conjugated BAs aggrandized the availability of organic sulphur utilized by Bilophila wadsworthia, which was associated with increased incidence of colitis in Il10−/− mice (26).
2.2.1 Deconjugation
In IBD patients, the low level of BSH activity in the microbiota was demonstrated by bioinformatic analysis of metagenomic data (6). BSH, which is oxygen insensitive, is generally located inside the cell, and the optimal pH is commonly between 5 and 6 (27). BSH catalyzes the hydrolysis of conjugated BAs at C24 N-acyl amide bond. A number of studies aimed to illustrate the key secondary structure element and amino acids that may be participated in the substrate binding in BSH. These reports exhibited that BSH can identify its substrates through hydrophobic interactions with steroid moiety (28–31). Functional BSH has been identified in all major gut bacteria divisions and archaeal species (32). The main bacteria involved in BAs deconjugation at genera level are Bifidobacterium, Lactobacillus, Bacteroides, Clostridium, and Listeria (25). Apart from the above five genera, Song and co-workers discovered that Parabacteroides, Bacillus, Mycobacterium, Staphylococcus, Enterococcus, Eubacterium, Blautia, Peptoclostridium, Fusobacterium, Rhodopseudomonas, Yersinia, and Vibrio possessed more than five BSHs (33). It is beneficial for the gut microbiota to modify conjugated BAs, since it may be relevant to the detoxication of conjugated BAs and the acquisition of nitrogen, carbon, and sulfur, which have effect on the growth of bacteria (27, 34). Catalysis of BSH enzymes is the first step in the transition from conjugated BAs to their unconjugated forms, followed by further conversion into secondary BAs. Due to the likely less easily absorbed of secondary BAs, the reabsorption of BAs in the enterohepatic circulation was reduced. Certain BAs may be more easily excreted from individuals, and BA neo-synthesis may be strengthened (35, 36). For instance, as the most hydrophobic BA, LCA is reabsorbed weakly back into enterohepatic cycle, which leads to higher amounts of LCA in feces (37). In the context of chronic inflammation, such as IBD, unconjugated BAs might be gradually exhausted, presumably on account of the depletion of BSH-rich gut bacteria (38). Replenishment of probiotics may improve the decreased BSH in IBD. More importantly, the BSH activity of probiotics may be desirable, because it maximizes its prospects for survival in the harsh environment of intestine, which is likely to enhance the overall beneficial effects related to the strains (27).
2.2.2 7α-dehydroxylation
Only a limited number of intestinal bacteria, including Clostridium and Eubacterium that encode bai genes, can accomplish this process of BAs dehydroxylation. The bai genes were significantly reduced in UC than familial adenomatous polyposis pouches (21). Furthermore, BA dehydroxylation can only occur after deconjugation because of the inaccessibility of the hydroxyl group, which is different to oxidation and epimerization, and the 7α-dehydroxylation is the most significant conversion of BAs from quantity and physiology aspects in humans (39). Taking the multi-step bai encoded pathway as an illustration, it contains BA import, modification, and export from bacteria. There exist noticeable species-specific discrepancies in the distribution of certain bai genes. Clostridium sordellii VPI 9048 carries merely baiA2, baiCD, baiE, baiH, and 7α-HSDH, whereas Clostridium hiranonis carries baiJ, baiH, baiBCDEA2FG, and 7α-HSDH (40). Nevertheless, it remains to be elucidated the true distributions of bai genes among bacterial species and is still poorly understanding the minimum bai gene set that secondary BAs synthesis requires. Moreover, in addition to one advantage that produces reduced nicotinamide adenine dinucleotide phosphate of the bai system, other evolutionary advantages are also the subject of speculation and should be further revealed (41).
2.2.3 Oxidation, epimerization, and esterification
HSDHs from intestinal bacteria catalyze the invertible oxidation of hydroxy to oxo groups. Epimerization occurs by stereospecific oxidation and reduction at 3-, 7-, and 12- hydroxyl groups of BAs (42). Epimerization requires the role of two different HSDHs and can be completed via a single species with both α- and β-HSDHs or through two species containing α-HSDH and β-HSDH, respectively (43). For instance, 3α/β-HSDH can epimerize DCA to 3-oxo/iso-DCA in Ruminococcus gnavus (44). Moreover, allo-BAs are generated by 5-β/α-epimerization (45). Esterified BAs in fecal samples of rodents and humans have been discovered by numerous reports. In addition, esterified BAs may account for more than 25% of total fecal BAs (46).
2.3 BAs and intestinal immunity
The disrupted composition, diversity, and/or functions of gut microbiome are closely associated with gut dysbiosis, which has deleterious influence on individuals through gut homeostatic imbalance and inappropriate immune activation (47–49). Although the member and abundance of intestinal microbiota have wide individual differences by taxonomic criteria, there exist relative consistent microbial patterns in the gut of IBD, such as decreased microbial diversity and relative abundance of Firmicutes, and increased Proteobacteria (50–53). Meanwhile, certain beneficial bacteria are reduced, whereas pathogens including Escherichia coli are elevated (54, 55). In the context of IBD, gut dysbiosis and BAs disturbance, especially the reduced secondary BAs, have an impact on intestinal immunity (55). IL-17 and interferon-γ (IFN-γ) are increased on account of the dysregulation of group 3 innate lymphoid cells (ILC3) and ILC1 as well as dysfunction of regulatory ILC (56). The gut microbiota from patients with IBD could decrease retinoic acid receptor related orphan receptor γt (RORγt)+ Treg cells that produce transforming growth factor-β (TGF-β) and IL-10, and elevate Th17 (T helper 17) cells with pro-inflammatory cytokines (e.g., IL-17) (57, 58). Bile acid–activated receptors (BARs), a family of nuclear and cell membrane receptors, encompass Takeda G-protein receptor 5 (TGR5), FXR, vitamin D receptor (VDR), pregnane X receptor (PXR), constitutive androstane receptor (CAR), sphingosine 1-phosphate receptor 2, liver-X-receptor α/β, and M2/3 muscarinic receptors (59). Deficiency or inactivation of FXR or TGR5 in macrophages and dendritic cells (DCs) augments the production of pro-inflammatory cytokines. In FXR−/− mice, infiltration of macrophages was enhanced. In addition, macrophages isolated from TNBS-treated FXR−/− mice rather than wild-type mice exhibited higher released inflammatory cytokines. The activation of TGR5 in lipopolysaccharide (LPS)–treated macrophages by DCA, LCA, and tauro-LCA was able to inhibit the generation of IL-12 and TNF-α. Simultaneously, it increased the ratio of IL-10/IL-12, indicating the phenotypic transformation of macrophages into anti-inflammatory forms (60). Recruitment of classically activated macrophages and intestinal inflammation was increased in TGR5−/− mice with colitis. On the contrary, TGR5 activation by agonist could reverse intestinal inflammation through decreasing the trafficking of monocytes from blood to gut (61).
BARs are discovered in intestinal epithelial cells, intestinal muscle and neurons, hepatocytes, biliary cells, liver sinusoidal cells, liver and intestinal endothelial cells, monocytes/macrophages cells, DCs, natural killer (NK) and NKT cells, ILC, Th1, and Th17 cells (62). CDCA (CA in mice) is recognized as the most potent FXR ligand in humans, followed by DCA, LCA, and CA, whereas secondary BAs, particularly LCA and DCA, are preferential ligands for the TGR5, whose other ligands are CDCA, UDCA, and CA (59). VDR is activated by LCA and its metabolites (isoallo-LCA and 3-oxo-LCA). Furthermore, isoallo-LCA and 3-oxo-LCA can act on RORγt as inverse agonists in DCs and T cells (63). We hereafter briefly summarized the main functions of BARs activation in different immune cells. In monocyte/macrophages cells, the activation of FXR reduces the generation of TNF-α and IL-1β under inflammatory conditions. TGR5 activation shifts intestinal macrophages that are treated with LPS from M1 to M2; decreases the expression of TNF-α, IL-1β, IFN-γ, IL-6, and IL-12; and elevates the level of IL-10. In addition, VDR activation inhibits the release of IL-1, IL-6, and TNF-α but enhances IL-10 production (61, 64, 65). As far as DCs, FXR activation inhibits the differentiation, activation, and maturation of intestinal DCs and downregulates TNF-α expression. In addition, as stated previously, the activation of TGR5 in DCs reduces the production of IL-12 and TNF-α. Suppression of DCs differentiation and maturation by activating VDR has been demonstrated (66–68). In ILC3, RORγt is required for its development and function (69, 70). In NKT cells, the activation of FXR could repress the production of IFN-γ and osteopontin. TGR5 activation redirects the NKT cells polarization toward NKT10, subsequently promoting the secretion of IL-10 whereas decreasing the TNF-α and IFN-γ (71, 72). Nonetheless, the effect of FXR/TGR5 activation in NKT cells has been investigated only in the liver. These results may indicate a similar effect of them on intestinal NKT cells, which needs to be further explored. As for T cells, by directly binding to the RORγt in Th17 cell, 3-oxo-LCA and isoLCA inhibit Th17 differentiation (73). Recently, LCA 3-sulfate, a synthesized sulfated product of LCA, suppresses Th17 cell differentiation through binding to RORγt (74). IsoalloLCA can increase the differentiation of Treg cells (75). IsoDCA acts on FXR in DCs and then results in the promotion of RORγt+FOXP3+ Treg cells (76). Beyond this, TGR5 agonism promotes T-cell differentiation toward the Treg phenotype (61). VDR activation facilitates a shift from Th1 to Th2 phenotype, inhibits proliferation of T cell and Th17 differentiation, and promotes the induction of Treg cells (77–81). Therefore, the anti-inflammatory and immunomodulatory effects of BAs-BARs-immune cell axis play essential roles in health and disease conditions.
3 Effectiveness of probiotics and their possible roles in gut microbiota-BAs axis
In accordance with the World Health Organization, probiotic bacteria are defined as live microorganisms that confer a health benefit to the host when administered in adequate amounts. Probiotic strains must have four criteria for selection and use in foods or dietary supplements. In brief, first, they must be sufficiently characterized. Second, safe use is an essential requirement. In addition, at least one human clinical trial can support their positive effectiveness by using probiotics. Last but not the least, it must be alive at an efficacious dose in the product during the quality guarantee period (82). The majority of probiotics used in research or commercial development are from limited bacteria, which mainly contain Bifidobacterium spp. and Lactobacillus spp. Currently, other probiotics, such as Saccharomyces, E. coli, Bacillus spp., Weissella spp., and Enterococci are available in the marketplace (83).
3.1 Effectiveness of probiotics on IBD/CAC
3.1.1 Probiotic effectiveness in clinical trials among IBD patients
As of now, some clinical trials have been presented the positive effects of probiotics on patients with IBD. Next, we listed some clinical trials and the altered fecal bacteria that might participate in the metabolism of BAs. In UC patients, administration of probiotic yogurt, which included Lactobacillus acidophilus La-5 and Bifidobacterium BB-12, led to increased numbers of Bifidobacterium, Lactobacillus and Bacteroides in feces (84). These three bacteria (genus level) might regulate the gut microbiota-BAs axis due to their BSH activities or other functions. BIFICO (Bifidobacterium longum, Lactobacillus acidophilus, and Enterococcus faecalis) prevented flare-ups of chronic UC by decreasing IL-1β and TNF-α levels, increasing IL-10 expression, and blocking Nuclear Factor kappa-light-chain-enhancer of activated B cells (NF-κB) activation. Moreover, it elevated fecal Lactobacillus and Bifidobacterium (85). Similar fecal bacteria variations were also found in another UC trials with BIFICO (86). Furthermore, mesalazine combined with BIFICO treatment reduced the adverse reactions (87). In a 2-year clinical study, mesalazine plus a probiotic mix improved the clinical response better than controls (88). Tsuda et al. showed that by making use of BIO-THREE (Streptococcus faecalis T-110, Clostridium butyricum TO-A, and Bacillus mesentericus TO-A) ameliorated endoscopic findings, clinical symptoms of UC patients, and increased fecal Bifidobacterium (89). In addition to these, many other studies also exhibited the effectiveness of the same or diverse probiotics on inducing remission or preventing recurrence of IBD patients (90–97). Moreover, application of Escherichia coli Nissle 1917 (EcN) by rectum was a well replacement treatment for moderate distal UC (98). Among CD patients, Saccharomyces boulardii was useful for maintenance treatment according to a clinical trial in 2000 (99).
Nevertheless, there exist some discrepant results of using probiotics in IBD. For example, Bifidobacterium breve fermented milk had no effect on relieving relapse in UC (100). In a small randomized double-blind placebo-controlled trial of UC, Lactobacillus acidophilus La-5 and Bifidobacterium animalis subsp. lactis BB-12 showed no significant clinical benefit (101). In another clinical trial, EcN did not improve the active UC followed by 7 weeks administration after ciprofloxacin or placebo intervention (102). With regard to CD, Bourreille et al. found that after steroid or salicylate treatment, Saccharomyces boulardii had no beneficial effect on CD patients in remission (103). Van Gossum et al. did not find that Lactobacillus johnsonii LA1 could prevent early endoscopic recurrence of CD patients after ileo-caecal resection (104). The reasons for why the diverse outcomes appear are intricate. It may attribute to strain-specific, dose, utility time, number of patients, individual differences, disease severity, microbiota composition, genetic aspects, inflammatory status, and so forth.
Hence, from the valid clinical trials of IBD, we can discover that the main elevated fecal bacteria (genus level) that may involve in modulating gut microbiota-BAs axis are Lactobacillus and Bifidobacterium. Only one clinical trial found the relative increased Bacteroides. Moreover, the outcomes and other effects including immune response of these clinical trials were also presented in Table 1.
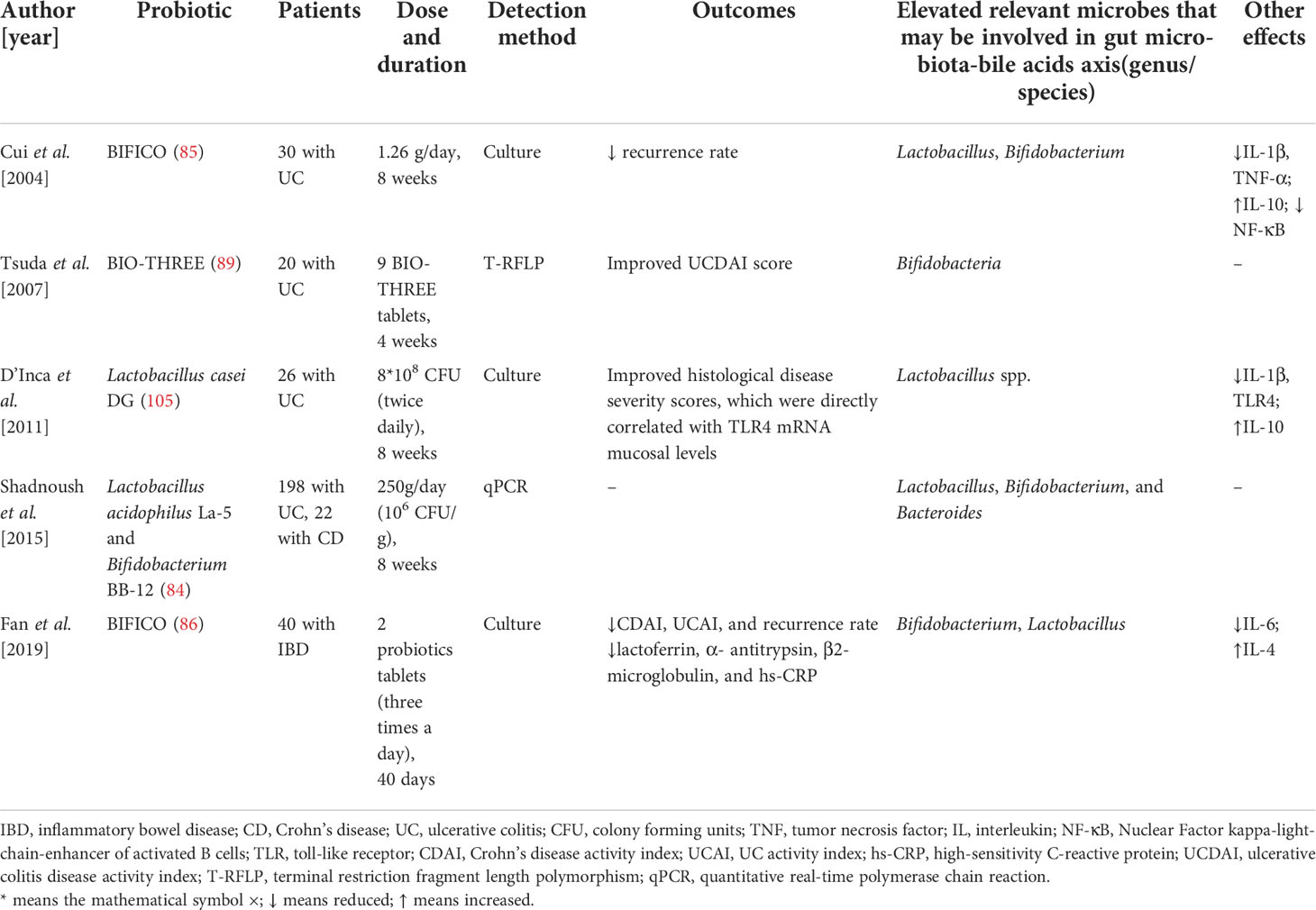
Table 1 Probiotics modulate immune response and the gut bacteria that may be involved in gut microbiota-bile acids axis in patients with inflammatory bowel disease.
3.1.2 Probiotic effectiveness in animal models with colitis
White and co-workers demonstrated that VisbiomeTM (Lactobacillus plantarum, Streptococcus thermophilus, Bifidobacterium breve, Lactobacillus paracasei, Lactobacillus delbrueckii subsp. bulgaricus, Lactobacillus acidophilus, Bifidobacterium longum, and Bifidobacterium infantis) plus prednisone therapy could accelerate clinical remission, upregulate the expression of E-cadherin, occludin, and zonulin but did not improve the histopathologic inflammation than prednisone alone group in canine with idiopathic IBD. In addition, the combination therapy increased fecal Bifidobacterium (106). In the TNBS-induced colitis model, Lactobacillus plantarum LC27 and Bifidobacterium longum LC67 inhibited Proteobacteria to Bacteroidetes ratio and NF-κB activation, elevated fecal Lactobacilli, Bifidobacteria and tight junction protein expression, simultaneously restored Th17/Treg balance (107). Another study using Lactobacillus acidophilus and Clostridium butyricum acquired similar results of elevated fecal Lactobacillus and Bifidobacterium. Furthermore, it alleviated reduced colon length and body weight, suppressed disease activity indices (DAI), and exerted an anti-inflammatory effect (108). VSL#3 (Lactobacillus paracasei, L. plantarum, L. acidophilus, L. delbrueckii subspecies bulgaricus, Bifidobacterium longum, B. breve, B. infantis, and Streptococcus thermophilus) lowered macroscopic and microscopic damage, reduced serum cytokine levels, but not dampening M1 macrophages. Of note, fecal Parabacteroides, Clostridium were increased in mice being given VSL#3 than in the TNBS colitis mice (109).
Within the DSS-induced colitis model, Lactobacillus rhamnosus GG (LGG) ameliorated decreased body weight and clinical features and simultaneously enhanced fecal Lactobacillus and Bacteroides (110). Three studies revealed the positive effect of Lactobacillus plantarum on DSS-induced colitis, Lactobacillus plantarum GIM17 and Lactobacillus plantarum-12 elevated the fecal Lactobacillus, whereas the Lactobacillus plantarum L15 could also increase other bacteria, such as Bifidobacterium and Bacteroides (111–113). LGG and Lactobacillus plantarum Q7–derived extracellular vesicles could also enhance the fecal Bifidobacterium_animalis or Lactobacillus and Bifidobacterium, respectively (114, 115). Jang et al. and Zhang et al. discovered that Lactobacillus fermentum KBL375 or Lactobacillus casei Zhang was capable of augmenting species levels of Lactobacillus in feces (116, 117). Lactobacillus M2S01 recovered decreased body weight and incremental DAI, promoted anti-inflammatory cytokines expression, and elevated fecal Bifidobacterium (118). Higher Lactobacilli, Bifidobacteria, or Bacteroides in feces were detected after the administration of Lactobacillus salivarius CPN60 or Lactobacillus brevis, respectively (111, 119). As for Bifidobacterium longum CCFM681, Chen et al. demonstrated the protective roles of it in mice with DSS-induced colitis. It significantly increased mucin 2, goblet cells, zonula occludens-1 (ZO-1), claudin-3 and α-catenin1, suppressed Toll-like receptor 4/NF-κB pathway and related pro-inflammatory cytokines. Moreover, it promoted the growth of Lactobacillus and Bifidobacterium in feces (120). The authors also discovered that Bifidobacterium pseudocatenulatum MY40C and CCFM680 were able to elevate fecal Lactobacillus as well (121). Oral delivery of Bacillus cereus or Bacillus subtilis alleviated DSS-induced colitis through anti-inflammation, protecting intestinal integrity, improving intestinal barrier function, and reshaping microbial composition, which comprised increased fecal Lactobacillus, Eubacterium, Bacillus, and Bacteroides (122–125). Rodriguez-Nogales et al. found that Saccharomyces boulardii CNCMI-745 improved the colitis-associated gut dysbiosis, including more Lactobacillus, Bifidobacterium in feces (126). Saccharomyces cerevisiae BR14 merely increased the fecal Lactobacillus (127), whereas an engineered strain reconstructed from Saccharomyces cerevisiae BY4741 could also enhance fecal Bacteroides apart from Lactobacillus in DSS-induced colitis (128). The aforementioned research revealed the potential functions and part mechanisms of single probiotic on animals with DSS-induced colitis. Noteworthy is that they are all involved in the alterations of fecal microbiota, which may modulate gut microbiota-BAs axis. Furthermore, numerous other researchers investigated the effectiveness of multi-strain or combined probiotics on the DSS model. Lactobacillus rhamnosus R0011 and Lactobacillus helveticus R0052 administration resulted in diminished colon disease and an increase in fecal Bacillus and Lactobacillus (129). Rodriguez-Nogales et al. uncovered the functional roles of Lactobacillus fermentum CECT5716 and Lactobacillus salivarius CECT5713 on DSS mouse colitis. Both probiotics contributed to anti-inflammatory effects, regulating immune response, improving microRNA (miR)-155 and miR-223 expression, and increasing fecal Parabacteroides, Bacillus (130). Lactobacillus sakei 07 and Bifidobacterium bifidum B10 combination decreased colonic IL-6, TNF-α, and intestinal permeability and increased intestinal flora biodiversity and the level of fecal Bifidobacterium in the colitis model (131). Quadruple probiotics were also showed benefit influences on colitis mice induced by DSS. Bifidobacterium infantis GMCC0460.1, Lactobacillus acidophilus GMCC0460.2, Enterococcus faecalis GMCC0460.3 and Bacillus cereus GMCC0460.4 repaired multi-barriers in the inflamed gut and enhanced Lactobacillus, Bifidobacterium, and Bacteroides in feces (132). Wang et al. uncovered that supplement of Lactobacillus reuteri RAM0101, Bacillus coagulans RAM1202, Bifidobacterium longum RAM0216, and Clostridium butyricum CICC6197 increased intestinal barrier function, IL-10 expression and fecal Bifidobacterium, Blautia, Lactobacillus, and Bacillus coagulans (133). Analogously, there exist some other new or potential probiotics, such as Faecalibacterium prausnitzii, Pediococcus pentosaceus, Ligilactobacillus salivarius, Lactiplantibacillus plantarum, and Akkermansia muciniphila have been certified effectively in DiNitroBenzene Sulfonic/DSS-induced colitis model. Lactobacillus, Bacteroides, Bifidobacterium, Parabacteroides, and Eubacterium_fissicatena_group were increased in feces after using these probiotics (134–138).
In short, the probiotics mentioned above bring forth favorable therapeutic effects on animal models with chemical-induced colitis. Probiotics may regulate gut microbiota-BAs axis by elevating the fecal concentration of Lactobacillus, Bifidobacterium, Bacteroides, Parabacteroides, Clostridium, Blautia, Bacillus coagulans, Eubacterium or Eubacterium_fissicatena_group at genus or species levels. Moreover, other effects involved in immune response were summarized in Table 2 as well.
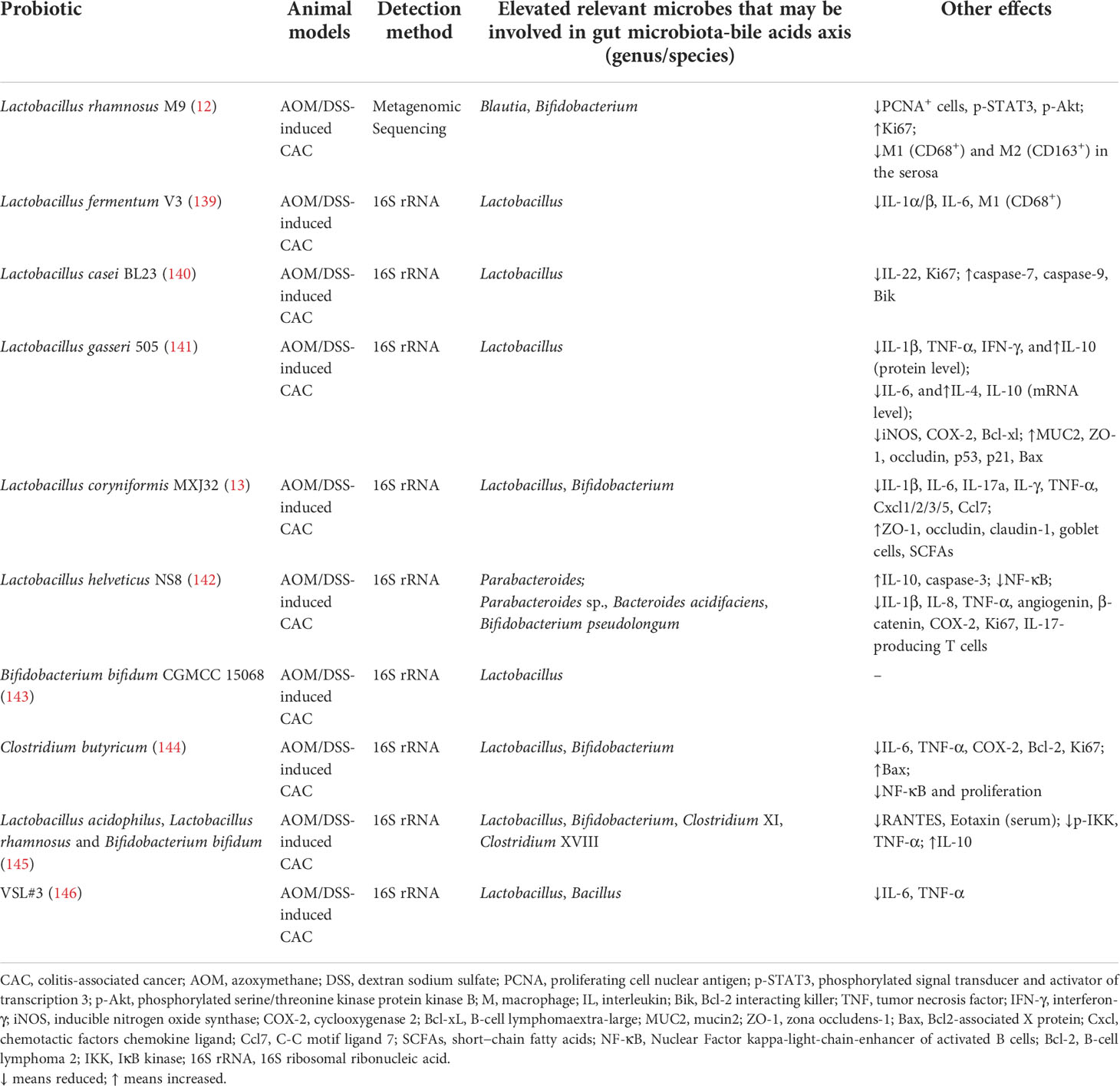
Table 2 Probiotics modulate immune response and the gut bacteria that may be involved in gut microbiota-bile acids axis in animal models with colitis-associated cancer.
3.1.3 Probiotic effectiveness in AOM/DSS induced CAC
Xu and co-workers indicated that Lactobacillus rhamnosus M9 suppressed the increased number and growth of colon tumors and expression of phosphorylated-signal transducer and activator of transcription 3 and phosphorylated-protein kinase B, ameliorated inflammatory damage and gut fibrosis in azoxymethane (AOM)/DSS-induced CAC. Furthermore, it changed fecal microbiota, which included elevated Blautia and Bifidobacterium at genus level (12). Lactobacillus fermentum V3 markedly inhibited colonic tumor formation and pro-inflammatory cytokines, accompanied by increased fecal Lactobacillus (139). The same fecal bacteria alteration that might participate in gut microbiota-BAs axis was also observed in AOM/DSS-induced CAC mice by using Lactobacillus casei BL23 or Lactobacillus gasseri 505. Other than this function, Lactobacillus casei BL23 reduced proliferation and histological scores, downregulated IL-22 cytokine, and upregulated caspase-7 and caspase-9 (140, 141). Wang et al. revealed that Lactobacillus coryniformis MXJ32 could significantly suppress the total number and average diameter of tumors, reinforce the expression of ZO-1, Claudin-1, Occludin, and recover the lesion of goblet cells. In addition, it lowered the expression of IL-1β, TNF-α, IL-6, IL-17a, C-C motif ligand 7, chemotactic factors chemokine ligand 5 (Cxcl5), Cxcl3, Cxcl2 and Cxcl1, increased the abundance of Lactobacillus, Bifidobacterium in the stool (13). Treatment with Lactobacillus helveticus NS8 overtly reduced the degree of hyperplasia and tumor number, suppressed enterocytes proliferation at the early stage of CAC, while it increased apoptosis level. Furthermore, NS8 significantly inhibited NF-κB activation, IL-17–producing T cells, and upregulated IL-10. Interestingly, at the genus level, fecal Parabacteroides was augmented after 80 days injection of AOM, while at the species level, Lactobacillus sp., Bacteroides acidifaciens or Parabacteroides sp., Bifidobacterium pseudolongum were increased at 14 or 80 days behind AOM injection (142). Wang and co-authors discovered the conducive role of Bifidobacterium bifidum CGMCC 15068 on the CAC mouse model. This probiotic was capable of attenuating tumorigenesis and shifting gut microbiota composition, which comprised a higher quantity of Lactobacillus in feces. In addition, the authors found the existence of differentially abundant metabolites between AOM/probiotic and AOM groups, which indicated that Bifidobacterium bifidum CGMCC 15068 was involved in multiple metabolic pathways, such as citrate cycle, galactose metabolism, butyrate metabolism, and so on (143). Clostridium butyricum, a relative new probiotic, has been identified to inhibit the incidence and size of tumors in CAC mice, decrease IL-6, TNF-α, cyclooxygenase-2, phosphorylation of NF-κB and B-cell lymphoma-2. Moreover, it aggrandized the expression of Bcl-2-associated X and fecal Lactobacillus, Bifidobacterium (144). Triple or multiple probiotics intervention still exhibited a satisfactory effect on the chemical-induced CAC model. Lactobacillus acidophilus, Lactobacillus rhamnosus, and Bifidobacterium bifidum mixture restrained the size and number of tumors, reduced colon inflammatory index, serum chemokines RANTES, eotaxin, phospho-IκB kinase, and TNF-α, whereas it enhanced IL-10 expression and fecal Lactobacillus, Bifidobacterium, Clostridium XI, and Clostridium XVIII (145). By making use of VSL#3 in AOM/DSS-induced CAC mice, Wang et al. revealed that this probiotic alone ameliorated oncogenesis and tumor load, and reduced the level of IL-6 and TNF-α in colon tissue. Furthermore, it increased fecal Bacillus and Lactobacillus as well, but the latter had no statistical significance (146).
As mentioned above, probiotics can improve the AOM/DSS-induced CAC model in a variety of ways, one of which is altering gut microbiota composition. In conclusion, the increased fecal bacteria that may involve in gut microbiota-BAs axis comprise Blautia, Bifidobacterium, Lactobacillus, Parabacteroides, Bacillus, Parabacteroides sp., Bacteroides acidifaciens, Bifidobacterium pseudolongum, Clostridium XI and Clostridium XVIII at genus/species level. In addition, other effects containing immune response after treatment with probiotics in CAC model were also showed in Table 3.

Table 3 Probiotics modulate immune response and the gut bacteria that may be involved in gut microbiota-bile acids axis in animal models with inflammatory bowel disease.
3.2 Possible mechanisms of probiotics on regulating gut microbiota-BAs axis in IBD/CAC
In the VSL#3-treated mice, the conjugated/unconjugated BAs ratio decreased, which was relevant to elevated fecal BSH expression and activity compared to the vehicle group. BSH activity was much higher after oral take of Lactobacillus acidophilus or Bifidobacterium infantis than Streptococcus thermophilus (149). Recently, a dual probiotics system including Lactobacillus delbrueckii subsp. bulgaricus and LGG was fabricated, and LGG could increase the abundance of BSH-containing gut bacteria (150). Recently, BSH-expressing engineered native E. coli could reduce primary conjugated fecal BAs and increase primary deconjugated BAs, consistent with increased BSH activity (151). BioPersist, a bioengineered live biotherapeutic product that enhanced persistence in the colitic mice could reduce the accumulation of BAs, suggesting that BioPersist facilitated efficient BA recycling via enterohepatic circulation or improved homeostasis of gut microbial growth (152). As a corollary, it is conceivable that probiotics with BSH can regulate gut microbiota-BAs axis through promoting deconjugation. Simultaneously, they decrease the accumulation of BAs in IBD. Sato and co-authors found that intestinal microbiota participated in 7α-dehydroxylation was restored in distal UC patients after 4 weeks of Clostridium butyricum intervention (153). Therefore, it can be inferred that probiotics may modulate gut microbiota-BAs axis by increasing intestinal bacteria involved in deconjugation and 7α-dehydroxylation over the course of BAs biotransformation. The possible elevated gut bacteria at the genus/species level after administration of diverse probiotics were discussed above. Collectively, after using probiotics in IBD and CAC, Lactobacillus, Bifidobacterium, Bacteroides, Parabacteroides, Clostridium, Blautia, Bacillus coagulans, Eubacterium or Eubacterium_fissicatena_group were increased in IBD, and Blautia, Bifidobacterium, Lactobacillus, Parabacteroides, Bacillus, Parabacteroides sp., Bacteroides acidifaciens, Bifidobacterium pseudolongum, Clostridium XI and XVIII were enhanced in CAC. Moreover, the altered BAs profile, especially the likely elevated secondary BAs, were able to bring forth anti-inflammatory and immunomodulatory effects through activating BARs on intestinal immune cells, as reviewed before. In addition to the altered gut microbes after administration of probiotics in these studies, other effects including beneficial immune response were also summarized in the Tables 1–3. Therefore, it could be speculated that the possible altered BA profiles through the administration of probiotics might ameliorate IBD/CAC via BAs-BARs–immune cell axis. However, whether probiotics alone and/or their elevated gut bacteria exert beneficial impacts on gut microbiota-BAs axis in the state of IBD or CAC need further explorations of clinical or animal studies. Also, the exact mechanisms are still not elaborated, which require to be appraised in depth. Lactobacilli and Bifidobacteria are representatives of the main probiotics, spontaneously amass primary, and possibly secondary unconjugated BAs or following intracellular BAs deconjugation in their cytoplasm (154, 155). Sanchez hypothesized that probiotic cytoplasm-sequestered primary BAs will escape being transformed into secondary BAs by other microorganisms. These unconverted primary BAs would be removed with the feces (156). Hence, this BAaccumulation mechanism may make a difference to chronic inflammation (such as IBD) and carcinogenesis (such as CAC).
In short, probiotics might involve in gut microbiota-BAs axis by several potential mechanisms, which we mainly focused on the increased gut bacteria containing BSH or BAI at genus/species level in IBD and CAC. We could speculate that probiotics treatment was able to enhance the activity of deconjugation and 7α-dehydroxylation by increasing the BSH- and BAI-containing bacteria in gut of IBD/CAC. Subsequently, the BAs profile was changed, which might include decreased conjugated primary BAs and increased unconjugated primary BAs and secondary BAs. Moreover, probiotics were possible to contribute to the absorption of these altered BAs. Afterwards, the absorbed BAs were likely to generate beneficial effects through activating BARs on intestinal immune cells, such as monocytes/macrophages cells, DCs, NKT cells, ILC and T cells. In addition, probiotic cytoplasm-sequestered primary BAs will escape being transformed into secondary BAs and then be excreted with the feces. Ultimately, the IBD may be improved by these mechanisms after using probiotics (Figure 1).
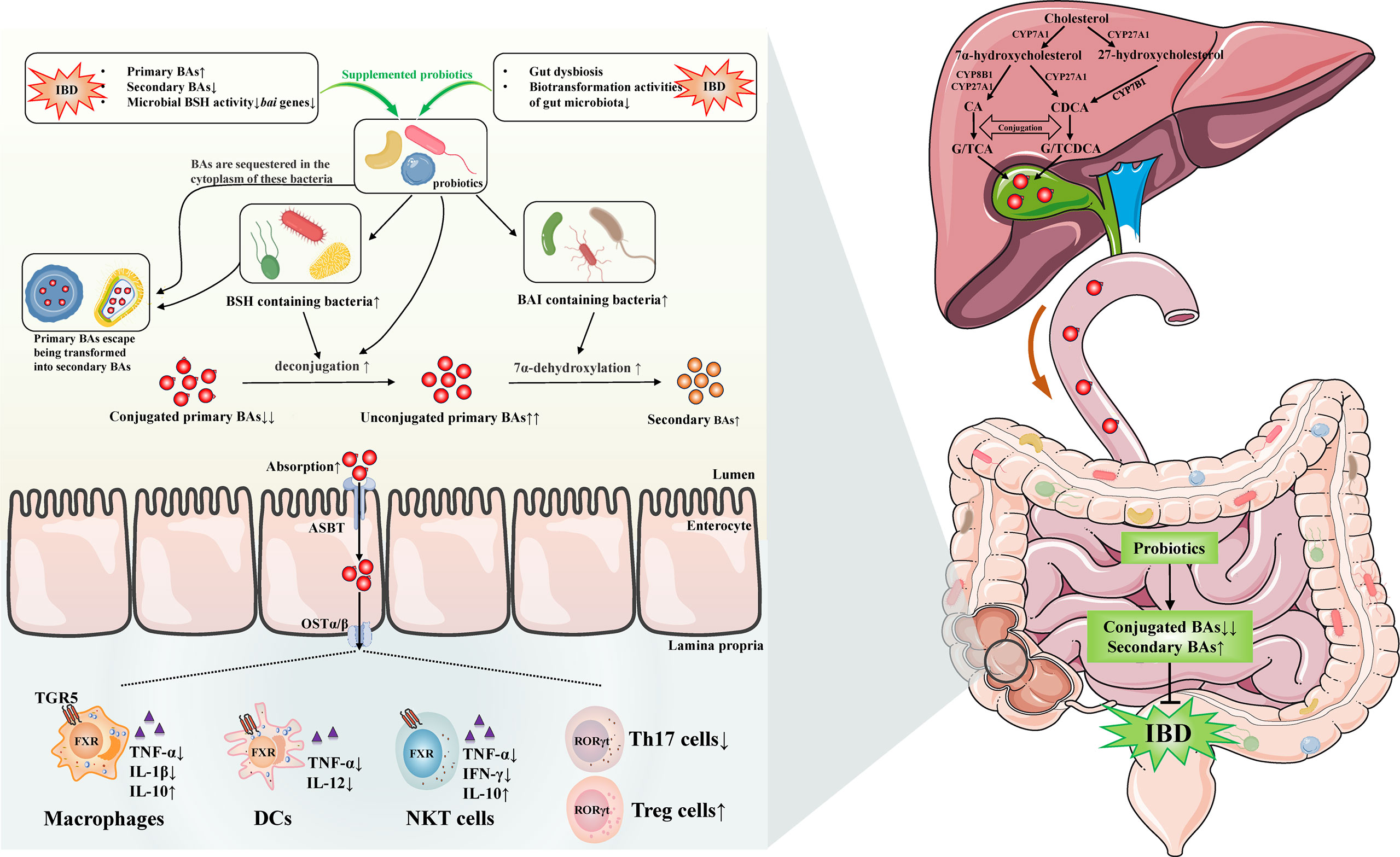
Figure 1 Possible mechanisms of probiotics on regulating gut microbiota-bile acids axis and related gut immunity in inflammatory bowel disease. Gut dysbiosis and BAs dysmetabolism existed in IBD has been discovered by many studies, the levels of secondary BAs are lower, primary BAs are elevated because of the impairment of microbiota biotransformation activities, and the microbial BSH activity and bai genes are decreased. The administration of probiotics are likely to increase the BSH and BAI containing bacteria in IBD. Therefore, the conjugated/unconjugated primary BAs ratio decreases and secondary BAs may increase by the elevated activities of deconjugation and 7α-dehydroxylation. Furthermore, probiotics may reduce the accumulation of BAs by facilitating the absorption of them, which are possible to exert beneficial effects by activating BARs on intestinal immune cells, such as monocytes/macrophages cells, DCs, NKT cells, ILC, and T cells. Beyond these, probiotic cytoplasm-sequestered primary BAs may escape being transformed into secondary BAs. These unconverted primary BAs are likely to be removed with the feces. Eventually, the IBD may be improved by these distinct mechanisms after using probiotics. IBD, inflammatory bowel disease; BAs, bile acids; TGR5, Takeda G-protein receptor 5; FXR, farnesoid X receptor; RORγt, retinoic acid receptor related orphan receptor γt; DCs, dendritic cells; NKT, natural killer T; ILC, innate lymphoid cells; Th17, T helper 17; Treg, regulatory T; TNF, tumor necrosis factor; IL, interleukin; IFN-γ, interferon-γ; CA, cholic acid; CDCA, chenodeoxycholic acid; GCA, glycocholic acid; TCA, taurocholic acid; GCDCA, glycochenodeoxycholic acid; TCDCA, taurochenodeoxycholic acid; CYP7A1, cholesterol-7α-hydroxylase; CYP8B1, sterol-12α-hydroxylase; CYP27A1, mitochondrial sterol-27-hydroxylase; CYP7B1, oxysterol 7α-hydroxylase; BSH, bile salt hydrolase; BAI, bile acid-inducible enzymes; ASBT, apical sodium-dependent bile acid transporter; OSTα/β, organic solute transporter subunit α/β.
4 Conclusions
A large number of studies have revealed the interaction between gut microbiota and BAs. Among them, the intestinal microbiota is involved in BAs synthesis and metabolism, thus influencing BAs composition. Both of which can facilitate the initiation and development of IBD and CAC. Additionally, growing evidence has indicated that certain probiotics exhibit beneficial effects on UC, CAC, and other diseases through multiple mechanisms. In the present review, the possible functional roles of probiotics on gut microbiota-BAs axis were discussed. We laid emphasis on elucidating the alterations of gut bacteria that may involve in gut microbiota-BAs axis in IBD/CAC patients or animal models treated with probiotics. These elevated bacteria at the genus/species level may include Lactobacillus, Bifidobacterium, Bacteroides, Parabacteroides, Clostridium, Blautia, Bacillus coagulans, Eubacterium, and Eubacterium_fissicatena_group. Some mentioned bacteria that may carry some functional genes seem to have the potential to become probiotic for the moment, but whether they can be used as probiotics still needs numerous and rigorous animal researches and clinical trials according to the four criteria for using as probiotics. Moreover, the activation of BARs, including FXR, TGR5, VDR in monocytes/macrophages cells, DCs, NKT cells, ILC, Th17, and Treg cells, exerts anti-inflammatory and immunomodulatory effects, which may partially explain the positive effects of probiotics on IBD. According to American Gastroenterological Association Institute’s advice, probiotics may be considered for the treatment of functional symptoms in IBD. The gut dysbiosis existed in IBD may be improved by using probiotics for a period of time. Due to the biological activity and characteristics of probiotics, there are potential risks in the treatment of IBD patients with probiotics, such as bacteremia, transfer of antibiotic resistance, and adverse reactions. Furthermore, the probiotics used for adjunctive therapy may not be effective in some patients on account of individual difference or strain specificity, so it should be discontinued at this time. Therefore, the duration of probiotics should be determined by different therapeutic goals, effects, and actual conditions. In the future, more clinical and animal studies are necessary to explore the direct changes of BAs and specific mechanisms by administration of probiotics in IBD and CAC.
Author contributions
LL, YG and HC contributed conception and prepared the manuscript. LL searched the literature and wrote the manuscript. XW, YS and YG prepared the figure. LL, TL, RX and HC revised the manuscript. BW and HC organized the framework. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the grants (81970477, 82070545 and 82100574) from the National Natural Science Foundation of China, the Key Project of Science and Technology Pillar Program of Tianjin (20YFZCSY00020).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
IBD, inflammatory bowel disease; BAs, bile acids; TGR5, Takeda G-protein receptor 5; FXR, farnesoid X receptor; RORγt, retinoic acid receptor related orphan receptor γt; DCs, dendritic cells; NKT, natural killer T; ILC, innate lymphoid cells; Th17, T helper 17; Treg, regulatory T; TNF, tumor necrosis factor; IL, interleukin; IFN-γ, interferon-γ; CA, cholic acid; CDCA, chenodeoxycholic acid; GCA, glycocholic acid; TCA, taurocholic acid; GCDCA, glycochenodeoxycholic acid; TCDCA, taurochenodeoxycholic acid; CYP7A1, cholesterol-7α-hydroxylase; CYP8B1, sterol-12α-hydroxylase; CYP27A1, mitochondrial sterol-27-hydroxylase; CYP7B1, oxysterol 7α-hydroxylase; BSH, bile salt hydrolase; BAI, bile acid-inducible enzymes; ASBT, apical sodium-dependent bile acid transporter; OSTα/β, organic solute transporter subunit α/β.
References
1. Mulder DJ, Noble AJ, Justinich CJ, Duffin JM. A tale of two diseases: The history of inflammatory bowel disease. J Crohns Colitis (2014) 8(5):341–8. doi: 10.1016/j.crohns.2013.09.009
2. Perri MR, Romano C, Marrelli M, Zicarelli L, Toma CC, Basta D, et al. Beneficial role of fruits, their juices, and freeze-dried powders on inflammatory bowel disease and related dysbiosis. Plants (Basel) (2021) 11(1):4. doi: 10.3390/plants11010004
3. Jakubczyk D, Leszczynska K, Gorska S. The effectiveness of probiotics in the treatment of inflammatory bowel disease (Ibd)-a critical review. Nutrients (2020) 12(7):1973. doi: 10.3390/nu12071973
4. Joyce SA, MacSharry J, Casey PG, Kinsella M, Murphy EF, Shanahan F, et al. Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut. Proc Natl Acad Sci U.S.A. (2014) 111(20):7421–6. doi: 10.1073/pnas.1323599111
5. Duboc H, Rajca S, Rainteau D, Benarous D, Maubert MA, Quervain E, et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut (2013) 62(4):531–9. doi: 10.1136/gutjnl-2012-302578
6. Ogilvie LA, Jones BV. Dysbiosis modulates capacity for bile acid modification in the gut microbiomes of patients with inflammatory bowel disease: A mechanism and marker of disease? Gut (2012) 61(11):1642–3. doi: 10.1136/gutjnl-2012-302137
7. Cai J, Sun L, Gonzalez FJ. Gut microbiota-derived bile acids in intestinal immunity, inflammation, and tumorigenesis. Cell Host Microbe (2022) 30(3):289–300. doi: 10.1016/j.chom.2022.02.004
8. Lu Q, Yang MF, Liang YJ, Xu J, Xu HM, Nie YQ, et al. Immunology of inflammatory bowel disease: Molecular mechanisms and therapeutics. J Inflammation Res (2022) 15:1825–44. doi: 10.2147/JIR.S353038
9. Sartor RB, Wu GD. Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology (2017) 152(2):327–39 e4. doi: 10.1053/j.gastro.2016.10.012
10. Bernstein CN, Blanchard JF, Kliewer E, Wajda A. Cancer risk in patients with inflammatory bowel disease: A population-based study. Cancer (2001) 91(4):854–62. doi: 10.1002/1097-0142(20010215)91:4<854::aid-cncr1073>3.0.co;2-z
11. Liu L, Yang M, Dong W, Liu T, Song X, Gu Y, et al. Gut dysbiosis and abnormal bile acid metabolism in colitis-associated cancer. Gastroenterol Res Pract (2021) 2021:6645970. doi: 10.1155/2021/6645970
12. Xu H, Hiraishi K, Kurahara LH, Nakano-Narusawa Y, Li X, Hu Y, et al. Inhibitory effects of breast milk-derived lactobacillus rhamnosus probio-M9 on colitis-associated carcinogenesis by restoration of the gut microbiota in a mouse model. Nutrients (2021) 13(4):1143. doi: 10.3390/nu13041143
13. Wang T, Zhang L, Wang P, Liu Y, Wang G, Shan Y, et al. Lactobacillus coryniformis Mxj32 administration ameliorates Azoxymethane/Dextran sulfate sodium-induced colitis-associated colorectal cancer Via reshaping intestinal microenvironment and alleviating inflammatory response. Eur J Nutr (2021) 61(1):85–99. doi: 10.1007/s00394-021-02627-8
14. Jia W, Xie G, Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol (2018) 15(2):111–28. doi: 10.1038/nrgastro.2017.119
15. Chiang JY. Bile acid metabolism and signaling. Compr Physiol (2013) 3(3):1191–212. doi: 10.1002/cphy.c120023
16. Gu Y, Li L, Yang M, Liu T, Song X, Qin X, et al. Bile acid-gut microbiota crosstalk in irritable bowel syndrome. Crit Rev Microbiol (2022) 1–20. doi: 10.1080/1040841X.2022.2058353
17. Vantrappen G, Ghoos Y, Rutgeerts P, Janssens J. Bile acid studies in uncomplicated crohn's disease. Gut (1977) 18(9):730–5. doi: 10.1136/gut.18.9.730
18. Heinken A, Ravcheev DA, Baldini F, Heirendt L, Fleming RMT, Thiele I. Systematic assessment of secondary bile acid metabolism in gut microbes reveals distinct metabolic capabilities in inflammatory bowel disease. Microbiome (2019) 7(1):75. doi: 10.1186/s40168-019-0689-3
19. Jahnel J, Fickert P, Hauer AC, Hogenauer C, Avian A, Trauner M. Inflammatory bowel disease alters intestinal bile acid transporter expression. Drug Metab Dispos (2014) 42(9):1423–31. doi: 10.1124/dmd.114.058065
20. Araki Y, Andoh A, Tsujikawa T, Fujiyama Y, Bamba T. Alterations in intestinal microflora, faecal bile acids and short chain fatty acids in dextran sulphate sodium-induced experimental acute colitis in rats. Eur J Gastroenterol Hepatol (2001) 13(2):107–12. doi: 10.1097/00042737-200102000-00004
21. Sinha SR, Haileselassie Y, Nguyen LP, Tropini C, Wang M, Becker LS, et al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe (2020) 27(4):659–70.e5. doi: 10.1016/j.chom.2020.01.021
22. Joyce SA, Gahan CG. Bile acid modifications at the microbe-host interface: Potential for nutraceutical and pharmaceutical interventions in host health. Annu Rev Food Sci Technol (2016) 7:313–33. doi: 10.1146/annurev-food-041715-033159
23. Midtvedt T. Microbial bile acid transformation. Am J Clin Nutr (1974) 27(11):1341–7. doi: 10.1093/ajcn/27.11.1341
24. Kriaa A, Bourgin M, Potiron A, Mkaouar H, Jablaoui A, Gerard P, et al. Microbial impact on cholesterol and bile acid metabolism: Current status and future prospects. J Lipid Res (2019) 60(2):323–32. doi: 10.1194/jlr.R088989
25. Gerard P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens (2013) 3(1):14–24. doi: 10.3390/pathogens3010014
26. Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, et al. Dietary-Fat-Induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature (2012) 487(7405):104–8. doi: 10.1038/nature11225
27. Begley M, Hill C, Gahan CG. Bile salt hydrolase activity in probiotics. Appl Environ Microbiol (2006) 72(3):1729–38. doi: 10.1128/AEM.72.3.1729-1738.2006
28. Yadav R, Singh PK, Puniya AK, Shukla P. Catalytic interactions and molecular docking of bile salt hydrolase (Bsh) from l. plantarum Rypr1 and its prebiotic utilization. Front Microbiol (2016) 7:2116. doi: 10.3389/fmicb.2016.02116
29. Xu F, Guo F, Hu XJ, Lin J. Crystal structure of bile salt hydrolase from lactobacillus salivarius. Acta Crystallogr F Struct Biol Commun (2016) 72(Pt 5):376–81. doi: 10.1107/S2053230X16005707
30. Lambert JM, Siezen RJ, de Vos WM, Kleerebezem M. Improved annotation of conjugated bile acid hydrolase superfamily members in gram-positive bacteria. Microbiol (Reading) (2008) 154(Pt 8):2492–500. doi: 10.1099/mic.0.2008/016808-0
31. Rossocha M, Schultz-Heienbrok R, von Moeller H, Coleman JP, Saenger W. Conjugated bile acid hydrolase is a tetrameric n-terminal thiol hydrolase with specific recognition of its cholyl but not of its tauryl product. Biochemistry (2005) 44(15):5739–48. doi: 10.1021/bi0473206
32. Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U.S.A. (2008) 105(36):13580–5. doi: 10.1073/pnas.0804437105
33. Song Z, Cai Y, Lao X, Wang X, Lin X, Cui Y, et al. Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (Bsh) genes based on worldwide human gut microbiome. Microbiome (2019) 7(1):9. doi: 10.1186/s40168-019-0628-3
34. De Smet I, Van Hoorde L, Vande Woestyne M, Christiaens H, Verstraete W. Significance of bile salt hydrolytic activities of lactobacilli. J Appl Bacteriol (1995) 79(3):292–301. doi: 10.1111/j.1365-2672.1995.tb03140.x
35. Costabile A, Buttarazzi I, Kolida S, Quercia S, Baldini J, Swann JR, et al. An in vivo assessment of the cholesterol-lowering efficacy of lactobacillus plantarum ecgc 13110402 in normal to mildly hypercholesterolaemic adults. PloS One (2017) 12(12):e0187964. doi: 10.1371/journal.pone.0187964
36. Costabile A, Klinder A, Fava F, Napolitano A, Fogliano V, Leonard C, et al. Whole-grain wheat breakfast cereal has a prebiotic effect on the human gut microbiota: A double-blind, placebo-controlled, crossover study. Br J Nutr (2008) 99(1):110–20. doi: 10.1017/S0007114507793923
37. Hanafi NI, Mohamed AS, Sheikh Abdul Kadir SH, Othman MHD. Overview of bile acids signaling and perspective on the signal of ursodeoxycholic acid, the most hydrophilic bile acid, in the heart. Biomolecules (2018) 8(4):159. doi: 10.3390/biom8040159
38. Labbe A, Ganopolsky JG, Martoni CJ, Prakash S, Jones ML. Bacterial bile metabolising gene abundance in crohn's, ulcerative colitis and type 2 diabetes metagenomes. PloS One (2014) 9(12):e115175. doi: 10.1371/journal.pone.0115175
39. Hamilton JP, Xie G, Raufman JP, Hogan S, Griffin TL, Packard CA, et al. Human cecal bile acids: Concentration and spectrum. Am J Physiol Gastrointest Liver Physiol (2007) 293(1):G256–63. doi: 10.1152/ajpgi.00027.2007
40. Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature (2013) 499(7456):97–101. doi: 10.1038/nature12347
41. Long SL, Gahan CGM, Joyce SA. Interactions between gut bacteria and bile in health and disease. Mol Aspects Med (2017) 56:54–65. doi: 10.1016/j.mam.2017.06.002
42. Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res (2006) 47(2):241–59. doi: 10.1194/jlr.R500013-JLR200
43. Lepercq P, Gerard P, Beguet F, Raibaud P, Grill JP, Relano P, et al. Epimerization of chenodeoxycholic acid to ursodeoxycholic acid by clostridium baratii isolated from human feces. FEMS Microbiol Lett (2004) 235(1):65–72. doi: 10.1016/j.femsle.2004.04.011
44. Devlin AS, Fischbach MA. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat Chem Biol (2015) 11(9):685–90. doi: 10.1038/nchembio.1864
45. Wahlstrom A, Sayin SI, Marschall HU, Backhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab (2016) 24(1):41–50. doi: 10.1016/j.cmet.2016.05.005
46. Ridlon JM, Harris SC, Bhowmik S, Kang DJ, Hylemon PB. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes (2016) 7(1):22–39. doi: 10.1080/19490976.2015.1127483
47. Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U.S.A. (2007) 104(34):13780–5. doi: 10.1073/pnas.0706625104
48. Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol (2012) 13(9):R79. doi: 10.1186/gb-2012-13-9-r79
49. Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis (2015) 26:26191. doi: 10.3402/mehd.v26.26191
50. Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Folsch UR, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut (2004) 53(5):685–93. doi: 10.1136/gut.2003.025403
51. Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset crohn's disease. Cell Host Microbe (2014) 15(3):382–92. doi: 10.1016/j.chom.2014.02.005
52. Hirano A, Umeno J, Okamoto Y, Shibata H, Ogura Y, Moriyama T, et al. Comparison of the microbial community structure between inflamed and non-inflamed sites in patients with ulcerative colitis. J Gastroenterol Hepatol (2018) 33(9):1590–97. doi: 10.1111/jgh.14129
53. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science (2005) 308(5728):1635–8. doi: 10.1126/science.1110591
54. Pittayanon R, Lau JT, Leontiadis GI, Tse F, Yuan Y, Surette M, et al. Differences in gut microbiota in patients with vs without inflammatory bowel diseases: A systematic review. Gastroenterology (2020) 158(4):930–46.e1. doi: 10.1053/j.gastro.2019.11.294
55. Yang M, Gu Y, Li L, Liu T, Song X, Sun Y, et al. Bile acid-gut microbiota axis in inflammatory bowel disease: From bench to bedside. Nutrients (2021) 13(9):3143. doi: 10.3390/nu13093143
56. Zeng B, Shi S, Ashworth G, Dong C, Liu J, Xing F. Ilc3 function as a double-edged sword in inflammatory bowel diseases. Cell Death Dis (2019) 10(4):315. doi: 10.1038/s41419-019-1540-2
57. Britton GJ, Contijoch EJ, Mogno I, Vennaro OH, Llewellyn SR, Ng R, et al. Microbiotas from humans with inflammatory bowel disease alter the balance of gut Th17 and rorgammat(+) regulatory T cells and exacerbate colitis in mice. Immunity (2019) 50(1):212–24.e4. doi: 10.1016/j.immuni.2018.12.015
58. Neurath MF. Targeting immune cell circuits and trafficking in inflammatory bowel disease. Nat Immunol (2019) 20(8):970–9. doi: 10.1038/s41590-019-0415-0
59. Fiorucci S, Biagioli M, Zampella A, Distrutti E. Bile acids activated receptors regulate innate immunity. Front Immunol (2018) 9:1853. doi: 10.3389/fimmu.2018.01853
60. Haselow K, Bode JG, Wammers M, Ehlting C, Keitel V, Kleinebrecht L, et al. Bile acids pka-dependently induce a switch of the il-10/Il-12 ratio and reduce proinflammatory capability of human macrophages. J Leukoc Biol (2013) 94(6):1253–64. doi: 10.1189/jlb.0812396
61. Biagioli M, Carino A, Cipriani S, Francisci D, Marchiano S, Scarpelli P, et al. The bile acid receptor Gpbar1 regulates the M1/M2 phenotype of intestinal macrophages and activation of Gpbar1 rescues mice from murine colitis. J Immunol (2017) 199(2):718–33. doi: 10.4049/jimmunol.1700183
62. Biagioli M, Marchiano S, Carino A, Di Giorgio C, Santucci L, Distrutti E, et al. Bile acids activated receptors in inflammatory bowel disease. Cells (2021) 10(6):1281. doi: 10.3390/cells10061281
63. Fiorucci S, Carino A, Baldoni M, Santucci L, Costanzi E, Graziosi L, et al. Bile acid signaling in inflammatory bowel diseases. Dig Dis Sci (2021) 66(3):674–93. doi: 10.1007/s10620-020-06715-3
64. Vavassori P, Mencarelli A, Renga B, Distrutti E, Fiorucci S. The bile acid receptor fxr is a modulator of intestinal innate immunity. J Immunol (2009) 183(10):6251–61. doi: 10.4049/jimmunol.0803978
65. Almerighi C, Sinistro A, Cavazza A, Ciaprini C, Rocchi G, Bergamini A. 1alpha,25-dihydroxyvitamin D3 inhibits Cd40l-induced pro-inflammatory and immunomodulatory activity in human monocytes. Cytokine (2009) 45(3):190–7. doi: 10.1016/j.cyto.2008.12.009
66. Gadaleta RM, van Erpecum KJ, Oldenburg B, Willemsen EC, Renooij W, Murzilli S, et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut (2011) 60(4):463–72. doi: 10.1136/gut.2010.212159
67. Massafra V, Ijssennagger N, Plantinga M, Milona A, Ramos Pittol JM, Boes M, et al. Splenic dendritic cell involvement in fxr-mediated amelioration of dss colitis. Biochim Biophys Acta (2016) 1862(2):166–73. doi: 10.1016/j.bbadis.2015.11.001
68. Ichikawa R, Takayama T, Yoneno K, Kamada N, Kitazume MT, Higuchi H, et al. Bile acids induce monocyte differentiation toward interleukin-12 hypo-producing dendritic cells Via a Tgr5-dependent pathway. Immunology (2012) 136(2):153–62. doi: 10.1111/j.1365-2567.2012.03554.x
69. Sawa S, Lochner M, Satoh-Takayama N, Dulauroy S, Berard M, Kleinschek M, et al. Rorgammat+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol (2011) 12(4):320–6. doi: 10.1038/ni.2002
70. Bassolas-Molina H, Raymond E, Labadia M, Wahle J, Ferrer-Picon E, Panzenbeck M, et al. An rorgammat oral inhibitor modulates il-17 responses in peripheral blood and intestinal mucosa of crohn's disease patients. Front Immunol (2018) 9:2307. doi: 10.3389/fimmu.2018.02307
71. Mencarelli A, Renga B, Migliorati M, Cipriani S, Distrutti E, Santucci L, et al. The bile acid sensor farnesoid X receptor is a modulator of liver immunity in a rodent model of acute hepatitis. J Immunol (2009) 183(10):6657–66. doi: 10.4049/jimmunol.0901347
72. Biagioli M, Carino A, Fiorucci C, Marchiano S, Di Giorgio C, Roselli R, et al. Gpbar1 functions as gatekeeper for liver nkt cells and provides counterregulatory signals in mouse models of immune-mediated hepatitis. Cell Mol Gastroenterol Hepatol (2019) 8(3):447–73. doi: 10.1016/j.jcmgh.2019.06.003
73. Paik D, Yao L, Zhang Y, Bae S, D'Agostino GD, Zhang M, et al. Human gut bacteria produce Taueta17-modulating bile acid metabolites. Nature (2022) 603(7903):907–912. doi: 10.1038/s41586-022-04480-z
74. Xiao R, Lei K, Kuok H, Deng W, Zhuang Y, Tang Y, et al. Synthesis and identification of lithocholic acid 3-sulfate as rorgammat ligand to inhibit Th17 cell differentiation. J Leukoc Biol (2022) 00:1–9. doi: 10.1002/JLB.1MA0122-513R
75. Hang S, Paik D, Yao L, Kim E, Trinath J, Lu J, et al. Bile acid metabolites control Th17 and treg cell differentiation. Nature (2019) 576(7785):143–8. doi: 10.1038/s41586-019-1785-z
76. Campbell C, McKenney PT, Konstantinovsky D, Isaeva OI, Schizas M, Verter J, et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature (2020) 581(7809):475–9. doi: 10.1038/s41586-020-2193-0
77. Bhalla AK, Amento EP, Serog B, Glimcher LH. 1,25-dihydroxyvitamin D3 inhibits antigen-induced T cell activation. J Immunol (1984) 133(4):1748–54.
78. Mattner F, Smiroldo S, Galbiati F, Muller M, Di Lucia P, Poliani PL, et al. Inhibition of Th1 development and treatment of chronic-relapsing experimental allergic encephalomyelitis by a non-hypercalcemic analogue of 1,25-dihydroxyvitamin D(3). Eur J Immunol (2000) 30(2):498–508. doi: 10.1002/1521-4141(200002)30:2<498::AID-IMMU498>3.0.CO;2-Q
79. Boonstra A, Barrat FJ, Crain C, Heath VL, Savelkoul HF, O'Garra A. 1alpha,25-dihydroxyvitamin D3 has a direct effect on naive Cd4(+) T cells to enhance the development of Th2 cells. J Immunol (2001) 167(9):4974–80. doi: 10.4049/jimmunol.167.9.4974
80. Tang J, Zhou R, Luger D, Zhu W, Silver PB, Grajewski RS, et al. Calcitriol suppresses antiretinal autoimmunity through inhibitory effects on the Th17 effector response. J Immunol (2009) 182(8):4624–32. doi: 10.4049/jimmunol.0801543
81. Penna G, Roncari A, Amuchastegui S, Daniel KC, Berti E, Colonna M, et al. Expression of the inhibitory receptor Ilt3 on dendritic cells is dispensable for induction of Cd4+Foxp3+ regulatory T cells by 1,25-dihydroxyvitamin D3. Blood (2005) 106(10):3490–7. doi: 10.1182/blood-2005-05-2044
82. Binda S, Hill C, Johansen E, Obis D, Pot B, Sanders ME, et al. Criteria to qualify microorganisms as "Probiotic" in foods and dietary supplements. Front Microbiol (2020) 11:1662. doi: 10.3389/fmicb.2020.01662
83. O'Toole PW, Marchesi JR, Hill C. Next-generation probiotics: The spectrum from probiotics to live biotherapeutics. Nat Microbiol (2017) 2:17057. doi: 10.1038/nmicrobiol.2017.57
84. Shadnoush M, Hosseini RS, Khalilnezhad A, Navai L, Goudarzi H, Vaezjalali M. Effects of probiotics on gut microbiota in patients with inflammatory bowel disease: A double-blind, placebo-controlled clinical trial. Korean J Gastroenterol (2015) 65(4):215–21. doi: 10.4166/kjg.2015.65.4.215
85. Cui HH, Chen CL, Wang JD, Yang YJ, Cun Y, Wu JB, et al. Effects of probiotic on intestinal mucosa of patients with ulcerative colitis. World J Gastroenterol (2004) 10(10):1521–5. doi: 10.3748/wjg.v10.i10.1521
86. Fan H, Du J, Liu X, Zheng WW, Zhuang ZH, Wang CD, et al. Effects of pentasa-combined probiotics on the microflora structure and prognosis of patients with inflammatory bowel disease. Turk J Gastroenterol (2019) 30(8):680–5. doi: 10.5152/tjg.2019.18426
87. Huang M, Chen Z, Lang C, Chen J, Yang B, Xue L, et al. Efficacy of mesalazine in combination with bifid triple viable capsules on ulcerative colitis and the resultant effect on the inflammatory factors. Pak J Pharm Sci (2018) 31(6(Special):2891–5.
88. Palumbo VD, Romeo M, Marino Gammazza A, Carini F, Damiani P, Damiano G, et al. The long-term effects of probiotics in the therapy of ulcerative colitis: A clinical study. BioMed Pap Med Fac Univ Palacky Olomouc Czech Repub (2016) 160(3):372–7. doi: 10.5507/bp.2016.044
89. Tsuda Y, Yoshimatsu Y, Aoki H, Nakamura K, Irie M, Fukuda K, et al. Clinical effectiveness of probiotics therapy (Bio-three) in patients with ulcerative colitis refractory to conventional therapy. Scand J Gastroenterol (2007) 42(11):1306–11. doi: 10.1080/00365520701396091
90. Kruis W, Schutz E, Fric P, Fixa B, Judmaier G, Stolte M. Double-blind comparison of an oral escherichia coli preparation and mesalazine in maintaining remission of ulcerative colitis. Aliment Pharmacol Ther (1997) 11(5):853–8. doi: 10.1046/j.1365-2036.1997.00225.x
91. Yoshimatsu Y, Yamada A, Furukawa R, Sono K, Osamura A, Nakamura K, et al. Effectiveness of probiotic therapy for the prevention of relapse in patients with inactive ulcerative colitis. World J Gastroenterol (2015) 21(19):5985–94. doi: 10.3748/wjg.v21.i19.5985
92. Kruis W, Fric P, Pokrotnieks J, Lukas M, Fixa B, Kascak M, et al. Maintaining remission of ulcerative colitis with the probiotic escherichia coli nissle 1917 is as effective as with standard mesalazine. Gut (2004) 53(11):1617–23. doi: 10.1136/gut.2003.037747
93. Tursi A, Brandimarte G, Papa A, Giglio A, Elisei W, Giorgetti GM, et al. Treatment of relapsing mild-to-Moderate ulcerative colitis with the probiotic Vsl3 as adjunctive to a standard pharmaceutical treatment: A double-blind, randomized, placebo-controlled study. Am J Gastroenterol (2010) 105(10):2218–27. doi: 10.1038/ajg.2010.218
94. Miele E, Pascarella F, Giannetti E, Quaglietta L, Baldassano RN, Staiano A. Effect of a probiotic preparation (Vsl#3) on induction and maintenance of remission in children with ulcerative colitis. Am J Gastroenterol (2009) 104(2):437–43. doi: 10.1038/ajg.2008.118
95. Tamaki H, Nakase H, Inoue S, Kawanami C, Itani T, Ohana M, et al. Efficacy of probiotic treatment with bifidobacterium longum 536 for induction of remission in active ulcerative colitis: A randomized, double-blinded, placebo-controlled multicenter trial. Dig Endosc (2016) 28(1):67–74. doi: 10.1111/den.12553
96. Sood A, Midha V, Makharia GK, Ahuja V, Singal D, Goswami P, et al. The probiotic preparation, Vsl3 induces remission in patients with mild-to-Moderately active ulcerative colitis. Clin Gastroenterol Hepatol (2009) 7(11):1202–9,9.e1. doi: 10.1016/j.cgh.2009.07.016
97. Bibiloni R, Fedorak RN, Tannock GW, Madsen KL, Gionchetti P, Campieri M, et al. Vsl3 probiotic-mixture induces remission in patients with active ulcerative colitis. Am J Gastroenterol (2005) 100(7):1539–46. doi: 10.1111/j.1572-0241.2005.41794.x
98. Matthes H, Krummenerl T, Giensch M, Wolff C, Schulze J. Clinical trial: Probiotic treatment of acute distal ulcerative colitis with rectally administered escherichia coli nissle 1917 (Ecn). BMC Complement Altern Med (2010) 10:13. doi: 10.1186/1472-6882-10-13
99. Guslandi M, Mezzi G, Sorghi M, Testoni PA. Saccharomyces boulardii in maintenance treatment of crohn's disease. Dig Dis Sci (2000) 45(7):1462–4. doi: 10.1023/a:1005588911207
100. Matsuoka K, Uemura Y, Kanai T, Kunisaki R, Suzuki Y, Yokoyama K, et al. Efficacy of bifidobacterium breve fermented milk in maintaining remission of ulcerative colitis. Dig Dis Sci (2018) 63(7):1910–9. doi: 10.1007/s10620-018-4946-2
101. Wildt S, Nordgaard I, Hansen U, Brockmann E, Rumessen JJ. A randomised double-blind placebo-controlled trial with lactobacillus acidophilus la-5 and bifidobacterium animalis subsp. lactis bb-12 for maintenance of remission in ulcerative colitis. J Crohns Colitis (2011) 5(2):115–21. doi: 10.1016/j.crohns.2010.11.004
102. Petersen AM, Mirsepasi H, Halkjaer SI, Mortensen EM, Nordgaard-Lassen I, Krogfelt KA. Ciprofloxacin and probiotic escherichia coli nissle add-on treatment in active ulcerative colitis: A double-blind randomized placebo controlled clinical trial. J Crohns Colitis (2014) 8(11):1498–505. doi: 10.1016/j.crohns.2014.06.001
103. Bourreille A, Cadiot G, Le Dreau G, Laharie D, Beaugerie L, Dupas JL, et al. Saccharomyces boulardii does not prevent relapse of crohn's disease. Clin Gastroenterol Hepatol (2013) 11(8):982–7. doi: 10.1016/j.cgh.2013.02.021
104. Van Gossum A, Dewit O, Louis E, de Hertogh G, Baert F, Fontaine F, et al. Multicenter randomized-controlled clinical trial of probiotics (Lactobacillus johnsonii, La1) on early endoscopic recurrence of crohn's disease after lleo-caecal resection. Inflammation Bowel Dis (2007) 13(2):135–42. doi: 10.1002/ibd.20063
105. D'Inca R, Barollo M, Scarpa M, Grillo AR, Brun P, Vettorato MG, et al. Rectal administration of lactobacillus casei dg modifies flora composition and toll-like receptor expression in colonic mucosa of patients with mild ulcerative colitis. Dig Dis Sci (2011) 56(4):1178–87. doi: 10.1007/s10620-010-1384-1
106. White R, Atherly T, Guard B, Rossi G, Wang C, Mosher C, et al. Randomized, controlled trial evaluating the effect of multi-strain probiotic on the mucosal microbiota in canine idiopathic inflammatory bowel disease. Gut Microbes (2017) 8(5):451–66. doi: 10.1080/19490976.2017.1334754
107. Jang SE, Jeong JJ, Kim JK, Han MJ, Kim DH. Simultaneous amelioratation of colitis and liver injury in mice by bifidobacterium longum Lc67 and lactobacillus plantarum Lc27. Sci Rep (2018) 8(1):7500. doi: 10.1038/s41598-018-25775-0
108. Wang Y, Gu Y, Fang K, Mao K, Dou J, Fan H, et al. Lactobacillus acidophilus and clostridium butyricum ameliorate colitis in murine by strengthening the gut barrier function and decreasing inflammatory factors. Benef Microbes (2018) 9(5):775–87. doi: 10.3920/BM2017.0035
109. Isidro RA, Lopez A, Cruz ML, Gonzalez Torres MI, Chompre G, Isidro AA, et al. The probiotic Vsl3 modulates colonic macrophages, inflammation, and microflora in acute trinitrobenzene sulfonic acid colitis. J Histochem Cytochem (2017) 65(8):445–61. doi: 10.1369/0022155417718542
110. Son SJ, Koh JH, Park MR, Ryu S, Lee WJ, Yun B, et al. Effect of the lactobacillus rhamnosus strain gg and tagatose as a synbiotic combination in a dextran sulfate sodium-induced colitis murine model. J Dairy Sci (2019) 102(4):2844–53. doi: 10.3168/jds.2018-15013
111. Ding S, Ma Y, Liu G, Yan W, Jiang H, Fang J. Lactobacillus brevis alleviates dss-induced colitis by reprograming intestinal microbiota and influencing serum metabolome in murine model. Front Physiol (2019) 10:1152. doi: 10.3389/fphys.2019.01152
112. Yu P, Ke C, Guo J, Zhang X, Li B. Lactobacillus plantarum L15 alleviates colitis by inhibiting lps-mediated nf-kappab activation and ameliorates dss-induced gut microbiota dysbiosis. Front Immunol (2020) 11:575173. doi: 10.3389/fimmu.2020.575173
113. Sun M, Liu Y, Song Y, Gao Y, Zhao F, Luo Y, et al. The ameliorative effect of lactobacillus plantarum-12 on dss-induced murine colitis. Food Funct (2020) 11(6):5205–22. doi: 10.1039/d0fo00007h
114. Tong L, Zhang X, Hao H, Liu Q, Zhou Z, Liang X, et al. Lactobacillus rhamnosus gg derived extracellular vesicles modulate gut microbiota and attenuate inflammatory in dss-induced colitis mice. Nutrients (2021) 13(10):3319. doi: 10.3390/nu13103319
115. Hao H, Zhang X, Tong L, Liu Q, Liang X, Bu Y, et al. Effect of extracellular vesicles derived from lactobacillus plantarum Q7 on gut microbiota and ulcerative colitis in mice. Front Immunol (2021) 12:777147. doi: 10.3389/fimmu.2021.777147
116. Zhang Y, Hou Q, Ma C, Zhao J, Xu H, Li W, et al. Lactobacillus casei protects dextran sodium sulfate- or rapamycin-induced colonic inflammation in the mouse. Eur J Nutr (2020) 59(4):1443–51. doi: 10.1007/s00394-019-02001-9
117. Jang YJ, Kim WK, Han DH, Lee K, Ko G. Lactobacillus fermentum species ameliorate dextran sulfate sodium-induced colitis by regulating the immune response and altering gut microbiota. Gut Microbes (2019) 10(6):696–711. doi: 10.1080/19490976.2019.1589281
118. Liu Y, Li Y, Yu X, Yu L, Tian F, Zhao J, et al. Physiological characteristics of lactobacillus casei strains and their alleviation effects against inflammatory bowel disease. J Microbiol Biotechnol (2021) 31(1):92–103. doi: 10.4014/jmb.2003.03041
119. Gupta M, Pattanaik AK, Singh A, Sharma S, Jadhav SE. An appraisal of the gut health modulatory effects of a calf faecal-origin probiotic lactobacillus salivarius Cpn60 using wistar rats with dextran sulfate sodium-induced colitis. J Sci Food Agric (2021) 101(4):1340–8. doi: 10.1002/jsfa.10744
120. Chen Y, Chen H, Ding J, Stanton C, Ross RP, Zhao J, et al. Bifidobacterium longum ameliorates dextran sulfate sodium-induced colitis by producing conjugated linoleic acid, protecting intestinal mechanical barrier, restoring unbalanced gut microbiota, and regulating the toll-like receptor-4/Nuclear factor-kappab signaling pathway. J Agric Food Chem (2021) 69(48):14593–608. doi: 10.1021/acs.jafc.1c06176
121. Chen Y, Yang B, Stanton C, Ross RP, Zhao J, Zhang H, et al. Bifidobacterium pseudocatenulatum ameliorates dss-induced colitis by maintaining intestinal mechanical barrier, blocking proinflammatory cytokines, inhibiting Tlr4/Nf-kappab signaling, and altering gut microbiota. J Agric Food Chem (2021) 69(5):1496–512. doi: 10.1021/acs.jafc.0c06329
122. Zhang X, Tong Y, Lyu X, Wang J, Wang Y, Yang R. Prevention and alleviation of dextran sulfate sodium salt-induced inflammatory bowel disease in mice with bacillus subtilis-fermented milk Via inhibition of the inflammatory responses and regulation of the intestinal flora. Front Microbiol (2020) 11:622354. doi: 10.3389/fmicb.2020.622354
123. Liu Y, Yin F, Huang L, Teng H, Shen T, Qin H. Long-term and continuous administration of bacillus subtilis during remission effectively maintains the remission of inflammatory bowel disease by protecting intestinal integrity, regulating epithelial proliferation, and reshaping microbial structure and function. Food Funct (2021) 12(5):2201–10. doi: 10.1039/d0fo02786c
124. Zhang HL, Li WS, Xu DN, Zheng WW, Liu Y, Chen J, et al. Mucosa-reparing and microbiota-balancing therapeutic effect of bacillus subtilis alleviates dextrate sulfate sodium-induced ulcerative colitis in mice. Exp Ther Med (2016) 12(4):2554–62. doi: 10.3892/etm.2016.3686
125. Sheng K, Xu Y, Kong X, Wang J, Zha X, Wang Y. Probiotic bacillus cereus alleviates dextran sulfate sodium-induced colitis in mice through improvement of the intestinal barrier function, anti-inflammation, and gut microbiota modulation. J Agric Food Chem (2021) 69(49):14810–23. doi: 10.1021/acs.jafc.1c03375
126. Rodriguez-Nogales A, Algieri F, Garrido-Mesa J, Vezza T, Utrilla MP, Chueca N, et al. Intestinal anti-inflammatory effect of the probiotic saccharomyces boulardii in dss-induced colitis in mice: Impact on micrornas expression and gut microbiota composition. J Nutr Biochem (2018) 61:129–39. doi: 10.1016/j.jnutbio.2018.08.005
127. Mu Z, Yang Y, Xia Y, Wang F, Sun Y, Yang Y, et al. Probiotic yeast Br14 ameliorates dss-induced colitis by restoring the gut barrier and adjusting the intestinal microbiota. Food Funct (2021) 12(18):8386–98. doi: 10.1039/d1fo01314a
128. Sun S, Xu X, Liang L, Wang X, Bai X, Zhu L, et al. Lactic acid-producing probiotic saccharomyces cerevisiae attenuates ulcerative colitis Via suppressing macrophage pyroptosis and modulating gut microbiota. Front Immunol (2021) 12:777665. doi: 10.3389/fimmu.2021.777665
129. Emge JR, Huynh K, Miller EN, Kaur M, Reardon C, Barrett KE, et al. Modulation of the microbiota-Gut-Brain axis by probiotics in a murine model of inflammatory bowel disease. Am J Physiol Gastrointest Liver Physiol (2016) 310(11):G989–98. doi: 10.1152/ajpgi.00086.2016
130. Rodriguez-Nogales A, Algieri F, Garrido-Mesa J, Vezza T, Utrilla MP, Chueca N, et al. Differential intestinal anti-inflammatory effects of lactobacillus fermentum and lactobacillus salivarius in dss mouse colitis: Impact on micrornas expression and microbiota composition. Mol Nutr Food Res (2017) 61(11). doi: 10.1002/mnfr.201700144
131. Wang W, Xing W, Wei S, Gao Q, Wei X, Shi L, et al. Semi-rational screening of probiotics from the fecal flora of healthy adults against dss-induced colitis mice by enhancing anti-inflammatory activity and modulating the gut microbiota. J Microbiol Biotechnol (2019) 29(9):1478–87. doi: 10.4014/jmb.1807.06061
132. Chen Y, Zhang L, Hong G, Huang C, Qian W, Bai T, et al. Probiotic mixtures with aerobic constituent promoted the recovery of multi-barriers in dss-induced chronic colitis. Life Sci (2020) 240:117089. doi: 10.1016/j.lfs.2019.117089
133. Wang Y, Xie Q, Zhang Y, Ma W, Ning K, Xiang JY, et al. Combination of probiotics with different functions alleviate dss-induced colitis by regulating intestinal microbiota, il-10, and barrier function. Appl Microbiol Biotechnol (2020) 104(1):335–49. doi: 10.1007/s00253-019-10259-6
134. Huang YY, Wu YP, Jia XZ, Lin J, Xiao LF, Liu DM, et al. Lactiplantibacillus plantarum dmdl 9010 alleviates dextran sodium sulfate (Dss)-induced colitis and behavioral disorders by facilitating microbiota-Gut-Brain axis balance. Food Funct (2022) 13(1):411–24. doi: 10.1039/d1fo02938j
135. Martin R, Chain F, Miquel S, Lu J, Gratadoux JJ, Sokol H, et al. The commensal bacterium faecalibacterium prausnitzii is protective in dnbs-induced chronic moderate and severe colitis models. Inflammation Bowel Dis (2014) 20(3):417–30. doi: 10.1097/01.MIB.0000440815.76627.64
136. Yao M, Lu Y, Zhang T, Xie J, Han S, Zhang S, et al. Improved functionality of ligilactobacillus salivarius Li01 in alleviating colonic inflammation by layer-by-Layer microencapsulation. NPJ Biofilms Microbiomes (2021) 7(1):58. doi: 10.1038/s41522-021-00228-1
137. Dong F, Xiao F, Li X, Li Y, Wang X, Yu G, et al. Pediococcus pentosaceus cect 8330 protects dss-induced colitis and regulates the intestinal microbiota and immune responses in mice. J Transl Med (2022) 20(1):33. doi: 10.1186/s12967-022-03235-8
138. Bian X, Wu W, Yang L, Lv L, Wang Q, Li Y, et al. Administration of akkermansia muciniphila ameliorates dextran sulfate sodium-induced ulcerative colitis in mice. Front Microbiol (2019) 10:2259. doi: 10.3389/fmicb.2019.02259
139. Chou YC, Ho PY, Chen WJ, Wu SH, Pan MH. Lactobacillus fermentum V3 ameliorates colitis-associated tumorigenesis by modulating the gut microbiome. Am J Cancer Res (2020) 10(4):1170–81.
140. Jacouton E, Chain F, Sokol H, Langella P, Bermudez-Humaran LG. Probiotic strain lactobacillus casei Bl23 prevents colitis-associated colorectal cancer. Front Immunol (2017) 8:1553. doi: 10.3389/fimmu.2017.01553
141. Oh NS, Lee JY, Kim YT, Kim SH, Lee JH. Cancer-protective effect of a synbiotic combination between lactobacillus gasseri 505 and a cudrania tricuspidata leaf extract on colitis-associated colorectal cancer. Gut Microbes (2020) 12(1):1785803. doi: 10.1080/19490976.2020.1785803
142. Rong J, Liu S, Hu C, Liu C. Single probiotic supplement suppresses colitis-associated colorectal tumorigenesis by modulating inflammatory development and microbial homeostasis. J Gastroenterol Hepatol (2019) 34(7):1182–92. doi: 10.1111/jgh.14516
143. Wang Q, Wang K, Wu W, Lv L, Bian X, Yang L, et al. Administration of bifidobacterium bifidum cgmcc 15068 modulates gut microbiota and metabolome in azoxymethane (Aom)/Dextran sulphate sodium (Dss)-induced colitis-associated colon cancer (Cac) in mice. Appl Microbiol Biotechnol (2020) 104(13):5915–28. doi: 10.1007/s00253-020-10621-z
144. Liu M, Xie W, Wan X, Deng T. Clostridium butyricum modulates gut microbiota and reduces colitis associated colon cancer in mice. Int Immunopharmacol (2020) 88:106862. doi: 10.1016/j.intimp.2020.106862
145. Mendes MCS, Paulino DS, Brambilla SR, Camargo JA, Persinoti GF, Carvalheira JBC. Microbiota modification by probiotic supplementation reduces colitis associated colon cancer in mice. World J Gastroenterol (2018) 24(18):1995–2008. doi: 10.3748/wjg.v24.i18.1995
146. Wang CS, Li WB, Wang HY, Ma YM, Zhao XH, Yang H, et al. Vsl3 can prevent ulcerative colitis-associated carcinogenesis in mice. World J Gastroenterol (2018) 24(37):4254–62. doi: 10.3748/wjg.v24.i37.4254
147. Ding S, Yan W, Fang J, Jiang H, Liu G. Potential role of lactobacillus plantarum in colitis induced by dextran sulfate sodium through altering gut microbiota and host metabolism in murine model. Sci China Life Sci (2021) 64(11):1906–16. doi: 10.1007/s11427-020-1835-4
148. Wang G, Huang S, Cai S, Yu H, Wang Y, Zeng X, et al. Lactobacillus reuteri ameliorates intestinal inflammation and modulates gut microbiota and metabolic disorders in dextran sulfate sodium-induced colitis in mice. Nutrients (2020) 12(8):2298. doi: 10.3390/nu12082298
149. Degirolamo C, Rainaldi S, Bovenga F, Murzilli S, Moschetta A. Microbiota modification with probiotics induces hepatic bile acid synthesis Via downregulation of the fxr-Fgf15 axis in mice. Cell Rep (2014) 7(1):12–8. doi: 10.1016/j.celrep.2014.02.032
150. Chen QW, Li QR, Cao MW, Yan JH, Zhang XZ. Hierarchy-assembled dual probiotics system ameliorates cholestatic drug-induced liver injury Via gut-liver axis modulation. Adv Sci (Weinh) (2022) 9:e2200986. doi: 10.1002/advs.202200986
151. Russell BJ, Brown SD, Siguenza N, Mai I, Saran AR, Lingaraju A, et al. Intestinal transgene delivery with native e. coli chassis allows persistent physiological changes. Cell (2022) 185(17):3263–77 doi: 10.1016/j.cell.2022.06.050
152. Ye J, Erland LAE, Gill SK, Bishop SL, Verdugo-Meza A, Murch SJ, et al. Metabolomics-guided hypothesis generation for mechanisms of intestinal protection by live biotherapeutic products. Biomolecules (2021) 11(5):738. doi: 10.3390/biom11050738
153. Sato S, Nagai H, Igarashi Y. Effect of probiotics on serum bile acids in patients with ulcerative colitis. Hepatogastroenterology (2012) 59(118):1804–8. doi: 10.5754/hge11789
154. Kurdi P, van Veen HW, Tanaka H, Mierau I, Konings WN, Tannock GW, et al. Cholic acid is accumulated spontaneously, driven by membrane deltaph, in many lactobacilli. J Bacteriol (2000) 182(22):6525–8. doi: 10.1128/JB.182.22.6525-6528.2000
155. Kurdi P, Tanaka H, van Veen HW, Asano K, Tomita F, Yokota A. Cholic acid accumulation and its diminution by short-chain fatty acids in bifidobacteria. Microbiol (Reading) (2003) 149(Pt 8):2031–7. doi: 10.1099/mic.0.26376-0
Keywords: inflammatory bowel disease, gut microbiota, bile acids, probiotics, bile salt hydrolase, bile acid-inducible enzymes, intestinal immunity
Citation: Li L, Liu T, Gu Y, Wang X, Xie R, Sun Y, Wang B and Cao H (2022) Regulation of gut microbiota-bile acids axis by probiotics in inflammatory bowel disease. Front. Immunol. 13:974305. doi: 10.3389/fimmu.2022.974305
Received: 21 June 2022; Accepted: 31 August 2022;
Published: 23 September 2022.
Edited by:
Xu Zhang, Health Canada, CanadaReviewed by:
Feng Zhang, Affiliated Hospital of Jiangnan University, ChinaJames Butcher, University of Ottawa, Canada
Copyright © 2022 Li, Liu, Gu, Wang, Xie, Sun, Wang and Cao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hailong Cao, Y2FvaGFpbG9uZ0B0bXUuZWR1LmNu
 Lingfeng Li
Lingfeng Li Tianyu Liu
Tianyu Liu Yu Gu
Yu Gu Runxiang Xie
Runxiang Xie Yue Sun
Yue Sun Bangmao Wang
Bangmao Wang Hailong Cao
Hailong Cao