
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol. , 12 September 2022
Sec. Molecular Innate Immunity
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.967055
This article is part of the Research Topic Hepatic Immune Response underlying Liver Cirrhosis and Portal Hypertension View all 19 articles
Tissue-resident memory (TRM) T cells are a unique subset of memory T cells that are critical for the first line of defense against pathogens or antigens in peripheral non-lymphoid tissues such as liver, gut, and skin. Generally, TRM cells are well adapted to the local environment in a tissue-specific manner and typically do not circulate but persist in tissues, distinguishing them from other memory T cell lineages. There is strong evidence that liver TRM cells provide a robust adaptive immune response to potential threats. Indeed, the potent effector function of hepatic TRM cells makes it essential for chronic liver diseases, including viral and parasite infection, autoimmune liver diseases (AILD), nonalcoholic fatty liver disease (NAFLD), hepatocellular carcinoma (HCC) and liver transplantation. Manipulation of hepatic TRM cells might provide novel promising strategies for precision immunotherapy of chronic liver diseases. Here, we provide insights into the phenotype of hepatic TRM cells through surface markers, transcriptional profiles and effector functions, discuss the development of hepatic TRM cells in terms of cellular origin and factors affecting their development, analyze the role of hepatic TRM cells in chronic liver diseases, as well as share our perspectives on the current status of hepatic TRM cell research.
T cells are essential for building an effective immune response against pathogens or antigens. Once the pathogen breaks through the barrier tissue and invades the body, antigen-presenting cells (APC) capture the foreign antigen and then migrate to the local draining lymph nodes to activate naive T cells. Primed naive T cells subsequently proliferate and differentiate into effector T cells that migrate into inflamed tissues to eliminate pathogens (1). Among these effector T cells, a minor fraction persists and develops into memory T cells precursors after the pathogens are cleared. According to their unique patrolling properties, proliferative potential, and effector function, these memory T cell precursors eventually develop into circulating memory T cells and tissue-resident memory T (TRM) cells (2, 3). Circulating memory T cells include central memory (TCM) cells that target and patrol in the lymph node and egress to the blood after infection, and effector memory (TEM) cells that survey nonlymphoid peripheral tissues and enter the peripheral circulation thorough the lymphatic system (4). By contrast, TRM cells almost not recirculate and are retained within tissues under homeostatic conditions (5).
Both CD8+ and CD4+ subpopulations of TRM cells are detected at different tissue sites (6–8). CD8+ TRM cells are well defined and enhance immune responses in peripheral tissues. However, the characteristics and functions of CD4+ TRM cells remain largely unclear (9, 10). In general, TRM cells primarily develop and persist in organs that are frequently exposed to pathogens or antigens, such as the liver, gut, skin and lung (11, 12). Among these organs, the liver is considered as a vital immune organ, and it is exposed to various pathogens and food antigens, that enter or re-enter the body via portal vein from the gastrointestinal tract and the systemic blood circulation.
Liver contains a large number of innate immune cells, including natural killer (NK) cells, NKT cells, γδ T cells, mucosal-associated invariant T cells, Kupffer cells, and dendritic cells (13). Interestingly, liver also include a number of liver-specific antigen-presenting cells, such as hepatic sinusoidal endothelial cells and hepatic stellate cells, which contribute to immune tolerance in the liver (13). Moreover, the hepatic specific immune microenvironment constructed by these immune cells promotes the generation of antigen-experienced T cells and TRM cells involved in pathogen clearance or autoimmune responses against self-antigens (14). Importantly, liver TRM cells perform an essential role in the first line of adaptive cellular defense while exposing to the cognate antigens in the liver (15, 16). Accordingly, the liver acts as an essential gatekeeper to prevent systemic infection and inflammation, while the liver TRM cells contribute to the efficient eradication of pathogens as well as immune responses.
In this review, we primarily focus on phenotype and development of hepatic TRM cells, mainly CD8+ TRM cells, with emphasis on their protective roles in viral and parasite infection, non-alcoholic fatty liver disease (NAFLD), hepatocellular carcinoma (HCC) and liver transplantation, as well as their pathogenic roles in autoimmune liver diseases (AILD) (Table 1).
The general characteristics of TRM cells include their strategic positioning in the tissues and effector functions. However, despite TRM cells share some similar features, the phenotype, such as surface markers and transcriptional profiles, and the underlying mechanisms for their generation and retention are highly heterogeneous in different tissues.
It is considered that the surface markers contribute to the identification and maintenance of hepatic TRM cells. Similar to other tissue-specific TRM cells, hepatic TRM cells downregulate the expression of tissue egression markers, like soingosine-1-phosphate 1 (S1PR1), and the homing receptors such as CD62L and CCR7 (43, 44). Furthermore, hepatic TRM cells usually express some adhesion molecule and chemokine receptors, including CD69 (44), CD103 (17, 45), CD49a (36), CXCR3 (17, 23) and CXCR6 (46, 47), which are involved in their localization and maintenance in the hepatic sinusoids and portal veins.
The lectin CD69 is constitutive expressed on the majority of liver TRM subsets. Upon exposure to antigens or pro-inflammatory mediators, the expression of CD69 is strongly upregulated on activated CD8+ T cells within peripheral tissues as a result of the downregulation of Krüppel-like factor 2(KLF2) (44, 48, 49). Meanwhile, as an antagonist of S1P1, CD69 complexs with S1P1 on the cell surface and leads to its internalization and degradation (50). Besides, CD69 also contributes to the retention status of hepatic TRM cells by downregulating sphingosine 1 phosphate receptor (S1PR1)-mediated tissue egress (44). Therefore, it is likely to that its primary role is to restrict the egress of TRM cells from the liver to the blood and lymphatic vessels.
CD103 is an α-chain of the integrin αEβ7. It is upregulated in activated peripheral CD8+ T lymphocytes upon exposure to TGFβ (51). CD103 is a receptor for E-cadherin, an adherens junction protein interlocking epithelial cells (52). Interestingly, E-cadherin is widely expressed by hepatocytes and cholangiocytes (36, 53, 54).The interaction of E-cadherin and CD103 expressing on the liver-infiltrating lymphocytes may be involved in positioning, adhesion and retention of hepatic TRM cells (36). Furthermore, CD103 may define two different functional subsets of TRM cells in human liver. The CD69+CD103+ subpopulations are antigen-specific autoreactive cytotoxic T cells in human liver, exhibiting more potent effector function than CD69+CD103- counterparts (45, 55, 56). Interestingly, there are differences between mouse and human liver TRM cells regarding CD103 expression. Interestingly, it appears that another liver-specific homing marker, lymphocyte function associated antigen 1 (LFA-1), rather than CD103, may be responsible for the retention of hepatic TRM cells in mice (16, 57).
CD49a, another adhesion molecule of TRM cells, is the α1 component of the integrin α1β1. CD49a pairs with integrin β1 to form the heterodimer VLA-1 which bind to collagen IV. This interaction is believed to be critical for retention of the resident population at the epithelium (58). In general, CD49a is upregulated following T cell activation and can be found on circulating T cells (59). Expression of CD49a contributes to protect cells from undergoing apoptosis (60). Importantly, blockade of CD49a with antibodies as well as genetic deletion of CD49a results in a diminution of TRM cells (59, 61). However, CD49a was not essential for the recruitment of CD8 T cells to the lung in mice, but for their persistence as memory cells (59). Therefore, CD49a may promote the survival, retention or proliferation of TRM cells. Moreover, CD49a may define different functional subsets of TRM cells. In the skin, CD49a expressing CD8+ TRM cells produce large amounts of IFN-γ, perforin and granzyme B, while CD49a negative counterparts prefer to produce IL17 (62). However, the effector function bias based on CD49a expression of liver TRM cells have not been comprehensively interrogated.
Chemokines and chemokine receptors have been extensively used to describe the correct localization, residence and effector function of immune cells within lymphoid organs and non-lymphoid tissues (63). Despite their expressions on TRM cells of different tissues have great heterogeneity, it is reported that the maintenance and effector function of TRM require constant chemokine stimulation (64–66). Chemokine receptors CXCR3 and CXCR6 have been extensively reported to be constitutively expressed on the surface of intrahepatic TRM cells (16, 17, 23, 46, 67). CXCR3 is a vital homing marker that may contributes to the retention of liver CD8+ TRM cells. It binds to multiple chemokines, such as CXCL9, CXCL10 and CXCL11, which are predominantly secreted by monocytes, liver sinusoidal endothelial cells and fibroblasts (17). On the other hand, CXCR6 also plays an important role in the maintenance of liver TRM cells (46, 68). CD8+ T cells lacking CXCR6 migrate to the liver normally after immunization, whereas perform a marked decrease capacity to form hepatic CD8+ TRM cells and severely impairs their effector functions against infection in the liver (46). In addition, CXCR6 also contributes to the maintenance of liver TRM cells via binding to CXCL16 secreted by liver sinusoidal endothelial cells (46, 68). These studies suggest that CXCR6 is essential for retention rather than recruitment of CD8+T cells to the liver. Additionally, deficiency of CXCR6 results in decreased survival of hepatic NKT cells patrolling the liver sinusoids, affecting hepatic intravascular immune surveillance (68).
Besides surface markers, multiple transcription factors are involved in the regulation of the distinct features of liver TRM cells.
The network of transcription factors underlies the unique features of TRM cells, including liver TRM cells (Figure 1). These transcription factors include B lymphocyte-induced maturation protein 1 (BLIMP1; also known as PRDM1), homologue of BLIMP1 in T cells (HOBIT; also known as ZFP683), runt-related transcription factor 3 (RUNX3), Notch, Peroxisome proliferator-activated receptor-γ (PPAR-γ), BHlHe40, TBX21(T-bet), aryl hydrocarbon receptor (AhR), Eomesodermin (EOMES), and NR4A family of orphan nuclear receptors (NR4As). The combined action of these transcription factors contributes to the residency status of liver TRM cells (64, 69).
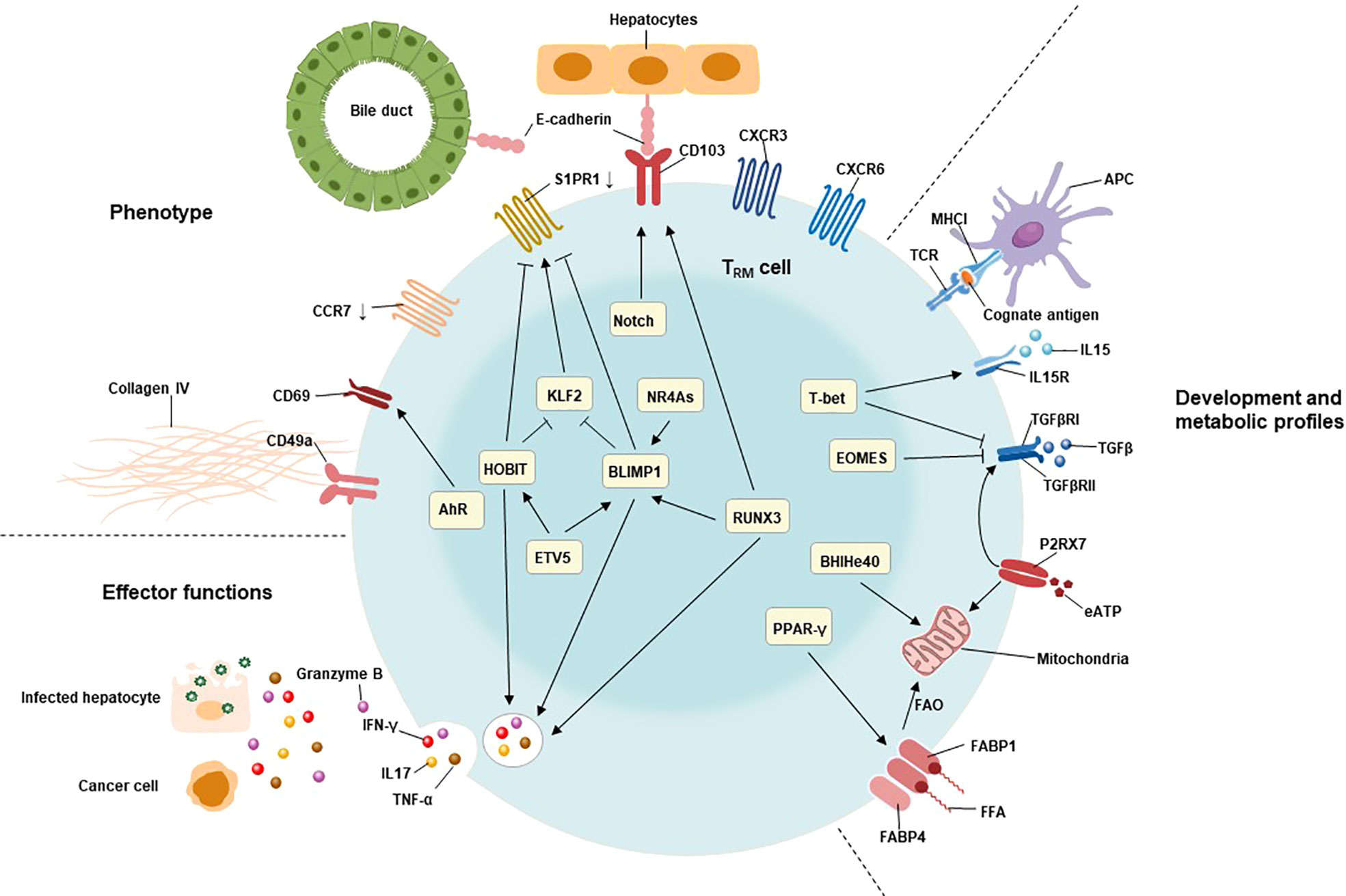
Figure 1 Characteristics of TRM cells include their tissue residency, long-term persistence, and effector function. The residency status of liver TRM cells is regulated by the combined action of B lymphocyte-induced maturation protein 1 (BLIMP1), BLIMP1 homolog in T cells (HOBIT), Notch, and runt-related transcription factor 3 (RUNX3). BLIMP1 and HOBIT downregulate CCR7, Krüppel-like factor 2 (KLF2) and tissue export pathway sphingosine 1-phosphate receptor 1 (S1PR1), while Notch directly upregulates the expression of CD103 on TRM cells. The interaction of CD103 and E-cadherin expressing on hepatocytes as well as cholangiocytes may be involved in adhesion and retention of hepatic TRM cells. Furthermore, the expression of BLIMP1 is regulated by the transcription factor runt-related transcription factor 3 (RUNX3) and NR4A family of orphan nuclear receptors (NR4As). The effector functions of liver TRM cells include direct killing of infected or malignant cells by secreting cytotoxic molecules and inflammatory cytokines, such as granzyme B, TNF-α, IFN-γ, and IL17. The expression of these cytotoxic components is regulated by HOBIT, BLIMP1 and RunX3. The development and maintenance of TRM cells require stimulation with IL15, and TGFβ, as well as cognate antigens presenting by antigen-presenting cells (APC). T-bet is essential for the sustain expression of IL15 receptor, albeit at low levels. Meanwhile, the expression of TGFβ receptor is also regulated by P2X purinreceptor 7 (P2RX7), a sensor for extracellular nucleotides that promotes mitochondrial homeostasis. Mitochondrial fatty acid β-oxidation (FAO) is an important energy source for TRM cells. Peroxisome proliferator-activated receptor-γ (PPARγ) drives the upregulation of FABP1 and FABP4 to promote free fatty acid uptake from the extracellular compartment, while the transcription factor BHlHe40 maintains mitochondrial fitness.
HOBIT is specifically up-regulated in TRM cells and, together with related Blimp1, mediates the development of TRM cells in lymphoid organs and non-lymphoid tissues (70). The co-expression of HOBIT and BLIMP1 instructs the downregulation of CCR7, transcription factor 7 (TCF7), KLF2, and S1PR1 in TRM cells (71). CCR7 is the receptor for chemokine ligand 19 (CCL19) and chemokine ligand 21 (CCL21) that responsible for cell migration to secondary lymphoid tissues (72).Meanwhile, TCF7, KLF2 and S1PR1 are involved in the tissue egression of lymphocytes (71). Interestingly, KLF2 regulates the expression of S1PR1 in lymphocytes of tissues, which directs them returning to circulation (44). Consequently, the Hobit-Blimp1 transcriptional module retains TRM cells within tissues through silencing the genes related to recirculation in addition to suppressing the markers related to egression. Furthermore, a murine study demonstrated that the transcriptional repressor Capicua (CIC) controls the development of liver TRM cells. Mechanistically, they found that CIC could regulate the expression of HOBIT by inhibiting the ETS variant transcription factor 5 (ETV5) (73). RUNX3 and Notch are essential for the maintenance of TRM cells by repressing the expression of genes involved in the formation of circulating memory T cells and inducing the expression of retention molecules, including CD103 (74). The collaboration of HOBIT, BLIMP1 and RUNX3 also drives immediate effector function in TRM cells by inducing and sustaining granzyme B production (75–77). Notch, predominantly expressed in newly developed TRM cells, not only regulates expression of IFN-γ upon restimulation but also contributes to the mitochondrial fatty acid β-oxidation (FAO) in TRM cells (74, 75). Importantly, exogenous free fatty acids uptake and their FAO are required for the survival and effector function of TRM cells (78). Meanwhile, PPAR-γ facilitate the uptake of free fatty acids by upregulating fatty acid binding proteins 1 and 4 (FABP1 and FABP4) in TRM cells (78, 79). Bhlhe40, a stress-responsive protein, promotes the survival and function of TRM cells under stress conditions by sustaining mitochondrial fitness (80).
T-bet is crucial to sustain the expression of the IL15 receptor β subunit (IL15Rβ) and therefore enable the long-term lineage stability of TRM cells, albeit at low levels (81). Activation of aryl hydrocarbon receptor (AhR) may be associated to the maintenance of liver TRM cells by increasing the expression of CD69 (82, 83).Recent studies reveal that EOMES directly inhibited expression of IFN-γ in vitro, while EOMES deletion in T cells led to substantially increased frequency and percentage of TRM precursor in the liver (84, 85). Therefore, the downregulation of EOMES in TRM cells is required to not only their formation, but also their effector function.
Additionally, the NR4As are composed of NR4A1 (Nur77), NR4A2 (Nurr1), and NR4A3 (Nor1). During the memory phase of influenza infection, Nur77 deficiency in CD8+ T cells reduces the frequency of CD8+ TRM cells in the liver without any effect on lung or bone marrow CD8+ TRM cells and other memory CD8+ T cells such as TCM and TEM (86), indicating a specific role of Nur77 on liver TRM cell differentiation. In addition, the expression of the transcription factors involving in TRM differentiation (BLIMP1 and T-bet) is decreased, while the expression of EOMES is increased in absence of Nor1 in CD8+ T cells (87). Interestingly, NR4As are particularly enriched in the highly functional CD28+ subset of CD8+ TRM cells. Importantly, deficiency of Nurr1 specifically reduces the percentage of these CD28+ TRM subsets (88). To conclude, NR4As are important regulators involved in the differentiation of CD8+ TRM cells. However, not all NR4As are comprehensively interrogated at the specific differentiation steps of CD8+TRM cells. Therefore, figure out which signals promote the expression of NR4As in addition the role of NR4As in CD8+ TRM cell differentiation await further investigation.
Although these transcription factors described above have been shown to be critical for TRM cells, it is difficult to determine which are the specific key regulators of TRM differentiation and maintenance, as they are also expressed in other CD8+ effector or memory subsets. Therefore, the differentiation and maintenance of TRM may be regulated by the cooperation of multiple transcription factors.
Similar to other tissue TRM cells, liver TRM cells also have timely, potent and durable effector functions. When pathogens enter the liver, TRM cells can take advantage of tissue residency to generate a rapid and effective protective immune response by secreting multiple chemokines and cytokines in a deployment-ready mode (75). The cytotoxic cytokines enable them to directly eliminate infected or malignant cells as well as control invading pathogens, while chemokines and pro-inflammatory cytokines recruit and activate other immune cells, thereby remodeling the local liver microenvironment for more potent effector functions. Furthermore, liver CD8+ TRM cells express high levels of Ki-67 and TCF1, showing their proliferative and self-renewal potential (89). Actually, TRM cells can persist in the liver for years and exert durable protective effect (17). In addition, TRM cells may help to significantly promote the repopulation of locally resident and circulating memory T cells after infection, suggesting their role in establishing secondary memory T cells to prevent future reinfection with the same pathogen (90, 91). Accordingly, TRM cells have been used to develop vaccines that generate stronger and longer-lasting immune responses than conventional vaccines (15, 28, 29). Meanwhile, CD8+ TRM cells are able to attract hepatic stellate cells (HSCs) in a CCR5-dependent manner and predispose activated HSCs to FasL-Fas-mediated apoptosis, thereby promoting liver fibrosis regression (39). However, every coin has two sides, as do liver TRM cells. Once TRM cells are interfered by cognate antigens and damage hepatocytes and cholangiocytes, it may lead to the occurrence of AILD. Meanwhile, auto-aggressive liver CXCR6+CD8+ TRM cells cause hepatic immune pathology in NASH in an MHC-class-I-independent manner (47). Therefore, clarifying the biological characteristics and development of liver TRM cells so as to accurately manipulate liver TRM cells can enhance the effector functions of TRM cells and avoid weaknesses.
Multiple factors including T cell-intrinsic and environmental factors are believed to be involved in the TRM cell differentiation. Thereinto, the first question to be addressed is the origin of TRM cells. Olivier, O et al. analyzed antigen-activated T cells from different tissues using TCR sequences. They found that TCM cells in the lymph nodes share a common clonal origin with TRM cells (92), indicating that these subsets derive from the same naïve T cell precursors. Moreover, the differ in TCR stimulation affinity, namely the strength of antigen binding of TCRs, affects the subsequent development of TRM cells (93). In this regard, high TCR affinity leads to TEM development, whereas a low TCR affinity results in short-lived memory cells with impaired secondary immune response (94, 95).It is reported that TRM cells have different TCR stimulation affinity compared to splenic memory T cells (93, 94). Furthermore, there is heterogeneity in the magnitude of TCR stimulation affinity required for the development of functional CD8+ TRM cells in different tissues (93, 96). For example, Maru, S et al. demonstrated that brain TRM cells stimulated with suboptimal stimulation strength respond more effectively to CNS infection than cognate antigen, suggesting that the strength of antigen stimulation affects the functional integrity of TRM cells in a persistent viral infection (93). However, the specific strength of TCR stimulation affinity required for inducing liver-adapted TRM cells has not been determined.
Additionally, the killer cell lectin-like receptor G1 (KLRG1) may contributes to figure out the source of TRM cells. KLRG1 is upregulated in short-lived effector cells (SLECs, KLRG1hi IL7Rαlo), whereas the memory precursor effector cells (MPECs) that turn into heterogenous populations of memory CD8+ T cells, bear negative or low expression of KLRG1 (9, 97). Adoptive transfer experiments have shown that MPECs could generate TRM cells after entering specific tissues (98). In addition, a portion of KLRG1+ CD8+ T cells can downregulate KLRG1 during the contraction phase of immune response and differentiated into TRM cells. The latter subset accounts for approximately half of the liver TRM cell population and has a stronger cytotoxic and proliferative capacity than those directly derived from KLRG1-CD8+ T cells (99). These findings suggest that liver TRM cell can originate from both KLRG1+ or KLRG1- lymphocytes.
On the other hand, studies have shown that cognate antigens and inflammatory cytokines also contribute to the development and maintenance of liver TRM cells (Figure 1).
Antigenic challenge induces and amplifies antigen-specific TRM cell proliferation, and maintained at low-level magnitude in the liver TRM pool after the clearance of infection. Actually, the capacity of hepatic TRM niches is large enough to lodge multiple TRM cells with different specificities without displacing previously established cells (14). Therefore, newly formed liver TRM cells do not displace existing TRM cell populations (14). Intriguingly, TRM cells induced by cognate antigen in secondary immune response are mainly developed from the pre-existing TRM populations, instead of circulating memory T cells (90, 100). Therefore, cognate antigens contribute to the immune response mediated by TRM cells and the construct of polyclonal TRM cell repertoire.
The differentiation and development of liver TRM cells can be mediated by multiple cytokines, including IL2, IL15, TGFβ and IL10. IL2 is mainly produced by activated T cells. It promotes the growth, proliferation and differentiation of lymphocytes, and is essential for the body’s immune response and antiviral infection. Interestingly, human liver CD8+ TRM cells express high levels of IL2 (17, 36). The unusually high IL2 production of hepatic CD8+ TRM may be important for their protective potential, as autocrine IL2 is needed to the persistence of memory responses to pathogens and secondary population expansion of CD8+ memory T cells (17, 101). In addition, IL15 is known to be involved in TRM development and longevity. Although shares a receptor subunit with IL2, IL15 has a perceptible difference in immunomodulatory properties. Generally, IL15 induces the proliferation and survival of circulating memory CD8+ T cells (102, 103). Nevertheless, the upregulation of the IL15 receptors in memory CD8+ T cells indicating that IL15 stimulation may be essential for TRM development (102). It was reported that IL15 was able to induce CD69, CXCR3 and CXCR6 expression on peripheral CD8 T cells in a dose-dependent manner, all of which were highly expressed on hepatic TRM cells (17). Consistently, IL15 knockout mice prevent CD8+ TRM cells development in the liver (14). Meanwhile, the expression of hepatic IL15 is positively correlated with TRM cells in AIH liver (36). Therefore, the presence of IL15 may be essential for the formation of liver TRM cells. Another important cytokine is the TGFβ. TGFβ is a pleiotropic cytokine that is produced in an inactive form, namely latency associated peptide (LAP). LAP can be activated by binding to integrin αvβ6 on epithelial cells and/or integrin αvβ8 on dendritic cells and endothelial cells (104). Activated-TGFβ induces CD8+ TRM cells to express CD103 as well as downregulate of EOMES (81, 98), which are mandatory for their generation, adhesion and long-term persistence in the liver. In fact, TGFβ is capable of inducing liver-adapted TRM cells, and importantly, hepatic TGFβ is significantly correlated with TRM cells infiltration in human liver (17, 18, 36). Actually, sequential exposure to IL-15 followed by TGFβ efficiently induced de novo CD69+CD103+CD8+ TRM cells, with similar frequencies to those found in healthy livers (17). These studies suggest that the expression of IL15 and TGFβ in the liver promotes the development and residency of CD103+ TRM cells in human. However, a recent mouse experiment showed that constitutive TGFβ signaling did not accelerate the development of liver TRM cells (105), indicating that TGFβ may have functional heterogeneity in liver TRM cells between human and mice. Meanwhile, monocyte-produced IL10 induced the release of surface-bound TGFβ of antigen-presenting cells, while blocking IL10 reduced CD103 expression on TRM cells (106). Therefore, IL10-mediated TGFβ signaling may have a critical role in the generation and retention of liver TRM cells.
Additionally, several cytokines have been reported to be involved in TRM development outside the liver. For example, the IFN-β and IL12 are described to positively influence TRM cells differentiation by regulating the expression of CD103 and CD69 in the intestine (107). Meanwhile, it is reported that hair follicle-derived IL7 is involved in CD4+ TRM cells generation and persistence in the skin (108, 109). Intriguingly, hepatocytes are the main source of IL7 in the liver, and the hepatocyte-derived IL7 can promote the survival of memory CD4+ and CD8+ T cells (110). However, the specific role of these cytokines on the development of liver TRM cells remains to be elucidated.
There are significant differences in the metabolic profiles of different T cell subsets. Several studied demonstrated that preferences for certain metabolic pathways for energy affect TRM cells generation, tissue retention, and effector functions.
Generally, highly proliferative and active cells prefer the glycolytic pathway, while quiescent cells primarily use oxidative phosphorylation and FAO to generate ATP. Thereinto, mammalian target of rapamycin (mTOR), including two subunits of mTOR complex 1 (mTORC1) and mTORC2, is a key regulator involved in regulating T cell nutrient metabolism, proliferation and activation (111). While activating, it induces glucose consumption to support T cell proliferation. There is strong evidence that mTOR plays an important role in the generation of TRM cells (112). Rapamycin, an mTORC1 inhibitor, has been reported to induce the formation of memory CD8+ T cells but reduce TRM production in the gut, thereby protecting mice from functional CD8+ TRM cell-mediated intestinal autoimmunity (113). However, the exact effects of rapamycin on the liver TRM cells are still under investigation.
Fatty acid binding proteins (FABPs) are a group of intracellular molecules that mediate the trafficking and metabolism of fatty acids (114). Reliance on FAO has recently been shown to be essential for the development and maturation of CD8+ TRM cells (78). For example, studies on skin TRM cells revealed that TRM cells upregulate FABP4 and FABP5 so as to uptake and utilize exogenous free fatty acid (FFA) as an energy source for their survival. Consistently, the deficiency of FABP4 and FABP5 results in impaired functional properties and longevity of skin CD8+ TRM cells, but not influence the survival of TCM cells in vivo (78). However, TRM cells from different tissues express distinct FABPs with selected in a tissue-specific fashion that is optimized for local fatty acid availability (78, 79). It has been demonstrated that liver TRM cells express high levels of FABP1 and a low concentration of FABP4, but do not express FABP5 (79). In a murine model of LCMV infection, FABP1 deficiency mice manifested impaired TRM cell development in the liver but not in the skin. Furthermore, the selective loss of liver TRM cells could be restored upon re-expression of FABP1 (79). Interestingly, bezafibrate, the PPAR agonists that promote FAO, has been confirmed to improve the effector function of memory T cells (115). Therefore, a unique FAO regulator, FABP1, driven by a liver-specific microenvironment may be a promising target for intervention in hepatic TRM cells.
Additionally, several studies revealed that P2X purinreceptor 7 (P2RX7) is required for the establishment, maintenance and functionality of TRM cells. P2RX7 is a sensor for extracellular nucleotides that promotes mitochondrial homeostasis and metabolic function of memory CD8+ T cells (116). Importantly, P2RX7 supports TRM development by enhancing CD8+ T cell sensing of TGFβ via upregulate the TGFβ receptor II (TGFβRII) through calcineurin signaling. Meanwhile, P2RX7-deficient TRM cells progressively decayed and expressed dysregulated TRM-specific markers such as CD103. Consistently, upregulation of TGFβRII expression rescued P2RX7-deficient TRM cell generation as well as mitochondrial function (116), indicating that sustained P2RX7 signaling is required for long-term TRM cell maintenance. However, another study demonstrated that P2RX7 activation in sterile tissue damage during acetaminophen-induced liver injury selectively enhanced the NAD-induced cell death of liver TRM cells compared with circulating T cells, whereas concurrent TCR engagement promoted survival of TRM cells (117).These studies suggest that differences in genetic background, microbiota as well as their metabolites might have caused discrepancies in the regulation of TRM differentiation and maintenance by P2RX7.
The porous epithelial layer is a unique feature of the liver, which not only enables the direct interaction of TRM cells with hepatocytes, but also facilitates the encounter of cognate antigens by TRM cells in the liver. TRM cells that reside in the unique microenvironments of the liver not only develop in response to infection, such as viral or parasite infection, but are also detected in AILD, NAFLD, HCC and liver allografts. Below, we discuss the unique characteristics of TRM cells in the local microenvironment of different chronic liver diseases, their role in disease progression, as well as their potential therapeutic value (Table 1).
Hepatoviral infection is mainly caused by the hepatitis B (HBV) and hepatitis C (HCV) viruses and the course can be acute or chronic. Chronic infection with hepatotropic virus can cause liver damage, cirrhosis, liver failure, development of HCC, and even liver transplantation. It has been demonstrated that hepatic TRM cells play a major antiviral immune response during chronic hepatic virus infections.
Pallett, J et al. were the first to report the virus-specific liver CD8+ T cells in chronic HBV infection, in which approximately 90% of them have a TRM cell-like phenotype (CD69+CD103+ or CD69+CD103−) (17). CD8+ TRM cells can persist in the liver for several years after primary infection and expand in patients with HBV. Importantly, virus-specific CD8+ TRM cells could still be detected in spontaneously recovered HBV patients, with effector functions equivalent to those from chronic HBV-infected patients (18), suggesting the long-term viral control of hepatic CD8+ TRM cells. Virus-specific CD8+ TRM are very efficient in their function. During HBV viral infection, PD-L1 expression is upregulated in hepatic sinusoidal endothelial cells and hepatocytes (118). PD-L1 on intrahepatic cells can interact with PD1 on TRM cells, thereby dampening pro-inflammatory TRM cell responses (19). Nevertheless, even though TRM cells express high levels of the PD1, they readily produce IFN-γ, TNF-α, perforin, and IL2 upon stimulation (17). IFN-γ and TNF-α mediated control of HBV replication, while perforin may contribute to the directly elimination of infected hepatocytes (20, 21). Furthermore, IL2 production is most strikingly enhanced within CD69+CD103+ TRM cells, which contributes to overcome PD-L1-mediated inhibition and exhaustion, stressing their ability for survival and maintenance (21, 119). Additionally, CD8+ TRM cells are enriched in HBV patients who achieved viral control, and their abundance is inversely correlated with HBV viral load, stressing that the virus-specific liver TRM cells can control viral replication and contribute to the functional cure for HBV patients (17, 22). Therefore, liver TRM cell expansion may be a potential therapeutic target for chronic HBV infection.
Additionally, a portion of HBV patients are co-infected with hepatitis D virus (HDV), which often indicates a poor prognosis. As the smallest known human virus, HDV has perfectly adapted to escape recognition by CD8+ T cells restricted by common human leukocyte antigen (HLA) class I alleles (120). A recent study suggested that antigen-nonspecific activation of hepatic CD8+ TRM cells may be involved in intrahepatic inflammation and disease progression in HDV infection (121).
CD8+ TRM cells also play an essential role in long-term antiviral response in chronic HCV infection (23–25). In the chimpanzee model of HCV reinfection, depletion of CD8+ T cells resulted in prolonged the virus persistence and prevented effective viral clearance, while recovery of CD8+ T cells lead to virus eradication (26). Meanwhile, a large number of CD69+CD8+ T cells were detected in the liver of animals recovered after HCV infection, but not in the peripheral blood. These subsets may be hepatic TRM cells, which are required for protection from persistent HCV Infection (26). Consistently, liver CD8+ TRM cells are highly increased in chronic HCV patients and possess a specific activation and cytolytic potential and are important in controlling chronic HCV infection (27).
Besides hepatotropic virus infection, liver CD8+ TRM cells contribute to the effective clearance of Lymphocytic choriomeningitis virus (LCMV) as well. In the murine model of LCMV infection, virus-specific TRM cells in the liver could be influenced by other liver-resident immune cells. For example, deficiency of liver-resident natural killer (LrNK) cells increased both the frequency and antiviral activity of hepatic TRM cells via the interaction of PD1 and PD-L1. Consistently, transfer of LrNK cells into LrNK-cell-deficient mice as well as PD-L1 inhibition restrain hepatic TRM cell function, resulting in impaired viral clearance (122). Furthermore, during LCMV infection, other liver-resident T cells, such as γδ T cells, also expand and promote viral clearance by producing IFN-γ and TNF-α (123).
Current studies suggest that hepatic TRM cells may be involved in the clearance of viral infection, protect patients from persistent viral infection, and improve disease prognosis. However, the role of TRM cells in different viral infections in the liver remains to be further elucidated.
Besides viral infections, several studies have investigated the role of liver TRM cells in parasitic infections, including malaria and leishmaniasis.
Malaria is an insect-borne infectious disease caused by the infection of Plasmodium through the bite of Anopheles mosquitoes or the transfusion of the blood of a person carrying Plasmodium (124). Plasmodium has a complex life cycle, including three stages in the liver, blood and mosquito. During infection of malaria, Plasmodium promotes the development of antigens-specific TRM cells (16, 125–127). These TRM cells could mediate protective immune responses through killing infected cells by producing pro-inflammatory cytokines, such as IFN-γ and TNF-α (16, 30). Additionally, TRM cell depletion abrogated an efficient immune response to a murine model of Plasmodium infection (31).Due to the protective immune response of TRM cells against malaria, vaccination strategies that maximize intrahepatic Plasmodium-specific TRM development have emerged (16, 28, 29, 32–34, 127). An example is the Plasmodium ribosomal protein vaccine (15). One of the antigens for this vaccine is PbRPL6120-127, a highly conserved H2-Kb-restricted epitope from the 60S ribosomal protein L6, expressed throughout the parasite life cycle, across Plasmodium species (15). It may be an optimal antigen for endogenous liver TRM development and protection against malaria. A single dose of this vaccine could provide effective and prolonged sterilizing immunity against high dose sporozoite challenges (15). Indeed, people living in malaria-endemic areas do not acquire effective protection against reinfection from malaria (128), while attenuated Plasmodium falciparum sporozoite (SPZ) vaccine is highly protective against controlled human malaria infection 3 weeks after immunization (129), suggesting multiple, complex factors are likely responsible for the lack of development of sterilizing immunity to malaria through natural infection. Furthermore, the protection and long-term efficacy of existing vaccines are not satisfactory. Accordingly, to improve the TRM-based vaccination against malaria in human, further investigation of the mechanisms that mediate Plasmodium-specific TRM generation and function, assessment of the feasibility of currently known antigens, as well as identification of novel target epitopes are required.
Recently, the role of TRM cells in Leishmaniasis was studied as well. Leishmaniasis is a zoonotic disease caused by Leishmania, which can cause cutaneous and visceral kala-azar in humans (130). There are various types of Leishmania in which Leishmania infantum (L. infantum) primarily infects the liver (131–133). During chronic L. infantum infection, liver TRM cells are generated and play a protective role. Importantly, induction by the Leishmania proteins LirCyP1 and LirSOD promotes the expansion of hepatic TRM cells, which could be a promising strategy for prophylactic or therapeutic vaccine formulations (131).
Taken together, hepatic TRM cells are critical in parasitic infections, and the TRM-based vaccination strategies could hold remarkable promise in providing long-term protection.
AILD is a group of liver inflammatory damage diseases mediated by abnormal autoimmunity, including autoimmune hepatitis (AIH), primary biliary cholangitis (PBC), primary sclerosing cholangitis (PSC), IgG4-related sclerosing cholangitis (IgG4-SC), etc. AIH is an inflammatory liver disease dominated by T cell-mediated hepatocyte injury. Antigen-specific CD8+ TRM cells have been reported to characterize the liver tissue of subjects with indeterminate pediatric acute liver failure (PALF) and may serve as a novel biomarker for PALF due to AIH (37, 38). Recently, our group demonstrated that CD69+CD103+CD8+ TRM cells play an important role in the pathogenesis of AIH, and histological remission is accompanied by decreased hepatic CD8+ TRM cells in AIH patients (36). In addition, hepatic CD8+ TRM cells from AIH patients expressed a higher level of PD-1, CXCR3 and granzyme B than those of healthy controls. Consistently, in AIH liver, both expression of IL15 and TGFβ, cytokines that induce TRM cells in vitro, were elevated, suggesting that the immunological microenvironment facilitates hepatic CD8+ TRM cells development and residency (36). Intriguingly, E-cadherin, the natural ligand of CD103, is widely expressed in hepatocytes of AIH patients, and located closely to CD8+ TRM cells, which may contribute to the residency of CD8+ TRM cells in the liver. Furthermore, E-cadherin is also widely expressed in cholangiocytes (53, 54), suggesting that CD103+ TRM cells may be involved in pathology of bile duct injury in cholestatic liver diseases, such as PBC and PSC. Interestingly, a recent study on biliary immune atlas revealed the presence of CD8+ TRM cells in areas of biliary inflammation in PSC patients (134).
Nonalcoholic fatty liver disease (NAFLD) is considered a hepatic manifestation of metabolic syndrome, hypertension and type 2 diabetes. Several studies have demonstrated that liver-resident T cells and the proinflammatory immune response they elicit are involved in NAFLD disease progression (135–138). Generally, liver-resident γδT cells induce chronic liver inflammation by producing proinflammatory cytokines such as IL17A, IFN-γ, and TNF-α, contributing to the pathogenic immune response to NAFLD (123, 137, 139). Furthermore, systemic inflammation in obese patients is associated with increased TRM cells in the liver and may be further involved in NAFLD disease progression. Importantly, activated TRM cells are significantly increased in the liver and visceral fat of obese patients. These activated TRM cells produce multiple pro-inflammatory cytokines, such as IL1β, IL2, IL12, and IL15 (140), further contributing to the generation of TRM cells in addition to the overall pro-inflammatory phenotype in obese patients.
Interestingly, a recent study revealed that CD69+CD103-CD8+ TRM cell may perform a protective role in resolving liver fibrosis of nonalcoholic steatohepatitis (NASH) (39). They demonstrated that the reduction of these CD8+ TRM cells significantly delayed fibrosis resolution via influencing predisposed HSCs apoptosis, while adoptive transfer of these cells protected mice from fibrosis progression in a CCR5-dependent manner (39). Therefore, the paradoxical roles of TRM cells in NAFLD and their specific mechanisms remain to be further investigated.
HCC accounts for the majority of primary liver cancers and is currently one of the leading causes of cancer-related deaths worldwide. The development of HCC is a complex multistep process caused by multiple risk factors, whereas the function of tumor-infiltrating T cells is important for moderating antitumor immunity in HCC development and determining the clinical fate of HCC patients (40). There are strong evidences that CD103+ TRM cells are enriched in HCC patients and associated with better prognosis (19, 41, 42).
In murine model of HCC, hepatic TRM cells were significantly expanded, and their frequencies decreased during HCC progression (141). Meanwhile, hepatic TRM cells in HCC have an exhausted phenotype, manifested by expression of PD1, LAG3, and TIM3 (40). Given that PD1 expression in TRM cells in HCC is associated with poor disease outcome (142), immunotherapy targeting checkpoint inhibition has been applied to HCC (143, 144). During immunotherapy for HCC, PD1high TRM cells are the most sensitive cells to anti-PD-1 therapy to overcome tumor growth and progression (145). Additionally, other markers of exhaustion and inhibition, such as TIM3 and CTLA4, and pro-inflammatory cytokines, such as IFN-γ and TNF-α, can also be simultaneously expressed on TRM cells in HCC patients (142), suggesting that hepatic TRM cells may be involved in direct killing of tumor cells. Overall, hepatic TRM cells might play an extremely important role in both HCC development and anti-tumor therapy.
Liver transplantation is the treatment of last option for end-stage liver disease of various causes and severe acute liver failure. It has been reported that donor-derived TRM cells are detectable in the liver allografts and that their abundancy could be correlated with organ survival and reduced rejection (146–148). Specifically, long-term persistence of lung donor-derived TRM cell is associated with reduced incidence of clinical events that precipitate allograft injury, including primary graft dysfunction (PGD) and acute cellular rejection (ACR) (149). However, the association of liver donor-derived TRM cells with the incidence of clinical events remains to be further elucidated (150). In liver allograft tissues, approximately 2-6% of CD8+ T cells had a donor-derived TRM phenotype at 11 years post-transplantation (18), well demonstrating the longevity of human liver TRM cells. Additionally, donor-derived TRM cells from an HBV-infected liver allograft could migrate to draining lymph nodes with down-regulation of some TRM-specific markers. However, they were not detectable in blood vessels (18). Interestingly, the same study demonstrated that a lower quantity of recipient-derived virus-specific T cells with a TRM-like phenotype were detected in the liver and blood (18), further revealing the extrahepatic origin of TRM cells in the liver. Nevertheless, CMV-specific TRM cells in human liver allografts did not acquire a TRM phenotype in the liver, possibly due to the lack of relevant antigens in the liver.
The tissue retention and longevity of hepatic TRM cells and their potent effector functions demonstrate their potential role in chronic liver diseases. The above studies have shown that hepatic TRM cells play a protective role in viral and parasitic infection, NAFLD, HCC, and liver transplantation, whereas they might be pathogenic in AILD such as AIH. However, further studies are needed to reveal more mechanisms of TRM cell biology, including the phenotype of TRM cells and the specific mechanisms that regulate their development and differentiation. Furthermore, there are several key points regarding hepatic TRM cells that remain to be investigated.
Firstly, TRM cells are heterogeneous, and the subsets of TRM cells that function in the liver under different conditions will differ in the expression of surface markers and biological behavior. For example, the predominant TRM cells associated with the pathogenesis of AIH are CD8+CD69+CD103+ TRM cells that highly express PD1, CXCR3 and granzyme B (36); whereas liver TRM cells of patients with acute hepatitis A are mainly CD8+CD69+CD103- TRM cells that express high levels of HIF-2α (55). TRM cells are essential for the adaptive immune response. While interfering different chronic liver diseases by hepatic TRM cells, the biological function and disease specificity of the corresponding TRM cells should be carefully considered. Therefore, identifying the specific subsets of hepatic TRM cells that play a major role in the chronic liver diseases will help to define precise future intervention strategies.
Secondly, since the liver is an immune organ, we should pay attention to the crosstalk of other immune cells in the liver to hepatic TRM cells. Clarify whether they are cooperative or antagonistic is of great significance. It has been shown that LrNK cells can reduce the frequency and antiviral activity of hepatic TRM cells through the interaction of PD1 and PD-L1 during LCMV infection (122). However, the interaction among other liver-resident cells remains to be further investigated. For example, liver-resident γδ T cells, participate in the pathogenic immune response to NAFLD by producing proinflammatory cytokines (123), are capable of form a long-lived resident memory-like subpopulation upon local inflammation or infection. Nevertheless, it is still unclear whether there is crosstalk between unconventional γδ TRM cells and conventional αβ TRM cells. Accordingly, clarifying these interactions will shed light on the overall immune homeostasis of the liver and lay the groundwork for developing holistic therapies.
Thirdly, given that the biliary system that communicates with the digestive tract and the portal blood that flows directly into the liver may contain various gut-derived microorganisms as well as their metabolites, hepatic TRM cells are chronically exposed to, and may be trained by them. Whether the composition of the gut microbiome, specific species of the gut microbiome or their metabolites would influence the phenotype and development of hepatic TRM cells are unknown yet. Elucidating these interactions may open up new avenues for the realization of therapeutic strategies for “enteric treatment of liver disease”.
To conclude, hepatic TRM cells are considered to play a crucial role in various chronic liver diseases. Elucidating and characterizing the underlying mechanisms of hepatic TRM cells will shed light on the control of chronic liver diseases and provide promising strategies for precision immunotherapy in different chronic liver diseases.
All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Sheridan BS, Lefrançois L. Regional and mucosal memory T cells. Nat Immunol (2011) 12:485–91. doi: 10.1038/ni.2029
2. Shin H, Iwasaki A. Tissue-resident memory T cells. Immunol Rev (2013) 255:165–81. doi: 10.1111/imr.12087
3. Watanabe R, Gehad A, Yang C, Scott LL, Teague JE, Schlapbach C, et al. Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Sci Transl Med (2015) 7(279):279ra39. doi: 10.1126/scitranslmed.3010302
4. Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature (1999) 401:708–12. doi: 10.1038/44385
5. Mueller SN, Mackay LK. Tissue-resident memory T cells: Local specialists in immune defence. Nat Rev Immunol (2016) 16:79–89. doi: 10.1038/nri.2015.3
6. Sun H, Sun C, Xiao W, Sun R. Tissue-resident lymphocytes: From adaptive to innate immunity. Cell Mol Immunol (2019) 16:205–15. doi: 10.1038/s41423-018-0192-y
7. Brummelman J, Pilipow K, Lugli E. The single-cell phenotypic identity of human CD8(+) and CD4(+) T cells. Int Rev Cell Mol Biol (2018) 341:63–124. doi: 10.1016/bs.ircmb.2018.05.007
8. Gebhardt T, Whitney PG, Zaid A, Mackay LK, Brooks AG, Heath WR, et al. Different patterns of peripheral migration by memory CD4+ and CD8+ T cells. Nature (2011) 477:216–9. doi: 10.1038/nature10339
9. Tokura Y, Phadungsaksawasdi P, Kurihara K, Fujiyama T, Honda T. Pathophysiology of skin resident memory T cells. Front Immunol (2020) 11:618897. doi: 10.3389/fimmu.2020.618897
10. Hirahara K, Kokubo K, Aoki A, Kiuchi M, Nakayama T. The role of CD4(+) resident memory T cells in local immunity in the mucosal tissue - protection versus pathology. Front Immunol (2021) 12:616309. doi: 10.3389/fimmu.2021.616309
11. Ariotti S, Haanen JB, Schumacher TN. Behavior and function of tissue-resident memory T cells. Adv Immunol (2012) 114:203–16. doi: 10.1016/b978-0-12-396548-6.00008-1
12. Li P, Zhang Y, Xu Y, Cao H, Li L. Characteristics of CD8+ and CD4+ tissue-resident memory lymphocytes in the gastrointestinal tract. Advanced Gut Microbiome Res (2022) 2022:9157455. doi: 10.1155/2022/9157455
13. Doherty DG. Immunity, tolerance and autoimmunity in the liver: A comprehensive review. J Autoimmun (2016) 66:60–75. doi: 10.1016/j.jaut.2015.08.020
14. Holz LE, Prier JE, Freestone D, Steiner TM, English K, Johnson DN, et al. CD8(+) T cell activation leads to constitutive formation of liver tissue-resident memory T cells that seed a Large and flexible niche in the liver. Cell Rep (2018) 25:68–79.e4. doi: 10.1016/j.celrep.2018.08.094
15. Valencia-Hernandez AM, Ng WY, Ghazanfari N, Ghilas S, de Menezes MN, Holz LE, et al. A natural peptide antigen within the plasmodium ribosomal protein RPL6 confers liver T(RM) cell-mediated immunity against malaria in mice. Cell Host Microbe (2020) 27:950–962.e7. doi: 10.1016/j.chom.2020.04.010
16. Fernandez-Ruiz D, Ng WY, Holz LE, Ma JZ, Zaid A, Wong YC, et al. Liver-resident memory CD8(+) T cells form a front-line defense against malaria liver-stage infection. Immunity (2016) 45:889–902. doi: 10.1016/j.immuni.2016.08.011
17. Pallett LJ, Davies J, Colbeck EJ, Robertson F, Hansi N, Easom NJW, et al. IL-2(high) tissue-resident T cells in the human liver: Sentinels for hepatotropic infection. J Exp Med (2017) 214:1567–80. doi: 10.1084/jem.20162115
18. Pallett LJ, Burton AR, Amin OE, Rodriguez-Tajes S, Patel AA, Zakeri N, et al. Longevity and replenishment of human liver-resident memory T cells and mononuclear phagocytes. J Exp Med (2020) 217(9):e20200050. doi: 10.1084/jem.20200050
19. Lim CJ, Lee YH, Pan L, Lai L, Chua C, Wasser M, et al. Multidimensional analyses reveal distinct immune microenvironment in hepatitis b virus-related hepatocellular carcinoma. Gut (2019) 68:916–27. doi: 10.1136/gutjnl-2018-316510
20. Guidotti LG, Rochford R, Chung J, Shapiro M, Purcell R, Chisari FV. Viral clearance without destruction of infected cells during acute HBV infection. Science (1999) 284:825–9. doi: 10.1126/science.284.5415.825
21. Bolte FJ, Rehermann B. Tissue-resident T cells in hepatitis b: A new target for cure? J Exp Med (2017) 214:1564–6. doi: 10.1084/jem.20170842
22. Dusséaux M, Masse-Ranson G, Darche S, Ahodantin J, Li Y, Fiquet O, et al. Viral load affects the immune response to HBV in mice with humanized immune system and liver. Gastroenterology (2017) 153:1647–1661.e9. doi: 10.1053/j.gastro.2017.08.034
23. Pirozyan MR, Nguyen N, Cameron B, Luciani F, Bull RA, Zekry A, et al. Chemokine-regulated recruitment of antigen-specific T-cell subpopulations to the liver in acute and chronic hepatitis c infection. J Infect Dis (2019) 219:1430–8. doi: 10.1093/infdis/jiy679
24. Stelma F, de Niet A, Sinnige MJ, van Dort KA, van Gisbergen K, Verheij J, et al. Human intrahepatic CD69 + CD8+ T cells have a tissue resident memory T cell phenotype with reduced cytolytic capacity. Sci Rep (2017) 7:6172. doi: 10.1038/s41598-017-06352-3
25. Abdel-Hakeem MS, Bédard N, Murphy D, Bruneau J, Shoukry NH. Signatures of protective memory immune responses during hepatitis c virus reinfection. Gastroenterology (2014) 147:870–881.e8. doi: 10.1053/j.gastro.2014.07.005
26. Shoukry NH, Grakoui A, Houghton M, Chien DY, Ghrayeb J, Reimann KA, et al. Memory CD8+ T cells are required for protection from persistent hepatitis c virus infection. J Exp Med (2003) 197:1645–55. doi: 10.1084/jem.20030239
27. Tonnerre P, Wolski D, Subudhi S, Aljabban J, Hoogeveen RC, Damasio M, et al. Differentiation of exhausted CD8(+) T cells after termination of chronic antigen stimulation stops short of achieving functional T cell memory. Nat Immunol (2021) 22:1030–41. doi: 10.1038/s41590-021-00982-6
28. Holz LE, Chua YC, de Menezes MN, Anderson RJ, Draper SL, Compton BJ, et al. Glycolipid-peptide vaccination induces liver-resident memory CD8(+) T cells that protect against rodent malaria. Sci Immunol (2020) 5(48):eaaz8035. doi: 10.1126/sciimmunol.aaz8035
29. Gola A, Silman D, Walters AA, Sridhar S, Uderhardt S, Salman AM, et al. Prime and target immunization protects against liver-stage malaria in mice. Sci Transl Med (2018) 10(460). doi: 10.1126/scitranslmed.aap9128
30. Grand M, Waqasi M, Demarta-Gatsi C, Wei Y, Peronet R, Commere PH, et al. Hepatic inflammation confers protective immunity against liver stages of malaria parasite. Front Immunol (2020) 11:585502. doi: 10.3389/fimmu.2020.585502
31. Fernandez-Ruiz D, de Menezes MN, Holz LE, Ghilas S, Heath WR, Beattie L. Harnessing liver-resident memory T cells for protection against malaria. Expert Rev Vaccines (2021) 20:127–41. doi: 10.1080/14760584.2021.1881485
32. Olsen TM, Stone BC, Chuenchob V, Murphy SC. Prime-and-Trap malaria vaccination to generate protective CD8(+) liver-resident memory T cells. J Immunol (2018) 201:1984–93. doi: 10.4049/jimmunol.1800740
33. Ghilas S, Enders MH, May R, Holz LE, Fernandez-Ruiz D, Cozijnsen A, et al. Development of plasmodium-specific liver-resident memory CD8(+) T cells after heat-killed sporozoite immunization in mice. Eur J Immunol (2021) 51:1153–65. doi: 10.1002/eji.202048757
34. Noé A, Datoo MS, Flaxman A, Husainy MA, Jenkin D, Bellamy D, et al. Deep immune phenotyping and single-cell transcriptomics allow identification of circulating TRM-like cells which correlate with liver-stage immunity and vaccine-induced protection from malaria. Front Immunol (2022) 13:795463. doi: 10.3389/fimmu.2022.795463
35. Lefebvre MN, Surette FA, Anthony SM, Vijay R, Jensen IJ, Pewe LL, et al. Expeditious recruitment of circulating memory CD8 T cells to the liver facilitates control of malaria. Cell Rep (2021) 37:109956. doi: 10.1016/j.celrep.2021.109956
36. You Z, Li Y, Wang Q, Zhao Z, Li Y, Qian Q, et al. The clinical significance of hepatic CD69(+) CD103(+) CD8(+) resident-memory T cells in autoimmune hepatitis. Hepatology (2021) 74:847–63. doi: 10.1002/hep.31739
37. Chapin CA, Burn T, Meijome T, Loomes KM, Melin-Aldana H, Kreiger PA, et al. Indeterminate pediatric acute liver failure is uniquely characterized by a CD103(+) CD8(+) T-cell infiltrate. Hepatology (2018) 68:1087–100. doi: 10.1002/hep.29901
38. Chapin CA, Melin-Aldana H, Kreiger PA, Burn T, Neighbors K, Taylor SA, et al. Activated CD8 T-cell hepatitis in children with indeterminate acute liver failure: Results from a multicenter cohort. J Pediatr Gastroenterol Nutr (2020) 71:713–9. doi: 10.1097/mpg.0000000000002893
39. Koda Y, Teratani T, Chu PS, Hagihara Y, Mikami Y, Harada Y, et al. CD8(+) tissue-resident memory T cells promote liver fibrosis resolution by inducing apoptosis of hepatic stellate cells. Nat Commun (2021) 12:4474. doi: 10.1038/s41467-021-24734-0
40. Chew V, Lai L, Pan L, Lim CJ, Li J, Ong R, et al. Delineation of an immunosuppressive gradient in hepatocellular carcinoma using high-dimensional proteomic and transcriptomic analyses. Proc Natl Acad Sci U.S.A. (2017) 114:E5900–e5909. doi: 10.1073/pnas.1706559114
41. Wang T, Shen Y, Luyten S, Yang Y, Jiang X. Tissue-resident memory CD8(+) T cells in cancer immunology and immunotherapy. Pharmacol Res (2020) 159:104876. doi: 10.1016/j.phrs.2020.104876
42. Wei J, Ishizuka JJ. Going viral: HBV-specific CD8(+) tissue-resident memory T cells propagate anti-tumor immunity. Immunity (2021) 54:1630–2. doi: 10.1016/j.immuni.2021.07.014
43. Hollingshead N, Wu X, Kenerson H, Chen A, Strickland I, Horton H, et al. Functional responses of resident human T cells in intact liver tissue. Cell Immunol (2021) 360:104275. doi: 10.1016/j.cellimm.2020.104275
44. Skon CN, Lee JY, Anderson KG, Masopust D, Hogquist KA, Jameson SC. Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat Immunol (2013) 14:1285–93. doi: 10.1038/ni.2745
45. Huang B, Lyu Z, Qian Q, Chen Y, Zhang J, Li B, et al. NUDT1 promotes the accumulation and longevity of CD103(+) TRM cells in primary biliary cholangitis. J Hepatol (2022) S0168-8278(22):00408–1. doi: 10.1016/j.jhep.2022.06.014
46. Tse SW, Radtke AJ, Espinosa DA, Cockburn IA, Zavala F. The chemokine receptor CXCR6 is required for the maintenance of liver memory CD8+ T cells specific for infectious pathogens. J Infect Dis (2014) 210:1508–16. doi: 10.1093/infdis/jiu281
47. Dudek M, Pfister D, Donakonda S, Filpe P, Schneider A, Laschinger M, et al. Auto-aggressive CXCR6(+) CD8 T cells cause liver immune pathology in NASH. Nature (2021) 592:444–9. doi: 10.1038/s41586-021-03233-8
48. Mackay LK, Braun A, Macleod BL, Collins N, Tebartz C, Bedoui S, et al. Cutting edge: CD69 interference with sphingosine-1-phosphate receptor function regulates peripheral T cell retention. J Immunol (2015) 194:2059–63. doi: 10.4049/jimmunol.1402256
49. Shiow LR, Rosen DB, Brdicková N, Xu Y, An J, Lanier LL, et al. CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature (2006) 440:540–4. doi: 10.1038/nature04606
50. Bankovich AJ, Shiow LR, Cyster JG. CD69 suppresses sphingosine 1-phosophate receptor-1 (S1P1) function through interaction with membrane helix 4. J Biol Chem (2010) 285:22328–37. doi: 10.1074/jbc.M110.123299
51. El-Asady R, Yuan R, Liu K, Wang D, Gress RE, Lucas PJ, et al. TGF-{beta}-dependent CD103 expression by CD8(+) T cells promotes selective destruction of the host intestinal epithelium during graft-versus-host disease. J Exp Med (2005) 201:1647–57. doi: 10.1084/jem.20041044
52. Cepek KL, Shaw SK, Parker CM, Russell GJ, Morrow JS, Rimm DL, et al. Adhesion between epithelial cells and T lymphocytes mediated by e-cadherin and the alpha e beta 7 integrin. Nature (1994) 372:190–3. doi: 10.1038/372190a0
53. Graham JJ, Mukherjee S, Yuksel M, Sanabria Mateos R, Si T, Huang Z, et al. Aberrant hepatic trafficking of gut-derived T cells is not specific to primary sclerosing cholangitis. Hepatology (2022) 75:518–30. doi: 10.1002/hep.32193
54. Li Y, Li B, You Z, Zhang J, Wei Y, Li Y, et al. Cytotoxic KLRG1 expressing lymphocytes invade portal tracts in primary biliary cholangitis. J Autoimmun (2019) 103:102293. doi: 10.1016/j.jaut.2019.06.004
55. Kim JH, Han JW, Choi YJ, Rha MS, Koh JY, Kim KH, et al. Functions of human liver CD69(+)CD103(-)CD8(+) T cells depend on HIF-2α activity in healthy and pathologic livers. J Hepatol (2020) 72:1170–81. doi: 10.1016/j.jhep.2020.01.010
56. Franciszkiewicz K, Le Floc’h A, Boutet M, Vergnon I, Schmitt A, Mami-Chouaib F. CD103 or LFA-1 engagement at the immune synapse between cytotoxic T cells and tumor cells promotes maturation and regulates T-cell effector functions. Cancer Res (2013) 73:617–28. doi: 10.1158/0008-5472.Can-12-2569
57. McNamara HA, Cai Y, Wagle MV, Sontani Y, Roots CM, Miosge LA, et al. Up-regulation of LFA-1 allows liver-resident memory T cells to patrol and remain in the hepatic sinusoids. Sci Immunol (2017) 2(9):eaaj1996. doi: 10.1126/sciimmunol.aaj1996
58. Conrad C, Boyman O, Tonel G, Tun-Kyi A, Laggner U, de Fougerolles A, et al. Alpha1beta1 integrin is crucial for accumulation of epidermal T cells and the development of psoriasis. Nat Med (2007) 13:836–42. doi: 10.1038/nm1605
59. Ray SJ, Franki SN, Pierce RH, Dimitrova S, Koteliansky V, Sprague AG, et al. The collagen binding alpha1beta1 integrin VLA-1 regulates CD8 T cell-mediated immune protection against heterologous influenza infection. Immunity (2004) 20:167–79. doi: 10.1016/s1074-7613(04)00021-4
60. Richter MV, Topham DJ. The alpha1beta1 integrin and TNF receptor II protect airway CD8+ effector T cells from apoptosis during influenza infection. J Immunol (2007) 179:5054–63. doi: 10.4049/jimmunol.179.8.5054
61. Meharra EJ, Schön M, Hassett D, Parker C, Havran W, Gardner H. Reduced gut intraepithelial lymphocytes in VLA1 null mice. Cell Immunol (2000) 201:1–5. doi: 10.1006/cimm.2000.1630
62. Cheuk S, Schlums H, Gallais Sérézal I, Martini E, Chiang SC, Marquardt N, et al. CD49a expression defines tissue-resident CD8(+) T cells poised for cytotoxic function in human skin. Immunity (2017) 46:287–300. doi: 10.1016/j.immuni.2017.01.009
63. Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol (2014) 32:659–702. doi: 10.1146/annurev-immunol-032713-120145
64. Kumar BV, Ma W, Miron M, Granot T, Guyer RS, Carpenter DJ, et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep (2017) 20:2921–34. doi: 10.1016/j.celrep.2017.08.078
65. Zaid A, Hor JL, Christo SN, Groom JR, Heath WR, Mackay LK, et al. Chemokine receptor-dependent control of skin tissue-resident memory T cell formation. J Immunol (2017) 199:2451–9. doi: 10.4049/jimmunol.1700571
66. Kumar BV, Kratchmarov R, Miron M, Carpenter DJ, Senda T, Lerner H, et al. Functional heterogeneity of human tissue-resident memory T cells based on dye efflux capacities. JCI Insight (2018) 3(22). doi: 10.1172/jci.insight.123568
67. Pichyangkul S, Spring MD, Yongvanitchit K, Kum-Arb U, Limsalakpetch A, Im-Erbsin R, et al. Chemoprophylaxis with sporozoite immunization in p. knowlesi rhesus monkeys confers protection and elicits sporozoite-specific memory T cells in the liver. PloS One (2017) 12:e0171826. doi: 10.1371/journal.pone.0171826
68. Geissmann F, Cameron TO, Sidobre S, Manlongat N, Kronenberg M, Briskin MJ, et al. Intravascular immune surveillance by CXCR6+ NKT cells patrolling liver sinusoids. PloS Biol (2005) 3:e113. doi: 10.1371/journal.pbio.0030113
69. Milner JJ, Goldrath AW. Transcriptional programming of tissue-resident memory CD8(+) T cells. Curr Opin Immunol (2018) 51:162–9. doi: 10.1016/j.coi.2018.03.017
70. Mackay LK, Minnich M, Kragten NA, Liao Y, Nota B, Seillet C, et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science (2016) 352:459–63. doi: 10.1126/science.aad2035
71. Bird L. Lymphocyte responses: Hunker down with HOBIT and BLIMP1. Nat Rev Immunol (2016) 16:338–9. doi: 10.1038/nri.2016.61
72. Förster R, Davalos-Misslitz AC, Rot A. CCR7 and its ligands: balancing immunity and tolerance. Nat Rev Immunol (2008) 8:362–71. doi: 10.1038/nri2297
73. Park S, Park J, Kim E, Lee Y. The Capicua/ETS translocation variant 5 axis regulates liver-resident memory CD8(+) T-cell development and the pathogenesis of liver injury. Hepatology (2019) 70:358–71. doi: 10.1002/hep.30594
74. Behr FM, Chuwonpad A, Stark R, van Gisbergen K. Armed and ready: Transcriptional regulation of tissue-resident memory CD8 T cells. Front Immunol (2018) 9:1770. doi: 10.3389/fimmu.2018.01770
75. Hombrink P, Helbig C, Backer RA, Piet B, Oja AE, Stark R, et al. Programs for the persistence, vigilance and control of human CD8(+) lung-resident memory T cells. Nat Immunol (2016) 17:1467–78. doi: 10.1038/ni.3589
76. Milner JJ, Toma C, Yu B, Zhang K, Omilusik K, Phan AT, et al. Runx3 programs CD8(+) T cell residency in non-lymphoid tissues and tumours. Nature (2017) 552:253–7. doi: 10.1038/nature24993
77. Kragten NAM, Behr FM, Vieira Braga FA, Remmerswaal EBM, Wesselink TH, Oja AE, et al. Blimp-1 induces and hobit maintains the cytotoxic mediator granzyme b in CD8 T cells. Eur J Immunol (2018) 48:1644–62. doi: 10.1002/eji.201847771
78. Pan Y, Tian T, Park CO, Lofftus SY, Mei S, Liu X, et al. Survival of tissue-resident memory T cells requires exogenous lipid uptake and metabolism. Nature (2017) 543:252–6. doi: 10.1038/nature21379
79. Frizzell H, Fonseca R, Christo SN, Evrard M, Cruz-Gomez S, Zanluqui NG, et al. Organ-specific isoform selection of fatty acid-binding proteins in tissue-resident lymphocytes. Sci Immunol (2020) 5(46):eaay9283. doi: 10.1126/sciimmunol.aay9283
80. Li C, Zhu B, Son YM, Wang Z, Jiang L, Xiang M, et al. The transcription factor Bhlhe40 programs mitochondrial regulation of resident CD8(+) T cell fitness and functionality. Immunity (2019) 51:491–507.e7. doi: 10.1016/j.immuni.2019.08.013
81. Mackay LK, Wynne-Jones E, Freestone D, Pellicci DG, Mielke LA, Newman DM, et al. T-Box transcription factors combine with the cytokines TGF-β and IL-15 to control tissue-resident memory T cell fate. Immunity (2015) 43:1101–11. doi: 10.1016/j.immuni.2015.11.008
82. Horras CJ, Lamb CL, King AL, Hanley JR, Mitchell KA. Consequences of TCDD treatment on intra-hepatic lymphocytes during liver regeneration. J Immunotoxicol (2012) 9:359–67. doi: 10.3109/1547691x.2012.664577
83. Hijnen D, Knol EF, Gent YY, Giovannone B, Beijn SJ, Kupper TS, et al. CD8(+) T cells in the lesional skin of atopic dermatitis and psoriasis patients are an important source of IFN-γ, IL-13, IL-17, and IL-22. J Invest Dermatol (2013) 133:973–9. doi: 10.1038/jid.2012.456
84. Parga-Vidal L, Behr FM, Kragten NAM, Nota B, Wesselink TH, Kavazović I, et al. Hobit identifies tissue-resident memory T cell precursors that are regulated by eomes. Sci Immunol (2021) 6(62):eabg3533. doi: 10.1126/sciimmunol.abg3533
85. Sun R, Wu Y, Zhou H, Wu Y, Yang Z, Gu Y, et al. Eomes impedes durable response to tumor immunotherapy by inhibiting stemness, tissue residency, and promoting the dysfunctional state of intratumoral CD8(+) T cells. Front Cell Dev Biol (2021) 9:640224. doi: 10.3389/fcell.2021.640224
86. Boddupalli CS, Nair S, Gray SM, Nowyhed HN, Verma R, Gibson JA, et al. ABC Transporters and NR4A1 identify a quiescent subset of tissue-resident memory T cells. J Clin Invest (2016) 126:3905–16. doi: 10.1172/jci85329
87. Odagiu L, Boulet S, Maurice De Sousa D, Daudelin JF, Nicolas S, Labrecque N. Early programming of CD8(+) T cell response by the orphan nuclear receptor NR4A3. Proc Natl Acad Sci U.S.A. (2020) 117:24392–402. doi: 10.1073/pnas.2007224117
88. Kurd NS, He Z, Louis TL, Milner JJ, Omilusik KD, Jin W, et al. Early precursors and molecular determinants of tissue-resident memory CD8(+) T lymphocytes revealed by single-cell RNA sequencing. Sci Immunol (2020) 5(47):eaaz6894. doi: 10.1126/sciimmunol.aaz6894
89. Bottois H, Ngollo M, Hammoudi N, Courau T, Bonnereau J, Chardiny V, et al. KLRG1 and CD103 expressions define distinct intestinal tissue-resident memory CD8 T cell subsets modulated in crohn’s disease. Front Immunol (2020) 11:896. doi: 10.3389/fimmu.2020.00896
90. Fonseca R, Beura LK, Quarnstrom CF, Ghoneim HE, Fan Y, Zebley CC, et al. Developmental plasticity allows outside-in immune responses by resident memory T cells. Nat Immunol (2020) 21:412–21. doi: 10.1038/s41590-020-0607-7
91. Behr FM, Parga-Vidal L, Kragten NAM, van Dam TJP, Wesselink TH, Sheridan BS, et al. Tissue-resident memory CD8(+) T cells shape local and systemic secondary T cell responses. Nat Immunol (2020) 21:1070–81. doi: 10.1038/s41590-020-0723-4
92. Gaide O, Emerson RO, Jiang X, Gulati N, Nizza S, Desmarais C, et al. Common clonal origin of central and resident memory T cells following skin immunization. Nat Med (2015) 21:647–53. doi: 10.1038/nm.3860
93. Maru S, Jin G, Schell TD, Lukacher AE. TCR stimulation strength is inversely associated with establishment of functional brain-resident memory CD8 T cells during persistent viral infection. PloS Pathog (2017) 13:e1006318. doi: 10.1371/journal.ppat.1006318
94. Frost EL, Kersh AE, Evavold BD, Lukacher AE. Cutting edge: Resident memory CD8 T cells express high-affinity TCRs. J Immunol (2015) 195:3520–4. doi: 10.4049/jimmunol.1501521
95. Krummey SM, Chen CW, Guasch SA, Liu D, Wagener M, Larsen CP, et al. Enhanced requirement for TNFR2 in graft rejection mediated by low-affinity memory CD8+ T cells during heterologous immunity. J Immunol (2016) 197:2009–15. doi: 10.4049/jimmunol.1502680
96. Yoshizawa A, Bi K, Keskin DB, Zhang G, Reinhold B, Reinherz EL. TCR-pMHC encounter differentially regulates transcriptomes of tissue-resident CD8 T cells. Eur J Immunol (2018) 48:128–50. doi: 10.1002/eji.201747174
97. Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity (2007) 27:281–95. doi: 10.1016/j.immuni.2007.07.010
98. Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon ML, et al. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat Immunol (2013) 14:1294–301. doi: 10.1038/ni.2744
99. Herndler-Brandstetter D, Ishigame H, Shinnakasu R, Plajer V, Stecher C, Zhao J, et al. KLRG1(+) effector CD8(+) T cells lose KLRG1, differentiate into all memory T cell lineages, and convey enhanced protective immunity. Immunity (2018) 48:716–729.e8. doi: 10.1016/j.immuni.2018.03.015
100. Park SL, Zaid A, Hor JL, Christo SN, Prier JE, Davies B, et al. Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat Immunol (2018) 19:183–91. doi: 10.1038/s41590-017-0027-5
101. Feau S, Arens R, Togher S, Schoenberger SP. Autocrine IL-2 is required for secondary population expansion of CD8(+) memory T cells. Nat Immunol (2011) 12:908–13. doi: 10.1038/ni.2079
102. Burkett PR, Koka R, Chien M, Chai S, Boone DL, Ma A. Coordinate expression and trans presentation of interleukin (IL)-15Ralpha and IL-15 supports natural killer cell and memory CD8+ T cell homeostasis. J Exp Med (2004) 200:825–34. doi: 10.1084/jem.20041389
103. Becker TC, Wherry EJ, Boone D, Murali-Krishna K, Antia R, Ma A, et al. Interleukin 15 is required for proliferative renewal of virus-specific memory CD8 T cells. J Exp Med (2002) 195:1541–8. doi: 10.1084/jem.20020369
104. Mohammed J, Beura LK, Bobr A, Astry B, Chicoine B, Kashem SW, et al. Stromal cells control the epithelial residence of DCs and memory T cells by regulated activation of TGF-β. Nat Immunol (2016) 17:414–21. doi: 10.1038/ni.3396
105. Christo SN, Evrard M, Park SL, Gandolfo LC, Burn TN, Fonseca R, et al. Discrete tissue microenvironments instruct diversity in resident memory T cell function and plasticity. Nat Immunol (2021) 22:1140–51. doi: 10.1038/s41590-021-01004-1
106. Thompson EA, Darrah PA, Foulds KE, Hoffer E, Caffrey-Carr A, Norenstedt S, et al. Monocytes acquire the ability to prime tissue-resident T cells via IL-10-Mediated TGF-β release. Cell Rep (2019) 28:1127–1135.e4. doi: 10.1016/j.celrep.2019.06.087
107. Bergsbaken T, Bevan MJ, Fink PJ. Local inflammatory cues regulate differentiation and persistence of CD8(+) tissue-resident memory T cells. Cell Rep (2017) 19:114–24. doi: 10.1016/j.celrep.2017.03.031
108. Adachi T, Kobayashi T, Sugihara E, Yamada T, Ikuta K, Pittaluga S, et al. Hair follicle-derived IL-7 and IL-15 mediate skin-resident memory T cell homeostasis and lymphoma. Nat Med (2015) 21:1272–9. doi: 10.1038/nm.3962
109. Barata JT, Durum SK, Seddon B. Flip the coin: IL-7 and IL-7R in health and disease. Nat Immunol (2019) 20:1584–93. doi: 10.1038/s41590-019-0479-x
110. Sawa Y, Arima Y, Ogura H, Kitabayashi C, Jiang JJ, Fukushima T, et al. Hepatic interleukin-7 expression regulates T cell responses. Immunity (2009) 30:447–57. doi: 10.1016/j.immuni.2009.01.007
111. Waickman AT, Powell JD. mTOR, metabolism, and the regulation of T-cell differentiation and function. Immunol Rev (2012) 249:43–58. doi: 10.1111/j.1600-065X.2012.01152.x
112. Sowell RT, Marzo AL. Resident-memory CD8 T cells and mTOR: Generation, protection, and clinical importance. Front Immunol (2015) 6:38. doi: 10.3389/fimmu.2015.00038
113. Sowell RT, Rogozinska M, Nelson CE, Vezys V, Marzo AL. Cutting edge: Generation of effector cells that localize to mucosal tissues and form resident memory CD8 T cells is controlled by mTOR. J Immunol (2014) 193:2067–71. doi: 10.4049/jimmunol.1400074
114. Chmurzyńska A. The multigene family of fatty acid-binding proteins (FABPs): function, structure and polymorphism. J Appl Genet (2006) 47:39–48. doi: 10.1007/bf03194597
115. Chowdhury PS, Chamoto K, Kumar A, Honjo T. PPAR-induced fatty acid oxidation in T cells increases the number of tumor-reactive CD8(+) T cells and facilitates anti-PD-1 therapy. Cancer Immunol Res (2018) 6:1375–87. doi: 10.1158/2326-6066.Cir-18-0095
116. Borges da Silva H, Beura LK, Wang H, Hanse EA, Gore R, Scott MC, et al. The purinergic receptor P2RX7 directs metabolic fitness of long-lived memory CD8(+) T cells. Nature (2018) 559:264–8. doi: 10.1038/s41586-018-0282-0
117. Stark R, Wesselink TH, Behr FM, Kragten NAM, Arens R, Koch-Nolte F, et al. T (RM) maintenance is regulated by tissue damage via P2RX7. Sci Immunol (2018) 3(30):eaau1022. doi: 10.1126/sciimmunol.aau1022
118. Mühlbauer M, Fleck M, Schütz C, Weiss T, Froh M, Blank C, et al. PD-L1 is induced in hepatocytes by viral infection and by interferon-alpha and -gamma and mediates T cell apoptosis. J Hepatol (2006) 45:520–8. doi: 10.1016/j.jhep.2006.05.007
119. Maini MK, Schurich A. The molecular basis of the failed immune response in chronic HBV: Therapeutic implications. J Hepatol (2010) 52:616–9. doi: 10.1016/j.jhep.2009.12.017
120. Oberhardt V, Hofmann M, Thimme R, Neumann-Haefelin C. Adaptive immune responses, immune escape and immune-mediated pathogenesis during HDV infection. Viruses (2022) 14(2):198. doi: 10.3390/v14020198
121. Kefalakes H, Horgan XJ, Jung MK, Amanakis G, Kapuria D, Bolte FJ, et al. Liver-resident bystander CD8(+) T cells contribute to liver disease pathogenesis in chronic hepatitis d virus infection. Gastroenterology (2021) 161:1567–1583.e9. doi: 10.1053/j.gastro.2021.07.027
122. Zhou J, Peng H, Li K, Qu K, Wang B, Wu Y, et al. Liver-resident NK cells control antiviral activity of hepatic T cells via the PD-1-PD-L1 axis. Immunity (2019) 50:403–417.e4. doi: 10.1016/j.immuni.2018.12.024
123. Hunter S, Willcox CR, Davey MS, Kasatskaya SA, Jeffery HC, Chudakov DM, et al. Human liver infiltrating γδ T cells are composed of clonally expanded circulating and tissue-resident populations. J Hepatol (2018) 69:654–65. doi: 10.1016/j.jhep.2018.05.007
124. Cockburn IA, Seder RA. Malaria prevention: from immunological concepts to effective vaccines and protective antibodies. Nat Immunol (2018) 19:1199–211. doi: 10.1038/s41590-018-0228-6
125. Berenzon D, Schwenk RJ, Letellier L, Guebre-Xabier M, Williams J, Krzych U. Protracted protection to plasmodium berghei malaria is linked to functionally and phenotypically heterogeneous liver memory CD8+ T cells. J Immunol (2003) 171:2024–34. doi: 10.4049/jimmunol.171.4.2024
126. Tse SW, Cockburn IA, Zhang H, Scott AL, Zavala F. Unique transcriptional profile of liver-resident memory CD8+ T cells induced by immunization with malaria sporozoites. Genes Immun (2013) 14:302–9. doi: 10.1038/gene.2013.20
127. Butler NS, Schmidt NW, Vaughan AM, Aly AS, Kappe SH, Harty JT. Superior antimalarial immunity after vaccination with late liver stage-arresting genetically attenuated parasites. Cell Host Microbe (2011) 9:451–62. doi: 10.1016/j.chom.2011.05.008
128. Tran TM, Li S, Doumbo S, Doumtabe D, Huang CY, Dia S, et al. An intensive longitudinal cohort study of malian children and adults reveals no evidence of acquired immunity to plasmodium falciparum infection. Clin Infect Dis (2013) 57:40–7. doi: 10.1093/cid/cit174
129. Ishizuka AS, Lyke KE, DeZure A, Berry AA, Richie TL, Mendoza FH, et al. Protection against malaria at 1 year and immune correlates following PfSPZ vaccination. Nat Med (2016) 22:614–23. doi: 10.1038/nm.4110
130. Rodrigues V, Cordeiro-da-Silva A, Laforge M, Silvestre R, Estaquier J. Regulation of immunity during visceral leishmania infection. Parasit Vectors (2016) 9:118. doi: 10.1186/s13071-016-1412-x
131. Rodrigues A, Claro M, Alexandre-Pires G, Santos-Mateus D, Martins C, Valério-Bolas A, et al. Leishmania infantum antigens modulate memory cell subsets of liver resident T lymphocyte. Immunobiology (2017) 222:409–22. doi: 10.1016/j.imbio.2016.08.009
132. Modabberi F, Ghadimi SN, Shahriarirad R, Nadimi E, Karbalay-Doust S, Rashidi S, et al. Stereological analysis of liver, spleen and bone of leishmania infantum-experimentally infected hamsters. Exp Parasitol (2021) 228:108137. doi: 10.1016/j.exppara.2021.108137
133. Zayats R, Uzonna JE, Murooka TT. Visualizing the In vivo dynamics of anti-leishmania immunity: Discoveries and challenges. Front Immunol (2021) 12:671582. doi: 10.3389/fimmu.2021.671582
134. Zimmer CL, von Seth E, Buggert M, Strauss O, Hertwig L, Nguyen S, et al. A biliary immune landscape map of primary sclerosing cholangitis reveals a dominant network of neutrophils and tissue-resident T cells. Sci Transl Med (2021) 13(599):eabb3107. doi: 10.1126/scitranslmed.abb3107
135. Hansel C, Erschfeld S, Baues M, Lammers T, Weiskirchen R, Trautwein C, et al. The inhibitory T cell receptors PD1 and 2B4 are differentially regulated on CD4 and CD8 T cells in a mouse model of non-alcoholic steatohepatitis. Front Pharmacol (2019) 10:244. doi: 10.3389/fphar.2019.00244
136. Diedrich T, Kummer S, Galante A, Drolz A, Schlicker V, Lohse AW, et al. Characterization of the immune cell landscape of patients with NAFLD. PloS One (2020) 15:e0230307. doi: 10.1371/journal.pone.0230307
137. Her Z, Tan JHL, Lim YS, Tan SY, Chan XY, Tan WWS, et al. CD4(+) T cells mediate the development of liver fibrosis in high fat diet-induced NAFLD in humanized mice. Front Immunol (2020) 11:580968. doi: 10.3389/fimmu.2020.580968
138. Fuchs A, Samovski D, Smith GI, Cifarelli V, Farabi SS, Yoshino J, et al. Associations among adipose tissue immunology, inflammation, exosomes and insulin sensitivity in people with obesity and nonalcoholic fatty liver disease. Gastroenterology (2021) 161:968–981.e12. doi: 10.1053/j.gastro.2021.05.008
139. Harley IT, Stankiewicz TE, Giles DA, Softic S, Flick LM, Cappelletti M, et al. IL-17 signaling accelerates the progression of nonalcoholic fatty liver disease in mice. Hepatology (2014) 59:1830–9. doi: 10.1002/hep.26746
140. Conroy MJ, Galvin KC, Doyle SL, Kavanagh ME, Mongan AM, Cannon A, et al. Parallel profiles of inflammatory and effector memory T cells in visceral fat and liver of obesity-associated cancer patients. Inflammation (2016) 39:1729–36. doi: 10.1007/s10753-016-0407-2
141. Williams M, Liu X, Zhang Y, Reske J, Bahal D, Gohl TG, et al. NCOA5 deficiency promotes a unique liver protumorigenic microenvironment through p21(WAF1/CIP1) overexpression, which is reversed by metformin. Oncogene (2020) 39:3821–36. doi: 10.1038/s41388-020-1256-x
142. Ma J, Zheng B, Goswami S, Meng L, Zhang D, Cao C, et al. PD1(Hi) CD8(+) T cells correlate with exhausted signature and poor clinical outcome in hepatocellular carcinoma. J Immunother Cancer (2019) 7:331. doi: 10.1186/s40425-019-0814-7
143. Okła K, Farber DL, Zou W. Tissue-resident memory T cells in tumor immunity and immunotherapy. J Exp Med (2021) 218(4):e20201605. doi: 10.1084/jem.20201605
144. Zakeri N, Hall A, Swadling L, Pallett LJ, Schmidt NM, Diniz MO, et al. Characterisation and induction of tissue-resident gamma delta T-cells to target hepatocellular carcinoma. Nat Commun (2022) 13:1372. doi: 10.1038/s41467-022-29012-1
145. Barsch M, Salié H, Schlaak AE, Zhang Z, Hess M, Mayer LS, et al. T Cell exhaustion and residency dynamics inform clinical outcomes in hepatocellular carcinoma. J Hepatol (2022) 77(2):397–409. doi: 10.1016/j.jhep.2022.02.032
146. Turner DL, Gordon CL, Farber DL. Tissue-resident T cells, in situ immunity and transplantation. Immunol Rev (2014) 258:150–66. doi: 10.1111/imr.12149
147. Fu J, Sykes M. Emerging concepts of tissue-resident memory T cells in transplantation. Transplantation (2021) 106(6):1132–42. doi: 10.1097/tp.0000000000004000
148. Taner T, Gustafson MP, Hansen MJ, Park WD, Bornschlegl S, Dietz AB, et al. Donor-specific hypo-responsiveness occurs in simultaneous liver-kidney transplant recipients after the first year. Kidney Int (2018) 93:1465–74. doi: 10.1016/j.kint.2018.01.022
149. Snyder ME, Finlayson MO, Connors TJ, Dogra P, Senda T, Bush E, et al. Generation and persistence of human tissue-resident memory T cells in lung transplantation. Sci Immunol (2019) 4(33):eaav5581. doi: 10.1126/sciimmunol.aav5581
Keywords: tissue-resident memory T cells, liver, chronic hepatitis B virus infection, malaria, autoimmune hepatitis, nonalcoholic fatty liver disease, hepatocellular carcinoma
Citation: Li Y, You Z, Tang R and Ma X (2022) Tissue-resident memory T cells in chronic liver diseases: Phenotype, development and function. Front. Immunol. 13:967055. doi: 10.3389/fimmu.2022.967055
Received: 12 June 2022; Accepted: 22 August 2022;
Published: 12 September 2022.
Edited by:
Martin Rottenberg, Karolinska Institutet (KI), SwedenReviewed by:
Hannah Kristin Drescher, Massachusetts General Hospital and Harvard Medical School, United StatesCopyright © 2022 Li, You, Tang and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiong Ma, bWF4aW9uZ21kQGhvdG1haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.