
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 16 August 2022
Sec. Cancer Immunity and Immunotherapy
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.955091
Background: Ovarian-tumor (OTU) domain-containing protein 6B (OTUD6B), one of newly identified OTU deubiquitylating enzyme families, is proved to be associated with tumor progression. However, whether it plays a key role in pan-cancer still remains unknown.
Methods: The profiles of OTUD6B expression in multiple cancers were analyzed using The Cancer Genome Atlas (TCGA) database. Information of protein expression was performed based on the HPA, GeneCards, and String databases. K-M plotter and survival data analysis were used to analyze the prognostic value of OTUD6B expression, including overall survival (OS), disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI). R package “clusterProfiler” was used for enrichment analysis of OTUD6B. Furthermore, we analyzed the correlation between the expression of OTUD6B, immune infiltration, and immune-related genes. Additionally, we preliminarily validated its tumorigenic effect in lung cancer cell lines.
Findings: OTUD6B expression was upregulated in most cancers, such as COAD, CHOL, and LUAD, and predicted poor prognosis in most cancers in TCGA. Results showed that OTUD6B expression was positively correlated with memory CD4+ T cells, Th1 CD4+ T cells, and CD8+ T cells. In terms of the immune-related genes, OTUD6B was found to be associated with most types of genes, such as immunostimulatory genes KDR, TGFBR1, and IL-10. Moreover, for most types of tumors, the immune score was found to be negatively correlated with OTUD6B expression. In addition, lung cancer cell lines with OTUD6B knockdown significantly inhibited proliferation and invasion ability of lung cancer cells.
Conclusions: The study indicated that OTUD6B is an oncogene and may serve as a new potential biomarker in various tumors. OTUD6B may play a part in TIME, which could be applied as a new target for cancer therapy.
Cancers are formidable in their breadth and scope of diversity. For decades, the primary focus of cancer research has been on malignant cancer cells, attempting to understand the carcinogenic mechanism of dominant oncogenes whose activation leads to the transformation from normal cells into cancer cells (1). Despite the fact that oncogenes are the core driving force of tumor development (2), the tumor microenvironment (TME), especially tumor immune microenvironment (TIME), is increasingly recognized as an important influence on tumor development.
Previous studies have demonstrated the synergistic interaction between cancer cells and the tumor immune microenvironment (TIME) together, which leads to tumor progression, metastasis, and even resistance to treatment (2–4). Moreover, cancer immunotherapy, especially immune checkpoint inhibitors (ICIs), has been shown to improve survival in cancer patients in recent years (5). Unfortunately, some patients are not sensitive to ICI treatment, because cancer cells have also developed sophisticated ways to evade from the immune system, which is a major obstacle to immunotherapy (6). Furthermore, the efficacy of immunotherapy is generally associated with the TIME. Hence, a powerful gene that plays key role in the TIME and tumor progression needs to be identified.
DUBs (deubiquitylating enzymes) contain enzymes that show linkage specificity when recognizing and cutting the diubiquitin moieties. DUBs have been proved to be active in diverse mechanisms to regulate a variety of cellular signalings, such as DNA repair and innate immune signaling pathways, which have been implicated in human diseases, including cancer (7, 8). OTUs, one of the most important members of DUBs, have been proved to modulate cellular cascade signaling and shown to be closely related to inflammation and cancer (9, 10). Moreover, quite a few OTUs such as DUBA, OTUB1, OTUD1, and OTUD5 are involved in the regulation of the TIME and cancer immunotherapy. For instance, OTUD5 was illustrated to positively regulate STING-mediated antitumor immune responses via deubiquitylation and maintain its stability (11). The tumor immunosuppression induced by PD-L1 has been implicated in OTUB1 (12). To date, only a few of studies have reported the role played by ovarian-tumor (OTU) domain-containing protein 6B (OTUD6B), the important one of the deubiquitylating enzyme family. Xinxin Liu (13) et al. reported that a low level of OTUD6B predicts poor survival and is correlated with a high recurrence rate in hepatocellular carcinoma. Meanwhile, in the present study we found that OTUD6B was closely associated with OTUB1, implying its potential role in immune response. Despite the fact that some researchers are illuminating the emerging functions of OTUD6B, most of the function and mechanism of OTUD6B remain elusive, especially its potential correlation with immune response; therefore, more research is needed to uncover the essential roles for OTUD6B in tumor development and progression.
In the present study, we evaluated the expression of OTUD6B in various cancers, as well as its role of prognosis and immunity value using multiple public databases. Furthermore, we explored the association between OTUD6B level and TMB, MSI, TME, immune checkpoint genes, immune infiltration cells, immune activating and immunosuppressive genes, chemokines, and chemokine receptors. What is more, we also conducted experiments in lung cancer cell lines to validate its oncogenic effect. Our results provide new insights into the remarkable functional role of OTUD6B and can be used as a potential target for future therapeutic development in various cancers.
RNA sequences and associated clinical data (including 33 types of cancer) were obtained from TCGA database using UCSC Xena (https://xena.ucsc.edu/). Gene mapping data of normal human tissue were downloaded from Genotype-Tissue Expression (GTEx) (https://commonfund.nih.gov/GTEx). Gene mutation and copy number alteration profiles of OTUD6B in pan-cancer were explored in cBioPortal for cancer genomics (http://www.cbioportal.org). We also downloaded the Simple Nucleotide Variation and Copy Number Variation dataset at level 4 of all TCGA samples from GDC (https://portal.gdc.cancer.gov/), and we integrated the samples’ mutation data, copy number data, and gene expression data of the samples. Protein domain information was obtained from the R package maftools (version 2.2.10). Box plots were developed by using the R package “ggplot2”.
The protein expression levels of OTUD6B in human tumors and normal tissues were acquired from the Human Protein Atlas database (HPA: https://www.proteinatlas.org/). The protein–protein interaction (PPI) network map of OTUD6B was conducted using the STRING (https://string-db.org/) open database. The subcellular localization information of OTUD6B was downloaded from GeneCards (https://www.genecards.org/).
Kaplan–Meier survival analysis was performed here to find different overall survival outcomes in TCGA cohort with high and low OTUD6B expressions, and it was realized using the Kaplan–Meier plotter (https://kmplot.com/analysis/). Univariate Cox regression analysis was performed to evaluate the prognosis value of OTUD6B in predicting OS, disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI) in pan-cancer. Forest plots for Cox regression were developed with the R packages “survival” and “forestplot”.
The TME is a microenvironment composed of different elements, such as tumor cells, immune cells surrounding tumor cells, and stromal cells, which is essential for the survival and progression of tumor cells. The intercellular equilibrium in the microenvironment affects the proliferation of tumor cells. We used the R software package ESTIMATE (version 1.0.13) to calculate the stromal and immune scores in each tumor based on gene expression in different cancers. The Tumor Immune Evaluation Resource (TIMER2) database (http://timer.comp-genomics.org/) was used to elucidate the association of OTUD6B levels with macrophage, CD4+ T-cell, and CD8+ T-cell infiltration. We analyzed the association of OTUD6B expression with immune-related genes, including those encoding chemokines (41), chemokine receptor proteins (14), major histocompatibility complex (MHC) (15), and immunoinhibitory (16) and immunostimulatory (46) genes. The Pearson correlation coefficient was also calculated, and the heat map of each type of cancer was displayed. Next, we extracted the OTUD6B gene and immune checkpoint pathway gene expression data from TCGA and GTEx (17) and then calculated the Pearson correlation between OTUD6B and 60 immune checkpoint-related genes.
MSI refers to the phenomenon that new microsatellite alleles appear at a certain microsatellite locus in tumors compared with normal tissues due to the insertion or deletion of repeat units. TMB refers to the number of somatic non-synonymous mutations in a specific genomic region. TMB can indirectly reflect the ability and degree of tumor production of neoantigens and predict the efficacy of immunotherapy for various tumors. They are both potential biomarkers for the efficacy prediction of immune checkpoint inhibitors (ICIs) (18, 19). We downloaded the standardized pan-cancer dataset from the UCSC database and extracted the expression data of the OTUD6B gene in each sample. We used the R package maftools (version 2.8.05) of the TMB and MSI and calculated the Pearson correlation of TMB and MSI for each tumor.
Gene set enrichment analysis (GSEA) was performed to investigate the potential biological pathways that OTU6B may be involved in pan-cancer. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways of this signature were mapped by using R packages “GSVA” and “clusterProfiler”. Different biological pathways of enrichment in different risk groups were identified.
Lung cancer cell lines A549 and Pc9 were cultured at 37°C in Dulbecco’s modified Eagle’s medium and RPMI-1640 with 10% fetal bovine serum (FBS), containing 1% penicillin and streptomycin, respectively.
Briefly, a 24-well Transwell plate was used to perform the cell migration assay. An 8-mm polyethylene terephthalate membrane filter (Corning) separates the lower and upper chambers, and cells were plated in the upper chamber at 2 × 104 cells per well in serum-free DMEM or RPMI-1640 medium. After 24 h of growing in a chamber with an atmosphere of 5% CO2 at 37°C, 4% formaldehyde was used to fixed the lower migrant cells for 20 min, followed by staining with crystal violet for another 10 min.
Human lung cancer tumor tissues were obtained from the Department of Thoracic Surgery, The First Affiliated Hospital of Xi’an Jiaotong University. Informed and signed contents were obtained from each patient. The samples were de-identified before being subjected to the described experiments in this study.
Cells were transfected with indicated constructs according to the manufacturer’s protocol using Lipofectamine 8000. The sequence for the control siRNA (NC) was as follows: 5′-UUCUCCGAACGUGUCACGU TT T-3′. The sequences for OTUD6B siRNA were as follows: 5′-GCUGACUACUAAGGAGAAUAATT-3′ and 5′-AAGGAGCGAGAAGAACGGAUA-3′. Western blot was performed to verify the transfection downregulating the level of OTUD6B protein.
The association between drug sensitivity and OTUD6B expression was evaluated using GSCALite (http://bioinfo.life.hust.edu.cn/GSCA). The drug sensitivity was presented through the half-maximal inhibitory concentration (IC50). Moreover, R package “pRRophetic” was performed to predict the potential drug responses in accordance with OTUD6B expression.
We used data from the GTEx database to analyze the profiles of OTUD6B expression in different tissues of healthy people. As shown in the histogram in Figure 1A, the OTUD6B levels vary across different tissue types and are particularly high in skeletal muscle. The expression pattern of OTUD6B in pan-cancer was subsequently evaluated. Figure 1B and Supplemental Figure 1A present the OTUD6B expression from TIMER and TCGA databases, respectively. The results showed that high OTUD6B mRNA expression was observed in 10 tumors in both methods, including breast invasive carcinoma (BRCA), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), rectum adenocarcinoma (READ), and stomach adenocarcinoma (STAD) (Figure 1B, Supplemental Figure 1A). Among the abovementioned, the highest expression levels of OTUD6B were found in LUSC, READ, and COAD in TCGA database. By contrast, it revealed a low expression in kidney renal papillary cell carcinoma (KIRP), kidney renal clear cell carcinoma (KIRC), and thyroid carcinoma (THCA), relative to normal tissues (Figure 1B, Supplemental Figure 1A). Moreover, we analyzed the protein level of OTUD6B by using the Human Protein Atlas (HPA) database. In normal tissue, a high level of OTUD6B protein was found in the lung, small intestine, colon, and rectum tissue, while the protein level was low in the hippocampus, oral mucosa, and liver (Supplemental Figure 2A). The protein level of OTUD6B was highest in thyroid tumor but lowest in liver and lung cancer (Supplemental Figure 2B).
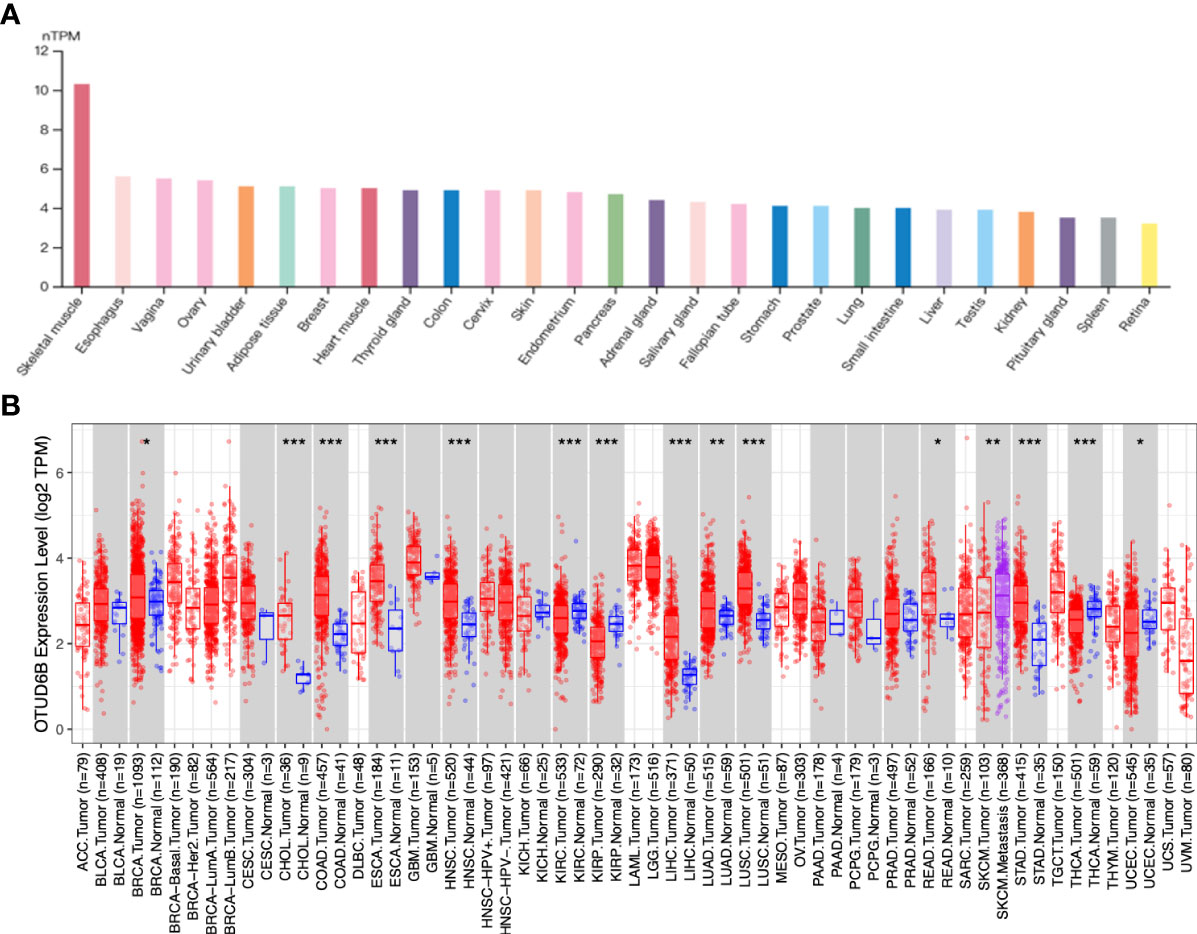
Figure 1 OTUD6B expression in various tissues and tumors. (A) The level of OTUD6B in different normal tissues from the HPA database. (B) The expression differences of OTUD6B between normal and tumor tissues from the TIMER database. (*p < 0.05; **p < 0.01; ***p < 0.001).
For paired tumors in TCGA, OTUD6B was upregulated in BLCA, BRCA, CHOL, COADREAD, ESCA, LIHC, HNSC, LUAD, LUSC, STAD, and OSCC, relative to adjacent normal tissues (Figures 2A–P); however, as indicated in Figures 2G, L it was downregulated in KIRC and THRA relative to adjacent normal tissues. Furthermore, we evaluated OTUD6B expression at different tumor stages according to the World Health Organization (WHO) and found that it was highly expressed in most advanced tumors including LUAD, ESAD, STAD, BLCA, BRCA, COADREAD, LIHC, and OSCC (Figures 3A–L). By contrast, a lower OTUD6B expression in advanced tumors was observed only in THCA (Figure 3K). From the abovementioned, we concluded that OTUD6B plays an important role in promoting tumor progression in most tumors.
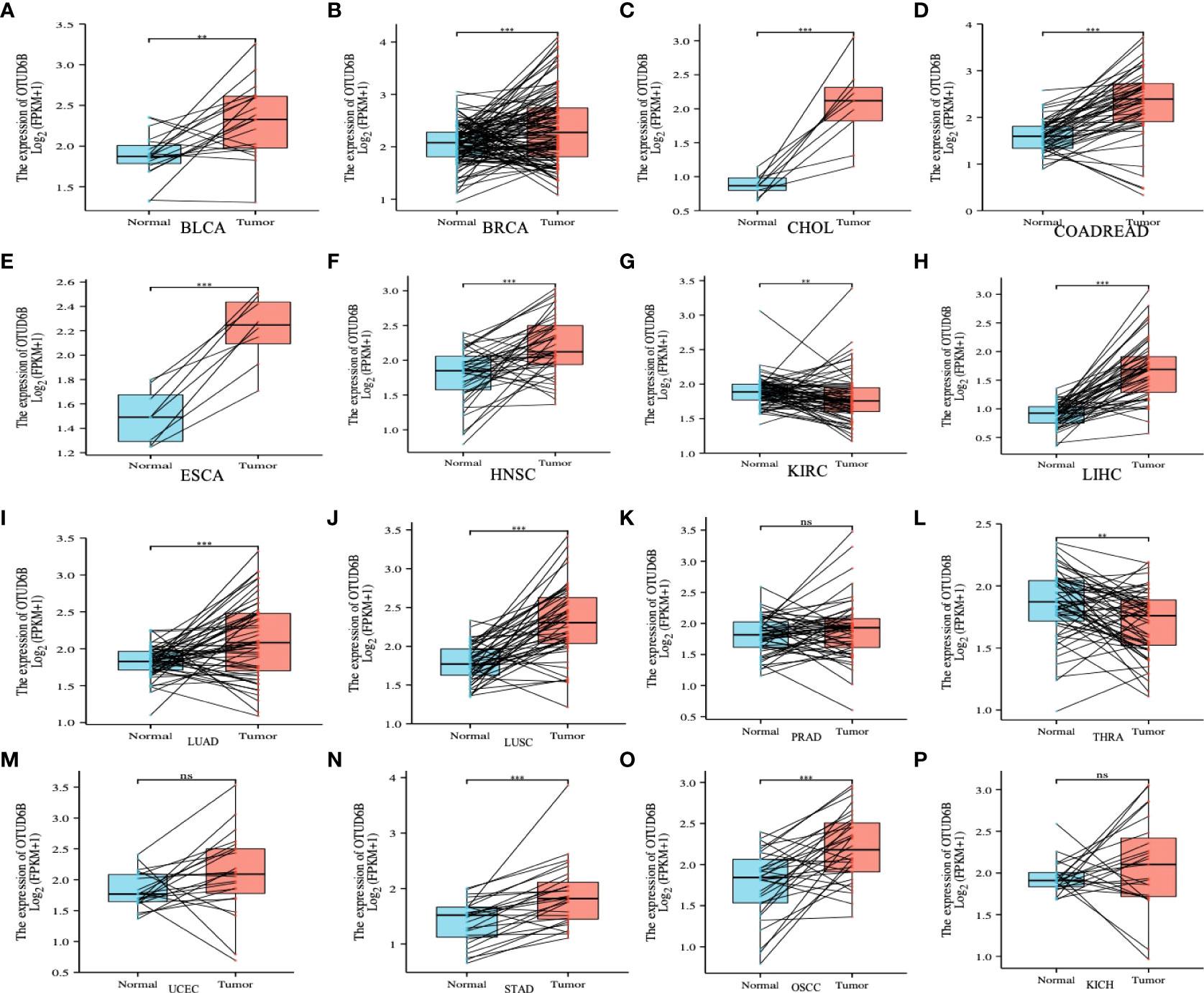
Figure 2 OTUD6B expression in pan-cancer. (A-P) Differential expression of OTUD6B in paired tumors and adjacent normal tissues from TCGA database (Ns, non-significant, **p < 0.01, ***p < 0.001).
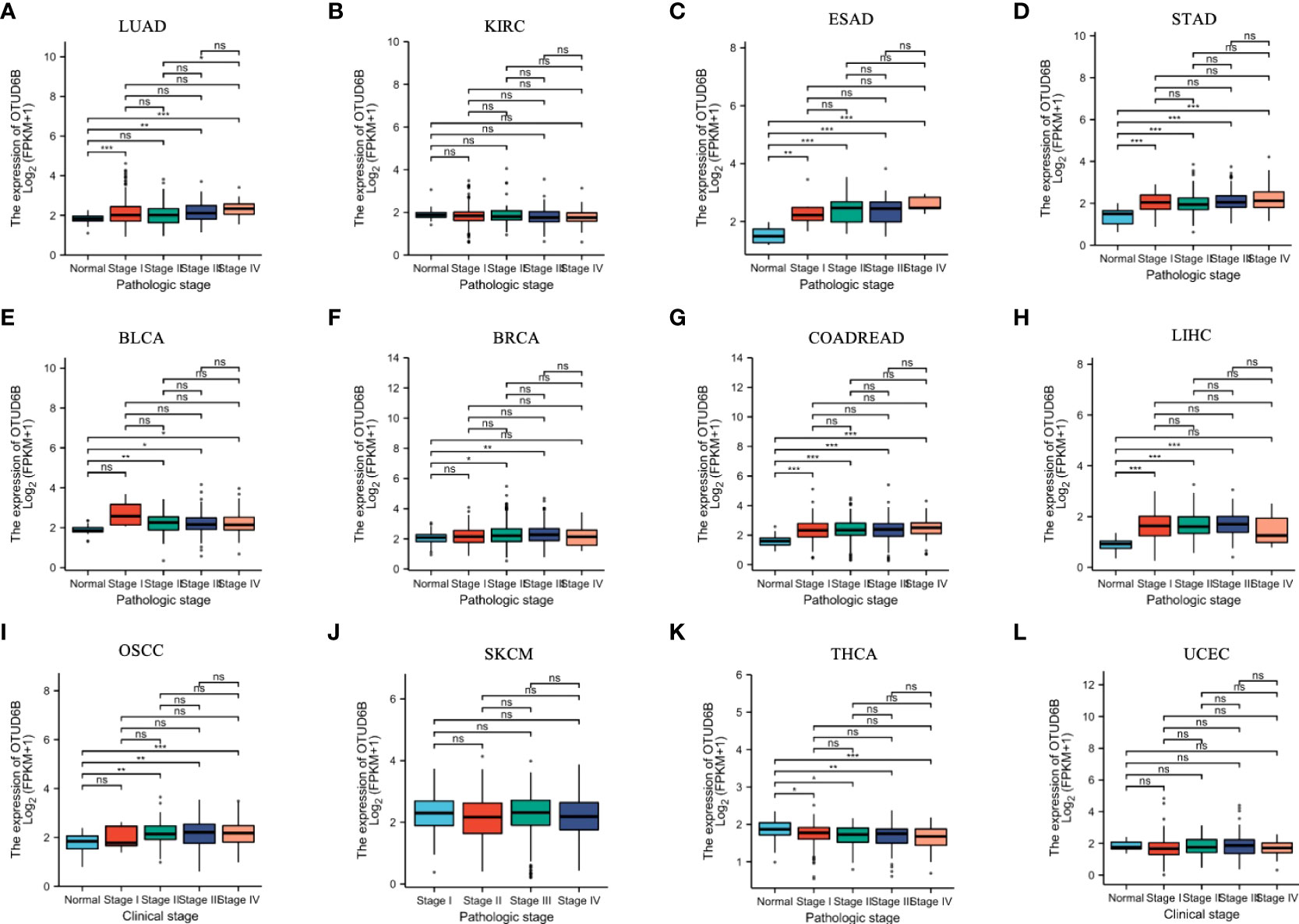
Figure 3 OTUD6B expression in different tumor stages. (A-L) Differential expression of OTUD6B in WHO stages in various tumors from TCGA database. (Ns, non-significant, *p < 0.05; **p < 0.01, ***p < 0.001).
As a newly identified OTU gene family member, we further used the GeneCards database to explore the subcellar location of OTUD6B for a better understanding of its function. OTUD6B is mainly distributed in the cytosol and nucleus, probably indicating a transfer or function between the two areas (Supplemental Figure 2C). Moreover, the PPI network of OTUD6B was constructed and we found that OTUD6B was closely associated with OTUB1, MYSM1, USP14, USP45, OTUD1, USP7, and UCHL3 proteins (Supplemental Figure 2C).
To further understand the structure and gene alteration of the OTUD6B gene, we used the cBioPortal database to explore the genetic alterations in OTUD6B and observed a high frequency of genetic alterations in patients with BRCA, UCS, PRAD, and LIHC, while patients with THCA, mesothelioma (MESO), KIRP, acute myeloid leukemia (ACC), and thymoma (THYM) possessed no gene alteration. The “amplification” type of CNA was the primary type in most cancers (Supplemental Figure 1B). The correlation between gene expression and copy number variation was also evaluated, and we observed significant differences in 16 tumors such as LGG (neutral = 490, gain = 14, loss = 4) (p = 0.04), CESC (neutral = 281, loss = 3, gain = 8) (p = 3.2e-3), and LUAD (neutral = 455, gain = 42, loss = 14) (p = 1.2e-3), as shown in Supplemental Figure 1C. The gene mutation landscape revealed that UCEC has the highest mutation frequency at 3.4%, followed by READ with a mutation rate of 2.2%. The predominant type of mutation in the OTU domain or other regions is “missense” mutation (Supplemental Figure 1D), indicating that this type of mutation may be responsible for gene function.
We then used Kaplan–Meier analysis to evaluate the overall survival (OS) of patients in TCGA pan-cancer. Our results showed that a high OTUD6B level was significantly associated with worse OS in patients with LUAD, BRCA, LIHC, THCA, and PAAD (Figures 4A, C, F, H, K). In contrast, a high OTUD6B expression was associated with better OS only in THYM (Figure 4L). While the OTUD6B expression showed no correlation in BLCA, UCEC, LUSC, STOD, HNSC and KIRC (Figures 4 B, D, E, G, I, J). Next, the forest map was generated by univariate Cox regression analysis of OS, disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI). The OS plot revealed that a high OTUD6B level was significantly linked to shorter OS times in patients with KICH, BRCA, GBM, UVM, LGG, LUAD, SRAC, MESO, and LIHC, and a better clinical outcome was observed in patients with KIRC (Figure 5A). DSS analysis showed that OTUD6B was associated with worse outcomes in patients with KICH, GBM, LGG, LUAD, UVM, MESO, and PRAD but was a protective factor in patients with KIRC only (Figure 5B). For DFI, a high OTUD6B expression was associated with significantly reduced DFI in patients with PAAD (Figure 5C).
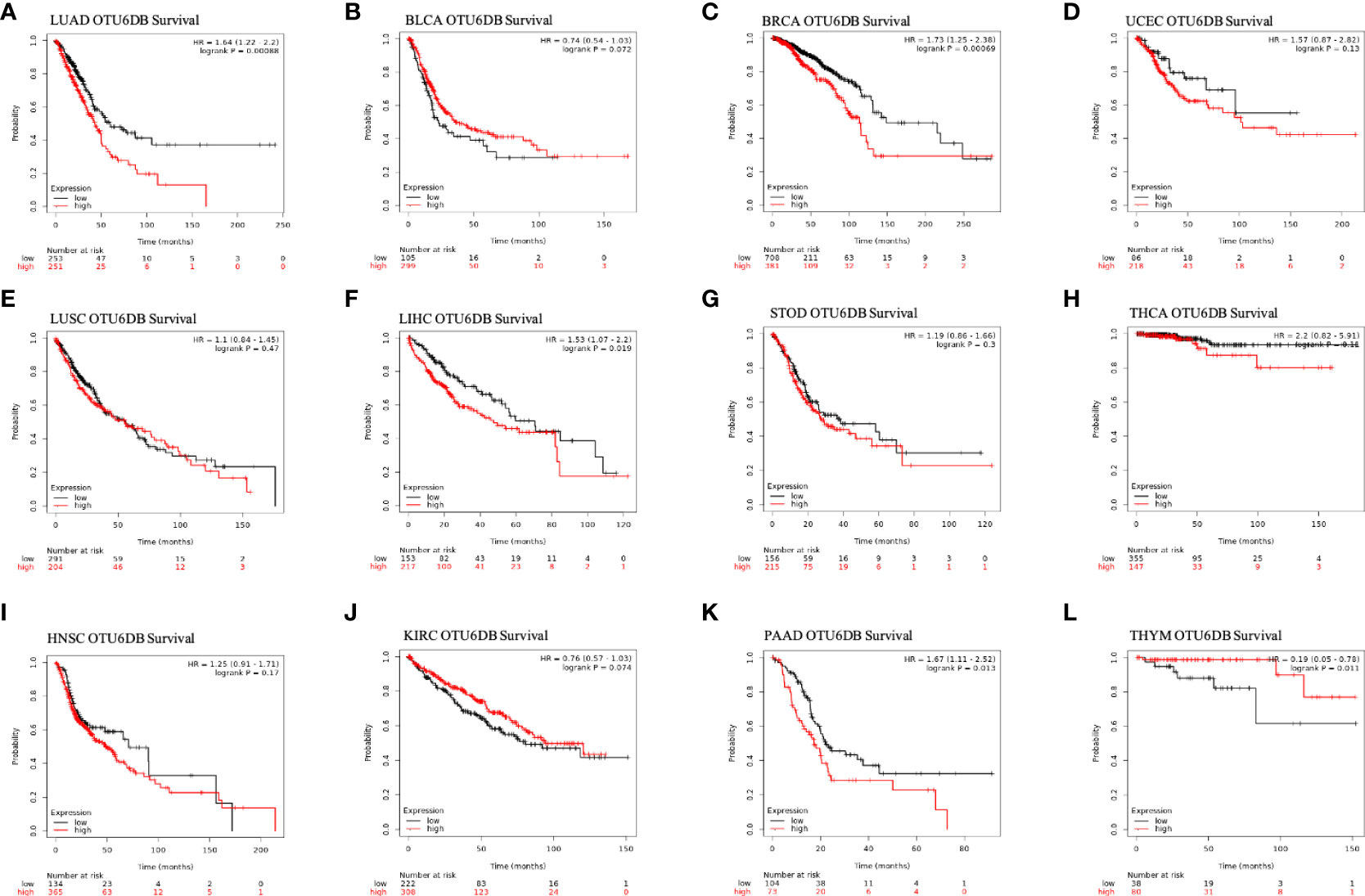
Figure 4 Overall survival and OTUD6B expression in the Kaplan–Meier database. (A-L) The relationship between OTUD6B levels and prognosis in indicated tumor types.
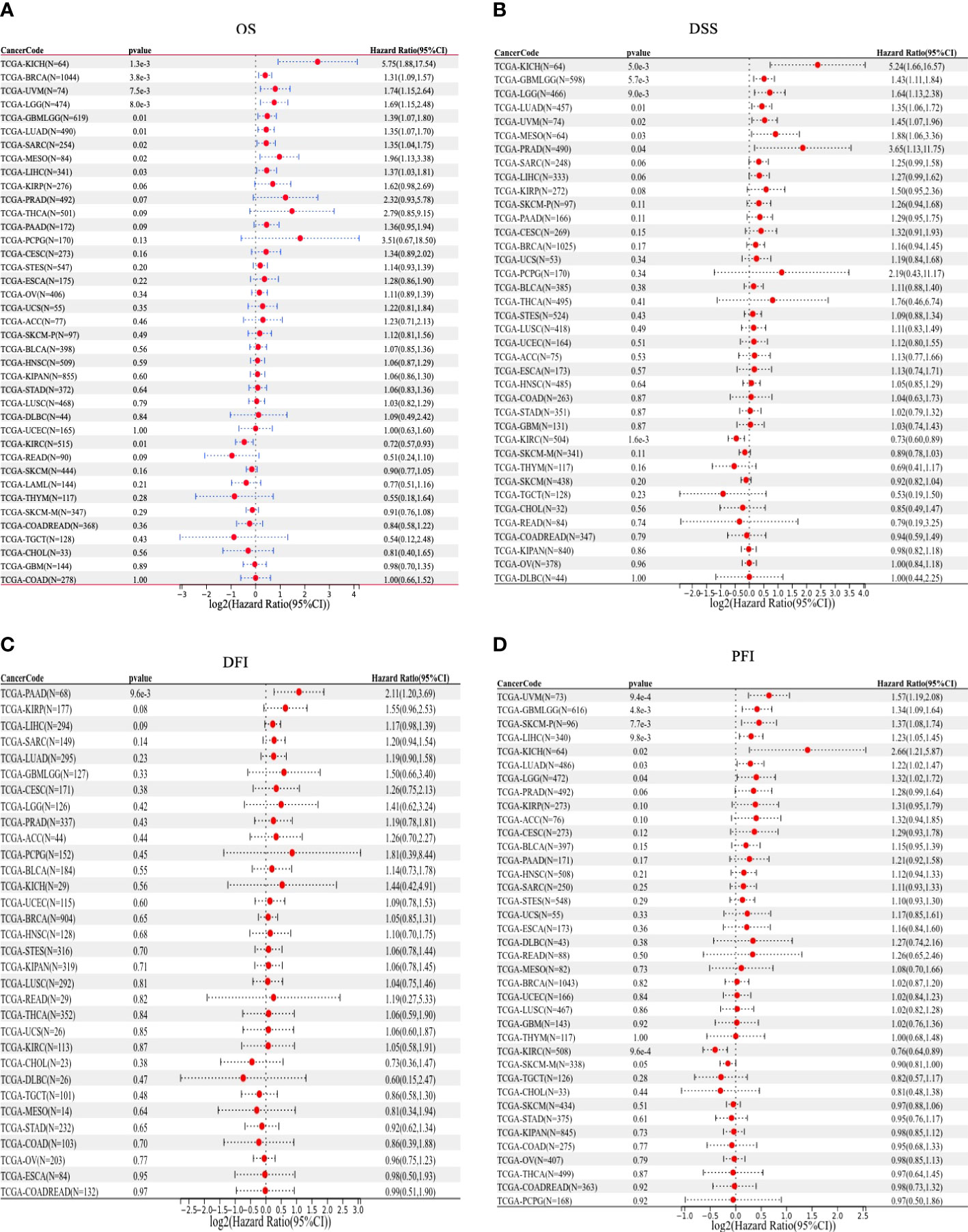
Figure 5 Forest plots showing results of univariate Cox regression analysis for (A) OS, (B) DSS, (C) DFI, and (D) PFI.
In addition, our results showed that a high OTUD6B level was significantly correlated with poor PFI in patients with UVM, GBM, SKCM, LIHC, KICH, LUAD, and LGG, while a low OTUD6B expression was correlated with better PFI in patients with KIRC (Figure 5D). These results indicated that OTUD6B is likely to be an oncogene in the majority of tumors.
To investigate the potential biological pathways in which OTUD6B may be involved in affecting tumor genesis and progression, we conducted GSEA in 33 tumors from TCGA. Eight tumors which are focused with significant enriched pathways were as presented in Figures 6A–H. We found that OTUD6B is involved in different enrichment pathways across these tumors and also plays the same functional role in the indicated tumors. The results showed that in PAAD, SKCM, and STAD, OTUD6B expression was significantly correlated with immune-related pathways, especially for the intestinal immune network (Figures 6B, D, E). Cycle-related pathways such as cytosolic DNA sensing and autophagy are closely associated with OTUD6B in LUSC and PAAD (Figures G, H). Interestingly, porphyrin and fatty acid metabolism was found correlated in LGG, PCPG, TGCT, and LGG (Figures 6A, E, F), probably meaning that OTUD6B may play roles in intracellular metal ion metabolism, such as iron metabolism.
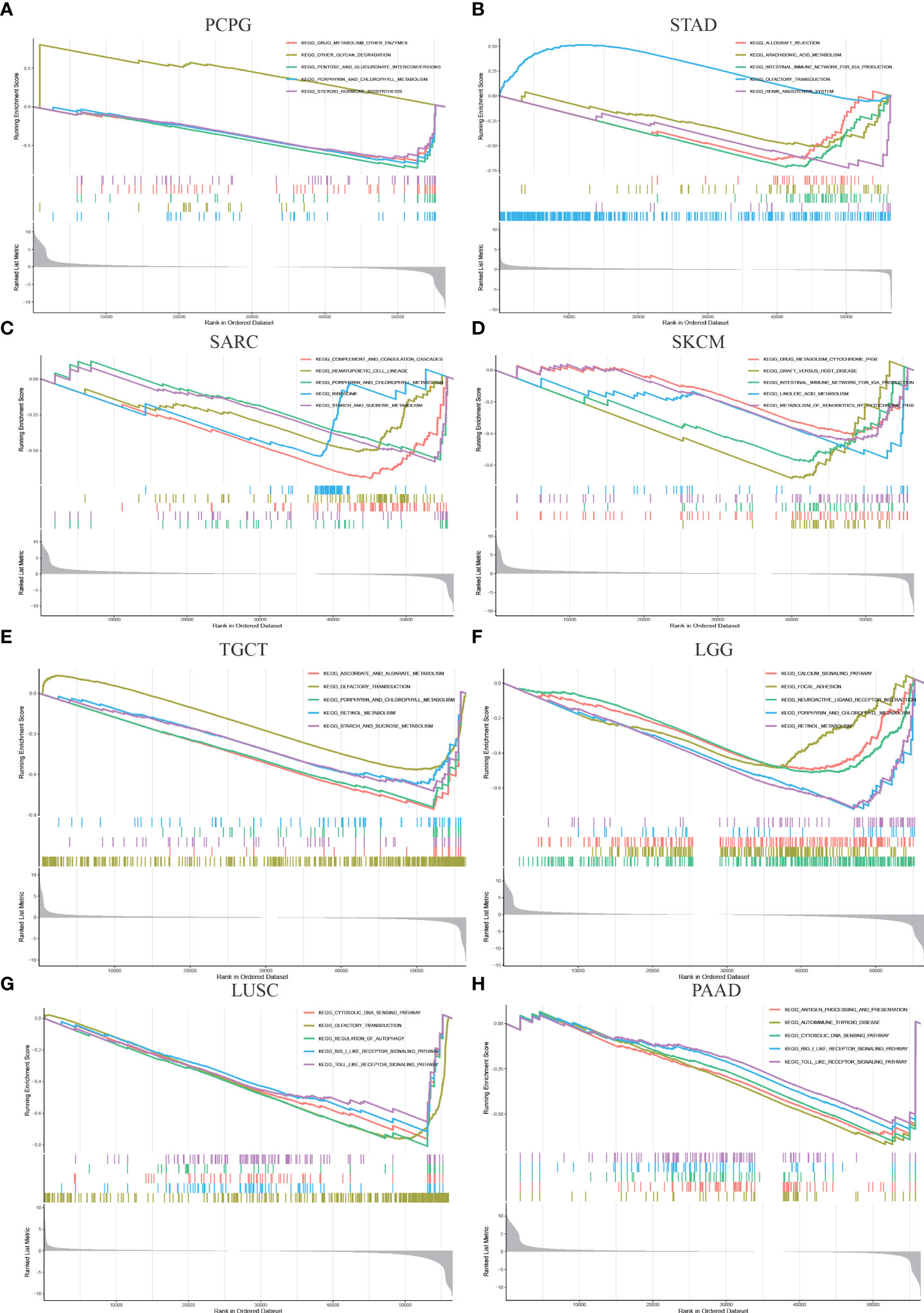
Figure 6 Gene enrichment analysis of OTUD6B in the KEGG database in PCPG, STAD, SARC, SKCM, TGCT, LUSC, LGG, and PAAD.
Due to immune dysfunction, immune cells cannot control tumor growth and may even promote the growth of tumors. We therefore performed immune correlation analysis through several immune-related databases. Correlation analyses using data from the TIMER2 database revealed that OTUD6B expression was positively associated with the level of M1-like macrophage infiltration in most tumor types while it was negatively correlated with M2-like macrophage. Interestingly, the results also showed that OTUD6B expression was positively correlated with memory CD4+ T cells, Th2 CD4+ T cells, and CD8+ T cells and negatively correlated with Th1 CD4+ T cells in TCGA pan-cancer (Figures 7A–C). Correlation analyses using data from the XCELL database showed that the OTUD6B level was significantly positively correlated with Th1 CD4+ T cells, CD8+ T cells, M1-like macrophage, and M2-like macrophage infiltration, while it was negatively linked with Th2 CD4+ T cells (Supplemental Figure 3A).
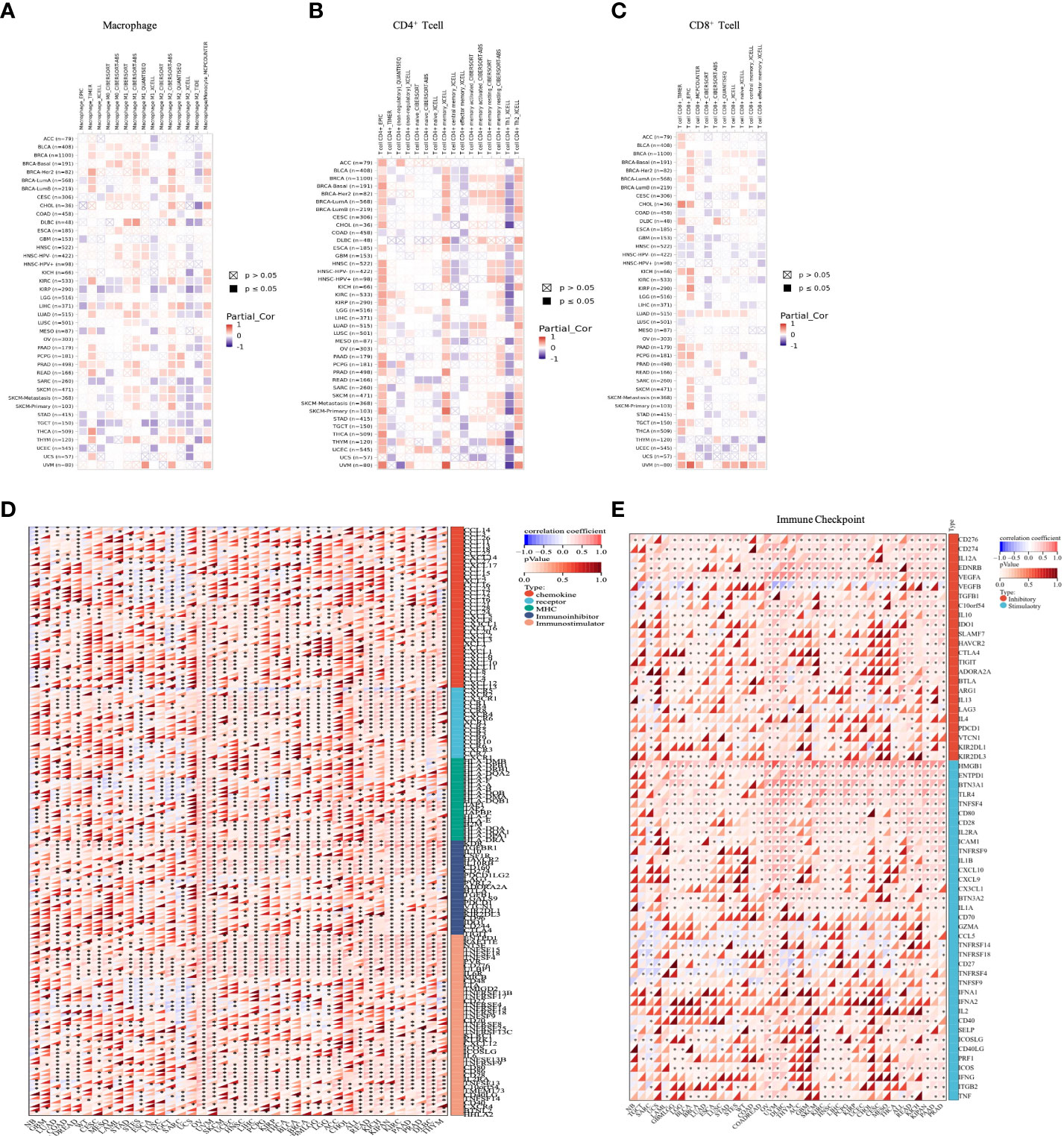
Figure 7 Analysis of different immune cell infiltrations. (A) Correlation between OTUD6B expression and macrophage, from the TIMER2 database. (B) Correlation between OTUD6B expression and CD4+ T cells from the TIMER2 database. (C) Correlation between OTUD6B expression and CD8+T cells from the TIMER2 database. (D) The heatmap reveals the correlation between immune-related gene expression such as chemokine genes, chemokine receptor genes, MHC, immunostimulatory genes, and immunoinhibitory renes. (E) Immune-checkpoint related genes. (*p < 0.05; **p < 0.01; ***p < 0.001 and ****p < 0.0001).
To further explore the association between OTUD6B expression and immune-related genes in pan-cancer, gene co-expression analyses of MHC genes, immunosuppressive and immunostimulant genes, chemokines, and chemokine receptors were conducted (Figure 7D). The results revealed a positive correlation between OTUD6B expression and these genes in most types of cancer, while in SARC, TGCT, and STES, a negative correlation was observed. In terms of immunostimulatory genes, OTUD6B was found to be associated with immunosuppressive marker genes, such as KDR, TGFBR1, and IL-10. This suggests that OTUD6B may be involved in immune regulation and possibly in the immune response.
To better explore the mechanism of OTUD6B involvement in immune regulation and its role in immune response, we investigated the relationship between OTUD6B and the immune-related gene. As presented in Figure 7E, we found that CD276 and VEGFA expression was significantly positively correlated with the expression of OTUD6B in nearly all types of cancer, so were the immune checkpoint inhibitor genes HMGB1 and TLR4, suggesting that OTUD6B may provide some help for tumor immunotherapy through these targets. Moreover, we analyzed the relationship between OTUD6B expression and RNA modifications including m1A, m5C, and m6A. A significant positive correlation between OTUD6B and the three types of RNA modification was found in almost all tumors, indicating the diversity of OTUD6B modification in tumors (Supplemental Figure 3B).
Moreover, correlations between OTUD6B and immune scores and stromal scores were measured; the results are presented in Figure 8 and Supplemental Figure 4. Immune scores were significantly associated with OTUD6B expression in 19 of 33 cancers, and stromal scores were significantly associated with OTUD6B expression in 12 of 33 cancers, respectively. In most types of tumor, the immune score was found to be negatively correlated with OTUD6B expression in UCEC, CESC, ESCA, LUAD, MESO, ACC, STES, SARC, LUSC, KIRP, STAD, THCA, THYM, SKCM, GBM, LGG, BLCA, HNSC, TGCT, and PCPG. On the other hand, THCA, BRCA, STAD, BLCA, LUSC, SKCM, TGCT, STES, and SARC were negatively correlated with OTUD6B expression in stromal scores. The stromal score was positively correlated with OTUD6B expression in LAML and KIRC (Supplemental Figure 4). Based on the results analyzed above, the immune score was consistent with analysis shown in supplemental Figure 3A showing that most tumors were negatively linked with common lymphoid progenitors.
Figure 9 displays the correlation of OTUD6B with TMB and MSI. OTUD6B expression was significantly associated with TMB in KICH, LUAD, STAD, OV, COADREAD, KIRP, and COAD. OTUD6B expression was positively related with KICH, LUAD, and STAD, while it was negatively related with the other five cancers. Among the abovementioned, KICH had the highest correlation coefficient with OTUD6B (Figure 9A). As indicated in Figure 9B, OTUD6B expression was significantly correlated with MSI in 10 out of 33 cancer types; it was positively associated with MSI in five (KIRC, SARC, CESC, STAD, LUSC) and negatively associated with MSI in five (DLBC, COAD, COADREAD, HNSC, PRAD) types of tumors respectively. In DLBC, the highest correlation was found between OTUD6B expression and MSI. Furthermore, Pearson’s correlation was performed to explore the relationship between OTUD6B expression and the status of promoter DNA methylation (Figure 9C). Results showed that DNA methylation had a significant positive correlation with OTUD6B expression in patients with TGCT, KIPAN, ACC, PCPG, CESC, THYM, LUSC, COAD, STES, PRAD, STAD, GBM, COADREAD, LUAD, and LGG.
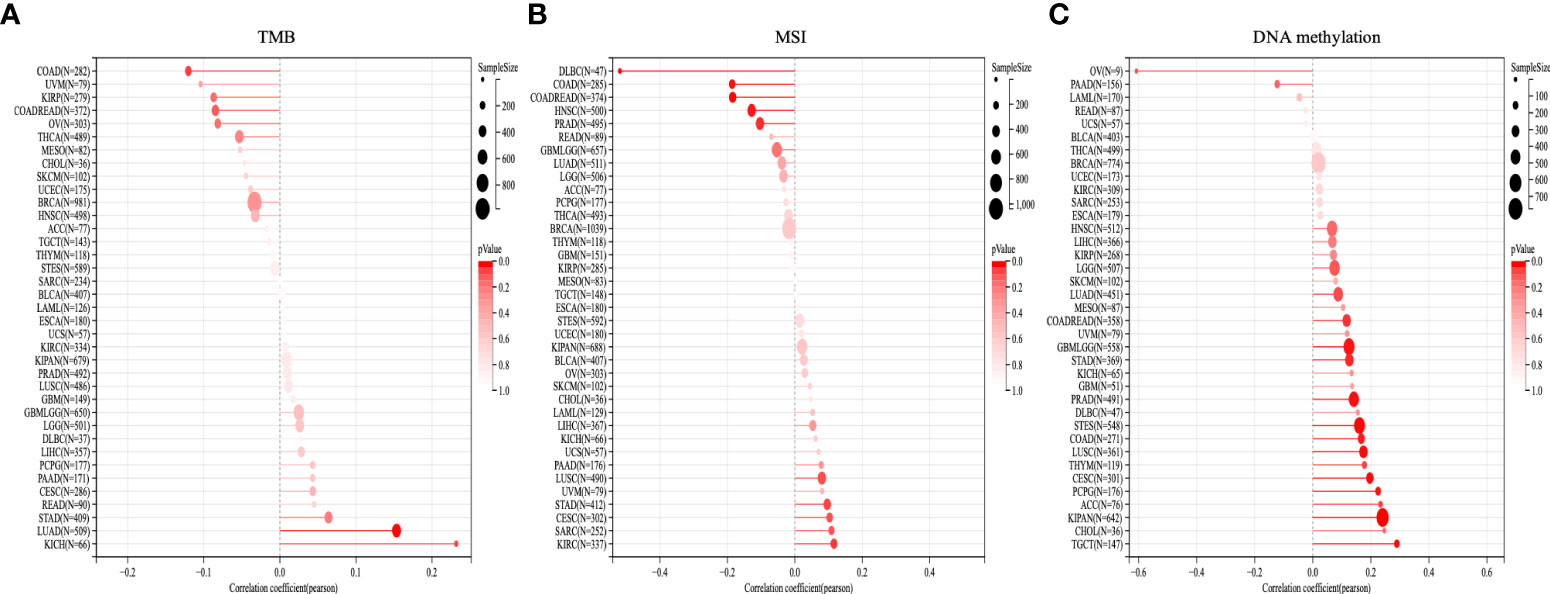
Figure 9 TMB, MSI, and DNA methylation analysis. The Lollipop plot represents the association between OTUD6B expression and TMB (A), MSI (B), and DNA methylation (C) in pan-cancer. The x-axis represents Spearman’s correlation coeficient. (TMB, tumor mutation burden; MSI, microsatellite instability).
To verify the carcinogenic and invasive capacity of OTUD6B, we generated stably silenced OTUD6B cell lines using A549 and Pc9 cells to assess its viability. Western blot analysis was performed to confirm OTUD6B and associated protein levels; E-cadherin was higher while N-cadherin was lower after knocking down OTUD6B (Figure 10A). Results showed that OTUD6B expression was highly expressed in lung cancer compared to normal tissues (Figure 10B). It is also obvious that OTUD6B silencing inhibited the proliferation, migration, and invasion of A549 and Pc9 cells (Figures 10C–E). The ability of OTUD6B to inhibit tumor proliferation was further verified in vivo in a nude mouse model (Figure 10F).
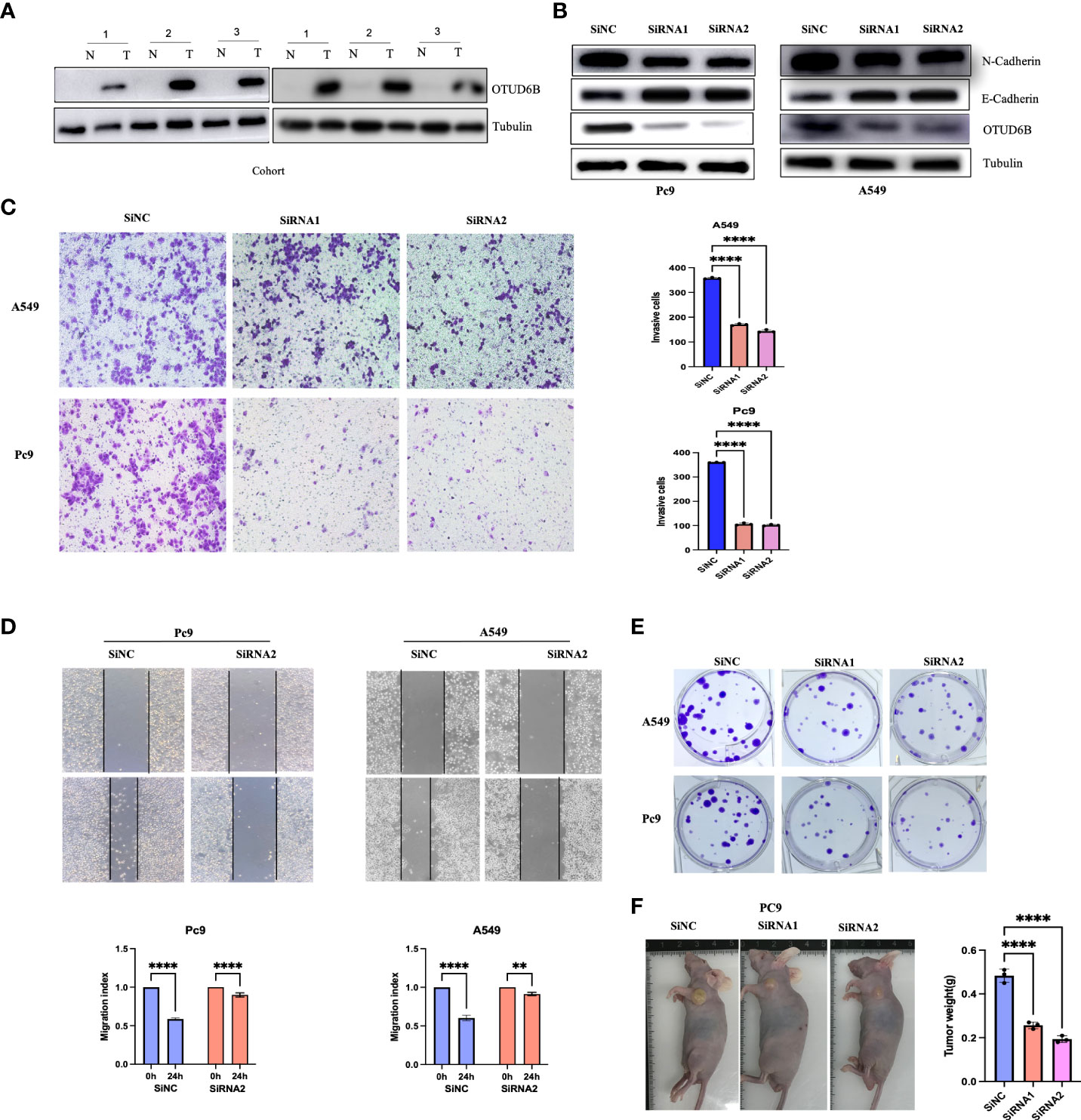
Figure 10 The knockdown of OTUD6B in lung cancer cells inhibits tumor self-renewal. (A) Immunoblot (IB) of OTUD6B expression in six randomly selected lung cancer samples (T) and their paired Normal (N) samples. n = 6 independent experiments. (B) Western blot validation of OTUD6B knockdown using siRNA in Pc9 and A549 cells. (C) Transwell migration assays in OTUD6B-knockdown Pc9 and A549 cells following transfection with the indicated siRNAs, typical images in the upper panels and invasive cell numbers in the lower panel. Scale bars, 70 μm. (D) OTUD6B knockdown inhibits Pc9 and A549 cell migration distance as seen in wound healing assays. (E) Cell proliferation detection of lung cancer cells was measured by clone formation. (F) Xenograft tumor models show that tumors grown from OTUD6B knockdown cells were smaller compared with those grown from control cells. (**p < 0.01; ****p < 0.0001).
Finally, we also investigated the response to targeted therapy and chemotherapy based on the level of OTUD6B. The results revealed a significant correlation between OTUD6B level and multiple-drug sensitivity (Figures 11A–K). For most targeted or chemotherapeutic agents including crizotinib, sorafenib, imatinib, etoposide, and paclitaxel, patients with a high OTUD6B level displayed higher sensitivity to these drugs, while only in erlotinib, higher sensitivity was observed in patients with a low OTUD6B expression (Figure 11C).
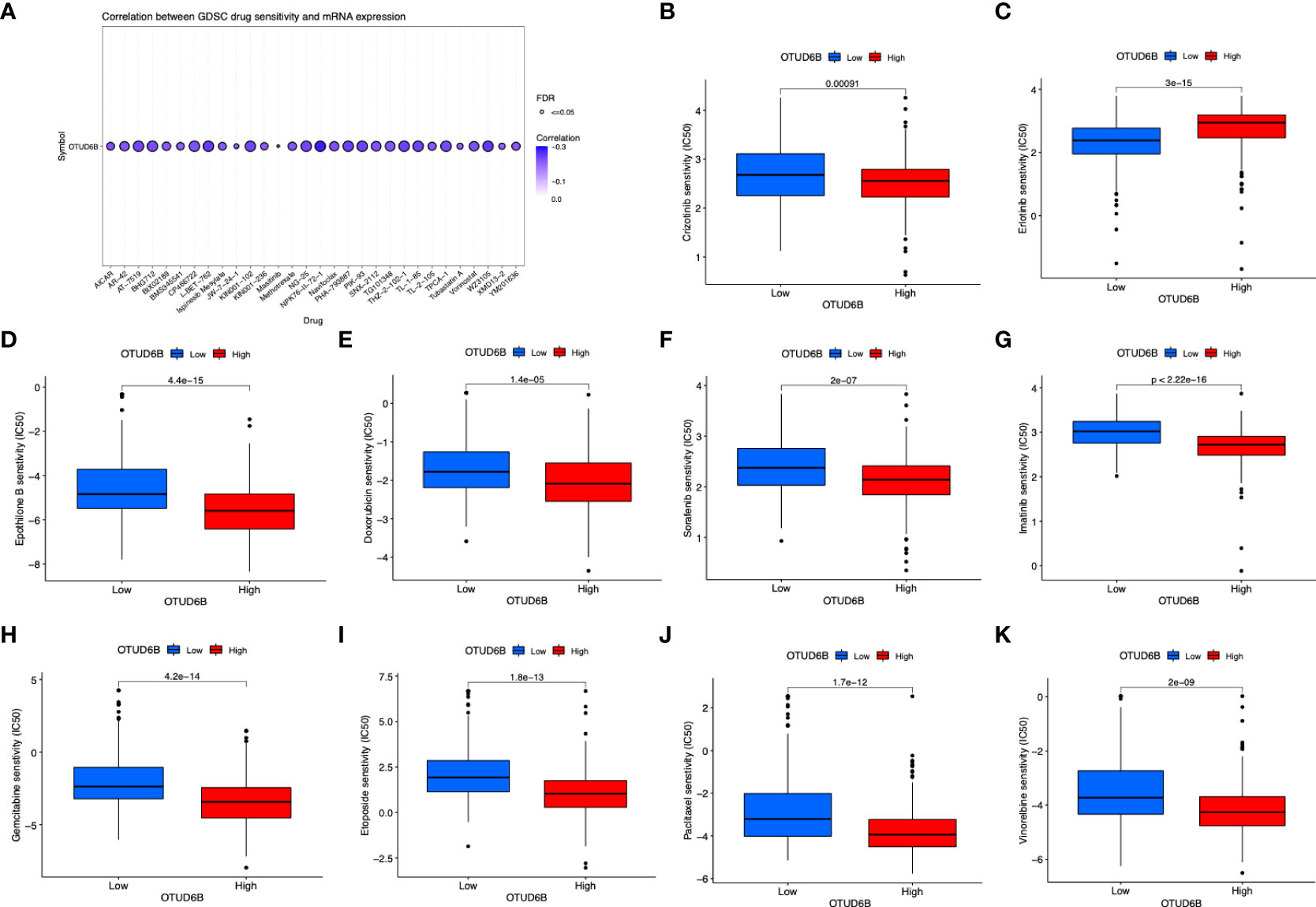
Figure 11 Relationship of OTUD6B expression with drug response. (A) The relationship between OTUD6B expression and GDSC drug sensitivity. Estimated IC50 indicating the efficiency of chemotherapy to ERGs in low- and high-risk patients of pazopanib (B), crizotinib (C), erlotinib (D), epothilone B (E), doxorubicin (F), sorafenib (G), imatinib (H), gemcitabine (I) etoposide (J), paclitaxel (K), vinorelbine.
OTUD6B, one of the subgroups of the OTU-containing family, does not have linkage specificity against diubiquitin which is different from most of other members (20). OTUD6B has been displayed to predict better patient survival in clear cell renal cell carcinoma and hepatocellular carcinoma through decreasing the ubiquitylation and proteasomal degradation of pVHL (13, 14). However, the functional role and specific mechanism of OTUD6B in other cancers remain a mystery. Here we conducted a pan-cancer analysis of OTUD6B to identify its impact on other cancer types.
Several databases were used to make a comprehensive analysis of OTUD6B expression and function. We firstly evaluated the OTUD6B expression in normal and cancer tissues in pan-cancer. Our results show that OTUD6B expression is relatively higher in most tumors than in normal tissues, which indicates that OTUD6B is an oncogenic gene in most tumors. In addition, based on Cox proportional hazard models and Kaplan–Meier survival curves, we found that a high OTUD6B expression was significantly correlated with poor prognosis of LUAD, BRCA, LIHC, THCA, and PAAD. Since we observed that OTUD6B has a potential cancer-promoting role in pan-cancers including LUAD, we further performed experiments in lung cancer cell lines to validate its carcinogenic role. We found alterations in E-cadherin and N-cadherin, which suggest that the invasive ability of OTUD6B in lung cancer may be related to EMT and may increase the risk of lung cancer metastasis. These results suggest that OTUD6B mainly plays an oncogenic role in tumor progression, which is consistent with previous studies. Furthermore, we evaluated the correlation between the OTUD6B level and drug sensitivity, indicating that in most circumstances, a low OTUD6B level was associated with better therapy response.
As one of the 18 DUB genes which contain the OTU domain, OTUD6B also has a common structure of these family members such as catalytic triad. DUBs remove Ub modifications and regulate nearly all Ub-dependent processes. Due to the complexity of Ub modifications, DuBs must exhibit different levels of specificity—they must distinguish between Ub and Ub-like modifications. Paul P. Geurink et al. have made a comparison of Ub and NEDD8-based (Ub-like modifier) peptide substrates and found that OTU DUBs hydrolyzed only Ub-based substrates but not NEDD8-based substrates under the same conditions, suggesting that human OTU DUBs are Ub specific (21). However, OTUD6B was affirmed to not have a linkage specificity against diubiquitin (20), indicating that it may function in its own way. Our results showed that the missense mutation occurs in the OTU domain which was accounted for almost half of the total mutations, suggesting its mutation tendency in OTU domain and its potential function sites.
Cancer treatments have changed dramatically in the last decade. To date, CTLA-4, PD-1, and PD-L1 are suggested to be the most widely targeted immune inhibitory receptors which had been verified to cause suppression of the T-cell immune response by binding to its corresponding ligands (15, 22, 23). It was indeed clinically beneficial in some patients, but unfortunately, a considerable number of patients showed resistance or non-response. Reports suggested that this may be related to the immune microenvironment which drew our attention to the importance of TIME.
The TIME includes stromal, fibroblastic, and endothelial cells and innate and adaptive immune cells, and the equilibrium of the TIME was based on these types of cells. By providing migration pathways that allow T cells to invade the tissue, the extracellular matrix can serve as both an inhibitor and a supporter for the adaptive immune response. Some studies have identified that OTUs are involved in the immune response. OTUB1 mediates the activation and metabolism of T cells through deubiquitinating AKT. OTUD1 can inhibit the transcriptome activity of IRF3, thus maintaining immune homeostasis in cells and hence dose OTUD3, PTUD4, and A20 (16, 24–26). In light of this, we explored the potential relationship between OTUD6B and immune response. Our study revealed that in most types of tumors, immune score was negatively correlated with OTUD6B expression including UCEC, CESC, ESCA, LUAD, MESO, and ACC. On the other hand, THCA, PCPG, BRCA, STAD, BLCA, LUSC, SKCM, TGCT, STES, and SARC were also found to be negatively correlated with OTUD6B expression in stromal scores. In LAML and KIRC, however, it showed a positive correlation between stromal score and OTUD6B expression. In the present study, we found that in most tumor types, OTUD6B expression was negatively linked with M1-like macrophage infiltration cells, while it was positively linked with M2-like macrophage infiltration cells, indicating that OTUD6B may play a role in macrophage polarization. In terms of T cells, we showed that OTUD6B expression was positively correlated with CD4+ memory T cells and Th2 CD4+ T cells but was negatively correlated with CD8+ T cells and Th1 CD4+ T cells. A previous study has verified that the level of M1 macrophage infiltration can be used as a reliable indicator of immunophenotype and is significantly associated with a good response (6). It is noteworthy to mention that immune cells can influence the tumor progression and prognosis (27). Various immune cells, such as CD8+ T cells, B cells, CD4+ T cells, neutrophils, and macrophages, secrete factors that influence the TIME, regulate tumor behavior, and have anticancer effects (28, 29). According to the abovementioned, OTUD6B may play an important role in stimulating human immune response and anti-immunotherapy.
ICIs is generally effective against a wide range of cancer types and is mostly not limited by the status of certain genetic mutations (30). In this study, OTUD6B expression has been unraveled to be positively correlated with most of the 47 immune checkpoint genes in almost all cancers, such as CD276, VEGFA, HMGB1, and TLR4. For instance, HMGB1 is associated with infectious and aseptic inflammatory disease states, and cancer. Studies have shown that HMGB1 is overexpressed in various solid carcinomas, and HMGB1 can stimulate innate immune cell chemotaxis and activation, thus mediating antitumor immune response and immunogenic cell death (31). Patients with a high level of CD276 are more sensitive to ICI treatment (32). From this, we can infer that OTUD6B was linked with better antitumor immune response. Therefore, OTUD6B may provide a pathway for tumor immunotherapy toward these targets.
In addition, we also investigated OTUD6B expression with TMB, MSI, and DVA methylation. We found that DNA methylation was significantly linked with OTUD6B in most tumors, indicating its potential effect on OTUD6B. ICI response has been shown to be associated with high TMB and MSI in various carcinomas and with good prognosis (33, 34). In this study, we showed that OTUD6B expression was significantly positively associated with TMB in KICH, LUAD, STAD, CESC, GBM, and LIHC and MSI in KIRC, SARC, CESC, STAD, LUSC, PAAD, and LIHC, respectively. The results indicated that OTUD6B may serve as a valuable biomarker for multiple cancers for their treatment and prognosis.
Several limitations also exist in this study. First, the majority of the patient data is derived from open databases, and only a few experiments have been verified. Second, OTUD6B is highly expressed and correlated with poor prognosis in many tumor types; while we have validated some mechanisms, its specific mechanism is still unknown. Therefore, further studies to evaluate the potential value of OTUD6B in diagnosis and prognosis in cancers need to be carried out.
To sum up, we detected an oncogenic effect of OTUD6B and suggested its potential as a prognostic biomarker in pan-cancer. An increased level of OTUD6B expression predicted a high level of TAM infiltration and generated an immunosuppressive microenvironment, which provides a possible target for tumor immune therapy. Our understanding of OTU is just the tip of the iceberg, and more functions, mechanisms, and potential therapeutic targets need to be explored and elucidate in the future.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The studies involving human participants were reviewed and approved by the ethics committee of the First Affiliated Hospital of Xi’an Jiaotong University. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements. The animal study was reviewed and approved by the ethics committee of the First Affiliated Hospital of Xi’an Jiaotong University. Written informed consent was not obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
GZ, DS, BZ, and HR designed the study. GZ and DS analyzed the data. GZ, JW, and SY wrote the manuscript and helped with the validation. SS and XC helped modify the manuscript. All authors contributed to the article and approved the submitted version.
The present study was supported by the National Natural Science Foundation of China (Program No. 82102801) and Natural Science Foundation of Shaanxi Province (Program No. 2022JQ-818&2019JM-559).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer ZJ declared a shared parent affiliation with the author SS to the handling editor at the time of the review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.955091/full#supplementary-material
Supplementary Figure 1 | Gene mutation profiles of OTUD6B in pan-cancer. (A) The expression differences of OTUD6B between normal and tumor tissues from TCGA database. (B) Gene alteration (C) Copy number variation of OTUD6B expression. (D) Mutant landscape of OTUD6B.
Supplementary Figure 2 | Protein expression and location of OTUD6B in normal and tumor tissues.
Supplementary Figure 3 | (A) Correlation between OTUD6B expression and different immune cells using data from XCell database. (B) RNA modification and OTUD6B level in multiple tumor tissues.
Supplementary Figure 4 | Association of OTUD6B expression level with Stromal Score across different types of cancers.
1. Hanahan D. Hallmarks of cancer: New dimensions. Cancer Discov (2022) 12(1):31–46. doi: 10.1158/2159-8290.CD-21-1059
2. Hanahan D, Coussens L. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell (2012) 21(3):309–22. doi: 10.1016/j.ccr.2012.02.022
3. Greten FR, Grivennikov SI. Inflammation and cancer: Triggers, mechanisms, and consequences. Immunity (2019) 51(1):27–41. doi: 10.1016/j.immuni.2019.06.025
4. Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res (2019) 79(18):4557–66. doi: 10.1158/0008-5472.CAN-18-3962
5. Bagchi S, Yuan R, Engleman EG. Immune checkpoint inhibitors for the treatment of cancer: Clinical impact and mechanisms of response and resistance. Annu Rev Pathol Mech Dis (2021) 16(1):223–49. doi: 10.1146/annurev-pathol-042020-042741
6. Zeng D, Ye Z, Wu J, Zhou R, Fan X, Wang G, et al. Macrophage correlates with immunophenotype and predicts anti-Pd-L1 response of urothelial cancer. Theranostics (2020) 10(15):7002–14. doi: 10.7150/thno.46176
7. Liu Q, Wu Y, Qin Y, Hu J, Cui J. Broad and diverse mechanisms used by deubiquitinase family members in regulating the type I interferon signaling pathway during antiviral responses. Sci Adv (2018) 4(5):eaar2824. doi: 10.1126/sciadv.aar2824
8. Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, Shiloh Y. Requirement of atm-dependent monoubiquitylation of histone H2b for timely repair of DNA double-strand breaks. Mol Cell (2011) 41(5):529–42. doi: 10.1016/j.molcel.2011.02.015
9. Sun SC. Deubiquitylation and regulation of the immune response. Nat Rev Immunol (2008) 8(7):501–11. doi: 10.1038/nri2337
10. Komander D, Rape M. The ubiquitin code. Annu Rev Biochem (2012) 81(7):203–29. doi: 10.1146/annurev-biochem-060310-170328
11. Guo Y, Jiang F, Kong L, Wu H, Zhang H, Chen X, et al. Otud5 promotes innate antiviral and antitumor immunity through deubiquitinating and stabilizing sting. Cell Mol Immunol (2021) 18(8):1945–55. doi: 10.1038/s41423-020-00531-5
12. Zhu D, Xu R, Huang X, Tang Z, Zheng X. Deubiquitinating enzyme Otub1 promotes cancer cell immunosuppression Via preventing er-associated degradation of immune checkpoint protein pd-L1. Cell Death Differentiation (2020) 28(6):1773–89. doi: 10.1038/s41418-020-00700-z
13. Liu X, Zhang X, Peng Z, Li C, Zhang L. Deubiquitylase Otud6b governs pvhl stability in an Enzyme£Ndependent manner and suppresses hepatocellular carcinoma metastasis. Advanced Sci (2020) 7(8):1902040. doi: 10.1002/advs.201902040
14. Guo K, Wei Y, Wang Z, Zhang X, Zhang X, Liu X, et al. Deubiquitylase Otud6b stabilizes the mutated pvhl and suppresses cell migration in clear cell renal cell carcinoma. Cell Death Dis (2022) 13(2):97. doi: 10.1038/s41419-021-04135-3
15. Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, et al. Engagement of the pd-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med (2000) 192(7):1027–34. doi: 10.1084/jem.192.7.1027
16. Zhou X, Yu J, Cheng X, Zhao B, Sun SC. The deubiquitinase Otub1 controls the activation of Cd8 T cells and nk cells by regulating il-15-Mediated priming. Nat Immunol (2019) 20(7):1–11. doi: 10.1038/s41590-019-0405-2
17. Thorsson V, Gibbs DL, Brown SD, Wolf D, Mariamidze A. The immune landscape of cancer. Immunity (2018) 48(4):812–30.e14. doi: 10.1016/j.immuni.2018.03.023
18. Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, et al. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic - sciencedirect. Ann Oncol (2019) 30(1):44–56. doi: 10.1093/annonc/mdy495
19. Cohen R, Rousseau B, Vidal J, Colle R, André T. Immune checkpoint inhibition in colorectal cancer: Microsatellite instability and beyond. Targeted Oncol (2019) 15(suppl):1–14. doi: 10.1007/s11523-019-00690-0
20. Mevissen TE, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N, et al. Otu deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell (2013) 154(1):169–84. doi: 10.1016/j.cell.2013.05.046
21. Geurink PP, Oualid FE, Jonker A, Hameed DS, Ovaa H. A general chemical ligation approach towards isopeptide-linked ubiquitin and ubiquitin-like assay reagents. Chembiochem (2012) 13(2):293–7. doi: 10.1002/cbic.201100706
22. Topalian S, Drake C, Pardoll D. Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell (2015) 27(4):450. doi: 10.1016/j.ccell.2015.03.001
23. Latchman Y, Wood CR, Chernova T, Chaudhary D, Borde M, Chernova I, et al. Pd-L2 is a second ligand for pd-1 and inhibits T cell activation. Nat Immunol (2001) 2(3):261–8. doi: 10.1038/85330
24. Zhang L, Jin L, Qian L, Qian F, Hui Z. Induction of Otud1 by rna viruses potently inhibits innate immune responses by promoting degradation of the Mavs/Traf3/Traf6 signalosome. PLoS Pathog (2018) 14(5):e1007067. doi: 10.1371/journal.ppat.1007067
25. Zhao Y, Mudge MC, Soll JM, Rodrigues RB, Byrum AK, Schwarzkopf EA, et al. Otud4 is a phospho-activated K63 deubiquitinase that regulates Myd88-dependent signaling. Mol Cell (2018) 69(3):505–16. doi: 10.1016/j.molcel.2018.01.009
26. Lin R, Yang L, Nakhaei P, Sun Q, Sharif-Askari E, Julkunen I, et al. Negative regulation of the retinoic acid-inducible gene I-induced antiviral state by the ubiquitin-editing protein A20. J Biol Chem (2006) 281(4):2095–103. doi: 10.1074/jbc.M510326200
27. Sokratous G, Polyzoidis S, Ashkan K. Immune infiltration of tumor microenvironment following immunotherapy for glioblastoma multiforme. Hum Vaccines Immunother (2017) 13(11):2575–82. doi: 10.1080/21645515.2017.1303582
28. DePeaux K, Delgoffe GM. Metabolic barriers to cancer immunotherapy. Nat Rev Immunol (2021) 21(12):785–97. doi: 10.1038/s41577-021-00541-y
29. Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, et al. Understanding the tumor immune microenvironment (Time) for effective therapy. Nat Med (2018) 24(5):541–50. doi: 10.1038/s41591-018-0014-x
30. Li B, Chan HL, Chen P. Immune checkpoint inhibitors: Basics and challenges. Curr Medicinal Chem (2017) 24(17):3009–25. doi: 10.2174/0929867324666170804143706
31. Hubert P, Roncarati P, Demoulin S, Pilard C, Ancion M, Reynders C, et al. Extracellular Hmgb1 blockade inhibits tumor growth through profoundly remodeling immune microenvironment and enhances checkpoint inhibitor-based immunotherapy. J Immunother Cancer (2021) 9(3). doi: 10.1136/jitc-2020-001966
32. Zhang G, Su L, Lv X, Yang Q. A novel tumor doubling time-related immune gene signature for prognosis prediction in hepatocellular carcinoma. Cancer Cell Int (2021) 21(1):1–15. doi: 10.1186/s12935-021-02227-w
33. Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to pd-1 inhibition. N Engl J Med (2017) 377(25):2500. doi: 10.1056/NEJMc1713444
Keywords: OTUD6B, pan-cancer, prognosis, immune microenvironment, TCGA
Citation: Zhao G, Song D, Wu J, Yang S, Shi S, Cui X, Ren H and Zhang B (2022) Identification of OTUD6B as a new biomarker for prognosis and immunotherapy by pan-cancer analysis. Front. Immunol. 13:955091. doi: 10.3389/fimmu.2022.955091
Received: 28 May 2022; Accepted: 15 July 2022;
Published: 16 August 2022.
Edited by:
Antonella Sistigu, Agostino Gemelli University Polyclinic (IRCCS), ItalyReviewed by:
Lele Zhu, University of Texas MD Anderson Cancer Center, United StatesCopyright © 2022 Zhao, Song, Wu, Yang, Shi, Cui, Ren and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong Ren, cmVob25nczIwMDBAMTYzLmNvbQ==; Boxiang Zhang, emhhbmdib3hpYW5nQHhqdHVmaC5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.