
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Immunol. , 14 July 2022
Sec. Comparative Immunology
Volume 13 - 2022 | https://doi.org/10.3389/fimmu.2022.927880
 Xiaomin Li1,2‡
Xiaomin Li1,2‡ Ting Chen1,3‡
Ting Chen1,3‡ Xiaofen Wu4
Xiaofen Wu4 Zhuobo Li1,2
Zhuobo Li1,2 Xin Zhang1,2
Xin Zhang1,2 Xiao Jiang1,3
Xiao Jiang1,3 Peng Luo1,3
Peng Luo1,3 Chaoqun Hu1,3
Chaoqun Hu1,3 Nai-Kei Wong5,6*†
Nai-Kei Wong5,6*† Chunhua Ren1,3*
Chunhua Ren1,3*Heavy-metal pollution has increasingly jeopardized the habitats of marine organisms including the sea cucumber, a seafloor scavenger vital to seawater bio-decontamination, ocean de-acidification and coral-reef protection. Normal physiology including immune functions of sea cucumbers is toxicologically modulated by marine metal pollutants such as cadmium (Cd). The processes underpinning Cd’s toxic effects on immune systems in the sea cucumber, Holothuria leucospilota, are still poorly understood. To this end, we cloned and characterized a full-length caspase-9 (Hl-CASP9) cDNA in the sea cucumber, Holothuria leucospilota. Hl-CASP9 mRNA levels evolved dynamically during embryonic development. Coelomocytes, a type of phagocytic immune effectors central to H. leucospilota immunity, were found to express Hl-CASP9 mRNA most abundantly. Hl-CASP9 protein structurally resembles caspases-2 and -9 in both invertebrate and vertebrate species, comprising a CARD domain and a CASc domain. Remarkably, Hl-CASP9 was transcriptionally sensitive to abiotic oxidative stress inducers including hydrogen peroxide (H2O2), nitric oxide (•NO) and cadmium (Cd), but insensitive to immunostimulants including lipopolysaccharide (LPS), and poly(I:C). Overexpression of Hl-CASP9 augmented mitochondria-dependent apoptosis in HEK293T cells, while knock-down of Hl-CASP9 blunted Cd-induced coelomocyte apoptosis in vivo. Overall, we illustrate that an evolutionarily ancient caspase-9-dependent pathway exists to sensitize coelomocytes to premature cell death precipitated by heavy metal pollutants, with important implications for negative modulation of organismal immune response in marine invertebrates.
Sea cucumbers (Echinodermata: Holothuroidea) constitute a major scavenging component integral to many marine ecosystems such as coral reefs, kelp grasslands and mangroves (1). Among these, Holothuria leucospilota is a tropical species endemic to the Indo-Pacific region, which dwells amidst coral reefs, mud or sand (2, 3). The sea cucumber typically feeds on sediments rich in organic carbons. It possesses a powerful digestive tract that efficiently degrades organic carbons, concomitantly reducing biocontamination and promoting local pH balance of seawater (4, 5). Thus, H. leucospilota serves as a seabed scavenger vital to the health and biodiversity of nearshore marine ecosystems, and has been prized for their roles in the restoration of injured coral reefs. However, the sea cucumber lacks an adaptive immune system as in other marine invertebrates; they rely overwhelmingly on innate immunity including humoral and cellular responses (6). In particular, the phagocytic coelomocytes (literally, cells derived from the coelom, a fluid-filled cavity occupying the space between the alimentary canal and body walls) have been recognized as the primary effector cells in the immune system of echinoderms (e.g., the sea cucumber), with indispensable roles in cell agglutination, encapsulation, phagocytosis and cell death (7, 8).
Within the increasingly volatile marine environments, physicochemical properties of seawater (e.g., pH, composition, etc.) can be adversely affected by anthropogenic pollutants from industrial discharges. Among the most deleterious pollutants are heavy metals, namely, cadmium (Cd), mercury, lead, and arsenic (9). The presence of Cd in ocean waters, for example, exerts global survival pressure on marine life, especially for species dwelling in the delicate coral-reef ecosystems, which ultimately leads to a profound loss of marine biodiversity (10, 11). In addition, Cd accumulates in tissues of marine organisms and could well cause harm to human health by entering or re-entering the food chain (12). Among marine benthos, echinoderms represent a simple but important model for assessing the effects of specific environmental pressure on biological growth and vitality (9). As heavy metals are generally toxic and difficult to remove, their accumulation may lead to acute or chronic oxidative stress in aquatic animals, which is accompanied by excessive ROS production, protein dysfunction, lipid peroxidation and DNA oxidation (12, 13).
As a key protein family involved with apoptosis, cysteine-dependent aspartate-directed proteases (caspases) are a family of cysteine proteases ubiquitously found in vertebrate and invertebrate species including echinoderms (14). Typically, apoptotic response is mediated through either an extrinsic or an intrinsic pathway. The extrinsic pathway is triggered by binding of extracellular ligands to death receptors on the cell membrane, whereas the intrinsic pathway is mainly turned on by different mitochondrial stressors, leading to the cytoplasmic release of mitochondrial cytochrome c, which then binds caspase-9 and apoptotic protease activating factor 1 (Apaf-1) to form an apoptosome (15). As an initiator caspase, caspase-9 is prerequisite for mitochondrial morphological changes and ROS production (16). Interestingly, despite its relevance to cell death, caspase-9 also plays pivotal roles in tissue differentiation (e.g., in muscles) in non-apoptotic signaling (17).
In recent years, there has been growing interest in the biology of apoptotic caspases in ecologically important echinoderms, including sea urchin (Paracentrotus lividus), sea cucumber (H. leucospilota and Apostichopus japonicus) and starfish (Asterina pectinifera) (18–22) (See a summary on this in Table 1). For example, exposure of P. lividus embryos to Cd triggered autophagy and concomitant apoptosis as assessed by TUNEL and cleaved caspase-3 (30). In previous studies on H. leucospilota by our group, HLcaspase-6 was shown to be an effector caspase in apoptosis (21), while HLcaspase-8 was found to operate in a receptor-related apoptotic pathway (22). Both HLcaspase-6 and HLcaspase-8 were transcriptionally up-regulated by immunostimulants such as lipopolysaccharide (LPS) or polyinosinic-polycytidylic acid [poly(I:C)] in H. leucospilota coelomocytes in vitro. Furthermore, both caspase-9 and Apaf-1 are present in the genome of Strongylocentrotus purpuratus (sea urchin), lending support to a conserved mechanism governing intrinsic apoptosis within deuterostomes (34). However, in echinoderms as in other marine invertebrates, the regulatory machinery for caspase-dependent pathways in Cd-induced apoptosis remains obscure.
In order to clarify the physiological roles of caspase-9 in echinoderms, we first cloned and characterized caspase-9 cDNA from tropical sea cucumber, H. leucospilota, an ecologically relevant and commercially valuable aquaculture species in southern China (35). To scrutinize the function of H. leucospilota caspase-9 (Hl-CASP9), we analyzed mRNA distribution profiles across developmental stages and various adult tissues of H. leucospilota, and compared transcriptional expression profiles following pharmacological challenges of immunostimulants and oxidative stressors including Cd and chemical donors of ROS/RNS (reactive oxygen/nitrogen species). Furthermore, the functional relevance of Hl-CASP9 to mitochondrial stress and apoptosis in response to Cd was also examined. Our current work thus provides fresh insights into the functional significance of sea cucumber caspase-9 in relation to environmental perturbations such as heavy metal contaminants and microbial infections, which should help stimulate interest in regulatory mechanisms underlying cell death and stress adaptation in marine invertebrates including echinoderms.
Healthy adult individuals of sea cucumbers (H. leucospilota) weighing 200 ± 10 g were obtained from Daya Bay (Shenzhen, China) and temporarily reared in a seawater aquarium with filtrated and aerated seawater with salinity of 35‰ and temperature of 32°C for a week before experiments. After being placed on ice for 30 min, sea cucumbers were dissected and the tissues were collected and frozen rapidly in liquid nitrogen for storage at −80°C, until RNA extraction. Meanwhile, coelomic fluids were filtered by 100 µm sterile nylon mesh and centrifuged immediately at 1,000 × g for 10 min at 4°C to harvest coelomocytes. Then, the harvested coelomocytes were then kept in 1 mL TRIzol reagent (Invitrogen, USA) at −80°C. All animal experiments have been approved by the Ethics Committees of the South China Sea Institute of Oceanology, Chinese Academy of Sciences, and were conducted in accordance with guidelines thereof.
Total RNA from coelomocytes of H. leucospilota was extracted by using TRIzol reagent and treated with RNase-free recombinant DNase I (Takara, Japan). First-strand cDNA was synthesized by using the PrimeScript™ II 1st Strand cDNA Synthesis Kit (Takara). Based on a transcriptomic library of H. leucospilota coelomocytes previously constructed by our lab, a partial sequence for the Hl-CASP9 homologue was verified with gene-specific primers Hl-CASP9-F and Hl-CASP9-R (Table S1). To obtain corresponding full-length cDNA sequences, the 3′- and 5′-rapid amplification of cDNA ends (RACE) was performed by using 3′ Full Race Core Set Ver. 2.0 and 5′ Full Race Kit (Takara) with gene-specific primers (3′ RACE1/3′ RACE2 and 5′ RACE1/5′ RACE2, see Table 1), respectively. Amplification conditions for verification of partial sequences and RACE were set as follows: 94°C for 60 s, 30 cycles of 98°C for 10 s, 55°C for 15 s and 68°C for 60 s. pMD™ 19-T vector (Takara) and DH5α competent cells (Takara) were used for subcloning of the amplicons. Three positive clones for each amplicon were subsequently sequenced.
Open reading frame (ORF) and amino acids sequences were predicted by using ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/). Structural domains of Hl-CASP9 were predicted by using the SMART (http://smart.embl-heidelberg.de/), ScanProsite (http://prosite.expasy.org/) and BLAST (http://blast.ncbi.nlm.nih.gov) programs. Molecular weight (M.W.) and theoretical isoelectric point were calculated according to deduced amino acids by ProtParam (http://web.expasy.org/protparam/). Alignment for amino acids sequences among various species was performed with the Clustal Omega program (http://www.ebi.ac.uk/Tools/msa/clustalo/) and presented by using the Jalview program (http://www.jalview.org/). Phylogenetic trees were constructed based on amino acid differences (p-distance) according to the neighbor-joining method (pairwise deletion) with 1,000 bootstrap replicates by using MEGA X. Three-dimensional (3-D) models were deduced with the I-TASSER server (http://zhanglab.ccmb.med.umich.edu/I-TASSER/) and visualized by the VDM program (http://www.ks.uiuc.edu/Research/vmd/).
Morphology of H. leucospilota embryos and larvae was studied by using an optical microscope, as described previously by our group (35). Samples were collected at different developmental stages including: fertilized egg, 2-cell, 4-cell, 8-cell, 16-cell, morula, blastula, rotated-blastula, early-gastrula, late-gastrula, early-auricularia, mid-auricularia, auricularia, doliolaria, pentactula, and juvenile stages. Tissue distribution of Hl-CASP9 mRNA was quantitatively detected in three adult individuals. The selected tissues include coelomocytes, intestine, body wall, respiratory tree, rete mirabile, transverse vessel, Polian vesicles, muscle, oesophagus, Cuvierian tubules and gonads. Total RNA was extracted with TRIzol reagent and digested with gDNA Eraser (Takara) and reverse transcription by using the PrimeScript™ RT reagent Kit (Takara) for quantitative PCR (qPCR). Specific primers QHl-CASP9-F and QHl-CASP9-R (Table S1) were designed based on the obtained Hl-CASP9 cDNA sequences. qPCR reactions were performed by using SYBR Premix Ex Taq™ II (Takara) in a final volume of 20 μL, with the conditions of 40 cycles of 95°C for 5 s and 60°C for 30 s. In this experiment, Hl-β-actin was used as an internal control to verify qPCR results with reference to previous modifications (22).
Coelomocytes were chosen as a cell model for investigating transcriptional responses of Hl-CASP9 following challenges of immunostimulants or oxidative stressors. Primary sea cucumber coelomocytes were prepared as previously described (36). After culture at 28°C for 18 h in Leiboviz’s L-15 culture medium (with 0.39 M NaCl) (Invitrogen), coelomocytes were challenged with immunostimulants including LPS (10 μg/mL), poly(I:C) (10 μg/mL) and heat-inactivated V. harveyi (107 CFU/mL), an important pathogen of many aquatic animals, or alternatively, with oxidative stress inducers including H2O2 (hydrogen peroxide; 2 μM), NOC-18 (a nitric oxide donor; 1μM) and CdCl2 (cadmium chloride; 20 μM), respectively. After these treatments for 0, 3, 6, 12 and 24 h, coelomocytes were harvested and lysed in TRIzol reagent, followed by procedures for RNA extraction and, first-strand cDNA synthesis. Quantitative changes in mRNA expressions of Hl-CASP9 after immune and oxidative challenges were analyzed quantitatively by qPCR assays, as described above.
Coding region of Hl-CASP9 was amplified by PCR by using the gene-specific primers PHl-CASP9-F and PHl-CASP9-R (Table S1), with restriction enzyme sites for BamH I and Xho I. PCR products and the pcDNA3.1(+) vector (Invitrogen) were digested with BamH I (Takara) and Xho I (Takara), and ligated by means of T4 DNA ligase (Fermentas, USA) at 16°C for 1 h and then used to transform DH5α competent cells. Accuracy of the constructed plasmid was confirmed by double-enzyme (BamH I and Xho I) digestion and sequencing. A positive clone was picked and cultured in lysogen broth (LB) containing ampicillin (100 μg/mL) at 37°C with shaking at 220 rpm overnight. The amplified pcDNA3.1(+)/Hl-CASP9 recombinant plasmid was purified by using the Plasmid MiniPrep DNA Kit (Axygen, USA).
Before transfection, HEK293T cells were seeded in a 6-well plate and cultured in Dulbecco’s Modified Eagle Medium (Hyclone, USA) containing 10% fetal calf serum (FCS), penicillin (100 μg/mL) and streptomycin (100 μg/mL) at 37°C with 5% CO2 for one day. Afterwards, HEK293T cells were transfected with 3 μg pcDNA3.1(+)/Hl-CASP9 recombinant plasmid by using 3 μL lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. By using the same methods, HEK293T cells were transfected with an empty pcDNA3.1(+) vector as a control.
HEK293T cells were incubated in the presence or absence of CCCP (carbonyl cyanide 3-chlorophenylhydrazone; 100 μM), a protonophore which uncouples oxidative phosphorylation in mitochondria, at 37°C for 30 min. Then, apoptosis was detected by an Annexin V-FITC/PI cell apoptosis detection kit (Solarbio, China). Cells were collected and washed twice with pre-cooled PBS (pH 7.4) and resuspended in 1×binding buffer, with a final cell suspension density of 1×106 cells/mL. Subsequently, 100 μL cell suspension mixed with 5 μL Annexin V-FITC and 5 μL propyl iodide (PI) was incubated in dark for 10 min at room temperature. Then, 400 μL of 1 × binding buffer was added prior to detection. Finally, cell apoptosis proportions were measured by flow cytometry (Beckman Coulter, USA) within 1 h. Annexin V-FITC was used to stain phospholipid serine on the cell membrane at the early stage of apoptosis, wherein FITC signals indicate the rate of early apoptosis. PI penetrates membranes of damaged cells, such as late apoptotic cells or necrotic cells, and binds nuclear DNA to give red staining. Cells doubly stained by PI and Annexin V-FITC are considered late apoptotic cells or necrotic cells.
Primary coelomocytes (1×106 cells/mL, survival rate > 95%) were seeded into 6-well plates pre-coated with poly D-lysine and grown overnight in Leiboviz’s L-15 culture medium (with 0.39 M NaCl). After treatment with different concentrations of CdCl2 (0, 5, 10 or 20 μM) for 3 h, the medium was removed. Then, 1 μM dichloro-dihydro-fluorescein diacetate (DCFH-DA) (Biosharp, China) diluted in serum-free medium was added. After 30-min incubation, coelomocytes were washed twice in PBS (with 0.53 M NaCl), followed by observation under a fluorescent microscope (Zeiss, Germany). Fluorescence intensity quantification was done with ImageJ.
Double-stranded RNA (dsRNA) targeting the Hl-CASP9 gene was synthesized by using the T7 RiboMAX™ Express RNAi System (Promega, USA) according to the manufacturer’s instructions. Similarly, a GFP cDNA fragment was amplified as the negative control. Specific primers used for the synthesis of dsRNA, as shown in Table S1. Concentration of synthesized dsRNA was verified by NanoDrop 2000 spectrophotometer (Thermo Scientific, USA), which was then diluted to 600 ng/μL in 150 mM RNase-free saline solution (RFSS). For in vivo dsRNA interference, nine healthy individuals of H. leucospilota (each weighing about 100 g) were randomly assigned into three groups (n = three individuals per group). The experimental group was injected with 100 μL dsCASP9, while the control group was injected with 100 μL RFSS and the negative control was injected with an equivalent amount of dsGFP. Following RNAi for 48 h, all sea cucumbers were dissected on the ice and their coelomocytes were isolated. Thereafter, a sample of coelomocytes was taken to assess interference efficiency of RNAi experiment by qPCR, with specific primers and reaction conditions identical to those listed in section 2.4.
Primary coelomocytes (1×106 cells/mL) from different groups (sea cucumbers injected with RFSS, dsGFP or dsCASP9) were plated into 3.5-cm glass-bottomed confocal dishes pre-coated with poly D-lysine, followed by incubation at 28°C for 18 h and subsequent treatment with CdCl2 (20 μM) for 3 h. Prior to imaging experiments, coelomocytes were loaded with the mitochondrial superoxide probe MitoSOX (2.5 μM; Invitrogen) for 30 min and then washed with HBSS (with 0.53 M NaCl) immediately before imaging. Images were obtained with a confocal fluorescence microscope (Leica, Germany). Fluorescence intensity quantification was done with ImageJ.
Mitochondrial membrane potential (Δψm) was measured by the commercial membrane potential probe JC-1 (2.5 μM; Invitrogen). Briefly, primary coelomocytes (1×106 cells/mL) from different groups (sea cucumbers injected with RFSS, dsGFP or dsCASP9) were seeded into 6-well plates pre-coated with poly D-lysine and grown in Leiboviz’s L-15 culture medium (with 0.39 M NaCl) overnight. Then, coelomocytes were treated with CdCl2 (20 μM; 3 h) or CCCP (100 μM; 30 min), respectively. After removing culture medium, coelomocytes were incubated with JC-1 for 20 min in dark and then washed twice with PBS (with 0.53 M NaCl) before acquisition. Δψm in coelomocytes from different groups was detected by flow cytometry (Beckman Coulter).
After RNAi for 48 h, primary coelomocytes from different groups (sea cucumbers injected with RFSS, dsGFP or dsCASP9) were harvested and cultured overnight in Leiboviz’s L-15 cell culture medium (with 0.39 M NaCl), and then exposed to oxidative stress inducers including H2O2 (2 μM), NOC-18 (1 μM) and CdCl2 (20 μM). After 24 h, the H. leucospilota coelomocytes were resuspended by proper pipetting and the coelomocytes proportions of undergoing apoptosis was detected by an Annexin V-FITC/PI cell apoptosis detection kit (Solarbio) and determined by flow cytometry (Beckman Coulter) as described above.
All data were analyzed by using one-way ANOVA and presented as mean ± SEM (n = 3), followed by Fisher’s least significant difference (LSD) test with SPSS 22.0 (IBM Software, USA). Statistical significance was determined at p > 0.05 (i.e., n.s.), *p < 0.05, **p < 0.01 or ***p < 0.001.
In this study, full-length cDNA sequence of caspase-9 was obtained from H. leucospilota by using 3’-/5’-RACE approaches, which has been deposited in GenBank under the accession number MW759843. The Hl-CASP9 cDNA is 1,841 bp in length, encompassing a 5’-untranslated region (UTR) of 180 bp, a 3’-UTR of 398 bp and an ORF of 1,263 bp encoding a protein of 420 a.a. (Figure S1A) with a deduced molecular weight of 47.25 kDa and a predicted isoelectric point of 6.44. Based on analysis with the SMART program, a caspase recruitment domain (CARD, residues 1-90) at the N-terminus, and a CASc domain (residues 146-415) at the C-terminus were predicted in the amino acid sequence of Hl-CASP9 (Figure 1A). Within its CASc domain, two active sites (His228 and Cys271), a substrate pocket (containing 12 conserved amino acids), a proteolytic cleavage site (containing 2 conserved amino acids) and a dimer interface (containing 15 conserved amino acids) were determined with the Blast program in NCBI (Figure S1B). By using ScanProsite prediction, the classical caspase subunits P20 (residues 153-275) and P10 (residues 326-415) within the CASc domain were identified. Moreover, the caspase family histidine active site motif (215HSNFDCVALAILTHG229) and the cysteine active site motif (262KPKIIILQACRG273) were also found in the P20 subunit (Figure S1A).
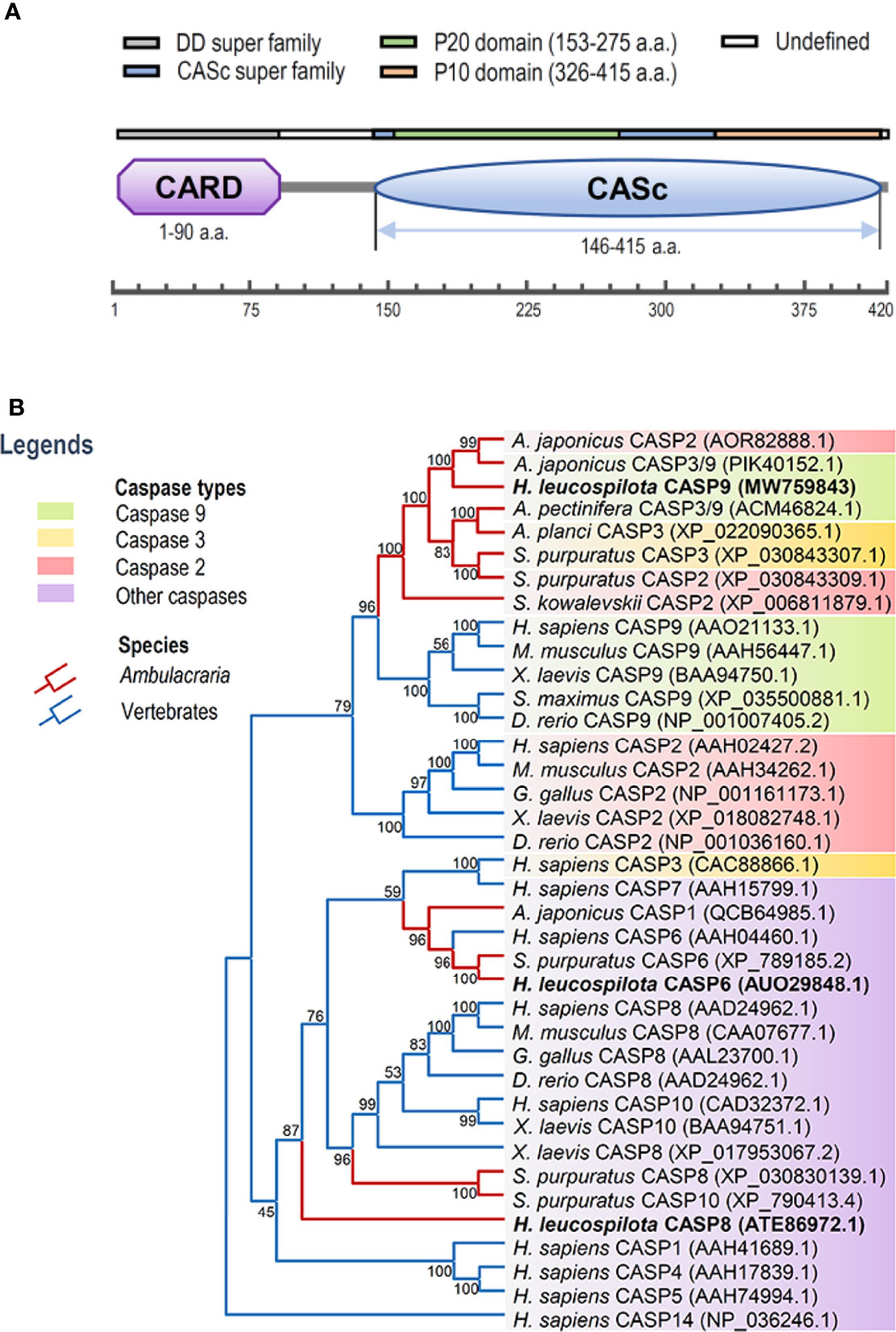
Figure 1 Functional domains of Hl-CASP9 and its evolutionary relationships with other caspases. (A) Structural domains of Hl-CASP9 predicted by using the SMART and ScanProsite programs. (B) Phylogenetic analysis of caspases among various species by using Neighbor-Joining method with bootstrap value of 1,000.
To clarify evolutionary relationships of caspases within deuterostomes, a phylogenetic tree was constructed. As shown in Figure 1B, caspase-9 and caspase-2 were clustered into one main branch, whereas other caspases were clustered into another main branch of the dendrogram. In the branch of caspase-2/9, Ambulacraria (represented by the phyla Echinodermata and Hemichordata) caspase-2/9 and vertebrate caspase-9 were clustered into one clade, whereas vertebrate caspase-2 was grouped into another clade. In addition, our newly identified Hl-CASP9 was highly similar in sequence to the sea cucumber A. japonicus caspase-2 and caspase-3/9, which have been named as Ajcaspase-2 and Ajcaspase-3/9 (19), respectively. Multiple alignments of amino acid sequences show that Hl-CASP9 proteins in different species share considerable consensus sequences among echinoderms and vertebrates. The catalytic CASc domain of caspase-9 and caspase-2 is remarkably conserved, especially in the P20 and P10 subunits. Unexpectedly, the Hl-CASP9 pentapeptide “QACRG” active sites were located within the P20 domain (Figure 2A), instead of the conserved pentapeptide “QACGG” found in the caspase-9 of vertebrates (37). In 3-D modeling, for caspase-9 CASc domains constructed with the amino acid sequences from human, zebrafish and sea cucumber H. leucospilota (Figure 2B), inter-species structural similarities in the P20 and P10 were confirmed. Likewise, the CARD domain and nestled active sites in the caspase-9 proteins are conserved (Figure 2C).
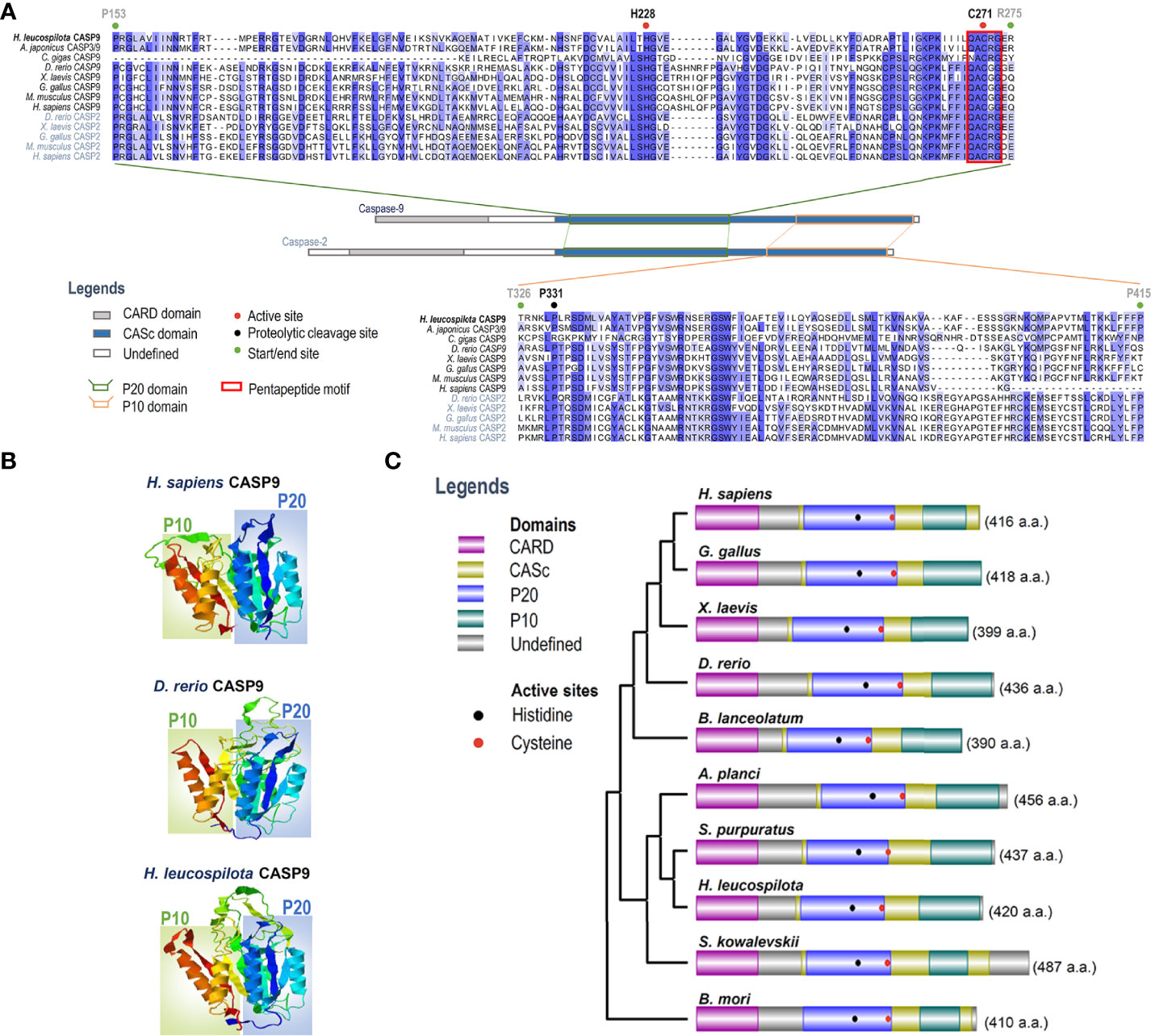
Figure 2 Analysis on amino acid sequence alignment, and organization of domains and 3-D structures of caspase-9. (A) Amino acid sequence alignment of caspase-2 and caspase-9 in various species. Conserved amino acid residues are boxed in dark blue, and similarities in amino acid residues are highlighted in light blue. (B) Comparison on 3-D structures of CASc domains in caspase-9 among three species. Positions of the P20 and P10 subunits are boxed. (C) Protein structural organization of caspase-9 among various species.
To characterize spatial and temporal expression patterns of Hl-CASP9, its transcript profiles in developing H. leucospilota embryos and larvae, as well as those in eleven selected adult tissues, were analyzed by qPCR. The representative developmental stages and anatomical architecture of the sea cucumber H. leucospilota are as shown in Figures 3A, B, respectively. Hl-CASP9 expression was found to be dynamic in all embryonic stages and fluctuated significantly, whereas Hl-CASP9 levels stayed relatively low in larval stages (Figure 3C). Specifically, Hl-CASP9 mRNA levels were high at the fertilized egg stage, but fell sharply at the early cleavage stage. As complexity of the embryos evolved, expression levels of Hl-CASP9 mRNA rose progressively, reaching a peak at the blastula stage, before subsiding again slowly in subsequent stages. Based on these observations, it could be speculated that Hl-CASP9 is implicated in some apoptosis- or caspase activity-dependent embryonic processes of H. leucospilota. As shown in Figure 3D, Hl-CASP9 transcripts were amply expressed in most adult tissues examined. Compared with the expression of rete mirabile, where the lowest expression levels were detected, the highest expression of Hl-CASP9 was found in the coelomocytes, followed by the body wall, muscle, oesophagus, Polian vesicles, intestine, gonads, respiratory tree, Cuvierian tubules and transverse vessel.
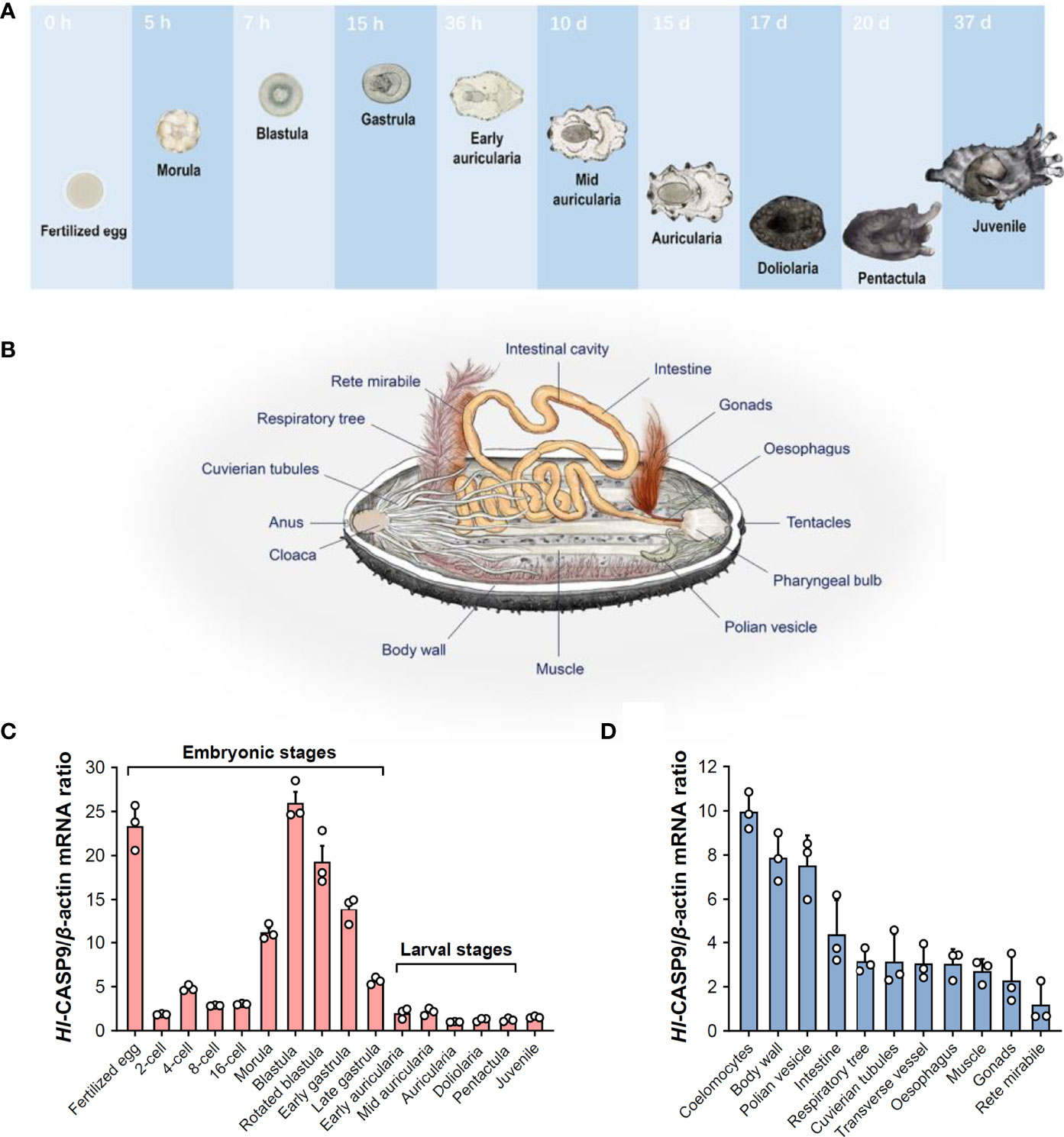
Figure 3 Spatial and temporal expression patterns of Hl-CASP9 mRNA in developing embryos and adult tissues. (A) Typical embryonic and larval development of H. leucospilota. Numbers indicate time lapsed post-fertilization. (B) Anatomy of H. leucospilota. (C) Hl-CASP9 mRNA expression profiles in developing embryos and larvae of H. leucospilota. (D) Hl-CASP9 mRNA expression profiles in different adult tissues of H. leucospilota.
Dynamic changes in Hl-CASP9 transcription in primary coelomocytes following challenges with immunostimulants LPS, poly(I:C) and heat-inactivated V. harveyi are as shown in Figures 4A-C. In this case, no significant changes were found in the expression of Hl-CASP9 mRNA from 0 to 24 h post-challenge. In contrast, Hl-CASP9 mRNA expression in primary coelomocytes were significantly upregulated under oxidative stressors including H2O2, NOC-18 and CdCl2 (Figures 4D-F). In response to H2O2 treatment, the expression of Hl-CASP9 was initially upregulated at 6 h and reach a peak of 2.87 folds (p < 0.001) relative to the untreated baseline at 24 h post-challenge. When treated with NOC-18, a well-established chemical donor of nitric oxide (•NO) (38), the expression of Hl-CASP9 was significantly elevated at 6 h and reached a peak of 2.84-fold (p < 0.001) increase relative to the basal levels at 24 h post-challenge. In the case of Cd, Hl-CASP9 expression was strongly stimulated: a 1.94-fold (p < 0.01) increase at 3 h following CdCl2 treatment, and a peak of 3.74-fold (p < 0.001) increase at 24 h. Taken together, these results suggest that Hl-CASP9 mRNA expression seems differentially regulated in response to oxidative stressors and immunostimulants.

Figure 4 Transcriptional expression patterns of Hl-CASP9 following challenges of immunostimulants or oxidative stress inducers. (A) Time-course study on the expression of Hl-CASP9 mRNA in coelomocytes treated with LPS (10 μg/mL). (B) Time-course study on the expression of Hl-CASP9 mRNA in coelomocytes treated with poly(I:C) (10 μg/mL). (C) Time-course study on the expression of Hl-CASP9 mRNA in coelomocytes treated with inactivated V. harveyi (107 cells/mL). (D) Time-course study on the expression of Hl-CASP9 mRNA in coelomocytes treated with H2O2 (2 μM). (E) Time-course study on the expression of Hl-CASP9 mRNA in coelomocytes treated with NOC-18 (1 μM). (F) Time-course study on the expression of Hl-CASP9 mRNA in coelomocytes treated with CdCl2 (20 μM).
ROS formation and its associated oxidative damage are recognized as hallmarks of heavy metals-induced cytotoxicity (12). To illustrate the relationship between ROS production and CdCl2 exposure in sea cucumber coelomocytes, intracellular ROS levels in coelomocytes were detected by means of the non-fluorescent probe DCFH-DA, which is switched on in a fluorescent state as DCF on reaction with ROS (39). As shown in Figure 5A, ROS formation in coelomocytes was dramatically elevated upon CdCl2. At 3 h post-Cd challenge, ROS levels were augmented by 4.73 folds (p < 0.001) relative to the untreated baseline (Figure 5B). Increase in ROS formation also showed a CdCl2 dose-dependent trend.
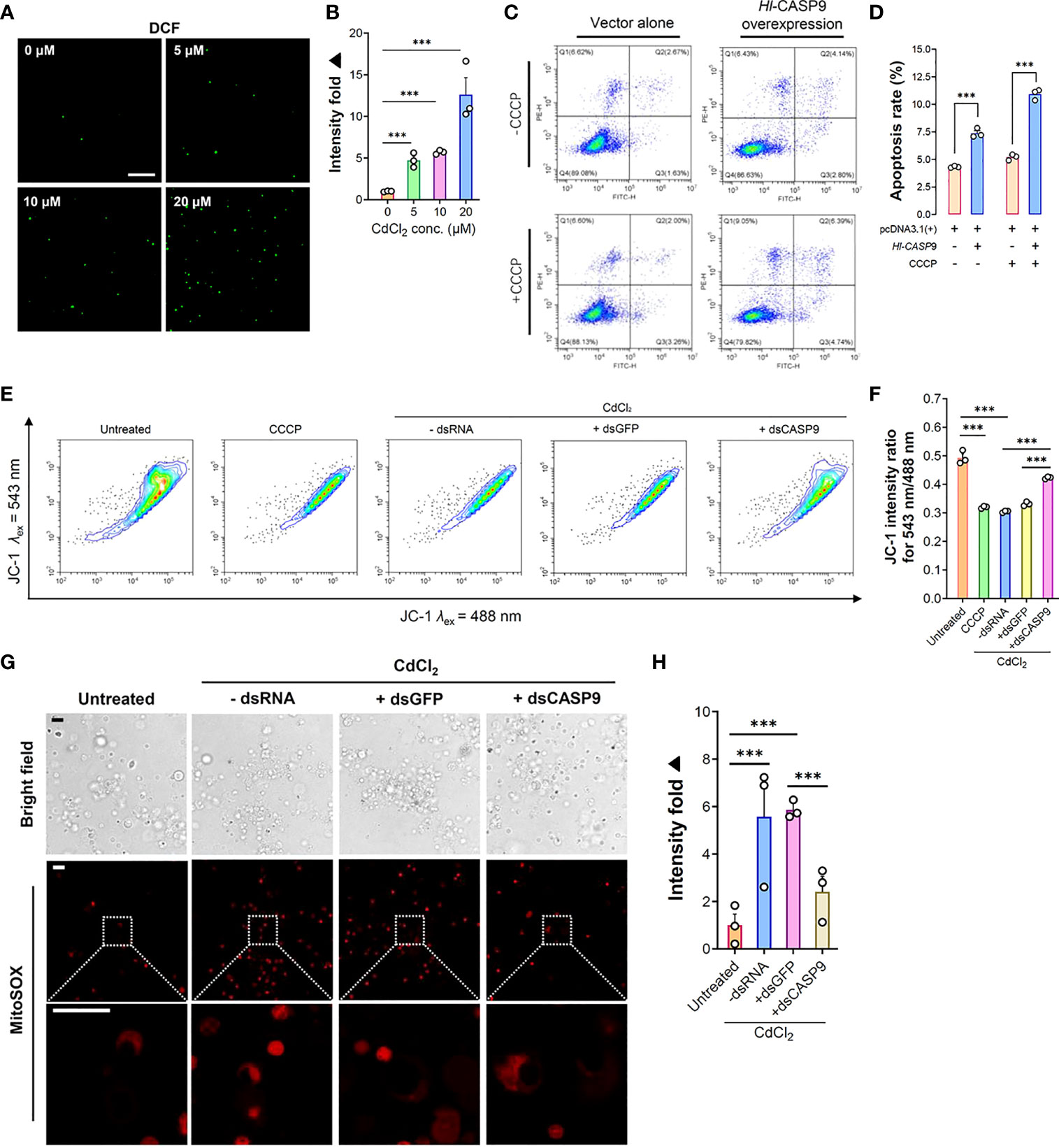
Figure 5 Formation of cellular ROS and associated mitochondrial stress in CdCl2-treated coelomocytes. (A) In vitro dose-dependent effects of CdCl2 (at 3 h) on ROS formation levels in primary coelomocytes as detected by the fluorescent DCF. Scale bar: 50 μm. (B) Quantification of DCF fluorescence intensities as reported in (A). (C) Effects of Hl-CASP9 overexpression on apoptosis in HEK293T cells, as analyzed by flow cytometry. HEK293T cells were treated with or without CCCP (100 μM). Comparisons are made between group with vector alone (HEK293T cells transfected with pcDNA3.1(+) blank plasmid) and group overexpressing Hl-CASP9 (HEK293T cells transfected with pcDNA3.1(+)/Hl-CASP9 recombinant plasmid). (D) Comparison on average apoptosis rates in HEK293T cells treated with or without CCCP in different groups. (E) Changes in mitochondrial membrane potential (Δψm) in CdCl2-treated primary coelomocytes with or without dsRNA (including dsGFP and dsCASP9) as assessed by JC-1 (2.5 μM) in flow cytometry. For comparison, coelomocytes were treated with or without CCCP (100 μM) or CdCl2 (20 μM). (F) Changes of Δψm in coelomocytes in response to CdCl2 were quantified by JC-1 intensity ratio for λex 543 nm/488 nm. (G) In vitro effects of CdCl2 (20 μM) on mitochondrial superoxide formation in primary coelomocytes with or without dsRNA (including dsGFP and dsCASP9), as visualized by MitoSOX (2.5 μM) in confocal imaging. Scale bar: 15 μm. (H) Quantification of MitoSOX fluorescence intensities in coelomocytes. Data were analyzed by using ImageJ.
In order to clarify the functional relevance of Hl-CASP9 in the mitochondria-dependent apoptosis pathway, pcDNA3.1(+)/Hl-CASP9 recombinant plasmid (rHl-CASP9) was transfected into HEK293T cells. CCCP was then used to test whether Hl-CASP9 could participate in apoptosis via the classical mitochondria-dependent pathway. As noted in Figure 5C, the number of cells from the Q3 quadrant (early apoptosis cells) and from the Q2 quadrant (later apoptosis cells) in flow cytometry was calculated. In the absence of CCCP, apoptosis rates were 4.32% and 7.29% in the untransfected control group (HEK293T cells transfected with pcDNA3.1(+) blank plasmid) and transfected control group (HEK293T cells transfected with rHl-CASP9), respectively (Figure 5C). The average apoptosis rates of HEK293T cells transfected with rHl-CASP9 significantly increased by 1.69 folds (p < 0.001) (Figure 5D). Upon treatment with CCCP, apoptosis rates in transfected treatment group became 10.92% while that of the untransfected treatment group were only 5.20% (Figure 5C). Evidently, apoptosis rate of the transfected treatment group was 2.10-fold (p < 0.001) higher than that of the untransfected treatment group (Figure 5D), thus implicating overexpressed Hl-CASP9 as a mediator of mitochondria-dependent apoptosis in HEK293T cells.
In general, Cd-induced oxidative stress often leads to mitochondrial dysfunction (40). We thus set out to assess any changes in mitochondrial membrane potential and mitochondrial superoxide formation in coelomocytes under Cd challenges. As anticipated, CdCl2 (20 μM; 3 h) rapidly induced a loss in mitochondrial membrane, as gauged by the membrane-potential sensitive dye JC-1 (2.5 μM) (Figures 5E, F). Similarly, mitochondrial superoxide formation was robustly induced by Cd (20 μM; 3 h) in coelomocytes, as measured by the mitochondrial superoxide probe MitoSOX (2.5 μM) (Figures 5G, H). To verify specific effects of Hl-CASP9 on CdCl2-treated coelomocytes, experiments of Hl-CASP9 knock-down by RNAi was carried out. After injection of dsCASP9 for 48 h, the expression of the Hl-CASP9 mRNA in sea cucumber coelomocytes dropped to 35% in the control group (with no dsRNA) (Figure S2). With RNAi by dsCASP9 (48 h), mitochondrial membrane potential was restored in CdCl2-treated coelomocytes (Figures 5E, F), while formation of mitochondrial superoxide was also lowered (Figures 5G, H).
Flow cytometry results for coelomocytes apoptosis show that exogenously applied oxidative stress inducers such as H2O2 (2 μM), NOC-18 (1 μM) and CdCl2 (20 μM) could efficiently induce apoptosis in sea cucumber coelomocytes. Early apoptosis rates of coelomocytes under the challenges of H2O2, NOC-18 and CdCl2 in the control group (with no dsRNA) were 32.71%, 33.97% and 47.26%, respectively, whereas corresponding rates in the untreated group was 24.27% (Figure 6A). With RNAi by dsCASP9 (48 h), average early apoptosis rates of coelomocytes under these oxidative stress inducers showed a 0.79-fold (p < 0.001) (Figure 6B), 0.88-fold (p < 0.05) (Figure 6C) and 0.79-fold (p < 0.001) (Figure 6D) reduction relative to the negative control group (with dsGFP), respectively. Collectively, these results indicate that coelomocyte apoptosis induced by oxidative stress inducers can be rescued by Hl-CASP9 knock-down, thus confirming Hl-CASP9 as a direct mediator of such environmental stress cues.
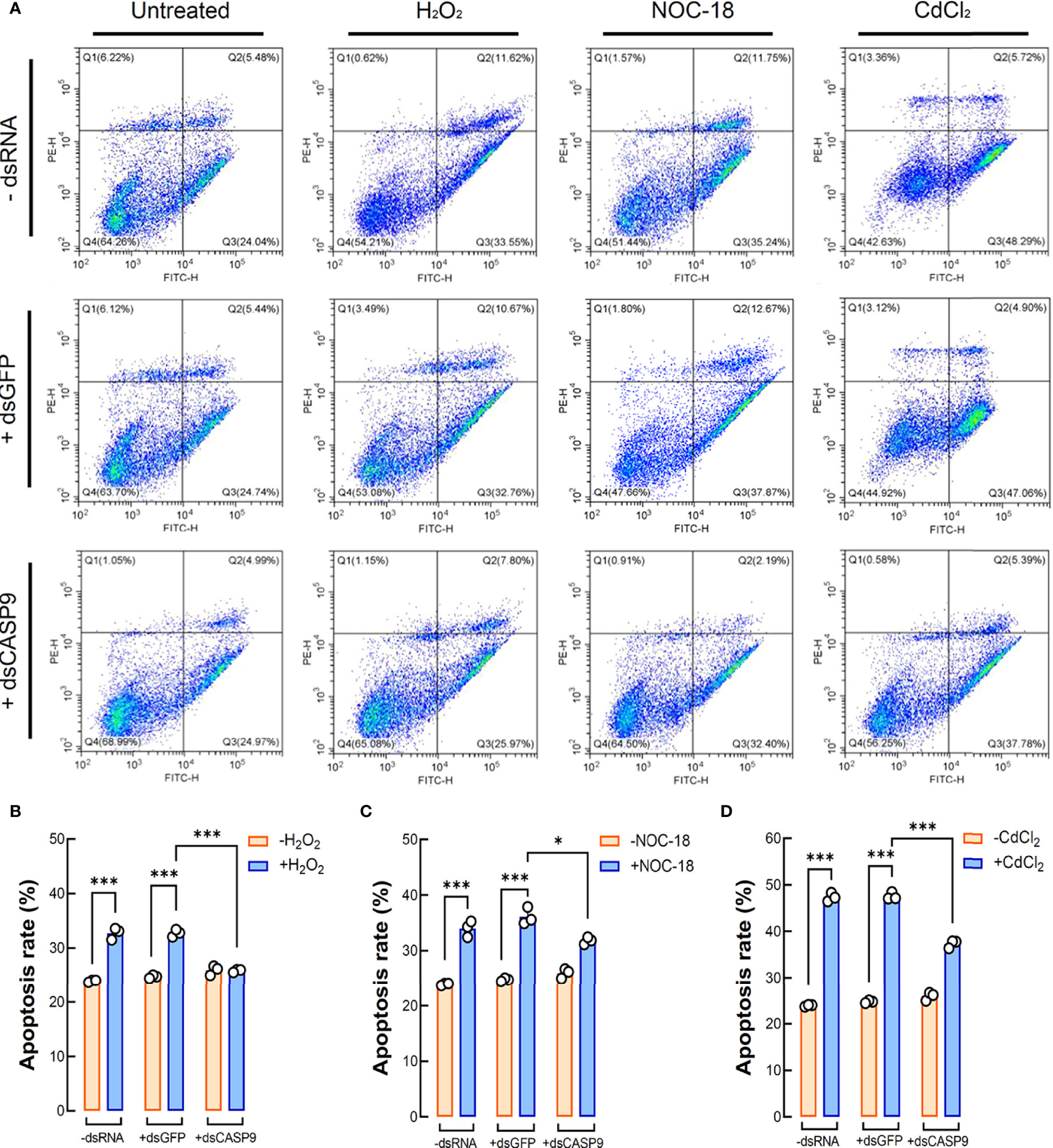
Figure 6 Coelomocyte apoptosis analysis by flow cytometry following Hl-CASP9 knock-down in vivo. (A) Coelomocytes treated with the indicated stimulants including H2O2 (2 μM), NOC-18 (1 μM) or CdCl2 (20 μM). Effects are compared among the “-dsRNA” group (sea cucumber injected of RNase-free saline solution), “+dsGFP” group (sea cucumber injected of dsGFP) and “+dsCASP9” group (sea cucumber injected of dsCASP9). (B) Statistical analysis on mean rates of early apoptosis in coelomocytes exposed to H2O2 (2 μM) for 24 h in different groups. (C) Statistical analysis on mean rates of early apoptosis in coelomocytes exposed to NOC-18 (1 μM) for 24 h in different groups. (D) Statistical analysis on mean rates of early apoptosis in coelomocytes exposed to CdCl2 (20 μM) for 24 h in different groups.
Apoptosis is an intricate process that crucially regulates tissue development, cellular homeostasis and immune defense of the host (26). For their diverse functions in cellular physiology, caspases have been exhaustively investigated in vertebrates, particularly in mammals (14). Several genetic homologues of these mammalian apoptosis effectors have been identified in bivalves, which are evolutionarily ancient invertebrates, hinting that a mitochondria-dependent apoptosis pathway may as well exist in marine invertebrates (41). In mammals, caspase-9 consists of three domains: an N-terminal pro-domain, i.e., caspase recruitment domain (CARD), a large catalytic subunit (P20; 20 kDa) and a small catalytic subunit (P10; 10 kDa) (42). Both P20 and P10 subunits are located in the CASc domain, as observed in zebrafish caspase-9 (43). Upon initiation of apoptosis, the P20 and P10 subunits are released and separated from the N-terminal region of pro-caspases to form an active heterotetramer (P20-P10)2 as an active caspase, which eventually commits irreversibly to apoptosis (44).
To date, the intrinsic mitochondrial apoptosis pathway involved in echinoderm remains scarcely investigated. This present work is the first to identify an echinoderm caspase-9. The deduced amino acid sequence of Hl-CASP9 contains a conserved caspase-9 architecture encompassing the CARD domain and CASc domain. Hl-CASP9 possesses two active sites rich in histidine and cysteine residues, which is consistent with previous reports that the active site is a catalytic dyad composed of a cysteine sulfohydryl group and a histidine imidazole ring (45). In most caspases, a typical pentapeptide active-site motif “QACRG” can be found, especially in executioner caspases or inflammatory caspases (46). In comparison, in initiator caspases, caspase-8 contains a novel sequence “QACQG” (43), while the pentapeptide “QACGG” is conserved for caspase-9 in vertebrates (37). Intriguingly, Hl-CASP9 contains an active site “QACRG” located in its P20 subunit. Despite low sequence similarity, evolutionary conserveness of protein structures is observed in caspase-9 and caspase-2 across multiple species from Ambulacraria to higher vertebrates. In multiple sequence alignments of caspase-9 and caspase-2 from human, zebrafish and sea cucumber, a degree of high similarity was revealed in their catalytic domains. Hl-CASP9 is sequence-wise most akin to the sea cucumber A. japonicus caspase-2 and caspase-3/9, suggesting that these caspases are phylogenetically related. As Hl-CASP9 and the vertebrate caspase-9 are grouped into one clade within the evolutionary tree, we proposed that Hl-CASP9 is a new member of the caspase-9 family.
It is noteworthy that only a restricted number of caspases (e.g., caspase-9) are amply expressed in the developing nervous system during embryonic development in zebrafish (43). The mitochondria-dependent apoptosis pathway can in turn be activated by multiple mechanisms during diverse processes of embryonic development and cell differentiation (47). In our study, Hl-CASP9 mRNA showed a dynamic expression pattern at various stages of embryonic and larval development in H. leucospilota, implying that Hl-CASP9 may play some essential roles in apoptosis-regulated embryonic development. Transcript expression of Hl-CASP9 was detected in all adult tissues examined, with the highest levels being observed in coelomocytes, which are deemed the primary effector cells in the immune system of echinoderms capable of phagocytosis (7, 8). Additionally, Hl-CASP9 was quite abundantly found in the body wall, which represents a first-line defense against invading microbes such as bacteria. Intuitively, the high mRNA expression levels of Hl-CASP9 observed in coelomocytes and body wall led us to speculate that Hl-CASP9 may be an immunity-related gene.
In innate immunity, Toll-like receptors (TLRs) respond to a variety of pathogen-associated molecular patterns (PAMP) and damage-associated molecular patterns (DAMP) to initiate or sustain a coordinated immune response during infection or inflammation. In our previous studies, transcriptional responses of HLcaspase-6 and HLcaspase-8 were positively stimulated by LPS or poly(I:C) in H. leucospilota coelomocytes (21, 22). To elucidate if a similar response occurs for Hl-CASP9 under immune challenges, we tested poly(I:C), LPS and heat-inactivated V. harveyi as immune stimulants. Surprisingly, Hl-CASP9 transcript expression was virtually unaltered after these treatments, suggesting that Hl-CASP9 may not be directly responsible for host responses to infiltrating pathogens. In vertebrates, caspase-9 has been recognized as a core initiator caspase in response to oxidative stress (48). Consequently, we picked H2O2, NOC-18 and CdCl2 as exogenous oxidant donors to test whether Hl-CASP9 mRNA responds to oxidative stress conditions. H2O2 is a long-lived major ROS that, in the range of 200-500 μM, can induce apoptosis mediated by the mitochondria-dependent pathway (49). In this study, expression of Hl-CASP9 in coelomocytes under H2O2 treatment was dramatically upregulated, in particular at 24 h post-challenge. This result is in agreement with previous studies reporting upregulation of caspase-9 mRNA by H2O2-induced oxidative stress in BmN-SWU1 cells from the ovary of silkworm and in Schwann cells from the peripheral nervous system of rat (38, 50). In addition to ROS, Hl-CASP9 transcript expression was also significantly upregulated under treatment with NOC-18, a well-documented •NO donor. When H. leucospilota coelomocytes were challenged with CdCl2, expression of Hl-CASP9 mRNA in coelomocytes was strongly elevated, in dose- and time-dependent manners. Previously, enhancement of caspase-9 transcript expression upon Cd challenge was reported in the rat glioma C6 cells (51). In brief, our results show that H. leucospilota coelomocytes are sensitive to oxidative stress instigated by H2O2, NO and Cd, as reflected by Hl-CASP9 transcriptional changes.
As CCCP is a protonophore capable of uncoupling mitochondrial oxidative phosphorylation leading to a loss of membrane potential on either side of the mitochondrial inner membrane (52), we used it to verify whether Hl-CASP9 participates in Cd-induced apoptosis via the mitochondria-dependent pathway. In HEK293T cells overexpressing Hl-CASP9, the results corroborate the idea that Hl-CASP9 is a mediator in mitochondria-dependent apoptosis, as in the case of intrinsic apoptosis in mammals (53). As a further test, apoptosis rates of sea cucumber coelomocytes were quantified by flow cytometry following pharmacological treatments with immunostimulants or oxidative stress inducers including Cd. In our previous studies, apoptosis of sea cucumber coelomocytes was significantly upregulated by challenges with LPS or poly(I:C) (22). Here, we showed that H2O2, NOC-18 and CdCl2 likewise potently induced apoptosis in sea cucumber coelomocytes, suggesting that coelomocytes are intrinsically sensitive to oxidative stress. Our findings support the proposition that oxidative stress can directly activate caspase-9, which has been described in vertebrates (54) and in invertebrates such as freshwater crab Sinopotamon yangtsekiense (55) and sea cucumber S. japonicus (31). Lastly, results for RNAi experiments show that Hl-CASP9 knock-down resulted in a significant reduction in coelomocyte apoptosis induced by exogenous oxidative stressors inducing Cd, thus implicating Hl-CASP9 as a direct and crucial mediator of coelomocyte apoptosis in Cd toxicity (See a scheme on concepts in Figure 7).
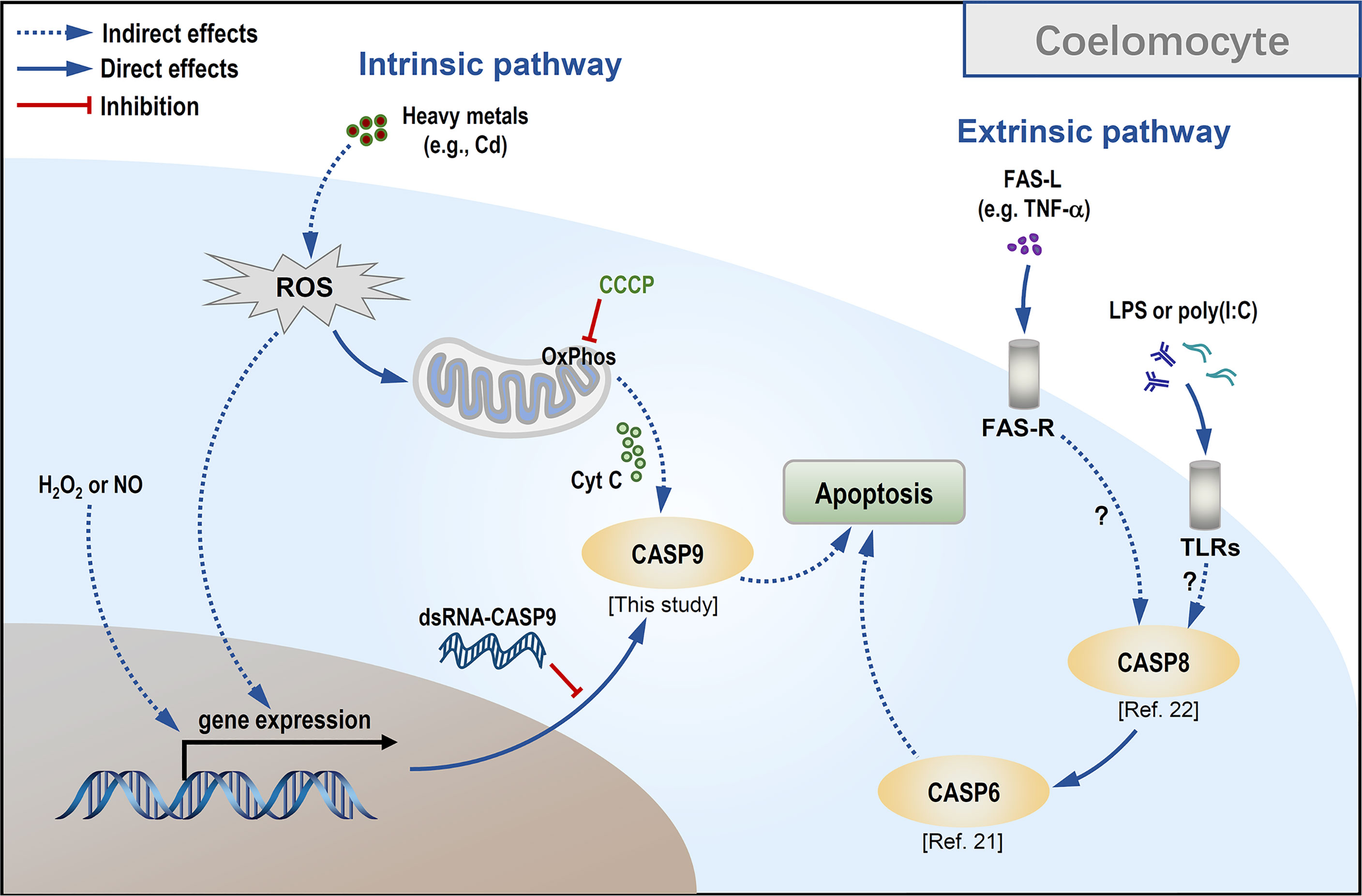
Figure 7 Conceptualization of Cd-induced mitochondria-dependent apoptosis in sea cucumber coelomocytes.
In recent studies, UV irradiation (31), Cd (56) and ZnO nanoparticles (57) were reported to drive oxidative stress by forcing cells to produce excessive oxidants such as H2O2, •NO, hydroxyl radical (•OH), peroxynitrite (ONOO-) and hypochlorite (OCl-). Unrestrained ROS generation in turn may directly or indirectly alter signal transduction, modulating the outcomes of apoptosis via the mitochondria-dependent pathway (58). In the present study, we produced evidence that Hl-CASP9 upregulation is sufficient to tone up mitochondrial stress in Cd toxicity, by depressing mitochondrial membrane potential and raising mitochondrial superoxide formation.
In summary, an echinoderm caspase-9 was cloned for the first time from the tropical sea cucumber H. leucospilota for functional characterization. Our findings give fresh insights into how this ancient caspase-9 mediates coelomocytic apoptosis in response to Cd toxicity via a mitochondria-dependent pathway. Evidently, Cd as an important heavy metal in marine pollution can profoundly impact the health and survival of benthic species. There is an urgent need to establish a mechanistic understanding of the organismal and cellular adaptive processes such as autophagy and apoptosis in toxic metal pollution (12). Echinoderms are ancient but useful models to systematically monitor and unveil the impact of environmental perturbations on marine organisms (9). Importantly, bioturbation as a fundamental function of holothurians (i.e., sea cucumbers) within marine benthic systems is intimately linked to the health of coral reefs (59). Our study here has fully demonstrated the utility of H. leucospilota as a robust model for the toxicology of heavy metal such as Cd. Further investigation on how Cd impacts processes such as embryogenesis, tissue development, and reproduction in relation to cell death regulation is warranted.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethical review and approval were not required for the animal study because all animal experiments were conducted in accordance with the guidelines of the South China Sea Institute of Oceanology, Chinese Academy of Sciences.
Conception: CR, TC, N-KW. Methodology and investigation: X-ML, XW, ZL, XZ. Resources: XJ, PL, CH, TC, CR. Writing-original draft: X-ML, N-KW. Writing-review and editing: N-KW, CR, X-ML, TC. Supervision and project administration: TC, CR. Funding acquisition: XJ, PL, CH. All authors read and provided inputs on the manuscript
This work was graciously supported by grants from the National Key R & D Program of China [2018YFD0901605, 2020YFD0901104], the Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) [GML2019ZD0402], the National Natural Science Foundation of China [42176132, 41906101, 32073002], Li Ka Shing Foundation [510858044 to N.W.] and the Key Deployment Project of Centre for Ocean Mega-Research of Science, Chinese Academy of Sciences [COMS2020Q03].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We are grateful to Dr. Zonghe Yu for providing the sea cucumber samples used in this study. We also thank Mr. Jinshang Liu for cultivation of the sea cucumbers.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.927880/full#supplementary-material
1. Collard M, Eeckhaut I, Dehairs F, Dubois P. Acid-Base Physiology Response to Ocean Acidification of Two Ecologically and Economically Important Holothuroids From Contrasting Habitats, Holothuria Scabra and Holothuria Parva. Environ Sci Pollut Res Int (2014) 21:13602–14. doi: 10.1007/s11356-014-3259-z
2. Bonham K, Held EE. Ecological Observations on the Sea Cucumbers Holothuria Atra and H. Leucospilota at Rongelap Atoll, Marshall Islands. Pac Sci (1963) 17:305–14. doi: 10.2307/1439679
4. Dzeroski S, Drumm D. Using Regression Trees to Identify the Habitat Preference of the Sea Cucumber (Holothuria Leucospilota) on Rarotonga, Cook Islands. Ecol Model (2003) 170:219–26. doi: 10.1016/s0304-3800(03)00229-1
5. Schneider K, Silverman J, Woolsey E, Eriksson H, Byrne M. Potential Influence of Sea Cucumbers on Coral Reef CaCO3 Budget: A Case Study at One Tree Reef. J Geophys Res-Biogeosci (2011) 116:G0403. doi: 10.1029/2011jg001755
6. Xue Z, Li H, Wang X, Li X, Liu Y, Sun J, et al. A Review of the Immune Molecules in the Sea Cucumber. Fish Shellfish Immunol (2015) 44:1–11. doi: 10.1016/j.fsi.2015.01.026
7. Dolmatova LS, Eliseikina MG, Romashina VV. Antioxidant Enzymatic Activity of Coelomocytes of the Far East Sea Cucumber Eupentacta Fraudatrix. J Evol Biochem Physiol (2004) 40:126–35. doi: 10.1023/b:joey.0000033803.35634.46
8. Gu M, Ma H, Mai K, Zhang W, Ai Q, Wang X, et al. Immune Response of Sea Cucumber Apostichopus Japonicus Coelomocytes to Several Immunostimulants In Vitro. Aquaculture (2010) 306:49–56. doi: 10.1016/j.aquaculture.2010.05.024
9. Chiarelli R, Martino C, Roccheri MC. Cadmium Stress Effects Indicating Marine Pollution in Different Species of Sea Urchin Employed as Environmental Bioindicators. Cell Stress Chaperones (2019) 24:675–87. doi: 10.1007/s12192-019-01010-1
10. Méndez S, Ruepert C, Mena F, Cortés J. Accumulation of Heavy Metals (Cd, Cr, Cu, Mn, Pb, Ni, Zn) in Sediments, Macroalgae (Cryptonemia Crenulata) and Sponge (Cinachyrella Kuekenthali) of a Coral Reef in Moín, Limón, Costa Rica: An Ecotoxicological Approach. Mar Pollut Bull (2021) 173:113159. doi: 10.1016/j.marpolbul.2021.113159
11. Mitchelmore CL, Verde EA, Weis VM. Uptake and Partitioning of Copper and Cadmium in the Coral Pocillopora Damicornis. Aquat Toxicol (2007) 85:48–56. doi: 10.1016/j.aquatox.2007.07.-015
12. Okereafor U, Makhatha M, Mekuto L, Uche-Okereafor N, Sebola T, Mavumengwana V. Toxic Metal Implications on Agricultural Soils, Plants, Animals, Aquatic Life and Human Health. Int J Environ Res Public Health (2020) 17:2204. doi: 10.3390/ijerph17072204
13. Wang W-X, Meng J, Weng N. Trace Metals in Oysters: Molecular and Cellular Mechanisms and Ecotoxicological Impacts. Environ Sci Process Impacts (2018) 20:892–912. doi: 10.1039/c8em00069g
14. Lamkanfi M, Declercq W, Kalai M, Saelens X, Vandenabeele P. Alice in Caspase Land. A Phylogenetic Analysis of Caspases From Worm to Man. Cell Death Differ (2002) 9:358–61. doi: 10.1038/sj.cdd.4400989
15. Rogers C, Erkes DA, Nardone A, Aplin AE, Fernandes-Alnemri T, Alnemri ES. Gasdermin Pores Permeabilize Mitochondria to Augment Caspase-3 Activation During Apoptosis and Inflammasome Activation. Nat Commun (2019) 10:1689. doi: 10.1038/s41467-019-09397-2
16. Brentnall M, Rodriguez-Menocal L, De Guevara RL, Cepero E, Boise LH. Caspase-9, Caspase-3 and Caspase-7 Have Distinct Roles During Intrinsic Apoptosis. BMC Cell Biol (2013) 14:32. doi: 10.1186/1471-2121-14-32
17. Murray TVA, McMahon JM, Howley BA, Stanley A, Ritter T, Mohr A, et al. A non-Apoptotic Role for Caspase-9 in Muscle Differentiation. J Cell Sci (2008) 121:3786–93. doi: 10.1242/jcs.024547
18. Ruocco N, Varrella S, Romano G, Ianora A, Bentley MG, Somma D, et al. Diatom-Derived Oxylipins Induce Cell Death in Sea Urchin Embryos Activating Caspase-8 and Caspase 3/7. Aquat Toxicol (2016) 176:128–40. doi: 10.1016/j.aquatox.2016.04.012
19. Shao Y, Li C, Zhang W, Duan X, Li Y, Jin C, et al. Molecular Cloning and Characterization of Four Caspases Members in Apostichopus Japonicus. Fish Shellfish Immunol (2016) 55:203–11. doi: 10.1016/j.fsi.2016.05.039
20. Tamura R, Takada M, Sakaue M, Yoshida A, Ohi S, Hirano K, et al. Starfish Apaf-1 Activates Effector Caspase-3/9 Upon Apoptosis of Aged Eggs. Sci Rep (2018) 8:1611. doi: 10.1038/s41598-018-19845-6
21. Yan A, Ren C, Chen T, Huo D, Jiang X, Sun H, et al. A Novel Caspase-6 From Sea Cucumber Holothuria Leucospilota: Molecular Characterization, Expression Analysis and Apoptosis Detection. Fish Shellfish Immunol (2018) 80:232–40. doi: 10.1016/j.fsi.2018.06.006
22. Yan A, Ren C, Chen T, Jiang X, Sun H, Huo D, et al. The First Tropical Sea Cucumber Caspase-8 From Holothuria Leucospilota: Molecular Characterization, Involvement of Apoptosis and Inducible Expression by Immune Challenge. Fish Shellfish Immunol (2018) 72:124–31. doi: 10.1016/j.fsi.2017.10.050
23. Shao Y, Che Z, Li C, Zhang W, Guo M. A Novel Caspase-1 Mediates Inflammatory Responses and Pyroptosis in Sea Cucumber Apostichopus Japonicus. Aquaculture (2019) 513:734399–. doi: 10.1016/j.aquaculture.2019.734399
24. Lu G, Yu Z, Lu M, Liu D, Wang F, Wu Y, et al. The Self-Activation and LPS Binding Activity of Executioner Caspase-1 in Oyster Crassostrea Gigas. Dev Comp Immunol (2017) 77:330–9. doi: 10.1016/j.dci.2017.09.002
25. Yang G, Wang J, Luo T, Zhang X. White Spot Syndrome Virus Infection Activates Caspase 1-Mediated Cell Death in Crustacean. Virology (2019) 528:37–47. doi: 10.1016/j.virol.2018.12.004
26. Wang Z, Li C, Xing R, Shao Y, Zhao X, Zhang W, et al. β-Integrin Mediates LPS-Induced Coelomocyte Apoptosis in Sea Cucumber Apostichopus Japonicus via the Integrin/Fak/caspase-3 Signaling Pathway. Dev Comp Immunol (2019) 91:26–36. doi: 10.1016/j.dci.2018.10.004
27. Qin Y, Zhang Y, Li X, Noor Z, Li J, Zhou Z, et al. Characterization and Functional Analysis of a Caspase 3 Gene: Evidence That ChCas 3 Participates in the Regulation of Apoptosis in Crassostrea Hongkongensis. Fish Shellfish Immunol (2020) 98:122–9. doi: 10.1016/j.fsi.2020.-01.007
28. Wang X, Li C, Jia Z, Xu T, Wang Y, Sun M, et al. Regulation of Apoptosis by Pacific Oyster Crassostrea Gigas Reveals Acclimation Strategy to CO2 Driven Acidification. Ecotoxicol Environ Saf (2021) 217:112235. doi: 10.1016/j.ecoenv.2021.112235
29. Havanapan P-O, Taengchaiyaphum S, Paemanee A, Phungthanom N, Roytrakul S, Sritunyalucksana K, et al. Caspase-3, a Shrimp Phosphorylated Hemocytic Protein is Necessary to Control YHV Infection. Fish Shellfish Immunol (2021) 114:36–48. doi: 10.1016/j.fsi.-2021.04.007
30. Chiarelli R, Agnello M, Bosco L, Roccheri MC. Sea Urchin Embryos Exposed to Cadmium as an Experimental Model for Studying the Relationship Between Autophagy and Apoptosis. Mar Environ Res (2014) 93:47–55. doi: 10.1016/j.marenvres.2013.06.001
31. Su L, Yang J, Fu X, Dong L, Zhou D, Sun L, et al. Ultraviolet Rays Induced Sea Cucumber (Stichopus Japonicus) Melting is Mediated Through the Caspase-Dependent Mitochondrial Apoptotic Pathway. J Agric Food Chem (2018) 66:45–52. doi: 10.1021/acs.jafc.7b03888
32. Li J, Dong L, Zhu D, Zhang M, Wang K, Chen F. An Effector Caspase Sp-Caspase First Identified in Mud Crab Scylla Paramamosain Exhibiting Immune Response and Cell Apoptosis. Fish Shellfish Immunol (2020) 103:442–53. doi: 10.1016/j.fsi.2020.05.045
33. Wang F, Yu Z, Wang W, Li Y, Lu G, Qu C, et al. A Novel Caspase-Associated Recruitment Domain (CARD) Containing Protein (CgCARDCP-1) Involved in LPS Recognition and NF-κb Activation in Oyster (Crassostrea Gigas). Fish Shellfish Immunol (2018) 79:120–9. doi: 10.1016/j.fsi.2018.05.018
34. Sakamaki K, Satou Y. Caspases: Evolutionary Aspects of Their Functions in Vertebrates. J Fish Biol (2009) 74:727–53. doi: 10.1111/j.1095-8649.2009.02184.x
35. Huang W, Huo D, Yu Z, Ren C, Jiang X, Luo P, et al. Spawning, Larval Development and Juvenile Growth of the Tropical Sea Cucumber Holothuria Leucospilota. Aquaculture (2018) 488:22–9. doi: 10.1016/j.aquaculture.2018.01.013
36. Wu X, Chen T, Huo D, Yu Z, Ruan Y, Cheng C, et al. Transcriptomic Analysis of Sea Cucumber (Holothuria Leucospilota) Coelomocytes Revealed the Echinoderm Cytokine Response During Immune Challenge. BMC Genomics (2020) 21:306. doi: 10.1186/s12864-020-6698-6
37. Li D, Ling S, Wu K, Luo Z. Identification of Five Key Genes Involved in Intrinsic Apoptotic Pathway From Yellow Catfish Pelteobagrus Fulvidraco and Their Transcriptional Responses to High Fat Diet (HFD). Front Physiol (2019) 10:921. doi: 10.3389/fphys.2019.00921
38. Chen P, Hu Y, Wang L, Xiao W, Bao X, Pan C, et al. Mitochondrial Apoptotic Pathway is Activated by H2O2-Mediated Oxidative Stress in BmN-SWU1 Cells From Bombyx Mori Ovary. PloS One (2015) 10:e0134694. doi: 10.1371/journal.pone.0134694
39. Eruslanov E, Kusmartsev S. Identification of ROS Using Oxidized DCFDA and Flow-Cytometry. Methods Mol Biol (2010) 594:57–72. doi: 10.1007/978-1-60761-411-1_4
40. Pan Y, Luo Z, Zhuo M, Wei C, Chen G, Song Y. Oxidative Stress and Mitochondrial Dysfunction Mediated Cd-Induced Hepatic Lipid Accumulation in Zebrafish Danio Rerio. Aquat Toxicol (2018) 199:12–20. doi: 10.1016/j.aquatox.2018.03.017
41. Li Y, Zhang L, Qu T, Tang X, Li L, Zhang G. Conservation and Divergence of Mitochondrial Apoptosis Pathway in the Pacific Oyster, Crassostrea Gigas. Cell Death Dis (2017) 8:e2915. doi: 10.1038/cddis.2017.307
43. Spead O, Verreet T, Donelson CJ, Poulain FE. Characterization of the Caspase Family in Zebrafish. PloS One (2018) 13:e0197966. doi: 10.1371/journal.pone.0197966
44. Zamaraev AV, Kopeina GS, Prokhorova EA, Zhivotovsky B, Lavrik IN. Post-Translational Modification of Caspases: The Other Side of Apoptosis Regulation. Trends Cell Biol (2017) 27:322–39. doi: 10.1016/j.tcb.2017.01.003
45. Würstle ML, Laussmann MA, Rehm M. The Central Role of Initiator Caspase-9 in Apoptosis Signal Transduction and the Regulation of its Activation and Activity on the Apoptosome. Exp Cell Res (2012) 318:1213–20. doi: 10.1016/j.yexcr.2012.02.013
46. Cohen GM. Caspases: The Executioners of Apoptosis. Biochem J (1997) 326:1–16. doi: 10.1042/bj3260001
47. Xu W, Guo G, Li J, Ding Z, Sheng J, Li J, et al. Activation of Bcl-2-Caspase-9 Apoptosis Pathway in the Testis of Asthmatic Mice. PloS One (2016) 11:e0149353. doi: 10.1371/journal.pone.-0149353
48. Kroemer G, Galluzzi L, Brenner C. Mitochondrial Membrane Permeabilization in Cell Death. Physiol Rev (2007) 87(1):99–163. doi: 10.1152/physrev.00013.2006
49. Lv R, Du L, Lu C, Wu J, Ding M, Wang C, et al. Allicin Protects Against H2O2-Induced Apoptosis of PC12 Cells via the Mitochondrial Pathway. Exp Ther Med (2017) 14:2053–9. doi: 10.3892/etm.2017.4725
50. Luo X, Chen B, Zheng R, Lin P, Li J, Chen H. Hydrogen Peroxide Induces Apoptosis Through the Mitochondrial Pathway in Rat Schwann Cells. Neurosci Lett (2010) 485:60–4. doi: 10.1016/-j.neulet.2010.08.063
51. Wtjen W, Cox M, Biagioli M, Beyersmann D. Cadmium-Induced Apoptosis in C6 Glioma Cells: Mediation by Caspase 9-Activation. Biometals (2002) 15:15–25. doi: 10.1023/a:1013141926228
52. Wang C, Cheng G, Yang S, Li L, Zhang Y, Zhao X, et al. Protective Effects of Platycodon Grandiflorus Polysaccharides Against Apoptosis Induced by Carbonyl Cyanide 3-Chlorophenylhydrazone in 3D4/21 Cells. Int J Biol Macromol (2019) 141:1220–7. doi: 10.1016/j.ijbiomac.2019.09.086
53. Yuan Y, Zhang Y, Zhao S, Chen J, Yang J, Wang T, et al. Cadmium-Induced Apoptosis in Neuronal Cells is Mediated by Fas/FasL-Mediated Mitochondrial Apoptotic Signaling Pathway. Sci Rep (2018) 8:1–11. doi: 10.1038/s41598-018-27106-9
54. Kannan K, Jain SK. Oxidative Stress and Apoptosis. Pathophysiology (2000) 7:153–63. doi: 10.1016/S0928-4680(00)00053-5
55. Liu D, Yan B, Yang J, Lei W, Wang L. Mitochondrial Pathway of Apoptosis in the Hepatopancreas of the Freshwater Crab Sinopotamon Yangtsekiense Exposed to Cadmium. Aquat Toxicol (2011) 105:394–402. doi: 10.1016/j.aquatox.2011.07.013
56. Wang J, Wang Q, Li J, Shen Q, Wang F, Wang L. Cadmium Induces Hydrogen Peroxide Production and Initiates Hydrogen Peroxide-Dependent Apoptosis in the Gill of Freshwater Crab, Sinopotamon Henanense. Comp Biochem Physiol C Toxicol Pharmacol (2012) 156:195–201. doi: 10.1016/j.cbpc.2012.05.006
57. Zhao X, Xin R, Rong Z, Luo Z, Ren B. Zinc Oxide Nanoparticles Induce Oxidative DNA Damage and ROS-Triggered Mitochondria-Mediated Apoptosis in Zebrafish Embryos. Aquat Toxicol (2016) 180:56–70. doi: 10.1016/j.aquatox.2016.09.013
58. Redza-Dutordoir M, Averill-Bates DA. Activation of Apoptosis Signalling Pathways by Reactive Oxygen Species. Biochim Biophys Acta (2016) 1863:2977–92. doi: 10.1016/j.bbamcr.2016.-09.012
Keywords: caspase-9, cadmium, reactive oxygen/nitrogen species (ROS/RNS), Holothuria leucospilota, invertebrate cell death
Citation: Li X, Chen T, Wu X, Li Z, Zhang X, Jiang X, Luo P, Hu C, Wong N-K and Ren C (2022) Evolutionarily Ancient Caspase-9 Sensitizes Immune Effector Coelomocytes to Cadmium-Induced Cell Death in the Sea Cucumber, Holothuria leucospilota. Front. Immunol. 13:927880. doi: 10.3389/fimmu.2022.927880
Received: 25 April 2022; Accepted: 24 June 2022;
Published: 14 July 2022.
Edited by:
Chris Hauton, University of Southampton, United KingdomCopyright © 2022 Li, Chen, Wu, Li, Zhang, Jiang, Luo, Hu, Wong and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nai-Kei Wong, d29uZ25rQHN0dS5lZHUuY24=; Chunhua Ren, cm9zZW1hcnkxNjZAc2luYS5jb20=
†ORCID: Nai-Kei Wong, orcid.org/0000-0003-1303-3170
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.