
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 13 September 2022
Sec. Mucosal Immunity
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.953196
This article is part of the Research TopicInterplay Between Diets, Microbiota, Bacterial Metabolites and Host for Intestinal Health and DiseaseView all 17 articles
The intestinal tract is an ecosystem in which the resident microbiota lives in symbiosis with its host. This symbiotic relationship is key to maintaining overall health, with dietary habits of the host representing one of the main external factors shaping the microbiome-host relationship. Diets high in fiber and low in fat and sugars, as opposed to Western and high-fat diets, have been shown to have a beneficial effect on intestinal health by promoting the growth of beneficial bacteria, improve mucus barrier function and immune tolerance, while inhibiting pro-inflammatory responses and their downstream effects. On the contrary, diets low in fiber and high in fat and sugars have been associated with alterations in microbiota composition/functionality and the subsequent development of chronic diseases such as food allergies, inflammatory bowel disease, and metabolic disease. In this review, we provided an updated overview of the current understanding of the connection between diet, microbiota, and health, with a special focus on the role of Western and high-fat diets in shaping intestinal homeostasis by modulating the gut microbiota.
The gastrointestinal (GI) tract is covered by a single layer of epithelial cells that act as a selectively permeable barrier allowing the absorption of dietary nutrients, microbial metabolites, electrolytes, and water from the lumen into the circulation, while maintaining an effective defense barrier against luminal microorganisms (1, 2). The GI tract harbors a complex and dynamic population of microbes encompassing bacteria, archaea, eukarya and viruses, collectively referred to as the gut microbiota, which has co-evolved with its host in a symbiotic relationship (3, 4). Although the gut microbiota comprises trillions of microbes, the relative sterility of blood and host tissue relies on an intact gut barrier, which acts as the gatekeeper of our health (5). Additionally, apart from acting as a selective barrier, the intestinal epithelium orchestrates the communication between intestinal microbes and the mucosal innate and adaptive immune system (1, 6). The microbiota and its metabolites regulate various aspects of gut immunity, and is thus critical for maintaining mucosal homeostasis (7, 8). The intestinal epithelial cells (IECs) protect the underlying tissues from commensal microbes and/or invading pathogenic microorganisms by secretion of a mucus layer which acts as an additional layer of physical defense of the host, and a habitat for bacteria providing binding sites and energy sources (9). The mucus also acts as a diffusion barrier for anti-microbial peptides (AMPs) released by Paneth cells and other epithelial cells, and immunoglobulin A (IgA) derived from mucosal B cells that prevents microbes from reaching the epithelium (10). Diet is one of the primary factors that influences gut microbiota composition, diversity, and functionality, which in turn have a strong impact on mucus properties, mucosal immunity, and thereby intestinal homeostasis (11, 12). The aim of this review is to summarize the current knowledge regarding the interplay between the diet, microbiota, mucus, and the intestinal immune system with a particular focus on the impact of Western and high-fat (HF) diets on the gut microbiota, and how shifts in the composition and functionality of the microbiota can compromise intestinal homeostasis.
The primary colonization of the GI tract begins at birth with the acquisition of microbes from the environment, mainly from the maternal vagina and the skin. The number of microorganisms that reside in the human gastrointestinal tract has been estimated to be 1013, a number that is similar to the number of human cells (13). However, the genes encoded by the human gut microbiota, known as the microbiome, are 100-fold more abundant than the genes of the human genome (14). 16S rRNA and metagenomics studies have revealed that the majority of gut microbiota sequences belong to the Bacteria, which is the predominant kingdom in the human adult gut microbiota (15–17). The human gut microbiota is mainly composed by two dominant bacterial phyla: gram-positive Firmicutes and gram-negative Bacteroidetes representing 85-90% of the total microbiota, whereas Actinobacteria, Proteobacteria, Fusobacteria, and Verrucomicrobia are minor costituents (15, 18, 19). Microbial density and diversity increase steadily along the GI tract from the proximal to the distal intestine, a process affected by host features and microbial community dynamics (20). The duodenum harbors 103 microbial cells per gram of intestinal content, and increasing densities/diversities are found in the jejunum (104 cells per gram), ileum (107 cells per gram), and colon (1012 cells per gram) (21). Gut microbiota composition also differs transversally from the lumen to the mucosa as demonstrated in both mice and human studies (15, 22, 23). In physiological conditions, the microbiota offers many benefits to the host, which include fortifying the intestinal epithelium, harvesting energy from undigested and unabsorbed nutrients, defending against pathogens, and regulating host immunity (24). However, several environmental (e.g., dietary patterns, antibiotics) and intrinsic (e.g., breast feeding, genetic background) factors can impact the gut microbiota composition and its structural, protective and metabolic functions (11). Additionally, other factors such as oxygen gradients, mucus properties, and the host immune system influence the transversal distribution pattern of the microbiota (23, 25, 26). Over the last years, the microbiota has emerged as a key regulator of host metabolism and health (25). There are several mechanisms by which the microbiota can regulate host metabolism and health, many of which can be ascribed to microbial metabolites (27). Among these bacterial metabolites, the most studied are the short-chain fatty acids (SCFAs) that are produced by bacterial fermentation of indigestible nutrients (i.e., dietary fibers and complex carbohydrates). The role of SCFAs in the regulation of metabolic function and intestinal homeostasis will be detailed in the next sections of this review. In addition to bacteria, another intricate kingdom that co-colonizes the human GI tract is composed of a substantial quantity of viruses collectively referred to as the gut virome (28). Viruses of bacteria, called bacteriophages (phages), are significantly more abundant than eukaryotic viruses, and the estimated phage-bacterial ratio in the human gut is believed to be 1:1 (29). Emerging studies have shown that phage-driven alterations of the microbiota composition by direct interactions or potentially via the human immune system have been associated with several diseases (e.g., inflammatory bowel disease, cancer, obesity) (28–30). However, little is known about the phage-mucus interactions, and therefore the present review only covers the mucus-bacteria feedback system of the gut.
Diet is a key discriminant in shaping health and aging trajectories with these effects being also mediated by the ability of nutrient quality and quantity to modulate gut microbiota composition and metabolic function (31). Indeed, in addition to providing the energy and the nutrients needed to sustain the cellular processes required for daily life (31), dietary components are also instrumental factors in modulating the microbial communities in the gut (32). In addition to the plethora of the effects exploited by the gut microbiota on host health, the intestinal microbiota community also regulates the mucus barrier function (12, 33). In light of this, dietary-driven modulations of the gut microbiota will also be reflected upon the gut mucus barrier function and the overall intestinal health (34) (Figures 1A, B). However, not all diets are equal, and it is established that different dietary patterns exert distinct effects on the gut microbiota. In agreement with this, a diet rich in fiber including galacto- and fructo-oligosaccharides (two important groups of prebiotics) and resistant starch strongly impacts the composition, diversity and metabolic function of the microbiota (35). To this end, dietary fiber provides a plethora of substrates for fermentation reactions carried out by specific species of microbes (e.g., Bifidobacterium, Faecalibacterium) that express the adequate enzymatic machinery to break down these complex carbohydrates and to produce SCFAs (e.g., acetate, butyrate, propionate). The SCFAs in turn exert beneficial effects on cardio-metabolic (36) and gut health (37), including promoting mucus barrier function (38, 39) (Figure 1A). Prompted by these evidences, a move towards a diet high in dietary fiber, low in glycemic index carbohydrates, long-chain saturated fatty acids, animal protein (i.e., red and processed meat), and sugar referred to as the Mediterranean diet (hereafter, MD), has been associated with the prevention of cardiovascular and metabolic diseases, and many other diseases (40, 41). The consumption of a MD has been shown to increase the levels of the fiber-degrading Faecalibacterium prausnitzii and genes associated with microbial carbohydrate degradation and butyrate metabolism in a population at risk for cardio-metabolic disease (42). An increased levels of fecal SCFAs, Prevotella and fiber degrading Firmicutes was also observed in healthy overweight and obese subjects with a high-level adherence to a MD (43). The importance of the dietary fiber (referred to as microbiota accessible carbohydrates, MACs) intake was also demonstrated in mice colonized with a human microbiota and showed that a low-MAC diet resulted in a reduction in microbial diversity in just three generations, which could not be brought back when mice were fed a normal MAC diet (44). Additionally, deprivation of dietary fiber was also associated with deleterious effects on the intestinal mucus layer by the gut microbiota (45–47), further highlighting the importance of dietary fiber for our health and gut microbiota ecosystem.
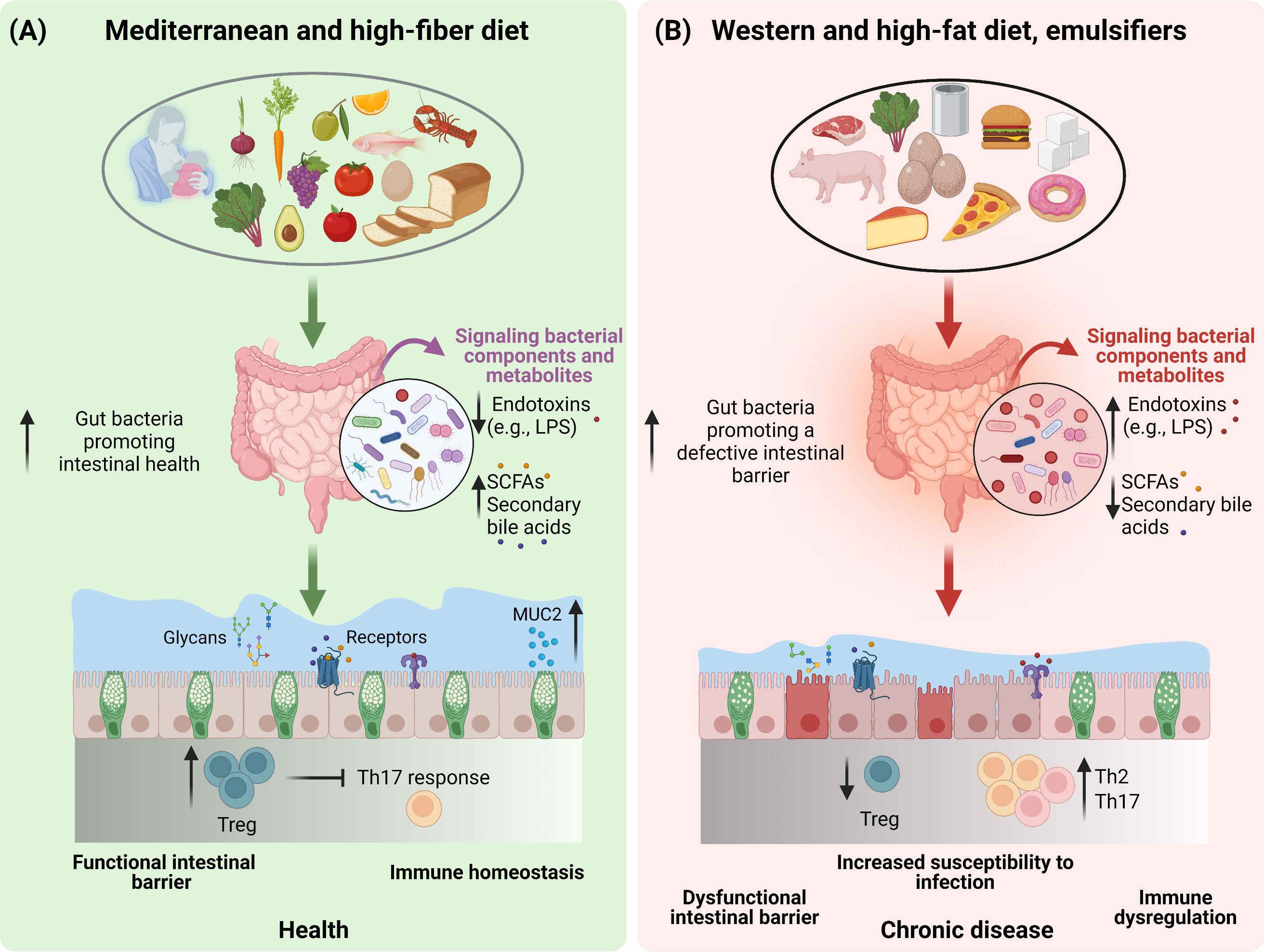
Figure 1 The impact of dietary patterns on gut microbiota and intestinal health. A schematic overview depicting the importance of diet and dietary constituents in discriminating between a healthy (A) or an unhealthy (B) state of the gut barrier function by modulating the composition and functionality of the gut microbiota.
In contrast to a fiber-rich diet, hypercaloric diets high in long-chain saturated fatty acids and ω-6, sugar and low in dietary fiber, referred to as the Western diet (WD) and HF diet in the case of animal models have not only been widely recognised for their detrimental effects on cardio-metabolic health, but also negatively affect the gut microbiota composition and functionality (11, 48–50) (Figure 1B). A key feature of both the WD and the HF-diet, is the high intake of dietary lipids especially in the form of triglycerides. Of note, dietary triglycerides differ in terms of their fatty acid composition (saturated vs unsaturated), which in turn represents a further discriminant dictating the effects of lipids on the gut microbiota. In mice, lard, which is rich in long-chain saturated fatty acids, promotes an increase in Bacteroides and Bilophila as compared to fish oil which is rich in unsaturated fatty acids and particularly ω-3 which promotes an increase in Bifidobacterium, Lactobacillus and Akkermansia muciniphila (51). Despite the majority of the lipids being digested and absorbed in the upper intestine, a high lipid intake, as in the case of HF-diets, in animal models promoted a decrease in bacteria count and a shift in microbiota species abundance (52). Indeed, when given to mice, HF-diets have been reported to increase the Firmicutes to Bacteroidetes ratio (52–54). Additionally, HF-diets have been shown to reduce gut bacteria promoting intestinal health, such as Akkermansia muciniphila, Bifidobacterium spp., Bacteroidetes spp., Lactobacillus spp. and Clostridiales spp., while increasing gut bacteria associated with defective gut barrier function, such as Oscillibacter spp. and Desulfovibrio spp (48) (Figure 1B). Disrupted gut barrier integrity results in increased gut permeability to luminal bacterial components such as lipopolysaccharide (LPS) resulting in chronic low-grade inflammation typical of obesity and related metabolic comorbidities (55–57). In addition to microbial activation of the intestinal immune system, the inflammation is also fuelled by saturated fatty acids overload, which by themselves are able to elicit pro-inflammatory responses (58). These effects translated in the development of metabolic aberrations, particularly given the role of low-grade chronic inflammation in impairing insulin sensitivity (59). Nevertheless, as already mentioned, WD and HF-diets are generally low in dietary fiber, therefore it is difficult to discern whether the observed effects on the gut microbiota are due to the deprivation of dietary fiber, or the high sugar and fat intake. In response to this conundrum, the effects of a WD and HF-diets on the gut microbiota were mitigated by the supplementation of dietary fibers, further supporting their prominent role in gut health (39, 60–62).
The metabolic activity and composition of the gut microbiota can also be modulated by dietary protein. Diets with a high protein/carbohydrate ratio may exert promising effects in preventing obesity and improving glycemic control as described in animal models (63, 64) and humans (65, 66), even though the effects of these dietary patterns on metabolic health in human remain controversial (67, 68). However, proteins, especially if consumed in excess are able to negatively influence the microbiota. In humans, a high-protein diet was found to decrease butyrate-producing bacteria and fecal butyrate levels (69), and decrease the abundance of beneficial bacteria like Bifidobacterium, Roseburia and Eubacterium rectale (70, 71). Despite these findings, it must be taken into consideration that the impact of protein on the gut microbiota is dictated by the amino acid composition and their relative abundance. Amino acids can in fact be metabolised by the microbiota resulting in the production of a wide array of metabolites which, in turn, affect the health of the host. In line with this, different amino acids exert different effects on the gut microbiota. For example, methionine restriction in mice results in an increase in SCFA-producing bacteria and inflammation-inhibiting bacteria with a concomitant decrease in inflammation-promoting bacteria (72). To the same extent, the protein source represents another variable underpinning the effects of dietary proteins on the gut microbiota. Indeed, vegetable proteins have been shown to increase Bifidobacterium and Lactobacillus, an effect which may also be dependent on the fact that vegetable source of proteins also represent a source of dietary fiber, as opposed to some animal proteins which combine a lack of fibers with high-levels saturated fatty acids (73).
Besides the role of nutrients, other food ingredients are also emerging as potent modulators of the gut microbiota. Of these, dietary emulsifiers like carboxymethylcellulose and polysorbate-80 have been shown to impact the gut microbiota composition increasing the susceptibility to colitis and the metabolic syndrome in animal models (74). Overall, the changes in the gut microbiota elicited by emulsifiers produced a shift in gut bacteria in a manner to promote and sustain intestinal inflammation (75) (Figure 1B). However, although numerous emulsifiers increased the pro-inflammatory potential of the gut microbiota ex vivo, these effects were not induced by all commonly used emulsifiers (76). In this regard, carrageenans and gums were shown to alter microbiota density, composition as well as the expression of pro-inflammatory molecules (76). Altogether, findings from both pre-clinical and clinical studies highlight the importance of diet, nutrients, and food ingredients in the modulation of the gut microbiota, resulting in either beneficial or detrimental health outcomes.
The intestinal barrier is multi-tiered, including the mucus layer, the epithelial layer and the underlying immune system (Figures 2A, B). At this interface, appropriate host-microbiome interactions play an important role in maintaining intestinal homeostasis throughout life (77). Disturbance to any of these layers by several factors such as dietary-driven changes in the gut microbiota composition, antibiotics usage, and genetic susceptibility, is associated with the onset of chronic disease including inflammatory bowel disease, extra intestinal autoimmune disease, metabolic disorders such as diabetes and obesity and allergic disease (78, 79). This section of the review will focus on the organization and composition of the different parts of the intestinal barrier in the steady state.
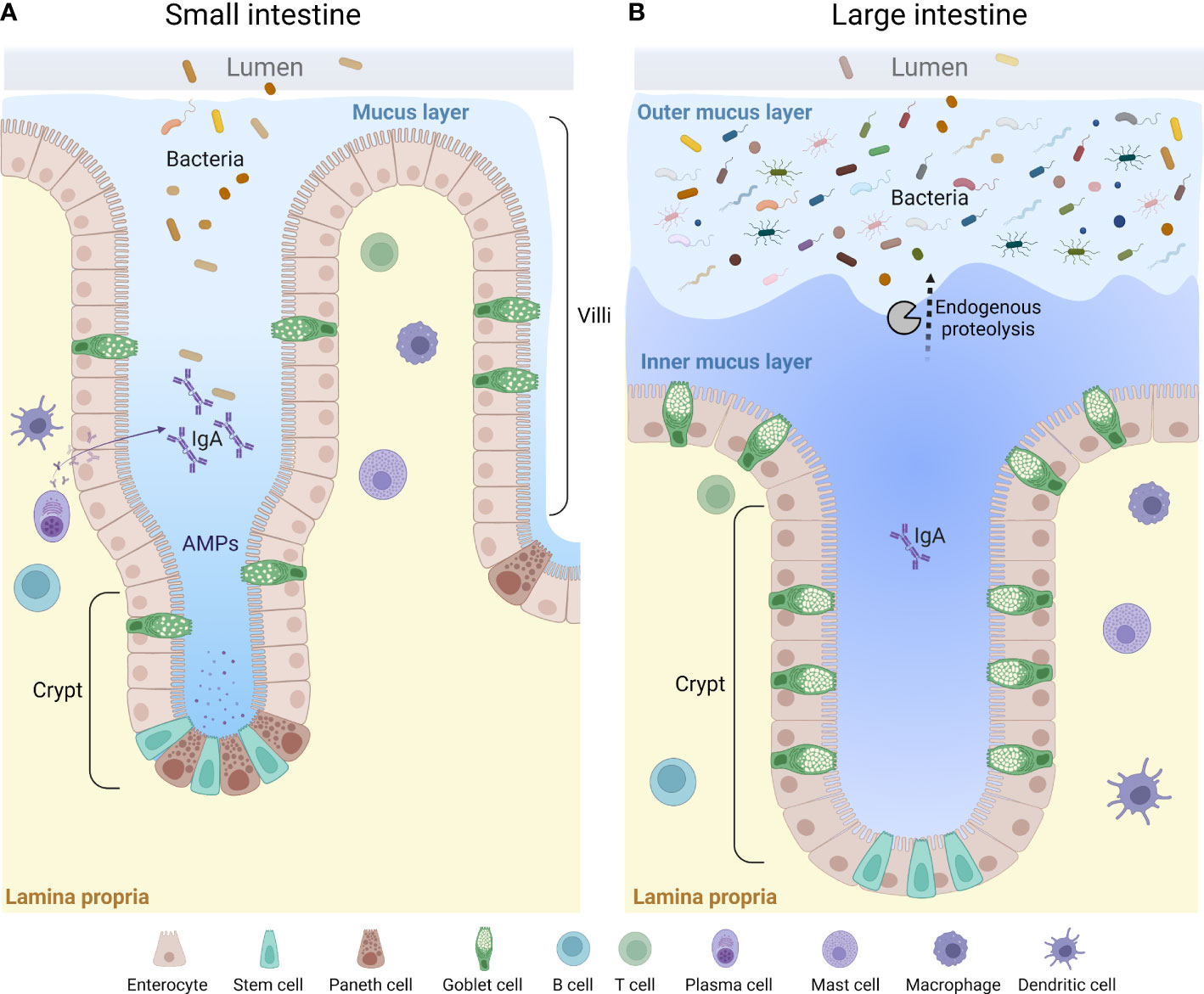
Figure 2 A general overview of the mucus layer in the small and large intestine. The mucus layer is a key component of the intestinal barrier. It is mainly composed of mucin glycoproteins produced and secreted by the intestinal goblet cells. The small intestine is characterized by a loose and unattached mucus layer containing antimicrobial products that limit penetration by bacteria (A), whereas the large intestine presents a dense and firmly attached mucus layer converted to a looser structure at the luminal side via endogenous proteolysis (B). AMPs, anti-microbial peptides.
The mucus layer, produced by specialised secretory goblet cells (GCs) in the epithelium covers the intestinal tissue to provide lubrication and protection against the luminal material, especially the microbiota. Mucus homeostasis is crucial for health and its dysregulation in either the small intestine or the colon causes or correlates with various diseases. Indeed, mucus accumulation and bacterial overgrowth due to loss of bicarbonate secretion is evident in cystic fibrosis, and lost mucus barrier properties and increased bacterial burden at the epithelium in the colon correlate with the inflammatory bowel disease ulcerative colitis (UC) (80–83). Thus, an understanding of factors that impact mucus homeostasis is needed in order to maintain a good “gut health”. External factors can alter mucus-related properties and as the major external factor affecting the intestine, diet can have profound effects on the mucus barrier. Mucus associated effects induced by the diet can be direct on the mucus or the epithelium (84–87) but perhaps more importantly indirect by influencing the microbiota which in turn can have strong effects on the intestinal mucus, which will be further discussed in the last part of this review (39, 45, 88–91).
The properties and thickness of the mucus layer varies along the GI tract to facilitate the physiological function in each location (33, 92, 93). The small intestinal mucus has been described as loosely organized to allow for efficient nutrient absorption along the full length of the villi protrusions (92, 94) (Figure 2A). To maintain a protective barrier against luminal bacteria and digestive enzymes, the small intestinal mucus is fortified by AMPs secreted from crypt-base residing Paneth cells and enterocytes at the base of the villi, IgA secreted by plasma B cells, and endogenous enzyme inhibitors (94–96) (Figure 2A). In the small intestine, the thickness of mucus layer covering the follicle associated epithelium of Peyer’s patches has been shown to be thinner as compared to the surrounding epithelium, which likely facilitates antigen and particle uptake by small intestinal M cells (97). Only a thin separating mucus layer covers the proximal colon of mice, and bacteria can be seen in close contact with the epithelium, as evident by histology (92, 98, 99). However, the mechanisms allowing the close proximity of bacteria to the epithelium in the proximal colon remains to be defined (100). As the luminal material solidifies during its distal transport, a separating mucus barrier is formed, distancing the fecal matter and the microbiota from the underlying tissue (98, 99). Thus, in the distal parts of the colon of both human and rodents, the epithelium is protected by a mucus barrier which physically separates the luminal microbiota from the tissue surface (93, 101, 102). However, mucus material can also be seen intermixed with the microbiota in the outer regions of fecal pellets, where it is thought to provide a nutrient rich ecological niche for some bacteria (20, 22, 101). This “inner” and “outer” mucus thus creates a two-layered mucus structure in the distal colon (80, 92, 93, 101) (Figure 2B). The conversion from the inner to the outer mucus layer is dependent on endogenous proteolytic activity, but bacterial proteolysis could play an additional role (101, 103) (Figure 2B).
The mucus is composed by a core set of about 30 proteins, most of which are produced by GCs, including mucin 2 (MUC2), chloride channel accessory-1 (CLCA1), Fc fragment of IgG binding protein (FCGBP) and zymogen granule protein 16 (ZG16) (83), as well as lipids, ions, and water (which makes up for approximately 95%) (104). The MUC2 glycoprotein is the main structural component and creates a net-like scaffolding backbone of the mucus along the GI-tract by oligomerization of its terminal ends (105, 106). MUC2 is a very large protein (>5000 amino acids) with two mucin characteristic PTS-domains, rich in proline, serine and threonine, which becomes heavily O-glycosylated by glycosyltransferases in the secretory pathway. The added glycans makes up for approximately 80% of the molecular weight in the mature protein (107). More than 100 different glycan structures have been identified in intestinal MUC2 (108). The distribution of different glycans is region and species specific, adding additional layers of complexity to the mucus barrier (109, 110). Apart from protecting the MUC2 protein backbone from proteolysis and giving MUC2 its gel properties by binding water, the glycans attached to MUC2 provide microbial adhesion sites and a nutritional source in the outer mucus layer (111–114). The glycan composition on MUC2 can thus provide a strategy for the host to select commensal bacteria, but can also be utilized by both commensal and pathogenic bacteria (115–117). Microbiota dependent degradation of mucin glycans induced by a low-fiber diet increases the susceptibility to Citrobacter rodentium infection in mice (45), indicating the importance of Muc2 glycosylation in protection against pathogenic bacteria. Shortened and less complex glycans have been identified in patients with active ulcerative colitis (118). It does, however, remain unknown whether the observed alterations are a cause, or an effect of the inflammation, and further investigations are needed to elucidate these mechanisms.
The bacterial diversity of the mucosa-associated bacteria is on par with that of the luminal microbiota, but the composition differs (39, 119). In general, mucosa-associated bacteria have a higher abundance of the phylum Firmicutes compared to Bacteroides both in humans and rodents (120). Although the inner mucus layer appears mostly sterile by histological examination, Bergström et al., detected appreciable levels of bacterial 16S in mucus collected in inter-pellet regions, indicating that some bacteria, especially so called mucus specialists, indeed can colonize this barrier, which was also evident by ex vivo imaging (119). Additionally, bacterial analyses of mucosal samples from laxative treated patients indicate their presence in mucus from the different sampling sites in the large intestine, as well as in the distal ileum (121). A couple of studies have demonstrated the presence of a crypt-specific microbiota in both mouse and human colon characterized by a low density of bacterial community dominated by Acinetobacter spp. in mouse and generally enriched for Proteobacteria capable of aerobic metabolism in both human and mouse (122, 123). It should however be noted that this was restricted to the proximal colon in mouse, and only affected a small number of colonic crypts in human. It is possible that the number of bacteria in the inner mucus layer and colonic crypts differ between strains/vivariums, reflecting differences in mucus quality controlled by the gut microbiota. Furthermore, in physiological conditions, mucin-degrading specialists (e.g., Akkermansia muciniphila and, Bacteroides thetaiotaomicron) live in mutual coexistence with host, and the rate of mucus degradation is balanced with the rate of mucus synthesis, resulting in a dynamic and stable mucus structure that is of fundamental importance for our “gut health”. Only few bacterial species have the enzymatic machinery for initiating partial or full mucin degradation, including A. muciniphila, Bacteroides thetaiotaomicron, B. bifidum, Bacteroides fragilis, Ruminococcus gnavus, and Ruminococcus torques (120). These mucin-specialist degrade the mucin protein, which possibly leads to the availability of oligosaccharides for other bacteria that do not harbour the correct enzymes for this process allowing bacterial cross-feeding and mucosal health (47, 120). An example is the interaction between the B. thetaiotaomicron (B.theta) and the Faecalibacterium prausnitzii. Wrzosek et al., have shown that B. theta, an acetate producer, increased goblet cell differentiation, expression of mucus-related genes, and mucus glycosylation in mono-colonized rats. In contrast when B.theta-mono-colonized rats were supplemented with Faecalibacterium prausnitzii, an acetate consumer and butyrate producer, the increase in goblet cell differentiation and the alteration in mucus glycan profile was reduced, thus maintaining an appropriate structure and composition on the gut epithelium (124). Moreover, additional studies are needed to elucidate the role of the mucosa-associated microbiota in regulation of intestinal homeostasis as this microbial community is located closely to the host.
Although the intestinal epithelium with its mucus layer act as the first layers of defense against the potentially harmful agents that pass through our GI tract, this is not an absolute barrier. Indeed, dietary and microbial antigens, and metabolites readily pass the mucus layer and epithelium and enter the lamina propria (LP) where they are sampled and sensed by the intestinal immune system (125, 126). In the intestine, the immunological challenge lies in how to accurately discriminate between harmless and harmful foreign antigens, and failure to respond adequately to the large variety of antigens that pass through the GI tract results in chronic inflammatory conditions such as food allergy and inflammatory bowel diseases (127, 128). The ability of the immune system to mount appropriate responses to the luminal content is to a large extent regulated by the composition of the content itself which influences the immune status of the host. The intestinal immune system can be divided into innate and adaptive immunity, where dendritic cells (DCs), macrophages, neutrophils, mast cells, eosinophils, basophils, natural killer T cells (NKT) and innate lymphoid cells (ILCs) make up the innate arm of immunity, and T and B cells make up the adaptive arm. During steady state, adaptive and innate immune responses in both the small intestine and colon promote tolerance and inhibit pro-inflammatory responses allowing us to live in symbiosis with our microbiota and tolerate the food we eat. Tolerance to the luminal content is to a large extent dependent on induction and maintenance of T regulatory cells (Tregs), a subset of CD4+ T helper cells (Th) that suppresses effector T cell responses, and by plasma cells that secrete large quantities of IgA directed towards luminal antigens (129). Innate immune cells such as macrophages and DCs contribute to maintaining a local environment promoting tolerance and induce adaptive immune responses. Tissue resident LP macrophages produce the anti-inflammatory interleukin (IL)-10, and prostaglandins that promote local tolerance in the LP, and migratory LP-DCs traffic to small intestine and colon draining lymph nodes following antigen acquisition where they promote de novo Treg induction (130, 131). Other innate cells such as eosinophils, more known for their role in pathological conditions such and allergic disease (132), were recently shown to contribute to intestinal homeostasis by stimulating and maintaining IgA producing plasma cells, and by regulating LP Treg and DC populations as mice lacking eosinophils were shown to have reduced number of LP Tregs and CD103+ tolerogenic DCs (133).
The role of the microbiota in regulation of the mucus layer is an area of continuous research, and a large proportion of the current knowledge comes from studies using germ free (GF) mice, and mono-colonization or conventionalization of GF mice. In this section we will describe how the absence or the re-introduction of microbes in murine models affect development and regulation of the mucus barrier, and intestinal immunity.
The first evidence on the importance of microbes in the regulation of the intestinal mucus layers was demonstrated in a conventionalization experiment using GF mice. The authors discovered that in the GF ileum, the mucus was attached to the epithelial surface in contrast to conventionally raised (Convr) mice in which the mucus is easily aspirated, a process shown to be regulated by microbial activation of meprin β, an enzyme involved in detachment and release of mucus in the small intestine (90, 134). In the GF colon, the mucus was thinner and more penetrable to bacterium sized fluorescent beads as compared to Convr mice, and the amount of Muc2 was lower in the GF colon as compared to Convr colon (90). Furthermore, the glycosylation pattern of Muc2, the overall expression levels of glycosyltransferases, and the length of the glycans were shown to differ between the two groups (90, 110). It has also been shown that single bacterial species (i.e., B. theta and Faecalibacterium prausnitzii) can promote colonic epithelial homeostasis by modifying goblet cell differentiation, expression of mucus-related genes, and mucin glycosylation (124). Johansson at al., have also shown that gut microbiota colonization of GF mice, and normalization of mucus properties is a slow process (90). Their results showed that it takes about 6 weeks for the colon inner mucus layer to become fully impenetrable to bacteria and bacteria sized beads, and 8 weeks for the microbiota to reach the composition of Convr mice (90). Noticeably, a normalizing change in several mucus parameters correlated with a shift in the ratio between Firmicutes and Bacteroides in the mucus (90). The importance of the microbiota in modulating the mucus phenotype was also illustrated in a comparison of two C57BL/6 mouse colonies with the same genetic background housed in different rooms of the same vivarium. The two colonies were characterized by a divergence in their gut microbiota composition as well as in their mucus phenotypes (88). One colony had mucus that was impenetrable to bacteria or bacterium sized fluorescent beads, whereas the other colony had an inner mucus layer penetrable to bacteria and beads (88). These divergences were traced back to the gut microbiota, since transfer of cecal microbiota from the two colonies to GF mice, was able to transfer the respective mucus properties. Analysis of the gut microbiota composition demonstrated that mice with an impenetrable mucus layer had increased amounts of the Erysipelotrichi class (mainly the genus Allobaculum), whereas mice with a penetrable mucus layer had increased levels of Proteobacteria and TM7 bacteria in the distal colon mucus (88). In a study of inflammasome-deficient mice, the mucus barrier function in Il18-/- mice was found to be dependent of the microbiota and could be either transferred or lost upon co-housing with different wild-type mice. Two fecal bacteria strains: the Bacteroidales family S24-7 (Muribaculaceae) and the genus Adlercrutzia were identified to be consistently and positively correlated with inner mucus layer function (89). Volk et al., have also provided further detailed information regarding correlative and causative relations between bacteria and mucus properties when pooling public dataset of different experiments (89). Additionally, feeding mice a WD induced a bloom of the Proteobacteria Helicobacter and a lower relative abundance of S24-7 family and Bifidobacteria, which correlated with a microbiota-dependent loss of mucus barrier function (39). Re-introduction of Bifidobacteria corrected certain aspects of the mucus dysfunction, but did not completely restore the mucus properties. These findings through a pre-clinical approach highlight three important points: (i) it is the selective increase in certain bacterial species and their specific functions rather than changes to the entire microbial community that regulates the properties of the inner colon mucus barrier, (ii) housing conditions are critical cofounding factors when investigating microbe-mucus interactions, and a standardized approach should be considered when comparing animal studies, (iii) time is of importance when analyzing bacterial-host interaction in GF and Convr mice.
The mechanisms by which microorganisms regulate mucus properties involve both bacterial metabolites such as SCFAs and secondary bile acids, and microbial components that bind pattern recognition receptors such as toll-like receptors (TLRs) expressed by GCs (135). Among the several SCFAs, butyrate has been shown to increase the production of MUC2 in cultured intestinal epithelial cells (136), and stimulates mucus release from the rat colon (86, 137). In addition to SCFAs, other classes of bacterial metabolites involved in the regulation of the mucus properties are the secondary bile acids. One example is deoxycholic acid (DCA), which has been shown to induce MUC2 expression in cultured colonic epithelial cells (138, 139). However, there are many controversial results associating bile acids with improvement of gut barrier function, and further investigations are required to elucidate their role in regulation of gut health (2). Like the bacteria metabolites, exposure of colonic tissues to high concentrations of bacterial products such as LPS and peptidoglycan (PGN), induces mucus secretion in both GF and Convr mice (140). Furthermore, mice lacking the TLR adaptor protein MyD88 present a decreased production of mucus, impaired goblet cell responses and reduced antimicrobial activity against Citrobacter rodentium infection (141). To the same extent, mice lacking the flagellin receptor, TLR5 deficient mice, have a disorganized mucus structure as compared to wild-type mice and an increased abundance of Proteobacteria in close contact with the epithelial surface (26). Altogether, these in vitro and in vivo studies indicate the importance of microbes or their metabolites and components, and host TLRs in maintaining mucosal health.
It is well established that adaptive immune responses are immature in both the small and large intestine of GF as compared to Convr mice. GF mice have reduced size of Peyer’s patches and reduced numbers of IgA producing plasma cells, LP T cells, and intra epithelial lymphocytes (IELs) (142–144). With respect to innate immunity, less is known about the role of the microbiota in regulation of the respective cell types, and observations differ between studies, possibly related to variations in the microbiota composition of the Convr control group. Colonic macrophages, DCs, and mast cells have been reported to be immature and reduced in numbers in GF mice as compared to Convr mice, suggesting a stimulatory role of the microbiota in driving proliferation and maturation of these cell types (145, 146). In contrast, eosinophil and NKT cell numbers have been reported to be increased in GF mice pointing towards a suppressive role of the microbiota in regulation of these cells (147). However, despite the observed increase in eosinophil numbers in GF mice, the cells appear inactive as they express less of eosinophil peroxidase (148, 149).
To further dissect the role of specific members of the microbiota in regulation of gut immunity, mono-colonization or colonization of GF mice with a limited consortium of bacteria have been used to study the role of the microbiota in regulation of both adaptive and innate immunity. In the context of microbial regulation of tolerance to the luminal content, studies have demonstrated that bacteria from the Clostridium genus cluster IV and XIVa promote induction of FoxP3+ Tregs in the colon (150). As mentioned previously, Clostridia species are well known for their ability to metabolize dietary fibers into SCFAs, and catabolize tryptophan into the ahryl carbon receptor (AhR) ligands indole and indole derivates, and it is considered that most effects of Clostridia on regulation of intestinal immunity is mediated via these metabolites. SCFAs regulate immune cell function by two main pathways 1) by activating G protein coupled receptors (GPRs): GPR41, 43 and 109A, and by acting as histone deacetylase (HDAC) inhibitors. HDACs are enzymes that regulate gene expression and thereby affect a variety of functions including proliferation and differentiation (151, 152). In the colon, SCFAs promote induction of Foxp3+ Tregs via HDAC inhibition, and by activation of GPR43 (153, 154). Feeding GF mice with SCFAs was shown to increase FoxP3 and Il10 expression, and increase the suppressive effect of Tregs (155). Butyrate has also been shown to inhibit the Th17 transcription factor RoRγt, and IL-17 production in vitro (156). Thus, in the colon, SCFAs promote tolerance via Treg induction and inhibition of Th17 responses. In contrast, SCFAs have not been shown to induce Treg responses in the small intestine, however other bacterial metabolites such as secondary bile acids (e.g., 3-oxolithocholic acid) and tryptophan catabolites (e.g., indole) that bind the AhR stimulate Treg induction and inhibit Th17 responses in both the small intestine and colon (157–159). SCFAs have also been shown to stimulate IgA production by intestinal B cells, and increase LP-DC expression of indoleamine 2,3-dioxygenase 1 (IDO1) and aldehyde dehydrogenase 1A2 (Aldh1A2) which promote conversion of naïve T cells into Tregs, thus, further promoting an environment favoring tolerance over inflammation (160, 161). Collectively these findings underscored the importance of the microbiota in regulation of maturation of intestinal immunity and immune homeostasis.
It is now well established that early nutrition can influence the development of the gut microbiota (162), and immediately after birth, breast milk or infant formula or a combination of both is our primary diet. Human milk oligosaccharides (HMOs) which are the most abundant components in breast milk cannot be digested by the human infant, but because of their structural similarity to mucin-glycans, they can be used as a primary carbon source by several bacterial strains (e.g., Bifidobacterium species, members from the genus Bacteroides) implicated in the initial colonization of our intestine, with beneficial effect on our mucosal, immune, and metabolic health during later life (163, 164). Both in vitro and in vivo studies have highlighted the ability of HMOs and HMO-compounds (i.e., 2′ -fucosyllactose) to modulate mucins expression and the secretory function of GCs (165, 166). HMOs can directly control intestinal immunity by decoying receptors of pathogenic bacteria and viruses, thereby preventing their binding on intestinal cells and the onset of a disease (167). Transitioning from milk based to solid food, and the introduction of fiber to our diet, the pivotal metabolic substrate for the gut microbiota, induce the production of SCFAs. As mentioned previously, these gut-microbiota-derived metabolites and especially butyrate can improve mucus barrier function via modulation of MUC2 production and expression (26) (Figure 1A). Moreover these bacterial metabolites act as ligands for GPR43, GPR41, and GPR109A that in addition to being expressed by immune cells also are expressed by epithelial cell, primarily cells of the secretory lineage (168). Activation of these receptors triggers the release of enteric peptide hormones including glucagon-like peptides 1 and 2 and peptide YY, which regulate several metabolic functions such as improvement of the gut barrier, metabolic inflammation, and gut transit time (25).
In contrast to diets high in dietary fiber that promote intestinal health, WD and HF-diets that often are low in dietary fiber, have been associated with loss of mucus barrier function, impaired immune homeostasis, and increased susceptibility to chronic inflammatory diseases including inflammatory bowel diseases and food allergies (46, 169). These effects have largely been related to loss of SCFAs production, but studies have also demonstrated a direct toxic effect of dietary fatty acids on T cells in vitro (170). As mentioned previously, high intake of WD and HF diet, is associated with an altered and less diverse gut microbiota composition (48, 171), which in turn contribute to an impaired mucus barrier (39, 172). Similar observations have been made in mice treated with dietary emulsifiers (74, 173) (Figure 1B). Lack of dietary fiber induces a shift in the gut microbiota composition toward the utilization of host-glycans such as those provided by mucins as energy source, with deleterious consequences on the mucus barrier (174, 175), and with an expansion and activity of colonic mucus degrading bacteria (i.e., Akkermansia muciniphila and Bacteroides caccae) and a decrease in fiber-degrading bacteria (i.e., Eubacterium rectale and Bacteroides ovatus) both in fecal samples, the colonic lumen, and the mucus layer, resulting in an increased susceptibility to gastrointestinal pathogen infections in mice (45) (Figure 1B). Nowadays, particular attention is given to the mucin specialist Akkermansia muciniphila, whose abundance is reduced in mice exposed to increased dietary fat content. This observation is of interest, since Akkermansia muciniphila supplementation in both mice and humans has been linked to improved health outcomes and gut barrier function (176, 177), further reinforcing the idea that the presence of certain bacteria in our gut is of fundamental importance for our health. With respect to the effect of WD and HF diets on intestinal immunity, HF diet has been associated with loss of both Treg and Th17 populations in the small intestine, while in the colon HF diet has been associated with Th2 skewing, and increased susceptibility to allergic disease (178–180) (Figure 1B). Altogether, these findings emphasize that dietary patterns, bacteria and bacterial components contribute to maintaining gut homeostasis.
In this review we have highlighted the importance of the interplay between the resident microbiota and the host in regulation of intestinal homeostasis, and how this interplay is influenced by the dietary habits of the host. It is well established that diets with a high fiber content, and low amounts of fat and sugar promote a microbiota that has beneficial effect on intestinal health by stimulating intestinal mucus barrier function and promoting immune tolerance over inflammation. On the contrary, diets low in fiber, and high in fat and sugar have been shown to promote a microbiota associated with development of intestinal and extra-intestinal diseases such as food allergy, inflammatory bowel disease and metabolic disease. Despite the established role of the diet and microbiota in regulation of intestinal health, many fundamental questions remain to be answered and the challenges ahead lies in 1) identify the molecular mechanisms involved in mucus impairment, 2) further characterization of the bacteria and bacterial metabolites that influence goblet cell function and mucus properties, 3) establish the importance of different type of goblet cells in the control of mucus production and intestinal immunity, 4) assessment of the role of peripheral organs such as the liver in regulation of the mucus barrier via production of bioactive compounds further metabolized by the microbiota, 5) characterization of the mechanisms by which specific dietary components influences intestinal homeostasis, and how diet induced changes in the microbiota influences the ability of the intestine to maintain tolerance to the luminal content, 6) evaluation of the therapeutical potential of dietary fiber/bacterial metabolite supplements in restoration of mucus barrier defects and loss of oral tolerance, and 7) deeper investigation of the phage-mucus interactions.
FS and JG conceived, supervised, and revised the first draft of the review. FS, EN, DS, and JG wrote and edited the review. All authors read and approved the final version of the manuscript.
This work was supported by the WoM Lundgren grant (2022-3916), the Swedish Research Council (2019-01134 and 2020-01588), the Crohn’s and Colitis foundation (580014), and the Swedish Society for Medical Research (PD20-0168).
We thank Malin E. V. Johansson for critically revising the review, and Ana Sofia de Jesus Vaz Luis for providing inputs and literature. Figures 1 and 2 are created with BioRender.com
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aldh1A2, Adehydrogenase 1A2; AhR, Ahryl carbon receptor; AGR, Anterior gradient; AMPs, Anti-microbial peptides; CLCA1, Chloride channel accessory; Convr, Conventionally raised; DCs, Dendritic cells; FCGBP, Fc fragment of IgG binding protein; GPRs, G protein coupled receptors; GI, Gastrointestinal; GF, Germ-free; GCs, Goblet cells; HF, High-fat; HDAC, Histone deacetylase; HMOs, Human milk oligosaccharides; IDO1, Indoleamine 2,3-dioxygenase 1; ILCs, Innate lymphoid cells; IL, Interleukin; IECs, Intestinal epithelial cells; IELs, intra epithelial lymphocytes; LP, Lamina propria; LPS, Lipopolysaccharide; MUC2, Mucin2; NKT, Natural killer T cells; PGN, Peptidoglycan; IgA, Immunoglobulin A; SCFAs, Short-chain fatty acids; Th, T helper cells; Tregs, T regulatory cells; TLRs, Toll-like receptors; UC, Ulcerative colitis; WD, Western diet; ZG16, Zymogen granule protein 16.
1. Turner JR. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol (2009) 9(11):799–809. doi: 10.1038/nri2653
2. Ghosh S, Whitley CS, Haribabu B, Jala VR. Regulation of intestinal barrier function by microbial metabolites. Cell Mol Gastroenterol Hepatol (2021) 11(5):1463–82. doi: 10.1016/j.jcmgh.2021.02.007
3. Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. (2005) 307(5717):1915–20. doi: 10.1126/science.1104816
4. Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. (2009) 136(1):65–80. doi: 10.1053/j.gastro.2008.10.080
5. Vancamelbeke M, Vermeire S. The intestinal barrier: A fundamental role in health and disease. Expert Rev Gastroenterol Hepatol (2017) 11(9):821–34. doi: 10.1080/17474124.2017.1343143
6. Soderholm AT, Pedicord VA. Intestinal epithelial cells: At the interface of the microbiota and mucosal immunity. Immunology. (2019) 158(4):267–80. doi: 10.1111/imm.13117
7. Hooper LV, Macpherson AJ. Immune adaptations that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol (2010) 10(3):159–69. doi: 10.1038/nri2710
8. Belkaid Y, Harrison OJ. Homeostatic immunity and the microbiota. Immunity. (2017) 46(4):562–76. doi: 10.1016/j.immuni.2017.04.008
9. Hansson GC. Mucins and the microbiome. Annu Rev Biochem (2020) 89:769–93. doi: 10.1146/annurev-biochem-011520-105053
10. Muniz LR, Knosp C, Yeretssian G. Intestinal antimicrobial peptides during homeostasis, infection, and disease. Front Immunol (2012) 3:310. doi: 10.3389/fimmu.2012.00310
11. Cani PD, Moens de Hase E, Van Hul M. Gut microbiota and host metabolism: From proof of concept to therapeutic intervention. Microorganisms. (2021) 9(6):1302. doi: 10.3390/microorganisms9061302
12. Alemao CA, Budden KF, Gomez HM, Rehman SF, Marshall JE, Shukla SD, et al. Impact of diet and the bacterial microbiome on the mucous barrier and immune disorders. Allergy. (2021) 76(3):714–34. doi: 10.1111/all.14548
13. Sender R, Fuchs S, Milo R. Are we really vastly outnumbered? revisiting the ratio of bacterial to host cells in humans. Cell. (2016) 164(3):337–40. doi: 10.1016/j.cell.2016.01.013
14. Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. (2006) 124(4):837–48. doi: 10.1016/j.cell.2006.02.017
15. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. (2005) 308(5728):1635–8. doi: 10.1126/science.1110591
16. Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. (2010) 464(7285):59–65. doi: 10.1038/nature08821
17. Mizrahi-Man O, Davenport ER, Gilad Y. Taxonomic classification of bacterial 16S rRNA genes using short sequencing reads: evaluation of effective study designs. PloS One (2013) 8(1):e53608. doi: 10.1371/journal.pone.0053608
18. Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, et al. Enterotypes of the human gut microbiome. Nature. (2011) 473(7346):174–80. doi: 10.1038/nature09944
19. Rajilic-Stojanovic M, Smidt H, de Vos WM. Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol (2007) 9(9):2125–36. doi: 10.1111/j.1462-2920.2007.01369.x
20. Tropini C, Earle KA, Huang KC, Sonnenburg JL. The gut microbiome: Connecting spatial organization to function. Cell Host Microbe (2017) 21(4):433–42. doi: 10.1016/j.chom.2017.03.010
21. Sommer F, Backhed F. The gut microbiota–masters of host development and physiology. Nat Rev Microbiol (2013) 11(4):227–38. doi: 10.1038/nrmicro2974
22. Li H, Limenitakis JP, Fuhrer T, Geuking MB, Lawson MA, Wyss M, et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat Commun (2015) 6:8292. doi: 10.1038/ncomms9292
23. Ringel Y, Maharshak N, Ringel-Kulka T, Wolber EA, Sartor RB, Carroll IM. High throughput sequencing reveals distinct microbial populations within the mucosal and luminal niches in healthy individuals. Gut Microbes (2015) 6(3):173–81. doi: 10.1080/19490976.2015.1044711
24. Thursby E, Juge N. Introduction to the human gut microbiota. Biochem J (2017) 474(11):1823–36. doi: 10.1042/BCJ20160510
25. de Vos WM, Tilg H, Van Hul M, Cani PD. Gut microbiome and health: mechanistic insights. Gut (2022) 71:1020–1032. doi: 10.1136/gutjnl-2021-326789
26. Etienne-Mesmin L, Chassaing B, Desvaux M, De Paepe K, Gresse R, Sauvaitre T, et al. Experimental models to study intestinal microbes-mucus interactions in health and disease. FEMS Microbiol Rev (2019) 43(5):457–89. doi: 10.1093/femsre/fuz013
27. Suriano F, Van Hul M, Cani PD. Gut microbiota and regulation of myokine-adipokine function. Curr Opin Pharmacol (2020) 52:9–17. doi: 10.1016/j.coph.2020.03.006
28. Cao Z, Sugimura N, Burgermeister E, Ebert MP, Zuo T, Lan P. The gut virome: A new microbiome component in health and disease. EBioMedicine. (2022) 81:104113. doi: 10.1016/j.ebiom.2022.104113
29. Shkoporov AN, Turkington CJ, Hill C. Mutualistic interplay between bacteriophages and bacteria in the human gut. Nat Rev Microbiol (2022). doi: 10.1038/s41579-022-00755-4
30. Guerin E, Hill C. Shining light on human gut bacteriophages. Front Cell Infect Microbiol (2020) 10:481. doi: 10.3389/fcimb.2020.00481
31. Cena H, Calder PC. Defining a healthy diet: Evidence for the role of contemporary dietary patterns in health and disease. Nutrients. (2020) 12(2):334. doi: 10.3390/nu12020334
32. Graf D, Di Cagno R, Fak F, Flint HJ, Nyman M, Saarela M, et al. Contribution of diet to the composition of the human gut microbiota. Microb Ecol Health Dis (2015) 26:26164. doi: 10.3402/mehd.v26.26164
33. Paone P, Cani PD. Mucus barrier, mucins and gut microbiota: The expected slimy partners? Gut (2020) 69(12):2232–43. doi: 10.1136/gutjnl-2020-322260
34. Singh RK, Chang H-W, Yan D, Lee KM, Ucmak D, Wong K, et al. Influence of diet on the gut microbiome and implications for human health. J Trans Med (2017) 15(1):73. doi: 10.1186/s12967-017-1175-y
35. Cronin P, Joyce SA, O’Toole PW, O’Connor EM. Dietary fibre modulates the gut microbiota. Nutrients. (2021) 13(5):1655. doi: 10.3390/nu13051655
36. Nogal A, Valdes AM, Menni C. The role of short-chain fatty acids in the interplay between gut microbiota and diet in cardio-metabolic health. Gut Microbes (2021) 13(1):1–24. doi: 10.1080/19490976.2021.1897212
37. Parada Venegas D, de la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol (2019) 10. doi: 10.3389/fimmu.2019.00277
38. Blaak EE, Canfora EE, Theis S, Frost G, Groen AK, Mithieux G, et al. Short chain fatty acids in human gut and metabolic health. Benef Microbes (2020) 11(5):411–55. doi: 10.3920/BM2020.0057
39. Schroeder BO, Birchenough GMH, Stahlman M, Arike L, Johansson MEV, Hansson GC, et al. Bifidobacteria or fiber protects against diet-induced microbiota-mediated colonic mucus deterioration. Cell Host Microbe (2018) 23(1):27–40 e7. doi: 10.1016/j.chom.2017.11.004
40. Nagpal R, Shively CA, Register TC, Craft S, Yadav H. Gut microbiome-Mediterranean diet interactions in improving host health. F1000Res. (2019) 8:699. doi: 10.12688/f1000research.18992.1
41. Sanz JM, Sergi D, Colombari S, Capatti E, Situlin R, Biolo G, et al. Dietary acid load but not Mediterranean diet adherence score is associated with metabolic and cardiovascular health state: A population observational study from northern Italy. Front Nutr (2022) 9:828587. doi: 10.3389/fnut.2022.828587
42. Meslier V, Laiola M, Roager HM, De Filippis F, Roume H, Quinquis B, et al. Mediterranean Diet intervention in overweight and obese subjects lowers plasma cholesterol and causes changes in the gut microbiome and metabolome independently of energy intake. Gut. (2020) 69(7):1258–68. doi: 10.1136/gutjnl-2019-320438
43. De Filippis F, Pellegrini N, Vannini L, Jeffery IB, La Storia A, Laghi L, et al. High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut. (2016) 65(11):1812–21. doi: 10.1136/gutjnl-2015-309957
44. Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, Sonnenburg JL. Diet-induced extinctions in the gut microbiota compound over generations. Nature. (2016) 529(7585):212–5. doi: 10.1038/nature16504
45. Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. (2016) 167(5):1339–53.e21. doi: 10.1016/j.cell.2016.10.043
46. Birchenough G, Schroeder BO, Backhed F, Hansson GC. Dietary destabilisation of the balance between the microbiota and the colonic mucus barrier. Gut Microbes (2019) 10(2):246–50. doi: 10.1080/19490976.2018.1513765
47. Schroeder BO. Fight them or feed them: How the intestinal mucus layer manages the gut microbiota. Gastroenterol Rep (2019) 7(1):3–12. doi: 10.1093/gastro/goy052
48. Malesza IJ, Malesza M, Walkowiak J, Mussin N, Walkowiak D, Aringazina R, et al. High-fat, Western-style diet, systemic inflammation, and gut microbiota: A narrative review. Cells (2021) 10(11):3164. doi: 10.3390/cells10113164
49. Cani PD, Osto M, Geurts L, Everard A. Involvement of gut microbiota in the development of low-grade inflammation and type 2 diabetes associated with obesity. Gut Microbes (2012) 3(4):279–88. doi: 10.4161/gmic.19625
50. Zinocker MK, Lindseth IA. The Western diet-Microbiome-Host interaction and its role in metabolic disease. Nutrients. (2018) 10(3):365. doi: 10.3390/nu10030365
51. Caesar R, Tremaroli V, Kovatcheva-Datchary P, Cani PD, Bäckhed F. Crosstalk between gut microbiota and dietary lipids aggravates WAT inflammation through TLR signaling. Cell Metab (2015) 22(4):658–68. doi: 10.1016/j.cmet.2015.07.026
52. Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen YY, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. (2009) 137(5):1716–24 e1-2. doi: 10.1053/j.gastro.2009.08.042
53. Murphy EF, Cotter PD, Healy S, Marques TM, O’Sullivan O, Fouhy F, et al. Composition and energy harvesting capacity of the gut microbiota: Relationship to diet, obesity and time in mouse models. Gut. (2010) 59(12):1635–42. doi: 10.1136/gut.2010.215665
54. Ravussin Y, Koren O, Spor A, LeDuc C, Gutman R, Stombaugh J, et al. Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obes (Silver Spring). (2012) 20(4):738–47. doi: 10.1038/oby.2011.111
55. Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. (2007) 56(7):1761–72. doi: 10.2337/db06-1491
56. Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. (2008) 57(6):1470–81. doi: 10.2337/db07-1403
57. Ghanim H, Abuaysheh S, Sia CL, Korzeniewski K, Chaudhuri A, Fernandez-Real JM, et al. Increase in plasma endotoxin concentrations and the expression of toll-like receptors and suppressor of cytokine signaling-3 in mononuclear cells after a high-fat, high-carbohydrate meal: Implications for insulin resistance. Diabetes Care (2009) 32(12):2281–7. doi: 10.2337/dc09-0979
58. Sergi D, Luscombe-Marsh N, Heilbronn LK, Birch-Machin M, Naumovski N, Lionetti L, et al. The inhibition of metabolic inflammation by EPA is associated with enhanced mitochondrial fusion and insulin signaling in human primary myotubes. J Nutr (2021) 151(4):810–9. doi: 10.1093/jn/nxaa430
59. Scheithauer TPM, Rampanelli E, Nieuwdorp M, Vallance BA, Verchere CB, van Raalte DH, et al. Gut microbiota as a trigger for metabolic inflammation in obesity and type 2 diabetes. Front Immunol (2020) 11. doi: 10.3389/fimmu.2020.571731
60. Zou J, Chassaing B, Singh V, Pellizzon M, Ricci M, Fythe MD, et al. Fiber-mediated nourishment of gut microbiota protects against diet-induced obesity by restoring IL-22-Mediated colonic health. Cell Host Microbe (2018) 23(1):41–53.e4. doi: 10.1016/j.chom.2017.11.003
61. Cani PD, Neyrinck AM, Fava F, Knauf C, Burcelin RG, Tuohy KM, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. (2007) 50(11):2374–83. doi: 10.1007/s00125-007-0791-0
62. Suriano F, Bindels LB, Verspreet J, Courtin CM, Verbeke K, Cani PD, et al. Fat binding capacity and modulation of the gut microbiota both determine the effect of wheat bran fractions on adiposity. Sci Rep (2017) 7(1):5621. doi: 10.1038/s41598-017-05698-y
63. Lacroix M, Gaudichon C, Martin A, Morens C, Mathe V, Tome D, et al. A long-term high-protein diet markedly reduces adipose tissue without major side effects in wistar male rats. Am J Physiol Regul Integr Comp Physiol (2004) 287(4):R934–42. doi: 10.1152/ajpregu.00100.2004
64. Freudenberg A, Petzke KJ, Klaus S. Dietary l-leucine and l-alanine supplementation have similar acute effects in the prevention of high-fat diet-induced obesity. Amino Acids (2013) 44(2):519–28. doi: 10.1007/s00726-012-1363-2
65. Drummen M, Tischmann L, Gatta-Cherifi B, Adam T, Westerterp-Plantenga M. Dietary protein and energy balance in relation to obesity and Co-morbidities. Front Endocrinol (Lausanne). (2018) 9:443. doi: 10.3389/fendo.2018.00443
66. Astrup A, Raben A, Geiker N. The role of higher protein diets in weight control and obesity-related comorbidities. Int J Obes (Lond). (2015) 39(5):721–6. doi: 10.1038/ijo.2014.216
67. Campos-Nonato I, Hernandez L, Barquera S. Effect of a high-protein diet versus standard-protein diet on weight loss and biomarkers of metabolic syndrome: A randomized clinical trial. Obes Facts. (2017) 10(3):238–51. doi: 10.1159/000471485
68. Lepe M, Bacardi Gascon M, Jimenez Cruz A. Long-term efficacy of high-protein diets: a systematic review. Nutr Hosp. (2011) 26(6):1256–9. doi: 10.1590/S0212-16112011000600010
69. Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol (2007) 73(4):1073–8. doi: 10.1128/AEM.02340-06
70. Salonen A, Lahti L, Salojarvi J, Holtrop G, Korpela K, Duncan SH, et al. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J (2014) 8(11):2218–30. doi: 10.1038/ismej.2014.63
71. Russell WR, Gratz SW, Duncan SH, Holtrop G, Ince J, Scobbie L, et al. High-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am J Clin Nutr (2011) 93(5):1062–72. doi: 10.3945/ajcn.110.002188
72. Yang Y, Zhang Y, Xu Y, Luo T, Ge Y, Jiang Y, et al. Dietary methionine restriction improves the gut microbiota and reduces intestinal permeability and inflammation in high-fat-fed mice. Food Funct (2019) 10(9):5952–68. doi: 10.1039/C9FO00766K
73. Singh RK, Chang HW, Yan D, Lee KM, Ucmak D, Wong K, et al. Influence of diet on the gut microbiome and implications for human health. J Transl Med (2017) 15(1):73. doi: 10.1186/s12967-017-1175-y
74. Chassaing B, Koren O, Goodrich JK, Poole AC, Srinivasan S, Ley RE, et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature. (2015) 519(7541):92–6. doi: 10.1038/nature14232
75. Chassaing B, Van de Wiele T, De Bodt J, Marzorati M, Gewirtz AT. Dietary emulsifiers directly alter human microbiota composition and gene expression ex vivo potentiating intestinal inflammation. Gut. (2017) 66(8):1414–27. doi: 10.1136/gutjnl-2016-313099
76. Naimi S, Viennois E, Gewirtz AT, Chassaing B. Direct impact of commonly used dietary emulsifiers on human gut microbiota. Microbiome. (2021) 9(1):66. doi: 10.1186/s40168-020-00996-6
77. Maloy KJ, Powrie F. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature. (2011) 474(7351):298–306. doi: 10.1038/nature10208
78. Chelakkot C, Ghim J, Ryu SH. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp Mol Med (2018) 50(8):1–9. doi: 10.1038/s12276-018-0126-x
79. Martini E, Krug SM, Siegmund B, Neurath MF, Becker C. Mend your fences: The epithelial barrier and its relationship with mucosal immunity in inflammatory bowel disease. Cell Mol Gastroenterol Hepatol (2017) 4(1):33–46. doi: 10.1016/j.jcmgh.2017.03.007
80. Gustafsson JK, Ermund A, Johansson ME, Schutte A, Hansson GC, Sjovall H. An ex vivo method for studying mucus formation, properties, and thickness in human colonic biopsies and mouse small and large intestinal explants. Am J Physiol Gastrointest Liver Physiol (2012) 302(4):G430–8. doi: 10.1152/ajpgi.00405.2011
81. Johansson ME, Gustafsson JK, Holmen-Larsson J, Jabbar KS, Xia L, Xu H, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. (2014) 63(2):281–91. doi: 10.1136/gutjnl-2012-303207
82. Nystrom EEL, Martinez-Abad B, Arike L, Birchenough GMH, Nonnecke EB, Castillo PA, et al. An intercrypt subpopulation of goblet cells is essential for colonic mucus barrier function. Science (2021) 372(6539):eabb1590. doi: 10.1126/science.abb1590
83. van der Post S, Jabbar KS, Birchenough G, Arike L, Akhtar N, Sjovall H, et al. Structural weakening of the colonic mucus barrier is an early event in ulcerative colitis pathogenesis. Gut. (2019) 68(12):2142–51. doi: 10.1136/gutjnl-2018-317571
84. Yildiz HM, McKelvey CA, Marsac PJ, Carrier RL. Size selectivity of intestinal mucus to diffusing particulates is dependent on surface chemistry and exposure to lipids. J Drug Target (2015) 23(7-8):768–74. doi: 10.3109/1061186X.2015.1086359
85. Burger-van Paassen N, Vincent A, Puiman PJ, van der Sluis M, Bouma J, Boehm G, et al. The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: Implications for epithelial protection. Biochem J (2009) 420(2):211–9. doi: 10.1042/BJ20082222
86. Barcelo A, Claustre J, Moro F, Chayvialle JA, Cuber JC, Plaisancié P. Mucin secretion is modulated by luminal factors in the isolated vascularly perfused rat colon. Gut. (2000) 46(2):218–24. doi: 10.1136/gut.46.2.218
87. Barcelo A, Claustre J, Toumi F, Burlet G, Chayvialle JA, Cuber JC, et al. Effect of bile salts on colonic mucus secretion in isolated vascularly perfused rat colon. Dig Dis Sci (2001) 46(6):1223–31. doi: 10.1023/A:1010607127822
88. Jakobsson HE, Rodriguez-Pineiro AM, Schutte A, Ermund A, Boysen P, Bemark M, et al. The composition of the gut microbiota shapes the colon mucus barrier. EMBO Rep (2015) 16(2):164–77. doi: 10.15252/embr.201439263
89. Volk JK, Nystrom EEL, van der Post S, Abad BM, Schroeder BO, Johansson A, et al. The Nlrp6 inflammasome is not required for baseline colonic inner mucus layer formation or function. J Exp Med (2019) 216(11):2602–18. doi: 10.1084/jem.20190679
90. Johansson ME, Jakobsson HE, Holmen-Larsson J, Schutte A, Ermund A, Rodriguez-Pineiro AM, et al. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe (2015) 18(5):582–92. doi: 10.1016/j.chom.2015.10.007
91. Schroeder BO, Birchenough GMH, Pradhan M, Nystrom EEL, Henricsson M, Hansson GC, et al. Obesity-associated microbiota contributes to mucus layer defects in genetically obese mice. J Biol Chem (2020) 295(46):15712–26. doi: 10.1074/jbc.RA120.015771
92. Ermund A, Schutte A, Johansson ME, Gustafsson JK, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. i. gastrointestinal mucus layers have different properties depending on location as well as over the peyer’s patches. Am J Physiol Gastrointest Liver Physiol (2013) 305(5):G341–7. doi: 10.1152/ajpgi.00046.2013
93. Atuma C, Strugala V, Allen A, Holm L. The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am J Physiol Gastrointest Liver Physiol (2001) 280(5):G922–9. doi: 10.1152/ajpgi.2001.280.5.G922
94. Moor AE, Harnik Y, Ben-Moshe S, Massasa EE, Rozenberg M, Eilam R, et al. Spatial reconstruction of single enterocytes uncovers broad zonation along the intestinal villus axis. Cell. (2018) 175(4):1156–67.e15. doi: 10.1016/j.cell.2018.08.063
95. Meyer-Hoffert U, Hornef MW, Henriques-Normark B, Axelsson LG, Midtvedt T, Putsep K, et al. Secreted enteric antimicrobial activity localises to the mucus surface layer. Gut. (2008) 57(6):764–71. doi: 10.1136/gut.2007.141481
96. Dupont A, Heinbockel L, Brandenburg K, Hornef MW. Antimicrobial peptides and the enteric mucus layer act in concert to protect the intestinal mucosa. Gut Microbes (2014) 5(6):761–5. doi: 10.4161/19490976.2014.972238
97. Ermund A, Gustafsson JK, Hansson GC, Keita AV. Mucus properties and goblet cell quantification in mouse, rat and human ileal peyer’s patches. PloS One (2013) 8(12):e83688. doi: 10.1371/journal.pone.0083688
98. Kamphuis JBJ, Mercier-Bonin M, Eutamène H, Theodorou V. Mucus organisation is shaped by colonic content; a new view. Sci Rep (2017) 7(1):8527. doi: 10.1038/s41598-017-08938-3
99. Bergstrom K, Shan X, Casero D, Batushansky A, Lagishetty V, Jacobs JP, et al. Proximal colon-derived O-glycosylated mucus encapsulates and modulates the microbiota. Science. (2020) 370(6515):467–72. doi: 10.1126/science.aay7367
100. Bergstrom K, Xia L. The barrier and beyond: Roles of intestinal mucus and mucin-type O-glycosylation in resistance and tolerance defense strategies guiding host-microbe symbiosis. Gut Microbes (2022) 14(1):2052699. doi: 10.1080/19490976.2022.2052699
101. Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A. (2008) 105(39):15064–9. doi: 10.1073/pnas.0803124105
102. Matsuo K, Ota H, Akamatsu T, Sugiyama A, Katsuyama T. Histochemistry of the surface mucous gel layer of the human colon. Gut. (1997) 40(6):782–9. doi: 10.1136/gut.40.6.782
103. Nystrom EEL, Birchenough GMH, van der Post S, Arike L, Gruber AD, Hansson GC, et al. Calcium-activated chloride channel regulator 1 (CLCA1) controls mucus expansion in colon by proteolytic activity. EBioMedicine. (2018) 33:134–43. doi: 10.1016/j.ebiom.2018.05.031
104. Barmpatsalou V, Dubbelboer IR, Rodler A, Jacobson M, Karlsson E, Pedersen BL, et al. Physiological properties, composition and structural profiling of porcine gastrointestinal mucus. Eur J Pharm Biopharm. (2021) 169:156–67. doi: 10.1016/j.ejpb.2021.10.008
105. Lidell ME, Johansson ME, Morgelin M, Asker N, Gum JR Jr., Kim YS, et al. The recombinant c-terminus of the human MUC2 mucin forms dimers in Chinese-hamster ovary cells and heterodimers with full-length MUC2 in LS 174T cells. Biochem J (2003) 372(Pt 2):335–45. doi: 10.1042/bj20030003
106. Godl K, Johansson ME, Lidell ME, Morgelin M, Karlsson H, Olson FJ, et al. The n terminus of the MUC2 mucin forms trimers that are held together within a trypsin-resistant core fragment. J Biol Chem (2002) 277(49):47248–56. doi: 10.1074/jbc.M208483200
107. Carlstedt I, Herrmann A, Karlsson H, Sheehan J, Fransson LA, Hansson GC. Characterization of two different glycosylated domains from the insoluble mucin complex of rat small intestine. J Biol Chem (1993) 268(25):18771–81. doi: 10.1016/S0021-9258(17)46696-8
108. Larsson JM, Karlsson H, Sjovall H, Hansson GC. A complex, but uniform O-glycosylation of the human MUC2 mucin from colonic biopsies analyzed by nanoLC/MSn. Glycobiology. (2009) 19(7):756–66. doi: 10.1093/glycob/cwp048
109. Holmen Larsson JM, Thomsson KA, Rodriguez-Pineiro AM, Karlsson H, Hansson GC. Studies of mucus in mouse stomach, small intestine, and colon. III. gastrointestinal Muc5ac and Muc2 mucin O-glycan patterns reveal a regiospecific distribution. Am J Physiol Gastrointest Liver Physiol (2013) 305(5):G357–63. doi: 10.1152/ajpgi.00048.2013
110. Arike L, Holmen-Larsson J, Hansson GC. Intestinal Muc2 mucin O-glycosylation is affected by microbiota and regulated by differential expression of glycosyltranferases. Glycobiology. (2017) 27(4):318–28. doi: 10.1093/glycob/cww134
111. Bergstrom K, Fu J, Johansson ME, Liu X, Gao N, Wu Q, et al. Core 1- and 3-derived O-glycans collectively maintain the colonic mucus barrier and protect against spontaneous colitis in mice. Mucosal Immunol (2017) 10(1):91–103. doi: 10.1038/mi.2016.45
112. Crouzier T, Boettcher K, Geonnotti AR, Kavanaugh NL, Hirsch JB, Ribbeck K, et al. Modulating mucin hydration and lubrication by deglycosylation and polyethylene glycol binding. Advanced Materials Interfaces. (2015) 2(18):1500308. doi: 10.1002/admi.201500308
113. van der Post S, Subramani DB, Backstrom M, Johansson MEV, Vester-Christensen MB, Mandel U, et al. Site-specific O-glycosylation on the MUC2 mucin protein inhibits cleavage by the porphyromonas gingivalis secreted cysteine protease (RgpB). J Biol Chem (2013) 288(20):14636–46. doi: 10.1074/jbc.M113.459479
114. Stanley RA, Ram SP, Wilkinson RK, Roberton AM. Degradation of pig gastric and colonic mucins by bacteria isolated from the pig colon. Appl Environ Microbiol (1986) 51(5):1104–9. doi: 10.1128/aem.51.5.1104-1109.1986
115. Juge N. Microbial adhesins to gastrointestinal mucus. Trends Microbiol (2012) 20(1):30–9. doi: 10.1016/j.tim.2011.10.001
116. Sommer F, Adam N, Johansson ME, Xia L, Hansson GC, Backhed F. Altered mucus glycosylation in core 1 O-glycan-deficient mice affects microbiota composition and intestinal architecture. PloS One (2014) 9(1):e85254. doi: 10.1371/journal.pone.0085254
117. Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol (2012) 10(5):323–35. doi: 10.1038/nrmicro2746
118. Larsson JM, Karlsson H, Crespo JG, Johansson ME, Eklund L, Sjovall H, et al. Altered O-glycosylation profile of MUC2 mucin occurs in active ulcerative colitis and is associated with increased inflammation. Inflammation Bowel Dis (2011) 17(11):2299–307. doi: 10.1002/ibd.21625
119. Bergström JH, Birchenough GM, Katona G, Schroeder BO, Schütte A, Ermund A, et al. Gram-positive bacteria are held at a distance in the colon mucus by the lectin-like protein ZG16. Proc Natl Acad Sci U S A. (2016) 113(48):13833–8. doi: 10.1073/pnas.1611400113
120. Ouwerkerk JP, de Vos WM, Belzer C. Glycobiome: bacteria and mucus at the epithelial interface. Best Pract Res Clin Gastroenterol (2013) 27(1):25–38. doi: 10.1016/j.bpg.2013.03.001
121. Vuik F, Dicksved J, Lam SY, Fuhler GM, van der Laan L, van de Winkel A, et al. Composition of the mucosa-associated microbiota along the entire gastrointestinal tract of human individuals. United Eur Gastroenterol J (2019) 7(7):897–907. doi: 10.1177/2050640619852255
122. Pedron T, Mulet C, Dauga C, Frangeul L, Chervaux C, Grompone G, et al. A crypt-specific core microbiota resides in the mouse colon. mBio (2012) 3(3):e00116-12. doi: 10.1128/mBio.00116-12
123. Saffarian A, Mulet C, Regnault B, Amiot A, Tran-Van-Nhieu J, Ravel J, et al. Crypt- and mucosa-associated core microbiotas in humans and their alteration in colon cancer patients. mBio (2019) 10(4):e01315-19. doi: 10.1128/mBio.01315-19
124. Wrzosek L, Miquel S, Noordine ML, Bouet S, Joncquel Chevalier-Curt M, Robert V, et al. Bacteroides thetaiotaomicron and faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol (2013) 11:61. doi: 10.1186/1741-7007-11-61
125. Knoop KA, McDonald KG, McCrate S, McDole JR, Newberry RD. Microbial sensing by goblet cells controls immune surveillance of luminal antigens in the colon. Mucosal Immunol (2015) 8(1):198–210. doi: 10.1038/mi.2014.58
126. Kulkarni DH, Gustafsson JK, Knoop KA, McDonald KG, Bidani SS, Davis JE, et al. Goblet cell associated antigen passages support the induction and maintenance of oral tolerance. Mucosal Immunol (2020) 13(2):271–82. doi: 10.1038/s41385-019-0240-7
127. Sicherer SH, Sampson HA. Food allergy: Epidemiology, pathogenesis, diagnosis, and treatment. J Allergy Clin Immunol (2014) 133(2):291–307. doi: 10.1016/j.jaci.2013.11.020
128. Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet (2015) 47(9):979–86. doi: 10.1038/ng.3359
129. Mestecky J, Strober W, Russell MW, Cheroutre H, Lambrecht BN, Kelsall BL. Mucosal immunology. Academic Press, Kidlington, Oxford (2015).
130. Denning TL, Wang YC, Patel SR, Williams IR, Pulendran B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat Immunol (2007) 8(10):1086–94. doi: 10.1038/ni1511
131. Veenbergen S, van Berkel LA, du Pre MF, He J, Karrich JJ, Costes LM, et al. Colonic tolerance develops in the iliac lymph nodes and can be established independent of CD103(+) dendritic cells. Mucosal Immunol (2016) 9(4):894–906. doi: 10.1038/mi.2015.118
132. Rothenberg ME, Mishra A, Brandt EB, Hogan SP. Gastrointestinal eosinophils. Immunol Rev (2001) 179:139–55. doi: 10.1034/j.1600-065X.2001.790114.x
133. Chu Van T, Beller A, Rausch S, Strandmark J, Zänker M, Arbach O, et al. Eosinophils promote generation and maintenance of immunoglobulin-A-Expressing plasma cells and contribute to gut immune homeostasis. Immunity. (2014) 40(4):582–93. doi: 10.1016/j.immuni.2014.02.014
134. Schutte A, Ermund A, Becker-Pauly C, Johansson ME, Rodriguez-Pineiro AM, Backhed F, et al. Microbial-induced meprin beta cleavage in MUC2 mucin and a functional CFTR channel are required to release anchored small intestinal mucus. Proc Natl Acad Sci U S A. (2014) 111(34):12396–401. doi: 10.1073/pnas.1407597111
135. Burgueño JF, Abreu MT. Epithelial toll-like receptors and their role in gut homeostasis and disease. Nat Rev Gastroenterol Hepatol (2020) 17(5):263–78. doi: 10.1038/s41575-019-0261-4
136. Willemsen LEM, Koetsier MA, van Deventer SJH, van Tol EAF. Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin E1 and E2 production by intestinal myofibroblasts. Gut. (2003) 52(10):1442–7. doi: 10.1136/gut.52.10.1442
137. Shimotoyodome A, Meguro S, Hase T, Tokimitsu I, Sakata T. Short chain fatty acids but not lactate or succinate stimulate mucus release in the rat colon. Comp Biochem Physiol Part A: Mol Integr Physiol (2000) 125(4):525–31. doi: 10.1016/S1095-6433(00)00183-5
138. Song S, Byrd JC, Koo JS, Bresalier RS. Bile acids induce MUC2 overexpression in human colon carcinoma cells. Cancer. (2005) 103(8):1606–14. doi: 10.1002/cncr.21015
139. Lee HY, Crawley S, Hokari R, Kwon S, Kim YS. Bile acid regulates MUC2 transcription in colon cancer cells via positive EGFR/PKC/Ras/ERK/CREB, PI3K/Akt/IkappaB/NF-kappaB and p38/MSK1/CREB pathways and negative JNK/c-Jun/AP-1 pathway. Int J Oncol (2010) 36(4):941–53. doi: 10.3892/ijo_00000573
140. Petersson J, Schreiber O, Hansson GC, Gendler SJ, Velcich A, Lundberg JO, et al. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am J Physiol Gastrointest Liver Physiol (2011) 300(2):G327–33. doi: 10.1152/ajpgi.00422.2010
141. Bhinder G, Stahl M, Sham HP, Crowley SM, Morampudi V, Dalwadi U, et al. Intestinal epithelium-specific MyD88 signaling impacts host susceptibility to infectious colitis by promoting protective goblet cell and antimicrobial responses. Infect Immun (2014) 82(9):3753–63. doi: 10.1128/IAI.02045-14
142. Macpherson Andrew J, Gatto D, Sainsbury E, Harriman Gregory R, Hengartner H, Zinkernagel Rolf M. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. (2000) 288(5474):2222–6. doi: 10.1126/science.288.5474.2222
143. Umesaki Y, Setoyama H, Matsumoto S, Okada Y. Expansion of alpha beta T-cell receptor-bearing intestinal intraepithelial lymphocytes after microbial colonization in germ-free mice and its independence from thymus. Immunology. (1993) 79(1):32–7.
144. Suzuki H, Jeong KI, Okutani T, Doi K. Regional variations in the number and subsets of intraepithelial lymphocytes in the mouse small intestine. Comp Med (2000) 50(1):39–42.
145. Bain CC, Bravo-Blas A, Scott CL, Perdiguero EG, Geissmann F, Henri S, et al. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nat Immunol (2014) 15(10):929–37. doi: 10.1038/ni.2967
146. Schwarzer M, Hermanova P, Srutkova D, Golias J, Hudcovic T, Zwicker C, et al. Germ-free mice exhibit mast cells with impaired functionality and gut homing and do not develop food allergy. Front Immunol (2019) 10:205. doi: 10.3389/fimmu.2019.00205
147. Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. (2012) 336(6080):489–93. doi: 10.1126/science.1219328
148. Olszak T, An D, Zeissig S, Vera Miguel P, Richter J, Franke A, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. (2012) 336(6080):489–93. doi: 10.1126/science.1219328
149. Jiménez-Saiz R, Anipindi VC, Galipeau H, Ellenbogen Y, Chaudhary R, Koenig JF, et al. Microbial regulation of enteric eosinophils and its impact on tissue remodeling and Th2 immunity. Front Immunol (2020) 11. doi: 10.3389/fimmu.2020.00155
150. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous clostridium species. Science. (2011) 331(6015):337–41. doi: 10.1126/science.1198469
151. Ellmeier W, Seiser C. Histone deacetylase function in CD4+ T cells. Nat Rev Immunol (2018) 18(10):617–34. doi: 10.1038/s41577-018-0037-z
152. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic treg cell homeostasis. Science. (2013) 341(6145):569–73. doi: 10.1126/science.1241165
153. Kespohl M, Vachharajani N, Luu M, Harb H, Pautz S, Wolff S, et al. Microbial metabolite butyrate induces expression of Th1-associated factors in CD4+ T cells. The Front Immunol (2017) 8:1036. doi: 10.3389/fimmu.2017.01036
154. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. (2013) 504(7480):446–50. doi: 10.1038/nature12721
155. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, et al. The microbial metabolites, short-chain fatty acids, regulate colonic treg cell homeostasis. Science (2013) 341(6145):569–73. doi: 10.1126/science.1241165
156. Chen L, Sun M, Wu W, Yang W, Huang X, Xiao Y, et al. Microbiota metabolite butyrate differentially regulates Th1 and Th17 cells’ differentiation and function in induction of colitis. Inflammatory Bowel Diseases. (2019) 25(9):1450–61. doi: 10.1093/ibd/izz046
157. Hang S, Paik D, Yao L, Kim E, Trinath J, Lu J, et al. Bile acid metabolites control TH17 and treg cell differentiation. Nature. (2019) 576(7785):143–8. doi: 10.1038/s41586-019-1785-z
158. Song X, Sun X, Oh SF, Wu M, Zhang Y, Zheng W, et al. Microbial bile acid metabolites modulate gut RORgamma(+) regulatory T cell homeostasis. Nature. (2020) 577(7790):410–5. doi: 10.1038/s41586-019-1865-0
159. Lee JS, Cella M, McDonald KG, Garlanda C, Kennedy GD, Nukaya M, et al. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of notch. Nat Immunol (2011) 13(2):144–51. doi: 10.1038/ni.2187
160. Isobe J, Maeda S, Obata Y, Iizuka K, Nakamura Y, Fujimura Y, et al. Commensal-bacteria-derived butyrate promotes the T-cell-independent IgA response in the colon. Int Immunol (2020) 32(4):243–58. doi: 10.1093/intimm/dxz078
161. Gurav A, Sivaprakasam S, Bhutia YD, Boettger T, Singh N, Ganapathy V. Slc5a8, a na+-coupled high-affinity transporter for short-chain fatty acids, is a conditional tumour suppressor in colon that protects against colitis and colon cancer under low-fibre dietary conditions. Biochem J (2015) 469(2):267–78. doi: 10.1042/BJ20150242
162. Wopereis H, Oozeer R, Knipping K, Belzer C, Knol J. The first thousand days - intestinal microbiology of early life: Establishing a symbiosis. Pediatr Allergy Immunol (2014) 25(5):428–38. doi: 10.1111/pai.12232
163. Wang BX, Wu CM, Ribbeck K. Home, sweet home: How mucus accommodates our microbiota. FEBS J (2021) 288(6):1789–99. doi: 10.1111/febs.15504
164. Belzer C. Nutritional strategies for mucosal health: The interplay between microbes and mucin glycans. Trends Microbiol (2022) 30(1):13–21. doi: 10.1016/j.tim.2021.06.003
165. Wu RY, Li B, Koike Y, Maattanen P, Miyake H, Cadete M, et al. Human milk oligosaccharides increase mucin expression in experimental necrotizing enterocolitis. Mol Nutr Food Res (2019) 63(3):e1800658. doi: 10.1002/mnfr.201800658
166. Figueroa-Lozano S, Akkerman R, Beukema M, van Leeuwen SS, Dijkhuizen L, de Vos P. 2′-fucosyllactose impacts the expression of mucus-related genes in goblet cells and maintains barrier function of gut epithelial cells. J Funct Foods. (2021) 85:104630. doi: 10.1016/j.jff.2021.104630
167. Singh RP, Niharika J, Kondepudi KK, Bishnoi M, Tingirikari JMR. Recent understanding of human milk oligosaccharides in establishing infant gut microbiome and roles in immune system. Food Res Int (2022) 151:110884. doi: 10.1016/j.foodres.2021.110884
168. Tabula Muris C, Overall C, Logistical C, Organ C, Processing, Library, et al. Single-cell transcriptomics of 20 mouse organs creates a tabula muris. Nature. (2018) 562(7727):367–72. doi: 10.1038/s41586-018-0590-4
169. Statovci D, Aguilera M, MacSharry J, Melgar S. The impact of Western diet and nutrients on the microbiota and immune response at mucosal interfaces. Front Immunol (2017) 8:838. doi: 10.3389/fimmu.2017.00838
170. Tanaka S, Nemoto Y, Takei Y, Morikawa R, Oshima S, Nagaishi T, et al. High-fat diet-derived free fatty acids impair the intestinal immune system and increase sensitivity to intestinal epithelial damage. Biochem Biophys Res Commun (2020) 522(4):971–7. doi: 10.1016/j.bbrc.2019.11.158
171. Everard A, Cani PD. Diabetes, obesity and gut microbiota. Best Pract Res Clin Gastroenterol (2013) 27(1):73–83. doi: 10.1016/j.bpg.2013.03.007
172. Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. (2013) 110(22):9066–71. doi: 10.1073/pnas.1219451110
173. Lock JY, Carlson TL, Wang CM, Chen A, Carrier RL. Acute exposure to commonly ingested emulsifiers alters intestinal mucus structure and transport properties. Sci Rep (2018) 8(1):10008. doi: 10.1038/s41598-018-27957-2
174. Sonnenburg ED, Sonnenburg JL. Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab (2014) 20(5):779–86. doi: 10.1016/j.cmet.2014.07.003
175. Earle KA, Billings G, Sigal M, Lichtman JS, Hansson GC, Elias JE, et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe (2015) 18(4):478–88. doi: 10.1016/j.chom.2015.09.002
176. Cani PD, de Vos WM. Next-generation beneficial microbes: The case of akkermansia muciniphila. Front Microbiol (2017) 8:1765. doi: 10.3389/fmicb.2017.01765
177. Depommier C, Everard A, Druart C, Plovier H, Van Hul M, Vieira-Silva S, et al. Supplementation with akkermansia muciniphila in overweight and obese human volunteers: A proof-of-concept exploratory study. Nat Med (2019) 25(7):1096–103. doi: 10.1038/s41591-019-0495-2
178. Garidou L, Pomié C, Klopp P, Waget A, Charpentier J, Aloulou M, et al. The gut microbiota regulates intestinal CD4 T cells expressing RORγt and controls metabolic disease. Cell Metab (2015) 22(1):100–12. doi: 10.1016/j.cmet.2015.06.001
179. Hussain M, Bonilla-Rosso G, Kwong Chung CKC, Bariswyl L, Rodriguez MP, Kim BS, et al. High dietary fat intake induces a microbiota signature that promotes food allergy. J Allergy Clin Immunol (2019) 144(1):157–70 e8. doi: 10.1016/j.jaci.2019.01.043
Keywords: Mediterranean diet, western diet, fibers, microbiota, bacterial metabolites, mucus layer, intestinal immune system, gut health
Citation: Suriano F, Nyström EEL, Sergi D and Gustafsson JK (2022) Diet, microbiota, and the mucus layer: The guardians of our health. Front. Immunol. 13:953196. doi: 10.3389/fimmu.2022.953196
Received: 25 May 2022; Accepted: 19 August 2022;
Published: 13 September 2022.
Edited by:
Silvia Melgar, University College Cork, IrelandReviewed by:
Johan Dabrosin Söderholm, Linköping University, SwedenCopyright © 2022 Suriano, Nyström, Sergi and Gustafsson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Francesco Suriano, ZnJhbmNlc2NvLnN1cmlhbm9AbWVka2VtLmd1LnNl; Jenny K. Gustafsson, amVubnkuZ3VzdGFmc3NvbkBndS5zZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.