
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 15 August 2022
Sec. T Cell Biology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.942465
 Yuan Luo1,2†
Yuan Luo1,2† Dan Li3†
Dan Li3† Luoyingzi Xie2
Luoyingzi Xie2 Shun Lei2
Shun Lei2 Xiangyu Chen2
Xiangyu Chen2 Cong Wang4
Cong Wang4 Dong Yao5
Dong Yao5 Lin Li1
Lin Li1 Jingyi Fang2
Jingyi Fang2 Cheng Chen2
Cheng Chen2 Shijie Yuan1
Shijie Yuan1 Fei Li1
Fei Li1 Xiaorong Xie1
Xiaorong Xie1 Yan Zhang2
Yan Zhang2 Zhirong Li2
Zhirong Li2 Li Hu2
Li Hu2 Jianfang Tang2
Jianfang Tang2 Lilin Ye2
Lilin Ye2 Zhengping Wei2,6*
Zhengping Wei2,6* Ran He1*
Ran He1*Follicular helper T (TFH) cells provide specialized help for B cells to ensure optimal humoral immunity. The histone methyltransferase EZH2, as a chromatin repressor, secures the TFH differentiation by promoting TFH lineage associated gene expression during acute viral infection, including Tcf7 and Bcl6. By using conditional deletion murine system, we observed that EZH2 ablation in CD4+ T cells was accompanied by aberrant accumulation of DNA methyltransferases (DNMTs) DNMT1 and DNMT3B in TFH cells. And the loss of EZH2 promoted aggravation of DNA methylation status at Tcf7 locus. Therefore, our findings suggested that EZH2 plays an important role in maintenance of hypomethylation at Tcf7 locus thus affecting TFH differentiation during acute viral infection.
Upon antigen engagement, antigen-specific naive CD4+ T cells differentiate into distinct effector populations to execute immune response under the regulation of specific transcription factors (1). Follicular helper T (TFH) cells are a subset of CD4+ T cells specialized in helping B cells by inducing the formation and maintenance of germinal centers (GCs), which are indispensable for differentiation of high-affinity antibody-producing plasma cells and production of memory B cells (2, 3). Abnormal TFH cell differentiation is closely related to antibody mediated autoimmune diseases, such as systemic lupus erythematosus and rheumatoid arthritis (4–6). Hence, dissecting the differentiation of TFH cells can help to modulate the humoral immunity for better control of infection or alleviation of autoimmune diseases (7).
TFH differentiation is characterized as a multi-stage process, which is precisely regulated by multiple transcription factors. The transcriptional repressor Bcl6 is the “master regulator” of TFH differentiation and is essential for the development of TFH cells (3). Moreover, induced ablation of Bcl6 converts “ex-TFH” cells into TH1 cells during acute lymphocytic choriomeningitis virus (LCMV-Armstrong) infection, suggesting that Bcl6 is critical for the integrity of TFH cells (8). On the contrary, Blimp1 (encoded by Prdm1), as the antagonist of Bcl6, promotes the differentiation of non-TFH effector cells by repressing Bcl6 expression (9, 10). Recent studies have shown that TCF1 (encoded by Tcf7) acts as upstream hub of the reciprocal antagonistic Bcl6-Blimp1 axis and secures TFH differentiation program by promoting Bcl6 expression but repressing Blimp1 expression, as manifested by the fact that the deficiency of TCF1 restricts the TFH differentiation and effector function (11, 12). Additionally, TCF1 maintains the transcriptional and metabolic signatures of TFH cells, it is not only necessary for adequate expansion of TFH cells, but also critical for TFH cell responses during LCMV infection (13). Other regulators, such as ICOS, Ascl2, Id2, STAT3, Klf2, and Foxo1 were also identified to regulate TFH differentiation (14–17). In addition to transcription mechanisms, epigenetic modification also plays a vital role in cell differentiation and plasticity by responding rapidly to external stimuli and incorporating a variety signals (18). For instance, the SUV39H1-dependent H3K9me3 is important for lineage integrity of TH2 cells (19), and the G9a-mediated H3K9me2 is involved in the control of TREG cell differentiation (20). Although the phenotypic and functional changes that occur during TFH differentiation have been well characterized, the detailed epigenetic mechanisms which control TFH differentiation remains little understood.
Enhancer of zeste homolog 2 (EZH2), the catalytic subunit of Polycomb complex 2 (PRC2), mediates the trimethylation at lysine 27 of histone H3 via its methyltransferase activity (HMT) (21). Generally, the EZH2-dependent H3K27me3 modification is associated with gene silencing through chromatin compaction (22). EZH2-mediated H3K27me3 has been demonstrated to restrict the differentiation and cytokine production through occupying Tbx21 and Ifng loci in TH1 cells, and the Gata3 and Il4 loci in TH2 cells (23). In TREG cells, the H3K27me3 deposition is also required for the repressive gene program, thus maintaining the lineage identity after activation (24). Moreover, EZH2 promotes TFH differentiation potentially by stabilizing the chromatin accessibility of TFH lineage associated genes, and the deletion of EZH2 caused reduced expression of TFH associated genes (25, 26). However, EZH2-mediated H3K27me3 deposition has not been observed at TFH lineage associated gene loci (25). Besides, it remains ambiguous whether and how the chromatin repressor EZH2 promotes TFH differentiation by regulating other modifiers that are negatively associated with TFH differentiation program.
In this study, we demonstrated that EZH2 restricts the expression of DNMT1 and DNMT3B, thus may help to maintain the hypomethylation status of Tcf7 locus. This study illustrated that EZH2 restrains the methylation status at Tcf7 locus and promotes the differentiation of TFH cells.
Ezh2fl/fl, Cd4-Cre transgenic mice, and wild type C57BL/6J (CD45.2 and CD45.1) mice were purchased from the Jackson Laboratory. SMARTA (CD45.1, expressing MHC II I-Ab-restricted TCR specific for LCMV glycoprotein amino acids 66–77 epitope) and LCMV-Armstrong strain were generously provided by Dr. Rafi Ahmed (Emory University). 2 × 105 plaque-forming units of LCMV-Armstrong strain with intraperitoneal injection to set up the acute viral infection model in mice. Both sexes were included without randomization or blinding to establish the experiments at the age of 6-10 weeks. All mice were house kept (3-5 mice per cage) in a specific pathogen-free facility with controlled environmental conditions. All experiments were performed according to the guidelines of the Institutional Animal Care and Use Committee of the Third Military Medical University.
Single-cell suspensions of spleens from the experimental animals were used for flow cytometry with a FACSCanto II (BD Biosciences). The surface staining was performed in FACS buffer, the anti-CD4 (RM4-5), anti-CD44 (IM7), anti-CD45.1 (A20), anti-CD45R (RA3-6B2) were obtained from Biolegend. For CXCR5 staining, all the surface antibodies were mixed in FACS buffer (PBS with 2% FBS) containing 2% normal mouse serum and 1% BSA. The CXCR5 staining was performed with a three-step protocol: firstly the cells were stained with purified rat anti-CXCR5 (2G8) at 4°C for 1h; then the cells were washed and stained with biotin-conjugated goat anti-rat IgG (Jackson ImmunoResearch) on ice for 30 min; lastly the cells were washed and stained with fluorescently-labeled streptavidin (Biolegend) and other surface antibodies on ice for 30 min. Ezh2 (11/EZH2) was obtained from BD Biosciences, the staining was performed with a Cytofix/Cytoperm Fixation/Permeabilization Kit (554714, BD Biosciences) according to the manufacturer’s instructions after surface staining. TCF1 (C46C7), DNMT1 (D63A6), DNMT3A (D23G1), DNMT3B (E4I4O) were obtained from Cell Signaling Technology, while Blimp1 (5E7), Foxp3 (3G3) were obtained from BD Biosciences. The staining was performed with a Foxp3/Transcription Factor Staining Buffer Set (eBioscience) according to the manufacturer’s instructions after surface staining. All data were analyzed by FlowJo (Treestar).
A total of 2 × 104 transgenic CD45.1+ SMARTA cells were harvested from naive mice and adoptively transferred intravenously to CD45.2+ C57BL/6 mice. On the following day, the recipient mice were intravenously infected with 2 × 105 pfu of LCMV-Armstrong strain.
The cell sorting was performed on a FACSAria II (BD Biosciences). The Naive SMARTA cells (CD25-CD44-CD62L+CD4+) were sorted from naive SMARTA mice, TFH cells (SLAM-CXCR5highCD4+) were sorted from mice infected with LCMV-Armstrong strain on day 4 and day 8 after CD4+ T cells enrichment. The biotin-conjugated antibodies: CD8 (53-6.7), CD45.2 (104), B220 (RA3-6B2), CD11b (M1/70), CD11c (N418), TER119 (TER), NK1.1 (PK136), F4/80 (BM8), CD25 (PC61) were used for the T cells enrichment. The purity of the sorted cells was >95% in all experiments.
For comparison the gene expression of target genes, total RNA was isolated from the cells sorted from mice infected with LCMV Armstrong strain. RNA was extracted with the RNeasy Mini Kit (74104, Qiagen) and reverse transcribed with the RevertAid H Minus First Strand cDNA Synthesis Kit (K1632, Thermo Scientific). Quantitative real-time PCR of target transcripts with appropriate primers (Supplementary Table 1) were carried out with SYBR Green PCR kit (208054, Qiagen) on a CFX96 Touch Real-Time System (Bio-Rad). Fold differences in expression levels were calculated according to the 2−ΔΔCT method.
The transfected 293T cells were washed with ice-cold PBS twice, and then lysed in RIPA buffer. Equal amounts of protein from each sample were separated with 10% SDS-PAGE and then transferred to PVDF membranes (IPVH00010, Millipore). The membranes were blocked with 5% bovine serum albumin (B2064, Sigma) for 1 hour at room temperature, and then incubated with appropriate antibodies overnight at 4°C. After four times washing, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody for another one hour at room temperature. Immunoblots were visualized with SuperSignal® West Pico Chemiluminescent Substrate (34080, Thermo Scientific) on a Bio-Rad XRS chemiluminescence detection system (Bio-Rad).
The full-length cDNA of Ezh2 was subcloned into pcDNA3.1-HA vector and Dnmt1, Dnmt3a, Dnmt3b were subcloned into pcDNA3.1-FLAG vector for expression. For protein stability studies, the pcDNA3.1-EZH2-HA vector was co-transfected with pcDNA3.1-FLAG-DNMT1 and pcDNA3.1-FLAG-DNMT3B vectors into 293T cells by using TranIT-293 Transfection Reagent (MIR 2705, Mirus), respectively. After 24 h, the transfected cells were treated with cycloheximide for 24 h, and the working solution of cycloheximide was 200 μg/mL. For immunoprecipitation studies, the 293T cells were transfected with pcDNA3.1-FLAG-DNMT1, pcDNA3.1-FLAG-DNMT3A, pcDNA3.1-FLAG-DNMT3B, pcDNA3.1-FLAG vectors. Cells were harvested 48 h after transfection, the extracted product were incubated with 2 μg of anti-Ezh2 (D2C9, Cell Signaling Technology), 2 μg of anti-FLAG (M2; Sigma-Aldrich) for 5 h, and then incubated with Dynabeads Protein G (10004D; Life Technologies) for 2 h. After washing, the protein stability and immunoprecipitated samples were analyzed by immunoblot analysis with appropriate antibodies.
The bisulfite sequencing of the target genomic region was used to measure the allelic frequency of methylated cytosines. Genomic DNA from the sorted cells were extracted with QIAamp® DNA Mini Kit (51304, Qiagen), and then bisulfate modified by EpiTect® Bisulfite Kit (59104, Qiagen). The bisulfite-modified DNA was amplified with locus-specific primers (Supplementary Table 1). The amplified product was cloned into the pMD19-T TA cloning Vector (6013, Takara), then transformed into DH5α competent cells. Independent colonies were sequenced to determine the methylation status from each sample. The CpG island was defined, and primer was designed by website of Li Lab (http://www.urogene.org/methprimer/) (27).
Statistical analysis was performed using Prism version 6.0 (GraphPad) software. Statistical significance was determined by unpaired two-tailed Student’s t test. A P value of less than 0.05 was considered statistically significant.
EZH2 is a subunit of PRC2, which acts as “writer” of the repressive H3K27me3 modification (21). We set out to elucidate the epigenetic role of EZH2 in TFH cells and identify the downstream regulators modulated by EZH2 during acute viral infection. To this end, we crossed mice containing loxP-flanked Ezh2 alleles with mice expressing Cre-recombinase under the control of Cd4 promoter, enhancer and silencer to generate the conditional knockout mice (Ezh2fl/flCd4-cre, hereafter called Ezh2-/-). The loss of Ezh2 mRNA in CD4+ T cells from Ezh2-/- mice was confirmed by quantitative RT-PCR (Figure 1A). We then infected the control (Ezh2fl/fl) mice and Ezh2-/- mice with LCMV-Armstrong strain, the deletion of EZH2 protein was validated in TFH cells from Ezh2-/- mice at day 8 after infection (Figure 1B). Moreover, the frequency and absolute number of TFH cells in Ezh2-/- mice were significantly diminished compared with those in the control mice (Supplemental Figure 1A, B), consistent with previous findings (25, 26). The transcript and protein levels of Tcf7 and Bcl6 were both decreased in TFH cells from Ezh2-/- mice, whereas that of Prdm1 and Blimp1 were increased (Figures 1C, D). These results suggested that EZH2 controls the expression of TFH lineage related regulators to secure TFH differentiation during acute viral infection.
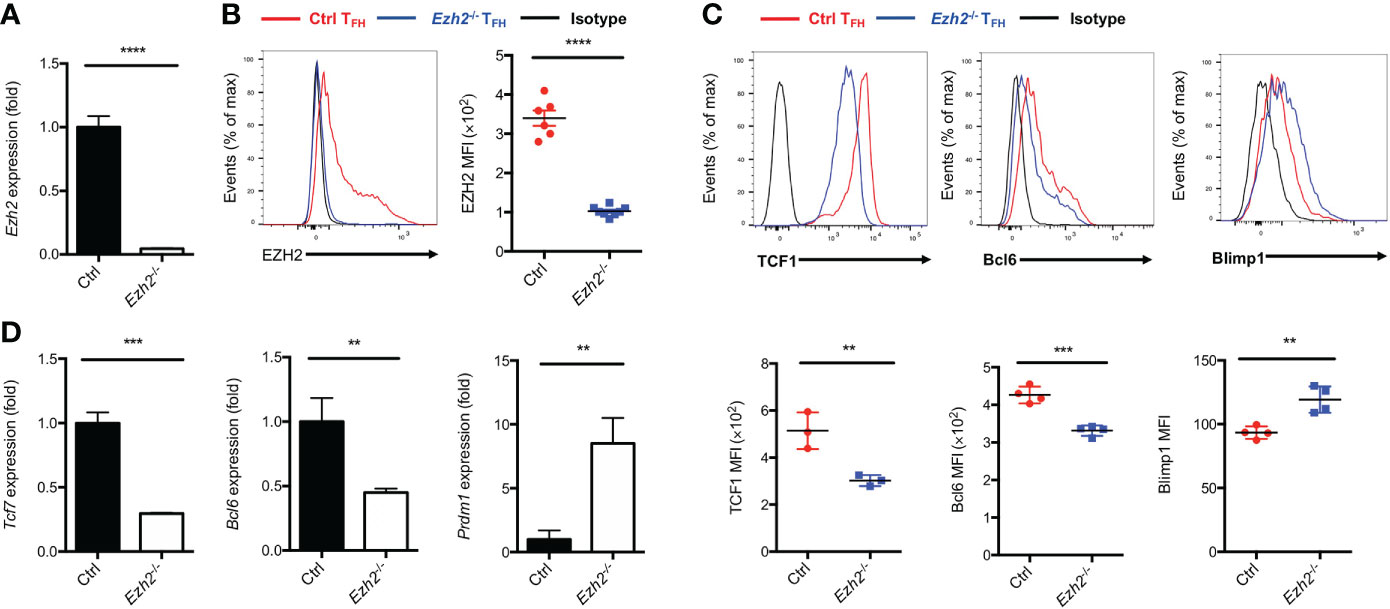
Figure 1 Epigenetic regulator EZH2 controls TFH differentiation. The control (Ezh2fl/fl) and Ezh2fl/flCd4-cre (Ezh2-/-) mice were infected with 2 × 105 plaque-forming units LCMV-Armstrong strain. After 8 days of infection, TFH (SLAM-CXCR5hiCD44+), sorted to perform the following experiments. (A) Analysis of mRNA of Ezh2 in naive splenic CD4+ T cells from control and Ezh2-/- mice via RT-PCR. Normalized to their expression in control CD4+ T cells. (B) Measurement of EZH2 expression in TFH cells via flow cytometry from control and Ezh2-/- mice. (C, D) Measurement of TCF1, Bcl6 and Blimp1 expression via flow cytometry, and the expression of Tcf7, Bcl6 and Prdm1 transcripts by RT-PCR in TFH cells from control and Ezh2-/- mice. Normalized to their expression in control TFH cells. P value was calculated by unpaired two-tailed Student’s t test from triplicate experiments. Error bars indicate mean ± SEM, **P < 0.01, ***P < 0.001, ****P < 0.0001.
The transcripts of Tcf7 and Bcl6 were decreased with EZH2 deletion in TFH cells, while no deposition of H3K27me3 marks were observed at those loci (25). Thus, EZH2 may regulate other chromatin modifiers to regulate the expression of those genes. It has been reported that EZH2 is involved in DNA methylation pathway to mediate gene expression through interacting with DNMT1, DNMT3A and DNMT3B (28). To determine whether the DNA methylation pathway in TFH cells is affected by EZH2 deletion, we measured the expression of DNA methyltransferases in TFH cells derived from control and Ezh2-/- mice on day 8 after acute viral infection. The DNMT1 and DNMT3B expression were increased with ablation of EZH2 in TFH cells, while the expression of DNMT3A was comparable between EZH2-intact and EZH2-deficient TFH cells (Figures 2A–C). When the protein synthesis inhibitor cycloheximide was applied to treat the transfected cells in vitro, the degradation of DNMT1 and DNMT3B were accelerated by forced expression of EZH2 (Figure 2D). These findings implied that DNMT1 and DNMT3B were regulated by EZH2-dependent signaling pathway. Moreover, the transcript level of demethyltransferases Tet1, Tet2, and Tet3 were not affected by the ablation of EZH2 in TFH cells (Figure 2E).
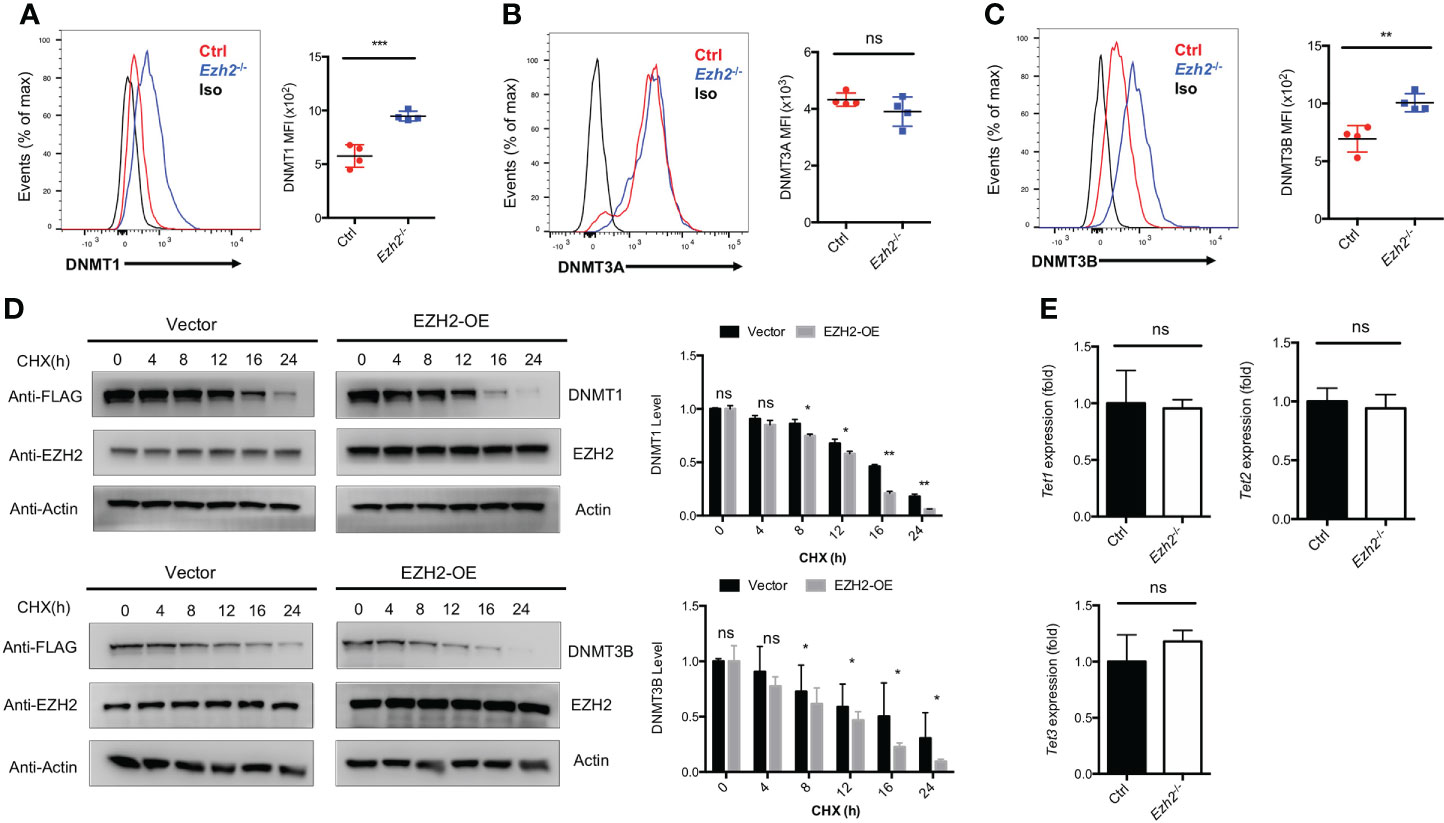
Figure 2 EZH2 regulates the expression of DNA Methyltransferases in TFH cells. (A–C) Analysis and summary of the expression of DNMT1, DNMT3A, DNMT3B in TFH cells via flow cytometry from control and Ezh2-/- mice at day 8 after infection, respectively. (D) The control and EZH2 overexpression-transfected 293T cells were treated with cycloheximide (CHX) at indicated times, followed by western blot analysis. Normalized the expression to the zero timepoint. (E) Real-Time PCR analysis of Tet1, Tet2, and Tet3 transcripts of TFH cells sorted from infected control and Ezh2-/- mice. Normalized to their expression in control TFH cells. P value was calculated by unpaired two-tailed Student’s t test from triplicate experiments. Error bars indicate mean ± SEM, ns not significant, *P < 0.05, **P < 0.01, ***P < 0.001.
These data indicated that EZH2 is associated with the DNA methylation pathway but not the DNA demethylation in TFH cells.
DNA methylation degree is inversely correlated with the expression of lineage-specific genes during T helper cell development (29, 30). As we know, TCF1 is intrinsically required for TFH cell differentiation (13). To determine whether the increased expression of DNMT1 and DNMT3B were associated with the impaired TFH differentiation, we measured the methylation status at CpG island of key regulator Tcf7 by bisulfite sequencing. Genomic DNA was isolated from TFH cells sorted from the spleen of control and Ezh2-/- mice at day 8 after infection. Strikingly, the methylation degree of CpG sites in the promoter of Tcf7 in Ezh2-/- TFH cells was 31.4%, which was three times higher than that of 10.7% in TFH cells from control mice (Figures 3A, B; Supplemental Figure 2A). Meanwhile, all the CpG sites became methylated in TFH cells from Ezh2-/- mice, and the Ezh2-/- TFH cells were 80% methylated at CpG site 1 and 6, while 3 of 7 CpG sites maintained unmethylated in the control TFH cells (Figure 3B). Moreover, the methylation status at Tcf7 locus was 23% in TFH cells from Ezh2-/- mice at early stage, which was twice higher as that of 9% in TFH cells from control mice (Figure 3C; Supplemental Figure 2B). Meanwhile, more than 85% of CpG sites became methylated in Ezh2-/- TFH cells, but the control TFH cells maintained 3 of 7 CpG sites unmethylated (Figure 3C). Additionally, the Tcf7 transcripts were diminished in early Ezh2-/- TFH cells, and thus accompanied by decreased expression of Bcl6 and increased expression of Prdm1 (Figure 3D).
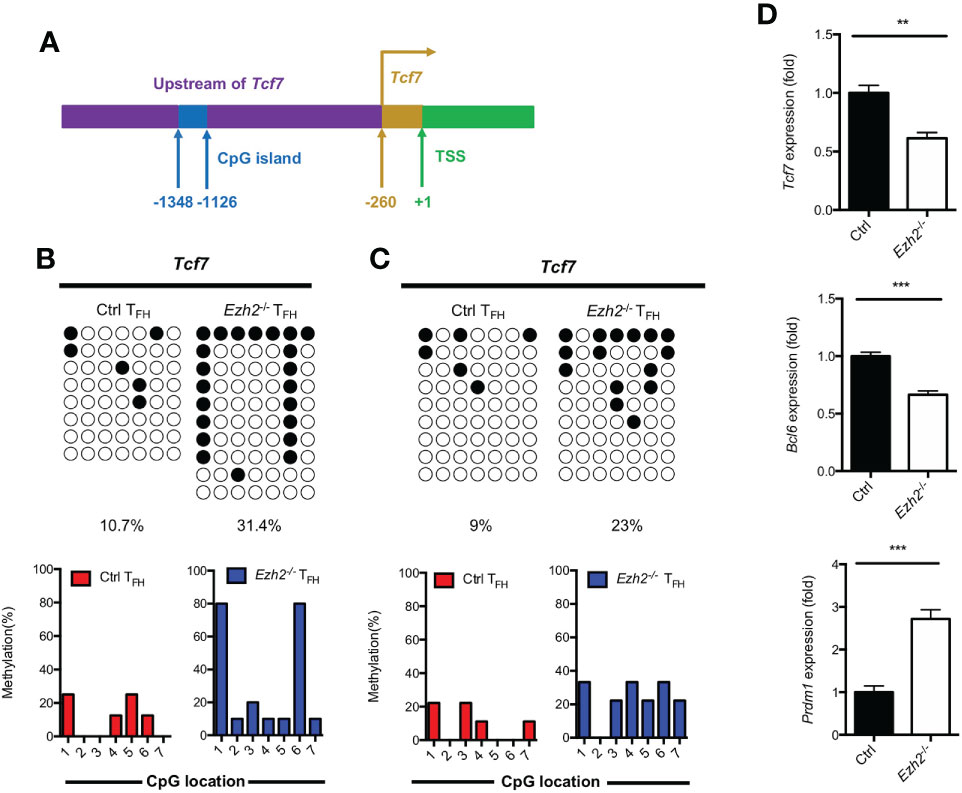
Figure 3 EZH2 deletion elevates the DNA methylation level at the Tcf7 locus. (A) Schematic diagram of CpG island in Tcf7 gene promoter. (B) Bisulfite sequencing analysis and graphical summary of conversed CpG island of Tcf7 promoter from sorted TFH cells from control and Ezh2-/- mice at day 8 after infection. (C) Bisulfite sequencing analysis and graphical summary of CpG island of Tcf7 promoter from sorted TFH cells from control and Ezh2-/- mice at day 4 after infection. (D) Real-Time PCR analysis of Tcf7, Bcl6, Prdm1 mRNA of TFH cells sorted from control and Ezh2-/- mice at day 4 after infection. Normalized to their expression in control TFH cells. The horizonal lines were corresponding to the colonies selected for sequencing. Filled black circles indicate methylated cytosine, open white circles indicate nonmethylated cytosine. P value was calculated by unpaired two-tailed Student’s t test. Error bars indicate mean ± SEM, **P < 0.01, ***P < 0.001.
Taken together, these results supported that EZH2 ensured the DNA hypomethylation degree at Tcf7 locus.
In addition to the Tcf7 locus, we also measured the methylation status of CpG islands at other TFH lineage related gene loci from both control and Ezh2-/- TFH cells at day 8 after infection. Bcl6 and Blimp1 are both downstream mediators of TCF1 (11, 12). The methylation status at Bcl6 locus were demethylated in TFH cells from both control mice and Ezh2-/- mice (Figures 4A, D), though its transcripts declined in TFH cells with EZH2 deletion (Figure 1D). Meanwhile, the CpG sites in the promoter region of Prdm1, were completely unmethylated in TFH cells from both control and Ezh2-/- mice (Figures 4B, E), but the expression of Prdm1 were much higher in TFH cells from Ezh2-/- mice than that from the control mice (Figure 1D). In addition, the CpG island of Id3, which is another TFH lineage associated factor (16), remained unmethylated in TFH cells from EZH2-intact and EZH2-deficient mice (Figures 4C, F).
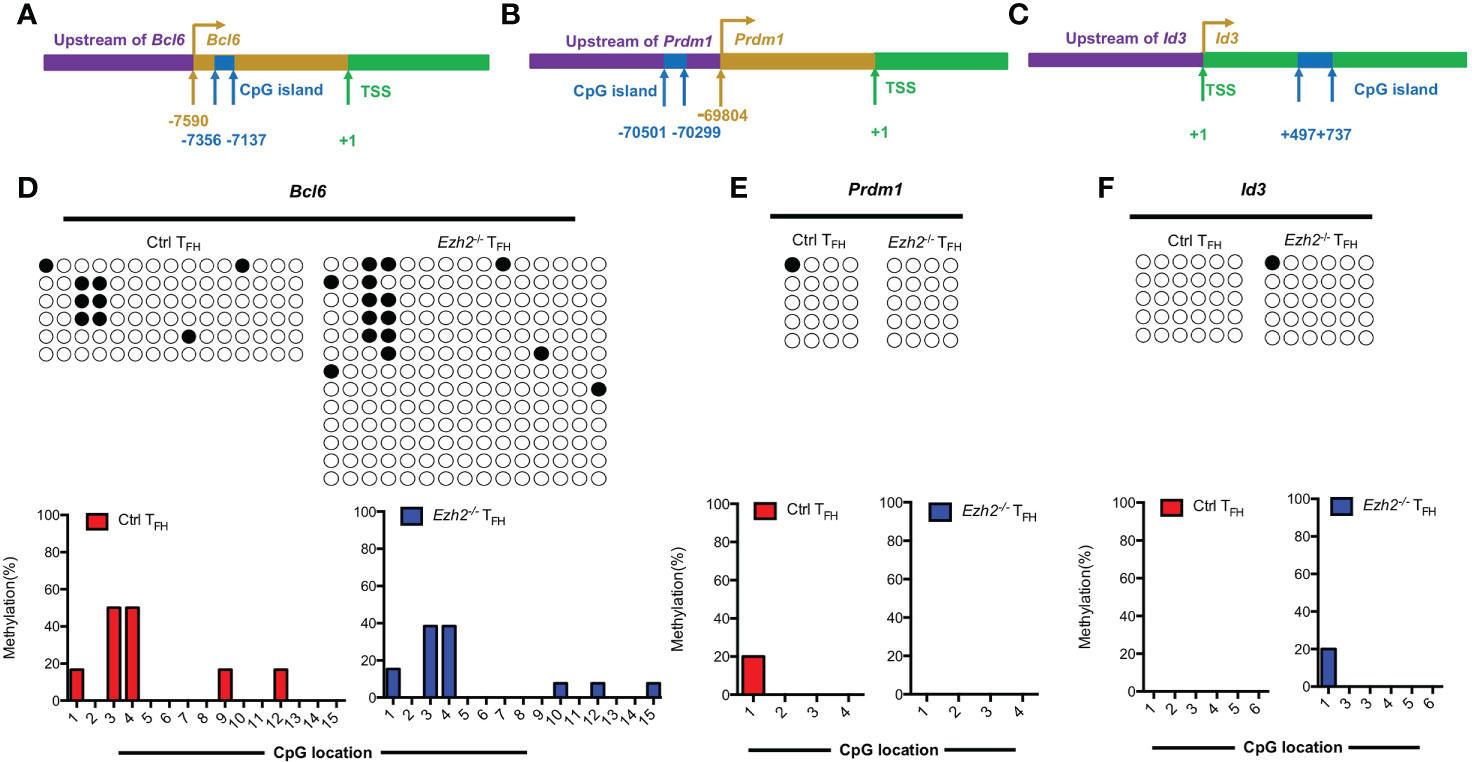
Figure 4 EZH2 displays inability to affect the methylation status at other gene loci. Genomic DNA was extracted from sorted TFH cells derived from control and Ezh2-/- mice at day 8 after LCMV Armstrong strain infection. (A–C) Schematic diagram of CpG island at Bcl6 locus (A), Prdm1 locus (B) and Id3 locus (C). (D–F) The DNA methylation status at Bcl6 locus (D), Prdm1 locus (E) and Id3 locus (F) were performed by bisulfite sequencing analysis with or without EZH2 ablation. The horizonal lines were corresponding to the colonies selected for sequencing. Filled black circles indicate methylated cytosine, open white circles indicate nonmethylated cytosine.
Taken together, these results together suggested that EZH2 displays inability to regulate the methylation status at Bcl6, Prdm1 and Id3 loci.
Results obtained from primary TFH cells demonstrated that the regulation of TCF1 expression is associated with methylation status of CpG sites in the promoter region of Tcf7 (Figure 3). To determine whether the methylation status at Tcf7 locus is associated with antigen specific TFH differentiation during acute viral infection, we transferred naive SMARTA cells into naive recipient mice, then the chimeras were subsequently infected with LCMV Armstrong strain (Figure 5A). At day 7 after infection, the CpG sites in Tcf7 promoter region was 27.1% methylated in antigen specific TH1 cells, which was nearly three times higher than that of 9.1% in antigen specific TFH cells (Figure 5B, Supplemental Figure 3A). And the expression of Tcf7 and TCF1 were remarkably higher in TFH cells compared with those in TH1 cells (Figure 5D, Supplemental Figure 3B). These results indicated that the methylation status at Tcf7 locus was inversely correlated with the transcripts and protein expression of Tcf7. Meanwhile, the Bcl6 locus maintained demethylated in TFH cells and TH1 cells (Figure 5C), while the transcripts and protein of Bcl6 were much higher in TFH cells compared with those in TH1 cells (Figure 5E, Supplemental Figure 3C).
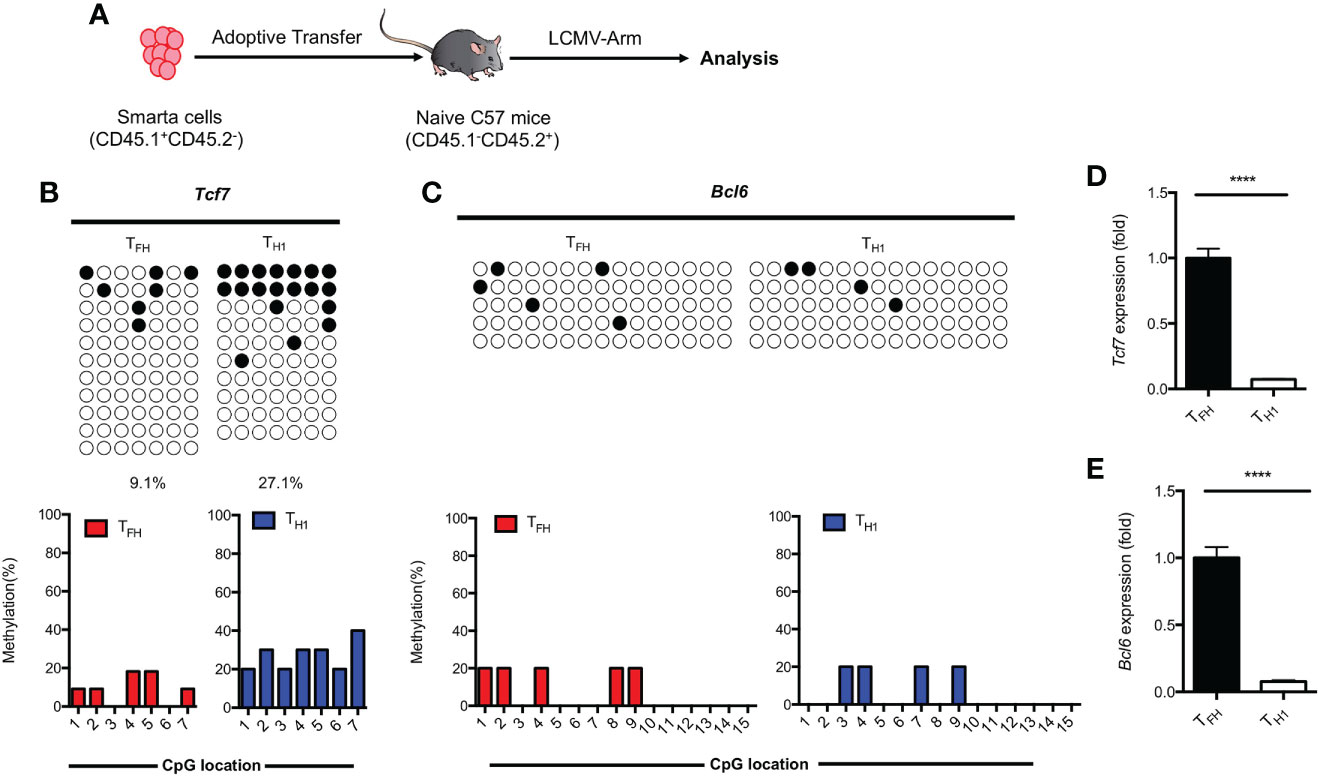
Figure 5 Methylation of Tcf7 Locus is associated with the differentiation of TFH cells during acute viral infection. (A) Procedure of experiment. 2 × 104 naive SMARTA cells were adoptively transferred into naive C57BL/6 mice, the chimeras were infected with LCMV Armstrong strain subsequently. (B, C) The virus experienced TFH (CD45.1+SLAMlowCXCR5+) and TH1 (CD45.1+SLAMhighCXCR5-) cells were sorted at day 7 after infection with LCMV Armstrong strain. The isolated genomic DNA purified from these two subtypes were subcloned into pMD19-T TA vectors for subsequent sequencing. The graphical summary of CpG islands of Tcf7 (B) and Bcl6 (C) were performed with bisulfite sequencing. (D, E) The transcriptional expression of Tcf7 (D) and Bcl6 (E) were measured by RT-PCR. Normalized to their expression in TFH cells. The horizonal lines were corresponding to the colonies selected for sequencing. Filled black circles indicate methylated cytosine, open white circles indicate nonmethylated cytosine. P value was calculated by unpaired two-tailed Student’s t test. Error bars indicate mean ± SEM. ****P < 0.0001.
These findings together suggested that hypomethylation of Tcf7 locus was positively correlated with the TFH differentiation during acute viral infection.
Besides transcription factors, epigenetic regulators have been demonstrated to involve in regulating TFH differentiation program recently (4, 25, 26, 31, 32). For instance, ablation of EZH2 led to less chromatin accessibility of TFH lineage associated genes (25), which reminded a unique function of EZH2 in transcriptional activation of TFH differentiation program (25). Moreover, EZH2 was associated with H3K27ac rather than H3K27me3 in TFH cells (26). EZH2 thus positively regulates TFH differentiation, despite generally being considered an epigenetic repressor per se. Our study further elucidates that EZH2 regulates the epigenetic modification of Tcf7 and promotes the differentiation of TFH cells. In TFH cells, EZH2 repressed the expression of DNMT1 and DNMT3B, which were associated with the methylation status at Tcf7 locus in TFH cells. Our study therefore uncovered a specific role for EZH2 in TFH differentiation from an epigenetic perspective and shed a new light on the mechanism of epigenetic modification associated with TFH differentiation program.
In this study, we provided the first evidence that ablation of EZH2 significantly increased the expression of DNMT1 and DNMT3B in TFH cells. We also observed that EZH2 could interact with DNMTs through direct protein-protein interaction (Supplemental Figure 4), which was consistent with a previous study (28). The H3K4me3 modification regulator histone methyltransferase SET7, was reported to trigger the degradation of DNMT1 by direct interaction (33), likely through the ubiquitin-proteasome system as shown by direct binding between EZH2 and the E3 ligase NP95 (34). Thus, it is plausible that EZH2 may similarly drive the degradation of DNMT1 and DNMT3B through direct interaction. It will be of great interest to verify the proper mechanism of how EZH2 mediates the expression of DNMT1 and DNMT3B in future studies.
DNA methylation is critical for the regulation of gene expression. We hence focused on the methylation status at TFH lineage related gene loci, including Tcf7, Bcl6, Id3 and Prdm1. The Tcf7 promoter was demonstrated as a target locus of DNMT3A, and the expression of DNMT3A was necessary to maintain the methylation status at Tcf7 promoter to restrict the frequency of memory precursor cells during acute viral infection (35). In this study, we found that Tcf7 locus was more selectively methylated in TFH cells with EZH2 deletion compared to other key loci were not affected. The inhibited transcriptional activity of Tcf7 led to less expression of Bcl6 and more Prdm1 expression. Additionally, the restricted TFH differentiation caused by EZH2-deficiency could be rescued by forced Bcl6 expression, which expression was assured by TCF1 (25). Although other functional target loci cannot yet be excluded, our observation demonstrated that the elevated methylation level of Tcf7 locus was associated with the increased expression of DNMTs in TFH cells. Further investigations are needed to determine whether DNMT1 or DNMT3B could act alone, or they coordinately associated with methylation level at Tcf7 locus.
It is still poorly understood what recruits DNMTs to regions of DNA loci that become methylated in T cells. In our study, the expression of DNMT1 and DNMT3B were elevated in TFH cells with EZH2 deletion, and the TFH lineage associated gene Tcf7 appears to be regulated by DNA methylation (Figure 3, Supplemental Figure 2). These findings prompt us to hypothesize that transcriptional repressor UHRF1 is a possible binding partner of DNMTs. UHRF1 is a PHD domain protein defined as the cooperator of DNMT1 (36–38). The PHD domain has been reported to interact with H3K18ac, which is associated with more accessible chromatin states (39). Thus, UHRF1 may serve as a link between DNMT1 and H3K18ac. Meanwhile, the expression of Kat2a and Kat2b (acetyltransferases of H3K18ac) were much higher in TFH cells than that in TH1 cells (data not shown). These facts suggest that H3K18ac might be associated with the expression of TFH lineage associated genes, including Tcf7. We suggest that DNMT1 may be driven to the H3K18ac associated Tcf7 locus by binding with UHRF1 for methylating the proper sites. However, it needs to be further disclosed.
In the scenario of acute viral infection, naive SMARTA cells differentiate into TFH and TH1 cells (40–43). The bifurcation of TFH and TH1 cells is mediated by Bcl6, and the expression of Bcl6 is ensured by TCF1. We observed that the methylation status at Tcf7 locus was more heavily in TH1 cells than that in TFH cells, while there was no detectable difference in the methylation status at Bcl6 locus between TFH and TH1 cells. Besides, the expression of DNMT1 and DNMT3B were comparable in TFH and TH1 cells, and the DNMT3A expression was much higher in TFH cells compared with that in TH1 cells (data not shown). These results indicated that Tcf7 locus may also be the target locus of DNA demethyltransferase. Although the precise mechanisms about the additional modifications affect the methylation status at Tcf7 locus are not clarified yet, our results suggest that the hypomethylation of Tcf7 locus but not Bcl6 locus was associated with the differentiation of TFH cells.
In summary, this study reveals for the first time that EZH2 regulates the epigenetic modification of Tcf7 during acute viral infection. EZH2 plays a crucial role in modulating the degree of DNA methylation at Tcf7 locus and promoting the differentiation of TFH cells. Since TFH cells are critical in humoral immunity responses and development of autoimmune diseases, the dissection of EZH2 function may provide substantial therapeutic benefits for the treatment of viral infection and autoimmune diseases.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by Institutional Animal Care and Use Committee of the Third Military Medical University.
YL, DL, ZW, LX, SL, XC, CW, LL, SY, FL, and XX performed the experiments. ZL, LH, JT, DY, JF, CC, and YZ provided reagents, materials and support. RH and LY designed the study, analyzed the data and wrote the paper with YL and ZW. RH and LY supervised the study. All authors contributed to the article and approved the submitted version.
This work was supported by National Key Research and Development Program of China (2020YFA0804400), National Natural Science Foundation of China (81871248 to RH, 32000619 to ZW), China Postdoctoral Science Foundation (2020M673647 to ZW), Chongqing Special Postdoctoral Science Foundation (cstc2020jcyj-bshX0039 to ZW) and Program of HUST Academic Frontier Youth Team (2018QYTD10).
We thank R. Ahmed (Emory University) for LCMV Armstrong virus and SMARTA transgenic mice. We thank the core facility center of Third Military Medical University for cell sorting.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.942465/full#supplementary-material
1. O'Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity. Helper Cd4+ T Cells Sci (2010) 327(5969):1098–102. doi: 10.1126/science.1178334
2. Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, et al. Bcl6 mediates the development of T follicular helper cells. Science (2009) 325(5943):1001–5. doi: 10.1126/science.1176676
3. Choi J, Crotty S. Bcl6-mediated transcriptional regulation of follicular helper T cells (Tfh). Trends Immunol (2021) 42(4):336–49. doi: 10.1016/j.it.2021.02.002
4. Liu L, Hu L, Yang L, Jia S, Du P, Min X, et al. Uhrf1 downregulation promotes T follicular helper cell differentiation by increasing Bcl6 expression in sle. Clin Epigenet (2021) 13(1):31. doi: 10.1186/s13148-021-01007-7
5. McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med (2011) 365(23):2205–19. doi: 10.1056/NEJMra1004965
6. Gabriel SE. The epidemiology of rheumatoid arthritis. Rheum Dis Clin North Am (2001) 27(2):269–81. doi: 10.1016/s0889-857x(05)70201-5
7. Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, et al. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell (2010) 143(4):592–605. doi: 10.1016/j.cell.2010.10.032
8. Alterauge D, Bagnoli JW, Dahlstrom F, Bradford BM, Mabbott NA, Buch T, et al. Continued Bcl6 expression prevents the transdifferentiation of established tfh cells into Th1 cells during acute viral infection. Cell Rep (2020) 33(1):108232. doi: 10.1016/j.celrep.2020.108232
9. Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and blimp-1 in T and b lymphocyte differentiation. Nat Immunol (2010) 11(2):114–20. doi: 10.1038/ni.1837
10. Choi J, Diao H, Faliti CE, Truong J, Rossi M, Belanger S, et al. Bcl-6 is the nexus transcription factor of T follicular helper cells via repressor-of-Repressor circuits. Nat Immunol (2020) 21(7):777–89. doi: 10.1038/s41590-020-0706-5
11. Choi YS, Gullicksrud JA, Xing S, Zeng Z, Shan Q, Li F, et al. Lef-1 and tcf-1 orchestrate T(Fh) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat Immunol (2015) 16(9):980–90. doi: 10.1038/ni.3226
12. Xu L, Cao Y, Xie Z, Huang Q, Bai Q, Yang X, et al. The transcription factor tcf-1 initiates the differentiation of T(Fh) cells during acute viral infection. Nat Immunol (2015) 16(9):991–9. doi: 10.1038/ni.3229
13. Wu T, Shin HM, Moseman EA, Ji Y, Huang B, Harly C, et al. Tcf1 is required for the T follicular helper cell response to viral infection. Cell Rep (2015) 12(12):2099–110. doi: 10.1016/j.celrep.2015.08.049
14. Choi YS, Kageyama R, Eto D, Escobar TC, Johnston RJ, Monticelli L, et al. Icos receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity (2011) 34(6):932–46. doi: 10.1016/j.immuni.2011.03.023
15. Liu X, Chen X, Zhong B, Wang A, Wang X, Chu F, et al. Transcription factor achaete-scute homologue 2 initiates follicular T-Helper-Cell development. Nature (2014) 507(7493):513–8. doi: 10.1038/nature12910
16. Shaw LA, Belanger S, Omilusik KD, Cho S, Scott-Browne JP, Nance JP, et al. Id2 reinforces Th1 differentiation and inhibits E2a to repress tfh differentiation. Nat Immunol (2016) 17(7):834–43. doi: 10.1038/ni.3461
17. Lee JY, Skon CN, Lee YJ, Oh S, Taylor JJ, Malhotra D, et al. The transcription factor Klf2 restrains Cd4(+) T follicular helper cell differentiation. Immunity (2015) 42(2):252–64. doi: 10.1016/j.immuni.2015.01.013
18. Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet (2016) 17(8):487–500. doi: 10.1038/nrg.2016.59
19. Allan RS, Zueva E, Cammas F, Schreiber HA, Masson V, Belz GT, et al. An epigenetic silencing pathway controlling T helper 2 cell lineage commitment. Nature (2012) 487(7406):249–53. doi: 10.1038/nature11173
20. Antignano F, Burrows K, Hughes MR, Han JM, Kron KJ, Penrod NM, et al. Methyltransferase G9a regulates T cell differentiation during murine intestinal inflammation. J Clin Invest (2014) 124(5):1945–55. doi: 10.1172/JCI69592
21. Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science (2002) 298(5595):1039–43. doi: 10.1126/science.1076997
22. Kanno Y, Vahedi G, Hirahara K, Singleton K, O'Shea JJ. Transcriptional and. epigenetic control of T helper cell specification: Molecular mechanisms underlying commitment and plasticity. Annu Rev Immunol (2012) 30:707–31. doi: 10.1146/annurev-immunol-020711-075058
23. Tumes DJ, Onodera A, Suzuki A, Shinoda K, Endo Y, Iwamura C, et al. The polycomb protein Ezh2 regulates differentiation and plasticity of Cd4(+) T helper type 1 and type 2 cells. Immunity (2013) 39(5):819–32. doi: 10.1016/j.immuni.2013.09.012
24. DuPage M, Chopra G, Quiros J, Rosenthal WL, Morar MM, Holohan D, et al. The chromatin-modifying enzyme Ezh2 is critical for the maintenance of regulatory T cell identity after activation. Immunity (2015) 42(2):227–38. doi: 10.1016/j.immuni.2015.01.007
25. Chen X, Cao G, Wu J, Wang X, Pan Z, Gao J, et al. The histone methyltransferase Ezh2 primes the early differentiation of follicular helper T cells during acute viral infection. Cell Mol Immunol (2020) 17(3):247–60. doi: 10.1038/s41423-019-0219-z
26. Li F, Zeng Z, Xing S, Gullicksrud JA, Shan Q, Choi J, et al. Ezh2 programs tfh differentiation by integrating phosphorylation-dependent activation of Bcl6 and polycomb-dependent repression of P19arf. Nat Commun (2018) 9(1):5452. doi: 10.1038/s41467-018-07853-z
27. Li LC, Dahiya R. Methprimer: Designing primers for methylation pcrs. Bioinformatics (2002) 18(11):1427–31. doi: 10.1093/bioinformatics/18.11.1427
28. Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The polycomb group protein Ezh2 directly controls DNA methylation. Nature (2006) 439(7078):871–4. doi: 10.1038/nature04431
29. Walsh CP, Chaillet JR, Bestor TH. Transcription of iap endogenous retroviruses. is constrained by cytosine methylation. Nat Genet (1998) 20(2):116–7. doi: 10.1038/2413
30. Walsh CP, Bestor TH. Cytosine methylation and mammalian development. Genes Dev (1999) 13(1):26–34. doi: 10.1101/gad.13.1.26
31. Yao Y, Yang Y, Guo W, Xu L, You M, Zhang YC, et al. Mettl3-dependent M(6)a. modification programs T follicular helper cell differentiation. Nat Commun (2021) 12(1):1333. doi: 10.1038/s41467-021-21594-6
32. Zhu Y, Zhao Y, Zou L, Zhang D, Aki D, Liu YC. The E3 ligase vhl promotes. follicular helper T cell differentiation Via glycolytic-epigenetic control. J Exp Med (2019) 216(7):1664–81. doi: 10.1084/jem.20190337
33. Esteve PO, Chin HG, Benner J, Feehery GR, Samaranayake M, Horwitz GA, et al. Regulation of Dnmt1 stability through Set7-mediated lysine methylation in mammalian cells. Proc Natl Acad Sci U.S.A. (2009) 106(13):5076–81. doi: 10.1073/pnas.0810362106
34. Babbio F, Pistore C, Curti L, Castiglioni I, Kunderfranco P, Brino L, et al. The sra. protein Uhrf1 promotes epigenetic crosstalks and is involved in prostate cancer progression. Oncogene (2012) 31(46):4878–87. doi: 10.1038/onc.2011.641
35. Ladle BH, Li KP, Phillips MJ, Pucsek AB, Haile A, Powell JD, et al. De novo DNA methylation by DNA methyltransferase 3a controls early effector Cd8+ T-cell fate decisions following activation. Proc Natl Acad Sci USA (2016) 113(38):10631–6. doi: 10.1073/pnas.1524490113
36. Guan D, Factor D, Liu Y, Wang Z, Kao HY. The epigenetic regulator Uhrf1. promotes ubiquitination-mediated degradation of the tumor-suppressor protein promyelocytic leukemia protein. Oncogene (2013) 32(33):3819–28. doi: 10.1038/onc.2012.406
37. Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, et al. The sra. protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature (2007) 450(7171):908–12. doi: 10.1038/nature06397
38. Rottach A, Frauer C, Pichler G, Bonapace IM, Spada F, Leonhardt H. The multi-domain protein Np95 connects DNA methylation and histone modification. Nucleic Acids Res (2010) 38(6):1796–804. doi: 10.1093/nar/gkp1152
39. He M, Xu Y, Chen J, Luo Y, Lv Y, Su J, et al. Mosnt2-dependent deacetylation of. histone H3 mediates motor-dependent autophagy and plant infection by the rice blast fungus magnaporthe oryzae. Autophagy (2018) 14(9):1543–61. doi: 10.1080/15548627.2018.1458171
40. Wang Y, Tian Q, Hao Y, Yao W, Lu J, Chen C, et al. The kinase complex Mtorc2 promotes the longevity of virus-specific memory Cd4(+) T cells by preventing ferroptosis. Nat Immunol (2022) 23(2):303–17. doi: 10.1038/s41590-021-01090-1
41. Hale JS, Youngblood B, Latner DR, Mohammed AU, Ye L, Akondy RS, et al. Distinct memory Cd4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity (2013) 38(4):805–17. doi: 10.1016/j.immuni.2013.02.020
42. Fahey LM, Wilson EB, Elsaesser H, Fistonich CD, McGavern DB, Brooks DG. Viral persistence redirects Cd4 T cell differentiation toward T follicular helper cells. J Exp Med (2011) 208(5):987–99. doi: 10.1084/jem.20101773
Keywords: LCMV, TFH cells, EZH2, DNA methylation, Tcf7
Citation: Luo Y, Li D, Xie L, Lei S, Chen X, Wang C, Yao D, Li L, Fang J, Chen C, Yuan S, Li F, Xie X, Zhang Y, Li Z, Hu L, Tang J, Ye L, Wei Z and He R (2022) EZH2 restricts Tcf7 DNA methylation and promotes TFH differentiation during acute viral infection. Front. Immunol. 13:942465. doi: 10.3389/fimmu.2022.942465
Received: 12 May 2022; Accepted: 28 July 2022;
Published: 15 August 2022.
Edited by:
Andrew D. Wells, Children’s Hospital of Philadelphia, United StatesReviewed by:
Gokhan Akkoyunlu, Akdeniz University, TurkeyCopyright © 2022 Luo, Li, Xie, Lei, Chen, Wang, Yao, Li, Fang, Chen, Yuan, Li, Xie, Zhang, Li, Hu, Tang, Ye, Wei and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ran He, cmFuaGVAaHVzdC5lZHUuY24=; Zhengping Wei, d2V6aHBpQDEyNi5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.