
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Immunol., 05 August 2022
Sec. Viral Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.947789
 Xizhen Zhang1,2
Xizhen Zhang1,2 Wei Yu1,2*
Wei Yu1,2*Heat shock proteins (HSPs) are a kind of proteins which mostly found in bacterial, plant and animal cells, in which they are involved in the monitoring and regulation of cellular life activities. HSPs protect other proteins under environmental and cellular stress by regulating protein folding and supporting the correctly folded structure of proteins as chaperones. During viral infection, some HSPs can have an antiviral effect by inhibiting viral proliferation through interaction and activating immune pathways to protect the host cell. However, although the biological function of HSPs is to maintain the homeostasis of cells, some HSPs will also be hijacked by viruses to help their invasion, replication, and maturation, thereby increasing the chances of viral survival in unfavorable conditions inside the host cell. In this review, we summarize the roles of the heat shock protein family in various stages of viral infection and the potential uses of these proteins in antiviral therapy.
Heat shock proteins (HSPs) were first discovered in the salivary glands of flies, where they are expressed under heat shock conditions. HSPs have a wide range of molecular weights from approximately 10 to 100 kDa and can be classified into different groups according to their molecular weight, including small heat shock proteins (sHSPs), HSP40, HSP60, HSP70, HSP90 and large heat shock proteins (1).
The sHSPs, most of which are heat-inducible, have a wide range of molecular weights from 12-43 kDa and are widely distributed in a variety of tissues. The ability to prevent the aggregation of proteins and polypeptides is the most important function of many sHSPs (2). Depending on the status of client proteins, sHSPs exert different molecular chaperone functions (3–5). HSP40, HSP60, HSP70 and HSP90 are well-studied heat shock proteins that often perform biological functions in cells as complexes. They are extensively involved in the lifecycle of proteins, including protein folding and refolding, transport, degradation, assembly, activity regulation, and translocation, as well as the depolymerization of protein aggregates. The heat shock protein family is also involved in many fundamental cellular processes including cell cycle control, cell survival, hormone signaling and response to cellular stress through the extensive regulation of intracellular proteins (6–11). Large HSPs, such as HSP100 and HSP110, contain a loop structure that gives them a high capacity of binding to polypeptide substrates or non-protein ligands such as pathogen-associated molecules (12). Both HSP100 and HSP110 have chaperone activity with HSP70, and they can regulate protein aggregation by forming HSP104-HSP70-HSP40 (13) and HSP110-HSP70-HSP40 (14) ternary complexes to maintain cellular homeostasis in a variety of cellular life activities.
The expression of HSPs is not only induced by heat or cold but is also responsive to a range of stressors including starvation (15), hypoxia (16), ultraviolet (UV) irradiation (17), exposure to heavy metals (18) and microbial infection (19). During viral infection, HSPs protect the host cells mainly by their chaperone functions. Small heat shock proteins are produced in large quantities in response to stress (20), partly activating immune signaling pathways (21), and partly assembling complexes to modulate apoptosis (22). The bigger members also assemble HSP complexes to fold host proteins correctly and refold aggregates of stress-denatured proteins (23). Importantly, some heat shock proteins are directly involved in the inhibition of viral replication and transcription (24).
Although the HSP family is a class of protective proteins, they can be hijacked by viruses to aid host cell invasion. Viruses lacking molecular chaperones can utilize the native HSPs of the host cell to help them invade cells and the nucleus (25), stabilize and regulate their own transcription and translation (26), assemble viral proteins, or alter the intracellular environment to promote viral proliferation (27, 28). The aim of this review is to collate relevant reports on the role of the HSP family in various phases of viral infection and pave the way for subsequent research on related treatments.
Based on the function of chaperones, HSPs widely participate in biomolecular networks by binding to proteins of various functions in space and time (Table 1). Under normal conditions, HSPs play a role in the regulation of the cellular life cycle and functions, while under stress, HSPs are one of the main systems to be activated and regulate stress resistance, thereby enhancing viability. The main stressors that organisms face can be roughly divided into three categories, including physical, chemical and biological factors (19). Here, we briefly summarize the responses of HSPs in response to the most common stressors such as cold, anoxia and pathogenic microorganisms.
In response to stressful environments, HSPs regulate transcription and translation by acting as accessory proteins. In a study on the cold adaptation of the Asiatic rice borer moth, Chilo suppressalis (20), small HSPs (sHSPs) were found to act as the first line of cellular defense against protein unfolding caused by the environmental stress, since their protein depolymerization activity is independent of ATP (29). They are capable of binding a large range of non-native substrate proteins to form sHSP-substrate complexes that prevent irreversible aggregation (30). Four sHSP genes were found in the genome of C. suppressalis, three of which are highly induced in response to cold stress and associated with HSP Beta-1 (HSPB1)-related protein (HSPB1AP). HSP70 and HSP90 are then synergistically upregulated at the transcriptional level, which requires the participation of HSP40 (31) and a protein called HSP90 ATPase homolog activator (HSP90aa) (32). Afterwards, HSP70 and HSP90 cooperatively refold proteins (Figure 1A) (33, 34).
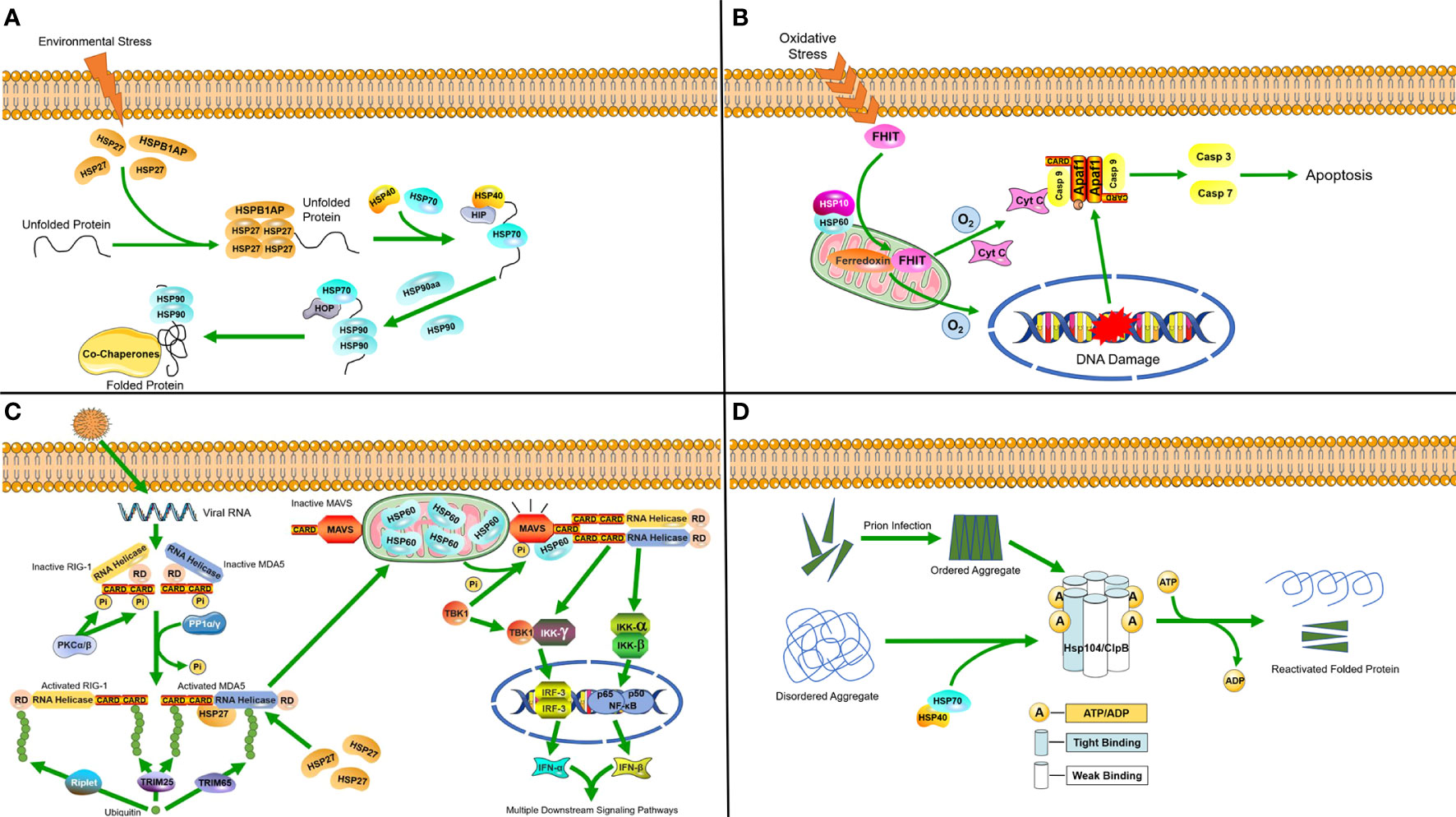
Figure 1 Molecular mechanisms of heat shock proteins induced under stress (A) Hsp27 binds to unfolded proteins that accumulate in the cytosol during stress, and then diverts unfolded proteins along the protein folding pathway, ultimately reaching HSP90. (B) In response to oxidative stress, the HSP60-10 complex helps to localize FHIT protein to the mitochondria, where it stabilizes ferredoxin reductase, leading to enhanced production of reactive oxygen species. This in turn triggers cytochrome c release and subsequent activation of the caspase cascade, ultimately causing apoptosis. (C) Following viral invasion, the RLR/MDA5 signaling pathway is activated. HSP27 can specifically stabilize MDA5 during expression to enhance the RLR/MDA5 signaling pathway. In mitochondria, HSP60 interacts with MAVS to increase MAVS-mediated IFN-β promoter activity and the transcriptional levels of IFN-β. Furthermore, it can upregulate MAVS-induced mRNA transcription of IFN-stimulated genes (ISGs). (D) Hsp104/ClpB complexes in host cells process disordered aggregates accumulated following cellular stress as well as ordered aggregates formed after prion infection with the help of the HSP70-40 partner system, dissociating them into component proteins and reactivating them.
At normal levels of oxygen, HSP60 forms a complex with the pro-apoptotic factor BCL2-associated X (Bax) in the cytosol and inhibits its translocation into the mitochondria, thereby preventing apoptosis. However, when cells are faced with hypoxia, the formation of complexes will be reduced and release Bax for translocation into the mitochondria, which results in the release of cytochrome c as an apoptotic signal (35). The HSP60-10 complex responds to oxidative stress and induces apoptosis when cells are under the dual stress of hypoxia and DNA damage (Figure 1B) (36).
HSPs are also induced when the host cell is infected by pathogenic microorganisms. A study on porcine reproductive and respiratory syndrome virus (PRRSV) identified HSP60 as a novel antiviral protein that inhibits viral replication (37). PRRSV infection activates PP1α/γ to dephosphorylate the originally phosphorylated MDA5 and RIG-1, after which MDA5 and RIG-1 are activated through ubiquitination. MAVS is phosphorylated and activated by TBK1, after which it interacts with RIG-I or MDA5, which triggers formation of a signaling synapse resulting in the formation of the canonical IFN-β enhanceosome complex that promotes IFN-β transcription. The RLR/MDA5 (RIG-I like receptor/melanoma differentiation-related gene 5) signaling pathway promotes the production of type I interferon to active the downstream signaling pathways (38). Upon the activation of mitochondrial antiviral signaling proteins (MAVS), HSP60 from the mitochondria binds to the MAVS protein and increases the expression of IFN-β, which can inhibit viral replication (37, 39). A recent study also found that the chaperone HSP27 positively regulates the RLR/MDA5 signaling pathway, which is triggered by encephalomyocarditis virus (EMCV) by stabilizing the expression of MDA5 to inhibit viral replication (Figure 1C) (40)
As molecular chaperones, heat shock proteins function by binding to client proteins in response to cellular or organismal stress. Small heat shock proteins bind directly to the target protein to prevent it from unfolding (2) or to transfer it to a complex transfer it to a complex which is inclined to be formed by larger heat shock proteins for further folding or refolding (20, 23). In response to stress, heat shock proteins either bind directly to protect the target protein (20), or affect factors that regulate cellular activities such as apoptosis (35) and immune signaling pathways (38). HSPs are activated by a wide variety of cellular stresses to maintain cellular homeostasis. However, it is also this characteristic that makes HSPs an easy target for viruses to break through host defenses. In this review, we will focus on the main functions of HSPs in viral infection.
There are a large number of studies reporting that HSPs are not only involved in antiviral responses, but also could be utilized by the virus to help cell entry, viral replication and virion assembly.
As a class of protective proteins, HSPs can inhibit viral proliferation by interacting with viral molecules and their related proteins. For example, HSC70/HSP90 has already been confirmed to be a driving force of the RNA-induced silencing complex (RISC) assembly pathway by providing ATP to load small RNA duplexes into argonaute protein, which can promote complex formation (24). Subsequently, RISC binds to viral mRNA, leading to the repression of viral translation. In the study of HPV, it was found that secreted HSP70 can effectively target dendritic cells with relevant antigens to enhance the antigen-specific immune response (41). The ClpB/HSP104 complex can disassemble disordered aggregates that accumulate due to cellular stress, as well as ordered aggregates formed by prions with the help of the HSP70 chaperone system, and reactivate their constituent proteins (Figure 1D) (23).
HSP can also regulate immune signaling pathways to resist viral infections. As mentioned above, HSP60 regulates the RLR/MDA5 signaling pathway to influence cellular immunity. In addition, HSP40 was also found to bind to MDA5 in the MDA5-MAVS pathway to disrupt the formation of MDA5 multimers, resulting in the suppression of type I IFN induction and protecting host cells from damage caused by excessive inflammation triggered by viral infection (42). Many studies have found that the upregulation of HSP27 inhibits the replication of porcine epidemic diarrhea virus (PEDV) and red spotted grouper neuro necrosis virus (RGNNV). HSP27 significantly increases the phosphorylation of NF-κB as an upstream regulator, which in turn upregulates interferon promoter activity and activates downstream interferon-stimulated genes. Viruses have also developed counteracting strategies to significantly downregulate HSP27 expression (43, 44). In general, HSP27 interacts with many different viral proteins to regulate the activity of IFN-1 and NF-κB signaling pathways (Figure 2A) (21, 45).
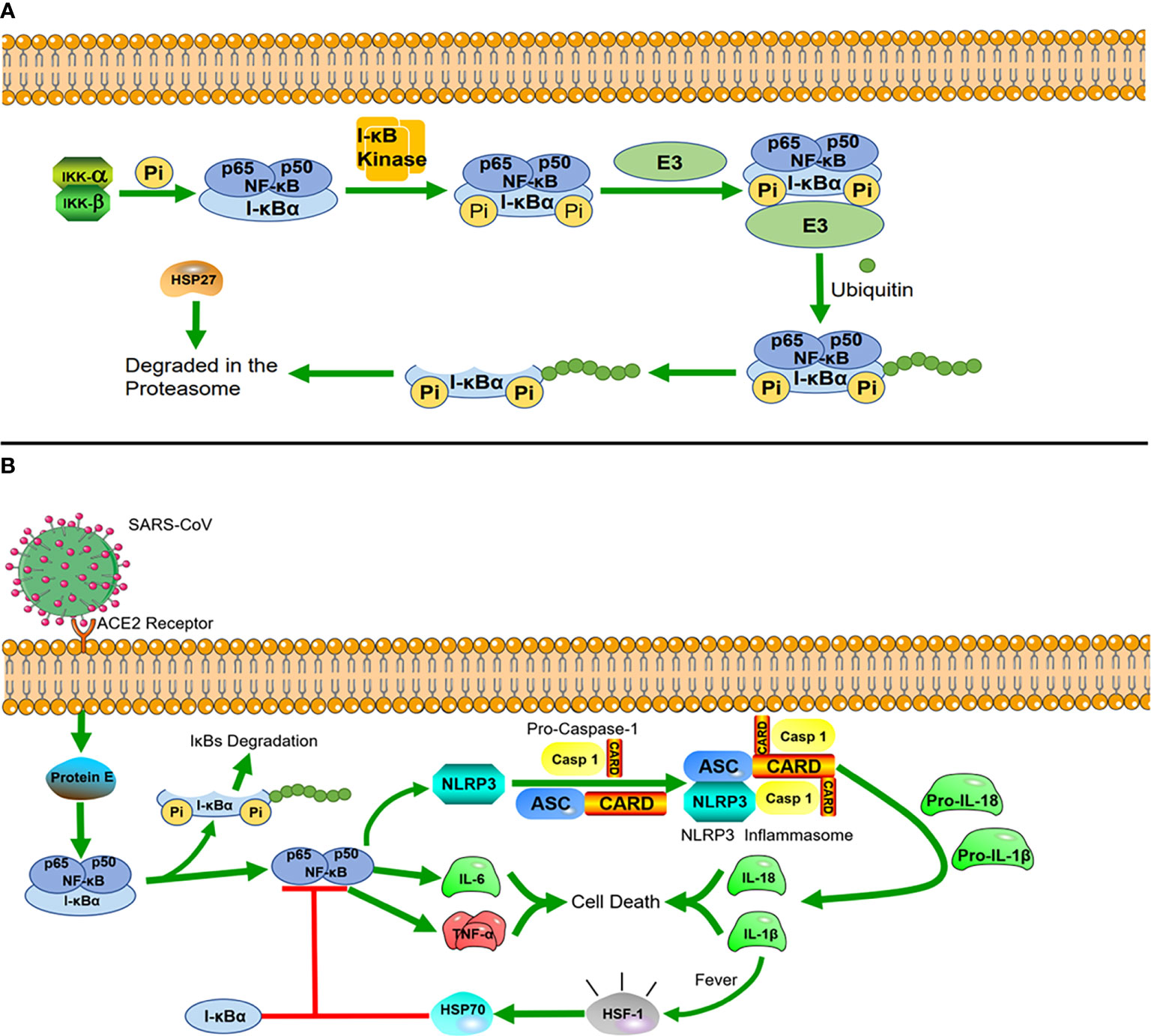
Figure 2 Heat shock proteins and immunological pathways (A) HSP27 regulates the NF-κB pathway. In the NF-κB signaling pathway, nuclear factor κB mainly exists as a heterodimer of p65 and P50, and I-κBα is a major inhibitor of NF-κB, which combines with them to form a complex in the resting state. The dimers are held inactive in the cytoplasm by their interaction with I-kBα proteins. I-κBα is phosphorylated when stimulated by external signals, and after phosphorylation, I-κBα proteins undergo ubiquitin-dependent degradation by the proteasome, after which NF-κB is translocated to the nucleus, where it acts as a transcription factor. The interaction of HSP27 with the 26S proteasome is necessary for the degradation of phosphorylated I-κBα, and overexpression of HSP27 enhances the proteasomal degradation of phosphorylated I-κBα. (B) HSP70 negatively regulates NLRP3 inflammatory vesicles. After SARS-CoV infects cells, the virus envelope E protein triggers the activation of the NF-κB inflammatory signaling cascade, which activates the NLRP3 inflammasome. Activation of NLRP3 induces the maturation of caspase-1, which in turn activates the secretion of interleukins IL-1β and IL-18. While IL-1β is an important factor in inducing a rise in core body temperature, HSP70, produced in response to the heat shock factor 1 (HSF-1), reduces the inflammatory response blocking NLRP3 and the articulator ASC to induce caspase-1 precursor maturation following a rise in body temperature.
During the two years of the COVID-19 epidemic, many studies on inflammation caused by coronavirus infection have been reported, in which we can also find new roles of HSPs. The evolutionarily conserved innate immune system is the first defense line against viral infection (46). The innate immune system is highly sensitive to stimuli, which rapidly recruit cells within minutes (neutrophils) to hours (monocytes/macrophages) to the site of injury. These rapid responses are orchestrated primarily by the expression of NF-κB, which drives inflammation during the early phase (47). There is a unique class of cytoplasmic receptors in the innate immune system called nucleotide-binding and oligomerization domain (NOD)-like receptors (NLRs), which constantly patrol for invading pathogens in the cytoplasm. At the heart of damaging inflammatory responses in many diseases is a multimolecular complex called the NOD-like receptor protein 3(NLRP3) inflammasome (48). In COVID-19, the viral envelope E protein triggers the activation of the NF-κB inflammatory signaling cascade and the interaction with inflammatory factors, such as tumor necrosis factor-alpha (TNF-α) and interleukin 6 (IL-6). These changes act as strong stimuli activating the cytosolic innate immune NLRP3 inflammasome. Once constituted, the NLRP3 inflammasome is secreted from the cells and can amplify the inflammatory response by activating the inflammasome and caspase-1 in neighboring cells. A recent study found that overexpression of HSP70 can inhibit the activation of the NLRP3 inflammasome, which in turn regulates the activation of caspase-1 (49)and the maturation of IL-1β (50) (Figure 2B). In related drug treatment studies, HSP90 inhibitors were found to block the initiation and activation of the NLRP3 inflammasome (51, 52).
Once the cells activate the inflammatory response, cyclooxygenase-2 (COX-2) is induced and starts producing proinflammatory arachidonic acid-derived prostaglandins (PGs) to promote the repair of inflammatory cells and tissues. Furthermore, PGs lead to an increase of the core body temperature (fever), which also triggers the heat shock response (HSR) (53). Under the influence of fever, structural changes in the plasma membrane directly activate heat shock factor 1 (HSF-1), whcih regulates the transcription of HSPs, expression of cytokines, and early response genes. The production of HSP70 in response to HSF-1 activation is correlated with complex formation between NF-κB and its inhibitor (I-κB) to prevent the translocation of NF-κB into the nucleus, which downregulates the acute inflammatory response (54). This avoids excessive protein damage or a cytokine storm induced by excessive inflammation (55).
Attachment is the first crucial step in the initiation of viral infection. It depends on the interaction between the viral attachment proteins and cellular receptors, which are key determinants of viral host specialization and pathogenesis. As a family of chaperone proteins widely distributed in cells, HSPs have been found to act as receptors for a variety of viruses in recent studies (56–58), and they are mainly involved in the viral contact and cell invasion in two ways.
Firstly, they participate in the process of viral entry into cells through endocytosis mediated by endocytosin and clathrin. A large number of helper proteins involved in endocytosis mediated by clathrin are present in various cells, and the HSP family is also represented among them. The D isoform of heat shock cognate protein 70 (HSC70) was found to help Japanese encephalitis virus (JEV) penetrate C6/36 cells via clathrin-mediated endocytosis (58). Another important chaperone, HSP90, was also recently found to form a complex with red spotted grouper neuronecrosis virus (RGNNV) on the cell surface and independently lead to RGNNV internalization through the clathrin endocytosis pathway (59).
Similarly, HSPs can also directly bind to virions as receptors on the cell surface. In existing reports, HSP70 was found to be involved in the invasion of various viruses in C6/36 cells. For example, HSC70 is involved in the process of dengue virus (DENV) invasion of cells by interacting with the DENV receptor complex (60, 61), while HSC70 interacts with the VP5 subunit of rotavirus spike protein to help it enter cells through endocytosis (24). Similar to HSP70, HSP90 is also an important component of the dengue virus receptor complex. In the available literature, HSP90 was found to be utilized directly as a cell surface to regulate receptor-mediated endocytosis pathways by many viruses, such as infectious bursal disease virus (62), dengue virus (63) and Japanese encephalitis virus (64). HSP90AA1 is a subtype of the HSP90 family and it was found to be involved in the cell entry of influenza A virus (IAV). IAV was reported to initiate the entry process via multiple endocytic pathways mediated by the viral hemagglutinin (HA) glycoprotein (65). HSP90AA1 is distributed on the cell surface and can regulate the entry of IAV directly by interacting with viral hemagglutinin (HA) (64).
Some viruses need to translocate viral molecules into the nucleus to interfere in the regulation of the cell’s internal environment or to advance replication of the viral own genome after invading a cell. HSPs are also involved in nuclear transport or the regulation of the intracellular environment to favor virion production, such as inducing tubulin acetylation to arrest the cell cycle (66) and so on. In IAV infection, HSP90 first exhibits downregulated acetylation levels along with enhanced nuclear transport to assist viral polymerase nuclear entry, after which the virus induces an upregulation of HSP90 acetylation levels, which indicates that HSPs play different roles at different phases of infection (67). Early in the IAV infection process, HSP40 (DnaJB1) can bind to the nucleoprotein (NP) of IAV with a nuclear localization signal and assists IAV viral ribonucleoprotein (vRNP) with nuclear trafficking through its interaction with nucleoproteins, which is also very important for viral protein entry (25). Similarly, HSP90 plays a role in enhancing the interaction between viral proteins and tubulin by binding to the acetylated α-tubulin to upregulate nuclear transport, which has been found in several viral infections, including mouse polyomavirus and herpes simplex virus 1 (68, 69).
HSPs not only assist in the nucleation of viral molecules, but are also intimately involved in the replication, transcription and translation of viruses, mainly in two ways. Since the HSP family is an important class of chaperones, they generally combine with virus-associated proteins to participate in their replication. Murine latency-associated nuclear antigen (mLANA) is a conserved protein of murine gammaherpesvirus 68 (MHV68) that is of great importance to latency maintenance and acute viral replication. In MHV68-infected 3T12 fibroblasts, mLANA directly interacts with HSC70 and recruits it to accumulate in the nucleus, which helps in the formation of viral replication complexes that can promote viral DNA replication, expression of late viral proteins, and ultimately lytic infection (70). Duck hepatitis B virus (DHBV) has been reported to rely on the recognition of RNA packaging signals by viral reverse transcriptase (RT), which can be efficiently activated by HSC70 and HSP40, thereby initiating downstream replication and nucleocapsid assembly (71). Enterovirus A71 (EV-71) is a positive-strand RNA virus in which the initiation of viral protein translation is guided by an internal ribosomal entry site (IRES), and HSC70 can upregulate the activity of IRES in cells to assist viral translation by interaction, thereby promoting the expression of viral proteins in RD cells (26). As mentioned before, IAV is a negative-sense single-stranded RNA virus that can utilize autophagy to facilitate its replication (72). Recent research has found that IAV induces autophagy through the binding of hemagglutinin (HA) to HSP90AA1 distributed on the cell surface. The interaction of HA1 and HSP90AA1 inhibits the phosphorylation of mTOR and AKT to induce autophagy through the AKT-MTOR pathway and thereby promote IAV replication (64).
In addition to protein-protein interactions, HSPs can also promote translation by binding to the viral genome. HSC70 can favor virus replication by binding regulator non-coding RNA (ncRNA). Studies have reported that many viruses, such as human immunodeficiency virus (HIV) (73), DENV (74), and West Nile virus (WNV) (75), encode microRNA-like ncRNA to regulate virus replication. Similarly, rabies virus (RABV) transcribes a small ncRNA, called leader RNA (leRNA). It was also found that HSC70 binds to leRNA to regulate viral replication during infection. Hepatitis C virus (HCV) is currently causing a worldwide epidemic. The nonstructural (NS) proteins are responsible for replication of HCV RNA as well as viral particle assembly, and are primary antiviral targets (76). In a recent study, Li et al. found that HSC70 co-precipitates with HCV NS proteins and RNA, interacting with the HCV replication complex and participating in HCV replication by regulating RNA translation from the HCV genome (77).
After completion of translation in the cell, the virus usually recruits several host factors to facilitate assembly and budding. Immunogold labeling revealed that HSC70 is attached to the surface of HCV particles by interacting with the HPD (His-Pro-Arg) motif on the E2 envelope protein of the virus. Then HSC70, HCV core, and E2 proteins were found to co-localize at the periphery of lipid droplets, an important site for HCV assembly and release (78). By using an allosteric HSC70 inhibitor and RNAi-mediated knockdown, Khachatoorian et al. (79)demonstrated that inactivation of HSC70 reduces the speed of HCV particle assembly, thus concluding that HSC70 plays a role in the assembly of viral particles during HCV infection. HSP90 was also found to play an important role in the maturation of viral proteins, including helping viral particle assembly, protein folding, and maintaining protein activity. HSP90 was found to be involved in multiple viral activities, including capsid precursor processing in coxsackieviruses, polioviruses and rhinoviruses (80), viral capsid assembly in early hepatitis E viral infection (81), maintenance of L protein stability in lacrosse virus (28), maintenance of reverse transcriptase activity in hepatitis B virus and NS2/3 protease in hepatitis C virus, as well as assistance in viral L polymerase folding in measles and Nipah virus (82).
In the context of the global coronavirus pandemic, research on HSPs and their roles in coronavirus infection is very popular. Here, we summarize the findings on the role of HSP90 in coronavirus (CoV) maturation. After CoVs invade cells, large numbers of proteins are translated in the endoplasmic reticulum (ER), which causes ER stress and triggers the unfolded protein response (UPR). HSP90 regulates the UPR by stabilizing the ER stress sensor transmembrane kinase IRE1α, which in turn contributes to viral protein folding and replication (27). In this regard, a recent analysis of RNA-sequencing data from COVID-19 patients also suggested that inhibition of HSP90 could reduce the replication rate of the novel coronavirus (preprint data) (83). This idea has been confirmed in numerous reports of HSP90 inhibitor experiments, which found that that HSP90 inhibitors such as 17-AAG and Luminespib trigger the activities of the unfolded protein response (UPR) in mice, which protected endothelial cells in the pulmonary aorta and pulmonary microvasculature (84). According to recent studies on coronaviruses, HSP90 is considered to be a host-dependent factor for human coronaviruses MERS-CoV, SARS- CoV and SARS-CoV-2. Li et al. found that the depletion of Hsp90β, the cytosolic isoform of HSP90, profoundly reduced viral growth as shown by both viral load quantification and virion titration (85). As confirmed by co-immunoprecipitation, MERS-CoV nucleocapsid protein (NP) is a substrate of HSP90β, which maintains the stability of NP by directly binding it and thereby preventing its degradation by the proteasome. Similarly, they also conducted experiments on the proliferation process of SARS-CoV and SARS-CoV-2, which revealed that the inhibition of HSP90 leads to a significant reduction of virion production. HSP70 and HSP90 are of great importance for viral gene expression since they play a key role in assembling the capsid of some viruses. Viruses utilize HSP70 and HSP90 to fold their proteins and increase their chances of survival under unfavorable host conditions (86).
In many studies on various viruses, the heat shock protein family has been shown to be extensively involved in the viral life cycle, and there have been many advances in the development of antiviral drugs targeting the heat shock protein family. Antiviral drugs targeting heat shock proteins work in three general ways, either by inhibiting the ATPase activity of HSPs, inhibiting the ability of HSPs to form complexes, or triggering modifications of HSPs such as phosphorylation and acetylation to reduce their activity (87). Hsp90 is thought to be the most abundant and evolutionarily conserved heat shock protein. There is also a wealth of research on HSP90-targeted drugs such as geldanamycin (GM) (88), tanespimycin (17-AAG) (89) and histone deacetylase inhibitors (90). Hsp90 inhibitors were demonstrated to protect cultured cells against infection by EV-A71 (91). Similarly, HSP70 is active in various phases of infection by HCV, Flavivirus and Enterovirus, while HSP70 inhibitors such as quercetin, VER155008 and JC40 also show great potential in the treatment of these viruses (92–94). In the treatment of COVID-19, the clinically approved HSP60 inhibitor mizoribine was found to exert an antiviral effect and is considered to be a potentially beneficial agent for hypertensive patients infected with the new coronavirus (95, 96). Quercetin is an inhibitor of HSP70 that also inhibits the activity of HSP40, and was found to decrease the intracellular accumulation of infectious particles when applied in the treatment of HCV infection (97). Among small heat shock proteins, HSP27 has been studied more frequently, and 1,3,5-trihydroxy-13,13-dimethyl-2H-pyran [7,6-b] xanthone (TDP), a compound isolated from a traditional Chinese herb, was found to inhibit HSP27 with significant anti-cytopathic effects, leading to the inhibition of EV-A71 infection (98, 99).
HSP family members participate in the promotion or inhibition of viral infection in many different ways. HSPs inhibit viral infection by acting on different client proteins, not only by activating immune pathways and regulating the cell cycle, but also by directly binding to proliferation-related factors of viruses to silence their replication. However, viruses also often hijack these molecular chaperones, the HSP family members are also extensively involved in all phases of viral proliferation (Table 2). The powerful regulatory ability of HSPs originates from numerous client proteins and more researches are required to explore the detailed mechanisms by which HSPs fight against viruses and help viral infections. In the process of viral infection, HSPs play different roles according to the different clients they serve, which makes them target proteins for the treatment of viral infections. Nowadays there are effective inhibitors, but few of them have become clinically approved drugs for complex reasons such as cell toxicity, side effects and drug stability.
In the background of the current global challenge of COVID-19, the development of drugs targeting heat shock proteins will be a new challenge and focus in this field. Therefore, a summary of the mechanisms of heat shock proteins in viral infection and the development of related inhibitor drugs offers a theoretical basis for future scientific exploration.
XZ: writing-original draft. WY: conceptualization, funding acquisition and writing-review and editing. All authors contributed to the article and approved the submitted version.
This work was financially supported by the National Natural Science Foundation of China (No. 31972623).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Kampinga HH, Hageman J, Vos MJ, Kubota H, Tanguay RM, Bruford EA, et al. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones (2009) 14(1):105–11. doi: 10.1007/s12192-008-0068-7
2. Bakthisaran R, Tangirala R, Rao CM. Small heat shock proteins: Role in cellular functions and pathology. Biochim Biophys Acta-Proteins Proteomics (2015) 1854(4):291–319. doi: 10.1016/j.bbapap.2014.12.019
3. Vos MJ, Zijlstra MP, Kanon B, van Waarde-Verhagen M, Brunt ERP, Oosterveld-Hut HMJ, et al. HSPB7 is the most potent polyQ aggregation suppressor within the HSPB family of molecular chaperones. Hum Mol Genet (2010) 19(23):4677–93. doi: 10.1093/hmg/ddq398
4. Vos MJ, Hageman J, Carra S, Kampinga HH. Structural and functional diversities between members of the human HSPB, HSPH, HSPA, and DNAJ chaperone families. Biochemistry (2008) 47(27):7001–11. doi: 10.1021/bi800639z
5. Wilhelmus MMM, Otte-Holler I, Wesseling P, de Waal RMW, Boelens WC, Verbeek MM. Specific association of small heat shock proteins with the pathological hallmarks of alzheimer's disease brains. Neuropathol Appl Neurobiol (2006) 32(2):119–30. doi: 10.1111/j.1365-2990.2006.00689.x
6. Liu QL, Liang C, Zhou L. Structural and functional analysis of the Hsp70/Hsp40 chaperone system. Protein Sci (2020) 29(2):378–90. doi: 10.1002/pro.3725
7. Rosenzweig R, Nillegoda NB, Mayer MP, Bukau B. The Hsp70 chaperone network. Nat Rev Mol Cell Biol (2019) 20(11):665–80. doi: 10.1038/s41580-019-0133-3
8. Schopf FH, Biebl MM, Buchner J. The HSP90 chaperone machinery. Nat Rev Mol Cell Biol (2017) 18(6):345–60. doi: 10.1038/nrm.2017.20
9. Bross P, Fernandez-Guerra P. Disease-associated mutations in the HSPD1 gene encoding the Large subunit of the mitochondrial HSP60/HSP10 chaperonin complex. Front Mol Biosci (2016) 3:49. doi: 10.3389/fmolb.2016.00049
10. McConnell JR, McAlpine SR. Heat shock proteins 27, 40, and 70 as combinational and dual therapeutic cancer targets. Bioorg Med Chem Lett (2013) 23(7):1923–8. doi: 10.1016/j.bmcl.2013.02.014
11. Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL. Not all J domains are created equal: implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci Publ Protein Soc (2005) 14(7):1697–709. doi: 10.1110/ps.051406805
12. Bolhassani A, Agi E. Heat shock proteins in infection. Clin Chim Acta (2019) 498:90–100. doi: 10.1016/j.cca.2019.08.015
13. Zietkiewicz S, Lewandowska A, Stocki P, Liberek K. Hsp70 chaperone machine remodels protein aggregates at the initial step of Hsp70-Hsp100-dependent disaggregation. J Biol Chem (2006) 281(11):7022–9. doi: 10.1074/jbc.M507893200
14. Gao XC, Carroni M, Nussbaum-Krammer C, Mogk A, Nillegoda NB, Szlachcic A, et al. Human Hsp70 disaggregase reverses parkinson's-linked alpha-synuclein amyloid fibrils. Mol Cell (2015) 59(5):781–93. doi: 10.1016/j.molcel.2015.07.012
15. Han D, Huang SSY, Wang WF, Deng DF, Hung SSO. Starvation reduces the heat shock protein responses in white sturgeon larvae. Environ Biol Fishes (2012) 93(3):333–42. doi: 10.1007/s10641-011-9918-8
16. Michaud MR, Teets NM, Peyton JT, Blobner BM, Denlinger DL. Heat shock response to hypoxia and its attenuation during recovery in the flesh fly, sarcophaga crassipalpis. J Insect Physiol (2011) 57(1):203–10. doi: 10.1016/j.jinsphys.2010.11.007
17. Wang ZY, Li A, Huang X, Bai GL, Jiang YX, Li RL, et al. HSP27 protects skin from ultraviolet b -induced photodamage by regulating autophagy and reactive oxygen species production. Front Cell Dev Biol (2022) 10:852244. doi: 10.3389/fcell.2022.852244
18. Wang H, Feng Y, Ming M, Song J, Chen Z, Xiao Z. Amelioration of cd-induced bioaccumulation, hematological parameters, and heat shock protein-related genes by vitamin c on common carp. Comp Biochem Physiol Toxicol Pharmacol CBP (2022) 258:109362. doi: 10.1016/j.cbpc.2022.109362
19. Hoekstra SP, Bishop NC, Leicht CA. Elevating body termperature to reduce low-grade inflammation: a welcome strategy for those unable to exercise? Exercise Immunol Rev (2020) 26:42–55.
20. Jiang F, Chang G, Li Z, Abouzaid M, Du X, Hull JJ, et al. The HSP/co-chaperone network in environmental cold adaptation of chilo suppressalis. Int J Biol Macromol (2021) 187:780–8. doi: 10.1016/j.ijbiomac.2021.07.113
21. Ling S, Luo M, Jiang S, Liu J, Ding C, Zhang Q, et al. Cellular Hsp27 interacts with classical swine fever virus NS5A protein and negatively regulates viral replication by the NF-kappaB signaling pathway. Virology (2018) 518:202–9. doi: 10.1016/j.virol.2018.02.020
22. Li PH, Cai YJ, Zhu XL, Yang JDH, Yang SQ, Huang W, et al. Epinephelus coioides Hsp27 negatively regulates innate immune response and apoptosis induced by Singapore grouper iridovirus (SGIV) infection. Fish Shellfish Immunol (2022) 120:470–80. doi: 10.1016/j.fsi.2021.12.016
23. Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol (2005) 6(7):519–29. doi: 10.1038/nrm1684
24. Wang Z, Li Y, Yang X, Zhao J, Cheng Y, Wang J. Mechanism and complex roles of HSC70 in viral infections. Front Microbiol (2020) 11:1577. doi: 10.3389/fmicb.2020.01577
25. Batra J, Tripathi S, Kumar A, Katz JM, Cox NJ, Lal RB, et al. Human heat shock protein 40 (Hsp40/DnaJB1) promotes influenza a virus replication by assisting nuclear import of viral ribonucleoproteins. Sci Rep (2016) 6:19063. doi: 10.1038/srep19063
26. Dong Q, Men R, Dan X, Chen Y, Li H, Chen G, et al. Hsc70 regulates the IRES activity and serves as an antiviral target of enterovirus A71 infection. Antiviral Res (2018) 150:39–46. doi: 10.1016/j.antiviral.2017.11.020
27. Wyler E, Mosbauer K, Franke V, Diag A, Gottula LT, Arsie R, et al. Transcriptomic profiling of SARS-CoV-2 infected human cell lines identifies HSP90 as target for COVID-19 therapy. Iscience (2021) 24(3):102151. doi: 10.1016/j.isci.2021.102151
28. Iyer K, Chand K, Mitra A, Trivedi J, Mitra D. Diversity in heat shock protein families: functional implications in virus infection with a comprehensive insight of their role in the HIV-1 life cycle. Cell Stress Chaperones (2021) 26(5):743–68. doi: 10.1007/s12192-021-01223-3
29. Mogk A, Ruger-Herreros C, Bukau B. Cellular functions and mechanisms of action of small heat shock proteins. Annu Rev Microbiol (2019) 73:89–110. doi: 10.1146/annurev-micro-020518-115515
30. Dabbaghizadeh A, Tanguay RM. Structural and functional properties of proteins interacting with small heat shock proteins. Cell Stress Chaperones (2020) 25(4):629–37. doi: 10.1007/s12192-020-01097-x
31. Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol (2010) 11(8):579–92. doi: 10.1038/nrm2941
32. Tesic M, Marsh JA, Cullinan SB, Gaber RF. Functional interactions between Hsp90 and the co-chaperones Cns1 and Cpr7 in saccharomyces cerevisiae. J Biol Chem (2003) 278(35):32692–701. doi: 10.1074/jbc.M304315200
33. Mayer MP. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem Sci (2013) 38(10):507–14. doi: 10.1016/j.tibs.2013.08.001
34. Karagoz GE, Rudiger SG. Hsp90 interaction with clients. Trends Biochem Sci (2015) 40(2):117–25. doi: 10.1016/j.tibs.2014.12.002
35. Gupta S, Knowlton AA. Cytosolic heat shock protein 60, hypoxia, and apoptosis. Circulation (2002) 106(21):2727–33. doi: 10.1161/01.cir.0000038112.64503.6e
36. Malik JA, Lone R. Heat shock proteins with an emphasis on HSP 60. Mol Biol Rep (2021) 48(10):6959–69. doi: 10.1007/s11033-021-06676-4
37. Chang X, Shi X, Zhang X, Chen J, Fan X, Yang Y, et al. miR-382-5p promotes porcine reproductive and respiratory syndrome virus (PRRSV) replication by negatively regulating the induction of type I interferon. FASEB J (2020) 34(3):4497–511. doi: 10.1096/fj.201902031RRR
38. Quicke KM, Diamond MS, Suthar MS. Negative regulators of the RIG-i-like receptor signaling pathway. Eur J Immunol (2017) 47(4):615–28. doi: 10.1002/eji.201646484
39. McNab F, Mayer-Barber K, Sher A, Wack A, O'Garra A. Type I interferons in infectious disease. Nat Rev Immunol (2015) 15(2):87–103. doi: 10.1038/nri3787
40. Li X, Ma R, Wu B, Niu Y, Li H, Li D, et al. HSP27 protein dampens encephalomyocarditis virus replication by stabilizing melanoma differentiation-associated gene 5. Front Microbiol (2021) 12:788870. doi: 10.3389/fmicb.2021.788870
41. Hauser H, Shen L, Gu Q, Krueger S, Chen S-Y. Secretory heat-shock protein as a dendritic cell-targeting molecule: a new strategy to enhance the potency of genetic vaccines. Gene Therapy (2004) 11(11):924–32. doi: 10.1038/sj.gt.3302160
42. Takashima K, Oshiumi H, Matsumoto M, Seya T. DNAJB1/HSP40 suppresses melanoma differentiation-associated gene 5-mitochondrial antiviral signaling protein function in conjunction with HSP70. J Innate Immun (2018) 10(1):44–55. doi: 10.1159/000480740
43. Sun M, Yu ZQ, Ma JL, Pan ZH, Lu CP, Yao HC. Down-regulating heat shock protein 27 is involved in porcine epidemic diarrhea virus escaping from host antiviral mechanism. Veterinary Microbiol (2017) 205:6–13. doi: 10.1016/j.vetmic.2017.04.031
44. Le Y, Jia P, Jin Y, Liu W, Jia K, Yi M. The antiviral role of heat shock protein 27 against red spotted grouper nervous necrosis virus infection in sea perch. Fish Shellfish Immunol (2017) 70:185–94. doi: 10.1016/j.fsi.2017.08.032
45. Sun P, Zhang S, Qin X, Chang X, Cui X, Li H, et al. Foot-and-mouth disease virus capsid protein VP2 activates the cellular EIF2S1-ATF4 pathway and induces autophagy via HSPB1. Autophagy (2018) 14(2):336–46. doi: 10.1080/15548627.2017.1405187
46. Netea MG, Schlitzer A, Placek K, Joosten LAB, Schultze JL. Innate and adaptive immune memory: an evolutionary continuum in the host's response to pathogens. Cell Host Microbe (2019) 25(1):13–26. doi: 10.1016/j.chom.2018.12.006
47. Oeckinghaus A, Ghosh S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harbor Perspect Biol (2009) 1(4):a000034. doi: 10.1101/cshperspect.a000034
48. Mangan MSJ, Olhava EJ, Roush WR, Seidel HM, Glick GD, Latz E. Targeting the NLRP3 inflammasome in inflammatory diseases. Nat Rev Drug Discovery (2018) 17(8):588–606. doi: 10.1038/nrd.2018.97
49. Martine P, Chevriaux A, Derangere V, Apetoh L, Garrido C, Ghiringhelli F, et al. HSP70 is a negative regulator of NLRP3 inflammasome activation. Cell Death Dis (2019) 10(4):256. doi: 10.1038/s41419-019-1491-7
50. Swanson KV, Deng M, Ting JP. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat Rev Immunol (2019) 19(8):477–89. doi: 10.1038/s41577-019-0165-0
51. Nizami S, Arunasalam K, Green J, Cook J, Lawrence CB, Zarganes-Tzitzikas T, et al. Inhibition of the NLRP3 inflammasome by HSP90 inhibitors. Immunology (2021) 162(1):84–91. doi: 10.1111/imm.13267
52. Shimp SK 3rd, Parson CD, Regna NL, Thomas AN, Chafin CB, Reilly CM, et al. HSP90 inhibition by 17-DMAG reduces inflammation in J774 macrophages through suppression of akt and nuclear factor-kappaB pathways. Inflammation Res (2012) 61(5):521–33. doi: 10.1007/s00011-012-0442-x
53. Lindquist S. The heat-shock response. Annu Rev Biochem (1986) 55:1151–91. doi: 10.1146/annurev.bi.55.070186.005443
54. Heck TG, Ludwig MS, Frizzo MN, Rasia AA, de Bittencourt PIH. Suppressed anti-inflammatory heat shock response in high-risk COVID-19 patients: lessons from basic research (inclusive bats), light on conceivable therapies. Clin Sci (2020) 134(15):1991–2017. doi: 10.1042/Cs20200596
55. Ruan Q, Yang K, Wang W, Jiang L, Song J. Correction to: Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from wuhan, China. Intensive Care Med (2020) 46(6):1294–7. doi: 10.1007/s00134-020-06028-z
56. Liu Q, Huang X, Zhao D, Han K, Liu Y, Yang J, et al. Identification of heat shock protein A9 as a tembusu virus binding protein on DF-1 cells. Virus Res (2017) 227:110–4. doi: 10.1016/j.virusres.2016.09.020
57. Wang Y, Li Y, Ding T. Heat shock protein 90beta in the vero cell membrane binds Japanese encephalitis virus. Int J Mol Med (2017) 40(2):474–82. doi: 10.3892/ijmm.2017.3041
58. Chuang CK, Yang TH, Chen TH, Yang CF, Chen WJ. Heat shock cognate protein 70 isoform d is required for clathrin-dependent endocytosis of Japanese encephalitis virus in C6/36 cells. J Gen Virol (2015) 96(Pt 4):793–803. doi: 10.1099/jgv.0.000015
59. Zhang W, Jia K, Jia P, Xiang Y, Lu X, Liu W, et al. Marine medaka heat shock protein 90ab1 is a receptor for red-spotted grouper nervous necrosis virus and promotes virus internalization through clathrin-mediated endocytosis. PloS Pathog (2020) 16(7):e1008668. doi: 10.1371/journal.ppat.1008668
60. Vega-Almeida TO, Salas-Benito M, De Nova-Ocampo MA, Del Angel RM, Salas-Benito JS. Surface proteins of C6/36 cells involved in dengue virus 4 binding and entry. Arch Virol (2013) 158(6):1189–207. doi: 10.1007/s00705-012-1596-0
61. Howe MK, Speer BL, Hughes PE, Loiselle DR, Vasudevan S, Haystead TAJ. An inducible heat shock protein 70 small molecule inhibitor demonstrates anti-dengue virus activity, validating Hsp70 as a host antiviral target. Antiviral Res (2016) 130:81–92. doi: 10.1016/j.antiviral.2016.03.017
62. Lin TW, Lo CW, Lai SY, Fan RJ, Lo CJ, Chou YM, et al. Chicken heat shock protein 90 is a component of the putative cellular receptor complex of infectious bursal disease virus. J Virol (2007) 81(16):8730–41. doi: 10.1128/JVI.00332-07
63. Reyes-Del Valle J, Chavez-Salinas S, Medina F, Del Angel RM. Heat shock protein 90 and heat shock protein 70 are components of dengue virus receptor complex in human cells. J Virol (2005) 79(8):4557–67. doi: 10.1128/JVI.79.8.4557-4567.2005
64. Wang X, Zheng T, Lin L, Zhang Y, Peng X, Yan Y, et al. Influenza a virus induces autophagy by its hemagglutinin binding to cell surface heat shock protein 90AA1. Front Microbiol (2020) 11:566348. doi: 10.3389/fmicb.2020.566348
65. Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem (2000) 69:531–69. doi: 10.1146/annurev.biochem.69.1.531
66. Wang J, Guo W, Long C, Zhou H, Wang H, Sun X. The split renilla luciferase complementation assay is useful for identifying the interaction of Epstein-Barr virus protein kinase BGLF4 and a heat shock protein Hsp90. Acta Virol (2016) 60(1):62–70. doi: 10.4149/av_2016_01_62
67. Panella S, Marcocci ME, Celestino I, Valente S, Zwergel C, Li Puma DD, et al. MC1568 inhibits HDAC6/8 activity and influenza a virus replication in lung epithelial cells: role of Hsp90 acetylation. Future Med Chem (2016) 8(17):2017–31. doi: 10.4155/fmc-2016-0073
68. Hornikova L, Fraiberk M, Man P, Janovec V, Forstova J. VP1, the major capsid protein of the mouse polyomavirus, binds microtubules, promotes their acetylation and blocks the host cell cycle. FEBS J (2017) 284(2):301–23. doi: 10.1111/febs.13977
69. Zhong M, Zheng K, Chen M, Xiang Y, Jin F, Ma K, et al. Heat-shock protein 90 promotes nuclear transport of herpes simplex virus 1 capsid protein by interacting with acetylated tubulin. PloS One (2014) 9(6):e99425. doi: 10.1371/journal.pone.0099425
70. Salinas E, Byrum SD, Moreland LE, Mackintosh SG, Tackett AJ, Forrest JC. Identification of viral and host proteins that interact with murine gammaherpesvirus 68 latency-associated nuclear antigen during lytic replication: a role for Hsc70 in viral replication. J Virol (2016) 90(3):1397–413. doi: 10.1128/JVI.02022-15
71. Beck J, Nassal M. Efficient Hsp90-independent in vitro activation by Hsc70 and Hsp40 of duck hepatitis b virus reverse transcriptase, an assumed Hsp90 client protein. J Biol Chem (2003) 278(38):36128–38. doi: 10.1074/jbc.M301069200
72. Wang R, Zhu Y, Zhao J, Ren C, Li P, Chen H, et al. Autophagy promotes replication of influenza a virus in vitro. J Virol (2019) 93(4):e01984–01918. doi: 10.1128/JVI.01984-18
73. Sun GH, Rossi JJ. MicroRNAs and their potential involvement in HIV infection. Trends Pharmacol Sci (2011) 32(11):675–81. doi: 10.1016/j.tips.2011.07.003
74. Hussain M, Asgari S. MicroRNA-like viral small RNA from dengue virus 2 autoregulates its replication in mosquito cells. Proc Natl Acad Sci United States America (2014) 111(7):2746–51. doi: 10.1073/pnas.1320123111
75. Hussain M, Torres S, Schnettler E, Funk A, Grundhoff A, Pijlman GP, et al. West Nile Virus encodes a microRNA-like small RNA in the 3' untranslated region which up-regulates GATA4 mRNA and facilitates virus replication in mosquito cells. Nucleic Acids Res (2012) 40(5):2210–23. doi: 10.1093/nar/gkr848
76. Wang Y, Lee S, Ha Y, Lam W, Chen SR, Dutschman GE, et al. Tylophorine analogs allosterically regulates heat shock cognate protein 70 and inhibits hepatitis c virus replication. Sci Rep (2017) 7(1):10037. doi: 10.1038/s41598-017-08815-z
77. Li HC, Yang CH, Lo SY. Cellular factors involved in the hepatitis c virus life cycle. World J Gastroenterol (2021) 27(28):4555–81. doi: 10.3748/wjg.v27.i28.4555
78. Parent R, Qu X, Petit MA, Beretta L. The heat shock cognate protein 70 is associated with hepatitis c virus particles and modulates virus infectivity. Hepatology (2009) 49(6):1798–809. doi: 10.1002/hep.22852
79. Khachatoorian R, French SW. Chaperones in hepatitis c virus infection. World J Hepatol (2016) 8(1):9–35. doi: 10.4254/wjh.v8.i1.9
80. Geller R, Vignuzzi M, Andino R, Frydman J. Evolutionary constraints on chaperone-mediated folding provide an antiviral approach refractory to development of drug resistance. Genes Dev (2007) 21(2):195–205. doi: 10.1101/gad.1505307
81. Zheng ZZ, Miao J, Zhao M, Tang M, Yeo AE, Yu H, et al. Role of heat-shock protein 90 in hepatitis e virus capsid trafficking. J Gen Virol (2010) 91(Pt 7):1728–36. doi: 10.1099/vir.0.019323-0
82. Bloyet LM, Welsch J, Enchery F, Mathieu C, de Breyne S, Horvat B, et al. HSP90 chaperoning in addition to phosphoprotein required for folding but not for supporting enzymatic activities of measles and nipah virus l polymerases. J Virol (2016) 90(15):6642–56. doi: 10.1128/JVI.00602-16
83. Sultan I, Howard S, Tbakhi A. Drug repositioning suggests a role for the heat shock protein 90 inhibitor geldanamycin in treating COVID-19 infection. ResearchGate (2020). doi: 10.21203/rs.3.rs-18714/v1
84. Kubra KT, Uddin MA, Akhter MS, Barabutis N. Hsp90 inhibitors induce the unfolded protein response in bovine and mice lung cells. Cell Signalling (2020) 67:109500. doi: 10.1016/j.cellsig.2019.109500
85. Li C, Chu H, Liu X, Chiu MC, Zhao X, Wang D, et al. Human coronavirus dependency on host heat shock protein 90 reveals an antiviral target. Emerg Microbes Infections (2020) 9(1):2663–72. doi: 10.1080/22221751.2020.1850183
86. Lubkowska A, Pluta W, Stronska A, Lalko A. Role of heat shock proteins (HSP70 and HSP90) in viral infection. Int J Mol Sci (2021) 22(17):9366. doi: 10.3390/ijms22179366
87. Wan QY, Song D, Li HC, He ML. Stress proteins: the biological functions in virus infection, present and challenges for target-based antiviral drug development. Signal Transduction Targeted Ther (2020) 5(1):40. doi: 10.1038/s41392-020-00233-4
88. Grenert JP, Sullivan WP, Fadden P, Haystead TA, Clark J, Mimnaugh E, et al. The amino-terminal domain of heat shock protein 90 (hsp90) that binds geldanamycin is an ATP/ADP switch domain that regulates hsp90 conformation. J Biol Chem (1997) 272(38):23843–50. doi: 10.1074/jbc.272.38.23843
89. Karkoulis PK, Stravopodis DJ, Margaritis LH, Voutsinas GE. 17-Allylamino-17-demethoxygeldanamycin induces downregulation of critical Hsp90 protein clients and results in cell cycle arrest and apoptosis of human urinary bladder cancer cells. BMC Cancer (2010) 10:15. doi: 10.1186/1471-2407-10-481
90. Park S, Park JA, Kim YE, Song S, Kwon HJ, Lee Y. Suberoylanilide hydroxamic acid induces ROS-mediated cleavage of HSP90 in leukemia cells. Cell Stress Chaperones (2015) 20(1):149–57. doi: 10.1007/s12192-014-0533-4
91. Tsou YL, Lin YW, Chang HW, Lin HY, Shao HY, Yu SL, et al. Heat shock protein 90: Role in enterovirus 71 entry and assembly and potential target for therapy. PloS One (2013) 8(10):13. doi: 10.1371/journal.pone.0077133
92. Su YS, Hsieh PY, Li JS, Pao YH, Chen CJ, Hwang LH. The heat shock protein 70 family of chaperones regulates all phases of the enterovirus A71 life cycle. Front Microbiol (2020) 11:1656. doi: 10.3389/fmicb.2020.01656
93. Taguwa S, Maringer K, Li X, Bernal-Rubio D, Rauch JN, Gestwicki JE, et al. Defining Hsp70 subnetworks in dengue virus replication reveals key vulnerability in flavivirus infection. Cell (2015) 163(5):1108–23. doi: 10.1016/j.cell.2015.10.046
94. Maeda Y, Yoshimura K, Matsui H, Shindo Y, Tamesa T, Tokumitsu Y, et al. Dendritic cells transfected with heat-shock protein 70 messenger RNA for patients with hepatitis c virus-related hepatocellular carcinoma: a phase 1 dose escalation clinical trial. Cancer Immunol Immunother (2015) 64(8):1047–56. doi: 10.1007/s00262-015-1709-1
95. Meng QL, Li BBX, Xiao XS. Toward developing chemical modulators of Hsp60 as potential therapeutics. Front Mol Biosci (2018) 5:35. doi: 10.3389/fmolb.2018.00035
96. Jakovac H. COVID-19 and hypertension: is the HSP60 culprit for the severe course and worse outcome? Am J Physiol-Heart Circulatory Physiol (2020) 319(4):H793–6. doi: 10.1152/ajpheart.00506.2020
97. Gonzalez O, Fontanes V, Raychaudhuri S, Loo R, Loo J, Arumugaswami V, et al. The heat shock protein inhibitor quercetin attenuates hepatitis c virus production. Hepatology (2009) 50(6):1756–64. doi: 10.1002/hep.23232
98. Fu WM, Zhang JF, Wang H, Xi ZC, Wang WM, Zhuang P, et al. Heat shock protein 27 mediates the effect of 1,3,5-trihydroxy-13,13-dimethyl-2H-pyran 7,6-b xanthone on mitochondrial apoptosis in hepatocellular carcinoma. J Proteomics (2012) 75(15):4833–43. doi: 10.1016/j.jprot.2012.05.032
99. Dan XL, Wan QY, Yi LN, Lu J, Jiao Y, Li HC, et al. Hsp27 responds to and facilitates enterovirus A71 replication by enhancing viral internal ribosome entry site-mediated translation. J Virol (2019) 93(9):17. doi: 10.1128/jvi.02322-18
100. Speth C, Prohaszka Z, Mair M, Stockl G, Zhu X, Jobstl B, et al. A 60 kD heat-shock protein-like molecule interacts with the HIV transmembrane glycoprotein gp41. Mol Immunol (1999) 36(9):619–28. doi: 10.1016/s0161-5890(99)00082-6
101. Ravindran MS, Bagchi P, Inoue T, Tsai B. A non-enveloped virus hijacks host disaggregation machinery to translocate across the endoplasmic reticulum membrane. PloS Pathog (2015) 11(8):e1005086. doi: 10.1371/journal.ppat.1005086
102. Cao M, Wei C, Zhao L, Wang J, Jia Q, Wang X, et al. DnaJA1/Hsp40 is co-opted by influenza a virus to enhance its viral RNA polymerase activity. J Virol (2014) 88(24):14078–89. doi: 10.1128/JVI.02475-14
103. Naito T, Momose F, Kawaguchi A, Nagata K. Involvement of Hsp90 in assembly and nuclear import of influenza virus RNA polymerase subunits. J Virol (2007) 81(3):1339–49. doi: 10.1128/JVI.01917-06
104. Park SG, Lim SO, Jung G. Binding site analysis of human HBV pol for molecular chaperonin, hsp60. Virology (2002) 298(1):116–23. doi: 10.1006/viro.2002.1496
105. Gurer C, Hoglund A, Hoglund S, Luban J. ATPgammaS disrupts human immunodeficiency virus type 1 virion core integrity. J Virol (2005) 79(9):5557–67. doi: 10.1128/JVI.79.9.5557-5567.2005
106. Radhakrishnan A, Yeo D, Brown G, Myaing MZ, Iyer LR, Fleck R, et al. Protein analysis of purified respiratory syncytial virus particles reveals an important role for heat shock protein 90 in virus particle assembly. Mol Cell Proteomics (2010) 9(9):1829–48. doi: 10.1074/mcp.M110.001651
107. Zhang SM, Sun DC, Lou S, Bo XC, Lu Z, Qian XH, et al. HBx protein of hepatitis b virus (HBV) can form complex with mitochondrial HSP60 and HSP70. Arch Virol (2005) 150(8):1579–90. doi: 10.1007/s00705-005-0521-1
Keywords: HSPs, viral infection, chaperones, protein folding, immunological pathways
Citation: Zhang X and Yu W (2022) Heat shock proteins and viral infection. Front. Immunol. 13:947789. doi: 10.3389/fimmu.2022.947789
Received: 19 May 2022; Accepted: 15 July 2022;
Published: 05 August 2022.
Edited by:
Esaki M. Shankar, Central University of Tamil Nadu, IndiaReviewed by:
Ji-Xin Tang, Guangdong Medical University, ChinaCopyright © 2022 Zhang and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Yu, bWlra3l1QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.